Supplementary Figures and Tables
|
|
|
- Joanna Benson
- 5 years ago
- Views:
Transcription
1 Supplementary Figures and Tables Supplementary Figure 1. Study design and sample collection. S.japonicum were harvested from C57 mice at 8 time points after infection. Total number of samples for RNA-Seq: 2 sexes per mouse x 3 replicates x 8 time points= 48.
2 Supplementary Figure 2. High throughput RNA Sequencing. Half of 48 libraries were sequenced for 3 times, generating 11.7 million reads per sample; while the other half were sequenced for 4 times, generating 15.6 million reads per sample. In all, 48 samples generated million reads with million reads per sample on average.
3 Supplementary Figure 3. Shared transcripts among three de novo reconstructed transcriptomes. M, transcriptome constructed from male reads. F, transcriptome constructed from female reads. MF, transcriptome constructed from male and female reads.
4 Supplementary Figure 4. Length distribution of the de novo reconstructed transcripts. Y-axis represents the number of transcripts in each size range and X-axis represents the size distribution.
5 Supplementary Figure 5. The expression patterns of female-specific transcripts at dpi. Y-axis represents the expression level (Log 2 RPKM) of transcript in female (red) and male (blue). X-axis represents 7 different time points after infection. (mean±s.e.m., n=3).
6 Supplementary Figure 6. The expression patterns of male-specific transcripts at dpi. Y-axis represents the expression level (Log 2 RPKM) of transcript in female (red) and male (blue). X-axis represents 7 different time points after infection. (mean±s.e.m., n=3).
7 Supplementary Figure 7. Clusters of co-expressed transcripts. 56 groups in all. X-axis represents 7 different time points after infection. The thin green line (male) and thin red line (female) represents the patterns of transcripts. Bold red line represents the average intensity of clustered transcripts.
8 Supplementary Figure 8. Verification of RNA-Seq results by qpcr. 24 genes in all. X-axis represents 7 different time points after infection. Y-axis represents the relative expression level (Log 2 Fold change) of transcript in female (red) and male (blue) evaluated by qpcr (solid line) and RNA-seq (dotted line). R, correlation.
9 Supplementary Figure 9. The pathway in Schistosoma japonicum of de novo synthesis of JH from Acetyl-CoA. The pathway map is the combination of terpenoid backbone biosynthesis and insect hormone biosynthesis (red box) from KEGG pathway database. Genes in green box indicated the identical genes found in de novo S. japonicum transcriptome.
10 Supplementary Figure 10. The relative expression levels of Sj AlstR after in vivo RNAi. Worms were perfused from infected mice (20 dpi) that either injected with control plasmid, scramble plasmid or si AlstR expression plasmid for 72h. Females worms (5~8) from each mouse were separated from male and taken as a replicate. The expression level of the Sj AlstR was evaluated by RT-PCR with the Sj PSMD gene as a reference calibrator. Five independent experiments were performed and the expression level of Sj AlstR was reduced by ~50% in Si AlstR group compared to control or scramble group (mean±s.e.m., n=5). Student s t-test: * p<0.05.
11 Supplementary Figure 11. The effect of 20E on the activities of Schistosoma japonicum. (mean±s.e.m., n=3).
12 Supplementary Figure 12. The profiles of elements involved in regulation of gene expression. Y-axis represents the expression level (Log 2 RPKM) of transcript in female (red) and male (blue). X-axis represents 7 different time points after infection. (mean± s.e.m., n=3).
13 Supplementary Figure 13. Schematic overview of male-female interplay in schistosomes. 1. Stimulus indicating the presence of the female is transferred to the nervous system of the male schistosome by receptor molecules on the surface of the gynecophoral canal of the male. 2. Biosynthesis of biogenic amines is enhanced by the over-expression of SjAADC. 3. Biogenic amines regulate copulatory activity. 4. The male transfers a tactile and/or chemical stimulus to the female during pairing. 5. The female receives the stimulus through sensory molecules on the tegument. 6. The female nervous system generates the neurotransmitter allatostatin. 7. Allatostatin is transferred to the ovary and gut through the nervous system. 8. Mediated by the allatostatin receptor, concentrations of insect-like hormones change, and this orchestrates the rapid development of the ovary and vitelline gland.
