Supplemental Table S1 Up-regulated genes in irjazh plants compared to WT plants by microarray.
|
|
|
- Opal Howard
- 5 years ago
- Views:
Transcription
1 Supplemental Table S1 Up-regulated genes in irjazh plants compared to WT plants by microarray. Classification Fold Change Bincode (TAIR) Annotation Probe number ACC number Cell wall. Degradation Glycosyl hydrolase family 3 protein CUST_146896_PI NP_ Lipid metabolism Putative trans-2-enoyl-coa reductase CUST_75436_PI NP_ Putative trans-2-enoyl-coa reductase CUST_33708_PI NP_ Lipase class 3 family protein CUST_160928_PI NP_ Squalene synthase CUST_163980_PI AAM Gamma-tocopherol methyltransferase CUST_112272_PI ABE Acyl-[acyl-carrier-protein] desaturase CUST_41928_PI P Glycerolipase A1 CUST_160212_PI ACZ Sphingolipid delta-8 desaturase CUST_80400_PI ABO31111 Amino acid metabolism Cyclase CUST_99572_PI AAS46038 Secondary metabolism epi-aristolochene synthase CUST_46640_PI AAO epi-aristolochene synthase (EAS) CUST_34180_PI Q Plastid 1,8-cineol synthase precursor CUST_12108_PI ABP epi-aristolochene synthase 34 CUST_173668_PI AAP05761
2 Hormone metabolism SAMT(salicylic acid methyltransferase) CUST_33280_PI AAW Short chain alcohol dehydrogenase CUST_42160_PI CAA Ethylene responsive element binding factor 1 CUST_115772_PI BAA32418 Tetrapyrrole synthesis ATCLH2; chlorophyllase CUST_94136_PI NP_ Stress Gamma-thionin (Plant defensins) CUST_57516_PI AAS Gamma-thionin (Plant defensins) CUST_8140_PI AAS Kunitz trypsin inhibitor CUST_14168_PI ACL domain trypsin inhibitor precursor CUST_9024_PI AAQ Proteinase inhibitor I-B (PI-IB) CUST_11524_PI Q Kunitz trypsin inhibitor CUST_79416_PI ACL Miraculin-like protein CUST_8752_PI ADK62529 Miscellaneous enzyme families CYP71D5v3 CUST_103900_PI ABC CYP71D47v1 CUST_102916_PI ABC Cytochrome P-450-like protein CUST_143016_PI BAB FAD-binding domain-containing protein CUST_127228_PI NP_ UDP-glucoronosyl/UDP-glucosyl transferase family protein CUST_43496_PI NP_ GDSL-motif lipase/hydrolase family protein CUST_64232_PI NP_563774
3 RNA. Regulation of transcription ATNAC2 (Arabidopsis NAC domain containing protein 2) CUST_14684_PI NP_ Basic helix-loop-helix (bhlh) family protein CUST_157324_PI NP_ ARR4 (Arabidospsis regulation regulator 4) CUST_56432_PI NP_ DNA.synthesis Histone H1 CUST_32040_PI AAC41651 Protein. Degradation AAA-type ATPase family protein CUST_76132_PI NP_ Putative metallocarboxypeptidase inhibitor precursor CUST_139476_PI BAJ Putative metallocarboxypeptidase inhibitor precursor CUST_31104_PI BAJ F-box family protein CUST_86652_PI NP_ RING/U-box domain-containing protein CUST_21448_PI AAG Putative metallocarboxypeptidase inhibitor precursor CUST_40216_PI BAJ AAA-type ATPase family protein CUST_78344_PI NP_ Cell.organisation AtPP2-A11 (Phloem protein 2-A11) CUST_73344_PI NP_ Development Tumor-related protein CUST_16488_PI BAA Patatin homolog CUST_168304_PI AAB Patatin CUST_1460_PI AAB Patatin homolog CUST_81764_PI AAB Patatin homolog CUST_142932_PI AAB Transducin family protein / WD-40 repeat family protein CUST_170740_PI NP_190535
4 Transport Mitochondrial carrier protein CUST_124384_PI ABO ATP-binding cassette transporter, putative CUST_87856_PI XP_ Nitrate transporter CUST_171856_PI BAC WBC11 (White-Brown Complex homolog protein 11) CUST_163156_PI NP_ Not assigned Phosphorylase family protein CUST_71848_PI NP_ FAD-binding domain-containing protein CUST_37768_PI NP_ JAZ8 (Jsmonate ZIM domain protein 8) CUST_80752_PI NP_ JAZ3 (Jasmonate ZIM-domain protein 3) CUST_52900_PI BAG Glycine-rich protein CUST_69052_PI BAA95941 * Microarray data is processed by SAM after 75% percentile normalization (FDR=4.82). * Up-regulated genes were determined by a greater than 3-fold induction of normalized signals in their expression ratio (irjazh / WT) of 2h-WOS treated leaves. The values are the average ratio of 3 biological replicates of the microarrays. * All changes in gene expression were statistically significant by t-test (P < 0.05) * Gene annotation is processed by Blast X (E-value < 1e-5) * Classification of genes is based on GO classification from TAIR (
5 Supplemental Table S2 Down-regulated genes in irjazh plants compared to WT plants by microarray. Calssification Fold Change Bincode (TAIR) Annotation Probe number ACC number Photosynthesis Photosystem I P700 apoprotein A2 CUST_32628_PI ABB Glycolate oxidase CUST_6068_PI ADM26718 Major Carbohydrates Alpha-amylase CUST_12856_PI ACZ26470 Lipid metabolism Acyltransferase-like protein CUST_12976_PI BAD Acyltransferase-like protein CUST_134584_PI BAD93693 Metal handling Vacuolar iron transporter-like protein CUST_26436_PI NP_ Ferric-chelate reductase CUST_64152_PI AAP Metallothionein-like protein type 2 CUST_144048_PI CAC12823 Secondary metabolism Sesquiterpene synthase CUST_166968_PI NP_ Plastid 1,8-cineol synthase precursor CUST_44188_PI ABP Plastid 1,8-cineol synthase precursor CUST_20468_PI ABP Plastid 1,8-cineol synthase precursor CUST_14120_PI ABP Monoterpene synthase 2 CUST_165928_PI AAX69064
