A new selectable marker for manipulating the simple. genomes of mycoplasma
|
|
|
- Opal Hart
- 5 years ago
- Views:
Transcription
1 AAC Accepts, published online ahead of print on 17 August 2009 Antimicrob. Agents Chemother. doi: /aac Copyright 2009, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights Reserved. 1 2 A new selectable marker for manipulating the simple genomes of mycoplasma running title: A new marker for manipulating mycoplasma Mikkel A. Algire, Carole Lartigue, David W. Thomas 1, Nacyra Assad-Garcia, John I. Glass and Chuck Merryman * The J. Craig Venter Institute 9704 Medical Center Drive Rockville, Maryland Present address: Biotechnology Industry Organization (BIO) 1201 Maryland Avenue SW Washington, DC * To whom correspondence should be addressed: 9704 Medical Center Drive Building 1, Room 322 Rockville, MD cmerryma@jcvi.org
2 Abstract Over the past several years significant advancements have been made on the molecular genetics of the Mollicutes (the simplest cells that can be grown in axenic culture). Nevertheless, a number of basic molecular tools are still required before genetic manipulations become routine. Here we describe the development of a new dominant selectable marker based on the enzyme puromycin-n-acetyltransferase from Streptomyces alboniger. Puromycin is an antibiotic that mimics the 3 -terminal end of aminoacylated trnas and attaches to the carboxyl-terminus of growing protein chains. This stops protein synthesis. Because puromycin conscripts rrnarecognition elements that are used by all of the various trnas in a cell, it is unlikely that spontaneous antibiotic resistance can be acquired via a simple point mutation an annoying issue with existing mycoplasma markers. Our codon-optimized cassette confers pronounced puromycin resistance in all five of the mycoplasma species we have tested so far. The resistance cassette was also designed to function in Escherichia coli which simplifies the construction of shuttle vectors and makes it trivial to produce the large quantities of DNA generally necessary for mycoplasma transformation. Due to these and other features, we expect the puromycin marker to be a widely applicable tool for studying these simple cells and pathogens. Introduction The mycoplasma, which is the generic name for members of the class Mollicutes, are wall-less bacteria that are often important pathogens in a variety of species (2, 20). In addition to their importance as pathogens, their small genomes and limited metabolic complexity make them platform organisms for studies on the universal elements of life itself. For example, transposon mutagenesis of the simplest mycoplasma (M. genitalium) showed that more than 100 of the 485 2
3 protein coding genes could be individually disrupted under laboratory conditions (5, 7, 18). Although it is unknown if the disrupted genes are simultaneously dispensable, the remaining 370 or so undisturbed genes provide an initial map of the minimum genetic requirements for life. The simplicity of mycoplasma also introduces some technical complications. For example, auxotrophic markers may be difficult to establish in organisms that utilize ill-defined media and that cannot synthesize most biological building blocks. Similarly, antibiotics that target the cell wall are not useful because this structure is completely absent. The use of other markers is not advisable because they are clinically relevant and the simplicity of mycoplasma naturally restricts clinical options in the first place. In our hands, fairly reliable markers include the tetracycline resistance protein (TetM), and to a lesser extent chloramphenicol acyltransferase (CAT) and aminoglycoside resistance protein (aaca/aphd). The tetm/tetracycline combination is usually preferable because of the potent anti-mycoplasma activity of tetracyclines and the low levels of spontaneous tetracycline resistance. In comparison, spontaneous resistance frequently arises with the other markers (presumably via a point mutation in an rrna). Such resistance is rarely insurmountable, but screening numerous colonies for authentic transformants wastes valuable resources unnecessarily. In spite of the limitations imposed by mycoplasma biology, the characteristics and mode of action of puromycin suggested that it was a likely candidate for marker development. Puromycin is readily available, relatively inexpensive, has no clinical value and it is a potent inhibitor of translation in both prokaryotes and eukaryotes. To our knowledge, no example of rrna based resistance to the antibiotic exists. Most likely this is a consequence of the universality of the acceptor end of trna and the mechanism of peptidyl tranferase the two translational features puromycin exploits as a near perfect trna mimic (25). Moreover, the PAC 3
4 gene from Streptomyces alboniger encodes a small protein (puromycin-n-acetyltransferase) which provides resistance to the antibiotic. This suggested that a codon optimized version of the enzyme might be efficiently expressed. The resistance genes for the usual markers have not been codon-optimized for production in mycoplasma and expression issues likely contribute to the use of low antibiotic concentrations and thus, to the establishment of spontaneous resistance. Here we describe the use of the PAC gene as a new selectable marker in several mycoplasma species. Materials and methods Mycoplasma strains. The mycoplasma species used were Mycoplasma mycoides subsp. mycoides large colony (LC) GM12 which has recently been renamed Mycoplasma mycoides subspecies capri GM12 (15), Mycoplasma capricolum subsp. capricolum California kid (ATCC 27343), Mycoplasma genitalium G37, Mycoplasma gallisepticum (ATCC 15302), and Mycoplasma pneumoniae Eaton (ATCC15531)and M. pneumoniae M129-B170 (ATCC 29343). M. mycoides and M. capricolum were grown at 37 ºC in SP4 media (8, 9, 23). M. genitalium and the M. pneumoniae strains were grown at 37 ºC in the presence of 5% CO 2 in SP4 media. M. gallisepticum was grown in Hayflick s media (23) in the presence of 5% CO 2 at 37 ºC. Puromycin N-acetlytransferase gene construction. The 597 bp puromycin N-acetlytransferase gene (PAC) was synthesized using overlapping oligonucleotides as described (22). Briefly, 5 phosphorylated oligonucleotides encoding both strands of a codon optimized version (for expression in M. genitalium) were ordered from IDT (Coralville, IA). The oligos were 48 bases long with an overlap of 24 bases. The top-strand and bottom-strand oligos were mixed, heated to 95 ºC, and slow cooled to allow annealing of the overlaps. The reactions were ligated for 12 hours and used as a template for PCR. The PCR amplicon was cloned into pgem-3zf(+) 4
5 (Promega, Madison, Wisconsin) and sequenced to identify correct PAC clones. The optimized PAC gene was then cloned under the control of the Spiroplasma citri spiralin promoter (Ps) (12) and used to replace the tetm gene in a derivative of Mini-Tn4001tet (17, 19) (GenBank accession no. GQ420676) as well as the tetm gene in pmyco1 (11). The new plasmid (Mini- Tn4001PsPuro; GenBank accession no. FJ872396) was used to transform M. genitalium, M. gallisepticum and M. pneumoniae, while the pmyco1 derivative (pmycopuro; GenBank accession no. GQ420675) was used to transform M. mycoides and M. capricolum. Mycoplasma transformations. M. genitalium and M. pneumoniae were transformed by electroporation as previously described (1, 6, 21). Polyethylene glycol (PEG) mediated transformation was used for M. mycoides, M. gallisepticum and M. capricolum (8, 9, 13, 17). Puromycin inhibitory concentration determination. M. capricolum wild-type cells and cells bearing the pmycopuro plasmid were grown in SP4 media containing various concentrations of puromycin to determine bacteriostatic activity. After 24 hours of exposure to the antibiotic, aliquots were removed, diluted 10,000-fold, and plated without antibiotic to evaluate bactericidal activity. A similar procedure was used for the other organisms but bactericidal activity was not determined. Results and discussion All five mycoplasma species were grown in SP4 or Hayflicks media supplemented with various concentrations of puromycin. All of the wild-type mycoplasmas that were tested were unable to grow in the presence of puromycin when the concentration exceeded 1-3 µg/ml. Thus, distantly related mycoplasmas from both the Mycoides group and the Pneumoniae group are sensitive to the antibiotic. This sensitivity may be commonplace within the Mollicutes as they 5