14 Supplementary Tables Supplementary Table 1. The S. japonicum transcripts that matched to JH synthesis genes in insects. Enzyme Accession# Accession# insects S. japonicum Identity/e-value Farnesol Oxidase D2WKD9 A. aegypti AAW %,4e-17 Farnesal AGI96742 dehydrogenase A. aegypti CAX %,1.2e-136 JH methyltransferase NA NA JH epoxidase NA NA
15 Supplementary Table 2. The cercarial infection doses at different time points. Days post infection Cercarial infection doses ~ 600~ 500~ 200~ 150~ 100~ 80~ 1, ~ 100
16 Supplementary Table 3. Primer sequences for probe synthesis. Gene name (Gene bank) allatostatin A receptor like (AY ) aromatic-l-amino-acid decarboxylase (AY ) Primer name Sense forward Sense reverse Antisense forward Antisense reverse Sense forward Sense reverse Antisense forward Antisense reverse Primer sequence 5 TAATACGACTCACTATAGGGAGTCATTTAGTT3 5 GACAAAATGGTAATGTAAATGTAGCGATTGTTTCA3 5 AGTCATTTAGTTGGAAGTTTTCGTGCATATATTTCA3 5 TAATACGACTCACTATAGGGATGTAGCGA3 5 TAATACGACTCACTATAGGGGATTTCACTTATT3 5 CATTTCAAAGTGACTTTTGTGTTGTTCAAATGC3 5 GATTTCACTTATTGGGGCAAACAAATGATTG3 5 TAATACGACTCACTATAGGGGACTTTTGTGTT3
17 Supplementary Table 4. The time point of each sample taken for RNA-seq Sample# Sj-male-1; Sj-male-2; Sj-male-3; Sj-female-1; Sj-female-2; Sj-female-3 Sj-male-4; Sj-male-5; Sj-male-6; Sj-female-4; Sj-female-5; Sj-female-6 Sj-male-7; Sj-male-8; Sj-male-9; Sj-female-7; Sj-female-8; Sj-female-9; Sj-male-10; Sj-male-11; Sj-male-12; Sj-female-10; Sj-female-11; Sj-female-12 Sj-male-13; Sj-male-14; Sj-male-15; Sj-female-13; Sj-female-14; Sj-female-15 Sj-male-16; Sj-male-17; Sj-male-18; Sj-female-16; Sj-female-17; Sj-female-18 Sj-male-19; Sj-male-20; Sj-male-21; Sj-female-19; Sj-female-20; Sj-female-21 Sj-male-22; Sj-male-23; Sj-male-24; Sj-female-22; Sj-female-23; Sj-female-24 Days post infection 14 dpi 16 dpi 18 dpi 20 dpi 22 dpi 24 dpi 26 dpi 28 dpi
RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays
 Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Nature Genetics: doi: /ng.3731
 Supplementary Figure 1 Circadian profiles of Adarb1 transcript and ADARB1 protein in mouse tissues. (a) Overlap of rhythmic transcripts identified in the previous transcriptome analyses. The mouse liver
Supplementary Figure 1 Circadian profiles of Adarb1 transcript and ADARB1 protein in mouse tissues. (a) Overlap of rhythmic transcripts identified in the previous transcriptome analyses. The mouse liver
Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion
 Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
SUPPLEMENTAL INFORMATIONS
 1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
Supplementary information
 Supplementary information High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss Majken Siersbæk, Lyuba Varticovski, Shutong Yang,
Supplementary information High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss Majken Siersbæk, Lyuba Varticovski, Shutong Yang,
Supplementary Information. Deciphering the Venomic Transcriptome of Killer- Wasp Vespa velutina
 Supplementary Information Deciphering the Venomic Transcriptome of Killer- Wasp Vespa velutina Zhirui Liu, Shuanggang Chen, You Zhou, Cuihong Xie, Bifeng Zhu, Huming Zhu, Shupeng Liu, Wei Wang, Hongzhuan
Supplementary Information Deciphering the Venomic Transcriptome of Killer- Wasp Vespa velutina Zhirui Liu, Shuanggang Chen, You Zhou, Cuihong Xie, Bifeng Zhu, Huming Zhu, Shupeng Liu, Wei Wang, Hongzhuan
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63.
 Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Supplementary Materials: Global Transcriptomic Analysis Reveals the Mechanism of Phelipanche Aegyptiaca Seed Germination
 Int. J. Mol. Sci. 2016, 17, 1139; doi:.3390/ijms17071139 S1 of S5 Supplementary Materials: Global Transcriptomic Analysis Reveals the Mechanism of Phelipanche Aegyptiaca Seed Germination Zhaoqun Yao, Fang
Int. J. Mol. Sci. 2016, 17, 1139; doi:.3390/ijms17071139 S1 of S5 Supplementary Materials: Global Transcriptomic Analysis Reveals the Mechanism of Phelipanche Aegyptiaca Seed Germination Zhaoqun Yao, Fang
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Functional genomics reveal that the serine synthesis pathway is essential in breast cancer
 Functional genomics reveal that the serine synthesis pathway is essential in breast cancer Results Presented by Stacey Lin Lloyd Lab http://www.amsbio.com/expression-ready-lentiviral-particles.aspx Overview
Functional genomics reveal that the serine synthesis pathway is essential in breast cancer Results Presented by Stacey Lin Lloyd Lab http://www.amsbio.com/expression-ready-lentiviral-particles.aspx Overview
The levels of mrna expression in the mouse brain were measured at 52 dpi after the brains were divided into five regions: cortex, hippocampus,
 Supplemental material Supplemental method RNA extraction, reverse transcription, and real-time PCR The levels of mrna expression in the mouse brain were measured at 52 dpi after the brains were divided
Supplemental material Supplemental method RNA extraction, reverse transcription, and real-time PCR The levels of mrna expression in the mouse brain were measured at 52 dpi after the brains were divided
Supplementary Figure 1: Experimental design. DISCOVERY PHASE VALIDATION PHASE (N = 88) (N = 20) Healthy = 20. Healthy = 6. Endometriosis = 33
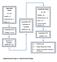 DISCOVERY PHASE (N = 20) Healthy = 6 Endometriosis = 7 EAOC = 7 Quantitative PCR (mirnas = 1113) a Quantitative PCR Verification of Candidate mirnas (N = 24) VALIDATION PHASE (N = 88) Healthy = 20 Endometriosis
