Systems Analysis of Immunity to Influenza Vaccination across Multiple Years and in Diverse Populations Reveals Shared Molecular Signatures
|
|
|
- Cordelia Perkins
- 6 years ago
- Views:
Transcription
1 Immunity Supplemental Information Systems Analysis of Immunity to Influenza Vaccination across Multiple Years and in Diverse Populations Reveals Shared Molecular Signatures Helder I. Nakaya, Thomas Hagan, Sai S. Duraisingham, Eva K. Lee, Marcin Kwissa, Nadine Rouphael, Daniela Frasca, Merril Gersten, Aneesh K. Mehta, Renaud Gaujoux, Gui-Mei Li, Shakti Gupta, Rafi Ahmed, Mark J. Mulligan, Shai Shen-Orr, Bonnie B. Blomberg, Shankar Subramaniam, and Bali Pulendran
2 Supplemental Figures Figure S1, Related to Figure 1. Study Demographics and HAI responses by phenotype. (a) Age distribution of subjects in this study. (b) Demographics of young and elderly subjects in this study. Subjects were only evaluated for type 2 diabetes in the 2011 season. (c) Antibody responses induced by TIV vaccination. The circos plot shows the log 2 fold-induction of HAI titers on day 28 post-vaccination for each one of the three Influenza virus strains (heat maps) or for the highest fold-induction among all 3 strains (i.e. the HAI response; histogram). High antibody responses are shown as red bars in the histogram, while low antibody responses are blue. The y-axis of the histogram ranges from 0-9 in log2, which represents HAI fold changes from Elderly antibody responses (in 2010 and 2011) are marked as pink in the inner circle of the plot. (d) HAI responses of young and elderly subjects. The box plots represent the median and first and third quartiles of HAI response in young (Y) (<65 years) and elderly (E) ( 65 years). *p < 0.03 (for t-test; two-tailed test). Data are from 58 subjects (2010) and 72 subjects (2011). (e) HAI responses by gender (all seasons). (f) HAI responses by type 2 diabetes status (2011 season).
3 Figure S2, Related to Figure 2. Comparison on transcriptional responses by phenotype. (a) Volcano plot of differentially expressed genes (p<0.01 and log 2 fold change>0.2) between male and female subjects on days 3 and 7 post-vaccination (using fold change expression compared to baseline). Green represents genes with increased expression in males, and red represents genes with increased expression in females. (b) Volcano plot of differentially expressed genes (p<0.01 and log 2 fold change>0.2) between type 2 diabetes postitve and negative subjects on day 7 post-vaccination (2011 season, day 3 was not analyzed due to the limited number of type 2 diabetes positive subjects with available day 3 expression data (n=4)). Green represents genes with increased expression in type 2 diabetes negative subjects, and red represents genes with increased expression in type 2 diabetes positive subjects. (c) Heat map of Blood Transcription Modules (BTMs, rows) whose activity at day 7 post-vaccination is associated with HAI response at day 28 after vaccination in type 2 diabetes negative (left column) or positive (right column) subjects (2011 season). Gene Set Enrichment Analysis (GSEA, nominal p<0.05; 1,000 permutations) was used to identify positive (red) or negative (blue) enrichment of BTMs (gene sets) within pre-ranked gene lists, where genes were ranked according to their correlation between expression and HAI response. Intensity of color represents the normalized enrichment score (NES).
4 Figure S3, Related to Figure 2. Multi-step DAMIP strategy. See methods below.
5 Figure S4, Related to Figure 2. BTMs associated with the HAI response in young and elderly subjects from the 2010 season. GSEA (nominal p<0.1; 1,000 permutations) was used to identify positive or negative enrichment of BTMs (gene sets) within pre-ranked gene lists, where genes were ranked according to their correlation between expression and HAI response in young and elderly subjects in the 2010 season. Modules represented are those consistently enriched for both young and elderly subjects, with Normalized Enrichment Score (NES) among Elderly subjects plotted on the y-axis and NES among young subjects plotted on the x-axis.
6 Figure S5, Related to Figure 3. Baseline signatures associated with the antibody response in each season. Heat map of Blood Transcription Modules (BTMs, rows) and TIV seasons (columns) whose activity at baseline is associated with HAI response at day 28 after vaccination. Gene Set Enrichment Analysis (GSEA, nominal p<0.05; 1,000 permutations) was used to identify positive (red), negative (blue) or no (grey) enrichment of BTMs (gene sets) within pre-ranked gene lists, where genes were ranked according to their correlation between expression and HAI response.
7 Figure S6, Related to Figure 5. Flow cytometry analysis of monocytes in young and elderly after TIV vaccination. (a) Blood monocytes were defined within the CD3-CD19-CD14+ PBMC. Dot-blot represents three distinct populations defined by CD14 and CD16 markers: CD14+CD16- monocytes, CD14+CD16+monocytes and CD14dimCD16++ monocytes. (b) Changes in total monocytes population after vaccination represented as % within all PBMC for young (red) and elderly (blue). Mean +/-SEM. (c) Kinetics of CD14+CD16- monocytes, CD14+CD16+monocytes and CD14dimCD16++ monocytes in young (left panel) and elderly (right panel) after vaccination. Mean +/-SEM. Areas Under Curve (AUC) in (b) and (c) were calculated to compare magnitudes of the monocyte subsets between young and elderly cohorts throughout the study duration (days 0-14) and compared by t-test. Changes of each of the monocytes subset on the indicated time-points after vaccination were compared to the day 0 (baseline) timepoint by t-test. (d) Activation of each of the monocyte subsets were assessed by CCR5 (left) and CD86 (right) staining and compared with day 0 (baseline) by t-test. Data are represented as the geometric mean fluorescence intensity (MFI) for young (left panel) and elderly (right panel) at each time point. Mean +/- SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<
8 Figure S7, Related to Figure 5. Correlation between estimated and FACS cell subset proportions on the 2010 trial. α, ρ, and R 2 represent Cronbach s alpha, Pearson correlation coefficient, and coefficient of determination (square of Pearson correlation coefficient), respectively.
