Supplementary Table 2b. List of spliced oncogenes and tumor suppressors identified in AML patients compared to normal donors
|
|
|
- Albert Sanders
- 5 years ago
- Views:
Transcription
1 1 Supplementary Table 2b. List of spliced oncogenes and tumor suppressors identified in AML patients compared to normal donors Gene Symbol Gene Name / Tumor Suppressors AKT1 v-akt murine thymoma viral oncogene homolog 1 v-raf v-raf murine sarcoma 3611 viral oncogene homolog BCL2 B-cell CLL/lymphoma 2 BCR breakpoint cluster region BRAF v-raf murine sarcoma viral oncogene homolog B1 BRCA1 breast cancer 1, early onset CASP8 caspase 8, apoptosis-related cysteine peptidase CBLB Cas-Br-M (murine) ecotropic retroviral transforming sequence b CCND2 cyclin D2 CDC42 cell division cycle 42 (GTP binding protein, 25kDa) CENTG1 centaurin, gamma 1 CSF1R colony stimulating factor 1 receptor, formerly McDonough feline sarcoma viral (v-fms) oncogene homolog DAPK1 death-associated protein kinase 1 ETS1 v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) FGFR1 fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) FOS v-fos FBJ murine osteosarcoma viral oncogene homolog HEXB hexosaminidase B (beta polypeptide) HIF1A hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) HSP90AA1 heat shock protein 90kDa alpha (cytosolic), class A member 1 IL8 interleukin 8 ITGB1 integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) JUN jun oncogene JUP junction plakoglobin KIT v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog LYN v-yes-1 Yamaguchi sarcoma viral related oncogene homolog MAP2K3 mitogen-activated protein kinase kinase 3 MCM3 minichromosome maintenance complex component 3 NCOA4 nuclear receptor coactivator 4 PAK1 p21/cdc42/rac1-activated kinase 1 (STE20 homolog, yeast) PIK3CG phosphoinositide-3-kinase, catalytic, gamma polypeptide PLCG2 phospholipase C, gamma 2 (phosphatidylinositol-specific) PML promyelocytic leukemia PTPN11 protein tyrosine phosphatase, non-receptor type 11 (Noonan syndrome 1)
2 2 RAB31 RAB31, member RAS oncogene family RAB5B RAB5B, member RAS oncogene family RAPGEF1 Rap guanine nucleotide exchange factor (GEF) 1 SNRPE small nuclear ribonucleoprotein polypeptide E SOS1 son of sevenless homolog 1 (Drosophila) THOC4 THO complex 4 TPR translocated promoter region (to activated MET oncogene) VAV1 vav 1 oncogene VAV3 vav 3 oncogene CXCR4 chemokine (C-X-C motif) receptor 4 NFKB1 nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105) PIK3CA phosphoinositide-3-kinase, catalytic, alpha polypeptide PRKAR1A protein kinase, camp-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) PTGS2 prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) RB1 retinoblastoma 1 (including osteosarcoma) RELA v-rel reticuloendotheliosis viral oncogene homolog A RUNX1 runt-related transcription factor 1 (acute myeloid leukemia 1) STAT3 signal transducer and activator of transcription 3 UBE3A ubiquitin protein ligase E3A TP53 tumor protein p53 (Li-Fraumeni syndrome) Tumor Suppressor ARRB2 arrestin, beta 2 Tumor Suppressors ATM ataxia telangiectasia mutated (includes complementation groups A, C and D) Tumor Suppressors BIRC5 baculoviral IAP repeat-containing 5 (survivin) Tumor Suppressors CD4 CD4 molecule Tumor Suppressors CD59 CD59 molecule, complement regulatory protein Tumor Suppressors CD81 CD81 molecule Tumor Suppressors CDC14A CDC14 cell division cycle 14 homolog A (S. cerevisiae) Tumor Suppressors CFLAR CASP8 and FADD-like apoptosis regulator Tumor Suppressors CREB1 camp responsive element binding protein 1 Tumor Suppressors CREBBP CREB binding protein (Rubinstein-Taybi syndrome) Tumor Suppressors CUL1 cullin 1 Tumor Suppressors DOT1L DOT1-like, histone H3 methyltransferase (S. cerevisiae) Tumor Suppressors DUSP6 dual specificity phosphatase 6 Tumor Suppressors ENO1 enolase 1, (alpha) Tumor Suppressors FLNA filamin A, alpha (actin binding protein 280) Tumor Suppressors FLT3 fms-related tyrosine kinase 3 Tumor Suppressors FRAP1 FK506 binding protein 12-rapamycin associated protein 1 Tumor Suppressors GSTP1 glutathione S-transferase pi Tumor Suppressors
3 3 HDAC2 histone deacetylase 2 Tumor Suppressors IGF1R insulin-like growth factor 1 receptor Tumor Suppressors INPP4B inositol polyphosphate-4-phosphatase, type II, 105kDa Tumor Suppressors ITGA5 integrin, alpha 5 (fibronectin receptor, alpha polypeptide) Tumor Suppressors ITGB3 integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) Tumor Suppressors JAK2 Janus kinase 2 (a protein tyrosine kinase) Tumor Suppressors MAP3K4 mitogen-activated protein kinase kinase kinase 4 Tumor Suppressors MAPK14 mitogen-activated protein kinase 14 Tumor Suppressors MAPK8 mitogen-activated protein kinase 8 Tumor Suppressors MSH2 muts homolog 2, colon cancer, nonpolyposis type 1 (E. coli) Tumor Suppressors NF1 neurofibromin 1 (neurofibromatosis, von Recklinghausen disease, Watson disease) Tumor Suppressors NR4A1 nuclear receptor subfamily 4, group A, member 1 Tumor Suppressors PECAM1 platelet/endothelial cell adhesion molecule (CD31 antigen) Tumor Suppressors PPM1A protein phosphatase 1A (formerly 2C), magnesium-dependent, alpha isoform Tumor Suppressors PRDM4 PR domain containing 4 Tumor Suppressors PRKCD protein kinase C, delta Tumor Suppressors PTPRJ protein tyrosine phosphatase, receptor type, J Tumor Suppressors RBL1 retinoblastoma-like 1 (p107) Tumor Suppressors SMAD2 SMAD family member 2 Tumor Suppressors SMAD3 SMAD family member 3 Tumor Suppressors SMAD4 SMAD family member 4 Tumor Suppressors STAT5A signal transducer and activator of transcription 5A Tumor Suppressors STK3 serine/threonine kinase 3 (STE20 homolog, yeast) Tumor Suppressors STK4 serine/threonine kinase 4 Tumor Suppressors SYK spleen tyrosine kinase Tumor Suppressors TRAF6 TNF receptor-associated factor 6 Tumor Suppressors TUSC4 tumor suppressor candidate 4 Tumor Suppressors UBE4B ubiquitination factor E4B (UFD2 homolog, yeast) Tumor Suppressors WASL Wiskott-Aldrich syndrome-like Tumor Suppressors YWHAE tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide Tumor Suppressors ZYX zyxin Tumor Suppressors Annotation analyses of the differentially spliced transcripts detected in both DFCI and UHN cohorts identified novel splice variants of 52 oncogenes and 50 tumor suppressor genes that are listed in this table. Splice variants of these genes were recurrently detected in AML patients as compared to normal donors.