DISCOVERY PHASE (N = 20) Healthy = 6 Endometriosis = 7 EAOC = 7 Quantitative PCR (mirnas = 1113) a Quantitative PCR Verification of Candidate mirnas (N = 24) VALIDATION PHASE (N = 88) Healthy = 20 Endometriosis
RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice
 SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
Hao D. H., Ma W. G., Sheng Y. L., Zhang J. B., Jin Y. F., Yang H. Q., Li Z. G., Wang S. S., GONG Ming*
 Comparison of transcriptomes and gene expression profiles of two chilling- and drought-tolerant and intolerant Nicotiana tabacum varieties under low temperature and drought stress Hao D. H., Ma W. G.,
Comparison of transcriptomes and gene expression profiles of two chilling- and drought-tolerant and intolerant Nicotiana tabacum varieties under low temperature and drought stress Hao D. H., Ma W. G.,
Supplementary Figure 1. Expression of phospho-sik3 in normal and osteoarthritic articular cartilage in the knee. (a) Semiserial histological sections
 Supplementary Figure 1. Expression of phospho-sik3 in normal and osteoarthritic articular cartilage in the knee. (a) Semiserial histological sections of normal cartilage were stained with safranin O-fast
Supplementary Figure 1. Expression of phospho-sik3 in normal and osteoarthritic articular cartilage in the knee. (a) Semiserial histological sections of normal cartilage were stained with safranin O-fast
Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent Protein.
 prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
Nature Structural & Molecular Biology: doi: /nsmb.2419
 Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
RNA-seq: filtering, quality control and visualisation. COMBINE RNA-seq Workshop
 RNA-seq: filtering, quality control and visualisation COMBINE RNA-seq Workshop QC and visualisation (part 1) Slide taken from COMBINE RNAseq workshop on 23/09/2016 RNA-seq of Mouse mammary gland Basal
RNA-seq: filtering, quality control and visualisation COMBINE RNA-seq Workshop QC and visualisation (part 1) Slide taken from COMBINE RNAseq workshop on 23/09/2016 RNA-seq of Mouse mammary gland Basal
RNA-Seq Preparation Comparision Summary: Lexogen, Standard, NEB
 RNA-Seq Preparation Comparision Summary: Lexogen, Standard, NEB CSF-NGS January 22, 214 Contents 1 Introduction 1 2 Experimental Details 1 3 Results And Discussion 1 3.1 ERCC spike ins............................................
RNA-Seq Preparation Comparision Summary: Lexogen, Standard, NEB CSF-NGS January 22, 214 Contents 1 Introduction 1 2 Experimental Details 1 3 Results And Discussion 1 3.1 ERCC spike ins............................................
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
f(x) = x R² = RPKM (M8.MXB) f(x) = x E-014 R² = 1 RPKM (M31.
 14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
Supporting Information
 Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
BIMM 143. RNA sequencing overview. Genome Informatics II. Barry Grant. Lecture In vivo. In vitro.
 RNA sequencing overview BIMM 143 Genome Informatics II Lecture 14 Barry Grant http://thegrantlab.org/bimm143 In vivo In vitro In silico ( control) Goal: RNA quantification, transcript discovery, variant
RNA sequencing overview BIMM 143 Genome Informatics II Lecture 14 Barry Grant http://thegrantlab.org/bimm143 In vivo In vitro In silico ( control) Goal: RNA quantification, transcript discovery, variant
The N-end rule pathway regulates pathogen responses. in plants
 SUPPLEMENTARY INFORMATION The N-end rule pathway regulates pathogen responses in plants Rémi de Marchi 1,2, Maud Sorel 1, Brian Mooney 1, Isabelle Fudal 3, Kevin Goslin 1, Kamila Kwaśniewska 4, Patrick
SUPPLEMENTARY INFORMATION The N-end rule pathway regulates pathogen responses in plants Rémi de Marchi 1,2, Maud Sorel 1, Brian Mooney 1, Isabelle Fudal 3, Kevin Goslin 1, Kamila Kwaśniewska 4, Patrick
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
Supplemental Data. Integrating omics and alternative splicing i reveals insights i into grape response to high temperature
 Supplemental Data Integrating omics and alternative splicing i reveals insights i into grape response to high temperature Jianfu Jiang 1, Xinna Liu 1, Guotian Liu, Chonghuih Liu*, Shaohuah Li*, and Lijun
Supplemental Data Integrating omics and alternative splicing i reveals insights i into grape response to high temperature Jianfu Jiang 1, Xinna Liu 1, Guotian Liu, Chonghuih Liu*, Shaohuah Li*, and Lijun
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
genomics for systems biology / ISB2020 RNA sequencing (RNA-seq)
 RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
SUPPLEMENTARY INFORMATION
 doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
Protein SD Units (P-value) Cluster order
 SUPPLEMENTAL TABLE AND FIGURES Table S1. Signature Phosphoproteome of CD22 E12 Transgenic Mouse BPL Cells. T-test vs. Other Protein SD Units (P-value) Cluster order ATPase (Ab-16) 1.41 0.000880 1 mtor
SUPPLEMENTAL TABLE AND FIGURES Table S1. Signature Phosphoproteome of CD22 E12 Transgenic Mouse BPL Cells. T-test vs. Other Protein SD Units (P-value) Cluster order ATPase (Ab-16) 1.41 0.000880 1 mtor
Supplementary Figure 1
 Supplementary Figure 1 A B mir-141, human cell lines mir-2c, human cell lines mir-141, hepatocytes mir-2c, hepatocytes Relative RNA.1.8.6.4.2 Relative RNA.3.2.1 Relative RNA 1.5 1..5 Relative RNA 2. 1.5
Supplementary Figure 1 A B mir-141, human cell lines mir-2c, human cell lines mir-141, hepatocytes mir-2c, hepatocytes Relative RNA.1.8.6.4.2 Relative RNA.3.2.1 Relative RNA 1.5 1..5 Relative RNA 2. 1.5
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
[U- 13 C5] glutamine. Glutamate. Acetyl-coA. Citrate. Citrate. Malate. Malate. Isocitrate OXIDATIVE METABOLISM. Oxaloacetate CO2.
