Abstract. Introduction
|
|
|
- Berenice Daniels
- 6 years ago
- Views:
Transcription
1 Activation of Cholesterol Synthesis in Preference to Fatty Acid Synthesis in Liver and Adipose Tissue of Transgenic Mice Overproducing Sterol Regulatory Element-binding Protein-2 Jay D. Horton,* Iichiro Shimomura,* Michael S. Brown,* Robert E. Hammer, Joseph L. Goldstein,* and Hitoshi Shimano* *Department of Molecular Genetics, Department of Biochemistry, and Howard Hughes Medical Institute, University of Texas Southwestern Medical Center, Dallas, Texas Abstract We produced transgenic mice that express a dominant-positive truncated form of sterol regulatory element-binding protein-2 (SREBP-2) in liver and adipose tissue. The encoded protein lacks the membrane-binding and COOH-terminal regulatory domains, and it is therefore not susceptible to negative regulation by cholesterol. Livers from the transgenic mice showed increases in mrnas encoding multiple enzymes of cholesterol biosynthesis, the LDL receptor, and fatty acid biosynthesis. The elevations in mrna for 3-hydroxy-3-methylglutaryl coenzyme A (HMG CoA) synthase and HMG CoA reductase were especially marked (13-fold and 75-fold, respectively). As a result, the transgenic livers showed a 28-fold increase in the rate of cholesterol synthesis and a lesser fourfold increase in fatty acid synthesis, as measured by intraperitoneal injection of [ 3 H]water. These results contrast with previously reported effects of dominant-positive SREBP-1a, which activated fatty acid synthesis more than cholesterol synthesis. In adipose tissue of the SREBP-2 transgenics, the mrnas for cholesterol biosynthetic enzymes were elevated, but the mrnas for fatty acid biosynthetic enzymes were not. We conclude that SREBP-2 is a relatively selective activator of cholesterol synthesis, as opposed to fatty acid synthesis, in liver and adipose tissue of mice. (J. Clin. Invest : ) Key words: cholesterol low density lipoprotein sterol regulatory element binding proteins fatty acids transgenic mice Introduction Cholesterol and fatty acids, the primary lipids synthesized in liver, are subject to different patterns of regulation (reviewed in references 1 and 2). Cholesterol synthesis is high when animals are fed a cholesterol-free diet, and it declines dramatically when cholesterol is fed. Cholesterol synthesis is also enhanced when the demand for cholesterol is increased, as when Address correspondence to Joseph L. Goldstein, Department of Molecular Genetics, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Room L5-238, Dallas, TX Phone: ; FAX: Received for publication 2 February 1998 and accepted in revised form 17 March J. Clin. Invest. The American Society for Clinical Investigation, Inc /98/06/2331/09 $2.00 Volume 101, Number 11, June 1998, animals consume bile acid binding resins. In contrast, fatty acid synthesis is driven primarily by the availability of carbohydrate and the stimulating action of insulin. Despite these different patterns of regulation, recent evidence suggests that both biosynthetic pathways can be regulated by a common family of transcription factors designated sterol regulatory element binding proteins (SREBPs) 1 (reviewed in reference 3). SREBPs are members of a broad class of transcription activators that use basic-helix-loop-helix-leucine zipper (bhlh-zip) motifs to dimerize and to bind DNA. SREBPs are unique in that they are synthesized as membrane-bound precursors that must be liberated by proteolysis in order to enter the nucleus (3). They are also unique in having a tyrosine in place of an otherwise conserved arginine, which allows them to bind to nonpalindromic sterol regulatory elements in preference to classic palindromic E boxes (4). The membrane-bound precursor forms of SREBPs contain 1,150 amino acids. The NH 2 -terminal segment of 480 amino acids includes the bhlh-zip domain and an acidic transcription activating domain at the extreme NH 2 terminus. The bhlh-zip domain is followed by a membrane attachment domain of 80 amino acids that consists of two membrane-spanning segments separated by a short hydrophilic loop. This is followed by a COOH-terminal regulatory domain of 590 amino acids. The SREBPs are oriented so that their NH 2 -terminal and COOH-terminal domains project into the cytoplasm, and the 31-amino acid loop projects into the lumen of the endoplasmic reticulum (3). The mechanism for regulation of SREBP activity was elucidated through studies of cultured fibroblasts. When such cells are deprived of sterols, a two-step proteolytic process releases the NH 2 -terminal domains of the SREBPs so that they can enter the nucleus (3). There they bind to sterol regulatory elements in the promoter regions of genes encoding enzymes of cholesterol biosynthesis (HMG CoA reductase, HMG CoA synthase, farnesyl diphosphate synthase, squalene synthase, and others) (5 8). The SREBPs also bind to regulatory sequences in the promoters of at least two genes involved in fatty acid synthesis, acetyl CoA carboxylase and fatty acid synthase (4, 9, 10), and at least one gene involved in triglyceride synthesis, glycerol-3-phosphate acyltransferase (11). Binding to all these promoters leads to transcriptional activation, which results in increased synthesis of cholesterol and fatty acids. When sterols overaccumulate in cells, the proteolytic release 1. Abbreviations used in this paper: bhlh-zip, basic-helix-loophelix-leucine zipper; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HMG CoA, 3-hydroxy-3-methylglutaryl coenzyme A; PEPCK, phosphoenolpyruvate carboxykinase; PPAR-, peroxisomal proliferator-activated receptor; SREBP, sterol regulatory element-binding protein. Transgenic Mice Expressing Truncated SREBP
2 of SREBPs is blocked, the NH 2 -terminal fragments remain bound to membranes, and transcription of all target genes declines (3). Three SREBPs are currently recognized (3). Two of these, designated SREBP-1a and SREBP-1c, are produced from a single gene through the use of alternate promoters (3, 12). SREBP-1c was cloned independently as a protein that binds to E-boxes and was named ADD-1 (13). SREBP-1a and -1c differ in the length of the first exon, which encodes a portion of the acidic transcription activating domain. This domain is much longer in SREBP-1a than it is in SREBP-1c (12). The other member of the family, designated SREBP-2, has a long transcription activation domain resembling that of SREBP-1a (6). Livers of mice, humans, and hamsters produce relatively large amounts of SREBP-1c and smaller amounts of SREBP-2 and -1a (12, 14). The reason for the three isoforms of SREBP is not yet known. The proteins appear to act primarily as homodimers, and heterodimerization is not required for their activity (reference 6 and Pai, J.-T., M.S. Brown, and J.L. Goldstein, unpublished observations). All three SREBPs are capable of activating all of the known target genes, but they do so with different efficiencies. In general, SREBP-1a and SREBP-2 are much more active than SREBP-1c (reference 15 and Pai, J.-T., M.S. Brown, and J.L. Goldstein, unpublished observations). In an attempt to decipher the roles of individual SREBPs in liver, we have produced transgenic mice that overexpress truncated dominant-positive forms of SREBP-1a (16) or SREBP-1c (15). The encoded proteins terminate at a point between the bhlh-zip domain and the membrane-attachment domain. Such proteins are never membrane bound, and they enter the nucleus directly without a requirement for proteolysis. As a result, these proteins are not down-regulated when cholesterol accumulates. Transgene expression is driven by the promoter for phosphoenolpyruvate carboxykinase (PEPCK), which is expressed in liver, adipose tissue, and kidney. Expression is induced to high levels in the liver when the mice consume a low carbohydrate/high protein diet (15, 16). Transgenic animals expressing dominant-positive SREBP-1a exhibited a striking phenotype. Their livers were enlarged up to fourfold, owing to the accumulation of massive amounts of triglyceride and cholesteryl ester (16). Despite the buildup of these end-products, the mrnas encoding multiple enzymes in the cholesterol and fatty acid biosynthetic pathways were markedly elevated. These include the mrnas for HMG CoA reductase, HMG CoA synthase, farnesyl diphosphate synthase, squalene synthase, acetyl CoA carboxylase, fatty acid synthase, and stearoyl CoA desaturase. The mrna encoding the LDL receptor was also increased. As a result, the livers of these animals massively overproduced cholesterol and fatty acids as measured by the rate of incorporation of intraperitoneally injected [ 3 H]water. The white adipose tissue of the SREBP-1a transgenic mice was normal in young animals, but it involuted as the animals became older (16). By the age of 4 mo, the amount of white adipose tissue had declined by 90%, as reflected by the amount of epididymal fat. In contrast to the striking phenotype of the SREBP-1a mice, we observed a much milder phenotype in animals expressing equivalent amounts of the truncated form of SREBP-1c (15). In these animals the liver was not significantly enlarged. The organ showed a modest accumulation of triglyceride and a modest increase in the mrnas encoding the fatty acid biosynthetic enzymes. The mrnas encoding the cholesterol biosynthetic enzymes were not elevated. In the current study we have extended these observations to SREBP-2. We have produced transgenic mice that overexpress a truncated dominant-positive form of SREBP-2 under control of the PEPCK promoter. The pathways of cholesterol biosynthesis and fatty acid biosynthesis are both activated in the SREBP-2 transgenic animals, but the ratio of cholesterol biosynthesis relative to fatty acid biosynthesis is much higher in these animals than in the SREBP-1a transgenics. This is attributable to a marked increase in the mrnas for HMG CoA synthase and HMG CoA reductase. These data indicate that the major function of SREBP-2 is to activate cholesterol biosynthesis in preference to fatty acid synthesis in liver and adipose tissue. Methods Materials and general methods. Sequencing reactions were performed on an Applied Biosystems 373A DNA Sequencer (Foster City, CA) by the dideoxy chain termination method. The content of cholesterol and triglyceride in plasma and liver was measured as described previously (16). Transgenic mice expressing SREBP-1c436 (encoding amino acids of human SREBP-1c) (15) and SREBP-1a460, line B (encoding amino acids of human SREBP-1a) (16) were described in the indicated reference. Measurement of hepatic unesterified and esterified cholesterol. Total lipids were extracted from mg aliquots of liver. Unesterified and esterified cholesterol were separated on 500-mg silica columns (Cat no ; Varian, Harbor City, CA) as described (17) and quantified by gas liquid chromatography (18). Plasmid construction. The expression plasmid ppepck-srebp- 2(1 468) contained the rat PEPCK promoter (2.4 kb) fused to a cdna encoding amino acids of human SREBP-2 (6) followed by two consecutive stop codons. It was constructed as previously described for ppepck-srebp-1a460 except that the cdna fragment was produced by PCR and ligated into the SmaI site of the pcmv- PEPCK promoter (16). The PCR reaction was performed using psrebp-2 (6) as a template, Pfu polymerase, a 5 primer corresponding to bp 74 to 45 in the 5 untranslated region of human SREBP-2 (5 -GGGCGGTGGCGACGGCACCGCCCCCG- CGTCT-3 ), and a 3 primer corresponding to amino acids followed by two consecutive stop codons (5 -GGGTCATCAG- TCTGGCTCATCTTTGACCTTTGCATCATC-3 ). The integrity of the construct was confirmed by DNA sequencing. Production and maintenance of transgenic mice. Techniques used for generating transgenic mice were previously described (16). A total of 1,502 fertilized eggs that were microinjected with the SalI-SphI fragment of ppepck-srebp-2(1 468) survived to the two-cell stage. Among the 184 offspring, 23% had integrated the transgene as determined by dot blot hybridization of DNA from tail homogenates. Of 42 DNA-positive mice subjected to partial hepatectomy, 30% produced truncated SREBP-2 as determined by immunoblot analysis. Mice with high levels of transgene expression in liver were bred to C57BL/6J SJL F1 mice, and two independent lines of SREBP- 2(1 468) mice were established, designated lines A and B. To obtain higher expression, hemizygotes of line B were mated to produce homozygotes for the transgene. Hemizygous mice from line A and homozygous mice from line B were used for all studies. Mice were housed in colony cages and maintained on a 14-h light/10-h dark cycle. Transgenic mice and nontransgenic littermates were fed a low carbohydrate/high protein diet (No. 5789C-3) from Purina Mills Inc. (St. Louis, MO) containing 71% (wt/wt) casein and 4.25% sucrose for 2 wk to induce maximal expression of the transgene (19). All mice were fasted 4 h before death during the early phase of the light cycle Horton et al.
