The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1
|
|
|
- Angela Lawson
- 5 years ago
- Views:
Transcription
1 The cytosolic exonuclease TREX inhibits the innate immune response to human immunodeficiency virus type Nan Yan, Ashton D Regalado-Magdos, Bart Stiggelbout, Min Ae Lee-Kirsch & Judy Lieberman Viral infection triggers innate immune sensors to produce type I interferon. However, infection of T cells and macrophages with human immunodeficiency virus (HIV) does not trip those alarms. How HIV avoids activating nucleic acid sensors is unknown. Here we found that the cytosolic exonuclease TREX suppressed interferon triggered by HIV. In Trex / mouse cells and human CD T cells and macrophages in which TREX was inhibited by RNA-mediated interference, cytosolic HIV DNA accumulated and HIV infection induced type I interferon that inhibited HIV replication and spreading. TREX bound to cytosolic HIV DNA and digested excess HIV DNA that would otherwise activate interferon expression via a pathway dependent on the kinase TBK, the adaptor STING and the transcription factor IRF. HIV-stimulated interferon production in cells deficient in TREX did not involve known nucleic acid sensors. Human immunodeficiency virus (HIV) introduces its single-stranded RNA (ssrna) genome into a cell in the reverse-transcription complex, which matures into the preintegration complex. This complex delivers reverse-transcribed HIV DNA to the nucleus for chromosomal integration. Few copies of HIV DNA integrate, leaving behind HIV DNA in the cytosol to be cleared by host enzymes. Although nucleic acids in the reverse-transcription complex might be shielded from nucleic acid sensors, viral DNA in the preintegration complex is accessible to exogenous endonucleases and thus is potentially accessible to cytosolic sensors of innate immunity. The endoplasmic reticulumassociated SET complex, which contains three DNases (APE, NM-H and TREX) and other proteins (SET, pp and HMGB), binds to the HIV preintegration complex and protects the integrase-activated DNA ends from self attack in suicidal autointegration. Suppressing the expression of any gene encoding a molecule of the SET complex increases autointegration and interferes with chromosomal integration. TREX is the most abundant - DNase in cells. Treatment with TREX-specific small interfering RNA (sirna) inhibits HIV replication more profoundly than do sirnas specific for other SET complex components, decreasing viral production by a log. TREX mutations are associated with inflammatory and autoimmune diseases, including Aicardi-Goutieres syndrome, chilblain lupus, and systemic lupus erythematosus, some of which have more type I interferon. TREX binds to transfected immunostimulatory DNA, and Trex / cells accumulate cytoplasmic DNA derived from endogenous retroelements, which activates interferon expression dependent on interferon-regulatory factor (IRF) 8. Like HIV, endogenous retroelements undergo cytoplasmic reverse transcription. We therefore investigated whether HIV might use TREX to avoid triggering antiviral innate immunity. RESULTS TREX inhibits interferon production in response to HIV We first compared the replication of HIV and the expression and secretion of type I interferons and inflammatory cytokines after infecting Trex / (wild-type) and Trex / mouse embryonic fibroblasts (MEFs) with vesicular stomatitis virus G (VSV-G)-pseudotyped HIV-luciferase virus, a single-round virus that does not produce infectious virus and has a nearly full-length HIV genome (lacking the gene encoding the envelope protein and with replacement of the gene encoding negative factor (Nef) with a gene encoding luciferase). This virus can infect MEFs, integrate and execute long terminal repeat (LTR)-driven expression of the luciferase reporter 9. After being infected with HIV-luciferase, Trex / MEFs had luciferase activity one tenth that of wild-type MEFs (Fig. a). Uninfected Trex / MEFs constitutively expressed slightly more interferon-β (IFN-β) mrna than did wild-type MEFs (Fig. b). In Trex / cells, HIV infection induced both IFN-β mrna (~-fold more than that of uninfected cells) and interleukin (IL-) mrna (~-fold more than that of uninfected cells; Fig. b,c). Neither IFN-β nor IL- was induced by HIV infection of wild-type MEFs. HIV did not induce IL-β, IFN-α or IFN-γ in wild-type or Trex / MEFs (data not shown). IFN-β was secreted, as assessed by enzyme-linked immunoassay (ELISA) of cultured supernatants (Fig. d). Nevirapine, which inhibits HIV Immune Disease Institute and Program in Cellular and Molecular Medicine, Children s Hospital, Boston, and Department of Pediatrics, Harvard Medical School, Boston, Massachusetts, USA. Children s Hospital, Technical University Dresden, Dresden, Germany. Correspondence should be addressed to J.L. (lieberman@idi.harvard.edu). Received 7 July; accepted 7 August; published online September ; doi:.8/ni.9 nature immunology VOLUME NUMBER NOVEMBER
2 a b c d Luc activity (% of ) IFN-β mrna (AU), HIV: Inhibitor: reverse transcription, inhibited the expression of IFN-β and IL- in response to HIV in Trex / cells, but the integrase inhibitor raltegravir, which acts after the synthesis of HIV DNA, did not (Fig. bf); this suggested that reverse-transcribed HIV DNA, rather than genomic RNA, was triggering the response. Treatment of the virus with DNase did not alter the IFN-β response to HIV-luciferase in Trex / cells (data not shown), which eliminated concerns that carryover of plasmid DNA was responsible for the induction of IFN-β. Virus-like particles lacking genomic RNA and heat-inactivated HIV also did not trigger IFN-β expression (Fig. g), which suggested that viral nucleic acid and an infectious virus were required. Because nonproductive autointegrated (autointegrant) DNA accumulates when TREX is inhibited by sirna, autointegrant DNA might have been triggering IFN-β. However, although only the reverse-transcription inhibitor suppressed IFN-β production, both the reverse-transcription inhibitor and integrase inhibitor blocked the production of autointegrants (Fig. h). These results suggest that the HIV-stimulated production of IFN-β in Trex / MEFs was activated by HIV DNA other than autointegrant DNA. HIV-stimulated interferon expression is IRF dependent IFN-β expression induced by transfected immunostimulatory DNA or endogenous retroelements in Trex / cells is mediated by the transcription factor IRF (refs.,7). To investigate whether IRF also activates HIV-induced expression of IFN-β, we compared IFN-β mrna and HIV infectivity in wild-type, Trex / and Trex / Irf / MEFs. Lack of IRF completely inhibited IFN-β induction (Fig. a). HIV-luciferase activity was also partially restored in Trex / Irf / cells (Fig. b). Therefore, the IFN-β induction in response to HIV in Trex / cells was mediated by IRF. Autointegration in the absence of Trex was indistinguishable in Trex / and Trex / Irf / cells (Fig. c), which suggested that autointegration is not altered by endogenous IFN-β production and that the two effects of TREX on HIV infection (blocking autointegration and inhibiting IFN-β induction) operate independently. Similarly, in human 9T embryonic kidney RT IN, HIV: Inhibitor: Figure TREX deficiency inhibits HIV replication and activates IFN-β in response to HIV infection. (a) Luciferase (Luc) activity in wild-type () and Trex / primary MEFs infected with VSV-Gpseudotyped HIV with luciferase reporter expression driven by the HIV LTR (as described in Results). (bd) Quantitative RT-PCR analysis (b,c) and ELISA (d) of cytokine induction in MEFs left uninfected (HIV ) or infected as described in a (HIV ) and left untreated ( ) or treated with inhibitors of reverse transcription (RT) or integrase (IN), assessed h after infection. Results are presented in arbitrary units (AU). (e,f) Luciferase activity (e) and late reverse transcripts (f) in primary MEFs infected as in a and left untreated (HIV) IL- mrna (AU) RT IN IFN-β protein (pg/ml) HIV: Inhibitor: f g, h Late RT (% of no drug) IFN-β mrna (AU) RT cells, TREX-specific sirna resulted in higher HIV-induced luciferase expression from the IFNB promoter, whereas sirnas specific for some other SET complex genes had no effect on IFN-β expression (Supplementary Fig. ). Therefore, protection from IFN-β activation involves TREX but not the entire SET complex. Activation of IRF triggers its nuclear translocation. To verify the role of IRF in HIV-induced IFN-β expression in Trex / cells, we monitored IRF localization by confocal microscopy. IRF was cytoplasmic in uninfected wild-type and Trex / MEFs (Fig. d). After infection with VSV-G-pseudotyped HIV expressing green fluorescent protein (HIV-GFP) at a multiplicity of infection (MOI) of, 8% of wild-type MEFs were infected, as assessed by GFP expression. At the same MOI, viral entry was similar in Trex / cells (assessed by quantitative RT-PCR for HIV genomic RNA; data not shown), but GFP expression was much lower (Fig. e). IRF remained cytoplasmic in % of wild-type cells but translocated into the nucleus in % of Trex / cells (Fig. d). These data confirm that the HIV-stimulated IFN-β response is IRF dependent and indicate the involvement of a cytosolic detection pathway. Likewise, these findings suggest that HIV DNA is not sensed by Toll-like receptor 9, which is contained in endosomes and activates type I interferon through IRF7 rather than IRF (refs.,). HIV reverse transcripts accumulate in Trex / cells To gain insight into how TREX suppresses HIV-stimulated IFN-β induction, we measured cytosolic HIV DNA and IFN-β mrna in wild-type and Trex / cells during infection with HIV-GFP (MOI = ; Fig. a). We also measured incoming HIV genomic RNA at h and h after infection to verify that wild-type and Trex / cells were infected with the same amount of virions (Fig. b). Cytosolic HIV DNA steadily increased for the first h after infection and then achieved a three- to fourfold higher plateau in Trex / cells than in wild-type cells. In wild-type cells, IFN-β mrna remained at baseline. In Trex / cells, IFN-β mrna was induced but lagged behind the accumulation of HIV DNA, first increasing h after infection. IN HIV HIV HIV HIV VLP Heat-inact RTin INin HIV e Autointegration (% of no drug) Luc activity (% of no drug) HIV HIV HIV RTin INin HIV HIV RTin or treated with inhibitors of reverse transcription (HIV RTin) or integrase (HIV INin), each added at the same time as HIV, assessed 8 h (e) or h (f) after infection. (g) IFN-β induction in MEFs infected with HIV (far left) or equivalent amounts (based on p ELISA) of virus-like particles (VLP) or HIV inactivated by heating for min at 9 C (Heat-inact HIV); results are presented in arbitrary units (AU). (h) HIV autointegration in primary MEFs infected and treated with inhibitors as in e,f. Data are representative of at least three independent experiments (error bars, s.d.). HIV INin VOLUME NUMBER NOVEMBER nature immunology
