An Atat1/Mec- 17- Myosin II Axis Controls Primary Ciliogenesis. Yanhua Rao. Department of Pharmacology and Cancer Biology Duke University
|
|
|
- Cleopatra Barker
- 6 years ago
- Views:
Transcription
1 An Atat1/Mec- 17- Myosin II Axis Controls Primary Ciliogenesis by Yanhua Rao Department of Pharmacology and Cancer Biology Duke University Date: Approved: Tso- Pang Yao, Supervisor Xiao- fan Wang Fan Wang Jeffrey Rathmell Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Pharmacology and Cancer Biology in the Graduate School of Duke University 2013
2 ABSTRACT An Atat1/Mec- 17- Myosin II Axis Controls Primary Ciliogenesis by Yanhua Rao Department of Pharmacology and Cancer Biology Duke University Date: Approved: Tso- Pang Yao, Supervisor Xiao- fan Wang Fan Wang Jeffrey Rathmell An abstract of a dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Pharmacology and Cancer Biology in the Graduate School of Duke University 2013
3 Copyright by Yanhua Rao 2013
4 An Abstract of a Dissertation Primary cilia are evolutionarily conserved, acetylated microtubule- based organelles that transduce mechanical and chemical signals. Primary cilium assembly is tightly controlled and its deregulation causes a spectrum of human diseases. Formation of primary cilium is a collaborative effort of multiple cellular machineries, including microtubule, actin network and membrane trafficking. How cells coordinate these components to construct the primary cilia remains unclear. In this dissertation research, we utilized a combination of cell biology, biochemistry and light microscopy technologies to tackle the enigma of primary cilia formation, with particular focus on isoform- specific roles of non- muscle myosin II family members. We found that myosin IIB (Myh10) is required for cilium formation. In contrast, myosin IIA (Myh9) suppresses cilium formation. In Myh10 deficient cells, Myh9 inactivation significantly restores cilia formation. Myh10 antagonizes Myh9 and increases actin dynamics, permitting pericentrosomal preciliary complex formation required for cilium assembly. Importantly, Myh10 is upregulated upon serum starvation- induced ciliogenesis and this induction requires Atat1/Mec- 17, the microtubule acetyltransferase. Our findings suggest that Atat1/Mec17- mediated microtubule acetylation is coupled to Myh10 induction, whose accumulation overcomes the Myh9- dependent actin cytoskeleton, iv
5 thereby activating cilium formation. Thus, Atat1/Mec17 and myosin II coordinate microtubules and the actin cytoskeleton to control primary cilium biogenesis. v
6 Contents An Abstract of a Dissertation... iv List of Tables... ix List of Figures... x Acknowledgements... xii Chapter 1. Introduction Structure and classification of the Cilia Primary cilia- related human diseases Primary cilium: a hub for signal transduction Primary cilia formation: a multiple- step process Basal body and Pericentrosomal Structures Actin cytoskeleton and primary cilia formation Non- muscle myosin II Microtubule Acetylation and Deacetylation HDAC6 and Atat1/Mec- 17 in cilia dynamics Rationales and aims Chapter 2. Materials and Methods Cell lines, constructs, sirnas, antibodies and chemical reagents Cell culture, transfection and cilia formation assay Immunofluorescence and F- actin staining Fluorescence Recovery After Photobleaching vi
7 2.5 Realtime PCR analysis of gene expression Data Processing and Statistical Analysis Chapter 3. A myosin II switch for primary cilia formation Myh10 is required for cilia formation Myh10 is required primary cilia- dependent sonic hedgehog signaling Myh9 and Myh10 exert opposite effects on cilia formation Myh10 promotes cilia formation by antagonizing Myh Myh10 is required to maintain actin network dynamics during cilia formation Myh10 is required to maintain branched actin dynamics to promote formation of pericentrosomal recycling endosome clusters Myh10- dependent actin dynamics regulates pericentrosomal PCM- 1/Cep290 localization Actin dynamics controls PCM- 1/Cep290 satellites organization Pericentrosomal Golgi apparatus is also regulated by actin dynamics Chapter 4. Atat1/Mec- 17 coordinates microtubule acetylation and actin dynamics during ciliogenesis Myh10 is upregulated during cilia formation Atat1/Mec- 17 controls Myh10 expression Atat1/Mec- 17 regulates myosin II switch to control the kinetics of ciliogenesis Tubulin acetylation activates Myh10 transcription Chapter 5. Conclusion, discussion and future directions Conclusions Discussions vii
8 5.2.1 Type II myosin motors are key regulators of cilium formation How do Myosin IIB promote actin dynamics? Actin cytoskeleton is the central regulator of pericentrosomal structures that sustain cilium growth The tubulin acetyltransferase Atat1/Mec- 17 coordinates actin dynamics and microtubule acetylation for proper cilium assembly Acetylated microtubules as possible intracellular signal amplifiers The mysterious microtubule lumen Acetylation and microtubule breathing The meaning of cilia to the cell Structural and functional heterogeneity of cilia Future directions Biography viii
9 List of Tables Table 1: Ciliopathies, causitive mutations and affected organs... 4 Table 2: sirna sequences used in this study Table 3 Realtime PCR Primers Used in This Study ix
10 List of Figures Figure 1: A primary cilium model Figure 2: Schematic presentation of primary cilia (a) and multiple cilia (b) Figure 3: Disease mutations targeting different functional complexes of cilia Figure 4: Primary cilium- dependent Sonic hedgehog transduction Figure 5: Effective knockdown of Myh9 and Myh10 in RPE cells 24 Figure 6: Myh9 and Myh10 play different roles in primary cilia formation Figure 7: Myh10 is also required for primary cilia formation in IMCD3 cells Figure 8: Reintroducing human Myh10 restored primary cilia formation in Myh10 knockdown IMCD3 cells Figure 11: Myh10 overexpression promotes primary cilia elongation Figure 12: Knocking down Myh9 restored ciliogenesis in Myh10 knockdown cells Figure 13: Blebbistatin treatment restored primary ciliogenesis in Myh10 knockdown cells Figure 14: Myh9 knockdown promotes spontaneous primary cilia formation Figure 15: Myh10 knockdown does not disrupt stress fiber formation Figure 17: Myh10 knockdown blocked fluorescence recovery of actin- GFP Figure 18: Myh10 is required to maintain actin network dynamics Figure 19: Myh10 knockdown enhanced branched actin network stabilization Figure 20: Myh10 knockdown reduced pericentrosomal recycling endosome cluster formation Figure 21: Myh10 knockdown impaired pericentrosomal Cep290 organization x
11 Figure 22: Myh10 knockdown does not affect centrosome- associated Cep Figure 23: Myh9 knockdown does not disrupt pericentrosomal Cep290 organization Figure 24: Pericentrosomal PCM- 1 organization is also dependent on Myh Figure 25: Myh10 knockdown causes PCM- 1 dispersion from pericentrosomal region.. 39 Figure 26: Myh10 knockdown does not affect Cep290 and PCM- 1 protein level Figure 27: Actin depolymerization reagents restored PCM- 1 pericentrosomal organization in Myh10 knockdown cells Figure 28: Golgi apparatus aggregation during cilia formation Figure 29: Myh9 and Myh10 affects Golgi apparatus morphology Figure 30: Actin depolymerization induces Golgi aggregation Figure 31: Myh10 protein expression is induced during primary cilia formation Figure 32: Atat1/Mec- 17 knockdown dramatically reduced Myh10 protein level Figure 33: Reduced expression of Myh10 in Atat1/Mec- 17 knockdown cells is not due to protein degradation Figure 34: Atat1/Mec- 17 controls Myh10 transcription Figure 35: Atat1/Mec- 17 is also induced during ciliogenesis Figure 36: Atat1/Mec- 17 knockdown induced PCM- 1 dispersion from pericentrosomal region Figure 37: Blebbistatin treatment restored ciliogenesis kinetics defects in Atat1/Mec- 17 knockdown cells Figure 38: Atat1/Mec- 17 controls Myh9- Myh10 switch to regulate ciliogenesis Figure 39: Atat1/Mec- 17 depends on its tubulin acetyltransferase activity to regulate Myh10 expression xi
12 Acknowledgements This work was completed under the supervision of my advisor and mentor Tso- Pang Yao, who provided me guidance and support throughout my time in his lab. This work could not have been possible without his vision and encouragement. His dedication for science set a unique example for me. I would like to express my sincere gratitude to my thesis committee (Xiao- fan Wang, Fan Wang, Jeffrey Rathmell) for their critical insights and constructive suggestions throughout. This piece of work could never be as good (at least not as bad) if not with their help and support. I also thank all the members in the Ehlers lab for their help and support during my time stay there and when Mike left. The friendship with them is an invaluable treasure for me. I also thank the Yao lab members, past and present, for their friendship, advice, assistance and for creating such a stimulating and cooperative work environment. My sincere gratitude goes to Rui Hao, Bin Wang, Meghan Woods, Christi Norris, Chun- Shiang Lai and Moon- Chang Choi. Finally, I thank my family for their devotion and support throughout graduate school. I am deeply grateful to my wife Hui for her unconditional love and embracement. Her gentle smile and graceful nature, even in the darkest time, will never fail to lighten my heart. xii
13 Chapter 1. Introduction Cilia are microtubule- based structures of the cell. The majority of cells in the human body have cilia. However, the function of cilia has long been ignored until the discovery of a cohort of cilia- related human diseases a decade ago(badano et al., 2006). Human genetic studies have identified a growing list of disease mutations tied to cilia dysfunctions. Cilia are now believed to be a critical structure for both developmental and homeostatic regulation. Recent functional studies have pointed to a pleiotropic role of cilia in regulating cellular functions, including mediating signal transduction of major signaling pathways(goetz and Anderson, 2010), cell cycle control(pan et al., 2012), cell polarization(vladar et al., 2012), energy homeostasis(davenport et al., 2007) and neuronal development(higginbotham et al., 2012). 1.1 Structure and classification of the Cilia Cilia are microtubule- based structures present in almost all vertebrate cells. It consists of a microtubule- based axoneme projecting from the basal body, which was transformed from one of the two centrioles. The microtubule axoneme is surrounded by the ciliary membrane which was the extension of, yet distinct from the plasma membrane in both lipid and protein composition. The ciliary and plasma membrane was physically segregated by a specialized structure called the transition zone, which forms a diffusion barrier to allow specific cargos to enter the cilium(badano et al., 2006; Gerdes et al., 2009) (Fig. 1). 1
14 Figure 1: Transmission electron microscopy imaging of a longitudinal sectioned flagellum. 1 Once passed the transition zone and entered the cilium, cargo proteins were mobilized bidirectional by specific motor proteins within the cilium. This transport process was called the intraflagellar transport (IFT) (Cole et al., 1998; Wang et al., 2006). Based on the organization of their axoneme microtubules, cilia can be classified as 9+0 and 9+2, depending on whether they have a pair of central microtubules (Gerdes et al., 2009). While most vertebrate cells develop single cilium, some other cells may grow multiple cilia (Fig. 2). 1 Modified from Allen, R.D., and P. Baumann Structure and arrangement of flagella in species of the genus Beneckea and Photobacterium fischeri. Journal of bacteriology. 107:
15 Figure 2: Scanning electron microscopy of primary cilium (a) and multiple cilia (b). 2 Multi- ciliated cells are often found in ventricles of the body, such as the ependymal cells lining the subventricular wall and the airway cells in the lung. Multiple cilia often beat rhythmically to generate force to drive the circulation of fluid within the cavity and they are often referred to as motile cilia for this reason. In contrast, single cilium is also called primary cilium or non- motile cilium. Primary cilia transduce extracellular signals to the interior of cells via specific receptors on the ciliary membrane, to regulate various cellular processes including proliferation, differentiation, polarity, morphogenesis and tissue patterning (Goetz and Anderson, 2010). They are also important mediators of sensory stimuli, including chemosensation, photosensation, thermosensation, olfaction and mechanosensation (Pazour and Witman, 2003). Motile cilia and primary cilia are functionally quite distinct but similar in in structure. Therefore, we will mainly focus our discussion on primary cilia in the following sections of this dissertation. 2 From by Charles Daghlian. 3
