MG132. GRXS17:V5-His MG132 GRXS17:HA. RuBisCo. Supplemental Data. Walton et al. (2015). Plant Cell /tpc
|
|
|
- Bruno Ray
- 5 years ago
- Views:
Transcription
1 MG132 GRXS17:V5-His MG132 GRXS17:HA RuBisCo Supplemental Figure 1. Degradation of GRXS17 by the 26S proteasome. (A) Cell-free degradation assay. Recombinant protein GRXS17:V5-HIS was incubated with total protein extract prepared from wild-type Arabidopsis seedlings in the presence or absence of MG132 for the indicated time. (B) Total proteins extracted from 35S:GRXS17:HA seedlings and incubated in the presence or absence of MG132. Samples were harvested at the indicated times. Coomassie Blue-stained ribulose-1,5- bisphosphate carboxylase/oxygenase (RuBisCo) was used as loading control. Proteins were detected with anti-v5 (1/2000) or anti-ha (1/1000) antibodies with their respective horseradish peroxidase (HRP)- conjugated secondary antibody.
2 Supplemental Figure 2. The Ubiquitination Site website page. In a query box, one or multiple AT numbers can be entered, returning the ubiquitination sites on corresponding proteins and information concerning the splice variant, modified sequence, sequence window, and the site position within the protein. The MAXQUANT score and delta score are provided as well. Through its link to PLAZA, access can be gained to ample information concerning the protein(s) in question. Digestion - LysC Tryp Acetylation - A - A - A Supplemental Figure 3. Acetylation efficiency testing. Silver-stained SDS-PAGE displaying the resistance of acetylated proteins to cleavage by endolysc. The used proteolytic enzyme [none (-), endolysc (LysC), or trypsin (Tryp)] and acetylation status [mock (-) or acetylated (A)] are indicated above the panels.
3 Supplemental Table 1. Identified ubiquitination sites on transcription factors and UPS members. Type Entry AGI code Description Modified peptide sequence Transcription factors bzip MYB WRKY bhlh TCP AT1G43700 Transcription factor VIP1 (Basic leucine zipper 51) (AtbZIP51) AT3G17609 Transcription factor HY5-like protein HYH (bzip transcription factor AtbZip64) AT5G11260 Transcription factor HY5 (Protein LONG HYPOCOTYL 5) AT1G72650 Myb family transcription factor TRFL6 AT2G04880 WRKY transcription factor 1 (Transcription factor ZAP1) AT4G26640 WRKY20 AT4G22070 WRKY31 AT4G30935 WRKY32 AT2G38470 WRKY33 AT3G01970 WRKY45 AT5G49520 WRKY48 AT4G36930 Transcription factor SPATULA (SPT) (Basic helix-loop-helix protein 24) AT4G02590 Transcription factor UNE12 (Basic helix-loophelix protein 59, bhlh 59) AT1G58100 TCP8 AT1G72010 TCP22 Linked to hormones ABA AT1G45249 Abscisic acid responsive elements-binding factor 2 ABF2 AT3G56850 ABSCISIC ACID-INSENSITIVE 5-like protein 2 DPBF3 (ABA-responsive element-binding protein 3) Auxin AT1G19220 Auxin response factor 19 ARF19 (Auxinresponsive protein IAA22) AT5G62000 Auxin response factor 2 ARF2 (ARF1-binding protein, ARF1-BP) Ethylene AT5G13330 Ethylene-responsive transcription factor ERF113 Brassinosteroids AT5G07310 Ethylene-responsive transcription factor ERF115 _GTSELNTENK(Ubi)HLKMR LAELALLDPK(Ubi)R KK(Ubi)VYVSDLESR VPEFGGEAVGK(Ubi)ETSGR TPAEK(Ubi)ENK(Ubi)R DSVEK(Ubi)SASR LSEK(Ubi)SEVR E(Ubi)VMEDGYNWR KK(Ubi)GGNIELSPVER SDVFTAVSKEK(Ubi)TSGSSVQTLR KYGQK(Ubi)VVR NSSQDHLLAQESK(Ubi)AEGR DEEK(Ubi)SLGADMEDLHDETVR ETLGK(Ubi)DQVQGVR K(Ubi)TSFSPR KYGQKQVK(Ubi)GSENPR LTEFHGVDNSAQPTTSSEEK(Ubi)PR SDTINTQTNEENK(Ubi)K AAEVHNLSEK(Ubi)R ALQELVPTVNK(Ubi)TDR STPPEDSTLATTSSTATATTTK(Ubi)R SVDLSK(Ubi)ENDDR EDYFK(ac)EPSSAAEPSESSQK(Ubi)ASQFQEQELAQGR KSGTVEK(Ubi)VVER VASGEVVEK(Ubi)TVER TYTKVQK(Ubi)R ASSEVSMK(Ubi)GNR DPKK(Ubi)AAR EEEEK(Ubi)NYGYNYYNYPR ANSGNYGK(Ubi)R_
4 Ubiquitin conjugation E1 AT5G06460 Ubiquitin-activating enzyme E1 2 UBA2 E2 AT5G50870 Ubiquitin-conjugating enzyme E2 27 UBC27 (Ubiquitin carrier protein 27) AT1G70660 Ubiquitin-conjugating enzyme E2 variant 1B UEV1B (Ubc enzyme variant 1B) AT2G36060 Ubiquitin-conjugating enzyme E2 variant 1C UEV1C (Ubc enzyme variant 1C) E3 UBOX ASK F-box 26S AT1G01680 U-box domain-containing protein 54 PUB54 AT5G05230 U-box domain-containing protein 62 PUB62 AT2G45950 SKP1-like protein 20 ASK20 AT3G61415 SKP1-like protein 21 ASK21 AT1G20140 SKP1-like protein 4 ASK4 _LEDVNSK(Ubi)LLR VCPK(Ubi)SDNLTR GSEEEK(Ubi)VVVPR GEK(Ubi)GIGDGTVSYGMDDGDDIYMR K(Ubi)LVQPPEGTFF KETIEK(Ubi)SKSNESDEDPR KETIEK(Ubi)SKSNESDEDPR VGEQDPK(Ubi)TR IIEGK(Ubi)NPEEIR LK(Ubi)NVEVEEHVDER IIEGK(Ubi)TPEEIR GK(Ubi)TPEQMR_ AT2G16365 F-box protein _K(Ubi)NESSAETNTLEMDR LQSLESSK(Ubi)DTQEDGPR_ At1g47765 Putative F-box protein _FELK(Ubi)EIADDQAR_ AT4G39756 Putative F-box/kelch-repeat protein _DIK(Ubi)GLATLNR_ AT5G S protease regulatory subunit 10B homolog A RPT4A AT3G S protease regulatory subunit 6A homolog A RPT5A AT1G S protease regulatory subunit 7 homolog A RPT1A AT5G S proteasome non-atpase regulatory subunit 12 homolog B RPN5B AT4G S proteasome regulatory subunit 4 homolog A RPT2A Deubiquitinating enzymes AT3G11910 Ubiquitin carboxyl-terminal hydrolase 13 UBP13 AT1G51710 Ubiquitin carboxyl-terminal hydrolase 6 UBP6 AT4G17510 Ubiquitin carboxyl-terminal hydrolase UCH3 _K(Ubi)IEIPLPNEQSR SKVDKEK(Ubi)LTSGTR K(Ubi)IEFPHPTEEAR DIEDEIRDEK(Ubi)NPR LLNEEK(Ubi)QMR LKPQEEK(Ubi)AEEDR AEEIPEEEK(Ubi)NIGPNDR KK(Ubi)LEAPR ATASESSSSK(Ubi)R_
5
Amino acids. Side chain. -Carbon atom. Carboxyl group. Amino group
