Trex1 regulates lysosomal biogenesis and interferon-independent activation of antiviral genes
|
|
|
- Silas Johns
- 5 years ago
- Views:
Transcription
1 Trex1 regulates lysosomal biogenesis and interferon-independent activation of antiviral genes Maroof Hasan 1,, James Koch 1,, Dinesh Rakheja 3, Asit K Pattnaik,, James Brugarolas 1,,7, Igor Dozmorov, Beth evine 1,,9,1, Edward K Wakeland, Min Ae ee-kirsch 11 & Nan Yan 1, npg 13 Nature America, Inc. All rights reserved. The sensing of viral nucleic acids by the innate immune system triggers the production of type I interferons, which activates interferon-stimulated genes (ISGs) and directs a multifaceted antiviral response. ISGs can also be activated through interferonindependent pathways, although the precise mechanisms remain elusive. Here we found that the cytosolic exonuclease Trex1 regulated the activation of a subset of ISGs independently of interferon. Both Trex1 / mouse cells and Trex1-mutant human cells had high expression of genes encoding antiviral molecules ( antiviral genes ) and were refractory to viral infection. The interferon-independent activation of antiviral genes in Trex1 / cells required the adaptor STING, the kinase TBK1 and the transcription factors IRF3 and IRF7. We also found that Trex1-deficient cells had an expanded lysosomal compartment, altered subcellular localization of the transcription factor TFEB and diminished activity of the regulator mtorc1. Together our data identify Trex1 as a regulator of lysosomal biogenesis and interferon-independent activation of antiviral genes and show that dysregulation of lysosomes can elicit innate immune responses. Vertebrates are constantly facing challenges from pathogenic microbes that introduce a variety of microbial proteins and nucleic acids into the host cell. To counter this, eukaryotic cells express many different pattern-recognition receptors that detect microbial pathogenassociated molecular patterns, which then activate antiviral interferon and proinflammatory responses 1. Mammalian pattern-recognition receptors include the Toll-like receptors (TRs), RIG-I-like receptors, Nod-like receptors, C-type lectin receptors and an emerging category of cytosolic receptors for DNA. During viral infection, viral nucleic acids are the main pathogen-associated molecular patterns detected by the receptors of the host innate immune system, which include RIG-I-like receptors and receptors of DNA in the cytosol and a subfamily of TRs that localize to the endosomal membrane 3. The central hub for the cytosolic sensing of DNA is the endoplasmic reticulum membrane adaptor STING (also known as MITA, MPYS or ERIS). Many proteins have been proposed to detect double-stranded DNA in the cytosol and signal through STING, including DAI, IFI1 and DDX1 (refs. ). STING can also directly recognize the cyclic dinucleotide c-di-gmp, which is usually associated with bacterial infection and activates interferon expression 7. Although interferon has a major role in controlling viral infections, interferon-independent pathways also exist and are vital for antiviral defense. For example, STING can activate the transcription factor STAT independently of the interferon pathway, and activated STAT induces chemokine expression that primes adaptive immune responses. Infection with enveloped viruses also triggers an interferon-independent pathway that involves the direct activation of a subset of interferon-stimulated genes (ISGs) by the transcription factor IRF3 (ref. 9). A study of innate immune responses to RNA viruses mediated by the signaling adaptor MAVS has demonstrated that interferon-independent induction of genes encoding antiviral molecules ( antiviral genes ) occurs rapidly after infection and is functionally important for the control of viral replication before the onset of more robust and sustained activation by interferon 1. The sensing pathways of the innate immune system are carefully designed to distinguish self ligands from non-self ligands, either by spatial separation (for example, TR7 and TR9 reside in endosomes, which are devoid of host nucleic acids) or by stringent ligand specificity (for example, TR9 recognizes CpG-containing DNA from bacteria, and the RNA heliase RIG-I recognizes RNA containing triphosphate from viruses). However, how cytosolic DNA-sensing pathways distinguish host DNA and viral DNA remains unclear. Trex1, an exonuclease that resides on the endoplasmic reticulum, is a negative regulator of the sensing of cytosolic human immunodeficiency virus (HIV) DNA by the innate immune system. In Trex1-deficient (Trex1 / ) mouse embryonic fibroblasts (MEFs) and in human CD + 1 Department of Internal Medicine, UT Southwestern Medical Center, Dallas, Texas, USA. Department of Microbiology, UT Southwestern Medical Center, Dallas, Texas, USA. 3 Department of Pathology, Children s Medical Center, UT Southwestern Medical Center, Dallas, Texas, USA. School of Veterinary Medicine and Biomedical Sciences, University of Nebraska, incoln, Nebraska, USA. Nebraska Center for Virology, University of Nebraska, incoln, Nebraska, USA. Department of Developmental Biology, UT Southwestern Medical Center, Dallas, Texas, USA. 7 Simmons Cancer Center, UT Southwestern Medical Center, Dallas, Texas, USA. Department of Immunology, UT Southwestern Medical Center, Dallas, Texas, USA. 9 Center for Autophagy Research, UT Southwestern Medical Center, Dallas, Texas, USA. 1 Howard Hughes Medical Institute, UT Southwestern Medical Center, Dallas, Texas, USA. 11 Children s Hospital, Technical University Dresden, Dresden, Germany. Correspondence should be addressed to N.Y. (nan.yan@utsouthwestern.edu). Received 7 June; accepted 19 October; published online 1 November 1; doi:1.13/ni.7 nature immunology VOUME 1 NUMBER 1 JANUARY 13 1
2 npg 13 Nature America, Inc. All rights reserved. T cells and macrophages depleted of Trex1 by RNA-mediated interference, cytosolic HIV DNA accumulates and triggers interferon through the STING kinase TBK1 IRF3 pathway 11. Those findings and other studies 1 suggest that cells rely on negative regulatory molecules such as Trex1 to keep cytosolic DNA-sensing pathways in check. Trex1 deficiency has been linked to the pathogenesis of autoimmunity. TREX1 mutations in humans are associated with a spectrum of autoimmune and inflammatory phenotypes, including Aicardi-Goutières syndrome (AGS; an inflammatory brain disease that mimics the symptoms of congenital viral infection 13,1 ), systemic lupus erythematosus (SE), familial chilbain lupus, and retinal vasculopathy with cerebral leukodystrophy TREX1 mutations have been found in up to % of patients with SE, with an extremely high odds ratio (of ) 1 ; thus, such mutations represent one of the highest disease risks recorded for a single susceptibility gene in complex polygenic SE 1. Studies of Trex1 / mice have shown that Trex1 / cells accumulate cytosolic single-stranded DNA that might be derived from DNA repair in the nucleus or from endogenous retroelements 19,. Genetic evidence has demonstrated that the STING-mediated DNAsensing pathway is essential for the pathogenesis of autoimmune disease in Trex1 / mice 1. The initiation of interferon expression is detected in only a subset of nonhematopoietic cells in Trex1 / mice, which raises the question of what happens to the majority of other cells that also lack Trex1 function. We also wondered whether Trex1 inhibits interferon responses to other viruses beyond HIV and/or if the mere loss of Trex1 function in a cell would elicit innate immune responses and establish an antiviral state. In this study, we found that Trex1-deficient and Trex1-mutant cells had broad antiviral activity against many RNA viruses. The antiviral activity was provided by higher expression of ISGs in cells that lacked Trex1 function and was mediated through an interferonindependent signaling pathway that involved STING, TBK1, IRF3 and IRF7. We also found that Trex1 regulated lysosomal biogenesis through the transcription factor TFEB and the regulator mtorc1 and provide evidence that dysregulation of lysosomes elicits innate immune responses. RESUTS Impaired replication in Trex1-deficient cells To investigate whether Trex1 is involved in the interferon response to RNA viruses, we infected wild-type and Trex1 / MEFs with the Indiana strain of vesicular stomatitis virus (; a negativestranded RNA virus), with glycoprotein (G protein) pseudotyped HIV 11 or treated them with mock infection, then measured mrna encoding interferon-β (IFN-β; Ifnb mrna) h after infection. As reported before 11, mock-infected wild-type and Trex1 / cells did not have detectable expression of Ifnb mrna, and infection with HIV stimulated the expression of Ifnb mrna only in Trex1 / cells, not in wild-type cells (Fig. 1a). In contrast, infection with stimulated similarly high Ifnb mrna expression in wild-type and Trex1 / cells (Fig. 1a), which suggested that Trex1 did not regulate the type I interferon response to. However, replication was considerably impaired in Trex1 / cells relative to its replication in wild-type cells, even though the induction of Ifnb was indistinguishable in the two cell types (Fig. 1b d). Specifically, the abundance of the two main forms of RNA, encoding the glycoprotein G and matrix protein M, in Trex1 / cells was 1% and 7%, respectively, that in wild-type cells (Fig. 1b). We also detected much lower abundance of proteins in Trex1 / cells than in wild-type cells after infection with two different doses of (multiplicity of infection (MOI), and 1; Fig. 1c). titers were also lower in infected Trex1 / cells than in wild-type cells (Fig. 1d). To better quantify and visualize replication, we infected wild-type and Trex1 / cells with -P-eGFP, a form of in which enhanced green fluorescent protein (egfp) is fused to the phosphoprotein P, which is usually associated with foci of viral RNA replication in the cell 1. We observed less replication of -P-eGFP in Trex1 / cells (3.% GFP) than in wild-type cells (.