T-Cell Network Example for GeneNet (August 2015) or later
|
|
|
- Jasper Osborn Young
- 6 years ago
- Views:
Transcription
1 T-Cell Network Example for GeneNet (August 2015) or later This note reproduces the T-Cell network example from R. Opgen-Rhein and K. Strimmer. 2006a. Using regularized dynamic correlation to infer gene dependency networks from time-series microarray data. Proceedings of WCSB 2006 (June 12-13, 2006, Tampere, Finland) and R. Opgen-Rhein and K. Strimmer. 2006b. Inferring gene dependency networks from genomic longitudinal data: a functional data approach. REVSTAT 4: ( Load GeneNet library library("genenet") Loading required package: corpcor Loading required package: longitudinal Loading required package: fdrtool get T cell data data(tcell) tc44 = combine.longitudinal(tcell.10, tcell.34) Estimate partial correlations pc1 = ggm.estimate.pcor(tc44, lambda=0) # static, no shrinkage Specified shrinkage intensity lambda (correlation matrix): 0 pc2 = ggm.estimate.pcor(tc44, method="dynamic", lambda=0) # dynamic, no shrinkage Specified shrinkage intensity lambda (correlation matrix): 0 pc3 = ggm.estimate.pcor(tc44) # static, with shrinkage Estimating optimal shrinkage intensity lambda (correlation matrix): pc4 = ggm.estimate.pcor(tc44, method="dynamic") # dynamic, with shrinkage Estimating optimal shrinkage intensity lambda (correlation matrix):
2 Find significant edges We use as selection criterion local fdr <= 0.2 static, no shrinkage t1.edges = network.test.edges(pc1) Type of Statistic: Correlation (kappa = 104, eta0 = ) (first row) and Distribution Function (second row) Fdr (tail area based) t1.net = extract.network(t1.edges) # prob > 0.8 Significant edges: 6 Corresponding to 0.36 % of possible edges t1.net pcor node1 node2 pval qval prob e e
3 e e e e dynamic, no shrinkage t2.edges = network.test.edges(pc2) Type of Statistic: Correlation (kappa = 110.7, eta0 = 0.964) (first row) and Distribution Function (second row) 0.5 fdr (density based) 0.5 t2.net = extract.network(t2.edges) # prob > 0.8 Significant edges: 9 Corresponding to 0.54 % of possible edges t2.net pcor node1 node2 pval qval prob 3
4 e e e e e e e e e e e e e e e e e e static, with shrinkage t3.edges = network.test.edges(pc3) Type of Statistic: Correlation (kappa = 151, eta0 = ) 0 6 (first row) and Distribution Function (second row) fdr (density based) t3.net = extract.network(t3.edges) # prob > 0.8 Significant edges: 10 Corresponding to 0.6 % of possible edges 4
5 t3.net pcor node1 node2 pval qval prob e e e e e e e e e e dynamic, with shrinkage t4.edges = network.test.edges(pc4) Type of Statistic: Correlation (kappa = 189.7, eta0 = ) 0 8 (first row) and Distribution Function (second row) fdr (density based) 5
6 t4.net = extract.network(t4.edges) # prob > 0.8 Significant edges: 21 Corresponding to 1.27 % of possible edges t4.net pcor node1 node2 pval qval prob e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e Produce plots using graph and Rgraphviz library("rgraphviz") Loading required package: graph Loading required package: grid node.labels = colnames(tc44) gr1 = network.make.graph( t1.net, node.labels) gr2 = network.make.graph( t2.net, node.labels) gr3 = network.make.graph( t3.net, node.labels) gr4 = network.make.graph( t4.net, node.labels) globalattrs = list() globalattrs$edge <- list(color = "blue", lty = "solid", lwd = 1, arrowsize=1) globalattrs$node <- list(fillcolor = "lightyellow", shape = "ellipse", fixedsize = FALSE) edgeattrs1 <- list() 6
7 edgeattrs1$dir = edge.info(gr1)$dir edgeattrs2 <- list() edgeattrs2$dir = edge.info(gr2)$dir edgeattrs3 <- list() edgeattrs3$dir = edge.info(gr3)$dir edgeattrs4 <- list() edgeattrs4$dir = edge.info(gr4)$dir par(mfrow=c(2,2)) plot(gr1, main="static, no shrinkage", attrs = globalattrs, edgeattrs = edgeattrs1, "fdp") plot(gr2, main="dynamic, no shrinkage", attrs = globalattrs, edgeattrs = edgeattrs2,"fdp") plot(gr3, main="static, with shrinkage", attrs = globalattrs, edgeattrs = edgeattrs3, "fdp") plot(gr4, main="dynamic, with shrinkage", attrs = globalattrs, edgeattrs = edgeattrs4, "fdp") Static, no shrinkage CD69 C3X1 TCF8 SOD1 MCL1 CDK4 IL4R IRAK1 TCF12 IFNAR1 GATA3 RB1 SCYA2 MAPK9 LAT TRAF5 CDC2 ID3 FYB IL2RG API1 SLA CASP8 ZNFN1A1 API2 NFKBIA SKIIP CYP19 JUND IL16 CCNG1 PIG3 JUNB CSF2RA EGR1 CCNA2 MPO RBL2 IL3RA CTNNB1 MAP2K4 RPS6KA1 AKT1 SIVA ITGAM MAP3K8 MYD88 CCNC CASP4 CASP7 PDE4B SMN1 PCNA CLU LCK APC E2F4 CIR Dynamic, no shrinkage ID3 MPO CD69 SIVA AKT1 JUNB RBL2 SMN1 PDE4B TRAF5 ITGAM SCYA2 RB1 TCF12 RPS6KA1 CLU MCL1 MAPK9 SLA IRAK1 ZNFN1A1 API2 GATA3 IL4R MAP3K8 SOD1 NFKBIA MYD88 CASP4 IL16 CDC2 JUND CCNA2 CIR CYP19 EGR1 CASP8 IFNAR1 CDK4 IL2RG API1 MAP2K4 E2F4 CSF2RA CTNNB1 LAT CCNG1 PCNA CCNC CASP7 IL3RA SKIIP LCK APC FYB PIG3 TCF8 C3X1 Static, with shrinkage JUNB AKT1 RBL2 SOD1 CDK4 CCNC PDE4B CD69 GATA3 MYD88 ITGAM SLA CASP4 CTNNB1 RPS6KA1 IL2RG API1 RB1 TRAF5 ZNFN1A1 JUND IL16 IL4R CLU CASP8 FYB EGR1 SIVA IFNAR1 E2F4 CDC2 MAPK9 MCL1 SCYA2 C3X1 MPO CCNA2 API2 CYP19 NFKBIA MAP2K4 TCF12 SKIIP PIG3 CSF2RA CCNG1 IRAK1 MAP3K8 SMN1 CASP7 PCNA IL3RA CIR LCK APC LAT ID3 TCF8 Dynamic, with shrinkage JUNB MPO TCF8 C3X1 CDK4 PDE4B SMN1 RPS6KA1 ID3 RBL2 TRAF5 MAP2K4 PCNA CTNNB1 SIVA MYD88 CIR IRAK1 CDC2 CCNA2 FYB GATA3 SOD1 IL4R CLU IL3RA IL16 API2 JUND ITGAM EGR1 MCL1 CCNG1 NFKBIA TCF12 CYP19 MAPK9 API1 CASP4 AKT1 CASP7 SLA IL2RG SCYA2 CASP8 IFNAR1 RB1 ZNFN1A1 LAT CCNC CSF2RA MAP3K8 E2F4 PIG3 CD69 LCK SKIIP APC par(mfrow=c(1,1)) 7
Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and
 Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and Promoter Motif Analysis Shisong Ma 1,2*, Michael Snyder 3, and Savithramma P Dinesh-Kumar 2* 1 School of Life Sciences, University
Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and Promoter Motif Analysis Shisong Ma 1,2*, Michael Snyder 3, and Savithramma P Dinesh-Kumar 2* 1 School of Life Sciences, University
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
R documentation. of GSCA/man/GSCA-package.Rd etc. June 8, GSCA-package. LungCancer metadi... 3 plotmnw... 5 plotnw... 6 singledc...
