SUPPLEMENTAL DATA. Table S1. Genes in each pathway selected for analysis.
|
|
|
- Liliana Jefferson
- 6 years ago
- Views:
Transcription
1 SUPPLEMENTAL DATA Table S1. Genes in each pathway selected for analysis. Gene Chr Start End bp Pathway 1 AKR1C COX 2 AKR1C COX 3 AKR1C COX 4 CXCL COX 5 CYP19A COX 6 FIGF COX 7 IL12A COX 8 IL12B COX 9 IL12RB COX 10 IL12RB COX 11 MGST COX 12 MGST COX 13 MGST COX 14 MMP COX 15 MMP COX 16 MMP COX 17 MMP COX 18 PPARA COX 19 PPARD COX 20 PPARG COX 21 PTGDR COX 22 PTGDR COX 23 PTGDS COX 24 PTGER COX 25 PTGER COX 26 PTGER COX 27 PTGER COX 28 PTGES COX 29 PTGES COX 30 PTGFR COX 31 PTGIR COX 32 PTGIS COX 33 PTGS COX 34 PTGS COX 35 TBXA2R COX 36 TBXAS COX 37 TIMP COX 38 VEGFA COX 39 VEGFB COX 40 VEGFC COX 1
2 ** No Omni1M SNPs available Gene Chr Start End bp Pathway 1 CCL cyto 2 CCL cyto 3 CCL cyto 4 CCL cyto 5 CCL cyto 6 CCL cyto 7 CCL cyto 8 CCL cyto 9 CCL cyto 10 CCL cyto 11 CCL cyto 12 CCL cyto 13 CCL cyto 14 CCL cyto 15 CCL cyto 16 CCL cyto 17 CCL cyto 18 CCL cyto 19 CCL cyto 20 CCL3L cyto 21 CCL3L cyto 22 CCL cyto 23 CCL4L cyto 24 CCL4L cyto 25 CCL cyto 26 CCL cyto 27 CCL cyto 28 CD40LG cyto 29 CD cyto 30 CRP cyto 31 CXCL cyto 32 CXCR cyto 33 CXCR cyto 34 EDA cyto 35 FASLG cyto 36 IFNA cyto 37 IFNA cyto 38 IFNA11P cyto 39 IFNA12P cyto 40 IFNA cyto 41 IFNA cyto 2
3 42 IFNA cyto 43 IFNA cyto 44 IFNA cyto 45 IFNA20P cyto 46 IFNA cyto 47 IFNA22P cyto 48 IFNA cyto 49 IFNA cyto 50 IFNA cyto 51 IFNA cyto 52 IFNA cyto 53 IFNAR cyto 54 IFNAR cyto 55 IFNB cyto 56 IFNG cyto 57 IFNGR cyto 58 IFNGR cyto 59 IFNK cyto 60 IFNL cyto 61 IFNL cyto 62 IFNL cyto 63 IFNLR cyto 64 IFNNP cyto 65 IFNW cyto 66 IFNWP cyto 67 IFNWP cyto 68 IFNWP cyto 69 IFNWP cyto 70 IFNWP cyto 71 IFNWP cyto 72 IL cyto 73 IL10RA cyto 74 IL10RB cyto 75 IL cyto 76 IL11RA cyto 77 IL12A cyto 78 IL12B cyto 79 IL12RB cyto 80 IL12RB cyto 81 IL cyto 82 IL13RA cyto 83 IL13RA cyto 84 IL cyto 85 IL15RA cyto 3
4 86 IL cyto 87 IL17A cyto 88 IL17B cyto 89 IL17C cyto 90 IL17D cyto 91 IL17F cyto 92 IL17RA cyto 93 IL17RB cyto 94 IL17RC cyto 95 IL17RD cyto 96 IL17RE cyto 97 IL cyto 98 IL18BP cyto 99 IL18R cyto 100 IL18RAP cyto 101 IL cyto 102 IL1A cyto 103 IL1B cyto 104 IL1F cyto 105 IL1R cyto 106 IL1R cyto 107 IL1RAP cyto 108 IL1RAPL cyto 109 IL1RAPL cyto 110 IL1RL cyto 111 IL1RL cyto 112 IL1RN cyto 113 IL cyto 114 IL cyto 115 IL20RA cyto 116 IL20RB cyto 117 IL cyto 118 IL21R cyto 119 IL cyto 120 IL22RA cyto 121 IL22RA cyto 122 IL23A cyto 123 IL23R cyto 124 IL cyto 125 IL cyto 126 IL cyto 127 IL cyto 128 IL27RA cyto 129 IL2RA cyto 4
5 130 IL2RB cyto 131 IL2RG cyto 132 IL cyto 133 IL cyto 134 IL31RA cyto 135 IL cyto 136 IL cyto 137 IL cyto 138 IL36A cyto 139 IL36B cyto 140 IL36G cyto 141 IL36RN cyto 142 IL cyto 143 IL3RA cyto 144 IL cyto 145 IL4R cyto 146 IL cyto 147 IL5RA cyto 148 IL cyto 149 IL6R cyto 150 IL6RP cyto 151 IL6ST cyto 152 IL6STP cyto 153 IL cyto 154 IL7R cyto 155 IL cyto 156 IL9R cyto 157 IL9RP cyto 158 IL9RP cyto 159 IL9RP cyto 160 IL9RP cyto 161 JAK cyto 162 JAK cyto 163 JAK cyto 164 LTA cyto 165 LTB cyto 166 SOCS cyto 167 SOCS cyto 168 SOCS cyto 169 SOCS cyto 170 SOCS cyto 171 SOCS cyto 172 SOCS cyto 173 STAT cyto 5
6 174 STAT cyto 175 STAT cyto 176 STAT cyto 177 STAT5A cyto 178 STAT5B cyto 179 STAT cyto 180 TGFB cyto 181 TGFB cyto 182 TGFB cyto 183 TGFBR cyto 184 TGFBR cyto 185 TMEM cyto 186 TNF cyto 187 TNFSF cyto 188 TNFSF cyto 189 TNFSF cyto 190 TNFSF cyto 191 TNFSF13B cyto 192 TNFSF cyto 193 TNFSF cyto 194 TNFSF cyto 195 TNFSF cyto 196 TNFSF cyto 197 TNFSF cyto 198 TRADD cyto 1 BTN1A ox 2 BTN2A ox 3 BTN2A ox 4 BTN2A3P ox 5 BTN3A ox 6 BTN3A ox 7 BTN3A ox 8 BTNL ox 9 BTNL ox 10 BTNL ox 11 BTNL ox 12 BTNL ox 13 CAT ox 14 CYBA ox 15 CYBB ox 16 CYBRD ox 17 CYP1A ox 18 CYP1A ox 6
7 19 FOS ox 20 FOSB ox 21 FOSL ox 22 FOSL ox 23 GCLC ox 24 GPX ox 25 GPX ox 26 GPX ox 27 GPX ox 28 GPX ox 29 GSR ox 30 GSTA ox 31 GSTA ox 32 GSTA ox 33 GSTA ox 34 GSTA ox 35 GSTA6P ox 36 GSTA7P ox 37 GSTM ox 38 GSTM ox 39 GSTM ox 40 GSTM ox 41 GSTM ox 42 GSTO ox 43 GSTO ox 44 GSTO3P ox 45 GSTP ox 46 GSTT ox 47 GSTT ox 48 GSTT2B ox 49 GSTZ ox 50 HIF1A ox 51 HMOX ox 52 HPGDS ox 53 JDP ox 54 JUN ox 55 JUNB ox 56 JUND ox 57 MAOA ox 58 MAPK ox 59 MAPK ox 60 MAPK ox 61 MAPK ox 62 MAPK ox 7
8 63 MAPK ox 64 MAPK ox 65 MAPK ox 66 MAPK ox 67 MAPK ox 68 MAPK ox 69 MAPK ox 70 MAPK ox 71 MGST ox 72 MGST ox 73 MGST ox 74 MT1X ox 75 NFE ox 76 NFE2L ox 77 NFE2L ox 78 NFE2L ox 79 NFIL ox 80 NFIX ox 81 NFKB ox 82 NFKB ox 83 NFKBIA ox 84 NFKBIB ox 85 NFKBID ox 86 NFKBIE ox 87 NFKBIZ ox 88 NOX ox 89 NQO ox 90 SOD ox 91 SOD ox 92 SOD ox 93 SP ox 94 SP ox 95 SP ox 96 SP ox 97 SP ox 98 SP ox 99 SP ox 100 SP ox 101 SP ox 102 STAP ox 103 STAT ox 104 STAT ox 105 STAT ox 106 STAT ox 8
9 107 STAT5A ox 108 STAT5B ox 109 STAT ox 110 TP ox 111 TXN ox 112 TXN ox 113 TXNRD ox 114 TXNRD ox 115 TXNRD ox 116 UGT1A ox 117 XDH ox 1 HLA-A immune 2 HLA-B immune 3 HLA-C immune 4 HLA-DMA immune 5 HLA-DMB immune 6 HLA-DOA immune 7 HLA-DOB immune 8 HLA-DPA immune 9 HLA-DPA immune 10 HLA-DPA immune 11 HLA-DPB immune 12 HLA-DPB immune 13 HLA-DQA immune 14 HLA-DQA immune 15 HLA-DQB immune 16 HLA-DQB immune 17 HLA-DRA immune 18 HLA-DRB immune 19 HLA-DRB immune 20 HLA-DRB immune 21 HLA-DRB immune 22 HLA-E immune 23 HLA-F immune 24 HLA-G immune 25 LTA immune 26 LTB immune 27 MICA immune 28 MICB immune 29 TAP immune 30 TAP immune 31 TAPBP immune 32 TNF immune 9