 PROTEINS Amino acids Side chain -Carbon atom Amino group Carboxyl group Amino acids Primary structure Amino acid monomers Peptide bond Peptide bond Amino group Carboxyl group Peptide bond N-terminal (
PROTEINS Amino acids Side chain -Carbon atom Amino group Carboxyl group Amino acids Primary structure Amino acid monomers Peptide bond Peptide bond Amino group Carboxyl group Peptide bond N-terminal (
Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis
 Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
Molecular Graphics Perspective of Protein Structure and Function
 Molecular Graphics Perspective of Protein Structure and Function VMD Highlights > 20,000 registered Users Platforms: Unix (16 builds) Windows MacOS X Display of large biomolecules and simulation trajectories
Molecular Graphics Perspective of Protein Structure and Function VMD Highlights > 20,000 registered Users Platforms: Unix (16 builds) Windows MacOS X Display of large biomolecules and simulation trajectories
WT siz1-2 siz1-3. WT siz1-2 siz1-3. -Pi, 0.05 M IAA. -Pi, 2.5 M NPA
 A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
Cleaning house is one way plants respond to inside and outside changes
 Cleaning house is one way plants respond to inside and outside changes Thank you! ADVANCE Nominators Colleagues Students Judy Callis, Department of MCB, UC-Davis Improving plant productivity impacts: Plants
Cleaning house is one way plants respond to inside and outside changes Thank you! ADVANCE Nominators Colleagues Students Judy Callis, Department of MCB, UC-Davis Improving plant productivity impacts: Plants
Proteasomes. When Death Comes a Knock n. Warren Gallagher Chem412, Spring 2001
 Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
Supplemental Table S1 Up-regulated genes in irjazh plants compared to WT plants by microarray.
 Supplemental Table S1 Up-regulated genes in irjazh plants compared to WT plants by microarray. Classification Fold Change Bincode (TAIR) Annotation Probe number ACC number Cell wall. Degradation 3.29 10.6.2
Supplemental Table S1 Up-regulated genes in irjazh plants compared to WT plants by microarray. Classification Fold Change Bincode (TAIR) Annotation Probe number ACC number Cell wall. Degradation 3.29 10.6.2
Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes
 Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes The major site of acetoacetate and 3-hydorxybutyrate production is in the liver. 3-hydorxybutyrate is the
Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes The major site of acetoacetate and 3-hydorxybutyrate production is in the liver. 3-hydorxybutyrate is the
Supplemental Data. Pick and Bräutgam et al. Plant Cell. (2011) /tpc Mean Adjustment FOM values (± SD) vs. Number of Clusters
 Mean Adjustment FOM values (± SD) vs. Number of Clusters Mean Adjustment FOM Number of Clusters Supplemental Figure 1. Figure of merit analysis of metabolite clustering. The algorithm tries 20 times to
Mean Adjustment FOM values (± SD) vs. Number of Clusters Mean Adjustment FOM Number of Clusters Supplemental Figure 1. Figure of merit analysis of metabolite clustering. The algorithm tries 20 times to
Supplementary Figures
 Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
There are approximately 30,000 proteasomes in a typical human cell Each proteasome is approximately 700 kda in size The proteasome is made up of 3
 Proteasomes Proteasomes Proteasomes are responsible for degrading proteins that have been damaged, assembled improperly, or that are of no profitable use to the cell. The unwanted protein is literally
Proteasomes Proteasomes Proteasomes are responsible for degrading proteins that have been damaged, assembled improperly, or that are of no profitable use to the cell. The unwanted protein is literally
REGULATORY MECHANISMS OF TRANSCRIPTION FACTOR FUNCTION
 Transcription Regulation And Gene Expression in Eukaryotes Cycle G2 (lecture 13709) RG Clerc 04.04.2012 REGULATORY MECHANISMS OF TRANSCRIPTION FACTOR FUNCTION Protein synthesized Protein phosphorylated
Transcription Regulation And Gene Expression in Eukaryotes Cycle G2 (lecture 13709) RG Clerc 04.04.2012 REGULATORY MECHANISMS OF TRANSCRIPTION FACTOR FUNCTION Protein synthesized Protein phosphorylated
The N-end rule pathway regulates pathogen responses. in plants
 SUPPLEMENTARY INFORMATION The N-end rule pathway regulates pathogen responses in plants Rémi de Marchi 1,2, Maud Sorel 1, Brian Mooney 1, Isabelle Fudal 3, Kevin Goslin 1, Kamila Kwaśniewska 4, Patrick
SUPPLEMENTARY INFORMATION The N-end rule pathway regulates pathogen responses in plants Rémi de Marchi 1,2, Maud Sorel 1, Brian Mooney 1, Isabelle Fudal 3, Kevin Goslin 1, Kamila Kwaśniewska 4, Patrick
Protein methylation CH 3
 Protein methylation CH 3 methionine S-adenosylmethionine (SAM or adomet) Methyl group of the methionine is activated by the + charge of the adjacent sulfur atom. SAM S-adenosylhomocysteine homocysteine
Protein methylation CH 3 methionine S-adenosylmethionine (SAM or adomet) Methyl group of the methionine is activated by the + charge of the adjacent sulfur atom. SAM S-adenosylhomocysteine homocysteine
NABIL KILLINY ASSISTANT PROFESSOR PLANT PATHOLOGY UNIVERSITY OF FLORIDA. Citrus responses to huanglongbing
 NABIL KILLINY ASSISTANT PROFESSOR PLANT PATHOLOGY UNIVERSITY OF FLORIDA Citrus responses to huanglongbing HLB is Vector-borne Disease! Vector psyllids Pathogen Ca. Liberibacter asiaticus. Host Citrus spp.
