Supplementary Data AtMYB12
|
|
|
- Alexia Owens
- 5 years ago
- Views:
Transcription
1 Supplementary Data AtMYB12 expression in tomato leads to large scale differential modulation in transcriptome and flavonoid content in leaf and fruit tissues Ashutosh Pandey 1,$,#, Prashant Misra 1,$,#, Dharmendra Choudhary 2, Reena Yadav 1, Ridhi Goel 1, Sweta Bhambhani 1, Indraneel Sanyal 1, Ritu Trivedi 2,*, Prabodh Kumar Trivedi 1,* 1 Council of Scientific and Industrial Research-National Botanical Research Institute (CSIR- NBRI), Rana Pratap Marg, Lucknow , INDIA 2 CSIR-Central Drug Research Institute (CSIR-CDRI), Endocrinology Division, Jankipuram Extension, Sitapur Road, Lucknow , INDIA $ Present address (AP): National Agri-Food Biotechnology Institute (NABI), Mohali , Punjab, INDIA $ Present address (PM): CSIR-Indian Institute of Integrative Medicine (IIIM), Canal Road, Jammu , INDIA #: Contributed equally to this study *Authors for correspondence PKT: prabodht@hotmail.com, prabodht@nbri.res.in RT: ritu_trivedi@nbri.res.in Running title: AtMYB12 differentially modulates flavonoid biosynthesis in tomato fruit and leaf
2 Supplementary Figure 1. Molecular characterization of transgenic lines. (a) Confirmation of the transgene by PCR amplification of genomic DNA with CaMV35S forward and gene specific reverse primers. (b) Expression analysis of transgene through semiquantitative RT- PCR using RNA from leaves.
3 Supplementary Figure 2. Effect of AtMYB12 expression on anthocyanin, polyphenol and flavonoid content in transgenic tomato lines as compared to WT. (a) Reduction in anthocyanin pigmentaion on stem and petiole in tomato transgenic line in comparison to wild type (WT) leaves. (b) Quantification of total polyphenolic content using methanolic extracts. Quantification has been carried out by gallic acid equivalent. (c) Quantification of total flavonoid content using methanolic extracts by quercetin equivalent. 1, 2 and 3 represent independent tomato transgenic lines expressing AtMYB12.
4 Supplementary Figure 3. Quantitative expression analysis of the structural genes upregulated in leaves of AtMYB12-expressing tomato transgenic lines. Expression of structural genes of phenylpropanoid pathway/flavonoid pathway in leaves of the WT and transgenic tomato lines. 1, 2 and 3 represent transgenic line 1, line 2 and line 3, respectively. PAL, Phenylalanine amonia lyase; CHI, Chalcone isomerase; CHS, Chalcone synthase; FLS, Flavonol synthase; F3H, Flavonone-3-hydroxylase; F3 H, flavonoid 3 -hydroxylase; 4CL, 4- coumaroyl CoA ligase; ANS, Anthocyanidine synthase HCT, Hydroxycinnamoyl transferase; GT, Glucosyltransferase.
5 Supplementary Figure 4. Distribution of common and unique genes with modulated expression among leaves and fruits of transgenic and WT tomato plants. (a) up-regulated unigenes and (b) down-regulated unigenes.
6 Secondary plant product stress responses a response to temperature stimulus response to heat response to wounding Down Up response to stress response to stimulus response to radiation response to light stimulus response to external stimulus response to abiotic stimulus flavonoid metabolic process flavonoid biosynthetic process secondary metabolic process phenylpropanoid metabolic process phenylpropanoid biosynthetic process b Percentage of gene count Supplementary Figure 5. Gene ontology of differentially regulated genes and functional network predicted. (a) Solanum lycopersicum homologues of the differentially-regulated genes in AtMYB12-expressing tomato leaves were identified and grouped with respect to their predicted role in different processes using agrigo (level 4). (b) Solanum lycopersicum homologues of the differentially expressed genes in AtMYB12-expressing tomato were identified to construct interactive network using Pathway Studio (Ariadne Genomics, USA).
7 Supplementary Figure 6. Validation of differential expression of genes involved in different processes in leaves of AtMYB12-expressing tomato transgenic lines. 1, 2 and 3 represent line 1, line 2 and line 3 respectively.acp-1-aminocyclopropane-1-carboxylate synthase (Solyc08g ); AQ, Aquaporin (Solyc12g ); AUX, Auxin efflux carrier family protein (Solyc02g ); CROT, Crotenase (Solyc12g ); CYT450, Cytochrome P450 (Solyc10g ); ERF, Ethylene-responsive transcription factor 4 (Solyc07g ); GST, Glutathione S-transferase (Solyc10g ); PDH, Prephenate dehydratase (Solyc06g ).
8 a b c d e Supplementary Figure 7. Differential expression of genes in leaves and fruits of tomato plants. (a) Differential expression of genes involved in flavonoid biosynthesis. (b) Differential expression of genes encoding glycosyltransferases. (c) Differential expression of genes encoding involved in phytohormone biosynthesis, signalling and response. (d) Differential expression of genes involved in aromatic amino acid biosynthesis. (e) Differential expression of genes involved in glutathione S transferase. The locus ids of various regulatory genes were searched from microarray data. These probe set ids were used for annotation using different databases. Heat maps denoting digital expression by using MeV version 4.3 program. WT and TR represent wild type and transgenic line.
9 Supplementary Figure 8. Differential expression of genes encoding enzymes involved in carbohydrate, lipid and organic acid metabolism in leaves and fruits of tomato plants. The locus ids of various regulatory genes were searched from microarray data. These probe set ids were used for annotation using different databases. Heat maps denoting digital expression by using MeV version 4.3 program. WT and TR represent wild type and transgenic line.
10 Table 1: List of up-regulated ( two-fold) genes in leaves of AtMYB12-expressing tomato lines S.No Locus ID SOLGEN locus id Annotation in SOLGEN 1 Les S1_at Solyc05g Chalcone synthase 2 Les S1_at Solyc09g Chalcone synthase 3 Les S1_at Solyc11g Anthocyanidin synthase 4 Les S1_at Solyc02g Flavanone 3-hydroxylase 5 LesAffx S1_at Solyc02g Dihydroflavonol-4-reductase 6 LesAffx S1_at Solyc02g Dihydroflavonol-4-reductase 7 LesAffx S1_at Solyc01g Ethylene responsive transcription factor 2b 8 LesAffx S1_at Solyc08g Ethylene-responsive transcription factor 10 9 LesAffx S1_at Solyc05g Chalcone--flavonone isomerase 10 LesAffx A1_at Solyc02g Dihydroflavonol-4-reductase 11 LesAffx S1_at Solyc12g coumarate-coa ligase 12 LesAffx A1_at Solyc02g Glycosyltransferase 13 LesAffx S1_at Solyc05g IAA-amino acid hydrolase 14 LesAffx S1_at Solyc07g Auxin-induced protein-like 15 LesAffx S1_at Solyc10g UDP flavonoid 3-O-glucosyltransferase (Fragment) 16 LesAffx A1_at Solyc03g Cinnamoyl-CoA reductase-like protein 17 LesAffx S1_at Solyc04g UDP-glucosyltransferase 18 Les S1_at Solyc02g Indole-3-acetic acid-amido synthetase GH LesAffx S1_at Solyc04g Ethylene responsive transcription factor 2b 20 Les S1_at Solyc02g aminocyclopropane-1-carboxylate oxidase 3 21 Les A1_at Solyc03g coumarate CoA ligase 22 LesAffx S1_at Solyc08g Ethylene-responsive transcription factor 1 23 RPTR-Les-U _s_at No Hit 24 Les A1_at No Hit 25 LesAffx S1_at Solyc08g Protein TIFY 5A 26 Les S1_at Solyc02g Proline dehydrogenase 27 Les S1_at Solyc01g Legumin 11S-globulin 28 Les S1_at Solyc03g Pectinesterase 29 LesAffx S1_at Solyc06g Ovarian cancer-associated gene 2 protein homolog 30 Les S1_at Solyc11g Proteinase inhibitor II 31 Les S1_at Solyc04g Monooxygenase FAD-binding 32 Les S1_at Solyc02g Proline dehydrogenase 33 Les A1_a_at No Hit 34 Les S1_x_at Solyc11g Calmodulin-like protein 35 LesAffx A1_at Solyc08g Laccase Les.13.1.S1_at Solyc04g cytochrome P450" 37 Les S1_a_at Solyc11g Calmodulin-like protein 38 Les S1_at Solyc09g Adenosine kinase 39 LesAffx S1_at Solyc02g Zinc finger AN1 domain-containing stress-associated protein Les S1_at Solyc03g Ammonium transporter 41 Les S1_s_at No Hit 42 Les S1_at Solyc06g Metallothionein-like protein type 2 43 LesAffx S1_at No Hit 44 Les A1_at No Hit 45 Les S1_at Solyc06g Bifunctional polymyxin resistance arna protein 46 Les S1_at Solyc07g Xyloglucan endotransglucosylase/hydrolase 7 47 LesAffx S1_at Solyc01g Gibberellin 2-oxidase 48 Les S1_at Solyc01g Receptor-like protein kinase At3g Les A1_at No Hit 50 LesAffx S1_at Solyc03g C4-dicarboxylate transporter/malic acid transport family protein 51 Les S1_at No Hit 52 Les S1_at Solyc04g Unknown Protein 53 Les A1_at No Hit 54 Les S1_at Solyc03g PPPDE peptidase domain-containing protein 1 55 Les S1_at Solyc01g Uncharacterized oxidoreductase Mb LesAffx S1_at Solyc08g ATP binding / serine-threonine kinase 57 Les S1_at Solyc10g Myb family transcription factor (Fragment) 58 LesAffx S1_at Solyc07g Solanesyl diphosphate synthase 59 Les S1_at Solyc03g CRT binding factor 2 60 LesAffx S1_at Solyc04g Hydrolase alpha/beta fold family protein expressed 61 Les A1_at No Hit 62 Les S1_at Solyc02g Xyloglucan endotransglucosylase/hydrolase 2 63 Les S1_at Solyc03g Major facilitator superfamily domain containing protein 5 64 Protease inhibitor/seed storage/lipid transfer protein family protein Les S1_at Solyc03g LesAffx S1_at Solyc04g Os03g protein (Fragment) 66 LesAffx A1_at Solyc01g COBRA-like protein
