An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors
|
|
|
- Imogen Joseph
- 5 years ago
- Views:
Transcription
1 377 Development 135, (2008) doi: /dev An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors Ophir D. Klein 1,2,3, David B. Lyons 1, Guive Balooch 4, Grayson W. Marshall 4, M. Albert Basson 5, Miroslav Peterka 6, Tomas Boran 6, Renata Peterkova 6 and Gail R. Martin 1, * Rodent incisors grow throughout adult life, but are prevented from becoming excessively long by constant abrasion, which is facilitated by the absence of enamel on one side of the incisor. Here we report that loss-of-function of sprouty genes, which encode antagonists of receptor tyrosine kinase signaling, leads to bilateral enamel deposition, thus impeding incisor abrasion and resulting in unchecked tooth elongation. We demonstrate that sprouty genes function to ensure that enamel-producing ameloblasts are generated on only one side of the tooth by inhibiting the formation of ectopic ameloblasts from self-renewing stem cells, and that they do so by preventing the establishment of an epithelial-mesenchymal FGF signaling loop. Interestingly, although inactivation of Spry4 alone initiates ectopic ameloblast formation in the embryo, the dosage of another sprouty gene must also be reduced to sustain it after birth. These data reveal that the generation of differentiated progeny from a particular stem cell population can be differently regulated in the embryo and adult. KEY WORDS: Ameloblast, Enamel, FGF signaling, Sprouty genes, Stem cells INTRODUCTION Rodent incisors are unusual among mammalian teeth in that they grow continuously throughout the life of the animal. This growth is fueled by stem cells in both the mesenchymal and epithelial compartments of the incisor, the progeny of which perpetually generate the various cell types in the tooth (Gronthos et al., 2002; Smith and Warshawsky, 1975). Incisor growth is counterbalanced by abrasion, without which the tooth would become excessively long and interfere with feeding. It would be difficult to abrade the incisors if enamel, the hardest component of the tooth, covered the entire surface of the incisor as it does in the molar. However, in rodent incisors enamel is normally present on the labial surface (facing the lip) and is absent on the lingual surface (facing the tongue) (see Fig. 1A). This asymmetry not only facilitates the abrasion that keeps incisor length relatively constant, but also ensures that the tooth will be filed down primarily on one side, thus generating a sharp tip (Addison and Appleton, 1915). Little is known about the stem cells that generate enamelproducing ameloblasts (ameloblast stem cells, or ASCs), because markers for them have not yet been identified. It has been proposed that ASCs reside in a niche located within a region called the cervical loop (CL) at the posterior end (base) of the incisor (Harada et al., 1999) (see Fig. 1A), but it is not known if ASCs give rise only to ameloblasts or also to the other epithelial cell types that must be continuously generated as the tooth grows. Based on models for the generation of differentiated progeny from stem cells in other tissues, such as the crypt of the intestinal villus (Fuchs et al., 2004), it has further been speculated that ameloblast formation begins when ASC 1 Department of Anatomy and Program in Developmental Biology, and 2 Department of Pediatrics, School of Medicine, University of California at San Francisco, San Francisco, CA , USA. 3 Department of Orofacial Sciences and 4 Department of Preventive and Restorative Dental Sciences, School of Dentistry, University of California at San Francisco, San Francisco, CA , USA. 5 Department of Craniofacial Development, King s College London, London, SE1 9RT, UK. 6 Department of Teratology, Institute of Experimental Medicine, Academy of Sciences of the Czech Republic, Prague, Czech Republic. *Author for correspondence ( gail.r.martin@ucsf.edu) Accepted 14 October 2007 progeny move out of the putative niche and develop into transitamplifying (T-A) cells that undergo a limited number of cell divisions (Harada et al., 1999; Wang et al., 2007). It is known that, once formed, ameloblasts move anteriorly along the length of the incisor (toward the tip) as they differentiate. After producing and depositing enamel, ameloblasts either undergo apoptosis or shrink in size (Smith and Warshawsky, 1975). Genetic analysis has shown that members of the fibroblast growth factor (FGF) family of secreted signaling molecules play a role in regulating ameloblast development or function, as Fgf3 / mice have defective enamel and Fgf3 / ;Fgf10 +/ mice have very thin or no enamel (Wang et al., 2007). Furthermore, based on data from studies of incisors developing in vitro it has been suggested that Fgf10 regulates epithelial stem cell survival (Harada et al., 2002; Yokohama-Tamaki et al., 2006). In wild-type incisors, the lack of lingual enamel is due to the absence of ameloblasts on that side (Smith and Warshawsky, 1975). However, it is not yet known whether lingual ameloblasts are absent because their formation from ASCs is blocked or because there are no ASCs in the lingual CL. Interestingly, the lingual CL differs in morphology from the labial CL (see Fig. 1A), which might reflect a lack of either T-A cells or ASCs. These morphological differences are correlated with asymmetries in the expression patterns of genes encoding members of the FGF family. For example, Fgf3 is detected in the mesenchyme surrounding the labial but not the lingual CL, and Fgf10 is expressed at a higher level on the labial side than on the lingual side (Harada et al., 1999). Furthermore, ectopic Fgf3 expression in lingual mesenchyme is associated with the formation of lingual ameloblasts in embryos homozygous for a null allele of follistatin (Fst), which encodes an extracellular inhibitor of signaling by transforming growth factor (TGF ) superfamily members (Wang et al., 2007; Wang et al., 2004). Together, the available data suggest that suppression of FGF signaling on the lingual side is necessary to prevent ameloblast formation. Here we identify sprouty (Spry) genes, which encode intracellular antagonists of FGF and other receptor-tyrosine kinase signaling pathways (Mason et al., 2006), as essential for establishing and sustaining the asymmetry of enamel deposition necessary for normal incisor length and shape. We provide genetic evidence that sprouty
2 378 Development 135 (2) genes prevent the generation of lingual ameloblasts by inhibiting an FGF-mediated epithelial-mesenchymal signaling loop on the lingual side. Furthermore, our data suggest that the earliest ameloblasts that form in the incisor do not arise from ASCs, but instead are derived from a transient embryonic ameloblast progenitor cell population that does not self-renew. MATERIALS AND METHODS Mouse lines Mouse lines carrying mutant alleles of Fgf9 (Colvin et al., 2001), Fgf10 (Min et al., 1998), Fst (Matzuk et al., 1995), Spry1 (Basson et al., 2005), Spry2 (Shim et al., 2005), Spry4 (Klein et al., 2006) and the K14-cre (Dassule et al., 2000) and Wnt1-cre (Danielian et al., 1998) transgenes were maintained and genotyped as reported, except that Fst wild-type and null alleles were genotyped using PCR rather than Southern blotting (primer sequences available on request). Age-matched CD1 embryos and adults were used as wild-type controls in all cases. Gene expression and histological analysis To stage embryos, noon of the day when a vaginal plug was detected was considered embryonic day (E) 0.5. RNA in situ hybridization was performed according to standard protocols on paraffin sections (10 m) using digoxigenin-labeled probes. Tissue was prepared for sectioning by fixing embryonic heads in 4% paraformaldehyde (PFA) or postnatal jaws in 4% PFA, and then decalcifying in RNAse-free EDTA for 2-4 days before sectioning. For postnatal histology, jaws were fixed in Bouin s solution, decalcified using a solution of 50% formic acid and 0.7 M sodium citrate (mixed 1:1), embedded in paraffin, sectioned at 7 m, and stained with Heidenhain s Azocarmine-aniline Blue (AZAN) stain. For photography of intact adult jaws, mouse heads were boiled for 30 minutes in distilled water and soft tissues were carefully removed. X-ray computed tomography X-ray computed tomography (XTM) was used to assess the degree of mineralization as previously described (Kinney et al., 2000). Briefly, mouse incisors (n=5 per group) were scanned at the Advanced Light Source (Lawrence Berkeley National Laboratory) and two-dimensional radiographs were obtained as the specimens were rotated through 180 in 0.5 increments. The radiographs were reconstructed into 1000 slices by Fourierfiltered back projection with a 10.5 m resolution. The attenuation coefficient (mm 1 ) of each pixel is represented by false colors and relates directly to mineral concentration. RESULTS Spry4 / ;Spry2 +/ mice have abnormal, tusk-like incisors due to ectopic enamel deposition on the lingual side of the tooth We found that Spry4 / ;Spry2 +/ mice, obtained by crossing mice carrying combinations of Spry4 (Klein et al., 2006) and Spry2 (Shim et al., 2005) null alleles, had excessively long and thick mandibular incisors that resembled tusks (Fig. 1B,F). Animals of this genotype will hereafter be referred to as tusk mutants. Abnormally long incisors can result from a lack of abrasion when the maxilla and mandible are misaligned. However, we detected no obvious craniofacial abnormalities in tusk mutants, and their incisors were thicker than those found in wild-type mice or in animals we sporadically found in our mouse colony in which there was overgrowth due to misalignment (not shown). Thus, we hypothesized that the excessive length and thickness of the tusk-like incisors might be due to deposition of lingual enamel, which would both thicken the mutant incisors and make them resistant to abrasion. We performed synchrotron XTM, which measures the relative mineral concentration in calcified tissues and distinguishes enamel from dentin and bone. In adult wild-type and Spry4 / (Spry4 null) incisors, which were of normal length and thickness, we detected enamel only on the labial surface (Fig. 1C,D). By contrast, tusk mutant incisors had enamel on both labial and lingual surfaces (Fig. 1E, and data not shown). Here we will focus on the phenotype of the mandibular incisors; the maxillary incisors have additional abnormalities, and will be described in a separate study. Sagittal histological sections of postnatal day (P) 14 tusk mutant incisors confirmed that they had ectopic enamel along much of the anteroposterior length of the lingual surface (Fig. 1G,H). Underlying this enamel we observed an ectopic layer of columnar cells, which resembled the ameloblasts that are normally found exclusively on the labial side (Fig. 1G-L). In addition to the presence of this ectopic layer of lingual ameloblasts, hereafter referred to as the lingual ameloblast phenotype, the morphology of the lingual CL was abnormal. In wild-type incisors the lingual CL is flattened and composed of a cuboidal epithelium (Fig. 1M, and data not shown), whereas in tusk mutants it resembled the labial CL in that it was more bulbous and composed of a columnar epithelium (Fig. 1O and data not shown; compare with Fig. 1N,P). These data indicate that sprouty genes function in the incisor to prevent ameloblast production on the lingual side, thus facilitating incisor abrasion and preventing excessive incisor length. Pre-ameloblasts are present on the lingual side of both Spry4 / ;Spry2 +/ and Spry4 / incisors at embryonic stages As ameloblasts are continuously lost at the anterior end of the rodent incisor as it grows (Smith and Warshawsky, 1977), and we found that lingual enamel was present throughout the life of the tusk mutants, it follows that ameloblasts must be continuously produced on the lingual side in these mice. To determine when lingual pre-ameloblasts (cells that have not yet begun to produce enamel matrix proteins) first appear in the tusk mutants we assayed for expression of sonic hedgehog (Shh), which is expressed in ameloblasts from a very early stage in their development and is downregulated as they mature (Bitgood and McMahon, 1995). On the labial side, we detected Shh-expressing cells in wild-type and tusk mutant incisors at E15.5 and 16.5, along the length of the epithelium anterior to the CL (Fig. 2A,B,D,E). At E16.5 we also detected expression of amelogenin (Amelx), a gene crucial for proper enamel formation (Zeichner-David et al., 1995), on the labial side near the anterior end of the incisor (Fig. 2C,F). Thus as expected, we found that cells in the labial epithelium were differentiating along the ameloblast lineage, with those at the anterior end of the incisor at a more advanced stage. On the lingual side, Shh-expressing cells were not detected in wild-type or tusk mutant incisors near the cervical loops at E15.5 (Fig. 2A,D), but by E16.5 Shh-expressing cells were observed in the lingual epithelium of the tusk mutant, in a small domain just anterior to the CL (Fig. 2B,E). These data suggest that lingual preameloblasts begin to form in tusk mutants between E15.5 and 16.5, at least one day after they are present on the labial side. These lingual pre-ameloblasts go on to differentiate, as Amelx expression was detected in the lingual epithelium of tusk mutants by E18.5 (not shown). We assumed that Spry4 null incisors would not have lingual ameloblasts, because the incisors in Spry4 null adults were neither excessively long nor thick (not shown) and had no enamel on the lingual side (Fig. 1D). Unexpectedly, we found that lingual Shhexpressing pre-ameloblasts were present in Spry4 null incisors at E16.5 (compare Fig. 2G,H and 2D,E), which matured into Amelxexpressing cells by E18.5 (data not shown). However, lingual ameloblasts ceased to be generated in Spry4 null animals after birth,
