Supplementary Materials for
|
|
|
- Kathleen Richards
- 6 years ago
- Views:
Transcription
1 Supplementary Materials for Controlling Long-Term Signaling: Receptor Dynamics Determine Attenuation and Refractory Behavior of the TGF-β Pathway Pedro Vizán, Daniel S. J. Miller, Ilaria Gori, Debipriya Das, Bernhard Schmierer,* Caroline S. Hill* *Corresponding author. (C.S.H.); (B.S.) Published 10 December 2013, Sci. Signal. 6, ra106 (2013) DOI: /scisignal The PDF file includes: Texts S1 to S3 Fig. S1. The refractory period is not unique to HaCaT cells. Fig. S2. Kinetics of Smad2 phosphorylation. Fig. S3. Blocking and depletion of ligand. Fig. S4. Specificity of the TβRII antibody in Western blotting. Fig. S5. Recovery from the refractory state. Fig. S6. Biotinylation of surface receptors in HaCaT cells at different time points. Fig. S7. Data fitting with previous data sets. Fig. S8. Comparison between simulations and experimental data in Figs. 1 and 2. Fig. S9. Comparison between simulations and experimental data in Fig. 3. Fig. S10. Restimulation of naïve cancer cell lines with medium from the D3 line. Table S1. Model parameters. Table S2. List of model species. Table S3. Ordinary differential equations and initial conditions. References (36, 37)
2 Supplementary Text S1. Model conception: the receptor module To model the findings of signal attenuation and the existence of a refractory period, the Smad model (4) was expanded to include a mathematical description of receptor biology that involved receptor maturation, receptor trafficking to the plasma membrane, ligand binding, receptor internalization, and receptor turnover. As described in the main text, binding of TGF-β is sufficient for signal attenuation, which does not require actively signaling receptors, but does require a functional proteasome. Because we excluded degradation of PSmad2 as the cause for attenuation, we concluded that degradation of a subpool of receptors that is enhanced by TGF-β binding attenuates the signal, which is consistent with previous experimental evidence (22, 23) and previous models (24, 36). The model has to account for signal attenuation and has to capture the refractory state. Although receptor turnover (synthesis and degradation) could, in principle, account for the observed refractory period, the measured half-lives of ALK5 and TβRII (~4 h and ~2 h, respectively; Fig. 3C) indicate that receptor turnover alone is too fast to account for the slow recovery from the refractory period (12 24 hours, Fig. 2C). These slow recovery kinetics point to at least one additional slow step distinct from receptor synthesis, such as posttranslational modification or trafficking of nascent receptors to the plasma membrane. As shown in Fig. 3D and fig. S6A, it is TβRII, rather than ALK5, that is rapidly depleted from the cell surface and thus responsible for loss of responsiveness. We used this finding to simplify the model, which contains only a single receptor species with the measured turnover rate for TβRII. TGF-β binds to competent surface receptors. We 1
3 know this step is fast, because a short time of TGF-β treatment is sufficient to induce a maximum response at 1 h (Fig. 2A). In contrast, the Smad2 phosphorylation time course data (fig. S7) exhibit a delay between TGF-β addition and the appearance of phosphorylated R-Smads. These findings suggest that receptor activation in response to ligand occurs in two steps: a fast TGF-β-binding step, which rapidly occupies the pool of available surface-exposed receptors, and a slower receptor activation step, which further converts TGF-β-bound receptors into fully active, signaling receptors. In the model, receptors mature from a precursor form () to a fully processed form ( ). has an intracellular form ( ) and a surface-exposed form ( ), which are in equilibrium. This equilibrium assumption holds if trafficking to and from the plasma membrane are fast compared with receptor maturation, which is likely, given the slow maturation rate. In the absence of TGF-β, the system is fully described by and. There is good evidence that the fraction of total cellular receptors that are competent and exposed on the cell surface is very small, and we set this fraction 0.05, that is 5% of total receptors are assumed to be at the surface in the absence of signal (6). We define the maturation efficiency α as the ratio of the maturation rate constant and the receptor degradation rate constant, and scale the system such that, in the absence of a signal, the total amount of receptors 1. We can now describe the system in the absence of signal by two ordinary differential equations (ODEs) and the three parameters,, and : The fractions of in the cytoplasm and the cell surface, respectively, are given by 2