11 67 Les A1_at No Hit 68 Les S1_at Solyc03g Xyloglucan endotransglucosylase/hydrolase 9 69 LesAffx S1_at Solyc05g Ribose-5-phosphate isomerase 70 Les S1_at Solyc02g Glycine rich protein 71 LesAffx S1_at Solyc01g Peptide methionine sulfoxide reductase msrb 72 LesAffx S1_at Solyc09g Acid phosphatase-like protein 73 Les S1_s_at Solyc04g Xyloglucan endotransglucosylase/hydrolase Les S1_at Solyc03g Lipoxygenase 75 LesAffx S1_at Solyc06g Methyltransferase like 7A 76 Les S1_a_at Solyc03g Phytoene synthase 1" 77 Les A1_at Solyc09g F-box family protein 78 LesAffx S1_at Solyc02g Peroxidase 1 79 LesAffx S1_at Solyc04g Ovarian cancer-associated gene 2 protein homolog 80 Les S1_at Solyc04g Phenylcoumaran benzylic ether reductase 81 Les S1_at Solyc12g Os07g protein (Fragment) 82 LesAffx S1_at Solyc07g Solanesyl diphosphate synthase 83 Les S1_at Solyc04g Carboxyl methyltransferase 84 Les S1_at Solyc01g Phospholipase D 85 Les S1_at Solyc12g Glyceraldehyde-3-phosphate dehydrogenase B 86 Les A1_at No Hit 87 LesAffx S1_at Solyc03g GCN5-related N-acetyltransferase 88 Les A1_at No Hit 89 LesAffx S1_at Solyc05g Cytidyltransferase-related 90 LesAffx S1_at Solyc06g Prephenate dehydrogenase family protein 91 LesAffx S1_at No Hit 92 LesAffx S1_at Solyc02g CBS domain containing protein 93 Les S1_at Solyc04g Xyloglucan endotransglucosylase/hydrolase Les A1_at Solyc04g Carboxyl methyltransferase 95 Les S1_at Solyc10g Late elongated hypocotyl and circadian clock associated-1-like" 96 LesAffx S1_at Solyc06g Conserved transmembrane protein 97 Les S1_at Solyc12g Glyceraldehyde-3-phosphate dehydrogenase B 98 Les S1_at Solyc02g Pectinesterase 99 LesAffx S1_at Solyc03g Oxalate oxidase-like germin LesAffx S1_at Solyc02g cdna clone J100026I16 full insert sequence 101 Les S1_at Solyc07g Xyloglucan endotransglucosylase/hydrolase LesAffx S1_at Solyc02g Zinc transporter protein 103 Les S1_at Solyc09g Ribulose-1 5-bisphosphate carboxylase/oxygenase activase LesAffx S1_at Solyc06g Bcl-2-associated athanogene-like protein
12 Table 2: List of down-regulated ( two-fold) genes in leaves of AtMYB12-expressing tomato lines S.No. Locus ID SOLGEN locus id Annotation in SOLGEN 1 LesAffx S1_at Solyc03g GDSL esterase/lipase At3g Les A1_at No Hit 3 Les A1_at No Hit 4 Les S1_a_at Solyc06g Unknown Protein 5 Les A1_at No Hit 6 Les S1_at Solyc02g B12D-like protein 7 Les A1_at No Hit 8 Les S1_at Solyc01g Cytochrome P450" 9 Les S1_at Solyc02g Cellular retinaldehyde-binding/triple function C-terminal 10 Les S1_at Solyc08g Osmotin-like protein (Fragment) 11 LesAffx S1_at Solyc10g Anthocyanidin synthase 12 Les A1_at No Hit 13 Les A1_at No Hit 14 LesAffx S1_at Solyc09g Os01g protein (Fragment) 15 Les A1_at No Hit 16 Les S2_at No Hit 17 LesAffx S1_at Solyc08g Heat stress transcription factor A3 18 Les S1_at Solyc03g Heat shock protein 19 Les A1_at No Hit 20 Les S1_at Solyc11g class IV heat shock protein 21 LesAffx S1_at Solyc10g Pyruvate decarboxylase 22 LesAffx S1_at Solyc01g LRR receptor-like serine/threonine-protein kinase, RLP" 23 Les S1_at Solyc04g Prostaglandin E synthase 3 24 LesAffx S1_at Solyc03g Nudix hydrolase 1 25 Les S1_at Solyc01g Unknown Protein 26 Les A1_at No Hit 27 LesAffx S1_at Solyc09g class I heat shock protein 3 28 LesAffx S1_at Solyc04g Pathogenesis-related protein-like protein 29 Les S1_at Solyc03g Kunitz trypsin inhibitor 30 Les S1_at Solyc06g Unknown Protein 31 Les S1_x_at Solyc00g Cysteine proteinase inhibitor 32 Les A1_at No Hit 33 Les S1_at Solyc05g Heat shock protein 34 LesAffx S1_at Solyc08g Anthocyanidin synthase (Fragment) 35 LesAffx S1_at Solyc04g class I heat shock protein 3 36 Les S1_at Solyc08g Auxin responsive protein 37 Les A1_at No Hit 38 Les S1_at Solyc07g aminocyclopropane-1-carboxylate oxidase 39 Les A1_at No Hit 40 Les S1_at Solyc11g Xyloglucan endotransglucosylase/hydrolase 9 41 Les S1_at Solyc07g Non-symbiotic hemoglobin protein
13 Table 3: List of up-regulated ( two-fold) genes in fruits of AtMYB12-expressing tomato lines S.No Locus ID SOLGEN locus id Annotation in SOLGEN 1 Les S1_at Solyc12g Aquaporin 2 Les S1_at Solyc08g aminocyclopropane-1-carboxylate synthase 3 Les S1_at Solyc09g Glutathione S-transferase-like protein 4 Les S1_at Solyc02g Indole-3-acetic acid-amido synthetase GH3.8 5 LesAffx S1_at Solyc08g UDP-glucose salicylic acid glucosyltransferase 6 Les S1_at Solyc05g Ethylene responsive transcription factor 1a 7 LesAffx S1_at Solyc12g Glucosyltransferase 8 Les S1_at Solyc02g Ethylene-responsive transcription factor 2 9 Les A1_at Solyc11g AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE-like protein 10 LesAffx S1_at Solyc12g Glucosyltransferase 11 Les A1_at Solyc02g WRKY transcription factor LesAffx S1_at Solyc03g BHLH transcription factor-like protein 13 LesAffx S1_at Solyc08g UDP-glucose glucosyltransferase 14 Les S1_at Solyc10g Glutathione S-transferase 15 LesAffx S1_at Solyc06g Prephenate dehydrogenase family protein 16 LesAffx S1_at Solyc02g Auxin efflux carrier family protein 17 Les S1_at Solyc05g hydroxyphenylpyruvate dioxygenase 18 LesAffx S1_at Solyc10g Cytochrome P450" 19 LesAffx S1_at Solyc02g Cytochrome P450" 20 Les S1_at Solyc07g Ethylene-responsive transcription factor 4 21 LesAffx S1_at Solyc06g Prephenate dehydratase 22 LesAffx S1_at Solyc10g Cytochrome P450" 23 Les S1_at Solyc08g Polyphenol oxidase 24 Les S1_at Solyc05g hydroxyphenylpyruvate dioxygenase 25 Les S1_at Solyc01g Glutathione S-transferase 26 Les S1_at Solyc01g UDP-glucosyltransferase family 1 protein 27 LesAffx S1_at Solyc09g aminocyclopropane-1-carboxylate oxidase-like protein 28 LesAffx S1_at Solyc05g Chalcone--flavonone isomerase 29 Les S1_at Solyc06g Ethylene responsive transcription factor 1b 30 LesAffx S1_at Solyc12g Enoyl-CoA-hydratase 31 Les S1_at Solyc09g Phenylalanine ammonia-lyase 32 LesAffx S1_at Solyc01g UDP-glucosyltransferase family 1 protein 33 Les S1_at Solyc03g Kunitz trypsin inhibitor 34 Les S1_at No Hit 35 Les S1_at Solyc01g Plant viral-response family protein 36 RPTR-Les-U _s_at No Hit 37 Les S1_s_at Solyc03g Ribulose bisphosphate carboxylase small chain 38 Les S1_at Solyc09g Heat stress transcription factor A3 39 Les A1_at No Hit 40 Les S1_at Solyc10g Heavy metal-associated domain containing protein expressed 41 Les S1_at Solyc09g Proteinase inhibitor I 42 Les S1_at Solyc01g Cyclin 43 Les S1_at No Hit 44 Les S1_at Solyc10g Aquaporin 45 Les S1_at Solyc08g Expansin-like protein 46 LesAffx A1_at Solyc07g Beta-glucosidase 47 Les A1_a_at Solyc03g Chlorophyll a-b binding protein 3C-like 48 Les S1_at Solyc09g Proteinase inhibitor I 49 Les A1_x_at Solyc03g Chlorophyll a-b binding protein 3C-like 50 Les A1_at Solyc09g F-box family protein 51 LesAffx S1_at Solyc03g Unknown Protein 52 Les S1_at Solyc09g Aldo/keto reductase family protein 53 Les S1_at Solyc05g Protein disulfide isomerase 54 Les S1_s_at Solyc09g Unknown Protein 55 Les S1_at Solyc02g Chlorophyll a/b binding protein 56 Les S1_x_at Solyc02g Chlorophyll a/b binding protein 57 LesAffx S1_at Solyc01g Xenotropic and polytropic retrovirus receptor 58 Les S1_at Solyc01g Xylanase inhibitor (Fragment) 59 Les S1_at Solyc11g Alcohol dehydrogenase 60 Les A1_at Solyc10g Aquaporin 61 Les A1_at No Hit 62 Les S1_at Solyc09g Proteinase inhibitor I 63 Les S1_a_at Solyc11g Proteinase inhibitor 64 LesAffx A1_at Solyc00g ATP synthase subunit a 65 Les S1_a_at Solyc11g Calmodulin-like protein 66 LesAffx S1_at Solyc09g Pyrimidine 5&apos-nucleotidase 67 Les S1_at No Hit
14 68 Les S1_at No Hit 69 LesAffx S1_at Solyc00g ATPase subunit 8 70 LesAffx S1_at Solyc02g Thaumatin-like protein 71 Les S1_at Solyc02g Ribulose bisphosphate carboxylase small chain 72 LesAffx S1_at Solyc07g Heat stress transcription factor A3 73 LesAffx S1_at Solyc08g Heat stress transcription factor A3 74 LesAffx S1_at Solyc02g Inositol trisphosphate 5-phosphatase-like protein 75 LesAffx S1_at Solyc01g Cell number regulator Les S1_at Solyc02g Carbonic anhydrase 77 LesAffx S1_at Solyc09g Heat shock protein 1 78 LesAffx S1_at Solyc01g Unknown Protein 79 Les S1_at Solyc05g Unknown Protein 80 Les A1_at No Hit 81 Les A1_at Solyc07g Metallocarboxypeptidase inhibitor 82 Les A1_at No Hit 83 Les S1_at Solyc03g Glyoxalase/bleomycin resistance protein/dioxygenase 84 Les S1_at Solyc09g Unknown Protein 85 Les S1_s_at Solyc10g Palmitoyltransferase PFA4 86 LesAffx S1_at Solyc06g Bcl-2-associated athanogene-like protein 87 Les A1_at No Hit 88 Les S1_at Solyc10g Arginine decarboxylase 89 Les S1_s_at Solyc07g Glutathione S-transferase-like protein 90 Les S1_at Solyc03g Proteinase inhibitor II 91 LesAffx S1_at Solyc10g F-box protein PP2-B1 92 LesAffx A1_at Solyc01g Serine/threonine protein kinase-like 93 LesAffx S1_at Solyc01g Glyoxalase/bleomycin resistance protein/dioxygenase 94 Les S1_at Solyc07g Endoribonuclease L-PSP family protein 95 LesAffx S1_at Solyc03g Chaperone protein dnaj 96 Les S1_at Solyc11g class IV heat shock protein 97 Les A1_at Solyc04g Pheophorbide a oxygenase 98 Les S1_at Solyc04g Calcium-dependent protein kinase 2 99 Les S1_at Solyc12g Chlorophyll a-b binding protein 13, chloroplastic 100 Les.13.1.S1_at Solyc04g cytochrome P450" 101 Les S1_x_at Solyc11g Calmodulin-like protein 102 Les S1_at Solyc03g Aluminum-induced protein-like 103 LesAffx S1_at Solyc06g Short-chain dehydrogenase/reductase family protein 104 LesAffx S1_at Solyc08g Unknown Protein 105 LesAffx S1_at Solyc01g Xylanase inhibitor (Fragment) 106 Les A1_at No Hit 107 LesAffx S1_at Solyc08g ChaC cation transport regulator-like Les S1_at Solyc12g Chlorophyll a-b binding protein 37, chloroplastic 109 LesAffx S1_at Solyc12g TBC1 domain family member LesAffx S1_at Solyc01g SRC2 homolog (Fragment) 111 Les S1_at Solyc11g DNA cross-link repair 1B-like protein 112 Les S1_at No Hit 113 LesAffx S1_at Solyc01g Hydrolase-like 114 Les A1_at No Hit 115 LesAffx S1_at Solyc03g Unknown Protein 116 Les S1_a_at No Hit 117 Les S1_at Solyc04g Lipase-like protein 118 Les S1_at Solyc07g Nuclear transcription factor Y subunit B Les S1_at Solyc03g Kunitz-type protease inhibitor 120 LesAffx S1_at Solyc08g ChaC cation transport regulator-like LesAffx S1_a_at Solyc03g AT1G52630-like protein (Fragment) 122 LesAffx S1_at Solyc02g Strictosidine synthase family protein 123 Les S1_at Solyc02g MYB transcription factor (Fragment) 124 LesAffx S1_at Solyc02g class I heat shock protein 125 Les S1_s_at Solyc06g Phosphatase 126 Les S1_s_at Solyc06g Sex-linked protein 9 (Fragment) 127 LesAffx S1_at Solyc08g WRKY transcription factor Les S1_s_at Solyc06g Sex-linked protein 9 (Fragment) 129 Les A1_at No Hit 130 LesAffx S1_at No Hit 131 Les S1_at Solyc06g WRKY transcription factor Les S1_at No Hit 133 LesAffx S1_at Solyc04g Alpha/beta hydrolase fold protein 134 Les A1_at No Hit 135 Les S1_at Solyc03g Transcription regulatory protein SNF5 136 Les A1_at No Hit 137 LesAffx S1_at Solyc02g Strictosidine synthase family protein 138 LesAffx S1_at Solyc06g Glutathione S-transferase 139 LesAffx S1_at Solyc02g Glycerophosphodiester phosphodiesterase gde1 140 Les S1_at Solyc09g Unknown Protein 141 Les A1_at No Hit 142 Les S1_at Solyc10g Arginine decarboxylase