3 Sprouty gene function in mouse incisors 379 Fig. 1. Spry4 / ;Spry2+/ mice develop a tusk -like incisor due to the presence of enamel on the lingual surface. (A) Schematic diagram of an adult incisor. Enamel and dentin, the calcified tissues of the tooth, are produced by ameloblasts and odontoblasts, respectively. Note that in the normal incisor, ameloblasts and enamel are present only on the labial surface. (B,F) Side views of mandibles from wild-type and Spry4 / ;Spry2+/ adult mice with soft tissue removed. Note the abnormal length and thickness (red bar) of the mutant incisor, as well as the absence of a sharp tip (asterisk). (C-E) Coronal sections from an XTM analysis of wild-type, Spry4 / (4 / ), and Spry4 / ;Spry2+/ (4 / ;2+/ ) incisors. The colors in the bar on the left indicate mineral content. In the XTM sections, enamel is false-colored blue and purple, and dentin and bone are false-colored green and yellow. The white arrow points to ectopic enamel. (G,H) Sagittal sections of the mandibular incisor from postnatal day 14 wild-type and Spry4 / ;Spry2+/ animals. Here, and in all other panels in this and subsequent figures, anterior is to the left and posterior to the right. Yellow arrowheads point to a layer of enamel. Black arrows point to the CLs at the posterior end of the lingual and labial sides of the incisor. (I-L) Higher magnification views of regions boxed in G and H. Note that in the wild-type and mutant incisors, dentin (stained pink and/or blue) and odontoblasts are present on both lingual and labial sides, whereas enamel and ameloblasts are found only on the labial side in wild-type and on both labial and lingual sides in the Spry4 / ;Spry2+/ incisor. (M-P) High magnification views showing lingual and labial CL morphology in wild-type and Spry4 / ;Spry2+/ incisors. Note that the mutant lingual CL is more similar to the labial CL than to the wild-type lingual CL. Scale bars: 500 m in H; 100 m in L; 50 m in P. A, anterior; Am, ameloblasts; CL, cervical loop; De, Dentin; En, enamel; Ep, epithelium; Mes, mesenchyme; M1 and M2, first and second molar; Od, odontoblasts; P, posterior; wt, wild type. epithelium, an anterior domain in which the formation of preameloblasts is normally suppressed by FST but not SPRY4, and a posterior domain in which it is suppressed by both genes. Loss of sprouty function leads to abnormal FGF gene expression on the lingual side of the incisor As sprouty genes normally function to suppress FGF signaling in different developmental settings (Klein et al., 2006; Shim et al., 2005), we investigated whether loss of sprouty function affects the expression of targets of FGF signaling, which in the molar include FGF genes themselves (Kettunen et al., 2000; Kratochwil et al., 2002). In wild-type incisors at E16.5, we found that Fgf3 RNA was localized in labial mesenchyme just anterior to the CL (Fig. 3A). Fgf10 RNA was also detected in labial mesenchyme, but was more broadly distributed around the CL, in a domain that extended further anterior and also posterior to the Fgf3 expression domain (Fig. 3D). Fgf9 RNA was localized in a small epithelial domain just anterior to the labial CL (Fig. 3G). By contrast, on the lingual side, only Fgf10 RNA was detected in lingual mesenchyme anterior to and surrounding the CL, but at a much lower level than on the labial side. In both tusk mutant and Spry4 null incisors, the FGF gene expression patterns were indistinguishable from what we observed in wild-type embryos at E15.5 and earlier (not shown). However, by as discussed below. Pre-ameloblasts were not detected on the lingual side in either Spry4 null heterozygotes, even when they were Spry2 null, or in Spry1 / ;Spry2 / embryos at E16.5 (not shown). Together, these data show that of the three sprouty genes known to be expressed in the embryo (Minowada et al., 1999), Spry4 is uniquely required to suppress the generation of preameloblasts on the lingual side of the embryonic incisor, and that both alleles of Spry4 must be inactivated to obtain a lingual ameloblast phenotype. Previous studies have shown that the incisors in Fst null embryos have lingual ameloblasts (Wang et al., 2004), although it is not known what the postnatal incisor phenotype would be because Fst null embryos die perinatally (Matzuk et al., 1995). Because Fst is the only other gene known to be necessary to suppress the formation of lingual ameloblasts, we were interested to compare preameloblast formation in Fst and Spry4 null embryos. When we assayed Fst null embryos at E16.5, we detected Shh-expressing cells in a domain adjacent to the lingual CL similar to the one we observed in tusk mutant and Spry4 null incisors. However, there was also a second lingual domain of Shh-expressing cells in Fst null incisors, which began a short distance anterior to the first domain and extended anteriorly toward the tip of the tooth (Fig. 2I). These data show that there are two discrete domains in the lingual
4 380 Development 135 (2) Fig. 2. Shh expression reveals the presence of ectopic pre-ameloblasts in Spry4 / ;Spry2+/, in Spry4 /, and in Fst / embryonic mouse incisors. (A-I) Gene expression analyzed by RNA in situ hybridization using the indicated probes on paraffin sections of wild-type and mutant embryonic incisors. The stage at which the embryos were collected is indicated. In this and subsequent figures, a dotted line outlines the incisor epithelium. In D, the Shh expression domain appears to extend slightly into the lingual epithelium, but as this was not reproducibly observed it is likely to be an artifact of the plane of section. The red arrowheads in E and H point to a domain in which Shh is ectopically expressed on the lingual side of Spry4 / ;Spry2+/ and Spry4 / incisors. A similar domain is detected in Fst null incisors (panel I), together with an additional anterior domain of ectopic Shh expression (open arrowheads). Scale bars: 100 m. alteration in the morphology of the lingual CL, as described for the tusk mutant incisor at P14 (see Fig. 1O); indeed, the lingual CL in the E16.5 tusk and Spry4 null mutants appeared to be a mirror image of the labial CL (see Fig. 3B,C). We next assayed for expression of Etv4 and Etv5 (previously known as Pea3 and Erm, respectively), which are considered to be direct targets of RTK signaling (O Hagan and Hassell, 1998; Roehl and Nusslein-Volhard, 2001). In wild-type incisors at E16.5, we detected Etv4 and Etv5 RNAs in both labial mesenchyme and epithelium, anterior to the CL, and on the lingual side, only in the Fig. 3. FGF genes are upregulated or ectopically expressed on the lingual side of Spry4 / ;Spry2+/ and Spry4 / embryonic incisors. (A-K) Gene expression was analyzed by RNA in situ hybridization using the probes indicated on paraffin sections of E16.5 incisors of the genotypes denoted. Yellow asterisks indicate gene expression domains in the mesenchyme, and arrows point to the anterior and posterior ends of gene expression domains in the epithelium. All samples are shown at the same magnification. E16.5, Fgf3, Fgf10 and Fgf9 were all abnormally expressed on the lingual side. For Fgf3 and Fgf10, the expression pattern on the lingual side appeared to be a mirror image of that in labial mesenchyme (Fig. 3B,C,E,F). For Fgf9, lingual expression was limited to a small domain localized slightly more posteriorly than the labial Fgf9 domain, extending into lingual CL epithelium (Fig. 3H,I). This domain was similar to that of the lingual Shh expression domain (see Fig. 2E,H), suggesting that Fgf9 is expressed in lingual pre-ameloblasts. The increase in FGF gene expression detected on the lingual side of the mutant incisors was correlated with a marked
5 Sprouty gene function in mouse incisors 381 epithelium, in a small domain at the posterior end of the CL (Fig. 3J, and data not shown). By contrast, in tusk mutant incisors, Etv4 and Etv5 RNAs were detected at a high level on both the labial and lingual sides, throughout the posterior epithelium and in the mesenchyme adjacent to the CL (Fig. 3K, and data not shown). These data indicate that loss of sprouty function results in an increase in RTK signaling in incisor mesenchyme and epithelium on both the labial and lingual sides. Together these results suggest that the normal function of sprouty genes is to prevent the establishment of an epithelial-mesenchymal FGF signaling loop on the lingual side of the incisor that can stimulate ameloblast formation from a stem cell population in the CL. We also assayed for expression of a number of other genes that are thought to play a role in tooth development, including activin, Bmp4, Dll1, Fgf4, Fst, Jag2, Lfng, Notch1, Notch2, Notch3, Runx2 and Twist1, but found no evidence that they were abnormally expressed in tusk mutant incisors (not shown). These observations further suggest that upregulation of FGF gene expression, and consequently FGF signaling, is the primary cause of the observed phenotype in sprouty loss-of-function mutants. Continuous formation of lingual ameloblasts in adult mice requires loss of Spry4 function and reduction in the dosage of another sprouty gene Although loss of Spry4 function alone results in the formation of ameloblasts on the lingual side of the incisor at E16.5 (Fig. 2), there is no lingual enamel in Spry4 null adults (Fig. 1D). To investigate when the generation of lingual ameloblasts ceases, we examined sections of Spry4 null incisors at P5. In sagittal sections, we observed a long swath of lingual enamel over a layer of ameloblasts (Fig. 4A) extending anteriorly from a point approximately 750 m anterior to the lingual CL. The region from this point posterior to the CL appeared devoid of ameloblasts (Fig. 4A,B). This absence of ameloblasts was confirmed by examining serial coronal sections (not shown). By contrast, the labial ameloblast layer extended along the entire length of the tooth up to the CL (Fig. 4A,C). These data confirm that the ectopic Shh-expressing pre-ameloblasts detected in Spry4 null incisors at E16.5 subsequently differentiated into functional, enamel-producing ameloblasts, and show that lingual ameloblasts ceased being produced before P5. To determine whether this change had occurred by P2, we assayed for the expression of two of the genes that we found were affected by loss of Spry4 function at E16.5. Shh was robustly expressed in labial epithelium in all samples (Fig. 4D,E), but very few Shh-expressing cells were detected in the lingual epithelium of Spry4 null incisors (Fig. 4D). By contrast, there was strong lingual expression of Shh in tusk mutant incisors (Fig. 4E). Concomitant with the absence of Shh expression in Spry4 null lingual epithelium, Fgf3 was no longer abnormally expressed in lingual mesenchyme (Fig. 4F), whereas ectopic Fgf3 expression was detected in lingual mesenchyme of tusk mutant incisors at this stage (Fig. 4G). The absence of lingual Fgf3 expression in Spry4 null postnatal incisors was accompanied by a morphological change in the lingual CL from labial-like to linguallike (Fig. 4D, compare with Fig. 3C). These data show that formation of lingual pre-ameloblasts ceases around the time of birth in Spry4 null incisors, but continues in Spry4 null mice that also lack one copy of Spry2. Interestingly, we found that Spry4 / animals that carried one null allele of Spry1 (Basson et al., 2005) likewise displayed a tusk mutant phenotype (not shown). Thus, the production of lingual ameloblasts that is initiated prenatally as a result of loss of Spry4 function continues after birth only when the dosage of another sprouty gene, either Spry1 or Spry2, is reduced. Fig. 4. Loss of Spry4 function alone is not sufficient to maintain continuous production of ameloblasts on the lingual side of the incisor. (A-C) Sagittal section of postnatal day (P) 5 Spry4 / incisor. The areas boxed in A are shown at higher magnification in B and C. Note that unlike on the labial side, the lingual ameloblast layer does not extend to the posterior end of the incisor. (D-G) RNA in situ hybridization assays for Shh and Fgf3 expression in paraffin sections of P2 incisors of the genotypes indicated. Yellow asterisks indicate mesenchymal expression, and black arrows demarcate the extent of epithelial expression in these panels. Note that no ectopic expression of either gene is detected in the Spry4 / incisors at this stage. (H) Frequency of the tusk phenotype (presence of lingual enamel) in Spry4 / ;Spry2 +/ adult mice, and the effects of reducing the dosage of Fgf9 or Fgf10. The Spry4 / ;Spry2 +/ animals evaluated were pooled from both crosses used to generate animals with reduced FGF gene dosage. P-values were calculated using Fisher s exact test. Am, ameloblasts; Od, odontoblasts. The lingual ameloblast phenotype can be rescued in the adult by reducing FGF gene dosage To determine the extent to which lingual ameloblast formation is sensitive to FGF signaling, we produced tusk mutants heterozygous for an Fgf9 null allele (Colvin et al., 2001) (Spry4 / ;Spry2 +/ ;Fgf9 +/ adults), and examined their incisors by XTM. We found that only 14% had lingual enamel, compared with 100% of their Spry4 / ;Spry2 +/ littermates (P<0.01). Likewise, heterozygosity for a null allele of Fgf10 (Min et al., 1998) had a similar, although less dramatic, effect, in that 37% of Spry4 / ;Spry2 +/ ;Fgf10 +/ adults had lingual enamel, compared with 93% of their Spry4 / ;Spry2 +/ littermates (P<0.01) Fig. 4H). Thus, continuous formation of ameloblasts on the lingual side in Spry4 / ;Spry2 +/ adults is dependent on persistent abnormal expression of Fgf9 and Fgf10 at high levels. By contrast, when we assayed for Shh expression at E16.5, all sprouty mutant embryos that were heterozygous for an Fgf9 null allele (Spry4 / ;Fgf9 +/, n=4; Spry4 / ;Spry2 +/ ;Fgf9 +/, n=3) had lingual Shh-expressing pre-ameloblasts (not shown), as was
