Nature Genetics: doi: /ng Supplementary Figure 1. Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms.
|
|
|
- Morris Nelson
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental stages grown at 20 C (Online Methods). The staining of H4 pan-acetyl (H4ac) served as a positive control. (a) Gonads (bar, 20 m). (b) Embryos and L2-stage larvae (bar, 20 m). 1
2 Supplementary Figure 2 Phenotypes of strains losing H3K9me in the germ line. (a) Number of viable progeny of wt and met-2 set-25 per worm at 26 C, over three generations, starting at generation 3 after transfer of the worms from 20 C (number of independent experiments (N) = 3, number of worms counted per experiment (n) = 60). (b) Number of viable progeny of wt, met-2 set-25 and mutants of the PIWI pathway per worm at 15 C, 20 C and 25 C by generation 3 after transfer from 20 C to the indicated temperature (N = 1, n = 25). (c) Percentage of worms developing full gonad arms at 20 C. mex-5:gfp-h2b was used to visualize gonad cells at all stages (N = 7, n = 2). (d) Analysis of RNA-seq data showing average fold change in expression of apoptosis response genes in the gonads of met-2 set-25 mutant versus wt (N = 3, P < 0.05 adjusted for multiple testing; FDR < 0.05). (e) Percentage of laid eggs hatching at 20 C. Strains deficient for CEP-1 (p53) additionally expressed the CED-1::GFP apoptosis reporter (N = 2, n = 80). (f) Analysis of RAD-51 foci detected by IF in the mitotic zone of the indicated genotypes to quantify the presence of resected DNA double-strand breaks. The percentage in images is equal to the frequency of mitotic tip cells with detectable RAD-51 foci. The bar graph further segregates positive cells by number of foci per cell (N = 3, n = 40 gonads). 2
3 3
4 Supplementary Figure 3 H3K9me distribution on genes and repeats. (a) H3K9me2 and H3K9me3 ChIP-seq were performed on early embryos at 20 C (N = 2) in a wild-type (N2) strain. Distribution along chr. I in relation to repetitive elements (REs) is shown. Quantification to the right shows the ratio of the coverage of the indicated histone modification on chromosome arms (outer two-thirds) versus the center (inner one-third). H3K27me3 and H3K4me3 mapping data are from the modencode project based on ChIP performed on mixed-population embryos, 20 C. (b) Metaplot and heat map showing log 2 enrichment of H3K9me2 and H3K9me3 (IP versus input) over gene bodies. Each row represents one gene, displaying the binned coding region plus 1 kb upstream of the transcription start site (TSS) and 1 kb downstream of the transcription termination sites (TES). Blue is most enriched, red is least enriched. (c) High-density scatterplot showing H3K9me2 and me3 enrichment over musclespecific genes and pseudogenes. The upper number indicates the percentage of genes that are H3K9me3 positive (including K9me2 positive and negative), and the lower number indicates the percentage of genes that are H3K9me2 positive but H3K9me3 negative (d,e) Distribution of H3K9me2 and H3K9me3 determined by ChIP-seq over the gene body of a gene with tissue-spec expression (d) neuronal unc-54, and a cluster of pseudogenes (e). 4
5 Supplementary Figure 4 Gene and repeat element derepression in the absence of H3K9me. (a,b) H3K9me2 and H3K9me3 ChIP-seq was performed on early embryos at 20 C (N = 2) in a wt strain, and gene expression was determined by two replicas of RNA-seq from embryos (20 C and 25 C) and from isolated gonads (20 C) from wt and met-2 set-25 strains. Scatterplots show H3K9me2 and H3K9me3 enrichment over the genes that are derepressed in early embryos at 20 C and 25 C, and in the gonads of met-2 set-25 animals, versus wt. Number indicates the percentage of derepressed genes enriched for either H3K9me2 or H3K9me3 in wt embryos. (b) Scatterplot of H3K9me2 and H3K9me3 enrichment over repeat subfamilies. Red dots mark repeat subfamilies that are derepressed in the indicated tissue and conditions; black dots represent non-affected repeat subfamilies. The red number indicates the percentage of derepressed repeat subfamilies that are either H3K9me2 or H3K9me3 positive. (c) qpcr verification of a subset of REs that were detected as derepressed (>2 fold, met-2 set-25/wt) in met-2 set-25 embryos at 20 C. Expression of the same REs was additionally analyzed in gonads and L1-stage larval RNA. REs of all three main classes are detected (N = 3), but clearly there are strong stage-specific expression differences, with larvae and embryos showing more similarity. 5