% GFP + ) by flow cytometry (Fig. 1e). Consistent with those data, analysis of infected wild-type cells by fluorescence microscopy showed bright replication foci marked by P-eGFP, whereas we detected very little green fluorescent signal in -P-eGFP infected Trex1 / cells (Fig. 1f). We also infected bone marrow derived macrophages (BMDMs) generated from wild-type, Trex1 +/ and Trex1 / mice and found that only Trex1 / cells were resistant to infection with (Fig. 1g,h). We next assessed whether the entry of was inhibited in Trex1 / cells. This seemed unlikely, as the entry of -G pseudotyped HIV into Trex1 / cells was not impaired 11, and infection with stimulated indistinguishable expression of Ifnb mrna in wild-type and Trex1 / cells (Fig. 1a). Nonetheless, to rule out the possibility of an entry defect, we labeled wild-type virions with the fluorescent dye Dil and monitored the infection of wild-type and Trex1 / cells with Dil-labeled by live cell fluorescence microscopy (Supplementary Fig. 1a). We observed no difference between wild-type and Trex1 / cells in intracellular Dil-labeled at 1 h after infection. We also observed similar amounts of RNA encoding G and M at 1 h after infection in both cell types (Supplementary Fig. 1b). These data suggested that replication was blocked at an early stage after entry, such as uncoating or RNA replication, in Trex1 / cells. We also found that in contrast to infected wild-type cells, -infected Trex1 / cells did not show detectable cytopathic effects (Supplementary Fig. ), consistent with the idea that Trex1 / cells were protected against viral infection. To investigate whether Trex1 was also required for replication in human cells, we studied wild-type skin fibroblasts and Trex1- mutant skin fibroblasts (homozygous for the TREX1 mutation that produces the R11H substitution of Trex1; called TREX1 R11H/R11H here) from a patient with AGS. Arg11 is a critical residue at the interface of the Trex1 dimer, and the R11H substitution substantially disrupts Trex1 function in vitro. R11H represents the most common Trex1 substitution in patients with AGS and has also been associated with SE 1. We infected the cells with or -P-eGFP and measured viral RNA. Infection with or -P-eGFP was lower in TREX1 R11H/R11H cells than in wild-type cells, as reflected by the lower abundance of viral RNA and viral proteins and fewer viral replication foci (Fig. 1i k). Given these collective results, we concluded that replication was impaired at an early after entry step in both mouse cells and human cells lacking Trex1 function. Broad antiviral resistance of Trex1-deficient cells To determine whether the replication block in Trex1 / and TREX1 R11H/R11H cells was unique for, we infected wild-type and Trex1 / MEFs and wild-type and TREX1 R11H/R11H human fibroblasts with the following three additional RNA viruses with negativeor postitive-stranded genomes: influenza virus (A/WSN/1933 strain), Sendai virus and West Nile virus (-TX strain). We then measured viral RNA and proteins, as well as viral titers in supernatants. None of the three viruses replicated efficiently in Trex1 / or TREX1 R11H/R11H cells, in contrast to their replication in wild-type cells (Fig. ). These results demonstrated that cells lacking Trex1 function were resistant to infection with several different types of RNA viruses. VOUME 1 NUMBER 1 JANUARY 13 nature immunology
3 npg 13 Nature America, Inc. All rights reserved. a Ifnb mrna (AU) f HIV -PeGFP DAPI b MEF ND ND : + + G RNA (AU) g ND : Trex1 regulates interferon-independent activation of ISGs Next we investigated the mechanism of antiviral resistance in Trex1 / cells. We first examined gene-expression profiles by infecting wildtype and Trex1 / MEFs with, influenza virus, Sendai virus or West Nile virus, or mock infecting the cells, followed by isolation of total RNA and analysis by high-throughput sequencing technologies (RNA-Seq) that offer quantitative measurement of both host and viral RNA simultaneously (Fig. 3a). The changes in gene expression confirmed by quantitative PCR were similar to those obtained by M RNA (AU) MEF BMDM -PeGFP DAPI ND + + Trex1 +/ c MOI: 1 1 HMGB1 Figure 1 Impaired replication in Trex1-deficient cells. (a) Quantitative RT-PCR analysis of Ifnb mrna in wild-type () and Trex1 / MEFs left uninfected ( ) or infected for h with -G pseudotyped HIV 11 or with (MOI, ). AU, arbitrary units. (b d) Quantitative RT-PCR analysis of RNA encoding the G and M proteins (b), immunoblot analysis of proteins (c) and viral titers in supernatants (d) of wild-type and Trex1 / MEFs mock infected or j TREX1 mut Time (h): h G RNA (AU) BMDM M RNA (AU) d titer ( 1 PFU/ml) P-eGFP i 7 TREX1 mut ND ND ND Time (h): Time (h): G RNA (AU) RNA-Seq (Fig. 3b, Supplementary Figs. 3 and and data not shown), which emphasized the quantitative power of our RNA-Seq analysis. We first analyzed the gene-expression profiles of uninfected wild-type and Trex1 / samples with Ingenuity pathway-analysis software and found that the gene network most represented in Trex1 / cells relative to its representation in wild-type cells was the network of antimicrobial response, inflammatory response, infectious diseases, which consists mostly of ISGs (Supplementary Fig. a,b). The networks of interferon signaling and cytosolic pattern-recognition receptors were also e SSC k 1, -P-eGFP DAPI M RNA (AU) Human fibroblast TREX1 mut infected for 1 h with. ND, not detectable; HMGB1, loading control (throughout); PFU, plaque-forming units. (e,f) Flow cytometry (e) and fluorescence microscopy (f) of wild-type and Trex1 / MEFs infected for 1 h with -P-eGFP 1. Numbers above outlined areas (e) indicate percent infected (GFP + ) cells. SSC, side scatter; DAPI, DNA-intercalating dye. Original magnification (f),. (g,h) Fluorescence microscopy (g) and quantitative RT-PCR analysis of RNA encoding G and M proteins (h) in wild-type, Trex1 +/ and Trex1 / BMDMs infected for 1 h with -P-eGFP (g) or (h). Original magnification,. (i,j) Quantitative RT-PCR analysis of RNA encoding G and M proteins (i) and immunoblot analysis of proteins (j) in wild-type and TREX1 R11H/R11H (TREX1-mut) primary human skin fibroblasts (isolated from a healthy donor and a patient with AGS, respectively), at 1 h after infection with. Arrowhead, nonspecific band;, tubulin (loading control throughout). (k) Fluorescence microscopy of wild-type and TREX1 R11H/R11H cells infected for 1 h with -P-eGFP. Original magnification,. Data are representative of at least three independent experiments (error bars (a,b,h,i), s.d.). a b c d e ND ND Influenza: + + Influenza NS1 RNA (AU) Influenza: Influenza Influenza Influenza: 37 TREX1 mut + + Figure Trex1-deficient cells have broad antiviral resistance. (a c) Quantitative RT-PCR analysis of RNA encoding the influenza virus nonstructural protein NS1 (a), immunoblot analysis of influenza virus proteins (b) and viral titers in supernatants (c) of wild-type and Trex1 / MEFs or wild-type and TREX1 R11H/R11H human fibroblasts left uninfected or infected with influenza virus (MOI, 1). (d f) Quantitative RT-PCR analysis of RNA encoding the Sendai virus phosphoprotein P (d), immunoblot analysis of Sendai virus proteins (e) and viral titers in the supernatants (f) of wild-type and Trex1 / MEFs or wild-type and TREX1 R11H/R11H human fibroblasts left uninfected or infected with Influenza virus titer ( 1 PFU/ml) f titer ( 1 PFU/ml) g Env RNA (AU) P RNA (AU) 1 ND ND : ND ND : + + h : MOI: 1 1 : i titer ( 1 PFU/ml) 1 1 TREX1 mut + + Sendai virus (; MOI, 1). (g i) Quantitative RT-PCR analysis of RNA encoding the West Nile virus envelope protein Env (g), immunoblot analysis of West Nile virus proteins (h) and viral titers in the supernatants (i) of wild-type and Trex1 / MEFs left uninfected or infected with West Nile virus (; MOI, 1 (h) or 1 (g i)). Data are representative of at least two independent experiments (error bars (a,d,g), s.d.). nature immunology VOUME 1 NUMBER 1 JANUARY 13 3
4 npg 13 Nature America, Inc. All rights reserved. a ow Expression Flu Flu Ifngr Ddx1 Oasl1 Cxcl1 nfkbia Nfkb Irf1 Isg Ddx1 Ccl Il Ifnb1 Ifit1 Ifih1 Nfkb1 Ptgs Cxcl1 Chh Ctsf Ifi7la Atpv1b1 lrf3 lrf Zbp1 lfib Nmi lfnar lfitm3 lfit lfit3 Parp9 Irf7 Usp1 lfi3 Trim1 Adar lrf9 lfi lfi3 Tlr Tlr Cxcr7 lfngr1 Ddx lfitm lfnar1 Rsad Gbp Ddx lsg1 lfi Tlr3 VSIVgp1 () VSIVgp () HA (influenza) PB1 (influenza) NP () P () E () High b mrna (fold) ND ND Ifit1 Ifit Ifit3 Med fifit1 mrna (fold) Iftm3 mrna (fold) g Ifit1 mrna (fold) MEF Ifnar1 / MEF 3 c d 1 e IFN-β (pg/ml) Ifit3 mrna (fold) Iftna mrna (fold) Irf7 mrna (fold) Ifnb1 mrna (AU) Irf7 mrna (fold) Iftnb1 mrna (fold) h infected (%) Flu Trex1 1 3 UI si-ifit1 si-ifitm3 si-stat1 si-stat Flu i Ifit1 mrna (fold) Ifit1 mrna (fold) Ifit1 mrna (fold) Ifit1 mrna (AU) Irf7 mrna (fold) Irf7 mrna (fold) Flu Ifnb mrna (fold) Ifnb mrna (fold) Flu Spleen Trex1 +/ Heart BMDM Figure 3 Interferon-independent activation of a subset of ISGs in Trex1-deficient cells. (a) RNA-Seq analysis of RNA expression in wild-type and Trex1 / MEFs left uninfected or infected for 1 h with, influenza virus (Flu), Sendai virus or West Nile virus (above columns). (b) Quantitative RT-PCR analysis of genes encoding members of the IFIT family in uninfected wild-type and Trex1 / MEFs; results are presented relative to those of the control gene Gapdh. (c) Enzyme-linked immunosorbent assay of IFN-β in supernatants of wild-type and Trex1 / MEFs left uninfected (Med) or infected with. (d,e) Quantitative RT-PCR analysis of Ifnb1 mrna (d) and Ifit1 mrna (e) in wild-type and Trex1 / MEFs mock infected or infected with, influenza virus, Sendai virus or West Nile virus. (f) Quantitative RT-PCR analysis of selected ISGs and interferon-encoding genes in wild-type MEFs 7 h after transfection with control sirna () or Trex1-specific sirna (, 1 3); results are presented relative to those obtained with control sirna, set as 1. Inset, immunoblot analysis of the knockdown of Trex1. P <. (Student s t-test). (g) Quantitative RT-PCR analysis of Ifit1 mrna in Ifnar1 / MEFs 7 h after transfection with sirna as in f (presented as in f). P <. (Student s t-test). (h) -P-eGFP infection in wild-type and Trex1 / MEFs transfected for h with control sirna or sirna specific for (si-) IFIT1, IFITM3, STAT1 or STAT (horizontal axis) and then mock infected (UI) or infected for 1 h with -P-eGFP, analyzed by flow cytometry; results are presented as the frequency of GFP + cells. P <. (Student s t-test). (i) Quantitative RT-PCR analysis of Ifit1, Irf7 and Ifnb1 mrna in spleen, heart and BMDMs isolated from wild-type, Trex1 +/ and Trex1 / mice (presented as in b). P <. (Student s t-test). Data are representative of one experiment (a), at least three independent experiments (b g) or two independent experiments (h,i; error bars (b i), s.d.). Irf7 mrna (fold) Ifnb mrna (fold) among the top-ranked canonical pathways with high hit ratio (determined by the frequency of genes in a pathway that are represented in a data set; Supplementary Fig. c). We then constructed a heat map of genes involved in the antimicrobial response network and representative genes from each virus based on the expression values (and standard deviation (s.d.)) of each gene across all samples (Fig. 3a). All four RNA viruses replicated less efficiently in Trex1 / cells than in wild-type cells (Fig. 3a and Supplementary Fig. ), with values ranging from % to % of those observed in wild-type cells. We also found that many ISGs, such as Ifit1, Ifit3, Isg1, Zbp1 and Usp1, showed considerable induction in uninfected Trex1 / cells (Fig. 3b and Supplementary Fig. 3). Notably, uninfected Trex1 / cells had an ISG-activation signature that resembled that of infected wild-type cells (Supplementary Fig. ), which suggested that the lack of Trex1 function alone was sufficient to initiate an antiviral state. The establishment of this antiviral state seemed to be independent of interferon, because we did not detect any activation of interferon-encoding genes or interferon proteins (Fig. 3c and Supplementary Fig. 3). Moreover, the patterns of induction of Ifnb1 by various viruses were indistinguishable in wild-type and Trex1 / cells (Fig. 3d; influenza virus is known to inhibit the activation of interferon 3 ). In contrast, the abundance of Ifit1 mrna was low in wild-type cells and increased after viral infection, whereas Trex1 / cells began with high expression of Ifit1 (that seemed to be equivalent to that observed in infected wild-type cells), and it remained high after viral infection (Fig. 3e). Trex1 / cells treated with increasing doses of recombinant IFN-β showed a further increase in Ifit1 expression (Supplementary Fig. 7), which suggested that Trex1 / cells were able to respond to interferon signaling. ISGs with substantial induction by Trex1 deficiency, such as those encoding members of the IFIT family, encode molecules with intrinsic antiviral activity against RNA viruses,. Of note, not all known ISGs were activated in Trex1 / cells; those encoding members VOUME 1 NUMBER 1 JANUARY 13 nature immunology
