RNP LOCALIZATION AND TRANSPORT IN YEAST
|
|
|
- Shawn Lamb
- 5 years ago
- Views:
Transcription
1 Annu. Rev. Cell Dev. Biol : Copyright c 2001 by Annual Reviews. All rights reserved RNP LOCALIZATION AND TRANSPORT IN YEAST Pascal Chartrand and Robert H. Singer Departments of Anatomy and Structural Biology, and Cell Biology, Albert-Einstein College of Medicine, Bronx, New York 10461; chartran@aecom.yu.edu; rhsinger@aecom.yu.edu Roy M. Long Department of Microbiology and Molecular Genetics, Medical College of Wisconsin, Milwaukee, Wisconsin ; rlong@mcw.edu Key Words ASH1 mrna, RNA localization, She proteins, locasome, myosin Abstract The localization of mrnas is used by various types of polarized cells to locally translate specific proteins, which restricts their distribution to a particular sub-region of the cytoplasm. This mechanism of protein sorting is involved in major biological processes such as asymmetric cell division, oogenesis, cellular motility, and synapse formation. With the finding of localized mrnas in the yeast Saccharomyces cerevisiae, it is now possible to benefit from the powerful yeast laboratory tools to explore the molecular basis of RNA localization. Because mrna transport and localization in yeast share many features with RNA localization in higher eukaryotes, including the formation of a large ribonucleoprotein (RNP) localization complex, the requirement of a polarized cytoskeleton and molecular motors, and the role of nuclear RNA-binding proteins in cytoplasmic localization, the yeast can be used as a paradigm for unraveling the molecular aspects of this process. This review summarizes the current knowledge on RNP transport and localization in yeast. CONTENTS INTRODUCTION LOCALIZED RNA IN YEAST: ROLES AND FUNCTIONS ASH IST ATM CIS-ACTING ELEMENTS THAT TARGET AN RNA TO ITS SPECIFIC LOCATION ASH1 mrna IST2 mrna ATM1 mrna TRANS-ACTING FACTORS INVOLVED IN RNA LOCALIZATION Myo4p/She1p /01/ $
2 298 CHARTRAND SINGER LONG She3p She2p A Nuclear Factor Involved in RNA Localization: Loc1p ROLE OF THE ACTIN CYTOSKELETON AND ASSOCIATED PROTEINS The Actin Cytoskeleton in Yeast She4p Bni1/She5p ANCHORING THE RNA AT THE LOCALIZATION SITE Role of mrna Translation in Anchoring Trans-Acting Factors Involved in Anchoring MECHANISM OF RNA TRANSPORT AND LOCALIZATION Kinetic Aspects of RNA Localization The Locasome: A Localization Machine PERSPECTIVES INTRODUCTION Establishing and maintaining cellular polarity requires the asymmetric segregation of specific proteins. A classic mechanism for segregating proteins relies on targeting sequences contained within the amino acid sequence of the protein. Alternatively, the localization of specific mrnas to the site of action of the protein is another mechanism employed by cells for intracellular sorting of proteins. At least three mechanisms have been identified for localizing mrnas: (a) direct transport of the mrna, (b) stabilization of the mrna at the site of localization, and (c) trapping the mrna at the site of localization (see review by Lipshitz & Smibert 2000). In the first pathway, mrnas destined for localization are packaged into ribonucleoprotein particles that are directly transported and anchored at the site of localization. Transport of the ribonucleoprotein (RNP) particles is dependent on motor proteins and cytoskeletal filaments. In a second pathway, localized mrnas are preferentially stabilized at the localization site and degraded at all other cellular positions. In a third pathway, localized mrnas are freely diffusible throughout the cell but upon reaching the site of localization are trapped at this position within the cell. In Drosophila, Xenopus, and zebrafish, mrna localization is responsible for the segregation of cell-fate determinants between daughter cells during development (Bashirullah et al. 1998). However, localization of mrna is not restricted to development. In polarized somatic cells of higher vertebrates such as fibroblasts and neurons, mrna localization is involved in the intracellular sorting of specific proteins within these cell types (Bassell et al. 1999). The yeast Saccharomyces cerevisiae also localizes specific mrnas to asymmetrically segregate proteins. The genetic and cell biological approaches available in yeast have allowed the identification and characterization of various factors involved in mrna localization at a pace unrivaled by other model systems. Consequently, yeast is an important route
3 mrna LOCALIZATION IN YEAST 299 toward understanding the molecular basis of mrna transport and localization in higher eukaryotes. This review highlights mrna localization in S. cerevisiae, focusing on the interplay between cis- and trans-acting localization factors. To date, three localized mrnas have been identified in S. cerevisiae: ASH1, IST2, and ATM1. LOCALIZED RNA IN YEAST: ROLES AND FUNCTIONS ASH1 ASH1 was identified in mutant selections designed to identify cell-type-specific regulators of HO expression, which is involved in mating-type switching in yeast (Bobola et al. 1996, Sil & Herskowitz 1996). In wild-type yeast cells, the HO endonuclease is expressed exclusively in mother cells. This results in mating-type switching in mother cells, whereas daughter cells maintain their original mating type. However, in ash1 cells, HO is active in both mother cells and daughter cells, demonstrating that Ash1p is a cell-type-specific regulator of HO expression. By immunofluorescence, Ash1p was observed exclusively in daughter cell nuclei where it functions as a transcriptional repressor of HO expression, restricting HO expression to mother cell nuclei (Bobola et al. 1996, Sil & Herskowitz 1996). It was observed by fluorescent in situ hybridization (FISH) (Figure 1) that during anaphase, ASH1 mrna localizes to the distal tip of daughter cells, which results in the asymmetric segregation of Ash1p (Long et al. 1997, Takizawa et al. 1997). Ash1p also functions in pseudohyphal growth (Chandarlapaty & Errede 1998, Pan & Heitman 2000). In response to nitrogen limitation, S. cerevisiae displays a dimorphic transition to a filamentous pseudohyphal morphology. In ash1 cells, pseudohyphal growth is not observed, whereas overexpression of ASH1 stimulates pseudohyphal growth. However, it remains to be determined if ASH1 mrna localization is required for pseudohyphal differentiation. IST2 In a genome-wide screen to identify additional mrnas that preferentially interact with the trans-acting factors required for ASH1 mrna localization, 11 mrnas, including IST2, were found to be associated with these RNA localization factors (Takizawa et al. 2000). From these 11 mrnas, only IST2 mrna was localized to daughter cells. In contrast to ASH1 mrna, IST2 mrna was localized to daughter cells throughout the cell cycle. IST2 mrna localization apparently functions to segregate Ist2p to the plasma membrane of medium- and large-budded cells. Ist2p, which has similarities to calcium and sodium channel proteins (Mannhaupt et al. 1994), has been reported to increase the sodium tolerance of yeast, but it remains to be determined why Ist2p is specifically segregated to daughter cells.
