Gene expression of muscular and neuronal pathways is cooperatively dysregulated in patients
|
|
|
- Edgar Robertson
- 5 years ago
- Views:
Transcription
1 Gene expression of muscular and neuronal pathways is cooperatively dysregulated in patients with idiopathic achalasia Orazio Palmieri 1, Tommaso Mazza 2, Antonio Merla 1, Caterina Fusilli 2, Antonello Cuttitta 3, Giuseppina Martino 1, Tiziana Latiano 1, Giuseppe Corritore 1, Fabrizio Bossa 1, Orazio Palumbo 4, Lucia Anna Muscarella 5, Massimo Carella 4, Paolo Graziano 6, Angelo Andriulli 1, Anna Latiano 1* 1. Division of Gastroenterology, IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo, Foggia, Italy; 2. Bioinformatics Unit, IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo, Foggia, Italy; 3. Unit of General Surgery 2nd and Thoracic Surgery, IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo, Foggia, Italy; 4. Medical Genetics Service, IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo, Foggia, Italy; 5. Laboratory of Oncology, IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo, Foggia, Italy: 6. Pathology Unit, IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo, Foggia, Italy *Correspondence to Anna Latiano BSc, PhD, Division of Gastroenterology, IRCCS Casa Sollievo della Sofferenza Hospital, San Giovanni Rotondo, 71013, Foggia, Italy. Telephone n.: ; fax n.:
2 Table S1. Differentially expressed genes involved in the smooth muscle contraction pathway (z-score = 1.455) NM_ CHRM2 Affected Affects (3) NM_ KCNMA1 Affected Affects (2) NM_ CNN1 Affected Affects (1) NM_ MYH11 Increased Increases (3) NM_ MYOCD Increased Increases (1) NM_ C3 Increased Increases (2) NM_ ATP1A2 Affected Affects (1) NM_ CACNA1C Affected Affects (2) NM_ ACTA2 Affected Affects (1) NM_ PTGS2 Increased Increases (1) NM_ CAV1 Affected Affects (3) NM_ SRF Increased Increases (1) NM_ ROCK1 Affected Affects (1) NM_ PTGS1 Increased Increases (1) NM_ C4A/C4B Increased Increases (1) ENST ROCK2 Affected Affects (1) NM_ NMU Decreased Increases (2) NM_ EDN1 Decreased Increases (3) Table S2. Differentially expressed genes involved in the smooth muscle contractility pathway (z-score = 2.623) NM_ SULF1 Increased Increases (2) NM_ MRVI1 Increased Increases (2) NM_ CAV1 Increased Increases (3) NM_ MYH11 Increased Increases (3) NM_ CHRM2 Increased Increases (4) NM_ PRKG1 Increased Increases (1) NM_ SULF2 Increased Increases (2) Table S3. Differentially expressed genes involved in the damage to nervous tissue pathway (z-score = 1.611) NM_ FGF2 Decreased Decreases (3) ENST TGM2 Decreased Decreases (7) NM_ PTGS2 Increased Increases (1) ENST EBF2 Affected Affects (1) NM_ IGF1 Decreased Decreases (23) NM_ ITPR1 Increased Increases (1) NM_ AGTR1 Increased Increases (1) ENST IL6 Increased Increases (6) NM_ PRNP Increased Increases (3) ENST CLU Affected Affects (2) NM_ NFE2L2 Increased Decreases (2) NM_ EIF2AK3 Increased Decreases (1) ENST MYRF Increased Decreases (2)
3 ENST PPARG Increased Decreases (5) NM_ MT1E Increased Decreases (1) ENST CHGA Affected Affects (1) ENST F2RL1 Decreased Increases (1) NM_ MT1H Increased Decreases (1) Table S4. Differentially expressed genes involved in the synaptic transmission of cells pathway (z-score = 1.062) NM_ GRPR Affected Affects (1) ENST BCHE Decreased Decreases (1) NM_ CHRM2 Increased Increases (2) NM_ NCS1 Increased Increases (1) NM_ KCNMA1 Affected Affects (2) NM_ CACNB2 Affected Affects (1) NM_ GABRA5 Affected Affects (1) NM_ DMD Affected Affects (1) NM_ DTNA Affected Affects (1) NM_ PCDHB5 Affected Affects (1) NM_ CACNA1C Affected Affects (1) NM_ CDH8 Affected Affects (1) NM_ FGF12 Affected Affects (1) NM_ NBEA Affected Affects (1) NM_ IGF1 Increased Increases (1) NM_ PMP22 Affected Affects (1) NM_ PCDHB16 Affected Affects (1) ENST HOMER1 Affected Affects (3) NM_ MYH10 Affected Affects (1) NM_ GRIK5 Affected Affects (1) NM_ PRNP Decreased Decreases (1) NM_ NPY1R Affected Affects (1) NM_ NPTX1 Affected Affects (1) NM_ PPP1R15A Increased Increases (1) NM_ NEDD4 Decreased Decreases (2) NM_ PPP1R9A Affected Affects (1) NM_ GLS Affected Affects (1) NM_ NRXN1 Affected Affects (1) NM_ NMU Decreased Increases (1) NM_ SST Affected Affects (1) NM_ CYTH1 Decreased Increases (1) NM_ CCKBR Decreased Increases (1) NM_ SLC1A1 Affected Affects (1) NM_ SSTR1 Affected Affects (2) NM_ MYO6 Affected Affects (1) NM_ XDH Decreased Increases (3)
4 Table S5. Differentially expressed genes involved in the leukocyte migration pathway (z-score = 2.474) NM_ BCL11B Increased Decreases (3) NM_ SST Increased Decreases (1) NM_ DCN Increased Increases (1) NM_ CYR61 Increased Increases (3) NM_ RGS1 Increased Decreases (4) NM_ PLA2G5 Increased Increases (5) ENST IL6 Increased Increases (12) NM_ THBS4 Increased Increases (1) ENST RARRES2 Increased Increases (15) NM_ PPBP Increased Increases (14) NM_ EDNRA Increased Increases (1) NM_ HGF Increased Increases (5) NM_ MCAM Increased Increases (3) NM_ PRDM1 Increased Decreases (1) NM_ VCAM1 Increased Increases (40) NM_ SCN9A Increased Increases (1) ENST CD36 Increased Increases (3) NM_ KITLG Increased Increases (12) NM_ PRNP Increased Increases (3) ENST ACKR3 Increased Increases (2) ENST THBS1 Increased Increases (17) NM_ ITGB3 Increased Increases (4) NM_ ENG Increased Increases (4) NM_ C5AR1 Increased Increases (11) NM_ C4A/C4B Increased Increases (1) NM_ GPR183 Increased Increases (2) ENST MYRF Increased Decreases (4) ENST CTGF Increased Increases (2) NM_ PTGS2 Increased Increases (6) NM_ TRPC6 Increased Increases (4) NM_ PIGR Increased Decreases (2) ENST PIP5K1C Increased Increases (5) NM_ MMP2 Increased Increases (4) NM_ IL1RN Increased Decreases (6) NM_ C3 Increased Increases (15) NM_ SEMA3E Increased Increases (1) ENST F13A1 Increased Increases (1) NM_ PIK3R1 Increased Increases (6) ENST LTF Increased Decreases (4) NM_ AOC3 Increased Increases (18) ENST BCL2 Increased Increases (1) ENST TGM2 Increased Increases (9) NM_ SERPINF1 Increased Increases (1) ENST LUM Increased Increases (9) NM_ SELE Increased Increases (57) NM_ GNAO1 Increased Increases (2) NM_ ITGAM Increased Increases (23) NM_ NPR1 Increased Increases (1) NM_ SERPINB3 Increased Decreases (1) NM_ CXCL12 Increased Increases (256)
5 ENST ANGPT1 Increased Increases (2) ENST ITGA9 Increased Increases (1) NM_ THBS2 Increased Increases (5) NM_ SWAP70 Increased Increases (9) ENST CYSLTR1 Increased Increases (3) AK IGHG1 Increased Decreases (2) ENST PIK3CA Increased Increases (1) NM_ TLN1 Increased Increases (3) NM_ ROCK1 Increased Increases (8) NM_ CCL2 Increased Increases (212) ENST INHBA Increased Increases (1) NM_ SLPI Increased Decreases (6) ENST FN1 Increased Increases (22) NM_ CTSG Increased Increases (15) NM_ TLR4 Increased Increases (25) ENST ROCK2 Increased Increases (1) NM_ RAP1GAP Increased Decreases (4) NM_ MUC1 Increased Decreases (1) NM_ RUNX3 Increased Decreases (3) NM_ JAM3 Increased Increases (7) NM_ SMPD2 Increased Decreases (1) NM_ FLNA Increased Increases (1) NM_ TPSAB1/TPSB2 Increased Increases (2) NM_ IL33 Increased Increases (8) ENST HSPA1A/HSPA1B Increased Increases (2) NM_ FUT8 Increased Decreases (1) NM_ LRRK2 Increased Increases (1) NM_ CCDC88A Increased Increases (1) NM_ ASB2 Increased Increases (1) NM_ HCAR2 Increased Decreases (1) ENST COL17A1 Increased Decreases (1) NM_ IRF6 Decreased Increases (2) ENST S100A7 Decreased Increases (8) NM_ RASGRP1 Decreased Increases (4) ENST GREM1 Decreased Decreases (2) NM_ EDN1 Decreased Increases (3) NM_ MMP1 Decreased Increases (1) NM_ CXCL14 Decreased Increases (3) NM_ TIMP1 Decreased Decreases (3) NM_ SDC1 Decreased Increases (3) ENST CCL24 Decreased Increases (13) NM_ CXCL13 Decreased Increases (42) NM_ CYTIP Decreased Increases (3) NM_ ICOS Decreased Increases (9) NM_ RORC Decreased Increases (1) NM_ AQP3 Decreased Increases (7) NM_ TIMP3 Decreased Decreases (2) NM_ IL18 Decreased Increases (13) NM_ GCNT1 Decreased Increases (11) NM_ SOD3 Decreased Decreases (1) NM_ PTPRJ Decreased Increases (1) NM_ DIAPH1 Decreased Increases (6) NM_ OCLN Decreased Increases (2)
