Telomerase RNA TLC1 Shuttling to the Cytoplasm Requires mrna Export Factors and Is Important for Telomere Maintenance
|
|
|
- Shanna Adams
- 6 years ago
- Views:
Transcription
1 Report Telomerase RNA TLC1 Shuttling to the Cytoplasm Requires mrna Export Factors and Is Important for Telomere Maintenance Graphical Abstract Authors Haijia Wu, Daniel Becker, Heike Krebber Correspondence de In Brief Telomerases protect the ends of linear chromosomes from shortening. They are composed of an RNA (TLC1 in S. cerevisiae) and several proteins. During telomerase maturation, TLC1 shuttles to the cytoplasm and recruit proteins. The mature complex is subsequently reimported into the nucleus to fulfill its function on telomeres. Wu et al. now uncover the export machinery for TLC1 and reveal the importance of this step by finding short telomere defects in export factor mutants. Highlights The telomerase RNA, TLC1, requires mrna export factors for nuclear exit TLC1 physically interacts with Mex67 and Dbp5 Nuclear retention of TLC1 leads to defects in telomerase formation Retention of TLC1 in the nucleus leads to telomere shortening Wu et al., 2014, Cell Reports 8, September 25, 2014 ª2014 The Authors
2 Cell Reports Report Telomerase RNA TLC1 Shuttling to the Cytoplasm Requires mrna Export Factors and Is Important for Telomere Maintenance Haijia Wu, 1 Daniel Becker, 1 and Heike Krebber 1, * 1 Abteilung für Molekulare Genetik, Institut für Mikrobiologie und Genetik, Göttinger Zentrum für Molekulare Biowissenschaften (GZMB), Georg-August Universität Göttingen, Göttingen, Germany *Correspondence: heike.krebber@biologie.uni-goettingen.de This is an open access article under the CC BY-NC-ND license ( SUMMARY Telomerases protect the ends of linear chromosomes from shortening. They are composed of an RNA (TLC1 in S. cerevisiae) and several proteins. TLC1 undergoes several maturation steps before it is exported into the cytoplasm to recruit the Est proteins for complete assembly. The mature telomerase is subsequently reimported into the nucleus, where it fulfills its function on telomeres. Here, we show that TLC1 export into the cytoplasm requires not only the Ran GTPase-dependent karyopherin Crm1/ Xpo1 but also the mrna export machinery. mrna export factor mutants accumulate mature and export-competent TLC1 RNAs in their nuclei. Moreover, TLC1 physically interacts with the mrna transport factors Mex67 and Dbp5/Rat8. Most importantly, we show that the nuclear export of TLC1 is an essential step for the formation of the functional RNA containing enzyme, because blocking TLC1 export in the mex67-5 xpo1-1 double mutant prevents its cytoplasmic maturation and leads to telomere shortening. INTRODUCTION Telomerases are enzymatic ribonucleoprotein complexes that antagonize the constant loss of the eukaryotic chromosome ends upon replication by the synthesis of repetitive DNA sequences. The telomerase of Saccharomyces cerevisiae is composed of an RNA molecule (TLC1), which provides a scaffold for the assembly of the protein components (Gallardo et al., 2008). The main catalytic enzyme is the reverse transcriptase Est2 (Lingner et al., 1997). In addition, regulatory subunits such as Est1 and Est3 are also required for the telomerase activity (Evans and Lundblad, 1999). All Est proteins are essential components of the telomerase as also reflected in their names: ever shorter telomeres. The mature nuclear telomerase at telomeres requires the Yku70/80 complex. In its absence, the telomerase shuttles in and out of the nucleus and is localized to the cytoplasm at steady state (Gallardo et al., 2008). The association of the Est proteins has been suggested to occur in the cytoplasm (Gallardo et al., 2008). The cytoplasmic assembly step of the telomerase, which might prevent immature complexes from occupying telomere ends, requires the nucleocytoplasmic shuttling of TLC1 that was reported to utilize the Ran GTPase-dependent exportin Xpo1/Crm1 and the importins Mtr10 and Kap122 (Gallardo et al., 2008). As a transcript of RNA polymerase II, TLC1 becomes polyadenylated like an mrna; however, the tail is removed during maturation (Chapon et al., 1997). At steady state, 5% 10% of all TLC1 transcripts contain an 80-nt-long poly(a) tail, which is accomplished by the same machinery that polyadenylates mrnas (Chapon et al., 1997). Moreover, maturation of the TLC1 RNA requires the entrance into the nucleolus, where hypermethylation of the cap structure by Tgs1 takes place (Gallardo et al., 2008; Seto et al., 1999). Furthermore, the Sm binding site is required for the nuclear exosome-mediated processing of TLC1 from different polyadenylated precursors into its mature form (Coy et al., 2013). The mature TLC1 is subsequently exported into the cytoplasm for full maturation. Other transcripts of RNA polymerase II (e.g., mrnas) recruit processing and mrna export factors cotranscriptionally, among them the mrna export receptor heterodimer Mex67-Mtr2, which mediates the nuclear export through the nuclear pore complex (NPC) (Niño et al., 2013; Strambio-De-Castillia et al., 2010). On the cytoplasmic side of the NPC, directionality of transport is generated by the displacement of Mex67-Mtr2 from the mrna by the DEAD box RNA helicase Dbp5 and its cofactors Gle1, IP 6, and Rat7/Nup159 (Tieg and Krebber, 2013). Although the nuclear export of TLC1 was reported to depend on the Ran GTPase-dependent Xpo1/Crm1 (Gallardo et al., 2008), no telomere shortening defects have been reported for mutants of Crm1/Xpo1, indicating that this export receptor might not be the only responsible transporter for the nuclear export of TLC1 RNAs. As a polymerase II transcript, it seems possible that TLC1 might also receive components of the mrna export machinery during transcription. Therefore, we addressed this question and found that indeed the nuclear export of TLC1 in addition to Xpo1/Crm1 requires the mrna export machinery. Strikingly, upon mutation of both export pathways, we find that the severely inhibited export of TLC1 subsequently leads to telomerase formation defects and consequently to telomere shortening Cell Reports 8, , September 25, 2014 ª2014 The Authors
3 RESULTS AND DISCUSSION The Nuclear Export of TLC1 Is Inhibited in mrna Export Mutants The mature TLC1 RNA is composed of 1,157 nt. This large size, which is similar to the size of a typical mrna in yeast, and its negative charge is a challenge for the passage through the hydrophobic NPC (Strambio-De-Castillia et al., 2010). Therefore, export receptors are required that coat the transport cargo to allow export. The nuclear export of TLC1 was shown to depend on Crm1/Xpo1 (Gallardo and Chartrand, 2008). Surprisingly, no telomere shortening defects have been reported for mutants of Crm1/Xpo1, which suggests that the nuclear export of TLC1 RNAs is not required for the assembly of functional telomerases or that other currently unknown export factors exist. As a polymerase II transcript, it seems possible that TLC1 might interact with the mrna export machinery. To investigate a requirement of mrna export factors in the transport of TLC1 to the cytoplasm, we compared the cellular localization of the TLC1 RNA and bulk mrnas by fluorescent in situ hybridization (FISH) experiments in wild-type, xpo1-1, and different mrna exportdefective mutants. In wild-type cells, TLC1 is localized within the nucleus in several discrete foci as reported earlier (Gallardo et al., 2008) (Figure S1A). In contrast, xpo1-1 cells shifted to the restrictive temperature showed a nuclear signal (Figure S1A). Strikingly, a similar accumulation was also detected in the mrna export factor mutants mex67-5, rat7-1, and rat8-2 (Figure S1A), suggesting defects in the nuclear export of TLC1. To better visualize this phenotype, we repeated this assay in the yku70 deletion strain in which the telomerase is cytoplasmic at steady state (Gallardo et al., 2008). In this background, 40% of TLC1 accumulates in the nuclei of all three mrna export mutants, as compared