Genome-wide association study dissects the genetic architecture of
|
|
|
- Esther Hensley
- 6 years ago
- Views:
Transcription
1 Supplementary Information for Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels Hui Li 1,6, Zhiyu Peng 2,6, Xiaohong Yang 1,6, Weidong Wang 1,6, Junjie Fu 3,6, Jianhua Wang 1,6, Yingjia Han 1, Yuchao Chai 1, Tingting Guo 1, Ning Yang 4, Jie Liu 4, Marilyn L. Warburton 5, Yanbing Cheng 2, Xiaomin Hao 1, Pan Zhang 1, Jinyang Zhao 2, Yunjun Liu 3, Guoying Wang 3, Jiansheng Li 1, Jianbing Yan 4 1 National Maize Improvement Center of China, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing , China. 2 BGI-Shenzhen, Shenzhen, , China 3 Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing , China 4 National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan , China. 5 United States Department of Agriculture-Agricultural Research Service: Corn Host Plant Resistance Research Unit, Box 9555, Mississippi State, MS, 39762, USA 6 These authors contributed equally to this work. Correspondence should be addressed to J.Y. (yjianbing@mail.hzau.edu.cn), J.L. (lijiansheng@cau.edu.cn) and G.W. (gywang@caas.net.cn). This file includes: Supplementary Figures 1-13 Supplementary Tables 1-11
2 Supplementary Figure 1 Percentage of 10 kernel oil components detected by gas chromatography and measured in 508 diverse inbred lines. Supplementary Figure 2 Distributions of maize kernel oil concentration and composition in 508 inbred lines. Supplementary Figure 3 Manhattan plots of each chromosome resulting from the GWAS results for oil concentration in maize kernels. Supplementary Figure 4 Manhattan and quantile-quantile plots resulting from the GWAS results for oil compositional traits in maize kernels. Supplementary Figure 5 The simplified lipid metabolic pathway in maize, modified from Arabidopsis and other plants. Supplementary Figure 6 Manhattan plots for the expression GWAS of proposed candidate genes with eqtl. Supplementary Figure 7 The co-expression network of the 54 most highly connected genes among 67 genes with expression data. Supplementary Figure 8 The validation of FAD2 via re-sequencing in an independent panel. Supplementary Figure 9 The validation of ACP via re-sequencing in an independent panel. Supplementary Figure 10 The validation of LACS via re-sequencing in an independent panel. Supplementary Figure 11 The validation of WRI1a via re-sequencing in an independent panel. Supplementary Figure 12 The co-expression network of WRI1a. Supplementary Figure 13 The validation of COPII via re-sequencing in an independent panel. Supplementary Table 1 Phenotypic analysis, correlations and heritability of all measured traits Supplementary Table 2 Correlations among all the measured traits Supplementary Table 3 SNPs and candidate genes significantly associated with oil compositional traits in two association panels of 368 and 508 maize inbred lines Supplementary Table 4 SNPs significantly associated with traits after conditioning upon lead SNPs of all oil traits from primary GWAS analysis Supplementary Table 5 List of possible additional candidate genes within 100 kb flanking region of the 74 lead SNPs Supplementary Table 6 Comparison of the associations of oil concentration between regular and high-oil lines Supplementary Table 7 Comparison of the associations of oil compositional traits between regular and high-oil lines Supplementary Table 8 Confirmation of significant candidate genes in three linkage populations Supplementary Table 9 Correlation analysis between the phenotype and the expression levels of proposed candidate genes with eqtls Supplementary Table 10 The full list of genes in the co-expression network of WRI1a Supplementary Table 11 List of the primers used in this study
3 Supplementary Figure 1 Percentage of 10 kernel oil components detected by gas chromatography and measured in 508 diverse inbred lines. Oil concentration was calculated as the sum of all the compositions. C16:0, palmitic acid; C16:1, palmitoleic acid; C18:0, stearic acid; C18:1, oleic acid; C18:2, linoleic acid; C18:3, linolenic acid; C20:0, arachidic acid; C20:1, gadoleic acid; C22:0, behenic acid; C24:0, lignoceric acid.
4 Supplementary Figure 2 Distributions of maize kernel oil concentration and composition in 508 inbred lines.the left plots are the frequency distributions and the right are the density distributions.
5 Supplementary Figure 3 Manhattan plots of each chromosome resulting from the GWAS results for oil concentration in maize kernels. The dashed horizontal line depicts the Bonferroni-adjusted significance threshold ( ). For each chromosome, the significant unique SNPs are indicated with red dots. The manhanttan plot of chromosome 9 is not shown as there were no significant loci detected on this chromosome. The full gene names are listed in Table 1.
6
7
8
9
10 Supplementary Figure 4 Manhattan and quantile-quantile plots resulting from the GWAS results for oil compositional traits in maize kernels. The dashed horizontal line in Manhattan plot depicts the Bonferroni-adjusted significance threshold ( ). The lead sites significantly associated with corresponding traits are indicated by red dots on the Manhattan plots. SFA: saturated fatty acid; USFA: Unsaturated fatty acid. Jianbing Yan 11/13/12 7:33 PM Deleted:
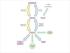
11 Supplementary Figure 5 The simplified lipid metabolic pathway in maize, modified from Arabidopsis and other plants 30. The candidate genes corresponding to significant loci detected by GWAS are in orange. The plasmid, endoplasmic reticulum (ER), oil body and peroxisome, are marked with light green, light blue, light orange and light purple, respectively. Abbreviations: PDHC, pyruvate dehydrogenase complex; ACC, acetyl-coa carboxylase; MCMT, malonyl-coa:acp malonyltransferase; FAS, fatty acid synthase; KAS, ketoacyl-acp synthase; SAD, stearoyl-acp desaturase; ACP, acyl carrier protein; FAT, acyl-acp thioesterase; FFA, free fatty acid; LACS, long chain acyl-coa synthase; FAD2, ER oleate desaturase; FAD3, ER linoleate desaturase; PC, phosphatidylcholine; G3P, glycerol 3-phosphate;GPAT, glycerol phosphate acyltransferase; LPAAT, lysophosphatidic acid (LPA) acyltransferase; PP, phosphatidic acid (PA) phosphatase; DGAT, acyl-coa: diacylglycerol (DAG) acyltransferase; FAE2, fatty acid elongase; KCS, ketoacyl-coa synthase; VLCFA, very long chain fatty acid; TAGL, triacylglycerol (TAG) lipase; MAGL: monoacylglycerol (MAG) lipase; CTS, COMATOSE (an ABC transporter); ACT, acyl-coa thioesterase; ACX, acyl-coa oxidase.
12
13
14
15 Supplementary Figure 6 Manhattan plots for the expression GWAS of proposed candidate genes with eqtl. A P) the top Manhattan plots of each panel are the GWAS results for oil compositional traits (A L) and the GWAS results after conditioning upon 63 lead SNPs (M P). The red dots are the proposed candidate genes with eqtls. The small Manhattan plots are the expression GWAS results for the candidate genes, and the red diamonds are the most significant sites associated with the expression level of each candidate gene.
16 Supplementary Figure 7 The co-expression network of the 54 most highly connected genes among 67 genes with expression data.
17 Supplementary Figure 8 The validation of FAD2 via re-sequencing in an independent panel. (a) Association between polymorphisms within the gene and the phenotypic variation in oleic acid levels in three environments. The x axis is the distance in base pairs along the gene; the first base of the start codon is defined as position 1. The y axis is the probability expressed as log 10 P from a mixed linear model. (b) The structure and functional domain of FAD2. The filled gray boxes represent the exons, open boxes indicate the UTRs, the black dashed lines mark the region re-sequenced in this study, and the orange ellipse represents the location of the conserved functional domain in gene. The red stars represent the lead polymorphisms (PZE ) detected by original GWAS and the proposed functional polymorphisms (S230A) detected by re-sequencing. R1, R2, and R3 are amplicons of
18 sequencing reaction; the primer sequences are given in Supplementary Table 11. (c) A representation of the pair-wise r 2 value(a measure of LD) among the significantly associated sites, where the color of each box corresponds to the r 2 value according to the legend. The purple star represents the proposed functional polymorphic locus. (d) Plots of the results from the correlation analysis. The x axis represents the normalized expression level of FAD2 in kernels collected at 15 DAP. The y axis represents the levels of oleic acid and linoleic acid, and the ratio of the two components. The r value is a Pearson correlation coefficient. (e) Box plot of normalized FAD2 expression for each allele of S230A site. The P value is based on a two-tailed t-test. (f) Linkage mapping result for chromosome 4 in a RIL population derived from By804 and B73. The x axis is the genetic map position, and the y axis is the LOD value. The purple line is the empirical significance threshold (LOD = 2.5). QTLs for C18:1 from two environments in Beijing 2005 (B 05) and Beijing 2006 (B 06) were identified; FAD2 is located within these QTLs. Jianbing Yan 11/13/12 7:34 PM Deleted: BY804
19 Supplementary Figure 9 The validation of ACP via re sequencing in an independent panel. (a) Association between polymorphisms within the gene and the phenotypic variation in oil concentration in three environments. The x axis is the distance in base pairs along the gene; the first base of the start codon is defined as position 1. The y axis is the probability expressed as log 10 P from a mixed linear model. (b) The structure and functional domain of ACP. Filled gray boxes represent exons, open boxes indicate the UTRs, the black dashed lines mark the region re-sequenced in this study and the orange ellipse represents the location of the conserved functional domain in gene. The red star represent the lead polymorphisms (M1c ) detected by original GWAS. The red triangle represents the proposed functional polymorphisms (InDel_8) by re-sequencing. The marker, InDel_8, represents the allele with 8-bp or without insertions. R1, R2, and R3 are amplicons of sequencing reaction; the primer sequences are given in Supplementary Table 11. (c) A representation of the pair-wise r 2 value (a measure of LD) among the significantly associated sites, where the color of each box corresponds to the r 2 value according to the legend. The purple star represents the proposed functional polymorphic locus. (d) Plots of the results from the correlation analysis. The x axis represents the normalized expression level of ACP in kernels collected at 15 DAP. The y axis represents the total oil concentration. The r value is a Pearson correlation coefficient. (e) Box plot of normalized ACP expression for each allele of InDel_8 site. The P value is based on a two-tailed t-test. The phenotypic variation explained (R 2 ) from ANOVA analysis.
