A Network Analysis of Chinese Medicine Lianhua-Qingwen Formula
|
|
|
- Lesley Booth
- 5 years ago
- Views:
Transcription
1 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Supplementary Information File A Network Analysis of Chinese Medicine Lianhua-Qingwen Formula to Identify its Main Effective Components Chun-Hua Wang, #a Yi Zhong, #b Yan Zhang, a Jin-Ping Liu, a Yue-Fei Wang, a Wei-Na Jia, a Guo-Cai Wang, * c Zheng Li* a, Yan Zhu a and Xiu-Mei Gao a a Tianjin Key Laboratory of Modern Chinese Medicine, Tianjin University of Traditional Chinese Medicine, Tianjin , China; b Pharmaceutical Informatics Institute, College of Pharmaceutical Sciences, Zhejiang University, Hangzhou , China; c Insitute of TCM and Natural Medicine, Jinan University, Guangzhou , China. # The authors contributed equally to this work. * Correspondence should be addressed to Dr. Zheng Li (lizheng1@gmail.com), phone: and Dr. Guo-Cai Wang (twangguocai@jnu.edu.cn), Phone:
2 Table S1. Relationship between candidate components and targets Candidate component Target Candidate component Target (+)-Pinoresinol-β-D-glucoside Chrysophanol glucoside F7 22-Acetoxyglycyrrhizin Chrysophanol glucoside PTGS2 22β-Acetoxy licorice saponin B2/uralsaponin F Chrysophanol glucoside TOP2A 3,4-Dicaffeoylquinic acid Chrysophanol glucoside TOP2B 3,5-Dicaffeoylquinic acid Citric acid AKR1B1 Amygdalin BAX Citric acid AKR1B1 0 Amygdalin CAT Citric acid CPB2 Amygdalin MTOR Citric acid CTSD Arctiin ADRB2 Citric acid F10 Arctiin F10 Citric acid GABRA1 Arctiin Arctiin HSP90AA 1 Citric acid GRIA2 HSP90AB 1 Citric acid GRIK2 Arctiin KCNH2 Citric acid GRIN1 Arctiin MUC1 Citric acid HDC Arctiin NCOA1 Citric acid IGHG1 Arctiin PTGS2 Citric acid NOS1 Arctiin PTON1 Citric acid PIPN1 Arctiin SCN5A Citric acid PLG Chlorogenic acid AKR1B1 Citric acid PTGS2 Chlorogenic acid CASP3 Citric acid SRC Chlorogenic acid PTGS2 Citric acid TPI1 Chlorogenic acid TYR Cryptochlorogenic acid
3 Chlorogenic acid UGT1A10 Emodin ACTA2 Chlorogenic acid UGT1A7 Emodin BTK Chlorogenic acid UGT1A8 Emodin CALM1 Chrysophanol CALM1 Emodin CASP3 Chrysophanol GABRA1 Emodin CDKN1A Chrysophanol Chrysophanol HSP90AA 1 Emodin CDKN1B HSP90AB 1 Emodin CDKN1C Chrysophanol IGHG1 Emodin CSF2 Chrysophanol NCOA2 Emodin CYP1A1 Chrysophanol PDE3A Emodin EGF Chrysophanol PIK3CG Emodin F10 Chrysophanol PKIA Emodin F7 Chrysophanol PRKACA Emodin Chrysophanol PTGS1 Emodin HSP90A A1 HSP90A B1 Chrysophanol PTGS2 Emodin IL1B Chrysophanol SCN5A Emodin MAOB Chrysophanol glucoside CALM1 Emodin MMP1 Chrysophanol glucoside F10 Emodin MMP9 Emodin NCOA1 Formononetin NDUFB6 Emodin NCOA2 Formononetin NOS2 Emodin PIK3CG Formononetin NOS3 Emodin PPARG Formononetin PDE3A Emodin PRKACA Formononetin PIM1 Emodin PRKCD Formononetin PPARG Emodin PRKCE Formononetin PPARG
4 Emodin PTGS1 Formononetin PPKACA Emodin PTGS2 Formononetin PRKAR1 A Emodin SLC2A1 Formononetin PRSS1 Emodin SLC2A4 Formononetin PTGS1 Emodin TGFB1 Formononetin PTGS2 Emodin TNF Formononetin RXRA Emodin TOP2A Formononetin SIRT1 Emodin TOP2B Forsythoside A AKR1B1 Emodin TP53 Forsythoside A PRKCA Emodin-8-O- glucoside TOP2A Forsythoside A PRKCB Emodin-8-O- glucoside TOP2B Forsythoside E Emodin-8-O- glucoside PTGS2 Gallic acid ABCB1 Formononetin ACHE Gallic acid AKT1 Formononetin ADRA1A Gallic acid ATM Formononetin ADRB2 Gallic acid CASP3 Formononetin AR Gallic acid CASP9 Formononetin ATP5B Gallic acid CHUK Formononetin ATR Gallic acid CYP3A4 3 Formononetin CALM1 Gallic acid EIF2AK3 Formononetin CCNA2 Gallic acid FASN Formononetin CDK2 Gallic acid Formononetin CHRM1 Gallic acid HSP90A A1 HSP90A B1 Formononetin DPP4 Gallic acid JUN Formononetin ESR1 Gallic acid MAOB Formononetin ESR2 Gallic acid MGST1
5 Formononetin F2 Gallic acid MMP2 Formononetin F2R Gallic acid MMP9 Formononetin GSK3B Gallic acid PGR Formononetin HSD3B1 Gallic acid PIK3CG Formononetin HSD3B2 Gallic acid PRKCA Formononetin Formononetin HSP90AA 1 Gallic acid PTGS1 HSP90AB 1 Gallic acid PTGS2 Formononetin IL4 Gallic acid PTPN1 Formononetin JUN Gallic acid SULT1A 1 Formononetin LACTB Gallic acid TNFSF6 Formononetin MAOB Gallic acid TOP2A Formononetin MAPK14 Gallic acid TOP2B Gallic acid TP53 Isoliquiritigenin AR glucoside ADPGK Isoliquiritigenin BAX glucoside AMY1C Isoliquiritigenin CA2 glucoside B4GALT1 Isoliquiritigenin CCNA2 glucoside F7 Isoliquiritigenin ESR1 glucoside GABRA1 Isoliquiritigenin ESR2 glucoside GALM Isoliquiritigenin FOS glucoside GBA Isoliquiritigenin GABBR1 glucoside GCK Isoliquiritigenin GSK3B glucoside HK1 Isoliquiritigenin glucoside IGHG1 Isoliquiritigenin HSP90A A1 HSP90A B1 glucoside LALBA Isoliquiritigenin JAK2
6 glucoside LCT Isoliquiritigenin LACTB glucoside PTGS1 Isoliquiritigenin MAOB glucoside PTGS2 Isoliquiritigenin MAPK14 Glycycoumarin Isoliquiritigenin MT2A Glycyrrhizic acid HS2ST1 Isoliquiritigenin NCOA2 Glycyrrhizic acid HSD11B1 Isoliquiritigenin NOS2 Glycyrrhizic acid HSD11B2 Isoliquiritigenin PIK3CG Glycyrrhizic acid HSD17B6 Isoliquiritigenin PIM1 Glycyrrhizic acid NR1I2 Isoliquiritigenin PKIA Glycyrrhizic acid NR3C1 Isoliquiritigenin PRKACA Glycyrrhizic acid PTGS1 Isoliquiritigenin PTGS1 Glycyrrhizic acid PTGS2 Isoliquiritigenin PTGS2 Glycyrrhizic acid PTPN2 Isoliquiritigenin SELE Glycyrrhizic acid UST Isoliquiritigenin SLC2A1 Hesperidin BAX Isoliquiritigenin TYR Hesperidin CASP3 Isoliquiritigenin VCAM1 Hesperidin ICAM1 Isoliquiritin AKR1B1 Hesperidin PTGS2 Kaempferol ACHE Hesperidin TIAM2 Kaempferol ADRA1B Hesperidin TP53 Kaempferol AHR Hyperin PTGS2 Kaempferol AHSA1 Hyperin TOP2A Kaempferol AKR1C3 Hyperin TOP2B Kaempferol ALOX5 Hyperin Aaur_2198 Kaempferol AR Hyperin ADRB2 Kaempferol BAX Hyperin AR Kaempferol BCL2 Hyperin BAX Kaempferol CALM1 Hyperin CA2 Kaempferol CASP3
7 Hyperin CCNA2 Kaempferol CHRM1 Hyperin TOP2A Kaempferol CHRM2 Hyperin TOP2B Kaempferol CYP1A1 Isoliquiritigenin ADRB2 Kaempferol CYP1A2 Kaempferol CYP1B1 Kaempferol VCAM1 Kaempferol CYP3A4 Kaempferol-3-O-rutinoside Kaempferol DIO1 Licorice saponin B2 Kaempferol DPP4 Licorice saponin E2 Kaempferol F2 Licorice saponin G2 Kaempferol F7 Licorice saponin H2 Kaempferol GABRA1 Liquiritigenin ADRB2 Kaempferol GABRA2 Liquiritigenin ESR1 Kaempferol GSTM1 Liquiritigenin ESR2 Kaempferol GSTM2 Liquiritigenin Kaempferol GSTP1 Liquiritigenin HSP90A A1 HSP90A B1 Kaempferol HAS2 Liquiritigenin LACTB Kaempferol HMOX1 Liquiritigenin MAOB Kaempferol HO1 Liquiritigenin PIK3CG Kaempferol Kaempferol HSP90AA 1 Liquiritigenin PKIA HSP90AB 1 Liquiritigenin PPKACA Kaempferol ICAM1 Liquiritigenin PTGS1 Kaempferol IKBKB Liquiritigenin PTGS2 Kaempferol IL1B Liquiritigenin RARA Kaempferol INSR Liquiritin Kaempferol MMP1 Liquiritin apioside
