RNA-seq: filtering, quality control and visualisation. COMBINE RNA-seq Workshop
|
|
|
- Samson Powers
- 5 years ago
- Views:
Transcription
1 RNA-seq: filtering, quality control and visualisation COMBINE RNA-seq Workshop
2 QC and visualisation (part 1)
3 Slide taken from COMBINE RNAseq workshop on 23/09/2016 RNA-seq of Mouse mammary gland Basal cells Virgin Pregnant Lactating n=2 n=2 n=2 Luminal cells Virgin Pregnant Lactating n=2 n=2 n=2 Fu et al. (2015) EGF-mediated induction of Mcl-1 at the switch to lactation is essential for alveolar cell survival Nat Cell Biol
4 Slide taken from COMBINE RNAseq workshop on 23/09/2016 (some) questions we can ask Which genes are differentially expressed between basal and luminal cells? between basal and luminal in virgin mice? between pregnant and lactating mice? between pregnant and lactating mice in basal cells?
5 Reading in the data counts data and sample information Formatting the data clean it up so we can look at it easily
6 Filtering out lowly expressed genes Genes with very low counts in all samples provide little evidence for differential expression Often samples have many genes with zero or very low counts A. Raw data B. Filtered data Density _6_5_11 9_6_5_11 purep53 JMS8 2 JMS8 3 JMS8 4 JMS8 5 JMS9 P7c JMS9 P8c Density _6_5_11 9_6_5_11 purep53 JMS8 2 JMS8 3 JMS8 4 JMS8 5 JMS9 P7c JMS9 P8c Log cpm Log cpm
7 Filtering out lowly expressed genes Testing for differential expression for many genes simultaneously adds to the multiple testing burden, reducing the power to detect DE genes. IT IS VERY IMPORTANT to filter out genes that have all zero counts or very low counts. We filter using CPM values rather than counts because they account for differences in sequencing depth between samples.
8 Filtering out lowly expressed genes CPM = counts per million, or how many counts would I get for a gene if the sample had a library size of 1M. For a given gene: Library size Count CPM 1M M M
9 Filtering out lowly expressed genes Use a CPM threshold to define expressed and unexpressed As a general rule, a good threshold can be chosen for a CPM value that corresponds to a count of 10. In our dataset, the samples have library sizes of 20 to 20 something million. Library size Count CPM 1M M M
10 Filtering out lowly expressed genes Use a CPM threshold to define expressed and unexpressed As a general rule, a good threshold can be chosen for a CPM value that corresponds to a count of 10. In our dataset, the samples have library sizes of 20 to 20 something million. Library size Count We CPM use a CPM 1M 1 threshold of 0.5! 1 10M M
11 Filtering out lowly expressed genes Use a CPM threshold to define expressed and unexpressed As a general rule, a good threshold can threshold be chosen of 1 works for a well in most cases. CPM value that corresponds to a count of 10. In our dataset, the samples have library sizes of 20 to 20 something million. Library size Count We CPM use a CPM 1M 1 threshold of 0.5! 1 10M 10 1 But if this is too hard to work out, a CPM 20M
12 Filtering out lowly expressed genes We keep any gene that is (roughly) expressed in at least one group. 12 samples, 6 groups, 2 replicates in each group. Keep if CPM > 0.5 in at least 2 out of 12 samples Basal cells Virgin Pregnant Lactating Luminal cells Virgin Pregnant Lactating
13 Filtering out lowly expressed genes We keep any gene that is (roughly) expressed in at least one group. 12 samples, 6 groups, 2 replicates in each group. Keep if CPM > 0.5 in at least 2 out of 12 samples Basal cells Virgin expressed Pregnant expressed Lactating expressed Luminal cells Virgin unexpressed Pregnant unexpressed Lactating unexpressed
14 Filtering out lowly expressed genes We keep any gene that is (roughly) expressed in at least one group. 12 samples, 6 groups, 2 replicates in each group. Keep if CPM > 0.5 in at least 2 out of 12 samples Basal cells Virgin unexpressed Pregnant expressed Lactating unexpressed Luminal cells Virgin unexpressed Pregnant unexpressed Lactating unexpressed
15 Filtering out lowly expressed genes We keep any gene that is (roughly) expressed in at least one group. 12 samples, 6 groups, 2 replicates in each group. Keep gene if CPM > 0.5 in at least 2 or more samples Basal cells Virgin unexpressed Pregnant expressed Lactating unexpressed Luminal cells Virgin unexpressed Pregnant unexpressed Lactating unexpressed
16 QC and visualisation (part 2)
17 MDS Plots A visualisation of a principle components analysis which looks at where the greatest sources of variation in the data come from. Distances represents the typical log2-fc observed between each pair of samples e.g. 6 units apart = 2^6 = 64-fold difference Unsupervised separation based on data, no prior knowledge of experimental design. Useful for an overview of the data. Do samples separate by experimental groups? Quality control Outliers?
18
19
20 QC and visualisation (part 3)
21 Normalisation for composition bias 10_6_5_11 9_6_5_11 purep53 JMS8 2 JMS8 3 JMS8 4 JMS8 5 JMS9 P7c JMS9 P8c A. Example: Unnormalised data Log cpm 10_6_5_11 9_6_5_11 purep53 JMS8 2 JMS8 3 JMS8 4 JMS8 5 JMS9 P7c JMS9 P8c B. Example: Normalised data Log cpm If we ran a DE analysis on Sample 1 and Sample 3, almost all genes will be downregulated in Sample 1!!
22 Normalisation for composition bias 10_6_5_11 9_6_5_11 purep53 JMS8 2 JMS8 3 JMS8 4 JMS8 5 JMS9 P7c JMS9 P8c A. Example: Unnormalised data Log cpm 10_6_5_11 9_6_5_11 purep53 JMS8 2 JMS8 3 JMS8 4 JMS8 5 JMS9 P7c JMS9 P8c B. Example: Normalised data Log cpm
23 Normalisation for composition bias TMM normalisation (Robinson and Oshlack, 2010) How do we make the expression of all the genes go UP in the one sample? Scaling factors E.g. scale library size by 0.1 so effective library size is 1M. Library size Count CPM 10M M Scaling factor <1 makes the CPM larger.
