Glycan fragment database: a database of PDB-based glycan 3D structures
|
|
|
- Stella McCormick
- 6 years ago
- Views:
Transcription
1 D470 D474 Nucleic Acids Research, 2013, Vol. 41, Database issue Published online 26 October 2012 doi: /nar/gks987 Glycan fragment database: a database of PDB-based glycan 3D structures Sunhwan Jo 1,2 and Wonpil Im 1,2, * 1 Department of Molecular Biosciences and 2 Center for Bioinformatics, The University of Kansas, 2030 Becker Drive, Lawrence, KS 66047, USA Received August 9, 2012; Revised September 14, 2012; Accepted September 27, 2012 ABSTRACT The glycan fragment database (GFDB), freely available at is a database of the glycosidic torsion angles derived from the glycan structures in the Protein Data Bank (PDB). Analogous to protein structure, the structure of an oligosaccharide chain in a glycoprotein, referred to as a glycan, can be characterized by the torsion angles of glycosidic linkages between relatively rigid carbohydrate monomeric units. Knowledge of accessible conformations of biologically relevant glycans is essential in understanding their biological roles. The GFDB provides an intuitive glycan sequence search tool that allows the user to search complex glycan structures. After a glycan search is complete, each glycosidic torsion angle distribution is displayed in terms of the exact match and the fragment match. The exact match results are from the PDB entries that contain the glycan sequence identical to the query sequence. The fragment match results are from the entries with the glycan sequence whose substructure (fragment) or entire sequence is matched to the query sequence, such that the fragment results implicitly include the influences from the nearby carbohydrate residues. In addition, clustering analysis based on the torsion angle distribution can be performed to obtain the representative structures among the searched glycan structures. INTRODUCTION An oligosaccharide moiety in a glycoprotein, referred to as a glycan, comes in a diversity of sequences and structures, and specific interactions between carbohydrates and proteins are essential in many cellular events (1 3). These events require molecular recognition of specific carbohydrate structures that seems to be sensitive to small differences in carbohydrate structure. For instance, the carbohydrate structures found on a host cell receptor, which only differ by the sequence of the terminal sugar residues, are believed to be a major factor in determining the host range (e.g. swine, avian or human) of influenza viruses (4,5). In addition, glycosyl transferases and glycosidases recognize specific sequences and spatially arranged oligosaccharide chains (6,7). Thus, understanding the carbohydrate conformations will provide insight into the role of glycans in modulating many cellular events. Analogous to protein structure, the structure of an oligosaccharide chain can be characterized by the torsion angles of glycosidic linkages between relatively rigid carbohydrate monomeric units. Considerable efforts have been already made to characterize the potential energy surface of the peptide bond conformation, and the accessible torsion angles of a peptide are well known (8 12). However, unlike proteins and peptides where the amino acid units are linearly linked together by the same peptide bonds, glycans can have branches, and each monosaccharide unit can be linked by different types of glycosidic linkages. In addition, the lack of experimentally derived atomic structures of oligosaccharides in aqueous solution makes it difficult to characterize the accessible torsion angles of a particular glycosidic linkage. Despite the difficulties involved in crystallization, the number of glycoprotein structures deposited in the Protein Data Bank (PDB) (13) has been steadily increasing (14,15). Although far from complete, glycan structures in the PDB can be used to study the accessible glycosidic torsion angles (16 19). Unfortunately, however, extracting structural information of glycans from the PDB is not trivial because of a lack of standardized nomenclature and the way the data are presented in the PDB (3,14). Recently, Sa we n et al. (19) analysed the accessible glycosidic torsion angles of the ð1! 2Þ linked mannose disaccharide using the PDB glycan structures, but they had to make considerable efforts to collect and filter out erroneous PDB entries. In this work, we present the glycan fragment database (GFDB), a database of the glycosidic torsion angles *To whom correspondence should be addressed. Tel: ; Fax: ; wonpil@ku.edu ß The Author(s) Published by Oxford University Press. This is an Open Access article distributed under the terms of the Creative Commons Attribution License ( which permits non-commercial reuse, distribution, and reproduction in any medium, provided the original work is properly cited. For commercial re-use, please contact journals.permissions@oup.com.
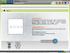
2 Nucleic Acids Research, 2013, Vol. 41, Database issue derived from the PDB glycan structures. Carbohydrate structures in the PDB are recognized by Glycan Reader, an automatic sugar identification algorithm that we developed (15), instead of using the nomenclature presented in the PDB entries. The GFDB provides an intuitive glycan sequence search tool that allows the user to search complex glycan structures. After a glycan search is complete, each glycosidic torsion angle distribution of the searched glycan structures is displayed. In addition, the torsion angle distributions can be clustered to generate representative structures using the clustering analysis facility on the GFDB interface. To facilitate the conformational analysis of glycosidic linkages, the GFDB also provides various filters. In the following sections, we discuss how the glycan structural information was collected, how to search a glycan sequence and how the search results are displayed. A stepwise guide about the GFDB is also provided in GLYCAN FRAGMENT DATABASE To recognize the PDB entries that contain carbohydrate molecules, we used Glycan Reader for automatic sugar identification (15). Briefly, in Glycan Reader, topologies of the molecules in the HETATM section of a PDB file are first generated using the atom connection information from the CONECT section. The carbohydrate candidate molecules (six-membered ring for a pyranose and five-membered ring for a furanose that are composed of only one oxygen and carbon atoms) are then identified. For each carbohydrate-like molecule, the chemical groups attached to each position of the ring and their orientations are compared with a pre-defined table to identify the correct chemical name for the carbohydrates. Glycan chains are constructed by examining the glycosidic linkages between the carbohydrate molecules that have chemical bonds between them. Identified carbohydrate molecules are further analysed and recorded in the GFDB. First, the residue name annotated in the PDB is compared with the molecular structure. The disparity of the residue name annotation in the PDB and the actual molecular structure is common (14). Although Glycan Reader returns the correct carbohydrate names according to the molecular structures, such disparity could be a sign of potential error. Second, because a distorted ring geometry could mislead the interpretation of the glycosidic torsion angles, the geometry of the carbohydrate ring is calculated by virtual torsion angle definition (20) and is recorded whether it is in a chair conformation (1C4 or 4C1). Finally, if the carbohydrate molecules have chemical groups (phosphate, sulphate, methyl and so forth) attached in one of the hydroxyl groups, the carbohydrates are marked as derived carbohydrates in the GFDB. The entries that belong to these cases can be excluded from the search using the filtering options, such as misassigned residues, distorted ring geometry and derived carbohydrates (see later in the text and also Figure 1). D471 WEB INTERFACE The GFDB provides a glycan sequence search interface that allows the user to search complex glycan sequences (Figure 1). The search interface provides a visual guide as the user builds a complex glycan query sequence, and the interface is compatible with any modern web browser with JavaScript capability. There is a report generation facility available to generate an archived report file that contains all the raw data for a given search and 3D structures based on the clustering analysis (see later in the text); the user also can get the archived report file by . There are several filtering functions available (Figure 1), which narrows the search results for specific needs, such as filters for only N-/O-glycosylated glycans, the resolution of the PDB entries and/or the aforementioned three structural features (misassigned residues, distorted ring geometry and derived carbohydrates). When analysing the glycosidic torsion angles in the PDB, it is important to understand that there are redundant PDB entries from the same or similar proteins. Without removing those redundant entries, it is possible to overestimate the preference of a certain conformation for a given glycan sequence. Although redundancies in the PDB can be removed by post-processing the data obtained by the GFDB, the GFDB provides a preliminary filter option for removing such redundant protein entries for N-linked or O-linked glycan chains based on the sequence similarity of the parent protein. Figure 1. The GFDB search interface.
