Exomes and Beyond Addressing the Genome with OneSeq. Madhuri Hegde, PhD, FACMG Emory University Emory Genetics Laboratory Atlanta, GA
|
|
|
- Annice Hutchinson
- 5 years ago
- Views:
Transcription
1 Exomes and Beyond Addressing the Genome with OneSeq Madhuri Hegde, PhD, FACMG Emory University Emory Genetics Laboratory Atlanta, GA
2 Technologies to Detect Various Types of Mutations RESOLUTION Targeted PCR assay Sequencing/ Next Gen Seq targeted array CGH 1 bp 1bp X 1000s CNVs 100s-1000s bp whole genome array CGH FISH G-banded karyotype CNVs 10,000s- 1,000,000s bp CNVs 100,000s- 1,000,000s bp CNVs 5,000,000+ bp Scherer et al. (2007) Nat Genet 39:s7-s15
3 Next Gen Sequencing Hybrid Capture and NGS are not 100% Efficient Need to fill-in low coverage gaps (regions with <15X coverage) ~1-5% of exons have low coverage GC rich regions typically have low coverage due to poor capture Repetitive sequences often have low coverage due to alignment issues Genes with pseudogenes have to be sequenced by specially designed Sanger assays CAN NOT DETECT GENOMIC COPY NUMBER AND INTRAGENIC DELETION AND DUPLICATIONS WHOLE EXOME HAS HOLES
4 Targeted panels/ exome / genome Targeted panels Exome Genome Smaller target region Allows interrogation of entire coding region of the genome(~92% coverage) Targets the Entire genome Highest depth of coverage (sanger fill in to complete is possible) Lower compared to targeted Lowest compared to targeted and exome Difficult for detection of CNVs, Trinucleotide repeats, pseudogenes No incidental findings (Allelic changes) Difficult for detection of CNVs, indels Trinucleotide repeats, Pseudogenes Need to address incidental findings Allows detection of CNVs, indels Difficult for detection Trinucleotide repeats, Pseudogenes Need to address incidental findings
5 EXOME AND GENOME SEQUENCING IN GENOMICS Basic Research Goals (Discovery): Clinical research (medical exome): Sequence a cohort to identify a new causative gene - Need to ensure all known disease causing genes have been covered and ruled out; follow up with functional studies Sequence large subset of genes to identify disease modifiers -Need to ensure adequate coverage of genes Identify novel genomic variants Sequence ambiguous cases to identify the cause of disease - Need to ensure all known disease causing genes have been covered Be able to detect genomic and intragenic CNVs Recommend cases for discovery projects based on phenotypes -Need to ensure adequate coverage of genes and known genes have been ruled out
6 PANELS OR EXOME/GENOME
7 Finishing the exome Gene: Technical aspects Gene: evidence Technically complete assay Medical Exome
8 THE MEDICAL EXOME PROJECT A highly curated gene resource and a technically optimized assay to provide a stepping stone for standardizing interpretation of genetic variation to fulfill the promise of genomic medicine
9 just "because three is better than one CHOP Emory Harvard LMM
10 The mendeliome : 4,600 genes Genome Exome Medical Exome v1 Exome ~1% of genome 22,000 genes ~4,600 genes implicated in disease 1998 genes w/pseudogenes ~1% genome 22,000 genes Enhanced coverage of 3,654 genes curated to be medically relevant Intronic mutations Medical Exome V1 Mitochondrial genome Genome ~complete genome with gaps
11 THE MEDICAL EXOME PROJECT STANDARDIZE MEDICAL EXOME SEQUENCING Develop a medically enhanced whole exome assay Develop complementary ancillary assays (pseudogenes) Determine performance metrics of the assay through intra- and inter-laboratory validation tests Define a framework for iterative curation of genes Support and integrate with existing efforts such as ClinGen, ClinVar, ICCG
12 ADDITIONAL CONTENTS OF ENHANCED EXOME CAPTURE ASSAY Ancestry Informative Marker panel* Able to differentiate main continental groups 0.4 Identify Informative Marker panel* 93 SNPs Match probability = 1 / Africa Asia Europe IndMex (J. Fagerness, R. Siburian) (J. Fagerness, R. Siburian) GWAS hits OR>5 n= 178; NHGRI GWAS catalog PGx targets Restricted to those with high evidence (PharmGKB)
13 TECHNICAL ENHANCEMENT
14 WHY THE BRICK WALL? MANY EXOME ASSAYS HAVE HOLES Cardiomyopathy Panel (51 genes) How much is covered at greater than 20x Targeted Capture Panel Exome (Agilent v6)
15 FILLING THE HOLES 2 types of performance issues (Agilent V6 exome kit) Exon not covered in design (no baits) Exon covered but performance suboptimal Worked with Agilent to create Version6-PLUS called the EmExome v2 ~3 Mb novel baits ~4 Mb boosted regions
16 BASE LINE EXOME DATA Whole Exome v6 (all genes) Medically Relevant Genes v6 11% of exons not covered ~10% of exons had a region not covered at greater than 20x
17 From v1 to v2 : Medical Exome v1 to Medical Exome v2 Progression in Design Medical Exome v1 (CREv1) 3654 genes curated to be medically relevant Medical Exome v2 (CREv2) 4753 genes curated to be medically relevant 475 genes removed from enhanced coverage from CREv1 to CREv2 but remain targeted by v remained in common from CREv1 to CREv2 Added curated deep intronic and promoter variants Breakpoint spanning baits for common events by precise mapping of breakpoints Non-coding splice sites OneSeq + CREv2 CREv CREv2 4753
18 Keeping Current on Curation Intensive process of reviewing the new literature Critical evaluation Example of deep intronic variant not in HGMD or LOVD Keeping an open door with clinician scientists
19 % of Exons Fully Covered MEDICAL EXOME v6 PLUS: UP TO 98% COVERAGE OF MEDICALLY RELEVANT HISEQ 2500 rapid ; 4-5 samples/lane GENES ~100X Coverage Across Samples
20 TOWARDS REPLACING TARGETED PANELS WITH EXOME SEQUENCING Cardiomyopathy Panel Targeted Capture Panel Medical Exome v2 (CREv2) = 99% of exons fully covered 99% of exons fully covered
21 WGS to EmExome v2: Coverage Comparison
22 ADDING CNV DETECTION USING AGILENT ONESEQ
23 Whole genome sequencing can find it all but at what cost? Cost of sequencing has defied Moore s law BUT. 1. $1000 genome only accessible with population-scale sequencer 2. And only if sequencer is run at full capacity WGS can cost up to $3,500 on a high throughput sequencer* -cost increase with increased depth Each WGS dataset (30x) is GB Enormous IT infrastructure needed to support data processing and storage Hidden cost of whole genome sequencing For Research Use Only.. Not for use in diagnostic procedures.
24 Knowing the Limitations Next Gen Sanger Single gene Sequencing panels Exome Genome Will not detect large deletions/duplications, repeat disorders, deep intronic variants NGS-based testing will have areas of low coverage
25 CNV Analysis from NGS Data - Benefits Detection of CNVs adds clinical value Can assay entire exome Other methods are reliable (qpcr, MPLA) but not scalable to a whole exome Retterer et al., Genet Med Dec 3. [epub ahead of print]; Poultney et al., Am J Hum Genet Oct 3;93(4): ; Fromer et al., Am J Hum Genet Oct 5;91(4):
26 Can whole exome sequencing detect CNVs? Detecting CNVs using read depth CNV Read Depth Read depth from WES is highly variable Duplication 1. Efficiency of target enrichment baits 2. Sequencing biases 3. Mappability 200+ computational methods to detect CNVs from whole exome sequencing No consensus on which method works best Same WES dataset analyzed by 5 different CNV yielded 10s-100s CNVs (Tan et al., 2014) For Research Use Only.. Not for use in diagnostic procedures.