16 1.2 Primary cilia-related human diseases Although the wide presence of primary cilia has been known for almost a century, its biological function remained elusive until the discovery of a cohort of primary cilia- related human disorders, including polycystic kidney disease, Bardet- Biedl syndrome (Katsanis et al., 2001), Joubert s syndrome (Louie and Gleeson, 2005), Meckel s syndrome and Nephronophthisis. These diseases, which later found to result from different gene mutations, shared strikingly similar disease manifestations involving multiple organ disorders (Table 1). Table 1: Ciliopathies, causitive mutations and affected organs Diseases Mutated Genes Affected Organs Alstrom syndrome ALMS1 Brain Bardet- Biedl syndrome BBS1, BBS2, ARL6, BBS4, BBS5, MKKS, BBS7, TTC8, BBS9, BBS10, TRIM32, BBS12 Joubert syndrome INPP5E, TMEM216, AHI1, NPHP1, CEP290, TMEM67, RPGRIP1L, ARL13B, CC2D2A, BRCC3 Meckel- Gruber syndrome MKS1, TMEM67, TMEM216, CEP290, RPGRIP1L, CC2D2A nephronophthisis NPHP1, INVS, NPHP3, NPHP4, IQCB1, CEP290, GLIS2, RPGRIP1L orofaciodigital syndrome 1 OFD1 Brain, liver, kidney, bone Brain liver, heart, bone kidney Senior- Loken syndrome NPHP1, NPHP4, IQCB1, CEP290, SDCCAG8 eye polycystic kidney disease PKD1, PKD2, PKHD1 kidney primary ciliary dyskinesia DNAI1, DNAH5, TXNDC3, DNAH11, DNAI2, KTU, RSPH4A, RSPH9, LRRC50 Patients often suffer from developmental as well as degenerative disorders such as cystic kidneys, retinal degeneration, anosmia, liver fibrosis, obesity, diabetes, situs 4
17 inversus, mental retardation and male infertility (Badano et al., 2006). Human genetic studies have identified multiple causative mutations for primary cilia- related diseases. Using combinatorial approaches including cell biology, biochemistry and electron microscopy, these disease mutations have been linked to different structures of the primary cilia, including IFT complex, basal body and transition zone (Fig 3). Figure 3: Disease mutations targeting different functional complexes of cilia. 3 3 Modified from Benzing, T., and B. Schermer Transition zone proteins and cilia dynamics. Nature genetics. 43:
18 Due to their genetic nature, there is currently no effective treatment for primary cilia- related diseases. Nevertheless, identification of these disease mutations has provided a powerful diagnostic tool to allow early detection of the diseases. 1.3 Primary cilium: a hub for signal transduction Studies of sonic hedgehog (Shh) signaling have lead to the discovery that several key components of sonic hedgehog signaling pathway, including Smoothened (Smo), Patched (Ptc), Sufu and Gli transcription factors, were localized to the primary cilium. Primary cilium was therefore found to be a critical mediator of sonic hedgehog signaling (Corbit et al., 2005). In various primary cilia- related diseases, sonic hedgehog signaling was disrupted which was reflected on organogenesis and body patterning defects (Quinlan et al., 2008). Under basal conditions, the sonic hedgehog receptor Smo was sequestered in the cytoplasmic vesicles and not entering the cilium. The Gli transcription activator Gli- 1 was sequestered at the tip of the primary cilium and was processed by Sufu to produce the repressive form GliR to inhibit Shh downstream gene expression. When stimulated with Shh protein, Smo translocates to enter the primary cilium to inhibit Sufu function and release Gli activators into the cytoplasm. Gli activators then enter the nuclear to activate downstream gene transcription (Fig 4). Shh signaling also activates the transcription of Gli- 1 itself by which to create a positive feedback. Gli- 1 mrna level could therefore serve as an indicator for Shh signaling (Jenkins, 2009). 6
19 Figure 4: Primary cilium- dependent Sonic hedgehog transduction. In addition to Shh, several other signaling pathways have also been found to associate with primary cilium, including PDGF (Schneider et al., 2005), Wnt (Lancaster et al., 2011) and Notch signaling (Lancaster et al., 2011). For example, PDGFα is localized to the ciliary membrane; β- catenin and presinilin- 2 are localized to the basal body; Notch- 3 is localized to the ciliary membrane. Primary cilia therefore function to compartmentalize and/or concentrate signaling components to spatially regulate transduction of these signal pathways. Disfunction of primary cilia will also disrupt this spatial arrangement and therefore cause deregulation of these signal pathways. 1.4 Primary cilia formation: a multiple-step process The construction of primary cilia is a multiple- step process and involves multiple intracellular organelles, structures and proteins. Although visualizing primary cilia 7
20 formation in vivo remains a difficult task, studies using in vitro cell culture models have revealed a step- wise process of primary cilia formation and have identified key regulators involved in each step. Most experiments are performed in primary or immortalized mouse or human cell lines. Typically, cells are grown as confluent monolayers and subjected to serum starvation to induce primary cilia formation. The earliest signals to initiating cilia formation under serum starvation still remain elusive but the downstream processes have been relatively well characterized. Upon serum starvation and cell cycle exit, one of the centriole pair translocate to the cortical plasma membrane where it was transformed to the basal body of the cilia. The basal body was then anchored to the plasma membrane by the transition fibers. The basal body provides an organization center for polarized growth of microtubules to form the axoneme of cilia. Meantime, intracellular membrane trafficking machineries, including the Golgi apparatus, recycling endosomes and motor protein complexes are also recruited to the vicinity of the basal body to deliver efficiently the membrane components and ciliary- specific proteins to support the outgrowth of the axoneme (Badano et al., 2006) (Fig 5). These structures surrounding the basal body will be referred to as pericentrosomal structures in the following discussions. 1.5 Basal body and Pericentrosomal Structures Centrosomes are organization center of the mitotic spindle during mitosis. In primary or immortalized cells, there is usually a pair of centrioles in G0 phase of cell 8
21 cycle. However, centrosome numbers could vary in cancer cells with abnormal cell cycle progression. When cells undergo mitosis, each one of the centrioles duplicates (Bornens, 2012)to form two new pairs of centrioles to keep centrosome numbers constant in daughter cells. When cells are about to form cilia, they exit cell cycle and one of the centrioles, the mother centriole translocates to the apical domain of the plasma membrane where it was anchored and becomes the basal body of the primary cilium. Microtubules polymerize at the basal body and further grow as the axoneme. The rapid outgrowth of microtubules further expedites intracellular trafficking toward the polarization region to create a local concentration of materials in the vicinity of the basal body. So far, the basal body region is known to enrich for various intracellular organelles and proteins, including the golgi apparatus, mitochondria, recycling endosomes and proteins (Bornens, 2012; Moser et al., 2010). The pericentrosomal area has attracted increasing attention srecently for its critical role in supporting cilia formation. Several recent reports have pointed to a critical role of pericentrosomal recycling endosome clusters in supporting membrane growth and protein trafficking into the cilia (Kim et al., 2008; Kim et al., 2012). For example, recycling endosome- localized Rab11 is required for Rab8, a ciliary Rab GTPase, to enter the cilia. PCM- 1 is a protein concentrated in the pericentrosomal region. Interestingly, PCM- 1 forms satellite structures around the basal body and these satellites are positive for Cep290, an important centrosomal protein required for cilia formation. PCM- 1 and 9
22 Cep290 depend on each other for their correct localization to the pericentrosomal satellites. Interestingly, although Cep290 is present on both centrosomes and pericentrosomal satellites, knocking down PCM- 1 only inhibited pericentrosomal Cep290 localization while leaving centrosome- associated PCM- 1 unaffected. Moreover, knocking down either PCM- 1 or Cep290 disrupted normal cilia formation, indicating a critical role of pericentrosomal satellites in regulating cilia formation. Organization of pericentrosomal materials is poorly understood. Evidence has shown that actin network may function to destabilize pericentrosomal recycling endosome clusters. Yet microtubules have been suggested to play a role in PCM- 1 trafficking to the pericentrosomal region (Kim et al., 2008). Although the pericentrosomal recycling endosome clusters and the PCM- 1 satellites are both concentrated to the pericentrosomal region, whether there is a common mechanism regulating their recruitment remains untested. 1.6 Actin cytoskeleton and primary cilia formation As the major building blocks of cilia, microtubule network in cilia formation has been well understood. In contrast, actin cytoskeleton is much less appreciated. In a sirna- based high- throughput screen, Kim et al. has identified several actin- destabilizing factor (gelsolin, AVIL ) as positive regulators of cilia formation and actin network stabilizing proteins (Actr3 and PARVA) as negative regulators of cilia formation (Kim et al., 2010). Their finding uncovered a restrictive role of actin network 10
23 in regulating cilia formation. A more recent report by Zhu et al. has identified microrna- 19-3p, to promote cilia formation by promoting branched actin dynamics (Cao et al., 2012). These two papers unambiguously confirmed the negative role of actin network and the necessity for cells to release actin inhibition in regulating cilia formation. It is therefore reasonable to infer that cells have to bypass the inhibitory role of actin network under cilia- inducing conditions to allow cilia formation. How cells accomplish this is unknown. However, both papers proposed that actin network inhibited ciliogenesis by inhibiting the pericentrosomal recycling endosome clusters formation. While this is a plausible explanation, additional mechanisms may also underlie the inhibitory role of actin network. 1.7 Non-muscle Myosin II Actin network is composed of filamentous actin polymers, also called F- actin. In living cells, F- actin also organizes to form different actin structures, including actin networks and actin bundles. When cells undergo morphological change or migration, actin network and actin bundles provide contractile force to facilitate morphogenesis or migration. Non- muscle myosin II family members are major force generator of the cell. They participate in various cellular processes, including formation and disassembly of major cellular actin structures, cell migration, morphogenesis and cell polarization (Even- Ram et al., 2007; Fukui et al., 1999; Haviv et al., 2008; Smutny et al., 2010; Vicente- 11
24 Manzanares et al., 2011; Wang et al., 2010; Wilson et al., 2010). The involvement of non- muscle myosin II in cilia formation has never been explored. Non- muscle myosin II family has three major family members: Myosin IIA (Myh9), Myosin IIB (Myh10) and Myosin IIC (Myh14). The former two are ubiquitously expressed and highly abundant. Myosin II family members share high sequence homology in their ATPase domain but differ significantly in their myosin tail domains, indicating the presence of isoform specific functions. Indeed, Adelstein and colleagues have proved that Myosin IIA and IIB are functionally irredundant in mice. Furthermore, swapping their ATPase domain did not change their functions to support mouse development whereas swapping their tail domains did (Wang et al., 2010). Loss of function experiments in tissue culture also suggested distinct functions of Myosin IIA and IIB in regulating F- actin network and cell migration. Knocking down Myosin IIA inhibited cellular stress fiber formation while knocking down Myosin IIB does not (Even- Ram et al., 2007; Vicente- Manzanares et al., 2011). Meanwhile, Myosin IIA inhibition promoted while Myosin IIB knockdown inhibited randomized cell migration(vicente- Manzanares et al., 2011). These two myosin isoforms also differ in their function in regulating cell- cell junctions, dendritic spine formation and cell polarization (Smutny et al., 2010; Vicente- Manzanares et al., 2011). Clearly, myosin IIA and IIB carry isoform specific roles. However, most early studies considering the function of Myosin II failed to distinguish these two isoforms. Instead, they were often 12
