CINCINNATA Controls Both Cell Differentiation and Growth in Petal Lobes and Leaves of Antirrhinum 1
|
|
|
- Hubert Briggs
- 5 years ago
- Views:
Transcription
1 CINCINNATA Controls Both Cell Differentiation and Growth in Petal Lobes and Leaves of Antirrhinum 1 Brian C.W. Crawford, Utpal Nath 2, Rosemary Carpenter, and Enrico S. Coen* John Innes Centre, Norwich NR4 7UH, United Kingdom To understand how differentiation and growth may be coordinated during development, we have studied the action of the CINCINNATA (CIN) gene of Antirrhinum. We show that in addition to affecting leaf lamina growth, CIN affects epidermal cell differentiation and growth of petal lobes. Strong alleles of cin give smaller petal lobes with flat instead of conical cells, correlating with lobe-specific expression of CIN in the wild type. Moreover, conical cells at the leaf margins are replaced by flatter cells, indicating that CIN has a role in cell differentiation of leaves as well as petals. A weak semidominant cin allele affects cell types mainly in the petal but does not affect leaf development, indicating these two effects can be separated. Expression of CIN correlates with expression of cell division markers, suggesting that CIN may influence petal growth, directly or indirectly, through effects on cell proliferation. For both leaves and petals, CIN affects growth and differentiation of the more distal and broadly extended domains (leaf lamina and petal lobe). However, while CIN promotes growth in petals, it promotes growth arrest in leaves, possibly because of different patterns of growth control in these systems. Development involves coordination of two interconnected processes: growth and cellular differentiation. The genetic control of each of these processes has been studied extensively in plants. For example, genes affecting leaf growth or epidermal cell fate have been isolated and analyzed (Masucci et al., 1996; Tsuge et al., 1996; Gu et al., 1998; Kim et al., 1998; Mizukami and Fischer, 2000; Kim et al., 2002). However, the way that growth and differentiation are coupled through the action of genes has been less well studied. To help address this, we have investigated the effects of the CINCINNATA (CIN) gene on growth and cell differentiation in Antirrhinum. CIN encodes a TCP transcription factor that promotes growth arrest, particularly in leaf margins (Nath et al., 2003). In cin mutants, leaves are larger and have an undulating edge due to excessive growth in marginal regions. In addition to these effects on leaf growth, cin mutants also show alterations in petal shape and cell types. This suggests that the CIN transcription factor may have targets involved in both growth and differentiation pathways. To determine how these pathways may be affected, we have analyzed the role of CIN in petal growth and differentiation. Flowers of Antirrhinum have five petals, which are united in their proximal region to form a corolla tube. 1 This work was supported by the Biotechnology and Biological Sciences Research Council. 2 Present address: Department of Microbiology and Cell Biology, Indian Institute of Science, Bangalore , India. * Corresponding author; enrico.coen@bbsrc.ac.uk; fax Article, publication date, and citation information can be found at The distal region of each petal forms a lobe, which itself can be subdivided into a more proximal region, termed the lip, and a more distal region (Fig. 1A; Keck et al., 2003). The flowers exhibit dorsoventral asymmetry and comprise two dorsal petals, two lateral petals, and one ventral petal. The lateral and ventral petal lobes form a platform for bees to land on and pry open the flower. A variety of cell types can be recognized in the flower, forming characteristic patterns along the proximodistal and dorsoventral axes (Keck et al., 2003). Several genes encoding transcription factors have been identified that influence these patterns. The CYCLOIDEA (CYC), DICHOTOMA (DICH), and DIVARICATA (DIV) genes encode TCP or MYB proteins that specify differences along the dorsoventral axis (Luo et al., 1996, 1999; Galego and Almeida, 2002). MIXTA (MIX) encodes a MYB-related protein that promotes conical cell differentiation in the lobes of the petal epidermis (Noda et al., 1994), while LIP genes encode APETALA2-like proteins that most strongly affect development in the lip region of the flower (Keck et al., 2003). However, none of these mutants have clear effects on leaf development. Here, we describe the effects of CIN on growth and differentiation. A weak semidominant cin allele carries a duplication of the CIN locus, most likely leading to posttranscriptional silencing of CIN. This allele affects cell types mainly in the lip region of the petal but does not affect leaf development. All strong alleles of cin have major disruptions in the CIN gene and give smaller petal lobes with flat instead of conical cells, correlating with lobe-specific expression of CIN in the wild type. In addition, conical cells normally observed at the margins of leaves are flatter than usual, indicating that CIN has a role in promoting conical cell development in leaves as well as petals. Expression 244 Plant Physiology, May 2004, Vol. 135, pp , Ó 2004 American Society of Plant Biologists
2 Growth and Cell Differentiation in Petals and Leaves Figure 1. Phenotype of wild-type and cin mutant flowers. Flowers of the wild type and cin mutants shown in side (left) or face view (right). Wild-type Stock 98 (A), cin-756 (B), cin-628 (C), wild-type Sippe 50 (D), and cin cin (E) are shown. The lobe and lip regions are indicated on the face view of wild-type Stock 98 (A). of CIN correlates with expression of cell division markers, suggesting that CIN may influence petal growth through effects on cell proliferation. However, while CIN promotes growth in petals, it promotes growth arrest in leaves, possibly because of different patterns of growth control in these systems. RESULTS Alleles of cincinnata To understand the role of CIN in petal development, we first characterized a range of cin alleles. Two alleles with indistinguishable mutant phenotypes, cin-755 Plant Physiol. Vol. 135,
3 Crawford et al. and cin-756, had previously been shown to carry deletions that extended into the CIN coding region (Nath et al., 2003). In addition to affecting leaf shape, these putative null alleles had a clear effect on the flower: the petal lobes were smaller, slightly curled, pale, and had altered cell types compared with their progenitor (Fig. 1, A and B). A broadly similar effect on both leaf and floral morphology was seen when comparing the classical allele, cin cin (Fig. 1E), with their progenitor (Fig. 1D). Nevertheless, there are some differences in detail. In particular, the degree to which the ventral and lateral petals bend at the tube-lobe boundary is reduced in the classical mutant. The cin cin phenotype is very similar to another mutant, subcrispa (Stubbe, 1966). An allelism test showed subcrispa is allelic to cin cin and was renamed cin suba. Genetic analysis indicated that many of the differences in appearance of the cin cin and cin suba alleles compared with cin-756 were a consequence of their distinct genetic backgrounds (the classical cin cin and cin suba alleles were produced in the Sippe 50 background, while the deletion alleles were produced in the John Innes Stock 98 background). Cloning and sequencing of cin cin showed that it had a 10-bp insertion 438 bp downstream of the start codon, resulting in a frameshift and a stop codon 444 bp downstream of the start (Fig. 2A). The predicted protein would be truncated at the C terminus by 256 amino acids compared to the wild type CIN protein and is expected to be inactive. PCR analysis on cin suba DNA revealed an insertion of a 4-kb DNA fragment in the CIN locus (Fig. 2A). Sequencing of the insertion site showed that the insertion was 63 bp downstream of the start codon. The insertion of this fragment disrupts the open reading frame (ORF) and is likely to disrupt CIN function. Another allele, cin-628, was also analyzed. Unlike other cin alleles, cin-628 was semidominant and had no clear effect on leaf development. The floral phenotype was different from the cin deletions: The effect of the mutation was primarily on the cell types within the adaxial epidermis of the petal lips (Fig. 1C). In cin-628/ CIN heterozygotes, the effect on cell types was weaker than in cin-628/cin-628 homozygotes and confined to more proximal regions of the petal lip. Crosses of cin-628 to cin cin gave a strong floral mutant phenotype in the F 1, consistent with their allelism. The cin-628 allele arose from a transposon mutagenesis screen. Southern blots of HindIII digests probed with CIN ORF revealed that in addition to the approximately 10-kb band seen in the wild-type progenitor, cin-628 had a novel band at approximately 15 kb of a similar intensity to the wild-type band (Fig. 2B, lanes 1 and 2). This indicated that cin-628 may carry a transposon-induced duplication of the CIN locus similar to a previously identified mutation of NIVEA in Antirrhinum (Bollmann et al., 1991). To determine whether the additional 15-kb band in cin-628 was responsible for the phenotype, stable homozygous revertants of cin-628 were analyzed (the Figure 2. Structure of cin alleles. A, Map of the cin locus. The rectangle represents the transcribed region of the CIN gene with the ORF shaded in gray. The thick black line represents the region deleted in cin-756. The triangle represents the insertion in cin suba. The star shows the position of the introduced stop codon in cin cin. E, B, and H are positions of restriction sites for the enzymes EcoRI, BamHI, and HindIII, respectively. B, DNA blots