27 Whole exome sequencing cannot detect non-coding CNVs Not all CNVs associated with constitutional diseases occur in genes Deletion of an intergenic regions upstream of SOX9 and NROB1 genes associated with male-to-female sex reversal in 46 X,Y individuals (Vetro et al., 2014, Smyk et al., 2007) Large duplication upstream of PMP22 associated with CMT1 neuropathy (Zhang et al., 2010, Weterman et al., 2010) Duplication downstream of PLP1 gene associated with spastic paraplegia and axonal neuropathy (Lee et al., 2006) Numerous more examples of CNVs in non-coding region can be found in the literature These CNVs would not be detected by WES! For Research Use Only.. Not for use in diagnostic procedures.
28 More information with just one capture Agilent OneSeq Target Enrichment: CNV, LOH, SNPs and indels in one assay Copy number and LOH determination Gene targets = SNPs and indels OneSeq CNV Backbone Focused Exome targets OneSeq Constitutional Research Panel All in one capture!
29 OneSeq Target Enrichment: One Assay, All Variants Med Exome associated regions Gene A Med Exome associated regions Gene B 1) Evenly spaced genome-wide baits 2) High density baits in Med Exome disease associated regions Copy number & LOH 3) User-defined baits in exonic regions SNVs & indels For Research Use Only.. Not for use in diagnostic procedures.
30 OneSeq is optimized for robust copy number calling OneSeq CNV backbone baits: Targeted to regions optimal GC content Best mappability Empirically validated Read depth from WES Read depth from OneSeq CNV backbone Dispersion Coefficient of variation = 65.8% Coefficient of variation = 36.1% For Research Use Only.. Not for use in diagnostic procedures.
31 OneSeq- Detecting Genomic CNV Genomic CNV EGL size OneSeq size Terminal deletion 21q 309 kb 272 kb Intragenic TCOF1 duplication 34.7 kb 34.9 kb Xq21 duplication 165 kb 181 kb Unbalanced translocation: 6p loss 5.89 Mb 5.73 Mb Chromothripsis: 7q21 loss 1.0 Mb 767 kb Chromothripsis: 11q14 loss 2.44 Mb 2.02 Mb Unbalanced translocation: 10q loss 1.49 Mb 1.29 Mb Unbalanced translocation: 17q gain 4.17 Mb 4.34 Mb
32 OneSeq 35-kb TCOF1duplication 309-kb 21q terminal deletion
33 Gene-specific Mutation Spectrum Familial Adenomatous Polyposis (APC) 80% point mutations 20% deletions/duplications RETT Syndrome (MECP2) 80% point mutations 20% deletions/duplications Point mutations Deletions/Duplications Cystic Fibrosis (CFTR) 90% point mutations 10% deletions/duplications Nuclear receptor binding SET domain protein 1 (NSD1) 75% point mutations 25% deletions/duplications Duchenne Muscular Dystrophy (DMD) 65% deletions/duplications 35% point mutations
34 Gene-specific Mutation Spectrum Familial Adenomatous Polyposis (APC) RETT Syndrome (MECP2) 80% point mutations 80% point mutations DMD gene : 65% del/dup 20% deletions/duplications 20% deletions/duplications Common intragenic deletion and duplications: GALC, FKTN Most known genes: 10-30% del/dup Point mutations Deep intronic/promoter/splice site targets Deletions/Duplications Common large events Cystic Fibrosis (CFTR) 90% point mutations Newly identified genes??? 10% deletions/duplications Nuclear receptor binding SET domain protein 1 (NSD1) 75% point mutations 25% deletions/duplications Duchenne Muscular Dystrophy (DMD) 65% deletions/duplications 35% point mutations
35 AJ Pathogenic Changes Disease ABCC8-Related Hyperinsulinism Bloom Syndrome Canavan Disease Cystic Fibrosis Familial Dysautonomia Fanconi Anemia Type C Gaucher Disease Glycogen Storage Disease Type 1a Joubert Syndrome Type 2 MSUD MSUD Type 3 (DLD) Mucolipidosis Type IV Nemaline Myopathy Niemann-Pick Disease Type A Tay-Sachs Disease Usher Syndrome Type IF Usher Syndrome Type III Mutations p.f1388del, c g>a (IVS32-9G>A), p.v187d c.2281del6ins7, c.2407inst c.433-2a>g, p.y231x, p.e285a, p.a305e 39 mutations c t>c (IVS20+6T>C), p.r696p c.322delg, IVS4+4A>T, p.q13x, p.r548x c.84gg, del55bp, IVS2+1G>A, p.n370, p.v394l, p.d409h, p.d409v, p.l444p, p.r463c, p.r463h, p.r496h c.79delc, c.378dupta, c.979del3, c.648g>t, p.r83c, p.r83h, p.g188r, p.e242x, p.g270v, p.q347x p.r12l p.r183p, p.g278s, p.e372x c.105insa (p.y35x), p.g229c Exon 1-exon 7del (Del6.4kb), IVS3-2A>G p.r2478_d2512del fsp330, p.r610del, p.l302p, p.t324i, p.h423y, p.r476w, p.r496l c.1278instatc, del7.6kb, G>C, IVS7+1G>A, IVS9+1G>A, p.r178h, p.r247w, p.r249w, p.g269s p.r245x c.459del3, p.n48k, p.l150p
36 Strategy for detection of large indels
37 NEXT STEPS TO ADDRESS LIMITATIONS OF NGS
38 MEDICALLY RELEVANT GENES WITH HOMOLOGY ISSUES The exome contains 1998 genes with at least one 250bp stretch of >98% homology Nearly 50% of these genes have issues in >3/4 of their exons Medical Exome 1998 homologous genes 286 homologous genes of likely/possible medical relevance ACMG Incidental gene list: PMS2, SMN Courtesy: Diana Mandelker
39 The Mendeliome: ~4,700 genes OneSeq Medical exome V2 Medical Exome V1 Exome Exome ~1% of genome 22,000 genes 4600 clinically relevant 1998 genes w/pseudogene(s) Medical Medical Exome Exome V2 V1 ~1% ~1% genome 22,000 genes Enhanced coverage of of ~4,700 4,600 clinically relevant genes Deep intronic mutations Mitochondrial genome Structural variants (including known targeted CNVs (eg 30kb GALC del) microrna OneSeq +Medical Exome V2 ~1% genome 22,000 genes Genome Enhanced coverage of ~4,700 clinically relevant genes Deep intronic mutations Mitochondrial genome Structural variants (including known targeted CNVs (eg 30kb GALC del) microrna PLUS copy number! Genome ~complete genome with gaps
Investigating rare diseases with Agilent NGS solutions
 Investigating rare diseases with Agilent NGS solutions Chitra Kotwaliwale, Ph.D. 1 Rare diseases affect 350 million people worldwide 7,000 rare diseases 80% are genetic 60 million affected in the US, Europe
Investigating rare diseases with Agilent NGS solutions Chitra Kotwaliwale, Ph.D. 1 Rare diseases affect 350 million people worldwide 7,000 rare diseases 80% are genetic 60 million affected in the US, Europe
Cytogenetics 101: Clinical Research and Molecular Genetic Technologies
 Cytogenetics 101: Clinical Research and Molecular Genetic Technologies Topics for Today s Presentation 1 Classical vs Molecular Cytogenetics 2 What acgh? 3 What is FISH? 4 What is NGS? 5 How can these
Cytogenetics 101: Clinical Research and Molecular Genetic Technologies Topics for Today s Presentation 1 Classical vs Molecular Cytogenetics 2 What acgh? 3 What is FISH? 4 What is NGS? 5 How can these
Identifying Mutations Responsible for Rare Disorders Using New Technologies
 Identifying Mutations Responsible for Rare Disorders Using New Technologies Jacek Majewski, Department of Human Genetics, McGill University, Montreal, QC Canada Mendelian Diseases Clear mode of inheritance
Identifying Mutations Responsible for Rare Disorders Using New Technologies Jacek Majewski, Department of Human Genetics, McGill University, Montreal, QC Canada Mendelian Diseases Clear mode of inheritance
DNA-seq Bioinformatics Analysis: Copy Number Variation
 DNA-seq Bioinformatics Analysis: Copy Number Variation Elodie Girard elodie.girard@curie.fr U900 institut Curie, INSERM, Mines ParisTech, PSL Research University Paris, France NGS Applications 5C HiC DNA-seq
DNA-seq Bioinformatics Analysis: Copy Number Variation Elodie Girard elodie.girard@curie.fr U900 institut Curie, INSERM, Mines ParisTech, PSL Research University Paris, France NGS Applications 5C HiC DNA-seq