6 have streamlined genomes unlikely to contain the sophisticated efflux pumps or the modifying enzymes typically required to establish puromycin resistance. Mini-Tn4001PsPuro (Figure 1) established resistance in M. genitalium, M. gallisepticum and M. pneumoniae by transposition. The pmycopuro plasmid replicates as a free DNA in M. capricolum, while in M. mycoides the plasmid integrates by single crossover at the OriC of the M. mycoides genome (data not shown). The puromycin constructs have not been used for directed mutagenesis although we presume it would work in mycoplasmas where such mutagenesis has been successful with a tetracycline marker. In general, expression of the PAC gene confers wild-type growth at concentrations beyond 50 µg/ml (M. capricolum had wild-type growth at 500 µg/ml). In practice, we typically selected for transformants on solid media at lower concentrations (3 µg/ml for M. genitalium, M. gallisepticum and M. pneumoniae and 8 µg/ml for M. capricolum and M. mycoides). Isolated colonies of M. capricolum or M. mycoides were subsequently grown in liquid culture at 8 µg/ml for the first passage and 50 µg/ml for later passages. M. genitalium, M. gallisepticum and M. pneumoniae were maintained at 3 µg/ml during passage. As shown in Figure 2, the inhibitory concentration for M. capricolum is approximately 3 µg/ml. We wanted to determine if the antibiotic action of puromycin on these mycoplasma species was bacteriocidal or bacteriostatic. After M. capricolum cells were exposed to 300 µg/ml of puromycin for 24 hours, they could recover when plated in the absence of the antibiotic (data not shown). Apparently, puromycin is not lethal even at high concentrations. It is possible that the stringent response, which is active in the organism, induces a persistent state and thereby, death is avoided (4, 10, 24). If this supposition is correct, the presence of stringent response proteins (rela/spot family proteins) in most sequenced mycoplasmas suggests that these bacteria 6
7 might also survive puromycin exposure even if they are incapable of growth. From a practical standpoint this issue does not affect the usefulness of the marker. For example, puromycin has become our marker of choice when working with the very slow growing organism M. genitalium. Over the three-week period required to produce clones, no special procedures are required to prevent the appearance of untransformed colonies. There are several minor features of the expression cassette that were designed to broaden its usefulness. Transcription of the PAC gene is driven by the promoter for spiralin (Ps), which is an abundant protein from Spiroplasma citri. We have found this promoter to be widely applicable. The efficacy of the spiralin promoter in the range of mycoplasmas examined here further documents its usefulness. Since the promoter also functions in E. coli (16), we designed the codon-optimized PAC gene so that it is free of TGA codons (TGA codes for tryptophan in most mycoplasma rather than a stop codon as in the standard genetic code). In the mycoplasmas that use TGA as a tryptophan codon, its use is heavily preferred over the other the tryptophan codon (TGG). Coding the Trp residues in the PAC gene with only TGG is a minor violation of codon-optimization rules but together with the Ps promoter it allows for expression in E. coli. This simplifies the construction of shuttle vectors as only a single marker is required. Puromycin sensitivity in E. coli begins between µg/ml and the cassette provides resistance beyond 200 µg/ml. We typically use 125 µg/ml of puromycin for selecting with E. coli. For puromycin expression, as well as the expression of numerous other proteins, we have used a large region upstream of the spiralin start codon (324 nucleotides). In the past such a large fragment could complicate its movement into various constructs. Now with the advent of new DNA assembly methods this is no longer a concern as we can seamlessly join 2-10 sequences without regard to restriction sites (3, 14). 7
8 Our main objective with this study was to identify a more consistent marker than tetracycline for our work on genome transplantation in M. genitalium (2). The finding that the puromycin/pac gene combination is broadly applicable for Mollicute research suggests that it may be generally preferable to the current antibiotic generally used by molecular mycoplasmologists, tetracycline. This is in part due to the recombinant DNA guidelines that regulate the insertion into pathogens of resistance genes for antibiotics in clinical or veterinary use. Tetracyclines remain a clinical option for some mycoplasma infections and are additives in some animal feeds. The efficacy of puromycin also introduces the possibility of generating double knockouts to further refine the minimal gene content able to support replication and life itself. Acknowledgments We thank Radha Krishnakumar for critically reading the manuscript. This work was supported by Synthetic Genomics, Inc. References 1. Cao, J., P. A. Kapke, and F. C. Minion Transformation of Mycoplasma gallisepticum with Tn916, Tn4001, and integrative plasmid vectors. J Bacteriol 176: Fadiel, A., K. D. Eichenbaum, N. El Semary, and B. Epperson Mycoplasma genomics: tailoring the genome for minimal life requirements through reductive evolution. Front Biosci 12:
9 Gibson, D. G., G. A. Benders, C. Andrews-Pfannkoch, E. A. Denisova, H. Baden- Tillson, J. Zaveri, T. B. Stockwell, A. Brownley, D. W. Thomas, M. A. Algire, C. Merryman, L. Young, V. N. Noskov, J. I. Glass, J. C. Venter, C. A. Hutchison, 3rd, and H. O. Smith Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319: Glaser, G., A. Razin, and S. Razin Stable RNA synthesis and its control in Mycoplasma capricolum. Nucleic Acids Res 9: Glass, J. I., N. Assad-Garcia, N. Alperovich, S. Yooseph, M. R. Lewis, M. Maruf, C. A. Hutchison, 3rd, H. O. Smith, and J. C. Venter Essential genes of a minimal bacterium. Proc Natl Acad Sci U S A 103: Hedreyda, C. T., K. K. Lee, and D. C. Krause Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid 30: Hutchison, C. A., S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286: King, K. W., and K. Dybvig Plasmid transformation of Mycoplasma mycoides subspecies mycoides is promoted by high concentrations of polyethylene glycol. Plasmid 26: King, K. W., and K. Dybvig Transformation of Mycoplasma capricolum and examination of DNA restriction modification in M. capricolum and Mycoplasma mycoides subsp. mycoides. Plasmid 31:
10 Korch, S. B., T. A. Henderson, and T. M. Hill Characterization of the hipa7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppgpp synthesis. Mol Microbiol 50: Lartigue, C., A. Blanchard, J. Renaudin, F. Thiaucourt, and P. Sirand-Pugnet Host specificity of mollicutes oric plasmids: functional analysis of replication origin. Nucleic Acids Res 31: Lartigue, C., S. Duret, M. Garnier, and J. Renaudin New plasmid vectors for specific gene targeting in Spiroplasma citri. Plasmid 48: Lartigue, C., J. I. Glass, N. Alperovich, R. Pieper, P. P. Parmar, C. A. Hutchison, 3rd, H. O. Smith, and J. C. Venter Genome transplantation in bacteria: changing one species to another. Science 317: Li, M. Z., and S. J. Elledge Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 4: Manso-Silvan, L., E. M. Vilei, K. Sachse, S. P. Djordjevic, F. Thiaucourt, and J. Frey Mycoplasma leachii sp. nov. as a new species designation for Mycoplasma sp. bovine group 7 of Leach, and reclassification of Mycoplasma mycoides subsp. mycoides LC as a serovar of Mycoplasma mycoides subsp. capri. Int J Syst Evol Microbiol 59: Mouches, C., T. Candresse, G. Barroso, C. Saillard, H. Wroblewski, and J. M. Bove Gene for spiralin, the major membrane protein of the helical mollicute Spiroplasma citri: cloning and expression in Escherichia coli. J Bacteriol 164:
11 Papazisi, L., K. E. Troy, T. S. Gorton, X. Liao, and S. J. Geary Analysis of cytadherence-deficient, GapA-negative Mycoplasma gallisepticum strain R. Infect Immun 68: Pich, O. Q., R. Burgos, R. Planell, E. Querol, and J. Pinol Comparative analysis of antibiotic resistance gene markers in Mycoplasma genitalium: application to studies of the minimal gene complement. Microbiology 152: Pour-El, I., C. Adams, and F. C. Minion Construction of mini-tn4001tet and its use in Mycoplasma gallisepticum. Plasmid 47: Razin, S., and R. Herrmann Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum, New York. 21. Reddy, S. P., W. G. Rasmussen, and J. B. Baseman Isolation and characterization of transposon Tn4001-generated, cytadherence-deficient transformants of Mycoplasma pneumoniae and Mycoplasma genitalium. FEMS Immunol Med Microbiol 15: Smith, H. O., C. A. Hutchison, 3rd, C. Pfannkoch, and J. C. Venter Generating a synthetic genome by whole genome assembly: phix174 bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci U S A 100: Tully, J. G., R. F. Whitcomb, H. F. Clark, and D. L. Williamson Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science 195: VanBogelen, R. A., and F. C. Neidhardt Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc Natl Acad Sci U S A 87:
12 Youngman, E. M., J. L. Brunelle, A. B. Kochaniak, and R. Green The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117: Downloaded from on September 25, 2018 by guest 12
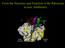
13 Figure 1. Maps of plasmids used in Mycoplamas. A. Map of the MiniTn4001PsTetM plasmid. The spiralin promoter is upstream of the TetM open reading frame (ORF). The IS256 elements flank the Tet resistance cassette with the transposase gene outside of the repeats. B. Map of the MiniTn4001PsPuro plasmid. The features are identical to the MiniTn4001PsTetM plasmid except that antibiotic resistance cassette is in the opposite orientation and the TetM ORF has been replaced with the puromycin N-acetlytransferase ORF. After the spiralin promoter was cloned upstream of the PAC gene in pgem-3zf(+) the entire cassette was cloned into the backbone of the MiniTn4001PsTetM plasmid that had the promoter and TetM ORF removed by digestion with PstI and BamHI. C. Map of the pmycopuro plasmid. The puromycin resistance unit was cloned into pmyco1 that had the promoter and TetM ORF removed by digestion with PstI. The M. mycoides OriC allows propagation in M. mycoides. The puc Ori and β-lactamase gene are used for propagation in E. coli. Figure 2. The pmycopuro plasmid confers puromycin resistance. M. capricolum was grown in SP4 media with a range of puromycin concentrations in the presence or absence of the pmycopuro plasmid. Without the plasmid M. capricolum grew slowly at puromycin concentrations of 1.2 µg/ml and no growth was observed at 3.7 µg/ml. In this color changing unit assay the change of the ph indicator phenol red in the media from red to yellow was used as an indicator of growth. In the absence of cell growth the SP4 media does not change color over the time course of the experiment (24 hr). M. capricolum cells containing the vector were resistant to puromycin at concentrations of at least 300 µg/ml. 13
14 A. IS256 Inner spiralin promoter puc Ori TetR B. C. AmpR MiniTn4001PsTetM 6637 bp puc Ori AmpR IS256 Inner puc Ori Transposase MiniTn4001PsPuro 5321 bp AmpR spiralin promoter pmycopuro 6202 bp PuroR M. mycoides OriC PuroR IS256 Outer spiralin promoter IS256 Outer Transposase
15 (-) vector µg/ml puromycin Downloaded from (+) vector on September 25, 2018 by guest
Objectives: Prof.Dr. H.D.El-Yassin
 Protein Synthesis and drugs that inhibit protein synthesis Objectives: 1. To understand the steps involved in the translation process that leads to protein synthesis 2. To understand and know about all
Protein Synthesis and drugs that inhibit protein synthesis Objectives: 1. To understand the steps involved in the translation process that leads to protein synthesis 2. To understand and know about all
Translation Activity Guide
 Translation Activity Guide Student Handout β-globin Translation Translation occurs in the cytoplasm of the cell and is defined as the synthesis of a protein (polypeptide) using information encoded in an
Translation Activity Guide Student Handout β-globin Translation Translation occurs in the cytoplasm of the cell and is defined as the synthesis of a protein (polypeptide) using information encoded in an
Lecture 2: Virology. I. Background
 Lecture 2: Virology I. Background A. Properties 1. Simple biological systems a. Aggregates of nucleic acids and protein 2. Non-living a. Cannot reproduce or carry out metabolic activities outside of a
Lecture 2: Virology I. Background A. Properties 1. Simple biological systems a. Aggregates of nucleic acids and protein 2. Non-living a. Cannot reproduce or carry out metabolic activities outside of a
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004
 Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Antibiotics and Ribosomes as Drug Targets
 Antibiotics and Ribosomes as Drug Targets www.biochemj.org/bj/330/0581/bj3300581.htm Professor Vassie Ware Bioscience in the 21 st Century November 5, 2012 PERSPECTIVE Widespread use of antibiotics after
Antibiotics and Ribosomes as Drug Targets www.biochemj.org/bj/330/0581/bj3300581.htm Professor Vassie Ware Bioscience in the 21 st Century November 5, 2012 PERSPECTIVE Widespread use of antibiotics after
Protein Synthesis
 Protein Synthesis 10.6-10.16 Objectives - To explain the central dogma - To understand the steps of transcription and translation in order to explain how our genes create proteins necessary for survival.
Protein Synthesis 10.6-10.16 Objectives - To explain the central dogma - To understand the steps of transcription and translation in order to explain how our genes create proteins necessary for survival.
One consequence of progress in the new field of synthetic
 Essential genes of a minimal bacterium John I. Glass, Nacyra Assad-Garcia, Nina Alperovich, Shibu Yooseph, Matthew R. Lewis, Mahir Maruf, Clyde A. Hutchison III, Hamilton O. Smith*, and J. Craig Venter
Essential genes of a minimal bacterium John I. Glass, Nacyra Assad-Garcia, Nina Alperovich, Shibu Yooseph, Matthew R. Lewis, Mahir Maruf, Clyde A. Hutchison III, Hamilton O. Smith*, and J. Craig Venter
LESSON 4.4 WORKBOOK. How viruses make us sick: Viral Replication
 DEFINITIONS OF TERMS Eukaryotic: Non-bacterial cell type (bacteria are prokaryotes).. LESSON 4.4 WORKBOOK How viruses make us sick: Viral Replication This lesson extends the principles we learned in Unit
DEFINITIONS OF TERMS Eukaryotic: Non-bacterial cell type (bacteria are prokaryotes).. LESSON 4.4 WORKBOOK How viruses make us sick: Viral Replication This lesson extends the principles we learned in Unit
AP Biology Reading Guide. Concept 19.1 A virus consists of a nucleic acid surrounded by a protein coat
 AP Biology Reading Guide Name Chapter 19: Viruses Overview Experimental work with viruses has provided important evidence that genes are made of nucleic acids. Viruses were also important in working out
AP Biology Reading Guide Name Chapter 19: Viruses Overview Experimental work with viruses has provided important evidence that genes are made of nucleic acids. Viruses were also important in working out
Genetic information flows from mrna to protein through the process of translation
 Genetic information flows from mrn to protein through the process of translation TYPES OF RN (RIBONUCLEIC CID) RN s job - protein synthesis (assembly of amino acids into proteins) Three main types: 1.
