Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function
|
|
|
- Juliet Matthews
- 6 years ago
- Views:
Transcription
1 Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL LrgA family BPSL hypothetical BPSL flagellar basal body BPSL flagellar motor switch BPSL probable flagellar motor switch BPSL flagellar BPSL flagellar biosynthetic BPSL flagellar biosynthesis BPSL flagellar biosynthetic 2 BPSL putative GMC oxidoreductase BPSL putative flagellar hook length control BPSL flagellar flij BPSL flagellum specific ATP synthase BPSL flagellar assembly BPSL flagellar motor BPSL flagellar M ring BPSL flagellar hook basal body complex BPSL flagellar BPSL hypothetical BPSL hypothetical BPSL putative export system BPSL hypothetical 3 BPSL hypothetical BPSL putative flagella synthesis BPSL putative negative regulator of flagellin synthesis BPSL flagellar basal body P ring biosynthesis BPSL putative flagellar basal body rod BPSL flagellar basal body rod BPSL putative basal body rod modification BPSL flagellar hook BPSL flagellar basal body rod BPSL flagellar basal body rod BPSL flagellar L ring precursor BPSL flagellar P ring precursor BPSL putative peptidoglycan hydrolase BPSL hypothetical
2 BPSL flagellar hook associated BPSL putative flagellar hook associated BPSL putative permease 4 BPSL metallo beta lactamase superfamily 5 BPSL DNA polymerase III subunit alpha BPSL hypothetical BPSL putative transporter BPSL putative fatty acid desaturase BPSL putative diaminobutyrate 2 oxoglutarate aminotransferase BPSL hypothetical BPSL hypothetical BPSL ABC transport system ATP binding BPSL hypothetical BPSL hypothetical BPSL hypothetical BPSL hypothetical BPSL hypothetical BPSL putative AMP binding enzyme BPSL putative pyridoxal dependent decarboxylase BPSL hypothetical BPSL hypothetical BPSL hypothetical BPSL hypothetical BPSL putative acyl carrier BPSL hypothetical BPSL putative AMP binding enzyme 6 BPSL hypothetical BPSL putative polysaccharide biosynthesis BPSL putative glycosyl transferase BPSL putative mannose 1 phosphate guanylyltransferase BPSL hypothetical BPSL putative glycosyl transferase BPSL PAP2 superfamily BPSL fis family regulatory BPSL hypothetical BPSL glycosyl hydrolase family
3 BPSL hypothetical BPSL hypothetical BPSL putative acyl CoA dehydrogenase BPSL acyl carrier BPSL hypothetical BPSL hypothetical BPSL putative sugar transferase BPSL putative polysaccharide biosynthesis/export BPSL glycosyl transferase group 1 BPSL hypothetical 7 BPSL putative RND family acriflavine resistance A precursor BPSL putative RND family acriflavine resistance BPSL outer membrane efflux 8 BPSL cobalamin biosynthesis CbiG BPSL cob(i)yrinic acid a,c diamide adenosyltransferase BPSL cobyrinic acid A,C diamide synthase BPSL putative siderophore biosynthesis related BPSL putative iron uptake receptor precursor BPSL putative L ornithine 5 monooxygenase BPSL putative siderophore related non ribosomal peptide synthase BPSL BPSL putative siderophore related no ribosomal peptide synthase putative siderophore biosynthesis related ABC transport BPSL hypothetical BPSL putative iron transport related exported BPSL putative iron transport related membrane BPSL BPSL putative iron transport related membrane putative iron transport related ATP bidning BPSL hypothetical BPSL hypothetical BPSL RNA polymerase sigma 70 factor
4 9 BPSL putative lipo BPSL putative fimbriae related BPSL putative fimbriae assembly chaperone BPSL hypothetical BPSL hypothetical BPSL hypothetical 10 BPSL putative multidrug resistance 11 BPSL putative acyltransferase BPSL hypothetical BPSL family M1 unassigned peptidase BPSL hypothetical BPSL hypothetical BPSL putative bacteriocin secretion BPSL colicin V processing peptidase BPSL putative outer membrane bacteriocin efflux 12 BPSL ,10 methylenetetrahydrofolate reductase BPSL hypothetical BPSL S adenosyl L homocysteine hydrolase BPSL flagellar biosynthesis sigma factor FliA BPSL flagellar biosynthesis FlhG BPSL flagellar biosynthesis BPSL flagellar biosynthesis BPSL flagellar biosynthetic FlhB BPSL hypothetical BPSL Gly/Ala/Ser rich lipo BPSL hypothetical BPSL chemotaxis CheZ BPSL chemotaxis CheY BPSL chemotaxis specific methylesterase BPSL putative chemotaxis BPSL chemotaxis methyltransferase BPSL methyl accepting chemotaxis I BPSL chemotaxis CheW BPSL chemotaxis two component sensor kinase CheA BPSL chemotaxis two component response regulator CheY1 BPSL flagellar motor
5 BPSL flagellar motor BPSL flagellar regulon master regulator subunit FlhC BPSL transcriptional activator FlhD BPSL S ribosomal S21 BPSL flagellin BPSL flagellar hook associated BPSL hypothetical BPSL putative TPR domain BPSL putative transferase BPSL putative keto/oxo acyl ACP synthase BPSL putative acyl carrier BPSL putative keto/oxo acyl ACP synthase BPSL putative short chain dehydrogenase BPSL putative acetyltransferase BPSL Rieske [2Fe 2S] domain BPSL hypothetical Table S9B: List of taurine regulated genes in Bp K96243 Chr 2 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSS alpha ketoglutarate permease BPSS C4 dicarboxylate transport sensor kinase BPSS C4 dicarboxylate transport transcriptional response regulator 2 BPSS hypothetical BPSS hemin ABC transport system, ATP binding BPSS hemin ABC transport system, membrane BPSS hemin transport system, substrate binding BPSS hemin ABC transport system related BPSS exported heme receptor 3 BPSS fatty acid CoA ligase BPSS malonyl CoA acyl carrier BPSS hypothetical BPSS fatty acid biosynthesis related CoA ligase BPSS diaminopimelate decarboxylase BPSS hypothetical BPSS ketol acid reductoisomerase BPSS multifunctional polyketide peptide syntase