6 Supplementary Figure 5 R-loop accumulation on repeat elements (REs). (a) RNA:DNA hybrids were detected on dot plots of genomic DNA isolated from the indicated genotypes grown at 20 C or 25 C (three blots at each temperature were quantified by scanning for Fig. 5a). Equal amounts of DNA (determined by OD 260/280 ) extracted from adult worms were spotted with or without RNase H treatment in decreasing concentrations (4, 2 and 1 g). The nitrocellulose membrane was probed with the RNA:DNA-specific S9.6 antibody (n(20 C) = 3, n(25 C) = 3). Quantification on the right side with signals normalized to the background of each blot. (b) DRIP-seq signals in wt embryos are shown for genes grouped on the basis of their transcriptional activity. The upper box blot shows the level of transcription of the separate groups (N = 1). This enhancement of R loops on very highly expressed genes has been observed in many organisms. (c,d) Graphs show the accumulation of RNA:DNA hybrids in wt and met-2 set-25 embryos relative to the distribution of H3K9me2 and me3 over the rdna cluster (c) or the right telomere of chr. I (d), in wt embryos. (e) DRIP-seq examples showing the R-loop signal over two RE clusters. The ChIP signal from antibody S9.6, which is specific for RNA:DNA hybrids, was normalized to input, and the RNase H control values were subtracted. 6
7 Supplementary Figure 6 Germline mutations in met-2 set-25 worms. Further verification and characterization of the mutations detected in the genome sequencing experiment described in Figure 6. (a) Number and distribution of single-nucleotide variants (SNVs) or polymorphisms observed in the sequencing experiment described in Figure 6a. No dinucleotide preferences were found among SNVs. (b) Sketch of a complex rearrangement involving a Tc3 transposon, found exclusively in the met-2 set-25 genome. The rearrangement was identified by genome sequencing. The graph to the left indicates the precise site of Tc3 insertion. Below are H3K9me2 and H3K9me3 ChIP-seq tracks for the region around the Tc3 transposon that provoked the inversion: it bears high levels of H3K9me3 and showed roughly a twofold change in expression in met-2 set-25 over wt gonads at 20 C. (c) Southern blotting with a probe against Tc3 shows a novel band detected in the met-2 set-25 mutant. (d) Depiction of the rearrangement shown in b indicating the position of the primer pairs used to verify the rearranged genomic context by PCR. PCR confirmation of the rearrangement is shown to the right. (e) Copy number ratios of met-2 set-25 worms relative to wt for the entire telomeric repeat subfamily, and for single telomere repeats. (f) Copy number ratios of met-2 set-25 worms relative to wt for rdna 7
8 repeats. 8
9 Size [bp] Type RE H3K9me3 Fold change enrichment expression Insertion PALTTAA Transposition Tc Transposition Tc Transposition Tc Transposion Tc Deletion Deletion AT-rich Deletion Trinucleotide-rep Deletion/insertion HELITRON Supplementary Table 1 Germline mutations identified in 1 month old met-2 set-25 substrains. Characteristics of the insertions (ins), deletions (del) and transpositions (trans) unique to one of the eight met-2 set-25 sub-strains. H3K9me3 enrichment and fold change in expression is given as the mean enrichment and mean fold change (log2) over the complete domain.
10 Name in manuscript Genotype International name Reference Wild-type (wt) N2 (bristol) met-2 set-25 met-2(n4256) III; set-25(n5021) III GW mex-5:h2b::gfp unc+119;rrrsi192[mex-5 prom::gfp: h2b::tbb-2 3'UTR;unc119(+)] II gift of R. Ciosk, FMI 82 met-2 set-25 mex- 5:h2b::gfp met-2(n4256) III; set-25(n5021) III; unc+119; rrrsi192[mex-5 prom::gfp: h2b::tbb-2 3'UTR;unc119(+)] II GW1028 ced-1::gfp bcis39 [Plim-7::ced-1::gfp; lin-15(+)] V MD met-2 set-25 ced-1::gfp met-2(n4256) III; set-25(n5021) III; bcis39 [Plim-7::ced-1::gfp; lin-15(+)] V GW1203 cep-1 ced-1::gfp cep-1(ep347) I; bcis39[(lim-7)ced- 1p::GFP + lin-15(+)] V GW1266 met-2 set-25 cep-1 ced- 1::gfp cep-1(ep347) I; met-2(n4256) III;set- 25(n5021) III; bcis39[(lim-7)ced- 1p::GFP + lin-15(+)] V GW1268 mut-7 mut-7(pk204) III NL mut-14 mut-14(pk738) V NL prg-1 prg-1(tm872) WM Hc array (heterochromatic array) pkls1604 [hsp-16.2 ATG(A)17::gfp/lacZ + prf4(rol-6(su1006))] NL met-2 set-25 hc array met-2(n4256) III; set-25(n5021) III; pkls1604 [hsp-16.2 ATG(A)17::gfp/lacZ + prf4(rol-6(su1006))] GW1198 msh-6 hc array msh-6(pk2504) I; pkls1604 [hsp-16.2 ATG(A)17::gfp/lacZ + prf4(rol- 6(su1006))] GW1280 ec array (Euchromatic array) pkls1604 [hsp-16.2 ATG(A)17::gfp/lacZ + prf4(rol-6(su1006))] GW1351 met-2 set-25 ec array met-2(n4256) III; set-25(n5021) III; pkls1604 [hsp-16.2 ATG(A)17::gfp/lacZ + prf4(rol-6(su1006))] GW1278