6 Terpene cyclase/mutase-related CUST_11104_PI NP_ Hormone metabolism SAUR-like auxin-responsive protein CUST_59948_PI NP_ Cytokinin-O-glucosyltransferase 2 CUST_173124_PI NP_ ERF transcription factor 5 CUST_116820_PI AAU81956 Stress Early dehydration inducible protein CUST_55152_PI AAR Salt responsive protein 1 CUST_45184_PI ACG CC-NBS-LRR putative disease resistance protein CUST_167984_PI ACB Pathogenesis-related protein CUST_15548_PI BAD15090 Miscellaneous enzyme families Elicitor-inducible cytochrome P450 CUST_25520_PI AAK Elicitor-inducible cytochrome P450 CUST_96052_PI AAK Short-chain dehydrogenase/reductase family protein CUST_122964_PI XP_ Beta-galactosidase STBG5 CUST_18760_PI ADO Beta-galactosidase STBG5 CUST_888_PI ADO CYP71D49v2 CUST_119780_PI ABC69401 RNA. regulation of transcription Zinc finger protein 4 CUST_73116_PI NP_ Class 2 knotted1-like protein CUST_139276_PI BAF bzip transcription factor BZI-2 CUST_107720_PI AAK Basic helix-loop-helix (bhlh) family protein CUST_56184_PI NP_181549
7 Transcription factor style2.1 CUST_33216_PI ABX Ribonuclease H-like protein CUST_79632_PI NP_ RNase H family protein CUST_121612_PI ABI Transcription factor style2.1 CUST_163752_PI ABX82930 Protein. degradation Class S F-box protein CUST_152204_PI ABR18786 Signaling Transducin/WD40 domain-containing protein CUST_21460_PI NP_ Calcineurin B-like protein CUST_107040_PI ABQ23353 Development Squamosa promoter-binding-like protein 12 CUST_109112_PI NP_ Nodulin MtN21 family protein CUST_29984_PI NP_ Senescence-associated protein-related CUST_6080_PI NP_ Fruitfull-like MADS-box protein CUST_42876_PI ABF NAC domain protein CUST_81900_PI AAU Putative gag polyprotein, identical CUST_81856_PI AAT Ripening regulated protein DDTFR18 CUST_97944_PI AAG49032 Transport Nitrate transporter (NTL1) CUST_131240_PI AAG Metal transporter CUST_159740_PI AAP21819
8 Not assigned Oxidoreductase family protei CUST_25856_PI NP_ Transducin family protein / WD-40 repeat family protein CUST_54908_PI NP_ Jasmonate ZIM-domain protein 1 CUST_40368_PI BAG GC-rich sequence DNA-binding factor CUST_122872_PI NP_ Serine-threonine protein kinase, plant-type, putative CUST_171656_PI XP_ * Microarray data is processed by SAM after 75% percentile normalization (FDR=4.82). * Down-regulated genes were determined by a greater than 3-fold repression of normalized signals in their expression ratio (irjazh / WT) of 2h-WOS treated leaves. The values are the average ratio of 3 biological replicates of the microarrays. * All changes in gene expression were statistically significant by t-test (P < 0.05) * Gene annotation is processed by Blast X (E-value < 1e-5) * Classification of genes is based on GO classification from TAIR (
9 Supplemental Table S3. Primer sequences used for cloning of JAZ genes from N. attenuata Gene name Primer sequences NtJAZa-FP NtJAZa-RP NtJAZb-FP NtJAZb-RP NtJAZc-FP NtJAZc-RP NaJAZc.1-FP & NaJAZc.2-RP & NtJAZd-FP-1* NtJAZd-FP-2* NtJAZe-FP NtJAZe-RP NtJAZf-FP NtJAZf-RP NtJAZp-FP NtJAZp-RP NtJAZh-FP NtJAZh-RP NtJAZj-FP NtJAZj-RP NtJAZk-FP NtJAZk-RP NaJAZc.1-FP & NaJAZc.2-RP & NtJAZl-FP-1* NtJAZl-FP-2* NtJAZm-FP-1* NtJAZm-FP-2* GACAGAAAAAGCAAAATATTTACAA ACAATAGGAAGTAAAAACTTAAGCC GTAATCCTAAATCTTTGAGAAAGTG AATTCAAAAATCCATTTGCCTTGAA TTTCTGGGGTTTGGTGTTTTGAGC GATGAAAGAGAATTAAGTACAGC CATTCAAAGCTCAATCCGC CGATGGAAGTATTGAGTGAGG TATCTTTCTCAAGCTTATTACAGATTCTTG ATTTAGTTTTTTGCCACCGGAAAACAAA TAAGTGAAAATAATGGAGAGAGATT TACAAAAGGCTCGTAGTGTTTTCAGATG TTGGCTGAAGTTGAATAAAG CTAGCATCATTCATCCATTATATTTCC ATTCTGTCAGATATATTTCAAGTTTCTG TCAATAGTTCACTCTCTTTGACACTA CAACTTGTTCAGTTGTAAAATTGTAG CATGGAATTGCAACTGGGGGACTCTTTA TTGTGGCTGAAGTTAAACGAAG CAAAATATATGTACAAATGGAACTA TTAAACGCCGGAACTTGACAGC GCAATATGGACAATGATACTGCC CAGAAACTCCATTGAATGCTAC ACCTGTCTTTTCGCTTCTCA GCACAGACTGAGAAAATGTATTGC ATGTATTGCAGCTCCAAAGTGGC AAGGAAGAAAACAATAGGCATAAAG ATAAGCTAAGCAATTCCCCCACATTG * cloned as 3'RACE product & Primers used to confirm alternative splicing form of NaJAZc and JAZk transcripts.
10 Supplemental Table S4. Primer sequences used in quantitative real time PCR of NaJAZ genes Gene name Primer sequences NaJAZa FP NaJAZa RP NaJAZb FP NaJAZb RP NaJAZc FP NaJAZc RP NaJAZd FP NaJAZd RP NaJAZe FP NaJAZe RP NaJAZf FP NaJAZf RP NaJAZg FP NaJAZg RP NaJAZh FP NaJAZh RP NaJAZj FP NaJAZj RP NaJAZk FP NaJAZk RP NaJAZl FP NaJAZl RP NaJAZm FP NaJAZm RP CAGTGAAAGCTGAGCAATTCTAGTACTC AGCCTTAGACGAATTGAATACCTACAC GGGAAAGCTAATTCAAGACAAATGG TGTATTCTTTAGCCACCAATCTAGC GGAAAGGGTGATGAACGCTGCA TATGGCAATAGGCTGCCTTCAGAC GCCTTGGCGTAACTGATGAAGTTG GGCAGCCCAGTGCACTAAGC CAATTTGGTCAAGGAGACGTGA GGCTCTGATCACAATTACAAGG CAAGTAGAGAAATGGAGGAG GCTAGTGATGATATGGAGAAG CAAAGCATCCTTGGCTCGGTTC CGCATCTACATTGACTTGCCATC TTCTGCTACGCCGCAAGTACTG GGTATGGCGCTCTAGCCGTTG CATCATCACCAATTTCAGAGCCTTC TCCAATTTTCCAATTTCTCCCAGCA TCTATGGTGATGTGCCTGCTGAC AACGGATATCCAAGCTAGCTGTTG TTGCCAGAAGGAAATCCCTGAAGAG TCCATCAAAAGCTAGCCCTACTTAGC AGTGCGTCAAATTTGAGAGCACCA GCTGCTTGAATCCTCCTTTCTCTTC
11 Supplemental Table S5. Primer sequences used in quantitative real time PCR experiments Gene name Primer sequences Hsr203 FP Hsr203 RP Hin1 FP Hin1 RP NaVPE361 FP NaVPE361 RP CAC TGT CTA CAC GCG CCT A GAG GAG GGC GGC GAA ACC GCGTCCAGTATTCAAAGGTCA CGCATGTAAAGCTTCACTTCC GCTGAAGCGTGTTCAACCTGTA GTGAGACTGCAAATTCCATGGCGT
Supplementary Tables 1
 Supplementary Tables 1 Supplementary Table 1: The 11 genes in the geneset for Case Study 1 At1g62570 At1g09350 At1g60470 At2g47180 At4g17090 At5g20830 At2g16890 At3g51240 At3g55120 At4g27560 At5g08640
Supplementary Tables 1 Supplementary Table 1: The 11 genes in the geneset for Case Study 1 At1g62570 At1g09350 At1g60470 At2g47180 At4g17090 At5g20830 At2g16890 At3g51240 At3g55120 At4g27560 At5g08640
WT siz1-2 siz1-3. WT siz1-2 siz1-3. -Pi, 0.05 M IAA. -Pi, 2.5 M NPA
 A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
Supplementary Data Transcriptome analysis in response to UV-B stress in soybean [Glycine max (L.) Merr.]