15 143 Les S1_at Solyc05g Chlorophyll a-b binding protein 6A, chloroplastic 144 Les S1_at No Hit 145 Les S1_at Solyc06g Sex-linked protein 9 (Fragment) 146 Les S1_at Solyc08g Photosystem I reaction center subunit 147 Les S1_at No Hit 148 Les S1_at No Hit 149 Les A1_at No Hit 150 Les A1_at No Hit 151 Les S1_at Solyc03g Protease inhibitor/seed storage/lipid transfer protein family protein 152 LesAffx A1_at No Hit 153 LesAffx S1_at Solyc03g AT1G52630-like protein (Fragment) 154 LesAffx S1_at Solyc04g TMV response-related protein 155 LesAffx S1_at Solyc05g Auxin-independent growth protein (Fragment) 156 Les S1_at Solyc03g Unknown Protein 157 LesAffx A1_at Solyc08g Anamorsin homolog 158 Les S1_at Solyc07g Xyloglucan endotransglucosylase/hydrolase Les S1_at No Hit 160 LesAffx S1_at Solyc02g MYB transcription factor 161 Les S1_at Solyc09g Threonine ammonia-lyase biosynthetic 162 Les A1_at Solyc10g Ferredoxin I 163 Les S1_at Solyc07g Ubiquitin fusion degradation protein LesAffx S1_at Solyc04g Ethylene-responsive transcription factor LesAffx S1_at Solyc10g Unknown Protein 166 Les A1_at No Hit 167 LesAffx S1_at Solyc01g Unknown Protein 168 Les S1_at Solyc10g Subtilisin-like protease 169 Les S1_at Solyc03g Chlorophyll a-b binding protein P4, chloroplastic 170 Les S1_at Solyc08g Stress-induced protein sti1-like protein 171 LesAffx S1_at Solyc06g Short-chain dehydrogenase/reductase family protein 172 Les S1_at Solyc06g Auxin responsive protein 173 Les S1_at No Hit 174 LesAffx A1_at Solyc02g Glycerophosphodiester phosphodiesterase gde1 175 LesAffx S1_at Solyc09g class I heat shock protein LesAffx S1_at Solyc02g Chorismate mutase Les A1_at No Hit 178 LesAffx.1.1.S1_at Solyc01g Agmatinase 179 Les S1_at No Hit 180 Les S1_at Solyc12g Photosystem II 5 kda protein, chloroplastic 181 Les A1_at No Hit 182 Les.84.1.S1_at Solyc12g Leucyl aminopeptidase 183 LesAffx S1_at Solyc06g Wound-induced protein RLK, Receptor like protein, putative resistance protein with an antifungal domain" LesAffx S1_at Solyc02g LesAffx S1_at No Hit 186 Les S1_at Solyc01g Glutamine synthetase 187 LesAffx A1_at Solyc04g Aspartate/glutamate/uridylate kinase family protein 188 Les S1_at Solyc03g Non-symbiotic hemoglobin LesAffx S1_at Solyc12g GRAS family transcription factor 190 Les A1_s_at No Hit 191 LesAffx S1_at Solyc10g LRR receptor-like serine/threonine-protein kinase, RLP" 192 LesAffx S1_at Solyc02g Macrophage erythroblast attacher 193 Les A1_at Solyc09g Proteinase inhibitor I 194 Les A1_s_at Solyc09g Unknown Protein 195 Les S1_at Solyc11g Calpain-2 catalytic subunit 196 Les S1_a_at No Hit 197 LesAffx S1_at Solyc05g F-box protein PP2-B1 198 Les A1_at Solyc02g MYB transcription factor (Fragment) 199 Les S1_at Solyc01g DNA polymerase 200 LesAffx S1_at Solyc01g Actin depolymerizing factor Les S1_at Solyc09g Unknown Protein 202 Les S1_at Solyc00g H-ATPase 203 Les S1_at Solyc04g Glyceraldehyde-3-phosphate dehydrogenase 204 Les S1_at Solyc04g Os01g protein (Fragment) 205 Les A1_at No Hit 206 LesAffx S1_at Solyc06g Prephenate dehydratase 207 LesAffx S1_at Solyc08g Os02g protein (Fragment) 208 LesAffx S1_at Solyc01g UBX domain protein LesAffx S1_at Solyc05g Poly polymerase catalytic domain containing protein expressed 210 Les S1_a_at No Hit 211 Les S1_at Solyc06g Integrin-linked kinase-associated serine/threonine phosphatase 2C 212 Les S1_at Solyc07g Saccharopine dehydrogenase (NAD(+) L-glutamate-forming) 213 Les S1_at Solyc10g Glutathione S-transferase 214 Les S1_at Solyc08g WRKY transcription factor Les S1_at No Hit 216 Les S1_at Solyc01g Cysteine-rich extensin-like protein-4
16 217 LesAffx S1_at No Hit 218 LesAffx S1_at Solyc05g Protein phosphatase-2c 219 LesAffx A1_at Solyc01g Unknown Protein 220 LesAffx S1_at Solyc03g Dof zinc finger protein LesAffx S1_at Solyc06g Cytochrome b5 reductase 222 Les S1_at Solyc09g Unknown Protein 223 LesAffx S1_at Solyc05g Chaperone protein dnaj LesAffx S1_at Solyc04g Phosphatidylserine synthase Les S1_at Solyc03g Pyridoxal biosynthesis lyase pdxs 226 Les S1_at Solyc06g Metallothionein-like protein type LesAffx S1_at Solyc02g Abhydrolase domain-containing protein FAM108B1 228 Les.13.1.S1_at Solyc04g cytochrome P450" 229 Les S1_at Solyc01g Zinc finger protein 230 Les A1_at Solyc11g ORF16-lacZ fusion protein 231 LesAffx S1_at Solyc01g Survival of motor neuron-related-splicing factor Les S1_at Solyc03g Chlorophyll a-b binding protein 3C-like 233 Les S1_at Solyc02g GDSL esterase/lipase At5g Les S1_at Solyc06g Os02g protein (Fragment) 235 LesAffx S1_at Solyc11g Zinc finger family protein 236 Les S1_at No Hit 237 Les S1_at Solyc04g NAC domain protein 238 Les A1_at No Hit 239 LesAffx A1_at Solyc03g Acid phosphatase 240 Les A1_at No Hit 241 Les S1_at No Hit 242 Les S1_at Solyc02g Plasma membrane associated protein 243 LesAffx S1_at Solyc07g Cotton fiber expressed protein Les S1_at Solyc07g Saccharopine dehydrogenase (NAD(+) L-glutamate-forming) 245 LesAffx S1_at Solyc01g Ubiquitin-conjugating enzyme E2 E3 246 LesAffx S1_at Solyc08g BHLH transcription factor 247 Les S1_at No Hit 248 Les A1_at No Hit 249 LesAffx S1_at Solyc00g Cytochrome c oxidase subunit LesAffx S1_at Solyc08g Zinc finger A20 and AN1 domain-containing stress-associated protein Les A1_at Solyc02g Peptidase trypsin-like serine and cysteine proteases (Fragment) 252 Les S1_at No Hit 253 LesAffx S1_at Solyc10g Response regulator Les S1_at Solyc09g PPPDE peptidase domain containing 2a 255 LesAffx S1_at Solyc02g Glycerophosphodiester phosphodiesterase gde1 256 LesAffx S1_at Solyc03g Unknown Protein 257 Les S1_at Solyc09g Ethylene responsive transcription factor 1a 258 Les A1_at No Hit 259 LesAffx S1_at Solyc01g Integral membrane protein 260 LesAffx S1_at Solyc05g RNA-binding protein Luc7-like LesAffx S1_at Solyc11g NADH-quinone oxidoreductase subunit 262 LesAffx S1_at Solyc02g Pantetheine-phosphate adenylyltransferase 263 LesAffx S1_at Solyc05g Poly polymerase catalytic domain containing protein expressed 264 Les S1_at Solyc03g Cellulose synthase 265 Les S1_at Solyc06g At1g72390-like protein (Fragment) 266 Les S1_a_at Solyc10g Ethylene-responsive transcription factor LesAffx A1_at Solyc04g Chaperone protein dnaj LesAffx S1_at Solyc10g Coiled-coil domain-containing protein 90B mitochondrial 269 LesAffx S1_at Solyc08g F-box family protein 270 Les S1_at Solyc06g Asparagine synthase (Glutamine-hydrolyzing) 271 LesAffx S1_at Solyc08g Avr9/Cf-9 rapidly elicited protein LesAffx S1_at Solyc00g Ring H2 finger protein 273 Les S1_at Solyc03g Rhodanese-like family protein-like protein (Fragment) 274 LesAffx S1_at Solyc02g Zinc finger AN1 domain-containing stress-associated protein Les A1_at No Hit 276 LesAffx S1_at Solyc00g ATP synthase subunit a 277 LesAffx S1_at Solyc07g FAD-dependent oxidoreductase family protein 278 Les S1_at Solyc10g Chlorophyll a-b binding protein 8, chloroplastic 279 Les S1_at Solyc07g aminocyclopropane-1-carboxylate oxidase 280 Les A1_at Solyc09g Unknown Protein 281 LesAffx S1_at Solyc07g Potassium channel tetramerization domain-containing protein 282 Les S1_at No Hit 283 Les A1_at No Hit 284 Les S1_at Solyc08g aminocyclopropane-1-carboxylate synthase 285 Les S1_at Solyc08g Phosphoribulokinase/uridine kinase 286 LesAffx S1_at Solyc02g Aldehyde dehydrogenase LesAffx S1_at Solyc02g Lrr, resistance protein fragment" 288 LesAffx S1_at Solyc01g Os05g protein (Fragment) 289 Les S1_s_at Solyc03g Ribulose bisphosphate carboxylase small chain 290 Les S1_at Solyc05g Acidic chitinase 291 LesAffx S1_at Solyc08g Zinc finger A20 and AN1 domain-containing stress-associated protein 7