Genomic structural variation
 Genomic structural variation Mario Cáceres The new genomic variation DNA sequence differs across individuals much more than researchers had suspected through structural changes A huge amount of structural
Genomic structural variation Mario Cáceres The new genomic variation DNA sequence differs across individuals much more than researchers had suspected through structural changes A huge amount of structural
Multiple Copy Number Variations in a Patient with Developmental Delay ASCLS- March 31, 2016
 Multiple Copy Number Variations in a Patient with Developmental Delay ASCLS- March 31, 2016 Marwan Tayeh, PhD, FACMG Director, MMGL Molecular Genetics Assistant Professor of Pediatrics Department of Pediatrics
Multiple Copy Number Variations in a Patient with Developmental Delay ASCLS- March 31, 2016 Marwan Tayeh, PhD, FACMG Director, MMGL Molecular Genetics Assistant Professor of Pediatrics Department of Pediatrics
Using the Bravo Liquid-Handling System for Next Generation Sequencing Sample Prep
 Using the Bravo Liquid-Handling System for Next Generation Sequencing Sample Prep Tom Walsh, PhD Division of Medical Genetics University of Washington Next generation sequencing Sanger sequencing gold
Using the Bravo Liquid-Handling System for Next Generation Sequencing Sample Prep Tom Walsh, PhD Division of Medical Genetics University of Washington Next generation sequencing Sanger sequencing gold
Benefits and pitfalls of new genetic tests
 Benefits and pitfalls of new genetic tests Amanda Krause Division of Human Genetics, NHLS and University of the Witwatersrand Definition of Genetic Testing the analysis of human DNA, RNA, chromosomes,
Benefits and pitfalls of new genetic tests Amanda Krause Division of Human Genetics, NHLS and University of the Witwatersrand Definition of Genetic Testing the analysis of human DNA, RNA, chromosomes,
PATIENT EDUCATION. carrier screening INFORMATION
 PATIENT EDUCATION carrier screening INFORMATION carrier screening AT A GLANCE Why is carrier screening recommended? Carrier screening is one of many tests that can help provide information to you and your
PATIENT EDUCATION carrier screening INFORMATION carrier screening AT A GLANCE Why is carrier screening recommended? Carrier screening is one of many tests that can help provide information to you and your
Merging single gene-level CNV with sequence variant interpretation following the ACMGG/AMP sequence variant guidelines
 Merging single gene-level CNV with sequence variant interpretation following the ACMGG/AMP sequence variant guidelines Tracy Brandt, Ph.D., FACMG Disclosure I am an employee of GeneDx, Inc., a wholly-owned
Merging single gene-level CNV with sequence variant interpretation following the ACMGG/AMP sequence variant guidelines Tracy Brandt, Ph.D., FACMG Disclosure I am an employee of GeneDx, Inc., a wholly-owned
Advance Your Genomic Research Using Targeted Resequencing with SeqCap EZ Library
 Advance Your Genomic Research Using Targeted Resequencing with SeqCap EZ Library Marilou Wijdicks International Product Manager Research For Life Science Research Only. Not for Use in Diagnostic Procedures.
Advance Your Genomic Research Using Targeted Resequencing with SeqCap EZ Library Marilou Wijdicks International Product Manager Research For Life Science Research Only. Not for Use in Diagnostic Procedures.
CURRENT GENETIC TESTING TOOLS IN NEONATAL MEDICINE. Dr. Bahar Naghavi
 2 CURRENT GENETIC TESTING TOOLS IN NEONATAL MEDICINE Dr. Bahar Naghavi Assistant professor of Basic Science Department, Shahid Beheshti University of Medical Sciences, Tehran,Iran 3 Introduction Over 4000
2 CURRENT GENETIC TESTING TOOLS IN NEONATAL MEDICINE Dr. Bahar Naghavi Assistant professor of Basic Science Department, Shahid Beheshti University of Medical Sciences, Tehran,Iran 3 Introduction Over 4000
A Lawyer s Perspective on Genetic Screening Performed by Cryobanks
 A Lawyer s Perspective on Genetic Screening Performed by Cryobanks As a lawyer practicing in the area of sperm bank litigation, I have, unfortunately, represented too many couples that conceived a child
A Lawyer s Perspective on Genetic Screening Performed by Cryobanks As a lawyer practicing in the area of sperm bank litigation, I have, unfortunately, represented too many couples that conceived a child
Breast and ovarian cancer in Serbia: the importance of mutation detection in hereditary predisposition genes using NGS
 Breast and ovarian cancer in Serbia: the importance of mutation detection in hereditary predisposition genes using NGS dr sc. Ana Krivokuća Laboratory for molecular genetics Institute for Oncology and
Breast and ovarian cancer in Serbia: the importance of mutation detection in hereditary predisposition genes using NGS dr sc. Ana Krivokuća Laboratory for molecular genetics Institute for Oncology and
Association for Molecular Pathology Promoting Clinical Practice, Basic Research, and Education in Molecular Pathology
 Association for Molecular Pathology Promoting Clinical Practice, Basic Research, and Education in Molecular Pathology 9650 Rockville Pike, Bethesda, Maryland 20814 Tel: 301-634-7939 Fax: 301-634-7990 Email:
Association for Molecular Pathology Promoting Clinical Practice, Basic Research, and Education in Molecular Pathology 9650 Rockville Pike, Bethesda, Maryland 20814 Tel: 301-634-7939 Fax: 301-634-7990 Email:
Golden Helix s End-to-End Solution for Clinical Labs
 Golden Helix s End-to-End Solution for Clinical Labs Steven Hystad - Field Application Scientist Nathan Fortier Senior Software Engineer 20 most promising Biotech Technology Providers Top 10 Analytics
Golden Helix s End-to-End Solution for Clinical Labs Steven Hystad - Field Application Scientist Nathan Fortier Senior Software Engineer 20 most promising Biotech Technology Providers Top 10 Analytics
QHerit Expanded Carrier Screen
 QHerit Expanded Carrier Screen Test Code: 94372 (X) Specimen Requirements: Preferred: 6 ml (4 ml minimum) room-temperature whole blood: 1.5 ml (1 ml minimum) in each of 4 lavender-top (EDTA) or yellow-top
QHerit Expanded Carrier Screen Test Code: 94372 (X) Specimen Requirements: Preferred: 6 ml (4 ml minimum) room-temperature whole blood: 1.5 ml (1 ml minimum) in each of 4 lavender-top (EDTA) or yellow-top
Applications of Chromosomal Microarray Analysis (CMA) in pre- and postnatal Diagnostic: advantages, limitations and concerns
 Applications of Chromosomal Microarray Analysis (CMA) in pre- and postnatal Diagnostic: advantages, limitations and concerns جواد کریمزاد حق PhD of Medical Genetics آزمايشگاه پاتوبيولوژي و ژنتيك پارسه
Applications of Chromosomal Microarray Analysis (CMA) in pre- and postnatal Diagnostic: advantages, limitations and concerns جواد کریمزاد حق PhD of Medical Genetics آزمايشگاه پاتوبيولوژي و ژنتيك پارسه
Nature Biotechnology: doi: /nbt.1904
 Supplementary Information Comparison between assembly-based SV calls and array CGH results Genome-wide array assessment of copy number changes, such as array comparative genomic hybridization (acgh), is
Supplementary Information Comparison between assembly-based SV calls and array CGH results Genome-wide array assessment of copy number changes, such as array comparative genomic hybridization (acgh), is
Expanded Carrier Screening: What s Best?