4 4 Supplementary Table 2c. List of genes spliced in both DFCI and UHN cohorts and absent in normal donors Differentially spliced gene symbol DFCI cohort (# of patients) UHN cohort (# of patients) RPS TRBV TNFSF12-TNFSF GOLGA CTNND PVRL HLA-C CHD HSPG UPK3B FLJ IGLV USP EEF1A C13ORF RPA NAIP 9 29 IGKC MAP3K MARVELD FLJ LTF HLA-DRB PDE4B MPL ARHGAP WDR MARK PRG PRKDC MLL TIMELESS TSC22D DDX MED12L RTN PRDM DOCK PDE4D DNTT AFF SEC31A KIAA
5 5 UBAP SNORD EDD JMJD MSX2P MLL TBL1X 9 22 PDE4DIP SRGN CAST CREBL ERC BAZ1B CBLB PTPN ATM 9 21 ITGA PKD RUNX FABP SLC44A SRRM BAT2D SF CFD APP MPO ARHGEF EPB STXBP LTBP BAZ1A PLXND SLC7A5P HDAC7A HDAC ZDHHC ANK HD HLA-DQA RBM ZUBR SPTA BMS TNS GOLGA8A LYST 10 18
6 6 C1ORF PTPRC HERC WHSC RARS HIPK IGLL ATRN ARHGAP HERC ZNF ZMYM BIRC VCAN NOTCH PDCD MACF KIAA DCTN ITGAX POLR2A MED ANAPC CA DPYSL PCAF GOLGA8B FOSB 9 16 MYCBP DOCK PTPRA TXNDC BPTF EIF4G MLL SORL SMARCA GTF2I NIPBL TTLL VCL FAM38A JARID1B CDK5RAP PTPRS HIST1H2AG DDEF
7 7 HIST1H3D LY DUT AGTPBP RNF FLT SFXN RERE 9 14 CD UBE2L SRBD ATXN LARS WNK DOCK PROM LEPROT JUP TOP2A FNDC3B PRAM TBCD MKI RABGAP1L SPTAN MAN1A MEF2C 9 13 WDR CD IDS 8 13 LGMN SYNE HIST1H2AC PHKB UBAC C2ORF FMNL NIN CHD FGR BNIP3L GUCY1A LILRB NCOR CHD COG EMR
8 8 ATP2C TRAK OAS GTF3C MDN CDC ILF NCAPD FCN UBE3C FNIP UBAP2L VAV SMARCA SLC7A5P APBB1IP EIF3S CLTC MEIS ME SIN3A EIF4G MCTP SIDT F11R GSTM SLC2A DDX WWP PPRC CPNE HLA-B 15 9 HIST1H1C 12 9 PRDX ASH1L 11 9 ZC3HAV ATE1 9 9 ZMYND MALAT UQCRC FANCI 12 8 HBA ATP2B OGT 10 8 ANXA BRPF ST7 9 8
9 9 DMXL1 9 8 CLASP1 9 8 MLL4 9 8 GIT2 9 8 PXDNL 18 7 TGFBI 16 7 CD TTC SMC1A 10 7 DNAJB CHST2 9 7 SNORA ACTN EHMT WDR40A 10 6 PDHA1 9 6 SETD3 9 6 SEL1L 9 6 HSPA1A 12 5 BCL FHL MBOAT SEPT CTSG 12 4 OGDH 9 4 SAMHD1 9 4 CDC42BPA 9 4 PCNT 11 3 SLC2A PRKACB 10 2 MLL3 9 2 RAB8B 16 1 FTH BPGM 10 1 NUP98 21 FLJ COL24A1 19 NCAPH 18 DDX3Y 17 CUL1 15 SFRS1 15 PFAS 15 ZBTB44 15 FAM120A 13 SAFB2 13 ABHD2 13 DOCK1 13
10 10 RBMX 13 BCL11A 12 PML 12 PPM1A 12 PNN 12 HNRPH3 12 TSC1 11 POMT1 11 TGFBR1 11 ANKRD17 11 SMC4 11 L3MBTL2 11 HM13 11 PPP3R1 11 RIOK3 11 FANCA 11 ARHGAP5 11 ZDHHC20 11 NLRP3 11 IARS 10 JMJD2C 10 SMURF1 10 TBRG4 10 EEF1A1 10 ARID1B 10 HERC5 10 DOPEY2 10 C20ORF19 10 SP3 10 PSD4 10 AFF3 10 SOS1 10 DOT1L 10 CTDP1 10 RALBP1 10 C14ORF ADRBK1 10 ZRANB1 10 SUPV3L1 10 CCDC7 10 USP33 10 EEF1A1 9 PHIP 9 MAN2A1 9 RICTOR 9 KLHL24 9 PLSCR1 9
11 11 DIRC2 9 NUP50 9 C21ORF59 9 SP100 9 CXCR4 9 KPNA2 9 MGC BIRC5 9 SUPT4H1 9 NFAT5 9 MT2A 9 SMG1 9 ZFAND6 9 KIAA CUL4A 9 EP400 9 PPHLN1 9 SNX15 9 PDCD4 9 ZDHHC16 9 SPOCK2 9 EIF2C3 9 FUSIP1 9 UTY 8 WWC3 8 This Table is summary of the analysis obtained by evaluating the extent of similarities between the DFCI and UHN patient cohorts with respect to splicing event frequency. We found that 75% of the transcripts that were recurrently spliced in 30% or more of DFCI patient cohort were also recurrently spliced in the UHN patient cohort.
Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction. Marker Sequence (5 3 ) Accession No.
 Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Developing a clinically feasible personalized medicine approach to pediatric
 Developing a clinically feasible personalized medicine approach to pediatric septic shock Hector R. Wong, Natalie Z. Cvijanovich, Nick Anas, Geoffrey L. Allen, Neal J. Thomas, Michael T. Bigham, Scott
Developing a clinically feasible personalized medicine approach to pediatric septic shock Hector R. Wong, Natalie Z. Cvijanovich, Nick Anas, Geoffrey L. Allen, Neal J. Thomas, Michael T. Bigham, Scott
TNFSF13B tumor necrosis factor (ligand) superfamily, member 13b NF-kB pathway cluster, Enrichment Score: 3.57
 Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
Supplementary Table S1: Gene expression targets
 Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
* Kyoto Encyclopedia of Genes and Genomes.
 Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array.
 Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Nature Genetics: doi: /ng Supplementary Figure 1. Depths and coverages in whole-exome and targeted deep sequencing data.