 R topics documented: R documentation of GSCA/man/GSCA-package.Rd etc. June 8, 2009 GSCA-package....................................... 1 LungCancer3........................................ 2 metadi...........................................
R topics documented: R documentation of GSCA/man/GSCA-package.Rd etc. June 8, 2009 GSCA-package....................................... 1 LungCancer3........................................ 2 metadi...........................................
Interpreting Diverse Genomic Data Using Gene Sets
 Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
Interpreting Diverse Genomic Data Using Gene Sets
 Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
* Kyoto Encyclopedia of Genes and Genomes.
 Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
blood mononuclear cells were stained with AAD, annexin, CD3, CD4, CD25, LAP and specific
 Supplementary figure legends Figure 1: Selective expression of regulatory markers on CD4 + LAP + T cells. Human peripheral blood mononuclear cells were stained with AAD, annexin, CD3, CD4, CD25, LAP and
Supplementary figure legends Figure 1: Selective expression of regulatory markers on CD4 + LAP + T cells. Human peripheral blood mononuclear cells were stained with AAD, annexin, CD3, CD4, CD25, LAP and
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study. February 2018
 Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
SUPPLEMENTARY FIGURES AND TABLES
 PD 0332991, a selective cyclin D kinase 4/6 inhibitor, sensitizes lung cancer cells to treatment with epidermal growth factor receptor tyrosine kinase inhibitors SUPPLEMENTARY FIGURES AND TABLES Supplementary
PD 0332991, a selective cyclin D kinase 4/6 inhibitor, sensitizes lung cancer cells to treatment with epidermal growth factor receptor tyrosine kinase inhibitors SUPPLEMENTARY FIGURES AND TABLES Supplementary
15. Supplementary Figure 9. Predicted gene module expression changes at 24hpi during HIV
 Supplementary Information Table of content 1. Supplementary Table 1. Summary of RNAseq data and mapping statistics 2. Supplementary Table 2. Biological functions enriched in 12 hpi DE genes, derived from
Supplementary Information Table of content 1. Supplementary Table 1. Summary of RNAseq data and mapping statistics 2. Supplementary Table 2. Biological functions enriched in 12 hpi DE genes, derived from
Protein SD Units (P-value) Cluster order
 SUPPLEMENTAL TABLE AND FIGURES Table S1. Signature Phosphoproteome of CD22 E12 Transgenic Mouse BPL Cells. T-test vs. Other Protein SD Units (P-value) Cluster order ATPase (Ab-16) 1.41 0.000880 1 mtor
SUPPLEMENTAL TABLE AND FIGURES Table S1. Signature Phosphoproteome of CD22 E12 Transgenic Mouse BPL Cells. T-test vs. Other Protein SD Units (P-value) Cluster order ATPase (Ab-16) 1.41 0.000880 1 mtor
SUPPLEMENTAL DATA. Table S1. Genes in each pathway selected for analysis.
