Identification of the LEDGF/p75 HIV-1 integraseinteraction domain and NLS reveals NLS-independent chromatin tethering
|
|
|
- Peter Stone
- 5 years ago
- Views:
Transcription
1 JCS epress online publication date 29 March 2005 Research Article 1733 Identification of the LEDGF/p75 HIV-1 integraseinteraction domain and NLS reveals NLS-independent chromatin tethering Maria Vanegas 1,2, Manuel Llano 1, Sharon Delgado 1, Daniah Thompson 1, Mary Peretz 1 and Eric Poeschla 1,2, * 1 Molecular Medicine Program and 2 Department of Immunology, Mayo Clinic College of Medicine, Rochester, MN 55905, USA *Author for correspondence ( poeschla.eric@mayo.edu) Accepted 17 January , Published by The Company of Biologists 2005 doi: /jcs Summary To investigate the basis for the LEDGF/p75 dependence of HIV-1 integrase (IN) nuclear localization and chromatin association, we used cell lines made stably deficient in endogenous LEDGF/p75 by RNAi to analyze determinants of its location in cells and its ability to interact with IN. Deletion of C-terminal LEDGF/p75 residues preserved nuclear and chromatin localization but abolished the interaction with IN and the tethering of IN to chromatin. Transfer of this IN-binding domain (IBD) was sufficient to confer HIV-1 IN interaction to GFP. HRP-2, the only other human protein with an identifiable IBD domain, was found to translocate IN to the nucleus of LEDGF/p75( ) cells. However, in contrast to LEDGF/p75, HRP-2 is not chromatin bound and does not tether IN to chromatin. A single classical nuclear localization signal (NLS) in the LEDGF/p75 N-terminal region ( 146 RRGRKRKAEKQ 156 ) was found by deletion mapping and was shown to be transferable to pyruvate kinase. Four central basic residues in the NLS are critical for its activity. Strikingly, however, stable expression studies with NLS(+/ ) and IBD(+/ ) mutants revealed that the NLS, although responsible for LEDGF/p75 nuclear import, is dispensable for stable, constitutive nuclear association of LEDGF/p75 and IN. Both wild-type LEDGF/p75 and NLSmutant LEDGF/p75 remain entirely chromatin associated throughout the cell cycle, and each tethers IN to chromatin. Thus, these experiments reveal stable nuclear sequestration of a transcriptional regulator by chromatin during the nuclear-cytosolic mixing of cell division, which additionally enables stable tethering of IN to chromatin. LEDGF/p75 is a multidomain adaptor protein that interacts with the nuclear import apparatus, lentiviral IN proteins and chromatin by means of an NLS, an IBD and additional chromatin-interacting domains. Key words: LEDGF/p75, HRP-2, Integrase, HIV, Chromatin, Nuclear localization Introduction Lens-epithelium-derived growth factor/p75 (LEDGF/p75) is a member of the hepatoma-derived growth factor (HDGF) family. The other six members are a smaller splicing variant, LEDGF/p52, as well as HDGF and four HDGF-related proteins (HRPs): HRP-1, HRP-2, HRP-3 and HRP-4 (Dietz et al., 2002; Ganapathy et al., 2003; Shinohara et al., 2002). Alternative names for LEDGF/p75 are PC4 and SFRS1 interacting protein 2 (PSIP2) and transcriptional coactivator p75. LEDGF/p75 was identified initially by its copurification with the transcriptional coactivator PC4 (Ge et al., 1998a). Interactions of LEDGF/p75 with components of the general transcription machinery and the transcription activation domain of VP16 indicate participation in transcriptional regulation (Ge et al., 1998a; Ge et al., 1998b). A role in activation of stress response genes has been suggested (Sharma et al., 2000; Shinohara et al., 2002). LEDGF/p75 is 530 amino acids in length. A 333 amino acid alternatively spliced product of the same gene, LEDGF/p52 (or PSIP1), has the same N-terminal 325 amino acid acids but a different C-terminus that is only 8 amino acids in length (Ge et al., 1998a; Ge et al., 1998b). Both p75 and p52 are nuclear proteins. LEDGF/p75 is less active as a coactivator than LEDGF/p52, but is much more abundant in cells (Llano et al., 2004a; Nishizawa et al., 2001). Since the discovery that LEDGF/p75 coprecipitates with human immunodeficiency virus type 1 (HIV-1) integrase (IN) (Cherepanov et al., 2003), LEDGF/p75 has drawn increasing interest from researchers studying HIV (Cherepanov et al., 2004; Llano et al., 2004a; Llano et al., 2004b; Maertens et al., 2003; Maertens et al., 2004). IN is a viral enzyme that catalyzes covalent insertion of the reverse-transcribed viral cdna into a host chromosome. HIV-1 and feline immunodeficiency virus (FIV) IN proteins are known to localize to cell nuclei when expressed in the absence of other viral components, as do some small IN fusions [e.g. to green fluorescence protein (GFP) (Bouyac-Bertoia et al., 2001; Cherepanov et al., 2000; Depienne et al., 2000; Gallay et al., 1997; Llano et al., 2004a; Petit et al., 2000; Pluymers et al., 1999; Woodward et al., 2003)]. This apparent karyophilia has led to the proposal of candidate nuclear localization signals (NLSs) in IN and attempts to implicate them in viral pre-integration complex (PIC) nuclear translocation (reviewed by Goff, 2001). However, some studies were inconsistent with an autonomous
2 (8) NLS in HIV-1 IN. For example, fusions of HIV-1 IN to larger indicator proteins such as pyruvate kinase and β-galactosidase are retained in the cytoplasm (Devroe et al., 2003; Kukolj et al., 1997; Llano et al., 2004a). Semi-permeabilized cell studies suggested that nuclear import of HIV-1 IN is facilitated by a limiting cellular factor that is not cytosolic (Depienne et al., 2001). An additional feature that remained to be explained was the tight association of IN with chromatin within the nucleus (Pluymers et al., 1999). Insights were provided into these issues by the findings that LEDGF/p75 co-immunoprecipitates and colocalizes with HIV-1 and FIV IN proteins, and determines their nuclear localization (Cherepanov et al., 2003; Llano et al., 2004a; Maertens et al., 2003). LEDGF/p75 was also found to be present in the functional PICS of HIV-1 and FIV (Llano et al., 2004a) and to protect these IN proteins from the ubiquitinproteasome (Llano et al., 2004b). RNA interference (RNAi) with stably expressed short-hairpin RNAs (shrna) caused highly effective stable knockdown of LEDGF/p75 (while preserving p52 expression) and produced a definitive phenotype: chromatin association of HIV-1 and FIV IN proteins was disrupted and they relocated permanently and completely to the cytoplasm of knocked down stable cell lines (Llano et al., 2004a). Nuclear/chromatin localization of IN was completely abrogated, consistent with a role for LEDGF/p75 as a lentiviral IN-to-chromatin tethering factor. Transient knockdown of LEDGF/p75 with sirna also disrupted chromatin association of a GFP-IN fusion protein, in this case shifting it from the nucleus to a diffuse distribution in both nucleus and cytoplasm (Maertens et al., 2003). The failure of HIV-1 IN karyophilia to be transferred by fusion of IN to some NLS reporter proteins (Devroe et al., 2003; Kukolj et al., 1997; Llano et al., 2004a) suggests that these fusion partners disrupt the IN-LEDGF/p75 interaction. So far, only one non-lentiviral IN, from murine leukemia virus (MLV), has been studied for interaction with LEDGF/p75. MLV IN did not interact with LEDGF/p75 and was cytoplasmic in its presence and absence, suggesting the interaction may be lentivirus specific (Llano et al., 2004a). Unlike LEDGF/p75, LEDGF/p52 does not interact with HIV-1 or FIV IN (Llano et al., 2004a; Maertens et al., 2003). These data suggested that the 205 amino acid LEDGF/p75 C- terminal domain contains the IN-binding domain (IBD), whereas the N-terminal 325 amino acids are likely to determine nuclear location. NLSs mediate nucleopore transit of proteins that exceed the approximately 60 kda upper limit for passive diffusion (Christophe et al., 2000; Jans et al., 2000; Moroianu, 1999). Several categories have been described (Christophe et al., 2000; Gorlich and Kutay, 1999). The best characterized classical NLSs (Kalderon et al., 1984; Lanford and Butel, 1984) are generally composed of a few contiguous basic residues, typically 4-6 lysines or arginines (monopartite signal), or of two such stretches separated by a non-conserved spacer of residues (bipartite signal). Several other kinds of NLSs also exist, such as the M9 sequence in the hnrnpa1 protein (Siomi and Dreyfuss, 1995); these are quite heterogeneous and generally non-basic in character (Christophe et al., 2000). Moreover, basic amino acid motifs that conform to the classical consensus can be found in nonnuclear proteins and many short basic sequences that are presumptive candidates for classical NLSs turn out not to have