Using Data from the Cancer Genome Atlas to Investigate Molecular Events Related to Ovarian Cancer. Abstract
|
|
|
- Bertina Spencer
- 5 years ago
- Views:
Transcription
1 Using Data from the Cancer Genome Atlas to Investigate Molecular Events Related to Ovarian Cancer Xiaoye Liu, Gang Fang, Michael Steinbach Abstract Ovarian Cancer is the most lethal gynecologic malignancy. Analyzing the molecular events related to ovarian cancer helps understand the pathogenesis of ovarian cancer from a genetic point of view. As mirna singletons have been found significantly related to ovarian cancer and many other cancers, mirnas have been recognized as an important riboregulator of gene expression. However, little is known about how pairs of mirna expression profiles associate with ovarian cancer. In our analysis, we explored the combinatorial effects of mirna pairs on regulating gene expression. We assessed the non-additive interaction between mirna pairs on gene expression of patients that carry high grade ovarian cancer. We demonstrate how different mirnas collectively contribute to ovarian cancer. We will illustrate two examples of mirna pairs, hsa.mir.937 & hsa.let.7b and hsa.mir.1277& hsa.mir.485.3b, that we found exhibit non-additive interaction pattern on affecting gene expression of the patients with high grade Ovarian Cancer. Introduction: Ovarian cancer is the second most common gynecologic cancer in the United States and sixth most common cancer in women worldwide[1,2,3]. In 2010 alone, there were 13,850 deaths from of ovarian cancer with an estimated 21,888 new cases diagnosed [4]. Although 90% of the patients with early-stage ovarian cancer could successfully survive 5 years or longer after initial diagnosis, only 21% of the patients who were diagnosed at advanced-stage could achieve that [2]. Due to the lack of robust methods for early detection, 19% of all ovarian cancer is diagnosed at early stage [5]. Thus, more understanding on the pathogenesis of ovarian cancer is needed for helping to improve the early stage detection diagnosis. Analyzing the molecular events related to ovarian cancer give the best help on understanding the pathogenesis of ovarian cancer from a genetic point of view. In order to identify the genetic alteration associates with the malignant phenotype of ovarian, many researchers start investigation on analyzing how mirna expression profiles contribute to ovarian cancer. mirna is a small and new class of noncoding RNAs that plays an important role in cellcycle progression, tissue differentiation and organ development [5,7]. Many mirna singletons have been found significantly related to many other cancers, such as breast cancer, pancreatic cancer and lung cancer. [7,8,9] Several recent studies have also successfully identified that particular mirna singletons such as mir-21, mir-125a, mir- 125b[3], mir-200a, mir-141, mir-199a [5], are significantly differentially or over expressed in the mirna expression profile of ovarian cancer patients. Although mirna has been recognized as a riboregulator of gene expression [3] and many mirna singletons have been discovered as essential cancer bio-makers, little is known about how pairs of mirna expression profiles associate with ovarian cancer. In this report, we represent the result of genomewide mirna expression profiles in a large set of high grade Ovarian Serous Cystadenocarcinoma (OSC) tissue, a type of highly aggressive epithelial ovarian tumor that accounts for 90% of all ovarian cancer [6]. We assessed the non-additive interaction between mirna pairs on gene expression of patients that carry OSC. We demonstrate how different mirnas collectively contributes to this high aggressive OSC. Material and Methods: An overview of the methods used can be found in Research Process Chart in Figure1. Data from The Cancer Genome Atlas (TCGA): We acquired the OSC mirna expression data, gene expression data and clinical data from TCGA data portal. (Available at: gov/tcga/) The level three OSC data from TCGA were analyzed for both mirna expression and gene expression. In TCGA, level three data represents the data which are background corrected, molecular abnormalities interpreted and normalized. mirna Expression Data: The mirna microarray platform of mirna expression data is Agilent 8*15K performed by University of North Carolina(UNC).The measurements for 799 mirnas were included in the mirna expression data in our analysis.
2 smaller set of biologically meaningful gene clusters. Table1: Data Information *clinical data were last updated on August 10 th 2010 Type Platform No. of Patients No. of Singletons mirna- Expressions Gene- Expressions UNC HmiRNA_8x15Kv BI HT_HG- U133A Clinical* _ 491 _ Figure 1: Research Process Chart. Gene Expression Data: The platform of gene expression data is Affymetrix HT Human Genome U133 Array Plate Set performed by Massachusetts Institute of Technology (MIT). We have included genes in our analysis. We have averaged the expression values of the duplicated samples from the same patients in mirna and gene expression data. We have also removed patients whose tumor stage information is null. 493 patients who have been annotated having OSC stage II to IV in the clinical data are finally selected in our analysis. (Table 1) Zscore Normalization: Before any methods are applied, both mirna and gene expression data are pre-processed by zscore normalization: subtracting the mean and dividing by standard deviation for data standardization. Construct Gene Co-expression Network: We performed Weighted Gene Co-expression Network Analysis (WGCNA) [10-13] on OSC gene expression data. The purpose of applying WGCNA is to attempt to identify co-expressed genes and to combine the high correlated genes into gene modules (clusters). Since gene modules might associate with biological pathways [14], combining genes into modules is an effective scheme for reducing the dimension of gene expression data from a level of thousands into a In WGCNA, each gene is considered a node in the network. The distance between a pair of gene nodes is first determined by their pair-wise Pearson correlation value. The gene network is constructed based on all pair-wise Pearson correlation values between the gene nodes in the network. The pair-wise Pearson correlation was calculated across all patients for all genes in the gene expression data matrix. In order to emphasize the large Pearson correlation value, a power β 1, known as soft threshold as against hard threshold in un-weighted network, needs to be chosen to raise the power of the absolute value of Pearson correlation by the formula: adjij = cor(xi, xj) ^β This adjij is defined as the adjacency of the genes in an unsigned weighted gene co-expression network. Picking a proper value for the soft threshold β is an essential step in WGCNA. In order to allow the network to keep the continuous nature of the gene co-expression information, β should be chosen obeying the criterion of scale-free topology. [10] In this way, a weighted network exhibits its advantages on the robustness in hierarchical clustering analysis [10,14]. As we applied WGCNA to gene expression data in our analysis, we chose soft power threshold β = 4, which is the smallest value reaching the level of 0.9 on the scale independence. (Figure 2)
3 Figure2 : the scale free topology and soft-power selection Taking the adjacency as input, the dissimilarity of a Topological Overlap Matrix (disstom) is computed. The disstom computation minimizes the effect of noise and unauthentic associations. disstom is the final input for the module detecting in WGCNA.[10] As we expect to analyze gene modules with a relatively large size, we set the minimum module size to be 30 when performing WGCNA. This allows all gene modules returned have a size no less than 30 genes. After combining dynamic gene modules with a correlation higher than 0.8, all genes have been assigned to 80 modules. The final results are module sizes ranging from 1530 to 33.(79 modules for signed genes and 1 module for unsigned ones). A graphic illustration is shown in figure 3. Module memberships were assigned to each gene in forms of numeric labels and color labels. In WGCNA, a module eigengene was computed for each gene module as the new representative of all genes in that module. It refers to the first principal component of a gene module. [10, 14] Its value summarizes the gene expression level of a certain gene module. We consider the new eigengene values as the new quantitative traits of genes in our analysis. Linear Regression Based Test for Finding Nonadditive Interaction: Figure3 : gene module detection. The color bar on the bottom level is the final module selection after merging the similar modules. In order to identify the level of non-additive interaction of each pair of mirnas, we performed linear least square regression fitting to each mirna pairs for each gene module. *y stands for the module eigengene data; gene expression *x1 stands for mirna1, x2 stands for mirna2 *x1*x2 stands for the interaction term * β 0 is the base,β1, β2 and β12 are the coefficient of each term *ε represents the error term Since our primary goal is assessing the nonadditive interaction between mirna pairs on the expressions of gene modules, the test of epistasis was performed in our analysis. We computed F- statistics to compare the full model (*) to the model which only includes the additive terms [17]. Such statistical computation allows us to assess significance of the non-additive interaction of each pair directly. (*) Ho: β12=0 purely additive model Ha: β12 0 full model (*) We have collected a matrix of all p-values from F- statistic computation in the model comparison for all possible mirna pair combinations on gene modules. There are 80 sets of p-value matrices returned and each set is a symmetric matrix with zeros on the diagonal and all 799 mirna as rows and columns. We have transformed all such matrices by taking log10 of the P-value in every cell (-log10pv). (Figure 4)
4 maximum log10pv. In this way, we see how the expression of the two mirnas affects the level of the gene module expressions. However, due to the high skewness and low variation of the eigengene value in some of the 80 gene modules, the non-additive pattern for some of the pairs we found is difficult to distinguish on their best gene module. Therefore, we have filtered out the gene modules that have IQR < in order to illustrate the non-additive pattern clearly. (Figure 5) There are 9 qualified modules selected. Figure4: An example for illustrating the symmetry log10pv matrix of all pairs on gene module 1. Since each matrix is symmetric, both the upper and the lower triangular contains the same - log10pv information. We selected the upper triangular for each of 80 log10pv matrices. We break every selected part into a vector, where each cell in that vector represents the p-value of corresponding pair of mirnas. We have combined all such vectors into a matrix with 80 columns and rows. (-log10pv matrix) Identify non-additive interaction mirna pairs: Since we are comparing the full model with the purely additive model, a lower P-value would suggest that the interaction effect is making a significant contribution. This is because we could not ignore the effect of the interaction if we reject the null hypothesis. Equivalently, after transforming PV to log10pv, a high logpv would support this conclusion as well. For identifying the significant mirnas, we took the row maximum of the log10pv matrix. We filtered out the low log10pv pairs and kept the ones that have relatively high (-log10pv>6) log10pv as target pairs for further graphical identification analysis. In the graphical analysis, for every target pair, we used scatter plot to show the pattern of normalized mirna expression for the two selected mirnas, one for each axis. We used color for all scatters to indicate the level of gene expressions of their selected best gene module. The best gene module refers to the module where the target pair has achieved their Figure 5: Histogram of IQR for each gene module False Discover Rate (FDR) computation: We used FDR computation to correct for the multiple comparisons. [15,16] Across all 9 gene modules, we found 942 pairs show significant to the gene expression trait. By setting the log10pv threshold to 6, there are 25 pairs left.(figure6) Figure 6: Brief graphic overview of 25 pairs.