to 20% in yku70d, clearly revealing nuclear export defects (Figures 1A and 1B). A nuclear, but not nucleolar, accumulation of TLC1 was detected (Figures S1B and S1C), suggesting that TLC1 is not affected in its nucleolar maturation steps. xpo1-1 mutants show also slight mrna export defects (Stade et al., 1997), raising the possibility that Xpo1 might not directly be involved in the TLC1 export. It has been reported earlier that overexpression of the mrna export factor DBP5 rescues the mrna export defects of xpo1-1 (Hodge et al., 1999). To investigate if this would also rescue the TLC1 mislocalization, we overexpressed DBP5 in xpo1-1 and found that while the mrna export defect was abrogated, TLC1 was still mislocalized to the nucleus, indicating that the nuclear export signal (NES) export function of Xpo1 has a direct impact on the TLC1 nuclear export (Figure S1D). To detect the TLC1 export defects in a distinct approach, we isolated total RNA of yeast cytoplasmic fractions and amplified TLC1. While the TLC1 RNA was clearly detectable in wild-type cells and in the unrelated translation termination factor mutant sup45-2, it was significantly reduced in mex67-5, rat7-1, and rat8-2 cells (Figure 1C). Together, these results suggest that TLC1 RNAs require the mrna export machinery for their transport into the cytoplasm. mrna Export Factors Physically Interact with TLC1 An involvement of the mrna export factors in TLC1 transport was further supported by the findings that TLC1 physically interacts with Mex67 and Dbp5/Rat8 in RNA coimmunoprecipitation (IP) analyses. Crm1/Xpo1, Est1, and Est2 served as a positive control and eif4g as a negative control (Figure 1D). This shows that not only Crm1/Xpo1 but also the mrna export factors physically bind to TLC1. Moreover, it suggests that the interaction period of TLC1 with mrna export factors is rather similar to that of Crm1/Xpo1 and possibly restricted to the export phase. In contrast to that, Est1 and Est2, although precipitated to a lesser extent, show a strong (1,000-fold enriched) interaction with TLC1 (Figure 1D), which one could speculate might be due to the fact that these proteins bind for a longer time period, as they are factors present on the matured telomerase, and that the export factors bind several transport cargoes while the Estproteins bind only TLC1. It is currently unclear if Mex67 for its interaction with TLC1 requires adaptor proteins such as Npl3 or Nab2, like in mrna export (Niño et al., 2013; Strambio-De- Castillia et al., 2010). However, it is equally possible that the interaction is rather direct, like in ribosomal subunit transport, where Mex67 contacts the 5S rrna directly (Yao et al., 2007). Taken together, the mislocalization of TLC1 in mrna export mutants and its physical interaction with the mrna export factors suggest their involvement in the nuclear export of TLC1. TLC1 Export Mutants Accumulate Fully Processed RNAs To exclude that defects in processing might be responsible for the nuclear export defects, we investigated the amount of the processed and export competent TLC1 RNA in export mutants. We observed that the ratio of the TLC1 to the U6 RNA, a transcript of RNA polymerase III, was comparable to wild-type in all export mutants, suggesting that the mrna export mutants are not defective in the transcription of TLC1 (Figure 2A). Analogous to mrnas, TLC1 RNAs are polyadenylated upon transcription termination. The poly(a) tail is, however, subsequently removed, so that in an average only 5% 10% of all TLC1 molecules contain a poly(a) tail (Chapon et al., 1997). To monitor potential defects in the maturation of TLC1, which would be reflected by an increase of the immature form of the TLC1 RNA, we determined the ratio of the total RNA and the unprocessed RNA by quantitative RT-PCRs (qrt-pcrs) with specific primers (Figure 2B) and found that significantly less immature TLC1 was detectable in all export mutants (Figure 2C), which is also visible in northern blot analyses (Figure S2), suggesting that only fully processed and export competent TLC1 RNAs accumulate in the nucleus, waiting to be exported. These data suggest that the processing of the immature form of TLC1 occurs prior to its export and that only the processed form is exported to the cytoplasm and accumulates in the export mutants. Also, since the amount is not reduced, it seems that the mature form is stable and not prone to degradation, possibly because in the export mutants, it is packaged for export and in this form not accessible to RNases. Both the NES and the mrna Export Pathways Act on One TLC1 RNA To investigate whether TLC1 can either be exported by the NES export pathway or the mrna export pathway or if both pathways contribute to the export of one TLC1 RNA, we performed IPs in wild-type cells and cells mutated either in XPO1 or in MEX67 Cell Reports 8, , September 25, 2014 ª2014 The Authors 1631
4 Figure 1. Nuclear Export of TLC1 Requires mrna Export Factors (A) FISH experiments in the yku70d background reveal TLC1 (in green) nuclear export defects. The indicated strains were grown to the logarithmic growth phase before they were shifted to 37 C for 1 hr. FISH experiments with a digoxigenin (DIG)-labeled TLC1 probe were performed and detected with fluorescein isothiocyanate-labeled DIG antibodies. The DNA was stained with Hoechst Single cells are shown enlarged in the framed area. Bulk mrna was detected with a Cy3-labeled oligo d(t) 50 probe. (B) Quantification of the signal intensity of 20 cells is shown. p values were calculated according to an unpaired two-tailed t test. (C) Reduced amounts of TLC1 are detected in the cytoplasm of mrna export factor mutants. Nucleocytoplasmic fractionation experiments were carried out with the indicated strains. Cells were grown to the log phase. Then they were either retained at 25 C or shifted to 37 C for 1 hr. The top shows a western blot from the total lysate (T) and the cytoplasmic fraction (C) probed with anti-zwf1 to stain a cytoplasmic protein and anti-nop1 to stain a nuclear protein. The bottom shows the results from three different qrt-pcr experiments in which TLC1 was amplified from the RNA extracted from the cytoplasmic fraction compared to the total RNA. The error bars show the SD. (D) Physical interactions of TLC1 and mrna export factors. Western blot analyses of the immunoprecipitated proteins from strains expressing the indicated tagged proteins are shown that were pulled with anti-myc and anti-gfp (top). The western blot was probed with a mixture of anti-myc and anti-gfp. The bottom shows RNA IP experiments that reveal a physical interaction of Xpo1, Est1, Est2, Mex67, and Rat8 with TLC1 via qrt-pcr. The translation factor eif4g served as a negative control. The binding of TLC1 to the proteins compared to the total RNA and the untagged wild-type control is shown. One representative western blot is shown. Three qrt-pcr experiments are shown in the bottom. The error bars shown in this figure show the SD of at least three experiments. p values shown in (C) and (D) were calculated according to a paired two-tailed t test compared to eif4g. and compared the amount of TLC1 associated with Xpo1 or Mex67. Interestingly, defects in either of these pathways, which both lead to the nuclear accumulation of TLC1 (Figure 1), increased the TLC1 association of both export factors (Figure 2D). This result shows that both export factors are connected via TLC1 and either export factor defect affects the other, which supports a model in which both export factors act on different positions of TLC1 to cooperatively export this large RNA Cell Reports 8, , September 25, 2014 ª2014 The Authors