6 NM_ SOCS3 Decreased Decreases (3) ENST SLIT2 Decreased Decreases (8) NM_ VAV3 Decreased Increases (1) NM_ P2RY2 Decreased Increases (2) ENST TPH1 Decreased Increases (3) NM_ SYK Decreased Increases (2) NM_ CTSE Decreased Increases (2) NM_ LDLR Decreased Increases (6) ENST CD2 Decreased Increases (2) NM_ TIAM1 Decreased Increases (1) NM_ LCN2 Decreased Increases (3) NM_ ANXA2 Decreased Increases (2) NM_ SEMA3A Decreased Decreases (4) NM_ DMBT1 Decreased Increases (1) NM_ MMP12 Decreased Increases (2) NM_ F3 Decreased Increases (3) ENST F2RL1 Decreased Increases (6) NM_ LGALS1 Decreased Decreases (7) ENST DEFB1 Decreased Increases (24) NM_ PRKG1 Decreased Decreases (3) NM_ YARS Decreased Increases (2) NM_ F11R Decreased Increases (11) NM_ TIMP2 Decreased Decreases (2) ENST PPARG Decreased Increases (6) ENST SERPINB1 Decreased Increases (1) NM_ XDH Decreased Increases (1) ENST S100A14 Decreased Increases (2) NM_ C1GALT1 Decreased Increases (3) NM_ BMPR2 Decreased Decreases (5) NM_ BMPR1A Decreased Decreases (1) NM_ PLXNC1 Decreased Decreases (1) NM_ DIAPH2 Affected Affects (2) NM_ EZR Affected Affects (2) NM_ PRKCA Affected Affects (1) NM_ CD34 Affected Affects (1) ENST ITGA1 Affected Affects (3) NM_ CDH1 Affected Affects (1) NM_ GPR34 Affected Affects (1) NM_ CAV1 Affected Affects (2) NM_ CXADR Affected Affects (1) ENST CRYAB Affected Affects (2) NM_ NFE2L2 Affected Affects (1) NM_ DDR2 Affected Affects (1) NM_ IL1A Affected Affects (2) NM_ COL4A1 Affected Affects (2) ENST CLU Affected Affects (1) ENST CD207 Affected Affects (1) ENST EDNRB Affected Affects (1) NM_ ELF4 Affected Affects (1) NM_ ILK Affected Affects (1) NM_ TGFB2 Affected Affects (1) NM_ DPYSL2 Affected Affects (2)
7 Table S6. Differentially expressed genes involved in the recruitment of phagocytes pathway (z-score = 2.416) NM_ CYR61 Increased Increases (1) ENST IL6 Increased Increases (2) NM_ VCAM1 Increased Increases (4) ENST CD36 Increased Increases (2) ENST THBS1 Increased Increases (2) NM_ C5AR1 Increased Increases (8) NM_ TRPC6 Increased Increases (1) ENST PIP5K1C Increased Increases (3) NM_ MMP2 Increased Increases (1) NM_ IL1RN Increased Decreases (3) ENST F13A1 Increased Increases (1) ENST LTF Increased Decreases (1) NM_ CAV1 Increased Increases (1) NM_ SELE Increased Increases (5) NM_ ITGAM Increased Increases (3) NM_ CXCL12 Increased Increases (3) NM_ THBS2 Increased Increases (1) ENST F2RL1 Increased Decreases (1) NM_ ROCK1 Increased Increases (1) NM_ CCL2 Increased Increases (31) NM_ SLPI Increased Decreases (4) NM_ CTSG Increased Increases (2) NM_ TLR4 Increased Increases (9) NM_ MUC1 Increased Decreases (1) NM_ IL33 Increased Increases (2) ENST HSPA1A/HSPA1B Increased Increases (2) NM_ FUT8 Increased Decreases (1) NM_ HCAR2 Increased Decreases (1) ENST COL17A1 Increased Decreases (1) NM_ EDN1 Decreased Increases (1) ENST RARRES2 Decreased Decreases (2) NM_ MMP1 Decreased Increases (1) NM_ IL18 Decreased Increases (5) NM_ GCNT1 Decreased Increases (1) NM_ IL1A Decreased Increases (1) ENST SLIT2 Decreased Decreases (1) NM_ VAV3 Decreased Increases (1) NM_ P2RY2 Decreased Increases (2) ENST CRYAB Decreased Decreases (1) NM_ LCN2 Decreased Increases (1) NM_ F3 Decreased Increases (1) ENST PPARG Decreased Increases (4) NM_ XDH Decreased Increases (1) NM_ NFE2L2 Affected Affects (1) ENST CLU Affected Affects (1) NM_ IRF6 Affected Affects (1)
8 Table S7. Differentially expressed genes involved in the adhesion of immune cells pathway (z-score = 2.461) NM_ RGS1 Increased Decreases (1) NM_ MRC1 Increased Increases (1) ENST IL6 Increased Increases (4) NM_ PPBP Increased Increases (6) NM_ HGF Increased Increases (1) NM_ MCAM Increased Increases (1) NM_ VCAM1 Increased Increases (36) NM_ ITGA7 Increased Increases (1) NM_ LRP1 Increased Increases (2) NM_ KITLG Increased Increases (4) NM_ ICOS Increased Decreases (4) ENST THBS1 Increased Increases (4) NM_ ITGB3 Increased Increases (2) ENST CLU Increased Increases (1) NM_ PTGS2 Increased Increases (3) NM_ C3 Increased Increases (2) ENST LTF Increased Decreases (3) NM_ AOC3 Increased Increases (7) NM_ SELE Increased Increases (22) NM_ ITGAM Increased Increases (44) NM_ CXCL12 Increased Increases (29) NM_ THBS2 Increased Increases (1) NM_ SWAP70 Increased Increases (4) NM_ CD34 Increased Increases (2) ENST PIK3CA Increased Increases (1) NM_ TLN1 Increased Increases (1) NM_ CCL2 Increased Increases (6) ENST FN1 Increased Increases (14) NM_ TLR4 Increased Increases (5) ENST PPARG Increased Decreases (1) NM_ CD97 Increased Increases (6) NM_ RAP1GAP Increased Decreases (3) NM_ MSN Increased Increases (2) NM_ JAM3 Increased Increases (3) NM_ IL33 Increased Increases (2) NM_ RASGRP1 Decreased Increases (3) NM_ DCN Decreased Decreases (4) NM_ IL1A Decreased Increases (4) NM_ DIAPH1 Decreased Increases (1) ENST PIP5K1C Decreased Decreases (1) NM_ VAV3 Decreased Increases (2) ENST TPH1 Decreased Increases (1) ENST CD2 Decreased Increases (4) NM_ CDH1 Decreased Increases (2) ENST ANGPT1 Decreased Decreases (3) NM_ ROCK1 Decreased Decreases (5) NM_ F11R Decreased Increases (2) ENST ADAM28 Decreased Increases (1) ENST A2M Decreased Decreases (1) NM_ GAS6 Decreased Decreases (5)
9 NM_ ALOX12 Decreased Increases (1) NM_ SERPING1 Decreased Decreases (5) NM_ PLXNC1 Decreased Decreases (1) ENST SLC35C1 Affected Affects (1) NM_ CTSG Affected Affects (4) NM_ EZR Affected Affects (1) NM_ F3 Affected Affects (3) NM_ AXL Affected Affects (2) NM_ NPTX1 Affected Affects (1) NM_ SYK Affected Affects (1) NM_ CXCL13 Affected Affects (1) ENST CD36 Affected Affects (1) Table S8. Differentially expressed genes involved in the regeneration of nerves pathway (z-score = 2.200) NM_ FGF2 Increased Increases (1) NM_ IL1RN Increased Decreases (1) ENST ITGA1 Increased Increases (1) NM_ CXCL12 Increased Increases (1) NM_ IGF1 Increased Increases (3) ENST CLU Affected Affects (1) Table S9. Differentially expressed genes involved in the synaptogenesis pathway (z-score = 2.164) NM_ NRXN1 Increased Increases (5) NM_ SPARCL1 Increased Increases (1) ENST THBS1 Increased Increases (1) NM_ ITGB3 Increased Increases (1) NM_ NCAM1 Increased Increases (3) NM_ CAV1 Increased Increases (1) NM_ THBS2 Increased Increases (1) NM_ IGF1 Increased Increases (2) ENST NLGN1 Increased Increases (3) NM_ DCLK1 Decreased Decreases (1) NM_ KLK8 Decreased Increases (1) NM_ PCDHB16 Affected Affects (1) NM_ PCDHB5 Affected Affects (1) NM_ IGSF9B Affected Affects (1) NM_ RGS2 Affected Affects (1) NM_ CNTN1 Affected Affects (1) NM_ CDH1 Affected Affects (1) NM_ MYO6 Affected Affects (1) NM_ NRN1 Affected Affects (2) NM_ HIPK2 Affected Affects (3) NM_ EPHA3 Affected Affects (1)