8 Kaempferol NCOA2 Loganic acid CA2 Kaempferol NOS2 Loganic acid DPP4 Kaempferol NOS3 Loganic acid F2 Kaempferol NR1I2 Loganic acid GABRA1 Kaempferol NR1I3 Loganic acid GABRA6 Kaempferol PGR Loganic acid GRID2 Kaempferol PIK3CG Loganic acid GRM2 Kaempferol PPARG Loganic acid PRSS1 Kaempferol PRKACA Loganic acid PTGS2 Kaempferol PRSS1 Loganin CA2 Kaempferol PSMD3 Loganin DPP4 Kaempferol PTGES Loganin F2 Kaempferol PTGS1 Loganin F2R Kaempferol PTGS2 Neochlorogenic acid C4H Kaempferol RPS6KA3 Neochlorogenic acid Kaempferol SELE Neochlorogenic acid CYP98A 3 CYP98A 8 Kaempferol SLC2A4 Neochlorogenic acid PTGS2 Kaempferol STAT1 Ononin AR Kaempferol TJP2 Ononin CALM1 Kaempferol TNF Ononin DPP4 Kaempferol TNFRSF1 1B Ononin ESR1 Kaempferol TOP2A Ononin ESR2 Kaempferol TOP2B Ononin F2 Ononin F2R Rutin ALOX5 Ononin F7 Rutin C5AR1 Ononin NOS2 Rutin C5AR2
9 Ononin PPARG Rutin CASP3 Ononin PTGS2 Rutin CAT Phillyrin CALM1 Rutin CHRM5 Phillyrin HSP90AA 1 Rutin CTGF Phillyrin NCOA2 Rutin CYP2C8 Phillyrin PIK3CG Rutin CYP2C9 Phillyrin PRKACA Rutin CYP2D6 Phillyrin PTGS1 Rutin CYP3A4 Phillyrin PTGS2 Rutin DIO1 Polydatin Polydatin Rutin FCER2 Polydatin Polydatin Rutin GSTP1 Polydatin Polydatin Rutin HAS2 Polydatin Polydatin Rutin HMGCR Quercetin NCOA2 Rutin HSPA4 Quercetin PRKACA Rutin IL1B Quinic acid IMPA1 Rutin IL6 Quinic acid IMPA2 Rutin IL8 Quinic acid PTGS2 Rutin INS Rhein AKR1B1 Rutin ITGB2 Rhein AKR1B10 Rutin POR Rhein BAX Rutin PRKCB Rhein CASP3 Rutin SOD1 Rhein CASP8 Rutin TBXA2R Rhein Rhein HSP90AA 1 Rutin TNF HSP90AB 1 Rutin TOP2A Rhein LOX Rutin TOP2B
10 Rhein NCOA2 Salidroside CASP3 Rhein NFKB1 Salidroside HIF1A Rhein PIK3CG Salidroside PARP1 Rhein PTGS1 Secoxyloganin CA2 Rhein PTGS2 Secoxyloganin DPP4 Rhein RARA Secoxyloganin F2 Rhein RELA S-suspensaside Rhein RXRA Sweroside CA2 Rhein SRD5A2 Sweroside F2 Rhein VEGFA Sweroside GABRA1 Rhodiosin Sweroside GABRA2 R-suspensaside Sweroside NQO2 Rutin ADRA2A Sweroside PRSS1 Rutin ADRA2C Tricin AR Rutin AKR1C3 Tricin CALM1 Tricin CDK2 Tricin PTGS1 Tricin DPP4 Tricin PTGS2 Tricin ESR1 Tricin SCN5A Tricin ESR2 Tricin SULT1A 1 Tricin F7 Isoliquiritin apioside Tricin GSK3B Isomer of liquiritin apioside Tricin HSP90AA 1 Isomer of Rengynic acid-1 -O-β-Dglucoside Tricin HSP90AB 1 Isomer1 of Chrysophanol Tricin MAPK14 Isomer1 of Forsythoside A Tricin NCOA2 Isomer2 of Chrysophanol Tricin NOS2 Physcion-8-O-β-D-glucopyranoside
11 Tricin PPARG Rhododendrol-4 -O-β-Dglucopyranoside Tricin PRSS1 Rengynicacid-1 -O-β-D-glucoside
12 Table S2. Interactions of targets in influenza-related PPI network Target1 Target2 Target1 Target2 Target1 Target2 Target1 Target CD86 DUSP9 MAPK1 LAPTM5 EPN1 PTPRE MAPK1 ABCF1 EIF6 DUSP9 MAPK14 LBP CD14 PTPRH MAPK1 ABL1 CDKN1B DYRK1B MAPK1 LCK CD247 PTPRJ IL2RG ACLY GSK3B DYRK1B MAPK14 LGALS2 LTA PTPRR MAPK1 ACP5 SPP1 E2F1 GSK3B LGALS3BP CD14 PTX3 C1QB ACP5 TNFSF10 E2F1 NFKB1 LRP2 TTR PTX3 C1QC ACTN4 CAMK2B EDAR TRAF1 LTA4H ARPC3 PTX3 CFH ACVR1 IKBKB EEF1A1 TTR LTA4H TMEM62 PTX3 FGF2 ADAMTS4 SERPINA3 EEF1D FOS LTB LTA PXN MAPK1 ADRB2 IL1B EEF1G CXCL13 LTBR LTA PXN SELE AES CCL7 EEF1G HDAC5 LUC7L3 CLK1 PYCARD CASP1 AFAP1L2 RASA1 EGFR ELF3 LY6E CD247 PYCARD MEFV AFAP1L2 SRC EGFR MAP2K1 LY96 TLR2 PYCARD NLRC4 AIMP1 AIMP2 EGFR MAPK1 LY96 TLR4 PYCARD NLRP3 AIMP1 ATP5A1 EGFR PIK3R2 LYN CDKN1B PYCARD SRF AIMP1 MAPK14 EIF2AK3 DNAJC3 MAF FOS PYDC2 PYCARD AIMP1 RARS EIF2AK3 IKBKB MAGEA11 ILF3 RAB27A GZMB AKT1 CDKN1B EIF2AK4 SMAD1 MALT1 BCL10 RAC1 TLR2 AKT1 EIF4EBP1 EIF2AK4 SMAD4 MALT1 SQSTM1 RAF1 MAP2K1 AKT1 GSK3B EIF2AK4 TGFBR1 MALT1 TRAF2 RAF1 MAP2K2 AKT1 IKBKB EIF4E2 EIF4EBP1 MALT1 TRAF6 RANBP2 HDAC4 ALOX5 ACTB EIF4EBP1 CHTF8 MAP1B GSK3B RANBP2 MDM2 ALOX5 GRB2 EIF4EBP1 EIF4E MAP2K1 CASP9 RANBP2 OPN1LW ALOX5AP ALOX5 EIF4EBP1 MVD MAP2K1 ELK1 RANBP2 OPN1MW ANKRD11 HDAC4 EIF4EBP2 EIF4E MAP2K1 EP300 RANBP2 RAN
13 ANKRD11 HDAC5 ELANE CXCR4 MAP2K1 MAPK14 RANGAP1 RANBP2 APC GSK3B ELANE SERPINA3 MAP2K1 PARVA RB1 FOS APOA1 TTR ELF1 NFKB1 MAP2K1 PRKCI RBP4 TTR AR GSK3B ELF3 EP300 MAP2K1 PRKCZ RBPJ MAML1 AR NFKB1 ELF3 NFKB1 MAP2K1 RPS6KA4 RELA IRF3 ARAF MAP2K1 ELK1 CEBPB MAP2K1 TRAF6 RET MAPK1 ARHGDIA CDKN1B ELK1 FOS MAP2K2 ARAF RET MAPK14 ARRB2 CXCR4 ELK1 MAPK1 MAP2K2 BRAF RFX5 CIITA ATF3 NFKB1 ELK4 FOS MAP2K2 CASP9 RFXANK CIITA ATF4 CEBPB ENO2 IL17B MAP2K2 MAPK1 RGS12 MAP2K2 ATF7 FOS EP300 CEBPB MAP2K2 MAPK3 RHEBL1 EIF4EBP1 ATM EIF4EBP1 EP300 IRF3 MAP2K3 MAPK14 RHOXF2 TOLLIP ATN1 PSME3 EP300 IRF7 MAP2K4 MAPK14 RIPK1 NFKB1 ATP5B PSME3 EPHA2 PIK3R2 MAP2K6 MAPK14 RIPK1 TICAM1 ATXN1 TOLLIP EPHB2 FOS MAP3K1 MAPK1 RIPK1 TRAF1 ATXN3 CASP1 EPN1 AP2A1 MAP3K13 IKBKB RIPK2 BCL10 AURKA GSK3B EPN1 AP2A2 MAP3K14 IKBKB RIPK2 BIRC2 AXL IL2RG EPN1 CLTC MAP3K14 TRAF1 RIPK2 CASP1 BAT5 PTGS2 EPN1 KIAA1377 MAP3K3 IKBKB RIPK2 CASP8 BATF FOS EPN1 MED31 MAP3K4 MAP2K1 RIPK2 CD40 BCAP31 CASP1 EPN1 REPS2 MAP3K7 FOS RIPK2 IKBKG BCL2 MAPK14 EPS15 ELF3 MAP3K7 IKBKB RIPK2 NGFR BCL3 GSK3B EPS15 EPN1 MAP3K7IP1 MAPK14 RIPK2 SMAD4 BCL6 MAPK1 EPS15L1 EPN1 MAP3K8 NFKB1 RIPK2 SMURF1 BCOR HDAC4 ERBB2 SERPINA3 MAP6 TRAF1 RIPK2 TNFRSF1A BIRC2 TRAF1 ERBB3 PIK3R2 MAPK1 AR RIPK2 TRAF1 BIRC3 TRAF1 ERBB4 PIK3R2 MAPK1 ARRB2 RIPK2 TRAF2 BIRC6 MAP2K1 ESR1 NFKB1 MAPK1 BCL2 RIPK2 TRAF5
14 BLM MX1 ETS1 NFKB1 MAPK1 CAD RIPK2 TRAF6 BRAF MAP2K1 ETV6 HDAC9 MAPK1 CALCOCO1 RIPK2 UBE2I BRAF MAPK1 F2 SPP1 MAPK1 CASP9 RIPK3 TICAM1 BRMS1 HDAC4 F5 SERPINA3 MAPK1 CAV1 RNF11 EPN1 BRMS1 HDAC5 FADD MYD88 MAPK1 CD19 RNF216 TLR3 BTK MYD88 FAM48A MAPK14 MAPK1 CEBPB RNF216 TLR4 BTK TLR4 FANCA IKBKB MAPK1 CMTM3 RNF216 TLR5 BTK TLR6 FCER2 CR2 MAPK1 DUSP1 RNF216 TLR9 BTK TLR9 FCGR3A CD247 MAPK1 DUSP16 RNF5 CFTR BUB1 EIF4EBP1 FGFR1 PIK3R2 MAPK1 DUSP2 RORC EIF4EBP1 C14orf166 NIN FHL3 MAPK1 MAPK1 DUSP5 RPS3 NFKB1 C1QBP MAPK1 FHOD1 CR2 MAPK1 DUSP6 RPS6KA1 CEBPB C2 C3 FKBP1A RYR1 MAPK1 EIF4EBP1 RPS6KA1 FOS C3 CR2 FLII MYD88 MAPK1 EPOR RPS6KA2 MAP2K1 C3 LRP1 FLNA SELE MAPK1 ERF RPS6KA2 MAPK1 C3AR1 C3 FOS ATF2 MAPK1 ESR1 RPS6KA3 MAPK1 C3AR1 C4A FOS BCL3 MAPK1 ETS1 RPS6KA4 FOS C5 C3 FOS CEBPG MAPK1 FCGR2B RPS6KA4 MAPK1 CAMK1 HDAC5 FOS CSNK2A1 MAPK1 FOS RPS6KA5 EIF4EBP1 CAMK2A CEBPB FOS CSNK2A2 MAPK1 FRS2 RPS6KB1 EIF4EBP1 CAMK2B CAMK2D FOS DDIT3 MAPK1 FRS3 RPS6KB1 MAPK1 CAMK2B COL4A3 FOS ETS1 MAPK1 GAB1 RPTOR EIF4EBP1 CAMK2B ETS1 FOS HNF1A MAPK1 GAB2 RSF1 NFKB1 CAMK2B GRIN2A FOS JUNB MAPK1 GABRR1 RUNX2 CEBPB CAMK2B ITPKA FOS LMNA MAPK1 GATA1 RUNX2 FOS CAMK2B PLCB3 FOS MITF MAPK1 GJA1 RXRA NFKB1 CAMK2G RYR1 FOS NFKB1 MAPK1 GORASP2 RXRG CAMK2B CAMK2N2 CAMK2B FOS RELA MAPK1 GRB10 RYR1 CACNA1S