17 292 Les A1_at No Hit 293 Les A1_at No Hit 294 Les S1_at Solyc07g Chlorophyll a-b binding protein 4, chloroplastic 298 Les S1_at Solyc09g Universal stress protein family protein 296 Les S1_at Solyc08g Zinc finger and SCAN domain containing 29 (Predicted) 297 LesAffx S1_at Solyc07g Decarboxylase family protein 298 Les S1_s_at Solyc09g Cell wall protein 299 Les S1_at Solyc03g MtN3-like protein 300 LesAffx S1_at Solyc07g Uncharacterized secreted protein 301 LesAffx S1_at Solyc01g Siroheme synthase 302 Les S1_at Solyc06g Heat shock protein 90 (Fragment) 303 Les S1_at Solyc12g Chlorophyll a-b binding protein 37, chloroplastic 304 LesAffx S1_at Solyc02g Protein serine/threonine kinase 305 LesAffx S1_s_at No Hit 306 Les A1_at No Hit 307 Les A1_at No Hit 308 Les S1_at Solyc01g ATPase AAA family protein expressed 309 Les A1_at No Hit 310 Les S1_at Solyc06g Vacuolar protein sorting-associated protein Les S1_at Solyc06g SET domain protein SUVR2 312 Les A1_at Solyc04g Water-stress inducible protein 3 (Fragment) 313 Les A1_at Solyc09g Unknown Protein 314 LesAffx S1_at Solyc01g Heavy metal-associated domain containing protein expressed 315 LesAffx S1_at Solyc06g N-acetyltransferase 316 LesAffx S1_at Solyc06g Mitochondrial import inner membrane translocase subunit TIM LesAffx S1_at Solyc01g Unknown Protein 318 Les S1_at Solyc06g Serine carboxypeptidase LesAffx S1_at Solyc07g NAC domain protein IPR LesAffx S1_at Solyc08g Avr9/Cf-9 rapidly elicited protein LesAffx S1_at Solyc10g MYB transcription factor (Fragment) 322 Les S1_at Solyc02g Pectinesterase 323 Les S1_at Solyc11g Mitogen-activated protein kinase 324 Les A1_at Solyc03g Glyoxal oxidase 325 Les A1_at No Hit 326 LesAffx A1_at Solyc01g UBX domain protein LesAffx S1_at Solyc03g Os05g protein (Fragment) 328 Les S1_at No Hit 329 LesAffx S1_at Solyc04g U-box domain-containing protein 330 Les S1_at Solyc01g Eukaryotic translation initiation factor 3 subunit B 331 Les A1_at No Hit 332 Les A1_at No Hit 333 Les S1_at Solyc04g Senescence-associated protein 334 Les S1_at Solyc01g Receptor-like kinase 335 LesAffx S1_at Solyc12g Dual specificity protein phosphatase Les A1_at No Hit 337 LesAffx A1_at Solyc10g Activator of heat shock protein ATPase homolog LesAffx S1_at Solyc03g Lipase-like protein 339 Les S1_at Solyc06g SNAP25 homologous protein SNAP Les S1_at Solyc04g Cell division protein kinase LesAffx S1_at Solyc03g Zinc finger protein-like protein 342 Les S1_at Solyc06g heat shock protein 343 Les S1_a_at Solyc01g Chitinase (Fragment) 344 Les S1_at Solyc12g TBC1 domain family member LesAffx S1_at Solyc04g UV excision repair protein RAD LesAffx S1_at Solyc01g Digalactosyldiacylglycerol synthase 2, chloroplastic 347 LesAffx S1_at Solyc02g Octanoyltransferase 348 LesAffx S1_at Solyc03g Hydrolase alpha/beta fold family protein 349 LesAffx S1_at Solyc09g Ras-related protein Rab LesAffx S1_at Solyc03g IBR finger domain protein 351 LesAffx S1_at Solyc01g Serine/threonine protein kinase-like 352 LesAffx S1_at Solyc07g ATP-dependent RNA helicase 353 Les S1_at Solyc06g Photosystem I reaction center subunit XI 354 LesAffx S1_at Solyc03g hydroxy-3-oxopropionate reductase 355 LesAffx S1_at Solyc04g MYB transcription factor 356 Les A1_at Solyc02g Succinate dehydrogenase iron-sulfur protein 357 LesAffx S1_at Solyc04g cdna clone J100026I16 full insert sequence 358 Les A1_at Solyc01g Unknown Protein 359 LesAffx S1_at Solyc07g UPF0431 protein C1orf66 homolog 360 Les S1_at No Hit 361 Les S1_at Solyc10g RING finger protein Les S1_at Solyc03g Alpha-hydroxynitrile lyase 363 LesAffx A1_at Solyc06g Nuclear SR-like RNA binding protein 364 Les S1_at Solyc01g Knotted-like homeobox protein 365 LesAffx A1_at Solyc12g F-box protein PP2-B1 366 LesAffx A1_at Solyc07g Decarboxylase family protein
18 367 Les S1_at Solyc08g Xenotropic and polytropic retrovirus receptor 368 Les S1_at Solyc06g Photosystem II reaction center W protein 369 Les A1_at No Hit 370 Les A1_at No Hit 371 LesAffx S1_at Solyc08g Globin 372 LesAffx S1_at Solyc02g C17orf95 protein (Fragment) 373 Les S1_x_at Solyc06g Phosphatase 374 Les S1_at Solyc07g SWIb domain-containing protein 375 Les A1_x_at No Hit 376 Les S1_at Solyc04g Unknown Protein 377 Les S1_at Solyc11g MYB transcription factor 378 LesAffx S1_at Solyc07g Serine/threonine protein kinase 379 LesAffx A1_at Solyc04g TMV response-related protein 380 Les S1_at Solyc07g ATP synthase I-like protein 381 Les A1_at No Hit 382 LesAffx A1_at Solyc01g COBRA-like protein 383 LesAffx A1_at Solyc11g Charged multivesicular body protein Les S1_at Solyc06g cdna clone C11 full insert sequence 385 LesAffx A1_at Solyc03g LYR motif-containing protein Les S1_at No Hit 387 Les S1_at Solyc05g Regulator of chromosome condensation RCC1 domain-containing protein 388 Les.47.1.S1_at Solyc02g MADS-box transcription factor 389 LesAffx S1_at Solyc03g E3 ubiquitin-protein ligase MARCH3 390 LesAffx S1_at Solyc07g Purine permease family protein 391 Les A1_at No Hit 392 Les S1_at No Hit 393 Les S1_at Solyc03g Polygalacturonase 394 Les S1_at Solyc09g PPPDE peptidase domain containing 2a 395 Les A1_at No Hit 396 LesAffx S1_at Solyc04g Tubby-like F-box protein Les S1_at No Hit 398 LesAffx S1_at Solyc03g Oleosin 399 Les S1_at Solyc01g Peroxidase 400 Les S1_at Solyc12g Exonuclease DNA polymerase III epsilon subunit family 401 LesAffx A1_at Solyc01g U-box domain-containing protein 402 LesAffx S1_at Solyc03g Unknown Protein 403 LesAffx S1_at Solyc09g Gibberellin receptor GID1L2 404 Les S1_at Solyc01g BZIP transcription factor 405 Les A1_at No Hit 406 Les S1_at Solyc06g Mitochondrial import inner membrane translocase subunit TIM LesAffx S1_at Solyc11g OTU domain-containing protein Les A1_at No Hit 409 LesAffx S1_at Solyc05g RING finger protein 410 LesAffx S1_at Solyc03g Ribulose bisphosphate carboxylase/oxygenase activase 411 LesAffx S1_at Solyc02g Splicing factor arginine/serine-rich Les S1_at Solyc05g Unknown Protein 413 LesAffx S1_at Solyc05g Unknown Protein 414 LesAffx S1_at Solyc03g RING finger protein Les A1_at No Hit 416 LesAffx S1_at Solyc03g Ring H2 finger protein 417 LesAffx S1_at Solyc01g U-box domain-containing protein 418 LesAffx S1_at Solyc04g Ras-related protein Rab LesAffx S1_at Solyc03g Hydrolase alpha/beta fold family protein 420 LesAffx S1_at Solyc01g AT4g33690/T16L1_180 (Fragment) 421 Les A1_at No Hit 422 Les S1_at No Hit 423 Les S1_at Solyc10g Chlorophyll a-b binding protein 7, chloroplastic 424 Les S1_at No Hit 425 Les S1_at Solyc01g Zinc finger A20 and AN1 domain-containing stress-associated protein LesAffx S1_at Solyc01g Os11g protein (Fragment) 427 LesAffx S1_at Solyc02g Ras-related protein Rab-2-A 428 Les S1_at Solyc10g Kelch-like protein Les S1_at Solyc10g Harpin-induced protein-like (Fragment) 430 Les A1_s_at Solyc09g Unknown Protein 431 Les S1_at Solyc07g L-lactate dehydrogenase 432 Les S1_at No Hit 433 LesAffx S1_at Solyc08g Short-chain dehydrogenase/reductase family protein 434 Les A1_at No Hit 435 LesAffx S1_at Solyc10g GDSL esterase/lipase At5g LesAffx S1_at Solyc11g Splicing factor 3B subunit Les S1_at Solyc09g Cobalt import ATP-binding protein CbiO Les S1_at Solyc02g Solute carrier family 2, facilitated glucose transporter member LesAffx S1_at Solyc07g RING finger protein 440 LesAffx A1_at Solyc05g Lipid phosphate phosphatase Les A1_at No Hit
19 442 LesAffx S1_at Solyc06g trna-dihydrouridine synthase 2-like protein 443 Les A1_at No Hit 444 Les A1_at No Hit 445 Les S1_at Solyc08g Unknown Protein 446 Les S1_at No Hit 447 LesAffx S1_at Solyc09g Short-chain dehydrogenase/reductase family protein 448 LesAffx A1_at Solyc10g CONSTANS-like zinc finger protein 449 Les A1_at Solyc12g Pectinesterase 450 Les S1_at Solyc01g Multiprotein bridging factor LesAffx S1_at Solyc12g F-box protein PP2-B1 452 Les S1_at Solyc05g Chloroplast photosystem II subunit X (Fragment) 453 Les S1_at Solyc10g Late elongated hypocotyl and circadian clock associated-1-like" 454 Les A1_at No Hit 455 LesAffx S1_at Solyc06g Golgi SNAP receptor complex member LesAffx S1_at Solyc07g MTD1 457 Les S1_at Solyc05g NAC domain transcription factor 458 Les S1_a_at No Hit 459 Les S1_at Solyc10g Heat shock protein Les A1_at No Hit 461 LesAffx S1_at Solyc07g Os06g protein (Fragment) 462 LesAffx S1_at No Hit 463 Les S1_at Solyc03g Guanylyl cyclase 464 LesAffx S1_at Solyc01g cdna clone J023121M11 full insert sequence 465 LesAffx A1_at Solyc01g S proteasome regulatory subunit 466 Les S1_at Solyc01g Vacuolar import and degradation protein VID LesAffx S1_at Solyc06g Transcription factor (Fragment) 468 Les S1_at Solyc03g Xyloglucan endotransglucosylase/hydrolase Les S1_at Solyc09g Selenium binding protein 470 LesAffx A1_at Solyc10g Eukaryotic translation initiation factor 4E 471 Les S1_at Solyc06g Protein tolb 472 LesAffx S1_at Solyc11g heat shock protein 473 Les S1_at Solyc12g Short-chain dehydrogenase/reductase family protein 474 Les S1_at Solyc05g Heat shock protein 475 Les S1_at Solyc01g S-adenosylmethionine decarboxylase proenzyme 476 Les A1_at No Hit 477 Les S1_at Solyc03g Heat shock protein 478 Les S1_at Solyc08g UDP-glucose 4-epimerase 479 LesAffx S1_at Solyc07g Yippee zinc-binding-like protein 480 Les S1_at Solyc01g SNARE associated Golgi protein 481 LesAffx S1_at Solyc08g Tasselseed2-like short-chain dehydrogenase/reductase (Fragment) 482 LesAffx S1_at Solyc01g Transporter major facilitator family 483 LesAffx A1_at Solyc06g Os11g protein (Fragment) 484 Les S1_at No Hit 485 Les S1_at Solyc03g Chlorophyll a-b binding protein P4, chloroplastic 486 Les S1_at Solyc12g ATP-dependent clp protease ATP-binding subunit 487 LesAffx A1_at Solyc10g SLT1 protein 488 LesAffx S1_at Solyc06g trna-dihydrouridine synthase 2-like protein 489 LesAffx S1_at Solyc04g Bet1-like protein At4g LesAffx S1_at Solyc05g Cytochrome P450" 491 Les A1_at No Hit 492 Les A1_at No Hit 493 Les S1_at Solyc08g Stress-induced protein sti1-like protein 494 LesAffx S1_at Solyc01g COBRA-like protein 495 Les S1_at Solyc01g Agmatinase 496 LesAffx S1_at Solyc11g Charged multivesicular body protein Les S1_at Solyc12g Protein phosphatase 2C 498 Les S1_at Solyc12g Aminotransferase-like protein 499 LesAffx S1_at No Hit 500 Les A1_at No Hit 501 LesAffx S1_at Solyc07g Oxalate oxidase-like germin LesAffx S1_at Solyc01g E3 ubiquitin-protein ligase MARCH6 503 Les S1_at Solyc12g Protochlorophyllide reductase 504 Les S1_at Solyc05g Unknown Protein 505 LesAffx S1_at Solyc01g Rubredoxin family protein 506 Les S1_at Solyc03g SPFH domain / Band 7 family protein 507 Les S1_at Solyc09g Cyclin dependent kinase inhibitor 508 LesAffx S1_at Solyc04g Prostaglandin E synthase 2-like 509 Les S1_at Solyc03g Transcription regulatory protein SNF5 510 LesAffx S1_at Solyc12g MADS box transcription factor 511 Les S1_s_at Solyc01g WD repeat-containing protein 512 Les S1_at No Hit 513 LesAffx S1_at Solyc04g U-box domain-containing protein 514 Les S1_at Solyc09g Tubby-like F-box protein LesAffx S1_at Solyc12g Density-regulated protein 516 LesAffx S1_at Solyc03g Cinnamoyl-CoA reductase-like protein