of wild-type, cin-628, cin-628 homozygous revertant, and an F 2 population segregating for cin-628 probed with CIN. DNA was digested with HindIII. Lane 1 contains DNA from wildtype Stock 98 (the background in which the cin-628 allele is maintained). Lane 2 contains DNA from cin-628. Lanes 3 and 4 contain DNA from two independent cin-628 homozygous revertants. Lanes 5 to 19 contain DNA from an F 2 population segregating for cin-628; the phenotypes of the plants are wild type (wt), heterozygous mutant (het), and homozygous mutant (hom). revertants were obtained by screening progeny of cin- 628 plants grown at 15 C, a temperature that favors transposon movement and rearrangements). The 15-kb band was absent in the revertants, although the 10-kb band was still present, indicating that the 15-kb band had been responsible for the cin-628 mutation (Fig. 2B, lanes 3 and 4). PCR analysis indicated that the 15-kb band carried a copy of the Tam3 transposon approximately 3.5 kb upstream of the ATG, indicating that the duplication was associated with a transposition event. To determine whether the 15-kb band was linked to the original 10-kb band, cin-628 was crossed to a wild-type line of a different genetic background (JI-7) with a distinct band of approximately 12 kb in HindIII digests. Analysis of the resulting F 2 showed that the approximately 15-kb and approximately 10-kb bands present in cin-628 were linked and cosegregated with the mutant phenotype (Fig. 2B, lanes 5 19). One explanation for these results is that cin-628 arose by a CIN duplication that led to repression of gene activity, possibly through posttranscriptional gene silencing (PTGS; Kusaba et al., 2003). PTGS would also account for the semidominance of cin-628. Similar transposon-induced semidominant alleles 246 Plant Physiol. Vol. 135, 2004
4 Growth and Cell Differentiation in Petals and Leaves Figure 3. SEM analysis of petals from the wild type compared to cin-756. Petals were dissected and flattened to illustrate the various areas used for comparisons of the wild type (i vii) and cin-756 (viii xiv). Scale bar 5 50 mm. dl, dorsal lobes; dt, dorsal tube; lp, lateral lobes; vp, ventral lobes; vt, ventral/lateral tube. carrying duplications have been described for the NIVEA locus of Antirrhinum and have subsequently been shown to contain small RNAs diagnostic of gene silencing (A. Hamilton and D. Baulcombe, personal communication; Bollmann et al., 1991). Cell Types in cin Alleles To investigate the effect of CIN on cell types, adaxial epidermal cells of petals from one of the deletion alleles, cin-756, and the wild type were analyzed by scanning electron microscopy (SEM). Four regions of the dorsal petal lobe and three of the ventral petal lobe were recorded (Fig. 3). Cells of the cin-756 were flatter than those of the wild type for all regions analyzed. Instead of conical cells typical of wild-type petal lobes, the cin mutant had nearly flat or weakly conical cells. The effect on conical cell formation was strongest in the lip region of the ventral petal lobe (Fig. 3, compare regions v and xii). Measurements of cell sizes showed that cells were also about 30% larger in area in cin mutants for three out of four regions of the dorsal lobes when compared to the wild type (Fig. 3, regions i iv and viii xi; Table I). Although, as these cells are flattened in the mutant, it is possible that the cell volume is not affected. The cin-628 allele had a slightly different effect on petal cell types. A mixture of flat cells and hair cells (Fig. 4, arrow) was produced in the lip region of the ventral and lateral petals instead of conical cells. The morphology of these hair cells was similar to that of wild-type hair cells from the adaxial surface of the petal tube (Fig. 4, arrowhead). The distal regions of the petal lobes in cin-628 did form conical cells indistinguishable Table I. Cell sizes in dorsal petals of cin-756 and the wild type Average size of petal conical cells (mm 2 ) from dorsal petals of cin-756 and the wild type. The sizes were obtained from different regions corresponding to Figure 3. Region i/viii Region ii/ix Region iii/x Region iv/xi cin-756 2, , , , Wild type 1, , , , Plant Physiol. Vol. 135,
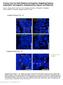
5 Crawford et al. from those found in wild-type petals, consistent with cin-628 being a weak rather than null allele. Conical cells are also found along the abaxialadaxial (dorsal-ventral) boundary (margin) of wildtype Antirrhinum leaves (Fig. 5A). To investigate whether these cells might also be affected by CIN, three regions along the leaf margin in the wild type and cin-756 allele were compared. The conical cells found in the wild type were replaced by flatter cells in cin-756 (Fig. 5B). Thus, CIN has an effect on conical cell development in both leaves and petals. Expression of CIN in Wild-Type Floral Tissue Figure 4. Adaxial epidermis surface of lateral petal in cin-628. Hair cells are produced on the lateral and ventral petal lips in cin-628 (arrow), resembling those normally found in the petal tube (arrowhead). The specific effect of CIN on petal lobe development might reflect restricted expression of CIN or factors that interact with CIN. To distinguish these possibilities, CIN expression in developing petals was studied by RNA in situ hybridization on longitudinal sections of floral buds. Antirrhinum floral development has been divided into 15 stages (Carpenter et al., 1995; Vincent and Coen, 2004). CIN expression in buds was first detected at stage 7, about halfway through floral development. Expression was in the distal regions of the growing petals (corresponding to the developing petal lobes) on both the adaxial and abaxial sides of the petal (Fig. 6A). Some signal was also observed in the distal region of the developing stamens. Figure 5. SEM analysis of leaf margins of the wild type compared to cin-756. Different areas along the proximal-distal axis of the leaf were compared in the wild type (A) and cin-756 (B). The arrows indicate conical cells found at the leaf margin in the wild type. Scale bar 5 50 mm. 248 Plant Physiol. Vol. 135, 2004
6 Growth and Cell Differentiation in Petals and Leaves Figure 6. Expression of CIN in wild-type Sippe 50 floral buds. Longitudinal sections of different stages were probed with digoxigenin-labeled CIN antisense RNA. Staining of RNA is dark blue, and tissue is counterstained with calcofluor (pale blue). In each case the dorsal side of the bud is on the left of the photograph. At stage 7, expression is found in the distal region of the developing petals (bracketed in A). At stage 9, expression is observed throughout the distal portion of the petals (B). At stage 10, expression is observed mainly in the epidermal layers of the distal region of the petal (C). At stage 11, expression is found only in the epidermal layers and vasculature of the lobes of all petals (D). Arrow indicates the position of the ventral furrow. s, stamen; c, carpel; vp, ventral petal; dp, dorsal petal. Scale bars mm. At stages 8 to 9, when the petal lobes cover the internal whorls, expression of CIN was observed in the lobes (Fig. 6B). Expression was stronger in, though not exclusive to, the adaxial side of each lobe. A slight kink could be observed at this stage in the ventral petal (Fig. 6B, arrow), corresponding to a furrow that forms between the tube and lobe. This region will give rise to the lip of the lower petals (Keck et al., 2003; Vincent and Coen, 2004). The expression domain of CIN included the region where the ventral furrow was forming. By stage 10, CIN expression was still in the petal lobes (Fig. 6C). Expression was now concentrated in the epidermal cell layers on both the adaxial and abaxial sides, although the expression was stronger in the adaxial epidermis. The CIN expression pattern included the kink in the ventral petal. Expression was also observed in regularly spaced groups of cells within the petal (Fig. 6C, arrowheads), most likely corresponding to the vascular tissue. At stages 11 to 12, when conical cells begin to form on the lobe, CIN expression was expressed strongly in the adaxial and abaxial epidermal layers of the lobe (Fig. 6D). CIN expression in the lobe stopped at a specific point on the ventral epidermis corresponding to the boundary between the tube and the lobe. CIN was expressed in both the cin-628 and cin cin mutants (data not shown). CIN expression in petals of cin-628 was reduced compared to wild type, although the pattern seemed unaltered (data not shown). The lowered expression in cin-628 is consistent with the hypothesis mentioned above that the effects of this allele may involve PTGS (Van der Krol et al., 1990). Early expression was also reduced in cin cin, although later expression was nearer to wild-type levels. Expression of CIN was not detected in mutants carrying the deletion allele, cin-756. Effect of CIN on Petal Growth In addition to the effect on cell types, petal lobes of cin-756 mutants were slightly smaller than those of the Plant Physiol. Vol. 135,
