Coordinated control of bile acids and lipogenesis through FXR-dependent regulation of fatty acid synthase 1
|
|
|
- Erica Randall
- 5 years ago
- Views:
Transcription
1 Coordinated control of bile acids and lipogenesis through FXR-dependent regulation of fatty acid synthase 1 Karen E. Matsukuma,* Mary K. Bennett,* Jiansheng Huang, Li Wang, Gregorio Gil, and Timothy F. Osborne 2, * Department of Molecular Biology and Biochemistry,* University of California, Irvine, CA; Departments of Medicine and Pharmacology, University of Kansas Medical Center, Kansas City, KS; and Department of Biochemistry, Medical College of Virginia, Virginia Commonwealth University, Richmond, VA Abstract We discovered a nuclear receptor element in the FAS promoter consisting of an inverted repeat spaced by one nucleotide (IR-1) and located 21 bases downstream of a direct repeat sequenced by 4 nucleotides (DR-4) oxysterol liver X receptor response element. An IR-1 is present in promoters of several genes of bile acid and lipid homeostasis and binds farnesoid X receptor/retinoid X receptor (FXR/ RXR) heterodimers to mediate bile acid-dependent transcription. We show that FXR/RXRa specifically binds to the FAS IR-1 and that the FAS promoter is activated z10-fold by the addition of a synthetic FXR agonist in transient transfection assays. We also demonstrate that endogenous FXR binds directly to the murine FAS promoter in the hepatic genome using a tissue-based chromatin immunoprecipitation procedure. Furthermore, we show that feeding wild-type mice a chow diet supplemented with the natural FXR agonist chenodeoxycholic acid results in a significant induction of FAS mrna expression. Thus, we have identified a novel IR-1 in the FAS promoter and demonstrate that it mediates FXR/bile acid regulation of the FAS gene. These findings provide the first evidence for direct regulation of lipogenesis by bile acids and also provide a mechanistic rationale for previously unexplained observations regarding bile acid control of FAS expression. Matsukuma, K.E.,M.K.Bennett,J.Huang,L.Wang,G.Gil,andT.F. Osborne. Coordinated control of bile acids and lipogenesis through FXR-dependent regulation of fatty acid synthase. J. Lipid Res : Supplementary key words farnesoid X receptor & nuclear receptors & small heterodimer partner In the intestine, bile acids serve to emulsify lipids from the diet, thereby facilitating efficient absorption of fatty acids, sterols, and fat-soluble vitamins. However, increased bile acid levels are cytotoxic (1, 2), so their levels must be tightly regulated. One of the key transcription factors responsible for the regulation of bile acid homeostasis is the nuclear receptor farnesoid X receptor (FXR) (3). Physiologic concentrations of select bile acids, such as chenodeoxycholic acid (CDCA) and cholic acid, are endogenous agonists for FXR (4 6). Activation of the FXR/retinoid X receptor (RXR) heterodimer by bile acids stimulates the synthesis of bile acid-regulatory proteins, including I-BABP (7), BSEP (8), and OATP1B3 (9), that modulate the flux of bile acids in the liver and intestine. Additionally, indirect regulation of the bile acid biosynthetic enzymes cytochrome P (CYP)7A1 and CYP8B1 is accomplished through FXR induction of the negative acting small heterodimer partner (SHP) protein in the liver, which binds to and represses the positive acting liver receptor homologue (LRH)- 1 nuclear receptor in the promoters for these genes (10 12). Another twist in the mechanism for bile acid-dependent repression of CYP7A1 was recently identified that involves the activation of FGF15 expression in the small intestine by bile acids and FGFR4 expression in the liver (13). Here, intestinal FGF15 is proposed to act through ligand-dependent stimulation of FGFR4 signaling in the liver. A second lipid-sensing nuclear receptor, liver X receptor (LXR), binds oxysterols and plays a key role in cholesterol homeostasis (14 18). As cholesterol is the direct precursor for bile acid biosynthesis, a complex interplay between the actions of LXR and FXR must exist to meet the metabolic needs of the organism. Serum cholesterol levels are in turn dependent on fatty acids as they are the organic substrate for cholesterol esterification, which is important for cholesterol storage. Thus, bile acids, cholesterol, and fatty acids are intimately tied to one another in the overall maintenance of lipid homeostasis. Several studies have also established that bile acids and FXR play important roles in both triglyceride and glucose metabolism. The triglyceride-lowering effect of bile acids was first described.20 years ago when clinicians ob- Manuscript received 26 July 2006 and in revised form 5 September Published, JLR Papers in Press, September 6, DOI /jlr.M JLR Journal of Lipid Research Volume 47, This paper is dedicated to the memory of Professor Edward K. Wagner, who died during the preparation of the manuscript. 2 To whom correspondence should be addressed. tim.osborne@uci.edu Copyright D 2006 by the American Society for Biochemistry and Molecular Biology, Inc. This article is available online at
2 served that bile acid therapy for the treatment of gallstone disease had the added effect of decreasing serum triglycerides (19 21). More recently, FXR induction of its target gene SHP was shown to correlate with decreased expression of the master lipogenic regulator sterolregulatory element binding protein (SREBP)-1c (as well as many of its gene targets), thus providing at least a partial mechanistic explanation for the triglyceride-lowering effect of bile acids (22). However, SHP modulation of SREBP-1c levels is unlikely to be the sole mechanism regulating the lipogenic response to bile acids because the repression of many of SREBP-1c s downstream targets (including FAS) is transient despite ongoing expression of SHP (22). Because FAS occupies a pivotal position for lipogenic flux, its activity is highly regulated. Significant regulation occurs at the transcriptional level, where major metabolic signals such as insulin and carbohydrates (23 26), thyroid hormone (27), fatty acids (28), and sterols (29) have all been shown to influence gene transcription. The promoter for the FAS gene contains a complex nuclear receptor response region at z2700. This region contains a classic direct repeat sequenced by 4 nucleotides (DR-4) nuclear receptor site that mediates the FAS response to thyroid hormone as well as LXR activators (27, 30). In this study, we report the identification of an inverted repeat spaced by one nucleotide (IR-1) located 21 bases downstream of the FAS DR-4 and demonstrate that it mediates bile acid regulation of the FAS gene. We thus establish FAS as an FXR target gene and provide the first example of the direct regulation of lipogenesis by bile acids. MATERIALS AND METHODS Cell culture HEK293T cells were obtained from the American Type Culture Collection, cultured in Dulbecco s Minimum Essential Medium (Irvine Scientific) containing penicillin/streptomycin (100 mg/ml), L-glutamine (10 mm), nonessential amino acids, (100 mm), 5 mm HEPES, ph 7.2, and 10% fetal bovine serum, and maintained at 378C and 5% CO 2. One day before transfection, cells were plated at a density of cells/well on a six-well plate in 3 ml of normal culture medium. Transient DNA transfections Twelve to 24 h after plating, cells were transfected by the standard calcium phosphate method as described (31). Cytomegalovirus X promoter (CMX)-rat FXRa (30 ng), CMX-human RXRa (100 ng), luciferase reporter (4 mg), CMV-b-galactosidase (2 mg), and salmon sperm DNA (to a final mass of 12 mg) were transfected per well as noted in the figure legends. Six to 8 h after transfection, cells were rinsed two times with sterile PBS. Medium was replaced with Defined Serum-Free Medium [Dulbecco s Minimum Essential Medium, 100 mg/ml penicillin/streptomycin, 2 mm L-glutamine, 100 mm nonessential amino acids, 5 mm HEPES, ph 7.2, insulin/transferrin/selenite (5 mg/ml; 5 mg/ml; 5 ng/ml; Sigma), 4% BSA (A-3803; Sigma), and 0.1 mg/ml 25- hydroxycholesterol] plus either DMSO (0.1%) as a vehicle control or the synthetic FXR agonist GW4064 (final concentration of 1 mm). (25-Hydroxycholesterol was added to suppress endogenous SREBP expression and thus minimize basal transcriptional activity.) Cells were allowed to incubate at 378C and5%co 2 for an additional h before harvesting. For each experiment, duplicate wells were plated for each condition. Transfection experiments were performed at least twice with similar results. Plasmids The FAS 2700/165 pgl2 luciferase reporter construct was subcloned by PCR from the rat FAS 21594/165 pgl2 described previously (29). All mutant reporter constructs were generated using the Quikchange site-directed mutagenesis kit (Stratagene). The remaining reporter constructs were generated by PCR amplification and subcloning. psyntataluc is a reporter vector containing the minimal promoter region of the hamster HMG- CoA synthase promoter (228/139) and has been described previously (32). The mouse SREBP-1c (2937/129) promoter reporter (33), the rat sterol 12-a hydroxylase promoter reporter (34), and the rat acetyl-coa carboxylase promoter reporter were described previously (35). The following expression vectors were generously provided by other laboratories: CMX-human thyroid receptor (TR)b (Barry Forman, City of Hope), CMXhuman RXRa (Ron Evans, Salk Institute), CMX-rat FXR (Bruce Blumberg, University of California, Irvine), and CMX-mouse LRH-1 (David Mangelsdorf, University of Texas Southwestern). Enzyme assays At the time of harvest, cells were rinsed once with PBS and then lysed in a reporter lysis buffer (25 mm Gly-Gly, 15 mm MgSO 4, 4 mm EGTA, and 0.25% Triton). Luciferase activity of the lysates was measured in an Analytical Luminescence Monolight 2010 luminometer using 5 20 ml of cell extract plus 100 ml of luciferase assay reagent (Promega), with data expressed in relative light units (RLUs). b-galactosidase activity was measured by a standard colorimetric assay at 420 nm absorbance using ml of cell lysate and 2-nitrophenyl b-galactopyranoside as the substrate. Luciferase activity for each sample was divided by the b-galactosidase activity to yield normalized RLUs. Fold activation was determined by dividing the normalized RLUs for a given sample by the normalized RLUs for the control sample (no activators plus vehicle). Each transfection was performed at least twice with similar results. Electrophoretic mobility shift assay In vitro transcribed and translated TRb (human), LXRa (human), FXR (rat), and RXRa (human) proteins were generated using the T7 TnT Rabbit Reticulocyte Lysate (Promega). Five microliters of each translation was added to each binding mixture [containing 10 mm HEPES, ph 7.6, 50 mm NaCl, 2.5 mm MgCl 2, 0.1 mg/ml poly di:dc, 0.05% Nonidet P-40 (v/v), and 10% glycerol] in a final volume of 20 ml. A double-stranded oligonucleotide DNA probe containing the wild-type sequence of IR-1 was 59 end-labeled using T 4 polynucleotide kinase (USB) and added to the binding mixtures at 1 ng per reaction. Binding mixtures were incubated at 48C for 1 2 h. Samples were then run on 5% polyacrylamide:bis-acrylamide (19:1) gels at room temperature for 1.5 h, fixed in a solution of 10% methanol/10% acetic acid, and dried onto 3MM chromatography paper at 808C for1h. Dried gels were exposed to X-ray film at 2208C for h. Chromatin immunoprecipitation B6/129 mice (6 week old males) were purchased from Taconic, allowed to adapt for 2 weeks to a 12 h light/12 h dark FXR regulation of fatty acid synthase gene 2755