NABIL KILLINY ASSISTANT PROFESSOR PLANT PATHOLOGY UNIVERSITY OF FLORIDA Citrus responses to huanglongbing HLB is Vector-borne Disease! Vector psyllids Pathogen Ca. Liberibacter asiaticus. Host Citrus spp.
Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene
 Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene YUELIN ZHANG, WEIHUA FAN, MARK KINKEMA, XIN LI, AND
Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene YUELIN ZHANG, WEIHUA FAN, MARK KINKEMA, XIN LI, AND
Cellular functions of protein degradation
 Protein Degradation Cellular functions of protein degradation 1. Elimination of misfolded and damaged proteins: Environmental toxins, translation errors and genetic mutations can damage proteins. Misfolded
Protein Degradation Cellular functions of protein degradation 1. Elimination of misfolded and damaged proteins: Environmental toxins, translation errors and genetic mutations can damage proteins. Misfolded
Ub-CHOP-Reporter Deubiquitination Assay Kit Catalog Number PR1001
 - 1 - MANUAL Ub-CHOP-Reporter Deubiquitination Assay Kit Catalog Number PR1001 all products are for research use only not intended for human or animal diagnostic or therapeutic uses LifeSensors, Inc.,
- 1 - MANUAL Ub-CHOP-Reporter Deubiquitination Assay Kit Catalog Number PR1001 all products are for research use only not intended for human or animal diagnostic or therapeutic uses LifeSensors, Inc.,
Functional mechanisms of drought tolerance in maize
 Functional mechanisms of drought tolerance in maize Nepolean Thirunavukkarasu Division of Genetics Indian Agricultural Research Institute New Delhi-110012 tnepolean@iari.resi.in tnepolean@gmail.com Mt
Functional mechanisms of drought tolerance in maize Nepolean Thirunavukkarasu Division of Genetics Indian Agricultural Research Institute New Delhi-110012 tnepolean@iari.resi.in tnepolean@gmail.com Mt
Biochemistry 2 Recita0on Amino Acid Metabolism
 Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
REGULATION OF ENZYME ACTIVITY. Medical Biochemistry, Lecture 25
 REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
Transcriptomic changes in Cucurbita pepo fruit after cold storage: differential response between two cultivars contrasting in chilling sensitivity
 Carvajal et al. BMC Genomics (2018) 19:125 https://doi.org/10.1186/s12864-018-4500-9 RESEARCH ARTICLE Open Access Transcriptomic changes in Cucurbita pepo fruit after cold storage: differential response
Carvajal et al. BMC Genomics (2018) 19:125 https://doi.org/10.1186/s12864-018-4500-9 RESEARCH ARTICLE Open Access Transcriptomic changes in Cucurbita pepo fruit after cold storage: differential response
PHRM 836 September 1, 2015
 PRM 836 September 1, 2015 Protein structure- function relationship: Catalysis example of serine proteases Devlin, section 9.3 Physiological processes requiring serine proteases Control of enzymatic activity
PRM 836 September 1, 2015 Protein structure- function relationship: Catalysis example of serine proteases Devlin, section 9.3 Physiological processes requiring serine proteases Control of enzymatic activity
Characterization of mirnas in Response to Short-Term Waterlogging in Three Inbred Lines of Zea mays
 Characterization of mirnas in Response to Short-Term Waterlogging in Three Inbred Lines of Zea mays Zhijie Liu 1,2, Sunita Kumari 2, Lifang Zhang 2, Yonglian Zheng 1 *, Doreen Ware 2,3 * 1 National Key
Characterization of mirnas in Response to Short-Term Waterlogging in Three Inbred Lines of Zea mays Zhijie Liu 1,2, Sunita Kumari 2, Lifang Zhang 2, Yonglian Zheng 1 *, Doreen Ware 2,3 * 1 National Key
Post-translational modifications of proteins in gene regulation under hypoxic conditions
 203 Review Article Post-translational modifications of proteins in gene regulation under hypoxic conditions 1, 2) Olga S. Safronova 1) Department of Cellular Physiological Chemistry, Tokyo Medical and
203 Review Article Post-translational modifications of proteins in gene regulation under hypoxic conditions 1, 2) Olga S. Safronova 1) Department of Cellular Physiological Chemistry, Tokyo Medical and
ab SREBP-2 Translocation Assay Kit (Cell-Based)
 ab133114 SREBP-2 Translocation Assay Kit (Cell-Based) Instructions for Use For analysis of translocation of SREBP-2 into nuclei. This product is for research use only and is not intended for diagnostic
ab133114 SREBP-2 Translocation Assay Kit (Cell-Based) Instructions for Use For analysis of translocation of SREBP-2 into nuclei. This product is for research use only and is not intended for diagnostic
Chemical Biology, Option II Mechanism Based Proteomic Tagging Case History CH1
 Proteome Wide Screening of Serine Protease Activity Proc Natl Acad Sci 1999, 97, 14694; Proteomics 2001, 1, 1067; Proc Natl Acad Sci 2002, 99, 10335; Biochemistry 2001, 40, 4005; J. Am. Chem. Soc., 2005,
Proteome Wide Screening of Serine Protease Activity Proc Natl Acad Sci 1999, 97, 14694; Proteomics 2001, 1, 1067; Proc Natl Acad Sci 2002, 99, 10335; Biochemistry 2001, 40, 4005; J. Am. Chem. Soc., 2005,
SUPPLEMENTARY INFORMATION
 a c e doi:10.1038/nature10407 b d f Supplementary Figure 1. SERCA2a complex analysis. (a) Two-dimensional SDS-PAGE gels of SERCA2a complexes. A silver-stained SDSPAGE gel is shown, which reveals a 12 kda
a c e doi:10.1038/nature10407 b d f Supplementary Figure 1. SERCA2a complex analysis. (a) Two-dimensional SDS-PAGE gels of SERCA2a complexes. A silver-stained SDSPAGE gel is shown, which reveals a 12 kda
Molecular Cell Biology - Problem Drill 10: Gene Expression in Eukaryotes
 Molecular Cell Biology - Problem Drill 10: Gene Expression in Eukaryotes Question No. 1 of 10 1. Which of the following statements about gene expression control in eukaryotes is correct? Question #1 (A)
