Jianqiang Shen Kabin Xie Lizhong Xiong
|
|
|
- Phillip Dennis
- 5 years ago
- Views:
Transcription
1 DOI /s ORIGINAL PAPER Global expression profiling of rice micrornas by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of micrornas in abiotic stress responses Jianqiang Shen Kabin Xie Lizhong Xiong Received: 14 May 2010 / Accepted: 26 September 2010 Ó Springer-Verlag 2010 Abstract MicroRNAs are a class of endogenous small RNA molecules (20 24 nucleotides) that have pivotal roles in regulating gene expression mostly at posttranscriptional levels in plants. Plant micrornas have been implicated in the regulation of diverse biological processes including growth and stress responses. However, the information about micrornas in regulating abiotic stress responses in rice is limited. We optimized a one-tube stem-loop reverse transcription quantitative PCR (ST-RT qpcr) for highthroughput expression profiling analysis of micrornas in rice under normal and stress conditions. The optimized ST- RT qpcr method was as accurate as small RNA gel blotting and was more convenient and time-saving than other methods in quantifying micrornas. With this method, 41 rice micrornas were quantified for their relative expression levels after drought, salt, cold, and abscisic acid (ABA) treatments. Thirty-two micrornas showed induced or suppressed expression after stress or Communicated by A. Tyagi. J. Shen and K. Xie contributed equally to this work. Electronic supplementary material The online version of this article (doi: /s ) contains supplementary material, which is available to authorized users. J. Shen K. Xie L. Xiong (&) National Key Laboratory of Crop Genetic Improvement and National Center of Plant Gene Research (Wuhan), Huazhong Agricultural University, Wuhan , China lizhongx@mail.hzau.edu.cn Present Address: K. Xie Department of Plant Pathology, State College, Pennsylvania State University, University Park, PA 16803, USA ABA treatment. Further analysis suggested that stressresponsive cis-elements were enriched in the promoters of stress-responsive microrna genes. The expressions of five and seven micrornas were significantly affected in the rice plant with defects in stress tolerance regulatory genes OsSKIPa and OsbZIP23, respectively. Some of the predicted target genes of these micrornas were also related to abiotic stresses. We conclude that ST-RT qpcr is an efficient and reliable method for expression profiling of micrornas and a significant portion of rice micrornas participate in abiotic stress response and regulation. Keywords PCR Introduction Oryza sativa mirna Stress Real-time MicroRNA (mirna) is a class of short (*22 nucleotides), endogenously expressed non-coding RNAs found in animals and plants. Plant mirnas are processed from primary transcripts by DICER-like proteins and mature mirnas are loaded into ARGONAUTE proteins to guide cleavage of target mrnas or translational repression (Baumberger and Baulcombe 2005; Liu et al. 2005; Henderson et al. 2006; Voinnet 2009). A number of mirnas have been identified by deep sequencing of small RNA fractions in rice (Sunkar et al. 2005, 2008; Johnson et al. 2007). The functions of plant mirnas are highly diverse and have essential roles in regulating plant growth, organogenesis, pattern formation, organ polarity, and hormone homeostasis (Voinnet 2009). Plants have evolved elaborate mechanisms to cope with adverse environmental changes including high or low temperature, high salinity, nutrition deficiency, and water deficit or drought stress. Plants can sense the signals
2 resulting from environmental changes and trigger activation or suppression of arrays of different genes to modulate hormone levels, metabolism, and developmental processes related to stress response and adaptation. Many transcription factors and their cognate cis-elements play essential roles during stress responses (Yamaguchi-Shinozaki and Shinozaki 2006). Evidence suggests that mirnas are also involved in abiotic stress responses (Jones-Rhoades and Bartel 2004; Sunkar and Zhu 2004; Lu et al. 2005; Li et al. 2008; Liu et al. 2008). A few Arabidopsis mirnas are induced by abiotic stresses (Jones-Rhoades and Bartel 2004; Sunkar and Zhu 2004; Liu et al. 2008). At-miR169 was recently characterized for its regulation role in drought tolerance (Li et al. 2008). However, little is known about mirnas during abiotic stress responses in economically important crops such as rice. High-throughput gene expression profiling is a powerful tool to identify stress-responsive genes. Unlike cdna microarray or DNA chip that have been routinely used for profiling of high-molecular-weight RNAs (especially mrnas), special techniques are required to quantify mirnas because of their small size and lack of common adaptor sequence as poly(a) in mrnas. Different methods have been developed to measure mirna levels (Chen et al. 2005; Castoldi et al. 2006; Hunt et al. 2009). The gold standard for mirna quantification is small RNA gel blotting, which is, however, not suitable for highthroughput assay. Microarray analysis is a powerful method for gene quantification and has recently been used extensively in mirna profiling (Barad et al. 2004; Liu et al. 2004, 2008; Castoldi et al. 2006). In plants, the mircury LNA array was used for mirna expression profiling (Castoldi et al. 2006; Liu et al. 2008). However, the sensitivity and reproducibility of the LNA array hybridization in plants have been questioned recently (Chambers and Shuai 2009). Real-time polymerase chain reaction (PCR) has been used as a robust, sensitive, and straightforward method for quantification of gene expression. By applying special techniques for cdna synthesis, real-time quantitative PCR can also be used to quantify mirna (Chen et al. 2005; Shi and Chiang 2005). The simplest method is stem-loop reverse transcription realtime quantitative PCR (ST-RT qpcr), in which mirna is converted to cdna using stem-loop primers with six nucleotides complementary to the 3 0 -end of mirna to guide the reverse transcription (Chen et al. 2005). Compared to other methods, ST-RT qpcr is one of the most convenient approaches for mirna quantification. Recently, the importance of following the correct experimental protocol in RT real-time qpcr to measure the mrna level was highlighted and included reference gene selection, experiment design, and careful statistical analysis (Udvardi et al. 2008). Similar principles were also recommended for mirna assays in animals using ST-RT qpcr (Peltier and Latham 2008; Mestdagh et al. 2009). Here, we optimized the ST-RT qpcr method and established a robust, straightforward mirna relative quantification approach in rice. The optimized ST-RT qpcr was as accurate as small RNA gel blotting but more sensitive and convenient for mirna quantification. By using this optimized ST-RT qpcr method, we analyzed the expression levels of rice mirnas under various stress conditions (drought, cold, and salt) and abscisic acid (ABA) treatment. Further analysis of the stress-responsive mirnas presented an overall view of mirna in abiotic stress responses in rice. Experimental procedures Plant materials and stress treatments Seeds of Oryza sativa L. ssp. japonica var. Zhonghua11 were sterilized and grown under controlled conditions at 28 C day/25 C night with a 12 h light/12 h dark photoperiod. After 15 days of germination, seedlings were exposed to drought or high-salinity stress (200 mm NaCl) or treated with 100 lm ABA. Leaf tissues were harvested at 0, 20, 40, and 180 min after stress treatment. For cold stress, the 15-day-old seedlings were exposed to 4 C, and the leaf tissues were harvested at 0, 40, 90, and 300 min. The samples were frozen in liquid nitrogen and stored at -80 C for further analysis. Total RNA was extracted from leaf tissues with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer s instruction. The RNA quality was checked by agarose gel electrophoresis. Primer design for mirna The rice mirna sequences were downloaded from the Sanger Institute mirbase Sequence Database (release 12.0) (Griffiths-Jones et al. 2008). The stem-loop RT primers (Supplemental Table 2) were designed according to Chen et al. (2005) with minor modification. Degenerated nucleotides were introduced to amplify mirna family members with single nucleotide difference at the 3 0 end. All DNA oligos were synthesized in Sangon (Shanghai, China). One-tube ST-RT qpcr The modified one-tube ST-RT qpcr was performed as follows. The stem-loop reverse transcription reaction was performed in 96-well plates in a volume of 5 ll containing 20 ng DNase I-treated RNA, 0.25 ll appropriate stemloop primers (10 lm), 30 U SuperScript III Reverse
3 Transcriptase (Invitrogen), and 6 U RNase inhibitor-hpri (Takara, Dalian, China). The cdna synthesis was conducted in an ABI PRISM 9700 thermal cycler using a pulse reverse transcription program (Varkonyi-Gasic et al. 2007). Real-time PCR analysis was carried out using SYBR Premix Ex Taq (Takara) on an ABI PRISM 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Briefly, 12.5 ll SYBR mixture, 1 ll universal reverse primer (Supplemental Table 2) and 1 ll mirna-specific forward primer were added to the 96-well plates. U6 control reactions were always included in each 96-well plate for each sample using U6-specific primers. All reactions were performed in triple replications. The amplification specificity was monitored by melting curve after PCR. Reverse transcription real-time qpcr The expression levels of OsSKIPa, OsbZIP23, and SNAC1 were quantified by real-time qpcr. Briefly, 1 lg total RNA was treated with 1 U DNase I (Invitrogen). Firststrand cdna were synthesized from 1 lg total RNA by Super Script III (Invitrogen) using poly (dt) oligos. About 1/20 of the first-strand cdna was used for realtime PCR using SYBR Premix Ex Taq (Takara) according to the manufacturer s instructions. U6 snrna was used as a reference gene. The RQ value was calculated as described by Pfaffl et al. (2002). Supplemental Table 2 lists the gene-specific primers for stress-responsive marker genes. Data analysis The Ct values were calculated by SDS software (Applied Biosystems). The threshold was determined manually and ranged from 0.01 to 0.1. PCR efficiency (E) of real-time PCR was calculated by absolute fluorescence using Lin- RegPCR (Ruijter et al. 2009). The RQ of mirnas was calculated with PCR efficiency correction using the following mathematical model (Pfaffl 2001): RQ ¼ E ðct:treatment Ct:controlÞ mirna E ðct:treatment Ct:controlÞ U6 In this study, untreated samples were used as controls for calibration. All statistical analysis was performed using R package. The putative target genes of rice mirnas were predicted on the website of WMD3 (Web microrna designer) (Warthmann et al. 2008). Genes with fewer than four mismatches to mirnas were considered as putative targets of mirnas. The sequences of promoter regions were downloaded from PlantGDB (Dong et al. 2004), and the cis-elements were predicted based on PLACE (Higo et al. 1999). Small RNA gel blotting Small RNA gel blotting was performed as described previously (Xie et al. 2006). Briefly, 20 lg total RNA was resolved in 20% urea denatured