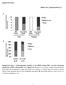
3 Immunoblotting. Nuclear extracts were prepared from pooled liver samples as previously described (16). Each sample (30 g protein) was subjected to 8% SDS-PAGE, electrophoretically transferred to Hybond C extra membranes (Amersham Corp., Arlington Heights, IL), and incubated with 5 g/ml of rabbit anti human SREBP-2 IgG, which was raised against amino acids of human SREBP-2 (6). Immunoblot analysis was carried out with the Enhanced Chemiluminescence (ECL) Western Blotting Detection System Kit (Amersham Corp.) with a horseradish peroxidase-labeled donkey anti rabbit IgG antibody (Amersham Corp.) as previously described (16). Blot hybridization of RNA. Total RNA was prepared from mouse liver and epididymal fat pad using the RNeasy Total RNA kit (Qiagen Inc., Chatsworth, CA) and the acid guanidinium thiocyanate-phenol-chloroform method (20), respectively. Equal aliquots of total RNA from each group of mice were pooled (total, 15 g) and subjected to Northern blot analysis with the indicated cdna probes prepared as previously described (15, 16, 21). A HindIII fragment (1.3 kb) of the human SREBP-2 cdna was used as a cdna probe for the transgene. A probe for mouse PPAR- (22) was prepared by reverse transcriptase-pcr using mouse liver poly(a) RNA as template as previously described for other probes (16). The 5 and 3 primers were 5 -ACAAGACTACCCTTTACTGAAATTACC- ATG-3 and 5 -GGCACTTCTGAAACCGACAGTACTGACAT- TTATTT-3, respectively. The bands on the Northern blots were quantified by exposure of the filter to a BAS1000 Fuji Phosphor- Imager (Tokyo, Japan), and the results were normalized to the signal generated from GAPDH mrna. RNase protection assay. crna probes were prepared from cdna fragments for human SREBP-1 and SREBP-2 that were produced by PCR amplification using the plasmids ppepck-srebp- 1a460 (16) and ppepck-srebp-2(1 468) (this paper) as templates, respectively. The following primers were used. Human SREBP-1: 5 primer, 5 -GGCACAGACCCTGCCAGCC-3 ; and 3 primer, 5 -CTGGGGTAGCCTAACACAGG-3 (5). Human SREBP-2: 5 primer, 5 -GTAGCAGCAGCGGCAGCAGTGG-3 ; and 3 primer, 5 -AAAGGCTGCTGGATGATCCTCG-3 (6). The cdna fragment for human SREBP-1 is identical in DNA sequence for isoforms SREBP-1a and -1c. HindIII and EcoRI sites were added to both 5 and 3 primers, respectively. The RNase protection assay was done as previously described (12, 14). For comparison of transgene expression levels in livers of SREBP-1a, -1c, and -2 transgenic mice, each probe was designed so that each protected fragment would contain the same number of 32 P atoms (i.e., 69). Cholesterol and fatty acid synthesis in vivo. Five 12-wk-old mice homozygous for the human SREBP-2 transgene (line B) and wildtype mice from the same litters (three females and one male for each group) were fasted for 4 h and killed during the early light cycle. The rates of incorporation of intraperitoneally injected [ 3 H]water into cholesterol and fatty acid were measured in liver as previously described (16). Results To produce transgenic mice expressing a dominant-positive form of SREBP-2, we constructed a plasmid encoding amino acids of human SREBP-2 under control of the PEPCK promoter. The encoded protein includes the bhlh-zip DNA binding domain and the NH 2 -terminal acidic transactivation domain, but it lacks the membrane-attachment domain and the COOH-terminal regulatory domain. Previous studies in tissue culture demonstrated that such truncated SREBPs enter Figure 1. Photograph of livers from 12-wk-old wild-type mice and transgenic mice expressing truncated dominant-positive SREBP-1a, -1c, and -2. The SREBP-1a (line B) and SREBP-1c mice were described in references 15 and 16. The SREBP-2 mouse was from line B. All animals were fed the low carbohydrate/high protein diet for 2 wk before killing. (Lower panel) schematic diagram of the domain structure of SREBP-1a, -1c, and -2, illustrating the site of truncation used for construction of the transgenes. TM, transmembrane segments. Transgenic Mice Expressing Truncated SREBP
4 Table I. Phenotypic Comparison of Wild-type and TgSREBP-2 Mice TgSREBP-2 mice Parameter Wild-type mice Line A Line B Number and sex Five males Five males Five males Body weight (grams) Liver weight (grams) Liver weight/body weight Epididymal fat pad weight (grams) Epididymal fat weight/body weight Liver cholesterol content (mg/gram) (P 0.001)* (P 0.01)* Liver triglyceride content (mg/gram) (P 0.05)* Total plasma cholesterol (mg/dl) (P 0.05)* (P 0.05)* Total plasma triglycerides (mg/dl) Each value represents the mean SEM of five 16-wk-old male mice. Wild-type and line A hemizygous transgenic mice were obtained from the same two litters. Line B transgenic mice were homozygous for the transgene and obtained from four littermates from hemizygote matings. All mice were fed the low carbohydrate/high protein diet for 2 wk and fasted 4 h before death. *Values in parentheses denote the level of statistical significance (Student s t test) between wild-type and the indicated line of transgenic mice. For parameters in which the level of significance is P 0.05, no values are shown. the nucleus directly without a requirement for proteolysis (23). The site of truncation in SREBP-2 is equivalent to the sites of truncation in the SREBP-1a and -1c proteins that have been studied previously in transgenic animals (Fig. 1, lower panel). The SREBP-2 transgene construct was injected into mouse eggs, and we derived two independent lines of transgenic mice, designated A and B. To increase expression, mice from line B were bred to homozygosity for the transgene. The line A mice were maintained in the hemizygous state. Fig. 1 (upper panel) compares the appearance of the liver of one representative mouse from SREBP-2, line B and livers from previously described transgenic mice expressing dominant-positive forms of SREBP-1a and -1c (15, 16). As reported previously, the livers from the SREBP-1a animals were grossly enlarged and pale, owing to massive accumulation of cholesteryl esters and triglycerides (16), whereas the SREBP-1c livers were generally indistinguishable from wild-type (15). Livers from the SREBP-2 animals were not visibly enlarged, but they exhibited a distinct pale color that was consistently different from that of wild-type mice (Fig. 1, upper panel). Table I compares various phenotypic parameters in wildtype mice and SREBP-2 transgenic mice of lines A and B. Statistically significant changes were confined to the liver and plasma. Livers from the SREBP-2 transgenic animals were normal in weight, but they contained increased amounts of cholesterol and triglyceride. The levels of both lipids were higher in line B than in line A. The plasma cholesterol level was increased slightly in the line A animals and decreased slightly in the line B animals, a finding of questionable biologic significance. Plasma triglyceride levels were reduced in the transgenic mice, but this was not statistically significant, owing to the large variation in triglyceride levels in the wild-type mice. Importantly, the weight of adipose tissue, as reflected in the epididymal fat pad, was normal in the SREBP-2 transgenic mice, a finding that contrasts with the marked decline observed in the SREBP-1a transgenics (16). Livers from line B mice expressed a somewhat higher level of truncated SREBP-2 protein as compared with livers from the line A animals (1.3-fold) as determined by immunoblotting (Fig. 2). Similar results were seen in one other experiment in which these levels were compared. In the Northern blotting experiment of Fig. 3, the livers of the line B mice contained about threefold more transgene SREBP-2 mrna than the livers of the line A mice (Fig. 3, upper left panel). A similar trend was seen when we examined levels of mrna for putative SREBP target genes. Elevations were observed in the levels of mrna for endogenous SREBP-1 and -2, the LDL receptor, and multiple genes of cholesterol biosynthesis, including HMG CoA synthase, HMG CoA reductase, farnesyl diphosphate (FPP) synthase, and squalene synthase. The magnitude of the increase in HMG CoA reductase mrna is particularly noteworthy (16-fold in line A and 75-fold in line B). Elevated levels of mrnas for fatty acid biosynthetic enzymes were also observed, including acetyl CoA carboxylase, fatty acid synthase, and stearoyl CoA desaturase. In each case the elevation was greater for the line B animals as compared with the line A animals. We also observed an elevation in the mrna encoding the microsomal triglyceride transfer protein (MTP), which participates in the synthesis of tri- Figure 2. Immunoblot analysis of truncated human SREBP-2 in nuclear extracts from livers of two lines of transgenic SREBP-2 mice (A and B). Livers from the mice described in Table I were pooled, and aliquots of the pooled nuclear extracts (30 g protein) were subjected to SDS-PAGE and transferred electrophoretically to a nitrocellulose filter. Immunoblot analysis was carried out with 5 g/ml rabbit anti human SREBP-2 IgG as the primary antibody, which does not recognize endogenous mouse SREBP-2. Bound antibodies were visualized as described in Methods. Filters were exposed to film for 15 s Horton et al.