3 cycle, and euthanized at the end of the dark cycle (8 AM). Livers from four mice were placed in 40 ml of ice-cold PBS containing a cocktail of protease inhibitors (1 mg/ml leupeptin, 1.4 mg/ml pepstatin, and 2 mg/ml PMSF) plus 1 mm EDTA and 1 mm EGTA. The tissue was disrupted in a Tekmar Tissumizer at the lowest setting. Formaldehyde was added from a 37% stock (v/v) to a final concentration of 1%, and samples were rotated on a shaker for 6 min followed by the addition of glycine to a final concentration of M. Samples were returned to the shaker for an additional 5 min, and then cells were collected by centrifugation (2,000 rpm in a Sorvall RC3B at 48C). The cell pellet was washed once with homogenization buffer A (10 mm HEPES, ph 7.6, 25 mm KCl, 1 mm EDTA, 1 mm EGTA, 2 M sucrose, 10% glycerol, and 0.15 mm spermine) plus protease inhibitors as described above. The final pellet was resuspended in buffer A and homogenized in a Dounce homogenizer with a B pestle to release nuclei. The solution was layered over buffer A and centrifuged in a Beckman ultracentrifuge (1 h at 26,000 rpm, 48C), and the nuclear pellet was resuspended in nuclei lysis buffer (1% SDS, 50 mm Tris, ph 7.6, and 10 mm EDTA). Nuclei were disrupted using an Ultrasonic model W-220F sonicator for s to shear chromatin. Chromatin size was checked by agarose electrophoresis to ensure that the average size was between 200 and 500 bp. Aliquots were used in immunoprecipitation experiments with an antibody to FXR (Santa Cruz H130X) and processed as described (36). Final DNA samples were analyzed by quantitative PCR in triplicate with a standard dilution curve of the input DNA performed in parallel. Oligonucleotide pairs for the FAS FXR binding region or exon 4 from the YY1 gene used in the quantitative PCR are as follows: FAS-700 (59) ATCCTGGTCTCCAAGGTG; FAS-534 (39) TAGGCAATAGGGT- GATGGG; YY1 (59) TCTGACGAGAGGATTGTGTGGAC; YY1 (39) CTGAAGGGCTTTTCTCCAGTATG. Animal feeding studies C57/BL6 mice (8 week old males) were maintained in a 12 h light/12 h dark controlled environment with free access to water and standard laboratory rodent chow. At the start of the feeding experiment, each group of mice (n 5 4) was fed either chow or chow mixed with 0.25% CDCA (w/w) for 6 days. Animals were euthanized and tissues harvested 6 h into the dark cycle. Tissues were homogenized and extracted for RNA using Trizol reagent (Invitrogen). For each liver sample, 8 mg of total RNA was separated on a 1% formaldehyde gel and transferred to a charged nylon membrane. FAS, L32, and SHP mrna levels were detected by Northern blotting using a- 32 P-labeled DNA probes. Densitometry was performed using Bio-Rad Quantity One software. FAS expression was normalized to L32 expression, and fold activation by CDCA feeding was determined by setting the normalized FAS mrna expression of the control fed animals to 1. The feeding study was performed twice with similar results. RESULTS We previously defined a DR-4 element in the rat FAS promoter as an LXR responsive site (30). In the course of these studies, we discovered an additional nuclear receptor half-site located near the DR-4 that was also indirectly required for LXR signaling through binding the monomeric nuclear receptor LRH-1 (K. E. Matsukuma et al., unpublished data). A closer inspection of the sequence surrounding this additional half-site suggested that it comprised half of a potential IR-1 nuclear receptor element (Fig. 1A). LXR/RXR heterodimers do not characteristi- Fig. 1. A: Alignment of the FAS promoter sequences from human, rat, and chicken. Nuclear receptor halfsites are shown in uppercase letters and denoted by arrows. The positions of the direct repeat sequenced by 4 nucleotides (DR-4) and inverted repeat spaced by one nucleotide (IR-1) elements are noted, and the liver receptor homologue (LRH)-1 binding site is denoted by a gray underline. The bases mutated in key electrophoretic mobility shift assay and reporter plasmids (IR-1 59 or 39) are also noted. B: Electrophoretic mobility shift assay demonstrating that the farnesoid X receptor/retinoid X receptor (FXR/RXR) heterodimer binds FAS IR-1. In vitro translated RXRa (R), thyroid receptor (TR)b (T), or FXR (F) were incubated alone or in combination as indicated with a 32 P-labeled sequence containing the FAS IR-1 [wild type (WT)] and analyzed by electrophoretic mobility shift assay. Fifty-fold excess cold competitor DNAs corresponding to the wild-type or mutated sequences (IR-1 59 or 39) were added along with the probe as noted at the top of the gel. C: FXR/RXR binding to human (hu) FAS IR-1 by electrophoretic mobility shift assay. In vitro translated RXRa (R), FXR (F), or both (F/R) were incubated with a 32 P-labeled sequence containing to either rat or human FAS IR-1 as indicated. Unlabeled competitor DNAs (50-fold molar excess) corresponding to the rat or human FAS probes or a mutated competitor (IR-1 59) were added along with the probe as noted at the top of the gel Journal of Lipid Research Volume 47, 2006
4 cally bind to this half-site configuration; however, it is a known high-affinity site for the FXR/RXR heterodimer (37). This site is conserved in the mouse promoter as well. To determine whether FXR/RXR heterodimers could bind this putative IR-1, we tested the binding of in vitro translated FXR and RXRa on a 32 P-labeled probe containing the sequence of the rat FAS IR-1 (Fig. 1B). We also tested the binding of TRb and LXRa (data not shown) to the IR-1. Individually, none of the nuclear receptors (including FXR) bound the DNA probe. In addition, when combined with RXRa, neither TRb (lanes 7 11) nor LXRa (data not shown) interacted efficiently with the DNA. In contrast, incubation of FXR and RXRa proteins with the radiolabeled probe resulted in a strong shifted band (lane 3). The binding was effectively competed off by 50-fold excess cold wild-type competitor DNA (lane 4) but not by cold competitor DNA containing minimal mutations in either half of the putative IR-1 (lanes 5, 6). Thus, the rat FAS IR-1 specifically binds FXR/RXRa and not TRb/ RXRa or LXRa/RXRa. Because the human FAS promoter diverges by one base from the rat promoter in the region of the putative IR-1 sequence, we investigated whether the site from the human FAS promoter could also bind the FXR/RXRa heterodimer (Fig. 1C). The FXR/RXRa heterodimer bound to the human FAS element (lane 9) at a level comparable to that seen with the rat sequence (lane 3). In addition, competition of FXR/RXRa binding to the rat FAS IR-1 element by 50-fold excess cold competitor DNA containing the human FAS sequence was equivalent to that seen with competitor DNA containing the rat FAS sequence (lanes 4, 5). To determine whether FXR/RXRa could activate FAS through the IR-1 site, we transfected HEK293T cells with expression vectors for FXR and RXRa and measured the activation of a FAS promoter construct containing the IR-1 (Fig. 2). We chose HEK293T cells because they do not show significant endogenous LRH-1 activity (K. E. Matsukuma et al., unpublished data). This is important because the LRH-1 binding site overlaps the FXR binding site in the FAS promoter, thus complicating a straightforward study of promoter activation by FXR. Cell lines derived from hepatocytes would be expected to express significant amounts of endogenous LRH-1 and therefore would not be appropriate model systems. When the synthetic FXR agonist GW4064 was added along with 10, 30, or 100 ng of the FXR expression vector, FAS activation increased significantly in a dose-dependent manner, and addition of the RXRa expression vector amplified this effect. In contrast, a point mutation in the putative IR-1 (FAS IR-1 59) extinguished the positive FXR response. Because the DR-4 that is required for the LXR activation of FAS is in close proximity to the IR-1 and because half of the IR-1 is an LRH-1 binding site essential for signaling through LXR, we analyzed the potential role of the DR-4 in FXR signaling (Fig. 3). In contrast to LXR activation of FAS, mutations in the DR-4 had little impact on activation by FXR. However, point mutations in either of the half-sites of the IR-1 significantly reduced the FXR response. Thus, FXR-dependent activation of FAS is specifically mediated through the IR-1 element. In other studies, we have shown that LXR activation of FAS is inhibited by SHP (K. E. Matsukuma et al., unpublished data); therefore, we investigated whether FXR activation of FAS was also affected by SHP expression in a cotransfection assay. Unlike LXR activation, however, FXR activation of FAS is independent of SHP expression (Fig. 4). It should be noted that in companion dishes from this same experiment, SHP addition did inhibit LXR activation of FAS (data not shown). To determine whether other LRH-1 activated genes might also contain dual-function binding elements for LRH-1 and FXR/RXR, we tested the effect of FXR activation on SREBP-1c and CYP8B1, two other genes known to be activated by LRH-1 overexpression (22, 34) (Fig. 5). In addition, the effect of FXR activation was tested on the promoter for acetyl-coa carboxylase, a lipogenic gene closely linked to FAS in the pathway of basic fatty acid biosynthesis (that has not been investigated for LRH-1 dependence). In this experiment, FAS was the lone promoter stimulated efficiently by FXR. Thus, FXR activation is neither a common feature of all LRH-1 gene targets nor a property shared by all lipogenic genes. To determine whether bile acid-dependent FAS activation could be recapitulated in an animal model, we fed C57/BL6 wild-type mice either a normal chow diet or a normal chow diet supplemented with 0.25% CDCA, a bile acid and potent activator of FXR, for 6 days and compared Fig. 2. FXR/RXR activates FAS through IR-1. HEK293T cells were transiently transfected with increasing amounts (10, 30, and 100 ng each) of cytomegalovirus X promoter (CMX)-RXRa (R) or CMX-FXR (F) and FAS 2700/165 wild type (WT) or FAS 2700/165 IR-1 59 mutant reporter construct. Cells were treated with or without the synthetic FXR agonist GW4064 (1 mm) for 16 h and harvested for measurement of luciferase and b-galactosidase activity as described in Materials and Methods. Error bars represent standard deviation. FXR regulation of fatty acid synthase gene 2757