5 From those 25 pairs, we have successfully found several that exhibit non-additive pattern on affecting eigengene value. We will be presenting two pairs that have a significant pattern as examples. Figure 7: hsa.mir.937 and hsa.let.7b on gene module 7 (Pair 1) expression values on their best eigengene, which best proves that their non-additive interaction exists on affecting module7. Pair2: hsa.mir.1277 and hsa.mir.485.3b on gene module 4. Pair1: hsa.mir.937 and hsa.let.7b on gene module 7 In the hsa.mir.937 and hsa.let.7b, the log10pv is on their best eigengene -- gene module 7 of the 9 qualified gene modules (The original gene module label is 75 before IQR filtering). This high log10pv indicates that the PV for the corresponding model comparison test is very small and indicates the significance of the interaction effect of the two mirnas. From figure 7, we can see that the corresponding expression value of the two mirnas over 493 patients shows a linear pattern. However, the nonadditive interaction is illustrated by color of figure 7. The color, which indicates the eigengene expression level, does not exhibit a linear trend related to the mirna expression values. In other words, we could not find the value of eigengene increasing or decreasing as the expression of mirnas monotonically changes. It means the mirnas are not additively controlling the Figure 8: hsa.mir.1277 and hsa.mir.485.3b on gene module 4 In this selected pair, hsa.mir.1277 and hsa.mir.485.3b, the log10pv is on their best eigengene -- gene module 4 of the 9 qualified gene modules (The original gene module label is 10 before IQR filtering). Similarly, the log10pv is large, which indicates that the PV for the corresponding model comparison test is very small. We fail to reject adding interaction term in the full model. In this pair, we do not find additive effects of the mirnas on their best eigengene, which proves
6 their non-additive interaction on the eigengene expression exists. Conclusion: We have successfully discovered the existence of non additive interactions between mirnas on affecting the gene expression of patients with ovarian cancer. Although the significance of mirna singletons on regulating gene expression is recognized today, there are seldom studies investigating on the pair effects of mirnas in ovarian cancer. Through our analysis, we have successfully discovered the existence of non additive interactions between mirnas that affect gene expression of patients with ovarian cancer. This discovery gives strong support for further analysis, such pathways analysis relates to nonadditive interaction effects of mirnas on ovarian cancer or other cancers. References: 1. "Ovarian Cancer- Get the Facts about Gynecologic Cancer." Centers for Disease Control and Prevention. Mar Web.04.Mar < 8.pdf>. 2. Douglas D. Taylor, Cicek Gercel-Taylor, MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer, Gynecologic Oncology, Volume 110, Issue 1, July 2008, Pages 13-21, ISSN , DOI: /j.ygyno ( ct.com/science/article/pii/s ) 3. Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, et al.microrna expression profiles in serous ovarian carcinoma. Clin Cancer Res 2008;14: < ull.html> 4. Altekruse SF, Kosary CL, Krapcho M, Neyman N, Aminou R, Waldron W, Ruhl J, Howlader N, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Cronin K, Chen HS, Feuer EJ, Stinchcomb DG, Edwards BK (eds). SEER Cancer Statistics Review, , National Cancer Institute. Bethesda, MD, _2007/, based on November 2009 SEER data submission, posted to the SEER web site, < > 6. Xiu-Qin Li, Shu-Lan Zhang, Zhen Cai, Yuan Zhou, Tian-Min Ye, Jen-Fu Chiu, Proteomic identification of tumorassociated protein in ovarian serous cystadenocarinoma, Cancer Letters, Volume 275, Issue 1, 8 March 2009, Pages , ISSN , DOI: /j.canlet < > 7. Marilena V. Iorio, Manuela Ferracin, Chang-Gong Liu, Angelo Veronese, et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer Cancer Res August 15, : ; doi: / can Mark Bloomston, Wendy L. Frankel, Fabio Petrocca, Stefano Volinia, Hansjuerg Alder, John P. Hagan, Chang-Gong Liu, Darshna Bhatt, Cristian Taccioli, Carlo M. Croce.Preliminary Communication MicroRNA Expression Patterns to Differentiate Pancreatic Adenocarcinoma From Normal Pancreas and Chronic Pancreatitis JAMA. 2007;297(17): doi: /jama Nozomu Yanaihara, Natasha Caplen, Elise Bowman, Masahiro Seike, Kensuke Kumamoto, Ming Yi, Robert M. Stephens, Aikou Okamoto, Jun Yokota, Tadao Tanaka, George Adrian Calin, Chang-Gong Liu, Carlo M. Croce, Curtis C. Harris, Unique microrna molecular profiles in lung cancer diagnosis and prognosis, Cancer Cell, Volume 9, Issue 3, March 2006, Pages , ISSN , DOI: /j.ccr < X> 10. Zhang B, Horvath S: A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 2005,4(1):. Article Horvath S, Zhang B, Carlson M, Lu KV, Zhu S, Felciano RM, Laurance MF, Zhao W, Qi S, Chen Z, et al.: Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci USA 2006, 103(46): Horvath S, Dong J: Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol 2008, 4(8):e Langfelder P, Horvath S: WGCNA: an R package for weighted gene co-expression network analysis. BMC Bioinformatics 2008, 9(1): Saris Christiaan Horvath, Steve van Vught, Paul van Es, Michael,Blauw, Hylke Fuller, Tova,et al / Weighted gene co-expression network analysis of the peripheral blood from Amyotrophic Lateral Sclerosis patients Storey JD (2002) A direct approach to false discovery rates. J R Stat Soc [SerB] 64: Storey JD, Tibshirani R (2003) Statistical significance for genome-wide studies. Proc Natl Acad Sci U S A Storey JD, Akey JM, Kruglyak L, 2005 Multiple Locus Linkage Analysis of Genomewide Expression in Yeast. PLoS Biol 3(8): e267. doi: /journal.pbio Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al.microrna signatures in human ovarian cancer. Cancer Res 2007;67: < ort>
Diet, Physical Activity, Weight & Cancer Survivorship: Perspectives from the NCI
 Diet, Physical Activity, Weight & Cancer Survivorship: Perspectives from the NCI American Institute for Cancer Research Meeting October 21, 2010 Catherine M. Alfano, Ph.D. Program Director & Behavioral
Diet, Physical Activity, Weight & Cancer Survivorship: Perspectives from the NCI American Institute for Cancer Research Meeting October 21, 2010 Catherine M. Alfano, Ph.D. Program Director & Behavioral
Lung and Bronchus Cancer Statistics in Virginia - Maps. July
 July 2011 4 July 2011 5 July 2011 6 July 2011 7 Lung and Bronchus Cancer Statistics in Virginia - Notes Sources: Incidence and percent local staging (VA Cancer Registry); mortality (VDH Division of Health
July 2011 4 July 2011 5 July 2011 6 July 2011 7 Lung and Bronchus Cancer Statistics in Virginia - Notes Sources: Incidence and percent local staging (VA Cancer Registry); mortality (VDH Division of Health
Colorectal Cancer Statistics in Virginia - Maps. July