5 Figure 3. Amounts of various mrnas in livers of wild-type (WT) mice and two lines of transgenic SREBP-2 mice (A and B) as measured by blot hybridization. Total liver RNA isolated from mice described in Table I was pooled (five mice per group), and 15- g aliquots were subjected to electrophoresis and blot hybridization with the indicated 32 P-labeled probe and with a control 32 P-labeled probe directed against GAPDH. The amount of radioactivity in each band was quantified as described in Methods. The fold increase in each mrna of transgenic mice, relative to that of wild-type control mice, was calculated after correction for loading differences with GAPDH. These values are shown below each blot. The probe for stearoyl CoA desaturase (SCD) was the mouse SCD-1 cdna fragment that can detect both SCD-1 and SCD-2 mrnas (16). FPP, farnesyl diphosphate; MTP, microsomal triglyceride transfer protein. Figure 4. Amount of various mrnas in white adipose tissues of wild-type (WT) and two lines of transgenic SREBP-2 mice (A and B) as measured by blot hybridization. Total RNA isolated from epididymal fat pads of mice described in Table I was pooled, and 15- g aliquots were subjected to electrophoresis and blot hybridization with the indicated 32 P-labeled probe and with a control 32 P-labeled probe directed against GAPDH. The amount of radioactivity in each band was quantified as described in Methods. The fold increase in each mrna of transgenic mice, relative to that of wild-type control mice, was calculated after correction for loading differences with GAPDH. These values are shown below each blot. The probe for stearoyl CoA desaturase (SCD) was the mouse SCD-1 cdna fragment that can detect both SCD-1 and SCD-2 mrnas (16). FPP, farnesyl diphosphate. Transgenic Mice Expressing Truncated SREBP
6 Table II. In Vivo Synthesis of Cholesterol and Fatty Acids in Livers from Wild-type and TgSREBP-2 Mice Incorporation of [ 3 H]water into Genotype of mice Liver weight Digitonin-precipitable sterols Fatty acids grams mol/h per gram mol/h per organ mol/h per gram mol/h per organ Wild-type (n 5) TgSREBP-2 (n 5) Each value represents the mean SEM of four female and one male 12-wk-old littermate mice of the indicated genotype. The transgenic SREBP-2 mice were from line B. All mice were fed the low carbohydrate/high protein diet for 2 wk and were fasted 4 h before the experiment. [ 3 H]water was injected intraperitoneally, and 1 h later the liver was removed for measurement of its content of 3 H-labeled digitonin-precipitable sterols and fatty acids as described in Methods. All differences between wild-type and transgenic SREBP-2 mice were statistically significant by the Student s t test (P 0.02). glyceride-rich VLDL (24). Inasmuch as this gene has not been studied previously as a target for SREBPs, we do not know whether the elevation is a direct effect of SREBP-2 action or whether it is secondary to the increased lipid content of the liver. The mrna for cholesterol 7- hydroxylase was elevated slightly in the line B animals. In agreement with previous observations with the other transgenic SREBP mice (15, 16), we found no elevation in the levels of mrna for apoproteins of the plasma lipoprotein transport system (namely, apo AI, apo B, and apo E) in livers of the mice expressing SREBP-2. The PEPCK promoter that drives expression of the SREBP-2 transgene is active in adipose tissue as well as in liver. In the experiment of Fig. 4, we compared the levels of various mrnas in epididymal fat pads of wild-type mice and transgenic lines A and B. The amount of transgene expression was higher in the line B animals than in the line A animals, and this was reflected in elevated levels of mrna for the LDL receptor and the cholesterol biosynthetic enzymes. Surprisingly, we did not observe consistent elevations in mrnas for acetyl CoA carboxylase, fatty acid synthase, or stearoyl CoA desaturase in adipose tissue. We also did not observe significant effects on mrnas for relatively fat-specific genes such as lipoprotein lipase, leptin, or PPAR-. Thus, in adipose tissue the truncated SREBP-2 has a relatively specific stimulatory effect on transcription of genes involved in cholesterol supply as opposed to fatty acid input. In livers of the SREBP-2 transgenic mice, the increases in mrnas encoding lipogenic enzymes were reflected in elevated rates of lipid synthesis (Table II). To make these measurements, wild-type and SREBP-2 transgenic mice (line B) were injected intraperitoneally with [ 3 H]water, and the incorporation into digitonin-precipitable sterols and fatty acids was measured in liver extracts 1 h later. These studies revealed a massive increase in sterol synthesis (28-fold) and a lesser increase in fatty acid synthesis (fourfold) in the SREBP-2 transgenics when expressed on the basis of liver weight. The increase in cholesterol synthesis was associated with an 11-fold increase in cholesteryl ester content in the SREBP-2 transgenic livers (Fig. 5). Discussion The current experiments complete a series in which dominantpositive truncated versions of all three SREBPs were expressed in liver and adipose tissue of transgenic mice under control of the PEPCK promoter. Figs. 6 9 compare the results with respect to the liver. The experiments in each strain were performed at different times, but the animals were all approximately the same age, and they were all fed the low carbohydrate/high protein diet for 2 wk to induce transgene expression before killing. Fig. 6 compares the levels of transgene expression as determined by RNase protection assays using probes that were adjusted to contain the same number of 32 P atoms in each protected fragment. The level of transgene-encoded mrna was higher for SREBP-1a than for SREBP-1c or SREBP-2. Although these differences preclude a direct comparison of each isoform s absolute effect on the absolute rate of lipid synthesis, they do allow a valid comparison of the relative effect of each isoform on cholesterol synthesis versus triglyceride synthesis. Although all three SREBP isoforms tended to stimulate expression of multiple genes involved in lipid synthesis, the relative degree of stimulation varied in ways that indicated differential effects on the pathways of cholesterol as opposed to fatty acid synthesis. As shown in Fig. 7, SREBP-2 was more effective than SREBP-1a in increasing the amounts of the three Figure 5. Content of unesterified (A) and esterified (B) cholesterol in livers of wild-type and transgenic SREBP-2, line B mice. Four 16-wkold wild-type female mice and five transgenic SREBP-2, line B littermate female mice were fed the low carbohydrate/high protein diet for 2 wk and fasted 4 h before study. Aliquots of liver ( mg) were taken for measurement of unesterified and esterified cholesterol as described in Methods. Each circle denotes the value from an individual animal, and the bar represents the mean Horton et al.
7 Figure 6. Relative amounts of mrna encoding truncated SREBP-1c (lane A), SREBP-1a (lane B), and SREBP-2 (lane C) in livers from transgenic mice as measured by RNase protection. Total RNA from the livers of mice expressing the truncated transgenes encoding SREBP-1c (15), SREBP-1a (16), and SREBP-2, line B (this paper) was isolated, and equal aliquots (10 g) were hybridized at 68 C to the appropriate 32 P-labeled crna probe as described in Methods. Protected fragments were separated by gel electrophoresis and exposed to film for 8 h at 80 C with an intensifying screen. The radioactivity in the gels was quantified as described in Methods, normalized to the -actin signal, and expressed as the fold change relative to the value in SREBP-1c mice, which was arbitrarily set at 1.0. mrnas of the cholesterol biosynthetic pathway (HMG CoA synthase, HMG CoA reductase, and squalene synthase), whereas SREBP-1a was more effective than SREBP-2 with respect to the two mrnas of fatty acid biosynthesis (acetyl CoA carboxylase and fatty acid synthase). Both SREBPs were approximately equipotent with respect to the LDL receptor. SREBP-1c was much less effective than either SREBP-1a or SREBP-2 in stimulating transcription of all of the genes. The differential effects of the three SREBPs on mrna transcription were reflected in differential rates of lipid synthesis from [ 3 H]water (Fig. 8). SREBP-2 stimulated sterol synthesis more profoundly than SREBP-1a (Fig. 8 A), and the reverse was observed with respect to fatty acid synthesis (Fig. 8 B). As shown in Fig. 9, SREBP-1a raised the hepatic content of triglycerides more than it did cholesterol, and therefore it raised the triglyceride/cholesterol ratio (Fig. 9 C). On the other hand, SREBP-2 raised the content of both lipids to approximately the same extent, and therefore the triglyceride/cholesterol ratio remained about the same as in the control animals (Fig. 9 C). Interestingly, although SREBP-1c was much less active than SREBP-1a or SREBP-2, it raised the triglyceride content of the livers while leaving the cholesterol content unchanged. Thus SREBP-1c, like SREBP-1a, increased the triglyceride/cholesterol ratio. The massive increase in cholesterol synthesis in the SREBP-2 transgenic mice (28-fold) correlates with the massive increase in the amount of mrna for HMG CoA reductase (75-fold in the line B mice). This was coupled with an 11-fold increase in the amount of mrna for HMG CoA synthase, which produces the substrate for the HMG CoA reductase reaction. These results emphasize the importance of these two enzymes as rate-controlling steps for the overall pathway of cholesterol biosynthesis. Considered together, the comparative results of Figs. 7 9 indicate that overexpression of dominant positive forms of SREBP-1a and SREBP-1c favors triglyceride synthesis as opposed to cholesterol synthesis, whereas expression of SREBP-2 tends to favor the synthesis of cholesterol over triglyceride in livers of transgenic mice. The experiments with dominant positive transgenes are conducted under highly artificial experimental conditions, yet they have implications for normal physiology. The results suggest that SREBP-1a, -1c, and -2 may have subtly distinct roles in the liver. Under normal conditions the mouse liver produces relatively large amounts of SREBP-1c relative to SREBP-1a (12). The same is true of the human (12) and the hamster (14). The function of the major SREBP-1c isoform may be to support fatty acid biosynthesis, and the smaller amounts of SREBP-1a may support low rates of cholesterol biosynthesis as well. On a low cholesterol chow diet, the mouse liver produces relatively little SREBP-2. The situation changes when the liver is deprived of cholesterol by treatment with diets con- Figure 7. Relative levels of various mrnas in livers of mice expressing transgenes encoding truncated SREBP-1c, -1a, and -2. Wild-type mrna levels in each experiment are arbitrarily set at 1. The mrna values for mice expressing truncated SREBP-1c (15) and SREBP-1a (16) were measured by Northern gel analysis and are taken from the indicated publication. mrna levels for the transgenic SREBP-2 mice are from this paper. All mice were fed the low carbohydrate/high protein diet for 2 wk. Transgenic SREBP-1a and SREBP-2 mice were fasted for 4 h before killing. Transgenic SREBP-1c mice were not fasted. Transgenic Mice Expressing Truncated SREBP