![Supplementary Data Transcriptome analysis in response to UV-B stress in soybean [Glycine max (L.) Merr.] Supplementary Data Transcriptome analysis in response to UV-B stress in soybean [Glycine max (L.) Merr.]](/thumbs/85/91947206.jpg) AJCS 6(9):195-1400 (2012) ISSN:185-27 Supplementary Data Transcriptome analysis in response to UV-B stress in soybean [Glycine max (L.) Merr.] Jun-Cheol Moon 1,2, Won Cheol Yim 1, Jae-Eun Lee, Young-Up
AJCS 6(9):195-1400 (2012) ISSN:185-27 Supplementary Data Transcriptome analysis in response to UV-B stress in soybean [Glycine max (L.) Merr.] Jun-Cheol Moon 1,2, Won Cheol Yim 1, Jae-Eun Lee, Young-Up
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function
 Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Supplemental Data. Shin et al. Plant Cell. (2012) /tpc YFP N
 MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
Mouse Meda-4 : chromosome 5G bp. EST (547bp) _at. 5 -Meda4 inner race (~1.8Kb)
 Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
 www.lessonplansinc.com Topic: Protein Synthesis - Sentence Activity Summary: Students will simulate transcription and translation by building a sentence/polypeptide from words/amino acids. Goals & Objectives:
www.lessonplansinc.com Topic: Protein Synthesis - Sentence Activity Summary: Students will simulate transcription and translation by building a sentence/polypeptide from words/amino acids. Goals & Objectives:
Supplementary Table 3. 3 UTR primer sequences. Primer sequences used to amplify and clone the 3 UTR of each indicated gene are listed.
 Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Chemistry 107 Exam 4 Study Guide
 Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
f(x) = x R² = RPKM (M8.MXB) f(x) = x E-014 R² = 1 RPKM (M31.
 14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
A basic helix loop helix transcription factor controls cell growth
 A basic helix loop helix transcription factor controls cell growth and size in root hairs Keke Yi 1,2, Benoît Menand 1,3, Elizabeth Bell 1, Liam Dolan 1,4 Supplementary note Low soil phosphate availability
A basic helix loop helix transcription factor controls cell growth and size in root hairs Keke Yi 1,2, Benoît Menand 1,3, Elizabeth Bell 1, Liam Dolan 1,4 Supplementary note Low soil phosphate availability
MG132. GRXS17:V5-His MG132 GRXS17:HA. RuBisCo. Supplemental Data. Walton et al. (2015). Plant Cell /tpc
 MG132 GRXS17:V5-His MG132 GRXS17:HA RuBisCo Supplemental Figure 1. Degradation of GRXS17 by the 26S proteasome. (A) Cell-free degradation assay. Recombinant protein GRXS17:V5-HIS was incubated with total
MG132 GRXS17:V5-His MG132 GRXS17:HA RuBisCo Supplemental Figure 1. Degradation of GRXS17 by the 26S proteasome. (A) Cell-free degradation assay. Recombinant protein GRXS17:V5-HIS was incubated with total
SSH fw: 5 TCGAGCGGCCGCCCGGGCAGGT 3 SSH rev: 5 AGCGTGGTCGCGGCCGAGGT 3. AY (Actin) 5 GAGAACTGGCATCACACCTT 3 5 AGCATGAGGAAGTGTGTATC 3
 ONLINE SUPPLEMENT 1: Oligonucleotide primers for reverse RNA gel blot: SSH fw: 5 TCGAGCGGCCGCCCGGGCAGGT 3 SSH rev: 5 AGCGTGGTCGCGGCCGAGGT 3 AY104722 (Actin) 5 GAGAACTGGCATCACACCTT 3 5 AGCATGAGGAAGTGTGTATC
ONLINE SUPPLEMENT 1: Oligonucleotide primers for reverse RNA gel blot: SSH fw: 5 TCGAGCGGCCGCCCGGGCAGGT 3 SSH rev: 5 AGCGTGGTCGCGGCCGAGGT 3 AY104722 (Actin) 5 GAGAACTGGCATCACACCTT 3 5 AGCATGAGGAAGTGTGTATC
Figure S1. Analysis of genomic and cdna sequences of the targeted regions in WT-KI and
 Figure S1. Analysis of genomic and sequences of the targeted regions in and indicated mutant KI cells, with WT and corresponding mutant sequences underlined. (A) cells; (B) K21E-KI cells; (C) D33A-KI cells;
Figure S1. Analysis of genomic and sequences of the targeted regions in and indicated mutant KI cells, with WT and corresponding mutant sequences underlined. (A) cells; (B) K21E-KI cells; (C) D33A-KI cells;
number Done by Corrected by Doctor Faisal Al-Khatibe
 number 24 Done by Mohammed tarabieh Corrected by Doctor Faisal Al-Khatibe 1 P a g e *Please look over the previous sheet about fatty acid synthesis **Oxidation(degradation) of fatty acids, occurs in the
number 24 Done by Mohammed tarabieh Corrected by Doctor Faisal Al-Khatibe 1 P a g e *Please look over the previous sheet about fatty acid synthesis **Oxidation(degradation) of fatty acids, occurs in the
Table S1. Oligonucleotides used for the in-house RT-PCR assays targeting the M, H7 or N9. Assay (s) Target Name Sequence (5 3 ) Comments
 SUPPLEMENTAL INFORMATION 2 3 Table S. Oligonucleotides used for the in-house RT-PCR assays targeting the M, H7 or N9 genes. Assay (s) Target Name Sequence (5 3 ) Comments CDC M InfA Forward (NS), CDC M
SUPPLEMENTAL INFORMATION 2 3 Table S. Oligonucleotides used for the in-house RT-PCR assays targeting the M, H7 or N9 genes. Assay (s) Target Name Sequence (5 3 ) Comments CDC M InfA Forward (NS), CDC M
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most
 Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
Chem 454, Spring Exam II F, Faraday's constant, = kcal/mol V
 Name Chem 454, Spring 2003 - Exam II F, Faraday's constant, = 23.06 kcal/mol V 1. Analysis of electron transport pathway in a pathogenic gram-negative bacterium reveals the presence of six electron transport
Name Chem 454, Spring 2003 - Exam II F, Faraday's constant, = 23.06 kcal/mol V 1. Analysis of electron transport pathway in a pathogenic gram-negative bacterium reveals the presence of six electron transport
ABA-dependant signalling of PR genes and potential involvement in the defence of lentil to Ascochyta lentis
 ABA-dependant signalling of PR genes and potential involvement in the defence of lentil to Ascochyta lentis Barkat Mustafa, David Tan, Paul WJ Taylor and Rebecca Ford Melbourne School of Land and Environment
ABA-dependant signalling of PR genes and potential involvement in the defence of lentil to Ascochyta lentis Barkat Mustafa, David Tan, Paul WJ Taylor and Rebecca Ford Melbourne School of Land and Environment