4 1 1 1 Where is the equilibrium constant of receptor trafficking to and from the membrane. The steady state of this system defines the initial conditions prior to the addition of TGF-β: S2. Model parameterization The model is expressed in dimensionless form with the total number of receptors, as well as total Smad2 and total Smad4, set to unity. In the model, 2 ng/ml TGF-β corresponds to a concentration of 4 (that is, there are 4 molecules of TGF-β for every molecule of receptor). This value was estimated by simulating the dose dependency of the response (fig. S2) and corresponds to a total number of roughly 15,000 receptor molecules per cell. TGF-β is depleted from the medium through active signaling and also by a constitutive, cell-dependent but signaling-independent TGF-β clearance (k cc ). Parameter values were either taken from previous work (4) or estimated from the data sets presented here. Three parameters, the receptor activation rate k act, the Smad2 phosphorylation rate k p, and the dissociation constant of the receptor inhibitor SB , K SBI, were optimized by curve fitting to the data shown in Fig. 2D of (4) (fig. S7). The fact that these data are derived from GFP-Smad2 overexpressing HaCaT cells 3
5 rather than the parental cell line used in this study does not cause significant differences and does not affect the fitting results. In summary, the comprehensive model presented here is not only consistent with the data sets shown in this manuscript, but also reproduces the data sets shown in (4). S3. Modeling autocrine signaling To simulate TGF-β-dependent autocrine TGF-β production, Equation 5 (table S3) was replaced by 1 24 Where is a small background synthesis rate ( 0.001), and is the rate of TGF-β-dependent synthesis, both expressed relative to k d. The model is available in the Biomodels database ( under the ID MODEL
6 Fig. S1. The refractory period is not unique to HaCaT cells. The mouse fibroblast cell line NIH-3T3, the transformed mouse mammary cell line EpRas, and the human breast cancer cell line MDA-MB-231 were treated for 1 h (b) or 8 h (c) with TGF-β (Tβ), after which cells were re-induced with fresh TGF-β for 1 h (e). Medium was also taken from cells after 8 h of ligand exposure and used to induce naïve cells for 1 h (d). Lysates were assayed by Western blot using antibodies recognizing PSmad2 and Smad2, with Tubulin or MCM6 as a loading control. In the experimental scheme shown, green arrows indicate addition of TGF-β; curved arrow, induction of naïve cells with conditioned media. Quantifications are the normalized average and ranges of two independent experiments. In all cases the amount of PSmad2 after a 1-h TGF-β stimulation was set to 1. 5
7 Fig. S2. Kinetics of Smad2 phosphorylation. Top panels: HaCaT cells were treated with 2 ng/ml or 0.1 ng/ml TGF-β for 60 min, harvesting samples every 10 minutes. PSmad2 was assayed by Western blot. Tubulin was used as a loading control. Representative panels of at least three independent experiments are shown. Bottom panel: Simulation of the amount of PSmad2 in cells treated with 2 ng/ml versus 0.1 ng/ml TGF-β. 6
8 Fig. S3. Blocking and depletion of ligand. (A) The ligand-neutralizing TGF-β antibody can be washed out. HaCaT cells were treated with ligand-neutralizing TGFβ antibody (block) or control antibody for 20 min. Cells were incubated with or without TGF-β for 1 h following washing, as indicated. PSmad2 was assayed by Western blot, using Tubulin as a loading control. (B) Time course of TGF-β depletion by the cells. HaCaT cells were treated with either 0.5 ng/ml or 2 ng/ml TGF-β for the times indicated. At each time point samples of media were taken and assayed in triplicate for TGF-β by ELISA. The values were normalized to the amount of TGF-β in the medium at the zero time point. Quantifications are the averages and standard deviations of triplicates. (C) Cell-dependent depletion of TGF-β. TGF-β was added to tissue culture plates with (+) or without (-) plated HaCaT cells and kept under normal culture conditions for 10 or 30 h (pre-incubation). The conditioned media was then used to induce naïve cells for 1 h. Induction capacity, as measured by ability to induce PSmad2, was assayed by Western blot as shown. Tubulin was used as a loading control. In A and C, representative blots of at least three independent experiments are shown. 7
9 Fig. S4. Specificity of the TβRII antibody in Western blotting. Whole-cell extracts were prepared from HaCaT cells or from T47D cells, which are negative for TβRII (37), and were assayed by Western blotting using antibodies recognizing TβRII and MCM7 as a loading control. Extracts were treated ± PNGase F to remove N-linked sugars. Representative blots of three independent experiments are shown. 8
10 Fig. S5. Recovery from the refractory state. HaCaT cells were either unstimulated (a) or stimulated with TGF-β for 1 h, 24 h, 30 h, 36 h or 48 h (b, c, e, g, i). At the latter four time points, cells were also restimulated with 2 ng/ml TGF-β for 1 h (d, f, h, j). Whole-cell extracts were assayed for PSmad2 and MCM7 by Western blotting. Quantifications are the normalized (relative to loading control) average and standard deviations of three replicates. The amount of PSmad2 after a 1-h TGF-β stimulation was set to 1. 9
11 Fig. S6. Biotinylation of surface receptors in HaCaT cells at different time points. (A) The experiment design was exactly as in Fig. 3D, except that cells were stimulated at time points between 0 and 8 h, or between 0 and 60 min. Quantifications are the normalized average and standard deviations of three independent experiments. (B) The experiment design was exactly as in Fig. 3D, except that cells were stimulated at time points between 0 and 48 h. Quantifications are the normalized average and standard deviations of three independent experiments. (C) Biotinylation of surface receptors in MDA-MB-231 cells. The experiment design was exactly as in Fig. 3D. Quantifications are the normalized average and ranges of two independent experiments. For each experiment, the amount of surface receptor in unstimulated cells was set to 1. 10