8 Figure 8. Rates of cholesterol and fatty acid synthesis in vivo in livers of mice expressing transgenes encoding truncated SREBP-1c, -1a, and -2. Values for SREBP-1a transgenic mice and their wild-type (WT) controls (mean SEM for 4 6 mice) are reproduced from experiments described in Table III in reference 16. Synthesis rates for SREBP-1c animals (7 mo of age) represent the mean SEM of values from three female and one male transgenic SREBP-1c mice as compared with four sex-matched littermate wild-type mice. Synthesis rates for transgenic SREBP-2, line B mice (mean SEM for five mice) are derived from the animals described in Table II. All mice were fed the low carbohydrate/high protein diet for 2 wk before the experiment. The SREBP-1a and -2 mice and their wild-type controls were fasted 2 and 4 h, respectively, before killing. The SREBP-1c mice were studied in the nonfasted state. All mice were injected with 50 mci of [ 3 H]water intraperitoneally, and 1 h later the liver was removed for measurement of its content of 3 H-labeled digitonin-precipitable sterols and fatty acids as described in Methods. taining an HMG CoA reductase inhibitor (lovastatin) and a bile acid binding resin (Colestipol). Under these conditions, livers of hamsters (25) and mice (Shimano, H., J. Horton, J.L. Goldstein, and M.S. Brown, unpublished observations) downregulate the production of the SREBP-1 isoforms and they upregulate production of SREBP-2. The proteolytic cleavage of the SREBP-2 precursor is also enhanced, leading to a marked increase in the content of the NH 2 -terminal segment of SREBP-2 in liver nuclei (reference 25 and Shimano, H., J. Horton, J.L. Goldstein, and M.S. Brown, unpublished observations). The current data suggest that the increase in SREBP-2, at the expense of the SREBP-1 isoforms, would lead to a selective increase in cholesterol biosynthesis as opposed to fatty acid biosynthesis. This would be an appropriate response to cholesterol deficiency. Figure 9. Comparison of cholesterol and triglyceride content in livers of mice expressing transgenes encoding truncated SREBP-1c, -1a, and -2. Values for the transgenic SREBP-1c mice and their wild-type (WT) controls (mean SEM for eight to nine mice) are taken from data of Table I in reference 15. Values for the transgenic SREBP-1a mice and their WT controls (mean SEM for six mice) are taken from data of Table I, Exp. B in reference 16. Values for the transgenic SREBP-2, line B mice are from this paper. All mice were fed the low carbohydrate/high protein diet for 2 wk before the experiment. Transgenic SREBP-1a and SREBP-2 mice were fasted for 4 h before killing. Transgenic SREBP-1c mice were not fasted. The selective effect of SREBP-2 in stimulating cholesterol synthesis was particularly apparent in adipose tissue. Here the SREBP-2 transgenic mice showed a distinct increase in mrnas encoding the LDL receptor and cholesterol biosynthetic enzymes yet no change in the mrnas of the fatty acid pathway (Fig. 4). It seems likely that the potential stimulating effects of SREBP-2 on fatty acid-related genes is counterbalanced by other regulatory proteins that tend to limit fatty acid biosynthesis. In addition to the differential effects of cholesterol and fatty acid biosynthesis, it is also possible that the three SREBP isoforms may have different effects on transcription of other genes whose existence is as yet unknown. Further elucidation of the effects of SREBPs on gene transcription will require detailed study of other genes that may be activated in the livers of the transgenic animals, as well as a more detailed comparison of the transcription-stimulating effects of the three iso Horton et al.
9 forms in tissue culture cells where a larger spectrum of quantitative measurements can be made. Studies along these lines are now underway. Acknowledgments We thank Yuri Bashmakov, Scott Clark, Shan Maika, and Robin Craddock for excellent technical assistance; Richard Gibson and Laura Quinlivan for invaluable help with the animals; and Dr. Laura Woollett for assistance with the [ 3 H]water experiments. This work was supported by grants from the National Institutes of Health (HL-20948), the Moss Heart Foundation, and the Perot Family Foundation. J.D. Horton is the recipient of a Postdoctoral Fellowship for Physicians from the Howard Hughes Medical Institute (Dallas, TX). I. Shimomura is the recipient of a Research Fellowship from the Manpei Suzuki Diabetes Foundation of Tokyo, Japan. References 1. Dietschy, J.M Overview of cholesterol and lipoprotein metabolism in the brain, liver and extrahepatic organs. Nutr. Metab. Cardiovasc. Dis. 7: Hillgartner, F.B., L.M. Salati, and A.G. Goodridge Physiological and molecular mechanisms involved in nutritional regulation of fatty acid synthesis. Physiol. Rev. 75: Brown, M.S., and J.L. Goldstein The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 89: Kim, J.B., G.D. Spotts, Y.-D. Halvorsen, H.-M. Shih, T. Ellenberger, H.C. Towle, and B.M. Spiegelman Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol. Cell. Biol. 15: Yokoyama, C., X. Wang, M.R. Briggs, A. Admon, J. Wu, X. Hua, J.L. Goldstein, and M.S. Brown SREBP-1, a basic helix-loop-helix leucine zipper protein that controls transcription of the LDL receptor gene. Cell. 75: Hua, X., C. Yokoyama, J. Wu, M.R. Briggs, M.S. Brown, J.L. Goldstein, and X. Wang SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc. Natl. Acad. Sci. USA. 90: Ericsson, J., S.M. Jackson, B.C. Lee, and P.A. Edwards Sterol regulatory element binding protein binds to a cis element in the promoter of the farnesyl diphosphate synthase gene. Proc. Natl. Acad. Sci. USA. 93: Guan, G., P.-H. Dai, T.F. Osborne, J.B. Kim, and I. Shechter Multiple sequence elements are involved in the transcriptional regulation of the human squalene synthase gene. J. Biol. Chem. 272: Kim, J.B., and B.M. Spiegelman ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes & Dev. 10: Lopez, J.M., M.K. Bennett, H.B. Sanchez, J.M. Rosenfeld, and T.F. Osborne Sterol regulation of acetyl CoA carboxylase: A mechanism for coordinate control of cellular lipid. Proc. Natl. Acad. Sci. USA. 93: Ericsson, J., S.M. Jackson, J.B. Kim, B.M. Spiegelman, and P.A. Edwards Identification of glycerol-3-phosphate acyltransferase as an adipocyte determination and differentiation factor 1 and sterol regulatory elementbinding protein-responsive gene. J. Biol. Chem. 272: Shimomura, I., H. Shimano, J.D. Horton, J.L. Goldstein, and M.S. Brown Differential expression of exons 1a and 1c in mrnas for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Invest. 99: Tontonoz, P., J.B. Kim, R.A. Graves, and B.M. Spiegelman ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol. Cell. Biol. 13: Shimomura, I., Y. Bashmakov, H. Shimano, J.D. Horton, J.L. Goldstein, and M.S. Brown Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver. Proc. Natl. Acad. Sci. USA. 94: Shimano, H., J.D. Horton, I. Shimomura, R.E. Hammer, M.S. Brown, and J.L. Goldstein Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J. Clin. Invest. 99: Shimano, H., J.D. Horton, R.E. Hammer, I. Shimomura, M.S. Brown, and J.L. Goldstein Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J. Clin. Invest. 98: Hamilton, J.G., and K. Comai Rapid separation of neutral lipids, free fatty acids and polar lipids using prepacked silica Sep-pak columns. Lipids. 23: Turley, S.D., M.W. Herndon, and J.M. Dietschy Reevaluation and application of the dual-isotope plasma ratio method for the measurement of intestinal cholesterol absorption in the hamster. J. Lipid Res. 35: Short, M.K., D.E. Clouthier, I.M. Schaefer, R.E. Hammer, M.A. Magnuson, and E.G. Beale Tissue-specific, developmental, hormonal, and dietary regulation of rat phosphoenolpyruvate carboxykinase-human growth hormone fusion genes in transgenic mice. Mol. Cell. Biol. 12: Chomczynski, P., and N. Sacchi Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: Shimano, H., I. Shimomura, R.E. Hammer, J. Herz, J.L. Goldstein, M.S. Brown, and J.D. Horton Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J. Clin. Invest. 100: Zhu, Y., K. Alvares, Q. Huang, M.S. Rao, and J.K. Reddy Cloning of a new member of the peroxisome proliferator activated receptor gene family from mouse liver. J. Biol. Chem. 268: Sato, R., J. Yang, X. Wang, M.J. Evans, Y.K. Ho, J.L. Goldstein, and M.S. Brown Assignment of the membrane attachment, DNA binding, and transcriptional activation domains of sterol regulatory element binding protein-1 (SREBP-1). J. Biol. Chem. 269: Sharp, D., L. Blinderman, K.A. Combs, B. Kienzle, B. Ricci, K. Wager- Smith, C.M. Gil, C.W. Turck, M.-E. Bouma, D.J. Rader, et al Cloning and gene defects in microsomal triglyceride transfer protein associated with abetalipoproteinaemia. Nature. 365: Sheng, Z., H. Otani, M.S. Brown, and J.L. Goldstein Independent regulation of sterol regulatory element binding proteins 1 and 2 in hamster liver. Proc. Natl. Acad. Sci. USA. 92: Transgenic Mice Expressing Truncated SREBP
Blunted Feedback Suppression of SREBP Processing by Dietary Cholesterol in Transgenic Mice Expressing Sterol-Resistant SCAP(D443N)
 Blunted Feedback Suppression of SREBP Processing by Dietary Cholesterol in Transgenic Mice Expressing Sterol-Resistant SCAP(D443N) Bobby S. Korn,* Iichiro Shimomura,* Yuriy Bashmakov,* Robert E. Hammer,
Blunted Feedback Suppression of SREBP Processing by Dietary Cholesterol in Transgenic Mice Expressing Sterol-Resistant SCAP(D443N) Bobby S. Korn,* Iichiro Shimomura,* Yuriy Bashmakov,* Robert E. Hammer,
Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver
 Proc. Natl. Acad. Sci. USA Vol. 94, pp. 12354 12359, November 1997 Biochemistry Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver (cholesterol biosynthesis
Proc. Natl. Acad. Sci. USA Vol. 94, pp. 12354 12359, November 1997 Biochemistry Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver (cholesterol biosynthesis
Jay D. Horton, Iichiro Shimomura, Shinji Ikemoto **, Yuriy Bashmakov, and Robert E. Hammer
 THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 278, No. 38, Issue of September 19, pp. 36652 36660, 2003 2003 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Overexpression
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 278, No. 38, Issue of September 19, pp. 36652 36660, 2003 2003 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Overexpression
Overexpression of SREBP-1a in Mouse Adipose Tissue Produces Adipocyte Hypertrophy, Increased Fatty Acid Secretion, and Fatty Liver
 JBC Papers in Press. Published on July 10, 2003 as Manuscript M306540200 1 Overexpression of SREBP-1a in Mouse Adipose Tissue Produces Adipocyte Hypertrophy, Increased Fatty Acid Secretion, and Fatty Liver
JBC Papers in Press. Published on July 10, 2003 as Manuscript M306540200 1 Overexpression of SREBP-1a in Mouse Adipose Tissue Produces Adipocyte Hypertrophy, Increased Fatty Acid Secretion, and Fatty Liver
Critical review. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver
 SPOTLIGHT Critical review SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver Jay D. Horton, 1,2 Joseph L. Goldstein, 1 and Michael S. Brown 1 1 Department of
SPOTLIGHT Critical review SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver Jay D. Horton, 1,2 Joseph L. Goldstein, 1 and Michael S. Brown 1 1 Department of
Fatty acid synthase (FAS) plays a central role in de novo
 Nutritional regulation of the fatty acid synthase promoter in vivo: Sterol regulatory element binding protein functions through an upstream region containing a sterol regulatory element Maria-Jesus Latasa,
Nutritional regulation of the fatty acid synthase promoter in vivo: Sterol regulatory element binding protein functions through an upstream region containing a sterol regulatory element Maria-Jesus Latasa,
The SCAP/SREBP Pathway: A Mediator of Hepatic Steatosis
 Review Article Endocrinol Metab 2017;32:6-10 https://doi.org/10.3803/enm.2017.32.1.6 pissn 2093-596X eissn 2093-5978 The SCAP/SREBP Pathway: A Mediator of Hepatic Steatosis Young-Ah Moon Department of
Review Article Endocrinol Metab 2017;32:6-10 https://doi.org/10.3803/enm.2017.32.1.6 pissn 2093-596X eissn 2093-5978 The SCAP/SREBP Pathway: A Mediator of Hepatic Steatosis Young-Ah Moon Department of
Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes
 Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes The major site of acetoacetate and 3-hydorxybutyrate production is in the liver. 3-hydorxybutyrate is the
Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes The major site of acetoacetate and 3-hydorxybutyrate production is in the liver. 3-hydorxybutyrate is the
Sterol regulatory element-binding proteins (SREBPs) are a
 Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mrna in rat hepatoma cells requires endogenous LXR ligands Russell A. DeBose-Boyd*, Jiafu Ou*, Joseph L. Goldstein, and Michael S.
Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mrna in rat hepatoma cells requires endogenous LXR ligands Russell A. DeBose-Boyd*, Jiafu Ou*, Joseph L. Goldstein, and Michael S.
Unit IV Problem 3 Biochemistry: Cholesterol Metabolism and Lipoproteins
 Unit IV Problem 3 Biochemistry: Cholesterol Metabolism and Lipoproteins - Cholesterol: It is a sterol which is found in all eukaryotic cells and contains an oxygen (as a hydroxyl group OH) on Carbon number
Unit IV Problem 3 Biochemistry: Cholesterol Metabolism and Lipoproteins - Cholesterol: It is a sterol which is found in all eukaryotic cells and contains an oxygen (as a hydroxyl group OH) on Carbon number
Supporting Information Table of content
 Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Hmgcoar AGCTTGCCCGAATTGTATGTG TCTGTTGTAACCATGTGACTTC. Cyp7α GGGATTGCTGTGGTAGTGAGC GGTATGGAATCAACCCGTTGTC
 Supplement Table I: primers for Real Time RT-PCR Gene Foward Reverse Hmgcoar AGCTTGCCCGAATTGTATGTG TCTGTTGTAACCATGTGACTTC Cyp7α GGGATTGCTGTGGTAGTGAGC GGTATGGAATCAACCCGTTGTC Cyp27a1 GTGGTCTTATTGGGTACTTGC
Supplement Table I: primers for Real Time RT-PCR Gene Foward Reverse Hmgcoar AGCTTGCCCGAATTGTATGTG TCTGTTGTAACCATGTGACTTC Cyp7α GGGATTGCTGTGGTAGTGAGC GGTATGGAATCAACCCGTTGTC Cyp27a1 GTGGTCTTATTGGGTACTTGC
Cholesterol metabolism. Function Biosynthesis Transport in the organism Hypercholesterolemia
 Cholesterol metabolism Function Biosynthesis Transport in the organism Hypercholesterolemia - component of all cell membranes - precursor of bile acids steroid hormones vitamin D Cholesterol Sources: dietary
Cholesterol metabolism Function Biosynthesis Transport in the organism Hypercholesterolemia - component of all cell membranes - precursor of bile acids steroid hormones vitamin D Cholesterol Sources: dietary
Cholesterol and its transport. Alice Skoumalová
 Cholesterol and its transport Alice Skoumalová 27 carbons Cholesterol - structure Cholesterol importance A stabilizing component of cell membranes A precursor of bile salts A precursor of steroid hormones
Cholesterol and its transport Alice Skoumalová 27 carbons Cholesterol - structure Cholesterol importance A stabilizing component of cell membranes A precursor of bile salts A precursor of steroid hormones
Dual regulation of mouse 5 - and 6 -desaturase gene expression by SREBP-1 and PPAR
 Dual regulation of mouse 5 - and 6 -desaturase gene expression by SREBP-1 and PPAR Takashi Matsuzaka, Hitoshi Shimano, 1, Naoya Yahagi,* Michiyo Amemiya-Kudo,* Tomohiro Yoshikawa,* Alyssa H. Hasty,* Yoshiaki
Dual regulation of mouse 5 - and 6 -desaturase gene expression by SREBP-1 and PPAR Takashi Matsuzaka, Hitoshi Shimano, 1, Naoya Yahagi,* Michiyo Amemiya-Kudo,* Tomohiro Yoshikawa,* Alyssa H. Hasty,* Yoshiaki
Regulating Hepatic Cellular Cholesterol
 Under circumstances of cholesterol deficiency, Sterol Regulatory Element Binding Proteins (SREBPs) via binding to DNA nuclear response elements set off genomic production of proteins and enzymes that induce
Under circumstances of cholesterol deficiency, Sterol Regulatory Element Binding Proteins (SREBPs) via binding to DNA nuclear response elements set off genomic production of proteins and enzymes that induce
ANSC/NUTR 601 GENERAL ANIMAL NUTRITION Stearoyl-CoA desaturase, VLDL metabolism, and obesity
 ANSC/NUTR 601 GENERAL ANIMAL NUTRITION Stearoyl-CoA desaturase, VLDL metabolism, and obesity I. Stearoyl coenzyme A desaturase and obesity in rodents A. Stearoyl coenzyme A desaturase (SCD) is the 9 desaturase.
ANSC/NUTR 601 GENERAL ANIMAL NUTRITION Stearoyl-CoA desaturase, VLDL metabolism, and obesity I. Stearoyl coenzyme A desaturase and obesity in rodents A. Stearoyl coenzyme A desaturase (SCD) is the 9 desaturase.