4 300 CHARTRAND SINGER LONG ATM1 ATM1 is a nuclear gene encoding a mitochondrial ATP-binding cassette (ABC) transporter (Leighton & Schatz 1995). Through biochemical fractionation, ATM1 mrna was observed to preferentially associate with mitochondria (Corral- Debrinski et al. 2000). Using a green fluorescent protein (GFP) reporter assay to monitor mrna in living yeast cells, two regions of ATM1 mrna were found to be sufficient to localize a heterologous reporter mrna to mitochondria. Transacting factors required for ATM1 mrna localization remain to be identified. These observations demonstrate that RNA localization is involved in targeting nuclearencoded proteins to mitochondria. CIS-ACTING ELEMENTS THAT TARGET AN RNA TO ITS SPECIFIC LOCATION Specific cis-acting element(s), known as zipcodes (Kislauskis & Singer 1992), must be present within the sequence of a localized mrna in order to target this mrna to a specific region in the cell cytoplasm. Usually present in the 3 untranslated region (3 UTR) of the mrna, the localization element(s) is recognized by the cell localization machinery. ASH1 mrna To identify cis-acting sequences responsible for the localization of the ASH1 mrna, fragments of ASH1 were inserted into a reporter mrna, and the cytoplasmic distribution of these chimeric mrnas was determined by FISH (Chartrand et al. 1999, Gonzalez et al. 1999). By this method, four ASH1 localization elements, each sufficient to localize a reporter mrna to the bud of dividing yeast cells, have been identified: three within the coding sequence (elements E1, E2A, and E2B) and one overlapping the stop codon (E3). Element E3 includes the last 15 nucleotides of the coding sequence and the first 100 nucleotides of the 3 UTR. Interestingly, in most known localized mrnas, the localization elements are located in the 3 UTR of the RNA, whereas all the ASH1 localization elements are located within or in part of the coding sequence. This unusual disposition of localization elements suggests an interaction between the RNA localization machinery and the translation machinery during transport of this mrna. A detailed mutagenesis analysis of the four elements reveals that although they perform a similar function, these elements have distinct sequences predicted to fold into different stem-loop structures (Chartrand et al. 1999, Gonzalez et al. 1999). These mutagenesis experiments also demonstrated that secondary and tertiary structures are important for recognition by the trans-acting factors involved in ASH1 mrna localization. Because all four elements appear to interact with the same RNA-binding protein, She2p (see below), it is likely that a structural motif shared between these four elements is recognized by She2p. In addition to She2p, RNA localization driven by each of these elements is dependent on She1p, She3p,
5 mrna LOCALIZATION IN YEAST 301 She4p, and She5p (see below) but is independent of Ash1p (Long et al. 1997, Takizawa et al. 1997). IST2 mrna Cis-acting localization elements for this RNA remain to be determined. ATM1 mrna Two independent cis-acting elements in the ATM1 mrna have been shown to localize a reporter mrna to mitochondria: one within the first 48 nucleotides of the N-terminal mitochondria-targeting sequence (mts) and one in the 3 UTR (Corral-Debrinsky et al. 2000). Interestingly, the localization function of the mts element appears to be translation independent, suggesting that the mts could perform two roles: mrna localization as a cis-acting RNA element and protein translocation into mitochondria as a peptide signal sequence. TRANS-ACTING FACTORS INVOLVED IN RNA LOCALIZATION A genetic selection for cell-type-specific regulators of HO expression identified five genes, SHE1 to 5, essential for asymmetric HO expression (Jansen et al. 1996). A mutation in any of these genes results in the repression of HO expression in both cell types (Bobola et al. 1996), and ASH1 mrna is symmetrically distributed between mother and daughter cells (Long et al. 1997, Takizawa et al. 1997). Numerous studies have now demonstrated that She1p, She2p, and She3p are components of a ribonucleoprotein particle that directly transports ASH1 and IST2 mrna to daughter cells. These observations are described below and are summarized in Table 1. Myo4p/She1p She3p SHE1 is identical to MYO4, which codes for a type V unconventional myosin that localizes to daughter cells (Haarer et al. 1994, Jansen et al. 1996). By immunoprecipitation, Myo4p associates with ASH1 mrna, which is dependent on She2p and She3p (Münchow et al. 1999, Takizawa & Vale 2000). Furthermore, it was observed that Myo4p colocalizes with ASH1 mrna-containing particles (Bertrand et al. 1998, Takizawa & Vale 2000), and in living yeast cells, Myo4p directly transports ASH1 mrna to daughter cells (Bertrand et al. 1998, Beach et al. 1999). Because Myo4p does not contain intrinsic RNA-binding activity, accessory proteins are necessary to interface the myosin with ASH1 mrna localization elements. She3p is a novel protein with no significant homology to any known proteins. She3p colocalizes with Myo4p and ASH1 mrna at the bud tip (Münchow et al. 1999). Furthermore, by two-hybrid, three-hybrid, and coimmunoprecipitation experiments,
6 302 CHARTRAND SINGER LONG TABLE 1 Trans-acting factors involved in ASH1 mrna localization Role in Location Name Function ASH1 localization Interactions in the cell She1p/ Type V Molecular motor She3p Bud tip Myo4p unconventional myosin She2p RNA-binding Binds ASH1 mrna She3p Cytoplasm/ protein localization elements bud tip She3p Myosin-binding Bridges She2p with the She1p Bud tip protein the myosin She1p She2p She4 Unknown Stabilizes the actin Unknown Cytoplasm cytoskeleton (?) She5p/ Scaffold protein Involved in the polarization Cdc42p, Bud6p Bud tip Bni1p of the actin cytoskeleton EF1α, Pfy1p Loc1p RNA-binding Binds ASH1 localization Unknown Nucleus protein elements; efficiency factor She2p the N-terminal half of the She3p was found to interact with the C-terminal tail of Myo4p (Münchow et al. 1999, Böhl et al. 2000, Long et al. 2000). It is hypothesized that the interaction between Myo4p and She3p occurs through coiled-coil domains located in both proteins (Böhl et al. 2000). By immunoprecipitation and threehybrid experiments, She3p association with ASH1 mrna was observed to be dependent on She2p (Münchow et al. 1999, Long et al. 2000, Takizawa & Vale 2000). These results suggested that the Myo4p-She3p complex interacts with the ASH1 mrna cis-acting RNA localization elements through She2p. She2p is also a novel protein with no homology to known proteins. Two-hybrid analysis and experiments using recombinant She2p and She3p demonstrate that She2p directly interacts with She3p through a domain in the C terminus of She3p (Böhl et al. 2000, Long et al. 2000). These results support the hypothesis that She2p functions to interface the Myo4p-She3p complex to ASH1 mrna (Böhl et al. 2000, Long et al. 2000). The hypothesis implies that She2p could be the RNA-binding protein required for ASH1 mrna localization. By electrophoretic gel mobility shift (EMSA) and UV cross-linking assays, it was observed that purified recombinant She2p could directly and specifically bind each of the ASH1 cis-acting localization elements (Böhl et al. 2000, Long et al. 2000). It is not known whether the simultaneous interaction of She2p with each of the cis-acting elements is additive or cooperative. The RNA-binding and She3p interaction domains of She2p remain to be identified. The function of She2p in ASH1 mrna localization is apparently restricted to bridge the Myo4p-She3p complex to cis-acting localization elements. When the
7 mrna LOCALIZATION IN YEAST 303 Myo4p-She3p complex was artificially tethered to a heterologous reporter mrna, Myo4p-She3p-dependent RNA localization was observed to be independent of She2p (Long et al. 2000). The function of She2p in yeast cells is apparently not limited to ASH1 and IST2 mrna localization. By immunofluorescence, She2p was observed to be uniformly distributed between mother and daughter cells (Böhl et al. 2000). However, when ASH1 mrna is overexpressed, She2p accumulates at the bud tip (Böhl et al. 2000). Furthermore, She2p can preferentially associates with at least 10 unlocalized mrnas (Takizawa et al. 2000). However, the function of She2p with these other mrnas remains to be determined. A Nuclear Factor Involved in RNA Localization: Loc1p In higher eukaryotic organisms, heterogeneous nuclear ribonucleoproteins (hnrnp) RNA-binding proteins such as Squid (Lall et al. 1999) and hnrnp A2 (Hoeck et al. 1998) have been shown to function in mrna localization. These proteins shuttle between the nucleus and cytoplasm, suggesting a role for a nuclear component in cytoplasmic mrna localization. In yeast, one trans-acting factor, Loc1p, was found to be a nuclear protein (Long et al. 2001). Loc1p is a very basic (pi 10), double-stranded RNA-binding protein identified in a three-hybrid screen for RNA-binding proteins specific for ASH1 cis-acting localization elements. Purified recombinant Loc1p was found to interact only with double-stranded RNA (Long et al. 2001). In loc1 cells, ASH1 mrna is predominantly delocalized, and in those cases, Ash1p is symmetrically distributed between mother and daughter nuclei. Loc1p is a nuclear protein, but in contrast to the proteins Squid and hnrnp A2, Loc1p does not shuttle between the nucleus and the cytoplasm. One possible function of Loc1p might be to interface the ASH1 mrna with a nucleo-cytoplasmic shuttling protein (perhaps She2p), which is required for ASH1 mrna localization. Another possibility is that localized mrna could be tagged by Loc1p in the nucleus after its synthesis in order to be recognized by the cytoplasmic localization machinery. Further studies are ongoing to determine if Loc1p interacts with nucleo-cytoplasmic shuttling proteins. ROLE OF THE ACTIN CYTOSKELETON AND ASSOCIATED PROTEINS The Actin Cytoskeleton in Yeast Transport of mrna by a molecular motor along the cytoskeleton is the main mechanism by which polarized cells localize mrnas (Lipshitz & Smibert 2000). The finding that a myosin motor (Myo4p) is involved in the localization of the ASH1 and IST2 mrnas points to a role for actin cytoskeleton being the track that directs mrna to the localization site. The yeast actin cytoskeleton is primarily organized in a network of F-actin cables that run parallel to the longitudinal axis of the cell. These cables are highly