20 517 LesAffx A1_at Solyc04g Genomic DNA chromosome 5 P1 clone MCA Les A1_at Solyc01g MADS-box transcription factor Les S1_at Solyc06g SCF ubiquitin ligase skp1 component 520 LesAffx S1_at Solyc10g SLT1 protein 521 LesAffx S1_at Solyc03g MYB transcription factor 522 LesAffx A1_at Solyc06g Pseudo response regulator 523 LesAffx S1_at Solyc07g S proteasome non-atpase regulatory subunit Les A1_at No Hit 525 LesAffx A1_at Solyc01g Carbohydrate kinase YjeF related protein 526 LesAffx S1_at Solyc01g Heterogeneous nuclear ribonucleoprotein A3-like protein LesAffx A1_at No Hit 528 Les S1_at Solyc06g Transmembrane BAX inhibitor motif-containing protein LesAffx S1_at Solyc08g Zinc finger family protein 530 Les S1_at No Hit 531 Les S1_at Solyc09g Arginine/serine-rich splicing factor 532 LesAffx S1_at Solyc12g CHCH domain containing protein 533 LesAffx S1_at Solyc02g Cysteine-rich PDZ-binding protein 534 Les A1_at No Hit 535 LesAffx S1_at Solyc09g C2 domain-containing protein-like 536 LesAffx S1_at Solyc01g Zinc finger family protein 537 LesAffx S1_at Solyc02g Dof zinc finger protein 538 Les A1_at No Hit 539 LesAffx S1_at Solyc10g F-box protein SKIP Les.32.2.S1_a_at Solyc04g Cryptochrome 1a" 541 LesAffx S1_at Solyc06g Trans-acting transcriptional protein ICP0 542 Les S1_at Solyc04g ABC-1 domain protein 543 Les S1_at Solyc01g Mitochondrial carrier protein 544 LesAffx S1_at Solyc10g Elicitor-responsive protein Les S1_at Solyc05g Transcription elongation factor A protein Les A1_at No Hit 547 LesAffx S1_at Solyc10g Unknown Protein 548 LesAffx S1_at Solyc03g Abscisic acid receptor PYL8 549 Les S1_at Solyc02g ATP synthase gamma chain 550 LesAffx S1_at Solyc12g Pre-mRNA-splicing factor ini1 551 LesAffx S1_at Solyc12g Unknown Protein 552 Les S1_at Solyc07g Receptor-like protein kinase At5g Les A1_at No Hit 554 LesAffx S1_at Solyc05g YIPF5 555 LesAffx S1_at Solyc01g UPF0468 protein C16orf80 homolog 556 LesAffx S1_at No Hit 557 Les A1_at No Hit 558 LesAffx S1_at Solyc02g Speckle-type POZ protein 559 Les S1_at Solyc06g Genomic DNA chromosome 5 TAC clone K15C Les A1_at No Hit 561 Les A1_at No Hit 562 LesAffx S1_at Solyc01g Abscisic acid receptor PYL8 563 LesAffx S1_at Solyc08g Coiled-coil domain-containing protein Les S1_at Solyc07g Acetolactate synthase 565 Les S1_at Solyc08g Class II small heat shock protein Le-HSP LesAffx A1_at Solyc09g Phytochrome A-associated F-box protein 567 Les S1_at Solyc10g Kelch-like protein Les S1_at No Hit 569 Les S1_at Solyc04g Unknown Protein 570 LesAffx S1_at Solyc03g WD-40 repeat family protein 571 Les S1_at Solyc02g Glucan endo-1 3-beta-glucosidase LesAffx S1_at Solyc02g Coiled-coil domain-containing protein 109A 573 LesAffx S1_at Solyc01g Cytochrome b6-f complex subunit LesAffx A1_at Solyc01g Digalactosyldiacylglycerol synthase 2, chloroplastic 575 LesAffx S1_at Solyc02g Tafazzin 576 LesAffx S1_at Solyc02g CBS domain containing protein 577 Les S1_at Solyc07g Xyloglucan endotransglucosylase/hydrolase LesAffx S1_at Solyc02g class I heat shock protein 579 Les S1_at Solyc04g Sucrose transporter 580 Les S1_at Solyc03g Lipoxygenase 581 LesAffx S1_at Solyc09g Actin-depolymerizing factor Les A1_at No Hit 583 Les A1_at No Hit 584 LesAffx S1_at No Hit 585 LesAffx S1_at Solyc08g Unknown Protein 586 Les A1_at Solyc10g CWC15 homolog 587 Les A1_at No Hit 588 Les A1_at No Hit 589 Les A1_at No Hit 590 LesAffx S1_at Solyc11g Inner membrane ALBINO3-like protein 591 Les S1_at Solyc09g Ammonium transporter
WT siz1-2 siz1-3. WT siz1-2 siz1-3. -Pi, 0.05 M IAA. -Pi, 2.5 M NPA
 A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
Supplementary Tables 1
 Supplementary Tables 1 Supplementary Table 1: The 11 genes in the geneset for Case Study 1 At1g62570 At1g09350 At1g60470 At2g47180 At4g17090 At5g20830 At2g16890 At3g51240 At3g55120 At4g27560 At5g08640
Supplementary Tables 1 Supplementary Table 1: The 11 genes in the geneset for Case Study 1 At1g62570 At1g09350 At1g60470 At2g47180 At4g17090 At5g20830 At2g16890 At3g51240 At3g55120 At4g27560 At5g08640
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. AtMYB12 antibody detects both Arabidopsis and tomato MYB12 protein. (a) AtMYB12 antibody detects both SlMYB12 and AtMYB12 in tomato fruit. Both WT and AtMYB12
Supplementary Figures Supplementary Figure 1. AtMYB12 antibody detects both Arabidopsis and tomato MYB12 protein. (a) AtMYB12 antibody detects both SlMYB12 and AtMYB12 in tomato fruit. Both WT and AtMYB12
0.5. Normalized 95% gray value interval h
 Normalized 95% gray value interval.5.4.3.2.1 h Supplemental Figure 1: Symptom score of root samples used in the proteomics study. For each time point, the normalized 95% gray value interval is an averaged
Normalized 95% gray value interval.5.4.3.2.1 h Supplemental Figure 1: Symptom score of root samples used in the proteomics study. For each time point, the normalized 95% gray value interval is an averaged
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function
 Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Supplemental Table S1 Up-regulated genes in irjazh plants compared to WT plants by microarray.
 Supplemental Table S1 Up-regulated genes in irjazh plants compared to WT plants by microarray. Classification Fold Change Bincode (TAIR) Annotation Probe number ACC number Cell wall. Degradation 3.29 10.6.2
Supplemental Table S1 Up-regulated genes in irjazh plants compared to WT plants by microarray. Classification Fold Change Bincode (TAIR) Annotation Probe number ACC number Cell wall. Degradation 3.29 10.6.2
Biological processes. Mitochondrion Metabolic process Catalytic activity Oxidoreductase
 Full name Glyceraldehyde3 phosphate dehydrogenase Succinatesemialdehyde Glutamate dehydrogenase 1, Alcohol dehydrogenase [NADP+] 2',3'cyclicnucleotide 3' phosphodiesterase Dihydropyrimidinaserelated 2
Full name Glyceraldehyde3 phosphate dehydrogenase Succinatesemialdehyde Glutamate dehydrogenase 1, Alcohol dehydrogenase [NADP+] 2',3'cyclicnucleotide 3' phosphodiesterase Dihydropyrimidinaserelated 2
Bacterial cellulose synthesis mechanism of facultative
 1 2 3 4 Bacterial cellulose synthesis mechanism of facultative anaerobe Kaihua Ji 1 +, Wei Wang 2, 3 +, Bing Zeng 1, Sibin Chen 1, Qianqian Zhao 4, Yueqing Chen 1, Guoqiang Li 1* and Ting Ma 1* 5 6 7 Figure
1 2 3 4 Bacterial cellulose synthesis mechanism of facultative anaerobe Kaihua Ji 1 +, Wei Wang 2, 3 +, Bing Zeng 1, Sibin Chen 1, Qianqian Zhao 4, Yueqing Chen 1, Guoqiang Li 1* and Ting Ma 1* 5 6 7 Figure
MG132. GRXS17:V5-His MG132 GRXS17:HA. RuBisCo. Supplemental Data. Walton et al. (2015). Plant Cell /tpc
 MG132 GRXS17:V5-His MG132 GRXS17:HA RuBisCo Supplemental Figure 1. Degradation of GRXS17 by the 26S proteasome. (A) Cell-free degradation assay. Recombinant protein GRXS17:V5-HIS was incubated with total
MG132 GRXS17:V5-His MG132 GRXS17:HA RuBisCo Supplemental Figure 1. Degradation of GRXS17 by the 26S proteasome. (A) Cell-free degradation assay. Recombinant protein GRXS17:V5-HIS was incubated with total
Lecture 10 - Protein Turnover and Amino Acid Catabolism
 Lecture 10 - Protein Turnover and Amino Acid Catabolism Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction 2 Proteins are degraded into amino acids. Protein
Lecture 10 - Protein Turnover and Amino Acid Catabolism Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction 2 Proteins are degraded into amino acids. Protein
Figure S3 Differentially expressed fungal and plant genes organised by catalytic activity ontology Organisation of E. festucae (A) and L.
 sak WT Number of branches 3 WT sak Figure S1 Vasculature of plants infected with the E. festucae sak mutant. Light micrographs of perennial ryegrass blade tissue showing branching between the vasculature
sak WT Number of branches 3 WT sak Figure S1 Vasculature of plants infected with the E. festucae sak mutant. Light micrographs of perennial ryegrass blade tissue showing branching between the vasculature
Supplementary Information
 Supplementary Information Figure S1 Sequencing quality. Sample screenshot of re-sequencing raw file quality generated by FastQC. For both the graphs, scores were based on the Illumina 1.5 quality score.
Supplementary Information Figure S1 Sequencing quality. Sample screenshot of re-sequencing raw file quality generated by FastQC. For both the graphs, scores were based on the Illumina 1.5 quality score.
Biochemistry 2 Recita0on Amino Acid Metabolism
 Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
f(x) = x R² = RPKM (M8.MXB) f(x) = x E-014 R² = 1 RPKM (M31.