3 Trex a b c d / e IFN-β mrna (AU), KO DKO mirf DAPI Merge No HIV HIV KO DKO KO DKO Luc activity (% of ) Autointegration (% of ) HIV HIV HIV HIV Events (% of max) GFP MEF, no HIV-GFP MEF HIV-GFP KO MEF HIV-GFP Figure HIV-stimulated expression of interferon is IRF dependent. (a) Quantitative RT-PCR analysis of IFN-β induction in wild-type (), Trex / (KO) and Trex / Irf / (DKO) primary MEFs infected with VSV-G-pseudotyped HIV-GFP, assessed h after infection; results are presented in arbitrary units. (b) Luciferase activity of primary MEFs infected with HIV-luciferase, assessed 8 h after infection. (c) HIV autointegration in primary MEFs infected as in a. (d) Epifluorescence microscopy of the translocation of IRF to the nucleus in wild-type and Trex / MEFs left uninfected ( HIV) or infected with HIV ( HIV) and stained h later for IRF (red) and with the DNA-intercalating dye DAPI (blue). Original magnification,. (e) Flow cytometry analysis of GFP expression in wild-type and Trex / MEFs infected as in a, assessed h after infection. Data are representative of three experiments (a,b), at least three independent experiments (c; error bars (ac), s.d.) or two experiments (d), or are from one of three independent experiments (e). IFN-β mrna peaked h after infection and then plummeted to close to baseline, even though cytosolic HIV DNA remained high. The induction of IFN-β and accumulation of cytosolic HIV DNA increased in tandem in Trex / cells as the amount of virus used for infection increased, but even the highest viral dose (MOI = 8) did not stimulate IFN-β expression in wild-type cells (Fig. c,d). Although cytosolic HIV DNA was about tenfold more abundant in Trex / MEFs than in wild-type MEFs, HIV DNA integration was lower in Trex / cells than in wild-type cells (Fig. e), which suggested that most of the HIV DNA that accumulated in Trex / cells did not contribute to productive infection. HIV-stimulated IFN- from Trex / cells inhibits HIV Type I interferons inhibit the replication of most viruses by both cell-autonomous and noncell-autonomous effects and block both early and late stages of the HIV life cycle. To determine whether IFN-β (and potentially other cytokines) secreted during IFN-β MRNA (fold) HIV infection of Trex / cells suppressed new HIV infection, we infected wild-type MEFs with VSV-G-pseudotyped HIVluciferase in the presence of conditioned medium collected from wild-type, Trex / or Trex / Irf / MEFs infected with HIV-GFP or mock infected. Only conditioned medium from HIV-infected Trex / MEFs inhibited luciferase activity (by a factor of ; Fig. f). Preincubating the conditioned medium from Trex / cells with neutralizing antibody to mouse IFN-β abrogated the antiviral effect (Fig. g). Infection with HIV-luciferase was inhibited similarly in cells incubated with mouse IFN-β at a concentration of pg/ml or with conditioned medium from HIV-infected Trex / MEFs, which contained IFN-β at a concentration of pg/ml (Figs. d and h). These findings suggest that IFN-β is the main secreted antiviral factor. To pinpoint which stage(s) of HIV replication IFN-β blocks, we treated wild-type MEFs with mouse IFN-β and measured HIV DNA synthesis, two-ltr circle formation and integration. To assess a, HIV DNA b c d e f, IFN-β mrna. h MOI,. MOI,. 8 MOI,., HIV DNA h MOI, MOI, MOI,. MOI, 8 MOI, 8 7, IFN-β mrna MOI, MEF: KO KODKO DKO HIV: Time (h) Conditioned medium g h Figure Cytosolic HIV DNA in Trex / cells is the trigger for interferon expression. (a) Quantitative PCR analysis of HIV DNA and quantitative RT-PCR analysis of IFN-β mrna in wild-type and Trex / cells infected with HIV-GFP, presented relative to the peak value in Trex / cells. (b) Quantitative RT-PCR analysis of HIV genomic RNA (grna) at h and h after infection as in a, presented relative to the value in wild-type cells at h. (c,d) Quantitative RT-PCR analysis of IFN-β mrna (c) and quantitative PCR analysis of cytosolic HIV DNA (d) in wild-type and Trex / MEFs infected with HIV at an MOI of., or 8; results relative to those of wild-type MEFs infected at an MOI of.. (e) Two-step semiquantitative PCR assay 9 of integrated DNA in MEFs infected as in c,d. MEF: KO KO (f) Luciferase activity of wild-type MEFs treated with conditioned medium from wild-type, Trex / mifn-β nab: Purified mifn-β (pg/ml) or Trex / Irf / cells left uninfected (HIV ) or infected with HIV-GFP (HIV ) (below graph) and then infected with HIV-luciferase, assessed 8 h after infection and presented relative to the activity of cells incubated with medium alone (far left). (g) Luciferase activity of wild-type and Trex / MEFs treated with medium alone (far left; MEF, mifn-β nab ) or with conditioned medium (preincubated with () or without ( ) neutralizing antibody to mouse IFN-β (mifn-β nab)) and infected with HIV-luciferase. (h) Luciferase activity of wild-type MEFs infected with HIV-luciferase and treated with various concentrations (horizontal axis) of purified recombinant mouse IFN-β (mifn-β). P <. (Student s t-test). Data are representative of three independent experiments (error bars, s.d.). DNA or mrna (%) Incoming HIV grna (normalized) HIV DNA (fold) Integrated DNA (fold) Luc activity (% of medium alone) Luc activity (% of medium alone) Luc activity (% of no mifn-β) nature immunology VOLUME NUMBER NOVEMBER 7
4 KO GFP-TREX KO GFP-D8N Figure Recognition of HIV products of reverse transcription by enzymatically active TREX suppresses interferon induction. (a) Synthetic nucleic acids used in b as generated during reverse transcription. (b) Quantitative RT-PCR analysis of IFN-β expression by wild-type and Trex / cells mock-transfected (Mock) or transfected with the synthetic nucleic acids in a and assessed h later. (c) Quantitative PCR analysis of cytosolic DNA in wild-type and Trex / MEFs (left) or wild-type and TREX- mutant (TREX(D8N)) human fibroblasts (right) infected with HIV or transfected with ssdna and dsdna oligonucleotides of Gag-encoding sequence ( base pairs), assessed with primers for sequence encoding Gag h after infection or h after transfection. P <. (Student s t-test). (d) Immunoprecipitation (IP), with anti-flag or immunoglobulin G (IgG), of cytosolic lysates of HeLa-CD cells expressing Flag-tagged TREX, left uninfected (HIV ) or infected for h with HIV IIIB (HIV ), followed by quantitative PCR (qpcr) or quantitative ssrna Polymerase RNA:DNA hybrid RNase H ssdna Polymerase RT-PCR (qrt-pcr), respectively, of DNA and RNA extracted from the immunoprecipitates. P <. (Student s t-test). (e) Luciferase activity (left) of wild-type and Trex / MEFs left untransfected ( ) or transfected to express GFP alone (GFP) or GFP-tagged TREX, either wild-type (GFP-TREX) or an enzymatically inactive mutant (GFP-D8N), and then infected with HIV-luciferase, assessed 8 h after infection. P <. (Student s t-test). Right, immunoblot analysis of TREX, GFP and tubulin (Tub; loading control). GFP-TREX FL, full-length TREX (wild type or D8N) fused to GFP. (f) Quantitative PCR analysis of HIV DNA in wild-type and Trex / MEFs treated as in e; results are presented relative to those of mock-transfected wild-type MEFs. P <. (Student s t-test). Data are representative of three independent experiments (b,c,e; error bars, s.d.) or two independent experiments (d,f; error bars, s.d. of triplicates). LTR-mediated transcription, we incubated the well characterized cell line TZM-bl, which is derived from human HeLa-CD cervical cancer cells that have an integrated copy of an LTR-driven luciferase reporter gene, with human IFN-β. At the IFN-β concentration in conditioned medium from infected Trex / cells, IFN-β substantially blocked HIV integration and LTR-mediated transcription (Supplementary Fig. ). Higher doses of IFN-β blocked nearly all early stages of HIV replication in single-round infection. Thus, secreted IFN-β inhibits multiple steps in the early phase of HIV infection. TREX digestion of HIV DNA blocks interferon induction To identify which HIV nucleic acids trigger IFN-β expression, we transfected wild-type and Trex / MEFs with synthetic base pair oligonucleotides containing sequences from the HIV gene encoding the group-associated antigen (Gag) protein, which corresponded to HIV nucleic acids in the cytosol during reverse transcription (ssrna to represent genomic RNA, RNA-DNA hybrids, single-stranded DNA (ssdna) and double-stranded DNA (dsdna)), and measured IFN-β mrna h later by quantitative RT-PCR (Fig. a,b). All DNAcontaining oligonucleotides induced more IFN-β in Trex / MEFs than in wild-type MEFs, but ssrna did not. We found that ssdna elicited the largest difference (~-fold more IFN-β for ssdna compared with - to -fold more for dsdna and - to fourfold for RNA-DNA duplexes). None of the oligonucleotides induced IFN-β in Trex / Irf / MEFs (data not shown), which supported the idea of IRF s role in signaling the presence of these cytosolic nucleic acids. Although synthetic oligonucleotides may not completely mimic native HIV products of reverse transcription, these data are consistent with the known enzymatic preference of TREX for ssdna substrates rather than dsdna substrates. To determine whether transfected oligonucleotides accumulated in Trex / cells as HIV DNA does during infection, we quantified cytosolic DNA after transfection of ssdna or dsdna containing the sequence encoding Gag or after HIV infection. Cytosolic DNA was four- to sixfold more abundant in Trex / MEFs a b c IFN-β mrna (AU) MEFs Human fibroblasts 7 TREX (D8N) 8 Mock ssrna RNA:DNA ssdna dsdna HIV ssdna dsdna HIV ssdna dsdna Stimulus 7 GFP- TREX FL GFP Tub IP: Flag Flag IgG Flag Flag IgG MEF: KO KO KO KO MEF: KO KO KO HIV: DNA: GFP GFP GFP- GFP- DNA: Mock GFP- GFP- Mock GFP- GFP- HIV DNA HIV RNA TREXD8N TREXD8N TREX D8N (qpcr) (qrt-pcr) HIV DNA or RNA (% of input) dsdna d e f Luc activity (% ) Gag DNA (fold) GFP KO GFP than in wild-type MEFs when measured h after transfection or h after infection (Fig. c). We repeated this experiment with human fibroblasts derived from a patient with chilblain lupus that expressed either wild-type TREX or mutant TREX (with substitution of asparagine for aspartic acid at position 8 (D8N)). D8N is a substitution in a highly conserved Mg -binding site in the Exo domain of TREX that eliminates exonuclease activity and interferes with the enzymatic activity of the wild-typemutant TREX heterodimer,7. Cells expressing the D8N mutant also accumulated all DNA species introduced by infection or transfection (Fig. c). The accumulation of HIV DNA in Trex / or TREX mutant cells is consistent with a published report showing that the cytosol of Trex / cells has more ssdna derived from endogenous retroelements 7. These results suggest that TREX suppresses IFN-β induction by digesting cytosolic DNA. For further evidence that TREX is responsible for removing extraneous cytosolic HIV DNA, we assessed whether TREX interacted with HIV DNA during infection with wild-type HIV strain IIIB. We infected HeLa-CD cells expressing Flag-tagged TREX with HIV IIIB for h, then obtained cytosolic extracts of these cells and immunoprecipitated proteins with antibody to Flag (anti-flag) or immunoglobulin G (control antibody). We assessed enrichment for HIV DNA and RNA encoding Gag in the precipitates by quantitative PCR or quantitative RT-PCR, respectively. Anti-Flag immunoprecipitated threefold more HIV DNA than did the immunoglobulin G control, but there was no enrichment for HIV RNA (Fig. d), which confirmed that HIV DNA binds to TREX and is its preferred target. To determine whether the enzymatic activity of TREX is needed to enhance HIV infection, we assessed whether expression of wild-type or D8N TREX in Trex / cells could restore HIV-luciferase infectivity (Fig. e). Wild-type TREX partially restored HIV infection, but the enzymatically inactive D8N mutant had no effect, which indicated that TREX s exonuclease activity is needed for both inhibiting autointegration and blocking IFN-β induction. We also measured the accumulation of HIV DNA in wild-type and Trex / cells transfected with GFP-tagged HIV DNA (fold) 8 VOLUME NUMBER NOVEMBER nature immunology
