A novel and universal method for microrna RT-qPCR data normalization
|
|
|
- Donald Sanders
- 5 years ago
- Views:
Transcription
1 A novel and universal method for microrna RT-qPCR data normalization Jo Vandesompele professor, Ghent University co-founder and CEO, Biogazelle 4 th International qpcr Symposium Weihenstephan, March 1, 29
2 outline megaplex stem-loop RT-PCR & PreAmp preamplification normalization of microrna gene expression levels
3 microrna quantification platform hybridisation based (microarray or beads in solution) Exiqon probeset mircury arrays flexmir beads Ambion mirvana probeset Invitrogen NCode probeset Agilent Human mirna Microarray Asuragen DiscovArray (service) PCR based Applied Biosystems stem-loop RT-PCR Exiqon mircury LNA microrna PCR System Invitrogen NCodemiRNA RT-PCR Qiagen miscript primer set miqpcr and other home brew protocols sequencing based (RNAseq)
4 microrna expression profiling stem-loop megaplex reverse transcription using 2 ng total RNA limited-cycle pre-amplification qpcr profiling 45 mirnas and controls higher sensitivity minimal amplification bias (Mestdagh et al., Nucleic Acids Research, 28) profiled > 1 samples
5 platform evaluation RT-qPCR qpcr plate setup: gene maximization (Hellemans et al., Genome Biology, 27) o a different mirna in each well of a 384 well plate (no replicates) o 1 sample per 384 well plate sample input o 2 ng total RNA (PreAmp) o 1.6 µg total RNA liquid handling Tecan Evo qpcr reactions on 79HT quality control o mean Cq for each 384 well plate o number of not expressed mirnas
6 Average Cq Average Cq Cq ( Cq NP - Cq P ) NBL-S, IMR-32 Cq ( Cq NP - Cq P ) NBL-S, IMR-32 minimal pre-amplification bias Average Cq NP (NBL-S, IMR-32) Average Cq NP (NBL-S, IMR-32)
7 Cq-value Cq-value Cq-value Cq-value single cell profiling A B mir-18a R 2 = mir-92 R 2 = total cell number total RNA input (pg) mir-2b R 2 = mir-19a R 2 = total cell number total RNA input (pg)
8 Mestdagh et al., Nucleic Acids Research, 28
9 normalization removal of experimentally induced noise input quantity: RNA quantity, cdna synthesis efficiency, input quality: RNA integrity, RNA purity, gold standard is the use of multiple stably expressed reference genes which genes? how many? how to do the calculations?
10 genorm normalisation framework for qpcr gene expression normalisation using the reference gene concept: quantified errors related to the use of a single reference gene (> 3 fold in 25% of the cases; > 6 fold in 1% of the cases) developed a robust algorithm for assessment of expression stability of candidate reference genes proposed the geometric mean of at least 3 reference genes for accurate and reliable normalisation Vandesompele et al., Genome Biology, 22 > 125 citations in PubMed > 8 software downloads
11 genorm software automated analysis ranking of candidate reference genes according to their stability determination of how many genes are required for reliable normalization
12 genorm validation robust insensitive to outliers maximal reduction of experimental variation accurate assessment of small expression differences statistically more significant results
13 microrna normalization small-rna controls classic normalization strategy small nuclear RNAs, small nucleolar RNAs 18 available from Applied Biosystems mean normalization method applied for microarray data universal: applicable for every mirna dataset many datapoints needed (megaplex vs. multiplex) mirnas/controls that resemble the mean minimal standard deviation when comparing mirna expression with mean ( genorm V value, st dev of log transformed ratios) compatible with multiplex asays need to determine mean
14 small RNA controls How stable is the mean compared to controls? genorm analysis using controls and mean as input variables exclusion of potentially co-regulated controls HY3 7q36 RNU19 5q31.2 RNU24 9q34 RNU38B 1p34.1-p32 RNU43 22q13 RNU44 1q25.1 RNU48 6p21.32 RNU49 17p11.2 RNU58A 18q21 RNU58B 18q21 RNU66 1p22.1 RNU6B 1p13 U18 15q22 U47 1q25.1 U54 8q12 U75 1q25.1 Z3 17q12 RPL21 13q12.2
15 mirna expression datasets neuroblastoma tumour samples T-ALL samples EVI1 deregulated leukemias retinoblastoma tumour samples normal tissues normal bone marrow
16 expression stability neuroblastoma 1,4 1,2 1,8,6,4,2
17 expression stability T-ALL 1,8 1,6 1,4 1,2 1,8,6,4,2
18 expression stability normal tissues 2,5 2 1,5 1,5
19 neuroblastoma not normalised stable controls mean mirnas
20 neuroblastoma not normalised stable controls mean mirnas
21 T-ALL not normalised stable controls mean mirnas
22 T-ALL not normalised stable controls mean mirnas
23 normal tissues not normalised stable controls mean mirnas
24 normal tissues not normalised stable controls mean mirnas
25 Fold enrichment CATGTG CATGTG CATGTG CACGTG CACGTG CATGTG CATGTG biological validation MYCN binds to the mir promoter -5 kb CpG island mir cluster +5 kb A B C IMR5 WAC2 A B C Amplicon
26 biological validation choice of normalization strategy influences differential mirna expression 3,5 Mir expression in neuroblastoma tumours 3 2,5 2 1,5 1,5 stable controls mean mirnas
27 biological validation choice of normalization strategy influences differential mirna expression 3,5 Mir expression in neuroblastoma tumours 3 2,5 2 1,5 1,5 stable controls mean mirnas
28 biological validation choice of normalization strategy influences differential mirna expression 3,5 Mir expression in neuroblastoma tumours 3 2,5 2 1,5 1,5 stable controls mean mirnas
29 fold change (MYCN amplified vs. MYCN single copy) balanced differential expression 3 2 controls mean average FC controls = -.44 average Fc mean =.5 average FC mirnas =
30 conclusions a highly sensitive mirna expression profiling platform novel and powerful mirna normalization strategy maximal reduction of technical noise improved identification of differentially expressed genes balancing of differential expression universally applicable o mean o multiple stable endogenous controls
31 acknowledgments mirna, T-UCR Pieter Mestdagh Frank Speleman Applied Biosystems qbase PLUS Jan Hellemans Stefaan Derveaux RNA QC, RNA amplification Nurten Yigit Justin Nuytens SHEP-tet Johannes Schulte (Essen) MYCN-ChIP Frank Westerman (Heidelberg)
From reference genes to global mean normalization