8 304 CHARTRAND SINGER LONG She4p dynamic and polarized, with their barbed ends directed toward the growing bud during anaphase (Pruyne & Bretscher 2000). The polarized growth of the actin cytoskeleton is responsible for the formation of the bud during mitosis (Adams & Pringle 1984), formation of the mating shmoo (Gehrung & Snyder 1990), polarized secretion (Finger & Novick 1998), and mitochondrial inheritance (for review, see Drubin et al. 1993, Botstein et al. 1997). Using a temperature-sensitive mutant allele act1-133, which affects the myosinbinding site on actin, Long et al. (1997) showed that ASH1 mrna is delocalized at the non-permissive temperature. Mutations in actin-associated proteins such as tropomyosin (tpm1) and profilin ( pfy1) also affect ASH1 mrna localization (Long et al. 1997). Because all these mutants affect the formation of actin cables that extend from the mother cell to the bud, these observations strongly suggest that the integrity of the actin cytoskeleton is essential for the proper transport of this mrna. In contrast, the localization of ASH1 mrna was not affected in a tubulin (tub2) mutant strain, which is defective in the formation of the astral microtubule, suggesting that the anaphase microtubule network is not required for mrna localization (Long et al. 1997). Another approach to study the role of the yeast cytoskeleton in mrna localization is to use drugs that specifically depolymerize either the actin or the microtubule cytoskeleton. In the presence of latrunculin A, an actin-depolymerizing drug, anaphase yeast cells fail to localize the ASH1 mrna (Takizawa et al. 1997). However, the addition of nocodazole, a microtubule-depolymerizing drug, to a yeast culture does not result in any defect in ASH1 mrna localization (Takizawa et al. 1997). Altogether, these results show that the transport of the ASH1 mrna to the bud tip of budding yeasts requires an intact actin cytoskeleton. Other proteins found to be essential for the localization of the ASH1 mrna are also involved in the formation and polarization of the actin cytoskeleton: SHE4 is among the five SHE genes essential for the proper localization of the ASH1 mrna (Jansen et al. 1996). SHE4 was also identified in a screen for yeast endocytosis mutants (Wendland et al. 1996). Although its specific function is not known (SHE4 is a non-essential gene), the actin cytoskeleton is altered in she4 cells, suggesting that She4p plays a role in actin polarization (Wendland et al. 1996). This result suggests that She4p functions indirectly in the localization of ASH1 mrna and indicates that She4p is probably not a direct member of the mrna localization machinery. Bni1/She5p Bni1p is a member of the formin family of proteins, which is defined by the presence of the proline-rich formin (FH) homology domains (Frazier et al. 1997). These proteins play an important role in polarization of the actin cytoskeleton and affect processes like cytokinesis, establishment of cell polarity, and polarized growth. A
9 mrna LOCALIZATION IN YEAST 305 bni1 strain is viable but shows defects in the organization of the actin cytoskeleton (Evangelista et al. 1997). Bni1p localizes at the site of bud growth, where the actin cytoskeleton is actively reorganized (Ozaki-Kuroda et al. 2001). One possible role of Bni1p is to act as a scaffold to recruit different factors involved in actin polymerization (Frazier et al. 1997). The formin domains of Bni1p have been shown to interact with profilin, an actin-binding protein implicated in actin polymerization (Imamura et al. 1997). Bni1p also interacts with EF1αp and Bud6p/Aip3p, which bind to and bundle actin filaments (Evangelista et al. 1997, Umikawa et al. 1998), and Rho1p and Cdc42, both Rho-related GTPase involved in the regulation of actin polymerization (Evangelista et al. 1997). Because of its role in the actin cytoskeleton organization, Bni1p probably functions indirectly in the localization of ASH1 mrna. However, some data suggest that it could play a role in the anchoring of this mrna at the bud tip (Beach et al. 1999; see below). Unlike the other she mutants, cells containing a SHE5 mutation accumulate ASH1 mrna at the bud neck between the mother and the daughter cells (Long et al. 1997, Takizawa et al. 1997). This particular phenotype may reflect a disruption of actin polarity in the bud. ANCHORING THE RNA AT THE LOCALIZATION SITE After the transport of mrna to its localization site, it must be anchored there in order to avoid diffusion throughout the cytoplasm. In the case of the ASH1 mrna, a tight localization of this mrna is observed at the bud tip during lateanaphase (Figure 1) (Long et al. 1997). IST2 mrna also localizes tightly at the bud tip (Takizawa et al. 2000). Several lines of evidence point to a role of various redundant factors involved in the anchoring process of mrna in yeast. Role of mrna Translation in Anchoring A potential factor involved in the anchoring of the ASH1 mrna could be the translated protein itself, Ash1p. Because Ash1p is translated at the bud tip and is a nucleic acid binding protein, it is reasonable to suggest that it may be involved in the anchoring of its own mrna at the bud tip. Such a case is known in Drosophila, where the Oskar protein is involved in the anchoring of its own messenger RNA at the posterior pole of the Drosophila oocyte (Rongo et al. 1995). Indeed, the insertion of a stop codon after the initiation codon results in a less tightly localized ASH1 mrna at the bud tip (Gonzalez et al. 1999). Moreover, the presence of a stop codon in the middle of the coding sequence also displays a partially localized phenotype, suggesting that the C-terminal half of the protein could be involved in the anchoring of the mrna. Interestingly, the C-terminal half of Ash1p contains a zinc-finger domain involved in DNA binding (Bobola et al. 1996, Sil & Herskowitz 1996). Because zinc fingers are also known to bind RNA (Burd & Dreyfuss 1994), one model could be that when the Ash1p is translated at the bud tip, it may bind
10 306 CHARTRAND SINGER LONG and anchor its own mrna at the cell cortex. Another possibility could be that the translation of the mrna is needed to reveal a cis-acting element involved in the anchoring of the RNA (Gonzalez et al. 1999). Trans-Acting Factors Involved in Anchoring Other trans-acting factors can also be involved in the anchoring of the ASH1 mrna. Obvious candidates are proteins present at the bud tip. These proteins are generally involved in bud site selection and polarized cell growth (Chant 1999), but some could potentially serve as anchors for messenger RNAs localized at the bud tip (Pruyne & Bretscher 2000). Several of these factors may have been identified by Beach et al. (1999) using a reporter RNA containing a single ASH1 localization element to follow the dynamics of localization and anchoring in living yeasts. They observed that in bni1/she5 and bud6/aip3 strains the reporter RNA was still asymmetrically distributed to the bud but had lost its cortical localization. Because both Bni1p/She5p and Bud6p/Aip3p are proteins localized to the cortex of the bud tip (Amberg et al. 1997, Ozaki-Kuroda et al. 2001), it is possible that these proteins are involved, directly or indirectly, in the anchoring of the localized ASH1 mrna. However, the endogenous ASH1 mrna appears not to be affected by a bud6 mutation (R.M. Long, unpublished observation; R.P. Jansen, personal communication), suggesting that redundant factors are involved in the anchoring of the full-length mrna. MECHANISM OF RNA TRANSPORT AND LOCALIZATION Kinetic Aspects of RNA Localization Since mrna localization is a dynamic process, the kinetic and temporal aspects of this mechanism remain to be revealed. Questions such as how fast the mrna is transported, how long the process takes, and when it is translated need to be answered. In the case of the ASH1 mrna, for instance, the timing of mrna transcription, localization, and translation must be carefully regulated in order to avoid contamination of Ash1p into the mother s nucleus before cytokinesis. Some of these questions were approached by the development of a new technique for the visualization of mrna in living yeast cells. GFP was fused to an RNA-binding protein (MS2 RNA-binding domain) and coexpressed with a reporter mrna containing RNA motifs (MS2-binding sites) recognized by this protein (Bertrand et al. 1998, Beach et al. 1999). A similar system was also developed with the U1A RNA-binding protein (Takizawa & Vale 2000, Brodsky & Silver 2000). When an ASH1 localization element is present in the reporter mrna, a single bright particle consisting of the GFP-MS2 fusion protein and the reporter mrna is seen localized at the bud tip of budding yeasts. The formation of this particle requires an intact localization element (Chartrand et al. 1999) and the proteins She2 and She3 (Bertrand et al. 1998), which suggests that it contains components of the localization machinery of the ASH1 mrna.
