Nature Genetics: doi: /ng Supplementary Figure 1
|
|
|
- Monica French
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1 Characterization of HPV Tg/+ ; Krt17 / mice. (a) Macroscopic images of HPV16 Tg/+ mouse ears at P40, P70 and P120. (b) Immunostaining of HPV16 Tg/+ ear tissue sections for K17 (red) and Hoechst (blue) at P20 and P40. (c) H&E staining of a wild-type P70 ear tissue section (this micrograph complements the data presented in Fig. 1b). (d) Immunostaining and quantification of vasculature in P70 ear tissue sections from wild-type (green bar), HPV16 Tg/+ (blue bar) and HPV16 Tg/+ ; Krt17 / (open bar) mice. The elements stained are PECAM (green), K14 (red) and Hoechst (blue). n = 9 biological replicates per genotype. (e) Flow cytometry analysis of total cells isolated from P70 ear tissue for four distinct genotypes. Cells double-positive for CD45 and CD4, CD207, CD11b or CD11c were analyzed. Number indicates the
2 percentage of double-positive cells per genotype. Data represent one of three biological replicates. (f) Immunoprecipitation of HPV16 E7 from P2 mouse skin keratinocytes. (g) p53 expression levels in P70 ear tissue. Actin was used as a loading control for total protein, and K14 serves as a loading control for epithelial cells. The K17 immunoblot establishes proper genotyping. PIS, preimmune sera. The blots in f and g represent one of six biological replicates. (h) TUNEL staining of P70 ear tissue, denoting apoptotic cells restricted to the suprabasal layers of epidermis, for both genotypes. Images represent one of six biological replicates. (i) Electron micrographs and aspect ratio quantification (box-and-whisker plot) of basal epithelial cells from 8-month-old ear tissue (8,400 magnification). n = 100 basal cells counted across 3 biological replicates per genotype.
3 Supplementary Figure 2 TPA- and HPV16-induced gene expression in skin tumor keratinocytes. (a) Normalized expression for target genes in intact A431 keratinocytes stimulated with TPA relative to DMSO (solid red bars) and in TPA-treated A431 cells expressing either shcontrol or shkrt17 plasmid (open bars). n = 5 biological replicates. (b) Normalized expression for gene transcripts restored upon Krt17 re-expression (n = 7 biological replicates; blue bars) relative to vector control and compared to Krt42 re-expression (n = 5 biological replicates; red bars). (c) Normalized expression for distinct regions of the Aire transcript (exon 2 or 10 or a region spanning exons 13 and 14) in TPA-treated ears (blue bars) or thymus (red bars) relative to liver, in two different genetic backgrounds (C57BL/6, left; FVB/N, right). n = 4 biological replicates. (d,e) Sense-strand RNA in situ hybridization controls in skin tissue sections (complements data reported in Fig. 2c,d). Scale bars, 200 µm. (f) Coimmunoprecipitation of K17 with RFP-Trap pulldown of mcherry-aire compared to unfused mcherry. GAPDH serves as a loading control for total protein. n = 1 of 7 biological replicates.
4
5 Supplementary Figure 3 Loss of Krt17 does not affect thymic Aire expression or function. (a) Normalized expression for Aire in P10 thymic tissue between wild-type (blue bar), HPV16 Tg/+ (red bar) and HPV16 Tg/+ ; Krt17 / (open bar) mice. *NS, no statistical significance. (b) RNA in situ hybridization for Aire in thymic tissue sections. Arrows denote regions of medullary thymus positive for hybridization. Scale bars, 200 µm. (c) H&E staining of lacrimal glands from aged (>4 month) C57BL/6 mice. Scale bars, 100 µm. (d) Histology scoring for the presence of lymphocytic infiltrate in multiple tissues from aged (>4 month) Krt17 +/+, Krt17 /, Aire +/+ and Aire / mice. *P values are from non-parametric ANOVA with Dunn s post-testing. n.s., no statistical significance. (e) Summary of histology scoring (each pie represents an individual mouse) with each tissue represented by a different color. A filled wedge indicates >10% of the tissue exhibiting inflammation.
6
7 Supplementary Figure 4 Characterization of Aire and nuclear K17 punctae in tumor epithelia. (a) A431 keratinocytes transfected with mcherry-aire and immunostained in green with markers for lysosomes (LAMP-1), stress granules (eif3η) or PML bodies (PML). Scale bars, 5 µm. (b) A431 keratinocytes stably expressing shrna targeting Krt17 (shkrt17) or shcontrol plasmids transiently transfected with mcherry-aire and immunostained for HSP70 (green). Scale bars, 20 µm. (c) Singleplane confocal images of HeLa cells stably expressing GFP-K17 and either mock treated or treated with LMB. Scale bars, 5 µm. (d) Single-plane confocal images through the nucleus of HeLa cells either mock treated or treated with LMB and immunostained for K17 (green) and Hoechst (blue) using mouse (left panels) or rabbit (right panels) anti-k17 antibodies. The z plane is on top. Arrows denote intranuclear punctae immunopositive for K17. Scale bars, 20 µm. (e,f) Single-plane confocal images of immortalized keratinocytes from wild-type (e) or Krt17 / (f) mouse epidermis immunostained for K17 and K5. Scale bars, 20 µm. (g) Immunoblots for subcellular fractions of DMSO- or TPA-treated A431 cells transfected with mcherry-aire compared to unfused mcherry control. Sol, soluble (GAPDH positive); NE, nuclear envelope enriched (nesprin 3 positive); Chr, chromatin enriched (histone H3 positive). (h) Immunoblots of subcellular fractions of DMSO- or TPA-treated A431 keratinocytes probed for various type I keratins, as indicated on the left. Sol, soluble (GAPDH positive); NE, nuclear envelope enriched (nesprin 3 positive); Chr, chromatin enriched (histone H3 positive). All images in a g are representative of three biological replicates. Blots in h are representative of five biological replicates.
8
9 Supplementary Figure 5 ChIP and EMSA assays. (a) Immunoblot for K17 after immunoprecipitation of K17, relative to preimmune sera (PIS), from the nuclear extract used for the ChIP analysis presented in Figure 4. Input represents the sample removed before immunoprecipitation. The image represents one of seven biological replicates. (b) DNA amplicons from the K17 ChIP assay (Fig. 4) observed after equal numbers of qpcr cycles by DNA electrophoresis. The images represent one of three biological replicates. (c,d) 5 upstream sequences of the CXCL10 (c) and CCL19 (d) transcripts. The locations of the forward and reverse primer sites used for the ChIP assays are depicted with colored arrows. NF- B consensus sequences are denoted with bold text. The ATG translation start site is italicized. (e) EMSA analysis of radiolabeled Oct1 consensus oligonucleotides using nuclear extracts from A431 keratinocytes treated with TPA, relative to DMSO control. Supershift analysis was conducted with antibodies against K17, relative to preimmune sera (PIS) control. Cold competition was conducted using 50-fold excess of non-labeled oligonucleotide. The image represents one of three biological replicates. (f) Reciprocal coimmunoprecipitation demonstrating p65 enrichment with K17 immunoprecipitation relative to preimmune sera (PIS) (left panel; n = 7 biological replicates) and K17 enrichment with p65 immunoprecipitation relative to IgG control (right panel; n = 3 biological replicates).