7 Crawford et al. wild type (Fig. 2). Measurements on several flowers showed that the dorsal petal lobes cin-756 were about 86% of the area of those in the wild type ( mm 2 compared to mm 2 ). Similarly, cin cin dorsal petals were about 62% of the area of their wild-type progenitor (68 6 8mm 2 compared to mm 2 ). The reduction in overall size contrasts with the observed increase in cell size in several regions of the petal lobes, suggesting that cin mutants had fewer cells in the dorsal petal lobes. To examine how CIN might be affecting growth of petals, expression of cell-cycle markers HISTONE4 (HIS4) and CYCLIND3B (CYCD3B) was analyzed in wild-type and cin mutant buds in the same genetic background. HIS4 is expressed only in the S phase of the cell cycle, and only a proportion of the cells give a strong signal in proliferating tissue. CYCD3B, a key regulator of the cell cycle in plants, is not restricted to one phase of the cell cycle and is expressed throughout proliferating tissue (Fobert et al., 1994; Riou-Khamlichi et al., 1999; Gaudin et al., 2000). In stage 9 buds, the spotty pattern of HIS4 expression was observed throughout the floral bud (Fig. 7, A and B). Spots of expression were most frequent in the region corresponding to the ventral furrow of the petal (Fig. 7B). Similarly, strongest CYCD3B expression was observed in the ventral furrow of the petals (Fig. 7, C and D). This indicates a high level of cell division activity and correlates with the highest level of CIN expression in this region (Figs. 7, E and F, and 6C). Figure 7. Expression of HIS4, CYCD3B, and CIN in stage 9 wild-type Sippe 50 floral buds. Consecutive longitudinal sections were probed with digoxigenin-labeled HIS4, CYCD3B, and CIN antisense RNA and viewed as in Figure 6. HIS4 expression is observed throughout the floral bud (A) but is more concentrated at the ventral furrow (shown enlarged in B). Strong CYCD3B expression is also found at the ventral furrow (C) and is concentrated on the adaxial side (shown enlarged in D). CIN expression is observed in the epidermal layers in the distal region of the petals (E), with stronger expression in the adaxial epidermal cell layer (shown enlarged in F). Rectangles indicate regions shown as enlargements. s, stamen; c, carpel; vp, ventral petal; dp, dorsal petal. Scale bars mm. 250 Plant Physiol. Vol. 135, 2004
8 Growth and Cell Differentiation in Petals and Leaves Expression analysis in the cin cin allele showed that CYCD3B expression was present in the region corresponding to the ventral furrow, although the prominence of the furrow was reduced (Fig. 8, A and B). The expression was mainly in the epidermal cell layer and was less intense than that in the wild type. Similarly, although HIS4 was expressed throughout floral buds of cin cin, the strong expression associated with the ventral furrow in the wild type was absent (Fig. 8, C and D). DISCUSSION We show that CIN affects cell types and growth of petal lobes in Antirrhinum, correlating with specific expression of CIN in these regions (no expression is detected in the petal tube region). There are parallels between these effects on petal development and the effect of CIN on leaves. Petal lobes can be compared to the leaf lamina, as both are broadly extended and positioned distally, while the petal tube can be compared to the petiole, as both are highly polarized and positioned proximally. Thus, for both petals and leaves, CIN plays a role in the development of the more distal and broadly extended regions of the organ. The parallel is further supported by the observation that some of the cell types in the leaf lamina are altered in cin mutants in a comparable way to cell-type alterations in the lobes. However, despite these broad parallels, many of the detailed effects of CIN on petals and leaves are different. Mature petal lobes of cin null mutant alleles have flat or nearly flat cells instead of the conical cells found in wild-type adaxial epidermis. Conical cells normally begin to form at late stages of floral bud development (Glover et al., 1998; Vincent and Coen, 2004) coinciding with a stage when CIN transcripts are present in the adaxial epidermis of the petal lobe. It is possible that CIN promotes conical cell formation through interactions with MIX, a gene encoding a MYB transcription factor that is also expressed in the adaxial epidermis (Noda et al., 1994); as with cin-756, mix mutants have flat cells instead of conical cells. However, the effect of MIX is more restricted than that of CIN, as organ shape and size are not affected in mix mutants. Moreover, cin mutants have altered conical cell development at the leaf margins while mix mutants did not. As CIN encodes a TCP transcription factor, one possibility is that it influences conical cell development in petals by directly switching on MIX specifically in petal lobes. However, some MIX expression is still detected by RT-PCR in cin-756 mutant petals, indicating that the interaction may be more indirect (our unpublished data). Interactions between TCP and MYB transcription factors have also been described for the control of dorsoventral asymmetry in Antirrhinum flowers. Two TCP transcription factors, CYC and DICH, are known to interact with the MYB transcription factor DIV to establish domains Figure 8. Expression of HIS4 and CYCD3B in stage 9 cin cin floral buds. Consecutive longitudinal sections were probed with digoxigenin-labeled HIS4 and CYC3B antisense RNA and viewed as in Figure 6. Expression of CYCD3B is observed in the petals (A) but is not greatly enhanced in the region where the ventral furrow normally forms (shown enlarged in B). Similarly, the frequency of spots of HIS4 expression (C) is not greatly enhanced in the ventral furrow region (shown enlarged in D). Arrows indicate the normal position of the ventral furrow and rectangles indicate regions chosen for enlargements. s, stamen; c, carpel; vp, ventral petal; dp, dorsal petal. Scale bars mm. Plant Physiol. Vol. 135,
9 Crawford et al. along the dorsal-ventral axis of the flower (Galego and Almeida, 2002). CIN may interact similarly with the family of MIX proteins to establish some of the distinctions within the petal. The effect of CIN on cell types varies for different regions of the petal lobes. Two results indicate that the lip region of the ventral petal lobe (i.e. the region nearest to the corolla tube) may be more sensitive to the loss of CIN. Firstly, in null cin alleles, the cells in this region are more severely affected (they are completely flat). Secondly, the weak cin-628 allele mainly affects cell types in this region. In this case, hair cells similar to those normally found inside the corolla tube are formed instead of conical cells. This suggests that one role of CIN may be to repress tube cell types while promoting the formation of conical cells. Other explanations are possible; for example, the hair cells in cin-628 may be the result of extended growth of conical cells (Glover et al., 1998). In addition, the difference between effects on ventral and dorsal petals indicates that the effects of CIN may depend on interactions with genes like CYC and DIV controlling dorsoventral asymmetry. CIN promotes growth in the petal lobes; mature lobes of the cin mutants are smaller than the wild type. The effect on growth depends on genetic background (cin-756 has a smaller effect on petal size than cin cin in their respective backgrounds). The promotion of growth by CIN involves extra cell divisions; cin mutants have fewer cells in the lobes. This correlates with reduced expression of cell cycle and histone genes in the developing lip region. CIN expression is strongest in the developing lip, suggesting that it may function in a region-specific manner to promote growth. Such regional specific effects on growth have also been described for other TCP transcription factors, such as CYC and DICH (Luo et al., 1996, 1999; Galego and Almeida, 2002). In both leaves and petals, CIN acts preferentially in particular regions, having its greatest effects in the petal lip and leaf margins. However, the effect of CIN on overall growth seems to be opposite for petals and leaves. In petals, CIN promotes growth, while in leaves CIN promotes arrest of growth and cell division (cin mutants have larger leaves). This difference may reflect a different mode of action for CIN in petals and leaves or different patterns of growth control in these systems. MATERIAL AND METHODS Plant Materials and Growth Conditions The insertion and deletion alleles of CIN were isolated in the John Innes Stock 98 background (Carpenter et al., 1987), which was used as the standard wild type for comparisons with the mutant. Both the classical mutants, cincinnata and subcrispa, originally came from the Gatersleben Collection (Hammer et al., 1990) and have been described by Stubbe (Stubbe, 1966). Plants were grown in a glasshouse, unless otherwise stated. For Tam3 mutagenesis, plants were grown in a growth cabinet at 15 C. For growth measurements, plants were grown at 25 C in growth cabinet with continuous light. DNA Analysis DNA extraction was conducted as described previously (Luo et al., 1996). PCR was carried out in a programmable thermal controller (PTC-100; MJ Research, Waltham, MA). Reactions were made in 10 mm Tris-HCl, ph 8.3; 50 mm KCl; 1.5 mm MgCl 2 ; 0.01% (w/v) gelatin; 0.2 mm each of datp, dctp, dgtp, and dttp; and 0.002% (w/v) Tween 20. Sixty nanograms of DNA and 2 units Taq polymerase were added per 50-mL reaction. Samples were heated to 95 C before addition of polymerase to minimize primer dimerization. Reactions were overlaid with 20 ml of mineral oil and 35 cycles of 30 s at 95 C, 2 min at 55 C, and 40 s at 72 C were performed. Samples were then heated to 72 C for 10 min before being stored at 4 C. Two primers were used to identify the cin cin allele: (1) GTA AGG TTG AAT GGC AGG AAC and (2) CCT TCT TTA TCA ACC AAT CAA C. The two primers used to clone the cin suba allele were (3) ATC CTC ACC ACC ATC ACC AT and primer 2. PCR products were electrophoresed in 0.8% (w/v) agarose gels, purified by Wizard columns (Promega, Madison, WI), and then sequenced with the ABI automatic sequencer (Perkin Elmer, Foster City, CA) using oligos B and D. DNA blots were conducted as described previously (Coen et al., 1990). The probe for Southern hybridization was