 14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
Supporting Information
 t t l l t l Supporting Information Zemach et al. 1.173/pnas.1969517.8 TE > 1 bp Leaf n o a Embryo h y C H H m e Endosperm Seedling root Seedling shoot -3-2 -1 1 2 3 4 3 2 1-1 -2-3 Fig. S1. Distribution
t t l l t l Supporting Information Zemach et al. 1.173/pnas.1969517.8 TE > 1 bp Leaf n o a Embryo h y C H H m e Endosperm Seedling root Seedling shoot -3-2 -1 1 2 3 4 3 2 1-1 -2-3 Fig. S1. Distribution
BIOL 158: BIOLOGICAL CHEMISTRY II
 BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
Chemistry 107 Exam 4 Study Guide
 Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
Figure 1 Original Advantages of biological reactions being catalyzed by enzymes:
 Enzyme basic concepts, Enzyme Regulation I III Carmen Sato Bigbee, Ph.D. Objectives: 1) To understand the bases of enzyme catalysis and the mechanisms of enzyme regulation. 2) To understand the role of
Enzyme basic concepts, Enzyme Regulation I III Carmen Sato Bigbee, Ph.D. Objectives: 1) To understand the bases of enzyme catalysis and the mechanisms of enzyme regulation. 2) To understand the role of
Coenzymes, vitamins and trace elements 209. Petr Tůma Eva Samcová
 Coenzymes, vitamins and trace elements 209 Petr Tůma Eva Samcová History and nomenclature of enzymes 1810, Gay-Lussac made an experiment with yeats alter saccharide to ethanol and CO 2 Fermentation From
Coenzymes, vitamins and trace elements 209 Petr Tůma Eva Samcová History and nomenclature of enzymes 1810, Gay-Lussac made an experiment with yeats alter saccharide to ethanol and CO 2 Fermentation From
Supplementary Data Transcriptome analysis in response to UV-B stress in soybean [Glycine max (L.) Merr.]
![Supplementary Data Transcriptome analysis in response to UV-B stress in soybean [Glycine max (L.) Merr.] Supplementary Data Transcriptome analysis in response to UV-B stress in soybean [Glycine max (L.) Merr.]](/thumbs/85/91947206.jpg) AJCS 6(9):195-1400 (2012) ISSN:185-27 Supplementary Data Transcriptome analysis in response to UV-B stress in soybean [Glycine max (L.) Merr.] Jun-Cheol Moon 1,2, Won Cheol Yim 1, Jae-Eun Lee, Young-Up
AJCS 6(9):195-1400 (2012) ISSN:185-27 Supplementary Data Transcriptome analysis in response to UV-B stress in soybean [Glycine max (L.) Merr.] Jun-Cheol Moon 1,2, Won Cheol Yim 1, Jae-Eun Lee, Young-Up
Table S1 Differentially expressed genes showing > 2 fold changes and p <0.01 for 0.01 mm NS-398, 0.1mM ibuprofen and COX-2 RNAi.
 Table S1 Differentially expressed genes showing > 2 fold changes and p
Table S1 Differentially expressed genes showing > 2 fold changes and p
NBCE Mock Board Questions Biochemistry
 1. Fluid mosaic describes. A. Tertiary structure of proteins B. Ribosomal subunits C. DNA structure D. Plasma membrane structure NBCE Mock Board Questions Biochemistry 2. Where in the cell does beta oxidation
1. Fluid mosaic describes. A. Tertiary structure of proteins B. Ribosomal subunits C. DNA structure D. Plasma membrane structure NBCE Mock Board Questions Biochemistry 2. Where in the cell does beta oxidation
Chem 280 Final Exam. Here is the summary of the total 150 points plus 6 points bonus. Carefully read the questions. Good luck!
 May 2 nd, 2012 Name: CLID: Score: Chem 280 Final Exam There are 32 multiple choices that are worth 3 points each. There are 5 problems and one bonus problem. Try to answer the questions, which you know
May 2 nd, 2012 Name: CLID: Score: Chem 280 Final Exam There are 32 multiple choices that are worth 3 points each. There are 5 problems and one bonus problem. Try to answer the questions, which you know
Table Ia. Differentially regulated genes in the arr2 loss-of-function mutant compared to wildtype
 i ANHANG Table Ia. Differentially regulated genes in the arr2 loss-of-function mutant compared to wildtype AGI No. Putative Function Fold Change a Abscisic acid-related AT4G24960 abscisic acid-induced
i ANHANG Table Ia. Differentially regulated genes in the arr2 loss-of-function mutant compared to wildtype AGI No. Putative Function Fold Change a Abscisic acid-related AT4G24960 abscisic acid-induced
Supporting Information
 Comparative Proteomic Study of Fatty Acid-treated Myoblasts Reveals Role of Cox-2 in Palmitate-induced Insulin Resistance Supporting Information Xiulan Chen 1#, Shimeng Xu 2,3#, Shasha Wei 1,3, Yaqin Deng
Comparative Proteomic Study of Fatty Acid-treated Myoblasts Reveals Role of Cox-2 in Palmitate-induced Insulin Resistance Supporting Information Xiulan Chen 1#, Shimeng Xu 2,3#, Shasha Wei 1,3, Yaqin Deng
number Done by Corrected by Doctor Nayef Karadsheh
 number 17 Done by Abdulrahman Alhanbali Corrected by Lara Abdallat Doctor Nayef Karadsheh 1 P a g e Pentose Phosphate Pathway (PPP) Or Hexose Monophosphate Shunt In this lecture We will talk about the
number 17 Done by Abdulrahman Alhanbali Corrected by Lara Abdallat Doctor Nayef Karadsheh 1 P a g e Pentose Phosphate Pathway (PPP) Or Hexose Monophosphate Shunt In this lecture We will talk about the
Lecture 11 - Biosynthesis of Amino Acids
 Lecture 11 - Biosynthesis of Amino Acids Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction Biosynthetic pathways for amino acids, nucleotides and lipids
Lecture 11 - Biosynthesis of Amino Acids Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction Biosynthetic pathways for amino acids, nucleotides and lipids
Protein Class/Name KEGG Pathways
 Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 4. Proteins Increased in Either Soil medium at 37 o C and 25 o C Table 4A. Proteins
Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 4. Proteins Increased in Either Soil medium at 37 o C and 25 o C Table 4A. Proteins
Lignin and the General Phenylpropanoid Pathway. Introduction and Importance:
 Lignin and the General Phenylpropanoid Pathway 13. Phenolics and Lignin p. 1 Introduction and Importance: Phenolic: a compound consisting of an aromatic ring plus at least one hydroxyl [= phenyl group],
Lignin and the General Phenylpropanoid Pathway 13. Phenolics and Lignin p. 1 Introduction and Importance: Phenolic: a compound consisting of an aromatic ring plus at least one hydroxyl [= phenyl group],
molecular function. The right hand y-axis indicates the number of annotated unigenes.
 Takemura et al. Supplementary Fig.S1 Supplementary Fig. S1. GO assignment of all unigenes. The unigenes were mapped to three main categories: iological process, cellular component and molecular function.
Takemura et al. Supplementary Fig.S1 Supplementary Fig. S1. GO assignment of all unigenes. The unigenes were mapped to three main categories: iological process, cellular component and molecular function.
Signal Transduction Cascades
 Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
SP GPI Found in exosomes
 Accession /Name/Functional group Spectr. counts SP GPI Found in exosomes Description Metabolism and energy production (21) CNAG_06770 fructose-bisphosphate aldolase 0.94 - - + gluconeogenesis and glycolysis
Accession /Name/Functional group Spectr. counts SP GPI Found in exosomes Description Metabolism and energy production (21) CNAG_06770 fructose-bisphosphate aldolase 0.94 - - + gluconeogenesis and glycolysis
BRIEF CONTENTS COPYRIGHTED MATERIAL III METABOLIC AND DEVELOPMENTAL INTEGRATION COMPARTMENTS CELL REPRODUCTION PLANT ENVIRONMENT AND AGRICULTURE
 BRIEF CONTENTS I COMPARTMENTS 1 Membrane Structure and Membranous Organelles 2 2 The Cell Wall 45 3 Membrane Transport 111 4 Protein Sorting and Vesicle Traffic 151 5 The Cytoskeleton 191 II CELL REPRODUCTION
BRIEF CONTENTS I COMPARTMENTS 1 Membrane Structure and Membranous Organelles 2 2 The Cell Wall 45 3 Membrane Transport 111 4 Protein Sorting and Vesicle Traffic 151 5 The Cytoskeleton 191 II CELL REPRODUCTION
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12864 Supplementary Table 1 1 2 3 4 5 6 7 Peak Gene code Screen Function or Read analysis AMP reads camp annotation reads minor Tb927.2.1810 AMP ISWI Confirmed
SUPPLEMENTARY INFORMATION doi:10.1038/nature12864 Supplementary Table 1 1 2 3 4 5 6 7 Peak Gene code Screen Function or Read analysis AMP reads camp annotation reads minor Tb927.2.1810 AMP ISWI Confirmed
Chapter 10. 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation)
 Chapter 10 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation) 1 Fueling Processes Respiration 1 Most respiration involves use of an electron transport chain As electrons pass through the electron
Chapter 10 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation) 1 Fueling Processes Respiration 1 Most respiration involves use of an electron transport chain As electrons pass through the electron
The Arabidopsis mir396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens
 The Arabidopsis mir396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens Mauricio Soto-Suárez, Patricia Baldrich, Detlef Weigel, Ignacio Rubio-Somoza and
The Arabidopsis mir396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens Mauricio Soto-Suárez, Patricia Baldrich, Detlef Weigel, Ignacio Rubio-Somoza and
Protein Class/Name. Pathways. bat00250, bat00280, bat00410, bat00640, bat bat00270, bat00330, bat00410, bat00480
 Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 1. Proteins Increased in Either Soil or Laboratory media Table 1A. Proteins Increased
Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 1. Proteins Increased in Either Soil or Laboratory media Table 1A. Proteins Increased
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the
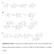 Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Lecture 13 (10/13/17)
 Lecture 13 (10/13/17) Reading: Ch6; 187-189, 204-205 Problems: Ch4 (text); 2, 3 NXT (after xam 2) Reading: Ch6; 190-191, 194-195, 197-198 Problems: Ch6 (text); 5, 6, 7, 24 OUTLIN NZYMS: Binding & Catalysis
Lecture 13 (10/13/17) Reading: Ch6; 187-189, 204-205 Problems: Ch4 (text); 2, 3 NXT (after xam 2) Reading: Ch6; 190-191, 194-195, 197-198 Problems: Ch6 (text); 5, 6, 7, 24 OUTLIN NZYMS: Binding & Catalysis
Protein Name. IFLENVIR,DSVTYTEHAK,TV TALDVVYALK,KTVTALDVV YALK,TVTALDVVYALKR,IF LENVIRDSVTYTEHAK gi Histone H2B
 Table 1. A functional category list of proteins (Lentinula edodes) identified by 1-DGE and nesi-lc-ms/ms. The table lists indicated fraction numbers, matching peptides, scores, accession numbers, protein
Table 1. A functional category list of proteins (Lentinula edodes) identified by 1-DGE and nesi-lc-ms/ms. The table lists indicated fraction numbers, matching peptides, scores, accession numbers, protein
SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS
 SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS No. Protein 1 serum albumin precursor gi 30794280 2 annexin A2 gi 27807289 3 Phosphatidylethanolamine-binding
SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS No. Protein 1 serum albumin precursor gi 30794280 2 annexin A2 gi 27807289 3 Phosphatidylethanolamine-binding
A CRISPR/Cas9-mediated gene editing to enhance the expression of the Solanum lycopersicum MYB12 gene and the nutritional value of tomato
 A CRISPR/Cas9-mediated gene editing to enhance the expression of the Solanum lycopersicum MYB12 gene and the nutritional value of tomato Dr. Aurelia Scarano Napoli, 22 Dicembre 2017 Phenylalanine Phenolic
A CRISPR/Cas9-mediated gene editing to enhance the expression of the Solanum lycopersicum MYB12 gene and the nutritional value of tomato Dr. Aurelia Scarano Napoli, 22 Dicembre 2017 Phenylalanine Phenolic
Front Plant Sci Mar 14
 Case Study Front Plant Sci. 2016 Mar 14 30 33 Study Background Cryptochromes (CRY) are blue-light photoreceptors that mediate various light responses in plants and animals. It is known that CRY1 and CRY2
Case Study Front Plant Sci. 2016 Mar 14 30 33 Study Background Cryptochromes (CRY) are blue-light photoreceptors that mediate various light responses in plants and animals. It is known that CRY1 and CRY2
De novo assembly of rubber transcriptome
 Project Title: De novo assembly of rubber transcriptome Name: Minami Matsui, Ng Kiaw Kiaw, Ong Wen Dee Laboratory at RIKEN: Synthetic Genomics Research Team, CSRS Background and Mission: Natural rubber
Project Title: De novo assembly of rubber transcriptome Name: Minami Matsui, Ng Kiaw Kiaw, Ong Wen Dee Laboratory at RIKEN: Synthetic Genomics Research Team, CSRS Background and Mission: Natural rubber
Catabolism of Carbon skeletons of Amino acids. Amino acid metabolism
 Catabolism of Carbon skeletons of Amino acids Amino acid metabolism Carbon skeleton Carbon Skeleton a carbon skeleton is the internal structure of organic molecules. Carbon Arrangements The arrangement
Catabolism of Carbon skeletons of Amino acids Amino acid metabolism Carbon skeleton Carbon Skeleton a carbon skeleton is the internal structure of organic molecules. Carbon Arrangements The arrangement
SI Materials Tomato fruit and treatment. Reference cited
 SI Materials Tomato fruit and treatment. Tomato Ailsa Craig, FLA 8059, Pearson and Nr mutant were grown in a heated greenhouse on the University of Florida campus. In order to evaluate the effect of cold
SI Materials Tomato fruit and treatment. Tomato Ailsa Craig, FLA 8059, Pearson and Nr mutant were grown in a heated greenhouse on the University of Florida campus. In order to evaluate the effect of cold
Validated Mouse Quantitative RT-PCR Genes Gene Gene Bank Accession Number Name