 Expanded Carrier Screening: What s Best? James D Goldberg, MD September 17, 2017 Disclosures James D. Goldberg, M.D. Chief Medical Officer, Counsyl 3 Learning Objectives Guidelines Data Design Practice
Expanded Carrier Screening: What s Best? James D Goldberg, MD September 17, 2017 Disclosures James D. Goldberg, M.D. Chief Medical Officer, Counsyl 3 Learning Objectives Guidelines Data Design Practice
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits
 AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
MEDICAL GENOMICS LABORATORY. Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG)
 Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
CHROMOSOMAL MICROARRAY (CGH+SNP)
 Chromosome imbalances are a significant cause of developmental delay, mental retardation, autism spectrum disorders, dysmorphic features and/or birth defects. The imbalance of genetic material may be due
Chromosome imbalances are a significant cause of developmental delay, mental retardation, autism spectrum disorders, dysmorphic features and/or birth defects. The imbalance of genetic material may be due
Implementation of the DDD/ClinGen OGT (CytoSure v3) Microarray
 Implementation of the DDD/ClinGen OGT (CytoSure v3) Microarray OGT UGM Birmingham 08/09/2016 Dom McMullan Birmingham Women's NHS Trust WM chromosomal microarray (CMA) testing Population of ~6 million (10%)
Implementation of the DDD/ClinGen OGT (CytoSure v3) Microarray OGT UGM Birmingham 08/09/2016 Dom McMullan Birmingham Women's NHS Trust WM chromosomal microarray (CMA) testing Population of ~6 million (10%)
Genetic Counselling in relation to genetic testing
 Genetic Counselling in relation to genetic testing Dr Julie Vogt Consultant Geneticist West Midlands Regional Genetics Service September 2016 Disclosures for Research Support/P.I. Employee Consultant Major
Genetic Counselling in relation to genetic testing Dr Julie Vogt Consultant Geneticist West Midlands Regional Genetics Service September 2016 Disclosures for Research Support/P.I. Employee Consultant Major
CNV Detection and Interpretation in Genomic Data
 CNV Detection and Interpretation in Genomic Data Benjamin W. Darbro, M.D., Ph.D. Assistant Professor of Pediatrics Director of the Shivanand R. Patil Cytogenetics and Molecular Laboratory Overview What
CNV Detection and Interpretation in Genomic Data Benjamin W. Darbro, M.D., Ph.D. Assistant Professor of Pediatrics Director of the Shivanand R. Patil Cytogenetics and Molecular Laboratory Overview What
Utility of Microarrays in Molecular Genetics
 Utility of Microarrays in Molecular Genetics Madhuri Hegde, Ph.D., FACMG Associate Professor Senior Director Department of Human Genetics Emory Genetics Laboratory Emory University School of Medicine Atlanta,
Utility of Microarrays in Molecular Genetics Madhuri Hegde, Ph.D., FACMG Associate Professor Senior Director Department of Human Genetics Emory Genetics Laboratory Emory University School of Medicine Atlanta,
Supplementary Table 2. Identified causative mutations and/or mutation candidates.
 Supplementary Table 2. Identified causative mutations and/or mutation candidates. Nonsense mutations base change aa change Average depth Result of next generation in 432 patient Hereditary form of the
Supplementary Table 2. Identified causative mutations and/or mutation candidates. Nonsense mutations base change aa change Average depth Result of next generation in 432 patient Hereditary form of the
NGS for Cancer Predisposition
 NGS for Cancer Predisposition Colin Pritchard MD, PhD University of Washington Dept. of Lab Medicine AMP Companion Society Meeting USCAP Boston March 22, 2015 Disclosures I am an employee of the University
NGS for Cancer Predisposition Colin Pritchard MD, PhD University of Washington Dept. of Lab Medicine AMP Companion Society Meeting USCAP Boston March 22, 2015 Disclosures I am an employee of the University
Multiplex target enrichment using DNA indexing for ultra-high throughput variant detection
 Multiplex target enrichment using DNA indexing for ultra-high throughput variant detection Dr Elaine Kenny Neuropsychiatric Genetics Research Group Institute of Molecular Medicine Trinity College Dublin
Multiplex target enrichment using DNA indexing for ultra-high throughput variant detection Dr Elaine Kenny Neuropsychiatric Genetics Research Group Institute of Molecular Medicine Trinity College Dublin
Genomics in Women s Health: Changing the Diagnostic and Therapeutic Paradigm
 Genomics in Women s Health: Changing the Diagnostic and Therapeutic Paradigm Lee P. Shulman MD The Anna Ross Lapham Professor in Obstetrics and Gynecology Feinberg School of Medicine of Northwestern University
Genomics in Women s Health: Changing the Diagnostic and Therapeutic Paradigm Lee P. Shulman MD The Anna Ross Lapham Professor in Obstetrics and Gynecology Feinberg School of Medicine of Northwestern University
GENETIC TESTING AND COUNSELING FOR HERITABLE DISORDERS
 Status Active Medical and Behavioral Health Policy Section: Laboratory Policy Number: VI-09 Effective Date: 03/17/2014 Blue Cross and Blue Shield of Minnesota medical policies do not imply that members
Status Active Medical and Behavioral Health Policy Section: Laboratory Policy Number: VI-09 Effective Date: 03/17/2014 Blue Cross and Blue Shield of Minnesota medical policies do not imply that members
Personalis ACE Clinical Exome The First Test to Combine an Enhanced Clinical Exome with Genome- Scale Structural Variant Detection
 Personalis ACE Clinical Exome The First Test to Combine an Enhanced Clinical Exome with Genome- Scale Structural Variant Detection Personalis, Inc. 1350 Willow Road, Suite 202, Menlo Park, California 94025
Personalis ACE Clinical Exome The First Test to Combine an Enhanced Clinical Exome with Genome- Scale Structural Variant Detection Personalis, Inc. 1350 Willow Road, Suite 202, Menlo Park, California 94025
CentoXome FUTURE'S KNOWLEDGE APPLIED TODAY
 CentoXome FUTURE'S KNOWLEDGE APPLIED TODAY More genetic information requires cutting-edge interpretation techniques Whole Exome Sequencing For certain patients the combination of symptoms does not allow
CentoXome FUTURE'S KNOWLEDGE APPLIED TODAY More genetic information requires cutting-edge interpretation techniques Whole Exome Sequencing For certain patients the combination of symptoms does not allow
CRISPR/Cas9 Enrichment and Long-read WGS for Structural Variant Discovery
 CRISPR/Cas9 Enrichment and Long-read WGS for Structural Variant Discovery PacBio CoLab Session October 20, 2017 For Research Use Only. Not for use in diagnostics procedures. Copyright 2017 by Pacific Biosciences
CRISPR/Cas9 Enrichment and Long-read WGS for Structural Variant Discovery PacBio CoLab Session October 20, 2017 For Research Use Only. Not for use in diagnostics procedures. Copyright 2017 by Pacific Biosciences
Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
 Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
CentoXome FUTURE'S KNOWLEDGE APPLIED TODAY
 CentoXome FUTURE'S KNOWLEDGE APPLIED TODAY More genetic information requires cutting-edge interpretation techniques Whole Exome Sequencing For some patients, the combination of symptoms does not allow
CentoXome FUTURE'S KNOWLEDGE APPLIED TODAY More genetic information requires cutting-edge interpretation techniques Whole Exome Sequencing For some patients, the combination of symptoms does not allow
MEDICAL GENOMICS LABORATORY. Peripheral Nerve Sheath Tumor Panel by Next-Gen Sequencing (PNT-NG)
 Peripheral Nerve Sheath Tumor Panel by Next-Gen Sequencing (PNT-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with receipt) Saliva (OGR-575 DNA Genotek;
Peripheral Nerve Sheath Tumor Panel by Next-Gen Sequencing (PNT-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with receipt) Saliva (OGR-575 DNA Genotek;
Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases.