24 Voom
25 Variance of log-cpm depends on mean of log-cpm SEQC (tech var only) Mouse (low bio var) Simulations (mod bio var) Nigerian (high bio var) D melanogaster (systematically dif) Average log 2 (count + 0.5) 25
26 RNA-seqdata is discrete has non-constant mean-variance trend Voom Transform to log-counts per million Remove mean-var dependence through the use of precision weights Normal dist. assumes that the data is continuous has constant variance
27 Variance weights Obtain variance estimates for each observation using mean-var trend. Assign inverse variance weights to each observation. Weights remove mean-variance trend from the data. Before: trended variance After: constant variance Qtr-root variance Log2- variance Mean log-count Mean log-cpm
Supplement to SCnorm: robust normalization of single-cell RNA-seq data
 Supplement to SCnorm: robust normalization of single-cell RNA-seq data Supplementary Note 1: SCnorm does not require spike-ins, since we find that the performance of spike-ins in scrna-seq is often compromised,
Supplement to SCnorm: robust normalization of single-cell RNA-seq data Supplementary Note 1: SCnorm does not require spike-ins, since we find that the performance of spike-ins in scrna-seq is often compromised,
Tutorial: RNA-Seq Analysis Part II: Non-Specific Matches and Expression Measures
 : RNA-Seq Analysis Part II: Non-Specific Matches and Expression Measures March 15, 2013 CLC bio Finlandsgade 10-12 8200 Aarhus N Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com support@clcbio.com
: RNA-Seq Analysis Part II: Non-Specific Matches and Expression Measures March 15, 2013 CLC bio Finlandsgade 10-12 8200 Aarhus N Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com support@clcbio.com
Nature Genetics: doi: /ng Supplementary Figure 1. Clinical timeline for the discovery WES cases.
 Supplementary Figure 1 Clinical timeline for the discovery WES cases. This illustrates the timeline of the disease events during the clinical course of each patient s disease, further indicating the available
Supplementary Figure 1 Clinical timeline for the discovery WES cases. This illustrates the timeline of the disease events during the clinical course of each patient s disease, further indicating the available
Predicting clinical outcomes in neuroblastoma with genomic data integration
 Predicting clinical outcomes in neuroblastoma with genomic data integration Ilyes Baali, 1 Alp Emre Acar 1, Tunde Aderinwale 2, Saber HafezQorani 3, Hilal Kazan 4 1 Department of Electric-Electronics Engineering,
Predicting clinical outcomes in neuroblastoma with genomic data integration Ilyes Baali, 1 Alp Emre Acar 1, Tunde Aderinwale 2, Saber HafezQorani 3, Hilal Kazan 4 1 Department of Electric-Electronics Engineering,
Exercises: Differential Methylation
 Exercises: Differential Methylation Version 2018-04 Exercises: Differential Methylation 2 Licence This manual is 2014-18, Simon Andrews. This manual is distributed under the creative commons Attribution-Non-Commercial-Share
Exercises: Differential Methylation Version 2018-04 Exercises: Differential Methylation 2 Licence This manual is 2014-18, Simon Andrews. This manual is distributed under the creative commons Attribution-Non-Commercial-Share
Elms, Hayes, Shelburne 1
 Elms, Hayes, Shelburne 1 Potential Influential Factors of Cognitive Decline and Engagement in Participants of Adult Day Services Lauren Elms, Cat Hayes, Will Shelburne Acknowledgements The authors would
Elms, Hayes, Shelburne 1 Potential Influential Factors of Cognitive Decline and Engagement in Participants of Adult Day Services Lauren Elms, Cat Hayes, Will Shelburne Acknowledgements The authors would
RNA-seq. Differential analysis
 RNA-seq Differential analysis Data transformations Count data transformations In order to test for differential expression, we operate on raw counts and use discrete distributions differential expression.
RNA-seq Differential analysis Data transformations Count data transformations In order to test for differential expression, we operate on raw counts and use discrete distributions differential expression.
Supplementary Figures and Tables
 Supplementary Figures and Tables Supplementary Figure 1. Study design and sample collection. S.japonicum were harvested from C57 mice at 8 time points after infection. Total number of samples for RNA-Seq:
Supplementary Figures and Tables Supplementary Figure 1. Study design and sample collection. S.japonicum were harvested from C57 mice at 8 time points after infection. Total number of samples for RNA-Seq:
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Unit 1 Exploring and Understanding Data
 Unit 1 Exploring and Understanding Data Area Principle Bar Chart Boxplot Conditional Distribution Dotplot Empirical Rule Five Number Summary Frequency Distribution Frequency Polygon Histogram Interquartile
Unit 1 Exploring and Understanding Data Area Principle Bar Chart Boxplot Conditional Distribution Dotplot Empirical Rule Five Number Summary Frequency Distribution Frequency Polygon Histogram Interquartile
SUPPLEMENTARY APPENDIX
 SUPPLEMENTARY APPENDIX 1) Supplemental Figure 1. Histopathologic Characteristics of the Tumors in the Discovery Cohort 2) Supplemental Figure 2. Incorporation of Normal Epidermal Melanocytic Signature
SUPPLEMENTARY APPENDIX 1) Supplemental Figure 1. Histopathologic Characteristics of the Tumors in the Discovery Cohort 2) Supplemental Figure 2. Incorporation of Normal Epidermal Melanocytic Signature
Small RNAs and how to analyze them using sequencing
 Small RNAs and how to analyze them using sequencing RNA-seq Course November 8th 2017 Marc Friedländer ComputaAonal RNA Biology Group SciLifeLab / Stockholm University Special thanks to Jakub Westholm for
Small RNAs and how to analyze them using sequencing RNA-seq Course November 8th 2017 Marc Friedländer ComputaAonal RNA Biology Group SciLifeLab / Stockholm University Special thanks to Jakub Westholm for
Clustering mass spectrometry data using order statistics
 Proteomics 2003, 3, 1687 1691 DOI 10.1002/pmic.200300517 1687 Douglas J. Slotta 1 Lenwood S. Heath 1 Naren Ramakrishnan 1 Rich Helm 2 Malcolm Potts 3 1 Department of Computer Science 2 Department of Wood
Proteomics 2003, 3, 1687 1691 DOI 10.1002/pmic.200300517 1687 Douglas J. Slotta 1 Lenwood S. Heath 1 Naren Ramakrishnan 1 Rich Helm 2 Malcolm Potts 3 1 Department of Computer Science 2 Department of Wood
ChIP-seq hands-on. Iros Barozzi, Campus IFOM-IEO (Milan) Saverio Minucci, Gioacchino Natoli Labs
 ChIP-seq hands-on Iros Barozzi, Campus IFOM-IEO (Milan) Saverio Minucci, Gioacchino Natoli Labs Main goals Becoming familiar with essential tools and formats Visualizing and contextualizing raw data Understand
ChIP-seq hands-on Iros Barozzi, Campus IFOM-IEO (Milan) Saverio Minucci, Gioacchino Natoli Labs Main goals Becoming familiar with essential tools and formats Visualizing and contextualizing raw data Understand
Author's response to reviews
 Author's response to reviews Title: Wnt1 is epistatic to Id2 in modeling mammary gland development and in causing mammary tumors Authors: Susan Marino (smarino@cmgm.stanford.edu) Claire Romelfanger (clairefrog@yahoo.com)
Author's response to reviews Title: Wnt1 is epistatic to Id2 in modeling mammary gland development and in causing mammary tumors Authors: Susan Marino (smarino@cmgm.stanford.edu) Claire Romelfanger (clairefrog@yahoo.com)
RNA-Seq Preparation Comparision Summary: Lexogen, Standard, NEB
 RNA-Seq Preparation Comparision Summary: Lexogen, Standard, NEB CSF-NGS January 22, 214 Contents 1 Introduction 1 2 Experimental Details 1 3 Results And Discussion 1 3.1 ERCC spike ins............................................