20 Supplementary Figure 10 The validation of LACS via re-sequencing in an independent panel. (a) Association between polymorphisms within the gene and the phenotypic variation in oil concentration in three environments. The x axis is the distance in base pairs along the gene; the first base of the start codon is defined as position 1. The y axis is the probability expressed as log 10P from a mixed linear model. (b) The structure and functional domain of LACS. Filled gray boxes represent exons, open boxes indicate the UTRs, the black dashed lines mark the region re-sequenced in this study and the location of the conserved functional domain in gene. The red star represent the lead polymorphisms (M2c ) detected by original GWAS. The red triangle represents the proposed functional polymorphisms (InDel_146/472) by re-sequencing. The marker, InDel_146/472, represents the allele with a 146 bp or 472 bp insertion. R1, R2, and R3 are amplicons of sequencing reaction; the primer sequences are given in Supplementary Table 11. (c) A representation of the pair wise r 2 value (a measure of LD) among the significantly associated sites, where the color of each box corresponds to the r 2 value according to the legend. The purple star represents the proposed functional polymorphic locus. (d) Plots of the results from the correlation analysis. The x axis represents the normalized expression of LACS in kernels collected at 15 DAP. The y axis represents the total oil concentration. The r value is a Pearson correlation coefficient. (e) Box plot of normalized LACS expression for each allele of InDel_146/472 site. The P value is based on a two-tailed t-test. The phenotypic variation explained (R 2 ) from ANOVA analysis.
21 Supplementary Figure 11 The validation of WRI1a via re sequencing in an independent panel. (a) Association between polymorphisms within the gene and the phenotypic variation in oil concentration in three environments. The x axis is the distance in base pairs along the gene; the first base of the start codon is defined as position 1. The y axis is the probability expressed as log 10P from a mixed linear model. (b) The structure and functional domain of WRI1a. Filled gray boxes represent exons, open boxes indicate the UTRs, the black dashed lines mark the region re-sequenced in this study and the orange ellipse represents the location of the conserved functional domains in gene. The red star represent the lead polymorphisms (M2c ) detected by original GWAS. The red triangle represents the proposed functional polymorphisms (InDel_2000) by re-sequencing. InDel_2000 represents the allele with 2000-bp or without insertions. R1, R2, and R3 are amplicons of sequencing reaction; the primer sequences are given in Supplementary Table 11. (c) A representation of the pair wise r 2 value (a measure of LD) among the significantly associated sites, where the color of each box corresponds to the r 2 value according to the legend. The purple star represents the proposed functional polymorphic locus. (d) Plots of the results from the correlation analysis. The x axis represents the normalized expression of WRI1a in kernels collected at 15 DAP. The y axis represents the total oil concentration. The r value is a Pearson correlation coefficient. (e) Box plot of normalized WRI1a expression for each allele of InDel_2000 site. The P value is based on a two tailed t-test. The phenotypic variation explained (R 2 ) from ANOVA analysis.
22 Supplementary Figure 12 The co expression network of WRI1a. Genes shown in light blue are annotated as transcription factors. Genes shown in yellow were previously reported as the putative target genes of WRI1a 33. Genes shown in orange are involved in late glycolysis and fatty acid biosynthesis in plastids. Genes shown in light green are mainly related to carbohydrate metabolism, amino acid metabolism and transmembrane transport. Genes shown in gray have an unknown function. The full list of genes can be found in Supplementary Table 10. Abbreviations: PDHE1, pyruvate dehydrogenase E1; PDHE2, pyruvate dehydrogenase E2; LT, lipoyltransferase; OAT, oleoyl ACP, thioesterase; KHT, ketopantoate hydroxymethyltransferase; PK, pyruvate kinase; ACAT2, acetoacetyl CoA thiolase 2; ACP, acyl carrier protein; OLE, oleosin; PPT, phosphoenolpyruvate/phosphate translocator; KAS II, ketoacyl ACP synthase II; OAS III, oxoacyl ACP synthase III; SAD, stearoyl ACP desaturase; FAD2, fatty acid desaturase 2; GH, glycosyl hydrolases; GP, ADP glucose pyrophosphorylase; PK, protein kinase; S/TPK, serine/ threonine kinase protein; P450, cytochrome P450; ZFP, zinc finger protein; GRF, growth regulating factor; UF, unknown function.
23 Supplementary Figure 13 The validation of COPII via re-sequencing in an independent panel. (a) Association between polymorphisms within the gene and the phenotypic variation in oil concentration in three environments. The x axis is the distance in base pairs along the gene; the first base of the start codon is defined as position 1. The y axis is the probability expressed as log 10P from a mixed linear model. (b) The structure and functional domain of COPII. Filled gray boxes represent exons, open boxes indicate the UTRs, the black dashed lines mark the region re-sequenced in this study and the orange ellipse represents the location of the conserved functional domain in gene. The red star represent the lead polymorphisms (M8c ) detected by original GWAS. The red triangle represents the proposed functional polymorphisms (InDel_20) by re-sequencing. InDel_20 represents the allele with 20-bp or without insertions. R1, R2, R3, and R4 are amplicons of sequencing reaction; the primer sequences are given in Supplementary Table 11. (c) A representation of the pair wise r 2 value (a measure of LD) among the significantly associated sites, where the color of each box corresponds to the r 2 value according to the legend. The purple star represents the proposed functional polymorphic locus. (d) Plots of the results from the correlation analysis. The x axis represents the normalized expression of COPII in kernels collected at 15 DAP. The y axis represents the total oil concentration. The r value is a Pearson correlation coefficient. (e) Box plot of normalized COPII expression for each allele of InDel_20 site. The P value is based on a two-tailed t-test. (f) Linkage mapping result for chromosome 8 in the F 2:3 population derived from Dan340 and K22. The x axis is the genetic map position, and the y axis is the LOD value. The gray line is the empirical significance threshold (LOD = 2.5). A QTL for total oil was identified; COPII is located within this QTL interval.
24 ARY TABLES 1 11 able 1 Phenotypic analysis, correlations and heritability of all measured traits of variation DF c Oil C16:0 C16:1 C18:0 C18:1 C18:2 C18:3 C20:0 C20:1 C22:0 C24:0 ment ** ** 0.09** 1.10** ** 79.23** 1.13** 0.56** 0.22** 0.16** 0.12** pe ** 9.96** 0.01** 0.98** ** ** 0.29** 0.03** 0.01** 0.01** 0.02** ** lity for H b s.d ± ± ± ± ± ± ± ± ± ± ± 0.09 of variation DF C16:0/C16:1 C16:0/C18:0 C18:0/C18:1 C18:1/C18:2 C18:2/C18:3 C18:0/C20:0 C20:0/C20:1 C20:0/C22:0 C22:0/C24:0 SFA/USFA ment ** 22.92** 0.01** 0.27** ** 22.46** 6.39** 15.66** 1.14** 0.05** otype ** 12.26** 0.00** 0.15** ** 2.56** 0.33** 0.77** 0.09** 0.00** r itability CI for H b ge n ± s.d ± ± ± ± ± ± ± ± ± ± 0.03 ce, showing the mean square and degrees of freedom. b 95% confidence interval (CI) for broad sense heritability. **, P < 0.01; *, P < 0.05; s.d., standard of freedom.
25 able 2 Correlations among all the measured traits C16:1 C18:0 C18:1 C18:2 C18:3 C20:0 C20:1 C22:0 C24:0 C16:0/C16:1 C16:0/C18:0 C18:0/C18:1 C18:1/C18:2 C18:2/C18:3 C18:0/C20:0 C20:0/C20:1 C20:0/C22:0 C22:0/C24:0 SFA/USFA 0.35** 0.19** 0.48** 0.33** 0.69** ** 0.60** 0.10* 0.36** 0.20** 0.40** 0.71** 0.23** ** 0.29** 0.43** 0.36** 0.12** 0.34** ** 0.16** ** 0.35** 0.20** 0.55** 0.13** 0.25** 0.41** 0.28** 0.16** 0.33** 0.13** 0.95** 0.13** ** ** 0.19** 0.30** 0.80** 0.23** ** 0.13** 0.15** 0.28** 0.17** 0.32** 0.08** 0.21** 0.27** 0.21** 0.47** 0.18** 0.09* 0.17** 0.10* 0.79** 0.65** 0.22** 0.09* 0.64** 0.62** 0.61** 0.12** 0.19** ** 0.95** 0.54** 0.21** 0.32** 0.19** 0.34** 0.16** 0.33** 0.56** 0.98** 0.12** ** 0.23** 0.27** 0.09** 0.27** 0.94** 0.43** 0.34** 0.34** ** 0.11* 0.24** 0.48** 0.96** ** 0.21** ** 0.19** 0.48** 0.39** 0.10* 0.13** 0.34** 0.48** ** 0.26** 0.47** 0.82** 0.14** ** 0.24** 0.28** ** 0.05* 0.47** 0.15** 0.31** 0.09** 0.41** 0.68** 0.37** 0.10* 0.29** 0.20** 0.23** 0.10* 0.35** 0.62** 0.13** 0.31** 0.38** ** 0.16** 0.16** 0.22** 0.27** 0.10** 0.40** 0.42** 0.28** 0.14** 0.19** 0.41** 0.30** ** 0.44** ** 0.01 ** 0.11** ** 0.07** 0.27** 0.67** 0.49** 0.72** ** ** 0.38** 0.67** 0.30** 0.60** 0.22** 0.37** ** 0.26** 0.12** 0.33** 0.19** 0.46** 0.41** 0.32** 0.69** 0.10* 0.30** 0.10* 0.29** 0.45** 0.49** 0.13** 0.61** 0.47** 0.37** ** 0.81** 0.08** 0.16** 0.11** 0.11** 0.09** 0.20** 0.07** 0.09** ** 0.15** 0.13** ** 0.10* 0.22** ** 0.16** 0.79** 0.32** 0.24** 0.29** 0.29** 0.21** 0.19** 0.25** 0.07** 0.44** 0.30** 0.26** 0.64** 0.45** 0.62** 0.15** 0.30** ** ** 0.54** 0.44** 0.20** 0.27** 0.31** 0.11** 0.13** 0.17** 0.46** 0.51** ** 0.56** 0.10* ** 5** ** 0.98** 0.96** 0.42** 0.18** 0.21** 0.16** 0.28** 0.16** 0.29** 0.48** ** 0.21** 0.17** 2** 0.32** 0.06* ** 0.81** 0.10** 0.05* 0.30** 0.41** 0.16** 0.20** ** ** 0.17** 0.39** 0** 0.12** 0.63** 0.08** ** 0.35** 0.52** 0.61** 0.46** ** 0.48** 0.07** 0.16** 0.14** 0.53** 0.13** 0.14** ** 0.20** 0.60** 0.06* 0.06* ** 0.46** 0.21** 0.11** 0.10** 0.45** 0.55** * 0.12** 0.20** 0.15** 0.37** 6** 0.12** 0.56** 0.46** 0.42** 0.50** 0.13** 0.19** 0.61** 0.52** ** 0.12** 0.43** 0.38** 0.46** 0.29** 0.11** 0.17** 8** 0.20** 0.09** 0.19** 0.18** 0.25** 0.27** 0.18** 0.28** 0.45** 0.16** 0.12** 0.05* 0.18** 0.18** 0.12** 0.09** * ** 0.29** 0.25** 0.27** 0.05* 0.21** 0.46** 0.11** 0.41** 0.38** 0.21** 0.23** 0.38** 0.17** 0.31** 0.15** 0.36** 0.11** 0.04 above and below the diagonal are the genetic and phenotypic correlation coefficients, respectively. **, P < 0.01; *, P < 0.05.