5 npg 13 Nature America, Inc. All rights reserved. of the IFITM family had similar expression in wild-type and Trex1 / cells (Supplementary Fig. 3). Together our data suggested substantial activation of a subset of ISGs in Trex1 / cells independently of the interferon response. To further confirm that the activation of ISGs was specific to the loss of Trex1 function, we knocked down Trex1 expression in wild-type MEFs through the use of three different small interfering RNA (sirna) constructs and noted significantly higher expression of Ifit1, Ifit3 and Irf7 (also an ISG), but not of Ifitm3, Ifna or Ifnb1 (Fig. 3f). We also knocked down Trex1 expression in MEFs deficient in the receptor for IFN-α and IFN-β (Ifnar1 / MEFs) and observed similarly higher expression of Ifit1 and Irf7 (Fig. 3g), which further suggested that the activation of ISGs regulated by Trex1 was interferon independent. To determine whether the activation of ISGs or the interferon pathway contributed to the control of viral infection in Trex1 / cells, we transfected wild-type and Trex1 / cells with a control sirna or sirna specific for the products of two ISGs (IFIT1 and IFITM3) or two key components of the interferon signaling pathway (STAT1 and STAT). We then infected cells with -P-eGFP and measured infectivity by flow cytometry (Fig. 3h). Knockdown of IFIT1 or IFITM3 in Trex1 / cells partially alleviated the blockade of replication, consistent with the known antiviral functions of these two moleules. In contrast, knockdown of STAT1 or STAT had no effect on replication, which further demonstrated that the interferon response was not required for the control of viral infection in Trex1 / cells. To determine whether this ISG-induction signature was present in primary cells of the immune response and tissues from Trex1 / mice, we isolated total RNA from whole spleen, heart and BMDMs from wild-type, Trex1 +/ and Trex1 / mice and measured Ifit1, Irf7 and Ifnb mrna. We observed an induction of ISGs of up to 3-fold in whole tissues and up to -fold in primary cells of the immune system only in Trex1 / mice, relative to their expression in wild-type mice (Fig. 3i). We also observed very low Ifnb expression in all samples from Trex1 / mice (Fig. 3i), consistent with published reports showing restriction of interferon expression to a subset of heart-muscle cells 1,19. We also used RNA-Seq to analyze total RNA from uninfected wild-type fibroblasts or fibroblasts from patients with AGS with the TREX1 R11H/R11H mutation or other mutations linked to AGS, including the RNASEHC D39Y/D11fs and SAMHD1 R9H/QX mutations 7. We again found substantial upregulation of a subset of ISGs, but not interferon-encoding genes, in TREX1 R11H/R11H cells (Fig. a). Notably, the ISG-activation signature was weak in RNASEHC D39Y/D11fs cells and was not present in SAMHD1 R9H/QX cells (Fig. a). To determine whether the same group of ISGs was activated in Trex1 / and TREX1 R11H/R11H cells, we selected 3 ISGs that are expressed in both mouse and human cells and compared their induction in Trex1 / and TREX1 R11H/R11H cells. ISGs that were induced in Trex1 / MEFs were also induced in TREX1 R11H/R11H fibroblasts, with a correlation r value of.9 (Fig. b). We observed weak correlation between gene induction in Trex1 / cells and that in RNASEHC D39Y/D11fs cells (r =.1) and no correlation for that in Trex1 / cells and SAMHD1 R9H/QX cells (r =.). Our data demonstrated that Trex1 also regulated the activation of ISGs in human fibroblasts. Factors for interferon-independent ISG activation We next sought to identify factors of the innate immune system required for the interferon-independent activation of ISGs in Trex1- deficient cells. We chose to measure Ifit1 mrna as an example of a Trex1-regulated ISGs because Ifit1 was the gene upregulated the a b 1 TREX1 mut RNASEHC mut SAMHD1 mut Tlr Ifngr Nmi Ifitm Irf7 Ifitm3 Isg Ifngr1 Chh Atpv1b1 Ddx1 Ifi3 Ifi Irf3 Parp9 Usp1 Irf9 Ifit Ifit1 Ifit3 Ddx Trim1 Rsad Ifih1 Adar Cxcl1 Ifnar1 Isg1 Ddx Cxcr7 Nfkb1 Ddx1 Nfkbia Tlr Irf1 Nfkb Ifnar ow High Expression Expression (fold) TREX1 mut induced Expression (fold) RNASEHC mut induced Expression (fold) SAMHD1 mut induced most in Trex1 deficiency. We first examined the effect of IRF3, which activates ISGs directly 9,. We measured Ifit1 mrna in wild-type, Trex1 / and Trex1 / Irf3 / MEFs and found induction of Ifit1 in Trex1 / single-deficiency cells and that the induction was inhibited by Trex1 / Irf3 / double deficiency (Fig. a), which suggested that IRF3 was required for activation of Ifit1. To determine whether IRF3 was also required for antiviral activity in the setting of Trex1 deficiency, we infected wild-type, Trex1 / and Trex1 / Irf3 / MEFs with or Sendai virus and measured viral proteins by immunoblot analysis or flow cytometry (Fig. b d). Infection with or Sendai virus was inhibited in Trex1 / cells, and infection with each was restored to close to wild-type amounts in Trex1 / Irf3 / cells. Therefore, IRF3 was a key component of the antiviral resistance in Trex1 / cells. We next explored which innate immune pathway upstream of IRF3 was involved in the activation of ISGs in Trex1 / cells. IRF3 is activated mainly by cytosolic DNA- or RNA-sensing pathways mediated by STING-TBK1 or RIG-I MAVS, respectively. Therefore, we knocked down key components of each pathway in Trex1 / cells through the use of sirna and measured Ifit1 expression. Knockdown r = Expression (fold) induced r = Expression (fold) induced r = Expression (fold) induced Figure Selective activation of ISGs in TREX1 mutant fibroblasts. (a) RNA-Seq analysis of RNA in skin fibroblasts from a healthy donor () or from patients with AGS with the TREX1 R11H/R11H (TREX1 mut), RNASEHC D39Y/D11fs (RNASEHC mut) or SAMHD1 R9H/QX (SAMHD1 mut) mutation. (b) Correlation analysis of ISGs (n = 3; expressed in both mouse and human cells) induced by Trex1 deficiency in MEFs versus AGS-related mutations in humans (as in a). Each symbol represents a gene; results are presented as expression in mutant cells relative to that in wild-type cells (fold); r values represent the quality of correlation obtained by the fitting of a power trend line through all data points. Data are from one experiment. nature immunology VOUME 1 NUMBER 1 JANUARY 13
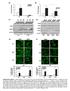
6 npg 13 Nature America, Inc. All rights reserved. a Ifit1 mrna (AU) Irf3 / b c / Trex1 Irf3 / UI Counts 3 UI GFP of IRF3, IRF7, TBK1, STING or IFI resulted in significantly lower Ifit1 expression, whereas knockdown of RIG-I or MAVS had no effect (Fig. e). We also did not observe any effect on Ifit1 expression in Trex1 / cells after knocking down TR7 or TR9 (data not shown). Knockdown of IRF3, IRF7 or TBK1 also diminished the -replication block in Trex1 / cells (Fig. f). These results suggested that the cytosolic DNA-sensing machinery was required for the interferon-independent activation of ISGs in Trex1-deficient cells, but the cytosolic RNA-sensing machinery was not. Knockdown of STING did not seem to enhance replication in Trex1 / cells, probably because the -replication assay measures the entire life cycle of, and many host factors may contribute to this, or because STING also regulates many other genes encoding molecules that could be required for replication. The same cytosolic DNA-sensing pathway is also involved in the activation of interferon-encoding genes during viral infection,11, whose products can then activate ISGs. Therefore, we did double knockdown of Trex1 plus components of the cytosolic DNA-sensing pathway in Ifnar1 / MEFs. Ifit1 expression was higher after knockdown of Trex1, and that was diminished by further knockdown of TBK1, IRF7 or STING (Fig. g). Knockdown of Trex1 in Ifnar1 / MEFs also inhibited replication, and further knockdown of TBK1 or IRF7 alleviated that inhibition (Fig. h). Together our data suggested that the core cytosolic DNA-sensing machinery (STING-TBK1-IRF3-IRF7) was involved in activating ISGs directly in cells with diminished or no Trex1 activity. Trex1 regulates lysosomal biogenesis via TFEB and mtorc1 We next sought to identify the underlying basis for the activation of ISGs in Trex1 / and TREX1 R11H/R11H cells. We first considered the possibility that Trex1 directly inhibits the cytosolic DNA-sensing machinery. To test this, we used 93T human embryonic kidney cells, in which the overexpression of STING induced sixfold more Ifit1 mrna (Supplementary Fig. ). We then coexpressed STING and increasing amounts of Trex1 to determine whether overexpression of Trex1 would inhibit the STING-mediated activation of Ifit Figure Interferon-independent activation of ISGs in Trex1 deficient cells requires STING, TBK1, IRF3 and IRF7. (a) Quantitative RT-PCR analysis of Ifit1 mrna in uninfected wild-type, Trex1 / and Trex1 / Irf3 / MEFs, presented relative to that of Gapdh. (b) Immunoblot analysis of proteins in wild-type, Trex1 / and Trex1 / Irf3 / MEFs left uninfected (UI) or infected for 1 h with. (c) Replication of in wild-type, Trex1 / and Trex1 / Irf3 / MEFs left uninfected or infected for 1 h with -P-eGFP, assessed by -P-eGFP 1 Irf3 / f MEF 7 3 infected (%) 1 Ctrl IRF IRF7 TBK1 STING IFI RIG-I MAVS g Ifit1 mrna (fold) d 3 1 Ifnar / MEF Ctrl Trex1 Trex1 + TBK1 e MEF 1 1 Ctrl IRF3 IRF7 TBK1 STING IFI RIG-I MAVS We did not observe any effect on ifit1 induction after overexpression of Trex1 (Supplementary Fig. ). The same degree of Trex1 overexpression inhibits the HIV-mediated activation of interferon-encoding genes 11. These results suggested that Trex1 did not directly inhibit the cytosolic DNA-sensing machinery. We then hypothesized that perhaps the accumulation of self ligands or a cellular abnormality in Trex1 / or TREX1 R11H/R11H cells might be detected by the STING-TBK1-IRF3-IRF7 pathway. We first assessed the abundance and morphology of cellular organelles in wild-type and Trex1 / cells by immunofluorescence staining of well-defined organelle markers (Fig. a). We did not observe substantial differences between those cells in their mitochondria, Golgi apparatus, endoplasmic reticulum or early endosomes. However, late endosomes (identified by staining with antibody to AMP-1) and lysosomes (identified by staining with the probe ysotracker Red) seemed to be more abundant in Trex1 / cells than in wildtype cells (Fig. a). We also observed a similarly greater abundance of the late endosome-lysosome compartment in Trex1 / BMDMs than in wild-type or Trex1 +/ BMDMs (Fig. b). To determine if this difference was also present in human cells, we stained wildtype and TREX1 R11H/R11H human fibroblasts, as well as control Hea human cervical cancer cells and Hea cells in which Trex1 was knocked down (by sirna), with ysotracker Red. In both cases, we observed much more ysotracker Red staining of Trex1-deficient cells than of wild-type cells (Fig. c and Supplementary Fig. 9), which indicated expansion of the late endosome-lysosome compartment in cells that lacked Trex1 function. We quantified the lysosomal expansion by flow cytometry of live cells with ysotracker