3 D472 Nucleic Acids Research, 2013, Vol. 41, Database issue Figure 3. An example of the exact and fragment matches based on the query sequence in Figure 1. (A) The glycan sequence for the exact match results. (B and C) Examples of the glycan sequences for the fragment match results. The matched substructure is highlighted in the red rectangles. The sequence in (A) is also included in the fragment match results. Figure 2. An example of the search result for the query sequence in Figure 1. The glycosidic torsion angle distribution of a particular glycosidic linkage can be displayed by clicking the glycosidic linkage in Sequence Graph in Figure 1. The clustering analysis of the glycan chain can be performed, and the top-five representative structures can be downloaded. The glycosidic torsion angle distribution of a selected cluster is shown in red. After a glycan search is finished, the interface shows two torsion angle distributions side by side (Figure 2), exact match and fragment match (Figure 3). For the exact match, the GFDB first performs a sequence search to find the PDB entries that contain the glycan sequence identical to the query sequence, and the resulting torsion angle values for each glycosidic linkage are displayed to the user. On the other hand, the fragment search performs a search against the substructures (hence, they are called fragments, Figure 3) and returns the entries having at least one substructure that matches to the query sequence. This provides more samples for the torsion angle analysis. The torsion angle values from the fragment match always contain the exact match results. However, the fragment search results may not be the same as the exact match results because part of a glycan structure can adopt a different structure when it has extra intra- and intermolecular interactions. Therefore, the fragment match results implicitly include the influences from the nearby carbohydrate residues and different protein carbohydrate interactions, such that one can assess the flexibility of a certain glycosidic linkage in the context of larger glycan chain by comparing the exact and fragment match results. The glycosidic torsion angle definition in the GFDB is adopted from the crystallographic definition; O5-C1-O1C0 x ( ), C1-O1-C0 x-c0 x 1 ( ), and O1-C0 6-C0 5-O0 5 (!). The torsion angle between the first residue of the N-glycan chain and the side chain of the asparagine residue is defined as O5-C1-N0 D2-C0 G ( ) and C1-N0 D2-C0 G-C0 B ( ). The torsion angle between the first residue of the O-glycan chain and the side chain of the serine residue is defined as O5-C1-O0 G-C0 B ( ) and C1-O0 G-C0 B-C0 A ( ). For threonine, OG1 is used instead of OG. The atom names are based on the CHARMM topology. CLUSTERING ANALYSIS Statistical analysis of the torsion angle values of a particular glycosidic linkage is useful to estimate the allowable conformations of glycan chains, but it is difficult to understand what would be the representative (or most probable) structures of the given glycan sequence among the available PDB glycan structures. To provide useful insight into the 3D glycan structure, the GFDB provides an option to perform clustering analysis of the torsion angle search results and produce the top-five most clustered glycanonly structures. The GFDB uses a simple clustering method to efficiently determine the members of each cluster. The pairwise torsion angle differences are first calculated by the following equation: vffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffi 2 2 up u t k ki kj + ki kj ð1þ dij ¼ N where N is the total number of glycosidic linkages in a glycan sequence, k and k are the torsion angle values of the k-th glycosidic linkage, and i and j represent two glycan structures.! torsion angle values are included only for
4 Nucleic Acids Research, 2013, Vol. 41, Database issue D473 glycosidic linkages that have three rotatable bonds such as 1 6 linkages. After the pairwise distance matrix of the searched glycan structures is calculated, the first cluster is identified with the maximum number of neighbours within a 30 cut-off radius; the cut-off value was empirically determined. The second cluster is identified in the same manner after excluding the members that belong to the first cluster. The result of the top-five clusters and the corresponding 3D glycan structure based on the centroid of each cluster is provided to the user along with the input files to generate the centroid glycan structures using the CHARMM biomolecular simulation program (21). CONCLUDING DISCUSSION There are several databases that provide information on glycan structures or sequences derived from the PDB (or from other experiments). Many of these databases, such as BCSDB (22), KEGG GLYCAN (23) and Glycoconjugate Data Bank (24), store only glycan sequence information, whereas the GFDB focuses on the 3D glycan structure. GlycoMaps DB (25) and GlyTorsion (26) provide torsion angle distributions of glycosidic linkages derived from computational calculations and from the PDB, respectively. Thus, the GlyTorsion database is the only database that can be directly compared with the GFDB. While the search interface of the GlyTorsion database is restricted to only one glycosidic linkage, the GFDB can search more complex glycan sequence with various filter functions and provide the clustering analysis and the top-five clustered structures. These unique features in the GFDB allow researchers to collect complex glycan structural information easily and reliably. As of August 2012, the GFDB contains 5360 PDB entries that contain at least one carbohydrate molecule and glycan chains. Among those glycan chains, (57%) are N-linked glycan chains and 788 (4%) are O-linked glycans. And the remaining 7944 (39%) exist as ligands. For the glycan structures with more than two carbohydrates, the hierarchical fragmentation identified a total of fragment structures with 4267 unique glycan sequences; a unique glycan sequence has more than two carbohydrates and is defined by the carbohydrate sequence and the glycosidic linkages. There are glycosidic torsion angle values available in the GFDB. By providing the straightforward search tool, the filtering functions and the clustering analysis for the representative structures, we hope that the GFDB can help conformational analysis of various oligosaccharide chains and glycosidic linkages. The database will be updated quarterly and is freely available at FUNDING University of Kansas General Research Fund [ ]; Kanas-COBRE NIH P20 [GM103420]; NSF [MCB ]; TeraGrid/XSEDE resources [TG-MCB070009]. Funding for open access charge: NSF [MCB ]. Conflict of interest statement. None declared. REFERENCES 1. Rudd,P.M., Wormald,M.R. and Dwek,R.A. (2004) Sugar-mediated ligand receptor interactions in the immune system. Trends Biotech., 22, Petrescu,A.-J., Wormald,M.R. and Dwek,R.A. (2006) Structural aspects of glycomes with a focus on N-glycosylation and glycoprotein folding. Curr. Opin. Struct. Biol., 16, Wormald,M.R., Petrescu,A.-J., Pao,Y., Glithero,A., Elliott,T. and Dwek,R.A. (2002) Conformational studies of oligosaccharides and glycopeptides: Complementarity of NMR, X-ray crystallography, and molecular modelling. Chem. Rev., 102, Shinya,K., Ebina,M., Yamada,S., Ono,M., Kasai,N. and Kawaoka,Y. (2006) Avian flu: influenza virus receptors in the human airway. Nature, 440, Skehel,J.J. and Wiley,D.C. (2000) Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem., 69, Shah,N., Kuntz,D.A. and Rose,D.R. (2008) Golgi alpha-mannosidase II cleaves two sugars sequentially in the same catalytic site. Proc. Natl Acad. Sci. USA, 105, Zhong,W., Kuntz,D.A., Ember,B., Singh,H., Moremen,K.W., Rose,D.R. and Boons,G.-J. (2008) Probing the substrate specificity of Golgi alpha-mannosidase II by use of synthetic oligosaccharides and a catalytic nucleophile mutant. J. Am. Chem. Soc., 130, Ramachandran,G.N., Ramakrishnan,C. and Sasisekharan,V. (1963) Stereochemistry of polypeptide chain configurations. J. Mol. Biol., 7, Ramachandran,G.N. and Sasisekharan,V. (1968) Conformation of polypeptides and proteins. Adv. Protein Chem., 23, Hovmoller,S., Zhou,T. and Ohlson,T. (2002) Conformations of amino acids in proteins. Acta Crystallogr. D Biol. Crystallogr., 58, Ho,B.K., Thomas,A. and Brasseur,R. (2003) Revisiting the Ramachandran plot: hard-sphere repulsion, electrostatics, and H-bonding in the alpha-helix. Protein Sci., 12, Porter,L.L. and Rose,G.D. (2011) Redrawing the Ramachandran plot after inclusion of hydrogen-bonding constraints. Proc. Natl Acad. Sci. USA, 108, Berman,H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, Lu tteke,t., Frank,M. and von der Lieth,C.-W. (2004) Data mining the protein data bank: automatic detection and assignment of carbohydrate structures. Carbohydr. Res., 339, Jo,S., Song,K.C., Desaire,H., Mackerell,A.D. Jr and Im,W. (2011) Glycan Reader: automated sugar identification and simulation preparation for carbohydrates and glycoproteins. J. Comput. Chem., 32, Petrescu,A.J., Petrescu,S.M., Dwek,R.A. and Wormald,M.R. (1999) A statistical analysis of N- and O-glycan linkage conformations from crystallographic data. Glycobiology, 9, Petrescu,A.-J., Milac,A.-L., Petrescu,S.M., Dwek,R.A. and Wormald,M.R. (2004) Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology, 14, Lu tteke,t. (2009) Analysis and validation of carbohydrate three-dimensional structures. Acta Crystallogr. D Biol. Crystallogr., 65, Sa we n,e., Massad,T., Landersjo,C., Damberg,P. and Widmalm,G.R. (2010) Population distribution of flexible molecules from maximum entropy analysis using different priors as background information: application to the, -conformational space of the a-(1 >2)-linked mannose disaccharide present in N- and O-linked glycoproteins. Org. Biomol. Chem., 8, Rao,V.S.R. (1998) Conformation of Carbohydrates. Harwood Academic Publishers, Australia. 21. Brooks,B.R., Brooks,C.L., Mackerell,A.D. Jr, Nilsson,L., Petrella,R.J., Roux,B., Won,Y., Archontis,G., Bartels,C., Boresch,S. et al. (2009) CHARMM: the biomolecular simulation program. J. Comput. Chem., 30,
5 D474 Nucleic Acids Research, 2013, Vol. 41, Database issue 22. Herget,S., Toukach,P.V., Ranzinger,R., Hull,W.E., Knirel,Y.A. and von der Lieth,C.W. (2008) Statistical analysis of the Bacterial Carbohydrate Structure Data Base (BCSDB): characteristics and diversity of bacterial carbohydrates in comparison with mammalian glycans. BMC Struct. Biol., 8, Hashimoto,K., Goto,S., Kawano,S., Aoki-Kinoshita,K.F., Ueda,N., Hamajima,M., Kawasaki,T. and Kanehisa,M. (2006) KEGG as a glycome informatics resource. Glycobiology, 16, 63R 70R. 24. Nakahara,T., Hashimoto,R., Nakagawa,H., Monde,K., Miura,N. and Nishimura,S.-I. (2008) Glycoconjugate Data Bank: structures an annotated glycan structure database and N-glycan primary structure verification service. Nucleic Acids Res., 36, D368 D Frank,M., Lutteke,T. and von der Lieth,C.W. (2007) GlycoMapsDB: a database of the accessible conformational space of glycosidic linkages. Nucleic Acids Res., 35, Lu tteke,t., Frank,M. and von der Lieth,C.-W. (2005) Carbohydrate Structure Suite (CSS): analysis of carbohydrate 3D structures derived from the PDB. Nucleic Acids Res., 33, D242 D246.