10 Table S10: regulatory networks explaining the gene expression changes in the dataset. Focus Molecules in Network Score Molecules Top Diseases and Functions ACPP,ARID5A,BLNK,CCND2,CD79A,CMA1,CPA3,DAB2,DDR1,EDN1,EDNRA,FGL2,FUT8,GFRA1,HBEGF, IGHM,IGKC,IL2RG,INHBA,ITGB3,LRRK2,MMP2,NEK7,PDGFRB,PRDM1,PTPRJ,RGS1,RGS2,SYK,TIMEL Cell Morphology, Humoral Immune Response, Cellular Movement ESS,TIMP2,TLR4,TPSAB1/TPSB2,USP12,ZC3H12C A2M,ALDOB,ATP1A2,ATP1B2,CNN1,DEF6,DES,EIF2S1,FOS,FOSB,FXYD1,GNAO1,HSPA1A/HSPA1B,IDH 1,KLK8,LCAT,LRP1,MEOX2,MFGE8,MT2A,MYH11,MYOCD,NR4A1,PLCB1,PLCB4,PPP1R15A,PPP1R3C, Organ Morphology, Skeletal and Muscular System Development and Function, Neurological Disease PRKAR2B,PRKG2,PRNP,ROCK2,SCN7A,SRF,SWAP70,TRPV4 ANTXR1,AP1B1,ATP11B,C5AR1,CAMK4,CCND2,CCR4,CD28,CEBPB,ERN1,FOXP3,GLI1,ICOS,ICOSLG/L OC ,IGFBP7,IL7R,KIF13A,mir-21,mir ,MMP2,NR2F2,NRN1,ODC1,PDE3B,PDZRN3,PPAP2C,PSIP1,PTGER4,RECK,SAT1,SERPINE1,SLC7 23 Cellular Development, Cellular Growth and Proliferation, Hematological System Development and Function A1,STOM,TIMP3,ZYX ASPM,B4GALNT2,CCNB2,CENPF,CEP55,DAG1,ECM1,GMNN,KIF20A,L-glutamic acid,mapk1,melk,mmp9,nusap1,pbk,plk1,prc1,ptger2,stil,timp1,tpx2,ttk Cell Cycle, Cellular Assembly and Organization, DNA Replication, Recombination, and Repair ADAM28,F13A1,FEZ1,GPNMB,IL2,IL4,IL13,KCNJ15,MN1,MS4A4A,MSMO1,MT1X,NID1,PLXNC1,RFTN1,S 11 PINT2,TFEC,TIMP4,TNS1,TRPS1 16 Cell-To-Cell Signaling and Interaction, Cellular Growth and Proliferation, Hematological System Development and Function C1GALT1,CD22,CD3D,Cdc42,CPT2,CRIM1,CRKL,CXCL10,CYR61,F11R,GLG1,HCK,IGHM,ITGB2,ITK,LT F,MS4A1,PIK3C2G,PIP5K1B,PLCG1,POU5F1,PRKD1,SELE,SELPLG,SETDB1,SH2D1B,SLAMF6,SLAMF Cellular Function and Maintenance, Hematological System Development and Function, Tissue Morphology,SLC35C1,STYK1,SULF2,SYK,THEMIS,VAV1,VDR C15orf48,C5AR1,CAMP,CD34,CD80,CSF3R,DAAM1,EFNB1,EGR3,ELANE,EPHA3,ERG,GFRA3,GINS2,GP ATCH2L,HPGDS,IGF1,IL17F,IL1A,IL1RN,KITLG,mir Cell-To-Cell Signaling and Interaction, Hematological System Development and Function, Immune Cell Trafficking 223,MPO,MREG,MUC13,MYO1D,NFIA,PDE3A,PRTN3,PTPN11,RCAN2,SLC16A7,SLIT2,TAC1,TLR7 ALOX12,BPTF,CACNA1H,CCND2,Cdkn1c,CYP2J2,EGR1,GNA15,HLTF,IKZF1,IKZF3,IL18R1,MAL,MBD2,M ECP2,MYO6,NAP1L2,NGFR,P2RY12,PCDH10,POLE2,PPP1R9A,PRDM1,PRNP,PROM1,RBBP4,RBBP7,R Cell Cycle, DNA Replication, Recombination, and Repair, Cellular Assembly and Organization EST,SELM,SGK1,SMARCA1,SMARCA4,SPIB,STAT6,XBP1 ACSL5,ALDOA,AQP1,ARNTL,ATF3,BHLHE22,CDH11,CDO1,COL1A2,COL4A1,CXCL12,DIXDC1,ENO3,Esr ra,fasn,fbp2,gapdh,htt,jup,kal1,kcnj8,lama4,lipe,map2,mcam,nr4a1,paqr3,peg10,phkg1, Organismal Functions, Cardiovascular System Development and Function, Organismal Development PRDM8,PYGM,RGL1,SKIL,THBS2,ZEB1 ACTN2,APP,ARL6IP5,ATP12A,ATP4A,C1R,CD28,ERMP1,F2RL3,FZD3,H19,HNRNPA1,HTT,KCND2,KCN 10 D3,MAGI1,MAPT,PRKAR2A,PRLR,PSEN1,RUNX1T1,TOP2A,TPD52L1 16 Nervous System Development and Function, Tissue Morphology, Cell-To-Cell Signaling and Interaction ABCA6,ABLIM3,APP,ARAP2,BRCA2,BRIP1,CEBPB,DDR2,DEFB1,HERC6,IFNB1,IFNG,IL10,LPAR4,NFE2L 9 3,SLC7A11,SOD3,STAT1,TGFB1,TGM1,TLR3,TMOD1,USP6NL,XIST 16 Cell Death and Survival, Cellular Development, Hematopoiesis C1S,CCL2,CCL20,CCR6,CD207,CDH1,CTGF,CYBB,Dglucose,EDNRB,F2R,FGF12,FN1,GCNT1,ITGA1,ITGA6,ITGA7,ITGA9,ITGAE,ITGB1,ITGBL1,JUN,KITLG,M 9 19 Cellular Movement, Hematological System Development and Function, Immune Cell Trafficking MP12,PKIG,prostaglandin E2,RGS10,RORA,RORC,RPTOR,TGFB1,TGM2,TNFRSF11A,TNFSF11,VEGFA ANXA2,CD300LD,EDN1,F2RL1,FPR2,GJC1,GLS,GSTA1,HSP90B1,HYOU1,IFIT1B,IRAK3,KLRB1,Map3k7, MMP1,MMP16,MST1R,nitrite,NR0B2,NUAK2,PI3,PPIF,PRDM1,PRTN3,RAB7B,S100A10,SLC29A1,SMAD4, 9 19 Inflammatory Response, Cell Death and Survival, Cell-To-Cell Signaling and Interaction TLR9,TNF,TNFAIP6,TNFRSF14,TNFSF14,TXK,XBP1 A2M,ADIPOR2,AIM1,ALDH1A1,CXCL1,DLL4,DUSP4,FN1,FPR2,FREM1,GDF15,GRIN1,IGHM,IL1B,IL1RN,I ns1,itga8,itga11,itgb1,itpkb,l1cam,lgals9b,lzts2,mapk1,myd88,pgf,pla2g5,pla2g4a,ppp1r9 19 Organismal Injury and Abnormalities, Cellular Movement, Cell-To-Cell Signaling and Interaction 12A,RAP2A,RPS6KA3,SELE,SRR,THBS1,ZC3H12A ADAM23,ADORA2A,APP,ARHGAP32,ATL2,ATP8A1,BDNF,CA12,COL4A2,ETNPPL,GRIA1,GRIN2A,GRIN2 9 B,HTT,MEIS2,MGP,mir-132,RASD2,SGTB,TESC,TFAP2C,TPH1,WISP1 15 Neurological Disease, Psychological Disorders, Skeletal and Muscular Disorders ATP,CCL3,CCL5,CENPU,CSF1,CXCL1,FABP3,GEM,GREM1,HDAC4,HTR2B,IFIT2,IFNA1/IFNA13,IL17A,IR F3,IRF7,LTBP1,MT1E,MT1F,MT1M,NEDD4L,PEA15,PIM2,PLAC8,PLD1,PRKCA,prostaglandin 8 18 Cell-To-Cell Signaling and Interaction, Hematological System Development and Function, Immune Cell Trafficking E2,SERPINB2,SOCS1,STARD4,STAT4,TBX21,TLR2,TLR4,TREM1 ADCYAP1,APP,ATP,B3GNT5,C1QL1,CKB,CRH,CTH,cyclic GMP,DCN,DHCR24,DUSP1,FOS,ganglioside GM1,GAP43,GLI3,GNRH1,HMOX2,MMP9,nitric 8 oxide,nos1,npc1,npr1,npr2,ogn,plat,plg,ptgs2,sdc1,serpine2,tgfb2,tlr2,ugcg,vcan,zf 18 Nucleic Acid Metabolism, Small Molecule Biochemistry, Molecular Transport HX3 BCL6,CCND2,CDH8,CRABP1,EBF1,EBI3,EOMES,EZH2,FAT3,IL2RB,IL2RG,IL4R,IL6R,IL7R,JAK3,MAP2K4,MAP3K14,MAPK8,MICAL2,mir ,PBX3,POU2AF1,PRDM1,PTPN14,RAB27B,SMARCC1,SOCS5,ST8SIA6,STAT5A,SYTL2,TCF3,TNFR 18 Cellular Development, Hematological System Development and Function, Hematopoiesis SF18,TRAF3,TRAF4,TRAT1 ASL,BAG2,CD28,CNTLN,FBXL7,FOXQ1,GALNT12,GBP2,GPX7,IFNB1,IFNG,IL4,IRF1,KMT2D,MIR17HG,M 7 YC,PCDH7,PHLPP2,RAB38,STAT1,STAT6,STXBP2,TOX3,WDR17 14 Cell-mediated Immune Response, Cellular Development, Cellular Function and Maintenance ascorbic acid,chn1,dll1,dpysl2,epha4,fgf2,fgfr1,fgfr2,fgfr3,gh1,ghrh,ghrhr,ghsr,grin2c,hes 7 1,HTR1A,IGF1,IGFBP2,IGFBP3,IGFBP4,IGFBP6,KATNA1,MAP1B,Nefm,NEUROG2,NF1,NRP1,OLIG2,SDC 17 Cellular Development, Nervous System Development and Function, Tissue Development 2,SEMA3A,SNX27,SST,SSTR1,TNIK,VIM ACAA2,ACP5,BTK,CASP8,CCND3,CD24,CD38,CD80,CFLAR,EGR3,ELL2,F8,HSPA6,ICOS,IGJ,IL5,IL18,IL2 7,IL2RA,KIAA1147,LIMK2,LMO7,MAF,MAPK8,MAPK9,MUC1,NFATC1,RBPMS,SCEL,SPI1,THY1,TMEM97, 7 17 Hematological System Development and Function, Tissue Morphology, Cell Death and Survival TRB,TSLP,ZNF25 ADCY5,ADCY9,APOA1,ARPP19,BMP3,CAMK4,CASP9,CDK5,CDK5R1,CHKA,CREBBP,CYP51A1,ELK1,F MR1,GNB5,GRIN2B,H2AFX,HAP1,HIP1,HTT,ITPR1,NOS1,OPRK1,OPRM1,PCP4,PGR,PPARGC1A,PPP Neurological Disease, Hereditary Disorder, Psychological Disorders R1A,PPP1R1B,PRKAA2,RAP1GAP,RGS9,SPTBN2,THRA,VCAM1 AGT,AKAP6,ANTXR2,ATP2B4,Ca2+,CACNA1C,CALCB,CAMLG,CAPN3,CCK,CCKAR,CCKBR,CCL24,CCN D2,CEBPD,CNR1,CXCL12,CXCL13,cyclic AMP,cyclic 7 GMP,EFNA5,FKBP1A,FPR1,GABA,GHRH,GPR126,HTR1A,IGHM,JPH2,NOS1,RYR1,S100A4,TRPC3,TRP 17 Cell Signaling, Molecular Transport, Vitamin and Mineral Metabolism C6,VAV3 ABI2,ADAM19,AICDA,ASB2,BATF,CCL20,CD180,COL8A1,EGR3,ESR1,ESR2,ICOS,IKZF2,IL6,IL23R,IRF4,J AK3,KCNE2,levothyroxine,MKI67,NCOA1,NCOA2,NOTCH1,PRDM1,RORA,RORC,SEMA3C,SULF1,SULT Inflammatory Disease, Inflammatory Response, Neurological Disease C2,TC2N,THRB,TIGIT,TNFSF13,TSHB,XBP1 APP,BTNL2,CD86,CD109,CDKN2B,CDKN2C,CEBPB,CSF2,CYP2S1,DAPL1,DAPP1,FOXO1,GJB2,GPD2, GPR34,IFITM3,IFNG,IKZF2,IL2,IL17A,IL1B,IRF5,MAVS,MYO10,NDN,RRAGD,SELL,SMPD2,ST14,TCF3,TG7 16 