 July 2011 5 July 2011 6 July 2011 7 July 2011 8 Colorectal Cancer Statistics in Virginia - Notes Sources: Incidence and percent local staging (VA Cancer Registry); mortality (VDH Division of Health Statistics);
July 2011 5 July 2011 6 July 2011 7 July 2011 8 Colorectal Cancer Statistics in Virginia - Notes Sources: Incidence and percent local staging (VA Cancer Registry); mortality (VDH Division of Health Statistics);
MicroRNA Cancer qpcr Array
 MicroRNA Cancer qpcr Array Array Layout Pre-designed set of 95 MicroRNA detection Primers U6 snrna Normalization transcript Selection of Candidate MicroRNAs for Cancer Array Sample References of MicroRNA
MicroRNA Cancer qpcr Array Array Layout Pre-designed set of 95 MicroRNA detection Primers U6 snrna Normalization transcript Selection of Candidate MicroRNAs for Cancer Array Sample References of MicroRNA
Overview of All SEER-Medicare Publications
 Overview of All SEER-Medicare Publications Outcomes Insights, Inc. Mark D. Danese, MHS, PhD Claire Cangialose February 22, 2017 Overview Information was extracted from the National Cancer Institute (NCI)
Overview of All SEER-Medicare Publications Outcomes Insights, Inc. Mark D. Danese, MHS, PhD Claire Cangialose February 22, 2017 Overview Information was extracted from the National Cancer Institute (NCI)
Gene-microRNA network module analysis for ovarian cancer
 Gene-microRNA network module analysis for ovarian cancer Shuqin Zhang School of Mathematical Sciences Fudan University Oct. 4, 2016 Outline Introduction Materials and Methods Results Conclusions Introduction
Gene-microRNA network module analysis for ovarian cancer Shuqin Zhang School of Mathematical Sciences Fudan University Oct. 4, 2016 Outline Introduction Materials and Methods Results Conclusions Introduction
developing new tools for diagnostics Join forces with IMGM Laboratories to make your mirna project a success
 micrornas developing new tools for diagnostics Join forces with IMGM Laboratories to make your mirna project a success Dr. Carola Wagner IMGM Laboratories GmbH Martinsried, Germany qpcr 2009 Symposium
micrornas developing new tools for diagnostics Join forces with IMGM Laboratories to make your mirna project a success Dr. Carola Wagner IMGM Laboratories GmbH Martinsried, Germany qpcr 2009 Symposium
Healthcare Reform and Cancer Survivorship: Implications for Care & Research
 Healthcare Reform and Cancer Survivorship: Implications for Care & Research Julia H. Rowland, Ph.D., Director Office of Cancer Survivorship National Cancer Institute National Institutes of Health DHHS
Healthcare Reform and Cancer Survivorship: Implications for Care & Research Julia H. Rowland, Ph.D., Director Office of Cancer Survivorship National Cancer Institute National Institutes of Health DHHS
South Asian Journal of Engineering and Technology Vol.3, No.9 (2017) 17 22
 South Asian Journal of Engineering and Technology Vol.3, No.9 (07) 7 Detection of malignant and benign Tumors by ANN Classification Method K. Gandhimathi, Abirami.K, Nandhini.B Idhaya Engineering College
South Asian Journal of Engineering and Technology Vol.3, No.9 (07) 7 Detection of malignant and benign Tumors by ANN Classification Method K. Gandhimathi, Abirami.K, Nandhini.B Idhaya Engineering College
Bootstrapped Integrative Hypothesis Test, COPD-Lung Cancer Differentiation, and Joint mirnas Biomarkers
 Bootstrapped Integrative Hypothesis Test, COPD-Lung Cancer Differentiation, and Joint mirnas Biomarkers Kai-Ming Jiang 1,2, Bao-Liang Lu 1,2, and Lei Xu 1,2,3(&) 1 Department of Computer Science and Engineering,
Bootstrapped Integrative Hypothesis Test, COPD-Lung Cancer Differentiation, and Joint mirnas Biomarkers Kai-Ming Jiang 1,2, Bao-Liang Lu 1,2, and Lei Xu 1,2,3(&) 1 Department of Computer Science and Engineering,
Cancer of the Breast (Female) - Cancer Stat Facts
 Page 1 of 9 Home Statistical Summaries Cancer Stat Facts Cancer of the Breast (Female) Cancer Stat Facts: Female Breast Cancer Statistics at a Glance At a Glance Estimated New Cases in 2016 % of All New
Page 1 of 9 Home Statistical Summaries Cancer Stat Facts Cancer of the Breast (Female) Cancer Stat Facts: Female Breast Cancer Statistics at a Glance At a Glance Estimated New Cases in 2016 % of All New
Breast Cancer Survivorship: Physical Activity
 WORKING EVERYDAY TO PROVIDE THE HIGHEST QUALITY OF LIFE FOR PEOPLE WITH CANCER Breast Cancer Survivorship: What is the Role of Diet and Physical Activity WENDY DEMARK-WAHNEFRIED, PHD, RD PROFESSOR AND
WORKING EVERYDAY TO PROVIDE THE HIGHEST QUALITY OF LIFE FOR PEOPLE WITH CANCER Breast Cancer Survivorship: What is the Role of Diet and Physical Activity WENDY DEMARK-WAHNEFRIED, PHD, RD PROFESSOR AND
National Surgical Adjuvant Breast and Bowel Project (NSABP) Foundation Annual Progress Report: 2009 Formula Grant
 National Surgical Adjuvant Breast and Bowel Project (NSABP) Foundation Annual Progress Report: 2009 Formula Grant Reporting Period July 1, 2011 June 30, 2012 Formula Grant Overview The National Surgical
National Surgical Adjuvant Breast and Bowel Project (NSABP) Foundation Annual Progress Report: 2009 Formula Grant Reporting Period July 1, 2011 June 30, 2012 Formula Grant Overview The National Surgical
Percent Surviving 5 Years 89.4%
 Page 1 of 8 Surveillance, Epidemiology, and End Results Program Turning Cancer Data Into Discovery Home Statistical Summaries Cancer Stat Fact Sheets Cancer of the Breast SEER Stat Fact Sheets: Breast
Page 1 of 8 Surveillance, Epidemiology, and End Results Program Turning Cancer Data Into Discovery Home Statistical Summaries Cancer Stat Fact Sheets Cancer of the Breast SEER Stat Fact Sheets: Breast
Patnaik SK, et al. MicroRNAs to accurately histotype NSCLC biopsies
 Patnaik SK, et al. MicroRNAs to accurately histotype NSCLC biopsies. 2014. Supplemental Digital Content 1. Appendix 1. External data-sets used for associating microrna expression with lung squamous cell
Patnaik SK, et al. MicroRNAs to accurately histotype NSCLC biopsies. 2014. Supplemental Digital Content 1. Appendix 1. External data-sets used for associating microrna expression with lung squamous cell
Nature Methods: doi: /nmeth.3115
 Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
The detection of differentially expressed micrornas from the serum of ovarian cancer patients using a novel real-time PCR platform
 Available online at www.sciencedirect.com Gynecologic Oncology 112 (2009) 55 59 www.elsevier.com/locate/ygyno The detection of differentially expressed micrornas from the serum of ovarian cancer patients
Available online at www.sciencedirect.com Gynecologic Oncology 112 (2009) 55 59 www.elsevier.com/locate/ygyno The detection of differentially expressed micrornas from the serum of ovarian cancer patients
Li-Xuan Qin 1* and Douglas A. Levine 2
 Qin and Levine BMC Medical Genomics (2016) 9:27 DOI 10.1186/s12920-016-0187-4 RESEARCH ARTICLE Open Access Study design and data analysis considerations for the discovery of prognostic molecular biomarkers:
Qin and Levine BMC Medical Genomics (2016) 9:27 DOI 10.1186/s12920-016-0187-4 RESEARCH ARTICLE Open Access Study design and data analysis considerations for the discovery of prognostic molecular biomarkers:
The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis
 The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis Tieliu Shi tlshi@bio.ecnu.edu.cn The Center for bioinformatics
The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis Tieliu Shi tlshi@bio.ecnu.edu.cn The Center for bioinformatics
Integration of Genetic and Genomic Approaches for the Analysis of Chronic Fatigue Syndrome Implicates Forkhead Box N1
 Integration of Genetic and Genomic Approaches for the Analysis of Chronic Fatigue Syndrome Implicates Forkhead Box N1 Angela Presson, Jeanette Papp, Eric Sobel, and Steve Horvath Biostatistics and Human
Integration of Genetic and Genomic Approaches for the Analysis of Chronic Fatigue Syndrome Implicates Forkhead Box N1 Angela Presson, Jeanette Papp, Eric Sobel, and Steve Horvath Biostatistics and Human
Improving Representation of Diverse Patients in Clinical Trials
 Improving Representation of Diverse Patients in Clinical Trials Coleman Obasaju, MD, PhD, Senior Medical Director US Medical Division, Oncology, Eli Lilly and Company Lilly s commitment to clinical diversity
Improving Representation of Diverse Patients in Clinical Trials Coleman Obasaju, MD, PhD, Senior Medical Director US Medical Division, Oncology, Eli Lilly and Company Lilly s commitment to clinical diversity
Introduction to Gene Sets Analysis
 Introduction to Svitlana Tyekucheva Dana-Farber Cancer Institute May 15, 2012 Introduction Various measurements: gene expression, copy number variation, methylation status, mutation profile, etc. Main
Introduction to Svitlana Tyekucheva Dana-Farber Cancer Institute May 15, 2012 Introduction Various measurements: gene expression, copy number variation, methylation status, mutation profile, etc. Main
Early Detection: Screening Guidelines
 Early Detection: Screening Guidelines National Cancer Statistics 2015 National Cancer Statistics 1. 6 Million Cases Expected 589,430 Cancer Deaths expected 2015 Virginia Cancer Statistics: Projected cases:
Early Detection: Screening Guidelines National Cancer Statistics 2015 National Cancer Statistics 1. 6 Million Cases Expected 589,430 Cancer Deaths expected 2015 Virginia Cancer Statistics: Projected cases:
On the Reproducibility of TCGA Ovarian Cancer MicroRNA Profiles
 On the Reproducibility of TCGA Ovarian Cancer MicroRNA Profiles Ying-Wooi Wan 1,2,4, Claire M. Mach 2,3, Genevera I. Allen 1,7,8, Matthew L. Anderson 2,4,5 *, Zhandong Liu 1,5,6,7 * 1 Departments of Pediatrics
On the Reproducibility of TCGA Ovarian Cancer MicroRNA Profiles Ying-Wooi Wan 1,2,4, Claire M. Mach 2,3, Genevera I. Allen 1,7,8, Matthew L. Anderson 2,4,5 *, Zhandong Liu 1,5,6,7 * 1 Departments of Pediatrics
Identification of Tissue Independent Cancer Driver Genes
 Identification of Tissue Independent Cancer Driver Genes Alexandros Manolakos, Idoia Ochoa, Kartik Venkat Supervisor: Olivier Gevaert Abstract Identification of genomic patterns in tumors is an important
Identification of Tissue Independent Cancer Driver Genes Alexandros Manolakos, Idoia Ochoa, Kartik Venkat Supervisor: Olivier Gevaert Abstract Identification of genomic patterns in tumors is an important
Cancer outlier differential gene expression detection
 Biostatistics (2007), 8, 3, pp. 566 575 doi:10.1093/biostatistics/kxl029 Advance Access publication on October 4, 2006 Cancer outlier differential gene expression detection BAOLIN WU Division of Biostatistics,
Biostatistics (2007), 8, 3, pp. 566 575 doi:10.1093/biostatistics/kxl029 Advance Access publication on October 4, 2006 Cancer outlier differential gene expression detection BAOLIN WU Division of Biostatistics,
Trends in Oncology Care. Challenges and Obstacles. Lawrence N. Shulman, MD Dana-Farber Cancer Institute Brigham and Women s Hospital
 Trends in Oncology Care Challenges and Obstacles Lawrence N. Shulman, MD Dana-Farber Cancer Institute Brigham and Women s Hospital Navigation Through The Continuum of Cancer Care Education Screening Early
Trends in Oncology Care Challenges and Obstacles Lawrence N. Shulman, MD Dana-Farber Cancer Institute Brigham and Women s Hospital Navigation Through The Continuum of Cancer Care Education Screening Early
Single SNP/Gene Analysis. Typical Results of GWAS Analysis (Single SNP Approach) Typical Results of GWAS Analysis (Single SNP Approach)
 High-Throughput Sequencing Course Gene-Set Analysis Biostatistics and Bioinformatics Summer 28 Section Introduction What is Gene Set Analysis? Many names for gene set analysis: Pathway analysis Gene set
High-Throughput Sequencing Course Gene-Set Analysis Biostatistics and Bioinformatics Summer 28 Section Introduction What is Gene Set Analysis? Many names for gene set analysis: Pathway analysis Gene set
Using CART to Mine SELDI ProteinChip Data for Biomarkers and Disease Stratification
 Using CART to Mine SELDI ProteinChip Data for Biomarkers and Disease Stratification Kenna Mawk, D.V.M. Informatics Product Manager Ciphergen Biosystems, Inc. Outline Introduction to ProteinChip Technology
Using CART to Mine SELDI ProteinChip Data for Biomarkers and Disease Stratification Kenna Mawk, D.V.M. Informatics Product Manager Ciphergen Biosystems, Inc. Outline Introduction to ProteinChip Technology
Bioimaging and Functional Genomics
 Bioimaging and Functional Genomics Elisa Ficarra, EPF Lausanne Giovanni De Micheli, EPF Lausanne Sungroh Yoon, Stanford University Luca Benini, University of Bologna Enrico Macii,, Politecnico di Torino
Bioimaging and Functional Genomics Elisa Ficarra, EPF Lausanne Giovanni De Micheli, EPF Lausanne Sungroh Yoon, Stanford University Luca Benini, University of Bologna Enrico Macii,, Politecnico di Torino
MicroRNA expression profiling and functional analysis in prostate cancer. Marco Folini s.c. Ricerca Traslazionale DOSL
 MicroRNA expression profiling and functional analysis in prostate cancer Marco Folini s.c. Ricerca Traslazionale DOSL What are micrornas? For almost three decades, the alteration of protein-coding genes
MicroRNA expression profiling and functional analysis in prostate cancer Marco Folini s.c. Ricerca Traslazionale DOSL What are micrornas? For almost three decades, the alteration of protein-coding genes
CoINcIDE: A framework for discovery of patient subtypes across multiple datasets
 Planey and Gevaert Genome Medicine (2016) 8:27 DOI 10.1186/s13073-016-0281-4 METHOD CoINcIDE: A framework for discovery of patient subtypes across multiple datasets Catherine R. Planey and Olivier Gevaert
Planey and Gevaert Genome Medicine (2016) 8:27 DOI 10.1186/s13073-016-0281-4 METHOD CoINcIDE: A framework for discovery of patient subtypes across multiple datasets Catherine R. Planey and Olivier Gevaert