10 1 AGER nfkb 2 BCL nfkb 3 BCL nfkb 4 BIRC nfkb 5 BIRC nfkb 6 CASP nfkb 7 CCL nfkb 8 CCL nfkb 9 CCL nfkb 10 CD nfkb 11 CD nfkb 12 CD nfkb 13 CD40LG nfkb 14 CD nfkb 15 CDC nfkb 16 CFB nfkb 17 CFLAR nfkb 18 CHUK nfkb 19 CXCL nfkb 20 CXCL nfkb 21 DDX nfkb 22 F nfkb 23 FAS nfkb 24 FASLG nfkb 25 FKBP nfkb 26 ICAM nfkb 27 IFNB nfkb 28 IKBKB nfkb 29 IKBKE nfkb 30 IKBKG nfkb 31 IL12B nfkb 32 IL1B nfkb 33 IL nfkb 34 IRAK nfkb 35 IRAK nfkb 36 IRAK nfkb 37 IRAK nfkb 38 IRF nfkb 39 IRF nfkb 40 IRF nfkb 41 LTA nfkb 42 LTB nfkb 43 MAP3K nfkb 10
11 44 MCC nfkb 45 MMP nfkb 46 MUL nfkb 47 MYD nfkb 48 NFKB nfkb 49 NFKB nfkb 50 NFKBIA nfkb 51 NFKBIB nfkb 52 NFKBID nfkb 53 NFKBIE nfkb 54 NFKBIZ nfkb 55 NGFR nfkb 56 NKAP nfkb 57 NKAPL nfkb 58 NKIRAS nfkb 59 NKIRAS nfkb 60 NKRF nfkb 61 NOS nfkb 62 RELA nfkb 63 RELB nfkb 64 RIPK nfkb 65 SELE nfkb 66 SELP nfkb 67 SELPLG nfkb 68 SIKE nfkb 69 SOD nfkb 70 SPAG nfkb 71 TAB nfkb 72 TAB nfkb 73 TAB nfkb 74 TANK nfkb 75 TBK nfkb 76 TBKBP nfkb 77 TICAM nfkb 78 TICAM nfkb 79 TIRAP nfkb 80 TLR nfkb 81 TLR nfkb 82 TLR nfkb 83 TLR nfkb 84 TLR nfkb 85 TLR nfkb 86 TLR nfkb 87 TLR nfkb 11
12 88 TNF nfkb 89 TNFAIP nfkb 90 TNFRSF10A nfkb 91 TNFRSF10B nfkb 92 TNFRSF10C nfkb 93 TNFRSF10D nfkb 94 TNFRSF11A nfkb 95 TNFRSF12A nfkb 96 TNFRSF13B nfkb 97 TNFRSF13C nfkb 98 TNFRSF nfkb 99 TNFRSF nfkb 100 TNFRSF nfkb 101 TNFRSF1A nfkb 102 TNFRSF1B nfkb 103 TNFRSF nfkb 104 TNFRSF nfkb 105 TNFRSF nfkb 106 TNFRSF nfkb 107 TNFSF nfkb 108 TNFSF nfkb 109 TNFSF nfkb 110 TNFSF13B nfkb 111 TNFSF nfkb 112 TNFSF nfkb 113 TNFSF nfkb 114 TOLLIP nfkb 115 TONSL nfkb 116 TRAF nfkb 117 TRAF nfkb 118 TRAF nfkb 119 TRAF nfkb 120 TRAF nfkb 121 TRAF nfkb 122 TRAF nfkb 123 TRAIP nfkb 124 TRIM nfkb 125 VCAM nfkb 12
13 Table S2. Gene-level associations with risk of EA. Gene Variants a PCs b P c q d 1 MGST1 Microsomal glutathione S-transferase TBXAS1 Thromboxane A synthase 1 (platelet) IL12RB2 Interleukin 12 receptor, beta PTGER1 Prostaglandin E receptor 1 (subtype EP1), 42kDa PTGER3 Prostaglandin E receptor 3 (subtype EP3) MMP2 Matrix metallopeptidase PPARG Peroxisome proliferator-activated receptor gamma PPARD Peroxisome proliferator-activated receptor delta IL12B Interleukin 12B CYP19A1 Cytochrome P450, family 19, subfamily A, polypeptide a Total number of SNPs selected for analysis of the indicated gene; b Gene-level principal components (PCs) included in the logistic regression model; c Likelihood ratio P value; d False discovery rate (FDR) q value. 13
14 Table S3. Assessment of MGST1 SNPs (n=14, Table 4) and risk of EA Controls EA cases SNP Chr Position Alleles a N MAF b N MAF b OR c 95% CI P q d 1 rs G/A ( ) rs G/C ( ) rs T/C ( ) rs C/T ( ) rs C/T ( ) rs T/C ( ) rs T/C ( ) rs T/C ( ) rs T/C ( ) rs A/G ( ) rs G/C ( ) rs G/A ( ) rs C/A ( ) rs A/G ( ) a Minor/major alleles, b Minor allele frequency, c Odds ratio, adjusted for age, sex, PC AIM,1 -PC AIM,4 using additive model (per-allele), d False discovery rate (FDR) 14
15 Table S4. Functional annotations for top MGST1 SNPs associated with risk of BE. SNP dbsnp a Motifs b eqtl c DNAse d rs INT Pou5f1, Sox Whole_blood (MGST1), Cerebellum (MPPE1), Adrenal (RP11-424M22.3) Fetal Adrenal Gland rs INT BRCA1, Foxp1, Hic1, NF-I, RREB-1, TATA Whole blood (MGST1), Cerebellum (MPPE1), Adrenal (RP11-424M22.3). rs INT. Whole blood (MGST1), Cerebellum (MPPE1), Adrenal (RP11-424M22.3) rs INT NRSF, Sin3Ak-20 Whole blood (MGST1), Cerebellum (MPPE1), Adrenal (RP11-424M22.3).. a dbsnp functional annotation (INT: intronic), b Predicted binding motifs altered, c Expression quantitative trait locus (eqtl), d DNAse hypersensitivity (ENCODE). Annotations extracted from HaploReg v4 ( 15
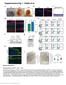
16 Figure S1. Overview of principal components-based approach for pathway-level and gene-level association analyses. 16
17 Figure S2. Regional association plot for n=36 genotyped SNPs at the MGST1 gene locus. The topranked variant among a cluster of six 5 SNPs associated with risk of BE (FDR q<0.05) is shown in solid purple. SNPs are ordered by genomic location. The color scheme indicates LD between the top-ranked SNP and other SNPs in the region using r 2 values calculated from the 1000 Genomes Project. The y axis shows log 10 (P) values computed from 3295 BE cases and 3204 controls. The recombination rate from CEU (Utah residents of Northern and Western European ancestry) HapMap data (right y axis) is shown in light blue. 17
18 Figure S3. NIH Roadmap Epigenome Atlas characterization of the MGST1 genomic locus. Multiple transcript variants for MGST1 are indicated (nc, non-coding). Patterns of DNA methylation (Bisulfite-Seq) and key histone modifications (H3K27me3, H3K4me3, H3K9me3) in esophageal tissue are displayed. The locus is segmented into regions of integrative chromatin states based on a Hidden Markov Model (HMM)-based algorithm trained on 127 epigenomes ( Red: Active TSS, Orange Red: Flanking Active TSS, LimeGreen: Transcr. at gene 5' and 3', Green: Strong transcription, DarkGreen: Weak transcription, GreenYellow: Genic enhancers, Yellow: Enhancers, Medium Aquamarine: ZNF genes & repeats, PaleTurquoise: Heterochromatin, IndianRed: Bivalent/Poised TSS, DarkSalmon: Flanking Bivalent TSS/Enh, DarkKhaki: Bivalent Enhancer, Silver: Repressed PolyComb, Gainsboro: Weak Repressed PolyComb, White: Quiescent/Low. 18