Red and found that Trex1 / cells and TREX1 R11H/R11H cells had three- to fivefold more lysosomes than did wild-type cells (Fig. d). We also detected more of the lysosomal membrane proteins AMP-1 and NPC1 in TREX1 R11H/R11H cells and Trex1 / cells than in wild-type cells by immunoblot analysis (Fig. e), which suggested enhanced lysosomal biogenesis in Trex1-deficient cells. To further confirm the expansion of the lysosome compartment, we analyzed wild-type and Ifit1 mrna (%) Irf3 / UI Trex1 + IRF7 Trex1 + STING h infected (%) 3 1 Ifnar / MEF Ctrl Trex1 Trex1 + TBK1 flow cytometry. (d) Immunoblot analysis of Sendai virus proteins in wild-type, Trex1 / and Trex1 / Irf3 / MEFs left uninfected or infected for 1 h with Sendai virus. (e) Quantitative RT-PCR analysis of Ifit1 mrna in Trex1 / MEFs transfected for 7 h with control sirna or sirna specific for components of DNA- or RNA-sensing pathways (horizontal axis), results are presented relative to those obtained with control sirna, set as 1%. (f) -P-eGFP infection in Trex1 / MEFs transfected with sirna as in e and infected for 1 h with -P-eGFP, assessed by flow cytometry. (g) Quantitative RT-PCR analysis of Ifit1 in Ifnar1 / MEFs transfected with control sirna or with various combinations (horizontal axis) of sirna specific for Trex1, TBK1, IRF7 or STING; results are presented relative to those of cells transfected with control sirna, set as 1. (h) -P-eGFP infection in Ifnar1 / MEFs transfected with sirna as in g and then infected with -P-eGFP. P <.1 and P <. (Student s t-test). Data are representative of at least two independent experiments (error bars (a,e h), s.d.). Trex1 + IRF7 Trex1 + STING VOUME 1 NUMBER 1 JANUARY 13 nature immunology
7 a Mito (Hsp) Golgi (GM13) MEF b ate endo-lyso (AMP1) ate endo-lyso (ysotracker) BMDM Trex1 +/ e FB AMP-1 FB NPC1 HMGB1 Mut Mut MEF NPC1 HMGB1 BMDM NPC1 HMGB1 npg 13 Nature America, Inc. All rights reserved. ER (Calreticulin) Early endo (EEA1) ate endo-lyso (AMP1) ate endo-lyso (ysotracker) Trex1 / cells by electron microscopy. Trex1 / cells had significantly more lysosome vacuolar structures (Fig. f,g). Those structures were surrounded by single-layer membranes, some of which contained electron-dense cellular materials commonly found in lysosomes (Fig. f, inset). ysosomes are important organelles for the breakdown and turnover of other cellular organelles (such as mitochondria), proteins and nucleic acids 9. Of note, we did not observe excessive accumulation of undigested cellular materials in these lysosomes; such accumulation is often found in cells associated with lysosomal storage diseases 3. We also did not detect more autolysosomes in Trex1 / cells than in wild-type cells, as measured by electron microscopy, the formation of dots of GFP-tagged autophagy marker C3, and immunoblot analysis of p, a ubiquitin-binding scaffold protein, and C3 (Supplementary Fig. 1 and data not shown). To determine whether the lysosome-expansion phenotype in Trex1 / cells was caused by the induction of genes encoding proteins of the lysosome, we measured the expression of Ctsa, Sgsh, amp1, Mcoln1 and Tpp1, which encode enzymes or structural proteins of the lysosome. All five genes were upregulated three- to fivefold in Trex1 / cells relative to their expression in wild-type cells, whereas other nonlysosomal genes were not (Fig. 7a). Many other genes encoding molecules involved in lysosomal biogenesis were also upregulated in Trex1 / cells relative to their expression in wild-type cells (Supplementary Fig. 11). ysosomal genes are regulated by TFEB through the recognition of conserved binding sites in their promoters. c d ysotracker Counts Counts TREX1 mut MEF Human fibroblast ysotracker TREX1 mut Figure Trex1 negatively regulates lysosomal biogenesis. (a,b) Fluorescence microscopy of wild-type and Trex1 / MEFs (a) and BMDMs (b) stained for various organelle markers (left margin; antibody or dye in parentheses): mito, mitochondria; ER, endoplasmic reticulum; endo, endosome; lyso, lysosome. Original magnification,. (c) Fluorescence microscopy of wild-type and TREX1 R11H/R11H human fibroblasts stained with ysotracker Red. Original magnification,. (d) Flow cytometry of live wild-type and Trex1 / MEFs or wild-type and TREX1 R11H/R11H human fibroblasts stained with ysotracker Red. (e) Immunoblot analysis of AMP-1 and NPC1 in wild-type and TREX1 R11H/R11H (Mut) human fibroblasts (FB) and wild-type and Trex1 / MEFs and BMDMs. (f) Electron microscopy of wild-type and Trex1 / MEFs: arrows indicate undigested (electron-dense) cellular materials in lysosome vacuoles; arrowheads in inset (enlargement of area outlined in main image) indicate single membrane surrounding a lysosome vacuole. N, nucleus; M, mitochondrion;, lysosome. Scale bar, 1 µm. (g) Quantification of lysosomal vacuoles in thin sections of MEFs (per cell; n = ). P <. (Student s t-test). Data are representative of at least three independent experiments (a e) or one experiment (f,g; error bars (g), s.d.). f M M M TFEB is a master regulator of the CEAR gene network ( coordinated lysosomal expression and regulation ) 31, and overexpression of TFEB results in higher lysosomal gene expression and promotes lysosome expansion 3. TFEB resides mostly in the cytoplasm and translocates to the nucleus after complex post-translational modifications 33,3. We did not observe any difference between wild-type and Trex1 / cells in their abundance of Tfeb mrna or TFEB protein (Fig. 7a and data not shown). To examine the subcellular localization of TFEB, we stained wild-type and Trex1 / cells with an antibody to TFEB and found that endogenous TFEB became mostly nuclear in Trex1 / cells (Fig. 7b). This result suggested that the higher lysosomal gene expression and expansion of the lysosomal compartment were connected to altered localization of TFEB in Trex1 / cells. We did not detect any interaction between Trex1 and TFEB by immunoprecipitation of proteins from wild-type MEFs (data not shown), which suggested that Trex1 did not regulate the translocation of TFEB through direct binding and retention in the cytosol. To determine whether TFEB function was required for the activation of ISGs and antiviral activity of Trex1 / cells, we knocked down TFEB expression in Trex1 / cells and measured Ifit1 and Ifit3 mrna in uninfected cells, as well as replication in those same cells. Knockdown of TFEB in Trex1 / cells resulted in lower expression of both Ifit1 and Ifit3 and more replication (Fig. 7c e). Knockdown of TFEB in wild-type MEFs did not affect Ifit1 or other genes encoding molecules of the innate immune system predicted to be targets N M M N g ysosome (per section) 3 1 nature immunology VOUME 1 NUMBER 1 JANUARY 13 7
8 npg 13 Nature America, Inc. All rights reserved. a mrna (fold) e infected (%) Ctsa Sgsh amp1 Mcoln1 Tpp1 Actb : sirna: 1 1+ si-tfeb Hmgb Set Tfeb flfit1 mrna (fold) of TFEB 31 (Supplementary Fig. 1), which suggested that TFEB did not regulate ISGs directly. Moreover, we found that Trex1 / Irf3 / cells had higher expression of lysosomal genes and AMP-1 protein, similar to that in Trex1 / cells, whereas ISG expression was much lower in Trex1 / Irf3 / cells than in Trex1 / cells (Supplementary Fig. 11), which suggested that lysosomal biogenesis (regulated by TFEB) acted upstream of ISG expression (regulated by IRF3 and IRF7). Overexpression of TFEB promotes lysosomal biogenesis 3. To determine whether manipulating the expression or nuclear translocation of TFEB in wild-type cells also induced the expression of ISGs, we overexpressed TFEB in wild-type MEFs and found that Ifit1 expression was increased in a dose-dependent manner (Fig. 7f). We also treated wild-type MEFs with chloroquine, which induces translocation of TFEB to the nucleus 3, and observed a dose-dependent increase in the expression of Mcoln1 (a lysosomal gene) and Ifit1 (Fig. 7g). These data further supported the proposal of a link between TFEB function in lysosomal biogenesis and induction of ISGs. Of note, the treatment of Trex1 / cells with chloroquine did not restore replication (Supplementary Fig. 13), probably because of the known antiviral effect of chloroquine 3 3. One of the upstream regulators of the translocation of TFEB to the nucleus is mtorc1, and inhibition of mtorc1 activity under many conditions promotes the transport of TFEB into the nucleus 3,39. b TFEB MEF TFEB 3 pcdna Plasmid (ng) g lfit1 mrna (fold) MEF Chloroquine Mock Mcoln1 mrna (fold) Mock c 1 TFEB Figure 7 Trex1 regulates lysosomal biogenesis via TFEB and mtorc1. (a) Quantitative RT-PCR analysis of lysosomal and nonlysosomal genes (horizontal axis) in wild-type and Trex1 / MEFs (presented as in Fig. 3b). (b) Fluorescence microscopy of wild-type and Trex1 / MEFs, assessing the endogenous localization of TFEB (left), and frequency of nuclear TFEB (right; n = 13 cells). Original magnification (left),. (c,d) Immunoblot analysis of the knockdown of TFEB (c) and quantitative RT-PCR analysis of Ifit1 and Ifit3 mrna (d; results presented as in Fig. 3f) in Trex1 / MEFs transfected with control sirna or TFEB-specific sirna (si-tfeb). (e) -P-eGFP infection in wild-type and Trex1 / MEFs transfected for 7 h with sirna as in d and mock infected or infected for 1 h with -P-eGFP, assessed by flow cytometry. (f) Quantitative RT-PCR analysis of Nuclear TFEB (%) h : + + mtorc1 p-sk (Thr39) p-sp (Ser3,Ser3) SP p-e-bp1 (Thr37,Thr) Chloroquine E-BP1 jlfit1 mrna (fold) MEF No sirna si-tfeb si-mtor d lfit1 mrna (% of Ctrl) MEF si-tfeb We thus examined mtorc1 activity in infected and uninfected wild-type and Trex1 / cells. We found that infection with induced mtorc1 activity in wild-type MEFs (Fig. 7h), consistent with the function of mtorc1 as a proviral factor. The activity of mtorc1 was much lower in uninfected and infected Trex1 / cells than in uninfected and infected wild-type cells (as assessed by phosphorylation of the kinase SK, the ribosomal protein SP and the translationinitiation inhibitor E-BP1; Fig. 7h,i). We also found that knockdown of mtor (via two independent sirna molecules) in wild-type MEFs resulted in higher Ifit1 expression (Fig. 7j). Moreover, expression of Flag-tagged Trex1 enhanced mtorc1 activity in wild-type cells and restored mtorc1 activity in Trex1 / cells relative to the activity achieved with the vector plasmid control (Fig. 7k). Our data suggested that Trex1 was important for maintaining mtorc1 activity and that a lower abundance of mtor led to the induction of ISGs. Consistent with our data, lower mtorc1 activity has been associated with antiviral effects. Collectively, our data suggested that Trex1 regulated lysosomal biogenesis through TFEB and mtorc1 and that lysosomal biogenesis had a critical role in innate immunity and antiviral defense (Supplementary Fig. 1). DISCUSSION It is well established that interferon has an important role in antiviral immunity. Cells are equipped with an extensive network of sensing k lfit3 mrna (% of Ctrl) p-sp (Ser3,Ser3) p-e-bp1 (Thr37,Thr) HMGB i Densitometry value MEF si-tfeb Vector F-Trex1 Vector F-Trex1 Ifit1 mrna in wild-type MEFs transfected for h with various concentrations (horizontal axis) of empty vector plasmid (pcdna) or plasmid encoding Myc-tagged TFEB; results are presented relative to those of mock-transfected cells ( ng), set as 1. (g) Quantitative RT-PCR analysis of the expression of Ifit1 and Mcoln1 in wild-type MEFs mock treated (Mock) or treated for 1 h with chloroquine (wedges: 1, or 1 µm); results are presented relative to those of mock-treated cells, set as 1. (h,i) Immunoblot analysis of phosphorylated (p-) and total mtorc1, SK, SP and E-BP1 in wild-type and Trex1 / MEFs left uninfected or infected for 1 h with (h; phosphorylated amino acids in parentheses), followed by densitometry analysis, presented relative to results obtained for wild-type cells, set as 1 (i). (j) Quantitative RT-PCR analysis of Ifit1 mrna in wild-type MEFs transfected for 7 h with control sirna or sirna specific for Trex1 or mtor (si-mtor, 1 and ); results are presented relative to those of cells transfected with control sirna, set as 1. (k) Immunoblot analysis of phosphorylated and total SP and E-BP1 in wild-type and Trex1 / MEFs transfected with for h with empty vector or vector encoding Flag-tagged Trex1 (F-Trex1). P <. (Student s t-test). Data are representative of at least three independent experiments (error bars (a,b,d,e,g,i,j), s.d.). p-sp p-e-bp1 VOUME 1 NUMBER 1 JANUARY 13 nature immunology