6 BPSS aldehyde dehydrogenase BPSS hypothetical BPSS peptide synthase regulatory BPSS hypothetical BPSS multifunctional polyketide peptide syntase BPSS LuxR family transcriptional regulator BPSS lipo BPSS monooxygenase BPSS ABC transport system, ATP binding BPSS ACB transport system, membrane BPSS monooxygenase BPSS hypothetical 4 BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS biopolymer transport BPSS bipolymer transport BPSS TonB like transport BPSS bacterioferritin ferredoxin BPSS glutamate racemase BPSS bacterioferritin 5 BPSS acetyl CoA hydrolase/transferase BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS ATPase BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS hypothetical
7 BPSS lipo BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS hypothetical BPSS hydroxydecanoyl ACP:CoA transacylase BPSS UDP glucoronosyl and UDP glucosyl transferase BPSS transport BPSS hypothetical BPSS outer membrane efflux BPSS HlyD family secretion BPSS glycosyl hydrolase 6 BPSS salicylate biosynthesis isochorismate synthase BPSS chorismate mutase BPSS pyochelin biosynthetic BPSS salicyl AMP ligase BPSS AraC family regulatory BPSS pyochelin synthetase BPSS pyochelin synthetase BPSS pyochelin biosynthetic 7 BPSS catalase precursor 8 BPSS HlyD family efflux pump BPSS outer membrane efflux BPSS dtdp rhamnosyl transferase BPSS efflux/sugar transport/multidrug resistance BPSS rhamnosyltransferase BPSS fatty acid biosynthetic 9 BPSS permease component of taurine ABC transporter BPSS ATP binding component of taurine ABC transporter BPSS periplasmic component of taurine ABC transporter alpha ketoglutarate dependent taurine BPSS dioxygenase
8 10 BPSS type IV pilus biosynthesis BPSS hypothetical BPSS major pilin subunit BPSS type IV pilus biosynthesis BPSS type IV pilus biosynthesis BPSS hypothetical BPSS type IV pilus biosynthesis BPSS type IV pilus biosynthesis BPSS type IV pilus biosynthesis BPSS twitching motility BPSS secretion BPSS probable two component system response regulator BPSS probable two component system sensor kinase 11 BPSS hypothetical BPSS probable non ribosomal peptide synthetase BPSS probable non ribosomal peptide synthetase BPSS probable non ribosomal peptide synthetase BPSS hypothetical 12 BPSS hypothetical BPSS hydroxy 2 ketovalerate aldolase BPSS acetaldehyde dehydrogenase BPSS thioesterase BPSS branched chain amino acid aminotransferase BPSS transferase BPSS non ribosomal peptide synthesis thioesterase BPSS non ribosomal peptide synthase related BPSS hypothetical BPSS non ribosomal peptide synthase 13 BPSS catalase HPII 14 BPSS permease BPSS ABC transporter ATP binding BPSS flavin binding monooxygenase like BPSS cytochrome P450 family BPSS multi domain beta keto acyl synthase BPSS acyl transferase
Protein Class/Name. Pathways. bat00250, bat00280, bat00410, bat00640, bat bat00270, bat00330, bat00410, bat00480
 Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 1. Proteins Increased in Either Soil or Laboratory media Table 1A. Proteins Increased
Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 1. Proteins Increased in Either Soil or Laboratory media Table 1A. Proteins Increased
Supplementary Table 8. Genes from supplementary table 4 ordered in the way in which they occur in the heatmap in Figure 4b of the main text.
 Supplementary Table 8. Genes from supplementary table 4 ordered in the way in which they occur in the heatmap in Figure 4b of the main text. gene name protein name reference name strand start positend
Supplementary Table 8. Genes from supplementary table 4 ordered in the way in which they occur in the heatmap in Figure 4b of the main text. gene name protein name reference name strand start positend
Figure S1: Abundance of Fe related proteins in oceanic Synechococcus sp. strain WH8102 in. Ferredoxin. Ferredoxin. Fe ABC transporter
 0.004 SW nm 0.04 0.03 nm nm 0.16 0.1 nm nm 0.40 0.3 nm nm 0.80 0.5 nm 1.6 1 nm 16 10 nm 160 100 nm 0 nm 0.04 nm 0.16 nm 0.40 nm 0.80 nm 1.6 nm 16 nm 160 nm Spectral counts 0 nm 0.04 nm 0.16 nm 0.40 nm
0.004 SW nm 0.04 0.03 nm nm 0.16 0.1 nm nm 0.40 0.3 nm nm 0.80 0.5 nm 1.6 1 nm 16 10 nm 160 100 nm 0 nm 0.04 nm 0.16 nm 0.40 nm 0.80 nm 1.6 nm 16 nm 160 nm Spectral counts 0 nm 0.04 nm 0.16 nm 0.40 nm
Module 1. Introduction. Microbial Physiology
 Module 1 Introduction to Microbial Physiology Module 1 INTRODUCTION TO MICROBIAL PHYSIOLOGY Page 2 Last time * Discussed the central dogma of molecular biology * Look at, in detail, the process of replication,
Module 1 Introduction to Microbial Physiology Module 1 INTRODUCTION TO MICROBIAL PHYSIOLOGY Page 2 Last time * Discussed the central dogma of molecular biology * Look at, in detail, the process of replication,
f(x) = x R² = RPKM (M8.MXB) f(x) = x E-014 R² = 1 RPKM (M31.
 14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
Protein Class/Name. Pathways. bat00250, bat00280, bat bat00640, bat00650 bat00270, bat00330, bat00410, bat00480 bat00270, bat00330,
 Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 2. Proteins Increased in Either Blood or Horizon media Table 2A. Proteins Increased
Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 2. Proteins Increased in Either Blood or Horizon media Table 2A. Proteins Increased
Genome-wide association studies for understanding pathogen evolution. Samuel K. Sheppard
 Genome-wide association studies for understanding pathogen evolution Samuel K. Sheppard Pathogenic bacteria Population genomics and evolution Sheppard et al. Science 2008; 11: 237-239. Sheppard et al.
Genome-wide association studies for understanding pathogen evolution Samuel K. Sheppard Pathogenic bacteria Population genomics and evolution Sheppard et al. Science 2008; 11: 237-239. Sheppard et al.
