Supplementary Methods
|
|
|
- Beverley April Stafford
- 5 years ago
- Views:
Transcription
1 Supplementary Methods Analysis of time course gene expression data. The time course data of the expression level of a representative gene is shown in the below figure. The trajectory of longitudinal expression of each gene was obtained by a cubic spline function, and the mean expression of controls was considered as the base line expression. The maximum deviation of the trajectory line from the base line was referred to as the maximum fold change, or simply the fold change of the gene between patients and healthy controls. For each gene, the time required for the gene to change to the half of its maximum fold change was defined as its response time, and the time required for the gene to return back to the half after reached maximum point was defined as its recovery time. The significance of the longitudinal gene expression change was estimated using EDGE (1) by 1,000 random permutations. Significant genes were selected by FDR<0.001 and fold change 2 for each data sets. 5,544 genes were identified as significant between patients and healthy subjects (4,389 in trauma, 3,250 in burns, and 2,251 in endotoxemia). Analysis of the longitudinal gene expression. Shown is the time course data of the measured expression level of a representative gene. On the x-axis, points of samples of controls are displayed at 0 hour, and points of patient samples are displayed according to their sampling time after injury. On the y-axis, each point shows the log2 fold change of the expression value of the gene in one sample compared with the mean expression of the controls. The longitudinal trend line (in green) was obtained by spline smoothing of the gene expression values. The maximum fold change is defined as the largest deviation of the trend line from the base level, i.e. the mean expression of the controls. The response time and recovery time are defined as the time when the trajectory reaches half of the maximum fold change before and after reaching the maximum respectively.
2 Human-mouse orthologs. 20,273 Entrez genes were assayed on Affymetrix human HU133 plus v2 arrays, among which 16,646 had mouse orthologs according to the Mouse Genome Database (2), and 15,686 were assayed on Affymetrix mouse MOE430 arrays. Among the 5,554 genes significantly changed in human conditions, 4,918 genes had mouse orthologs assayed on the mouse arrays, which were included in the subsequent analysis. Comparison of gene response between datasets. The maximum fold changes of gene expression were measured in log-scale between patients and healthy subjects for each data set of human burn, trauma and endotoxemia, and between treated group and control group for the corresponding murine models. Between two datasets, the agreements of the maximum gene fold changes (R 2 ) as well as directionality of the changes (%) were compared. R 2 represents the square of Pearson s correlation. Similar results were seen when the rank correlation was calculated as in Fig. S1. % represents the percentages of genes changed to the same direction between the two datasets. In Fig. S9, gene expression changes of patients subjected to irradiation were compared with those of mouse models (GSE10640) (3, 4). Gene expression data was obtained from GSE10640, which includes two independent sets of patients under irradiation as part of their pretransplantation conditioning (n=24 and 10, each set was assayed on a different microarray platform) and 11 mouse models. 487 significantly changed genes (FDR<0.05) between patients under irradiation and controls were identified. R 2 and % were calculated in these genes between the first data set of patients (n=24) and the second set of patients (n=10) and the mouse models.
3 Fig. S1. Rank correlations (R 2 ) and percentages of genes changed to the same direction (%) between the human and mouse studies. The lower left half of the figure shows the scatter plots of the log2 fold changes of 4,918 human genes responsive to trauma, burns or endotoxemia (FDR<0.001; fold change >2), the same as shown in Fig. 1. The upper right half lists the rank correlations (R 2 ) and percentages of genes changed to the same direction (%) between the corresponding studies. Note that by random chance 50% of the genes between two uncorrelated conditions are expected to change in the same direction. The results show that the genomic responses to human trauma and burns are highly correlated, while the murine models correlate poorly with human conditions and each other.
4 Fig. S2. Correlations of gene changes between the human and murine studies in subsets of genes significantly changed in human injury and showed a greater magnitude of fold changes (FC). Among 4,918 human genes significantly changed in human trauma, burns or endotoxemia (FDR<0.001; FC 2), 336 genes had FC 4, 171 had FC 5, and 105 had FC 6 in human burn. Shown are bar graphs of Pearson correlations (R 2 ) between human trauma, endotoxemia, three murine models versus human burns as a reference. While the murine models correlate poorly among the 4,918 genes significantly changed in human injuries (FDR<0.001; FC 2) with the corresponding human conditions (R 2 = ), the correlations increase to (R 2 = ) among the top 105 genes with the greatest magnitude of changes (FDR<0.001; FC 6).
5 Fig. S3. Scatter plots and Pearson correlations (R 2 ) of genes significantly changed in the murine models. In the mouse models, 1,519 genes were significant at FDR<0.001 and 2 fold changes between the treated and control groups in either one of the three murine models, among which 87% (1,318) have human orthologs. In contrast to the 4,918 genes responsive to one of the human conditions as shown in Fig. 1, here the scatter plots depict fold changes of these 1,318 genes. The axes represent log2 fold changes. The high correlation was maintained in these genes between human trauma and burn (R 2 =0.88), whereas correlation between models was low (R ).
6 A. B. Fig. S4. Robustness of the analysis with respect to the sample sizes. The two human injury studies included 167 trauma patients sampled over the first 28 days after injury, 244 burn patients over the first year after injury, and 37 healthy controls, while the human endotoxemia study included 4 healthy volunteers received bacterial endotoxin and 4 controls each measured at four time points. The mouse models of trauma, burns, and endotoxemia each included 16 mice as the treated group and 16 as the control group (four time points, and 4 treated and 4 controls at each time point). To address the potential confounding feature of different sample sizes between the studies, comparable numbers of humans and mice were compared by random resampling. Since significant genes in the mouse burn and trauma models were identified from the time
7 course changes over four time points after injury, to simulate a similar environment in the human injury studies, we randomly selected four burn subjects at 3 days (0-3d), 7 days (3d-7d), 14 days (7d-14d), and 21 days (14d-21d) as well as four healthy controls at each time point, and then measured the significance of each gene with the same method applied to the mouse data. For human trauma, four trauma subjects were randomly selected at 1 day (0-1d), 4 days (2d-4d), 7 days (4d-7d), and 14 days (7d-14d) as well as the controls. The comparison was repeated by 1,000 times. (A) Box plots of the correlations (R 2 ) between burn injury and the other human conditions and murine models. The maximum fold changes of the randomly selected burn patients were compared with those of randomly selected trauma patients as well as the maximum fold changes from human endotoxemia and murine models. The correlations remained high between human injuries (R 2 =0.85). (B) The number of significantly changed genes detected in each human condition and murine model. For the human injuries, the time course of the randomly selected burn and trauma patients and controls were analyzed and the number of significant genes were identified (FDR<0.001 & FC 2). The median of the results from the 1,000 repeats is shown for human burns and trauma. Human conditions cause much more profound genomic response ( 3x more genes) than the corresponding mouse models.
8 Fig. S5. Box plots of the response and recovery times of gene expression in human burns, trauma and endotoxemia as well as corresponding mouse models. The response time is the time when the gene fold change reaches half of the maximum change. The recovery time is the time when the gene fold change returns back to half after it reached the maximum. While most of the gene response (75% percentile) occurred within the first 12 hours in all of the human injuries (human burns and trauma) and models (human endotoxemia and the three mouse models), the recovery differed dramatically in the time scale. The genomic disturbance in the models recovered within hours to 4 days (75% percentile), but lasted for 1-6 months in trauma and burn patients.