 SUPPLEMENTAL DATA Table S1. Genes in each pathway selected for analysis. Gene Chr Start End bp Pathway 1 AKR1C1 10 5000044 5022159 22115 COX 2 AKR1C2 10 5029967 5060207 30240 COX 3 AKR1C3 10 5077549 5149878
SUPPLEMENTAL DATA Table S1. Genes in each pathway selected for analysis. Gene Chr Start End bp Pathway 1 AKR1C1 10 5000044 5022159 22115 COX 2 AKR1C2 10 5029967 5060207 30240 COX 3 AKR1C3 10 5077549 5149878
Analysis of DNA methylation patterns associated with the gastric cancer genome
 ONCOLOGY LETTERS 7: 1021-1026, 2014 Analysis of DNA methylation patterns associated with the gastric cancer genome YI CHENG 1,2, ZHI YAN 2, YIN LIU 1, CHENGBAI LIANG 1, HONG XIA 1, JUNMING FENG 3, GUORONG
ONCOLOGY LETTERS 7: 1021-1026, 2014 Analysis of DNA methylation patterns associated with the gastric cancer genome YI CHENG 1,2, ZHI YAN 2, YIN LIU 1, CHENGBAI LIANG 1, HONG XIA 1, JUNMING FENG 3, GUORONG
The LiquidAssociation Package
 The LiquidAssociation Package Yen-Yi Ho October 30, 2018 1 Introduction The LiquidAssociation package provides analytical methods to study three-way interactions. It incorporates methods to examine a particular
The LiquidAssociation Package Yen-Yi Ho October 30, 2018 1 Introduction The LiquidAssociation package provides analytical methods to study three-way interactions. It incorporates methods to examine a particular
CPM (x 10-3 ) Tregs +Teffs. Tregs alone ICOS CLTA-4
 A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
Phosphorylation Site Company Cat #
 Supplemental Table 1. Antibodies used for RPPA analysis. Label Protein Phosphorylation Site Company Cat # Used on MDA_CLSS Used on MDA_Pilot 4EBP1 4EBP1 Cell Signaling 9452 No 4EBP1.pS65 4EBP1 S65 Cell
Supplemental Table 1. Antibodies used for RPPA analysis. Label Protein Phosphorylation Site Company Cat # Used on MDA_CLSS Used on MDA_Pilot 4EBP1 4EBP1 Cell Signaling 9452 No 4EBP1.pS65 4EBP1 S65 Cell
An Integrative Pharmacogenomic Approach Identifies Two-drug. Combination Therapies for Personalized Cancer Medicine
 An Integrative Pharmacogemic Approach Identifies Two-drug Combination Therapies for Personalized Cancer Medicine Yin Liu 1,2,3, Teng Fei 4,5, Xiaoqi Zheng 6, Myles Brown 4,5, Peng Zhang 7*, X. Shirley
An Integrative Pharmacogemic Approach Identifies Two-drug Combination Therapies for Personalized Cancer Medicine Yin Liu 1,2,3, Teng Fei 4,5, Xiaoqi Zheng 6, Myles Brown 4,5, Peng Zhang 7*, X. Shirley
Supplemental Information. High-Throughput Microfluidic Labyrinth for the. Label-free Isolation of Circulating Tumor Cells
 Cell Systems, Volume 5 Supplemental Information High-Throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells Eric Lin, Lianette Rivera-Báez, Shamileh Fouladdel, Hyeun
Cell Systems, Volume 5 Supplemental Information High-Throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells Eric Lin, Lianette Rivera-Báez, Shamileh Fouladdel, Hyeun
Supplementary Material. Part I: Sample Information. Part II: Pathway Information
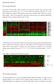 Supplementary Material Part I: Sample Information Three NPC cell lines, CNE1, CNE2, and HK1 were treated with CYC202. Gene expression of 380 selected genes were collected at 0, 2, 4, 6, 12 and 24 hours
Supplementary Material Part I: Sample Information Three NPC cell lines, CNE1, CNE2, and HK1 were treated with CYC202. Gene expression of 380 selected genes were collected at 0, 2, 4, 6, 12 and 24 hours
RchyOptimyx: Gating Hierarchy Optimization for Flow Cytometry
 RchyOptimyx: Gating Hierarchy Optimization for Flow Cytometry Nima Aghaeepour and Adrin Jalali April 4, 2013 naghaeep@bccrc.ca Contents 1 Licensing 1 2 Introduction 1 3 First Example: Preparing Raw Data
RchyOptimyx: Gating Hierarchy Optimization for Flow Cytometry Nima Aghaeepour and Adrin Jalali April 4, 2013 naghaeep@bccrc.ca Contents 1 Licensing 1 2 Introduction 1 3 First Example: Preparing Raw Data
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature89 IFN- (ng ml ) 5 4 3 1 Splenocytes NS IFN- (ng ml ) 6 4 Lymph node cells NS Nfkbiz / Nfkbiz / Nfkbiz / Nfkbiz / IL- (ng ml ) 3 1 Splenocytes IL- (ng ml ) 1 8 6 4 *** ** Lymph node cells
doi: 1.138/nature89 IFN- (ng ml ) 5 4 3 1 Splenocytes NS IFN- (ng ml ) 6 4 Lymph node cells NS Nfkbiz / Nfkbiz / Nfkbiz / Nfkbiz / IL- (ng ml ) 3 1 Splenocytes IL- (ng ml ) 1 8 6 4 *** ** Lymph node cells
Supplementary Figure S1. Flow cytometric analysis of the expression of Thy1 in NH cells. Flow cytometric analysis of the expression of T1/ST2 and
 Supplementary Figure S1. Flow cytometric analysis of the expression of Thy1 in NH cells. Flow cytometric analysis of the expression of T1/ST2 and Thy1 in NH cells derived from the lungs of naïve mice.
Supplementary Figure S1. Flow cytometric analysis of the expression of Thy1 in NH cells. Flow cytometric analysis of the expression of T1/ST2 and Thy1 in NH cells derived from the lungs of naïve mice.
Bronchoalveolar lavage fluid analysis showed increased eosinophil in 30% of subjects with IrIa BAL
 Bronchoalveolar lavage fluid analysis showed increased eosinophil in 30% of subjects with IrIa BAL UBC Chan-Yeung Centre for Occupational and Environmental Respiratory Disease Understanding exposure effects
Bronchoalveolar lavage fluid analysis showed increased eosinophil in 30% of subjects with IrIa BAL UBC Chan-Yeung Centre for Occupational and Environmental Respiratory Disease Understanding exposure effects
Supplementary Figures. T Cell Factor-1 initiates T helper 2 fate by inducing GATA-3 and repressing Interferon-γ
 Supplementary Figures T Cell Factor-1 initiates T helper 2 fate by inducing GATA-3 and repressing Interferon-γ Qing Yu, Archna Sharma, Sun Young Oh, Hyung-Geun Moon, M. Zulfiquer Hossain, Theresa M. Salay,
Supplementary Figures T Cell Factor-1 initiates T helper 2 fate by inducing GATA-3 and repressing Interferon-γ Qing Yu, Archna Sharma, Sun Young Oh, Hyung-Geun Moon, M. Zulfiquer Hossain, Theresa M. Salay,
Vega: Variational Segmentation for Copy Number Detection
 Vega: Variational Segmentation for Copy Number Detection Sandro Morganella Luigi Cerulo Giuseppe Viglietto Michele Ceccarelli Contents 1 Overview 1 2 Installation 1 3 Vega.RData Description 2 4 Run Vega
Vega: Variational Segmentation for Copy Number Detection Sandro Morganella Luigi Cerulo Giuseppe Viglietto Michele Ceccarelli Contents 1 Overview 1 2 Installation 1 3 Vega.RData Description 2 4 Run Vega
HTG EdgeSeq Immuno-Oncology Assay Gene List
 A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
Supplementary figures
 Supplementary figures Supplementary figure 1: Pathway enrichment for each comparison. The first column shows enrichment with differentially expressed genes between the diet groups at t = 0, the second
Supplementary figures Supplementary figure 1: Pathway enrichment for each comparison. The first column shows enrichment with differentially expressed genes between the diet groups at t = 0, the second
Lung Reperfusion Injury
 Lung Reperfusion Injury Michael S. Mulligan, M.D. Professor of Surgery Chief, Section of Thoracic Surgery Director, Lung Transplant Program University of Washington Medical Center Guiding Principles in
Lung Reperfusion Injury Michael S. Mulligan, M.D. Professor of Surgery Chief, Section of Thoracic Surgery Director, Lung Transplant Program University of Washington Medical Center Guiding Principles in
1,000 in silico simulated alpha, beta, gamma and delta TCR repertoires were created.