generated using pjam193. Microscopy Samples for SEM were prepared on plastic replicas as described earlier (Carpenter et al., 1995). Young leaves were dissected from the plants, and a plastic mold and cast were taken (Green and Linstead, 1990). The cast was gold coated, and micrographs were taken using a CT1500 HF microscope (Oxford Instruments, Concord, MA). In Situ Hybridization The methods used for tissue preparation, digoxigenin-labeling of RNA probes, and in situ hybridization were as described previously (Coen et al., 1990). The probe used to detect the CIN transcript was a 1048-bp fragment from the cdna clone, covering the entire ORF. For HIS4, the probe consisted of the entire cdna (Fobert et al., 1994). For CYCD3B,a3#-terminal fragment of the cdna lacking the poly(a) tail was used (Gaudin et al., 2000). Petal Measurements Petals were dissected under a microscope, flattened between glass plates, and photographs taken with a Nikon Coolpix 995 digital camera (Tokyo), either fitted on a microscope or a tripod. Quantitative parameters such as area, perimeter, length, and width were measured in the digital images using programs written in Matlab (The MathWorks, Natick, MA; program written by A.G. Rolland-Lagan). The average epidermal cell size was determined by counting the number of cells in SEM of 0.1 to 1 square mm area of the leaf surface, then dividing the area by the number of cells. Sequence data from this article have been deposited with the EMBL/ GenBank data libraries under accession number AY ACKNOWLEDGMENTS We thank Anne-Gaelle Rolland-Lagan for the Matlab program. In addition, we thank Annabel Whibley for helpful comments on the manuscript. Received November 19, 2003; returned for revision February 18, 2004; accepted February 21, LITERATURE CITED Bollmann J, Carpenter R, Coen ES (1991) Allelic interactions at the nivea locus of Antirrhinum. Plant Cell 3: Carpenter R, Copsey L, Vincent C, Doyle S, Magrath R, Coen E (1995) Control of flower development and phyllotaxy by meristem identity genes in Antirrhinum. Plant Cell 7: Carpenter R, Martin C, Coen ES (1987) Comparison of genetic behaviour of the transposable element Tam3 at two unlinked pigment loci in Antirrhinum majus. Mol Gen Genet 207: Plant Physiol. Vol. 135, 2004
10 Growth and Cell Differentiation in Petals and Leaves Coen ES, Romero JM, Doyle S, Elliott R, Murphy G, Carpenter R (1990) floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell63: Fobert PR, Coen ES, Murphy GJ, Doonan JH (1994) Patterns of cell division revealed by transcriptional regulation of genes during the cell cycle in plants. EMBO J 13: Galego L, Almeida J (2002) Role of DIVARICATA in the control of dorsoventral asymmetry in Antirrhinum flowers. Genes Dev 16: Gaudin V, Lunness PA, Fobert PR, Towers M, Riou-Khamlichi C, Murray JA,CoenE,DoonanJH(2000) The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiol 122: Glover BJ, Perez-Rodriguez M, Martin C (1998) Development of several epidermal cell types can be specified by the same MYB-related plant transcription factor. Development 125: Green PB, Linstead P (1990) A procedure for SEM of complex shoot structures applied to the inflorescence of snapdragon (Antirrhinum). Protoplasma 158: Gu Q, Ferrandiz C, Yanofsky MF, Martienssen R (1998) The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125: Hammer K, Knüpffer S, Knüpffer H (1990) Das Gaterslebener Antirrhinum-sortiment. Kulturpflanze 38: Keck E, McSteen P, Carpenter R, Coen E (2003) Separation of genetic functions controlling organ identity in flowers. EMBO J 22: Kim GT, Shoda K, Tsuge T, Cho KH, Uchimiya H, Yokoyama R, Nishitani K, Tsukaya H (2002) The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO J 21: Kim GT, Tsukaya H, Uchimiya H (1998) The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev 12: Kusaba M, Miyahara K, Iida S, Fukuoka H, Takano T, Sassa H, Nishimura M, Nishio T (2003) Low glutelin content1: a dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. Plant Cell 15: Luo D, Carpenter R, Copsey L, Vincent C, Clark J, Coen E (1999) Control of organ asymmetry in flowers of Antirrhinum. Cell 99: Luo D, Carpenter R, Vincent C, Copsey L, Coen E (1996) Origin of floral asymmetry in Antirrhinum. Nature 383: Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW (1996) The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: Mizukami Y, Fischer RL (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97: Nath U, Crawford BC, Carpenter R, Coen E (2003) Genetic control of surface curvature. Science 299: Noda K, Glover BJ, Linstead P, Martin C (1994) Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature 369: Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JA (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: Stubbe H (1966) Genetik und Zytologie von Antirrhinum L. sect. Antirrhinum. Veb Gustav Fischer Verlag, Jena, Germany Tsuge T, Tsukaya H, Uchimiya H (1996) Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122: VanderKrolAR,MurLA,BeldM,MolJN,StuitjeAR(1990) Flavonoid genes in petunia: Addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2: Vincent C, Coen ES (2004) A temporal and morphological framework for flower development in Antirrhinum majus. Can J Bot (in press) Plant Physiol. Vol. 135,
Testing the ABC floral-organ identity model: expression of A and C function genes
 Objectives: Testing the ABC floral-organ identity model: expression of A and C function genes To test the validity of the ABC model for floral organ identity we will: 1. Use the model to make predictions
Objectives: Testing the ABC floral-organ identity model: expression of A and C function genes To test the validity of the ABC model for floral organ identity we will: 1. Use the model to make predictions
Regulation of Floral Organ Identity. Dr. Chloe Diamond Mara
 Regulation of Floral Organ Identity Dr. Chloe Diamond Mara Flower Development Angiosperms (flowering plants) are the most widespread group of land plants Flowers are the reproductive organs that consist
Regulation of Floral Organ Identity Dr. Chloe Diamond Mara Flower Development Angiosperms (flowering plants) are the most widespread group of land plants Flowers are the reproductive organs that consist
Supplemental Data. Müller-Xing et al. (2014). Plant Cell /tpc
 Supplemental Figure 1. Phenotypes of iclf (clf-28 swn-7 CLF pro :CLF-GR) plants. A, Late rescue of iclf plants by renewed DEX treatment; senescent inflorescence with elongated siliques (arrow; 90 DAG,
Supplemental Figure 1. Phenotypes of iclf (clf-28 swn-7 CLF pro :CLF-GR) plants. A, Late rescue of iclf plants by renewed DEX treatment; senescent inflorescence with elongated siliques (arrow; 90 DAG,
Supplemental Figure S1. The number of hydathodes is reduced in the as2-1 rev-1
 Supplemental Data Supplemental Figure S1. The number of hydathodes is reduced in the as2-1 rev-1 and kan1-11 kan2-5 double mutants. A, The numbers of hydathodes in different leaves of Col-0, as2-1 rev-1,
Supplemental Data Supplemental Figure S1. The number of hydathodes is reduced in the as2-1 rev-1 and kan1-11 kan2-5 double mutants. A, The numbers of hydathodes in different leaves of Col-0, as2-1 rev-1,
Regulation of Floral-Organ- Type by SUPERMAN
 Regulation of Floral-Organ- Type by SUPERMAN 1. Need for regulators of the organ-identity genes. 2. The Superman mutant phenotype-predicting the role of SUPERMAN. 3. Testing our hypothesis of the role
Regulation of Floral-Organ- Type by SUPERMAN 1. Need for regulators of the organ-identity genes. 2. The Superman mutant phenotype-predicting the role of SUPERMAN. 3. Testing our hypothesis of the role
Cell lineage patterns and homeotic gene activity during Antirrhinum flower development
 Cell lineage patterns and homeotic gene activity during Antirrhinum flower development Coral A. Vincent, Rosemary Carpenter and Enrico S. Coen Genetics Department, John Innes Centre, Colney Lane, Norwich
Cell lineage patterns and homeotic gene activity during Antirrhinum flower development Coral A. Vincent, Rosemary Carpenter and Enrico S. Coen Genetics Department, John Innes Centre, Colney Lane, Norwich
Floral Organ Mutants and the Study of Organ Morphogenesis
 Floral Organ Mutants and the Study of Organ Morphogenesis Objectives: 1. How does one use mutants to understand floral organ morphogenesis? 2. What are the phenotypes of some floral organ mutants? 3. What
Floral Organ Mutants and the Study of Organ Morphogenesis Objectives: 1. How does one use mutants to understand floral organ morphogenesis? 2. What are the phenotypes of some floral organ mutants? 3. What
Arabidopsis: Flower Development and Patterning
 Arabidopsis: Flower Development and Patterning John L Bowman, University of California, Davis, California, USA The development of flowers and floral organs is directed by genetic programmes likely to be
Arabidopsis: Flower Development and Patterning John L Bowman, University of California, Davis, California, USA The development of flowers and floral organs is directed by genetic programmes likely to be
FILAMENTOUS FLOWER Controls the Formation and Development of Arabidopsis Inflorescences and Floral Meristems
 The Plant Cell, Vol. 11, 69 86, January 1999, www.plantcell.org 1999 American Society of Plant Physiologists FILAMENTOUS FLOWER Controls the Formation and Development of Arabidopsis Inflorescences and
The Plant Cell, Vol. 11, 69 86, January 1999, www.plantcell.org 1999 American Society of Plant Physiologists FILAMENTOUS FLOWER Controls the Formation and Development of Arabidopsis Inflorescences and
Genetics of Floral Development An Evo-Devo Approach
 Genetics of Floral Development An Evo-Devo Approach Biology 317 Kelsey Galimba 8.5.2013 Why are flowering plants so diverse? Why are flowering plants so diverse? http://www.botanicalgarden.ubc.ca/potd/