![[U- 13 C5] glutamine. Glutamate. Acetyl-coA. Citrate. Citrate. Malate. Malate. Isocitrate OXIDATIVE METABOLISM. Oxaloacetate CO2. [U- 13 C5] glutamine. Glutamate. Acetyl-coA. Citrate. Citrate. Malate. Malate. Isocitrate OXIDATIVE METABOLISM. Oxaloacetate CO2.](/thumbs/76/74112154.jpg) Supplementary Figures a. Relative mrna levels Supplementary Figure 1 (Christofk) 3.0 2.5 2.0 1.5 1.0 0.5 0.0 LAT1 Fumarate Succinate Palmitate Acetyl-coA Oxaloacetate OXIDATIVE METABOLISM α-ketoglutarate
Supplementary Figures a. Relative mrna levels Supplementary Figure 1 (Christofk) 3.0 2.5 2.0 1.5 1.0 0.5 0.0 LAT1 Fumarate Succinate Palmitate Acetyl-coA Oxaloacetate OXIDATIVE METABOLISM α-ketoglutarate
Supplementary Figure 1. Using DNA barcode-labeled MHC multimers to generate TCR fingerprints
 Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
PBZ FT01_PBZ FT01_TZ FT01_NZ. interface zone (I) tumor zone (TZ) necrotic zone (NZ)
 Oncotarget, Supplementary Materials www.impactjournals.com/oncotarget/ SUPPLEMENTRY FLES ndividuals factor map (P) FT_ FT_ FT_ Dim (.%) Dim (.%) >% peripheral brain zone () around % interface zone () FT
Oncotarget, Supplementary Materials www.impactjournals.com/oncotarget/ SUPPLEMENTRY FLES ndividuals factor map (P) FT_ FT_ FT_ Dim (.%) Dim (.%) >% peripheral brain zone () around % interface zone () FT
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols.
 Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 )
 770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
Tutorial: RNA-Seq Analysis Part II: Non-Specific Matches and Expression Measures
 : RNA-Seq Analysis Part II: Non-Specific Matches and Expression Measures March 15, 2013 CLC bio Finlandsgade 10-12 8200 Aarhus N Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com support@clcbio.com
: RNA-Seq Analysis Part II: Non-Specific Matches and Expression Measures March 15, 2013 CLC bio Finlandsgade 10-12 8200 Aarhus N Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com support@clcbio.com
Nature Biotechnology: doi: /nbt Supplementary Figure 1. PL gene expression in tomato fruit.