 From reference genes to global mean normalization Jo Vandesompele professor, Ghent University co-founder and CEO, Biogazelle qpcr Symposium USA November 9, 2009 Millbrae, CA outline what is normalization
From reference genes to global mean normalization Jo Vandesompele professor, Ghent University co-founder and CEO, Biogazelle qpcr Symposium USA November 9, 2009 Millbrae, CA outline what is normalization
Chapter 18. mirna Expression Profiling: From Reference Genes to Global Mean Normalization
 Chapter 18 mirna Expression Profiling: From Reference Genes to Global Mean Normalization Barbara D haene, Pieter Mestdagh, Jan Hellemans, and Jo Vandesompele Abstract MicroRNAs (mirnas) are an important
Chapter 18 mirna Expression Profiling: From Reference Genes to Global Mean Normalization Barbara D haene, Pieter Mestdagh, Jan Hellemans, and Jo Vandesompele Abstract MicroRNAs (mirnas) are an important
A two-microrna signature in urinary exosomes for diagnosis of prostate cancer
 Poster # B4 A two-microrna signature in urinary exosomes for diagnosis of prostate cancer Anne Karin Ildor Rasmussen 1, Peter Mouritzen 1, Karina Dalsgaard Sørensen 3, Thorarinn Blondal 1, Jörg Krummheuer
Poster # B4 A two-microrna signature in urinary exosomes for diagnosis of prostate cancer Anne Karin Ildor Rasmussen 1, Peter Mouritzen 1, Karina Dalsgaard Sørensen 3, Thorarinn Blondal 1, Jörg Krummheuer
microrna PCR System (Exiqon), following the manufacturer s instructions. In brief, 10ng of
 SUPPLEMENTAL MATERIALS AND METHODS Quantitative RT-PCR Quantitative RT-PCR analysis was performed using the Universal mircury LNA TM microrna PCR System (Exiqon), following the manufacturer s instructions.
SUPPLEMENTAL MATERIALS AND METHODS Quantitative RT-PCR Quantitative RT-PCR analysis was performed using the Universal mircury LNA TM microrna PCR System (Exiqon), following the manufacturer s instructions.
Extracellular Vesicle RNA isolation Kits
 Extracellular Vesicle RNA isolation Kits Summary Section 4 Introduction 42 Exo-TotalRNA and TumorExo-TotalRNA isolation kits 43 Extracellular Vesicle RNA extraction kits Ordering information Products can
Extracellular Vesicle RNA isolation Kits Summary Section 4 Introduction 42 Exo-TotalRNA and TumorExo-TotalRNA isolation kits 43 Extracellular Vesicle RNA extraction kits Ordering information Products can
Benchmark study: Exiqon mircury LNA microrna arrays vs. Supplier A s DNA based capture probes. Includes comparison of:
 Benchmark study: Exiqon mircury LNA microrna arrays vs. Supplier A s DNA based capture probes Includes comparison of: I. II. III. Signal levels Relative quantification False differences For complete experimental
Benchmark study: Exiqon mircury LNA microrna arrays vs. Supplier A s DNA based capture probes Includes comparison of: I. II. III. Signal levels Relative quantification False differences For complete experimental
Assaying micrornas in biofluids for detection of drug induced cardiac injury. HESI Annual Meeting State of the Science Session June 8, 2011
 Assaying micrornas in biofluids for detection of drug induced cardiac injury HESI Annual Meeting State of the Science Session June 8, 2011 Karol Thompson, PhD Center for Drug Evaluation & Research US Food
Assaying micrornas in biofluids for detection of drug induced cardiac injury HESI Annual Meeting State of the Science Session June 8, 2011 Karol Thompson, PhD Center for Drug Evaluation & Research US Food
Pieter Mestdagh *, Pieter Van Vlierberghe *, An De Weer *, Daniel Muth, Frank Westermann, Frank Speleman * and Jo Vandesompele *
 2009 Mestdagh Volume al. 10, Issue 6, Article R64 Open Access Method A novel and universal method for microrna RT-qPCR data normalization Pieter Mestdagh *, Pieter Van Vlierberghe *, An De Weer *, Daniel
2009 Mestdagh Volume al. 10, Issue 6, Article R64 Open Access Method A novel and universal method for microrna RT-qPCR data normalization Pieter Mestdagh *, Pieter Van Vlierberghe *, An De Weer *, Daniel
Supplementary webappendix
 Supplementary webappendix This webappendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Kratz JR, He J, Van Den Eeden SK, et
Supplementary webappendix This webappendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Kratz JR, He J, Van Den Eeden SK, et
Profiles of gene expression & diagnosis/prognosis of cancer. MCs in Advanced Genetics Ainoa Planas Riverola
 Profiles of gene expression & diagnosis/prognosis of cancer MCs in Advanced Genetics Ainoa Planas Riverola Gene expression profiles Gene expression profiling Used in molecular biology, it measures the
Profiles of gene expression & diagnosis/prognosis of cancer MCs in Advanced Genetics Ainoa Planas Riverola Gene expression profiles Gene expression profiling Used in molecular biology, it measures the
A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis
 APPLICATION NOTE Cell-Free DNA Isolation Kit A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis Abstract Circulating cell-free DNA (cfdna) has been shown
APPLICATION NOTE Cell-Free DNA Isolation Kit A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis Abstract Circulating cell-free DNA (cfdna) has been shown
developing new tools for diagnostics Join forces with IMGM Laboratories to make your mirna project a success
 micrornas developing new tools for diagnostics Join forces with IMGM Laboratories to make your mirna project a success Dr. Carola Wagner IMGM Laboratories GmbH Martinsried, Germany qpcr 2009 Symposium
micrornas developing new tools for diagnostics Join forces with IMGM Laboratories to make your mirna project a success Dr. Carola Wagner IMGM Laboratories GmbH Martinsried, Germany qpcr 2009 Symposium
Deploying the full transcriptome using RNA sequencing. Jo Vandesompele, CSO and co-founder The Non-Coding Genome May 12, 2016, Leuven
 Deploying the full transcriptome using RNA sequencing Jo Vandesompele, CSO and co-founder The Non-Coding Genome May 12, 2016, Leuven Roadmap Biogazelle the power of RNA reasons to study non-coding RNA
Deploying the full transcriptome using RNA sequencing Jo Vandesompele, CSO and co-founder The Non-Coding Genome May 12, 2016, Leuven Roadmap Biogazelle the power of RNA reasons to study non-coding RNA
Single Cell Quantitative Polymer Chain Reaction (sc-qpcr)