Schoenheimer effect explained feedback regulation of cholesterol synthesis in mice mediated by Insig proteins
 Research article Schoenheimer effect explained feedback regulation of cholesterol synthesis in mice mediated by Insig proteins Luke J. Engelking, 1 Guosheng Liang, 1 Robert E. Hammer, 2 Kiyosumi Takaishi,
Research article Schoenheimer effect explained feedback regulation of cholesterol synthesis in mice mediated by Insig proteins Luke J. Engelking, 1 Guosheng Liang, 1 Robert E. Hammer, 2 Kiyosumi Takaishi,
Cleavage of Sterol Regulatory Element-binding Proteins (SREBPs) at Site-1 Requires Interaction with SREBP Cleavage-activating Protein
 THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 273, No. 10, Issue of March 6, pp. 5785 5793, 1998 1998 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Cleavage of Sterol
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 273, No. 10, Issue of March 6, pp. 5785 5793, 1998 1998 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Cleavage of Sterol
Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes
 Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes Michiyo Amemiya-Kudo,* Hitoshi Shimano, 1, Alyssa H. Hasty,* Naoya Yahagi,*
Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes Michiyo Amemiya-Kudo,* Hitoshi Shimano, 1, Alyssa H. Hasty,* Naoya Yahagi,*
Oxidation of Long Chain Fatty Acids
 Oxidation of Long Chain Fatty Acids Dr NC Bird Oxidation of long chain fatty acids is the primary source of energy supply in man and animals. Hibernating animals utilise fat stores to maintain body heat,
Oxidation of Long Chain Fatty Acids Dr NC Bird Oxidation of long chain fatty acids is the primary source of energy supply in man and animals. Hibernating animals utilise fat stores to maintain body heat,
ab SREBP-2 Translocation Assay Kit (Cell-Based)
 ab133114 SREBP-2 Translocation Assay Kit (Cell-Based) Instructions for Use For analysis of translocation of SREBP-2 into nuclei. This product is for research use only and is not intended for diagnostic
ab133114 SREBP-2 Translocation Assay Kit (Cell-Based) Instructions for Use For analysis of translocation of SREBP-2 into nuclei. This product is for research use only and is not intended for diagnostic
Synergistic activation of transcription by nuclear factor Y and sterol regulatory element binding protein
 Synergistic activation of transcription by nuclear factor Y and sterol regulatory element binding protein Simon M. Jackson,* Johan Ericsson,* Roberto Mantovani, and Peter A. Edwards 1, *, Departments of
Synergistic activation of transcription by nuclear factor Y and sterol regulatory element binding protein Simon M. Jackson,* Johan Ericsson,* Roberto Mantovani, and Peter A. Edwards 1, *, Departments of
Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR
 Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) cadl CTTGGGGGCGCGTCT CTGTTCTTTTGTGCCGTTTCG cyl-coenzyme Dehydrogenase, very
Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) cadl CTTGGGGGCGCGTCT CTGTTCTTTTGTGCCGTTTCG cyl-coenzyme Dehydrogenase, very
Gene Polymorphisms and Carbohydrate Diets. James M. Ntambi Ph.D
 Gene Polymorphisms and Carbohydrate Diets James M. Ntambi Ph.D Fatty Acids that Flux into Tissue Lipids are from Dietary Sources or are Made De novo from Glucose or Fructose Glucose Fructose Acetyl-CoA
Gene Polymorphisms and Carbohydrate Diets James M. Ntambi Ph.D Fatty Acids that Flux into Tissue Lipids are from Dietary Sources or are Made De novo from Glucose or Fructose Glucose Fructose Acetyl-CoA
Supplementary Figure S1
 Lipidomic-based investigation into the regulatory effect of Schisandrin B on palmitic acid level in non-alcoholic steatotic livers Hiu Yee Kwan 1,2, Xuyan Niu 3, Wenlin Dai 4, Tiejun Tong 4, Xiaojuan Chao
Lipidomic-based investigation into the regulatory effect of Schisandrin B on palmitic acid level in non-alcoholic steatotic livers Hiu Yee Kwan 1,2, Xuyan Niu 3, Wenlin Dai 4, Tiejun Tong 4, Xiaojuan Chao
Investigations on the mechanism of hypercholesterolemia observed in copper deficiency in rats
 J. Biosci., Vol. 12, Number 2, June 1987, pp. 137 142. Printed in India. Investigations on the mechanism of hypercholesterolemia observed in copper deficiency in rats P. VALSALA and P. A. KURUP Department
J. Biosci., Vol. 12, Number 2, June 1987, pp. 137 142. Printed in India. Investigations on the mechanism of hypercholesterolemia observed in copper deficiency in rats P. VALSALA and P. A. KURUP Department
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies
 Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Lipoprotein Formation, Structure and Metabolism: Cholesterol Balance and the Regulation of Plasma Lipid Levels
 Lipoprotein Formation, Structure and Metabolism: Balance and the Regulation of Plasma Lipid Levels David E. Cohen, MD, PhD Director of Hepatology, Gastroenterology Division, Brigham and Women s Hospital
Lipoprotein Formation, Structure and Metabolism: Balance and the Regulation of Plasma Lipid Levels David E. Cohen, MD, PhD Director of Hepatology, Gastroenterology Division, Brigham and Women s Hospital
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at
 Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
2.5. AMPK activity
 Supplement Fig. A 3 B phos-ampk 2.5 * Control AICAR AMPK AMPK activity (Absorbance at 45 nm) 2.5.5 Control AICAR Supplement Fig. Effects of AICAR on AMPK activation in macrophages. J774. macrophages were
Supplement Fig. A 3 B phos-ampk 2.5 * Control AICAR AMPK AMPK activity (Absorbance at 45 nm) 2.5.5 Control AICAR Supplement Fig. Effects of AICAR on AMPK activation in macrophages. J774. macrophages were
MILK BIOSYNTHESIS PART 3: FAT
 MILK BIOSYNTHESIS PART 3: FAT KEY ENZYMES (FROM ALL BIOSYNTHESIS LECTURES) FDPase = fructose diphosphatase Citrate lyase Isocitrate dehydrogenase Fatty acid synthetase Acetyl CoA carboxylase Fatty acyl
MILK BIOSYNTHESIS PART 3: FAT KEY ENZYMES (FROM ALL BIOSYNTHESIS LECTURES) FDPase = fructose diphosphatase Citrate lyase Isocitrate dehydrogenase Fatty acid synthetase Acetyl CoA carboxylase Fatty acyl
General Laboratory methods Plasma analysis: Gene Expression Analysis: Immunoblot analysis: Immunohistochemistry:
 General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
MCB130 Midterm. GSI s Name:
 1. Peroxisomes are small, membrane-enclosed organelles that function in the degradation of fatty acids and in the degradation of H 2 O 2. Peroxisomes are not part of the secretory pathway and peroxisomal
1. Peroxisomes are small, membrane-enclosed organelles that function in the degradation of fatty acids and in the degradation of H 2 O 2. Peroxisomes are not part of the secretory pathway and peroxisomal
Suppression of fatty acid synthase promoter by polyunsaturated fatty acids
 Suppression of fatty acid synthase promoter by polyunsaturated fatty acids Yang Soo Moon, 1 Maria-Jesus Latasa, Michael J. Griffin, and Hei Sook Sul 2 Department of Nutritional Sciences, University of
Suppression of fatty acid synthase promoter by polyunsaturated fatty acids Yang Soo Moon, 1 Maria-Jesus Latasa, Michael J. Griffin, and Hei Sook Sul 2 Department of Nutritional Sciences, University of
Lipid Metabolism Prof. Dr. rer physiol. Dr.h.c. Ulrike Beisiegel
 Lipid Metabolism Department of Biochemistry and Molecular Biology II Medical Center Hamburg-ppendorf 1 Lipids. visceral fat. nutritional lipids 0 1.5 3 4.5 9 h. serum lipids. lipid accumulation in the
Lipid Metabolism Department of Biochemistry and Molecular Biology II Medical Center Hamburg-ppendorf 1 Lipids. visceral fat. nutritional lipids 0 1.5 3 4.5 9 h. serum lipids. lipid accumulation in the
Is it really that simple? Alyssa Hasty, PhD Associate Professor Molecular Physiology and Biophysics
 Alyssa Hasty, PhD Associate Professor Molecular Physiology and Biophysics Why we care about hepatic lipogenesis Control of lipid synthesis What can go wrong in humans Animal models dlto study lipoprotein
Alyssa Hasty, PhD Associate Professor Molecular Physiology and Biophysics Why we care about hepatic lipogenesis Control of lipid synthesis What can go wrong in humans Animal models dlto study lipoprotein
Plasma lipoproteins & atherosclerosis by. Prof.Dr. Maha M. Sallam
 Biochemistry Department Plasma lipoproteins & atherosclerosis by Prof.Dr. Maha M. Sallam 1 1. Recognize structures,types and role of lipoproteins in blood (Chylomicrons, VLDL, LDL and HDL). 2. Explain
Biochemistry Department Plasma lipoproteins & atherosclerosis by Prof.Dr. Maha M. Sallam 1 1. Recognize structures,types and role of lipoproteins in blood (Chylomicrons, VLDL, LDL and HDL). 2. Explain
Insulin-induced gene 1 (insig-1) mrna increases dramatically in
 Insig-1 brakes lipogenesis in adipocytes and inhibits differentiation of preadipocytes Jinping Li*, Kiyosumi Takaishi*, William Cook*, Sara Kay McCorkle*, and Roger H. Unger* *Touchstone Center for Diabetes
Insig-1 brakes lipogenesis in adipocytes and inhibits differentiation of preadipocytes Jinping Li*, Kiyosumi Takaishi*, William Cook*, Sara Kay McCorkle*, and Roger H. Unger* *Touchstone Center for Diabetes
Niacin Metabolism: Effects on Cholesterol
 Niacin Metabolism: Effects on Cholesterol By Julianne R. Edwards For Dr. William R. Proulx, PhD, RD Associate Professor of Nutrition and Dietetics In partial fulfillments for the requirements of NUTR342
Niacin Metabolism: Effects on Cholesterol By Julianne R. Edwards For Dr. William R. Proulx, PhD, RD Associate Professor of Nutrition and Dietetics In partial fulfillments for the requirements of NUTR342
control kda ATGL ATGLi HSL 82 GAPDH * ** *** WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi iwat gwat ibat
 body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
THE LDL RECEPTOR AND THE REGULATION OF CELLULAR CHOLESTEROL METABOLISM
 J. Cell Set. Suppl. 3, 131-137 (1985) Printed in Great Britain The Company of Biologists Limited 1985 131 THE LDL RECEPTOR AND THE REGULATION OF CELLULAR CHOLESTEROL METABOLISM JO SEPH L. G O L D S T E
J. Cell Set. Suppl. 3, 131-137 (1985) Printed in Great Britain The Company of Biologists Limited 1985 131 THE LDL RECEPTOR AND THE REGULATION OF CELLULAR CHOLESTEROL METABOLISM JO SEPH L. G O L D S T E
Supplementary Information. MicroRNA-33b knock-in mice for an intron of sterol regulatory
 Supplementary Information MicroRNA-33b knock-in mice for an intron of sterol regulatory element-binding factor 1 (Srebf1) exhibit reduced HDL-C in vivo Takahiro Horie, Tomohiro Nishino, Osamu Baba, Yasuhide
Supplementary Information MicroRNA-33b knock-in mice for an intron of sterol regulatory element-binding factor 1 (Srebf1) exhibit reduced HDL-C in vivo Takahiro Horie, Tomohiro Nishino, Osamu Baba, Yasuhide
ANSC/NUTR 618 Lipids & Lipid Metabolism
 I. Overall concepts A. Definitions ANC/NUTR 618 Lipids & Lipid Metabolism 1. De novo synthesis = synthesis from non-fatty acid precursors a. Carbohydrate precursors (glucose, lactate, and pyruvate) b.
I. Overall concepts A. Definitions ANC/NUTR 618 Lipids & Lipid Metabolism 1. De novo synthesis = synthesis from non-fatty acid precursors a. Carbohydrate precursors (glucose, lactate, and pyruvate) b.
7.06 Cell Biology EXAM #3 April 24, 2003
 7.06 Spring 2003 Exam 3 Name 1 of 8 7.06 Cell Biology EXAM #3 April 24, 2003 This is an open book exam, and you are allowed access to books and notes. Please write your answers to the questions in the
7.06 Spring 2003 Exam 3 Name 1 of 8 7.06 Cell Biology EXAM #3 April 24, 2003 This is an open book exam, and you are allowed access to books and notes. Please write your answers to the questions in the
Materials and Methods , The two-hybrid principle.