polyacrylamide gel and blotted into Hybond N? membrane (Amersham, Piscataway, NJ, USA) using MINI PROTEAN system III (Bio- Rad). DNA oligo (mir156-probe: 5 0 -GTGCTCACTCTCT TCTGTCA-3 0, mir167-probe: 5 0 -AGATCATGCTGGCA GCTTCA-3 0, mir171-probe: 5 0 -GATATTGGCACGGCT CAATCA-3 0 and U6-probe: 5 0 -TGTATCGTTCCAATTT TATCGGATGT-3 0 ) labeling and hybridization were performed as described by Xie et al. (2006). The signal intensity was captured by the FUJIFLIM system (FLA5100, Fuji). The RQ of mirna was obtained by normalizing the mirna to U6 and calibrated to nonstressed samples. Results Optimization of ST-RT PCR for mirna quantification Traditional ST-RT qpcr requires multiple steps performed in different tubes, which is inconvenient for highthroughput analysis (Chen et al. 2005; Varkonyi-Gasic et al. 2007). Therefore, we set up a one-tube ST-RT qpcr approach in which DNase I treatment, stem-loop reverse transcription, and real-time qpcr were conducted in one tube (Fig. 1a, detailed in Experimental procedures ). We tested the modified method by profiling the expression levels of mir156 and mir164 in rice seedlings. There was no difference in Ct (threshold cycle) numbers obtained by the modified and traditional ST-RT qpcr (data not shown). We also tested the sensitivity of one-tube ST-RT qpcr and the result indicated that mir156 and mir164 could be detected in as little as 20 pg total RNA. We further checked 23 plant conserved mirnas using sequential dilutions of total RNA as templates. The results showed that the Ct values were highly correlated to the amount of input RNA (averaged correlation efficiency R 2 =-0.95, Fig. 1b). These results suggested that the onetube ST-RT qpcr could be efficiently used for mirna quantification. Proper data normalization using a stable reference gene is critical for qpcr assays. Different reference genes have been proposed in animal mirna qpcr assays, including stable mirnas (mir-191 and mir-103) (Peltier and Latham 2008), other small RNAs (Bandres et al. 2006), and mean mirna expression, which combined expression of several stable mirnas (Mestdagh et al. 2009). To find a suitable reference gene for ST-RT qpcr in rice, we tested the stability of U6 small nuclear RNA (snrna),
4 Fig. 1 Optimization of ST-RT qpcr assay to quantify mirnas. a Schematic diagram of one-tube ST-RT qpcr assay. b Relation of Ct (threshold = 0.1) and input total RNAs in ST-RT qpcr. c Diagram of plate design for high-throughput mirna assays. The calibration sample and internal reference gene are always included in each plate with seven genes of interest (GOI) in one plate. d Typical one-tube ST-RT real-time qpcr amplification plots of eight 96-well plates showing increase in SYBR green fluorescence (DRn, log scale) over PCR cycle number OsActin1 (AK060893), and seven conserved plant mirnas (mir156, mir160, mir162, mir166, mir168, and mir319; Table 1). The primers of U6 snrna were designed in the loop region to eliminate the potential inhibiting effect of the RNA secondary structure on cdna synthesis. OsActin1 and U6 snrna were detected by following the same RT PCR procedure as used for mirna qualification in which reverse PCR primers were used instead of stem-loop primers for cdna synthesis. The stability of these genes was assessed in drought, salinity, and cold conditions and also in ABA-treated samples. The results indicated that the Ct values of U6 snrna had the least standard deviation in these samples among all the candidate reference genes (Table 1). Stability values by pair-wise comparison (Vandesompele et al. 2002) also suggested that U6 snrna had minimal M values, which indicated the highest stability (Table 1). Further validation of U6 snrna using diluted total RNA suggested that U6 snrna was extremely stable in a large range of input RNAs (Supplemental Fig. 1). These results suggest that U6 snrna can be used as a stable reference gene for ST-RT qpcr data normalization.
5 Table 1 Stability of candidate internal reference genes in ST-RT qpcr MiRNA or Gene name Accession no. Function Ct SD M U6 snrna AC Small nuclear RNA mir168b MI mirna processing mir156 MI Flowering mir166 MI Root development mir162 MI Pre-miRNA processing mir168a MI Small RNA effector mir160 MI Auxin homeostasis mir319 MI Auxin signaling OsActin-1 AK Cytoskeleton The amplified product of U6 is located at 96,034 96,094 bp of AC Accession numbers of mirnas are from mirbase and their functions were predicted according to their targets M stability value calculated by pair-wise comparison of these genes as described by Vandesompele et al. (2002), SD standard deviation of Ct value across 16 samples The efficiency of PCR has a major impact on the accuracy of gene expression quantification. Therefore, the PCR efficiency of all detected mirnas was determined by absolute fluorescence signal method using LinRegPCR (version 11.0) (Ruijter et al. 2009). The results showed that PCR efficiency for all primers was higher than 1.5, except for mir169d-p and mir439 (Supplemental Table 1). However, the PCR efficiency of these mirnas was less than 1.9 (except for mir395), which is significantly different from U6 snrna (PCR efficiency = 1.987). These results suggest that PCR efficiency correction is required at the mirna level quantification in ST-RT qpcr data analysis. Detection of mirnas by one-tube ST-RT qpcr in rice seedlings Two primers, stem-loop primer and PCR forward primer, were designed for each of the mirnas representing all 60 rice mirna families in mirbase (release 12.0) (Griffiths- Jones et al. 2006). Degenerated nucleotides were introduced in six stem-loop primers (mir159, mir160, mir167, mir168, mir169, and mir399) because they have only one or two different nucleotides (Supplemental Table 2). All the primers were tested for ST-RT qpcr using rice seedling total RNA. The results suggested that 41 mirnas were detectable with a cut-off Ct value of 35 (threshold = 0.1; Fig. 2) at which nonspecific/primer dimer amplification was not detected. Among the 24 undetected mirnas, 8 (mir413, mir414, mir416, mir417, mir418, mir419, mir420, and mir426) were predicted by conserved sequences without experimental support (Wang et al. 2004), and 7 (mir807, mir811, mir813, mir817, mir818, mir819, and mir821) showed organ-specific expression (Luo et al. 2006) and therefore may not be expressed in the seedlings. It should be also noted that mirna-specific regions in forward primers of these undetectable mirnas tend to have either lower annealing temperatures or longer A/T regions than the detectable ones. For instance, mir818 and mir420 have as long as nine continued A/T in primers (Supplemental Table 2). These results suggested that the sequence of mirnas may affect the efficiency of ST-RT qpcr even though most mirnas could be quantified by ST-RT qpcr. The Ct values of the 41 mirnas detected by one-tube ST-RT qpcr in rice seedlings ranged from 14 to 33 (threshold 0.1, Fig. 2). These mirnas can be classified into high (Ct \ 20), medium (Ct \ 30), and low expression (Ct [ 30) groups (Fig. 2). The top two abundant mirnas in the seedlings are mir528 and mir168. Thirtyfive of these 41 mirnas were identified in the rice seedling small RNA library (Sunkar et al. 2008) and 25 were found in the MPSS small RNA signature library of rice seedlings (Nakano et al. 2006). However, the correlation efficiency between Ct and sequence frequency is low (Fig. 3). These results suggest that ST-RT qpcr may have high sensitivity in quantification of mirna abundance. Profiling rice mirna expression under stress by one-tube ST-RT qpcr The one-tube ST-RT qpcr was applied for mirna profiling in rice seedlings after stress treatment. RNA samples from rice seedlings after drought, salt, cold, and ABA treatments were collected and the treatments were confirmed by checking the expression of SNAC1 and Osb- ZIP23, two well-characterized stress-induced genes in rice (Hu et al. 2006; Xiang et al. 2008). The results showed that both genes were dramatically induced in the samples (Supplemental Fig. 2), suggesting that the stress treatments
6 Fig. 2 The Ct values of mirnas detected in rice seedlings. The raw Ct value at threshold 0.1 was calculated in rice seedlings. The rice mirnas were classified into high, medium, and low groups according their Ct value Fig. 3 Correlation between Ct and sequence frequency of mirnas. The Ct value was compared to sequence frequency (log transferred) in rice seedling samples. The sequence frequency were from Sunkar et al. (2008) were successfully applied for these samples. According to the suggestions for RT-qPCR of mrna (Udvardi et al. 2008; Rieu and Powers 2009), a one-tube ST-RT qpcr experiment was set up (Fig. 1c): (1) U6 snrna reference is included for each sample in all reactions, (2) calibration samples (untreated samples) are always included, (3) mirna expression is relatively quantified with PCR efficiency correction. Figure 1d illustrates typical amplification curves, which combined eight plates for profiling 14 mirnas. To validate the accuracy of one-tube ST-RT qpcr for quantification of mirnas under stress conditions, the expression levels of three mirnas (mir156, mir167, and mir171) were compared by using one-tube ST-RT qpcr and small RNA gel blotting methods with the same RNA samples (Fig. 4a). The small RNA gel blotting clearly shows that mir156 expression was altered after stress and ABA treatments. In contrast, the level of U6 was very stable in these samples (Fig. 4a). The relative quantity (RQ [gel blotting], or fold-change) of the three mirnas was normalized over the U6 signal and calibrated to the nonstress samples (Fig. 4b). In the ST-RT qpcr assays, the relative quantity (RQ [ST-RT qpcr] ) was also normalized over U6 snrna and calibrated to the non-stress samples with PCR efficiency corrected (detailed in Experimental procedures ). The results indicated that RQ [ST-RT qpcr] agreed with RQ [gel blotting] very well (Fig. 4b). The maximal difference in RQ between the two methods was less than 0.3 (Fig. 4b). These results suggest that the optimized ST-RT qpcr method is reliable and efficient in profiling mirnas under stress conditions. Among the mirnas detectable in rice seedlings, 35 mirnas (belonging to 31 mirna families) were induced (RQ [ 1.5) or suppressed (RQ \ 0.67; Supplemental Fig. 3) after stress treatments (drought, salt, or cold), suggesting that most mirnas are responsive to abiotic stresses in rice. Three mirnas (mir530, mir408, and mir435) were induced more than threefold in one of the stresses. The highest induced mirna was mir530, which was induced 6.8-fold under high-salinity stress. The mir530 was also found in Populus (but not in Arabidopsis), but it is not induced by abiotic stress and has distinct targets in Populus (Lu et al. 2008). Notably, most mirnas showed dynamic induction or suppression patterns after stress treatments. For example, mir408 was induced at 20 min after drought stress and then reduced to a level comparable to non-stressed conditions. Several mirnas showed constitutive induction/suppression during the stress treatments: mir319 (cold and drought), mir395 (salinity), mir396 (salinity), mir399 (salinity), and mir530 (salinity).
