Probing a Role for TRAP150 in Gene Regulation
|
|
|
- Emma Pierce
- 5 years ago
- Views:
Transcription
1 Wright State University CORE Scholar Browse all Theses and Dissertations Theses and Dissertations 2009 Probing a Role for TRAP150 in Gene Regulation Divya Potabathula Wright State University Follow this and additional works at: Part of the Biology Commons Repository Citation Potabathula, Divya, "Probing a Role for TRAP150 in Gene Regulation" (2009). Browse all Theses and Dissertations This Thesis is brought to you for free and open access by the Theses and Dissertations at CORE Scholar. It has been accepted for inclusion in Browse all Theses and Dissertations by an authorized administrator of CORE Scholar. For more information, please contact corescholar@ library-corescholar@wright.edu.
2 PROBING A ROLE FOR TRAP15O IN GENE REGULATION A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science By DIVYA POTABATHULA B.Tech., Jawaharlal Nehru Technological University, Wright State University
3 WRIGHT STATE UNIVERSITY SCHOOL OF GRADUATE STUDIES January 20, 2009 I HEREBY RECOMMEND THAT THE THESIS PREPARED UNDER MY SUPERVISION BY Divya Potabathula ENTITLED Probing a role for TRAP150 in gene regulation BE ACCEPTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Master of Science. Paula A. Bubulya, Ph. D. Thesis Director Committee on Final Examination David L. Goldstein, Ph. D. Department Chair Paula A. Bubulya, Ph.D. Barbara Hull, Ph. D. Scott E. Baird, Ph.D. Joseph F. Thomas, Jr., Ph.D. Dean, School of Graduate Studies ii
4 ABSTRACT: Potabathula, Divya. M.S., Biological Sciences, Wright State University Probing a Role for TRAP150 in Gene Regulation. The mammalian cell nucleus is a highly organized cellular compartment. The nucleus is surrounded by the nuclear envelope and contains distinct domains such as nucleoli, nuclear speckles and Cajal bodies that may reflect nuclear functions such as transcription and RNA processing. Nuclear speckles are regions in the nucleus that are enriched in splicing factors and other RNA processing factors, as well as some transcription factors and the large subunit of RNA polymerase II. Mass spectrometry analysis of purified nuclear speckles revealed 146 nuclear speckle proteins of which 33 are novel nuclear speckle components. One such novel nuclear speckle protein is thyroid hormone receptor associated protein (TRAP150/THRAP3). Immunofluorescence of TRAP150 in Hela cells shows that it localizes predominantly in a diffuse distribution throughout the nucleoplasm that resembles the localization of transcription factors. Phylogenetics showed that TRAP150 and Btf (another novel speckle protein) are members of the same gene family. The amino acid sequence alignment of TRAP150 and Btf showed that they share 39.5% sequence identity and 62% similarity. High sequence similarity was observed at the C-termini, suggesting a second functional domain of unknown function. TRAP150 and Btf associate with affinity-purified in vitro-spliced human mrnps. Hence, I hypothesized that TRAP150 might have a role in transcription and/or premrna processing. Inhibition of RNA polymerase II transcription caused extensive iii
5 enrichment of TRAP150 in large, rounded nuclear speckles. I observed the association of TRAP150 and Btf with a reporter gene locus in vivo. Results showed that TRAP150 and Btf associate more frequently with the activated reporter locus than with the inactive locus. TRAP150 and Btf also localized in the vicinity of the reporter RNA. These results are all consistent with a role for TRAP150 and Btf in gene expression. iv
6 TABLE OF CONTENTS Page I. INTRODUCTION 1 TRAP150 3 Phylogenetics of TRAP150 4 Transcription factories 6 U2OS locus 7 II. SPECIFIC AIMS 10 III. MATERIALS and METHODS 13 cdna constructs 13 Cell culture 13 Transient transfections using the calcium phosphate method 13 Transient transfection by Electroporation (for U2OS cells) 14 Immunofluorescence 14 Microscopy 15 Scoring the cells 15 Sequence alignments 16 IV. RESULTS 17 Specific aim 1 17 v
7 TABLE OF CONTENTS (Continued) Page Specific aim 2 20 Specific aim 2A 22 Specific aim 2B 24 Specific aim 2B-i 26 Specific aim 2B-ii 34 V. DISCUSSION 37 VI. REFERENCES 43 vi
8 LIST OF FIGURES Figure Page 1. Tree fam of TRAP150 gene Schematic representation of the U2OS transgene array Sequence alignment of Btf and TRAP A graphical representation of local alignments between TRAP150 and Btf Immunoflourescence of TRAP150 and nuclear speckles Localization of YFP-TRAP Localization of TRAP150 in transcription inhibited nuclei Different morphologies of the U2OS locus Localization of TRAP150 in ON vs OFF gene locus Localization of Btf in ON vs OFF gene locus Localization of TRAP150, Btf and SF2/ASF at reporter RNA. 35 vii
9 LIST OF TABLES Table Page 1. TRAP150 at the reporter locus Btf at the reporter locus. 33 viii
10 LIST OF ABBREVIATIONS Br-UTP- Bromo-Uridine triphosphate. Btf- Bcl2 associated transcription factor. CBC- Cap binding complex. CXorf23- Chromosome X open reading frame 23. DEAD box- D-E-A-D (Aspartic acid-glutamic acid-alanine-aspartic acid) box. DMEM- Dulbecco s Modified Eagle Medium. Dox- Doxycycline. HDACs- Histone deacetylases. HP1- Heterochromatin protein1. IGCs- Interchromatin granule clusters. NLS- Nuclear localization signal. RS domain- Arginine-Serine rich domain. SF2/ASF- Splicing factor 2/Alternative splicing factor. snrnp- small nuclear ribonucleoprotein. TR- Thyroid hormone receptor. TRAPs- Thyroid hormone receptor associated proteins. TRAP150- Thyroid hormone receptor associated protein 150. ix
11 ACKNOWLEDGEMENTS I wish to express my gratitude to my advisor, Dr. Paula Bubulya, for her training, supervision, and immense support throughout this project. I would also like to thank Dr. Barbara Hull and Dr. Scott Baird for their suggestions, guidance, support and most of all for giving me their valuable time. I would like to thank Dr.Athanasios Bubulya for his assistance and insight. x
12 I. INTRODUCTION The mammalian cell nucleus is a highly organized cellular compartment. The nucleus is surrounded by the nuclear envelope and is arranged into distinct domains that may reflect functions such as transcription, pre-mrna processing, and DNA replication (Spector, 1993). Some of these domains include the nucleolus, chromosome territories, nuclear pore complexes, Cajal bodies, Gems and nuclear speckles. (Spector, 1993; Lamond and Earnshaw, 1998; Misteli, 2001). These large regions in mammalian nuclei are functionally distinct from their surroundings and are unique because they are nonmembrane bound nuclear organelles. One idea is that these domains may serve as storage sites to supply functional complexes to regions of activity else where in the nucleus (Misteli, 2005). Nuclear speckles are regions in the nucleus that are enriched in splicing factors and other RNA processing factors, as well as some transcription factors and the large subunit of RNA polymerase II (Misteli et al., Mintz et al., 1999; Saitoh, 2004). Nuclear speckles are not the sites of splicing and transcription, rather they are storage areas and/or assembly sites for these factors (Misteli and Spector, 1999; Caceres et al., 1997). Nuclear speckles are also called interchromatin granule clusters (IGCs) based upon their ultratructure, with clusters of ribonucleoprotein particles seen interspersed between regions of chromatin. IGCs are one of the largest non-chromatin structures of the nucleoplasm. A proteomic analysis of IGCs purified from mouse liver showed that 81% of the proteins are involved in RNA metabolism among which 54% are involved in 1
13 pre-mrna splicing (Mintz et al., 1999; Saitoh et al., 2004). Other components are thought to have a role in apoptosis, mrna export, transcription and cleavage and polyadenylation (Saitoh et al., 2004). Mass spectrometry analysis of purified IGCs revealed 146 nuclear speckle proteins of which 33 are novel proteins that had not previously been identified as nuclear speckle proteins (Mintz et al., 1999; Saitoh et al., 2004). Assignment of these proteins to distinct functional groups was based upon motif analysis. For RNA processing factors, this included motifs such as RNA binding motifs, RNA helicase motifs, RNA recognition motifs, arginine serine (RS) rich domain motif and the D-E-A-D (Asp-Glu-Ala-Asp) box (DEAD box) helicase domain (Saitoh et al., 2004). These domains all serve specific functions in proteins that are involved in different aspects of RNA metabolism such as pre-mrna processing. Serine-arginine (SR)-rich splicing factors called SR proteins play a key role during constitutive and alternative splicing and are one of the major families of proteins found in nuclear speckles (Li and Bingham, 1991). During transcription, SR proteins and other RNA processing factors are recruited to transcription sites for co-transcriptional splicing (Misteli et al., 1997; Caceres et al., 1997). The arginine/serine rich (RS), domain found in pre-mrna splicing factors is a speckle-targeting signal (Li and Bingham, 1991; Hedley et al., 1995; Caceres et al., 1997). Phosphorylation of serines in the RS domain regulates the release of SR protein complexes from the nuclear speckles and their subsequent assembly into spliceosomes at the transcription site (Hedley et al., 1995; Misteli et al., 1998). The dynamic movements of speckle components observed by expressing fluorescently tagged splicing factors in living cells may reflect the 2
14 recruitment of RNA processing machinery from nuclear speckles to transcription sites (Misteli et al., 1998; Caceres and Krainer, 1997). SR splicing factors are divided into 3 groups according to the arrangement of the RS domain with respect to their other domains. Type I SR proteins have a RS domain and one to three RNA recognition motifs. Examples of type I SR proteins include classical SR proteins such as SF2/ASF, SC35 and 9G8. Type II SR proteins have a RS domain but do not have a RNA recognition motif. Examples of type II SR proteins include thyroid hormone receptor associated protein 150 (TRAP 150) and Bcl-2 associated transcription factor (Btf). Type III SR proteins are high molecular weight proteins that contain repetitive sequences in addition to the RS domain. Son and NP220 belong to this class of SR proteins (Saitoh et al., 2004). TRAP150 TRAP150 was initially identified as a component of thyroid hormone receptor associated proteins. Thyroid hormone receptor (TR) is a nuclear receptor. In the absence of a thyroid hormone, thyroid hormone receptor binds to DNA and acts as a transcription repressor (Ito et al., 1999). In the presence of thyroid hormone, a conformational change occurs in the receptor, and it becomes a transcriptional activator (Ito et al., 1999). Thyroid hormone receptor associated proteins (TRAPs) co-activate TR-mediated transcription in a TATA-box-binding-independent manner. Some TRAPs have functions similar to the yeast mediator proteins that are responsible for transcription activation (Ito et al., 1999). 3