 The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
Print: ISSN Online: ISSN
 Review Article Interplay between Cholesterol, SREBPs, MicroRNA-33 in Dyslipidemia Abdul Hafeez Kandhro Abdul Hafeez Kandhro, Healthcare Molecular & Diagnostic Laboratory, Hyderabad, Pakistan, Email of
Review Article Interplay between Cholesterol, SREBPs, MicroRNA-33 in Dyslipidemia Abdul Hafeez Kandhro Abdul Hafeez Kandhro, Healthcare Molecular & Diagnostic Laboratory, Hyderabad, Pakistan, Email of
Mouse Meda-4 : chromosome 5G bp. EST (547bp) _at. 5 -Meda4 inner race (~1.8Kb)
 Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
Compensatory increase in fatty acid synthesis in adipose tissue of mice with conditional deficiency of SCAP in liver
 Compensatory increase in fatty acid synthesis in adipose tissue of mice with conditional deficiency of SCAP in liver Hiroshi Kuriyama, 1 Guosheng Liang, 1 Luke J. Engelking, 1 Jay D. Horton, 1,2 Joseph
Compensatory increase in fatty acid synthesis in adipose tissue of mice with conditional deficiency of SCAP in liver Hiroshi Kuriyama, 1 Guosheng Liang, 1 Luke J. Engelking, 1 Jay D. Horton, 1,2 Joseph
LIPID METABOLISM. Sri Widia A Jusman Department of Biochemistry & Molecular Biology FMUI
 LIPID METABOLISM Sri Widia A Jusman Department of Biochemistry & Molecular Biology FMUI Lipid metabolism is concerned mainly with fatty acids cholesterol Source of fatty acids from dietary fat de novo
LIPID METABOLISM Sri Widia A Jusman Department of Biochemistry & Molecular Biology FMUI Lipid metabolism is concerned mainly with fatty acids cholesterol Source of fatty acids from dietary fat de novo
Quantitative Real-Time PCR was performed as same as Materials and Methods.
 Supplemental Material Quantitative Real-Time PCR Quantitative Real-Time PCR was performed as same as Materials and Methods. Expression levels in the aorta were normalized to peptidylprolyl isomerase B
Supplemental Material Quantitative Real-Time PCR Quantitative Real-Time PCR was performed as same as Materials and Methods. Expression levels in the aorta were normalized to peptidylprolyl isomerase B
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM. Fatty Acid Elongation and Desaturation
 ANSC/NUTR 618 LIPIDS & LIPID METABOLISM I. Fatty acid elongation A. General 1. At least 60% of fatty acids in triacylglycerols are C18. 2. Free palmitic acid (16:0) synthesized in cytoplasm is elongated
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM I. Fatty acid elongation A. General 1. At least 60% of fatty acids in triacylglycerols are C18. 2. Free palmitic acid (16:0) synthesized in cytoplasm is elongated
review Genetically modified mice for the study of apolipoprotein B Edward Kim 1 and Stephen G. Young
 review Genetically modified mice for the study of apolipoprotein B Edward Kim 1 and Stephen G. Young Gladstone Institute of Cardiovascular Disease, San Francisco, CA 94141-9100, and Cardiovascular Research
review Genetically modified mice for the study of apolipoprotein B Edward Kim 1 and Stephen G. Young Gladstone Institute of Cardiovascular Disease, San Francisco, CA 94141-9100, and Cardiovascular Research
Lipid Metabolism. Remember fats?? Triacylglycerols - major form of energy storage in animals
 Remember fats?? Triacylglycerols - major form of energy storage in animals Your energy reserves: ~0.5% carbs (glycogen + glucose) ~15% protein (muscle, last resort) ~85% fat Why use fat for energy? 1 gram
Remember fats?? Triacylglycerols - major form of energy storage in animals Your energy reserves: ~0.5% carbs (glycogen + glucose) ~15% protein (muscle, last resort) ~85% fat Why use fat for energy? 1 gram
Doctor of Philosophy
 Regulation of Gene Expression of the 25-Hydroxyvitamin D la-hydroxylase (CYP27BI) Promoter: Study of A Transgenic Mouse Model Ivanka Hendrix School of Molecular and Biomedical Science The University of
Regulation of Gene Expression of the 25-Hydroxyvitamin D la-hydroxylase (CYP27BI) Promoter: Study of A Transgenic Mouse Model Ivanka Hendrix School of Molecular and Biomedical Science The University of
Intramembrane glycine mediates multimerization of Insig-2, a requirement for sterol regulation in Chinese hamster ovary cells
 Intramembrane glycine mediates multimerization of Insig-2, a requirement for sterol regulation in Chinese hamster ovary cells Peter C. W. Lee * and Russell A. DeBose-Boyd 1, Howard Hughes Medical Institute
Intramembrane glycine mediates multimerization of Insig-2, a requirement for sterol regulation in Chinese hamster ovary cells Peter C. W. Lee * and Russell A. DeBose-Boyd 1, Howard Hughes Medical Institute
Membrane Lipids & Cholesterol Metabolism
 Membrane Lipids & Cholesterol Metabolism Learning Objectives 1. How Are Acylglycerols and Compound Lipids Produced? 2. The synthesis of Sphingolipids from Ceramide 3. Diseases due to Disruption of Lipid
Membrane Lipids & Cholesterol Metabolism Learning Objectives 1. How Are Acylglycerols and Compound Lipids Produced? 2. The synthesis of Sphingolipids from Ceramide 3. Diseases due to Disruption of Lipid
GPR120 *** * * Liver BAT iwat ewat mwat Ileum Colon. UCP1 mrna ***
 a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
GASTROENTEROLOGY 1983;84:253-64
 GASTROENTEROLOGY 1983;84:253-64 Alteration of the Degree of Biliary Cholesterol Saturation in the Hamster and Rat by Manipulation of the Pools of Preformed and Newly Synthesized Cholesterol STEPHEN D.
GASTROENTEROLOGY 1983;84:253-64 Alteration of the Degree of Biliary Cholesterol Saturation in the Hamster and Rat by Manipulation of the Pools of Preformed and Newly Synthesized Cholesterol STEPHEN D.
cholesterol structure Cholesterol FAQs Cholesterol promotes the liquid-ordered phase of membranes Friday, October 15, 2010
 cholesterol structure most plasma cholesterol is in the esterified form (not found in cells or membranes) cholesterol functions in all membranes (drives formation of lipid microdomains) cholesterol is
cholesterol structure most plasma cholesterol is in the esterified form (not found in cells or membranes) cholesterol functions in all membranes (drives formation of lipid microdomains) cholesterol is
Srebp2: A Master Regulator of Sterol and Fatty Acid Synthesis
 Srebp2: A Master Regulator of Sterol and Fatty Acid Synthesis Blair B. Madison Division of Gastroenterology, Washington University School of Medicine, Saint Louis, MO 63110. Corresponding Author: Blair
Srebp2: A Master Regulator of Sterol and Fatty Acid Synthesis Blair B. Madison Division of Gastroenterology, Washington University School of Medicine, Saint Louis, MO 63110. Corresponding Author: Blair
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Cellular control of cholesterol. Peter Takizawa Department of Cell Biology
 Cellular control of cholesterol Peter Takizawa Department of Cell Biology Brief overview of cholesterol s biological role Regulation of cholesterol synthesis Dietary and cellular uptake of cholesterol
Cellular control of cholesterol Peter Takizawa Department of Cell Biology Brief overview of cholesterol s biological role Regulation of cholesterol synthesis Dietary and cellular uptake of cholesterol
The SREBP Pathway: Regulation of Cholesterol Metabolism by Proteolysis. SREBPs: Membrane-Bound Transcription Factors
 Cell, Vol. 89, 331 340, May 2, 1997, Copyright 1997 by Cell Press The SREBP Pathway: Regulation of Cholesterol Metabolism by Proteolysis of a Membrane-Bound Transcription Factor Review Michael S. Brown
Cell, Vol. 89, 331 340, May 2, 1997, Copyright 1997 by Cell Press The SREBP Pathway: Regulation of Cholesterol Metabolism by Proteolysis of a Membrane-Bound Transcription Factor Review Michael S. Brown
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna
 Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
Amplification of the gene for SCAP, coupled with Insig-1 deficiency, confers sterol resistance in mutant Chinese hamster ovary cells
 Amplification of the gene for SCAP, coupled with Insig-1 deficiency, confers sterol resistance in mutant Chinese hamster ovary cells Peter C. W. Lee,* Pingsheng Liu, Wei-Ping Li, and Russell A. DeBose-Boyd
Amplification of the gene for SCAP, coupled with Insig-1 deficiency, confers sterol resistance in mutant Chinese hamster ovary cells Peter C. W. Lee,* Pingsheng Liu, Wei-Ping Li, and Russell A. DeBose-Boyd
activity through redistribution of intracellular cholesterol pools
 Proc. Natl. Acad. Sci. USA Vol. 89, pp. 1797-181, November 1992 Medical Sciences Fatty acids regulate hepatic low density lipoprotein receptor activity through redistribution of intracellular cholesterol
Proc. Natl. Acad. Sci. USA Vol. 89, pp. 1797-181, November 1992 Medical Sciences Fatty acids regulate hepatic low density lipoprotein receptor activity through redistribution of intracellular cholesterol
Gene expression of sterol regulatory element-binding
 Gene expression of sterol regulatory element-binding proteins in hamster small intestine F. Jeffrey Field, 1 Ella Born, Shubha Murthy, and Satya N. Mathur Department of Internal Medicine and Veterans Administration,
Gene expression of sterol regulatory element-binding proteins in hamster small intestine F. Jeffrey Field, 1 Ella Born, Shubha Murthy, and Satya N. Mathur Department of Internal Medicine and Veterans Administration,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
Supplemental Figures
 Supplemental Figures Supplemental Figure 1. Fasting-dependent regulation of the SREBP ortholog SBP-1 and lipid homeostasis mediated by the SIRT1 ortholog SIR-2.1 in C. elegans. (A) Wild-type or sir-2.1(lof)
Supplemental Figures Supplemental Figure 1. Fasting-dependent regulation of the SREBP ortholog SBP-1 and lipid homeostasis mediated by the SIRT1 ortholog SIR-2.1 in C. elegans. (A) Wild-type or sir-2.1(lof)
SUPPLEMENTARY DATA. Supplementary Table 1. Primers used in qpcr
 Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
Commentary. New mechanisms by which statins lower plasma cholesterol. Henri Brunengraber
 Commentary New mechanisms by which statins lower plasma cholesterol Henri Brunengraber Department of Nutrition Case Western Reserve University Cleveland, OH 44109 Phone: 216 368 6429 Fax: 216 368 6560
Commentary New mechanisms by which statins lower plasma cholesterol Henri Brunengraber Department of Nutrition Case Western Reserve University Cleveland, OH 44109 Phone: 216 368 6429 Fax: 216 368 6560
Supplemental Table 1. List of primers used for real time PCR.
