Synergistic activation of transcription by nuclear factor Y and sterol regulatory element binding protein
|
|
|
- Sharlene Sparks
- 5 years ago
- Views:
Transcription
1 Synergistic activation of transcription by nuclear factor Y and sterol regulatory element binding protein Simon M. Jackson,* Johan Ericsson,* Roberto Mantovani, and Peter A. Edwards 1, *, Departments of Biological Chemistry and Medicine* and the Molecular Biology Institute, University of California-Los Angeles, Los Angeles, CA 90095, and Departimento di Genetica e Biologia dei Microorganismi, Universita di Milano, Milano, Italy Abstract The current studies define the role of three distinct cis-elements in the proximal promoter of the rat farnesyl diphosphate (FPP) synthase gene. The three cis-elements, a sterol regulatory element 3 (SRE-3) flanked by an ATTGG motif (inverted CCAAT box), and a CCAAT box, form a sterol regulatory unit that is necessary and sufficient for sterol-regulated expression of FPP synthase promoter-reporter genes. FPP synthase promoter-reporter genes, that contain promoters with either wild-type nucleotide sequences or mutations in one or more of the three cis-elements, were transiently transfected into CV-1 cells. The activity of the wild-type promoterreporter gene increased when the cells were incubated in steroldepleted media or when the cells were co-transfected with a plasmid encoding the mature form of SRE binding protein (SREBP-1a). The results with the mutant promoter-reporter genes demonstrated that all three cis-elements were necessary for normal expression/regulation of the reporter gene by either sterols or by co-expressed SREBP-1a. Gel mobility shift assays demonstrated that the synergistic binding of SREBP-1a to SRE-3 was dependent on the binding of recombinant nuclear factor Y (NF-Y) to the DNA, consistent with the in vivo regulation studies. Jackson, S. M., J. Ericsson, R. Mantovani, and P. A. Edwards. Synergistic activation of transcription by nuclear factor Y and sterol regulatory element binding protein. J. Lipid Res : Supplementary key words SREBP-1a SRE-3 In previous studies we identified a 61 bp sequence in the proximal promoter of FPP synthase (nucleotides 293 to 233) that was necessary and sufficient to impart sterol-regulated expression to a reporter gene (1). These studies also identified four domains (defined by mutants 2, A, B and C in ref. 1) within this 61 bp sequence that, when mutated, impaired sterol-regulated expression of reporter genes. Mutant 2, the most 5 domain, identified a 5 bp motif (ATTGG) termed an inverted CCAAT box (1 3). The high expression of FPP synthase promoter-reporter genes observed in cells incubated in sterol-depleted media was attenuated by either mutation of the ATTGG motif within the FPP synthase promoter or by co-expression of a dominant negative form of NF-YA (2). Based on these studies, we proposed that the induced expression of FPP synthase was dependent upon the binding of the heterotrimeric transcription factor NF-Y to the inverted CCAAT box motif (2). Mutants A and B defined a novel motif termed SRE-3 (CTCACACGAG) that was shown to function as a binding site for SREBP-1a (1, 4). SRE-3 has 60% identity with SRE-1 (ATCACCCCAC), an element originally identified as an SREBP-binding site in the promoters of the LDL receptor and HMG-CoA synthase genes (5, 6). SRE-3 also has 50% identity with the nucleotides surrounding the E-box motif in the promoter of the fatty acid synthase gene (7). This latter motif was originally shown to bind ADD1, the rat homologue of SREBP-1 (7, 8). More recent studies have demonstrated that the fatty acid synthase promoter contains four half-sites that overlap the E-box motif and appear to function as two distinct binding sites for SREBP (9). Electromobility shift assays that utilized oligonucleotides containing the SRE-3 and the 5 inverted CCAAT box of the FPP synthase promoter demonstrated that binding of SREBP-1a to SRE-3 in vitro was enhanced over 20-fold when partially purified NF-Y bound to the inverted CCAAT box, located 20 bp 5 of SRE-3 (4). A different but somewhat analogous situation has been reported in studies utilizing the promoter of the LDL receptor gene (10, 11). In those studies, the binding of SREBP to SRE-1 was shown to stimulate the binding Abbreviations: FPP, farnesyl diphosphate; SRE, sterol regulatory element; SREBP, sterol regulatory element binding protein; NF-Y, nuclear factor Y; HMG-CoA, 3-hydroxy-3-methylglutaryl coenzyme A; CAT, chloramphenicol acetyltransferase; LDL, low density lipoprotein; bp, base pair; CHO, Chinese hamster ovary. 1 To whom correspondence should be addressed. Journal of Lipid Research Volume 39,
2 of Sp1 to an adjacent low affinity Sp1-binding motif. Thus, the requirement for an additional transcription factor(s), in addition to SREBP, appears to be a common theme in achieving optimal regulation of sterol-sensitive genes; studies with the LDL receptor (10, 11), fatty acid synthase (9, 12), and acetyl CoA carboxylase (13) promoters indicate that Sp1 fulfills this function in these promoters. Other transcription factors (NF-1, AP1, or Oct1) were unable to substitute fully for Sp1 in the sterol-regulated expression of LDL receptor promoterreporter constructs (10). In contrast, high expression of reporter genes under the control of promoters derived from the FPP synthase (1, 2), HMG-CoA synthase (2), SREBP-2 (14), glycerol-3-phosphate acyltransferase (15), and possibly squalene synthase (16, 17) genes requires NF-Y, in addition to SREBP. SREBP (5, 18), Sp1 (11, 19), and NF-Y (20) all contain transcriptional activation domains. Yieh, Sanchez, and Osborne (11) demonstrated that two distinct domains of Sp1 are involved both in transcriptional activation and SREBP-dependent DNA binding to the LDL receptor gene/promoter. However, the mechanism by which synergistic stimulation of transcription occurs when SREBP and either NF-Y or Sp1 bind to DNA is poorly understood. A fourth mutation, termed mutant C, defined a 12 bp sequence in the FPP synthase promoter that was necessary for the increased expression observed in cells incubated in sterol-depleted media (1). These changes in activity and regulation of the mutant construct were similar to those exhibited by FPP synthase promoter-reporter genes that contained a mutation in SRE-3 (1, 4). The results are consistent with the presence of important regulatory sequences in the 12 bp defined by mutant C. The wild-type nucleotides in this region contained a CCAAT box motif located 10 bp 3 of the SRE-3 motif (1). Several proteins have been identified that bind to CCAAT or related sequences (21 23). NF-Y, a heterotrimeric protein that binds to a CCAAT sequence, has been given many names including CP1, CBF, -CP1, EF1, and CBPtk (reviewed in ref. 24). All three NF-Y subunits (A, B, C) are required for DNA binding (25), which results in distortion of the DNA (24). The mechanism by which NF-Y regulates transcription of different genes is unknown. It may depend on the glutamineand serine/threonine-rich domains of NF-YA (20), the recruitment of other transcription factors to adjacent sites on the DNA (4), or recruitment of other non-dna binding proteins. The current studies were performed to determine the role of the nucleotides within the region defined by mutant C in the rat FPP synthase promoter (1). First, we demonstrate that NF-Y binds to the CCAAT box motif defined by mutant C. Second, we utilized recombinant NF-YA, NF-YB, and NF-YC to demonstrate that the enhanced binding of recombinant SREBP-1a to SRE-3 within the FPP synthase promoter requires that the heterotrimeric NF-Y binds to the adjacent ATTGG and/or CCAAT motifs. This NF-Y-dependent binding of SREBP-1a to DNA occurs in the absence of other nuclear proteins. Surprisingly, the increased formation of the SREBP-1a:DNA complex did not require that NF-Y remain bound to the DNA. We conclude that the normal regulation/expression of FPP synthase promoter-reporter genes, either in response to cellular sterol deprivation or after co-expression of mature SREBP, requires the binding of NF-Y to two motifs; one motif (CCAAT) is present within the sequence defined by mutant C (10 bp 3 of the SRE-3) and the second (ATTGG) lies 20 bp 5 of the SRE-3. EXPERIMENTAL PROCEDURES Materials DNA restriction and modification enzymes were obtained from Gibco BRL. 32 P-labeled nucleotide triphosphates were obtained from Amersham Corp. prsetb (Invitrogen) containing both a partial sequence of SREBP-1a (amino acids 1 490) and T7 and polyhistidine tags and pcmv-csa10, which encodes amino acids of SREBP-1a were kindly provided by Dr. T. Osborne (Department of Molecular Biology and Biochemistry, UC Irvine). Lipoprotein-deficient fetal calf serum was purchased from PerImmune. Antibodies to CBF-A, that also recognizes human NF-YB, were a generous gift from Dr. B. de Crombrugghe (University of Texas). Antibodies to CBF-HSP were a generous gift from Dr. B. Wu (Northwestern University) (26). The T7 antibody was from Invitrogen. The sources of all other reagents and plasmids have been given (1, 2, 27). Cell culture, transient transfections, and reporter gene assays CV-1 cells were cultured as previously described (4). Cells were transiently transfected with promoterreporter genes, a plasmid encoding -galactosidase and, where indicated, pcmv-csa10 (encoding mature SREBP-1a) as previously described (1, 2). After transfection, the cells were incubated for 20 h in media supplemented with either 10% lipoprotein-deficient calf serum in the absence (inducing media) or presence (repressing media) of sterols (10 g/ml cholesterol and 1 g/ml 25-hydroxycholesterol) (1, 2) or 10% fetal bovine serum, as indicated in the legends. Cells were then lysed and the CAT and -galactosidase activities 768 Journal of Lipid Research Volume 39, 1998