this function, making empirical mapping essential (Christophe et al., 2000; Gorlich and Kutay, 1999). To determine the basis for the regulation of lentiviral IN trafficking by LEDGF/p75, we systematically analyzed this protein for domains mediating two functions: (1) interaction with HIV-1 IN and (2) nuclear localization. To facilitate the analyses, we engineered stable cell lines by RNAi to lack endogenous LEDGF/p75. We then studied mutant protein phenotypes with respect to the nuclear and chromatin association of LEDGF/p75 and IN. LEDGF/p75 NLSindependent nuclear location of LEDGF/p75 and IN that persists through cell division in stable cell lines is shown. To extend our previous comparative analyses, we cloned and sequenced the feline LEDGF/p75 ortholog, and investigated whether LEDGF/p75 interacts with the IN protein of a second oncoretrovirus, avian leukosis virus (ALV). Materials and Methods Plasmids phin encodes a C-terminally Myc-epitope-tagged version of HIV-1 IN and has been previously described (Llano et al., 2004a). ppk- LEDGF encodes a fusion of pyruvate kinase (Myc-epitope-tagged) to the amino terminus of LEDGF/p75; it was constructed from the pcdna3-based version of pmyc-pk (Siomi and Dreyfuss, 1995). LEDGF/p75 fusions were inserted as NheI/NotI fragments into pmyc- PK. Overlap-extension PCR was used for alanine substitution of residues in LEDGF/p75 using outer primers 5 -AAATA- AGGAAAAGTATGGC-3 and 5 -AAGCAAGTTCATCCAAGGCC- 3, and inner primers 5 -GGGGGGCAGCGGCAGCGGCAGAA- AACAAGTAGAAACTGAGAGG-3 and 5 -CTGCCGCTGCCGC- TGCCCCCCTTCTGGCAGCTTTTGG-3. To allow expression in the presence of stable RNAi against endogenous LEDGF/p75, this and other mutations used pledgf/p75simut, a LEDGF/p75 cdna in which seven synonymous nucleotide changes were introduced at the shrna target site (Llano et al., 2004a). LEDGF/p was constructed by overlap-extension PCR using outer primers 5 - CCCGGGTCGACTCTAGAGGTACC-3 and 5 -TAGGCACCTATT- GGTCTTACTGAC-3 and inner primers 5 -AACATTGTTTTCTT- AACTTCTGGC-3 and 5 -GTTAAGAAAACAATGTTGTATAAC- 3. To enable stable expression, an internal ribosome entry-site-linked puromycin resistance gene (pac) was inserted immediately downstream of LEDGF/p75 genes. A nuclear GFP protein, NLS-GFP, was constructed by inserting the SV40 T antigen NLS (Lanford and Butel, 1984) in frame at the N-terminus of egfp using oligonucleotide adaptors. LEDGF/p75 segments were fused to the NLS-GFP C-terminus as insertions between BamHI and XbaI of pnls-gfp. ALV IN was amplified by PCR from RCASBP (Petropoulos and Hughes, 1991) with primers that incorporate a C- terminal Myc epitope tag as previously described for HIV and FIV IN (Llano et al., 2004a). pflag-hrp-2 was constructed by PCR amplification of HRP-2 from a human cdna library (ATCC Mammalian Genome Collection), followed by addition of an N- terminal FLAG epitope tag and insertion between PstI and NotI in pcmv (Llano et al., 2004a) and verification by DNA sequencing of identity with the published sequence of Izumoto and colleagues (Izumoto et al., 1997). Cell culture, transfections and selection of stable cell lines 293T HEK, QT6, Vero and NIH 3T3 cells from the American Type Tissue Culture Collection were grown in Dulbecco s modified Eagle s medium (Gibco BRL) supplemented with 10% fetal calf serum, penicillin and streptomycin. si1340 cells, abbreviated here as L cells, stably express a highly effective anti-ledgf/p75 short-hairpin RNA
3 HIV-1 integrase and functional domains of LEDGF/p (shrna) (Llano et al., 2004a). siscram cells, abbreviated here as S cells, express a control shrna. The derivation of this cell line is described elsewhere (Llano et al., 2004a). Transfections were performed by the calcium phosphate coprecipitation method with a total of 2 µg of DNA per well of a six-well plate or 1µg of DNA per chamber in a two-chamber LabTek II glass chamber slide (Nalge Nunc). Briefly, cells were transfected 24 hours after being plated in 2 ml of medium at cells/well or 1 ml of medium at cells/chamber. After hours, the transfection mix was replaced with fresh culture medium. Cells were harvested or used for indirect immunofluorescence hours after the transfection mix was added. For LEDGF/p75.NLS and LEDGF/p stable cell lines, L cells were plated in 75 cm 2 flasks and cotransfected the next day with 7 µg of DNA that had been linearized at a restriction site in the prokaryotic backbone. After hours, the transfection mix was replaced with fresh culture medium. 48 hours after transfection, cells were selected in 3.0 µg/ml puromycin. Immunoblotting and laser-scanning confocal immunofluorescence microscopy For Western blotting, cells were lysed in 150 mm NaCl, 0.5% DOC, 0.1% SDS, 1% NP-40, and 150 mm Tris-HCl ph 8.0 plus a protease inhibitor cocktail (Complete-mini; Boehringer). Proteins (30 µg/lane) were resolved in SDS-10% polyacrylamide gels and transferred to Immobilon P membranes (Millipore). Blocked membranes were incubated overnight at 4 C with anti-myc mab (clone 9E10; Covance), anti-alpha-tubulin mab (clone B-5-1-2; Sigma), anti- LEDGF/p75 monoclonal antibody (BD Transduction Laboratories), or anti-flag mab (Sigma) in Tris-buffered saline with 5% nonfat milk plus 0.05% Tween 20. After washing, membranes were incubated with the appropriate horseradish peroxidase-tagged secondary antibody. ECL (Amersham Pharmacia Biotech) detected bound antibodies. Indirect immunofluorescence detection of Myc-epitope-tagged proteins was performed by laser-scanning confocal fluorescence microscopy with a monoclonal anti-myc epitope antibody (Covance; clone 9E10) or a rabbit anti-myc antibody (Santa Cruz) when costained with anti-ledgf/p75 mab. Cells grown in LabTek II chamber slides were fixed with 4% formaldehyde in PBS for 10 minutes at 37 C, washed with PBS, and then permeabilized with ice-cold methanol for two minutes at room temperature. Fixed cells were blocked in 10% fetal calf serum, 20 mm ammonium chloride, and PBS for 30 minutes at room temperature, then incubated with the appropriate antibodies, followed by Alexa or Texas Red-conjugated goat anti-mouse antibody or goat anti-rabbit antibody (Molecular Probes). Nuclear DNA was stained with DAPI (Molecular Probes). Triton X extraction of cells was done as described by Kannouche et al.; briefly, unfixed cells were extracted in 1% Triton X-100 at 37 C prior to fixation in 4% paraformaldehyde and immunofluorescence, with control cells unexposed to the detergent (Kannouche et al., 2004). 40 and 400 mm NaCl. The cell lysates were then centrifuged at 6000 g for 2 minutes. 20 µl supernatant was used for immunoblotting of the total cellular fraction, whereas the rest was incubated with 6 µg of anti-gfp mab (BD Transduction Laboratories) for 15 minutes, followed by addition of 30 µl sheep anti-mouse IgG magnetic beads (Dynal) and incubated at 4 C for one hour on a rotating platform. Beads were washed three times in CSK buffer containing 0.5% NP- 40 and boiled in Laemmli buffer plus β-mercaptoethanol. Samples were separated by SDS-PAGE and analyzed by western blotting with an anti-myc mab and anti-gfp mab as described above. Feline LEDGF/p75 ortholog cloning cdna was prepared from cat peripheral blood mononuclear cell (PBMC) RNA using ProSTAR High fidelity RT-PCR system (Stratagen). The total cdna was used in a PCR reaction with human LEDGF/p75-specific primers: 5 -AGCTAGCTCTAGAATGACTC- GCGATTTCAAACCTGG-3 and 5 -ATATGCGGCCGCCTAGTTA- TCTAGTGTAGAATCC-3. The product was inserted into pcdna3 as an Nhe/Not fragment and sequenced. Results Characterization of an NLS in LEDGF/p75 Interactions of IN with mutant LEDGF/p75 proteins are optimally studied in the absence of the endogenous protein. Therefore, most studies in the present work were carried out in L cells, which stably express a highly effective anti- LEDGF/p75 short-hairpin RNA (shrna) and lack detectable endogenous LEDGF/p75 (Llano et al., 2004a). A control line (S cells) expresses a control shrna. Both L and S cells were derived from 293T cells for analyses of subcellular location (Llano et al., 2004a). To override the stable RNAi for experimental purposes in the present work, a LEDGF/p75 cdna in which seven synonymous nucleotide changes were Immunoprecipitation 48 hours after transfection, cells were lysed for 10 minutes on ice in 350 µl of modified CSK buffer [10 mm Pipes ph 6.8, 10% w/v sucrose, 1 mm DTT, 1 mm MgCl 2 plus an EDTA-free protease inhibitors mixture (Roche Molecular Biochemicals)] supplemented with 0.5% NP- Fig. 1. PK-LEDGF/p75 fusion proteins. Transfections were in L cells. (A) Confocal microscopy. Lower panels show DAPI staining. (B) Immunoblotting. The primary antibody was an anti-myc epitope mab.