5 wild-type or D8N TREX. As shown above (Fig. ), Trex / cells accumulated about five times more cytosolic HIV DNA than did wild-type cells. The excess HIV DNA was completely eliminated by expression of GFP-tagged wild-type TREX but not by expression of GFP-tagged D8N TREX (Fig. f). These results suggest that TREX metabolizes cytosolic HIV DNA. Because overexpressing GFP-tagged wild-type TREX did not diminish HIV DNA abundance below that in an infected wild-type cell, TREX may not have access to all HIV DNA products. HIV activates interferon in TREX-deficient human cells Most experiments in this study reported above used mouse Trex / cells to take advantage of their complete lack of TREX, in contrast to the incomplete inhibition afforded by sirnas. To investigate whether our findings were physiologically relevant during HIV infection of primary human immune cells, we used sirna to suppress TREX alone or both TREX and IRF in monocyte-derived macrophages (MDMs) from two human donors. At d after sirna treatment, we infected sirna-treated cells, which had % less TREX and/or IRF mrna than did cells treated with control sirna, with the BaL strain of HIV (Fig. ac). At h after infection, cytosolic HIV DNA was about fourfold higher in samples treated with sirna specific for TREX or for both TREX plus IRF than in samples treated with control sirna. HIV replication and spreading in cultures of macrophages treated with TREX-specific sirna was one fourth to one half that in control cells, as assessed by measurement of HIV Gag p antigen in the medium. When expression of both IRF a TREX mrna MDM b TREX TREX IRF sirna HIV BaL transfection infection Days 7 Confirm knockdown Donor Donor sirna IRF mrna TREX TREX IRF sirna Donor Donor c HIV DNA TREX TREX IRF sirna d p released (ng/ml) ND ND ND TREX TREX IRF TREX TREX IRF TREX TREX IRF TREX TREX IRF Day Day Day 7 Day Figure TREX-specific sirna treatment induces IFN-α and IFN-β and inhibits HIV replication in primary human immune cells. (a) Experimental procedure: human MDMs were transfected with control sirna or sirna specific for TREX alone or TREX plus IRF and infected d later with HIV (BaL strain). (b) Quantitative RT-PCR analysis of TREX and IRF mrna in MDMs from two different donors ( and ), treated as outlined in a and assessed d after sirna transfection; results are relative to those of cells treated with control () sirna. (c) Cytosolic HIV DNA in MDMs treated as outlined in a, presented relative to results of cells treated with control sirna. (d) ELISA of p released into culture supernatants of MDMs treated as outlined in a, assessed on days after infection. ND, not determined (not measured on day ). (e) Quantitative RT-PCR analysis of IFN-α and IFN-β mrna in MDMs left uninfected (left) or treated as outlined in a, assessed on days, or 7 after infection and presented relative to the control values on day. Flow cytometry analysis of p immunostaining indicated that % and TREX was suppressed, HIV replication was partially restored (Fig. d), which suggested that IRF-dependent induction of IFN-β contributed to the inhibition of HIV replication caused by TREX- specific sirna. Consistent with that idea, expression of both IFN-α and IFN-β mrna increased up to tenfold in macrophages treated with TREX-specific sirna, but not in those treated with control sirna or with TREX-specific and IRF-specific sirnas (Fig. e). We obtained similar results after transfection with TREX-specific sirna into human peripheral blood CD T cells infected with HIV IIIB (Fig. fl). In T cells treated with TREX-specific sirna, HIV DNA accumulated in the cytosol and the expression of both IFN-α and IFN-β was induced. TREX-specific sirna similarly resulted in less HIV production, as measured by flow cytometry analysis of intracellular p. Both the number of p cells and p mean fluorescence intensity were lower. Therefore, TREX deficiency resulted in less HIV replication and spreading in culture. The magnitude of these effects increased with the amount of sirna transfected and the extent of TREX suppression. Therefore, during infection of primary human cells with wild-type HIV, TREX suppresses HIV-induced IFN-β activation through IRF to promote HIV replication. HIV DNA signals via STING, TBK and IRF To investigate the pathway triggered by HIV DNA, we treated Trex / MEFs with sirnas targeting selected genes linked to DNA-stimulated induction of interferon and examined the effect on HIV-stimulated IFN-β expression (Fig. a,b). Inhibiting a gene e IFN-α mrna IFN-β mrna h 8 7 HIV: sirna: IFN-α mrna (fold) TREX TREX IRF Uninfected p (%) 7 TREX Mock k TREX TREX IRF Day IFN-β mrna (fold) i HIV: sirna: TREX TREX TREX IRF Day 8 TREX TREX IRF HIV: sirna: Day 7 l Mock f TREX mrna Mock TREX HIV: sirna: TREX p MFI Cells TREX sirna j g HIV DNA (fold) 8 Mock. No HIV.7 sirna p-fitc TREX sirna TREX sirna of cells were infected by day 7 (data not shown). P <. (Student s t-test). (f) TREX mrna in CD T cells positively selected from peripheral blood mononuclear cells, transfected with no sirna (Mock), control sirna ( pmol) or TREX-specific sirna (, or pmol per cells) and infected d later with HIV IIIB; mrna measured d after transfection is presented relative to that in cells treated with control sirna. (g) Cytosolic HIV DNA in the cells in f, assessed h after infection. (h,i) IFN-α mrna (h) and IFN-β mrna (i) in the cells in f, measured h after infection and presented relative to results obtained with control cells. (jl) Flow cytometry analysis of HIV IIIB replication in CD T cells treated with control or TREX-specific sirna and then left uninfected (No HIV) or infected with HIV IIIB, and assessed d after infection as percent p cells (j; numbers above bracketed lines), frequency of p cells (k) and the mean fluorescence intensity (MFI) of p (l). FITC, fluorescein isothiocyanate. P <. (Student s t-test). Data are representative of two experiments ((be); error bars, s.d. (b) or average and s.d. of two replicates from two independent donors (d,e)) or experiments (d; error bars, s.d.) or are from two independent experiments (fl; error bars, s.d. of six replicates). nature immunology VOLUME NUMBER NOVEMBER 9
6 Figure HIV-stimulated interferon induction requires IRF, TBK and STING. (a) Quantitative RT-PCR analysis of genes related to innate immunity in Trex / MEFs 8 h after transfection with control or gene-specific sirna. Ddx8 encodes RIG-I; Tmem7 encodes STING. P <. (Student s t-test). (b) Quantitative RT-PCR analysis of IFN-β mrna in wild-type and Trex / MEFs left untransfected (sirna ) or transfected with control or gene-specific sirna (sirna ), a sirna b c Gene sirna HIV: HIV: sirna: sirna: Target mrna inhibition (% of ctrl) Tlr9 Ddx8 Aim Lrrfip Mavs Tmem7 Irf Tbk Hmgb Tlr9 Ddx8 Aim Lrrfip Mavs Tmem7 Irf Tbk Hmgb Ddx8 Sting Irf Tbk Hmgb and left uninfected (HIV ) or infected with VSV-G-pseudotyped HIV (HIV ), assessed h after infection; results are presented relative to those of untransfected wild-type MEFs. P <. (Student s t-test). (c) Quantitative PCR analysis of cytosolic HIV DNA in Trex / MEFs transfected with control or gene-specific sirna, assessed h after infection with HIV-GFP; results are presented relative to those of infected Trex / MEFs transfected with control sirna. Data are representative of three independent experiments (error bars, s.d.). IFN-β mrna (fold) HIV DNA (fold) required for HIV-stimulated interferon signaling in Trex / cells should result in lower IFN-β expression. As expected, treatment with Irf-specific sirna resulted in less HIV-stimulated IFN-β mrna. Similarly, sirna targeting Tbk, which encodes an IRF kinase, also inhibited HIV-stimulated expression of IFN-β (Fig. b). We found that sirna specific for genes encoding the DNA sensors TLR9 (ref. 8), AIM (ref. 9), LRRFIP (ref. ) or HMGB (ref. ), or encoding RIG-I (which recognizes RNA transcribed from cytosolic DNA, ) or its adaptor MAVS (also known as IPS-, VISA and CARDIF) 7, did not suppress HIV-stimulated expression of IFN-β, which suggested that an unknown sensor detects HIV DNA or that multiple known DNA sensors might function redundantly. Additional experiments confirmed that the RNA polymerase III RIG-IMAVS DNA-detection pathway, was not involved in HIV-stimulated induction of interferon (Supplementary Fig. ). However, sirna specific for the gene encoding another membraneassociated adaptor, STING (also known as MITA and ERIS 8 ), which has been identified as mediating innate immune responses to cytosolic DNA 8,9, inhibited HIV-stimulated expression of IFN-β (Fig. b). STING is phosphorylated by the kinase TBK (refs. 9,). We found that sirna specific for the genes encoding STING, IRF or TBK did not affect cytosolic accumulation of HIV DNA in Trex / MEFs (Fig. c), which suggested that they act downstream of DNA sensing. Thus, HIV DNA is detected by a pathway that signals through STING, TBK and IRF and does not involve any known DNA sensor (Supplementary Fig. ). HMGB, but not its homolog HMGB, associates with TREX in the cytosolic SET complex. Treatment of HIV-infected wild-type and Trex / MEFs with Hmgb-specific sirna enhanced IFN-β expression (Fig. b), which suggested that HMGB inhibits the response to cytosolic HIV DNA. However, treatment with Hmgb-specific sirna did not result in more cytosolic HIV DNA in Trex / MEFs than in those treated with control sirna (Fig. c), which suggested that HMGB might act downstream of the recognition of HIV DNA. HMGB proteins repress the transcription of many genes, including proinflammatory genes, such as TNF. To determine whether HMGB regulates IFNB transcription, we transfected HMGB- specific sirna into 9T cells expressing a luciferase reporter plasmid driven by the IFNB promoter. IFN-βluciferase expression was about twofold higher in cells treated with HMGB-specific sirna than in those treated with control sirna in response to poly(da:dt) or MAVS overexpression (Supplementary Fig. ), which suggested that HMGB inhibits IFNB expression, either directly or indirectly, though its promoter, but acts downstream of DNA sensing. To determine whether HMGB has a role in the removal of cytosolic HIV DNA or HIV-stimulated expression of interferon in primary human MDMs, we suppressed HMGB alone or together with TREX. HMGB-specific sirna on its own did not result in more cytosolic HIV DNA. However, unlike results obtained with Trex / MEFs, treatment of human macrophages with HMGB- and TREX-specific sirna resulted in significantly more accumulation of cytosolic HIV DNA than did treatment with TREX-specific sirna alone (P <.). Treatment with HMGB-specific sirna led to HIV-stimulated expression of IFN-α and IFN-β, but did not induce interferon expression in uninfected cells (data not shown); this was significantly greater than that induced by TREX-specific sirna alone and was not enhanced by treatment with both sirnas (P <.). These results suggest that HMGB may act together with TREX to remove HIV DNA from the cytoplasm (model, Supplementary Fig. ). Further work is needed to elucidate the role of HMGB in suppressing HIV DNAstimulated induction of interferon, but these results suggest that HMGB might act at multiple points (by recognizing and/or helping eliminate cytosolic DNA and suppressing the IFNB