3 were determined (1, 2). The -galactosidase activity was used to normalize for transfection efficiencies (1). FPP synthase promoter-reporter gene constructs The wild type (wt) ptkciii promoter-reporter plasmid used in these studies is a promoter-chloramphenicol acetyltransferase (CAT) reporter construct containing 61 bp of the FPP synthase proximal promoter ( 293 to 233) (see Fig. 1). This 61 bp fragment was flanked 5 by 24 bp containing a HindIII restriction site and 3 by 9 bp containing a BamHI site. The double digested fragment was cloned into the HindIII/BamHI sites of ptkciii (1). The mutant promoter-reporter plasmids were constructed as follows. The Sculptor in vitro mutagenesis kit (Amersham) was used to produce a 3 bp transversion (nucleotides 242 to 238; CCAAT to CACCT) in pfpps A 61 bp fragment ( 293 to 233) was then generated utilizing the mutated pfpps and polymerase chain reaction (PCR) utilizing primers containing restriction sites (HindIII/BamHI) compatible with ptkciii to produce the Mut w construct utilized in this study. Mut 2-5 (ATTGG to ATTGT; a mutation in the inverted CCAAT box motif) and Mut q (CAC to ACA; a mutation in the SRE-3 motif) (Fig. 1) were generated in a similar manner and the 61 bp fragments ( 293 to 233) were subcloned into ptkciii. Mut 2-5:w was generated by digesting gel-purified HindIII/BamHI fragments of Mut 2-5 and Mut w with MaeIII. The gel-purified upstream fragment derived from the digestion of Mut 2-5 was subsequently religated with the downstream fragment derived from Mut w and subcloned into ptkciii to produce the double mutant construct. All PCR products were sequenced after subcloning in order to verify the sequence. Purification of recombinant SREBP-1a, NF-YA, NF-YB, and NF-YC Recombinant SREBP-1a, containing the N-terminal T7 and polyhistidine (His 6 ) tags was purified to homogeneity from E. coli extracts by nickel affinity chromatography as described by Sanchez, Yieh, and Osborne (10). Nucleotides corresponding to amino acids 262 to 317 of mouse NF-YA (28), 51 to 140 of mouse NF-YB (CBF-A) (29), and 37 to 120 of human NF-YC (30), respectively, were generated by polymerase chain reaction and cloned into parent plasmids to produce PET15YA9, PET29YB4, and PET29YC5. The polyhistidine-tagged proteins were purified from E. coli lysates by nickel affinity chromatography, as described above. Oligonucleotides Oligonucleotides used in gel mobility shift assays were synthesized by Gibco BRL. They include a 27 bp wild-type oligonucleotide ( 298 to 272) (27) containing the ATTGG motif of the FPP synthase promoter, a 21 bp oligonucleotide ( 250 to 230) containing the CCAAT motif and a 27 bp oligonucleotide ( 298 to 272) containing a mutated ATTGG motif (CGGTT; Mut 2-0). Electrobility shift assay CHO nuclear extracts were isolated as described (1, 4). Complementary single-stranded DNA, corresponding to the indicated nucleotides of the FPP synthase promoter (Fig. 1) and containing either the wild-type sequence or the indicated mutations, were annealed, and the double-stranded probe was isolated from acrylamide gels (31). Radiolabeled probes were generated in the same manner after an initial radiolabeling of a single-stranded oligonucleotide with 32 P (31). Doublestranded probes were also generated by HindIII/ BamHI restriction enzyme digestion of the wt and mutant ptkciii constructs. The fragments, containing nucleotides 293 to 233 of the FPP synthase promoter and HindIII/BamHI flanking sequences, were gel-purified and end-radiolabeled with 32 P. The radiolabeled probes (20,000 cpm; 1.5 fmol) were used in gel mobility shift assays in the presence of 2 or 5 g of CHO nuclear extract and non-fat milk (2.5 mg/ml) or in the presence of recombinant purified histidine-tagged NF- YA, NF-YB, NF-YC, and/or SREBP-1a in the absence of non-fat milk. The concentration of the recombinant proteins, based on silver-stained gels, was less than 50 ng/ l. Where indicated, antibody (0.2 g) to NF-YB or CBF-HSP was added to the incubations on ice for 30 min before gel electrophoresis was performed (1, 2). RESULTS Mutational analysis of the FPP synthase promoter identifies three cis-elements required for sterol-regulated expression Figure 1 shows the nucleotide sequence of the rat FPP synthase proximal promoter between nucleotides 293 and 233 (1). The ATTGG (inverted CCAAT box), SRE-3, and CCAAT box motifs are indicated, as are the mutations that were introduced to produce the mutant promoters (Fig. 1). We have previously demonstrated that the expression of a reporter gene under the control of nucleotides 293 to 233 from the FPP synthase promoter was regulated by sterols and by coexpressed SREBP-1a (1). We concluded that these 61 bp of the FPP synthase promoter were sufficient to confer sterol-regulated expression to an otherwise sterolunresponsive reporter gene (1). Jackson et al. Synergistic activation of transcription by NF-Y and SREBP 769
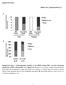
4 Fig. 1. A 61-bp sequence (nucleotides 293 to 233) of the FPP synthase promoter contains three cis-elements critical for sterol-regulated transcription. The ATTGG motif (heavy underline), SRE-3 motif (heavy overline), and CCAAT motif (light underline) are shown. Nucleotides shown below the wild-type sequences represent the changes that were introduced to produce the mutant constructs Mut 2-0, Mut 2-5, Mut q, and Mut w. A series of reporter genes were constructed in which the wild-type 61 bp FPP synthase promoter sequence ( 293 to 233) or the same sequence containing the indicated mutations (Mut 2-5, Mut q, Mut w, and Mut 2-5:w, Fig. 1) was placed 5 of a minimal thymidine kinase promoter-cat gene. CV-1 cells were transiently transfected with these constructs and the cells were then incubated for 20 h in media either depleted of sterols (inducing media) or supplemented with cholesterol and 25-hydroxycholesterol (repressing media). As expected, the expression of the reporter gene under the control of the wild-type FPP synthase promoter was induced approximately 14-fold when the cells were deprived of sterols (Fig. 2). The expression of a reporter gene that contained a 3 bp mutation in the CCAAT box motif (Mut w) was severely crippled; the reporter gene activity was low and was induced less than 1.5-fold after incubation of cells in the inducing media (Fig. 2). Similar results were observed when the promoter contained mutations in both the CCAAT box and the ATTGG motif (Mut w:2-5) (Fig. 2). Reporter genes that contained a 3 bp mutation in the SRE-3 motif (Mut q) were expressed at levels that approximated the repressed levels of the wild-type construct (Fig. 2). As expected, the activity of this mutant reporter gene was not induced when the cells were incubated in sterol-deficient medium (Fig. 2), consistent with the inability of endogenous SREBP to bind to the mutated SRE-3 (1). Finally, reporter genes that contained a point mutation in the ATTGG motif (Mut 2-5) also exhibited impaired regulation in response to sterol deprivation; reporter gene activity was regulated 3.6- Fig. 2. Mutations within three distinct cis-elements in the 61-bp FPP synthase promoter impair sterolregulated transcription. CV-1 cells were transfected in duplicate with plasmids expressing -galactosidase and either wild-type ptkciii (wt), the same construct but containing a mutation in the CCAAT motif (Mut w), the ATTGG motif (Mut 2-5), the SRE-3 motif (Mut q), or a combined ATTGG and CCAAT motif mutant (Mut 2-5:w). M signifies the mutated motif. After transfection, cells were incubated for 20 h in medium containing 10% lipoprotein-deficient fetal calf serum in the presence ( ) of absence ( ) of sterols (10 g of cholesterol and 1 g of 25-hydroxycholesterol per ml). Chloramphenicol acetyltransferase activities were normalized to -galactosidase activities in order to account for differences in transfection efficiencies. The normalized activity obtained from the wild-type construct expressed in cells incubated in the presence of sterols was given a value of 1.0. The results are representative of three separate studies, each performed in duplicate. 770 Journal of Lipid Research Volume 39, 1998
5 fold versus 14.1-fold in controls (Fig. 2). The data in Fig. 2 also demonstrate that, under inducing conditions, the absolute activity of the wild-type reporter construct exceeds the additive activities of the Mut q (two functional NF-Y binding sites) and Mut 2-5:w (functional SREBP binding site). Thus, synergistic activation of the FPP synthase promoter-reporter gene requires the presence of three motifs (ATTGG, SRE-3, and CCAAT). Figure 3 shows that co-expression of low levels of a plasmid encoding mature SREBP-1a, together with the wild-type FPP synthase promoter-reporter gene, resulted in a 6.7-fold increase in CAT activity. In the absence of co-expressed SREBP-1a, the FPP synthase promoter-reporter genes, containing mutations in the ATTGG, CCAAT, or SRE-3 motifs, were expressed at lower levels than the control (Fig. 3). In the presence of co-expressed SREBP-1a, there was a variable increase in reporter gene activity, although the maximal levels obtained were significantly less than the control (Fig. 3). Figure 3 demonstrates that co-expressed SREBP-1a does stimulate reporter genes containing an intact SRE-3 and either the wild-type ATTGG or CCAAT motif. However, mutation of both of these motifs (Mut 2-5:w) results in a reporter gene that is the least responsive to the co-expressed SREBP-1a (Fig. 3). The small stimulation of the Mutant q reporter gene by coexpressed SREBP-1a (Fig. 3) is likely the result of the relatively high levels of nuclear SREBP-1a that persist under these conditions. Gel mobility shift assays have shown that SREBP-1a binds weakly to DNA containing the q mutation, under specific conditions that include high concentration of the recombinant protein (data not shown). The observation that the maximal reporter gene activity obtained with the wild-type construct was greater than the added activities