15 One among the potential nuclear speckle proteins is thyroid hormone receptor associated protein of 150 kda (TRAP150). It was identified in proteomic analysis of purified nuclear speckles as a Type II RS domain containing protein (Mintz et al., 1999; Saitoh et al., 2004). Initially, TRAP150 was reported to have a role in transcription regulation acting as a co-transcriptional activator (Ito et al., 1999) but further work suggests that TRAP150 is not a bonafide component of the mediator complex (Dr. Robert Roeder, personal communication). Phylogenetics of TRAP150 TRAP150 is a member of a small gene family that also includes Btf and Chromosome X open reading frame 23 (CXorf23) and their orthologs. A phylogeny of this gene family is shown in Figure 1. Orthologs of TRAP150 gene family are present in vertebrates including mammals, birds and fish (Figure 1). The lineage is not seen in invertebrates. One TRAP150-related protein Btf, is predicted to have roles in transcription regulation (Kasof et al., 1999). CXorf23 is a TRAP150 related protein that is uncharacterized. Within the gene family, TRAP150 is more closely related to Btf and its orthologs than to CXorf23. 4
16 Btf TRAP150 CXorf23 Uncharacterized proteins Figure 1: Treefam phylogeny of TRAP150 gene. The three clusters of genes Btf, TRAP150 and CXorf23 hypothetical proteins are grouped together under one family of genes. Genes from the same species are denoted in the same color, the blue boxes denote nodes of the tree from which a specific gene is branched. The numbers at each node indicate the boot strap value of each clade. 5
17 Research from the Luhrman lab suggests that TRAP 150 and Btf might possibly be associated with affinity purified in vitro-spliced human mrnps (Merz et al., 2007). For that study, mrnps were synthesized in vitro under different conditions. For example, the mrnps were synthesized either in the presence or absence of capping, splicing and polyadenylation and then purified for mass spectrometry analysis (Merz et al., 2007). The results show that TRAP150 and Btf loaded onto mrnps during pre-mrna processing. Btf and TRAP150 associate with mrna in a way that is independent of pre-mrna splicing events (Merz et al., 2007). While Btf remains as a stable component of affinity purified mrnps, TRAP150 (and some RNA-binding proteins like YB-1 and NFAR1) dissociated from mrnps under stringent conditions. One contrast between TRAP150 and Btf is that Btf was not found on purified mrnps that lack a cap-binding complex (CBC), suggesting that the binding of Btf to mrnps may be cap-dependent. It is unclear, whether Btf directly interacts with the CBC or the CBC indirectly influences Btf s association with mrnps (Merz et al., 2007). Transcription factories Nuclear compartments are large regions in the nucleus that are functionally distinct from their surroundings. The distribution and spatial organization of transcription complexes correspond to sub nuclear transcription centers or transcription factories (Cook, 1998, Chakalova et al., 2005). To visualize global transcription sites in cells, transcription sites can be labeled when nascent transcripts are extended in the presence of the nucleotide analog bromo-utp, thus tagging the nascent RNA. Each focus of Br-UTP incorporation seen by microscopy would then represent a transcription 6
18 site. Many such nascent RNAs or transcription units are packed into a focus to form a transcription factory. The labeling is not from a single gene locus, but rather from a cluster of genes. In this study, we were interested to know if TRAP150 localization was consistent with localization of transcription sites. RNA polymerase II, transcription factors and splicing factors are also seen in a pattern of dispersed foci all over the nucleoplasm in an actively transcribing nucleus that is similar to the pattern seen by Br- UTP incorporation. However, when transcription is inhibited, the transcription factors and splicing factors accumulate in nuclear speckles. One idea here is that TRAP150 might be associated with transcription factories if its localization resembles that of transcription factors and RNA polymerase II. Determining the localization of TRAP150 with respect to RNA polymerase II or nascent mrna would help determine the possibility of such a role. U2OS locus: Labeling global transcription sites with BrUTP would be expected to show hundreds of sites throughout the nucleus. Co-labeling of TRAP150 would also show hundreds of foci throughout the nucleus. Spatial overlap between the two signals may not give a definitive conclusion whether TRAP150 is at every transcription site. The U20S locus is a special gene array that allows gene expression to be observed at three different levels by directly visualizing DNA, RNA and protein in the cell (Janicki et al., 2004). A diagram of the transgene array is shown in Figure 2. 7
19 Figure 2: Adapted from Janicki et al. (2004). Schematic representation of the U2OS transgene array. The plasmid is composed of 256 copies of the lac operator, 96 tetracycline response elements, a CMV promoter, CFP fused to the peroxisomal targeting signal SKL, 24 MS2 translational operators (MS2 repeats), a rabbit β globin intron/exon module, and a cleavage/polyadenylation signal. Expression of CFP-lac repressor allows the DNA to be visualized and expression of ptet-on (rtta) in the presence of doxycycline (dox) drives expression from the CMV minimal promoter. When MS2-YFP (YFP fused to the MS2 coat protein) dimerizes and interacts with the stem loop structure of the translational operator, it allows the transcribed RNA to be visualized. The labeling of peroxisomes with CFP in the cytoplasm confirm the translation and expression of the reporter protein. The U2OS transgene array contains approximately 200 copies of this plasmid construct (Janicki et al., 2004). The U2OS cell line has ~200 copies of stably integrated transgene plasmid as estimated by Southern blotting (Janicki et al., 2004). Each copy contains a transcription unit that has 256 copies of lac operator, which serves a binding site for CFP lac repressor or YFP lac repressor. This region of the DNA is fluorescently labeled when plasmids encoding these fusion proteins are transfected into the cell line (Janicki et al., 2004). The locus also has 96 tetracycline regulatory elements that allow inducible regulation of transcription. To induce transcription of the array, a fusion of VP16 activation domain and tetracycline controlled transactivation domain (pteton) are transfected into the U2OS cells. Doxycycline (Dox) addition to the culture medium induces binding of pteton (transcription activator) binds to tetracycline response elements and activates transcription starting from the CMV minimal promoter (Janicki et 8
20 al., 2004). Transcription at the locus is switched ON and reporter gene expression takes place in the presence of doxycycline. The locus can be visualized by fluorescence microscopy because of CFP-LacI that accumulates on lac operator repeats. In the OFF state, the chromatin at the locus appears as highly condensed as a single dot. The condensed locus is heterochromatic, as evident from its association with HP1 and methylated histone 3 at lysine 9 (H3MeK9) (Janicki et al., 2004). Upon induction of transcription, HP1 is removed and exchanged with histone H3.3 variant. The chromatin becomes highly decondensed and U2OS locus becomes transcriptionally active. The U2OS reporter mrna encodes a peroxisomal targeting signal-1 (Serine-Lysine-Leucine) fused to CFP (CFP-SKL). In the presence of dox, transcription of the reporter RNA begins. Accumulation of the nascent pre-mrna containing MS2 stem loops can be visualized at the locus when YFP-tagged MS2 binding protein is expressed in the cells (Janicki et al., 2004). The appearance of CFP-SKL protein (that is targeted to persoxisomes) in the cytoplasm shows that all steps of reporter gene expression are complete. The localization of TRAP150 with respect to the U2OS locus in its ON and OFF state was used to test for a possible role of TRAP150 in gene expression. 9
21 II. SPECIFIC AIMS: The goal of this study is to determine the role of TRAP150 in gene regulation. Hypothesis: TRAP150 is a transcription activator or a splicing factor. It is likely to be shuttling between its site of action (transcription sites) and storage areas (nuclear speckles). Specific Aim 1: To identify potentially functional domains of TRAP150 and Btf. Amino acid sequence alignment of TRAP150 and Btf gives percentage similarity and also highlights conserved regions, which could lead to understanding the functionality of both the proteins. Specific Aim 2: To determine if TRAP150 is associated with transcription. a) To determine if the localization of TRAP150 is altered after inhibition of transcription. Immunoflourescence labeling of TRAP150 in Hela cells after inhibition of RNA polymerase II by α-amanitin treatment. b) To determine if TRAP150 and Btf are recruited to transcription sites in vivo. (i) Localization of TRAP150 and Btf at a specific gene locus For this aim, I employed a specialized synthetic gene locus called the U2OS locus. The condensed heterochromatic (or transcriptionally off) locus was compared to a completely decondensed and transcriptionally active gene locus to 10
22 determine if TRAP150 or Btf accumulated at the transcription site in either case. (ii) Localization of TRAP150 and Btf with respect to nascent reporter RNA. The U2OS cells were transfected with a plasmid encoding YFP-MS2 to label nascent reporter RNA, and immunofluorescence was performed to label TRAP150 or Btf to determine if these proteins associate with the active gene locus. EXPECTED RESULTS: For Specific Aim 1: The percent identity/similarity is expected to be high at the amino termini because of the presence of the arginine-serine (RS) domain at the N-terminus. If stretches of identical amino acids are seen in both the proteins, perhaps conserved domains perform similar functions and homologs may be functional substitutes for each other. Conserved motifs could also serve as a binding site for some other protein involved in a process e.g. a phosphorylation site that binds to GSK3-β. For Specific Aim 2: Upon transcription inhibition by α-amanitin, nuclear speckles would be enlarged and rounded. This is because all the speckle proteins will accumulate at nuclear speckles since the transcription sites will not actively produce pre-mrna substrate that normally recruits splicing factors away from nuclear speckles. TRAP150 is expected to distinctly co-localize with the nuclear speckles since it is hypothesized to be a transcription and/or splicing factor. When looking for localization of TRAP150 at transcription sites, it is expected to co-localize with the decondensed U20S gene locus just like any other transcription factor or pre-mrna processing factor. It is also 11
23 predicted to co-localize with the reporter RNA. TRAP150 is not expected to associate with the transcriptionally inactive gene locus. 12