 938 939 940 941 942 Figure S1 Schematic of the in silico TCRminer and MiXCR validation. 1,000 in silico simulated alpha, beta, gamma and delta TCR repertoires were created. Then, 100,000 simulated 80 bp
938 939 940 941 942 Figure S1 Schematic of the in silico TCRminer and MiXCR validation. 1,000 in silico simulated alpha, beta, gamma and delta TCR repertoires were created. Then, 100,000 simulated 80 bp
Fig. 3 Terms and their Frequencies across documents
 SUPPLEMENTAL MATERIALS Application Notes Supplemental Materials Data and text mining NLPCORE Computer Aided Discovery of Annotations of Bioentities and their Relationships in Articles Varun Mittal, Alexander
SUPPLEMENTAL MATERIALS Application Notes Supplemental Materials Data and text mining NLPCORE Computer Aided Discovery of Annotations of Bioentities and their Relationships in Articles Varun Mittal, Alexander
Nature Genetics: doi: /ng Supplementary Figure 1
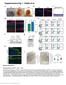 Supplementary Figure 1 Characterization of HPV Tg/+ ; Krt17 / mice. (a) Macroscopic images of HPV16 Tg/+ mouse ears at P40, P70 and P120. (b) Immunostaining of HPV16 Tg/+ ear tissue sections for K17 (red)
Supplementary Figure 1 Characterization of HPV Tg/+ ; Krt17 / mice. (a) Macroscopic images of HPV16 Tg/+ mouse ears at P40, P70 and P120. (b) Immunostaining of HPV16 Tg/+ ear tissue sections for K17 (red)
Doing Thousands of Hypothesis Tests at the Same Time. Bradley Efron Stanford University
 Doing Thousands of Hypothesis Tests at the Same Time Bradley Efron Stanford University 1 Simultaneous Hypothesis Testing 1980: Simultaneous Statistical Inference (Rupert Miller) 2, 3,, 20 simultaneous
Doing Thousands of Hypothesis Tests at the Same Time Bradley Efron Stanford University 1 Simultaneous Hypothesis Testing 1980: Simultaneous Statistical Inference (Rupert Miller) 2, 3,, 20 simultaneous
Introduction to Cancer Biology
 Introduction to Cancer Biology Robin Hesketh Multiple choice questions (choose the one correct answer from the five choices) Which ONE of the following is a tumour suppressor? a. AKT b. APC c. BCL2 d.
Introduction to Cancer Biology Robin Hesketh Multiple choice questions (choose the one correct answer from the five choices) Which ONE of the following is a tumour suppressor? a. AKT b. APC c. BCL2 d.
Fasting selectively blocks development of acute lymphoblastic leukemia via leptin-receptor upregulation
 a r t i c l e s Fasting selectively blocks development of acute lymphoblastic leukemia via leptin-receptor upregulation 7 Nature America, Inc., part of Springer Nature. All rights reserved. Zhigang Lu,,
a r t i c l e s Fasting selectively blocks development of acute lymphoblastic leukemia via leptin-receptor upregulation 7 Nature America, Inc., part of Springer Nature. All rights reserved. Zhigang Lu,,
Innate immune regulation of T-helper (Th) cell homeostasis in the intestine
 Innate immune regulation of T-helper (Th) cell homeostasis in the intestine Masayuki Fukata, MD, Ph.D. Research Scientist II Division of Gastroenterology, Department of Medicine, F. Widjaja Foundation,
Innate immune regulation of T-helper (Th) cell homeostasis in the intestine Masayuki Fukata, MD, Ph.D. Research Scientist II Division of Gastroenterology, Department of Medicine, F. Widjaja Foundation,
Models of cell signalling uncover molecular mechanisms of high-risk neuroblastoma and predict outcome
 Models of cell signalling uncover molecular mechanisms of high-risk neuroblastoma and predict outcome Marta R. Hidalgo 1, Alicia Amadoz 2, Cankut Çubuk 1, José Carbonell-Caballero 2 and Joaquín Dopazo
Models of cell signalling uncover molecular mechanisms of high-risk neuroblastoma and predict outcome Marta R. Hidalgo 1, Alicia Amadoz 2, Cankut Çubuk 1, José Carbonell-Caballero 2 and Joaquín Dopazo
Package ORIClust. February 19, 2015
 Type Package Package ORIClust February 19, 2015 Title Order-restricted Information Criterion-based Clustering Algorithm Version 1.0-1 Date 2009-09-10 Author Maintainer Tianqing Liu
Type Package Package ORIClust February 19, 2015 Title Order-restricted Information Criterion-based Clustering Algorithm Version 1.0-1 Date 2009-09-10 Author Maintainer Tianqing Liu
The Center for PERSONALIZED DIAGNOSTICS
 The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
A mathematical model for short-term vs. long-term survival in patients with. glioma
 SUPPLEMENTARY DATA A mathematical model for short-term vs. long-term survival in patients with glioma Jason B. Nikas 1* 1 Genomix, Inc., Minneapolis, MN 55364, USA * Correspondence: Dr. Jason B. Nikas
SUPPLEMENTARY DATA A mathematical model for short-term vs. long-term survival in patients with glioma Jason B. Nikas 1* 1 Genomix, Inc., Minneapolis, MN 55364, USA * Correspondence: Dr. Jason B. Nikas
fl/+ KRas;Atg5 fl/+ KRas;Atg5 fl/fl KRas;Atg5 fl/fl KRas;Atg5 Supplementary Figure 1. Gene set enrichment analyses. (a) (b)
 KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Expression deviation of the genes mapped to gene-wise recurrent mutations in the TCGA breast cancer cohort (top) and the TCGA lung cancer cohort (bottom). For each gene (each pair
Supplementary Figure 1 Expression deviation of the genes mapped to gene-wise recurrent mutations in the TCGA breast cancer cohort (top) and the TCGA lung cancer cohort (bottom). For each gene (each pair
Statistical Issues in Translational Cancer Research
 Statistical Issues in Translational Cancer Research Martin Bøgsted Department of Haematology Aalborg University Hospital and Department of Clinical Medicine Aalborg University The only useful function
Statistical Issues in Translational Cancer Research Martin Bøgsted Department of Haematology Aalborg University Hospital and Department of Clinical Medicine Aalborg University The only useful function
Package CorMut. January 21, 2019
 Type Package Package CorMut January 21, 2019 Title Detect the correlated mutations based on selection pressure Version 1.24.0 Date 2018-02-21 Author Maintainer Depends methods,seqinr,igraph