Molecular Cell Biology - Problem Drill 10: Gene Expression in Eukaryotes Question No. 1 of 10 1. Which of the following statements about gene expression control in eukaryotes is correct? Question #1 (A)
Supplemental Information. Spatial Auxin Signaling. Controls Leaf Flattening in Arabidopsis
 Current Biology, Volume 27 Supplemental Information Spatial Auxin Signaling Controls Leaf Flattening in Arabidopsis Chunmei Guan, Binbin Wu, Ting Yu, Qingqing Wang, Naden T. Krogan, Xigang Liu, and Yuling
Current Biology, Volume 27 Supplemental Information Spatial Auxin Signaling Controls Leaf Flattening in Arabidopsis Chunmei Guan, Binbin Wu, Ting Yu, Qingqing Wang, Naden T. Krogan, Xigang Liu, and Yuling
Plant Signal Transduction
 Plant Signal Transduction EDITED BY Dierk Scheel Institute of Plant Biochemistry, Halle, Germany and Claus Wasternack Institute of Plant Biochemistry, Halle, Germany OXTORD UNIVERSITY PRESS Contents List
Plant Signal Transduction EDITED BY Dierk Scheel Institute of Plant Biochemistry, Halle, Germany and Claus Wasternack Institute of Plant Biochemistry, Halle, Germany OXTORD UNIVERSITY PRESS Contents List
Supporting Information
 Supporting Information Supplemental Figure Legends Supplementary Figure 1. Experimental Design. A. To benchmark the performance of software parameter sets proteins were extracted from a pool of Arabidopsis
Supporting Information Supplemental Figure Legends Supplementary Figure 1. Experimental Design. A. To benchmark the performance of software parameter sets proteins were extracted from a pool of Arabidopsis
Short-Term and Continuing Stresses Differentially Interplay with Multiple Hormones to Regulate Plant Survival and Growth
 Molecular Plant Volume 7 Number 5 Pages 841 855 May 2014 RESEARCH ARTICLE Short-Term and Continuing Stresses Differentially Interplay with Multiple Hormones to Regulate Plant Survival and Growth Cangjing
Molecular Plant Volume 7 Number 5 Pages 841 855 May 2014 RESEARCH ARTICLE Short-Term and Continuing Stresses Differentially Interplay with Multiple Hormones to Regulate Plant Survival and Growth Cangjing
The Nobel Prize in Chemistry 2004
 The Nobel Prize in Chemistry 2004 Ubiquitous Quality Control of Life C S Karigar and K R Siddalinga Murthy The Nobel Prize in Chemistry for 2004 is shared by Aaron Ciechanover, Avram Hershko and Irwin
The Nobel Prize in Chemistry 2004 Ubiquitous Quality Control of Life C S Karigar and K R Siddalinga Murthy The Nobel Prize in Chemistry for 2004 is shared by Aaron Ciechanover, Avram Hershko and Irwin
Front Plant Sci Mar 14
 Case Study Front Plant Sci. 2016 Mar 14 30 33 Study Background Cryptochromes (CRY) are blue-light photoreceptors that mediate various light responses in plants and animals. It is known that CRY1 and CRY2
Case Study Front Plant Sci. 2016 Mar 14 30 33 Study Background Cryptochromes (CRY) are blue-light photoreceptors that mediate various light responses in plants and animals. It is known that CRY1 and CRY2
Comparative Transcriptome Profiling of Two Tomato Genotypes in Response to Potassium-Deficiency Stress
 Article Comparative Transcriptome Profiling of Two Tomato Genotypes in Response to Potassium-Deficiency Stress Xiaoming Zhao 1,2, Yang Liu 1, Xin Liu 1, * and Jing Jiang 1, * 1 The Key Laboratory of Protected
Article Comparative Transcriptome Profiling of Two Tomato Genotypes in Response to Potassium-Deficiency Stress Xiaoming Zhao 1,2, Yang Liu 1, Xin Liu 1, * and Jing Jiang 1, * 1 The Key Laboratory of Protected
Protein Identification and Phosphorylation Site Determination by de novo sequencing using PepFrag TM MALDI-Sequencing kit
 Application Note Tel: +82-54-223-2463 Fax : +82-54-223-2460 http://www.genomine.com venture ldg 306 Pohang techno park Pohang, kyungbuk, Korea(ROK) Protein Identification and Phosphorylation Site Determination
Application Note Tel: +82-54-223-2463 Fax : +82-54-223-2460 http://www.genomine.com venture ldg 306 Pohang techno park Pohang, kyungbuk, Korea(ROK) Protein Identification and Phosphorylation Site Determination
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.10/nature10195 NCBI gene: Tagged Subunit(s: HA-Vpx; FLAG-Cul4 HA-DCAF1 FLAG-Cul4 HA-FLAG-Vpx Mock Vpx (SIVmac 100 (a ; 0.159 (b ; 0.05 DCAF1 DDB1 DDA1 Cul4A 1; 0.024591
SUPPLEMENTARY INFORMATION doi:10.10/nature10195 NCBI gene: Tagged Subunit(s: HA-Vpx; FLAG-Cul4 HA-DCAF1 FLAG-Cul4 HA-FLAG-Vpx Mock Vpx (SIVmac 100 (a ; 0.159 (b ; 0.05 DCAF1 DDB1 DDA1 Cul4A 1; 0.024591
Signal Transduction Cascades
 Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
Arabidopsis Transcriptome Analysis Reveals Key Roles of Melatonin in Plant Defense Systems
 Virginia Commonwealth University VCU Scholars Compass Study of Biological Complexity Publications Center for the Study of Biological Complexity 2014 Arabidopsis Transcriptome Analysis Reveals Key Roles
Virginia Commonwealth University VCU Scholars Compass Study of Biological Complexity Publications Center for the Study of Biological Complexity 2014 Arabidopsis Transcriptome Analysis Reveals Key Roles
Supplements. Figure S1. B Phalloidin Alexa488
 Supplements A, DMSO, PP2, PP3 Crk-myc Figure S1. (A) Src kinase activity is necessary for recruitment of Crk to Nephrin cytoplasmic domain. Human podocytes expressing /7-NephrinCD () were treated with
Supplements A, DMSO, PP2, PP3 Crk-myc Figure S1. (A) Src kinase activity is necessary for recruitment of Crk to Nephrin cytoplasmic domain. Human podocytes expressing /7-NephrinCD () were treated with
Mass Spectrometry and Proteomics - Lecture 4 - Matthias Trost Newcastle University
 Mass Spectrometry and Proteomics - Lecture 4 - Matthias Trost Newcastle University matthias.trost@ncl.ac.uk previously Peptide fragmentation Hybrid instruments 117 The Building Blocks of Life DNA RNA Proteins