15 CAMK4 HDAC4 FOS RUNX1 MAPK1 GRB2 RYR1 CALM1 CAMTA2 HDAC5 FOS SMAD3 MAPK1 HSF4 RYR1 S100A1 CAPN1 IL1A FOS STAT1 MAPK1 HSP90AA1 RYR2 RYR1 CARD16 CASP1 FOXK2 IL2 MAPK1 IER3 S100A13 IL1A CARD16 RIPK2 FOXO3 IKBKB MAPK1 IFNAR1 SCHIP1 FOS CARD17 CASP1 FRAT2 GSK3B MAPK1 IQGAP1 SDC1 IL8 CARD6 NOD1 G3BP2 NFKB1 MAPK1 IRS1 SDC4 CCL5 CARD6 RIPK2 GAB2 CD247 MAPK1 KHDRBS1 SELE ACTN2 CARD8 CASP1 GADD45A MAPK14 MAPK1 KRT8 SELE CD44 CARD8 NLRP3 GAPDH CAMK2B MAPK1 LCK SELL SELE CASP1 AR GATA2 MAPK1 MAPK1 LIFR SELPLG SELE CASP1 ATN1 GATA4 FOS MAPK1 LIPE SERF2 PSME3 CASP1 BCL2L1 GCM1 HDAC4 MAPK1 LRPAP1 SERPINA3 APP CASP1 BID GCM1 HDAC5 MAPK1 MAFA SERPINA3 HOXB1 CASP1 CDC2L2 GDF9 ILF3 MAPK1 MAP2K1 SERPINA3 SERPINA3 CASP1 CDK11B GLG1 SELE MAPK1 MBP SERPINB9 GZMB CASP1 EGFR GMFB MAPK1 MAPK1 MCL1 SERPING1 SELE CASP1 FGF2 GMFB MAPK14 MAPK1 MYB SETDB1 TOLLIP CASP1 HTT GNA14 CCR1 MAPK1 NCOA1 SFTPD DCN CASP1 IL18 GNB1 HDAC5 MAPK1 NCOA3 SGK1 GSK3B CASP1 IL1A GOPC CFTR MAPK1 NDE1 SGK3 GSK3B CASP1 IL1B GPR77 C3 MAPK1 NEFH SGK3 ITCH CASP1 LMNA GRB10 MAP2K1 MAPK1 NR3C1 SH2B3 CD247 CASP1 MAPT GRB2 CD247 MAPK1 NR4A1 SHB IL2RG CASP1 NEDD4 GRIN1 MAP2K2 MAPK1 NR5A1 SHC1 CD247 CASP1 PARP1 GRIN2B CAMK2B MAPK1 PAK1 SHC1 IL2 CASP1 PLA2G4A GRIN2D MAP2K2 MAPK1 PAK2 SHC1 IL2RG CASP1 PSEN1 GRIP1 IRF3 MAPK1 PLAT SHC1 MAPK1
16 CASP1 PSEN2 GSK3B AKAP11 MAPK1 PLCB1 SHC1 MAPK14 CASP1 TFAP2A GSK3B AKT2 MAPK1 PPARA SIGIRR TLR4 CASP10 TRAF1 GSK3B APP MAPK1 PPARG SIGIRR TLR5 CASP14 CASP1 GSK3B AXIN1 MAPK1 PPP1CA SIGIRR TLR9 CASP3 IL18 GSK3B AXIN2 MAPK1 PPP1R9B SIGLEC12 CMAS CASP3 PSME3 GSK3B BICD1 MAPK1 PPP2CA SIN3A HDAC9 CASP3 TRAF1 GSK3B BICD2 MAPK1 PRKCD SIN3A NFKB1 CASP4 IL18 GSK3B CABYR MAPK1 PRKCZ SIN3B HDAC9 CASP4 IL1B GSK3B CCND1 MAPK1 PTPN5 SIRPA IL1RAP CASP6 PSME3 GSK3B CCNE1 MAPK1 PTPN7 SKP2 CDKN1B CASP6 TRAF1 GSK3B CDH1 MAPK1 RAF1 SLA CD247 CASP7 PSME3 GSK3B CDK5 MAPK1 RB1 SLA2 CD247 CASP8 CDKN1B GSK3B CEBPA MAPK1 RGS19 SLAMF7 SH2D1A CASP8 MAPK1 GSK3B CREB1 MAPK1 RPS6KA1 SLC12A9 CPSF4 CASP8 TRAF1 GSK3B CREM MAPK1 RXRA SLC35A2 MDFI CAV1 PTGS2 GSK3B CTNNB1 MAPK1 SCNN1G SLC35A2 PLSCR1 CBL ITCH GSK3B CTNND1 MAPK1 SH2D3C SLC4A7 CFTR CBLC ITCH GSK3B DCTN1 MAPK1 SLC9A1 SLC4A8 CFTR CCBP2 CCL13 GSK3B DCTN2 MAPK1 SMAD1 SLC9A3R1 CFTR CCBP2 CCL19 GSK3B DPYSL2 MAPK1 SMAD2 SLC9A3R2 CFTR CCBP2 CCL2 GSK3B DYNC1I1 MAPK1 SMAD3 SMAD3 CEBPB CCBP2 CCL21 GSK3B EIF2B5 MAPK1 SMAD4 SMAD3 MECOM CCBP2 CCL3 GSK3B FOXO1 MAPK1 SNCA SMAD4 CEBPB CCBP2 CCL4 GSK3B FRAT1 MAPK1 SORBS3 SMAD7 MAPK14 CCBP2 CCL5 GSK3B GYS1 MAPK1 SOS1 SMARCA4 CEBPB CCBP2 CCL7 GSK3B HNRNPD MAPK1 SP1 SMARCB1 CEBPB CCL13 CCR2 GSK3B ILK MAPK1 SREBF1 SMARCC1 CEBPB CCL13 CCR3 GSK3B LRP6 MAPK1 STAT3 SMARCD1 FOS
17 CCL13 CCR5 GSK3B MAPT MAPK1 TCF3 SNAI1 GSK3B CCL14 CCR1 GSK3B MARK2 MAPK1 TGIF1 SNAP23 CFTR CCL14 CCR3 GSK3B MCL1 MAPK1 TNFRSF1A SNCG MAPK1 CCL15 CCR1 GSK3B MITF MAPK1 TNFRSF25 SOCS1 CXCR4 CCL15 CCR3 GSK3B MUC1 MAPK1 TOB1 SOCS3 CXCR4 CCL16 CCR1 GSK3B MYOCD MAPK1 TP53 SOCS6 PIK3R2 CCL17 VCAM1 GSK3B NFKB1 MAPK1 TSC2 SON PRPF40A CCL19 CCR10 GSK3B NOTCH1 MAPK1 UBTF SON SNIP1 CCL19 CCR7 GSK3B NOTCH2 MAPK1 VAV1 SOS1 PIK3R2 CCL19 CCRL1 GSK3B OGT MAPK1 ZFP36 SPAG9 MAPK14 CCL2 DARC GSK3B PPP1R2 MAPK14 ATF2 SPAG9 NFKB1 CCL2 VCAN GSK3B PRKCB MAPK14 CDC25C SPI1 NFKB1 CCL20 CCR6 GSK3B PSEN1 MAPK14 CREB1 SPIB CEBPB CCL20 VCAN GSK3B PTK2 MAPK14 CSNK2A2 SPOP TRAF1 CCL21 CCR7 GSK3B PTPN1 MAPK14 CSNK2B SPP1 ABCF3 CCL21 CCRL1 GSK3B PXN MAPK14 DDIT3 SPP1 CTNNBL1 CCL21 UBQLN4 GSK3B RCAN1 MAPK14 EEA1 SPP1 IGFBP5 CCL21 VCAN GSK3B SNCAIP MAPK14 EEF2K SPP1 ITGA5 CCL22 CCL19 GSK3B TP53 MAPK14 EIF4EBP1 SPP1 ITGB1 CCL23 CCR1 GTF2F2 FOS MAPK14 ELK1 SPP1 PDLIM7 CCL24 CCR3 GZMB BID MAPK14 ELK3 SPP1 SGTA CCL26 CCR3 GZMB CASP3 MAPK14 ESR1 SPP1 UBQLN4 CCL28 CCR3 GZMB CASP7 MAPK14 FKBP8 SRC CDKN1B CCL3 SRGN GZMB CASP9 MAPK14 FLNA SRC IKBKB CCL3L1 CCR1 GZMB CHRM3 MAPK14 GRB2 SRC TRAF1 CCL3L1 CCR3 GZMB DFFA MAPK14 HNF4A SREBF2 MAPK1 CCL4 CCL3 GZMB GRIA3 MAPK14 HSF4 SRF CEBPB CCL4 CCR3 GZMB LMNB1 MAPK14 JDP2 SRGN GZMB
18 CCL4 IL8 GZMB PARP1 MAPK14 JUNB ST3GAL3 TTR CCL5 DARC GZMB PRKDC MAPK14 KRT18 STAT1 CXCR4 CCL5 GRB2 GZMB PTK2 MAPK14 KRT8 STAT1 IL2RG CCL5 PIGR GZMB UBE4A MAPK14 MAFA STAT1 MAPK14 CCL5 SDC1 GZMB UBE4B MAPK14 MAP3K7 STAT2 CXCR4 CCL5 VCAN GZMB XRCC4 MAPK14 MAPK1 STAT3 CXCR4 CCL7 CCL7 HAX1 IL1A MAPK14 MAPKAPK2 STAT5A CD247 CCL7 FEZ1 HDAC1 MECOM MAPK14 MAPKAPK5 STAT5A CEBPB CCL8 CCR1 HDAC1 NFKB1 MAPK14 MARS STAT5A IL2RG CCL8 CCR3 HDAC3 HDAC4 MAPK14 MAX STAT5A MAPK1 CCNA1 CDKN1B HDAC3 HDAC9 MAPK14 MEF2A STAT5B CD247 CCND2 CDKN1B HDAC4 ANKRA2 MAPK14 MEF2C STAT5B CXCR4 CCNE2 CDKN1B HDAC4 ATRX MAPK14 NCF1 STAT5B MAPK1 CCR1 CCL2 HDAC4 BCL6 MAPK14 NCOA3 STAT6 CEBPB CCR1 CCL26 HDAC4 CBX5 MAPK14 NFATC1 STAT6 NFKB1 CCR1 CCL3 HDAC4 CIITA MAPK14 NFATC4 STK39 MAPK14 CCR1 CCL4 HDAC4 MAP1S MAPK14 NUP153 SUMO1 FOS CCR1 CCL5 HDAC4 MAPK1 MAPK14 PLA2G4A SUMO1 HDAC4 CCR1 CCL7 HDAC4 MAPK3 MAPK14 PML SUMO2 FOS CCR1 CREB3 HDAC4 MECOM MAPK14 PPARGC1A SUMO3 FOS CCR1 JAK1 HDAC4 MEF2A MAPK14 PTPN7 SUMO4 FOS CCR1 PLP2 HDAC4 MEF2C MAPK14 RPS6KA4 SYK TLR4 CCR1 STAT1 HDAC4 MEF2D MAPK14 SCN8A TAF1 FOS CCR1 STAT3 HDAC4 NCOR2 MAPK14 SLC9A1 TAF6 CIITA CCR10 CCL2 HDAC4 NR2C1 MAPK14 SMAD3 TAF9 CEBPB CCR10 CCL7 HDAC4 RBBP4 MAPK14 STAT4 TAF9 ELF3 CCR2 CCL2 HDAC4 REV3L MAPK14 ZFP36L1 TANK IKBKB CCR2 CCL7 HDAC4 RFXANK MAPK14 ZFYVE20 TANK TRAF1
19 CCR3 CCL11 HDAC4 RUNX2 MAPK3 MAP2K1 TBK1 TICAM1 CCR3 CCL3 HDAC4 RUNX3 MAPK3 MAPK14 TBP FOS CCR3 CCL5 HDAC4 RUVBL2 MAPK8 MAPK1 TFRC CD247 CCR3 CCL7 HDAC4 SUMO2 MAPK8 MAPK14 TGFB1 CCL3 CCR3 CXCL10 HDAC4 TNFRSF14 MAPK8IP3 MAP2K1 TGFBR2 PIK3R2 CCR3 FGR HDAC4 YWHAE MAPK9 NFATC3 TGFBRAP1 ALOX5 CCR3 HCK HDAC4 YWHAZ MAPKAPK2 ALOX5 TH MAPK1 CCR4 CCL17 HDAC5 ANKRA2 MAPKAPK3 MAPK14 THRB CAMK2B CCR4 CCL3 HDAC5 BCL6 MAPKAPK5 EIF4EBP1 TICAM1 AZI2 CCR4 CCL5 HDAC5 BCOR MAPKAPK5 MAPK1 TICAM1 IRF3 CCR5 CCL2 HDAC5 CBX5 MAPKSP1 MAP2K1 TICAM1 IRF7 CCR5 CCL3 HDAC5 CIITA MAPKSP1 MAP2K2 TICAM1 MAVS CCR5 CCL4 HDAC5 DDX20 MAPKSP1 MAPK1 TICAM1 PIAS4 CCR5 CCL5 HDAC5 GABARAP MARK3 CPSF4 TICAM1 TLR3 CCR5 CCL7 HDAC5 GATA1 MAVS DDX58 TICAM1 TRAF1 CCR5 CXCR4 HDAC5 GATA2 MBP MAPK14 TICAM1 TRAF6 CCR7 CCR10 HDAC5 HDAC3 MED23 CEBPB TICAM1 TRAM1 CCR8 CCL17 HDAC5 HR MED23 ELF3 TICAM2 IL1RAP CCR8 CCL4 HDAC5 MEF2A MEF2B HDAC4 TICAM2 IRAK2 CCRL1 CCL13 HDAC5 MEF2C MEFV PSTPIP1 TICAM2 IRF3 CCRL1 CCL2 HDAC5 MEF2D MEGF10 HDAC4 TICAM2 IRF7 CCRL1 CCL5 HDAC5 NCOR1 METAP2 MAPK1 TICAM2 TICAM1 CCRL1 CCL7 HDAC5 NCOR2 MFAP4 SFTPD TICAM2 TLR3 CCRL1 CXCL13 HDAC5 RFXANK MITF MAPK1 TICAM2 TLR4 CD14 CD55 HDAC5 RUNX3 MITF MAPK14 TIRAP CD247 CD14 LTF HDAC5 SFN MKNK1 MAPK1 TIRAP IRAK2 CD14 TLR3 HDAC5 SUV39H1 MKNK1 MAPK14 TIRAP MYD88 CD2 CD247 HDAC5 UBC MKNK2 MAPK1 TIRAP TICAM1