10 R.P. Hobbs et al. Keratin 17 regulates Aire to promote skin tumorigenesis Supplementary Table 1. Summary of "Inflammatory Cytokine and Receptors" array Wildtype HPV16 tg/+ ;Krt17 -/- vs. HPV16 tg vs. HPV16 tg Cxcl Cxcl Hsp90a 1.41 Ccr Il1b Lta 1.31 Il Ccl Tnfrsf1b 1.05 Ccl Cxcl Cxcl Ccl Il1f Il13ra Il8rb Cxcl Mif Ccl Cxcl C Xcr Ccl Cxcr Ccr Ccl Il1r Ccl Ccl Il10rb Ccr Ccl Abcf Il2rg Il1a Ltb Ccr Il Gapdh Ccl Casp Il2rb Ifng Il1f Il1r Ccr Cd40lg Gusb Ccl Itgb Il17b Cxcl Spp Ccr Tnf Il Il Il5ra Hprt Crp Itgam Ccl Il10ra Il Ccr Il HPV16 tg -induced, Krt17-dependent Wildtype HPV16 tg/+ ;Krt17 -/- Cxcr Cx3cl Ccl Ccr Il Pf Ccr Tollip Cxcl Tgfb Scye Actb Il6st Il6ra Ccl Ccl Il Tnfrsf1a Ccl Ccr Ccl Bcl Il HPV16 tg -induced, Krt17-independent Wildtype Not induced by HPV16 tg
11 R.P. Hobbs et al. Keratin 17 regulates Aire to promote skin tumorigenesis Supplementary Table 2. Summary of "NfκB Signaling Pathway" array HPV16 tg/+ ;Krt17 -/- vs. HPV16 tg Il HPV16 tg -induced, Tnfrsf1a 1.09 Chuk Gja Krt17-dependent Ltbr 1.08 Tlr Nfkb Htr2b Tnf Raf Irf Relb Traf Ikbkb HPV16 tg/+ ;Krt17 -/- Rela Traf Akt Tlr vs. HPV16 tg Il1b Hsp90ab Ikbke Csf Rel Tollip Il1a Tnfrsf1b Irak Bcl Icam Ikbkg Il Zap Crebbp Ifng HPV16 tg -induced, Bcl Tgfbr Gapdh Krt17-independent Nod Ripk Casp Eif2ak Ripk Stat Nfkbia Tgfbr Tnfsf Tbk Tlr Nlrp Tnfaip Card Cd Atf Kat2b Mapk Tnfrsf10b Cflar Casp Tradd F2r Egr Lta Jun Il1r Nfkb Elk Fasl 1.49 Tlr Atf Csf C Tlr Hprt Actb Map3k Myd Cd Tlr Fadd 1.33 Tlr Fos Not induced by HPV16 tg Tlr Irak Smad Tnfsf Ccl Lpar Slc20a Gusb Not induced by HPV16 tg Not induced by HPV16 tg
12 R.P. Hobbs et al. Keratin 17 regulates Aire to promote skin tumorigenesis Supplementary Table 3. Summary of "T h 17 for Autoimmunity and Inflammation" Array HPV16 tg/+ ;Krt17 -/- vs. HPV16 tg Il12rb Hprt Isg Socs Socs Cd Ccl Cebpb 1.25 Cx3cl Il Il Gata Il Cacybp 1.16 Il Mmp Cd Mmp Cxcl Myd Cxcl Cxcl Syk 1.08 Mmp Traf Tlr Ccl Cd Nfkb Il17rb Stat S1pr HPV16 tg/+ ;Krt17 -/- Hsp90ab Cd3d vs. HPV16 tg Il17rc Nfatc Il23a Tnf Tbx Csf Il Il Il1b Il17c Il6ra Il Il23r Il17d Ccl Cd Il17rd Il Icam Cd Il12b Il7r Rorc Il17a Yy Cd3e Clec7a Il12rb Cd3g Ifng Stat5a Il Icos Actb Il Il Stat Cxcl Tirap Csf Ccl Cd8a Jak Il17f Gusb Foxp Il17re Cd40lg Jak Gapdh Stat Il Il Ccl Tgfb HPV16 tg -induced, HPV16 tg -induced, Krt17-independent Krt17-dependent Not induced by HPV16 tg Not induced by HPV16 tg
13 R.P. Hobbs et al. Keratin 17 regulates Aire to promote skin tumorigenesis Supplementary Table 4. Summary of "Signal Transduction PathwayFinder" array HPV16 tg/+ ;Krt17 -/- vs. HPV16 tg Cdkn2a Rbp Cdh Cxcl Brca Icam Fos Hprt Vegfa Ccl Hsp90ab Pparg Selp Cdk Lta Fas Birc Hhip Il4ra Mmp Wnt Tnf Mmp Fasn Tfrc 1.20 Naip Vcam Birc Myc 1.14 Ikbkb HPV16 tg/+ ;Krt17 -/- Greb Bax vs. HPV16 tg Igfbp Nab Csf Tcf Hsf Fasl Wisp Tert Ccl Gapdh 1.10 Ei Cxcl Gadd45a 1.03 Cdkn1b Il1a Nos Ccnd Il2ra En Lef Ptgs Atf Bmp Birc Il Hoxa Cd Irf Wnt Cebpb Trp Ptch Egr Actb Hspb Sele Foxa Pmepa Hk Nfkbia Fn Tank Cdkn2b Bmp Odc Fgf Cdkn1a Igfbp Nrip Gys Mdm Bcl2l Jun Bcl Gusb Lep Cyp19a HPV16 tg -induced, HPV16 tg -induced, Krt17-independent Krt17-dependent Not induced by HPV16 tg Not induced by HPV16 tg
14 R.P. Hobbs et al. Keratin 17 regulates Aire to promote skin tumorigenesis Supplementary Table 5. Summary of "Innate and Adaptive Immune Response" array HPV16 tg/+ ;Krt17 -/- vs. HPV16 tg Ccr C8a 1.47 Mapk Pglyrp Tlr Cfp Prg Il12rb Pglyrp Tnf Pglyrp Tgfb Hc Irak Actb Ltf 1.05 Nos HPV16 tg/+ ;Krt17 -/- Ptafr 1.03 Traf Nfkb Crp vs. HPV16 tg HPV16 tg - induced, Krt17- dependent Defb Ppbp Adora2a Trem Cxcr Hmox Il1b Ncf Lalba Il Serpina1a Colec Il1f Tlr Ikbkb Clec7a Irf Tnfrsf1a Il1f Gusb Ifngr Ifnb Il1r Ifngr Tlr Cybb Camp Il Nfkbia Cd1d Il1a Cd Il1f Cd Mif Myd Il1f Mapk Stab Casp Tlr Fn Casp Gapdh Il1rapl2 N/A Il1f Serpine Il1rl2 N/A Ccl Nlrc Lyz Il1r Hprt Tlr Il1rn Nfkb Ly Irak Hsp90ab Tlr Chuk Lbp Dmbt N/A Tollip Proc 1.59 N/A Il1rap Sftpd Tlr HPV16 tg -induced, Krt17-independent Not induced by HPV16 tg Not induced by HPV16 tg
15 R.P. Hobbs et al. Keratin 17 regulates Aire to promote skin tumorigenesis Supplementary Table 6. List of qrt-pcr target genes and primers sets MOUSE HUMAN Primer Sequence (5'-->3') Primer Sequence (5'-->3') Actb Forward GGCTGTATTCCCCTCCATCG ACTB Forward CATGTACGTTGCTATCCAGGC Actb Reverse CCAGTTGGTAACAATGCCATGT ACTB Reverse CTCCTTAATGTCACGCACGAT Aire Forward AGGTCAGCTTCAGAGAAAACCA AIRE Forward CCAGGCTCTCAACTGAAGGC Aire Reverse TCATTCCCAGCACTCAGTAGA AIRE Reverse GAATCCCGTTCCCGAGTGG Anxa2 Forward TGCTCTGAACATTGAGACAGC ANXA2 Forward GAGCGGGATGCTTTGAACATT Anxa2 Reverse AATAGGCCCAAAATCACCGTC ANXA2 Reverse TAGGCGAAGGCAATATCCTGT Axin2 Forward ATGAGTAGCGCCGTGTTAGTG AXIN2 Forward CAACACCAGGCGGAACGAA Axin2 Reverse GGGCATAGGTTTGGTGGACT AXIN2 Reverse GCCCAATAAGGAGTGTAAGGACT Ccl19 Forward CCTGGGAACATCGTGAAAGC CCL19 Forward TACATCGTGAGGAACTTCCACT Ccl19 Reverse TAGTGTGGTGAACACAACAGC CCL19 Reverse CTGGATGATGCGTTCTACCCA Ccl2 Forward TTAAAAACCTGGATCGGAACCAA CCL2 Forward CAGCCAGATGCAATCAATGCC Ccl2 Reverse GCATTAGCTTCAGATTTACGGGT CCL2 Reverse TGGAATCCTGAACCCACTTCT Ccl22 Forward AAGCCTGGCGTTGTTTTGATA CCL22 Forward ATCGCCTACAGACTGCACTC Ccl22 Reverse CCTGGGATCGGCACAGATA CCL22 Reverse GACGGTAACGGACGTAATCAC Ccl25 Forward GAGGGCGATGAGAATCTTGAC CCL25 Forward CACCACAACACGCAGACCTT Ccl25 Reverse TCCTCACGCTTGTACTGTTGG CCL25 Reverse TGCTGATGGGATTGCTAAACTT Ccl5 Forward GCTGCTTTGCCTACCTCTCC CCL5 Forward TCCAACCCAGCAGTCGTCT Ccl5 Reverse TCGAGTGACAAACACGACTGC CCL5 Reverse TTGGCGGTTCTTTCGGGTG Ccr4 