Genetic information flows from mrn to protein through the process of translation TYPES OF RN (RIBONUCLEIC CID) RN s job - protein synthesis (assembly of amino acids into proteins) Three main types: 1.
Course Title Form Hours subject
 Course Title Form Hours subject Types, and structure of chromosomes L 1 Histology Karyotyping and staining of human chromosomes L 2 Histology Chromosomal anomalies L 2 Histology Sex chromosomes L 1 Histology
Course Title Form Hours subject Types, and structure of chromosomes L 1 Histology Karyotyping and staining of human chromosomes L 2 Histology Chromosomal anomalies L 2 Histology Sex chromosomes L 1 Histology
Certificate of Analysis
 Certificate of Analysis plvx-ef1α-ires-puro Vector Table of Contents Product Information... 1 Description... 2 Location of Features... 3 Additional Information... 3 Quality Control Data... 4 Catalog No.
Certificate of Analysis plvx-ef1α-ires-puro Vector Table of Contents Product Information... 1 Description... 2 Location of Features... 3 Additional Information... 3 Quality Control Data... 4 Catalog No.
TRANSLATION. Translation is a process where proteins are made by the ribosomes on the mrna strand.
 TRANSLATION Dr. Mahesha H B, Yuvaraja s College, University of Mysore, Mysuru. Translation is a process where proteins are made by the ribosomes on the mrna strand. Or The process in the ribosomes of a
TRANSLATION Dr. Mahesha H B, Yuvaraja s College, University of Mysore, Mysuru. Translation is a process where proteins are made by the ribosomes on the mrna strand. Or The process in the ribosomes of a
Supplemental Material for. Figure S1. Identification of TetR responsive promoters in F. novicida and E. coli.
 Supplemental Material for Synthetic promoters functional in Francisella novicida and Escherichia coli Ralph L. McWhinnie and Francis E. Nano Department of Biochemistry and Microbiology, University of Victoria,
Supplemental Material for Synthetic promoters functional in Francisella novicida and Escherichia coli Ralph L. McWhinnie and Francis E. Nano Department of Biochemistry and Microbiology, University of Victoria,
Viral Genetics. BIT 220 Chapter 16
 Viral Genetics BIT 220 Chapter 16 Details of the Virus Classified According to a. DNA or RNA b. Enveloped or Non-Enveloped c. Single-stranded or double-stranded Viruses contain only a few genes Reverse
Viral Genetics BIT 220 Chapter 16 Details of the Virus Classified According to a. DNA or RNA b. Enveloped or Non-Enveloped c. Single-stranded or double-stranded Viruses contain only a few genes Reverse
Pre-mRNA has introns The splicing complex recognizes semiconserved sequences
 Adding a 5 cap Lecture 4 mrna splicing and protein synthesis Another day in the life of a gene. Pre-mRNA has introns The splicing complex recognizes semiconserved sequences Introns are removed by a process
Adding a 5 cap Lecture 4 mrna splicing and protein synthesis Another day in the life of a gene. Pre-mRNA has introns The splicing complex recognizes semiconserved sequences Introns are removed by a process
Practice Problems 8. a) What do we define as a beneficial or advantageous mutation to the virus? Why?
 Life Sciences 1a Practice Problems 8 1. You have two strains of HIV one is a wild type strain of HIV and the second has acquired a mutation in the gene encoding the protease. This mutation has a dual effect
Life Sciences 1a Practice Problems 8 1. You have two strains of HIV one is a wild type strain of HIV and the second has acquired a mutation in the gene encoding the protease. This mutation has a dual effect
INTRODUCTION PRODUCT DESCRIPTION
 INTRODUCTION Mycoplasma are known as important contaminants of biological products derived from cell lines in the biopharmaceutical industry affecting every parameter of a cell culture system. Contaminated
INTRODUCTION Mycoplasma are known as important contaminants of biological products derived from cell lines in the biopharmaceutical industry affecting every parameter of a cell culture system. Contaminated
Bio 111 Study Guide Chapter 17 From Gene to Protein
 Bio 111 Study Guide Chapter 17 From Gene to Protein BEFORE CLASS: Reading: Read the introduction on p. 333, skip the beginning of Concept 17.1 from p. 334 to the bottom of the first column on p. 336, and
Bio 111 Study Guide Chapter 17 From Gene to Protein BEFORE CLASS: Reading: Read the introduction on p. 333, skip the beginning of Concept 17.1 from p. 334 to the bottom of the first column on p. 336, and
Chapter 19: The Genetics of Viruses and Bacteria
 Chapter 19: The Genetics of Viruses and Bacteria What is Microbiology? Microbiology is the science that studies microorganisms = living things that are too small to be seen with the naked eye Microorganisms
Chapter 19: The Genetics of Viruses and Bacteria What is Microbiology? Microbiology is the science that studies microorganisms = living things that are too small to be seen with the naked eye Microorganisms
Gene Vaccine Dr. Sina Soleimani
 Gene Vaccine Dr. Sina Soleimani Human Viral Vaccines Quality Control Laboratory (HVVQC) Titles 1. A short Introduction of Vaccine History 2. First Lineage of Vaccines 3. Second Lineage of Vaccines 3. New
Gene Vaccine Dr. Sina Soleimani Human Viral Vaccines Quality Control Laboratory (HVVQC) Titles 1. A short Introduction of Vaccine History 2. First Lineage of Vaccines 3. Second Lineage of Vaccines 3. New
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation
 Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
From the Structure and Function of the Ribosome to new Antibiotics
 From the Structure and Function of the Ribosome to new Antibiotics Crick s central dogma of molecular biology: DNA makes DNA makes RNA makes protein Jim Watson, 1964 J.A. Lake, 1976 (J.M.B. 105, 131)
From the Structure and Function of the Ribosome to new Antibiotics Crick s central dogma of molecular biology: DNA makes DNA makes RNA makes protein Jim Watson, 1964 J.A. Lake, 1976 (J.M.B. 105, 131)
Bordeaux, France, from 1999 to 2002 and Description of Two tet(m)-positive Isolates of ACCEPTED
 AAC Accepts, published online ahead of print on 19 November 2007 Antimicrob. Agents Chemother. doi:10.1128/aac.00960-07 Copyright 2007, American Society for Microbiology and/or the Listed Authors/Institutions.
AAC Accepts, published online ahead of print on 19 November 2007 Antimicrob. Agents Chemother. doi:10.1128/aac.00960-07 Copyright 2007, American Society for Microbiology and/or the Listed Authors/Institutions.