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 10 Carbohydrates 2013 W. H. Freeman and Company Chapter 10 Outline Monosaccharides are aldehydes or ketones that contain two or
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 10 Carbohydrates 2013 W. H. Freeman and Company Chapter 10 Outline Monosaccharides are aldehydes or ketones that contain two or
PHARMACOPHORE MODELING
 CHAPTER 9 PHARMACOPHORE MODELING 9.1 PHARMACOPHORES AS VIRTUAL SCREENING TOOLS The pharmacophore concept has been widely used over the past decades for rational drug design approaches and has now been
CHAPTER 9 PHARMACOPHORE MODELING 9.1 PHARMACOPHORES AS VIRTUAL SCREENING TOOLS The pharmacophore concept has been widely used over the past decades for rational drug design approaches and has now been
Carbohydrates. Learning Objective
 , one of the four major classes of biomolecules, are aldehyde or ketone compounds with multiple hydroxyl groups. They function as energy stores, metabolic intermediates and important fuels for the body.
, one of the four major classes of biomolecules, are aldehyde or ketone compounds with multiple hydroxyl groups. They function as energy stores, metabolic intermediates and important fuels for the body.
Chapter 11. Learning objectives: Structure and function of monosaccharides, polysaccharide, glycoproteins lectins.
 Chapter 11 Learning objectives: Structure and function of monosaccharides, polysaccharide, glycoproteins lectins. Carbohydrates Fuels Structural components Coating of cells Part of extracellular matrix
Chapter 11 Learning objectives: Structure and function of monosaccharides, polysaccharide, glycoproteins lectins. Carbohydrates Fuels Structural components Coating of cells Part of extracellular matrix
GlycoFragment and GlycoSearchMS: web tools to support the interpretation of mass spectra of complex carbohydrates
 GlycoFragment and GlycoSearchMS: web tools to support the interpretation of mass spectra of complex carbohydrates Klaus Karl Lohmann* and Claus-W. von der Lieth Nucleic Acids Research, 2004, Vol. 32, Web
GlycoFragment and GlycoSearchMS: web tools to support the interpretation of mass spectra of complex carbohydrates Klaus Karl Lohmann* and Claus-W. von der Lieth Nucleic Acids Research, 2004, Vol. 32, Web
GLYCOSYLATION OF PROTEINS: A COMPUTER BASED METHOD FOR THE RAPID EXPLORATION OF COMFORMATIONAL SPACE OF N-GLYCANS
 GLYCOSYLATION OF PROTEINS: A COMPUTER BASED METHOD FOR THE RAPID EXPLORATION OF COMFORMATIONAL SPACE OF N-GLYCANS A BOHNE, C. -W. VON DER LIETH DKFZ (German Cancer Research Centre), Molecular Modelling,
GLYCOSYLATION OF PROTEINS: A COMPUTER BASED METHOD FOR THE RAPID EXPLORATION OF COMFORMATIONAL SPACE OF N-GLYCANS A BOHNE, C. -W. VON DER LIETH DKFZ (German Cancer Research Centre), Molecular Modelling,
A Graphic User Interface for the Glyco-MGrid Portal System for Collaboration among Scientists
 Proceedings of the 6th WSEAS Int. Conference on TELECOMMUNICATIONS and INFORMATICS, Dallas, Texas, USA, March 22-24, 2007 82 A Graphic User Interface for the Glyco-MGrid Portal System for Collaboration
Proceedings of the 6th WSEAS Int. Conference on TELECOMMUNICATIONS and INFORMATICS, Dallas, Texas, USA, March 22-24, 2007 82 A Graphic User Interface for the Glyco-MGrid Portal System for Collaboration
Topic 4 - #2 Carbohydrates Topic 2
 Topic 4 - #2 Carbohydrates Topic 2 Biologically Important Monosaccharide Derivatives There are a large number of monosaccharide derivatives. A variety of chemical and enzymatic reactions produce these
Topic 4 - #2 Carbohydrates Topic 2 Biologically Important Monosaccharide Derivatives There are a large number of monosaccharide derivatives. A variety of chemical and enzymatic reactions produce these
CS612 - Algorithms in Bioinformatics
 Spring 2016 Protein Structure February 7, 2016 Introduction to Protein Structure A protein is a linear chain of organic molecular building blocks called amino acids. Introduction to Protein Structure Amine
Spring 2016 Protein Structure February 7, 2016 Introduction to Protein Structure A protein is a linear chain of organic molecular building blocks called amino acids. Introduction to Protein Structure Amine
Integrated use of Databases in Structural Investigation of Complex Carbohydrates. Roberta Marchetti, Ph.D. University of Naples Federico II
 Integrated use of Databases in Structural Investigation of Complex Carbohydrates Roberta Marchetti, Ph.D. University of Naples Federico II Carbohydrates Proteins Nucleic Acids Lipids Glycans are made up
Integrated use of Databases in Structural Investigation of Complex Carbohydrates Roberta Marchetti, Ph.D. University of Naples Federico II Carbohydrates Proteins Nucleic Acids Lipids Glycans are made up
A BEGINNER S GUIDE TO BIOCHEMISTRY
 A BEGINNER S GUIDE TO BIOCHEMISTRY Life is basically a chemical process Organic substances: contain carbon atoms bonded to other carbon atom 4 classes: carbohydrates, lipids, proteins, nucleic acids Chemical
A BEGINNER S GUIDE TO BIOCHEMISTRY Life is basically a chemical process Organic substances: contain carbon atoms bonded to other carbon atom 4 classes: carbohydrates, lipids, proteins, nucleic acids Chemical
Carbohydrates are aldehyde or ketone compounds with multiple hydroxyl groups Have multiple roles in all forms of life
 Carbohydrates 1 Carbohydrates are aldehyde or ketone compounds with multiple hydroxyl groups Have multiple roles in all forms of life Classification Serve as energy stores, fuels, and metabolic intermediates
Carbohydrates 1 Carbohydrates are aldehyde or ketone compounds with multiple hydroxyl groups Have multiple roles in all forms of life Classification Serve as energy stores, fuels, and metabolic intermediates
Glycosaminoglycans: Anionic polysaccharide chains made of repeating disaccharide units
 Glycosaminoglycans: Anionic polysaccharide chains made of repeating disaccharide units Glycosaminoglycans present on the animal cell surface and in the extracellular matrix. Glycoseaminoglycans (mucopolysaccharides)
Glycosaminoglycans: Anionic polysaccharide chains made of repeating disaccharide units Glycosaminoglycans present on the animal cell surface and in the extracellular matrix. Glycoseaminoglycans (mucopolysaccharides)
Chapter 1. Chemistry of Life - Advanced TABLE 1.2: title
 Condensation and Hydrolysis Condensation reactions are the chemical processes by which large organic compounds are synthesized from their monomeric units. Hydrolysis reactions are the reverse process.