 Supplementary Figure 1 PL gene expression in tomato fruit. Relative expression of five PL-coding genes measured in at least three fruit of each genotype (cv. Alisa Craig) at four stages of development,
Supplementary Figure 1 PL gene expression in tomato fruit. Relative expression of five PL-coding genes measured in at least three fruit of each genotype (cv. Alisa Craig) at four stages of development,
Endocrine Physiology
 Endocrine Physiology 1. Introduction to Endocrine Principles Endocrine cell Amount of hormone Receptor levels Chemical messenger Target cell Half life of hormones Carrier proteins Metabolism: activation
Endocrine Physiology 1. Introduction to Endocrine Principles Endocrine cell Amount of hormone Receptor levels Chemical messenger Target cell Half life of hormones Carrier proteins Metabolism: activation
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS
 Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Supplemental Information. Metabolic Maturation during Muscle Stem Cell. Differentiation Is Achieved by mir-1/133a-mediated
 Cell Metabolism, Volume 27 Supplemental Information Metabolic Maturation during Muscle Stem Cell Differentiation Is Achieved by mir-1/133a-mediated Inhibition of the Dlk1-Dio3 Mega Gene Cluster Stas Wüst,
Cell Metabolism, Volume 27 Supplemental Information Metabolic Maturation during Muscle Stem Cell Differentiation Is Achieved by mir-1/133a-mediated Inhibition of the Dlk1-Dio3 Mega Gene Cluster Stas Wüst,
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH
 EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
Hormones and the Endocrine System
 Chapter 45 Hormones and the Endocrine System PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions
Chapter 45 Hormones and the Endocrine System PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions
Cellecta Overview. Started Operations in 2007 Headquarters: Mountain View, CA
 Cellecta Overview Started Operations in 2007 Headquarters: Mountain View, CA Focus: Development of flexible, scalable, and broadly parallel genetic screening assays to expedite the discovery and characterization
Cellecta Overview Started Operations in 2007 Headquarters: Mountain View, CA Focus: Development of flexible, scalable, and broadly parallel genetic screening assays to expedite the discovery and characterization
PREPARED FOR: U.S. Army Medical Research and Materiel Command Fort Detrick, MD
 AD Award Number: W81XWH-11-1-0126 TITLE: Chemical strategy to translate genetic/epigenetic mechanisms to breast cancer therapeutics PRINCIPAL INVESTIGATOR: Xiang-Dong Fu, PhD CONTRACTING ORGANIZATION:
AD Award Number: W81XWH-11-1-0126 TITLE: Chemical strategy to translate genetic/epigenetic mechanisms to breast cancer therapeutics PRINCIPAL INVESTIGATOR: Xiang-Dong Fu, PhD CONTRACTING ORGANIZATION:
Supplementary Information
 Supplementary Information Astrocytes regulate adult hippocampal neurogenesis through ephrin-b signaling Randolph S. Ashton, Anthony Conway, Chinmay Pangarkar, Jamie Bergen, Kwang-Il Lim, Priya Shah, Mina
Supplementary Information Astrocytes regulate adult hippocampal neurogenesis through ephrin-b signaling Randolph S. Ashton, Anthony Conway, Chinmay Pangarkar, Jamie Bergen, Kwang-Il Lim, Priya Shah, Mina
Dioxin induces Ahr-dependent robust DNA demethylation of the Cyp1a1 promoter via Tdg in the mouse liver
 Dioxin induces Ahr-dependent robust DNA demethylation of the Cypa promoter via Tdg in the mouse liver Hesbon Z. Amenya, Chiharu Tohyama, Seiichiroh Ohsako Laboratory of Environmental Health Sciences, Center
Dioxin induces Ahr-dependent robust DNA demethylation of the Cypa promoter via Tdg in the mouse liver Hesbon Z. Amenya, Chiharu Tohyama, Seiichiroh Ohsako Laboratory of Environmental Health Sciences, Center
Supplemental Information. Genomic Characterization of Murine. Monocytes Reveals C/EBPb Transcription. Factor Dependence of Ly6C Cells
 Immunity, Volume 46 Supplemental Information Genomic Characterization of Murine Monocytes Reveals C/EBPb Transcription Factor Dependence of Ly6C Cells Alexander Mildner, Jörg Schönheit, Amir Giladi, Eyal
Immunity, Volume 46 Supplemental Information Genomic Characterization of Murine Monocytes Reveals C/EBPb Transcription Factor Dependence of Ly6C Cells Alexander Mildner, Jörg Schönheit, Amir Giladi, Eyal
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature8645 Physical coverage (x haploid genomes) 11 6.4 4.9 6.9 6.7 4.4 5.9 9.1 7.6 125 Neither end mapped One end mapped Chimaeras Correct Reads (million ns) 1 75 5 25 HCC1187 HCC1395 HCC1599
doi: 1.138/nature8645 Physical coverage (x haploid genomes) 11 6.4 4.9 6.9 6.7 4.4 5.9 9.1 7.6 125 Neither end mapped One end mapped Chimaeras Correct Reads (million ns) 1 75 5 25 HCC1187 HCC1395 HCC1599
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Figure 1: Additional metabolic parameters of obesity mouse models and controls. (a) Body weight, (b) blood glucose and (c) insulin
 Supplementary Figure 1: Additional metabolic parameters of obesity mouse models and controls. (a) Body weight, (b) blood glucose and (c) insulin resistance index of homeostatic model assessment (HOMA IR)
Supplementary Figure 1: Additional metabolic parameters of obesity mouse models and controls. (a) Body weight, (b) blood glucose and (c) insulin resistance index of homeostatic model assessment (HOMA IR)
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Quantification of myelin fragments in the aging brain (a) Electron microscopy on corpus callosum is shown for a 18-month-old wild type mice. Myelin fragments (arrows) were detected
Supplementary Figure 1 Quantification of myelin fragments in the aging brain (a) Electron microscopy on corpus callosum is shown for a 18-month-old wild type mice. Myelin fragments (arrows) were detected
Supplemental Information
 Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
Molecular Profiling of Tumor Microenvironment Alex Chenchik, Ph.D. Cellecta, Inc.