Page 32 AP Biology: 2013 Exam Review CONCEPT 6 REGULATION
 Page 32 AP Biology: 2013 Exam Review CONCEPT 6 REGULATION 1. Feedback a. Negative feedback mechanisms maintain dynamic homeostasis for a particular condition (variable) by regulating physiological processes,
Page 32 AP Biology: 2013 Exam Review CONCEPT 6 REGULATION 1. Feedback a. Negative feedback mechanisms maintain dynamic homeostasis for a particular condition (variable) by regulating physiological processes,
Biochemistry 2000 Sample Question Transcription, Translation and Lipids. (1) Give brief definitions or unique descriptions of the following terms:
 (1) Give brief definitions or unique descriptions of the following terms: (a) exon (b) holoenzyme (c) anticodon (d) trans fatty acid (e) poly A tail (f) open complex (g) Fluid Mosaic Model (h) embedded
(1) Give brief definitions or unique descriptions of the following terms: (a) exon (b) holoenzyme (c) anticodon (d) trans fatty acid (e) poly A tail (f) open complex (g) Fluid Mosaic Model (h) embedded
Certificate of Analysis
 Certificate of Analysis Catalog No. Amount Lot Number 631987 10 μg Specified on product label. Product Information plvx-ef1α-ires-mcherry is a bicistronic lentiviral expression vector that can be used
Certificate of Analysis Catalog No. Amount Lot Number 631987 10 μg Specified on product label. Product Information plvx-ef1α-ires-mcherry is a bicistronic lentiviral expression vector that can be used
Mycoplasma Total Solution. Mycoplasma Detection Mycoplasma Elimination Mycoplasma Prevention Cell Freezing Medium (Mycoplasma Free)
 Mycoplasma Total Solution Mycoplasma Detection Mycoplasma Elimination Mycoplasma Prevention Cell Freezing Medium (Mycoplasma Free) Mycoplasma Detection Mycoplasma Detection CellSafe s Mycoplasma Detection
Mycoplasma Total Solution Mycoplasma Detection Mycoplasma Elimination Mycoplasma Prevention Cell Freezing Medium (Mycoplasma Free) Mycoplasma Detection Mycoplasma Detection CellSafe s Mycoplasma Detection
RNA (Ribonucleic acid)
 RNA (Ribonucleic acid) Structure: Similar to that of DNA except: 1- it is single stranded polunucleotide chain. 2- Sugar is ribose 3- Uracil is instead of thymine There are 3 types of RNA: 1- Ribosomal
RNA (Ribonucleic acid) Structure: Similar to that of DNA except: 1- it is single stranded polunucleotide chain. 2- Sugar is ribose 3- Uracil is instead of thymine There are 3 types of RNA: 1- Ribosomal
Multi-clonal origin of macrolide-resistant Mycoplasma pneumoniae isolates. determined by multiple-locus variable-number tandem-repeat analysis
 JCM Accepts, published online ahead of print on 30 May 2012 J. Clin. Microbiol. doi:10.1128/jcm.00678-12 Copyright 2012, American Society for Microbiology. All Rights Reserved. 1 2 Multi-clonal origin
JCM Accepts, published online ahead of print on 30 May 2012 J. Clin. Microbiol. doi:10.1128/jcm.00678-12 Copyright 2012, American Society for Microbiology. All Rights Reserved. 1 2 Multi-clonal origin
The Blueprint of Life: DNA to Protein. What is genetics? DNA Structure 4/27/2011. Chapter 7
 The Blueprint of Life: NA to Protein Chapter 7 What is genetics? The science of heredity; includes the study of genes, how they carry information, how they are replicated, how they are expressed NA Structure
The Blueprint of Life: NA to Protein Chapter 7 What is genetics? The science of heredity; includes the study of genes, how they carry information, how they are replicated, how they are expressed NA Structure
The Blueprint of Life: DNA to Protein
 The Blueprint of Life: NA to Protein Chapter 7 What is genetics? The science of heredity; includes the y; study of genes, how they carry information, how they are replicated, how they are expressed 1 NA
The Blueprint of Life: NA to Protein Chapter 7 What is genetics? The science of heredity; includes the y; study of genes, how they carry information, how they are replicated, how they are expressed 1 NA
Molecular mechanisms of antibiotic resistance in Neisseria gonorrhoeae
 medicaldaily.com http://en.vircell.com/diseases ppcorn.com Molecular mechanisms of antibiotic resistance in Neisseria gonorrhoeae Robert Nicholas University of North Carolina at Chapel Hill cdc.gov Timeline
medicaldaily.com http://en.vircell.com/diseases ppcorn.com Molecular mechanisms of antibiotic resistance in Neisseria gonorrhoeae Robert Nicholas University of North Carolina at Chapel Hill cdc.gov Timeline
Human Genome: Mapping, Sequencing Techniques, Diseases
 Human Genome: Mapping, Sequencing Techniques, Diseases Lecture 4 BINF 7580 Fall 2005 1 Let us review what we talked about at the previous lecture. Please,... 2 The central dogma states that the transfer
Human Genome: Mapping, Sequencing Techniques, Diseases Lecture 4 BINF 7580 Fall 2005 1 Let us review what we talked about at the previous lecture. Please,... 2 The central dogma states that the transfer
Introduction retroposon
 17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
Computational Biology I LSM5191
 Computational Biology I LSM5191 Aylwin Ng, D.Phil Lecture Notes: Transcriptome: Molecular Biology of Gene Expression II TRANSLATION RIBOSOMES: protein synthesizing machines Translation takes place on defined
Computational Biology I LSM5191 Aylwin Ng, D.Phil Lecture Notes: Transcriptome: Molecular Biology of Gene Expression II TRANSLATION RIBOSOMES: protein synthesizing machines Translation takes place on defined
TRANSCRIPTION. DNA à mrna
 TRANSCRIPTION DNA à mrna Central Dogma Animation DNA: The Secret of Life (from PBS) http://www.youtube.com/watch? v=41_ne5ms2ls&list=pl2b2bd56e908da696&index=3 Transcription http://highered.mcgraw-hill.com/sites/0072507470/student_view0/
TRANSCRIPTION DNA à mrna Central Dogma Animation DNA: The Secret of Life (from PBS) http://www.youtube.com/watch? v=41_ne5ms2ls&list=pl2b2bd56e908da696&index=3 Transcription http://highered.mcgraw-hill.com/sites/0072507470/student_view0/
Introduction 1/3. The protagonist of our story: the prokaryotic ribosome
 Introduction 1/3 The protagonist of our story: the prokaryotic ribosome Introduction 2/3 The core functional domains of the ribosome (the peptidyl trasferase center and the decoding center) are composed
Introduction 1/3 The protagonist of our story: the prokaryotic ribosome Introduction 2/3 The core functional domains of the ribosome (the peptidyl trasferase center and the decoding center) are composed
8 Suppression Analysis
 Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 8 Suppression Analysis OVERVIEW Suppression
Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 8 Suppression Analysis OVERVIEW Suppression
Genetics. Instructor: Dr. Jihad Abdallah Transcription of DNA
 Genetics Instructor: Dr. Jihad Abdallah Transcription of DNA 1 3.4 A 2 Expression of Genetic information DNA Double stranded In the nucleus Transcription mrna Single stranded Translation In the cytoplasm
Genetics Instructor: Dr. Jihad Abdallah Transcription of DNA 1 3.4 A 2 Expression of Genetic information DNA Double stranded In the nucleus Transcription mrna Single stranded Translation In the cytoplasm
DNA codes for RNA, which guides protein synthesis.