promoter). DISCUSSION HIV infection of its main target cells, macrophages and CD T cells, does not induce cell-autonomous interferons. We have shown here that the host cytoplasmic exonuclease TREX helps HIV evade innate immunity by digesting reverse transcripts that are not imported into the nucleus and would otherwise induce interferons. When TREX was inhibited by RNAi, HIV infection of primary cells triggered expression and secretion of type I interferon. The HIV-stimulated interferon response in cells deficient in TREX, like the response to endogenous retroelement DNA and transfected DNA 7, was IRF dependent. We found that interferon induction in TREX-deficient cells was blocked by the expression of enzymatically active TREX or by cosuppression of IRF expression to interrupt interferon signaling. HIV-stimulated innate immune signaling also required STING and TBK. On the basis of our sirna experiments, we conclude that none of the known DNA sensors was involved. Therefore, our working model of the innate immune pathway activated by cytosolic HIV DNA starts with an unknown sensor (that may preferentially recognize ssdna) that signals through STING, TBK and IRF to activate interferon expression. Type I interferons inhibit HIV replication at multiple steps in the early phase of its life cycle and thereby suppress viral spreading. Failure to induce antiviral interferons in infected T cells and macrophages may promote transmission by allowing the virus to spread from the initial nidus of infection to neighboring cells in genital tissue. However, assessing the importance of TREX in transmission will require efficient methods for inhibiting TREX expression in vivo in the immune cells that HIV infects. Such methods are not yet available but are being developed. We used an MOI of to achieve a reasonable frequency of infected cells. As cytoplasmic accumulation VOLUME NUMBER NOVEMBER nature immunology
7 of and interferon triggering by HIV DNA depend on the MOI, the physiological relevance of our results to HIV transmission hinges on what viral concentrations are achieved in vivo, which is unknown. Viral concentrations may reach a high MOI locally during the replicative burst that occurs in the genital tract during transmission, when a strong intrinsic antiviral immune response might prevent dissemination 7. Other settings of high viral concentration might be activated lymph nodes or gut-associated lymphoid tissues. We did not determine whether TREX affects interferon production by plasmacytoid DCs, the main source of type I interferon during HIV infection. HIV replication is inefficient in DCs. Interferon stimulation in plasmacytoid DCs seems to be triggered mostly by endocytosed virus, whose genomic RNA is recognized by Toll-like receptor 7 in endosomes 8. Productive HIV infection of macrophages and T cells, however, involves fusion of the viral membrane with the cell membrane and direct uncoating of the viral capsid into the cytosol, bypassing the endosomal compartment and Toll-like receptor signaling. Nonetheless, it will be important to determine whether TREX modulates interferon signaling in plasmacytoid DCs. We found that HIV-stimulated activation of interferon in Trex / cells was eliminated by treatment of infected cells with an inhibitor of reverse transcription but not by treatment with an integrase inhibitor, which suggests that HIV DNA, not genomic RNA, is the nucleic acid that triggers innate immunity. The nucleic acid most sensitive to TREX activity is ssdna, and therefore ssdna is probably its main substrate. IFN-β mrna is normally detected 8 h after transfection of immunostimulatory DNA or infection with a DNA virus,,9. After HIV infection, interferon mrna is not detected until h after infection; the lag in IFN-β expression is probably due to the time needed to complete reverse transcription. The rapid decrease in IFN-β mrna expression after it reaches its peak value suggests that a cell-autonomous secondary mechanism tempers the innate immune response that, if unchecked, could be harmful to the host. Some HIV DNA accumulated in the cytoplasm of HIV-infected cells, even when TREX was normally expressed, but it did not activate interferon expression. A cytoplasmic DNA threshold, which might vary in different cell types, may need to be exceeded to trigger innate immunity. HMGB proteins have been proposed as innate immune sentinel proteins that facilitate nucleic acid recognition by sensors of RNA and DNA. Here, experiments with HMGB-specific sirna demonstrated the opposite effect: HMGB helped suppress interferon induction by HIV in human cells. Although RNAi of Hmgb in Trex / MEFs did not result in more cytosolic HIV DNA, RNAi of HMGB in human macrophages that were also treated with TREX-specific sirna resulted in enhanced cytosolic HIV DNA and induction of IFN-α and IFN-β. Therefore, HMGB proteins may have a more complex role in innate immunity than originally suggested. In their role as sentinels for foreign nucleic acids, they may facilitate the recognition of nucleic acids both by sensors that trigger innate immune responses and by proteins, such as TREX, that inhibit interferon induction. Therefore, the net effect of a lower abundance of HMGB proteins could be either to inhibit interferon induction (as reported before ) or to enhance it, as shown here. We also found that HMGB acted downstream of nucleic acid sensing at the IFNB promoter to suppress IFNB transcription, which adds another layer of complexity. This transcriptional effect extended to non-hiv innate immune stimuli (the synthetic dsdna poly(da:dt) and overexpression of MAVS). In the study noted above, IFN-β expression stimulated by poly(da:dt) was lower in Hmgb / MEFs than in wild-type MEFs, whereas we found the opposite effect with RNAi of Hmgb. The apparent discrepancy between those results and ours could be due to a difference in the consequences of complete or partial Hmgb elimination, especially if HMGB operates at multiple steps in interferon induction. In many of our experiments we used genetically deficient mouse cells to demonstrate that TREX helps HIV evade detection by the innate immune system and to define the HIV DNAstimulated interferon signaling pathway. Knockout mouse cells are powerful tools for HIV research 9,,. Once the block in the entry into mouse cells is overcome by VSV-G pseudotyping, most early steps of HIV replication, including reverse transcription, integration and LTR-driven transcription, are similar in human and mouse cells. Furthermore, human TREX is 7.% identical to its mouse homolog in amino acid sequence, and is 7.%, % and 8.7% identical to its mouse homolog in its three exonuclease motifs. Human and mouse TREX have the same enzymatic activity and can substitute for each other. Therefore, mouse cells are well suited for the study of TREX function in HIV replication. Nonetheless, human immune cells susceptible to HIV can differ from MEFs in their ability to activate innate immune pathways. For example, IFN-α was induced by HIV in human immune cells but not in MEFs. The differences in the role of HMGB in the accumulation of HIV DNA in human macrophages and MEFs may be another case in point. We confirmed all key findings in primary human cells, including HIV DNA accumulation, interferon induction and inhibition of HIV replication when TREX was inhibited by RNAi, and efficient rescue by cosuppression of IRF expression. Our data shed light on the fate of nonproductive or nonintegrated HIV DNA in the cell. At an MOI of, HIV infection produced many reverse transcripts (although only one per incoming genomic RNA), but very few copies managed to integrate into the host chromosome. The remaining HIV DNA was cleared by TREX, as cytosolic HIV DNA built up when TREX function was deficient or inhibited and was removed after expression of enzymatically active TREX. As a consequence, wild-type TREX fully restored HIV infectivity in Trex / MEFs, and the D8N mutant failed to do so. Other host nucleases might also help digest cytosolic HIV DNA. It is unclear why the excess HIV DNA that accumulated in Trex / cells did not lead to more chromosomal integration. Sequencing these excess HIV DNAs may show whether they are able to integrate and what prevents them from integrating. These excess HIV DNAs may be mostly nonproductive products of reverse transcription. TREX promotes HIV replication in the following two ways: it inhibits autointegration, and it suppresses the interferon response. Several models might explain the dual effects of TREX on HIV DNA. One possibility is that TREX might sort productive versus nonproductive HIV products of reverse transcription. Reverse transcription of HIV is error prone and often produces incomplete products. TREX recognizes ssdna or dsdna with single-strand overhangs, the kind of DNA in failed products of reverse transcription. TREX might bind to HIV DNA nonspecifically in the cytosol but as an exonuclease can only efficiently digest HIV DNA that contains broken ends or single-strand overhangs. HIV integrase binds to the ends of reverse transcripts that are capable of chromosomal integration and might protect them from digestion by TREX. Incomplete products of reverse transcription, however, would not bind integrase and therefore would be susceptible to degradation by TREX. Autointegration requires the full-length product of reverse transcription and active DNA ends bound by integrase, which catalyzes autointegration. Although TREX probably binds to full-length integration-competent products, as well as to transcripts that are not competent for integration, its exonuclease activity might be inhibited nature immunology VOLUME NUMBER NOVEMBER
8 in the full-length transcript by lack of some DNA feature that facilitates digestion (such as shielding by integrase). Another possibility is that TREX is inhibited by components of the SET complex that also bind to the HIV preintegration complex. Another DNase in the SET complex, NM-H, is inhibited by SET protein and is activated only when granzyme A cleaves SET. TREX is an abundant protein that is not exclusively present in the SET complex. Two subpopulations of TREX could be involved in different actions: the SET complexassociated TREX inhibits autointegration, whereas TREX outside the SET complex is enzymatically active and removes excess HIV DNA. This model would also explain why sirnas directed against genes encoding most other molecules of the SET complex do not induce interferon but do protect against autointegration. Further studies are needed to test these ideas. Mutations in TREX that interfere with the enzymatic function or localization of TREX are associated with systemic lupus erythematosus and other autoimmune and/or inflammatory diseases. Patients with systemic lupus erythematosus are underrepresented in HIV-infected populations. It would be worth evaluating whether TREX polymorphisms or autoimmune diseases are associated with less HIV transmission or a more benign disease course. The innate immune pathway identified in this study will improve understanding of how HIV intersects with innate immunity and may also shed light on autoimmune and inflammatory syndromes linked to TREX mutation. Methods Methods and any associated references are available in the online version of the paper at Note: Supplementary information is available on the Nature Immunology website. Acknowledgments We thank D. Stetson (University of Washington) for wild-type, Trex / and Trex / Irf / primary MEFs, under agreement with D. Barnes and T. Lindahl (Cancer Research UK); J. Jung (University of Southern California) for RIG-Ideficient (Ddx8 / ) MEFs; S. Harvey and F. Perrino (Wake Forest University) for wild-type and Trex / transformed MEFs; A. Engelman (Dana-Farber Cancer Institute) for the HIV-luciferase plasmid (pnl-/env ); D. Gabuzda (Dana- Farber Cancer Institute) for the HIV-GFP plasmid (pnl-/env ); T. Fujita (Kyoto University) for antiserum to mouse IRF; S. Nagata (Kyoto University) for the hemagglutinin-tagged MAVS plasmid; K. Fitzgerald (University of Massachusetts) for sirna and inhibitors of RNA polymerase III; J. Hiscott (McGill University) for IFNB-luciferase plasmid; L. Gehrke (Harvard Medical School) for CMV renilla luciferase plasmid; and members of the Lieberman lab for discussions. Supported by the US National Institutes of Health (AI87 to J.L., and T HL987 to N.Y.), the Harvard Center for AIDS Research (N.Y.), Harvard Summer Honors Undergraduate Research Program (A.D.R.