 Supplementary Figure 1 Depths and coverages in whole-exome and targeted deep sequencing data. Depth (top) and coverage (bottom) of whole-exome sequencing for 38 independent JPN cases (mean depth = 130)
Supplementary Figure 1 Depths and coverages in whole-exome and targeted deep sequencing data. Depth (top) and coverage (bottom) of whole-exome sequencing for 38 independent JPN cases (mean depth = 130)
Fetal-maternal alignment of regulatory T cells correlates with Interleukin 10 mediated Bcl-
 Fetal-maternal alignment of regulatory T cells correlates with Interleukin 10 mediated Bcl- 2 upregulation in pregnancy B. Santner-Nanan, K. Straubinger, P. Hsu, G. Parnell, B. Tang, B. Xu, A. Makris,
Fetal-maternal alignment of regulatory T cells correlates with Interleukin 10 mediated Bcl- 2 upregulation in pregnancy B. Santner-Nanan, K. Straubinger, P. Hsu, G. Parnell, B. Tang, B. Xu, A. Makris,
U118MG. Supplementary Figure 1 U373MG U118MG 3.5 A CCF-SSTG
 A172 CCF-SSTG1 15 - - - 1 1 1 2 2 3 4 6 7 8 10101112131617192022-1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22 T98G U373MG - - - 1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22-1 1 1 2 2 3 4 6 7
A172 CCF-SSTG1 15 - - - 1 1 1 2 2 3 4 6 7 8 10101112131617192022-1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22 T98G U373MG - - - 1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22-1 1 1 2 2 3 4 6 7
Supplementary Figure 1 A. Relative mrna level for cells. Number of Wells. treated with different sirnas
 Supplementary Figure 1 A Relative mrna level for cells treated with different sirnas 1 0.9 0.8 0.7 0.6 0.5 0.4 0.3 0.2 0.1 0 Control FRS2 PTEN PTPR FGFR1 CDH5 ACTN1 RAC2 Gene targeted and probed C Number
Supplementary Figure 1 A Relative mrna level for cells treated with different sirnas 1 0.9 0.8 0.7 0.6 0.5 0.4 0.3 0.2 0.1 0 Control FRS2 PTEN PTPR FGFR1 CDH5 ACTN1 RAC2 Gene targeted and probed C Number
q 2017 by The University of Chicago. All rights reserved. DOI: /690105
 q 2017 by The University of Chicago. All rights reserved. DOI: 10.1086/690105 Appendix from R. M. Cox et al., Hormonally Mediated Increases in Sex-Biased Gene Expression Accompany the Breakdown of Between-
q 2017 by The University of Chicago. All rights reserved. DOI: 10.1086/690105 Appendix from R. M. Cox et al., Hormonally Mediated Increases in Sex-Biased Gene Expression Accompany the Breakdown of Between-
Supplementary Material. Table S1. Summary of mapping results
 Supplementary Material Table S1. Summary of mapping results Sample Total reads Mapped (%) HNE0-1 99687516 80786083 (81.0%) HNE0-2 94318720 77047792 (81.7%) HNE0-3 104033900 84348102 (81.1%) HNE15-1 94426598
Supplementary Material Table S1. Summary of mapping results Sample Total reads Mapped (%) HNE0-1 99687516 80786083 (81.0%) HNE0-2 94318720 77047792 (81.7%) HNE0-3 104033900 84348102 (81.1%) HNE15-1 94426598
RAS Genes. The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes.
 ۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
1. Activated receptor tyrosine kinases (RTKs) phosphorylates themselves
 Enzyme-coupled receptors Transmembrane proteins Ligand-binding domain on the outer surface Cytoplasmic domain acts as an enzyme itself or forms a complex with enzyme 1. Activated receptor tyrosine kinases
Enzyme-coupled receptors Transmembrane proteins Ligand-binding domain on the outer surface Cytoplasmic domain acts as an enzyme itself or forms a complex with enzyme 1. Activated receptor tyrosine kinases
Receptor mediated Signal Transduction
 Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
RT 2 Profiler PCR Array:
 RT 2 Profiler PCR Array: Rat Cell Cycle Catalog Number For Real-Time Instruments: PARN-020A ABI Standard Blocks; Bio-Rad icycler, MyiQ, and (MJ Research) Chromo 4; and Stratagene Mx3005p, Mx3000p PARN-020C
RT 2 Profiler PCR Array: Rat Cell Cycle Catalog Number For Real-Time Instruments: PARN-020A ABI Standard Blocks; Bio-Rad icycler, MyiQ, and (MJ Research) Chromo 4; and Stratagene Mx3005p, Mx3000p PARN-020C
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study. February 2018
 Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Molecular Cell Biology. Prof. D. Karunagaran. Department of Biotechnology. Indian Institute of Technology Madras
 Molecular Cell Biology Prof. D. Karunagaran Department of Biotechnology Indian Institute of Technology Madras Module 9 Molecular Basis of Cancer, Oncogenes and Tumor Suppressor Genes Lecture 2 Genes Associated
Molecular Cell Biology Prof. D. Karunagaran Department of Biotechnology Indian Institute of Technology Madras Module 9 Molecular Basis of Cancer, Oncogenes and Tumor Suppressor Genes Lecture 2 Genes Associated
I. Top scoring maps. 1.IFN gamma signaling
 I. Top scoring maps 1.IFN gamma signaling Interferon-gamma signaling Interferons (IFNs) are pleiotropic cytokines that mediate anti-viral responses, inhibit proliferation and participate in immune surveillance
I. Top scoring maps 1.IFN gamma signaling Interferon-gamma signaling Interferons (IFNs) are pleiotropic cytokines that mediate anti-viral responses, inhibit proliferation and participate in immune surveillance
Cell cycle, signaling to cell cycle, and molecular basis of oncogenesis
 Cell cycle, signaling to cell cycle, and molecular basis of oncogenesis MUDr. Jiří Vachtenheim, CSc. CELL CYCLE - SUMMARY Basic terminology: Cyclins conserved proteins with homologous regions; their cellular
Cell cycle, signaling to cell cycle, and molecular basis of oncogenesis MUDr. Jiří Vachtenheim, CSc. CELL CYCLE - SUMMARY Basic terminology: Cyclins conserved proteins with homologous regions; their cellular
devseek (Sequence(Analysis(Panel(for(Neurodevelopmental(Disorders
 ACSL4 Mental/retardation,/XBlinked/63 XLBR ADSL Adenylosuccinase/Deficiency AR AFF2 Mental/retardation,/XBlinked,/FRAXE/type XLBR ALG6 Congenital/disorder/of/glycosylation/type/Ic AR ANK3 Mental/retardation,/autosomal/recessive/37
ACSL4 Mental/retardation,/XBlinked/63 XLBR ADSL Adenylosuccinase/Deficiency AR AFF2 Mental/retardation,/XBlinked,/FRAXE/type XLBR ALG6 Congenital/disorder/of/glycosylation/type/Ic AR ANK3 Mental/retardation,/autosomal/recessive/37
 Table S1. Metadata for transcriptome interaction network and pathway analysis of 5448 intracelluarly infected TEpi cells in comparison to mock TEpi cells. Gene symbol Full name Log2FC gene expression in
Table S1. Metadata for transcriptome interaction network and pathway analysis of 5448 intracelluarly infected TEpi cells in comparison to mock TEpi cells. Gene symbol Full name Log2FC gene expression in