Protein Class/Name KEGG Pathways
 Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 4. Proteins Increased in Either Soil medium at 37 o C and 25 o C Table 4A. Proteins
Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 4. Proteins Increased in Either Soil medium at 37 o C and 25 o C Table 4A. Proteins
Figure S3 Differentially expressed fungal and plant genes organised by catalytic activity ontology Organisation of E. festucae (A) and L.
 sak WT Number of branches 3 WT sak Figure S1 Vasculature of plants infected with the E. festucae sak mutant. Light micrographs of perennial ryegrass blade tissue showing branching between the vasculature
sak WT Number of branches 3 WT sak Figure S1 Vasculature of plants infected with the E. festucae sak mutant. Light micrographs of perennial ryegrass blade tissue showing branching between the vasculature
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 28 Fatty Acid Synthesis 2013 W. H. Freeman and Company Chapter 28 Outline 1. The first stage of fatty acid synthesis is transfer
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 28 Fatty Acid Synthesis 2013 W. H. Freeman and Company Chapter 28 Outline 1. The first stage of fatty acid synthesis is transfer
Bacterial cellulose synthesis mechanism of facultative
 1 2 3 4 Bacterial cellulose synthesis mechanism of facultative anaerobe Kaihua Ji 1 +, Wei Wang 2, 3 +, Bing Zeng 1, Sibin Chen 1, Qianqian Zhao 4, Yueqing Chen 1, Guoqiang Li 1* and Ting Ma 1* 5 6 7 Figure
1 2 3 4 Bacterial cellulose synthesis mechanism of facultative anaerobe Kaihua Ji 1 +, Wei Wang 2, 3 +, Bing Zeng 1, Sibin Chen 1, Qianqian Zhao 4, Yueqing Chen 1, Guoqiang Li 1* and Ting Ma 1* 5 6 7 Figure
Supplementary information
 Supplementary information Crosstalk between sugarcane and a plant-growth promoting Burkholderia species Chanyarat Paungfoo-Lonhienne, Thierry G. A. Lonhienne, Yun Kit Yeoh, Bogdan C. Donose, Richard I.
Supplementary information Crosstalk between sugarcane and a plant-growth promoting Burkholderia species Chanyarat Paungfoo-Lonhienne, Thierry G. A. Lonhienne, Yun Kit Yeoh, Bogdan C. Donose, Richard I.
BIOL 158: BIOLOGICAL CHEMISTRY II
 BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site.
 Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site. Still having trouble understanding the material? Check
Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site. Still having trouble understanding the material? Check
Lecture 29: Membrane Transport and metabolism
 Chem*3560 Lecture 29: Membrane Transport and metabolism Insulin controls glucose uptake Adipose tissue and muscles contain a passive glucose transporter GluT4 which takes up glucose from blood. (This is
Chem*3560 Lecture 29: Membrane Transport and metabolism Insulin controls glucose uptake Adipose tissue and muscles contain a passive glucose transporter GluT4 which takes up glucose from blood. (This is
WT siz1-2 siz1-3. WT siz1-2 siz1-3. -Pi, 0.05 M IAA. -Pi, 2.5 M NPA
 A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
Supplementary Information
 Supplementary Information Assembly and Clustering of Natural Antibiotics Guides Target Identification Chad W. Johnston 1,2, Michael A. Skinnider 1,2, Chris A. Dejong 1,2, Philip N. Rees 1,2, Gregory M.
Supplementary Information Assembly and Clustering of Natural Antibiotics Guides Target Identification Chad W. Johnston 1,2, Michael A. Skinnider 1,2, Chris A. Dejong 1,2, Philip N. Rees 1,2, Gregory M.
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the
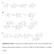 Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Lecture 11 - Biosynthesis of Amino Acids
 Lecture 11 - Biosynthesis of Amino Acids Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction Biosynthetic pathways for amino acids, nucleotides and lipids
Lecture 11 - Biosynthesis of Amino Acids Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction Biosynthetic pathways for amino acids, nucleotides and lipids
Syllabus for BASIC METABOLIC PRINCIPLES
 Syllabus for BASIC METABOLIC PRINCIPLES The video lecture covers basic principles you will need to know for the lectures covering enzymes and metabolism in Principles of Metabolism and elsewhere in the
Syllabus for BASIC METABOLIC PRINCIPLES The video lecture covers basic principles you will need to know for the lectures covering enzymes and metabolism in Principles of Metabolism and elsewhere in the
SUPPLEMENTARY INFORMATION. A comparative study of outer membrane proteome between a paired colistinsusceptible
 SUPPLEMENTARY INFORMATION A comparative study of outer membrane proteome between a paired colistinsusceptible and extremely colistin-resistant Klebsiella pneumoniae strains Raad Jasim, Mark A. Baker, 2
SUPPLEMENTARY INFORMATION A comparative study of outer membrane proteome between a paired colistinsusceptible and extremely colistin-resistant Klebsiella pneumoniae strains Raad Jasim, Mark A. Baker, 2
PAPER No. : 16 Bioorganic and biophysical chemistry MODULE No. : 25 Coenzyme-I Coenzyme A, TPP, B12 and biotin
 Subject Paper No and Title Module No and Title Module Tag 16, Bio organic and Bio physical chemistry 25, Coenzyme-I : Coenzyme A, TPP, B12 and CHE_P16_M25 TABLE OF CONTENTS 1. Learning Outcomes 2. Introduction
Subject Paper No and Title Module No and Title Module Tag 16, Bio organic and Bio physical chemistry 25, Coenzyme-I : Coenzyme A, TPP, B12 and CHE_P16_M25 TABLE OF CONTENTS 1. Learning Outcomes 2. Introduction
Electron Transport Chain and Oxidative phosphorylation
 Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
Coenzymes, vitamins and trace elements 209. Petr Tůma Eva Samcová
 Coenzymes, vitamins and trace elements 209 Petr Tůma Eva Samcová History and nomenclature of enzymes 1810, Gay-Lussac made an experiment with yeats alter saccharide to ethanol and CO 2 Fermentation From
Coenzymes, vitamins and trace elements 209 Petr Tůma Eva Samcová History and nomenclature of enzymes 1810, Gay-Lussac made an experiment with yeats alter saccharide to ethanol and CO 2 Fermentation From
Coenzymes. Coenzymes 9/11/2018. BCMB 3100 Introduction to Coenzymes & Vitamins
 BCMB 3100 Introduction to Coenzymes & Vitamins Cofactors Essential ions Coenzymes Cosubstrates Prosthetic groups Coenzymes structure/function/active group Vitamins 1 Coenzymes Some enzymes require for
BCMB 3100 Introduction to Coenzymes & Vitamins Cofactors Essential ions Coenzymes Cosubstrates Prosthetic groups Coenzymes structure/function/active group Vitamins 1 Coenzymes Some enzymes require for
0.5. Normalized 95% gray value interval h
 Normalized 95% gray value interval.5.4.3.2.1 h Supplemental Figure 1: Symptom score of root samples used in the proteomics study. For each time point, the normalized 95% gray value interval is an averaged