Type Package Package CorMut January 21, 2019 Title Detect the correlated mutations based on selection pressure Version 1.24.0 Date 2018-02-21 Author Maintainer Depends methods,seqinr,igraph
HD-SNP Microarray Analysis of the Study 9 High Risk ALL Patients -
 HD-SNP Microarray Analysis of the Study 9 High Risk ALL Patients - Increased Yield of Important Prognostic Information Nadine K Berry 1,2,4, Rodney J. Scott 1,4, Rosemary Sutton 5, Toby N Trahair 5, Philip
HD-SNP Microarray Analysis of the Study 9 High Risk ALL Patients - Increased Yield of Important Prognostic Information Nadine K Berry 1,2,4, Rodney J. Scott 1,4, Rosemary Sutton 5, Toby N Trahair 5, Philip
CNV PCA Search Tutorial
 CNV PCA Search Tutorial Release 8.1 Golden Helix, Inc. March 18, 2014 Contents 1. Data Preparation 2 A. Join Log Ratio Data with Phenotype Information.............................. 2 B. Activate only
CNV PCA Search Tutorial Release 8.1 Golden Helix, Inc. March 18, 2014 Contents 1. Data Preparation 2 A. Join Log Ratio Data with Phenotype Information.............................. 2 B. Activate only
TNFSF13B tumor necrosis factor (ligand) superfamily, member 13b NF-kB pathway cluster, Enrichment Score: 3.57
 Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was
 Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Fluxion Biosciences and Swift Biosciences Somatic variant detection from liquid biopsy samples using targeted NGS
 APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
Genomic and transcriptomic landscapes of acute promyelocy3c leukemia Kankan Wang
 Genomic and transcriptomic landscapes of acute promyelocy3c leukemia Kankan Wang State Key Laboratory of Medical Genomics Rui-Jin Hospital Shanghai Jiaotong University School of Medicine Understanding
Genomic and transcriptomic landscapes of acute promyelocy3c leukemia Kankan Wang State Key Laboratory of Medical Genomics Rui-Jin Hospital Shanghai Jiaotong University School of Medicine Understanding
Multiplicity Considerations in Confirmatory Subgroup Analyses
 Multiplicity Considerations in Confirmatory Subgroup Analyses Frank Bretz European Statistical Meeting on Subgroup Analyses Brussels, November 30, 2012 Subgroup analyses Exploratory subgroup analyses are
Multiplicity Considerations in Confirmatory Subgroup Analyses Frank Bretz European Statistical Meeting on Subgroup Analyses Brussels, November 30, 2012 Subgroup analyses Exploratory subgroup analyses are
SureSelect Cancer All-In-One Custom and Catalog NGS Assays
 SureSelect Cancer All-In-One Custom and Catalog NGS Assays Detect all cancer-relevant variants in a single SureSelect assay SNV Indel TL SNV Indel TL Single DNA input Single AIO assay Single data analysis
SureSelect Cancer All-In-One Custom and Catalog NGS Assays Detect all cancer-relevant variants in a single SureSelect assay SNV Indel TL SNV Indel TL Single DNA input Single AIO assay Single data analysis
Statistics and Probability
 Statistics and a single count or measurement variable. S.ID.1: Represent data with plots on the real number line (dot plots, histograms, and box plots). S.ID.2: Use statistics appropriate to the shape
Statistics and a single count or measurement variable. S.ID.1: Represent data with plots on the real number line (dot plots, histograms, and box plots). S.ID.2: Use statistics appropriate to the shape
Lung Met 1 Lung Met 2 Lung Met Lung Met H3K4me1. Lung Met H3K27ac Primary H3K4me1
 a Gained Met-VELs 1.5 1.5 -.5 Lung Met 1 Lung Met Lung Met 3 1. Lung Met H3K4me1 Lung Met H3K4me1 1 Lung Met H3K4me1 Lung Met H3K7ac 1.5 Lung Met H3K7ac Lung Met H3K7ac.8 Primary H3K4me1 Primary H3K7ac
a Gained Met-VELs 1.5 1.5 -.5 Lung Met 1 Lung Met Lung Met 3 1. Lung Met H3K4me1 Lung Met H3K4me1 1 Lung Met H3K4me1 Lung Met H3K7ac 1.5 Lung Met H3K7ac Lung Met H3K7ac.8 Primary H3K4me1 Primary H3K7ac
Supplementary Material
 Supplementary Material Taghon, Yui, and Rothenberg: Mast cell diversion of T-lineage precursor cells by the essential T-lineage transcription factor GATA Supplemental Table Supplemental Figures 1-6 Supplemental
Supplementary Material Taghon, Yui, and Rothenberg: Mast cell diversion of T-lineage precursor cells by the essential T-lineage transcription factor GATA Supplemental Table Supplemental Figures 1-6 Supplemental
Causal Protein Signalling Networks
 Hunting for the Lost Pathways m.scutari@ucl.ac.uk Genetics Institute January 25, 2013 Graphical Models Graphical Models Graphical Models Graphical models are defined by: a network structure, G = (V, E),
Hunting for the Lost Pathways m.scutari@ucl.ac.uk Genetics Institute January 25, 2013 Graphical Models Graphical Models Graphical Models Graphical models are defined by: a network structure, G = (V, E),
Department of International Health (Vaccine science track) Johns Hopkins University Bloomberg School of Public Health
 Acute Respiratory Distress Syndrome is a TH17 and Treg immune disease By Wan-Jiung Hu Postdoctorate Genomics Research Center Academia Sinica No 128 Academia Road section2 Nangang 115, Taipei, Taiwan Previous
Acute Respiratory Distress Syndrome is a TH17 and Treg immune disease By Wan-Jiung Hu Postdoctorate Genomics Research Center Academia Sinica No 128 Academia Road section2 Nangang 115, Taipei, Taiwan Previous
Network-assisted data analysis
 Network-assisted data analysis Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Protein identification in shotgun proteomics Protein digestion LC-MS/MS Protein
Network-assisted data analysis Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Protein identification in shotgun proteomics Protein digestion LC-MS/MS Protein
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Item Response Theory for Polytomous Items Rachael Smyth
 Item Response Theory for Polytomous Items Rachael Smyth Introduction This lab discusses the use of Item Response Theory (or IRT) for polytomous items. Item response theory focuses specifically on the items
Item Response Theory for Polytomous Items Rachael Smyth Introduction This lab discusses the use of Item Response Theory (or IRT) for polytomous items. Item response theory focuses specifically on the items
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols.
 Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Identifying the Zygosity Status of Twins Using Bayes Network and Estimation- Maximization Methodology
 Identifying the Zygosity Status of Twins Using Bayes Network and Estimation- Maximization Methodology Yicun Ni (ID#: 9064804041), Jin Ruan (ID#: 9070059457), Ying Zhang (ID#: 9070063723) Abstract As the
Identifying the Zygosity Status of Twins Using Bayes Network and Estimation- Maximization Methodology Yicun Ni (ID#: 9064804041), Jin Ruan (ID#: 9070059457), Ying Zhang (ID#: 9070063723) Abstract As the
Biomedical Informatics Insights. Differential Transcriptional Changes in Mice Exposed to Chemically Distinct Diesel Samples
 Biomedical Informatics Insights Original Research Open Access Full open access to this and thousands of other papers at http://www.la-press.com. Differential Transcriptional Changes in Mice Exposed to
Biomedical Informatics Insights Original Research Open Access Full open access to this and thousands of other papers at http://www.la-press.com. Differential Transcriptional Changes in Mice Exposed to
A Systems Biology Approach to the Analysis of Subset- Specific Responses to Lipopolysaccharide in Dendritic Cells
 A Systems Biology Approach to the Analysis of Subset- Specific Responses to Lipopolysaccharide in Dendritic Cells David G. Hancock 1,2, Elena Shklovskaya 1,2, Thomas V. Guy 1,2, Reza Falsafi 3, Chris D.
A Systems Biology Approach to the Analysis of Subset- Specific Responses to Lipopolysaccharide in Dendritic Cells David G. Hancock 1,2, Elena Shklovskaya 1,2, Thomas V. Guy 1,2, Reza Falsafi 3, Chris D.
Supplementary Materials
 1 Supplementary Materials Rotger et al. Table S1A: Demographic characteristics of study participants. VNP RP EC CP (n=6) (n=66) (n=9) (n=5) Male gender, n(%) 5 (83) 54 (82) 5 (56) 3 (60) White ethnicity,
1 Supplementary Materials Rotger et al. Table S1A: Demographic characteristics of study participants. VNP RP EC CP (n=6) (n=66) (n=9) (n=5) Male gender, n(%) 5 (83) 54 (82) 5 (56) 3 (60) White ethnicity,
Gene expression profiling predicts clinical outcome of prostate cancer. Gennadi V. Glinsky, Anna B. Glinskii, Andrew J. Stephenson, Robert M.
 SUPPLEMENTARY DATA Gene expression profiling predicts clinical outcome of prostate cancer Gennadi V. Glinsky, Anna B. Glinskii, Andrew J. Stephenson, Robert M. Hoffman, William L. Gerald Table of Contents
SUPPLEMENTARY DATA Gene expression profiling predicts clinical outcome of prostate cancer Gennadi V. Glinsky, Anna B. Glinskii, Andrew J. Stephenson, Robert M. Hoffman, William L. Gerald Table of Contents
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/3/114/ra23/dc1 Supplementary Materials for Regulation of Zap70 Expression During Thymocyte Development Enables Temporal Separation of CD4 and CD8 Repertoire Selection
www.sciencesignaling.org/cgi/content/full/3/114/ra23/dc1 Supplementary Materials for Regulation of Zap70 Expression During Thymocyte Development Enables Temporal Separation of CD4 and CD8 Repertoire Selection
Comparing Multifunctionality and Association Information when Classifying Oncogenes and Tumor Suppressor Genes
 000 001 002 003 004 005 006 007 008 009 010 011 012 013 014 015 016 017 018 019 020 021 022 023 024 025 026 027 028 029 030 031 032 033 034 035 036 037 038 039 040 041 042 043 044 045 046 047 048 049 050
000 001 002 003 004 005 006 007 008 009 010 011 012 013 014 015 016 017 018 019 020 021 022 023 024 025 026 027 028 029 030 031 032 033 034 035 036 037 038 039 040 041 042 043 044 045 046 047 048 049 050
This is a free sample of content from Immune Tolerance. Click here for more information or to buy the book.