-M.) and Deutsche Forschungsgemeinschaft (Le 7/- to M.A.L.-K.). AUTHOR CONTRIBUTIONS N.Y. conceived of the study, designed and did most experiments and helped write the paper; A.D.R.-M. and B.S. helped do the experiments; M.A.L.-K. provided human cell lines and scientific advice; and J.L. conceived of and supervised the study and helped write the paper. COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests. Published online at Reprints and permissions information is available online at reprintsandpermissions/.. Bowerman, B., Brown, N., Bishop, K. & Varmus, H. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev., 978 (989).. Yan, N., Cherepanov, P., Daigle, J.E., Engelman, A. & Lieberman, J. The SET complex acts as a barrier to autointegration of HIV-. PLoS Pathog., e7 (9).. Lee-Kirsch, M.A. et al. A mutation in TREX that impairs susceptibility to granzyme A- mediated cell death underlies familial chilblain lupus. J. Mol. Med. 8, 7 (7).. Lee-Kirsch, M.A. et al. Mutations in the gene encoding the - DNA exonuclease TREX are associated with systemic lupus erythematosus. Nat. Genet. 9, 7 (7).. Crow, Y. et al. Mutations in the gene encoding the - DNA exonuclease TREX cause Aicardi-Goutieres syndrome at the AGS locus. Nat. Genet. 8, 979 ().. Stetson, D.B. & Medzhitov, R. Recognition of cytosolic DNA activates an IRF- dependent innate immune response. Immunity, 9 (). 7. Stetson, D.B., Ko, J.S., Heidmann, T. & Medzhitov, R. Trex prevents cell-intrinsic initiation of autoimmunity. Cell, 8798 (8). 8. Yang, Y., Lindahl, T. & Barnes, D. Trex exonuclease degrades ssdna to prevent chronic checkpoint activation and autoimmune disease. Cell, 8788 (7). 9. Shun, M. et al. LEDGF/p7 functions downstream from preintegration complex formation to effect gene-specific HIV- integration. Genes Dev., (7).. Honda, K. et al. IRF-7 is the master regulator of type-i interferon-dependent immune responses. Nature, ().. Kawai, T. et al. Interferon-α induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF. Nat. Immunol., 8 ().. Agy, M.B., Acker, R.L., Sherbert, C.H. & Katze, M.G. Interferon treatment inhibits virus replication in HIV-- and SIV-infected CD T-cell lines by distinct mechanisms: evidence for decreased stability and aberrant processing of HIV- proteins. Virology, 798 (99).. Coccia, E.M., Krust, B. & Hovanessian, A.G. Specific inhibition of viral protein synthesis in HIV-infected cells in response to interferon treatment. J. Biol. Chem. 9, 879 (99).. Shirazi, Y. & Pitha, P.M. Alpha interferon inhibits early stages of the human immunodeficiency virus type replication cycle. J. Virol., 8 (99).. Baca-Regen, L., Heinzinger, N., Stevenson, M. & Gendelman, H.E. Alpha interferoninduced antiretroviral activities: restriction of viral nucleic acid synthesis and progeny virion production in human immunodeficiency virus type -infected monocytes. J. Virol. 8, 797 (99).. Mazur, D. & Perrino, F. Identification and expression of the TREX and TREX cdna sequences encoding mammalian exonucleases. J. Biol. Chem. 7, 99 (999). 7. Lehtinen, D.A., Harvey, S., Mulcahy, M.J., Hollis, T. & Perrino, F.W. The TREX double-stranded DNA degradation activity is defective in dominant mutations associated with autoimmune disease. J. Biol. Chem. 8, 9 (8). 8. Hornung, V. & Latz, E. Intracellular DNA recognition. Nat. Rev. Immunol., (). 9. Schroder, K., Muruve, D.A. & Tschopp, J. Innate immunity: cytoplasmic DNA sensing by the AIM inflammasome. Curr. Biol. 9, RR (9).. Yang, P. et al. The cytosolic nucleic acid sensor LRRFIP mediates the production of type I interferon via a β-catenin-dependent pathway. Nat. Immunol., 879 ().. Yanai, H. et al. HMGB proteins function as universal sentinels for nucleic-acidmediated innate immune responses. Nature, 99 (9).. Chiu, Y.-H., Macmillan, J.B. & Chen, Z.J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 8, 79 (9).. Ablasser, A. et al. RIG-I-dependent sensing of poly(da:dt) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol., 7 (9).. Seth, R.B., Sun, L., Ea, C.-K. & Chen, Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF. Cell, 98 ().. Kawai, T. et al. IPS-, an adaptor triggering RIG-I- and Mda-mediated type I interferon induction. Nat. Immunol., ().. Xu, L.-G. VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 9, 777 (). 7. Meylan, E. et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 7, 77 (). 8. Ishikawa, H. & Barber, G. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature, 778 (8). 9. Zhong, B. et al. The adaptor protein MITA links virus-sensing receptors to IRF transcription factor activation. Immunity 9, 8 (8).. Sun, W. et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA, 888 (9).. Ishikawa, H., Ma, Z. & Barber, G.N. STING regulates intracellular DNAmediated, type I interferon-dependent innate immunity. Nature, (9).. Fan, Z., Beresford, P., Zhang, D. & Lieberman, J. HMG interacts with the nucleosome assembly protein SET and is a target of the cytotoxic T-lymphocyte protease granzyme A. Mol. Cell. Biol., 88 (). VOLUME NUMBER NOVEMBER nature immunology
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Intrinsic cellular defenses against virus infection
 Intrinsic cellular defenses against virus infection Detection of virus infection Host cell response to virus infection Interferons: structure and synthesis Induction of antiviral activity Viral defenses
Intrinsic cellular defenses against virus infection Detection of virus infection Host cell response to virus infection Interferons: structure and synthesis Induction of antiviral activity Viral defenses
Nature Immunology doi: /ni Supplementary Figure 1. Raf-1 inhibition does not affect TLR4-induced type I IFN responses.
 Supplementary Figure 1 Raf-1 inhibition does not affect TLR4-induced type I IFN responses. Real-time PCR analyses of IFNB, ISG15, TRIM5, TRIM22 and APOBEC3G mrna in modcs 6 h after stimulation with TLR4
Supplementary Figure 1 Raf-1 inhibition does not affect TLR4-induced type I IFN responses. Real-time PCR analyses of IFNB, ISG15, TRIM5, TRIM22 and APOBEC3G mrna in modcs 6 h after stimulation with TLR4
Supplementary Figure 1
 Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Innate Immunity & Inflammation
 Innate Immunity & Inflammation The innate immune system is an evolutionally conserved mechanism that provides an early and effective response against invading microbial pathogens. It relies on a limited
Innate Immunity & Inflammation The innate immune system is an evolutionally conserved mechanism that provides an early and effective response against invading microbial pathogens. It relies on a limited
Intracellular MHC class II molecules promote TLR-triggered innate. immune responses by maintaining Btk activation
 Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
October 26, Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh-
 1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
MedChem 401~ Retroviridae. Retroviridae
 MedChem 401~ Retroviridae Retroviruses plus-sense RNA genome (!8-10 kb) protein capsid lipid envelop envelope glycoproteins reverse transcriptase enzyme integrase enzyme protease enzyme Retroviridae The
MedChem 401~ Retroviridae Retroviruses plus-sense RNA genome (!8-10 kb) protein capsid lipid envelop envelope glycoproteins reverse transcriptase enzyme integrase enzyme protease enzyme Retroviridae The
Fayth K. Yoshimura, Ph.D. September 7, of 7 HIV - BASIC PROPERTIES
 1 of 7 I. Viral Origin. A. Retrovirus - animal lentiviruses. HIV - BASIC PROPERTIES 1. HIV is a member of the Retrovirus family and more specifically it is a member of the Lentivirus genus of this family.
1 of 7 I. Viral Origin. A. Retrovirus - animal lentiviruses. HIV - BASIC PROPERTIES 1. HIV is a member of the Retrovirus family and more specifically it is a member of the Lentivirus genus of this family.
Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay
 Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay Background ImQuest BioSciences has developed and qualified a single-plate method to expedite the screening of antiviral agents against
Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay Background ImQuest BioSciences has developed and qualified a single-plate method to expedite the screening of antiviral agents against
Reviewers' comments: Reviewer #1 (Remarks to the Author):
 Reviewers' comments: Reviewer #1 (Remarks to the Author): In this manuscript, Song et al. identified FBXW7 as a new positive regulator for RIG-Itriggered type I IFN signaling pathway. The authors observed
Reviewers' comments: Reviewer #1 (Remarks to the Author): In this manuscript, Song et al. identified FBXW7 as a new positive regulator for RIG-Itriggered type I IFN signaling pathway. The authors observed
Crucial role for human Toll-like receptor 4 in the development of contact allergy to nickel
 Supplementary Figures 1-8 Crucial role for human Toll-like receptor 4 in the development of contact allergy to nickel Marc Schmidt 1,2, Badrinarayanan Raghavan 1,2, Verena Müller 1,2, Thomas Vogl 3, György
Supplementary Figures 1-8 Crucial role for human Toll-like receptor 4 in the development of contact allergy to nickel Marc Schmidt 1,2, Badrinarayanan Raghavan 1,2, Verena Müller 1,2, Thomas Vogl 3, György
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information. Supplementary Figure 1
 Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Introduction retroposon
 17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
7.012 Quiz 3 Answers
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
Medical Virology Immunology. Dr. Sameer Naji, MB, BCh, PhD (UK) Head of Basic Medical Sciences Dept. Faculty of Medicine The Hashemite University
 Medical Virology Immunology Dr. Sameer Naji, MB, BCh, PhD (UK) Head of Basic Medical Sciences Dept. Faculty of Medicine The Hashemite University Human blood cells Phases of immune responses Microbe Naïve
Medical Virology Immunology Dr. Sameer Naji, MB, BCh, PhD (UK) Head of Basic Medical Sciences Dept. Faculty of Medicine The Hashemite University Human blood cells Phases of immune responses Microbe Naïve
S1a S1b S1c. S1d. S1f S1g S1h SUPPLEMENTARY FIGURE 1. - si sc Il17rd Il17ra bp. rig/s IL-17RD (ng) -100 IL-17RD
 SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
AGAINST VIRAL INFECTIONS. Identify the types of immunity involve in the mechanisms of protection against viral infections.