RNA-Seq Preparation Comparision Summary: Lexogen, Standard, NEB CSF-NGS January 22, 214 Contents 1 Introduction 1 2 Experimental Details 1 3 Results And Discussion 1 3.1 ERCC spike ins............................................
Supplementary Materials Extracting a Cellular Hierarchy from High-dimensional Cytometry Data with SPADE
 Supplementary Materials Extracting a Cellular Hierarchy from High-dimensional Cytometry Data with SPADE Peng Qiu1,4, Erin F. Simonds2, Sean C. Bendall2, Kenneth D. Gibbs Jr.2, Robert V. Bruggner2, Michael
Supplementary Materials Extracting a Cellular Hierarchy from High-dimensional Cytometry Data with SPADE Peng Qiu1,4, Erin F. Simonds2, Sean C. Bendall2, Kenneth D. Gibbs Jr.2, Robert V. Bruggner2, Michael
Understandable Statistics
 Understandable Statistics correlated to the Advanced Placement Program Course Description for Statistics Prepared for Alabama CC2 6/2003 2003 Understandable Statistics 2003 correlated to the Advanced Placement
Understandable Statistics correlated to the Advanced Placement Program Course Description for Statistics Prepared for Alabama CC2 6/2003 2003 Understandable Statistics 2003 correlated to the Advanced Placement
Exercise Verify that the term on the left of the equation showing the decomposition of "total" deviation in a two-factor experiment.
 Exercise 2.2.1 Verify that the term on the left of the equation showing the decomposition of "total" deviation in a two-factor experiment y ijk y = ( y i y ) + ( y j y ) + [( y ij y ) ( y i y ) ( y j y
Exercise 2.2.1 Verify that the term on the left of the equation showing the decomposition of "total" deviation in a two-factor experiment y ijk y = ( y i y ) + ( y j y ) + [( y ij y ) ( y i y ) ( y j y
Statistics 202: Data Mining. c Jonathan Taylor. Final review Based in part on slides from textbook, slides of Susan Holmes.
 Final review Based in part on slides from textbook, slides of Susan Holmes December 5, 2012 1 / 1 Final review Overview Before Midterm General goals of data mining. Datatypes. Preprocessing & dimension
Final review Based in part on slides from textbook, slides of Susan Holmes December 5, 2012 1 / 1 Final review Overview Before Midterm General goals of data mining. Datatypes. Preprocessing & dimension
Statistics: A Brief Overview Part I. Katherine Shaver, M.S. Biostatistician Carilion Clinic
 Statistics: A Brief Overview Part I Katherine Shaver, M.S. Biostatistician Carilion Clinic Statistics: A Brief Overview Course Objectives Upon completion of the course, you will be able to: Distinguish
Statistics: A Brief Overview Part I Katherine Shaver, M.S. Biostatistician Carilion Clinic Statistics: A Brief Overview Course Objectives Upon completion of the course, you will be able to: Distinguish
APPENDIX D REFERENCE AND PREDICTIVE VALUES FOR PEAK EXPIRATORY FLOW RATE (PEFR)
 APPENDIX D REFERENCE AND PREDICTIVE VALUES FOR PEAK EXPIRATORY FLOW RATE (PEFR) Lung function is related to physical characteristics such as age and height. In order to assess the Peak Expiratory Flow
APPENDIX D REFERENCE AND PREDICTIVE VALUES FOR PEAK EXPIRATORY FLOW RATE (PEFR) Lung function is related to physical characteristics such as age and height. In order to assess the Peak Expiratory Flow
Framework for Defining Evidentiary Standards for Biomarker Qualification: Minimal Residual Disease (MRD) in Multiple Myeloma (MM)
 Framework for Defining Evidentiary Standards for Biomarker Qualification: Minimal Residual Disease (MRD) in Multiple Myeloma (MM) MRD in Multiple Myeloma Team Biomarker Qualification Workshop Framework
Framework for Defining Evidentiary Standards for Biomarker Qualification: Minimal Residual Disease (MRD) in Multiple Myeloma (MM) MRD in Multiple Myeloma Team Biomarker Qualification Workshop Framework
Abstract. Optimization strategy of Copy Number Variant calling using Multiplicom solutions APPLICATION NOTE. Introduction
 Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Mostly Harmless Simulations? On the Internal Validity of Empirical Monte Carlo Studies
 Mostly Harmless Simulations? On the Internal Validity of Empirical Monte Carlo Studies Arun Advani and Tymon Sªoczy«ski 13 November 2013 Background When interested in small-sample properties of estimators,
Mostly Harmless Simulations? On the Internal Validity of Empirical Monte Carlo Studies Arun Advani and Tymon Sªoczy«ski 13 November 2013 Background When interested in small-sample properties of estimators,
STATISTICAL METHODS FOR DIAGNOSTIC TESTING: AN ILLUSTRATION USING A NEW METHOD FOR CANCER DETECTION XIN SUN. PhD, Kansas State University, 2012
 STATISTICAL METHODS FOR DIAGNOSTIC TESTING: AN ILLUSTRATION USING A NEW METHOD FOR CANCER DETECTION by XIN SUN PhD, Kansas State University, 2012 A THESIS Submitted in partial fulfillment of the requirements
STATISTICAL METHODS FOR DIAGNOSTIC TESTING: AN ILLUSTRATION USING A NEW METHOD FOR CANCER DETECTION by XIN SUN PhD, Kansas State University, 2012 A THESIS Submitted in partial fulfillment of the requirements
Peak-calling for ChIP-seq and ATAC-seq
 Peak-calling for ChIP-seq and ATAC-seq Shamith Samarajiwa CRUK Autumn School in Bioinformatics 2017 University of Cambridge Overview Peak-calling: identify enriched (signal) regions in ChIP-seq or ATAC-seq
Peak-calling for ChIP-seq and ATAC-seq Shamith Samarajiwa CRUK Autumn School in Bioinformatics 2017 University of Cambridge Overview Peak-calling: identify enriched (signal) regions in ChIP-seq or ATAC-seq
Technical note on seasonal adjustment for Gross domestic product (Expenditure)
 Technical note on seasonal adjustment for Gross domestic product (Expenditure) July 1, 2013 Contents 1 Gross domestic product (Expenditure) 2 1.1 Additive versus multiplicative seasonality.....................
