ACBD2/ECI2-mediated peroxisome-mitochondria interactions in Leydig cell steroid
|
|
|
- Jonas Copeland
- 5 years ago
- Views:
Transcription
1 Supplemental data ACBD2/ECI2-mediated peroxisome-mitochondria interactions in Leydig cell steroid biosynthesis Jinjiang Fan 1,2, Xinlu Li 1,3, Leeyah Issop 1,2, Martine Culty 1,2,4 and Vassilios Papadopoulos 1,2,3,4 1 The Research Institute of the McGill University Health Centre; Departments of 2 Medicine, 3 Biochemistry, and 4 Pharmacology & Therapeutics, McGill University, Montréal, Québec, H4A 3J1, Canada Supplemental Figures: 9 Supplemental Tables: 2 1
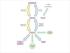
2 A B 99 Gene family and subcellular localization (P, peroxisomes; M, mitochondria) 99 hpeci peci2 reci2 ACBD2/ECI2/PECI meci2-isoa reci3 meci3 geci2 seci1 100 pechs1 hechs1 rechs1 59 mechs1 hech1 pech1 100 rech1 mech1 heci1 peci1 reci1 100 meci1 0.2 geci1 P, M, or both ECHS1 M ECH1/DCI* P, M, or both ECI1/DCI**/mECI M Figure S1. Phylogenetic analysis of ACBD2/ECI2 proteins and their closely related proteins. A, Neighbor-joining phylogenetic tree of ACBD2/ECI2 proteins with their closest functional homologous proteins. Corresponding subcellular localizations of each gene family are indicated at the right of the tree. h, human; p, chimpanzee; r, rat; m, mouse; g, chicken; s, yeast; P, peroxisomes; M, mitochondria; both, both P and M. *DCI/ECH1, dienoyl-coa isomerase; **DCI/ECI1, dodecenoyl- CoA isomerase. meci, mitochondrial ECI. The number at each note indicates the bootstrap support values over 50%, a statistical approach to estimate re-sampling distribution. B, Circle phylogenetic tree of putative ACBD2/ECI2 homologues with their evolutionary origin. Unrooted neighbor-joining phylogenetic tree depicting evolutionary relationships among ACBD2/ECI2 and related sequences, resulting from BLAST searches with mouse ACBD2/ECI2 as a query. Three main clades are indicated: ACBD2/ECI2, CDYL (testis-specific chromodomain Y-like protein), and bacterial enoyl CoA hydrolases. 2
3 BSC SLC PLC ILC ALC GENE SYMBOL GENE TITLE CORE ENRICHMENT EHHADH enoyl-coenzyme A, hydratase Yes ADHFE1 alcohol dehydrogenase, iron containing, 1 Yes CPT1B carnitine palmitoyltransferase 1B (muscle) Yes ADH1A alcohol dehydrogenase 1A (class I), alpha polypeptide Yes CPT2 carnitine palmitoyltransferase II Yes ALDH2 aldehyde dehydrogenase 2 family (mitochondrial) Yes ECHS1 enoylcoenzyme A hydratase, short chain, 1, mitochondrial Yes ACAA2 acetyl-coenzyme A acyltransferase 2 (mitochondrial) Yes ACADS acyl-coenzyme A dehydrogenase, C-2 to C-3 short chain Yes ECI1 dodecenoyl-coenzyme A delta isomerase (ECI1/DCI/mECI) Yes ACAT1 acetyl-coenzyme A acetyltransferase 1 Yes CPT1A carnitine palmitoyltransferase 1A (liver) Yes ECI2 peroxisomal D3,D2-enoyl-CoA isomerase (ACBD2/ECI2/PECI) Yes HADH hydroxyacyl-coenzyme A dehydrogenase Yes ACSL6 acyl-coa synthetase long-chain family member 6 Yes ACADVL acyl-coenzyme A dehydrogenase, very long chain Yes ACOX1 acyl-coenzyme A oxidase 1, palmitoyl Yes ACADM acyl-coenzyme A dehydrogenase, C-4 to C-12 straight chain Yes HADHA hydroxyacyl-coenzyme A dehydrogenase Yes ACADL acyl-coenzyme A dehydrogenase, long chain Yes HSD17B4 hydroxysteroid (17-beta) dehydrogenase 4 Yes ALDH7A1 aldehyde dehydrogenase 7 family, member A1 Yes ALDH9A1 aldehyde dehydrogenase 9 family, member A1 Yes ALDH3A2 aldehyde dehydrogenase 3 family, member A2 Yes ACAT2 acetyl-coenzyme A acetyltransferase 2 Yes HADHB hydroxyacyl-coenzyme A dehydrogenase No ACOX3 acyl-coenzyme A oxidase 3, pristanoyl No ACADSB acyl-coenzyme A dehydrogenase, short/branched chain No ACSL3 acyl-coa synthetase long-chain family member 3 No ADH4 alcohol dehydrogenase 4 (class II), pi polypeptide No ADH7 alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide No ALDH3A1 aldehyde dehydrogenase 3 family, membera1 No ACSL1 acyl-coa synthetase long-chain family member 1 No ACSL5 acyl-coa synthetase long-chain family member 5 No ACSL4 acyl-coa synthetase long-chain family member 4 No CPT1C carnitine palmitoyltransferase 1C No Figure S2. Heat map displaying the gene set enrichment analysis (GSEA) of fatty acid metabolism during rat Leydig cell development and the melting curve profiles of three known Acbd2/Eci2 mrna isoforms. Confirmation of the fatty acid metabolism in ALC via GSEA. The HSA00071_FATTY_ ACID_METABOLISM was enriched on the top among 28 enriched gene sets with NOM p-value < and FDR q-value = ( The range of colors (red, pink, light blue, dark blue) in the heat map shows the range of expression values (high, moderate, low, lowest). SLC, PLC, ILC, and ALC represent the stem, putative, immature, and adult rat Leydig cells, respectively; BSC, bone stem cells as control. The microarray dataset used herein is from the GEO: GSE The Acbd2/Eci2 and Eci1 genes used in this study are highlighted. 3
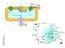
4 A B Single colony Colony expansion C D Mito-H E Mito-L F Progesterone (ng/mg protein) *** MA-10 Mito-H 1 mm dbcamp, 2 hr Mito-L Figure S3. Establishment and characterization of a stable sub-cell line, Mito-H, from MA-10 mouse Leydig tumor cells. A, Isolation of a single colony of MA-10 cells transfected with the plasmid mito-rogfp with a mitochondrial targeting sequence in pegfp-n1 for mammalian expression (pra306). Selective growth occurred under the presence of 500 µg/ml of G418 (Invitrogen) in DMEM/F-12 medium supplemented with 5% heat-inactivated FBS and 2.5% horse serum. Representative GFP fluorescence detected in a single colony using an Olympus 1X51 fluorescence microscope at 40 original magnification. B, The expansion of the single colony under selective growth conditions: DMEM/F-12 medium supplemented with 5% heat-inactivated FBS and 2.5% horse serum at 37 C and 5% CO 2, with an initial supplement of G418 (500 μg/ml). C, Cell sorting of the expanded single colony of MA-10 cells with the stable expression of rogfp1 by flow cytometry. Wild-type MA-10 cells were used as controls (Speciment_001_sample.fcs), and transfected MA-10 cells (Specifiment_001_WT.fcs) were further divided into two groups that were 4
5 cultured under the same conditions: Mito-H (GFP hi) with higher GFP expression and Mito-L (GFP low) with low GFP expression. The remaining cells were abandoned, including the cells with GFP expression between these two groups (GFP+) and the cells without GFP or lower than the Mito-L group (Live). D, Representative GFP fluorescence of Mito-H cells. Scale bars in red = 5 μm. E, Representative GFP fluorescence of Mito-L cells. Scale bars in red = 5 μm. F, Steroid production in the Mito-H cell line. To provide initial data about steroid production in the newly established Mito-H cell line, progesterone production before and after dbcamp stimulation ( or +, as indicated) were measured using RIA, with MA-10 cells used as controls. Steroid production was three times higher in Mito-H cells than that in the parent MA-10 cells. Statistical analysis was performed using Student s t- tests (n = 4; ***p < 0.001). 5