4 (8) introduced at the shrna target site (pledgf/p75simut) was used for all transfected LEDGF/p75 constructs. First, all 530 amino acids of LEDGF/p75 were fused to the Fig. 2. Basic residues are required for nuclear localization. Transfections were in L cells. (A) Confocal microscopy. (B) Immunoblotting. Lane 1: Mock; lane 2: PK; lane 3: PK-LEDGF/ ; lane 4: PK-LEDGF/RR AA; lane 5: PK-LEDGF/ RKRK AAAA. C-terminus of Myc-epitope-tagged pyruvate kinase (PK), yielding PK-LEDGF/ PK is frequently used as an NLS reporter because it is a large (60 kda), monomeric, exclusively cytoplasmic protein that exceeds the size limit for diffusion through nucleopores (Bouyac-Bertoia et al., 2001; Fouchier et al., 1997; Llano et al., 2004a; Siomi and Dreyfuss, 1995). We used a flexible linker in all fusions (Ser-Gly 4 -Ser). PK was exclusively cytoplasmic as expected (data not shown), but PK- LEDGF/1-530 localized to the nucleus, which is consistent with the existence of a transferable NLS in LEDGF/p75 (Fig. 1A,i). Portions of LEDGF/p75 were then fused to PK. Fusions of the most N-terminal 200 and 325 amino acids to PK resulted in nuclear localization identical to that of LEDGF/p75 (data not shown). By contrast, fusions of the first 100 amino acids (PK-LEDGF/1-100) or of two C-terminal regions (PK- LEDGF/ and PK-LEDGF/ ) led to exclusively cytoplasmic location (data not shown). PK-LEDGF/1-160 and PK-LEDGF/ were exclusively nuclear, whereas PK- LEDGF was cytoplasmic (Fig. 1A,ii-iv). Expression of single, full-length proteins of predicted molecular mass was verified by immunoblotting for these and all mutant proteins in this study (Fig. 1B). From these experiments, amino acid residues required for LEDGF/p75 nuclear localization could be clearly deduced to reside within interval 146 RRGRKRKAEKQVETE 160, which contains the basic, classical NLS-consistent stretch 146 RRGRKRKAEKQ 156. To confirm this, only amino acids were fused to PK (PK-LEDGF/ ). This fusion protein was exclusively nuclear, identifying this sequence as a transferable NLS (Fig. 2A,i; immunoblotting shown in Fig. 2B). When arginine residues 146 and 147 in PK-LEDGF/ were substituted with alanine (RR146-47AA), the protein was predominantly nuclear in location, but some cytoplasmic immunolabeling was observed (Fig. 2A,ii). When the four central basic residues ( 149 RKRK 152 ) in PK-LEDGF/ were substituted with alanine, the protein was exclusively cytoplasmic (Fig. 2A,iii). Thus, these four basic residues are critical to the NLS, whereas the upstream two basic amino acids 146 RR 147 appear to be needed for optimal function. To test this hypothesis further, we alaninesubstituted 149 RKRK 152 in full-length LEDGF/p75, yielding LEDGF/p75.NLS (Fig. 3A,B). Wild-type LEDGF/p75 was nuclear as expected (Fig. 3A,ii), but LEDGF/p75.NLS was cytoplasmic in interphase L cells, confirming the results of the PK fusion experiments (Fig. 3A,iii; Fig. 3A,iv is discussed further below). Fig. 3. LEDGF/p75 and LEDGF/p75.NLS ; subcellular location and interactions with chromatin, in different phases of the cell cycle. (A) Transient transfection into L cells. (B) Immunoblotting in L cells, using anti-ledgf/p75 mab. Endogenous p52 is also detected by the N-terminus antibody. LEDGF/p75.NLS interacts with HIV-1 IN and sequesters it in the cytoplasm in interphase yet tethers IN to chromosomes in dividing cells We then proceeded to use L cells to test whether LEDGF/p75.NLS colocalizes with
5 HIV-1 integrase and functional domains of LEDGF/p IN (Fig. 4A,B). As expected, IN was cytoplasmic in L cells (Fig. 4A,a-d), but was directed to the nucleus by coexpressed wild-type LEDGF/p75 (Fig. 4A,e-h). By contrast, IN and LEDGF/p75.NLS were excluded from the nuclei of interphase L cells (Fig. 4A,il). In both cases, the transcriptional coactivator and HIV-1 IN precisely colocalized (Fig. 4A,g,k). Moreover, over-expression of IN and LEDGF/ p75.nls in 293T cells (i.e. in the presence of endogenous LEDGF/p75), also resulted in sequestration of IN in the cytoplasm, yielding localizations in these cells opposite to that seen with wild-type LEDGF/p75 and indistinguishable from those in panels i-k of Fig. 4A (data not shown). However, we then found that, although this NLS mediates nuclear import, it is not required for tethering to chromatin and is thus dispensable for stable residence in nuclei of proliferating cells. We were first intrigued by the observation that intense LEDGF/p75.NLS -specific immunolabeling of metaphase and anaphase chromosomes could be found in some transiently transfected L cells that had proceeded into mitosis after transfection (see anaphase cell in Fig. 3A,iv, and mitotic cells in Fig. 4A,m-p). This is the same appearance that wild-type LEDGF/p75 has in mitotic cells (Llano et al., 2004a) (data not shown). However, it was difficult to interpret the finding since the great majority of LEDGF/p75.NLS - transfected cells showed cytoplasmic location (Fig. 3A,iii; Fig. 4A,j). To pursue the issue with greater clarity, we stably expressed LEDGF/p75.NLS in L cells (Fig. 5A,B). Remarkably, in all cells of this cell line (which we derived twice to verify the result), LEDGF/p75.NLS has a wild-type, nuclear location (Fig, 5A) moreover, it mediates precise colocalization and chromatin tethering of IN regardless of position in the cell cycle (Fig. 5B). Thus, mutation of the classical NLS prevents nuclear import of this protein when the nucleus is intact, but does not abrogate its ability to bind to nuclear DNA. We infer that other domains in the NLS-mutant LEDGF/p75 protein are able to mediate binding to chromatin and nuclear retention when it is exposed to cytoplasmic proteins after nuclear envelope breakdown. Identification and characterization of the IBD: residues are necessary and sufficient to mediate IN interaction As noted, p52 does not interact with IN, suggesting that the C- terminal domain of LEDGF/p75 (residues ) contains Fig. 4. Co-expression of IN with LEDGF/p75 or LEDGF/p75.NLS in L cells. (A) Panels a- d: HIV-1 IN was transfected alone. Panels e-h: cotransfection of HIV-1 IN with LEDGF/p75 (using pledgf/p75simut). Panels i-l: cotransfection of HIV-1 IN with pledgf/p75.nls. (B) Immunoblotting of lysates from cells in (A). an IBD. We also noted from BLAST and other sequence alignment tool analyses (data not shown) that the region corresponding to residues of human LEDGF/p75 is well conserved in different species, including Xenopus (92% conserved, 85% identical). We therefore used a method of fusing segments of the p75-unique C-terminus (residues and ) to the C-terminus of a nuclear-targeted GFP test protein (NLS-GFP). We constructed the latter protein by incorporating the SV40 T antigen NLS (Lanford and Butel, 1984) at the GFP N-terminus. The experimental strategy is that the NLS-GFP moiety in the fusions is predicted to confine them to the nucleus (substituting functionally for the N- terminal NLS of LEDGF/p75), allowing potential interactions with untethered, cytoplasmic HIV-1 IN in L cells to be detected by confocal microscopy. Use of L cells both eliminates confounding effects of endogenous LEDGF/p75 and permits
6 (8) Fig. 5. Stable expression of LEDGF/p75.NLS in L cells; subcellular localization and interaction with IN. Stable cell lines were derived by puromycin selection. (A) Confocal immunofluorescence with anti- LEDGF/p75 mab. DAPI staining (right) indicates chromatin (approximately 50% of the cells in this stable L cell line express the LEDGF/p75 mutant). (B) The same cells after transfection of HIV-1 IN. LEDGF/p75.NLS and IN colocalize (overlay, right). cytoplasmically located IN for testing. We initially analyzed their subcellular locations in the absence of IN (Fig. 6A) and verified expression of full-length fusions by immunoblotting (Fig. 6B). As predicted, NLS-GFP and NLS-GFP/ were nuclear (Fig. 6A). NLS-GFP/ was also nuclear, with discrete intra-nuclear foci apparent (Fig. 6A). We then cotransfected L cells with plasmids encoding HIV- 1 IN and the NLS-GFP fusion proteins (Fig. 7). NLS-GFP had no effect on IN, which remained cytoplasmic (Fig. 7A,a-d). By contrast, the GFP protein with the smaller LEDGF/p75 C- terminal fragment (NLS-GFP/ ) caused a diagnostic relocation and colocalization with IN. IN was re-directed from the cytoplasm to the nucleus (Fig. 7A,i-k). Co-expression of HIV-1 IN with NLS-GFP/ also caused total relocation/ colocalization of the fusion protein, but from the nucleus to the cytoplasm (Fig. 7A,e-h). This intriguing opposite polarity is probably due to masking of the NLS of NLS-GFP/ by the interaction with IN or induction of additional proteinprotein interactions by the IN NLS-GFP/ complex. To confirm the interaction between the putative IBD and IN, lysates were also immunoprecipitated. HIV IN coimmunoprecipitated with NLS-GFP/ , but not with NLS-GFP (Fig. 7B). To test the model, we deleted amino