24 III. Methods: cdna constructs: Dr. Robert Roeder, Rockfeller University, kindly provided cdna of TRAP150. For sub-cloning, oligonucleotide primers were designed to amplify the open reading frame. The 5 203StartKpnI primer introduces a KpnI site immediately upstream 203StartKpnI 3070StopBamHI GGGGTACCTCAAAAACAAACAAATCCAA GGGATCCCTACTCGGTTGTGGGCTGTA of the start codon for translation. The StopBamHI primer introduces a BamHI site immediately downstream of the stop codon. After PCR amplification, the product was sub-cloned into the peyfp plasmid vector to create the YFP-TRAP150 fusion protein. Cell Culture: Hela cells were cultured in DMEM (Dulbecco s Modified Eagle Medium) supplemented with 10% fetal bovine serum (Hyclone) and penicillin-streptomycin. The cultures were washed with 1X Phospahate buffered saline (PBS) trypsinized and seeded onto 100 mm plates with 10 ml of medium. Passage number was maintained. U2OS cells were cultured in DMEM containing 10% Tet System Approved FBS (BD Biosciences Clontech, Palo Alto, CA) and penicillin-streptomycin. The cells were maintained in the same manner as Hela cells. Transient Transfections using the calcium phosphate method: 1x10 5 Hela cells were plated on the previous day in a 35 mm dish in 2 ml of medium. After 24 hours, a mixture of 2M Calcium Chloride, dh 2 O and 3 µg of plasmid 13
25 DNA of interest plus 1.5 µg of carrier pbluescript was added drop by drop to 1X Hepes buffer saline. The transfection mixture was then added to the cells in drop-wise fashion. 24 hrs post-transfection cells were washed with 1X PBS and fresh medium was added. Cells were fixed after 48 hrs to get maximum transfection efficiency. Transient transfection by Electroporation (for U2OS cells): Cells that reached ~70-80% confluency in 100 mm plates were trypsinized and centrifuged to pellet the cells. The pellet was resuspended in 600 µl of medium. 200 µl of cells were added to 4 mm gap cuvettes containing 40 µg of sheared salmon sperm DNA, 2 µg of pteton, and 2 µg of either CFP-lacI or psv2-yfp-laci. In some experiments, such as for observing nascent reporter RNA, 0.5 µg of YFP-MS2 binding protein/yfp-trap150/yfp-btf was also added. Electroporation was done using a Gene Pulser II (BioRad, Hercules, CA) (170 V, 950 µf). The electroporated cells were resuspended in 2 ml of medium, transferred to a 15 ml conical tube and centrifuged to pellet the cells. The cells were then resuspended in 5 ml of medium and plated 1 ml per well in a 6-well dish containing sterilized glass cover slips (rinsed with 70% EtOH and flamed). After 6 hrs of incubation at 37 o C, doxycycline (dox) was added to the cells in half of the wells to activate transcription of the reporter locus. Immunofluorescence was performed 2h after dox addition. Immunofluorescence: Cells were rinsed briefly in PBS fixed for 15 min in freshly prepared 2% formaldehyde in PBS (ph 7.4) and washed three times for 5 minutes each in 1X PBS. 14
26 Cells were permeabilized in PBS +0.2% Triton X-100 and washed three times for 5 minutes each in 1X PBS+0.5% normal goat serum. Primary antibodies were added for 1 h at room temperature: anti-thrap3 956A (anti-trap150 Bethyl Laboratories, Inc.), (1:2000), anti-sf2/asf (1:2500). Cells were rinsed in PBS + 0.5% goat serum, then secondary anti-species-specific antibodies were added for 1 h at room temperature: donkey anti mouse Texas red (1:500), donkey anti-rabbit FITC (1:500). Microscopy: U2OS cells were examined using a DeltaVision RT Microscope (Applied Precision) using Olympus IX70 inverted microscope, a 60X 1.4 n.a objective and a SONY ICX285 ER Progressive Scan CCD equipped with live cell imaging filter and CFP/YFP/mRFP and EGFP excitation and emission filters. SoftWoRx Explorer Image Viewer software was used to collect digital images. Images were acquired by collecting a stack of 0.28 µm increments in the Z-axis in all the required filters. Such a stack consisted of 4.2 µm µm 0.(15-36 sections) total depth and it contained sections, depending upon the transcription status and size of the locus. Scrolling through the stack confirms that the co-localization even in the individual images within the stack. Images were deconvolved using 10 iterations of the conservative algorithm based on the point-spread function of the 60X lens. Stacked images were then compressed for display. The images were saved as TIFF images. Individual Z-images were also examined to confirm results obtained from projected images. Scoring the association of TRAP150 or Btf with the U2OS locus: Starting from one corner of the cover slip, the cells were scored for having an open versus closed locus and also for the absence or presence of TRAP150 or Btf. The 15
27 morphology of locus was observed first under the microscope to determine the status of the locus (closed, partially open, open) by using the filter specific for the lac repressor then the localization of TRAP150/Btf was observed with respect to the locus by using the other filter. The cell was then scored accordingly. For example: # 45 Open locus TRAP150 absent Sequence Alignments: Full-length amino acid alignment of TRAP150 and Btf was done using the SIM alignment tool at The amino acid sequences of TRAP150 and Btf were used to perform the alignment. The resulting alignments can be viewed under LALNView that gives color-coded regions of similarity. 16
28 IV. RESULTS: AIM 1: To identify the potential functional domains of TRAP150. Proteins with similar amino acid sequences/conserved regions often share similar function. Hence, the higher the similarity in amino acid sequences between 2 proteins, the greater is the probability of their functional similarities. By comparing the protein sequence of newly identified speckle proteins, one can begin to understand the functions of the novel nuclear speckle proteins. In order to find potential functional domains of TRAP150, the amino acid sequence was compared with Btf (nuclear speckle component and a homolog of TRAP150) (Figure 3). Regions containing high sequence similarity suggest conserved domains, which in turn suggest regions of functionality of the protein. The alignment in Figure 3 shows 39.5% identities, 62% similarities and 5% gaps. Several stretches of identical amino acids are seen at the N-terminus and the C-terminus. High sequence identity ( 50%) is seen at amino acids (which is the RS domain for both of the proteins), 40% similarity is seen from amino acids (Figure 4). Less similarity was seen in the middle region. 40% similarity was seen from amino acid , and from amino acid there is a similarity range of 50%-80% (Figure 4). In this context, it is clear that TRAP150 and Btf share highest similarity at the N- and C- termini. Both TRAP150 and Btf have a RS domain at the N-terminus, which is a nuclear speckle-targeting signal. High similarity in the C-terminus suggests that this is a second functional domain that is not yet characterized. Motif analysis did not show up any results except for the RS domain and nuclear localization signal. 17
29 BCLF1_HUMA TR150_HUMA BCLF1_HUMA TR150_HUMA BCLF1_HUMA TR150_HUMA BCLF1_HUMA TR150_HUMA BCLF1_HUMA TR150_HUMA BCLF1_HUMA TR150_HUMA BCLF1_HUMA TR150_HUMA BCLF1_HUMA TR150_HUMA BCLF1_HUMA TR150_HUMA 4 SNSRSHS-SRSKSRSQSSSRSRSRSHSRKKRYSSRSRSRTYSRSRSRDRMYSRDYR-RDY 14 SRSRSASRSRSRSFSKSRSRSRSLSRSRKRRLSSRSRSRSYSPAHNRERNHPRVYQNRDF * *** * *** * * * ***** * *** * ******* ** * * * * ** 62 R-NNRGMRRPYGYRGRGRGYYQGGG-GRYHRGGYRPVW-NRRHSRSPRRGRSRSRSPKRR 74 RGHNRGYRRPYYFRGRNRGFYPWGQYNRGGYGNYRSNWQNYRQAYSPRRGRSRSRSPKRR * *** **** *** ** * * * * ** * * * *************** 119 SVSSQRSRSRSRRSYRSS--RSPRSSSSRSSSPYSKSPVSKRRGSQEKQTKKAEGEPQEE 134 S-PSPRSRSHSRNSDKSSSDRSRRSSSSRSSSNHSRVESSKRKSAKEKKSSSKDSRPSQA * * **** ** * ** ** ********* * *** ** * 177 SPLKSKSQEEPKDTFEHDPSESIDEFNKSSATSGDIWPGLSAYDNSPRSPHSPSPIATPP 193 AG-DNQGDEAKEQTFSGGTSQDTKASESSKPWPDATYGTGSASRASAVSELSPRERSPAL * ** * * ** * * ** 237 SQSSSCSDAPMLSTVHSAKNTPSQHSHSIQHSPERSGSGSVGNGSSRYSPSQNSPIHHIP 252 KSPLQSVVVRRRSPRPSPVPKPSPPLSSTSQMGSTLPSGAGYQSGTHQGQFDHGSGSLSP * * ** * ** * 297 SRRSPAKTIAPQNAPRDESRGRSSFYPDGGDQETAKTGKFLKRFTDEESRVFLLDRGNTR 312 SKKSPVGKSPPSTGSTYGSSQKEESAASGG---AAYTKRYLEEQKTENGKDKEQKQTNT- * ** * * ** * * * * ** 357 DKEASKEKGSEKGRAEGEWEDQEALDYFSDKESGKQKFNDSEGDDTEETED-----YRQF 368 DKEKIKEKGSFSDTGLGDGKMKS--DSFAPKTDSEKPFRGSQSPKRYKLRDDFEKKMADF *** ***** * * * * * * * * 412 RKSVLADQGKSFATASHRNTEEEGLKYKSKV---SLKGNRESDGFREEKNY------KLK 426 HKEEMDDQDKDKAKGRKESEFDDEPKFMSKVIGANKNQEEEKSGKWEGLVYAPPGKEKQR * ** * * * *** * * * * * 463 ETGYVVERPSTTKDKHKEEDKNSERITVKKETQSPEQVKSEKLKDLFDYSPPLHKNLDAR 486 KTEELEEESFPERSKKEDRGKRSEGGHRGFVPEKNFRVTAYKAVQEKSSSPPPRKTSESR * * * * ** * * *** * * BCLF1_HUMA TR150_HUMA BCLF1_HUMA TR150_HUMA BCLF1_HUMA TR150_HUMA BCLF1_HUMA TR150_HUMA BCLF1_HUMA TR150_HUMA BCLF1_HUMA TR150_HUMA BCLF1_HUMA TR150_HUMA 523 EKSTFREESPLRIKMIASDSHRPEVKLKMAPVPLDDSNRPASLTKDRLLASTLVHSVKKE 546 DKLGAKGDFPTG-KSSFSITREAQVNVRMDSFDEDLARPSGLLAQERKLCRDLVHSNKKE * * * * * * * * * * **** *** 583 QEFRSIFDHIKLPQASKSTSESFIQHIVSLVHHVKEQYFKSAAMTLNERFTSYQK--ATE 605 QEFRSIFQHIQSAQSQRSPSELFAQHIVTIVHHVKEHHFGSSGMTLHERFTKYLKRGTEQ ******* ** * * ** * **** ****** * * *** **** * * 641 EHSTRQKSPEIHRRIDISPSTLRKHTRLAGEERVFKEENQKGDKKLRCDSADLRHDIDRR 665 EAAKNKKSPEIHRRIDISPSTFRKHGLAHDEMKSPREPGYKAEGKYKDDPVDLRLDIERR * *************** *** * * * * * *** ** ** 701 RK--ERSKERGDSKGSRES--SGSRKQEKTPKDYKEYKSYKDDSKHKREQDHSRSSSSSA 725 KKHKERDLKRGKSRESVDSRDSSHSRERSAEKTEKTHKGSKKQKKHRRARDRSRSSSSSS * ** ** * * * * * * * * ** * * ******* 757 SPSSPSSREEKESKKEREEEFKTHHEM-----KEYSGFA--GVSRPRGTF-FRIRGRGRA 785 QSSHSYKAEEYTEETEEREESTTGFDKSRLGTKDFVGPSERGGGRARGTFQFRARGRGWG * ** * ** * * * * * **** ** **** 809 RGVFAGTNTGPNNSNTTFQKRPKEEEWDPEYTPKSKKYFLHDDRD-DGVDYWAKRGRGRG 845 RGNYSGNNN--NNSNNDFQKRNREEEWDPEYTPKSKKYYLHDDREGEGSDKWVSRGRGRG ** * * **** **** *************** ***** * * * ****** 868 TFQRGRGRFNFKKSGSSPKWTHDKYQGD-GIVEDEEETMENNEEK 903 AFPRGRGRFMFRKSSTSPKWAHDKFSGEEGEIEDDESGTENREEK * ****** * ** **** *** * * ** * ** *** Figure 3: BCLF-1 is Bcl-2 associated transcription factor and TR150 is Thyroid hormone receptor associated transcription factor (TRAP150). The boxes in red and blue border a GSK3β site and a CHD1 interaction region respectively which are conserved in both the proteins. 39.5% identity in 945 residues overlap; Score: ; Gap frequency: 5.1%. 18
30 19
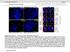
31 AIM 2: To determine if TRAP150 is associated with transcription. Localization of TRAP150 TRAP150 was identified as a novel nuclear speckle protein (Mintz et al., 1999; Saitoh et al., 2004). In order to observe at the localization of TRAP150 with respect to nuclear speckles, dual-labeling immunofluorescence was performed using anti-rabbit anti-trap A antibody from Bethyl Laboratories and anti-sf2/asf (splicing factor2/alternative splicing factor is a bonafide nuclear speckle protein). The results showed that endogenous TRAP150 localizes all over the nucleoplasm in foci or punctate pattern that resembles the localization pattern of transcription sites (Figure 5). In some cases, enrichment of TRAP150 is also seen near the periphery of nuclear speckles. These are also sites that have been previously reported as regions of high transcription activity (Wei et al., 1999; Xing et al., 1993). 20
