SUPPLEMENTARY INFORMATION
|
|
|
- Winifred King
- 5 years ago
- Views:
Transcription
1 Supplementary igure 1. The expression level of PP4 (At4g16860) is diminished in the rpp4 mutant (SK017569). The mna was extracted to analyze the expression level of PP4 using quantitative PC (qpc). Gene expression was normalized against constitutively expressed UBQ5. Error bars represent SD (n = 3). WT, wild-type. WWW NATUE.COM/NATUE 1
2 Supplementary igure 2. Identification of PP4-regulated genes through expression profiling. a, In planta growth of Hpa Emwa1 and plant responses were examined under a microscope following lactophenol trypan blue (LTB) staining. ed arrows indicate H cell death and blue arrows point at hyphae. dpi, days post inoculation. b, Expression levels of the marker gene P1 were measured using qpc and normalized with UBQ5. Error bars represent SD (n = 3 biological replicates). c, Expression profiles of candidate genes that were significantly induced (q-values < 0.01) more than 2-fold in WT at 2 dpi. WWW NATUE.COM/NATUE 2
3 WWW NATUE.COM/NATUE 3
4 Supplementary igure 3-1. T-DNA insertion diminished the expression of candidate genes as measured by qpc. Gene expression levels were normalized against UBQ5. Error bars represent SD (n = 3 biological replicates). ull numbers for the T-DNA insertion mutant lines are shown in Supplementary Table 1. Primers used in this analysis are shown in Supplementary Table 2. WWW NATUE.COM/NATUE 4
5 WWW NATUE.COM/NATUE 5
6 Supplementary igure 3-2. T-DNA insertion diminished the expression of candidate genes as measured by qpc. Gene expression levels were normalized against UBQ5. Error bars represent SD (n = 3 biological replicates). ull numbers for the T-DNA insertion mutant lines are shown in Supplementary Table 1. Primers used in this analysis are shown in Supplementary Table 2. WWW NATUE.COM/NATUE 6
7 Supplementary igure 4. Characterization of mutants in PP4-mediated resistance against Hpa Emwa1. a, The seven phenotypes scored in WT, rpp4, and 22 mutants 7 dpi with Hpa Emwa1. SPP, Sporangiophore; TN, Trailing necrosis; OOS, Oospore; H, ree hypha; HI, ree hyphal intermediate; EXH, Expanding H; DIH, Discrete H. b, Bar graphs showing distributions of phenotype (PT) scores (% of occurrence in 40 leaves/genotype) of the mutants (black bars) compared with WT (blue bar) and rpp4 (red bar). Mutants are ranked from the highest to the lowest with no correspondence from panel to panel. The error bars, 95% confidence intervals based on binomial distribution. WWW NATUE.COM/NATUE 7
8 Supplementary igure 5. Phenotypic clustering analysis using data from 3 biological replicates. Hierarchical clustering performed using the mean phenotype scores of the three biological replicates with bootstrap (100,000 times) showed that mutants were divided into 2 groups similar to ig. 1b (Mutants were coloured according to ig. 1b). Distance was measured by the standard correlation coefficient (average linkage; scale 0-1). WWW NATUE.COM/NATUE 8
9 Supplementary igure 6. Defects in gene-mediated resistance in Group 2 mutants. a, Phenolic compound accumulation at 7 dpi with Hpa Emwa1. Arrows point at sites of spore penetration. b, Callose deposition 7 dpi with Hpa Emwa1. ed arrows indicate callose produced by plants (WT) and along the hyphae of the pathogen (mutants). WWW NATUE.COM/NATUE 9
10 Supplementary igure 7. Some of the mutants compromised in PP4-mediated resistance against Hpa Emwa1 are also defective in immune responses against Pseudomonas. a, Mutants 2, 6, 8 and 12 are compromised in PS2-mediated resistance against Psm ES4326/Avrpt2. Error bars represent 95% confidence interval (n = 8). cfu, colony-forming unit. This experiment was performed three times with similar results. Asterisks indicate p-values < b, Mutants 2, 8, and 12 showed enhanced susceptibility to Psm ES4326. WWW NATUE.COM/NATUE 10
11 WWW NATUE.COM/NATUE 11
12 Supplementary igure 8. Luciferase-based diurnal and free running tests. a, Luciferase measurement of CCA1:LUC under the conditions used for the ASL-seq experiment (ig. 3c, Col CK). Two biological replicates were included. The white bars, the day; black bar, the night. b, ree running test of CCA1:LUC and LHY:LUC expression with/without Hpa Emwa1 treatment. CK, control; Emwa1, Hpa Emwa1 treated; ZT, zeitgeber time. Error bars represent SEM (n=18). This experiment has been repeated three times with similar results. WWW NATUE.COM/NATUE 12
13 Supplementary igure 9. Determination of the cluster number used in nonnegative matrix factorization (NM). A range of cluster numbers from 2 to 22 were implemented in NM with 300 runs (10,000 iterations/run) and the corresponding cophenetic correlation coefficients were calculated. When cluster number equalled 2, the highest cophenetic correlation coefficient was achieved. WWW NATUE.COM/NATUE 13
14 Supplementary igure 10. Convergence of NM. 300 runs (10,000 iterations/run) were performed using 2 clusters and the corresponding costs were calculated for each run. The NM algorithm converged with fewer than 50 runs. WWW NATUE.COM/NATUE 14
15 WWW NATUE.COM/NATUE 15