 LECTURE: 02 Title: THE IMMUNOLOGICAL PROTECTIVE MECHANISMS AGAINST VIRAL INFECTIONS LEARNING OBJECTIVES: The student should be able to: Identify the types of immunity involve in the mechanisms of protection
LECTURE: 02 Title: THE IMMUNOLOGICAL PROTECTIVE MECHANISMS AGAINST VIRAL INFECTIONS LEARNING OBJECTIVES: The student should be able to: Identify the types of immunity involve in the mechanisms of protection
Under the Radar Screen: How Bugs Trick Our Immune Defenses
 Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 7: Cytokines Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research HHV-8 Discovered in the 1980 s at the
Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 7: Cytokines Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research HHV-8 Discovered in the 1980 s at the
7.012 Problem Set 6 Solutions
 Name Section 7.012 Problem Set 6 Solutions Question 1 The viral family Orthomyxoviridae contains the influenza A, B and C viruses. These viruses have a (-)ss RNA genome surrounded by a capsid composed
Name Section 7.012 Problem Set 6 Solutions Question 1 The viral family Orthomyxoviridae contains the influenza A, B and C viruses. These viruses have a (-)ss RNA genome surrounded by a capsid composed
Trex1 regulates lysosomal biogenesis and interferon-independent activation of antiviral genes
 Trex1 regulates lysosomal biogenesis and interferon-independent activation of antiviral genes Maroof Hasan 1,, James Koch 1,, Dinesh Rakheja 3, Asit K Pattnaik,, James Brugarolas 1,,7, Igor Dozmorov, Beth
Trex1 regulates lysosomal biogenesis and interferon-independent activation of antiviral genes Maroof Hasan 1,, James Koch 1,, Dinesh Rakheja 3, Asit K Pattnaik,, James Brugarolas 1,,7, Igor Dozmorov, Beth
Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
Viral Genetics. BIT 220 Chapter 16
 Viral Genetics BIT 220 Chapter 16 Details of the Virus Classified According to a. DNA or RNA b. Enveloped or Non-Enveloped c. Single-stranded or double-stranded Viruses contain only a few genes Reverse
Viral Genetics BIT 220 Chapter 16 Details of the Virus Classified According to a. DNA or RNA b. Enveloped or Non-Enveloped c. Single-stranded or double-stranded Viruses contain only a few genes Reverse
Trim29 gene-targeting strategy. (a) Genotyping of wildtype mice (+/+), Trim29 heterozygous mice (+/ ) and homozygous mice ( / ).
 Supplementary Figure 1 Trim29 gene-targeting strategy. (a) Genotyping of wildtype mice (+/+), Trim29 heterozygous mice (+/ ) and homozygous mice ( / ). (b) Immunoblot analysis of TRIM29 in lung primary
Supplementary Figure 1 Trim29 gene-targeting strategy. (a) Genotyping of wildtype mice (+/+), Trim29 heterozygous mice (+/ ) and homozygous mice ( / ). (b) Immunoblot analysis of TRIM29 in lung primary
Problem Set 5 KEY
 2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
Newly Recognized Components of the Innate Immune System
 Newly Recognized Components of the Innate Immune System NOD Proteins: Intracellular Peptidoglycan Sensors NOD-1 NOD-2 Nod Protein LRR; Ligand Recognition CARD RICK I-κB p50 p65 NF-κB Polymorphisms in Nod-2
Newly Recognized Components of the Innate Immune System NOD Proteins: Intracellular Peptidoglycan Sensors NOD-1 NOD-2 Nod Protein LRR; Ligand Recognition CARD RICK I-κB p50 p65 NF-κB Polymorphisms in Nod-2
Recombinant Protein Expression Retroviral system
 Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Section 6. Junaid Malek, M.D.
 Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
General information. Cell mediated immunity. 455 LSA, Tuesday 11 to noon. Anytime after class.
 General information Cell mediated immunity 455 LSA, Tuesday 11 to noon Anytime after class T-cell precursors Thymus Naive T-cells (CD8 or CD4) email: lcoscoy@berkeley.edu edu Use MCB150 as subject line
General information Cell mediated immunity 455 LSA, Tuesday 11 to noon Anytime after class T-cell precursors Thymus Naive T-cells (CD8 or CD4) email: lcoscoy@berkeley.edu edu Use MCB150 as subject line
Basis and Clinical Applications of Interferon
 Interferon Therapy Basis and Clinical Applications of Interferon JMAJ 47(1): 7 12, 2004 Jiro IMANISHI Professor, Kyoto Prefectural University of Medicine Abstract: Interferon (IFN) is an antiviral substance
Interferon Therapy Basis and Clinical Applications of Interferon JMAJ 47(1): 7 12, 2004 Jiro IMANISHI Professor, Kyoto Prefectural University of Medicine Abstract: Interferon (IFN) is an antiviral substance
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/366/ra25/dc1 Supplementary Materials for Viral entry route determines how human plasmacytoid dendritic cells produce type I interferons Daniela Bruni, Maxime
www.sciencesignaling.org/cgi/content/full/8/366/ra25/dc1 Supplementary Materials for Viral entry route determines how human plasmacytoid dendritic cells produce type I interferons Daniela Bruni, Maxime
CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION
 CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION What is Cytokine? Secreted popypeptide (protein) involved in cell-to-cell signaling. Acts in paracrine or autocrine fashion through specific cellular receptors.
CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION What is Cytokine? Secreted popypeptide (protein) involved in cell-to-cell signaling. Acts in paracrine or autocrine fashion through specific cellular receptors.
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature05732 SUPPLEMENTARY INFORMATION Supplemental Data Supplement Figure Legends Figure S1. RIG-I 2CARD undergo robust ubiquitination a, (top) At 48 h posttransfection with a GST, GST-RIG-I-2CARD
doi: 10.1038/nature05732 SUPPLEMENTARY INFORMATION Supplemental Data Supplement Figure Legends Figure S1. RIG-I 2CARD undergo robust ubiquitination a, (top) At 48 h posttransfection with a GST, GST-RIG-I-2CARD
Title: Cytosolic DNA-mediated, STING-dependent pro-inflammatory gene. Fig. S1. STING ligands-mediated signaling response in MEFs. (A) Primary MEFs (1
 1 Supporting Information 2 3 4 Title: Cytosolic DNA-mediated, STING-dependent pro-inflammatory gene induction necessitates canonical NF-κB activation through TBK1 5 6 Authors: Abe et al. 7 8 9 Supporting
1 Supporting Information 2 3 4 Title: Cytosolic DNA-mediated, STING-dependent pro-inflammatory gene induction necessitates canonical NF-κB activation through TBK1 5 6 Authors: Abe et al. 7 8 9 Supporting
Nature Immunology doi: /ni.3268
 Supplementary Figure 1 Loss of Mst1 and Mst2 increases susceptibility to bacterial sepsis. (a) H&E staining of colon and kidney sections from wild type and Mst1 -/- Mst2 fl/fl Vav-Cre mice. Scale bar,
Supplementary Figure 1 Loss of Mst1 and Mst2 increases susceptibility to bacterial sepsis. (a) H&E staining of colon and kidney sections from wild type and Mst1 -/- Mst2 fl/fl Vav-Cre mice. Scale bar,
Supplementary information
 Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
OCCUPATIONAL HEALTH CONSIDERATIONS FOR WORK WITH VIRAL VECTORS
 OCCUPATIONAL HEALTH CONSIDERATIONS FOR WORK WITH VIRAL VECTORS GARY R. FUJIMOTO, M.D. PALO ALTO MEDICAL FOUNDATION ADJUNCT ASSOCIATE CLINICAL PROFESSOR OF MEDICINE DIVISION OF INFECTIOUS DISEASES AND GEOGRAPHIC
OCCUPATIONAL HEALTH CONSIDERATIONS FOR WORK WITH VIRAL VECTORS GARY R. FUJIMOTO, M.D. PALO ALTO MEDICAL FOUNDATION ADJUNCT ASSOCIATE CLINICAL PROFESSOR OF MEDICINE DIVISION OF INFECTIOUS DISEASES AND GEOGRAPHIC
Stochastic Expression of the Interferon-b Gene
 Mingwei Zhao 1, Jiangwen Zhang 2., Hemali Phatnani 3., Stefanie Scheu 4, Tom Maniatis 1,3 * 1 Department of Molecular and Cellular Biology, Harvard University, Cambridge, Massachusetts, United States of
Mingwei Zhao 1, Jiangwen Zhang 2., Hemali Phatnani 3., Stefanie Scheu 4, Tom Maniatis 1,3 * 1 Department of Molecular and Cellular Biology, Harvard University, Cambridge, Massachusetts, United States of
HIV 101: Fundamentals of HIV Infection
 HIV 101: Fundamentals of HIV Infection David H. Spach, MD Professor of Medicine University of Washington Seattle, Washington Learning Objectives After attending this presentation, learners will be able
HIV 101: Fundamentals of HIV Infection David H. Spach, MD Professor of Medicine University of Washington Seattle, Washington Learning Objectives After attending this presentation, learners will be able
Peli1 negatively regulates T-cell activation and prevents autoimmunity
 Peli1 negatively regulates T-cell activation and prevents autoimmunity Mikyoung Chang 1,*, Wei Jin 1,5,*, Jae-Hoon Chang 1, Yi-chuan Xiao 1, George Brittain 1, Jiayi Yu 1, Xiaofei Zhou 1, Yi-Hong Wang
Peli1 negatively regulates T-cell activation and prevents autoimmunity Mikyoung Chang 1,*, Wei Jin 1,5,*, Jae-Hoon Chang 1, Yi-chuan Xiao 1, George Brittain 1, Jiayi Yu 1, Xiaofei Zhou 1, Yi-Hong Wang
Immunity to Viruses. Patricia Fitzgerald-Bocarsly September 25, 2008