Technical note on seasonal adjustment for Gross domestic product (Expenditure) July 1, 2013 Contents 1 Gross domestic product (Expenditure) 2 1.1 Additive versus multiplicative seasonality.....................
Knowledge Discovery and Data Mining I
 Ludwig-Maximilians-Universität München Lehrstuhl für Datenbanksysteme und Data Mining Prof. Dr. Thomas Seidl Knowledge Discovery and Data Mining I Winter Semester 2018/19 Introduction What is an outlier?
Ludwig-Maximilians-Universität München Lehrstuhl für Datenbanksysteme und Data Mining Prof. Dr. Thomas Seidl Knowledge Discovery and Data Mining I Winter Semester 2018/19 Introduction What is an outlier?
High-Throughput Sequencing Course
 High-Throughput Sequencing Course Introduction Biostatistics and Bioinformatics Summer 2017 From Raw Unaligned Reads To Aligned Reads To Counts Differential Expression Differential Expression 3 2 1 0 1
High-Throughput Sequencing Course Introduction Biostatistics and Bioinformatics Summer 2017 From Raw Unaligned Reads To Aligned Reads To Counts Differential Expression Differential Expression 3 2 1 0 1
Discussion Meeting for MCP-Mod Qualification Opinion Request. Novartis 10 July 2013 EMA, London, UK
 Discussion Meeting for MCP-Mod Qualification Opinion Request Novartis 10 July 2013 EMA, London, UK Attendees Face to face: Dr. Frank Bretz Global Statistical Methodology Head, Novartis Dr. Björn Bornkamp
Discussion Meeting for MCP-Mod Qualification Opinion Request Novartis 10 July 2013 EMA, London, UK Attendees Face to face: Dr. Frank Bretz Global Statistical Methodology Head, Novartis Dr. Björn Bornkamp
Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and
 Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and Promoter Motif Analysis Shisong Ma 1,2*, Michael Snyder 3, and Savithramma P Dinesh-Kumar 2* 1 School of Life Sciences, University
Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and Promoter Motif Analysis Shisong Ma 1,2*, Michael Snyder 3, and Savithramma P Dinesh-Kumar 2* 1 School of Life Sciences, University
User Guide. Association analysis. Input
 User Guide TFEA.ChIP is a tool to estimate transcription factor enrichment in a set of differentially expressed genes using data from ChIP-Seq experiments performed in different tissues and conditions.
User Guide TFEA.ChIP is a tool to estimate transcription factor enrichment in a set of differentially expressed genes using data from ChIP-Seq experiments performed in different tissues and conditions.
A Statistical Framework for Classification of Tumor Type from microrna Data
 DEGREE PROJECT IN MATHEMATICS, SECOND CYCLE, 30 CREDITS STOCKHOLM, SWEDEN 2016 A Statistical Framework for Classification of Tumor Type from microrna Data JOSEFINE RÖHSS KTH ROYAL INSTITUTE OF TECHNOLOGY
DEGREE PROJECT IN MATHEMATICS, SECOND CYCLE, 30 CREDITS STOCKHOLM, SWEDEN 2016 A Statistical Framework for Classification of Tumor Type from microrna Data JOSEFINE RÖHSS KTH ROYAL INSTITUTE OF TECHNOLOGY
Basic Statistics 01. Describing Data. Special Program: Pre-training 1
 Basic Statistics 01 Describing Data Special Program: Pre-training 1 Describing Data 1. Numerical Measures Measures of Location Measures of Dispersion Correlation Analysis 2. Frequency Distributions (Relative)
Basic Statistics 01 Describing Data Special Program: Pre-training 1 Describing Data 1. Numerical Measures Measures of Location Measures of Dispersion Correlation Analysis 2. Frequency Distributions (Relative)
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
CHAPTER 9: Producing Data: Experiments
 CHAPTER 9: Producing Data: Experiments The Basic Practice of Statistics 6 th Edition Moore / Notz / Fligner Lecture PowerPoint Slides Chapter 9 Concepts 2 Observation vs. Experiment Subjects, Factors,
CHAPTER 9: Producing Data: Experiments The Basic Practice of Statistics 6 th Edition Moore / Notz / Fligner Lecture PowerPoint Slides Chapter 9 Concepts 2 Observation vs. Experiment Subjects, Factors,
Digitizing the Proteomes From Big Tissue Biobanks
 Digitizing the Proteomes From Big Tissue Biobanks Analyzing 24 Proteomes Per Day by Microflow SWATH Acquisition and Spectronaut Pulsar Analysis Jan Muntel 1, Nick Morrice 2, Roland M. Bruderer 1, Lukas
Digitizing the Proteomes From Big Tissue Biobanks Analyzing 24 Proteomes Per Day by Microflow SWATH Acquisition and Spectronaut Pulsar Analysis Jan Muntel 1, Nick Morrice 2, Roland M. Bruderer 1, Lukas
DeconRNASeq: A Statistical Framework for Deconvolution of Heterogeneous Tissue Samples Based on mrna-seq data
 DeconRNASeq: A Statistical Framework for Deconvolution of Heterogeneous Tissue Samples Based on mrna-seq data Ting Gong, Joseph D. Szustakowski April 30, 2018 1 Introduction Heterogeneous tissues are frequently
DeconRNASeq: A Statistical Framework for Deconvolution of Heterogeneous Tissue Samples Based on mrna-seq data Ting Gong, Joseph D. Szustakowski April 30, 2018 1 Introduction Heterogeneous tissues are frequently
Experimental Design For Microarray Experiments. Robert Gentleman, Denise Scholtens Arden Miller, Sandrine Dudoit
 Experimental Design For Microarray Experiments Robert Gentleman, Denise Scholtens Arden Miller, Sandrine Dudoit Copyright 2002 Complexity of Genomic data the functioning of cells is a complex and highly
Experimental Design For Microarray Experiments Robert Gentleman, Denise Scholtens Arden Miller, Sandrine Dudoit Copyright 2002 Complexity of Genomic data the functioning of cells is a complex and highly
Catherine A. Welch 1*, Séverine Sabia 1,2, Eric Brunner 1, Mika Kivimäki 1 and Martin J. Shipley 1
 Welch et al. BMC Medical Research Methodology (2018) 18:89 https://doi.org/10.1186/s12874-018-0548-0 RESEARCH ARTICLE Open Access Does pattern mixture modelling reduce bias due to informative attrition
Welch et al. BMC Medical Research Methodology (2018) 18:89 https://doi.org/10.1186/s12874-018-0548-0 RESEARCH ARTICLE Open Access Does pattern mixture modelling reduce bias due to informative attrition
Nature Structural & Molecular Biology: doi: /nsmb.2419
 Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
C-1: Variables which are measured on a continuous scale are described in terms of three key characteristics central tendency, variability, and shape.