11 mrna LOCALIZATION IN YEAST 307 Using video microscopy, it was possible to follow the movement of this particle from the mother cell cytoplasm to the bud tip (Bertrand et al. 1998). The particle moved at velocities that varied between 200 and 440 nm/s, which is consistent with the velocity expected from a myosin V motor, e.g., Myo4p, which is reported to move between 200 and 400 nm/s (Cheney et al. 1993). Overall, it took 128 s for the particle to be transported over a net linear distance of 4.4 µm, from the mother cell cytoplasm to the bud tip. When the GFP-MS2 construct was expressed with the reporter mrna in a strain deleted of the MYO4/SHE1 gene, the particle exhibited diffusion only within the cytoplasm of the mother cell (Bertrand et al. 1998). Altogether, these results indicate that Myo4p directly transports the ASH1 particle to the bud tip. Moreover, it shows that the ASH1 mrna can be transported and localized within 2 min after its export from the nucleus. The Locasome: A Localization Machine The transport of an mrna to its site of localization usually requires the assembly of a multiprotein complex, the localization machine, on the RNA localization element(s). The resulting RNP complex, called a locasome (Bertrand et al. 1998, Bassell et al. 1999), consists minimally of a protein motor, an RNA-binding protein, and the localizing mrna (Long et al. 2000). The formation of a localization complex appears to be a characteristic of several localized mrnas in various organisms. It has been observed as particles or granules in different systems: in vivo using fluorophore-labeled mrna (Ainger et al. 1993) or GFP-labeled localization trans-acting factors (Wang & Hazelrigg 1994, Kohrmann et al. 1999) or in vitro by biochemical purification (Wilhelm et al. 2000). In living yeast, using the GFP-MS2 fusion protein and the lacz-ash1 mrna system, the ASH1 locasome is seen as a single large particle in the cell cytoplasm (see above; Bertrand et al. 1998, Beach et al. 1999). This phenotype is possibly created by the aggregation of multiple small particles into a single large one. This particle was found by in situ hybridization to contain the ASH1 mrna, and its formation requires the presence of intact SHE2 and SHE3 genes (Bertrand et al. 1998). Moreover, the proteins She1p, She2p, and She3p coimmunoprecipitated with the GFP-MS2/lacZ-ASH1 mrna complex (W. Gu & R.H. Singer, unpublished observations), suggesting that this particle contains the main trans-acting factors involved in ASH1 mrna localization. Because the same trans-acting factors are involved in the localization of the IST2 mrna (Takizawa et al. 2000), a single locasome could possibly contain different mrnas. From all the genetic, cell biology, and biochemical studies discussed above on the various cis- and trans-acting factors involved in ASH1 mrna localization, we propose the following model for the assembly of the ASH1 mrna locasome in the yeast cytoplasm (Figure 2). As shown previously, (1) the localization elements in the ASH1 mrna sequence are bound by the She2 protein. (2) She2p interacts with the C-terminal domain of She3p. (3) The N-terminal coiled-coil domain of She3p then associates with a similar coiled-coil domain at the C-terminal end of the She1p myosin. (4) Together, this complex interacts
12 308 CHARTRAND SINGER LONG with the actin cytoskeleton via the myosin. Although this model is based on the core components of the localization machinery (She1p, She2p, and She3p), other unidentified trans-acting factors would also be expected to be part of this complex, e.g., factors involved in the translation or the stability of the mrna. A general working model for the localization of the ASH1 mrna in yeast can also be proposed (Figure 3), where this mrna is first recognized in the nucleus by nuclear proteins, such as Loc1p, which act as tags for the localization machinery (1). Once in the cytoplasm, the localization machinery completes its assembly on the tagged mrna and forms the locasome (3). The locasome is transported along the actin cytoskeleton up to the bud tip (5) where it becomes anchored (7). Once localized, the mrna is translated (8), and the Ash1 protein is synthesized at the bud tip juxtanuclearly and then is imported into the daughter cell nucleus. PERSPECTIVES Remarkable progress has been made in the characterization of the molecular process behind RNA localization in yeast. Although several features of this process are specific to yeast, the broad outlines of the mechanism are certainly conserved across the eukaryotic kingdom: for instance, the role of nuclear factors, the formation of a locasome by the localization machinery, and the importance of the cytoskeleton and associated proteins in this process. These aspects are also present in RNA localization pathways in other biological systems. However, several aspects of this process are still poorly understood: for example, the role of nuclear factors in RNA localization, the mechanism of mrna anchoring, and the assembly of the various trans-acting factors and the mrna into a locasome. Filling in the details of the mrna localization mechanism will occupy a new generation of investigators over the next years. Visit the Annual Reviews home page at LITERATURE CITED Adams A, Pringle J Relationship of actin and tubulin distribution to bud-growth in wild-type and morphogenetic mutant Saccharomyce cerevisiae. J. Cell Biol. 98: Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, et al Transport and localization of exogenous MBP mrna microinjected into oligodendrocytes. J. Cell. Biol. 123: Amberg DC, Zahner JE, Mulholland JW, Pringle JR, Botstein D Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol. Biol. Cell 8: Bashirullah A, Cooperstock RL, Lipshitz HD RNA localization in development. Annu. Rev. Biochem. 67: Bassell GJ, Oleynikov Y, Singer RH The travels of mrnas through all cells large and small. FASEB J. 13: Beach DL, Salmon ED, Bloom K Localization and anchoring of mrna in budding yeast. Curr. Biol. 9:569 78
13 mrna LOCALIZATION IN YEAST 309 Bertrand E, Chartrand P, Schaeffer M, Shenoy SM, Singer RH, Long RM Localization of ASH1 mrna particles in living yeast. Mol. Cell 2: Bobola N, Jansen RP, Shin TH, Nasmyth K Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell 84: Böhl F, Kruse C, Frank A, Ferring D, Jansen RP She2p, a novel RNA-binding protein tethers ASH1 mrna to the Myo4p myosin motor via She3p. EMBO J. 19: Botstein D, Amberg D, Mulholland J The yeast cytoskeleton. In The Molecular and Cellular Biology of the Yeast Saccharomyces, ed. JR Pringle, JR Broach, EW Jones, 3:1 90. New York: Cold Spring Harbor Lab. Press pp. Brodsky AS, Silver PA Pre-mRNA processing factors are required for nuclear export. RNA 6: Burd CG, Dreyfuss G Conserved structures and diversity of functions of RNAbinding proteins. Science 265: Chandarlapaty S, Errede B Ash1, a daughter cell-specific protein, is required for pseudohyphal growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: Chant J Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 15: Chartrand P, Meng XH, Singer RH, Long RM Structural elements required for the localization of ASH1 mrna and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 9: Corral-Debrinski M, Bludgeon C, Jacq C In yeast, the 3 untranslated region or the presequence of ATM1 is required for the exclusive localization of its mrna to the vicinity of mitochondria. Mol. Cell. Biol. 20: Drubin DG, Jones HD, Wertman KF Actin structure and function: roles of mitochondria reorganization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol. Biol. Cell 4: Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, et al Bni1p, a yeast formin linking Cdc42 and the actin cytoskeleton during polarized morphogenesis. Science 276: Finger F, Novick P Spatial regulation of exocytosis: lessons from yeast. J. Cell. Biol. 142: Frazier JA, Field CM Actin cytoskeleton: Are FH proteins local organizers? Curr. Biol. 7:R Gehrung S, Snyder M The SPA2 gene of Saccharomyces cerevisiae is important for pheromone-induced morphogenesis and efficient mating. J. Cell. Biol. 111: Gonzalez I, Buonomo SBC, Nasmyth K, von Ahsen U ASH1 mrna localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr. Biol. 9: Haarer BK, Petzold A, Lillie SH, Brown SS Identification of MYO4, a second classv myosin gene in yeast. J. Cell. Sci. 107: Hoeck KS, Kidd GJ, Carson JH, Smith R hnrnp A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mrna. Biochemistry 37: Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, et al Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 16: Jansen RP, Dowzer C, Michaelis C, Galova M, Nasmyth K Mother cell-specific HO expression in budding yeast depends on the unconventional myosin Myo4p and other cytoplasmic proteins. Cell 84: Kislauskis EH, Singer RH Determinants of mrna localization. Curr. Opin. Cell. Biol. 4: Kohrmann M, Luo M, Kaether C, DesGroseillers L, Dotti CG, Kiebler MA Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal
14 310 CHARTRAND SINGER LONG neurons. Mol. Biol. Cell. 10: Lall S, Francis-Lang H, Flament A, Norvell A, Schüpbach T, Ish-Horowicz D Squid hnrnp protein promotes apical cytoplasmic transport and localization of Drosophila pairrule transcripts. Cell 98: Leighton J, Schatz G An ABC transporter in the mitochondrial inner membrane is required for normal growth of yeast. EMBO J. 14: Lipshitz HD, Smibert CA Mechanisms of RNA localization and translational regulation. Curr. Opin. Gen. Dev. 10: Long RM, Gu W, Lorimer E, Singer RH, Chartrand P She2p is a novel RNA-binding protein that recruits the Myo4p/She3p complex to ASH1 mrna. EMBO J. 19: Long RM, Gu W, Meng X-H, Gonsalvez G, Singer RH, Chartrand P An exclusively nuclear RNA binding protein affects asymmetric localization of ASH1 mrna and Ash1p in yeast. J. Cell Biol. 153: Long RM, Singer RH, Meng X-H, Gonzalez I, Nasmyth K, Jansen RP Mating type switching in yeast controlled by asymmetric localization of ASH1 mrna. Science 277: Mannhaupt G, Stucka R, Ehnle S, Vetter I, Feldmann H Analysis of a 70 Kb region of the right arm of yeast chromosome II. Yeast 10: Münchow S, Sauter C, Jansen RP Association of the class V myosin Myo4p with a localized messenger RNA in budding yeast depends on She proteins. J. Cell Sci. 112: Ozaki-Kuroda K, Yamamoto Y, Nohara H, Kinoshita M, Fujiwara T, et al Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: Pan X, Heitman J Sok2 regulates yeast pseudohyphal differentiation via a transcriptional factor cascade that regulates cell-cell adhesion. Mol. Cell Biol. 22: Pruyne D, Bretscher A Polarization of cell growth in yeast. II. The role of the cortical actin cytoskeleton. J. Cell Sci. 113: Rongo C, Davis ER, Lehmann R Localization of oskar RNA regulates oskar translation and requires Oskar protein. Development 121: Sil A, Herskowitz I Identification of an asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell 84: Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290: Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature 389:90 93 Takizawa PA, Vale RD The myosin motor, Myo4p, binds Ash1 mrna via the adapter protein, She3p. Proc. Natl. Acad. Sci. USA 97: Umikawa M, Tanaka K, Kamei T, Shimizu K, Imamura H, et al Interaction of Rho1p target Bni1p with F-actin-binding elongation factor 1α: implication in Rho1p-regulated reorganization of the actin cytoskeleton in Saccharomyces cerevisiae. Oncogene 16: Wang S, Hazelrigg T Implications for bcd mrna localization from spatial distribution of exu protein in Drosophila oogenesis. Nature 369:400 3 Wendland B, McCaffery JM, Xiao Q, Emr SD A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J. Cell Biol. 135: Wilhelm JE, Mansfield J, Hom-Booher N, Wang S, Turck CW, et al Isolation of a ribonucleoprotein complex involved in mrna localization in Drosophila oocytes. J. Cell. Biol. 148:427 40
15 Figure 1 Fluorescent in situ hybridization on a late-anaphase budding yeast to detect the ASH1 mrna. Note the tight crescent localization of the mrna at the bud tip. DAPI: DNA staining. Size bar: 10 µm.