 Single Cell Quantitative Polymer Chain Reaction (sc-qpcr) Analyzing gene expression profiles from a bulk population of cells provides an average profile which may obscure important biological differences
Single Cell Quantitative Polymer Chain Reaction (sc-qpcr) Analyzing gene expression profiles from a bulk population of cells provides an average profile which may obscure important biological differences
Cellecta Overview. Started Operations in 2007 Headquarters: Mountain View, CA
 Cellecta Overview Started Operations in 2007 Headquarters: Mountain View, CA Focus: Development of flexible, scalable, and broadly parallel genetic screening assays to expedite the discovery and characterization
Cellecta Overview Started Operations in 2007 Headquarters: Mountain View, CA Focus: Development of flexible, scalable, and broadly parallel genetic screening assays to expedite the discovery and characterization
Phosphate buffered saline (PBS) for washing the cells TE buffer (nuclease-free) ph 7.5 for use with the PrimePCR Reverse Transcription Control Assay
 Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Simple, rapid, and reliable RNA sequencing
 Simple, rapid, and reliable RNA sequencing RNA sequencing applications RNA sequencing provides fundamental insights into how genomes are organized and regulated, giving us valuable information about the
Simple, rapid, and reliable RNA sequencing RNA sequencing applications RNA sequencing provides fundamental insights into how genomes are organized and regulated, giving us valuable information about the
Patnaik SK, et al. MicroRNAs to accurately histotype NSCLC biopsies
 Patnaik SK, et al. MicroRNAs to accurately histotype NSCLC biopsies. 2014. Supplemental Digital Content 1. Appendix 1. External data-sets used for associating microrna expression with lung squamous cell
Patnaik SK, et al. MicroRNAs to accurately histotype NSCLC biopsies. 2014. Supplemental Digital Content 1. Appendix 1. External data-sets used for associating microrna expression with lung squamous cell
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer. Application Note
 Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Obstacles and challenges in the analysis of microrna sequencing data
 Obstacles and challenges in the analysis of microrna sequencing data (mirna-seq) David Humphreys Genomics core Dr Victor Chang AC 1936-1991, Pioneering Cardiothoracic Surgeon and Humanitarian The ABCs
Obstacles and challenges in the analysis of microrna sequencing data (mirna-seq) David Humphreys Genomics core Dr Victor Chang AC 1936-1991, Pioneering Cardiothoracic Surgeon and Humanitarian The ABCs
# of Normal brain # of controls Upregulated Downregulated Normalizer # of mirnas in Platform profile
 Profiling studies comparing mirna expression in glioblastoma or glioblastoma cell lines to normal brain. MiRNAs were only included if fold change was greater than or equal to twofold Author Year Type of
Profiling studies comparing mirna expression in glioblastoma or glioblastoma cell lines to normal brain. MiRNAs were only included if fold change was greater than or equal to twofold Author Year Type of
Reference mirnas for mirnaome Analysis of Urothelial Carcinomas
 Reference mirnas for mirnaome Analysis of Urothelial Carcinomas Nadine Ratert 1,2, Hellmuth-Alexander Meyer 1,3, Monika Jung 1, Hans-Joachim Mollenkopf 4, Ina Wagner 4, Kurt Miller 1, Ergin Kilic 5, Andreas
Reference mirnas for mirnaome Analysis of Urothelial Carcinomas Nadine Ratert 1,2, Hellmuth-Alexander Meyer 1,3, Monika Jung 1, Hans-Joachim Mollenkopf 4, Ina Wagner 4, Kurt Miller 1, Ergin Kilic 5, Andreas
mirna Whole Transcriptome Assay
 mirna Whole Transcriptome Assay HTG EdgeSeq mirna Whole Transciptome Assay The HTG EdgeSeq mirna Whole Transcriptome Assay (WTA) is a next generation sequencing (NGS) application that measures the expression
mirna Whole Transcriptome Assay HTG EdgeSeq mirna Whole Transciptome Assay The HTG EdgeSeq mirna Whole Transcriptome Assay (WTA) is a next generation sequencing (NGS) application that measures the expression
Identification of mirna expression profiles for diagnosis and prognosis of prostate cancer
 Identification of mirna expression profiles for diagnosis and prognosis of prostate cancer To my grandfather Curt "Cula" Carlsson "Research is to see what everybody else has seen, and to think what nobody
Identification of mirna expression profiles for diagnosis and prognosis of prostate cancer To my grandfather Curt "Cula" Carlsson "Research is to see what everybody else has seen, and to think what nobody
Identification of suitable reference genes for the relative quantification of micrornas in pleural effusion
 ONCOLOGY LETTERS 8: 1889-1895, 2014 Identification of suitable reference genes for the relative quantification of micrornas in pleural effusion HYE SUK HAN 1, YEONG NANG JO 2, JIN YONG LEE 3, SONG YI CHOI
ONCOLOGY LETTERS 8: 1889-1895, 2014 Identification of suitable reference genes for the relative quantification of micrornas in pleural effusion HYE SUK HAN 1, YEONG NANG JO 2, JIN YONG LEE 3, SONG YI CHOI
Identification of Suitable Reference Genes for qpcr Analysis of Serum microrna in Gastric Cancer Patients
 DOI 10.1007/s10620-011-1981-7 ORIGINAL ARTICLE Identification of Suitable Reference Genes for qpcr Analysis of Serum microrna in Gastric Cancer Patients Jianning Song Zhigang Bai Wei Han Jun Zhang Hua
DOI 10.1007/s10620-011-1981-7 ORIGINAL ARTICLE Identification of Suitable Reference Genes for qpcr Analysis of Serum microrna in Gastric Cancer Patients Jianning Song Zhigang Bai Wei Han Jun Zhang Hua
SUPPLEMENTARY FIGURES AND TABLES
 SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: CaSR expression in neuroblastoma models. A. Proteins were isolated from three neuroblastoma cell lines and from the liver metastasis of a MYCN-non
SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: CaSR expression in neuroblastoma models. A. Proteins were isolated from three neuroblastoma cell lines and from the liver metastasis of a MYCN-non
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers
 Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Ambient Temperature Stabilization of RNA derived from Jurkat, HeLa and HUVEC Cell Lines for Use in RT-qPCR Assays