9 npg 13 Nature America, Inc. All rights reserved. mechanisms in the innate immune system for the detection of invading pathogens through recognition by pattern-recognition receptors. When such receptors are engaged, they trigger signaling pathways that often lead to the activation of interferon expression 3. Infection with enveloped viruses also triggers an interferon-independent pathway that involves the direct activation by IRF3 of a subset of ISGs. In fact, IRF3 can bind to the promoters of many ISGs in addition to those of interferon-encoding genes 1. The promoters of interferonencoding genes (such as Ifnb1) are complex, containing both positive and negative regulatory elements for transcripion factors such as members of the IRF family, NF-κB and AP-1, and a concerted effort by multiple transcription factors is often required for their stimulation. In contrast, the promoters of many ISGs (such as Ifit1) are simpler and can be easily turned on by IRF proteins independently of interferon 9,1. Direct activation of antiviral genes is important for nonprofessional interferon-producing cells such as fibroblasts to effectively defend themselves against viral infection or for cells to defend themselves against viruses that have evolved mechanisms to disrupt the interferon response. It is also advantageous for cells to rapidly induce some ISGs after viral infection before a stronger and more sustained response can be established by interferon signaling pathways. A study of a cytosolic RNA-sensing pathway has provided evidence that interferon-independent activation of ISGs mediated by peroxisomal MAVS is functionally important for defense against infection with RNA viruses 1. Very little is known about whether interferon-independent activation of ISGs occurs in the absence of infection and how it is regulated. Here we have identified Trex1, a cytosolic protein associated with the endoplasmic reticulum, as a key negative regulator of interferonindependent activation of Ifit1 and other ISGs in uninfected cells. When the function of Trex1 was disrupted, either by genetic deficiency in mice or by a homozygous mutation in humans, or by sirnamediated knockdown in a variety of cell types, a subset of ISGs were activated independently of interferon, which led to an antiviral state. Notably, the induction of ISGs in Trex1-deficient cells was sustained at a very high degree and achieved an antiviral state similar to that caused by the interferon-dependent pathway. That was in contrast to the interferon-independent response induced by viral infection in wild-type cells that seems to be temporary and less robust 1. We also challenged wild-type and Trex1 / mouse cells and TREX1 R11H/R11H human cells with a variety of RNA viruses, including, influenza virus, Sendai virus and West Nile virus, and all these viruses failed to replicate in cells that lacked Trex1 function. We have also identified a pathway in the innate immune system, involving STING, TBK1, IRF3 and IRF7, that was important for the interferon-independent activation of ISGs in Trex1-deficient cells. STING is a critical factor for the sensing of pathogen-associated DNA or cyclic di-gmp in the cytosol and subsequent induction of interferon expression,7. Our data have expanded the function of the STING-TBK1-IRF3-IRF7 pathway to include both interferon-dependent and interferon-independent branches as downstream pathways. A published study has also shown that STING activates the phosphorylation of STAT after viral infection, which then induces the expression of chemokines such as CC, CC and CC and homing of cells of the immune system. We did not observe induction of the expression of those chemokines in Trex1 / or TREX1 R11H/R11H cells relative to their expression in wild-type cells (data not shown). Collectively, our study has expanded the understanding of STING and associated factors of the innate immune system as a versatile machinery that can activate multiple distinct downstream pathways. Our data have also shed some light on the potential endogenous trigger of interferon-independent activation of ISGs. We found that Trex1-deficient or Trex1-mutant cells had excessive amounts of lysosomal vacuoles and expanded lysosomal compartments, as determined by immunofluorescence and immunoblot analysis of lysosomal markers, quantitative RT-PCR analysis of lysosomal genes, and electron microscopy. Consistent with such enhanced lysosome biogenesis, the master regulator of lysosome genes TFEB translocated to become predominantly nuclear in Trex1 / cells. We also found lower mtorc1 activity in Trex1 / cells and restoration of mtorc1 activity after expression of Flag-tagged Trex1 in Trex1 / cells; this suggested an important role for Trex1 in maintaining the activity of mtorc1, which regulates the translocation of TFEB to the nucleus 3,39. We also provided the following lines of evidence that demonstrated a functional linkage of TFEB-regulated lysosomal biogenesis to the activation of ISGs: knockdown of TFEB in Trex1- deficient cells tempered the activation of ISGs and antiviral immunity; overexpression of TFEB in wild-type cells, which promotes lysosomal biogenesis 3, resulted in higher Ifit1 expression; treatment of wildtype cells with chloroquine, which induces translocation of TFEB to the nucleus 3 and has antiviral activity 3 3, resulted in higher Ifit1 expression, up to 1-fold; and knockdown of mtor by sirna in wild-type cells also resulted in higher Ifit1 expression. Furthermore, given our observation of higher expression of lysosomal genes and a greater abundance of lysosomal proteins and lack of excess accumulation of undigested contents, Trex1 / cells probably have enhanced lysosomal function. The release of abnormally large amounts of processed peptides or nucleic acids into the cytosol or into the extracellular space (via exocytosis ) might break cellular homeostasis or immunotolerance or exceed the threshold for cytosolic sensing of DNA. The exact identity of such cytosolic DNA remains unclear; published studies have indicated that it might be DNA-replication debris or endogenous retroelements 19. Aberrant functions of lysosomes have been indicated in lupus nephritis, in which lysosomal contents mimic viral particles and activate innate immunity 3. It is also possible that a greater abundance of lysosome vacuoles could result in a membrane perturbation that would elicit an interferon-dependent or interferon-independent antiviral response 9,. Further studies are needed to distinguish among these possibilities. Collectively, our work has demonstrated a link between lysosomal biogenesis and activation of ISGs by the innate immune system, as well as a previously unknown role for Trex1 in regulating lysosomal biogenesis through TFEB and mtorc1. Trex1 inhibits HIV-mediated activation of interferons 11. Here we confirmed that finding and further identified a previously unknown function of Trex1 in the regulation of interferon-independent activation of the innate immune response through lysosomal biogenesis in uninfected cells, which resulted in a broad-spectrum antiviral state in which the replication of several different RNA viruses was inhibited. Both functions of Trex1 shared a similar signaling pathway of the innate immune system that involved STING-TBK1-IRF3, which was able to activate multiple downstream pathways. The upstream stimulus for HIV-mediated interferon activation is HIV DNA from nonproductive reverse transcription 11, whereas the upstream stimulus for the interferon-independent pathway probably involves lysosome function. Our work has also provided further insight into pathogenetic mechanisms underlying systemic autoimmunity associated with TREX1 mutation, such as SE, a prototypical autoimmune disease. Central to SE pathogenesis is that ineffective waste disposal due to impaired apoptosis or defective clearance of cellular debris leads to nature immunology VOUME 1 NUMBER 1 JANUARY 13 9
Trex1 regulates lysosomal biogenesis and interferon-independent activation of antiviral genes
 University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Papers in Veterinary and Biomedical Science Veterinary and Biomedical Sciences, Department of 2013 Trex1 regulates lysosomal
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Papers in Veterinary and Biomedical Science Veterinary and Biomedical Sciences, Department of 2013 Trex1 regulates lysosomal
Supplementary Figure 1
 Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Title: Cytosolic DNA-mediated, STING-dependent pro-inflammatory gene. Fig. S1. STING ligands-mediated signaling response in MEFs. (A) Primary MEFs (1
 1 Supporting Information 2 3 4 Title: Cytosolic DNA-mediated, STING-dependent pro-inflammatory gene induction necessitates canonical NF-κB activation through TBK1 5 6 Authors: Abe et al. 7 8 9 Supporting
1 Supporting Information 2 3 4 Title: Cytosolic DNA-mediated, STING-dependent pro-inflammatory gene induction necessitates canonical NF-κB activation through TBK1 5 6 Authors: Abe et al. 7 8 9 Supporting
Intrinsic cellular defenses against virus infection
 Intrinsic cellular defenses against virus infection Detection of virus infection Host cell response to virus infection Interferons: structure and synthesis Induction of antiviral activity Viral defenses
Intrinsic cellular defenses against virus infection Detection of virus infection Host cell response to virus infection Interferons: structure and synthesis Induction of antiviral activity Viral defenses
Intracellular MHC class II molecules promote TLR-triggered innate. immune responses by maintaining Btk activation
 Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
FIG S1 Examination of eif4b expression after virus infection. (A) A549 cells
 Supplementary Figure Legends FIG S1 Examination of expression after virus infection. () 549 cells were infected with herpes simplex virus (HSV) (MOI = 1), and harvested at the indicated times, followed
Supplementary Figure Legends FIG S1 Examination of expression after virus infection. () 549 cells were infected with herpes simplex virus (HSV) (MOI = 1), and harvested at the indicated times, followed
Trim29 gene-targeting strategy. (a) Genotyping of wildtype mice (+/+), Trim29 heterozygous mice (+/ ) and homozygous mice ( / ).