6 382 Development 135 (2) observed in all Spry4 null and tusk mutant incisors in which Fgf9 dosage was normal. Preliminary results on Spry4 null embryos with reduced dosage of Fgf10 were similar. Thus reducing FGF gene dosage in sprouty mutant embryos did not rescue the prenatal lingual ameloblast phenotype. These data raise the possibility that loss of sprouty function causes lingual ameloblast formation in embryonic incisors as a consequence of effects on non-fgf-mediated RTK signaling. Alternatively, there may be higher levels of FGF signaling in the embryonic than in the adult incisor, and therefore reducing FGF gene dosage by one copy does not sufficiently decrease FGF signaling in the embryo to prevent lingual ameloblast formation in the absence of sprouty gene function. Spry4 is required in both mesenchyme and epithelium to suppress the initiation of lingual ameloblast formation To determine where in the embryonic incisor Spry4 might function to suppress the initiation of lingual ameloblast formation, we assessed the expression pattern of Spry4 as well as Spry1 and Spry2 in wild-type embryos. At E14.5, we found significant differences in the expression domains of the three sprouty genes: Spry1 RNA was detected in both epithelium and mesenchyme (Fig. 5A); Spry2 RNA was detected exclusively in the epithelium (Fig. 5B); and Spry4 RNA was detected exclusively in the mesenchyme (Fig. 5C). However, by E16.5 not only Spry1 and Spry2, but also Spry4, expression was detected in the epithelium on both the labial and lingual sides of the incisor. In the mesenchyme, Spry1 and Spry4, but not Spry2, expression was detected on the labial side, and only Spry4 expression was detected, albeit at a low level, on the lingual side (Fig. 5D-F). As Spry4 expression was detected in both lingual epithelium and mesenchyme, we next sought to determine in which tissue(s) Spry4 function is required. To test whether Spry4 function is required in the epithelium, we performed Cre-mediated conditional loss-of-function experiments using a K14-cre transgene that has previously been used to inactivate floxed alleles in dental epithelium from early stages of tooth development (Dassule et al., 2000). First, we produced K14-cre; Spry4fl/ ;Spry2fl/+ embryos, and assayed for Shh, Fgf3, Fgf10 and Fgf9 expression at E17.5, to determine the effect of epitheliumspecific inactivation of Spry4 on lingual gene expression. As expected, Spry4 RNA was detected in incisor mesenchyme but not epithelium (Fig. 5G, compare with Fig. 5F). However, no abnormal lingual gene expression was observed in such embryos (Fig. 5I and data not shown), indicating that a complete loss of Spry4 function in the epithelium, even when the mesenchyme is heterozygous for Spry4, is not sufficient to allow the formation of lingual preameloblasts. To test whether Spry4 function is required in the mesenchyme, we employed a Wnt1-cre transgene (Danielian et al., 1998) that has previously been used to inactivate floxed alleles in dental mesenchyme from early stages of tooth development (Chai et al., 2000). We produced Wnt1-cre; Spry4fl/fl;Spry2fl/fl embryos, and assayed to determine the effect on lingual gene expression at E16.5. As expected, Spry4 expression was detected in incisor epithelium but not mesenchyme (Fig. 5H, compare with Fig. 5F). However, despite inactivation of both alleles of Spry4 in the mesenchyme, we Fig. 5. sprouty gene expression and tissue-specific inactivation in the developing mouse incisor. Gene expression was analyzed by RNA in situ hybridization using the probes indicated on paraffin sections of embryonic incisors of the genotypes denoted at E16.5 or (A-F) A comparison of the expression domains of sprouty gene family members in the incisor at the stages indicated. Yellow asterisks indicate mesenchymal expression. (G-K) Tissue-specific inactivation of Spry4. The absence of Spry4 expression is indicated by red circles in the epithelium of an incisor carrying K14-cre, one Spry4fl and one Spry4 allele (G) and by red asterisks in the mesenchyme of an incisor carrying Wnt1-cre and two Spry4fl alleles (H). For each genotype shown in I-K, the diagram illustrates the tissue in which Cre-mediated recombination occurred (green fill), the sprouty alleles that were inactivated by Cre (white lettering), or that were inherited as nulls (black lettering). The photograph shows Shh expression, which marks cells that are differentiating along the ameloblast lineage. The red arrowheads point to the ectopic lingual Shh expression domain.
7 Sprouty gene function in mouse incisors 383 detected no lingual expression of Shh or the other genes assayed in these embryos (Fig. 5J and data not shown). Thus, elimination of Spry4 function in only the mesenchyme is not sufficient to cause the formation of lingual pre-ameloblasts. We next assayed Wnt1-cre; Spry4 fl/ ;Spry2 fl/+ embryos, which inherited one Spry4 null allele and one floxed allele. In these embryos, in which the mesenchyme was null for Spry4 as a consequence of Wnt1-cre activity, and the epithelium was heterozygous for Spry4, we observed abnormal lingual gene expression (Fig. 5K and data not shown). These data indicate that inactivation of both Spry4 alleles in the mesenchyme and one Spry4 allele in the epithelium is required, when there are two functional Spry2 alleles (floxed and wild-type) in the epithelium, for the generation of Shh-expressing pre-ameloblasts on the lingual side of the embryonic incisor. Because these animals do have two functional Spry2 alleles in the epithelium, where Spry2 is exclusively expressed they would presumably cease producing ameloblasts after birth and thus would not display the tusk mutant phenotype. DISCUSSION In a few mammalian species, teeth have evolved the ability to grow throughout adult life (Tummers and Thesleff, 2003). One such tooth is the rodent incisor, in which continuous growth is coupled with a lack of enamel deposition on the lingual side, thereby providing a means of sharpening the tip and limiting incisor length. Here we demonstrate that in mice, sprouty genes are essential for ensuring that this vital asymmetry in enamel deposition occurs. Loss of Spry4 function results in the generation of enamel-producing ameloblasts on both the lingual and labial sides of the embryonic incisor. We present evidence that this phenotype is due to the establishment of an ectopic epithelial-mesenchymal FGF signaling loop on the lingual side. Interestingly, we found that this lingual ameloblast phenotype, which is robust in the embryo, is not sustained after birth unless the dosage of an additional sprouty gene, either Spry1 or Spry2, is reduced. The additive loss of sprouty function results in tusk-like adult incisors that are resistant to abrasion because enamel is deposited on both the lingual and labial sides. FGF signaling regulates the generation of lingual ameloblasts from stem cells It is now well established that loss of sprouty function causes hypersensitivity to FGF and other receptor tyrosine kinase signaling (Mason et al., 2006). Here we provide evidence that in the incisor, sprouty genes antagonize FGF signaling, and that loss of sprouty function results in upregulation of FGF gene expression on the lingual side. Three lines of evidence indicate that this increase in FGF gene expression is responsible for the generation of lingual ameloblasts. First, an increase in the level of lingual FGF gene expression is closely correlated with the initiation of Shh expression, a marker for pre-ameloblasts, in the lingual epithelium of late gestation embryos. Second, the reversal of the increase in lingual Fgf3 expression that we observed in Spry4 null incisors shortly after birth is closely correlated with cessation of lingual Shh expression. Third, reducing the dosage of either Fgf9 or Fgf10 substantially rescues the postnatal lingual ameloblast phenotype in tusk mutants, thus providing genetic evidence that high levels of FGF signaling are required to sustain the generation of lingual ameloblasts in adult sprouty mutants. Based on the results of our tissue-specific knockout experiments and gene expression analyses, we propose the following model to explain how sprouty genes normally function to prevent an increase in FGF gene expression and the consequent establishment of an FGF A E16.5 B Wild type E15.5 lingual labial SPRY2 SPRY4 FGF9 Wild type FGF10 SPRY2 SPRY4 FGF3 SPRY4 FGF10 FGF9 SPRY4 EAP Ameloblasts (differentiation in situ ) CL Ameloblast formation from EAPs inhibited by FST E16.5 Spry4 null SPRY2 FGF9 FGF3 FGF10 Ameloblast formation from ASCs inhibited by FST and Sprouty ASC ASC-derived Ameloblasts Fig. 6. Models for the role of sprouty genes in controlling FGF signaling in the mouse incisor and for the generation of embryonic ameloblasts. (A) Functions of sprouty genes in inhibiting the establishment of a lingual FGF epithelial-mesenchymal signaling loop. Arrows indicate a stimulatory effect and the symbol indicates an inhibitory effect of one signaling molecule on the expression of another. In wild type, sprouty genes are expressed on the labial side, but they do not prevent (dashed symbol) reciprocal signaling between FGF9 in epithelium and FGF3/FGF10 in mesenchyme. On the lingual side, sprouty genes inhibit signaling to adjacent tissues by the low levels of FGF9 in the epithelium and of FGF10 in the mesenchyme, and consequently there is no upregulation of FGF gene expression in either tissue. However, in Spry4 null incisors a reciprocal signaling loop between epithelium and mesenchyme is established because, in the absence of SPRY4, these tissues are hypersensitive to the low level of FGF signaling. In turn, the increase in FGF signaling on the lingual side results in the generation of ameloblasts from self-renewing stem cells in the CL. (B) A proposal for how ameloblasts develop in the embryonic incisor. At E15.5, wild-type incisor epithelium contains embryonic ameloblast progenitor (EAP) cells capable of limited proliferation. On the labial side, their descendants (in the domain colored pink) differentiate in situ into enamel-producing cells. Similar cells are present on the lingual side (in the domain colored lighter pink), but their differentiation is inhibited by Follistatin. Between E15.5 and 16.5, an ameloblast stem cell (ASC) population is established in the labial CL. Unlike EAP cells, ASCs have the capacity to self-renew (circular arrow), as well as give rise to ameloblasts. ASC descendants that will develop into enamel-producing cells may first form transit-amplifying (T-A) cells in the anterior CL. After several divisions, their descendants move out of the CL, and begin differentiating. In the E16.5 incisor, these ASC-derived pre-ameloblasts are found in a domain in the labial epithelium (colored dark brown), between the EAP domain and the CL. The diagram illustrates the possibility that ASCs are normally also present on the lingual side. However, no pre-ameloblasts derived from these ASCs are present in the lingual epithelium anterior to the CL (in the domain colored light brown), because the generation of ameloblasts from lingual ASCs is blocked due to the inhibitory effects of sprouty as well as FST function on lingual FGF gene expression. signaling loop on the lingual side of the incisor between E15.5 and 16.5 (see Fig. 6A). In the epithelium, SPRY4 normally prevents an FGF signal produced in the mesenchyme from inducing/ A P T-A