25 studied together under the nomenclature Myosin II. Straight et al. have developed a highly specific inhibitor, blebbistatin, for Myosin IIA and IIB, which have been widely used to study the function of Myosin II (Straight et al., 2003). However, because blebbistatin does not distinguish Myosin IIA and IIB, it is often difficult to tell apart the effects of Myosin IIA vs. IIB in a blebbistatin treatment experiment. 1.8 Microtubule Acetylation and Deacetylation Microtubule has long been known to undergo various chemical modifications, including phosphorylation, polyglutamylation and acetylation. Ciliary microtubules are heavily acetylated and polyglutamylated. Glutamylated tubulins are often present in ultrastable microtubule structures. Glutamylase C.elegans mutants are defective in cilia motility, indicating a critical role of glutamylation in proper function of ciliary microtubules(kubo et al., 2010; Suryavanshi et al., 2010). Similarly, deglutamylase mutants are defective in intra- ciliary trafficking of cilia- localized proteins (O'ʹHagan et al., 2011). Ciliary microtubules are also heavily acetylated. In fact, acetylated microtubule is the most commonly used marker for cilia. Tubulins could be acetylated on multiple sites but the most prominent one is K40 (L'ʹHernault and Rosenbaum, 1985). Acetylation on K40 of microtubules could be reversed by deacetylation reactions catalyzed by the microtubule deacetylase HDAC6, which removes the acetyl group from the K40 of microtubules (Hubbert et al., 2002). Sirt2 has also been reported to carry tubulin 13
26 deacetylase activity (North et al., 2003) but so far HDAC6 remains the major and more powerful microtubule deacetylase. HDAC6 has a preference over polymerized microtubules rather than tubulin monomers or dimers (Hubbert et al., 2002). However, very little is known about the biological functions of microtubule acetylation. Reed et al. proposed that acetylated microtubules guides kinesn- 1- dependent directional trafficking along microtubules but this theory is later challenged by others (Reed et al., 2006; Walter et al., 2012). Others also proposed that microtubule acetylation regulated microtubule stability but this theory seems to only apply in a few cases (Sudo and Baas, 2010). Interestingly, microtubule is also linked to p38/mapk signal transduction. Cheung et al. showed that in both cultured mammalian cells, destabilizing microtubules inhibited p38/mapk signaling (Cheung et al., 2004). Chalfie et al. also discovered similar microtubule- MAPK link in C. elegans using genetic approaches (Bounoutas et al., 2011). Mec- 17 was first identified in a mechanosensation mutant C.elegans. Recently, two groups reported independently that Mec- 17 and Atat1 (α- tubulin N- acetyltransferase 1), the mammalian homolog of Mec- 17, are bona fide microtubule acetyltransferases (Akella et al., 2010; Shida et al., 2010). The discovery of Mec- 17 and Atat1 brings about new hope to tackle the function of acetylated microtubules. Chalfie et al. and Goodman et al. reported a similar function of Mec- 17 in regulating microtubule length and protofilament organization (Cueva et al., 2012; Topalidou et al., 2012). By using an acetylation defective K40R tubulin mutant, Goodman and colleagues reached a 14
27 conclusion that tubulin acetylation is required to maintain the 15- protofilament structure and the lumen size of microtubules. They further showed by molecular dynamics simulation that K40 acetylation affects salt bridge formation mediated by K40 residues and thereby changed the overall conformation of microtubules. Interestingly, Goodman et al. also revealed a catalytic activity- independent function of Mec- 17. They showed that in Mec- 17 deficient mutant, microtubule lumen content was disappeared. Unexpectedly, a catalytic- deficient Mec- 17 mutant transgene can restore the lumen content, indicative of a structural role of Mec- 17 in regulating microtubule lumen structure. However, due to limitations of their approaches, they did not identify the components of the lumen materials, which left room for further research. 1.9 HDAC6 and Atat1/Mec-17 in cilia dynamics Although cilia are heavily acetylated, microtubule acetylation is not required for cilia formation. Ptk2, a kangaroo cell line devoid of microtubule acetylation, forms cilia normally (Piperno et al., 1987; Shida et al., 2010). Consistently, Atat1 knockdown cells also form cilia at normal percentage after serum starvation for 24 hours. Interestingly, cilia formation was dramatically delayed at 12 hours in Atat1 knockdown cells (Shida et al., 2010). This data suggests that Atat1, although not required for cilia formation, do play a role in regulating the kinetics of cilia formation. The underlying mechanism of this kinetics delay was not well understood. 15
28 When cells receive mitogenic stimuli, they disassemble their cilia to allow cell cycle re- entry. HDAC6 is an important mediator of this process. Upon serum stimulation, HDAC6, which was mainly localized to the cytoplasm and basal body, entered the cilia. HDAC6 entry into the cilia is a critical step of cilia resorption because inhibiting HDAC6 by either sirna knockdown or specific inhibitors blocked serum induced cilia resorption (Pugacheva et al., 2007). The downstream target of HDAC6 in this context is unclear but recent report suggested that HSP90 might mediate HDAC6 effect during cilia resorption (Prodromou et al., 2012) Rationales and aims Primary cilia underlie many developmental and homeostatic processes. Dysfunction of primary cilia causes many human diseases involving multiple organs. Therefore, a deeper knowledge about the regulation of primary cilia formation promotes our understanding of primary cilia- related human diseases and may help develop effective treatment of these diseases. Actin network has been proposed as a negative regulator of primary cilia formation. The mechanism by which cells bypass the inhibition of actin network remains poorly understood. In addition, although actin network regulates pericentrosomal recycling endosome cluster formation, whether other pericentrosomal structures are also controlled under similar mechanisms is unclear. To obtain a better understanding of this 16
29 question, I focus on non- muscle myosin II, the major actin network regulator and aim to address the following questions: 1) How does myosin II regulate actin network during cilia formation? 2) How are pericentrosomal structures regulated? 3) How do cells coordinate microtubule, actin network and membrane trafficking to promote cilia formation? 17
30 Chapter 2. Materials and Methods 2.1 Cell lines, constructs, sirnas, antibodies and chemical reagents Three cell lines were used in this study: RPE- Mchr1 GFP cells were a kind gift from Dr. Peter Jackson s laboratory at Genentech, Inc. IMCD3 cells were obtained from Dr. Nicholas Katsanis laboratory at Duke University Medical Center. ARPE- 19 cells (ATCC #CRL- 2302) were purchased from Duke University Cell Culture Facility. All cells were maintained and passaged according to provider s instructions. Myh9- GFP (Addgene plasmid 11347) and Myh10- GFP (Addgene plasmid 11348) constructs were purchased from addgene and was originally constructed by Dr. Robert S. Adelstein s laboratory. GFP- actin was obtained from Dr. Michael D. Ehlers laboratory. MSCV- Puro retroviral vector was a kind gift from Dr. Xiao- fan Wang s lab at Duke University. Human Myh10 cdna sequence was amplified by PCR reactions and cloned into a modified MSCV- puro vector to generate MSCV- Myh10- HA. Human plko.1 Atat1/Mec- 17 MISSION shrna constructs were purchased from Sigma (#1 shrna: TRCN ; #3 shrna: TRCN ). plko.1 non- target control constructs was also a gift from Dr. Xiao- fan Wang s lab at Duke University. Antibodies used in this study includes: rabbit anti- glu- tubulin (Millipore #AB3201, 1:200), mouse anti- acetylated- tubulin (sigma #T7451, 1:1000 for IF and 1:3000 for WB), mouse anti- γ- tubulin (sigma #T5326, 1:2000), rabbit anti- γ- tubulin (sigma #T3559, 1:500), mouse anti- Rab11 (BD Transduction Laboratories #610656, 1:200), rabbit anti- Cep290 (Bethyl Laboratories, Inc. #IHC , 18
31 1:300), rabbit anti- PCM- 1 (Epitomics #T2443, 1:200), mouse anti- Myh10 (Developmental Studies Hybridoma Bank #CMII 23, 1:1000 for western blot), rabbit anti- Myh9 (Santa Cruz #sc , 1:1000 for western blot). sirna duplex sequence used in this study are listed in supplemental table S1. Chemical reagents used in this study includes: Blebbistatin (Sigma #B0560), Latrunculin A (Sigma #L5163), Jasp (Santa Cruz #sc ), MG132 (Sigma #C2211), Tubastatin A (BioVision #1724-1), Nicotinamide (Sigma #N0636). Table 2: sirna sequences used in this study Genes Species Vendor Sequences MYH10 #1 Human Invitrogen UUCUGUACAAGUUUGGGUCCAAUUC MYH10 #2 Human Sigma GCAAUACAGUGGGACAGUU[dT][dT] MYH9 Human Invitrogen GAGUCUGAGCGUGCUUCCAGGAAUA 2.2 Cell culture, transfection and cilia formation assay RPE- Mchr1GFP cells were maintained in DMEM (Gibco) containing 10% FBS (Fetal Bovine Serum) in a 37 C humidified incubator with 5% CO2. ARPE- 19 and IMCD3 cells were maintained in DMEM:F12 medium (Gibco) containing 10% FBS. Cells were passaged at 80%- 90% confluence at a regular basis. To induce cilia formation, cells were allow to grow confluent on a coverslip before switching to DMEM or DMEM: F12 containing 0.2% FBS. For the time course study of ciliogenesis, confluent cell layers were 19
32 first stimulated with 20% FBS for 6 hours to disassemble spontaneously formed cilia and then switched to medium containing 0.2% FBS. Transient transfections of plasmids or sirna duplexes were performed with lipofectamine 2000 or RNAiMAX (Life Technologies) following manufacturer s guide. For stable transfection using lentivirus or retrovirus, virus was packaged with appropriate packaging plasmids (Δ8.9 and pcmv- VSVG for lentivirus; pcl- Ampho for retrovirus) in 293T cells. Viruses were collected 48 hours after transfection, filtered through 0.45µμm sterile cellulose acetate membrane filters. Viruses were added to cell culture medium and selected with puromycin (1µμg/ml) for a week. Survived cells were then collected or passaged for analysis. Multiple rounds of infection were performed in a need basis. For non- virus based stable transfection, plasmids were delivered into the cell by Lipofectamine 2000 transfection. Transfected cells were then split at 1:10 and selected with corresponding antibiotics (1µμg/ml puromycin or 0.5mg/ml G418) for 3 weeks. Individual cell colonies were picked and expanded for further analysis. To maintain all stable cell lines, culture medium contained antibiotics (1µμg/ml puromycin or 0.5mg/ml G418). 2.3 Immunofluorescence and F-actin staining For immunofluorescence, cells were fixated with ice- cold methanol (for antibodies targeting acetylated- tubulin, γ- tubulin, Cep290 and PCM- 1) or 4% paraformaldehyde in PBS (for Rab11), washed with PBS, blocked with 10% normal goat serum, then incubated with primary antibodies diluted in 10% normal goat serum 20
33 overnight in 4 C. Primary antibodies were rinsed with PBS and incubated with secondary antibody in 10% normal goat serum for 1 hour and then rinsed and mounted on glass slides for microscopy imaging. For F- actin staining, cells were fixated in pre- warmed (37 C) 4% paraformaldehyde and rinsed with PBS, then incubated with phalloidin- 555 (Cytoskeleton #PHDH1- A) for 1 hour. Coverslips were then mounted on glass slides for imaging on a Zeiss 780 microscopy. 2.4 Fluorescence Recovery After Photobleaching ARPE- 19 cells were first transfected with Actin- GFP plasmid and then with control, Myh9 and Myh10 sirna. 48 hours after sirna transfection, cells were switched to warm phenol- free DMEM for FRAP analysis. Cells were imaged for 10 seconds, then photobleached at selected regions and imaged for 3 minutes to allow fluorescence recovery. For each knockdown group, a total of five cells were used for analysis. Intensity data from 5 cells were analyzed using imagej FRAP profiler plugin, averaged and plotted. The fluorescence recovery curve fitting was performed with Prism 6 software (GraphPad Software) under the exponential association function f(t) = A(1- e - τt ) 2.5 Realtime PCR analysis of gene expression Total RNA was extracted from cells using RNAeasy mini kit (Qiagen), quantified using a spectrometer. 1 ug of total RNA from each sample was reverse transcribed using Promega M- MLV Reverse Transcriptase to be used as template for realtime PCR 21
34 analysis. cdna and primers for specific genes were mixed in realtime PCR master mix (Qiagen). Realtime PCR reaction was performed on a Realplex Mastercycler (Eppendorf). Data was analysed with Excel. PCR primers used for realtime PCR analysis are listed in table 2. Table 3 Realtime PCR Primers Used in This Study Genes Species Forward Primer Reverse Primer Gli- 1 Human GGGTGCCGGAAGTCATACTC GCTAGGATCTGTATAGCGTTTGG Myh9 Human CAGCAAGCTGCCGATAAGTAT CTTGTCGGAAGGCACCCAT Myh10 Human TGGTTTTGAGGCAGCTAGTATCA AGTCCTGAATAGTAGCGATCCTT Atat1/Mec- 17 Human GGCCCAGAATCTTTCCGCTC GATGCAAAGTGGTTCTACCTCAT Myh9 Mouse GGCCCTGCTAGATGAGGAGT CTTGGGCTTCTGGAACTTGG Myh10 Mouse GGAATCCTTTGGAAATGCGAAGA GCCCCAACAATATAGCCAGTTAC 2.6 Data Processing and Statistical Analysis All the quantifications were from three independent experiments. To quantify the percentage of ciliated cells, cells in randomly chosen light fields under the microscopy were counted. Only cells with acetylated tubulin staining longer than 1µμm were considered as bearing mature cilia and counted as positive. For cilia length analysis, cilia lengths were determined using the line scan function in Fiji imaging processing software. Lengths of cilia were plotted as cumulative probability distribution curve. Data are demonstrated as mean±sd. Two- group hypothesis testing were 22
35 performed by student t- test. Results were considered statistically significant when p<