26
27 able 3 SNPs and candidate genes significantly associated with oil compositional traits in two association panels of 368 and 508 Chr. Position b SNPs Lead trait c Other trait d Alleles e MAF f P value g QTL h j QTL eqtl Annotation k direction i PZE C20:1 A/G N # Ketoacyl CoA Synthase, KCS S1,S2,S3,S M1c C16:1 G/C Y + (D/C) Haem peroxidase M1c C16:1 A/C Y + (D/C) Unknown M1c C20:0 G/A N # Unknown M2c C18:3 A/C N # N.S* A cytochrome b5 containing fusion protein, CYTOCHROMB5 30,S5,S6,S7,S8,S M2c C18:0 C/T N # N.S* Unknown M3c C18:0/C18:1 T/A Phosphatidylinositol/phosphat idylcholine transfer protein, PITP S10,S M3c C16:1 C/G N # N.S* Unknown PZE C18:1 C18:2,C18:0/C A/C Y + (C) N.S* Oleate desaturase, FAD2 18:1, (refs.22,30,71,72,73,74) C18:1/C18: M4c C180/C181 C/A N # Sequence specific DNA binding transcription factor activity M4c C18:0 C/G N # N.S* Unknown M4c C16:0 SFA/USFA T/C N # Nutrient reservoir activity PUT 163a C18:0 C/A N # Zinc finger Jianbing Yan Formatted: S
28 Chr. Position b SNPs Lead trait c Other trait d Alleles e MAF f P value g QTL h j QTL eqtl Annotation k direction i PZA C16:0 A/G Y + (D) N.S* COPII coated vesicles, COPII 30, M5c C20:0/C22:0 A/G N # N.A* Unknown M6c C18:0/C20:0 T/C Y + (D/C) N.S* WD 40 repeats M6c C20:0 A/G Y + (B) Acyl ACP thioesterases, FATB.a $ 24,30,S12,S13,S M6c C22:0 C20:0,C24:0 T/A N # Unknown M6c C24:0 G/T Y + (B) Protein binding M6c C20:1 G/A Y + (B/C) Vegetative storage protein M6c C22:0 C/A N # Structural constituent of ribosome M6c C20:0 C/T Y + (B) N.S* Zinc finger PZE C20:0 A/G N # Unknown M6c C18:0/C18:1 G/T N # N.S* Unknown M6c C18:2 G/A N # N.S* Unknown M6c C18:0 G/C N # Fasciclin like arabinogalactan protein M6c C20:1 T/C N # N.S* Unknown M8c C18:1 G/C Y + (C) N.S* Unknown M8c C20:0 A/G Y + (C) Unknown M8c C18:0 T/C Y + (C) Unknown M9c C16:0 A/G Y + (D/C) Unknown M9c SFA/USFA G/A Y + (B/D/C) Acyl transferase M9c C16:0 C16:0/C16:1,C G/T Y + (D/C) Acyl ACP thioesterases, 16:0/C18:0,SF FATB.b $ 24,30,S12,S13,S14 Jianbing Yan Formatted: S
29 Chr. Position b SNPs Lead trait c Other trait d Alleles e MAF f P value g QTL h j QTL eqtl Annotation k direction i A/USFA,C18:0 /C18: PZB C18:0/C20:0 A/G N # N.S* Fatty acid elongase, FAE2 30,S15,S16,S PZE C18:2/C18:3 C18:3 C/A Y + (B) Unknown M10c C18:1/C18:2 C18:1 C/G N # Hydrolase activity M10c C18:0 T/C Y + (C) N.S* RNA polymerase II candidate gene in the locus or the nearest annotated gene to the lead SNP. b Position in base pairs for the lead SNP according to B73 reference sequence.60, c The oil trait associated with the highest P value among oil compositional traits. d Additional oil trait with a high probability of 10 6 ). e Major allele (first) and minor allele. The underlined base is the favorable allele. f Minor allele frequency (MAF). g P values of the lead trait only. h The ated in one of the QTL intervals reported previously (see supplementary references indicated in brackets) or in the BY804/B73 RIL population (B) or in one populations K22/Dan340 (D) and CI7/K22 (C). # The candidate was not located in any QTL interval. + The candidate gene was located in one or more QTL nd C populations. i The allele effect direction in B, D and C populations. High parent. Low parent. Not segregated. j SNP located within 200 kb of the ene, that was most significantly associated with the expression level of this gene. N.S., not significant (P > ); N.A., not available (no expression e gene). k Each candidate gene is annotated according to Interproscan ( The supplementary references for these genes are t the end of the annotation. $ There are two copies of this gene in the maize genome.
30
31 able 4 SNPs significantly associated with traits after conditioning upon lead SNPs of all oil traits from primary GWAS analysis Chr. Position b SNP Lead trait c Other trait d e Alleles MAF f P value g QTL h QTL direction i j eqtl Annotation k M1c C22:0/C24:0 C/T Y + (D) N.A. Acyl CoA oxidase, M1c C22:0/C24:0 A/G Y + (B) ACX S18,S19,S Acetyl CoA synthetase 30,S SYN27357 C18:1/C18:2 C/A N # Unknown M2c C22:0/C24:0 C/A N # N.S.* Protein tyrosine phosphatase M4c C22:0/C24:0 T/C N # Nutrient reservoir activity M4c C18:3 C/G N # Unknown M4c C16:0/C16:1 C/T N # Protein phosphatase 2C, PP2C M5c C20:0/C20:1 G/T N # N.A.* 4A nuclear receptor M6c C22:0/C24:0 C/A N # N.A.* Epoxide hydrolase, EH 51, M8c C18:1/C18:2 C18:1 C/G Y + (C) Translation elongation factor, EF1B M8c C22:0/C24:0 T/C N # Unknown ical candidate gene either in the locus or the nearest annotated gene to the lead SNP. b Position in base pairs for the lead SNP according to version 5b.60 of e sequence ( c The lipid traits associated with the highest P value among all traits. d Additional lipid traits with P < e Major or allele. The underlined base indicates the favorable allele. f Minor allele frequency (MAF). g P values of the lead trait only. h The candidate gene is located in ervals reported previously (see supplementary references indicated in brackets) or in the BY804/B73 RIL population (B) or in one or both of the F 2:3/F 2:4 an340 (D) and CI7/K22 (C). # The candidate was not located in any QTL interval. + The candidate gene was located in one or more QTL intervals in the B, D. i The allele effect direction in B, D and C populations. Not segregated. j SNP located within 200 kb of the candidate gene, that was most significantly e expression of this gene. N.S., not significant (P > ); V. A., not available. k Each candidate gene is annotated according to InterProScan k/interpro). The supplementary references for these genes are included in brackets at the end of the annotation.
32 Table 5 List of possible additional candidate genes within a 100 kb flanking region of the 74 lead SNPs Chr. Position Gene list b GRMZM2G031718,GRMZM2G172448,GRMZM2G172410,GRMZM2G GRMZM5G855347,GRMZM2G064663,GRMZM2G064732,GRMZM2G GRMZM2G496991,GRMZM2G153393,GRMZM5G815358,GRMZM2G GRMZM2G144995,GRMZM2G GRMZM2G None GRMZM2G005887,GRMZM2G009530,GRMZM2G GRMZM5G839017,GRMZM2G059064,GRMZM2G GRMZM2G110328,GRMZM2G110304,GRMZM2G110294,GRMZM2G GRMZM2G062394,GRMZM2G062377,GRMZM2G062289,GRMZM2G047299,GRMZM2G047223,GRMZM2G349062,GRMZM2G046231,GR MZM2G046267,GRMZM2G GRMZM2G051675,GRMZM2G None GRMZM2G GRMZM2G455491,GRMZM2G GRMZM2G358619,GRMZM2G GRMZM5G881605,GRMZM2G082974,GRMZM2G083094,GRMZM2G GRMZM2G471529,GRMZM2G GRMZM2G176570,GRMZM2G GRMZM2G044055,GRMZM2G044092,GRMZM2G118362,GRMZM2G GRMZM2G003682,GRMZM2G GRMZM2G148532,GRMZM2G GRMZM2G088273,GRMZM2G088365,GRMZM2G088441,GRMZM2G388461,GRMZM2G045387,GRMZM2G346895,GRMZM2G346897,GR MZM2G044104,GRMZM2G044152,GRMZM2G GRMZM2G054651,GRMZM2G133756,GRMZM2G GRMZM5G828945,GRMZM2G055520,GRMZM2G055499,GRMZM5G890820,GRMZM2G152573
33 Chr. Position Gene list b GRMZM2G059021,GRMZM2G GRMZM2G064960,GRMZM2G365298,GRMZM2G065030,GRMZM2G GRMZM2G122846,GRMZM2G122843,GRMZM2G122811,GRMZM2G GRMZM2G GRMZM2G166713,GRMZM2G125378,GRMZM2G125387,GRMZM2G125455,GRMZM2G125512,GRMZM2G GRMZM2G008839,GRMZM2G008819,GRMZM2G008691,GRMZM2G GRMZM2G441541,GRMZM2G GRMZM2G173903,GRMZM2G AC _FG002,GRMZM2G345055,GRMZM2G GRMZM2G GRMZM2G339018,GRMZM2G041310,GRMZM2G041159,GRMZM2G580389,GRMZM2G GRMZM2G147809,GRMZM2G147814,GRMZM2G147862,GRMZM2G GRMZM2G007810,AC _FG003,GRMZM2G GRMZM2G064852,GRMZM2G GRMZM2G363437,GRMZM2G GRMZM2G GRMZM2G149452,GRMZM2G GRMZM2G GRMZM2G139796,GRMZM2G079080,GRMZM2G GRMZM2G335618,GRMZM2G567373,GRMZM2G GRMZM2G055209,GRMZM2G GRMZM2G GRMZM2G GRMZM2G367631,GRMZM2G331283,GRMZM2G GRMZM2G030583,GRMZM2G116986,GRMZM2G GRMZM2G442215,GRMZM2G442244
34 Chr. Position Gene list b GRMZM5G826194,GRMZM5G GRMZM2G None GRMZM2G366778,GRMZM2G065757,GRMZM2G GRMZM2G142068,GRMZM2G167786,GRMZM2G167860,GRMZM2G010953,GRMZM2G GRMZM2G GRMZM2G527017,GRMZM2G GRMZM2G097141,GRMZM2G097103,GRMZM2G003103,GRMZM5G GRMZM2G052926,GRMZM2G GRMZM2G GRMZM2G None GRMZM2G043737,GRMZM5G800211,GRMZM5G847358,GRMZM2G None GRMZM2G098721,GRMZM2G098750,GRMZM2G GRMZM2G103179,GRMZM2G005461,GRMZM2G005339,GRMZM2G485304,GRMZM2G104269,GRMZM2G GRMZM2G391473,GRMZM5G899300,GRMZM2G GRMZM2G GRMZM2G None GRMZM2G028307,GRMZM2G091819,AC _FG None None GRMZM2G043737,GRMZM5G800211,GRMZM5G847358,GRMZM2G according to chromosome and physical position, which are listed in Table 1, Supplementary Tables 3 and 4. b Other possible candidate genes besides
35 able 6 Comparison of the associations of oil concentration between regular and high-oil lines P value* Alleles* MAF* P value (2) a Alleles (2) b MAF (2) c P value (3) a Alleles (3) b MAF(3) c T/C T/C T/C T/A T/A T/A C/A C/A C/A T/C T/C C/T A/G A/G A/G T/G T/G T/G C/G C/G C/G C/T C/T C/T G/C G/C G/C A/G A/G A/G A/G A/G A/G G/C G/C G/C G/A G/A G/A C/T C/T C/T G/C G/C G/C A/G A/G A/G G/C G/C G/C C/T C/T C/T C/T C/T C/T C/A C/A C/A G/T G/T G/T G/T G/T G/T G/A G/A G/A A/C A/C A/C T/C T/C T/C 0.13
36 P value* Alleles* MAF* P value (2) a Alleles (2) b MAF (2) c P value (3) a Alleles (3) b MAF(3) c G/A G/A G/A 0.22 ed in Table1. (2) Associations of oil concentration in regular lines excluding high-oil lines. (3) Associations of oil concentration in high-oil lines excluding of association. b Major allele (first), minor allele. The underlined bases are the favorable alleles. c Minor allele frequency (MAF).