16 Supplementary igure 11. Membership distance for Cluster 1 and Cluster 2 genes determined by their expression patterns. a, The two expression clusters overlap with the phenotypic grouping (as in ig. 1b; Group 1 genes in red and Group 2 in blue). or each gene, its membership distance in the expression cluster is represented by the radius of the circle. The calculation is detailed in Methods. b, Timecourse expression of the NM Cluster 2 genes with/without Hpa Emwa1 infection. The white bars, the day; black bar, the night. CK, control; EMWA1, Hpa Emwa1 inoculated. WWW NATUE.COM/NATUE 16
17 Supplementary igure 12. Enhanced disease resistance to Hpa Noco2 observed in the CCA1 OE transgenic line based on sporangiospore count 7 dpi. Error bars, SD (n = 3, One-way ANOVA and Bonferroni's Multiple Comparison Test); **, p- values < 0.01, *** p-values < This experiment has been repeated twice with similar results. WWW NATUE.COM/NATUE 17
18 WWW NATUE.COM/NATUE 18
19 WWW NATUE.COM/NATUE 19
20 Supplementary Table 1. A list of all mutant alleles and their phenotypes. SK, Salk line; CS, Sail line; S, susceptible;, resistant; +, WT level of callose or phenolic deposition observed; +/-, lower than WT level of callose observed. NT, not tested; EDS, enhanced disease symptoms; Ins, insensitive. Vir, virulent; MAMP, microbial-associated molecular pattern; W/T, fresh weight assay and root length assay. Note (*) that the infection phenotype against Hpa Emwa1 was determined based on microscopic observations of the symptoms; not all alleles were quantitatively measured as in ig. 1a. WWW NATUE.COM/NATUE 20
21 # Description Mutant Sequence 1 L-kinase SK SK GGCTCACGCTTTATCG TGTCCCCGTGGTTCTA GGCTCACGCTTTATCG TGTCCCCGTGGTTCTA 2 Calcineurin-like phosphoesterase SK SK CCCAACTCAGGATCGT ACGGTGAGCGAGGAATA CCCAACTCAGGATCGT ACGGTGAGCGAGGAATA 3 AD1-like 1 (AD1-L1) CS SK TCTCGTGGCACTACCT CAACGTCGGTATCGCA TCTCGTGGCACTACCT CAACGTCGGTATCGCA 4 Aspartic protease 38 (ATASP38) SK UDP-glucosyl transferase 72E1 (UGT72E1) SK Ankyrin (ANK) SK GTACTCAGTTTACTTTCCTGC GTTCTGCTGATGATGATGTC TGCAGTGCATATTTATCCG GCCCTCGTAATCACTCC AGGTGACGTAGACCTT AAAACCGTGTTGCCTC 7 Disease resistance protein-related SK SK ATTGTGTTTGATGATGTCC AGCCAACAGTCTAGCA ACGAATGGCTAGGGAC CACCGTATTCTCCGACA 8 Protein kinase family protein SK SK CAGAACATCGCCCTAC GTGAGGCATTAGTGGTTT CAGAACATCGCCCTAC GTGAGGCATTAGTGGTTT 9 ABC transporter family protein SK SK TGCCCCAAATTACATTACG TGGAGGCTAAGCACGA CCGATGCTACTGCTCT GGCGAACATACCGAAT 10 ATPase-like domain-containing protein CS SK TTCGTACCGCTATCGAC CTCCCATAACACCATTGC TTCGTACCGCTATCGAC CTCCCATAACACCATTGC 11 Ankyrin repeat family protein SK TTGGGAATTAACAGCC GCAACCGTATTACCTC WWW NATUE.COM/NATUE 21
22 SK GCACTGTTCACGTCCT AGTACGGTCAACGCTC 12 Lipase class 3 family protein SK CS ACTCAATAGGCTCCGC CTCTGCTCCCCTCGAA ACTCAATAGGCTCCGC CTCTGCTCCCCTCGAA 13 Constitutive shade-avoidance 1 (CSA1) SK AACTGTAAAAGTCATGCC CATTCCTTGGGGATGGAAC 14 Cyclic nucleotide gated channel 3 (CNGC3) SK CS AGGACATTAAACGCCATC ATAGGGAATTGGGAACTG AGGACATTAAACGCCATC ATAGGGAATTGGGAACTG 15 Lipid transfer family protein (LTP) SK CS ATGGTCTCTGCTCAGT CTGGTACGGTTTTCGAT ATGGTCTCTGCTCAGT CTGGTACGGTTTTCGAT 16 Tyrosine aminotransferase 3 (TAT3) SK SK AATACCGTCGCAATGG CTTGAACTACCCCTGTAGAT AATACCGTCGCAATGG CTTGAACTACCCCTGTAGAT 17 Glycoside hydrolase SK CS GATCGAGTAGTTGGGA GCCAAAGGTATTGTCA GGAGTGGAAGTCAGGT GTAGGTGCTCGCCAAA 18 Cold-responsive protein (CO15B) SK TCGTTGATATACGCCG CTCTTCTGCTTTACCCTC 19 Aspartyl protease family protein SK CS TAGCGCCATGTACTGC AGGGACTAGATTGGCTG TGCACTACGCCAATGT GCTCTAAACCCTCGTCC 20 BON association protein 1 (BAP1) SK SK CTGAGATGATGGCGGT GCAATTGAAGTAGCAGAAACA CTGAGATGATGGCGGT GCAATTGAAGTAGCAGAAACA 21 L-kinase SK SK TGCTGGTACTTACGGC AGAAAAGGCAGTGGAG TGAAAGGTGTGGCTCAT GCCGTAAGTACCAGCA WWW NATUE.COM/NATUE 22
23 22 Ser/Thr protein kinase family protein SK SK GTGGCTTCGGTACTGT CGGGTTGACCAGCAGA GTGGCTTCGGTACTGT CGGGTTGACCAGCAGA ecognition of Peronospora parasitica 4 (PP4) SK GCATTTGAAGTTGCCGAGC GGGCATCCAGTGAGATTGAGG Supplementary Table 2. Primers used in qpc characterization of gene expression in T- DNA insertion mutants in Supplementary ig. 3. SK, Salk line; CS, Sail line. WWW NATUE.COM/NATUE 23
24 Description Name Sequence 1 2 L-kinase Calcineurin-like phosphoesterase family protein 3 AD1-like 1 (AD1-L1) 4 Aspartic protease 38 (ATASP38) 5 UDP-glucosyl transferase 72E1 (UGT72E1) 6 Ankyrin (ANK) 7 Disease resistance protein-related 8 Protein kinase family protein 9 ABC transporter family protein 10 ATPase-like domain-containing protein 11 Ankyrin repeat family protein 12 Lipase class 3 family protein Constitutive shade-avoidance 1 (CSA1) Cyclic nucleotide gated channel 3 (CNGC3) 15 Lipid transfer family protein (LTP) 16 Tyrosine aminotransferase 3 (TAT3) 17 Glycoside hydrolase family 1G35710AD1 1G35710pA1 1G13750AD1 1G13750pA1 4G33300AD1 4G33300pA1 5G02190AD2 5G02190pA2 3G50740AD1 3G50740pA1 5G54610AD2 5G54610pA2 5G45440AD1 5G45440pA1 1G16260AD1 1G16260pA1 5G52860AD1 5G52860pA1 4G36290AD1 4G36290pA1 1G10340AD2 1G10340pA2 3G48080AD1 3G48080pA1 5G17880AD2 5G17880pA2 2G46430AD1 2G46430pA1 3G22600AD2 3G22600pA2 2G24850AD2 2G24850pA2 3G54440AD2 3G54440pA2 CTGGTGCAACGTAGCCGTAGCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATCTTCATCGTGTAAGCAAACT CTGGTAAACCGGGCATGTTCCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATTCATGACTCGTGCATACACT CAGTATGAATAGAGACAATCCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATTTGCATTTCATTCATTTCCC TGATGATGTCCAATCACGTACCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATCGATCCACATGTTCTGCTGA TGACACGCTCCAAAAGATGCCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATGGCACCACGTGCCATGCACC CTCCTCTACCACGAATACGACCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATAAGATGCAATGGTGTCATAC CTTCTGAATTGGGGTTGTCGCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATCAGAGGTTTAGTTTTACTCT TTAAAATGGCTGGCCAAGTTCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATTCCAGAGCTTAGAGTTCCAC CAACACACGATAAAGTACGACCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATCTAAGGAGAGCAAGAAAACA CGTCTGTTCTACTTCATTCTCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATTCTTTTTCTAAGCTCTTTAC TAGTGGATCCACTTGATATGCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATCAGTATAGTCCCTACGATAC CATTCCAAGGTCGTCGAGAGCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATCACTGTCTGGCTCTTGTGCT CTCATCGCATCGTTTAAGCGCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATAAAATCTCAGGAACACAACT CTGAACATGTGTCGGAGTTGCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATGCCATTGCACTGAATAAAAC CGGTATTACAGCGACTGACACCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATAGAACCACCACCACCACCAC CCATAAATAAGAACAAGATTCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATATTGATGTGTCAAGCTTCAA CTTCCAACCTTCAATAGTCCCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATATAAGGAGAACTCCTTGAAT WWW NATUE.COM/NATUE 24
25 18 Cold-responsive protein (CO15B) 19 Aspartyl protease family protein 20 BON association protein 1 (BAP1) 21 L-kinase 22 Ser/Thr protein kinase family protein Ubiquitin 5 (UBQ5) Late elongated hypocotyl (LHY) Circadian clock associated 1 (CCA1) 2G42530AD1 2G42530pA1 3G51330AD1 3G51330pA1 3G61190AD1 3G61190pA1 4G08850AD1 4G08850pA1 1G66880AD1 1G66880pA1 3G62250AD1 3G62250pA1 1G01060AD2 1G01060pA2 2G46830AD1 2G46830pA1 GTGGCTTCGTTGAGGTCATCCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATCGAAATCAGAAGCTTTCTTT AATCAGACCGTTTCCAACCCCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATCAAGCTTTCATCTTCAAAAC CCCAAACCGGAACTCCGGTACCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATAAATTGGCGTTGATACAGAC CTGGAGCAACATAGCCGTAACCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATTTTCATCGCATAAGCTAGTT CTTTTATGTGGAGGAATGATCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATGATGTCTCTGTGTATGATTC GAACCTTTCCAGATCCATCGCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATGCACTCCTTCCTCAAACGCT TTTCGTTGGTAAGGGATACTCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATTCAATGTCGCCACTTACCTG CTCGTCAGACACAGACTTCCCCTGTGGTCGTAGCATCAGC AATGATACGGCGACCACCGAGATGCTTGGAAGGCAATTCGACC Supplementary Table 3. Primers used in ASL-seq in ig. 3c. WWW NATUE.COM/NATUE 25
Lecture 3-4. Identification of Positive Regulators downstream of SA
 Lecture 3-4 Identification of Positive Regulators downstream of SA Positive regulators between SA and resistance/pr Systemic resistance PR gene expression? SA Local infection Necrosis SA What could they
Lecture 3-4 Identification of Positive Regulators downstream of SA Positive regulators between SA and resistance/pr Systemic resistance PR gene expression? SA Local infection Necrosis SA What could they
Supplementary Figures
 Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first
 Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Table S1. Relative abundance of AGO1/4 proteins in different organs. Table S2. Summary of smrna datasets from various samples.
 Supplementary files Table S1. Relative abundance of AGO1/4 proteins in different organs. Table S2. Summary of smrna datasets from various samples. Table S3. Specificity of AGO1- and AGO4-preferred 24-nt
Supplementary files Table S1. Relative abundance of AGO1/4 proteins in different organs. Table S2. Summary of smrna datasets from various samples. Table S3. Specificity of AGO1- and AGO4-preferred 24-nt
Supplementary Material
 Supplementary Material Supplementary Figure 1. NOS2 -/- mice develop an analogous Ghon complex after infection in the ear dermis and show dissemination of Mtb to the lung. (A) WT and NOS2 -/- mice were
Supplementary Material Supplementary Figure 1. NOS2 -/- mice develop an analogous Ghon complex after infection in the ear dermis and show dissemination of Mtb to the lung. (A) WT and NOS2 -/- mice were
Supplemental Data. Shin et al. Plant Cell. (2012) /tpc YFP N
 MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
Supplemental Data. Wang et al. (2013). Plant Cell /tpc
 Supplemental Data. Wang et al. (2013). Plant Cell 10.1105/tpc.112.108993 Supplemental Figure 1. 3-MA Treatment Reduces the Growth of Seedlings. Two-week-old Nicotiana benthamiana seedlings germinated on
Supplemental Data. Wang et al. (2013). Plant Cell 10.1105/tpc.112.108993 Supplemental Figure 1. 3-MA Treatment Reduces the Growth of Seedlings. Two-week-old Nicotiana benthamiana seedlings germinated on
Supplemental Data. Hiruma et al. Plant Cell. (2010) /tpc Col-0. pen2-1
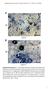 A Ch B Col-0 Cg pen2-1 Supplemental Figure 1. Trypan Blue Staining of Leaves Inoculated with Adapted and Nonadapted Colletotrichum Species.(A) Conidial suspensions of C. higginsianum MAFF305635 (Ch) were
A Ch B Col-0 Cg pen2-1 Supplemental Figure 1. Trypan Blue Staining of Leaves Inoculated with Adapted and Nonadapted Colletotrichum Species.(A) Conidial suspensions of C. higginsianum MAFF305635 (Ch) were
Supplemental Figure 1. Genes showing ectopic H3K9 dimethylation in this study are DNA hypermethylated in Lister et al. study.
 mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
doi: /nature10632
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10632 Supplementary Figure 1 Lyn mediates neutrophil wound responses as a redox sensor. a, A schematic model. Wounded epithelial cells release H 2 O 2 by an
SUPPLEMENTARY INFORMATION doi:10.1038/nature10632 Supplementary Figure 1 Lyn mediates neutrophil wound responses as a redox sensor. a, A schematic model. Wounded epithelial cells release H 2 O 2 by an
Nature Neuroscience: doi: /nn Supplementary Figure 1. Large-scale calcium imaging in vivo.
 Supplementary Figure 1 Large-scale calcium imaging in vivo. (a) Schematic illustration of the in vivo camera imaging set-up for large-scale calcium imaging. (b) High-magnification two-photon image from
Supplementary Figure 1 Large-scale calcium imaging in vivo. (a) Schematic illustration of the in vivo camera imaging set-up for large-scale calcium imaging. (b) High-magnification two-photon image from
Supplemental data. Uppalapati et al. (2012) Plant Cell /tpc
 PAL Control P. emaculata 0 8 24 48 8 24 48 OPR Control P. emaculata 0 8 24 48 8 24 48 CHS PR3 CHR PR5 CHI PR10 IFS IFR Supplemental Figure 1. Expression profiles for selected genes in phenylpropanoid pathway
PAL Control P. emaculata 0 8 24 48 8 24 48 OPR Control P. emaculata 0 8 24 48 8 24 48 CHS PR3 CHR PR5 CHI PR10 IFS IFR Supplemental Figure 1. Expression profiles for selected genes in phenylpropanoid pathway
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols.
 Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
SUPPLEMENTARY INFORMATION
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 2 ARTICLE NUMBER: 17084 Metabolic anticipation in Mycobacterium tuberculosis Hyungjin Eoh, Zhe Wang, Emilie Layre,
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 2 ARTICLE NUMBER: 17084 Metabolic anticipation in Mycobacterium tuberculosis Hyungjin Eoh, Zhe Wang, Emilie Layre,
Nature Immunology: doi: /ni eee Supplementary Figure 1
 eee Supplementary Figure 1 Hyphae induce NET release, but yeast do not. (a) NET release by human peripheral neutrophils stimulated with a hgc1 yeast-locked C. albicans mutant (yeast) or pre-formed WT C.
eee Supplementary Figure 1 Hyphae induce NET release, but yeast do not. (a) NET release by human peripheral neutrophils stimulated with a hgc1 yeast-locked C. albicans mutant (yeast) or pre-formed WT C.
Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 )
 770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis
 Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
Supplementary Information and Figure legends
 Supplementary Information and Figure legends Table S1. Primers for quantitative RT-PCR Target Sequence (5 -> 3 ) Target Sequence (5 -> 3 ) DAB2IP F:TGGACGATGTGCTCTATGCC R:GGATGGTGATGGTTTGGTAG Snail F:CCTCCCTGTCAGATGAGGAC
Supplementary Information and Figure legends Table S1. Primers for quantitative RT-PCR Target Sequence (5 -> 3 ) Target Sequence (5 -> 3 ) DAB2IP F:TGGACGATGTGCTCTATGCC R:GGATGGTGATGGTTTGGTAG Snail F:CCTCCCTGTCAGATGAGGAC
Supplemental Data. Hao et al. (2014). Plant Cell /tpc
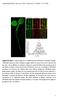 Supplemental Figure 1. Confocal Images and VA-TIRFM Analysis of GFP-RbohD in Arabidopsis Seedlings. (A) RbohD expression in whole Arabidopsis seedlings. RbohD was expressed in the leaves, hypocotyl, and
Supplemental Figure 1. Confocal Images and VA-TIRFM Analysis of GFP-RbohD in Arabidopsis Seedlings. (A) RbohD expression in whole Arabidopsis seedlings. RbohD was expressed in the leaves, hypocotyl, and
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
WT siz1-2 siz1-3. WT siz1-2 siz1-3. -Pi, 0.05 M IAA. -Pi, 2.5 M NPA
 A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
Supplementary Figure 1. Repression of hepcidin expression in the liver of mice treated with
 Supplementary Figure 1. Repression of hepcidin expression in the liver of mice treated with DMN Immunohistochemistry for hepcidin and H&E staining (left). qrt-pcr assays for hepcidin in the liver (right).
Supplementary Figure 1. Repression of hepcidin expression in the liver of mice treated with DMN Immunohistochemistry for hepcidin and H&E staining (left). qrt-pcr assays for hepcidin in the liver (right).
Supplementary Figure 1. Using DNA barcode-labeled MHC multimers to generate TCR fingerprints
 Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the
 Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the cardiac glycoside target, ATP1A1. (a) The percentage
Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the cardiac glycoside target, ATP1A1. (a) The percentage
Nature Genetics: doi: /ng.3731
 Supplementary Figure 1 Circadian profiles of Adarb1 transcript and ADARB1 protein in mouse tissues. (a) Overlap of rhythmic transcripts identified in the previous transcriptome analyses. The mouse liver
Supplementary Figure 1 Circadian profiles of Adarb1 transcript and ADARB1 protein in mouse tissues. (a) Overlap of rhythmic transcripts identified in the previous transcriptome analyses. The mouse liver
Supplementary Figures
 Supplementary Figures 9 10 11 Supplementary Figure 1. Old plants are more resistant to insect herbivores than young plants. (a) Image of young (1-day-old, 1D) and old (-day-old, D) plants of Arabidopsis
Supplementary Figures 9 10 11 Supplementary Figure 1. Old plants are more resistant to insect herbivores than young plants. (a) Image of young (1-day-old, 1D) and old (-day-old, D) plants of Arabidopsis
Nature Immunology: doi: /ni Supplementary Figure 1. DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells.
 Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
Nature Medicine doi: /nm.3957
 Supplementary Fig. 1. p38 alternative activation, IL-21 expression, and T helper cell transcription factors in PDAC tissue. (a) Tissue microarrays of pancreatic tissue from 192 patients with pancreatic
Supplementary Fig. 1. p38 alternative activation, IL-21 expression, and T helper cell transcription factors in PDAC tissue. (a) Tissue microarrays of pancreatic tissue from 192 patients with pancreatic
7.012 Quiz 3 Answers
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
Supplemental Data. Beck et al. (2010). Plant Cell /tpc
 Supplemental Figure 1. Phenotypic comparison of the rosette leaves of four-week-old mpk4 and Col-0 plants. A mpk4 vs Col-0 plants grown in soil. Note the extreme dwarfism of the mpk4 plants (white arrows)
Supplemental Figure 1. Phenotypic comparison of the rosette leaves of four-week-old mpk4 and Col-0 plants. A mpk4 vs Col-0 plants grown in soil. Note the extreme dwarfism of the mpk4 plants (white arrows)
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
Supplementary Fig. S1. Schematic diagram of minigenome segments.
 open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
Open Flower. Juvenile leaf Flowerbud. Carpel 35 NA NA NA NA 61 NA 95 NA NA 15 NA 41 3 NA
 PaxDB Root Juvenile leaf Flowerbud Open Flower Carpel Mature Pollen Silique Seed Sec3a Sec3b Sec5a Sec5b Sec6 Sec8 Sec10a/b Sec15a Sec15b Exo84a Exo84b Exo84c Exo70A1 Exo70A2 Exo70A3 49 47 8 75 104 79
PaxDB Root Juvenile leaf Flowerbud Open Flower Carpel Mature Pollen Silique Seed Sec3a Sec3b Sec5a Sec5b Sec6 Sec8 Sec10a/b Sec15a Sec15b Exo84a Exo84b Exo84c Exo70A1 Exo70A2 Exo70A3 49 47 8 75 104 79
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated
 Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation
 Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplementary Tables 1
 Supplementary Tables 1 Supplementary Table 1: The 11 genes in the geneset for Case Study 1 At1g62570 At1g09350 At1g60470 At2g47180 At4g17090 At5g20830 At2g16890 At3g51240 At3g55120 At4g27560 At5g08640
Supplementary Tables 1 Supplementary Table 1: The 11 genes in the geneset for Case Study 1 At1g62570 At1g09350 At1g60470 At2g47180 At4g17090 At5g20830 At2g16890 At3g51240 At3g55120 At4g27560 At5g08640
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
Testing the ABC floral-organ identity model: expression of A and C function genes
 Objectives: Testing the ABC floral-organ identity model: expression of A and C function genes To test the validity of the ABC model for floral organ identity we will: 1. Use the model to make predictions
Objectives: Testing the ABC floral-organ identity model: expression of A and C function genes To test the validity of the ABC model for floral organ identity we will: 1. Use the model to make predictions
Gene Ontology and Functional Enrichment. Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein
 Gene Ontology and Functional Enrichment Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein The parsimony principle: A quick review Find the tree that requires the fewest
Gene Ontology and Functional Enrichment Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein The parsimony principle: A quick review Find the tree that requires the fewest
A. Generation and characterization of Ras-expressing autophagycompetent
 Supplemental Material Supplemental Figure Legends Fig. S1 A. Generation and characterization of Ras-expressing autophagycompetent and -deficient cell lines. HA-tagged H-ras V12 was stably expressed in
Supplemental Material Supplemental Figure Legends Fig. S1 A. Generation and characterization of Ras-expressing autophagycompetent and -deficient cell lines. HA-tagged H-ras V12 was stably expressed in
The N-end rule pathway regulates pathogen responses. in plants
 SUPPLEMENTARY INFORMATION The N-end rule pathway regulates pathogen responses in plants Rémi de Marchi 1,2, Maud Sorel 1, Brian Mooney 1, Isabelle Fudal 3, Kevin Goslin 1, Kamila Kwaśniewska 4, Patrick
SUPPLEMENTARY INFORMATION The N-end rule pathway regulates pathogen responses in plants Rémi de Marchi 1,2, Maud Sorel 1, Brian Mooney 1, Isabelle Fudal 3, Kevin Goslin 1, Kamila Kwaśniewska 4, Patrick
effects on organ development. a-f, Eye and wing discs with clones of ε j2b10 show no
 Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Nature Genetics: doi: /ng Supplementary Figure 1. Clinical timeline for the discovery WES cases.
 Supplementary Figure 1 Clinical timeline for the discovery WES cases. This illustrates the timeline of the disease events during the clinical course of each patient s disease, further indicating the available
Supplementary Figure 1 Clinical timeline for the discovery WES cases. This illustrates the timeline of the disease events during the clinical course of each patient s disease, further indicating the available
A quick review. The clustering problem: Hierarchical clustering algorithm: Many possible distance metrics K-mean clustering algorithm:
 The clustering problem: partition genes into distinct sets with high homogeneity and high separation Hierarchical clustering algorithm: 1. Assign each object to a separate cluster. 2. Regroup the pair
The clustering problem: partition genes into distinct sets with high homogeneity and high separation Hierarchical clustering algorithm: 1. Assign each object to a separate cluster. 2. Regroup the pair
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Frequency of alternative-cassette-exon engagement with the ribosome is consistent across data from multiple human cell types and from mouse stem cells. Box plots showing AS frequency
Supplementary Figure 1 Frequency of alternative-cassette-exon engagement with the ribosome is consistent across data from multiple human cell types and from mouse stem cells. Box plots showing AS frequency
Supplementary Fig. 1 No relative growth advantage of Foxp3 negative cells.