 Ambient Temperature Stabilization of RNA derived from Jurkat, HeLa and HUVEC Cell Lines for Use in RT-qPCR Assays C. Litterst 1, H. Martinez, B. Iverson and R. Nuňez 1 Bio-Rad Laboratories, Life Science
Ambient Temperature Stabilization of RNA derived from Jurkat, HeLa and HUVEC Cell Lines for Use in RT-qPCR Assays C. Litterst 1, H. Martinez, B. Iverson and R. Nuňez 1 Bio-Rad Laboratories, Life Science
microrna analysis for the detection of autologous blood transfusion in doping control
 microrna analysis for the detection of autologous blood transfusion in doping control Application note Doping Control Authors Francesco Donati*, Fiorella Boccia, Xavier de la Torre, Alessandra Stampella,
microrna analysis for the detection of autologous blood transfusion in doping control Application note Doping Control Authors Francesco Donati*, Fiorella Boccia, Xavier de la Torre, Alessandra Stampella,
Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay
 Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay Background ImQuest BioSciences has developed and qualified a single-plate method to expedite the screening of antiviral agents against
Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay Background ImQuest BioSciences has developed and qualified a single-plate method to expedite the screening of antiviral agents against
Upcoming Webinars. Profiling genes by pathways and diseases. Sample & Assay Technologies -1-
 Upcoming Webinars -1- Keep up to date: Follow Pathway focused biology on Facebook www.facebook.com/pathwaycentral Latest information on, pathway focused research and demos. -2- Understanding Gene Expression
Upcoming Webinars -1- Keep up to date: Follow Pathway focused biology on Facebook www.facebook.com/pathwaycentral Latest information on, pathway focused research and demos. -2- Understanding Gene Expression
York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
 MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
DETECTING DISEASE IN BLOOD
 Science Webinar Series DETECTING DISEASE IN BLOOD What mirna Biomarkers Can Tell Us 30 May, 2012 Change the size of any window by dragging the lower left corner. Use controls in top right corner to close
Science Webinar Series DETECTING DISEASE IN BLOOD What mirna Biomarkers Can Tell Us 30 May, 2012 Change the size of any window by dragging the lower left corner. Use controls in top right corner to close
Screening for novel oncology biomarker panels using both DNA and protein microarrays. John Anson, PhD VP Biomarker Discovery
 Screening for novel oncology biomarker panels using both DNA and protein microarrays John Anson, PhD VP Biomarker Discovery Outline of presentation Introduction to OGT and our approach to biomarker studies
Screening for novel oncology biomarker panels using both DNA and protein microarrays John Anson, PhD VP Biomarker Discovery Outline of presentation Introduction to OGT and our approach to biomarker studies
Products for cfdna and mirna isolation. Subhead Circulating Cover nucleic acids from plasma
 MACHEREY-NAGEL Products for cfdna and mirna isolation Bioanalysis Subhead Circulating Cover nucleic acids from plasma n Flexible solutions for small and large blood plasma volumes n Highly efficient recovery
MACHEREY-NAGEL Products for cfdna and mirna isolation Bioanalysis Subhead Circulating Cover nucleic acids from plasma n Flexible solutions for small and large blood plasma volumes n Highly efficient recovery
STAT1 regulates microrna transcription in interferon γ stimulated HeLa cells
 CAMDA 2009 October 5, 2009 STAT1 regulates microrna transcription in interferon γ stimulated HeLa cells Guohua Wang 1, Yadong Wang 1, Denan Zhang 1, Mingxiang Teng 1,2, Lang Li 2, and Yunlong Liu 2 Harbin
CAMDA 2009 October 5, 2009 STAT1 regulates microrna transcription in interferon γ stimulated HeLa cells Guohua Wang 1, Yadong Wang 1, Denan Zhang 1, Mingxiang Teng 1,2, Lang Li 2, and Yunlong Liu 2 Harbin
Abstract. Optimization strategy of Copy Number Variant calling using Multiplicom solutions APPLICATION NOTE. Introduction
 Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Genome-wide promoter methylation analysis in neuroblastoma identifies prognostic methylation biomarkers
 RESEARCH Open Access Genome-wide promoter methylation analysis in neuroblastoma identifies prognostic methylation biomarkers Anneleen Decock 1, Maté Ongenaert 1, Jasmien Hoebeeck 1,2, Katleen De Preter
RESEARCH Open Access Genome-wide promoter methylation analysis in neuroblastoma identifies prognostic methylation biomarkers Anneleen Decock 1, Maté Ongenaert 1, Jasmien Hoebeeck 1,2, Katleen De Preter
ncounter TM Analysis System
 ncounter TM Analysis System Molecules That Count TM www.nanostring.com Agenda NanoString Technologies History Introduction to the ncounter Analysis System CodeSet Design and Assay Principals System Performance
ncounter TM Analysis System Molecules That Count TM www.nanostring.com Agenda NanoString Technologies History Introduction to the ncounter Analysis System CodeSet Design and Assay Principals System Performance
Analysis of extracellular RNA in cerebrospinal fluid
 JOURNAL OF EXTRACELLULAR VESICLES, 217 VOL. 6, 1317577 https://doi.org/1.18/21378.217.1317577 TRANSFERRED ARTICLE Analysis of extracellular RNA in cerebrospinal fluid Julie A. Saugstad a *, Theresa A.
JOURNAL OF EXTRACELLULAR VESICLES, 217 VOL. 6, 1317577 https://doi.org/1.18/21378.217.1317577 TRANSFERRED ARTICLE Analysis of extracellular RNA in cerebrospinal fluid Julie A. Saugstad a *, Theresa A.
RECENT ADVANCES IN THE MOLECULAR DIAGNOSIS OF BREAST CANCER
 Technology Transfer in Diagnostic Pathology. 6th Central European Regional Meeting. Cytopathology. Balatonfüred, Hungary, April 7-9, 2011. RECENT ADVANCES IN THE MOLECULAR DIAGNOSIS OF BREAST CANCER Philippe
Technology Transfer in Diagnostic Pathology. 6th Central European Regional Meeting. Cytopathology. Balatonfüred, Hungary, April 7-9, 2011. RECENT ADVANCES IN THE MOLECULAR DIAGNOSIS OF BREAST CANCER Philippe
Advance Your Genomic Research Using Targeted Resequencing with SeqCap EZ Library
 Advance Your Genomic Research Using Targeted Resequencing with SeqCap EZ Library Marilou Wijdicks International Product Manager Research For Life Science Research Only. Not for Use in Diagnostic Procedures.