11 msh-6 ec array msh-6(pk2504) I; pkls1604 [hsp-16.2 ATG(A)17::gfp/lacZ + prf4(rol- 6(su1006))] GW1279 thoc-2 thoc-2(tm1310) III/hT2[bli-4(e937) let-?(q782) qis48](i;iii) GIN Supplementary Table 2 Worm strains used in this study
12 Repeat qpcr: Forward Reverse Name Sequence Name Sequence DNA1 SG-7490 AGGCCCATTCCATAGCTTTT SG-7491 TTTCTGGGATTTCATGCACA DNA2 SG-7494 CCGTTATCGTGACAAGCAGA SG-7495 AAGCTTGAGCCGCTGATTAC DNA3 SG-7521 CACGTTCCCAGCTTCAATTT SG-7522 TGCAAATCTACAGTAACCCAGAA RNA1 SG-7498 TACCAACGAGCCGAGTCTTC SG-7499 TCTTCAGTTTCCTCGCCTGT RNA2 SG-7502 GAAGCAGTCATTGTGGCTCA SG-7503 GATTCTCCCTGTGCTCTTGC RNA3 SG-7509 TACCAACGAGCCGAGTCTTC SG-7510 TCTTCAGTTTCCTCGCCTGT RNA4 SG-7523 TGCATGCAAGACTAATTTTCAA SG-7524 CCAAAAGTTATTGGGCTTTCG RNA5 SG-7527 GGCACGATGTTTTTGTTGAA SG-7528 TCTATGAATTTCCCGCAGAA RNA6 SG-7533 GCCGCTAGACACCTAACGAG SG-7534 ATTATGGGGACGCAGAAAAA Tan1 SG-7537 GGATGGATTGGGATGGATG SG-7536 AACCATGCCAATCCTTTGTT Tan2 SG-7541 GTGTAGCCGTGGATTGTGTG SG-7542 ATTTGCCTGCTGGTCCATAG Tan3 SG-7543 GTTGGGGCGGCTGTAGTT SG-7544 AGCACCAAAGACGACAACAA Tan4 SG-7545 ATCGGGATGGCTCGGTAT SG-7546 ATCCCGATCCTTTGTTGTTG Tan5 SG-7548 CAGCCACAACTACCACAACG SG-7550 CAGTCTGTTGGACACGGAAC Tan6 SG-7551 ACCTACGTGCCTGCCTACAT SG-7552 TTTTGTCAGGGACATGCGTA Tan7 SG-7555 CCTGGCACTTACCAGTCCAT SG-7556 ATCGGGATGGATGGCTTC DRIP qpcr: Tc4 SG-7513 GAGTTTCCGTCCCGATTACA SG-7514 AGAAACGGTTCGAACAATGC CEMUDR-1 SG-7490 AGGCCCATTCCATAGCTTTT SG-7491 TTTCTGGGATTTCATGCACA Tc3 SG-6909 GAAGGATCCGGTGAGCTACG SG-6910 TACAGGAGTTGGAGGCAGCA CER10-I_CE SG-7498 TACCAACGAGCCGAGTCTTC SG-7499 TCTTCAGTTTCCTCGCCTGT Helitron 2 SG-8080 GATGTTTGGAGAGATAGTGG SG-8082 GACAACATCCATCCACTAACC G MSAT-1 SG-8076 GGAATGTTCCAGAACTTTCTA SG-8077 ACATTCCAGACTTTTCCCAA G (TATCG)n SG-8078 CGTATCGTTTCTAGCTATATCG SG-8079 CGATACGATATAGCTTTTTACG unc-119 SG-2869 CCACACCACCTCTAATCTCC SG-2870 TCATTTCTCTGCGTCTTCCT lmn-1 SG-2797 CAAGAGAACAACAGACTCCA SG-2798 TAATAAGACCACCGCATCAG G eef-1a.1 SG-8975 AGGAATGGTCGTTACCTTCGC SG-8976 CAGACGGATCCACGACGAATA Repeat copy number: HELITRON Y4 SG-8074 TGTTCTGCCAATTTATTTACTC SG-8075 GGCAGAACATTAGAGTAAATAT HELITRON 2 SG-8080 GATGTTTGGAGAGATAGTGGG SG-8082 GACAACATCCATCCACTAACC MSAT-1 SG-8076 GGAATGTTCCAGAACTTTCTAG SG-8077 ACATTCCAGACTTTTCCCAA (TATCG)n SG-8078 CGTATCGTTTCTAGCTATATCG SG-8079 CGATACGATATAGCTTTTTACG CEREP 58 SG-8071 CGGGCAAATCAACAATTGAA SG-8073 GAAGATGAGGAATTTAATTG lmn-1 SG-2797 CAAGAGAACAACAGACTCCAG SG-2798 TAATAAGACCACCGCATCAG Rearrangement:
13 PP1 SG-7680 CAAGGTGGAAGACATGTA SG-7681 TACGAGCAATTCAACTGG PP2 SG-7682 TTTATATCGGGTCCAACC SG-7683 GAAGCACGTCGACTGAAG PP3 SG-7684 AGGGGGGAAGAATTTAAC SG-7685 TCAAATTAGGGGGGGTCC PP4 SG-7685 TCAAATTAGGGGGGGTCC SG-7686 GTGTGGATACTGTCGTTC PP5 SG-7680 CAAGGTGGAAGACATGTA SG-7685 TCAAATTAGGGGGGGTCC PP6 SG-7682 TTTATATCGGGTCCAACC SG-7684 AGGGGGGAAGAATTTAAC Tc3-probe SG-7201 CTGTAAGACGGCAAGAGA SG-7202 TCTTGTTCTGAGCATACACG H3K9me2 and H3K9me3 ChIP on LacZ mutator arrays Tc4 SG-7513 GAGTTTCCGTCCCGATTACA SG-7514 AGAAACGGTTCGAACAATGC gfp (array) SG-6987 GGAGAGGGTGAAGGTGATGC SG-6988 CATAACCTTCGGGCATGGCA lmn-1 SG-2797 CAAGAGAACAACAGACTCCAG SG-2798 TAATAAGACCACCGCATCAG Cloning of array parts Part 1 SG-8150 CTACTTTTCCATGTACCG SG-8258 GTTTCTTTCATCTCACTGGATCCCC Part 2 SG-8151 TCATCTCACTGGATCCCC SG-8259 GTTTCTTGTGTCCAAGAATGTTTCC Part 3 SG-6987 GGAGAGGGTGAAGGTGATGC SG-2039 GCAGCTGTTACAAACTCAAG Supplementary Table 3 Primers used in this study
14 supplement-only references 83. Tocchini, C. et al. The TRIM-NHL protein LIN-41 controls the onset of developmental plasticity in Caenorhabditis elegans. PLoS Genet 10, e (2014). 84. Lu, N., Yu, X., He, X. & Zhou, Z. Detecting apoptotic cells and monitoring their clearance in the nematode Caenorhabditis elegans. Methods Mol Biol 559, (2009). 85. Castellano-Pozo, M., Garcia-Muse, T. & Aguilera, A. R-loops cause replication impairment and genome instability during meiosis. EMBO Rep 13, (2012).