6 A ECI1-DsRed Mito-roGFP Colocalization/ Hoechst M B ECI3-DsRed Mito-roGFP Colocalization/ Hoechst C C DsRed-ECI1 Mito-roGFP Colocalization/ Hoechst C D DsRed-ECI3 Mito-roGFP Colocalization/ Hoechst P Figure S4. Confocal live cell imaging of subcellular targeting of two control proteins: ECI1 and ECI3. Mitochondrial localization of ECI1-DsRed fusion protein (A) compared with peroxisomal localization of DsRed-ECI3 (D), whereas ECI3-DsRed and DsRed-ECI1 are mislocated in the cytocol as shown in (B) and (C), respectively. Both fusion proteins (red) served as controls in comparison with the subcellular localization of ACBD2/ECI2 in the main text. Mito-roGFP (green) was used to stain the mitochondria in Mito-H cells, and Hoechst (blue) was used to stain the nuclei. Magnification of the overlap between the red and green channels and the nuclear DNA staining is shown in the right panels and the inset. M, mitochondria; C, cytosol; P, peroxisomes. Scale bar, 2 μm. 6
7 Figure S5. ACBD2/ECI2 isoform B targets peroxisomes in mouse Leydig cells mltc1. A. Subcellular localization of DsRed-ACBD2/ECI2 (isoform B). B. Subcellular localization of DsRed-ACBD2/ECI2 (isoform B) with Mito-BFP (mitochondria in blue). C. Subcellular localization of DsRed-ACBD2/ECI2 (isoform B) with AcGFP-pero (peroxisomes). D. Overlaps of the three channels with DsRed- ACBD2/ECI2 (isoform B), mito-bfp, and AcGFP-pero. E-G, A magnified view of the highlighted area to show the distribution of DsRed-ACBD2/ECI2 (isoform B) with overlap of peroxisomes (F). M, mitochondria; P*, peroxisomal cargo; P, peroxisomes. 7
8 Figure S6. ACBD2/ECI2 with blocked PTS1 targets mitochondria in mouse Leydig cells mltc1. A. Subcellular localization of ACBD2/ECI2-DsRed (with blocked PTS1). B. Subcellular localization of ACBD2/ECI2-DsRed with Mito-BFP (mitochondria in blue). C. Subcellular localization of ACBD2/ECI2- DsRed with AcGFP-pero (peroxisomes). D. Overlaps of the three channels with ACBD2/ECI2-DsRed, mito-bfp, and AcGFP-pero. E-G, A magnified view of the highlighted area to show the distribution of ACBD2/ECI2-DsRed with overlap of mitochondria (G). M, mitochondria; P*, peroxisomal cargo; P, peroxisomes. 8
9 Figure S7. In-cell Co-IP of endogenous ACBD2/ECI2 with TOMM20 in mouse Leydig cells mltc1. The signals of PLA Duolink- In-cell co-ip (red) were collected in cultures without (A) and at present of 1 mm dbcamp for 2 hr (B). C and E, A representative magnified area shows the red blobs as the PLA signals from A and B, respectively. D, Bar graphs of the quantification of colocalized ACBD2/ECI2 and TOMM20 puncta or contact sites revealed before and after 1 mm dbcamp treatment. Mean + SD; **, student t-test, p < 0.01; n = 299 (control), and 304 (1 mm dbcamp). 9
10 Figure S8. ACBD2/ECI2 isoform A targets mitochondria in mouse Leydig cells mltc1. A. Subcellular localization of mito-dsred-acbd2/eci2 (isoform A). B. Subcellular localization of mito- DsRed-ACBD2/ECI2 (isoform A) with Mito-BFP (mitochondria in blue). C. Subcellular localization of mito-dsred-acbd2/eci2 (isoform A) with AcGFP-pero (peroxisomes). D. Overlaps of the three channels with mito-dsred-acbd2/eci2 (isoform A), mito-bfp, and AcGFP-pero. E-G, A magnified view of the highlighted area to show the distribution of mito-dsred-acbd2/eci2 (isoform A) with overlap of mitochondria (G). M, mitochndria; P*, peroxisomal cargo; P, peroxisomes. 10
11 Figure S9. The tissue/organ distribution of ACBD2/ECI2 isoform A revealed by expressed sequence tags (NCBI). Using the mrna sequence of Acbd2/Eci2 isoform A from MA-10 cell as reference, shown under the red dotted line, the Acbd2/Eci2 gene was found to be expressed in embryo, blastocyst, diaphragm, eye, fibroblast, joints, kidney, liver, tonge, neurosphere, placenta, thymus, TIB BB88 cell line, tumor, uterus, and pooled tissues, where. The conserved mrna sequences among the different sequence varieties are indicated as conserved, and the first ATG encoding the mitochondrial presequence is shown. The mrna expression of the Acbd2/Eci2 isoform A suggests that the findings reported herein on MA-10 Leydig cells may apply to other non-steroidogenic tissues. 11
12 Table S1. Oligonucleotides used in PCR and plasmid construction. Primer Sequence Plasmid/Purpose Eci2-R-XhoI GTCCTCGAGATGGCGGCAGTGACCTGGAGTCGGGCTCGATGCTGGTGTCCG pacbd2/eci2-dsred Eci2-F-SacII CAGCCGCGGCAGCTTTGGTTTTCTGGAGACGAAGC pacbd2/eci2-dsred Eci1-R-NheI CTAGCTAGCATGGCGCTGGCTGCTGCGCGTCG peci1-dsred Eci1-F-SacII CAGCCGCGGGCCCTTCTTTTGCTTGAGC peci1-dsred Eci3-R-NheI CTAGCTAGCATGCCTAAGCCTGGTGTGTTTAACTTTG peci3-dsred Eci3-F-SacII CAGCCGCGGTAGCTTAGCCTTTGCCTTTCTG peci3-dsred Eci2-R-XhoI GTCCTCGAGCTGCGGCAGTGACCTGGAGTCGGGCTCG pdsred-acbd2/eci2 Eci2-F-SacII CAGCCGCGGTCACAGCTTTGGTTTTCTGGAGACGAAGC pdsred-acbd2/eci2 Eci1-R-XholI GTCCTCGAGCTGCGCTGGCTGCTGCGCGTCG pdsred-eci1 Eci1-F-SacII CAGCCGCGGTTAGCCCTTCTTTTGCTTGAGC pdsred-eci1 Eci3-R-XhoI GTCCTCGAGCTCCTAAGCCTGGTGTGTTTAACTTTG pdsred-eci3 Eci3-F-SacIII CAGCCGCGGTCATAGCTTAGCCTTTGCCTTTCTG pdsred-eci3 mpex11b-r-xhol AGATCTCGAGCTGACGCCTGGGTCCGCTTCAGTGC pamcyan-pex11 mpex11b-f-sacii GGCCCGCGGTCAGGGCTTGAGTCGTAGCC pamcyan-pex11 Eci2isoB-R-XhoI ATCTCGAGCTAGAGCCAGTCAGCAGGACTTTG pmito-dsred-eci2 Eci2noACBDR-XhoI AGATCTCGAGCTTCATCTGAAGCCCCGAGCCAGG pmito-dsred-eci2 (noacbp) Exon-lR AGATGCTTCTCTTTGGGAAGAAGC 3 -end mrna Exon-lF TGCTCCCTATGAGGAGTCTAGC 3 -end mrna NcoI-R ACCATGGACAACACCGAGGACG pmito-dsred-eci2 NheI-F TGCTAGCTGCCTCCTCTATCC pmito-dsred-eci2 NcoI-NheI-mitoR GACCATGGAGCTAGCATGGCGGCAGTGACCTGG pmito-dsred-eci2 NcoI-mitoF GTCCATGGTTGTTGGTCTGCCCAAGTGC pmito-dsred-eci2 R TGACCACAGTATCTAGCTATGGC PCR for isoform R TTGACGATTTCCCAGCGAGAGAGC PCR for isoform F TGGCTCTCATTGTTGGTCTGCC PCR for isoform DsRed-pR TCGAGGAAAACCAAAGCTGTGACCGC pdsred-pero/amcyan-pero DsRed-pF GGTCACAGCTTTGGTTTTCC pdsred-pero/amcyan-pero Mito-DsRed-sR TCGAGTGTGATGACCGC pmito-dsred Mito-DsRed-sF GGTCATCACAC pmito-dsred mtomm20-r-nhei GCGGCTAGCATGGTGGGCCGGAACAGCGCCATCG pmtomm20-cfp mtomm20-f-kpni CGCGGTACCGTTCCACATCATCTTCAGC pmtomm20-cfp mpmp70-r-xhoi AGATCTCGAGCTGCGGCCTTCAGCAAGTACTTGACG pamcyan-mpmp70 mpmp70-f-sacii GGCCCGCGGCTATGATCCGAACTCAACTGTATCTTCTGTG pamcyan-mpmp70 12
