The N-end rule pathway regulates pathogen responses. in plants
|
|
|
- Sibyl Townsend
- 5 years ago
- Views:
Transcription
1 SUPPLEMENTARY INFORMATION The N-end rule pathway regulates pathogen responses in plants Rémi de Marchi 1,2, Maud Sorel 1, Brian Mooney 1, Isabelle Fudal 3, Kevin Goslin 1, Kamila Kwaśniewska 4, Patrick T. Ryan 4, Marina Pfalz 5, Juergen Kroymann 5, Stephan Pollmann 6, Angela Feechan 7, Frank Wellmer 4, Susana Rivas 2*, and Emmanuelle Graciet 1* 1
2 Supplementary figures Supplementary Figure 1: Comparison of transcriptomics datasets obtained with ate1 ate2 seedlings. a, Overlap between our transcriptomics dataset obtained and the results of a microarray analysis using seedlings grown under normal oxygen conditions 1. For the latter dataset, DEGs with log 2 (fold change) > 0.5 and adjusted P-value < 0.05 were considered. b, Overlap between our transcriptomics dataset and core hypoxia response genes defined by Mustroph et al. 2. 2
3 Supplementary Figure 2: Results of glucosinolate measurements. a, Results of glucosinolate measurements. The biosynthesis pathways of methionine-derived aliphatic glucosinolates and tryptophan-derived indolic glucosinolates (inset) are shown. The biosynthesis pathway of aliphatic glucosinolates includes reactions that occur in the chloroplast (represented by a green rectangle). The number of methylene groups in the chain of different precursors is indicated (2C, 3C, etc ). The level of the different glucosinolates in wild-type and ate1 ate2 seedlings is represented by blue and yellow 3
4 circles, respectively. The diameter of the circles is proportional to the quantity of the different glucosinolates measured in nine independent sets of samples. Only protein names encoded by genes that were found to be differentially expressed in ate1 ate2 are indicated, with black and red fonts corresponding to genes that were down and upregulated in the mutant, respectively. n.d.: not detected. b, Estimated total levels of aliphatic glucosinolates. c, Estimated total levels of indole glucosinolates. Estimated minimum and maximum are indicated for each genotype. 3BZOP (aliphatic glucosinolate) and 1MOI3M (indole glucosinolate) could not be separated, hence minimum and maximum levels of the two types of glucosinolates were calculated using ratios between 4BZOB and 3BZOP of 1.27:1 and 1.92:1, respectively 3-5 (see Methods for details). 4
5 Supplementary Figure 3: Response of JA and SA related genes in ate1 ate2 and prt6-5 mutant seedlings. The regulation of selected JA (a-c) and SA response genes (d-f) in ate1 ate2, prt6-5 and wild-type seedlings was monitored after treatment with 20 µm MeJA (a, d), 0.5 mm SA (b, e) and 10 µm ABA (c, f). Solid and dashed bars represent mock- or hormone-treated samples, respectively. Error bars correspond to SEM of at least four independent biological replicates. The results of Student s t-tests are shown with a indicating P-values < 0.05 and b P-values between 0.05 and 0.1. VSP1: VEGETATIVE STORAGE PROTEIN1; VSP2: VEGETATIVE STORAGE PROTEIN2; LOX2: LIPOXYGENASE2; AOS: ALLENE OXIDE SYNTHASE; PR1: PATHOGENESIS- RELATED1; NDR1: NON RACE-SPECIFIC DISEASE RESISTANCE1; PAD4: PHYTOALEXIN DEFICIENT4; EDS1: ENHANCED DISEASE SUSCEPTIBILITY1; ABI5: ABA INSENSITIVE5. 5
6 Supplementary Figure 4: Representative symptoms exhibited following inoculation with S. sclerotiorum. Wild-type and N-end rule mutant plants were photographed 7 days following inoculation with S. sclerotiorum. The Shahdara (Sha) and Rubezhnoe (Rbz) accessions were used as susceptible and resistant control plants, respectively. 6
7 Supplementary Figure 5: Expression of N-end rule genes following inoculation with Pst AvrRpm1. Four-week-old plants were inoculated with Pst AvrRpm1 (5x10 7 cfu/ml) and tissue was collected at the indicated time points. Gene expression was assessed using RT-qPCRs and oligonucleotides described in Supplementary Table 3. Gene expression levels were first normalized using the MON1 reference gene. Results are presented relative to the expression level of a given gene at the t0 time point. 7
8 Supplementary Figure 6: Comparison of transcriptomics datasets obtained following inoculation with Pst AvrRpm1. a, Overlap between our transcriptomics datasets obtained with wild-type (Col-0) plants following inoculation with Pst AvrRpm1 and the dataset presented by Truman et al. 6. The latter was generated using wild-type Col-0 plants inoculated with Pst AvrRpm1 and total RNA isolated at 4 hpi. b, Overlap between our transcriptomics datasets obtained with ate1 ate2 mutant plants following inoculation with Pst AvrRpm1 and the dataset presented by Truman et al. 6. 8
9 Supplementary Figure 7: Expression profile of genes associated with GO categories enriched in both the wild type and the ate1 ate2 mutant. Gene expression changes in wild type and ate1 ate2 plants following inoculation with Pst AvrRpm1 were extracted from the microarray datasets. ICS1 and PAD4 are SA-related genes; AOS and VSP2 are JA-response genes; MYB30 and MPK3 are known regulators of Arabidopsis response to Pst AvrRpm1. 9
10 Supplementary Figure 8: Expression profile of genes associated with GO categories that are only enriched among DEGs identified in the wild type. Gene expression changes in wild type and ate1 ate2 plants following inoculation with Pst AvrRpm1 were extracted from the microarray datasets. MAM3 and FMO-GSox1 are glucosinolate-related genes; ADH1 (ALCOHOL DEHYDROGENASE 1) and GLB1 (HEMOGLOBIN 1) are hypoxia-related genes; VPS45 (VACUOLAR PROTEIN SORTING 45) and RABA6b (RAB GTPase HOMOLOG A6B) are involved in intracellular protein movement. 10
11 Supplementary Figure 9: Gene expression changes of selected genes in prt6-5 mutant plants inoculated with Pst AvrRpm1. Gene expression changes of JA (VSP1 and VSP2) and SA (ICS1 and PR1) response genes, as well as of known regulators of the response to Pst AvrRpm1 (MPK3 and MYB30), were monitored following inoculation of wild type (Col-0) and prt6-5 plants with Pst AvrRpm1 at a density of 5x10 7 cfu/ml. Expression levels relative to those of the REF1 reference gene are presented. Error bars indicate SEM of four independent experiments. The results of Student s t-tests are shown with a indicating P-values < 0.05 and b P-values between 0.05 and
12 Supplementary Table 3: List of oligonucleotides used for this study, including gene name and AGI. Oligonucleotide name and gene AGI REF1 fwd (AT1G13320) REF1 rev (AT1G13320) REF2 fwd (AT4G34270) REF2 rev (AT4G34270) MON1-F (AT2G28390) MON1-R (AT2G28390) DNA contamination control fwd DNA contamination control rev EG_qPCR143 (MYB34; AT5G60890) EG_qPCR144 (MYB34; AT5G60890) EG_qPCR147 (CYP79F2; AT1G16400) EG_qPCR148 (CYP79F1 & CYP79F2; AT1G16410 & AT1G16400) EG_qPCR149 (CYP79F1; AT1G16410) EG_qPCR38 (MAM3; AT5G23020) EG_qPCR39 (MAM3; AT5G23020) EG_qPCR122 (FMO GS-ox1; AT1G65860) EG_qPCR123 (FMO GS-ox1; AT1G65860) EG_qPCR124 (BCAT4; AT3G19710) EG_qPCR125 (BCAT4; AT3G19710) PR1 fwd (AT2G14610) PR1 rev (AT2G14610) WRKY70 fwd (AT3G56400) WRKY70 rev (AT3G56400) NDR1 rev (AT3G20600) NDR1 fwd AT3G20600) PAD4 rev (AT3G52430) PAD4 fwd (AT3G52430) EDS1 rev (AT3G48090) EDS1 fwd (AT3G48090) Oligonucleotide sequence (5 to 3 ) AAGCGGTTGTGGAGAACATGATACG TGGAGAGCTTGATTTGCGAAATACCG ATCTGCGAAAGGGTATCCAGTTGAC TGGAAGCCTCTGACTGATGGAGC AACTCTATGCAGCATTTGATCCACT TGATTGCATATCTTTATCGCCATC TCATCAGCGGTAGATGGAACTCTGAG ATGACGTAGAAGCATCATCCTGCG CATGCTCTTAAGGGTAACAAGTGGGC CCTAGCGGAACCGGATGAATATACCG CATCGTAGAGTCCATTGGAGACAATTACAA CCGACCGTTGATACATCGAGTGAATG TCATCATGGAGACAATCGGAGACAATTACA TGGTGAACGGTGCTGAAATCTCATC AATGTCTTCCCAAACTTATACAACAGCGG GGTTCATTCCGAGATAGACTTCGCC AGACATGCTTGTATAGAGGTTCGACGC CGGCTACCAGGTCGAGGAAC CTAGTCACAATGGAAGCAGTGCCAG GAGAAGGCTAACTACAACTACGCTGC TTGGCACATCCGAGTCTCACTGA TACTTGAGGACGCATTTTCTTGGAGG GGACTTGCTTTGTTGCCTTGCAC CTGGTTGTTTAGCGGCTTTACTTGACC CACCATCAACACGACCAAGATCAATTCATC TTCCGAGCAGAGGAGAGCCAA CCTCCGATGAACCTCTACCTATGGTC ACCTCTCTTGCTCGATCACCTGA GAGGCAGACAGTACGTTCAAGCT 12
13 VSP1 fwd (AT5G24780) VSP1 rev (AT5G24780) VSP2 rev (AT5G24770) VSP2 fwd (AT5G24770) LOX2 fwd (AT3G45140) LOX2 rev (AT3G45140) AOS fwd (AT5G42650) AOS rev (AT5G42650) ABI5 rev (AT2G36270) ABI5 fwd (AT2G36270) MYC2 rev (AT1G32640) MYC2 fwd (AT1G32640) ICS1 rev (AT1G74710) ICS1 fwd (AT1G74710) MPK3 fwd (AT3G45640) MPK3 rev (AT3G45640) MYB30 fwd (AT3G28910) MYB30 rev (AT3G28910) RCA fwd (AT2G39730) RCA rev (AT2G39730) ATE1 fwd (AT5G05700) ATE1 rev (AT5G05700) ATE2 fwd (AT3G11240) ATE2 rev (AT3G11240) NTAQ1 fwd (AT2G41760) NTAQ1 rev (AT2G41760) PRT1 fwd (AT3G24800) PRT1 rev (AT3G24800) PRT6 fwd (AT5G02310) PRT6 rev (AT5G02310) GACCTAGACGACACTCTCCTCTCTAGT ACTCTAACCACGACCAGTACGCC CACGAGACTCTTCCTCACCTTTGACT AAGCTGCTGGCGTGACCTAC ACGGAGGTGGAATCATTGAGACTTGTT CGGTCTTATCTTCCTCAGCCAACC TTGGTGGCGAGGTTGTTTGTGAT ACAGATGGACTACACAGGTGCGA CAATGTCCGCAATCTCCCGTTCG CGG AGT TGG AGA GGA AGA GGA AGC CCATCTTCACCGTCGCTTGTTGA GAGGTTGATGTCGGCGTTGATGG AGATGGGTCACTTCCAGCTACTATCC CGAGGAGAGTGAATTTGCAGTCGG CTGTTGAACAAGCTCTGAATCACCAGT GTGCTATGGCTTCTTGGTAGATCATCTC AAGGGAGTTCAAGATCATAATGGTGAGG GCCCAGTATTGGTAGGAACAGCTC AGAGAGTCCAACTTGCCGAGACC TTGCTGGGCTCCTTTTCCGTAGAA TACAAGGAAATGCGGCAGATC TGGAGCTTGGTATTGTTGAAGC TGTATAACTCTGATGAAGACTCAGACTC CGGTATATCCTTGTATCGAAGTCGA TTCGCAGAGTACCAGAGGTTCTTCC GGTGGAGGTTGAGCAGTCCAAC TCAAAGCTATCTCAAAGAAGGCC ATGATTGGATACACCCCACAAGA CCTTGTTGCAGAGAAAGTGGTT CCAGGTAAGGAGATGGCCA 13
14 Supplementary references 1. Gibbs, D.J. et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, (2011). 2. Mustroph, A. et al. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci U S A 106, (2009). 3. Kliebenstein, D.J. et al. Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol 126, (2001). 4. Petersen, B.L., Chen, S., Hansen, C.H., Olsen, C.E. & Halkier, B.A. Composition and content of glucosinolates in developing Arabidopsis thaliana. Planta 214, (2002). 5. Brown, P.D., Tokuhisa, J.G., Reichelt, M. & Gershenzon, J. Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62, (2003). 6. Truman, W., de Zabala, M.T. & Grant, M. Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance. Plant J 46, (2006). 14
Supplemental Data. Shin et al. Plant Cell. (2012) /tpc YFP N
 MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
Supplementary Figures
 Supplementary Figures 9 10 11 Supplementary Figure 1. Old plants are more resistant to insect herbivores than young plants. (a) Image of young (1-day-old, 1D) and old (-day-old, D) plants of Arabidopsis
Supplementary Figures 9 10 11 Supplementary Figure 1. Old plants are more resistant to insect herbivores than young plants. (a) Image of young (1-day-old, 1D) and old (-day-old, D) plants of Arabidopsis
A Complex Interplay of Three R2R3 MYB Transcription Factors Determines the Profile of Aliphatic Glucosinolates in Arabidopsis 1[C][W][OA]
![A Complex Interplay of Three R2R3 MYB Transcription Factors Determines the Profile of Aliphatic Glucosinolates in Arabidopsis 1[C][W][OA] A Complex Interplay of Three R2R3 MYB Transcription Factors Determines the Profile of Aliphatic Glucosinolates in Arabidopsis 1[C][W][OA]](/thumbs/93/112961649.jpg) A Complex Interplay of Three R2R3 MYB Transcription Factors Determines the Profile of Aliphatic Glucosinolates in Arabidopsis 1[C][W][OA] Ida Elken Sønderby, Meike Burow, Heather C. Rowe 2, Daniel J. Kliebenstein,
A Complex Interplay of Three R2R3 MYB Transcription Factors Determines the Profile of Aliphatic Glucosinolates in Arabidopsis 1[C][W][OA] Ida Elken Sønderby, Meike Burow, Heather C. Rowe 2, Daniel J. Kliebenstein,
Supplementary Figures
 Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
Supplemental Data. Wang et al. (2013). Plant Cell /tpc
 Supplemental Data. Wang et al. (2013). Plant Cell 10.1105/tpc.112.108993 Supplemental Figure 1. 3-MA Treatment Reduces the Growth of Seedlings. Two-week-old Nicotiana benthamiana seedlings germinated on
Supplemental Data. Wang et al. (2013). Plant Cell 10.1105/tpc.112.108993 Supplemental Figure 1. 3-MA Treatment Reduces the Growth of Seedlings. Two-week-old Nicotiana benthamiana seedlings germinated on
Københavns Universitet
 university of copenhagen Københavns Universitet Linking metabolic QTLs with network and cis-eqtls controlling biosynthetic pathways Wentzell, Adam M.; Rowe, Heather C.; Hansen, Bjarne Gram; Ticconi, Carla;
university of copenhagen Københavns Universitet Linking metabolic QTLs with network and cis-eqtls controlling biosynthetic pathways Wentzell, Adam M.; Rowe, Heather C.; Hansen, Bjarne Gram; Ticconi, Carla;
Supplemental data. Uppalapati et al. (2012) Plant Cell /tpc
 PAL Control P. emaculata 0 8 24 48 8 24 48 OPR Control P. emaculata 0 8 24 48 8 24 48 CHS PR3 CHR PR5 CHI PR10 IFS IFR Supplemental Figure 1. Expression profiles for selected genes in phenylpropanoid pathway
PAL Control P. emaculata 0 8 24 48 8 24 48 OPR Control P. emaculata 0 8 24 48 8 24 48 CHS PR3 CHR PR5 CHI PR10 IFS IFR Supplemental Figure 1. Expression profiles for selected genes in phenylpropanoid pathway
WT siz1-2 siz1-3. WT siz1-2 siz1-3. -Pi, 0.05 M IAA. -Pi, 2.5 M NPA
 A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
Genomic Analysis of QTLs and Genes Altering Natural Variation in Stochastic Noise
 Genomic Analysis of QTLs and Genes Altering Natural Variation in Stochastic Noise Jose M. Jimenez-Gomez 1., Jason A. Corwin 2., Bindu Joseph 2., Julin N. Maloof 1, Daniel J. Kliebenstein 2 * 1 Department
Genomic Analysis of QTLs and Genes Altering Natural Variation in Stochastic Noise Jose M. Jimenez-Gomez 1., Jason A. Corwin 2., Bindu Joseph 2., Julin N. Maloof 1, Daniel J. Kliebenstein 2 * 1 Department
Supplementary Figures and Tables
 Supplementary Figures and Tables Supplementary Figure 1. Study design and sample collection. S.japonicum were harvested from C57 mice at 8 time points after infection. Total number of samples for RNA-Seq:
Supplementary Figures and Tables Supplementary Figure 1. Study design and sample collection. S.japonicum were harvested from C57 mice at 8 time points after infection. Total number of samples for RNA-Seq:
Supplemental Data. Hiruma et al. Plant Cell. (2010) /tpc Col-0. pen2-1
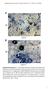 A Ch B Col-0 Cg pen2-1 Supplemental Figure 1. Trypan Blue Staining of Leaves Inoculated with Adapted and Nonadapted Colletotrichum Species.(A) Conidial suspensions of C. higginsianum MAFF305635 (Ch) were
A Ch B Col-0 Cg pen2-1 Supplemental Figure 1. Trypan Blue Staining of Leaves Inoculated with Adapted and Nonadapted Colletotrichum Species.(A) Conidial suspensions of C. higginsianum MAFF305635 (Ch) were
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated
 Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplemental Data. Beck et al. (2010). Plant Cell /tpc
 Supplemental Figure 1. Phenotypic comparison of the rosette leaves of four-week-old mpk4 and Col-0 plants. A mpk4 vs Col-0 plants grown in soil. Note the extreme dwarfism of the mpk4 plants (white arrows)
Supplemental Figure 1. Phenotypic comparison of the rosette leaves of four-week-old mpk4 and Col-0 plants. A mpk4 vs Col-0 plants grown in soil. Note the extreme dwarfism of the mpk4 plants (white arrows)
Oxi1 mutant plays an important role in Arabidopsis resistance against aphid (Myzus persicae)
 Oxi1 mutant plays an important role in Arabidopsis resistance against aphid (Myzus persicae) Dr. Tahsin Shoala Assistant professor - College of Biotechnology - Misr University for Science and Technology
Oxi1 mutant plays an important role in Arabidopsis resistance against aphid (Myzus persicae) Dr. Tahsin Shoala Assistant professor - College of Biotechnology - Misr University for Science and Technology
Supplemental Figure S1. Sequence feature and phylogenetic analysis of GmZF351. (A) Amino acid sequence alignment of GmZF351, AtTZF1, SOMNUS, AtTZF5,
 Supplemental Figure S. Sequence feature and phylogenetic analysis of GmZF35. (A) Amino acid sequence alignment of GmZF35, AtTZF, SOMNUS, AtTZF5, and OsTZF. Characters with black background indicate conserved
Supplemental Figure S. Sequence feature and phylogenetic analysis of GmZF35. (A) Amino acid sequence alignment of GmZF35, AtTZF, SOMNUS, AtTZF5, and OsTZF. Characters with black background indicate conserved
Molecular mechanism of the priming by jasmonic acid of specific dehydration stress response genes in Arabidopsis
 University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications in the Biological Sciences Papers in the Biological Sciences 2016 Molecular mechanism of the priming
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications in the Biological Sciences Papers in the Biological Sciences 2016 Molecular mechanism of the priming
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
MOLECULAR BIOLOGY AND GENETICS OF PROGRAMMED CELL DEATH IN ARABIDOPSIS THALIANA
 PROCEEDINGS OF THE BALKAN SCIENTIFIC CONFERENCE OF BIOLOGY IN PLOVDIV (BULGARIA) FROM 19 TH TILL 21 ST OF MAY 2005 (EDS B. GRUEV, M. NIKOLOVA AND A. DONEV), 2005 (P. 157 161) MOLECULAR BIOLOGY AND GENETICS
PROCEEDINGS OF THE BALKAN SCIENTIFIC CONFERENCE OF BIOLOGY IN PLOVDIV (BULGARIA) FROM 19 TH TILL 21 ST OF MAY 2005 (EDS B. GRUEV, M. NIKOLOVA AND A. DONEV), 2005 (P. 157 161) MOLECULAR BIOLOGY AND GENETICS
MG132. GRXS17:V5-His MG132 GRXS17:HA. RuBisCo. Supplemental Data. Walton et al. (2015). Plant Cell /tpc
 MG132 GRXS17:V5-His MG132 GRXS17:HA RuBisCo Supplemental Figure 1. Degradation of GRXS17 by the 26S proteasome. (A) Cell-free degradation assay. Recombinant protein GRXS17:V5-HIS was incubated with total
MG132 GRXS17:V5-His MG132 GRXS17:HA RuBisCo Supplemental Figure 1. Degradation of GRXS17 by the 26S proteasome. (A) Cell-free degradation assay. Recombinant protein GRXS17:V5-HIS was incubated with total
Biosynthesis of glucosinolates gene discovery and beyond
 Review Biosynthesis of glucosinolates gene discovery and beyond Ida E. Sønderby, Fernando Geu-Flores and Barbara A. Halkier Plant Biochemistry Laboratory, VKR Research Centre Pro-Active Plants, Department
Review Biosynthesis of glucosinolates gene discovery and beyond Ida E. Sønderby, Fernando Geu-Flores and Barbara A. Halkier Plant Biochemistry Laboratory, VKR Research Centre Pro-Active Plants, Department
High coverage in planta RNA sequencing identifies Fusarium oxysporum effectors and Medicago truncatularesistancemechanisms
 High coverage in planta RNA sequencing identifies Fusarium oxysporum effectors and Medicago truncatularesistancemechanisms Louise Thatcher Gagan Garg, Angela Williams, Judith Lichtenzveig and Karam Singh
High coverage in planta RNA sequencing identifies Fusarium oxysporum effectors and Medicago truncatularesistancemechanisms Louise Thatcher Gagan Garg, Angela Williams, Judith Lichtenzveig and Karam Singh
Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at
 Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at different concentrations for 30 min and analyzed for
Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at different concentrations for 30 min and analyzed for
fl/+ KRas;Atg5 fl/+ KRas;Atg5 fl/fl KRas;Atg5 fl/fl KRas;Atg5 Supplementary Figure 1. Gene set enrichment analyses. (a) (b)
 KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
Table S1. Relative abundance of AGO1/4 proteins in different organs. Table S2. Summary of smrna datasets from various samples.
 Supplementary files Table S1. Relative abundance of AGO1/4 proteins in different organs. Table S2. Summary of smrna datasets from various samples. Table S3. Specificity of AGO1- and AGO4-preferred 24-nt
Supplementary files Table S1. Relative abundance of AGO1/4 proteins in different organs. Table S2. Summary of smrna datasets from various samples. Table S3. Specificity of AGO1- and AGO4-preferred 24-nt
Biochemical Networks and Epistasis Shape the Plant Metabolome. Kliebenstein 1
 Biochemical Networks and Epistasis Shape the Plant Metabolome Heather C. Rowe 1, Bjarne Gram Hansen 2, Barbara Ann Halkier 2 and Daniel J. Kliebenstein 1 1 Genetics Graduate Group and Department of Plant
Biochemical Networks and Epistasis Shape the Plant Metabolome Heather C. Rowe 1, Bjarne Gram Hansen 2, Barbara Ann Halkier 2 and Daniel J. Kliebenstein 1 1 Genetics Graduate Group and Department of Plant
Supplemental Information. Spatial Auxin Signaling. Controls Leaf Flattening in Arabidopsis
 Current Biology, Volume 27 Supplemental Information Spatial Auxin Signaling Controls Leaf Flattening in Arabidopsis Chunmei Guan, Binbin Wu, Ting Yu, Qingqing Wang, Naden T. Krogan, Xigang Liu, and Yuling
Current Biology, Volume 27 Supplemental Information Spatial Auxin Signaling Controls Leaf Flattening in Arabidopsis Chunmei Guan, Binbin Wu, Ting Yu, Qingqing Wang, Naden T. Krogan, Xigang Liu, and Yuling
A -GLS Arabidopsis Cuscuta gronovii
 -GLS Arabidopsis Cuscuta gronovii internal standard 4MS Wildtype Arabidopsis Cuscuta gronovii base Cuscuta gronovii apex 3MSP internal standard MSP 4MT 8MSO 4MO 1MO Figure S1. Representative HPLC-DAD chromatograms
-GLS Arabidopsis Cuscuta gronovii internal standard 4MS Wildtype Arabidopsis Cuscuta gronovii base Cuscuta gronovii apex 3MSP internal standard MSP 4MT 8MSO 4MO 1MO Figure S1. Representative HPLC-DAD chromatograms
Effects of jasmonic acid signalling on the wheat microbiome differ between
 Supplementary Information: Effects of jasmonic acid signalling on the wheat microbiome differ between body sites Hongwei Liu 1, Lilia C. Carvalhais 1, Peer M. Schenk 1, Paul G. Dennis 1* 1 School of Agriculture
Supplementary Information: Effects of jasmonic acid signalling on the wheat microbiome differ between body sites Hongwei Liu 1, Lilia C. Carvalhais 1, Peer M. Schenk 1, Paul G. Dennis 1* 1 School of Agriculture
The radish genome and comprehensive gene expression profile of tuberous root formation
 The radish genome and comprehensive gene expression profile of tuberous root formation and development Yuki Mitsui 1*, Michihiko Shimomura 2*, Kenji Komatsu 1*, Nobukazu Namiki 2, Mari Shibata-Hatta 1,
The radish genome and comprehensive gene expression profile of tuberous root formation and development Yuki Mitsui 1*, Michihiko Shimomura 2*, Kenji Komatsu 1*, Nobukazu Namiki 2, Mari Shibata-Hatta 1,
Genomics of plant defense against insects. Dr. Philippe Reymond, MER
 Genomics of plant defense against insects Dr. Philippe Reymond, MER Herbivorous insects are abundant on earth and cause severe damage to crops Biomass of ants compared to biomass of vertebrates (4 to 1)
Genomics of plant defense against insects Dr. Philippe Reymond, MER Herbivorous insects are abundant on earth and cause severe damage to crops Biomass of ants compared to biomass of vertebrates (4 to 1)
m 6 A mrna methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
Production of the Cancer-Preventive Glucoraphanin in Tobacco
 Molecular Plant Volume 3 Number 4 Pages 751 759 July 2010 RESEARCH ARTICLE Production of the Cancer-Preventive Glucoraphanin in Tobacco Michael Dalgaard Mikkelsen a, Carl Erik Olsen b and Barbara Ann Halkier
Molecular Plant Volume 3 Number 4 Pages 751 759 July 2010 RESEARCH ARTICLE Production of the Cancer-Preventive Glucoraphanin in Tobacco Michael Dalgaard Mikkelsen a, Carl Erik Olsen b and Barbara Ann Halkier
SUPPLEMENTARY INFORMATION
 Supplementary igure 1. The expression level of PP4 (At4g16860) is diminished in the rpp4 mutant (SK017569). The mna was extracted to analyze the expression level of PP4 using quantitative PC (qpc). Gene
Supplementary igure 1. The expression level of PP4 (At4g16860) is diminished in the rpp4 mutant (SK017569). The mna was extracted to analyze the expression level of PP4 using quantitative PC (qpc). Gene
Nature Genetics: doi: /ng Supplementary Figure 1. Assessment of sample purity and quality.
 Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
A quantitative genetics and ecological model system: Understanding the. aliphatic glucosinolate biosynthetic network via QTLs
 A quantitative genetics and ecological model system: Understanding the aliphatic glucosinolate biosynthetic network via QTLs Daniel J. Kliebenstein 1 1 Department of Plant Sciences University of California,
A quantitative genetics and ecological model system: Understanding the aliphatic glucosinolate biosynthetic network via QTLs Daniel J. Kliebenstein 1 1 Department of Plant Sciences University of California,
Supplemental Information. Figures. Figure S1
 Supplemental Information Figures Figure S1 Identification of JAGGER T-DNA insertions. A. Positions of T-DNA and Ds insertions in JAGGER are indicated by inverted triangles, the grey box represents the
Supplemental Information Figures Figure S1 Identification of JAGGER T-DNA insertions. A. Positions of T-DNA and Ds insertions in JAGGER are indicated by inverted triangles, the grey box represents the
Analysis of Plant Resistance against Herbivorous Insects with Molecular and Quantitative Genetics
 Analysis of Plant Resistance against Herbivorous Insects with Molecular and Quantitative Genetics Dissertation zur Erlangung des akademischen Grades doctor rerum naturalium (Dr. rer. nat.) vorgelegt dem
Analysis of Plant Resistance against Herbivorous Insects with Molecular and Quantitative Genetics Dissertation zur Erlangung des akademischen Grades doctor rerum naturalium (Dr. rer. nat.) vorgelegt dem
A basic helix loop helix transcription factor controls cell growth
 A basic helix loop helix transcription factor controls cell growth and size in root hairs Keke Yi 1,2, Benoît Menand 1,3, Elizabeth Bell 1, Liam Dolan 1,4 Supplementary note Low soil phosphate availability
A basic helix loop helix transcription factor controls cell growth and size in root hairs Keke Yi 1,2, Benoît Menand 1,3, Elizabeth Bell 1, Liam Dolan 1,4 Supplementary note Low soil phosphate availability
Jyoti Shah, Vamsi Nalam, Sujon Sarowar, Syeda Alam, Sumita Behera, Hyeonju Lee, Delia Burdan and Harold N. Trick
 Targeting Host Defense and Susceptibility Mechanisms for Engineering FHB Resistance Jyoti Shah, Vamsi Nalam, Sujon Sarowar, Syeda Alam, Sumita Behera, Hyeonju Lee, Delia Burdan and Harold N. Trick General
Targeting Host Defense and Susceptibility Mechanisms for Engineering FHB Resistance Jyoti Shah, Vamsi Nalam, Sujon Sarowar, Syeda Alam, Sumita Behera, Hyeonju Lee, Delia Burdan and Harold N. Trick General
Supplementary Materials: Global Transcriptomic Analysis Reveals the Mechanism of Phelipanche Aegyptiaca Seed Germination
 Int. J. Mol. Sci. 2016, 17, 1139; doi:.3390/ijms17071139 S1 of S5 Supplementary Materials: Global Transcriptomic Analysis Reveals the Mechanism of Phelipanche Aegyptiaca Seed Germination Zhaoqun Yao, Fang
Int. J. Mol. Sci. 2016, 17, 1139; doi:.3390/ijms17071139 S1 of S5 Supplementary Materials: Global Transcriptomic Analysis Reveals the Mechanism of Phelipanche Aegyptiaca Seed Germination Zhaoqun Yao, Fang
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
Supplementary Table 3. 3 UTR primer sequences. Primer sequences used to amplify and clone the 3 UTR of each indicated gene are listed.
 Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Lecture 3-4. Identification of Positive Regulators downstream of SA
 Lecture 3-4 Identification of Positive Regulators downstream of SA Positive regulators between SA and resistance/pr Systemic resistance PR gene expression? SA Local infection Necrosis SA What could they
Lecture 3-4 Identification of Positive Regulators downstream of SA Positive regulators between SA and resistance/pr Systemic resistance PR gene expression? SA Local infection Necrosis SA What could they
A smart acid nanosystem for ultrasensitive. live cell mrna imaging by the target-triggered intracellular self-assembly
 Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2017 A smart ZnO@polydopamine-nucleic acid nanosystem for ultrasensitive live cell mrna imaging
Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2017 A smart ZnO@polydopamine-nucleic acid nanosystem for ultrasensitive live cell mrna imaging
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. H3F3B expression in lung cancer. a. Comparison of H3F3B expression in relapsed and non-relapsed lung cancer patients. b. Prognosis of two groups of lung cancer
Supplementary Figures Supplementary Figure 1. H3F3B expression in lung cancer. a. Comparison of H3F3B expression in relapsed and non-relapsed lung cancer patients. b. Prognosis of two groups of lung cancer
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
A putative MYB35 ortholog is a candidate for the sex-determining genes in Asparagus
 Supplementary figures for: A putative MYB35 ortholog is a candidate for the sex-determining genes in Asparagus officinalis Daisuke Tsugama, Kohei Matsuyama, Mayui Ide, Masato Hayashi, Kaien Fujino, and
Supplementary figures for: A putative MYB35 ortholog is a candidate for the sex-determining genes in Asparagus officinalis Daisuke Tsugama, Kohei Matsuyama, Mayui Ide, Masato Hayashi, Kaien Fujino, and
Supplemental Figure 1. Genes showing ectopic H3K9 dimethylation in this study are DNA hypermethylated in Lister et al. study.
 mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
Supplementary Figure 1. ALVAC-protein vaccines and macaque immunization. (A) Maximum likelihood
 Supplementary Figure 1. ALVAC-protein vaccines and macaque immunization. (A) Maximum likelihood tree illustrating CRF01_AE gp120 protein sequence relationships between 107 Envs sampled in the RV144 trial
Supplementary Figure 1. ALVAC-protein vaccines and macaque immunization. (A) Maximum likelihood tree illustrating CRF01_AE gp120 protein sequence relationships between 107 Envs sampled in the RV144 trial
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
A bts-1 (SALK_016526)
 Electronic Supplementary Material (ESI) for Metallomics. This journal is The Royal Society of Chemistry 217 A ts-1 (SALK_16526) BTS (At3g1829) ATG tsl1 (SALK_1554) 2 p BTSL1 (At1g7477) ATG tsl2 (SAIL_615_HO1)
Electronic Supplementary Material (ESI) for Metallomics. This journal is The Royal Society of Chemistry 217 A ts-1 (SALK_16526) BTS (At3g1829) ATG tsl1 (SALK_1554) 2 p BTSL1 (At1g7477) ATG tsl2 (SAIL_615_HO1)
Figure S1 Expression of AHL gene family members in diploid (Ler Col) and triploid (Ler
 Supplemental material Supplemental figure legends Figure S Expression of AHL gene family members in diploid (Ler ) and triploid (Ler osd) seeds. AHLs from clade B are labelled with (I), and AHLs from clade
Supplemental material Supplemental figure legends Figure S Expression of AHL gene family members in diploid (Ler ) and triploid (Ler osd) seeds. AHLs from clade B are labelled with (I), and AHLs from clade
Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 )
 770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
Molecular Plant Volume 5 Number 6 Pages November 2012
 Molecular Plant Volume 5 Number 6 Pages 1389 1402 November 2012 RESEARCH ARTICLE Transcriptional Activation and Production of Tryptophan-Derived Secondary Metabolites in Arabidopsis Roots Contributes to
Molecular Plant Volume 5 Number 6 Pages 1389 1402 November 2012 RESEARCH ARTICLE Transcriptional Activation and Production of Tryptophan-Derived Secondary Metabolites in Arabidopsis Roots Contributes to
Supporting Information
 Supporting Information Lee et al. 10.1073/pnas.0910950106 Fig. S1. Fe (A), Zn (B), Cu (C), and Mn (D) concentrations in flag leaves from WT, osnas3-1, and OsNAS3-antisense (AN-2) plants. Each measurement
Supporting Information Lee et al. 10.1073/pnas.0910950106 Fig. S1. Fe (A), Zn (B), Cu (C), and Mn (D) concentrations in flag leaves from WT, osnas3-1, and OsNAS3-antisense (AN-2) plants. Each measurement
RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays
 Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana
 Biochemistry: Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana 38 CAMERON E. NIENABER ʻ04 Abstract Long recognized as essential plant nutrients and metabolites, inorganic and organic
Biochemistry: Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana 38 CAMERON E. NIENABER ʻ04 Abstract Long recognized as essential plant nutrients and metabolites, inorganic and organic
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation
 Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplemental Data. Jones et al. (2012). Plant Cell /tpc
 Supplemental Data. Jones et al. (). Plant Cell.5/tpc..88 Relative ioluminescence C Relative ioluminescence E Relative ioluminescence 8 7 96 8 7 96 STIPL:STIPL-GFP 8 7 96 Relative ioluminescence Relative
Supplemental Data. Jones et al. (). Plant Cell.5/tpc..88 Relative ioluminescence C Relative ioluminescence E Relative ioluminescence 8 7 96 8 7 96 STIPL:STIPL-GFP 8 7 96 Relative ioluminescence Relative
Nature Immunology: doi: /ni Supplementary Figure 1. Transcriptional program of the TE and MP CD8 + T cell subsets.
 Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Evaluation of Changes in Glucosinolates Profile and Biosynthetic Genes Expression in Turnip (Brassica rapa L.)
 Evaluation of Changes in Glucosinolates Profile and Biosynthetic Genes Expression in Turnip (Brassica rapa L.) Supervisors Dr. ir. Guusje Bonnema Dr. Jules Beekwilder Dr.ir. Chris Maliepaard Ir. Johan
Evaluation of Changes in Glucosinolates Profile and Biosynthetic Genes Expression in Turnip (Brassica rapa L.) Supervisors Dr. ir. Guusje Bonnema Dr. Jules Beekwilder Dr.ir. Chris Maliepaard Ir. Johan
Supplemental Figure S1. The number of hydathodes is reduced in the as2-1 rev-1
 Supplemental Data Supplemental Figure S1. The number of hydathodes is reduced in the as2-1 rev-1 and kan1-11 kan2-5 double mutants. A, The numbers of hydathodes in different leaves of Col-0, as2-1 rev-1,
Supplemental Data Supplemental Figure S1. The number of hydathodes is reduced in the as2-1 rev-1 and kan1-11 kan2-5 double mutants. A, The numbers of hydathodes in different leaves of Col-0, as2-1 rev-1,
Sbarrato_Supplementary_Fig1
 Sbarrato_Supplementary_Fig1 Supplementary Figure 1. Translatome analysis of CLL patients based on IGVH mutational status ) Profile for the translatome of unmutated IGVH CLL versus mutated IGVH CLL pa9ents.
Sbarrato_Supplementary_Fig1 Supplementary Figure 1. Translatome analysis of CLL patients based on IGVH mutational status ) Profile for the translatome of unmutated IGVH CLL versus mutated IGVH CLL pa9ents.
Influenza virus exploits tunneling nanotubes for cell-to-cell spread
 Supplementary Information Influenza virus exploits tunneling nanotubes for cell-to-cell spread Amrita Kumar 1, Jin Hyang Kim 1, Priya Ranjan 1, Maureen G. Metcalfe 2, Weiping Cao 1, Margarita Mishina 1,
Supplementary Information Influenza virus exploits tunneling nanotubes for cell-to-cell spread Amrita Kumar 1, Jin Hyang Kim 1, Priya Ranjan 1, Maureen G. Metcalfe 2, Weiping Cao 1, Margarita Mishina 1,
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Folate stability in GA9.15
 Supplementary Figure 1 Folate stability in GA9.15 Total folate levels in GA9.15 until 7 months of storage at 28 C. Seeds of the sixth (T5) generation (upon harvest stored at -80 C) were used to confirm
Supplementary Figure 1 Folate stability in GA9.15 Total folate levels in GA9.15 until 7 months of storage at 28 C. Seeds of the sixth (T5) generation (upon harvest stored at -80 C) were used to confirm
12-Oxo-Phytodienoic Acid Accumulation during Seed Development Represses Seed Germination in Arabidopsis C W OA
 The Plant Cell, Vol. 23: 583 599, February 2011, www.plantcell.org ã 2011 American Society of Plant Biologists 12-Oxo-Phytodienoic Acid Accumulation during Seed Development Represses Seed Germination in
The Plant Cell, Vol. 23: 583 599, February 2011, www.plantcell.org ã 2011 American Society of Plant Biologists 12-Oxo-Phytodienoic Acid Accumulation during Seed Development Represses Seed Germination in
Short-Term and Continuing Stresses Differentially Interplay with Multiple Hormones to Regulate Plant Survival and Growth
 Molecular Plant Volume 7 Number 5 Pages 841 855 May 2014 RESEARCH ARTICLE Short-Term and Continuing Stresses Differentially Interplay with Multiple Hormones to Regulate Plant Survival and Growth Cangjing
Molecular Plant Volume 7 Number 5 Pages 841 855 May 2014 RESEARCH ARTICLE Short-Term and Continuing Stresses Differentially Interplay with Multiple Hormones to Regulate Plant Survival and Growth Cangjing
Role of 9-Lipoxygenase and a-dioxygenase Oxylipin Pathways as Modulators of Local and Systemic Defense
 RESEARCH ARTICLE Role of 9-Lipoxygenase and a-dioxygenase Oxylipin Pathways as Modulators of Local and Systemic Defense Jorge Vicente a, Tomás Cascón a, Begonya Vicedo b, Pilar García-Agustín b, Mats Hamberg
RESEARCH ARTICLE Role of 9-Lipoxygenase and a-dioxygenase Oxylipin Pathways as Modulators of Local and Systemic Defense Jorge Vicente a, Tomás Cascón a, Begonya Vicedo b, Pilar García-Agustín b, Mats Hamberg
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Biosynthesis and functions of free and combined fatty alcohols associated with suberin
 Laboratoire Biosynthesis and functions of free and combined fatty alcohols associated with suberin Collaborating Laboratories: Dr. Owen Rowland Sollapura Vishwanath PhD Candidate Department of Biology
Laboratoire Biosynthesis and functions of free and combined fatty alcohols associated with suberin Collaborating Laboratories: Dr. Owen Rowland Sollapura Vishwanath PhD Candidate Department of Biology
SUPPLEMENTAL INFORMATIONS
 1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
4. Th1-related gene expression in infected versus mock-infected controls from Fig. 2 with gene annotation.
 List of supplemental information 1. Graph of mouse weight loss during course of infection- Line graphs showing mouse weight data during course of infection days 1 to 10 post-infections (p.i.). 2. Graph
List of supplemental information 1. Graph of mouse weight loss during course of infection- Line graphs showing mouse weight data during course of infection days 1 to 10 post-infections (p.i.). 2. Graph
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH
 EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53
 1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most
 Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
Open Flower. Juvenile leaf Flowerbud. Carpel 35 NA NA NA NA 61 NA 95 NA NA 15 NA 41 3 NA
 PaxDB Root Juvenile leaf Flowerbud Open Flower Carpel Mature Pollen Silique Seed Sec3a Sec3b Sec5a Sec5b Sec6 Sec8 Sec10a/b Sec15a Sec15b Exo84a Exo84b Exo84c Exo70A1 Exo70A2 Exo70A3 49 47 8 75 104 79
PaxDB Root Juvenile leaf Flowerbud Open Flower Carpel Mature Pollen Silique Seed Sec3a Sec3b Sec5a Sec5b Sec6 Sec8 Sec10a/b Sec15a Sec15b Exo84a Exo84b Exo84c Exo70A1 Exo70A2 Exo70A3 49 47 8 75 104 79
Expression constructs
 Gene expressed in bebe3 ZmBEa Expression constructs 35S ZmBEa Pnos:Hygromycin r 35S Pnos:Hygromycin r 35S ctp YFP Pnos:Hygromycin r B -1 Chl YFP- Merge Supplemental Figure S1: Constructs Used for the Expression
Gene expressed in bebe3 ZmBEa Expression constructs 35S ZmBEa Pnos:Hygromycin r 35S Pnos:Hygromycin r 35S ctp YFP Pnos:Hygromycin r B -1 Chl YFP- Merge Supplemental Figure S1: Constructs Used for the Expression
Supplemental Table 1 Molecular Profile of the SCLC Cell Line Panel
 Supplemental Table 1 Molecular Profile of the SCLC Cell Line Panel p53 RB Myc Cell Line Mutation A Mutation A Amplification B COR-L103 C p.y234c p.d584e L-Myc NCI-H526 p.s33_splice None N-Myc NCI-H1048
Supplemental Table 1 Molecular Profile of the SCLC Cell Line Panel p53 RB Myc Cell Line Mutation A Mutation A Amplification B COR-L103 C p.y234c p.d584e L-Myc NCI-H526 p.s33_splice None N-Myc NCI-H1048
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1164627/dc1 Supporting Online Material for Glucosinolate Metabolites Required for an Arabidopsis Innate Immune Response Nicole K. Clay, Adewale M. Adio, Carine Denoux,
www.sciencemag.org/cgi/content/full/1164627/dc1 Supporting Online Material for Glucosinolate Metabolites Required for an Arabidopsis Innate Immune Response Nicole K. Clay, Adewale M. Adio, Carine Denoux,
Table S1. Total and mapped reads produced for each ChIP-seq sample
 Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
Nature Structural & Molecular Biology: doi: /nsmb.3218
 Supplementary Figure 1 Endogenous EGFR trafficking and responses depend on biased ligands. (a) Lysates from HeLa cells stimulated for 2 min. with increasing concentration of ligands were immunoblotted
Supplementary Figure 1 Endogenous EGFR trafficking and responses depend on biased ligands. (a) Lysates from HeLa cells stimulated for 2 min. with increasing concentration of ligands were immunoblotted
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Generation and validation of mtef4-knockout mice.
 Supplementary Figure 1 Generation and validation of mtef4-knockout mice. (a) Alignment of EF4 (E. coli) with mouse, yeast and human EF4. (b) Domain structures of mouse mtef4 compared to those of EF4 (E.
Supplementary Figure 1 Generation and validation of mtef4-knockout mice. (a) Alignment of EF4 (E. coli) with mouse, yeast and human EF4. (b) Domain structures of mouse mtef4 compared to those of EF4 (E.
Supplementary figure 1
 Supplementary figure 1 Nature Medicine: doi:1.138/nm.275 CLUSTER BY SELF-ORGANIZING MAPS SELECTED PATHWAY ANALISYS TERMS Cluster : up-regulated genes in acute patients Cell cycle/dna repair Fatty acid
Supplementary figure 1 Nature Medicine: doi:1.138/nm.275 CLUSTER BY SELF-ORGANIZING MAPS SELECTED PATHWAY ANALISYS TERMS Cluster : up-regulated genes in acute patients Cell cycle/dna repair Fatty acid
Toluidin-Staining of mast cells Ear tissue was fixed with Carnoy (60% ethanol, 30% chloroform, 10% acetic acid) overnight at 4 C, afterwards
 Toluidin-Staining of mast cells Ear tissue was fixed with Carnoy (60% ethanol, 30% chloroform, 10% acetic acid) overnight at 4 C, afterwards incubated in 100 % ethanol overnight at 4 C and embedded in
Toluidin-Staining of mast cells Ear tissue was fixed with Carnoy (60% ethanol, 30% chloroform, 10% acetic acid) overnight at 4 C, afterwards incubated in 100 % ethanol overnight at 4 C and embedded in
Supplementary Information. Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase
 Journal: Nature Medicine Supplementary Information Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase 1,2 Peng Wang PhD, 1,2 Juan-Carlos Alvarez-Perez
Journal: Nature Medicine Supplementary Information Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase 1,2 Peng Wang PhD, 1,2 Juan-Carlos Alvarez-Perez
Studies on arabiopsides using two Arabidopsis thaliana ecotypes
 Studies on arabiopsides using two Arabidopsis thaliana ecotypes Nilay Peker Practical work, July - August 2010, 15 hp Department of Plant and Environmental Sciences University of Gothenburg Abstract In
Studies on arabiopsides using two Arabidopsis thaliana ecotypes Nilay Peker Practical work, July - August 2010, 15 hp Department of Plant and Environmental Sciences University of Gothenburg Abstract In
Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types.
 Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Nature Immunology: doi: /ni Supplementary Figure 1. Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice.
 Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Nature Getetics: doi: /ng.3471
 Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
Nicotine biosynthesis pathway: beyond tobacco
 Nicotine biosynthesis pathway: beyond tobacco Masataka Kajikawa, Nicolas Sierro, Haruhiko Kawaguchi, Nicolas Bakaher, Nikolai V Ivanov, Takashi Hashimoto, Tsubasa Shoji Nicotiana tabacum Cultivated as
Nicotine biosynthesis pathway: beyond tobacco Masataka Kajikawa, Nicolas Sierro, Haruhiko Kawaguchi, Nicolas Bakaher, Nikolai V Ivanov, Takashi Hashimoto, Tsubasa Shoji Nicotiana tabacum Cultivated as
Involvement of phytohormones and transcription factors in tissue - reunion in the cut tissues of plant
 奨励賞 植物の生長調節 Regulation of Plant Growth & Development Vol.48, No.1, 17-23, 2013 Involvement of phytohormones and transcription factors in tissue - reunion in the cut tissues of plant Masashi Asahina Department
奨励賞 植物の生長調節 Regulation of Plant Growth & Development Vol.48, No.1, 17-23, 2013 Involvement of phytohormones and transcription factors in tissue - reunion in the cut tissues of plant Masashi Asahina Department
Potential use of High iron and low phytate GM rice and their Bio-safety Assessment
 Potential use of High iron and low phytate GM rice and their Bio-safety Assessment Dr. Karabi Datta University of Calcutta, India Background High iron rice and iron bioavailability Micronutrient deficiency
Potential use of High iron and low phytate GM rice and their Bio-safety Assessment Dr. Karabi Datta University of Calcutta, India Background High iron rice and iron bioavailability Micronutrient deficiency
Culture Density (OD600) 0.1. Culture Density (OD600) Culture Density (OD600) Culture Density (OD600) Culture Density (OD600)
 A. B. C. D. E. PA JSRI JSRI 2 PA DSAM DSAM 2 DSAM 3 PA LNAP LNAP 2 LNAP 3 PAO Fcor Fcor 2 Fcor 3 PAO Wtho Wtho 2 Wtho 3 Wtho 4 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB
A. B. C. D. E. PA JSRI JSRI 2 PA DSAM DSAM 2 DSAM 3 PA LNAP LNAP 2 LNAP 3 PAO Fcor Fcor 2 Fcor 3 PAO Wtho Wtho 2 Wtho 3 Wtho 4 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB
Metabolic Engineering in Nicotiana benthamiana Reveals Key Enzyme Functions in Arabidopsis Indole Glucosinolate Modification W
 The Plant Cell, Vol. 23: 716 729, February 2011, www.plantcell.org ã 2011 American Society of Plant Biologists Metabolic Engineering in Nicotiana benthamiana Reveals Key Enzyme Functions in Arabidopsis
The Plant Cell, Vol. 23: 716 729, February 2011, www.plantcell.org ã 2011 American Society of Plant Biologists Metabolic Engineering in Nicotiana benthamiana Reveals Key Enzyme Functions in Arabidopsis
Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition
 In the format provided y the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 217.15 Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition Chloé
In the format provided y the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 217.15 Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition Chloé
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato
 The Plant Journal (2010) 63, 443 457 doi: 10.1111/j.1365-313X.2010.04254.x The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato Jane L.
The Plant Journal (2010) 63, 443 457 doi: 10.1111/j.1365-313X.2010.04254.x The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato Jane L.