obtained with Mut q and Mut 2-5:w (Fig. 3) is consistent with synergistic activation of the reporter gene under conditions where nuclear proteins bind to all three motifs. Taken together, the data in Figs. 2 and 3 identify a third motif (CCAAT), in addition to the previously defined ATTGG and SRE-3 motifs (1, 2, 27), that is important for sterol- and SREBP-1a-regulated expression of FPP synthase promoter-reporter genes. Role of CCAAT- and ATTGG-binding proteins in the synergistic binding of SREBP-1a to SRE-3 Electromobility shift assays were carried out with oligonucleotides corresponding to different motifs within the FPP synthase promoter; a 27 bp 5 probe ( 298 to 272) containing either the wild-type 5 ATTGG sequences or 5 bp transversion (CGGTT; Mut 2-0) and a 21 bp 3 probe ( 250 to 230) containing the CCAAT motif (Fig. 1). Incubation of the 32 P end-labeled 5 probe with nuclear extract derived from CHO cells resulted in the formation of one major complex (Fig. 4A, lanes 2 and 7) that was competed by unlabeled competitor DNA Fig. 3. Mutations within the ATTGG, CCAAT, and SRE-3 motifs impair transcriptional activation of an FPP synthase promoter-reporter construct by co-expressed SREBP-1a. CV-1 cells were transfected with various constructs as described in the legend to Fig. 2, in the absence ( ) or presence ( ) of 20 ng of pcmv- CSA10 (SREBP-1a). The cells were then incubated for 20 h in medium containing 10% fetal bovine serum. The chloramphenicol acetyltransferase activity obtained from the wild-type promoter-reporter gene construct expressed in cells in the absence of co-expressed SREBP-1a was given a value of 1.0. The fold induction of chloramphenicol acetyltransferase activity, as a result of the co-expressed SREBP-1a, was 6.7, 8.5, 8,5, 2.5, and 1.5 for the wt, Mut w, Mut 2-5, Mut q, and Mut 2-5:w constructs, respectively. Jackson et al. Synergistic activation of transcription by NF-Y and SREBP 771
6 Fig. 4. NF-Y binds to the CCAAT motif in the FPP synthase promoter. A: A 32 P-labeled double-stranded oligonucleotide probe (20,000 cpm, 1.5 fmol), corresponding to nucleotides 298 to 272 of the FPP synthase promoter containing the ATTGG motif was used in a gel mobility shift assay either in the absence (lane 1) or the presence of 5 g of CHO nuclear extract (lanes 2 15) and either zero (lanes 1, 2, and 7), or increasing molar excess of competitor oligonucleotides. The competitor DNA contained 100-, 1,000-, 5,000-, and 10,000-fold molar excess, respectively, of wild-type sequence (lanes 3 6); the same sequence except ATTGG was mutated to CGGTT (Mut 2-0) (lanes 8 11) or an oligonucleotide (nucleotides 250 to 230) containing the wild-type CCAAT sequence (lanes 12 15). The NF-Y:DNA complex is indicated. The free probe is not shown. B: Radiolabeled probes corresponding to the wild-type ATTGG sequence (lanes 1 4), mutated ATTGG sequence (Mut 2-0; lanes 5 8) and wild-type CCAAT sequence (lanes 9 12) were incubated at 20 C with 5 g CHO nuclear extract for 60 min. After the addition of 0.2 g of antibody to the indicated CCAAT box-binding protein, the assay mixture was placed on ice for 30 min. The bound NF-Y/DNA complex (NF-Y), supershifted (SS) complexes, and a non-specific shifted complex (NS) are indicated. The free probe is not shown. corresponding to either the 5 probe (Fig. 4A, lanes 3 6) or the 3 oligonucleotide containing the CCAAT motif (lanes 12 15). The competition by these two probes was similar at equivalent molar concentrations (Fig. 4A, lanes 3 6 vs ). In contrast, the formation of the shifted complex was unaffected by inclusion of the 5 mutant oligonucleotide over the same molar concentration range (Fig. 4A, lanes 8 11). Incubation of radiolabeled 5 ATTGG- or 3 CCAATcontaining probes with CHO nuclear extract produced a supershifted complex when antibody to NF-YA was also included in the assay (Fig. 4B, lanes 4 and 12 vs. lanes 2 and 10, respectively). This latter effect was specific, as inclusion of antibody to CBF-HSP, a distinct CCAAT box binding protein (26), did not affect the formation or migration of the single complex (Fig. 4B, lanes 3 and 11). The mutant probe (Mut 2-0) formed a non-specific shifted complex that was unaffected when antibodies to either CBF-HSP or NF-YA were included in the assay (Fig. 4B, lanes 6 8). We previously utilized nuclear extracts, enriched in NF-Y, to demonstrate that the binding of NF-Y to the 5 ATTGG motif within the FPP synthase promoter, stimulated by more than 20-fold the binding of SREBP-1a to the adjacent SRE-3 motif (4). Immuno-depletion of this nuclear fraction with anti-nf-y antibodies provided direct evidence of the importance of NF-Y in enhancing the binding of SREBP-1a to SRE-3 (4). However, we were unable to assess whether other nuclear proteins, in addition to NF-Y, played a role in the stimulated binding of SREBP-1a to SRE-3. The data shown in Figs. 2 and 3 suggested that the binding of NF-Y to the 3 CCAAT motif, in addition to binding to the 5 ATTGG motif, might influence the binding of SREBP-1a to the adjacent SRE-3 (Figs. 2 and 3). The availability of recombinant NF-YA, NF-YB, NF- YC, and SREBP-1a permitted us to determine more exactly the role of the three NF-Y proteins per se in stimulating the binding of SREBP-1a to SRE-3 and to determine the function of the 3 CCAAT motif in this latter process. Recombinant truncated NF-YA, NF-YB, and NF-YC, containing N-terminal T7 and polyhistidine tags, were purified from Escherichia coli extracts by nickel affinity chromatography. Each of the proteins migrated as a single major band when analyzed by polyacrylamide gel electrophoresis (data not shown). Figure 5A demonstrates that, under these conditions, recombinant trimeric NF-Y (lane 2) and recombinant SREBP-1a (lane 3) bound poorly to the wild-type probe containing the ATTGG, SRE-3, and CCAAT motifs. Addition of increasing amounts of the three recombinant 772 Journal of Lipid Research Volume 39, 1998
7 Fig. 5. Stimulated binding of SREBP-1a to SRE-3 within the FPP synthase promoter requires that NF-Y binds to the CCAAT and/or ATTGG adjacent motifs. 32 P end-labeled double-stranded DNA oligonucleotides (nucleotides 293 to 233) containing (A) wild-type sequences (lanes 1 7) or mutations in both the ATTGG and CCAAT motifs (Mut 2-0:Mut w) (lanes 8 14) or (B) mutations in the ATTGG (Mut 2-0) (lanes 1 7) or CCAAT (Mut w) (lanes 8 14) motifs were used in gel electrophoresis mobility studies as described in Materials and Methods. The probes containing wild-type ATTGG (A), SRE-3 (S), CCAAT (C), or mutant (m) motifs are indicated. The probes were incubated with 1 l recombinant SREBP-1a and recombinant trimeric NF-Y, as indicated. Equal volumes of recombinant NF-Y A, B, and C were premixed and the amounts added in A and B was as follows; lanes 2, 7, 9, and 14, (3.6 l); lanes 4 and 11, (0.6 l); lanes 5 and 12, (1.2 l); lanes 6 and 13, (1.8 l). * indicates a multimeric SREBP-1a:DNA complex. In C the probe, containing ATTGG, SRE-3, and a mutated CCAAT motif, was incubated with 1.0 l of each recombinant NF-Y subunit, as indicated, and the DNA-protein shifted complexes are identified as described above. The free probe and shifted complexes containing SREBP-1a or NF-Y are shown. The NF-Y:DNA complex was also observed in the presence of high levels of NF-Y A, B, and C and non-fat milk but in the absence of SREBP-1a (data not shown). subunits of NF-Y together with SREBP-1a resulted in a large increase in the formation of both an SREBP- 1a:DNA complex and an NF-Y:DNA complex (Fig. 5A, lanes 4 7). Under these conditions, an NF-Y:SREBP- 1a:DNA complex was not observed. A slower migrating complex (indicated by * in Fig. 5A and B) corresponds to multimeric SREBP-1a bound to DNA. This complex was observed in some, but not all, experiments in which NF-Y was included and in other gel mobility shift assays where high levels of SREBP-1a and non-fat milk were used in the absence of NF-Y (data not shown). Formation of the SREBP-1a:DNA complex required Jackson et al. Synergistic activation of transcription by NF-Y and SREBP 773
8 that the DNA contain binding sites for the added recombinant NF-Y; a probe that contained mutations in both the ATTGG and CCAAT motifs bound neither SREBP-1a nor NF-Y (Fig. 5A, lanes 9 14). Figure 5B shows that recombinant NF-Y also stimulated, in a dosedependent manner, the formation of an SREBP:DNA complex when the radiolabeled probe contained a mutation in either the 5 ATTGG (compare lane 3 with lanes 4 7) or 3 CCAAT (compare lane 10 with lanes 11 14) motifs. Figure 5B also demonstrates that the heterotrimeric NF-Y forms a more stable complex with the DNA when bound to the 5 ATTGG motif as compared to the 3 CCAAT motif (compare lanes 7 and 14). However, when the probe contained both the ATTGG and CCAAT motifs, the NF-Y:DNA complex was observed at lower concentrations of NF-Y than when the probe contained only one of these motifs (compare Fig. 5A with 5B). This enhanced binding of NF-Y to the DNA may be important for the subsequent binding of SREBP-1a to the SRE-3 motif. Previous studies have demonstrated that the AT TGG/CCAAT nucleotides are absolutely required for the binding of the heterotrimeric NF-Y to DNA but that there is a strong preference for additional flanking nucleotides (21, 23, 32). We hypothesize that, under the DNA binding and electrophoretic conditions used in these studies, the stability and/or affinity of truncated recombinant NF-YA, B, and C for the DNA is reduced, when compared to non-truncated NF-Y, and results in impaired formation of stable SREBP:NF-Y:DNA or NF- Y:DNA complexes. Nonetheless, the results demonstrate that the formation of a stable SREBP-1a:DNA complex is markedly enhanced by NF-Y by a process that requires either the ATTGG and/or CCAAT motifs. Figure 5C demonstrates that the formation of the SREBP-1a:DNA or NF-Y:DNA complex is dependent on the presence of all three NF-Y subunits (compare