13 Table S2. Peroxisomal proteins with potential targeting to mitochondria Swiss-Prot Protein Name Gene Name Q8JZV9 3-hydroxybutyrate dehydrogenase type 2 Bdh2 Q921H8 3-ketoacyl-CoA thiolase A, peroxisomal Acaa1a Q80XL6 Acyl-CoA dehydrogenase family member 11 Acad11 Q99L15 Acyl-coenzyme A thioesterase 1 Acot1 Q544M5 Acyl-coenzyme A thioesterase 12 Acot12 Q8BWN8 Acyl-coenzyme A thioesterase 4 Acot4 Q8VCR7 Alpha/beta hydrolase domain-containing protein 14B Abhd14b O09174 Alpha-methylacyl-CoA racemase Amacr ENSMUSP Argininosuccinate synthase Ass1 Q3UD91 Aspartate aminotransferase, mitochondrial Got2 Q99KR3 Beta-lactamase-like protein 2 Lactb2 Q3TCG3 Carnitine O-acetyltransferase Crat Q544Z9 Cytochrome b-5, isoform CRA_d Cyb5 Q9DB77 Cytochrome b-c1 complex subunit 2, mitochondrial Uqcrc2 O35459 Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial Ech1 O54734 Dolichyl-diphosphooligosaccharide-protein glycosyltransferase 48 kda subunit Ddost Q9CQJ4 E3 ubiquitin-protein ligase RING2 Rnf2 Q9WUR2 Enoyl-CoA delta isomerase 2, mitochondrial* ACBD2/ECI2 Q9DCM2 Glutathione S-transferase kappa 1 Gstk1 P13707 Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic Gpd1 Q9D0C9 Histone H4 Hist4h4 Q9NYQ2 Hydroxyacid oxidase 2 Hao2 Q3TT11 Hydroxysteroid(17-Beta)Dehydrogenase 4 Hsd17b4 Q3TJ51 Isocitrate dehydrogenase [NADP] Idh1 Q3TEG8 Lon protease homolog 2, peroxisomal Lonp2 P41216 Long-chain-fatty-acid--CoA ligase 1 Acsl1 P14152 Malate dehydrogenase, cytoplasmic Mdh1 Q9CQ92 Mitochondrial fission 1 protein Fis1 Q922Q1 MOSC domain-containing protein 2, mitochondrial Marc2 Q9CY59 NADH-cytochrome b5 reductase (ec ) homolog Cyb5r3 P32020 Non-specific lipid-transfer protein Scp2 Q3U7H9 Peroxiredoxin-5 Prdx5 Q9WV68 Peroxisomal 2,4-dienoyl-CoA reductase Decr2 Q3TDG0 Peroxisomal acyl-coenzyme A oxidase 1 Acox1 Q9QXD1 Peroxisomal acyl-coenzyme A oxidase 2 Acox2 Q9Z211 Peroxisomal membrane protein 11A Pex11a Q3TF72 Prolyl 4-hydroxylase P4hb Q569N4 Ribonuclease UK114 Hrsp12 P20108 Thioredoxin-dependent peroxide reductase, mitochondrial Prdx3 Q8R164 Valacyclovir hydrolase Bphl O35488 Very long-chain acyl-coa synthetase Slc27a2 Q3U4P9 von Willebrand factor A domain-containing protein 8 Vwa8 Q8R0Z8 Zinc-binding alcohol dehydrogenase domain-containing protein 2 Zadh2 *Acbd2/Eci2/Peci, predicted as peroxisomal protein only (Wiese, et al., 2007). The black font highlights the actual literature report on the dual targets in peroxisomes and mitochondria. 13
Supplementary Figure 1. Mitochondrial function in skeletal muscle and plasma parameters of STZ mice A D-F
 Supplementary Figure 1. Mitochondrial function in skeletal muscle and plasma parameters of STZ mice A Expression of representative subunits of the mitochondrial respiratory chain was analyzed by real time
Supplementary Figure 1. Mitochondrial function in skeletal muscle and plasma parameters of STZ mice A Expression of representative subunits of the mitochondrial respiratory chain was analyzed by real time
CFSE Thy1.2 + CD4 + cells
 Time after immunization (d) IFN-g Spleen Lymph nodes Rbpj +/+ ;OT-II Rbpj -/- ;OT-II Rbpj +/+ ;OT-II Rbpj -/- ;OT-II 2.3.2 22.9 5.6 3 23.3 3. 21.3 32.7 8.7 18.5 11.6 13.5 7 84.2 74.2 85. 83.8 CFSE Thy1.2
Time after immunization (d) IFN-g Spleen Lymph nodes Rbpj +/+ ;OT-II Rbpj -/- ;OT-II Rbpj +/+ ;OT-II Rbpj -/- ;OT-II 2.3.2 22.9 5.6 3 23.3 3. 21.3 32.7 8.7 18.5 11.6 13.5 7 84.2 74.2 85. 83.8 CFSE Thy1.2
Lecture: 26 OXIDATION OF FATTY ACIDS
 Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at
 Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/2/e1602038/dc1 Supplementary Materials for Mitochondrial metabolic regulation by GRP78 Manoj Prasad, Kevin J. Pawlak, William E. Burak, Elizabeth E. Perry, Brendan
advances.sciencemag.org/cgi/content/full/3/2/e1602038/dc1 Supplementary Materials for Mitochondrial metabolic regulation by GRP78 Manoj Prasad, Kevin J. Pawlak, William E. Burak, Elizabeth E. Perry, Brendan
Lehninger 5 th ed. Chapter 17
 Lehninger 5 th ed. Chapter 17 December 26, 2010 Prof. Shimon Schuldiner Email: Shimon.Schuldiner@huji.ac.il Phone: 6585992 CHAPTER 17 Fatty Acid Catabolism Key topics: How fats are digested in animals
Lehninger 5 th ed. Chapter 17 December 26, 2010 Prof. Shimon Schuldiner Email: Shimon.Schuldiner@huji.ac.il Phone: 6585992 CHAPTER 17 Fatty Acid Catabolism Key topics: How fats are digested in animals
Mitochondrial Fatty Acid Oxidation Deficiencies Prof. Niels Gregersen
 Mitochondrial Fatty Acid Oxidation Deficiencies Research Unit for Molecular Medicine Clinical Institute Aarhus University Hospital and Faculty of Health Sciences Aarhus University, Aarhus, Denmark 1 Menu
Mitochondrial Fatty Acid Oxidation Deficiencies Research Unit for Molecular Medicine Clinical Institute Aarhus University Hospital and Faculty of Health Sciences Aarhus University, Aarhus, Denmark 1 Menu
Summary of fatty acid synthesis
 Lipid Metabolism, part 2 1 Summary of fatty acid synthesis 8 acetyl CoA + 14 NADPH + 14 H+ + 7 ATP palmitic acid (16:0) + 8 CoA + 14 NADP + + 7 ADP + 7 Pi + 7 H20 1. The major suppliers of NADPH for fatty
Lipid Metabolism, part 2 1 Summary of fatty acid synthesis 8 acetyl CoA + 14 NADPH + 14 H+ + 7 ATP palmitic acid (16:0) + 8 CoA + 14 NADP + + 7 ADP + 7 Pi + 7 H20 1. The major suppliers of NADPH for fatty
Biosynthesis of Fatty Acids
 Biosynthesis of Fatty Acids Fatty acid biosynthesis takes place in the cytosol rather than the mitochondria and requires a different activation mechanism and different enzymes and coenzymes than fatty
Biosynthesis of Fatty Acids Fatty acid biosynthesis takes place in the cytosol rather than the mitochondria and requires a different activation mechanism and different enzymes and coenzymes than fatty
Tala Saleh. Razi Kittaneh ... Nayef Karadsheh
 Tala Saleh Razi Kittaneh... Nayef Karadsheh β-oxidation of Fatty Acids The oxidation of fatty acids occurs in 3 steps: Step 1: Activation of the Fatty acid FA + HS-CoA + ATP FA-CoA + AMP + PPi - The fatty
Tala Saleh Razi Kittaneh... Nayef Karadsheh β-oxidation of Fatty Acids The oxidation of fatty acids occurs in 3 steps: Step 1: Activation of the Fatty acid FA + HS-CoA + ATP FA-CoA + AMP + PPi - The fatty
Synthesis and degradation of fatty acids Martina Srbová
 Synthesis and degradation of fatty acids Martina Srbová martina.srbova@lfmotol.cuni.cz Fatty acids (FA) mostly an even number of carbon atoms and linear chain in esterified form as component of lipids
Synthesis and degradation of fatty acids Martina Srbová martina.srbova@lfmotol.cuni.cz Fatty acids (FA) mostly an even number of carbon atoms and linear chain in esterified form as component of lipids
Voet Biochemistry 3e John Wiley & Sons, Inc.