HTG EdgeSeq Immuno-Oncology Assay Gene List
 A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Characterization of HPV Tg/+ ; Krt17 / mice. (a) Macroscopic images of HPV16 Tg/+ mouse ears at P40, P70 and P120. (b) Immunostaining of HPV16 Tg/+ ear tissue sections for K17 (red)
Supplementary Figure 1 Characterization of HPV Tg/+ ; Krt17 / mice. (a) Macroscopic images of HPV16 Tg/+ mouse ears at P40, P70 and P120. (b) Immunostaining of HPV16 Tg/+ ear tissue sections for K17 (red)
* Kyoto Encyclopedia of Genes and Genomes.
 Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
1,000 in silico simulated alpha, beta, gamma and delta TCR repertoires were created.
 938 939 940 941 942 Figure S1 Schematic of the in silico TCRminer and MiXCR validation. 1,000 in silico simulated alpha, beta, gamma and delta TCR repertoires were created. Then, 100,000 simulated 80 bp
938 939 940 941 942 Figure S1 Schematic of the in silico TCRminer and MiXCR validation. 1,000 in silico simulated alpha, beta, gamma and delta TCR repertoires were created. Then, 100,000 simulated 80 bp
qpcr-array Analysis Service
 qpcr-array Analysis Service Customer Name Institute Telephone Address E-mail PO Number Service Code Report Date Service Laboratory Department Phalanx Biotech Group, Inc 6 Floor, No.6, Technology Road 5,
qpcr-array Analysis Service Customer Name Institute Telephone Address E-mail PO Number Service Code Report Date Service Laboratory Department Phalanx Biotech Group, Inc 6 Floor, No.6, Technology Road 5,
Bronchoalveolar lavage fluid analysis showed increased eosinophil in 30% of subjects with IrIa BAL
 Bronchoalveolar lavage fluid analysis showed increased eosinophil in 30% of subjects with IrIa BAL UBC Chan-Yeung Centre for Occupational and Environmental Respiratory Disease Understanding exposure effects
Bronchoalveolar lavage fluid analysis showed increased eosinophil in 30% of subjects with IrIa BAL UBC Chan-Yeung Centre for Occupational and Environmental Respiratory Disease Understanding exposure effects
A genome-wide association study identifies vitiligo
 A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
Supplementary Table 1: OCAC study location, type, and number of cases and controls
 Supplementary Table 1: OCAC study location, type, and number of cases and controls Study Name Abbreviation Study Location Study Type N Cases N Controls Australian Ovarian Cancer Study/Australian Cancer
Supplementary Table 1: OCAC study location, type, and number of cases and controls Study Name Abbreviation Study Location Study Type N Cases N Controls Australian Ovarian Cancer Study/Australian Cancer
Eosinophils! 40! 30! 20! 10! 0! NS!
 A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
To compare the relative amount of of selected gene expression between sham and
 Supplementary Materials and Methods Gene Expression Analysis To compare the relative amount of of selected gene expression between sham and mice given renal ischemia-reperfusion injury (IRI), ncounter
Supplementary Materials and Methods Gene Expression Analysis To compare the relative amount of of selected gene expression between sham and mice given renal ischemia-reperfusion injury (IRI), ncounter
Transduction of lentivirus to human primary CD4+ T cells
 Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
 Table S1. Metadata for transcriptome interaction network and pathway analysis of 5448 intracelluarly infected TEpi cells in comparison to mock TEpi cells. Gene symbol Full name Log2FC gene expression in
Table S1. Metadata for transcriptome interaction network and pathway analysis of 5448 intracelluarly infected TEpi cells in comparison to mock TEpi cells. Gene symbol Full name Log2FC gene expression in
Supplementary Figure 1. Quantile-quantile (Q-Q) plot of the log 10 p-value association results from logistic regression models for prostate cancer
 Supplementary Figure 1. Quantile-quantile (Q-Q) plot of the log 10 p-value association results from logistic regression models for prostate cancer risk in stage 1 (red) and after removing any SNPs within
Supplementary Figure 1. Quantile-quantile (Q-Q) plot of the log 10 p-value association results from logistic regression models for prostate cancer risk in stage 1 (red) and after removing any SNPs within
Molecular basis of age-associated cytokine dysregulation in LPS-stimulated macrophages
 Uncorrected Version. Published on April 6, 2006 as DOI:10.1189/jlb.0106024 Molecular basis of age-associated cytokine dysregulation in LPS-stimulated macrophages R. Lakshman Chelvarajan,*, Yushu Liu, Diana
Uncorrected Version. Published on April 6, 2006 as DOI:10.1189/jlb.0106024 Molecular basis of age-associated cytokine dysregulation in LPS-stimulated macrophages R. Lakshman Chelvarajan,*, Yushu Liu, Diana
Figure 1: Effects of cisplatin on survival of lung cancer cells.