1. What substance could be represented by the letter X in the diagram below?
 1. What substance could be represented by the letter X in the diagram below? A) carbohydrates B) ozone C) carbon dioxide D) water 2. Base your answer to the following question on the diagram below. For
1. What substance could be represented by the letter X in the diagram below? A) carbohydrates B) ozone C) carbon dioxide D) water 2. Base your answer to the following question on the diagram below. For
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
BCM 221 LECTURES OJEMEKELE O.
 BCM 221 LECTURES BY OJEMEKELE O. OUTLINE INTRODUCTION TO LIPID CHEMISTRY STORAGE OF ENERGY IN ADIPOCYTES MOBILIZATION OF ENERGY STORES IN ADIPOCYTES KETONE BODIES AND KETOSIS PYRUVATE DEHYDROGENASE COMPLEX
BCM 221 LECTURES BY OJEMEKELE O. OUTLINE INTRODUCTION TO LIPID CHEMISTRY STORAGE OF ENERGY IN ADIPOCYTES MOBILIZATION OF ENERGY STORES IN ADIPOCYTES KETONE BODIES AND KETOSIS PYRUVATE DEHYDROGENASE COMPLEX
Figure S1: Abundance of Fe related proteins in oceanic Synechococcus sp. strain WH8102 in. Ferredoxin. Ferredoxin. Fe ABC transporter
 0.004 SW nm 0.04 0.03 nm nm 0.16 0.1 nm nm 0.40 0.3 nm nm 0.80 0.5 nm 1.6 1 nm 16 10 nm 160 100 nm 0 nm 0.04 nm 0.16 nm 0.40 nm 0.80 nm 1.6 nm 16 nm 160 nm Spectral counts 0 nm 0.04 nm 0.16 nm 0.40 nm
0.004 SW nm 0.04 0.03 nm nm 0.16 0.1 nm nm 0.40 0.3 nm nm 0.80 0.5 nm 1.6 1 nm 16 10 nm 160 100 nm 0 nm 0.04 nm 0.16 nm 0.40 nm 0.80 nm 1.6 nm 16 nm 160 nm Spectral counts 0 nm 0.04 nm 0.16 nm 0.40 nm
sesquiterpenes monoterpenes mg gfw -1 fruit skin peel seed whole fruit
 mg gfw -1 14 12 1 8 6 4 2 sesquiterpenes monoterpenes fruit skin peel seed whole fruit Supplementary Figure S1. Terpene amount in fruit skin, seeds, and whole fruits of Japanese pepper. Monoterpenes (β-phellandrene,
mg gfw -1 14 12 1 8 6 4 2 sesquiterpenes monoterpenes fruit skin peel seed whole fruit Supplementary Figure S1. Terpene amount in fruit skin, seeds, and whole fruits of Japanese pepper. Monoterpenes (β-phellandrene,
Supplementary Materials
 Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
Abbreviations: P- paraffin-embedded section; C, cryosection; Bio-SA, biotin-streptavidin-conjugated fluorescein amplification.
 Supplementary Table 1. Sequence of primers for real time PCR. Gene Forward primer Reverse primer S25 5 -GTG GTC CAC ACT ACT CTC TGA GTT TC-3 5 - GAC TTT CCG GCA TCC TTC TTC-3 Mafa cds 5 -CTT CAG CAA GGA
Supplementary Table 1. Sequence of primers for real time PCR. Gene Forward primer Reverse primer S25 5 -GTG GTC CAC ACT ACT CTC TGA GTT TC-3 5 - GAC TTT CCG GCA TCC TTC TTC-3 Mafa cds 5 -CTT CAG CAA GGA
c Tuj1(-) apoptotic live 1 DIV 2 DIV 1 DIV 2 DIV Tuj1(+) Tuj1/GFP/DAPI Tuj1 DAPI GFP
 Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Biology 12 January 2004 Provincial Examination
 Biology 12 January 2004 Provincial Examination ANSWER KEY / SCORING GUIDE CURRICULUM: Organizers 1. Cell Biology 2. Cell Processes and Applications 3. Human Biology Sub-Organizers A, B, C, D E, F, G, H
Biology 12 January 2004 Provincial Examination ANSWER KEY / SCORING GUIDE CURRICULUM: Organizers 1. Cell Biology 2. Cell Processes and Applications 3. Human Biology Sub-Organizers A, B, C, D E, F, G, H
Citation for published version (APA): Oosterveer, M. H. (2009). Control of metabolic flux by nutrient sensors Groningen: s.n.
 University of Groningen Control of metabolic flux by nutrient sensors Oosterveer, Maaike IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it.
University of Groningen Control of metabolic flux by nutrient sensors Oosterveer, Maaike IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it.