 * * Voet Biochemistry 3e Lipid Metabolism Part I: (Chap. 25, sec.1-3) Glucose C 6 H 12 O 6 + 6 O 2 6 CO 2 + 6 H 2 O G o = -2823 kj/mol Fats (palmitic acid) C 16 H 32 O 2 + 23 O 2 16 CO 2 + 16 H 2 O G o
* * Voet Biochemistry 3e Lipid Metabolism Part I: (Chap. 25, sec.1-3) Glucose C 6 H 12 O 6 + 6 O 2 6 CO 2 + 6 H 2 O G o = -2823 kj/mol Fats (palmitic acid) C 16 H 32 O 2 + 23 O 2 16 CO 2 + 16 H 2 O G o
Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR
 Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) cadl CTTGGGGGCGCGTCT CTGTTCTTTTGTGCCGTTTCG cyl-coenzyme Dehydrogenase, very
Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) cadl CTTGGGGGCGCGTCT CTGTTCTTTTGTGCCGTTTCG cyl-coenzyme Dehydrogenase, very
by E. Lucchinetti, C. Hofer, L. Bestmann, M. Hersberger, J. Feng, M. Zhu, L. Furrer, M.C. Schaub, R. Tavakoli, M. Genoni, A. Zollinger, and M.
 Lucchinetti et al., Supplementary Material Supplementary material to: Gene Regulatory Control of Myocardial Energy Metabolism Predicts Postoperative Cardiac Function in Patients Undergoing Off-pump Coronary
Lucchinetti et al., Supplementary Material Supplementary material to: Gene Regulatory Control of Myocardial Energy Metabolism Predicts Postoperative Cardiac Function in Patients Undergoing Off-pump Coronary
Dietary Lipid Utilization by Haddock (Melanogrammus aeglefinus)
 Dietary Lipid Utilization by Haddock (Melanogrammus aeglefinus) Santosh P. Lall & Dominic A. Nanton National Research Council of Canada Halifax, Canada vis, California ne 23, 2003 Body Components of Wild
Dietary Lipid Utilization by Haddock (Melanogrammus aeglefinus) Santosh P. Lall & Dominic A. Nanton National Research Council of Canada Halifax, Canada vis, California ne 23, 2003 Body Components of Wild
Expanded View Figures
 PEX13 functions in selective autophagy Ming Y Lee et al Expanded View Figures Figure EV1. PEX13 is required for Sindbis virophagy. A, B Quantification of mcherry-capsid puncta per cell (A) and GFP-LC3
PEX13 functions in selective autophagy Ming Y Lee et al Expanded View Figures Figure EV1. PEX13 is required for Sindbis virophagy. A, B Quantification of mcherry-capsid puncta per cell (A) and GFP-LC3
Global gene expression analysis of liver for androstenone and skatole production in young boars
 Global gene expression analysis of liver for androstenone and skatole production in young boars Christiane Neuhoff Session 36 Institute of Animal Science Animal Breeding and Husbandry/ Animal Genetics
Global gene expression analysis of liver for androstenone and skatole production in young boars Christiane Neuhoff Session 36 Institute of Animal Science Animal Breeding and Husbandry/ Animal Genetics
Mouse Meda-4 : chromosome 5G bp. EST (547bp) _at. 5 -Meda4 inner race (~1.8Kb)
 Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
LIPID METABOLISM
 LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
Biochemistry - I SPRING Mondays and Wednesdays 9:30-10:45 AM (MR-1307) Lectures Based on Profs. Kevin Gardner & Reza Khayat
 Biochemistry - I Mondays and Wednesdays 9:30-10:45 AM (MR-1307) SPRING 2017 Lectures 21-22 Based on Profs. Kevin Gardner & Reza Khayat 1 Outline Vertebrate processing of dietary lipids Mobilization of
Biochemistry - I Mondays and Wednesdays 9:30-10:45 AM (MR-1307) SPRING 2017 Lectures 21-22 Based on Profs. Kevin Gardner & Reza Khayat 1 Outline Vertebrate processing of dietary lipids Mobilization of
Fatty acid breakdown
 Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
Lipid metabolism. Degradation and biosynthesis of fatty acids Ketone bodies
 Lipid metabolism Degradation and biosynthesis of fatty acids Ketone bodies Fatty acids (FA) primary fuel molecules in the fat category main use is for long-term energy storage high level of energy storage:
Lipid metabolism Degradation and biosynthesis of fatty acids Ketone bodies Fatty acids (FA) primary fuel molecules in the fat category main use is for long-term energy storage high level of energy storage:
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 28 Fatty Acid Synthesis 2013 W. H. Freeman and Company Chapter 28 Outline 1. The first stage of fatty acid synthesis is transfer
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 28 Fatty Acid Synthesis 2013 W. H. Freeman and Company Chapter 28 Outline 1. The first stage of fatty acid synthesis is transfer
Biology 638 Biochemistry II Exam-3. (Note that you are not allowed to use any calculator)
 Biology 638 Biochemistry II Exam-3 (Note that you are not allowed to use any calculator) 1. In the non-cyclic pathway, electron pathway is. Select the most accurate one. a. PSII PC Cyt b 6 f PC PSI Fd-NADP
Biology 638 Biochemistry II Exam-3 (Note that you are not allowed to use any calculator) 1. In the non-cyclic pathway, electron pathway is. Select the most accurate one. a. PSII PC Cyt b 6 f PC PSI Fd-NADP
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM. Fatty Acid Elongation and Desaturation
 ANSC/NUTR 618 LIPIDS & LIPID METABOLISM I. Fatty acid elongation A. General 1. At least 60% of fatty acids in triacylglycerols are C18. 2. Free palmitic acid (16:0) synthesized in cytoplasm is elongated
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM I. Fatty acid elongation A. General 1. At least 60% of fatty acids in triacylglycerols are C18. 2. Free palmitic acid (16:0) synthesized in cytoplasm is elongated
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease using MetPA analysis (panel A).