7 Fig. 4 Validation of the accuracy of ST-RT qpcr by small RNA gel blotting. a Small RNA gel blot measuring mir156, mir167, mir171, and U6. Ethidium bromide-stained RNA is shown as loading control. b Comparison of the RQ values from small RNA gel blots and onetube ST-RT qpcr on the basis of 16 treated samples We noticed that different members of mir159 showed distinct expression patterns after cold treatment. Based on the ST-RT qpcr detection, 19, 18, and 23 mirnas were responsive to drought, salt, and cold stress, respectively (Fig. 5a). Except for mir394 and mir400 (does not exist in rice), all the stress-induced mirnas identified by microarray in Arabidopsis (Liu et al. 2008) were also induced in rice. Most mirnas were responsive to more than one stress condition (Fig. 5a). Our data indicate that 13 mirnas are responsive (induced or suppressed) to all the three stresses and 8 mirnas are responsive to two of the three stresses. These results suggest that many mirnas may be commonly regulated in the responses to different stresses.
8 Diverse function of stress-regulated mirnas Fig. 5 Stress- and ABA-responsive mirnas in rice. a Venn diagram of stress-responsive mirnas in rice. The number of mirnas responsive to each class is indicated. b Cluster analysis of mirnas and three stress-related regulatory genes based on RQ values. The RQ value was changed to gray, as color key indicates ABA is one of most important mediators of stress signal transduction (Zhu 2002). Therefore, we also quantified the relative expression changes of mirnas in rice seedlings after ABA treatment. The results showed that 34 mirnas were significantly induced (RQ [ 1.5) or suppressed (RQ \ 0.67) after ABA treatment (Supplemental Fig. 3). Except for mir398 and mir535, all the ABA-responsive mirnas were also induced or suppressed by at least one of the stress treatments. In fact, mir398 and mir535 showed slight stress-inducible expression although the RQ values of these two mirnas were within the arbitrary cut-off range (0.75 \ RQ \ 1.5). This result suggested that most of the stress-regulated mirna expressions may be ABA-dependent. Cluster analysis according to RQs of mirnas and a few reported stress-responsive genes (OsbZIP23, OsSKIPa, and SNAC1) revealed two groups in terms of stress-responsive patterns (Fig. 5b). Group I includes 30 mirnas and OsSKIPa that have less RQ change than those in group II after stress/aba treatment. Group II includes five mirnas and two stress-responsive transcription factor genes (OsbZIP23 and SNAC1), whose expressions were dramatically changed after stress/aba treatment. Interestingly, most of the mirnas in group I are important for developmental regulation in Arabidopsis (Voinnet 2009). In rice, mir156 and OsSKIPa have been shown to have essential regulation roles in plant growth (Xie et al. 2006; Hou et al. 2009). Because abiotic stresses have strong impacts on rice development, it is possible that the stress-caused developmental changes may be mediated by some of these mirnas. To investigate putative function of stress-responsive mirnas, the targets of these mirnas were predicted by searching the mirna sequence against the rice genome annotation (Ouyang et al. 2007). According to the annotation of putative mirna targets, stress-responsive mir- NAs target a large number of genes with diverse annotations including transcription factors, kinase, transporters, zinc fingers (Supplemental Table 3). The putative functions of these targets were also diverse including flowering time, pollination, phytohormones homeostasis, and stress responses (Supplemental Table 3). Very few of these targets have been confirmed for their functions in stress responses. By searching public microarray data, at least 26 predicted target genes changed their expression more than twofold in drought- or salt stress-treated leaves (Zhou et al. 2007). However, the target genes of the same mirna tend to control different biologic processes. Therefore, it is possible that only portions of these target genes are involved in stress responses. Consistent with this hypothesis, only portions of these target genes were responsive to stresses. For example, among the 11 SPL genes targeted by mir156, only 4 were induced by drought or salt stress. Stress-responsive mirnas may be regulated by stress-related transcription factors Interactions between trans-factors and cis-elements are major processes to induce or suppress gene expression in stress responses (Yamaguchi-Shinozaki and Shinozaki 2006). Some stress-related cis-elements have been identified in the promoters of stress-inducible mirnas in Arabidopsis (Megraw et al. 2006; Liu et al. 2008). To further investigate the rice stress-responsive mirnas, we analyzed the distribution of putative stress- and ABA-related
9 Table 2 Stress-responsive mirnas with changed expression levels in the mutants of regulatory genes OsSKIPa and OsbZIP23 mirna osskipa osbzip23 cis-elements in their promoters. All the stress-responsive mirna promoters contained more than one stress/abarelated cis-element (Supplemental Table 4). The promoters of 11 mirnas (mir156, mir169, mir171, mir393, mir399, mir439, mir535, mir806, mir809, mir812, and mir815) contained all the stress-related cis-elements investigated including ABA-responsive element, droughtresponsive element, low temperature-induced element, MYC-binding site, and MYB-binding site (Supplemental Table 4). This result suggested that stress-regulated mirnas may be regulated by stress-related transcription factors. To further confirm this speculation, we checked seven stress-responsive mirnas in the osskipa knockdown plants (OsSKIPa-RNAi) and osbzip23 knock-out mutant by ST-RT qpcr. OsSKIPa is a transcription regulator that regulates both development and stress tolerance (Hou et al. 2009). OsbZIP23 is a bzip transcription factor mediating ABA-dependent stress responses (Xiang et al. 2008). Relative quantification identified five and six mir- NAs with altered expression in the osskipa and osbzip23 plants, respectively. OsSKIPa may positively regulate the expression of mir172, mir399, mir442, and mir815, but negatively regulate mir156 (Table 2). OsbZIP23 may positively regulate mir156 and mir530, but negatively regulate mir172, mir399, mir442, and mir810 (Table 2). Although the direct regulatory relationship of these mirnas and stress-related transcription regulators requires more detailed experimental support, these preliminary data imply that some of the stress-responsive mirnas may be regulated by key stress-related transcriptional regulators such as OsbZIP23 and OsSKIPa. Discussion RQ 95% CI RQ 95% CI mir mir mir * * mir mir mir * mir * The RQ of mirnas were calibrated to wild type CI confidence interval High-throughput qpcr is a robust and convenient method for profiling gene expression. The accuracy of qpcr relies on correct experimental protocol, careful experimental design, and statistical analysis (Udvardi et al. 2008; Rieu and Powers 2009). Here, we outlined a one-tube ST-RT qpcr to quantify the mirna expression level. Performing all reactions in a single tube can minimize manipulation errors and is convenient for high-throughput experiments. According to our experience and results, the following principles should be considered for the accurate measurement of mirna levels by one-tube ST-RT qpcr: (1) Always include a reference gene and calibrate samples in a single-reaction plate to reduce variations among the plates. (2) Calculate RQ with PCR efficiency correction. (3) Use stable reference genes. In rice, U6 snrna is an ideal reference gene for small RNA quantification. We validated that an optimized ST-RT qpcr assay can quantify the mirna level as precisely as small RNA gel blotting (Fig. 2). The ST-RT qpcr assay requires mirna-specific primers. Some mirnas have high A/U content or long A/U repeat that may make the specific amplification difficult. Unlike regular RT-qPCR, the choice of ST-RT qpcr primers is rather limited because of the short length of mirnas. Therefore, some mirnas could not be detected using ST-RT qpcr. For those mirnas with high A/U content or long A/U regions, we performed qpcr with a lower annealing temperature, but nonspecific amplification increased dramatically. Such problems may be addressed by using locked nucleic acid-modified primers in qpcr (Latorra et al. 2003). High-throughput gene profiling has been successfully used to study stress responses and identify putative drought-tolerance genes. For example, a few genes such as SNAC1, OsSKIPa, and OsbZIP23 that confer stress tolerance in rice were identified based on global gene expression profiling and functional testing (Hu et al. 2006; Xiang et al. 2008; Hou et al. 2009). In this study, the expression profiles of mirnas under abiotic stresses were surveyed using the one-tube ST-RT qpcr method. As a result, about 88% (35 of 40) of the detected mirnas were induced or suppressed by drought, cold, or salt stress. Many of the stress-responsive mirnas are conserved between rice (this study) and Arabidopsis (Liu et al. 2008). So far, only few Arabidopsis mirnas have been investigated for their roles in the regulation of stress response. By suppressing the drought tolerance gene NFYA5, transgenic Arabidopsis overexpressing mir169 was more sensitive to drought stress than wild type (Li et al. 2008). ABA induced accumulation of mir159 direct degradation of MYB33 and MYB101, which are positive regulators of the ABA response. Overexpression of mir159 in Arabidopsis showed hypersensitivity to ABA (Reyes and Chua 2007). Recently, members of mir169 family, targeting the NF-YA transcript factor in rice, were identified to be