acids in LEDGF/p75 and tested its interaction with IN in L cells (Fig. 8). LEDGF/p was nuclear, with wild-type intranuclear distribution (Fig. 8A, top row) and displayed the predicted reduction in molecular weight (Fig. 8B). This protein Fig. 6. C-terminal segments of LEDGF/p75 fused to NLS-GFP: expression and intracellular location. (A) Confocal microscopy of NLS-GFP-LEDGF fusions. (B) Immunoblotting with anti-gfp mab. Lane 1: Mock; lane 2: NLS-GFP; lane 3: NLS-GFP/ ; lane 4: NLS-GFP/ had the same appearance in the presence of endogenous LEDGF/p75 (data not shown). However, in contrast to the wild-type protein, did not interact with HIV-1 IN or sequester it to the nucleus of L cells, in any stage of the cell cycle (Fig. 8A). Taken together, the experiments indicate residues in the p75 unique C-terminal domain mediate interaction with HIV-1 IN. All proteins we test that have this IBD colocalize with HIV-1 IN, whether they are LEDGF/p75 proteins or chimeric GFP test proteins. All proteins that lack the IBD do not colocalize with HIV-1 IN. Comparative virological aspects of the IN-LEDGF/p75 interaction We have been interested in whether the LEDGF/p75 interaction with IN is lenti-retrovirus specific. FIV IN also displays LEDGF/p75-dependent nuclear localization (Llano et al., 2004a), whereas murine leukemia virus IN does not. In both L and S cells, MLV IN is stably cytoplasmic (Llano et al., 2004a). By contrast, in the present work, we found that ALV IN is stably nuclear in L cells, S cells and Quail (QT6) cells (data not shown), a result that is consistent with the transferable NLS that exists in this protein (Kukolj et al., 1997). FIV IN
7 HIV-1 integrase and functional domains of LEDGF/p subcellular localization is the same in both feline and human cells (data not shown). To extend our comparative data with both HIV-1 and FIV IN, we cloned and sequenced the feline LEDGF/p75 ortholog and compared the domains we have mapped. A cdna was isolated by RT-PCR from Felis catus PBMC mrna using human-specific primers, and sequenced (GenBank accession no. AY705213). Human and feline LEDGF/p75 show overall 4% nonidentity. The NLS is fully conserved. The IBD is also identical in the two proteins except for a conservative valine to isoleucine change at residue 411. Nine of nineteen total variant residues (and six of the nine total non-conservative changes) cluster in the extreme C-terminus between amino acids 485 and 530. HRP-2 co-expression translocates HIV-1 IN from the cytoplasm to the nucleus in the LEDGF/p75( ) background BLAST analyses with LEDGF/p75 and LEDGF/p75 fragments identified only one other human protein with a region similar to the IBD. This is hepatoma-derived growth factor (HDGF)-related protein 2 (HRP-2) (Izumoto et al., 1997). The HDGF gene family, of which HRP-2 is a member, is reviewed in Dietz et al. (Dietz et al., 2002). ClustalW alignment showed that residues of human HRP-2 possess 54% identity and 83% similarity to the LEDGF IBD ( ), suggesting a conserved functional domain. We therefore cloned a human HRP-2 cdna by PCR from a human cdna library and expressed the protein in L cells (Fig. 9). Although 670 amino acids in length, HRP- 2 displayed a molecular mass of approximately 140 kda (Fig. 9B), which is larger than the predicted mass of 74 kda. A similarly sized HRP-2 band was observed by Engelman and colleagues (Cherepanov et al., 2004). We speculate that this size shift is due to post-translational modifications (e.g. phosphorylation at serine-acidic clusters) (Meier and Blobel, 1992) that are prominent in HRP-2. HRP-2 was nuclear in the presence and absence of LEDGF/p75 (i.e. in L cells; Fig. 9A, top row) and 293T cells (data not shown). To determine if HRP-2 interacts with HIV-1 IN and can substitute functionally for the nuclear-translocating function displayed by exogenously expressed LEDGF/p75 in L cells, we coexpressed HIV-1 IN with and without HRP-2 in these cells. Similar to the action of LEDGF/p75 (Fig. 4A,e-h), HRP-2 relocated HIV-IN from the cytoplasm into the nucleus where both proteins colocalized (Fig. 9A). Notably however, multiple attempts to co-immunoprecipitate IN with HRP-2 and vice Fig. 7. C-terminal segments of LEDGF/p75 fused to NLS-GFP: IN interaction. L cells were cotransfected with HIV-1 IN and either NLS-GFP, NLS-GFP/ , or NLS-GFP/ shrna target-site-mutated (simut) versions of LEDGF/p75 were used in all expression plasmids. (A) Confocal microscopy. (B) Co-immunoprecipitation of NLS-GFP/ and IN. Immunoprecipitation was carried out with a mab to GFP, followed by immunoblotting for GFP (lower) and IN (upper). versa from these cells were unsuccessful (data not shown), a result that strongly contrasts with the abundant coprecipitation we observe under the same conditions for LEDGF/p75 and IN (Llano et al., 2004a). HRP-2 is not a chromatin-tethering protein Since LEDGF/p75 is tightly associated with chromatin and this forms the basis for its IN-tethering function, we examined whether HRP-2 also displays this property by expressing HRP- 2 in L cells, in the presence and absence of exogenous LEDGF/p75 (Fig. 10). The intranuclear distribution of HRP-2 in interphase cells was more homogeneous than that of LEDGF/p75. In addition, the HRP-2 and LEDGF/p75 did not precisely overlap in interphase nuclei (note the red-yellow variegation in the two such nuclei with both proteins in Fig. 10p). However, the striking difference between the proteins is observed in actively dividing cells. HRP-2 was seen to be unassociated with condensed chromatin in cells in various phases of mitosis [e.g. metaphase (Fig. 10p) or telophase (Fig. 10l)]. Note the complete lack of overlap of LEDGF/p75, which is chromosome bound, and HRP-2, which is distributed away
8 (8) Fig. 8. LEDGF/p75 residues are required for nuclear import of HIV IN. Transfections were done in L cells. (A) Confocal microscopy. (B) Immunoblotting. from the chromosomes. The distribution of HRP-2 was the same when LEDGF/p75 was absent (Fig. 10a-h). In addition, Triton X-100 extraction of unfixed cells (Kannouche et al., 2004) removed detectable HRP-2 but did not extract LEDGF/p75 from cell nuclei (data not shown). Finally, coexpression of HIV-1 IN and HRP-2 in L cells showed they colocalized precisely in all cellcycle phases, with the same distribution seen for HRP-2 alone in Fig. 10; however, when all three proteins were co-expressed in L cells, HRP-2 over-expression did not affect the colocalization of IN, LEDGF/p75 and chromatin (data not shown). These results suggest a dominant role for LEDGF/p75 over HRP-2 for IN localization, which is consistent with a weaker interaction of HRP-2 with IN as discussed above. observed subcellular locations of lentiviral IN proteins. First, we determined that a discrete region in the LEDGF/p75 C- terminus is responsible for IN interaction. Second, in the course of identifying and characterizing a single NLS in the N- terminus that determines the nuclear import of the protein, our data revealed novel processes that result in LEDGF/p75 and HIV-1 IN chromatin association. The experiments provide clear evidence for NLS-independent stable nuclear sequestering of a transcriptional regulator by chromatin during the nuclear-cytosolic mixing of cell division. This process in turn enables sequestration of IN through a LEDGF/p75 tether. Overall, LEDGF/p75 can be considered a multi-domain adaptor protein that interacts with the nuclear import apparatus, lentiviral IN proteins, and chromatin by means of an NLS, an IBD and additional chromatin-interacting domains. Two complementary lines of evidence identify residues in LEDGF/p75 as a functional domain necessary and sufficient for interaction with HIV-1 IN. This segment of the p75-unique C terminus is transferable and acts autonomously within the transferred context. In addition, its deletion abrogates IN interaction, in all phases of the cell cycle. There is complete correlation between the presence of the IBD in a protein and IN interaction as assessed by coimmunoprecipitation and confocal immunofluorescence. The location of this IBD in the C-terminus of LEDGF/p75 is consistent with the prior observation that LEDGF/p52 does not interact with IN proteins and lends further support to the chromatin-tethering model. After this paper was first submitted, Engelman and colleagues published data that identified the NLS and IBD domains using different methods (Cherepanov et al., 2004). Our data are consistent with their identifications of these Discussion Here we address two problems that concern LEDGF/p75 domain structure and illuminate the process by which LEDGF/p75 governs the Fig. 9. Expression and subcellular localization of HRP-2 and HIV-1 IN. (A) Confocal microscopy of L cells cotransfected with pflag-hrp-2 (top), HIV-1 IN (middle), or both (bottom). (B) Immunoblotting of L cells transfected with pflag-hrp-2.