 Figure 1 Figure 1: Effects of cisplatin on survival of lung cancer cells. To determine the IC 50 concentration of cisplatin, cells were treated with various concentrations of cisplatin and cell survival
Figure 1 Figure 1: Effects of cisplatin on survival of lung cancer cells. To determine the IC 50 concentration of cisplatin, cells were treated with various concentrations of cisplatin and cell survival
The innate immune system: Contributions to Immunogenicity of Therapeutic Proteins
 The innate immune system: Contributions to Immunogenicity of Therapeutic Proteins Daniela Verthelyi, M.D., Ph.D. Laboratory of Immunology Division of Therapeu8c Proteins OBP, CDER, FDA Map to the talk:
The innate immune system: Contributions to Immunogenicity of Therapeutic Proteins Daniela Verthelyi, M.D., Ph.D. Laboratory of Immunology Division of Therapeu8c Proteins OBP, CDER, FDA Map to the talk:
Late regulation of immune genes and micrornas in circulating leukocytes in a pig model of
 1 Supplementary material for: 2 3 4 5 6 Late regulation of immune genes and micrornas in circulating leukocytes in a pig model of influenza A (H1N2) infection Louise Brogaard, Peter M. H. Heegaard, Lars
1 Supplementary material for: 2 3 4 5 6 Late regulation of immune genes and micrornas in circulating leukocytes in a pig model of influenza A (H1N2) infection Louise Brogaard, Peter M. H. Heegaard, Lars
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supporting Information
 Supporting Information Rothhammer et al. 1.173/pnas.161413114 SI Materials and Methods EAE Clinical Scoring and Agents. Clinical sig of EAE were assessed as follows:, no sig of disease; 1, loss of tone
Supporting Information Rothhammer et al. 1.173/pnas.161413114 SI Materials and Methods EAE Clinical Scoring and Agents. Clinical sig of EAE were assessed as follows:, no sig of disease; 1, loss of tone
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature89 IFN- (ng ml ) 5 4 3 1 Splenocytes NS IFN- (ng ml ) 6 4 Lymph node cells NS Nfkbiz / Nfkbiz / Nfkbiz / Nfkbiz / IL- (ng ml ) 3 1 Splenocytes IL- (ng ml ) 1 8 6 4 *** ** Lymph node cells
doi: 1.138/nature89 IFN- (ng ml ) 5 4 3 1 Splenocytes NS IFN- (ng ml ) 6 4 Lymph node cells NS Nfkbiz / Nfkbiz / Nfkbiz / Nfkbiz / IL- (ng ml ) 3 1 Splenocytes IL- (ng ml ) 1 8 6 4 *** ** Lymph node cells
CPM (x 10-3 ) Tregs +Teffs. Tregs alone ICOS CLTA-4
 A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.
 Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.32 PCOS locus after conditioning for the lead SNP rs10993397;
Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.32 PCOS locus after conditioning for the lead SNP rs10993397;
Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types.
 Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array.
 Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplemental Table 1. Eighty-nine single nucleotide polymorphisms of 49 immune response genes analyzed in the study
 Supplemental Table 1. Eighty-nine single nucleotide polymorphisms of 49 immune response genes analyzed in the study Gene Symbol Gene ID Gene Full Name SNP ID (alphabetical order) CARD15/NOD2 64127 nucleotide-binding
Supplemental Table 1. Eighty-nine single nucleotide polymorphisms of 49 immune response genes analyzed in the study Gene Symbol Gene ID Gene Full Name SNP ID (alphabetical order) CARD15/NOD2 64127 nucleotide-binding
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/310/ra11/dc1 Supplementary Materials for STAT3 Induction of mir-146b Forms a Feedback Loop to Inhibit the NF-κB to IL-6 Signaling Axis and STAT3-Driven Cancer
www.sciencesignaling.org/cgi/content/full/7/310/ra11/dc1 Supplementary Materials for STAT3 Induction of mir-146b Forms a Feedback Loop to Inhibit the NF-κB to IL-6 Signaling Axis and STAT3-Driven Cancer
Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases
 Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Supplementary Table S1: Gene expression targets
 Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
The circulating T helper subsets and regulatory T cells in patients with common variable immunodeficiency with no monogenic disease
 1 Supplementary data The circulating T helper subsets and regulatory T cells in patients with common variable immunodeficiency with no monogenic disease Azizi et al. 2 Table S1. Excluded 373 genes variants
1 Supplementary data The circulating T helper subsets and regulatory T cells in patients with common variable immunodeficiency with no monogenic disease Azizi et al. 2 Table S1. Excluded 373 genes variants
TNFSF13B tumor necrosis factor (ligand) superfamily, member 13b NF-kB pathway cluster, Enrichment Score: 3.57
 Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
MathIOmica: Dynamic Transcriptome
 Printed from the Complete Wolfram Language Documentation 1 MathIOmica: Dynamic Transcriptome Loading the MathIOmica Package Classification, Clustering and Visualization of Transcriptome Time eries Importing
Printed from the Complete Wolfram Language Documentation 1 MathIOmica: Dynamic Transcriptome Loading the MathIOmica Package Classification, Clustering and Visualization of Transcriptome Time eries Importing
An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action
 An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action Supplementary Figure : Expression levels of toll-like receptor 4 (Tlr4) in muse lung does not change throughout the
An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action Supplementary Figure : Expression levels of toll-like receptor 4 (Tlr4) in muse lung does not change throughout the
Use of Molecular Assays to Assess Cellular Therapies
 Use of Molecular Assays to Assess Cellular Therapies D. Stroncek, P. Jin, M. Sabatino, J. Ren, L. Castiello, E. Wang, and F.M. Marincola. Cellular Therapies Section Clinical Center, NIH Methods for the
Use of Molecular Assays to Assess Cellular Therapies D. Stroncek, P. Jin, M. Sabatino, J. Ren, L. Castiello, E. Wang, and F.M. Marincola. Cellular Therapies Section Clinical Center, NIH Methods for the
Biomedical Informatics Insights. Differential Transcriptional Changes in Mice Exposed to Chemically Distinct Diesel Samples
 Biomedical Informatics Insights Original Research Open Access Full open access to this and thousands of other papers at http://www.la-press.com. Differential Transcriptional Changes in Mice Exposed to
Biomedical Informatics Insights Original Research Open Access Full open access to this and thousands of other papers at http://www.la-press.com. Differential Transcriptional Changes in Mice Exposed to
Interpreting Diverse Genomic Data Using Gene Sets
 Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
Supplementary appendix
 Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Kennedy GA, Varelias A, Vuckovic S, et al.
Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Kennedy GA, Varelias A, Vuckovic S, et al.
Department of International Health (Vaccine science track) Johns Hopkins University Bloomberg School of Public Health
 Acute Respiratory Distress Syndrome is a TH17 and Treg immune disease By Wan-Jiung Hu Postdoctorate Genomics Research Center Academia Sinica No 128 Academia Road section2 Nangang 115, Taipei, Taiwan Previous
Acute Respiratory Distress Syndrome is a TH17 and Treg immune disease By Wan-Jiung Hu Postdoctorate Genomics Research Center Academia Sinica No 128 Academia Road section2 Nangang 115, Taipei, Taiwan Previous
A Systems Biology Approach to the Analysis of Subset- Specific Responses to Lipopolysaccharide in Dendritic Cells
 A Systems Biology Approach to the Analysis of Subset- Specific Responses to Lipopolysaccharide in Dendritic Cells David G. Hancock 1,2, Elena Shklovskaya 1,2, Thomas V. Guy 1,2, Reza Falsafi 3, Chris D.
A Systems Biology Approach to the Analysis of Subset- Specific Responses to Lipopolysaccharide in Dendritic Cells David G. Hancock 1,2, Elena Shklovskaya 1,2, Thomas V. Guy 1,2, Reza Falsafi 3, Chris D.
T-Cell Network Example for GeneNet (August 2015) or later
 T-Cell Network Example for GeneNet 1.2.13 (August 2015) or later This note reproduces the T-Cell network example from R. Opgen-Rhein and K. Strimmer. 2006a. Using regularized dynamic correlation to infer
T-Cell Network Example for GeneNet 1.2.13 (August 2015) or later This note reproduces the T-Cell network example from R. Opgen-Rhein and K. Strimmer. 2006a. Using regularized dynamic correlation to infer
Bezzi et al., Supplementary Figure 1 *** Nature Medicine: doi: /nm Pten pc-/- ;Zbtb7a pc-/- Pten pc-/- ;Pml pc-/- Pten pc-/- ;Trp53 pc-/-
 Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Abstract. Introduction
 Lack of Clinical Manifestations in Asymptomatic Dengue Infection Is Attributed to Broad Down-Regulation and Selective Up-Regulation of Host Defence Response Genes Adeline S. L. Yeo 1, Nur Atiqah Azhar
Lack of Clinical Manifestations in Asymptomatic Dengue Infection Is Attributed to Broad Down-Regulation and Selective Up-Regulation of Host Defence Response Genes Adeline S. L. Yeo 1, Nur Atiqah Azhar
BD Rhapsody Immune Response Panel Hs
 Doc ID: 55665 Rev. 2.0 PRODUCT INSERT BD Rhapsody Immune Response Panel Hs BD Biosciences Kit Part Number: 633750 Purpose The BD Rhapsody Immune Response Panel Hs is a set of ready-to-use primer pools
Doc ID: 55665 Rev. 2.0 PRODUCT INSERT BD Rhapsody Immune Response Panel Hs BD Biosciences Kit Part Number: 633750 Purpose The BD Rhapsody Immune Response Panel Hs is a set of ready-to-use primer pools
The epigenetic landscape of T cell subsets in SLE identifies known and potential novel drivers of the autoimmune response
 Abstract # 319030 Poster # F.9 The epigenetic landscape of T cell subsets in SLE identifies known and potential novel drivers of the autoimmune response Jozsef Karman, Brian Johnston, Sofija Miljovska,
Abstract # 319030 Poster # F.9 The epigenetic landscape of T cell subsets in SLE identifies known and potential novel drivers of the autoimmune response Jozsef Karman, Brian Johnston, Sofija Miljovska,
Interpreting Diverse Genomic Data Using Gene Sets
 Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
Biomarkers of cancer immunotherapy Jason Luke, MD, FACP
 Biomarkers of cancer immunotherapy Jason Luke, MD, FACP Assistant Professor of Medicine Disclosures Consultancy: 7 Hills, Actym, Amgen, Array, AstraZeneca, BeneVir, Bristol-Myers Squibb, Castle, CheckMate,
Biomarkers of cancer immunotherapy Jason Luke, MD, FACP Assistant Professor of Medicine Disclosures Consultancy: 7 Hills, Actym, Amgen, Array, AstraZeneca, BeneVir, Bristol-Myers Squibb, Castle, CheckMate,
Genes, Genes, Everywhere! The key to a cure for IBD
 Genes, Genes, Everywhere! The key to a cure for IBD Dermot McGovern MD, PhD, FRCP Director, Translational Medicine Professor of Medicine Joshua L and Lisa Z Greer Endowed Chair in IBD Genetics F. Widjaja
Genes, Genes, Everywhere! The key to a cure for IBD Dermot McGovern MD, PhD, FRCP Director, Translational Medicine Professor of Medicine Joshua L and Lisa Z Greer Endowed Chair in IBD Genetics F. Widjaja
Development of an In Vitro Human Liver System for Interrogating Non-Alcoholic. Brian R. Wamhoff, 501 Locust Ave., Charlottesville, VA 22902
 Supplemental Information Development of an In Vitro Human Liver System for Interrogating Non-Alcoholic Steatohepatitis Ryan Feaver, Banumathi K Cole, Mark J Lawson, Stephen A Hoang, Svetlana Marukian,
Supplemental Information Development of an In Vitro Human Liver System for Interrogating Non-Alcoholic Steatohepatitis Ryan Feaver, Banumathi K Cole, Mark J Lawson, Stephen A Hoang, Svetlana Marukian,
Supplemental Table 1: Enriched GO categories per cluster
 Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Accessing and Using ENCODE Data Dr. Peggy J. Farnham
 1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
CPT Codes for Pharmacogenomic Tests
 CPT s for Pharmacogenomic Tests The table below lists CPT codes and lab fee information for pharmacogenomic tests as established by the Centers for Medicare and Medicaid Services. It was compiled by the
CPT s for Pharmacogenomic Tests The table below lists CPT codes and lab fee information for pharmacogenomic tests as established by the Centers for Medicare and Medicaid Services. It was compiled by the
Genetics of Pediatric Inflammatory Bowel Disease
 Genetics of Pediatric Inflammatory Bowel Disease Judith Kelsen MD Assistant Professor of Pediatrics Division of Gastroenterology, Hepatology, and Nutrition IBD Education Day 2/9/2014 Objectives Brief overview
Genetics of Pediatric Inflammatory Bowel Disease Judith Kelsen MD Assistant Professor of Pediatrics Division of Gastroenterology, Hepatology, and Nutrition IBD Education Day 2/9/2014 Objectives Brief overview
Molecular mechanisms underlying anti-inflammatory phenotype of neonatal splenic macrophages
 Molecular mechanisms underlying anti-inflammatory phenotype of neonatal splenic macrophages Lakshman Chelvarajan,*, Diana Popa,* Yushu Liu, Thomas V. Getchell,*, Arnold J. Stromberg, and Subbarao Bondada*,,1
Molecular mechanisms underlying anti-inflammatory phenotype of neonatal splenic macrophages Lakshman Chelvarajan,*, Diana Popa,* Yushu Liu, Thomas V. Getchell,*, Arnold J. Stromberg, and Subbarao Bondada*,,1
The Epigenome Tools 2: ChIP-Seq and Data Analysis
 The Epigenome Tools 2: ChIP-Seq and Data Analysis Chongzhi Zang zang@virginia.edu http://zanglab.com PHS5705: Public Health Genomics March 20, 2017 1 Outline Epigenome: basics review ChIP-seq overview
The Epigenome Tools 2: ChIP-Seq and Data Analysis Chongzhi Zang zang@virginia.edu http://zanglab.com PHS5705: Public Health Genomics March 20, 2017 1 Outline Epigenome: basics review ChIP-seq overview
Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction. Marker Sequence (5 3 ) Accession No.
 Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Differential association of HLA with three subtypes of type 1 diabetes: fulminant, slowly progressive and acute-onset
 Diabetologia (2009) 52:2513 2521 DOI 10.1007/s00125-009-1539-9 ARTICLE Differential association of HLA with three subtypes of type 1 diabetes: fulminant, slowly progressive and acute-onset Y. Kawabata
Diabetologia (2009) 52:2513 2521 DOI 10.1007/s00125-009-1539-9 ARTICLE Differential association of HLA with three subtypes of type 1 diabetes: fulminant, slowly progressive and acute-onset Y. Kawabata
Supplemental Figure S1. Tertiles of FKBP5 promoter methylation and internal regulatory region
 Supplemental Figure S1. Tertiles of FKBP5 promoter methylation and internal regulatory region methylation in relation to PSS and fetal coupling. A, PSS values for participants whose placentas showed low,
Supplemental Figure S1. Tertiles of FKBP5 promoter methylation and internal regulatory region methylation in relation to PSS and fetal coupling. A, PSS values for participants whose placentas showed low,
HLA Mismatches. Professor Steven GE Marsh. Anthony Nolan Research Institute EBMT Anthony Nolan Research Institute
 HLA Mismatches Professor Steven GE Marsh HLA Mismatches HLA Genes, Structure, Polymorphism HLA Nomenclature HLA Mismatches in HSCT Defining a mismatch HLA Mismatches HLA Genes, Structure, Polymorphism
HLA Mismatches Professor Steven GE Marsh HLA Mismatches HLA Genes, Structure, Polymorphism HLA Nomenclature HLA Mismatches in HSCT Defining a mismatch HLA Mismatches HLA Genes, Structure, Polymorphism
A Genome-wide Association Study in Han Chinese Identifies Multiple. Susceptibility loci for IgA Nephropathy. Supplementary Material
 A Genome-wide Association Study in Han Chinese Identifies Multiple Susceptibility loci for IgA Nephropathy Supplementary Material Xue-Qing Yu 1,13, Ming Li 1,13, Hong Zhang 2,13, Hui-Qi Low 3, Xin Wei
A Genome-wide Association Study in Han Chinese Identifies Multiple Susceptibility loci for IgA Nephropathy Supplementary Material Xue-Qing Yu 1,13, Ming Li 1,13, Hong Zhang 2,13, Hui-Qi Low 3, Xin Wei
Nature Structural & Molecular Biology: doi: /nsmb.2419
 Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
A Network Analysis of Chinese Medicine Lianhua-Qingwen Formula
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Supplementary Information File A Network Analysis of Chinese Medicine Lianhua-Qingwen
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Supplementary Information File A Network Analysis of Chinese Medicine Lianhua-Qingwen
For focused group profiling of human HIV infection and host response genes expression
 ExProfile TM Human HIV Infection and Host Response Related Gene qpcr Array For focused group profiling of human HIV infection and host response genes expression Cat. No. QG023-A (1 x 96-well plate, Format
ExProfile TM Human HIV Infection and Host Response Related Gene qpcr Array For focused group profiling of human HIV infection and host response genes expression Cat. No. QG023-A (1 x 96-well plate, Format
SUPPLEMENTARY FIG. S2. b-galactosidase staining of
 SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 LD (r 2 ) between the A3AB deletion and all markers in a 400-kb APOBEC3 region in 1000 Genomes Project populations. Populations: CEU, individuals of European ancestry from Utah,
Supplementary Figure 1 LD (r 2 ) between the A3AB deletion and all markers in a 400-kb APOBEC3 region in 1000 Genomes Project populations. Populations: CEU, individuals of European ancestry from Utah,
The Genetic Epidemiology of Rheumatoid Arthritis. Lindsey A. Criswell AURA meeting, 2016
 The Genetic Epidemiology of Rheumatoid Arthritis Lindsey A. Criswell AURA meeting, 2016 Overview Recent successes in gene identification genome wide association studies (GWAS) clues to etiologic pathways
The Genetic Epidemiology of Rheumatoid Arthritis Lindsey A. Criswell AURA meeting, 2016 Overview Recent successes in gene identification genome wide association studies (GWAS) clues to etiologic pathways
Applied Biosystems models 7500 (Fast block), 7900HT (Fast block), StepOnePlus, ViiA 7 (Fast block)
 RT² Profiler PCR Array (96-Well Format and 384-Well [4 x 96] Format) Human HIV Infection and Host Response Cat. no. 330231 PAHS-051YA For pathway expression analysis Format Format A Format C Format D Format
RT² Profiler PCR Array (96-Well Format and 384-Well [4 x 96] Format) Human HIV Infection and Host Response Cat. no. 330231 PAHS-051YA For pathway expression analysis Format Format A Format C Format D Format
Immunological characterization of a recipient of a cross-match positive. Thet Su Win, Naoka Murakami, Thiago J. Borges, Anil Chandraker, George
 Supplementary Materials Immunological characterization of a recipient of a cross-match positive face transplant Thet Su Win, Naoka Murakami, Thiago J. Borges, Anil Chandraker, George Murphy, Christine
Supplementary Materials Immunological characterization of a recipient of a cross-match positive face transplant Thet Su Win, Naoka Murakami, Thiago J. Borges, Anil Chandraker, George Murphy, Christine
BMJ Open. IL-6 pathway driven investigation of response to IL-6 receptor inhibition in rheumatoid arthritis
 IL- pathway driven investigation of response to IL- receptor inhibition in rheumatoid arthritis Journal: Manuscript ID: bmjopen-0-00 Article Type: Research Date Submitted by the Author: 0-May-0 Complete
IL- pathway driven investigation of response to IL- receptor inhibition in rheumatoid arthritis Journal: Manuscript ID: bmjopen-0-00 Article Type: Research Date Submitted by the Author: 0-May-0 Complete
Supplementary Material
 Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
HLA-Cw*16:02. rs Supplementary Figure 1: Genetic association of imputed markers and HLA types in the MHC region.