20 CD247 CD244 HDAC5 YWHAB MKNK2 MAPK14 TIRAP TLR4 CD247 CD28 HDAC5 YWHAE MMP1 CCL13 TLR1 DHX36 CD247 CD48 HDAC9 BCL6 MMP1 CCL2 TLR1 TLR10 CD247 CD5 HDAC9 CBX5 MMP1 CCL7 TLR1 TLR5 CD247 CSK HDAC9 CTBP1 MMP1 SERPINA3 TLR1 TLR9 CD247 FYN HDAC9 HDAC1 MMP13 CCL7 TLR1 TRAP1 CD247 PTPN3 HDAC9 HDAC4 MMP14 CCL7 TLR2 CD14 CD247 PTPRC HDAC9 MEF2A MMP2 CCL7 TLR2 HMGB1 CD247 ZAP70 HDAC9 NCOR1 MMP2 IL1B TLR2 HSP90B1 CD28 PIK3R2 HDAC9 SUMO1 MMP3 CCL13 TLR2 IRAK1 CD3D CD247 HDAC9 SUMO2 MMP3 CCL2 TLR2 MAPK8IP3 CD4 CXCR4 HIVEP3 TRAF1 MMP3 CCL7 TLR2 RIPK2 CD4 HLA-DPA1 HLA-DPA1 HLA-DPB1 MMP3 SPP1 TLR2 SFTPA1 CD40 BIRC2 HMGA1 CEBPB MMP7 SPP1 TLR2 TICAM1 CD40 BIRC3 HMGA2 NFKB1 MMP8 CCL2 TLR2 TLR1 CD40 CAV1 HMGB1 TLR4 MMP9 IL8 TLR2 TLR10 CD40 CBL HNRNPH3 CCL13 MS4A1 CD40 TLR3 CSK CD40 HSPA4 HNRNPH3 TTR MST1 SERPINA3 TLR3 MYD88 CD40 HSPA8 HNRNPK CEBPB MTOR EIF4EBP1 TLR3 TLR3 CD40 IL4R HOMER1 RYR1 MTPN NFKB1 TLR4 BCL10 CD40 JAK3 HOMER2 RYR1 MX1 ACTB TLR4 CD14 CD40 MAP3K8 HOMER3 RYR1 MX1 TRPC1 TLR4 CNPY4 CD40 PIK3R1 HRAS MAP2K1 MX1 TRPC3 TLR4 HSP90B1 CD40LG CD40 HRAS TLR2 MX1 TRPC4 TLR4 IRAK2 CD53 IL2 HRAS TLR9 MX1 TRPC5 TLR4 MAPK8IP3 CD53 IL4 HSP90B1 TLR1 MX1 TRPC6 TLR4 MYD88 CD74 HLA-DPA1 HSPA5 PSME3 MX1 TRPC7 TLR4 NOX4 CD86 CD28 HSPA8 CXCR4 MYB CEBPB TLR4 RIPK2
21 CD86 CD80 HSPD1 TLR1 MYC CEBPB TLR4 TLR1 CD86 CTLA4 HTRA2 MAPK14 MYC ELF3 TLR4 TLR5 CDC25B MAPK14 IFITM1 CR2 MYC GSK3B TLR6 TLR2 CDC37 IKBKB IFNA1 CR2 MYC MAP2K1 TLR6 TRAP1 CDK1 EPN1 IFNA2 IFNAR1 MYD88 IL1R1 TLR7 MYD88 CDK2 CDKN1B IFNAR2 IFNA2 MYD88 IRAK1 TLR8 MYD88 CDK5 MAP2K1 IFNAR2 IFNB1 MYD88 IRF5 TLR9 MAPK8IP3 CDK8 MAML1 IFNB1 IFNAR1 MYD88 IRF7 TMEM132A GSK3B CDK9 NFKB1 IFNG IFNGR1 MYD88 SPOP TNFAIP3 TICAM1 CDKN1B CCNA2 IFNG IFNGR2 MYD88 STAP2 TNFRSF10A TNFSF10 CDKN1B CCNB1 IFNG TNF MYD88 TLR10 TNFRSF10B TNFSF10 CDKN1B CCND1 IGF1R PIK3R2 MYD88 TLR2 TNFRSF10C TNFSF10 CDKN1B CCND3 IGF2R GZMB MYD88 TLR5 TNFRSF10D TNFSF10 CDKN1B CDK3 IGFBP7 CCL21 MYOC IGLL1 TNFRSF11A TRAF1 CDKN1B CDK4 IGFBP7 CCL5 NACC1 HDAC4 TNFRSF12A TRAF1 CDKN1B CDK5 IGFBP7 CXCL10 NCF1 NFKB1 TNFRSF14 LTA CDKN1B COPS5 IKBKAP IKBKB NCOA1 CIITA TNFRSF14 TRAF1 CDKN1B GRB2 IKBKB CASP8 NCOA1 FOS TNFRSF18 TRAF1 CDKN1B KPNA3 IKBKB CTNNB1 NCOA1 NFKB1 TNFRSF19 TRAF1 CDKN1B KPNA5 IKBKB EIF2AK2 NCOA3 PSME3 TNFRSF1A IKBKB CDKN1B MCM7 IKBKB HSP90AA1 NCOR2 FOS TNFRSF1A LTA CDKN1B SPDYA IKBKB HSP90AB1 NCOR2 NFKB1 TNFRSF1B LTA CDKN1B STMN1 IKBKB IRS1 NCR1 CD247 TNFRSF4 SIVA1 CDKN1B TRAF2 IKBKB KIAA1967 NCR1 CD59 TNFRSF4 TNFRSF9 CDKN1B UCHL1 IKBKB MAP3K11 NCR1 FCER1G TNFRSF4 TRAF1 CDKN1B YWHAB IKBKB NCOA3 NCR3 CD247 TNFRSF4 TRAF4 CDKN1B YWHAG IKBKB NFKB1 NDFIP2 ITCH TNFRSF9 TRAF1 CDKN1B YWHAH IKBKB NFKBIA NDN IL1A TNFRSF9 TRAF2
22 CDKN1B YWHAQ IKBKB NFKBIB NDRG1 DDX1 TNFRSF9 TRAF3 CDX2 GSK3B IKBKB PEBP1 NDRG1 ILF3 TNFSF10 C7orf64 CDX2 MAPK1 IKBKB PRKCQ NEDD9 ITCH TNFSF10 TNFRSF11B CDX2 MAPK14 IKBKB RELA NEK2 MAPK1 TNFSF10 TNFSF10 CEBPB ATF2 IKBKB STAP2 NFATC3 FOS TNFSF11 MAPK1 CEBPB CCL3 IKBKB TBK1 NFATC3 TTF1 TNFSF11 NFKB1 CEBPB CEBPA IKBKB TGFBR1 NFATC4 CREBBP TNFSF4 TNFRSF4 CEBPB CEBPD IKBKB TNFAIP3 NFATC4 GATA4 TNFSF4 TRAF2 CEBPB CEBPG IKBKB TRAF2 NFATC4 YWHAQ TNFSF9 TNFRSF9 CEBPB CREB1 IKBKB YWHAB NFATC4 YWHAZ TNFSF9 TRAF1 CEBPB CREBBP IKBKE TICAM1 NFE2L2 CASP1 TNIP1 MAPK1 CEBPB DDIT3 IKBKG GSK3B NFKB1 BCL3 TNIP2 NFKB1 CEBPB EGFR IKBKG IKBKB NFKB1 BRCA1 TNPO1 RANBP2 CEBPB EGR1 IL10 A2M NFKB1 BTRC TOLLIP CEBPB ESR1 IL10 IL10RB NFKB1 CFLAR TOLLIP DAZAP2 CEBPB FOXO1 IL10RA IL10 NFKB1 CHUK TOLLIP IL1RAP CEBPB HMGB1 IL12A IL8 NFKB1 FBXW11 TOLLIP IRAK1 CEBPB HOMER3 IL13RA1 IL13 NFKB1 HMGB1 TOLLIP LDB1 CEBPB HSF1 IL13RA2 IL13 NFKB1 IKBKG TOLLIP RBPMS CEBPB MAPK3 IL13RA2 IL4 NFKB1 IL2RA TOLLIP TLR2 CEBPB NFKB1 IL15 IL2RG NFKB1 IRF1 TOLLIP TLR4 CEBPB NR3C1 IL15RA IL2RG NFKB1 IRF2 TOLLIP TOM1 CEBPB PTGES2 IL17RA IL17A NFKB1 KPNA3 TOLLIP TOM1L1 CEBPB RB1 IL17RB IL17B NFKB1 LYL1 TP53 CR2 CEBPB RELA IL18BP IL18 NFKB1 MEN1 TP53BP1 NFKB1 CEBPB RPS6KA5 IL18R1 IL18 NFKB1 NFKB2 TP73 ITCH CEBPB RUNX1 IL18R1 IL1RAP NFKB1 NFKBIA TPPP GSK3B CEBPB SP1 IL18RAP IL18 NFKB1 NFKBIB TPR MAPK1
23 CEBPB SPI1 IL1A IL1R1 NFKB1 NFKBIE TPST1 CCR1 CEBPZ LAG3 IL1B A2M NFKB1 NR3C1 TPST1 CCR2 CELA1 SERPINA3 IL1R1 IL1B NFKB1 PDCD11 TPST1 CCR8 CENPJ LAG3 IL1R1 IL1RN NFKB1 REL TPST1 CMKLR1 CEP55 MAPK1 IL1R1 TOLLIP NFKB1 RELA TPST1 GP1BA CFHR4 C3 IL1R2 IL1A NFKB1 RELB TPST1 GPR1 CFHR5 C3 IL1R2 IL1B NFKB1 STAT3 TPST1 SELPLG CFI C3 IL1R2 IL1RN NFKB1 UBE2D3 TPST2 CCR1 CFLAR IKBKB IL1RAP IL1A NFKB2 HDAC4 TRA CD247 CFLAR RIPK2 IL1RAP IL1B NFKB2 HDAC5 TRA2B IVNS1ABP CFLAR TRAF1 IL1RAP IL1R1 NFKB2 IKBKB TRADD TRAF1 CFTR CANX IL1RAP IL1R2 NFKBIE IL1A TRAF1 CD40 CFTR EZR IL1RAP IRAK1 NFKBIZ NFKB1 TRAF1 GFI1B CFTR PDZD3 IL1RAP MYD88 NFRKB NFKB1 TRAF1 TNFAIP3 CFTR PRKAR2A IL1RAP PIK3R1 NGFR IL2 TRAF1 TNFRSF17 CFTR PRKCE IL1RAP PRPF40A NGFR MAPK1 TRAF1 TNFRSF1A CFTR SLC9A6 IL1RAP RAC1 NGFRAP1 TRAF1 TRAF1 TNFRSF1B CFTR STX1A IL1RAP STAT3 NIN GSK3B TRAF1 TNFRSF8 CHD3 IVNS1ABP IL1RL1 MYD88 NKRF NFKB1 TRAF1 TRAF2 CHGB MOGS IL1RL2 IL18 NKX2-1 MAPK1 TRAF1 TRAF3IP2 CHUK IKBKB IL2 IFNA1 NLRC4 CASP1 TRAF2 CD40 CIITA NFYB IL2 IL2RB NLRP1 CASP1 TRAF2 TNFRSF4 CIITA ZXDC IL22 IL10RB NLRP1 NOD1 TRAF3 CD40 CKS1B CDKN1B IL22RA1 IL22 NLRP1 PYCARD TRAF3 MAP2K1 CLK1 GOLM1 IL22RA2 IL22 NLRP12 PYCARD TRAF3 TICAM1 CLK1 MBP IL23A IL12RB1 NOD1 CASP1 TRAF3 TNFRSF4 CLK1 PIAS4 IL23R IL23A NOD1 CASP2 TRAF3IP2 CD40 CLK1 PPIG IL28RA IL29 NOD1 CASP4 TRAF3IP2 IKBKB
24 CLK1 PRPF4 IL2RA IL2 NOD1 CASP8 TRAF4 TICAM1 CLK1 PTPN1 IL2RG CAPNS1 NOD1 CASP9 TRAF5 CD40 CLK1 SFRS1 IL2RG ICAM1 NOD1 RIPK2 TRAF5 TNFRSF4 CLK1 SFRS6 IL2RG IL2 NOD2 RIPK2 TRAF6 CD40 CLK1 UBL5 IL2RG IL2RB NOLC1 CEBPB TRAF6 IRF7 CLK1 YWHAG IL2RG IL4 NOTCH1 NFKB1 TRAF6 MAPK14 CMA1 SERPINA3 IL2RG IL4R NR2C2 IKBKB TRAF6 TLR3 CNKSR1 MAP2K2 IL2RG IL7 NRIP1 HDAC5 TRAIP TRAF1 COBRA1 FOS IL2RG IL7R NSMAF CD40 TRAT1 CD247 COG5 MEFV IL2RG IL9R NTRK3 MAPK1 TRB CD247 COIL PSME3 IL2RG JAK1 NUCB1 PTGS2 TRDN RYR1 COTL1 ALOX5 IL3 CSF2RB NUP50 CDKN1B TRIB1 MAP2K1 CPN1 C3 IL3 HRAS NUP62 RANBP2 TRIM28 CEBPB CPNE1 MAP2K1 IL3 PIK3CA OLFM4 C3 TRIM37 TRAF1 CPNE4 MAP2K1 IL3RA IL3 OPTN FOS TRIP4 NFKB1 CPSF4 FIP1L1 IL4R IL13 PAK1 CASP1 TRIP6 ILF3 CR2 ANXA6 IL4R IL4 PAK1 MAP2K1 TRIP6 SON CR2 CD19 IL5RA IL5 PARK2 CASP1 TRIP6 TLR2 CR2 CR1 IL6 SH3GL2 PARK2 RANBP2 TRO FOS CR2 HNRNPU IL6 ZBTB16 PARP1 NFKB1 TRPC4AP IKBKB CREBBP ELF3 IL6R IL6 PDGFRB PIK3R2 TRPV4 ITCH CREBBP IRF7 IL6ST IL6 PDIA3 IFNG TSC2 CDKN1B CREBBP MECOM IL7R IL7 PDZK1 CFTR TSC2 GSK3B CRMP1 EPN1 IL8 CXCR1 PEA15 MAPK1 TSC22D3 FOS CSDA GSK3B IL8 CXCR2 PEBP1 MAP2K1 TSC22D3 NFKB1 CSDA MAPK1 IL8 DARC PEBP1 MAPK1 TTR AGER CSF2RA IKBKB IL8 GNAI2 PEBP4 MAP2K1 TTR ATF4 CSNK1A1 NFATC3 IL8 PF4 PELI2 MYD88 TTR C7orf20