Genetics of Floral Development An Evo-Devo Approach Biology 317 Kelsey Galimba 8.5.2013 Why are flowering plants so diverse? Why are flowering plants so diverse? http://www.botanicalgarden.ubc.ca/potd/
BIOL2005 WORKSHEET 2008
 BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
UFO and LEAFY in Arabidopsis
 Development 128, 2735-2746 (2001) Printed in Great Britain The Company of Biologists Limited 2001 DEV0360 2735 The ASK1 gene regulates B function gene expression in cooperation with UFO and LEAFY in Arabidopsis
Development 128, 2735-2746 (2001) Printed in Great Britain The Company of Biologists Limited 2001 DEV0360 2735 The ASK1 gene regulates B function gene expression in cooperation with UFO and LEAFY in Arabidopsis
A genetic and molecular model for flower development in Arabidopsis thaliana
 Development Supplement I, 1991, 157-167 Printed in Great Britain The Company of Biologists Limited 1991 157 A genetic and molecular model for flower development in Arabidopsis thaliana ELLIOT M. MEYEROWITZ*,
Development Supplement I, 1991, 157-167 Printed in Great Britain The Company of Biologists Limited 1991 157 A genetic and molecular model for flower development in Arabidopsis thaliana ELLIOT M. MEYEROWITZ*,
The Role of Lipids in Flowering Development of Arabidopsis Enhanced pah1pah2 Plants. Toshiro Ito 1 & Lee Lishi 2
 The Role of Lipids in Flowering Development of Arabidopsis Enhanced pah1pah2 Plants Toshiro Ito 1 & Lee Lishi 2 Department of Biological Sciences, Faculty of Science, National University of Singapore,
The Role of Lipids in Flowering Development of Arabidopsis Enhanced pah1pah2 Plants Toshiro Ito 1 & Lee Lishi 2 Department of Biological Sciences, Faculty of Science, National University of Singapore,
CLAVATA3 is a specific regulator of shoot and floral meristem development
 Development 2, 20572067 (995) Printed in Great Britain The Company of Biologists Limited 995 2057 CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes
Development 2, 20572067 (995) Printed in Great Britain The Company of Biologists Limited 995 2057 CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes
PLENA and FARINELLI: redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development
 The EMBO Journal Vol.18 No.14 pp.4023 4034, 1999 PLENA and FARINELLI: redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development Brendan Davies 1, Patrick
The EMBO Journal Vol.18 No.14 pp.4023 4034, 1999 PLENA and FARINELLI: redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development Brendan Davies 1, Patrick
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Figure 1. Generation of a conditional allele of the Kindlin-2 gene. (A) A restriction map of the relevant genomic region of Kindlin-2 (top), the targeting construct
SUPPLEMENTARY INFORMATION Supplementary Figure 1. Generation of a conditional allele of the Kindlin-2 gene. (A) A restriction map of the relevant genomic region of Kindlin-2 (top), the targeting construct
A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS Ilha Lee, Diana S. Wolfe, Ove Nilsson and Detlef Weigel
 Research Paper 95 A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS Ilha Lee, Diana S. Wolfe, Ove Nilsson and Detlef Weigel Background: Development of petals and stamens in Arabidopsis flowers requires
Research Paper 95 A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS Ilha Lee, Diana S. Wolfe, Ove Nilsson and Detlef Weigel Background: Development of petals and stamens in Arabidopsis flowers requires
A genetic framework for fruit patterning in Arabidopsis thaliana
 Research article 4687 A genetic framework for fruit patterning in Arabidopsis thaliana José R. Dinneny 1,2, Detlef Weigel 2,3 and Martin F. Yanofsky 1, * 1 Division of Biological Sciences, University of
Research article 4687 A genetic framework for fruit patterning in Arabidopsis thaliana José R. Dinneny 1,2, Detlef Weigel 2,3 and Martin F. Yanofsky 1, * 1 Division of Biological Sciences, University of
Supplemental Data. Shin et al. Plant Cell. (2012) /tpc YFP N
 MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
Supplemental Data. Wu et al. (2010). Plant Cell /tpc
 Supplemental Figure 1. FIM5 is preferentially expressed in stamen and mature pollen. The expression data of FIM5 was extracted from Arabidopsis efp browser (http://www.bar.utoronto.ca/efp/development/),
Supplemental Figure 1. FIM5 is preferentially expressed in stamen and mature pollen. The expression data of FIM5 was extracted from Arabidopsis efp browser (http://www.bar.utoronto.ca/efp/development/),
The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function
 University of South Carolina Scholar Commons Faculty Publications Biological Sciences, Department of 1-1-1996 The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class
University of South Carolina Scholar Commons Faculty Publications Biological Sciences, Department of 1-1-1996 The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class
Functional Diversification of the Two C-Class MADS Box Genes OSMADS3 and OSMADS58 in Oryza sativa W OA
 The Plant Cell, Vol. 18, 15 28, January 2006, www.plantcell.org ª 2005 American Society of Plant Biologists RESEARCH ARTICLES Functional Diversification of the Two C-Class MADS Box Genes OSMADS3 and OSMADS58
The Plant Cell, Vol. 18, 15 28, January 2006, www.plantcell.org ª 2005 American Society of Plant Biologists RESEARCH ARTICLES Functional Diversification of the Two C-Class MADS Box Genes OSMADS3 and OSMADS58
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
Activation of the Arabidopsis B Class Homeotic Genes by APETALA1
 The Plant Cell, Vol. 13, 739 753, April 2001, www.plantcell.org 2001 American Society of Plant Physiologists RESEARCH ARTICLE Activation of the Arabidopsis B Class Homeotic Genes by APETALA1 Medard Ng
The Plant Cell, Vol. 13, 739 753, April 2001, www.plantcell.org 2001 American Society of Plant Physiologists RESEARCH ARTICLE Activation of the Arabidopsis B Class Homeotic Genes by APETALA1 Medard Ng
a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53
 1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
ACURIOUS malformation in one of the flowers on a raceme of
 ON AN ABNORMALITY IN PURPUREA By VIOLET L. ANDERSON. DIGITALIS Quain Student of Botany, University College, London. (With 6 figures in the text.) ACURIOUS malformation in one of the flowers on a raceme
ON AN ABNORMALITY IN PURPUREA By VIOLET L. ANDERSON. DIGITALIS Quain Student of Botany, University College, London. (With 6 figures in the text.) ACURIOUS malformation in one of the flowers on a raceme
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated
 Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Problem Set 5 KEY
 2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
The SEP4 Gene of Arabidopsis thaliana Functions in Floral Organ and Meristem Identity
 Current Biology, Vol. 14, 1935 1940, November 9, 2004, 2004 Elsevier Ltd. All rights reserved. DOI 10.1016/j.cub.2004.10.028 The SEP4 Gene of Arabidopsis thaliana Functions in Floral Organ and Meristem
Current Biology, Vol. 14, 1935 1940, November 9, 2004, 2004 Elsevier Ltd. All rights reserved. DOI 10.1016/j.cub.2004.10.028 The SEP4 Gene of Arabidopsis thaliana Functions in Floral Organ and Meristem
Supplemental Data. Wang et al. (2013). Plant Cell /tpc
 Supplemental Data. Wang et al. (2013). Plant Cell 10.1105/tpc.112.108993 Supplemental Figure 1. 3-MA Treatment Reduces the Growth of Seedlings. Two-week-old Nicotiana benthamiana seedlings germinated on
Supplemental Data. Wang et al. (2013). Plant Cell 10.1105/tpc.112.108993 Supplemental Figure 1. 3-MA Treatment Reduces the Growth of Seedlings. Two-week-old Nicotiana benthamiana seedlings germinated on
Patterns of Cell Division, Cell Differentiation and Cell Elongation in Epidermis and Cortex of Arabidopsis pedicels in the Wild Type and in erecta
 Patterns of Cell Division, Cell Differentiation and Cell Elongation in Epidermis and Cortex of Arabidopsis pedicels in the Wild Type and in erecta Mark G. R. Bundy, Olivia A. Thompson, Matthew T. Sieger,
Patterns of Cell Division, Cell Differentiation and Cell Elongation in Epidermis and Cortex of Arabidopsis pedicels in the Wild Type and in erecta Mark G. R. Bundy, Olivia A. Thompson, Matthew T. Sieger,
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
AINTEGUMENTA Contributes to Organ Polarity and Regulates Growth of Lateral Organs in Combination with YABBY Genes 1
 AINTEGUMENTA Contributes to Organ Polarity and Regulates Growth of Lateral Organs in Combination with YABBY Genes 1 Staci Nole-Wilson 2 and Beth A. Krizek* Department of Biological Sciences, University
AINTEGUMENTA Contributes to Organ Polarity and Regulates Growth of Lateral Organs in Combination with YABBY Genes 1 Staci Nole-Wilson 2 and Beth A. Krizek* Department of Biological Sciences, University
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Ectopic Expression of AINTEGUMENTA in Arabidopsis Plants Results in Increased Growth of Floral Organs
 DEVELOPMENTAL GENETICS 25:224 236 (1999) Ectopic Expression of AINTEGUMENTA in Arabidopsis Plants Results in Increased Growth of Floral Organs BETH ALLYN KRIZEK* Department of Biological Sciences, University
DEVELOPMENTAL GENETICS 25:224 236 (1999) Ectopic Expression of AINTEGUMENTA in Arabidopsis Plants Results in Increased Growth of Floral Organs BETH ALLYN KRIZEK* Department of Biological Sciences, University
Probe. Hind III Q,!?R'!! /0!!!!D1"?R'! vector. Homologous recombination
 Supple-Zhang Page 1 Wild-type locus Targeting construct Targeted allele Exon Exon3 Exon Probe P1 P P3 FRT FRT loxp loxp neo vector amh I Homologous recombination neo P1 P P3 FLPe recombination Q,!?R'!!