Condensation and Hydrolysis Condensation reactions are the chemical processes by which large organic compounds are synthesized from their monomeric units. Hydrolysis reactions are the reverse process.
A. B. C. D. E. F. G. H. I. J. K. Ser/Thr. Ser/Thr. Ser/Thr. Ser/Thr. Ser/Thr. Asn. Asn. Asn. Asn. Asn. Asn
 A. B. C. D. E. F. "3 "3!4!3 Ser/Thr "3!4!3!4 Asn Asn Ser/Thr Asn!3!6 Ser/Thr G. H. I. J. K.!3 Ser/Thr Ser/Thr 4 4 2 2 6 3 6 Asn Asn Asn Glycosidases and Glycosyltransferases Introduction to Inverting/Retaining
A. B. C. D. E. F. "3 "3!4!3 Ser/Thr "3!4!3!4 Asn Asn Ser/Thr Asn!3!6 Ser/Thr G. H. I. J. K.!3 Ser/Thr Ser/Thr 4 4 2 2 6 3 6 Asn Asn Asn Glycosidases and Glycosyltransferases Introduction to Inverting/Retaining
FurinDB: A Database of 20-Residue Furin Cleavage Site Motifs, Substrates and Their Associated Drugs
 Int. J. Mol. Sci. 2011, 12, 1060-1065; doi:10.3390/ijms12021060 OPEN ACCESS Technical Note International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms FurinDB: A Database of 20-Residue
Int. J. Mol. Sci. 2011, 12, 1060-1065; doi:10.3390/ijms12021060 OPEN ACCESS Technical Note International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms FurinDB: A Database of 20-Residue
A. Incorrect! No, this is not the description of this type of molecule. B. Incorrect! No, this is not the description of this type of molecule.
 Biochemistry - Problem Drill 08: Carbohydrates No. 1 of 10 1. have one aldehyde (-CHO) or one keto (-C=O) group and many hydroxyl (-OH) groups. (A) Amino acids (B) Proteins (C) Nucleic Acids (D) Carbohydrates
Biochemistry - Problem Drill 08: Carbohydrates No. 1 of 10 1. have one aldehyde (-CHO) or one keto (-C=O) group and many hydroxyl (-OH) groups. (A) Amino acids (B) Proteins (C) Nucleic Acids (D) Carbohydrates
Lecture Series 2 Macromolecules: Their Structure and Function
 Lecture Series 2 Macromolecules: Their Structure and Function Reading Assignments Read Chapter 4 (Protein structure & Function) Biological Substances found in Living Tissues The big four in terms of macromolecules
Lecture Series 2 Macromolecules: Their Structure and Function Reading Assignments Read Chapter 4 (Protein structure & Function) Biological Substances found in Living Tissues The big four in terms of macromolecules
Abdullah zurayqat. Bahaa Najjar. Mamoun Ahram
 9 Abdullah zurayqat Bahaa Najjar Mamoun Ahram Polysaccharides Polysaccharides Definition and Structure [Greek poly = many; sacchar = sugar] are complex carbohydrates, composed of 10 to up to several thousand
9 Abdullah zurayqat Bahaa Najjar Mamoun Ahram Polysaccharides Polysaccharides Definition and Structure [Greek poly = many; sacchar = sugar] are complex carbohydrates, composed of 10 to up to several thousand
Lecture Series 2 Macromolecules: Their Structure and Function
 Lecture Series 2 Macromolecules: Their Structure and Function Reading Assignments Read Chapter 4 (Protein structure & Function) Biological Substances found in Living Tissues The big four in terms of macromolecules
Lecture Series 2 Macromolecules: Their Structure and Function Reading Assignments Read Chapter 4 (Protein structure & Function) Biological Substances found in Living Tissues The big four in terms of macromolecules
A. Lipids: Water-Insoluble Molecules
 Biological Substances found in Living Tissues Lecture Series 3 Macromolecules: Their Structure and Function A. Lipids: Water-Insoluble Lipids can form large biological molecules, but these aggregations
Biological Substances found in Living Tissues Lecture Series 3 Macromolecules: Their Structure and Function A. Lipids: Water-Insoluble Lipids can form large biological molecules, but these aggregations
-can be classified by the number of sugars that constitute the molecules: -how to differentiate between glucose and galactose?
 Carbohydrates (Also called: saccharides) -can be classified by the number of sugars that constitute the molecules: 1- monosaccharides: -General formula: (CH2O)n -Contain one sugar molecule -Contain two
Carbohydrates (Also called: saccharides) -can be classified by the number of sugars that constitute the molecules: 1- monosaccharides: -General formula: (CH2O)n -Contain one sugar molecule -Contain two
A look at macromolecules (Text pages 38-54) What is the typical chemical composition of a cell? (Source of figures to right: Madigan et al.
 A look at macromolecules (Text pages 38-54) What is the typical chemical composition of a cell? (Source of figures to right: Madigan et al. 2002 Chemical Bonds Ionic Electron-negativity differences cause
A look at macromolecules (Text pages 38-54) What is the typical chemical composition of a cell? (Source of figures to right: Madigan et al. 2002 Chemical Bonds Ionic Electron-negativity differences cause
Questions- Carbohydrates. A. The following structure is D-sorbose. (Questions 1 7) CH 2 OH C = O H C OH HO C H H C OH
 Questions- Carbohydrates A. The following structure is D-sorbose. (Questions 1 7) CH 2 C = O H C HO C H H C CH 2 1. 2. 3. 4. 5. Which characteristic is different when comparing the open-chain forms of
Questions- Carbohydrates A. The following structure is D-sorbose. (Questions 1 7) CH 2 C = O H C HO C H H C CH 2 1. 2. 3. 4. 5. Which characteristic is different when comparing the open-chain forms of
Glycosaminoglycans, Proteoglycans, and Glycoproteins
 Glycosaminoglycans, Proteoglycans, and Glycoproteins Presented by Dr. Mohammad Saadeh The requirements for the Pharmaceutical Biochemistry I Philadelphia University Faculty of pharmacy I. OVERVIEW OF GLYCOSAMINOGLYCANS
Glycosaminoglycans, Proteoglycans, and Glycoproteins Presented by Dr. Mohammad Saadeh The requirements for the Pharmaceutical Biochemistry I Philadelphia University Faculty of pharmacy I. OVERVIEW OF GLYCOSAMINOGLYCANS
Problem Set 2 September 18, 2009
 September 18, 2009 General Instructions: 1. You are expected to state all your assumptions and provide step-by-step solutions to the numerical problems. Unless indicated otherwise, the computational problems
September 18, 2009 General Instructions: 1. You are expected to state all your assumptions and provide step-by-step solutions to the numerical problems. Unless indicated otherwise, the computational problems
Amino Acids. Review I: Protein Structure. Amino Acids: Structures. Amino Acids (contd.) Rajan Munshi
 Review I: Protein Structure Rajan Munshi BBSI @ Pitt 2005 Department of Computational Biology University of Pittsburgh School of Medicine May 24, 2005 Amino Acids Building blocks of proteins 20 amino acids
Review I: Protein Structure Rajan Munshi BBSI @ Pitt 2005 Department of Computational Biology University of Pittsburgh School of Medicine May 24, 2005 Amino Acids Building blocks of proteins 20 amino acids
Glycosidic bond cleavage
 Glycosidases and Glycosyltransferases Introduction to Inverting/Retaining Mechanisms Inhibitor design Chemical Reaction Proposed catalytic mechanisms Multiple slides courtesy of Harry Gilbert with Wells
Glycosidases and Glycosyltransferases Introduction to Inverting/Retaining Mechanisms Inhibitor design Chemical Reaction Proposed catalytic mechanisms Multiple slides courtesy of Harry Gilbert with Wells
Objectives. Carbon Bonding. Carbon Bonding, continued. Carbon Bonding
 Biochemistry Table of Contents Objectives Distinguish between organic and inorganic compounds. Explain the importance of carbon bonding in biological molecules. Identify functional groups in biological
Biochemistry Table of Contents Objectives Distinguish between organic and inorganic compounds. Explain the importance of carbon bonding in biological molecules. Identify functional groups in biological
Carbohydrate Structure and Nomenclature. Essentials of Glycobiology 1 April 2004
 1 Carbohydrate Structure and Nomenclature Essentials of Glycobiology 1 April 2004 Nathaniel Finney Dept. of Chemistry and Biochemistry UCSD nfinney@chem.ucsd.edu 2 Lecture utline 1. Carbohydrates - definition
1 Carbohydrate Structure and Nomenclature Essentials of Glycobiology 1 April 2004 Nathaniel Finney Dept. of Chemistry and Biochemistry UCSD nfinney@chem.ucsd.edu 2 Lecture utline 1. Carbohydrates - definition
Chemical Formulas. Chemical Formula CH 3 COCHCHOCHClCHNH Lewis Dot Structure
 Biochemistry . Chemical Formulas A chemical formula represents the chemical makeup of a compound. It shows the numbers and kinds of atoms present in a compound. It is a kind of shorthand that scientists
Biochemistry . Chemical Formulas A chemical formula represents the chemical makeup of a compound. It shows the numbers and kinds of atoms present in a compound. It is a kind of shorthand that scientists
Molecular Graphics Perspective of Protein Structure and Function
 Molecular Graphics Perspective of Protein Structure and Function VMD Highlights > 20,000 registered Users Platforms: Unix (16 builds) Windows MacOS X Display of large biomolecules and simulation trajectories
Molecular Graphics Perspective of Protein Structure and Function VMD Highlights > 20,000 registered Users Platforms: Unix (16 builds) Windows MacOS X Display of large biomolecules and simulation trajectories
Biological Sciences 4087 Exam I 9/20/11
 Name: Biological Sciences 4087 Exam I 9/20/11 Total: 100 points Be sure to include units where appropriate. Show all calculations. There are 5 pages and 11 questions. 1.(20pts)A. If ph = 4.6, [H + ] =
Name: Biological Sciences 4087 Exam I 9/20/11 Total: 100 points Be sure to include units where appropriate. Show all calculations. There are 5 pages and 11 questions. 1.(20pts)A. If ph = 4.6, [H + ] =
BIOCHEMISTRY. How Are Macromolecules Formed? Dehydration Synthesis or condensation reaction Polymers formed by combining monomers and removing water.