 A ACPAs. See Antibodies to citrullinated peptide antigens Activation-induced cell death (AICD), 25 AICD. See Activation-induced cell death AIRE, 3, 18 19, 24, 88, 103 104 AKT, 35 Alefacept, 136 ALPS. See
A ACPAs. See Antibodies to citrullinated peptide antigens Activation-induced cell death (AICD), 25 AICD. See Activation-induced cell death AIRE, 3, 18 19, 24, 88, 103 104 AKT, 35 Alefacept, 136 ALPS. See
Dynamic Gene Regulatory Networks of Human Myeloid Differentiation
 Article Dynamic Gene Regulatory Networks of Human Myeloid Differentiation Graphical Abstract Authors Ricardo N. Ramirez, Nicole C. El-Ali, Mikayla Anne Mager, Dana Wyman, Ana Conesa, Ali Mortazavi Correspondence
Article Dynamic Gene Regulatory Networks of Human Myeloid Differentiation Graphical Abstract Authors Ricardo N. Ramirez, Nicole C. El-Ali, Mikayla Anne Mager, Dana Wyman, Ana Conesa, Ali Mortazavi Correspondence
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Zhi-Ping Liu 1, Hulin Wu 2, Jian Zhu 3* and Hongyu Miao 2*
 Liu et al. BMC Bioinformatics 2014, 15:336 METHODOLOGY ARTICLE Open Access Systematic identification of transcriptional and post-transcriptional regulations in human respiratory epithelial cells during
Liu et al. BMC Bioinformatics 2014, 15:336 METHODOLOGY ARTICLE Open Access Systematic identification of transcriptional and post-transcriptional regulations in human respiratory epithelial cells during
Bezzi et al., Supplementary Figure 1 *** Nature Medicine: doi: /nm Pten pc-/- ;Zbtb7a pc-/- Pten pc-/- ;Pml pc-/- Pten pc-/- ;Trp53 pc-/-
 Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Cancer outlier differential gene expression detection
 Biostatistics (2007), 8, 3, pp. 566 575 doi:10.1093/biostatistics/kxl029 Advance Access publication on October 4, 2006 Cancer outlier differential gene expression detection BAOLIN WU Division of Biostatistics,
Biostatistics (2007), 8, 3, pp. 566 575 doi:10.1093/biostatistics/kxl029 Advance Access publication on October 4, 2006 Cancer outlier differential gene expression detection BAOLIN WU Division of Biostatistics,
cis-regulatory enrichment analysis in human, mouse and fly
 cis-regulatory enrichment analysis in human, mouse and fly Zeynep Kalender Atak, PhD Laboratory of Computational Biology VIB-KU Leuven Center for Brain & Disease Research Laboratory of Computational Biology
cis-regulatory enrichment analysis in human, mouse and fly Zeynep Kalender Atak, PhD Laboratory of Computational Biology VIB-KU Leuven Center for Brain & Disease Research Laboratory of Computational Biology
Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice
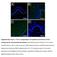 Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice undergoing AD- and psoriasis-like disease. Immunofluorescence staining for Fn14 (green) and DAPI (blue) in skin of naïve
Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice undergoing AD- and psoriasis-like disease. Immunofluorescence staining for Fn14 (green) and DAPI (blue) in skin of naïve
Supplementary Figure 1. The overlap between statistically-significant and highconfidence
 Supplementary figures Supplementary Figure 1. The overlap between statistically-significant and highconfidence PPIs with previously reported PPIs in public databases. Venn diagrams showing the overlap
Supplementary figures Supplementary Figure 1. The overlap between statistically-significant and highconfidence PPIs with previously reported PPIs in public databases. Venn diagrams showing the overlap
Review II: Cell Biology
 Review II: Cell Biology Rajan Munshi BBSI @ Pitt 2006 Department of Computational Biology University of Pittsburgh School of Medicine May 24, 2006 Outline Cell Cycle Signal Transduction 1 Cell Cycle Four
Review II: Cell Biology Rajan Munshi BBSI @ Pitt 2006 Department of Computational Biology University of Pittsburgh School of Medicine May 24, 2006 Outline Cell Cycle Signal Transduction 1 Cell Cycle Four
The Research Roadmap Checklist
 1/5 The Research Roadmap Checklist Version: December 1, 2007 All enquires to bwhitworth@acm.org This checklist is at http://brianwhitworth.com/researchchecklist.pdf The element details are explained at
1/5 The Research Roadmap Checklist Version: December 1, 2007 All enquires to bwhitworth@acm.org This checklist is at http://brianwhitworth.com/researchchecklist.pdf The element details are explained at
VAP Are strict diagnostic criteria advisable?
 VAP Are strict diagnostic criteria advisable? Javier Garau, MD, PhD 18th Infection and Sepsis Symposium, Porto, 27th February 2013 Limitations of current definitions Alternatives -Streamlined definition
VAP Are strict diagnostic criteria advisable? Javier Garau, MD, PhD 18th Infection and Sepsis Symposium, Porto, 27th February 2013 Limitations of current definitions Alternatives -Streamlined definition
Supplementary Methods
 Supplementary Methods Analysis of time course gene expression data. The time course data of the expression level of a representative gene is shown in the below figure. The trajectory of longitudinal expression
Supplementary Methods Analysis of time course gene expression data. The time course data of the expression level of a representative gene is shown in the below figure. The trajectory of longitudinal expression
Metabolomic Data Analysis with MetaboAnalyst
 Metabolomic Data Analysis with MetaboAnalyst User ID: guest6501 April 16, 2009 1 Data Processing and Normalization 1.1 Reading and Processing the Raw Data MetaboAnalyst accepts a variety of data types
Metabolomic Data Analysis with MetaboAnalyst User ID: guest6501 April 16, 2009 1 Data Processing and Normalization 1.1 Reading and Processing the Raw Data MetaboAnalyst accepts a variety of data types
CENTRAL LABORATORY SERVICES
 CENTRAL LABORATORY SERVICES Bridging the Gaps in Biomarker Development and Testing for Global Clinical Trials Biomarkers, Digital Pathology & the Central Lab Presentation Title Presenter Name July 16,
CENTRAL LABORATORY SERVICES Bridging the Gaps in Biomarker Development and Testing for Global Clinical Trials Biomarkers, Digital Pathology & the Central Lab Presentation Title Presenter Name July 16,
Expanded View Figures
 Solip Park & Ben Lehner Epistasis is cancer type specific Molecular Systems Biology Expanded View Figures A B G C D E F H Figure EV1. Epistatic interactions detected in a pan-cancer analysis and saturation
Solip Park & Ben Lehner Epistasis is cancer type specific Molecular Systems Biology Expanded View Figures A B G C D E F H Figure EV1. Epistatic interactions detected in a pan-cancer analysis and saturation
Transduction of lentivirus to human primary CD4+ T cells
 Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
HEDIS 2018 Medications List Directory User Manual
 HEDIS 2018 Medications List Directory User Manual 2017-11-01 Table of Contents Table of Contents What Is the HEDIS 2018 Medications List Directory?... 1 Included in the 2018 Medications List Directory...
HEDIS 2018 Medications List Directory User Manual 2017-11-01 Table of Contents Table of Contents What Is the HEDIS 2018 Medications List Directory?... 1 Included in the 2018 Medications List Directory...