Oncogenes and Tumor Suppressors MCB 5068 November 12, 2013 Jason Weber
 Oncogenes and Tumor Suppressors MCB 5068 November 12, 2013 Jason Weber jweber@dom.wustl.edu Oncogenes & Cancer DNA Tumor Viruses Simian Virus 40 p300 prb p53 Large T Antigen Human Adenovirus p300 E1A
Oncogenes and Tumor Suppressors MCB 5068 November 12, 2013 Jason Weber jweber@dom.wustl.edu Oncogenes & Cancer DNA Tumor Viruses Simian Virus 40 p300 prb p53 Large T Antigen Human Adenovirus p300 E1A
SUPPLEMENTARY FIG. S2. b-galactosidase staining of
 SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
SIRT6 histone deacetylase functions as a potential oncogene in human melanoma -
 SIRT6 histone deacetylase functions as a potential oncogene in human melanoma - Garcia-Peterson et al Supplementary Figure S:Tissue microarray (TMA) staining and organization. A) Diagram of the TMA indicating
SIRT6 histone deacetylase functions as a potential oncogene in human melanoma - Garcia-Peterson et al Supplementary Figure S:Tissue microarray (TMA) staining and organization. A) Diagram of the TMA indicating
Supporting Information
 Supporting Information Gorgun et al. 10.1073/pnas.0901166106 SI Text Animals, Disease Detection and Infusion of CLL Cells. Mice were examined for lymphadenopathy and splenomegaly and at specific ages or
Supporting Information Gorgun et al. 10.1073/pnas.0901166106 SI Text Animals, Disease Detection and Infusion of CLL Cells. Mice were examined for lymphadenopathy and splenomegaly and at specific ages or
Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion
 Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
Table S1 Differentially expressed genes showing > 2 fold changes and p <0.01 for 0.01 mm NS-398, 0.1mM ibuprofen and COX-2 RNAi.
 Table S1 Differentially expressed genes showing > 2 fold changes and p
Table S1 Differentially expressed genes showing > 2 fold changes and p
Enzyme-coupled Receptors. Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors
 Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
Sample Metrics. Allele Frequency (%) Read Depth Ploidy. Gene CDS Effect Protein Effect. LN Metastasis Tumor Purity Computational Pathology 80% 60%
 Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
Supplementary Table 1. Information on the 174 single nucleotide variants identified by whole-exome sequencing
 Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
Cover Page. The handle holds various files of this Leiden University dissertation.
 Cover Page The handle http://hdl.handle.net/1887/20177 holds various files of this Leiden University dissertation. Author: Kester, Maria Sophia van (Marloes) Title: Molecular aspects of cutaneous T-cell
Cover Page The handle http://hdl.handle.net/1887/20177 holds various files of this Leiden University dissertation. Author: Kester, Maria Sophia van (Marloes) Title: Molecular aspects of cutaneous T-cell
Determination Differentiation. determinated precursor specialized cell
 Biology of Cancer -Developmental Biology: Determination and Differentiation -Cell Cycle Regulation -Tumor genes: Proto-Oncogenes, Tumor supressor genes -Tumor-Progression -Example for Tumor-Progression:
Biology of Cancer -Developmental Biology: Determination and Differentiation -Cell Cycle Regulation -Tumor genes: Proto-Oncogenes, Tumor supressor genes -Tumor-Progression -Example for Tumor-Progression:
SUPPLEMENTARY DATA. Assessment of cell survival (MTT) Assessment of early apoptosis and cell death
 Overexpression of a functional calcium-sensing receptor dramatically increases osteolytic potential of MDA-MB-231 cells in a mouse model of bone metastasis through epiregulinmediated osteoprotegerin downregulation
Overexpression of a functional calcium-sensing receptor dramatically increases osteolytic potential of MDA-MB-231 cells in a mouse model of bone metastasis through epiregulinmediated osteoprotegerin downregulation
KEY CONCEPT QUESTIONS IN SIGNAL TRANSDUCTION
 Signal Transduction - Part 2 Key Concepts - Receptor tyrosine kinases control cell metabolism and proliferation Growth factor signaling through Ras Mutated cell signaling genes in cancer cells are called
Signal Transduction - Part 2 Key Concepts - Receptor tyrosine kinases control cell metabolism and proliferation Growth factor signaling through Ras Mutated cell signaling genes in cancer cells are called
Signaling Through Immune System Receptors (Ch. 7)
 Signaling Through Immune System Receptors (Ch. 7) 1. General principles of signal transduction and propagation. 2. Antigen receptor signaling and lymphocyte activation. 3. Other receptors and signaling
Signaling Through Immune System Receptors (Ch. 7) 1. General principles of signal transduction and propagation. 2. Antigen receptor signaling and lymphocyte activation. 3. Other receptors and signaling
Direct Intracellular Regulators of Apoptosis Array 1
 Supplement Table A Direct Intracellular Regulators of Apoptosis 2 Mean Name Description.5 2.2.9 ATM ataxia telangiectasia mutated homolog.9.8.9 Bcl2aa B-cell leukemia/lymphoma 2 related protein Aa.6 2..9
Supplement Table A Direct Intracellular Regulators of Apoptosis 2 Mean Name Description.5 2.2.9 ATM ataxia telangiectasia mutated homolog.9.8.9 Bcl2aa B-cell leukemia/lymphoma 2 related protein Aa.6 2..9
Principles of Genetics and Molecular Biology
 Cell signaling Dr. Diala Abu-Hassan, DDS, PhD School of Medicine Dr.abuhassand@gmail.com Principles of Genetics and Molecular Biology www.cs.montana.edu Modes of cell signaling Direct interaction of a
Cell signaling Dr. Diala Abu-Hassan, DDS, PhD School of Medicine Dr.abuhassand@gmail.com Principles of Genetics and Molecular Biology www.cs.montana.edu Modes of cell signaling Direct interaction of a
Chapter 6: Cancer Pathways. Other Pathways. Cancer Pathways
 Chapter 6: Cancer Pathways Limited number of pathways control proliferation and differentiation Transmit signals from growth factors, hormones, cell-to-cell communications/interactions Pathways turn into
Chapter 6: Cancer Pathways Limited number of pathways control proliferation and differentiation Transmit signals from growth factors, hormones, cell-to-cell communications/interactions Pathways turn into
Supplementary Material. Part I: Sample Information. Part II: Pathway Information
 Supplementary Material Part I: Sample Information Three NPC cell lines, CNE1, CNE2, and HK1 were treated with CYC202. Gene expression of 380 selected genes were collected at 0, 2, 4, 6, 12 and 24 hours