Forward TGCACCAAGGAAGGTATCAAGG CCR4 Forward AGAAGGCATCAAGGCATTTGG Ccr4 Reverse GTACACGTCCGTCATGGACTT CCR4 Reverse ACACATCAGTCATGGACCTGAG Ccr7 Forward GTACGAGTCGGTGTGCTTCAA CCR7 Forward TGAGGTCACGGACGATTACAT Ccr7 Reverse GGTAGGTATCCGTCATGGTCT CCR7 Reverse GTAGGCCCACGAAACAAATGAT Cd40 Forward TTGTTGACAGCGGTCCATCTA CD40 Forward CACCTGGAACAGAGAGACACA Cd40 Reverse CCATCGTGGAGGTACTGTTTG CD40 Reverse CCTCACTCGTACAGTGCCA Cflar Forward GCTCCAGAATGGGCGAAGTAA CFLAR Forward TGCTCTTTTTGTGCCGGGAT Cflar Reverse ACGGATGTGCGGAGGTAAAAA CFLAR Reverse CGACAGACAGCTTACCTCTTTC Cxcl10 Forward CCAAGTGCTGCCGTCATTTTC CXCL10 Forward GTGGCATTCAAGGAGTACCTC Cxcl10 Reverse GGCTCGCAGGGATGATTTCAA CXCL10 Reverse TGATGGCCTTCGATTCTGGATT Cxcl11 Forward GGCTTCCTTATGTTCAAACAGGG CXCL11 Forward GACGCTGTCTTTGCATAGGC Cxcl11 Reverse GCCGTTACTCGGGTAAATTACA CXCL11 Reverse GGATTTAGGCATCGTTGTCCTTT Cxcl5 Forward TCCAGCTCGCCATTCATGC CXCL5 Forward AGCTGCGTTGCGTTTGTTTAC Cxcl5 Reverse TTGCGGCTATGACTGAGGAAG CXCL5 Reverse GCGAACACTTGCAGATTACTGA Cxcl9 Forward TCCTTTTGGGCATCATCTTCC CXCL9 Forward GTAGTGAGAAAGGGTCGCTGT Cxcl9 Reverse TTTGTAGTGGATCGTGCCTCG CXCL9 Reverse AGGGCTTGGGGCAAATTGTT Cxcr3 Forward TACCTTGAGGTTAGTGAACGTCA CXCR3 Forward CCACCTAGCTGTAGCAGACAC Cxcr3 Reverse CGCTCTCGTTTTCCCCATAATC CXCR3 Reverse AGGGCTCCTGCGTAGAAGTT Deptor Forward GATAGACGGCACCATCTCAAAA DEPTOR Forward CCTACCCAAACTGTTTTGTCGC Deptor Reverse GGCCTCCTTATGTTCAATGAGC DEPTOR Reverse CGGTCTGCTAATTTCTGCATGAG Egr1 Forward TCGGCTCCTTTCCTCACTCA EGR1 Forward GGTCAGTGGCCTAGTGAGC Egr1 Reverse CTCATAGGGTTGTTCGCTCGG EGR1 Reverse GTGCCGCTGAGTAAATGGGA Eif4e Forward AACCGGAAACCACCCCTAC EIF4E Forward TGCGGCTGATCTCCAAGTTTG Eif4e Reverse CTCTGGGTTAGCAACCTCCT EIF4E Reverse CCCACATAGGCTCAATACCATC Eif4ebp1 Forward GGGGACTACAGCACCACTC EIF4EBP1 Forward CTATGACCGGAAATTCCTGATGG Eif4ebp1 Reverse GTTCCGACACTCCATCAGAAAT EIF4EBP1 Reverse CCCGCTTATCTTCTGGGCTA Fn1 Forward TTCAAGTGTGATCCCCATGAAG FN1 Forward AGGAAGCCGAGGTTTTAACTG Fn1 Reverse CAGGTCTACGGCAGTTGTCA FN1 Reverse AGGACGCTCATAAGTGTCACC
16 Foxp3 Forward ACCATTGGTTTACTCGCATGT FOXP3 Forward CTTCAAGTTCCACAACATGCG Foxp3 Reverse TCCACTCGCACAAAGCACTT FOXP3 Reverse CGTGGCGTAGGTGAAAGGG GAPDH Forward AAATGGTGAAGGTCGGTGT GAPDH Forward AAGGTGAAGGTCGGAGTCAAC GAPDH Reverse ACTCCACGACATACTCAGCAC GAPDH Reverse GGGGTCATTGATGGCAACAATA Hnrnpa2b1 Forward GTTGAGCCAAAACGTGCTGTA HNRNPA2B1 Forward TGGAGGTAGCCCCGGTTATG Hnrnpa2b1 Reverse TTTCCAGACTGCCTATCGGTA HNRNPA2B1 Reverse GGACCGTAGTTAGAAGGTTGCT Hnrnpd Forward CCCCACGACACACTGAAGC HNRNPD Forward GCGTGGGTTCTGCTTTATTACC Hnrnpd Reverse CGCCCTGTGATAGGATCTAACT HNRNPD Reverse TTGCTGATATTGTTCCTTCGACA Hnrnpk Forward ACTGATGAGATGGTTGAATTGCG HNRNPK Forward GCAGGAGGAATTATTGGGGTC Hnrnpk Reverse CTGGCATTGTAGTCTGTACGG HNRNPK Reverse TGCACTCTACAACCCTATCGG Hspa1a/1b Forward TGGTGCAGTCCGACATGAAG HSPA1A/1B Forward TTTGAGGGCATCGACTTCTACA Hspa1a/1b Reverse GCTGAGAGTCGTTGAAGTAGGC HSPA1A/1B Reverse CCAGGACCAGGTCGTGAATC Hspa5 Forward ACTTGGGGACCACCTATTCCT HSPA5 Forward CATCACGCCGTCCTATGTCG Hspa5 Reverse ATCGCCAATCAGACGCTCC HSPA5 Reverse CGTCAAAGACCGTGTTCTCG Hspa8 Forward TCTCGGCACCACCTACTCC HSPA8 Forward ATGCCAAACGTCTGATTGGAC Hspa8 Reverse CTACGCCCGATCAGACGTTT HSPA8 Reverse AGCATCATTCACCACCATAAAGG Hspa9 Forward ATGGCTGGAATGGCCTTAGC HSPA9 Forward CTTGTTTCAAGGCGGGATTATGC Hspa9 Reverse ACCCAAATCAATACCAACCACTG HSPA9 Reverse GCAGGAGTTGGTAGTACCCAAA Icam1 Forward GTGATGCTCAGGTATCCATCCA ICAM1 Forward ATGCCCAGACATCTGTGTCC Icam1 Reverse CACAGTTCTCAAAGCACAGCG ICAM1 Reverse GGGGTCTCTATGCCCAACAA Ido1 Forward GCTTTGCTCTACCACATCCAC IDO1 Forward TTGCACGTCTAGTTCTGGGAT Ido1 Reverse CAGGCGCTGTAACCTGTGT IDO1 Reverse GCAAGACCTTACGGACATCTCC Ido2 Forward AAGGCCAACCCCAAAAGGTG IDO2 Forward CCACAGACCGAATGTGAAGAC Ido2 Reverse ACCAGGATAGGCGGGAGTC IDO2 Reverse TGTTGGCAATTTCCATCCAAGG Ifng Forward ATGAACGCTACACACTGCATC IFNG Forward TCGGTAACTGACTTGAATGTCCA Ifng Reverse CCATCCTTTTGCCAGTTCCTC IFNG Reverse TCGCTTCCCTGTTTTAGCTGC Ikbkb Forward GACATCGCATCGGCTCTTAGA IKBKB Forward GTCTTTGCACATCATTCGTGGG Ikbkb Reverse AACGGTCACGGTGTACTTCTG IKBKB Reverse GTGCCGAAGCTCCAGTAGTC Il17a Forward TTTAACTCCCTTGGCGCAAAA IL17A Forward TCCCACGAAATCCAGGATGC Il17a Reverse CTTTCCCTCCGCATTGACAC IL17A Reverse TGTTCAGGTTGACCATCACAGT Il17c Forward CTCCTGCTTCTAGGCTGGTTG IL17C Forward CCACACTGCTACTCGGCTG Il17c Reverse CCACCTGGCACTTCGAGTTAG IL17C Reverse CACACGGTATCTCCAGGGTGA Il20 Forward TCTTGCCTTTGGACTGTTCTCC IL20 Forward TTTTCTGAGATACGGGGCAGT Il20 Reverse GTTTGCAGTAATCACACAGCTTC IL20 Reverse GTCTTAGCAAATGGCGCAGGA Il4 Forward GGTCTCAACCCCCAGCTAGT IL4 Forward CGGCAACTTTGTCCACGGA Il4 Reverse GCCGATGATCTCTCTCAAGTGAT IL4 Reverse TCTGTTACGGTCAACTCGGTG Il6 Forward CCAAGAGGTGAGTGCTTCCC IL6 Forward AATTCGGTACATCCTCGACGG Il6 Reverse CTGTTGTTCAGACTCTCTCCCT IL6 Reverse TTGGAAGGTTCAGGTTGTTTTCT Irf1 Forward ATGCCAATCACTCGAATGCG IRF1 Forward TTCACACAGGCCGATACAAAG Irf1 Reverse TTGTATCGGCCTGTGTGAATG IRF1 Reverse CATGGCACAGCGAAAGTTGG Krt14 Forward AGCGGCAAGAGTGAGATTTCT KRT14 Forward TGAGCCGCATTCTGAACGAG Krt14 Reverse CCTCCAGGTTATTCTCCAGGG KRT14 Reverse GATGACTGCGATCCAGAGGA Krt16 Forward GGTGGCCTCTAACAGTGATCT KRT16 Forward GACCGGCGGAGATGTGAAC Krt16 Reverse TGCATACAGTATCTGCCTTTGG KRT16 Reverse CTGCTCGTACTGGTCACGC Krt17 Forward ACCATCCGCCAGTTTACCTC KRT17 Forward GGTGGGTGGTGAGATCAATGT Krt17 Reverse CTACCCAGGCCACTAGCTGA KRT17 Reverse CGCGGTTCAGTTCCTCTGTC Mapkap1 Forward GAGACGCAGGGCTACATATACG MAPKAP1 Forward GGTGGACACCGATTTCCCC Mapkap1 Reverse GCGGAGTCGTTCTAATCTTTGA MAPKAP1 Reverse CGCTTCACTGCCTTCAGTAAGA Mlst8 Forward ATCAGTGGGCTTTCACGAGG MLST8 Forward TGTGGGCTTCCACGAAGAC Mlst8 Reverse GGTGCGTTCACCTGGAAGAT MLST8 Reverse AGTTAATGGGTGCGTTCACCT Mmp2 Forward ACCTGAACACTTTCTATGGCTG MMP2 Forward GATACCCCTTTGACGGTAAGGA Mmp2 Reverse CTTCCGCATGGTCTCGATG MMP2 Reverse CCTTCTCCCAAGGTCCATAGC