Supplemental Figure 1. Genes showing ectopic H3K9 dimethylation in this study are DNA hypermethylated in Lister et al. study.
 mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Nature Genetics: doi: /ng Supplementary Figure 1. Assessment of sample purity and quality.
 Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/8/e1701143/dc1 Supplementary Materials for Impaired DNA replication derepresses chromatin and generates a transgenerationally inherited epigenetic memory Adam
advances.sciencemag.org/cgi/content/full/3/8/e1701143/dc1 Supplementary Materials for Impaired DNA replication derepresses chromatin and generates a transgenerationally inherited epigenetic memory Adam
High Throughput Sequence (HTS) data analysis. Lei Zhou
 High Throughput Sequence (HTS) data analysis Lei Zhou (leizhou@ufl.edu) High Throughput Sequence (HTS) data analysis 1. Representation of HTS data. 2. Visualization of HTS data. 3. Discovering genomic
High Throughput Sequence (HTS) data analysis Lei Zhou (leizhou@ufl.edu) High Throughput Sequence (HTS) data analysis 1. Representation of HTS data. 2. Visualization of HTS data. 3. Discovering genomic
Patterns of Histone Methylation and Chromatin Organization in Grapevine Leaf. Rachel Schwope EPIGEN May 24-27, 2016
 Patterns of Histone Methylation and Chromatin Organization in Grapevine Leaf Rachel Schwope EPIGEN May 24-27, 2016 What does H3K4 methylation do? Plant of interest: Vitis vinifera Culturally important
Patterns of Histone Methylation and Chromatin Organization in Grapevine Leaf Rachel Schwope EPIGEN May 24-27, 2016 What does H3K4 methylation do? Plant of interest: Vitis vinifera Culturally important
An epigenetic approach to understanding (and predicting?) environmental effects on gene expression
 www.collaslab.com An epigenetic approach to understanding (and predicting?) environmental effects on gene expression Philippe Collas University of Oslo Institute of Basic Medical Sciences Stem Cell Epigenetics
www.collaslab.com An epigenetic approach to understanding (and predicting?) environmental effects on gene expression Philippe Collas University of Oslo Institute of Basic Medical Sciences Stem Cell Epigenetics
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
Supplementary Table 1. List of primers used in this study
 Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Not IN Our Genes - A Different Kind of Inheritance.! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014
 Not IN Our Genes - A Different Kind of Inheritance! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014 Epigenetics in Mainstream Media Epigenetics *Current definition:
Not IN Our Genes - A Different Kind of Inheritance! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014 Epigenetics in Mainstream Media Epigenetics *Current definition:
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
BIO360 Fall 2013 Quiz 1
 BIO360 Fall 2013 Quiz 1 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function mutation.
BIO360 Fall 2013 Quiz 1 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function mutation.
klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach
![klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach](/thumbs/91/104639484.jpg) DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
Structural Variation and Medical Genomics
 Structural Variation and Medical Genomics Andrew King Department of Biomedical Informatics July 8, 2014 You already know about small scale genetic mutations Single nucleotide polymorphism (SNPs) Deletions,
Structural Variation and Medical Genomics Andrew King Department of Biomedical Informatics July 8, 2014 You already know about small scale genetic mutations Single nucleotide polymorphism (SNPs) Deletions,
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
MIR retrotransposon sequences provide insulators to the human genome
 Supplementary Information: MIR retrotransposon sequences provide insulators to the human genome Jianrong Wang, Cristina Vicente-García, Davide Seruggia, Eduardo Moltó, Ana Fernandez- Miñán, Ana Neto, Elbert
Supplementary Information: MIR retrotransposon sequences provide insulators to the human genome Jianrong Wang, Cristina Vicente-García, Davide Seruggia, Eduardo Moltó, Ana Fernandez- Miñán, Ana Neto, Elbert
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq
 Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
p.r623c p.p976l p.d2847fs p.t2671 p.d2847fs p.r2922w p.r2370h p.c1201y p.a868v p.s952* RING_C BP PHD Cbp HAT_KAT11
 ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
Nature Immunology: doi: /ni Supplementary Figure 1. Characteristics of SEs in T reg and T conv cells.
 Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
C. elegans Embryonic Development
 Autonomous Specification in Tunicate development Autonomous & Conditional Specification in C. elegans Embryonic Development Figure 8.36 Bilateral Symmetry in the Egg of the Tunicate Styela partita Fig.
Autonomous Specification in Tunicate development Autonomous & Conditional Specification in C. elegans Embryonic Development Figure 8.36 Bilateral Symmetry in the Egg of the Tunicate Styela partita Fig.