5 Fig. 3. Both IR-1 half-sites contribute to the FAS FXR response. HEK293T cells were transiently transfected with 100 ng of CMX- RXRa and 30 ng of CMX-FXR and one of four FAS reporter constructs as indicated. Cells were then treated with the synthetic FXR agonist GW4064 (1 mm) for 16 h and harvested for measurement of luciferase and b-galactosidase activity as described in Materials and Methods. Error bars represent standard deviation. Fig. 5. FXR/RXR activation is specific to the FAS promoter. HEK293T cells were transiently transfected with 100 ng of CMX- RXRa, 100 ng of CMX-FXR, and reporter plasmids as indicated. Cells were then treated with the synthetic FXR agonist GW4064 (1 mm) or DMSO for 16 h and harvested for measurement of luciferase and b-galactosidase activities as described in Materials and Methods. ACC PII, promoter for acetyl-coa carboxylase; SREBP, sterol-regulatory element binding protein. Error bars represent standard deviation. hepatic FAS gene expression (Fig. 6A, B). FAS mrna increased z2-fold in livers of animals fed the 0.25% CDCA diet with respect to the control fed animals. In this same experiment, the expression of SHP, a known FXR target gene, increased by z3-fold. Thus, bile acids induce FAS expression in vivo, and induction of FAS occurs at a level comparable to that of other FXR target genes. Unfortunately, feeding bile acids to FXR-deficient mice is highly toxic, and the animals become cachetic and begin to die after a few days of bile acid feeding (38). Thus, a knockout mouse model was not available for further verification of the role of FXR in FAS activation. Fig. 4. LRH-1 interferes with FXR/RXR activation of FAS; SHP does not. HEK293T cells were transiently transfected with 100 ng of CMX-RXRa, 30 ng of CMX-FXR, 100 ng of CMX-SHP, or combinations as noted. The reporter plasmid was the FAS promoter construct with mutations in both halves of the DR-4 (DR-4M). Transfected cells were then treated with the synthetic FXR agonist GW4064 (1 mm) for 16 h and harvested for measurement of luciferase and b-galactosidase as described in Materials and Methods. Error bars represent standard deviation. Because the bile acid feeding study by itself did not provide direct evidence for FXR regulation of FAS expression in animals, we performed chromatin immunoprecipitation studies on livers of wild-type mice expressing endogenous levels of FXR to determine whether FXR is directly associated with the endogenous FAS promoter. Chromatin was analyzed for FXR binding to the FAS promoter using an antibody to FXR and PCR primers specific for the relevant region of the FAS promoter or a genomic region from the mouse YY1 gene as a negative control (Fig. 7). The FAS promoter DNA was enriched.100-fold relative to an equivalent sample in which a control IgG fraction was used. In addition, FXR binding to the negative control YY1 locus was negligible (Fig. 7). Thus, endogenous FXR is specifically recruited to the native FAS promoter. DISCUSSION Although FXR was originally identified as a key regulator of cholesterol and bile acid homeostasis (38), it is now understood to also play an important role in fatty acid metabolism. The first evidence that bile acids might be involved in fatty acid metabolism derived from the incidental observation.20 years ago that administration of bile acids decreased serum triglycerides in patients with gallstone disease (19 21). An FXR-dependent mechanism for this effect was proposed by Watanabe et al. (22), who observed that SREBP-1c levels decreased z50% but SHP levels increased by 300% after 1 day of treatment of wildtype or prediabetic (KK-A y ) mice with 0.5% cholic acid. In in vitro experiments, these authors demonstrated that LXR activation of the SREBP-1c promoter was significantly inhibited by the expression of SHP or the addition of bile acids. However, in this same study, levels of several key lipogenic targets of SREBP-1c did not correlate with SREBP-1c levels when the feeding study was extended to 2758 Journal of Lipid Research Volume 47, 2006
6 Fig. 7. FXR binds FAS promoter in hepatic chromatin. Endogenous hepatic FXR binding to the FAS promoter was analyzed by a chromatin immunoprecipitation (ChIP) experiment as described in Materials and Methods. The level of DNA precipitated by the FXR or control IgG antibody is presented for the FAS promoter or exon 4 from the YY1 gene as a control. The amount of DNA precipitated was calculated using a serial dilution curve of the input DNA from native chromatin that was quantified by optical density at 260 nm. Error bars represent standard deviation. Fig. 6. Six day chenodeoxycholic acid (CDCA) feeding (0.25%) increases FAS mrna in livers of wild-type mice. Wild-type mice were fed ad libitum either a normal chow diet (control) or a normal chow diet supplemented with 0.25% CDCA for 6 days. RNA was extracted from livers of mice and individually analyzed by Northern blotting. A: Autoradiogram of Northern blot probed for FAS and L32 or SHP. Each lane was loaded with 20 mg of RNA extracted from an individual animal in each group. B: Densitometric analysis of the data in A with the ratios of FAS/L32 and SHP/L32 represented in the graphs. Error bars represent standard deviation. 7 days. In particular, FAS mrna decreased by z50% after 1 day of cholic acid feeding but returned to control levels at 1 week, despite ongoing SREBP-1c repression and continued SHP elevation. Thus, only the transient inhibitory effect of bile acids is consistent with SHP inhibition of SREBP-1c-dependent lipogenesis, so additional mechanisms for the bile acid regulation of lipogenesis under more long-term bile acid-feeding conditions must exist. Therefore, we designed our feeding study to reflect this more chronic increase of bile acids. In this study, we identified an IR-1 element in the FAS promoter that mediates the bile acid activation of the FAS gene. Although neither FAS nor other lipogenic gene targets have been identified as a result of FXR microarray experiments, analysis of these microarray data sets reveals that whereas some well-established FXR target genes (such as SHP) are activated by the synthetic ligand GW4064, they are not significantly activated by CDCA, or vice versa (39, 40). Other studies have reported opposing effects of a single FXR ligand on different known FXR target genes (41) and differential patterns for the recruitment of coactivators by the various endogenous bile acid agonists (42). These findings suggest that an exhaustive list of FXR gene targets may not be achieved simply by use of broad microarray analyses. Physiologically speaking, it seems likely that the discrepancies in FXR target gene activation observed in these experiments underlie complex regulatory mechanisms in which the specific ligand and the unique architecture of the nuclear receptor response regions in each promoter influence the final gene output response. Demonstration of a direct activating role for FXR on the FAS promoter was unexpected given the wellestablished triglyceride-lowering effect of bile acids. However, an important role for fatty acid synthesis may be revealed when cholesterol levels are chronically increased. High cellular cholesterol levels increase oxysterols and therefore LXR signaling, leading to the upregulation of fatty acid synthase (30). Increased fatty acid production facilitates cholesterol storage by increasing the availability of the cosubstrate for cholesterol esterification. In the rodent, increased LXR activity resulting from the presence of oxysterols also results in the upregulation of CYP7A1 and the conversion of the excess cholesterol to bile acids (14, 15). If this process is allowed to continue, however, bile acids eventually accumulate to levels that activate the endogenous bile acid receptor FXR and result in the activation of FXR target genes such as SHP (10, 11). Because SHP negatively regulates both CYP7A1 (11) and FAS (22) (K. E. Matsukuma et al., unpublished data), activation of SHP would both limit the flow of cholesterol into the bile acid synthetic pathway and simultaneously decrease fatty acid synthesis, resulting in a shift in the balance of cholesterol and fatty acids to a state of cholesterol excess. This would limit the ability of the liver to FXR regulation of fatty acid synthase gene 2759