Supplementary Material Part I: Sample Information Three NPC cell lines, CNE1, CNE2, and HK1 were treated with CYC202. Gene expression of 380 selected genes were collected at 0, 2, 4, 6, 12 and 24 hours
Signaling. Dr. Sujata Persad Katz Group Centre for Pharmacy & Health research
 Signaling Dr. Sujata Persad 3-020 Katz Group Centre for Pharmacy & Health research E-mail:sujata.persad@ualberta.ca 1 Growth Factor Receptors and Other Signaling Pathways What we will cover today: How
Signaling Dr. Sujata Persad 3-020 Katz Group Centre for Pharmacy & Health research E-mail:sujata.persad@ualberta.ca 1 Growth Factor Receptors and Other Signaling Pathways What we will cover today: How
A particular set of insults induces apoptosis (part 1), which, if inhibited, can switch to autophagy. At least in some cellular settings, autophagy se
 A particular set of insults induces apoptosis (part 1), which, if inhibited, can switch to autophagy. At least in some cellular settings, autophagy serves as a defence mechanism that prevents or retards
A particular set of insults induces apoptosis (part 1), which, if inhibited, can switch to autophagy. At least in some cellular settings, autophagy serves as a defence mechanism that prevents or retards
Deregulation of signal transduction and cell cycle in Cancer
 Deregulation of signal transduction and cell cycle in Cancer Tuangporn Suthiphongchai, Ph.D. Department of Biochemistry Faculty of Science, Mahidol University Email: tuangporn.sut@mahidol.ac.th Room Pr324
Deregulation of signal transduction and cell cycle in Cancer Tuangporn Suthiphongchai, Ph.D. Department of Biochemistry Faculty of Science, Mahidol University Email: tuangporn.sut@mahidol.ac.th Room Pr324
Biol403 MAP kinase signalling
 Biol403 MAP kinase signalling The mitogen activated protein kinase (MAPK) pathway is a signalling cascade activated by a diverse range of effectors. The cascade regulates many cellular activities including
Biol403 MAP kinase signalling The mitogen activated protein kinase (MAPK) pathway is a signalling cascade activated by a diverse range of effectors. The cascade regulates many cellular activities including
Oncogenes and Tumor. supressors
 Oncogenes and Tumor supressors From history to therapeutics Serge ROCHE Neoplastic transformation TUMOR SURESSOR ONCOGENE ONCOGENES History 1911 1960 1980 2001 Transforming retrovirus RSV v-src is an oncogene
Oncogenes and Tumor supressors From history to therapeutics Serge ROCHE Neoplastic transformation TUMOR SURESSOR ONCOGENE ONCOGENES History 1911 1960 1980 2001 Transforming retrovirus RSV v-src is an oncogene
Table 1: NFkB Target Gene Sets 1.Genes included in the subset of 15k genes selected according to a MAD-based variation filter
 Supplementary Information NFkB target gene sets The NFkB target gene sets are listed below (Table1). In addition to the previously performed mapping of Affymetrix probesets and corresponding Lymphochip
Supplementary Information NFkB target gene sets The NFkB target gene sets are listed below (Table1). In addition to the previously performed mapping of Affymetrix probesets and corresponding Lymphochip
Integrative Radiation Biology
 Dr. Kristian Unger Integrative Biology Group Research Unit of Radiation Cytogenetics Department of Radiation Sciences Helmholtz-Zentrum München 1 Key Questions Molecular mechanisms of radiation-induced
Dr. Kristian Unger Integrative Biology Group Research Unit of Radiation Cytogenetics Department of Radiation Sciences Helmholtz-Zentrum München 1 Key Questions Molecular mechanisms of radiation-induced
Cell Signaling part 2
 15 Cell Signaling part 2 Functions of Cell Surface Receptors Other cell surface receptors are directly linked to intracellular enzymes. The largest family of these is the receptor protein tyrosine kinases,
15 Cell Signaling part 2 Functions of Cell Surface Receptors Other cell surface receptors are directly linked to intracellular enzymes. The largest family of these is the receptor protein tyrosine kinases,
Signal Transduction Pathways. Part 2
 Signal Transduction Pathways Part 2 GPCRs G-protein coupled receptors > 700 GPCRs in humans Mediate responses to senses taste, smell, sight ~ 1000 GPCRs mediate sense of smell in mouse Half of all known
Signal Transduction Pathways Part 2 GPCRs G-protein coupled receptors > 700 GPCRs in humans Mediate responses to senses taste, smell, sight ~ 1000 GPCRs mediate sense of smell in mouse Half of all known
Supplementary Figure 1: Func8onal Network Analysis of Kinases Significantly Modulated by MERS CoV Infec8on and Conserved Across All Time Points
 A. B. 8 4 Supplementary Figure : Func8onal Network Analysis of Kinases Significantly Modulated by MERS CoV Infec8on and Conserved Across All Time Points Examined. A) Venn diagram analysis of kinases significantly
A. B. 8 4 Supplementary Figure : Func8onal Network Analysis of Kinases Significantly Modulated by MERS CoV Infec8on and Conserved Across All Time Points Examined. A) Venn diagram analysis of kinases significantly
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature12912 Figure S1. Distribution of mutation rates and spectra across 4,742 tumor-normal (TN) pairs from 21 tumor types, as in Figure 1 from Lawrence et al. Nature
SUPPLEMENTARY INFORMATION doi:1.138/nature12912 Figure S1. Distribution of mutation rates and spectra across 4,742 tumor-normal (TN) pairs from 21 tumor types, as in Figure 1 from Lawrence et al. Nature
Quantitative Real Time PCR (RT-qPCR) Differentiation of human preadipocytes to adipocytes
 Quantitative Real Time PCR (RT-qPCR) First strand cdna was synthesized and RT-qPCR was performed using RT2 first strand kits (Cat#330401) and RT2 qpcr Master Mix (SABioscience, Frederick, MD). Experiments
Quantitative Real Time PCR (RT-qPCR) First strand cdna was synthesized and RT-qPCR was performed using RT2 first strand kits (Cat#330401) and RT2 qpcr Master Mix (SABioscience, Frederick, MD). Experiments
Computational Biology I LSM5191
 Computational Biology I LSM5191 Aylwin Ng, D.Phil Lecture 6 Notes: Control Systems in Gene Expression Pulling it all together: coordinated control of transcriptional regulatory molecules Simple Control:
Computational Biology I LSM5191 Aylwin Ng, D.Phil Lecture 6 Notes: Control Systems in Gene Expression Pulling it all together: coordinated control of transcriptional regulatory molecules Simple Control:
Precision Oncology: Experience at UW
 Precision Oncology: Experience at UW Colin Pritchard MD, PhD University of Washington, Department of Lab Medicine WSMOS Meeting November 1, 2013 Conflict of Interest Disclosures I declare the following,