 5HT1a NM_008308.4 Serotonin Receptor 1a 5HT1b NM_010482.1 Serotonin Receptor 1b 5HT2a NM_172812.2 Serotonin Receptor 2a ACO-2 NM_080633.2 Aconitase 2 Adora2A NM_009630.02 Adenosine A2a Receptor Aif1 (IbaI)
5HT1a NM_008308.4 Serotonin Receptor 1a 5HT1b NM_010482.1 Serotonin Receptor 1b 5HT2a NM_172812.2 Serotonin Receptor 2a ACO-2 NM_080633.2 Aconitase 2 Adora2A NM_009630.02 Adenosine A2a Receptor Aif1 (IbaI)
Amino acid metabolism I
 Amino acid metabolism I Jana Novotná Department of the Medical Chemistry and Clinical Biochemistry The 2nd Faculty of Medicine, Charles Univ. Metabolic relationship of amino acids DIETARY PROTEINS GLYCOLYSIS
Amino acid metabolism I Jana Novotná Department of the Medical Chemistry and Clinical Biochemistry The 2nd Faculty of Medicine, Charles Univ. Metabolic relationship of amino acids DIETARY PROTEINS GLYCOLYSIS
University of Guelph Department of Chemistry and Biochemistry Structure and Function In Biochemistry
 University of Guelph Department of Chemistry and Biochemistry 19-356 Structure and Function In Biochemistry Final Exam, April 21, 1997. Time allowed, 120 min. Answer questions 1-30 on the computer scoring
University of Guelph Department of Chemistry and Biochemistry 19-356 Structure and Function In Biochemistry Final Exam, April 21, 1997. Time allowed, 120 min. Answer questions 1-30 on the computer scoring
Supporting Information
 Supporting Information Bar-Even et al. 10.1073/pnas.0907176107 Fig. S1. A scheme of the reductive pentose phosphate cycle, as inspired from (81). Dashed line corresponds to RUBISCO's oxygenase reaction
Supporting Information Bar-Even et al. 10.1073/pnas.0907176107 Fig. S1. A scheme of the reductive pentose phosphate cycle, as inspired from (81). Dashed line corresponds to RUBISCO's oxygenase reaction
Lecture 16. Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III
 Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
Supplemental Information
 Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
MBB 115:511 and 694:407 Final Exam Niederman/Deis
 MBB 115:511 and 694:407 Final Exam Niederman/Deis Tue. Dec. 19, 2006 Name Row Letter Seat Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is worth two points.
MBB 115:511 and 694:407 Final Exam Niederman/Deis Tue. Dec. 19, 2006 Name Row Letter Seat Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is worth two points.
Protein Class/Name. Pathways. bat00250, bat00280, bat bat00640, bat00650 bat00270, bat00330, bat00410, bat00480 bat00270, bat00330,
 Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 2. Proteins Increased in Either Blood or Horizon media Table 2A. Proteins Increased
Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 2. Proteins Increased in Either Blood or Horizon media Table 2A. Proteins Increased
Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease using MetPA analysis (panel A).
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease
Lecture: 26 OXIDATION OF FATTY ACIDS
 Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Name. The following exam contains 44 questions, valued at 2.6 points/question. 2. Which of the following is not a principal use of proteins?
 Chemistry 131 Exam 3 Practice Proteins, Enzymes, and Carbohydrates Spring 2018 Name The following exam contains 44 questions, valued at 2.6 points/question 1. Which of the following is a protein? a. Amylase
Chemistry 131 Exam 3 Practice Proteins, Enzymes, and Carbohydrates Spring 2018 Name The following exam contains 44 questions, valued at 2.6 points/question 1. Which of the following is a protein? a. Amylase
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 30 Amino Acid Degradation and the Urea Cycle 2013 W. H. Freeman and Company Chapter 30 Outline Amino acids are obtained from the
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 30 Amino Acid Degradation and the Urea Cycle 2013 W. H. Freeman and Company Chapter 30 Outline Amino acids are obtained from the
PCB 3023 Exam 4 - Form A First and Last Name
 PCB 3023 Exam 4 - Form A First and Last Name Student ID # (U Number) A Before beginning this exam, please complete the following instructions: 1) Write your name and U number on the first page of this
PCB 3023 Exam 4 - Form A First and Last Name Student ID # (U Number) A Before beginning this exam, please complete the following instructions: 1) Write your name and U number on the first page of this
Biochemistry 423 Final Examination NAME:
 Biochemistry 423 Final Examination NAME: 1 Circle the single BEST answer (3 points each) 1. At equilibrium the free energy of a reaction G A. depends only on the temperature B. is positive C. is 0 D. is
Biochemistry 423 Final Examination NAME: 1 Circle the single BEST answer (3 points each) 1. At equilibrium the free energy of a reaction G A. depends only on the temperature B. is positive C. is 0 D. is
Short polymer. Dehydration removes a water molecule, forming a new bond. Longer polymer (a) Dehydration reaction in the synthesis of a polymer
 HO 1 2 3 H HO H Short polymer Dehydration removes a water molecule, forming a new bond Unlinked monomer H 2 O HO 1 2 3 4 H Longer polymer (a) Dehydration reaction in the synthesis of a polymer HO 1 2 3
HO 1 2 3 H HO H Short polymer Dehydration removes a water molecule, forming a new bond Unlinked monomer H 2 O HO 1 2 3 4 H Longer polymer (a) Dehydration reaction in the synthesis of a polymer HO 1 2 3
NABIL KILLINY ASSISTANT PROFESSOR PLANT PATHOLOGY UNIVERSITY OF FLORIDA. Citrus responses to huanglongbing
 NABIL KILLINY ASSISTANT PROFESSOR PLANT PATHOLOGY UNIVERSITY OF FLORIDA Citrus responses to huanglongbing HLB is Vector-borne Disease! Vector psyllids Pathogen Ca. Liberibacter asiaticus. Host Citrus spp.
NABIL KILLINY ASSISTANT PROFESSOR PLANT PATHOLOGY UNIVERSITY OF FLORIDA Citrus responses to huanglongbing HLB is Vector-borne Disease! Vector psyllids Pathogen Ca. Liberibacter asiaticus. Host Citrus spp.
AMINO ACIDS NON-ESSENTIAL ESSENTIAL
 Edith Frederika Introduction A major component of food is PROTEIN The protein ingested as part of our diet are not the same protein required by the body Only 40 to 50 gr of protein is required by a normal
Edith Frederika Introduction A major component of food is PROTEIN The protein ingested as part of our diet are not the same protein required by the body Only 40 to 50 gr of protein is required by a normal
Biochemical networks related to carbon accumulation in sugarcane
 Biochemical networks related to carbon accumulation in sugarcane Marcos Buckeridge Department of Botany, Laboratory of Plant Physiological Ecology IB USP How are we approaching the sugarcane metabolic
Biochemical networks related to carbon accumulation in sugarcane Marcos Buckeridge Department of Botany, Laboratory of Plant Physiological Ecology IB USP How are we approaching the sugarcane metabolic
Proteins are sometimes only produced in one cell type or cell compartment (brain has 15,000 expressed proteins, gut has 2,000).
 Lecture 2: Principles of Protein Structure: Amino Acids Why study proteins? Proteins underpin every aspect of biological activity and therefore are targets for drug design and medicinal therapy, and in
Lecture 2: Principles of Protein Structure: Amino Acids Why study proteins? Proteins underpin every aspect of biological activity and therefore are targets for drug design and medicinal therapy, and in
Comparative proteomic analysis provides novel insight into the interaction between resistant vs susceptible tomato cultivars and TYLCV infection
 Huang et al. BMC Plant Biology (2016) 16:162 DOI 10.1186/s12870-016-0819-z RESEARCH ARTICLE Comparative proteomic analysis provides novel insight into the interaction between resistant vs susceptible tomato
Huang et al. BMC Plant Biology (2016) 16:162 DOI 10.1186/s12870-016-0819-z RESEARCH ARTICLE Comparative proteomic analysis provides novel insight into the interaction between resistant vs susceptible tomato
BOT 6516 Plant Metabolism
 BOT 6516 Plant Metabolism Lecture 22 Natural Products Slide sets available at: http://hort.ifas.ufl.edu/teach/guyweb/bot6516/index.html Some Big Ideas and Aspirations for Plant Natural Products Why is
BOT 6516 Plant Metabolism Lecture 22 Natural Products Slide sets available at: http://hort.ifas.ufl.edu/teach/guyweb/bot6516/index.html Some Big Ideas and Aspirations for Plant Natural Products Why is
Comparative Transcriptome Profiling of Two Tomato Genotypes in Response to Potassium-Deficiency Stress
 Article Comparative Transcriptome Profiling of Two Tomato Genotypes in Response to Potassium-Deficiency Stress Xiaoming Zhao 1,2, Yang Liu 1, Xin Liu 1, * and Jing Jiang 1, * 1 The Key Laboratory of Protected
Article Comparative Transcriptome Profiling of Two Tomato Genotypes in Response to Potassium-Deficiency Stress Xiaoming Zhao 1,2, Yang Liu 1, Xin Liu 1, * and Jing Jiang 1, * 1 The Key Laboratory of Protected
1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled
 Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
If you ate a clown, would it taste funny? Oh, wait, that s cannibalism . Anabolism
 If you ate a clown, would it taste funny? Oh, wait, that s cannibalism. Anabolism is about putting things together. Anabolism: The Use of Energy in Biosynthesis Anabolism energy from catabolism is used
If you ate a clown, would it taste funny? Oh, wait, that s cannibalism. Anabolism is about putting things together. Anabolism: The Use of Energy in Biosynthesis Anabolism energy from catabolism is used
Cell Signaling part 2
 15 Cell Signaling part 2 Functions of Cell Surface Receptors Other cell surface receptors are directly linked to intracellular enzymes. The largest family of these is the receptor protein tyrosine kinases,
15 Cell Signaling part 2 Functions of Cell Surface Receptors Other cell surface receptors are directly linked to intracellular enzymes. The largest family of these is the receptor protein tyrosine kinases,
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 31 Amino Acid Synthesis 2013 W. H. Freeman and Company Chapter 31 Outline Although the atmosphere is approximately 80% nitrogen,
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 31 Amino Acid Synthesis 2013 W. H. Freeman and Company Chapter 31 Outline Although the atmosphere is approximately 80% nitrogen,
Fatty acid breakdown
 Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
Chapter 23 Enzymes 1
 Chapter 23 Enzymes 1 Enzymes Ribbon diagram of cytochrome c oxidase, the enzyme that directly uses oxygen during respiration. 2 Enzyme Catalysis Enzyme: A biological catalyst. With the exception of some
Chapter 23 Enzymes 1 Enzymes Ribbon diagram of cytochrome c oxidase, the enzyme that directly uses oxygen during respiration. 2 Enzyme Catalysis Enzyme: A biological catalyst. With the exception of some
Microbial Metabolism. PowerPoint Lecture Presentations prepared by Bradley W. Christian, McLennan Community College C H A P T E R
 PowerPoint Lecture Presentations prepared by Bradley W. Christian, McLennan Community College C H A P T E R 5 Microbial Metabolism Big Picture: Metabolism Metabolism is the buildup and breakdown of nutrients
PowerPoint Lecture Presentations prepared by Bradley W. Christian, McLennan Community College C H A P T E R 5 Microbial Metabolism Big Picture: Metabolism Metabolism is the buildup and breakdown of nutrients
endomembrane system internal membranes origins transport of proteins chapter 15 endomembrane system
 endo system chapter 15 internal s endo system functions as a coordinated unit divide cytoplasm into distinct compartments controls exocytosis and endocytosis movement of molecules which cannot pass through
endo system chapter 15 internal s endo system functions as a coordinated unit divide cytoplasm into distinct compartments controls exocytosis and endocytosis movement of molecules which cannot pass through
Syllabus for BASIC METABOLIC PRINCIPLES
 Syllabus for BASIC METABOLIC PRINCIPLES The video lecture covers basic principles you will need to know for the lectures covering enzymes and metabolism in Principles of Metabolism and elsewhere in the
Syllabus for BASIC METABOLIC PRINCIPLES The video lecture covers basic principles you will need to know for the lectures covering enzymes and metabolism in Principles of Metabolism and elsewhere in the
Supplemental Data. Pick and Bräutgam et al. Plant Cell. (2011) /tpc Mean Adjustment FOM values (± SD) vs. Number of Clusters
 Mean Adjustment FOM values (± SD) vs. Number of Clusters Mean Adjustment FOM Number of Clusters Supplemental Figure 1. Figure of merit analysis of metabolite clustering. The algorithm tries 20 times to
Mean Adjustment FOM values (± SD) vs. Number of Clusters Mean Adjustment FOM Number of Clusters Supplemental Figure 1. Figure of merit analysis of metabolite clustering. The algorithm tries 20 times to
Metabolism. Metabolic pathways. BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 11: Metabolic Pathways
 BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 11: Metabolic Pathways http://compbio.uchsc.edu/hunter/bio5099 Larry.Hunter@uchsc.edu Metabolism Metabolism is the chemical change of
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 11: Metabolic Pathways http://compbio.uchsc.edu/hunter/bio5099 Larry.Hunter@uchsc.edu Metabolism Metabolism is the chemical change of
Glycolysis. Cellular Respiration
 Glucose is the preferred carbohydrate of cells. In solution, it can change from a linear chain to a ring. Energy is stored in the bonds of the carbohydrates. Breaking these bonds releases that energy.