8 384 Development 135 (2) upregulating FGF gene expression. FGF10 is a good candidate for the mesenchymal signal that is antagonized by SPRY4 in the epithelium, as Fgf10 expression is normally detected at a low level in the mesenchyme surrounding the wild-type lingual CL at E16.5. Moreover, FGF10 is known to signal via the b isoform of FGFR2, which is expressed in lingual (and labial) epithelium (Peters et al., 1992). We propose that inactivation of a single allele of Spry4 in the epithelium renders it sufficiently hypersensitive to the small amount of FGF10 normally produced in the mesenchyme, such that the expression of Fgf9, a direct or indirect target of FGF10 signaling, is slightly upregulated in the lingual epithelium. However, if the mesenchyme is not sufficiently hypersensitive, then this increase in epithelial Fgf9 expression is presumably transitory, because the mesenchymal FGFs are not upregulated in response to the increase in epithelial FGF signal, and a positive-feedback signaling loop is therefore not established on the lingual side. Likewise, in the mesenchyme, SPRY4 prevents an FGF signal produced in the lingual epithelium from upregulating the expression of mesenchymal FGF gene(s). FGF9, which is normally produced in the lingual epithelium at such a low level that its expression is detected only with a radiolabeled probe [compare Fig. 3G with Wang et al. (Wang et al., 2007)], presumably signals via FGFR1, which is expressed throughout the incisor mesenchyme (Peters et al., 1992). Inactivating two alleles of Spry4 in the mesenchyme renders it hypersensitive to the small amount of FGF9 produced in the epithelium, such that the expression of Fgf3 and Fgf10, two direct or indirect targets of FGF9 signaling, is induced/upregulated in the lingual mesenchyme. Thus a positive-feedback FGF loop between lingual epithelium and mesenchyme is initiated and maintained for a few days in late-gestation Spry4 mutant incisors. However, if the epithelium is not also sufficiently hypersensitive, then it will not respond to increases in the level of mesenchymal FGF signals, and a positive-feedback signaling loop will not be established. Loss of sprouty function did not appear to cause any gross abnormalities on the labial side, where sprouty genes are highly expressed and presumably antagonize the FGF signaling that is required for normal ameloblast development and function. Analogous observations have been made in other developmental settings, where loss of sprouty function apparently affects only cells that do not normally respond to RTK signaling, but has little or no effect on cells that normally respond to high levels of RTK signaling (Basson et al., 2005; Klein et al., 2006; Shim et al., 2005). Precisely how the dynamics of the negative feedback between RTK signaling and sprouty-mediated inhibition of that signaling impacts such developmental systems remains to be elucidated. Comparison of sprouty and follistatin mutant incisors suggests that ameloblasts form from two different progenitor populations The data reported here must also be considered in the context of a larger genetic network that includes TGF family members and their antagonists, such as follistatin. Previous studies of the incisor phenotype in Fst null embryos have provided evidence that FST inhibits lingual ameloblast differentiation in the anterior incisor via a direct negative effect on BMP4 signaling (Wang et al., 2004), whereas in the lingual CL region it may do so by negatively regulating Fgf3 expression indirectly via effects on signaling by other TGF family members (Wang et al., 2007). Our analysis of Shh expression in Fst null mutants revealed that there are two physically separate domains in which pre-ameloblasts are found in the lingual epithelium at E16.5 (see Fig. 2I). We suggest that these anterior and posterior domains correspond to those in which ameloblast formation is thought to be inhibited by direct and indirect effects on TGF signaling, respectively. Significantly, we found that in embryonic Spry4 null incisors, lingual pre-ameloblasts are detected only in the posterior domain (see Fig. 2H), thus providing strong evidence that when ameloblasts form in mutant embryonic lingual epithelium, they do so in two domains that respond differently to increased signaling. To explain these observations, we propose the model illustrated in Fig. 6B. In the anterior domain, ameloblasts are generated from an embryonic ameloblast progenitor (EAP) population capable of giving rise to a limited number of progeny that subsequently differentiate in situ into enamel-producing cells. This process is analogous to that by which all ameloblasts in molars are thought to form (Zeichner-David et al., 1995). Development of ameloblasts from EAP cells in the anterior lingual domain is normally inhibited by FST acting to suppress BMP4 signaling, and is not suppressed by sprouty-mediated inhibition of FGF signaling. In contrast, in the posterior lingual domain, ameloblasts are derived from a selfrenewing ameloblast stem cell population located within the nearby CL. The formation of these ameloblasts depends on a high level of FGF signaling, which is normally suppressed by Spry4 function in the epithelium and mesenchyme. However, ameloblast formation in this domain is also suppressed by FST, most likely by an indirect negative effect on Fgf3 expression. One possible explanation for the observation that loss of function of either Fst or Spry4 leads to the induction of Fgf3 expression in lingual mesenchyme is that Fst and Spry4 act in the same genetic pathway. However, we found that loss of Fst function does not result in a decrease in Spry4 expression or vice versa (not shown), indicating that these genes may instead act in parallel pathways that converge on downstream targets and that can each affect Fgf3 expression. The increase in lingual Fgf3 expression that has been detected in Fst null mutants could sufficiently increase the level of FGF signaling from the mesenchyme to the epithelium, such that it overcomes the antagonism by sprouty genes that normally prevents the establishment of an FGF-signaling loop. We further suggest that ameloblast formation on the labial side likewise occurs by two different mechanisms: the first ameloblasts to form in the embryonic incisor develop from EAP cells; their formation requires BMP4 signaling but may occur independent of FGF signaling. Subsequently, ameloblasts are generated from ASCs in the labial CL, and this process depends on signaling via other TGF family members as well as high levels of FGF signaling. Based on these hypotheses, we speculate that a key event in incisor development is the establishment of the ASC population on the labial side between E15.5 and Formation of such stem cells would involve the acquisition, perhaps by a subpopulation of EAP cells, of the ability to self-renew as well as to produce progeny that give rise to ameloblasts (and perhaps other epithelial cell types) throughout the life of the animal. Possible mechanisms by which FGF signaling controls ameloblast formation Perhaps the most important question raised by our data is: how does an increase in FGF signaling on the lingual side of the incisor result in the formation of ameloblasts? One possibility is that ASCs are not normally present in the lingual CL, and that ectopic FGF signaling functions to induce their formation. Another possibility is that ASCs are present in the lingual CL, but normally they do not give rise to progeny that develop into ameloblasts. If so, then increased FGF signaling might stimulate them to form such
9 Sprouty gene function in mouse incisors 385 progeny, perhaps by promoting the formation and/or expansion of T-A cells that subsequently develop into ameloblasts. With respect to the latter suggestion, it is tempting to speculate that the transformation to a more labial CL-like morphology that occurs in the mutant lingual CL when it begins producing ameloblasts might reflect the presence of an expanded T-A cell population. At present, we are unable to explore these and other possibilities because markers for ASCs and T-A cells have not been identified. However, we are inclined to favor the hypothesis that FGF signaling affects T-A cells, if only because the domains in which ectopic Fgf3 and Fgf9 expression are detected in Spry4 null incisors are localized just anterior to the region in the CL where T-A cells are speculated to reside. One of our most intriguing findings is that a phenotype that appears to be very robust in the embryo the generation of an ectopic ameloblast population from stem cells on the lingual side of the incisor due to loss of Spry4 function is reversed just after birth unless an additional sprouty gene is inactivated. Based on our genetic rescue data, a likely explanation for this phenomenon is that the normal level of FGF signaling on the lingual side, albeit low, is higher in the embryo than in the adult. Thus the removal of an additional sprouty gene is required to render the adult epithelium and mesenchyme sufficiently sensitive to the adult level of FGF signaling to sustain the generation of lingual ameloblasts. These data reveal that the generation of differentiated progeny from a particular stem cell population can be differently regulated in the embryo and adult, and illustrate how manipulating levels of signals or their antagonists can provide a mechanism for enhancing the production of differentiated progeny from adult stem cells. We thank Drs M. Matzuk, D. Ornitz, L. Reichardt, D. Sheppard and D. Srivastava for helping us to obtain the mutant mouse lines used in this study, and Drs P. Denbesten and I. Thesleff for in situ probes. We are grateful to P. Ghatpande, I. Koppova, Z. Markova, A. Nemati, G. Nonomura, L. Prentice, H. Schmidt and E. Yu for excellent technical assistance. We also thank Drs J. Jernvall, B. Kraatz, and I. Thesleff for helpful discussion and advice, as well as M. Kumar, R. Metzger and our laboratory colleagues for critical reading of the manuscript. The Advanced Light Source is supported by the Director, Office of Basic Energy Sciences, of the U.S. Department of Energy (under Contract No. DE-AC02-05CH11231). O.K. was supported by a Pediatric Scientist Development Program award from the NIH (K12-HD00850). This work was supported by grants from the GACR (304/07/0223) and MSMT CR (project COST B23.002) to R.P. and M.P., the Medical Research Council (G ) and the Wellcome Trust (080470) to M.A.B., and the US National Institutes of Health (P01 DE09859) to G.W.M. and (R01 DE17744) to G.R.M. References Addison, W. H. and Appleton, J. L. (1915). The structure and growth of the incisor teeth of the albino rat. J. Morphol. 26, Basson, M. A., Akbulut, S., Watson-Johnson, J., Simon, R., Carroll, T. J., Shakya, R., Gross, I., Martin, G. R., Lufkin, T., McMahon, A. P. et al. (2005). Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev. Cell 8, Bitgood, M. J. and McMahon, A. P. (1995). Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev. Biol. 172, Chai, Y., Jiang, X., Ito, Y., Bringas, P., Jr, Han, J., Rowitch, D. H., Soriano, P., McMahon, A. P. and Sucov, H. M. (2000). Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127, Colvin, J. S., White, A. C., Pratt, S. J. and Ornitz, D. M. (2001). Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development 128, Danielian, P. S., Muccino, D., Rowitch, D. H., Michael, S. K. and McMahon, A. P. (1998). Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8, Dassule, H. R., Lewis, P., Bei, M., Maas, R. and McMahon, A. P. (2000). Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 127, Fuchs, E., Tumbar, T. and Guasch, G. (2004). Socializing with the neighbors: stem cells and their niche. Cell 116, Gronthos, S., Brahim, J., Li, W., Fisher, L. W., Cherman, N., Boyde, A., DenBesten, P., Robey, P. G. and Shi, S. (2002). Stem cell properties of human dental pulp stem cells. J. Dent. Res. 81, Harada, H., Kettunen, P., Jung, H. S., Mustonen, T., Wang, Y. A. and Thesleff, I. (1999). Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J. Cell Biol. 147, Harada, H., Toyono, T., Toyoshima, K., Yamasaki, M., Itoh, N., Kato, S., Sekine, K. and Ohuchi, H. (2002). FGF10 maintains stem cell compartment in developing mouse incisors. Development 129, Kettunen, P., Laurikkala, J., Itaranta, P., Vainio, S., Itoh, N. and Thesleff, I. (2000). Associations of FGF-3 and FGF-10 with signaling networks regulating tooth morphogenesis. Dev. Dyn. 219, Kinney, J. H., Haupt, D. L., Balooch, M., Ladd, A. J., Ryaby, J. T. and Lane, N. E. (2000). Three-dimensional morphometry of the L6 vertebra in the ovariectomized rat model of osteoporosis: biomechanical implications. J. Bone Miner. Res. 15, Klein, O. D., Minowada, G., Peterkova, R., Kangas, A., Yu, B. D., Lesot, H., Peterka, M., Jernvall, J. and Martin, G. R. (2006). Sprouty genes control diastema tooth development via bidirectional antagonism of epithelialmesenchymal FGF signaling. Dev. Cell 11, Kratochwil, K., Galceran, J., Tontsch, S., Roth, W. and Grosschedl, R. (2002). FGF4, a direct target of LEF1 and Wnt signaling, can rescue the arrest of tooth organogenesis in Lef1( / ) mice. Genes Dev. 16, Mason, J. M., Morrison, D. J., Basson, M. A. and Licht, J. D. (2006). Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 16, Matzuk, M. M., Lu, N., Vogel, H., Sellheyer, K., Roop, D. R. and Bradley, A. (1995). Multiple defects and perinatal death in mice deficient in follistatin. Nature 374, Min, H., Danilenko, D. M., Scully, S. A., Bolon, B., Ring, B. D., Tarpley, J. E., DeRose, M. and Simonet, W. S. (1998). Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 12, Minowada, G., Jarvis, L. A., Chi, C. L., Neubuser, A., Sun, X., Hacohen, N., Krasnow, M. A. and Martin, G. R. (1999). Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development 126, O Hagan, R. C. and Hassell, J. A. (1998). The PEA3 Ets transcription factor is a downstream target of the HER2/Neu receptor tyrosine kinase. Oncogene 16, Peters, K. G., Werner, S., Chen, G. and Williams, L. T. (1992). Two FGF receptor genes are differentially expressed in epithelial and mesenchymal tissues during limb formation and organogenesis in the mouse. Development 114, Roehl, H. and Nusslein-Volhard, C. (2001). Zebrafish pea3 and erm are general targets of FGF8 signaling. Curr. Biol. 11, Shim, K., Minowada, G., Coling, D. E. and Martin, G. R. (2005). Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev. Cell 8, Smith, C. E. and Warshawsky, H. (1975). Cellular renewal in the enamel organ and the odontoblast layer of the rat incisor as followed by radioautography using 3H-thymidine. Anat. Rec. 183, Smith, C. E. and Warshawsky, H. (1977). Quantitative analysis of cell turnover in the enamel organ of the rat incisor. Evidence for ameloblast death immediately after enamel matrix secretion. Anat. Rec. 187, Tummers, M. and Thesleff, I. (2003). Root or crown: a developmental choice orchestrated by the differential regulation of the epithelial stem cell niche in the tooth of two rodent species. Development 130, Wang, X. P., Suomalainen, M., Jorgez, C. J., Matzuk, M. M., Werner, S. and Thesleff, I. (2004). Follistatin regulates enamel patterning in mouse incisors by asymmetrically inhibiting BMP signaling and ameloblast differentiation. Dev. Cell 7, Wang, X. P., Suomalainen, M., Felszeghy, S., Zelarayan, L. C., Alonso, M. T., Plikus, M. V., Maas, R. L., Chuong, C. M., Schimmang, T. and Thesleff, I. (2007). An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 5, e159. Yokohama-Tamaki, T., Ohshima, H., Fujiwara, N., Takada, Y., Ichimori, Y., Wakisaka, S., Ohuchi, H. and Harada, H. (2006). Cessation of Fgf10 signaling, resulting in a defective dental epithelial stem cell compartment, leads to the transition from crown to root formation. Development 133, Zeichner-David, M., Diekwisch, T., Fincham, A., Lau, E., MacDougall, M., Moradian-Oldak, J., Simmer, J., Snead, M. and Slavkin, H. C. (1995). Control of ameloblast differentiation. Int. J. Dev. Biol. 39,
BCL11B Regulates Epithelial Proliferation and Asymmetric Development of the Mouse Mandibular Incisor
 BCL11B Regulates Epithelial Proliferation and Asymmetric Development of the Mouse Mandibular Incisor Kateryna Kyrylkova 1, Sergiy Kyryachenko 1, Brian Biehs 2 *, Ophir Klein 2, Chrissa Kioussi 1 *, Mark
BCL11B Regulates Epithelial Proliferation and Asymmetric Development of the Mouse Mandibular Incisor Kateryna Kyrylkova 1, Sergiy Kyryachenko 1, Brian Biehs 2 *, Ophir Klein 2, Chrissa Kioussi 1 *, Mark
Temporal Analysis of Ectopic Enamel Production in Incisors From Sprouty Mutant Mice
 JOURNAL OF EXPERIMENTAL ZOOLOGY (MOL DEV EVOL) 312B (2009) Temporal Analysis of Ectopic Enamel Production in Incisors From Sprouty Mutant Mice TOMAS BORAN 1,2, RENATA PETERKOVA 1, HERVE LESOT 3,4,5, DAVID
JOURNAL OF EXPERIMENTAL ZOOLOGY (MOL DEV EVOL) 312B (2009) Temporal Analysis of Ectopic Enamel Production in Incisors From Sprouty Mutant Mice TOMAS BORAN 1,2, RENATA PETERKOVA 1, HERVE LESOT 3,4,5, DAVID
The eternal tooth germ is formed at the apical end of continuously growing teeth*
 The eternal tooth germ is formed at the apical end of continuously growing teeth* Hayato Ohshima 1, Naohiro Nakasone 1, 2, Emi Hashimoto 1, Hideo Sakai 1, Kuniko Nakakura-Ohshima 3 and Hidemitsu Harada
The eternal tooth germ is formed at the apical end of continuously growing teeth* Hayato Ohshima 1, Naohiro Nakasone 1, 2, Emi Hashimoto 1, Hideo Sakai 1, Kuniko Nakakura-Ohshima 3 and Hidemitsu Harada
Sprouty Genes Control Diastema Tooth Development via Bidirectional Antagonism of Epithelial-Mesenchymal FGF Signaling
 Developmental Cell 11, 181 190, August, 2006 ª2006 Elsevier Inc. DOI 10.1016/j.devcel.2006.05.014 Sprouty Genes Control Diastema Tooth Development via Bidirectional Antagonism of Epithelial-Mesenchymal
Developmental Cell 11, 181 190, August, 2006 ª2006 Elsevier Inc. DOI 10.1016/j.devcel.2006.05.014 Sprouty Genes Control Diastema Tooth Development via Bidirectional Antagonism of Epithelial-Mesenchymal
The Epithelial-Mesenchymal Interaction Plays a Role in the Maintenance of the Stem Cell Niche of Mouse Incisors via Fgf10 and Fgf9 Signaling
 The Open Biotechnology Journal, 2008, 2, 111-115 111 The Epithelial-Mesenchymal Interaction Plays a Role in the Maintenance of the Stem Cell Niche of Mouse Incisors via Fgf10 and Fgf9 Signaling Tamaki
The Open Biotechnology Journal, 2008, 2, 111-115 111 The Epithelial-Mesenchymal Interaction Plays a Role in the Maintenance of the Stem Cell Niche of Mouse Incisors via Fgf10 and Fgf9 Signaling Tamaki
Supplementary Figure 1: Signaling centers contain few proliferating cells, express p21, and
 Supplementary Figure 1: Signaling centers contain few proliferating cells, express p21, and exclude YAP from the nucleus. (a) Schematic diagram of an E10.5 mouse embryo. (b,c) Sections at B and C in (a)
Supplementary Figure 1: Signaling centers contain few proliferating cells, express p21, and exclude YAP from the nucleus. (a) Schematic diagram of an E10.5 mouse embryo. (b,c) Sections at B and C in (a)
Tooth development in the 'crooked' mouse
 /. Embryo!, exp. Morph. Vol. 41, pp. 279-287, 1977 279 Printed in Great Britain Company of Biologists Limited 1977 Tooth development in the 'crooked' mouse By J. A. SOFAER 1 From the University of Edinburgh,
/. Embryo!, exp. Morph. Vol. 41, pp. 279-287, 1977 279 Printed in Great Britain Company of Biologists Limited 1977 Tooth development in the 'crooked' mouse By J. A. SOFAER 1 From the University of Edinburgh,
Signaling by FGFR2b controls the regenerative capacity of adult mouse incisors
 AND STEM CELLS RESEARCH ARTICLE 3743 Development 137, 3743-3752 (2010) doi:10.1242/dev.051672 2010. Published by The Company of Biologists Ltd Signaling by FGFR2b controls the regenerative capacity of
AND STEM CELLS RESEARCH ARTICLE 3743 Development 137, 3743-3752 (2010) doi:10.1242/dev.051672 2010. Published by The Company of Biologists Ltd Signaling by FGFR2b controls the regenerative capacity of
Cooperative and independent functions of FGF and Wnt signaling during early inner ear development
 Wright et al. BMC Developmental Biology (2015) 15:33 DOI 10.1186/s12861-015-0083-8 RESEARCH ARTICLE Open Access Cooperative and independent functions of FGF and Wnt signaling during early inner ear development
Wright et al. BMC Developmental Biology (2015) 15:33 DOI 10.1186/s12861-015-0083-8 RESEARCH ARTICLE Open Access Cooperative and independent functions of FGF and Wnt signaling during early inner ear development
Revitalization of a Diastemal Tooth Primordium in Spry2 Null Mice Results From Increased Proliferation and Decreased Apoptosis
 JOURNAL OF EXPERIMENTAL ZOOLOGY (MOL DEV EVOL) 312B:292 308 (2009) Revitalization of a Diastemal Tooth Primordium in Spry2 Null Mice Results From Increased Proliferation and Decreased Apoptosis RENATA
JOURNAL OF EXPERIMENTAL ZOOLOGY (MOL DEV EVOL) 312B:292 308 (2009) Revitalization of a Diastemal Tooth Primordium in Spry2 Null Mice Results From Increased Proliferation and Decreased Apoptosis RENATA
Ahtiainen et al., http :// /cgi /content /full /jcb /DC1
 Supplemental material JCB Ahtiainen et al., http ://www.jcb.org /cgi /content /full /jcb.201512074 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Distinct distribution of different cell cycle phases in the
Supplemental material JCB Ahtiainen et al., http ://www.jcb.org /cgi /content /full /jcb.201512074 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Distinct distribution of different cell cycle phases in the
An Integrated Gene Regulatory Network Controls Stem Cell Proliferation in Teeth
 An Integrated Gene Regulatory Network Controls Stem Cell Proliferation in Teeth The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters.
An Integrated Gene Regulatory Network Controls Stem Cell Proliferation in Teeth The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters.
CHAPTER 6 SUMMARIZING DISCUSSION
 CHAPTER 6 SUMMARIZING DISCUSSION More than 20 years ago the founding member of the Wnt gene family, Wnt-1/Int1, was discovered as a proto-oncogene activated in mammary gland tumors by the mouse mammary
CHAPTER 6 SUMMARIZING DISCUSSION More than 20 years ago the founding member of the Wnt gene family, Wnt-1/Int1, was discovered as a proto-oncogene activated in mammary gland tumors by the mouse mammary
Distinct Roles Of CCN1 And CCN2 In Limb Development
 Distinct Roles Of CCN1 And CCN2 In Limb Development Jie Jiang, PhD 1, Jessica Ong, BS 1, Faith Hall-Glenn, PhD 2, Teni Anbarchian, BS 1, Karen M. Lyons, PhD 1. 1 University of California, Los Angeles,
Distinct Roles Of CCN1 And CCN2 In Limb Development Jie Jiang, PhD 1, Jessica Ong, BS 1, Faith Hall-Glenn, PhD 2, Teni Anbarchian, BS 1, Karen M. Lyons, PhD 1. 1 University of California, Los Angeles,
Development of teeth. 5.DM - Pedo
 Development of teeth 5.DM - Pedo Tooth development process of continuous changes in predetermined order starts from dental lamina A band of ectodermal cells growing from the epithelium of the embryonic
Development of teeth 5.DM - Pedo Tooth development process of continuous changes in predetermined order starts from dental lamina A band of ectodermal cells growing from the epithelium of the embryonic
DEVELOPMENT AND DISEASE FGF10 maintains stem cell compartment in developing mouse incisors
 Development 129, 1533-1541 (2002) Printed in Great Britain The Company of Biologists Limited 2002 DEV14510 1533 DEVELOPMENT AND DISEASE FGF10 maintains stem cell compartment in developing mouse incisors
Development 129, 1533-1541 (2002) Printed in Great Britain The Company of Biologists Limited 2002 DEV14510 1533 DEVELOPMENT AND DISEASE FGF10 maintains stem cell compartment in developing mouse incisors
Branching morphogenesis of the lung: new molecular insights into an old problem
 86 Review TRENDS in Cell Biology Vol.13 No.2 February 2003 Branching morphogenesis of the lung: new molecular insights into an old problem Pao-Tien Chuang 1 and Andrew P. McMahon 2 1 Cardiovascular Research
86 Review TRENDS in Cell Biology Vol.13 No.2 February 2003 Branching morphogenesis of the lung: new molecular insights into an old problem Pao-Tien Chuang 1 and Andrew P. McMahon 2 1 Cardiovascular Research
Inhibition of Wnt signaling by Wise (Sostdc1) and negative feedback from Shh controls tooth number and patterning
 RESEARCH ARTICLE 3221 Development 137, 3221-3231 (2010) doi:10.1242/dev.054668 2010. Published by The Company of Biologists Ltd Inhibition of Wnt signaling by Wise (Sostdc1) and negative feedback from
RESEARCH ARTICLE 3221 Development 137, 3221-3231 (2010) doi:10.1242/dev.054668 2010. Published by The Company of Biologists Ltd Inhibition of Wnt signaling by Wise (Sostdc1) and negative feedback from
Vertebrate Limb Patterning
 Vertebrate Limb Patterning What makes limb patterning an interesting/useful developmental system How limbs develop Key events in limb development positioning and specification initiation of outgrowth establishment
Vertebrate Limb Patterning What makes limb patterning an interesting/useful developmental system How limbs develop Key events in limb development positioning and specification initiation of outgrowth establishment
Teeth, orofacial development and
 Teeth, orofacial development and cleft anomalies Miroslav Peterka Variability of jaws in vertebrates. (A) cartilaginous fish shark; (B) an example of a bone fish; (C ) amphibian frog; (D) reptile - turtle;
Teeth, orofacial development and cleft anomalies Miroslav Peterka Variability of jaws in vertebrates. (A) cartilaginous fish shark; (B) an example of a bone fish; (C ) amphibian frog; (D) reptile - turtle;
ODONTOGENESIS- A HIGHLY COMPLEX CELL-CELL INTERACTION PROCESS
 ODONTOGENESIS- A HIGHLY COMPLEX CELL-CELL INTERACTION PROCESS AMBRISH KAUSHAL, MALA KAMBOJ Department of Oral and Maxillofacial Pathology Career Post Graduate Institute of Dental Sciences and Hospital
ODONTOGENESIS- A HIGHLY COMPLEX CELL-CELL INTERACTION PROCESS AMBRISH KAUSHAL, MALA KAMBOJ Department of Oral and Maxillofacial Pathology Career Post Graduate Institute of Dental Sciences and Hospital
Emx2 patterns the neocortex by regulating FGF positional signaling
 Emx2 patterns the neocortex by regulating FGF positional signaling Tomomi Fukuchi-Shimogori and Elizabeth A Grove Presented by Sally Kwok Background Cerebral cortex has anatomically and functionally distinct
Emx2 patterns the neocortex by regulating FGF positional signaling Tomomi Fukuchi-Shimogori and Elizabeth A Grove Presented by Sally Kwok Background Cerebral cortex has anatomically and functionally distinct
Developmental Biology
 Developmental Biology 328 (2009) 493 505 Contents lists available at ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology Enamel-free teeth: Tbx1 deletion affects
Developmental Biology 328 (2009) 493 505 Contents lists available at ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology Enamel-free teeth: Tbx1 deletion affects
06 Tooth Development and Eruption
 + 06 Tooth Development and Eruption Tooth development Root development PDL and alveolar bone development Primary tooth eruption and shedding Permanent tooth eruption Q. Where and how tooth starts to form?