Inflammatory Response, Cell Death and Survival, Cell-To-Cell Signaling and Interaction FB1,TNF,XDH AGL,ARHGAP21,CREM,CRYAB,CYP27A1,DNASE2,DOCK8,GAS1,GBP6,GLP2R,HMGCR,HMGCS1,IFIT3,I FNAR1,IFNB1,IRF3,IRF5,IRF7,MAPK13,MAVS,MPZ,NFATC2,NFIX,NPTX1,OAS1,PKP2,SAMSN1,SASH1,SI6 16 Infectious Disease, Antimicrobial Response, Inflammatory Response RT2,SOCS1,SQLE,STAT2,THBS2,TLR3,VCL AQP3,CAMK4,CCL2,CD14,CD55,CYP2E1,DIO2,DMBT1,DRD2,DYSF,EGR1,EGR2,ethanol,FOS,GABA,GA D2,GRIN2A,GZMB,Gzmc,HMCN1,ID4,IFIT1,IL15,IL1R2,JUN,MAP2,MT1G,NCAM1,NLGN2,PLCL1,PRF1,SE 6 16 Neurological Disease, Skeletal and Muscular Disorders, Hereditary Disorder C23A,SRGN,TFRC,YARS BTBD11,CAMK2A,CAMK2B,CASK,CCNE2,CDCA8,corticosterone,DLG3,DLG4,DLGAP2,DSTN,FMR1,FYN, GRIK2,GRIK5,GUCY1A2,GUCY1B3,IQSEC1,KCNJ10,KCNJ16,MECP2,MSRB2,NGF,NLGN1,NRXN1,NTRK 6 16 Cell-To-Cell Signaling and Interaction, Nervous System Development and Function, Behavior 2,PSEN1,SLC25A4,SLC4A4,SNRPN,SOX9,SRC,SYT1,TUBB6,UBE3A ACKR3,AQP1,AR,ATP1B1,CASQ2,CD28,CD38,CD40LG,CDH5,Cdkn1c,CELF2,CLCN3,CSF2,CSGALNACT 2,HN1,HTT,IFNG,IGFBP5,IL2,IL4,IL5,IL15,JUNB,MMD,NR3C1,ODC1,PRKAG3,RNF128,SOD1,TGFB1,TRE 6 16 Cell-To-Cell Signaling and Interaction, Hematological System Development and Function, Immune Cell Trafficking M1,TSPAN8,UGT1A9 (includes others),vamp8,wnk2 CA9,CCNG2,CD40LG,CD79A,CD79B,CDC6,CDK2,CYTH1,CYTIP,E2F2,E2F3,E2F4,EBI3,GOT1,GPR183, HIF1A,HK2,IGHA1,IGHG1,MYC,POU2AF1,PTPRG,RASSF2,RBL1,RBL2,RGS1,RRM2,RUVBL1,RUVBL2,S 6 16 Cell Morphology, Hematological System Development and Function, Tissue Morphology EMA4D,SLC38A1,TNFAIP2,TNFRSF13C,TNFSF13,UBD AKT3,CD247,CLEC4G,CYBA,DICER1,ELF4,ERBB2,FPR2,GNB4,IFNG,IL4I1,ITCH,KLF4,KLK7,KLRB1,LPA R3,mir-21,mir ,NCR1,norepinephrine,NR2F6,PDE5A,PTPRN2,RNF128,RUNX3,SEMA7A,SLC16A2,SLC7A5,SLC7A8,S 16 Cellular Development, Cellular Growth and Proliferation, Hematological System Development and Function ST,TARDBP,TNFRSF18,TP63,TRD,TRG Ccl2,CD40,CD69,CD86,CEACAM1,CEACAM5,CSF2,CTNNB1,GADD45B,GPT2,ICAM1,IL5,IL6,IL18,IL17A,IL 2RA,LPIN2,LRG1,LRRN3,MYD88,MYRF,NTN1,PAX6,PDCD1,PTGS2,SAMHD1,SATB1,SELL,SLC27A2,ST 6 16 Cellular Movement, Hematological System Development and Function, Immune Cell Trafficking AT1,STAT6,TBC1D9,TICAM1,TMEM2,UNC5B ACTN1,AKT1,APC,Ccl2,CD44,CD3E,CDK5,CORO1C,CTNNB1,DIAPH1,EZR,FZD7,GPR37,IL23A,IVNS1AB P,KIF11,LCN2,MRC1,MSN,NF2,NOS2,PARK2,PTEN,PTPN22,RAC1,RDX,RELA,SATB1,SLC6A3,SLC9A3R 6 16 Cell Death and Survival, Cellular Movement, Cancer 1,SNCA,SPN,TARDBP,TBC1D1,VCP AGT,AGTR1,ALDH1A2,betaestradiol,BIRC5,BMP2,BTC,C4A/C4B,CPT1B,DKK1,DRD1,DRD2,EFEMP1,ESCO2,FRMD6,GAL,GHRH,HO 6 XA4,HOXD4,HSD11B1,IGF1,IGF2,IGFBP2,NBN,OCLN,PDK4,POU1F1,PRL,PTGFRN,PTGIS,PTTG1,RARB 16 Tissue Morphology, Embryonic Development, Organismal Development,RASSF8,TP53,WNT7A APBB1,APP,AVPR1A,Ca2+,CAP2,CD27,CHRDL1,CNKSR2,DGKB,DLG4,ETS1,GABRA5,GRM5,HDAC1,HD AC2,HDAC3,HDAC4,HDAC9,HOMER1,HOMER2,HTT,IL4,IL13,K+,L-glutamic 6 15 Amino Acid Metabolism, Cell-To-Cell Signaling and Interaction, Molecular Transport acid,myog,nbea,ppp3ca,prkcb,ptgfr,tnf,trpc1,tshz3,tspan13 ANXA6,CA2,CD38,CD40,CD83,CD40LG,CEBPB,COL6A3,CSF1,CYSLTR1,CYSLTR2,F2RL2,FGF7,FOXP3,GLIPR2,GPM6B,IGHG3,IL4,IL5,IL13,IL19,IL17A,IL1RL2,NFATC1,PKP3,PNP,PRKG1,PROS1,PSEN1,PTBP5 15 Cell-To-Cell Signaling and Interaction, Hematological System Development and Function, Cellular Growth and Proliferation 2,S1PR1,STAT6,TFEC,TNFSF8,TP53 ADAMTS1,ATP1A1,BMPR1B,BOC,BUB1,Cacnb1,CACNB2,CALCB,CARTPT,CD209,CDK5,CLU,dopamine, DSG3,FGF8,FGF19,FTH1,GDNF,glutathione,GUCY1A3,HSF1,HSF2,JUN,KCNK1,KCNN3,MAOA,MAOB,mir 5 15 Cell-To-Cell Signaling and Interaction, Drug Metabolism, Molecular Transport 506,MYOM1,NMU,NPY,NR4A2,SHH,SOD1,XIAP ACADM,ARG1,BCL11B,BMP1,CFD,EIF4EBP1,EMP3,ESRRG,Foxp1,GFAP,HIP1,HTT,IER3,ILK,inosine,KC NIP2,KLF5,MAP3K8,mir- 5 29,MPZ,MYL9,NGEF,PMP22,Pou3f1,PPARGC1B,PPP1R1B,PPP3CA,SAMD4A,SATB2,SERPING1,SKI,TS 15 Hereditary Disorder, Neurological Disease, Psychological Disorders PO,UCP2,WFS1,ZC3H6 ACHE,ADAM10,APLP1,BACE1,BDH1,CDH2,CHAT,CNTF,CNTFR,DCLK1,DPYSL2,FOXA1,GJA1,GJB6,HT R2C,INADL,LRP1,MAP6,MBP,MPDZ,MYH10,NOS1,NR4A2,PER3,PSEN1,QKI,RGS4,STX2,STXBP1,SYP,T 5 15 Cellular Development, Nervous System Development and Function, Tissue Development GFBR2,TH,Tpm1,TPM3,YWHAZ ANK3,CDK5,CNTN1,CNTNAP1,CTNNB1,CTNNBIP1,CTSF,DKK1,FGF5,FRZB,GAP43,HDAC4,HDAC9,HE PH,HOOK1,JUN,MBP,MPZ,NFASC,NGFR,NRCAM,NTF3,NTRK1,PTN,PTPRZ1,quinolinic acid,rtn4,sox2,sptbn4,tnc,tspan2,ugt8,ulk2,vegfa,wnt3a 5 15 Cellular Development, Nervous System Development and Function, Tissue Development ADCYAP1,ADCYAP1R1,AGER,AKAP12,ANGPT1,BMPR1A,CD80,CD247,CNTNAP2,CTSE,CXCL3,EBI3,EP B41,GAP43,GFAP,GYS2,IL10,IL25,IL17RB,IL23A,IRF1,ITGAM,JAK1,LAMP2,MAN1A1,MOG,NEDD4,nitric 5 14 Cellular Movement, Immune Cell Trafficking, Hematological System Development and Function oxide,notch1,p2ry2,scarb1,serpinb1,smpdl3b,snca,superoxide ACTG1,AQP4,BMPR2,CAV3,DAG1,Dmd,DMD,DTNA,EGR2,ERBB2,ERBB3,FBXO32,FKRP,FOXO3,KCNJ1 4 0,LAMA2,LAMC1,MBP,mir- 13 Developmental Disorder, Skeletal and Muscular Disorders, Hereditary Disorder 130,MYOC,NOS1,PRX,SGCA,SGCB,SGCD,SGCE,SGCG,SNTA1,SNTB1,SNTB2,SSPN,TRIM63,UTRN acetylcholine,ache,adenosine,bche,bdnf,cadps2,cdon,chat,chga,chrm2,cnr1,cntf,dlg1,dn AJB5,DRD1,DRD2,DUSP1,GABA,GDNF,GHRL,GPRASP1,GRB14,HCN1,IGF1R,MAP3K3,MAPK14,NCS1,N3 12 Behavior, Cellular Compromise, Neurological Disease GF,PLG,PPP1R1B,PRIMA1,SLC17A6,SLC6A4,TRPA1,TUBB3
11 Figure legends Figure S1: Heatmap of differentially expressed genes between achalasic and control sample tissues. Samples are grouped by hierarchical clustering. Figure S2: Graph connecting network-eligible molecules as calculated by IPA and overrepresenting the cell morphology, humoral immune response, and cellular movement processes. The achieved score of the graph was 33, while the sparseness ρ value was Figure S3: Graph connecting network-eligible molecules as calculated by IPA and overrepresenting the organ morphology, skeletal and muscular system development processes, and neurological disease processes. The achieved score of the graph was 33, while the sparseness ρ value was 6.01.
12
13 MYOCD CNN1 DES PPP1R3C MYH11 muscle contraction NR4A1 ALDOB contraction of smooth muscle SRF formation of myosin stress fibers ROCK2 FOSB FXYD1 ATP1A2 GNAO1 MT2A DEF6 SCN7A PLCB4 SWAP70 PLCB1 FOS MEOX2 disorder of basal ganglia LRP1 PRKG2 loss of neurons ATP1B2 KLK8 LCAT HSPA1A/HSPA1B PRKAR2B IDH1 A2M PPP1R15A EIF2S1 PRNP motor function TRPV4 MFGE8 prion disease neuritogenesis of neurons
14 IL2RG CD79A FUT8 PDGFRB IGHM TIMP2 MMP2 IGKC morphology of B lymphocytes migration of lymphatic system cells ITGB3 BLNK SYK PRDM1 leukocyte migration DDR1 RGS2 CCND2 HBEGF inflammatory response EDNRA CMA1 INHBA TIMELESS FGL2 PTPRJ EDN1 TPSAB1/TPSB2 CPA3 RGS1 GFRA1 ARID5A TLR4 LRRK2 USP12 ZC3H12C ACPP NEK7 DAB2
B16F1 B16F10 Supplemental Figure S1
 B16F1 B16F1 Supplemental Figure S1 Representative microangiography images of B16F1 and B16F1 tumors grown in the cranial windows. FITC-dextran (2 million MW) was injected systemically to visualize blood
B16F1 B16F1 Supplemental Figure S1 Representative microangiography images of B16F1 and B16F1 tumors grown in the cranial windows. FITC-dextran (2 million MW) was injected systemically to visualize blood
Eosinophils! 40! 30! 20! 10! 0! NS!