Supplemental Information. Figures. Figure S1
 Supplemental Information Figures Figure S1 Identification of JAGGER T-DNA insertions. A. Positions of T-DNA and Ds insertions in JAGGER are indicated by inverted triangles, the grey box represents the
Supplemental Information Figures Figure S1 Identification of JAGGER T-DNA insertions. A. Positions of T-DNA and Ds insertions in JAGGER are indicated by inverted triangles, the grey box represents the
Breast cancer. Risk factors you cannot change include: Treatment Plan Selection. Inferring Transcriptional Module from Breast Cancer Profile Data
 Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
An Unexpected Function of the Prader-Willi Syndrome Imprinting Center in Maternal Imprinting in Mice
 An Unexpected Function of the Prader-Willi Syndrome Imprinting Center in Maternal Imprinting in Mice Mei-Yi Wu 1 *, Ming Jiang 1, Xiaodong Zhai 2, Arthur L. Beaudet 2, Ray-Chang Wu 1 * 1 Department of
An Unexpected Function of the Prader-Willi Syndrome Imprinting Center in Maternal Imprinting in Mice Mei-Yi Wu 1 *, Ming Jiang 1, Xiaodong Zhai 2, Arthur L. Beaudet 2, Ray-Chang Wu 1 * 1 Department of
Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy
 Kavya Puchhalapalli CALS Honors Project Report Spring 2017 Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy Abstract Malignant brain tumors including medulloblastomas and primitive neuroectodermal
Kavya Puchhalapalli CALS Honors Project Report Spring 2017 Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy Abstract Malignant brain tumors including medulloblastomas and primitive neuroectodermal
Nature Genetics: doi: /ng Supplementary Figure 1. HOX fusions enhance self-renewal capacity.
 Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University
 Role of Chemical lexposure in Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University CNV Discovery Reference Genetic
Role of Chemical lexposure in Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University CNV Discovery Reference Genetic
Programmed Cell Death (apoptosis)
 Programmed Cell Death (apoptosis) Stereotypic death process includes: membrane blebbing nuclear fragmentation chromatin condensation and DNA framentation loss of mitochondrial integrity and release of
Programmed Cell Death (apoptosis) Stereotypic death process includes: membrane blebbing nuclear fragmentation chromatin condensation and DNA framentation loss of mitochondrial integrity and release of
pirna pathway targets active LINE1 elements to establish the repressive H3K9me3markingermcells
 pirna pathway targets active LINE1 elements to establish the repressive H3K9me3markingermcells Dubravka Pezic, 1,3 Sergei A. Manakov, 1,3 Ravi Sachidanandam, 2 and Alexei A. Aravin 1,4 1 Division of Biology
pirna pathway targets active LINE1 elements to establish the repressive H3K9me3markingermcells Dubravka Pezic, 1,3 Sergei A. Manakov, 1,3 Ravi Sachidanandam, 2 and Alexei A. Aravin 1,4 1 Division of Biology
Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63.
 Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Nature Structural & Molecular Biology: doi: /nsmb.2419
 Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Variant Classification. Author: Mike Thiesen, Golden Helix, Inc.
 Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Supplementary Materials and Methods
 Supplementary Materials and Methods Whole Mount X-Gal Staining Whole tissues were collected, rinsed with PBS and fixed with 4% PFA. Tissues were then rinsed in rinse buffer (100 mm Sodium Phosphate ph
Supplementary Materials and Methods Whole Mount X-Gal Staining Whole tissues were collected, rinsed with PBS and fixed with 4% PFA. Tissues were then rinsed in rinse buffer (100 mm Sodium Phosphate ph
Abstract. Optimization strategy of Copy Number Variant calling using Multiplicom solutions APPLICATION NOTE. Introduction
 Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
CANCER. Inherited Cancer Syndromes. Affects 25% of US population. Kills 19% of US population (2nd largest killer after heart disease)
 CANCER Affects 25% of US population Kills 19% of US population (2nd largest killer after heart disease) NOT one disease but 200-300 different defects Etiologic Factors In Cancer: Relative contributions
CANCER Affects 25% of US population Kills 19% of US population (2nd largest killer after heart disease) NOT one disease but 200-300 different defects Etiologic Factors In Cancer: Relative contributions
Genetics and Genomics in Medicine Chapter 6 Questions
 Genetics and Genomics in Medicine Chapter 6 Questions Multiple Choice Questions Question 6.1 With respect to the interconversion between open and condensed chromatin shown below: Which of the directions
Genetics and Genomics in Medicine Chapter 6 Questions Multiple Choice Questions Question 6.1 With respect to the interconversion between open and condensed chromatin shown below: Which of the directions
Zhu et al, page 1. Supplementary Figures
 Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
ddm1a (PFG_3A-51065) ATG ddm1b (PFG_2B-60109) ATG osdrm2 (PFG_3A-04110) osdrm2 osdrm2 osdrm2
 Relative expression.6.5.4.3.2.1 OsDDM1a OsDDM1b OsDRM2 TG TG TG ddm1a (PFG_3-5165) P1 P3 ddm1b (PFG_2-619) P2 531bp 5233bp P1 P4 P3 P2 F1 R1 F1 TG TG TG F1 R1 C ddm1a -/- -/- ddm1b -/- +/- +/- -/- D (PFG_3-411)
Relative expression.6.5.4.3.2.1 OsDDM1a OsDDM1b OsDRM2 TG TG TG ddm1a (PFG_3-5165) P1 P3 ddm1b (PFG_2-619) P2 531bp 5233bp P1 P4 P3 P2 F1 R1 F1 TG TG TG F1 R1 C ddm1a -/- -/- ddm1b -/- +/- +/- -/- D (PFG_3-411)
Introduction to Cancer Biology
 Introduction to Cancer Biology Robin Hesketh Multiple choice questions (choose the one correct answer from the five choices) Which ONE of the following is a tumour suppressor? a. AKT b. APC c. BCL2 d.
Introduction to Cancer Biology Robin Hesketh Multiple choice questions (choose the one correct answer from the five choices) Which ONE of the following is a tumour suppressor? a. AKT b. APC c. BCL2 d.