 MODULE 02: DESCRIBING DT SECTION C: KEY POINTS C-1: Variables which are measured on a continuous scale are described in terms of three key characteristics central tendency, variability, and shape. C-2:
MODULE 02: DESCRIBING DT SECTION C: KEY POINTS C-1: Variables which are measured on a continuous scale are described in terms of three key characteristics central tendency, variability, and shape. C-2:
Supplementary Figure 1: Experimental design. DISCOVERY PHASE VALIDATION PHASE (N = 88) (N = 20) Healthy = 20. Healthy = 6. Endometriosis = 33
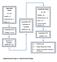 DISCOVERY PHASE (N = 20) Healthy = 6 Endometriosis = 7 EAOC = 7 Quantitative PCR (mirnas = 1113) a Quantitative PCR Verification of Candidate mirnas (N = 24) VALIDATION PHASE (N = 88) Healthy = 20 Endometriosis
DISCOVERY PHASE (N = 20) Healthy = 6 Endometriosis = 7 EAOC = 7 Quantitative PCR (mirnas = 1113) a Quantitative PCR Verification of Candidate mirnas (N = 24) VALIDATION PHASE (N = 88) Healthy = 20 Endometriosis
Types of Data. Systematic Reviews: Data Synthesis Professor Jodie Dodd 4/12/2014. Acknowledgements: Emily Bain Australasian Cochrane Centre
 Early Nutrition Workshop, December 2014 Systematic Reviews: Data Synthesis Professor Jodie Dodd 1 Types of Data Acknowledgements: Emily Bain Australasian Cochrane Centre 2 1 What are dichotomous outcomes?
Early Nutrition Workshop, December 2014 Systematic Reviews: Data Synthesis Professor Jodie Dodd 1 Types of Data Acknowledgements: Emily Bain Australasian Cochrane Centre 2 1 What are dichotomous outcomes?
Profiling of the Exosomal Cargo of Bovine Milk Reveals the Presence of Immune- and Growthmodulatory Non-coding RNAs (ncrna)
 Animal Industry Report AS 664 ASL R3235 2018 Profiling of the Exosomal Cargo of Bovine Milk Reveals the Presence of Immune- and Growthmodulatory Non-coding RNAs (ncrna) Eric D. Testroet Washington State
Animal Industry Report AS 664 ASL R3235 2018 Profiling of the Exosomal Cargo of Bovine Milk Reveals the Presence of Immune- and Growthmodulatory Non-coding RNAs (ncrna) Eric D. Testroet Washington State
Package AbsFilterGSEA
 Type Package Package AbsFilterGSEA September 21, 2017 Title Improved False Positive Control of Gene-Permuting GSEA with Absolute Filtering Version 1.5.1 Author Sora Yoon Maintainer
Type Package Package AbsFilterGSEA September 21, 2017 Title Improved False Positive Control of Gene-Permuting GSEA with Absolute Filtering Version 1.5.1 Author Sora Yoon Maintainer
RNA-seq Introduction
 RNA-seq Introduction DNA is the same in all cells but which RNAs that is present is different in all cells There is a wide variety of different functional RNAs Which RNAs (and sometimes then translated
RNA-seq Introduction DNA is the same in all cells but which RNAs that is present is different in all cells There is a wide variety of different functional RNAs Which RNAs (and sometimes then translated
Exploring chromatin regulation by ChIP-Sequencing
 Exploring chromatin regulation by ChIP-Sequencing From datasets quality assessment, enrichment patterns identification and multi-profiles integration to the reconstitution of gene regulatory wires describing
Exploring chromatin regulation by ChIP-Sequencing From datasets quality assessment, enrichment patterns identification and multi-profiles integration to the reconstitution of gene regulatory wires describing
# Assessment of gene expression levels between several cell group types is a common application of the unsupervised technique.
 # Aleksey Morozov # Microarray Data Analysis Using Hierarchical Clustering. # The "unsupervised learning" approach deals with data that has the features X1,X2...Xp, but does not have an associated response
# Aleksey Morozov # Microarray Data Analysis Using Hierarchical Clustering. # The "unsupervised learning" approach deals with data that has the features X1,X2...Xp, but does not have an associated response
DNA Sequence Bioinformatics Analysis with the Galaxy Platform
 DNA Sequence Bioinformatics Analysis with the Galaxy Platform University of São Paulo, Brazil 28 July - 1 August 2014 Dave Clements Johns Hopkins University Robson Francisco de Souza University of São
DNA Sequence Bioinformatics Analysis with the Galaxy Platform University of São Paulo, Brazil 28 July - 1 August 2014 Dave Clements Johns Hopkins University Robson Francisco de Souza University of São
International Journal of Research in Science and Technology. (IJRST) 2018, Vol. No. 8, Issue No. IV, Oct-Dec e-issn: , p-issn: X
 CLOUD FILE SHARING AND DATA SECURITY THREATS EXPLORING THE EMPLOYABILITY OF GRAPH-BASED UNSUPERVISED LEARNING IN DETECTING AND SAFEGUARDING CLOUD FILES Harshit Yadav Student, Bal Bharati Public School,
CLOUD FILE SHARING AND DATA SECURITY THREATS EXPLORING THE EMPLOYABILITY OF GRAPH-BASED UNSUPERVISED LEARNING IN DETECTING AND SAFEGUARDING CLOUD FILES Harshit Yadav Student, Bal Bharati Public School,
UNIVERSITY of PENNSYLVANIA CIS 520: Machine Learning Midterm, 2016
 UNIVERSITY of PENNSYLVANIA CIS 520: Machine Learning Midterm, 2016 Exam policy: This exam allows one one-page, two-sided cheat sheet; No other materials. Time: 80 minutes. Be sure to write your name and
UNIVERSITY of PENNSYLVANIA CIS 520: Machine Learning Midterm, 2016 Exam policy: This exam allows one one-page, two-sided cheat sheet; No other materials. Time: 80 minutes. Be sure to write your name and
Supplementary information for: Human micrornas co-silence in well-separated groups and have different essentialities
 Supplementary information for: Human micrornas co-silence in well-separated groups and have different essentialities Gábor Boross,2, Katalin Orosz,2 and Illés J. Farkas 2, Department of Biological Physics,
Supplementary information for: Human micrornas co-silence in well-separated groups and have different essentialities Gábor Boross,2, Katalin Orosz,2 and Illés J. Farkas 2, Department of Biological Physics,
A novel and universal method for microrna RT-qPCR data normalization
 A novel and universal method for microrna RT-qPCR data normalization Jo Vandesompele professor, Ghent University co-founder and CEO, Biogazelle 4 th International qpcr Symposium Weihenstephan, March 1,
A novel and universal method for microrna RT-qPCR data normalization Jo Vandesompele professor, Ghent University co-founder and CEO, Biogazelle 4 th International qpcr Symposium Weihenstephan, March 1,
Predominant contribution of cis-regulatory divergence in the evolution of mouse alternative splicing
 Molecular Systems Biology Peer Review Process File Predominant contribution of cis-regulatory divergence in the evolution of mouse alternative splicing Mr. Qingsong Gao, Wei Sun, Marlies Ballegeer, Claude
Molecular Systems Biology Peer Review Process File Predominant contribution of cis-regulatory divergence in the evolution of mouse alternative splicing Mr. Qingsong Gao, Wei Sun, Marlies Ballegeer, Claude
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplemental Figure S1. PLAG1 kidneys contain fewer glomeruli (A) Quantitative PCR for Igf2 and PLAG1 in whole kidneys taken from mice at E15.