32 TRAP150 SF2/ASF Merge DAPI Figure 5. 1 X 10 5 Hela cells were plated in a 6-well dish and processed for IF after 24 hours using rabbit anti-trap150/thrap3 956A polyclonal antibody (Bethyl labs) and mouse anti- SF2/ASF monoclonal antibody (AK103, A. Krainer). TRAP150 showed some overlap with SF2/ASF in nuclear speckles, and showed additional small foci throughout the nucleoplasm. DNA was stained with DAPI. Scale Bar= 5µm. with DAPI. Scale Bar= 5µm. In order to further study TRAP150 localization, TRAP150 cdna was PCR amplified sub-cloned into peyfp-c1. The sub-cloned DNA was then expressed in HeLa cells and its localization was observed. TRAP150 localization was strictly nuclear. Some co-localization of TRAP150 with splicing factor SF2/ASF was observed in nuclear speckles (Figures 5 and 6). The localization of YFP-TRAP150 and endogenous TRAP150 was observed in order to validate the localization of YFP-TRAP150 with respect to nuclear speckles. 21
33 YFP-TRAP150 SF2/ASF Merge DAPI Figure 6: YFP-TRAP150 was transfected into Hela cells using the Calcium phosphate method and IF was performed to label nuclear speckles using SF2/ASF antibody. The localization of YFP-TRAP is the same as endogenous TRAP150. DNA was stained with DAPI. Scale Bar= 5µm. AIM 2A: To examine if the localization of TRAP150 is altered after inhibition of RNA polymerase II transcription. Since I hypothesized that TRAP150 may have a role in pre-mrna transcription, I examined the localization of TRAP150 upon transcription inhibition. Alpha-amanitin arrests transcription of pre-mrna by inhibiting RNA polymerase II (Kedinger et al., 1970; Lindell et al., 1970). α-amanitin is a cyclic octapeptide that binds with a high affinity to the largest subunit of RNA pol II, RPBI. It blocks the binding among subunits, preventing them from forming the RNA polymerase II complex during the initiation of transcription (Cochet-Meilhac and Chambon, 1974; Lutter, 1982). After treating Hela cells with α-amanitin for 6h, immunofluorescence was performed to label U1-70K, a component of the U1 SnRNP that localizes to nuclear speckles (Figure 7). 22
34 + Amanitin A B C D U1-70K TRAP150 Merge DAPI - Amanitin - U1-70K TRAP150 Merge DAPI Figure 7. 1X 10 5 Hela cells were seeded onto cover slips for 24 hours, and 50µg/ml α- amanitin was added for 6h prior to processing for immunofluorescence localization of TRAP150 and U1-70K. After transcription inhibition, TRAP150 accumulates in enlarged nuclear speckles. DNA was stained with DAPI. Scale Bar= 5µm. In amanitin-treated cells, nuclear speckles appear larger and rounder than untreated cells, as indicated by arrows in the insets in Figure 7 E and M. It appears that TRAP150 was targeted to nuclear speckles in the amanitin-treated cells contrary to its punctate or foci-like localization in the nucleoplasm in the untreated cells (shown by arrow-heads in the insets in Figure 7 G and O respectively). Hence, in a transcriptionally active nucleus, most of the components of nuclear speckles including TRAP150 are dynamic and localize throughout the nucleoplasm (Misteli et al., 1994). In transcription inhibited nuclei that are devoid of any splicing factor substrates, pre-mrna processing factors are devoid of their sites of function, they tend to accumulate at their storage sites 23
35 in the nuclear speckles. Here, TRAP150 exhibited similar enrichment in nuclear speckles as U-170K snrnp upon inhibition of RNA pol II. AIM 2B: To determine if TRAP150 and Btf are recruited to transcription sites in vivo. Since the previous results are consistent with a possible role for TRAP150 in gene regulation, we employed a more direct approach to study the localization of TRAP150 at a single gene locus. A synthetic gene locus called the locus stably incorporated in human U2OS cells was used to determine if TRAP150 is present at the inactive and/or the active locus. A range of morphologies is seen upon induction of transcription of the locus. These morphologies are illustrated in Figure 8. The U2OS cells were transfected with YFP/CFP-tagged Lac repressor (YFP-LacI or CFP-LacI) and the tetracyclineinducible transcriptional activator pteton. Doxycycline was added 6h post-transfection to induce transcription at the reporter locus. After treating the cells with doxycycline for 1.5-2h, the cells were fixed and processed for microscopy. The closed locus is highly condensed and heterochromatic (Janicki et al., 2004) and it appears like a single tightly condensed spot in the nucleus as seen in Figure 8A. 24
36 A B C Locus Morphology Closed Partially Open Open D E F G DAPI stained nucleus Figure 8: Different morphologies of the U2OS locus. The YFP-LacI repressor labeled gene locus has 3 different morphologies. Panel A shows a condensed locus that is heterochromatic. B is a partially open locus and C shows a transcriptionally active completely decondensed locus. Panels D,E and F show DAPI staining of these nuclei. Scale bar = 5 µm. The transcriptionally active loci showed a range from partially decondensed (Figure 8B) to completely open and decondensed (Figure 8C). The locus in each individual nucleus is potentially induced or activated at different times following dox addition in the living cells. This is one explanation for why different morphologies of open loci were observed in fixed cells. The partially open morphology is likely a transitional state that might be either a transcriptionally active locus beginning to decondense or an open locus beginning to condense. Because it takes hrs for this reporter locus to completely to decondense, the second possibility is unlikely only 2 hrs after inducing transcription (Janicki et al., 2004). 25
37 AIM 2B-i: TRAP150 and Btf at U2OS gene locus: Specific aim 2B of this project was to examine if TRAP150 and Btf localizes at the transcriptionally inactive or activated gene locus. U2OS cells were transfected with YFP-LacI along with ptet-on. Doxycycline was added to half of the coverslips 6h after transfection and cells were processed for microscopy 2h after dox addition. In the nuclei of cells treated with doxycycline, TRAP150 was enriched at the completely decondensed locus, where it distinctly localized at the vicinity of the locus as shown in Figure 9C. A B C A B C D TRAP150 LacI Merge DAPI E F G H I J K I J K L M N O TRAP150 LacI Merge DAPI M N O P Figure 9: U2OS cells were transiently transfected with psv2-yfp-laci repressor and ptet-on (rtta). Doxycycline was added 6h post transfection. Cells were fixed 2h after dox addition and immunolabeled for TRAP150. Panels A-D show dox-induced cells with an activated locus. Panels I-L show the inactive locus. In panel B, the locus is decondensed and open in comparison to panel J where the locus is closed (arrowheads indicate the position of the locus in B and J). Panels A and I show endogenous TRAP150. The overlay images in C and K show the localization of TRAP150 with respect to the lac repressor that labels the DNA at the locus. TRAP150 is enriched at the decondensed 26
38 active locus (panel C, arrow), but is not enriched at the condensed inactive locus (panel K, arrow). Panels (E-H) and (M-P) are enlarged insets to panels (A-D) and (I-L), respectively. DNA was stained with DAPI. Scale Bar = 5µm. In contrast, at the heterochromatic locus (i.e., nuclei transfected with pteton and YFPlacI but without dox addition), no such enrichment of TRAP150 was seen (Figure 9K). Similar results were seen with TRAP150 homolog Btf. In the nuclei of cells treated with doxycyline, Btf was localized at the vicinity of the locus (Figure 10C), while Btf enrichment was not seen at the transcriptionally inactive locus (Figure 10K). A B C D +DO Btf LacR Merge DAPI E F G H I J K L - Btf LacR Merge DAPI M N O P Figure 10: U2OS cells were transiently transfected with psv2-yfp-laci, and ptet-on (rtta). Doxycycline was added 6h post transfection. Cells were fixed 2h after Dox addition and Btf was immuno-labeled with rabbit anti-btf WU10 polyclonal antibody. Arrowheads indicate the position of the locus in panels B and J. Panels A-D show Btf enrichment at the activated locus, while panels I-L do not show enrichment of Btf at the inactive locus (arrow in I). E-H and M-P are insets of A-D and I-L respectively. DNA is stained with DAPI. Scale Bar = 5µm. 27
39 Mammalian gene regulation is controlled by regulatory mechanims that modulate chromatin structure. The first level of chromatin condensation looks like beads on a string, which consists of DNA wound around histone octamers into the basic unit of chromatin called a nucleosome. There are eight histone proteins - two molecules each of H2A, H2B, H3 and H4 (Alberts et al., 2002). Two H2A-H2B dimers form a tetramer, which bind with a H3-H4 tetramer to form a core histone octamer (Alberts et al., 2002). The DNA wraps 1.65 times around the histone core to form nucleosomes that are separated by a short stretch of linker DNA. Formation of nucleosomes reduces the length of the DNA to 1/3 of its original length. Nucleosomes are further packed into a 30-nm fiber as histone NH 2 -terminal tails interact with neighboring nucleosomes. Regulation of these interactions by chromatin remodeling enzyme complexes controls the accessibility of promoter and enhancer elements, and ultimately the activity of genes. The U2OS transgene array is stably integrated into a single region on chromosome 1. The large number of plasmid copies in the array results in makes it a huge gene locus. This transgene array is heterochromatic during its inactive state (OFF), when it exibhits higher order compaction and is inaccessible to the transcription machinery. The inactive locus is associated with histone methyl transferases and heterochromatin protein 1 (HP1) (Janicki et al., 2004). The methylation of lysine 9 on histone H3 creates a binding site for the chromodomain of HP1 hence silencing the gene (Jacobs and Khorasanizadeh, 2002). Observations of the inactive locus show that it appears as a single tight spot in the nucleus. When the gene locus is turned ON upon transcription activation the methylated Histone 3 is exchanged with H3.3 variant. This exchange removes methylated histone H3, replacing it with H3.3 whose tail is not 28