BRIEF CONTENTS COPYRIGHTED MATERIAL III METABOLIC AND DEVELOPMENTAL INTEGRATION COMPARTMENTS CELL REPRODUCTION PLANT ENVIRONMENT AND AGRICULTURE
 BRIEF CONTENTS I COMPARTMENTS 1 Membrane Structure and Membranous Organelles 2 2 The Cell Wall 45 3 Membrane Transport 111 4 Protein Sorting and Vesicle Traffic 151 5 The Cytoskeleton 191 II CELL REPRODUCTION
BRIEF CONTENTS I COMPARTMENTS 1 Membrane Structure and Membranous Organelles 2 2 The Cell Wall 45 3 Membrane Transport 111 4 Protein Sorting and Vesicle Traffic 151 5 The Cytoskeleton 191 II CELL REPRODUCTION
Supplementary Figure 1 a
 Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Regulation of P450. b. competitive-inhibitor but not substrate. c. non-competitive inhibition. 2. Covalent binding to heme or protein
 10. Inhibition of P450 Regulation of P450 a. competitive-2 chemicals metabolized by same isoform b. competitive-inhibitor but not substrate c. non-competitive inhibition 11. Induction of P450 1. Non-covalent
10. Inhibition of P450 Regulation of P450 a. competitive-2 chemicals metabolized by same isoform b. competitive-inhibitor but not substrate c. non-competitive inhibition 11. Induction of P450 1. Non-covalent
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
FATTY ACID SYNTHESIS
 FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
Supplementary Document
 Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
Supporting Information. Mutational analysis of a phenazine biosynthetic gene cluster in
 Supporting Information for Mutational analysis of a phenazine biosynthetic gene cluster in Streptomyces anulatus 9663 Orwah Saleh 1, Katrin Flinspach 1, Lucia Westrich 1, Andreas Kulik 2, Bertolt Gust
Supporting Information for Mutational analysis of a phenazine biosynthetic gene cluster in Streptomyces anulatus 9663 Orwah Saleh 1, Katrin Flinspach 1, Lucia Westrich 1, Andreas Kulik 2, Bertolt Gust
Lecture: 26 OXIDATION OF FATTY ACIDS
 Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Metabolism. Topic 11&12 (ch8) Microbial Metabolism. Metabolic Balancing Act. Topics. Catabolism Anabolism Enzymes
 Topic 11&12 (ch8) Microbial Metabolism Topics Metabolism Energy Pathways Biosynthesis 1 Catabolism Anabolism Enzymes Metabolism 2 Metabolic Balancing Act Catabolism Enzymes involved in breakdown of complex
Topic 11&12 (ch8) Microbial Metabolism Topics Metabolism Energy Pathways Biosynthesis 1 Catabolism Anabolism Enzymes Metabolism 2 Metabolic Balancing Act Catabolism Enzymes involved in breakdown of complex
a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53
 1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
Figure 1 Original Advantages of biological reactions being catalyzed by enzymes:
 Enzyme basic concepts, Enzyme Regulation I III Carmen Sato Bigbee, Ph.D. Objectives: 1) To understand the bases of enzyme catalysis and the mechanisms of enzyme regulation. 2) To understand the role of
Enzyme basic concepts, Enzyme Regulation I III Carmen Sato Bigbee, Ph.D. Objectives: 1) To understand the bases of enzyme catalysis and the mechanisms of enzyme regulation. 2) To understand the role of
Metabolism Energy Pathways Biosynthesis. Catabolism Anabolism Enzymes
 Topics Microbial Metabolism Metabolism Energy Pathways Biosynthesis 2 Metabolism Catabolism Catabolism Anabolism Enzymes Breakdown of complex organic molecules in order to extract energy and dform simpler
Topics Microbial Metabolism Metabolism Energy Pathways Biosynthesis 2 Metabolism Catabolism Catabolism Anabolism Enzymes Breakdown of complex organic molecules in order to extract energy and dform simpler
NABIL KILLINY ASSISTANT PROFESSOR PLANT PATHOLOGY UNIVERSITY OF FLORIDA. Citrus responses to huanglongbing
 NABIL KILLINY ASSISTANT PROFESSOR PLANT PATHOLOGY UNIVERSITY OF FLORIDA Citrus responses to huanglongbing HLB is Vector-borne Disease! Vector psyllids Pathogen Ca. Liberibacter asiaticus. Host Citrus spp.
NABIL KILLINY ASSISTANT PROFESSOR PLANT PATHOLOGY UNIVERSITY OF FLORIDA Citrus responses to huanglongbing HLB is Vector-borne Disease! Vector psyllids Pathogen Ca. Liberibacter asiaticus. Host Citrus spp.
Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at
 Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at different concentrations for 30 min and analyzed for
Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at different concentrations for 30 min and analyzed for
Metabolism. Chapter 8 Microbial Metabolism. Metabolic balancing act. Catabolism Anabolism Enzymes. Topics. Metabolism Energy Pathways Biosynthesis
 Chapter 8 Microbial Metabolism Topics Metabolism Energy Pathways Biosynthesis Catabolism Anabolism Enzymes Metabolism 1 2 Metabolic balancing act Catabolism and anabolism simple model Catabolism Enzymes
Chapter 8 Microbial Metabolism Topics Metabolism Energy Pathways Biosynthesis Catabolism Anabolism Enzymes Metabolism 1 2 Metabolic balancing act Catabolism and anabolism simple model Catabolism Enzymes
Ch 07. Microbial Metabolism
 Ch 07 Microbial Metabolism SLOs Differentiate between metabolism, catabolism, and anabolism. Fully describe the structure and function of enzymes. Differentiate between constitutive and regulated enzymes.
Ch 07 Microbial Metabolism SLOs Differentiate between metabolism, catabolism, and anabolism. Fully describe the structure and function of enzymes. Differentiate between constitutive and regulated enzymes.
Enzymes what are they?
 Topic 11 (ch8) Microbial Metabolism Topics Metabolism Energy Pathways Biosynthesis 1 Catabolism Anabolism Enzymes Metabolism 2 Metabolic balancing act Catabolism Enzymes involved in breakdown of complex
Topic 11 (ch8) Microbial Metabolism Topics Metabolism Energy Pathways Biosynthesis 1 Catabolism Anabolism Enzymes Metabolism 2 Metabolic balancing act Catabolism Enzymes involved in breakdown of complex
Phosphorus Deficiency in Potatoes. Helen Bowen, Rory Hayden & Will Spracklen
 A Molecular Diagnostic for Phosphorus Deficiency in Potatoes Philip J. White John P. Hammond, Martin R. Broadley, Helen Bowen, Rory Hayden & Will Spracklen (Warwick, Nottingham, SCRI) IPNC XVI, Sacramento,
A Molecular Diagnostic for Phosphorus Deficiency in Potatoes Philip J. White John P. Hammond, Martin R. Broadley, Helen Bowen, Rory Hayden & Will Spracklen (Warwick, Nottingham, SCRI) IPNC XVI, Sacramento,
Biosynthesis of Fatty Acids
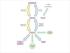 Biosynthesis of Fatty Acids Fatty acid biosynthesis takes place in the cytosol rather than the mitochondria and requires a different activation mechanism and different enzymes and coenzymes than fatty
Biosynthesis of Fatty Acids Fatty acid biosynthesis takes place in the cytosol rather than the mitochondria and requires a different activation mechanism and different enzymes and coenzymes than fatty
Glycolysis - Plasmodium
 Apicomplexan Biochemistry Basics Toxoplasma Good cell biology model Genome sequencing not completed Virtual pathways Cryptosporidium The strange one Genome sequence completed Virtual pathways Plasmodium
Apicomplexan Biochemistry Basics Toxoplasma Good cell biology model Genome sequencing not completed Virtual pathways Cryptosporidium The strange one Genome sequence completed Virtual pathways Plasmodium
Very-Long Chain Fatty Acid Biosynthesis
 Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes
 Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes The major site of acetoacetate and 3-hydorxybutyrate production is in the liver. 3-hydorxybutyrate is the
Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes The major site of acetoacetate and 3-hydorxybutyrate production is in the liver. 3-hydorxybutyrate is the
MBB 694:407, 115:511 Name Third Exam Niederman, Deis. Please use BLOCK CAPITAL letters like this --- A, B, C, D, E. Not lowercase!