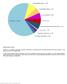 Supplementary Figure 1 Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases. A total of 85 somatically acquired
Supplementary Figure 1 Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases. A total of 85 somatically acquired
AVENIO ctdna Analysis Kits The complete NGS liquid biopsy solution EMPOWER YOUR LAB
 Analysis Kits The complete NGS liquid biopsy solution EMPOWER YOUR LAB Analysis Kits Next-generation performance in liquid biopsies 2 Accelerating clinical research From liquid biopsy to next-generation
Analysis Kits The complete NGS liquid biopsy solution EMPOWER YOUR LAB Analysis Kits Next-generation performance in liquid biopsies 2 Accelerating clinical research From liquid biopsy to next-generation
Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University
 Role of Chemical lexposure in Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University CNV Discovery Reference Genetic
Role of Chemical lexposure in Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University CNV Discovery Reference Genetic
Lab Prior Authorization
 Lab Prior Authorization On July 22, 2015, BlueCross BlueShield of South Carolina announced that it will partner with Avalon Healthcare Solutions (Avalon) to administer a comprehensive suite of laboratory
Lab Prior Authorization On July 22, 2015, BlueCross BlueShield of South Carolina announced that it will partner with Avalon Healthcare Solutions (Avalon) to administer a comprehensive suite of laboratory
Whole exome sequencing as a first line test: Is there even a role for metabolic biochemists in the future?
 Whole exome sequencing as a first line test: Is there even a role for metabolic biochemists in the future? Tony Marinakii Purine Research Laboratory Biochemical Sciences Whole exome sequencing No doubt
Whole exome sequencing as a first line test: Is there even a role for metabolic biochemists in the future? Tony Marinakii Purine Research Laboratory Biochemical Sciences Whole exome sequencing No doubt
Challenges of CGH array testing in children with developmental delay. Dr Sally Davies 17 th September 2014
 Challenges of CGH array testing in children with developmental delay Dr Sally Davies 17 th September 2014 CGH array What is CGH array? Understanding the test Benefits Results to expect Consent issues Ethical
Challenges of CGH array testing in children with developmental delay Dr Sally Davies 17 th September 2014 CGH array What is CGH array? Understanding the test Benefits Results to expect Consent issues Ethical
p.r623c p.p976l p.d2847fs p.t2671 p.d2847fs p.r2922w p.r2370h p.c1201y p.a868v p.s952* RING_C BP PHD Cbp HAT_KAT11
 ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
LIST OF INVESTIGATIONS
 Karyotyping: K001 K002 LIST OF INVESTIGATIONS SAMPLE CONTAINER TYPE cells For Karyotyping [Single] cells For Karyotyping [Couple] Vacutainer Vacutainer 7-8 7-8 K003 Fetal Blood Sample For Karyotyping Vacutainer
Karyotyping: K001 K002 LIST OF INVESTIGATIONS SAMPLE CONTAINER TYPE cells For Karyotyping [Single] cells For Karyotyping [Couple] Vacutainer Vacutainer 7-8 7-8 K003 Fetal Blood Sample For Karyotyping Vacutainer
PALB2 c g>c is. VARIANT OF UNCERTAIN SIGNIFICANCE (VUS) CGI s summary of the available evidence is in Appendices A-C.
 Consultation sponsor (may not be the patient): First LastName [Patient identity withheld] Date received by CGI: 2 Sept 2017 Variant Fact Checker Report ID: 0000001.5 Date Variant Fact Checker issued: 12
Consultation sponsor (may not be the patient): First LastName [Patient identity withheld] Date received by CGI: 2 Sept 2017 Variant Fact Checker Report ID: 0000001.5 Date Variant Fact Checker issued: 12
Sharan Goobie, MD, MSc, FRCPC
 Sharan Goobie, MD, MSc, FRCPC Chromosome testing in 2014 Presenter Disclosure: Sharan Goobie has no potential for conflict of interest with this presentation Objectives Review of standard genetic investigations
Sharan Goobie, MD, MSc, FRCPC Chromosome testing in 2014 Presenter Disclosure: Sharan Goobie has no potential for conflict of interest with this presentation Objectives Review of standard genetic investigations
Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research
 Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research Application Note Authors John McGuigan, Megan Manion,
Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research Application Note Authors John McGuigan, Megan Manion,
Next Generation Sequencing as a tool for breakpoint analysis in rearrangements of the globin-gene clusters
 Next Generation Sequencing as a tool for breakpoint analysis in rearrangements of the globin-gene clusters XXXth International Symposium on Technical Innovations in Laboratory Hematology Honolulu, Hawaii
Next Generation Sequencing as a tool for breakpoint analysis in rearrangements of the globin-gene clusters XXXth International Symposium on Technical Innovations in Laboratory Hematology Honolulu, Hawaii
Andrew Parrish, Richard Caswell, Garan Jones, Christopher M. Watson, Laura A. Crinnion 3,4, Sian Ellard 1,2
 METHOD ARTICLE An enhanced method for targeted next generation sequencing copy number variant detection using ExomeDepth [version 1; referees: 1 approved, 1 approved with reservations] 1 2 1 3,4 Andrew
METHOD ARTICLE An enhanced method for targeted next generation sequencing copy number variant detection using ExomeDepth [version 1; referees: 1 approved, 1 approved with reservations] 1 2 1 3,4 Andrew
Clinical Policy Title: Genetic tests for Duchenne muscular dystrophy
 Clinical Policy Title: Genetic tests for Duchenne muscular dystrophy Clinical Policy Number: 02.01.23 Effective Date: February 1, 2017 Initial Review Date: January 18, 2017 Most Recent Review Date: January
Clinical Policy Title: Genetic tests for Duchenne muscular dystrophy Clinical Policy Number: 02.01.23 Effective Date: February 1, 2017 Initial Review Date: January 18, 2017 Most Recent Review Date: January
CNV detection. Introduction and detection in NGS data. G. Demidov 1,2. NGSchool2016. Centre for Genomic Regulation. CNV detection. G.