 Section 3: DNA codes for RNA, which guides protein synthesis. K What I Know W What I Want to Find Out L What I Learned Vocabulary Review synthesis New RNA messenger RNA ribosomal RNA transfer RNA transcription
Section 3: DNA codes for RNA, which guides protein synthesis. K What I Know W What I Want to Find Out L What I Learned Vocabulary Review synthesis New RNA messenger RNA ribosomal RNA transfer RNA transcription
Chapter 32: Translation
 Chapter 32: Translation Voet & Voet: Pages 1343-1385 (Parts of sections 1-3) Slide 1 Genetic code Translates the genetic information into functional proteins mrna is read in 5 to 3 direction Codons are
Chapter 32: Translation Voet & Voet: Pages 1343-1385 (Parts of sections 1-3) Slide 1 Genetic code Translates the genetic information into functional proteins mrna is read in 5 to 3 direction Codons are
Host-parasite interactions: Evolutionary genetics of the House Finch- Mycoplasma epizootic
 Host-parasite interactions: Evolutionary genetics of the House Finch- Mycoplasma epizootic Scott V. Edwards Department of Organismic and Evolutionary Biology Harvard University Cambridge, MA USA http://www.oeb.harvard.edu/faculty/edwards
Host-parasite interactions: Evolutionary genetics of the House Finch- Mycoplasma epizootic Scott V. Edwards Department of Organismic and Evolutionary Biology Harvard University Cambridge, MA USA http://www.oeb.harvard.edu/faculty/edwards
1) DNA unzips - hydrogen bonds between base pairs are broken by special enzymes.
 Biology 12 Cell Cycle To divide, a cell must complete several important tasks: it must grow, during which it performs protein synthesis (G1 phase) replicate its genetic material /DNA (S phase), and physically
Biology 12 Cell Cycle To divide, a cell must complete several important tasks: it must grow, during which it performs protein synthesis (G1 phase) replicate its genetic material /DNA (S phase), and physically
Synthetic Genomics and Its Application to Viral Infectious Diseases. Timothy Stockwell (JCVI) David Wentworth (JCVI)
 Synthetic Genomics and Its Application to Viral Infectious Diseases Timothy Stockwell (JCVI) David Wentworth (JCVI) Outline Using informatics to predict drift (strain selection) Synthetic Genomics: Preparedness
Synthetic Genomics and Its Application to Viral Infectious Diseases Timothy Stockwell (JCVI) David Wentworth (JCVI) Outline Using informatics to predict drift (strain selection) Synthetic Genomics: Preparedness
Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins.
 Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins. The RNA transcribed from a complex transcription unit
Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins. The RNA transcribed from a complex transcription unit
An Introduction to Carbohydrates
 An Introduction to Carbohydrates Carbohydrates are a large class of naturally occurring polyhydroxy aldehydes and ketones. Monosaccharides also known as simple sugars, are the simplest carbohydrates containing
An Introduction to Carbohydrates Carbohydrates are a large class of naturally occurring polyhydroxy aldehydes and ketones. Monosaccharides also known as simple sugars, are the simplest carbohydrates containing
Overview: Chapter 19 Viruses: A Borrowed Life
 Overview: Chapter 19 Viruses: A Borrowed Life Viruses called bacteriophages can infect and set in motion a genetic takeover of bacteria, such as Escherichia coli Viruses lead a kind of borrowed life between
Overview: Chapter 19 Viruses: A Borrowed Life Viruses called bacteriophages can infect and set in motion a genetic takeover of bacteria, such as Escherichia coli Viruses lead a kind of borrowed life between
Chapter 7 Conclusions
 VII-1 Chapter 7 Conclusions VII-2 The development of cell-based therapies ranging from well-established practices such as bone marrow transplant to next-generation strategies such as adoptive T-cell therapy
VII-1 Chapter 7 Conclusions VII-2 The development of cell-based therapies ranging from well-established practices such as bone marrow transplant to next-generation strategies such as adoptive T-cell therapy
PROTEIN SYNTHESIS. It is known today that GENES direct the production of the proteins that determine the phonotypical characteristics of organisms.
 PROTEIN SYNTHESIS It is known today that GENES direct the production of the proteins that determine the phonotypical characteristics of organisms.» GENES = a sequence of nucleotides in DNA that performs
PROTEIN SYNTHESIS It is known today that GENES direct the production of the proteins that determine the phonotypical characteristics of organisms.» GENES = a sequence of nucleotides in DNA that performs
Text Mycoplasmas -general structure
 Text Mycoplasmas -general structure The mycoplasmas are the smallest (0.15-0.30μm) and the simplest self replicating prokaryotes. Mycoplasma is a genus of over 100 species of cell wall lacking bacteria.
Text Mycoplasmas -general structure The mycoplasmas are the smallest (0.15-0.30μm) and the simplest self replicating prokaryotes. Mycoplasma is a genus of over 100 species of cell wall lacking bacteria.
BCH Graduate Survey of Biochemistry
 BCH 5045 Graduate Survey of Biochemistry Instructor: Charles Guy Producer: Ron Thomas Director: Glen Graham Lecture 7 Slide sets available at: http://hort.ifas.ufl.edu/teach/guyweb/bch5045/index.html David
BCH 5045 Graduate Survey of Biochemistry Instructor: Charles Guy Producer: Ron Thomas Director: Glen Graham Lecture 7 Slide sets available at: http://hort.ifas.ufl.edu/teach/guyweb/bch5045/index.html David
Post-PKS tailoring steps of the spiramycin. macrolactone ring in Streptomyces ambofaciens
 Post-PKS tailoring steps of the spiramycin macrolactone ring in Streptomyces ambofaciens Hoang-Chuong NGUYEN, Emmanuelle DARBN, Robert THAI, Jean-Luc PERNDET, Sylvie LAUTRU SUPPLEMENTARY DATA 1 Figure
Post-PKS tailoring steps of the spiramycin macrolactone ring in Streptomyces ambofaciens Hoang-Chuong NGUYEN, Emmanuelle DARBN, Robert THAI, Jean-Luc PERNDET, Sylvie LAUTRU SUPPLEMENTARY DATA 1 Figure
Figure S1. Molecular confirmation of the precise insertion of the AsMCRkh2 cargo into the kh w locus.
 Supporting Information Appendix Table S1. Larval and adult phenotypes of G 2 progeny of lines 10.1 and 10.2 G 1 outcrosses to wild-type mosquitoes. Table S2. List of oligonucleotide primers. Table S3.
Supporting Information Appendix Table S1. Larval and adult phenotypes of G 2 progeny of lines 10.1 and 10.2 G 1 outcrosses to wild-type mosquitoes. Table S2. List of oligonucleotide primers. Table S3.
Viral vaccines. Lec. 3 أ.د.فائزة عبد هللا مخلص
 Lec. 3 أ.د.فائزة عبد هللا مخلص Viral vaccines 0bjectives 1-Define active immunity. 2-Describe the methods used for the preparation of attenuated live & killed virus vaccines. 3- Comparison of Characteristics
Lec. 3 أ.د.فائزة عبد هللا مخلص Viral vaccines 0bjectives 1-Define active immunity. 2-Describe the methods used for the preparation of attenuated live & killed virus vaccines. 3- Comparison of Characteristics
INFECTION AND IMMUNITY, Sept. 1999, p Vol. 67, No. 9. Copyright 1999, American Society for Microbiology. All Rights Reserved.