 MBB 694:407, 115:511 Name Third Exam Niederman, Deis Wed. Nov. 22, 2006 Row Letter Seat Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is worth two points.
MBB 694:407, 115:511 Name Third Exam Niederman, Deis Wed. Nov. 22, 2006 Row Letter Seat Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is worth two points.
Toluidin-Staining of mast cells Ear tissue was fixed with Carnoy (60% ethanol, 30% chloroform, 10% acetic acid) overnight at 4 C, afterwards
 Toluidin-Staining of mast cells Ear tissue was fixed with Carnoy (60% ethanol, 30% chloroform, 10% acetic acid) overnight at 4 C, afterwards incubated in 100 % ethanol overnight at 4 C and embedded in
Toluidin-Staining of mast cells Ear tissue was fixed with Carnoy (60% ethanol, 30% chloroform, 10% acetic acid) overnight at 4 C, afterwards incubated in 100 % ethanol overnight at 4 C and embedded in
LIPID METABOLISM
 LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
BIOL 158: BIOLOGICAL CHEMISTRY II
 BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
Oxidation of Long Chain Fatty Acids
 Oxidation of Long Chain Fatty Acids Dr NC Bird Oxidation of long chain fatty acids is the primary source of energy supply in man and animals. Hibernating animals utilise fat stores to maintain body heat,
Oxidation of Long Chain Fatty Acids Dr NC Bird Oxidation of long chain fatty acids is the primary source of energy supply in man and animals. Hibernating animals utilise fat stores to maintain body heat,
Protein sequence alignment using binary string
 Available online at www.scholarsresearchlibrary.com Scholars Research Library Der Pharmacia Lettre, 2015, 7 (5):220-225 (http://scholarsresearchlibrary.com/archive.html) ISSN 0975-5071 USA CODEN: DPLEB4
Available online at www.scholarsresearchlibrary.com Scholars Research Library Der Pharmacia Lettre, 2015, 7 (5):220-225 (http://scholarsresearchlibrary.com/archive.html) ISSN 0975-5071 USA CODEN: DPLEB4
3) How many different amino acids are proteogenic in eukaryotic cells? A) 12 B) 20 C) 25 D) 30 E) None of the above
 Suggesting questions for Biochemistry 1 and 2 and clinical biochemistry 1) Henderson Hasselbalch Equation shows: A) The relationship between ph and the concentration of an acid and its conjugate base B)
Suggesting questions for Biochemistry 1 and 2 and clinical biochemistry 1) Henderson Hasselbalch Equation shows: A) The relationship between ph and the concentration of an acid and its conjugate base B)
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 27 Fatty Acid Degradation Dietary Lipid (Triacylglycerol) Metabolism - In the small intestine, fat particles are coated with bile
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 27 Fatty Acid Degradation Dietary Lipid (Triacylglycerol) Metabolism - In the small intestine, fat particles are coated with bile
The Arabidopsis mir396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens
 The Arabidopsis mir396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens Mauricio Soto-Suárez, Patricia Baldrich, Detlef Weigel, Ignacio Rubio-Somoza and
The Arabidopsis mir396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens Mauricio Soto-Suárez, Patricia Baldrich, Detlef Weigel, Ignacio Rubio-Somoza and
Post-translational modifications of proteins in gene regulation under hypoxic conditions
 203 Review Article Post-translational modifications of proteins in gene regulation under hypoxic conditions 1, 2) Olga S. Safronova 1) Department of Cellular Physiological Chemistry, Tokyo Medical and
203 Review Article Post-translational modifications of proteins in gene regulation under hypoxic conditions 1, 2) Olga S. Safronova 1) Department of Cellular Physiological Chemistry, Tokyo Medical and
cholesterol structure Cholesterol FAQs Cholesterol promotes the liquid-ordered phase of membranes Friday, October 15, 2010
 cholesterol structure most plasma cholesterol is in the esterified form (not found in cells or membranes) cholesterol functions in all membranes (drives formation of lipid microdomains) cholesterol is
cholesterol structure most plasma cholesterol is in the esterified form (not found in cells or membranes) cholesterol functions in all membranes (drives formation of lipid microdomains) cholesterol is
Summary of fatty acid synthesis
 Lipid Metabolism, part 2 1 Summary of fatty acid synthesis 8 acetyl CoA + 14 NADPH + 14 H+ + 7 ATP palmitic acid (16:0) + 8 CoA + 14 NADP + + 7 ADP + 7 Pi + 7 H20 1. The major suppliers of NADPH for fatty
Lipid Metabolism, part 2 1 Summary of fatty acid synthesis 8 acetyl CoA + 14 NADPH + 14 H+ + 7 ATP palmitic acid (16:0) + 8 CoA + 14 NADP + + 7 ADP + 7 Pi + 7 H20 1. The major suppliers of NADPH for fatty
Lignin and the General Phenylpropanoid Pathway. Introduction and Importance:
 Lignin and the General Phenylpropanoid Pathway 13. Phenolics and Lignin p. 1 Introduction and Importance: Phenolic: a compound consisting of an aromatic ring plus at least one hydroxyl [= phenyl group],
Lignin and the General Phenylpropanoid Pathway 13. Phenolics and Lignin p. 1 Introduction and Importance: Phenolic: a compound consisting of an aromatic ring plus at least one hydroxyl [= phenyl group],
Lipid Metabolism. Catabolism Overview
 Lipid Metabolism Pratt & Cornely, Chapter 17 Catabolism Overview Lipids as a fuel source from diet Beta oxidation Mechanism ATP production Ketone bodies as fuel 1 High energy More reduced Little water
Lipid Metabolism Pratt & Cornely, Chapter 17 Catabolism Overview Lipids as a fuel source from diet Beta oxidation Mechanism ATP production Ketone bodies as fuel 1 High energy More reduced Little water
Bacterial Gene Finding CMSC 423
 Bacterial Gene Finding CMSC 423 Finding Signals in DNA We just have a long string of A, C, G, Ts. How can we find the signals encoded in it? Suppose you encountered a language you didn t know. How would
Bacterial Gene Finding CMSC 423 Finding Signals in DNA We just have a long string of A, C, G, Ts. How can we find the signals encoded in it? Suppose you encountered a language you didn t know. How would
Functional mechanisms of drought tolerance in maize
 Functional mechanisms of drought tolerance in maize Nepolean Thirunavukkarasu Division of Genetics Indian Agricultural Research Institute New Delhi-110012 tnepolean@iari.resi.in tnepolean@gmail.com Mt
Functional mechanisms of drought tolerance in maize Nepolean Thirunavukkarasu Division of Genetics Indian Agricultural Research Institute New Delhi-110012 tnepolean@iari.resi.in tnepolean@gmail.com Mt
Please use BLOCK CAPITAL letters like this --- A, B, C, D, E. Not lowercase! 1. E 10. D 19. E 2. D 11. A 20. A 3. D 12. D 21. A 4. D 13. A 22.