17 Mmp3 Forward ACATGGAGACTTTGTCCCTTTTG MMP3 Forward CTGGACTCCGACACTCTGGA Mmp3 Reverse TTGGCTGAGTGGTAGAGTCCC MMP3 Reverse CAGGAAAGGTTCTGAAGTGACC Mmp9 Forward CTGGACAGCCAGACACTAAAG MMP9 Forward TGTACCGCTATGGTTACACTCG Mmp9 Reverse CTCGCGGCAAGTCTTCAGAG MMP9 Reverse GGCAGGGACAGTTGCTTCT Mpz Forward CGGACAGGGAAATCTATGGTGC MPZ Forward CATCGTGGTTTACACCGACAG Mpz Reverse GCGCCAGGTAAAAGAGATGTCA MPZ Reverse TGGAAGATCGAAATGGCATCTCT Mpzl1 Forward TGGATGGTTGACCACGGTCT MPZL1 Forward ACGCCAAAAGAAATCTTCGTGG Mpzl1 Reverse CCCAGGTGACTCGGTCTTT MPZL1 Reverse TCAACCCGCCAGTCGTACTA Mpzl2 Forward GATGCGCTAACTGTGACGTG MPZL2 Forward TTAATGGGACAGATGCTCGGT Mpzl2 Reverse GGTCTTTGAACCGTCCGCT MPZL2 Reverse AAGACACCCGGTCCTTAAACC Mpzl3 Forward CCTTGGAGATAAGTGCCGATG MPZL3 Forward ATGCCCATGTCCGAGGTTATG Mpzl3 Reverse CGGTATGTCCAGTCTATGGTCA MPZL3 Reverse GGAGGGCGATATGTCCAGTCT Mtor Forward CAGTTCGCCAGTGGACTGAAG MTOR Forward ACTCGCTTCTATGACCAACTGA Mtor Reverse GCTGGTCATAGAAGCGAGTAGAC MTOR Reverse CACCTTCCACTCCTATGAGGC Myd88 Forward TCATGTTCTCCATACCCTTGGT MYD88 Forward GGCTGCTCTCAACATGCGA Myd88 Reverse AAACTGCGAGTGGGGTCAG MYD88 Reverse CTGTGTCCGCACGTTCAAGA Nfkbia Forward TGAAGGACGAGGAGTACGAGC NFKBIA Forward CTCCGAGACTTTCGAGGAAATAC Nfkbia Reverse TTCGTGGATGATTGCCAAGTG NFKBIA Reverse GCCATTGTAGTTGGTAGCCTTCA Prkca Forward GTTTACCCGGCCAACGACT PRKCA Forward AAGGGACCCGACACTGATGA Prkca Reverse GGGCGATGAATTTGTGGTCTT PRKCA Reverse AAGGTGGGGCTTCCGTAAGT Ptgs2 Forward TGCACTATGGTTACAAAAGCTGG PTGS2 Forward CCAGTATAAGTGCGATTGTACCC Ptgs2 Reverse TCAGGAAGCTCCTTATTTCCCTT PTGS2 Reverse TCAAAAATTCCGGTGTTGAGCA Rela Forward AGGCTTCTGGGCCTTATGTG RELA Forward GTGGGGACTACGACCTGAATG Rela Reverse TGCTTCTCTCGCCAGGAATAC RELA Reverse GGGGCACGATTGTCAAAGATG Rictor Forward GCTGCGCTATCTCATCCAAGA RICTOR Forward TCCAAAGACTCGACAGTATGTGC Rictor Reverse CTTTCTGACTAAGCGAAGGGC RICTOR Reverse GGCTAGAAATCGTGCTTCTCTG Rn18s Forward CCTGTGCCTTCCTTGGA RPS18 Forward GCGGCGGAAAATAGCCTTTG Rn18s Reverse CATTCGAACGTCTGCCCTATC RPS18 Reverse GATCACACGTTCCACCTCATC Rps6kb1 Forward GGGGCTATGGAAAGGTTTTTCA RPS6KB1 Forward AGAACTTCTGGCTCGAAAGGT Rps6kb1 Reverse CGTGTCCTTAGCATTCCTCACT RPS6KB1 Reverse CGACAGGTGTCTGACGTGTAA Rptor Forward TGTGAGAAAATCGAAGGCTCC RPTOR Forward AATGCTGCAATCGCCTCTTCT Rptor Reverse TTTGCACCGATGGTTTCCAGA RPTOR Reverse GCCAAAGGTAGGTTCCAGTCTG Runx1 Forward GCAGGCAACGATGAAAACTACT RUNX1 Forward CTGCCCATCGCTTTCAAGGT Runx1 Reverse GCAACTTGTGGCGGATTTGTA RUNX1 Reverse GCCGAGTAGTTTTCATCATTGCC S100a10 Forward TGGAAACCATGATGCTTACGTT S100A10 Forward GGCTACTTAACAAAGGAGGACC S100a10 Reverse GAAGCCCACTTTGCCATCTC S100A10 Reverse GAGGCCCGCAATTAGGGAAA S100a4 Forward TGAGCAACTTGGACAGCAACA S100A4 Forward GATGAGCAACTTGGACAGCAA S100a4 Reverse TTCCGGGGTTCCTTATCTGGG S100A4 Reverse CTGGGCTGCTTATCTGGGAAG S100a5 Forward ATTCAGGGAGAGAGGGTAGCA S100A5 Forward GGAGAGAGGGTAGCAAACTGA S100a5 Reverse CTCTGCAAGACTCAGCTCTGT S100A5 Reverse TCCCCAAGACACAGCTCTTTC S100a7a Forward AGGAGTTGAAAGCTCTGCTCT S100A7 Forward ACGTGATGACAAGATTGACAAGC S100a7a Reverse GCTCTGTGATGTAGTATGGCTG S100A7 Reverse GCGAGGTAATTTGTGCCCTTT S100a8 Forward AAATCACCATGCCCTCTACAAG S100A8 Forward ATGCCGTCTACAGGGATGAC S100a8 Reverse CCCACTTTTATCACCATCGCAA S100A8 Reverse ACACTCGGTCTCTAGCAATTTCT S100a9 Forward ATACTCTAGGAAGGAAGGACACC S100A9 Forward GGTCATAGAACACATCATGGAGG S100a9 Reverse TCCATGATGTCATTTATGAGGGC S100A9 Reverse GGCCTGGCTTATGGTGGTG Sdc1 Forward CTTTGTCACGGCAGACACCTT SDC1 Forward CTGCCGCAAATTGTGGCTAC Sdc1 Reverse GACAGAGGTAAAAGCAGTCTCG SDC1 Reverse TGAGCCGGAGAAGTTGTCAGA Sfn Forward GTGTGTGCGACACCGTACT SFN Forward TGACGACAAGAAGCGCATCAT Sfn Reverse CTCGGCTAGGTAGCGGTAG SFN Reverse GTAGTGGAAGACGGAAAAGTTCA Socs1 Forward CTGCGGCTTCTATTGGGGAC SOCS1 Forward TTTTCGCCCTTAGCGTGAAGA Socs1 Reverse AAAAGGCAGTCGAAGGTCTCG SOCS1 Reverse GAGGCAGTCGAAGCTCTCG
18 Stat1 Forward GCTGCCTATGATGTCTCGTTT STAT1 Forward GCAGGTTCACCAGCTTTATGA Stat1 Reverse TGCTTTTCCGTATGTTGTGCT STAT1 Reverse TGAAGATTACGCTTGCTTTTCCT Stat2 Forward TTTGGCTACCTGGATTGAAGAC STAT2 Forward CCAGCTTTACTCGCACAGC Stat2 Reverse GGCTGAATTTTCGCAAGTTATGC STAT2 Reverse AGCCTTGGAATCATCACTCCC Stat3 Forward CACCTTGGATTGAGAGTCAAGAC STAT3 Forward CAGCAGCTTGACACACGGTA Stat3 Reverse AGGAATCGGCTATATTGCTGGT STAT3 Reverse AAACACCAAAGTGGCATGTGA Stat4 Forward GCACTCAGTAAGATGACGCAG STAT4 Forward TGTTGGCCCAATGGATTGAAA Stat4 Reverse CCAGTAGGGTAAAGCAGTTCTG STAT4 Reverse GGAAACACGACCTAACTGTTCAT Stat5a Forward CAGCCGTGGGATGCTATTGA STAT5A Forward GCAGAGTCCGTGACAGAGG Stat5a Reverse GGGACAGCGGTCATACGTG STAT5A Reverse CCACAGGTAGGGACAGAGTCT Stat5b Forward TGTCCCTGAAACGAATCAAGAG STAT5B Forward GAACACCCGCAATGATTACAGT Stat5b Reverse CTGAACTGTGAGTCAAACAGGAT STAT5B Reverse ACGGTCTGACCTCTTAATTCGT Stat6 Forward TGGAGAGCATCTATCAGAGGGA STAT6 Forward GTTCCGCCACTTGCCAATG Stat6 Reverse GCGGAACTCTTCTATAACAGCTT STAT6 Reverse TGGATCTCCCCTACTCGGTG Tcf4 Forward GATGGGACTCCCTATGACCAC TCF4 Forward AAAAATGGACCAACTTCTTTGGC Tcf4 Reverse GAAAGGGTTCCTGGATTGCCC TCF4 Reverse GGAATTGACAAAAGGTGGAGAGA Thbs1 Forward CCTGCCAGGGAAGCAACAA THBS1 Forward AGACTCCGCATCGCAAAGG Thbs1 Reverse ACAGTCTATGTAGAGTTGAGCCC THBS1 Reverse TCACCACGTTGTTGTCAAGGG Thbs2 Forward GGGGACACTTTGGACCTCAAC THBS2 Forward GACACGCTGGATCTCACCTAC Thbs2 Reverse GCAGCCCACATACAGGCTA THBS2 Reverse GAAGCTGTCTATGAGGTCGCA Tnfrsf11a Forward CAGATGTCTTTTCGTCCACAGA