+ 06 Tooth Development and Eruption Tooth development Root development PDL and alveolar bone development Primary tooth eruption and shedding Permanent tooth eruption Q. Where and how tooth starts to form?
evolution and development of primate teeth
 evolution and development of primate teeth diversity of mammalian teeth upper left molars buccal mesial distal lingual Jernvall & Salazar-Ciudad 07 trends in dental evolution many similar single-cusped
evolution and development of primate teeth diversity of mammalian teeth upper left molars buccal mesial distal lingual Jernvall & Salazar-Ciudad 07 trends in dental evolution many similar single-cusped
Regulation of tooth number by fine-tuning levels of receptor-tyrosine kinase signaling
 RESEARCH ARTICLE 4063 Development 138, 4063-4073 (2011) doi:10.1242/dev.069195 2011. Published by The Company of Biologists Ltd Regulation of tooth number by fine-tuning levels of receptor-tyrosine kinase
RESEARCH ARTICLE 4063 Development 138, 4063-4073 (2011) doi:10.1242/dev.069195 2011. Published by The Company of Biologists Ltd Regulation of tooth number by fine-tuning levels of receptor-tyrosine kinase
Sonic hedgehog regulates growth and morphogenesis of the tooth
 Development 127, 4775-4785 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV3251 4775 Sonic hedgehog regulates growth and morphogenesis of the tooth Hélène R. Dassule 1, Paula
Development 127, 4775-4785 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV3251 4775 Sonic hedgehog regulates growth and morphogenesis of the tooth Hélène R. Dassule 1, Paula
Journal Club. 03/04/2012 Lama Nazzal
 Journal Club 03/04/2012 Lama Nazzal NOTCH and the kidneys Is an evolutionarily conserved cell cell communication mechanism. Is a regulator of cell specification, differentiation, and tissue patterning.
Journal Club 03/04/2012 Lama Nazzal NOTCH and the kidneys Is an evolutionarily conserved cell cell communication mechanism. Is a regulator of cell specification, differentiation, and tissue patterning.
Early cell death (FGF) B No RunX transcription factor produced Yes No differentiation
 Solution Key - Practice Questions Question 1 a) A recent publication has shown that the fat stem cells (FSC) can act as bone stem cells to repair cavities in the skull, when transplanted into immuno-compromised
Solution Key - Practice Questions Question 1 a) A recent publication has shown that the fat stem cells (FSC) can act as bone stem cells to repair cavities in the skull, when transplanted into immuno-compromised
Tooth organogenesis and regeneration
 Tooth organogenesis and regeneration Irma Thesleff and Mark Tummers, Developmental Biology Program, Institute of Biotechnology, PO Box 56, University of Helsinki, FIN-00014, Helsinki, Finland Table of
Tooth organogenesis and regeneration Irma Thesleff and Mark Tummers, Developmental Biology Program, Institute of Biotechnology, PO Box 56, University of Helsinki, FIN-00014, Helsinki, Finland Table of
The morphological studies of root r maxillary primary canines and their Title the position of successive permanen Micro-CT
 The morphological studies of root r maxillary primary canines and their Title the position of successive permanen Micro-CT Author(s) Saka, H; Koyama, T; Tamatsu, Y; Usa Alternative Journal Pediatric dental
The morphological studies of root r maxillary primary canines and their Title the position of successive permanen Micro-CT Author(s) Saka, H; Koyama, T; Tamatsu, Y; Usa Alternative Journal Pediatric dental
TITLE: A PSCA Promoter Based Avian Retroviral Transgene Model of Normal and Malignant Prostate
 AD Award Number: DAMD17-03-1-0163 TITLE: A PSCA Promoter Based Avian Retroviral Transgene Model of Normal and Malignant Prostate PRINCIPAL INVESTIGATOR: Robert Reiter, M.D. CONTRACTING ORGANIZATION: The
AD Award Number: DAMD17-03-1-0163 TITLE: A PSCA Promoter Based Avian Retroviral Transgene Model of Normal and Malignant Prostate PRINCIPAL INVESTIGATOR: Robert Reiter, M.D. CONTRACTING ORGANIZATION: The
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at
 Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
Nature Neuroscience: doi: /nn Supplementary Figure 1. MADM labeling of thalamic clones.
 Supplementary Figure 1 MADM labeling of thalamic clones. (a) Confocal images of an E12 Nestin-CreERT2;Ai9-tdTomato brain treated with TM at E10 and stained for BLBP (green), a radial glial progenitor-specific
Supplementary Figure 1 MADM labeling of thalamic clones. (a) Confocal images of an E12 Nestin-CreERT2;Ai9-tdTomato brain treated with TM at E10 and stained for BLBP (green), a radial glial progenitor-specific
Supplemental Information. Otic Mesenchyme Cells Regulate. Spiral Ganglion Axon Fasciculation. through a Pou3f4/EphA4 Signaling Pathway
 Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Signaling Vascular Morphogenesis and Maintenance
 Signaling Vascular Morphogenesis and Maintenance Douglas Hanahan Science 277: 48-50, in Perspectives (1997) Blood vessels are constructed by two processes: vasculogenesis, whereby a primitive vascular
Signaling Vascular Morphogenesis and Maintenance Douglas Hanahan Science 277: 48-50, in Perspectives (1997) Blood vessels are constructed by two processes: vasculogenesis, whereby a primitive vascular
INTRODUCTION. Developmental Biology 229, (2001) doi: /dbio , available online at
 Developmental Biology 229, 443 455 (2001) doi:10.1006/dbio.2000.9955, available online at http://www.idealibrary.com on TNF Signaling via the Ligand Receptor Pair Ectodysplasin and Edar Controls the Function
Developmental Biology 229, 443 455 (2001) doi:10.1006/dbio.2000.9955, available online at http://www.idealibrary.com on TNF Signaling via the Ligand Receptor Pair Ectodysplasin and Edar Controls the Function
Lecture 3: Skeletogenesis and diseases
 Jilin University School of Stomatology Skeletogenesis Lecture 3: Skeletogenesis and diseases Aug. 21, 2015 Yuji Mishina, Ph.D. mishina@umich.edu Bone Development Mouse embryo, E14.5 Mouse embryo, E18.0
Jilin University School of Stomatology Skeletogenesis Lecture 3: Skeletogenesis and diseases Aug. 21, 2015 Yuji Mishina, Ph.D. mishina@umich.edu Bone Development Mouse embryo, E14.5 Mouse embryo, E18.0
EMBO REPORT SUPPLEMENTARY SECTION. Quantitation of mitotic cells after perturbation of Notch signalling.
 EMBO REPORT SUPPLEMENTARY SECTION Quantitation of mitotic cells after perturbation of Notch signalling. Notch activation suppresses the cell cycle indistinguishably both within and outside the neural plate
EMBO REPORT SUPPLEMENTARY SECTION Quantitation of mitotic cells after perturbation of Notch signalling. Notch activation suppresses the cell cycle indistinguishably both within and outside the neural plate
Children's Hospital of Pittsburgh Annual Progress Report: 2008 Formula Grant
 Children's Hospital of Pittsburgh Annual Progress Report: 2008 Formula Grant Reporting Period July 1, 2011 June 30, 2012 Formula Grant Overview The Children's Hospital of Pittsburgh received $958,038 in
Children's Hospital of Pittsburgh Annual Progress Report: 2008 Formula Grant Reporting Period July 1, 2011 June 30, 2012 Formula Grant Overview The Children's Hospital of Pittsburgh received $958,038 in
Restriction of sonic hedgehog signalling during early tooth development
 Research article 2875 Restriction of sonic hedgehog signalling during early tooth development Martyn T. Cobourne 1, Isabelle Miletich 2 and Paul T. Sharpe 2, * 1 Department of Craniofacial Development
Research article 2875 Restriction of sonic hedgehog signalling during early tooth development Martyn T. Cobourne 1, Isabelle Miletich 2 and Paul T. Sharpe 2, * 1 Department of Craniofacial Development
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
Developing Facial Symmetry Using an Intraoral Device: A Case Report
 Developing Facial Symmetry Using an Intraoral Device: A Case Report by Theodore R. Belfor, D.D.S.; and G. Dave Singh, D.D.Sc., Ph.D., B.D.S. Dr. Theodore Belfor graduated from New York University College
Developing Facial Symmetry Using an Intraoral Device: A Case Report by Theodore R. Belfor, D.D.S.; and G. Dave Singh, D.D.Sc., Ph.D., B.D.S. Dr. Theodore Belfor graduated from New York University College
SUPPLEMENTARY INFORMATION
 b 350 300 250 200 150 100 50 0 E0 E10 E50 E0 E10 E50 E0 E10 E50 E0 E10 E50 Number of organoids per well 350 300 250 200 150 100 50 0 R0 R50 R100 R500 1st 2nd 3rd Noggin 100 ng/ml Noggin 10 ng/ml Noggin
b 350 300 250 200 150 100 50 0 E0 E10 E50 E0 E10 E50 E0 E10 E50 E0 E10 E50 Number of organoids per well 350 300 250 200 150 100 50 0 R0 R50 R100 R500 1st 2nd 3rd Noggin 100 ng/ml Noggin 10 ng/ml Noggin
Patterning the Embryo
 Patterning the Embryo Anteroposterior axis Regional Identity in the Vertebrate Neural Tube Fig. 2.2 1 Brain and Segmental Ganglia in Drosophila Fig. 2.1 Genes that create positional and segment identity
Patterning the Embryo Anteroposterior axis Regional Identity in the Vertebrate Neural Tube Fig. 2.2 1 Brain and Segmental Ganglia in Drosophila Fig. 2.1 Genes that create positional and segment identity
Contribution of the Tooth Bud Mesenchyme to Alveolar Bone
 JOURNAL OF EXPERIMENTAL ZOOLOGY (MOL DEV EVOL) 312B (2009) Contribution of the Tooth Bud Mesenchyme to Alveolar Bone LISA DIEP 1, EVA MATALOVA 2,3, THIMIOS A. MITSIADIS 4, AND ABIGAIL S. TUCKER 1 1 Department
JOURNAL OF EXPERIMENTAL ZOOLOGY (MOL DEV EVOL) 312B (2009) Contribution of the Tooth Bud Mesenchyme to Alveolar Bone LISA DIEP 1, EVA MATALOVA 2,3, THIMIOS A. MITSIADIS 4, AND ABIGAIL S. TUCKER 1 1 Department
Genotype analysis by Southern blots of nine independent recombinated ES cell clones by
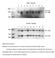 Supplemental Figure 1 Selected ES cell clones show a correctly recombined conditional Ngn3 allele Genotype analysis by Southern blots of nine independent recombinated ES cell clones by hybridization with
Supplemental Figure 1 Selected ES cell clones show a correctly recombined conditional Ngn3 allele Genotype analysis by Southern blots of nine independent recombinated ES cell clones by hybridization with
Src-INACTIVE / Src-INACTIVE
 Biology 169 -- Exam 1 February 2003 Answer each question, noting carefully the instructions for each. Repeat- Read the instructions for each question before answering!!! Be as specific as possible in each
Biology 169 -- Exam 1 February 2003 Answer each question, noting carefully the instructions for each. Repeat- Read the instructions for each question before answering!!! Be as specific as possible in each
Problem Set 8 Key 1 of 8
 7.06 2003 Problem Set 8 Key 1 of 8 7.06 2003 Problem Set 8 Key 1. As a bright MD/PhD, you are interested in questions about the control of cell number in the body. Recently, you've seen three patients
7.06 2003 Problem Set 8 Key 1 of 8 7.06 2003 Problem Set 8 Key 1. As a bright MD/PhD, you are interested in questions about the control of cell number in the body. Recently, you've seen three patients
Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate
 Research article Related Commentary, page 1676 Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate Ritva Rice, 1,2 Bradley Spencer-Dene, 3 Elaine C. Connor, 1
Research article Related Commentary, page 1676 Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate Ritva Rice, 1,2 Bradley Spencer-Dene, 3 Elaine C. Connor, 1
Supporting Information
 Supporting Information Franco et al. 10.1073/pnas.1015557108 SI Materials and Methods Drug Administration. PD352901 was dissolved in 0.5% (wt/vol) hydroxyl-propyl-methylcellulose, 0.2% (vol/vol) Tween
Supporting Information Franco et al. 10.1073/pnas.1015557108 SI Materials and Methods Drug Administration. PD352901 was dissolved in 0.5% (wt/vol) hydroxyl-propyl-methylcellulose, 0.2% (vol/vol) Tween
First posted online on 22 September 2016 as /dev
 First posted online on 22 September 2016 as 10.1242/dev.138883 Access the most recent version at http://dev.biologists.org/lookup/doi/10.1242/dev.138883 Sox2 and Lef-1 interact with Pitx2 to regulate incisor
First posted online on 22 September 2016 as 10.1242/dev.138883 Access the most recent version at http://dev.biologists.org/lookup/doi/10.1242/dev.138883 Sox2 and Lef-1 interact with Pitx2 to regulate incisor
a b c periosteum parietal bone bone marrow dura periosteum suture mesenchyme osteogenic front suture mesenchyme 1
 coronary suture sagittal suture DOI: 10.1038/ncb3139 a b c e parietal bone suture mesenchyme parietal bone bone marrow ura ura ura f parietal bone ura suture mesenchyme bone g ura osteogenic front suture
coronary suture sagittal suture DOI: 10.1038/ncb3139 a b c e parietal bone suture mesenchyme parietal bone bone marrow ura ura ura f parietal bone ura suture mesenchyme bone g ura osteogenic front suture
DENTIN It a hard vital tissue, surrounds the pulp & underlies the enamel on the crown & the cementum on the roots of the teeth.