9 Fig. S6. Centroid expression of each gene cluster in human as shown in Fig. 2A. Each panel describes the centroid expression of a gene cluster for the three human conditions. The longitudinal trend lines were obtained by spline smoothing. Clusters 1-3 depict the common early activation (35% of the genes in Fig. 2A), clusters 5-8 are for the common early suppression (60%), and cluster 4 presents the late activation specific to burn and trauma (5%). Most of the genes changed in the same direction between burns and trauma (97%) and between the two injuries and endotoxemia (88%). In addition, the gene response time was within hours to burns, trauma and endotoxemia, while gene recovery differed dramatically between hours for endotoxemia, one month for trauma, and six months for burns.
10 A. B. Fig. S7. Gene changes over time for the genes responsive in the murine models. 1,519 genes were differentially expressed between the treated and control groups (FDR< and 2-fold change). (A) K-means clustering of genes over time. Red indicates increased and blue indicates decreased expression relative to the mean (white). (B) Centroid expression of each gene cluster in the murine models. Each panel describes the centroid expression of a gene cluster for the three murine models. The longitudinal trend lines were obtained by spline smoothing. There was great variability among the three murine models within each cluster for the time span, suggesting that mouse gene responses differed from one another.
11 A. Toll-like receptor expression in human burns Fold Change Log 2 (2) Log 2 (1.5) 0 -Log 2 (1.5) -Log 2 (2) FDR B. Toll-like receptor expression in the murine burns model
12 C. Toll-like receptor expression in human trauma D. Toll-like receptor expression in the murine trauma model
13 E. Toll-like receptor expression in human endotoxemia F. Toll-like receptor expression in the murine endotoxemia model
14 Fig. S8. The changes of genes in the TLR pathways for human injuries and the mouse models. Genes with FDR<0.01 are colored, and the rest genes are in white. Genes with significant changes (FDR<0.001 and fold change 2) are colored in red if activated or blue if suppressed. Genes with mild changes (FDR<0.01 and fold change 1.5) are colored in light red or light blue. Other genes (FDR<0.01 but fold change<1.5) are colored in gray. Toll-like receptor expression in (A) human burns, (B) the murine burns model, (C) human trauma, (D) the murine trauma model, (E) human endotoxemia, and (F) the murine endotoxemia model.
15 Fig. S9. Comparison of gene expression changes by radiation exposure in mice and patients. Gene expression data was obtained from GSE10640 (3, 4), which includes two independent sets of patients under irradiation as part of their pre-transplantation conditioning (n=24 and 10) and 11 mouse models. 487 significantly changed genes (FDR<0.05) between patients under irradiation and controls were identified. Plotted are the correlations (R 2, left, pink) of the fold changes of these genes and the percentages of genes changed in the same direction (%, right, green) between the first data set of patients (n=24) and the second set of patients (n=10) and the mouse models.
16 Table S1. Reproducibility of human and mouse studies. Standard deviations (SD) and coefficients of variation (CoV) were calculated from the biological replicates at each time point and averaged over the time points for mouse experiments and human endotoxemia. For human trauma and burn patients, the SD and CoV were calculated from samples of each binned time as described in Fig. S4 and averaged over the bins. Table S1. SD CoV Human Controls Human Burn Human Trauma Human Endotoxemia Mouse Controls Mouse Burn Model Mouse Trauma-Hemorrhage Model Mouse Endotoxemia Model
17 Table S2. Pathway comparisons between human injuries and mouse models. Shown are Pearson correlations (R 2 ) and percentages of genes changed to the same direction (%) for (A) the 20 most up-regulated and (B) the 20 most down-regulated pathways between the four model systems (human endotoxemia and the three murine models) versus human burn injury. Negative correlations are shown as negative numbers (-R 2 ). Human trauma is shown as reference. In every pathway, human endotoxemia had much higher similarity to human injury than mouse models had. Table S2A # Genes Human Trauma (R 2 ) Human Trauma (%) Human Endotoxemia (R 2 ) Fcγ Receptor-mediated Phagocytosis in Macrophages % % % % % and Monocytes IL-10 Signaling % % % % % Integrin Signaling % % % % % B Cell Receptor Signaling % % % % % Toll-like Receptor Signaling % % % % % Production of Nitric Oxide and Reactive Oxygen Species in % % % % % Macrophages Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses % % % % % TREM1 Signaling % % % % % GM-CSF Signaling % % % % % IL-6 Signaling % % % % % IL-15 Signaling % % % % % IL-8 Signaling % % % % % IL-1 Signaling % % % % % IL-3 Signaling % % % % % PI3K/AKT Signaling % % % % % p38 MAPK Signaling % % % % % PPARα/RXRα Activation % % % % % Leukocyte Extravasation Signaling % % % % % JAK/Stat Signaling % % % % % Role of PKR in Interferon Induction and Antiviral Response % % % % % Summary median minimum maximum % 96% 100% Human Endotoxemia (%) 95% 82% 100% Mouse Burn (R 2 ) Mouse Burn (%) 65% 48% 79% Mouse Trauma (R 2 ) Mouse Trauma (%) 51% 35% 71% Mouse Endotoxemia (R 2 ) Mouse Endotoxemia (%) 61% 43% 80%
18 Table S2B # Genes Human Trauma (R 2 ) Human Trauma (%) Human Endotoxemia (R 2 ) icos-icosl Signaling in T Helper Cells % % % % % CD28 Signaling in T Helper Cells % % % % % Calcium-induced T Lymphocyte Apoptosis % % % % % PKCθ Signaling in T Lymphocytes % % % % % T Cell Receptor Signaling % % % % % Role of NFAT in Regulation of the Immune Response % % % % % Primary Immunodeficiency Signaling % % % % % B Cell Development % % % % % CTLA4 Signaling in Cytotoxic T Lymphocytes % % % % % Antigen Presentation Pathway % % % % % EIF2 Signaling % % % % % IL-4 Signaling % % % % % NF-κB Activation by Viruses % % % % % Type I Diabetes Mellitus Signaling % % % % % Cytotoxic T Lymphocytemediated Apoptosis of Target Cells % % % % % T Helper Cell Differentiation % % % % % Purine Metabolism % % % % % Regulation of eif4 and p70s6k Signaling % % % % % mtor Signaling % % % % % OX40 Signaling Pathway % % % % % Summary median minimum maximum % 89% 100% % 80% 100% % 53% 75% % 47% 77% % 30% 60% Human Endotoxemia (%) Mouse Burn (R 2 ) Mouse Burn (%) Mouse Trauma (R 2 ) Mouse Trauma (%) Mouse Endotoxemia (R 2 ) Mouse Endotoxemia (%)
19 Acknowledgements List of additional participants and affiliations of the Inflammation and Host Response to Injury Large Scale Collaborative Research Program: Amer Abouhamze, Dept. of Surgery, University of Florida College of Medicine, Gainesville, FL Ulysses G.J. Balis, Dept. of Pathology, University of Michigan Medical School, Ann Arbor, MI David G. Camp II, Pacific Northwest National Laboratory, Richland, WA Asit K. De, Dept. of Surgery, University of Rochester Medical Center, Rochester, NY Brian G. Harbrecht, Dept. of Surgery, University of Louisville, Louisville, KY Douglas L. Hayden, Biostatistics Center, Massachusetts General Hospital, Boston, MA Amit Kaushal, Stanford Genome Technology Center, Stanford University, Palo Alto, CA Grant E. O'Keefe, Dept. of Surgery, University of Washington, Seattle, WA Kenneth T. Kotz, Dept. of Surgery, Massachusetts General Hospital, Boston, MA Weijun Qian, Pacific Northwest National Laboratory, Richland, WA David A. Schoenfeld, Biostatistics Center, Massachusetts General Hospital, Boston, MA Michael B. Shapiro, Dept. of Surgery, Northwestern University, Chicago, IL Geoffrey M. Silver, Dept. of Surgery, Loyola University School of Medicine, Maywood, IL Richard D. Smith, Pacific Northwest National Laboratory, Richland, WA John D. Storey, Dept. of Molecular Biology, Princeton University, Princeton, NJ Robert Tibshirani, Dept. of Statistics, Stanford University, Stanford, CA Mehmet Toner, Dept. of Surgery, Massachusetts General Hospital, Boston, MA Julie Wilhelmy, Stanford Genome Technology Center, Stanford University, Palo Alto, CA Bram Wispelwey, Biostatistics Center, Massachusetts General Hospital, Boston, MA Wing H Wong, Dept. of Statistics, Stanford University, Stanford, CA Supplemental References: 1. Storey JD, et al. (2005) Significance analysis of time course microarray experiments. Proc Natl Acad Sci U S A., 102(36): Blake JA, et al. (2011) The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res., 39(Database issue):d Dressman HK, et al. (2007) Gene expression signatures that predict radiation exposure in mice and humans. PLoS Med., 4(4):e Meadows SK, et al. (2008) Gene expression signatures of radiation response are specific, durable and accurate in mice and humans. PLoS One, 3(5):e1912.