 Introduction and detection in NGS data 1,2 1 Genomic and Epigenomic Variation in Disease group, Centre for Genomic Regulation 2 Universitat Pompeu Fabra NGSchool2016 methods: methods Outline methods: methods
Introduction and detection in NGS data 1,2 1 Genomic and Epigenomic Variation in Disease group, Centre for Genomic Regulation 2 Universitat Pompeu Fabra NGSchool2016 methods: methods Outline methods: methods
Genetics and Genomics: Applications to Developmental Disability
 Tuesday, 12:30 2:00, B1 Objective: Genetics and Genomics: Applications to Developmental Disability Helga Toriello 616-234-2712 toriello@msu.edu Identify advances in clinical assessment and management of
Tuesday, 12:30 2:00, B1 Objective: Genetics and Genomics: Applications to Developmental Disability Helga Toriello 616-234-2712 toriello@msu.edu Identify advances in clinical assessment and management of
What s the Human Genome Project Got to Do with Developmental Disabilities?
 What s the Human Genome Project Got to Do with Developmental Disabilities? Disclosures Neither speaker has anything to disclose. Phase Two: Interpretation Officially started in October 1990 Goals of the
What s the Human Genome Project Got to Do with Developmental Disabilities? Disclosures Neither speaker has anything to disclose. Phase Two: Interpretation Officially started in October 1990 Goals of the
variant led to a premature stop codon p.k316* which resulted in nonsense-mediated mrna decay. Although the exact function of the C19L1 is still
 157 Neurological disorders primarily affect and impair the functioning of the brain and/or neurological system. Structural, electrical or metabolic abnormalities in the brain or neurological system can
157 Neurological disorders primarily affect and impair the functioning of the brain and/or neurological system. Structural, electrical or metabolic abnormalities in the brain or neurological system can
The Deciphering Development Disorders (DDD) project: What a genomic approach can achieve
 The Deciphering Development Disorders (DDD) project: What a genomic approach can achieve RCP ADVANCED MEDICINE, LONDON FEB 5 TH 2018 HELEN FIRTH DM FRCP DCH, SANGER INSTITUTE 3,000,000,000 bases in each
The Deciphering Development Disorders (DDD) project: What a genomic approach can achieve RCP ADVANCED MEDICINE, LONDON FEB 5 TH 2018 HELEN FIRTH DM FRCP DCH, SANGER INSTITUTE 3,000,000,000 bases in each
Detection of aneuploidy in a single cell using the Ion ReproSeq PGS View Kit
 APPLICATION NOTE Ion PGM System Detection of aneuploidy in a single cell using the Ion ReproSeq PGS View Kit Key findings The Ion PGM System, in concert with the Ion ReproSeq PGS View Kit and Ion Reporter
APPLICATION NOTE Ion PGM System Detection of aneuploidy in a single cell using the Ion ReproSeq PGS View Kit Key findings The Ion PGM System, in concert with the Ion ReproSeq PGS View Kit and Ion Reporter
IHCP bulletin INDIANA HEALTH COVERAGE PROGRAMS BT MARCH 13, 2012
 IHCP bulletin INDIANA HEALTH COVERAGE PROGRAMS BT201208 MARCH 13, 2012 Updates to the 2012 Healthcare Common Coding System This bulletin updates information published by the Indiana Health Coverage Programs
IHCP bulletin INDIANA HEALTH COVERAGE PROGRAMS BT201208 MARCH 13, 2012 Updates to the 2012 Healthcare Common Coding System This bulletin updates information published by the Indiana Health Coverage Programs
SNP Array NOTE: THIS IS A SAMPLE REPORT AND MAY NOT REFLECT ACTUAL PATIENT DATA. FORMAT AND/OR CONTENT MAY BE UPDATED PERIODICALLY.
 SAMPLE REPORT SNP Array NOTE: THIS IS A SAMPLE REPORT AND MAY NOT REFLECT ACTUAL PATIENT DATA. FORMAT AND/OR CONTENT MAY BE UPDATED PERIODICALLY. RESULTS SNP Array Copy Number Variations Result: LOSS,
SAMPLE REPORT SNP Array NOTE: THIS IS A SAMPLE REPORT AND MAY NOT REFLECT ACTUAL PATIENT DATA. FORMAT AND/OR CONTENT MAY BE UPDATED PERIODICALLY. RESULTS SNP Array Copy Number Variations Result: LOSS,
SNP Array NOTE: THIS IS A SAMPLE REPORT AND MAY NOT REFLECT ACTUAL PATIENT DATA. FORMAT AND/OR CONTENT MAY BE UPDATED PERIODICALLY.
 SAMPLE REPORT SNP Array NOTE: THIS IS A SAMPLE REPORT AND MAY NOT REFLECT ACTUAL PATIENT DATA. FORMAT AND/OR CONTENT MAY BE UPDATED PERIODICALLY. RESULTS SNP Array Copy Number Variations Result: GAIN,
SAMPLE REPORT SNP Array NOTE: THIS IS A SAMPLE REPORT AND MAY NOT REFLECT ACTUAL PATIENT DATA. FORMAT AND/OR CONTENT MAY BE UPDATED PERIODICALLY. RESULTS SNP Array Copy Number Variations Result: GAIN,
Characterisation of structural variation in breast. cancer genomes using paired-end sequencing on. the Illumina Genome Analyser
 Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
NGS in tissue and liquid biopsy
 NGS in tissue and liquid biopsy Ana Vivancos, PhD Referencias So, why NGS in the clinics? 2000 Sanger Sequencing (1977-) 2016 NGS (2006-) ABIPrism (Applied Biosystems) Up to 2304 per day (96 sequences
NGS in tissue and liquid biopsy Ana Vivancos, PhD Referencias So, why NGS in the clinics? 2000 Sanger Sequencing (1977-) 2016 NGS (2006-) ABIPrism (Applied Biosystems) Up to 2304 per day (96 sequences
Back to the Basics: Methyl-Seq 101
 Back to the Basics: Methyl-Seq 101 Presented By: Alex Siebold, Ph.D. October 9, 2013 Field Applications Scientist Agilent Technologies Life Sciences & Diagnostics Group Life Sciences & Diagnostics Group
Back to the Basics: Methyl-Seq 101 Presented By: Alex Siebold, Ph.D. October 9, 2013 Field Applications Scientist Agilent Technologies Life Sciences & Diagnostics Group Life Sciences & Diagnostics Group
Home Brewed Personalized Genomics
 Home Brewed Personalized Genomics The Quest for Meaningful Analysis Results of a 23andMe Exome Pilot Trio of Myself, Wife, and Son February 22, 2013 Gabe Rudy, Vice President of Product Development Exome
Home Brewed Personalized Genomics The Quest for Meaningful Analysis Results of a 23andMe Exome Pilot Trio of Myself, Wife, and Son February 22, 2013 Gabe Rudy, Vice President of Product Development Exome
Corporate Medical Policy
 Corporate Medical Policy Carrier Screening for Genetic Disease File Name: Origination: Last CAP Review: Next CAP Review: Last Review: carrier_screening_for_genetic_disease 12/2013 7/2017 7/2018 7/2017
Corporate Medical Policy Carrier Screening for Genetic Disease File Name: Origination: Last CAP Review: Next CAP Review: Last Review: carrier_screening_for_genetic_disease 12/2013 7/2017 7/2018 7/2017
MRC-Holland MLPA. Description version 08; 30 March 2015
 SALSA MLPA probemix P351-C1 / P352-D1 PKD1-PKD2 P351-C1 lot C1-0914: as compared to the previous version B2 lot B2-0511 one target probe has been removed and three reference probes have been replaced.
SALSA MLPA probemix P351-C1 / P352-D1 PKD1-PKD2 P351-C1 lot C1-0914: as compared to the previous version B2 lot B2-0511 one target probe has been removed and three reference probes have been replaced.