 Supplementary Figure 1 Trim29 gene-targeting strategy. (a) Genotyping of wildtype mice (+/+), Trim29 heterozygous mice (+/ ) and homozygous mice ( / ). (b) Immunoblot analysis of TRIM29 in lung primary
Supplementary Figure 1 Trim29 gene-targeting strategy. (a) Genotyping of wildtype mice (+/+), Trim29 heterozygous mice (+/ ) and homozygous mice ( / ). (b) Immunoblot analysis of TRIM29 in lung primary
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1
 The cytosolic exonuclease TREX inhibits the innate immune response to human immunodeficiency virus type Nan Yan, Ashton D Regalado-Magdos, Bart Stiggelbout, Min Ae Lee-Kirsch & Judy Lieberman Viral infection
The cytosolic exonuclease TREX inhibits the innate immune response to human immunodeficiency virus type Nan Yan, Ashton D Regalado-Magdos, Bart Stiggelbout, Min Ae Lee-Kirsch & Judy Lieberman Viral infection
Nature Immunology doi: /ni Supplementary Figure 1. Raf-1 inhibition does not affect TLR4-induced type I IFN responses.
 Supplementary Figure 1 Raf-1 inhibition does not affect TLR4-induced type I IFN responses. Real-time PCR analyses of IFNB, ISG15, TRIM5, TRIM22 and APOBEC3G mrna in modcs 6 h after stimulation with TLR4
Supplementary Figure 1 Raf-1 inhibition does not affect TLR4-induced type I IFN responses. Real-time PCR analyses of IFNB, ISG15, TRIM5, TRIM22 and APOBEC3G mrna in modcs 6 h after stimulation with TLR4
October 26, Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
SUPPLEMENTAL INFORMATIONS
 1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
Innate Immunity & Inflammation
 Innate Immunity & Inflammation The innate immune system is an evolutionally conserved mechanism that provides an early and effective response against invading microbial pathogens. It relies on a limited
Innate Immunity & Inflammation The innate immune system is an evolutionally conserved mechanism that provides an early and effective response against invading microbial pathogens. It relies on a limited
Lecture 2: Virology. I. Background
 Lecture 2: Virology I. Background A. Properties 1. Simple biological systems a. Aggregates of nucleic acids and protein 2. Non-living a. Cannot reproduce or carry out metabolic activities outside of a
Lecture 2: Virology I. Background A. Properties 1. Simple biological systems a. Aggregates of nucleic acids and protein 2. Non-living a. Cannot reproduce or carry out metabolic activities outside of a
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS
 Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
T-cell activation T cells migrate to secondary lymphoid tissues where they interact with antigen, antigen-presenting cells, and other lymphocytes:
 Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
T-cell activation T cells migrate to secondary lymphoid tissues where they interact with antigen, antigen-presenting cells, and other lymphocytes:
 Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh-
 1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
SUPPLEMENTARY INFORMATION
 sirna pool: Control Tetherin -HA-GFP HA-Tetherin -Tubulin Supplementary Figure S1. Knockdown of HA-tagged tetherin expression by tetherin specific sirnas. HeLa cells were cotransfected with plasmids expressing
sirna pool: Control Tetherin -HA-GFP HA-Tetherin -Tubulin Supplementary Figure S1. Knockdown of HA-tagged tetherin expression by tetherin specific sirnas. HeLa cells were cotransfected with plasmids expressing
Irf1 fold changes (D) 24h 48h. p-p65. t-p65. p-irf3. t-irf3. β-actin SKO TKO 100% 80% 60% 40% 20%
 Irf7 Fold changes 3 1 Irf1 fold changes 3 1 8h h 8h 8h h 8h p-p6 p-p6 t-p6 p-irf3 β-actin p-irf3 t-irf3 β-actin TKO TKO STKO (E) (F) TKO TKO % of p6 nuclear translocation % % 1% 1% % % p6 TKO % of IRF3
Irf7 Fold changes 3 1 Irf1 fold changes 3 1 8h h 8h 8h h 8h p-p6 p-p6 t-p6 p-irf3 β-actin p-irf3 t-irf3 β-actin TKO TKO STKO (E) (F) TKO TKO % of p6 nuclear translocation % % 1% 1% % % p6 TKO % of IRF3
Reviewers' comments: Reviewer #1 (Remarks to the Author):
 Reviewers' comments: Reviewer #1 (Remarks to the Author): In this manuscript, Song et al. identified FBXW7 as a new positive regulator for RIG-Itriggered type I IFN signaling pathway. The authors observed
Reviewers' comments: Reviewer #1 (Remarks to the Author): In this manuscript, Song et al. identified FBXW7 as a new positive regulator for RIG-Itriggered type I IFN signaling pathway. The authors observed
Innate immunity. Abul K. Abbas University of California San Francisco. FOCiS
 1 Innate immunity Abul K. Abbas University of California San Francisco FOCiS 2 Lecture outline Components of innate immunity Recognition of microbes and dead cells Toll Like Receptors NOD Like Receptors/Inflammasome
1 Innate immunity Abul K. Abbas University of California San Francisco FOCiS 2 Lecture outline Components of innate immunity Recognition of microbes and dead cells Toll Like Receptors NOD Like Receptors/Inflammasome
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
General information. Cell mediated immunity. 455 LSA, Tuesday 11 to noon. Anytime after class.
 General information Cell mediated immunity 455 LSA, Tuesday 11 to noon Anytime after class T-cell precursors Thymus Naive T-cells (CD8 or CD4) email: lcoscoy@berkeley.edu edu Use MCB150 as subject line
General information Cell mediated immunity 455 LSA, Tuesday 11 to noon Anytime after class T-cell precursors Thymus Naive T-cells (CD8 or CD4) email: lcoscoy@berkeley.edu edu Use MCB150 as subject line
MISSION: understanding the mechanisms of therapeutic strategies
 Telethon Institute of Genetics and Medicine MISSION: understanding the mechanisms of genetic diseases to develop preventive and therapeutic strategies G E N O T Y P E G E Researcher N 1 3 5 O T Y P E 8
Telethon Institute of Genetics and Medicine MISSION: understanding the mechanisms of genetic diseases to develop preventive and therapeutic strategies G E N O T Y P E G E Researcher N 1 3 5 O T Y P E 8
Chapter 6. Antigen Presentation to T lymphocytes
 Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature05732 SUPPLEMENTARY INFORMATION Supplemental Data Supplement Figure Legends Figure S1. RIG-I 2CARD undergo robust ubiquitination a, (top) At 48 h posttransfection with a GST, GST-RIG-I-2CARD
doi: 10.1038/nature05732 SUPPLEMENTARY INFORMATION Supplemental Data Supplement Figure Legends Figure S1. RIG-I 2CARD undergo robust ubiquitination a, (top) At 48 h posttransfection with a GST, GST-RIG-I-2CARD
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
Crucial role for human Toll-like receptor 4 in the development of contact allergy to nickel
 Supplementary Figures 1-8 Crucial role for human Toll-like receptor 4 in the development of contact allergy to nickel Marc Schmidt 1,2, Badrinarayanan Raghavan 1,2, Verena Müller 1,2, Thomas Vogl 3, György
Supplementary Figures 1-8 Crucial role for human Toll-like receptor 4 in the development of contact allergy to nickel Marc Schmidt 1,2, Badrinarayanan Raghavan 1,2, Verena Müller 1,2, Thomas Vogl 3, György
Nature Immunology doi: /ni.3268
 Supplementary Figure 1 Loss of Mst1 and Mst2 increases susceptibility to bacterial sepsis. (a) H&E staining of colon and kidney sections from wild type and Mst1 -/- Mst2 fl/fl Vav-Cre mice. Scale bar,
Supplementary Figure 1 Loss of Mst1 and Mst2 increases susceptibility to bacterial sepsis. (a) H&E staining of colon and kidney sections from wild type and Mst1 -/- Mst2 fl/fl Vav-Cre mice. Scale bar,
S1a S1b S1c. S1d. S1f S1g S1h SUPPLEMENTARY FIGURE 1. - si sc Il17rd Il17ra bp. rig/s IL-17RD (ng) -100 IL-17RD
 SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
Nature Immunology: doi: /ni Supplementary Figure 1. Production of cytokines and chemokines after vaginal HSV-2 infection.