lane 4 with lanes 3, 5, 6, and 7). DISCUSSION In the current studies we demonstrate that the regulated expression of FPP synthase promoter-reporter genes by sterols is dependent on three cis-elements in the proximal promoter. We have termed this 61 bp fragment a sterol regulatory unit because mutation in any of the three elements interferes with the regulation of reporter genes by sterols. The stimulated expression of FPP synthase promoter-reporter genes that occurs when cells are incubated either under conditions of sterol deprivation or when the cells are co-transfected with a plasmid encoding mature SREBP-1a, is dependent on the binding of SREBP-1a to SRE-3. However, the binding of SREBP-1a to SRE-3 is itself dependent on the binding of NF-Y to the ATTGG and CCAAT motifs. These latter two motifs lie 20 bp and 10 bp 5 and 3, respectively, of SRE-3. The current studies demonstrate that the CCAAT motif, 3 of SRE-3, is also critical for the sterol- and SREBP-1a-regulated expression of FPP synthase promoter-reporter genes (Figs. 2 and 3), presumably because of its role in stimulating the binding of SREBP to SRE-3 (Fig. 5). The in vitro gel mobility shift studies utilized truncated recombinant NF-YA, B, and C proteins that are devoid of activation domains but are able to form a heterotrimeric core complex that binds DNA. The binding of this heterotrimeric NF-Y core to either the ATTGG or CCAAT motifs in the FPP synthase promoter is required and sufficient to stimulate the binding of SREBP-1a to its cognate DNA binding site (Fig. 5). Other nuclear proteins are not necessary for the observed synergism. We hypothesize that the NF-Y-dependent increase in SREBP-1a:DNA complex formation is a result of altered conformation of the SREBP-1a bound to the DNA. This hypothesis is based on previous studies in which both the heterotrimeric full length and truncated NF-Y subunits were shown to bend DNA (24) (R. Mantovani, unpublished results). Further experiments will be necessary to determine whether the synergistic binding of SREBP-1a to the SRE-3 motif is dependent on this NF-Y-induced bending of the DNA. Studies with other transcriptional factors, including Ets-1 (33), p53 (34), and the TATA binding protein (35) demonstrate that truncated forms of these proteins exhibit increased DNA binding activity relative to the full-length proteins. Such results are consistent with the presence of inhibitory protein domains that are deleted from the truncated proteins. In more recent studies, BamHI endonuclease was shown to undergo a conformational change upon binding to DNA that disordered an inhibitory domain of the protein (36). The protein:dna interactions in these latter studies did not require the presence of a second DNA binding protein. In contrast, the current studies demonstrate that SREBP-1a conformation/affinity for DNA is altered in an NF-Y:DNA-dependent manner. Further studies will be required to determine whether SREBP-1a also contains an inhibitory domain that modulates the interaction with DNA. Induced expression of reporter genes in steroldeprived cells has been observed for a number of different constructs; reporter genes containing promoters derived from the genes encoding either FPP synthase, HMG-CoA synthase (2, 37), SREBP-2 (14), squalene synthase (17), or glycerol-3-phosphate acyltransferase (15) are dependent on the presence of binding sites 774 Journal of Lipid Research Volume 39, 1998
9 for both SREBP and NF-Y. Based on the current studies, we hypothesize that SREBP and NF-Y will bind synergistically to the promoters of the latter four genes and result in transcriptional activation. In contrast, under similar conditions of cellular sterol deprivation, high levels of expression of reporter genes under the control of promoters derived from either the LDL receptor (10, 11, 38, 39), fatty acid synthase (12) or acetyl CoA carboxylase (13) genes, require that the promoters contain binding sites for SREBP and Sp1. It is possible that transcriptional activation of the FPP synthase gene observed in sterol-depleted cells is dependent in part on the bending of DNA that has been shown to occur when NF-Y binds to its cognate site (24) and in part on the interaction of SREBP with other nuclear proteins, including CBP (40). Further studies should determine whether CBP and/or other associated proteins are involved in the NF-Y- and SREBP-dependent expression of genes encoding FPP synthase, HMG-CoA synthase, squalene synthase, SREBP-2, and glycerol-3-phosphate acyltransferase. We thank Dr. T. Osborne of the University of California, Irvine for providing the plasmids encoding SREBP-1a and Dr. B. de Crombrugghe of The University of Texas for antibody to CBF-A. We also acknowledge Bernard Lee for his technical contributions. The work was supported by Grant HL from the National Institutes of Health, and a grant from the Laubisch fund. J. E. is the recipient of an American Heart Association, Greater Los Angeles Affiliate Post-doctoral Fellowship. Manuscript received 19 September 1997 and in revised form 17 December REFERENCES 1. Ericsson, J., S. M. Jackson, B. C. Lee, and P. A. Edwards Sterol regulatory element binding protein binds to a novel cis element in the promoter of the farnesyl diphosphate synthase gene. Proc. Natl. Acad. Sci. USA. 93: Jackson, S. M., J. Ericsson, T. F. Osborne, and P. A. Edwards NF-Y has a novel role in sterol-dependent transcription of two cholesterogenic genes. J. Biol. Chem. 270: Spear, D. H., J. Ericsson, S. M. Jackson, and P. A. Edwards Identification of a 6-base pair element involved in the sterol-mediated transcriptional regulation of farnesyl diphosphate synthase. J. Biol. Chem. 269: Ericsson, J., S. M. Jackson, and P. A. Edwards Synergistic binding of sterol regulatory element-binding protein and NF-Y to the farnesyl diphosphate synthase promoter is critical for sterol-regulated expression of the gene. J. Biol. Chem. 271: Hua, X., C. Yokoyama, J. Wu, M. R. Briggs, M. S. Brown, and J. L. Goldstein SREBP-2, a second basic-helixloop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc. Natl. Acad. Sci. USA. 90: Wang, X., M. R. Briggs, X. Hua, C. Yokoyama, J. L. Goldstein, and M. S. Brown Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. J. Biol. Chem. 268: Tontonoz, P., J. B. Kim, R. A. Graves, and B. M. Spiegelman ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol. Cell. Biol. 13: Kim, J. B., G. D. Spotts, Y-D. Halvorsen, H-M. Shih, T. Ellenberger, H. C. Towle, and B. M. Spiegelman Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol. Cell. Biol. 15: Magana, M. M., and T. F. Osborne Two tandem binding sites for sterol regulatory element binding proteins are required for sterol regulation of fatty-acid synthase promoter. J. Biol. Chem. 271: Sanchez, H. B., L. Yieh, and T. F. Osborne Cooperation by sterol regulatory element-binding protein and Sp1 in sterol regulation of low density lipoprotein receptor gene. J. Biol. Chem. 270: Yieh, L., H. B. Sanchez, and T. F. Osborne Domains of transcription factor Sp1 required for synergistic activation with sterol regulatory element binding protein 1 of low density lipoprotein receptor promoter. Proc. Natl. Acad. Sci. USA. 92: Bennett, M. K., J. M. Lopez, H. B. Sanchez, and T. F. Osborne Sterol regulation of fatty acid synthase promoter: coordinate feedback regulation of two major lipid pathways. J. Biol. Chem. 270: Lopez, J. M., M. K. Bennett, H. B. Sanchez, and T. F. Osborne Sterol regulation of acetyl coenzyme A carboxylase: a mechanism for coordinate control of cellular lipid. Proc. Natl. Acad. Sci. USA. 93: Sato, R., J. Inoue, Y. Kawabe, T. Kodama, T. Takano, and M. Maeda Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J. Biol. Chem. 271: Ericsson, J., S. M. Jackson, J. B. Kim, B. M. Spiegelman, and P. A. Edwards Identification of glycerol-3-phosphate acyltransferase as an adipocyte determination and differentiation factor 1- and sterol regulatory elementbinding protein-responsive gene. J. Biol. Chem. 272: Guan, G., G. Jiang, R. L. Koch, and I. Shechter Molecular cloning and functional analysis of the promoter of the human squalene synthase gene. J. Biol. Chem. 270: Guan, G., P-H. Dai, T. F. Osborne, J. B. Kim, and I. Shechter Multiple sequence elements are involved in the transcriptional regulation of the human squalene synthase gene. J. Biol. Chem. 272: Yokoyama, C., X. Wang, M. R. Briggs, A. Admon, J. Wu, X. Hua, J. L. Goldstein, and M. S. Brown SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 75: Pascal, E., and R. Tjian Different activation domains of Sp1 govern formation of multimers and mediate transcriptional synergism. Genes Dev. 5: Coustry, F., S. N. Maity, and B. de Crombrugghe Jackson et al. Synergistic activation of transcription by NF-Y and SREBP 775