 Immunity to Viruses Patricia Fitzgerald-Bocarsly September 25, 2008 The Immune System Deals with a Huge Range of Pathogens Roitt, 2003 Immune Responses to Viruses Viruses are dependent on the host cell
Immunity to Viruses Patricia Fitzgerald-Bocarsly September 25, 2008 The Immune System Deals with a Huge Range of Pathogens Roitt, 2003 Immune Responses to Viruses Viruses are dependent on the host cell
How HIV Causes Disease Prof. Bruce D. Walker
 How HIV Causes Disease Howard Hughes Medical Institute Massachusetts General Hospital Harvard Medical School 1 The global AIDS crisis 60 million infections 20 million deaths 2 3 The screen versions of
How HIV Causes Disease Howard Hughes Medical Institute Massachusetts General Hospital Harvard Medical School 1 The global AIDS crisis 60 million infections 20 million deaths 2 3 The screen versions of
19 Viruses BIOLOGY. Outline. Structural Features and Characteristics. The Good the Bad and the Ugly. Structural Features and Characteristics
 9 Viruses CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Viruses A. Structure of viruses B. Common Characteristics of Viruses C. Viral replication D. HIV Lecture Presentation
9 Viruses CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Viruses A. Structure of viruses B. Common Characteristics of Viruses C. Viral replication D. HIV Lecture Presentation
VIROLOGY. Engineering Viral Genomes: Retrovirus Vectors
 VIROLOGY Engineering Viral Genomes: Retrovirus Vectors Viral vectors Retrovirus replicative cycle Most mammalian retroviruses use trna PRO, trna Lys3, trna Lys1,2 The partially unfolded trna is annealed
VIROLOGY Engineering Viral Genomes: Retrovirus Vectors Viral vectors Retrovirus replicative cycle Most mammalian retroviruses use trna PRO, trna Lys3, trna Lys1,2 The partially unfolded trna is annealed
RAISON D ETRE OF THE IMMUNE SYSTEM:
 RAISON D ETRE OF THE IMMUNE SYSTEM: To Distinguish Self from Non-Self Thereby Protecting Us From Our Hostile Environment. Innate Immunity Acquired Immunity Innate immunity: (Antigen nonspecific) defense
RAISON D ETRE OF THE IMMUNE SYSTEM: To Distinguish Self from Non-Self Thereby Protecting Us From Our Hostile Environment. Innate Immunity Acquired Immunity Innate immunity: (Antigen nonspecific) defense
Anti-infectious Immunity
 Anti-infectious Immunity innate immunity barrier structures Secretory molecules Phagocytes NK cells Anatomical barriers 1. Skin and mucosa barrier 2.hemo-Spinal Fluid barrier 3. placental barrier Phagocytic
Anti-infectious Immunity innate immunity barrier structures Secretory molecules Phagocytes NK cells Anatomical barriers 1. Skin and mucosa barrier 2.hemo-Spinal Fluid barrier 3. placental barrier Phagocytic
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS
 Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
10th International Rotavirus Symposium Bangkok, Thailand
 Rotavirus Host Range Restriction and Innate Immunity: Mechanisms of Vaccine Attenuation Harry Greenberg MD Stanford University 10th International Rotavirus Symposium Bangkok, Thailand 09/19/12 B dsrna
Rotavirus Host Range Restriction and Innate Immunity: Mechanisms of Vaccine Attenuation Harry Greenberg MD Stanford University 10th International Rotavirus Symposium Bangkok, Thailand 09/19/12 B dsrna
number Done by Corrected by Doctor Ashraf
 number 4 Done by Nedaa Bani Ata Corrected by Rama Nada Doctor Ashraf Genome replication and gene expression Remember the steps of viral replication from the last lecture: Attachment, Adsorption, Penetration,
number 4 Done by Nedaa Bani Ata Corrected by Rama Nada Doctor Ashraf Genome replication and gene expression Remember the steps of viral replication from the last lecture: Attachment, Adsorption, Penetration,
Supplementary information for: Community detection for networks with. unipartite and bipartite structure. Chang Chang 1, 2, Chao Tang 2
 Supplementary information for: Community detection for networks with unipartite and bipartite structure Chang Chang 1, 2, Chao Tang 2 1 School of Life Sciences, Peking University, Beiing 100871, China
Supplementary information for: Community detection for networks with unipartite and bipartite structure Chang Chang 1, 2, Chao Tang 2 1 School of Life Sciences, Peking University, Beiing 100871, China
Chapter 6. Antigen Presentation to T lymphocytes
 Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
Follow this and additional works at: Part of the Medicine and Health Sciences Commons
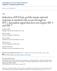 Washington University School of Medicine Digital Commons@Becker Open Access Publications 2009 Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2009 Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent
Pre-made Reporter Lentivirus for NF-κB Signal Pathway
 Pre-made Reporter for NF-κB Signal Pathway Cat# Product Name Amounts LVP965-P or: LVP965-P-PBS NFKB-GFP (Puro) LVP966-P or: LVP966-P-PBS NFKB-RFP (Puro) LVP967-P or: LVP967-P-PBS NFKB-Luc (Puro) LVP968-P
Pre-made Reporter for NF-κB Signal Pathway Cat# Product Name Amounts LVP965-P or: LVP965-P-PBS NFKB-GFP (Puro) LVP966-P or: LVP966-P-PBS NFKB-RFP (Puro) LVP967-P or: LVP967-P-PBS NFKB-Luc (Puro) LVP968-P
RAS Genes. The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes.
 ۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
Chapter13 Characterizing and Classifying Viruses, Viroids, and Prions
 Chapter13 Characterizing and Classifying Viruses, Viroids, and Prions 11/20/2017 MDufilho 1 Characteristics of Viruses Viruses Minuscule, acellular, infectious agent having either DNA or RNA Cause infections
Chapter13 Characterizing and Classifying Viruses, Viroids, and Prions 11/20/2017 MDufilho 1 Characteristics of Viruses Viruses Minuscule, acellular, infectious agent having either DNA or RNA Cause infections
Nature Medicine: doi: /nm.2109
 HIV 1 Infects Multipotent Progenitor Cells Causing Cell Death and Establishing Latent Cellular Reservoirs Christoph C. Carter, Adewunmi Onafuwa Nuga, Lucy A. M c Namara, James Riddell IV, Dale Bixby, Michael
HIV 1 Infects Multipotent Progenitor Cells Causing Cell Death and Establishing Latent Cellular Reservoirs Christoph C. Carter, Adewunmi Onafuwa Nuga, Lucy A. M c Namara, James Riddell IV, Dale Bixby, Michael
L I F E S C I E N C E S
 1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 OCTOBER 31, 2006
1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 OCTOBER 31, 2006
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.10/nature10195 NCBI gene: Tagged Subunit(s: HA-Vpx; FLAG-Cul4 HA-DCAF1 FLAG-Cul4 HA-FLAG-Vpx Mock Vpx (SIVmac 100 (a ; 0.159 (b ; 0.05 DCAF1 DDB1 DDA1 Cul4A 1; 0.024591
SUPPLEMENTARY INFORMATION doi:10.10/nature10195 NCBI gene: Tagged Subunit(s: HA-Vpx; FLAG-Cul4 HA-DCAF1 FLAG-Cul4 HA-FLAG-Vpx Mock Vpx (SIVmac 100 (a ; 0.159 (b ; 0.05 DCAF1 DDB1 DDA1 Cul4A 1; 0.024591
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
Antiviral Drugs Lecture 5
 Antiviral Drugs Lecture 5 Antimicrobial Chemotherapy (MLAB 366) 1 Dr. Mohamed A. El-Sakhawy 2 Introduction Viruses are microscopic organisms that can infect all living cells. They are parasitic and multiply
Antiviral Drugs Lecture 5 Antimicrobial Chemotherapy (MLAB 366) 1 Dr. Mohamed A. El-Sakhawy 2 Introduction Viruses are microscopic organisms that can infect all living cells. They are parasitic and multiply
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Human Immunodeficiency Virus
 Human Immunodeficiency Virus Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Viruses and hosts Lentivirus from Latin lentis (slow), for slow progression of disease
Human Immunodeficiency Virus Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Viruses and hosts Lentivirus from Latin lentis (slow), for slow progression of disease
1. TLR. TLR Toll-like receptors. Toll Toll-like receptor, TLR TLR TLR TLR. type I TLR TLR. Toll
 54pp.145 152 2004 1. TLR T B TLR Toll-like receptors TLR TLR I IFN TLR T B B T Toll NF- B 1996 565-0871 3-1 TEL 06-6879-8303 FAX 06-6879-8305 E-mail uemattsu@biken.osaka-u.ac.jp Toll Toll-like receptor,
54pp.145 152 2004 1. TLR T B TLR Toll-like receptors TLR TLR I IFN TLR T B B T Toll NF- B 1996 565-0871 3-1 TEL 06-6879-8303 FAX 06-6879-8305 E-mail uemattsu@biken.osaka-u.ac.jp Toll Toll-like receptor,
Toll-like Receptors (TLRs): Biology, Pathology and Therapeutics
 Toll-like Receptors (TLRs): Biology, Pathology and Therapeutics Dr Sarah Sasson SydPATH Registrar 23 rd June 2014 TLRs: Introduction Discovered in 1990s Recognise conserved structures in pathogens Rely
Toll-like Receptors (TLRs): Biology, Pathology and Therapeutics Dr Sarah Sasson SydPATH Registrar 23 rd June 2014 TLRs: Introduction Discovered in 1990s Recognise conserved structures in pathogens Rely
Allergy and Immunology Review Corner: Chapter 13 of Immunology IV: Clinical Applications in Health and Disease, by Joseph A. Bellanti, MD.