Precision Oncology: Experience at UW Colin Pritchard MD, PhD University of Washington, Department of Lab Medicine WSMOS Meeting November 1, 2013 Conflict of Interest Disclosures I declare the following,
Molecular Hematopathology Leukemias I. January 14, 2005
 Molecular Hematopathology Leukemias I January 14, 2005 Chronic Myelogenous Leukemia Diagnosis requires presence of Philadelphia chromosome t(9;22)(q34;q11) translocation BCR-ABL is the result BCR on chr
Molecular Hematopathology Leukemias I January 14, 2005 Chronic Myelogenous Leukemia Diagnosis requires presence of Philadelphia chromosome t(9;22)(q34;q11) translocation BCR-ABL is the result BCR on chr
VIII Curso Internacional del PIRRECV. Some molecular mechanisms of cancer
 VIII Curso Internacional del PIRRECV Some molecular mechanisms of cancer Laboratorio de Comunicaciones Celulares, Centro FONDAP Estudios Moleculares de la Celula (CEMC), ICBM, Facultad de Medicina, Universidad
VIII Curso Internacional del PIRRECV Some molecular mechanisms of cancer Laboratorio de Comunicaciones Celulares, Centro FONDAP Estudios Moleculares de la Celula (CEMC), ICBM, Facultad de Medicina, Universidad
Diagnostic test Suggested website label Description Hospitals available
 Diagnostic test Suggested website label Description Hospitals available Abbott Molecular Inc, PATHVYSION HER-2 DNA Probe Kit (FISH) PathVysion kit A diagnostic tool used to determine whether a particular
Diagnostic test Suggested website label Description Hospitals available Abbott Molecular Inc, PATHVYSION HER-2 DNA Probe Kit (FISH) PathVysion kit A diagnostic tool used to determine whether a particular
Jocelyn Chapman, MD Division of Gynecologic Oncology. Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic
 Jocelyn Chapman, MD Division of Gynecologic Oncology Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic Genetics underlies all cancers Somatic or tumor genetics Germline or inherited genetics
Jocelyn Chapman, MD Division of Gynecologic Oncology Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic Genetics underlies all cancers Somatic or tumor genetics Germline or inherited genetics
Next Generation Sequencing in Haematological Malignancy: A European Perspective. Wolfgang Kern, Munich Leukemia Laboratory
 Next Generation Sequencing in Haematological Malignancy: A European Perspective Wolfgang Kern, Munich Leukemia Laboratory Diagnostic Methods Cytomorphology Cytogenetics Immunophenotype Histology FISH Molecular
Next Generation Sequencing in Haematological Malignancy: A European Perspective Wolfgang Kern, Munich Leukemia Laboratory Diagnostic Methods Cytomorphology Cytogenetics Immunophenotype Histology FISH Molecular
CELL CYCLE MOLECULAR BASIS OF ONCOGENESIS
 CELL CYCLE MOLECULAR BASIS OF ONCOGENESIS Summary of the regulation of cyclin/cdk complexes during celll cycle Cell cycle phase Cyclin-cdk complex inhibitor activation Substrate(s) G1 Cyclin D/cdk 4,6
CELL CYCLE MOLECULAR BASIS OF ONCOGENESIS Summary of the regulation of cyclin/cdk complexes during celll cycle Cell cycle phase Cyclin-cdk complex inhibitor activation Substrate(s) G1 Cyclin D/cdk 4,6
Patricia Aoun MD, MPH Professor and Vice-Chair for Clinical Affairs Medical Director, Clinical Laboratories Department of Pathology City of Hope
 Patricia Aoun MD, MPH Professor and Vice-Chair for Clinical Affairs Medical Director, Clinical Laboratories Department of Pathology City of Hope National Medical Center Disclosures I have no disclosures
Patricia Aoun MD, MPH Professor and Vice-Chair for Clinical Affairs Medical Director, Clinical Laboratories Department of Pathology City of Hope National Medical Center Disclosures I have no disclosures
CHAPTER INTRODUCTION
 CHAPTER 6 IDENTIFICATION OF RHEUMATOID ARTHRITIS SIGNIFICANT TARGET PROTEINS AND THEIR PATHWAYS THROUGH THE COMBINATORIAL APPROACH (RA-GIM, RA-DTP AND MICROARRAY DATANALYSIS OF RA) 6.1 INTRODUCTION The
CHAPTER 6 IDENTIFICATION OF RHEUMATOID ARTHRITIS SIGNIFICANT TARGET PROTEINS AND THEIR PATHWAYS THROUGH THE COMBINATORIAL APPROACH (RA-GIM, RA-DTP AND MICROARRAY DATANALYSIS OF RA) 6.1 INTRODUCTION The
Out-Patient Billing CPT Codes
 Out-Patient Billing CPT Codes Updated Date: August 3, 08 Client Billed Molecular Tests HPV DNA Tissue Testing 8764 No Medicare Billed - Molecular Tests NeoARRAY NeoARRAY SNP/Cytogenetic No 89 NeoLAB NeoLAB
Out-Patient Billing CPT Codes Updated Date: August 3, 08 Client Billed Molecular Tests HPV DNA Tissue Testing 8764 No Medicare Billed - Molecular Tests NeoARRAY NeoARRAY SNP/Cytogenetic No 89 NeoLAB NeoLAB
Cancer genetics
 Cancer genetics General information about tumorogenesis. Cancer induced by viruses. The role of somatic mutations in cancer production. Oncogenes and Tumor Suppressor Genes (TSG). Hereditary cancer. 1
Cancer genetics General information about tumorogenesis. Cancer induced by viruses. The role of somatic mutations in cancer production. Oncogenes and Tumor Suppressor Genes (TSG). Hereditary cancer. 1
The Center for PERSONALIZED DIAGNOSTICS
 The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
G-Protein Signaling. Introduction to intracellular signaling. Dr. SARRAY Sameh, Ph.D
 G-Protein Signaling Introduction to intracellular signaling Dr. SARRAY Sameh, Ph.D Cell signaling Cells communicate via extracellular signaling molecules (Hormones, growth factors and neurotransmitters
G-Protein Signaling Introduction to intracellular signaling Dr. SARRAY Sameh, Ph.D Cell signaling Cells communicate via extracellular signaling molecules (Hormones, growth factors and neurotransmitters
A genome-wide association study identifies vitiligo
 A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
BL 424 Chapter 15: Cell Signaling; Signal Transduction
 BL 424 Chapter 15: Cell Signaling; Signal Transduction All cells receive and respond to signals from their environments. The behavior of each individual cell in multicellular plants and animals must be
BL 424 Chapter 15: Cell Signaling; Signal Transduction All cells receive and respond to signals from their environments. The behavior of each individual cell in multicellular plants and animals must be
Designer Affinity Reagents. Brian Kay
 Designer Affinity Reagents Brian Kay bkay@uic.edu Types of Affinity Reagents Src SH3 domain Lysozyme Src SH3 domain FN3 monobody Peptide Ligand Antibody Fragment Scaffold M13 Bacteriophage 900 nm x 10
Designer Affinity Reagents Brian Kay bkay@uic.edu Types of Affinity Reagents Src SH3 domain Lysozyme Src SH3 domain FN3 monobody Peptide Ligand Antibody Fragment Scaffold M13 Bacteriophage 900 nm x 10