9 HIV-1 integrase and functional domains of LEDGF/p Fig. 10. Differential HRP-2 and LEDGF/p75 location in L cells. L cells were cotransfected with pflag-hrp-2 without (a-h) or with (i-p) LEDGF/p75 (using pledgf/p75simut). motifs, although some differences are apparent, and additional findings about subcellular trafficking of LEDGF/p75 and HRP- 2 are presented here. They localized the IBD to residues using in vitro pull-downs, and this is in good agreement with our mapping to as the minimal domain fragment that caused GFP to interact with HIV-1 IN in colocalization and co-immunoprecipitation assays. In addition, we showed that deletion of this region ( ) from LEDGF/p75 resulted in loss of IN interaction. Thus, considering both studies together, it is reasonable to conclude that residues represent the minimal IBD. The function of HRP-2 is currently unknown. Like other members of the HDGF family, it contains a PWWP domain in its N-terminus (Izumoto, 1997; Diezt et al., 2002). Within this family, only LEDGF/p75 and HRP-2 contain a basic C- terminus, in which the IBD resides. GST pull-downs showed that the homologous IBD region in HRP-2 (amino acids ) interact with IN, and IN could be co-immunoprecipitated from 293T cells with HA-tagged HRP-2 (Cherepanov et al., 2004). We have also shown here that when IN and HRP-2 are co-expressed in L cells, which have been thoroughly documented to exclude lentiviral integrases from the nucleus (Llano et al., 2004a), HIV-1 IN translocates from the cytoplasm to the nucleus. These data suggest that HIV-1 integrase can utilize both LEDGF/p75 and HRP-2 for nuclear localization. However, a key distinction emerged in our experiments: HRP- 2 clearly does not associate with chromatin-like LEDGF/p75 and does not tether IN to chromatin (Fig. 10). In contrast to Cherepanov et al. (Cherepanov et al., 2004), we were unable to co-immunoprecipitate HRP-2 and IN using a variety of extraction and precipitation conditions, although we are able to reciprocally co-immunoprecipitate IN and LEDGF/p75 easily (Llano et al., 2004a; Llano et al., 2004b). In this regard, considerably less HRP-2 than LEDGF/p75 was coprecipitable by IN in the experiments reported (Cherepanov et al., 2004). Thus, the IN HRP-2 interaction appears to be considerably weaker than the IN- LEDGF/p75 interaction. In dividing cells, the nuclear envelope breaks down in early prophase with microtubule-induced tearing of the nuclear lamina, allowing mixing of cytoplasmic and nuclear contents, and it reassembles at the end of mitosis after sister chromatid separation (Beaudouin et al., 2002; Foisner, 2003). The only apparent requirement for stable chromatin association of LEDGF/ p75.nls is that nuclear contents become accessible during cell division. This aspect was clearly established by the LEDGF/p75.NLS stable cell line, where cytoplasmic LEDGF/p75.NLS was never visualized in confocal microscopy. In transiently transfected cells, LEDGF/p75.NLS, which is identical to the wild-type protein except for the four amino acids changed to alanine, was excluded from the nucleus until the advent of cell division, an exclusion that is not seen with wild-type LEDGF/p75. Thus, four basic amino acids in the identified LEDGF/p75 NLS are required for IN to be imported into an intact nucleus and become chromatin associated. The LEDGF/p75 NLS conforms well to consensus sequences for classical NLSs of the SV40 T antigen-type (Christophe et al., 2000), and functions as a discrete, transferable, linear element. Maertens et al. also analyzed LEDGF/p75 for NLSs, using transiently transfected GFP- LEDGF/p75 and HcRed1-IN fusion proteins (Maertens et al., 2004). Our data agree with Maertens et al. on the identity of the NLS and in the conclusion that NLS-mutant LEDGF/p75 can colocalize with HIV-1 IN in the cytoplasm. Our NLS experiments were different methodologically in that we used immunofluorescence, allowing us to assess directly the location of LEDGF/p75 that is unfused to any protein and is wild type except for the NLS mutation, and to assess its interactions with unfused IN. Because fluorescent proteins (in particular red fluorescent protein) tend to oligomerize and may aggregate (Rizzo and Piston, 2005), this methodological difference might explain the absence of apparent large cytoplasmic co-aggregates or inclusions of colocalized proteins in our study (compare each in Fig. 4). In addition, we extended our analyses to stable expression, which allowed us to follow cells through multiple generations, revealing that this NLS is totally dispensable for stable nuclear localization if cells are dividing. Confocal immunofluorescence shows LEDGF/p75.NLS to be always and entirely chromatin associated in all cells of the population. These data form an interesting parallel with the results of Devroe et al., who found that the addition of an NES to integrase caused IN to be
10 (8) cytoplasmic after transient transfection, but nuclear after stable expression (Devroe et al., 2003). The finding that ALV IN is nuclear in the presence and absence of LEDGF/p75 shows that stable nuclear residence of a retroviral IN protein does not necessarily imply LEDGF/p75 interaction. This result with a fourth retroviral IN protein also adds support to our hypothesis that the interaction of IN with LEDGF/p75 is lentivirus specific and might therefore be implicated in aspects of replication distinctive to the lentiviridae. Because the present data involve expression of HIV-1 integrase protein outside the viral context, they do not yet demonstrate a functional role for LEDGF/p75 in HIV-1 replication. The precise role of the IN-LEDGF/p75 interaction in the viral life cycle remains uncertain. A role in determining the predilection of HIV-1 for integration into active genes (Schroder et al., 2002) merits investigation. Our experiments are consistent with a model in which LEDGF/p75 is a multidomain adaptor protein that can mediate either nuclear import of IN or chromatin sequestration of IN, both of which would be IBD dependent. Retention of both proteins, with LEDGF/ p75 tethering IN, is dependent on the binding of LEDGF/p75 to chromatin. No nuclear import is required for stable chromatin association if cells are cycling. The ability of both LEDGF/p75 and HRP-2 to function in translocating IN to the nucleus in L cells suggests that these proteins mediate nucleopore transit of IN. Alternatively, IN may undergo nuclear import in nondividing cells independently of LEDGF/p75, and then be subjected to nuclear sequestration through interaction with intra-nuclear LEDGF/p75 or HRP-2. However, the latter model would not seem to explain the exclusion of IN from the nuclei of L cells (Fig. 4A,a-c), unless net active export of IN is occurring in these cells. Thus, whereas it is clear that LEDGF/p75 and IN can interact in the cytoplasm, whether IN-LEDGF/p75 complexes are the principal means for the nucleopore transit of IN cannot be stated definitively based on presently available data. Semi-permeabilized cell assays suggest LEDGF/p75 import occurs through the importin alpha/beta pathway (Maertens et al., 2004), which is plausible since this has been the case for all classical NLSs yet studied (Christophe et al., 2000). However, IN nuclear import in the in vitro assay was apparently not enhanced by LEDGF/p75 (Maertens et al., 2004). Furthermore, Depienne et al., using the same digitoninpermeabilization method, reported that HIV-1 IN nuclear import did not involve the importin alpha/beta pathway (Depienne et al., 2001). Deletion analyses focused on identifying the chromatininteracting domains in LEDGF/p75 will be useful. Both LEDGF/p75 and HRP-2 contain an N-terminal PWWP domain upstream of the NLS. In other proteins, PWWP domain involvement in protein-protein interactions (Stec et al., 2000) and chromatin association (Ge et al., 2004) has been suggested. Downstream of the NLS, AT-Hook motifs are present in both LEDGF/p75 and HRP-2. Proteins containing AT hooks bind AT-rich DNA and are thought to coregulate transcription by modifying the architecture of DNA to enhance the accessibility of promoters to transcription factors (Aravind and Landsman, 1998; Reeves and Nissen, 1990; Siddiqa et al., 2001). Both of these elements are candidates for mediating chromatin interaction of LEDGF/p75, perhaps cooperatively, and might also influence interactions with the transcription machinery. We thank G. Dreyfuss (University of Pennsylvania, PA) for pyruvate kinase plasmids, Z. Debyser (Rega Institute, Belgium) for a human LEDGF/p75 cdna, and G. Bren (Mayo Clinic College of Medicine, MN) for pcdna3.gfp. References Aravind, L. and Landsman, D. (1998). AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids. Res. 26, Beaudouin, J., Gerlich, D., Daigle, N., Eils, R. and Ellenberg, J. (2002). Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108, Bouyac-Bertoia, M., Dvorin, J., Fouchier, R., Jenkins, Y., Meyer, B., Wu, L., Emerman, M. and Malim, M. H. (2001). HIV-1 infection requires a functional integrase NLS. Mol. Cell 7, Cherepanov, P., Pluymers, W., Claeys, A., Proost, P., de Clercq, E. and Debyser, Z. (2000). High-level expression of active HIV-1 integrase from a synthetic gene in human cells. FASEB J. 14, Cherepanov, P., Maertens, G., Proost, P., Devreese, B., van Beeumen, J., Engelborghs, Y., de Clercq, E. and Debyser, Z. (2003). HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278, Cherepanov, P., Devroe, E., Silver, P. A. and Engelman, A. (2004). Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 279, Christophe, D., Christophe-Hobertus, C. and Pichon, B. (2000). Nuclear targeting of proteins: how many different signals? Cell Signal. 