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease
Fatty acid synthesis. Dr. Nalini Ganesan M.Sc., Ph.D Associate Professor Department of Biochemistry SRMC & RI (DU) Porur, Chennai - 116
 Fatty acid synthesis Dr. Nalini Ganesan M.Sc., Ph.D Associate Professor Department of Biochemistry SRMC & RI (DU) Porur, Chennai 116 Harper s biochemistry 24 th ed, Pg 218 Fatty acid Synthesis Known as
Fatty acid synthesis Dr. Nalini Ganesan M.Sc., Ph.D Associate Professor Department of Biochemistry SRMC & RI (DU) Porur, Chennai 116 Harper s biochemistry 24 th ed, Pg 218 Fatty acid Synthesis Known as
Oxidation of Long Chain Fatty Acids
 Oxidation of Long Chain Fatty Acids Dr NC Bird Oxidation of long chain fatty acids is the primary source of energy supply in man and animals. Hibernating animals utilise fat stores to maintain body heat,
Oxidation of Long Chain Fatty Acids Dr NC Bird Oxidation of long chain fatty acids is the primary source of energy supply in man and animals. Hibernating animals utilise fat stores to maintain body heat,
Chapter 14. Energy conversion: Energy & Behavior
 Chapter 14 Energy conversion: Energy & Behavior Why do you Eat and Breath? To generate ATP Foods, Oxygen, and Mitochodria Cells Obtain Energy by the Oxidation of Organic Molecules Food making ATP making
Chapter 14 Energy conversion: Energy & Behavior Why do you Eat and Breath? To generate ATP Foods, Oxygen, and Mitochodria Cells Obtain Energy by the Oxidation of Organic Molecules Food making ATP making
number Done by Corrected by Doctor Faisal Al- Khateeb
 number 21 Done by Omar Sami Corrected by حسام أبو عوض Doctor Faisal Al- Khateeb 1 P a g e (Only one or two marks are allocated for this sheetin the exam). Through this lecture we are going to cover the
number 21 Done by Omar Sami Corrected by حسام أبو عوض Doctor Faisal Al- Khateeb 1 P a g e (Only one or two marks are allocated for this sheetin the exam). Through this lecture we are going to cover the
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified
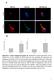 Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Generation and validation of mtef4-knockout mice.
 Supplementary Figure 1 Generation and validation of mtef4-knockout mice. (a) Alignment of EF4 (E. coli) with mouse, yeast and human EF4. (b) Domain structures of mouse mtef4 compared to those of EF4 (E.
Supplementary Figure 1 Generation and validation of mtef4-knockout mice. (a) Alignment of EF4 (E. coli) with mouse, yeast and human EF4. (b) Domain structures of mouse mtef4 compared to those of EF4 (E.
Rapid parallel measurements of macroautophagy and mitophagy in
 Supplemental Figures Rapid parallel measurements of macroautophagy and mitophagy in mammalian cells using a single fluorescent biosensor Sargsyan A, Cai J, Fandino LB, Labasky ME, Forostyan T, Colosimo
Supplemental Figures Rapid parallel measurements of macroautophagy and mitophagy in mammalian cells using a single fluorescent biosensor Sargsyan A, Cai J, Fandino LB, Labasky ME, Forostyan T, Colosimo
Supplemental Table 1. Cardiac mitochondrial acetyl proteoforms in mouse heart. Uniprot ID Gene Symbol Acetyl Proteoform Q8BWT1 Acaa2 K137 Q8BWT1
 Supplemental Table 1. Cardiac mitochondrial acetyl proteoforms in mouse heart. Uniprot ID Gene Symbol Acetyl Proteoform Q8BWT1 Acaa2 K137 Q8BWT1 Acaa2 K171 Q8BWT1 Acaa2 K234 Q8BWT1 Acaa2 K240 D3Z7X0 Acad12
Supplemental Table 1. Cardiac mitochondrial acetyl proteoforms in mouse heart. Uniprot ID Gene Symbol Acetyl Proteoform Q8BWT1 Acaa2 K137 Q8BWT1 Acaa2 K171 Q8BWT1 Acaa2 K234 Q8BWT1 Acaa2 K240 D3Z7X0 Acad12
GENERAL FEATURES OF FATTY ACIDS BIOSYNTHESIS
 1 GENERAL FEATURES OF FATTY ACIDS BIOSYNTHESIS 1. Fatty acids may be synthesized from dietary glucose via pyruvate. 2. Fatty acids are the preferred fuel source for the heart and the primary form in which
1 GENERAL FEATURES OF FATTY ACIDS BIOSYNTHESIS 1. Fatty acids may be synthesized from dietary glucose via pyruvate. 2. Fatty acids are the preferred fuel source for the heart and the primary form in which
Triose-P isomerase Enolase
 Select the single best answer. 1 onsider the catabolism of glucose to carbon dioxide and water. In this metabolic direction, which of these enzymes catalyzes a reaction where the PRUTS have one more "high-energy"
Select the single best answer. 1 onsider the catabolism of glucose to carbon dioxide and water. In this metabolic direction, which of these enzymes catalyzes a reaction where the PRUTS have one more "high-energy"
f(x) = x R² = RPKM (M8.MXB) f(x) = x E-014 R² = 1 RPKM (M31.
 14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
Synthesis of Fatty Acids and Triacylglycerol
 Synthesis of Fatty Acids and Triacylglycerol Lippincott s Chapter 16 Fatty Acid Synthesis Mainly in the Liver Requires Carbon Source: Acetyl CoA Reducing Power: NADPH 8 CH 3 COO C 15 H 33 COO Energy Input:
Synthesis of Fatty Acids and Triacylglycerol Lippincott s Chapter 16 Fatty Acid Synthesis Mainly in the Liver Requires Carbon Source: Acetyl CoA Reducing Power: NADPH 8 CH 3 COO C 15 H 33 COO Energy Input:
Biological processes. Mitochondrion Metabolic process Catalytic activity Oxidoreductase
 Full name Glyceraldehyde3 phosphate dehydrogenase Succinatesemialdehyde Glutamate dehydrogenase 1, Alcohol dehydrogenase [NADP+] 2',3'cyclicnucleotide 3' phosphodiesterase Dihydropyrimidinaserelated 2
Full name Glyceraldehyde3 phosphate dehydrogenase Succinatesemialdehyde Glutamate dehydrogenase 1, Alcohol dehydrogenase [NADP+] 2',3'cyclicnucleotide 3' phosphodiesterase Dihydropyrimidinaserelated 2
Electron Transport Chain and Oxidative phosphorylation
 Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
* * Days after darkness transition
 Protein (mg g -1 FW) 20 15 10 5 * * * * * * WT ivdh-1 d2hgdh1-2 etfqo-1 etfqo-2 35S:IVDH 35S:D2HGDH * * * * 0 0 3 7 10 Days after darkness transition Supplemental Figure 1. Protein content in Arabidopsis
Protein (mg g -1 FW) 20 15 10 5 * * * * * * WT ivdh-1 d2hgdh1-2 etfqo-1 etfqo-2 35S:IVDH 35S:D2HGDH * * * * 0 0 3 7 10 Days after darkness transition Supplemental Figure 1. Protein content in Arabidopsis
Proteomic Analysis of Glomeruli in Diabetes 1
 Proteomic Analysis of Glomeruli in Diabetes 1 Table 1: Identified Proteins from Mouse Glomerular Proteome Map spot# ; Regulation Functional swiss prot mass with group Protein entry (kda) pi Diabetes* Cytoskeletal
Proteomic Analysis of Glomeruli in Diabetes 1 Table 1: Identified Proteins from Mouse Glomerular Proteome Map spot# ; Regulation Functional swiss prot mass with group Protein entry (kda) pi Diabetes* Cytoskeletal
Fatty acid oxidation. doc. Ing. Zenóbia Chavková, CSc.