Mass Spectrometry and Proteomics - Lecture 4 - Matthias Trost Newcastle University matthias.trost@ncl.ac.uk previously Peptide fragmentation Hybrid instruments 117 The Building Blocks of Life DNA RNA Proteins
Supplementary Tables 1
 Supplementary Tables 1 Supplementary Table 1: The 11 genes in the geneset for Case Study 1 At1g62570 At1g09350 At1g60470 At2g47180 At4g17090 At5g20830 At2g16890 At3g51240 At3g55120 At4g27560 At5g08640
Supplementary Tables 1 Supplementary Table 1: The 11 genes in the geneset for Case Study 1 At1g62570 At1g09350 At1g60470 At2g47180 At4g17090 At5g20830 At2g16890 At3g51240 At3g55120 At4g27560 At5g08640
Enzyme-coupled Receptors. Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors
 Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Regulation of Enzymatic Activity. Lesson 4
 Regulation of Enzymatic Activity Lesson 4 Regulation of Enzymatic Activity no real regulation: - regulation of enzyme expression and turnover - control of enzyme trafficking - supply of cofactors real
Regulation of Enzymatic Activity Lesson 4 Regulation of Enzymatic Activity no real regulation: - regulation of enzyme expression and turnover - control of enzyme trafficking - supply of cofactors real
Table S1. Sequence of human and mouse primers used for RT-qPCR measurements.
 Table S1. Sequence of human and mouse primers used for RT-qPCR measurements. Ca9, carbonic anhydrase IX; Ndrg1, N-myc downstream regulated gene 1; L28, ribosomal protein L28; Hif1a, hypoxia inducible factor
Table S1. Sequence of human and mouse primers used for RT-qPCR measurements. Ca9, carbonic anhydrase IX; Ndrg1, N-myc downstream regulated gene 1; L28, ribosomal protein L28; Hif1a, hypoxia inducible factor
Histone acetyltransferase inhibitors block neuroblastoma cell growth in vivo
 Histone acetyltransferase inhibitors block neuroblastoma cell growth in vivo 1 JM Gajer #, 1,2 SD Furdas #, 3 A Gründer, 3 M Gothwal, 4 U Heinicke, 1 K. Keller, 5,10 F Colland, 4 S Fulda, 3 HL Pahl, 6
Histone acetyltransferase inhibitors block neuroblastoma cell growth in vivo 1 JM Gajer #, 1,2 SD Furdas #, 3 A Gründer, 3 M Gothwal, 4 U Heinicke, 1 K. Keller, 5,10 F Colland, 4 S Fulda, 3 HL Pahl, 6
Chem Lecture 2 Protein Structure
 Chem 452 - Lecture 2 Protein Structure 110923 Proteins are the workhorses of a living cell and involve themselves in nearly all of the activities that take place in a cell. Their wide range of structures
Chem 452 - Lecture 2 Protein Structure 110923 Proteins are the workhorses of a living cell and involve themselves in nearly all of the activities that take place in a cell. Their wide range of structures
Supplementary Data AtMYB12
 Supplementary Data AtMYB12 expression in tomato leads to large scale differential modulation in transcriptome and flavonoid content in leaf and fruit tissues Ashutosh Pandey 1,$,#, Prashant Misra 1,$,#,
Supplementary Data AtMYB12 expression in tomato leads to large scale differential modulation in transcriptome and flavonoid content in leaf and fruit tissues Ashutosh Pandey 1,$,#, Prashant Misra 1,$,#,
The Arabidopsis mir396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens
 The Arabidopsis mir396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens Mauricio Soto-Suárez, Patricia Baldrich, Detlef Weigel, Ignacio Rubio-Somoza and
The Arabidopsis mir396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens Mauricio Soto-Suárez, Patricia Baldrich, Detlef Weigel, Ignacio Rubio-Somoza and
Day Date Title Instructor 5 th Ed 6 th Ed. Protein digestion and AA absorption
 Day Date Title Instructor 5 th Ed 6 th Ed 1 Tuesday 18 April 2017 Protein digestion and AA absorption D S Jairajpuri 250 256 250 256 2 Wednesday 19 April 2017 Removal of nitrogen and urea cycle D S Jairajpuri
Day Date Title Instructor 5 th Ed 6 th Ed 1 Tuesday 18 April 2017 Protein digestion and AA absorption D S Jairajpuri 250 256 250 256 2 Wednesday 19 April 2017 Removal of nitrogen and urea cycle D S Jairajpuri
Transcription and chromatin. General Transcription Factors + Promoter-specific factors + Co-activators
 Transcription and chromatin General Transcription Factors + Promoter-specific factors + Co-activators Cofactor or Coactivator 1. work with DNA specific transcription factors to make them more effective
Transcription and chromatin General Transcription Factors + Promoter-specific factors + Co-activators Cofactor or Coactivator 1. work with DNA specific transcription factors to make them more effective
Hands-On Ten The BRCA1 Gene and Protein
 Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated
 Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
THE UBIQUITIN-PROTEASOME SYSTEM DURING PROTEOTOXIC STRESS
 Cell and Molecular Biology (CMB) Karolinska Institutet Stockholm, Sweden THE UBIQUITIN-PROTEASOME SYSTEM DURING PROTEOTOXIC STRESS Victoria Menéndez Benito Stockholm 2006 Front cover: Ub-R-YFP MelJuSo
Cell and Molecular Biology (CMB) Karolinska Institutet Stockholm, Sweden THE UBIQUITIN-PROTEASOME SYSTEM DURING PROTEOTOXIC STRESS Victoria Menéndez Benito Stockholm 2006 Front cover: Ub-R-YFP MelJuSo
DSB. Double-Strand Breaks causate da radiazioni stress ossidativo farmaci
 DSB Double-Strand Breaks causate da radiazioni stress ossidativo farmaci METODI DDR foci formation in irradiated (2 Gy) cells fixed 2 h later IRIF IRradiation Induced Focus Laser micro-irradiation DDR
DSB Double-Strand Breaks causate da radiazioni stress ossidativo farmaci METODI DDR foci formation in irradiated (2 Gy) cells fixed 2 h later IRIF IRradiation Induced Focus Laser micro-irradiation DDR
Supplemental Figure 1. Genes showing ectopic H3K9 dimethylation in this study are DNA hypermethylated in Lister et al. study.
 mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is worth 2 points.
 MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
ubiquitin, protein degradation, proteasome, proteolysis SIGNALS IN PROTEINS FOR UBIQUITINYLATION AND DEGRADATION...438
 Annu. Rev. Biochem. 1998. 67:425 79 Copyright c 1998 by Annual Reviews. All rights reserved THE UBIQUITIN SYSTEM Avram Hershko and Aaron Ciechanover Unit of Biochemistry, Faculty of Medicine and the Rappaport
Annu. Rev. Biochem. 1998. 67:425 79 Copyright c 1998 by Annual Reviews. All rights reserved THE UBIQUITIN SYSTEM Avram Hershko and Aaron Ciechanover Unit of Biochemistry, Faculty of Medicine and the Rappaport
Previous Class. Today. Detection of enzymatic intermediates: Protein tyrosine phosphatase mechanism. Protein Kinase Catalytic Properties
 Previous Class Detection of enzymatic intermediates: Protein tyrosine phosphatase mechanism Today Protein Kinase Catalytic Properties Protein Phosphorylation Phosphorylation: key protein modification
Previous Class Detection of enzymatic intermediates: Protein tyrosine phosphatase mechanism Today Protein Kinase Catalytic Properties Protein Phosphorylation Phosphorylation: key protein modification
SUPPLEMENTARY INFORMATION
 Figure S1. Silver staining and immunoblotting of the purified TAK1 kinase complex. The TAK1 kinase complex was purified through tandem affinity methods (Protein A and FLAG), and aliquots of the purified
Figure S1. Silver staining and immunoblotting of the purified TAK1 kinase complex. The TAK1 kinase complex was purified through tandem affinity methods (Protein A and FLAG), and aliquots of the purified
Effects of Sucrose on Germination and Seedling Development of Brassica Napus
 Effects of Sucrose on Germination and Seedling Development of Brassica Napus Feng Xu E-mail: xufeng198677@yahoo.com.cn Xiaoli Tan (Corresponding author) Tel: 86-511-8291-4326 E-mail: xltan@ujs.edu.cn Zheng
Effects of Sucrose on Germination and Seedling Development of Brassica Napus Feng Xu E-mail: xufeng198677@yahoo.com.cn Xiaoli Tan (Corresponding author) Tel: 86-511-8291-4326 E-mail: xltan@ujs.edu.cn Zheng
Regulation of Gene Expression in Eukaryotes
 Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Expression constructs
 Gene expressed in bebe3 ZmBEa Expression constructs 35S ZmBEa Pnos:Hygromycin r 35S Pnos:Hygromycin r 35S ctp YFP Pnos:Hygromycin r B -1 Chl YFP- Merge Supplemental Figure S1: Constructs Used for the Expression
Gene expressed in bebe3 ZmBEa Expression constructs 35S ZmBEa Pnos:Hygromycin r 35S Pnos:Hygromycin r 35S ctp YFP Pnos:Hygromycin r B -1 Chl YFP- Merge Supplemental Figure S1: Constructs Used for the Expression
Supplemental Data. Hao et al. (2014). Plant Cell /tpc
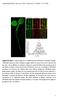 Supplemental Figure 1. Confocal Images and VA-TIRFM Analysis of GFP-RbohD in Arabidopsis Seedlings. (A) RbohD expression in whole Arabidopsis seedlings. RbohD was expressed in the leaves, hypocotyl, and
Supplemental Figure 1. Confocal Images and VA-TIRFM Analysis of GFP-RbohD in Arabidopsis Seedlings. (A) RbohD expression in whole Arabidopsis seedlings. RbohD was expressed in the leaves, hypocotyl, and
CKIP-1 couples Smurf1 ubiquitin ligase with Rpt6 subunit of proteasome to promote substrate degradation
 Manuscript EMBO-2012-36312 CKIP-1 couples Smurf1 ubiquitin ligase with Rpt6 subunit of proteasome to promote substrate degradation Yifang Wang, Jing Nie, Yiwu Wang, Luo Zhang, Guichun Xing, Ping Xie, Fuchu
Manuscript EMBO-2012-36312 CKIP-1 couples Smurf1 ubiquitin ligase with Rpt6 subunit of proteasome to promote substrate degradation Yifang Wang, Jing Nie, Yiwu Wang, Luo Zhang, Guichun Xing, Ping Xie, Fuchu
De novo assembly of rubber transcriptome
 Project Title: De novo assembly of rubber transcriptome Name: Minami Matsui, Ng Kiaw Kiaw, Ong Wen Dee Laboratory at RIKEN: Synthetic Genomics Research Team, CSRS Background and Mission: Natural rubber
Project Title: De novo assembly of rubber transcriptome Name: Minami Matsui, Ng Kiaw Kiaw, Ong Wen Dee Laboratory at RIKEN: Synthetic Genomics Research Team, CSRS Background and Mission: Natural rubber
This PDF file includes: Supplementary Figures 1 to 6 Supplementary Tables 1 to 2 Supplementary Methods Supplementary References
 Structure of the catalytic core of p300 and implications for chromatin targeting and HAT regulation Manuela Delvecchio, Jonathan Gaucher, Carmen Aguilar-Gurrieri, Esther Ortega, Daniel Panne This PDF file
Structure of the catalytic core of p300 and implications for chromatin targeting and HAT regulation Manuela Delvecchio, Jonathan Gaucher, Carmen Aguilar-Gurrieri, Esther Ortega, Daniel Panne This PDF file
A Novel Inhibitor of SUMOylation Pathway: Understanding the Mechanism of Action
 A Novel Inhibitor of SUMOylation Pathway: Understanding the Mechanism of Action, Hilda Wiryawan, and Jiayu Liao Department of Bioengineering ABSTRACT SUMOylation, one of the many important post-translational
A Novel Inhibitor of SUMOylation Pathway: Understanding the Mechanism of Action, Hilda Wiryawan, and Jiayu Liao Department of Bioengineering ABSTRACT SUMOylation, one of the many important post-translational
Where Splicing Joins Chromatin And Transcription. 9/11/2012 Dario Balestra
 Where Splicing Joins Chromatin And Transcription 9/11/2012 Dario Balestra Splicing process overview Splicing process overview Sequence context RNA secondary structure Tissue-specific Proteins Development