9 Supplemental Experimental Procedures Human subjects and influenza vaccines This study is comprised of subjects vaccinated with TIV during the 5 consecutive influenza seasons in, (9 subjects), (28 subjects), (28 subjects), (58 subjects), and (72 subjects). The age of vaccinees ranged from 20 to 86, with 39 and 15 subjects in the and seasons >65 years old, respectively. Subjects were vaccinated with one dose of TIV (Fluarix, GlaxoSmithKline Biologicals, in , , and seasons, Fluvirin, Novartis Vaccines and Diagnostics Limited, in season, and Fluzone, Sanofi Pasteur Inc., in season) following current guidelines for influenza vaccination. Written informed consent was obtained from each subject with institutional review and approval from the Emory University Institutional Review Board. Season Vaccine Brand H1N1 strain H3N2 strain B strain Fluarix-GSK A/Solomon Islands/3/2006 A/Wisconsin/67/2005 B/Malaysia/2506/ Fluarix-GSK A/Brisbane/59/2007 A/Uruguay/716/2007 X- B/Brisbane/3/ Fluvirin-Novartis A/Brisbane/59/2007 A/Brisbane/10/2007 B/Brisbane/60/ Fluzone-Sanofi A/California/7/2009 A/Perth/16/2009 B/Brisbane/60/ Fluarix-GSK & A/California/7/2009 A/Victoria/210/2009 B/Wisconsin/01/ Fluvirin-Novartis Influenza virus strains contained in the vaccines. Cells, plasma and RNA isolation Four blood samples were obtained from influenza vaccinees: at baseline, and at days 3, 7 and 28 (range, day 28 to 32) post-vaccination. Additional samples were collected at days 1, 14, and 180 for selected subjects. Peripheral blood mononuclear cells (PBMCs) and plasma were isolated from fresh blood (CPTs; Vacutainer with Sodium Citrate; BD), following the manufacturer s protocol. PBMCs were frozen in DMSO with 10% FBS and stored at 80 C and then transferred on the next day to liquid nitrogen freezers ( 210 C). Plasma samples from CPTs were stored at 80 C. Trizol (Invitrogen) was used to lyse fresh PBMCs (1 ml of Trizol to ~1.5 x 10 6 cells) and to protect RNA from degradation. Trizol samples were stored at 80 C. Hemagglutination inhibition assays Hemagglutination inhibition (HAI) titers were determined based on the standard WHO protocol, as previously described (Chen et al., 2010). Briefly, plasma samples were treated with receptor destroying enzyme (RDE; Denka Seiken Co.) by adding 1 part plasma to 3 parts RDE and incubating at 37 C overnight. The following morning, the RDE was inactivated by incubating the samples at 56 C for one hour. The samples were then serially diluted with PBS in 96 well v-bottom plates (Nunc, Rochester, NY) and 4 HA units of either the H1N1, H3N2, or influenza B virus was added to each well. After 30 minutes at room temperature, 50 ul of 0.5% turkey RBCs (Rockland Immunochemicals) suspended in PBS with 0.5% BSA was added to each well and the plates were shaken manually. After an additional 30 minutes at room temperature, the plasma titers were read as the reciprocal of the final dilution for which a button was observed. Negative and positive control plasma samples for each virus were used for reference (data not shown). Microarray experiments Total RNA from fresh PBMCs (~1.5 x 10 6 cells) was purified using Trizol (Invitrogen, Life Technologies Corporation) according to the manufacturer's instructions. All RNA samples were checked for purity using a ND-1000 spectrophotometer (NanoDrop Technologies) and for integrity by electrophoresis on a 2100 BioAnalyzer (Agilent Technologies). Two-round in vitro transcription amplification and labeling was performed starting with 50 ng intact, total RNA per sample, following the Affymetrix protocol. After hybridization on Human U133 Plus 2.0 Arrays (using GeneTitan platform, Affymetrix, or individual
10 cartridges) for 16 h at 45 C and 60 r.p.m. in a Hybridization Oven 640 (Affymetrix), slides were washed and stained with a Fluidics Station 450 (Affymetrix). Scanning was performed on a seventh-generation GeneChip Scanner 3000 (Affymetrix), and Affymetrix GCOS software was used to perform image analysis and generate raw intensity data. Initial data quality was assessed by background level, 3 labeling bias, and pairwise correlation among samples. CEL files from outlier samples were excluded and the remaining CEL files of all the samples belonging to the same influenza season trial were grouped, and the microarray intensity data of probe sets (hereafter referred to genes ) were normalized by RMA, which includes global background adjustment and quantile normalization. mirna array experiments Fresh PBMCs (~1.5 x 10 6 cells) were lysed using mirna Lysis Buffer (2ml per subject, HTG Molecular Diagnostics). HTG Molecular Diagnostics core facility extracted and processed the total RNA, and performed the mirna expression profiling according to the manufacturer's instructions. Data was normalized using 40% trimmed mean and log2 transformed. Blood Transcriptional Module Enrichment Probes were first ranked by Pearson correlation with either the day 28 antibody fold change, day 180/ day 28 residual, or subject age using fold change microarray data from days 0, 1, 3, and 7 post-vaccination. Separate rankings were generated for each flu season for the day 28 antibody and age correlations. Probes were then collapsed to Entrez gene IDs, selecting the probe with the smallest correlation p value in cases where multiple probes mapped to a single Entrez ID. Gene set enrichment analysis software (GSEA) was then run in pre-ranked list mode with 1000 permutations to generate enrichment scores for 346 blood transcriptional modules (BTMs) based on the distribution of member genes of each module in the ranked list. Enrichment scores were visualized using the corrplot package in R (Wei, 2012). Individual modules and associated gene coexpression relationships were represented using Cytoscape software (Shannon et al., 2003). Baseline Transcriptional Analysis For analysis of baseline signatures of vaccine response (Fig. 3), ComBat software (Johnson et al., 2007) was applied to the RMA normalized baseline expression values to remove batch effects from each flu season within the study. Probes were then ranked by Pearson correlation between their ComBat normalized day 0 expression (across all 5 seasons combined) and the day 28 antibody fold change. This ranking was used with GSEA as described above to generate a list of enriched BTMs. As validation, this procedure was repeated separately in two independent datasets, from Franco et al. (Franco et al., 2013) and Furman et al. (Furman et al., 2013). The Franco et al. study included two separate cohorts that were analyzed separately. BTMs consistently enriched in 3 out of 4 of the datasets were identified and represented in Fig. 3. Flow Cytometry PBMCs were thawed and stained with the live/dead marker Alexa Fluor 430 (Lifetechnologies) to exclude dead cells. 2-3 x 10 6 cells were stained with an appropriate antibody cocktail. Cells were washed in PBS with 5% fetal bovine serum and fixed with the Cytofix buffer (BD) and analyzed on the LSRII flow cytometer (BD). All flow cytometry analysis was done using the FlowJo (Treestar). Blood NK cells were defined within the live singlets gate as CD3 - CD4 - CD19 - CD14 - cells as the CD56 ++ NK, CD56 ++ CD16 + NK and CD56dim CD16 ++ NK. Monocytes were defined within the live singlet CD3 - CD19 - PBMC as the CD14 + CD16 -, CD14 + CD16 + and CD14 dim CD16 ++ monocytes. Geometric mean fluorescence intensity (MFI) was reported for expression of CD69 on NK cell subsets and CCR5 and CD86 on monocytes. Antibodies used for staining of human PBMCs: BD: CCR5 (2D7/CCR5), CD14 (MQp9 and M5E2), CD16 (3G8), CD56 (NCAM16.2), CD3 (SK7), CD19 (SJ25C1), CD69 (FN50). Biolegend: CD16 (3G8), CD56 (HCD56), CD86 (IT2.2). Molecular Probes: CD14 (TUK4). Statistical analyses were performed with GraphPad Prism to identify significant change relevant to the baseline (day 0), mean ± SEM, t test. Areas Under Curve (AUC) were calculated to compare magnitudes of the cell subsets between cohorts throughout the study duration (days 0, 1, 3, 7 and 14) and compared by t- test.