10 induced by high salinity and drought (Zhao et al. 2007, 2009). We also found that most members of the mir169 family were induced by multiple stresses (Fig. 5b). Overexpression of mir396c in rice and Arabidopsis exhibited reduced salt and alkali tolerance (Gao et al. 2010), but we observed that only mir396d was slightly induced by the stresses. By using microarray, mir156k, mir167a/b/c, mir169e/f/h and mir319 were found to be down-regulated under cold stress (Jian et al. 2009; Lv et al. 2010), which were confirmed in this study. No significant change was detected for some of the previously identified droughtresponsive mirnas such as mir531 (Zhao et al. 2007), but they still showed a trend of induction by drought in our analysis. In the other hand, some mirnas (such as mir393) showed prominent induction by the stresses but they were not identified as stress-responsive mirnas in the microarray analysis. Besides the sensitivity difference of the different mirna quantification methods, such discrepancies might be also explained by the different temporal and spatial stages at which stress was applied and the difference of rice varieties used. We noticed that most mirnas are responsive to at least one of the abiotic stresses. Considering the functional diversity of the target genes of these mirnas, our results suggest that mirnas may have a wide range of roles in the regulation of stress responses in plants. Although many stress-responsive mirnas have been identified in plants, the regulation of stress-responsive mirnas by stress stimuli remains an unanswered question. Genetic and genomic analyses suggest that abiotic stress signals are perceived and transduced by internal signaling processes that lead to specific regulation of gene expression involving many transcriptional regulators (Zhu 2002; Yamaguchi-Shinozaki and Shinozaki 2006). Since the premirnas are transcribed from genomic DNA (mirna genes) in a way similar to encoding mrna transcription, the alteration of mirna levels under stress conditions is most likely regulated at the transcriptional level of the mirna gene. This can be supported by the fact that quite a few stress-responsive mirnas exhibit expressional level changes in the knock-down or knock-out rice plants of stress-related transcription regulators such as OsSKIPa and OsbZIP23. However, we cannot exclude that the alteration of mirna level may be regulated during pre-mirna processing. It has been reported that plant pathologic effector proteins could suppress mirna processing (Navarro et al. 2008). It is possible that mirna processing may be also regulated by abiotic stress stimuli. Compared to the stress-responsive expression patterns of the well-characterized stress-related regulatory genes OsSKIPa, OsbZIP23, and SNAC1, most of the stressresponsive mirnas have more similar patterns to OsSKIPa than to others (Fig. 5b; Supplemental Fig. 3), i.e., transiently induced or suppressed less than threefold. This is consistent with the functions of OsSKIPa and most mirnas (especially conserved mirnas in plants) that have essential regulatory roles in plant growth and development. Transient and slight alteration of the expression levels of such genes under stress can fine-tune the stress response without significantly affecting growth and development. In contrast, some stressspecific regulatory genes such as OsbZIP23 and SNAC1 showed dramatic changes in expression levels to respond to the stresses. Interestingly, very few mirnas showed dramatic changes in expression levels under stress conditions. Only mir435, mir530, and mir809 were dramatically induced. Unlike other mirnas, each is conserved among different species and target genes belong to the same family; both mir530 and mir809 showed less conservation among different plant species and target mrnas encoding diverse function proteins. These two mirnas may be involved in species-specific stress responses. Stress responses and adaptations involve a series of complex processes including developmental, metabolic, and physiologic changes. Functional characterization of the stress-responsive mirnas may provide new clues for a complete understanding of the molecular basis of stress tolerance in plants. Acknowledgments This work was supported by grants from the National Natural Science Foundation of China ( , , and ), the National Special Key Project of China on Functional Genomics of Major Plants and Animals (2006AA10A103), and the National Special Key Project of China on Transgenic Research (2008ZX001). References Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzo M, Garcia-Foncillas J (2006) Identification by real-time PCR of 13 mature micrornas differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer 5:29 Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, Einav U, Gilad S, Hurban P, Karov Y, Lobenhofer EK, Sharon E, Shiboleth YM, Shtutman M, Bentwich Z, Einat P (2004) MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res 14: Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits micrornas and short interfering RNAs. Proc Natl Acad Sci USA 102: Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU (2006) A sensitive array for microrna expression profiling (michip) based on locked nucleic acids (LNA). RNA 12: Chambers C, Shuai B (2009) Profiling microrna expression in Arabidopsis pollen using microrna array and real-time PCR. BMC Plant Biol 9:87 Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ (2005) Real-time quantification of micrornas by stem-loop RT-PCR. Nucleic Acids Res 33:e179
11 Dong Q, Schlueter SD, Brendel V (2004) PlantGDB, plant genome database and analysis tools. Nucleic Acids Res 32:D354 D359 Gao P, Bai X, Yang L, Lv D, Li Y, Cai H, Ji W, Guo D, Zhu Y (2010) Over-expression of osa-mir396c decreases salt and alkali stress tolerance. Planta 231: Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) mirbase: microrna sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140 D144 Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) mirbase: tools for microrna genomics. Nucleic Acids Res 36:D154 D158 Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE (2006) Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38: Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: Nucleic Acids Res 27: Hou X, Xie K, Yao J, Qi Z, Xiong L (2009) A homolog of human skiinteracting protein in rice positively regulates cell viability and stress tolerance. Proc Natl Acad Sci USA 106: Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103: Hunt EA, Goulding AM, Deo SK (2009) Direct detection and quantification of micrornas. Anal Biochem 387:1 12 Jian X, Zhang L, Li G, Zhang L, Wang X, Cao X, Fang X, Chen F (2009) Identification of novel stress-regulated micrornas from Oryza sativa L. Genomics 95:47 55 Johnson C, Bowman L, Adai AT, Vance V, Sundaresan V (2007) CSRDB: a small RNA integrated database and browser resource for cereals. Nucleic Acids Res 35:D829 D833 Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant micrornas and their targets, including a stress-induced mirna. Mol Cell 14: Latorra D, Arar K, Hurley JM (2003) Design considerations and effects of LNA in PCR primers. Mol Cell Probes 17: Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20: Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, Negrini M, Croce CM (2004) An oligonucleotide microchip for genome-wide microrna profiling in human and mouse tissues. Proc Natl Acad Sci USA 101: Liu B, Li P, Li X, Liu C, Cao S, Chu C, Cao X (2005) Loss of function of OsDCL1 affects microrna accumulation and causes developmental defects in rice. Plant Physiol 139: Liu HH, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated micrornas in Arabidopsis thaliana. RNA 14: Lu S, Sun Y-H, Shi R, Clark C, Li L, Chiang VL (2005) Novel and mechanical stress-responsive micrornas in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17: Lu S, Sun YH, Chiang VL (2008) Stress-responsive micrornas in Populus. Plant J 55: Luo YC, Zhou H, Li Y, Chen JY, Yang JH, Chen YQ, Qu LH (2006) Rice embryogenic calli express a unique set of micrornas, suggesting regulatory roles of micrornas in plant postembryogenic development. FEBS Lett 580: Lv DK, Bai X, Li Y, Ding XD, Ge Y, Cai H, Ji W, Wu N, Zhu YM (2010) Profiling of cold-stress-responsive mirnas in rice by microarrays. Gene 459:39 47 Megraw M, Baev V, Rusinov V, Jensen ST, Kalantidis K, Hatzigeorgiou AG (2006) MicroRNA promoter element discovery in Arabidopsis. RNA 12: Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J (2009) A novel and universal method for microrna RT-qPCR data normalization. Genome Biol 10:R64 Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC (2006) Plant MPSS databases: signature-based transcriptional resources for analyses of mrna and small RNA. Nucleic Acids Res 34:D731 D735 Navarro L, Jay F, Nomura K, He SY, Voinnet O (2008) Suppression of the microrna pathway by bacterial effector proteins. Science 321: Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, Orvis J, Haas B, Wortman J, Buell CR (2007) The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res 35:D883 D887 Peltier HJ, Latham GJ (2008) Normalization of microrna expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 14: Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45 Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36 Reyes JL, Chua NH (2007) ABA induction of mir159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J 49: Rieu I, Powers SJ (2009) Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell 21: Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:e45 Shi R, Chiang VL (2005) Facile means for quantifying microrna expression by real-time PCR. Biotechniques 39: Sunkar R, Zhu JK (2004) Novel and stress-regulated micrornas and other small RNAs from Arabidopsis. Plant Cell 16: Sunkar R, Girke T, Jain PK, Zhu JK (2005) Cloning and characterization of micrornas from rice. Plant Cell 17: Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK (2008) Identification of novel and candidate mirnas in rice by high throughput sequencing. BMC Plant Biol 8:25 Udvardi MK, Czechowski T, Scheible WR (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20: Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034 Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of micrornas. Plant Methods 3:12 Voinnet O (2009) Origin, biogenesis, and activity of plant micror- NAs. Cell 136: Wang XJ, Reyes JL, Chua NH, Gaasterland T (2004) Prediction and identification of Arabidopsis thaliana micrornas and their mrna targets. Genome Biol 5:R65 Warthmann N, Chen H, Ossowski S, Weigel D, Herve P (2008) Highly specific gene silencing by artificial mirnas in rice. PLoS One 3:e1829 Xiang Y, Tang N, Du H, Ye H, Xiong L (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper
12 transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148: Xie K, Wu C, Xiong L (2006) Genomic organization, differential expression, and interaction of SQUAMOSA promoter-bindinglike transcription factors and microrna156 in rice. Plant Physiol 142: Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57: Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, Ruan K, Jin Y (2007) Identification of drought-induced micrornas in rice. Biochem Biophys Res Commun 354: Zhao B, Ge L, Liang R, Li W, Ruan K, Lin H, Jin Y (2009) Members of mir-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol Biol 10:29 Zhou J, Wang X, Jiao Y, Qin Y, Liu X, He K, Chen C, Ma L, Wang J, Xiong L, Zhang Q, Fan L, Deng XW (2007) Global genome expression analysis of rice in response to drought and highsalinity stresses in shoot, flag leaf, and panicle. Plant Mol Biol 63: Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:
Chapter 18. mirna Expression Profiling: From Reference Genes to Global Mean Normalization
 Chapter 18 mirna Expression Profiling: From Reference Genes to Global Mean Normalization Barbara D haene, Pieter Mestdagh, Jan Hellemans, and Jo Vandesompele Abstract MicroRNAs (mirnas) are an important
Chapter 18 mirna Expression Profiling: From Reference Genes to Global Mean Normalization Barbara D haene, Pieter Mestdagh, Jan Hellemans, and Jo Vandesompele Abstract MicroRNAs (mirnas) are an important
RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
 Xu et al. Molecular Cancer (2019) 18:8 https://doi.org/10.1186/s12943-018-0932-8 LETTER TO THE EDITOR RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
Xu et al. Molecular Cancer (2019) 18:8 https://doi.org/10.1186/s12943-018-0932-8 LETTER TO THE EDITOR RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
High AU content: a signature of upregulated mirna in cardiac diseases
 https://helda.helsinki.fi High AU content: a signature of upregulated mirna in cardiac diseases Gupta, Richa 2010-09-20 Gupta, R, Soni, N, Patnaik, P, Sood, I, Singh, R, Rawal, K & Rani, V 2010, ' High