25 CSNK2A1 EIF4EBP1 ILF3 DHX9 PF4 CCL5 TTR CHD3 CSNK2A1 SPP1 ILF3 EIF2AK2 PFN2 IVNS1ABP TTR DDR1 CSNK2A2 EIF4EBP1 ILF3 FUS PHB2 HDAC5 TTR FEZ1 CSNK2A2 SPP1 ILF3 KRTAP4-12 PI4K2B CD247 TTR HSPG2 CTBP1 MECOM ILF3 MDFI PIAS1 DDX21 TTR IKBKAP CTBP2 MECOM ILF3 PLSCR1 PIK3CA TICAM1 TTR MARK3 CTNNB1 NFKB1 ILF3 PRMT1 PIK3CD IL3 TTR PHYHIP CTRB1 SERPINA3 ILF3 SMN1 PIK3CD IL4 TTR SETDB1 CTRC SERPINA3 ILF3 XPO5 PIK3R1 IL13 TTR VIM CTRL SERPINA3 IPO5 RANBP2 PIK3R1 TLR2 TTRAP CD40 CTSD CCL21 IQGAP1 AIMP1 PIK3R TUBA4A LTA CTSG CXCR4 IRAK1 IRF7 PIK3R2 AXL TUBB MX1 CTSG SERPINA3 IRAK1 MAPK14 PIK3R2 CBL TXN NFKB1 CTSL1 IL8 IRAK1 RIPK2 PIK3R2 CRKL UBB CDKN1B CUL4A CDKN1B IRAK1 TLR4 PIK3R2 CSF1R UBB IKBKB CXCL1 DARC IRAK2 HRAS PIK3R2 EGF UBE2I RANBP2 CXCL10 CXCL10 IRAK2 IL1R1 PIK3R2 ERBB2 UBE2K NFKB1 CXCL10 VCAN IRAK2 IRAK1 PIK3R2 FYN UBR5 MAPK1 CXCL11 CCR3 IRAK2 MYD88 PIK3R2 GRB2 UMOD LTA CXCL11 CXCR3 IRAK2 PELI1 PIK3R2 HCK UMOD MMP8 CXCL12 CXCR4 IRAK2 PELI2 PIK3R2 IRS1 UNC119 CD247 CXCL13 CCL19 IRAK2 PELI3 PIK3R2 KIT UNC5CL NFKB1 CXCL13 CCL21 IRAK2 SMAD2 PIK3R2 PIK3CD UPF3A GSK3B CXCL13 CCR10 IRAK2 SMURF1 PIK3R2 SHC1 USF1 HDAC9 CXCL13 CXCR3 IRAK2 TRAF6 PIK3R2 SOCS1 USF2 FOS CXCL13 CXCR5 IRAK3 IRAK2 PIK3R2 SOCS7 USP7 TRAF1 CXCL13 FGF2 IRAK3 MYD88 PIK3R2 SYK VCAM1 IL13 CXCL2 CXCL2 IRAK4 MYD88 PIK3R2 TGFBR1 VCL SELE
26 CXCL9 CCR3 IRF3 CREBBP PLCG1 SELE VIM IVNS1ABP CXCL9 CXCR3 IRF3 IKBKE PLG CXCL2 VSIG4 C3 CXCR1 CXCL2 IRF3 MAVS PLK3 MAP2K1 WASF1 IL8 CXCR1 CXCL3 IRF3 PIAS4 PLK3 MAPK1 WASF3 PIK3R2 CXCR1 CXCL6 IRF3 SERPINB2 POLA1 SERPINA3 WDR65 FOS CXCR2 CXCL2 IRF3 SMURF2 POP1 PYCARD WWP1 IL2RG CXCR2 CXCL3 IRF3 SYT1 PPARGC1A GSK3B WWP2 IL2RG CXCR2 CXCL6 IRF3 TBK1 PPIB IRF3 XBP1 FOS CXCR3 CXCL10 IRF4 MYD88 PPIL5 TNFRSF9 XPO1 CDKN1B CXCR4 ADRBK2 IRF5 IRF3 PPM1B IKBKB XPO1 CIITA CXCR4 GNA13 IRF7 IRF3 PPP2CA EIF4EBP1 XPO1 RANBP2 CXCR4 GNAI1 IRF7 MAVS PPP2R1B CFTR XPOT RANBP2 CXCR4 JAK2 IRF7 PIAS4 PPP4C NFKB1 XRCC5 CD40 CXCR4 MYH9 IRS2 PIK3R2 PRDX1 IRF3 XRCC6 CD40 CXCR4 PTK2 ITCH ATN1 PRF1 GZMB XRN2 TOLLIP CXCR4 PTPN11 ITCH CPSF6 PRKAA1 CFTR YBX1 GSK3B CXCR4 PTPRC ITCH DTX1 PRKACA CFTR YBX1 MAPK1 CXCR4 SDC4 ITCH JUN PRKACA CIITA YES1 CDKN1B CXCR4 VAV1 ITCH NOTCH1 PRKACA FOS YWHAB PIK3R2 CYLD DDX58 ITCH NUMB PRKACA GSK3B YWHAB SON DAPK1 MAPK1 ITCH OCLN PRKACA NFKB1 YWHAE CDKN1B DARC CCL17 ITCH POLR2A PRKACA RYR1 YWHAG HDAC4 DARC CCL7 ITCH RIPK1 PRKAR1A PYCARD YWHAG SON DARC CCL8 ITCH RNF11 PRKCA CFTR ZBTB16 EPN1 DARC CXCL5 ITCH SH3GL2 PRKCA SPP1 ZBTB16 HDAC4 DAXX MX1 ITCH TRPC4 PRKCB IKBKB ZBTB16 HDAC5 DDX1 CSTF2 ITCH TRPV1 PRKCE MAPK1 ZBTB16 HDAC9 DDX1 RELA ITGA9 SPP1 PRKCZ GSK3B ZBTB16 TOLLIP
27 DDX21 JUN ITGAV SPP1 PRKCZ IKBKB ZHX1 TOLLIP DEFB4 TLR4 ITGB2 CD14 PRKD1 HDAC5 ZNF346 ILF3 DERL1 CFTR ITGB3BP NFKB1 PRKG1 CFTR ZNHIT1 MAPK14 DMBT1 SFTPD ITPKB PSME3 PRKG1 RYR1 TNFRSF7 DNAJA1 CFTR IVNS1ABP SYNCRIP PRKG1 SPP1 STAU DNAJB1 DNAJC3 JAK1 CXCR4 PRKRIR DNAJC3 SIAT1 DNAJC1 SERPINA3 JAK3 CD247 PRTN3 IL1B PFC DNAJC3 EIF2AK2 JAK3 CXCR4 PSMB9 NCOA1 PAF1 DNAJC3 HSPA1A JAK3 IL2RG PSMB9 NCOA2 MX2 DNAJC3 TIRAP JUN ELF3 PSMB9 NCOA3 MOX2 DNAJC5 CFTR JUN FOS PSMB9 PSMB10 IFITM3 DNM1L GSK3B JUND FOS PSMC5 FOS IFI30 DOCK2 CD247 JUND MAPK1 PSME3 ADAP1 IFIH1 DPP4 CCL5 KAT2A IRF7 PSME3 BBS2 IL17F DPP4 CXCL10 KAT2B CIITA PSME3 FXR2 CLEC4C DPP4 CXCL11 KAT2B IRF7 PSME3 KIAA1267 CMAHP DPP4 CXCL9 KAT2B MECOM PSME3 MDM2 CPT2 DPP4 CXCR4 KAT5 NFKB1 PSME3 NUDT18 CRTAM DUSP1 MAPK14 KLF11 MAPK1 PSME3 TP53 APG5L DUSP10 MAPK14 KLF4 HDAC5 PSME3 WDR25 DUSP16 MAPK14 KLF5 NFKB1 PSME3 ZCCHC10 DUSP2 MAPK14 KLK2 SERPINA3 PTGS2 TP53 DUSP22 MAPK14 KLK3 SERPINA3 PTHLH IL6 DUSP3 MAP2K2 KLK4 SERPINA3 PTK2 SELE DUSP3 MAPK1 KPNB1 RANBP2 PTPN1 MAPK1 DUSP4 MAPK1 KSR1 MAP2K1 PTPN11 SELE DUSP4 MAPK14 KSR1 MAPK1 PTPN22 CD247 DUSP7 MAPK1 KSR2 MAP2K1 PTPN6 CD247
28 DUSP7 MAPK14 KSR2 MAPK1 PTPN6 CXCR4
HTG EdgeSeq Immuno-Oncology Assay Gene List
 A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
SUPPLEMENTAL DATA. Table S1. Genes in each pathway selected for analysis.
 SUPPLEMENTAL DATA Table S1. Genes in each pathway selected for analysis. Gene Chr Start End bp Pathway 1 AKR1C1 10 5000044 5022159 22115 COX 2 AKR1C2 10 5029967 5060207 30240 COX 3 AKR1C3 10 5077549 5149878
SUPPLEMENTAL DATA Table S1. Genes in each pathway selected for analysis. Gene Chr Start End bp Pathway 1 AKR1C1 10 5000044 5022159 22115 COX 2 AKR1C2 10 5029967 5060207 30240 COX 3 AKR1C3 10 5077549 5149878
Nature Genetics: doi: /ng Supplementary Figure 1
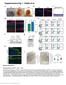 Supplementary Figure 1 Characterization of HPV Tg/+ ; Krt17 / mice. (a) Macroscopic images of HPV16 Tg/+ mouse ears at P40, P70 and P120. (b) Immunostaining of HPV16 Tg/+ ear tissue sections for K17 (red)
Supplementary Figure 1 Characterization of HPV Tg/+ ; Krt17 / mice. (a) Macroscopic images of HPV16 Tg/+ mouse ears at P40, P70 and P120. (b) Immunostaining of HPV16 Tg/+ ear tissue sections for K17 (red)
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study. February 2018
 Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases
 Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Assessing the Economics of Genomic Medicine. Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center
 Assessing the Economics of Genomic Medicine Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center DISCLOSURES No conflicts Patents unenforced Consultancies unpaid
Assessing the Economics of Genomic Medicine Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center DISCLOSURES No conflicts Patents unenforced Consultancies unpaid
Jocelyn Chapman, MD Division of Gynecologic Oncology. Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic
 Jocelyn Chapman, MD Division of Gynecologic Oncology Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic Genetics underlies all cancers Somatic or tumor genetics Germline or inherited genetics
Jocelyn Chapman, MD Division of Gynecologic Oncology Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic Genetics underlies all cancers Somatic or tumor genetics Germline or inherited genetics
Eosinophils! 40! 30! 20! 10! 0! NS!