10 Studies on transcription activation by the multimeric CCAAT-binding factor CBF. J. Biol. Chem. 270: Bucher, P Weight matrix descriptions of four eukaryotic RNA polymerase promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 212: Chodosh, L. A., A. S. Baldwin, R. W. Carthew, and P. Sharp Human CCAAT-binding proteins have heterologous subunits. Cell. 53: Dorn, A., J. Bollekens, A. Staub, C. Benoist, and D. Mathis A multiplicity of CCAAT box-binding proteins. Cell. 50: Ronchi, A., M. Bellorini, N. Mongelli, and R. Mantovani CAAT-box binding protein NF-Y (CBF, CP1) recognizes the minor groove and distorts DNA. Nucleic Acids Res. 23: Sinha, S., S. N. Maity, J. Lu, and B. de Crombrugghe Recombinant rat CBF-C, the third subunit of CBF/ NFY, allows formation of a protein-dna complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc. Natl. Acad. Sci. USA. 92: Lum, L. S. Y., S. Hsu, M. Vaewhongs, and B. Wu The hsp70 gene CCAAT-binding factor mediates transcriptional activation by the adenovirus Ela protein. Mol. Cell. Biol. 12: Spear, D. H., S. Y. Kutsunai, C. C. Correll, and P. A. Edwards Molecular cloning and promoter analysis of the rat farnesyl diphosphate synthase gene. J. Biol. Chem. 267: Mantovani, R., X-Y. Li, U. Pessara, R. H. van Huisjduijnen, C. Benoist, and D. Mathis Dominant negative analogs of NF-YA. J. Biol. Chem. 269: Sohn, K. Y., S. N. Maity, and B. de Crombrugghe Studies on the structure of the mouse CBF-A gene and properties of a truncated CBF-A isoform generated from an alternatively spliced RNA. Gene. 139: Bellorini, M., K. Zemzoumi, A. Farina, J. Berthelsen, G. Piaggio, and R. Mantovani Cloning and expression of human NF-YC. Gene. 193: Sambrook, J., E. F. Fritsch, and T. Maniatis Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Plainview, NY. 32. van Huijsduijnen, R. H., J. Bollekens, A. Dorn, C. Benoist, and D. Mathis Properties of a CCAAT box-binding protein. Nucleic Acids Res. 15: Petersen, J. M., J. J. Skalicky, L. W. Donaldson, L. P. McIntosh, T. Alber, and B. J. Graves Modulation of transcription factor Ets-1 DNA binding: DNA-induced unfolding of an alpha helix. Science. 269: Hupp, T. R., D. W. Meek, C. A. Midgley, and D. P. Lane Regulation of the specific DNA binding function of p53. Cell. 71: Lieberman, P. M., M. C. Schmidt, C. C. Kao, and A. J. Berke Two distinct domains in the yeast transcription factor IID and evidence for a TATA box-induced conformational change. Mol. Cell. Biol. 11: Newman, M., T. S. Strzelecka, L. F. Dorner, I. Schildkraut, and A. K. Aggarwal Structure of Bam H1 endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science. 269: Smith, J. R., T. F. Osborne, M. S. Brown, J. L. Goldstein, and G. Gil Multiple sterol regulatory elements in promoter for hamster 3-hydroxy-3-methylglutaryl-coenzyme A synthase. J. Biol. Chem. 263: Briggs, M. R., C. Yokoyama, X. Wang, M. S. Brown, and J. L. Goldstein Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. J. Biol. Chem. 268: Sudhof, T. C., D. R. Van der Westhuyzen, J. L. Goldstein, M. S. Brown, and D. W. Russell Three direct repeats and a TATA-like sequence are required for regulated expression of the human low density lipoprotein receptor gene. J. Biol. Chem. 262: Oliner, J. D., J. M. Andresen, S. K. Hansen, S. Zhou, and R. Tjian SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes Dev. 10: Journal of Lipid Research Volume 39, 1998
In vitro DNase I foot printing. In vitro DNase I footprinting was performed as described
 Supplemental Methods In vitro DNase I foot printing. In vitro DNase I footprinting was performed as described previously 1 2 using 32P-labeled 211 bp fragment from 3 HS1. Footprinting reaction mixes contained
Supplemental Methods In vitro DNase I foot printing. In vitro DNase I footprinting was performed as described previously 1 2 using 32P-labeled 211 bp fragment from 3 HS1. Footprinting reaction mixes contained
Fatty acid synthase (FAS) plays a central role in de novo
 Nutritional regulation of the fatty acid synthase promoter in vivo: Sterol regulatory element binding protein functions through an upstream region containing a sterol regulatory element Maria-Jesus Latasa,
Nutritional regulation of the fatty acid synthase promoter in vivo: Sterol regulatory element binding protein functions through an upstream region containing a sterol regulatory element Maria-Jesus Latasa,
Abstract. Introduction
 Activation of Cholesterol Synthesis in Preference to Fatty Acid Synthesis in Liver and Adipose Tissue of Transgenic Mice Overproducing Sterol Regulatory Element-binding Protein-2 Jay D. Horton,* Iichiro
Activation of Cholesterol Synthesis in Preference to Fatty Acid Synthesis in Liver and Adipose Tissue of Transgenic Mice Overproducing Sterol Regulatory Element-binding Protein-2 Jay D. Horton,* Iichiro
SREBP-1 integrates the actions of thyroid hormone, insulin, camp, and medium-chain fatty acids on ACC transcription in hepatocytes
 SREBP-1 integrates the actions of thyroid hormone, insulin, camp, and medium-chain fatty acids on ACC transcription in hepatocytes Yanqiao Zhang, Liya Yin, and F. Bradley Hillgartner 1 Department of Biochemistry
SREBP-1 integrates the actions of thyroid hormone, insulin, camp, and medium-chain fatty acids on ACC transcription in hepatocytes Yanqiao Zhang, Liya Yin, and F. Bradley Hillgartner 1 Department of Biochemistry
CTP:phosphocholine cytidylyltransferase, a new steroland SREBP-responsive gene
 CTP:phosphocholine cytidylyltransferase, a new steroland SREBP-responsive gene Heidi Rachelle Kast,* Catherine M. Nguyen,* Andrew M. Anisfeld,* Johan Ericsson, and Peter A. Edwards 1, *, Departments of
CTP:phosphocholine cytidylyltransferase, a new steroland SREBP-responsive gene Heidi Rachelle Kast,* Catherine M. Nguyen,* Andrew M. Anisfeld,* Johan Ericsson, and Peter A. Edwards 1, *, Departments of
Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver
 Proc. Natl. Acad. Sci. USA Vol. 94, pp. 12354 12359, November 1997 Biochemistry Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver (cholesterol biosynthesis
Proc. Natl. Acad. Sci. USA Vol. 94, pp. 12354 12359, November 1997 Biochemistry Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver (cholesterol biosynthesis
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation
 Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes
 Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes Michiyo Amemiya-Kudo,* Hitoshi Shimano, 1, Alyssa H. Hasty,* Naoya Yahagi,*
Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes Michiyo Amemiya-Kudo,* Hitoshi Shimano, 1, Alyssa H. Hasty,* Naoya Yahagi,*
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
Sensitization of the HIV-1-LTR upon Long Term Low Dose Oxidative Stress*
 THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 271, No. 36, Issue of September 6, pp. 21798 21802, 1996 1996 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Sensitization
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 271, No. 36, Issue of September 6, pp. 21798 21802, 1996 1996 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Sensitization
Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene
 Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene YUELIN ZHANG, WEIHUA FAN, MARK KINKEMA, XIN LI, AND
Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene YUELIN ZHANG, WEIHUA FAN, MARK KINKEMA, XIN LI, AND
Life Sciences 1A Midterm Exam 2. November 13, 2006
 Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
/05/$15.00/0 Molecular Endocrinology 19(5): Copyright 2005 by The Endocrine Society doi: /me
 0888-8809/05/$15.00/0 Molecular Endocrinology 19(5):1343 1360 Printed in U.S.A. Copyright 2005 by The Endocrine Society doi: 10.1210/me.2003-0493 Elevated Glucose Attenuates Human Insulin Gene Promoter
0888-8809/05/$15.00/0 Molecular Endocrinology 19(5):1343 1360 Printed in U.S.A. Copyright 2005 by The Endocrine Society doi: 10.1210/me.2003-0493 Elevated Glucose Attenuates Human Insulin Gene Promoter
AP Biology Summer Assignment Chapter 3 Quiz
 AP Biology Summer Assignment Chapter 3 Quiz 2016-17 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. All of the following are found in a DNA nucleotide
AP Biology Summer Assignment Chapter 3 Quiz 2016-17 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. All of the following are found in a DNA nucleotide
Received 20 August 1997/Accepted 17 November 1997
 JOURNAL OF VIROLOGY, Mar. 1998, p. 2113 2124 Vol. 72, No. 3 0022-538X/98/$04.00 0 Copyright 1998, American Society for Microbiology Overexpression of C/EBP Represses Human Papillomavirus Type 18 Upstream
JOURNAL OF VIROLOGY, Mar. 1998, p. 2113 2124 Vol. 72, No. 3 0022-538X/98/$04.00 0 Copyright 1998, American Society for Microbiology Overexpression of C/EBP Represses Human Papillomavirus Type 18 Upstream
Cleavage of Sterol Regulatory Element-binding Proteins (SREBPs) at Site-1 Requires Interaction with SREBP Cleavage-activating Protein
 THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 273, No. 10, Issue of March 6, pp. 5785 5793, 1998 1998 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Cleavage of Sterol
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 273, No. 10, Issue of March 6, pp. 5785 5793, 1998 1998 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Cleavage of Sterol
The nuclear pre-mrna introns of eukaryotes are removed by
 U4 small nuclear RNA can function in both the major and minor spliceosomes Girish C. Shukla and Richard A. Padgett* Department of Molecular Biology, Lerner Research Institute, Cleveland Clinic Foundation,
U4 small nuclear RNA can function in both the major and minor spliceosomes Girish C. Shukla and Richard A. Padgett* Department of Molecular Biology, Lerner Research Institute, Cleveland Clinic Foundation,
Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes
 Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes The major site of acetoacetate and 3-hydorxybutyrate production is in the liver. 3-hydorxybutyrate is the
Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes The major site of acetoacetate and 3-hydorxybutyrate production is in the liver. 3-hydorxybutyrate is the
Activation of Gene Expression by Human Herpes Virus 6
 Activation of Gene Expression by Human Herpes Virus 6 M. E. M. Campbell and S. McCorkindale 1 Introduction Human herpes virus type 6 (HHV-6) was first detected by Salahuddin et al. [6] and has been isolated
Activation of Gene Expression by Human Herpes Virus 6 M. E. M. Campbell and S. McCorkindale 1 Introduction Human herpes virus type 6 (HHV-6) was first detected by Salahuddin et al. [6] and has been isolated
REGULATORY MECHANISMS OF TRANSCRIPTION FACTOR FUNCTION
 Transcription Regulation And Gene Expression in Eukaryotes Cycle G2 (lecture 13709) RG Clerc 04.04.2012 REGULATORY MECHANISMS OF TRANSCRIPTION FACTOR FUNCTION Protein synthesized Protein phosphorylated
Transcription Regulation And Gene Expression in Eukaryotes Cycle G2 (lecture 13709) RG Clerc 04.04.2012 REGULATORY MECHANISMS OF TRANSCRIPTION FACTOR FUNCTION Protein synthesized Protein phosphorylated
Materials and Methods , The two-hybrid principle.