Nature Genetics: doi: /ng Supplementary Figure 1. SEER data for male and female cancer incidence from
 Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Chapter 1. Introduction
 Chapter 1 Introduction 1.1 Motivation and Goals The increasing availability and decreasing cost of high-throughput (HT) technologies coupled with the availability of computational tools and data form a
Chapter 1 Introduction 1.1 Motivation and Goals The increasing availability and decreasing cost of high-throughput (HT) technologies coupled with the availability of computational tools and data form a
Statistical Analysis of Single Nucleotide Polymorphism Microarrays in Cancer Studies
 Statistical Analysis of Single Nucleotide Polymorphism Microarrays in Cancer Studies Stanford Biostatistics Workshop Pierre Neuvial with Henrik Bengtsson and Terry Speed Department of Statistics, UC Berkeley
Statistical Analysis of Single Nucleotide Polymorphism Microarrays in Cancer Studies Stanford Biostatistics Workshop Pierre Neuvial with Henrik Bengtsson and Terry Speed Department of Statistics, UC Berkeley
Package AbsFilterGSEA
 Type Package Package AbsFilterGSEA September 21, 2017 Title Improved False Positive Control of Gene-Permuting GSEA with Absolute Filtering Version 1.5.1 Author Sora Yoon Maintainer
Type Package Package AbsFilterGSEA September 21, 2017 Title Improved False Positive Control of Gene-Permuting GSEA with Absolute Filtering Version 1.5.1 Author Sora Yoon Maintainer
Inferring condition-specific mirna activity from matched mirna and mrna expression data
 Inferring condition-specific mirna activity from matched mirna and mrna expression data Junpeng Zhang 1, Thuc Duy Le 2, Lin Liu 2, Bing Liu 3, Jianfeng He 4, Gregory J Goodall 5 and Jiuyong Li 2,* 1 Faculty
Inferring condition-specific mirna activity from matched mirna and mrna expression data Junpeng Zhang 1, Thuc Duy Le 2, Lin Liu 2, Bing Liu 3, Jianfeng He 4, Gregory J Goodall 5 and Jiuyong Li 2,* 1 Faculty
Mercy s 2013 Cancer Program Annual Report
 Mercy s 2013 Cancer Program Annual Report Mercy Hospital & Medical Center is accredited by the Commission on Cancer as an Academic Comprehensive Cancer Program. This study directed by the Mercy s Cancer
Mercy s 2013 Cancer Program Annual Report Mercy Hospital & Medical Center is accredited by the Commission on Cancer as an Academic Comprehensive Cancer Program. This study directed by the Mercy s Cancer
HALLA KABAT * Outreach Program, mircore, 2929 Plymouth Rd. Ann Arbor, MI 48105, USA LEO TUNKLE *
 CERNA SEARCH METHOD IDENTIFIED A MET-ACTIVATED SUBGROUP AMONG EGFR DNA AMPLIFIED LUNG ADENOCARCINOMA PATIENTS HALLA KABAT * Outreach Program, mircore, 2929 Plymouth Rd. Ann Arbor, MI 48105, USA Email:
CERNA SEARCH METHOD IDENTIFIED A MET-ACTIVATED SUBGROUP AMONG EGFR DNA AMPLIFIED LUNG ADENOCARCINOMA PATIENTS HALLA KABAT * Outreach Program, mircore, 2929 Plymouth Rd. Ann Arbor, MI 48105, USA Email:
The LiquidAssociation Package
 The LiquidAssociation Package Yen-Yi Ho October 30, 2018 1 Introduction The LiquidAssociation package provides analytical methods to study three-way interactions. It incorporates methods to examine a particular
The LiquidAssociation Package Yen-Yi Ho October 30, 2018 1 Introduction The LiquidAssociation package provides analytical methods to study three-way interactions. It incorporates methods to examine a particular
Weighted Gene Co-expression Network Analysis (WGCNA) R Tutorial, Part C Summary The data and biological implications are described in
 Weighted Gene Co-expression Network Analysis (WGCNA) R Tutorial, Part C Breast Cancer Microarray Data. Steve Horvath, Paul Mischel Correspondence: shorvath@mednet.ucla.edu, http://www.ph.ucla.edu/biostat/people/horvath.htm
Weighted Gene Co-expression Network Analysis (WGCNA) R Tutorial, Part C Breast Cancer Microarray Data. Steve Horvath, Paul Mischel Correspondence: shorvath@mednet.ucla.edu, http://www.ph.ucla.edu/biostat/people/horvath.htm
Kidney Cancer Causes, Risk Factors, and Prevention
 Kidney Cancer Causes, Risk Factors, and Prevention Risk Factors A risk factor is anything that affects your chance of getting a disease such as cancer. Learn more about the risk factors for kidney cancer.
Kidney Cancer Causes, Risk Factors, and Prevention Risk Factors A risk factor is anything that affects your chance of getting a disease such as cancer. Learn more about the risk factors for kidney cancer.
SUPPLEMENTARY FIGURE LEGENDS
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Negative correlation between mir-375 and its predicted target genes, as demonstrated by gene set enrichment analysis (GSEA). 1 The correlation between
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Negative correlation between mir-375 and its predicted target genes, as demonstrated by gene set enrichment analysis (GSEA). 1 The correlation between
Profiles of gene expression & diagnosis/prognosis of cancer. MCs in Advanced Genetics Ainoa Planas Riverola
 Profiles of gene expression & diagnosis/prognosis of cancer MCs in Advanced Genetics Ainoa Planas Riverola Gene expression profiles Gene expression profiling Used in molecular biology, it measures the
Profiles of gene expression & diagnosis/prognosis of cancer MCs in Advanced Genetics Ainoa Planas Riverola Gene expression profiles Gene expression profiling Used in molecular biology, it measures the
What you should know before you collect data. BAE 815 (Fall 2017) Dr. Zifei Liu
 What you should know before you collect data BAE 815 (Fall 2017) Dr. Zifei Liu Zifeiliu@ksu.edu Types and levels of study Descriptive statistics Inferential statistics How to choose a statistical test
What you should know before you collect data BAE 815 (Fall 2017) Dr. Zifei Liu Zifeiliu@ksu.edu Types and levels of study Descriptive statistics Inferential statistics How to choose a statistical test
Supplementary Figure 1: Experimental design. DISCOVERY PHASE VALIDATION PHASE (N = 88) (N = 20) Healthy = 20. Healthy = 6. Endometriosis = 33
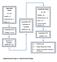 DISCOVERY PHASE (N = 20) Healthy = 6 Endometriosis = 7 EAOC = 7 Quantitative PCR (mirnas = 1113) a Quantitative PCR Verification of Candidate mirnas (N = 24) VALIDATION PHASE (N = 88) Healthy = 20 Endometriosis
DISCOVERY PHASE (N = 20) Healthy = 6 Endometriosis = 7 EAOC = 7 Quantitative PCR (mirnas = 1113) a Quantitative PCR Verification of Candidate mirnas (N = 24) VALIDATION PHASE (N = 88) Healthy = 20 Endometriosis
Simple Linear Regression the model, estimation and testing
 Simple Linear Regression the model, estimation and testing Lecture No. 05 Example 1 A production manager has compared the dexterity test scores of five assembly-line employees with their hourly productivity.
Simple Linear Regression the model, estimation and testing Lecture No. 05 Example 1 A production manager has compared the dexterity test scores of five assembly-line employees with their hourly productivity.