 BIOCHEMISTRY Organic compounds Compounds that contain carbon are called organic. Inorganic compounds do not contain carbon. Carbon has 4 electrons in outer shell. Carbon can form covalent bonds with as
BIOCHEMISTRY Organic compounds Compounds that contain carbon are called organic. Inorganic compounds do not contain carbon. Carbon has 4 electrons in outer shell. Carbon can form covalent bonds with as
189,311, , ,561, ,639, ,679, Ch13; , Carbohydrates
 Lecture 31 (12/8/17) Reading: Ch7; 258-267 Ch10; 371-373 Problems: Ch7 (text); 26,27,28 Ch7 (study-guide: applying); 2,5 Ch7 (study-guide: facts); 6 NEXT (LAST!) Reading: Chs4,6,8,10,14,16,17,18; 128-129,
Lecture 31 (12/8/17) Reading: Ch7; 258-267 Ch10; 371-373 Problems: Ch7 (text); 26,27,28 Ch7 (study-guide: applying); 2,5 Ch7 (study-guide: facts); 6 NEXT (LAST!) Reading: Chs4,6,8,10,14,16,17,18; 128-129,
The. Crash Course. Basically, almost all living things are made up of these 4 Elements: - Carbon (C) - Nitrogen (N) - Hydrogen (H) - Oxygen (O)
 The Biochemistry Crash Course Basically, almost all living things are made up of these 4 Elements: - Carbon (C) - Nitrogen (N) - Hydrogen (H) - Oxygen (O) This exercise is designed to familiarize you with
The Biochemistry Crash Course Basically, almost all living things are made up of these 4 Elements: - Carbon (C) - Nitrogen (N) - Hydrogen (H) - Oxygen (O) This exercise is designed to familiarize you with
BIO 311C Spring Lecture 15 Friday 26 Feb. 1
 BIO 311C Spring 2010 Lecture 15 Friday 26 Feb. 1 Illustration of a Polypeptide amino acids peptide bonds Review Polypeptide (chain) See textbook, Fig 5.21, p. 82 for a more clear illustration Folding and
BIO 311C Spring 2010 Lecture 15 Friday 26 Feb. 1 Illustration of a Polypeptide amino acids peptide bonds Review Polypeptide (chain) See textbook, Fig 5.21, p. 82 for a more clear illustration Folding and
Ch. 5 The S & F of Macromolecules. They may be extremely small but they are still macro.
 Ch. 5 The S & F of Macromolecules They may be extremely small but they are still macro. Background Information Cells join small molecules together to form larger molecules. Macromolecules may be composed
Ch. 5 The S & F of Macromolecules They may be extremely small but they are still macro. Background Information Cells join small molecules together to form larger molecules. Macromolecules may be composed
Significance and Functions of Carbohydrates. Bacterial Cell Walls
 Biochemistry 462a - Carbohydrate Function Reading - Chapter 9 Practice problems - Chapter 9: 2, 4a, 4b, 6, 9, 10, 13, 14, 15, 16a, 17; Carbohydrate extra problems Significance and Functions of Carbohydrates
Biochemistry 462a - Carbohydrate Function Reading - Chapter 9 Practice problems - Chapter 9: 2, 4a, 4b, 6, 9, 10, 13, 14, 15, 16a, 17; Carbohydrate extra problems Significance and Functions of Carbohydrates
Workshop on Analysis and prediction of contacts in proteins
 Workshop on Analysis and prediction of contacts in proteins 1.09.09 Eran Eyal 1, Vladimir Potapov 2, Ronen Levy 3, Vladimir Sobolev 3 and Marvin Edelman 3 1 Sheba Medical Center, Ramat Gan, Israel; Departments
Workshop on Analysis and prediction of contacts in proteins 1.09.09 Eran Eyal 1, Vladimir Potapov 2, Ronen Levy 3, Vladimir Sobolev 3 and Marvin Edelman 3 1 Sheba Medical Center, Ramat Gan, Israel; Departments
Final Report. Identification of Residue Mutations that Increase the Binding Affinity of LSTc to HA RBD. Wendy Fong ( 方美珍 ) CNIC; Beijing, China
 Final Report Identification of Residue Mutations that Increase the Binding Affinity of LSTc to HA RBD Wendy Fong ( 方美珍 ) CNIC; Beijing, China Influenza s Viral Life Cycle 1. HA on virus binds to sialic
Final Report Identification of Residue Mutations that Increase the Binding Affinity of LSTc to HA RBD Wendy Fong ( 方美珍 ) CNIC; Beijing, China Influenza s Viral Life Cycle 1. HA on virus binds to sialic
Life on earth is based on carbon. A carbon atom forms four covalent bonds It can join with other carbon atoms to make chains or rings
 Some atoms share outer shell electrons with other atoms, forming covalent bonds Atoms joined together by covalent bonds form molecules Molecules can be represented in many ways Table 2.8 Life on earth
Some atoms share outer shell electrons with other atoms, forming covalent bonds Atoms joined together by covalent bonds form molecules Molecules can be represented in many ways Table 2.8 Life on earth
Introduction to proteins and protein structure
 Introduction to proteins and protein structure The questions and answers below constitute an introduction to the fundamental principles of protein structure. They are all available at [link]. What are
Introduction to proteins and protein structure The questions and answers below constitute an introduction to the fundamental principles of protein structure. They are all available at [link]. What are
Honors Biology Chapter 3: Macromolecules PPT Notes
 Honors Biology Chapter 3: Macromolecules PPT Notes 3.1 I can explain why carbon is unparalleled in its ability to form large, diverse molecules. Diverse molecules found in cells are composed of carbon
Honors Biology Chapter 3: Macromolecules PPT Notes 3.1 I can explain why carbon is unparalleled in its ability to form large, diverse molecules. Diverse molecules found in cells are composed of carbon