 Supplementary Figure 1 Production of cytokines and chemokines after vaginal HSV-2 infection. C57BL/6 mice were (a) treated intravaginally with 20 µl of PBS or infected with 6.7x10 4 pfu of HSV-2 in the
Supplementary Figure 1 Production of cytokines and chemokines after vaginal HSV-2 infection. C57BL/6 mice were (a) treated intravaginally with 20 µl of PBS or infected with 6.7x10 4 pfu of HSV-2 in the
Stochastic Expression of the Interferon-b Gene
 Mingwei Zhao 1, Jiangwen Zhang 2., Hemali Phatnani 3., Stefanie Scheu 4, Tom Maniatis 1,3 * 1 Department of Molecular and Cellular Biology, Harvard University, Cambridge, Massachusetts, United States of
Mingwei Zhao 1, Jiangwen Zhang 2., Hemali Phatnani 3., Stefanie Scheu 4, Tom Maniatis 1,3 * 1 Department of Molecular and Cellular Biology, Harvard University, Cambridge, Massachusetts, United States of
Under the Radar Screen: How Bugs Trick Our Immune Defenses
 Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 7: Cytokines Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research HHV-8 Discovered in the 1980 s at the
Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 7: Cytokines Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research HHV-8 Discovered in the 1980 s at the
Scott Abrams, Ph.D. Professor of Oncology, x4375 Kuby Immunology SEVENTH EDITION
 Scott Abrams, Ph.D. Professor of Oncology, x4375 scott.abrams@roswellpark.org Kuby Immunology SEVENTH EDITION CHAPTER 11 T-Cell Activation, Differentiation, and Memory Copyright 2013 by W. H. Freeman and
Scott Abrams, Ph.D. Professor of Oncology, x4375 scott.abrams@roswellpark.org Kuby Immunology SEVENTH EDITION CHAPTER 11 T-Cell Activation, Differentiation, and Memory Copyright 2013 by W. H. Freeman and
NOTES. The Naturally Attenuated Kunjin Strain of West Nile Virus Shows Enhanced Sensitivity to the Host Type I Interferon Response
 JOURNAL OF VIROLOGY, June 2011, p. 5664 5668 Vol. 85, No. 11 0022-538X/11/$12.00 doi:10.1128/jvi.00232-11 Copyright 2011, American Society for Microbiology. All Rights Reserved. NOTES The Naturally Attenuated
JOURNAL OF VIROLOGY, June 2011, p. 5664 5668 Vol. 85, No. 11 0022-538X/11/$12.00 doi:10.1128/jvi.00232-11 Copyright 2011, American Society for Microbiology. All Rights Reserved. NOTES The Naturally Attenuated
IFNγ Inhibits the Cytosolic Replication of Shigella flexneri via the Cytoplasmic RNA Sensor RIG-I
 IFNγ Inhibits the Cytosolic Replication of Shigella flexneri via the Cytoplasmic RNA Sensor RIG-I The Harvard community has made this article openly available. Please share how this access benefits you.
IFNγ Inhibits the Cytosolic Replication of Shigella flexneri via the Cytoplasmic RNA Sensor RIG-I The Harvard community has made this article openly available. Please share how this access benefits you.
2. Innate immunity 2013
 1 Innate Immune Responses 3 Innate immunity Abul K. Abbas University of California San Francisco The initial responses to: 1. Microbes: essential early mechanisms to prevent, control, or eliminate infection;
1 Innate Immune Responses 3 Innate immunity Abul K. Abbas University of California San Francisco The initial responses to: 1. Microbes: essential early mechanisms to prevent, control, or eliminate infection;
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/366/ra25/dc1 Supplementary Materials for Viral entry route determines how human plasmacytoid dendritic cells produce type I interferons Daniela Bruni, Maxime
www.sciencesignaling.org/cgi/content/full/8/366/ra25/dc1 Supplementary Materials for Viral entry route determines how human plasmacytoid dendritic cells produce type I interferons Daniela Bruni, Maxime
Structure and Function of Antigen Recognition Molecules
 MICR2209 Structure and Function of Antigen Recognition Molecules Dr Allison Imrie allison.imrie@uwa.edu.au 1 Synopsis: In this lecture we will examine the major receptors used by cells of the innate and
MICR2209 Structure and Function of Antigen Recognition Molecules Dr Allison Imrie allison.imrie@uwa.edu.au 1 Synopsis: In this lecture we will examine the major receptors used by cells of the innate and
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
7.012 Quiz 3 Answers
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
Newly Recognized Components of the Innate Immune System
 Newly Recognized Components of the Innate Immune System NOD Proteins: Intracellular Peptidoglycan Sensors NOD-1 NOD-2 Nod Protein LRR; Ligand Recognition CARD RICK I-κB p50 p65 NF-κB Polymorphisms in Nod-2
Newly Recognized Components of the Innate Immune System NOD Proteins: Intracellular Peptidoglycan Sensors NOD-1 NOD-2 Nod Protein LRR; Ligand Recognition CARD RICK I-κB p50 p65 NF-κB Polymorphisms in Nod-2
Nature Medicine: doi: /nm.2109
 HIV 1 Infects Multipotent Progenitor Cells Causing Cell Death and Establishing Latent Cellular Reservoirs Christoph C. Carter, Adewunmi Onafuwa Nuga, Lucy A. M c Namara, James Riddell IV, Dale Bixby, Michael
HIV 1 Infects Multipotent Progenitor Cells Causing Cell Death and Establishing Latent Cellular Reservoirs Christoph C. Carter, Adewunmi Onafuwa Nuga, Lucy A. M c Namara, James Riddell IV, Dale Bixby, Michael
Identification of Host Cytosolic Sensors and Bacterial Factors Regulating the Type I Interferon Response to Legionella pneumophila
 Identification of Host Cytosolic Sensors and Bacterial Factors Regulating the Type I Interferon Response to Legionella pneumophila Kathryn M. Monroe, Sarah M. McWhirter, Russell E. Vance* Division of Immunology
Identification of Host Cytosolic Sensors and Bacterial Factors Regulating the Type I Interferon Response to Legionella pneumophila Kathryn M. Monroe, Sarah M. McWhirter, Russell E. Vance* Division of Immunology
AP Biology Reading Guide. Concept 19.1 A virus consists of a nucleic acid surrounded by a protein coat
 AP Biology Reading Guide Name Chapter 19: Viruses Overview Experimental work with viruses has provided important evidence that genes are made of nucleic acids. Viruses were also important in working out
AP Biology Reading Guide Name Chapter 19: Viruses Overview Experimental work with viruses has provided important evidence that genes are made of nucleic acids. Viruses were also important in working out
THE ROLE OF ALTERED CALCIUM AND mtor SIGNALING IN THE PATHOGENESIS OF CYSTINOSIS
 Research Foundation, 18 month progress report THE ROLE OF ALTERED CALCIUM AND mtor SIGNALING IN THE PATHOGENESIS OF CYSTINOSIS Ekaterina Ivanova, doctoral student Elena Levtchenko, MD, PhD, PI Antonella
Research Foundation, 18 month progress report THE ROLE OF ALTERED CALCIUM AND mtor SIGNALING IN THE PATHOGENESIS OF CYSTINOSIS Ekaterina Ivanova, doctoral student Elena Levtchenko, MD, PhD, PI Antonella
Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent Protein.
 prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
Dr. Ahmed K. Ali Attachment and entry of viruses into cells
 Lec. 6 Dr. Ahmed K. Ali Attachment and entry of viruses into cells The aim of a virus is to replicate itself, and in order to achieve this aim it needs to enter a host cell, make copies of itself and
Lec. 6 Dr. Ahmed K. Ali Attachment and entry of viruses into cells The aim of a virus is to replicate itself, and in order to achieve this aim it needs to enter a host cell, make copies of itself and
Innate Immunity. Hathairat Thananchai, DPhil Department of Microbiology Faculty of Medicine Chiang Mai University 2 August 2016
 Innate Immunity Hathairat Thananchai, DPhil Department of Microbiology Faculty of Medicine Chiang Mai University 2 August 2016 Objectives: Explain how innate immune system recognizes foreign substances
Innate Immunity Hathairat Thananchai, DPhil Department of Microbiology Faculty of Medicine Chiang Mai University 2 August 2016 Objectives: Explain how innate immune system recognizes foreign substances
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous
 Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Dynamic Interaction of Stress Granule, DDX3X and IKK-α Mediates Multiple Functions in
 Dynamic Interaction of Stress Granule, and Mediates Multiple Functions in Hepatitis C Virus Infection Véronique Pène, Qisheng Li#, Catherine Sodroski, Ching-Sheng Hsu, T. Jake Liang# Liver Diseases Branch,
Dynamic Interaction of Stress Granule, and Mediates Multiple Functions in Hepatitis C Virus Infection Véronique Pène, Qisheng Li#, Catherine Sodroski, Ching-Sheng Hsu, T. Jake Liang# Liver Diseases Branch,
Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication
 Manuscript EMBO-2010-74756 Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication Tsai-Ling Liao, Chung-Yi Wu, Wen-Chi Su, King-Song Jeng and Michael Lai Corresponding
Manuscript EMBO-2010-74756 Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication Tsai-Ling Liao, Chung-Yi Wu, Wen-Chi Su, King-Song Jeng and Michael Lai Corresponding
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
Supplementary Figure 1
 Supplementary Figure 1 Constitutive EGFR signaling does not activate canonical EGFR signals (a) U251EGFRInd cells with or without tetracycline exposure (24h, 1µg/ml) were treated with EGF for 15 minutes
Supplementary Figure 1 Constitutive EGFR signaling does not activate canonical EGFR signals (a) U251EGFRInd cells with or without tetracycline exposure (24h, 1µg/ml) were treated with EGF for 15 minutes
Supplementary Information. Supplementary Figure 1
 Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Toll-like Receptors (TLRs): Biology, Pathology and Therapeutics
 Toll-like Receptors (TLRs): Biology, Pathology and Therapeutics Dr Sarah Sasson SydPATH Registrar 23 rd June 2014 TLRs: Introduction Discovered in 1990s Recognise conserved structures in pathogens Rely
Toll-like Receptors (TLRs): Biology, Pathology and Therapeutics Dr Sarah Sasson SydPATH Registrar 23 rd June 2014 TLRs: Introduction Discovered in 1990s Recognise conserved structures in pathogens Rely
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table Title of file for HTML: Peer Review File Description: Innate Scavenger Receptor-A regulates
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table Title of file for HTML: Peer Review File Description: Innate Scavenger Receptor-A regulates
Follow this and additional works at: Part of the Medicine and Health Sciences Commons
 Washington University School of Medicine Digital Commons@Becker Open Access Publications 2009 Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2009 Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent
Influenza virus exploits tunneling nanotubes for cell-to-cell spread
 Supplementary Information Influenza virus exploits tunneling nanotubes for cell-to-cell spread Amrita Kumar 1, Jin Hyang Kim 1, Priya Ranjan 1, Maureen G. Metcalfe 2, Weiping Cao 1, Margarita Mishina 1,
Supplementary Information Influenza virus exploits tunneling nanotubes for cell-to-cell spread Amrita Kumar 1, Jin Hyang Kim 1, Priya Ranjan 1, Maureen G. Metcalfe 2, Weiping Cao 1, Margarita Mishina 1,
LESSON 4.4 WORKBOOK. How viruses make us sick: Viral Replication
 DEFINITIONS OF TERMS Eukaryotic: Non-bacterial cell type (bacteria are prokaryotes).. LESSON 4.4 WORKBOOK How viruses make us sick: Viral Replication This lesson extends the principles we learned in Unit
DEFINITIONS OF TERMS Eukaryotic: Non-bacterial cell type (bacteria are prokaryotes).. LESSON 4.4 WORKBOOK How viruses make us sick: Viral Replication This lesson extends the principles we learned in Unit
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
Fayth K. Yoshimura, Ph.D. September 7, of 7 HIV - BASIC PROPERTIES
 1 of 7 I. Viral Origin. A. Retrovirus - animal lentiviruses. HIV - BASIC PROPERTIES 1. HIV is a member of the Retrovirus family and more specifically it is a member of the Lentivirus genus of this family.