Mir-595 is a significant indicator of poor patient prognosis in epithelial ovarian cancer
 European Review for Medical and Pharmacological Sciences 2017; 21: 4278-4282 Mir-595 is a significant indicator of poor patient prognosis in epithelial ovarian cancer Q.-H. ZHOU 1, Y.-M. ZHAO 2, L.-L.
European Review for Medical and Pharmacological Sciences 2017; 21: 4278-4282 Mir-595 is a significant indicator of poor patient prognosis in epithelial ovarian cancer Q.-H. ZHOU 1, Y.-M. ZHAO 2, L.-L.
SubLasso:a feature selection and classification R package with a. fixed feature subset
 SubLasso:a feature selection and classification R package with a fixed feature subset Youxi Luo,3,*, Qinghan Meng,2,*, Ruiquan Ge,2, Guoqin Mai, Jikui Liu, Fengfeng Zhou,#. Shenzhen Institutes of Advanced
SubLasso:a feature selection and classification R package with a fixed feature subset Youxi Luo,3,*, Qinghan Meng,2,*, Ruiquan Ge,2, Guoqin Mai, Jikui Liu, Fengfeng Zhou,#. Shenzhen Institutes of Advanced
Classification of cancer profiles. ABDBM Ron Shamir
 Classification of cancer profiles 1 Background: Cancer Classification Cancer classification is central to cancer treatment; Traditional cancer classification methods: location; morphology, cytogenesis;
Classification of cancer profiles 1 Background: Cancer Classification Cancer classification is central to cancer treatment; Traditional cancer classification methods: location; morphology, cytogenesis;
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Association of mir-21 with esophageal cancer prognosis: a meta-analysis
 Association of mir-21 with esophageal cancer prognosis: a meta-analysis S.-W. Wen 1, Y.-F. Zhang 1, Y. Li 1, Z.-X. Liu 2, H.-L. Lv 1, Z.-H. Li 1, Y.-Z. Xu 1, Y.-G. Zhu 1 and Z.-Q. Tian 1 1 Department of
Association of mir-21 with esophageal cancer prognosis: a meta-analysis S.-W. Wen 1, Y.-F. Zhang 1, Y. Li 1, Z.-X. Liu 2, H.-L. Lv 1, Z.-H. Li 1, Y.-Z. Xu 1, Y.-G. Zhu 1 and Z.-Q. Tian 1 1 Department of
Analysis of paired mirna-mrna microarray expression data using a stepwise multiple linear regression model
 Analysis of paired mirna-mrna microarray expression data using a stepwise multiple linear regression model Yiqian Zhou 1, Rehman Qureshi 2, and Ahmet Sacan 3 1 Pure Storage, 650 Castro Street, Suite #260,
Analysis of paired mirna-mrna microarray expression data using a stepwise multiple linear regression model Yiqian Zhou 1, Rehman Qureshi 2, and Ahmet Sacan 3 1 Pure Storage, 650 Castro Street, Suite #260,
Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and
 Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and Promoter Motif Analysis Shisong Ma 1,2*, Michael Snyder 3, and Savithramma P Dinesh-Kumar 2* 1 School of Life Sciences, University
Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and Promoter Motif Analysis Shisong Ma 1,2*, Michael Snyder 3, and Savithramma P Dinesh-Kumar 2* 1 School of Life Sciences, University
Nature Medicine: doi: /nm.3967
 Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Undertaking statistical analysis of
 Descriptive statistics: Simply telling a story Laura Delaney introduces the principles of descriptive statistical analysis and presents an overview of the various ways in which data can be presented by
Descriptive statistics: Simply telling a story Laura Delaney introduces the principles of descriptive statistical analysis and presents an overview of the various ways in which data can be presented by
ASMS 2015 ThP 459 Glioblastoma Multiforme Subtype Classification: Integrated Analysis of Protein and Gene Expression Data
 ASMS 2015 ThP 459 Glioblastoma Multiforme Subtype Classification: Integrated Analysis of Protein and Gene Expression Data Durairaj Renu 1, Vadiraja Bhat 2, Mona Al-Gizawiy 3, Carolina B. Livi 2, Stephen
ASMS 2015 ThP 459 Glioblastoma Multiforme Subtype Classification: Integrated Analysis of Protein and Gene Expression Data Durairaj Renu 1, Vadiraja Bhat 2, Mona Al-Gizawiy 3, Carolina B. Livi 2, Stephen
Aspects of Statistical Modelling & Data Analysis in Gene Expression Genomics. Mike West Duke University
 Aspects of Statistical Modelling & Data Analysis in Gene Expression Genomics Mike West Duke University Papers, software, many links: www.isds.duke.edu/~mw ABS04 web site: Lecture slides, stats notes, papers,
Aspects of Statistical Modelling & Data Analysis in Gene Expression Genomics Mike West Duke University Papers, software, many links: www.isds.duke.edu/~mw ABS04 web site: Lecture slides, stats notes, papers,
Identifying Relevant micrornas in Bladder Cancer using Multi-Task Learning
 Identifying Relevant micrornas in Bladder Cancer using Multi-Task Learning Adriana Birlutiu (1),(2), Paul Bulzu (1), Irina Iereminciuc (1), and Alexandru Floares (1) (1) SAIA & OncoPredict, Cluj-Napoca,
Identifying Relevant micrornas in Bladder Cancer using Multi-Task Learning Adriana Birlutiu (1),(2), Paul Bulzu (1), Irina Iereminciuc (1), and Alexandru Floares (1) (1) SAIA & OncoPredict, Cluj-Napoca,
Diagnosis of multiple cancer types by shrunken centroids of gene expression
 Diagnosis of multiple cancer types by shrunken centroids of gene expression Robert Tibshirani, Trevor Hastie, Balasubramanian Narasimhan, and Gilbert Chu PNAS 99:10:6567-6572, 14 May 2002 Nearest Centroid
Diagnosis of multiple cancer types by shrunken centroids of gene expression Robert Tibshirani, Trevor Hastie, Balasubramanian Narasimhan, and Gilbert Chu PNAS 99:10:6567-6572, 14 May 2002 Nearest Centroid
Comparison of Gene Set Analysis with Various Score Transformations to Test the Significance of Sets of Genes
 Comparison of Gene Set Analysis with Various Score Transformations to Test the Significance of Sets of Genes Ivan Arreola and Dr. David Han Department of Management of Science and Statistics, University
Comparison of Gene Set Analysis with Various Score Transformations to Test the Significance of Sets of Genes Ivan Arreola and Dr. David Han Department of Management of Science and Statistics, University
Profile Analysis. Intro and Assumptions Psy 524 Andrew Ainsworth
 Profile Analysis Intro and Assumptions Psy 524 Andrew Ainsworth Profile Analysis Profile analysis is the repeated measures extension of MANOVA where a set of DVs are commensurate (on the same scale). Profile
Profile Analysis Intro and Assumptions Psy 524 Andrew Ainsworth Profile Analysis Profile analysis is the repeated measures extension of MANOVA where a set of DVs are commensurate (on the same scale). Profile
Bayes Linear Statistics. Theory and Methods
 Bayes Linear Statistics Theory and Methods Michael Goldstein and David Wooff Durham University, UK BICENTENNI AL BICENTENNIAL Contents r Preface xvii 1 The Bayes linear approach 1 1.1 Combining beliefs
Bayes Linear Statistics Theory and Methods Michael Goldstein and David Wooff Durham University, UK BICENTENNI AL BICENTENNIAL Contents r Preface xvii 1 The Bayes linear approach 1 1.1 Combining beliefs
Classification. Methods Course: Gene Expression Data Analysis -Day Five. Rainer Spang
 Classification Methods Course: Gene Expression Data Analysis -Day Five Rainer Spang Ms. Smith DNA Chip of Ms. Smith Expression profile of Ms. Smith Ms. Smith 30.000 properties of Ms. Smith The expression
Classification Methods Course: Gene Expression Data Analysis -Day Five Rainer Spang Ms. Smith DNA Chip of Ms. Smith Expression profile of Ms. Smith Ms. Smith 30.000 properties of Ms. Smith The expression
Reveal Relationships in Categorical Data
 SPSS Categories 15.0 Specifications Reveal Relationships in Categorical Data Unleash the full potential of your data through perceptual mapping, optimal scaling, preference scaling, and dimension reduction
SPSS Categories 15.0 Specifications Reveal Relationships in Categorical Data Unleash the full potential of your data through perceptual mapping, optimal scaling, preference scaling, and dimension reduction
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Illustration of the working of network-based SVM to confidently predict a new (and now confirmed) ASD gene. Gene CTNND2 s brain network neighborhood that enabled its prediction by
Supplementary Figure 1 Illustration of the working of network-based SVM to confidently predict a new (and now confirmed) ASD gene. Gene CTNND2 s brain network neighborhood that enabled its prediction by