 Fatty acid oxidation doc. Ing. Zenóbia Chavková, CSc. Physiological functions of fatty acids 1. Structural components of cell membranes (phospholipids and sphingolipids) 2. Energy storage (triacylglycerols)
Fatty acid oxidation doc. Ing. Zenóbia Chavková, CSc. Physiological functions of fatty acids 1. Structural components of cell membranes (phospholipids and sphingolipids) 2. Energy storage (triacylglycerols)
BCH 4054 Spring 2001 Chapter 24 Lecture Notes
 BCH 4054 Spring 2001 Chapter 24 Lecture Notes 1 Chapter 24 Fatty Acid Catabolism 2 Fatty Acids as Energy Source Triglycerides yield 37 kj/g dry weight Protein 17 kj/g Glycogen 16 kj/g (even less wet weight)
BCH 4054 Spring 2001 Chapter 24 Lecture Notes 1 Chapter 24 Fatty Acid Catabolism 2 Fatty Acids as Energy Source Triglycerides yield 37 kj/g dry weight Protein 17 kj/g Glycogen 16 kj/g (even less wet weight)
OVERVIEW M ET AB OL IS M OF FR EE FA TT Y AC ID S
 LIPOLYSIS LIPOLYSIS OVERVIEW CATABOLISM OF FREE FATTY ACIDS Nonesterified fatty acids Source:- (a) breakdown of TAG in adipose tissue (b) action of Lipoprotein lipase on plasma TAG Combined with Albumin
LIPOLYSIS LIPOLYSIS OVERVIEW CATABOLISM OF FREE FATTY ACIDS Nonesterified fatty acids Source:- (a) breakdown of TAG in adipose tissue (b) action of Lipoprotein lipase on plasma TAG Combined with Albumin
Lipid Metabolism. Catabolism Overview
 Lipid Metabolism Pratt & Cornely, Chapter 17 Catabolism Overview Lipids as a fuel source from diet Beta oxidation Mechanism ATP production Ketone bodies as fuel 1 High energy More reduced Little water
Lipid Metabolism Pratt & Cornely, Chapter 17 Catabolism Overview Lipids as a fuel source from diet Beta oxidation Mechanism ATP production Ketone bodies as fuel 1 High energy More reduced Little water
nature methods Organelle-specific, rapid induction of molecular activities and membrane tethering
 nature methods Organelle-specific, rapid induction of molecular activities and membrane tethering Toru Komatsu, Igor Kukelyansky, J Michael McCaffery, Tasuku Ueno, Lidenys C Varela & Takanari Inoue Supplementary
nature methods Organelle-specific, rapid induction of molecular activities and membrane tethering Toru Komatsu, Igor Kukelyansky, J Michael McCaffery, Tasuku Ueno, Lidenys C Varela & Takanari Inoue Supplementary
Synthesis and elongation of fatty acids
 Synthesis and elongation of fatty acids A molecular caliper mechanism for determining very long-chain fatty acid length Vladimir Denic and Jonathan S. Weissman (2007) Cell 130, 663-677 February 28, 2008
Synthesis and elongation of fatty acids A molecular caliper mechanism for determining very long-chain fatty acid length Vladimir Denic and Jonathan S. Weissman (2007) Cell 130, 663-677 February 28, 2008
Supplemental Data. Article. Serotonin Regulates C. elegans Fat and Feeding. through Independent Molecular Mechanisms
 Cell Metabolism, Volume 7 Supplemental Data Article Serotonin Regulates C. elegans Fat and Feeding through Independent Molecular Mechanisms Supriya Srinivasan, Leila Sadegh, Ida C. Elle, Anne G.L. Christensen,
Cell Metabolism, Volume 7 Supplemental Data Article Serotonin Regulates C. elegans Fat and Feeding through Independent Molecular Mechanisms Supriya Srinivasan, Leila Sadegh, Ida C. Elle, Anne G.L. Christensen,
Total RNA was isolated and reverse transcribed with AffinityScript QPCR cdna Synthesis Kit
 SUPPLEMENTAL MATERIAL RNA analyses Total RNA was isolated and reverse transcribed with AffinityScript QPCR cdna Synthesis Kit (Agilent Technologies). PCR reactions were performed in triplicate in a 96-well
SUPPLEMENTAL MATERIAL RNA analyses Total RNA was isolated and reverse transcribed with AffinityScript QPCR cdna Synthesis Kit (Agilent Technologies). PCR reactions were performed in triplicate in a 96-well
Supplementary information
 Supplementary information Supplementary Figure 1: Components of Arabidopsis tricarboxylic acid (TCA) cycle. Schematic summary of the TCA cycle and the enzymes related to the reactions. The large text and
Supplementary information Supplementary Figure 1: Components of Arabidopsis tricarboxylic acid (TCA) cycle. Schematic summary of the TCA cycle and the enzymes related to the reactions. The large text and
6. How Are Fatty Acids Produced? 7. How Are Acylglycerols and Compound Lipids Produced? 8. How Is Cholesterol Produced?
 Lipid Metabolism Learning bjectives 1 How Are Lipids Involved in the Generationand Storage of Energy? 2 How Are Lipids Catabolized? 3 What Is the Energy Yield from the xidation of Fatty Acids? 4 How Are
Lipid Metabolism Learning bjectives 1 How Are Lipids Involved in the Generationand Storage of Energy? 2 How Are Lipids Catabolized? 3 What Is the Energy Yield from the xidation of Fatty Acids? 4 How Are
Biologic Oxidation BIOMEDICAL IMPORTAN
 Biologic Oxidation BIOMEDICAL IMPORTAN Chemically, oxidation is defined as the removal of electrons and reduction as the gain of electrons. Thus, oxidation is always accompanied by reduction of an electron
Biologic Oxidation BIOMEDICAL IMPORTAN Chemically, oxidation is defined as the removal of electrons and reduction as the gain of electrons. Thus, oxidation is always accompanied by reduction of an electron
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the
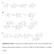 Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
FREE ENERGY Reactions involving free energy: 1. Exergonic 2. Endergonic
 BIOENERGETICS FREE ENERGY It is the portion of the total energy change in a system that is available for doing work at constant temperature and pressure; it is represented as ΔG. Reactions involving free
BIOENERGETICS FREE ENERGY It is the portion of the total energy change in a system that is available for doing work at constant temperature and pressure; it is represented as ΔG. Reactions involving free
Roles of Lipids. principal form of stored energy major constituents of cell membranes vitamins messengers intra and extracellular
 Roles of Lipids principal form of stored energy major constituents of cell membranes vitamins messengers intra and extracellular = Oxidation of fatty acids Central energy-yielding pathway in animals. O
Roles of Lipids principal form of stored energy major constituents of cell membranes vitamins messengers intra and extracellular = Oxidation of fatty acids Central energy-yielding pathway in animals. O
Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site.
 Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site. Still having trouble understanding the material? Check
Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site. Still having trouble understanding the material? Check
Part III => METABOLISM and ENERGY. 3.4 Lipid Catabolism 3.4a Fatty Acid Degradation 3.4b Ketone Bodies
 Part III => METABOLISM and ENERGY 3.4 Lipid Catabolism 3.4a Fatty Acid Degradation 3.4b Ketone Bodies Section 3.4a: Fatty Acid Degradation Synopsis 3.4a - Triglycerides (or fats) in the diet or adipose
Part III => METABOLISM and ENERGY 3.4 Lipid Catabolism 3.4a Fatty Acid Degradation 3.4b Ketone Bodies Section 3.4a: Fatty Acid Degradation Synopsis 3.4a - Triglycerides (or fats) in the diet or adipose
8 th NUGO week Wageningen. #nugo. Michael Müller Conclusions What is health?