12 Fig. S7. Data fitting with previous data sets. The parameter set for the extended model described here fits the set of data used in (4). Nuclear accumulation of Smad2 upon TGF-β stimulation and its clearance from the nucleus upon SB addition is shown, as are the amounts of PSmad2 during a 45-min stimulation with TGF-β. 11
13 Fig. S8. Comparison between simulations and experimental data in Figs. 1 and 2. Lines represent time course simulations, points and bars represent the experimentally determined values +/- their standard deviations or ranges. For a detailed description refer to the corresponding figures (indicated at the top of each graph) in the main text. 12
14 Fig. S9. Comparison between simulations and experimental data in Fig. 3. (A) Time course simulation in cells treated with TGF-β ± cycloheximide (CHX) ± MG-132 or MG-132 followed by SB Left panel, replotting of the simulation shown in Fig. 5E. Right panel, experimental data from Figs 3A and B. (B) Time course of receptor degradation in cells treated with CHX ± TGF-β. Left panel, replotting of parts of the simulation shown in Fig. 5F. Right panel, experimental data for TβRII from Fig. 3C. 13
15 Fig. S10. Restimulation of naïve cancer cell lines with medium from the D3 line. Medium from D3 cells, the cancer cell line with the most autocrine TGF-β production, was added to naïve cancer cell lines and PSmad2 was detected by Western Blot after 1 h. Quantifications are the normalized (relative to the cell line, CN5) average and standard deviations of three independent experiments. 14
16 Table S1. Model parameters. Parameters of the receptor module Value Source receptor degradation 0.32 h -1 estimate from Fig. 3C competent surface receptors in the absence of a signal 0.05 Ref (6) receptor synthesis rate, defined such that in the absence of TGF-β, h -1 1 maturation efficiency 0.08 estimate from Fig. 2C ligand-induced to constitutive degradation ratio 4 estimate from Fig. 1A equilibrium constant for receptor trafficking, defined such that in the absence of TGF-β, TGF-β binding relative to 100 estimate from Fig. 2A receptor activation relative to optimized by data fitting dissociation constant of SB optimized by data fitting Molecules of TGF-β per receptor if TGF-β = 1 ng/ml 2 estimate from fig. S2 constitutive TGF-β clearance relative to 0.35 Parameters of the Smad module Value Source estimate from Fig. 2D and fig. S4B Smad2 phosphorylation rate constant h -1 optimized by data fitting Smad2 dephosphorylation rate constant 23.6 h -1 Ref (4) nuclear import of Smad2 and Smad h -1 Ref (4) nuclear export of Smad2 20 h -1 Ref (4) nuclear export of Smad h -1 Ref (4) on-rate of Smad complex association 350 h -1 Ref (4) off-rate of Smad complex dissociation 60 h -1 Ref (4) % % of Smad2 phosphorylated at t = 0.75 h 0.31 Ref (4) Complex import factor 5.7 Ref (4) cytonucleoplasmic volume ratio 2.27 Ref (4) 15
17 Table S2. List of model species. See table S3 for the equations. Ligand and Receptors Compartment Definition Nascent receptors Cytoplasm Eq. 1 Competent receptors Cytoplasm Eq. 2 1 Competent, intracellular receptors Cytoplasm 1 Competent surface receptors Cytoplasm 1 TGF-β bound receptors, not yet active Cytoplasm Eq. 3 Actively signaling receptors Cytoplasm Eq. 4 TGF-β Cytoplasm Eq. 5 Unphosphorylated Smad2 Compartment Definition 2 Cytoplasmic unphosphorylated Smad2 Cytoplasm Eq. 6 2 Nuclear unphosphorylated Smad2 Nucleus Smad4; sum of free and complexed Compartment Definition 4 Total cytoplasmic Smad4 Cytoplasm Eq. 7 4 Total nuclear Smad4 Nucleus psmad2; sum of free and complexed Compartment Definition 2 Total cellular psmad2 Cell Eq. 8 2 Total cytoplasmic psmad2 Cytoplasm Eq. 9 2 Total nuclear psmad2 Nucleus Smad complexes Compartment Definition 24 Total cellular heteromeric complexes Cell Eq Cytoplasmic heteromeric complexes Cytoplasm Eq Nuclear heteromeric complexes Nucleus Total cellular homomeric complexes Cell Eq Cytoplasmic homomeric complexes Cytoplasm Eq Nuclear homomeric complexes Nucleus Free, monomeric forms of psmad2 and Smad4 Compartment Definition 2 Cytoplasmic monomeric psmad2 Cytoplasm Nuclear monomeric psmad2 Nucleus Cytoplasmic monomeric Smad4 Cytoplasm Nuclear monomeric Smad4 Nucleus
18 Table S3. Ordinary differential equations and initial conditions. Receptor Module Eq. 1 1 Eq. 2 Initial condition Eq Eq Eq. 5./ Smad Module Eq. 6 Initial condition Eq Eq Eq Eq Eq Eq Eq
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Part I. Boolean modelling exercises
 Part I. Boolean modelling exercises. Glucose repression of Icl in yeast In yeast Saccharomyces cerevisiae, expression of enzyme Icl (isocitrate lyase-, involved in the gluconeogenesis pathway) is important
Part I. Boolean modelling exercises. Glucose repression of Icl in yeast In yeast Saccharomyces cerevisiae, expression of enzyme Icl (isocitrate lyase-, involved in the gluconeogenesis pathway) is important
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR-2B cells untreated (-) or stimulated (+) for 45 min
 Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR2B cells untreated () or stimulated () for 45 min with 5 ng/ml TGFβ or 10 ng/ml BMP4 were incubated with
Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR2B cells untreated () or stimulated () for 45 min with 5 ng/ml TGFβ or 10 ng/ml BMP4 were incubated with
Supplementary Fig. 1 p38 MAPK negatively regulates DC differentiation. (a) Western blot analysis of p38 isoform expression in BM cells, immature DCs
 Supplementary Fig. 1 p38 MAPK negatively regulates DC differentiation. (a) Western blot analysis of p38 isoform expression in BM cells, immature DCs (idcs) and mature DCs (mdcs). A myeloma cell line expressing
Supplementary Fig. 1 p38 MAPK negatively regulates DC differentiation. (a) Western blot analysis of p38 isoform expression in BM cells, immature DCs (idcs) and mature DCs (mdcs). A myeloma cell line expressing
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/310/ra11/dc1 Supplementary Materials for STAT3 Induction of mir-146b Forms a Feedback Loop to Inhibit the NF-κB to IL-6 Signaling Axis and STAT3-Driven Cancer
www.sciencesignaling.org/cgi/content/full/7/310/ra11/dc1 Supplementary Materials for STAT3 Induction of mir-146b Forms a Feedback Loop to Inhibit the NF-κB to IL-6 Signaling Axis and STAT3-Driven Cancer
Figure S1: Effects on haptotaxis are independent of effects on cell velocity A)
 Supplemental Figures Figure S1: Effects on haptotaxis are independent of effects on cell velocity A) Velocity of MV D7 fibroblasts expressing different GFP-tagged Ena/VASP family proteins in the haptotaxis
Supplemental Figures Figure S1: Effects on haptotaxis are independent of effects on cell velocity A) Velocity of MV D7 fibroblasts expressing different GFP-tagged Ena/VASP family proteins in the haptotaxis
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/308/ra4/dc1 Supplementary Materials for Antipsychotics Activate mtorc1-dependent Translation to Enhance Neuronal Morphological Complexity Heather Bowling, Guoan
www.sciencesignaling.org/cgi/content/full/7/308/ra4/dc1 Supplementary Materials for Antipsychotics Activate mtorc1-dependent Translation to Enhance Neuronal Morphological Complexity Heather Bowling, Guoan
Supplemental Information. CD4 + CD25 + Foxp3 + Regulatory T Cells Promote. Th17 Cells In Vitro and Enhance Host Resistance
 Immunity, Volume 34 Supplemental Information D4 + D25 + + Regulatory T ells Promote Th17 ells In Vitro and Enhance Host Resistance in Mouse andida albicans Th17 ell Infection Model Pushpa Pandiyan, Heather
Immunity, Volume 34 Supplemental Information D4 + D25 + + Regulatory T ells Promote Th17 ells In Vitro and Enhance Host Resistance in Mouse andida albicans Th17 ell Infection Model Pushpa Pandiyan, Heather
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/418/ra27/dc1 Supplementary Materials for The adhesion molecule PECAM-1 enhances the TGF- mediated inhibition of T cell function Debra K. Newman,* Guoping Fu,
www.sciencesignaling.org/cgi/content/full/9/418/ra27/dc1 Supplementary Materials for The adhesion molecule PECAM-1 enhances the TGF- mediated inhibition of T cell function Debra K. Newman,* Guoping Fu,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/381/ra59/dc1 Supplementary Materials for Analysis of single-cell cytokine secretion reveals a role for paracrine signaling in coordinating macrophage responses
www.sciencesignaling.org/cgi/content/full/8/381/ra59/dc1 Supplementary Materials for Analysis of single-cell cytokine secretion reveals a role for paracrine signaling in coordinating macrophage responses
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Figure 1. The mir-182 binding site of SMAD7 3 UTR and the. mutated sequence.
 Supplementary Figure 1. The mir-182 binding site of SMAD7 3 UTR and the mutated sequence. 1 Supplementary Figure 2. Expression of mir-182 and SMAD7 in various cell lines. (A) Basal levels of mir-182 expression
Supplementary Figure 1. The mir-182 binding site of SMAD7 3 UTR and the mutated sequence. 1 Supplementary Figure 2. Expression of mir-182 and SMAD7 in various cell lines. (A) Basal levels of mir-182 expression
LPS LPS P6 - + Supplementary Fig. 1.
 P6 LPS - - - + + + - LPS + + - - P6 + Supplementary Fig. 1. Pharmacological inhibition of the JAK/STAT blocks LPS-induced HMGB1 nuclear translocation. RAW 267.4 cells were stimulated with LPS in the absence
P6 LPS - - - + + + - LPS + + - - P6 + Supplementary Fig. 1. Pharmacological inhibition of the JAK/STAT blocks LPS-induced HMGB1 nuclear translocation. RAW 267.4 cells were stimulated with LPS in the absence
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/278/rs11/dc1 Supplementary Materials for In Vivo Phosphoproteomics Analysis Reveals the Cardiac Targets of β-adrenergic Receptor Signaling Alicia Lundby,* Martin
www.sciencesignaling.org/cgi/content/full/6/278/rs11/dc1 Supplementary Materials for In Vivo Phosphoproteomics Analysis Reveals the Cardiac Targets of β-adrenergic Receptor Signaling Alicia Lundby,* Martin
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the T H 17 cell it reg cell balance Rajatava Basu 1,5, Sarah K.
SUPPLEMENTARY MATERIAL IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the T H 17 cell it reg cell balance Rajatava Basu 1,5, Sarah K.