TNFRSF11A Forward TCCTCCACGGACAAATGCAG Tnfrsf11a Reverse AGACTGGGCAGGTAAGCCT TNFRSF11A Reverse CAAACCGCATCGGATTTCTCT Tnfsf10 Forward ATGGTGATTTGCATAGTGCTCC TNFSF10 Forward TGCGTGCTGATCGTGATCTTC Tnfsf10 Reverse GCAAGCAGGGTCTGTTCAAGA TNFSF10 Reverse GCTCGTTGGTAAAGTACACGTA Traf2 Forward AGAGAGTAGTTCGGCCTTTCC TRAF2 Forward TCCCTGGAGTTGCTACAGC Traf2 Reverse GTGCATCCATCATTGGGACAG TRAF2 Reverse AGGCGGAGCACAGGTACTT Trim28 Forward CCCTGTCTACATTCGGCCTG TRIM28 Forward TTTCATGCGTGATAGTGGCAG Trim28 Reverse ACGATGTCTTTGGAGTAGCACT TRIM28 Reverse GCCTCTACACAGGTCTCACAC Wnt1 Forward CGACTGATCCGACAGAACCC WNT1 Forward GGTGGGGTATTGTGAACGTAG Wnt1 Reverse CCATTTGCACTCTCGCACA WNT1 Reverse CGTATCAGACGCCGCTGTTT Wnt10a Forward CATGCTCGAATGAGACTCCAC WNT10A Forward AAGCCTGGAGACTCGCAAC Wnt10a Reverse CCCTACTGTGCGGAACTCAG WNT10A Reverse CGATGGCGTAGGCAAAAGC Wnt10b Forward ATCGCCGTTCACGAGTGTC WNT10B Forward GTGAGCGAGACCCCACTATG Wnt10b Reverse GGAAACCGCGCTTGAGGAT WNT10B Reverse CACTCTGTAACCTTGCACTCATC Wnt11 Forward CAGCCTTCGTGTATGCCCTG WNT11 Forward GGAGTCGGCCTTCGTGTATG Wnt11 Reverse CCGTAGCTGAGGTTGTCCG WNT11 Reverse GCTGAGGTTGTCCGCACAT Wnt2 Forward CCTCCGAAGTAGTCGGGAATC WNT2 Forward GCCTTTGTTTATGCCATCTCCT Wnt2 Reverse GCAGGACTTTAATTCTCCTTGGC WNT2 Reverse CTTGGCGCTTCCCATCTTCTT Wnt2b Forward GACACGTCCTGGTGGTACATA WNT2 Reverse GGGTGGTACAGTTCCAGCG Wnt2b Reverse GTCTGGGTAGCGTTGACACA WNT2B Forward GTTACCCAGACATCATGCGTT Wnt3 Forward TGGCTACCCAATTTGGTGGTC WNT3 Forward GGAGAGGGACCTGGTCTACTA Wnt3 Reverse CTTCACACCTTCTGCTACGCT WNT3 Reverse CTTGTGCCAAAGGAACCCGT Wnt4 Forward TGCGAGGTAAAGACGTGCTG WNT4 Forward CTCCACACTCGACTCCTTGC Wnt4 Reverse CTTGAACTGTGCATTCCGAGG WNT4 Reverse CCGAAGAGATGGCGTACACG Wnt5a Forward GGACCACATGCAGTACATTGG WNT5A Forward ATTCTTGGTGGTCGCTAGGTA Wnt5a Reverse CGTCTCTCGGCTGCCTATTT WNT5A Reverse CGCCTTCTCCGATGTACTGC Wnt7b Forward CTTCACCTATGCCATCACGG WNT7B Forward GAAGCAGGGCTACTACAACCA Wnt7b Reverse TGGTTGTAGTAGCCTTGCTTCT WNT7B Reverse CGGCCTCATTGTTATGCAGGT
19 R.P. Hobbs et al. Keratin 17 regulates Aire to promote skin tumorigenesis Supplementary Table 7. List of primer sets used for ChIP and mutagenesis Primer set Forward Sequence (5'-->3') Reverse sequence (5'-->3') hcxcl11 1 TCTTTAGGAACTCATTTACAGGT CCTTTCACCCACCCTTCAT 2 TGTATAGATATGTGAAGGATGAAGGG GATAAGAATGTGAAAGCACTGTGAA 3 GCTGGTTACCATCTGGGTTT GGCACAGGAATTTCCCTCTT 4 GGAAAGGTGCATGACTRCAAAG CAGCTTTGCTGCTCTTCTTG hcxcl10 1 GAATGGATTGCAACCTTTGTT ATTTCCCTCTGCTCCTCTTT 2 AAAGAGGAGCAGAGGGAAAT CTGATTGGATAAGGGCATTACAG 3 AGTCAACTGTAATGCCCTTATCC TGGAGGTTCCTCTGCTGTA hmmp9 1 GGAAGGCATTTACTCCAGGTTA CAAGGATACAGTGAGCCATGAT 2 GCTGGAGTGCAGTGACATAA CTAAAGCCAAGACCCAAGATT 3 TTACAGGAATGAGCCACCATAC TGTCCCTAAATTGATGTGAGGATTA 4 CCTCACATCAATTTAGGGACAAAG GCTGAATCTTCAGGGCAGTA 5 TCTAGAGGCTGCTACTGTCC CTCCCTGACAGCCTTCTTTG 6 AGGGAGAGGAAGCTGAGT CCTGAGACTTCTTTCTGAAACTAATG 7 GGAGGATATCTGACCTGGGA GTGGGAACTGTATGAAAGGGA 8 CTGACCACTGGAGGCTTTC CAGTCTCCACTGCCAAGTC 9 GGTTTCCTGCGGGTCTG TGTGTGTGTGTGTGTGTGT 10 GTGGTGTAAGCCCTTTCTCA TGAGGGCAGAGGTGTCT hccl19 1 AGGGCAATCCCTCTCTCT CCCTTTCCTTCTGCTTTCCT 2 AGAGCAGGAGCCAAGGA GCCAGAGCTTCACTTTCTCTT 3 TCACAGAATGGGACATGAAGG GAAATGCAAGGTGTGAGTGATG 4 CATCACTCACACCTTGCATTTC AACAGAGGCAGGCCAAC hgapdh 1 TGAGCAGTCCGGTGTCACTA GGACTTTGGGAACGACTGAGATG 2 AACTTTCCCGCCTCTCA CAGGACACTAGGGAGTCAAG 3 TCCTTGACTCCCTAGTGTCCTG TAGTAGCCGGGCCCTACTTT 4 ATCGGGCCAATCTCAGT TCTTGAGGCCTGAGCTA K17 Lys399Ala mutagenesis ACTCAGTACGCGAAAGAACC CAGGTGGGCATCCTCTC
HTG EdgeSeq Immuno-Oncology Assay Gene List
 A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
Transduction of lentivirus to human primary CD4+ T cells
 Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Eosinophils! 40! 30! 20! 10! 0! NS!
 A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
SUPPLEMENTAL DATA. Table S1. Genes in each pathway selected for analysis.
 SUPPLEMENTAL DATA Table S1. Genes in each pathway selected for analysis. Gene Chr Start End bp Pathway 1 AKR1C1 10 5000044 5022159 22115 COX 2 AKR1C2 10 5029967 5060207 30240 COX 3 AKR1C3 10 5077549 5149878
SUPPLEMENTAL DATA Table S1. Genes in each pathway selected for analysis. Gene Chr Start End bp Pathway 1 AKR1C1 10 5000044 5022159 22115 COX 2 AKR1C2 10 5029967 5060207 30240 COX 3 AKR1C3 10 5077549 5149878
Bezzi et al., Supplementary Figure 1 *** Nature Medicine: doi: /nm Pten pc-/- ;Zbtb7a pc-/- Pten pc-/- ;Pml pc-/- Pten pc-/- ;Trp53 pc-/-
 Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action
 An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action Supplementary Figure : Expression levels of toll-like receptor 4 (Tlr4) in muse lung does not change throughout the
An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action Supplementary Figure : Expression levels of toll-like receptor 4 (Tlr4) in muse lung does not change throughout the
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis
 Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplementary Material
 Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
qpcr-array Analysis Service
 qpcr-array Analysis Service Customer Name Institute Telephone Address E-mail PO Number Service Code Report Date Service Laboratory Department Phalanx Biotech Group, Inc 6 Floor, No.6, Technology Road 5,
qpcr-array Analysis Service Customer Name Institute Telephone Address E-mail PO Number Service Code Report Date Service Laboratory Department Phalanx Biotech Group, Inc 6 Floor, No.6, Technology Road 5,
SUPPLEMENTAL INFORMATIONS
 1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
* Kyoto Encyclopedia of Genes and Genomes.
 Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature89 IFN- (ng ml ) 5 4 3 1 Splenocytes NS IFN- (ng ml ) 6 4 Lymph node cells NS Nfkbiz / Nfkbiz / Nfkbiz / Nfkbiz / IL- (ng ml ) 3 1 Splenocytes IL- (ng ml ) 1 8 6 4 *** ** Lymph node cells
doi: 1.138/nature89 IFN- (ng ml ) 5 4 3 1 Splenocytes NS IFN- (ng ml ) 6 4 Lymph node cells NS Nfkbiz / Nfkbiz / Nfkbiz / Nfkbiz / IL- (ng ml ) 3 1 Splenocytes IL- (ng ml ) 1 8 6 4 *** ** Lymph node cells
Figure 1: Effects of cisplatin on survival of lung cancer cells.
 Figure 1 Figure 1: Effects of cisplatin on survival of lung cancer cells. To determine the IC 50 concentration of cisplatin, cells were treated with various concentrations of cisplatin and cell survival
Figure 1 Figure 1: Effects of cisplatin on survival of lung cancer cells. To determine the IC 50 concentration of cisplatin, cells were treated with various concentrations of cisplatin and cell survival
1,000 in silico simulated alpha, beta, gamma and delta TCR repertoires were created.
 938 939 940 941 942 Figure S1 Schematic of the in silico TCRminer and MiXCR validation. 1,000 in silico simulated alpha, beta, gamma and delta TCR repertoires were created. Then, 100,000 simulated 80 bp
938 939 940 941 942 Figure S1 Schematic of the in silico TCRminer and MiXCR validation. 1,000 in silico simulated alpha, beta, gamma and delta TCR repertoires were created. Then, 100,000 simulated 80 bp
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was
 Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
For focused group profiling of human HIV infection and host response genes expression
 ExProfile TM Human HIV Infection and Host Response Related Gene qpcr Array For focused group profiling of human HIV infection and host response genes expression Cat. No. QG023-A (1 x 96-well plate, Format
ExProfile TM Human HIV Infection and Host Response Related Gene qpcr Array For focused group profiling of human HIV infection and host response genes expression Cat. No. QG023-A (1 x 96-well plate, Format
Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases
 Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Irf1 fold changes (D) 24h 48h. p-p65. t-p65. p-irf3. t-irf3. β-actin SKO TKO 100% 80% 60% 40% 20%
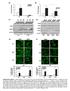 Irf7 Fold changes 3 1 Irf1 fold changes 3 1 8h h 8h 8h h 8h p-p6 p-p6 t-p6 p-irf3 β-actin p-irf3 t-irf3 β-actin TKO TKO STKO (E) (F) TKO TKO % of p6 nuclear translocation % % 1% 1% % % p6 TKO % of IRF3
Irf7 Fold changes 3 1 Irf1 fold changes 3 1 8h h 8h 8h h 8h p-p6 p-p6 t-p6 p-irf3 β-actin p-irf3 t-irf3 β-actin TKO TKO STKO (E) (F) TKO TKO % of p6 nuclear translocation % % 1% 1% % % p6 TKO % of IRF3
Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice
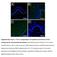 Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice undergoing AD- and psoriasis-like disease. Immunofluorescence staining for Fn14 (green) and DAPI (blue) in skin of naïve
Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice undergoing AD- and psoriasis-like disease. Immunofluorescence staining for Fn14 (green) and DAPI (blue) in skin of naïve
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array.
 Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
To compare the relative amount of of selected gene expression between sham and
 Supplementary Materials and Methods Gene Expression Analysis To compare the relative amount of of selected gene expression between sham and mice given renal ischemia-reperfusion injury (IRI), ncounter
Supplementary Materials and Methods Gene Expression Analysis To compare the relative amount of of selected gene expression between sham and mice given renal ischemia-reperfusion injury (IRI), ncounter
CPM (x 10-3 ) Tregs +Teffs. Tregs alone ICOS CLTA-4
 A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
Comparison of in vitro transfection efficiency of HBV plasmids between genotypes HBV expressing plasmids (5
 Supplemental Materials Figures legend Supplemental Table 1, 2 and 3 Supplemental Figure 1, Figure 2 and Figure 3 Supplemental Figure 1 Comparison of in vitro transfection efficiency of HBV plasmids between
Supplemental Materials Figures legend Supplemental Table 1, 2 and 3 Supplemental Figure 1, Figure 2 and Figure 3 Supplemental Figure 1 Comparison of in vitro transfection efficiency of HBV plasmids between
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/310/ra11/dc1 Supplementary Materials for STAT3 Induction of mir-146b Forms a Feedback Loop to Inhibit the NF-κB to IL-6 Signaling Axis and STAT3-Driven Cancer
www.sciencesignaling.org/cgi/content/full/7/310/ra11/dc1 Supplementary Materials for STAT3 Induction of mir-146b Forms a Feedback Loop to Inhibit the NF-κB to IL-6 Signaling Axis and STAT3-Driven Cancer
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Late regulation of immune genes and micrornas in circulating leukocytes in a pig model of
 1 Supplementary material for: 2 3 4 5 6 Late regulation of immune genes and micrornas in circulating leukocytes in a pig model of influenza A (H1N2) infection Louise Brogaard, Peter M. H. Heegaard, Lars
1 Supplementary material for: 2 3 4 5 6 Late regulation of immune genes and micrornas in circulating leukocytes in a pig model of influenza A (H1N2) infection Louise Brogaard, Peter M. H. Heegaard, Lars
Supplementary. presence of the. (c) mrna expression. Error. in naive or
 Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
Bronchoalveolar lavage fluid analysis showed increased eosinophil in 30% of subjects with IrIa BAL
 Bronchoalveolar lavage fluid analysis showed increased eosinophil in 30% of subjects with IrIa BAL UBC Chan-Yeung Centre for Occupational and Environmental Respiratory Disease Understanding exposure effects
Bronchoalveolar lavage fluid analysis showed increased eosinophil in 30% of subjects with IrIa BAL UBC Chan-Yeung Centre for Occupational and Environmental Respiratory Disease Understanding exposure effects
Nature Immunology: doi: /ni Supplementary Figure 1. Production of cytokines and chemokines after vaginal HSV-2 infection.
 Supplementary Figure 1 Production of cytokines and chemokines after vaginal HSV-2 infection. C57BL/6 mice were (a) treated intravaginally with 20 µl of PBS or infected with 6.7x10 4 pfu of HSV-2 in the
Supplementary Figure 1 Production of cytokines and chemokines after vaginal HSV-2 infection. C57BL/6 mice were (a) treated intravaginally with 20 µl of PBS or infected with 6.7x10 4 pfu of HSV-2 in the
pplementary Figur Supplementary Figure 1. a.
 pplementary Figur Supplementary Figure 1. a. Quantification by RT-qPCR of YFV-17D and YFV-17D pol- (+) RNA in the supernatant of cultured Huh7.5 cells following viral RNA electroporation of respective
pplementary Figur Supplementary Figure 1. a. Quantification by RT-qPCR of YFV-17D and YFV-17D pol- (+) RNA in the supernatant of cultured Huh7.5 cells following viral RNA electroporation of respective
Supplementary Figure 1. NAFL enhanced immunity of other vaccines (a) An over-the-counter, hand-held non-ablative fractional laser (NAFL).
 Supplementary Figure 1. NAFL enhanced immunity of other vaccines (a) An over-the-counter, hand-held non-ablative fractional laser (NAFL). (b) Depiction of a MTZ array generated by NAFL. (c-e) IgG production
Supplementary Figure 1. NAFL enhanced immunity of other vaccines (a) An over-the-counter, hand-held non-ablative fractional laser (NAFL). (b) Depiction of a MTZ array generated by NAFL. (c-e) IgG production
Supplementary Information. Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase
 Journal: Nature Medicine Supplementary Information Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase 1,2 Peng Wang PhD, 1,2 Juan-Carlos Alvarez-Perez
Journal: Nature Medicine Supplementary Information Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase 1,2 Peng Wang PhD, 1,2 Juan-Carlos Alvarez-Perez
Supplemental Table 1. Primers used for RT-PCR analysis of inflammatory cytokines Gene Primer Sequence
 Supplemental Table 1. Primers used for RT-PCR analysis of inflammatory cytokines Gene Primer Sequence IL-1α Forward primer 5 -CAAGATGGCCAAAGTTCGTGAC-3' Reverse primer 5 -GTCTCATGAAGTGAGCCATAGC-3 IL-1β
Supplemental Table 1. Primers used for RT-PCR analysis of inflammatory cytokines Gene Primer Sequence IL-1α Forward primer 5 -CAAGATGGCCAAAGTTCGTGAC-3' Reverse primer 5 -GTCTCATGAAGTGAGCCATAGC-3 IL-1β
Supplementary Table 1 Clinicopathological characteristics of 35 patients with CRCs
 Supplementary Table Clinicopathological characteristics of 35 patients with CRCs Characteristics Type-A CRC Type-B CRC P value Sex Male / Female 9 / / 8.5 Age (years) Median (range) 6. (9 86) 6.5 (9 76).95
Supplementary Table Clinicopathological characteristics of 35 patients with CRCs Characteristics Type-A CRC Type-B CRC P value Sex Male / Female 9 / / 8.5 Age (years) Median (range) 6. (9 86) 6.5 (9 76).95
Supplementary Figure 1. Dynamic Response of WT and mir-21 -/- mice to caerulein. (a) Representative histological sections of mouse pancreas stained
 Supplementary Figure 1. Dynamic Response of WT and mir-21 -/- mice to caerulein. (a) Representative histological sections of mouse pancreas stained with hematoxylin from caerulein-treated WT and mir-21
Supplementary Figure 1. Dynamic Response of WT and mir-21 -/- mice to caerulein. (a) Representative histological sections of mouse pancreas stained with hematoxylin from caerulein-treated WT and mir-21
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
blood mononuclear cells were stained with AAD, annexin, CD3, CD4, CD25, LAP and specific
 Supplementary figure legends Figure 1: Selective expression of regulatory markers on CD4 + LAP + T cells. Human peripheral blood mononuclear cells were stained with AAD, annexin, CD3, CD4, CD25, LAP and
Supplementary figure legends Figure 1: Selective expression of regulatory markers on CD4 + LAP + T cells. Human peripheral blood mononuclear cells were stained with AAD, annexin, CD3, CD4, CD25, LAP and
Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction. Marker Sequence (5 3 ) Accession No.
 Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Applied Biosystems models 7500 (Fast block), 7900HT (Fast block), StepOnePlus, ViiA 7 (Fast block)
 RT² Profiler PCR Array (96-Well Format and 384-Well [4 x 96] Format) Human HIV Infection and Host Response Cat. no. 330231 PAHS-051YA For pathway expression analysis Format Format A Format C Format D Format
RT² Profiler PCR Array (96-Well Format and 384-Well [4 x 96] Format) Human HIV Infection and Host Response Cat. no. 330231 PAHS-051YA For pathway expression analysis Format Format A Format C Format D Format
The innate immune system: Contributions to Immunogenicity of Therapeutic Proteins
 The innate immune system: Contributions to Immunogenicity of Therapeutic Proteins Daniela Verthelyi, M.D., Ph.D. Laboratory of Immunology Division of Therapeu8c Proteins OBP, CDER, FDA Map to the talk:
The innate immune system: Contributions to Immunogenicity of Therapeutic Proteins Daniela Verthelyi, M.D., Ph.D. Laboratory of Immunology Division of Therapeu8c Proteins OBP, CDER, FDA Map to the talk:
well for 2 h at rt. Each dot represents an individual mouse and bar is the mean ±
 Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
CD Marker Antibodies. atlasantibodies.com
 CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
B16F1 B16F10 Supplemental Figure S1
 B16F1 B16F1 Supplemental Figure S1 Representative microangiography images of B16F1 and B16F1 tumors grown in the cranial windows. FITC-dextran (2 million MW) was injected systemically to visualize blood
B16F1 B16F1 Supplemental Figure S1 Representative microangiography images of B16F1 and B16F1 tumors grown in the cranial windows. FITC-dextran (2 million MW) was injected systemically to visualize blood
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.
 Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
a. b. c. d. e. f. g. h. i. j. k. l. m. n. o. p.
 a. b. c. d. e. f. g. h. i. j. k. l. 2.5 2 1.5 1.5 IL-1β 12 8 6 4 2 IL-1β 9 8 7 6 4 3 3 2.9 IL-1β m. n. o. p. 1.8 1.6 1.4 1.2 1.8.6.4.2 6h LPS 2 15 1 5 6h LPS 2 6h LPS 6 4 3 6h LPS Supplementary Figure
a. b. c. d. e. f. g. h. i. j. k. l. 2.5 2 1.5 1.5 IL-1β 12 8 6 4 2 IL-1β 9 8 7 6 4 3 3 2.9 IL-1β m. n. o. p. 1.8 1.6 1.4 1.2 1.8.6.4.2 6h LPS 2 15 1 5 6h LPS 2 6h LPS 6 4 3 6h LPS Supplementary Figure
Supplementary Information
 Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was
 Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity.
 Supplementary Figure 1 Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity. Male C57BL/6J mice were fed a normal chow (NC, 10% fat) or a high-fat diet (HFD,
Supplementary Figure 1 Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity. Male C57BL/6J mice were fed a normal chow (NC, 10% fat) or a high-fat diet (HFD,
Supplementary Figure 1: TSLP receptor skin expression in dcssc. A: Healthy control (HC) skin with TSLP receptor expression in brown (10x
 Supplementary Figure 1: TSLP receptor skin expression in dcssc. A: Healthy control (HC) skin with TSLP receptor expression in brown (10x magnification). B: Second HC skin stained for TSLP receptor in brown
Supplementary Figure 1: TSLP receptor skin expression in dcssc. A: Healthy control (HC) skin with TSLP receptor expression in brown (10x magnification). B: Second HC skin stained for TSLP receptor in brown
Supplementary information CD4 T cells are required for both development and maintenance of disease in a new model of reversible colitis
 Supplementary information CD4 T cells are required for both development and maintenance of disease in a new model of reversible colitis rasseit and Steiner et al. .. Supplementary Figure 1 % of initial
Supplementary information CD4 T cells are required for both development and maintenance of disease in a new model of reversible colitis rasseit and Steiner et al. .. Supplementary Figure 1 % of initial
Supplementary Figure 1. Repression of hepcidin expression in the liver of mice treated with
 Supplementary Figure 1. Repression of hepcidin expression in the liver of mice treated with DMN Immunohistochemistry for hepcidin and H&E staining (left). qrt-pcr assays for hepcidin in the liver (right).
Supplementary Figure 1. Repression of hepcidin expression in the liver of mice treated with DMN Immunohistochemistry for hepcidin and H&E staining (left). qrt-pcr assays for hepcidin in the liver (right).
Supplementary Figure 1
 Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
S1a S1b S1c. S1d. S1f S1g S1h SUPPLEMENTARY FIGURE 1. - si sc Il17rd Il17ra bp. rig/s IL-17RD (ng) -100 IL-17RD
 SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
Nature Immunology: doi: /ni Supplementary Figure 1. Cytokine pattern in skin in response to urushiol.
 Supplementary Figure 1 Cytokine pattern in skin in response to urushiol. Wild-type (WT) and CD1a-tg mice (n = 3 per group) were sensitized and challenged with urushiol (uru) or vehicle (veh). Quantitative
Supplementary Figure 1 Cytokine pattern in skin in response to urushiol. Wild-type (WT) and CD1a-tg mice (n = 3 per group) were sensitized and challenged with urushiol (uru) or vehicle (veh). Quantitative
SUPPLEMENTARY FIGURES AND TABLE
 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh-
 1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
Supplementary Material. Part I: Sample Information. Part II: Pathway Information
 Supplementary Material Part I: Sample Information Three NPC cell lines, CNE1, CNE2, and HK1 were treated with CYC202. Gene expression of 380 selected genes were collected at 0, 2, 4, 6, 12 and 24 hours
Supplementary Material Part I: Sample Information Three NPC cell lines, CNE1, CNE2, and HK1 were treated with CYC202. Gene expression of 380 selected genes were collected at 0, 2, 4, 6, 12 and 24 hours
Regulation of inflammation by histone deacetylases in rheumatoid arthritis: beyond epigenetics Grabiec, A.M.
 UvA-DARE (Digital Academic Repository) Regulation of inflammation by histone deacetylases in rheumatoid arthritis: beyond epigenetics Grabiec, A.M. Link to publication Citation for published version (APA):
UvA-DARE (Digital Academic Repository) Regulation of inflammation by histone deacetylases in rheumatoid arthritis: beyond epigenetics Grabiec, A.M. Link to publication Citation for published version (APA):
Supplementary Figure 1. Flow cytometry panels used for BD Canto (A) and BD Fortessa (B).
 Intra Immune nalysis Surface Supplementary Figure 1. Flow cytometry panels used for D Canto () and D Fortessa (). Name Fluorochrome ID F488 PE PerCp-Cy5.5 PC Paclue PE-Cy7 PC-H7 Lympho* 1 CD56 CD8 CD16
Intra Immune nalysis Surface Supplementary Figure 1. Flow cytometry panels used for D Canto () and D Fortessa (). Name Fluorochrome ID F488 PE PerCp-Cy5.5 PC Paclue PE-Cy7 PC-H7 Lympho* 1 CD56 CD8 CD16
Table S1. Primer sequences used for qrt-pcr. CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT ACTB AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT LCOR
 Table S1. Primer sequences used for qrt-pcr. ACTB LCOR KLF6 CTBP1 CDKN1A CDH1 ATF3 PLAU MMP9 TFPI2 CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT CGGCTGCAGGAAAGTTTACA
Table S1. Primer sequences used for qrt-pcr. ACTB LCOR KLF6 CTBP1 CDKN1A CDH1 ATF3 PLAU MMP9 TFPI2 CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT CGGCTGCAGGAAAGTTTACA
Title: Cytosolic DNA-mediated, STING-dependent pro-inflammatory gene. Fig. S1. STING ligands-mediated signaling response in MEFs. (A) Primary MEFs (1
 1 Supporting Information 2 3 4 Title: Cytosolic DNA-mediated, STING-dependent pro-inflammatory gene induction necessitates canonical NF-κB activation through TBK1 5 6 Authors: Abe et al. 7 8 9 Supporting
1 Supporting Information 2 3 4 Title: Cytosolic DNA-mediated, STING-dependent pro-inflammatory gene induction necessitates canonical NF-κB activation through TBK1 5 6 Authors: Abe et al. 7 8 9 Supporting
Supplementary Figure 1. ETBF activate Stat3 in B6 and Min mice colons
 Supplementary Figure 1 ETBF activate Stat3 in B6 and Min mice colons a pstat3 controls Pos Neg ETBF 1 2 3 4 b pstat1 pstat2 pstat3 pstat4 pstat5 pstat6 Actin Figure Legend: (a) ETBF induce predominantly
Supplementary Figure 1 ETBF activate Stat3 in B6 and Min mice colons a pstat3 controls Pos Neg ETBF 1 2 3 4 b pstat1 pstat2 pstat3 pstat4 pstat5 pstat6 Actin Figure Legend: (a) ETBF induce predominantly
Loss of RhoA promotes skin tumor formation. Supplementary Figure 1. Loss of RhoA does not impair F-actin organization.