https://helda.helsinki.fi High AU content: a signature of upregulated mirna in cardiac diseases Gupta, Richa 2010-09-20 Gupta, R, Soni, N, Patnaik, P, Sood, I, Singh, R, Rawal, K & Rani, V 2010, ' High
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer. Application Note
 Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
A novel and universal method for microrna RT-qPCR data normalization
 A novel and universal method for microrna RT-qPCR data normalization Jo Vandesompele professor, Ghent University co-founder and CEO, Biogazelle 4 th International qpcr Symposium Weihenstephan, March 1,
A novel and universal method for microrna RT-qPCR data normalization Jo Vandesompele professor, Ghent University co-founder and CEO, Biogazelle 4 th International qpcr Symposium Weihenstephan, March 1,
MicroRNA and Male Infertility: A Potential for Diagnosis
 Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
Identification of mirnas in Eucalyptus globulus Plant by Computational Methods
 International Journal of Pharmaceutical Science Invention ISSN (Online): 2319 6718, ISSN (Print): 2319 670X Volume 2 Issue 5 May 2013 PP.70-74 Identification of mirnas in Eucalyptus globulus Plant by Computational
International Journal of Pharmaceutical Science Invention ISSN (Online): 2319 6718, ISSN (Print): 2319 670X Volume 2 Issue 5 May 2013 PP.70-74 Identification of mirnas in Eucalyptus globulus Plant by Computational
Downregulation of serum mir-17 and mir-106b levels in gastric cancer and benign gastric diseases
 Brief Communication Downregulation of serum mir-17 and mir-106b levels in gastric cancer and benign gastric diseases Qinghai Zeng 1 *, Cuihong Jin 2 *, Wenhang Chen 2, Fang Xia 3, Qi Wang 3, Fan Fan 4,
Brief Communication Downregulation of serum mir-17 and mir-106b levels in gastric cancer and benign gastric diseases Qinghai Zeng 1 *, Cuihong Jin 2 *, Wenhang Chen 2, Fang Xia 3, Qi Wang 3, Fan Fan 4,
Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana
 Biochemistry: Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana 38 CAMERON E. NIENABER ʻ04 Abstract Long recognized as essential plant nutrients and metabolites, inorganic and organic
Biochemistry: Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana 38 CAMERON E. NIENABER ʻ04 Abstract Long recognized as essential plant nutrients and metabolites, inorganic and organic
he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003)
 MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
From reference genes to global mean normalization
 From reference genes to global mean normalization Jo Vandesompele professor, Ghent University co-founder and CEO, Biogazelle qpcr Symposium USA November 9, 2009 Millbrae, CA outline what is normalization
From reference genes to global mean normalization Jo Vandesompele professor, Ghent University co-founder and CEO, Biogazelle qpcr Symposium USA November 9, 2009 Millbrae, CA outline what is normalization
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated
 Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Bi 8 Lecture 17. interference. Ellen Rothenberg 1 March 2016
 Bi 8 Lecture 17 REGulation by RNA interference Ellen Rothenberg 1 March 2016 Protein is not the only regulatory molecule affecting gene expression: RNA itself can be negative regulator RNA does not need
Bi 8 Lecture 17 REGulation by RNA interference Ellen Rothenberg 1 March 2016 Protein is not the only regulatory molecule affecting gene expression: RNA itself can be negative regulator RNA does not need
Expression of mir-146a-5p in patients with intracranial aneurysms and its association with prognosis
 European Review for Medical and Pharmacological Sciences 2018; 22: 726-730 Expression of mir-146a-5p in patients with intracranial aneurysms and its association with prognosis H.-L. ZHANG 1, L. LI 2, C.-J.
European Review for Medical and Pharmacological Sciences 2018; 22: 726-730 Expression of mir-146a-5p in patients with intracranial aneurysms and its association with prognosis H.-L. ZHANG 1, L. LI 2, C.-J.
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Detection of let-7a MicroRNA by Real-time PCR in Colorectal Cancer: a Single-centre Experience from China
 The Journal of International Medical Research 2007; 35: 716 723 Detection of let-7a MicroRNA by Real-time PCR in Colorectal Cancer: a Single-centre Experience from China W-J FANG 1, C-Z LIN 2, H-H ZHANG
The Journal of International Medical Research 2007; 35: 716 723 Detection of let-7a MicroRNA by Real-time PCR in Colorectal Cancer: a Single-centre Experience from China W-J FANG 1, C-Z LIN 2, H-H ZHANG
Small RNA existed in commercial reverse transcriptase: primary evidence of functional small RNAs
 Protein Cell 15, 6(1):1 5 DOI 1.17/s13238-14-116-2 Small RNA existed in commercial reverse transcriptase: primary evidence of functional small RNAs Jie Xu, Xi Chen, Donghai Li, Qun Chen, Zhen Zhou, Dongxia
Protein Cell 15, 6(1):1 5 DOI 1.17/s13238-14-116-2 Small RNA existed in commercial reverse transcriptase: primary evidence of functional small RNAs Jie Xu, Xi Chen, Donghai Li, Qun Chen, Zhen Zhou, Dongxia
Genome-wide microrna profiling in human fetal nervous tissues by oligonucleotide microarray
 Childs Nerv Syst (2006) 22:1419 1425 DOI 10.1007/s00381-006-0173-9 ORIGINAL PAPER Genome-wide microrna profiling in human fetal nervous tissues by oligonucleotide microarray Jian-Jun Zhao & You-Jia Hua
Childs Nerv Syst (2006) 22:1419 1425 DOI 10.1007/s00381-006-0173-9 ORIGINAL PAPER Genome-wide microrna profiling in human fetal nervous tissues by oligonucleotide microarray Jian-Jun Zhao & You-Jia Hua
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Phosphate buffered saline (PBS) for washing the cells TE buffer (nuclease-free) ph 7.5 for use with the PrimePCR Reverse Transcription Control Assay
 Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Profiles of gene expression & diagnosis/prognosis of cancer. MCs in Advanced Genetics Ainoa Planas Riverola
 Profiles of gene expression & diagnosis/prognosis of cancer MCs in Advanced Genetics Ainoa Planas Riverola Gene expression profiles Gene expression profiling Used in molecular biology, it measures the
Profiles of gene expression & diagnosis/prognosis of cancer MCs in Advanced Genetics Ainoa Planas Riverola Gene expression profiles Gene expression profiling Used in molecular biology, it measures the
Supplementary Figures
 Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
Santosh Patnaik, MD, PhD! Assistant Member! Department of Thoracic Surgery! Roswell Park Cancer Institute!
 Santosh Patnaik, MD, PhD Assistant Member Department of Thoracic Surgery Roswell Park Cancer Institute MicroRNA biology, techniques and applications History Biogenesis Nomenclature Tissue specificity Mechanisms
Santosh Patnaik, MD, PhD Assistant Member Department of Thoracic Surgery Roswell Park Cancer Institute MicroRNA biology, techniques and applications History Biogenesis Nomenclature Tissue specificity Mechanisms
MicroRNA-mediated incoherent feedforward motifs are robust
 The Second International Symposium on Optimization and Systems Biology (OSB 8) Lijiang, China, October 31 November 3, 8 Copyright 8 ORSC & APORC, pp. 62 67 MicroRNA-mediated incoherent feedforward motifs
The Second International Symposium on Optimization and Systems Biology (OSB 8) Lijiang, China, October 31 November 3, 8 Copyright 8 ORSC & APORC, pp. 62 67 MicroRNA-mediated incoherent feedforward motifs
Supplementary information
 Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Decreased expression of mir-490-3p in osteosarcoma and its clinical significance
 European Review for Medical and Pharmacological Sciences Decreased expression of mir-490-3p in osteosarcoma and its clinical significance B. TANG, C. LIU, Q.-M. ZHANG, M. NI Department of Orthopedics,
European Review for Medical and Pharmacological Sciences Decreased expression of mir-490-3p in osteosarcoma and its clinical significance B. TANG, C. LIU, Q.-M. ZHANG, M. NI Department of Orthopedics,
Circular RNAs (circrnas) act a stable mirna sponges
 Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Phenomena first observed in petunia
 Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
Differentially expressed microrna cohorts in seed development may contribute to poor grain filling of inferior spikelets in rice
 Peng et al. BMC Plant Biology 2014, 14:196 RESEARCH ARTICLE Open Access Differentially expressed microrna cohorts in seed development may contribute to poor grain filling of inferior spikelets in rice
Peng et al. BMC Plant Biology 2014, 14:196 RESEARCH ARTICLE Open Access Differentially expressed microrna cohorts in seed development may contribute to poor grain filling of inferior spikelets in rice
RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays
 Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
WHO Prequalification of In Vitro Diagnostics PUBLIC REPORT. Product: Alere q HIV-1/2 Detect WHO reference number: PQDx
 WHO Prequalification of In Vitro Diagnostics PUBLIC REPORT Product: Alere q HIV-1/2 Detect WHO reference number: PQDx 0226-032-00 Alere q HIV-1/2 Detect with product codes 270110050, 270110010 and 270300001,
WHO Prequalification of In Vitro Diagnostics PUBLIC REPORT Product: Alere q HIV-1/2 Detect WHO reference number: PQDx 0226-032-00 Alere q HIV-1/2 Detect with product codes 270110050, 270110010 and 270300001,
Widespread Long Noncoding RNAs as Endogenous Target Mimics for MicroRNAs in Plants 1[W]
![Widespread Long Noncoding RNAs as Endogenous Target Mimics for MicroRNAs in Plants 1[W] Widespread Long Noncoding RNAs as Endogenous Target Mimics for MicroRNAs in Plants 1[W]](/thumbs/96/127589351.jpg) Widespread Long Noncoding RNAs as Endogenous Target Mimics for MicroRNAs in Plants 1[W] Hua-Jun Wu 2, Zhi-Min Wang 2, Meng Wang, and Xiu-Jie Wang* State Key Laboratory of Plant Genomics, Institute of Genetics
Widespread Long Noncoding RNAs as Endogenous Target Mimics for MicroRNAs in Plants 1[W] Hua-Jun Wu 2, Zhi-Min Wang 2, Meng Wang, and Xiu-Jie Wang* State Key Laboratory of Plant Genomics, Institute of Genetics
AIDS - Knowledge and Dogma. Conditions for the Emergence and Decline of Scientific Theories Congress, July 16/ , Vienna, Austria
 AIDS - Knowledge and Dogma Conditions for the Emergence and Decline of Scientific Theories Congress, July 16/17 2010, Vienna, Austria Reliability of PCR to detect genetic sequences from HIV Juan Manuel
AIDS - Knowledge and Dogma Conditions for the Emergence and Decline of Scientific Theories Congress, July 16/17 2010, Vienna, Austria Reliability of PCR to detect genetic sequences from HIV Juan Manuel
Blocking the expression of the hepatitis B virus S gene in hepatocellular carcinoma cell lines with an anti-gene locked nucleic acid in vitro
 Blocking the expression of the hepatitis B virus S gene in hepatocellular carcinoma cell lines with an anti-gene locked nucleic acid in vitro Y.-B. Deng, H.-J. Qin, Y.-H. Luo, Z.-R. Liang and J.-J. Zou
Blocking the expression of the hepatitis B virus S gene in hepatocellular carcinoma cell lines with an anti-gene locked nucleic acid in vitro Y.-B. Deng, H.-J. Qin, Y.-H. Luo, Z.-R. Liang and J.-J. Zou
RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using
 Supplementary Information Materials and Methods RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using Trizol reagent (Invitrogen,Carlsbad, CA) according to the manufacturer's instructions.