1 of 7 I. Viral Origin. A. Retrovirus - animal lentiviruses. HIV - BASIC PROPERTIES 1. HIV is a member of the Retrovirus family and more specifically it is a member of the Lentivirus genus of this family.
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Cellular compartments
 Cellular compartments 1. Cellular compartments and their function 2. Evolution of cellular compartments 3. How to make a 3D model of cellular compartment 4. Cell organelles in the fluorescent microscope
Cellular compartments 1. Cellular compartments and their function 2. Evolution of cellular compartments 3. How to make a 3D model of cellular compartment 4. Cell organelles in the fluorescent microscope
Chapter 19: The Genetics of Viruses and Bacteria
 Chapter 19: The Genetics of Viruses and Bacteria What is Microbiology? Microbiology is the science that studies microorganisms = living things that are too small to be seen with the naked eye Microorganisms
Chapter 19: The Genetics of Viruses and Bacteria What is Microbiology? Microbiology is the science that studies microorganisms = living things that are too small to be seen with the naked eye Microorganisms
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Protein Trafficking in the Secretory and Endocytic Pathways
 Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was
 Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Toll-like Receptor Signaling
 Toll-like Receptor Signaling 1 Professor of Medicine University of Massachusetts Medical School, Worcester, MA, USA Why do we need innate immunity? Pathogens multiply very fast We literally swim in viruses
Toll-like Receptor Signaling 1 Professor of Medicine University of Massachusetts Medical School, Worcester, MA, USA Why do we need innate immunity? Pathogens multiply very fast We literally swim in viruses
Human type I interferonopathies
 Human type I interferonopathies Yanick Crow Institut Imagine, Paris University of Manchester Human type I interferonopathies 1 Inborn errors of enhanced type I interferon signaling Type I interferons 2
Human type I interferonopathies Yanick Crow Institut Imagine, Paris University of Manchester Human type I interferonopathies 1 Inborn errors of enhanced type I interferon signaling Type I interferons 2
Peli1 negatively regulates T-cell activation and prevents autoimmunity
 Peli1 negatively regulates T-cell activation and prevents autoimmunity Mikyoung Chang 1,*, Wei Jin 1,5,*, Jae-Hoon Chang 1, Yi-chuan Xiao 1, George Brittain 1, Jiayi Yu 1, Xiaofei Zhou 1, Yi-Hong Wang
Peli1 negatively regulates T-cell activation and prevents autoimmunity Mikyoung Chang 1,*, Wei Jin 1,5,*, Jae-Hoon Chang 1, Yi-chuan Xiao 1, George Brittain 1, Jiayi Yu 1, Xiaofei Zhou 1, Yi-Hong Wang
Tbk1-TKO! DN cells (%)! 15! 10!
 a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting
 Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
System Biology analysis of innate and adaptive immune responses during HIV infection
 System Biology analysis of innate and adaptive immune responses during HIV infection Model of T cell memory persistence and exhaustion Naive Ag+APC Effector TEM (Pfp, Gr.B, FasL, TNF) Ag stim. IL-2, IL-7,
System Biology analysis of innate and adaptive immune responses during HIV infection Model of T cell memory persistence and exhaustion Naive Ag+APC Effector TEM (Pfp, Gr.B, FasL, TNF) Ag stim. IL-2, IL-7,
Nature Medicine: doi: /nm.4322
 1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
Overview: Chapter 19 Viruses: A Borrowed Life
 Overview: Chapter 19 Viruses: A Borrowed Life Viruses called bacteriophages can infect and set in motion a genetic takeover of bacteria, such as Escherichia coli Viruses lead a kind of borrowed life between
Overview: Chapter 19 Viruses: A Borrowed Life Viruses called bacteriophages can infect and set in motion a genetic takeover of bacteria, such as Escherichia coli Viruses lead a kind of borrowed life between
Cell Quality Control. Peter Takizawa Department of Cell Biology
 Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Chapter13 Characterizing and Classifying Viruses, Viroids, and Prions
 Chapter13 Characterizing and Classifying Viruses, Viroids, and Prions 11/20/2017 MDufilho 1 Characteristics of Viruses Viruses Minuscule, acellular, infectious agent having either DNA or RNA Cause infections
Chapter13 Characterizing and Classifying Viruses, Viroids, and Prions 11/20/2017 MDufilho 1 Characteristics of Viruses Viruses Minuscule, acellular, infectious agent having either DNA or RNA Cause infections
10th International Rotavirus Symposium Bangkok, Thailand
 Rotavirus Host Range Restriction and Innate Immunity: Mechanisms of Vaccine Attenuation Harry Greenberg MD Stanford University 10th International Rotavirus Symposium Bangkok, Thailand 09/19/12 B dsrna
Rotavirus Host Range Restriction and Innate Immunity: Mechanisms of Vaccine Attenuation Harry Greenberg MD Stanford University 10th International Rotavirus Symposium Bangkok, Thailand 09/19/12 B dsrna
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells
 Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
ISG15 sirna # Ctrl sirna+ifn+wt. Virus titer (Pfu/ml) hours post infection. d USP18 sirna #2 IFN
 a ISG15 sirna Ctrl #1 #2 ISG15 conjugates IB: ISG15 Virus titer (Pfu/ml) b ISG15 sirna #2 10 7 Ctrl sirna+ifn+wt Ctrl sirna+ifn+67 10 6 ISG15 sirna+ifn+wt ISG15 sirna+ifn+67 10 5 10 4 10 3 Free ISG15 10
a ISG15 sirna Ctrl #1 #2 ISG15 conjugates IB: ISG15 Virus titer (Pfu/ml) b ISG15 sirna #2 10 7 Ctrl sirna+ifn+wt Ctrl sirna+ifn+67 10 6 ISG15 sirna+ifn+wt ISG15 sirna+ifn+67 10 5 10 4 10 3 Free ISG15 10
VIROLOGY. Engineering Viral Genomes: Retrovirus Vectors
 VIROLOGY Engineering Viral Genomes: Retrovirus Vectors Viral vectors Retrovirus replicative cycle Most mammalian retroviruses use trna PRO, trna Lys3, trna Lys1,2 The partially unfolded trna is annealed
VIROLOGY Engineering Viral Genomes: Retrovirus Vectors Viral vectors Retrovirus replicative cycle Most mammalian retroviruses use trna PRO, trna Lys3, trna Lys1,2 The partially unfolded trna is annealed
Supplementary information
 Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Expanded View Figures
 PEX13 functions in selective autophagy Ming Y Lee et al Expanded View Figures Figure EV1. PEX13 is required for Sindbis virophagy. A, B Quantification of mcherry-capsid puncta per cell (A) and GFP-LC3
PEX13 functions in selective autophagy Ming Y Lee et al Expanded View Figures Figure EV1. PEX13 is required for Sindbis virophagy. A, B Quantification of mcherry-capsid puncta per cell (A) and GFP-LC3
CELL PART OF THE DAY. Chapter 7: Cell Structure and Function
 CELL PART OF THE DAY Chapter 7: Cell Structure and Function Cell Membrane Cell membranes are composed of two phospholipid layers. Cell membrane is flexible, not rigid The cell membrane has two major functions.
CELL PART OF THE DAY Chapter 7: Cell Structure and Function Cell Membrane Cell membranes are composed of two phospholipid layers. Cell membrane is flexible, not rigid The cell membrane has two major functions.
Summary of Endomembrane-system
 Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
Lecture on Innate Immunity and Inflammation
 Lecture on Innate Immunity and Inflammation Evolutionary View Epithelial barriers to infection Four main types of innate recognition molecules:tlrs, CLRs, NLRs, RLRs NF-κB, the master transcriptional regulator
Lecture on Innate Immunity and Inflammation Evolutionary View Epithelial barriers to infection Four main types of innate recognition molecules:tlrs, CLRs, NLRs, RLRs NF-κB, the master transcriptional regulator
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature89 IFN- (ng ml ) 5 4 3 1 Splenocytes NS IFN- (ng ml ) 6 4 Lymph node cells NS Nfkbiz / Nfkbiz / Nfkbiz / Nfkbiz / IL- (ng ml ) 3 1 Splenocytes IL- (ng ml ) 1 8 6 4 *** ** Lymph node cells
doi: 1.138/nature89 IFN- (ng ml ) 5 4 3 1 Splenocytes NS IFN- (ng ml ) 6 4 Lymph node cells NS Nfkbiz / Nfkbiz / Nfkbiz / Nfkbiz / IL- (ng ml ) 3 1 Splenocytes IL- (ng ml ) 1 8 6 4 *** ** Lymph node cells
Supplementary information
 Supplementary information Exosomes mediate the cell-to-cell transmission of interferon alpha-induced antiviral activity Jianhua Li, Kuancheng Liu, Yang Liu, Yan Xu, Fei Zhang, Huijuan Yang, Jiangxia Liu,
Supplementary information Exosomes mediate the cell-to-cell transmission of interferon alpha-induced antiviral activity Jianhua Li, Kuancheng Liu, Yang Liu, Yan Xu, Fei Zhang, Huijuan Yang, Jiangxia Liu,
Antigen Presentation and T Lymphocyte Activation. Abul K. Abbas UCSF. FOCiS
 1 Antigen Presentation and T Lymphocyte Activation Abul K. Abbas UCSF FOCiS 2 Lecture outline Dendritic cells and antigen presentation The role of the MHC T cell activation Costimulation, the B7:CD28 family
1 Antigen Presentation and T Lymphocyte Activation Abul K. Abbas UCSF FOCiS 2 Lecture outline Dendritic cells and antigen presentation The role of the MHC T cell activation Costimulation, the B7:CD28 family
CPM (x 10-3 ) Tregs +Teffs. Tregs alone ICOS CLTA-4
 A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled
 Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
RAS Genes. The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes.
 ۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
Oncolytic virus strategy
 Oncolytic viruses Oncolytic virus strategy normal tumor NO replication replication survival lysis Oncolytic virus strategy Mechanisms of tumor selectivity of several, some of them naturally, oncolytic
Oncolytic viruses Oncolytic virus strategy normal tumor NO replication replication survival lysis Oncolytic virus strategy Mechanisms of tumor selectivity of several, some of them naturally, oncolytic