Identification of influential proteins in the classical retinoic acid signaling pathway
 Ghaffari and Petzold Theoretical Biology and Medical Modelling (2018) 15:16 https://doi.org/10.1186/s12976-018-0088-7 RESEARCH Open Access Identification of influential proteins in the classical retinoic
Ghaffari and Petzold Theoretical Biology and Medical Modelling (2018) 15:16 https://doi.org/10.1186/s12976-018-0088-7 RESEARCH Open Access Identification of influential proteins in the classical retinoic
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous
 Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/364/ra19/dc1 Supplementary Materials for Plasma membrane Ca 2+ -ATPases can shape the pattern of Ca 2+ transients induced by store-operated Ca 2+ entry Katalin
www.sciencesignaling.org/cgi/content/full/8/364/ra19/dc1 Supplementary Materials for Plasma membrane Ca 2+ -ATPases can shape the pattern of Ca 2+ transients induced by store-operated Ca 2+ entry Katalin
Figure S1. PMVs from THP-1 cells expose phosphatidylserine and carry actin. A) Flow
 SUPPLEMENTARY DATA Supplementary Figure Legends Figure S1. PMVs from THP-1 cells expose phosphatidylserine and carry actin. A) Flow cytometry analysis of PMVs labelled with annexin-v-pe (Guava technologies)
SUPPLEMENTARY DATA Supplementary Figure Legends Figure S1. PMVs from THP-1 cells expose phosphatidylserine and carry actin. A) Flow cytometry analysis of PMVs labelled with annexin-v-pe (Guava technologies)
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Analysis of Wnt signaling β-catenin spatial dynamics in HEK293T cells
 Tan et al. BMC Systems Biology 2014, 8:44 RESEARCH ARTICLE Analysis of Wnt signaling β-catenin spatial dynamics in HEK293T cells Chin Wee Tan 1,2,3,4*, Bruce S Gardiner 5, Yumiko Hirokawa 1,3, David W
Tan et al. BMC Systems Biology 2014, 8:44 RESEARCH ARTICLE Analysis of Wnt signaling β-catenin spatial dynamics in HEK293T cells Chin Wee Tan 1,2,3,4*, Bruce S Gardiner 5, Yumiko Hirokawa 1,3, David W
hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This
 SUPPLEMENTAL FIGURE LEGEND Fig. S1. Generation and characterization of. (A) Coomassie staining of soluble hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This protein was expressed
SUPPLEMENTAL FIGURE LEGEND Fig. S1. Generation and characterization of. (A) Coomassie staining of soluble hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This protein was expressed
Control of Cell Proliferation by Peptide Growth Factors. Autocrine Growth Factor Production Causes Malignant Transformation?
 Control of Cell Proliferation by Peptide Growth Factors Autocrine Growth Factor Production Causes Malignant Transformation? Transforming Activities From Condition Media from a Tumor Cell Line Condition
Control of Cell Proliferation by Peptide Growth Factors Autocrine Growth Factor Production Causes Malignant Transformation? Transforming Activities From Condition Media from a Tumor Cell Line Condition
W/T Itgam -/- F4/80 CD115. F4/80 hi CD115 + F4/80 + CD115 +
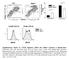 F4/8 % in the peritoneal lavage 6 4 2 p=.15 n.s p=.76 CD115 F4/8 hi CD115 + F4/8 + CD115 + F4/8 hi CD115 + F4/8 + CD115 + MHCII MHCII Supplementary Figure S1. CD11b deficiency affects the cellular responses
F4/8 % in the peritoneal lavage 6 4 2 p=.15 n.s p=.76 CD115 F4/8 hi CD115 + F4/8 + CD115 + F4/8 hi CD115 + F4/8 + CD115 + MHCII MHCII Supplementary Figure S1. CD11b deficiency affects the cellular responses
Intracellular MHC class II molecules promote TLR-triggered innate. immune responses by maintaining Btk activation
 Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis
 Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells
 Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Use of a camp BRET Sensor to Characterize a Novel Regulation of camp by the Sphingosine-1-phosphate/G 13 Pathway
 Use of a camp BRET Sensor to Characterize a Novel Regulation of camp by the Sphingosine-1-phosphate/G 13 Pathway SUPPLEMENTAL DATA Characterization of the CAMYEL sensor and calculation of intracellular
Use of a camp BRET Sensor to Characterize a Novel Regulation of camp by the Sphingosine-1-phosphate/G 13 Pathway SUPPLEMENTAL DATA Characterization of the CAMYEL sensor and calculation of intracellular
genome edited transient transfection, CMV promoter
 Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
5K ALDEFLUOR-positive/ CXCR1-negative. 5K ALDEFLUOR-positive/ CXCR1-positive BAAA BAAA CXCR1-APC BAAA BAAA CXCR1-APC
 A +DEAB -DEAB K ALDEFLUOR-positive/ CXCR-negative BAAA BAAA CXCR-APC B +DEAB -DEAB K ALDEFLUOR-positive/ CXCR-positive BAAA BAAA CXCR-APC C Supplemental Figure. Tumorigenicity of the ALDEFLUOR-positive/CXCR-positive
A +DEAB -DEAB K ALDEFLUOR-positive/ CXCR-negative BAAA BAAA CXCR-APC B +DEAB -DEAB K ALDEFLUOR-positive/ CXCR-positive BAAA BAAA CXCR-APC C Supplemental Figure. Tumorigenicity of the ALDEFLUOR-positive/CXCR-positive
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human
 Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
SUPPLEMENTARY INFORMATION
 sirna pool: Control Tetherin -HA-GFP HA-Tetherin -Tubulin Supplementary Figure S1. Knockdown of HA-tagged tetherin expression by tetherin specific sirnas. HeLa cells were cotransfected with plasmids expressing
sirna pool: Control Tetherin -HA-GFP HA-Tetherin -Tubulin Supplementary Figure S1. Knockdown of HA-tagged tetherin expression by tetherin specific sirnas. HeLa cells were cotransfected with plasmids expressing