 A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
Bronchoalveolar lavage fluid analysis showed increased eosinophil in 30% of subjects with IrIa BAL
 Bronchoalveolar lavage fluid analysis showed increased eosinophil in 30% of subjects with IrIa BAL UBC Chan-Yeung Centre for Occupational and Environmental Respiratory Disease Understanding exposure effects
Bronchoalveolar lavage fluid analysis showed increased eosinophil in 30% of subjects with IrIa BAL UBC Chan-Yeung Centre for Occupational and Environmental Respiratory Disease Understanding exposure effects
Precision Oncology: Experience at UW
 Precision Oncology: Experience at UW Colin Pritchard MD, PhD University of Washington, Department of Lab Medicine WSMOS Meeting November 1, 2013 Conflict of Interest Disclosures I declare the following,
Precision Oncology: Experience at UW Colin Pritchard MD, PhD University of Washington, Department of Lab Medicine WSMOS Meeting November 1, 2013 Conflict of Interest Disclosures I declare the following,
An Integrative Pharmacogenomic Approach Identifies Two-drug. Combination Therapies for Personalized Cancer Medicine
 An Integrative Pharmacogemic Approach Identifies Two-drug Combination Therapies for Personalized Cancer Medicine Yin Liu 1,2,3, Teng Fei 4,5, Xiaoqi Zheng 6, Myles Brown 4,5, Peng Zhang 7*, X. Shirley
An Integrative Pharmacogemic Approach Identifies Two-drug Combination Therapies for Personalized Cancer Medicine Yin Liu 1,2,3, Teng Fei 4,5, Xiaoqi Zheng 6, Myles Brown 4,5, Peng Zhang 7*, X. Shirley
Genomic and transcriptomic landscapes of acute promyelocy3c leukemia Kankan Wang
 Genomic and transcriptomic landscapes of acute promyelocy3c leukemia Kankan Wang State Key Laboratory of Medical Genomics Rui-Jin Hospital Shanghai Jiaotong University School of Medicine Understanding
Genomic and transcriptomic landscapes of acute promyelocy3c leukemia Kankan Wang State Key Laboratory of Medical Genomics Rui-Jin Hospital Shanghai Jiaotong University School of Medicine Understanding
The circulating T helper subsets and regulatory T cells in patients with common variable immunodeficiency with no monogenic disease
 1 Supplementary data The circulating T helper subsets and regulatory T cells in patients with common variable immunodeficiency with no monogenic disease Azizi et al. 2 Table S1. Excluded 373 genes variants
1 Supplementary data The circulating T helper subsets and regulatory T cells in patients with common variable immunodeficiency with no monogenic disease Azizi et al. 2 Table S1. Excluded 373 genes variants
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Frampton et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing Supplementary Figure 1A Base substitution detection
SUPPLEMENTARY INFORMATION Frampton et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing Supplementary Figure 1A Base substitution detection
Changing the Culture of Cancer Care II. Eric Holland Fred Hutchinson Cancer Research Center University of Washington Seattle
 Changing the Culture of Cancer Care II Eric Holland Fred Hutchinson Cancer Research Center University of Washington Seattle Transforming Cancer Therapy Eric Holland Fred Hutchinson Cancer Research Center
Changing the Culture of Cancer Care II Eric Holland Fred Hutchinson Cancer Research Center University of Washington Seattle Transforming Cancer Therapy Eric Holland Fred Hutchinson Cancer Research Center
APPLICATIONS OF NEXT GENERATION SEQUENCING IN SOLID TUMORS - PATHOLOGIST PROSPECTIVE
 AMP COMPANION MEETING SYMPOSIUM AT USCAP 2015 NEXT-GENERATION OF PATHOLOGY: ROLE OF PATHOLOGIST IN NGS-BASED PERSONALIZED MEDICINE APPLICATIONS OF NEXT GENERATION SEQUENCING IN SOLID TUMORS - PATHOLOGIST
AMP COMPANION MEETING SYMPOSIUM AT USCAP 2015 NEXT-GENERATION OF PATHOLOGY: ROLE OF PATHOLOGIST IN NGS-BASED PERSONALIZED MEDICINE APPLICATIONS OF NEXT GENERATION SEQUENCING IN SOLID TUMORS - PATHOLOGIST
# 1 India s Best Selling Cancer Genomics Test Precision treatment for every Cancer Patient Patient Guide
 # 1 India s Best Selling Cancer Genomics Test Precision treatment for every Cancer Patient Patient Guide PositiveSelect is India s widely used Cancer Genomics test with every 8 out of 10 patients opting
# 1 India s Best Selling Cancer Genomics Test Precision treatment for every Cancer Patient Patient Guide PositiveSelect is India s widely used Cancer Genomics test with every 8 out of 10 patients opting
Jennifer Hauenstein Oncology Cytogenetics Emory University Hospital Atlanta, GA
 Comparison of Genomic Coverage using Affymetrix OncoScan Array and Illumina TruSight Tumor 170 NGS Panel for Detection of Copy Number Abnormalities in Clinical GBM Specimens Jennifer Hauenstein Oncology
Comparison of Genomic Coverage using Affymetrix OncoScan Array and Illumina TruSight Tumor 170 NGS Panel for Detection of Copy Number Abnormalities in Clinical GBM Specimens Jennifer Hauenstein Oncology
Nature Genetics: doi: /ng Supplementary Figure 1. Depths and coverages in whole-exome and targeted deep sequencing data.
 Supplementary Figure 1 Depths and coverages in whole-exome and targeted deep sequencing data. Depth (top) and coverage (bottom) of whole-exome sequencing for 38 independent JPN cases (mean depth = 130)
Supplementary Figure 1 Depths and coverages in whole-exome and targeted deep sequencing data. Depth (top) and coverage (bottom) of whole-exome sequencing for 38 independent JPN cases (mean depth = 130)
Supplementary Figure 1. The overlap between statistically-significant and highconfidence
 Supplementary figures Supplementary Figure 1. The overlap between statistically-significant and highconfidence PPIs with previously reported PPIs in public databases. Venn diagrams showing the overlap
Supplementary figures Supplementary Figure 1. The overlap between statistically-significant and highconfidence PPIs with previously reported PPIs in public databases. Venn diagrams showing the overlap
A genome-wide association study identifies vitiligo
 A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
Supplementary Table 1: OCAC study location, type, and number of cases and controls
 Supplementary Table 1: OCAC study location, type, and number of cases and controls Study Name Abbreviation Study Location Study Type N Cases N Controls Australian Ovarian Cancer Study/Australian Cancer
Supplementary Table 1: OCAC study location, type, and number of cases and controls Study Name Abbreviation Study Location Study Type N Cases N Controls Australian Ovarian Cancer Study/Australian Cancer
What is the status of the technologies of "precision medicine?
 Session 2: What is the status of the technologies of "precision medicine? Gideon Blumenthal, MD, Clinical Team Leader, Thoracic and Head/Neck Oncology, Center for Drug Evaluation and Research (CDER), U.S.
Session 2: What is the status of the technologies of "precision medicine? Gideon Blumenthal, MD, Clinical Team Leader, Thoracic and Head/Neck Oncology, Center for Drug Evaluation and Research (CDER), U.S.
A computational approach to identify microrna (mirna) based biomarker from the regulation of disease pathology.
 Research Article http://www.alliedacademies.org/biology-and-medicine-case-report/ A computational approach to identify microrna (mirna) based biomarker from the regulation of disease pathology. Anandaram
Research Article http://www.alliedacademies.org/biology-and-medicine-case-report/ A computational approach to identify microrna (mirna) based biomarker from the regulation of disease pathology. Anandaram
Figure 1: Effects of cisplatin on survival of lung cancer cells.
 Figure 1 Figure 1: Effects of cisplatin on survival of lung cancer cells. To determine the IC 50 concentration of cisplatin, cells were treated with various concentrations of cisplatin and cell survival
Figure 1 Figure 1: Effects of cisplatin on survival of lung cancer cells. To determine the IC 50 concentration of cisplatin, cells were treated with various concentrations of cisplatin and cell survival
Supplementary Materials
 Supplementary Materials Using Multi objective Optimization to Identify Dynamical Network Biomarkers as Early warning Signals of Complex Diseases Fatemeh Vafaee 1, 2 1 Charles Perkins Centre, University
Supplementary Materials Using Multi objective Optimization to Identify Dynamical Network Biomarkers as Early warning Signals of Complex Diseases Fatemeh Vafaee 1, 2 1 Charles Perkins Centre, University
SI Table. SI Table 3. SI Table 4. SI Table 5A. SI Table 5B. SI Table 5C. SI Table 6A. SI Table 6B. SI Table 6C
 SI Table SI Table 1 SI Table 2 SI Table 3 SI Table 4 SI Table 5A SI Table 5B SI Table 5C SI Table 6A SI Table 6B SI Table 6C Title List of primers used to clone HCV genes and gene fragments HCV genotype
SI Table SI Table 1 SI Table 2 SI Table 3 SI Table 4 SI Table 5A SI Table 5B SI Table 5C SI Table 6A SI Table 6B SI Table 6C Title List of primers used to clone HCV genes and gene fragments HCV genotype
Clinically Useful Next Generation Sequencing and Molecular Testing in Gliomas MacLean P. Nasrallah, MD PhD
 Clinically Useful Next Generation Sequencing and Molecular Testing in Gliomas MacLean P. Nasrallah, MD PhD Neuropathology Fellow Division of Neuropathology Center for Personalized Diagnosis (CPD) Glial
Clinically Useful Next Generation Sequencing and Molecular Testing in Gliomas MacLean P. Nasrallah, MD PhD Neuropathology Fellow Division of Neuropathology Center for Personalized Diagnosis (CPD) Glial
Sample Metrics. Allele Frequency (%) Read Depth Ploidy. Gene CDS Effect Protein Effect. LN Metastasis Tumor Purity Computational Pathology 80% 60%
 Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits.
 Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits. All With Fst in the With long With SNP(s) at the 5' top 5% bracket haplotypes regulatory region
Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits. All With Fst in the With long With SNP(s) at the 5' top 5% bracket haplotypes regulatory region
Illumina s Cancer Research Portfolio and Dedicated Workflows
 Illumina s Cancer Research Portfolio and Dedicated Workflows Michael Sohn Clinical Sales Specialist Spain&Italy 2017 2017 Illumina, Inc. All rights reserved. Illumina, 24sure, BaseSpace, BeadArray, BlueFish,
Illumina s Cancer Research Portfolio and Dedicated Workflows Michael Sohn Clinical Sales Specialist Spain&Italy 2017 2017 Illumina, Inc. All rights reserved. Illumina, 24sure, BaseSpace, BeadArray, BlueFish,
Exons from 137 genes were targeted for hybrid selection and massively parallel sequencing.
 SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Material. Part I: Sample Information. Part II: Pathway Information
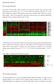 Supplementary Material Part I: Sample Information Three NPC cell lines, CNE1, CNE2, and HK1 were treated with CYC202. Gene expression of 380 selected genes were collected at 0, 2, 4, 6, 12 and 24 hours
Supplementary Material Part I: Sample Information Three NPC cell lines, CNE1, CNE2, and HK1 were treated with CYC202. Gene expression of 380 selected genes were collected at 0, 2, 4, 6, 12 and 24 hours
Case Study - Informatics
 bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature12912 Figure S1. Distribution of mutation rates and spectra across 4,742 tumor-normal (TN) pairs from 21 tumor types, as in Figure 1 from Lawrence et al. Nature
SUPPLEMENTARY INFORMATION doi:1.138/nature12912 Figure S1. Distribution of mutation rates and spectra across 4,742 tumor-normal (TN) pairs from 21 tumor types, as in Figure 1 from Lawrence et al. Nature
Fig. 3 Terms and their Frequencies across documents
 SUPPLEMENTAL MATERIALS Application Notes Supplemental Materials Data and text mining NLPCORE Computer Aided Discovery of Annotations of Bioentities and their Relationships in Articles Varun Mittal, Alexander
SUPPLEMENTAL MATERIALS Application Notes Supplemental Materials Data and text mining NLPCORE Computer Aided Discovery of Annotations of Bioentities and their Relationships in Articles Varun Mittal, Alexander
Table S1. Demographics of patients and tumor characteristics.
 Supplemental Tables for: A Clinical Actionability of Comprehensive Genomic Profiling for Management of Rare or Refractory Cancers Kim Hirshfield et al. Table S1. Demographics of patients and tumor characteristics.
Supplemental Tables for: A Clinical Actionability of Comprehensive Genomic Profiling for Management of Rare or Refractory Cancers Kim Hirshfield et al. Table S1. Demographics of patients and tumor characteristics.
Supplementary Data Figure S1. A) PKI-587 suppression of p-akt in A498 and (+/- 10
 Supplementary Data Figure S1. A) PKI-587 suppression of p-akt in A498 and 786-0 (+/- 10 μm Verapamil), and B) PKI-587 suppression of p-akt in BT474, & U87MG. 3 1 0.3 0.1 0.03 0 [μm] A) 786-0 - + p-akt
Supplementary Data Figure S1. A) PKI-587 suppression of p-akt in A498 and 786-0 (+/- 10 μm Verapamil), and B) PKI-587 suppression of p-akt in BT474, & U87MG. 3 1 0.3 0.1 0.03 0 [μm] A) 786-0 - + p-akt
Provide your cancer patients personalized treatment options with ClariFind
 ONCOLOGY ClariFind employs next-generation sequencing (NGS) for comprehensive genomic profiling of a patient s tumor sample to identify potential clinically actionable targets and associated therapies.
ONCOLOGY ClariFind employs next-generation sequencing (NGS) for comprehensive genomic profiling of a patient s tumor sample to identify potential clinically actionable targets and associated therapies.