37 able 7 Comparison of the associations of oil compositional traits between regular and high-oil lines P value * Alleles * MAF * P value (2) a Alleles (2) b MAF (2) b P value (3) a Alleles (3) b MAF (3) c A/G A/G A # G/C G/C G # A/C A/C A # G/A G/A A/G A/C A/C A # C/T C/T T/C T/A T/A A/T C/G C/G G/C A/C A/C C/A C/A C/A C/A C/G C/G G/C T/C T/C C/T C/A C/A C # A/G A/G G/A A/G A/G A/G T/C T/C C/T A/G A/G A # T/A T/A T # G/T G/T G* G/A G/A A/G C/A C/A C # C/T C/T T/C A/G A/G G/A G/T G/T T/G 0.17
38 P value * Alleles * MAF * P value (2) a Alleles (2) b MAF (2) b P value (3) a Alleles (3) b MAF (3) c G/A G/A A/G G/C G/C G # T/C T/C T # G/C G/C G # A/G A/G G/A T/C T/C T # A/G A/G A/G G/A G/A A/G G/T G/T T/G A/G A/G G/A C/A C/A A/G C/G C/G C # T/C T/C C/T 0.09 listed in Supplementary Table 3. (2) Associations of oil concentration in regular lines excluding high-oil lines. (3) Associations of oil concentration in high-oil ular lines. a P value of association. b Major allele (first), minor allele. The underlined bases are the favorable alleles. C Minor allele frequency (MAF). # Not
39 able 8 Confirmation of significant candidate genes in three linkage populations Chr. Confidence Interval (Mb) a Population b Trait A c D d R 2 (%) e 1 B C22:0/C24: D (BJ $ ) C 1 B C16:1 D / C(BJ, WH*) 0.01/ / / / B (05BJ, 06BJ # ) C18:0/C18:1 0.01/ / /20.30 D C 1 B C16: D (BJ $ ) C (WH*) B (06BJ # ) C22:0/C24: D C 3 B Oil D (BJ $ ) / C(BJ, WH*) 0.20/ / / / B (05BJ, 06BJ # ) C18:1 2.35/ / /10.29 D / C (BJ, WH*) 0.94/ / / B C16: D (BJ $ ) C 6 B C18:0/C20: D (BJ $ ) C / B (05BJ, 06BJ # ) C20:0 0.05/ / /30.36 D C / B(05BJ, 06BJ # ) C24:0 0.02/ / /24.65 D C / B (05BJ, 06BJ # ) C20:1 0.03/ / /21.18 D C (HN*) / B (05BJ, 06BJ # ) C18:1/C18:2 0.17/ / /39.48 D C / B (05BJ/06BJ # ) C20:0 0.05/ / /29.37 D C
40 Chr. Confidence Interval (Mb) a Population b Trait A c D d R 2 (%) e 8 B Oil / D (BJ, WH $ ) 0.13/ / / C (BJ*) B Oil / D (WH $ ) C 8 B Oil / D(BJ, WH $ ) 0.03/ / /17.06 C 8 B C18:1/C18:2 D / / C (BJ, HN, WH*) 0.03/0.02/ /0.00/ /6.16/ B C18:1 D / / C (BJ, HN, WH*) 1.34/0.94/ /0.15/ /7.54/ B C20:0 D C(WH*) B C18:0 D / / C (BJ, HN, WH*) 0.19/0.12/ /0.11/ /9.94/ / B (05BJ, 06BJ # ) C16:0 1.28/ / / / D (BJ, HN $ ) 1.88/ / / / C (BJ, HN*) 1.56/ / / / B (05BJ, 06BJ # ) SFA/USFA 0.02/ / / / D (BJ,WH $ ) 0.02/ / / / / C (BJ, HN, WH*) 0.02/0.02/ /0.00/ /28.76/ / B (05BJ, 06BJ # ) C16:0 1.28/ / / / / D (BJ, HN, WH $ ) 1.88/0.99/ / 0.08/ /6.41/ / / C (BJ, HN, WH*) 1.56/1.33/ / 0.06/ /31.78/ / B (05BJ, 06BJ # ) C18:2/C18:3 4.60/ / /17.49 D C / B (05BJ, 06BJ # ) Oil 0.31/ / /10.53 D C 10 B C18:0 D C (HN*) ion of the candidate gene refers to version 5b.60 of the maize reference sequence, flanked by the confidence interval of the QTL to the left and right of the ulations B, D and C correspond to the By804 B73 RIL, K22 D340 and CI7 K22 populations, respectively. # Phenotypic data were collected in year 2005 (05) ijing (BJ) for QTL analysis in B population. $ F 2:3 families of the D population were harvested for phenotyping in Beijing 2009, and F 2:4 families were obtained
41 s, Hainan in 2009 and Wuhan in 2010 (HN and WH, respectively). *F2:3 families of the C population were harvested for phenotyping in Beijing, and F2:4 d from Hainan and Wuhan. c Additive effect. d Dominant effect. e The phenotypic variation explained by this QTL.
42 able 9 Correlation analysis between the phenotype and the expression of proposed candidate genes with eqtls Lead trait Oil C16:0 C16:1 C18:0 C18:1 C18:2 C18:3 C20:0 C20:1 C22:0 C24:0 C16:0/C16:1 C16:0/C18:0 C18:0/C18:1 C18:1/C18:2 C18:2/C18:3 C18:0/C20:0 C20:0/C20:1 C20:0/C22:0 C22:0/C24:0 SFA/USFA C20:1 r p < Oil r p C16:1 r p C16:1 r p < < < C20:0 r p Oil r p C18:3 r p < < C18:0/C18:1 r p < < < < < < Oil r p Oil r p C18:0/C18:1 r p Oil r p C16:0 r p C18:0 r p Oil r p < < Oil r p Oil r p Oil r p Oil r p C22:0 r p < < < < < C24:0 r p < < C20:1 r p < < < < < < < C22:0 r p < <
43 Lead trait Oil C16:0 C16:1 C18:0 C18:1 C18:2 C18:3 C20:0 C20:1 C22:0 C24:0 C16:0/C16:1 C16:0/C18:0 C18:0/C18:1 C18:1/C18:2 C18:2/C18:3 C18:0/C20:0 C20:0/C20:1 C20:0/C22:0 C22:0/C24:0 SFA/USFA C20:0 r p C18:0 r p < C20:1 r p Oil r p C20:0 r p C18:0 r p < < C16:0 r p SFA/USFA r p C16:0 r p C18:1/C18:2 r p Oil r p C22:0/C24:0 r p C18:1/C18:2 r p C22:0/C24:0 r p C18:3 r p C16:0/C16:1 r p C18:1/C18:2 r p C22:0/C24:0 r p xpression of candidate gene correlates with target trait at P < 0.01; blue highlighting, expression of candidate gene does not correlate with target trait but with other traits at P <, expression of candidate does not correlate with any trait at P < 0.01.
Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels
 Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels Hui Li 1,, Zhiyu Peng,, Xiaohong Yang 1,, Weidong Wang 1,, Junjie Fu,, Jianhua Wang 1,, Yingjia Han
Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels Hui Li 1,, Zhiyu Peng,, Xiaohong Yang 1,, Weidong Wang 1,, Junjie Fu,, Jianhua Wang 1,, Yingjia Han
Very-Long Chain Fatty Acid Biosynthesis
 Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Very-Long Chain Fatty Acid Biosynthesis
 Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
LIPID METABOLISM
 LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM. Fatty Acid Elongation and Desaturation
 ANSC/NUTR 618 LIPIDS & LIPID METABOLISM I. Fatty acid elongation A. General 1. At least 60% of fatty acids in triacylglycerols are C18. 2. Free palmitic acid (16:0) synthesized in cytoplasm is elongated
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM I. Fatty acid elongation A. General 1. At least 60% of fatty acids in triacylglycerols are C18. 2. Free palmitic acid (16:0) synthesized in cytoplasm is elongated
Synthesis of Fatty Acids and Triacylglycerol
 Synthesis of Fatty Acids and Triacylglycerol Lippincott s Chapter 16 Fatty Acid Synthesis Mainly in the Liver Requires Carbon Source: Acetyl CoA Reducing Power: NADPH 8 CH 3 COO C 15 H 33 COO Energy Input:
Synthesis of Fatty Acids and Triacylglycerol Lippincott s Chapter 16 Fatty Acid Synthesis Mainly in the Liver Requires Carbon Source: Acetyl CoA Reducing Power: NADPH 8 CH 3 COO C 15 H 33 COO Energy Input:
BCM 221 LECTURES OJEMEKELE O.