5 Figure 2. Mutations in mrna Export Factors Result in an Increased Amount of Processed TLC1 (A) All TLC1 export-defective mutants are not impaired in the total level of TLC1 RNA. The indicated strains were grown to log phase before shifted to 37 C for 1 hr. The total RNA was isolated from the cells, and qrt-pcr experiments revealed the ratio of the total TLC1 in comparison to the amount of the U6 small nuclear RNA. (B) Primers used to detect the total TLC1 (mature and immature) and the unprocessed TLC1. A schematic representation of the TLC1 RNA is shown that includes the mature TLC1 (pink area) and the immature TLC1 (pink and blue area). The primer pairs (in black) that amplify either all TLC1 molecules present in the cell (total TLC1) or the immature forms (unprocessed TLC1) are indicated. (C) Fully processed TLC1 RNAs accumulate in the TLC1 export mutants. The indicated strains were grown to log phase before they were shifted to 37 C for 1 hr. The total RNA was isolated from the cells and qrt-pcr experiments revealed the decreased ratio of the unprocessed TLC1 in comparison to the total TLC1 in the TLC1 export mutants. (D) IP analyses reveal connected export activities of Xpo1 and Mex67. Western blots of immunoprecipitates of Xpo1-GFP or Mex67-GFP in wild-type (WT) or mex67-5 or xpo1-1 are show (top). qrt-pcr experiments were performed to determine the amount of coprecipitated TLC1 RNA (bottom). All experiments were done at least three times, and the error bars show the SD. The error bars depicted in this figure show the SD. p values were calculated according to a paired two-tailed t test. Defects in mrna Export Factors Prevent Proper Telomerase Formation Upon entry of TLC1 in the cytoplasm, maturation of the telomerase continues and proteins including Est1 and Est2 associate. However, because its export is affected in xpo1-1 and the mrna export mutants, TLC1 is depleted from the cytoplasm, and one would expect that lesser amounts of Est1 and Est2 should be tethered to the telomerase in the nucleus. Indeed, we found that the wild typical nuclear localization of Est1 and Est2 is significantly decreased in all export mutants (Figure 3A) to similar amounts of 30% (Figure 3B). In other words, 70% of Est1 and Est2 are localized to the cytoplasm as compared to 40% in the wild-type, indicating defects in the cytoplasmic assembly of the telomerase. Interestingly, none of the export mutants showed a comparably low percentage of nuclear signals as detected in tlc1d, which might be an indication that both export pathways, the Ran-dependent Crm1/Xpo1 and the mrna export pathway, contribute to the transport of TLC1 to the cytoplasm. Cell Reports 8, , September 25, 2014 ª2014 The Authors 1633
6 1634 Cell Reports 8, , September 25, 2014 ª2014 The Authors (legend on next page)
7 A similar result was also obtained in nucleocytoplasmic fractionation experiments in which the cytoplasmic level of Est1 and Est2 clearly increased (4-fold) in all TLC1 export mutants (Figure 3C). To analyze these potential defects in telomerase assembly in the TLC1 export mutants directly, we determined the amount of TLC1 that is bound to Est2 by RNA IP experiments and found a 40% reduction of TLC1 RNA bound to Est2 in all export mutants relative to wild-type, indicating a requirement for TLC1 translocation to the cytoplasm for its binding to Est2 (Figure 3D). These assembly defects are further confirmed by interaction studies of Est1 with Est2. Both proteins interact with each other in the cytoplasm upon binding to TLC1 (Gallardo and Chartrand, 2008). We coimmunoprecipitated Est1 bound to Est2 in wildtype cells and compared the precipitates from wild-type to the export factor mutants (Figure 3E). Quantification of these interactions revealed a 40% reduced interaction of Est1 and Est2 in all export mutants (Figure 3F), reflecting the cytoplasmic depletion of TLC1. Even stronger results were obtained when cells were either treated with RNase A, which destroys the TLC1 scaffold, or when the IP analyses were done in cells lacking TLC1 (tlc1d) (Figures 3E and 3F). Notably, when related to the loading control Hem15, it became apparent that the cellular amounts of Est1 and Est2 are relatively stable in all analyzed mutants (Figure S3). However, some degradation seems to occur in tlc1d and might suggest that Est1 and Est2 are possibly more susceptible to degradation when not incorporated into the telomerase complex. These findings suggest that less telomerase is reimported into the nucleus to bind to the telomere ends for telomere maintenance. To finally show that directly, we investigated the incorporation of Est2 into the mature telomere at the telomere ends by IP of Est2 with the telomere protein Yku70 (Gallardo et al., 2008). In contrast to wild-type cells, in which both proteins show a clear interaction, this interaction is reduced to 40% 60% in xpo1-1, rat8-2, and mex67-5 mutants (Figure 3G). These results clearly show that less functional telomerase is generated in the TLC1 export mutants. Taken together, these experiments reinforce the requirement of the mrna export machinery for the nuclear export of TLC1 and support the requirement for the cytoplasmic translocation of TLC1 for further maturation steps. Blocking the Export of TLC1 Results in Shorter Telomere Ends Our data suggest that TLC1 RNAs are transported in a combined action of two cellular transport pathways, the Ran GTPasedependent Crm1/Xpo1 pathway and the mrna export pathway. The inhibition of one of the export pathways still allows enough export of this RNA into the cytoplasm to support the formation of sufficient intact telomerases to prevent telomere shortening. Inhibiting both TLC1 export pathways should prevent sufficient export and result in a dysfunctional telomerase leading to severe telomere shortening. Indeed, xpo1-1 mex67-5 yku70d cells show a significant further increase of the nuclear TLC1 RNA signal as compared to the single mutants in the presence of yku70d and to yku70d (Figure 4A), suggesting that both pathways cooperate to transport TLC1. This strong nuclear export block of TLC1 in the double mutant is also reflected by a further increase of the fully processed TLC1 RNA compared to the amount detected in the single mutants (Figure 4B). This effect is not due to a decrease of the amount of TLC1 (Figure S4A). Furthermore, the amount of mislocalized Est1 and Est2 is increased in xpo1-1 mex67-5 double mutant (60% cytoplasmic) as compared to the single mutants (40% cytoplasmic) (Figure 4C). These additive effects of defects in XPO1 and MEX67 on the export of TLC1 further support a model in which both pathways join forces to collectively transfer TLC1 to the cytoplasm. Consequently, subsequent Southern blot analyses in which telomere shortening can be visualized revealed severe defects in telomere maintenance, reflected in the shortening of the telomere, for the double mutant xpo1-1 mex67-5, but not the single mutants (Figure 4D). Wild-type and yku70 deletion strains served as negative and positive controls, respectively, for the progressive shortening of the telomeres. All strains were grown at the semipermissive temperature of 32 C for xpo1-1 mex67-5 for 30 and 60 generations. This effect is not due to a decrease of the amount of TLC1 and reduced level of the Est proteins at this temperature (Figures S4B and S4C). Figure 3. Mutations in mrna Export Factors Result in Telomerase Formation Defects (A) Est1 and Est2 are mislocalized to the cytoplasm in TLC1 export mutants. Immunofluorescence experiments were performed with the indicated strains either retained at 25 C or shifted to 37 C for 1 hr. Hoechst stains the DNA and indicates the nucleus. (B) Comparison of cells in the permissive and the restrictive situation (25 C versus 37 C and WT versus tlc1d) reveal a highly significant increase of mislocalized telomerase components Est1 and Est2. Quantification of the nuclear signal compared to the total signal of at lest 50 cells per strain and condition from Figure 3A are shown. p values were calculated according to an unpaired two-tailed t test. (C) Nucleocytoplasmic fractionation experiments reveal increased cytoplasmic level of Est1 and Est2. Nucleocytoplasmic fractionation experiments were carried out with the indicated strains. Cells were grown to the log phase. Then, they were