40 methylated. This first step inlocus activation decondenses the chromatin for it to be accessible for transcription machinery (Ahmad and Henikoff, 2002). The transcription activator (rtta) binds to the tetracycline response elements (upon addition of doxycycline) and recruits gene expression machinery to the promoter region. When the locus is turned on completely, it has a large and irregular morphology with many lobes. Because not all copies of the transgene are simultaneously activated, there is usually a delay in the total activation of the locus. Some copies of the transgene persist in the inactive state hence, the intermediate morphologies of the locus that had only 2-3 lobes scored as partially open were observed. When transcription is activated and the locus is decondensed (either completely or partially), TRAP150 was frequently seen in the vicinity of the locus. In order to confirm that TRAP150 is recruited to the transcriptionally active reporter gene locus, it was imperative to score a significant number of cells with enrichment of TRAP150 at the partially open or open locus. Also, to conclude that TRAP150 is recruited to the transcriptionally active reporter gene locus, it should be absent from the inactive locus. Cells were scored according to the status of the locus (active versus inactive) and the association with TRAP150 or Btf. Results are shown in Tables 1 and 2. For scoring, TRAP150 association with the U2OS locus, +Dox/+pTet-ON (active) and -Dox/-pTet-ON (inactive) were scored for TRAP150 and Btf in separate experiments. In a +Dox experiment, to examine TRAP150 association, a total of 87 cells were scored. Of these 15 cells had a completely open locus and 72 had a partially open locus (Table 1B). Out of the 15 nuclei with completely open/decondensed loci, 11 (73.3%) of 29
41 them had TRAP150 enrichment and 4 did not have TRAP150 enrichment. Out of 72 nuclei with partially open loci, 45 (62.5%) of these loci had TRAP150 enrichment while 27 nuclei (37.5%) of the loci had no TRAP150 association. In a Dox experiment to examine TRAP150 association, 98 cells were scored. 30 of these loci were closed and 68 were partially open. Only 6.6% of these 30 closed loci had TRAP150 association, while 93.4% did not show TRAP150 association. A Condition of the experiment Status of the Locus Number of cells % of loci associated with TRAP150 % of loci with TRAP150 absent +DOX Open % 35.6% -DOX Closed % 93.4% B Condition of the experiment Status of Locus Number of cells/number of cells with TRAP150 enrichment % Recruitment +DOX Open Partially Open 15/ 11 72/ % 62.5% -DOX Close Partially Open 30/2 68/18 6.6% 26.6% Table 1: TRAP150 at the reporter locus. A. Comparison between +Dox and -DOX experiments regarding loci association with TRAP150. The number of cells scored with such association is shown for both cases. The percentages of the presence or absence of TRAP150 for each condition are also given. B. Further analysis of Table A. In a +Dox experiment, the numbers for an open locus containing cells is again divided into partially open and open loci. Also, for Dox experiment, the types of loci observed were close and partially open. The number of cells scored and the percentages given in the Table 1.A are classified accordingly in Table 1.B. TRAP150 was enriched on 26.6% of the 68 partially open loci. Comprehensively, 64.36% of nuclei with an open locus in a +Dox experiment have TRAP150 enrichment at 30
42 the locus, while 6.6% of nuclei in a Dox experiment had TRAP150 association with a closed locus (Table 1A). While 35.6% of open loci containing nuclei in a +Dox experiment did not have TRAP150 at the vicinity of the locus, 93.4% of the nuclei with closed loci in a Dox experiment were without TRAP150 enrichment. In an ideal experiment, when transcription is activated, the gene locus is decondensed and acquires an open conformation. If TRAP150 is predicted to be a transcription activator or a premrna processing factor, it would be expected to always co-localize at the gene locus. If TRAP150 is not involved in transcription/pre-mrna processing, there would be no such enrichment. If TRAP150 is associated with transcription repression, it would be expected to co-localize with an OFF/condensed locus. Our experiment showed that there was a higher percentage of cells with TRAP150 enrichment to open loci than cells with closed loci. However, 35.6% of the cells with open loci were not seen with TRAP150 association. The reason could be that most of the cells counted had partially open conformation (Table 1B), suggesting that some of the (45 out of 72) partially open loci might not have been activated enough to associate with TRAP150 at the time of fixation. A higher percentage of nuclei with completely open loci associated with TRAP150 (11 out of 15) also reinforces the idea. If TRAP150 is predicted to be a transcription repressor, a high percentage of cells would have had TRAP150 enrichment at an OFF/inactive/closed locus. However, the finding that only 6.6% of closed loci had TRAP150 association would suggest that TRAP150 does not associate with inactive genes. While the locus should be completely turned off when its activity has not been intentionally induced by addition of Dox to the culture medium. The relatively small number of closed loci (6.6%) that showed TRAP150 association could be explained by 31
43 leaky transcription from the inducible promoter. I observed a significant difference in the percentage of partially open loci in a DOX experiment versus a +Dox experiment with regard to TRAP150 association 62.5% of loci in a +DOX experiment had TRAP150 association with partially open loci, versus only 26.6% of nuclei in a DOX experiment. Once again this suggests a difference in the extent of activation of the locus at the time of fixation, or could more importantly reflect that the intentionally activated loci were in some way more competent for recruiting gene expression machinery. A Condition of the experiment Status of the Locus Number of cells % of loci associated with Btf % of loci with Btf absent +DOX Open % 38.9% -DOX Close 72 7% 93% B Condition of the experiment Status of Locus Number of cells/number of cells with Btf enrichment % Recruitment +DOX Open Partially Open 18/ 12 72/ 43 67% 60% -DOX Close Partially Open 72/5 28/10 7% 36% Table 2: Btf at the reporter locus. A. Comparison of lociʼs association with Btf between +Dox and Dox experiments. The number of cells scored with such association is shown for both cases. The percentages of the presence or absence of Btf for each condition are also given. B. Further analysis of Table A. In a +Dox experiment, the numbers for an open locus containing cells is again divided into partially open and open loci. Likewise, for Dox experiment, the types of loci observed were close and partially open. Accordingly, percentages of cells with Btf enrichment at each of the conditions of the loci in both +/- Dox experiments were noted. 32
44 Cell counts for the absence/presence of Btf were done in order to examine Btf association with the U2OS locus. In a +Dox experiment, 67% of the nuclei scored showed Btf association with the open locus, while in a Dox experiment, only 7% of the nuclei showed Btf recruitment at the closed locus. Also, in a Dox experiment, Btf was absent from 93% of the closed loci while it was absent from 39% of the open/partially open loci. Ideally, if Btf functions as a transcription activator/pre-mrna processing factor, we would expect it to always co-localize with the open/on/decondensed locus. Our result showed 61.1% of open loci (scored in 90 nuclei) with Btf association. Of those 90 nuclei with open loci, most of them (72) were partially open. Considering the fact that partially open loci are not completely decondensed, the probability of transcription factors/pre-mrna processing factors accessing the DNA is less. Hence, this may explain why fewer open loci than expected with Btf. 7% of nuclei in a Dox experiment showed Btf association suggesting that there is possibly a leakiness in the system. Partially open loci are generally not expected in a Dox experiment yet they were observed. The reason could be once again basal activity that could have decondensed the loci partially. While 60% of nuclei in a +Dox experiment with partially open loci had Btf association, 36% of the nuclei in a Dox experiment with partially open loci had Btf association. This suggests once again that the partially open loci that had intentionally been activated are somehow more competent for gene activation. 33
45 AIM 2B-ii: TRAP150 and Btf at RNA: Next, the localization of TRAP150 and Btf with respect to U2OS reporter RNA was examined. U2OS cells were transfected with plasmids encoding YFP- MS2 and ptet-on (rtta). YFP-MS2 binding protein reporter labels the RNA upon induction of transcription by doxycycline. YFP-MS2 binding protein has a diffuse pattern all over the nucleoplasm in cells with inactive loci, but it accumulates at the activated RNA of the reporter gene locus. No such accumulation of YFP-MS2 at the reporter locus was seen in a Dox experiment (data not shown), and was not expected since transcription was not induced. The localization of TRAP150, Btf and SF2/ASF (a splicing factor) with respect to the reporter RNA was the objective of this sub-aim. SF2/ASF is an essential splicing factor that associates with nascent pre-mrna and it was used as a positive control for these experiments. We would expect to see SF2/ASF exactly colocalized with nascent reporter RNA on the activated reporter locus. 34
46 A TRAP150 YFP MS2 Merge DAPI B C mcherryms2 Merge YFP-SF2 Figure 11. U2OS cells were transiently transfected with pteton (rtta), YFP- or Cherry- MS2 binding protein. Doxycycline was added 6 hours post transfection and the cells were processed 2 hours later for immunofluorescence localization of A) Btf, B) TRAP150 and C) SF2/ASF. While all three proteins are recruited to the immediate vicinity of the RNA, there is only partial overlap between these proteins and the MS2-binding protein. DNA was stained with DAPI. Scale bar = 5µm. 35
Eukaryotic transcription (III)
 Eukaryotic transcription (III) 1. Chromosome and chromatin structure Chromatin, chromatid, and chromosome chromatin Genomes exist as chromatins before or after cell division (interphase) but as chromatids
Eukaryotic transcription (III) 1. Chromosome and chromatin structure Chromatin, chromatid, and chromosome chromatin Genomes exist as chromatins before or after cell division (interphase) but as chromatids
supplementary information
 DOI: 10.1038/ncb2157 Figure S1 Immobilization of histone pre-mrna to chromatin leads to formation of histone locus body with associated Cajal body. Endogenous histone H2b(e) pre-mrna is processed with
DOI: 10.1038/ncb2157 Figure S1 Immobilization of histone pre-mrna to chromatin leads to formation of histone locus body with associated Cajal body. Endogenous histone H2b(e) pre-mrna is processed with
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression. Chromatin Array of nuc
 Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
The In Vivo Kinetics of RNA Polymerase II Elongation during Co-Transcriptional Splicing
 The In Vivo Kinetics of RNA Polymerase II Elongation during Co-Transcriptional Splicing Yehuda Brody 1, Noa Neufeld 1, Nicole Bieberstein 2, Sebastien Z. Causse 3, Eva-Maria Böhnlein 2, Karla M. Neugebauer
The In Vivo Kinetics of RNA Polymerase II Elongation during Co-Transcriptional Splicing Yehuda Brody 1, Noa Neufeld 1, Nicole Bieberstein 2, Sebastien Z. Causse 3, Eva-Maria Böhnlein 2, Karla M. Neugebauer