Supplementary Information Materials and Methods RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using Trizol reagent (Invitrogen,Carlsbad, CA) according to the manufacturer's instructions.
Expression of mir-1294 is downregulated and predicts a poor prognosis in gastric cancer
 European Review for Medical and Pharmacological Sciences 2018; 22: 5525-5530 Expression of mir-1294 is downregulated and predicts a poor prognosis in gastric cancer Y.-X. SHI, B.-L. YE, B.-R. HU, X.-J.
European Review for Medical and Pharmacological Sciences 2018; 22: 5525-5530 Expression of mir-1294 is downregulated and predicts a poor prognosis in gastric cancer Y.-X. SHI, B.-L. YE, B.-R. HU, X.-J.
Supplemental Figure 1. Small RNA size distribution from different soybean tissues.
 Supplemental Figure 1. Small RNA size distribution from different soybean tissues. The size of small RNAs was plotted versus frequency (percentage) among total sequences (A, C, E and G) or distinct sequences
Supplemental Figure 1. Small RNA size distribution from different soybean tissues. The size of small RNAs was plotted versus frequency (percentage) among total sequences (A, C, E and G) or distinct sequences
mirna Dr. S Hosseini-Asl
 mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
genomics for systems biology / ISB2020 RNA sequencing (RNA-seq)
 RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
Mir-595 is a significant indicator of poor patient prognosis in epithelial ovarian cancer
 European Review for Medical and Pharmacological Sciences 2017; 21: 4278-4282 Mir-595 is a significant indicator of poor patient prognosis in epithelial ovarian cancer Q.-H. ZHOU 1, Y.-M. ZHAO 2, L.-L.
European Review for Medical and Pharmacological Sciences 2017; 21: 4278-4282 Mir-595 is a significant indicator of poor patient prognosis in epithelial ovarian cancer Q.-H. ZHOU 1, Y.-M. ZHAO 2, L.-L.
Small RNAs and how to analyze them using sequencing
 Small RNAs and how to analyze them using sequencing RNA-seq Course November 8th 2017 Marc Friedländer ComputaAonal RNA Biology Group SciLifeLab / Stockholm University Special thanks to Jakub Westholm for
Small RNAs and how to analyze them using sequencing RNA-seq Course November 8th 2017 Marc Friedländer ComputaAonal RNA Biology Group SciLifeLab / Stockholm University Special thanks to Jakub Westholm for
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
m 6 A mrna methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
mirna in Plasma Exosome is Stable under Different Storage Conditions
 Molecules 2014, 19, 1568-1575; doi:10.3390/molecules19021568 Article OPEN ACCESS molecules ISSN 1420-3049 www.mdpi.com/journal/molecules mirna in Plasma Exosome is Stable under Different Storage Conditions
Molecules 2014, 19, 1568-1575; doi:10.3390/molecules19021568 Article OPEN ACCESS molecules ISSN 1420-3049 www.mdpi.com/journal/molecules mirna in Plasma Exosome is Stable under Different Storage Conditions
microrna analysis for the detection of autologous blood transfusion in doping control
 microrna analysis for the detection of autologous blood transfusion in doping control Application note Doping Control Authors Francesco Donati*, Fiorella Boccia, Xavier de la Torre, Alessandra Stampella,
microrna analysis for the detection of autologous blood transfusion in doping control Application note Doping Control Authors Francesco Donati*, Fiorella Boccia, Xavier de la Torre, Alessandra Stampella,
CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer
 European Review for Medical and Pharmacological Sciences 2018; 22: 3713-3718 CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer N. LIU 1, J. ZHANG 1, L.-Y. ZHANG 1, L.
European Review for Medical and Pharmacological Sciences 2018; 22: 3713-3718 CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer N. LIU 1, J. ZHANG 1, L.-Y. ZHANG 1, L.
Effects of Soil Copper Concentration on Growth, Development and Yield Formation of Rice (Oryza sativa)
 Rice Science, 05, 12(2): 125-132 125 http://www.ricescience.org Effects of Soil Copper Concentration on Growth, Development and Yield Formation of Rice (Oryza sativa) XU Jia-kuan 1, 2, YANG Lian-xin 1,
Rice Science, 05, 12(2): 125-132 125 http://www.ricescience.org Effects of Soil Copper Concentration on Growth, Development and Yield Formation of Rice (Oryza sativa) XU Jia-kuan 1, 2, YANG Lian-xin 1,
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
STAT1 regulates microrna transcription in interferon γ stimulated HeLa cells
 CAMDA 2009 October 5, 2009 STAT1 regulates microrna transcription in interferon γ stimulated HeLa cells Guohua Wang 1, Yadong Wang 1, Denan Zhang 1, Mingxiang Teng 1,2, Lang Li 2, and Yunlong Liu 2 Harbin
CAMDA 2009 October 5, 2009 STAT1 regulates microrna transcription in interferon γ stimulated HeLa cells Guohua Wang 1, Yadong Wang 1, Denan Zhang 1, Mingxiang Teng 1,2, Lang Li 2, and Yunlong Liu 2 Harbin
Single Cell Quantitative Polymer Chain Reaction (sc-qpcr)
 Single Cell Quantitative Polymer Chain Reaction (sc-qpcr) Analyzing gene expression profiles from a bulk population of cells provides an average profile which may obscure important biological differences
Single Cell Quantitative Polymer Chain Reaction (sc-qpcr) Analyzing gene expression profiles from a bulk population of cells provides an average profile which may obscure important biological differences
Oncolytic Adenovirus Complexes Coated with Lipids and Calcium Phosphate for Cancer Gene Therapy
 Oncolytic Adenovirus Complexes Coated with Lipids and Calcium Phosphate for Cancer Gene Therapy Jianhua Chen, Pei Gao, Sujing Yuan, Rongxin Li, Aimin Ni, Liang Chu, Li Ding, Ying Sun, Xin-Yuan Liu, Yourong
Oncolytic Adenovirus Complexes Coated with Lipids and Calcium Phosphate for Cancer Gene Therapy Jianhua Chen, Pei Gao, Sujing Yuan, Rongxin Li, Aimin Ni, Liang Chu, Li Ding, Ying Sun, Xin-Yuan Liu, Yourong
PREPARED FOR: U.S. Army Medical Research and Materiel Command Fort Detrick, MD
 AD Award Number: W81XWH-11-1-0126 TITLE: Chemical strategy to translate genetic/epigenetic mechanisms to breast cancer therapeutics PRINCIPAL INVESTIGATOR: Xiang-Dong Fu, PhD CONTRACTING ORGANIZATION:
AD Award Number: W81XWH-11-1-0126 TITLE: Chemical strategy to translate genetic/epigenetic mechanisms to breast cancer therapeutics PRINCIPAL INVESTIGATOR: Xiang-Dong Fu, PhD CONTRACTING ORGANIZATION:
Supplementary Fig. 1. Identification of acetylation of K68 of SOD2
 Supplementary Fig. 1. Identification of acetylation of K68 of SOD2 A B H. sapiens 54 KHHAAYVNNLNVTEEKYQEALAK 75 M. musculus 54 KHHAAYVNNLNATEEKYHEALAK 75 X. laevis 55 KHHATYVNNLNITEEKYAEALAK 77 D. rerio
Supplementary Fig. 1. Identification of acetylation of K68 of SOD2 A B H. sapiens 54 KHHAAYVNNLNVTEEKYQEALAK 75 M. musculus 54 KHHAAYVNNLNATEEKYHEALAK 75 X. laevis 55 KHHATYVNNLNITEEKYAEALAK 77 D. rerio
Serum mirna expression profile as a prognostic biomarker of stage II/III
 Serum mirna expression profile as a prognostic biomarker of stage II/III colorectal adenocarcinoma Jialu Li a,b,, Yang Liu c,, Cheng Wang d,, Ting Deng a,, Hongwei Liang b, Yifei Wang e, Dingzhi Huang
Serum mirna expression profile as a prognostic biomarker of stage II/III colorectal adenocarcinoma Jialu Li a,b,, Yang Liu c,, Cheng Wang d,, Ting Deng a,, Hongwei Liang b, Yifei Wang e, Dingzhi Huang
Iso-Seq Method Updates and Target Enrichment Without Amplification for SMRT Sequencing
 Iso-Seq Method Updates and Target Enrichment Without Amplification for SMRT Sequencing PacBio Americas User Group Meeting Sample Prep Workshop June.27.2017 Tyson Clark, Ph.D. For Research Use Only. Not
Iso-Seq Method Updates and Target Enrichment Without Amplification for SMRT Sequencing PacBio Americas User Group Meeting Sample Prep Workshop June.27.2017 Tyson Clark, Ph.D. For Research Use Only. Not
MicroRNA expression profiling and functional analysis in prostate cancer. Marco Folini s.c. Ricerca Traslazionale DOSL
 MicroRNA expression profiling and functional analysis in prostate cancer Marco Folini s.c. Ricerca Traslazionale DOSL What are micrornas? For almost three decades, the alteration of protein-coding genes
MicroRNA expression profiling and functional analysis in prostate cancer Marco Folini s.c. Ricerca Traslazionale DOSL What are micrornas? For almost three decades, the alteration of protein-coding genes
BIO360 Fall 2013 Quiz 1
 BIO360 Fall 2013 Quiz 1 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function mutation.