 Allergy and Immunology Review Corner: Chapter 13 of Immunology IV: Clinical Applications in Health and Disease, by Joseph A. Bellanti, MD. Chapter 13: Mechanisms of Immunity to Viral Disease Prepared by
Allergy and Immunology Review Corner: Chapter 13 of Immunology IV: Clinical Applications in Health and Disease, by Joseph A. Bellanti, MD. Chapter 13: Mechanisms of Immunity to Viral Disease Prepared by
Irf1 fold changes (D) 24h 48h. p-p65. t-p65. p-irf3. t-irf3. β-actin SKO TKO 100% 80% 60% 40% 20%
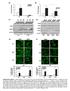 Irf7 Fold changes 3 1 Irf1 fold changes 3 1 8h h 8h 8h h 8h p-p6 p-p6 t-p6 p-irf3 β-actin p-irf3 t-irf3 β-actin TKO TKO STKO (E) (F) TKO TKO % of p6 nuclear translocation % % 1% 1% % % p6 TKO % of IRF3
Irf7 Fold changes 3 1 Irf1 fold changes 3 1 8h h 8h 8h h 8h p-p6 p-p6 t-p6 p-irf3 β-actin p-irf3 t-irf3 β-actin TKO TKO STKO (E) (F) TKO TKO % of p6 nuclear translocation % % 1% 1% % % p6 TKO % of IRF3
Name Section Problem Set 6
 Name Section 7.012 Problem Set 6 Question 1 The viral family Orthomyxoviridae contains the influenza A, B and C viruses. These viruses have a (-)ss RNA genome surrounded by a capsid composed of lipids
Name Section 7.012 Problem Set 6 Question 1 The viral family Orthomyxoviridae contains the influenza A, B and C viruses. These viruses have a (-)ss RNA genome surrounded by a capsid composed of lipids
Toll-like Receptor Signaling
 Toll-like Receptor Signaling 1 Professor of Medicine University of Massachusetts Medical School, Worcester, MA, USA Why do we need innate immunity? Pathogens multiply very fast We literally swim in viruses
Toll-like Receptor Signaling 1 Professor of Medicine University of Massachusetts Medical School, Worcester, MA, USA Why do we need innate immunity? Pathogens multiply very fast We literally swim in viruses
ECM1 controls T H 2 cell egress from lymph nodes through re-expression of S1P 1
 ZH, Li et al, page 1 ECM1 controls T H 2 cell egress from lymph nodes through re-expression of S1P 1 Zhenhu Li 1,4,Yuan Zhang 1,4, Zhiduo Liu 1, Xiaodong Wu 1, Yuhan Zheng 1, Zhiyun Tao 1, Kairui Mao 1,
ZH, Li et al, page 1 ECM1 controls T H 2 cell egress from lymph nodes through re-expression of S1P 1 Zhenhu Li 1,4,Yuan Zhang 1,4, Zhiduo Liu 1, Xiaodong Wu 1, Yuhan Zheng 1, Zhiyun Tao 1, Kairui Mao 1,
HIV INFECTION: An Overview
 HIV INFECTION: An Overview UNIVERSITY OF PAPUA NEW GUINEA SCHOOL OF MEDICINE AND HEALTH SCIENCES DIVISION OF BASIC MEDICAL SCIENCES DISCIPLINE OF BIOCHEMISTRY & MOLECULAR BIOLOGY PBL MBBS II SEMINAR VJ
HIV INFECTION: An Overview UNIVERSITY OF PAPUA NEW GUINEA SCHOOL OF MEDICINE AND HEALTH SCIENCES DIVISION OF BASIC MEDICAL SCIENCES DISCIPLINE OF BIOCHEMISTRY & MOLECULAR BIOLOGY PBL MBBS II SEMINAR VJ
LESSON 4.6 WORKBOOK. Designing an antiviral drug The challenge of HIV
 LESSON 4.6 WORKBOOK Designing an antiviral drug The challenge of HIV In the last two lessons we discussed the how the viral life cycle causes host cell damage. But is there anything we can do to prevent
LESSON 4.6 WORKBOOK Designing an antiviral drug The challenge of HIV In the last two lessons we discussed the how the viral life cycle causes host cell damage. But is there anything we can do to prevent
Size nm m m
 1 Viral size and organization Size 20-250nm 0.000000002m-0.000000025m Virion structure Capsid Core Acellular obligate intracellular parasites Lack organelles, metabolic activities, and reproduction Replicated
1 Viral size and organization Size 20-250nm 0.000000002m-0.000000025m Virion structure Capsid Core Acellular obligate intracellular parasites Lack organelles, metabolic activities, and reproduction Replicated
Polyomaviridae. Spring
 Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Innate Sensing of HIV-Infected Cells
 Innate Sensing of HIV-Infected Cells Alice Lepelley 1", Stéphanie Louis 1", Marion Sourisseau 1", Helen K. W. Law 2, Julien Pothlichet 3, Clémentine Schilte 4, Laurence Chaperot 5, Joël Plumas 5, Richard
Innate Sensing of HIV-Infected Cells Alice Lepelley 1", Stéphanie Louis 1", Marion Sourisseau 1", Helen K. W. Law 2, Julien Pothlichet 3, Clémentine Schilte 4, Laurence Chaperot 5, Joël Plumas 5, Richard
Coronaviruses cause acute, mild upper respiratory infection (common cold).
 Coronaviruses David A. J. Tyrrell Steven H. Myint GENERAL CONCEPTS Clinical Presentation Coronaviruses cause acute, mild upper respiratory infection (common cold). Structure Spherical or pleomorphic enveloped
Coronaviruses David A. J. Tyrrell Steven H. Myint GENERAL CONCEPTS Clinical Presentation Coronaviruses cause acute, mild upper respiratory infection (common cold). Structure Spherical or pleomorphic enveloped
Supplemental Materials and Methods Plasmids and viruses Quantitative Reverse Transcription PCR Generation of molecular standard for quantitative PCR
 Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Phenomena first observed in petunia
 Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
FOR OPTIMAL GUT HEALTH KEMIN.COM/GUTHEALTH
 FOR OPTIMAL GUT HEALTH KEMIN.COM/GUTHEALTH ALETA A SOURCE OF 1,3-BETA GLUCANS Aleta is highly bioavailable, offering a concentration greater than 5% of 1,3-beta glucans. Aleta provides a consistent response
FOR OPTIMAL GUT HEALTH KEMIN.COM/GUTHEALTH ALETA A SOURCE OF 1,3-BETA GLUCANS Aleta is highly bioavailable, offering a concentration greater than 5% of 1,3-beta glucans. Aleta provides a consistent response
Viruses Tomasz Kordula, Ph.D.
 Viruses Tomasz Kordula, Ph.D. Resources: Alberts et al., Molecular Biology of the Cell, pp. 295, 1330, 1431 1433; Lehninger CD Movie A0002201. Learning Objectives: 1. Understand parasitic life cycle of
Viruses Tomasz Kordula, Ph.D. Resources: Alberts et al., Molecular Biology of the Cell, pp. 295, 1330, 1431 1433; Lehninger CD Movie A0002201. Learning Objectives: 1. Understand parasitic life cycle of
Reoviruses. Virion. Genome. Genes and proteins. Viruses and hosts. Diseases. Distinctive characteristics
 Reoviruses Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Virion Naked icosahedral capsid (T=13), diameter 60-85 nm Capsid consists of two or three concentric protein
Reoviruses Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Virion Naked icosahedral capsid (T=13), diameter 60-85 nm Capsid consists of two or three concentric protein
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Oncolytic Viruses as a Potential Approach to Eliminate the HIV Reservoir
 Oncolytic Viruses as a Potential Approach to Eliminate the HIV Reservoir Costiniuk CT, Côté SC, Al-Ghazawi FM, Carrasco-Medina L,Young CD, & Angel JB University of Ottawa, November 12 th, 2012 HIV Reservoirs
Oncolytic Viruses as a Potential Approach to Eliminate the HIV Reservoir Costiniuk CT, Côté SC, Al-Ghazawi FM, Carrasco-Medina L,Young CD, & Angel JB University of Ottawa, November 12 th, 2012 HIV Reservoirs
VIRUSES. Biology Applications Control. David R. Harper. Garland Science Taylor & Francis Group NEW YORK AND LONDON
 VIRUSES Biology Applications Control David R. Harper GS Garland Science Taylor & Francis Group NEW YORK AND LONDON vii Chapter 1 Virus Structure and 2.2 VIRUS MORPHOLOGY 26 Infection 1 2.3 VIRAL CLASSIFICATION
VIRUSES Biology Applications Control David R. Harper GS Garland Science Taylor & Francis Group NEW YORK AND LONDON vii Chapter 1 Virus Structure and 2.2 VIRUS MORPHOLOGY 26 Infection 1 2.3 VIRAL CLASSIFICATION
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were
 Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
Identification of Mutation(s) in. Associated with Neutralization Resistance. Miah Blomquist
 Identification of Mutation(s) in the HIV 1 gp41 Subunit Associated with Neutralization Resistance Miah Blomquist What is HIV 1? HIV-1 is an epidemic that affects over 34 million people worldwide. HIV-1
Identification of Mutation(s) in the HIV 1 gp41 Subunit Associated with Neutralization Resistance Miah Blomquist What is HIV 1? HIV-1 is an epidemic that affects over 34 million people worldwide. HIV-1
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/1/8/e1500296/dc1 Supplementary Materials for Transcriptional regulation of APOBEC3 antiviral immunity through the CBF- /RUNX axis This PDF file includes: Brett
advances.sciencemag.org/cgi/content/full/1/8/e1500296/dc1 Supplementary Materials for Transcriptional regulation of APOBEC3 antiviral immunity through the CBF- /RUNX axis This PDF file includes: Brett
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
RAISON D ETRE OF THE IMMUNE SYSTEM:
 RAISON D ETRE OF THE IMMUNE SYSTEM: To Distinguish Self from Non-Self Thereby Protecting Us From Our Hostile Environment. Innate Immunity Adaptive Immunity Innate immunity: (Antigen - nonspecific) defense
RAISON D ETRE OF THE IMMUNE SYSTEM: To Distinguish Self from Non-Self Thereby Protecting Us From Our Hostile Environment. Innate Immunity Adaptive Immunity Innate immunity: (Antigen - nonspecific) defense
1. Overview of Adaptive Immunity
 Chapter 17A: Adaptive Immunity Part I 1. Overview of Adaptive Immunity 2. T and B Cell Production 3. Antigens & Antigen Presentation 4. Helper T cells 1. Overview of Adaptive Immunity The Nature of Adaptive
Chapter 17A: Adaptive Immunity Part I 1. Overview of Adaptive Immunity 2. T and B Cell Production 3. Antigens & Antigen Presentation 4. Helper T cells 1. Overview of Adaptive Immunity The Nature of Adaptive
Supplementary Figure 1 Induction of cellular senescence and isolation of exosome. a to c, Pre-senescent primary normal human diploid fibroblasts
 Supplementary Figure 1 Induction of cellular senescence and isolation of exosome. a to c, Pre-senescent primary normal human diploid fibroblasts (TIG-3 cells) were rendered senescent by either serial passage
Supplementary Figure 1 Induction of cellular senescence and isolation of exosome. a to c, Pre-senescent primary normal human diploid fibroblasts (TIG-3 cells) were rendered senescent by either serial passage
Antigen Presentation and T Lymphocyte Activation. Abul K. Abbas UCSF. FOCiS
 1 Antigen Presentation and T Lymphocyte Activation Abul K. Abbas UCSF FOCiS 2 Lecture outline Dendritic cells and antigen presentation The role of the MHC T cell activation Costimulation, the B7:CD28 family
1 Antigen Presentation and T Lymphocyte Activation Abul K. Abbas UCSF FOCiS 2 Lecture outline Dendritic cells and antigen presentation The role of the MHC T cell activation Costimulation, the B7:CD28 family
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Differential expression of mirnas from the pri-mir-17-92a locus.
 Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
RIG-I dependent sensing of poly(da-dt) via the induction of an RNA
 RI-I dependent sensing of via the induction of an RNA polymerase III transcribed RNA intermediate Andrea Ablasser 1*, Franz Bauernfeind 1*, unther Hartmann 1, Eicke Latz 2, Katherine A. Fitzgerald 2* and
RI-I dependent sensing of via the induction of an RNA polymerase III transcribed RNA intermediate Andrea Ablasser 1*, Franz Bauernfeind 1*, unther Hartmann 1, Eicke Latz 2, Katherine A. Fitzgerald 2* and
Identification of Host Cytosolic Sensors and Bacterial Factors Regulating the Type I Interferon Response to Legionella pneumophila
 Identification of Host Cytosolic Sensors and Bacterial Factors Regulating the Type I Interferon Response to Legionella pneumophila Kathryn M. Monroe, Sarah M. McWhirter, Russell E. Vance* Division of Immunology
Identification of Host Cytosolic Sensors and Bacterial Factors Regulating the Type I Interferon Response to Legionella pneumophila Kathryn M. Monroe, Sarah M. McWhirter, Russell E. Vance* Division of Immunology
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