Normalized 95% gray value interval.5.4.3.2.1 h Supplemental Figure 1: Symptom score of root samples used in the proteomics study. For each time point, the normalized 95% gray value interval is an averaged
Catabolism of Carbon skeletons of Amino acids. Amino acid metabolism
 Catabolism of Carbon skeletons of Amino acids Amino acid metabolism Carbon skeleton Carbon Skeleton a carbon skeleton is the internal structure of organic molecules. Carbon Arrangements The arrangement
Catabolism of Carbon skeletons of Amino acids Amino acid metabolism Carbon skeleton Carbon Skeleton a carbon skeleton is the internal structure of organic molecules. Carbon Arrangements The arrangement
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 31 Amino Acid Synthesis 2013 W. H. Freeman and Company Chapter 31 Outline Although the atmosphere is approximately 80% nitrogen,
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 31 Amino Acid Synthesis 2013 W. H. Freeman and Company Chapter 31 Outline Although the atmosphere is approximately 80% nitrogen,
BCMB 3100 Introduction to Coenzymes & Vitamins
 BCMB 3100 Introduction to Coenzymes & Vitamins Cofactors Essential ions Coenzymes Cosubstrates Prosthetic groups Coenzymes structure/function/active group Vitamins 1 Coenzymes Some enzymes require for
BCMB 3100 Introduction to Coenzymes & Vitamins Cofactors Essential ions Coenzymes Cosubstrates Prosthetic groups Coenzymes structure/function/active group Vitamins 1 Coenzymes Some enzymes require for
Chapter 10. 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation)
 Chapter 10 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation) 1 Fueling Processes Respiration 1 Most respiration involves use of an electron transport chain As electrons pass through the electron
Chapter 10 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation) 1 Fueling Processes Respiration 1 Most respiration involves use of an electron transport chain As electrons pass through the electron
Supplemental Information
 Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
MITOCHONDRIA LECTURES OVERVIEW
 1 MITOCHONDRIA LECTURES OVERVIEW A. MITOCHONDRIA LECTURES OVERVIEW Mitochondrial Structure The arrangement of membranes: distinct inner and outer membranes, The location of ATPase, DNA and ribosomes The
1 MITOCHONDRIA LECTURES OVERVIEW A. MITOCHONDRIA LECTURES OVERVIEW Mitochondrial Structure The arrangement of membranes: distinct inner and outer membranes, The location of ATPase, DNA and ribosomes The
Glycolysis - Plasmodium
 Apicomplexan Biochemistry Basics Toxoplasma Good cell biology model Genome sequencing not completed Virtual pathways Cryptosporidium The strange one Genome sequence completed Virtual pathways Plasmodium
Apicomplexan Biochemistry Basics Toxoplasma Good cell biology model Genome sequencing not completed Virtual pathways Cryptosporidium The strange one Genome sequence completed Virtual pathways Plasmodium
MITOCW watch?v=ddt1kusdoog
 MITOCW watch?v=ddt1kusdoog The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high-quality educational resources for free. To
MITOCW watch?v=ddt1kusdoog The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high-quality educational resources for free. To
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting
 Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
CELLS. Cells. Basic unit of life (except virus)
 Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
FATTY ACID SYNTHESIS
 FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
SDR families. SDR10E Fatty acyl-coa reductase FACR1_HUMAN 544 x x SDR11E 3 beta-hydroxysteroid dehydrogenase 3BHS1_HUMAN 254 x x
 SDR1E UDP-glucose 4-epimerase GALE_HUMAN 5305 x x x x SDR2E dtdp-d-glucose 4,6-dehydratase TGDS_HUMAN 4194 x x x x SDR3E GDP-mannose 4,6 dehydratase GMDS_HUMAN 2314 x x x x SDR4E GDP-L-fucose synthetase
SDR1E UDP-glucose 4-epimerase GALE_HUMAN 5305 x x x x SDR2E dtdp-d-glucose 4,6-dehydratase TGDS_HUMAN 4194 x x x x SDR3E GDP-mannose 4,6 dehydratase GMDS_HUMAN 2314 x x x x SDR4E GDP-L-fucose synthetase
LIPID METABOLISM
 LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
Electron transport chain, oxidative phosphorylation, mitochondrial transport systems
 Electron transport chain, oxidative phosphorylation, mitochondrial transport systems JAN ILLNER Respiratory chain & oxidative phosphorylation INTERMEMBRANE SPACE ubiquinone cytochrome c ATPase Production
Electron transport chain, oxidative phosphorylation, mitochondrial transport systems JAN ILLNER Respiratory chain & oxidative phosphorylation INTERMEMBRANE SPACE ubiquinone cytochrome c ATPase Production
BCM 221 LECTURES OJEMEKELE O.
 BCM 221 LECTURES BY OJEMEKELE O. OUTLINE INTRODUCTION TO LIPID CHEMISTRY STORAGE OF ENERGY IN ADIPOCYTES MOBILIZATION OF ENERGY STORES IN ADIPOCYTES KETONE BODIES AND KETOSIS PYRUVATE DEHYDROGENASE COMPLEX
BCM 221 LECTURES BY OJEMEKELE O. OUTLINE INTRODUCTION TO LIPID CHEMISTRY STORAGE OF ENERGY IN ADIPOCYTES MOBILIZATION OF ENERGY STORES IN ADIPOCYTES KETONE BODIES AND KETOSIS PYRUVATE DEHYDROGENASE COMPLEX
2. What is molecular oxygen directly converted into? a. Carbon Dioxide b. Water c. Glucose d. None of the Above
 Biochem 1 Mock Exam 3 Chapter 11: 1. What is glucose completely oxidized into? a. Carbon Dioxide and Water b. Carbon Dioxide and Oxygen c. Oxygen and Water d. Water and Glycogen 2. What is molecular oxygen
Biochem 1 Mock Exam 3 Chapter 11: 1. What is glucose completely oxidized into? a. Carbon Dioxide and Water b. Carbon Dioxide and Oxygen c. Oxygen and Water d. Water and Glycogen 2. What is molecular oxygen
CELLULAR METABOLISM. Metabolic pathways can be linear, branched, cyclic or spiral
 CHM333 LECTURE 24 & 25: 3/27 29/13 SPRING 2013 Professor Christine Hrycyna CELLULAR METABOLISM What is metabolism? - How cells acquire, transform, store and use energy - Study reactions in a cell and how
CHM333 LECTURE 24 & 25: 3/27 29/13 SPRING 2013 Professor Christine Hrycyna CELLULAR METABOLISM What is metabolism? - How cells acquire, transform, store and use energy - Study reactions in a cell and how
Citric acid cycle and respiratory chain. Pavla Balínová
 Citric acid cycle and respiratory chain Pavla Balínová Mitochondria Structure of mitochondria: Outer membrane Inner membrane (folded) Matrix space (mtdna, ribosomes, enzymes of CAC, β-oxidation of FA,
Citric acid cycle and respiratory chain Pavla Balínová Mitochondria Structure of mitochondria: Outer membrane Inner membrane (folded) Matrix space (mtdna, ribosomes, enzymes of CAC, β-oxidation of FA,
Supplementary figure legends
 Supplementary figure legends Fig. S1. Lineweaver-Burk plot of putrescine uptake by YeeF. An overnight culture of SK629 was inoculated in 100-mL LBG medium in 500-mL Erlenmeyer flasks. The medium was supplemented