7 store excess cholesterol as cholesteryl ester and result in cholesterol toxicity (43). The direct mechanism for the bile acid activation of FAS presented in this study provides a plausible means by which FAS may circumvent SHP inhibition to maintain an adequate fatty acid pool for cholesterol esterification when both cholesterol and bile acid levels are chronically high. Finally, it is interesting that the FXR/RXR binding site in the FAS promoter overlaps an LRH-1 binding site that is required for efficient LXR activation (K. E. Matsukuma et al., unpublished data). Although the significance of this binding site arrangement is not known, overlapping nuclear receptor binding sites have been found in many other promoters, including CYP7A1, CYP8B1, and apolipoprotein C-III (12, 44, 45). In the promoters of CYP7A1 and CYP8B1, binding sites for the nuclear receptors HNF- 4a and FTF overlap. Using the technique of chromatin immunoprecipitation, investigators found that in the absence of bile acids, HNF4a preferentially bound to the overlapping site in the CYP7A1 promoter. In the presence of bile acids, however, only FTF bound this site. Given that each nuclear receptor has a unique transcriptional activation potential, the differential binding of these two nuclear receptors may function to ensure that the precise amount of gene product accumulates for optimal physiologic responses. In this way, overlapping binding sites may be an effective mechanism by which dynamic physiologic responses are achieved. The authors thank Drs. Bruce Blumberg, Ronald Evans, Barry Forman, Peter Tontonoz, and David Mangelsdorf for plasmid reagents and Timothy Willson for the GW4064. The authors gratefully acknowledge support from the National Institutes of Health to T.F.O. (Grants HL and DK-71021), G.G. (Grant DK-38030), and L.W. (Grant P20 RR from the IdeA Network of Biomedical Research Excellence (INBRE) Program of the National Center for Research Resources). L.W. also was supported in part through a grant from Kansas Masonic Cancer Research. K.E.M. was supported by fellowships from the Achievement Rewards for College Scientists Foundation and the American Diabetes Association and is a member of the Medical Scientist Training Program in the University of California Irvine School of Medicine. REFERENCES 1. Schmucker, D. L., M. Ohta, S. Kanai, Y. Sato, and K. Kitani Hepatic injury induced by bile salts: correlation between biochemical and morphological events. Hepatology. 12: Spivey, J. R., S. F. Bronk, and G. J. Gores Glycochenodeoxycholate-induced lethal hepatocellular injury in rat hepatocytes. Role of ATP depletion and cytosolic free calcium. J. Clin. Invest. 92: Forman, B. M., E. Goode, J. Chen, A. E. Oro, D. J. Bradley, T. Perlmann, D. J. Noonan, L. T. Burka, T. McMorris, W. W. Lamph, et al Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 81: Makishima, M., A. Y. Okamoto, J. J. Repa, H. Tu, R. M. Learned, A. Luk, M. V. Hull, K. D. Lustig, D. J. Mangelsdorf, and B. Shan Identification of a nuclear receptor for bile acids. Science. 284: Parks, D. J., S. G. Blanchard, R. K. Bledsoe, G. Chandra, T. G. Consler, S. A. Kliewer, J. B. Stimmel, T. M. Willson, A. M. Zavacki, D. D. Moore, et al Bile acids: natural ligands for an orphan nuclear receptor. Science. 284: Wang, H., J. Chen, K. Hollister, L. C. Sowers, and B. M. Forman Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 3: Grober, J., I. Zaghini, H. Fujii, S. A. Jones, S. A. Kliewer, T. M. Willson, T. Ono, and P. Besnard Identification of a bile acidresponsive element in the human ileal bile acid-binding protein gene. Involvement of the farnesoid X receptor/9-cis-retinoic acid receptor heterodimer. J. Biol. Chem. 274: Ananthanarayanan, M., N. Balasubramanian, M. Makishima, D. J. Mangelsdorf, and F. J. Suchy Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J. Biol. Chem. 276: Jung, D., M. Podvinec, U. A. Meyer, D. J. Mangelsdorf, M. Fried, P. J. Meier, and G. A. Kullak-Ublick Human organic anion transporting polypeptide 8 promoter is transactivated by the farnesoid X receptor/bile acid receptor. Gastroenterology. 122: Lu, T. T., M. Makishima, J. J. Repa, K. Schoonjans, T. A. Kerr, J. Auwerx, and D. J. Mangelsdorf Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 6: Goodwin, B., S. A. Jones, R. R. Price, M. A. Watson, D. D. McKee, L. B. Moore, C. Galardi, J. G. Wilson, M. C. Lewis, M. E. Roth, et al A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 6: del Castillo-Olivares, A., and G. Gil Suppression of sterol 12alpha-hydroxylase transcription by the short heterodimer partner: insights into the repression mechanism. Nucleic Acids Res. 29: Inagaki, T., M. Choi, A. Moschetta, L. Peng, C. L. Cummins, J. G. McDonald, G. Luo, S. A. Jones, B. Goodwin, J. A. Richardson, et al Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2: Lehmann, J. M., S. A. Kliewer, L. B. Moore, T. A. Smith-Oliver, B. B. Oliver, D. A. Winegar, D. E. Blanchard, T. A. Spencer, and T. M. Willson Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J. Biol. Chem. 272: Peet, D. J., S. D. Turley, W. Ma, B. A. Janowski, J-M. A. Lobaccard, R. E. Hammer, and D. J. Mangelsdorf Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXRa. Cell. 93: Repa, J. J., S. D. Turley, J. A. Lobaccaro, J. Medina, L. Li, K. Lustig, B. Shan, R. A. Heyman, J. M. Dietschy, and D. J. Mangelsdorf Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science. 289: Laffitte, B. A., J. J. Repa, S. B. Joseph, D. C. Wilpitz, H. R. Kast, D. J. Mangelsdorf, and P. Tontonoz LXRs control lipidinducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc. Natl. Acad. Sci. USA. 98: Luo, Y., and A. R. Tall Sterol upregulation of human CETP expression in vitro and in transgenic mice by an LXR element. J. Clin. Invest. 105: Angelin, B., K. Einarsson, K. Hellstrom, and B. Leijd Effects of cholestyramine and chenodeoxycholic acid on the metabolism of endogenous triglyceride in hyperlipoproteinemia. J. Lipid Res. 19: Bateson, M. C., M. D. Evans, Jr., and I. A. Bouchier Chenodeoxycholic acid therapy for hypertriglyceridaemia in men. Br. J. Clin. Pharmacol. 5: Bell,G.,B.Lewis,A.Petrie,andR.Dowling.1973.Serumlipidsincholelithiasis: effect of chenodeoxycholic acid therapy. BMJ. 3: Watanabe, M., S. M. Houten, L. Wang, A. Moschetta, D. J. Mangelsdorf, R. A. Heyman, D. D. Moore, and J. Auwerx Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J. Clin. Invest. 113: Moustaid, N., K. Sakamoto, S. Clarke, R. S. Beyer, and H. S. Sul Regulation of fatty acid synthase gene transcription. Biochem. J. 292: Moustaid, N., R. S. Beyer, and H. S. Sul Identification of an insulin response element in the fatty acid synthase promoter. J. Biol. Chem. 269: Wolf, S. S., G. Hofer, K. F. Beck, K. Roder, and M. Schweizer Insulin-responsive regions of the rat fatty acid synthase gene promoter. Biochem. Biophys. Res. Commun. 203: Journal of Lipid Research Volume 47, 2006
8 26. Ishii, S., K. Iizuka, B. C. Miller, and K. Uyeda Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc. Natl. Acad. Sci. USA. 101: Xiong, S., S. S. Chirala, M. H. Hsu, and S. J. Wakil Identification of thyroid hormone response elements in the human fatty acid synthase promoter. Proc. Natl. Acad. Sci. USA. 95: Clarke, S. D., M. K. Armstrong, and D. B. Jump Dietary polyunsaturated fats uniquely suppress rat liver fatty acid synthase and S14 mrna content. J. Nutr. 120: Bennett, M. K., J. M. Lopez, H. B. Sanchez, and T. F. Osborne Sterol regulation of fatty acid synthase promoter. J. Biol. Chem. 270: Joseph, S. B., B. A. Laffitte, P. H. Patel, M. A. Watson, K. E. Matsukuma, R. Walczak, J. L. Collins, T. F. Osborne, and P. Tontonoz Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J. Biol. Chem. 277: Yieh, L., H. B. Sanchez, and T. F. Osborne Domains of transcription factor Sp1 required for synergistic activation with sterol regulatory element binding protein 1 of low density lipoprotein receptor promoter. Proc. Natl. Acad. Sci. USA. 92: Sanchez, H. B., L. Yieh, and T. F. Osborne Cooperation by SREBP and Sp1 in sterol regulation of low density lipoprotein receptor gene. J. Biol. Chem. 270: Zhang, C., D. J. Shin, and T. F. Osborne A simple promoter containing two Sp1 sites controls the expression of sterol-regulatoryelement-binding protein 1a (SREBP-1a). Biochem. J. 386: del Castillo-Olivares, A., and G. Gil Alpha 1-fetoprotein transcription factor is required for the expression of sterol 12alpha -hydroxylase, the specific enzyme for cholic acid synthesis. Potential role in the bile acid-mediated regulation of gene transcription. J. Biol. Chem. 275: Lopez, J. M., M. K. Bennett, H. B. Sanchez, J. M. Rosenfeld, and T. F. Osborne Sterol regulation of acetyl CoA carboxylase: a mechanism for coordinate control of cellular lipid. Proc. Natl. Acad. Sci. USA. 93: Bennett, M., J. I. Toth, and T. F. Osborne Selective association of sterol regulatory element binding protein isoforms with target promoters in vivo. J. Biol. Chem. 279: Laffitte, B. A., H. R. Kast, C. M. Nguyen, A. M. Zavacki, D. D. Moore, and P. A. Edwards Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J. Biol. Chem. 275: Sinal, C. J., M. Tohkin, M. Miyata, J. M. Ward, G. Lambert, and F. J. Gonzalez Targeted disruption of the nuclear receptor FXR/ BAR impairs bile acid and lipid homeostasis. Cell. 102: Pircher, P. C., J. L. Kitto, M. L. Petrowski, R. K. Tangirala, E. D. Bischoff, I. G. Schulman, and S. K. Westin Farnesoid X receptor regulates bile acid-amino acid conjugation. J. Biol. Chem. 278: Downes, M., M. A. Verdecia, A. J. Roecker, R. Hughes, J. B. Hogenesch, H. R. Kast-Woelbern, M. E. Bowman, J. L. Ferrer, A. M. Anisfeld, P. A. Edwards, et al A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol. Cell. 11: Dussault, I., R. Beard, M. Lin, K. Hollister, J. Chen, J. H. Xiao, R. Chandraratna, and B. M. Forman Identification of geneselective modulators of the bile acid receptor FXR. J. Biol. Chem. 278: Lew, J. L., A. Zhao, J. Yu, L. Huang, N. De Pedro, F. Pelaez, S. D. Wright, and J. Cui The farnesoid X receptor controls gene expression in a ligand- and promoter-selective fashion. J. Biol. Chem. 279: Warner, G. J., G. Stoudt, M. Bamberger, W. J. Johnson, and G. H. Rothblat Cell toxicity induced by inhibition of acyl coenzyme A:cholesterol acyltransferase and accumulation of unesterified cholesterol. J. Biol. Chem. 270: Stroup, D., and J. Y. L. Chiang HNF4 and COUP-TFII interact to modulate transcription of the cholesterol 7{alpha}- hydroxylase gene (CYP7A1). J. Lipid Res. 41: Claudel, T., Y. Inoue, O. Barbier, D. Duran-Sandoval, V. Kosykh, J. Fruchart, J. C. Fruchart, F. J. Gonzalez, and B. Staels Farnesoid X receptor agonists suppress hepatic apolipoprotein CIII expression. Gastroenterology. 125: FXR regulation of fatty acid synthase gene 2761
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
Identification of PLTP as an LXR target gene and apoe as an FXR target gene reveals overlapping targets for the two nuclear receptors
 rapid communication Identification of PLTP as an LXR target gene and apoe as an FXR target gene reveals overlapping targets for the two nuclear receptors Puiying A. Mak,* Heidi R. Kast-Woelbern,* Andrew
rapid communication Identification of PLTP as an LXR target gene and apoe as an FXR target gene reveals overlapping targets for the two nuclear receptors Puiying A. Mak,* Heidi R. Kast-Woelbern,* Andrew
Peroxisome proliferator-activated receptor- coactivator 1 (PGC-1 ) regulates triglyceride metabolism by activation of the nuclear receptor FXR
 Peroxisome proliferator-activated receptor- coactivator 1 (PGC-1 ) regulates triglyceride metabolism by activation of the nuclear receptor FXR Yanqiao Zhang, 1,2 Lawrence W. Castellani, 2 Christopher J.