 The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
Intramembrane glycine mediates multimerization of Insig-2, a requirement for sterol regulation in Chinese hamster ovary cells
 Intramembrane glycine mediates multimerization of Insig-2, a requirement for sterol regulation in Chinese hamster ovary cells Peter C. W. Lee * and Russell A. DeBose-Boyd 1, Howard Hughes Medical Institute
Intramembrane glycine mediates multimerization of Insig-2, a requirement for sterol regulation in Chinese hamster ovary cells Peter C. W. Lee * and Russell A. DeBose-Boyd 1, Howard Hughes Medical Institute
Supplemental Figure 1
 1 Supplemental Figure 1 Effects of DATE shortening on HGF promoter activity. The HGF promoter region (-1037 to +56) containing wild-type (30As) or truncated DATE (26As, 27As, 28A, 29As) from breast cancer
1 Supplemental Figure 1 Effects of DATE shortening on HGF promoter activity. The HGF promoter region (-1037 to +56) containing wild-type (30As) or truncated DATE (26As, 27As, 28A, 29As) from breast cancer
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004
 Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Structural vs. nonstructural proteins
 Why would you want to study proteins associated with viruses or virus infection? Receptors Mechanism of uncoating How is gene expression carried out, exclusively by viral enzymes? Gene expression phases?
Why would you want to study proteins associated with viruses or virus infection? Receptors Mechanism of uncoating How is gene expression carried out, exclusively by viral enzymes? Gene expression phases?
Direct Repression of Splicing by transformer-2
 Direct Repression of Splicing by transformer-2 Dawn S. Chandler, Junlin Qi and William Mattox Mol. Cell. Biol. 2003, 23(15):5174. DOI: 10.1128/MCB.23.15.5174-5185.2003. REFERENCES CONTENT ALERTS Updated
Direct Repression of Splicing by transformer-2 Dawn S. Chandler, Junlin Qi and William Mattox Mol. Cell. Biol. 2003, 23(15):5174. DOI: 10.1128/MCB.23.15.5174-5185.2003. REFERENCES CONTENT ALERTS Updated
/06/$15.00/0 Molecular Endocrinology 20(9): Copyright 2006 by The Endocrine Society doi: /me
 0888-8809/06/$15.00/0 Molecular Endocrinology 20(9):2062 2079 Printed in U.S.A. Copyright 2006 by The Endocrine Society doi: 10.1210/me.2005-0316 Androgens, Progestins, and Glucocorticoids Induce Follicle-Stimulating
0888-8809/06/$15.00/0 Molecular Endocrinology 20(9):2062 2079 Printed in U.S.A. Copyright 2006 by The Endocrine Society doi: 10.1210/me.2005-0316 Androgens, Progestins, and Glucocorticoids Induce Follicle-Stimulating
2.5. AMPK activity
 Supplement Fig. A 3 B phos-ampk 2.5 * Control AICAR AMPK AMPK activity (Absorbance at 45 nm) 2.5.5 Control AICAR Supplement Fig. Effects of AICAR on AMPK activation in macrophages. J774. macrophages were
Supplement Fig. A 3 B phos-ampk 2.5 * Control AICAR AMPK AMPK activity (Absorbance at 45 nm) 2.5.5 Control AICAR Supplement Fig. Effects of AICAR on AMPK activation in macrophages. J774. macrophages were
Received 26 January 1996/Returned for modification 28 February 1996/Accepted 15 March 1996
 MOLECULAR AND CELLULAR BIOLOGY, June 1996, p. 3012 3022 Vol. 16, No. 6 0270-7306/96/$04.00 0 Copyright 1996, American Society for Microbiology Base Pairing at the 5 Splice Site with U1 Small Nuclear RNA
MOLECULAR AND CELLULAR BIOLOGY, June 1996, p. 3012 3022 Vol. 16, No. 6 0270-7306/96/$04.00 0 Copyright 1996, American Society for Microbiology Base Pairing at the 5 Splice Site with U1 Small Nuclear RNA
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
L I F E S C I E N C E S
 1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 OCTOBER 31, 2006
1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 OCTOBER 31, 2006
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Design of isolated protein and RNC constructs, and homogeneity of purified RNCs. (a) Schematic depicting the design and nomenclature used for all the isolated proteins and RNCs used
Supplementary Figure 1 Design of isolated protein and RNC constructs, and homogeneity of purified RNCs. (a) Schematic depicting the design and nomenclature used for all the isolated proteins and RNCs used
ZHIHONG JIANG, 1 JOCELYN COTE, 1 JENNIFER M. KWON, 2 ALISON M. GOATE, 3
 MOLECULAR AND CELLULAR BIOLOGY, June 2000, p. 4036 4048 Vol. 20, No. 11 0270-7306/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Aberrant Splicing of tau Pre-mRNA Caused
MOLECULAR AND CELLULAR BIOLOGY, June 2000, p. 4036 4048 Vol. 20, No. 11 0270-7306/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Aberrant Splicing of tau Pre-mRNA Caused
RayBio Human PPAR-alpha Transcription Factor Activity Assay Kit
 RayBio Human PPAR-alpha Transcription Factor Activity Assay Kit Catalog #: TFEH-PPARa User Manual Jan 5, 2018 3607 Parkway Lane, Suite 200 Norcross, GA 30092 Tel: 1-888-494-8555 (Toll Free) or 770-729-2992,
RayBio Human PPAR-alpha Transcription Factor Activity Assay Kit Catalog #: TFEH-PPARa User Manual Jan 5, 2018 3607 Parkway Lane, Suite 200 Norcross, GA 30092 Tel: 1-888-494-8555 (Toll Free) or 770-729-2992,
ADD1 SREBP1 activates PPAR through the production of endogenous ligand (fatty acid metabolism nuclear hormone receptor adipocyte differentiation)
 Proc. Natl. Acad. Sci. USA Vol. 95, pp. 4333 4337, April 1998 Cell Biology ADD1 SREBP1 activates PPAR through the production of endogenous ligand (fatty acid metabolism nuclear hormone receptor adipocyte
Proc. Natl. Acad. Sci. USA Vol. 95, pp. 4333 4337, April 1998 Cell Biology ADD1 SREBP1 activates PPAR through the production of endogenous ligand (fatty acid metabolism nuclear hormone receptor adipocyte
Polyomaviridae. Spring
 Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced to express
 Supplementary Figure 1. VapC-mt4 cleaves trna Ala2 in E. coli. Histograms representing the fold change in reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced
Supplementary Figure 1. VapC-mt4 cleaves trna Ala2 in E. coli. Histograms representing the fold change in reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced
Acetyl-CoA or BC acyl-coa. Int1-ACP. FabH. FabF. Anti-FabF Platensimycin ACP. Fak
 Initiation Acetyl-CoA AccBCDA FabZ Int-ACP Elongation [N+] FabG Int1-ACP Acetyl-CoA or BC acyl-coa FabH FabD CoA Malonyl-ACP Malonyl CoA ACP Anti-FabI Triclosan AFN-15 FabI acyl-acp PlsX PlsY Pls C Lipoic
Initiation Acetyl-CoA AccBCDA FabZ Int-ACP Elongation [N+] FabG Int1-ACP Acetyl-CoA or BC acyl-coa FabH FabD CoA Malonyl-ACP Malonyl CoA ACP Anti-FabI Triclosan AFN-15 FabI acyl-acp PlsX PlsY Pls C Lipoic
Supplementary Figures. T Cell Factor-1 initiates T helper 2 fate by inducing GATA-3 and repressing Interferon-γ
 Supplementary Figures T Cell Factor-1 initiates T helper 2 fate by inducing GATA-3 and repressing Interferon-γ Qing Yu, Archna Sharma, Sun Young Oh, Hyung-Geun Moon, M. Zulfiquer Hossain, Theresa M. Salay,
Supplementary Figures T Cell Factor-1 initiates T helper 2 fate by inducing GATA-3 and repressing Interferon-γ Qing Yu, Archna Sharma, Sun Young Oh, Hyung-Geun Moon, M. Zulfiquer Hossain, Theresa M. Salay,
RECAP (1)! In eukaryotes, large primary transcripts are processed to smaller, mature mrnas.! What was first evidence for this precursorproduct
 RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
MCB130 Midterm. GSI s Name:
 1. Peroxisomes are small, membrane-enclosed organelles that function in the degradation of fatty acids and in the degradation of H 2 O 2. Peroxisomes are not part of the secretory pathway and peroxisomal
1. Peroxisomes are small, membrane-enclosed organelles that function in the degradation of fatty acids and in the degradation of H 2 O 2. Peroxisomes are not part of the secretory pathway and peroxisomal
7.06 Cell Biology EXAM #3 April 24, 2003
 7.06 Spring 2003 Exam 3 Name 1 of 8 7.06 Cell Biology EXAM #3 April 24, 2003 This is an open book exam, and you are allowed access to books and notes. Please write your answers to the questions in the
7.06 Spring 2003 Exam 3 Name 1 of 8 7.06 Cell Biology EXAM #3 April 24, 2003 This is an open book exam, and you are allowed access to books and notes. Please write your answers to the questions in the
RECAP (1)! In eukaryotes, large primary transcripts are processed to smaller, mature mrnas.! What was first evidence for this precursorproduct
 RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
Loss of Calreticulin Uncovers a Critical Role for Calcium in Regulating Cellular Lipid Homeostasis
 SUPPLEMENTARY MATERIAL Loss of Calreticulin Uncovers a Critical Role for Calcium in Regulating Cellular Lipid Homeostasis Wen-An Wang 1, Wen-Xin Liu 1, Serpen Durnaoglu 2, Sun-Kyung Lee 2, Jihong Lian