SUPPLEMENTARY FIGURE S1: nlp-22 is expressed in the RIA interneurons and is secreted. (a) An animal expressing both the RIA specific reporter
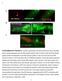 1 SUPPLEMENTARY FIGURE S1: nlp-22 is expressed in the RIA interneurons and is secreted. (a) An animal expressing both the RIA specific reporter Pglr-3:mCherry (red) and Pnlp-22:gfp (green) shows co-localization
1 SUPPLEMENTARY FIGURE S1: nlp-22 is expressed in the RIA interneurons and is secreted. (a) An animal expressing both the RIA specific reporter Pglr-3:mCherry (red) and Pnlp-22:gfp (green) shows co-localization
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols.
 Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Expression deviation of the genes mapped to gene-wise recurrent mutations in the TCGA breast cancer cohort (top) and the TCGA lung cancer cohort (bottom). For each gene (each pair
Supplementary Figure 1 Expression deviation of the genes mapped to gene-wise recurrent mutations in the TCGA breast cancer cohort (top) and the TCGA lung cancer cohort (bottom). For each gene (each pair
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells
 Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Alpha thalassemia mental retardation X-linked. Acquired alpha-thalassemia myelodysplastic syndrome
 Alpha thalassemia mental retardation X-linked Acquired alpha-thalassemia myelodysplastic syndrome (Alpha thalassemia mental retardation X-linked) Acquired alpha-thalassemia myelodysplastic syndrome Schematic
Alpha thalassemia mental retardation X-linked Acquired alpha-thalassemia myelodysplastic syndrome (Alpha thalassemia mental retardation X-linked) Acquired alpha-thalassemia myelodysplastic syndrome Schematic
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Monitoring for Drug Resistance by Genotyping. Urvi M Parikh, PhD MTN Virology Core Lab
 Monitoring for Drug Resistance by Genotyping Urvi M Parikh, PhD MTN Virology Core Lab Outline What is Drug Resistance? Genotyping Algorithm Standard vs Sensitive Resistance Testing Sequencing Protocols
Monitoring for Drug Resistance by Genotyping Urvi M Parikh, PhD MTN Virology Core Lab Outline What is Drug Resistance? Genotyping Algorithm Standard vs Sensitive Resistance Testing Sequencing Protocols
DNA-seq Bioinformatics Analysis: Copy Number Variation
 DNA-seq Bioinformatics Analysis: Copy Number Variation Elodie Girard elodie.girard@curie.fr U900 institut Curie, INSERM, Mines ParisTech, PSL Research University Paris, France NGS Applications 5C HiC DNA-seq
DNA-seq Bioinformatics Analysis: Copy Number Variation Elodie Girard elodie.girard@curie.fr U900 institut Curie, INSERM, Mines ParisTech, PSL Research University Paris, France NGS Applications 5C HiC DNA-seq
he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003)
 MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
Genomic structural variation
 Genomic structural variation Mario Cáceres The new genomic variation DNA sequence differs across individuals much more than researchers had suspected through structural changes A huge amount of structural
Genomic structural variation Mario Cáceres The new genomic variation DNA sequence differs across individuals much more than researchers had suspected through structural changes A huge amount of structural
Nature Biotechnology: doi: /nbt.1904
 Supplementary Information Comparison between assembly-based SV calls and array CGH results Genome-wide array assessment of copy number changes, such as array comparative genomic hybridization (acgh), is
Supplementary Information Comparison between assembly-based SV calls and array CGH results Genome-wide array assessment of copy number changes, such as array comparative genomic hybridization (acgh), is
Supplementary Figure 1. Spitzoid Melanoma with PPFIBP1-MET fusion. (a) Histopathology (4x) shows a domed papule with melanocytes extending into the
 Supplementary Figure 1. Spitzoid Melanoma with PPFIBP1-MET fusion. (a) Histopathology (4x) shows a domed papule with melanocytes extending into the deep dermis. (b) The melanocytes demonstrate abundant
Supplementary Figure 1. Spitzoid Melanoma with PPFIBP1-MET fusion. (a) Histopathology (4x) shows a domed papule with melanocytes extending into the deep dermis. (b) The melanocytes demonstrate abundant
Supplementary Information
 Supplementary Information The fate of W chromosomes in hybrids between wild silkmoths, Samia cynthia ssp.: no role in sex determination and reproduction Atsuo Yoshido, Frantisek Marec, Ken Sahara Supplementary
Supplementary Information The fate of W chromosomes in hybrids between wild silkmoths, Samia cynthia ssp.: no role in sex determination and reproduction Atsuo Yoshido, Frantisek Marec, Ken Sahara Supplementary
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
BIO360 Fall 2013 Quiz 1
 BIO360 Fall 2013 Quiz 1 Name: Key 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function
BIO360 Fall 2013 Quiz 1 Name: Key 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function
Nature Genetics: doi: /ng Supplementary Figure 1. Clinical timeline for the discovery WES cases.