36 Chapter 3. A myosin II switch for primary cilia formation Actin cytoskeleton has been proposed as a negative regulator of cilia formation. Cells have to release the inhibitory brake enforced by actin network to allow cilia formation. Although several important actin stabilization and destabilization factors have been found to control cilia formation, these findings did not address the question how cells bypass the actin brake during ciliogenesis. As introduced in Chapter 2, non- muscle myosin II proteins are important actin network regulators. By understanding how non- muscle myosins IIs regulate cilia formation, we should be able to gain more knowledge in how actin network was regulated during ciliogenesis. 3.1 Myh10 is required for cilia formation To investigate the role of the actin network in ciliogenesis, we focused on non- muscle myosin II motors, which are known to regulate actin network dynamics and actinomyosin- microtubule crosstalk. We adopted the serum starvation protocol to Figure 5: Effective knockdown of Myh9 and Myh10 in RPE cells. 24
37 induce cilium formation in RPE- Mchr1 GFP retina epithelial cells. To study the role of myosin IIA (Myh9) and IIB (Myh10) in cilium formation, we first designed sirna duplexes specifically targeting Myh10 or Myh9. Western blot analysis and Q- PCR confirmed the effective knockdown of Myh10 and Myh9 by sirnas (Fig 5). When RPE- Mchr1 GFP cells were subjected to serum deprivation for 24 hours to induce ciliogenesis, Myh9 sirna- treated cells were able to form cilia as efficiently as control sirna group. In stark contrast, two different Myh10 sirna duplexes both dramatically inhibited cilium formation (Fig 6). Figure 6: Myh9 and Myh10 play different roles in primary cilia formation Knocking down Myh10, but not Myh9, in mouse IMCD3 cells also potently inhibited cilium formation in IMCD3 cells (Fig 7). As shown in Fig 7, Myh10 knockdown in IMCD3 cells reduced percentage of ciliated cells to ~25%, while Myh9 knockdown does not have a dramatic effect on cilia formation, further suggesting that Myh9 and Myh10 play different roles in regulating cilia formation. And this effect is conserved between mouse and human. 25
38 Figure 7: Myh10 is also required for primary cilia formation in IMCD3 cells. Moreover, re- introducing a wild type human Myh10 in Myh10- knockdown- IMCD3 cells restored cilium formation (Fig 8). These findings show that Myh10 is required for efficient ciliogenesis. Figure 8: Reintroducing human Myh10 restored primary cilia formation in Myh10 knockdown IMCD3 cells 26
39 3.2 Myh10 is required primary cilia-dependent sonic hedgehog signaling Cilia are important for sonic hedgehog signaling. To confirm that Myh10 is required for cilia- mediated signal transduction, we stimulated cells with SAG, a small Figure 9: Myh10 is required for cilia- mediated sonic hedgehog signaling. molecule agonist of Smoothened, to induce sonic hedgehog signaling in serum- starved cells. We measured Gli- 1 mrna induction, a downstream target of hedgehog signaling, by RT- PCR. As shown in Fig 9, Myh10 (red bar) but not Myh9 (green bar) knockdown strongly inhibited Gli- 1 induction upon SAG stimulation, indicating that Myh10 is required for cilium- mediated sonic hedgehog signal transduction. 3.3 Myh9 and Myh10 exert opposite effects on cilia formation To further ascertain the role of Mhy10 in cilium formation, we determined whether an increase of Myh10 expression is sufficient to promote the growth of cilia. To this end, we stably expressed Myh10- GFP in IMCD3 cells. Myh9- GFP was included as a 27
40 control. We then labeled acetylated tubulin to examine cilium formation in these two stable cell lines. Myh10- GFP IMCD3 cells form cilia at a modestly higher percentage than control cells (Fig 10, left panel, middle row, and right panel, red bar); however, cilium length in Myh10- GFP expressing cells is significantly longer (Fig 11). Figure 10: Myh9 and Myh10 overexpression have opposite effects on cilia formation. This data indicate that increasing Myh10 expression stimulates cilia elongation. Unexpectedly, cilium formation was dramatically repressed in Myh9- GFP expressing IMCD3 cells (Fig 10, left panel, bottom row, and right panel, green bar). Thus Myh9, in contrast to Myh10, appears to be a negative regulator of cilium formation. This data, when consistent with the loss of function experiments (Fig 6-9), also consolidated existing theory that actin network rigidity is a negative regulator of cilia formation and elongation. 28
41 Figure 11: Myh10 overexpression promotes primary cilia elongation 3.4 Myh10 promotes cilia formation by antagonizing Myh9 As overexpression of Myh9 and Myh10 caused opposite phenotypes on cilia morphology, we tested if Myh10 promotes cilium formation by antagonizing Myh9 activity. To test this possibility, we knocked down both Myh9 and Myh10 by sirna in RPE- Mchr1GFP cells and assessed cilium formation. In contrast to Myh10 single knockdown, where few cilia were observed, concurrent knockdown of Myh9 restored cilium formation in more than 40% of the cells (Fig 12). Figure 12: Knocking down Myh9 restored ciliogenesis in Myh10 knockdown cells 29
42 To further support this conclusion, we treated Myh10 knockdown cells with a myosin II- specific inhibitor, Blebbistatin, to inhibit Myh9 activity in the cell. If Myh10 is indeed antagonizing Myh9 activity, blebbistatin treatment should be able to rescue cilia formation defect in Myh10 knockdown cells. As expected, blebbistatin treatment dramatically restored cilia formation in Myh10 knockdown cells (Fig 13). Figure 13: Blebbistatin treatment restored primary ciliogenesis in Myh10 knockdown cells. We also noted that the cilia formed under these conditions were generally shorter than those in wild type control cells. These data indicate that Myh9 activity is, at least partially, responsible for cilium inhibition caused by Myh10 knockdown. We next asked whether Myh9 activity might function to inhibit ciliogenesis in the presence of high serum when ciliogenesis is typically low. As shown in Fig 14, knocking down Myh9 expression indeed significantly increased ciliogenesis. 30
43 Figure 14: Myh9 knockdown promotes spontaneous primary cilia formation. These experiments indicate that Myh9 suppresses ciliogenesis under non- permissive conditions and Myh10 can counteract this activity. Importantly, the counteracting effects of Myh9 and Myh10 suggested a plausible mechanism by which Myh10 promotes ciliogenesis by inhibiting Myh9 activity. Thus, Myh9 and Myh10 formed a molecular switch to control cilia formation. 3.5 Myh10 is required to maintain actin network dynamics during cilia formation Myh9 is known to stabilize actin bundles and the filament network. We therefore asked whether Myh10 might oppose Myh9 and increase actin network dynamics. To test this possibility, we first performed Phalloidin staining in ARPE- 19 cells to examine the influence of Myh10 knockdown upon the F- actin network. As reported previously, Myh10 knockdown did not disrupt stress fiber formation (Fig 15), which is controlled by 31
44 Myh9. In fact, there appeared to be more and bigger stress fibers in Myh10 knockdown cells, which resembles Myh10 knockout phenotype (Even- Ram et al., 2007). Figure 15: Myh10 knockdown does not disrupt stress fiber formation. Myh10 knockdown, however, dramatically increased F- actin staining intensity at the cell edge and the cell cortex (Fig 16), indicative of stabilization of F- actin network. On the cell edge, Myh10 knockdown cells seem to accumulate both lamellapodia- and stress fiber- like structures. Particularly, there seems to be a redistribution of stress fiber actin bundles from other parts of the cell to the cell periphery, which could be owing to changes in contractility of actin network in the cell. Interestingly, previous reports have indicated that Myh10 could be localized to stress fibers, leading edge and cell cortex, 32
45 which is consistent with the actin cytoskeleton changes observed in Myh10 knockdown cells. Figure 16: Myh10 knockdown enhanced F- actin on the cell edge and the cortex. To quantitatively measure actin dynamics, we performed a fluorescence recovery after photobleaching (FRAP) assay using β- actin- GFP in ARPE- 19 cells. We found that in contrast to Myh9 knockdown, which accelerated the initial phase of fluorescence recovery after photobleaching, Myh10 knockdown dramatically blocked the recovery (Fig 17). We then averaged FRAP data from 5 different cells in each experimental group and fitted the curve using exponential association function f(t) = A(1- e - τt ). Results showed that Myh9 knockdown significantly increased the rate of fluorescence recovery and the 33
46 ratio of mobile actin- GFP fraction, indicative of a more flexible actin network. Figure 17: Myh10 knockdown blocked fluorescence recovery of actin- GFP. In contrast, Myh10 knockdown dramatically reduced the mobile fraction and recovery rate, suggesting that Myh10 knockdown stabilized cellular actin network (Fig 18). Figure 18: Myh10 is required to maintain actin network dynamics. 34
47 3.6 Myh10 is required to maintain branched actin dynamics to promote formation of pericentrosomal recycling endosome clusters Previous reports have suggested an critical role of branched actin network in regulating ciliogenesis (Cao et al., 2012; Kim et al., 2010), we therefore tested whether the enhanced F- actin on the cell periphery in Myh10 knockdown cells contains branched actin components. P34/Arc is a component of the arp2/3 complex of actin nucleation factor. The areas with enhanced F- actin staining in Myh10- knockdown cells were enriched for p34/arc, indicating that these actin regions contain branched actin (Fig 19). Figure 19: Myh10 knockdown enhanced branched actin network stabilization. Branched actin network has been proposed to prevent the concentration of pericentrosomal recycling endosomes that provide membranes for primary cilium growth (Cao et al., 2012; Kim et al., 2010). We therefore examined whether Myh10 regulates pericentrosomal recycling endosome cluster formation. This could serve as indirect evidence whether Myh10 actually regulates actin stability during ciliogenesis. Rab11 is a Rab small GTPase required for recycling endosome function and it is also localized to recycling endosomes. 35
48 Figure 20: Myh10 knockdown reduced pericentrosomal recycling endosome cluster formation As shown in Fig 20, Myh10 knockdown dramatically reduced the percentage of cells with Rab11- positive recycling endosomes clusters (dropped to ~5% in Myh10 knockdown cells), consistent with the function of Myh10 in maintaining actin dynamics. Taken together, Myh10, contrary to Myh9, functions to maintain a dynamic actin network and supports pericentrosomal recycling endosome cluster formation. 3.7 Myh10-dependent actin dynamics regulates pericentrosomal PCM-1/Cep290 localization The requirement of Myh10 for the assembly of pericentrosomal recycling endosome pool prompted us to examine whether Myh10- dependent actin dynamics are also necessary for clustering other pericentrosomal components required for cilium formation. We examined Cep290 and PCM- 1, which are co- localized to centriolar satellites and recruit Rab8 to facilitate the fusion of preciliary vesicles to the ciliary membrane. Pericentrosomal Cep290 staining showed a marked decrease of signal intensity in Myh10- knockdown cells (Fig 21). To measure the intensity of 36
49 pericentrosomal Cep290, we drew a 30- pixel diameter circle around γ- tubulin staining of each cell and quantified the corresponding Cep290 staining within the circle. Figure 21: Myh10 knockdown impaired pericentrosomal Cep290 organization. More detailed examination revealed a specific loss of Cep290 at the centriolar satellite (Fig 21, left panel, bottom, yellow boxes) whereas centrosome- associated Cep290 is retained (Fig 22). Figure 22: Myh10 knockdown does not affect centrosome- associated Cep
50 In contrast, Myh9 knockdown did not disrupt Cep290 concentration to centriolar satellites (Fig 23), suggesting that the effect observed in Myh10 knockdown cells is specific. Figure 23: Myh9 knockdown does not disrupt pericentrosomal Cep290 organization. Similarly, pericentrosomal PCM- 1 staining intensity was also dramatically reduced in Myh10 knockdown cells (Fig 24). Figure 24: Pericentrosomal PCM- 1 organization is also dependent on Myh10. Careful examination of subcellular PCM- 1 localization revealed that PCM- 1 satellite structures surrounding the centrosome was dispersed in Myh10 knockdown 38
51 cells (Fig 25), suggesting that Myh10 is required for proper pericentrisomal organization of Cep290/PCM- 1 complex. Figure 25: Myh10 knockdown causes PCM- 1 dispersion from pericentrosomal region. To examine whether the reduction of PCM- 1 staining intensity is a consequence of enhanced protein degradation, we evaluated the total protein level in Myh10 knockdown cells by western blot. The overall protein levels of Cep290 and PCM- 1 remained unaltered in Myh10 knockdown cells (Fig 26), supporting the conclusion that the Cep290- PCM- 1 complex is mis- localized in Myh10 deficient cells. Figure 26: Myh10 knockdown does not affect Cep290 and PCM- 1 protein level. 39