Effector T Cells and
 1 Effector T Cells and Cytokines Andrew Lichtman, MD PhD Brigham and Women's Hospital Harvard Medical School 2 Lecture outline Cytokines Subsets of CD4+ T cells: definitions, functions, development New
1 Effector T Cells and Cytokines Andrew Lichtman, MD PhD Brigham and Women's Hospital Harvard Medical School 2 Lecture outline Cytokines Subsets of CD4+ T cells: definitions, functions, development New
Protein SD Units (P-value) Cluster order
 SUPPLEMENTAL TABLE AND FIGURES Table S1. Signature Phosphoproteome of CD22 E12 Transgenic Mouse BPL Cells. T-test vs. Other Protein SD Units (P-value) Cluster order ATPase (Ab-16) 1.41 0.000880 1 mtor
SUPPLEMENTAL TABLE AND FIGURES Table S1. Signature Phosphoproteome of CD22 E12 Transgenic Mouse BPL Cells. T-test vs. Other Protein SD Units (P-value) Cluster order ATPase (Ab-16) 1.41 0.000880 1 mtor
RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays
 Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Animal Models to Understand Immunity
 Animal Models to Understand Immunity Hussein El Saghire hesaghir@sckcen.be Innate Adaptive immunity Immunity MAPK and NF-kB TLR pathways receptors Fast Slow Non-specific Specific NOD-like receptors T-cell
Animal Models to Understand Immunity Hussein El Saghire hesaghir@sckcen.be Innate Adaptive immunity Immunity MAPK and NF-kB TLR pathways receptors Fast Slow Non-specific Specific NOD-like receptors T-cell
ACTIVATION AND EFFECTOR FUNCTIONS OF CELL-MEDIATED IMMUNITY AND NK CELLS. Choompone Sakonwasun, MD (Hons), FRCPT
 ACTIVATION AND EFFECTOR FUNCTIONS OF CELL-MEDIATED IMMUNITY AND NK CELLS Choompone Sakonwasun, MD (Hons), FRCPT Types of Adaptive Immunity Types of T Cell-mediated Immune Reactions CTLs = cytotoxic T lymphocytes
ACTIVATION AND EFFECTOR FUNCTIONS OF CELL-MEDIATED IMMUNITY AND NK CELLS Choompone Sakonwasun, MD (Hons), FRCPT Types of Adaptive Immunity Types of T Cell-mediated Immune Reactions CTLs = cytotoxic T lymphocytes
4. Th1-related gene expression in infected versus mock-infected controls from Fig. 2 with gene annotation.
 List of supplemental information 1. Graph of mouse weight loss during course of infection- Line graphs showing mouse weight data during course of infection days 1 to 10 post-infections (p.i.). 2. Graph
List of supplemental information 1. Graph of mouse weight loss during course of infection- Line graphs showing mouse weight data during course of infection days 1 to 10 post-infections (p.i.). 2. Graph
Supplemental Information. Systems Scale Interactive Exploration. Reveals Quantitative and Qualitative Differences
 Immunity, Volume 38 Supplemental Information Systems Scale Interactive Exploration Reveals Quantitative and Qualitative Differences in Response to Influenza and Pneumococcal Vaccines Gerlinde Obermoser,
Immunity, Volume 38 Supplemental Information Systems Scale Interactive Exploration Reveals Quantitative and Qualitative Differences in Response to Influenza and Pneumococcal Vaccines Gerlinde Obermoser,
Expanded View Figures
 Solip Park & Ben Lehner Epistasis is cancer type specific Molecular Systems Biology Expanded View Figures A B G C D E F H Figure EV1. Epistatic interactions detected in a pan-cancer analysis and saturation
Solip Park & Ben Lehner Epistasis is cancer type specific Molecular Systems Biology Expanded View Figures A B G C D E F H Figure EV1. Epistatic interactions detected in a pan-cancer analysis and saturation
Parallels between cancer and infectious disease
 Washington University School of Medicine Digital Commons@Becker Open Access Publications 2014 Parallels between cancer and infectious disease Richard S. Hotchkiss Washington University School of Medicine
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2014 Parallels between cancer and infectious disease Richard S. Hotchkiss Washington University School of Medicine
Statistical Assessment of the Global Regulatory Role of Histone. Acetylation in Saccharomyces cerevisiae. (Support Information)
 Statistical Assessment of the Global Regulatory Role of Histone Acetylation in Saccharomyces cerevisiae (Support Information) Authors: Guo-Cheng Yuan, Ping Ma, Wenxuan Zhong and Jun S. Liu Linear Relationship
Statistical Assessment of the Global Regulatory Role of Histone Acetylation in Saccharomyces cerevisiae (Support Information) Authors: Guo-Cheng Yuan, Ping Ma, Wenxuan Zhong and Jun S. Liu Linear Relationship
chapter 1 - fig. 2 Mechanism of transcriptional control by ppar agonists.
 chapter 1 - fig. 1 The -omics subdisciplines. chapter 1 - fig. 2 Mechanism of transcriptional control by ppar agonists. 201 figures chapter 1 chapter 2 - fig. 1 Schematic overview of the different steps
chapter 1 - fig. 1 The -omics subdisciplines. chapter 1 - fig. 2 Mechanism of transcriptional control by ppar agonists. 201 figures chapter 1 chapter 2 - fig. 1 Schematic overview of the different steps
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded
 Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
Nature Neuroscience: doi: /nn Supplementary Figure 1. Behavioral training.
 Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Supplementary Figures
 Supplementary Figures Figure S1. Urodynamic recording of BOO-induced LUTD patients. (A) DO group. BOO patients with increased detrusor pressure and reduced urine flow during pressure flow in combination
Supplementary Figures Figure S1. Urodynamic recording of BOO-induced LUTD patients. (A) DO group. BOO patients with increased detrusor pressure and reduced urine flow during pressure flow in combination
Supplementary Figure 1. ALVAC-protein vaccines and macaque immunization. (A) Maximum likelihood
 Supplementary Figure 1. ALVAC-protein vaccines and macaque immunization. (A) Maximum likelihood tree illustrating CRF01_AE gp120 protein sequence relationships between 107 Envs sampled in the RV144 trial
Supplementary Figure 1. ALVAC-protein vaccines and macaque immunization. (A) Maximum likelihood tree illustrating CRF01_AE gp120 protein sequence relationships between 107 Envs sampled in the RV144 trial
Computational Identification and Prediction of Tissue-Specific Alternative Splicing in H. Sapiens. Eric Van Nostrand CS229 Final Project
 Computational Identification and Prediction of Tissue-Specific Alternative Splicing in H. Sapiens. Eric Van Nostrand CS229 Final Project Introduction RNA splicing is a critical step in eukaryotic gene
Computational Identification and Prediction of Tissue-Specific Alternative Splicing in H. Sapiens. Eric Van Nostrand CS229 Final Project Introduction RNA splicing is a critical step in eukaryotic gene
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/278/rs11/dc1 Supplementary Materials for In Vivo Phosphoproteomics Analysis Reveals the Cardiac Targets of β-adrenergic Receptor Signaling Alicia Lundby,* Martin
www.sciencesignaling.org/cgi/content/full/6/278/rs11/dc1 Supplementary Materials for In Vivo Phosphoproteomics Analysis Reveals the Cardiac Targets of β-adrenergic Receptor Signaling Alicia Lundby,* Martin
SUPPLEMENTARY DATA Expression of complement factors is induced in DBA/2 mice following administration of C. albicans
 SUPPLEMENTARY DATA Expression of complement factors is induced in DBA/2 mice following administration of C. albicans water-soluble mannoprotein-beta-glucan complex (CAWS) Noriko Nagi-Miura 1,, Daisuke
SUPPLEMENTARY DATA Expression of complement factors is induced in DBA/2 mice following administration of C. albicans water-soluble mannoprotein-beta-glucan complex (CAWS) Noriko Nagi-Miura 1,, Daisuke
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 A β-strand positions consistently places the residues at CDR3β P6 and P7 within human and mouse TCR-peptide-MHC interfaces. (a) E8 TCR containing V β 13*06 carrying with an 11mer
Supplementary Figure 1 A β-strand positions consistently places the residues at CDR3β P6 and P7 within human and mouse TCR-peptide-MHC interfaces. (a) E8 TCR containing V β 13*06 carrying with an 11mer
Supplementary Figure 1. Biological characteris=cs of Smarcb1 flox/flox ; Rosa26- Cre ERT2 ; lymphomas
 Supplementary Figure 1. Biological characteris=cs of Smarcb1 flox/flox ; Rosa26- Cre ERT2 ; lymphomas a Tec Kinase Signaling CD28 Signaling in T Helper Cells Role of Macrophages in Rheumatoid Arthri>s
Supplementary Figure 1. Biological characteris=cs of Smarcb1 flox/flox ; Rosa26- Cre ERT2 ; lymphomas a Tec Kinase Signaling CD28 Signaling in T Helper Cells Role of Macrophages in Rheumatoid Arthri>s
Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types.
 Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplemental Information. Genomic Characterization of Murine. Monocytes Reveals C/EBPb Transcription. Factor Dependence of Ly6C Cells
 Immunity, Volume 46 Supplemental Information Genomic Characterization of Murine Monocytes Reveals C/EBPb Transcription Factor Dependence of Ly6C Cells Alexander Mildner, Jörg Schönheit, Amir Giladi, Eyal
Immunity, Volume 46 Supplemental Information Genomic Characterization of Murine Monocytes Reveals C/EBPb Transcription Factor Dependence of Ly6C Cells Alexander Mildner, Jörg Schönheit, Amir Giladi, Eyal
Expanded View Figures
 EMO Molecular Medicine Proteomic map of squamous cell carcinomas Hanibal ohnenberger et al Expanded View Figures Figure EV1. Technical reproducibility. Pearson s correlation analysis of normalised SILC
EMO Molecular Medicine Proteomic map of squamous cell carcinomas Hanibal ohnenberger et al Expanded View Figures Figure EV1. Technical reproducibility. Pearson s correlation analysis of normalised SILC
Supplementary Figure 1:
 Supplementary Figure 1: (A) Whole aortic cross-sections stained with Hematoxylin and Eosin (H&E), 7 days after porcine-pancreatic-elastase (PPE)-induced AAA compared to untreated, healthy control aortas
Supplementary Figure 1: (A) Whole aortic cross-sections stained with Hematoxylin and Eosin (H&E), 7 days after porcine-pancreatic-elastase (PPE)-induced AAA compared to untreated, healthy control aortas
Supplementary Figure 1
 Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer.
 Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Supplementary Materials
 Supplementary Materials Supplementary figure 1. Taxonomic representation summarized at genus level. Fecal microbiota from a separate set of Jackson and Harlan mice prior to irradiation. A taxon was included
Supplementary Materials Supplementary figure 1. Taxonomic representation summarized at genus level. Fecal microbiota from a separate set of Jackson and Harlan mice prior to irradiation. A taxon was included
The Human Microbiome in Chronic Obstructive Pulmonary Disease (COPD)
 The Human Microbiome in Chronic Obstructive Pulmonary Disease (COPD) Jeffrey L. Curtis, M.D. University of Michigan & Ann Arbor VA Healthsystem Ning Shen, M.D. Peking University Third Hospital Outline
The Human Microbiome in Chronic Obstructive Pulmonary Disease (COPD) Jeffrey L. Curtis, M.D. University of Michigan & Ann Arbor VA Healthsystem Ning Shen, M.D. Peking University Third Hospital Outline
Interferon-gamma pathway is activated in a chronically HBV infected chimpanzee that controls HBV following ARC-520 RNAi treatment
 Interferon-gamma pathway is activated in a chronically HBV infected chimpanzee that controls HBV following ARC-520 RNAi treatment Zhao Xu, Deborah Chavez, Jason E. Goetzmann, Robert E. Lanford and Christine
Interferon-gamma pathway is activated in a chronically HBV infected chimpanzee that controls HBV following ARC-520 RNAi treatment Zhao Xu, Deborah Chavez, Jason E. Goetzmann, Robert E. Lanford and Christine
T-cell activation T cells migrate to secondary lymphoid tissues where they interact with antigen, antigen-presenting cells, and other lymphocytes:
 Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
T-cell activation T cells migrate to secondary lymphoid tissues where they interact with antigen, antigen-presenting cells, and other lymphocytes:
 Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
Chapter 35 Active Reading Guide The Immune System
 Name: AP Biology Mr. Croft Chapter 35 Active Reading Guide The Immune System Section 1 Phagocytosis plays an important role in the immune systems of both invertebrates and vertebrates. Review the process
Name: AP Biology Mr. Croft Chapter 35 Active Reading Guide The Immune System Section 1 Phagocytosis plays an important role in the immune systems of both invertebrates and vertebrates. Review the process
Journal: Nature Methods
 Journal: Nature Methods Article Title: Network-based stratification of tumor mutations Corresponding Author: Trey Ideker Supplementary Item Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure
Journal: Nature Methods Article Title: Network-based stratification of tumor mutations Corresponding Author: Trey Ideker Supplementary Item Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure
The Immune System. A macrophage. ! Functions of the Immune System. ! Types of Immune Responses. ! Organization of the Immune System
 The Immune System! Functions of the Immune System! Types of Immune Responses! Organization of the Immune System! Innate Defense Mechanisms! Acquired Defense Mechanisms! Applied Immunology A macrophage
The Immune System! Functions of the Immune System! Types of Immune Responses! Organization of the Immune System! Innate Defense Mechanisms! Acquired Defense Mechanisms! Applied Immunology A macrophage
Lymphatic System. Chapter 14. Introduction. Main Channels of Lymphatics. Lymphatics. Lymph Tissue. Major Lymphatic Vessels of the Trunk
 Lymphatic System Chapter 14 Components Lymph is the fluid Vessels lymphatics Structures & organs Functions Return tissue fluid to the bloodstream Transport fats from the digestive tract to the bloodstream
Lymphatic System Chapter 14 Components Lymph is the fluid Vessels lymphatics Structures & organs Functions Return tissue fluid to the bloodstream Transport fats from the digestive tract to the bloodstream
Innate Immunity. Connection Between Innate and Adaptive Immunity. Know Differences and Provide Examples Chapter 3. Antimicrobial peptide psoriasin
 Know Differences and Provide Examples Chapter * Innate Immunity * kin and Epithelial Barriers * Antimicrobial peptide psoriasin -Activity against Gram (-) E. coli Connection Between Innate and Adaptive
Know Differences and Provide Examples Chapter * Innate Immunity * kin and Epithelial Barriers * Antimicrobial peptide psoriasin -Activity against Gram (-) E. coli Connection Between Innate and Adaptive
Titrations in Cytobank
 The Premier Platform for Single Cell Analysis (1) Titrations in Cytobank To analyze data from a titration in Cytobank, the first step is to upload your own FCS files or clone an experiment you already
The Premier Platform for Single Cell Analysis (1) Titrations in Cytobank To analyze data from a titration in Cytobank, the first step is to upload your own FCS files or clone an experiment you already
Supplemental Figure S1. RANK expression on human lung cancer cells.
 Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
System Biology analysis of innate and adaptive immune responses during HIV infection
 System Biology analysis of innate and adaptive immune responses during HIV infection Model of T cell memory persistence and exhaustion Naive Ag+APC Effector TEM (Pfp, Gr.B, FasL, TNF) Ag stim. IL-2, IL-7,
System Biology analysis of innate and adaptive immune responses during HIV infection Model of T cell memory persistence and exhaustion Naive Ag+APC Effector TEM (Pfp, Gr.B, FasL, TNF) Ag stim. IL-2, IL-7,
Innate immunity. Abul K. Abbas University of California San Francisco. FOCiS
 1 Innate immunity Abul K. Abbas University of California San Francisco FOCiS 2 Lecture outline Components of innate immunity Recognition of microbes and dead cells Toll Like Receptors NOD Like Receptors/Inflammasome
1 Innate immunity Abul K. Abbas University of California San Francisco FOCiS 2 Lecture outline Components of innate immunity Recognition of microbes and dead cells Toll Like Receptors NOD Like Receptors/Inflammasome
Comments on Significance of candidate cancer genes as assessed by the CaMP score by Parmigiani et al.
 Comments on Significance of candidate cancer genes as assessed by the CaMP score by Parmigiani et al. Holger Höfling Gad Getz Robert Tibshirani June 26, 2007 1 Introduction Identifying genes that are involved
Comments on Significance of candidate cancer genes as assessed by the CaMP score by Parmigiani et al. Holger Höfling Gad Getz Robert Tibshirani June 26, 2007 1 Introduction Identifying genes that are involved
Structure and Function of Antigen Recognition Molecules
 MICR2209 Structure and Function of Antigen Recognition Molecules Dr Allison Imrie allison.imrie@uwa.edu.au 1 Synopsis: In this lecture we will examine the major receptors used by cells of the innate and
MICR2209 Structure and Function of Antigen Recognition Molecules Dr Allison Imrie allison.imrie@uwa.edu.au 1 Synopsis: In this lecture we will examine the major receptors used by cells of the innate and
10th International Rotavirus Symposium Bangkok, Thailand
 Rotavirus Host Range Restriction and Innate Immunity: Mechanisms of Vaccine Attenuation Harry Greenberg MD Stanford University 10th International Rotavirus Symposium Bangkok, Thailand 09/19/12 B dsrna