 Supplementary Figure 1 Clinical timeline for the discovery WES cases. This illustrates the timeline of the disease events during the clinical course of each patient s disease, further indicating the available
Supplementary Figure 1 Clinical timeline for the discovery WES cases. This illustrates the timeline of the disease events during the clinical course of each patient s disease, further indicating the available
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first
 Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease
 1 Supplemental Materials 2 3 Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease 4 5 6 Authors: Madhumita Basu, 1 Jun-Yi Zhu, 2 Stephanie LaHaye 1,3, Uddalak
1 Supplemental Materials 2 3 Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease 4 5 6 Authors: Madhumita Basu, 1 Jun-Yi Zhu, 2 Stephanie LaHaye 1,3, Uddalak
Results. Abstract. Introduc4on. Conclusions. Methods. Funding
 . expression that plays a role in many cellular processes affecting a variety of traits. In this study DNA methylation was assessed in neuronal tissue from three pigs (frontal lobe) and one great tit (whole
. expression that plays a role in many cellular processes affecting a variety of traits. In this study DNA methylation was assessed in neuronal tissue from three pigs (frontal lobe) and one great tit (whole
Polyomaviridae. Spring
 Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
SUPPLEMENTARY INFORMATION
 doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
Global variation in copy number in the human genome
 Global variation in copy number in the human genome Redon et. al. Nature 444:444-454 (2006) 12.03.2007 Tarmo Puurand Study 270 individuals (HapMap collection) Affymetrix 500K Whole Genome TilePath (WGTP)
Global variation in copy number in the human genome Redon et. al. Nature 444:444-454 (2006) 12.03.2007 Tarmo Puurand Study 270 individuals (HapMap collection) Affymetrix 500K Whole Genome TilePath (WGTP)
Nature Medicine: doi: /nm.4322
 1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
pplementary Figur Supplementary Figure 1. a.
 pplementary Figur Supplementary Figure 1. a. Quantification by RT-qPCR of YFV-17D and YFV-17D pol- (+) RNA in the supernatant of cultured Huh7.5 cells following viral RNA electroporation of respective
pplementary Figur Supplementary Figure 1. a. Quantification by RT-qPCR of YFV-17D and YFV-17D pol- (+) RNA in the supernatant of cultured Huh7.5 cells following viral RNA electroporation of respective
Table S1. Relative abundance of AGO1/4 proteins in different organs. Table S2. Summary of smrna datasets from various samples.
 Supplementary files Table S1. Relative abundance of AGO1/4 proteins in different organs. Table S2. Summary of smrna datasets from various samples. Table S3. Specificity of AGO1- and AGO4-preferred 24-nt
Supplementary files Table S1. Relative abundance of AGO1/4 proteins in different organs. Table S2. Summary of smrna datasets from various samples. Table S3. Specificity of AGO1- and AGO4-preferred 24-nt
Table S1. Total and mapped reads produced for each ChIP-seq sample
 Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
CTCF-Mediated Functional Chromatin Interactome in Pluripotent Cells
 SUPPLEMENTARY INFORMATION CTCF-Mediated Functional Chromatin Interactome in Pluripotent Cells Lusy Handoko 1,*, Han Xu 1,*, Guoliang Li 1,*, Chew Yee Ngan 1, Elaine Chew 1, Marie Schnapp 1, Charlie Wah
SUPPLEMENTARY INFORMATION CTCF-Mediated Functional Chromatin Interactome in Pluripotent Cells Lusy Handoko 1,*, Han Xu 1,*, Guoliang Li 1,*, Chew Yee Ngan 1, Elaine Chew 1, Marie Schnapp 1, Charlie Wah
CHAPTER 17 CHROMOSOME REARRANGEMENTS
 CHROMOSOME REARRANGEMENTS CHAPTER 17 Figure 1. Comparing an ideogram of the human chromosome 2 to the equivalent chromosomes in chimpanzees, we notice that the human chromosome 2 likely came from a fusion
CHROMOSOME REARRANGEMENTS CHAPTER 17 Figure 1. Comparing an ideogram of the human chromosome 2 to the equivalent chromosomes in chimpanzees, we notice that the human chromosome 2 likely came from a fusion
Nature Genetics: doi: /ng Supplementary Figure 1. SEER data for male and female cancer incidence from
 Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Nature Immunology: doi: /ni Supplementary Figure 1. DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells.
 Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
Raymond Auerbach PhD Candidate, Yale University Gerstein and Snyder Labs August 30, 2012
 Elucidating Transcriptional Regulation at Multiple Scales Using High-Throughput Sequencing, Data Integration, and Computational Methods Raymond Auerbach PhD Candidate, Yale University Gerstein and Snyder
Elucidating Transcriptional Regulation at Multiple Scales Using High-Throughput Sequencing, Data Integration, and Computational Methods Raymond Auerbach PhD Candidate, Yale University Gerstein and Snyder
Accessing and Using ENCODE Data Dr. Peggy J. Farnham
 1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1: Quality assessment of formalin-fixed paraffin-embedded (FFPE)-derived DNA and nuclei. (a) Multiplex PCR analysis of unrepaired and repaired bulk FFPE gdna from
Supplementary Figure 1 Supplementary Fig. 1: Quality assessment of formalin-fixed paraffin-embedded (FFPE)-derived DNA and nuclei. (a) Multiplex PCR analysis of unrepaired and repaired bulk FFPE gdna from
Supplementary Fig. S1. Schematic diagram of minigenome segments.