 INFECTION AND IMMUNITY, Sept. 1999, p. 4557 4562 Vol. 67, No. 9 0019-9567/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Identification of a New Variable Sequence in
INFECTION AND IMMUNITY, Sept. 1999, p. 4557 4562 Vol. 67, No. 9 0019-9567/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Identification of a New Variable Sequence in
18.2 Viruses and Prions
 KEY CONCEPT Infections can be caused in several ways. Viruses, bacteria, viroids, and prions can all cause infection. Any disease-causing agent is called a pathogen. 1 nanometer (nm) = one billionth of
KEY CONCEPT Infections can be caused in several ways. Viruses, bacteria, viroids, and prions can all cause infection. Any disease-causing agent is called a pathogen. 1 nanometer (nm) = one billionth of
L I F E S C I E N C E S
 1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 NOVEMBER 2, 2006
1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 NOVEMBER 2, 2006
Supplementary Information. Supplementary Figure 1
 Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Materials and Methods , The two-hybrid principle.
 The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
SelenoMet Dream TM Media Kit (MD12-506)
 SelenoMet Dream TM Media Kit (MD12-506) For the Overnight Expression of Recombinant Proteins About the Kit: MD12-506 contains: (*each packet contains enough to make up 1L of media) Description For the
SelenoMet Dream TM Media Kit (MD12-506) For the Overnight Expression of Recombinant Proteins About the Kit: MD12-506 contains: (*each packet contains enough to make up 1L of media) Description For the
Complete Student Notes for BIOL2202
 Complete Student Notes for BIOL2202 Revisiting Translation & the Genetic Code Overview How trna molecules interpret a degenerate genetic code and select the correct amino acid trna structure: modified
Complete Student Notes for BIOL2202 Revisiting Translation & the Genetic Code Overview How trna molecules interpret a degenerate genetic code and select the correct amino acid trna structure: modified
Life Sciences 1A Midterm Exam 2. November 13, 2006
 Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Review of Biochemistry
 Review of Biochemistry Chemical bond Functional Groups Amino Acid Protein Structure and Function Proteins are polymers of amino acids. Each amino acids in a protein contains a amino group, - NH 2,
Review of Biochemistry Chemical bond Functional Groups Amino Acid Protein Structure and Function Proteins are polymers of amino acids. Each amino acids in a protein contains a amino group, - NH 2,
Crl and RpoS May Not Be Involved in Cross Protection against Tetracycline in Escherichia coli in Response to Heat Stress
 rl and RpoS May Not Be Involved in ross Protection against Tetracycline in Escherichia coli in Response to Heat Stress Irina han, hi Wing heng, Jasmine hin, and Veronica how Department of Microbiology
rl and RpoS May Not Be Involved in ross Protection against Tetracycline in Escherichia coli in Response to Heat Stress Irina han, hi Wing heng, Jasmine hin, and Veronica how Department of Microbiology
Polyomaviridae. Spring
 Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
CELLS. Cells. Basic unit of life (except virus)
 Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Supplemental Information
 Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
Microsart Calibration Reagent
 Instructions for Use Microsart Calibration Reagent Prod. No. SMB95-2021 Mycoplasma arginini Prod. No. SMB95-2022 Mycoplasma orale Prod. No. SMB95-2023 Mycoplasma gallisepticum Prod. No. SMB95-2024 Mycoplasma
Instructions for Use Microsart Calibration Reagent Prod. No. SMB95-2021 Mycoplasma arginini Prod. No. SMB95-2022 Mycoplasma orale Prod. No. SMB95-2023 Mycoplasma gallisepticum Prod. No. SMB95-2024 Mycoplasma
Problem-solving Test: The Mechanism of Protein Synthesis
 Q 2009 by The International Union of Biochemistry and Molecular Biology BIOCHEMISTRY AND MOLECULAR BIOLOGY EDUCATION Vol. 37, No. 1, pp. 58 62, 2009 Problem-based Learning Problem-solving Test: The Mechanism
Q 2009 by The International Union of Biochemistry and Molecular Biology BIOCHEMISTRY AND MOLECULAR BIOLOGY EDUCATION Vol. 37, No. 1, pp. 58 62, 2009 Problem-based Learning Problem-solving Test: The Mechanism
PhysicsAndMathsTutor.com. Question Number. 1. prevents viruses attaching to {uninfected / eq} host cells / eq ; 2. by binding to receptors / eq ;
 1(a) 1. prevents viruses attaching to {uninfected / eq} host cells / eq ; 2. by binding to receptors / eq ; 3. (therefore) preventing virus from entering cell / eq ; 4. (therefore) viruses cannot replicate
1(a) 1. prevents viruses attaching to {uninfected / eq} host cells / eq ; 2. by binding to receptors / eq ; 3. (therefore) preventing virus from entering cell / eq ; 4. (therefore) viruses cannot replicate
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10913 Supplementary Figure 1 2F o -F c electron density maps of cognate and near-cognate trna Leu 2 in the A site of the 70S ribosome. The maps are contoured at 1.2 sigma and some of
doi:10.1038/nature10913 Supplementary Figure 1 2F o -F c electron density maps of cognate and near-cognate trna Leu 2 in the A site of the 70S ribosome. The maps are contoured at 1.2 sigma and some of
Antibiotics for Research
 Antibiotics for Research Natural antibiotics have existed for centuries prior to scientists identifying and isolating active moieties responsible for antibacterial activity. Today antibiotics are widely
Antibiotics for Research Natural antibiotics have existed for centuries prior to scientists identifying and isolating active moieties responsible for antibacterial activity. Today antibiotics are widely
Section 1 Proteins and Proteomics
 Section 1 Proteins and Proteomics Learning Objectives At the end of this assignment, you should be able to: 1. Draw the chemical structure of an amino acid and small peptide. 2. Describe the difference
Section 1 Proteins and Proteomics Learning Objectives At the end of this assignment, you should be able to: 1. Draw the chemical structure of an amino acid and small peptide. 2. Describe the difference
Affinity of Doripenem and Comparators to Penicillin-Binding Proteins in Escherichia coli and ACCEPTED
 AAC Accepts, published online ahead of print on February 00 Antimicrob. Agents Chemother. doi:./aac.01-0 Copyright 00, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights
AAC Accepts, published online ahead of print on February 00 Antimicrob. Agents Chemother. doi:./aac.01-0 Copyright 00, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights
A.Kavitha Assistant professor Department of Botany RBVRR Womens college
 A.Kavitha Assistant professor Department of Botany RBVRR Womens college The Ultrastructure Of A Typical Bacterial Cell The Bacterial Cell This is a diagram of a typical bacterial cell, displaying all of
A.Kavitha Assistant professor Department of Botany RBVRR Womens college The Ultrastructure Of A Typical Bacterial Cell The Bacterial Cell This is a diagram of a typical bacterial cell, displaying all of
numbe r Done by Corrected by Doctor
 numbe r 5 Done by Mustafa Khader Corrected by Mahdi Sharawi Doctor Ashraf Khasawneh Viral Replication Mechanisms: (Protein Synthesis) 1. Monocistronic Method: All human cells practice the monocistronic
numbe r 5 Done by Mustafa Khader Corrected by Mahdi Sharawi Doctor Ashraf Khasawneh Viral Replication Mechanisms: (Protein Synthesis) 1. Monocistronic Method: All human cells practice the monocistronic
Central Dogma. Central Dogma. Translation (mrna -> protein)
 Central Dogma Central Dogma Translation (mrna -> protein) mrna code for amino acids 1. Codons as Triplet code 2. Redundancy 3. Open reading frames 4. Start and stop codons 5. Mistakes in translation 6.