Supple-Zhang Page 1 Wild-type locus Targeting construct Targeted allele Exon Exon3 Exon Probe P1 P P3 FRT FRT loxp loxp neo vector amh I Homologous recombination neo P1 P P3 FLPe recombination Q,!?R'!!
8 Suppression Analysis
 Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 8 Suppression Analysis OVERVIEW Suppression
Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 8 Suppression Analysis OVERVIEW Suppression
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Genetic Specification of floral organ identity. Initiating floral development. Deciding when to initiate flowering - induced mutations -in Nature
 Genetic Specification of floral organ identity Initiating floral development Deciding when to initiate flowering - induced mutations -in Nature Flower structure of rabidopsis Stamens arpels Petals Sepals
Genetic Specification of floral organ identity Initiating floral development Deciding when to initiate flowering - induced mutations -in Nature Flower structure of rabidopsis Stamens arpels Petals Sepals
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
c Tuj1(-) apoptotic live 1 DIV 2 DIV 1 DIV 2 DIV Tuj1(+) Tuj1/GFP/DAPI Tuj1 DAPI GFP
 Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
Genetic suppressors and enhancers provide clues to gene regulation and genetic pathways
 Genetic suppressors and enhancers provide clues to gene regulation and genetic pathways Suppressor mutation: a second mutation results in a less severe phenotype than the original mutation Suppressor mutations
Genetic suppressors and enhancers provide clues to gene regulation and genetic pathways Suppressor mutation: a second mutation results in a less severe phenotype than the original mutation Suppressor mutations
Supplemental Information. Spatial Auxin Signaling. Controls Leaf Flattening in Arabidopsis
 Current Biology, Volume 27 Supplemental Information Spatial Auxin Signaling Controls Leaf Flattening in Arabidopsis Chunmei Guan, Binbin Wu, Ting Yu, Qingqing Wang, Naden T. Krogan, Xigang Liu, and Yuling
Current Biology, Volume 27 Supplemental Information Spatial Auxin Signaling Controls Leaf Flattening in Arabidopsis Chunmei Guan, Binbin Wu, Ting Yu, Qingqing Wang, Naden T. Krogan, Xigang Liu, and Yuling
Arabidopsis fruit development
 Development 125, 1509-1517 (1998) Printed in Great Britain The Company of Biologists Limited 1998 DEV155 1509 The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development
Development 125, 1509-1517 (1998) Printed in Great Britain The Company of Biologists Limited 1998 DEV155 1509 The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development
Supplemental Data. Beck et al. (2010). Plant Cell /tpc
 Supplemental Figure 1. Phenotypic comparison of the rosette leaves of four-week-old mpk4 and Col-0 plants. A mpk4 vs Col-0 plants grown in soil. Note the extreme dwarfism of the mpk4 plants (white arrows)
Supplemental Figure 1. Phenotypic comparison of the rosette leaves of four-week-old mpk4 and Col-0 plants. A mpk4 vs Col-0 plants grown in soil. Note the extreme dwarfism of the mpk4 plants (white arrows)
Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities
 Research article 2997 Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities Yuval Eshed 1,2, *,, Anat Izhaki 1, *, Stuart F. Baum 1,, Sandra K. Floyd
Research article 2997 Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities Yuval Eshed 1,2, *,, Anat Izhaki 1, *, Stuart F. Baum 1,, Sandra K. Floyd
Gwyneth C. Ingram,a Justin Goodrich,a Mark D. Wilkinson,b Rüdiger Simon,a George W. Haughn,b and Enrico S. Coena,
 The Plant Cell, Vol. 7, 1501-1510, September 1995 O 1995 American Society of Plant Physiologists Parallels between UNUSUAL FLORAL ORGANS and FMBRATA, Genes Controlling Flower Development in Arabidopsis
The Plant Cell, Vol. 7, 1501-1510, September 1995 O 1995 American Society of Plant Physiologists Parallels between UNUSUAL FLORAL ORGANS and FMBRATA, Genes Controlling Flower Development in Arabidopsis
Luminescent platforms for monitoring changes in the solubility of amylin and huntingtin in living cells
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
HANABA TARANU Is a GATA Transcription Factor That Regulates Shoot Apical Meristem and Flower Development in Arabidopsis W
 The Plant Cell, Vol. 16, 2586 2600, October 2004, www.plantcell.org ª 2004 American Society of Plant Biologists HANABA TARANU Is a GATA Transcription Factor That Regulates Shoot Apical Meristem and Flower
The Plant Cell, Vol. 16, 2586 2600, October 2004, www.plantcell.org ª 2004 American Society of Plant Biologists HANABA TARANU Is a GATA Transcription Factor That Regulates Shoot Apical Meristem and Flower
Variations in Chromosome Structure & Function. Ch. 8
 Variations in Chromosome Structure & Function Ch. 8 1 INTRODUCTION! Genetic variation refers to differences between members of the same species or those of different species Allelic variations are due
Variations in Chromosome Structure & Function Ch. 8 1 INTRODUCTION! Genetic variation refers to differences between members of the same species or those of different species Allelic variations are due
An Enhancer Trap Line Associated with a D-Class Cyclin Gene in Arabidopsis 1
 An Enhancer Trap Line Associated with a D-Class Cyclin Gene in Arabidopsis 1 Kankshita Swaminathan, Yingzhen Yang, Natasha Grotz, Lauren Campisi, and Thomas Jack* Department of Biological Sciences, Dartmouth
An Enhancer Trap Line Associated with a D-Class Cyclin Gene in Arabidopsis 1 Kankshita Swaminathan, Yingzhen Yang, Natasha Grotz, Lauren Campisi, and Thomas Jack* Department of Biological Sciences, Dartmouth
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Supplemental Information. Figures. Figure S1
 Supplemental Information Figures Figure S1 Identification of JAGGER T-DNA insertions. A. Positions of T-DNA and Ds insertions in JAGGER are indicated by inverted triangles, the grey box represents the
Supplemental Information Figures Figure S1 Identification of JAGGER T-DNA insertions. A. Positions of T-DNA and Ds insertions in JAGGER are indicated by inverted triangles, the grey box represents the
Nature Neuroscience: doi: /nn Supplementary Figure 1. Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites.
 Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana
 Biochemistry: Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana 38 CAMERON E. NIENABER ʻ04 Abstract Long recognized as essential plant nutrients and metabolites, inorganic and organic
Biochemistry: Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana 38 CAMERON E. NIENABER ʻ04 Abstract Long recognized as essential plant nutrients and metabolites, inorganic and organic
Supplemental Material for. Figure S1. Identification of TetR responsive promoters in F. novicida and E. coli.