Supplementary figure legends Fig. S1. Lineweaver-Burk plot of putrescine uptake by YeeF. An overnight culture of SK629 was inoculated in 100-mL LBG medium in 500-mL Erlenmeyer flasks. The medium was supplemented
number Done by Corrected by Doctor Faisal Al-Khatibe
 number 24 Done by Mohammed tarabieh Corrected by Doctor Faisal Al-Khatibe 1 P a g e *Please look over the previous sheet about fatty acid synthesis **Oxidation(degradation) of fatty acids, occurs in the
number 24 Done by Mohammed tarabieh Corrected by Doctor Faisal Al-Khatibe 1 P a g e *Please look over the previous sheet about fatty acid synthesis **Oxidation(degradation) of fatty acids, occurs in the
Lecture 16. Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III
 Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 30 Amino Acid Degradation and the Urea Cycle 2013 W. H. Freeman and Company Chapter 30 Outline Amino acids are obtained from the
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 30 Amino Acid Degradation and the Urea Cycle 2013 W. H. Freeman and Company Chapter 30 Outline Amino acids are obtained from the
Cell Structure. Morphology of Prokaryotic Cell. Cytoplasmic Membrane 4/6/2011. Chapter 3. Cytoplasmic membrane
 Cell Structure Chapter 3 Morphology of Prokaryotic Cell Cytoplasmic membrane Delicate thin fluid structure Surrounds cytoplasm of cell Defines boundary Defines boundary Serves as a selectively permeable
Cell Structure Chapter 3 Morphology of Prokaryotic Cell Cytoplasmic membrane Delicate thin fluid structure Surrounds cytoplasm of cell Defines boundary Defines boundary Serves as a selectively permeable
Coenzymes. Coenzymes 9/15/2014. BCMB 3100 Introduction to Coenzymes & Vitamins
 BCMB 3100 Introduction to Coenzymes & Vitamins Cofactors Essential ions Coenzymes Cosubstrates Prosthetic groups Coenzymes structure/function/active group Vitamins 1 Coenzymes Some enzymes require for
BCMB 3100 Introduction to Coenzymes & Vitamins Cofactors Essential ions Coenzymes Cosubstrates Prosthetic groups Coenzymes structure/function/active group Vitamins 1 Coenzymes Some enzymes require for
9/16/2015. Coenzymes. Coenzymes. BCMB 3100 Introduction to Coenzymes & Vitamins. Types of cofactors
 BCMB 3100 Introduction to Coenzymes & Vitamins Cofactors Essential ions Coenzymes Cosubstrates Prosthetic groups Coenzymes structure/function/active group Vitamins 1 Coenzymes Some enzymes require for
BCMB 3100 Introduction to Coenzymes & Vitamins Cofactors Essential ions Coenzymes Cosubstrates Prosthetic groups Coenzymes structure/function/active group Vitamins 1 Coenzymes Some enzymes require for
respiration mitochondria mitochondria metabolic pathways reproduction can fuse or split DRP1 interacts with ER tubules chapter DRP1 ER tubule
 mitochondria respiration chapter 3-4 shape highly variable can fuse or split structure outer membrane inner membrane cristae intermembrane space mitochondrial matrix free ribosomes respiratory enzymes
mitochondria respiration chapter 3-4 shape highly variable can fuse or split structure outer membrane inner membrane cristae intermembrane space mitochondrial matrix free ribosomes respiratory enzymes
Figure 1 Original Advantages of biological reactions being catalyzed by enzymes:
 Enzyme basic concepts, Enzyme Regulation I III Carmen Sato Bigbee, Ph.D. Objectives: 1) To understand the bases of enzyme catalysis and the mechanisms of enzyme regulation. 2) To understand the role of
Enzyme basic concepts, Enzyme Regulation I III Carmen Sato Bigbee, Ph.D. Objectives: 1) To understand the bases of enzyme catalysis and the mechanisms of enzyme regulation. 2) To understand the role of
Carbohydrate Metabolism I
 Carbohydrate Metabolism I Outline Glycolysis Stages of glycolysis Regulation of Glycolysis Carbohydrate Metabolism Overview Enzyme Classification Dehydrogenase - oxidizes substrate using cofactors as
Carbohydrate Metabolism I Outline Glycolysis Stages of glycolysis Regulation of Glycolysis Carbohydrate Metabolism Overview Enzyme Classification Dehydrogenase - oxidizes substrate using cofactors as
Physiological Adaptation. Microbial Physiology Module 4
 Physiological Adaptation Microbial Physiology Module 4 Topics Coordination of Metabolic Reactions Regulation of Enzyme Activity Regulation of Gene Expression Global Control, Signal Transduction and Twocomponent
Physiological Adaptation Microbial Physiology Module 4 Topics Coordination of Metabolic Reactions Regulation of Enzyme Activity Regulation of Gene Expression Global Control, Signal Transduction and Twocomponent
Summary of fatty acid synthesis
 Lipid Metabolism, part 2 1 Summary of fatty acid synthesis 8 acetyl CoA + 14 NADPH + 14 H+ + 7 ATP palmitic acid (16:0) + 8 CoA + 14 NADP + + 7 ADP + 7 Pi + 7 H20 1. The major suppliers of NADPH for fatty
Lipid Metabolism, part 2 1 Summary of fatty acid synthesis 8 acetyl CoA + 14 NADPH + 14 H+ + 7 ATP palmitic acid (16:0) + 8 CoA + 14 NADP + + 7 ADP + 7 Pi + 7 H20 1. The major suppliers of NADPH for fatty
Biosynthesis of Fatty Acids
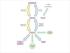 Biosynthesis of Fatty Acids Fatty acid biosynthesis takes place in the cytosol rather than the mitochondria and requires a different activation mechanism and different enzymes and coenzymes than fatty
Biosynthesis of Fatty Acids Fatty acid biosynthesis takes place in the cytosol rather than the mitochondria and requires a different activation mechanism and different enzymes and coenzymes than fatty
Nitrogen Metabolism. Overview
 Nitrogen Metabolism Pratt and Cornely Chapter 18 Overview Nitrogen assimilation Amino acid biosynthesis Nonessential aa Essential aa Nucleotide biosynthesis Amino Acid Catabolism Urea Cycle Juicy Steak
Nitrogen Metabolism Pratt and Cornely Chapter 18 Overview Nitrogen assimilation Amino acid biosynthesis Nonessential aa Essential aa Nucleotide biosynthesis Amino Acid Catabolism Urea Cycle Juicy Steak
Bacterial Structures. Capsule or Glycocalyx TYPES OF FLAGELLA FLAGELLA. Average size: µm 2-8 µm Basic shapes:
 PROKARYOTIC One circular chromosome, not in a membrane No histones No organelles Peptidoglycan cell walls Binary fission EUKARYOTIC Paired chromosomes, in nuclear membrane Histones Organelles Polysaccharide
PROKARYOTIC One circular chromosome, not in a membrane No histones No organelles Peptidoglycan cell walls Binary fission EUKARYOTIC Paired chromosomes, in nuclear membrane Histones Organelles Polysaccharide
Dental Students Biochemistry Exam V Questions ( Note: In all cases, the only correct answer is the best answer)
 Dental Students Biochemistry Exam V Questions - 2006 ( Note: In all cases, the only correct answer is the best answer) 1. Essential fatty acids are: A. precursors of biotin B. precursors of tyrosine C.