Rotavirus Host Range Restriction and Innate Immunity: Mechanisms of Vaccine Attenuation Harry Greenberg MD Stanford University 10th International Rotavirus Symposium Bangkok, Thailand 09/19/12 B dsrna
Supplementary Figure 1: TSLP receptor skin expression in dcssc. A: Healthy control (HC) skin with TSLP receptor expression in brown (10x
 Supplementary Figure 1: TSLP receptor skin expression in dcssc. A: Healthy control (HC) skin with TSLP receptor expression in brown (10x magnification). B: Second HC skin stained for TSLP receptor in brown
Supplementary Figure 1: TSLP receptor skin expression in dcssc. A: Healthy control (HC) skin with TSLP receptor expression in brown (10x magnification). B: Second HC skin stained for TSLP receptor in brown
Version A. AP* Biology: Immune System. Name: Period
 Name: Period Version A AP* Biology: Immune System Directions: Each of the questions or incomplete statements below is followed by four suggested answers or completions. Select the one that is best in each
Name: Period Version A AP* Biology: Immune System Directions: Each of the questions or incomplete statements below is followed by four suggested answers or completions. Select the one that is best in each
Regulation of cell signaling cascades by influenza A virus
 The Second International Symposium on Optimization and Systems Biology (OSB 08) Lijiang, China, October 31 November 3, 2008 Copyright 2008 ORSC & APORC, pp. 389 394 Regulation of cell signaling cascades
The Second International Symposium on Optimization and Systems Biology (OSB 08) Lijiang, China, October 31 November 3, 2008 Copyright 2008 ORSC & APORC, pp. 389 394 Regulation of cell signaling cascades
Chromatin marks identify critical cell-types for fine-mapping complex trait variants
 Chromatin marks identify critical cell-types for fine-mapping complex trait variants Gosia Trynka 1-4 *, Cynthia Sandor 1-4 *, Buhm Han 1-4, Han Xu 5, Barbara E Stranger 1,4#, X Shirley Liu 5, and Soumya
Chromatin marks identify critical cell-types for fine-mapping complex trait variants Gosia Trynka 1-4 *, Cynthia Sandor 1-4 *, Buhm Han 1-4, Han Xu 5, Barbara E Stranger 1,4#, X Shirley Liu 5, and Soumya
Chronic HIV-1 Infection Frequently Fails to Protect against Superinfection
 Chronic HIV-1 Infection Frequently Fails to Protect against Superinfection Anne Piantadosi 1,2[, Bhavna Chohan 1,2[, Vrasha Chohan 3, R. Scott McClelland 3,4,5, Julie Overbaugh 1,2* 1 Division of Human
Chronic HIV-1 Infection Frequently Fails to Protect against Superinfection Anne Piantadosi 1,2[, Bhavna Chohan 1,2[, Vrasha Chohan 3, R. Scott McClelland 3,4,5, Julie Overbaugh 1,2* 1 Division of Human
A quick review. The clustering problem: Hierarchical clustering algorithm: Many possible distance metrics K-mean clustering algorithm:
 The clustering problem: partition genes into distinct sets with high homogeneity and high separation Hierarchical clustering algorithm: 1. Assign each object to a separate cluster. 2. Regroup the pair
The clustering problem: partition genes into distinct sets with high homogeneity and high separation Hierarchical clustering algorithm: 1. Assign each object to a separate cluster. 2. Regroup the pair
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Study Guide 23, 24 & 47
 Study Guide 23, 24 & 47 STUDY GUIDE SECTION 23-3 Bacteria and Humans Name Period Date 1. One bacterial disease that is transmitted by contaminated drinking water is a. Lyme disease b. gonorrhea c. tuberculosis
Study Guide 23, 24 & 47 STUDY GUIDE SECTION 23-3 Bacteria and Humans Name Period Date 1. One bacterial disease that is transmitted by contaminated drinking water is a. Lyme disease b. gonorrhea c. tuberculosis
Bayesian Inference for Single-cell ClUstering and ImpuTing (BISCUIT) Elham Azizi
 Bayesian Inference for Single-cell ClUstering and ImpuTing (BISCUIT) Elham Azizi BioC 2017: Where Software and Biology Connect Profiling Tumor-Immune Ecosystem in Breast Cancer Immunotherapy treatments
Bayesian Inference for Single-cell ClUstering and ImpuTing (BISCUIT) Elham Azizi BioC 2017: Where Software and Biology Connect Profiling Tumor-Immune Ecosystem in Breast Cancer Immunotherapy treatments
Standard Scores. Richard S. Balkin, Ph.D., LPC-S, NCC
 Standard Scores Richard S. Balkin, Ph.D., LPC-S, NCC 1 Normal Distributions While Best and Kahn (2003) indicated that the normal curve does not actually exist, measures of populations tend to demonstrate
Standard Scores Richard S. Balkin, Ph.D., LPC-S, NCC 1 Normal Distributions While Best and Kahn (2003) indicated that the normal curve does not actually exist, measures of populations tend to demonstrate
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
Protein Biomarker Biomarker Discover y y in Organ Organ Transplantation A A Proteomics Proteomics Approach Tara r Sig del, Sig PhD 9/26/2011
 Protein Biomarker Discovery in Organ Transplantation A Proteomics Approach Tara Sigdel, PhD 9/26/2011 Presented at the 2011 Stanford Mass Spectrometry Users Meeting For personal use only. Please do not
Protein Biomarker Discovery in Organ Transplantation A Proteomics Approach Tara Sigdel, PhD 9/26/2011 Presented at the 2011 Stanford Mass Spectrometry Users Meeting For personal use only. Please do not
OVARIAN CANCER POSSIBLE TUMOR-SUPPRESSIVE ROLE OF BATF2 IN OVARIAN CANCER
 POSSIBLE TUMOR-SUPPRESSIVE ROLE OF BATF2 IN OVARIAN CANCER Rissa Fedora, OMS-III OVARIAN CANCER 5 th leading cause of death among American women Risk factors: Age Gender female Greater lifetime ovulations
POSSIBLE TUMOR-SUPPRESSIVE ROLE OF BATF2 IN OVARIAN CANCER Rissa Fedora, OMS-III OVARIAN CANCER 5 th leading cause of death among American women Risk factors: Age Gender female Greater lifetime ovulations
Supplementary Material
 Supplementary Material Fig. S1. Gpa33 -/- mice do not express Gpa33 mrna or GPA33 protein. (A) The Gpa33 null allele contains a premature translational stop codon (STOP) in exon 2, and almost all of the
Supplementary Material Fig. S1. Gpa33 -/- mice do not express Gpa33 mrna or GPA33 protein. (A) The Gpa33 null allele contains a premature translational stop codon (STOP) in exon 2, and almost all of the
Intrinsic cellular defenses against virus infection
 Intrinsic cellular defenses against virus infection Detection of virus infection Host cell response to virus infection Interferons: structure and synthesis Induction of antiviral activity Viral defenses
Intrinsic cellular defenses against virus infection Detection of virus infection Host cell response to virus infection Interferons: structure and synthesis Induction of antiviral activity Viral defenses
Cancer outlier differential gene expression detection
 Biostatistics (2007), 8, 3, pp. 566 575 doi:10.1093/biostatistics/kxl029 Advance Access publication on October 4, 2006 Cancer outlier differential gene expression detection BAOLIN WU Division of Biostatistics,
Biostatistics (2007), 8, 3, pp. 566 575 doi:10.1093/biostatistics/kxl029 Advance Access publication on October 4, 2006 Cancer outlier differential gene expression detection BAOLIN WU Division of Biostatistics,
Nature Immunology: doi: /ni Supplementary Figure 1. Transcriptional program of the TE and MP CD8 + T cell subsets.
 Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Computational Approaches to Transcriptome Signatures in the Human Brain
 Computational Approaches to Transcriptome Signatures in the Human Brain Agilent Technologies eseminar February 18, 2016 Mike Hawrylycz, Ph.D. ALLEN Adult Human Atlas Online tools An Anatomic Transcriptional
Computational Approaches to Transcriptome Signatures in the Human Brain Agilent Technologies eseminar February 18, 2016 Mike Hawrylycz, Ph.D. ALLEN Adult Human Atlas Online tools An Anatomic Transcriptional
Models for HSV shedding must account for two levels of overdispersion
 UW Biostatistics Working Paper Series 1-20-2016 Models for HSV shedding must account for two levels of overdispersion Amalia Magaret University of Washington - Seattle Campus, amag@uw.edu Suggested Citation
UW Biostatistics Working Paper Series 1-20-2016 Models for HSV shedding must account for two levels of overdispersion Amalia Magaret University of Washington - Seattle Campus, amag@uw.edu Suggested Citation
CHAPTER I INTRODUCTION. for both infectious diseases and malignancies. Immunity is known as the innate
 CHAPTER I INTRODUCTION 1.1. Background of Study The immune system s function is to provide defense of the human body for both infectious diseases and malignancies. Immunity is known as the innate immunity
CHAPTER I INTRODUCTION 1.1. Background of Study The immune system s function is to provide defense of the human body for both infectious diseases and malignancies. Immunity is known as the innate immunity
MONTGOMERY COUNTY COMMUNITY COLLEGE Department of Science LECTURE OUTLINE CHAPTERS 16, 17, 18 AND 19
 MONTGOMERY COUNTY COMMUNITY COLLEGE Department of Science LECTURE OUTLINE CHAPTERS 16, 17, 18 AND 19 CHAPTER 16: NONSPECIFIC DEFENSES OF THE HOST I. THE FIRST LINE OF DEFENSE A. Mechanical Barriers (Physical
MONTGOMERY COUNTY COMMUNITY COLLEGE Department of Science LECTURE OUTLINE CHAPTERS 16, 17, 18 AND 19 CHAPTER 16: NONSPECIFIC DEFENSES OF THE HOST I. THE FIRST LINE OF DEFENSE A. Mechanical Barriers (Physical
Nature Genetics: doi: /ng Supplementary Figure 1. SEER data for male and female cancer incidence from
 Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
CPM (x 10-3 ) Tregs +Teffs. Tregs alone ICOS CLTA-4
 A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
Immunity. Skin. Mucin-containing mucous membranes. Desmosome (attaches keratincontaining. Fig. 43.2
 Immunity 1 Fig. 43.2 2 Skin Mucin-containing mucous membranes Desmosome (attaches keratincontaining skin cells together) 1 http://www.mhhe.com/biosci/ap/histology_mh/pseudos2l.jpg http://training.seer.cancer.gov/module_anatomy/images/
Immunity 1 Fig. 43.2 2 Skin Mucin-containing mucous membranes Desmosome (attaches keratincontaining skin cells together) 1 http://www.mhhe.com/biosci/ap/histology_mh/pseudos2l.jpg http://training.seer.cancer.gov/module_anatomy/images/
Supplementary. properties of. network types. randomly sampled. subsets (75%
 Supplementary Information Gene co-expression network analysis reveals common system-level prognostic genes across cancer types properties of Supplementary Figure 1 The robustness and overlap of prognostic
Supplementary Information Gene co-expression network analysis reveals common system-level prognostic genes across cancer types properties of Supplementary Figure 1 The robustness and overlap of prognostic
Basis and Clinical Applications of Interferon
 Interferon Therapy Basis and Clinical Applications of Interferon JMAJ 47(1): 7 12, 2004 Jiro IMANISHI Professor, Kyoto Prefectural University of Medicine Abstract: Interferon (IFN) is an antiviral substance
Interferon Therapy Basis and Clinical Applications of Interferon JMAJ 47(1): 7 12, 2004 Jiro IMANISHI Professor, Kyoto Prefectural University of Medicine Abstract: Interferon (IFN) is an antiviral substance
Supplementary Materials
 1 Supplementary Materials Rotger et al. Table S1A: Demographic characteristics of study participants. VNP RP EC CP (n=6) (n=66) (n=9) (n=5) Male gender, n(%) 5 (83) 54 (82) 5 (56) 3 (60) White ethnicity,
1 Supplementary Materials Rotger et al. Table S1A: Demographic characteristics of study participants. VNP RP EC CP (n=6) (n=66) (n=9) (n=5) Male gender, n(%) 5 (83) 54 (82) 5 (56) 3 (60) White ethnicity,
TD-BF01: Innate immunity to microorganisms
 TD-BF01: Innate immunity to microorganisms I. Toll receptors (adapted from Takeuchi, O. et al. (1999) Immunity 11:443; Kawai, T. et al. (1999) Immunity 11:115; Hemmi, H. et al. (2000) Nature 408:740; Muzio,
TD-BF01: Innate immunity to microorganisms I. Toll receptors (adapted from Takeuchi, O. et al. (1999) Immunity 11:443; Kawai, T. et al. (1999) Immunity 11:115; Hemmi, H. et al. (2000) Nature 408:740; Muzio,
chapter 17: specific/adaptable defenses of the host: the immune response
 chapter 17: specific/adaptable defenses of the host: the immune response defense against infection & illness body defenses innate/ non-specific adaptable/ specific epithelium, fever, inflammation, complement,
chapter 17: specific/adaptable defenses of the host: the immune response defense against infection & illness body defenses innate/ non-specific adaptable/ specific epithelium, fever, inflammation, complement,
T Cell Activation. Patricia Fitzgerald-Bocarsly March 18, 2009
 T Cell Activation Patricia Fitzgerald-Bocarsly March 18, 2009 Phases of Adaptive Immune Responses Phases of T cell responses IL-2 acts as an autocrine growth factor Fig. 11-11 Clonal Expansion of T cells
T Cell Activation Patricia Fitzgerald-Bocarsly March 18, 2009 Phases of Adaptive Immune Responses Phases of T cell responses IL-2 acts as an autocrine growth factor Fig. 11-11 Clonal Expansion of T cells
Skin. Mucin-containing mucous membranes. Desmosome (attaches keratincontaining
 Immunity 1 Fig. 43.2 2 Skin Mucin-containing mucous membranes Desmosome (attaches keratincontaining skin cells together) http://www.mhhe.com/biosci/ap/histology_mh/pseudos2l.jpg http://training.seer.cancer.gov/module_anatomy/images/
Immunity 1 Fig. 43.2 2 Skin Mucin-containing mucous membranes Desmosome (attaches keratincontaining skin cells together) http://www.mhhe.com/biosci/ap/histology_mh/pseudos2l.jpg http://training.seer.cancer.gov/module_anatomy/images/
L1 on PyMT tumor cells but Py117 cells are more responsive to IFN-γ. (A) Flow
 A MHCI B PD-L1 Fold expression 8 6 4 2 Fold expression 3 2 1 No tx 1Gy 2Gy IFN Py117 Py117 Supplementary Figure 1. Radiation and IFN-γ enhance MHCI expression and PD- L1 on PyMT tumor cells but Py117 cells
A MHCI B PD-L1 Fold expression 8 6 4 2 Fold expression 3 2 1 No tx 1Gy 2Gy IFN Py117 Py117 Supplementary Figure 1. Radiation and IFN-γ enhance MHCI expression and PD- L1 on PyMT tumor cells but Py117 cells
Nature Getetics: doi: /ng.3471
 Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
The Innate Immune Response
 The Innate Immune Response FUNCTIONS OF THE IMMUNE SYSTEM: Recognize, destroy and clear a diversity of pathogens. Initiate tissue and wound healing processes. Recognize and clear damaged self components.
The Innate Immune Response FUNCTIONS OF THE IMMUNE SYSTEM: Recognize, destroy and clear a diversity of pathogens. Initiate tissue and wound healing processes. Recognize and clear damaged self components.
Colon cancer subtypes from gene expression data
 Colon cancer subtypes from gene expression data Nathan Cunningham Giuseppe Di Benedetto Sherman Ip Leon Law Module 6: Applied Statistics 26th February 2016 Aim Replicate findings of Felipe De Sousa et
Colon cancer subtypes from gene expression data Nathan Cunningham Giuseppe Di Benedetto Sherman Ip Leon Law Module 6: Applied Statistics 26th February 2016 Aim Replicate findings of Felipe De Sousa et
SUPPLEMENTARY INFORMATION
 doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
Overview: The immune responses of animals can be divided into innate immunity and acquired immunity.
 GUIDED READING - Ch. 43 - THE IMMUNE SYSTEM NAME: Please print out these pages and HANDWRITE the answers directly on the printouts. Typed work or answers on separate sheets of paper will not be accepted.