 Molecular Profiling of Tumor Microenvironment Alex Chenchik, Ph.D. Cellecta, Inc. Cellecta, Inc. Founded: April 2006 Headquarters: Mountain View, CA 12 SBIR Grants Custom Service Provider for Functional
Molecular Profiling of Tumor Microenvironment Alex Chenchik, Ph.D. Cellecta, Inc. Cellecta, Inc. Founded: April 2006 Headquarters: Mountain View, CA 12 SBIR Grants Custom Service Provider for Functional
SUPPLEMENTARY INFORMATION
 Supplementary text Collectively, we were able to detect ~14,000 expressed genes with RPKM (reads per kilobase per million) > 1 or ~16,000 with RPKM > 0.1 in at least one cell type from oocyte to the morula
Supplementary text Collectively, we were able to detect ~14,000 expressed genes with RPKM (reads per kilobase per million) > 1 or ~16,000 with RPKM > 0.1 in at least one cell type from oocyte to the morula
Supporting Information Table of content
 Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
microrna PCR System (Exiqon), following the manufacturer s instructions. In brief, 10ng of
 SUPPLEMENTAL MATERIALS AND METHODS Quantitative RT-PCR Quantitative RT-PCR analysis was performed using the Universal mircury LNA TM microrna PCR System (Exiqon), following the manufacturer s instructions.
SUPPLEMENTAL MATERIALS AND METHODS Quantitative RT-PCR Quantitative RT-PCR analysis was performed using the Universal mircury LNA TM microrna PCR System (Exiqon), following the manufacturer s instructions.
Introduction to Systems Biology of Cancer Lecture 2
 Introduction to Systems Biology of Cancer Lecture 2 Gustavo Stolovitzky IBM Research Icahn School of Medicine at Mt Sinai DREAM Challenges High throughput measurements: The age of omics Systems Biology
Introduction to Systems Biology of Cancer Lecture 2 Gustavo Stolovitzky IBM Research Icahn School of Medicine at Mt Sinai DREAM Challenges High throughput measurements: The age of omics Systems Biology
Transcription factor Foxp3 and its protein partners form a complex regulatory network
 Supplementary figures Resource Paper Transcription factor Foxp3 and its protein partners form a complex regulatory network Dipayan Rudra 1, Paul deroos 1, Ashutosh Chaudhry 1, Rachel Niec 1, Aaron Arvey
Supplementary figures Resource Paper Transcription factor Foxp3 and its protein partners form a complex regulatory network Dipayan Rudra 1, Paul deroos 1, Ashutosh Chaudhry 1, Rachel Niec 1, Aaron Arvey
Profiling of the Exosomal Cargo of Bovine Milk Reveals the Presence of Immune- and Growthmodulatory Non-coding RNAs (ncrna)
 Animal Industry Report AS 664 ASL R3235 2018 Profiling of the Exosomal Cargo of Bovine Milk Reveals the Presence of Immune- and Growthmodulatory Non-coding RNAs (ncrna) Eric D. Testroet Washington State
Animal Industry Report AS 664 ASL R3235 2018 Profiling of the Exosomal Cargo of Bovine Milk Reveals the Presence of Immune- and Growthmodulatory Non-coding RNAs (ncrna) Eric D. Testroet Washington State
Figure S1A. Blood glucose levels in mice after glucose injection
 ## Figure S1A. Blood glucose levels in mice after glucose injection Blood glucose (mm/l) 25 2 15 1 5 # 15 3 6 3+3 Time after glucose injection (min) # Figure S1B. α-kg levels in mouse livers after glucose
## Figure S1A. Blood glucose levels in mice after glucose injection Blood glucose (mm/l) 25 2 15 1 5 # 15 3 6 3+3 Time after glucose injection (min) # Figure S1B. α-kg levels in mouse livers after glucose
Supplementary figure legends
 Supplementary figure legends Fig. S1. Lineweaver-Burk plot of putrescine uptake by YeeF. An overnight culture of SK629 was inoculated in 100-mL LBG medium in 500-mL Erlenmeyer flasks. The medium was supplemented
Supplementary figure legends Fig. S1. Lineweaver-Burk plot of putrescine uptake by YeeF. An overnight culture of SK629 was inoculated in 100-mL LBG medium in 500-mL Erlenmeyer flasks. The medium was supplemented
MII. Supplement Figure 1. CapZ β2. Merge. 250ng. 500ng DIC. Merge. Journal of Cell Science Supplementary Material. GFP-CapZ β2 DNA
 A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
Introduction to metabolic regulation. Prof K Syed Department of Biochemistry & Microbiology University of Zululand Room no. 247
 Introduction to metabolic regulation Prof K Syed Department of Biochemistry & Microbiology University of Zululand Room no. 247 SyedK@unizulu.ac.za Topics Metabolism Metabolism: Categories Important metabolic
Introduction to metabolic regulation Prof K Syed Department of Biochemistry & Microbiology University of Zululand Room no. 247 SyedK@unizulu.ac.za Topics Metabolism Metabolism: Categories Important metabolic
Summary of behavioral performances for mice in imaging experiments.
 Supplementary Figure 1 Summary of behavioral performances for mice in imaging experiments. (a) Task performance for mice during M2 imaging experiments. Open triangles, individual experiments. Filled triangles,
Supplementary Figure 1 Summary of behavioral performances for mice in imaging experiments. (a) Task performance for mice during M2 imaging experiments. Open triangles, individual experiments. Filled triangles,
Nature Immunology: doi: /ni Supplementary Figure 1. RNA-Seq analysis of CD8 + TILs and N-TILs.
 Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Ayako Suzuki 1, Koutatsu Matsushima 2, Hideki Makinoshima 2, Sumio Sugano 1, Takashi Kohno 3,4, Katsuya Tsuchihara 2 and Yutaka Suzuki 1,5*
 Suzuki et al. Genome Biology (2015) 16:66 DOI 10.1186/s13059-015-0636-y RESEARCH Open Access Single-cell analysis of lung adenocarcinoma cell lines reveals diverse expression patterns of individual cells
Suzuki et al. Genome Biology (2015) 16:66 DOI 10.1186/s13059-015-0636-y RESEARCH Open Access Single-cell analysis of lung adenocarcinoma cell lines reveals diverse expression patterns of individual cells
Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease
 1 Supplemental Materials 2 3 Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease 4 5 6 Authors: Madhumita Basu, 1 Jun-Yi Zhu, 2 Stephanie LaHaye 1,3, Uddalak
1 Supplemental Materials 2 3 Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease 4 5 6 Authors: Madhumita Basu, 1 Jun-Yi Zhu, 2 Stephanie LaHaye 1,3, Uddalak
Figure S2. Distribution of acgh probes on all ten chromosomes of the RIL M0022
 96 APPENDIX B. Supporting Information for chapter 4 "changes in genome content generated via segregation of non-allelic homologs" Figure S1. Potential de novo CNV probes and sizes of apparently de novo
96 APPENDIX B. Supporting Information for chapter 4 "changes in genome content generated via segregation of non-allelic homologs" Figure S1. Potential de novo CNV probes and sizes of apparently de novo
Figure 1. Effects of FGF21 on adipose tissue. (A) Representative histological. findings of epididymal adipose tissue (B) mrna expression of
 SUPPLEMENTAL MATERIAL EN-12-2276 Figure 1. Effects of FGF21 on adipose tissue. (A) Representative histological findings of epididymal adipose tissue (B) mrna expression of adipocytokines in adipose tissue.
SUPPLEMENTAL MATERIAL EN-12-2276 Figure 1. Effects of FGF21 on adipose tissue. (A) Representative histological findings of epididymal adipose tissue (B) mrna expression of adipocytokines in adipose tissue.
SUPPLEMENTARY INFORMATION
 -. -. SUPPLEMENTARY INFORMATION DOI: 1.1/ncb86 a WAT-1 WAT- BAT-1 BAT- sk-muscle-1 sk-muscle- mir-133b mir-133a mir-6 mir-378 mir-1 mir-85 mir-378 mir-6a mir-18 mir-133a mir- mir- mir-341 mir-196a mir-17
-. -. SUPPLEMENTARY INFORMATION DOI: 1.1/ncb86 a WAT-1 WAT- BAT-1 BAT- sk-muscle-1 sk-muscle- mir-133b mir-133a mir-6 mir-378 mir-1 mir-85 mir-378 mir-6a mir-18 mir-133a mir- mir- mir-341 mir-196a mir-17
Simple, rapid, and reliable RNA sequencing
 Simple, rapid, and reliable RNA sequencing RNA sequencing applications RNA sequencing provides fundamental insights into how genomes are organized and regulated, giving us valuable information about the
Simple, rapid, and reliable RNA sequencing RNA sequencing applications RNA sequencing provides fundamental insights into how genomes are organized and regulated, giving us valuable information about the
Solid-Phase Purification of Synthetic DNA Sequences. Serge L. Beaucage, Ph.D. Chief, Laboratory of Biological Chemistry FDA-CDER
 Solid-Phase Purification of Synthetic DNA Sequences Serge L. Beaucage, Ph.D. Chief, Laboratory of Biological Chemistry FDA-CDER The one who follows the crowd will usually go no further than the crowd.
Solid-Phase Purification of Synthetic DNA Sequences Serge L. Beaucage, Ph.D. Chief, Laboratory of Biological Chemistry FDA-CDER The one who follows the crowd will usually go no further than the crowd.
Lecture 8 Understanding Transcription RNA-seq analysis. Foundations of Computational Systems Biology David K. Gifford
 Lecture 8 Understanding Transcription RNA-seq analysis Foundations of Computational Systems Biology David K. Gifford 1 Lecture 8 RNA-seq Analysis RNA-seq principles How can we characterize mrna isoform
Lecture 8 Understanding Transcription RNA-seq analysis Foundations of Computational Systems Biology David K. Gifford 1 Lecture 8 RNA-seq Analysis RNA-seq principles How can we characterize mrna isoform
Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay
 Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay Background ImQuest BioSciences has developed and qualified a single-plate method to expedite the screening of antiviral agents against
Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay Background ImQuest BioSciences has developed and qualified a single-plate method to expedite the screening of antiviral agents against
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation
 Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplementary information
 Supplementary information Supplementary Figure 1: Components of Arabidopsis tricarboxylic acid (TCA) cycle. Schematic summary of the TCA cycle and the enzymes related to the reactions. The large text and
Supplementary information Supplementary Figure 1: Components of Arabidopsis tricarboxylic acid (TCA) cycle. Schematic summary of the TCA cycle and the enzymes related to the reactions. The large text and
Supplementary Figure 1: High-throughput profiling of survival after exposure to - radiation. (a) Cells were plated in at least 7 wells in a 384-well
 Supplementary Figure 1: High-throughput profiling of survival after exposure to - radiation. (a) Cells were plated in at least 7 wells in a 384-well plate at cell densities ranging from 25-225 cells in
Supplementary Figure 1: High-throughput profiling of survival after exposure to - radiation. (a) Cells were plated in at least 7 wells in a 384-well plate at cell densities ranging from 25-225 cells in
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at
 Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Supplemental Fig. 1. Relative mrna Expression. Relative mrna Expression WT KO WT KO RT 4 0 C
 Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative
Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative
Nature Genetics: doi: /ng Supplementary Figure 1. Assessment of sample purity and quality.
 Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figures
 J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity.
 Supplementary Figure 1 Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity. Male C57BL/6J mice were fed a normal chow (NC, 10% fat) or a high-fat diet (HFD,
Supplementary Figure 1 Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity. Male C57BL/6J mice were fed a normal chow (NC, 10% fat) or a high-fat diet (HFD,
Lung Met 1 Lung Met 2 Lung Met Lung Met H3K4me1. Lung Met H3K27ac Primary H3K4me1
 a Gained Met-VELs 1.5 1.5 -.5 Lung Met 1 Lung Met Lung Met 3 1. Lung Met H3K4me1 Lung Met H3K4me1 1 Lung Met H3K4me1 Lung Met H3K7ac 1.5 Lung Met H3K7ac Lung Met H3K7ac.8 Primary H3K4me1 Primary H3K7ac
a Gained Met-VELs 1.5 1.5 -.5 Lung Met 1 Lung Met Lung Met 3 1. Lung Met H3K4me1 Lung Met H3K4me1 1 Lung Met H3K4me1 Lung Met H3K7ac 1.5 Lung Met H3K7ac Lung Met H3K7ac.8 Primary H3K4me1 Primary H3K7ac
Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types.
 Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Nature Genetics: doi: /ng Supplementary Figure 1. Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms.
 Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
In glycolysis, glucose is converted to pyruvate. If the pyruvate is reduced to lactate, the pathway does not require O 2 and is called anaerobic
 Glycolysis 1 In glycolysis, glucose is converted to pyruvate. If the pyruvate is reduced to lactate, the pathway does not require O 2 and is called anaerobic glycolysis. If this pyruvate is converted instead
Glycolysis 1 In glycolysis, glucose is converted to pyruvate. If the pyruvate is reduced to lactate, the pathway does not require O 2 and is called anaerobic glycolysis. If this pyruvate is converted instead
P. Tang ( 鄧致剛 ); PJ Huang ( 黄栢榕 ) g( ); g ( ) Bioinformatics Center, Chang Gung University.
 Databases and Tools for High Throughput Sequencing Analysis P. Tang ( 鄧致剛 ); PJ Huang ( 黄栢榕 ) g( ); g ( ) Bioinformatics Center, Chang Gung University. HTseq Platforms Applications on Biomedical Sciences
Databases and Tools for High Throughput Sequencing Analysis P. Tang ( 鄧致剛 ); PJ Huang ( 黄栢榕 ) g( ); g ( ) Bioinformatics Center, Chang Gung University. HTseq Platforms Applications on Biomedical Sciences
Supplemental data. Uppalapati et al. (2012) Plant Cell /tpc
 PAL Control P. emaculata 0 8 24 48 8 24 48 OPR Control P. emaculata 0 8 24 48 8 24 48 CHS PR3 CHR PR5 CHI PR10 IFS IFR Supplemental Figure 1. Expression profiles for selected genes in phenylpropanoid pathway
PAL Control P. emaculata 0 8 24 48 8 24 48 OPR Control P. emaculata 0 8 24 48 8 24 48 CHS PR3 CHR PR5 CHI PR10 IFS IFR Supplemental Figure 1. Expression profiles for selected genes in phenylpropanoid pathway
2.28E E AKT/CAT P-3-4:
 Supplemental Table 1: IPA network analysis of the microarray data presented in Figure 6 showing the most significant molecular and cellular function classifications along with the respective number of
Supplemental Table 1: IPA network analysis of the microarray data presented in Figure 6 showing the most significant molecular and cellular function classifications along with the respective number of
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Binding capacity of DNA-barcoded MHC multimers and recovery of antigen specificity
 Supplementary Figure 1 Binding capacity of DNA-barcoded MHC multimers and recovery of antigen specificity (a, b) Fluorescent-based determination of the binding capacity of DNA-barcoded MHC multimers (+barcode)
Supplementary Figure 1 Binding capacity of DNA-barcoded MHC multimers and recovery of antigen specificity (a, b) Fluorescent-based determination of the binding capacity of DNA-barcoded MHC multimers (+barcode)
Potential use of High iron and low phytate GM rice and their Bio-safety Assessment
 Potential use of High iron and low phytate GM rice and their Bio-safety Assessment Dr. Karabi Datta University of Calcutta, India Background High iron rice and iron bioavailability Micronutrient deficiency
Potential use of High iron and low phytate GM rice and their Bio-safety Assessment Dr. Karabi Datta University of Calcutta, India Background High iron rice and iron bioavailability Micronutrient deficiency