11 Deconvolution Cell subset proportions for the 2011 trial were estimated in R using the CellMix package and the approach proposed by Abbas et al., which performs a nonnegative linear regression of the total gene expression on a reference basis matrix composed of gene expression profiles from sorted cells. We used a custom basis matrix built from publicly available expression profiles of sorted cell subsets (Allantaz et al., 2012) (GSE28490), which we validated on the 2010 trial data. More precisely, we looked for groups of probe sets whose total gene expression profiles in mixed samples are able to track the proportion of a given cell subset. In order to identify such probe sets, we simulated 500 blood samples by mixing in silico -- all cell subset average expression profiles (mean across replicate of the same cell subset), using random mixing proportions drawn from uniform distributions over realistic ranges for each cell subset (Table S4). We then selected, for each subset, the top 20 probe sets whose total mixed expression profile was highly correlated (Pearson correlation above 0.9) with its corresponding simulated proportions. A second filtering step consisted in removing probe sets whose maximum average gene expression was not achieved on its assigned cell subset. This procedure retained a total of 164 probe sets, whose average gene expression profiles were combined into a reference basis matrix (Table S4). We evaluated its performance on the 2010 trial data, by comparing the estimated proportions with the available FACS proportions, which showed very good overall correlation for CD4+ and CD8+ T cells, CD19+ B cells, CD14+ monocytes and NK cells (Figure S7). DAMIP method Discriminant Analysis via Mixed Integer Programming (DAMIP) was used to identify the signatures that predict the antibody response to vaccination. In our previous work, we described in detail how DAMIP can be used to identify signatures that predict the immunogenicity of the yellow fever (Querec et al., 2009) and influenza vaccines (Nakaya et al., 2011). Briefly, the entire dataset is separated into training and blind testing sets. Feature selection and classification are performed on the training set to obtain a set of discriminatory gene signatures that satisfies a pre-set level of unbiased estimate of classification accuracy (e.g., 80%). The resulting classification rules are then used to predict the blind test set, resulting in an estimate of the predictive accuracy. In this 5-year analysis, we utilized a multi-step DAMIP strategy (Fig. S3). We divided the vaccinees in each season into training and blind test sets containing roughly 85% and 15% of the subjects, respectively (Fig. S3). Then for each season separately, we used DAMIP to train on the training set and identified classification rules which contained predictive gene sets of size 10, 15, 20, 25, 30, and 50, using day 7 fold change expression. The vaccinees from the 2007 season were pooled with those from 2008 due to their low numbers. For each season, among the classification rules generated, we selected those with unbiased estimate of at least 70% for blind prediction of HAI responses for all other seasons. For example, when using to train, we blind predicted the responses in 2009, 2010, and 2011, and selected those rules with predictive accuracies of at least 70%. Similarly we used 2009, 2010, and 2011 for training, predicting the remaining seasons. The frequencies of representation of genes in these selected classification rules were calculated, and the top 250 most frequent genes were selected (among the results from each season). We ranked these genes by average frequency and selected the 66 genes that appeared in all 4 lists of top 250 most frequent genes. In the next step, combinatorial feature selection was run using these 66 genes for each season. Classification rules associated with 3, 4, or 5 discriminatory genes were identified. We reported the classification rules in which the predictive accuracies of HAI response in all other seasons are at least 70%. This resulted in a total of 175 predictive rules, with accuracy ranging between % (Table S1, Fig. S3). For each of these rules, a second layer of blind prediction was then performed on the remaining blind test set (15% of subjects from each season) (Table S2). Module-based responder classification via artificial neural network Artificial neural networks (ANNs) are computational algorithms whose structure is based on the connectivity between neurons in biological nervous systems. ANNs are capable of a wide variety of tasks including machine learning and pattern recognition. As nonlinear models, ANNs have advantages over linear approaches such as logistic regression in situations where the relationship between the inputs and outputs of the system is not linear, which is often the case in biological systems. In order to use an ANN to predict a high/low antibody response based on module-level features, blood transcription module
12 enrichment scores were generated for each subject using single sample Gene Set Enrichment Analysis (ssgsea) based on a ranking of the genes by fold change on days 3 and 7 post-vaccination. In order to reduce the computational complexity and reduce overfitting during feature selection, a pre-filter was applied by ranking the modules by independent two-sample t-test and selecting the top 25 modules with the smallest t-test p values for inclusion as possible features. A forward selection approach was then implemented for feature selection, in which the network is trained using each module separately as the sole input. The module that achieves the highest average accuracy across 10 trials is included in the final input set, and then modules are iteratively added to this set. Each iteration, all remaining modules are tested one at a time as an addition to the input set, and only the module that improves the average accuracy the most is kept. This process is stopped when the accuracy fails to increase in a given iteration. Once the final input set was selected, the final classifier was generated using a bagging approach (Breiman, 1996), in which a committee of networks is created by training single networks using randomized partitioning of the training data. The output of the ensemble classifier is then determined by a majority vote of the output of the member networks of the committee. By averaging over the variation caused by initial weighting and training/validation partitioning, this method has been shown to increase the accuracy and reduce the variability of classification when using neural networks and other methods (Bauer and Kohavi, 1999). Based on preliminary analysis of the classifier performance with varying network and committee sizes, a committee of 20 two-layer neural networks with 5 first-layer neurons each was used in the final approach. All coding was done in MATLAB using the Neural Network Toolbox. In order to evaluate the performance of the algorithm, a bootstrapping approach was implemented. The subjects across all seasons were randomly divided into one training and one testing group in an 85%/15% ratio, respectively. Elderly subjects ( 65 years old) were separated and included as an independent test set. To prevent overfitting, feature selection and training of the network weights were performed using only the training subjects, and then the performance of the trained network in blind prediction of the testing subjects was assessed. This process was repeated for 100 trials in order to ensure that the performance was robust across many random divisions of the data. The performance of the algorithm as well as the frequency with which each module was selected as an input is shown in Table S3. Differential expression analysis and WGCNA For significance assessment of each season and time point, expression differences between baseline and the day post-vaccination were assessed by pairwise Student s t-test (p-value < 0.01 and log 2 fold-change > 0.2). Weighted correlation network analysis (WGCNA) was used to find clusters of highly correlated genes among the TIV regulated genes in young (power = 9) and elderly (power = 4). Identification of signatures of antibody persistence Day 180 HAI titers were measured for subjects in the cohorts, using the same strain that achieved the maximum fold change on day 28 on a per subject basis. As the day 180 samples were collected and analyzed separately from the day 28 samples, the baseline HAI titer was also remeasured in order to provide accurate normalization during fold change calculation. Subjects whose baseline titer measurements differed by more than 2-fold were omitted from further analysis. Next, a simple linear regression was performed using the day 180 titer fold change as the dependent variable and the day 28 titer fold change as the independent variable. The distance to this regression line (residual) was computed for each subject and used as a measure of relative antibody response persistence, with a greater value representing a higher day 180 response relative to the original day 28 response. Blood transcriptional module enrichment analysis was then performed using genes ranked by correlation with the day 180/day 28 residual as described above. Integrated mrna-mirna expression analysis Differentially expressed mirna were identified using pairwise Student s t-test (p-value <0.05) between baseline and post-vaccination expression values (days 1, 3, and 7). Correlated gene module-mirna pairs were identified by performing Pearson correlation between normalized mirna expression and single sample blood transcription module enrichment scores. Next, for modules of interest, each significantly negatively correlating (p<0.05) mirna was mapped using Targetscan database to identify any predicted
13 target genes (context score < 0) within the module. Pearson correlation was then used to validate the interactions of each mirna with its predicted target genes within our dataset. References Allantaz, F., Cheng, D.T., Bergauer, T., Ravindran, P., Rossier, M.F., Ebeling, M., Badi, L., Reis, B., Bitter, H., D'Asaro, M., et al. (2012). Expression profiling of human immune cell subsets identifies mirna-mrna regulatory relationships correlated with cell type specific expression. PLoS One 7, e Bauer, E., and Kohavi, R. (1999). An empirical comparison of voting classification algorithms: Bagging, boosting, and variants. Mach Learn 36, Breiman, L. (1996). Bagging predictors. Mach Learn 24, Chen, G.L., Lamirande, E.W., Jin, H., Kemble, G., and Subbarao, K. (2010). Safety, immunogencity, and efficacy of a cold-adapted A/Ann Arbor/6/60 (H2N2) vaccine in mice and ferrets. Virology 398, Johnson, W.E., Li, C., and Rabinovic, A. (2007). Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, Nakaya, H.I., Wrammert, J., Lee, E.K., Racioppi, L., Marie-Kunze, S., Haining, W.N., Means, A.R., Kasturi, S.P., Khan, N., Li, G.M., et al. (2011). Systems biology of vaccination for seasonal influenza in humans. Nat Immunol 12, Querec, T.D., Akondy, R.S., Lee, E.K., Cao, W., Nakaya, H.I., Teuwen, D., Pirani, A., Gernert, K., Deng, J., Marzolf, B., et al. (2009). Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol 10, Shannon, P., Markiel, A., Ozier, O., Baliga, N.S., Wang, J.T., Ramage, D., Amin, N., Schwikowski, B., and Ideker, T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 13, Wei, T. (2012). Package corrplot Visualization of a correlation matrix. (cran.r-project.org.).