 Lec. 7 Dr. Ali H.Murad DENTIN It a hard vital tissue, surrounds the pulp & underlies the enamel on the crown & the cementum on the roots of the teeth. Physical properties: 1-Dentin is pale yellow in color,
Lec. 7 Dr. Ali H.Murad DENTIN It a hard vital tissue, surrounds the pulp & underlies the enamel on the crown & the cementum on the roots of the teeth. Physical properties: 1-Dentin is pale yellow in color,
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Supporting Information
 Supporting Information Creuzet 10.1073/pnas.0906072106 Fig. S1. Migrating CNC cells express Bmp inhibitors. (A) HNK1 immunolabeling of migrating CNC cells at 7 ss. (B) At this stage, Fgf8 is strongly expressed
Supporting Information Creuzet 10.1073/pnas.0906072106 Fig. S1. Migrating CNC cells express Bmp inhibitors. (A) HNK1 immunolabeling of migrating CNC cells at 7 ss. (B) At this stage, Fgf8 is strongly expressed
AP VP DLP H&E. p-akt DLP
 A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
To the root of the stem cell problem. The evolutionary importance of the epithelial stem cell niche during tooth development
 To the root of the stem cell problem The evolutionary importance of the epithelial stem cell niche during tooth development Mark Tummers Developmental Biology Programme Institute of Biotechnology University
To the root of the stem cell problem The evolutionary importance of the epithelial stem cell niche during tooth development Mark Tummers Developmental Biology Programme Institute of Biotechnology University
Index. Note: Page numbers of article titles are in boldface type.
 Index Note: Page numbers of article titles are in boldface type. A Alginate, tooth-shaped, for constructs, encapsulated pulp cells in, 589 590 Antibiotic paste, triple, change in root length and width
Index Note: Page numbers of article titles are in boldface type. A Alginate, tooth-shaped, for constructs, encapsulated pulp cells in, 589 590 Antibiotic paste, triple, change in root length and width
Interactions of the endocrine system, bone and oral health
 Interactions of the endocrine system, bone and oral health All bones are not equal! Dense high proportion of cortical bone High proportion of trabecular bone Mandible Functions: mastication, respiration,
Interactions of the endocrine system, bone and oral health All bones are not equal! Dense high proportion of cortical bone High proportion of trabecular bone Mandible Functions: mastication, respiration,
Regionalization of the nervous system. Paul Garrity 7.68J/9.013J February 25, 2004
 Regionalization of the nervous system Paul Garrity 7.68J/9.013J February 25, 2004 Patterning along: Rostral/Caudal (AP) axis Dorsal/Ventral (DV) axis Start with DV axial patterning in Spinal Cord Dorsal/Ventral
Regionalization of the nervous system Paul Garrity 7.68J/9.013J February 25, 2004 Patterning along: Rostral/Caudal (AP) axis Dorsal/Ventral (DV) axis Start with DV axial patterning in Spinal Cord Dorsal/Ventral
Alveolar Growth in Japanese Infants: A Comparison between Now and 40 Years ago
 Bull Tokyo Dent Coll (2017) 58(1): 9 18 Original Article doi:10.2209/tdcpublication.2016-0500 Alveolar Growth in Japanese Infants: A Comparison between Now and 40 Years ago Hiroki Imai 1), Tetsuhide Makiguchi
Bull Tokyo Dent Coll (2017) 58(1): 9 18 Original Article doi:10.2209/tdcpublication.2016-0500 Alveolar Growth in Japanese Infants: A Comparison between Now and 40 Years ago Hiroki Imai 1), Tetsuhide Makiguchi
Corporate Medical Policy
 Corporate Medical Policy File Name: Origination: Last CAP Review: Next CAP Review: Last Review: orthodontics_for_pediatric_patients 2/2014 10/2017 10/2018 10/2017 Description of Procedure or Service Children
Corporate Medical Policy File Name: Origination: Last CAP Review: Next CAP Review: Last Review: orthodontics_for_pediatric_patients 2/2014 10/2017 10/2018 10/2017 Description of Procedure or Service Children
Developmental Biology
 Developmental Biology 362 (2012) 141 153 Contents lists available at SciVerse ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology Dual function of suppressor of
Developmental Biology 362 (2012) 141 153 Contents lists available at SciVerse ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology Dual function of suppressor of
Bone Marrow Pop. (% Total) Mature Pool (Absolute %) Immature Pool (Absolute %) A10 EC Control A10 EC Control A10 EC Control
 Bone Marrow Pop. (% Total) Mature Pool (Asolute %) Immature Pool (Asolute %) A10 EC A10 EC A10 EC Myeloid 50.7 57.5 37.5 46.2 13.2 11.3 Erythroid 38.3 23.2 33.3 16.8 9.3 6.3 Lymphocytes 13.8 19.0 - - -
Bone Marrow Pop. (% Total) Mature Pool (Asolute %) Immature Pool (Asolute %) A10 EC A10 EC A10 EC Myeloid 50.7 57.5 37.5 46.2 13.2 11.3 Erythroid 38.3 23.2 33.3 16.8 9.3 6.3 Lymphocytes 13.8 19.0 - - -
CAP STAGE. Ans 1 The following are the stages of tooth development :
 Ans 1 The following are the stages of tooth development : 1. Bud stage 2. Cap stage 3. Bell stage 4. Advanced bell stage 5. Formation of Hertwig s epithelial root sheath BUD STAGE 1. Around the eighth
Ans 1 The following are the stages of tooth development : 1. Bud stage 2. Cap stage 3. Bell stage 4. Advanced bell stage 5. Formation of Hertwig s epithelial root sheath BUD STAGE 1. Around the eighth
Supplemental Data Macrophage Migration Inhibitory Factor MIF Interferes with the Rb-E2F Pathway
 Supplemental Data Macrophage Migration Inhibitory Factor MIF Interferes with the Rb-E2F Pathway S1 Oleksi Petrenko and Ute M. Moll Figure S1. MIF-Deficient Cells Have Reduced Transforming Ability (A) Soft
Supplemental Data Macrophage Migration Inhibitory Factor MIF Interferes with the Rb-E2F Pathway S1 Oleksi Petrenko and Ute M. Moll Figure S1. MIF-Deficient Cells Have Reduced Transforming Ability (A) Soft
Primary cilia regulate Shh activity in the control of molar tooth number
 RESEARCH REPORT 897 Development 136, 897-903 (2009) doi:10.1242/dev.027979 Primary cilia regulate Shh activity in the control of molar tooth number Atsushi Ohazama 1, Courtney J. Haycraft 2, Maisa Seppala
RESEARCH REPORT 897 Development 136, 897-903 (2009) doi:10.1242/dev.027979 Primary cilia regulate Shh activity in the control of molar tooth number Atsushi Ohazama 1, Courtney J. Haycraft 2, Maisa Seppala
The Histology of Dentin
 The Histology of Dentin Pauline Hayes Garrett, D.D.S. Department of Endodontics, Prosthodontics, and Operative Dentistry University of Maryland, Baltimore This material was taken from: Essentials of Oral
The Histology of Dentin Pauline Hayes Garrett, D.D.S. Department of Endodontics, Prosthodontics, and Operative Dentistry University of Maryland, Baltimore This material was taken from: Essentials of Oral
Mucous and ciliated cell metaplasia in epithelial linings of odontogenic inflammatory and developmental cysts
 77 Journal of Oral Science, Vol. 47, No. 2, 77-81, 2005 Original Mucous and ciliated cell metaplasia in epithelial linings of odontogenic inflammatory and developmental cysts Yasunori Takeda, Yuko Oikawa,
77 Journal of Oral Science, Vol. 47, No. 2, 77-81, 2005 Original Mucous and ciliated cell metaplasia in epithelial linings of odontogenic inflammatory and developmental cysts Yasunori Takeda, Yuko Oikawa,
Only 40% of the Story
 X-RAY, X-RAY, READ ALL ABOUT IT! The Use and Utility of Dental Radiographs in Practice Lisa Fink, DVM, DAVDC Dentistry & Oral Surgery Service October 4, 2015 Only 40% of the Story Radiographs of teeth
X-RAY, X-RAY, READ ALL ABOUT IT! The Use and Utility of Dental Radiographs in Practice Lisa Fink, DVM, DAVDC Dentistry & Oral Surgery Service October 4, 2015 Only 40% of the Story Radiographs of teeth
Diseases of the skeleton
 Diseases of the skeleton Flexibility Protection of vital organs Strength Skeletal defects impact human health Developmental diseases Degenerative diseases Fins regenerate and grow rapidly following amputation
Diseases of the skeleton Flexibility Protection of vital organs Strength Skeletal defects impact human health Developmental diseases Degenerative diseases Fins regenerate and grow rapidly following amputation
Attachment G. Orthodontic Criteria Index Form Comprehensive D8080. ABBREVIATIONS CRITERIA for Permanent Dentition YES NO
 First Review IL HFS Dental Program Models Second Review Ortho cad Attachment G Orthodontic Criteria Index Form Comprehensive D8080 Ceph Film X-Rays Photos Narrative Patient Name: DOB: ABBREVIATIONS CRITERIA
First Review IL HFS Dental Program Models Second Review Ortho cad Attachment G Orthodontic Criteria Index Form Comprehensive D8080 Ceph Film X-Rays Photos Narrative Patient Name: DOB: ABBREVIATIONS CRITERIA
The Beauty of the Skin
 The Beauty of the Skin Rose-Anne Romano, Ph.D Assistant Professor Department of Oral Biology School of Dental Medicine State University of New York at Buffalo The Big Question How do approximately 50 trillion
The Beauty of the Skin Rose-Anne Romano, Ph.D Assistant Professor Department of Oral Biology School of Dental Medicine State University of New York at Buffalo The Big Question How do approximately 50 trillion
Formation of Urine: Formation of Urine
 The Urinary outflow tract: monitors and regulates extra-cellular fluids excretes harmful substances in urine, including nitrogenous wastes (urea) returns useful substances to bloodstream maintain balance
The Urinary outflow tract: monitors and regulates extra-cellular fluids excretes harmful substances in urine, including nitrogenous wastes (urea) returns useful substances to bloodstream maintain balance
Cancer. The fundamental defect is. unregulated cell division. Properties of Cancerous Cells. Causes of Cancer. Altered growth and proliferation
 Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
GENETIC ANALYSIS OF RAS SIGNALING PATHWAYS IN CELL PROLIFERATION, MIGRATION AND SURVIVAL
 Manuscript EMBO-2009-72496 GENETIC ANALYSIS OF RAS SIGNALING PATHWAYS IN CELL PROLIFERATION, MIGRATION AND SURVIVAL Matthias Drosten, Alma Dhawahir, Eleanor Sum, Jelena Urosevic, Carmen Lechuga, Luis Esteban,
Manuscript EMBO-2009-72496 GENETIC ANALYSIS OF RAS SIGNALING PATHWAYS IN CELL PROLIFERATION, MIGRATION AND SURVIVAL Matthias Drosten, Alma Dhawahir, Eleanor Sum, Jelena Urosevic, Carmen Lechuga, Luis Esteban,
Root Proximity Characteristics and Type of Alveolar Bone Loss: A Case-control Study
 Journal of the International Academy of Periodontology 011 13/3: 73-79 Root Proximity Characteristics and Type of Alveolar Bone Loss: A Case-control Study 1 1 1 Maria-Anna Loukideli, Alexandra Tsami, Eudoxie
Journal of the International Academy of Periodontology 011 13/3: 73-79 Root Proximity Characteristics and Type of Alveolar Bone Loss: A Case-control Study 1 1 1 Maria-Anna Loukideli, Alexandra Tsami, Eudoxie