 A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases
 Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
CD Marker Antibodies. atlasantibodies.com
 CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
HTG EdgeSeq Immuno-Oncology Assay Gene List
 A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
Supplemental Data for Toischer et al., Cardiomyocyte proliferation prevents failure in
 Supplemental Data, Page 1 Supplemental Data for Toischer et al., Cardiomyocyte proliferation prevents failure in pressure but not volume overload. Contents: Page 3: Supplemental Figure 1 - Echocardiographic
Supplemental Data, Page 1 Supplemental Data for Toischer et al., Cardiomyocyte proliferation prevents failure in pressure but not volume overload. Contents: Page 3: Supplemental Figure 1 - Echocardiographic
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature15260 Supplementary Data 1: Gene expression in individual basal/stem, luminal, and luminal progenitor cells. Box plots show expression levels for each gene from the 49-gene differentiation
doi:10.1038/nature15260 Supplementary Data 1: Gene expression in individual basal/stem, luminal, and luminal progenitor cells. Box plots show expression levels for each gene from the 49-gene differentiation
Supplementary Figure 1!
 a!!gt-cre x Twist LoxP/LoxP!!GT-Cre x Twist LoxP/LoxP! x RosaLSLEYFP!!GT-Cre x Snail LoxP/LoxP!!GT-Cre x Snail LoxP/LoxP! x RosaLSLEYFP! Supplementary Figure!!GT-Cre x Twist LoxP/LoxP! x RosaLSLEYFP x
a!!gt-cre x Twist LoxP/LoxP!!GT-Cre x Twist LoxP/LoxP! x RosaLSLEYFP!!GT-Cre x Snail LoxP/LoxP!!GT-Cre x Snail LoxP/LoxP! x RosaLSLEYFP! Supplementary Figure!!GT-Cre x Twist LoxP/LoxP! x RosaLSLEYFP x
Supplementary Table S1: Gene expression targets
 Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array.
 Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Screening of potential biomarkers in uterine leiomyomas disease via gene expression profiling analysis
 MOLECULAR MEDICINE REPORTS 17: 6985-6996, 2018 Screening of potential biomarkers in uterine leiomyomas disease via gene expression profiling analysis XUHUI LIU 1*, YANFEI LIU 2*, JINGRONG ZHAO 3 and YAN
MOLECULAR MEDICINE REPORTS 17: 6985-6996, 2018 Screening of potential biomarkers in uterine leiomyomas disease via gene expression profiling analysis XUHUI LIU 1*, YANFEI LIU 2*, JINGRONG ZHAO 3 and YAN
SUPPLEMENTAL TABLE AND FIGURES
 SUPPLEMENTAL TABLE AND FIGURES Zhang et al. (29) - Enzymes in the NAD + Salvage Pathway Regulate SIRT1 Activity at Target Gene Promoters This document contains supplemental data (1 table and 6 figures)
SUPPLEMENTAL TABLE AND FIGURES Zhang et al. (29) - Enzymes in the NAD + Salvage Pathway Regulate SIRT1 Activity at Target Gene Promoters This document contains supplemental data (1 table and 6 figures)
Cardiovascular Disease Products
 Cardiovascular Disease Products For more information, visit: www.bosterbio.com Cardiovascular Disease Research Cardiovascular disease is the leading cause of death in developed nations. Boster Bio aims
Cardiovascular Disease Products For more information, visit: www.bosterbio.com Cardiovascular Disease Research Cardiovascular disease is the leading cause of death in developed nations. Boster Bio aims
TITLE: Genetic and epigenetic biomarkers for recurrent prostate cancer after radiotherapy
 AD Award Number: W81XWH-12-1-0113 TITLE: Genetic and epigenetic biomarkers for recurrent prostate cancer after radiotherapy PRINCIPAL INVESTIGATOR: Jong Park CONTRACTING ORGANIZATION: H. Lee Moffitt Cancer
AD Award Number: W81XWH-12-1-0113 TITLE: Genetic and epigenetic biomarkers for recurrent prostate cancer after radiotherapy PRINCIPAL INVESTIGATOR: Jong Park CONTRACTING ORGANIZATION: H. Lee Moffitt Cancer
Supplemental Figure 1: Anti-GDF15 antibody blocks binding of GDF15 to hgfral.
 Supplemental Figure 1: Anti-GDF15 antibody blocks binding of GDF15 to hgfral. Anti-GDF15 antibody (mab26, a neutralizing monoclonal antibody to wtgdf15) blocks binding of GDF15 to human GFRAL in a dose-dependent
Supplemental Figure 1: Anti-GDF15 antibody blocks binding of GDF15 to hgfral. Anti-GDF15 antibody (mab26, a neutralizing monoclonal antibody to wtgdf15) blocks binding of GDF15 to human GFRAL in a dose-dependent
The expression of genes contributing to pancreatic adenocarcinoma progression is influenced by the respective environment Sagini et al
 The expression of genes contributing to pancreatic adenocarcinoma progression is influenced by the respective environment Sagini et al Supplementary Figure 1: Target genes regulated by TGM2. Figure represents
The expression of genes contributing to pancreatic adenocarcinoma progression is influenced by the respective environment Sagini et al Supplementary Figure 1: Target genes regulated by TGM2. Figure represents
Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction. Marker Sequence (5 3 ) Accession No.
 Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Table 1: Motor-clutch gene mrna expression Myosin Motors
 Supplementary Table 1: Motor-clutch gene mrna expression Myosin Motors Symbol Description Expression Rank MYL6 myosin, light chain 6, alkali, smooth muscle and non-muscle 11903 79 MYH9 myosin, heavy chain
Supplementary Table 1: Motor-clutch gene mrna expression Myosin Motors Symbol Description Expression Rank MYL6 myosin, light chain 6, alkali, smooth muscle and non-muscle 11903 79 MYH9 myosin, heavy chain
Supplementary Figures
 Supplementary Figures Figure S1. Urodynamic recording of BOO-induced LUTD patients. (A) DO group. BOO patients with increased detrusor pressure and reduced urine flow during pressure flow in combination
Supplementary Figures Figure S1. Urodynamic recording of BOO-induced LUTD patients. (A) DO group. BOO patients with increased detrusor pressure and reduced urine flow during pressure flow in combination
Contribution of the intronic mir-338-3p and its Hosting Gene AATK to Compensatory β-cell Mass Expansion
 Contribution of the intronic mir-338-3p and its Hosting Gene AATK to Compensatory β-cell Mass Expansion Cécile Jacovetti 1, Veronica Jimenez 2, Eduard Ayuso 2#, Ross Laybutt 3, Marie-Line Peyot 4, Marc
Contribution of the intronic mir-338-3p and its Hosting Gene AATK to Compensatory β-cell Mass Expansion Cécile Jacovetti 1, Veronica Jimenez 2, Eduard Ayuso 2#, Ross Laybutt 3, Marie-Line Peyot 4, Marc
The Genetics of Alcoholism. Robert Hitzemann, Ph.D. Departments of Behavioral Neuroscience & Psychiatry Oregon Health & Science University
 The Genetics of Alcoholism Robert Hitzemann, Ph.D. Departments of Behavioral Neuroscience & Psychiatry Oregon Health & Science University Genetics, Genomics and Alcoholism Robert Hitzemann, Ph.D. Departments
The Genetics of Alcoholism Robert Hitzemann, Ph.D. Departments of Behavioral Neuroscience & Psychiatry Oregon Health & Science University Genetics, Genomics and Alcoholism Robert Hitzemann, Ph.D. Departments
antibody and ELISA experts Cancer Research Products
 Cancer Research Products WWW.BOSTERBIO.COM Cancer Research Cancer is a leading cause of death in developed nations, affecting millions of people around the world. Research into the molecular basis of these
Cancer Research Products WWW.BOSTERBIO.COM Cancer Research Cancer is a leading cause of death in developed nations, affecting millions of people around the world. Research into the molecular basis of these
1. Jing Wang, Ph.D., Key Laboratory of Mental Health, Institute of Psychology,
 Network analysis reveals a stress-affected common gene module among seven stress-related diseases/systems which provide potential targets for mechanism research Liyuan Guo 1 *, Yang Du 1, 2 and Jing Wang
Network analysis reveals a stress-affected common gene module among seven stress-related diseases/systems which provide potential targets for mechanism research Liyuan Guo 1 *, Yang Du 1, 2 and Jing Wang
BD Rhapsody Immune Response Panel Hs
 Doc ID: 55665 Rev. 2.0 PRODUCT INSERT BD Rhapsody Immune Response Panel Hs BD Biosciences Kit Part Number: 633750 Purpose The BD Rhapsody Immune Response Panel Hs is a set of ready-to-use primer pools
Doc ID: 55665 Rev. 2.0 PRODUCT INSERT BD Rhapsody Immune Response Panel Hs BD Biosciences Kit Part Number: 633750 Purpose The BD Rhapsody Immune Response Panel Hs is a set of ready-to-use primer pools
Control Histogram Log 2 Fold Change
 300 Control Histogram 250 200 Frequency 150 100 50 0-2 -1.5-1 -0.5 0 0.5 1 1.5 2 Log 2 Fold Change SI-Figure 1: Control Histogram. Histogram represents biological and experimental variability of itraq
300 Control Histogram 250 200 Frequency 150 100 50 0-2 -1.5-1 -0.5 0 0.5 1 1.5 2 Log 2 Fold Change SI-Figure 1: Control Histogram. Histogram represents biological and experimental variability of itraq
Supplementary information. Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls
 Supplementary information Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls angiogenesis in breast cancer Ali Flores-Pérez, Laurence A. Marchat, Sergio Rodríguez-Cuevas, Verónica Bautista-Piña,
Supplementary information Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls angiogenesis in breast cancer Ali Flores-Pérez, Laurence A. Marchat, Sergio Rodríguez-Cuevas, Verónica Bautista-Piña,
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Characterization of HPV Tg/+ ; Krt17 / mice. (a) Macroscopic images of HPV16 Tg/+ mouse ears at P40, P70 and P120. (b) Immunostaining of HPV16 Tg/+ ear tissue sections for K17 (red)
Supplementary Figure 1 Characterization of HPV Tg/+ ; Krt17 / mice. (a) Macroscopic images of HPV16 Tg/+ mouse ears at P40, P70 and P120. (b) Immunostaining of HPV16 Tg/+ ear tissue sections for K17 (red)
ONLINE DATA SUPPLEMENT. Single Cell RNA-Sequencing Identifies Diverse Roles of Epithelial Cells in. Idiopathic Pulmonary Fibrosis
 ONLINE DATA SUPPLEMENT Single Cell RNA-Sequencing Identifies Diverse Roles of Epithelial Cells in Idiopathic Pulmonary Fibrosis Yan Xu Takako Mizuno Anusha Sridharan Yina Du Minzhe Guo Jie Tang Kathryn
ONLINE DATA SUPPLEMENT Single Cell RNA-Sequencing Identifies Diverse Roles of Epithelial Cells in Idiopathic Pulmonary Fibrosis Yan Xu Takako Mizuno Anusha Sridharan Yina Du Minzhe Guo Jie Tang Kathryn
Aberrant gene expression in mucosa adjacent to tumor reveals a molecular crosstalk in colon cancer
 Sanz-Pamplona et al. Molecular Cancer 2014, 13:46 RESEARCH Open Access Aberrant gene expression in mucosa adjacent to tumor reveals a molecular crosstalk in colon cancer Rebeca Sanz-Pamplona 1, Antoni
Sanz-Pamplona et al. Molecular Cancer 2014, 13:46 RESEARCH Open Access Aberrant gene expression in mucosa adjacent to tumor reveals a molecular crosstalk in colon cancer Rebeca Sanz-Pamplona 1, Antoni
Supplementary Figures and Tables. An inflammatory gene signature distinguishes neurofibroma Schwann cells and
 Supplementary Figures and Tables An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system Kwangmin Choi 1, Kakajan Komurov
Supplementary Figures and Tables An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system Kwangmin Choi 1, Kakajan Komurov
Supplementary Information for: Predictors of chronic kidney disease in type 1 diabetes: a longitudinal study from the AMD Annals initiative.