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
Supplementary Figure 1
 Supplementary Figure 1 6 HE-50 HE-116 E-1 HE-108 Supplementary Figure 1. Targeted drug response curves of endometrial cancer cells. Endometrial cancer cell lines were incubated with serial dilutions of
Supplementary Figure 1 6 HE-50 HE-116 E-1 HE-108 Supplementary Figure 1. Targeted drug response curves of endometrial cancer cells. Endometrial cancer cell lines were incubated with serial dilutions of
Supplementary Table 1. Metabolic parameters in GFP and OGT-treated mice
 Supplementary Table 1. Metabolic parameters in GFP and OGT-treated mice Fasted Refed GFP OGT GFP OGT Liver G6P (mmol/g) 0.03±0.01 0.04±0.02 0.60±0.04 0.42±0.10 A TGs (mg/g of liver) 20.08±5.17 16.29±0.8
Supplementary Table 1. Metabolic parameters in GFP and OGT-treated mice Fasted Refed GFP OGT GFP OGT Liver G6P (mmol/g) 0.03±0.01 0.04±0.02 0.60±0.04 0.42±0.10 A TGs (mg/g of liver) 20.08±5.17 16.29±0.8
Cell isolation. Spleen and lymph nodes (axillary, inguinal) were removed from mice
 Supplementary Methods: Cell isolation. Spleen and lymph nodes (axillary, inguinal) were removed from mice and gently meshed in DMEM containing 10% FBS to prepare for single cell suspensions. CD4 + CD25
Supplementary Methods: Cell isolation. Spleen and lymph nodes (axillary, inguinal) were removed from mice and gently meshed in DMEM containing 10% FBS to prepare for single cell suspensions. CD4 + CD25
Supplementary Figure 1. BMS enhances human T cell activation in vitro in a
 Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
MATHEMATICAL MODELING OF PERIFUSION CELL CULTURE EXPERIMENTS ON GNRH SIGNALING
 MATHEMATICAL MODELING OF PERIFUSION CELL CULTURE EXPERIMENTS ON GNRH SIGNALING N Ezgi Temamogullari a,, H Frederik Nijhout b, Michael C Reed a arxiv:1606.00463v1 [q-bio.cb] 9 Dec 2015 Abstract a Department
MATHEMATICAL MODELING OF PERIFUSION CELL CULTURE EXPERIMENTS ON GNRH SIGNALING N Ezgi Temamogullari a,, H Frederik Nijhout b, Michael C Reed a arxiv:1606.00463v1 [q-bio.cb] 9 Dec 2015 Abstract a Department
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Figure 1 Induction of cellular senescence and isolation of exosome. a to c, Pre-senescent primary normal human diploid fibroblasts
 Supplementary Figure 1 Induction of cellular senescence and isolation of exosome. a to c, Pre-senescent primary normal human diploid fibroblasts (TIG-3 cells) were rendered senescent by either serial passage
Supplementary Figure 1 Induction of cellular senescence and isolation of exosome. a to c, Pre-senescent primary normal human diploid fibroblasts (TIG-3 cells) were rendered senescent by either serial passage
TEB. Id4 p63 DAPI Merge. Id4 CK8 DAPI Merge
 a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
SD-1 SD-1: Cathepsin B levels in TNF treated hch
 SD-1 SD-1: Cathepsin B levels in TNF treated hch. A. RNA and B. protein extracts from TNF treated and untreated human chondrocytes (hch) were analyzed via qpcr (left) and immunoblot analyses (right) for
SD-1 SD-1: Cathepsin B levels in TNF treated hch. A. RNA and B. protein extracts from TNF treated and untreated human chondrocytes (hch) were analyzed via qpcr (left) and immunoblot analyses (right) for
Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening
 Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening Introduction: Vimentin (VIM) intermediate filament (IF) proteins are associated with EMT in lung cancer and its metastatic
Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening Introduction: Vimentin (VIM) intermediate filament (IF) proteins are associated with EMT in lung cancer and its metastatic
Supplementary Material
 Supplementary Material accompanying the manuscript Interleukin 37 is a fundamental inhibitor of innate immunity Marcel F Nold, Claudia A Nold-Petry, Jarod A Zepp, Brent E Palmer, Philip Bufler & Charles
Supplementary Material accompanying the manuscript Interleukin 37 is a fundamental inhibitor of innate immunity Marcel F Nold, Claudia A Nold-Petry, Jarod A Zepp, Brent E Palmer, Philip Bufler & Charles
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/4/199/ra75/dc1 Supplementary Materials for Signaling by the Matrix Proteoglycan Decorin Controls Inflammation and Cancer Through PDCD4 and MicroRNA-21 Rosetta
www.sciencesignaling.org/cgi/content/full/4/199/ra75/dc1 Supplementary Materials for Signaling by the Matrix Proteoglycan Decorin Controls Inflammation and Cancer Through PDCD4 and MicroRNA-21 Rosetta
Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured
 Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured under Th0, Th1, Th2, Th17, and Treg conditions. mrna
Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured under Th0, Th1, Th2, Th17, and Treg conditions. mrna
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
Supplemental Materials. STK16 regulates actin dynamics to control Golgi organization and cell cycle
 Supplemental Materials STK16 regulates actin dynamics to control Golgi organization and cell cycle Juanjuan Liu 1,2,3, Xingxing Yang 1,3, Binhua Li 1, Junjun Wang 1,2, Wenchao Wang 1, Jing Liu 1, Qingsong
Supplemental Materials STK16 regulates actin dynamics to control Golgi organization and cell cycle Juanjuan Liu 1,2,3, Xingxing Yang 1,3, Binhua Li 1, Junjun Wang 1,2, Wenchao Wang 1, Jing Liu 1, Qingsong
Nature Neuroscience: doi: /nn Supplementary Figure 1. PICALM expression in brain capillary endothelium in human brain and in mouse brain.