Peroxisome proliferator-activated receptor- coactivator 1 (PGC-1 ) regulates triglyceride metabolism by activation of the nuclear receptor FXR Yanqiao Zhang, 1,2 Lawrence W. Castellani, 2 Christopher J.
FXR agonists and FGF15 reduce fecal bile acid excretion in a mouse model of bile acid malabsorption
 FXR agonists and FGF15 reduce fecal bile acid excretion in a mouse model of bile acid malabsorption Diana Jung,* Takeshi Inagaki,*, Robert D. Gerard,, Paul A. Dawson,** Steven A. Kliewer,*, David J. Mangelsdorf,
FXR agonists and FGF15 reduce fecal bile acid excretion in a mouse model of bile acid malabsorption Diana Jung,* Takeshi Inagaki,*, Robert D. Gerard,, Paul A. Dawson,** Steven A. Kliewer,*, David J. Mangelsdorf,
Regulating Hepatic Cellular Cholesterol
 Under circumstances of cholesterol deficiency, Sterol Regulatory Element Binding Proteins (SREBPs) via binding to DNA nuclear response elements set off genomic production of proteins and enzymes that induce
Under circumstances of cholesterol deficiency, Sterol Regulatory Element Binding Proteins (SREBPs) via binding to DNA nuclear response elements set off genomic production of proteins and enzymes that induce
Protocol for Gene Transfection & Western Blotting
 The schedule and the manual of basic techniques for cell culture Advanced Protocol for Gene Transfection & Western Blotting Schedule Day 1 26/07/2008 Transfection Day 3 28/07/2008 Cell lysis Immunoprecipitation
The schedule and the manual of basic techniques for cell culture Advanced Protocol for Gene Transfection & Western Blotting Schedule Day 1 26/07/2008 Transfection Day 3 28/07/2008 Cell lysis Immunoprecipitation
FXR regulates organic solute transporters a and b in the adrenal gland, kidney, and intestine
 FXR regulates organic solute transporters a and b in the adrenal gland, kidney, and intestine Hans Lee,* Yanqiao Zhang,* Florence Y. Lee,* Stanley F. Nelson, Frank J. Gonzalez, and Peter A. Edwards, 1,
FXR regulates organic solute transporters a and b in the adrenal gland, kidney, and intestine Hans Lee,* Yanqiao Zhang,* Florence Y. Lee,* Stanley F. Nelson, Frank J. Gonzalez, and Peter A. Edwards, 1,
2.5. AMPK activity
 Supplement Fig. A 3 B phos-ampk 2.5 * Control AICAR AMPK AMPK activity (Absorbance at 45 nm) 2.5.5 Control AICAR Supplement Fig. Effects of AICAR on AMPK activation in macrophages. J774. macrophages were
Supplement Fig. A 3 B phos-ampk 2.5 * Control AICAR AMPK AMPK activity (Absorbance at 45 nm) 2.5.5 Control AICAR Supplement Fig. Effects of AICAR on AMPK activation in macrophages. J774. macrophages were
SREBP-1 integrates the actions of thyroid hormone, insulin, camp, and medium-chain fatty acids on ACC transcription in hepatocytes
 SREBP-1 integrates the actions of thyroid hormone, insulin, camp, and medium-chain fatty acids on ACC transcription in hepatocytes Yanqiao Zhang, Liya Yin, and F. Bradley Hillgartner 1 Department of Biochemistry
SREBP-1 integrates the actions of thyroid hormone, insulin, camp, and medium-chain fatty acids on ACC transcription in hepatocytes Yanqiao Zhang, Liya Yin, and F. Bradley Hillgartner 1 Department of Biochemistry
The Schedule and the Manual of Basic Techniques for Cell Culture
 The Schedule and the Manual of Basic Techniques for Cell Culture 1 Materials Calcium Phosphate Transfection Kit: Invitrogen Cat.No.K2780-01 Falcon tube (Cat No.35-2054:12 x 75 mm, 5 ml tube) Cell: 293
The Schedule and the Manual of Basic Techniques for Cell Culture 1 Materials Calcium Phosphate Transfection Kit: Invitrogen Cat.No.K2780-01 Falcon tube (Cat No.35-2054:12 x 75 mm, 5 ml tube) Cell: 293
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism SUPPLEMENTARY FIGURES, LEGENDS AND METHODS
 A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
Lecithin Cholesterol Acyltransferase (LCAT) ELISA Kit
 Product Manual Lecithin Cholesterol Acyltransferase (LCAT) ELISA Kit Catalog Number STA-616 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Cholesterol is a lipid sterol
Product Manual Lecithin Cholesterol Acyltransferase (LCAT) ELISA Kit Catalog Number STA-616 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Cholesterol is a lipid sterol
Supporting Information Table of content
 Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Activation of the nuclear receptor FXR induces fibrinogen expression: a new role for bile acid signaling
 Activation of the nuclear receptor FXR induces fibrinogen expression: a new role for bile acid signaling Andrew M. Anisfeld,* Heidi R. Kast-Woelbern,* Hans Lee,* Yanqiao Zhang,* Florence Y. Lee,* and Peter
Activation of the nuclear receptor FXR induces fibrinogen expression: a new role for bile acid signaling Andrew M. Anisfeld,* Heidi R. Kast-Woelbern,* Hans Lee,* Yanqiao Zhang,* Florence Y. Lee,* and Peter
FXR-mediated down-regulation of CYP7A1 dominates LXR in long-term cholesterol-fed NZW rabbits
 FXR-mediated down-regulation of CYP7A1 dominates LXR in long-term cholesterol-fed NZW rabbits Guorong Xu, 1, *, Hai Li, Lu-xing Pan, Quan Shang, Akira Honda, M. Ananthanarayanan,** Sandra K. Erickson,
FXR-mediated down-regulation of CYP7A1 dominates LXR in long-term cholesterol-fed NZW rabbits Guorong Xu, 1, *, Hai Li, Lu-xing Pan, Quan Shang, Akira Honda, M. Ananthanarayanan,** Sandra K. Erickson,
Chromatin Immunoprecipitation (ChIPs) Protocol (Mirmira Lab)
 Chromatin Immunoprecipitation (ChIPs) Protocol (Mirmira Lab) Updated 12/3/02 Reagents: ChIP sonication Buffer (1% Triton X-100, 0.1% Deoxycholate, 50 mm Tris 8.1, 150 mm NaCl, 5 mm EDTA): 10 ml 10 % Triton
Chromatin Immunoprecipitation (ChIPs) Protocol (Mirmira Lab) Updated 12/3/02 Reagents: ChIP sonication Buffer (1% Triton X-100, 0.1% Deoxycholate, 50 mm Tris 8.1, 150 mm NaCl, 5 mm EDTA): 10 ml 10 % Triton
Supplementary data Supplementary Figure 1 Supplementary Figure 2
 Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue
 Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue Bryan A. Laffitte*, Lily C. Chao, Jing Li, Robert Walczak*, Sarah
Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue Bryan A. Laffitte*, Lily C. Chao, Jing Li, Robert Walczak*, Sarah
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
/06/$15.00/0 Molecular Endocrinology 20(9): Copyright 2006 by The Endocrine Society doi: /me
 0888-8809/06/$15.00/0 Molecular Endocrinology 20(9):2062 2079 Printed in U.S.A. Copyright 2006 by The Endocrine Society doi: 10.1210/me.2005-0316 Androgens, Progestins, and Glucocorticoids Induce Follicle-Stimulating
0888-8809/06/$15.00/0 Molecular Endocrinology 20(9):2062 2079 Printed in U.S.A. Copyright 2006 by The Endocrine Society doi: 10.1210/me.2005-0316 Androgens, Progestins, and Glucocorticoids Induce Follicle-Stimulating
Chenodeoxycholic acid suppresses the activation of acetyl-coenzyme A carboxylase-a gene transcription by the liver X receptor agonist T
 Chenodeoxycholic acid suppresses the activation of acetyl-coenzyme A carboxylase-a gene transcription by the liver X receptor agonist T0-901317 Saswata Talukdar, Sushant Bhatnagar, Sami Dridi, and F. Bradley
Chenodeoxycholic acid suppresses the activation of acetyl-coenzyme A carboxylase-a gene transcription by the liver X receptor agonist T0-901317 Saswata Talukdar, Sushant Bhatnagar, Sami Dridi, and F. Bradley
Decreased serum triglyceride levels have long been. Farnesoid X Receptor Agonists Suppress Hepatic Apolipoprotein CIII Expression
 GASTROENTEROLOGY 2003;125:544 555 Farnesoid X Receptor Agonists Suppress Hepatic Apolipoprotein CIII Expression THIERRY CLAUDEL,*, YUSUKE INOUE, OLIVIER BARBIER,*, DANIEL DURAN-SANDOVAL,*, VLADIMIR KOSYKH,
GASTROENTEROLOGY 2003;125:544 555 Farnesoid X Receptor Agonists Suppress Hepatic Apolipoprotein CIII Expression THIERRY CLAUDEL,*, YUSUKE INOUE, OLIVIER BARBIER,*, DANIEL DURAN-SANDOVAL,*, VLADIMIR KOSYKH,
TFEB-mediated increase in peripheral lysosomes regulates. Store Operated Calcium Entry
 TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
FGF15/19 protein levels in the portal blood do not reflect changes in the ileal FGF15/19 or hepatic CYP7A1 mrna levels
 FGF15/19 protein levels in the portal blood do not reflect changes in the ileal FGF15/19 or hepatic CYP7A1 mrna levels Quan Shang, *, Grace L. Guo, Akira Honda, ** Monica Saumoy, Gerald Salen, * and Guorong
FGF15/19 protein levels in the portal blood do not reflect changes in the ileal FGF15/19 or hepatic CYP7A1 mrna levels Quan Shang, *, Grace L. Guo, Akira Honda, ** Monica Saumoy, Gerald Salen, * and Guorong
Ligand-Dependent Coactivation of the Human Bile Acid Receptor FXR by the Peroxisome Proliferator-Activated Receptor Coactivator-1
 0022-3565/05/3121-170 178$20.00 THE JOURNAL OF PHARMACOLOGY AND EXPERIMENTAL THERAPEUTICS Vol. 312, No. 1 Copyright 2005 by The American Society for Pharmacology and Experimental Therapeutics 72124/1182236
0022-3565/05/3121-170 178$20.00 THE JOURNAL OF PHARMACOLOGY AND EXPERIMENTAL THERAPEUTICS Vol. 312, No. 1 Copyright 2005 by The American Society for Pharmacology and Experimental Therapeutics 72124/1182236
RayBio Human PPAR-alpha Transcription Factor Activity Assay Kit
 RayBio Human PPAR-alpha Transcription Factor Activity Assay Kit Catalog #: TFEH-PPARa User Manual Jan 5, 2018 3607 Parkway Lane, Suite 200 Norcross, GA 30092 Tel: 1-888-494-8555 (Toll Free) or 770-729-2992,
RayBio Human PPAR-alpha Transcription Factor Activity Assay Kit Catalog #: TFEH-PPARa User Manual Jan 5, 2018 3607 Parkway Lane, Suite 200 Norcross, GA 30092 Tel: 1-888-494-8555 (Toll Free) or 770-729-2992,
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Requires Signaling though Akt2 Independent of the. Transcription Factors FoxA2, FoxO1, and SREBP1c
 Cell Metabolism, Volume 14 Supplemental Information Postprandial Hepatic Lipid Metabolism Requires Signaling though Akt2 Independent of the Transcription Factors FoxA2, FoxO1, and SREBP1c Min Wan, Karla
Cell Metabolism, Volume 14 Supplemental Information Postprandial Hepatic Lipid Metabolism Requires Signaling though Akt2 Independent of the Transcription Factors FoxA2, FoxO1, and SREBP1c Min Wan, Karla
Total Histone H3 Acetylation Detection Fast Kit (Colorimetric)
 Total Histone H3 Acetylation Detection Fast Kit (Colorimetric) Catalog Number KA1538 48 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use...