2.3 Carbon-Based Molecules. KEY CONCEPT Carbon-based molecules are the foundation of life.
 KEY CONCEPT Carbon-based molecules are the foundation of life. Carbon atoms have unique bonding properties. Carbon forms covalent bonds with up to four other atoms, including other carbon atoms. Carbon-based
KEY CONCEPT Carbon-based molecules are the foundation of life. Carbon atoms have unique bonding properties. Carbon forms covalent bonds with up to four other atoms, including other carbon atoms. Carbon-based
Carbohydrate Chemistry
 Carbohydrate Chemistry edited by Geert-Jan Boons University of Birmingham Birmingham UK m BLACKIE ACADEMIC & PROFESSIONAL An Imprint of Chapman & Hall London Weinheim New York * Tokyo Melbourne Madras
Carbohydrate Chemistry edited by Geert-Jan Boons University of Birmingham Birmingham UK m BLACKIE ACADEMIC & PROFESSIONAL An Imprint of Chapman & Hall London Weinheim New York * Tokyo Melbourne Madras
Carbohydrates. What are they? What do cells do with carbs? Where do carbs come from? O) n. Formula = (CH 2
 Carbohydrates What are they? Formula = (C 2 O) n where n > 3 Also called sugar Major biomolecule in body What do cells do with carbs? Oxidize them for energy Store them to oxidize later for energy Use
Carbohydrates What are they? Formula = (C 2 O) n where n > 3 Also called sugar Major biomolecule in body What do cells do with carbs? Oxidize them for energy Store them to oxidize later for energy Use
Bioinformatics for molecular biology
 Bioinformatics for molecular biology Structural bioinformatics tools, predictors, and 3D modeling Structural Biology Review Dr Research Scientist Department of Microbiology, Oslo University Hospital -
Bioinformatics for molecular biology Structural bioinformatics tools, predictors, and 3D modeling Structural Biology Review Dr Research Scientist Department of Microbiology, Oslo University Hospital -
We are going to talk about two classifications of proteins: fibrous & globular.
 Slide # 13 (fibrous proteins) : We are going to talk about two classifications of proteins: fibrous & globular. *fibrous proteins: (dense fibers) *Their structures are mainly formed of the secondary structure
Slide # 13 (fibrous proteins) : We are going to talk about two classifications of proteins: fibrous & globular. *fibrous proteins: (dense fibers) *Their structures are mainly formed of the secondary structure
Biosynthesis of N and O Glycans
 TechNote #TNGL101 Biosynthesis of N and O Glycans These suggestions and data are based on information we believe to be reliable. They are offered in good faith, but without guarantee, as conditions and
TechNote #TNGL101 Biosynthesis of N and O Glycans These suggestions and data are based on information we believe to be reliable. They are offered in good faith, but without guarantee, as conditions and
Biology. Slide 1 of 37. End Show. Copyright Pearson Prentice Hall
 Biology 1 of 37 2 of 37 The Chemistry of Carbon The Chemistry of Carbon Organic chemistry is the study of all compounds that contain bonds between carbon atoms. 3 of 37 Macromolecules Macromolecules Macromolecules
Biology 1 of 37 2 of 37 The Chemistry of Carbon The Chemistry of Carbon Organic chemistry is the study of all compounds that contain bonds between carbon atoms. 3 of 37 Macromolecules Macromolecules Macromolecules
Protein Trafficking in the Secretory and Endocytic Pathways
 Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Chapter Three (Biochemistry)
 Chapter Three (Biochemistry) 1 SECTION ONE: CARBON COMPOUNDS CARBON BONDING All compounds can be classified in two broad categories: organic compounds and inorganic compounds. Organic compounds are made
Chapter Three (Biochemistry) 1 SECTION ONE: CARBON COMPOUNDS CARBON BONDING All compounds can be classified in two broad categories: organic compounds and inorganic compounds. Organic compounds are made
Carbohydrates 1. Steven E. Massey, Ph.D. Assistant Professor Bioinformatics Department of Biology University of Puerto Rico Río Piedras
 Carbohydrates 1 Steven E. Massey, Ph.D. Assistant Professor Bioinformatics Department of Biology University of Puerto Rico Río Piedras Office & Lab: NCN#343B Tel: 787-764-0000 ext. 7798 E-mail: stevenemassey@gmail.com
Carbohydrates 1 Steven E. Massey, Ph.D. Assistant Professor Bioinformatics Department of Biology University of Puerto Rico Río Piedras Office & Lab: NCN#343B Tel: 787-764-0000 ext. 7798 E-mail: stevenemassey@gmail.com
AP BIOLOGY: READING ASSIGNMENT FOR CHAPTER 5
 1) Complete the following table: Class Monomer Functions Carbohydrates 1. 3. Lipids 1. 3. Proteins 1. 3. 4. 5. 6. Nucleic Acids 1. 2) Circle the atoms of these two glucose molecules that will be removed
1) Complete the following table: Class Monomer Functions Carbohydrates 1. 3. Lipids 1. 3. Proteins 1. 3. 4. 5. 6. Nucleic Acids 1. 2) Circle the atoms of these two glucose molecules that will be removed
A Getting-It-On Review and Self-Test. . Carbohydrates are
 A Getting-It-n Review and Self-Test arbohydrates arbohydrates, one of the three principal classes of foods, contain only three elements: (1), (2), and (3). The name carbohydrate is derived from the French
A Getting-It-n Review and Self-Test arbohydrates arbohydrates, one of the three principal classes of foods, contain only three elements: (1), (2), and (3). The name carbohydrate is derived from the French
Chapter 3. Table of Contents. Section 1 Carbon Compounds. Section 2 Molecules of Life. Biochemistry
 Biochemistry Table of Contents Section 1 Carbon Compounds Section 2 Molecules of Life Section 1 Carbon Compounds Objectives Distinguish between organic and inorganic compounds. Explain the importance of
Biochemistry Table of Contents Section 1 Carbon Compounds Section 2 Molecules of Life Section 1 Carbon Compounds Objectives Distinguish between organic and inorganic compounds. Explain the importance of
MITOCW watch?v=kl2kpdlb8sq
 MITOCW watch?v=kl2kpdlb8sq The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high quality educational resources for free. To
MITOCW watch?v=kl2kpdlb8sq The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high quality educational resources for free. To
Dr. Entedhar Carbohydrates Carbohydrates are carbon compounds that have aldehyde (C-H=0) or ketone (C=O) moiety and comprises polyhyroxyl alcohol
 Dr. Entedhar Carbohydrates Carbohydrates are carbon compounds that have aldehyde (C-H=0) or ketone (C=O) moiety and comprises polyhyroxyl alcohol (polyhydroxyaldehyde or polyhyroxyketone); their polymers,which
Dr. Entedhar Carbohydrates Carbohydrates are carbon compounds that have aldehyde (C-H=0) or ketone (C=O) moiety and comprises polyhyroxyl alcohol (polyhydroxyaldehyde or polyhyroxyketone); their polymers,which
CARBOHYDRATES. Produce energy for living things Atoms? Monomer Examples? Carbon, hydrogen, and oxygen in 1:2:1 ratio.
 CARBOHYDRATES Produce energy for living things Atoms? Carbon, hydrogen, and oxygen in 1:2:1 ratio Monomer Examples? Sugars, starches MONOSACCHARIDES--- main source of energy for cells Glucose Know formula?
CARBOHYDRATES Produce energy for living things Atoms? Carbon, hydrogen, and oxygen in 1:2:1 ratio Monomer Examples? Sugars, starches MONOSACCHARIDES--- main source of energy for cells Glucose Know formula?
molecules ISSN
 Molecules 2000, 5, 1399-1407 molecules ISSN 1420-3049 http://www.mdpi.org Substitution Effects on Reactivity of N-Acyl-2-amino-2-desoxyglucopyranoses. Quantum Chemical Study Aušra Vektariene 1, *, Arvydas
Molecules 2000, 5, 1399-1407 molecules ISSN 1420-3049 http://www.mdpi.org Substitution Effects on Reactivity of N-Acyl-2-amino-2-desoxyglucopyranoses. Quantum Chemical Study Aušra Vektariene 1, *, Arvydas
DEPARTMENT OF SCIENCE
 DEPARTMENT OF SCIENCE COURSE OUTLINE WINTER 2012-13 BC 3200 STRUCTURE & CATALYSIS INSTRUCTOR: Philip Johnson PHONE: 780-539-2863 OFFICE: J224 E-MAIL: PJohnson@gprc.ab.ca OFFICE HOURS: Mondays 1000-1120
DEPARTMENT OF SCIENCE COURSE OUTLINE WINTER 2012-13 BC 3200 STRUCTURE & CATALYSIS INSTRUCTOR: Philip Johnson PHONE: 780-539-2863 OFFICE: J224 E-MAIL: PJohnson@gprc.ab.ca OFFICE HOURS: Mondays 1000-1120