 -log10 P HLA-B*51 rs1050502 rs116799036 HLA-Cw*16:02 rs12525170 rs114854070 Uncondition Condition on HLA-B*51 HLA-F HLA-A HLA-B HLA-G HLA-C MICA Supplementary Figure 1: Genetic association of imputed markers
-log10 P HLA-B*51 rs1050502 rs116799036 HLA-Cw*16:02 rs12525170 rs114854070 Uncondition Condition on HLA-B*51 HLA-F HLA-A HLA-B HLA-G HLA-C MICA Supplementary Figure 1: Genetic association of imputed markers
Gene Expression Multiplexing Menu
 Gene Expression Multiplexing Menu Mouse Rat Human Table of CONTENTS Mouse Inflammatory Plex 2 Rat Stress Plex 3 Growth Regulation Plex 4 Multitox Plex 5 Pharma III Plex 6 Reference Plex 7 Human Transporter
Gene Expression Multiplexing Menu Mouse Rat Human Table of CONTENTS Mouse Inflammatory Plex 2 Rat Stress Plex 3 Growth Regulation Plex 4 Multitox Plex 5 Pharma III Plex 6 Reference Plex 7 Human Transporter
Cancer, inflammation, and immunity crosstalk
 Contact Technical Support BRCsupport@QIAGEN.COM 1-800-362-7737 Cancer, inflammation, and immunity crosstalk Jesse Liang, Ph.D. Legal disclaimer QIAGEN products shown here are intended for molecular biology
Contact Technical Support BRCsupport@QIAGEN.COM 1-800-362-7737 Cancer, inflammation, and immunity crosstalk Jesse Liang, Ph.D. Legal disclaimer QIAGEN products shown here are intended for molecular biology
Genome-Wide Associations between Genetic and Epigenetic Variation Influence mrna Expression and Insulin Secretion in Human Pancreatic Islets
 Genome-Wide Associations between Genetic and Epigenetic Variation Influence mrna Expression and Insulin Secretion in Human Pancreatic Islets Anders H. Olsson 1, Petr Volkov 1, Karl Bacos 1, Tasnim Dayeh
Genome-Wide Associations between Genetic and Epigenetic Variation Influence mrna Expression and Insulin Secretion in Human Pancreatic Islets Anders H. Olsson 1, Petr Volkov 1, Karl Bacos 1, Tasnim Dayeh
Transcriptional profiling provides insights into metronomic cyclophosphamide-activated, innate immune-dependent regression of brain tumor xenografts
 Doloff and Waxman BMC Cancer (2015) 15:375 DOI 10.1186/s12885-015-1358-y RESEARCH ARTICLE Open Access Transcriptional profiling provides insights into metronomic cyclophosphamide-activated, innate immune-dependent
Doloff and Waxman BMC Cancer (2015) 15:375 DOI 10.1186/s12885-015-1358-y RESEARCH ARTICLE Open Access Transcriptional profiling provides insights into metronomic cyclophosphamide-activated, innate immune-dependent
Inflammation and extracellular proteinases
 Inflammation and extracellular proteinases Plaque rupture 75% of MI (heart attack) Foamy macrophages Thin cap No collagen Thin fibrous cap (
Inflammation and extracellular proteinases Plaque rupture 75% of MI (heart attack) Foamy macrophages Thin cap No collagen Thin fibrous cap (
The Changing Diagnostic Landscape of Primary Immune Deficiency
 The Changing Diagnostic Landscape of Primary Immune Deficiency Dr. Sabeena Selvarajah Paediatric Immunology Fellow, Princess Margaret Hospital for Children, Term infant girl of non-consanguineous parents
The Changing Diagnostic Landscape of Primary Immune Deficiency Dr. Sabeena Selvarajah Paediatric Immunology Fellow, Princess Margaret Hospital for Children, Term infant girl of non-consanguineous parents
Supplemental Figure 1. Mother. Son. Brother. Nephew. Sister (deceased)
 Supplemental Figure 1 IRF8:NM_002163:c.C602T IRF8:NM_002163:c.C671T M Son Br Nephew Sister (deceased) Supplemental Figure 1. Confirmation of family member IRF8 variants by Sanger sequencing. The proband
Supplemental Figure 1 IRF8:NM_002163:c.C602T IRF8:NM_002163:c.C671T M Son Br Nephew Sister (deceased) Supplemental Figure 1. Confirmation of family member IRF8 variants by Sanger sequencing. The proband
B16F1 B16F10 Supplemental Figure S1
 B16F1 B16F1 Supplemental Figure S1 Representative microangiography images of B16F1 and B16F1 tumors grown in the cranial windows. FITC-dextran (2 million MW) was injected systemically to visualize blood
B16F1 B16F1 Supplemental Figure S1 Representative microangiography images of B16F1 and B16F1 tumors grown in the cranial windows. FITC-dextran (2 million MW) was injected systemically to visualize blood
Human genetic susceptibility to infectious disease
 GENOME-WIDE ASSOCIATION STUDIES Human genetic susceptibility to infectious disease Stephen J. Chapman and Adrian V. S. Hill Abstract Recent genome-wide studies have reported novel associations between
GENOME-WIDE ASSOCIATION STUDIES Human genetic susceptibility to infectious disease Stephen J. Chapman and Adrian V. S. Hill Abstract Recent genome-wide studies have reported novel associations between
Statistical Issues in Translational Cancer Research
 Statistical Issues in Translational Cancer Research Martin Bøgsted Department of Haematology Aalborg University Hospital and Department of Clinical Medicine Aalborg University The only useful function
Statistical Issues in Translational Cancer Research Martin Bøgsted Department of Haematology Aalborg University Hospital and Department of Clinical Medicine Aalborg University The only useful function
Nature Immunology: doi: /ni.3745
 Supplementary Figure 1 Experimental settings. (a) Experimental setup and bioinformatic data analysis of transcriptomic changes induced in neonatal and adult monocytes by stimulation with 10 ng/ml LPS for
Supplementary Figure 1 Experimental settings. (a) Experimental setup and bioinformatic data analysis of transcriptomic changes induced in neonatal and adult monocytes by stimulation with 10 ng/ml LPS for
Chromatin marks identify critical cell-types for fine-mapping complex trait variants
 Chromatin marks identify critical cell-types for fine-mapping complex trait variants Gosia Trynka 1-4 *, Cynthia Sandor 1-4 *, Buhm Han 1-4, Han Xu 5, Barbara E Stranger 1,4#, X Shirley Liu 5, and Soumya
Chromatin marks identify critical cell-types for fine-mapping complex trait variants Gosia Trynka 1-4 *, Cynthia Sandor 1-4 *, Buhm Han 1-4, Han Xu 5, Barbara E Stranger 1,4#, X Shirley Liu 5, and Soumya
Epigenetics. Jenny van Dongen Vrije Universiteit (VU) Amsterdam Boulder, Friday march 10, 2017
 Epigenetics Jenny van Dongen Vrije Universiteit (VU) Amsterdam j.van.dongen@vu.nl Boulder, Friday march 10, 2017 Epigenetics Epigenetics= The study of molecular mechanisms that influence the activity of
Epigenetics Jenny van Dongen Vrije Universiteit (VU) Amsterdam j.van.dongen@vu.nl Boulder, Friday march 10, 2017 Epigenetics Epigenetics= The study of molecular mechanisms that influence the activity of
OVARIAN CANCER POSSIBLE TUMOR-SUPPRESSIVE ROLE OF BATF2 IN OVARIAN CANCER
 POSSIBLE TUMOR-SUPPRESSIVE ROLE OF BATF2 IN OVARIAN CANCER Rissa Fedora, OMS-III OVARIAN CANCER 5 th leading cause of death among American women Risk factors: Age Gender female Greater lifetime ovulations
POSSIBLE TUMOR-SUPPRESSIVE ROLE OF BATF2 IN OVARIAN CANCER Rissa Fedora, OMS-III OVARIAN CANCER 5 th leading cause of death among American women Risk factors: Age Gender female Greater lifetime ovulations