Cancer. The fundamental defect is. unregulated cell division. Properties of Cancerous Cells. Causes of Cancer. Altered growth and proliferation
 Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Assessing the Economics of Genomic Medicine. Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center
 Assessing the Economics of Genomic Medicine Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center DISCLOSURES No conflicts Patents unenforced Consultancies unpaid
Assessing the Economics of Genomic Medicine Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center DISCLOSURES No conflicts Patents unenforced Consultancies unpaid
Genomic Medicine: What every pathologist needs to know
 Genomic Medicine: What every pathologist needs to know Stephen P. Ethier, Ph.D. Professor, Department of Pathology and Laboratory Medicine, MUSC Director, MUSC Center for Genomic Medicine Genomics and
Genomic Medicine: What every pathologist needs to know Stephen P. Ethier, Ph.D. Professor, Department of Pathology and Laboratory Medicine, MUSC Director, MUSC Center for Genomic Medicine Genomics and
Principles of cell signaling Lecture 4
 Principles of cell signaling Lecture 4 Johan Lennartsson Molecular Cell Biology (1BG320), 2014 Johan.Lennartsson@licr.uu.se 1 Receptor tyrosine kinase-induced signal transduction Erk MAP kinase pathway
Principles of cell signaling Lecture 4 Johan Lennartsson Molecular Cell Biology (1BG320), 2014 Johan.Lennartsson@licr.uu.se 1 Receptor tyrosine kinase-induced signal transduction Erk MAP kinase pathway
Growth and Differentiation Phosphorylation Sampler Kit
 Growth and Differentiation Phosphorylation Sampler Kit E 0 5 1 0 1 4 Kits Includes Cat. Quantity Application Reactivity Source Akt (Phospho-Ser473) E011054-1 50μg/50μl IHC, WB Human, Mouse, Rat Rabbit
Growth and Differentiation Phosphorylation Sampler Kit E 0 5 1 0 1 4 Kits Includes Cat. Quantity Application Reactivity Source Akt (Phospho-Ser473) E011054-1 50μg/50μl IHC, WB Human, Mouse, Rat Rabbit
Revision. camp pathway
 االله الرحمن الرحيم بسم Revision camp pathway camp pathway Revision camp pathway Adenylate cyclase Adenylate Cyclase enzyme Adenylate cyclase catalyses the formation of camp from ATP. Stimulation or inhibition
االله الرحمن الرحيم بسم Revision camp pathway camp pathway Revision camp pathway Adenylate cyclase Adenylate Cyclase enzyme Adenylate cyclase catalyses the formation of camp from ATP. Stimulation or inhibition
Phospho-AKT Sampler Kit
 Phospho-AKT Sampler Kit E 0 5 1 0 0 3 Kits Includes Cat. Quantity Application Reactivity Source Akt (Ab-473) Antibody E021054-1 50μg/50μl IHC, WB Human, Mouse, Rat Rabbit Akt (Phospho-Ser473) Antibody
Phospho-AKT Sampler Kit E 0 5 1 0 0 3 Kits Includes Cat. Quantity Application Reactivity Source Akt (Ab-473) Antibody E021054-1 50μg/50μl IHC, WB Human, Mouse, Rat Rabbit Akt (Phospho-Ser473) Antibody
Cancer. The fundamental defect is. unregulated cell division. Properties of Cancerous Cells. Causes of Cancer. Altered growth and proliferation
 Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Supplementary Information
 Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
Cell Cell Communication
 IBS 8102 Cell, Molecular, and Developmental Biology Cell Cell Communication January 29, 2008 Communicate What? Why do cells communicate? To govern or modify each other for the benefit of the organism differentiate
IBS 8102 Cell, Molecular, and Developmental Biology Cell Cell Communication January 29, 2008 Communicate What? Why do cells communicate? To govern or modify each other for the benefit of the organism differentiate
Designer Affinity Reagents. Reagents. Types of Affinity. M13 Bacteriophage. 900 nm x 10 nm. Brian Kay Src SH3 domain.
 Designer Affinity Reagents Brian Kay bkay@uic.edu Types of Affinity Reagents Src SH3 domain Lysozyme Src SH3 domain FN3 monobody Peptide Ligand Antibody Fragment Scaffold M13 Bacteriophage 900 nm 10 nm
Designer Affinity Reagents Brian Kay bkay@uic.edu Types of Affinity Reagents Src SH3 domain Lysozyme Src SH3 domain FN3 monobody Peptide Ligand Antibody Fragment Scaffold M13 Bacteriophage 900 nm 10 nm
Additional file 2 List of pathway from PID
 Additional file 2 List of pathway from PID Pathway ID Pathway name # components # enriched GO terms a4b1_paxdep_pathway Paxillin-dependent events mediated by a4b1 20 179 a4b1_paxindep_pathway Paxillin-independent
Additional file 2 List of pathway from PID Pathway ID Pathway name # components # enriched GO terms a4b1_paxdep_pathway Paxillin-dependent events mediated by a4b1 20 179 a4b1_paxindep_pathway Paxillin-independent
Gene Expression Multiplexing Menu
 Gene Expression Multiplexing Menu Mouse Rat Human Table of CONTENTS Mouse Inflammatory Plex 2 Rat Stress Plex 3 Growth Regulation Plex 4 Multitox Plex 5 Pharma III Plex 6 Reference Plex 7 Human Transporter
Gene Expression Multiplexing Menu Mouse Rat Human Table of CONTENTS Mouse Inflammatory Plex 2 Rat Stress Plex 3 Growth Regulation Plex 4 Multitox Plex 5 Pharma III Plex 6 Reference Plex 7 Human Transporter
Cell Cell Communication
 IBS 8102 Cell, Molecular, and Developmental Biology Cell Cell Communication January 29, 2008 Communicate What? Why do cells communicate? To govern or modify each other for the benefit of the organism differentiate
IBS 8102 Cell, Molecular, and Developmental Biology Cell Cell Communication January 29, 2008 Communicate What? Why do cells communicate? To govern or modify each other for the benefit of the organism differentiate
Case Study - Informatics
 bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
The Yeast Two-Hybrid System
 The Yeast Two-Hybrid System What is the yeast two-hybrid system used for? Identifies novel protein-protein interactions Can identify protein cascades Identifies mutations that affect protein-protein binding
The Yeast Two-Hybrid System What is the yeast two-hybrid system used for? Identifies novel protein-protein interactions Can identify protein cascades Identifies mutations that affect protein-protein binding
Supplementary Material Correlation matrices on FP and FN profiles
 Supplementary Material Correlation matrices on FP and FN profiles The following two tables give the correlation coefficients for the FP profiles and the FN profiles of a single tagging solutions against
Supplementary Material Correlation matrices on FP and FN profiles The following two tables give the correlation coefficients for the FP profiles and the FN profiles of a single tagging solutions against
BL 424 Test pts name Multiple choice have one choice each and are worth 3 points.
 BL 424 Test 3 2010 150 pts name Multiple choice have one choice each and are worth 3 points. 1. The plasma membrane functions as a a. selective barrier to the passage of molecules. b. sensor through which
BL 424 Test 3 2010 150 pts name Multiple choice have one choice each and are worth 3 points. 1. The plasma membrane functions as a a. selective barrier to the passage of molecules. b. sensor through which
Applied Biosystems models 7500 (Fast block), 7900HT (Fast block), StepOnePlus, ViiA 7 (Fast block)
 RT² Profiler PCR Array (96-Well Format and 384-Well [4 x 96] Format) Human HIV Infection and Host Response Cat. no. 330231 PAHS-051YA For pathway expression analysis Format Format A Format C Format D Format
RT² Profiler PCR Array (96-Well Format and 384-Well [4 x 96] Format) Human HIV Infection and Host Response Cat. no. 330231 PAHS-051YA For pathway expression analysis Format Format A Format C Format D Format
Next Generation Sequencing in Clinical Practice: Impact on Therapeutic Decision Making
 Next Generation Sequencing in Clinical Practice: Impact on Therapeutic Decision Making November 20, 2014 Capturing Value in Next Generation Sequencing Symposium Douglas Johnson MD, MSCI Vanderbilt-Ingram
Next Generation Sequencing in Clinical Practice: Impact on Therapeutic Decision Making November 20, 2014 Capturing Value in Next Generation Sequencing Symposium Douglas Johnson MD, MSCI Vanderbilt-Ingram
Changing the Culture of Cancer Care II. Eric Holland Fred Hutchinson Cancer Research Center University of Washington Seattle
 Changing the Culture of Cancer Care II Eric Holland Fred Hutchinson Cancer Research Center University of Washington Seattle Transforming Cancer Therapy Eric Holland Fred Hutchinson Cancer Research Center
Changing the Culture of Cancer Care II Eric Holland Fred Hutchinson Cancer Research Center University of Washington Seattle Transforming Cancer Therapy Eric Holland Fred Hutchinson Cancer Research Center
Blastic Plasmacytoid Dendritic Cell Neoplasm with DNMT3A and TET2 mutations (SH )
 Blastic Plasmacytoid Dendritic Cell Neoplasm with DNMT3A and TET2 mutations (SH2017-0314) Habibe Kurt, Joseph D. Khoury, Carlos E. Bueso-Ramos, Jeffrey L. Jorgensen, Guilin Tang, L. Jeffrey Medeiros, and
Blastic Plasmacytoid Dendritic Cell Neoplasm with DNMT3A and TET2 mutations (SH2017-0314) Habibe Kurt, Joseph D. Khoury, Carlos E. Bueso-Ramos, Jeffrey L. Jorgensen, Guilin Tang, L. Jeffrey Medeiros, and
Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice
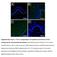 Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice undergoing AD- and psoriasis-like disease. Immunofluorescence staining for Fn14 (green) and DAPI (blue) in skin of naïve
Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice undergoing AD- and psoriasis-like disease. Immunofluorescence staining for Fn14 (green) and DAPI (blue) in skin of naïve
CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION
 CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION What is Cytokine? Secreted popypeptide (protein) involved in cell-to-cell signaling. Acts in paracrine or autocrine fashion through specific cellular receptors.
CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION What is Cytokine? Secreted popypeptide (protein) involved in cell-to-cell signaling. Acts in paracrine or autocrine fashion through specific cellular receptors.
Supplementary Table 1: Motor-clutch gene mrna expression Myosin Motors
 Supplementary Table 1: Motor-clutch gene mrna expression Myosin Motors Symbol Description Expression Rank MYL6 myosin, light chain 6, alkali, smooth muscle and non-muscle 11903 79 MYH9 myosin, heavy chain
Supplementary Table 1: Motor-clutch gene mrna expression Myosin Motors Symbol Description Expression Rank MYL6 myosin, light chain 6, alkali, smooth muscle and non-muscle 11903 79 MYH9 myosin, heavy chain
Exons from 137 genes were targeted for hybrid selection and massively parallel sequencing.
 SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
Structure and Function of Fusion Gene Products in. Childhood Acute Leukemia
 Structure and Function of Fusion Gene Products in Childhood Acute Leukemia Chromosomal Translocations Chr. 12 Chr. 21 der(12) der(21) A.T. Look, Science 278 (1997) Distribution Childhood ALL TEL-AML1 t(12;21)
Structure and Function of Fusion Gene Products in Childhood Acute Leukemia Chromosomal Translocations Chr. 12 Chr. 21 der(12) der(21) A.T. Look, Science 278 (1997) Distribution Childhood ALL TEL-AML1 t(12;21)
Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies
 Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies Dr. Maricel G. Kann Assistant Professor Dept of Biological Sciences UMBC 2 The term protein domain
Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies Dr. Maricel G. Kann Assistant Professor Dept of Biological Sciences UMBC 2 The term protein domain
The Tissue Engineer s Toolkit
 The Tissue Engineer s Toolkit Stimuli Detection and Response Ken Webb, Ph. D. Assistant Professor Dept. of Bioengineering Clemson University Environmental Stimulus-Cellular Response Environmental Stimuli
The Tissue Engineer s Toolkit Stimuli Detection and Response Ken Webb, Ph. D. Assistant Professor Dept. of Bioengineering Clemson University Environmental Stimulus-Cellular Response Environmental Stimuli
Signal Transduction: G-Protein Coupled Receptors
 Signal Transduction: G-Protein Coupled Receptors Federle, M. (2017). Lectures 4-5: Signal Transduction parts 1&2: nuclear receptors and GPCRs. Lecture presented at PHAR 423 Lecture in UIC College of Pharmacy,
Signal Transduction: G-Protein Coupled Receptors Federle, M. (2017). Lectures 4-5: Signal Transduction parts 1&2: nuclear receptors and GPCRs. Lecture presented at PHAR 423 Lecture in UIC College of Pharmacy,
Supplementary Information. Table of contents
 Supplementary Information Table of contents Fig. S1. Inhibition of specific upstream kinases affects the activity of the analyzed readouts Fig. S2. Down-regulation of INCENP gene induces the formation
Supplementary Information Table of contents Fig. S1. Inhibition of specific upstream kinases affects the activity of the analyzed readouts Fig. S2. Down-regulation of INCENP gene induces the formation