Mechanisms of alternative splicing regulation
 Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
Eukaryotic Gene Regulation
 Eukaryotic Gene Regulation Chapter 19: Control of Eukaryotic Genome The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different,
Eukaryotic Gene Regulation Chapter 19: Control of Eukaryotic Genome The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different,
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004
 Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Chapter 10 - Post-transcriptional Gene Control
 Chapter 10 - Post-transcriptional Gene Control Chapter 10 - Post-transcriptional Gene Control 10.1 Processing of Eukaryotic Pre-mRNA 10.2 Regulation of Pre-mRNA Processing 10.3 Transport of mrna Across
Chapter 10 - Post-transcriptional Gene Control Chapter 10 - Post-transcriptional Gene Control 10.1 Processing of Eukaryotic Pre-mRNA 10.2 Regulation of Pre-mRNA Processing 10.3 Transport of mrna Across
Where Splicing Joins Chromatin And Transcription. 9/11/2012 Dario Balestra
 Where Splicing Joins Chromatin And Transcription 9/11/2012 Dario Balestra Splicing process overview Splicing process overview Sequence context RNA secondary structure Tissue-specific Proteins Development
Where Splicing Joins Chromatin And Transcription 9/11/2012 Dario Balestra Splicing process overview Splicing process overview Sequence context RNA secondary structure Tissue-specific Proteins Development
Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression
 Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Polyomaviridae. Spring
 Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
MOLECULAR CELL BIOLOGY
 1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
MCB Chapter 11. Topic E. Splicing mechanism Nuclear Transport Alternative control modes. Reading :
 MCB Chapter 11 Topic E Splicing mechanism Nuclear Transport Alternative control modes Reading : 419-449 Topic E Michal Linial 14 Jan 2004 1 Self-splicing group I introns were the first examples of catalytic
MCB Chapter 11 Topic E Splicing mechanism Nuclear Transport Alternative control modes Reading : 419-449 Topic E Michal Linial 14 Jan 2004 1 Self-splicing group I introns were the first examples of catalytic
Chapter 11 How Genes Are Controlled
 Chapter 11 How Genes Are Controlled PowerPoint Lectures for Biology: Concepts & Connections, Sixth Edition Campbell, Reece, Taylor, Simon, and Dickey Copyright 2009 Pearson Education, Inc. Lecture by Mary
Chapter 11 How Genes Are Controlled PowerPoint Lectures for Biology: Concepts & Connections, Sixth Edition Campbell, Reece, Taylor, Simon, and Dickey Copyright 2009 Pearson Education, Inc. Lecture by Mary
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system
 Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
RNA Processing in Eukaryotes *
 OpenStax-CNX module: m44532 1 RNA Processing in Eukaryotes * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you
OpenStax-CNX module: m44532 1 RNA Processing in Eukaryotes * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you
Genetics. Instructor: Dr. Jihad Abdallah Transcription of DNA
 Genetics Instructor: Dr. Jihad Abdallah Transcription of DNA 1 3.4 A 2 Expression of Genetic information DNA Double stranded In the nucleus Transcription mrna Single stranded Translation In the cytoplasm
Genetics Instructor: Dr. Jihad Abdallah Transcription of DNA 1 3.4 A 2 Expression of Genetic information DNA Double stranded In the nucleus Transcription mrna Single stranded Translation In the cytoplasm
Optimization of the Fuse-It-mRNA Protocol for L929 Cells in the µ-plate 24 Well
 Optimization of the Fuse-It-mRNA Protocol for L929 Cells in the µ-plate 24 Well 1. General Information... 1 2. Background... 1 3. Material and Equipment Required... 2 4. Experimental Procedure and Results...
Optimization of the Fuse-It-mRNA Protocol for L929 Cells in the µ-plate 24 Well 1. General Information... 1 2. Background... 1 3. Material and Equipment Required... 2 4. Experimental Procedure and Results...
Supplementary Information. Preferential associations between co-regulated genes reveal a. transcriptional interactome in erythroid cells
 Supplementary Information Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells Stefan Schoenfelder, * Tom Sexton, * Lyubomira Chakalova, * Nathan
Supplementary Information Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells Stefan Schoenfelder, * Tom Sexton, * Lyubomira Chakalova, * Nathan
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Chromatin position in HepG2 cells: Although being non-random, significantly changed in daughter cells
 Chromatin position in HepG2 cells: Although being non-random, significantly changed in daughter cells Ivan Raška mother cell nucleus Charles University in Prague, First Faculty of Medicine, and Institute
Chromatin position in HepG2 cells: Although being non-random, significantly changed in daughter cells Ivan Raška mother cell nucleus Charles University in Prague, First Faculty of Medicine, and Institute
Alpha thalassemia mental retardation X-linked. Acquired alpha-thalassemia myelodysplastic syndrome
 Alpha thalassemia mental retardation X-linked Acquired alpha-thalassemia myelodysplastic syndrome (Alpha thalassemia mental retardation X-linked) Acquired alpha-thalassemia myelodysplastic syndrome Schematic
Alpha thalassemia mental retardation X-linked Acquired alpha-thalassemia myelodysplastic syndrome (Alpha thalassemia mental retardation X-linked) Acquired alpha-thalassemia myelodysplastic syndrome Schematic
Genetics and Genomics in Medicine Chapter 6 Questions
 Genetics and Genomics in Medicine Chapter 6 Questions Multiple Choice Questions Question 6.1 With respect to the interconversion between open and condensed chromatin shown below: Which of the directions
Genetics and Genomics in Medicine Chapter 6 Questions Multiple Choice Questions Question 6.1 With respect to the interconversion between open and condensed chromatin shown below: Which of the directions
MODULE 3: TRANSCRIPTION PART II
 MODULE 3: TRANSCRIPTION PART II Lesson Plan: Title S. CATHERINE SILVER KEY, CHIYEDZA SMALL Transcription Part II: What happens to the initial (premrna) transcript made by RNA pol II? Objectives Explain
MODULE 3: TRANSCRIPTION PART II Lesson Plan: Title S. CATHERINE SILVER KEY, CHIYEDZA SMALL Transcription Part II: What happens to the initial (premrna) transcript made by RNA pol II? Objectives Explain
Transcription and RNA processing
 Transcription and RNA processing Lecture 7 Biology 3310/4310 Virology Spring 2018 It is possible that Nature invented DNA for the purpose of achieving regulation at the transcriptional rather than at the
Transcription and RNA processing Lecture 7 Biology 3310/4310 Virology Spring 2018 It is possible that Nature invented DNA for the purpose of achieving regulation at the transcriptional rather than at the
BIO 5099: Molecular Biology for Computer Scientists (et al)
 BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
Biology. Lectures winter term st year of Pharmacy study
 Biology Lectures winter term 2008 1 st year of Pharmacy study 3 rd Lecture Chemical composition of living matter chemical basis of life. Atoms, molecules, organic compounds carbohydrates, lipids, proteins,
Biology Lectures winter term 2008 1 st year of Pharmacy study 3 rd Lecture Chemical composition of living matter chemical basis of life. Atoms, molecules, organic compounds carbohydrates, lipids, proteins,
L6 GLUT4myc Cell Growth Protocol
 L6 GLUT4myc Cell Growth Protocol Background: Parental L6 cells selected for high fusion (2, 3) were stably transfected with a rat GLUT4 cdna carrying a myc epitope (recognized by the commercially available
L6 GLUT4myc Cell Growth Protocol Background: Parental L6 cells selected for high fusion (2, 3) were stably transfected with a rat GLUT4 cdna carrying a myc epitope (recognized by the commercially available
Molecular Cell Biology - Problem Drill 10: Gene Expression in Eukaryotes
 Molecular Cell Biology - Problem Drill 10: Gene Expression in Eukaryotes Question No. 1 of 10 1. Which of the following statements about gene expression control in eukaryotes is correct? Question #1 (A)
Molecular Cell Biology - Problem Drill 10: Gene Expression in Eukaryotes Question No. 1 of 10 1. Which of the following statements about gene expression control in eukaryotes is correct? Question #1 (A)
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment
 Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
Lecture 10. Eukaryotic gene regulation: chromatin remodelling
 Lecture 10 Eukaryotic gene regulation: chromatin remodelling Recap.. Eukaryotic RNA polymerases Core promoter elements General transcription factors Enhancers and upstream activation sequences Transcriptional
Lecture 10 Eukaryotic gene regulation: chromatin remodelling Recap.. Eukaryotic RNA polymerases Core promoter elements General transcription factors Enhancers and upstream activation sequences Transcriptional
L I F E S C I E N C E S
 1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 NOVEMBER 2, 2006
1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 NOVEMBER 2, 2006
Instructions. Fuse-It-mRNA easy. Shipping and Storage. Overview. Kit Contents. Specifications. Note: Important Guidelines
 Membrane fusion is a highly efficient method for transfecting various molecules and particles into mammalian cells, even into sensitive and primary cells. The Fuse-It reagents are cargo-specific liposomal
Membrane fusion is a highly efficient method for transfecting various molecules and particles into mammalian cells, even into sensitive and primary cells. The Fuse-It reagents are cargo-specific liposomal
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Epigenetics. Lyle Armstrong. UJ Taylor & Francis Group. f'ci Garland Science NEW YORK AND LONDON
 ... Epigenetics Lyle Armstrong f'ci Garland Science UJ Taylor & Francis Group NEW YORK AND LONDON Contents CHAPTER 1 INTRODUCTION TO 3.2 CHROMATIN ARCHITECTURE 21 THE STUDY OF EPIGENETICS 1.1 THE CORE
... Epigenetics Lyle Armstrong f'ci Garland Science UJ Taylor & Francis Group NEW YORK AND LONDON Contents CHAPTER 1 INTRODUCTION TO 3.2 CHROMATIN ARCHITECTURE 21 THE STUDY OF EPIGENETICS 1.1 THE CORE
SR Proteins. The SR Protein Family. Advanced article. Brenton R Graveley, University of Connecticut Health Center, Farmington, Connecticut, USA
 s s Brenton R Graveley, University of Connecticut Health Center, Farmington, Connecticut, USA Klemens J Hertel, University of California Irvine, Irvine, California, USA Members of the serine/arginine-rich
s s Brenton R Graveley, University of Connecticut Health Center, Farmington, Connecticut, USA Klemens J Hertel, University of California Irvine, Irvine, California, USA Members of the serine/arginine-rich
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
Annexin V-PE Apoptosis Detection Kit
 Annexin V-PE Apoptosis Detection Kit Catalog Number KA0716 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information...