12, Depienne, C., Roques, P., Creminon, C., Fritsch, L., Casseron, R., Dormont, D., Dargemont, C. and Benichou, S. (2000). Cellular distribution and karyophilic properties of matrix, integrase, and Vpr proteins from the human and simian immunodeficiency viruses. Exp. Cell Res. 260, Depienne, C., Mousnier, A., Leh, H., le Rouzic, E., Dormont, D., Benichou, S. and Dargemont, C. (2001). Characterization of the nuclear import pathway for HIV-1 integrase. J. Biol. Chem. 276, Devroe, E., Engelman, A. and Silver, P. A. (2003). Intracellular transport of human immunodeficiency virus type 1 integrase. J. Cell Sci. 116, Dietz, F., Franken, S., Yoshida, K., Nakamura, H., Kappler, J. and Gieselmann, V. (2002). The family of hepatoma-derived growth factor proteins: characterization of a new member HRP-4 and classification of its subfamilies. Biochem. J. 366, Foisner, R. (2003). Cell cycle dynamics of the nuclear envelope. ScientificWorldJournal 3, Fouchier, R. A., Meyer, B. E., Simon, J. H., Fischer, U. and Malim, M. H. (1997). HIV-1 infection of non-dividing cells: evidence that the aminoterminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16, Gallay, P., Hope, T., Chin, D. and Trono, D. (1997). HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94, Ganapathy, V., Daniels, T. and Casiano, C. A. (2003). LEDGF/p75: a novel nuclear autoantigen at the crossroads of cell survival and apoptosis. Autoimmun. Rev. 2, Ge, H., Si, Y. and Roeder, R. G. (1998a). Isolation of cdnas encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 17, Ge, H., Si, Y. and Wolffe, A. P. (1998b). A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol. Cell 2, Ge, Y. Z., Pu, M. T., Gowher, H., Wu, H. P., Ding, J. P., Jeltsch, A. and Xu, G. L. (2004). Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 279, Goff, S. P. (2001). Intracellular trafficking of retroviral genomes during the early phase of infection: viral exploitation of cellular pathways. J. Gene Med. 3, Gorlich, D. and Kutay, U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, Izumoto, Y., Kuroda, T., Harada, H., Kishimoto, T. and Nakamura, H. (1997). Hepatoma-derived growth factor belongs to a gene family in mice showing significant homology in the amino terminus. Biochem. Biophys. Res. Commun. 238,
11 HIV-1 integrase and functional domains of LEDGF/p Jans, D. A., Xiao, C. Y. and Lam, M. H. (2000). Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22, Kalderon, D., Richardson, W. D., Markham, A. F. and Smith, A. E. (1984). Sequence requirements for nuclear location of simian virus 40 large-t antigen. Nature 311, Kannouche, P. L., Wing, J. and Lehmann, A. R. (2004). Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14, Kukolj, G., Jones, K. S. and Skalka, A. M. (1997). Subcellular localization of avian sarcoma virus and human immunodeficiency virus type 1 integrases. J. Virol. 71, Lanford, R. E. and Butel, J. S. (1984). Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell 37, Llano, M., Vanegas, M., Fregoso, O., Saenz, D., Chung, S., Peretz, M. and Poeschla, E. M. (2004a). LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral pre-integration complexes. J. Virol. 78, Llano, M., Delgado, S., Vanegas, M. and Poeschla, E. M. (2004b). LEDGF/p75 prevents proteasomal degradation of HIV-1 integrase. J. Biol. Chem. 279, Maertens, G., Cherepanov, P., Pluymers, W., Busschots, K., de Clercq, E., Debyser, Z. and Engelborghs, Y. (2003). LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278, Maertens, G., Cherepanov, P., Debyser, Z., Engelborghs, Y. and Engelman, A. (2004). Identification and characterization of a functional nuclear localization signal in the HIV-1 integrase (IN) interactor LEDGF/p75. J. Biol. Chem. 279, Meier, U. T. and Blobel, G. (1992). Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell 70, Moroianu, J. (1999). Nuclear import and export pathways. J. Cell Biochem. Suppl , Nishizawa, Y., Usukura, J., Singh, D. P., Chylack, L. T., Jr and Shinohara, T. (2001). Spatial and temporal dynamics of two alternatively spliced regulatory factors, lens epithelium-derived growth factor (ledgf/p75) and p52, in the nucleus. Cell Tissue Res. 305, Petit, C., Schwartz, O. and Mammano, F. (2000). The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA. J. Virol. 74, Petropoulos, C. J. and Hughes, S. H. (1991). Replication-competent retrovirus vectors for the transfer and expression of gene cassettes in avian cells. J. Virol. 65, Pluymers, W., Cherepanov, P., Schols, D., de Clercq, E. and Debyser, Z. (1999). Nuclear localization of human immunodeficiency virus type 1 integrase expressed as a fusion protein with green fluorescent protein. Virology 258, Reeves, R. and Nissen, M. S. (1990). The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem. 265, Rizzo, M. and Piston, D. (2005). Fluorescent protein trafficking and detection. In Live Cell Imaging: A Laboratory Manual (eds R. D. Goldman and D. L. Spector), pp Cold Spring Harbor: Cold Spring Harbor Press. Schroder, A. R., Shinn, P., Chen, H., Berry, C., Ecker, J. R. and Bushman, F. (2002). HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110, Sharma, P., Singh, D. P., Fatma, N., Chylack, L. T., Jr and Shinohara, T. (2000). Activation of LEDGF gene by thermal- and oxidative-stresses. Biochem. Biophys. Res. Commun. 276, Shinohara, T., Singh, D. P. and Fatma, N. (2002). LEDGF, a survival factor, activates stress-related genes. Prog. Retin. Eye Res. 21, Siddiqa, A., Sims-Mourtada, J. C., Guzman-Rojas, L., Rangel, R., Guret, C., Madrid-Marina, V., Sun, Y. and Martinez-Valdez, H. (2001). Regulation of CD40 and CD40 ligand by the AT-hook transcription factor AKNA. Nature 410, Siomi, H. and Dreyfuss, G. (1995). A nuclear localization domain in the hnrnp A1 protein. J. Cell Biol. 129, Stec, I., Nagl, S. B., van Ommen, G. J. and den Dunnen, J. T. (2000). The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 473, 1-5. Woodward, C. L., Wang, Y., Dixon, W. J., Htun, H. and Chow, S. A. (2003). Subcellular localization of feline immunodeficiency virus integrase and mapping of its karyophilic determinant. J. Virol. 77,
Recombinant Protein Expression Retroviral system
 Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Supplementary Material
 Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Intracellular transport of human immunodeficiency virus type 1 integrase
 Research Article 4401 Intracellular transport of human immunodeficiency virus type 1 integrase Eric Devroe 1,2, Alan Engelman 3,4 and Pamela A. Silver 1,2, * 1 Department of Biological Chemistry and Molecular
Research Article 4401 Intracellular transport of human immunodeficiency virus type 1 integrase Eric Devroe 1,2, Alan Engelman 3,4 and Pamela A. Silver 1,2, * 1 Department of Biological Chemistry and Molecular
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Regulators of Cell Cycle Progression
 Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
VIROLOGY. Engineering Viral Genomes: Retrovirus Vectors
 VIROLOGY Engineering Viral Genomes: Retrovirus Vectors Viral vectors Retrovirus replicative cycle Most mammalian retroviruses use trna PRO, trna Lys3, trna Lys1,2 The partially unfolded trna is annealed
VIROLOGY Engineering Viral Genomes: Retrovirus Vectors Viral vectors Retrovirus replicative cycle Most mammalian retroviruses use trna PRO, trna Lys3, trna Lys1,2 The partially unfolded trna is annealed
Supplementary data Supplementary Figure 1 Supplementary Figure 2
 Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates
 HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates Department of Microbiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA
HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates Department of Microbiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA
Cell Cycle, Mitosis, and Microtubules. LS1A Final Exam Review Friday 1/12/07. Processes occurring during cell cycle
 Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting. protein3) regulate autophagy and mitophagy in renal tubular cells in. acute kidney injury
 Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
The Lentiviral Integrase Binding Protein LEDGF/p75 and HIV-1 Replication
 Review The Lentiviral Integrase Binding Protein LEDGF/p75 and HIV-1 Replication Alan Engelman 1 *, Peter Cherepanov 2 * 1 Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, Division
Review The Lentiviral Integrase Binding Protein LEDGF/p75 and HIV-1 Replication Alan Engelman 1 *, Peter Cherepanov 2 * 1 Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, Division
Bio 401 Sp2014. Multiple Choice (Circle the letter corresponding to the correct answer)
 MIDTERM EXAM KEY (60 pts) You should have 5 pages. Please put your name on every page. You will have 70 minutes to complete the exam. You may begin working as soon as you receive the exam. NOTE: the RED
MIDTERM EXAM KEY (60 pts) You should have 5 pages. Please put your name on every page. You will have 70 minutes to complete the exam. You may begin working as soon as you receive the exam. NOTE: the RED
Eukaryotic transcription (III)
 Eukaryotic transcription (III) 1. Chromosome and chromatin structure Chromatin, chromatid, and chromosome chromatin Genomes exist as chromatins before or after cell division (interphase) but as chromatids
Eukaryotic transcription (III) 1. Chromosome and chromatin structure Chromatin, chromatid, and chromosome chromatin Genomes exist as chromatins before or after cell division (interphase) but as chromatids
Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression
 Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplementary Information
 Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
Supplementary Information. Supplementary Figure 1
 Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
7.012 Problem Set 6 Solutions
 Name Section 7.012 Problem Set 6 Solutions Question 1 The viral family Orthomyxoviridae contains the influenza A, B and C viruses. These viruses have a (-)ss RNA genome surrounded by a capsid composed
Name Section 7.012 Problem Set 6 Solutions Question 1 The viral family Orthomyxoviridae contains the influenza A, B and C viruses. These viruses have a (-)ss RNA genome surrounded by a capsid composed
Supplemental Information
 Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
Problem Set 5 KEY
 2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer
 Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Protocol for Gene Transfection & Western Blotting
 The schedule and the manual of basic techniques for cell culture Advanced Protocol for Gene Transfection & Western Blotting Schedule Day 1 26/07/2008 Transfection Day 3 28/07/2008 Cell lysis Immunoprecipitation
The schedule and the manual of basic techniques for cell culture Advanced Protocol for Gene Transfection & Western Blotting Schedule Day 1 26/07/2008 Transfection Day 3 28/07/2008 Cell lysis Immunoprecipitation
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
Materials and Methods , The two-hybrid principle.