 Supplemental Material for Synthetic promoters functional in Francisella novicida and Escherichia coli Ralph L. McWhinnie and Francis E. Nano Department of Biochemistry and Microbiology, University of Victoria,
Supplemental Material for Synthetic promoters functional in Francisella novicida and Escherichia coli Ralph L. McWhinnie and Francis E. Nano Department of Biochemistry and Microbiology, University of Victoria,
Vertebrate Limb Patterning
 Vertebrate Limb Patterning What makes limb patterning an interesting/useful developmental system How limbs develop Key events in limb development positioning and specification initiation of outgrowth establishment
Vertebrate Limb Patterning What makes limb patterning an interesting/useful developmental system How limbs develop Key events in limb development positioning and specification initiation of outgrowth establishment
Genes Directing Flower Development in Arabidopsis
 The Plant Cell, Vol. 1,37-52, January 1989, 1989 American Society of Plant Physiologists Genes Directing Flower Development in Arabidopsis John L. Bowman, David R. Smyth, 1 and Elliot M. Meyerowitz 2 Division
The Plant Cell, Vol. 1,37-52, January 1989, 1989 American Society of Plant Physiologists Genes Directing Flower Development in Arabidopsis John L. Bowman, David R. Smyth, 1 and Elliot M. Meyerowitz 2 Division
Life Sciences 1A Midterm Exam 2. November 13, 2006
 Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
The Ff.010 Gene Product Regulates the Expression Domain of Homeotic Genes AP3 and PI in Arabidopsis Flowers
 The Plant Cell, Vol. 3, 1221-1237, November 1991 1991 American Society of Plant Physiologists The Ff.010 Gene Product Regulates the Expression Domain of Homeotic Genes AP3 and PI in Arabidopsis Flowers
The Plant Cell, Vol. 3, 1221-1237, November 1991 1991 American Society of Plant Physiologists The Ff.010 Gene Product Regulates the Expression Domain of Homeotic Genes AP3 and PI in Arabidopsis Flowers
Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal
 Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus Fadel TISSIR, Yibo QU, Mireille MONTCOUQUIOL, Libing ZHOU, Kouji KOMATSU, Dongbo SHI, Toshihiko FUJIMORI,
Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus Fadel TISSIR, Yibo QU, Mireille MONTCOUQUIOL, Libing ZHOU, Kouji KOMATSU, Dongbo SHI, Toshihiko FUJIMORI,
Supplemental Figures Legends and Supplemental Figures. for. pirna-guided slicing of transposon transcripts enforces their transcriptional
 Supplemental Figures Legends and Supplemental Figures for pirn-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear pirn repertoire Kirsten-ndré
Supplemental Figures Legends and Supplemental Figures for pirn-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear pirn repertoire Kirsten-ndré
CHAPTER 4 RESULTS. showed that all three replicates had similar growth trends (Figure 4.1) (p<0.05; p=0.0000)
 CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
CLAVATA1, a regulator of meristem and flower development in Arabidopsis
 Development 119, 397-418 (1993) Printed in Great Britain The Company of Biologists Limited 1993 397 CLAVATA1, a regulator of meristem and flower development in Arabidopsis Steven E. Clark, Mark P. Running
Development 119, 397-418 (1993) Printed in Great Britain The Company of Biologists Limited 1993 397 CLAVATA1, a regulator of meristem and flower development in Arabidopsis Steven E. Clark, Mark P. Running
Transcriptional repression of Xi
 Transcriptional repression of Xi Xist Transcription of Xist Xist RNA Spreading of Xist Recruitment of repression factors. Stable repression Translocated Xic cannot efficiently silence autosome regions.
Transcriptional repression of Xi Xist Transcription of Xist Xist RNA Spreading of Xist Recruitment of repression factors. Stable repression Translocated Xic cannot efficiently silence autosome regions.
Primary Cilia Can Both Mediate and Suppress Hedgehog Pathway- Dependent Tumorigenesis (Supplementary Figures and Materials)
 Primary Cilia Can Both Mediate and Suppress Hedgehog Pathway- Dependent Tumorigenesis (Supplementary Figures and Materials) Sunny Y. Wong, Allen D. Seol, Po-Lin So, Alexandre N. Ermilov, Christopher K.
Primary Cilia Can Both Mediate and Suppress Hedgehog Pathway- Dependent Tumorigenesis (Supplementary Figures and Materials) Sunny Y. Wong, Allen D. Seol, Po-Lin So, Alexandre N. Ermilov, Christopher K.
Expression of acid base transporters in the kidney collecting duct in Slc2a7 -/-
 Supplemental Material Results. Expression of acid base transporters in the kidney collecting duct in Slc2a7 -/- and Slc2a7 -/- mice. The expression of AE1 in the kidney was examined in Slc26a7 KO mice.
Supplemental Material Results. Expression of acid base transporters in the kidney collecting duct in Slc2a7 -/- and Slc2a7 -/- mice. The expression of AE1 in the kidney was examined in Slc26a7 KO mice.
Mendel s Methods: Monohybrid Cross
 Mendel s Methods: Monohybrid Cross Mendel investigated whether the white-flowered form disappeared entirely by breeding the F1 purple flowers with each other. Crossing two purple F1 monohybrid plants is
Mendel s Methods: Monohybrid Cross Mendel investigated whether the white-flowered form disappeared entirely by breeding the F1 purple flowers with each other. Crossing two purple F1 monohybrid plants is
Tetrapod Limb Development
 IBS 8102 Cell, Molecular and Developmental Biology Tetrapod Limb Development February 11, 2008 Tetrapod Limbs Merlin D. Tuttle Vicki Lockard and Paul Barry Father Alejandro Sanchez Anne Fischer Limb Patterning
IBS 8102 Cell, Molecular and Developmental Biology Tetrapod Limb Development February 11, 2008 Tetrapod Limbs Merlin D. Tuttle Vicki Lockard and Paul Barry Father Alejandro Sanchez Anne Fischer Limb Patterning
Supporting Information
 Supporting Information Franco et al. 10.1073/pnas.1015557108 SI Materials and Methods Drug Administration. PD352901 was dissolved in 0.5% (wt/vol) hydroxyl-propyl-methylcellulose, 0.2% (vol/vol) Tween
Supporting Information Franco et al. 10.1073/pnas.1015557108 SI Materials and Methods Drug Administration. PD352901 was dissolved in 0.5% (wt/vol) hydroxyl-propyl-methylcellulose, 0.2% (vol/vol) Tween
Supporting Information
 Supporting Information Harries et al. 1.173/pnas.9923916 A Fig. S1. Disruption of microfilaments within epidermal cells after treatment with 5 M Lat. Images of N. benthamiana cells are from plants expressing
Supporting Information Harries et al. 1.173/pnas.9923916 A Fig. S1. Disruption of microfilaments within epidermal cells after treatment with 5 M Lat. Images of N. benthamiana cells are from plants expressing
SUPPLEMENTARY INFORMATION. Otx2 controls neuron subtype identity in ventral tegmental area and antagonizes
 Di Salvio et al. 1 SUPPLEMENTARY INFORMATION Otx2 controls neuron subtype identity in ventral tegmental area and antagonizes vulnerability to MPTP Michela Di Salvio, Luca Giovanni Di Giovannantonio, Dario
Di Salvio et al. 1 SUPPLEMENTARY INFORMATION Otx2 controls neuron subtype identity in ventral tegmental area and antagonizes vulnerability to MPTP Michela Di Salvio, Luca Giovanni Di Giovannantonio, Dario
The Rab GTPase RabA4d Regulates Pollen Tube Tip Growth in Arabidopsis thaliana W
 The Plant Cell, Vol. 21: 526 544, February 2009, www.plantcell.org ã 2009 American Society of Plant Biologists The Rab GTPase RabA4d Regulates Pollen Tube Tip Growth in Arabidopsis thaliana W Amy L. Szumlanski
The Plant Cell, Vol. 21: 526 544, February 2009, www.plantcell.org ã 2009 American Society of Plant Biologists The Rab GTPase RabA4d Regulates Pollen Tube Tip Growth in Arabidopsis thaliana W Amy L. Szumlanski
Identification of a novel duplication mutation in the VHL gene in a large Chinese family with Von Hippel-Lindau (VHL) syndrome
 Identification of a novel duplication mutation in the VHL gene in a large Chinese family with Von Hippel-Lindau (VHL) syndrome L.H. Cao 1, B.H. Kuang 2, C. Chen 1, C. Hu 2, Z. Sun 1, H. Chen 2, S.S. Wang
Identification of a novel duplication mutation in the VHL gene in a large Chinese family with Von Hippel-Lindau (VHL) syndrome L.H. Cao 1, B.H. Kuang 2, C. Chen 1, C. Hu 2, Z. Sun 1, H. Chen 2, S.S. Wang
Unit D Notebook Directions
 DO NOT PUT THIS FIRST PAGE IN YOUR NOTEBOOK!! Unit D Notebook Directions Immediately following the last page of Unit C, do the following: (Page numbers are not important, but the order needs to be exact)
DO NOT PUT THIS FIRST PAGE IN YOUR NOTEBOOK!! Unit D Notebook Directions Immediately following the last page of Unit C, do the following: (Page numbers are not important, but the order needs to be exact)