Where Splicing Joins Chromatin And Transcription 9/11/2012 Dario Balestra Splicing process overview Splicing process overview Sequence context RNA secondary structure Tissue-specific Proteins Development
Molecular mechanism of the priming by jasmonic acid of specific dehydration stress response genes in Arabidopsis
 University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications in the Biological Sciences Papers in the Biological Sciences 2016 Molecular mechanism of the priming
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications in the Biological Sciences Papers in the Biological Sciences 2016 Molecular mechanism of the priming
Chapter 31. Completing the Protein Life Cycle: Folding, Processing and Degradation. Biochemistry by Reginald Garrett and Charles Grisham
 Chapter 31 Completing the Protein Life Cycle: Folding, Processing and Degradation Biochemistry by Reginald Garrett and Charles Grisham Essential Question How are newly synthesized polypeptide chains transformed
Chapter 31 Completing the Protein Life Cycle: Folding, Processing and Degradation Biochemistry by Reginald Garrett and Charles Grisham Essential Question How are newly synthesized polypeptide chains transformed
Supplementary Figures
 Supplementary Figures a b c d PDI activity in % ERp72 activity in % 4 3 2 1 1 1 ERp activity in % e ΔRFU min -1 1 1 ERp7 activity in % 1 1 Supplementary Figure 1. Selectivity of the bepristat-mediated
Supplementary Figures a b c d PDI activity in % ERp72 activity in % 4 3 2 1 1 1 ERp activity in % e ΔRFU min -1 1 1 ERp7 activity in % 1 1 Supplementary Figure 1. Selectivity of the bepristat-mediated
Proteins. Amino acids, structure and function. The Nobel Prize in Chemistry 2012 Robert J. Lefkowitz Brian K. Kobilka
 Proteins Amino acids, structure and function The Nobel Prize in Chemistry 2012 Robert J. Lefkowitz Brian K. Kobilka O O HO N N HN OH Ser65-Tyr66-Gly67 The Nobel prize in chemistry 2008 Osamu Shimomura,
Proteins Amino acids, structure and function The Nobel Prize in Chemistry 2012 Robert J. Lefkowitz Brian K. Kobilka O O HO N N HN OH Ser65-Tyr66-Gly67 The Nobel prize in chemistry 2008 Osamu Shimomura,
Brassinosteroids (BRs) are a class of steroid hormones that
 The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis Jun-Xian He*, Joshua M. Gendron*, Yanli Yang*, Jianming Li,
The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis Jun-Xian He*, Joshua M. Gendron*, Yanli Yang*, Jianming Li,
Improve Protein Analysis with the New, Mass Spectrometry- Compatible ProteasMAX Surfactant
 Improve Protein Analysis with the New, Mass Spectrometry- Compatible Surfactant ABSTRACT Incomplete solubilization and digestion and poor peptide recovery are frequent limitations in protein sample preparation
Improve Protein Analysis with the New, Mass Spectrometry- Compatible Surfactant ABSTRACT Incomplete solubilization and digestion and poor peptide recovery are frequent limitations in protein sample preparation
Supplemental Data. Wang et al. (2013). Plant Cell /tpc
 Supplemental Data. Wang et al. (2013). Plant Cell 10.1105/tpc.112.108993 Supplemental Figure 1. 3-MA Treatment Reduces the Growth of Seedlings. Two-week-old Nicotiana benthamiana seedlings germinated on
Supplemental Data. Wang et al. (2013). Plant Cell 10.1105/tpc.112.108993 Supplemental Figure 1. 3-MA Treatment Reduces the Growth of Seedlings. Two-week-old Nicotiana benthamiana seedlings germinated on
Involvement of phytohormones and transcription factors in tissue - reunion in the cut tissues of plant
 奨励賞 植物の生長調節 Regulation of Plant Growth & Development Vol.48, No.1, 17-23, 2013 Involvement of phytohormones and transcription factors in tissue - reunion in the cut tissues of plant Masashi Asahina Department
奨励賞 植物の生長調節 Regulation of Plant Growth & Development Vol.48, No.1, 17-23, 2013 Involvement of phytohormones and transcription factors in tissue - reunion in the cut tissues of plant Masashi Asahina Department
Supplementary information
 Supplementary information Supplementary Figure 1: Components of Arabidopsis tricarboxylic acid (TCA) cycle. Schematic summary of the TCA cycle and the enzymes related to the reactions. The large text and
Supplementary information Supplementary Figure 1: Components of Arabidopsis tricarboxylic acid (TCA) cycle. Schematic summary of the TCA cycle and the enzymes related to the reactions. The large text and
Ubiquitinated proteins promote the association of proteasomes with the deubiquitinating enzyme Usp14 and the ubiquitin ligase Ube3c
 Ubiquitinated proteins promote the association of proteasomes with the deubiquitinating enzyme Usp14 and the ubiquitin ligase Ube3c Chueh-Ling Kuo a and Alfred Lewis Goldberg a,1 a Department of Cell Biology,
Ubiquitinated proteins promote the association of proteasomes with the deubiquitinating enzyme Usp14 and the ubiquitin ligase Ube3c Chueh-Ling Kuo a and Alfred Lewis Goldberg a,1 a Department of Cell Biology,
SUPPLEMENTARY INFORMATION
 Supplementary igure 1. The expression level of PP4 (At4g16860) is diminished in the rpp4 mutant (SK017569). The mna was extracted to analyze the expression level of PP4 using quantitative PC (qpc). Gene
Supplementary igure 1. The expression level of PP4 (At4g16860) is diminished in the rpp4 mutant (SK017569). The mna was extracted to analyze the expression level of PP4 using quantitative PC (qpc). Gene
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression. Chromatin Array of nuc
 Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
BIO360 Fall 2013 Quiz 1
 BIO360 Fall 2013 Quiz 1 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function mutation.