Analysis with SureCall 2.1
 Analysis with SureCall 2.1 Danielle Fletcher Field Application Scientist July 2014 1 Stages of NGS Analysis Primary analysis, base calling Control Software FASTQ file reads + quality 2 Stages of NGS Analysis
Analysis with SureCall 2.1 Danielle Fletcher Field Application Scientist July 2014 1 Stages of NGS Analysis Primary analysis, base calling Control Software FASTQ file reads + quality 2 Stages of NGS Analysis
No mutations were identified.
 Hereditary High Cholesterol Test ORDERING PHYSICIAN PRIMARY CONTACT SPECIMEN Report date: Aug 1, 2017 Dr. Jenny Jones Sample Medical Group 123 Main St. Sample, CA Kelly Peters Sample Medical Group 123
Hereditary High Cholesterol Test ORDERING PHYSICIAN PRIMARY CONTACT SPECIMEN Report date: Aug 1, 2017 Dr. Jenny Jones Sample Medical Group 123 Main St. Sample, CA Kelly Peters Sample Medical Group 123
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers
 Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Abstract. Optimization strategy of Copy Number Variant calling using Multiplicom solutions APPLICATION NOTE. Introduction
 Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Nature Genetics: doi: /ng Supplementary Figure 1. PCA for ancestry in SNV data.
 Supplementary Figure 1 PCA for ancestry in SNV data. (a) EIGENSTRAT principal-component analysis (PCA) of SNV genotype data on all samples. (b) PCA of only proband SNV genotype data. (c) PCA of SNV genotype
Supplementary Figure 1 PCA for ancestry in SNV data. (a) EIGENSTRAT principal-component analysis (PCA) of SNV genotype data on all samples. (b) PCA of only proband SNV genotype data. (c) PCA of SNV genotype
Genetic Testing for Single-Gene and Multifactorial Conditions
 Clinical Appropriateness Guidelines Genetic Testing for Single-Gene and Multifactorial Conditions EFFECTIVE DECEMBER 1, 2017 Appropriate.Safe.Affordable 2017 AIM Specialty Health 2069-1217 Table of Contents
Clinical Appropriateness Guidelines Genetic Testing for Single-Gene and Multifactorial Conditions EFFECTIVE DECEMBER 1, 2017 Appropriate.Safe.Affordable 2017 AIM Specialty Health 2069-1217 Table of Contents
The Focused Exome service at Bristol Genetics Laboratory
 The Focused Exome service at Bristol Genetics Laboratory Chris Buxton Maggie Williams July 2016 Bristol Clinical exome Service to mid July Validation: Agilent FE kit, NextSeq 500 and new pipeline 1st reports
The Focused Exome service at Bristol Genetics Laboratory Chris Buxton Maggie Williams July 2016 Bristol Clinical exome Service to mid July Validation: Agilent FE kit, NextSeq 500 and new pipeline 1st reports
Managing Moderate Penetrance
 Managing Moderate Penetrance Thomas Slavin, MD, FACMG Assistant Clinical Professor, Department of Medical Oncology, Division of Clinical Cancer Genetics Program Member, Cancer Control and Population Sciences
Managing Moderate Penetrance Thomas Slavin, MD, FACMG Assistant Clinical Professor, Department of Medical Oncology, Division of Clinical Cancer Genetics Program Member, Cancer Control and Population Sciences
PERSONALIZED GENETIC REPORT CLIENT-REPORTED DATA PURPOSE OF THE X-SCREEN TEST
 INCLUDED IN THIS REPORT: REVIEW OF YOUR GENETIC INFORMATION RELEVANT TO ENDOMETRIOSIS PERSONAL EDUCATIONAL INFORMATION RELEVANT TO YOUR GENES INFORMATION FOR OBTAINING YOUR ENTIRE X-SCREEN DATA FILE PERSONALIZED
INCLUDED IN THIS REPORT: REVIEW OF YOUR GENETIC INFORMATION RELEVANT TO ENDOMETRIOSIS PERSONAL EDUCATIONAL INFORMATION RELEVANT TO YOUR GENES INFORMATION FOR OBTAINING YOUR ENTIRE X-SCREEN DATA FILE PERSONALIZED
MRC-Holland MLPA. Description version 29; 31 July 2015
 SALSA MLPA probemix P081-C1/P082-C1 NF1 P081 Lot C1-0114. As compared to the previous B2 version (lot 0813 and 0912), 11 target probes are replaced or added, and 10 new reference probes are included. P082
SALSA MLPA probemix P081-C1/P082-C1 NF1 P081 Lot C1-0114. As compared to the previous B2 version (lot 0813 and 0912), 11 target probes are replaced or added, and 10 new reference probes are included. P082
BWA alignment to reference transcriptome and genome. Convert transcriptome mappings back to genome space
 Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
Detection of copy number variations in PCR-enriched targeted sequencing data
 Detection of copy number variations in PCR-enriched targeted sequencing data German Demidov Parseq Lab, Saint-Petersburg University of Russian Academy of Sciences, current: Center for Genomic Regulation
Detection of copy number variations in PCR-enriched targeted sequencing data German Demidov Parseq Lab, Saint-Petersburg University of Russian Academy of Sciences, current: Center for Genomic Regulation
Agilent s Copy Number Variation (CNV) Portfolio
 Technical Overview Agilent s Copy Number Variation (CNV) Portfolio Abstract Copy Number Variation (CNV) is now recognized as a prevalent form of structural variation in the genome contributing to human
Technical Overview Agilent s Copy Number Variation (CNV) Portfolio Abstract Copy Number Variation (CNV) is now recognized as a prevalent form of structural variation in the genome contributing to human
New: P077 BRCA2. This new probemix can be used to confirm results obtained with P045 BRCA2 probemix.
 SALSA MLPA KIT P045-B2 BRCA2/CHEK2 Lot 0410, 0609. As compared to version B1, four reference probes have been replaced and extra control fragments at 100 and 105 nt (X/Y specific) have been included. New:
SALSA MLPA KIT P045-B2 BRCA2/CHEK2 Lot 0410, 0609. As compared to version B1, four reference probes have been replaced and extra control fragments at 100 and 105 nt (X/Y specific) have been included. New:
Illuminating the genetics of complex human diseases
 Illuminating the genetics of complex human diseases Michael Schatz Sept 27, 2012 Beyond the Genome @mike_schatz / #BTG2012 Outline 1. De novo mutations in human diseases 1. Autism Spectrum Disorder 2.
Illuminating the genetics of complex human diseases Michael Schatz Sept 27, 2012 Beyond the Genome @mike_schatz / #BTG2012 Outline 1. De novo mutations in human diseases 1. Autism Spectrum Disorder 2.
QUESTION. Personal Behavior History. Donor Genetic History. Donor Medical History. Family Medical History PERSONAL BEHAVIOR HISTORY. Never N/A.