52 3.8 Actin dynamics controls PCM-1/Cep290 satellites organization Next, we tested if actin dynamics underlie Myh10- dependent pericentrosomal structure organization. We treated Myh10 knockdown RPE- 1- Mchr1 GFP line with a low dosage (20nM) of Latrunculin A (Lat), an actin depolymerization reagent, to increase actin dynamics in Myh10- knockdown cells. 24 hours after Latrunculin A treatment, pericentrosomal PCM- 1 was partially restored (Fig 27, LAT panels). Inhibiting Myh9 by the myosin II- specific inhibitor, Blebbistatin (25µμM), also restored PCM- 1 clusters in Myh10 knockdown cells (Fig 27, Blebbi panels). These data indicate that Myh10 promotes pericentrosomal concentration of the Cep290- PCM- 1 complexes by opposing Myh9 and maintaining a dynamic actin cytoskeleton. Figure 27: Actin depolymerization reagents restored PCM- 1 pericentrosomal organization in Myh10 knockdown cells 40
53 3.9 Pericentrosomal Golgi apparatus is also regulated by actin dynamics Our data clearly showed that Myh10- dependent actin dynamics underlies both pericentrosomal recycling endosome cluster formation and Cep290/PCM- 1 satellites organization. Whether this Myh10- actin axis represents a broad mechanism for other pericentrosomal structure organization becomes an interesting question to test. Golgi apparatus is another important structure for primary cilium formation and it is also localized adjacent to the basal body. We first monitored Golgi apparatus morphology change during the time course of primary cilium formation. We serum starved RPE- Mchr1 GFP cells and immunostained GM130, a cis- golgi marker, to assess the morphological change of Golgi apparatus at different time points of cilia formation. Surprisingly, we observed an obvious aggregation of golgi apparatus around the basal body (Fig 28). Figure 28: Golgi apparatus aggregation during cilia formation. 41
54 As expected, Myh9 and Myh10 knockdown displayed very different phenotypes on Golgi apparatus. Inhibiting Myh9 induced a marked golgi aggregation while Myh10 knockdown dispersed golgi (Fig 29). Figure 29: Myh9 and Myh10 affects Golgi apparatus morphology. Similarly, chemical reagents influencing actin dynamics also changed Golgi morphology. Latrunculin A, the actin depolymerization reagent, promotes Golgi aggregation while jaspkinolide, an actin stabilizer, induces Golgi dispersion (Fig 30). Figure 30: Actin depolymerization induces Golgi aggregation. This evidence suggests that Golgi apparatus, yet another pericentrosomal structure, is also regulated by actin dynamics during cilia formation. This data, together 42
55 with pericentrosomal recycling endosomes and Cep290- PCM- 1 satellites, revealed an interesting regulation of pericentrosomal structure organization by actin dynamics. 43
56 Chapter 4. Atat1/Mec-17 coordinates microtubule acetylation and actin dynamics during ciliogenesis Primary cilia formation involves actin remodeling, microtubule reorganization and membrane trafficking. Obviously, these cellular processes must be tightly coordinated to construct such an exquisite organelle as the primary cilium. However, how these cellular processes are coordinated remains an enigma. Non- muscle myosin II provides such an opportunity since they have been suggested to regulate actin- microtubule crosstalk. To study this question, we started by understanding how myosin II genes are regulated during cilia formation. 4.1 Myh10 is upregulated during cilia formation The fact that Myh10 overexpression promoted cilium elongation prompted us to examine whether Myh10 expression is regulated during cilium formation. We monitored the expression of Myh10 during cilium formation induced by serum starvation. Interestingly, Myh10 protein levels gradually increased during the early phase (0-12h after starvation) of cilium formation (Fig 31). In contrast, Myh9 protein levels remained relatively constant. Real- time PCR analysis showed that Myh10 mrna level increased within 2 hours of serum starvation whereas Myh9 mrna level remained relatively unchanged. This data reveal that relative abundance of Myh10 vs. Mhy9 increases in response to serum starvation, resulting in conditions favorable for primary cilia formation. Furthermore, the upregulation of Myh10 is at least partially attributable 44
57 to transcriptional level of regulation. But how Myh10 is regulated on the transcription level is unknown. Figure 31: Myh10 protein expression is induced during primary cilia formation. 4.2 Atat1/Mec-17 controls Myh10 expression Next, we sought to explore the mechanism by which Myh10 is induced during the induction of cilium formation. As cilia are heavily enriched for acetylated microtubules, we determined whether microtubule acetyltransferase Atat1/Mec- 17 affects Myh10 expression. We transduced RPE- Mchr1 GFP cells with several independent Atat1/Mec- 17 shrna lentiviral particles. Two (#1 and #3) out of 5 independent shrna constructs effectively reduced acetylated tubulin levels (Fig 32a) and Atat1/Mec
58 mrna (Fig 32b). In both Atat1/Mec- 17 shrna knockdown cell lines, Myh10 protein level was dramatically reduced (Fig 32a) whereas Myh9 remained relatively unchanged. level. Figure 32: Atat1/Mec- 17 knockdown dramatically reduced Myh10 protein The decrease in Myh10 protein abundance was not due to protein degradation as inhibiting proteasome function by MG132 did not restore Myh10 level (Fig 33). Figure 33: Reduced expression of Myh10 in Atat1/Mec- 17 knockdown cells is not due to protein degradation 46
59 Quantitative PCR detected a significant reduction of Myh10 mrna (~60-70%) in Atat1/Mec- 17- knockdown cells (Fig 34), suggesting that Atat1/Mec- 17 regulates Myh10 protein expression via a transcriptional mechanism. Please note that Myh9 mrna level was also decreased in Atat1/Mec- 17 knockdown cells, but this decrease was much less than Myh10. Importantly, this decrease in Myh9 mrna does not seem to affect the protein level of Myh9, an effect probably owing to compensation in protein stability or posttranscriptional regulation. Figure 34: Atat1/Mec- 17 controls Myh10 transcription. Together, Atat1/Mec- 17 seems to regulate Myh10 via a transcription- dependent mechanism. We know that acetylated tubulin level increases during ciliogenesis, which could be due to inhibition of HDAC6 activity or upregulation of Atat1/Mec- 17 activity. We address this question, we performed a Q- PCR to measure Atat1/Mec- 17 mrna at different time points of ciliogenesis. Interestingly, Atat1/Mec- 17 mrna level is also 47
60 upregulated by serum starvation and this pattern is strikingly similar to Myh10 mrna change during ciliogenesis(fig 35). Figure 35: Atat1/Mec- 17 is also induced during ciliogenesis. Collectively, these results suggest that Atat1/Mec- 17 is required for Myh10 upregulation in response to serum starvation. Furthermore, Atat1/Mec- 17 knockdown also caused PCM- 1 dispersion as observed in Myh10 knockdown (Fig 36). Figure 36: Atat1/Mec- 17 knockdown induced PCM- 1 dispersion from pericentrosomal region. 48
61 4.3 Atat1/Mec-17 regulates myosin II switch to control the kinetics of ciliogenesis If Atat1/Mec- 17 controls Myh10 expression, one would expect a ciliogenesis defect in Atat1/Mec- 17 knockdown cells. Consistent with the previous report, we found that ciliogenesis was significantly reduced in the early phase (0-12 hours after serum withdrawal) (Fig 37, 12 hours). Similar to Myh10 knockdown cells, this deficiency in cilium formation can be reversed by Blebbistatin (Fig 37, red at 12 hours). However, cilium formation in Atat1/Mec- 17 knockdown cells eventually recovered (Fig 37, green). Figure 37: Blebbistatin treatment restored ciliogenesis kinetics defects in Atat1/Mec- 17 knockdown cells. Protein analysis revealed that Myh9 protein level decreased in Atat1/Mec- 17- knockdown cells in the late (12-24 hours), but not early (0-12 hours), phase of the ciliogenesis (Fig 38). Given the inhibitory activity of Myh9 on cilium formation, the loss of Myh9 could explain the eventual recovery of cilium formation in Atat1/Mec- 17 knockdown cells, although the mechanism is unclear. Our data reveal that activation of ciliogenesis is preceded by Atat1/Mec- 17- dependent Myh10 induction, suggesting 49
62 transcriptional upregulation of Atat1/Mec- 17 and Myh10 are key molecular events leading to starvation- induced cilium formation. Figure 38: Atat1/Mec- 17 controls Myh9- Myh10 switch to regulate ciliogenesis. 4.4 Tubulin acetylation activates Myh10 transcription To further explore the mechanism by which Atat1/Mec- 17 regulates Myh10 transcription, we focused on the dependence of catalytic activity of Atat1/Mec- 17. The only substrate so far found for Atat1/Mec- 17 is tubulin. Chalfie and colleagues have discovered an interesting link between microtubule and gene transcription (Bounoutas et al., 2011). It is therefore of great interest to test whether tubulin acetylation also regulates gene transcription, Myh10 transcription in this case in particular. To this end, I generated a catalytic- dead mutant of mouse Atat1/Mec- 17, which is incapable of acetylating microtubules. I expressed this mutant Atat1/Mec- 17 as well as the wild- type mouse Atat1/Mec- 17 in Atat1/Mec- 17 knockdown ARPE- 19 cell lines to rescue the knockdown of Atat1/Mec- 17. Western blot showed good expression of HA- tagged Atat1/Mec- 17 and its mutant in knockdown cells, indicating successful rescue of gene expression. We the blotted acetylated- tubulin and showed that exogenously expressed 50
63 Atat1/Mec- 17 is functional in acetylating microtubules. Interestingly, Myh10 protein expression could only be restored by wild- type Atat1/Mec- 17 but not catalytic- dead form of Atat1/Mec- 17 (Fig 39). Figure 39: Atat1/Mec- 17 depends on its tubulin acetyltransferase activity to regulate Myh10 expression. 51
Supplemental Information. Supernumerary Centrosomes. Nucleate Extra Cilia and Compromise. Primary Cilium Signaling. Current Biology, Volume 22
 Current Biology, Volume 22 Supplemental Information Supernumerary Centrosomes Nucleate Extra Cilia and Compromise Primary Cilium Signaling Moe R. Mahjoub and Tim Stearns Supplemental Inventory 1. Supplemental
Current Biology, Volume 22 Supplemental Information Supernumerary Centrosomes Nucleate Extra Cilia and Compromise Primary Cilium Signaling Moe R. Mahjoub and Tim Stearns Supplemental Inventory 1. Supplemental
CilioPathy panel. 3-Jul-2018 (102 genen) Centrum voor Medische Genetica Gent. versie. OMIM gene ID
 versie 3-Jul-2018 (102 genen) CilioPathy panel Centrum voor Medische Genetica Gent Gene OMIM gene ID Associated phenotype, OMIM phenotype ID, phenotype mapping key and inheritance pattern AHI1 608894 Joubert
versie 3-Jul-2018 (102 genen) CilioPathy panel Centrum voor Medische Genetica Gent Gene OMIM gene ID Associated phenotype, OMIM phenotype ID, phenotype mapping key and inheritance pattern AHI1 608894 Joubert
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
Utilization of the MiSeq in a clinical lab. Tony Krentz, PhD PreventionGenetics
 Utilization of the MiSeq in a clinical lab Tony Krentz, PhD PreventionGenetics PreventionGenetics Founded in 2004 in Marshfield, Wisconsin by James Weber ~90 employees Largest test menu in US Vision: Disease
Utilization of the MiSeq in a clinical lab Tony Krentz, PhD PreventionGenetics PreventionGenetics Founded in 2004 in Marshfield, Wisconsin by James Weber ~90 employees Largest test menu in US Vision: Disease
CMB621: Cytoskeleton. Also known as How the cell plays with LEGOs to ensure order, not chaos, is temporally and spatially achieved
 CMB621: Cytoskeleton Also known as How the cell plays with LEGOs to ensure order, not chaos, is temporally and spatially achieved Lecture(s) Overview Lecture 1: What is the cytoskeleton? Membrane interaction
CMB621: Cytoskeleton Also known as How the cell plays with LEGOs to ensure order, not chaos, is temporally and spatially achieved Lecture(s) Overview Lecture 1: What is the cytoskeleton? Membrane interaction
Centrosomes and Cilia
 Centrosomes and Cilia SMC6052/BIM6028/EXMD604 William Y. Tsang Research Unit Director, Cell Division and Centrosome Biology, IRCM Chercheur Adjoint, Faculté de medécine, Université de Montréal Adjunct
Centrosomes and Cilia SMC6052/BIM6028/EXMD604 William Y. Tsang Research Unit Director, Cell Division and Centrosome Biology, IRCM Chercheur Adjoint, Faculté de medécine, Université de Montréal Adjunct
Fixation methods can differentially affect ciliary protein immunolabeling
 DOI 10.1186/s13630-017-0045-9 Cilia METHODOLOGY Open Access Fixation methods can differentially affect ciliary protein immunolabeling Kiet Hua 1 and Russell J. Ferland 1,2* Abstract Background: Primary
DOI 10.1186/s13630-017-0045-9 Cilia METHODOLOGY Open Access Fixation methods can differentially affect ciliary protein immunolabeling Kiet Hua 1 and Russell J. Ferland 1,2* Abstract Background: Primary
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous
 Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Centrosome & Centrioles,It s Structure,Function!!