Dental Students Biochemistry Exam V Questions - 2006 ( Note: In all cases, the only correct answer is the best answer) 1. Essential fatty acids are: A. precursors of biotin B. precursors of tyrosine C.
E.coli Core Model: Metabolic Core
 1 E.coli Core Model: Metabolic Core 2 LEARNING OBJECTIVES Each student should be able to: Describe the glycolysis pathway in the core model. Describe the TCA cycle in the core model. Explain gluconeogenesis.
1 E.coli Core Model: Metabolic Core 2 LEARNING OBJECTIVES Each student should be able to: Describe the glycolysis pathway in the core model. Describe the TCA cycle in the core model. Explain gluconeogenesis.
Supplementary Tables 1
 Supplementary Tables 1 Supplementary Table 1: The 11 genes in the geneset for Case Study 1 At1g62570 At1g09350 At1g60470 At2g47180 At4g17090 At5g20830 At2g16890 At3g51240 At3g55120 At4g27560 At5g08640
Supplementary Tables 1 Supplementary Table 1: The 11 genes in the geneset for Case Study 1 At1g62570 At1g09350 At1g60470 At2g47180 At4g17090 At5g20830 At2g16890 At3g51240 At3g55120 At4g27560 At5g08640
Oxidative Phosphorylation
 Electron Transport Chain (overview) The NADH and FADH 2, formed during glycolysis, β- oxidation and the TCA cycle, give up their electrons to reduce molecular O 2 to H 2 O. Electron transfer occurs through
Electron Transport Chain (overview) The NADH and FADH 2, formed during glycolysis, β- oxidation and the TCA cycle, give up their electrons to reduce molecular O 2 to H 2 O. Electron transfer occurs through
number Done by Corrected by Doctor F. Al-Khateeb
 number 23 Done by A. Rawajbeh Corrected by Doctor F. Al-Khateeb Ketone bodies Ketone bodies are used by the peripheral tissues like the skeletal and cardiac muscles, where they are the preferred source
number 23 Done by A. Rawajbeh Corrected by Doctor F. Al-Khateeb Ketone bodies Ketone bodies are used by the peripheral tissues like the skeletal and cardiac muscles, where they are the preferred source
Ahmad Ulnar. Faisal Nimri ... Dr.Faisal
 24 Ahmad Ulnar Faisal Nimri... Dr.Faisal Fatty Acid Synthesis - Occurs mainly in the Liver (to store excess carbohydrates as triacylglycerols(fat)) and in lactating mammary glands (for the production of
24 Ahmad Ulnar Faisal Nimri... Dr.Faisal Fatty Acid Synthesis - Occurs mainly in the Liver (to store excess carbohydrates as triacylglycerols(fat)) and in lactating mammary glands (for the production of
Microbiology: A Systems Approach
 Microbiology: A Systems Approach First Edition Cowan & Talaro Chapter 4 Prokaryotic Profiles: the Bacteria and the Archaea Chapter 4 Fig. 4.1 3 3 parts flagella filament long, thin, helical structure composed
Microbiology: A Systems Approach First Edition Cowan & Talaro Chapter 4 Prokaryotic Profiles: the Bacteria and the Archaea Chapter 4 Fig. 4.1 3 3 parts flagella filament long, thin, helical structure composed
Synthesis and degradation of fatty acids Martina Srbová
 Synthesis and degradation of fatty acids Martina Srbová martina.srbova@lfmotol.cuni.cz Fatty acids (FA) mostly an even number of carbon atoms and linear chain in esterified form as component of lipids
Synthesis and degradation of fatty acids Martina Srbová martina.srbova@lfmotol.cuni.cz Fatty acids (FA) mostly an even number of carbon atoms and linear chain in esterified form as component of lipids
Lecture 10 - Protein Turnover and Amino Acid Catabolism
 Lecture 10 - Protein Turnover and Amino Acid Catabolism Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction 2 Proteins are degraded into amino acids. Protein
Lecture 10 - Protein Turnover and Amino Acid Catabolism Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction 2 Proteins are degraded into amino acids. Protein
Amino Acid Metabolism
 Amino Acid Metabolism Last Week Most of the Animal Kingdom = Lazy - Most higher organisms in the animal kindom don t bother to make all of the amino acids. - Instead, we eat things that make the essential
Amino Acid Metabolism Last Week Most of the Animal Kingdom = Lazy - Most higher organisms in the animal kindom don t bother to make all of the amino acids. - Instead, we eat things that make the essential
Lecture: 26 OXIDATION OF FATTY ACIDS
 Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Table removed due to copyright reasons.