 Supplementary Information for: Predictors of chronic kidney disease in type 1 diabetes: a longitudinal study from the AMD Annals initiative. Authors: Pamela Piscitelli 1, Francesca Viazzi 2 ; Paola Fioretto
Supplementary Information for: Predictors of chronic kidney disease in type 1 diabetes: a longitudinal study from the AMD Annals initiative. Authors: Pamela Piscitelli 1, Francesca Viazzi 2 ; Paola Fioretto
Lions and Tigers and Bears.Oh My!
 Lions and Tigers and Bears.Oh My! TAKING THE FEAR OUT OF GENETIC TESTING Lisa Butterfield, MS, CGC Genetic Counselor Clinical Assistant Professor University of Kansas Health System Why is Genetic Testing
Lions and Tigers and Bears.Oh My! TAKING THE FEAR OUT OF GENETIC TESTING Lisa Butterfield, MS, CGC Genetic Counselor Clinical Assistant Professor University of Kansas Health System Why is Genetic Testing
Glia in Alzheimer's disease and aging: Molecular mechanisms underlying astrocyte and microglia reactivity Orre, A.M.
 UvA-DARE (Digital Academic Repository) Glia in Alzheimer's disease and aging: Molecular mechanisms underlying astrocyte and microglia reactivity Orre, A.M. Link to publication Citation for published version
UvA-DARE (Digital Academic Repository) Glia in Alzheimer's disease and aging: Molecular mechanisms underlying astrocyte and microglia reactivity Orre, A.M. Link to publication Citation for published version
Case Study - Informatics
 bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
The Human Body: An Orientation
 PowerPoint Lecture Slide Presentation by Patty Bostwick-Taylor, Florence-Darlington Technical College The Human Body: An Orientation 1 PART A The Human Body An Orientation Anatomy Study of the structure
PowerPoint Lecture Slide Presentation by Patty Bostwick-Taylor, Florence-Darlington Technical College The Human Body: An Orientation 1 PART A The Human Body An Orientation Anatomy Study of the structure
 Supplementary Table 1. Annotated candidate SNV and indels in melanoma tumors. Patient C1 Patient C2 Patient C3 Patient C4 Patient C5 Patient C6 Patient C7 Patient C10 Chr Position Reference Allele Sample
Supplementary Table 1. Annotated candidate SNV and indels in melanoma tumors. Patient C1 Patient C2 Patient C3 Patient C4 Patient C5 Patient C6 Patient C7 Patient C10 Chr Position Reference Allele Sample
Characterization of mirna-regulated networks, hubs of signaling, and biomarkers in obstruction-induced bladder dysfunction
 Characterization of mirna-regulated networks, hubs of signaling, and biomarkers in obstruction-induced bladder dysfunction Ali Hashemi Gheinani, 1 Bernhard Kiss, 2 Felix Moltzahn, 2 Irene Keller, 3 Rémy
Characterization of mirna-regulated networks, hubs of signaling, and biomarkers in obstruction-induced bladder dysfunction Ali Hashemi Gheinani, 1 Bernhard Kiss, 2 Felix Moltzahn, 2 Irene Keller, 3 Rémy
Regulation of inflammation by histone deacetylases in rheumatoid arthritis: beyond epigenetics Grabiec, A.M.
 UvA-DARE (Digital Academic Repository) Regulation of inflammation by histone deacetylases in rheumatoid arthritis: beyond epigenetics Grabiec, A.M. Link to publication Citation for published version (APA):
UvA-DARE (Digital Academic Repository) Regulation of inflammation by histone deacetylases in rheumatoid arthritis: beyond epigenetics Grabiec, A.M. Link to publication Citation for published version (APA):
LIX1 APOD RRM2 FAIM2 MGST1 UHRF1 PPIA BEX1
 C2orf80 1 C2orf80 A2M ANXA1 APOD ARL4A BEX1 C1S C3 CD44 CHI3L1 CHI3L2 CLU DPYD DSCAM EMP3 FN1 GBP1 GEM GLIPR1 GRIA2 IDH1 LIX1 MAP2 MDK MGST1 MIR3917 NAMPT PCMTD2 PPIA PTN PTPRZ1 S100B SCG3 SERPINA3 TCF12
C2orf80 1 C2orf80 A2M ANXA1 APOD ARL4A BEX1 C1S C3 CD44 CHI3L1 CHI3L2 CLU DPYD DSCAM EMP3 FN1 GBP1 GEM GLIPR1 GRIA2 IDH1 LIX1 MAP2 MDK MGST1 MIR3917 NAMPT PCMTD2 PPIA PTN PTPRZ1 S100B SCG3 SERPINA3 TCF12
In vivo bromodeoxyuridine (BrdU) incorporation was performed to analyze cell
 Supplementary Methods BrdU incorporation in vivo In vivo bromodeoxyuridine (BrdU) incorporation was performed to analyze cell proliferation in the heart. Mice were subjected to LI-TAC, and 5 days later
Supplementary Methods BrdU incorporation in vivo In vivo bromodeoxyuridine (BrdU) incorporation was performed to analyze cell proliferation in the heart. Mice were subjected to LI-TAC, and 5 days later
Table 1 Demographic data of PD-patients. Data are represented as counts or median and full range.
 Table 1 Demographic data of PD-patients. Data are represented as counts or median and full range. Parameter Number Patients 5 Diabetes mellitus 0 Previous NTX 0 Steroids 0 Patients with residual renal
Table 1 Demographic data of PD-patients. Data are represented as counts or median and full range. Parameter Number Patients 5 Diabetes mellitus 0 Previous NTX 0 Steroids 0 Patients with residual renal
EMT and ECM. Jesse Liang, Ph.D. Sample & Assay Technologies
 EMT and ECM Jesse Liang, Ph.D. 1 Epithelial Cells.1. Closely adjoined.2. Polarized.Epithelial Markers: E-Cadherin (adherens junctions) Claudins (tight junctions) Occludin (tight junctions) Desmoplakin
EMT and ECM Jesse Liang, Ph.D. 1 Epithelial Cells.1. Closely adjoined.2. Polarized.Epithelial Markers: E-Cadherin (adherens junctions) Claudins (tight junctions) Occludin (tight junctions) Desmoplakin
Xiaodan Wu 1, Xiaoru Sun 2, Chengshui Chen 2*, Chunxue Bai 3 and Xiangdong Wang 2,3*
 Wu et al. Critical Care (2014) 18:508 DOI 10.1186/s13054-014-0508-y RESEARCH Open Access Dynamic gene expressions of peripheral blood mononuclear cells in patients with acute exacerbation of chronic obstructive
Wu et al. Critical Care (2014) 18:508 DOI 10.1186/s13054-014-0508-y RESEARCH Open Access Dynamic gene expressions of peripheral blood mononuclear cells in patients with acute exacerbation of chronic obstructive
The Human Body: An Orientation
 PowerPoint Lecture Slide Presentation by Patty Bostwick-Taylor, Florence-Darlington Technical College The Human Body: An Orientation 1PART A The Human Body An Orientation Anatomy Study of the structure
PowerPoint Lecture Slide Presentation by Patty Bostwick-Taylor, Florence-Darlington Technical College The Human Body: An Orientation 1PART A The Human Body An Orientation Anatomy Study of the structure
Jocelyn Chapman, MD Division of Gynecologic Oncology. Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic
 Jocelyn Chapman, MD Division of Gynecologic Oncology Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic Genetics underlies all cancers Somatic or tumor genetics Germline or inherited genetics
Jocelyn Chapman, MD Division of Gynecologic Oncology Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic Genetics underlies all cancers Somatic or tumor genetics Germline or inherited genetics
Cytokines, adhesion molecules and apoptosis markers. A comprehensive product line for human and veterinary ELISAs
 Cytokines, adhesion molecules and apoptosis markers A comprehensive product line for human and veterinary ELISAs IBL International s cytokine product line... is extremely comprehensive. The assays are
Cytokines, adhesion molecules and apoptosis markers A comprehensive product line for human and veterinary ELISAs IBL International s cytokine product line... is extremely comprehensive. The assays are
Natural and Synthetic lipid membrane nanovesiclesto repair and regenerate skin. Sai Kiang Lim, PhD
 Natural and Synthetic lipid membrane nanovesiclesto repair and regenerate skin Sai Kiang Lim, PhD Research Focus of Lab Nano Lipid membrane vesicles Natural Extracellular Vesicles: microvesicles, exosomes,
Natural and Synthetic lipid membrane nanovesiclesto repair and regenerate skin Sai Kiang Lim, PhD Research Focus of Lab Nano Lipid membrane vesicles Natural Extracellular Vesicles: microvesicles, exosomes,
under the aegis of cover picture: Chiostro d Onore University of Milan
 under the aegis of cover picture: Chiostro d Onore University of Milan Milan. 2,3 July 2012 Palazzo Visconti MEETING AIM In the last year the therapeutic scenario of chronic hepatitis C has been subverted
under the aegis of cover picture: Chiostro d Onore University of Milan Milan. 2,3 July 2012 Palazzo Visconti MEETING AIM In the last year the therapeutic scenario of chronic hepatitis C has been subverted
Robot acceptability in caregivers and elderly subjects with dementia
 Robot acceptability in caregivers and elderly subjects with dementia -------------- Antonio Greco and Grazia D Onofrio Complex Unit of Geriatrics, Department of Medical Sciences, IRCCS Casa Sollievo della
Robot acceptability in caregivers and elderly subjects with dementia -------------- Antonio Greco and Grazia D Onofrio Complex Unit of Geriatrics, Department of Medical Sciences, IRCCS Casa Sollievo della
 Table S1. Metadata for transcriptome interaction network and pathway analysis of 5448 intracelluarly infected TEpi cells in comparison to mock TEpi cells. Gene symbol Full name Log2FC gene expression in
Table S1. Metadata for transcriptome interaction network and pathway analysis of 5448 intracelluarly infected TEpi cells in comparison to mock TEpi cells. Gene symbol Full name Log2FC gene expression in
Deconvolution of Heterogeneous Wound Tissue Samples into Relative Macrophage Phenotype Composition via Models based on Gene Expression
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2017 Supplementary Information for: Deconvolution of Heterogeneous Wound Tissue Samples into
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2017 Supplementary Information for: Deconvolution of Heterogeneous Wound Tissue Samples into
Supporting Information
 Supporting Information Rothhammer et al. 1.173/pnas.161413114 SI Materials and Methods EAE Clinical Scoring and Agents. Clinical sig of EAE were assessed as follows:, no sig of disease; 1, loss of tone
Supporting Information Rothhammer et al. 1.173/pnas.161413114 SI Materials and Methods EAE Clinical Scoring and Agents. Clinical sig of EAE were assessed as follows:, no sig of disease; 1, loss of tone
Supplement Results, Figures and Tables
 Supplement Results, Figures and Tables Results PET imaging The SUV was significantly higher in tumours of the parental HSC-3 cell line than tumours of the EV and the MMP-8 overexpressing cell lines (Figure
Supplement Results, Figures and Tables Results PET imaging The SUV was significantly higher in tumours of the parental HSC-3 cell line than tumours of the EV and the MMP-8 overexpressing cell lines (Figure
Nature Genetics: doi: /ng Supplementary Figure 1. Depths and coverages in whole-exome and targeted deep sequencing data.