 8 th NUGO week Wageningen #nugo Michael Müller Conclusions What is health? 1 1 You are what you eat What's healthy? 4 4 What is health? WHO 1946:..a state of complete physical, mental, and social well-being
8 th NUGO week Wageningen #nugo Michael Müller Conclusions What is health? 1 1 You are what you eat What's healthy? 4 4 What is health? WHO 1946:..a state of complete physical, mental, and social well-being
A role for Peroxisome Proliferator-Activated Receptor Beta in T cell development
 SUPPLEMENTARY INFORMATION A role for Peroxisome Proliferator-Activated Receptor Beta in T cell development Isabelle Mothe-Satney, Joseph Murdaca, Brigitte Sibille, Anne-Sophie Rousseau, Raphaëlle Squillace,
SUPPLEMENTARY INFORMATION A role for Peroxisome Proliferator-Activated Receptor Beta in T cell development Isabelle Mothe-Satney, Joseph Murdaca, Brigitte Sibille, Anne-Sophie Rousseau, Raphaëlle Squillace,
Chemistry 3503 Final exam April 17, Student s name:
 Chemistry 3503 Final exam April 17, 2008 Student s name: THIS EXAM IS FOR STUDENTS IN D. CRAIG S SECTION. IF YOU ARE IN M. EZE S SECTION THIS EXAM IS NOT FOR YOU. Part I /40 Part II Question 1 /4 Question
Chemistry 3503 Final exam April 17, 2008 Student s name: THIS EXAM IS FOR STUDENTS IN D. CRAIG S SECTION. IF YOU ARE IN M. EZE S SECTION THIS EXAM IS NOT FOR YOU. Part I /40 Part II Question 1 /4 Question
Role of mitochondrial beta-oxidation in ethanol response: A candidate gene study using Caenorhabditis elegans
 Virginia Commonwealth University VCU Scholars Compass Theses and Dissertations Graduate School 2017 Role of mitochondrial beta-oxidation in ethanol response: A candidate gene study using Caenorhabditis
Virginia Commonwealth University VCU Scholars Compass Theses and Dissertations Graduate School 2017 Role of mitochondrial beta-oxidation in ethanol response: A candidate gene study using Caenorhabditis
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
WT siz1-2 siz1-3. WT siz1-2 siz1-3. -Pi, 0.05 M IAA. -Pi, 2.5 M NPA
 A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
Biochemistry of cellular organelles
 Kontinkangas, L101A Biochemistry of cellular organelles Lectures: 1. Membrane channels; 2. Membrane transporters; 3. Soluble lipid/metabolite-transfer proteins; 4. Mitochondria as cellular organelles;
Kontinkangas, L101A Biochemistry of cellular organelles Lectures: 1. Membrane channels; 2. Membrane transporters; 3. Soluble lipid/metabolite-transfer proteins; 4. Mitochondria as cellular organelles;
Name Animal source Vendor Cat # Dilutions
 Supplementary data Table S1. Primary and Secondary antibody sources Devi et al, TXNIP in mitophagy A. Primary Antibodies Name Animal source Vendor Cat # Dilutions 1. TXNIP mouse MBL KO205-2 1:2000 (WB)
Supplementary data Table S1. Primary and Secondary antibody sources Devi et al, TXNIP in mitophagy A. Primary Antibodies Name Animal source Vendor Cat # Dilutions 1. TXNIP mouse MBL KO205-2 1:2000 (WB)
Ahmad Ulnar. Faisal Nimri ... Dr.Faisal
 24 Ahmad Ulnar Faisal Nimri... Dr.Faisal Fatty Acid Synthesis - Occurs mainly in the Liver (to store excess carbohydrates as triacylglycerols(fat)) and in lactating mammary glands (for the production of
24 Ahmad Ulnar Faisal Nimri... Dr.Faisal Fatty Acid Synthesis - Occurs mainly in the Liver (to store excess carbohydrates as triacylglycerols(fat)) and in lactating mammary glands (for the production of
ANSC/NUTR 618 Lipids & Lipid Metabolism
 I. Overall concepts A. Definitions ANC/NUTR 618 Lipids & Lipid Metabolism 1. De novo synthesis = synthesis from non-fatty acid precursors a. Carbohydrate precursors (glucose, lactate, and pyruvate) b.
I. Overall concepts A. Definitions ANC/NUTR 618 Lipids & Lipid Metabolism 1. De novo synthesis = synthesis from non-fatty acid precursors a. Carbohydrate precursors (glucose, lactate, and pyruvate) b.
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function
 Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Lecture 29: Membrane Transport and metabolism
 Chem*3560 Lecture 29: Membrane Transport and metabolism Insulin controls glucose uptake Adipose tissue and muscles contain a passive glucose transporter GluT4 which takes up glucose from blood. (This is
Chem*3560 Lecture 29: Membrane Transport and metabolism Insulin controls glucose uptake Adipose tissue and muscles contain a passive glucose transporter GluT4 which takes up glucose from blood. (This is
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Identification of peroxisomal targeting signals in cholesterol biosynthetic enzymes: AA-CoA thiolase, HMG-CoA synthase, MPPD, and FPP synthase
 Identification of peroxisomal targeting signals in cholesterol biosynthetic enzymes: AA-CoA thiolase, HMG-CoA synthase, MPPD, and FPP synthase Lisa M. Olivier, Werner Kovacs, Kim Masuda, Gilbert-Andre
Identification of peroxisomal targeting signals in cholesterol biosynthetic enzymes: AA-CoA thiolase, HMG-CoA synthase, MPPD, and FPP synthase Lisa M. Olivier, Werner Kovacs, Kim Masuda, Gilbert-Andre
Synthesis of Fatty Acids and Triacylglycerol
 Fatty Acid Synthesis Synthesis of Fatty Acids and Triacylglycerol Requires Carbon Source: Reducing Power: NADPH Energy Input: ATP Why Energy? Why Energy? Fatty Acid Fatty Acid + n(atp) ΔG o : -ve Fatty
Fatty Acid Synthesis Synthesis of Fatty Acids and Triacylglycerol Requires Carbon Source: Reducing Power: NADPH Energy Input: ATP Why Energy? Why Energy? Fatty Acid Fatty Acid + n(atp) ΔG o : -ve Fatty
FATTY ACID SYNTHESIS
 FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
Fatty Acid Degradation. Catabolism Overview. TAG and FA 11/11/2015. Chapter 27, Stryer Short Course. Lipids as a fuel source diet Beta oxidation
 Fatty Acid Degradation Chapter 27, Stryer Short Course Catabolism verview Lipids as a fuel source diet Beta oxidation saturated Unsaturated dd chain Ketone bodies as fuel Physiology High energy More reduced
Fatty Acid Degradation Chapter 27, Stryer Short Course Catabolism verview Lipids as a fuel source diet Beta oxidation saturated Unsaturated dd chain Ketone bodies as fuel Physiology High energy More reduced
incorporation into fructose-6-phosphate and glucose-6-phosphate (F6P/G6P); 2-
 Relative abundance (%) Relative abundance (%) Relative abundance (%) Relative abundance (%) 100% 80% 60% 40% F6P / G6P M+0 M+1 M+2 M+3 M+4 M+5 M+6 100% 80% 60% 40% 2PG / 3PG M+0 M+1 M+2 M+3 20% 20% 0%
Relative abundance (%) Relative abundance (%) Relative abundance (%) Relative abundance (%) 100% 80% 60% 40% F6P / G6P M+0 M+1 M+2 M+3 M+4 M+5 M+6 100% 80% 60% 40% 2PG / 3PG M+0 M+1 M+2 M+3 20% 20% 0%
number Done by Corrected by Doctor Faisal Al-Khatibe
 number 24 Done by Mohammed tarabieh Corrected by Doctor Faisal Al-Khatibe 1 P a g e *Please look over the previous sheet about fatty acid synthesis **Oxidation(degradation) of fatty acids, occurs in the
number 24 Done by Mohammed tarabieh Corrected by Doctor Faisal Al-Khatibe 1 P a g e *Please look over the previous sheet about fatty acid synthesis **Oxidation(degradation) of fatty acids, occurs in the
MITOCHONDRIA LECTURES OVERVIEW
 1 MITOCHONDRIA LECTURES OVERVIEW A. MITOCHONDRIA LECTURES OVERVIEW Mitochondrial Structure The arrangement of membranes: distinct inner and outer membranes, The location of ATPase, DNA and ribosomes The
1 MITOCHONDRIA LECTURES OVERVIEW A. MITOCHONDRIA LECTURES OVERVIEW Mitochondrial Structure The arrangement of membranes: distinct inner and outer membranes, The location of ATPase, DNA and ribosomes The
Electron transport chain, oxidative phosphorylation, mitochondrial transport systems
 Electron transport chain, oxidative phosphorylation, mitochondrial transport systems JAN ILLNER Respiratory chain & oxidative phosphorylation INTERMEMBRANE SPACE ubiquinone cytochrome c ATPase Production
Electron transport chain, oxidative phosphorylation, mitochondrial transport systems JAN ILLNER Respiratory chain & oxidative phosphorylation INTERMEMBRANE SPACE ubiquinone cytochrome c ATPase Production
SDR families. SDR10E Fatty acyl-coa reductase FACR1_HUMAN 544 x x SDR11E 3 beta-hydroxysteroid dehydrogenase 3BHS1_HUMAN 254 x x
 SDR1E UDP-glucose 4-epimerase GALE_HUMAN 5305 x x x x SDR2E dtdp-d-glucose 4,6-dehydratase TGDS_HUMAN 4194 x x x x SDR3E GDP-mannose 4,6 dehydratase GMDS_HUMAN 2314 x x x x SDR4E GDP-L-fucose synthetase
SDR1E UDP-glucose 4-epimerase GALE_HUMAN 5305 x x x x SDR2E dtdp-d-glucose 4,6-dehydratase TGDS_HUMAN 4194 x x x x SDR3E GDP-mannose 4,6 dehydratase GMDS_HUMAN 2314 x x x x SDR4E GDP-L-fucose synthetase
Lecture 16. Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III
 Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
7 Cellular Respiration and Fermentation
 CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 7 Cellular Respiration and Fermentation Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION
CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 7 Cellular Respiration and Fermentation Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION
Dietary Lipid Metabolism
 Dietary Lipid Metabolism Presented by Dr. Mohammad Saadeh The requirements for the Pharmaceutical Biochemistry II Philadelphia University Faculty of pharmacy OVERVIEW Lipids are a heterogeneous group.