Total Histone H3 Acetylation Detection Fast Kit (Colorimetric) Catalog Number KA1538 48 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use...
Western Immunoblotting Preparation of Samples:
 Western Immunoblotting Preparation of Samples: Total Protein Extraction from Culture Cells: Take off the medium Wash culture with 1 x PBS 1 ml hot Cell-lysis Solution into T75 flask Scrap out the cells
Western Immunoblotting Preparation of Samples: Total Protein Extraction from Culture Cells: Take off the medium Wash culture with 1 x PBS 1 ml hot Cell-lysis Solution into T75 flask Scrap out the cells
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
General Laboratory methods Plasma analysis: Gene Expression Analysis: Immunoblot analysis: Immunohistochemistry:
 General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
Human Leptin ELISA Kit
 Product Manual Human Leptin ELISA Kit Catalog Numbers MET-5057 MET-5057-5 96 assays 5 x 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Leptin is a polypeptide hormone
Product Manual Human Leptin ELISA Kit Catalog Numbers MET-5057 MET-5057-5 96 assays 5 x 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Leptin is a polypeptide hormone
Chromatin IP (Isw2) Fix soln: 11% formaldehyde, 0.1 M NaCl, 1 mm EDTA, 50 mm Hepes-KOH ph 7.6. Freshly prepared. Do not store in glass bottles.
 Chromatin IP (Isw2) 7/01 Toshi last update: 06/15 Reagents Fix soln: 11% formaldehyde, 0.1 M NaCl, 1 mm EDTA, 50 mm Hepes-KOH ph 7.6. Freshly prepared. Do not store in glass bottles. 2.5 M glycine. TBS:
Chromatin IP (Isw2) 7/01 Toshi last update: 06/15 Reagents Fix soln: 11% formaldehyde, 0.1 M NaCl, 1 mm EDTA, 50 mm Hepes-KOH ph 7.6. Freshly prepared. Do not store in glass bottles. 2.5 M glycine. TBS:
Mammalian Membrane Protein Extraction Kit
 Mammalian Membrane Protein Extraction Kit Catalog number: AR0155 Boster s Mammalian Membrane Protein Extraction Kit is a simple, rapid and reproducible method to prepare cellular protein fractions highly
Mammalian Membrane Protein Extraction Kit Catalog number: AR0155 Boster s Mammalian Membrane Protein Extraction Kit is a simple, rapid and reproducible method to prepare cellular protein fractions highly
Human LDL Receptor / LDLR ELISA Pair Set
 Human LDL Receptor / LDLR ELISA Pair Set Catalog Number : SEK10231 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized in
Human LDL Receptor / LDLR ELISA Pair Set Catalog Number : SEK10231 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized in
Total Phosphatidic Acid Assay Kit
 Product Manual Total Phosphatidic Acid Assay Kit Catalog Number MET- 5019 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Phosphatidic Acid (PA) is a critical precursor
Product Manual Total Phosphatidic Acid Assay Kit Catalog Number MET- 5019 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Phosphatidic Acid (PA) is a critical precursor
SREBF2 Transcription Factor Assay Kit ( Cat # KA1379 V.01 ) 1
 Background Lipid homeostasis in vertebrate cells is regulated by a family of transcription factors called sterol regulatory element binding proteins (SREBP s). SREBP s directly activate the expression
Background Lipid homeostasis in vertebrate cells is regulated by a family of transcription factors called sterol regulatory element binding proteins (SREBP s). SREBP s directly activate the expression
Bile acid receptor FXR: metabolic regulator in the gut. Sungsoon Fang
 Bile acid receptor FXR: metabolic regulator in the gut Sungsoon Fang Nuclear hormone receptor 1905: Ernest Starling coined hormone 1929: Estrogen structure 1958: Estrogen receptor by Elwood Jensen 1985:
Bile acid receptor FXR: metabolic regulator in the gut Sungsoon Fang Nuclear hormone receptor 1905: Ernest Starling coined hormone 1929: Estrogen structure 1958: Estrogen receptor by Elwood Jensen 1985:
Antibodies: LB1 buffer For 50 ml For 10ml For 30 ml Final 1 M HEPES, ph 2.5 ml 0.5 ml 1.5 ml 50mM. 5 M NaCl 1.4 ml 280 µl 0.
 Experiment: Date: Tissue: Purpose: ChIP-Seq Antibodies: 11x cross-link buffer: Regent Stock Solution Final Vol for 10 ml of 11xstock concentration 5 M NaCl 0.1M 0.2 ml 0.5 M EDTA 1 mm 20 ul 0.5 M EGTA,
Experiment: Date: Tissue: Purpose: ChIP-Seq Antibodies: 11x cross-link buffer: Regent Stock Solution Final Vol for 10 ml of 11xstock concentration 5 M NaCl 0.1M 0.2 ml 0.5 M EDTA 1 mm 20 ul 0.5 M EGTA,
MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands)
 Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
PREPARATION OF IF- ENRICHED CYTOSKELETAL PROTEINS
 TMM,5-2011 PREPARATION OF IF- ENRICHED CYTOSKELETAL PROTEINS Ice-cold means cooled in ice water. In order to prevent proteolysis, make sure to perform all steps on ice. Pre-cool glass homogenizers, buffers
TMM,5-2011 PREPARATION OF IF- ENRICHED CYTOSKELETAL PROTEINS Ice-cold means cooled in ice water. In order to prevent proteolysis, make sure to perform all steps on ice. Pre-cool glass homogenizers, buffers
EpiQuik Total Histone H3 Acetylation Detection Fast Kit (Colorimetric)
 EpiQuik Total Histone H3 Acetylation Detection Fast Kit (Colorimetric) Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Total Histone H3 Acetylation Detection Fast Kit (Colorimetric)
EpiQuik Total Histone H3 Acetylation Detection Fast Kit (Colorimetric) Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Total Histone H3 Acetylation Detection Fast Kit (Colorimetric)
Farnesoid X receptor (FXR)-mediated down-regulation of CYP7A1 dominates LXRα in
 JLR Papers In Press. Published on August 1, 2003 as Manuscript M300182-JLR200 Farnesoid X receptor (FXR)-mediated down-regulation of CYP7A1 dominates LXRα in long term cholesterol fed NZW rabbits Guorong
JLR Papers In Press. Published on August 1, 2003 as Manuscript M300182-JLR200 Farnesoid X receptor (FXR)-mediated down-regulation of CYP7A1 dominates LXRα in long term cholesterol fed NZW rabbits Guorong
Farnesoid X receptor represses hepatic human APOA gene expression
 Research article Farnesoid X receptor represses hepatic human APOA gene expression Indumathi Chennamsetty, 1 Thierry Claudel, 2 Karam M. Kostner, 3 Anna Baghdasaryan, 2 Dagmar Kratky, 1 Sanja Levak-Frank,
Research article Farnesoid X receptor represses hepatic human APOA gene expression Indumathi Chennamsetty, 1 Thierry Claudel, 2 Karam M. Kostner, 3 Anna Baghdasaryan, 2 Dagmar Kratky, 1 Sanja Levak-Frank,
In vitro DNase I foot printing. In vitro DNase I footprinting was performed as described
 Supplemental Methods In vitro DNase I foot printing. In vitro DNase I footprinting was performed as described previously 1 2 using 32P-labeled 211 bp fragment from 3 HS1. Footprinting reaction mixes contained
Supplemental Methods In vitro DNase I foot printing. In vitro DNase I footprinting was performed as described previously 1 2 using 32P-labeled 211 bp fragment from 3 HS1. Footprinting reaction mixes contained
HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation
 SUPPLEMENTARY INFORMATION Materials and Methods Human cell lines and culture conditions HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation in exon 20 of BRCA1
SUPPLEMENTARY INFORMATION Materials and Methods Human cell lines and culture conditions HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation in exon 20 of BRCA1
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Bile Acid Receptor Farnesoid X Receptor: A Novel Therapeutic Target for Metabolic Diseases
 Review J Lipid Atheroscler 2017 June;6(1):1-7 https://doi.org/10.12997/jla.2017.6.1.1 pissn 2287-2892 eissn 2288-2561 JLA Bile Acid Receptor Farnesoid X Receptor: A Novel Therapeutic Target for Metabolic
Review J Lipid Atheroscler 2017 June;6(1):1-7 https://doi.org/10.12997/jla.2017.6.1.1 pissn 2287-2892 eissn 2288-2561 JLA Bile Acid Receptor Farnesoid X Receptor: A Novel Therapeutic Target for Metabolic
Serum Amyloid A3 Gene Expression in Adipocytes is an Indicator. of the Interaction with Macrophages
 Serum Amyloid A3 Gene Expression in Adipocytes is an Indicator of the Interaction with Macrophages Yohei Sanada, Takafumi Yamamoto, Rika Satake, Akiko Yamashita, Sumire Kanai, Norihisa Kato, Fons AJ van
Serum Amyloid A3 Gene Expression in Adipocytes is an Indicator of the Interaction with Macrophages Yohei Sanada, Takafumi Yamamoto, Rika Satake, Akiko Yamashita, Sumire Kanai, Norihisa Kato, Fons AJ van
2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein
![2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein](/thumbs/86/94743397.jpg) Supplementary Information 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked amino-modification products by acrolein Ayumi Tsutsui and Katsunori Tanaka* Biofunctional Synthetic Chemistry Laboratory, RIKEN
Supplementary Information 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked amino-modification products by acrolein Ayumi Tsutsui and Katsunori Tanaka* Biofunctional Synthetic Chemistry Laboratory, RIKEN