BIO360 Fall 2013 Quiz 1 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function mutation.
Expression of mirnas of Hevea brasiliensis under drought stress is altered in clones with varying levels of drought tolerance
 Indian Journal of Biotechnology Vol 15, April 2016, pp 153-160 Expression of mirnas of Hevea brasiliensis under drought stress is altered in clones with varying levels of drought tolerance Linu Kuruvilla,
Indian Journal of Biotechnology Vol 15, April 2016, pp 153-160 Expression of mirnas of Hevea brasiliensis under drought stress is altered in clones with varying levels of drought tolerance Linu Kuruvilla,
microrna Presented for: Presented by: Date:
 microrna Presented for: Presented by: Date: 2 micrornas Non protein coding, endogenous RNAs of 21-22nt length Evolutionarily conserved Regulate gene expression by binding complementary regions at 3 regions
microrna Presented for: Presented by: Date: 2 micrornas Non protein coding, endogenous RNAs of 21-22nt length Evolutionarily conserved Regulate gene expression by binding complementary regions at 3 regions
Research Article Base Composition Characteristics of Mammalian mirnas
 Journal of Nucleic Acids Volume 2013, Article ID 951570, 6 pages http://dx.doi.org/10.1155/2013/951570 Research Article Base Composition Characteristics of Mammalian mirnas Bin Wang Department of Chemistry,
Journal of Nucleic Acids Volume 2013, Article ID 951570, 6 pages http://dx.doi.org/10.1155/2013/951570 Research Article Base Composition Characteristics of Mammalian mirnas Bin Wang Department of Chemistry,
Identification of UV-B-induced micrornas in wheat
 Identification of UV-B-induced micrornas in wheat B. Wang*, Y.F. Sun*, N. Song, X.J. Wang, H. Feng, L.L. Huang and Z.S. Kang College of Plant Protection and State Key Laboratory of Crop Stress Biology
Identification of UV-B-induced micrornas in wheat B. Wang*, Y.F. Sun*, N. Song, X.J. Wang, H. Feng, L.L. Huang and Z.S. Kang College of Plant Protection and State Key Laboratory of Crop Stress Biology
microrna PCR System (Exiqon), following the manufacturer s instructions. In brief, 10ng of
 SUPPLEMENTAL MATERIALS AND METHODS Quantitative RT-PCR Quantitative RT-PCR analysis was performed using the Universal mircury LNA TM microrna PCR System (Exiqon), following the manufacturer s instructions.
SUPPLEMENTAL MATERIALS AND METHODS Quantitative RT-PCR Quantitative RT-PCR analysis was performed using the Universal mircury LNA TM microrna PCR System (Exiqon), following the manufacturer s instructions.
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Instructions for Use. RealStar Influenza Screen & Type RT-PCR Kit /2017 EN
 Instructions for Use RealStar Influenza Screen & Type RT-PCR Kit 4.0 05/2017 EN RealStar Influenza Screen & Type RT-PCR Kit 4.0 For research use only! (RUO) 164003 INS-164000-EN-S01 96 05 2017 altona
Instructions for Use RealStar Influenza Screen & Type RT-PCR Kit 4.0 05/2017 EN RealStar Influenza Screen & Type RT-PCR Kit 4.0 For research use only! (RUO) 164003 INS-164000-EN-S01 96 05 2017 altona
NOVEL FUNCTION OF mirnas IN REGULATING GENE EXPRESSION. Ana M. Martinez
 NOVEL FUNCTION OF mirnas IN REGULATING GENE EXPRESSION Ana M. Martinez Switching from Repression to Activation: MicroRNAs can Up-Regulate Translation. Shoba Vasudevan, Yingchun Tong, Joan A. Steitz AU-rich
NOVEL FUNCTION OF mirnas IN REGULATING GENE EXPRESSION Ana M. Martinez Switching from Repression to Activation: MicroRNAs can Up-Regulate Translation. Shoba Vasudevan, Yingchun Tong, Joan A. Steitz AU-rich
Nature Genetics: doi: /ng Supplementary Figure 1. Assessment of sample purity and quality.
 Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
The Need for a PARP in vivo Pharmacodynamic Assay
 The Need for a PARP in vivo Pharmacodynamic Assay Jay George, Ph.D. Chief Scientific Officer Trevigen, Inc. Gaithersburg, MD Poly(ADP-ribose) polymerases are promising therapeutic targets. In response
The Need for a PARP in vivo Pharmacodynamic Assay Jay George, Ph.D. Chief Scientific Officer Trevigen, Inc. Gaithersburg, MD Poly(ADP-ribose) polymerases are promising therapeutic targets. In response
ANALYSIS OF SOYBEAN SMALL RNA POPULATIONS HLAING HLAING WIN THESIS
 ANALYSIS OF SOYBEAN SMALL RNA POPULATIONS BY HLAING HLAING WIN THESIS Submitted in partial fulfillment of the requirements for the degree of Master of Science in Bioinformatics in the Graduate College
ANALYSIS OF SOYBEAN SMALL RNA POPULATIONS BY HLAING HLAING WIN THESIS Submitted in partial fulfillment of the requirements for the degree of Master of Science in Bioinformatics in the Graduate College
For in vitro Veterinary Diagnostics only. Kylt Rotavirus A. Real-Time RT-PCR Detection.
 For in vitro Veterinary Diagnostics only. Kylt Rotavirus A Real-Time RT-PCR Detection www.kylt.eu DIRECTION FOR USE Kylt Rotavirus A Real-Time RT-PCR Detection A. General Kylt Rotavirus A products are
For in vitro Veterinary Diagnostics only. Kylt Rotavirus A Real-Time RT-PCR Detection www.kylt.eu DIRECTION FOR USE Kylt Rotavirus A Real-Time RT-PCR Detection A. General Kylt Rotavirus A products are
Original Article Identification of hsa-mir-21 as a target gene of tumorigenesis in neuroblastoma
 Int J Clin Exp Med 2016;9(2):2718-2727 www.ijcem.com /ISSN:1940-5901/IJCEM0016181 Original Article Identification of hsa-mir-21 as a target gene of tumorigenesis in neuroblastoma Zuopeng Wang *, Wei Yao
Int J Clin Exp Med 2016;9(2):2718-2727 www.ijcem.com /ISSN:1940-5901/IJCEM0016181 Original Article Identification of hsa-mir-21 as a target gene of tumorigenesis in neuroblastoma Zuopeng Wang *, Wei Yao
Molecular mechanism of the priming by jasmonic acid of specific dehydration stress response genes in Arabidopsis
 University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications in the Biological Sciences Papers in the Biological Sciences 2016 Molecular mechanism of the priming
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications in the Biological Sciences Papers in the Biological Sciences 2016 Molecular mechanism of the priming
Supporting Information
 Supporting Information Hu et al..73/pnas.3489 SI Materials and Methods Histone Extraction and Western Blot Analysis. Rice histone proteins were extracted from -d-old seedlings. After being washed with
Supporting Information Hu et al..73/pnas.3489 SI Materials and Methods Histone Extraction and Western Blot Analysis. Rice histone proteins were extracted from -d-old seedlings. After being washed with
Instructions for Use. RealStar Influenza S&T RT-PCR Kit /2017 EN
 Instructions for Use RealStar Influenza S&T RT-PCR Kit 3.0 01/2017 EN RealStar Influenza S&T RT-PCR Kit 3.0 For research use only! (RUO) 163003 INS-163000-EN-S02 96 01 2017 altona Diagnostics GmbH Mörkenstr.
Instructions for Use RealStar Influenza S&T RT-PCR Kit 3.0 01/2017 EN RealStar Influenza S&T RT-PCR Kit 3.0 For research use only! (RUO) 163003 INS-163000-EN-S02 96 01 2017 altona Diagnostics GmbH Mörkenstr.
Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins.
 Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins. The RNA transcribed from a complex transcription unit
Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins. The RNA transcribed from a complex transcription unit
Analysis of mirna expression under stress in Arabidopsis thaliana
 Analysis of mirna expression under stress in Arabidopsis thaliana Aida Hajdarpašić*, Pia Ruggenthaler Max F. Perutz Laboratories, Medical University of Vienna, Department of Medical Biochemistry, Dr. Bohrgasse
Analysis of mirna expression under stress in Arabidopsis thaliana Aida Hajdarpašić*, Pia Ruggenthaler Max F. Perutz Laboratories, Medical University of Vienna, Department of Medical Biochemistry, Dr. Bohrgasse
Low levels of serum mir-99a is a predictor of poor prognosis in breast cancer
 Low levels of serum mir-99a is a predictor of poor prognosis in breast cancer J. Li 1, Z.J. Song 2, Y.Y. Wang 1, Y. Yin 1, Y. Liu 1 and X. Nan 1 1 Tumor Research Department, Shaanxi Provincial Tumor Hospital,
Low levels of serum mir-99a is a predictor of poor prognosis in breast cancer J. Li 1, Z.J. Song 2, Y.Y. Wang 1, Y. Yin 1, Y. Liu 1 and X. Nan 1 1 Tumor Research Department, Shaanxi Provincial Tumor Hospital,
CHAPTER 4 RESULTS. showed that all three replicates had similar growth trends (Figure 4.1) (p<0.05; p=0.0000)
 CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
Association of mir-21 with esophageal cancer prognosis: a meta-analysis
 Association of mir-21 with esophageal cancer prognosis: a meta-analysis S.-W. Wen 1, Y.-F. Zhang 1, Y. Li 1, Z.-X. Liu 2, H.-L. Lv 1, Z.-H. Li 1, Y.-Z. Xu 1, Y.-G. Zhu 1 and Z.-Q. Tian 1 1 Department of
Association of mir-21 with esophageal cancer prognosis: a meta-analysis S.-W. Wen 1, Y.-F. Zhang 1, Y. Li 1, Z.-X. Liu 2, H.-L. Lv 1, Z.-H. Li 1, Y.-Z. Xu 1, Y.-G. Zhu 1 and Z.-Q. Tian 1 1 Department of
Marta Puerto Plasencia. microrna sponges
 Marta Puerto Plasencia microrna sponges Introduction microrna CircularRNA Publications Conclusions The most well-studied regions in the human genome belong to proteincoding genes. Coding exons are 1.5%
Marta Puerto Plasencia microrna sponges Introduction microrna CircularRNA Publications Conclusions The most well-studied regions in the human genome belong to proteincoding genes. Coding exons are 1.5%
Expression of mirnas confers enhanced tolerance to drought and salt stress in Finger millet (Eleusine coracona)
 Journal of Stress Physiology & Biochemistry, Vol. 9 No. 3 2013, pp. 220-231 ISSN 1997-0838 Original Text Copyright 2013 by Nageshbabu, Usha, Jyothi, Sharadamma, Rai, Devaraj ORIGINAL ARTICLE Expression
Journal of Stress Physiology & Biochemistry, Vol. 9 No. 3 2013, pp. 220-231 ISSN 1997-0838 Original Text Copyright 2013 by Nageshbabu, Usha, Jyothi, Sharadamma, Rai, Devaraj ORIGINAL ARTICLE Expression
Original Article MicroRNA-27a acts as a novel biomarker in the diagnosis of patients with laryngeal squamous cell carcinoma
 Int J Clin Exp Pathol 2016;9(2):2049-2053 www.ijcep.com /ISSN:1936-2625/IJCEP0014841 Original Article MicroRNA-27a acts as a novel biomarker in the diagnosis of patients with laryngeal squamous cell carcinoma
Int J Clin Exp Pathol 2016;9(2):2049-2053 www.ijcep.com /ISSN:1936-2625/IJCEP0014841 Original Article MicroRNA-27a acts as a novel biomarker in the diagnosis of patients with laryngeal squamous cell carcinoma
Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54
 CORRECTION NOTICE Nat. Genet. 42, 759 763 (2010); published online 22 August 2010; corrected online 27 August 2014 Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects
CORRECTION NOTICE Nat. Genet. 42, 759 763 (2010); published online 22 August 2010; corrected online 27 August 2014 Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects
Supplementary webappendix
 Supplementary webappendix This webappendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Kratz JR, He J, Van Den Eeden SK, et
Supplementary webappendix This webappendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Kratz JR, He J, Van Den Eeden SK, et
In silico identification of mirnas and their targets from the expressed sequence tags of Raphanus sativus
 www.bioinformation.net Hypothesis Volume 8(2) In silico identification of mirnas and their targets from the expressed sequence tags of Raphanus sativus Charuvaka Muvva 1 *, Lata Tewari 2, Kasoju Aruna
www.bioinformation.net Hypothesis Volume 8(2) In silico identification of mirnas and their targets from the expressed sequence tags of Raphanus sativus Charuvaka Muvva 1 *, Lata Tewari 2, Kasoju Aruna
MicroRNA in Cancer Karen Dybkær 2013
 MicroRNA in Cancer Karen Dybkær RNA Ribonucleic acid Types -Coding: messenger RNA (mrna) coding for proteins -Non-coding regulating protein formation Ribosomal RNA (rrna) Transfer RNA (trna) Small nuclear
MicroRNA in Cancer Karen Dybkær RNA Ribonucleic acid Types -Coding: messenger RNA (mrna) coding for proteins -Non-coding regulating protein formation Ribosomal RNA (rrna) Transfer RNA (trna) Small nuclear
Ling Xu, Wei-Qi Dai, Xuan-Fu Xu, Fan Wang, Lei He, Chuan-Yong Guo*
 DOI:http://dx.doi.org/10.7314/APJCP.2012.13.7.3203 RESEARCH ARTICLE Effects of Multiple-target Anti-microRNA Antisense Oligodeoxyribonucleotides on Proliferation and Migration of Gastric Cancer Cells Ling
DOI:http://dx.doi.org/10.7314/APJCP.2012.13.7.3203 RESEARCH ARTICLE Effects of Multiple-target Anti-microRNA Antisense Oligodeoxyribonucleotides on Proliferation and Migration of Gastric Cancer Cells Ling
Rice Mutation Breeding for Various Grain Qualities in Thailand
 8. Thailand Rice Mutation Breeding for Various Grain Qualities in Thailand S. Taprab, W. Sukviwat, D. Chettanachit, S. Wongpiyachon and W. Rattanakarn Bureau of Rice Research and Development, Rice Department,
8. Thailand Rice Mutation Breeding for Various Grain Qualities in Thailand S. Taprab, W. Sukviwat, D. Chettanachit, S. Wongpiyachon and W. Rattanakarn Bureau of Rice Research and Development, Rice Department,
High-resolution identification and abundance profiling of cassava (Manihot esculenta Crantz) micrornas
 Khatabi et al. BMC Genomics (216) 17:85 DOI 1.1186/s12864-16-2391-1 RESEARCH ARTICLE High-resolution identification and abundance profiling of cassava (Manihot esculenta Crantz) micrornas Behnam Khatabi
Khatabi et al. BMC Genomics (216) 17:85 DOI 1.1186/s12864-16-2391-1 RESEARCH ARTICLE High-resolution identification and abundance profiling of cassava (Manihot esculenta Crantz) micrornas Behnam Khatabi
Milk micro-rna information and lactation
 Christophe Lefèvre, BioDeakin,. Deakin University, Geelong VIC Australia Milk micro-rna information and lactation New Signals in milk? - Markers of milk and lactation status? - Signal infant development?
Christophe Lefèvre, BioDeakin,. Deakin University, Geelong VIC Australia Milk micro-rna information and lactation New Signals in milk? - Markers of milk and lactation status? - Signal infant development?
Supporting Information
 Supporting Information Lee et al. 10.1073/pnas.0910950106 Fig. S1. Fe (A), Zn (B), Cu (C), and Mn (D) concentrations in flag leaves from WT, osnas3-1, and OsNAS3-antisense (AN-2) plants. Each measurement
Supporting Information Lee et al. 10.1073/pnas.0910950106 Fig. S1. Fe (A), Zn (B), Cu (C), and Mn (D) concentrations in flag leaves from WT, osnas3-1, and OsNAS3-antisense (AN-2) plants. Each measurement
Cellular MicroRNA and P Bodies Modulate Host-HIV-1 Interactions. 指導教授 : 張麗冠博士 演講者 : 黃柄翰 Date: 2009/10/19
 Cellular MicroRNA and P Bodies Modulate Host-HIV-1 Interactions 指導教授 : 張麗冠博士 演講者 : 黃柄翰 Date: 2009/10/19 1 MicroRNA biogenesis 1. Pri mirna: primary mirna 2. Drosha: RNaseIII 3. DCR 1: Dicer 1 RNaseIII
Cellular MicroRNA and P Bodies Modulate Host-HIV-1 Interactions 指導教授 : 張麗冠博士 演講者 : 黃柄翰 Date: 2009/10/19 1 MicroRNA biogenesis 1. Pri mirna: primary mirna 2. Drosha: RNaseIII 3. DCR 1: Dicer 1 RNaseIII
Paternal exposure and effects on microrna and mrna expression in developing embryo. Department of Chemical and Radiation Nur Duale
 Paternal exposure and effects on microrna and mrna expression in developing embryo Department of Chemical and Radiation Nur Duale Our research question Can paternal preconceptional exposure to environmental
Paternal exposure and effects on microrna and mrna expression in developing embryo Department of Chemical and Radiation Nur Duale Our research question Can paternal preconceptional exposure to environmental
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation
 Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
95% CI: , P=0.037).
 Biomedical Research 2017; 28 (21): 9248-9252 Reduced expression level of mir-205 predicts poor prognosis in Qiang Yang 1*#, Peng Sun 2#, Haotian Feng 1, Xin Li 1, Jianmin Li 1 1 Department of Orthopedics,
Biomedical Research 2017; 28 (21): 9248-9252 Reduced expression level of mir-205 predicts poor prognosis in Qiang Yang 1*#, Peng Sun 2#, Haotian Feng 1, Xin Li 1, Jianmin Li 1 1 Department of Orthopedics,
MicroRNAs, RNA Modifications, RNA Editing. Bora E. Baysal MD, PhD Oncology for Scientists Lecture Tue, Oct 17, 2017, 3:30 PM - 5:00 PM
 MicroRNAs, RNA Modifications, RNA Editing Bora E. Baysal MD, PhD Oncology for Scientists Lecture Tue, Oct 17, 2017, 3:30 PM - 5:00 PM Expanding world of RNAs mrna, messenger RNA (~20,000) trna, transfer
MicroRNAs, RNA Modifications, RNA Editing Bora E. Baysal MD, PhD Oncology for Scientists Lecture Tue, Oct 17, 2017, 3:30 PM - 5:00 PM Expanding world of RNAs mrna, messenger RNA (~20,000) trna, transfer
Supplementary Figure 1: Experimental design. DISCOVERY PHASE VALIDATION PHASE (N = 88) (N = 20) Healthy = 20. Healthy = 6. Endometriosis = 33
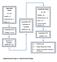 DISCOVERY PHASE (N = 20) Healthy = 6 Endometriosis = 7 EAOC = 7 Quantitative PCR (mirnas = 1113) a Quantitative PCR Verification of Candidate mirnas (N = 24) VALIDATION PHASE (N = 88) Healthy = 20 Endometriosis
DISCOVERY PHASE (N = 20) Healthy = 6 Endometriosis = 7 EAOC = 7 Quantitative PCR (mirnas = 1113) a Quantitative PCR Verification of Candidate mirnas (N = 24) VALIDATION PHASE (N = 88) Healthy = 20 Endometriosis