Synergy of radiotherapy and PD-1 blockade in Kras-mutant lung cancer
 Supplementary Information Synergy of radiotherapy and PD-1 blockade in Kras-mutant lung cancer Grit S. Herter-Sprie, Shohei Koyama, Houari Korideck, Josephine Hai, Jiehui Deng, Yvonne Y. Li, Kevin A. Buczkowski,
Supplementary Information Synergy of radiotherapy and PD-1 blockade in Kras-mutant lung cancer Grit S. Herter-Sprie, Shohei Koyama, Houari Korideck, Josephine Hai, Jiehui Deng, Yvonne Y. Li, Kevin A. Buczkowski,
An integrated transcriptomic and functional genomic approach identifies a. type I interferon signature as a host defense mechanism against Candida
 Supplementary information for: An integrated transcriptomic and functional genomic approach identifies a type I interferon signature as a host defense mechanism against Candida albicans in humans Sanne
Supplementary information for: An integrated transcriptomic and functional genomic approach identifies a type I interferon signature as a host defense mechanism against Candida albicans in humans Sanne
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Association-heterogeneity mapping identifies an Asian-specific association of the GTF2I locus with rheumatoid arthritis
 Supplementary Material Association-heterogeneity mapping identifies an Asian-specific association of the GTF2I locus with rheumatoid arthritis Kwangwoo Kim 1,, So-Young Bang 1,, Katsunori Ikari 2,3, Dae
Supplementary Material Association-heterogeneity mapping identifies an Asian-specific association of the GTF2I locus with rheumatoid arthritis Kwangwoo Kim 1,, So-Young Bang 1,, Katsunori Ikari 2,3, Dae
Using Ozone to Validate a. Toxicity Testing
 Using Ozone to Validate a Systems Biology Approach to Toxicity Testing Robert Devlin Environmental Public Health Division US EPA Systems Biology Informed Risk Assessment Meeting NationalAcademy of Sciences
Using Ozone to Validate a Systems Biology Approach to Toxicity Testing Robert Devlin Environmental Public Health Division US EPA Systems Biology Informed Risk Assessment Meeting NationalAcademy of Sciences
Molecular profile of cochlear immunity in the resident cells of the organ of Corti
 Cai et al. Journal of Neuroinflammation 2014, 11:173 JOURNAL OF NEUROINFLAMMATION RESEARCH Open Access Molecular profile of cochlear immunity in the resident cells of the organ of Corti Qunfeng Cai 1,
Cai et al. Journal of Neuroinflammation 2014, 11:173 JOURNAL OF NEUROINFLAMMATION RESEARCH Open Access Molecular profile of cochlear immunity in the resident cells of the organ of Corti Qunfeng Cai 1,
Supplementary information. Supplementary figure 1. Flow chart of study design
 Supplementary information Supplementary figure 1. Flow chart of study design Supplementary Figure 2. Quantile-quantile plot of stage 1 results QQ plot of the observed -log10 P-values (y axis) versus the
Supplementary information Supplementary figure 1. Flow chart of study design Supplementary Figure 2. Quantile-quantile plot of stage 1 results QQ plot of the observed -log10 P-values (y axis) versus the
Immunogenetics of juvenile idiopathic arthritis: A comprehensive review
 Immunogenetics of juvenile idiopathic arthritis: A comprehensive review Aimee O. Hersh, University of Utah School of Medicine Sampath Prahalad, Emory University Journal Title: Journal of Autoimmunity Volume:
Immunogenetics of juvenile idiopathic arthritis: A comprehensive review Aimee O. Hersh, University of Utah School of Medicine Sampath Prahalad, Emory University Journal Title: Journal of Autoimmunity Volume:
Integrative analysis of genetic data sets reveals a shared innate immune component in autism spectrum disorder and its co-morbidities
 Integrative analysis of genetic data sets reveals a shared innate immune component in autism spectrum disorder and its co-morbidities The MIT Faculty has made this article openly available. Please share
Integrative analysis of genetic data sets reveals a shared innate immune component in autism spectrum disorder and its co-morbidities The MIT Faculty has made this article openly available. Please share
Significance of the MHC
 CHAPTER 7 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
CHAPTER 7 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
Regulation of inflammation by histone deacetylases in rheumatoid arthritis: beyond epigenetics Grabiec, A.M.
 UvA-DARE (Digital Academic Repository) Regulation of inflammation by histone deacetylases in rheumatoid arthritis: beyond epigenetics Grabiec, A.M. Link to publication Citation for published version (APA):
UvA-DARE (Digital Academic Repository) Regulation of inflammation by histone deacetylases in rheumatoid arthritis: beyond epigenetics Grabiec, A.M. Link to publication Citation for published version (APA):
indicated in shaded lowercase letters (hg19, Chr2: 217,955, ,957,266).
 Legend for Supplementary Figures Figure S1: Sequence of 2q35 encnv. The DNA sequence of the 1,375bp 2q35 encnv is indicated in shaded lowercase letters (hg19, Chr2: 217,955,892-217,957,266). Figure S2:
Legend for Supplementary Figures Figure S1: Sequence of 2q35 encnv. The DNA sequence of the 1,375bp 2q35 encnv is indicated in shaded lowercase letters (hg19, Chr2: 217,955,892-217,957,266). Figure S2:
CD Marker Antibodies. atlasantibodies.com
 CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
Profiling micrornas in lung tissue from pigs infected with Actinobacillus pleuropneumoniae
 Podolska et al. BMC Genomics 2012, 13:459 RESEARCH ARTICLE Open Access Profiling micrornas in lung tissue from pigs infected with Actinobacillus pleuropneumoniae Agnieszka Podolska 1,2, Christian Anthon
Podolska et al. BMC Genomics 2012, 13:459 RESEARCH ARTICLE Open Access Profiling micrornas in lung tissue from pigs infected with Actinobacillus pleuropneumoniae Agnieszka Podolska 1,2, Christian Anthon
Effects of age-at-diagnosis and duration of diabetes on GADA and IA-2A positivity
 Effects of age-at-diagnosis and duration of diabetes on GADA and IA-2A positivity Duration of diabetes was inversely correlated with age-at-diagnosis (ρ=-0.13). However, as backward stepwise regression
Effects of age-at-diagnosis and duration of diabetes on GADA and IA-2A positivity Duration of diabetes was inversely correlated with age-at-diagnosis (ρ=-0.13). However, as backward stepwise regression
Multi-omics integration reveals molecular networks and regulators of psoriasis
 Zhao et al. BMC Systems Biology (2019) 13:8 https://doi.org/10.1186/s12918-018-0671-x RESEARCH ARTICLE Multi-omics integration reveals molecular networks and regulators of psoriasis Yuqi Zhao 1, Deepali
Zhao et al. BMC Systems Biology (2019) 13:8 https://doi.org/10.1186/s12918-018-0671-x RESEARCH ARTICLE Multi-omics integration reveals molecular networks and regulators of psoriasis Yuqi Zhao 1, Deepali