BIO360 Fall 2013 Quiz 1 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function mutation.
Chem 280 Final Exam. Here is the summary of the total 150 points plus 6 points bonus. Carefully read the questions. Good luck!
 May 2 nd, 2012 Name: CLID: Score: Chem 280 Final Exam There are 32 multiple choices that are worth 3 points each. There are 5 problems and one bonus problem. Try to answer the questions, which you know
May 2 nd, 2012 Name: CLID: Score: Chem 280 Final Exam There are 32 multiple choices that are worth 3 points each. There are 5 problems and one bonus problem. Try to answer the questions, which you know
Receptor mediated Signal Transduction
 Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
FATTY ACID SYNTHESIS
 FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
Minjia Tan, Ph.D Shanghai Institute of Materia Medica, Chinese Academy of Sciences
 Lysine Glutarylation Is a Protein Posttranslational Modification Regulated by SIRT5 Minjia Tan, Ph.D Shanghai Institute of Materia Medica, Chinese Academy of Sciences Lysine: the most frequently modified
Lysine Glutarylation Is a Protein Posttranslational Modification Regulated by SIRT5 Minjia Tan, Ph.D Shanghai Institute of Materia Medica, Chinese Academy of Sciences Lysine: the most frequently modified
Complexity DNA. Genome RNA. Transcriptome. Protein. Proteome. Metabolites. Metabolome
 DNA Genome Complexity RNA Transcriptome Systems Biology Linking all the components of a cell in a quantitative and temporal manner Protein Proteome Metabolites Metabolome Where are the functional elements?
DNA Genome Complexity RNA Transcriptome Systems Biology Linking all the components of a cell in a quantitative and temporal manner Protein Proteome Metabolites Metabolome Where are the functional elements?
SHORT LINEAR MOTIFS. Holger Dinkel EMBO Practical Course Computational analysis of protein-protein interactions From sequences to networks
 Holger Dinkel EMBO Practical Course Computational analysis of protein-protein interactions From sequences to networks IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19
Holger Dinkel EMBO Practical Course Computational analysis of protein-protein interactions From sequences to networks IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19
Supplemental Figure 1. Small RNA size distribution from different soybean tissues.
 Supplemental Figure 1. Small RNA size distribution from different soybean tissues. The size of small RNAs was plotted versus frequency (percentage) among total sequences (A, C, E and G) or distinct sequences
Supplemental Figure 1. Small RNA size distribution from different soybean tissues. The size of small RNAs was plotted versus frequency (percentage) among total sequences (A, C, E and G) or distinct sequences
Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism- 2015
 Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism- 2015 The major site of acetoacetate and 3-hydorxybutyrate production is the liver. They are preferred substrates for myocardiocytes
Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism- 2015 The major site of acetoacetate and 3-hydorxybutyrate production is the liver. They are preferred substrates for myocardiocytes
Overview of the Expressway Cell-Free Expression Systems. Expressway Mini Cell-Free Expression System
 Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Introduction Session 1 and 2 (Ubiquitin, proteasome and human disease)
 Introduction Session 1 and 2 (Ubiquitin, proteasome and human disease) Courtesy of Sam Griffiths-Jones. Used with permission. Source: "Peptide models for protein beta-sheets." PhD thesis, University of
Introduction Session 1 and 2 (Ubiquitin, proteasome and human disease) Courtesy of Sam Griffiths-Jones. Used with permission. Source: "Peptide models for protein beta-sheets." PhD thesis, University of
Cancer. The fundamental defect is. unregulated cell division. Properties of Cancerous Cells. Causes of Cancer. Altered growth and proliferation
 Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Ubiquitination of Keap1, a BTB-Kelch Substrate Adaptor Protein for Cul3, Targets Keap1 for Degradation by a Proteasome-independent Pathway*
 THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 280, No. 34, Issue of August 26, pp. 30091 30099, 2005 2005 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Ubiquitination
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 280, No. 34, Issue of August 26, pp. 30091 30099, 2005 2005 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Ubiquitination
supplementary information
 DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
The Molecular Mechanism of Ethylene- Mediated Root Hair Development Induced by Phosphate Starvation
 RESEARCH ARTICLE The Molecular Mechanism of Ethylene- Mediated Root Hair Development Induced by Phosphate Starvation Li Song 1, Haopeng Yu 2, Jinsong Dong 1, Ximing Che 1, Yuling Jiao 2, Dong Liu 1 * a11111
RESEARCH ARTICLE The Molecular Mechanism of Ethylene- Mediated Root Hair Development Induced by Phosphate Starvation Li Song 1, Haopeng Yu 2, Jinsong Dong 1, Ximing Che 1, Yuling Jiao 2, Dong Liu 1 * a11111