2. Which of the following amino acids is most likely to be found on the outer surface of a properly folded protein?
 Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
a) The statement is true for X = 400, but false for X = 300; b) The statement is true for X = 300, but false for X = 200;
 1. Consider the following statement. To produce one molecule of each possible kind of polypeptide chain, X amino acids in length, would require more atoms than exist in the universe. Given the size of
1. Consider the following statement. To produce one molecule of each possible kind of polypeptide chain, X amino acids in length, would require more atoms than exist in the universe. Given the size of
Chapter 16: Carbohydrates
 Vocabulary Aldose: a sugar that contains an aldehyde group as part of its structure Amylopectin: a form of starch; a branched chain polymer of glucose Amylose: a form of starch; a linear polymer of glucose
Vocabulary Aldose: a sugar that contains an aldehyde group as part of its structure Amylopectin: a form of starch; a branched chain polymer of glucose Amylose: a form of starch; a linear polymer of glucose
Haworth structure for fructose
 BIOMOLECULES carbohydrates may be defined as optically active polyhydroxy aldehydes or ketones or the compounds which produce such units on hydrolysis. Some of the carbohydrates, which are sweet in taste,
BIOMOLECULES carbohydrates may be defined as optically active polyhydroxy aldehydes or ketones or the compounds which produce such units on hydrolysis. Some of the carbohydrates, which are sweet in taste,
Lecture Series 2 Macromolecules: Their Structure and Function
 Lecture Series 2 Macromolecules: Their Structure and Function Reading Assignments Read Chapter 4 (Protein structure & Function) Biological Substances found in Living Tissues The big four in terms of macromolecules
Lecture Series 2 Macromolecules: Their Structure and Function Reading Assignments Read Chapter 4 (Protein structure & Function) Biological Substances found in Living Tissues The big four in terms of macromolecules
H C. C α. Proteins perform a vast array of biological function including: Side chain
 Topics The topics: basic concepts of molecular biology elements on Python overview of the field biological databases and database searching sequence alignments phylogenetic trees microarray data analysis
Topics The topics: basic concepts of molecular biology elements on Python overview of the field biological databases and database searching sequence alignments phylogenetic trees microarray data analysis
Visualizing Biopolymers and Their Building Blocks
 Visualizing Biopolymers and Their Building Blocks By Sharlene Denos (UIUC) & Kathryn Hafner (Danville High) Living things are primarily composed of carbon-based (organic) polymers. These are made up many
Visualizing Biopolymers and Their Building Blocks By Sharlene Denos (UIUC) & Kathryn Hafner (Danville High) Living things are primarily composed of carbon-based (organic) polymers. These are made up many
Details of Organic Chem! Date. Carbon & The Molecular Diversity of Life & The Structure & Function of Macromolecules
 Details of Organic Chem! Date Carbon & The Molecular Diversity of Life & The Structure & Function of Macromolecules Functional Groups, I Attachments that replace one or more of the hydrogens bonded to
Details of Organic Chem! Date Carbon & The Molecular Diversity of Life & The Structure & Function of Macromolecules Functional Groups, I Attachments that replace one or more of the hydrogens bonded to
CLASS 11th. Biomolecules
 CLASS 11th 01. Carbohydrates These are the compound of carbon, hydrogen and oxygen having hydrogen and oxygen in the same ratio as that of water, i.e. 2 : 1. They are among the most widely distributed
CLASS 11th 01. Carbohydrates These are the compound of carbon, hydrogen and oxygen having hydrogen and oxygen in the same ratio as that of water, i.e. 2 : 1. They are among the most widely distributed
Chapter-8 Saccharide Chemistry
 Chapter-8 Saccharide Chemistry Page 217-228 Carbohydrates (Saccharides) are most abundant biological molecule, riginally produced through C 2 fixation during photosynthesis I (C 2 ) n or - C - I where
Chapter-8 Saccharide Chemistry Page 217-228 Carbohydrates (Saccharides) are most abundant biological molecule, riginally produced through C 2 fixation during photosynthesis I (C 2 ) n or - C - I where
Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site.
 Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site. Still having trouble understanding the material? Check
Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site. Still having trouble understanding the material? Check
Bio 100 Serine Proteases 9/26/11
 Assigned Reading: 4th ed. 6.4.1 The Chymotrypsin Mechanism Involves Acylation And Deacylation Of A Ser Residue p. 213 BOX 20-1 Penicillin and β-lactamase p. 779 6.5.7 Some Enzymes Are Regulated By Proteolytic
Assigned Reading: 4th ed. 6.4.1 The Chymotrypsin Mechanism Involves Acylation And Deacylation Of A Ser Residue p. 213 BOX 20-1 Penicillin and β-lactamase p. 779 6.5.7 Some Enzymes Are Regulated By Proteolytic
Downloaded from
 Biomolecules Section A (One Mark Question) 1. Name the sugar present in milk. A: Lactose, 2. How many monosaccharide units are present in it? A: two monosaccharide units are present. 3.What are such oligosaccharides
Biomolecules Section A (One Mark Question) 1. Name the sugar present in milk. A: Lactose, 2. How many monosaccharide units are present in it? A: two monosaccharide units are present. 3.What are such oligosaccharides
Carbohydrates - Chemical Structure
 Carbohydrates - Chemical Structure Carbohydrates consist of the elements carbon (C), hydrogen (H) and oxygen (O) with a ratio of hydrogen twice that of carbon and oxygen. Carbohydrates include sugars,
Carbohydrates - Chemical Structure Carbohydrates consist of the elements carbon (C), hydrogen (H) and oxygen (O) with a ratio of hydrogen twice that of carbon and oxygen. Carbohydrates include sugars,
Computer modelling studies on the modes of binding of some of the α- glycosidically linked disaccharides to concanavalin A
 Proc. Int. Symp. Biomol. Struct. Interactions, Suppl. J. Biosci., Vol. 8, Nos 1 & 2, August 1985, pp. 389 401. Printed in India. Computer modelling studies on the modes of binding of some of the α- glycosidically
Proc. Int. Symp. Biomol. Struct. Interactions, Suppl. J. Biosci., Vol. 8, Nos 1 & 2, August 1985, pp. 389 401. Printed in India. Computer modelling studies on the modes of binding of some of the α- glycosidically
Lecture 10 More about proteins
 Lecture 10 More about proteins Today we're going to extend our discussion of protein structure. This may seem far-removed from gene cloning, but it is the path to understanding the genes that we are cloning.
Lecture 10 More about proteins Today we're going to extend our discussion of protein structure. This may seem far-removed from gene cloning, but it is the path to understanding the genes that we are cloning.
Downloaded from
 Chapter 14. Biomolecules Section A (One Mark Question) 1.Name the sugar present in milk. A: Lactose, 2.How many monosaccharide units are present in it? A: two monosaccharide units are present. 3. What
Chapter 14. Biomolecules Section A (One Mark Question) 1.Name the sugar present in milk. A: Lactose, 2.How many monosaccharide units are present in it? A: two monosaccharide units are present. 3. What
Macro molecule = is all the reactions that take place in cells, the sum of all chemical reactions that occur within a living organism Anabolism:
 Macromolecule Macro molecule = molecule that is built up from smaller units The smaller single subunits that make up macromolecules are known as Joining two or more single units together form a M is all
Macromolecule Macro molecule = molecule that is built up from smaller units The smaller single subunits that make up macromolecules are known as Joining two or more single units together form a M is all
Biochemistry: Macromolecules
 1 Biology: Macromolecules 2 Carbohydrates Carbohydrate organic compound containing carbon, hydrogen, & oxygen in a 1:2:1 ratio Meaning: hydrated carbon ratio of h:0 is 2:1 (same as in water) Source: plants
1 Biology: Macromolecules 2 Carbohydrates Carbohydrate organic compound containing carbon, hydrogen, & oxygen in a 1:2:1 ratio Meaning: hydrated carbon ratio of h:0 is 2:1 (same as in water) Source: plants
CHAPTER 3. Carbon & the Molecular Diversity of Life
 CHAPTER 3 Carbon & the Molecular Diversity of Life Carbon: The Organic Element Compounds that are synthesized by cells and contain carbon are organic So what is inorganic? Why are carbon compounds so prevalent?
CHAPTER 3 Carbon & the Molecular Diversity of Life Carbon: The Organic Element Compounds that are synthesized by cells and contain carbon are organic So what is inorganic? Why are carbon compounds so prevalent?