Advance Your Genomic Research Using Targeted Resequencing with SeqCap EZ Library Marilou Wijdicks International Product Manager Research For Life Science Research Only. Not for Use in Diagnostic Procedures.
Genomic explorations have offered us a great deal. Buried Treasure
 18 Buried Treasure Noncoding DNA makes up the majority of our genomes, much of it being conserved and transcribed. Though the functions of small regulatory RNA molecules are well known, what about so-called
18 Buried Treasure Noncoding DNA makes up the majority of our genomes, much of it being conserved and transcribed. Though the functions of small regulatory RNA molecules are well known, what about so-called
Prognostic value of microrna expression pattern in upper tract urothelial carcinoma
 Prognostic value of microrna expression pattern in upper tract urothelial carcinoma Laura Izquierdo 1, Mercedes Ingelmo-Torres 1, Carmen Mallofré 2, Juan José Lozano 3, Marie Verhasselt-Crinquette 4, Xavier
Prognostic value of microrna expression pattern in upper tract urothelial carcinoma Laura Izquierdo 1, Mercedes Ingelmo-Torres 1, Carmen Mallofré 2, Juan José Lozano 3, Marie Verhasselt-Crinquette 4, Xavier
QIAsymphony DSP Circulating DNA Kit
 QIAsymphony DSP Circulating DNA Kit February 2017 Performance Characteristics 937556 Sample to Insight Contents Performance Characteristics... 4 Basic performance... 4 Run precision... 6 Equivalent performance
QIAsymphony DSP Circulating DNA Kit February 2017 Performance Characteristics 937556 Sample to Insight Contents Performance Characteristics... 4 Basic performance... 4 Run precision... 6 Equivalent performance
Monitor TM assay: comparison with traditional RT-qPCR methods
 Preliminary evaluation of the GeneXpert Dx System for CML patients monitoring through the Xpert BCR-ABL Monitor TM assay: comparison with traditional RT-qPCR methods Silvia Calatroni Division of Hematology
Preliminary evaluation of the GeneXpert Dx System for CML patients monitoring through the Xpert BCR-ABL Monitor TM assay: comparison with traditional RT-qPCR methods Silvia Calatroni Division of Hematology
Next Generation Sequencing as a tool for breakpoint analysis in rearrangements of the globin-gene clusters
 Next Generation Sequencing as a tool for breakpoint analysis in rearrangements of the globin-gene clusters XXXth International Symposium on Technical Innovations in Laboratory Hematology Honolulu, Hawaii
Next Generation Sequencing as a tool for breakpoint analysis in rearrangements of the globin-gene clusters XXXth International Symposium on Technical Innovations in Laboratory Hematology Honolulu, Hawaii
The value of Omics to chemical risk assessment
 The value of Omics to chemical risk assessment Timothy W Gant There is a focus on transcriptomics in this talk but for example only. All omics are useful in risk assessment Outline What are we aiming to
The value of Omics to chemical risk assessment Timothy W Gant There is a focus on transcriptomics in this talk but for example only. All omics are useful in risk assessment Outline What are we aiming to
Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63.
 Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
WHO Prequalification of In Vitro Diagnostics PUBLIC REPORT. Product: Alere q HIV-1/2 Detect WHO reference number: PQDx
 WHO Prequalification of In Vitro Diagnostics PUBLIC REPORT Product: Alere q HIV-1/2 Detect WHO reference number: PQDx 0226-032-00 Alere q HIV-1/2 Detect with product codes 270110050, 270110010 and 270300001,
WHO Prequalification of In Vitro Diagnostics PUBLIC REPORT Product: Alere q HIV-1/2 Detect WHO reference number: PQDx 0226-032-00 Alere q HIV-1/2 Detect with product codes 270110050, 270110010 and 270300001,
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
NGS in Cancer Pathology After the Microscope: From Nucleic Acid to Interpretation
 NGS in Cancer Pathology After the Microscope: From Nucleic Acid to Interpretation Michael R. Rossi, PhD, FACMG Assistant Professor Division of Cancer Biology, Department of Radiation Oncology Department
NGS in Cancer Pathology After the Microscope: From Nucleic Acid to Interpretation Michael R. Rossi, PhD, FACMG Assistant Professor Division of Cancer Biology, Department of Radiation Oncology Department
iplex genotyping IDH1 and IDH2 assays utilized the following primer sets (forward and reverse primers along with extension primers).
 Supplementary Materials Supplementary Methods iplex genotyping IDH1 and IDH2 assays utilized the following primer sets (forward and reverse primers along with extension primers). IDH1 R132H and R132L Forward:
Supplementary Materials Supplementary Methods iplex genotyping IDH1 and IDH2 assays utilized the following primer sets (forward and reverse primers along with extension primers). IDH1 R132H and R132L Forward:
Gene-microRNA network module analysis for ovarian cancer
 Gene-microRNA network module analysis for ovarian cancer Shuqin Zhang School of Mathematical Sciences Fudan University Oct. 4, 2016 Outline Introduction Materials and Methods Results Conclusions Introduction
Gene-microRNA network module analysis for ovarian cancer Shuqin Zhang School of Mathematical Sciences Fudan University Oct. 4, 2016 Outline Introduction Materials and Methods Results Conclusions Introduction
Multiplex-PCR in clinical virology benefits and limitations. Weihenstephan Multiplex-PCR the concept:
 Multiplex-PCR in clinical virology benefits and limitations ans itschko, elga Mairhofer, Anna-Lena Winkler Max von Pettenkofer-Institute Department of Virology Ludwig-Maximilians-University, Munich Weihenstephan
Multiplex-PCR in clinical virology benefits and limitations ans itschko, elga Mairhofer, Anna-Lena Winkler Max von Pettenkofer-Institute Department of Virology Ludwig-Maximilians-University, Munich Weihenstephan
Supplementary Figure 1: Experimental design. DISCOVERY PHASE VALIDATION PHASE (N = 88) (N = 20) Healthy = 20. Healthy = 6. Endometriosis = 33
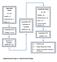 DISCOVERY PHASE (N = 20) Healthy = 6 Endometriosis = 7 EAOC = 7 Quantitative PCR (mirnas = 1113) a Quantitative PCR Verification of Candidate mirnas (N = 24) VALIDATION PHASE (N = 88) Healthy = 20 Endometriosis
DISCOVERY PHASE (N = 20) Healthy = 6 Endometriosis = 7 EAOC = 7 Quantitative PCR (mirnas = 1113) a Quantitative PCR Verification of Candidate mirnas (N = 24) VALIDATION PHASE (N = 88) Healthy = 20 Endometriosis
Circulating Cell-Free DNA Pre-analytics: Importance of ccfdna Stabilization and Extraction for Liquid Biopsy Applications
 Circulating Cell-Free DNA Pre-analytics: Importance of ccfdna Stabilization and Extraction for Liquid Biopsy Applications Martin Schlumpberger, Ass.Director R&D, QIAGEN GmbH IQNpath 2017 - Circulating
Circulating Cell-Free DNA Pre-analytics: Importance of ccfdna Stabilization and Extraction for Liquid Biopsy Applications Martin Schlumpberger, Ass.Director R&D, QIAGEN GmbH IQNpath 2017 - Circulating