CRITERIA FOR USE. A GRAPHICAL EXPLANATION OF BI-VARIATE (2 VARIABLE) REGRESSION ANALYSISSys
 Multiple Regression Analysis 1 CRITERIA FOR USE Multiple regression analysis is used to test the effects of n independent (predictor) variables on a single dependent (criterion) variable. Regression tests
Multiple Regression Analysis 1 CRITERIA FOR USE Multiple regression analysis is used to test the effects of n independent (predictor) variables on a single dependent (criterion) variable. Regression tests
Unit 1 Exploring and Understanding Data
 Unit 1 Exploring and Understanding Data Area Principle Bar Chart Boxplot Conditional Distribution Dotplot Empirical Rule Five Number Summary Frequency Distribution Frequency Polygon Histogram Interquartile
Unit 1 Exploring and Understanding Data Area Principle Bar Chart Boxplot Conditional Distribution Dotplot Empirical Rule Five Number Summary Frequency Distribution Frequency Polygon Histogram Interquartile
Feature Vector Denoising with Prior Network Structures. (with Y. Fan, L. Raphael) NESS 2015, University of Connecticut
 Feature Vector Denoising with Prior Network Structures (with Y. Fan, L. Raphael) NESS 2015, University of Connecticut Summary: I. General idea: denoising functions on Euclidean space ---> denoising in
Feature Vector Denoising with Prior Network Structures (with Y. Fan, L. Raphael) NESS 2015, University of Connecticut Summary: I. General idea: denoising functions on Euclidean space ---> denoising in
Statistical Assessment of the Global Regulatory Role of Histone. Acetylation in Saccharomyces cerevisiae. (Support Information)
 Statistical Assessment of the Global Regulatory Role of Histone Acetylation in Saccharomyces cerevisiae (Support Information) Authors: Guo-Cheng Yuan, Ping Ma, Wenxuan Zhong and Jun S. Liu Linear Relationship
Statistical Assessment of the Global Regulatory Role of Histone Acetylation in Saccharomyces cerevisiae (Support Information) Authors: Guo-Cheng Yuan, Ping Ma, Wenxuan Zhong and Jun S. Liu Linear Relationship
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Figure S2. Distribution of acgh probes on all ten chromosomes of the RIL M0022
 96 APPENDIX B. Supporting Information for chapter 4 "changes in genome content generated via segregation of non-allelic homologs" Figure S1. Potential de novo CNV probes and sizes of apparently de novo
96 APPENDIX B. Supporting Information for chapter 4 "changes in genome content generated via segregation of non-allelic homologs" Figure S1. Potential de novo CNV probes and sizes of apparently de novo
About diffcoexp. Wenbin Wei, Sandeep Amberkar, Winston Hide
 About diffcoexp Wenbin Wei, Sandeep Amberkar, Winston Hide Sheffield Institute of Translational Neuroscience, University of Sheffield, Sheffield, United Kingdom April 11, 2018 Contents 1 Description 1
About diffcoexp Wenbin Wei, Sandeep Amberkar, Winston Hide Sheffield Institute of Translational Neuroscience, University of Sheffield, Sheffield, United Kingdom April 11, 2018 Contents 1 Description 1
Method Comparison Report Semi-Annual 1/5/2018
 Method Comparison Report Semi-Annual 1/5/2018 Prepared for Carl Commissioner Regularatory Commission 123 Commission Drive Anytown, XX, 12345 Prepared by Dr. Mark Mainstay Clinical Laboratory Kennett Community
Method Comparison Report Semi-Annual 1/5/2018 Prepared for Carl Commissioner Regularatory Commission 123 Commission Drive Anytown, XX, 12345 Prepared by Dr. Mark Mainstay Clinical Laboratory Kennett Community
A Biclustering Based Classification Framework for Cancer Diagnosis and Prognosis
 A Biclustering Based Classification Framework for Cancer Diagnosis and Prognosis Baljeet Malhotra and Guohui Lin Department of Computing Science, University of Alberta, Edmonton, Alberta, Canada T6G 2E8
A Biclustering Based Classification Framework for Cancer Diagnosis and Prognosis Baljeet Malhotra and Guohui Lin Department of Computing Science, University of Alberta, Edmonton, Alberta, Canada T6G 2E8
PANCREATIC CANCER IS A LEthal
 PRELIMINARY COMMUNICATION MicroRNA Expression Patterns to Differentiate Pancreatic Adenocarcinoma From Normal Pancreas and Chronic Pancreatitis Mark Bloomston, MD Wendy L. Frankel, MD Fabio Petrocca, MD
PRELIMINARY COMMUNICATION MicroRNA Expression Patterns to Differentiate Pancreatic Adenocarcinoma From Normal Pancreas and Chronic Pancreatitis Mark Bloomston, MD Wendy L. Frankel, MD Fabio Petrocca, MD
Head and neck cancer disparities, Genetics, Environment, Behavior: What really matters?
 Head and neck cancer disparities, Genetics, Environment, Behavior: What really matters? Camille Ragin, PhD, MPH Associate Professor Cancer Prevention and Control Program No financial relationships with
Head and neck cancer disparities, Genetics, Environment, Behavior: What really matters? Camille Ragin, PhD, MPH Associate Professor Cancer Prevention and Control Program No financial relationships with
Introduction to Discrimination in Microarray Data Analysis
 Introduction to Discrimination in Microarray Data Analysis Jane Fridlyand CBMB University of California, San Francisco Genentech Hall Auditorium, Mission Bay, UCSF October 23, 2004 1 Case Study: Van t
Introduction to Discrimination in Microarray Data Analysis Jane Fridlyand CBMB University of California, San Francisco Genentech Hall Auditorium, Mission Bay, UCSF October 23, 2004 1 Case Study: Van t
LAB ASSIGNMENT 4 INFERENCES FOR NUMERICAL DATA. Comparison of Cancer Survival*
 LAB ASSIGNMENT 4 1 INFERENCES FOR NUMERICAL DATA In this lab assignment, you will analyze the data from a study to compare survival times of patients of both genders with different primary cancers. First,
LAB ASSIGNMENT 4 1 INFERENCES FOR NUMERICAL DATA In this lab assignment, you will analyze the data from a study to compare survival times of patients of both genders with different primary cancers. First,
2.75: 84% 2.5: 80% 2.25: 78% 2: 74% 1.75: 70% 1.5: 66% 1.25: 64% 1.0: 60% 0.5: 50% 0.25: 25% 0: 0%
 Capstone Test (will consist of FOUR quizzes and the FINAL test grade will be an average of the four quizzes). Capstone #1: Review of Chapters 1-3 Capstone #2: Review of Chapter 4 Capstone #3: Review of
Capstone Test (will consist of FOUR quizzes and the FINAL test grade will be an average of the four quizzes). Capstone #1: Review of Chapters 1-3 Capstone #2: Review of Chapter 4 Capstone #3: Review of
Machine Learning! Robert Stengel! Robotics and Intelligent Systems MAE 345,! Princeton University, 2017
 Machine Learning! Robert Stengel! Robotics and Intelligent Systems MAE 345,! Princeton University, 2017 A.K.A. Artificial Intelligence Unsupervised learning! Cluster analysis Patterns, Clumps, and Joining
Machine Learning! Robert Stengel! Robotics and Intelligent Systems MAE 345,! Princeton University, 2017 A.K.A. Artificial Intelligence Unsupervised learning! Cluster analysis Patterns, Clumps, and Joining
Binary Diagnostic Tests Two Independent Samples
 Chapter 537 Binary Diagnostic Tests Two Independent Samples Introduction An important task in diagnostic medicine is to measure the accuracy of two diagnostic tests. This can be done by comparing summary
Chapter 537 Binary Diagnostic Tests Two Independent Samples Introduction An important task in diagnostic medicine is to measure the accuracy of two diagnostic tests. This can be done by comparing summary
Package ORIClust. February 19, 2015
 Type Package Package ORIClust February 19, 2015 Title Order-restricted Information Criterion-based Clustering Algorithm Version 1.0-1 Date 2009-09-10 Author Maintainer Tianqing Liu
Type Package Package ORIClust February 19, 2015 Title Order-restricted Information Criterion-based Clustering Algorithm Version 1.0-1 Date 2009-09-10 Author Maintainer Tianqing Liu
Preliminary Report on Simple Statistical Tests (t-tests and bivariate correlations)
 Preliminary Report on Simple Statistical Tests (t-tests and bivariate correlations) After receiving my comments on the preliminary reports of your datasets, the next step for the groups is to complete
Preliminary Report on Simple Statistical Tests (t-tests and bivariate correlations) After receiving my comments on the preliminary reports of your datasets, the next step for the groups is to complete
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
bivariate analysis: The statistical analysis of the relationship between two variables.