 Supplementary Figure 1 Depths and coverages in whole-exome and targeted deep sequencing data. Depth (top) and coverage (bottom) of whole-exome sequencing for 38 independent JPN cases (mean depth = 130)
Supplementary Figure 1 Depths and coverages in whole-exome and targeted deep sequencing data. Depth (top) and coverage (bottom) of whole-exome sequencing for 38 independent JPN cases (mean depth = 130)
Supplementary Material: Exploring wound-healing genomic machinery with a network-based approach
 Supplementary Material: Exploring wound-healing genomic machinery with a network-based approach Francesca Vitali, Simone Marini, Martina Balli, Hanne Grosemans, Maurilio Sampaolesi, Yves A. Lussier, Maria
Supplementary Material: Exploring wound-healing genomic machinery with a network-based approach Francesca Vitali, Simone Marini, Martina Balli, Hanne Grosemans, Maurilio Sampaolesi, Yves A. Lussier, Maria
Study on the Biological Basis of Hypertension and Syndrome with Liver Fire Hyperactivity Based on Data Mining Technology
 Download free from www.wjtcm.net Original Article Study on the Biological Basis of Hypertension and Syndrome with Liver Fire Hyperactivity Based on Data Mining Technology Xue-Ling Ma a, Xing Zhai b, Jing-Wei
Download free from www.wjtcm.net Original Article Study on the Biological Basis of Hypertension and Syndrome with Liver Fire Hyperactivity Based on Data Mining Technology Xue-Ling Ma a, Xing Zhai b, Jing-Wei
Casa Sollievo della Sofferenza - San Giovanni Rotondo, Italy
 - San Giovanni Rotondo, Italy General Information New breast cancer cases treated per year 358 Breast multidisciplinarity team members 20 Radiologists, surgeons, pathologists, medical oncologists, radiotherapists
- San Giovanni Rotondo, Italy General Information New breast cancer cases treated per year 358 Breast multidisciplinarity team members 20 Radiologists, surgeons, pathologists, medical oncologists, radiotherapists
PSK4U THE NEUROMUSCULAR SYSTEM
 PSK4U THE NEUROMUSCULAR SYSTEM REVIEW Review of muscle so we can see how the neuromuscular system works This is not on today's note Skeletal Muscle Cell: Cellular System A) Excitation System Electrical
PSK4U THE NEUROMUSCULAR SYSTEM REVIEW Review of muscle so we can see how the neuromuscular system works This is not on today's note Skeletal Muscle Cell: Cellular System A) Excitation System Electrical
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study. February 2018
 Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Tissue-specific Differentiation Potency of Mesenchymal Stromal Cells from Perinatal
 Tissue-specific Differentiation Potency of Mesenchymal Stromal Cells from Perinatal Tissues Ahlm Kwon 1,*, Yonggoo Kim 1,2,*, Myungshin Kim 1,2,*, Jiyeon Kim 1, Hayoung Choi 1, Dong Wook Jekarl 1,2, Seungok
Tissue-specific Differentiation Potency of Mesenchymal Stromal Cells from Perinatal Tissues Ahlm Kwon 1,*, Yonggoo Kim 1,2,*, Myungshin Kim 1,2,*, Jiyeon Kim 1, Hayoung Choi 1, Dong Wook Jekarl 1,2, Seungok
Supplementary Table 2. Identified causative mutations and/or mutation candidates.
 Supplementary Table 2. Identified causative mutations and/or mutation candidates. Nonsense mutations base change aa change Average depth Result of next generation in 432 patient Hereditary form of the
Supplementary Table 2. Identified causative mutations and/or mutation candidates. Nonsense mutations base change aa change Average depth Result of next generation in 432 patient Hereditary form of the
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation
 Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
SCIENCE CHINA Life Sciences
 SCIENCE CHINA Life Sciences RESEARCH PAPER April 2013 Vol.56 No.4: 1 7 doi: 10.1007/s11427-013-4460-x Table S1 Human paired-end RNA-Seq data sets from the Sequence Read Archive (SRA) database Experiment
SCIENCE CHINA Life Sciences RESEARCH PAPER April 2013 Vol.56 No.4: 1 7 doi: 10.1007/s11427-013-4460-x Table S1 Human paired-end RNA-Seq data sets from the Sequence Read Archive (SRA) database Experiment
Chapter 8 11/1/2012. Synaptic Components are Ancient. Syncytium or Synapses? Synapse Formation and Function. Early Calcium Spikes
 Chapter 8 Synaptic Components are Ancient Synapse Formation and Function Fig 8.1 Syncytium or Synapses? Electrical Development Synapses Improve in Function with Time Fig 8.2 Fig 8.3 Early Calcium Spikes
Chapter 8 Synaptic Components are Ancient Synapse Formation and Function Fig 8.1 Syncytium or Synapses? Electrical Development Synapses Improve in Function with Time Fig 8.2 Fig 8.3 Early Calcium Spikes
Adenocarcinoma of the pancreas
 Adenocarcinoma of the pancreas SEMINARS IN DIAGNOSTIC PATHOLOGY 31 (2014) 443 451 Ralph H.Hruban, MD, David S. Klimstra, MD Paola Parente Anatomia Patologica Casa Sollievo della Sofferenza San Giovanni
Adenocarcinoma of the pancreas SEMINARS IN DIAGNOSTIC PATHOLOGY 31 (2014) 443 451 Ralph H.Hruban, MD, David S. Klimstra, MD Paola Parente Anatomia Patologica Casa Sollievo della Sofferenza San Giovanni
Elaine N. Marieb Katja N. Hoehn Ninth Edition
 Human Anatomy & Physiology Elaine N. Marieb Katja N. Hoehn Ninth Edition Pearson Education Limited Edinburgh Gate Harlow Essex CM20 2JE England and Associated Companies throughout the world Visit us on
Human Anatomy & Physiology Elaine N. Marieb Katja N. Hoehn Ninth Edition Pearson Education Limited Edinburgh Gate Harlow Essex CM20 2JE England and Associated Companies throughout the world Visit us on
HIBBING COMMUNITY COLLEGE COURSE OUTLINE
 HIBBING COMMUNITY COLLEGE COURSE OUTLINE COURSE NUMBER & TITLE: BIOL 2151: Human Physiology CREDITS: 4 (Lecture 3 / Lab 1) PREREQUISITES: Human Anatomy or Integrated Science recommended CATALOG DESCRIPTION:
HIBBING COMMUNITY COLLEGE COURSE OUTLINE COURSE NUMBER & TITLE: BIOL 2151: Human Physiology CREDITS: 4 (Lecture 3 / Lab 1) PREREQUISITES: Human Anatomy or Integrated Science recommended CATALOG DESCRIPTION:
Performance Objective Critical Attributes Benchmarks/Assessment. Can the student identify structures and functions of whole systems?
 Curriculum Standard One: Organization of the Human Body The student will understand how structure and function compliment each other, the nature of homeostasis, and the language of Anatomy. 1A. The student
Curriculum Standard One: Organization of the Human Body The student will understand how structure and function compliment each other, the nature of homeostasis, and the language of Anatomy. 1A. The student
The Muscular System PART A
 6 The Muscular System PART A PowerPoint Lecture Slide Presentation by Jerry L. Cook, Sam Houston University ESSENTIALS OF HUMAN ANATOMY & PHYSIOLOGY EIGHTH EDITION ELAINE N. MARIEB The Muscular System
6 The Muscular System PART A PowerPoint Lecture Slide Presentation by Jerry L. Cook, Sam Houston University ESSENTIALS OF HUMAN ANATOMY & PHYSIOLOGY EIGHTH EDITION ELAINE N. MARIEB The Muscular System
! BIOL 2401! Week 5. Nervous System. Nervous System
 Collin County Community College! BIOL 2401! Week 5 Nervous System 1 Nervous System The process of homeostasis makes sure that the activities that occur in the body are maintained within normal physiological
Collin County Community College! BIOL 2401! Week 5 Nervous System 1 Nervous System The process of homeostasis makes sure that the activities that occur in the body are maintained within normal physiological
Postmortem brain transcriptomes in Dup15q and idiopathic autism. T Grant Belgard, DPhil Dan Geschwind s Lab UCLA Neuropsychiatric Institute
 Postmortem brain transcriptomes in Dup15q and idiopathic autism T Grant Belgard, DPhil Dan Geschwind s Lab UCLA Neuropsychiatric Institute Many genetic causes of autism, none particularly dominant Is there
Postmortem brain transcriptomes in Dup15q and idiopathic autism T Grant Belgard, DPhil Dan Geschwind s Lab UCLA Neuropsychiatric Institute Many genetic causes of autism, none particularly dominant Is there
The pluripotency factor NANOG promotes the formation of squamous cell carcinomas
 SUPPLEMENTAL DATA The pluripotency factor NANOG promotes the formation of squamous cell carcinomas Adelaida R. Palla, Daniela Piazzolla, Noelia Alcazar, Marta Cañamero, Osvaldo Graña, Gonzalo Gómez-López,
SUPPLEMENTAL DATA The pluripotency factor NANOG promotes the formation of squamous cell carcinomas Adelaida R. Palla, Daniela Piazzolla, Noelia Alcazar, Marta Cañamero, Osvaldo Graña, Gonzalo Gómez-López,
Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK
 Supplemental legends Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK Table SП. List of genes in cluster 1 identified using two-dimensional hierarchical clustering analysis Table SШ.
Supplemental legends Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK Table SП. List of genes in cluster 1 identified using two-dimensional hierarchical clustering analysis Table SШ.
Cover Page. The handle holds various files of this Leiden University dissertation.
 Cover Page The handle http://hdl.handle.net/1887/43190 holds various files of this Leiden University dissertation. Author: Raeven, R.H.M. Title: Systems vaccinology : molecular signatures of immunity to
Cover Page The handle http://hdl.handle.net/1887/43190 holds various files of this Leiden University dissertation. Author: Raeven, R.H.M. Title: Systems vaccinology : molecular signatures of immunity to
ptglab.com PECAM1: Customer Testimonial
 ptglab.com 1 CD MARKER ANTIBODIES www.ptglab.com Introduction The cluster of differentiation (abbreviated as CD) is a protocol used for the identification and investigation of cell surface molecules. So-called
ptglab.com 1 CD MARKER ANTIBODIES www.ptglab.com Introduction The cluster of differentiation (abbreviated as CD) is a protocol used for the identification and investigation of cell surface molecules. So-called
الكيمياء احليوية لألنسجة املتخصصة. Biochemistry of Specialized Tissues
 الكيمياء احليوية لألنسجة املتخصصة Biochemistry of Specialized Tissues BCH 443 Biochemistry of specialized Tissues Course Symbol & No. : BCH 443 Credit Hours : 2 (2+0) Prerequisite : BCH 347 Class schedule
الكيمياء احليوية لألنسجة املتخصصة Biochemistry of Specialized Tissues BCH 443 Biochemistry of specialized Tissues Course Symbol & No. : BCH 443 Credit Hours : 2 (2+0) Prerequisite : BCH 347 Class schedule
Biomarcatori per la immunoterapia: cosa e come cercare Paolo Graziano
 Biomarcatori per la immunoterapia: cosa e come cercare Paolo Graziano Unit of Pathology Fondazione IRCCS Casa Sollievo della Sofferenza San Giovanni Rotondo, Foggia,Italy p.graziano@operapadrepio.it Disclosure
Biomarcatori per la immunoterapia: cosa e come cercare Paolo Graziano Unit of Pathology Fondazione IRCCS Casa Sollievo della Sofferenza San Giovanni Rotondo, Foggia,Italy p.graziano@operapadrepio.it Disclosure
Investigation into differential RNA expression between diabetic and non-diabetic patients in acute ankle fractures.