An aldose contains an aldehyde functionality A ketose contains a ketone functionality
 RCT Chapter 7 Aldoses and Ketoses; Representative monosaccharides. (a)two trioses, an aldose and a ketose. The carbonyl group in each is shaded. An aldose contains an aldehyde functionality A ketose contains
RCT Chapter 7 Aldoses and Ketoses; Representative monosaccharides. (a)two trioses, an aldose and a ketose. The carbonyl group in each is shaded. An aldose contains an aldehyde functionality A ketose contains
The Structure and Function of Macromolecules
 The Structure and Function of Macromolecules Macromolecules are polymers Polymer long molecule consisting of many similar building blocks. Monomer the small building block molecules. Carbohydrates, proteins
The Structure and Function of Macromolecules Macromolecules are polymers Polymer long molecule consisting of many similar building blocks. Monomer the small building block molecules. Carbohydrates, proteins
BIOLOGICAL MOLECULES. Although many inorganic compounds are essential to life, the vast majority of substances in living things are organic compounds.
 BIOLOGY 12 BIOLOGICAL MOLECULES NAME: Although many inorganic compounds are essential to life, the vast majority of substances in living things are organic compounds. ORGANIC MOLECULES: Organic molecules
BIOLOGY 12 BIOLOGICAL MOLECULES NAME: Although many inorganic compounds are essential to life, the vast majority of substances in living things are organic compounds. ORGANIC MOLECULES: Organic molecules
Macromolecules. Note: If you have not taken Chemistry 11 (or if you ve forgotten some of it), read the Chemistry Review Notes on your own.
 Macromolecules Note: If you have not taken Chemistry 11 (or if you ve forgotten some of it), read the Chemistry Review Notes on your own. Macromolecules are giant molecules made up of thousands or hundreds
Macromolecules Note: If you have not taken Chemistry 11 (or if you ve forgotten some of it), read the Chemistry Review Notes on your own. Macromolecules are giant molecules made up of thousands or hundreds
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10913 Supplementary Figure 1 2F o -F c electron density maps of cognate and near-cognate trna Leu 2 in the A site of the 70S ribosome. The maps are contoured at 1.2 sigma and some of
doi:10.1038/nature10913 Supplementary Figure 1 2F o -F c electron density maps of cognate and near-cognate trna Leu 2 in the A site of the 70S ribosome. The maps are contoured at 1.2 sigma and some of
2.2 Cell Construction
 2.2 Cell Construction Elemental composition of typical bacterial cell C 50%, O 20%, N 14%, H 8%, P 3%, S 1%, and others (K +, Na +, Ca 2+, Mg 2+, Cl -, vitamin) Molecular building blocks Lipids Carbohydrates
2.2 Cell Construction Elemental composition of typical bacterial cell C 50%, O 20%, N 14%, H 8%, P 3%, S 1%, and others (K +, Na +, Ca 2+, Mg 2+, Cl -, vitamin) Molecular building blocks Lipids Carbohydrates
GLYCAN STRUCTURES, CLUES TO THE ORIGIN OF SACCHARIDES
 GLYCAN STRUCTURES, CLUES TO THE ORIGIN OF SACCHARIDES Jun Hirabayashi Department of Biological Chemistry, Faculty of Pharmaceutical Sciences, Teikyo University Sagamiko, Kanagawa 199-0195, Japan Tel: 0426-85-3741
GLYCAN STRUCTURES, CLUES TO THE ORIGIN OF SACCHARIDES Jun Hirabayashi Department of Biological Chemistry, Faculty of Pharmaceutical Sciences, Teikyo University Sagamiko, Kanagawa 199-0195, Japan Tel: 0426-85-3741
Biological Sciences B. CHEM. ENGG. Part 1. Chemicals of life: Water, Amino acids, Carbohydrates, Lipids. Dr. Ratnesh Jain
 Biological Sciences B. CHEM. ENGG. Part 1. Chemicals of life: Water, Amino acids, Carbohydrates, Lipids Dr. Ratnesh Jain The Foundations Content Cellular Foundations Chemical Foundations Physical Foundations
Biological Sciences B. CHEM. ENGG. Part 1. Chemicals of life: Water, Amino acids, Carbohydrates, Lipids Dr. Ratnesh Jain The Foundations Content Cellular Foundations Chemical Foundations Physical Foundations
KEY NAME (printed very legibly) UT-EID
 BIOLOGY 311C - Brand Spring 2007 KEY NAME (printed very legibly) UT-EID EXAMINATION II Before beginning, check to be sure that this exam contains 7 pages (including front and back) numbered consecutively,
BIOLOGY 311C - Brand Spring 2007 KEY NAME (printed very legibly) UT-EID EXAMINATION II Before beginning, check to be sure that this exam contains 7 pages (including front and back) numbered consecutively,
Guided Inquiry Skills Lab. Additional Lab 1 Making Models of Macromolecules. Problem. Introduction. Skills Focus. Materials.
 Additional Lab 1 Making Models of Macromolecules Guided Inquiry Skills Lab Problem How do monomers join together to form polymers? Introduction A small number of elements make up most of the mass of your
Additional Lab 1 Making Models of Macromolecules Guided Inquiry Skills Lab Problem How do monomers join together to form polymers? Introduction A small number of elements make up most of the mass of your
Introduction to Macromolecules. If you were to look at the nutrition label of whole milk, what main items stick out?
 Introduction to Macromolecules Macromolecules are a set of molecules that are found in living organisms. Macromolecules essentially mean big molecules as the word macro means large. The functions of these
Introduction to Macromolecules Macromolecules are a set of molecules that are found in living organisms. Macromolecules essentially mean big molecules as the word macro means large. The functions of these
2.1.1 Biological Molecules
 2.1.1 Biological Molecules Relevant Past Paper Questions Paper Question Specification point(s) tested 2013 January 4 parts c and d p r 2013 January 6 except part c j k m n o 2012 June 1 part ci d e f g
2.1.1 Biological Molecules Relevant Past Paper Questions Paper Question Specification point(s) tested 2013 January 4 parts c and d p r 2013 January 6 except part c j k m n o 2012 June 1 part ci d e f g
Sheet #10 Dr. Mamoun Ahram Sec 1,2,3 15/07/2014. Carbohydrates 2
 Carbohydrates 2 A study Guide: Kindly,refer to the slide number,look at the structures and read the sheet notes well,most of the slides content besides all what the doctor said are mentioned here,good
Carbohydrates 2 A study Guide: Kindly,refer to the slide number,look at the structures and read the sheet notes well,most of the slides content besides all what the doctor said are mentioned here,good
BIOCHEMISTRY I HOMEWORK III DUE 10/15/03 66 points total + 2 bonus points = 68 points possible Swiss-PDB Viewer Exercise Attached
 BIOCHEMISTRY I HOMEWORK III DUE 10/15/03 66 points total + 2 bonus points = 68 points possible Swiss-PDB Viewer Exercise Attached 1). 20 points total T or F (2 points each; if false, briefly state why
BIOCHEMISTRY I HOMEWORK III DUE 10/15/03 66 points total + 2 bonus points = 68 points possible Swiss-PDB Viewer Exercise Attached 1). 20 points total T or F (2 points each; if false, briefly state why
Biochemistry - I. Prof. S. Dasgupta Department of Chemistry Indian Institute of Technology, Kharagpur Lecture 1 Amino Acids I
 Biochemistry - I Prof. S. Dasgupta Department of Chemistry Indian Institute of Technology, Kharagpur Lecture 1 Amino Acids I Hello, welcome to the course Biochemistry 1 conducted by me Dr. S Dasgupta,
Biochemistry - I Prof. S. Dasgupta Department of Chemistry Indian Institute of Technology, Kharagpur Lecture 1 Amino Acids I Hello, welcome to the course Biochemistry 1 conducted by me Dr. S Dasgupta,
Why Carbon? What does a carbon atom look like?
 Biomolecules Organic Chemistry In the 1800 s it was believed to be impossible to recreate molecules in a lab Thus, the study of organic chemistry was originally the study of molecules in living organisms
Biomolecules Organic Chemistry In the 1800 s it was believed to be impossible to recreate molecules in a lab Thus, the study of organic chemistry was originally the study of molecules in living organisms
Molecular Dynamics of Sialic Acid Analogues and their Interaction with Influenza Hemagglutinin
 Research Papers www.ijpsonline.com Molecular Dynamics of Sialic Acid Analogues and their Interaction with Influenza Hemagglutinin T. FEMLIN BLESSIA, V. S. RAPHEAL 1 AND D. J. S. SHARMILA* Department of
Research Papers www.ijpsonline.com Molecular Dynamics of Sialic Acid Analogues and their Interaction with Influenza Hemagglutinin T. FEMLIN BLESSIA, V. S. RAPHEAL 1 AND D. J. S. SHARMILA* Department of