Annexin V-PE Apoptosis Detection Kit Catalog Number KA0716 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information...
INTERACTION DRUG BODY
 INTERACTION DRUG BODY What the drug does to the body What the body does to the drug Receptors - intracellular receptors - membrane receptors - Channel receptors - G protein-coupled receptors - Tyrosine-kinase
INTERACTION DRUG BODY What the drug does to the body What the body does to the drug Receptors - intracellular receptors - membrane receptors - Channel receptors - G protein-coupled receptors - Tyrosine-kinase
Supplemental Information. 3D-CLEM Reveals that a Major Portion. of Mitotic Chromosomes Is Not Chromatin
 Molecular Cell, Volume 64 Supplemental Information 3D-CLEM Reveals that a Major Portion of Mitotic Chromosomes Is Not Chromatin Daniel G. Booth, Alison J. Beckett, Oscar Molina, Itaru Samejima, Hiroshi
Molecular Cell, Volume 64 Supplemental Information 3D-CLEM Reveals that a Major Portion of Mitotic Chromosomes Is Not Chromatin Daniel G. Booth, Alison J. Beckett, Oscar Molina, Itaru Samejima, Hiroshi
TFEB-mediated increase in peripheral lysosomes regulates. Store Operated Calcium Entry
 TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
Eukaryotic mrna is covalently processed in three ways prior to export from the nucleus:
 RNA Processing Eukaryotic mrna is covalently processed in three ways prior to export from the nucleus: Transcripts are capped at their 5 end with a methylated guanosine nucleotide. Introns are removed
RNA Processing Eukaryotic mrna is covalently processed in three ways prior to export from the nucleus: Transcripts are capped at their 5 end with a methylated guanosine nucleotide. Introns are removed
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature05473 SUPPLEMENTARY FIGURES SUPPLEMENTARY INFORMATION Supplementary Figure 1: Association of Runx2 with mitotic chromosomes Mitotic chromosome spreads were prepared for a. Human Saos-2
doi: 10.1038/nature05473 SUPPLEMENTARY FIGURES SUPPLEMENTARY INFORMATION Supplementary Figure 1: Association of Runx2 with mitotic chromosomes Mitotic chromosome spreads were prepared for a. Human Saos-2
EPIGENTEK. EpiQuik Global Histone H4 Acetylation Assay Kit. Base Catalog # P-4009 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik Global Histone H4 Acetylation Assay Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Global Histone H4 Acetylation Assay Kit is suitable for specifically measuring global
EpiQuik Global Histone H4 Acetylation Assay Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Global Histone H4 Acetylation Assay Kit is suitable for specifically measuring global
Transcription and chromatin. General Transcription Factors + Promoter-specific factors + Co-activators
 Transcription and chromatin General Transcription Factors + Promoter-specific factors + Co-activators Cofactor or Coactivator 1. work with DNA specific transcription factors to make them more effective
Transcription and chromatin General Transcription Factors + Promoter-specific factors + Co-activators Cofactor or Coactivator 1. work with DNA specific transcription factors to make them more effective
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being A Eukaryote. Eukaryotic Cells. Basic eukaryotes have:
 BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
Supplementary Fig. S1. Schematic diagram of minigenome segments.
 open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
Regulation of Gene Expression in Eukaryotes
 Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Identification and characterization of multiple splice variants of Cdc2-like kinase 4 (Clk4)
 Identification and characterization of multiple splice variants of Cdc2-like kinase 4 (Clk4) Vahagn Stepanyan Department of Biological Sciences, Fordham University Abstract: Alternative splicing is an
Identification and characterization of multiple splice variants of Cdc2-like kinase 4 (Clk4) Vahagn Stepanyan Department of Biological Sciences, Fordham University Abstract: Alternative splicing is an
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion
 Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
U3.2.3: Eukaryotic chromosomes are linear DNA molecules associated with histone proteins. (Oxford Biology Course Companion page 151).
 Cell Division Study Guide U3.2.3: Eukaryotic chromosomes are linear DNA molecules associated with histone proteins. (Oxford Biology Course Companion page 151). 1. Describe the structure of eukaryotic DNA
Cell Division Study Guide U3.2.3: Eukaryotic chromosomes are linear DNA molecules associated with histone proteins. (Oxford Biology Course Companion page 151). 1. Describe the structure of eukaryotic DNA
Organization of genetic material in eukaryotes
 Organization of genetic material in eukaryotes biologiemoleculara.usmf.md pass.: bmgu e.usmf.md 1 DNA in eukaryotes Location: In nucleus In mitochondria biologiemoleculara.usmf.md e.usmf.md pass.: bmgu
Organization of genetic material in eukaryotes biologiemoleculara.usmf.md pass.: bmgu e.usmf.md 1 DNA in eukaryotes Location: In nucleus In mitochondria biologiemoleculara.usmf.md e.usmf.md pass.: bmgu
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
General Biology 1004 Chapter 11 Lecture Handout, Summer 2005 Dr. Frisby
 Slide 1 CHAPTER 11 Gene Regulation PowerPoint Lecture Slides for Essential Biology, Second Edition & Essential Biology with Physiology Presentation prepared by Chris C. Romero Neil Campbell, Jane Reece,
Slide 1 CHAPTER 11 Gene Regulation PowerPoint Lecture Slides for Essential Biology, Second Edition & Essential Biology with Physiology Presentation prepared by Chris C. Romero Neil Campbell, Jane Reece,
RECAP (1)! In eukaryotes, large primary transcripts are processed to smaller, mature mrnas.! What was first evidence for this precursorproduct
 RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
Breast cancer. Risk factors you cannot change include: Treatment Plan Selection. Inferring Transcriptional Module from Breast Cancer Profile Data
 Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
Trafficking of mrnas containing ALREXpromoting elements through nuclear speckles
 Research paper Nucleus 4:4, 326 340; July/August 2013; 2013 Landes Bioscience Trafficking of mrnas containing ALREXpromoting elements through nuclear speckles Abdalla Akef, 1 Hui Zhang, 1 Seiji Masuda
Research paper Nucleus 4:4, 326 340; July/August 2013; 2013 Landes Bioscience Trafficking of mrnas containing ALREXpromoting elements through nuclear speckles Abdalla Akef, 1 Hui Zhang, 1 Seiji Masuda
Chapter 3. Expression of α5-megfp in Mouse Cortical Neurons. on the β subunit. Signal sequences in the M3-M4 loop of β nachrs bind protein factors to
 22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
Instructions for Use. APO-AB Annexin V-Biotin Apoptosis Detection Kit 100 tests
 3URGXFW,QIRUPDWLRQ Sigma TACS Annexin V Apoptosis Detection Kits Instructions for Use APO-AB Annexin V-Biotin Apoptosis Detection Kit 100 tests For Research Use Only. Not for use in diagnostic procedures.
3URGXFW,QIRUPDWLRQ Sigma TACS Annexin V Apoptosis Detection Kits Instructions for Use APO-AB Annexin V-Biotin Apoptosis Detection Kit 100 tests For Research Use Only. Not for use in diagnostic procedures.