Central Dogma Central Dogma Translation (mrna -> protein) mrna code for amino acids 1. Codons as Triplet code 2. Redundancy 3. Open reading frames 4. Start and stop codons 5. Mistakes in translation 6.
RNA Processing in Eukaryotes *
 OpenStax-CNX module: m44532 1 RNA Processing in Eukaryotes * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you
OpenStax-CNX module: m44532 1 RNA Processing in Eukaryotes * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you
ASSESMENT OF CRYOPRESERVATION SYSTEMS INFLUENCE ON THE SURVAVIAL OF E. COLI RECOMBINANT STRAINS
 Lucrări ştiinńifice Zootehnie şi Biotehnologii, vol. 41(1) (2008), Timişoara ASSESMENT OF CRYOPRESERVATION SYSTEMS INFLUENCE ON THE SURVAVIAL OF E. COLI RECOMBINANT STRAINS TESTAREA INFLUENłEI SISTEMELOR
Lucrări ştiinńifice Zootehnie şi Biotehnologii, vol. 41(1) (2008), Timişoara ASSESMENT OF CRYOPRESERVATION SYSTEMS INFLUENCE ON THE SURVAVIAL OF E. COLI RECOMBINANT STRAINS TESTAREA INFLUENłEI SISTEMELOR
Aspirin antagonism in isonizaid treatment of tuberculosis in mice ACCEPTED. Department of Molecular Microbiology & Immunology, Bloomberg School of
 AAC Accepts, published online ahead of print on 4 December 2006 Antimicrob. Agents Chemother. doi:10.1128/aac.01145-06 Copyright 2006, American Society for Microbiology and/or the Listed Authors/Institutions.
AAC Accepts, published online ahead of print on 4 December 2006 Antimicrob. Agents Chemother. doi:10.1128/aac.01145-06 Copyright 2006, American Society for Microbiology and/or the Listed Authors/Institutions.
* these authors contributed equally to the preparation of this report
 AAC Accepts, published online ahead of print on June 00 Antimicrob. Agents Chemother. doi:0./aac.000-0 Copyright 00, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights
AAC Accepts, published online ahead of print on June 00 Antimicrob. Agents Chemother. doi:0./aac.000-0 Copyright 00, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights
Auxin-Inducible Degron (AID) System Total Set
 Catalog No. BRS-APC011A Auxin-Inducible Degron (AID) System Total Set This AID System Charactoristics Principle of Auxin-Inducible Degron (AID ) System AID plasmid protein 1. Transfection of AID plasmid
Catalog No. BRS-APC011A Auxin-Inducible Degron (AID) System Total Set This AID System Charactoristics Principle of Auxin-Inducible Degron (AID ) System AID plasmid protein 1. Transfection of AID plasmid
Genome-wide association studies for understanding pathogen evolution. Samuel K. Sheppard
 Genome-wide association studies for understanding pathogen evolution Samuel K. Sheppard Pathogenic bacteria Population genomics and evolution Sheppard et al. Science 2008; 11: 237-239. Sheppard et al.
Genome-wide association studies for understanding pathogen evolution Samuel K. Sheppard Pathogenic bacteria Population genomics and evolution Sheppard et al. Science 2008; 11: 237-239. Sheppard et al.
Chemistry 107 Exam 4 Study Guide
 Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
ACCEPTED. Proteins P24 and P41 Function in the Regulation of Terminal Organelle. Benjamin M. Hasselbring and Duncan C. Krause*
 JB Accepts, published online ahead of print on 10 August 2007 J. Bacteriol. doi:10.1128/jb.00867-07 Copyright 2007, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights
JB Accepts, published online ahead of print on 10 August 2007 J. Bacteriol. doi:10.1128/jb.00867-07 Copyright 2007, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights
Gene expression scaled by distance to the genome replication site. Ying et al. Supporting Information. Contents. Supplementary note I p.
 Gene expression scaled by distance to the genome replication site Ying et al Supporting Information Contents Supplementary note I p. 2 Supplementary note II p. 3-5 Supplementary note III p. 5-7 Supplementary
Gene expression scaled by distance to the genome replication site Ying et al Supporting Information Contents Supplementary note I p. 2 Supplementary note II p. 3-5 Supplementary note III p. 5-7 Supplementary
7.012 Quiz 3 Answers
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
RNA and Protein Synthesis Guided Notes
 RNA and Protein Synthesis Guided Notes is responsible for controlling the production of in the cell, which is essential to life! o DNARNAProteins contain several thousand, each with directions to make
RNA and Protein Synthesis Guided Notes is responsible for controlling the production of in the cell, which is essential to life! o DNARNAProteins contain several thousand, each with directions to make
Effect of Lincomycin and Clindamycin on Peptide
 ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, Jan. 1975, p. 32-37 Copyright 0 1975 American Society for Microbiology Vol. 7, No. 1 Printed in U.S.A. Effect of Lincomycin and Clindamycin on Peptide Chain Initiation
ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, Jan. 1975, p. 32-37 Copyright 0 1975 American Society for Microbiology Vol. 7, No. 1 Printed in U.S.A. Effect of Lincomycin and Clindamycin on Peptide Chain Initiation
CHAPTER 4 RESULTS. showed that all three replicates had similar growth trends (Figure 4.1) (p<0.05; p=0.0000)
 CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
October 26, Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is worth 2 points.
 MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
HUMAN BIOLOGY (SPECIFICATION A) Unit 9 (Written Synoptic)
 Surname Other Names Leave blank Centre Number Candidate Number Candidate Signature General Certificate of Education June 2003 Advanced Level Examination HUMAN BIOLOGY (SPECIFICATION A) Unit 9 (Written
Surname Other Names Leave blank Centre Number Candidate Number Candidate Signature General Certificate of Education June 2003 Advanced Level Examination HUMAN BIOLOGY (SPECIFICATION A) Unit 9 (Written
Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University
 Role of Chemical lexposure in Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University CNV Discovery Reference Genetic
Role of Chemical lexposure in Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University CNV Discovery Reference Genetic
Molecular Biology (BIOL 4320) Exam #2 April 22, 2002
 Molecular Biology (BIOL 4320) Exam #2 April 22, 2002 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Molecular Biology (BIOL 4320) Exam #2 April 22, 2002 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
HEREDITY SAMPLE TOURNAMENT
 HEREDITY SAMPLE TOURNAMENT PART 1 - BACKGROUND: 1. Heterozygous means. A. Information about heritable traits B. Unique/ different molecular forms of a gene that are possible at a given locus C. Having
HEREDITY SAMPLE TOURNAMENT PART 1 - BACKGROUND: 1. Heterozygous means. A. Information about heritable traits B. Unique/ different molecular forms of a gene that are possible at a given locus C. Having
BIOLOGY 111. CHAPTER 3: The Cell: The Fundamental Unit of Life
 BIOLOGY 111 CHAPTER 3: The Cell: The Fundamental Unit of Life The Cell: The Fundamental Unit of Life Learning Outcomes 3.1 Explain the similarities and differences between prokaryotic and eukaryotic cells
BIOLOGY 111 CHAPTER 3: The Cell: The Fundamental Unit of Life The Cell: The Fundamental Unit of Life Learning Outcomes 3.1 Explain the similarities and differences between prokaryotic and eukaryotic cells