Deconvolution of Heterogeneous Wound Tissue Samples into Relative Macrophage Phenotype Composition via Models based on Gene Expression
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2017 Supplementary Information for: Deconvolution of Heterogeneous Wound Tissue Samples into
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2017 Supplementary Information for: Deconvolution of Heterogeneous Wound Tissue Samples into
Supplementary Information
 Supplementary Information Title: Role of Cytochrome P450 (CYP)1A in Hyperoxic Lung Injury: Analysis of the Transcriptome and Proteome Authors: Krithika Lingappan a, Suman Maity b, Weiwu Jiang a, Lihua
Supplementary Information Title: Role of Cytochrome P450 (CYP)1A in Hyperoxic Lung Injury: Analysis of the Transcriptome and Proteome Authors: Krithika Lingappan a, Suman Maity b, Weiwu Jiang a, Lihua
Supplemental Table 1: Enriched GO categories per cluster
 Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
CD Marker Antibodies. atlasantibodies.com
 CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
Department of International Health (Vaccine science track) Johns Hopkins University Bloomberg School of Public Health
 Acute Respiratory Distress Syndrome is a TH17 and Treg immune disease By Wan-Jiung Hu Postdoctorate Genomics Research Center Academia Sinica No 128 Academia Road section2 Nangang 115, Taipei, Taiwan Previous
Acute Respiratory Distress Syndrome is a TH17 and Treg immune disease By Wan-Jiung Hu Postdoctorate Genomics Research Center Academia Sinica No 128 Academia Road section2 Nangang 115, Taipei, Taiwan Previous
Gene expression of muscular and neuronal pathways is cooperatively dysregulated in patients
 Gene expression of muscular and neuronal pathways is cooperatively dysregulated in patients with idiopathic achalasia Orazio Palmieri 1, Tommaso Mazza 2, Antonio Merla 1, Caterina Fusilli 2, Antonello
Gene expression of muscular and neuronal pathways is cooperatively dysregulated in patients with idiopathic achalasia Orazio Palmieri 1, Tommaso Mazza 2, Antonio Merla 1, Caterina Fusilli 2, Antonello
Next generation sequencing on FFPE tumor tissues for lung and colorectal carcinoma diagnostics and research Sakari Knuutila
 Next generation sequencing on FFPE tumor tissues for lung and colorectal carcinoma diagnostics and research Sakari Knuutila Faculty of Medicine / Haartman Institute / Department of Pathology / Sakari Knuutila
Next generation sequencing on FFPE tumor tissues for lung and colorectal carcinoma diagnostics and research Sakari Knuutila Faculty of Medicine / Haartman Institute / Department of Pathology / Sakari Knuutila
Transduction of lentivirus to human primary CD4+ T cells
 Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
BD Rhapsody Immune Response Panel Hs
 Doc ID: 55665 Rev. 2.0 PRODUCT INSERT BD Rhapsody Immune Response Panel Hs BD Biosciences Kit Part Number: 633750 Purpose The BD Rhapsody Immune Response Panel Hs is a set of ready-to-use primer pools
Doc ID: 55665 Rev. 2.0 PRODUCT INSERT BD Rhapsody Immune Response Panel Hs BD Biosciences Kit Part Number: 633750 Purpose The BD Rhapsody Immune Response Panel Hs is a set of ready-to-use primer pools
MathIOmica: Dynamic Transcriptome
 Printed from the Complete Wolfram Language Documentation 1 MathIOmica: Dynamic Transcriptome Loading the MathIOmica Package Classification, Clustering and Visualization of Transcriptome Time eries Importing
Printed from the Complete Wolfram Language Documentation 1 MathIOmica: Dynamic Transcriptome Loading the MathIOmica Package Classification, Clustering and Visualization of Transcriptome Time eries Importing
Supplementary Table S1: Gene expression targets
 Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
The expression of genes contributing to pancreatic adenocarcinoma progression is influenced by the respective environment Sagini et al
 The expression of genes contributing to pancreatic adenocarcinoma progression is influenced by the respective environment Sagini et al Supplementary Figure 1: Target genes regulated by TGM2. Figure represents
The expression of genes contributing to pancreatic adenocarcinoma progression is influenced by the respective environment Sagini et al Supplementary Figure 1: Target genes regulated by TGM2. Figure represents
Nature Immunology: doi: /ni.3745
 Supplementary Figure 1 Experimental settings. (a) Experimental setup and bioinformatic data analysis of transcriptomic changes induced in neonatal and adult monocytes by stimulation with 10 ng/ml LPS for
Supplementary Figure 1 Experimental settings. (a) Experimental setup and bioinformatic data analysis of transcriptomic changes induced in neonatal and adult monocytes by stimulation with 10 ng/ml LPS for
Supplementary Table S1. Histopathologic findings for multi-region analysis.
 SUPPLEMENTARY INFORMATION SUPPLEMENTARY TABLES Supplementary Table S1. Histopathologic findings for multi-region analysis. Patient Histology R1: clear cell RCC, Fuhrman 2-3, acinar pattern 1 R2: clear
SUPPLEMENTARY INFORMATION SUPPLEMENTARY TABLES Supplementary Table S1. Histopathologic findings for multi-region analysis. Patient Histology R1: clear cell RCC, Fuhrman 2-3, acinar pattern 1 R2: clear
Use of Molecular Assays to Assess Cellular Therapies
 Use of Molecular Assays to Assess Cellular Therapies D. Stroncek, P. Jin, M. Sabatino, J. Ren, L. Castiello, E. Wang, and F.M. Marincola. Cellular Therapies Section Clinical Center, NIH Methods for the
Use of Molecular Assays to Assess Cellular Therapies D. Stroncek, P. Jin, M. Sabatino, J. Ren, L. Castiello, E. Wang, and F.M. Marincola. Cellular Therapies Section Clinical Center, NIH Methods for the
q 2017 by The University of Chicago. All rights reserved. DOI: /690105
 q 2017 by The University of Chicago. All rights reserved. DOI: 10.1086/690105 Appendix from R. M. Cox et al., Hormonally Mediated Increases in Sex-Biased Gene Expression Accompany the Breakdown of Between-
q 2017 by The University of Chicago. All rights reserved. DOI: 10.1086/690105 Appendix from R. M. Cox et al., Hormonally Mediated Increases in Sex-Biased Gene Expression Accompany the Breakdown of Between-
Oncology Update: The 10 Most Talked About Breast Cancer Topics of The Top 10 10/24/2013. The Awareness Debate
 Oncology Update: The 10 Most Talked About Breast Cancer Topics of 2013 Michaela Tsai, MD Martha Bacon Stimpson Chair of Breast Oncology Virginia Piper Cancer Institute Minnesota Oncology The Top 10 1.
Oncology Update: The 10 Most Talked About Breast Cancer Topics of 2013 Michaela Tsai, MD Martha Bacon Stimpson Chair of Breast Oncology Virginia Piper Cancer Institute Minnesota Oncology The Top 10 1.
103.5 Membrane metalloendopeptidase TGA GGG GTC ACG ATT TTA GG ATG ATG GTG AGG AGC AGG AC
 Supplementary Table SA Primers used for real-time RT-PCR gene regulation and their confirmation Gene Name Accession no Size Region Forward Primer Reverse Primer Efficiency% AKT v-akt oncogene homolog NM_00563
Supplementary Table SA Primers used for real-time RT-PCR gene regulation and their confirmation Gene Name Accession no Size Region Forward Primer Reverse Primer Efficiency% AKT v-akt oncogene homolog NM_00563
Developing a clinically feasible personalized medicine approach to pediatric
 Developing a clinically feasible personalized medicine approach to pediatric septic shock Hector R. Wong, Natalie Z. Cvijanovich, Nick Anas, Geoffrey L. Allen, Neal J. Thomas, Michael T. Bigham, Scott
Developing a clinically feasible personalized medicine approach to pediatric septic shock Hector R. Wong, Natalie Z. Cvijanovich, Nick Anas, Geoffrey L. Allen, Neal J. Thomas, Michael T. Bigham, Scott
* Kyoto Encyclopedia of Genes and Genomes.
 Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
SUPPLEMENTAL MATERIAL. Characterization and discovery of novel mirnas and mornas in JAK2V617F. mutated SET2 cells
 SUPPLEMENTAL MATERIAL Characterization and discovery of novel mirnas and mornas in JAK2V617F mutated SET2 cells Stefania Bortoluzzi 1*, Andrea Bisognin 1*, et al. 1 Supplemental methods Short RNA library
SUPPLEMENTAL MATERIAL Characterization and discovery of novel mirnas and mornas in JAK2V617F mutated SET2 cells Stefania Bortoluzzi 1*, Andrea Bisognin 1*, et al. 1 Supplemental methods Short RNA library
For focused group profiling of human HIV infection and host response genes expression
 ExProfile TM Human HIV Infection and Host Response Related Gene qpcr Array For focused group profiling of human HIV infection and host response genes expression Cat. No. QG023-A (1 x 96-well plate, Format
ExProfile TM Human HIV Infection and Host Response Related Gene qpcr Array For focused group profiling of human HIV infection and host response genes expression Cat. No. QG023-A (1 x 96-well plate, Format
Gene List HTG EdgeSeq Oncology Biomarker Panel
 Gene List HTG EdgeSeq Oncology Biomarker Panel 1 Gene List HTG EdgeSeq Oncology Biomarker Panel ACACA ACACB ACTB ADIPOR1 ADIPOR2 ADRA1A ADRA1B ADRA1D ADRA2A ADRA2B ADRA2C AK1 AK2 AK3 ATG13 CAMKK1 CAMKK2
Gene List HTG EdgeSeq Oncology Biomarker Panel 1 Gene List HTG EdgeSeq Oncology Biomarker Panel ACACA ACACB ACTB ADIPOR1 ADIPOR2 ADRA1A ADRA1B ADRA1D ADRA2A ADRA2B ADRA2C AK1 AK2 AK3 ATG13 CAMKK1 CAMKK2
Editorial Type 2 Diabetes and More Gene Panel: A Predictive Genomics Approach for a Polygenic Disease
 Cronicon OPEN ACCESS EC DIABETES AND METABOLIC RESEARCH Editorial Type 2 Diabetes and More Gene Panel: A Predictive Genomics Approach for a Polygenic Disease Amr TM Saeb* University Diabetes Center, College
Cronicon OPEN ACCESS EC DIABETES AND METABOLIC RESEARCH Editorial Type 2 Diabetes and More Gene Panel: A Predictive Genomics Approach for a Polygenic Disease Amr TM Saeb* University Diabetes Center, College
SureSelect Cancer All-In-One Custom and Catalog NGS Assays
 SureSelect Cancer All-In-One Custom and Catalog NGS Assays Detect all cancer-relevant variants in a single SureSelect assay SNV Indel TL SNV Indel TL Single DNA input Single AIO assay Single data analysis
SureSelect Cancer All-In-One Custom and Catalog NGS Assays Detect all cancer-relevant variants in a single SureSelect assay SNV Indel TL SNV Indel TL Single DNA input Single AIO assay Single data analysis
SUPPLEMENTARY FIG. S2. b-galactosidase staining of
 SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
15. Supplementary Figure 9. Predicted gene module expression changes at 24hpi during HIV
 Supplementary Information Table of content 1. Supplementary Table 1. Summary of RNAseq data and mapping statistics 2. Supplementary Table 2. Biological functions enriched in 12 hpi DE genes, derived from
Supplementary Information Table of content 1. Supplementary Table 1. Summary of RNAseq data and mapping statistics 2. Supplementary Table 2. Biological functions enriched in 12 hpi DE genes, derived from
Supplemental Information. High-Throughput Microfluidic Labyrinth for the. Label-free Isolation of Circulating Tumor Cells
 Cell Systems, Volume 5 Supplemental Information High-Throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells Eric Lin, Lianette Rivera-Báez, Shamileh Fouladdel, Hyeun
Cell Systems, Volume 5 Supplemental Information High-Throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells Eric Lin, Lianette Rivera-Báez, Shamileh Fouladdel, Hyeun
Supplemental Data for Toischer et al., Cardiomyocyte proliferation prevents failure in
 Supplemental Data, Page 1 Supplemental Data for Toischer et al., Cardiomyocyte proliferation prevents failure in pressure but not volume overload. Contents: Page 3: Supplemental Figure 1 - Echocardiographic
Supplemental Data, Page 1 Supplemental Data for Toischer et al., Cardiomyocyte proliferation prevents failure in pressure but not volume overload. Contents: Page 3: Supplemental Figure 1 - Echocardiographic
Integrative Radiation Biology
 Dr. Kristian Unger Integrative Biology Group Research Unit of Radiation Cytogenetics Department of Radiation Sciences Helmholtz-Zentrum München 1 Key Questions Molecular mechanisms of radiation-induced
Dr. Kristian Unger Integrative Biology Group Research Unit of Radiation Cytogenetics Department of Radiation Sciences Helmholtz-Zentrum München 1 Key Questions Molecular mechanisms of radiation-induced
Supplementary Material: Exploring wound-healing genomic machinery with a network-based approach
 Supplementary Material: Exploring wound-healing genomic machinery with a network-based approach Francesca Vitali, Simone Marini, Martina Balli, Hanne Grosemans, Maurilio Sampaolesi, Yves A. Lussier, Maria
Supplementary Material: Exploring wound-healing genomic machinery with a network-based approach Francesca Vitali, Simone Marini, Martina Balli, Hanne Grosemans, Maurilio Sampaolesi, Yves A. Lussier, Maria
Hepatocellular Carcinoma (HCC): What Treatment Modality for Which Tumor?
 K. Rajender Reddy, MD, CG Hepatocellular Carcinoma (HCC): What Treatment Modality for Which Tumor? K. Rajender Reddy, MD, CG Ruimy Family President s Distinguished Professor of Medicine Director of Hepatology
K. Rajender Reddy, MD, CG Hepatocellular Carcinoma (HCC): What Treatment Modality for Which Tumor? K. Rajender Reddy, MD, CG Ruimy Family President s Distinguished Professor of Medicine Director of Hepatology
Diagnostica Molecolare!