either retained at 25 C or shifted to 37 C for 1 hr. The top shows a western blot from the total lysate (T) and the cytoplasmic fraction (C) probed with anti-zwf1 to stain a cytoplasmic protein and anti-nop1 to stain a nuclear protein. The bottom shows the results from three different qrt-pcr experiments in which TLC1 was amplified from the RNA extracted from the cytoplasmic fraction compared to the total RNA. (D) The interaction of the TLC1 RNA and Est2 is reduced in TLC1 export mutants. RNA IP experiments were performed with Est2 in the indicated strains grown at 25 C to the log phase and then shifted to 37 C for 1 hr. The Est2-bound TLC1 RNA was then determined by qrt-pcr and compared to the total TLC1 and to 25 C. (E) The interaction of Est1 and Est2 is disturbed in TLC1 export mutants. Anti-FLAG IP experiments with FLAG-myc-tagged Est2 and myc-tagged Est1 are shown in western blot analyses with anti-myc. (F) Quantification of three independent experiments, one of which is shown in (E). (G) Less mature telomerase is assembled in the TLC1 export mutants. Western blots of a representative FLAG-IP analysis of Yku70 with FLAG-myc-tagged Est2 is shown from the indicated strains that were grown to log phase and then shifted to 37 C for 1 hr (top). Zwf1 served as a negative control. All error bars depicted in this figure show the SD of at least three different experiments. p values were calculated according to a paired two-tailed t test. Cell Reports 8, , September 25, 2014 ª2014 The Authors 1635
8 Figure 4. Mutations in mrna Export Factors Lead to the Shortening of Telomeres (A) FISH experiments reveal an export block of TLC1 RNA in the double mutant xpo1-1 mex67-5. The experiment was performed in the yku70d background. All indicated strains were grown to the logarithmic growth phase at 25 C before the cells were shifted to 37 C for 1 hr. TLC1 was detected with Cy3-labeled 1636 Cell Reports 8, , September 25, 2014 ª2014 The Authors (legend continued on next page)
9 Interestingly, while mex67-5 after 60 generations shows no effect on telomere shortening, a very slight effect was visible for xpo1-1 (Figure 4D). This might be due to the fact that this mutant has also slight mrna export defects, which might be caused by the mislocalized mrna export factor Dbp5 (Hodge et al., 1999). Strikingly, although these mrna export defects are rescued by the overexpression of DBP5, the TLC1 export defects remain (Figure S1D). Also, the telomere shortening defects remain in the presence of high-copy DBP5 (Figure S4D), which clearly shows that both the NES and the mrna export pathways participate in the TLC1 export. In sum, these data reveal the dependence of TLC1 to exit the nucleus for the maturation of the telomerase, which requires that several export factors join forces to produce a functional telomerase (Figure 4E). It has been suggested earlier that the cytoplasmic assembly step of the telomerase might have evolved to prevent that immature complexes occupy the telomere ends (Gallardo et al., 2008). However, if the shuttling is indeed essential for telomere maintenance was not clear, because Crm1/ Xpo1 mutants did not affect telomere length. Our findings that telomere ends cannot be maintained in the double mutant mex67-5 xpo1-1, in which the nuclear export of TLC1 is prevented (Figure 4), clearly show a necessity for the cytoplasmic shuttling of TLC1 and indicate that the nuclear maturation is not sufficient to create functional telomerases that support telomere maintenance. Thus, our results support a model in which upon completion of the nuclear maturation the TLC1 RNA is exported into the cytoplasm (Figure 4E). Its export is mediated by the Ran GTPase-dependent karyopherin Crm1/Xpo1 and the mrna export factors Mex67, Dbp5/Rat8, and Nup159/Rat7. Both pathways contribute to the nuclear export of TLC1 and when mutated prevent its export, which ultimately leads to telomerase formation defects and subsequently to telomere shortening. EXPERIMENTAL PROCEDURES Oligonucleotides, Strains, and Plasmids Oligonucleotides, yeast strains, and plasmids used in this study are listed in the Supplemental Experimental Procedures. Detailed descriptions on how strains were constructed as well as detailed growth conditions can be found in the Supplemental Experimental Procedures. FISH and Immunofluorescence The experiments were essentially performed as described earlier (Hackmann et al., 2014). For further details, see the Supplemental Experimental Procedures. IP Analyses, RNA Co-IP, and qrt-pcr The experiments were essentially performed as published earlier (Hackmann et al., 2014). Homogenized cell lysates were incubated with the indicated antibodies and immobilized on matrix beads. The protein interaction partners were detected by western blot analyses. The RNA interaction partners were further quantified by using qrt-pcr. For further details, see the Supplemental Experimental Procedures. Signal Detection and Quantification Detailed description of western blot and Southern blot assays, fluorescent signals, and qrt-pcr analyses can be found in the Supplemental Experimental Procedures. p values are indicated as follows: ****p < , *** < p < 0.001, **0.001 < p < 0.01, *0.01 < p < Detection of Telomere Shortening Southern blot analyses were performed to detect the length of the telomeres. For additional details, see the Supplemental Experimental Procedures. SUPPLEMENTAL INFORMATION Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at /j.celrep AUTHOR CONTRIBUTIONS H.W. and H.K. designed the study. H.W. and D.B. performed the experiments. H.W., D.B., and H.K. analyzed the data. H.K. wrote the manuscript. ACKNOWLEDGMENTS We are grateful to P. Chartrand, R. Lill, V. Lundblad, P.A. Silver, and K. Weis for providing strains or antibodies. We thank H. Bastians and the members of the H.K. lab for discussions. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) awarded to H.K. and the SFB 860. oligonucleotide probes and the DNA was stained with Hoechst Quantification of the nuclear signals of 20 cells/mutant in comparison to yku70d is shown (bottom). (B) Increased accumulation of the processed TLC1 RNAs in the xpo1-1 mex67-5 double mutant. The indicated strains were grown to log phase before they were shifted to 37 C for 1 hr. The total RNA was isolated from the cells and qrt-pcr experiments revealed the decreased ratio of the unprocessed TLC1 in comparison to the total TLC1 in the TLC1 export mutants. p values were calculated according to a paired two-tailed t test. (C) The double mutant xpo1-1 mex67-5 shows an increased cytoplasmic accumulation of Est1. Colocalization experiments of Est1-myc (green) and the DNA (blue) in the logarithmic growing indicated strains either retained at 25 C or shifted to 37 C for 1 hr are shown (top). Quantification of the nuclear signals of 20 cells/ condition is shown and set into relation (bottom). p values shown in (A) and (C) were calculated according to an unpaired two-tailed t test. All error bars depicted in this figure show the SD of at least three different experiments. (D) Southern blot analyses reveal the progressive shortening of telomeres in mrna export factor mutants. The indicated single and double mutant strains were grown for the indicated generations at the semipermissive temperature of 32 C. The DNA was extracted, digested, and separated on an agarose gel. The length of the Y 0 telomere was determined by Southern blot analyses with a DIG-labeled probe. (E) Model for the life cycle of TLC1. TLC1 is generated in the nucleus by RNA polymerase II (RNAP II). Upon association of the Sm ring, the RNA is processed and the poly(a) tail removed by the nuclear exosome. The CAP is hypermethylated and a trimethylguanine (TMG) CAP is generated. The Ran-dependent nuclear export signal (NES) export receptor Crm1/Xpo1 associates via a currently unknown NES containing protein (X). Proper nuclear export additionally requires the mrna export machinery; the export receptor heterodimer Mex67-Mtr2 associates and supports transport through the nuclear pore complex (NPC). Upon transit Mex67-Mtr2 is displaced by the DEAD box RNA helicase Dbp5 that is located at the cytoplasmic filaments of the NPC, where it interacts with Rat7/Nup159. The subsequent cytoplasmic maturation of the telomerase requires the association of several proteins including Est1, Est2, and Est3. The mature telomerase is reimported into the nucleus via the importins Mtr10 and Kap122, where it finally associates with the telomere ends. Cell Reports 8, , September 25, 2014 ª2014 The Authors 1637