 BCM 221 LECTURES BY OJEMEKELE O. OUTLINE INTRODUCTION TO LIPID CHEMISTRY STORAGE OF ENERGY IN ADIPOCYTES MOBILIZATION OF ENERGY STORES IN ADIPOCYTES KETONE BODIES AND KETOSIS PYRUVATE DEHYDROGENASE COMPLEX
BCM 221 LECTURES BY OJEMEKELE O. OUTLINE INTRODUCTION TO LIPID CHEMISTRY STORAGE OF ENERGY IN ADIPOCYTES MOBILIZATION OF ENERGY STORES IN ADIPOCYTES KETONE BODIES AND KETOSIS PYRUVATE DEHYDROGENASE COMPLEX
ANSC/NUTR 618 Lipids & Lipid Metabolism
 I. Overall concepts A. Definitions ANC/NUTR 618 Lipids & Lipid Metabolism 1. De novo synthesis = synthesis from non-fatty acid precursors a. Carbohydrate precursors (glucose, lactate, and pyruvate) b.
I. Overall concepts A. Definitions ANC/NUTR 618 Lipids & Lipid Metabolism 1. De novo synthesis = synthesis from non-fatty acid precursors a. Carbohydrate precursors (glucose, lactate, and pyruvate) b.
Synthesis of Fatty Acids and Triacylglycerol
 Fatty Acid Synthesis Synthesis of Fatty Acids and Triacylglycerol Requires Carbon Source: Reducing Power: NADPH Energy Input: ATP Why Energy? Why Energy? Fatty Acid Fatty Acid + n(atp) ΔG o : -ve Fatty
Fatty Acid Synthesis Synthesis of Fatty Acids and Triacylglycerol Requires Carbon Source: Reducing Power: NADPH Energy Input: ATP Why Energy? Why Energy? Fatty Acid Fatty Acid + n(atp) ΔG o : -ve Fatty
Very-Long Chain Fatty Acid Biosynthesis
 Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Very-Long Chain Fatty Acid Biosynthesis
 Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM. Triacylglycerol and Fatty Acid Metabolism
 ANSC/NUTR 618 LIPIDS & LIPID METABOLISM II. Triacylglycerol synthesis A. Overall pathway Glycerol-3-phosphate + 3 Fatty acyl-coa à Triacylglycerol + 3 CoASH B. Enzymes 1. Acyl-CoA synthase 2. Glycerol-phosphate
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM II. Triacylglycerol synthesis A. Overall pathway Glycerol-3-phosphate + 3 Fatty acyl-coa à Triacylglycerol + 3 CoASH B. Enzymes 1. Acyl-CoA synthase 2. Glycerol-phosphate
Leen Alsahele. Razan Al-zoubi ... Faisal
 25 Leen Alsahele Razan Al-zoubi... Faisal last time we started talking about regulation of fatty acid synthesis and degradation *regulation of fatty acid synthesis by: 1- regulation of acetyl CoA carboxylase
25 Leen Alsahele Razan Al-zoubi... Faisal last time we started talking about regulation of fatty acid synthesis and degradation *regulation of fatty acid synthesis by: 1- regulation of acetyl CoA carboxylase
number Done by Corrected by Doctor Faisal Al-Khatibe
 number 24 Done by Mohammed tarabieh Corrected by Doctor Faisal Al-Khatibe 1 P a g e *Please look over the previous sheet about fatty acid synthesis **Oxidation(degradation) of fatty acids, occurs in the
number 24 Done by Mohammed tarabieh Corrected by Doctor Faisal Al-Khatibe 1 P a g e *Please look over the previous sheet about fatty acid synthesis **Oxidation(degradation) of fatty acids, occurs in the
Biochemistry Sheet 27 Fatty Acid Synthesis Dr. Faisal Khatib
 Page1 بسم رلاهللا On Thursday, we discussed the synthesis of fatty acids and its regulation. We also went on to talk about the synthesis of Triacylglycerol (TAG). Last time, we started talking about the
Page1 بسم رلاهللا On Thursday, we discussed the synthesis of fatty acids and its regulation. We also went on to talk about the synthesis of Triacylglycerol (TAG). Last time, we started talking about the
Synthesis and elongation of fatty acids
 Synthesis and elongation of fatty acids A molecular caliper mechanism for determining very long-chain fatty acid length Vladimir Denic and Jonathan S. Weissman (2007) Cell 130, 663-677 February 28, 2008
Synthesis and elongation of fatty acids A molecular caliper mechanism for determining very long-chain fatty acid length Vladimir Denic and Jonathan S. Weissman (2007) Cell 130, 663-677 February 28, 2008
Biosynthesis of Fatty Acids. By Dr.QUTAIBA A. QASIM
 Biosynthesis of Fatty Acids By Dr.QUTAIBA A. QASIM Fatty Acids Definition Fatty acids are comprised of hydrocarbon chains terminating with carboxylic acid groups. Fatty acids and their associated derivatives
Biosynthesis of Fatty Acids By Dr.QUTAIBA A. QASIM Fatty Acids Definition Fatty acids are comprised of hydrocarbon chains terminating with carboxylic acid groups. Fatty acids and their associated derivatives
Synthesis and degradation of fatty acids Martina Srbová
 Synthesis and degradation of fatty acids Martina Srbová martina.srbova@lfmotol.cuni.cz Fatty acids (FA) mostly an even number of carbon atoms and linear chain in esterified form as component of lipids
Synthesis and degradation of fatty acids Martina Srbová martina.srbova@lfmotol.cuni.cz Fatty acids (FA) mostly an even number of carbon atoms and linear chain in esterified form as component of lipids
MPS Advanced Plant Biochemistry Course. Fall Semester Lecture 11. Lipids III
 MPS 587 - Advanced Plant Biochemistry Course Fall Semester 2011 Lecture 11 Lipids III 9. Triacylglycerol synthesis 10. Engineering triacylglycerol fatty acid composition Today s topics on the Arabidopsis
MPS 587 - Advanced Plant Biochemistry Course Fall Semester 2011 Lecture 11 Lipids III 9. Triacylglycerol synthesis 10. Engineering triacylglycerol fatty acid composition Today s topics on the Arabidopsis
Table S1. Trait summary for Northern Leaf Blight resistance in the maize nested association mapping (NAM) population and individual NAM families
 SUPPLEMENTAL MATERIAL Supplemental Tables: Table S1. Trait summary for Northern Leaf Blight resistance in the maize nested association mapping (NAM) population and individual NAM families "#$%&' (')*'%$+,-'
SUPPLEMENTAL MATERIAL Supplemental Tables: Table S1. Trait summary for Northern Leaf Blight resistance in the maize nested association mapping (NAM) population and individual NAM families "#$%&' (')*'%$+,-'
BIOSYNTHESIS OF FATTY ACIDS. doc. Ing. Zenóbia Chavková, CSc.
 BIOSYNTHESIS OF FATTY ACIDS doc. Ing. Zenóbia Chavková, CSc. The pathway for the of FAs is not the reversal of the oxidation pathway Both pathways are separated within different cellular compartments In
BIOSYNTHESIS OF FATTY ACIDS doc. Ing. Zenóbia Chavková, CSc. The pathway for the of FAs is not the reversal of the oxidation pathway Both pathways are separated within different cellular compartments In
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Lipids and Fatty Acids
 Lipids and Fatty Acids Objectives: 1. What are Lipids? properties glycerolipids vs. isoprenoids glycerolipid structure glycerolipid nomenclature 2. Fatty acid biosynthesis ellular localization Substrate
Lipids and Fatty Acids Objectives: 1. What are Lipids? properties glycerolipids vs. isoprenoids glycerolipid structure glycerolipid nomenclature 2. Fatty acid biosynthesis ellular localization Substrate
Fatty acids synthesis
 Fatty acids synthesis The synthesis start from Acetyl COA the first step requires ATP + reducing power NADPH! even though the oxidation and synthesis are different pathways but from chemical part of view
Fatty acids synthesis The synthesis start from Acetyl COA the first step requires ATP + reducing power NADPH! even though the oxidation and synthesis are different pathways but from chemical part of view
Energy storage in cells
 Energy storage in cells Josef Fontana EC - 58 Overview of the lecture Introduction to the storage substances of human body Overview of storage compounds in the body Glycogen metabolism Structure of glycogen
Energy storage in cells Josef Fontana EC - 58 Overview of the lecture Introduction to the storage substances of human body Overview of storage compounds in the body Glycogen metabolism Structure of glycogen
Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.
 Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.32 PCOS locus after conditioning for the lead SNP rs10993397;
Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.32 PCOS locus after conditioning for the lead SNP rs10993397;
6. How Are Fatty Acids Produced? 7. How Are Acylglycerols and Compound Lipids Produced? 8. How Is Cholesterol Produced?
 Lipid Metabolism Learning bjectives 1 How Are Lipids Involved in the Generationand Storage of Energy? 2 How Are Lipids Catabolized? 3 What Is the Energy Yield from the xidation of Fatty Acids? 4 How Are
Lipid Metabolism Learning bjectives 1 How Are Lipids Involved in the Generationand Storage of Energy? 2 How Are Lipids Catabolized? 3 What Is the Energy Yield from the xidation of Fatty Acids? 4 How Are
Anabolism of Fatty acids (Anabolic Lynen spiral) Glycerol and Triglycerides
 Anabolism of Fatty acids (Anabolic Lynen spiral) Glycerol and Triglycerides Anabolism of fatty acids Fatty acids are not stored in the body free. They are a source of energy in the form of triglycerides
Anabolism of Fatty acids (Anabolic Lynen spiral) Glycerol and Triglycerides Anabolism of fatty acids Fatty acids are not stored in the body free. They are a source of energy in the form of triglycerides
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 28 Fatty Acid Synthesis 2013 W. H. Freeman and Company Chapter 28 Outline 1. The first stage of fatty acid synthesis is transfer
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 28 Fatty Acid Synthesis 2013 W. H. Freeman and Company Chapter 28 Outline 1. The first stage of fatty acid synthesis is transfer
FATTY ACID SYNTHESIS
 FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
Biosynthesis of Fatty Acids
 Biosynthesis of Fatty Acids Fatty acid biosynthesis takes place in the cytosol rather than the mitochondria and requires a different activation mechanism and different enzymes and coenzymes than fatty
Biosynthesis of Fatty Acids Fatty acid biosynthesis takes place in the cytosol rather than the mitochondria and requires a different activation mechanism and different enzymes and coenzymes than fatty
Optimising. membranes
 Optimising Neuronal membranes Lipids are classified as 1. Simple lipids oils and fats 2. Complex lipids a) Phospholipids b) Glycosphingolipids containing a fatty acid, sphingosine and a CHO c) Lipoproteins
Optimising Neuronal membranes Lipids are classified as 1. Simple lipids oils and fats 2. Complex lipids a) Phospholipids b) Glycosphingolipids containing a fatty acid, sphingosine and a CHO c) Lipoproteins
Summary of fatty acid synthesis
 Lipid Metabolism, part 2 1 Summary of fatty acid synthesis 8 acetyl CoA + 14 NADPH + 14 H+ + 7 ATP palmitic acid (16:0) + 8 CoA + 14 NADP + + 7 ADP + 7 Pi + 7 H20 1. The major suppliers of NADPH for fatty
Lipid Metabolism, part 2 1 Summary of fatty acid synthesis 8 acetyl CoA + 14 NADPH + 14 H+ + 7 ATP palmitic acid (16:0) + 8 CoA + 14 NADP + + 7 ADP + 7 Pi + 7 H20 1. The major suppliers of NADPH for fatty
Dietary α-linolenic acid-rich flaxseed oil prevents against alcoholic hepatic steatosis
 Dietary α-linolenic acid-rich flaxseed oil prevents against alcoholic hepatic steatosis via ameliorating lipid homeostasis at adipose tissue-liver axis in mice Meng Wang a, Xiao-Jing Zhang a, Kun Feng
Dietary α-linolenic acid-rich flaxseed oil prevents against alcoholic hepatic steatosis via ameliorating lipid homeostasis at adipose tissue-liver axis in mice Meng Wang a, Xiao-Jing Zhang a, Kun Feng
Figure S2. Distribution of acgh probes on all ten chromosomes of the RIL M0022
 96 APPENDIX B. Supporting Information for chapter 4 "changes in genome content generated via segregation of non-allelic homologs" Figure S1. Potential de novo CNV probes and sizes of apparently de novo
96 APPENDIX B. Supporting Information for chapter 4 "changes in genome content generated via segregation of non-allelic homologs" Figure S1. Potential de novo CNV probes and sizes of apparently de novo
LIPID METABOLISM. Sri Widia A Jusman Department of Biochemistry & Molecular Biology FMUI
 LIPID METABOLISM Sri Widia A Jusman Department of Biochemistry & Molecular Biology FMUI Lipid metabolism is concerned mainly with fatty acids cholesterol Source of fatty acids from dietary fat de novo
LIPID METABOLISM Sri Widia A Jusman Department of Biochemistry & Molecular Biology FMUI Lipid metabolism is concerned mainly with fatty acids cholesterol Source of fatty acids from dietary fat de novo
Application of NIR spectroscopy for seed composition improvement in soybean. Jason D. Gillman USDA-ARS/PGRU
 Application of NIR spectroscopy for seed composition improvement in soybean Jason D. Gillman USDA-ARS/PGRU 2-14-2017 Soybean fatty acid profile P o l y - u n s a t u SATURATED FATS Mono-unsaturated 16:0
Application of NIR spectroscopy for seed composition improvement in soybean Jason D. Gillman USDA-ARS/PGRU 2-14-2017 Soybean fatty acid profile P o l y - u n s a t u SATURATED FATS Mono-unsaturated 16:0
IDENTIFICATION OF QTLS FOR STARCH CONTENT IN SWEETPOTATO (IPOMOEA BATATAS (L.) LAM.)
 Journal of Integrative Agriculture Advanced Online Publication: 2013 Doi: 10.1016/S2095-3119(13)60357-3 IDENTIFICATION OF QTLS FOR STARCH CONTENT IN SWEETPOTATO (IPOMOEA BATATAS (L.) LAM.) YU Xiao-xia
Journal of Integrative Agriculture Advanced Online Publication: 2013 Doi: 10.1016/S2095-3119(13)60357-3 IDENTIFICATION OF QTLS FOR STARCH CONTENT IN SWEETPOTATO (IPOMOEA BATATAS (L.) LAM.) YU Xiao-xia
MILK BIOSYNTHESIS PART 3: FAT
 MILK BIOSYNTHESIS PART 3: FAT KEY ENZYMES (FROM ALL BIOSYNTHESIS LECTURES) FDPase = fructose diphosphatase Citrate lyase Isocitrate dehydrogenase Fatty acid synthetase Acetyl CoA carboxylase Fatty acyl
MILK BIOSYNTHESIS PART 3: FAT KEY ENZYMES (FROM ALL BIOSYNTHESIS LECTURES) FDPase = fructose diphosphatase Citrate lyase Isocitrate dehydrogenase Fatty acid synthetase Acetyl CoA carboxylase Fatty acyl
GENERAL FEATURES OF FATTY ACIDS BIOSYNTHESIS
 1 GENERAL FEATURES OF FATTY ACIDS BIOSYNTHESIS 1. Fatty acids may be synthesized from dietary glucose via pyruvate. 2. Fatty acids are the preferred fuel source for the heart and the primary form in which
1 GENERAL FEATURES OF FATTY ACIDS BIOSYNTHESIS 1. Fatty acids may be synthesized from dietary glucose via pyruvate. 2. Fatty acids are the preferred fuel source for the heart and the primary form in which
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies
 Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Roles of Lipids. principal form of stored energy major constituents of cell membranes vitamins messengers intra and extracellular
 Roles of Lipids principal form of stored energy major constituents of cell membranes vitamins messengers intra and extracellular = Oxidation of fatty acids Central energy-yielding pathway in animals. O
Roles of Lipids principal form of stored energy major constituents of cell membranes vitamins messengers intra and extracellular = Oxidation of fatty acids Central energy-yielding pathway in animals. O
Huang et al. BMC Plant Biology (2018) 18:140
 Huang et al. BMC Plant Biology (2018) 18:140 https://doi.org/10.1186/s12870-018-1356-8 RESEARCH ARTICLE Temporal transcriptome profiling of developing seeds reveals a concerted gene regulation in relation
Huang et al. BMC Plant Biology (2018) 18:140 https://doi.org/10.1186/s12870-018-1356-8 RESEARCH ARTICLE Temporal transcriptome profiling of developing seeds reveals a concerted gene regulation in relation
26.1 Acetyl Coenzyme A
 Chapter 26 Lipids Lipids Lipids are naturally occurring substances grouped together on the basis of a common property they they are more soluble in nonpolar solvents than in water. Some of the most important
Chapter 26 Lipids Lipids Lipids are naturally occurring substances grouped together on the basis of a common property they they are more soluble in nonpolar solvents than in water. Some of the most important
Supplemental Information. Genetic Regulation of Plasma Lipid Species. and Their Association with Metabolic Phenotypes
 Cell Systems, Volume 6 Supplemental Information Genetic Regulation of Plasma Lipid Species and Their Association with Metabolic Phenotypes Pooja Jha, Molly T. McDevitt, Emina Halilbasic, Evan G. Williams,
Cell Systems, Volume 6 Supplemental Information Genetic Regulation of Plasma Lipid Species and Their Association with Metabolic Phenotypes Pooja Jha, Molly T. McDevitt, Emina Halilbasic, Evan G. Williams,
Lipid metabolism. Degradation and biosynthesis of fatty acids Ketone bodies
 Lipid metabolism Degradation and biosynthesis of fatty acids Ketone bodies Fatty acids (FA) primary fuel molecules in the fat category main use is for long-term energy storage high level of energy storage:
Lipid metabolism Degradation and biosynthesis of fatty acids Ketone bodies Fatty acids (FA) primary fuel molecules in the fat category main use is for long-term energy storage high level of energy storage:
Fatty Acid and Triacylglycerol Metabolism 1
 Fatty Acid and Triacylglycerol Metabolism 1 Mobilization of stored fats and oxidation of fatty acids Lippincott s Chapter 16 What is the first lecture about What is triacylglycerol Fatty acids structure
Fatty Acid and Triacylglycerol Metabolism 1 Mobilization of stored fats and oxidation of fatty acids Lippincott s Chapter 16 What is the first lecture about What is triacylglycerol Fatty acids structure
Lecture: 26 OXIDATION OF FATTY ACIDS
 Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Chemistry 107 Exam 4 Study Guide
 Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
f(x) = x R² = RPKM (M8.MXB) f(x) = x E-014 R² = 1 RPKM (M31.
 14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
Supplementary Figure S1A
 Supplementary Figure S1A-G. LocusZoom regional association plots for the seven new cross-cancer loci that were > 1 Mb from known index SNPs. Genes up to 500 kb on either side of each new index SNP are
Supplementary Figure S1A-G. LocusZoom regional association plots for the seven new cross-cancer loci that were > 1 Mb from known index SNPs. Genes up to 500 kb on either side of each new index SNP are
number Done by Corrected by Doctor F. Al-Khateeb
 number 23 Done by A. Rawajbeh Corrected by Doctor F. Al-Khateeb Ketone bodies Ketone bodies are used by the peripheral tissues like the skeletal and cardiac muscles, where they are the preferred source
number 23 Done by A. Rawajbeh Corrected by Doctor F. Al-Khateeb Ketone bodies Ketone bodies are used by the peripheral tissues like the skeletal and cardiac muscles, where they are the preferred source
Determination of genetic effects of ATF3 and CDKN1A genes on milk yield and compositions in Chinese Holstein population
 Han et al. BMC Genetics (2017) 18:47 DOI 10.1186/s12863-017-0516-4 RESEARCH ARTICLE Open Access Determination of genetic effects of ATF3 and CDKN1A genes on milk yield and compositions in Chinese Holstein
Han et al. BMC Genetics (2017) 18:47 DOI 10.1186/s12863-017-0516-4 RESEARCH ARTICLE Open Access Determination of genetic effects of ATF3 and CDKN1A genes on milk yield and compositions in Chinese Holstein
Lecture 16. Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III
 Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
A REVIEW ON BIOCHEMICAL MECHANISM OF FATTY ACIDS SYNTHESIS AND OIL DEPOSITION IN BRASSICA AND ARABIDOPSIS
 American Journal of Agricultural and Biological Sciences 9 (4): 534-545, 2014 ISSN: 1557-4989 2014 M. Rahman, This open access article is distributed under a Creative Commons Attribution (CC-BY) 3.0 license
American Journal of Agricultural and Biological Sciences 9 (4): 534-545, 2014 ISSN: 1557-4989 2014 M. Rahman, This open access article is distributed under a Creative Commons Attribution (CC-BY) 3.0 license
Hand in the Test Sheets (with the checked multiple choice answers) and your Sheets with written answers.