 Supplementary Figure Legends Supplementary Figure 1. Loss of RhoA does not impair F-actin organization. a. Representative IF images of F-actin staining of big and small control (left) and RhoA ko tumors
Supplementary Figure Legends Supplementary Figure 1. Loss of RhoA does not impair F-actin organization. a. Representative IF images of F-actin staining of big and small control (left) and RhoA ko tumors
SUPPLEMENTARY FIGURE LEGENDS
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Hippocampal sections from new-born Pten+/+ and PtenFV/FV pups were stained with haematoxylin and eosin (H&E) and were imaged at (a) low and (b) high
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Hippocampal sections from new-born Pten+/+ and PtenFV/FV pups were stained with haematoxylin and eosin (H&E) and were imaged at (a) low and (b) high
Supplementary Information. Induction of p53-independent apoptosis by ectopic expression of HOXA5
 Supplementary Information Induction of p53-independent apoptosis by ectopic expression of in human liposarcomas Dhong Hyun Lee 1, *, Charles Forscher 1, Dolores Di Vizio 2, 3, and H. Phillip Koeffler 1,
Supplementary Information Induction of p53-independent apoptosis by ectopic expression of in human liposarcomas Dhong Hyun Lee 1, *, Charles Forscher 1, Dolores Di Vizio 2, 3, and H. Phillip Koeffler 1,
fl/+ KRas;Atg5 fl/+ KRas;Atg5 fl/fl KRas;Atg5 fl/fl KRas;Atg5 Supplementary Figure 1. Gene set enrichment analyses. (a) (b)
 KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
Nature Immunology doi: /ni Supplementary Figure 1. Raf-1 inhibition does not affect TLR4-induced type I IFN responses.
 Supplementary Figure 1 Raf-1 inhibition does not affect TLR4-induced type I IFN responses. Real-time PCR analyses of IFNB, ISG15, TRIM5, TRIM22 and APOBEC3G mrna in modcs 6 h after stimulation with TLR4
Supplementary Figure 1 Raf-1 inhibition does not affect TLR4-induced type I IFN responses. Real-time PCR analyses of IFNB, ISG15, TRIM5, TRIM22 and APOBEC3G mrna in modcs 6 h after stimulation with TLR4
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the
 Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Molecular basis of age-associated cytokine dysregulation in LPS-stimulated macrophages
 Uncorrected Version. Published on April 6, 2006 as DOI:10.1189/jlb.0106024 Molecular basis of age-associated cytokine dysregulation in LPS-stimulated macrophages R. Lakshman Chelvarajan,*, Yushu Liu, Diana
Uncorrected Version. Published on April 6, 2006 as DOI:10.1189/jlb.0106024 Molecular basis of age-associated cytokine dysregulation in LPS-stimulated macrophages R. Lakshman Chelvarajan,*, Yushu Liu, Diana
Nature Immunology: doi: /ni.3745
 Supplementary Figure 1 Experimental settings. (a) Experimental setup and bioinformatic data analysis of transcriptomic changes induced in neonatal and adult monocytes by stimulation with 10 ng/ml LPS for
Supplementary Figure 1 Experimental settings. (a) Experimental setup and bioinformatic data analysis of transcriptomic changes induced in neonatal and adult monocytes by stimulation with 10 ng/ml LPS for
Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion
 Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A.
 Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Case Study - Informatics
 bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
Supplemental Information. High-Throughput Microfluidic Labyrinth for the. Label-free Isolation of Circulating Tumor Cells
 Cell Systems, Volume 5 Supplemental Information High-Throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells Eric Lin, Lianette Rivera-Báez, Shamileh Fouladdel, Hyeun
Cell Systems, Volume 5 Supplemental Information High-Throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells Eric Lin, Lianette Rivera-Báez, Shamileh Fouladdel, Hyeun
Supporting Information
 Supporting Information Rothhammer et al. 1.173/pnas.161413114 SI Materials and Methods EAE Clinical Scoring and Agents. Clinical sig of EAE were assessed as follows:, no sig of disease; 1, loss of tone
Supporting Information Rothhammer et al. 1.173/pnas.161413114 SI Materials and Methods EAE Clinical Scoring and Agents. Clinical sig of EAE were assessed as follows:, no sig of disease; 1, loss of tone
SUPPLEMENTARY INFORMATION
 Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supporting Information
 Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
A Systems Biology Approach to the Analysis of Subset- Specific Responses to Lipopolysaccharide in Dendritic Cells
 A Systems Biology Approach to the Analysis of Subset- Specific Responses to Lipopolysaccharide in Dendritic Cells David G. Hancock 1,2, Elena Shklovskaya 1,2, Thomas V. Guy 1,2, Reza Falsafi 3, Chris D.
A Systems Biology Approach to the Analysis of Subset- Specific Responses to Lipopolysaccharide in Dendritic Cells David G. Hancock 1,2, Elena Shklovskaya 1,2, Thomas V. Guy 1,2, Reza Falsafi 3, Chris D.
Cancer, inflammation, and immunity crosstalk
 Contact Technical Support BRCsupport@QIAGEN.COM 1-800-362-7737 Cancer, inflammation, and immunity crosstalk Jesse Liang, Ph.D. Legal disclaimer QIAGEN products shown here are intended for molecular biology
Contact Technical Support BRCsupport@QIAGEN.COM 1-800-362-7737 Cancer, inflammation, and immunity crosstalk Jesse Liang, Ph.D. Legal disclaimer QIAGEN products shown here are intended for molecular biology
Interpreting Diverse Genomic Data Using Gene Sets
 Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
Transcription factor Foxp3 and its protein partners form a complex regulatory network
 Supplementary figures Resource Paper Transcription factor Foxp3 and its protein partners form a complex regulatory network Dipayan Rudra 1, Paul deroos 1, Ashutosh Chaudhry 1, Rachel Niec 1, Aaron Arvey
Supplementary figures Resource Paper Transcription factor Foxp3 and its protein partners form a complex regulatory network Dipayan Rudra 1, Paul deroos 1, Ashutosh Chaudhry 1, Rachel Niec 1, Aaron Arvey
Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3
 Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Trim29 gene-targeting strategy. (a) Genotyping of wildtype mice (+/+), Trim29 heterozygous mice (+/ ) and homozygous mice ( / ).
 Supplementary Figure 1 Trim29 gene-targeting strategy. (a) Genotyping of wildtype mice (+/+), Trim29 heterozygous mice (+/ ) and homozygous mice ( / ). (b) Immunoblot analysis of TRIM29 in lung primary
Supplementary Figure 1 Trim29 gene-targeting strategy. (a) Genotyping of wildtype mice (+/+), Trim29 heterozygous mice (+/ ) and homozygous mice ( / ). (b) Immunoblot analysis of TRIM29 in lung primary
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary fig. 1. Crystals induce necroptosis does not involve caspases, TNF receptor or NLRP3. A. Mouse tubular epithelial cells were pretreated
 Supplementary fig. 1. Crystals induce necroptosis does not involve caspases, TNF receptor or NLRP3. A. Mouse tubular epithelial cells were pretreated with zvad-fmk (10µM) and exposed to calcium oxalate
Supplementary fig. 1. Crystals induce necroptosis does not involve caspases, TNF receptor or NLRP3. A. Mouse tubular epithelial cells were pretreated with zvad-fmk (10µM) and exposed to calcium oxalate
Supplementary Table 1. List of primers used in this study
 Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
p.r623c p.p976l p.d2847fs p.t2671 p.d2847fs p.r2922w p.r2370h p.c1201y p.a868v p.s952* RING_C BP PHD Cbp HAT_KAT11
 ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
Interpreting Diverse Genomic Data Using Gene Sets
 Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients.
 Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent Protein.
 prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
Supplementary Information
 Supplementary Information Supplementary Figure 1. Short-term coreceptor and costimulation blockade induces tolerance to pluripotent human ESCs. (A) Schematic diagram showing experimental approaches and
Supplementary Information Supplementary Figure 1. Short-term coreceptor and costimulation blockade induces tolerance to pluripotent human ESCs. (A) Schematic diagram showing experimental approaches and
Supplementary Figures. T Cell Factor-1 initiates T helper 2 fate by inducing GATA-3 and repressing Interferon-γ
 Supplementary Figures T Cell Factor-1 initiates T helper 2 fate by inducing GATA-3 and repressing Interferon-γ Qing Yu, Archna Sharma, Sun Young Oh, Hyung-Geun Moon, M. Zulfiquer Hossain, Theresa M. Salay,
Supplementary Figures T Cell Factor-1 initiates T helper 2 fate by inducing GATA-3 and repressing Interferon-γ Qing Yu, Archna Sharma, Sun Young Oh, Hyung-Geun Moon, M. Zulfiquer Hossain, Theresa M. Salay,