Specification of Arabidopsis floral meristem identity by repression of flowering time genes
 RESEARCH ARTICLE 1901 Development 134, 1901-1910 (2007) doi:10.1242/dev.003103 Specification of Arabidopsis floral meristem identity by repression of flowering time genes Chang Liu 1, *, Jing Zhou 1, *,
RESEARCH ARTICLE 1901 Development 134, 1901-1910 (2007) doi:10.1242/dev.003103 Specification of Arabidopsis floral meristem identity by repression of flowering time genes Chang Liu 1, *, Jing Zhou 1, *,
See pedigree info on extra sheet 1. ( 6 pts)
 Biol 321 Quiz #2 Spring 2010 40 pts NAME See pedigree info on extra sheet 1. ( 6 pts) Examine the pedigree shown above. For each mode of inheritance listed below indicate: E = this mode of inheritance
Biol 321 Quiz #2 Spring 2010 40 pts NAME See pedigree info on extra sheet 1. ( 6 pts) Examine the pedigree shown above. For each mode of inheritance listed below indicate: E = this mode of inheritance
Phylogenetic analysis of human and chicken importins. Only five of six importins were studied because
 Supplementary Figure S1 Phylogenetic analysis of human and chicken importins. Only five of six importins were studied because importin-α6 was shown to be testis-specific. Human and chicken importin protein
Supplementary Figure S1 Phylogenetic analysis of human and chicken importins. Only five of six importins were studied because importin-α6 was shown to be testis-specific. Human and chicken importin protein
Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene
 Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene YUELIN ZHANG, WEIHUA FAN, MARK KINKEMA, XIN LI, AND
Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene YUELIN ZHANG, WEIHUA FAN, MARK KINKEMA, XIN LI, AND
Structural Variation and Medical Genomics
 Structural Variation and Medical Genomics Andrew King Department of Biomedical Informatics July 8, 2014 You already know about small scale genetic mutations Single nucleotide polymorphism (SNPs) Deletions,
Structural Variation and Medical Genomics Andrew King Department of Biomedical Informatics July 8, 2014 You already know about small scale genetic mutations Single nucleotide polymorphism (SNPs) Deletions,
Floral organogenesis in Antirrhinum majus (Scrophulariaceae)
 Proc. Indian Acad. Sci., Vol. 88 B, Part II, Number 3, May 1979, pp. 183-188, printed in India. Floral organogenesis in Antirrhinum majus (Scrophulariaceae) V SINGH and D K JAIN* Department of Botany,
Proc. Indian Acad. Sci., Vol. 88 B, Part II, Number 3, May 1979, pp. 183-188, printed in India. Floral organogenesis in Antirrhinum majus (Scrophulariaceae) V SINGH and D K JAIN* Department of Botany,
Supplemental Figure 1. Small RNA size distribution from different soybean tissues.
 Supplemental Figure 1. Small RNA size distribution from different soybean tissues. The size of small RNAs was plotted versus frequency (percentage) among total sequences (A, C, E and G) or distinct sequences
Supplemental Figure 1. Small RNA size distribution from different soybean tissues. The size of small RNAs was plotted versus frequency (percentage) among total sequences (A, C, E and G) or distinct sequences
Nature Genetics: doi: /ng Supplementary Figure 1. Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms.
 Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy
 Kavya Puchhalapalli CALS Honors Project Report Spring 2017 Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy Abstract Malignant brain tumors including medulloblastomas and primitive neuroectodermal
Kavya Puchhalapalli CALS Honors Project Report Spring 2017 Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy Abstract Malignant brain tumors including medulloblastomas and primitive neuroectodermal
Flowering Plant Reproduction
 Lab Exercise Flowering Plant Reproduction Objectives - To be able to identify the parts of a flower - Be able to distinguish between dicots and monocots based on flower morphology - Become familiar with
Lab Exercise Flowering Plant Reproduction Objectives - To be able to identify the parts of a flower - Be able to distinguish between dicots and monocots based on flower morphology - Become familiar with
Genetics 275 Examination February 10, 2003.
 Genetics 275 Examination February 10, 2003. Do all questions in the spaces provided. The value for this examination is twenty marks (20% of the grade for the course). The value for individual questions
Genetics 275 Examination February 10, 2003. Do all questions in the spaces provided. The value for this examination is twenty marks (20% of the grade for the course). The value for individual questions
Open Flower. Juvenile leaf Flowerbud. Carpel 35 NA NA NA NA 61 NA 95 NA NA 15 NA 41 3 NA
 PaxDB Root Juvenile leaf Flowerbud Open Flower Carpel Mature Pollen Silique Seed Sec3a Sec3b Sec5a Sec5b Sec6 Sec8 Sec10a/b Sec15a Sec15b Exo84a Exo84b Exo84c Exo70A1 Exo70A2 Exo70A3 49 47 8 75 104 79
PaxDB Root Juvenile leaf Flowerbud Open Flower Carpel Mature Pollen Silique Seed Sec3a Sec3b Sec5a Sec5b Sec6 Sec8 Sec10a/b Sec15a Sec15b Exo84a Exo84b Exo84c Exo70A1 Exo70A2 Exo70A3 49 47 8 75 104 79
The Biology and Genetics of Cells and Organisms The Biology of Cancer
 The Biology and Genetics of Cells and Organisms The Biology of Cancer Mendel and Genetics How many distinct genes are present in the genomes of mammals? - 21,000 for human. - Genetic information is carried
The Biology and Genetics of Cells and Organisms The Biology of Cancer Mendel and Genetics How many distinct genes are present in the genomes of mammals? - 21,000 for human. - Genetic information is carried
SALSA MLPA probemix P185-C2 Intersex Lot C2-1015: As compared to the previous version C1 (lot C1-0611), the lengths of four probes have been adjusted.
 mix P185-C2 Intersex Lot C2-1015: As compared to the previous version C1 (lot C1-0611), the lengths of four s have been adjusted. The sex-determining region on chromosome Y (SRY) is the most important
mix P185-C2 Intersex Lot C2-1015: As compared to the previous version C1 (lot C1-0611), the lengths of four s have been adjusted. The sex-determining region on chromosome Y (SRY) is the most important
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Class XII Chapter 5 Principles of Inheritance and Variation Biology
 Question 1: Mention the advantages of selecting pea plant for experiment by Mendel. Mendel selected pea plants to carry out his study on the inheritance of characters from parents to offspring. He selected
Question 1: Mention the advantages of selecting pea plant for experiment by Mendel. Mendel selected pea plants to carry out his study on the inheritance of characters from parents to offspring. He selected
Chapter 02 Mendelian Inheritance
 Chapter 02 Mendelian Inheritance Multiple Choice Questions 1. The theory of pangenesis was first proposed by. A. Aristotle B. Galen C. Mendel D. Hippocrates E. None of these Learning Objective: Understand
Chapter 02 Mendelian Inheritance Multiple Choice Questions 1. The theory of pangenesis was first proposed by. A. Aristotle B. Galen C. Mendel D. Hippocrates E. None of these Learning Objective: Understand
Phenomena first observed in petunia
 Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
Developmental Biology
 Developmental Biology 362 (2012) 141 153 Contents lists available at SciVerse ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology Dual function of suppressor of
Developmental Biology 362 (2012) 141 153 Contents lists available at SciVerse ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology Dual function of suppressor of
s u p p l e m e n ta ry i n f o r m at i o n
 Figure S1 Characterization of tet-off inducible cell lines expressing GFPprogerin and GFP-wt lamin A. a, Western blot analysis of GFP-progerin- or GFP-wt lamin A- expressing cells before induction (0d)
Figure S1 Characterization of tet-off inducible cell lines expressing GFPprogerin and GFP-wt lamin A. a, Western blot analysis of GFP-progerin- or GFP-wt lamin A- expressing cells before induction (0d)
NO APICAL MERISTEM (MtNAM) regulates floral organ identity and lateral organ separation in Medicago truncatula
 Research NO APICAL MERISTEM (MtNAM) regulates floral organ identity and lateral organ separation in Medicago truncatula Xiaofei Cheng, Jianling Peng, Junying Ma, Yuhong Tang, Rujin Chen, Kirankumar S.
Research NO APICAL MERISTEM (MtNAM) regulates floral organ identity and lateral organ separation in Medicago truncatula Xiaofei Cheng, Jianling Peng, Junying Ma, Yuhong Tang, Rujin Chen, Kirankumar S.