 Donor 4576 Medical Profile S Personal Behavior History Donor Genetic History Donor Medical History Family Medical History PERSONAL BEHAVIOR HISTORY Current alcohol use: If yes, oz./week and type of alcohol:
Donor 4576 Medical Profile S Personal Behavior History Donor Genetic History Donor Medical History Family Medical History PERSONAL BEHAVIOR HISTORY Current alcohol use: If yes, oz./week and type of alcohol:
Preparent Standard Panel
 Preparent Standard Panel Disorder Gene Variants Ethnicity Alpha-thalassemia HBA1 HBA2 -alpha3.7, -alpha4.2, -alpha20.5, --SEA, --MED, --FIL, --THAI Mediterranean 1 in 20 1 in 191 1 in 291 African American
Preparent Standard Panel Disorder Gene Variants Ethnicity Alpha-thalassemia HBA1 HBA2 -alpha3.7, -alpha4.2, -alpha20.5, --SEA, --MED, --FIL, --THAI Mediterranean 1 in 20 1 in 191 1 in 291 African American
MutationTaster & RegulationSpotter
 Practical MutationTaster & RegulationSpotter Daniela Hombach & Jana Marie Schwarz Exzellenzcluster NeuroCure Charité Universitätsmedizin Berlin AG Translational Genomics 08.11.2017 Rare diseases 7000 8000rare
Practical MutationTaster & RegulationSpotter Daniela Hombach & Jana Marie Schwarz Exzellenzcluster NeuroCure Charité Universitätsmedizin Berlin AG Translational Genomics 08.11.2017 Rare diseases 7000 8000rare
Understanding genetics, mutation and other details. Stanley F. Nelson, MD 6/29/18
 Understanding genetics, mutation and other details Stanley F. Nelson, MD 6/29/18 1 6 11 16 21 Duchenne muscular dystrophy 26 31 36 41 46 51 56 61 66 71 76 81 86 91 96 600 500 400 300 200 100 0 Duchenne/Becker
Understanding genetics, mutation and other details Stanley F. Nelson, MD 6/29/18 1 6 11 16 21 Duchenne muscular dystrophy 26 31 36 41 46 51 56 61 66 71 76 81 86 91 96 600 500 400 300 200 100 0 Duchenne/Becker
Iowa Wellstone Center Muscle Tissue and Cell Culture Repository
 Iowa Wellstone Center Muscle Tissue and Cell Culture Repository Steven A. Moore, M.D., Ph.D. The University of Iowa Department of Pathology and Iowa Wellstone Muscular Dystrophy Cooperative Research Center
Iowa Wellstone Center Muscle Tissue and Cell Culture Repository Steven A. Moore, M.D., Ph.D. The University of Iowa Department of Pathology and Iowa Wellstone Muscular Dystrophy Cooperative Research Center
MSI positive MSI negative
 Pritchard et al. 2014 Supplementary Figure 1 MSI positive MSI negative Hypermutated Median: 673 Average: 659.2 Non-Hypermutated Median: 37.5 Average: 43.6 Supplementary Figure 1: Somatic Mutation Burden
Pritchard et al. 2014 Supplementary Figure 1 MSI positive MSI negative Hypermutated Median: 673 Average: 659.2 Non-Hypermutated Median: 37.5 Average: 43.6 Supplementary Figure 1: Somatic Mutation Burden
Variant Classification. Author: Mike Thiesen, Golden Helix, Inc.
 Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
MRC-Holland MLPA. Description version 30; 06 June 2017
 SALSA MLPA probemix P081-C1/P082-C1 NF1 P081 Lot C1-0517, C1-0114. As compared to the previous B2 version (lot B2-0813, B2-0912), 11 target probes are replaced or added, and 10 new reference probes are
SALSA MLPA probemix P081-C1/P082-C1 NF1 P081 Lot C1-0517, C1-0114. As compared to the previous B2 version (lot B2-0813, B2-0912), 11 target probes are replaced or added, and 10 new reference probes are
Nature Genetics: doi: /ng Supplementary Figure 1. TCGA data set on HNSCCs reanalyzed in this study.
 Supplementary Figure 1 TCGA data set on HNSCCs reanalyzed in this study. Summary of the TCGA dataset on HNSCCs re-analyzed in this study and the respective numbers of samples available within each. Supplementary
Supplementary Figure 1 TCGA data set on HNSCCs reanalyzed in this study. Summary of the TCGA dataset on HNSCCs re-analyzed in this study and the respective numbers of samples available within each. Supplementary
MLPA SAMPLES USER GUIDE
 MLPA SAMPLES USER GUIDE MLPA microdeletion studies Requirements: 1-2 mls (minimum) of peripheral blood in a lithium heparin tube (green or orange top) AND 1-2 mls of peripheral blood in a EDTA tube (purple
MLPA SAMPLES USER GUIDE MLPA microdeletion studies Requirements: 1-2 mls (minimum) of peripheral blood in a lithium heparin tube (green or orange top) AND 1-2 mls of peripheral blood in a EDTA tube (purple
Supplementary appendix
 Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Wells JM, Farris RF, Gosdin TA, et al. Pulmonary
Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Wells JM, Farris RF, Gosdin TA, et al. Pulmonary
Whole Exome Sequencing (WES) Whole Exome Sequencing. What Is Whole Exome Sequencing?
 Whole Exome Sequencing (WES) Procedure(s) addressed by this policy: Exome (e.g., unexplained constitutional or heritable disorder or syndrome); sequence analysis Sequence analysis, each comparator exome
Whole Exome Sequencing (WES) Procedure(s) addressed by this policy: Exome (e.g., unexplained constitutional or heritable disorder or syndrome); sequence analysis Sequence analysis, each comparator exome
BROADENING YOUR PATIENT S OPTIONS FOR GENETIC CARRIER SCREENING.
 BROADENING YOUR PATIENT S OPTIONS FOR GENETIC CARRIER SCREENING. The Inheritest SM Carrier Screen provides relevant genetic screening for many inherited diseases found throughout the pan-ethnic US population.
BROADENING YOUR PATIENT S OPTIONS FOR GENETIC CARRIER SCREENING. The Inheritest SM Carrier Screen provides relevant genetic screening for many inherited diseases found throughout the pan-ethnic US population.
Reporting TP53 gene analysis results in CLL
 Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
Welcome to the Genetic Code: An Overview of Basic Genetics. October 24, :00pm 3:00pm
 Welcome to the Genetic Code: An Overview of Basic Genetics October 24, 2016 12:00pm 3:00pm Course Schedule 12:00 pm 2:00 pm Principles of Mendelian Genetics Introduction to Genetics of Complex Disease
Welcome to the Genetic Code: An Overview of Basic Genetics October 24, 2016 12:00pm 3:00pm Course Schedule 12:00 pm 2:00 pm Principles of Mendelian Genetics Introduction to Genetics of Complex Disease
Centers for Medicare and Medicaid Services
 Centers for Medicare and Medicaid Services Clinical Laboratory Fee Schedule Annual Laboratory Public Meeting June 25, 2018 Anthony Sireci, MD, Msc Association for Molecular Pathology Outline Germline Procedures
Centers for Medicare and Medicaid Services Clinical Laboratory Fee Schedule Annual Laboratory Public Meeting June 25, 2018 Anthony Sireci, MD, Msc Association for Molecular Pathology Outline Germline Procedures
TumorNext-HRD with OvaNext: Paired Germline and Tumor Analyses of Genes Involved in
 SAMPLE REPORT Ordered By Contact ID:1251298 Example, Doctor, MD MOCKORG44 (10829) 123 Somewhere LaneSuite 4 Heaven NV 78872 US Ph:123-123-1234 Fx:123-123-1223 Org ID:8141 Normal Specimen Accession #: 00-086947
SAMPLE REPORT Ordered By Contact ID:1251298 Example, Doctor, MD MOCKORG44 (10829) 123 Somewhere LaneSuite 4 Heaven NV 78872 US Ph:123-123-1234 Fx:123-123-1223 Org ID:8141 Normal Specimen Accession #: 00-086947
SALSA MLPA probemix P185-C2 Intersex Lot C2-1015: As compared to the previous version C1 (lot C1-0611), the lengths of four probes have been adjusted.
 mix P185-C2 Intersex Lot C2-1015: As compared to the previous version C1 (lot C1-0611), the lengths of four s have been adjusted. The sex-determining region on chromosome Y (SRY) is the most important
mix P185-C2 Intersex Lot C2-1015: As compared to the previous version C1 (lot C1-0611), the lengths of four s have been adjusted. The sex-determining region on chromosome Y (SRY) is the most important