 Molecular Biology and Biochemistry 694:407/115:511 Third Exam Deis Niederman Wednesday, November 22, 2006 Name Row Letter Seat Number This exam consists of two parts. Part I is multiple choice. Each of
Molecular Biology and Biochemistry 694:407/115:511 Third Exam Deis Niederman Wednesday, November 22, 2006 Name Row Letter Seat Number This exam consists of two parts. Part I is multiple choice. Each of
ANSC 619 PHYSIOLOGICAL CHEMISTRY OF LIVESTOCK SPECIES. Carbohydrate Metabolism
 ANSC 619 PHYSIOLOGICAL CHEMISTRY OF LIVESTOCK SPECIES I. Glycolysis A. Pathway Regulation of glycolysis Hexokinase: Activated by glucose. Inhibited by G6P. 6-Phosphofructokinase: Inhibited by ATP, especially
ANSC 619 PHYSIOLOGICAL CHEMISTRY OF LIVESTOCK SPECIES I. Glycolysis A. Pathway Regulation of glycolysis Hexokinase: Activated by glucose. Inhibited by G6P. 6-Phosphofructokinase: Inhibited by ATP, especially
CD31 5'-AGA GAC GGT CTT GTC GCA GT-3' 5 ' -TAC TGG GCT TCG AGA GCA GT-3'
 Table S1. The primer sets used for real-time RT-PCR analysis. Gene Forward Reverse VEGF PDGFB TGF-β MCP-1 5'-GTT GCA GCA TGA ATC TGA GG-3' 5'-GGA GAC TCT TCG AGG AGC ACT T-3' 5'-GAA TCA GGC ATC GAG AGA
Table S1. The primer sets used for real-time RT-PCR analysis. Gene Forward Reverse VEGF PDGFB TGF-β MCP-1 5'-GTT GCA GCA TGA ATC TGA GG-3' 5'-GGA GAC TCT TCG AGG AGC ACT T-3' 5'-GAA TCA GGC ATC GAG AGA
BIOSYNTHESIS OF FATTY ACIDS. doc. Ing. Zenóbia Chavková, CSc.
 BIOSYNTHESIS OF FATTY ACIDS doc. Ing. Zenóbia Chavková, CSc. The pathway for the of FAs is not the reversal of the oxidation pathway Both pathways are separated within different cellular compartments In
BIOSYNTHESIS OF FATTY ACIDS doc. Ing. Zenóbia Chavková, CSc. The pathway for the of FAs is not the reversal of the oxidation pathway Both pathways are separated within different cellular compartments In
Journal of Cell Science Supplementary information. Arl8b +/- Arl8b -/- Inset B. electron density. genotype
 J. Cell Sci. : doi:.4/jcs.59: Supplementary information E9. A Arl8b /- Arl8b -/- Arl8b Arl8b non-specific band Gapdh Tbp E7.5 HE Inset B D Control al am hf C E Arl8b -/- al am hf E8.5 F low middle high
J. Cell Sci. : doi:.4/jcs.59: Supplementary information E9. A Arl8b /- Arl8b -/- Arl8b Arl8b non-specific band Gapdh Tbp E7.5 HE Inset B D Control al am hf C E Arl8b -/- al am hf E8.5 F low middle high
Chem 280 Final Exam. Here is the summary of the total 150 points plus 6 points bonus. Carefully read the questions. Good luck!
 May 2 nd, 2012 Name: CLID: Score: Chem 280 Final Exam There are 32 multiple choices that are worth 3 points each. There are 5 problems and one bonus problem. Try to answer the questions, which you know
May 2 nd, 2012 Name: CLID: Score: Chem 280 Final Exam There are 32 multiple choices that are worth 3 points each. There are 5 problems and one bonus problem. Try to answer the questions, which you know
Electron Transport Chain and Oxidative phosphorylation
 Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM. Fatty Acid Elongation and Desaturation
 ANSC/NUTR 618 LIPIDS & LIPID METABOLISM I. Fatty acid elongation A. General 1. At least 60% of fatty acids in triacylglycerols are C18. 2. Free palmitic acid (16:0) synthesized in cytoplasm is elongated
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM I. Fatty acid elongation A. General 1. At least 60% of fatty acids in triacylglycerols are C18. 2. Free palmitic acid (16:0) synthesized in cytoplasm is elongated
Tala Saleh. Razi Kittaneh ... Nayef Karadsheh
 Tala Saleh Razi Kittaneh... Nayef Karadsheh β-oxidation of Fatty Acids The oxidation of fatty acids occurs in 3 steps: Step 1: Activation of the Fatty acid FA + HS-CoA + ATP FA-CoA + AMP + PPi - The fatty
Tala Saleh Razi Kittaneh... Nayef Karadsheh β-oxidation of Fatty Acids The oxidation of fatty acids occurs in 3 steps: Step 1: Activation of the Fatty acid FA + HS-CoA + ATP FA-CoA + AMP + PPi - The fatty
Supplementary Data AtMYB12
 Supplementary Data AtMYB12 expression in tomato leads to large scale differential modulation in transcriptome and flavonoid content in leaf and fruit tissues Ashutosh Pandey 1,$,#, Prashant Misra 1,$,#,
Supplementary Data AtMYB12 expression in tomato leads to large scale differential modulation in transcriptome and flavonoid content in leaf and fruit tissues Ashutosh Pandey 1,$,#, Prashant Misra 1,$,#,
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. H3F3B expression in lung cancer. a. Comparison of H3F3B expression in relapsed and non-relapsed lung cancer patients. b. Prognosis of two groups of lung cancer
Supplementary Figures Supplementary Figure 1. H3F3B expression in lung cancer. a. Comparison of H3F3B expression in relapsed and non-relapsed lung cancer patients. b. Prognosis of two groups of lung cancer
Table S1 Differentially expressed genes showing > 2 fold changes and p <0.01 for 0.01 mm NS-398, 0.1mM ibuprofen and COX-2 RNAi.
 Table S1 Differentially expressed genes showing > 2 fold changes and p
Table S1 Differentially expressed genes showing > 2 fold changes and p
Supplementary Information
 Supplementary Information Figure S1 Sequencing quality. Sample screenshot of re-sequencing raw file quality generated by FastQC. For both the graphs, scores were based on the Illumina 1.5 quality score.
Supplementary Information Figure S1 Sequencing quality. Sample screenshot of re-sequencing raw file quality generated by FastQC. For both the graphs, scores were based on the Illumina 1.5 quality score.
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at
 Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Residues (aa) MW pi GRAVY
 Supplementary Table 1. Table showing the calculated properties based on the primary nucleotide and protein sequences of the 39 rubber tree chitinases. Highlights: green denotes lower GRAVY, orange denotes
Supplementary Table 1. Table showing the calculated properties based on the primary nucleotide and protein sequences of the 39 rubber tree chitinases. Highlights: green denotes lower GRAVY, orange denotes
L I F E S C I E N C E S
 1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 OCTOBER 31, 2006
1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 OCTOBER 31, 2006
Lecture 16. Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III
 Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
BCMB 3100 Fall 2013 Exam III
 BCMB 3100 Fall 2013 Exam III 1. (10 pts.) (a.) Briefly describe the purpose of the glycerol dehydrogenase phosphate shuttle. (b.) How many ATPs can be made when electrons enter the electron transport chain
BCMB 3100 Fall 2013 Exam III 1. (10 pts.) (a.) Briefly describe the purpose of the glycerol dehydrogenase phosphate shuttle. (b.) How many ATPs can be made when electrons enter the electron transport chain
TetR repressor-based bioreporters for the detection of doxycycline using Escherichia
 Supplementary materials TetR repressor-based bioreporters for the detection of doxycycline using Escherichia coli and Acinetobacter oleivorans Hyerim Hong and Woojun Park * Department of Environmental
Supplementary materials TetR repressor-based bioreporters for the detection of doxycycline using Escherichia coli and Acinetobacter oleivorans Hyerim Hong and Woojun Park * Department of Environmental
Fatty Acid and Triacylglycerol Metabolism 1
 Fatty Acid and Triacylglycerol Metabolism 1 Mobilization of stored fats and oxidation of fatty acids Lippincott s Chapter 16 What is the first lecture about What is triacylglycerol Fatty acids structure
Fatty Acid and Triacylglycerol Metabolism 1 Mobilization of stored fats and oxidation of fatty acids Lippincott s Chapter 16 What is the first lecture about What is triacylglycerol Fatty acids structure
Chemistry 5.07 Problem Set
 Chemistry 5.07 Problem Set 8 2013 Problem 1. All oxidation steps in the pathway from glucose to CO 2 result in the production of NADH, except the succinate dehydrogenase (SDH) step in the TCA cycle, which
Chemistry 5.07 Problem Set 8 2013 Problem 1. All oxidation steps in the pathway from glucose to CO 2 result in the production of NADH, except the succinate dehydrogenase (SDH) step in the TCA cycle, which
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature05883 SUPPLEMENTARY INFORMATION Supplemental Figure 1 Prostaglandin agonists and antagonists alter runx1/cmyb expression. a-e, Embryos were exposed to (b) PGE2 and (c) PGI2 (20μM) and
doi: 10.1038/nature05883 SUPPLEMENTARY INFORMATION Supplemental Figure 1 Prostaglandin agonists and antagonists alter runx1/cmyb expression. a-e, Embryos were exposed to (b) PGE2 and (c) PGI2 (20μM) and
Very-Long Chain Fatty Acid Biosynthesis
 Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
III. Metabolism The Citric Acid Cycle
 Department of Chemistry and Biochemistry University of Lethbridge III. Metabolism The Citric Acid Cycle Slide 1 The Eight Steps of the Citric Acid Cycle Enzymes: 4 dehydrogenases (2 decarboxylation) 3
Department of Chemistry and Biochemistry University of Lethbridge III. Metabolism The Citric Acid Cycle Slide 1 The Eight Steps of the Citric Acid Cycle Enzymes: 4 dehydrogenases (2 decarboxylation) 3
Fatty acid breakdown
 Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
Student name ID # 2. (4 pts) What is the terminal electron acceptor in respiration? In photosynthesis?
 1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? 2. (4 pts) What is the terminal electron acceptor
1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? 2. (4 pts) What is the terminal electron acceptor
Transcriptional regulatory network in potato potato virus Y interaction signaling
 Transcriptional regulatory network in potato potato virus Y interaction signaling Tjaša Lukan National Institute of Biology, Ljubljana, Slovenia Department of Biotechnology and Systems biology August 2016
Transcriptional regulatory network in potato potato virus Y interaction signaling Tjaša Lukan National Institute of Biology, Ljubljana, Slovenia Department of Biotechnology and Systems biology August 2016
Fatty acids synthesis
 Fatty acids synthesis The synthesis start from Acetyl COA the first step requires ATP + reducing power NADPH! even though the oxidation and synthesis are different pathways but from chemical part of view
Fatty acids synthesis The synthesis start from Acetyl COA the first step requires ATP + reducing power NADPH! even though the oxidation and synthesis are different pathways but from chemical part of view
Synthesis of Fatty Acids and Triacylglycerol
 Synthesis of Fatty Acids and Triacylglycerol Lippincott s Chapter 16 Fatty Acid Synthesis Mainly in the Liver Requires Carbon Source: Acetyl CoA Reducing Power: NADPH 8 CH 3 COO C 15 H 33 COO Energy Input:
Synthesis of Fatty Acids and Triacylglycerol Lippincott s Chapter 16 Fatty Acid Synthesis Mainly in the Liver Requires Carbon Source: Acetyl CoA Reducing Power: NADPH 8 CH 3 COO C 15 H 33 COO Energy Input:
Beta Thalassemia Sami Khuri Department of Computer Science San José State University Spring 2015
 Bioinformatics in Medical Product Development SMPD 287 Three Beta Thalassemia Sami Khuri Department of Computer Science San José State University Hemoglobin Outline Anatomy of a gene Hemoglobinopathies
Bioinformatics in Medical Product Development SMPD 287 Three Beta Thalassemia Sami Khuri Department of Computer Science San José State University Hemoglobin Outline Anatomy of a gene Hemoglobinopathies
Biosynthesis of Triacylglycerides (TG) in liver. Mobilization of stored fat and oxidation of fatty acids
 Biosynthesis of Triacylglycerides (TG) in liver Mobilization of stored fat and oxidation of fatty acids Activation of hormone sensitive lipase This enzyme is activated when phosphorylated (3,5 cyclic AMPdependent
Biosynthesis of Triacylglycerides (TG) in liver Mobilization of stored fat and oxidation of fatty acids Activation of hormone sensitive lipase This enzyme is activated when phosphorylated (3,5 cyclic AMPdependent
Physiological Chemistry II Exam IV Dr. Melissa Kelley April 13, 2004
 Name Write your name on the back of the exam Physiological Chemistry II Exam IV Dr. Melissa Kelley April 13, 2004 This examination consists of forty-four questions, each having 2 points. The remaining
Name Write your name on the back of the exam Physiological Chemistry II Exam IV Dr. Melissa Kelley April 13, 2004 This examination consists of forty-four questions, each having 2 points. The remaining
Very-Long Chain Fatty Acid Biosynthesis
 Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