Glucose is the preferred carbohydrate of cells. In solution, it can change from a linear chain to a ring. Energy is stored in the bonds of the carbohydrates. Breaking these bonds releases that energy.
III. Metabolism The Citric Acid Cycle
 Department of Chemistry and Biochemistry University of Lethbridge III. Metabolism The Citric Acid Cycle Slide 1 The Eight Steps of the Citric Acid Cycle Enzymes: 4 dehydrogenases (2 decarboxylation) 3
Department of Chemistry and Biochemistry University of Lethbridge III. Metabolism The Citric Acid Cycle Slide 1 The Eight Steps of the Citric Acid Cycle Enzymes: 4 dehydrogenases (2 decarboxylation) 3
BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001
 BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) The
BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) The
Bio Metabolism. Metabolism Life is a bag of biochemistry. Chloroplasts and mitochondria. What is food?
 Metabolism Life is a bag of biochemistry 1 Chloroplasts and mitochondria Heat Carbohydrate O 2 CO 2 + H 2 O Heat Chloroplast Mitochondria 2 What is food? Proteins - polymers of amino acids Carbohydrates
Metabolism Life is a bag of biochemistry 1 Chloroplasts and mitochondria Heat Carbohydrate O 2 CO 2 + H 2 O Heat Chloroplast Mitochondria 2 What is food? Proteins - polymers of amino acids Carbohydrates
Cells. Variation and Function of Cells
 Cells Variation and Function of Cells Plasma Membrane= the skin of a cell, it protects and nourishes the cell while communicating with other cells at the same time. Lipid means fat and they are hydrophobic
Cells Variation and Function of Cells Plasma Membrane= the skin of a cell, it protects and nourishes the cell while communicating with other cells at the same time. Lipid means fat and they are hydrophobic
Integration of Metabolism
 Integration of Metabolism Metabolism is a continuous process. Thousands of reactions occur simultaneously in order to maintain homeostasis. It ensures a supply of fuel, to tissues at all times, in fed
Integration of Metabolism Metabolism is a continuous process. Thousands of reactions occur simultaneously in order to maintain homeostasis. It ensures a supply of fuel, to tissues at all times, in fed
MBBS First Professional (Part-I Examination) BIOCHEMISTRY Model Paper
 MBBS FIRST PROFESSIONAL (PART I) Biochemistry (MCQs) Maximum Marks: 35 Time Allowed: 45 minutes Q 1. What is not correct about cell membrane? A. Contains two layers of phospholipids B. May contain receptor
MBBS FIRST PROFESSIONAL (PART I) Biochemistry (MCQs) Maximum Marks: 35 Time Allowed: 45 minutes Q 1. What is not correct about cell membrane? A. Contains two layers of phospholipids B. May contain receptor
PROTEIN METABOLISM: NITROGEN CYCLE; DIGESTION OF PROTEINS. Red meat is an important dietary source of protein nitrogen
 PROTEIN METABOLISM: NITROGEN CYCLE; DIGESTION OF PROTEINS Red meat is an important dietary source of protein nitrogen The Nitrogen Cycle and Nitrogen Fixation Nitrogen is needed for amino acids, nucleotides,
PROTEIN METABOLISM: NITROGEN CYCLE; DIGESTION OF PROTEINS Red meat is an important dietary source of protein nitrogen The Nitrogen Cycle and Nitrogen Fixation Nitrogen is needed for amino acids, nucleotides,
Endoplasmic Reticulum
 Endoplasmic Reticulum What s ER? How is ER? Why is ER? definition description functions Nissl s bodies neurons Berg s bodies hepatocytes Organelle structure histocytochemical evidences Ergastoplasm pancreatic
Endoplasmic Reticulum What s ER? How is ER? Why is ER? definition description functions Nissl s bodies neurons Berg s bodies hepatocytes Organelle structure histocytochemical evidences Ergastoplasm pancreatic
Points 1. Following is the overall reaction catalyzed by the Calvin-Benson cycle:
 BCH 4054 February 22, 2002 HOUR TEST 2 NAME_ Points 1. Following is the overall reaction catalyzed by the Calvin-Benson cycle: CO 2 + 3ATP + 2NADPH 1/3 glyceraldehyde-3-p + 3ADP + 2NADP + Give the structures
BCH 4054 February 22, 2002 HOUR TEST 2 NAME_ Points 1. Following is the overall reaction catalyzed by the Calvin-Benson cycle: CO 2 + 3ATP + 2NADPH 1/3 glyceraldehyde-3-p + 3ADP + 2NADP + Give the structures
SDR families. SDR10E Fatty acyl-coa reductase FACR1_HUMAN 544 x x SDR11E 3 beta-hydroxysteroid dehydrogenase 3BHS1_HUMAN 254 x x
 SDR1E UDP-glucose 4-epimerase GALE_HUMAN 5305 x x x x SDR2E dtdp-d-glucose 4,6-dehydratase TGDS_HUMAN 4194 x x x x SDR3E GDP-mannose 4,6 dehydratase GMDS_HUMAN 2314 x x x x SDR4E GDP-L-fucose synthetase
SDR1E UDP-glucose 4-epimerase GALE_HUMAN 5305 x x x x SDR2E dtdp-d-glucose 4,6-dehydratase TGDS_HUMAN 4194 x x x x SDR3E GDP-mannose 4,6 dehydratase GMDS_HUMAN 2314 x x x x SDR4E GDP-L-fucose synthetase
Dental Students Biochemistry Exam V Questions ( Note: In all cases, the only correct answer is the best answer)
 Dental Students Biochemistry Exam V Questions - 2006 ( Note: In all cases, the only correct answer is the best answer) 1. Essential fatty acids are: A. precursors of biotin B. precursors of tyrosine C.
Dental Students Biochemistry Exam V Questions - 2006 ( Note: In all cases, the only correct answer is the best answer) 1. Essential fatty acids are: A. precursors of biotin B. precursors of tyrosine C.
Proteins consist in whole or large part of amino acids. Simple proteins consist only of amino acids.
 Today we begin our discussion of the structure and properties of proteins. Proteins consist in whole or large part of amino acids. Simple proteins consist only of amino acids. Conjugated proteins contain
Today we begin our discussion of the structure and properties of proteins. Proteins consist in whole or large part of amino acids. Simple proteins consist only of amino acids. Conjugated proteins contain
Hand in the Test Sheets (with the checked multiple choice answers) and your Sheets with written answers.
 Page 1 of 13 IMPORTANT INFORMATION Hand in the Test Sheets (with the checked multiple choice answers) and your Sheets with written answers. THE exam has an 'A' and 'B' section SECTION A (based on Dykyy's
Page 1 of 13 IMPORTANT INFORMATION Hand in the Test Sheets (with the checked multiple choice answers) and your Sheets with written answers. THE exam has an 'A' and 'B' section SECTION A (based on Dykyy's
Lecture 16. Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III
 Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
SUPPLEMENTARY INFORMATION. A comparative study of outer membrane proteome between a paired colistinsusceptible
 SUPPLEMENTARY INFORMATION A comparative study of outer membrane proteome between a paired colistinsusceptible and extremely colistin-resistant Klebsiella pneumoniae strains Raad Jasim, Mark A. Baker, 2
SUPPLEMENTARY INFORMATION A comparative study of outer membrane proteome between a paired colistinsusceptible and extremely colistin-resistant Klebsiella pneumoniae strains Raad Jasim, Mark A. Baker, 2
BIOLOGY 103 Spring 2001 MIDTERM LAB SECTION
 BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
Insulin mrna to Protein Kit
 Insulin mrna to Protein Kit A 3DMD Paper BioInformatics and Mini-Toober Folding Activity Student Handout www.3dmoleculardesigns.com Insulin mrna to Protein Kit Contents Becoming Familiar with the Data...
Insulin mrna to Protein Kit A 3DMD Paper BioInformatics and Mini-Toober Folding Activity Student Handout www.3dmoleculardesigns.com Insulin mrna to Protein Kit Contents Becoming Familiar with the Data...
U118MG. Supplementary Figure 1 U373MG U118MG 3.5 A CCF-SSTG
 A172 CCF-SSTG1 15 - - - 1 1 1 2 2 3 4 6 7 8 10101112131617192022-1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22 T98G U373MG - - - 1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22-1 1 1 2 2 3 4 6 7
A172 CCF-SSTG1 15 - - - 1 1 1 2 2 3 4 6 7 8 10101112131617192022-1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22 T98G U373MG - - - 1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22-1 1 1 2 2 3 4 6 7
Eukaryotic mrna is covalently processed in three ways prior to export from the nucleus:
 RNA Processing Eukaryotic mrna is covalently processed in three ways prior to export from the nucleus: Transcripts are capped at their 5 end with a methylated guanosine nucleotide. Introns are removed
RNA Processing Eukaryotic mrna is covalently processed in three ways prior to export from the nucleus: Transcripts are capped at their 5 end with a methylated guanosine nucleotide. Introns are removed
Under aerobic conditions, pyruvate enters the mitochondria where it is converted into acetyl CoA.
 Under aerobic conditions, pyruvate enters the mitochondria where it is converted into acetyl CoA. Acetyl CoA is the fuel for the citric acid cycle, which processes the two carbon acetyl unit to two molecules
Under aerobic conditions, pyruvate enters the mitochondria where it is converted into acetyl CoA. Acetyl CoA is the fuel for the citric acid cycle, which processes the two carbon acetyl unit to two molecules