Cellular Reproduction, Part 1: Mitosis Lecture 10 Fall 2008
 Cell Theory 1 Cellular Reproduction, Part 1: Mitosis Lecture 10 Fall 2008 Cell theory: All organisms are made of cells All cells arise from preexisting cells How do new cells arise? Cell division the reproduction
Cell Theory 1 Cellular Reproduction, Part 1: Mitosis Lecture 10 Fall 2008 Cell theory: All organisms are made of cells All cells arise from preexisting cells How do new cells arise? Cell division the reproduction
ab SREBP-2 Translocation Assay Kit (Cell-Based)
 ab133114 SREBP-2 Translocation Assay Kit (Cell-Based) Instructions for Use For analysis of translocation of SREBP-2 into nuclei. This product is for research use only and is not intended for diagnostic
ab133114 SREBP-2 Translocation Assay Kit (Cell-Based) Instructions for Use For analysis of translocation of SREBP-2 into nuclei. This product is for research use only and is not intended for diagnostic
Ch. 18 Regulation of Gene Expression
 Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Biochemistry 673 Lecture 2 Jason Kahn, UMCP Introduction to steroid hormone receptor (nuclear receptor) signalling
 Biochemistry 673 Lecture 2 Jason Kahn, UMCP Introduction to steroid hormone receptor (nuclear receptor) signalling Resources: Latchman Lodish chapter 10, 20 Helmreich, chapter 11 http://www.nursa.org,
Biochemistry 673 Lecture 2 Jason Kahn, UMCP Introduction to steroid hormone receptor (nuclear receptor) signalling Resources: Latchman Lodish chapter 10, 20 Helmreich, chapter 11 http://www.nursa.org,
Circular RNAs (circrnas) act a stable mirna sponges
 Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
RayBio Annexin V-FITC Apoptosis Detection Kit
 RayBio Annexin V-FITC Apoptosis Detection Kit User Manual Version 1.0 May 25, 2014 (Cat#: 68FT-AnnV-S) RayBiotech, Inc. We Provide You With Excellent Support And Service Tel:(Toll Free)1-888-494-8555 or
RayBio Annexin V-FITC Apoptosis Detection Kit User Manual Version 1.0 May 25, 2014 (Cat#: 68FT-AnnV-S) RayBiotech, Inc. We Provide You With Excellent Support And Service Tel:(Toll Free)1-888-494-8555 or
How Cells Divide. Chapter 10
 How Cells Divide Chapter 10 Bacterial Cell Division Bacteria divide by binary fission. -the single, circular bacterial chromosome is replicated -replication begins at the origin of replication and proceeds
How Cells Divide Chapter 10 Bacterial Cell Division Bacteria divide by binary fission. -the single, circular bacterial chromosome is replicated -replication begins at the origin of replication and proceeds
REGULATED SPLICING AND THE UNSOLVED MYSTERY OF SPLICEOSOME MUTATIONS IN CANCER
 REGULATED SPLICING AND THE UNSOLVED MYSTERY OF SPLICEOSOME MUTATIONS IN CANCER RNA Splicing Lecture 3, Biological Regulatory Mechanisms, H. Madhani Dept. of Biochemistry and Biophysics MAJOR MESSAGES Splice
REGULATED SPLICING AND THE UNSOLVED MYSTERY OF SPLICEOSOME MUTATIONS IN CANCER RNA Splicing Lecture 3, Biological Regulatory Mechanisms, H. Madhani Dept. of Biochemistry and Biophysics MAJOR MESSAGES Splice
nature methods Organelle-specific, rapid induction of molecular activities and membrane tethering
 nature methods Organelle-specific, rapid induction of molecular activities and membrane tethering Toru Komatsu, Igor Kukelyansky, J Michael McCaffery, Tasuku Ueno, Lidenys C Varela & Takanari Inoue Supplementary
nature methods Organelle-specific, rapid induction of molecular activities and membrane tethering Toru Komatsu, Igor Kukelyansky, J Michael McCaffery, Tasuku Ueno, Lidenys C Varela & Takanari Inoue Supplementary
Life Sciences 1A Midterm Exam 2. November 13, 2006
 Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS
 Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Molecular Biology (BIOL 4320) Exam #2 April 22, 2002
 Molecular Biology (BIOL 4320) Exam #2 April 22, 2002 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Molecular Biology (BIOL 4320) Exam #2 April 22, 2002 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Nature Methods: doi: /nmeth.4257
 Supplementary Figure 1 Screen for polypeptides that affect cellular actin filaments. (a) Table summarizing results from all polypeptides tested. Source shows organism, gene, and amino acid numbers used.
Supplementary Figure 1 Screen for polypeptides that affect cellular actin filaments. (a) Table summarizing results from all polypeptides tested. Source shows organism, gene, and amino acid numbers used.
Transcription and RNA processing
 Transcription and RNA processing Lecture 7 Biology W3310/4310 Virology Spring 2016 It is possible that Nature invented DNA for the purpose of achieving regulation at the transcriptional rather than at
Transcription and RNA processing Lecture 7 Biology W3310/4310 Virology Spring 2016 It is possible that Nature invented DNA for the purpose of achieving regulation at the transcriptional rather than at
Supplementary Material
 Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Electron micrograph of phosphotungstanic acid-stained exosomes derived from murine
 1 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Physical properties of murine DC-derived exosomes. a, Electron micrograph of phosphotungstanic acid-stained exosomes derived from
1 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Physical properties of murine DC-derived exosomes. a, Electron micrograph of phosphotungstanic acid-stained exosomes derived from
Supplementary Material and Methods
 Online Supplement Kockx et al, Secretion of Apolipoprotein E from Macrophages 1 Supplementary Material and Methods Cloning of ApoE-GFP Full-length human apoe3 cdna (pcdna3.1/zeo + -apoe) was kindly provided
Online Supplement Kockx et al, Secretion of Apolipoprotein E from Macrophages 1 Supplementary Material and Methods Cloning of ApoE-GFP Full-length human apoe3 cdna (pcdna3.1/zeo + -apoe) was kindly provided
ab CytoPainter Golgi/ER Staining Kit
 ab139485 CytoPainter Golgi/ER Staining Kit Instructions for Use Designed to detect Golgi bodies and endoplasmic reticulum by microscopy This product is for research use only and is not intended for diagnostic
ab139485 CytoPainter Golgi/ER Staining Kit Instructions for Use Designed to detect Golgi bodies and endoplasmic reticulum by microscopy This product is for research use only and is not intended for diagnostic
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the
 Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Total Histone H3 Acetylation Detection Fast Kit (Colorimetric)
 Total Histone H3 Acetylation Detection Fast Kit (Colorimetric) Catalog Number KA1538 48 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use...
Total Histone H3 Acetylation Detection Fast Kit (Colorimetric) Catalog Number KA1538 48 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use...
Nature Biotechnology: doi: /nbt.3828
 Supplementary Figure 1 Development of a FRET-based MCS. (a) Linker and MA2 modification are indicated by single letter amino acid code. indicates deletion of amino acids and N or C indicate the terminus
Supplementary Figure 1 Development of a FRET-based MCS. (a) Linker and MA2 modification are indicated by single letter amino acid code. indicates deletion of amino acids and N or C indicate the terminus
Nature Genetics: doi: /ng Supplementary Figure 1. Assessment of sample purity and quality.
 Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Mechanism of splicing
 Outline of Splicing Mechanism of splicing Em visualization of precursor-spliced mrna in an R loop Kinetics of in vitro splicing Analysis of the splice lariat Lariat Branch Site Splice site sequence requirements
Outline of Splicing Mechanism of splicing Em visualization of precursor-spliced mrna in an R loop Kinetics of in vitro splicing Analysis of the splice lariat Lariat Branch Site Splice site sequence requirements
EPIGENTEK. EpiQuik HDAC2 Assay Kit (Colorimetric) Base Catalog # P-4006 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik HDAC2 Assay Kit (Colorimetric) Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik HDAC2 Assay Kit is very suitable for measuring HDAC2 levels from various fresh tissues and
EpiQuik HDAC2 Assay Kit (Colorimetric) Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik HDAC2 Assay Kit is very suitable for measuring HDAC2 levels from various fresh tissues and
RECAP (1)! In eukaryotes, large primary transcripts are processed to smaller, mature mrnas.! What was first evidence for this precursorproduct
 RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
The clathrin adaptor Numb regulates intestinal cholesterol. absorption through dynamic interaction with NPC1L1
 The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
Regulation of Gene Expression
 18 Regulation of Gene Expression KEY CONCEPTS Figure 18.1 What regulates the precise pattern of gene expression in the developing wing of a fly embryo? 18.1 Bacteria often respond to environmental change
18 Regulation of Gene Expression KEY CONCEPTS Figure 18.1 What regulates the precise pattern of gene expression in the developing wing of a fly embryo? 18.1 Bacteria often respond to environmental change
Translation. Host Cell Shutoff 1) Initiation of eukaryotic translation involves many initiation factors
 Translation Questions? 1) How does poliovirus shutoff eukaryotic translation? 2) If eukaryotic messages are not translated how can poliovirus get its message translated? Host Cell Shutoff 1) Initiation
Translation Questions? 1) How does poliovirus shutoff eukaryotic translation? 2) If eukaryotic messages are not translated how can poliovirus get its message translated? Host Cell Shutoff 1) Initiation
Instructions. Fuse-It-Color. Overview. Specifications
 Membrane fusion is a novel and highly superior method for incorporating various molecules and particles into mammalian cells. Cargo-specific liposomal carriers are able to attach and rapidly fuse with
Membrane fusion is a novel and highly superior method for incorporating various molecules and particles into mammalian cells. Cargo-specific liposomal carriers are able to attach and rapidly fuse with
Influenza virus exploits tunneling nanotubes for cell-to-cell spread
 Supplementary Information Influenza virus exploits tunneling nanotubes for cell-to-cell spread Amrita Kumar 1, Jin Hyang Kim 1, Priya Ranjan 1, Maureen G. Metcalfe 2, Weiping Cao 1, Margarita Mishina 1,
Supplementary Information Influenza virus exploits tunneling nanotubes for cell-to-cell spread Amrita Kumar 1, Jin Hyang Kim 1, Priya Ranjan 1, Maureen G. Metcalfe 2, Weiping Cao 1, Margarita Mishina 1,
B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small cell lung cancer
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Experimental Methods Cell culture B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Experimental Methods Cell culture B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small
mirna Dr. S Hosseini-Asl
 mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
Chapter 3: Cells. I. Overview
 Chapter 3: Cells I. Overview A. Characteristics 1. Basic structural/functional unit 2. Diameter is too small to see by the naked eye 3. Can be over 3 feet long 4. Trillions of cells in over 200 basic types
Chapter 3: Cells I. Overview A. Characteristics 1. Basic structural/functional unit 2. Diameter is too small to see by the naked eye 3. Can be over 3 feet long 4. Trillions of cells in over 200 basic types
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting. protein3) regulate autophagy and mitophagy in renal tubular cells in. acute kidney injury
 Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Human Anatomy & Physiology
 PowerPoint Lecture Slides prepared by Barbara Heard, Atlantic Cape Community College Ninth Edition Human Anatomy & Physiology C H A P T E R 3 Annie Leibovitz/Contact Press Images 2013 Pearson Education,
PowerPoint Lecture Slides prepared by Barbara Heard, Atlantic Cape Community College Ninth Edition Human Anatomy & Physiology C H A P T E R 3 Annie Leibovitz/Contact Press Images 2013 Pearson Education,
'''''''''''''''''Fundamental'Biology' BI'1101' ' an'interdisciplinary'approach'to'introductory'biology' Five'Levels'of'Organiza-on' Molecular'
 '''''''''''''''''Fundamental'Biology' BI'1101' ' an'interdisciplinary'approach'to'introductory'biology' Anggraini'Barlian,' Iriawa-' Tjandra'Anggraeni' SITH4ITB' Five'Levels'of'Organiza-on' Molecular'
'''''''''''''''''Fundamental'Biology' BI'1101' ' an'interdisciplinary'approach'to'introductory'biology' Anggraini'Barlian,' Iriawa-' Tjandra'Anggraeni' SITH4ITB' Five'Levels'of'Organiza-on' Molecular'
Supplementary Information
 Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