 Centrosome & Centrioles,It s Structure,Function!! CENTROSOME In cell biology, the centrosome (Latin centrum center and Greek soma body ) is an organelle that serves as the main microtubule organizing center
Centrosome & Centrioles,It s Structure,Function!! CENTROSOME In cell biology, the centrosome (Latin centrum center and Greek soma body ) is an organelle that serves as the main microtubule organizing center
HOMEWORK RUBRICS MECHANOTRANSDUCTION UNIT: HOMEWORK #1 (20 pts towards your grade)
 HOMEWORK RUBRICS MECHANOTRANSDUCTION UNIT: HOMEWORK #1 (20 pts towards your grade) 1. Mesenchymal stem cells (MSC) cultured on extracellular matrices with different stiffness exhibit diverse lineage commitment
HOMEWORK RUBRICS MECHANOTRANSDUCTION UNIT: HOMEWORK #1 (20 pts towards your grade) 1. Mesenchymal stem cells (MSC) cultured on extracellular matrices with different stiffness exhibit diverse lineage commitment
cell movement and neuronal migration
 cell movement and neuronal migration Paul Letourneau letou001@umn.edu Chapter 16; The Cytoskeleton; Molecular Biology of the Cell, Alberts et al. 1 Cell migration Cell migration in 3 steps; protrusion,
cell movement and neuronal migration Paul Letourneau letou001@umn.edu Chapter 16; The Cytoskeleton; Molecular Biology of the Cell, Alberts et al. 1 Cell migration Cell migration in 3 steps; protrusion,
04_polarity. The formation of synaptic vesicles
 Brefeldin prevents assembly of the coats required for budding Nocodazole disrupts microtubules Constitutive: coatomer-coated Selected: clathrin-coated The formation of synaptic vesicles Nerve cells (and
Brefeldin prevents assembly of the coats required for budding Nocodazole disrupts microtubules Constitutive: coatomer-coated Selected: clathrin-coated The formation of synaptic vesicles Nerve cells (and
* * A3027. A4623 e A3507 A3507 A3507
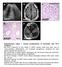 a c L A327 d e A37 A37 A37 Supplementary Figure 1. Clinical manifestations of individuals with mutations. (a) Renal ultrasound of right kidney in A327 reveals small renal cysts, loss of corticomedullary
a c L A327 d e A37 A37 A37 Supplementary Figure 1. Clinical manifestations of individuals with mutations. (a) Renal ultrasound of right kidney in A327 reveals small renal cysts, loss of corticomedullary
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Nature Medicine doi: /nm.2860
 Supplemental Figure Legends Supplemental Figure 1: Hypomorphic expression of IFT88 results in olfactory signaling proteins no longer localizing to the ciliary layer. (a) ACIII localizes to the cilia and
Supplemental Figure Legends Supplemental Figure 1: Hypomorphic expression of IFT88 results in olfactory signaling proteins no longer localizing to the ciliary layer. (a) ACIII localizes to the cilia and
Molecular Cell Biology - Problem Drill 20: Cytoskeleton and Cellular Mobility
 Molecular Cell Biology - Problem Drill 20: Cytoskeleton and Cellular Mobility Question No. 1 of 10 1. Which of the following statements about cytoskeletal filaments is correct? Question #1 (A) The Cytoskeleton
Molecular Cell Biology - Problem Drill 20: Cytoskeleton and Cellular Mobility Question No. 1 of 10 1. Which of the following statements about cytoskeletal filaments is correct? Question #1 (A) The Cytoskeleton
A Role for Intraflagellar Transport Proteins in Mitosis: A Dissertation
 University of Massachusetts Medical School escholarship@umms GSBS Dissertations and Theses Graduate School of Biomedical Sciences 6-18-2013 A Role for Intraflagellar Transport Proteins in Mitosis: A Dissertation
University of Massachusetts Medical School escholarship@umms GSBS Dissertations and Theses Graduate School of Biomedical Sciences 6-18-2013 A Role for Intraflagellar Transport Proteins in Mitosis: A Dissertation
Cell Cycle, Mitosis, and Microtubules. LS1A Final Exam Review Friday 1/12/07. Processes occurring during cell cycle
 Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Supporting Information
 Supporting Information Kim et al. 10.1073/pnas.0912180106 SI Materials and Methods DNA Constructs. The N-terminally-tagged mouse Gli2 expression construct, pcefl/3 HA-Gli2, was constructed by ligating
Supporting Information Kim et al. 10.1073/pnas.0912180106 SI Materials and Methods DNA Constructs. The N-terminally-tagged mouse Gli2 expression construct, pcefl/3 HA-Gli2, was constructed by ligating
Microtubule Forces Kevin Slep
 Microtubule Forces Kevin Slep Microtubules are a Dynamic Scaffold Microtubules in red, XMA215 family MT polymerase protein in green Some Microtubule Functions Cell Structure Polarized Motor Track (kinesins
Microtubule Forces Kevin Slep Microtubules are a Dynamic Scaffold Microtubules in red, XMA215 family MT polymerase protein in green Some Microtubule Functions Cell Structure Polarized Motor Track (kinesins
Structures in Cells. Lecture 5, EH1008: Biology for Public Health, Biomolecules.
 Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Tubulointerstitial Renal Disease. Anna Vinnikova, MD Division of Nephrology
 Tubulointerstitial Renal Disease Anna Vinnikova, MD Division of Nephrology Part I: Cystic Renal Disease www.pathguy.com Simple cysts Simple cysts May be multiple Usually 1 5cm, may be bigger Translucent,
Tubulointerstitial Renal Disease Anna Vinnikova, MD Division of Nephrology Part I: Cystic Renal Disease www.pathguy.com Simple cysts Simple cysts May be multiple Usually 1 5cm, may be bigger Translucent,
Chapter 2. Investigation into mir-346 Regulation of the nachr α5 Subunit
 15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Structures in Cells. Cytoplasm. Lecture 5, EH1008: Biology for Public Health, Biomolecules
 Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question.
 Exam Name MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) All of the following are synthesized along various sites of the endoplasmic reticulum
Exam Name MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) All of the following are synthesized along various sites of the endoplasmic reticulum
m 6 A mrna methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
APGRU4L1 Chap 12 Extra Reading Cell Cycle and Mitosis
 APGRU4L1 Chap 12 Extra Reading Cell Cycle and Mitosis Dr. Ramesh Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008
APGRU4L1 Chap 12 Extra Reading Cell Cycle and Mitosis Dr. Ramesh Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008
Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling
 Developmental Cell Supplemental Information Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling Francesc R. Garcia-Gonzalo, Siew Cheng Phua, Elle C. Roberson, Galo Garcia
Developmental Cell Supplemental Information Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling Francesc R. Garcia-Gonzalo, Siew Cheng Phua, Elle C. Roberson, Galo Garcia
Supplemental Figure 1. Western blot analysis indicated that MIF was detected in the fractions of
 Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Centrosomes and Cilia
 Centrosomes and Cilia SMC6052/BIM6027/516-604D William Y. Tsang Research Unit Director, Cell Division and Centrosome Biology, IRCM Chercheur Adjoint, Faculté de medécine, Université de Montréal Adjunct
Centrosomes and Cilia SMC6052/BIM6027/516-604D William Y. Tsang Research Unit Director, Cell Division and Centrosome Biology, IRCM Chercheur Adjoint, Faculté de medécine, Université de Montréal Adjunct
SUPPLEMENTARY INFORMATION
 Figure S1. Loss of Ena/VASP proteins inhibits filopodia and neuritogenesis. (a) Bar graph of filopodia number per stage 1 control and mmvvee (Mena/ VASP/EVL-null) neurons at 40hrs in culture. Loss of all
Figure S1. Loss of Ena/VASP proteins inhibits filopodia and neuritogenesis. (a) Bar graph of filopodia number per stage 1 control and mmvvee (Mena/ VASP/EVL-null) neurons at 40hrs in culture. Loss of all
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division The Cell Cycle: Cell Growth, Cell Division 2007-2008 2007-2008 Getting from there to here Going from egg to baby. the original
Biology is the only subject in which multiplication is the same thing as division The Cell Cycle: Cell Growth, Cell Division 2007-2008 2007-2008 Getting from there to here Going from egg to baby. the original
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
MCB Topic 19 Regulation of Actin Assembly- Prof. David Rivier
 MCB 252 -Topic 19 Regulation of Actin Assembly- Prof. David Rivier MCB 252 Spring 2017 MCB 252 Cell Biology Topic 19 Regulation of Actin Assembly Reading: Lodish 17.2-17.3, 17.7 MCB 252 Actin Cytoskeleton
MCB 252 -Topic 19 Regulation of Actin Assembly- Prof. David Rivier MCB 252 Spring 2017 MCB 252 Cell Biology Topic 19 Regulation of Actin Assembly Reading: Lodish 17.2-17.3, 17.7 MCB 252 Actin Cytoskeleton
Supplemental Information. Induction of Expansion and Folding. in Human Cerebral Organoids
 Cell Stem Cell, Volume 20 Supplemental Information Induction of Expansion and Folding in Human Cerebral Organoids Yun Li, Julien Muffat, Attya Omer, Irene Bosch, Madeline A. Lancaster, Mriganka Sur, Lee
Cell Stem Cell, Volume 20 Supplemental Information Induction of Expansion and Folding in Human Cerebral Organoids Yun Li, Julien Muffat, Attya Omer, Irene Bosch, Madeline A. Lancaster, Mriganka Sur, Lee
Cell division functions in 1. reproduction, 2. growth, and 3. repair
 Cell division functions in 1. reproduction, 2. growth, and 3. repair What do you think you are looking at here??? Can something like you or I do this??? Fig. 12.1 How did you start out? How did you grow?