16 Figure 2 The yeast locasome. Only the elements E3 in the 3 UTR of the ASH1 mrna is depicted. A similar complex should also form on the remaining three localization elements. She2p is represented in red, She3p is shown in dark and light blue, and She1p is in gray.
17 Figure 3 Model of ASH1 mrna localization. See text for details.
mrna cis Elements Located in Non-Protein-Coding Regions
 mrna cis Elements Located in Non-Protein-Coding Regions IRP Repressor mirnas Bcd/4EHP Repressor cap 5 Leader ORF 3 UTR (A) n eif4e/4e-bp 2 o structure uorf eif2α kinases Localization Transcript-independent
mrna cis Elements Located in Non-Protein-Coding Regions IRP Repressor mirnas Bcd/4EHP Repressor cap 5 Leader ORF 3 UTR (A) n eif4e/4e-bp 2 o structure uorf eif2α kinases Localization Transcript-independent
MOLECULAR CELL BIOLOGY
 1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
BIO 5099: Molecular Biology for Computer Scientists (et al)
 BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
Cell Cycle, Mitosis, and Microtubules. LS1A Final Exam Review Friday 1/12/07. Processes occurring during cell cycle
 Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
CELLS. Cells. Basic unit of life (except virus)
 Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
04_polarity. The formation of synaptic vesicles
 Brefeldin prevents assembly of the coats required for budding Nocodazole disrupts microtubules Constitutive: coatomer-coated Selected: clathrin-coated The formation of synaptic vesicles Nerve cells (and
Brefeldin prevents assembly of the coats required for budding Nocodazole disrupts microtubules Constitutive: coatomer-coated Selected: clathrin-coated The formation of synaptic vesicles Nerve cells (and
Regulators of Cell Cycle Progression
 Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
MCB Chapter 11. Topic E. Splicing mechanism Nuclear Transport Alternative control modes. Reading :
 MCB Chapter 11 Topic E Splicing mechanism Nuclear Transport Alternative control modes Reading : 419-449 Topic E Michal Linial 14 Jan 2004 1 Self-splicing group I introns were the first examples of catalytic
MCB Chapter 11 Topic E Splicing mechanism Nuclear Transport Alternative control modes Reading : 419-449 Topic E Michal Linial 14 Jan 2004 1 Self-splicing group I introns were the first examples of catalytic
CMB621: Cytoskeleton. Also known as How the cell plays with LEGOs to ensure order, not chaos, is temporally and spatially achieved
 CMB621: Cytoskeleton Also known as How the cell plays with LEGOs to ensure order, not chaos, is temporally and spatially achieved Lecture(s) Overview Lecture 1: What is the cytoskeleton? Membrane interaction
CMB621: Cytoskeleton Also known as How the cell plays with LEGOs to ensure order, not chaos, is temporally and spatially achieved Lecture(s) Overview Lecture 1: What is the cytoskeleton? Membrane interaction
Polyomaviridae. Spring
 Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being A Eukaryote. Eukaryotic Cells. Basic eukaryotes have:
 BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
Molecular Cell Biology 5068 In Class Exam 1 October 3, 2013
 Molecular Cell Biology 5068 In Class Exam 1 October 3, 2013 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
Molecular Cell Biology 5068 In Class Exam 1 October 3, 2013 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
Neuronal plasma membrane
 ORGANELLES ORGANELLES Neuronal plasma membrane The neuronal plasma membrane contains several local domains with unique properties Presynaptic terminal Endoplasmic Reticulum In neurons the Nissl bodies
ORGANELLES ORGANELLES Neuronal plasma membrane The neuronal plasma membrane contains several local domains with unique properties Presynaptic terminal Endoplasmic Reticulum In neurons the Nissl bodies
Neuronal plasma membrane
 ORGANELLES ORGANELLES Neuronal plasma membrane The neuronal plasma membrane contains several local domains with unique properties Presynaptic terminal Endoplasmic Reticulum In neurons the Nissl bodies
ORGANELLES ORGANELLES Neuronal plasma membrane The neuronal plasma membrane contains several local domains with unique properties Presynaptic terminal Endoplasmic Reticulum In neurons the Nissl bodies
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division The Cell Cycle: Cell Growth, Cell Division 2007-2008 2007-2008 Getting from there to here Going from egg to baby. the original
Biology is the only subject in which multiplication is the same thing as division The Cell Cycle: Cell Growth, Cell Division 2007-2008 2007-2008 Getting from there to here Going from egg to baby. the original
Zool 3200: Cell Biology Exam 4 Part II 2/3/15
 Name:Key Trask Zool 3200: Cell Biology Exam 4 Part II 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Name:Key Trask Zool 3200: Cell Biology Exam 4 Part II 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
APGRU4L1 Chap 12 Extra Reading Cell Cycle and Mitosis
 APGRU4L1 Chap 12 Extra Reading Cell Cycle and Mitosis Dr. Ramesh Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008
APGRU4L1 Chap 12 Extra Reading Cell Cycle and Mitosis Dr. Ramesh Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008
Anatomy Chapter 2 - Cells
 Cells Cells are the basic living structural, functional unit of the body Cytology is the branch of science that studies cells The human body has 100 trillion cells 200 different cell types with a variety
Cells Cells are the basic living structural, functional unit of the body Cytology is the branch of science that studies cells The human body has 100 trillion cells 200 different cell types with a variety
A. Incorrect! All the cells have the same set of genes. (D)Because different types of cells have different types of transcriptional factors.