Subject Index. AXIN2, cleft defects 24, 26
 Subject Index ADAMTS, mouse mutants and palate development 37, 38 Africa, cleft lip and palate prevalence 6, 7 Alcohol dependence, pregnancy risks for cleft 25, 61 Altitude, pregnancy risks for cleft 25,
Subject Index ADAMTS, mouse mutants and palate development 37, 38 Africa, cleft lip and palate prevalence 6, 7 Alcohol dependence, pregnancy risks for cleft 25, 61 Altitude, pregnancy risks for cleft 25,
Tooth eruption and movement
 Tooth eruption and movement Dr. Krisztián Nagy Diphydont dentition Deciduous dentition primary dentition Diphydont dentition Permanent dentition secondary dentition Mixed Dentition: Presence of both dentitions
Tooth eruption and movement Dr. Krisztián Nagy Diphydont dentition Deciduous dentition primary dentition Diphydont dentition Permanent dentition secondary dentition Mixed Dentition: Presence of both dentitions
Treatment planning of nonskeletal problems. in preadolescent children
 In the name of GOD Treatment planning of nonskeletal problems in preadolescent children Presented by: Dr Somayeh Heidari Orthodontist Reference: Contemporary Orthodontics Chapter 7 William R. Proffit,
In the name of GOD Treatment planning of nonskeletal problems in preadolescent children Presented by: Dr Somayeh Heidari Orthodontist Reference: Contemporary Orthodontics Chapter 7 William R. Proffit,
Hepatogenesis I Liver development
 Hepatogenesis I Liver development HB 308 George Yeoh Room 2.59 MCS Building yeoh@cyllene.uwa.edu.au Topics Early liver development Tissue interaction - role of morphogens and cytokines Liver enriched transcription
Hepatogenesis I Liver development HB 308 George Yeoh Room 2.59 MCS Building yeoh@cyllene.uwa.edu.au Topics Early liver development Tissue interaction - role of morphogens and cytokines Liver enriched transcription
Interactions between Shh, Sostdc1 and Wnt signaling and a new feedback loop for spatial patterning of the teeth
 RESEARCH ARTICLE 1807 Development 138, 1807-1816 (2011) doi:10.1242/dev.056051 2011. Published by The Company of Biologists Ltd Interactions between Shh, Sostdc1 and Wnt signaling and a new feedback loop
RESEARCH ARTICLE 1807 Development 138, 1807-1816 (2011) doi:10.1242/dev.056051 2011. Published by The Company of Biologists Ltd Interactions between Shh, Sostdc1 and Wnt signaling and a new feedback loop
A Cxcl12-Cxcr4 Chemokine Signaling Pathway Defines
 Supplemental Data A Cxcl12-Cxcr4 Chemokine Signaling Pathway Defines the Initial Trajectory of Mammalian Motor Axons Ivo Lieberam, Dritan Agalliu, Takashi Nagasawa, Johan Ericson, and Thomas M. Jessell
Supplemental Data A Cxcl12-Cxcr4 Chemokine Signaling Pathway Defines the Initial Trajectory of Mammalian Motor Axons Ivo Lieberam, Dritan Agalliu, Takashi Nagasawa, Johan Ericson, and Thomas M. Jessell
Cell Birth and Death. Chapter Three
 Cell Birth and Death Chapter Three Neurogenesis All neurons and glial cells begin in the neural tube Differentiated into neurons rather than ectoderm based on factors we have already discussed If these
Cell Birth and Death Chapter Three Neurogenesis All neurons and glial cells begin in the neural tube Differentiated into neurons rather than ectoderm based on factors we have already discussed If these
Dental Morphology and Vocabulary
 Dental Morphology and Vocabulary Palate Palate Palate 1 2 Hard Palate Rugae Hard Palate Palate Palate Soft Palate Palate Palate Soft Palate 4 Palate Hard Palate Soft Palate Maxillary Arch (Maxilla) (Uppers)
Dental Morphology and Vocabulary Palate Palate Palate 1 2 Hard Palate Rugae Hard Palate Palate Palate Soft Palate Palate Palate Soft Palate 4 Palate Hard Palate Soft Palate Maxillary Arch (Maxilla) (Uppers)
POSTGRADUATE INSTITUTE OF MEDICINE UNIVERSITY OF COLOMBO SELECTION EXAMINATION IN MD (ORAL SURGERY) OCTOBER 2009 PAPER 1.1. Part A (General Anatomy)
 POSTGRADUATE INSTITUTE OF MEDICINE UNIVERSITY OF COLOMBO SELECTION EXAMINATION IN MD (ORAL SURGERY) OCTOBER 2009 Date : 5 th October 2009 Time : 2.00 p.m. 5.00p.m. PAPER 1.1 Answer three (03) questions
POSTGRADUATE INSTITUTE OF MEDICINE UNIVERSITY OF COLOMBO SELECTION EXAMINATION IN MD (ORAL SURGERY) OCTOBER 2009 Date : 5 th October 2009 Time : 2.00 p.m. 5.00p.m. PAPER 1.1 Answer three (03) questions
Lecture 2 Maxillary central incisor
 Lecture 2 Maxillary central incisor Generally The deciduous tooth appears in the mouth at 3 18 months of age, with 6 months being the average and is replaced by the permanent tooth around 7 8 years of
Lecture 2 Maxillary central incisor Generally The deciduous tooth appears in the mouth at 3 18 months of age, with 6 months being the average and is replaced by the permanent tooth around 7 8 years of
Supplemental Data. Wnt/β-Catenin Signaling in Mesenchymal Progenitors. Controls Osteoblast and Chondrocyte
 Supplemental Data Wnt/β-Catenin Signaling in Mesenchymal Progenitors Controls Osteoblast and Chondrocyte Differentiation during Vertebrate Skeletogenesis Timothy F. Day, Xizhi Guo, Lisa Garrett-Beal, and
Supplemental Data Wnt/β-Catenin Signaling in Mesenchymal Progenitors Controls Osteoblast and Chondrocyte Differentiation during Vertebrate Skeletogenesis Timothy F. Day, Xizhi Guo, Lisa Garrett-Beal, and
Canonical Wnt Signaling in Osteoblasts Is Required for Osteoclast Differentiation
 Canonical Wnt Signaling in Osteoblasts Is Required for Osteoclast Differentiation DONALD A. GLASS, II AND GERARD KARSENTY Department of Molecular and Human Genetics, Bone Disease Program of Texas, Baylor
Canonical Wnt Signaling in Osteoblasts Is Required for Osteoclast Differentiation DONALD A. GLASS, II AND GERARD KARSENTY Department of Molecular and Human Genetics, Bone Disease Program of Texas, Baylor
Supplemental Figure 1: Lrig1-Apple expression in small intestine. Lrig1-Apple is observed at the crypt base and in insterstial cells of Cajal, but is
 Supplemental Figure 1: Lrig1-Apple expression in small intestine. Lrig1-Apple is observed at the crypt base and in insterstial cells of Cajal, but is not co-expressed in DCLK1-positive tuft cells. Scale
Supplemental Figure 1: Lrig1-Apple expression in small intestine. Lrig1-Apple is observed at the crypt base and in insterstial cells of Cajal, but is not co-expressed in DCLK1-positive tuft cells. Scale
Biology Developmental Biology Spring Quarter Midterm 1 Version A
 Biology 411 - Developmental Biology Spring Quarter 2013 Midterm 1 Version A 75 Total Points Open Book Choose 15 out the 20 questions to answer (5 pts each). Only the first 15 questions that are answered
Biology 411 - Developmental Biology Spring Quarter 2013 Midterm 1 Version A 75 Total Points Open Book Choose 15 out the 20 questions to answer (5 pts each). Only the first 15 questions that are answered
Postnatal Growth. The study of growth in growing children is for two reasons : -For health and nutrition assessment
 Growth of The Soft Tissues Postnatal Growth Postnatal growth is defined as the first 20 years of growth after birth krogman 1972 The study of growth in growing children is for two reasons : -For health
Growth of The Soft Tissues Postnatal Growth Postnatal growth is defined as the first 20 years of growth after birth krogman 1972 The study of growth in growing children is for two reasons : -For health
Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease
 1 Supplemental Materials 2 3 Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease 4 5 6 Authors: Madhumita Basu, 1 Jun-Yi Zhu, 2 Stephanie LaHaye 1,3, Uddalak
1 Supplemental Materials 2 3 Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease 4 5 6 Authors: Madhumita Basu, 1 Jun-Yi Zhu, 2 Stephanie LaHaye 1,3, Uddalak
Dental Anatomy and Occlusion
 CHAPTER 53 Dental Anatomy and Occlusion Ma Lou C. Sabino DDS, and Emily G. Smythe, DDS What numerical system is used most commonly in the United States for designating the adult dentition? Pediatric dentition?
CHAPTER 53 Dental Anatomy and Occlusion Ma Lou C. Sabino DDS, and Emily G. Smythe, DDS What numerical system is used most commonly in the United States for designating the adult dentition? Pediatric dentition?
Molecular and genetic analysis of stromal fibroblasts in prostate cancer
 Final report ESMO Translational Research Fellowship 2010-2011 Molecular and genetic analysis of stromal fibroblasts in prostate cancer Michalis Karamouzis Host Institute Department of Biological Chemistry,
Final report ESMO Translational Research Fellowship 2010-2011 Molecular and genetic analysis of stromal fibroblasts in prostate cancer Michalis Karamouzis Host Institute Department of Biological Chemistry,
Urogenital Development
 2-5-03 Urogenital Development Greg Dressler Assoc. Professor Dept. of Pathology x46490 Dressler@umich.edu The Origin of the Kidney In the vertebrate embryo, the first stage of kidney development occurs
2-5-03 Urogenital Development Greg Dressler Assoc. Professor Dept. of Pathology x46490 Dressler@umich.edu The Origin of the Kidney In the vertebrate embryo, the first stage of kidney development occurs
Benefits of conducting research while completing the DDS program Critical thinking skills and opportunity to publish scientific papers NIH/NIDCR
 Benefits of conducting research while completing the DDS program Critical thinking skills and opportunity to publish scientific papers NIH/NIDCR training opportunities (basic and clinical research) Presentation
Benefits of conducting research while completing the DDS program Critical thinking skills and opportunity to publish scientific papers NIH/NIDCR training opportunities (basic and clinical research) Presentation
Dentin Formation(Dentinogenesis)
 Lecture four Dr. Wajnaa Oral Histology Dentin Formation(Dentinogenesis) Dentinogenesis begins at the cusp tips after the odontoblasts have differentiated and begin collagen production. Dentinogenesis growth
Lecture four Dr. Wajnaa Oral Histology Dentin Formation(Dentinogenesis) Dentinogenesis begins at the cusp tips after the odontoblasts have differentiated and begin collagen production. Dentinogenesis growth
Institution : College of Dentistry Academic Department : Maxillofacial surgery and Diagnostic sciences
 Institution : College of Dentistry Academic Department : Maxillofacial surgery and Diagnostic sciences [MDS] Program : Bachelor of Dentistry [ BDS ] Course : Oral Biology Course Coordinator : Saleem Shaikh
Institution : College of Dentistry Academic Department : Maxillofacial surgery and Diagnostic sciences [MDS] Program : Bachelor of Dentistry [ BDS ] Course : Oral Biology Course Coordinator : Saleem Shaikh