10 Received: April 11, 2014 Revised: July 9, 2014 Accepted: August 8, 2014 Published: September 11, 2014 REFERENCES Chapon, C., Cech, T.R., and Zaug, A.J. (1997). Polyadenylation of telomerase RNA in budding yeast. RNA 3, Coy, S., Volanakis, A., Shah, S., and Vasiljeva, L. (2013). The Sm complex is required for the processing of non-coding RNAs by the exosome. PLoS ONE 8, e Evans, S.K., and Lundblad, V. (1999). Est1 and Cdc13 as comediators of telomerase access. Science 286, Gallardo, F., and Chartrand, P. (2008). Telomerase biogenesis: The long road before getting to the end. RNA Biol. 5, Gallardo, F., Olivier, C., Dandjinou, A.T., Wellinger, R.J., and Chartrand, P. (2008). TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 27, Hackmann, A., Wu, H., Schneider, U.M., Meyer, K., Jung, K., and Krebber, H. (2014). Quality control of spliced mrnas requires the shuttling SR proteins Gbp2 and Hrb1. Nat Commun 5, Hodge, C.A., Colot, H.V., Stafford, P., and Cole, C.N. (1999). Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mrna export defect of xpo1-1 cells. EMBO J. 18, Lingner, J., Hughes, T.R., Shevchenko, A., Mann, M., Lundblad, V., and Cech, T.R. (1997). Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276, Niño, C.A., Hérissant, L., Babour, A., and Dargemont, C. (2013). mrna nuclear export in yeast. Chem. Rev. 113, Seto, A.G., Zaug, A.J., Sobel, S.G., Wolin, S.L., and Cech, T.R. (1999). Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401, Stade, K., Ford, C.S., Guthrie, C., and Weis, K. (1997). Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90, Strambio-De-Castillia, C., Niepel, M., and Rout, M.P. (2010). The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11, Tieg, B., and Krebber, H. (2013). Dbp5 - from nuclear export to translation. Biochim. Biophys. Acta 1829, Yao, W., Roser, D., Köhler, A., Bradatsch, B., Bassler, J., and Hurt, E. (2007). Nuclear export of ribosomal 60S subunits by the general mrna export receptor Mex67-Mtr2. Mol. Cell 26, Cell Reports 8, , September 25, 2014 ª2014 The Authors
RECAP FROM MONDAY AND TUESDAY LECTURES
 RECAP FROM MONDAY AND TUESDAY LECTURES Nucleo-cytoplasmic transport factors: interact with the target macromolecule (signal recognition) interact with the NPC (nucleoporin Phe - Gly repeats) NPC cytoplasmic
RECAP FROM MONDAY AND TUESDAY LECTURES Nucleo-cytoplasmic transport factors: interact with the target macromolecule (signal recognition) interact with the NPC (nucleoporin Phe - Gly repeats) NPC cytoplasmic
MOLECULAR CELL BIOLOGY
 1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
L I F E S C I E N C E S
 1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 NOVEMBER 2, 2006
1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 NOVEMBER 2, 2006
Bio 401 Sp2014. Multiple Choice (Circle the letter corresponding to the correct answer)
 MIDTERM EXAM KEY (60 pts) You should have 5 pages. Please put your name on every page. You will have 70 minutes to complete the exam. You may begin working as soon as you receive the exam. NOTE: the RED
MIDTERM EXAM KEY (60 pts) You should have 5 pages. Please put your name on every page. You will have 70 minutes to complete the exam. You may begin working as soon as you receive the exam. NOTE: the RED
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Edens and Levy, http://www.jcb.org/cgi/content/full/jcb.201406004/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nuclear shrinking does not depend on the cytoskeleton
Supplemental material Edens and Levy, http://www.jcb.org/cgi/content/full/jcb.201406004/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nuclear shrinking does not depend on the cytoskeleton
MCB Chapter 11. Topic E. Splicing mechanism Nuclear Transport Alternative control modes. Reading :
 MCB Chapter 11 Topic E Splicing mechanism Nuclear Transport Alternative control modes Reading : 419-449 Topic E Michal Linial 14 Jan 2004 1 Self-splicing group I introns were the first examples of catalytic
MCB Chapter 11 Topic E Splicing mechanism Nuclear Transport Alternative control modes Reading : 419-449 Topic E Michal Linial 14 Jan 2004 1 Self-splicing group I introns were the first examples of catalytic
Organization of genetic material in eukaryotes
 Organization of genetic material in eukaryotes biologiemoleculara.usmf.md pass.: bmgu e.usmf.md 1 DNA in eukaryotes Location: In nucleus In mitochondria biologiemoleculara.usmf.md e.usmf.md pass.: bmgu
Organization of genetic material in eukaryotes biologiemoleculara.usmf.md pass.: bmgu e.usmf.md 1 DNA in eukaryotes Location: In nucleus In mitochondria biologiemoleculara.usmf.md e.usmf.md pass.: bmgu
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Genetics. Instructor: Dr. Jihad Abdallah Transcription of DNA
 Genetics Instructor: Dr. Jihad Abdallah Transcription of DNA 1 3.4 A 2 Expression of Genetic information DNA Double stranded In the nucleus Transcription mrna Single stranded Translation In the cytoplasm
Genetics Instructor: Dr. Jihad Abdallah Transcription of DNA 1 3.4 A 2 Expression of Genetic information DNA Double stranded In the nucleus Transcription mrna Single stranded Translation In the cytoplasm
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
RNA Processing in Eukaryotes *
 OpenStax-CNX module: m44532 1 RNA Processing in Eukaryotes * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you
OpenStax-CNX module: m44532 1 RNA Processing in Eukaryotes * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you
Life Sciences 1A Midterm Exam 2. November 13, 2006
 Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
October 26, Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
Reviewers' comments: Reviewer #1 (Remarks to the Author):
 Reviewers' comments: Reviewer #1 (Remarks to the Author): In this manuscript, Song et al. identified FBXW7 as a new positive regulator for RIG-Itriggered type I IFN signaling pathway. The authors observed
Reviewers' comments: Reviewer #1 (Remarks to the Author): In this manuscript, Song et al. identified FBXW7 as a new positive regulator for RIG-Itriggered type I IFN signaling pathway. The authors observed
TRANSCRIPTION. DNA à mrna
 TRANSCRIPTION DNA à mrna Central Dogma Animation DNA: The Secret of Life (from PBS) http://www.youtube.com/watch? v=41_ne5ms2ls&list=pl2b2bd56e908da696&index=3 Transcription http://highered.mcgraw-hill.com/sites/0072507470/student_view0/
TRANSCRIPTION DNA à mrna Central Dogma Animation DNA: The Secret of Life (from PBS) http://www.youtube.com/watch? v=41_ne5ms2ls&list=pl2b2bd56e908da696&index=3 Transcription http://highered.mcgraw-hill.com/sites/0072507470/student_view0/
Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
Polyomaviridae. Spring
 Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Eukaryotic transcription (III)
 Eukaryotic transcription (III) 1. Chromosome and chromatin structure Chromatin, chromatid, and chromosome chromatin Genomes exist as chromatins before or after cell division (interphase) but as chromatids
Eukaryotic transcription (III) 1. Chromosome and chromatin structure Chromatin, chromatid, and chromosome chromatin Genomes exist as chromatins before or after cell division (interphase) but as chromatids
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Protein Synthesis
 Protein Synthesis 10.6-10.16 Objectives - To explain the central dogma - To understand the steps of transcription and translation in order to explain how our genes create proteins necessary for survival.