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Accepted Contact information for corresponding author Lourde
 1 Accepted 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 PROGNOSTIC VALUE OF microrna EXPRESSION PATTERN IN UPPER TRACT UROTHELIAL CARCINOMA 1 Laura Izquierdo 1,
1 Accepted 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 PROGNOSTIC VALUE OF microrna EXPRESSION PATTERN IN UPPER TRACT UROTHELIAL CARCINOMA 1 Laura Izquierdo 1,
MicroRNA expression profiling and functional analysis in prostate cancer. Marco Folini s.c. Ricerca Traslazionale DOSL
 MicroRNA expression profiling and functional analysis in prostate cancer Marco Folini s.c. Ricerca Traslazionale DOSL What are micrornas? For almost three decades, the alteration of protein-coding genes
MicroRNA expression profiling and functional analysis in prostate cancer Marco Folini s.c. Ricerca Traslazionale DOSL What are micrornas? For almost three decades, the alteration of protein-coding genes
In Situ Hybridization: Market Strategies and Forecasts, US,
 In Situ Hybridization: Market Strategies and Forecasts, US, 2018-2024 Table of Contents In Situ Hybridization: Executive Summary The study is designed to give a comprehensive overview of the In Situ Hybridization
In Situ Hybridization: Market Strategies and Forecasts, US, 2018-2024 Table of Contents In Situ Hybridization: Executive Summary The study is designed to give a comprehensive overview of the In Situ Hybridization
Detection of copy number variations in PCR-enriched targeted sequencing data
 Detection of copy number variations in PCR-enriched targeted sequencing data German Demidov Parseq Lab, Saint-Petersburg University of Russian Academy of Sciences, current: Center for Genomic Regulation
Detection of copy number variations in PCR-enriched targeted sequencing data German Demidov Parseq Lab, Saint-Petersburg University of Russian Academy of Sciences, current: Center for Genomic Regulation
Circular RNAs (circrnas) act a stable mirna sponges
 Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Title: Dysregulated mir-183 Inhibits Migration in Breast Cancer
 Author's response to reviews Title: Dysregulated mir-183 Inhibits Migration in Breast Cancer Authors: Aoife J Lowery (aoife.lowery@gmail.com) Nicola Miller (nicola.miller@nuigalway.ie) Roisin M Dwyer (roisin.dwyer@nuigalway.ie)
Author's response to reviews Title: Dysregulated mir-183 Inhibits Migration in Breast Cancer Authors: Aoife J Lowery (aoife.lowery@gmail.com) Nicola Miller (nicola.miller@nuigalway.ie) Roisin M Dwyer (roisin.dwyer@nuigalway.ie)
Department of Pharmaceutical Sciences, School of Pharmacy, Northeastern University, Boston, MA 02115, USA 2
 Pancreatic Cancer Cell Exosome-Mediated Macrophage Reprogramming and the Role of MicroRNAs 155 and 125b2 Transfection using Nanoparticle Delivery Systems Mei-Ju Su 1, Hibah Aldawsari 2, and Mansoor Amiji
Pancreatic Cancer Cell Exosome-Mediated Macrophage Reprogramming and the Role of MicroRNAs 155 and 125b2 Transfection using Nanoparticle Delivery Systems Mei-Ju Su 1, Hibah Aldawsari 2, and Mansoor Amiji
MRC-Holland MLPA. Description version 13;
 SALSA MLPA probemix P027-C1 Uveal Melanoma Lot C1-0211: A large number of probes have been replaced by other probes in the same chromosomal regions as compared to previous lots, and several reference probes
SALSA MLPA probemix P027-C1 Uveal Melanoma Lot C1-0211: A large number of probes have been replaced by other probes in the same chromosomal regions as compared to previous lots, and several reference probes
Identification of aberrantly expressed mirnas in rectal cancer
 ONCOLOGY REPORTS 28: 77-84, 2012 Identification of aberrantly expressed mirnas in rectal cancer XINHUA LI 1*, GUIYING ZHANG 1*, FEIJUN LUO 2, JINDE RUAN 3, DAMAO HUANG 4, DEYUN FENG 3, DESHENG XIAO 3,
ONCOLOGY REPORTS 28: 77-84, 2012 Identification of aberrantly expressed mirnas in rectal cancer XINHUA LI 1*, GUIYING ZHANG 1*, FEIJUN LUO 2, JINDE RUAN 3, DAMAO HUANG 4, DEYUN FENG 3, DESHENG XIAO 3,
micrornas (mirna) and Biomarkers
 micrornas (mirna) and Biomarkers Small RNAs Make Big Splash mirnas & Genome Function Biomarkers in Cancer Future Prospects Javed Khan M.D. National Cancer Institute EORTC-NCI-ASCO November 2007 The Human
micrornas (mirna) and Biomarkers Small RNAs Make Big Splash mirnas & Genome Function Biomarkers in Cancer Future Prospects Javed Khan M.D. National Cancer Institute EORTC-NCI-ASCO November 2007 The Human
Droplet Digital PCR, the new tool in HIV reservoir quantification? Ward De Spiegelaere
 Droplet Digital PCR, the new tool in HIV reservoir quantification? Ward De Spiegelaere Droplet Digital PCR, the new tool in HIV reservoir quantification? Content: - Digital PCR - Applications - Total HIV
Droplet Digital PCR, the new tool in HIV reservoir quantification? Ward De Spiegelaere Droplet Digital PCR, the new tool in HIV reservoir quantification? Content: - Digital PCR - Applications - Total HIV
Mature microrna identification via the use of a Naive Bayes classifier
 Mature microrna identification via the use of a Naive Bayes classifier Master Thesis Gkirtzou Katerina Computer Science Department University of Crete 13/03/2009 Gkirtzou K. (CSD UOC) Mature microrna identification
Mature microrna identification via the use of a Naive Bayes classifier Master Thesis Gkirtzou Katerina Computer Science Department University of Crete 13/03/2009 Gkirtzou K. (CSD UOC) Mature microrna identification
Iso-Seq Method Updates and Target Enrichment Without Amplification for SMRT Sequencing
 Iso-Seq Method Updates and Target Enrichment Without Amplification for SMRT Sequencing PacBio Americas User Group Meeting Sample Prep Workshop June.27.2017 Tyson Clark, Ph.D. For Research Use Only. Not
Iso-Seq Method Updates and Target Enrichment Without Amplification for SMRT Sequencing PacBio Americas User Group Meeting Sample Prep Workshop June.27.2017 Tyson Clark, Ph.D. For Research Use Only. Not
genomics for systems biology / ISB2020 RNA sequencing (RNA-seq)
 RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
MPS for translocations
 MPS for translocations Filip Van Nieuwerburgh, Ph.D. Lab of Pharmaceutical Biotechnology NXTGNT massively parallel sequencing facility, Ghent University In collaboration with: Center for Medical Genetics,
MPS for translocations Filip Van Nieuwerburgh, Ph.D. Lab of Pharmaceutical Biotechnology NXTGNT massively parallel sequencing facility, Ghent University In collaboration with: Center for Medical Genetics,
Supplementary Table 2: ALK mutations in neuroblastoma cell lines and comparison
 Supplementary data Supplementary Table 2: ALK mutations in neuroblastoma cell lines and comparison with published studies. (MNA = MYCN amplification, A = MYCN amplified, NA = not MYCN amplified, wt = wild-type).