 atural Product Biosynthesis Relevance of natural products in the pharmaceutical industry (see Table 2 in Butler J. at. Prod. 2004, 67, 2141-2153.)) Table removed due to copyright reasons. 2 * a phosphopantetheinyl
atural Product Biosynthesis Relevance of natural products in the pharmaceutical industry (see Table 2 in Butler J. at. Prod. 2004, 67, 2141-2153.)) Table removed due to copyright reasons. 2 * a phosphopantetheinyl
Supplemental Information:
 SupplementalInformation: Content: FigureS1.AccessibilityoftheL.interrogansproteomebyLC MSanalysis. FigureS2.Functionalannotationoftheidentifiedproteins. FigureS3.DistributionofMS1 featureintensities. FigureS4.ComparisonoftheDDA
SupplementalInformation: Content: FigureS1.AccessibilityoftheL.interrogansproteomebyLC MSanalysis. FigureS2.Functionalannotationoftheidentifiedproteins. FigureS3.DistributionofMS1 featureintensities. FigureS4.ComparisonoftheDDA
Oxidation of Long Chain Fatty Acids
 Oxidation of Long Chain Fatty Acids Dr NC Bird Oxidation of long chain fatty acids is the primary source of energy supply in man and animals. Hibernating animals utilise fat stores to maintain body heat,
Oxidation of Long Chain Fatty Acids Dr NC Bird Oxidation of long chain fatty acids is the primary source of energy supply in man and animals. Hibernating animals utilise fat stores to maintain body heat,
BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001
 BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) The
BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) The
III. Metabolism The Citric Acid Cycle
 Department of Chemistry and Biochemistry University of Lethbridge III. Metabolism The Citric Acid Cycle Slide 1 The Eight Steps of the Citric Acid Cycle Enzymes: 4 dehydrogenases (2 decarboxylation) 3
Department of Chemistry and Biochemistry University of Lethbridge III. Metabolism The Citric Acid Cycle Slide 1 The Eight Steps of the Citric Acid Cycle Enzymes: 4 dehydrogenases (2 decarboxylation) 3
Tala Saleh. Razi Kittaneh ... Nayef Karadsheh
 Tala Saleh Razi Kittaneh... Nayef Karadsheh β-oxidation of Fatty Acids The oxidation of fatty acids occurs in 3 steps: Step 1: Activation of the Fatty acid FA + HS-CoA + ATP FA-CoA + AMP + PPi - The fatty
Tala Saleh Razi Kittaneh... Nayef Karadsheh β-oxidation of Fatty Acids The oxidation of fatty acids occurs in 3 steps: Step 1: Activation of the Fatty acid FA + HS-CoA + ATP FA-CoA + AMP + PPi - The fatty
Energy storage in cells
 Energy storage in cells Josef Fontana EC - 58 Overview of the lecture Introduction to the storage substances of human body Overview of storage compounds in the body Glycogen metabolism Structure of glycogen
Energy storage in cells Josef Fontana EC - 58 Overview of the lecture Introduction to the storage substances of human body Overview of storage compounds in the body Glycogen metabolism Structure of glycogen
Lecture 13 (10/13/17)
 Lecture 13 (10/13/17) Reading: Ch6; 187-189, 204-205 Problems: Ch4 (text); 2, 3 NXT (after xam 2) Reading: Ch6; 190-191, 194-195, 197-198 Problems: Ch6 (text); 5, 6, 7, 24 OUTLIN NZYMS: Binding & Catalysis
Lecture 13 (10/13/17) Reading: Ch6; 187-189, 204-205 Problems: Ch4 (text); 2, 3 NXT (after xam 2) Reading: Ch6; 190-191, 194-195, 197-198 Problems: Ch6 (text); 5, 6, 7, 24 OUTLIN NZYMS: Binding & Catalysis
Fatty acid breakdown
 Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
NBCE Mock Board Questions Biochemistry
 1. Fluid mosaic describes. A. Tertiary structure of proteins B. Ribosomal subunits C. DNA structure D. Plasma membrane structure NBCE Mock Board Questions Biochemistry 2. Where in the cell does beta oxidation
1. Fluid mosaic describes. A. Tertiary structure of proteins B. Ribosomal subunits C. DNA structure D. Plasma membrane structure NBCE Mock Board Questions Biochemistry 2. Where in the cell does beta oxidation
Nafith Abu Tarboush DDS, MSc, PhD
 Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush Why do we need it? Location & where is it in the picture? Electron (energy) carrying molecules Components Enzymes & cofactors
Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush Why do we need it? Location & where is it in the picture? Electron (energy) carrying molecules Components Enzymes & cofactors
BIOSYNTHESIS OF FATTY ACIDS. doc. Ing. Zenóbia Chavková, CSc.
 BIOSYNTHESIS OF FATTY ACIDS doc. Ing. Zenóbia Chavková, CSc. The pathway for the of FAs is not the reversal of the oxidation pathway Both pathways are separated within different cellular compartments In
BIOSYNTHESIS OF FATTY ACIDS doc. Ing. Zenóbia Chavková, CSc. The pathway for the of FAs is not the reversal of the oxidation pathway Both pathways are separated within different cellular compartments In
10/25/2010 CHAPTER 9 CELLULAR RESPIRATION. Life is Work. Types of cellular respiration. Catabolic pathways = oxidizing fuels
 CHAPTER 9 CELLULAR RESPIRATION Life is Work Living cells require transfusions of energy from outside sources to perform their many tasks: Chemical work Transport work Mechanical work Energy stored in the
CHAPTER 9 CELLULAR RESPIRATION Life is Work Living cells require transfusions of energy from outside sources to perform their many tasks: Chemical work Transport work Mechanical work Energy stored in the
BY: RASAQ NURUDEEN OLAJIDE
 BY: RASAQ NURUDEEN OLAJIDE LECTURE CONTENT INTRODUCTION CITRIC ACID CYCLE (T.C.A) PRODUCTION OF ACETYL CoA REACTIONS OF THE CITIRC ACID CYCLE THE AMPHIBOLIC NATURE OF THE T.C.A CYCLE THE GLYOXYLATE CYCLE
BY: RASAQ NURUDEEN OLAJIDE LECTURE CONTENT INTRODUCTION CITRIC ACID CYCLE (T.C.A) PRODUCTION OF ACETYL CoA REACTIONS OF THE CITIRC ACID CYCLE THE AMPHIBOLIC NATURE OF THE T.C.A CYCLE THE GLYOXYLATE CYCLE
number Done by Corrected by Doctor Nayef Karadsheh
 number 11 Done by حسام أبو عوض Corrected by Moayyad Al-Shafei Doctor Nayef Karadsheh 1 P a g e General Regulatory Aspects in Metabolism: We can divide all pathways in metabolism to catabolicand anabolic.
number 11 Done by حسام أبو عوض Corrected by Moayyad Al-Shafei Doctor Nayef Karadsheh 1 P a g e General Regulatory Aspects in Metabolism: We can divide all pathways in metabolism to catabolicand anabolic.