Transcriptional Profiles from Paired Normal Samples Offer Complementary Information on Cancer Patient Survival -- Evidence from TCGA Pan-Cancer Data
 Transcriptional Profiles from Paired Normal Samples Offer Complementary Information on Cancer Patient Survival -- Evidence from TCGA Pan-Cancer Data Supplementary Materials Xiu Huang, David Stern, and
Transcriptional Profiles from Paired Normal Samples Offer Complementary Information on Cancer Patient Survival -- Evidence from TCGA Pan-Cancer Data Supplementary Materials Xiu Huang, David Stern, and
Abstract. Introduction
 Lack of Clinical Manifestations in Asymptomatic Dengue Infection Is Attributed to Broad Down-Regulation and Selective Up-Regulation of Host Defence Response Genes Adeline S. L. Yeo 1, Nur Atiqah Azhar
Lack of Clinical Manifestations in Asymptomatic Dengue Infection Is Attributed to Broad Down-Regulation and Selective Up-Regulation of Host Defence Response Genes Adeline S. L. Yeo 1, Nur Atiqah Azhar
Single SNP/Gene Analysis. Typical Results of GWAS Analysis (Single SNP Approach) Typical Results of GWAS Analysis (Single SNP Approach)
 High-Throughput Sequencing Course Gene-Set Analysis Biostatistics and Bioinformatics Summer 28 Section Introduction What is Gene Set Analysis? Many names for gene set analysis: Pathway analysis Gene set
High-Throughput Sequencing Course Gene-Set Analysis Biostatistics and Bioinformatics Summer 28 Section Introduction What is Gene Set Analysis? Many names for gene set analysis: Pathway analysis Gene set
Late regulation of immune genes and micrornas in circulating leukocytes in a pig model of
 1 Supplementary material for: 2 3 4 5 6 Late regulation of immune genes and micrornas in circulating leukocytes in a pig model of influenza A (H1N2) infection Louise Brogaard, Peter M. H. Heegaard, Lars
1 Supplementary material for: 2 3 4 5 6 Late regulation of immune genes and micrornas in circulating leukocytes in a pig model of influenza A (H1N2) infection Louise Brogaard, Peter M. H. Heegaard, Lars
Characteriza*on of Soma*c Muta*ons in Cancer Genomes
 Characteriza*on of Soma*c Muta*ons in Cancer Genomes Ben Raphael Department of Computer Science Center for Computa*onal Molecular Biology Soma*c Muta*ons and Cancer Clonal Theory (Nowell 1976) Passenger
Characteriza*on of Soma*c Muta*ons in Cancer Genomes Ben Raphael Department of Computer Science Center for Computa*onal Molecular Biology Soma*c Muta*ons and Cancer Clonal Theory (Nowell 1976) Passenger
Supplementary information. Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls
 Supplementary information Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls angiogenesis in breast cancer Ali Flores-Pérez, Laurence A. Marchat, Sergio Rodríguez-Cuevas, Verónica Bautista-Piña,
Supplementary information Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls angiogenesis in breast cancer Ali Flores-Pérez, Laurence A. Marchat, Sergio Rodríguez-Cuevas, Verónica Bautista-Piña,
Supplementary Figure 1. Successful excision of genes from WBM lysates and
 Supplementary Information: Supplementary Figure 1. Successful excision of genes from WBM lysates and survival of mice with different genotypes. (a) The proper excision of Pten, p110α, p110α and p110δ was
Supplementary Information: Supplementary Figure 1. Successful excision of genes from WBM lysates and survival of mice with different genotypes. (a) The proper excision of Pten, p110α, p110α and p110δ was
Supplement Results, Figures and Tables
 Supplement Results, Figures and Tables Results PET imaging The SUV was significantly higher in tumours of the parental HSC-3 cell line than tumours of the EV and the MMP-8 overexpressing cell lines (Figure
Supplement Results, Figures and Tables Results PET imaging The SUV was significantly higher in tumours of the parental HSC-3 cell line than tumours of the EV and the MMP-8 overexpressing cell lines (Figure
Clinical Grade Genomic Profiling: The Time Has Come
 Clinical Grade Genomic Profiling: The Time Has Come Gary Palmer, MD, JD, MBA, MPH Senior Vice President, Medical Affairs Foundation Medicine, Inc. Oct. 22, 2013 1 Why We Are Here A Shared Vision At Foundation
Clinical Grade Genomic Profiling: The Time Has Come Gary Palmer, MD, JD, MBA, MPH Senior Vice President, Medical Affairs Foundation Medicine, Inc. Oct. 22, 2013 1 Why We Are Here A Shared Vision At Foundation
Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester
 Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester dsg6@le.ac.uk CFDNA/CTDNA Circulating-free AS A LIQUID DNA BIOPSY (cfdna) Tumour Biopsy Liquid Biopsy
Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester dsg6@le.ac.uk CFDNA/CTDNA Circulating-free AS A LIQUID DNA BIOPSY (cfdna) Tumour Biopsy Liquid Biopsy
Supplement of Co-occurrence patterns in aquatic bacterial communities across changing permafrost landscapes
 Supplement of Biogeosciences, 13, 175 190, 2016 http://www.biogeosciences.net/13/175/2016/ doi:10.5194/bg-13-175-2016-supplement Author(s) 2016. CC Attribution 3.0 License. Supplement of Co-occurrence
Supplement of Biogeosciences, 13, 175 190, 2016 http://www.biogeosciences.net/13/175/2016/ doi:10.5194/bg-13-175-2016-supplement Author(s) 2016. CC Attribution 3.0 License. Supplement of Co-occurrence
Supplemental Figure S1. RANK expression on human lung cancer cells.
 Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
Supplementary Figure 1. Flow cytometry panels used for BD Canto (A) and BD Fortessa (B).
 Intra Immune nalysis Surface Supplementary Figure 1. Flow cytometry panels used for D Canto () and D Fortessa (). Name Fluorochrome ID F488 PE PerCp-Cy5.5 PC Paclue PE-Cy7 PC-H7 Lympho* 1 CD56 CD8 CD16
Intra Immune nalysis Surface Supplementary Figure 1. Flow cytometry panels used for D Canto () and D Fortessa (). Name Fluorochrome ID F488 PE PerCp-Cy5.5 PC Paclue PE-Cy7 PC-H7 Lympho* 1 CD56 CD8 CD16
What s in a Tipping Point?
 What s in a Tipping Point? Using Systems Biology to Characterize Adverse Oxidative Responses in Human Lung Cells Jenna M. Currier, Ph.D. ORISE Postdoctoral Fellow at U.S. EPA Mentors: Brian Chorley, Ph.D.
What s in a Tipping Point? Using Systems Biology to Characterize Adverse Oxidative Responses in Human Lung Cells Jenna M. Currier, Ph.D. ORISE Postdoctoral Fellow at U.S. EPA Mentors: Brian Chorley, Ph.D.