 Supplemental Table 1. List of primers used for real time PCR. Primer Sequence Primer Sequence Mouse Pcsk9-F TTGCAGCAGCTGGGAACTT Mouse Scd1-F CATCATTCTCATGGTCCTGCT Mouse Pcsk9-R CCGACTGTGATGACCTCTGGA Mouse
Supplemental Table 1. List of primers used for real time PCR. Primer Sequence Primer Sequence Mouse Pcsk9-F TTGCAGCAGCTGGGAACTT Mouse Scd1-F CATCATTCTCATGGTCCTGCT Mouse Pcsk9-R CCGACTGTGATGACCTCTGGA Mouse
Upregulation of hepatic LDL transport by n-3 fatty acids in LDL receptor knockout mice
 Upregulation of hepatic LDL transport by n-3 fatty acids in LDL receptor knockout mice Chandna Vasandani,* Abdallah I. Kafrouni,* Antonella Caronna,* Yuriy Bashmakov, Michael Gotthardt, Jay D. Horton,*,
Upregulation of hepatic LDL transport by n-3 fatty acids in LDL receptor knockout mice Chandna Vasandani,* Abdallah I. Kafrouni,* Antonella Caronna,* Yuriy Bashmakov, Michael Gotthardt, Jay D. Horton,*,
Chapter 26 Biochemistry 5th edition. phospholipids. Sphingolipids. Cholesterol. db=books&itool=toolbar
 http://www.ncbi.nlm.nih.gov/sites/entrez? db=books&itool=toolbar 1 The surface of a soap bubble is a bilayer formed by detergent molecules 2 Chapter 26 Biochemistry 5th edition phospholipids Sphingolipids
http://www.ncbi.nlm.nih.gov/sites/entrez? db=books&itool=toolbar 1 The surface of a soap bubble is a bilayer formed by detergent molecules 2 Chapter 26 Biochemistry 5th edition phospholipids Sphingolipids
PLASMA LIPOPROTEINS AND LIPIDS DETERMINATION OF PLASMA CHOLESTEROL AND TRIGLICERIDE LEVEL
 PLASMA LIPOPROTEINS AND LIPIDS DETERMINATION OF PLASMA CHOLESTEROL AND TRIGLICERIDE LEVEL Lipids are characterized by low polarity and limited solubility in water. Their plasma concentration is about 500-600
PLASMA LIPOPROTEINS AND LIPIDS DETERMINATION OF PLASMA CHOLESTEROL AND TRIGLICERIDE LEVEL Lipids are characterized by low polarity and limited solubility in water. Their plasma concentration is about 500-600
Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene
 Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene YUELIN ZHANG, WEIHUA FAN, MARK KINKEMA, XIN LI, AND
Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene YUELIN ZHANG, WEIHUA FAN, MARK KINKEMA, XIN LI, AND
Sterol regulatory element binding protein-1c (SREBP-1c) (1),
 Unsaturated fatty acids inhibit transcription of the sterol regulatory element-binding protein-1c (SREBP-1c) gene by antagonizing liganddependent activation of the LXR Jiafu Ou*, Hua Tu, Bei Shan, Alvin
Unsaturated fatty acids inhibit transcription of the sterol regulatory element-binding protein-1c (SREBP-1c) gene by antagonizing liganddependent activation of the LXR Jiafu Ou*, Hua Tu, Bei Shan, Alvin
For pair feeding, mice were fed 2.7g of HFD containing tofogliflozin
 Materials and Methods Pair Feeding Experiment For pair feeding, mice were fed 2.7g of HFD containing tofogliflozin (0.005%), which is average daily food intake of mice fed control HFD ad libitum at week
Materials and Methods Pair Feeding Experiment For pair feeding, mice were fed 2.7g of HFD containing tofogliflozin (0.005%), which is average daily food intake of mice fed control HFD ad libitum at week
FATTY ACID SYNTHESIS
 FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
The clathrin adaptor Numb regulates intestinal cholesterol. absorption through dynamic interaction with NPC1L1
 The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
Week 3 The Pancreas: Pancreatic ph buffering:
 Week 3 The Pancreas: A gland with both endocrine (secretion of substances into the bloodstream) & exocrine (secretion of substances to the outside of the body or another surface within the body) functions
Week 3 The Pancreas: A gland with both endocrine (secretion of substances into the bloodstream) & exocrine (secretion of substances to the outside of the body or another surface within the body) functions
Hepatic cholesterol and bile acid metabolism and intestinal cholesterol absorption in scavenger receptor class B type I-deficient mice
 Hepatic cholesterol and bile acid metabolism and intestinal cholesterol absorption in scavenger receptor class B type I-deficient mice Pablo Mardones,* Verónica Quiñones,* Ludwig Amigo,* Mauricio Moreno,*
Hepatic cholesterol and bile acid metabolism and intestinal cholesterol absorption in scavenger receptor class B type I-deficient mice Pablo Mardones,* Verónica Quiñones,* Ludwig Amigo,* Mauricio Moreno,*
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
BCH 447. Triglyceride Determination in Serum
 BCH 447 Triglyceride Determination in Serum Introduction: Triglycerides are esters of fatty acids and are hydrolyzed by lipase to glycerol and free fatty acids. Triglyceride determinations when performed
BCH 447 Triglyceride Determination in Serum Introduction: Triglycerides are esters of fatty acids and are hydrolyzed by lipase to glycerol and free fatty acids. Triglyceride determinations when performed
Chapter 2 The Chemistry of Life Part 2
 Chapter 2 The Chemistry of Life Part 2 Carbohydrates are Polymers of Monosaccharides Three different ways to represent a monosaccharide Carbohydrates Carbohydrates are sugars and starches and provide
Chapter 2 The Chemistry of Life Part 2 Carbohydrates are Polymers of Monosaccharides Three different ways to represent a monosaccharide Carbohydrates Carbohydrates are sugars and starches and provide
ADD-1/SREBP-1 is a major determinant of tissue. differential lipogenic capacity in mammalian and avian species.
 ADD-1/SREBP-1 is a major determinant of tissue differential lipogenic capacity in mammalian and avian species Florence Gondret, 1 Pascal Ferré, and Isabelle Dugail INSERM Unité 465, Institut Biomédical
ADD-1/SREBP-1 is a major determinant of tissue differential lipogenic capacity in mammalian and avian species Florence Gondret, 1 Pascal Ferré, and Isabelle Dugail INSERM Unité 465, Institut Biomédical
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
in vivo promoter analysis on refeeding response of hepatic sterol regulatory element-binding protein-1c expression.
 * Manuscript in vivo promoter analysis on refeeding response of hepatic sterol regulatory element-binding protein-1c expression. Yoshinori Takeuchi,1,2 Naoya Yahagi,1,3 Yoshimi Nakagawa,2,3 Takashi Matsuzaka,2,3
* Manuscript in vivo promoter analysis on refeeding response of hepatic sterol regulatory element-binding protein-1c expression. Yoshinori Takeuchi,1,2 Naoya Yahagi,1,3 Yoshimi Nakagawa,2,3 Takashi Matsuzaka,2,3
Problem Set #5 4/3/ Spring 02
 Question 1 Chloroplasts contain six compartments outer membrane, intermembrane space, inner membrane, stroma, thylakoid membrane, and thylakoid lumen each of which is populated by specific sets of proteins.
Question 1 Chloroplasts contain six compartments outer membrane, intermembrane space, inner membrane, stroma, thylakoid membrane, and thylakoid lumen each of which is populated by specific sets of proteins.
Figure 1. Effects of FGF21 on adipose tissue. (A) Representative histological. findings of epididymal adipose tissue (B) mrna expression of
 SUPPLEMENTAL MATERIAL EN-12-2276 Figure 1. Effects of FGF21 on adipose tissue. (A) Representative histological findings of epididymal adipose tissue (B) mrna expression of adipocytokines in adipose tissue.
SUPPLEMENTAL MATERIAL EN-12-2276 Figure 1. Effects of FGF21 on adipose tissue. (A) Representative histological findings of epididymal adipose tissue (B) mrna expression of adipocytokines in adipose tissue.
GROWTH HORMONE AND PPARα IN THE REGULATION OF GENES INVOLVED IN HEPATIC LIPID METABOLISM. Caroline Améen
 GROWTH HORMONE AND PPARα IN THE REGULATION OF GENES INVOLVED IN HEPATIC LIPID METABOLISM Caroline Améen Department of Physiology and Wallenberg Laboratory for Cardiovascular Research Sahlgrenska Academy
GROWTH HORMONE AND PPARα IN THE REGULATION OF GENES INVOLVED IN HEPATIC LIPID METABOLISM Caroline Améen Department of Physiology and Wallenberg Laboratory for Cardiovascular Research Sahlgrenska Academy
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original
 Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
23.1 Lipid Metabolism in Animals. Chapter 23. Micelles Lipid Metabolism in. Animals. Overview of Digestion Lipid Metabolism in
 Denniston Topping Caret Copyright! The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Chapter 23 Fatty Acid Metabolism Triglycerides (Tgl) are emulsified into fat droplets
Denniston Topping Caret Copyright! The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Chapter 23 Fatty Acid Metabolism Triglycerides (Tgl) are emulsified into fat droplets
ADD1 SREBP1 activates PPAR through the production of endogenous ligand (fatty acid metabolism nuclear hormone receptor adipocyte differentiation)
 Proc. Natl. Acad. Sci. USA Vol. 95, pp. 4333 4337, April 1998 Cell Biology ADD1 SREBP1 activates PPAR through the production of endogenous ligand (fatty acid metabolism nuclear hormone receptor adipocyte
Proc. Natl. Acad. Sci. USA Vol. 95, pp. 4333 4337, April 1998 Cell Biology ADD1 SREBP1 activates PPAR through the production of endogenous ligand (fatty acid metabolism nuclear hormone receptor adipocyte
Bio 366: Biological Chemistry II Test #1, 100 points (7 pages)
 Bio 366: Biological Chemistry II Test #1, 100 points (7 pages) READ THIS: Take a numbered test and sit in the seat with that number on it. Remove the numbered sticker from the desk, and stick it on the
Bio 366: Biological Chemistry II Test #1, 100 points (7 pages) READ THIS: Take a numbered test and sit in the seat with that number on it. Remove the numbered sticker from the desk, and stick it on the
Triglyceride determination
 Triglyceride determination Introduction: - Triglycerides are esters of fatty acids and are hydrolyzed to glycerol and free fatty acids (by lipase) - Triglyceride determinations when performed in conjunction
Triglyceride determination Introduction: - Triglycerides are esters of fatty acids and are hydrolyzed to glycerol and free fatty acids (by lipase) - Triglyceride determinations when performed in conjunction