 Page 1 of 13 IMPORTANT INFORMATION Hand in the Test Sheets (with the checked multiple choice answers) and your Sheets with written answers. THE exam has an 'A' and 'B' section SECTION A (based on Dykyy's
Page 1 of 13 IMPORTANT INFORMATION Hand in the Test Sheets (with the checked multiple choice answers) and your Sheets with written answers. THE exam has an 'A' and 'B' section SECTION A (based on Dykyy's
Increasing erucic acid content in the seed oil of rapeseed (Brassica napus L.) by combining selection for natural variation and transgenic approaches
 Increasing erucic acid content in the seed oil of rapeseed (Brassica napus L.) by combining selection for natural variation and transgenic approaches Dissertation to obtain the Ph. D. degree in the Faculty
Increasing erucic acid content in the seed oil of rapeseed (Brassica napus L.) by combining selection for natural variation and transgenic approaches Dissertation to obtain the Ph. D. degree in the Faculty
Cong Li 1, Dongxiao Sun 1*, Shengli Zhang 1*, Shaohua Yang 1, M. A. Alim 1, Qin Zhang 1, Yanhua Li 2 and Lin Liu 2
 Li et al. BMC Genetics (2016) 17:110 DOI 10.1186/s12863-016-0418-x RESEARCH ARTICLE Open Access Genetic effects of FASN, PPARGC1A, ABCG2 and IGF1 revealing the association with milk fatty acids in a Chinese
Li et al. BMC Genetics (2016) 17:110 DOI 10.1186/s12863-016-0418-x RESEARCH ARTICLE Open Access Genetic effects of FASN, PPARGC1A, ABCG2 and IGF1 revealing the association with milk fatty acids in a Chinese
Clinical Genetics. Functional validation in a diagnostic context. Robert Hofstra. Leading the way in genetic issues
 Clinical Genetics Leading the way in genetic issues Functional validation in a diagnostic context Robert Hofstra Clinical Genetics Leading the way in genetic issues Future application of exome sequencing
Clinical Genetics Leading the way in genetic issues Functional validation in a diagnostic context Robert Hofstra Clinical Genetics Leading the way in genetic issues Future application of exome sequencing
Supplementary Figures
 Supplementary Figures Supplementary Fig 1. Comparison of sub-samples on the first two principal components of genetic variation. TheBritishsampleisplottedwithredpoints.The sub-samples of the diverse sample
Supplementary Figures Supplementary Fig 1. Comparison of sub-samples on the first two principal components of genetic variation. TheBritishsampleisplottedwithredpoints.The sub-samples of the diverse sample
Tala Saleh. Razi Kittaneh ... Nayef Karadsheh
 Tala Saleh Razi Kittaneh... Nayef Karadsheh β-oxidation of Fatty Acids The oxidation of fatty acids occurs in 3 steps: Step 1: Activation of the Fatty acid FA + HS-CoA + ATP FA-CoA + AMP + PPi - The fatty
Tala Saleh Razi Kittaneh... Nayef Karadsheh β-oxidation of Fatty Acids The oxidation of fatty acids occurs in 3 steps: Step 1: Activation of the Fatty acid FA + HS-CoA + ATP FA-CoA + AMP + PPi - The fatty
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Replicability of blood eqtl effects in ileal biopsies from the RISK study. eqtls detected in the vicinity of SNPs associated with IBD tend to show concordant effect size and direction
Supplementary Figure 1 Replicability of blood eqtl effects in ileal biopsies from the RISK study. eqtls detected in the vicinity of SNPs associated with IBD tend to show concordant effect size and direction
Supplementary Information. Supplementary Figures
 Supplementary Information Supplementary Figures.8 57 essential gene density 2 1.5 LTR insert frequency diversity DEL.5 DUP.5 INV.5 TRA 1 2 3 4 5 1 2 3 4 1 2 Supplementary Figure 1. Locations and minor
Supplementary Information Supplementary Figures.8 57 essential gene density 2 1.5 LTR insert frequency diversity DEL.5 DUP.5 INV.5 TRA 1 2 3 4 5 1 2 3 4 1 2 Supplementary Figure 1. Locations and minor
Algal Biofuels Research: Using basic science to maximize fuel output. Jacob Dums, PhD candidate, Heike Sederoff Lab March 9, 2015
 Algal Biofuels Research: Using basic science to maximize fuel output Jacob Dums, PhD candidate, jtdums@ncsu.edu Heike Sederoff Lab March 9, 2015 Outline Research Approach Dunaliella Increase Oil Content
Algal Biofuels Research: Using basic science to maximize fuel output Jacob Dums, PhD candidate, jtdums@ncsu.edu Heike Sederoff Lab March 9, 2015 Outline Research Approach Dunaliella Increase Oil Content
Fatty acid breakdown
 Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
Biosynthesis of Triacylglycerides (TG) in liver. Mobilization of stored fat and oxidation of fatty acids
 Biosynthesis of Triacylglycerides (TG) in liver Mobilization of stored fat and oxidation of fatty acids Activation of hormone sensitive lipase This enzyme is activated when phosphorylated (3,5 cyclic AMPdependent
Biosynthesis of Triacylglycerides (TG) in liver Mobilization of stored fat and oxidation of fatty acids Activation of hormone sensitive lipase This enzyme is activated when phosphorylated (3,5 cyclic AMPdependent
Fatty Acid and Triacylglycerol Metabolism 1
 Fatty Acid and Triacylglycerol Metabolism 1 Mobilization of stored fats and oxidation of fatty acids Lippincott s Chapter 16 What is the first lecture about What is triacylglycerol Fatty acids structure
Fatty Acid and Triacylglycerol Metabolism 1 Mobilization of stored fats and oxidation of fatty acids Lippincott s Chapter 16 What is the first lecture about What is triacylglycerol Fatty acids structure
indicated in shaded lowercase letters (hg19, Chr2: 217,955, ,957,266).
 Legend for Supplementary Figures Figure S1: Sequence of 2q35 encnv. The DNA sequence of the 1,375bp 2q35 encnv is indicated in shaded lowercase letters (hg19, Chr2: 217,955,892-217,957,266). Figure S2:
Legend for Supplementary Figures Figure S1: Sequence of 2q35 encnv. The DNA sequence of the 1,375bp 2q35 encnv is indicated in shaded lowercase letters (hg19, Chr2: 217,955,892-217,957,266). Figure S2:
Complex Trait Genetics in Animal Models. Will Valdar Oxford University
 Complex Trait Genetics in Animal Models Will Valdar Oxford University Mapping Genes for Quantitative Traits in Outbred Mice Will Valdar Oxford University What s so great about mice? Share ~99% of genes
Complex Trait Genetics in Animal Models Will Valdar Oxford University Mapping Genes for Quantitative Traits in Outbred Mice Will Valdar Oxford University What s so great about mice? Share ~99% of genes
Fatty acid synthesis. Dr. Nalini Ganesan M.Sc., Ph.D Associate Professor Department of Biochemistry SRMC & RI (DU) Porur, Chennai - 116
 Fatty acid synthesis Dr. Nalini Ganesan M.Sc., Ph.D Associate Professor Department of Biochemistry SRMC & RI (DU) Porur, Chennai 116 Harper s biochemistry 24 th ed, Pg 218 Fatty acid Synthesis Known as
Fatty acid synthesis Dr. Nalini Ganesan M.Sc., Ph.D Associate Professor Department of Biochemistry SRMC & RI (DU) Porur, Chennai 116 Harper s biochemistry 24 th ed, Pg 218 Fatty acid Synthesis Known as
SUPPLEMENTARY DATA. 1. Characteristics of individual studies
 1. Characteristics of individual studies 1.1. RISC (Relationship between Insulin Sensitivity and Cardiovascular disease) The RISC study is based on unrelated individuals of European descent, aged 30 60
1. Characteristics of individual studies 1.1. RISC (Relationship between Insulin Sensitivity and Cardiovascular disease) The RISC study is based on unrelated individuals of European descent, aged 30 60
Summary of Endomembrane-system
 Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
MITOCW watch?v=ddt1kusdoog
 MITOCW watch?v=ddt1kusdoog The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high-quality educational resources for free. To
MITOCW watch?v=ddt1kusdoog The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high-quality educational resources for free. To
Supplemental Figure 1. Genes showing ectopic H3K9 dimethylation in this study are DNA hypermethylated in Lister et al. study.
 mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
A genome-wide association study of seed composition traits in wild soybean (Glycine soja)
 Leamy et al. BMC Genomics (2017) 18:18 DOI 10.1186/s12864-016-3397-4 RESEARCH ARTICLE A genome-wide association study of seed composition traits in wild soybean (Glycine soja) Larry J. Leamy 1, Hengyou
Leamy et al. BMC Genomics (2017) 18:18 DOI 10.1186/s12864-016-3397-4 RESEARCH ARTICLE A genome-wide association study of seed composition traits in wild soybean (Glycine soja) Larry J. Leamy 1, Hengyou
BIOLOGY 103 Spring 2001 MIDTERM LAB SECTION
 BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
Lipid metabolism I Triacylglycerols
 Lipid metabolism I Triacylglycerols Biochemistry I Lecture 8 2008 (J.S.) Major classes of lipids Simple lipids Triacylglycerols serve as energy-providing nutrients, the turnover about 100 g per day in
Lipid metabolism I Triacylglycerols Biochemistry I Lecture 8 2008 (J.S.) Major classes of lipids Simple lipids Triacylglycerols serve as energy-providing nutrients, the turnover about 100 g per day in
MALDI Activity 4 MALDI-TOF Mass Spectrometry: Data Analysis
 MALDI Activity 4 MALDI-TOF Mass Spectrometry: Data Analysis Model 1: Introduction to Triacylglycerides (TAGs) In MALDI Activity 3 you learned how to open your raw mass spectrum data and use the MMass program
MALDI Activity 4 MALDI-TOF Mass Spectrometry: Data Analysis Model 1: Introduction to Triacylglycerides (TAGs) In MALDI Activity 3 you learned how to open your raw mass spectrum data and use the MMass program
CHEM/MBIO 2370 Biochemistry 2: Catabolism, Synthesis and Information Pathways--Syllabus
 An introductory course dealing with the basic metabolic processes that occur in living cells including the production and use of metabolic energy, the breakdown and synthesis of biomolecules, the synthesis
An introductory course dealing with the basic metabolic processes that occur in living cells including the production and use of metabolic energy, the breakdown and synthesis of biomolecules, the synthesis
Skeletal muscle metabolism was studied by measuring arterio-venous concentration differences
 Supplemental Data Dual stable-isotope experiment Skeletal muscle metabolism was studied by measuring arterio-venous concentration differences across the forearm, adjusted for forearm blood flow (FBF) (1).
Supplemental Data Dual stable-isotope experiment Skeletal muscle metabolism was studied by measuring arterio-venous concentration differences across the forearm, adjusted for forearm blood flow (FBF) (1).
Integration & Hormone Regulation
 Integration Branchpoints in metabolism where metabolites can go several directions 1. Glucose 6-phosphate Energy needed (low energy charge): glycolysis Low blood sugar: high [glucagon], low [insulin] glycogen
Integration Branchpoints in metabolism where metabolites can go several directions 1. Glucose 6-phosphate Energy needed (low energy charge): glycolysis Low blood sugar: high [glucagon], low [insulin] glycogen
Genetic regulation of bovine milk fatty acid composition: Improving the healthfulness of milk through selection
 Graduate Theses and Dissertations Graduate College 2010 Genetic regulation of bovine milk fatty acid composition: Improving the healthfulness of milk through selection Rafael Nafikov Iowa State University
Graduate Theses and Dissertations Graduate College 2010 Genetic regulation of bovine milk fatty acid composition: Improving the healthfulness of milk through selection Rafael Nafikov Iowa State University
Gene Polymorphisms and Carbohydrate Diets. James M. Ntambi Ph.D
 Gene Polymorphisms and Carbohydrate Diets James M. Ntambi Ph.D Fatty Acids that Flux into Tissue Lipids are from Dietary Sources or are Made De novo from Glucose or Fructose Glucose Fructose Acetyl-CoA
Gene Polymorphisms and Carbohydrate Diets James M. Ntambi Ph.D Fatty Acids that Flux into Tissue Lipids are from Dietary Sources or are Made De novo from Glucose or Fructose Glucose Fructose Acetyl-CoA
Lecture 36. Key Concepts. Overview of lipid metabolism. Reactions of fatty acid oxidation. Energy yield from fatty acid oxidation
 Lecture 36 Lipid Metabolism 1 Fatty Acid Oxidation Ketone Bodies Key Concepts Overview of lipid metabolism Reactions of fatty acid oxidation Energy yield from fatty acid oxidation Formation of ketone bodies
Lecture 36 Lipid Metabolism 1 Fatty Acid Oxidation Ketone Bodies Key Concepts Overview of lipid metabolism Reactions of fatty acid oxidation Energy yield from fatty acid oxidation Formation of ketone bodies
Fats and Lipids (Ans570)
 Fats and Lipids (Ans570) Outlines Fats and Lipids Structure, nomenclature Phospholipids, Sterols, and Lipid Derivatives Lipid Oxidation Roles of fat in food processing and dietary fat Lipid and fat analysis:
Fats and Lipids (Ans570) Outlines Fats and Lipids Structure, nomenclature Phospholipids, Sterols, and Lipid Derivatives Lipid Oxidation Roles of fat in food processing and dietary fat Lipid and fat analysis:
23.1 Lipid Metabolism in Animals. Chapter 23. Micelles Lipid Metabolism in. Animals. Overview of Digestion Lipid Metabolism in
 Denniston Topping Caret Copyright! The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Chapter 23 Fatty Acid Metabolism Triglycerides (Tgl) are emulsified into fat droplets
Denniston Topping Caret Copyright! The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Chapter 23 Fatty Acid Metabolism Triglycerides (Tgl) are emulsified into fat droplets
Lecture 16. Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III
 Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
ANSC/NUTR 618 Lipids & Lipid Metabolism
 I. verall concepts A. Definitions ANSC/NUTR 618 Lipids & Lipid Metabolism 1. De novo synthesis = synthesis from non-fatty acid precursors a. Carbohydrate precursors (glucose and lactate) 1) Uses glucose
I. verall concepts A. Definitions ANSC/NUTR 618 Lipids & Lipid Metabolism 1. De novo synthesis = synthesis from non-fatty acid precursors a. Carbohydrate precursors (glucose and lactate) 1) Uses glucose
Acetyl-CoA or BC acyl-coa. Int1-ACP. FabH. FabF. Anti-FabF Platensimycin ACP. Fak
 Initiation Acetyl-CoA AccBCDA FabZ Int-ACP Elongation [N+] FabG Int1-ACP Acetyl-CoA or BC acyl-coa FabH FabD CoA Malonyl-ACP Malonyl CoA ACP Anti-FabI Triclosan AFN-15 FabI acyl-acp PlsX PlsY Pls C Lipoic
Initiation Acetyl-CoA AccBCDA FabZ Int-ACP Elongation [N+] FabG Int1-ACP Acetyl-CoA or BC acyl-coa FabH FabD CoA Malonyl-ACP Malonyl CoA ACP Anti-FabI Triclosan AFN-15 FabI acyl-acp PlsX PlsY Pls C Lipoic
Fatty Acid Desaturation
 Fatty Acid Desaturation Objectives: 1. Isolation of desaturase mutants 2. Substrates for fatty acid desaturation 3. ellular localization of desaturases References: Buchanan et al. 2000. Biochemistry and
Fatty Acid Desaturation Objectives: 1. Isolation of desaturase mutants 2. Substrates for fatty acid desaturation 3. ellular localization of desaturases References: Buchanan et al. 2000. Biochemistry and
Fatty Acids, lipolysis and goat flavour (1) Specificities of lipid metabolim in goats :
 Specificities of lipid metabolim in goats : Yves Chilliard INRA Clermont-Ferrand / Theix France - milk FA profile (high C8 & C1, and B-CFA) - lipolytic system (LPL regulations; flavour) (e.g. Chilliard
Specificities of lipid metabolim in goats : Yves Chilliard INRA Clermont-Ferrand / Theix France - milk FA profile (high C8 & C1, and B-CFA) - lipolytic system (LPL regulations; flavour) (e.g. Chilliard
Supplemental data. Uppalapati et al. (2012) Plant Cell /tpc
 PAL Control P. emaculata 0 8 24 48 8 24 48 OPR Control P. emaculata 0 8 24 48 8 24 48 CHS PR3 CHR PR5 CHI PR10 IFS IFR Supplemental Figure 1. Expression profiles for selected genes in phenylpropanoid pathway
PAL Control P. emaculata 0 8 24 48 8 24 48 OPR Control P. emaculata 0 8 24 48 8 24 48 CHS PR3 CHR PR5 CHI PR10 IFS IFR Supplemental Figure 1. Expression profiles for selected genes in phenylpropanoid pathway
7/11/17. Cell Function & Chemistry. Molecular and Cellular Biology. 2. Bio-Chemical Foundations & Key Molecules of a Cell
 Molecular and Cellular Biology Cell Function & Chemistry 2. Bio-Chemical Foundations & Key Molecules of a Cell Prof. Dr. Klaus Heese Interaction Molecular Bonds Define Cellular Functions Water H 2 O Interactions
Molecular and Cellular Biology Cell Function & Chemistry 2. Bio-Chemical Foundations & Key Molecules of a Cell Prof. Dr. Klaus Heese Interaction Molecular Bonds Define Cellular Functions Water H 2 O Interactions
Dietary Lipid Metabolism
 Dietary Lipid Metabolism Presented by Dr. Mohammad Saadeh The requirements for the Pharmaceutical Biochemistry II Philadelphia University Faculty of pharmacy OVERVIEW Lipids are a heterogeneous group.
Dietary Lipid Metabolism Presented by Dr. Mohammad Saadeh The requirements for the Pharmaceutical Biochemistry II Philadelphia University Faculty of pharmacy OVERVIEW Lipids are a heterogeneous group.
Supplementary information
 Supplementary information Hepatitis B virus genotype, mutations, human leukocyte antigen polymorphisms and their interactions in hepatocellular carcinoma: a multi-centre case-control study Juan Wen, Ci
Supplementary information Hepatitis B virus genotype, mutations, human leukocyte antigen polymorphisms and their interactions in hepatocellular carcinoma: a multi-centre case-control study Juan Wen, Ci
MCQS ON LIPIDS. Dr. RUCHIKA YADU
 MCQS ON LIPIDS Dr. RUCHIKA YADU Q1. THE FATS AND OILS ARE RESPECTIVELY RICH IN a) Unsaturated fatty acids b) Saturated fatty acids c) Saturated and unsaturated fatty acids d) None of these Q2. ESSENTIAL
MCQS ON LIPIDS Dr. RUCHIKA YADU Q1. THE FATS AND OILS ARE RESPECTIVELY RICH IN a) Unsaturated fatty acids b) Saturated fatty acids c) Saturated and unsaturated fatty acids d) None of these Q2. ESSENTIAL
Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54
 CORRECTION NOTICE Nat. Genet. 42, 759 763 (2010); published online 22 August 2010; corrected online 27 August 2014 Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects
CORRECTION NOTICE Nat. Genet. 42, 759 763 (2010); published online 22 August 2010; corrected online 27 August 2014 Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects
A total of 2,822 Mexican dyslipidemic cases and controls were recruited at INCMNSZ in
 Supplemental Material The N342S MYLIP polymorphism is associated with high total cholesterol and increased LDL-receptor degradation in humans by Daphna Weissglas-Volkov et al. Supplementary Methods Mexican
Supplemental Material The N342S MYLIP polymorphism is associated with high total cholesterol and increased LDL-receptor degradation in humans by Daphna Weissglas-Volkov et al. Supplementary Methods Mexican
Journal of Chemical and Pharmaceutical Research, 2013, 5(12): Research Article
 Available online www.jocpr.com Journal of Chemical and Pharmaceutical Research, 2013, 5(12):1252-1257 Research Article ISSN : 0975-7384 CODEN(USA) : JCPRC5 An analysis and research on biosynthesis pathway
Available online www.jocpr.com Journal of Chemical and Pharmaceutical Research, 2013, 5(12):1252-1257 Research Article ISSN : 0975-7384 CODEN(USA) : JCPRC5 An analysis and research on biosynthesis pathway