 Genetics - Problem Drill 21: Cytogenetics and Chromosomal Mutation No. 1 of 10 1. Why do some cells express one set of genes while other cells express a different set of genes during development? (A) Because
Genetics - Problem Drill 21: Cytogenetics and Chromosomal Mutation No. 1 of 10 1. Why do some cells express one set of genes while other cells express a different set of genes during development? (A) Because
Lecture 2: Virology. I. Background
 Lecture 2: Virology I. Background A. Properties 1. Simple biological systems a. Aggregates of nucleic acids and protein 2. Non-living a. Cannot reproduce or carry out metabolic activities outside of a
Lecture 2: Virology I. Background A. Properties 1. Simple biological systems a. Aggregates of nucleic acids and protein 2. Non-living a. Cannot reproduce or carry out metabolic activities outside of a
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
Overview of the Cellular Basis of Life. Copyright 2009 Pearson Education, Inc., publishing as Benjamin Cummings
 Overview of the Cellular Basis of Life Cells and Tissues Cells: Carry out all chemical activities needed to sustain life Cells are the building blocks of all living things Tissues Cells vary in length,
Overview of the Cellular Basis of Life Cells and Tissues Cells: Carry out all chemical activities needed to sustain life Cells are the building blocks of all living things Tissues Cells vary in length,
Structures in Cells. Cytoplasm. Lecture 5, EH1008: Biology for Public Health, Biomolecules
 Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Chapter 3: Cells. I. Overview
 Chapter 3: Cells I. Overview A. Characteristics 1. Basic structural/functional unit 2. Diameter is too small to see by the naked eye 3. Can be over 3 feet long 4. Trillions of cells in over 200 basic types
Chapter 3: Cells I. Overview A. Characteristics 1. Basic structural/functional unit 2. Diameter is too small to see by the naked eye 3. Can be over 3 feet long 4. Trillions of cells in over 200 basic types
Protein Trafficking in the Secretory and Endocytic Pathways
 Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
Mitosis vs. microtubule
 Mitosis vs. microtubule Anaphase-promoting complex/cyclosome (APC/C) Duplicated centrosomes align and begin separating in prophase Relation of centrosome duplication to the cell cycle. Parent centrioles
Mitosis vs. microtubule Anaphase-promoting complex/cyclosome (APC/C) Duplicated centrosomes align and begin separating in prophase Relation of centrosome duplication to the cell cycle. Parent centrioles
Cell Overview. Hanan Jafar BDS.MSc.PhD
 Cell Overview Hanan Jafar BDS.MSc.PhD THE CELL is made of: 1- Nucleus 2- Cell Membrane 3- Cytoplasm THE CELL Formed of: 1. Nuclear envelope 2. Chromatin 3. Nucleolus 4. Nucleoplasm (nuclear matrix) NUCLEUS
Cell Overview Hanan Jafar BDS.MSc.PhD THE CELL is made of: 1- Nucleus 2- Cell Membrane 3- Cytoplasm THE CELL Formed of: 1. Nuclear envelope 2. Chromatin 3. Nucleolus 4. Nucleoplasm (nuclear matrix) NUCLEUS
RECAP FROM MONDAY AND TUESDAY LECTURES
 RECAP FROM MONDAY AND TUESDAY LECTURES Nucleo-cytoplasmic transport factors: interact with the target macromolecule (signal recognition) interact with the NPC (nucleoporin Phe - Gly repeats) NPC cytoplasmic
RECAP FROM MONDAY AND TUESDAY LECTURES Nucleo-cytoplasmic transport factors: interact with the target macromolecule (signal recognition) interact with the NPC (nucleoporin Phe - Gly repeats) NPC cytoplasmic
Name: Multiple choice questions. Pick the BEST answer (2 pts ea)
 Exam 1 202 Oct. 5, 1999 Multiple choice questions. Pick the BEST answer (2 pts ea) 1. The lipids of a red blood cell membrane are all a. phospholipids b. amphipathic c. glycolipids d. unsaturated 2. The
Exam 1 202 Oct. 5, 1999 Multiple choice questions. Pick the BEST answer (2 pts ea) 1. The lipids of a red blood cell membrane are all a. phospholipids b. amphipathic c. glycolipids d. unsaturated 2. The
Structures in Cells. Lecture 5, EH1008: Biology for Public Health, Biomolecules.
 Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
The Cell Organelles. Eukaryotic cell. The plasma membrane separates the cell from the environment. Plasma membrane: a cell s boundary
 Eukaryotic cell The Cell Organelles Enclosed by plasma membrane Subdivided into membrane bound compartments - organelles One of the organelles is membrane bound nucleus Cytoplasm contains supporting matrix
Eukaryotic cell The Cell Organelles Enclosed by plasma membrane Subdivided into membrane bound compartments - organelles One of the organelles is membrane bound nucleus Cytoplasm contains supporting matrix
RECAP (1)! In eukaryotes, large primary transcripts are processed to smaller, mature mrnas.! What was first evidence for this precursorproduct
 RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Getting from there to here Going from egg to baby. the original
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Getting from there to here Going from egg to baby. the original
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis
 Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Chromosomes Chromosomes were first observed by the German embryologist Walther Fleming in 1882. Chromosome number varies among organisms most
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Chromosomes Chromosomes were first observed by the German embryologist Walther Fleming in 1882. Chromosome number varies among organisms most
Cytoskelet Prednáška 6 Mikrotubuly a mitóza
 Cytoskelet Prednáška 6 Mikrotubuly a mitóza Polarity of tubulin polymerization Nuclei Tubulin > C C Preferential addition of tubulin ar (+) ends Tubulin < C C Preferential loss of tubulin ar (+) ends Kinesin
Cytoskelet Prednáška 6 Mikrotubuly a mitóza Polarity of tubulin polymerization Nuclei Tubulin > C C Preferential addition of tubulin ar (+) ends Tubulin < C C Preferential loss of tubulin ar (+) ends Kinesin
cell movement and neuronal migration
 cell movement and neuronal migration Paul Letourneau letou001@umn.edu Chapter 16; The Cytoskeleton; Molecular Biology of the Cell, Alberts et al. 1 Cell migration Cell migration in 3 steps; protrusion,
cell movement and neuronal migration Paul Letourneau letou001@umn.edu Chapter 16; The Cytoskeleton; Molecular Biology of the Cell, Alberts et al. 1 Cell migration Cell migration in 3 steps; protrusion,
Chapter 3 Part 2! Pages (10 th and 11 th eds.)! The Cellular Level of Organization! Cellular Organelles and Protein Synthesis!
 Chapter 3 Part 2! Pages 65 89 (10 th and 11 th eds.)! The Cellular Level of Organization! Cellular Organelles and Protein Synthesis! The Cell Theory! Living organisms are composed of one or more cells.!
Chapter 3 Part 2! Pages 65 89 (10 th and 11 th eds.)! The Cellular Level of Organization! Cellular Organelles and Protein Synthesis! The Cell Theory! Living organisms are composed of one or more cells.!
10/13/11. Cell Theory. Cell Structure
 Cell Structure Grade 12 Biology Cell Theory All organisms are composed of one or more cells. Cells are the smallest living units of all living organisms. Cells arise only by division of a previously existing
Cell Structure Grade 12 Biology Cell Theory All organisms are composed of one or more cells. Cells are the smallest living units of all living organisms. Cells arise only by division of a previously existing
MCB Topic 19 Regulation of Actin Assembly- Prof. David Rivier
 MCB 252 -Topic 19 Regulation of Actin Assembly- Prof. David Rivier MCB 252 Spring 2017 MCB 252 Cell Biology Topic 19 Regulation of Actin Assembly Reading: Lodish 17.2-17.3, 17.7 MCB 252 Actin Cytoskeleton
MCB 252 -Topic 19 Regulation of Actin Assembly- Prof. David Rivier MCB 252 Spring 2017 MCB 252 Cell Biology Topic 19 Regulation of Actin Assembly Reading: Lodish 17.2-17.3, 17.7 MCB 252 Actin Cytoskeleton
BIOL 4374/BCHS 4313 Cell Biology Exam #2 March 22, 2001
 BIOL 4374/BCHS 4313 Cell Biology Exam #2 March 22, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) In the
BIOL 4374/BCHS 4313 Cell Biology Exam #2 March 22, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) In the
2013 John Wiley & Sons, Inc. All rights reserved. PROTEIN SORTING. Lecture 10 BIOL 266/ Biology Department Concordia University. Dr. S.
 PROTEIN SORTING Lecture 10 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Introduction Membranes divide the cytoplasm of eukaryotic cells into distinct compartments. The endomembrane
PROTEIN SORTING Lecture 10 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Introduction Membranes divide the cytoplasm of eukaryotic cells into distinct compartments. The endomembrane
Bio10 Cell Structure SRJC
 3.) Cell Structure and Function Structure of Cell Membranes Fluid mosaic model Mixed composition: Phospholipid bilayer Glycolipids Sterols Proteins Fluid Mosaic Model Phospholipids are not packed tightly
3.) Cell Structure and Function Structure of Cell Membranes Fluid mosaic model Mixed composition: Phospholipid bilayer Glycolipids Sterols Proteins Fluid Mosaic Model Phospholipids are not packed tightly
Nucleic acids. Nucleic acids are information-rich polymers of nucleotides
 Nucleic acids Nucleic acids are information-rich polymers of nucleotides DNA and RNA Serve as the blueprints for proteins and thus control the life of a cell RNA and DNA are made up of very similar nucleotides.
Nucleic acids Nucleic acids are information-rich polymers of nucleotides DNA and RNA Serve as the blueprints for proteins and thus control the life of a cell RNA and DNA are made up of very similar nucleotides.