Dietary Lipid Metabolism Presented by Dr. Mohammad Saadeh The requirements for the Pharmaceutical Biochemistry II Philadelphia University Faculty of pharmacy OVERVIEW Lipids are a heterogeneous group.
Citric acid cycle and respiratory chain. Pavla Balínová
 Citric acid cycle and respiratory chain Pavla Balínová Mitochondria Structure of mitochondria: Outer membrane Inner membrane (folded) Matrix space (mtdna, ribosomes, enzymes of CAC, β-oxidation of FA,
Citric acid cycle and respiratory chain Pavla Balínová Mitochondria Structure of mitochondria: Outer membrane Inner membrane (folded) Matrix space (mtdna, ribosomes, enzymes of CAC, β-oxidation of FA,
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
BCMB 3100 Fall 2013 Exam III
 BCMB 3100 Fall 2013 Exam III 1. (10 pts.) (a.) Briefly describe the purpose of the glycerol dehydrogenase phosphate shuttle. (b.) How many ATPs can be made when electrons enter the electron transport chain
BCMB 3100 Fall 2013 Exam III 1. (10 pts.) (a.) Briefly describe the purpose of the glycerol dehydrogenase phosphate shuttle. (b.) How many ATPs can be made when electrons enter the electron transport chain
III. Metabolism The Citric Acid Cycle
 Department of Chemistry and Biochemistry University of Lethbridge III. Metabolism The Citric Acid Cycle Slide 1 The Eight Steps of the Citric Acid Cycle Enzymes: 4 dehydrogenases (2 decarboxylation) 3
Department of Chemistry and Biochemistry University of Lethbridge III. Metabolism The Citric Acid Cycle Slide 1 The Eight Steps of the Citric Acid Cycle Enzymes: 4 dehydrogenases (2 decarboxylation) 3
Dynamic Interaction of Stress Granule, DDX3X and IKK-α Mediates Multiple Functions in
 Dynamic Interaction of Stress Granule, and Mediates Multiple Functions in Hepatitis C Virus Infection Véronique Pène, Qisheng Li#, Catherine Sodroski, Ching-Sheng Hsu, T. Jake Liang# Liver Diseases Branch,
Dynamic Interaction of Stress Granule, and Mediates Multiple Functions in Hepatitis C Virus Infection Véronique Pène, Qisheng Li#, Catherine Sodroski, Ching-Sheng Hsu, T. Jake Liang# Liver Diseases Branch,
Name Class Date. 1. Cellular respiration is the process by which the of "food"
 Name Class Date Cell Respiration Introduction Cellular respiration is the process by which the chemical energy of "food" molecules is released and partially captured in the form of ATP. Carbohydrates,
Name Class Date Cell Respiration Introduction Cellular respiration is the process by which the chemical energy of "food" molecules is released and partially captured in the form of ATP. Carbohydrates,
MITOCW watch?v=ddt1kusdoog
 MITOCW watch?v=ddt1kusdoog The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high-quality educational resources for free. To
MITOCW watch?v=ddt1kusdoog The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high-quality educational resources for free. To
ARTICLE IN PRESS. Peroxisomal β-oxidation A metabolic pathway with multiple functions
 BBAMCR-15482; No. of pages: 14; 4C: + model Biochimica et Biophysica Acta xx (2006) xxx xxx www.elsevier.com/locate/bbamcr Review Peroxisomal β-oxidation A metabolic pathway with multiple functions Yves
BBAMCR-15482; No. of pages: 14; 4C: + model Biochimica et Biophysica Acta xx (2006) xxx xxx www.elsevier.com/locate/bbamcr Review Peroxisomal β-oxidation A metabolic pathway with multiple functions Yves
2-more complex molecules (fatty acyl esters) as triacylglycerols.
 ** Fatty acids exist in two forms:- 1-free fatty acids (unesterified) 2-more complex molecules (fatty acyl esters) as triacylglycerols. ** most tissues might use fatty acids as source of energy during
** Fatty acids exist in two forms:- 1-free fatty acids (unesterified) 2-more complex molecules (fatty acyl esters) as triacylglycerols. ** most tissues might use fatty acids as source of energy during
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies
 Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Oxidative Phosphorylation
 Electron Transport Chain (overview) The NADH and FADH 2, formed during glycolysis, β- oxidation and the TCA cycle, give up their electrons to reduce molecular O 2 to H 2 O. Electron transfer occurs through
Electron Transport Chain (overview) The NADH and FADH 2, formed during glycolysis, β- oxidation and the TCA cycle, give up their electrons to reduce molecular O 2 to H 2 O. Electron transfer occurs through
7 Pathways That Harvest Chemical Energy
 7 Pathways That Harvest Chemical Energy Pathways That Harvest Chemical Energy How Does Glucose Oxidation Release Chemical Energy? What Are the Aerobic Pathways of Glucose Metabolism? How Is Energy Harvested
7 Pathways That Harvest Chemical Energy Pathways That Harvest Chemical Energy How Does Glucose Oxidation Release Chemical Energy? What Are the Aerobic Pathways of Glucose Metabolism? How Is Energy Harvested
Lipid Metabolism. Remember fats?? Triacylglycerols - major form of energy storage in animals
 Remember fats?? Triacylglycerols - major form of energy storage in animals Your energy reserves: ~0.5% carbs (glycogen + glucose) ~15% protein (muscle, last resort) ~85% fat Why use fat for energy? 1 gram
Remember fats?? Triacylglycerols - major form of energy storage in animals Your energy reserves: ~0.5% carbs (glycogen + glucose) ~15% protein (muscle, last resort) ~85% fat Why use fat for energy? 1 gram
Fatty acids synthesis
 Fatty acids synthesis The synthesis start from Acetyl COA the first step requires ATP + reducing power NADPH! even though the oxidation and synthesis are different pathways but from chemical part of view
Fatty acids synthesis The synthesis start from Acetyl COA the first step requires ATP + reducing power NADPH! even though the oxidation and synthesis are different pathways but from chemical part of view
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS
 Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