Molecular Basis for Feedback Regulation of Bile Acid Synthesis by Nuclear Receptors
 Molecular Cell, Vol. 6, 507 515, September, 2000, Copyright 2000 by Cell Press Molecular Basis for Feedback Regulation of Bile Acid Synthesis by Nuclear Receptors Timothy T. Lu, Makoto Makishima,* Joyce
Molecular Cell, Vol. 6, 507 515, September, 2000, Copyright 2000 by Cell Press Molecular Basis for Feedback Regulation of Bile Acid Synthesis by Nuclear Receptors Timothy T. Lu, Makoto Makishima,* Joyce
ab SREBP-2 Translocation Assay Kit (Cell-Based)
 ab133114 SREBP-2 Translocation Assay Kit (Cell-Based) Instructions for Use For analysis of translocation of SREBP-2 into nuclei. This product is for research use only and is not intended for diagnostic
ab133114 SREBP-2 Translocation Assay Kit (Cell-Based) Instructions for Use For analysis of translocation of SREBP-2 into nuclei. This product is for research use only and is not intended for diagnostic
Luminescent platforms for monitoring changes in the solubility of amylin and huntingtin in living cells
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Gene Polymorphisms and Carbohydrate Diets. James M. Ntambi Ph.D
 Gene Polymorphisms and Carbohydrate Diets James M. Ntambi Ph.D Fatty Acids that Flux into Tissue Lipids are from Dietary Sources or are Made De novo from Glucose or Fructose Glucose Fructose Acetyl-CoA
Gene Polymorphisms and Carbohydrate Diets James M. Ntambi Ph.D Fatty Acids that Flux into Tissue Lipids are from Dietary Sources or are Made De novo from Glucose or Fructose Glucose Fructose Acetyl-CoA
Total Bile Acid Assay Kit (Fluorometric)
 Product Manual Total Bile Acid Assay Kit (Fluorometric) Catalog Number MET-5005 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Bile is a complex mixture of lipids, protein,
Product Manual Total Bile Acid Assay Kit (Fluorometric) Catalog Number MET-5005 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Bile is a complex mixture of lipids, protein,
Trident Membrane Protein Extraction Kit
 Cat. No. Size Shelf life GTX16373 5/ 20 tests 12 months at the appropriate storage temperatures (see below) Contents Component Storage Amount for 5 tests Amount for 20 tests Buffer A -20 o C 2.5 ml 10
Cat. No. Size Shelf life GTX16373 5/ 20 tests 12 months at the appropriate storage temperatures (see below) Contents Component Storage Amount for 5 tests Amount for 20 tests Buffer A -20 o C 2.5 ml 10
Functional Specificities of Brm and Brg-1 Swi/Snf ATPases in the Feedback Regulation of Hepatic Bile Acid Biosynthesis
 MOLECULAR AND CELLULAR BIOLOGY, Dec. 2009, p. 6170 6181 Vol. 29, No. 23 0270-7306/09/$12.00 doi:10.1128/mcb.00825-09 Copyright 2009, American Society for Microbiology. All Rights Reserved. Functional Specificities
MOLECULAR AND CELLULAR BIOLOGY, Dec. 2009, p. 6170 6181 Vol. 29, No. 23 0270-7306/09/$12.00 doi:10.1128/mcb.00825-09 Copyright 2009, American Society for Microbiology. All Rights Reserved. Functional Specificities
Niacin Metabolism: Effects on Cholesterol
 Niacin Metabolism: Effects on Cholesterol By Julianne R. Edwards For Dr. William R. Proulx, PhD, RD Associate Professor of Nutrition and Dietetics In partial fulfillments for the requirements of NUTR342
Niacin Metabolism: Effects on Cholesterol By Julianne R. Edwards For Dr. William R. Proulx, PhD, RD Associate Professor of Nutrition and Dietetics In partial fulfillments for the requirements of NUTR342
The antiparasitic drug ivermectin is a novel FXR ligand that regulates metabolism
 Supplementary Information The antiparasitic drug ivermectin is a novel FXR ligand that regulates metabolism Address correspondence to Yong Li (yongli@xmu.edu.cn, Tel: 86-592-218151) GW464 CDCA Supplementary
Supplementary Information The antiparasitic drug ivermectin is a novel FXR ligand that regulates metabolism Address correspondence to Yong Li (yongli@xmu.edu.cn, Tel: 86-592-218151) GW464 CDCA Supplementary
(Stratagene, La Jolla, CA) (Supplemental Fig. 1A). A 5.4-kb EcoRI fragment
 SUPPLEMENTAL INFORMATION Supplemental Methods Generation of RyR2-S2808D Mice Murine genomic RyR2 clones were isolated from a 129/SvEvTacfBR λ-phage library (Stratagene, La Jolla, CA) (Supplemental Fig.
SUPPLEMENTAL INFORMATION Supplemental Methods Generation of RyR2-S2808D Mice Murine genomic RyR2 clones were isolated from a 129/SvEvTacfBR λ-phage library (Stratagene, La Jolla, CA) (Supplemental Fig.
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna
 Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
Glycerol- 3- Phosphate (G3P) Assay Kit (Colorimetric)
 Product Manual Glycerol- 3- Phosphate (G3P) Assay Kit (Colorimetric) Catalog Number MET- 5075 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Glycerol-3-phosphate (G3P)
Product Manual Glycerol- 3- Phosphate (G3P) Assay Kit (Colorimetric) Catalog Number MET- 5075 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Glycerol-3-phosphate (G3P)
PRODUCT INFORMATION & MANUAL
 PRODUCT INFORMATION & MANUAL Mitochondrial Extraction Kit NBP2-29448 Research use only. Not for diagnostic or therapeutic procedures www.novusbio.com P: 303.760.1950 P: 888.506.6887 F: 303.730.1966 technical@novusbio.com
PRODUCT INFORMATION & MANUAL Mitochondrial Extraction Kit NBP2-29448 Research use only. Not for diagnostic or therapeutic procedures www.novusbio.com P: 303.760.1950 P: 888.506.6887 F: 303.730.1966 technical@novusbio.com
Peroxisome proliferator-activated receptor (PPAR ) and agonist inhibit cholesterol 7 -hydroxylase gene (CYP7A1) transcription
 Peroxisome proliferator-activated receptor (PPAR ) and agonist inhibit cholesterol 7 -hydroxylase gene (CYP7A1) transcription Maria Marrapodi and John Y. L. Chiang 1 Department of Biochemistry and Molecular
Peroxisome proliferator-activated receptor (PPAR ) and agonist inhibit cholesterol 7 -hydroxylase gene (CYP7A1) transcription Maria Marrapodi and John Y. L. Chiang 1 Department of Biochemistry and Molecular
Bile-acid synthesis is the major mechanism to
 AUTOIMMUNE, CHOLESTATIC AND BILIARY DISEASE Mechanism of Tissue-Specific Farnesoid X Receptor in Suppressing the Expression of Genes in Bile-Acid Synthesis in Mice Bo Kong, 1 Li Wang, 2 John Y.L. Chiang,
AUTOIMMUNE, CHOLESTATIC AND BILIARY DISEASE Mechanism of Tissue-Specific Farnesoid X Receptor in Suppressing the Expression of Genes in Bile-Acid Synthesis in Mice Bo Kong, 1 Li Wang, 2 John Y.L. Chiang,
FOCUS SubCell. For the Enrichment of Subcellular Fractions. (Cat. # ) think proteins! think G-Biosciences
 169PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name FOCUS SubCell For the Enrichment of Subcellular Fractions (Cat. # 786 260) think
169PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name FOCUS SubCell For the Enrichment of Subcellular Fractions (Cat. # 786 260) think
EPIGENTEK. EpiQuik Global Acetyl Histone H3K27 Quantification Kit (Colorimetric) Base Catalog # P-4059 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik Global Acetyl Histone H3K27 Quantification Kit (Colorimetric) Base Catalog # P-4059 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Global Acetyl Histone H3K27 Quantification Kit (Colorimetric)
EpiQuik Global Acetyl Histone H3K27 Quantification Kit (Colorimetric) Base Catalog # P-4059 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Global Acetyl Histone H3K27 Quantification Kit (Colorimetric)
Fatty acid synthase (FAS) plays a central role in de novo
 Nutritional regulation of the fatty acid synthase promoter in vivo: Sterol regulatory element binding protein functions through an upstream region containing a sterol regulatory element Maria-Jesus Latasa,
Nutritional regulation of the fatty acid synthase promoter in vivo: Sterol regulatory element binding protein functions through an upstream region containing a sterol regulatory element Maria-Jesus Latasa,
Supplementary information
 Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Doctor of Philosophy
 Regulation of Gene Expression of the 25-Hydroxyvitamin D la-hydroxylase (CYP27BI) Promoter: Study of A Transgenic Mouse Model Ivanka Hendrix School of Molecular and Biomedical Science The University of
Regulation of Gene Expression of the 25-Hydroxyvitamin D la-hydroxylase (CYP27BI) Promoter: Study of A Transgenic Mouse Model Ivanka Hendrix School of Molecular and Biomedical Science The University of
AP Biology Summer Assignment Chapter 3 Quiz
 AP Biology Summer Assignment Chapter 3 Quiz 2016-17 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. All of the following are found in a DNA nucleotide
AP Biology Summer Assignment Chapter 3 Quiz 2016-17 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. All of the following are found in a DNA nucleotide
Transcriptional regulation of the human hepatic lipase (LIPC) gene promoter
 Transcriptional regulation of the human hepatic lipase (LIPC) gene promoter Laura E. Rufibach, 1, * Stephen A. Duncan, Michele Battle, and Samir S. Deeb* Departments of Medical Genetics and Genome Sciences,*
Transcriptional regulation of the human hepatic lipase (LIPC) gene promoter Laura E. Rufibach, 1, * Stephen A. Duncan, Michele Battle, and Samir S. Deeb* Departments of Medical Genetics and Genome Sciences,*
Islet viability assay and Glucose Stimulated Insulin Secretion assay RT-PCR and Western Blot
 Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
SensoLyte pnpp Alkaline Phosphatase Assay Kit *Colorimetric*
 SensoLyte pnpp Alkaline Phosphatase Assay Kit *Colorimetric* Catalog # 72146 Kit Size 500 Assays (96-well plate) Optimized Performance: This kit is optimized to detect alkaline phosphatase activity Enhanced
SensoLyte pnpp Alkaline Phosphatase Assay Kit *Colorimetric* Catalog # 72146 Kit Size 500 Assays (96-well plate) Optimized Performance: This kit is optimized to detect alkaline phosphatase activity Enhanced
CTP:phosphocholine cytidylyltransferase, a new steroland SREBP-responsive gene
 CTP:phosphocholine cytidylyltransferase, a new steroland SREBP-responsive gene Heidi Rachelle Kast,* Catherine M. Nguyen,* Andrew M. Anisfeld,* Johan Ericsson, and Peter A. Edwards 1, *, Departments of
CTP:phosphocholine cytidylyltransferase, a new steroland SREBP-responsive gene Heidi Rachelle Kast,* Catherine M. Nguyen,* Andrew M. Anisfeld,* Johan Ericsson, and Peter A. Edwards 1, *, Departments of
Retinoic acid represses CYP7A1 expression in human hepatocytes and HepG2 cells by FXR/RXR-dependent and independent mechanisms
 Retinoic acid represses CYP7A1 expression in human hepatocytes and HepG2 cells by FXR/RXR-dependent and independent mechanisms Shi-Ying Cai, 1 Hongwei He, Trong Nguyen, Albert Mennone, and James L. Boyer
Retinoic acid represses CYP7A1 expression in human hepatocytes and HepG2 cells by FXR/RXR-dependent and independent mechanisms Shi-Ying Cai, 1 Hongwei He, Trong Nguyen, Albert Mennone, and James L. Boyer
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies
 Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Supplemental Figures
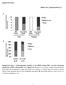 Supplemental Figures Supplemental Figure 1. Fasting-dependent regulation of the SREBP ortholog SBP-1 and lipid homeostasis mediated by the SIRT1 ortholog SIR-2.1 in C. elegans. (A) Wild-type or sir-2.1(lof)
Supplemental Figures Supplemental Figure 1. Fasting-dependent regulation of the SREBP ortholog SBP-1 and lipid homeostasis mediated by the SIRT1 ortholog SIR-2.1 in C. elegans. (A) Wild-type or sir-2.1(lof)
ab Adipogenesis Assay Kit (Cell-Based)
 ab133102 Adipogenesis Assay Kit (Cell-Based) Instructions for Use For the study of induction and inhibition of adipogenesis in adherent cells. This product is for research use only and is not intended
ab133102 Adipogenesis Assay Kit (Cell-Based) Instructions for Use For the study of induction and inhibition of adipogenesis in adherent cells. This product is for research use only and is not intended
Human Apolipoprotein A1 EIA Kit
 A helping hand for your research Product Manual Human Apolipoprotein A1 EIA Kit Catalog Number: 83901 96 assays 1 Table of Content Product Description 3 Assay Principle 3 Kit Components 3 Storage 4 Reagent
A helping hand for your research Product Manual Human Apolipoprotein A1 EIA Kit Catalog Number: 83901 96 assays 1 Table of Content Product Description 3 Assay Principle 3 Kit Components 3 Storage 4 Reagent
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer. Application Note
 Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Farnesoid X receptor (FXR, NR1H4) belongs to
 Genomic Analysis of Hepatic Farnesoid X Receptor Binding Sites Reveals Altered Binding in Obesity and Direct Gene Repression by Farnesoid X Receptor in Mice Jiyoung Lee, 1 * Sunmi Seok, 1 * Pengfei Yu,
Genomic Analysis of Hepatic Farnesoid X Receptor Binding Sites Reveals Altered Binding in Obesity and Direct Gene Repression by Farnesoid X Receptor in Mice Jiyoung Lee, 1 * Sunmi Seok, 1 * Pengfei Yu,
Serum Triglyceride Quantification Kit (Colorimetric)
 Product Manual Serum Triglyceride Quantification Kit (Colorimetric) Catalog Number STA-396 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Triglycerides (TAG) are a type
Product Manual Serum Triglyceride Quantification Kit (Colorimetric) Catalog Number STA-396 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Triglycerides (TAG) are a type
Supplementary Material
 Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates
 HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates Department of Microbiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA
HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates Department of Microbiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA
Prevention of cholesterol gallstone disease by FXR agonists in a mouse model
 24 Nature Publishing Group http://www.nature.com/naturemedicine Prevention of cholesterol gallstone disease by FXR agonists in a mouse model Antonio Moschetta, Angie L Bookout & David J Mangelsdorf Cholesterol
24 Nature Publishing Group http://www.nature.com/naturemedicine Prevention of cholesterol gallstone disease by FXR agonists in a mouse model Antonio Moschetta, Angie L Bookout & David J Mangelsdorf Cholesterol
Chapter 2. Investigation into mir-346 Regulation of the nachr α5 Subunit
 15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
Free Fatty Acid Assay Kit (Fluorometric)
 Product Manual Free Fatty Acid Assay Kit (Fluorometric) Catalog Number STA-619 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Triglycerides (TAG) are a type of lipid
Product Manual Free Fatty Acid Assay Kit (Fluorometric) Catalog Number STA-619 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Triglycerides (TAG) are a type of lipid
Spherical Nucleic Acids For Advanced Wound Healing Applications Chad A. Mirkin
 Spherical Nucleic Acids For Advanced Wound Healing Applications Chad A. Mirkin Departments of Chemistry, Infectious Disease, Materials Science & Engineering, Chemical & Biological Engineering, and Biomedical
Spherical Nucleic Acids For Advanced Wound Healing Applications Chad A. Mirkin Departments of Chemistry, Infectious Disease, Materials Science & Engineering, Chemical & Biological Engineering, and Biomedical
OxiSelect Human Oxidized LDL ELISA Kit (OxPL-LDL Quantitation)
 Product Manual OxiSelect Human Oxidized LDL ELISA Kit (OxPL-LDL Quantitation) Catalog Number STA-358 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Lipoproteins are submicroscopic
Product Manual OxiSelect Human Oxidized LDL ELISA Kit (OxPL-LDL Quantitation) Catalog Number STA-358 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Lipoproteins are submicroscopic
Mammalian Tissue Protein Extraction Reagent
 Mammalian Tissue Protein Extraction Reagent Catalog number: AR0101 Boster s Mammalian Tissue Protein Extraction Reagent is a ready-to-use Western blot related reagent solution used for efficient extraction
Mammalian Tissue Protein Extraction Reagent Catalog number: AR0101 Boster s Mammalian Tissue Protein Extraction Reagent is a ready-to-use Western blot related reagent solution used for efficient extraction
RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice
 SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
ab Histone Deacetylase (HDAC) Activity Assay Kit (Fluorometric)
 ab156064 Histone Deacetylase (HDAC) Activity Assay Kit (Fluorometric) Instructions for Use For the quantitative measurement of Histone Deacetylase activity in cell lysates This product is for research
ab156064 Histone Deacetylase (HDAC) Activity Assay Kit (Fluorometric) Instructions for Use For the quantitative measurement of Histone Deacetylase activity in cell lysates This product is for research
Constitutive Regulation of P450s by Endocrine Factors
 References: Constitutive Regulation of P450s by Endocrine Factors Meyer UA. Endo-xenobiotic crosstalk and the regulation of cytochromes P450. Drug Metab Rev 39:639-46, 2007. Waxman DJ and O Connor C. Growth
References: Constitutive Regulation of P450s by Endocrine Factors Meyer UA. Endo-xenobiotic crosstalk and the regulation of cytochromes P450. Drug Metab Rev 39:639-46, 2007. Waxman DJ and O Connor C. Growth
Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION
 Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION X. Shawn Liu 1, 3, Bing Song 2, 3, Bennett D. Elzey 3, 4, Timothy L. Ratliff 3, 4, Stephen F. Konieczny
Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION X. Shawn Liu 1, 3, Bing Song 2, 3, Bennett D. Elzey 3, 4, Timothy L. Ratliff 3, 4, Stephen F. Konieczny