 Supplemental Figure S1. PLAG1 kidneys contain fewer glomeruli (A) Quantitative PCR for Igf2 and PLAG1 in whole kidneys taken from mice at E15.5, E18.5, P4, and P8. Values shown are means from four technical
Supplemental Figure S1. PLAG1 kidneys contain fewer glomeruli (A) Quantitative PCR for Igf2 and PLAG1 in whole kidneys taken from mice at E15.5, E18.5, P4, and P8. Values shown are means from four technical
Identification of Tissue Independent Cancer Driver Genes
 Identification of Tissue Independent Cancer Driver Genes Alexandros Manolakos, Idoia Ochoa, Kartik Venkat Supervisor: Olivier Gevaert Abstract Identification of genomic patterns in tumors is an important
Identification of Tissue Independent Cancer Driver Genes Alexandros Manolakos, Idoia Ochoa, Kartik Venkat Supervisor: Olivier Gevaert Abstract Identification of genomic patterns in tumors is an important
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits
 AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
Results & Statistics: Description and Correlation. I. Scales of Measurement A Review
 Results & Statistics: Description and Correlation The description and presentation of results involves a number of topics. These include scales of measurement, descriptive statistics used to summarize
Results & Statistics: Description and Correlation The description and presentation of results involves a number of topics. These include scales of measurement, descriptive statistics used to summarize
Georgina Salas. Topics EDCI Intro to Research Dr. A.J. Herrera
 Homework assignment topics 51-63 Georgina Salas Topics 51-63 EDCI Intro to Research 6300.62 Dr. A.J. Herrera Topic 51 1. Which average is usually reported when the standard deviation is reported? The mean
Homework assignment topics 51-63 Georgina Salas Topics 51-63 EDCI Intro to Research 6300.62 Dr. A.J. Herrera Topic 51 1. Which average is usually reported when the standard deviation is reported? The mean
Sensory Cue Integration
 Sensory Cue Integration Summary by Byoung-Hee Kim Computer Science and Engineering (CSE) http://bi.snu.ac.kr/ Presentation Guideline Quiz on the gist of the chapter (5 min) Presenters: prepare one main
Sensory Cue Integration Summary by Byoung-Hee Kim Computer Science and Engineering (CSE) http://bi.snu.ac.kr/ Presentation Guideline Quiz on the gist of the chapter (5 min) Presenters: prepare one main
Chapter 20: Test Administration and Interpretation
 Chapter 20: Test Administration and Interpretation Thought Questions Why should a needs analysis consider both the individual and the demands of the sport? Should test scores be shared with a team, or
Chapter 20: Test Administration and Interpretation Thought Questions Why should a needs analysis consider both the individual and the demands of the sport? Should test scores be shared with a team, or
Observational studies; descriptive statistics
 Observational studies; descriptive statistics Patrick Breheny August 30 Patrick Breheny University of Iowa Biostatistical Methods I (BIOS 5710) 1 / 38 Observational studies Association versus causation
Observational studies; descriptive statistics Patrick Breheny August 30 Patrick Breheny University of Iowa Biostatistical Methods I (BIOS 5710) 1 / 38 Observational studies Association versus causation
Appendix B1. Details of Sampling and Weighting Procedures
 Appendix B1 Details of Sampling and Weighting Procedures Appendix B1 Details of Sampling and Weighting Procedures B1.1 Selection of WIC Sites The WIC service sites were selected using a stratified two-stage
Appendix B1 Details of Sampling and Weighting Procedures Appendix B1 Details of Sampling and Weighting Procedures B1.1 Selection of WIC Sites The WIC service sites were selected using a stratified two-stage
Effective Implementation of Bayesian Adaptive Randomization in Early Phase Clinical Development. Pantelis Vlachos.
 Effective Implementation of Bayesian Adaptive Randomization in Early Phase Clinical Development Pantelis Vlachos Cytel Inc, Geneva Acknowledgement Joint work with Giacomo Mordenti, Grünenthal Virginie
Effective Implementation of Bayesian Adaptive Randomization in Early Phase Clinical Development Pantelis Vlachos Cytel Inc, Geneva Acknowledgement Joint work with Giacomo Mordenti, Grünenthal Virginie
On the Possible Pitfalls in the Evaluation of Brain Computer Interface Mice
 On the Possible Pitfalls in the Evaluation of Brain Computer Interface Mice Riccardo Poli and Mathew Salvaris Brain-Computer Interfaces Lab, School of Computer Science and Electronic Engineering, University
On the Possible Pitfalls in the Evaluation of Brain Computer Interface Mice Riccardo Poli and Mathew Salvaris Brain-Computer Interfaces Lab, School of Computer Science and Electronic Engineering, University
4. Model evaluation & selection
 Foundations of Machine Learning CentraleSupélec Fall 2017 4. Model evaluation & selection Chloé-Agathe Azencot Centre for Computational Biology, Mines ParisTech chloe-agathe.azencott@mines-paristech.fr
Foundations of Machine Learning CentraleSupélec Fall 2017 4. Model evaluation & selection Chloé-Agathe Azencot Centre for Computational Biology, Mines ParisTech chloe-agathe.azencott@mines-paristech.fr
Tutorial 3: MANOVA. Pekka Malo 30E00500 Quantitative Empirical Research Spring 2016
 Tutorial 3: Pekka Malo 30E00500 Quantitative Empirical Research Spring 2016 Step 1: Research design Adequacy of sample size Choice of dependent variables Choice of independent variables (treatment effects)
Tutorial 3: Pekka Malo 30E00500 Quantitative Empirical Research Spring 2016 Step 1: Research design Adequacy of sample size Choice of dependent variables Choice of independent variables (treatment effects)
MULTIPLE REGRESSION OF CPS DATA
 MULTIPLE REGRESSION OF CPS DATA A further inspection of the relationship between hourly wages and education level can show whether other factors, such as gender and work experience, influence wages. Linear
MULTIPLE REGRESSION OF CPS DATA A further inspection of the relationship between hourly wages and education level can show whether other factors, such as gender and work experience, influence wages. Linear
Methods for assessing consistency in Mixed Treatment Comparison Meta-analysis
 Methods for assessing consistency in Mixed Treatment Comparison Meta-analysis Sofia Dias, NJ Welton, AE Ades RSS 2009, Edinburgh Department of Community Based Medicine Overview Mixed Treatment Comparison
Methods for assessing consistency in Mixed Treatment Comparison Meta-analysis Sofia Dias, NJ Welton, AE Ades RSS 2009, Edinburgh Department of Community Based Medicine Overview Mixed Treatment Comparison
Serum mirna signature diagnoses and discriminates murine colitis subtypes and predicts ulcerative colitis in humans
 Serum mirna signature diagnoses and discriminates murine colitis subtypes and predicts ulcerative colitis in humans Emilie Viennois 1*, Yuan Zhao 1, 2, Moon Kwon Han 1, Bo Xiao 1, 3, Mingzhen Zhang 1,
Serum mirna signature diagnoses and discriminates murine colitis subtypes and predicts ulcerative colitis in humans Emilie Viennois 1*, Yuan Zhao 1, 2, Moon Kwon Han 1, Bo Xiao 1, 3, Mingzhen Zhang 1,
Human breast milk mirna, maternal probiotic supplementation and atopic dermatitis in offsrping
 Human breast milk mirna, maternal probiotic supplementation and atopic dermatitis in offsrping Melanie Rae Simpson PhD candidate Department of Public Health and General Practice Norwegian University of
Human breast milk mirna, maternal probiotic supplementation and atopic dermatitis in offsrping Melanie Rae Simpson PhD candidate Department of Public Health and General Practice Norwegian University of
War and Relatedness Enrico Spolaore and Romain Wacziarg September 2015
 War and Relatedness Enrico Spolaore and Romain Wacziarg September 2015 Online Appendix Supplementary Empirical Results, described in the main text as "Available in the Online Appendix" 1 Table AUR0 Effect
War and Relatedness Enrico Spolaore and Romain Wacziarg September 2015 Online Appendix Supplementary Empirical Results, described in the main text as "Available in the Online Appendix" 1 Table AUR0 Effect
Nature Neuroscience: doi: /nn Supplementary Figure 1. Behavioral training.
 Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Bayesian Inference for Single-cell ClUstering and ImpuTing (BISCUIT) Elham Azizi
 Bayesian Inference for Single-cell ClUstering and ImpuTing (BISCUIT) Elham Azizi BioC 2017: Where Software and Biology Connect Profiling Tumor-Immune Ecosystem in Breast Cancer Immunotherapy treatments
Bayesian Inference for Single-cell ClUstering and ImpuTing (BISCUIT) Elham Azizi BioC 2017: Where Software and Biology Connect Profiling Tumor-Immune Ecosystem in Breast Cancer Immunotherapy treatments
Discontinuous Traits. Chapter 22. Quantitative Traits. Types of Quantitative Traits. Few, distinct phenotypes. Also called discrete characters
 Discontinuous Traits Few, distinct phenotypes Chapter 22 Also called discrete characters Quantitative Genetics Examples: Pea shape, eye color in Drosophila, Flower color Quantitative Traits Phenotype is
Discontinuous Traits Few, distinct phenotypes Chapter 22 Also called discrete characters Quantitative Genetics Examples: Pea shape, eye color in Drosophila, Flower color Quantitative Traits Phenotype is
QUANTIFYING THE EFFECT OF SETTING QUALITY CONTROL STANDARD DEVIATIONS GREATER THAN ACTUAL STANDARD DEVIATIONS ON WESTGARD RULES
 AACC24 QUANTIFYING THE EFFECT OF SETTING QUALITY CONTROL STANDARD DEVIATIONS GREATER THAN ACTUAL STANDARD DEVIATIONS ON WESTGARD RULES Graham Jones Department of Chemical Pathology, St Vincent s Hospital,
AACC24 QUANTIFYING THE EFFECT OF SETTING QUALITY CONTROL STANDARD DEVIATIONS GREATER THAN ACTUAL STANDARD DEVIATIONS ON WESTGARD RULES Graham Jones Department of Chemical Pathology, St Vincent s Hospital,
Lecture 8 Understanding Transcription RNA-seq analysis. Foundations of Computational Systems Biology David K. Gifford
 Lecture 8 Understanding Transcription RNA-seq analysis Foundations of Computational Systems Biology David K. Gifford 1 Lecture 8 RNA-seq Analysis RNA-seq principles How can we characterize mrna isoform
Lecture 8 Understanding Transcription RNA-seq analysis Foundations of Computational Systems Biology David K. Gifford 1 Lecture 8 RNA-seq Analysis RNA-seq principles How can we characterize mrna isoform
World Congress on Breast Cancer
 World Congress on Breast Cancer 05.08.2015 How pregnancy at early age protects against breast cancer Fabienne Meier-Abt, MD PhD Background: Early age pregnancy protects against breast cancer. MacMahon,
World Congress on Breast Cancer 05.08.2015 How pregnancy at early age protects against breast cancer Fabienne Meier-Abt, MD PhD Background: Early age pregnancy protects against breast cancer. MacMahon,
Chapter 1: Explaining Behavior
 Chapter 1: Explaining Behavior GOAL OF SCIENCE is to generate explanations for various puzzling natural phenomenon. - Generate general laws of behavior (psychology) RESEARCH: principle method for acquiring
Chapter 1: Explaining Behavior GOAL OF SCIENCE is to generate explanations for various puzzling natural phenomenon. - Generate general laws of behavior (psychology) RESEARCH: principle method for acquiring
BIMM 143. RNA sequencing overview. Genome Informatics II. Barry Grant. Lecture In vivo. In vitro.