Fatty acid synthesis. Dr. Nalini Ganesan M.Sc., Ph.D Associate Professor Department of Biochemistry SRMC & RI (DU) Porur, Chennai - 116
 Fatty acid synthesis Dr. Nalini Ganesan M.Sc., Ph.D Associate Professor Department of Biochemistry SRMC & RI (DU) Porur, Chennai 116 Harper s biochemistry 24 th ed, Pg 218 Fatty acid Synthesis Known as
Fatty acid synthesis Dr. Nalini Ganesan M.Sc., Ph.D Associate Professor Department of Biochemistry SRMC & RI (DU) Porur, Chennai 116 Harper s biochemistry 24 th ed, Pg 218 Fatty acid Synthesis Known as
Module No. # 01 Lecture No. # 19 TCA Cycle
 Biochemical Engineering Prof. Dr. Rintu Banerjee Department of Agricultural and Food Engineering Asst. Prof. Dr. Saikat Chakraborty Department of Chemical Engineering Indian Institute of Technology, Kharagpur
Biochemical Engineering Prof. Dr. Rintu Banerjee Department of Agricultural and Food Engineering Asst. Prof. Dr. Saikat Chakraborty Department of Chemical Engineering Indian Institute of Technology, Kharagpur
23.1 Lipid Metabolism in Animals. Chapter 23. Micelles Lipid Metabolism in. Animals. Overview of Digestion Lipid Metabolism in
 Denniston Topping Caret Copyright! The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Chapter 23 Fatty Acid Metabolism Triglycerides (Tgl) are emulsified into fat droplets
Denniston Topping Caret Copyright! The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Chapter 23 Fatty Acid Metabolism Triglycerides (Tgl) are emulsified into fat droplets
Scantron Instructions
 BIOLOGY 1A MIDTERM # 1 February 17 th, 2012 NAME SECTION # DISCUSSION GSI 1. Sit every other seat and sit by section number. Place all books and paper on the floor. Turn off all phones, pagers, etc. and
BIOLOGY 1A MIDTERM # 1 February 17 th, 2012 NAME SECTION # DISCUSSION GSI 1. Sit every other seat and sit by section number. Place all books and paper on the floor. Turn off all phones, pagers, etc. and
The diagram below summarizes the conversion of the twenty standard amino acids. Copyright Mark Brandt, Ph.D. 23
 Amino acid breakdown Amino acids comprise one of the three major energy sources for animals. They are an especially important energy source for carnivorous animals, and for all animals during early starvation
Amino acid breakdown Amino acids comprise one of the three major energy sources for animals. They are an especially important energy source for carnivorous animals, and for all animals during early starvation
FREE ENERGY Reactions involving free energy: 1. Exergonic 2. Endergonic
 BIOENERGETICS FREE ENERGY It is the portion of the total energy change in a system that is available for doing work at constant temperature and pressure; it is represented as ΔG. Reactions involving free
BIOENERGETICS FREE ENERGY It is the portion of the total energy change in a system that is available for doing work at constant temperature and pressure; it is represented as ΔG. Reactions involving free
Marah Bitar. Faisal Nimri ... Nafeth Abu Tarboosh
 8 Marah Bitar Faisal Nimri... Nafeth Abu Tarboosh Summary of the 8 steps of citric acid cycle Step 1. Acetyl CoA joins with a four-carbon molecule, oxaloacetate, releasing the CoA group and forming a six-carbon
8 Marah Bitar Faisal Nimri... Nafeth Abu Tarboosh Summary of the 8 steps of citric acid cycle Step 1. Acetyl CoA joins with a four-carbon molecule, oxaloacetate, releasing the CoA group and forming a six-carbon
Six Types of Enzyme Catalysts
 Six Types of Enzyme Catalysts Although a huge number of reactions occur in living systems, these reactions fall into only half a dozen types. The reactions are: 1. Oxidation and reduction. Enzymes that
Six Types of Enzyme Catalysts Although a huge number of reactions occur in living systems, these reactions fall into only half a dozen types. The reactions are: 1. Oxidation and reduction. Enzymes that
Lecture 17: Nitrogen metabolism 1. Urea cycle detoxification of NH 3 2. Amino acid degradation
 Lecture 17: Nitrogen metabolism 1. Urea cycle detoxification of NH 3 2. Amino acid degradation Reference material Biochemistry 4 th edition, Mathews, Van Holde, Appling, Anthony Cahill. Pearson ISBN:978
Lecture 17: Nitrogen metabolism 1. Urea cycle detoxification of NH 3 2. Amino acid degradation Reference material Biochemistry 4 th edition, Mathews, Van Holde, Appling, Anthony Cahill. Pearson ISBN:978
Metabolism III. Aim: understand gluconeogenesis, pentose phosphate pathway, photosynthesis and amino acid synthesis
 Metabolism III Aim: understand gluconeogenesis, pentose phosphate pathway, photosynthesis and amino acid synthesis Anabolism From a carbon source and inorganic molecules, microbes synthesize new organelles
Metabolism III Aim: understand gluconeogenesis, pentose phosphate pathway, photosynthesis and amino acid synthesis Anabolism From a carbon source and inorganic molecules, microbes synthesize new organelles
number Done by Corrected by Doctor Nayef Karadsheh
 number 17 Done by Abdulrahman Alhanbali Corrected by Lara Abdallat Doctor Nayef Karadsheh 1 P a g e Pentose Phosphate Pathway (PPP) Or Hexose Monophosphate Shunt In this lecture We will talk about the
number 17 Done by Abdulrahman Alhanbali Corrected by Lara Abdallat Doctor Nayef Karadsheh 1 P a g e Pentose Phosphate Pathway (PPP) Or Hexose Monophosphate Shunt In this lecture We will talk about the
Protein Name. IFLENVIR,DSVTYTEHAK,TV TALDVVYALK,KTVTALDVV YALK,TVTALDVVYALKR,IF LENVIRDSVTYTEHAK gi Histone H2B
 Table 1. A functional category list of proteins (Lentinula edodes) identified by 1-DGE and nesi-lc-ms/ms. The table lists indicated fraction numbers, matching peptides, scores, accession numbers, protein
Table 1. A functional category list of proteins (Lentinula edodes) identified by 1-DGE and nesi-lc-ms/ms. The table lists indicated fraction numbers, matching peptides, scores, accession numbers, protein
Synthesis of Fatty Acids and Triacylglycerol
 Synthesis of Fatty Acids and Triacylglycerol Lippincott s Chapter 16 Fatty Acid Synthesis Mainly in the Liver Requires Carbon Source: Acetyl CoA Reducing Power: NADPH 8 CH 3 COO C 15 H 33 COO Energy Input:
Synthesis of Fatty Acids and Triacylglycerol Lippincott s Chapter 16 Fatty Acid Synthesis Mainly in the Liver Requires Carbon Source: Acetyl CoA Reducing Power: NADPH 8 CH 3 COO C 15 H 33 COO Energy Input:
