Supplementary Table S1. List of PTPRK-RSPO3 gene fusions in TCGA's colon cancer cohort. Chr. # of Gene 2. Chr. # of Gene 1
|
|
|
- Jacob Phelps
- 5 years ago
- Views:
Transcription
1 Supplementary Tale S1. List of PTPRK-RSPO3 gene fusions in TCGA's colon cancer cohort TCGA Case ID Gene-1 Gene-2 Chr. # of Gene 1 Chr. # of Gene 2 Genomic coordiante of Gene 1 at fusion junction Genomic coordinate of Gene 2 at fusion junction Numer of spanning reads found in RNAseq TCGA-AA-3842 PTPRK RSPO3 chr6 chr TCGA-AA-3664 PTPRK RSPO3 chr6 chr TCGA-AA-3518 PTPRK RSPO3 chr6 chr TCGA-A PTPRK RSPO3 chr6 chr
2 Wnt receptors Wnt responders Wnt modulators Wnt ligands Wnt signaling RSPO-LGR Suppplementary Tale S2. Expression profile of Wnt and RSPO signalings genes in three lung cancer cell lines. The data in log2(microarray signal) were downloaded from CCLE's gene expression dataase. Category Cell line A549 H46 H29 Category Cell line A549 H46 H29 PORCN RSPO Norrin RSPO WNT RSPO WNT RSPO WNT2B LGR WNT LGR WNT LGR WNT DVL WNT5A DVL WNT5B DVL WNT IQGAP WNT7A IQGAP WNT7B IQGAP WNT8A DKK WNT8B DKK WNT9A DKK WNT9B E-cadherin WNT1A RNF WNT1B ZNRF WNT SFRP WNT SFRP FZD SFRP FZD SFRP FZD AXIN FZD WIF FZD catenin FZD CD FZD FZD PTPRK FZD FZD LRP LRP ROR ROR Ryk
3 15 1 Gene Exp (log2[rsem]) 1 5 Gene Exp (log2[signal[) c LGR4 LGR5 LGR6 d LGR4 LGR5 LGR Gene Exp (log2[rsem]) 1 5 Gene Exp (log2[signal[) RSPO1 RSPO2 RSPO3 RSPO4 RSPO1 RSPO2 RSPO3 RSPO4 Supplementary Figure S1. RSPO3 and LGR4 were the predominant ligand receptor pair expressed in lung AD and lung cancer cell lines. (a ) Expression level and distriution of LGR4 6 in the 23 samples of TCGA s LUAD cohort (a) and CCLE s 186 lung cancer cell lines (). (c d) Expression level and distriution of RSPO1 4 in the same TCGA cohort (c) and CCLE lung cancer cell line collection (d).
4 15 Normal Tumor RSPO3 Exp (log2[signal]) 1 5 TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA TCGA Supplementary Figure S2. High expression of RSPO3 in selected tumors was due to upregulation of gene expression instead of repression. Log2[RSEM] values of RSPO3 from tumor and matched normal tissues in TCGA s 55 cases were plotted side y side for each tumor.
5 % Surviving RSPO3-high RSPO3-norm (Logrank test p =.7) N = 84 2 N = Months of Survival 1 % Surviving N = 5 N = RSPO3-norm RSPO3-high (Logrank test p =.59) Months of Survival Supplementary Figure S3. High expression of RSPO3 was associated with poor survival in two other lung cancer studies. Kaplan Meier survival plot of RSPO3 expression vs. overall survival in two lung cancer cohorts, GSE3141 (a) and GSE3121 (). RSPO3 high were defined as those samples at 2 SDs aove the population mean (Z = 2) for oth GSE3141 and GSE3121.
6 25 2 TCGA-AA-3664 RPKM TCGA-AA-3842 TCGA-AA-3518 TCGA-A Sample Numer Supplementary Figure S4. High levels of RSPO3 expression in colon cancers were all driven y recurrent fusions with PTPRK. The RNA seq data in RPKM (reads per kiloase per million reads) of RSPO3 from TCGA s 244 colorectal samples were sorted from low to high and plotted. Samples with PTPRK RSPO3 gene fusions were marked y arrows with case IDs.
7 RSPO3 Keap1 ABCC2 K Ras TP53 EGFR mrna Downregulation mrna Upregulation Mutation RSPO3 Keap1 ABCC2 AKR1C1 AKR1C2 -Expression high -Expression low Supplementary Figure S5. High expression of RSPO3 is strongly associated with Keap1 deficiency in oth primary lung AD tumors and lung cancer cell lines. (a) Parallel view of gene expression and mutation status of RSPO3 vs. Keap1, ABCC2 ( a target gene of the Keap1 NRF2 pathway), K Ras, TP53, and EGFR in the 23 samples of TCGA s LUAD cohort. The diagram was generated using the OncoPrint function of cbioportal. Down and up regulation are defined as those samples with log2[rsem] values at 1 SD elow or aove, respectively, the population mean. () A heat map of the expression profile of RSPO3, Keap1, ABCC2, AKR1C1, and AKR1C2 in CCLE s 186 lung cancer cell line set. Mutation status of Keap1 in CCLE cell lines is unknown and thus not included.
8 GFP PLVX GFP Keap1 GFP PLVX GFP Keap1 Keap1 Actin NRF2 Actin c anti Keap1 anti NRF GFP GFP-Keap1 % Control Exp AKR1C2 RSPO3 Supplementary Figure S6. Expression of GFP Keap1 in lung cancer cell line H1944 cells led to decreased levels of NRF2 and reduced expression of RSPO3 and AKR1C2. (a) WB results of H1944 cells expressing GFP control or Keap1 GFP proed with anti Keap1 antiody. () WB results of the same cells proed with anti NRF2 antiody. These experiments were repeated once with representative data shown. (c) RT qpcr analysis of AKR1C2 and RSPO3 in H1944 cells expressing GFP control or GFP Keap1. The data are normalized to 18S RNA and the values are the mean of three replicate experiments. Error ars are S.E.M.
9 Supplementary Figure S7. Nine putative NRF2 inding sites are located upstream of the promoter region of RSPO3. The sequences and positions of the 9 putative NRF inding sites located upstream of the transcription start site (TSS) of RSPO3 are listed. The profile of NRF2 inding sites determined from ChIP Seq (Malhotra et al., 21) is shown for comparison. ntgannnngcn is the consensus sequence of NRF inding sites.
10 15 15 RSPO3 Exp (log2[rsem]) 1 5 RSPO3 Exp (log2[rsem]) 1 5 c Log2[RSPO3 copy numer] d RSPO3-GISTIC 2 15 P =.65 (One way ANOVA) 1..8 P =.55 (One way ANOVA) # of Mutations 1 Fraction RSPO3-high, Keap1-def RSPO3-low, Keap1-def RSPO3-low, Keap1-norm. RSPO3-high, Keap1-def RSPO3-low, Keap1-def RSPO3-low, Keap1-norm Supplementary Figure S8. RSPO3 high and norm tumors were not significantly different in gene copy numer, mutation urden, and the fraction of copy numer altered genome in the 23 samples of TCGA s LUAD set. (a) RSPO3 Expression vs. RSPO3 gene copy numer as determined y Affymetrix SNP6. () RSPO3 expression vs. gene copy numer as determined y the GISTIC assay. (c) RSPO3 expression vs. mutation urden. (d) RSPO3 expression vs. fraction of copy numer altered genome.
11 c Methylaiton eta at Keap1-norm Keap1-def Methylaiton eta at Keap-norm Keap1-def Methylaiton eta at Keap1-norm Keap1-def RSPO3 Exp (log2[rsem]) RSPO3 Exp (log2[rsem]) RSPO3 Exp (log2[rsem]) Supplementary Figure S9. All RSPO3 high tumors had lost methylation in the three CpG sequences located downstream of the RSPO3 promoter region. (a c) Scatter plot of the methylation eta values at the three CpG sites, (a), (), and (c) vs. RSPO3 expression in relation to Keap1 status. Keap1 norm = no Keap1 mutation or down regulation. Keap1 def = Keap1 deficient.
12 P LGR4 p LRP6 tlrp6 Catenin Actin Expression of protein (%) LGR4 plrp6 LRP6 -Catenin Parent shlgr4-#39 shlgr4-#4 shlgr4-#43 c Cell Index Parental shlgr4-39 shlgr4-4 shlgr4-43 d Migration cell numer P <.5 (One way ANOVA) * * e Invasion cell numer P <.5 (One way ANOVA) * * Growth Time (Hour) Parental shlgr4-39 shlgr4-4 shlgr4-43 Parental shlgr4-39 shlgr4-4 shlgr4-43 Supplementary Figure S1 Confirmation of the effect of LGR4 KD y a second shrna (#43). (a) WB results of LGR4 expression and Wnt signaling in A549 parental cells (P) and those expressing control shrna (C), shlgr4 4 (4), or shlgr4 43 (43). () Quantification of the WB signals in (a). (c e) Growth (c), migration (d), and invasion (e) of A549 parental cells and those expressing control shrna, shlgr4 4, or shlgr4 43. The experiments were repeated twice and errors ars are S.E.M. *p <.5 vs. control shrna, Dunnett s multiple comparison test.
13 Parental shrna-cont shlgr4-4 O.D. (57 nm) Growth Time (day) O.D. (57 nm) Parental shrna-cont shrspo3-63 shrspo Growth Time (day) Supplementary Figure S11. MTT assay confirmed the negative effect of KD of LGR4 or RSPO3 on cell growth of A549 cells. (a) Growth curve of A549 cells with KD of LGR4 (shlgr4 4) vs. parental A549 cells and those expressing control shrna. () Growth curve of A549 cells with KD of RSPO3 (shrna 63, 67) vs. parental A549 cells and those expressing control shrna. The experiments were repeated once with representative data shown here.
14 Expression of LGR4 (%) Parental shrna-cont shlgr4-4 Expression of IQGAP1 (%) Parental shrna-cont shiqgap1 Supplementary Figure S12. Quantified results of the WB signals in Figure 3e. (a), Levels of LGR4 as normalized to actin. () Levels of IQGAP1 as normalized to actin.
15 Migration cell numer RSPO3 +RSPO3 Parental Vector LGR4-KD IQGAP1-KD Supplementary Figure S13. The actual numer of migrated cells of the experiment in Fig. 3f.
16 Parental shrna-cont LGR4-KD Supplementary Figure S14. KD of LGR4 in A549 cells led to change from a mesenchymal like morphology to an epithelial (cole stone) like morphology. Representative right field micrographs of parental A549 cells, cells expressing control shrna or LGR4 KD shrna (shlgr4 4).
17 15 Expression of protein (%) 1 5 A549-Parental A549-shLGR4-39 A549-shLGR4-4 H46-Parental H46-shLGR4-39 H46-shLGR4-4 LGR4 E-cadherin Fironectin N-cadherin Vimentin 3 Expression of protein (%) 2 1 Parental Vector-cont shrspo3-63 shrspo3-67 Ze1 E-cadherin Fironectin N-cadherin Vimentin Supplementary Figure S15. Quantified results of the WB signals in Figure 4 c. (a) results of the WB signals in Figure 4 as normalized to actin. () Results of the WB signals in Figure 4c as normalized to actin.
18 Parental shrna-control LGR4-KD Parental shrna-control LGR4-KD -LGR4 -Actin Supplementary Figure S16. KD of LGR4 in A549 cells led to reduced tumor growth. (a) Photographs of the sucutaneous tumors from parental A549 cells and those expressing control shrna or the LGR4 KD shrna (shlgr4 4). () WB analysis of LGR4 of the tumors from sucutaneous study using the LGR4 antiody 7E7. Actin was used as loading control.
19 #96 #93 #97 #94 #98 #95 #99 #97 Supplementary Figure S17. H &E staining of lung sections from the animals injected with A549 cell expressing control shrna (a) or LGR4 KD (shlgr 4) (). The numers denote the codes used for each animal. Representative micrographs of the remaining two animals are shown in Figure 6C.
20 % Surviving Axin2-norm Axin2-high (Z > +1) Axin2-low (Z < -1) (logrank test p =.16 N = 126 RSPO3 Exp (log2[rsem]) 2 1 N = 41 N = Months Survival 15 RSPO3 (R =.4, p =.5) AXIN2 Exp (log2[rsem]) Supplementary Figure S18. Expression level of Axin2 was not correlated with survival nor with that of RSPO3. (a) Kaplan Meier survival curves of patients with tumors of either high, low, or norm levels of expression of Axin2. High and low expressions are defined as 1 standard deviation aove (Z > +1) or elow (Z < 1) the population mean of log2[resm] of RNA seq data. The rest of the samples are define as norm. () Plot of the RNA seq data of Axin2 vs. RSPO3. Correlation efficiency (R) and p values are y Spearmen test (non parametric).
SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer.
 Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
Table S1. Total and mapped reads produced for each ChIP-seq sample
 Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Nature Medicine: doi: /nm.3967
 Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Supplementary Figure 1: High-throughput profiling of survival after exposure to - radiation. (a) Cells were plated in at least 7 wells in a 384-well
 Supplementary Figure 1: High-throughput profiling of survival after exposure to - radiation. (a) Cells were plated in at least 7 wells in a 384-well plate at cell densities ranging from 25-225 cells in
Supplementary Figure 1: High-throughput profiling of survival after exposure to - radiation. (a) Cells were plated in at least 7 wells in a 384-well plate at cell densities ranging from 25-225 cells in
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Materials and Methods
 DD2 suppresses tumorigenicity of ovarian cancer cells by limiting cancer stem cell population Chunhua Han et al. Supplementary Materials and Methods Analysis of publicly available datasets: To analyze
DD2 suppresses tumorigenicity of ovarian cancer cells by limiting cancer stem cell population Chunhua Han et al. Supplementary Materials and Methods Analysis of publicly available datasets: To analyze
Supplemental Information. NRF2 Is a Major Target of ARF. in p53-independent Tumor Suppression
 Molecular Cell, Volume 68 Supplemental Information NRF2 Is a Major Target of ARF in p53-independent Tumor Suppression Delin Chen, Omid Tavana, Bo Chu, Luke Erber, Yue Chen, Richard Baer, and Wei Gu Figure
Molecular Cell, Volume 68 Supplemental Information NRF2 Is a Major Target of ARF in p53-independent Tumor Suppression Delin Chen, Omid Tavana, Bo Chu, Luke Erber, Yue Chen, Richard Baer, and Wei Gu Figure
Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs.
 Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs. (a) CNA analysis of expression microarray data obtained from 15 tumors in the SV40Tag
Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs. (a) CNA analysis of expression microarray data obtained from 15 tumors in the SV40Tag
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.
 Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
a Anti-Dab2 RGD Merge
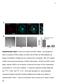 a Anti-Da Merge *** ** Percentage of cells adhered 8 6 4 RGE RAD RGE RAD Supplementary Figure Da are localized at clusters. (a) Endogenous Da is localized at clusters. (), ut not RGE nor RAD peptides can
a Anti-Da Merge *** ** Percentage of cells adhered 8 6 4 RGE RAD RGE RAD Supplementary Figure Da are localized at clusters. (a) Endogenous Da is localized at clusters. (), ut not RGE nor RAD peptides can
Supplementary Figure 1. MLN8237 treatments show an unusual camel-back response pattern.
 Supplementary Figure 1. MLN8237 treatments show an unusual camel-ack response pattern. NCI-H1819 cells were treated with serial concentrations of MLN8237 and four days after treatment, cell viaility was
Supplementary Figure 1. MLN8237 treatments show an unusual camel-ack response pattern. NCI-H1819 cells were treated with serial concentrations of MLN8237 and four days after treatment, cell viaility was
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Nature Genetics: doi: /ng Supplementary Figure 1. Phenotypic characterization of MES- and ADRN-type cells.
 Supplementary Figure 1 Phenotypic characterization of MES- and ADRN-type cells. (a) Bright-field images showing cellular morphology of MES-type (691-MES, 700-MES, 717-MES) and ADRN-type (691-ADRN, 700-
Supplementary Figure 1 Phenotypic characterization of MES- and ADRN-type cells. (a) Bright-field images showing cellular morphology of MES-type (691-MES, 700-MES, 717-MES) and ADRN-type (691-ADRN, 700-
mir-509-5p and mir-1243 increase the sensitivity to gemcitabine by inhibiting
 mir-509-5p and mir-1243 increase the sensitivity to gemcitabine by inhibiting epithelial-mesenchymal transition in pancreatic cancer Hidekazu Hiramoto, M.D. 1,3, Tomoki Muramatsu, Ph.D. 1, Daisuke Ichikawa,
mir-509-5p and mir-1243 increase the sensitivity to gemcitabine by inhibiting epithelial-mesenchymal transition in pancreatic cancer Hidekazu Hiramoto, M.D. 1,3, Tomoki Muramatsu, Ph.D. 1, Daisuke Ichikawa,
SUPPLEMENTARY FIGURES: Supplementary Figure 1
 SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
TEB. Id4 p63 DAPI Merge. Id4 CK8 DAPI Merge
 a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
m 6 A mrna methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
Supplemental Data. Integrating omics and alternative splicing i reveals insights i into grape response to high temperature
 Supplemental Data Integrating omics and alternative splicing i reveals insights i into grape response to high temperature Jianfu Jiang 1, Xinna Liu 1, Guotian Liu, Chonghuih Liu*, Shaohuah Li*, and Lijun
Supplemental Data Integrating omics and alternative splicing i reveals insights i into grape response to high temperature Jianfu Jiang 1, Xinna Liu 1, Guotian Liu, Chonghuih Liu*, Shaohuah Li*, and Lijun
Supplemental information
 Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
SUPPLEMENTARY INFORMATION
 -. -. SUPPLEMENTARY INFORMATION DOI: 1.1/ncb86 a WAT-1 WAT- BAT-1 BAT- sk-muscle-1 sk-muscle- mir-133b mir-133a mir-6 mir-378 mir-1 mir-85 mir-378 mir-6a mir-18 mir-133a mir- mir- mir-341 mir-196a mir-17
-. -. SUPPLEMENTARY INFORMATION DOI: 1.1/ncb86 a WAT-1 WAT- BAT-1 BAT- sk-muscle-1 sk-muscle- mir-133b mir-133a mir-6 mir-378 mir-1 mir-85 mir-378 mir-6a mir-18 mir-133a mir- mir- mir-341 mir-196a mir-17
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Color Key PCA. mir- 15a let-7c 106b let-7b let-7a 16 10b 99a 26a 20b 374b 19b 135b 125b a-5p 199b-5p 93 92b MES PN.
 1123 83 528 816 84 2 17 718 Comp3 3 2 1 1 2 3 4 Comp2 a b Color Key MES PN PCA 3 2 1 1 2 3 4 Comp1 1 2 3 1 2 3 4-2 -1 1 2 mi- 15a let-7c 16b let-7b let-7a 16 1b 99a 26a 2b 374b 19b 135b 125b 9 34 125a-5p
1123 83 528 816 84 2 17 718 Comp3 3 2 1 1 2 3 4 Comp2 a b Color Key MES PN PCA 3 2 1 1 2 3 4 Comp1 1 2 3 1 2 3 4-2 -1 1 2 mi- 15a let-7c 16b let-7b let-7a 16 1b 99a 26a 2b 374b 19b 135b 125b 9 34 125a-5p
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 DOT1L regulates the expression of epithelial and mesenchymal markers. (a) The expression levels and cellular localizations of EMT markers were confirmed by
Supplementary Figures Supplementary Figure 1 DOT1L regulates the expression of epithelial and mesenchymal markers. (a) The expression levels and cellular localizations of EMT markers were confirmed by
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded
 Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays
 Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Inhibition of Gastric Cancer Invasion and Metastasis by PLA2G2A, a Novel β-catenin/tcf Target Gene
 Supplementary Information Inhibition of Gastric Cancer Invasion and Metastasis by PLA2G2A, a Novel β-catenin/tcf Target Gene Kumaresan Ganesan, 1 Tatiana Ivanova, 1 Yonghui Wu, 1 Vikneswari Rajasegaran,
Supplementary Information Inhibition of Gastric Cancer Invasion and Metastasis by PLA2G2A, a Novel β-catenin/tcf Target Gene Kumaresan Ganesan, 1 Tatiana Ivanova, 1 Yonghui Wu, 1 Vikneswari Rajasegaran,
Supplementary Figure 1. IHC and proliferation analysis of pten-deficient mammary tumors
 Wang et al LEGENDS TO SUPPLEMENTARY INFORMATION Supplementary Figure 1. IHC and proliferation analysis of pten-deficient mammary tumors A. Induced expression of estrogen receptor α (ERα) in AME vs PDA
Wang et al LEGENDS TO SUPPLEMENTARY INFORMATION Supplementary Figure 1. IHC and proliferation analysis of pten-deficient mammary tumors A. Induced expression of estrogen receptor α (ERα) in AME vs PDA
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
Guangdong Medical University, Zhanjiang, China; 5 Guangxi Medical University, Nanning, China; 6 Department of Pathology, University of Michigan
 Overexpression of FAM83H-AS1 indicates poor patient survival and knockdown impairs cell proliferation and invasion via MET/EGFR signaling in lung cancer Jie Zhang 1,2, Shumei Feng 3, Wenmei Su 4, Shengbin
Overexpression of FAM83H-AS1 indicates poor patient survival and knockdown impairs cell proliferation and invasion via MET/EGFR signaling in lung cancer Jie Zhang 1,2, Shumei Feng 3, Wenmei Su 4, Shengbin
Nature Genetics: doi: /ng Supplementary Figure 1. Assessment of sample purity and quality.
 Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
HALLA KABAT * Outreach Program, mircore, 2929 Plymouth Rd. Ann Arbor, MI 48105, USA LEO TUNKLE *
 CERNA SEARCH METHOD IDENTIFIED A MET-ACTIVATED SUBGROUP AMONG EGFR DNA AMPLIFIED LUNG ADENOCARCINOMA PATIENTS HALLA KABAT * Outreach Program, mircore, 2929 Plymouth Rd. Ann Arbor, MI 48105, USA Email:
CERNA SEARCH METHOD IDENTIFIED A MET-ACTIVATED SUBGROUP AMONG EGFR DNA AMPLIFIED LUNG ADENOCARCINOMA PATIENTS HALLA KABAT * Outreach Program, mircore, 2929 Plymouth Rd. Ann Arbor, MI 48105, USA Email:
Nature Genetics: doi: /ng.2995
 Supplementary Figure 1 Kaplan-Meier survival curves of patients with brainstem tumors. (a) Comparison of patients with PPM1D mutation versus wild-type PPM1D. (b) Comparison of patients with PPM1D mutation
Supplementary Figure 1 Kaplan-Meier survival curves of patients with brainstem tumors. (a) Comparison of patients with PPM1D mutation versus wild-type PPM1D. (b) Comparison of patients with PPM1D mutation
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
days days and gbt-i.cd Recipient 20
 gbt-i. GFP+ Resident memory cells: gbt-i.gfp+ Recruited memory cells: gbt-i.cd45.1+ 1 2-3 gbt-i. flu.gb sc. CD45.1+ Graft with gbt-i.gfp+ 1 Recipient 1 re- 3 36 Graft with gbt-i.gfp+ and gbt-i.cd45.1+
gbt-i. GFP+ Resident memory cells: gbt-i.gfp+ Recruited memory cells: gbt-i.cd45.1+ 1 2-3 gbt-i. flu.gb sc. CD45.1+ Graft with gbt-i.gfp+ 1 Recipient 1 re- 3 36 Graft with gbt-i.gfp+ and gbt-i.cd45.1+
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
BIMM 143. RNA sequencing overview. Genome Informatics II. Barry Grant. Lecture In vivo. In vitro.
 RNA sequencing overview BIMM 143 Genome Informatics II Lecture 14 Barry Grant http://thegrantlab.org/bimm143 In vivo In vitro In silico ( control) Goal: RNA quantification, transcript discovery, variant
RNA sequencing overview BIMM 143 Genome Informatics II Lecture 14 Barry Grant http://thegrantlab.org/bimm143 In vivo In vitro In silico ( control) Goal: RNA quantification, transcript discovery, variant
Metastasis regulation by PPARD expression in cancer cells
 Metastasis regulation by PPARD expression in cancer cells Xiangsheng Zuo, 1 Weiguo Xu, 1,2 Min Xu, 1 Rui Tian, 1 Micheline J. Moussalli, 3 Fei Mao, 1 Xiaofeng Zheng, 4 Jing Wang, 4 Jeffrey S. Morris, 5
Metastasis regulation by PPARD expression in cancer cells Xiangsheng Zuo, 1 Weiguo Xu, 1,2 Min Xu, 1 Rui Tian, 1 Micheline J. Moussalli, 3 Fei Mao, 1 Xiaofeng Zheng, 4 Jing Wang, 4 Jeffrey S. Morris, 5
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the
 Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
RNA SEQUENCING AND DATA ANALYSIS
 RNA SEQUENCING AND DATA ANALYSIS Length of mrna transcripts in the human genome 5,000 5,000 4,000 3,000 2,000 4,000 1,000 0 0 200 400 600 800 3,000 2,000 1,000 0 0 2,000 4,000 6,000 8,000 10,000 Length
RNA SEQUENCING AND DATA ANALYSIS Length of mrna transcripts in the human genome 5,000 5,000 4,000 3,000 2,000 4,000 1,000 0 0 200 400 600 800 3,000 2,000 1,000 0 0 2,000 4,000 6,000 8,000 10,000 Length
Computer Science, Biology, and Biomedical Informatics (CoSBBI) Outline. Molecular Biology of Cancer AND. Goals/Expectations. David Boone 7/1/2015
 Goals/Expectations Computer Science, Biology, and Biomedical (CoSBBI) We want to excite you about the world of computer science, biology, and biomedical informatics. Experience what it is like to be a
Goals/Expectations Computer Science, Biology, and Biomedical (CoSBBI) We want to excite you about the world of computer science, biology, and biomedical informatics. Experience what it is like to be a
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Supplementary Information and Figure legends
 Supplementary Information and Figure legends Table S1. Primers for quantitative RT-PCR Target Sequence (5 -> 3 ) Target Sequence (5 -> 3 ) DAB2IP F:TGGACGATGTGCTCTATGCC R:GGATGGTGATGGTTTGGTAG Snail F:CCTCCCTGTCAGATGAGGAC
Supplementary Information and Figure legends Table S1. Primers for quantitative RT-PCR Target Sequence (5 -> 3 ) Target Sequence (5 -> 3 ) DAB2IP F:TGGACGATGTGCTCTATGCC R:GGATGGTGATGGTTTGGTAG Snail F:CCTCCCTGTCAGATGAGGAC
Nature Genetics: doi: /ng Supplementary Figure 1. Details of sequencing analysis.
 Supplementary Figure 1 Details of sequencing analysis. (a) Flow chart showing which patients fall into each category and were used for analysis. (b) Graph showing the average and median coverage for all
Supplementary Figure 1 Details of sequencing analysis. (a) Flow chart showing which patients fall into each category and were used for analysis. (b) Graph showing the average and median coverage for all
WNT signalling pathways as therapeutic targets in cancer
 WNT signalling pathways as therapeutic targets in cancer Jamie N. Anastas 1,2,3,4 and Randall T. Moon 1,2,4 Abstract Since the initial discovery of the oncogenic activity of WNT1 in mouse mammary glands,
WNT signalling pathways as therapeutic targets in cancer Jamie N. Anastas 1,2,3,4 and Randall T. Moon 1,2,4 Abstract Since the initial discovery of the oncogenic activity of WNT1 in mouse mammary glands,
microrna-200b and microrna-200c promote colorectal cancer cell proliferation via
 Supplementary Materials microrna-200b and microrna-200c promote colorectal cancer cell proliferation via targeting the reversion-inducing cysteine-rich protein with Kazal motifs Supplementary Table 1.
Supplementary Materials microrna-200b and microrna-200c promote colorectal cancer cell proliferation via targeting the reversion-inducing cysteine-rich protein with Kazal motifs Supplementary Table 1.
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
Nature Genetics: doi: /ng Supplementary Figure 1. Somatic coding mutations identified by WES/WGS for 83 ATL cases.
 Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
Protein Kinase C Modulates Wnt Signaling In Colon Tumoral Cell Lines
 Protein Kinase C Modulates Wnt Signaling In Colon Tumoral Cell Lines Dra Martha Robles Flores Department of Biochemistry Facultad de Medicina Universidad Nacional Autónoma de México Tissue anatomy of the
Protein Kinase C Modulates Wnt Signaling In Colon Tumoral Cell Lines Dra Martha Robles Flores Department of Biochemistry Facultad de Medicina Universidad Nacional Autónoma de México Tissue anatomy of the
Lung Met 1 Lung Met 2 Lung Met Lung Met H3K4me1. Lung Met H3K27ac Primary H3K4me1
 a Gained Met-VELs 1.5 1.5 -.5 Lung Met 1 Lung Met Lung Met 3 1. Lung Met H3K4me1 Lung Met H3K4me1 1 Lung Met H3K4me1 Lung Met H3K7ac 1.5 Lung Met H3K7ac Lung Met H3K7ac.8 Primary H3K4me1 Primary H3K7ac
a Gained Met-VELs 1.5 1.5 -.5 Lung Met 1 Lung Met Lung Met 3 1. Lung Met H3K4me1 Lung Met H3K4me1 1 Lung Met H3K4me1 Lung Met H3K7ac 1.5 Lung Met H3K7ac Lung Met H3K7ac.8 Primary H3K4me1 Primary H3K7ac
Dominic J Smiraglia, PhD Department of Cancer Genetics. DNA methylation in prostate cancer
 Dominic J Smiraglia, PhD Department of Cancer Genetics DNA methylation in prostate cancer Overarching theme Epigenetic regulation allows the genome to be responsive to the environment Sets the tone for
Dominic J Smiraglia, PhD Department of Cancer Genetics DNA methylation in prostate cancer Overarching theme Epigenetic regulation allows the genome to be responsive to the environment Sets the tone for
Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion
 Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/308/ra4/dc1 Supplementary Materials for Antipsychotics Activate mtorc1-dependent Translation to Enhance Neuronal Morphological Complexity Heather Bowling, Guoan
www.sciencesignaling.org/cgi/content/full/7/308/ra4/dc1 Supplementary Materials for Antipsychotics Activate mtorc1-dependent Translation to Enhance Neuronal Morphological Complexity Heather Bowling, Guoan
fl/+ KRas;Atg5 fl/+ KRas;Atg5 fl/fl KRas;Atg5 fl/fl KRas;Atg5 Supplementary Figure 1. Gene set enrichment analyses. (a) (b)
 KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
Supplementary Table S1. Tumor samples used for analysis Tumor size (cm) BNG (grade) ERα PR. pn-
 Supplementary Table S1. Tumor samples used for analysis Sample# Age Tumor size (cm) pn- Stage Stage BNG (grade) ERα PR HER2 (FISH) Triple negative T1 46 3 N1a III 2 Pos Neg N T2 58 1 N(i-) I 3 Pos Neg
Supplementary Table S1. Tumor samples used for analysis Sample# Age Tumor size (cm) pn- Stage Stage BNG (grade) ERα PR HER2 (FISH) Triple negative T1 46 3 N1a III 2 Pos Neg N T2 58 1 N(i-) I 3 Pos Neg
p.r623c p.p976l p.d2847fs p.t2671 p.d2847fs p.r2922w p.r2370h p.c1201y p.a868v p.s952* RING_C BP PHD Cbp HAT_KAT11
 ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
Nature Immunology: doi: /ni Supplementary Figure 1. Id2 and Id3 define polyclonal T H 1 and T FH cell subsets.
 Supplementary Figure 1 Id2 and Id3 define polyclonal T H 1 and T FH cell subsets. Id2 YFP/+ (a) or Id3 GFP/+ (b) mice were analyzed 7 days after LCMV infection. T H 1 (SLAM + CXCR5 or CXCR5 PD-1 ), T FH
Supplementary Figure 1 Id2 and Id3 define polyclonal T H 1 and T FH cell subsets. Id2 YFP/+ (a) or Id3 GFP/+ (b) mice were analyzed 7 days after LCMV infection. T H 1 (SLAM + CXCR5 or CXCR5 PD-1 ), T FH
Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier
 Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) in a cohort of patients (N=52) with stage III primary ovarian
Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) in a cohort of patients (N=52) with stage III primary ovarian
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH
 EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
underlying metastasis and recurrence in HNSCC, we analyzed two groups of patients. The
 Supplementary Figures Figure S1. Patient cohorts and study design. To define and interrogate the genetic alterations underlying metastasis and recurrence in HNSCC, we analyzed two groups of patients. The
Supplementary Figures Figure S1. Patient cohorts and study design. To define and interrogate the genetic alterations underlying metastasis and recurrence in HNSCC, we analyzed two groups of patients. The
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature19360 Supplementary Tables Supplementary Table 1. Number of monoclonal reads in each sample Sample Number of cells Total reads Aligned reads Monoclonal reads
SUPPLEMENTARY INFORMATION doi:10.1038/nature19360 Supplementary Tables Supplementary Table 1. Number of monoclonal reads in each sample Sample Number of cells Total reads Aligned reads Monoclonal reads
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous
 Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Introduction. Introduction
 Introduction We are leveraging genome sequencing data from The Cancer Genome Atlas (TCGA) to more accurately define mutated and stable genes and dysregulated metabolic pathways in solid tumors. These efforts
Introduction We are leveraging genome sequencing data from The Cancer Genome Atlas (TCGA) to more accurately define mutated and stable genes and dysregulated metabolic pathways in solid tumors. These efforts
Expert-guided Visual Exploration (EVE) for patient stratification. Hamid Bolouri, Lue-Ping Zhao, Eric C. Holland
 Expert-guided Visual Exploration (EVE) for patient stratification Hamid Bolouri, Lue-Ping Zhao, Eric C. Holland Oncoscape.sttrcancer.org Paul Lisa Ken Jenny Desert Eric The challenge Given - patient clinical
Expert-guided Visual Exploration (EVE) for patient stratification Hamid Bolouri, Lue-Ping Zhao, Eric C. Holland Oncoscape.sttrcancer.org Paul Lisa Ken Jenny Desert Eric The challenge Given - patient clinical
Social deficits in Shank3-deficient mouse models of autism are rescued by histone deacetylase (HDAC) inhibition
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41593-018-0110-8 In the format provided by the authors and unedited. Social deficits in Shank3-deficient mouse models of autism are rescued by
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41593-018-0110-8 In the format provided by the authors and unedited. Social deficits in Shank3-deficient mouse models of autism are rescued by
IPA Advanced Training Course
 IPA Advanced Training Course October 2013 Academia sinica Gene (Kuan Wen Chen) IPA Certified Analyst Agenda I. Data Upload and How to Run a Core Analysis II. Functional Interpretation in IPA Hands-on Exercises
IPA Advanced Training Course October 2013 Academia sinica Gene (Kuan Wen Chen) IPA Certified Analyst Agenda I. Data Upload and How to Run a Core Analysis II. Functional Interpretation in IPA Hands-on Exercises
Nature Genetics: doi: /ng Supplementary Figure 1. HOX fusions enhance self-renewal capacity.
 Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding
 Supplementary Information A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding Qilai Huang 1,2*, Thomas Whitington 3,4*, Ping Gao 1,2, Johan
Supplementary Information A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding Qilai Huang 1,2*, Thomas Whitington 3,4*, Ping Gao 1,2, Johan
Supporting Information online
 Supporting Information online THE RIVER BLINDNESS DRUG IVERMECTIN AND RELATED MACROCYCLIC LACTONES INHIBIT WNT-TCF PATHWAY RESPONSES IN HUMAN CANCER Alice Melotti, Christophe Mas, Monika Kuciak, Aiala
Supporting Information online THE RIVER BLINDNESS DRUG IVERMECTIN AND RELATED MACROCYCLIC LACTONES INHIBIT WNT-TCF PATHWAY RESPONSES IN HUMAN CANCER Alice Melotti, Christophe Mas, Monika Kuciak, Aiala
RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice
 SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
Results. Abstract. Introduc4on. Conclusions. Methods. Funding
 . expression that plays a role in many cellular processes affecting a variety of traits. In this study DNA methylation was assessed in neuronal tissue from three pigs (frontal lobe) and one great tit (whole
. expression that plays a role in many cellular processes affecting a variety of traits. In this study DNA methylation was assessed in neuronal tissue from three pigs (frontal lobe) and one great tit (whole
Nature Immunology: doi: /ni Supplementary Figure 1. Characteristics of SEs in T reg and T conv cells.
 Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Cancer Informatics Lecture
 Cancer Informatics Lecture Mayo-UIUC Computational Genomics Course June 22, 2018 Krishna Rani Kalari Ph.D. Associate Professor 2017 MFMER 3702274-1 Outline The Cancer Genome Atlas (TCGA) Genomic Data Commons
Cancer Informatics Lecture Mayo-UIUC Computational Genomics Course June 22, 2018 Krishna Rani Kalari Ph.D. Associate Professor 2017 MFMER 3702274-1 Outline The Cancer Genome Atlas (TCGA) Genomic Data Commons
CHAPTER 6 SUMMARIZING DISCUSSION
 CHAPTER 6 SUMMARIZING DISCUSSION More than 20 years ago the founding member of the Wnt gene family, Wnt-1/Int1, was discovered as a proto-oncogene activated in mammary gland tumors by the mouse mammary
CHAPTER 6 SUMMARIZING DISCUSSION More than 20 years ago the founding member of the Wnt gene family, Wnt-1/Int1, was discovered as a proto-oncogene activated in mammary gland tumors by the mouse mammary
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Supplementary Figure 1. IDH1 and IDH2 mutation site sequences on WHO grade III
 Supplementary Materials: Supplementary Figure 1. IDH1 and IDH2 mutation site sequences on WHO grade III patient samples. Genomic DNA samples extracted from punch biopsies from either FFPE or frozen tumor
Supplementary Materials: Supplementary Figure 1. IDH1 and IDH2 mutation site sequences on WHO grade III patient samples. Genomic DNA samples extracted from punch biopsies from either FFPE or frozen tumor
Figure S1: Effects on haptotaxis are independent of effects on cell velocity A)
 Supplemental Figures Figure S1: Effects on haptotaxis are independent of effects on cell velocity A) Velocity of MV D7 fibroblasts expressing different GFP-tagged Ena/VASP family proteins in the haptotaxis
Supplemental Figures Figure S1: Effects on haptotaxis are independent of effects on cell velocity A) Velocity of MV D7 fibroblasts expressing different GFP-tagged Ena/VASP family proteins in the haptotaxis
Pathologic Stage. Lymph node Stage
 ASC ASC a c Patient ID BMI Age Gleason score Non-obese PBMC 1 22.1 81 6 (3+3) PBMC 2 21.9 6 6 (3+3) PBMC 3 22 84 8 (4+4) PBMC 4 24.6 68 7 (3+4) PBMC 24. 6 (3+3) PBMC 6 24.7 73 7 (3+4) PBMC 7 23. 67 7 (3+4)
ASC ASC a c Patient ID BMI Age Gleason score Non-obese PBMC 1 22.1 81 6 (3+3) PBMC 2 21.9 6 6 (3+3) PBMC 3 22 84 8 (4+4) PBMC 4 24.6 68 7 (3+4) PBMC 24. 6 (3+3) PBMC 6 24.7 73 7 (3+4) PBMC 7 23. 67 7 (3+4)
genomics for systems biology / ISB2020 RNA sequencing (RNA-seq)
 RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
Inhibition of TGFβ enhances chemotherapy action against triple negative breast cancer by abrogation of
 SUPPLEMENTAL DATA Inhibition of TGFβ enhances chemotherapy action against triple negative breast cancer by abrogation of cancer stem cells and interleukin-8 Neil E. Bhola 1, Justin M. Balko 1, Teresa C.
SUPPLEMENTAL DATA Inhibition of TGFβ enhances chemotherapy action against triple negative breast cancer by abrogation of cancer stem cells and interleukin-8 Neil E. Bhola 1, Justin M. Balko 1, Teresa C.
Supplementary Figure 1: Neuregulin 1 increases the growth of mammary organoids compared to EGF. (a) Mammary epithelial cells were freshly isolated,
 1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1: Neuregulin 1 increases the growth of mammary organoids compared to EGF. (a) Mammary epithelial cells were freshly isolated, embedded in matrigel and exposed
1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1: Neuregulin 1 increases the growth of mammary organoids compared to EGF. (a) Mammary epithelial cells were freshly isolated, embedded in matrigel and exposed
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung
 Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Exploring TCGA Pan-Cancer Data at the UCSC Cancer Genomics Browser
 Exploring TCGA Pan-Cancer Data at the UCSC Cancer Genomics Browser Melissa S. Cline 1*, Brian Craft 1, Teresa Swatloski 1, Mary Goldman 1, Singer Ma 1, David Haussler 1, Jingchun Zhu 1 1 Center for Biomolecular
Exploring TCGA Pan-Cancer Data at the UCSC Cancer Genomics Browser Melissa S. Cline 1*, Brian Craft 1, Teresa Swatloski 1, Mary Goldman 1, Singer Ma 1, David Haussler 1, Jingchun Zhu 1 1 Center for Biomolecular
Tutorial: RNA-Seq Analysis Part II: Non-Specific Matches and Expression Measures
 : RNA-Seq Analysis Part II: Non-Specific Matches and Expression Measures March 15, 2013 CLC bio Finlandsgade 10-12 8200 Aarhus N Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com support@clcbio.com
: RNA-Seq Analysis Part II: Non-Specific Matches and Expression Measures March 15, 2013 CLC bio Finlandsgade 10-12 8200 Aarhus N Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com support@clcbio.com
Supplementary Information
 Supplementary Information Supplementary Figure 1. Effect of mir mimics and anti-mirs on DTPs a, Representative fluorescence microscopy images of GFP vector control or mir mimicexpressing parental and DTP
Supplementary Information Supplementary Figure 1. Effect of mir mimics and anti-mirs on DTPs a, Representative fluorescence microscopy images of GFP vector control or mir mimicexpressing parental and DTP
Supplementary Figure 1
 Supplementary Figure 1 Constitutive EGFR signaling does not activate canonical EGFR signals (a) U251EGFRInd cells with or without tetracycline exposure (24h, 1µg/ml) were treated with EGF for 15 minutes
Supplementary Figure 1 Constitutive EGFR signaling does not activate canonical EGFR signals (a) U251EGFRInd cells with or without tetracycline exposure (24h, 1µg/ml) were treated with EGF for 15 minutes
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
<10. IL-1β IL-6 TNF + _ TGF-β + IL-23
 3 ns 25 ns 2 IL-17 (pg/ml) 15 1 ns ns 5 IL-1β IL-6 TNF
3 ns 25 ns 2 IL-17 (pg/ml) 15 1 ns ns 5 IL-1β IL-6 TNF
* * A3027. A4623 e A3507 A3507 A3507
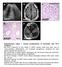 a c L A327 d e A37 A37 A37 Supplementary Figure 1. Clinical manifestations of individuals with mutations. (a) Renal ultrasound of right kidney in A327 reveals small renal cysts, loss of corticomedullary
a c L A327 d e A37 A37 A37 Supplementary Figure 1. Clinical manifestations of individuals with mutations. (a) Renal ultrasound of right kidney in A327 reveals small renal cysts, loss of corticomedullary
Supplementary Figure 1
 Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