Cell division functions in 1. reproduction, 2. growth, and 3. repair What do you think you are looking at here??? Can something like you or I do this??? Fig. 12.1 How did you start out? How did you grow?
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Getting from there to here Going from egg to baby. the original
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Getting from there to here Going from egg to baby. the original
SUPPLEMENTARY INFORMATION
 Supplementary Information included with Nature MS 2008-02-01484B by Colantonio et al., entitled The dynein regulatory complex is required for ciliary motility and otolith biogenesis in the inner ear. This
Supplementary Information included with Nature MS 2008-02-01484B by Colantonio et al., entitled The dynein regulatory complex is required for ciliary motility and otolith biogenesis in the inner ear. This
(a) TEM of a plasma. Fimbriae. Nucleoid. Ribosomes. Plasma membrane. Cell wall Capsule. Bacterial chromosome
 0 m m 0. m cm mm 00 µm 0 µm 00 nm 0 nm Human height Length of some nerve and muscle cells Chicken egg Frog egg Most plant and animal cells Most bacteria Smallest bacteria Viruses Proteins Unaided eye Light
0 m m 0. m cm mm 00 µm 0 µm 00 nm 0 nm Human height Length of some nerve and muscle cells Chicken egg Frog egg Most plant and animal cells Most bacteria Smallest bacteria Viruses Proteins Unaided eye Light
A study of cilia density in sea urchin embryos during gastrulation
 A study of cilia density in sea urchin embryos during gastrulation Steve Das Independent Research Project Report Bio 254 Developmental Biology May 3, 2012 Introduction Embryo growth and development is
A study of cilia density in sea urchin embryos during gastrulation Steve Das Independent Research Project Report Bio 254 Developmental Biology May 3, 2012 Introduction Embryo growth and development is
Mitosis vs. microtubule
 Mitosis vs. microtubule Anaphase-promoting complex/cyclosome (APC/C) Duplicated centrosomes align and begin separating in prophase Relation of centrosome duplication to the cell cycle. Parent centrioles
Mitosis vs. microtubule Anaphase-promoting complex/cyclosome (APC/C) Duplicated centrosomes align and begin separating in prophase Relation of centrosome duplication to the cell cycle. Parent centrioles
CELL PART OF THE DAY. Chapter 7: Cell Structure and Function
 CELL PART OF THE DAY Chapter 7: Cell Structure and Function Cell Membrane Cell membranes are composed of two phospholipid layers. Cell membrane is flexible, not rigid The cell membrane has two major functions.
CELL PART OF THE DAY Chapter 7: Cell Structure and Function Cell Membrane Cell membranes are composed of two phospholipid layers. Cell membrane is flexible, not rigid The cell membrane has two major functions.
Supplementary Information
 Supplementary Information CEP41 is mutated in Joubert syndrome and is required for tubulin glutamylation at the cilium Ji Eun Lee, Jennifer L. Silhavy, Maha S. Zaki, Jana Schroth, Stephanie L. Bielas,
Supplementary Information CEP41 is mutated in Joubert syndrome and is required for tubulin glutamylation at the cilium Ji Eun Lee, Jennifer L. Silhavy, Maha S. Zaki, Jana Schroth, Stephanie L. Bielas,
The cytoskeleton and cell movement. (Actin microfilaments)
 The cytoskeleton and cell movement (Actin microfilaments) What is the cytoskeleton? A dynamic network of protein filaments extending throughout the cytoplasm Three types: microfilaments (actin), microtubules
The cytoskeleton and cell movement (Actin microfilaments) What is the cytoskeleton? A dynamic network of protein filaments extending throughout the cytoplasm Three types: microfilaments (actin), microtubules
Human height. Length of some nerve and muscle cells. Chicken egg. Frog egg. Most plant and animal cells Nucleus Most bacteria Mitochondrion
 10 m 1 m 0.1 m 1 cm Human height Length of some nerve and muscle cells Chicken egg Unaided eye 1 mm Frog egg 100 µm 10 µm 1 µm 100 nm 10 nm Most plant and animal cells Nucleus Most bacteria Mitochondrion
10 m 1 m 0.1 m 1 cm Human height Length of some nerve and muscle cells Chicken egg Unaided eye 1 mm Frog egg 100 µm 10 µm 1 µm 100 nm 10 nm Most plant and animal cells Nucleus Most bacteria Mitochondrion
Chapter 3: Cells 3-1
 Chapter 3: Cells 3-1 Introduction: A. Human body consists of 75 trillion cells B. About 260 types of cells that vary in shape & size yet have much in common B. Differences in cell shape make different
Chapter 3: Cells 3-1 Introduction: A. Human body consists of 75 trillion cells B. About 260 types of cells that vary in shape & size yet have much in common B. Differences in cell shape make different
~Lentivirus production~
 ~Lentivirus production~ May 30, 2008 RNAi core R&D group member Lentivirus Production Session Lentivirus!!! Is it health threatening to lab technician? What s so good about this RNAi library? How to produce
~Lentivirus production~ May 30, 2008 RNAi core R&D group member Lentivirus Production Session Lentivirus!!! Is it health threatening to lab technician? What s so good about this RNAi library? How to produce
Influenza virus exploits tunneling nanotubes for cell-to-cell spread
 Supplementary Information Influenza virus exploits tunneling nanotubes for cell-to-cell spread Amrita Kumar 1, Jin Hyang Kim 1, Priya Ranjan 1, Maureen G. Metcalfe 2, Weiping Cao 1, Margarita Mishina 1,
Supplementary Information Influenza virus exploits tunneling nanotubes for cell-to-cell spread Amrita Kumar 1, Jin Hyang Kim 1, Priya Ranjan 1, Maureen G. Metcalfe 2, Weiping Cao 1, Margarita Mishina 1,
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Figure S1
 Supplementary Figure S1 Supplementary Figure S1. PARP localization patterns using GFP-PARP and PARP-specific antibody libraries GFP-PARP localization in non-fixed (A) and formaldehyde fixed (B) GFP-PARPx
Supplementary Figure S1 Supplementary Figure S1. PARP localization patterns using GFP-PARP and PARP-specific antibody libraries GFP-PARP localization in non-fixed (A) and formaldehyde fixed (B) GFP-PARPx
Chapter 12 The Cell Cycle: Cell Growth, Cell Division
 Chapter 12 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a period at the end of a sentence And now look at you How did you get from there to
Chapter 12 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a period at the end of a sentence And now look at you How did you get from there to
PCB 3023 Exam 4 - Form A First and Last Name
 PCB 3023 Exam 4 - Form A First and Last Name Student ID # (U Number) A Before beginning this exam, please complete the following instructions: 1) Write your name and U number on the first page of this
PCB 3023 Exam 4 - Form A First and Last Name Student ID # (U Number) A Before beginning this exam, please complete the following instructions: 1) Write your name and U number on the first page of this
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division Ch. 10 Where it all began You started as a cell smaller than a period
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division Ch. 10 Where it all began You started as a cell smaller than a period
Myelin suppresses axon regeneration by PIR-B/SHPmediated inhibition of Trk activity
 Manuscript EMBO-2010-76298 Myelin suppresses axon regeneration by PIR-B/SHPmediated inhibition of Trk activity Yuki Fujita, Shota Endo, Toshiyuki Takai and Toshihide Yamashita Corresponding author: Toshihide
Manuscript EMBO-2010-76298 Myelin suppresses axon regeneration by PIR-B/SHPmediated inhibition of Trk activity Yuki Fujita, Shota Endo, Toshiyuki Takai and Toshihide Yamashita Corresponding author: Toshihide
Regulators of Cell Cycle Progression
 Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
Supplemental Materials. STK16 regulates actin dynamics to control Golgi organization and cell cycle
 Supplemental Materials STK16 regulates actin dynamics to control Golgi organization and cell cycle Juanjuan Liu 1,2,3, Xingxing Yang 1,3, Binhua Li 1, Junjun Wang 1,2, Wenchao Wang 1, Jing Liu 1, Qingsong
Supplemental Materials STK16 regulates actin dynamics to control Golgi organization and cell cycle Juanjuan Liu 1,2,3, Xingxing Yang 1,3, Binhua Li 1, Junjun Wang 1,2, Wenchao Wang 1, Jing Liu 1, Qingsong
The Role of Sorting Nexins in Antigen Presentation
 The Role of Sorting Nexins in Antigen Presentation Chng X.R.J 1 and Wong S.H. 2 Department of Microbiology Yong Loo Lin School of Medicine, National University of Singapore Block MD4, 5 Science Drive 2,
The Role of Sorting Nexins in Antigen Presentation Chng X.R.J 1 and Wong S.H. 2 Department of Microbiology Yong Loo Lin School of Medicine, National University of Singapore Block MD4, 5 Science Drive 2,
BIOLOGY - CLUTCH CH.12 - CELL DIVISION.
 !! www.clutchprep.com CONCEPT: CELL DIVISION Cell division is the process by which one cell splits into two or more daughter cells. Cell division generally requires that cells produce enough materials,
!! www.clutchprep.com CONCEPT: CELL DIVISION Cell division is the process by which one cell splits into two or more daughter cells. Cell division generally requires that cells produce enough materials,
Supplementary Information
 1 Supplementary Information A role for primary cilia in glutamatergic synaptic integration of adult-orn neurons Natsuko Kumamoto 1,4,5, Yan Gu 1,4, Jia Wang 1,4, Stephen Janoschka 1,2, Ken-Ichi Takemaru
1 Supplementary Information A role for primary cilia in glutamatergic synaptic integration of adult-orn neurons Natsuko Kumamoto 1,4,5, Yan Gu 1,4, Jia Wang 1,4, Stephen Janoschka 1,2, Ken-Ichi Takemaru
ANALYSIS OF THE CILIARY GENES GAS8 AND MKS6 REVEAL CONSERVED ROLES IN CILIA MOTILITY AND TRANSITION ZONE FUNCTION. by: WESLEY R.
 ANALYSIS OF THE CILIARY GENES GAS8 AND MKS6 REVEAL CONSERVED ROLES IN CILIA MOTILITY AND TRANSITION ZONE FUNCTION by: WESLEY R. LEWIS JAMES F. COLLAWN: COMMITTEE CHAIR BRADLEY K. YODER: MENTOR JOHN M.
ANALYSIS OF THE CILIARY GENES GAS8 AND MKS6 REVEAL CONSERVED ROLES IN CILIA MOTILITY AND TRANSITION ZONE FUNCTION by: WESLEY R. LEWIS JAMES F. COLLAWN: COMMITTEE CHAIR BRADLEY K. YODER: MENTOR JOHN M.