GUIDED READING - Ch. 43 - THE IMMUNE SYSTEM NAME: Please print out these pages and HANDWRITE the answers directly on the printouts. Typed work or answers on separate sheets of paper will not be accepted.
Gene Ontology 2 Function/Pathway Enrichment. Biol4559 Thurs, April 12, 2018 Bill Pearson Pinn 6-057
 Gene Ontology 2 Function/Pathway Enrichment Biol4559 Thurs, April 12, 2018 Bill Pearson wrp@virginia.edu 4-2818 Pinn 6-057 Function/Pathway enrichment analysis do sets (subsets) of differentially expressed
Gene Ontology 2 Function/Pathway Enrichment Biol4559 Thurs, April 12, 2018 Bill Pearson wrp@virginia.edu 4-2818 Pinn 6-057 Function/Pathway enrichment analysis do sets (subsets) of differentially expressed
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Illustration of the working of network-based SVM to confidently predict a new (and now confirmed) ASD gene. Gene CTNND2 s brain network neighborhood that enabled its prediction by
Supplementary Figure 1 Illustration of the working of network-based SVM to confidently predict a new (and now confirmed) ASD gene. Gene CTNND2 s brain network neighborhood that enabled its prediction by
Nature Neuroscience: doi: /nn Supplementary Figure 1. Trial structure for go/no-go behavior
 Supplementary Figure 1 Trial structure for go/no-go behavior a, Overall timeline of experiments. Day 1: A1 mapping, injection of AAV1-SYN-GCAMP6s, cranial window and headpost implantation. Water restriction
Supplementary Figure 1 Trial structure for go/no-go behavior a, Overall timeline of experiments. Day 1: A1 mapping, injection of AAV1-SYN-GCAMP6s, cranial window and headpost implantation. Water restriction
Innate Immunity. Bởi: OpenStaxCollege
 Innate Immunity Bởi: OpenStaxCollege The vertebrate, including human, immune system is a complex multilayered system for defending against external and internal threats to the integrity of the body. The
Innate Immunity Bởi: OpenStaxCollege The vertebrate, including human, immune system is a complex multilayered system for defending against external and internal threats to the integrity of the body. The
Principles of Adaptive Immunity
 Principles of Adaptive Immunity Chapter 3 Parham Hans de Haard 17 th of May 2010 Agenda Recognition molecules of adaptive immune system Features adaptive immune system Immunoglobulins and T-cell receptors
Principles of Adaptive Immunity Chapter 3 Parham Hans de Haard 17 th of May 2010 Agenda Recognition molecules of adaptive immune system Features adaptive immune system Immunoglobulins and T-cell receptors
Numerous hypothesis tests were performed in this study. To reduce the false positive due to
 Two alternative data-splitting Numerous hypothesis tests were performed in this study. To reduce the false positive due to multiple testing, we are not only seeking the results with extremely small p values
Two alternative data-splitting Numerous hypothesis tests were performed in this study. To reduce the false positive due to multiple testing, we are not only seeking the results with extremely small p values
Effector mechanisms of cell-mediated immunity: Properties of effector, memory and regulatory T cells
 ICI Basic Immunology course Effector mechanisms of cell-mediated immunity: Properties of effector, memory and regulatory T cells Abul K. Abbas, MD UCSF Stages in the development of T cell responses: induction
ICI Basic Immunology course Effector mechanisms of cell-mediated immunity: Properties of effector, memory and regulatory T cells Abul K. Abbas, MD UCSF Stages in the development of T cell responses: induction
Table of Contents. Plots. Essential Statistics for Nursing Research 1/12/2017
 Essential Statistics for Nursing Research Kristen Carlin, MPH Seattle Nursing Research Workshop January 30, 2017 Table of Contents Plots Descriptive statistics Sample size/power Correlations Hypothesis
Essential Statistics for Nursing Research Kristen Carlin, MPH Seattle Nursing Research Workshop January 30, 2017 Table of Contents Plots Descriptive statistics Sample size/power Correlations Hypothesis
Innate Immunity. Chapter 3. Connection Between Innate and Adaptive Immunity. Know Differences and Provide Examples. Antimicrobial peptide psoriasin
 Chapter Know Differences and Provide Examples Innate Immunity kin and Epithelial Barriers Antimicrobial peptide psoriasin -Activity against Gram (-) E. coli Connection Between Innate and Adaptive Immunity
Chapter Know Differences and Provide Examples Innate Immunity kin and Epithelial Barriers Antimicrobial peptide psoriasin -Activity against Gram (-) E. coli Connection Between Innate and Adaptive Immunity
2. Innate immunity 2013
 1 Innate Immune Responses 3 Innate immunity Abul K. Abbas University of California San Francisco The initial responses to: 1. Microbes: essential early mechanisms to prevent, control, or eliminate infection;
1 Innate Immune Responses 3 Innate immunity Abul K. Abbas University of California San Francisco The initial responses to: 1. Microbes: essential early mechanisms to prevent, control, or eliminate infection;
Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and
 Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and Promoter Motif Analysis Shisong Ma 1,2*, Michael Snyder 3, and Savithramma P Dinesh-Kumar 2* 1 School of Life Sciences, University
Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and Promoter Motif Analysis Shisong Ma 1,2*, Michael Snyder 3, and Savithramma P Dinesh-Kumar 2* 1 School of Life Sciences, University
ONCOS-102 in melanoma Dr. Alexander Shoushtari. 4. ONCOS-102 in mesothelioma 5. Summary & closing
 ONCOS-102 in melanoma Dr. Alexander Shoushtari 4. ONCOS-102 in mesothelioma 5. Summary & closing 1 Preliminary data from C824 Activating the Alexander Shoushtari, MD Assistant Attending Physician Melanoma
ONCOS-102 in melanoma Dr. Alexander Shoushtari 4. ONCOS-102 in mesothelioma 5. Summary & closing 1 Preliminary data from C824 Activating the Alexander Shoushtari, MD Assistant Attending Physician Melanoma
Gene Ontology and Functional Enrichment. Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein
 Gene Ontology and Functional Enrichment Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein The parsimony principle: A quick review Find the tree that requires the fewest
Gene Ontology and Functional Enrichment Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein The parsimony principle: A quick review Find the tree that requires the fewest
Scientific Research. The Scientific Method. Scientific Explanation
 Scientific Research The Scientific Method Make systematic observations. Develop a testable explanation. Submit the explanation to empirical test. If explanation fails the test, then Revise the explanation
Scientific Research The Scientific Method Make systematic observations. Develop a testable explanation. Submit the explanation to empirical test. If explanation fails the test, then Revise the explanation
Supplementary Figure 1 Information on transgenic mouse models and their recording and optogenetic equipment. (a) 108 (b-c) (d) (e) (f) (g)
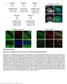 Supplementary Figure 1 Information on transgenic mouse models and their recording and optogenetic equipment. (a) In four mice, cre-dependent expression of the hyperpolarizing opsin Arch in pyramidal cells
Supplementary Figure 1 Information on transgenic mouse models and their recording and optogenetic equipment. (a) In four mice, cre-dependent expression of the hyperpolarizing opsin Arch in pyramidal cells