 open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
Course Title Form Hours subject
 Course Title Form Hours subject Types, and structure of chromosomes L 1 Histology Karyotyping and staining of human chromosomes L 2 Histology Chromosomal anomalies L 2 Histology Sex chromosomes L 1 Histology
Course Title Form Hours subject Types, and structure of chromosomes L 1 Histology Karyotyping and staining of human chromosomes L 2 Histology Chromosomal anomalies L 2 Histology Sex chromosomes L 1 Histology
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
Genetic suppressors and enhancers provide clues to gene regulation and genetic pathways
 Genetic suppressors and enhancers provide clues to gene regulation and genetic pathways Suppressor mutation: a second mutation results in a less severe phenotype than the original mutation Suppressor mutations
Genetic suppressors and enhancers provide clues to gene regulation and genetic pathways Suppressor mutation: a second mutation results in a less severe phenotype than the original mutation Suppressor mutations
Cytogenetics 101: Clinical Research and Molecular Genetic Technologies
 Cytogenetics 101: Clinical Research and Molecular Genetic Technologies Topics for Today s Presentation 1 Classical vs Molecular Cytogenetics 2 What acgh? 3 What is FISH? 4 What is NGS? 5 How can these
Cytogenetics 101: Clinical Research and Molecular Genetic Technologies Topics for Today s Presentation 1 Classical vs Molecular Cytogenetics 2 What acgh? 3 What is FISH? 4 What is NGS? 5 How can these
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical
 Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
BIOL2005 WORKSHEET 2008
 BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
Variations in Chromosome Structure & Function. Ch. 8
 Variations in Chromosome Structure & Function Ch. 8 1 INTRODUCTION! Genetic variation refers to differences between members of the same species or those of different species Allelic variations are due
Variations in Chromosome Structure & Function Ch. 8 1 INTRODUCTION! Genetic variation refers to differences between members of the same species or those of different species Allelic variations are due
Nature Immunology: doi: /ni Supplementary Figure 1 33,312. Aire rep 1. Aire rep 2 # 44,325 # 44,055. Aire rep 1. Aire rep 2.
 a 33,312 b rep 1 rep 1 # 44,325 rep 2 # 44,055 [0-84] rep 2 [0-84] 1810043G02Rik Pfkl Dnmt3l Icosl rep 1 [0-165] rep 2 [0-165] Rps14 Cd74 Mir5107 Tcof1 rep 1 [0-69] rep 2 [0-68] Id3 E2f2 Asap3 rep 1 [0-141]
a 33,312 b rep 1 rep 1 # 44,325 rep 2 # 44,055 [0-84] rep 2 [0-84] 1810043G02Rik Pfkl Dnmt3l Icosl rep 1 [0-165] rep 2 [0-165] Rps14 Cd74 Mir5107 Tcof1 rep 1 [0-69] rep 2 [0-68] Id3 E2f2 Asap3 rep 1 [0-141]
fl/+ KRas;Atg5 fl/+ KRas;Atg5 fl/fl KRas;Atg5 fl/fl KRas;Atg5 Supplementary Figure 1. Gene set enrichment analyses. (a) (b)
 KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Information. Supplementary Figures
 Supplementary Information Supplementary Figures.8 57 essential gene density 2 1.5 LTR insert frequency diversity DEL.5 DUP.5 INV.5 TRA 1 2 3 4 5 1 2 3 4 1 2 Supplementary Figure 1. Locations and minor
Supplementary Information Supplementary Figures.8 57 essential gene density 2 1.5 LTR insert frequency diversity DEL.5 DUP.5 INV.5 TRA 1 2 3 4 5 1 2 3 4 1 2 Supplementary Figure 1. Locations and minor
DAPI ASY1 DAPI/ASY1 DAPI RAD51 DAPI/RAD51. Supplementary Figure 1. Additional information on meiosis in R. pubera. a) The
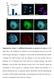 a % 10 Number of crossover per bivalent b 0 1 c DAPI/telomere 80 1 60 40 1 2 20 d 0 0 1 2 >=3 DAPI ASY1 DAPI/ASY1 e DAPI RAD51 DAPI/RAD51 Supplementary Figure 1. Additional information on meiosis in R.
a % 10 Number of crossover per bivalent b 0 1 c DAPI/telomere 80 1 60 40 1 2 20 d 0 0 1 2 >=3 DAPI ASY1 DAPI/ASY1 e DAPI RAD51 DAPI/RAD51 Supplementary Figure 1. Additional information on meiosis in R.
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
Peak-calling for ChIP-seq and ATAC-seq
 Peak-calling for ChIP-seq and ATAC-seq Shamith Samarajiwa CRUK Autumn School in Bioinformatics 2017 University of Cambridge Overview Peak-calling: identify enriched (signal) regions in ChIP-seq or ATAC-seq
Peak-calling for ChIP-seq and ATAC-seq Shamith Samarajiwa CRUK Autumn School in Bioinformatics 2017 University of Cambridge Overview Peak-calling: identify enriched (signal) regions in ChIP-seq or ATAC-seq
Challenges of CGH array testing in children with developmental delay. Dr Sally Davies 17 th September 2014
 Challenges of CGH array testing in children with developmental delay Dr Sally Davies 17 th September 2014 CGH array What is CGH array? Understanding the test Benefits Results to expect Consent issues Ethical
Challenges of CGH array testing in children with developmental delay Dr Sally Davies 17 th September 2014 CGH array What is CGH array? Understanding the test Benefits Results to expect Consent issues Ethical
Supplementary figures
 Supplementary figures Supplementary Figure 1. B cells stimulated with pokeweed mitogen display normal mitotic figures but not cells infected with B95-8. The figures show cells stimulated with pokeweed
Supplementary figures Supplementary Figure 1. B cells stimulated with pokeweed mitogen display normal mitotic figures but not cells infected with B95-8. The figures show cells stimulated with pokeweed
Supplemental Information For: The genetics of splicing in neuroblastoma
 Supplemental Information For: The genetics of splicing in neuroblastoma Justin Chen, Christopher S. Hackett, Shile Zhang, Young K. Song, Robert J.A. Bell, Annette M. Molinaro, David A. Quigley, Allan Balmain,
Supplemental Information For: The genetics of splicing in neuroblastoma Justin Chen, Christopher S. Hackett, Shile Zhang, Young K. Song, Robert J.A. Bell, Annette M. Molinaro, David A. Quigley, Allan Balmain,