SUPPLEMENTARY MATERIAL Loss of Calreticulin Uncovers a Critical Role for Calcium in Regulating Cellular Lipid Homeostasis Wen-An Wang 1, Wen-Xin Liu 1, Serpen Durnaoglu 2, Sun-Kyung Lee 2, Jihong Lian
Overview of the Expressway Cell-Free Expression Systems. Expressway Mini Cell-Free Expression System
 Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
SREBF2 Transcription Factor Assay Kit ( Cat # KA1379 V.01 ) 1
 Background Lipid homeostasis in vertebrate cells is regulated by a family of transcription factors called sterol regulatory element binding proteins (SREBP s). SREBP s directly activate the expression
Background Lipid homeostasis in vertebrate cells is regulated by a family of transcription factors called sterol regulatory element binding proteins (SREBP s). SREBP s directly activate the expression
Blunted Feedback Suppression of SREBP Processing by Dietary Cholesterol in Transgenic Mice Expressing Sterol-Resistant SCAP(D443N)
 Blunted Feedback Suppression of SREBP Processing by Dietary Cholesterol in Transgenic Mice Expressing Sterol-Resistant SCAP(D443N) Bobby S. Korn,* Iichiro Shimomura,* Yuriy Bashmakov,* Robert E. Hammer,
Blunted Feedback Suppression of SREBP Processing by Dietary Cholesterol in Transgenic Mice Expressing Sterol-Resistant SCAP(D443N) Bobby S. Korn,* Iichiro Shimomura,* Yuriy Bashmakov,* Robert E. Hammer,
Luminescent platforms for monitoring changes in the solubility of amylin and huntingtin in living cells
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Chapter 2 Gene and Promoter Structures of the Dopamine Receptors
 Chapter 2 Gene and Promoter Structures of the Dopamine Receptors Ursula M. D Souza Abstract The dopamine receptors have been classified into two groups, the D 1 - like and D 2 -like dopamine receptors,
Chapter 2 Gene and Promoter Structures of the Dopamine Receptors Ursula M. D Souza Abstract The dopamine receptors have been classified into two groups, the D 1 - like and D 2 -like dopamine receptors,
BIO360 Fall 2013 Quiz 1
 BIO360 Fall 2013 Quiz 1 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function mutation.
BIO360 Fall 2013 Quiz 1 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function mutation.
Eukaryotic transcription (III)
 Eukaryotic transcription (III) 1. Chromosome and chromatin structure Chromatin, chromatid, and chromosome chromatin Genomes exist as chromatins before or after cell division (interphase) but as chromatids
Eukaryotic transcription (III) 1. Chromosome and chromatin structure Chromatin, chromatid, and chromosome chromatin Genomes exist as chromatins before or after cell division (interphase) but as chromatids
Supplemental Figures
 Supplemental Figures Supplemental Figure 1. Fasting-dependent regulation of the SREBP ortholog SBP-1 and lipid homeostasis mediated by the SIRT1 ortholog SIR-2.1 in C. elegans. (A) Wild-type or sir-2.1(lof)
Supplemental Figures Supplemental Figure 1. Fasting-dependent regulation of the SREBP ortholog SBP-1 and lipid homeostasis mediated by the SIRT1 ortholog SIR-2.1 in C. elegans. (A) Wild-type or sir-2.1(lof)
Supplemental Figure 1 ELISA scheme to measure plasma total, mature and furin-cleaved
 1 Supplemental Figure Legends Supplemental Figure 1 ELISA scheme to measure plasma total, mature and furin-cleaved PCSK9 concentrations. 4 Plasma mature and furin-cleaved PCSK9s were measured by a sandwich
1 Supplemental Figure Legends Supplemental Figure 1 ELISA scheme to measure plasma total, mature and furin-cleaved PCSK9 concentrations. 4 Plasma mature and furin-cleaved PCSK9s were measured by a sandwich
Differentiation-induced Changes of Mediterranean Fever Gene (MEFV) Expression in HL-60 Cell
 Differentiation-induced Changes of Mediterranean Fever Gene (MEFV) Expression in HL-60 Cell Wenxin Li Department of Biological Sciences Fordham University Abstract MEFV is a human gene that codes for an
Differentiation-induced Changes of Mediterranean Fever Gene (MEFV) Expression in HL-60 Cell Wenxin Li Department of Biological Sciences Fordham University Abstract MEFV is a human gene that codes for an
7.012 Quiz 3 Answers
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
Zool 3200: Cell Biology Exam 4 Part II 2/3/15
 Name:Key Trask Zool 3200: Cell Biology Exam 4 Part II 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Name:Key Trask Zool 3200: Cell Biology Exam 4 Part II 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Mammalian Tissue Protein Extraction Reagent
 Mammalian Tissue Protein Extraction Reagent Catalog number: AR0101 Boster s Mammalian Tissue Protein Extraction Reagent is a ready-to-use Western blot related reagent solution used for efficient extraction
Mammalian Tissue Protein Extraction Reagent Catalog number: AR0101 Boster s Mammalian Tissue Protein Extraction Reagent is a ready-to-use Western blot related reagent solution used for efficient extraction
Supplementary information
 Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Regulation of expression of glial filament acidic protein
 Regulation of expression of glial filament acidic protein SRIJATA SARKAR and NICHOLAS J. COWAN Department of Biochemistry, NYU Medical Center, 550 First Avenue, New York, NY 10016, USA Summary The regulation
Regulation of expression of glial filament acidic protein SRIJATA SARKAR and NICHOLAS J. COWAN Department of Biochemistry, NYU Medical Center, 550 First Avenue, New York, NY 10016, USA Summary The regulation
Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression
 Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Use of double- stranded DNA mini- circles to characterize the covalent topoisomerase- DNA complex
 SUPPLEMENTARY DATA Use of double- stranded DNA mini- circles to characterize the covalent topoisomerase- DNA complex Armêl Millet 1, François Strauss 1 and Emmanuelle Delagoutte 1 1 Structure et Instabilité
SUPPLEMENTARY DATA Use of double- stranded DNA mini- circles to characterize the covalent topoisomerase- DNA complex Armêl Millet 1, François Strauss 1 and Emmanuelle Delagoutte 1 1 Structure et Instabilité
Spliceosome Pathway. is required for a stable interaction between U2 snrnp and
 MOLECULAR AND CELLULAR BIOLOGY, Sept. 1988, p. 3755-3760 Vol. 8, No. 9 0270-7306/88/093755-06$02.00/0 Copyright 1988, American Society for Microbiology Early Commitment of Yeast Pre-mRNA to the Spliceosome
MOLECULAR AND CELLULAR BIOLOGY, Sept. 1988, p. 3755-3760 Vol. 8, No. 9 0270-7306/88/093755-06$02.00/0 Copyright 1988, American Society for Microbiology Early Commitment of Yeast Pre-mRNA to the Spliceosome
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer. Application Note
 Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
The clathrin adaptor Numb regulates intestinal cholesterol. absorption through dynamic interaction with NPC1L1
 The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
Cholesterol metabolism. Function Biosynthesis Transport in the organism Hypercholesterolemia
 Cholesterol metabolism Function Biosynthesis Transport in the organism Hypercholesterolemia - component of all cell membranes - precursor of bile acids steroid hormones vitamin D Cholesterol Sources: dietary
Cholesterol metabolism Function Biosynthesis Transport in the organism Hypercholesterolemia - component of all cell membranes - precursor of bile acids steroid hormones vitamin D Cholesterol Sources: dietary
Supporting Information Table of content
 Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Transient Ribosomal Attenuation Coordinates Protein Synthesis and Co-translational Folding
 SUPPLEMENTARY INFORMATION: Transient Ribosomal Attenuation Coordinates Protein Synthesis and Co-translational Folding Gong Zhang 1,2, Magdalena Hubalewska 1 & Zoya Ignatova 1,2 1 Department of Cellular
SUPPLEMENTARY INFORMATION: Transient Ribosomal Attenuation Coordinates Protein Synthesis and Co-translational Folding Gong Zhang 1,2, Magdalena Hubalewska 1 & Zoya Ignatova 1,2 1 Department of Cellular
Blocking the expression of the hepatitis B virus S gene in hepatocellular carcinoma cell lines with an anti-gene locked nucleic acid in vitro
 Blocking the expression of the hepatitis B virus S gene in hepatocellular carcinoma cell lines with an anti-gene locked nucleic acid in vitro Y.-B. Deng, H.-J. Qin, Y.-H. Luo, Z.-R. Liang and J.-J. Zou
Blocking the expression of the hepatitis B virus S gene in hepatocellular carcinoma cell lines with an anti-gene locked nucleic acid in vitro Y.-B. Deng, H.-J. Qin, Y.-H. Luo, Z.-R. Liang and J.-J. Zou
JBC Papers in Press. Published on January 14, 2002 as Manuscript M
 JBC Papers in Press. Published on January 14, 2002 as Manuscript M109976200 Pig Liver Carnitine Palmitoyltransferase: Chimera Studies Show That Both the N- and C-Terminal Regions of the Enzyme Are Important