 bivariate analysis: The statistical analysis of the relationship between two variables. cell frequency: The number of cases in a cell of a cross-tabulation (contingency table). chi-square (χ 2 ) test for
bivariate analysis: The statistical analysis of the relationship between two variables. cell frequency: The number of cases in a cell of a cross-tabulation (contingency table). chi-square (χ 2 ) test for
micrornas (mirna) and Biomarkers
 micrornas (mirna) and Biomarkers Small RNAs Make Big Splash mirnas & Genome Function Biomarkers in Cancer Future Prospects Javed Khan M.D. National Cancer Institute EORTC-NCI-ASCO November 2007 The Human
micrornas (mirna) and Biomarkers Small RNAs Make Big Splash mirnas & Genome Function Biomarkers in Cancer Future Prospects Javed Khan M.D. National Cancer Institute EORTC-NCI-ASCO November 2007 The Human
Psychology Research Process
 Psychology Research Process Logical Processes Induction Observation/Association/Using Correlation Trying to assess, through observation of a large group/sample, what is associated with what? Examples:
Psychology Research Process Logical Processes Induction Observation/Association/Using Correlation Trying to assess, through observation of a large group/sample, what is associated with what? Examples:
Problem #1 Neurological signs and symptoms of ciguatera poisoning as the start of treatment and 2.5 hours after treatment with mannitol.
 Ho (null hypothesis) Ha (alternative hypothesis) Problem #1 Neurological signs and symptoms of ciguatera poisoning as the start of treatment and 2.5 hours after treatment with mannitol. Hypothesis: Ho:
Ho (null hypothesis) Ha (alternative hypothesis) Problem #1 Neurological signs and symptoms of ciguatera poisoning as the start of treatment and 2.5 hours after treatment with mannitol. Hypothesis: Ho:
Doing Thousands of Hypothesis Tests at the Same Time. Bradley Efron Stanford University
 Doing Thousands of Hypothesis Tests at the Same Time Bradley Efron Stanford University 1 Simultaneous Hypothesis Testing 1980: Simultaneous Statistical Inference (Rupert Miller) 2, 3,, 20 simultaneous
Doing Thousands of Hypothesis Tests at the Same Time Bradley Efron Stanford University 1 Simultaneous Hypothesis Testing 1980: Simultaneous Statistical Inference (Rupert Miller) 2, 3,, 20 simultaneous
Downregulation of serum mir-17 and mir-106b levels in gastric cancer and benign gastric diseases
 Brief Communication Downregulation of serum mir-17 and mir-106b levels in gastric cancer and benign gastric diseases Qinghai Zeng 1 *, Cuihong Jin 2 *, Wenhang Chen 2, Fang Xia 3, Qi Wang 3, Fan Fan 4,
Brief Communication Downregulation of serum mir-17 and mir-106b levels in gastric cancer and benign gastric diseases Qinghai Zeng 1 *, Cuihong Jin 2 *, Wenhang Chen 2, Fang Xia 3, Qi Wang 3, Fan Fan 4,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63.
 Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
1) What is the independent variable? What is our Dependent Variable?
 1) What is the independent variable? What is our Dependent Variable? Independent Variable: Whether the font color and word name are the same or different. (Congruency) Dependent Variable: The amount of
1) What is the independent variable? What is our Dependent Variable? Independent Variable: Whether the font color and word name are the same or different. (Congruency) Dependent Variable: The amount of
MicroRNAs (mirnas) are highly conserved small
 MINERVA BIOTEC 2008;20:23-30 Using mirna expression data for the study of human cancer N. MASCELLANI 1, L. TAGLIAVINI 1, G. GAMBERONI 1, S. ROSSI 1, J. MARCHESINI 1, C. TACCIOLI 1, G. DI LEVA 1, M. NEGRINI
MINERVA BIOTEC 2008;20:23-30 Using mirna expression data for the study of human cancer N. MASCELLANI 1, L. TAGLIAVINI 1, G. GAMBERONI 1, S. ROSSI 1, J. MARCHESINI 1, C. TACCIOLI 1, G. DI LEVA 1, M. NEGRINI
MEASURES OF ASSOCIATION AND REGRESSION
 DEPARTMENT OF POLITICAL SCIENCE AND INTERNATIONAL RELATIONS Posc/Uapp 816 MEASURES OF ASSOCIATION AND REGRESSION I. AGENDA: A. Measures of association B. Two variable regression C. Reading: 1. Start Agresti
DEPARTMENT OF POLITICAL SCIENCE AND INTERNATIONAL RELATIONS Posc/Uapp 816 MEASURES OF ASSOCIATION AND REGRESSION I. AGENDA: A. Measures of association B. Two variable regression C. Reading: 1. Start Agresti
A COMBINATORY ALGORITHM OF UNIVARIATE AND MULTIVARIATE GENE SELECTION
 5-9 JATIT. All rights reserved. A COMBINATORY ALGORITHM OF UNIVARIATE AND MULTIVARIATE GENE SELECTION 1 H. Mahmoodian, M. Hamiruce Marhaban, 3 R. A. Rahim, R. Rosli, 5 M. Iqbal Saripan 1 PhD student, Department
5-9 JATIT. All rights reserved. A COMBINATORY ALGORITHM OF UNIVARIATE AND MULTIVARIATE GENE SELECTION 1 H. Mahmoodian, M. Hamiruce Marhaban, 3 R. A. Rahim, R. Rosli, 5 M. Iqbal Saripan 1 PhD student, Department
Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD
 Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD Department of Biomedical Informatics Department of Computer Science and Engineering The Ohio State University Review
Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD Department of Biomedical Informatics Department of Computer Science and Engineering The Ohio State University Review
SUPPLEMENTARY APPENDIX
 SUPPLEMENTARY APPENDIX 1) Supplemental Figure 1. Histopathologic Characteristics of the Tumors in the Discovery Cohort 2) Supplemental Figure 2. Incorporation of Normal Epidermal Melanocytic Signature
SUPPLEMENTARY APPENDIX 1) Supplemental Figure 1. Histopathologic Characteristics of the Tumors in the Discovery Cohort 2) Supplemental Figure 2. Incorporation of Normal Epidermal Melanocytic Signature
Understanding DNA Copy Number Data
 Understanding DNA Copy Number Data Adam B. Olshen Department of Epidemiology and Biostatistics Helen Diller Family Comprehensive Cancer Center University of California, San Francisco http://cc.ucsf.edu/people/olshena_adam.php
Understanding DNA Copy Number Data Adam B. Olshen Department of Epidemiology and Biostatistics Helen Diller Family Comprehensive Cancer Center University of California, San Francisco http://cc.ucsf.edu/people/olshena_adam.php