 RNA sequencing overview BIMM 143 Genome Informatics II Lecture 14 Barry Grant http://thegrantlab.org/bimm143 In vivo In vitro In silico ( control) Goal: RNA quantification, transcript discovery, variant
RNA sequencing overview BIMM 143 Genome Informatics II Lecture 14 Barry Grant http://thegrantlab.org/bimm143 In vivo In vitro In silico ( control) Goal: RNA quantification, transcript discovery, variant
Introduction to SPSS. Katie Handwerger Why n How February 19, 2009
 Introduction to SPSS Katie Handwerger Why n How February 19, 2009 Overview Setting up a data file Frequencies/Descriptives One-sample T-test Paired-samples T-test Independent-samples T-test One-way ANOVA
Introduction to SPSS Katie Handwerger Why n How February 19, 2009 Overview Setting up a data file Frequencies/Descriptives One-sample T-test Paired-samples T-test Independent-samples T-test One-way ANOVA
Overview Detection epileptic seizures
 Overview Detection epileptic seizures Overview Problem Goal Method Detection with accelerometers Detection with video data Result Problem Detection of epileptic seizures with EEG (Golden Standard) Not
Overview Detection epileptic seizures Overview Problem Goal Method Detection with accelerometers Detection with video data Result Problem Detection of epileptic seizures with EEG (Golden Standard) Not
NEW METHODS FOR SENSITIVITY TESTS OF EXPLOSIVE DEVICES
 NEW METHODS FOR SENSITIVITY TESTS OF EXPLOSIVE DEVICES Amit Teller 1, David M. Steinberg 2, Lina Teper 1, Rotem Rozenblum 2, Liran Mendel 2, and Mordechai Jaeger 2 1 RAFAEL, POB 2250, Haifa, 3102102, Israel
NEW METHODS FOR SENSITIVITY TESTS OF EXPLOSIVE DEVICES Amit Teller 1, David M. Steinberg 2, Lina Teper 1, Rotem Rozenblum 2, Liran Mendel 2, and Mordechai Jaeger 2 1 RAFAEL, POB 2250, Haifa, 3102102, Israel
TEB. Id4 p63 DAPI Merge. Id4 CK8 DAPI Merge
 a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
Diurnal Pattern of Reaction Time: Statistical analysis
 Diurnal Pattern of Reaction Time: Statistical analysis Prepared by: Alison L. Gibbs, PhD, PStat Prepared for: Dr. Principal Investigator of Reaction Time Project January 11, 2015 Summary: This report gives
Diurnal Pattern of Reaction Time: Statistical analysis Prepared by: Alison L. Gibbs, PhD, PStat Prepared for: Dr. Principal Investigator of Reaction Time Project January 11, 2015 Summary: This report gives
NGS ONCOPANELS: FDA S PERSPECTIVE
 NGS ONCOPANELS: FDA S PERSPECTIVE CBA Workshop: Biomarker and Application in Drug Development August 11, 2018 Rockville, MD You Li, Ph.D. Division of Molecular Genetics and Pathology Food and Drug Administration
NGS ONCOPANELS: FDA S PERSPECTIVE CBA Workshop: Biomarker and Application in Drug Development August 11, 2018 Rockville, MD You Li, Ph.D. Division of Molecular Genetics and Pathology Food and Drug Administration
RNA-seq. Design of experiments
 RNA-seq Design of experiments Experimental design Introduction An experiment is a process or study that results in the collection of data. Statistical experiments are conducted in situations in which researchers
RNA-seq Design of experiments Experimental design Introduction An experiment is a process or study that results in the collection of data. Statistical experiments are conducted in situations in which researchers
omiras: MicroRNA regulation of gene expression
 omiras: MicroRNA regulation of gene expression Sören Müller, Goethe University of Frankfurt am Main Molecular Bioinformatics Group, Institute of Computer Science Plant Molecular Biology Group, Institute
omiras: MicroRNA regulation of gene expression Sören Müller, Goethe University of Frankfurt am Main Molecular Bioinformatics Group, Institute of Computer Science Plant Molecular Biology Group, Institute
Comments on Significance of candidate cancer genes as assessed by the CaMP score by Parmigiani et al.
 Comments on Significance of candidate cancer genes as assessed by the CaMP score by Parmigiani et al. Holger Höfling Gad Getz Robert Tibshirani June 26, 2007 1 Introduction Identifying genes that are involved
Comments on Significance of candidate cancer genes as assessed by the CaMP score by Parmigiani et al. Holger Höfling Gad Getz Robert Tibshirani June 26, 2007 1 Introduction Identifying genes that are involved
DiffVar: a new method for detecting differential variability with application to methylation in cancer and aging
 Genome Biology This Provisional PDF corresponds to the article as it appeared upon acceptance. Fully formatted PDF and full text (HTML) versions will be made available soon. DiffVar: a new method for detecting
Genome Biology This Provisional PDF corresponds to the article as it appeared upon acceptance. Fully formatted PDF and full text (HTML) versions will be made available soon. DiffVar: a new method for detecting
Dr. Allen Back. Sep. 30, 2016
 Dr. Allen Back Sep. 30, 2016 Extrapolation is Dangerous Extrapolation is Dangerous And watch out for confounding variables. e.g.: A strong association between numbers of firemen and amount of damge at
Dr. Allen Back Sep. 30, 2016 Extrapolation is Dangerous Extrapolation is Dangerous And watch out for confounding variables. e.g.: A strong association between numbers of firemen and amount of damge at
Stats 95. Statistical analysis without compelling presentation is annoying at best and catastrophic at worst. From raw numbers to meaningful pictures
 Stats 95 Statistical analysis without compelling presentation is annoying at best and catastrophic at worst. From raw numbers to meaningful pictures Stats 95 Why Stats? 200 countries over 200 years http://www.youtube.com/watch?v=jbksrlysojo
Stats 95 Statistical analysis without compelling presentation is annoying at best and catastrophic at worst. From raw numbers to meaningful pictures Stats 95 Why Stats? 200 countries over 200 years http://www.youtube.com/watch?v=jbksrlysojo
Iso-Seq Method Updates and Target Enrichment Without Amplification for SMRT Sequencing
 Iso-Seq Method Updates and Target Enrichment Without Amplification for SMRT Sequencing PacBio Americas User Group Meeting Sample Prep Workshop June.27.2017 Tyson Clark, Ph.D. For Research Use Only. Not
Iso-Seq Method Updates and Target Enrichment Without Amplification for SMRT Sequencing PacBio Americas User Group Meeting Sample Prep Workshop June.27.2017 Tyson Clark, Ph.D. For Research Use Only. Not
SUPPLEMENTAL INFORMATION
 SUPPLEMENTAL INFORMATION GO term analysis of differentially methylated SUMIs. GO term analysis of the 458 SUMIs with the largest differential methylation between human and chimp shows that they are more
SUPPLEMENTAL INFORMATION GO term analysis of differentially methylated SUMIs. GO term analysis of the 458 SUMIs with the largest differential methylation between human and chimp shows that they are more
genomics for systems biology / ISB2020 RNA sequencing (RNA-seq)
 RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
Chapter 11: Advanced Remedial Measures. Weighted Least Squares (WLS)
 Chapter : Advanced Remedial Measures Weighted Least Squares (WLS) When the error variance appears nonconstant, a transformation (of Y and/or X) is a quick remedy. But it may not solve the problem, or it
Chapter : Advanced Remedial Measures Weighted Least Squares (WLS) When the error variance appears nonconstant, a transformation (of Y and/or X) is a quick remedy. But it may not solve the problem, or it
MicroRNA roles in signaling during lactation: an insight from differential expression, time course and pathway analyses of deep sequence data
 MicroRNA roles in signaling during lactation: an insight from differential expression, time course and pathway analyses of deep sequence data Duy N. Do 1, 2, Ran Li 1, 3, Pier-Luc Dudemaine 1 and Eveline
MicroRNA roles in signaling during lactation: an insight from differential expression, time course and pathway analyses of deep sequence data Duy N. Do 1, 2, Ran Li 1, 3, Pier-Luc Dudemaine 1 and Eveline