 Diagnostica Molecolare! Aldo Scarpa Unità Diagnostica Molecolare Azienda Ospedaliera Universitaria Integrata di Verona e ARC-NET Centro di Ricerca Applicata sul Cancro PDTA CARCINOMA POLMONARE - IL PAZIENTE
Diagnostica Molecolare! Aldo Scarpa Unità Diagnostica Molecolare Azienda Ospedaliera Universitaria Integrata di Verona e ARC-NET Centro di Ricerca Applicata sul Cancro PDTA CARCINOMA POLMONARE - IL PAZIENTE
Hematology Fusion/Expression Profile
 Hematology Fusion/Expression Profile Patient Name: Ordered By Date of Birth: Ordering Physician: Gender (M/F): Physician ID: Client: Accession #: Case #: Specimen Type: Body Site: Specimen ID: Ethnicity:
Hematology Fusion/Expression Profile Patient Name: Ordered By Date of Birth: Ordering Physician: Gender (M/F): Physician ID: Client: Accession #: Case #: Specimen Type: Body Site: Specimen ID: Ethnicity:
Clinical implication of MEN1 mutation in surgically resected thymic carcinoid patients
 Case Report Clinical implication of MEN1 in surgically resected thymic carcinoid patients Xiongfei Li 1 *, Mingbiao Li 1 *, Tao Shi 2 *, Renwang Liu 1, Dian Ren 1, Fan Yang 1, Sen Wei 1, Gang Chen 1, Jun
Case Report Clinical implication of MEN1 in surgically resected thymic carcinoid patients Xiongfei Li 1 *, Mingbiao Li 1 *, Tao Shi 2 *, Renwang Liu 1, Dian Ren 1, Fan Yang 1, Sen Wei 1, Gang Chen 1, Jun
ShinyGO: a web application for in-depth analysis of gene sets
 ShinyGO: a web application for in-depth analysis of gene sets Steven Xijin Ge 1,*, and Dongmin Jung 1,2 1 Department of Mathematics and Statistics, Box 2225, Brookings, SD, USA 57007 2 Avison Biomedical
ShinyGO: a web application for in-depth analysis of gene sets Steven Xijin Ge 1,*, and Dongmin Jung 1,2 1 Department of Mathematics and Statistics, Box 2225, Brookings, SD, USA 57007 2 Avison Biomedical
SUPPLEMENTARY INFORMATION
 Meta-Analysis Supplementary Fig.. Schematic of experimental design. Large scale shrna screens for proliferation/viability were performed in 9 cell lines using a library of 52 shrnas targeting 957 genes
Meta-Analysis Supplementary Fig.. Schematic of experimental design. Large scale shrna screens for proliferation/viability were performed in 9 cell lines using a library of 52 shrnas targeting 957 genes
Supplementary Materials for
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 217 Supplementary Materials for Chemical genomic analysis of GPR35 signaling Heidi (Haibei)
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 217 Supplementary Materials for Chemical genomic analysis of GPR35 signaling Heidi (Haibei)
Statistical Issues in Translational Cancer Research
 Statistical Issues in Translational Cancer Research Martin Bøgsted Department of Haematology Aalborg University Hospital and Department of Clinical Medicine Aalborg University The only useful function
Statistical Issues in Translational Cancer Research Martin Bøgsted Department of Haematology Aalborg University Hospital and Department of Clinical Medicine Aalborg University The only useful function
Interpreting Diverse Genomic Data Using Gene Sets
 Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
T-Cell Network Example for GeneNet (August 2015) or later
 T-Cell Network Example for GeneNet 1.2.13 (August 2015) or later This note reproduces the T-Cell network example from R. Opgen-Rhein and K. Strimmer. 2006a. Using regularized dynamic correlation to infer
T-Cell Network Example for GeneNet 1.2.13 (August 2015) or later This note reproduces the T-Cell network example from R. Opgen-Rhein and K. Strimmer. 2006a. Using regularized dynamic correlation to infer
Zhi-Ping Liu 1, Hulin Wu 2, Jian Zhu 3* and Hongyu Miao 2*
 Liu et al. BMC Bioinformatics 2014, 15:336 METHODOLOGY ARTICLE Open Access Systematic identification of transcriptional and post-transcriptional regulations in human respiratory epithelial cells during
Liu et al. BMC Bioinformatics 2014, 15:336 METHODOLOGY ARTICLE Open Access Systematic identification of transcriptional and post-transcriptional regulations in human respiratory epithelial cells during
Supplemental Table unique genes
 Supplemental Table 1 Inflammation KEGG ID Title Gene Count KEGG:mmu04662 B cell receptor signaling pathway 20 KEGG:mmu04060 Cytokine-cytokine receptor interaction 44 KEGG:mmu04010 MAPK signaling pathway
Supplemental Table 1 Inflammation KEGG ID Title Gene Count KEGG:mmu04662 B cell receptor signaling pathway 20 KEGG:mmu04060 Cytokine-cytokine receptor interaction 44 KEGG:mmu04010 MAPK signaling pathway
Brigatinib overcomes ALK resistance mechanisms preclinically Zhang et al Supplementary information
 Supplementary information The potent ALK inhibitor brigatinib (AP26113) overcomes mechanisms of resistance to first- and second-generation ALK inhibitors in preclinical models Sen Zhang, Rana Anjum, Rachel
Supplementary information The potent ALK inhibitor brigatinib (AP26113) overcomes mechanisms of resistance to first- and second-generation ALK inhibitors in preclinical models Sen Zhang, Rana Anjum, Rachel
Next-generation sequencing of Chinese stage IV lung cancer patients reveals an association between EGFR mutation status and survival outcome
 Clin Genet 2017: 91: 488 493 Printed in Singapore. All rights reserved Short Report 2016 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd CLINICAL GENETICS doi: 10.1111/cge.12809 Next-generation
Clin Genet 2017: 91: 488 493 Printed in Singapore. All rights reserved Short Report 2016 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd CLINICAL GENETICS doi: 10.1111/cge.12809 Next-generation
devseek (Sequence(Analysis(Panel(for(Neurodevelopmental(Disorders
 ACSL4 Mental/retardation,/XBlinked/63 XLBR ADSL Adenylosuccinase/Deficiency AR AFF2 Mental/retardation,/XBlinked,/FRAXE/type XLBR ALG6 Congenital/disorder/of/glycosylation/type/Ic AR ANK3 Mental/retardation,/autosomal/recessive/37
ACSL4 Mental/retardation,/XBlinked/63 XLBR ADSL Adenylosuccinase/Deficiency AR AFF2 Mental/retardation,/XBlinked,/FRAXE/type XLBR ALG6 Congenital/disorder/of/glycosylation/type/Ic AR ANK3 Mental/retardation,/autosomal/recessive/37
Supplementary Materials
 Supplementary Materials Recurrent SMARCA4 Mutations in Small Cell Carcinoma of the Ovary Petar Jelinic, Jennifer J. Mueller, Narciso Olvera, Fanny Dao, Sasinya N. Scott, Ronak Shah, JianJiong Gao, Nikolaus
Supplementary Materials Recurrent SMARCA4 Mutations in Small Cell Carcinoma of the Ovary Petar Jelinic, Jennifer J. Mueller, Narciso Olvera, Fanny Dao, Sasinya N. Scott, Ronak Shah, JianJiong Gao, Nikolaus
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature15260 Supplementary Data 1: Gene expression in individual basal/stem, luminal, and luminal progenitor cells. Box plots show expression levels for each gene from the 49-gene differentiation
doi:10.1038/nature15260 Supplementary Data 1: Gene expression in individual basal/stem, luminal, and luminal progenitor cells. Box plots show expression levels for each gene from the 49-gene differentiation
Supporting Information
 Supporting Information Rothhammer et al. 1.173/pnas.161413114 SI Materials and Methods EAE Clinical Scoring and Agents. Clinical sig of EAE were assessed as follows:, no sig of disease; 1, loss of tone
Supporting Information Rothhammer et al. 1.173/pnas.161413114 SI Materials and Methods EAE Clinical Scoring and Agents. Clinical sig of EAE were assessed as follows:, no sig of disease; 1, loss of tone
Supplementary information. Supplementary Table 1. Kinase Type Format N Average (IC 50 )
 Supplementary information Supplementary Table 1 Kinase Type Format N Average (IC 50 ) SD ALK Y Lantha 6 > 10 n.d. AURORA_A S/T Caliper 6 > 10 n.d. AXL Y Lantha 3 > 10 n.d. BTK Y Caliper 6 > 10 n.d. cabl
Supplementary information Supplementary Table 1 Kinase Type Format N Average (IC 50 ) SD ALK Y Lantha 6 > 10 n.d. AURORA_A S/T Caliper 6 > 10 n.d. AXL Y Lantha 3 > 10 n.d. BTK Y Caliper 6 > 10 n.d. cabl
Supporting Information
 Supporting Information Lin et al. 10.1073/pnas.1408759111 Fig. S1. Safety and potential toxicity evaluation of M1 in immunocompetent mice. Immunocompetent BALB/c mice received two doses of i.v. infusion
Supporting Information Lin et al. 10.1073/pnas.1408759111 Fig. S1. Safety and potential toxicity evaluation of M1 in immunocompetent mice. Immunocompetent BALB/c mice received two doses of i.v. infusion
A brain tumor and NGS/multiplexing
 A brain tumor and NGS/multiplexing Santiago Ramón y Cajal Jefe de Servicio. Hospital Vall d Hebron Catedrático de Anatomía Patológica U.A.B. Académico de Número de la Real Academia Nacional de Medicina
A brain tumor and NGS/multiplexing Santiago Ramón y Cajal Jefe de Servicio. Hospital Vall d Hebron Catedrático de Anatomía Patológica U.A.B. Académico de Número de la Real Academia Nacional de Medicina
Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic
 Supplementary Information for Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic signals in endothelial cells Kazuhiro Ohkubo 1,, Yasunari Sakai 1,*,, Hirosuke Inoue 1, Satoshi
Supplementary Information for Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic signals in endothelial cells Kazuhiro Ohkubo 1,, Yasunari Sakai 1,*,, Hirosuke Inoue 1, Satoshi
Phosphorylation Site Company Cat #
 Supplemental Table 1. Antibodies used for RPPA analysis. Label Protein Phosphorylation Site Company Cat # Used on MDA_CLSS Used on MDA_Pilot 4EBP1 4EBP1 Cell Signaling 9452 No 4EBP1.pS65 4EBP1 S65 Cell
Supplemental Table 1. Antibodies used for RPPA analysis. Label Protein Phosphorylation Site Company Cat # Used on MDA_CLSS Used on MDA_Pilot 4EBP1 4EBP1 Cell Signaling 9452 No 4EBP1.pS65 4EBP1 S65 Cell
Applied Biosystems models 7500 (Fast block), 7900HT (Fast block), StepOnePlus, ViiA 7 (Fast block)
 RT² Profiler PCR Array (96-Well Format and 384-Well [4 x 96] Format) Human HIV Infection and Host Response Cat. no. 330231 PAHS-051YA For pathway expression analysis Format Format A Format C Format D Format
RT² Profiler PCR Array (96-Well Format and 384-Well [4 x 96] Format) Human HIV Infection and Host Response Cat. no. 330231 PAHS-051YA For pathway expression analysis Format Format A Format C Format D Format
 Supplementary Table 1. Annotated candidate SNV and indels in melanoma tumors. Patient C1 Patient C2 Patient C3 Patient C4 Patient C5 Patient C6 Patient C7 Patient C10 Chr Position Reference Allele Sample
Supplementary Table 1. Annotated candidate SNV and indels in melanoma tumors. Patient C1 Patient C2 Patient C3 Patient C4 Patient C5 Patient C6 Patient C7 Patient C10 Chr Position Reference Allele Sample
Supplementary Information
 Supplementary Information Index of Supplementary Materials Supplementary Figure. Distribution of methylation levels, characteristics of cis-meqtls and association plots for meqtls in methylation-related
Supplementary Information Index of Supplementary Materials Supplementary Figure. Distribution of methylation levels, characteristics of cis-meqtls and association plots for meqtls in methylation-related
Patricia Aoun MD, MPH Professor and Vice-Chair for Clinical Affairs Medical Director, Clinical Laboratories Department of Pathology City of Hope
 Patricia Aoun MD, MPH Professor and Vice-Chair for Clinical Affairs Medical Director, Clinical Laboratories Department of Pathology City of Hope National Medical Center Disclosures I have no disclosures
Patricia Aoun MD, MPH Professor and Vice-Chair for Clinical Affairs Medical Director, Clinical Laboratories Department of Pathology City of Hope National Medical Center Disclosures I have no disclosures
The lung cancer program of Alliance Against Cancer. Ruggero De Maria, President
 The lung cancer program of Alliance Against Cancer Ruggero De Maria, President Alliance Against Cancer (ACC) was established in 2002 by the Italian Ministry of Health with the task of promoting an active
The lung cancer program of Alliance Against Cancer Ruggero De Maria, President Alliance Against Cancer (ACC) was established in 2002 by the Italian Ministry of Health with the task of promoting an active
Research Article Differential Expression of MicroRNAs between Eutopic and Ectopic Endometrium in Ovarian Endometriosis
 Hindawi Publishing Corporation Journal of Biomedicine and Biotechnology Volume 2010, Article ID 369549, 29 pages doi:10.1155/2010/369549 Research Article Differential Expression of MicroRNAs between Eutopic
Hindawi Publishing Corporation Journal of Biomedicine and Biotechnology Volume 2010, Article ID 369549, 29 pages doi:10.1155/2010/369549 Research Article Differential Expression of MicroRNAs between Eutopic