Supplementary data Supplementary Table 2: ALK mutations in neuroblastoma cell lines and comparison with published studies. (MNA = MYCN amplification, A = MYCN amplified, NA = not MYCN amplified, wt = wild-type).
EPIGENOMICS PROFILING SERVICES
 EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
mirna Expression Profiling Enables Risk Stratification in Archived and Fresh Neuroblastoma Tumor Samples
 Imaging, Diagnosis, Prognosis Clinical Cancer Research mirna Expression Profiling Enables Risk Stratification in Archived and Fresh Neuroblastoma Tumor Samples Katleen De Preter 1, Pieter Mestdagh 1,Jo
Imaging, Diagnosis, Prognosis Clinical Cancer Research mirna Expression Profiling Enables Risk Stratification in Archived and Fresh Neuroblastoma Tumor Samples Katleen De Preter 1, Pieter Mestdagh 1,Jo
Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer
 Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer Avrum Spira, Jennifer E Beane, Vishal Shah, Katrina Steiling, Gang Liu, Frank Schembri, Sean Gilman, Yves-Martine
Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer Avrum Spira, Jennifer E Beane, Vishal Shah, Katrina Steiling, Gang Liu, Frank Schembri, Sean Gilman, Yves-Martine
Profiles of gene expression & diagnosis/prognosis of cancer Lorena Roa de la Cruz
 Genomics Profiles of gene expression & diagnosis/prognosis of cancer Lorena Roa de la Cruz Gene expression profiling Measurement of the activity of thousands of genes at once Techniques used for gene expression
Genomics Profiles of gene expression & diagnosis/prognosis of cancer Lorena Roa de la Cruz Gene expression profiling Measurement of the activity of thousands of genes at once Techniques used for gene expression
mirna in Plasma Exosome is Stable under Different Storage Conditions
 Molecules 2014, 19, 1568-1575; doi:10.3390/molecules19021568 Article OPEN ACCESS molecules ISSN 1420-3049 www.mdpi.com/journal/molecules mirna in Plasma Exosome is Stable under Different Storage Conditions
Molecules 2014, 19, 1568-1575; doi:10.3390/molecules19021568 Article OPEN ACCESS molecules ISSN 1420-3049 www.mdpi.com/journal/molecules mirna in Plasma Exosome is Stable under Different Storage Conditions
medoct group, Medical University Vienna, Jack Ballantyne, PhD National Center for Forensic Science, Orlando, FL USA
 medoct group, Medical University Vienna, Jack Ballantyne, PhD National Center for Forensic Science, Orlando, FL USA Association of Forensic DNA Analysts and Administrators San Antonio, TX June 2012 Perceived
medoct group, Medical University Vienna, Jack Ballantyne, PhD National Center for Forensic Science, Orlando, FL USA Association of Forensic DNA Analysts and Administrators San Antonio, TX June 2012 Perceived
Circulating Microrna Expression As A Biomarker For Early Diagnosis Of Severity In Spinal Cord Injury
 Circulating Microrna Expression As A Biomarker For Early Diagnosis Of Severity In Spinal Cord Injury Susumu Hachisuka 1, Naosuke Kamei, MD, PhD 2, Satoshi Ujigo 3, Shigeru Miyaki 3, Mitsuo Ochi 3. 1 Hiroshima
Circulating Microrna Expression As A Biomarker For Early Diagnosis Of Severity In Spinal Cord Injury Susumu Hachisuka 1, Naosuke Kamei, MD, PhD 2, Satoshi Ujigo 3, Shigeru Miyaki 3, Mitsuo Ochi 3. 1 Hiroshima
ARTICLE IN PRESS. Critical Reviews in Oncology/Hematology xxx (2010) xxx xxx
 Critical Reviews in Oncology/Hematology xxx (2010) xxx xxx Circulating micrornas: Association with disease and potential use as biomarkers Glen Reid, Michaela B. Kirschner, Nico van Zandwijk Asbestos Diseases
Critical Reviews in Oncology/Hematology xxx (2010) xxx xxx Circulating micrornas: Association with disease and potential use as biomarkers Glen Reid, Michaela B. Kirschner, Nico van Zandwijk Asbestos Diseases
SALSA MLPA probemix P315-B1 EGFR
 SALSA MLPA probemix P315-B1 EGFR Lot B1-0215 and B1-0112. As compared to the previous A1 version (lot 0208), two mutation-specific probes for the EGFR mutations L858R and T709M as well as one additional
SALSA MLPA probemix P315-B1 EGFR Lot B1-0215 and B1-0112. As compared to the previous A1 version (lot 0208), two mutation-specific probes for the EGFR mutations L858R and T709M as well as one additional
SALSA MS-MLPA KIT ME011-A1 Mismatch Repair genes (MMR) Lot 0609, 0408, 0807, 0407
 SALSA MS-MLPA KIT ME011-A1 Mismatch Repair genes (MMR) Lot 0609, 0408, 0807, 0407 The Mismatch Repair (MMR) system is critical for the maintenance of genomic stability. MMR increases the fidelity of DNA
SALSA MS-MLPA KIT ME011-A1 Mismatch Repair genes (MMR) Lot 0609, 0408, 0807, 0407 The Mismatch Repair (MMR) system is critical for the maintenance of genomic stability. MMR increases the fidelity of DNA
Non-Invasive Array Based Screening for Cancer. Dr. Amit Kumar President and CEO CombiMatrix Corporation
 Non-Invasive Array Based Screening for Cancer Dr. Amit Kumar President and CEO CombiMatrix Corporation www.combimatrix.com May 13, 2009 This presentation contains forward-looking statements within the