BIOL 4374/BCHS 4313 Cell Biology Exam #2 March 22, 2001
 BIOL 4374/BCHS 4313 Cell Biology Exam #2 March 22, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) In the
BIOL 4374/BCHS 4313 Cell Biology Exam #2 March 22, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) In the
Molecular Cell Biology 5068 In Class Exam 1 October 3, 2013
 Molecular Cell Biology 5068 In Class Exam 1 October 3, 2013 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
Molecular Cell Biology 5068 In Class Exam 1 October 3, 2013 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
Human Epithelial Cells
 The Cell Human Epithelial Cells Plant Cells Cells have an internal structure Eukaryotic cells are organized Protective membrane around them that communicates with other cells Organelles have specific jobs
The Cell Human Epithelial Cells Plant Cells Cells have an internal structure Eukaryotic cells are organized Protective membrane around them that communicates with other cells Organelles have specific jobs
10/13/11. Cell Theory. Cell Structure
 Cell Structure Grade 12 Biology Cell Theory All organisms are composed of one or more cells. Cells are the smallest living units of all living organisms. Cells arise only by division of a previously existing
Cell Structure Grade 12 Biology Cell Theory All organisms are composed of one or more cells. Cells are the smallest living units of all living organisms. Cells arise only by division of a previously existing
MBios 401/501: Lecture 12.1 Signaling IV. Slide 1
 MBios 401/501: Lecture 12.1 Signaling IV Slide 1 Pathways that require regulated proteolysis 1. Notch and Delta 2. Wnt/ b-catenin 3. Hedgehog 4. NFk-B Our last topic on cell signaling are pathways that
MBios 401/501: Lecture 12.1 Signaling IV Slide 1 Pathways that require regulated proteolysis 1. Notch and Delta 2. Wnt/ b-catenin 3. Hedgehog 4. NFk-B Our last topic on cell signaling are pathways that
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human
 Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
TFEB-mediated increase in peripheral lysosomes regulates. Store Operated Calcium Entry
 TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
PHSI3009 Frontiers in Cellular Physiology 2017
 Overview of PHSI3009 L2 Cell membrane and Principles of cell communication L3 Signalling via G protein-coupled receptor L4 Calcium Signalling L5 Signalling via Growth Factors L6 Signalling via small G-protein
Overview of PHSI3009 L2 Cell membrane and Principles of cell communication L3 Signalling via G protein-coupled receptor L4 Calcium Signalling L5 Signalling via Growth Factors L6 Signalling via small G-protein
Overview of the Cellular Basis of Life. Copyright 2009 Pearson Education, Inc., publishing as Benjamin Cummings
 Overview of the Cellular Basis of Life Cells and Tissues Cells: Carry out all chemical activities needed to sustain life Cells are the building blocks of all living things Tissues Cells vary in length,
Overview of the Cellular Basis of Life Cells and Tissues Cells: Carry out all chemical activities needed to sustain life Cells are the building blocks of all living things Tissues Cells vary in length,
Supplementary data Supplementary Figure 1 Supplementary Figure 2
 Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Erzsebet Kokovay, Susan Goderie, Yue Wang, Steve Lotz, Gang Lin, Yu Sun, Badrinath Roysam, Qin Shen,
 Cell Stem Cell, Volume 7 Supplemental Information Adult SVZ Lineage Cells Home to and Leave the Vascular Niche via Differential Responses to SDF1/CXCR4 Signaling Erzsebet Kokovay, Susan Goderie, Yue Wang,
Cell Stem Cell, Volume 7 Supplemental Information Adult SVZ Lineage Cells Home to and Leave the Vascular Niche via Differential Responses to SDF1/CXCR4 Signaling Erzsebet Kokovay, Susan Goderie, Yue Wang,
MII. Supplement Figure 1. CapZ β2. Merge. 250ng. 500ng DIC. Merge. Journal of Cell Science Supplementary Material. GFP-CapZ β2 DNA
 A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
CELLS. Cells. Basic unit of life (except virus)
 Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
CELL PARTS TYPICAL ANIMAL CELL
 AP BIOLOGY CText Reference, Campbell v.8, Chapter 6 ACTIVITY1.12 NAME DATE HOUR CELL PARTS TYPICAL ANIMAL CELL ENDOMEMBRANE SYSTEM TYPICAL PLANT CELL QUESTIONS: 1. Write the name of the cell part in the
AP BIOLOGY CText Reference, Campbell v.8, Chapter 6 ACTIVITY1.12 NAME DATE HOUR CELL PARTS TYPICAL ANIMAL CELL ENDOMEMBRANE SYSTEM TYPICAL PLANT CELL QUESTIONS: 1. Write the name of the cell part in the
7.2 Cell Structure. Sunday, December 16, :13 PM. Chapter 7 Cell Strucutre and Function Page 1
 7.2 Cell Structure Sunday, December 16, 2012 5:13 PM Vocabulary: Cytoplasm: in eukaryotic cells, all cellular contents outside the nucleus; in prokaryotic cells, all of the cells' contents Organelle: specialized
7.2 Cell Structure Sunday, December 16, 2012 5:13 PM Vocabulary: Cytoplasm: in eukaryotic cells, all cellular contents outside the nucleus; in prokaryotic cells, all of the cells' contents Organelle: specialized
Actin structure. Actin a highly conserved gene. Molecular Cell Biology
 Harvey Lodish Arnold Berk Paul Matsudaira Chris A. Kaiser Monty Krieger Matthew P. Scott Lawrence Zipursky James Darnell Molecular Cell Biology Fifth Edition Chapter 19: Cytoskeleton I: Microfilaments
Harvey Lodish Arnold Berk Paul Matsudaira Chris A. Kaiser Monty Krieger Matthew P. Scott Lawrence Zipursky James Darnell Molecular Cell Biology Fifth Edition Chapter 19: Cytoskeleton I: Microfilaments
Anatomy Chapter 2 - Cells
 Cells Cells are the basic living structural, functional unit of the body Cytology is the branch of science that studies cells The human body has 100 trillion cells 200 different cell types with a variety
Cells Cells are the basic living structural, functional unit of the body Cytology is the branch of science that studies cells The human body has 100 trillion cells 200 different cell types with a variety
Fungal cell walls are rigid with less flexibility due to a combination of more sugar (more chitin) and protein flexibility.
 Cell Structure Assignment Score. Name Sec.. Date. Working by yourself or in a group, answer the following questions about the Cell Structure material. This assignment is worth 40 points with the possible
Cell Structure Assignment Score. Name Sec.. Date. Working by yourself or in a group, answer the following questions about the Cell Structure material. This assignment is worth 40 points with the possible
8/7/18. UNIT 2: Cells Chapter 3: Cell Structure and Function. I. Cell Theory (3.1) A. Early studies led to the development of the cell theory
 8/7/18 UNIT 2: Cells Chapter 3: Cell Structure and Function I. Cell Theory (3.1) A. Early studies led to the development of the cell theory 1. Discovery of Cells a. Robert Hooke (1665)-Used compound microscope
8/7/18 UNIT 2: Cells Chapter 3: Cell Structure and Function I. Cell Theory (3.1) A. Early studies led to the development of the cell theory 1. Discovery of Cells a. Robert Hooke (1665)-Used compound microscope
BIO 5099: Molecular Biology for Computer Scientists (et al)
 BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
Campbell Biology in Focus (Urry) Chapter 9 The Cell Cycle. 9.1 Multiple-Choice Questions
 Campbell Biology in Focus (Urry) Chapter 9 The Cell Cycle 9.1 Multiple-Choice Questions 1) Starting with a fertilized egg (zygote), a series of five cell divisions would produce an early embryo with how
Campbell Biology in Focus (Urry) Chapter 9 The Cell Cycle 9.1 Multiple-Choice Questions 1) Starting with a fertilized egg (zygote), a series of five cell divisions would produce an early embryo with how
Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy
 Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy Norann A. Zaghloul, Nicholas Katsanis J Clin Invest. 2009;119(3):428-437. https://doi.org/10.1172/jci37041. Science in Medicine Bardet-Biedl
Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy Norann A. Zaghloul, Nicholas Katsanis J Clin Invest. 2009;119(3):428-437. https://doi.org/10.1172/jci37041. Science in Medicine Bardet-Biedl
Chapter 3. Expression of α5-megfp in Mouse Cortical Neurons. on the β subunit. Signal sequences in the M3-M4 loop of β nachrs bind protein factors to
 22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
Lecture 10. G1/S Regulation and Cell Cycle Checkpoints. G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint
 Lecture 10 G1/S Regulation and Cell Cycle Checkpoints Outline: G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint Paper: The roles of Fzy/Cdc20 and Fzr/Cdh1
Lecture 10 G1/S Regulation and Cell Cycle Checkpoints Outline: G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint Paper: The roles of Fzy/Cdc20 and Fzr/Cdh1
Signaling. Dr. Sujata Persad Katz Group Centre for Pharmacy & Health research
 Signaling Dr. Sujata Persad 3-020 Katz Group Centre for Pharmacy & Health research E-mail:sujata.persad@ualberta.ca 1 Growth Factor Receptors and Other Signaling Pathways What we will cover today: How
Signaling Dr. Sujata Persad 3-020 Katz Group Centre for Pharmacy & Health research E-mail:sujata.persad@ualberta.ca 1 Growth Factor Receptors and Other Signaling Pathways What we will cover today: How
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Supporting Information
 Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Organelles in EUKARYOTIC CELLS. Pages 4 5: cell membrane, cytoplasm/cytosol, cytoskeleton, nucleus, chromatin/chromosomes, and centrioles
 Organelles in EUKARYOTIC CELLS Pages 4 5: cell membrane, cytoplasm/cytosol, cytoskeleton, nucleus, chromatin/chromosomes, and centrioles Use the info on the following slides to complete pages 4 5 in your
Organelles in EUKARYOTIC CELLS Pages 4 5: cell membrane, cytoplasm/cytosol, cytoskeleton, nucleus, chromatin/chromosomes, and centrioles Use the info on the following slides to complete pages 4 5 in your
By Sally Pobojewski Photos by Scott Galvin. Secrets of the. Cilia. { long-overlooked organelles hold some hefty keys to human health }
 By Sally Pobojewski Photos by Scott Galvin Secrets of the Cilia { long-overlooked organelles hold some hefty keys to human health } Photo: Dr. G. Moscoso/Photo Researchers, Inc. 20 Fall 2007 Medicine at
By Sally Pobojewski Photos by Scott Galvin Secrets of the Cilia { long-overlooked organelles hold some hefty keys to human health } Photo: Dr. G. Moscoso/Photo Researchers, Inc. 20 Fall 2007 Medicine at
Supplemental Information. Otic Mesenchyme Cells Regulate. Spiral Ganglion Axon Fasciculation. through a Pou3f4/EphA4 Signaling Pathway
 Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Chapter 3 Part 2! Pages (10 th and 11 th eds.)! The Cellular Level of Organization! Cellular Organelles and Protein Synthesis!
 Chapter 3 Part 2! Pages 65 89 (10 th and 11 th eds.)! The Cellular Level of Organization! Cellular Organelles and Protein Synthesis! The Cell Theory! Living organisms are composed of one or more cells.!
Chapter 3 Part 2! Pages 65 89 (10 th and 11 th eds.)! The Cellular Level of Organization! Cellular Organelles and Protein Synthesis! The Cell Theory! Living organisms are composed of one or more cells.!
International Graduate Research Programme in Cardiovascular Science
 1 International Graduate Research Programme in Cardiovascular Science This work has been supported by the European Community s Sixth Framework Programme under grant agreement n LSHM-CT-2005-01883 EUGeneHeart.
1 International Graduate Research Programme in Cardiovascular Science This work has been supported by the European Community s Sixth Framework Programme under grant agreement n LSHM-CT-2005-01883 EUGeneHeart.
The Complexity of Simple Genetics
 The Complexity of Simple Genetics? The ciliopathies: a journey into variable penetrance and expressivity Bardet-Biedl Syndrome Allelism at a single locus is insufficient to explain phenotypic variability
The Complexity of Simple Genetics? The ciliopathies: a journey into variable penetrance and expressivity Bardet-Biedl Syndrome Allelism at a single locus is insufficient to explain phenotypic variability