 The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
The Interaction of LEDGF/p75 with Integrase Is Lentivirus-specific and Promotes DNA Binding*
 THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 280, No. 18, Issue of May 6, pp. 17841 17847, 2005 2005 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. The Interaction of
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 280, No. 18, Issue of May 6, pp. 17841 17847, 2005 2005 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. The Interaction of
Structural vs. nonstructural proteins
 Why would you want to study proteins associated with viruses or virus infection? Receptors Mechanism of uncoating How is gene expression carried out, exclusively by viral enzymes? Gene expression phases?
Why would you want to study proteins associated with viruses or virus infection? Receptors Mechanism of uncoating How is gene expression carried out, exclusively by viral enzymes? Gene expression phases?
Supplemental Materials and Methods Plasmids and viruses Quantitative Reverse Transcription PCR Generation of molecular standard for quantitative PCR
 Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Subcellular Localization of Feline Immunodeficiency Virus Integrase and Mapping of Its Karyophilic Determinant
 JOURNAL OF VIROLOGY, Apr. 2003, p. 4516 4527 Vol. 77, No. 8 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.8.4516 4527.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Subcellular
JOURNAL OF VIROLOGY, Apr. 2003, p. 4516 4527 Vol. 77, No. 8 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.8.4516 4527.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Subcellular
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
SUPPLEMENTARY INFORMATION
 sirna pool: Control Tetherin -HA-GFP HA-Tetherin -Tubulin Supplementary Figure S1. Knockdown of HA-tagged tetherin expression by tetherin specific sirnas. HeLa cells were cotransfected with plasmids expressing
sirna pool: Control Tetherin -HA-GFP HA-Tetherin -Tubulin Supplementary Figure S1. Knockdown of HA-tagged tetherin expression by tetherin specific sirnas. HeLa cells were cotransfected with plasmids expressing
MOLECULAR CELL BIOLOGY
 1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
The Schedule and the Manual of Basic Techniques for Cell Culture
 The Schedule and the Manual of Basic Techniques for Cell Culture 1 Materials Calcium Phosphate Transfection Kit: Invitrogen Cat.No.K2780-01 Falcon tube (Cat No.35-2054:12 x 75 mm, 5 ml tube) Cell: 293
The Schedule and the Manual of Basic Techniques for Cell Culture 1 Materials Calcium Phosphate Transfection Kit: Invitrogen Cat.No.K2780-01 Falcon tube (Cat No.35-2054:12 x 75 mm, 5 ml tube) Cell: 293
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION
 Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION X. Shawn Liu 1, 3, Bing Song 2, 3, Bennett D. Elzey 3, 4, Timothy L. Ratliff 3, 4, Stephen F. Konieczny
Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION X. Shawn Liu 1, 3, Bing Song 2, 3, Bennett D. Elzey 3, 4, Timothy L. Ratliff 3, 4, Stephen F. Konieczny
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation
 Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
TFEB-mediated increase in peripheral lysosomes regulates. Store Operated Calcium Entry
 TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
The HIV life cycle. integration. virus production. entry. transcription. reverse transcription. nuclear import
 The HIV life cycle entry reverse transcription transcription integration virus production nuclear import Hazuda 2012 Integration Insertion of the viral DNA into host chromosomal DNA, essential step in
The HIV life cycle entry reverse transcription transcription integration virus production nuclear import Hazuda 2012 Integration Insertion of the viral DNA into host chromosomal DNA, essential step in
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
~Lentivirus production~
 ~Lentivirus production~ May 30, 2008 RNAi core R&D group member Lentivirus Production Session Lentivirus!!! Is it health threatening to lab technician? What s so good about this RNAi library? How to produce
~Lentivirus production~ May 30, 2008 RNAi core R&D group member Lentivirus Production Session Lentivirus!!! Is it health threatening to lab technician? What s so good about this RNAi library? How to produce
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
supplementary information
 DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
SUPPLEMENT. Materials and methods
 SUPPLEMENT Materials and methods Cell culture and reagents Cell media and reagents were from Invitrogen unless otherwise indicated. Antibiotics and Tet-certified serum were from Clontech. In experiments
SUPPLEMENT Materials and methods Cell culture and reagents Cell media and reagents were from Invitrogen unless otherwise indicated. Antibiotics and Tet-certified serum were from Clontech. In experiments
Name Section Problem Set 6
 Name Section 7.012 Problem Set 6 Question 1 The viral family Orthomyxoviridae contains the influenza A, B and C viruses. These viruses have a (-)ss RNA genome surrounded by a capsid composed of lipids
Name Section 7.012 Problem Set 6 Question 1 The viral family Orthomyxoviridae contains the influenza A, B and C viruses. These viruses have a (-)ss RNA genome surrounded by a capsid composed of lipids
TSH Receptor Monoclonal Antibody (49) Catalog Number MA3-218 Product data sheet
 Website: thermofisher.com Customer Service (US): 1 800 955 6288 ext. 1 Technical Support (US): 1 800 955 6288 ext. 441 TSH Receptor Monoclonal Antibody (49) Catalog Number MA3-218 Product data sheet Details
Website: thermofisher.com Customer Service (US): 1 800 955 6288 ext. 1 Technical Support (US): 1 800 955 6288 ext. 441 TSH Receptor Monoclonal Antibody (49) Catalog Number MA3-218 Product data sheet Details
The clathrin adaptor Numb regulates intestinal cholesterol. absorption through dynamic interaction with NPC1L1
 The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
Fayth K. Yoshimura, Ph.D. September 7, of 7 HIV - BASIC PROPERTIES
 1 of 7 I. Viral Origin. A. Retrovirus - animal lentiviruses. HIV - BASIC PROPERTIES 1. HIV is a member of the Retrovirus family and more specifically it is a member of the Lentivirus genus of this family.
1 of 7 I. Viral Origin. A. Retrovirus - animal lentiviruses. HIV - BASIC PROPERTIES 1. HIV is a member of the Retrovirus family and more specifically it is a member of the Lentivirus genus of this family.
Life Sciences 1A Midterm Exam 2. November 13, 2006
 Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were
 Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
7.012 Quiz 3 Answers
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
Supplementary Information
 Supplementary Information Supplementary Figure 1. EBV-gB 23-431 mainly exists as trimer in HEK 293FT cells. (a) Western blotting analysis for DSS crosslinked FLAG-gB 23-431. HEK 293FT cells transfected
Supplementary Information Supplementary Figure 1. EBV-gB 23-431 mainly exists as trimer in HEK 293FT cells. (a) Western blotting analysis for DSS crosslinked FLAG-gB 23-431. HEK 293FT cells transfected
Mechanisms of alternative splicing regulation
 Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Materials and Methods
 Supplementary Materials and Methods Reagents and antibodies was purchased from iaffin GmbH & Co KG. Cisplatin (ristol-myers Squibb Co.) and etoposide (Sandoz Pharma Ltd.) were used. Antibodies recognizing
Supplementary Materials and Methods Reagents and antibodies was purchased from iaffin GmbH & Co KG. Cisplatin (ristol-myers Squibb Co.) and etoposide (Sandoz Pharma Ltd.) were used. Antibodies recognizing
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS
 Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Under the Radar Screen: How Bugs Trick Our Immune Defenses
 Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 7: Cytokines Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research HHV-8 Discovered in the 1980 s at the
Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 7: Cytokines Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research HHV-8 Discovered in the 1980 s at the
supplementary information
 DOI: 10.1038/ncb2153 Figure S1 Ectopic expression of HAUSP up-regulates REST protein. (a) Immunoblotting showed that ectopic expression of HAUSP increased REST protein levels in ENStemA NPCs. (b) Immunofluorescent
DOI: 10.1038/ncb2153 Figure S1 Ectopic expression of HAUSP up-regulates REST protein. (a) Immunoblotting showed that ectopic expression of HAUSP increased REST protein levels in ENStemA NPCs. (b) Immunofluorescent
supplementary information
 Figure S1 Nucleotide binding status of RagA mutants. Wild type and mutant forms of MycRagA was transfected into HEK293 cells and the transfected cells were labeled with 32 Pphosphate. MycRagA was immunoprecipitated
Figure S1 Nucleotide binding status of RagA mutants. Wild type and mutant forms of MycRagA was transfected into HEK293 cells and the transfected cells were labeled with 32 Pphosphate. MycRagA was immunoprecipitated
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh-
 1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
Supporting Information
 Supporting Information Kim et al. 10.1073/pnas.0912180106 SI Materials and Methods DNA Constructs. The N-terminally-tagged mouse Gli2 expression construct, pcefl/3 HA-Gli2, was constructed by ligating
Supporting Information Kim et al. 10.1073/pnas.0912180106 SI Materials and Methods DNA Constructs. The N-terminally-tagged mouse Gli2 expression construct, pcefl/3 HA-Gli2, was constructed by ligating
Reviewers' comments: Reviewer #1 (Remarks to the Author):
 Reviewers' comments: Reviewer #1 (Remarks to the Author): Nature Communications manuscript number NCOMMS-16-15882, by Miyakawa et al. presents an intriguing analysis of the effects of the tumor suppressor