Protein Synthesis 10.6-10.16 Objectives - To explain the central dogma - To understand the steps of transcription and translation in order to explain how our genes create proteins necessary for survival.
1. Identify and characterize interesting phenomena! 2. Characterization should stimulate some questions/models! 3. Combine biochemistry and genetics
 1. Identify and characterize interesting phenomena! 2. Characterization should stimulate some questions/models! 3. Combine biochemistry and genetics to gain mechanistic insight! 4. Return to step 2, as
1. Identify and characterize interesting phenomena! 2. Characterization should stimulate some questions/models! 3. Combine biochemistry and genetics to gain mechanistic insight! 4. Return to step 2, as
number Done by Corrected by Doctor Ashraf
 number 4 Done by Nedaa Bani Ata Corrected by Rama Nada Doctor Ashraf Genome replication and gene expression Remember the steps of viral replication from the last lecture: Attachment, Adsorption, Penetration,
number 4 Done by Nedaa Bani Ata Corrected by Rama Nada Doctor Ashraf Genome replication and gene expression Remember the steps of viral replication from the last lecture: Attachment, Adsorption, Penetration,
The Human Nuclear Poly(A)-Binding Protein Promotes RNA Hyperadenylation and Decay
 The Human Nuclear Poly(A)-Binding Protein Promotes RNA Hyperadenylation and Decay Stefan M. Bresson, Nicholas K. Conrad* Department of Microbiology, University of Texas Southwestern Medical Center, Dallas,
The Human Nuclear Poly(A)-Binding Protein Promotes RNA Hyperadenylation and Decay Stefan M. Bresson, Nicholas K. Conrad* Department of Microbiology, University of Texas Southwestern Medical Center, Dallas,
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
SUPPLEMENTARY INFORMATION
 Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
Translation Activity Guide
 Translation Activity Guide Student Handout β-globin Translation Translation occurs in the cytoplasm of the cell and is defined as the synthesis of a protein (polypeptide) using information encoded in an
Translation Activity Guide Student Handout β-globin Translation Translation occurs in the cytoplasm of the cell and is defined as the synthesis of a protein (polypeptide) using information encoded in an
Homework Hanson section MCB Course, Fall 2014
 Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Cellular MicroRNA and P Bodies Modulate Host-HIV-1 Interactions. 指導教授 : 張麗冠博士 演講者 : 黃柄翰 Date: 2009/10/19
 Cellular MicroRNA and P Bodies Modulate Host-HIV-1 Interactions 指導教授 : 張麗冠博士 演講者 : 黃柄翰 Date: 2009/10/19 1 MicroRNA biogenesis 1. Pri mirna: primary mirna 2. Drosha: RNaseIII 3. DCR 1: Dicer 1 RNaseIII
Cellular MicroRNA and P Bodies Modulate Host-HIV-1 Interactions 指導教授 : 張麗冠博士 演講者 : 黃柄翰 Date: 2009/10/19 1 MicroRNA biogenesis 1. Pri mirna: primary mirna 2. Drosha: RNaseIII 3. DCR 1: Dicer 1 RNaseIII
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
(A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939 (5 µm)
 Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
7.012 Quiz 3 Answers
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
Minor point: In Table S1 there seems to be a typo - multiplicity of the data for the Sm3+ structure 134.8?
 PEER REVIEW FILE Reviewers' comments: Reviewer #1 (Remarks to the Author): Finogenova and colleagues investigated aspects of the structural organization of the Nuclear Exosome Targeting (NEXT) complex,
PEER REVIEW FILE Reviewers' comments: Reviewer #1 (Remarks to the Author): Finogenova and colleagues investigated aspects of the structural organization of the Nuclear Exosome Targeting (NEXT) complex,
Eukaryotic Gene Regulation
 Eukaryotic Gene Regulation Chapter 19: Control of Eukaryotic Genome The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different,
Eukaryotic Gene Regulation Chapter 19: Control of Eukaryotic Genome The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different,
Supplementary Fig. S1. Schematic diagram of minigenome segments.
 open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
MicroRNA dysregulation in cancer. Systems Plant Microbiology Hyun-Hee Lee
 MicroRNA dysregulation in cancer Systems Plant Microbiology Hyun-Hee Lee Contents 1 What is MicroRNA? 2 mirna dysregulation in cancer 3 Summary What is MicroRNA? What is MicroRNA? MicroRNAs (mirnas) -
MicroRNA dysregulation in cancer Systems Plant Microbiology Hyun-Hee Lee Contents 1 What is MicroRNA? 2 mirna dysregulation in cancer 3 Summary What is MicroRNA? What is MicroRNA? MicroRNAs (mirnas) -
Chapter 10 - Post-transcriptional Gene Control
 Chapter 10 - Post-transcriptional Gene Control Chapter 10 - Post-transcriptional Gene Control 10.1 Processing of Eukaryotic Pre-mRNA 10.2 Regulation of Pre-mRNA Processing 10.3 Transport of mrna Across
Chapter 10 - Post-transcriptional Gene Control Chapter 10 - Post-transcriptional Gene Control 10.1 Processing of Eukaryotic Pre-mRNA 10.2 Regulation of Pre-mRNA Processing 10.3 Transport of mrna Across
Last time we talked about the few steps in viral replication cycle and the un-coating stage:
 Zeina Al-Momani Last time we talked about the few steps in viral replication cycle and the un-coating stage: Un-coating: is a general term for the events which occur after penetration, we talked about
Zeina Al-Momani Last time we talked about the few steps in viral replication cycle and the un-coating stage: Un-coating: is a general term for the events which occur after penetration, we talked about
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Generation and validation of mtef4-knockout mice.
 Supplementary Figure 1 Generation and validation of mtef4-knockout mice. (a) Alignment of EF4 (E. coli) with mouse, yeast and human EF4. (b) Domain structures of mouse mtef4 compared to those of EF4 (E.
Supplementary Figure 1 Generation and validation of mtef4-knockout mice. (a) Alignment of EF4 (E. coli) with mouse, yeast and human EF4. (b) Domain structures of mouse mtef4 compared to those of EF4 (E.