JBC Papers in Press. Published on January 14, 2002 as Manuscript M109976200 Pig Liver Carnitine Palmitoyltransferase: Chimera Studies Show That Both the N- and C-Terminal Regions of the Enzyme Are Important
Chemistry 107 Exam 4 Study Guide
 Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Homologous DNA Sequences and Cellular Factors Are Implicated in the Control of Glucagon and Insulin Gene Expression
 MOLECULAR AND CELLULAR BIOLOGY, July 1995, p. 3904 3916 Vol. 15, No. 7 0270-7306/95/$04.00 0 Copyright 1995, American Society for Microbiology Homologous DNA Sequences and Cellular Factors Are Implicated
MOLECULAR AND CELLULAR BIOLOGY, July 1995, p. 3904 3916 Vol. 15, No. 7 0270-7306/95/$04.00 0 Copyright 1995, American Society for Microbiology Homologous DNA Sequences and Cellular Factors Are Implicated
Lipid Metabolism. Catabolism Overview
 Lipid Metabolism Pratt & Cornely, Chapter 17 Catabolism Overview Lipids as a fuel source from diet Beta oxidation Mechanism ATP production Ketone bodies as fuel 1 High energy More reduced Little water
Lipid Metabolism Pratt & Cornely, Chapter 17 Catabolism Overview Lipids as a fuel source from diet Beta oxidation Mechanism ATP production Ketone bodies as fuel 1 High energy More reduced Little water
ARV Mode of Action. Mode of Action. Mode of Action NRTI. Immunopaedia.org.za
 ARV Mode of Action Mode of Action Mode of Action - NRTI Mode of Action - NNRTI Mode of Action - Protease Inhibitors Mode of Action - Integrase inhibitor Mode of Action - Entry Inhibitors Mode of Action
ARV Mode of Action Mode of Action Mode of Action - NRTI Mode of Action - NNRTI Mode of Action - Protease Inhibitors Mode of Action - Integrase inhibitor Mode of Action - Entry Inhibitors Mode of Action
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Genetics. Instructor: Dr. Jihad Abdallah Transcription of DNA
 Genetics Instructor: Dr. Jihad Abdallah Transcription of DNA 1 3.4 A 2 Expression of Genetic information DNA Double stranded In the nucleus Transcription mrna Single stranded Translation In the cytoplasm
Genetics Instructor: Dr. Jihad Abdallah Transcription of DNA 1 3.4 A 2 Expression of Genetic information DNA Double stranded In the nucleus Transcription mrna Single stranded Translation In the cytoplasm
Data Sheet. Notch Pathway Reporter Kit Catalog # 60509
 Data Sheet Notch Pathway Reporter Kit Catalog # 60509 6042 Cornerstone Court W, Ste B Background The Notch signaling pathway controls cell fate decisions in vertebrate and invertebrate tissues. NOTCH signaling
Data Sheet Notch Pathway Reporter Kit Catalog # 60509 6042 Cornerstone Court W, Ste B Background The Notch signaling pathway controls cell fate decisions in vertebrate and invertebrate tissues. NOTCH signaling
TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer
 Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
1. Identify and characterize interesting phenomena! 2. Characterization should stimulate some questions/models! 3. Combine biochemistry and genetics
 1. Identify and characterize interesting phenomena! 2. Characterization should stimulate some questions/models! 3. Combine biochemistry and genetics to gain mechanistic insight! 4. Return to step 2, as
1. Identify and characterize interesting phenomena! 2. Characterization should stimulate some questions/models! 3. Combine biochemistry and genetics to gain mechanistic insight! 4. Return to step 2, as
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism SUPPLEMENTARY FIGURES, LEGENDS AND METHODS
 A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Segment-specific and common nucleotide sequences in the
 Proc. Nati. Acad. Sci. USA Vol. 84, pp. 2703-2707, May 1987 Biochemistry Segment-specific and common nucleotide sequences in the noncoding regions of influenza B virus genome RNAs (viral transcription/viral
Proc. Nati. Acad. Sci. USA Vol. 84, pp. 2703-2707, May 1987 Biochemistry Segment-specific and common nucleotide sequences in the noncoding regions of influenza B virus genome RNAs (viral transcription/viral
RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice
 SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
Part-4. Cell cycle regulatory protein 5 (Cdk5) A novel target of ERK in Carb induced cell death
 Part-4 Cell cycle regulatory protein 5 (Cdk5) A novel target of ERK in Carb induced cell death 95 1. Introduction The process of replicating DNA and dividing cells can be described as a series of coordinated
Part-4 Cell cycle regulatory protein 5 (Cdk5) A novel target of ERK in Carb induced cell death 95 1. Introduction The process of replicating DNA and dividing cells can be described as a series of coordinated
Supplementary Material
 Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Course Title Form Hours subject
 Course Title Form Hours subject Types, and structure of chromosomes L 1 Histology Karyotyping and staining of human chromosomes L 2 Histology Chromosomal anomalies L 2 Histology Sex chromosomes L 1 Histology
Course Title Form Hours subject Types, and structure of chromosomes L 1 Histology Karyotyping and staining of human chromosomes L 2 Histology Chromosomal anomalies L 2 Histology Sex chromosomes L 1 Histology
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system
 Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
TECHNICAL BULLETIN. R 2 GlcNAcβ1 4GlcNAcβ1 Asn
 GlycoProfile II Enzymatic In-Solution N-Deglycosylation Kit Product Code PP0201 Storage Temperature 2 8 C TECHNICAL BULLETIN Product Description Glycosylation is one of the most common posttranslational
GlycoProfile II Enzymatic In-Solution N-Deglycosylation Kit Product Code PP0201 Storage Temperature 2 8 C TECHNICAL BULLETIN Product Description Glycosylation is one of the most common posttranslational
7.06 Cell Biology Exam #3 April 23, 2002
 RECITATION TA: NAME: 7.06 Cell Biology Exam #3 April 23, 2002 This is an open book exam and you are allowed access to books, notes, and calculators. Please limit your answers to the spaces allotted after
RECITATION TA: NAME: 7.06 Cell Biology Exam #3 April 23, 2002 This is an open book exam and you are allowed access to books, notes, and calculators. Please limit your answers to the spaces allotted after
Unit IV Problem 3 Biochemistry: Cholesterol Metabolism and Lipoproteins
 Unit IV Problem 3 Biochemistry: Cholesterol Metabolism and Lipoproteins - Cholesterol: It is a sterol which is found in all eukaryotic cells and contains an oxygen (as a hydroxyl group OH) on Carbon number
Unit IV Problem 3 Biochemistry: Cholesterol Metabolism and Lipoproteins - Cholesterol: It is a sterol which is found in all eukaryotic cells and contains an oxygen (as a hydroxyl group OH) on Carbon number
A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis
 APPLICATION NOTE Cell-Free DNA Isolation Kit A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis Abstract Circulating cell-free DNA (cfdna) has been shown
APPLICATION NOTE Cell-Free DNA Isolation Kit A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis Abstract Circulating cell-free DNA (cfdna) has been shown
Breast cancer. Risk factors you cannot change include: Treatment Plan Selection. Inferring Transcriptional Module from Breast Cancer Profile Data
 Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Cholesterol and its transport. Alice Skoumalová
 Cholesterol and its transport Alice Skoumalová 27 carbons Cholesterol - structure Cholesterol importance A stabilizing component of cell membranes A precursor of bile salts A precursor of steroid hormones
Cholesterol and its transport Alice Skoumalová 27 carbons Cholesterol - structure Cholesterol importance A stabilizing component of cell membranes A precursor of bile salts A precursor of steroid hormones
hemagglutinin and the neuraminidase genes (RNA/recombinant viruses/polyacrylamide gel electrophoresis/genetics)
 Proc. Natl. Acad. Sci. USA Vol. 73, No. 6, pp. 242-246, June 976 Microbiology Mapping of the influenza virus genome: Identification of the hemagglutinin and the neuraminidase genes (RNA/recombinant viruses/polyacrylamide
Proc. Natl. Acad. Sci. USA Vol. 73, No. 6, pp. 242-246, June 976 Microbiology Mapping of the influenza virus genome: Identification of the hemagglutinin and the neuraminidase genes (RNA/recombinant viruses/polyacrylamide
LS1a Fall 2014 Problem Set #4 Due Monday 11/3 at 6 pm in the drop boxes on the Science Center 2 nd Floor
 LS1a Fall 2014 Problem Set #4 Due Monday 11/3 at 6 pm in the drop boxes on the Science Center 2 nd Floor Note: Adequate space is given for each answer. Questions that require a brief explanation should
LS1a Fall 2014 Problem Set #4 Due Monday 11/3 at 6 pm in the drop boxes on the Science Center 2 nd Floor Note: Adequate space is given for each answer. Questions that require a brief explanation should