Systems Analysis of Immunity to Influenza Vaccination across Multiple Years and in Diverse Populations Reveals Shared Molecular Signatures
 Systems Analysis of Immunity to Influenza Vaccination across Multiple Years and in Diverse Populations Reveals Shared Molecular Signatures Helder Nakaya, Emory University Thomas Hagan, University of California
Systems Analysis of Immunity to Influenza Vaccination across Multiple Years and in Diverse Populations Reveals Shared Molecular Signatures Helder Nakaya, Emory University Thomas Hagan, University of California
Nature Neuroscience: doi: /nn Supplementary Figure 1. Behavioral training.
 Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Supplemental Information. Systems Scale Interactive Exploration. Reveals Quantitative and Qualitative Differences
 Immunity, Volume 38 Supplemental Information Systems Scale Interactive Exploration Reveals Quantitative and Qualitative Differences in Response to Influenza and Pneumococcal Vaccines Gerlinde Obermoser,
Immunity, Volume 38 Supplemental Information Systems Scale Interactive Exploration Reveals Quantitative and Qualitative Differences in Response to Influenza and Pneumococcal Vaccines Gerlinde Obermoser,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
ADCC Assay Protocol Vikram Srivastava 1, Zheng Yang 1, Ivan Fan Ngai Hung 2, Jianqing Xu 3, Bojian Zheng 3 and Mei- Yun Zhang 3*
 ADCC Assay Protocol Vikram Srivastava 1, Zheng Yang 1, Ivan Fan Ngai Hung 2, Jianqing Xu 3, Bojian Zheng 3 and Mei- Yun Zhang 3* 1 Department of Microbiology, Li Ka Shing Faculty of Medicine, University
ADCC Assay Protocol Vikram Srivastava 1, Zheng Yang 1, Ivan Fan Ngai Hung 2, Jianqing Xu 3, Bojian Zheng 3 and Mei- Yun Zhang 3* 1 Department of Microbiology, Li Ka Shing Faculty of Medicine, University
Application of Artificial Neural Networks in Classification of Autism Diagnosis Based on Gene Expression Signatures
 Application of Artificial Neural Networks in Classification of Autism Diagnosis Based on Gene Expression Signatures 1 2 3 4 5 Kathleen T Quach Department of Neuroscience University of California, San Diego
Application of Artificial Neural Networks in Classification of Autism Diagnosis Based on Gene Expression Signatures 1 2 3 4 5 Kathleen T Quach Department of Neuroscience University of California, San Diego
Supplementary Figure 1. Using DNA barcode-labeled MHC multimers to generate TCR fingerprints
 Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplementary Fig. 1: Ex vivo tetramer enrichment with anti-c-myc beads
 Supplementary Fig. 1: Ex vivo tetramer enrichment with anti-c-myc beads Representative example of comparative ex vivo tetramer enrichment performed in three independent experiments with either conventional
Supplementary Fig. 1: Ex vivo tetramer enrichment with anti-c-myc beads Representative example of comparative ex vivo tetramer enrichment performed in three independent experiments with either conventional
SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer.
 Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Nature Methods: doi: /nmeth.3115
 Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Supplementary Figure 1. ALVAC-protein vaccines and macaque immunization. (A) Maximum likelihood
 Supplementary Figure 1. ALVAC-protein vaccines and macaque immunization. (A) Maximum likelihood tree illustrating CRF01_AE gp120 protein sequence relationships between 107 Envs sampled in the RV144 trial
Supplementary Figure 1. ALVAC-protein vaccines and macaque immunization. (A) Maximum likelihood tree illustrating CRF01_AE gp120 protein sequence relationships between 107 Envs sampled in the RV144 trial
EXPression ANalyzer and DisplayER
 EXPression ANalyzer and DisplayER Tom Hait Aviv Steiner Igor Ulitsky Chaim Linhart Amos Tanay Seagull Shavit Rani Elkon Adi Maron-Katz Dorit Sagir Eyal David Roded Sharan Israel Steinfeld Yossi Shiloh
EXPression ANalyzer and DisplayER Tom Hait Aviv Steiner Igor Ulitsky Chaim Linhart Amos Tanay Seagull Shavit Rani Elkon Adi Maron-Katz Dorit Sagir Eyal David Roded Sharan Israel Steinfeld Yossi Shiloh
Supplementary Figure 1
 Supplementary Figure 1 FMO control (no HLA-DR antibody) FMO control (no HLA-DR antibody) Supplementary Figure 1. Gating strategy for HLA-DR+ T-cells. A gate was drawn around lymphocytes based on cell size
Supplementary Figure 1 FMO control (no HLA-DR antibody) FMO control (no HLA-DR antibody) Supplementary Figure 1. Gating strategy for HLA-DR+ T-cells. A gate was drawn around lymphocytes based on cell size
Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary Materials and Methods
 Silva et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary
Silva et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/278/rs11/dc1 Supplementary Materials for In Vivo Phosphoproteomics Analysis Reveals the Cardiac Targets of β-adrenergic Receptor Signaling Alicia Lundby,* Martin
www.sciencesignaling.org/cgi/content/full/6/278/rs11/dc1 Supplementary Materials for In Vivo Phosphoproteomics Analysis Reveals the Cardiac Targets of β-adrenergic Receptor Signaling Alicia Lundby,* Martin
Supplementary Figure 1
 Supplementary Figure 1 Identification of IFN-γ-producing CD8 + and CD4 + T cells with naive phenotype by alternative gating and sample-processing strategies. a. Contour 5% probability plots show definition
Supplementary Figure 1 Identification of IFN-γ-producing CD8 + and CD4 + T cells with naive phenotype by alternative gating and sample-processing strategies. a. Contour 5% probability plots show definition
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h)
 Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h) after feeding. A small slice (~5-1 mm 3 ) was taken
Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h) after feeding. A small slice (~5-1 mm 3 ) was taken
Supplementary Methods
 Supplementary Methods Analysis of time course gene expression data. The time course data of the expression level of a representative gene is shown in the below figure. The trajectory of longitudinal expression
Supplementary Methods Analysis of time course gene expression data. The time course data of the expression level of a representative gene is shown in the below figure. The trajectory of longitudinal expression
Nature Immunology: doi: /ni Supplementary Figure 1. Transcriptional program of the TE and MP CD8 + T cell subsets.
 Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and
 Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and Promoter Motif Analysis Shisong Ma 1,2*, Michael Snyder 3, and Savithramma P Dinesh-Kumar 2* 1 School of Life Sciences, University
Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and Promoter Motif Analysis Shisong Ma 1,2*, Michael Snyder 3, and Savithramma P Dinesh-Kumar 2* 1 School of Life Sciences, University
Unit 1 Exploring and Understanding Data
 Unit 1 Exploring and Understanding Data Area Principle Bar Chart Boxplot Conditional Distribution Dotplot Empirical Rule Five Number Summary Frequency Distribution Frequency Polygon Histogram Interquartile
Unit 1 Exploring and Understanding Data Area Principle Bar Chart Boxplot Conditional Distribution Dotplot Empirical Rule Five Number Summary Frequency Distribution Frequency Polygon Histogram Interquartile
Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes
 Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes Kaifu Chen 1,2,3,4,5,10, Zhong Chen 6,10, Dayong Wu 6, Lili Zhang 7, Xueqiu Lin 1,2,8,
Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes Kaifu Chen 1,2,3,4,5,10, Zhong Chen 6,10, Dayong Wu 6, Lili Zhang 7, Xueqiu Lin 1,2,8,
Supplemental Information. Aryl Hydrocarbon Receptor Controls. Monocyte Differentiation. into Dendritic Cells versus Macrophages
 Immunity, Volume 47 Supplemental Information Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages Christel Goudot, Alice Coillard, Alexandra-Chloé Villani,
Immunity, Volume 47 Supplemental Information Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages Christel Goudot, Alice Coillard, Alexandra-Chloé Villani,
Human B Cell Responses to Influenza Virus Vaccination
 Human B Cell Responses to Influenza Virus Vaccination Nucleoprotein (RNA) Influenza Virus Neuraminidase (NA) Hemagglutinin (HA) Enveloped, single-stranded, negative-sense RNA virus with segmented genome.
Human B Cell Responses to Influenza Virus Vaccination Nucleoprotein (RNA) Influenza Virus Neuraminidase (NA) Hemagglutinin (HA) Enveloped, single-stranded, negative-sense RNA virus with segmented genome.
User Guide. Association analysis. Input
 User Guide TFEA.ChIP is a tool to estimate transcription factor enrichment in a set of differentially expressed genes using data from ChIP-Seq experiments performed in different tissues and conditions.
User Guide TFEA.ChIP is a tool to estimate transcription factor enrichment in a set of differentially expressed genes using data from ChIP-Seq experiments performed in different tissues and conditions.
Statistics 202: Data Mining. c Jonathan Taylor. Final review Based in part on slides from textbook, slides of Susan Holmes.
 Final review Based in part on slides from textbook, slides of Susan Holmes December 5, 2012 1 / 1 Final review Overview Before Midterm General goals of data mining. Datatypes. Preprocessing & dimension
Final review Based in part on slides from textbook, slides of Susan Holmes December 5, 2012 1 / 1 Final review Overview Before Midterm General goals of data mining. Datatypes. Preprocessing & dimension
Pearson r = P (one-tailed) = n = 9
 8F4-Specific Lysis, % 1 UPN1 UPN3 8 UPN7 6 Pearson r =.69 UPN2 UPN5 P (one-tailed) =.192 4 UPN8 n = 9 2 UPN9 UPN4 UPN6 5 1 15 2 25 8 8F4, % Max MFI Supplementary Figure S1. AML samples UPN1-UPN9 show variable
8F4-Specific Lysis, % 1 UPN1 UPN3 8 UPN7 6 Pearson r =.69 UPN2 UPN5 P (one-tailed) =.192 4 UPN8 n = 9 2 UPN9 UPN4 UPN6 5 1 15 2 25 8 8F4, % Max MFI Supplementary Figure S1. AML samples UPN1-UPN9 show variable
Supplemental Figure S1
 Supplemental Figure S1 Children from Nandi (low pathogen burden area) and Kisumu (high pathogen burden area) have significantly different pathogen-specific serological profiles. Serum antibody titers for
Supplemental Figure S1 Children from Nandi (low pathogen burden area) and Kisumu (high pathogen burden area) have significantly different pathogen-specific serological profiles. Serum antibody titers for
Study No.: Title: Rationale: Phase: Study Period: Study Design: Centers: Indication: Treatment: Objectives:
 The study listed may include approved and non-approved uses, formulations or treatment regimens. The results reported in any single study may not reflect the overall results obtained on studies of a product.
The study listed may include approved and non-approved uses, formulations or treatment regimens. The results reported in any single study may not reflect the overall results obtained on studies of a product.
Detailed step-by-step operating procedures for NK cell and CTL degranulation assays
 Supplemental methods Detailed step-by-step operating procedures for NK cell and CTL degranulation assays Materials PBMC isolated from patients, relatives and healthy donors as control K562 cells (ATCC,
Supplemental methods Detailed step-by-step operating procedures for NK cell and CTL degranulation assays Materials PBMC isolated from patients, relatives and healthy donors as control K562 cells (ATCC,
Mapping the Antigenic and Genetic Evolution of Influenza Virus
 Mapping the Antigenic and Genetic Evolution of Influenza Virus Derek J. Smith, Alan S. Lapedes, Jan C. de Jong, Theo M. Bestebroer, Guus F. Rimmelzwaan, Albert D. M. E. Osterhaus, Ron A. M. Fouchier Science
Mapping the Antigenic and Genetic Evolution of Influenza Virus Derek J. Smith, Alan S. Lapedes, Jan C. de Jong, Theo M. Bestebroer, Guus F. Rimmelzwaan, Albert D. M. E. Osterhaus, Ron A. M. Fouchier Science
AbSeq on the BD Rhapsody system: Exploration of single-cell gene regulation by simultaneous digital mrna and protein quantification
 BD AbSeq on the BD Rhapsody system: Exploration of single-cell gene regulation by simultaneous digital mrna and protein quantification Overview of BD AbSeq antibody-oligonucleotide conjugates. High-throughput
BD AbSeq on the BD Rhapsody system: Exploration of single-cell gene regulation by simultaneous digital mrna and protein quantification Overview of BD AbSeq antibody-oligonucleotide conjugates. High-throughput
Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD
 Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD Department of Biomedical Informatics Department of Computer Science and Engineering The Ohio State University Review
Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD Department of Biomedical Informatics Department of Computer Science and Engineering The Ohio State University Review
Nature Immunology: doi: /ni Supplementary Figure 1. RNA-Seq analysis of CD8 + TILs and N-TILs.
 Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer. Application Note
 Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Colon cancer subtypes from gene expression data
 Colon cancer subtypes from gene expression data Nathan Cunningham Giuseppe Di Benedetto Sherman Ip Leon Law Module 6: Applied Statistics 26th February 2016 Aim Replicate findings of Felipe De Sousa et
Colon cancer subtypes from gene expression data Nathan Cunningham Giuseppe Di Benedetto Sherman Ip Leon Law Module 6: Applied Statistics 26th February 2016 Aim Replicate findings of Felipe De Sousa et
RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays
 Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
List of Figures. List of Tables. Preface to the Second Edition. Preface to the First Edition
 List of Figures List of Tables Preface to the Second Edition Preface to the First Edition xv xxv xxix xxxi 1 What Is R? 1 1.1 Introduction to R................................ 1 1.2 Downloading and Installing
List of Figures List of Tables Preface to the Second Edition Preface to the First Edition xv xxv xxix xxxi 1 What Is R? 1 1.1 Introduction to R................................ 1 1.2 Downloading and Installing
Modeling Sentiment with Ridge Regression
 Modeling Sentiment with Ridge Regression Luke Segars 2/20/2012 The goal of this project was to generate a linear sentiment model for classifying Amazon book reviews according to their star rank. More generally,
Modeling Sentiment with Ridge Regression Luke Segars 2/20/2012 The goal of this project was to generate a linear sentiment model for classifying Amazon book reviews according to their star rank. More generally,
Supplementary Figure 1. mrna targets were found in exosomes and absent in free-floating supernatant. Serum exosomes and exosome-free supernatant were
 Supplementary Figure 1. mrna targets were found in exosomes and absent in free-floating supernatant. Serum exosomes and exosome-free supernatant were separated via ultracentrifugation and lysed to analyze
Supplementary Figure 1. mrna targets were found in exosomes and absent in free-floating supernatant. Serum exosomes and exosome-free supernatant were separated via ultracentrifugation and lysed to analyze
Understanding Poor Vaccine Responses: Transcriptomics of Vaccine Failure
 Final Report - Summer Research Project 1 Introduction Understanding Poor Vaccine Responses: Transcriptomics of Vaccine Failure Katherine Miller, Year 3, Dalhousie University (Supervisor: Dr. Lisa Barrett,
Final Report - Summer Research Project 1 Introduction Understanding Poor Vaccine Responses: Transcriptomics of Vaccine Failure Katherine Miller, Year 3, Dalhousie University (Supervisor: Dr. Lisa Barrett,
Nature Medicine: doi: /nm.2109
 HIV 1 Infects Multipotent Progenitor Cells Causing Cell Death and Establishing Latent Cellular Reservoirs Christoph C. Carter, Adewunmi Onafuwa Nuga, Lucy A. M c Namara, James Riddell IV, Dale Bixby, Michael
HIV 1 Infects Multipotent Progenitor Cells Causing Cell Death and Establishing Latent Cellular Reservoirs Christoph C. Carter, Adewunmi Onafuwa Nuga, Lucy A. M c Namara, James Riddell IV, Dale Bixby, Michael
Identification of Tissue Independent Cancer Driver Genes
 Identification of Tissue Independent Cancer Driver Genes Alexandros Manolakos, Idoia Ochoa, Kartik Venkat Supervisor: Olivier Gevaert Abstract Identification of genomic patterns in tumors is an important
Identification of Tissue Independent Cancer Driver Genes Alexandros Manolakos, Idoia Ochoa, Kartik Venkat Supervisor: Olivier Gevaert Abstract Identification of genomic patterns in tumors is an important
Pandemic Preparedness Team Immunology and Pathogenesis Branch Influenza Division Centers for Disease Control and Prevention USA VERSION 1
 MODIFIED HEMAGGLUTINATION INHIBITION (HI) ASSAY USING HORSE RBCS FOR SEROLOGIC DETECTION OF ANTIBODIES TO H7 SUBTYPE AVIAN INFLUENZA VIRUS IN HUMAN SERA Pandemic Preparedness Team Immunology and Pathogenesis
MODIFIED HEMAGGLUTINATION INHIBITION (HI) ASSAY USING HORSE RBCS FOR SEROLOGIC DETECTION OF ANTIBODIES TO H7 SUBTYPE AVIAN INFLUENZA VIRUS IN HUMAN SERA Pandemic Preparedness Team Immunology and Pathogenesis
Supplementary Material. for. A cell-centered meta-analysis reveals baseline predictors of anti-tnf nonresponse. in biopsy and blood of IBD patients
 Supplementary Material for A cell-centered meta-analysis reveals baseline predictors of anti-tnf nonresponse in biopsy and blood of IBD patients This document includes Supplementary Methods, Figures and
Supplementary Material for A cell-centered meta-analysis reveals baseline predictors of anti-tnf nonresponse in biopsy and blood of IBD patients This document includes Supplementary Methods, Figures and
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded
 Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
OncoPPi Portal A Cancer Protein Interaction Network to Inform Therapeutic Strategies
 OncoPPi Portal A Cancer Protein Interaction Network to Inform Therapeutic Strategies 2017 Contents Datasets... 2 Protein-protein interaction dataset... 2 Set of known PPIs... 3 Domain-domain interactions...
OncoPPi Portal A Cancer Protein Interaction Network to Inform Therapeutic Strategies 2017 Contents Datasets... 2 Protein-protein interaction dataset... 2 Set of known PPIs... 3 Domain-domain interactions...
ncounter TM Analysis System
 ncounter TM Analysis System Molecules That Count TM www.nanostring.com Agenda NanoString Technologies History Introduction to the ncounter Analysis System CodeSet Design and Assay Principals System Performance
ncounter TM Analysis System Molecules That Count TM www.nanostring.com Agenda NanoString Technologies History Introduction to the ncounter Analysis System CodeSet Design and Assay Principals System Performance
Supporting Information
 Supporting Information Valkenburg et al. 10.1073/pnas.1403684111 SI Materials and Methods ELISA and Microneutralization. Sera were treated with Receptor Destroying Enzyme II (RDE II, Accurate) before ELISA
Supporting Information Valkenburg et al. 10.1073/pnas.1403684111 SI Materials and Methods ELISA and Microneutralization. Sera were treated with Receptor Destroying Enzyme II (RDE II, Accurate) before ELISA
From Biostatistics Using JMP: A Practical Guide. Full book available for purchase here. Chapter 1: Introduction... 1
 From Biostatistics Using JMP: A Practical Guide. Full book available for purchase here. Contents Dedication... iii Acknowledgments... xi About This Book... xiii About the Author... xvii Chapter 1: Introduction...
From Biostatistics Using JMP: A Practical Guide. Full book available for purchase here. Contents Dedication... iii Acknowledgments... xi About This Book... xiii About the Author... xvii Chapter 1: Introduction...
Supplementary Materials
 Supplementary Materials July 2, 2015 1 EEG-measures of consciousness Table 1 makes explicit the abbreviations of the EEG-measures. Their computation closely follows Sitt et al. (2014) (supplement). PE
Supplementary Materials July 2, 2015 1 EEG-measures of consciousness Table 1 makes explicit the abbreviations of the EEG-measures. Their computation closely follows Sitt et al. (2014) (supplement). PE
Supplementary. properties of. network types. randomly sampled. subsets (75%
 Supplementary Information Gene co-expression network analysis reveals common system-level prognostic genes across cancer types properties of Supplementary Figure 1 The robustness and overlap of prognostic
Supplementary Information Gene co-expression network analysis reveals common system-level prognostic genes across cancer types properties of Supplementary Figure 1 The robustness and overlap of prognostic
Supplemental Figure S1. RANK expression on human lung cancer cells.
 Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
Predicting Breast Cancer Survival Using Treatment and Patient Factors
 Predicting Breast Cancer Survival Using Treatment and Patient Factors William Chen wchen808@stanford.edu Henry Wang hwang9@stanford.edu 1. Introduction Breast cancer is the leading type of cancer in women
Predicting Breast Cancer Survival Using Treatment and Patient Factors William Chen wchen808@stanford.edu Henry Wang hwang9@stanford.edu 1. Introduction Breast cancer is the leading type of cancer in women
In vitro human regulatory T cell expansion
 - 1 - Human CD4 + CD25 + CD127 dim/- regulatory T cell Workflow isolation, in vitro expansion and analysis In vitro human regulatory T cell expansion Introduction Regulatory T (Treg) cells are a subpopulation
- 1 - Human CD4 + CD25 + CD127 dim/- regulatory T cell Workflow isolation, in vitro expansion and analysis In vitro human regulatory T cell expansion Introduction Regulatory T (Treg) cells are a subpopulation
Influenza A H1N1 (Swine Flu 2009) Hemagglutinin / HA ELISA Pair Set
 Influenza A H1N1 (Swine Flu 2009) Hemagglutinin / HA ELISA Pair Set Catalog Number : SEK001 To achieve the best assay results, this manual must be read carefully before using this product and the assay
Influenza A H1N1 (Swine Flu 2009) Hemagglutinin / HA ELISA Pair Set Catalog Number : SEK001 To achieve the best assay results, this manual must be read carefully before using this product and the assay
Comparison of discrimination methods for the classification of tumors using gene expression data
 Comparison of discrimination methods for the classification of tumors using gene expression data Sandrine Dudoit, Jane Fridlyand 2 and Terry Speed 2,. Mathematical Sciences Research Institute, Berkeley
Comparison of discrimination methods for the classification of tumors using gene expression data Sandrine Dudoit, Jane Fridlyand 2 and Terry Speed 2,. Mathematical Sciences Research Institute, Berkeley
Christopher P. Corkum 1, Danielle P. Ings 1, Christopher Burgess 2, Sylwia Karwowska 2, Werner Kroll 2,3 and Tomasz I. Michalak 1*
 Corkum et al. BMC Immunology (2015) 16:48 DOI 10.1186/s12865-015-0113-0 METHODOLOGY Open Access Immune cell subsets and their gene expression profiles from human PBMC isolated by Vacutainer Cell Preparation
Corkum et al. BMC Immunology (2015) 16:48 DOI 10.1186/s12865-015-0113-0 METHODOLOGY Open Access Immune cell subsets and their gene expression profiles from human PBMC isolated by Vacutainer Cell Preparation
Gene expression analysis. Roadmap. Microarray technology: how it work Applications: what can we do with it Preprocessing: Classification Clustering
 Gene expression analysis Roadmap Microarray technology: how it work Applications: what can we do with it Preprocessing: Image processing Data normalization Classification Clustering Biclustering 1 Gene
Gene expression analysis Roadmap Microarray technology: how it work Applications: what can we do with it Preprocessing: Image processing Data normalization Classification Clustering Biclustering 1 Gene
Supplementary Figure 1. Nature Neuroscience: doi: /nn.4547
 Supplementary Figure 1 Characterization of the Microfetti mouse model. (a) Gating strategy for 8-color flow analysis of peripheral Ly-6C + monocytes from Microfetti mice 5-7 days after TAM treatment. Living
Supplementary Figure 1 Characterization of the Microfetti mouse model. (a) Gating strategy for 8-color flow analysis of peripheral Ly-6C + monocytes from Microfetti mice 5-7 days after TAM treatment. Living
Technical Resources. BD Immunocytometry Systems. FastImmune Intracellular Cytokine Staining Procedures
 FastImmune Intracellular Cytokine Staining Procedures BD has developed protocols for the detection of intracellular cytokines in activated lymphocytes and in activated monocytes. The procedures have been
FastImmune Intracellular Cytokine Staining Procedures BD has developed protocols for the detection of intracellular cytokines in activated lymphocytes and in activated monocytes. The procedures have been
Influenza A H1N1 HA ELISA Pair Set
 Influenza A H1N1 HA ELISA Pair Set for H1N1 ( A/Puerto Rico/8/1934 ) HA Catalog Number : SEK11684 To achieve the best assay results, this manual must be read carefully before using this product and the
Influenza A H1N1 HA ELISA Pair Set for H1N1 ( A/Puerto Rico/8/1934 ) HA Catalog Number : SEK11684 To achieve the best assay results, this manual must be read carefully before using this product and the
Immunological Data Processing & Analysis
 Immunological Data Processing & Analysis Hongmei Yang Center for Biodefence Immune Modeling Department of Biostatistics and Computational Biology University of Rochester June 12, 2012 Hongmei Yang (CBIM
Immunological Data Processing & Analysis Hongmei Yang Center for Biodefence Immune Modeling Department of Biostatistics and Computational Biology University of Rochester June 12, 2012 Hongmei Yang (CBIM
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
NASDAQ:NVAX Novavax, Inc. All rights reserved.
 Novavax vaccine induced improved immune responses against homologous and drifted A(H3N2) viruses in older adults compared to egg-based, high-dose, influenza vaccine World Vaccine Congress April 4, 2018
Novavax vaccine induced improved immune responses against homologous and drifted A(H3N2) viruses in older adults compared to egg-based, high-dose, influenza vaccine World Vaccine Congress April 4, 2018
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/3/114/ra23/dc1 Supplementary Materials for Regulation of Zap70 Expression During Thymocyte Development Enables Temporal Separation of CD4 and CD8 Repertoire Selection
www.sciencesignaling.org/cgi/content/full/3/114/ra23/dc1 Supplementary Materials for Regulation of Zap70 Expression During Thymocyte Development Enables Temporal Separation of CD4 and CD8 Repertoire Selection
Limited efficacy of inactivated influenza vaccine in elderly individuals is associated with decreased production of vaccine-specific antibodies
 Related Commentary, page 2981 Research article Limited efficacy of inactivated influenza vaccine in elderly individuals is associated with decreased production of vaccine-specific antibodies Sanae Sasaki,
Related Commentary, page 2981 Research article Limited efficacy of inactivated influenza vaccine in elderly individuals is associated with decreased production of vaccine-specific antibodies Sanae Sasaki,
White Paper Estimating Complex Phenotype Prevalence Using Predictive Models
 White Paper 23-12 Estimating Complex Phenotype Prevalence Using Predictive Models Authors: Nicholas A. Furlotte Aaron Kleinman Robin Smith David Hinds Created: September 25 th, 2015 September 25th, 2015
White Paper 23-12 Estimating Complex Phenotype Prevalence Using Predictive Models Authors: Nicholas A. Furlotte Aaron Kleinman Robin Smith David Hinds Created: September 25 th, 2015 September 25th, 2015
Nature Medicine: doi: /nm.3967
 Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Supplementary information for: Human micrornas co-silence in well-separated groups and have different essentialities
 Supplementary information for: Human micrornas co-silence in well-separated groups and have different essentialities Gábor Boross,2, Katalin Orosz,2 and Illés J. Farkas 2, Department of Biological Physics,
Supplementary information for: Human micrornas co-silence in well-separated groups and have different essentialities Gábor Boross,2, Katalin Orosz,2 and Illés J. Farkas 2, Department of Biological Physics,
To test the possible source of the HBV infection outside the study family, we searched the Genbank
 Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplemental Data Figure S1 Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells.
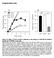 Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
Supplemental Information. Genomic Characterization of Murine. Monocytes Reveals C/EBPb Transcription. Factor Dependence of Ly6C Cells
 Immunity, Volume 46 Supplemental Information Genomic Characterization of Murine Monocytes Reveals C/EBPb Transcription Factor Dependence of Ly6C Cells Alexander Mildner, Jörg Schönheit, Amir Giladi, Eyal
Immunity, Volume 46 Supplemental Information Genomic Characterization of Murine Monocytes Reveals C/EBPb Transcription Factor Dependence of Ly6C Cells Alexander Mildner, Jörg Schönheit, Amir Giladi, Eyal
2009 H1N1 Influenza ( Swine Flu ) Hemagglutinin ELISA kit
 2009 H1N1 Influenza ( Swine Flu ) Hemagglutinin ELISA kit Catalog Number : SEK001 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as
2009 H1N1 Influenza ( Swine Flu ) Hemagglutinin ELISA kit Catalog Number : SEK001 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Primary Adult Naïve CD4+ CD45RA+ Cells. Prepared by: David Randolph at University of Alabama, Birmingham
 Primary Adult Naïve CD4+ CD45RA+ Cells Prepared by: David Randolph (drdrdr@uab.edu) at University of Alabama, Birmingham Goal: To obtain large numbers of highly pure primary CD4+ CD45RO- CD25- cells from
Primary Adult Naïve CD4+ CD45RA+ Cells Prepared by: David Randolph (drdrdr@uab.edu) at University of Alabama, Birmingham Goal: To obtain large numbers of highly pure primary CD4+ CD45RO- CD25- cells from
Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types.
 Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Low Avidity CMV + T Cells accumulate in Old Humans
 Supplementary Figure Legends Supplementary Figure 1. CD45RA expressing CMVpp65-specific T cell populations accumulate within HLA-A*0201 and HLA-B*0701 individuals Pooled data showing the size of the NLV/HLA-A*0201-specific
Supplementary Figure Legends Supplementary Figure 1. CD45RA expressing CMVpp65-specific T cell populations accumulate within HLA-A*0201 and HLA-B*0701 individuals Pooled data showing the size of the NLV/HLA-A*0201-specific
Suppl Video: Tumor cells (green) and monocytes (white) are seeded on a confluent endothelial
 Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
Influenza Clinical Bulletin # 3: October 8, 2009 Vaccination Guidelines for Patients for Influenza
 The purpose of this document is to provide NYP providers with the most current recommendations regarding influenza vaccination for their patients. It is important to recognize that guidance reflects optimal
The purpose of this document is to provide NYP providers with the most current recommendations regarding influenza vaccination for their patients. It is important to recognize that guidance reflects optimal
Statistical Assessment of the Global Regulatory Role of Histone. Acetylation in Saccharomyces cerevisiae. (Support Information)
 Statistical Assessment of the Global Regulatory Role of Histone Acetylation in Saccharomyces cerevisiae (Support Information) Authors: Guo-Cheng Yuan, Ping Ma, Wenxuan Zhong and Jun S. Liu Linear Relationship
Statistical Assessment of the Global Regulatory Role of Histone Acetylation in Saccharomyces cerevisiae (Support Information) Authors: Guo-Cheng Yuan, Ping Ma, Wenxuan Zhong and Jun S. Liu Linear Relationship
38 Int'l Conf. Bioinformatics and Computational Biology BIOCOMP'16
 38 Int'l Conf. Bioinformatics and Computational Biology BIOCOMP'16 PGAR: ASD Candidate Gene Prioritization System Using Expression Patterns Steven Cogill and Liangjiang Wang Department of Genetics and
38 Int'l Conf. Bioinformatics and Computational Biology BIOCOMP'16 PGAR: ASD Candidate Gene Prioritization System Using Expression Patterns Steven Cogill and Liangjiang Wang Department of Genetics and
In vitro human regulatory T cell expansion
 - 1 - Human CD4 + CD25 + regulatory T cell isolation, Workflow in vitro expansion and analysis In vitro human regulatory T cell expansion Introduction Regulatory T (Treg) cells are a subpopulation of T
- 1 - Human CD4 + CD25 + regulatory T cell isolation, Workflow in vitro expansion and analysis In vitro human regulatory T cell expansion Introduction Regulatory T (Treg) cells are a subpopulation of T
Computational purification of individual tumor gene expression profiles leads to significant improvements in prognostic prediction
 METHOD Open Access Computational purification of individual tumor gene expression profiles leads to significant improvements in prognostic prediction Gerald Quon 1,9, Syed Haider 2,3, Amit G Deshwar 4,
METHOD Open Access Computational purification of individual tumor gene expression profiles leads to significant improvements in prognostic prediction Gerald Quon 1,9, Syed Haider 2,3, Amit G Deshwar 4,
SUPPLEMENTARY INFORMATION
 doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
Supplemental figures
 Supplemental figures Supplemental Figure 1. Impact of immunogenic-llc tumors (LLC-OVA) on peripheral OVA-specific CD8 T cells. Tetramer analysis showing percent OVA-specific CD8 T cells in peripheral blood
Supplemental figures Supplemental Figure 1. Impact of immunogenic-llc tumors (LLC-OVA) on peripheral OVA-specific CD8 T cells. Tetramer analysis showing percent OVA-specific CD8 T cells in peripheral blood
fl/+ KRas;Atg5 fl/+ KRas;Atg5 fl/fl KRas;Atg5 fl/fl KRas;Atg5 Supplementary Figure 1. Gene set enrichment analyses. (a) (b)
 KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
HD1 (FLU) HD2 (EBV) HD2 (FLU)
 ramer staining + anti-pe beads ramer staining a HD1 (FLU) HD2 (EBV) HD2 (FLU).73.11.56.46.24 1.12 b CD127 + c CD127 + d CD127 - e CD127 - PD1 - PD1 + PD1 + PD1-1 1 1 1 %CD127 + PD1-8 6 4 2 + anti-pe %CD127
ramer staining + anti-pe beads ramer staining a HD1 (FLU) HD2 (EBV) HD2 (FLU).73.11.56.46.24 1.12 b CD127 + c CD127 + d CD127 - e CD127 - PD1 - PD1 + PD1 + PD1-1 1 1 1 %CD127 + PD1-8 6 4 2 + anti-pe %CD127
Cell subset prediction for blood genomic studies
 METHODOLOGY ARTICLE Open Access Cell subset prediction for blood genomic studies Christopher R Bolen 1, Mohamed Uduman 1 and Steven H Kleinstein 1,2* Abstract Background: Genome-wide transcriptional profiling
METHODOLOGY ARTICLE Open Access Cell subset prediction for blood genomic studies Christopher R Bolen 1, Mohamed Uduman 1 and Steven H Kleinstein 1,2* Abstract Background: Genome-wide transcriptional profiling
Generation of ST2-GFP reporter mice and characterization of ILC1 cells following infection
 Supplementary Figure 1 Generation of ST2-GFP reporter mice and characterization of ILC1 cells following infection with influenza virus. (a) ST2-GFP reporter mice were generated as described in Methods.
Supplementary Figure 1 Generation of ST2-GFP reporter mice and characterization of ILC1 cells following infection with influenza virus. (a) ST2-GFP reporter mice were generated as described in Methods.
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Study No.: Title: Rationale: Phase: Study Period: Study Design: Centers: Indication: Treatment: Objectives: Primary Outcome/Efficacy Variable:
 The study listed may include approved and non-approved uses, formulations or treatment regimens. The results reported in any single study may not reflect the overall results obtained on studies of a product.
The study listed may include approved and non-approved uses, formulations or treatment regimens. The results reported in any single study may not reflect the overall results obtained on studies of a product.
Supplement to SCnorm: robust normalization of single-cell RNA-seq data
 Supplement to SCnorm: robust normalization of single-cell RNA-seq data Supplementary Note 1: SCnorm does not require spike-ins, since we find that the performance of spike-ins in scrna-seq is often compromised,
Supplement to SCnorm: robust normalization of single-cell RNA-seq data Supplementary Note 1: SCnorm does not require spike-ins, since we find that the performance of spike-ins in scrna-seq is often compromised,
HIV-1 p24 ELISA Pair Set Cat#: orb54951 (ELISA Manual)
 HIV-1 p24 ELISA Pair Set Cat#: orb54951 (ELISA Manual) BACKGROUND Human Immunodeficiency Virus ( HIV ) can be divided into two major types, HIV type 1 (HIV-1) and HIV type 2 (HIV-2). HIV-1 is related to
HIV-1 p24 ELISA Pair Set Cat#: orb54951 (ELISA Manual) BACKGROUND Human Immunodeficiency Virus ( HIV ) can be divided into two major types, HIV type 1 (HIV-1) and HIV type 2 (HIV-2). HIV-1 is related to
Simple, rapid, and reliable RNA sequencing
 Simple, rapid, and reliable RNA sequencing RNA sequencing applications RNA sequencing provides fundamental insights into how genomes are organized and regulated, giving us valuable information about the
Simple, rapid, and reliable RNA sequencing RNA sequencing applications RNA sequencing provides fundamental insights into how genomes are organized and regulated, giving us valuable information about the