 Investigation into differential RNA expression between diabetic and non-diabetic patients in acute ankle fractures. Brian A Silvia MD, PhD brian.silvia@gmail.com Lorenzo Gamez MD lorenzo.gamez@gmail.com
Investigation into differential RNA expression between diabetic and non-diabetic patients in acute ankle fractures. Brian A Silvia MD, PhD brian.silvia@gmail.com Lorenzo Gamez MD lorenzo.gamez@gmail.com
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06030 SUPPLEMENTARY INFORMATION Whole Mount eno-empty eno-cre Original Magnification 200X eno-cre eno-empty Ji, Ramsey et al., Supplementary Figure 1 www.nature.com/nature 1 a Percent
doi: 10.1038/nature06030 SUPPLEMENTARY INFORMATION Whole Mount eno-empty eno-cre Original Magnification 200X eno-cre eno-empty Ji, Ramsey et al., Supplementary Figure 1 www.nature.com/nature 1 a Percent
Integrative analysis of genetic data sets reveals a shared innate immune component in autism spectrum disorder and its co-morbidities
 Integrative analysis of genetic data sets reveals a shared innate immune component in autism spectrum disorder and its co-morbidities The MIT Faculty has made this article openly available. Please share
Integrative analysis of genetic data sets reveals a shared innate immune component in autism spectrum disorder and its co-morbidities The MIT Faculty has made this article openly available. Please share
THE BIOLOGY OF PLATELET-GEL THERAPY
 THE BIOLOGY OF PLATELET-GEL THERAPY The synopsis of normal healing includes a well known sequence of coordinated phases. The unique process leading to healing is ontologically partitioned in three sequential
THE BIOLOGY OF PLATELET-GEL THERAPY The synopsis of normal healing includes a well known sequence of coordinated phases. The unique process leading to healing is ontologically partitioned in three sequential
The Amazing Brain Webinar Series: Select Topics in Neuroscience and Child Development for the Clinician
 The Amazing Brain Webinar Series: Select Topics in Neuroscience and Child Development for the Clinician Part VII Recent Advances in the Genetics of Autism Spectrum Disorders Abha R. Gupta, MD, PhD Jointly
The Amazing Brain Webinar Series: Select Topics in Neuroscience and Child Development for the Clinician Part VII Recent Advances in the Genetics of Autism Spectrum Disorders Abha R. Gupta, MD, PhD Jointly
MicroRNA expression signature in atrial fibrillation with mitral stenosis
 Physiol Genomics 43: 655 664, 2011. First published February 15, 2011; doi:10.1152/physiolgenomics.00139.2010. CALL FOR PAPERS MicroRNA and Tissue Injury MicroRNA expression signature in atrial fibrillation
Physiol Genomics 43: 655 664, 2011. First published February 15, 2011; doi:10.1152/physiolgenomics.00139.2010. CALL FOR PAPERS MicroRNA and Tissue Injury MicroRNA expression signature in atrial fibrillation
g. RA Bardot et al., 2016 Supplementary Figure 1 RV LV E15.5 E6.5 E18.5 E7.5 VE RV LV E8.5 RV LV E10.5 Brightfield Foxa2Cre:YFP Brightfield
 Bardot et al., 216 Supplementary Figure 1 a. VE e. RA LA LA RV LV E15.5 E6.5 b. E7.5 VE f. RA E18.5 LA c. E8.5 ML N CM g. RA RV LV LA P7 PHT RV LV d. E1.5 Brightfield Foxa2Cre:YFP H Brightfield Foxa2Cre:YFP
Bardot et al., 216 Supplementary Figure 1 a. VE e. RA LA LA RV LV E15.5 E6.5 b. E7.5 VE f. RA E18.5 LA c. E8.5 ML N CM g. RA RV LV LA P7 PHT RV LV d. E1.5 Brightfield Foxa2Cre:YFP H Brightfield Foxa2Cre:YFP
Supplemental Table 1: Enriched GO categories per cluster
 Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
Functions of Nervous System Neuron Structure
 Chapter 10 Nervous System I Divisions of the Nervous System Cell Types of Neural Tissue neurons neuroglial cells Central Nervous System brain spinal cord Peripheral Nervous System nerves cranial nerves
Chapter 10 Nervous System I Divisions of the Nervous System Cell Types of Neural Tissue neurons neuroglial cells Central Nervous System brain spinal cord Peripheral Nervous System nerves cranial nerves
Proteome Analysis of Human Perilymph using an Intraoperative Sampling Method
 Supporting Information Article Title Proteome Analysis of Human Perilymph using an Intraoperative Sampling Method Authors Heike A. Schmitt 1,3, Andreas Pich 2, Anke Schröder 2, Verena Scheper 1,3, Giorgio
Supporting Information Article Title Proteome Analysis of Human Perilymph using an Intraoperative Sampling Method Authors Heike A. Schmitt 1,3, Andreas Pich 2, Anke Schröder 2, Verena Scheper 1,3, Giorgio
A blood based 12-miRNA signature of Alzheimer disease patients
 RESEARCH Open Access A blood based 12-miRNA signature of Alzheimer disease patients Petra Leidinger 1, Christina Backes 1, Stephanie Deutscher 1, Katja Schmitt 1, Sabine C Mueller 1, Karen Frese 2, Jan
RESEARCH Open Access A blood based 12-miRNA signature of Alzheimer disease patients Petra Leidinger 1, Christina Backes 1, Stephanie Deutscher 1, Katja Schmitt 1, Sabine C Mueller 1, Karen Frese 2, Jan
Supplementary Figure S1: Pial and periventricular vascular networks and the dual GABA neuron streams of the early embryonic mouse telencephalon
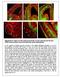 Supplementary Figure S1: Pial and periventricular vascular networks and the dual GABA neuron streams of the early embryonic mouse telencephalon a) The profile of GABA neurons arriving at the pallial-subpallial
Supplementary Figure S1: Pial and periventricular vascular networks and the dual GABA neuron streams of the early embryonic mouse telencephalon a) The profile of GABA neurons arriving at the pallial-subpallial
Identification by genetics of. expressed in the brain as potential novel
 Identification by genetics of neurotransmitter and other transporters expressed in the brain as potential novel targets for schizophrenia 13 rd November 2014, Drug Transporters symposium, target of avoid?
Identification by genetics of neurotransmitter and other transporters expressed in the brain as potential novel targets for schizophrenia 13 rd November 2014, Drug Transporters symposium, target of avoid?
Anatomy & Physiology Muscular System Worksheet
 Anatomy & Physiology Muscular System Worksheet 1. What are the three categories of muscle tissue? a) b) c) 2. The smallest functional unit of a muscle fiber is called a. 3. What are the four characteristics
Anatomy & Physiology Muscular System Worksheet 1. What are the three categories of muscle tissue? a) b) c) 2. The smallest functional unit of a muscle fiber is called a. 3. What are the four characteristics
Skeletal Muscle and the Molecular Basis of Contraction. Lanny Shulman, O.D., Ph.D. University of Houston College of Optometry
 Skeletal Muscle and the Molecular Basis of Contraction Lanny Shulman, O.D., Ph.D. University of Houston College of Optometry Like neurons, all muscle cells can be excited chemically, electrically, and
Skeletal Muscle and the Molecular Basis of Contraction Lanny Shulman, O.D., Ph.D. University of Houston College of Optometry Like neurons, all muscle cells can be excited chemically, electrically, and
HTAD PATIENT PATHWAY
 HTAD PATIENT PATHWAY Strategy for Diagnosis and Initial Management of patients and families with (suspected) Heritable Thoracic Aortic Disease (HTAD) DISCLAIMER This document is an opinion statement reflecting
HTAD PATIENT PATHWAY Strategy for Diagnosis and Initial Management of patients and families with (suspected) Heritable Thoracic Aortic Disease (HTAD) DISCLAIMER This document is an opinion statement reflecting
Annotation SNPEff_ Annotation SNPEff_. SNPEff_. Annotation. SNPEff_. Annotation. SNPEff_. Annotation. SNPEff_. Annotation
 Chr Gene p-value Binned_ variants Phenotype Biological_Feature Note 11 DRD2 0.001179727 35 Asthma Gene 16 CES2 0.010970381 40 Asthma Gene 19 LDLR 0.023074213 43 Asthma Gene 10 ARID5B 0.025870289 44 Asthma
Chr Gene p-value Binned_ variants Phenotype Biological_Feature Note 11 DRD2 0.001179727 35 Asthma Gene 16 CES2 0.010970381 40 Asthma Gene 19 LDLR 0.023074213 43 Asthma Gene 10 ARID5B 0.025870289 44 Asthma
TEB. Id4 p63 DAPI Merge. Id4 CK8 DAPI Merge
 a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
Expression levels of b-pix mrna in ALS-MSCs and C-MSCs.
 Supplementary Data SUPPLEMENTARY FIG. S1. The neurological functioning of ischemic rats depends on the origin of the transplanted MSCs. The scores of healthy rats in tests on forelimb placing, forelimb
Supplementary Data SUPPLEMENTARY FIG. S1. The neurological functioning of ischemic rats depends on the origin of the transplanted MSCs. The scores of healthy rats in tests on forelimb placing, forelimb
Sample Metrics. Allele Frequency (%) Read Depth Ploidy. Gene CDS Effect Protein Effect. LN Metastasis Tumor Purity Computational Pathology 80% 60%
 Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
ACTIVATION AND EFFECTOR FUNCTIONS OF CELL-MEDIATED IMMUNITY AND NK CELLS. Choompone Sakonwasun, MD (Hons), FRCPT
 ACTIVATION AND EFFECTOR FUNCTIONS OF CELL-MEDIATED IMMUNITY AND NK CELLS Choompone Sakonwasun, MD (Hons), FRCPT Types of Adaptive Immunity Types of T Cell-mediated Immune Reactions CTLs = cytotoxic T lymphocytes
ACTIVATION AND EFFECTOR FUNCTIONS OF CELL-MEDIATED IMMUNITY AND NK CELLS Choompone Sakonwasun, MD (Hons), FRCPT Types of Adaptive Immunity Types of T Cell-mediated Immune Reactions CTLs = cytotoxic T lymphocytes
The Human Body: An Orientation
 1 The Human Body: An Orientation Yong Jeong, MD, PhD Department of Bio and Brain Engineering The Human Body An Orientation Anatomy Study of the structure and shape of the body and its parts Physiology
1 The Human Body: An Orientation Yong Jeong, MD, PhD Department of Bio and Brain Engineering The Human Body An Orientation Anatomy Study of the structure and shape of the body and its parts Physiology