 Supplementary Fig. 1 Supplementary Figure S1: No relative growth advantage of Foxp3 negative cells. itreg were induced from WT (A) or FIR (B) CD4 + T cells. FIR itregs were then removed from the TCR signal
Supplementary Fig. 1 Supplementary Figure S1: No relative growth advantage of Foxp3 negative cells. itreg were induced from WT (A) or FIR (B) CD4 + T cells. FIR itregs were then removed from the TCR signal
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
Loss of Calreticulin Uncovers a Critical Role for Calcium in Regulating Cellular Lipid Homeostasis
 SUPPLEMENTARY MATERIAL Loss of Calreticulin Uncovers a Critical Role for Calcium in Regulating Cellular Lipid Homeostasis Wen-An Wang 1, Wen-Xin Liu 1, Serpen Durnaoglu 2, Sun-Kyung Lee 2, Jihong Lian
SUPPLEMENTARY MATERIAL Loss of Calreticulin Uncovers a Critical Role for Calcium in Regulating Cellular Lipid Homeostasis Wen-An Wang 1, Wen-Xin Liu 1, Serpen Durnaoglu 2, Sun-Kyung Lee 2, Jihong Lian
Nature Genetics: doi: /ng Supplementary Figure 1. SEER data for male and female cancer incidence from
 Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
p.r623c p.p976l p.d2847fs p.t2671 p.d2847fs p.r2922w p.r2370h p.c1201y p.a868v p.s952* RING_C BP PHD Cbp HAT_KAT11
 ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
Nature Neuroscience: doi: /nn Supplementary Figure 1. Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites.
 Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Supplementary Material
 Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supporting Information
 Supporting Information Plikus et al. 10.1073/pnas.1215935110 SI Text Movies S1, S2, S3, and S4 are time-lapse recordings from individually cultured Period2 Luc vibrissa follicles show that circadian cycles
Supporting Information Plikus et al. 10.1073/pnas.1215935110 SI Text Movies S1, S2, S3, and S4 are time-lapse recordings from individually cultured Period2 Luc vibrissa follicles show that circadian cycles
Supplemental Data. Müller-Xing et al. (2014). Plant Cell /tpc
 Supplemental Figure 1. Phenotypes of iclf (clf-28 swn-7 CLF pro :CLF-GR) plants. A, Late rescue of iclf plants by renewed DEX treatment; senescent inflorescence with elongated siliques (arrow; 90 DAG,
Supplemental Figure 1. Phenotypes of iclf (clf-28 swn-7 CLF pro :CLF-GR) plants. A, Late rescue of iclf plants by renewed DEX treatment; senescent inflorescence with elongated siliques (arrow; 90 DAG,
Supporting Information
 Supporting Information Harries et al. 1.173/pnas.9923916 A Fig. S1. Disruption of microfilaments within epidermal cells after treatment with 5 M Lat. Images of N. benthamiana cells are from plants expressing
Supporting Information Harries et al. 1.173/pnas.9923916 A Fig. S1. Disruption of microfilaments within epidermal cells after treatment with 5 M Lat. Images of N. benthamiana cells are from plants expressing
Supplemental Information. Increased 4E-BP1 Expression Protects. against Diet-Induced Obesity and Insulin. Resistance in Male Mice
 Cell Reports, Volume 16 Supplemental Information Increased 4E-BP1 Expression Protects against Diet-Induced Obesity and Insulin Resistance in Male Mice Shih-Yin Tsai, Ariana A. Rodriguez, Somasish G. Dastidar,
Cell Reports, Volume 16 Supplemental Information Increased 4E-BP1 Expression Protects against Diet-Induced Obesity and Insulin Resistance in Male Mice Shih-Yin Tsai, Ariana A. Rodriguez, Somasish G. Dastidar,
Supplementary Information. Supplementary Figures
 Supplementary Information Supplementary Figures.8 57 essential gene density 2 1.5 LTR insert frequency diversity DEL.5 DUP.5 INV.5 TRA 1 2 3 4 5 1 2 3 4 1 2 Supplementary Figure 1. Locations and minor
Supplementary Information Supplementary Figures.8 57 essential gene density 2 1.5 LTR insert frequency diversity DEL.5 DUP.5 INV.5 TRA 1 2 3 4 5 1 2 3 4 1 2 Supplementary Figure 1. Locations and minor
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/393/rs9/dc1 Supplementary Materials for Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts
www.sciencesignaling.org/cgi/content/full/8/393/rs9/dc1 Supplementary Materials for Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts
Supplemental Data. Wu et al. (2010). Plant Cell /tpc
 Supplemental Figure 1. FIM5 is preferentially expressed in stamen and mature pollen. The expression data of FIM5 was extracted from Arabidopsis efp browser (http://www.bar.utoronto.ca/efp/development/),
Supplemental Figure 1. FIM5 is preferentially expressed in stamen and mature pollen. The expression data of FIM5 was extracted from Arabidopsis efp browser (http://www.bar.utoronto.ca/efp/development/),
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Jyoti Shah, Vamsi Nalam, Sujon Sarowar, Syeda Alam, Sumita Behera, Hyeonju Lee, Delia Burdan and Harold N. Trick
 Targeting Host Defense and Susceptibility Mechanisms for Engineering FHB Resistance Jyoti Shah, Vamsi Nalam, Sujon Sarowar, Syeda Alam, Sumita Behera, Hyeonju Lee, Delia Burdan and Harold N. Trick General
Targeting Host Defense and Susceptibility Mechanisms for Engineering FHB Resistance Jyoti Shah, Vamsi Nalam, Sujon Sarowar, Syeda Alam, Sumita Behera, Hyeonju Lee, Delia Burdan and Harold N. Trick General
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A.
 Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary Figure 1) GABAergic enhancement by leptin hyperpolarizes POMC neurons A) Representative recording samples showing the membrane
 Supplementary Figure 1) GABAergic enhancement by leptin hyperpolarizes POMC neurons A) Representative recording samples showing the membrane potential recorded from POMC neurons following treatment with
Supplementary Figure 1) GABAergic enhancement by leptin hyperpolarizes POMC neurons A) Representative recording samples showing the membrane potential recorded from POMC neurons following treatment with
Supporting Information
 Supporting Information Lee et al. 10.1073/pnas.0910950106 Fig. S1. Fe (A), Zn (B), Cu (C), and Mn (D) concentrations in flag leaves from WT, osnas3-1, and OsNAS3-antisense (AN-2) plants. Each measurement
Supporting Information Lee et al. 10.1073/pnas.0910950106 Fig. S1. Fe (A), Zn (B), Cu (C), and Mn (D) concentrations in flag leaves from WT, osnas3-1, and OsNAS3-antisense (AN-2) plants. Each measurement
NC bp. b 1481 bp
 Kcna3 NC 11 178 p 1481 p 346 p c *** *** d relative expression..4.3.2.1 CD4 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna Kcna6 Kcna7 Kcnn4 relative expression..4.3.2.1 CD8 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna
Kcna3 NC 11 178 p 1481 p 346 p c *** *** d relative expression..4.3.2.1 CD4 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna Kcna6 Kcna7 Kcnn4 relative expression..4.3.2.1 CD8 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Nature Genetics: doi: /ng Supplementary Figure 1. HOX fusions enhance self-renewal capacity.
 Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
TIME FOR COFFEE Encodes a Nuclear Regulator in the Arabidopsis thaliana Circadian Clock W
 The Plant Cell, Vol. 19: 1522 1536, May 2007, www.plantcell.org ª 2007 American Society of Plant Biologists TIME FOR COFFEE Encodes a Nuclear Regulator in the Arabidopsis thaliana Circadian Clock W Zhaojun
The Plant Cell, Vol. 19: 1522 1536, May 2007, www.plantcell.org ª 2007 American Society of Plant Biologists TIME FOR COFFEE Encodes a Nuclear Regulator in the Arabidopsis thaliana Circadian Clock W Zhaojun
Supplemental Figure 1. (A) The localization of Cre DNA recombinase in the testis of Cyp19a1-Cre mice was detected by immunohistchemical analyses
 Supplemental Figure 1. (A) The localization of Cre DNA recombinase in the testis of Cyp19a1-Cre mice was detected by immunohistchemical analyses using an anti-cre antibody; testes at 1 week (left panel),
Supplemental Figure 1. (A) The localization of Cre DNA recombinase in the testis of Cyp19a1-Cre mice was detected by immunohistchemical analyses using an anti-cre antibody; testes at 1 week (left panel),
Balancing intestinal and systemic inflammation through cell type-specific expression of
 Supplementary Information Balancing intestinal and systemic inflammation through cell type-specific expression of the aryl hydrocarbon receptor repressor Olga Brandstätter 1,2,6, Oliver Schanz 1,6, Julia
Supplementary Information Balancing intestinal and systemic inflammation through cell type-specific expression of the aryl hydrocarbon receptor repressor Olga Brandstätter 1,2,6, Oliver Schanz 1,6, Julia
s u p p l e m e n ta ry i n f o r m at i o n
 Figure S1 Characterization of tet-off inducible cell lines expressing GFPprogerin and GFP-wt lamin A. a, Western blot analysis of GFP-progerin- or GFP-wt lamin A- expressing cells before induction (0d)
Figure S1 Characterization of tet-off inducible cell lines expressing GFPprogerin and GFP-wt lamin A. a, Western blot analysis of GFP-progerin- or GFP-wt lamin A- expressing cells before induction (0d)
Supplemental Figure S1. Sequence feature and phylogenetic analysis of GmZF351. (A) Amino acid sequence alignment of GmZF351, AtTZF1, SOMNUS, AtTZF5,
 Supplemental Figure S. Sequence feature and phylogenetic analysis of GmZF35. (A) Amino acid sequence alignment of GmZF35, AtTZF, SOMNUS, AtTZF5, and OsTZF. Characters with black background indicate conserved
Supplemental Figure S. Sequence feature and phylogenetic analysis of GmZF35. (A) Amino acid sequence alignment of GmZF35, AtTZF, SOMNUS, AtTZF5, and OsTZF. Characters with black background indicate conserved
Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma.
 Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Nature Biotechnology: doi: /nbt Supplementary Figure 1. PL gene expression in tomato fruit.
 Supplementary Figure 1 PL gene expression in tomato fruit. Relative expression of five PL-coding genes measured in at least three fruit of each genotype (cv. Alisa Craig) at four stages of development,
Supplementary Figure 1 PL gene expression in tomato fruit. Relative expression of five PL-coding genes measured in at least three fruit of each genotype (cv. Alisa Craig) at four stages of development,
Supplementary Figure 1. Double-staining immunofluorescence analysis of invasive colon and breast cancers. Specimens from invasive ductal breast
 Supplementary Figure 1. Double-staining immunofluorescence analysis of invasive colon and breast cancers. Specimens from invasive ductal breast carcinoma (a) and colon adenocarcinoma (b) were staining
Supplementary Figure 1. Double-staining immunofluorescence analysis of invasive colon and breast cancers. Specimens from invasive ductal breast carcinoma (a) and colon adenocarcinoma (b) were staining
Figure S1. Standard curves for amino acid bioassays. (A) The standard curve for leucine concentration versus OD600 values for L. casei.
 Figure S1. Standard curves for amino acid bioassays. (A) The standard curve for leucine concentration versus OD600 values for L. casei. (B) The standard curve for lysine concentrations versus OD600 values
Figure S1. Standard curves for amino acid bioassays. (A) The standard curve for leucine concentration versus OD600 values for L. casei. (B) The standard curve for lysine concentrations versus OD600 values
Student name ID # 2. (4 pts) What is the terminal electron acceptor in respiration? In photosynthesis?
 1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? 2. (4 pts) What is the terminal electron acceptor
1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? 2. (4 pts) What is the terminal electron acceptor
Supplementary Figure 1. Ent inhibits LPO activity in a dose- and time-dependent fashion.