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004
 Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Schwarz et al. Activity-Dependent Ubiquitination of GluA1 Mediates a Distinct AMPAR Endocytosis
 Schwarz et al Activity-Dependent Ubiquitination of GluA1 Mediates a Distinct AMPAR Endocytosis and Sorting Pathway Supplemental Data Supplemental Fie 1: AMPARs undergo activity-mediated ubiquitination
Schwarz et al Activity-Dependent Ubiquitination of GluA1 Mediates a Distinct AMPAR Endocytosis and Sorting Pathway Supplemental Data Supplemental Fie 1: AMPARs undergo activity-mediated ubiquitination
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTAL DATA RESULTS
 SUPPLEMENTAL DATA RESULTS Ded1-mediated ribosomal scanning is less leaky than scanning promoted by eifs 4A/4B/4F. The efficiency of leaky scanning in the presence of Ded1 or of eifs 4A/4B/4F was investigated
SUPPLEMENTAL DATA RESULTS Ded1-mediated ribosomal scanning is less leaky than scanning promoted by eifs 4A/4B/4F. The efficiency of leaky scanning in the presence of Ded1 or of eifs 4A/4B/4F was investigated
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Molecular Cell Biology - Problem Drill 10: Gene Expression in Eukaryotes
 Molecular Cell Biology - Problem Drill 10: Gene Expression in Eukaryotes Question No. 1 of 10 1. Which of the following statements about gene expression control in eukaryotes is correct? Question #1 (A)
Molecular Cell Biology - Problem Drill 10: Gene Expression in Eukaryotes Question No. 1 of 10 1. Which of the following statements about gene expression control in eukaryotes is correct? Question #1 (A)
RNA-quality control by the exosome
 RNA-quality control by the exosome Jonathan Houseley, John LaCava and David Tollervey Abstract The exosome complex of 3 5 exonucleases is an important component of the RNA-processing machinery in eukaryotes.
RNA-quality control by the exosome Jonathan Houseley, John LaCava and David Tollervey Abstract The exosome complex of 3 5 exonucleases is an important component of the RNA-processing machinery in eukaryotes.
GW182 proteins cause PABP dissociation from silenced mirna targets in the absence of deadenylation
 Manuscript EMBO-2012-83275 GW182 proteins cause PABP dissociation from silenced mirna targets in the absence of deadenylation Latifa Zekri, Duygu Kuzuoğlu-Öztürk and Elisa Izaurralde Corresponding author:
Manuscript EMBO-2012-83275 GW182 proteins cause PABP dissociation from silenced mirna targets in the absence of deadenylation Latifa Zekri, Duygu Kuzuoğlu-Öztürk and Elisa Izaurralde Corresponding author:
Practice Exam 2 MCBII
 1. Which feature is true for signal sequences and for stop transfer transmembrane domains (4 pts)? A. They are both 20 hydrophobic amino acids long. B. They are both found at the N-terminus of the protein.
1. Which feature is true for signal sequences and for stop transfer transmembrane domains (4 pts)? A. They are both 20 hydrophobic amino acids long. B. They are both found at the N-terminus of the protein.
Fayth K. Yoshimura, Ph.D. September 7, of 7 HIV - BASIC PROPERTIES
 1 of 7 I. Viral Origin. A. Retrovirus - animal lentiviruses. HIV - BASIC PROPERTIES 1. HIV is a member of the Retrovirus family and more specifically it is a member of the Lentivirus genus of this family.
1 of 7 I. Viral Origin. A. Retrovirus - animal lentiviruses. HIV - BASIC PROPERTIES 1. HIV is a member of the Retrovirus family and more specifically it is a member of the Lentivirus genus of this family.
BIO 5099: Molecular Biology for Computer Scientists (et al)
 BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated
 Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
RNA (Ribonucleic acid)
 RNA (Ribonucleic acid) Structure: Similar to that of DNA except: 1- it is single stranded polunucleotide chain. 2- Sugar is ribose 3- Uracil is instead of thymine There are 3 types of RNA: 1- Ribosomal
RNA (Ribonucleic acid) Structure: Similar to that of DNA except: 1- it is single stranded polunucleotide chain. 2- Sugar is ribose 3- Uracil is instead of thymine There are 3 types of RNA: 1- Ribosomal
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Introduction retroposon
 17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being A Eukaryote. Eukaryotic Cells. Basic eukaryotes have:
 BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
10/13/11. Cell Theory. Cell Structure
 Cell Structure Grade 12 Biology Cell Theory All organisms are composed of one or more cells. Cells are the smallest living units of all living organisms. Cells arise only by division of a previously existing
Cell Structure Grade 12 Biology Cell Theory All organisms are composed of one or more cells. Cells are the smallest living units of all living organisms. Cells arise only by division of a previously existing
Repair of Broken Chromosomes and Maintenance of Chromosome Stability
 Repair of Broken Chromosomes and Maintenance of Chromosome Stability Jim Haber Brandeis University Genome instability in tumor cells Truncations Translocations Inversions Duplications Amplifications Deletions
Repair of Broken Chromosomes and Maintenance of Chromosome Stability Jim Haber Brandeis University Genome instability in tumor cells Truncations Translocations Inversions Duplications Amplifications Deletions
MODULE 3: TRANSCRIPTION PART II
 MODULE 3: TRANSCRIPTION PART II Lesson Plan: Title S. CATHERINE SILVER KEY, CHIYEDZA SMALL Transcription Part II: What happens to the initial (premrna) transcript made by RNA pol II? Objectives Explain
MODULE 3: TRANSCRIPTION PART II Lesson Plan: Title S. CATHERINE SILVER KEY, CHIYEDZA SMALL Transcription Part II: What happens to the initial (premrna) transcript made by RNA pol II? Objectives Explain
9/3/2009 DNA i DNA n euk euk yotes Organizatio Organ izatio n of o f gen ge e n tic Locati t on: In n ucleu e s material mater in e ial
 DNA in eukaryotes Organization of genetic material in eukaryotes Location: In nucleus In mitochondria DNA in eukaryotes Nuclear DNA: Long, linear molecules; Chromatin chromosomes; 10% of DNA in genes,
DNA in eukaryotes Organization of genetic material in eukaryotes Location: In nucleus In mitochondria DNA in eukaryotes Nuclear DNA: Long, linear molecules; Chromatin chromosomes; 10% of DNA in genes,
MicroRNA and Male Infertility: A Potential for Diagnosis
 Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Processing of RNA II Biochemistry 302. February 18, 2004 Bob Kelm
 Processing of RNA II Biochemistry 302 February 18, 2004 Bob Kelm What s an intron? Transcribed sequence removed during the process of mrna maturation Discovered by P. Sharp and R. Roberts in late 1970s
Processing of RNA II Biochemistry 302 February 18, 2004 Bob Kelm What s an intron? Transcribed sequence removed during the process of mrna maturation Discovered by P. Sharp and R. Roberts in late 1970s
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
L I F E S C I E N C E S
 1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 OCTOBER 31, 2006
1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 OCTOBER 31, 2006
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression. Chromatin Array of nuc
 Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
7.06 Cell Biology EXAM #3 April 24, 2003
 7.06 Spring 2003 Exam 3 Name 1 of 8 7.06 Cell Biology EXAM #3 April 24, 2003 This is an open book exam, and you are allowed access to books and notes. Please write your answers to the questions in the
7.06 Spring 2003 Exam 3 Name 1 of 8 7.06 Cell Biology EXAM #3 April 24, 2003 This is an open book exam, and you are allowed access to books and notes. Please write your answers to the questions in the
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS
 Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
BIO360 Fall 2013 Quiz 1
 BIO360 Fall 2013 Quiz 1 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function mutation.