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system
 Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Lecture 6 9/17 Dr. Hirsh Organization of Cells, continued
 Cell structure of Eukaryotic cells Lecture 6 9/17 Dr. Hirsh Organization of Cells, continued Lots of double-membraned organelles Existence of an Endo-membrane system separation of areas of cell, transport
Cell structure of Eukaryotic cells Lecture 6 9/17 Dr. Hirsh Organization of Cells, continued Lots of double-membraned organelles Existence of an Endo-membrane system separation of areas of cell, transport
Chapter 12 The Cell Cycle: Cell Growth, Cell Division
 Chapter 12 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a period at the end of a sentence And now look at you How did you get from there to
Chapter 12 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a period at the end of a sentence And now look at you How did you get from there to
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment
 Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
Lecture 10. G1/S Regulation and Cell Cycle Checkpoints. G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint
 Lecture 10 G1/S Regulation and Cell Cycle Checkpoints Outline: G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint Paper: The roles of Fzy/Cdc20 and Fzr/Cdh1
Lecture 10 G1/S Regulation and Cell Cycle Checkpoints Outline: G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint Paper: The roles of Fzy/Cdc20 and Fzr/Cdh1
Cellular compartments
 Cellular compartments 1. Cellular compartments and their function 2. Evolution of cellular compartments 3. How to make a 3D model of cellular compartment 4. Cell organelles in the fluorescent microscope
Cellular compartments 1. Cellular compartments and their function 2. Evolution of cellular compartments 3. How to make a 3D model of cellular compartment 4. Cell organelles in the fluorescent microscope
Principles of Anatomy and Physiology
 Principles of Anatomy and Physiology 14 th Edition CHAPTER 3 The Cellular Level of Organization Introduction The purpose of the chapter is to: 1. Introduce the parts of a cell 2. Discuss the importance
Principles of Anatomy and Physiology 14 th Edition CHAPTER 3 The Cellular Level of Organization Introduction The purpose of the chapter is to: 1. Introduce the parts of a cell 2. Discuss the importance
RAS Genes. The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes.
 ۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
Life Sciences 1A Midterm Exam 2. November 13, 2006
 Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Intracellular Compartments and Protein Sorting
 Intracellular Compartments and Protein Sorting Intracellular Compartments A eukaryotic cell is elaborately subdivided into functionally distinct, membrane-enclosed compartments. Each compartment, or organelle,
Intracellular Compartments and Protein Sorting Intracellular Compartments A eukaryotic cell is elaborately subdivided into functionally distinct, membrane-enclosed compartments. Each compartment, or organelle,
October 26, Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
Campbell Biology in Focus (Urry) Chapter 9 The Cell Cycle. 9.1 Multiple-Choice Questions
 Campbell Biology in Focus (Urry) Chapter 9 The Cell Cycle 9.1 Multiple-Choice Questions 1) Starting with a fertilized egg (zygote), a series of five cell divisions would produce an early embryo with how
Campbell Biology in Focus (Urry) Chapter 9 The Cell Cycle 9.1 Multiple-Choice Questions 1) Starting with a fertilized egg (zygote), a series of five cell divisions would produce an early embryo with how
Cell Signaling (III) Cell Cycle (I)
 BME 42-620 Engineering Molecular Cell Biology Lecture 22: Cell Signaling (III) Cell Cycle (I) Chapter 15 BME42-620 Lecture 22, December 01, 2011 1 Comments on Reading Assignment 5 (I) I assume that this
BME 42-620 Engineering Molecular Cell Biology Lecture 22: Cell Signaling (III) Cell Cycle (I) Chapter 15 BME42-620 Lecture 22, December 01, 2011 1 Comments on Reading Assignment 5 (I) I assume that this
Mechanisms of alternative splicing regulation
 Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis
 Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Prokaryotes Have a Simpler Cell Cycle Cell division in prokaryotes takes place in two stages, which together make up a simple cell cycle 1. Copy
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Prokaryotes Have a Simpler Cell Cycle Cell division in prokaryotes takes place in two stages, which together make up a simple cell cycle 1. Copy
Molecular Cell Biology 5068 In Class Exam 1 September 29, Please print your name:
 Molecular Cell Biology 5068 In Class Exam 1 September 29, 2015 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your
Molecular Cell Biology 5068 In Class Exam 1 September 29, 2015 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your
Molecular Trafficking
 SCBM 251 Molecular Trafficking Assoc. Prof. Rutaiwan Tohtong Department of Biochemistry Faculty of Science rutaiwan.toh@mahidol.ac.th Lecture outline 1. What is molecular trafficking? Why is it important?
SCBM 251 Molecular Trafficking Assoc. Prof. Rutaiwan Tohtong Department of Biochemistry Faculty of Science rutaiwan.toh@mahidol.ac.th Lecture outline 1. What is molecular trafficking? Why is it important?
Cell Physiology Final Exam Fall 2008
 Cell Physiology Final Exam Fall 2008 Guys, The average on the test was 69.9. Before you start reading the right answers please do me a favor and remember till the end of your life that GLUCOSE TRANSPORT
Cell Physiology Final Exam Fall 2008 Guys, The average on the test was 69.9. Before you start reading the right answers please do me a favor and remember till the end of your life that GLUCOSE TRANSPORT
Problem Set 5 KEY
 2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
The Cell Cycle and How Cells Divide
 The Cell Cycle and How Cells Divide 1 Phases of the Cell Cycle The cell cycle consists of Interphase normal cell activity The mitotic phase cell divsion INTERPHASE Growth G 1 (DNA synthesis) Growth G 2
The Cell Cycle and How Cells Divide 1 Phases of the Cell Cycle The cell cycle consists of Interphase normal cell activity The mitotic phase cell divsion INTERPHASE Growth G 1 (DNA synthesis) Growth G 2
AP Biology Book Notes Chapter 4: Cells v Cell theory implications Ø Studying cell biology is in some sense the same as studying life Ø Life is
 AP Biology Book Notes Chapter 4: Cells v Cell theory implications Ø Studying cell biology is in some sense the same as studying life Ø Life is continuous v Small cell size is becoming more necessary as
AP Biology Book Notes Chapter 4: Cells v Cell theory implications Ø Studying cell biology is in some sense the same as studying life Ø Life is continuous v Small cell size is becoming more necessary as
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
Cells. Variation and Function of Cells
 Cells Variation and Function of Cells Cell Theory states that: 1. All living things are made of cells 2. Cells are the basic unit of structure and function in living things 3. New cells are produced from
Cells Variation and Function of Cells Cell Theory states that: 1. All living things are made of cells 2. Cells are the basic unit of structure and function in living things 3. New cells are produced from
Homework Hanson section MCB Course, Fall 2014
 Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Alpha thalassemia mental retardation X-linked. Acquired alpha-thalassemia myelodysplastic syndrome
 Alpha thalassemia mental retardation X-linked Acquired alpha-thalassemia myelodysplastic syndrome (Alpha thalassemia mental retardation X-linked) Acquired alpha-thalassemia myelodysplastic syndrome Schematic
Alpha thalassemia mental retardation X-linked Acquired alpha-thalassemia myelodysplastic syndrome (Alpha thalassemia mental retardation X-linked) Acquired alpha-thalassemia myelodysplastic syndrome Schematic
Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
Chapter 3. Expression of α5-megfp in Mouse Cortical Neurons. on the β subunit. Signal sequences in the M3-M4 loop of β nachrs bind protein factors to
 22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson
 Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson General principles Properties of lysosomes Delivery of enzymes to lysosomes Endocytic uptake clathrin, others Endocytic pathways recycling vs.
Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson General principles Properties of lysosomes Delivery of enzymes to lysosomes Endocytic uptake clathrin, others Endocytic pathways recycling vs.
Biology 218 Human Anatomy
 Chapter 2 Adapted from Tortora 10 th ed LECTURE OUTLINE A. A Generalized Cell (p. 25) 1. A human cell consists of three major parts (see Table 2.2 on p. 42): a. Plasma membrane b. Cytoplasm which includes
Chapter 2 Adapted from Tortora 10 th ed LECTURE OUTLINE A. A Generalized Cell (p. 25) 1. A human cell consists of three major parts (see Table 2.2 on p. 42): a. Plasma membrane b. Cytoplasm which includes
Chapter 10 - Post-transcriptional Gene Control
 Chapter 10 - Post-transcriptional Gene Control Chapter 10 - Post-transcriptional Gene Control 10.1 Processing of Eukaryotic Pre-mRNA 10.2 Regulation of Pre-mRNA Processing 10.3 Transport of mrna Across
Chapter 10 - Post-transcriptional Gene Control Chapter 10 - Post-transcriptional Gene Control 10.1 Processing of Eukaryotic Pre-mRNA 10.2 Regulation of Pre-mRNA Processing 10.3 Transport of mrna Across
Supplementary information. The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic. Spindle Orientation
 Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
1. endoplasmic reticulum This is the location where N-linked oligosaccharide is initially synthesized and attached to glycoproteins.
 Biology 4410 Name Spring 2006 Exam 2 A. Multiple Choice, 2 pt each Pick the best choice from the list of choices, and write it in the space provided. Some choices may be used more than once, and other
Biology 4410 Name Spring 2006 Exam 2 A. Multiple Choice, 2 pt each Pick the best choice from the list of choices, and write it in the space provided. Some choices may be used more than once, and other
Capu and Spire Assemble a Cytoplasmic Actin Mesh
 Developmental Cell 13 Supplemental Data Capu and Spire Assemble a Cytoplasmic Actin Mesh that Maintains Microtubule Organization in the Drosophila Oocyte Katja Dahlgaard, Alexandre A.S.F. Raposo, Teresa
Developmental Cell 13 Supplemental Data Capu and Spire Assemble a Cytoplasmic Actin Mesh that Maintains Microtubule Organization in the Drosophila Oocyte Katja Dahlgaard, Alexandre A.S.F. Raposo, Teresa
Eukaryotic cell. Premedical IV Biology
 Eukaryotic cell Premedical IV Biology The size range of organisms Light microscopes visible light is passed through the specimen and glass lenses the resolution is limited by the wavelength of the visible
Eukaryotic cell Premedical IV Biology The size range of organisms Light microscopes visible light is passed through the specimen and glass lenses the resolution is limited by the wavelength of the visible
Section 6. Junaid Malek, M.D.
 Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
Introduction retroposon
 17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
The Cell. Biology 105 Lecture 4 Reading: Chapter 3 (pages 47 62)
 The Cell Biology 105 Lecture 4 Reading: Chapter 3 (pages 47 62) Outline I. Prokaryotic vs. Eukaryotic II. Eukaryotic A. Plasma membrane transport across B. Main features of animal cells and their functions
The Cell Biology 105 Lecture 4 Reading: Chapter 3 (pages 47 62) Outline I. Prokaryotic vs. Eukaryotic II. Eukaryotic A. Plasma membrane transport across B. Main features of animal cells and their functions
Main differences between plant and animal cells: Plant cells have: cell walls, a large central vacuole, plastids and turgor pressure.
 Main differences between plant and animal cells: Plant cells have: cell walls, a large central vacuole, plastids and turgor pressure. Animal cells have a lysosome (related to vacuole) and centrioles (function
Main differences between plant and animal cells: Plant cells have: cell walls, a large central vacuole, plastids and turgor pressure. Animal cells have a lysosome (related to vacuole) and centrioles (function
8/7/18. UNIT 2: Cells Chapter 3: Cell Structure and Function. I. Cell Theory (3.1) A. Early studies led to the development of the cell theory
 8/7/18 UNIT 2: Cells Chapter 3: Cell Structure and Function I. Cell Theory (3.1) A. Early studies led to the development of the cell theory 1. Discovery of Cells a. Robert Hooke (1665)-Used compound microscope
8/7/18 UNIT 2: Cells Chapter 3: Cell Structure and Function I. Cell Theory (3.1) A. Early studies led to the development of the cell theory 1. Discovery of Cells a. Robert Hooke (1665)-Used compound microscope
Cell wall components:
 Main differences between plant and animal cells: Plant cells have: cell walls, a large central vacuole, plastids and turgor pressure. The Cell Wall The primary cell wall is capable of rapid expansion during
Main differences between plant and animal cells: Plant cells have: cell walls, a large central vacuole, plastids and turgor pressure. The Cell Wall The primary cell wall is capable of rapid expansion during
Chapter 4 Genetics and Cellular Function. The Nucleic Acids (medical history) Chromosome loci. Organization of the Chromatin. Nucleotide Structure
 Chapter 4 Genetics and Cellular Function The Nucleic Acids (medical history) Nucleus and nucleic acids Protein synthesis and secretion DNA replication and the cell cycle Chromosomes and heredity Organization
Chapter 4 Genetics and Cellular Function The Nucleic Acids (medical history) Nucleus and nucleic acids Protein synthesis and secretion DNA replication and the cell cycle Chromosomes and heredity Organization
Microtubule Forces Kevin Slep
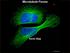 Microtubule Forces Kevin Slep Microtubules are a Dynamic Scaffold Microtubules in red, XMA215 family MT polymerase protein in green Some Microtubule Functions Cell Structure Polarized Motor Track (kinesins
Microtubule Forces Kevin Slep Microtubules are a Dynamic Scaffold Microtubules in red, XMA215 family MT polymerase protein in green Some Microtubule Functions Cell Structure Polarized Motor Track (kinesins
Introduction. Biochemistry: It is the chemistry of living things (matters).
 Introduction Biochemistry: It is the chemistry of living things (matters). Biochemistry provides fundamental understanding of the molecular basis for the function and malfunction of living things. Biochemistry
Introduction Biochemistry: It is the chemistry of living things (matters). Biochemistry provides fundamental understanding of the molecular basis for the function and malfunction of living things. Biochemistry
REGULATED SPLICING AND THE UNSOLVED MYSTERY OF SPLICEOSOME MUTATIONS IN CANCER
 REGULATED SPLICING AND THE UNSOLVED MYSTERY OF SPLICEOSOME MUTATIONS IN CANCER RNA Splicing Lecture 3, Biological Regulatory Mechanisms, H. Madhani Dept. of Biochemistry and Biophysics MAJOR MESSAGES Splice
REGULATED SPLICING AND THE UNSOLVED MYSTERY OF SPLICEOSOME MUTATIONS IN CANCER RNA Splicing Lecture 3, Biological Regulatory Mechanisms, H. Madhani Dept. of Biochemistry and Biophysics MAJOR MESSAGES Splice
Dendritic RNA Transport: Dynamic Spatio-Temporal Control of Neuronal Gene Expression 437
 Dendritic RNA Transport: Dynamic Spatio-Temporal Control of Neuronal Gene Expression 437 Dendritic RNA Transport: Dynamic Spatio-Temporal Control of Neuronal Gene Expression J B Dictenberg Hunter College
Dendritic RNA Transport: Dynamic Spatio-Temporal Control of Neuronal Gene Expression 437 Dendritic RNA Transport: Dynamic Spatio-Temporal Control of Neuronal Gene Expression J B Dictenberg Hunter College
Regulation of Gene Expression in Eukaryotes
 Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Lecture 10. Eukaryotic gene regulation: chromatin remodelling
 Lecture 10 Eukaryotic gene regulation: chromatin remodelling Recap.. Eukaryotic RNA polymerases Core promoter elements General transcription factors Enhancers and upstream activation sequences Transcriptional
Lecture 10 Eukaryotic gene regulation: chromatin remodelling Recap.. Eukaryotic RNA polymerases Core promoter elements General transcription factors Enhancers and upstream activation sequences Transcriptional
Lecture 1, Fall 2014 The structure of the eukaryotic cell, as it relates to cells chemical and biological functions.
 Lecture 1, Fall 2014 The structure of the eukaryotic cell, as it relates to cells chemical and biological functions. There are several important themes that transcends just the chemistry and bring the
Lecture 1, Fall 2014 The structure of the eukaryotic cell, as it relates to cells chemical and biological functions. There are several important themes that transcends just the chemistry and bring the
Watson-Crick Model of B-DNA
 Reading: Ch8; 285-290 Ch24; 963-978 Problems: Ch8 (text); 9 Ch8 (study-guide: facts); 3 Ch24 (text); 5,7,9,10,14,16 Ch24 (study-guide: applying); 1 Ch24 (study-guide: facts); 1,2,4 NEXT Reading: Ch1; 29-34
Reading: Ch8; 285-290 Ch24; 963-978 Problems: Ch8 (text); 9 Ch8 (study-guide: facts); 3 Ch24 (text); 5,7,9,10,14,16 Ch24 (study-guide: applying); 1 Ch24 (study-guide: facts); 1,2,4 NEXT Reading: Ch1; 29-34
Circular RNAs (circrnas) act a stable mirna sponges
 Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Chapter 3: Cells 3-1
 Chapter 3: Cells 3-1 Introduction: A. Human body consists of 75 trillion cells B. About 260 types of cells that vary in shape & size yet have much in common B. Differences in cell shape make different
Chapter 3: Cells 3-1 Introduction: A. Human body consists of 75 trillion cells B. About 260 types of cells that vary in shape & size yet have much in common B. Differences in cell shape make different
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
Chapt. 10 Cell Biology and Biochemistry. The cell: Student Learning Outcomes: Describe basic features of typical human cell
 Chapt. 10 Cell Biology and Biochemistry Cell Chapt. 10 Cell Biology and Biochemistry The cell: Lipid bilayer membrane Student Learning Outcomes: Describe basic features of typical human cell Integral transport
Chapt. 10 Cell Biology and Biochemistry Cell Chapt. 10 Cell Biology and Biochemistry The cell: Lipid bilayer membrane Student Learning Outcomes: Describe basic features of typical human cell Integral transport
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division Ch. 10 Where it all began You started as a cell smaller than a period
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division Ch. 10 Where it all began You started as a cell smaller than a period
Cell Quality Control. Peter Takizawa Department of Cell Biology
 Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Mitosis and the Cell Cycle
 Mitosis and the Cell Cycle Chapter 12 The Cell Cycle: Cell Growth & Cell Division Where it all began You started as a cell smaller than a period at the end of a sentence Getting from there to here Cell
Mitosis and the Cell Cycle Chapter 12 The Cell Cycle: Cell Growth & Cell Division Where it all began You started as a cell smaller than a period at the end of a sentence Getting from there to here Cell