Reviewers' comments: Reviewer #1 (Remarks to the Author): Nature Communications manuscript number NCOMMS-16-15882, by Miyakawa et al. presents an intriguing analysis of the effects of the tumor suppressor
SUPPLEMENTARY INFORMATION
 Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
Supplemental information contains 7 movies and 4 supplemental Figures
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
Supplementary Figure S1
 Supplementary Figure S1 Supplementary Figure S1. PARP localization patterns using GFP-PARP and PARP-specific antibody libraries GFP-PARP localization in non-fixed (A) and formaldehyde fixed (B) GFP-PARPx
Supplementary Figure S1 Supplementary Figure S1. PARP localization patterns using GFP-PARP and PARP-specific antibody libraries GFP-PARP localization in non-fixed (A) and formaldehyde fixed (B) GFP-PARPx
Rabbit Polyclonal antibody to NFkB p65 (v-rel reticuloendotheliosis viral oncogene homolog A (avian))
 Datasheet GeneTex, Inc : Toll Free 1-877-GeneTex (1-877-436-3839) Fax:1-949-309-2888 info@genetex.com GeneTex International Corporation : Tel:886-3-6208988 Fax:886-3-6208989 infoasia@genetex.com Date :
Datasheet GeneTex, Inc : Toll Free 1-877-GeneTex (1-877-436-3839) Fax:1-949-309-2888 info@genetex.com GeneTex International Corporation : Tel:886-3-6208988 Fax:886-3-6208989 infoasia@genetex.com Date :
Supporting Information
 Supporting Information Valkenburg et al. 10.1073/pnas.1403684111 SI Materials and Methods ELISA and Microneutralization. Sera were treated with Receptor Destroying Enzyme II (RDE II, Accurate) before ELISA
Supporting Information Valkenburg et al. 10.1073/pnas.1403684111 SI Materials and Methods ELISA and Microneutralization. Sera were treated with Receptor Destroying Enzyme II (RDE II, Accurate) before ELISA
condition. Left panel, the HCT-116 cells were lysed with RIPA buffer containing 0.1%
 FIGURE LEGENDS Supplementary Fig 1 (A) sumoylation pattern detected under denaturing condition. Left panel, the HCT-116 cells were lysed with RIPA buffer containing 0.1% SDS in the presence and absence
FIGURE LEGENDS Supplementary Fig 1 (A) sumoylation pattern detected under denaturing condition. Left panel, the HCT-116 cells were lysed with RIPA buffer containing 0.1% SDS in the presence and absence
Polyomaviridae. Spring
 Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
SUPPLEMENTARY INFORMATION
 Figure S1 Treatment with both Sema6D and Plexin-A1 sirnas induces the phenotype essentially identical to that induced by treatment with Sema6D sirna alone or Plexin-A1 sirna alone. (a,b) The cardiac tube
Figure S1 Treatment with both Sema6D and Plexin-A1 sirnas induces the phenotype essentially identical to that induced by treatment with Sema6D sirna alone or Plexin-A1 sirna alone. (a,b) The cardiac tube
Alternative splicing. Biosciences 741: Genomics Fall, 2013 Week 6
 Alternative splicing Biosciences 741: Genomics Fall, 2013 Week 6 Function(s) of RNA splicing Splicing of introns must be completed before nuclear RNAs can be exported to the cytoplasm. This led to early
Alternative splicing Biosciences 741: Genomics Fall, 2013 Week 6 Function(s) of RNA splicing Splicing of introns must be completed before nuclear RNAs can be exported to the cytoplasm. This led to early
The Process of Cell Division
 Lesson Overview 10.2 The Process of Cell Division THINK ABOUT IT What role does cell division play in your life? Does cell division stop when you are finished growing? Chromosomes What is the role of chromosomes
Lesson Overview 10.2 The Process of Cell Division THINK ABOUT IT What role does cell division play in your life? Does cell division stop when you are finished growing? Chromosomes What is the role of chromosomes
RAS Genes. The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes.
 ۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
Pre-made Reporter Lentivirus for NF-κB Signal Pathway
 Pre-made Reporter for NF-κB Signal Pathway Cat# Product Name Amounts LVP965-P or: LVP965-P-PBS NFKB-GFP (Puro) LVP966-P or: LVP966-P-PBS NFKB-RFP (Puro) LVP967-P or: LVP967-P-PBS NFKB-Luc (Puro) LVP968-P
Pre-made Reporter for NF-κB Signal Pathway Cat# Product Name Amounts LVP965-P or: LVP965-P-PBS NFKB-GFP (Puro) LVP966-P or: LVP966-P-PBS NFKB-RFP (Puro) LVP967-P or: LVP967-P-PBS NFKB-Luc (Puro) LVP968-P
Nature Medicine: doi: /nm.4322
 1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
SUPPLEMENTARY INFORMATION
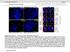 doi: 10.1038/nature05473 SUPPLEMENTARY FIGURES SUPPLEMENTARY INFORMATION Supplementary Figure 1: Association of Runx2 with mitotic chromosomes Mitotic chromosome spreads were prepared for a. Human Saos-2
doi: 10.1038/nature05473 SUPPLEMENTARY FIGURES SUPPLEMENTARY INFORMATION Supplementary Figure 1: Association of Runx2 with mitotic chromosomes Mitotic chromosome spreads were prepared for a. Human Saos-2
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism SUPPLEMENTARY FIGURES, LEGENDS AND METHODS
 A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
LESSON 4.4 WORKBOOK. How viruses make us sick: Viral Replication
 DEFINITIONS OF TERMS Eukaryotic: Non-bacterial cell type (bacteria are prokaryotes).. LESSON 4.4 WORKBOOK How viruses make us sick: Viral Replication This lesson extends the principles we learned in Unit
DEFINITIONS OF TERMS Eukaryotic: Non-bacterial cell type (bacteria are prokaryotes).. LESSON 4.4 WORKBOOK How viruses make us sick: Viral Replication This lesson extends the principles we learned in Unit
Mammalian Membrane Protein Extraction Kit
 Mammalian Membrane Protein Extraction Kit Catalog number: AR0155 Boster s Mammalian Membrane Protein Extraction Kit is a simple, rapid and reproducible method to prepare cellular protein fractions highly
Mammalian Membrane Protein Extraction Kit Catalog number: AR0155 Boster s Mammalian Membrane Protein Extraction Kit is a simple, rapid and reproducible method to prepare cellular protein fractions highly
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Jewell et al., http://www.jcb.org/cgi/content/full/jcb.201007176/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. IR Munc18c association is independent of IRS-1. (A and
Supplemental material Jewell et al., http://www.jcb.org/cgi/content/full/jcb.201007176/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. IR Munc18c association is independent of IRS-1. (A and
Supplementary Figure 1. The CagA-dependent wound healing or transwell migration of gastric cancer cell. AGS cells transfected with vector control or
 Supplementary Figure 1. The CagA-dependent wound healing or transwell migration of gastric cancer cell. AGS cells transfected with vector control or 3xflag-CagA expression vector were wounded using a pipette
Supplementary Figure 1. The CagA-dependent wound healing or transwell migration of gastric cancer cell. AGS cells transfected with vector control or 3xflag-CagA expression vector were wounded using a pipette
Essential Medium, containing 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin. Huvec were cultured in
 Supplemental data Methods Cell culture media formulations A-431 and U-87 MG cells were maintained in Dulbecco s Modified Eagle s Medium. FaDu cells were cultured in Eagle's Minimum Essential Medium, containing
Supplemental data Methods Cell culture media formulations A-431 and U-87 MG cells were maintained in Dulbecco s Modified Eagle s Medium. FaDu cells were cultured in Eagle's Minimum Essential Medium, containing
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Certificate of Analysis
 Certificate of Analysis plvx-ef1α-ires-puro Vector Table of Contents Product Information... 1 Description... 2 Location of Features... 3 Additional Information... 3 Quality Control Data... 4 Catalog No.
Certificate of Analysis plvx-ef1α-ires-puro Vector Table of Contents Product Information... 1 Description... 2 Location of Features... 3 Additional Information... 3 Quality Control Data... 4 Catalog No.
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
Supplementary Figure 1
 Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figure 1 Induction of cellular senescence and isolation of exosome. a to c, Pre-senescent primary normal human diploid fibroblasts
 Supplementary Figure 1 Induction of cellular senescence and isolation of exosome. a to c, Pre-senescent primary normal human diploid fibroblasts (TIG-3 cells) were rendered senescent by either serial passage
Supplementary Figure 1 Induction of cellular senescence and isolation of exosome. a to c, Pre-senescent primary normal human diploid fibroblasts (TIG-3 cells) were rendered senescent by either serial passage
Mitosis. AND Cell DiVISION
 Mitosis AND Cell DiVISION Cell Division Characteristic of living things: ability to reproduce their own kind. Cell division purpose: When unicellular organisms such as amoeba divide to form offspring reproduction
Mitosis AND Cell DiVISION Cell Division Characteristic of living things: ability to reproduce their own kind. Cell division purpose: When unicellular organisms such as amoeba divide to form offspring reproduction
Figure S1. Schematic presentation of genomic replication of idsiv after transfection and infection. After transfection of idsiv plasmid DNA into 293T
 Figure S1. Schematic presentation of genomic replication of idsiv after transfection and infection. After transfection of idsiv plasmid DNA into 293T cells, the RNA genomes with all modifications are generated
Figure S1. Schematic presentation of genomic replication of idsiv after transfection and infection. After transfection of idsiv plasmid DNA into 293T cells, the RNA genomes with all modifications are generated
Ch. 18 Regulation of Gene Expression
 Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