BIO360 Fall 2013 Quiz 1 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function mutation.
Received 26 January 1996/Returned for modification 28 February 1996/Accepted 15 March 1996
 MOLECULAR AND CELLULAR BIOLOGY, June 1996, p. 3012 3022 Vol. 16, No. 6 0270-7306/96/$04.00 0 Copyright 1996, American Society for Microbiology Base Pairing at the 5 Splice Site with U1 Small Nuclear RNA
MOLECULAR AND CELLULAR BIOLOGY, June 1996, p. 3012 3022 Vol. 16, No. 6 0270-7306/96/$04.00 0 Copyright 1996, American Society for Microbiology Base Pairing at the 5 Splice Site with U1 Small Nuclear RNA
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells
 Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Supplementary Information
 Supplementary Information Precursors of trnas are stabilized by methylguanosine cap structures Takayuki Ohira and Tsutomu Suzuki Department of Chemistry and Biotechnology, Graduate School of Engineering,
Supplementary Information Precursors of trnas are stabilized by methylguanosine cap structures Takayuki Ohira and Tsutomu Suzuki Department of Chemistry and Biotechnology, Graduate School of Engineering,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/305/ra106/dc1 Supplementary Materials for Controlling Long-Term Signaling: Receptor Dynamics Determine Attenuation and Refractory Behavior of the TGF-β Pathway
www.sciencesignaling.org/cgi/content/full/6/305/ra106/dc1 Supplementary Materials for Controlling Long-Term Signaling: Receptor Dynamics Determine Attenuation and Refractory Behavior of the TGF-β Pathway
Recruitment of the Complete htrex Complex Is Required for Kaposi s Sarcoma Associated Herpesvirus Intronless mrna Nuclear Export and Virus Replication
 Recruitment of the Complete htrex Complex Is Required for Kaposi s Sarcoma Associated Herpesvirus Intronless mrna Nuclear Export and Virus Replication James R. Boyne 1, Kevin J. Colgan 1, Adrian Whitehouse
Recruitment of the Complete htrex Complex Is Required for Kaposi s Sarcoma Associated Herpesvirus Intronless mrna Nuclear Export and Virus Replication James R. Boyne 1, Kevin J. Colgan 1, Adrian Whitehouse
Section 6. Junaid Malek, M.D.
 Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
L I F E S C I E N C E S
 1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 NOVEMBER 2, 2006
1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 NOVEMBER 2, 2006
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human
 Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
SUPPLEMENTARY LEGENDS...
 TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
AP Biology
 Tour of the Cell (1) 2007-2008 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Cell Size Why organelles? Specialized structures - specialized
Tour of the Cell (1) 2007-2008 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Cell Size Why organelles? Specialized structures - specialized
Supplementary Table 1. List of primers used in this study
 Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
mirna Dr. S Hosseini-Asl
 mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
Protein Trafficking in the Secretory and Endocytic Pathways
 Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Pol II - mrna synthesis
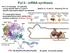 Pol II - mrna synthesis Pol II, S. cerevisiae (12 subunits) - core by specific Rpb1-3, 9 and 11 - Rpb5-6, 8, 10 and 12 - shared by Pol I-III - specific subcomplex Rpb4/7 not essential - TF: transcription
Pol II - mrna synthesis Pol II, S. cerevisiae (12 subunits) - core by specific Rpb1-3, 9 and 11 - Rpb5-6, 8, 10 and 12 - shared by Pol I-III - specific subcomplex Rpb4/7 not essential - TF: transcription
Circular RNAs (circrnas) act a stable mirna sponges
 Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
Nature Genetics: doi: /ng Supplementary Figure 1. Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms.
 Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Processing of RNA II Biochemistry 302. February 13, 2006
 Processing of RNA II Biochemistry 302 February 13, 2006 Precursor mrna: introns and exons Intron: Transcribed RNA sequence removed from precursor RNA during the process of maturation (for class II genes:
Processing of RNA II Biochemistry 302 February 13, 2006 Precursor mrna: introns and exons Intron: Transcribed RNA sequence removed from precursor RNA during the process of maturation (for class II genes:
Supplementary Figure 1
 Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Prof. R. V. Skibbens. BIOS 10 and BIOS 90: BioScience in the 21 st Century. Cell Cycle, Cell Division and intro to Cancer.
 Prof. R. V. Skibbens August 31, 2015 BIOS 10 and BIOS 90: BioScience in the 21 st Century Cell Cycle, Cell Division and intro to Cancer Cell Cycle Why a cell cycle? What is the goal? trauma growth development
Prof. R. V. Skibbens August 31, 2015 BIOS 10 and BIOS 90: BioScience in the 21 st Century Cell Cycle, Cell Division and intro to Cancer Cell Cycle Why a cell cycle? What is the goal? trauma growth development
LESSON 4.4 WORKBOOK. How viruses make us sick: Viral Replication
 DEFINITIONS OF TERMS Eukaryotic: Non-bacterial cell type (bacteria are prokaryotes).. LESSON 4.4 WORKBOOK How viruses make us sick: Viral Replication This lesson extends the principles we learned in Unit
DEFINITIONS OF TERMS Eukaryotic: Non-bacterial cell type (bacteria are prokaryotes).. LESSON 4.4 WORKBOOK How viruses make us sick: Viral Replication This lesson extends the principles we learned in Unit
The Cell Organelles. Eukaryotic cell. The plasma membrane separates the cell from the environment. Plasma membrane: a cell s boundary
 Eukaryotic cell The Cell Organelles Enclosed by plasma membrane Subdivided into membrane bound compartments - organelles One of the organelles is membrane bound nucleus Cytoplasm contains supporting matrix
Eukaryotic cell The Cell Organelles Enclosed by plasma membrane Subdivided into membrane bound compartments - organelles One of the organelles is membrane bound nucleus Cytoplasm contains supporting matrix
Zool 3200: Cell Biology Exam 4 Part II 2/3/15
 Name:Key Trask Zool 3200: Cell Biology Exam 4 Part II 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Name:Key Trask Zool 3200: Cell Biology Exam 4 Part II 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Objectives: Prof.Dr. H.D.El-Yassin
 Protein Synthesis and drugs that inhibit protein synthesis Objectives: 1. To understand the steps involved in the translation process that leads to protein synthesis 2. To understand and know about all
Protein Synthesis and drugs that inhibit protein synthesis Objectives: 1. To understand the steps involved in the translation process that leads to protein synthesis 2. To understand and know about all
Types of cells. Cell size comparison. The Jobs of Cells 10/5/2015. Cells & Cell Organelles. Doing Life s Work
 Types of cells Prokaryote Cells & Cell Organelles bacteria cells Doing Life s Work Eukaryotes 2009-2010 animal cells plant cells Cell size comparison Animal cell Bacterial cell most bacteria (prokaryotic)
Types of cells Prokaryote Cells & Cell Organelles bacteria cells Doing Life s Work Eukaryotes 2009-2010 animal cells plant cells Cell size comparison Animal cell Bacterial cell most bacteria (prokaryotic)