 Supplementary Information Supplementary Figure 1. Ent inhibits LPO activity in a dose- and time-dependent fashion. Various concentrations of Ent, DHBA or ABAH were pre-incubated for 10 min with LPO (50
Supplementary Information Supplementary Figure 1. Ent inhibits LPO activity in a dose- and time-dependent fashion. Various concentrations of Ent, DHBA or ABAH were pre-incubated for 10 min with LPO (50
Supplemental Figure S1. The number of hydathodes is reduced in the as2-1 rev-1
 Supplemental Data Supplemental Figure S1. The number of hydathodes is reduced in the as2-1 rev-1 and kan1-11 kan2-5 double mutants. A, The numbers of hydathodes in different leaves of Col-0, as2-1 rev-1,
Supplemental Data Supplemental Figure S1. The number of hydathodes is reduced in the as2-1 rev-1 and kan1-11 kan2-5 double mutants. A, The numbers of hydathodes in different leaves of Col-0, as2-1 rev-1,
Supplementary Information. Conformational states of Lck regulate clustering in early T cell signaling
 Supplementary Information Conformational states of Lck regulate clustering in early T cell signaling Jérémie Rossy, Dylan M. Owen, David J. Williamson, Zhengmin Yang and Katharina Gaus Centre for Vascular
Supplementary Information Conformational states of Lck regulate clustering in early T cell signaling Jérémie Rossy, Dylan M. Owen, David J. Williamson, Zhengmin Yang and Katharina Gaus Centre for Vascular
Supplementary Figure 1
 VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
Supplementary Figure 1. Biological characteris=cs of Smarcb1 flox/flox ; Rosa26- Cre ERT2 ; lymphomas
 Supplementary Figure 1. Biological characteris=cs of Smarcb1 flox/flox ; Rosa26- Cre ERT2 ; lymphomas a Tec Kinase Signaling CD28 Signaling in T Helper Cells Role of Macrophages in Rheumatoid Arthri>s
Supplementary Figure 1. Biological characteris=cs of Smarcb1 flox/flox ; Rosa26- Cre ERT2 ; lymphomas a Tec Kinase Signaling CD28 Signaling in T Helper Cells Role of Macrophages in Rheumatoid Arthri>s
Supplementary information. Supplementary figure 1. Flow chart of study design
 Supplementary information Supplementary figure 1. Flow chart of study design Supplementary Figure 2. Quantile-quantile plot of stage 1 results QQ plot of the observed -log10 P-values (y axis) versus the
Supplementary information Supplementary figure 1. Flow chart of study design Supplementary Figure 2. Quantile-quantile plot of stage 1 results QQ plot of the observed -log10 P-values (y axis) versus the
Nature Immunology: doi: /ni Supplementary Figure 1. Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice.
 Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Nature Immunology: doi: /ni Supplementary Figure 1. Transcriptional program of the TE and MP CD8 + T cell subsets.
 Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Figure 1
 Supplementary Figure 1 Miniature microdrive, spike sorting and sleep stage detection. a, A movable recording probe with 8-tetrodes (32-channels). It weighs ~1g. b, A mouse implanted with 8 tetrodes in
Supplementary Figure 1 Miniature microdrive, spike sorting and sleep stage detection. a, A movable recording probe with 8-tetrodes (32-channels). It weighs ~1g. b, A mouse implanted with 8 tetrodes in
Supplementary Information. Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase
 Journal: Nature Medicine Supplementary Information Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase 1,2 Peng Wang PhD, 1,2 Juan-Carlos Alvarez-Perez
Journal: Nature Medicine Supplementary Information Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase 1,2 Peng Wang PhD, 1,2 Juan-Carlos Alvarez-Perez
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Cheng MH, Fan U, Grewal N, et al. Acquired autoimmune polyglandular
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Cheng MH, Fan U, Grewal N, et al. Acquired autoimmune polyglandular
Supplementary Figure 1. Successful excision of genes from WBM lysates and
 Supplementary Information: Supplementary Figure 1. Successful excision of genes from WBM lysates and survival of mice with different genotypes. (a) The proper excision of Pten, p110α, p110α and p110δ was
Supplementary Information: Supplementary Figure 1. Successful excision of genes from WBM lysates and survival of mice with different genotypes. (a) The proper excision of Pten, p110α, p110α and p110δ was
Host-Induced Gene Silencing in the Fusarium-Wheat interaction. Wanxin Chen and Patrick Schweizer
 Host-Induced Gene Silencing in the Fusarium-Wheat interaction Wanxin Chen and Patrick Schweizer What is Host-Induced Gene Silencing (HIGS)? Fungus Bgh Trujillo et al. (2004) MPMI HIGS: HI reduced GAME
Host-Induced Gene Silencing in the Fusarium-Wheat interaction Wanxin Chen and Patrick Schweizer What is Host-Induced Gene Silencing (HIGS)? Fungus Bgh Trujillo et al. (2004) MPMI HIGS: HI reduced GAME
El Azzouzi et al., http ://www.jcb.org /cgi /content /full /jcb /DC1
 Supplemental material JCB El Azzouzi et al., http ://www.jcb.org /cgi /content /full /jcb.201510043 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Acquisition of -phluorin correlates negatively with podosome
Supplemental material JCB El Azzouzi et al., http ://www.jcb.org /cgi /content /full /jcb.201510043 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Acquisition of -phluorin correlates negatively with podosome
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
A putative MYB35 ortholog is a candidate for the sex-determining genes in Asparagus
 Supplementary figures for: A putative MYB35 ortholog is a candidate for the sex-determining genes in Asparagus officinalis Daisuke Tsugama, Kohei Matsuyama, Mayui Ide, Masato Hayashi, Kaien Fujino, and
Supplementary figures for: A putative MYB35 ortholog is a candidate for the sex-determining genes in Asparagus officinalis Daisuke Tsugama, Kohei Matsuyama, Mayui Ide, Masato Hayashi, Kaien Fujino, and
Color Key PCA. mir- 15a let-7c 106b let-7b let-7a 16 10b 99a 26a 20b 374b 19b 135b 125b a-5p 199b-5p 93 92b MES PN.
 1123 83 528 816 84 2 17 718 Comp3 3 2 1 1 2 3 4 Comp2 a b Color Key MES PN PCA 3 2 1 1 2 3 4 Comp1 1 2 3 1 2 3 4-2 -1 1 2 mi- 15a let-7c 16b let-7b let-7a 16 1b 99a 26a 2b 374b 19b 135b 125b 9 34 125a-5p
1123 83 528 816 84 2 17 718 Comp3 3 2 1 1 2 3 4 Comp2 a b Color Key MES PN PCA 3 2 1 1 2 3 4 Comp1 1 2 3 1 2 3 4-2 -1 1 2 mi- 15a let-7c 16b let-7b let-7a 16 1b 99a 26a 2b 374b 19b 135b 125b 9 34 125a-5p