Non-Invasive Array Based Screening for Cancer Dr. Amit Kumar President and CEO CombiMatrix Corporation www.combimatrix.com May 13, 2009 This presentation contains forward-looking statements within the
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Set the stage: Genomics technology. Jos Kleinjans Dept of Toxicogenomics Maastricht University, the Netherlands
 Set the stage: Genomics technology Jos Kleinjans Dept of Toxicogenomics Maastricht University, the Netherlands Amendment to the latest consolidated version of the REACH legislation REACH Regulation 1907/2006:
Set the stage: Genomics technology Jos Kleinjans Dept of Toxicogenomics Maastricht University, the Netherlands Amendment to the latest consolidated version of the REACH legislation REACH Regulation 1907/2006:
KAPA Stranded RNA-Seq Library Preparation Kit
 KAPA Stranded RNA-Seq Illumina platforms KR0934 v1.13 This provides product information and a detailed protocol for the KAPA Stranded RNA- Seq (Illumina platforms), product codes KK8400 and KK8401. Contents
KAPA Stranded RNA-Seq Illumina platforms KR0934 v1.13 This provides product information and a detailed protocol for the KAPA Stranded RNA- Seq (Illumina platforms), product codes KK8400 and KK8401. Contents
Human influenza A virus subtype (H3)
 PCRmax Ltd TM qpcr test Human influenza A virus subtype (H3) Haemoglutinin H3 gene 150 tests For general laboratory and research use only 1 Introduction to Human influenza A virus subtype (H3) Influenza,
PCRmax Ltd TM qpcr test Human influenza A virus subtype (H3) Haemoglutinin H3 gene 150 tests For general laboratory and research use only 1 Introduction to Human influenza A virus subtype (H3) Influenza,
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
CMV DNA Quantification Using an Automated Platform for Nucleic Acid Extraction and Real- time PCR Assay Set-up
 JCM Accepts, published online ahead of print on 11 May 2011 J. Clin. Microbiol. doi:10.1128/jcm.00721-11 Copyright 2011, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights
JCM Accepts, published online ahead of print on 11 May 2011 J. Clin. Microbiol. doi:10.1128/jcm.00721-11 Copyright 2011, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights
EXOSOMES & MICROVESICLES
 The Sample Preparation Experts EXOSOMES & MICROVESICLES The New Standard in Exosome Purification and RNA Isolation Best-in-Class, Pure & Simple Exosome Purification, Fractionation of Exosomal Free & Circulating
The Sample Preparation Experts EXOSOMES & MICROVESICLES The New Standard in Exosome Purification and RNA Isolation Best-in-Class, Pure & Simple Exosome Purification, Fractionation of Exosomal Free & Circulating
On the Reproducibility of TCGA Ovarian Cancer MicroRNA Profiles
 On the Reproducibility of TCGA Ovarian Cancer MicroRNA Profiles Ying-Wooi Wan 1,2,4, Claire M. Mach 2,3, Genevera I. Allen 1,7,8, Matthew L. Anderson 2,4,5 *, Zhandong Liu 1,5,6,7 * 1 Departments of Pediatrics
On the Reproducibility of TCGA Ovarian Cancer MicroRNA Profiles Ying-Wooi Wan 1,2,4, Claire M. Mach 2,3, Genevera I. Allen 1,7,8, Matthew L. Anderson 2,4,5 *, Zhandong Liu 1,5,6,7 * 1 Departments of Pediatrics
Supplementary Online Content
 Supplementary Online Content Fumagalli D, Venet D, Ignatiadis M, et al. RNA Sequencing to predict response to neoadjuvant anti-her2 therapy: a secondary analysis of the NeoALTTO randomized clinical trial.
Supplementary Online Content Fumagalli D, Venet D, Ignatiadis M, et al. RNA Sequencing to predict response to neoadjuvant anti-her2 therapy: a secondary analysis of the NeoALTTO randomized clinical trial.
MicroRNA expression profiling in endometrial adenocarcinoma
 MicroRNA expression profiling in endometrial adenocarcinoma To my parents Örebro Studies in Medicine 118 SANJA JURCEVIC MicroRNA expression profiling in endometrial adenocarcinoma Sanja Jurcevic, 2015
MicroRNA expression profiling in endometrial adenocarcinoma To my parents Örebro Studies in Medicine 118 SANJA JURCEVIC MicroRNA expression profiling in endometrial adenocarcinoma Sanja Jurcevic, 2015
Detection of let-7a MicroRNA by Real-time PCR in Colorectal Cancer: a Single-centre Experience from China
 The Journal of International Medical Research 2007; 35: 716 723 Detection of let-7a MicroRNA by Real-time PCR in Colorectal Cancer: a Single-centre Experience from China W-J FANG 1, C-Z LIN 2, H-H ZHANG
The Journal of International Medical Research 2007; 35: 716 723 Detection of let-7a MicroRNA by Real-time PCR in Colorectal Cancer: a Single-centre Experience from China W-J FANG 1, C-Z LIN 2, H-H ZHANG
Hypoxia represses microrna biogenesis proteins in breast cancer cells
 Bandara et al. BMC Cancer 2014, 14:533 RESEARCH ARTICLE Open Access Hypoxia represses microrna biogenesis proteins in breast cancer cells Veronika Bandara 1,2, Michael Z Michael 2 and Jonathan M Gleadle
Bandara et al. BMC Cancer 2014, 14:533 RESEARCH ARTICLE Open Access Hypoxia represses microrna biogenesis proteins in breast cancer cells Veronika Bandara 1,2, Michael Z Michael 2 and Jonathan M Gleadle