 Supplementary Figure 1 PICALM expression in brain capillary endothelium in human brain and in mouse brain. a, Double immunostaining for PICALM (red, left) and lectin positive endothelial profiles (blue,
Supplementary Figure 1 PICALM expression in brain capillary endothelium in human brain and in mouse brain. a, Double immunostaining for PICALM (red, left) and lectin positive endothelial profiles (blue,
Supplemental Information. Menin Deficiency Leads to Depressive-like. Behaviors in Mice by Modulating. Astrocyte-Mediated Neuroinflammation
 Neuron, Volume 100 Supplemental Information Menin Deficiency Leads to Depressive-like Behaviors in Mice by Modulating Astrocyte-Mediated Neuroinflammation Lige Leng, Kai Zhuang, Zeyue Liu, Changquan Huang,
Neuron, Volume 100 Supplemental Information Menin Deficiency Leads to Depressive-like Behaviors in Mice by Modulating Astrocyte-Mediated Neuroinflammation Lige Leng, Kai Zhuang, Zeyue Liu, Changquan Huang,
In situ drug-receptor binding kinetics in single cells: a quantitative label-free study of anti-tumor drug resistance
 Supplementary Information for In situ drug-receptor binding kinetics in single cells: a quantitative label-free study of anti-tumor drug resistance Wei Wang 1, Linliang Yin 2,3, Laura Gonzalez-Malerva
Supplementary Information for In situ drug-receptor binding kinetics in single cells: a quantitative label-free study of anti-tumor drug resistance Wei Wang 1, Linliang Yin 2,3, Laura Gonzalez-Malerva
Stewart et al. CD36 ligands promote sterile inflammation through assembly of a TLR 4 and 6 heterodimer
 NFκB (fold induction) Stewart et al. ligands promote sterile inflammation through assembly of a TLR 4 and 6 heterodimer a. mrna (fold induction) 5 4 3 2 1 LDL oxldl Gro1a MIP-2 RANTES mrna (fold induction)
NFκB (fold induction) Stewart et al. ligands promote sterile inflammation through assembly of a TLR 4 and 6 heterodimer a. mrna (fold induction) 5 4 3 2 1 LDL oxldl Gro1a MIP-2 RANTES mrna (fold induction)
colorimetric sandwich ELISA kit datasheet
 colorimetric sandwich ELISA kit datasheet For the quantitative detection of human IL5 in serum, plasma, cell culture supernatants and urine. general information Catalogue Number Product Name Species cross-reactivity
colorimetric sandwich ELISA kit datasheet For the quantitative detection of human IL5 in serum, plasma, cell culture supernatants and urine. general information Catalogue Number Product Name Species cross-reactivity
Nature Structural & Molecular Biology: doi: /nsmb.3218
 Supplementary Figure 1 Endogenous EGFR trafficking and responses depend on biased ligands. (a) Lysates from HeLa cells stimulated for 2 min. with increasing concentration of ligands were immunoblotted
Supplementary Figure 1 Endogenous EGFR trafficking and responses depend on biased ligands. (a) Lysates from HeLa cells stimulated for 2 min. with increasing concentration of ligands were immunoblotted
Inhibition of TGFβ enhances chemotherapy action against triple negative breast cancer by abrogation of
 SUPPLEMENTAL DATA Inhibition of TGFβ enhances chemotherapy action against triple negative breast cancer by abrogation of cancer stem cells and interleukin-8 Neil E. Bhola 1, Justin M. Balko 1, Teresa C.
SUPPLEMENTAL DATA Inhibition of TGFβ enhances chemotherapy action against triple negative breast cancer by abrogation of cancer stem cells and interleukin-8 Neil E. Bhola 1, Justin M. Balko 1, Teresa C.
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical
 Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
SDC (Supplemental Digital Content) Figure S1. Percentages of Granzyme B expressing CD8 + T lymphocytes
 Figure S1. Percentages of Granzyme expressing D8 + T lymphocytes 13.5% 1.4%.6% 77% 21% 24% 76% 8 8 Granzyme + (%) 6 4 2 Granzyme + (%) 6 4 2 D8 + Tnaive Tcm Tem Temra D8 + D8 + D28 + D8 + D28 null We analyzed
Figure S1. Percentages of Granzyme expressing D8 + T lymphocytes 13.5% 1.4%.6% 77% 21% 24% 76% 8 8 Granzyme + (%) 6 4 2 Granzyme + (%) 6 4 2 D8 + Tnaive Tcm Tem Temra D8 + D8 + D28 + D8 + D28 null We analyzed
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/271/ra25/dc1 Supplementary Materials for Phosphoproteomic Analysis Implicates the mtorc2-foxo1 Axis in VEGF Signaling and Feedback Activation of Receptor Tyrosine
www.sciencesignaling.org/cgi/content/full/6/271/ra25/dc1 Supplementary Materials for Phosphoproteomic Analysis Implicates the mtorc2-foxo1 Axis in VEGF Signaling and Feedback Activation of Receptor Tyrosine
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Nair S, Branagan AR, Liu J, Boddupalli CS, Mistry PK, Dhodapkar
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Nair S, Branagan AR, Liu J, Boddupalli CS, Mistry PK, Dhodapkar
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 DOT1L regulates the expression of epithelial and mesenchymal markers. (a) The expression levels and cellular localizations of EMT markers were confirmed by
Supplementary Figures Supplementary Figure 1 DOT1L regulates the expression of epithelial and mesenchymal markers. (a) The expression levels and cellular localizations of EMT markers were confirmed by
Protein tyrosine phosphatase 1B targets PITX1/p120RasGAP. thus showing therapeutic potential in colorectal carcinoma
 Protein tyrosine phosphatase 1B targets PITX1/p120RasGAP thus showing therapeutic potential in colorectal carcinoma Hao-Wei Teng, Man-Hsin Hung, Li-Ju Chen, Mao-Ju Chang, Feng-Shu Hsieh, Ming-Hsien Tsai,
Protein tyrosine phosphatase 1B targets PITX1/p120RasGAP thus showing therapeutic potential in colorectal carcinoma Hao-Wei Teng, Man-Hsin Hung, Li-Ju Chen, Mao-Ju Chang, Feng-Shu Hsieh, Ming-Hsien Tsai,
SUPPLEMENTARY LEGENDS...
 TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
supplementary information
 DOI: 10.1038/ncb2133 Figure S1 Actomyosin organisation in human squamous cell carcinoma. (a) Three examples of actomyosin organisation around the edges of squamous cell carcinoma biopsies are shown. Myosin
DOI: 10.1038/ncb2133 Figure S1 Actomyosin organisation in human squamous cell carcinoma. (a) Three examples of actomyosin organisation around the edges of squamous cell carcinoma biopsies are shown. Myosin
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system
 Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Crystallization-grade After D After V3 cocktail. Time (s) Time (s) Time (s) Time (s) Time (s) Time (s)
 Ligand Type Name 6 Crystallization-grade After 447-52D After V3 cocktail Receptor CD4 Resonance Units 5 1 5 1 5 1 Broadly neutralizing antibodies 2G12 VRC26.9 Resonance Units Resonance Units 3 1 15 1 5
Ligand Type Name 6 Crystallization-grade After 447-52D After V3 cocktail Receptor CD4 Resonance Units 5 1 5 1 5 1 Broadly neutralizing antibodies 2G12 VRC26.9 Resonance Units Resonance Units 3 1 15 1 5
Supplementary Figure 1. Deletion of Smad3 prevents B16F10 melanoma invasion and metastasis in a mouse s.c. tumor model.
 A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Relative Rates. SUM159 CB- 839-Resistant *** n.s Intracellular % Labeled by U- 13 C-Asn 0.
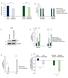 A Relative Growth Rates 1.2 1.8.6.4.2 B Relative Rates 1.6 1.4 1.2 1.8.6.4.2 LPS2 Parental LPS2 Q-Independent SUM159 Parental SUM159 CB-839-Resistant LPS2 Parental LPS2 Q- Independent SUM159 Parental SUM159
A Relative Growth Rates 1.2 1.8.6.4.2 B Relative Rates 1.6 1.4 1.2 1.8.6.4.2 LPS2 Parental LPS2 Q-Independent SUM159 Parental SUM159 CB-839-Resistant LPS2 Parental LPS2 Q- Independent SUM159 Parental SUM159
Supplementary Figure 1 Protease allergens induce IgE and IgG1 production. (a-c)
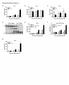 1 Supplementary Figure 1 Protease allergens induce IgE and IgG1 production. (a-c) Serum IgG1 (a), IgM (b) and IgG2 (c) concentrations in response to papain immediately before primary immunization (day
1 Supplementary Figure 1 Protease allergens induce IgE and IgG1 production. (a-c) Serum IgG1 (a), IgM (b) and IgG2 (c) concentrations in response to papain immediately before primary immunization (day
mir-509-5p and mir-1243 increase the sensitivity to gemcitabine by inhibiting
 mir-509-5p and mir-1243 increase the sensitivity to gemcitabine by inhibiting epithelial-mesenchymal transition in pancreatic cancer Hidekazu Hiramoto, M.D. 1,3, Tomoki Muramatsu, Ph.D. 1, Daisuke Ichikawa,
mir-509-5p and mir-1243 increase the sensitivity to gemcitabine by inhibiting epithelial-mesenchymal transition in pancreatic cancer Hidekazu Hiramoto, M.D. 1,3, Tomoki Muramatsu, Ph.D. 1, Daisuke Ichikawa,
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure S1: Fibroblast-induced elongation of cancer cells requires direct contact with living fibroblasts. A. Representative images of HT29-GFP cultured in the presence
SUPPLEMENTARY FIGURES Supplementary Figure S1: Fibroblast-induced elongation of cancer cells requires direct contact with living fibroblasts. A. Representative images of HT29-GFP cultured in the presence
ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP
 ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP Peera Jaru-Ampornpan 1, Kuang Shen 1,3, Vinh Q. Lam 1,3, Mona Ali 2, Sebastian Doniach 2, Tony Z. Jia 1, Shu-ou Shan 1 Supplementary
ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP Peera Jaru-Ampornpan 1, Kuang Shen 1,3, Vinh Q. Lam 1,3, Mona Ali 2, Sebastian Doniach 2, Tony Z. Jia 1, Shu-ou Shan 1 Supplementary
Fang et al. NMuMG. PyVmT unstained Anti-CCR2-PE MDA-MB MCF MCF10A
 A NMuMG PyVmT 16.5+.5 47.+7.2 Fang et al. unstained Anti-CCR2-PE 4T1 Control 37.6+6.3 56.1+.65 MCF1A 16.1+3. MCF-7 3.1+5.4 MDA-M-231 42.1+5.5 unstained Secondary antibody only Anti-CCR2 SUPPLEMENTAL FIGURE
A NMuMG PyVmT 16.5+.5 47.+7.2 Fang et al. unstained Anti-CCR2-PE 4T1 Control 37.6+6.3 56.1+.65 MCF1A 16.1+3. MCF-7 3.1+5.4 MDA-M-231 42.1+5.5 unstained Secondary antibody only Anti-CCR2 SUPPLEMENTAL FIGURE
hemodynamic stress. A. Echocardiographic quantification of cardiac dimensions and function in
 SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Fbn1 C1039G/+ hearts display normal cardiac function in the absence of hemodynamic stress. A. Echocardiographic quantification of cardiac dimensions and
SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Fbn1 C1039G/+ hearts display normal cardiac function in the absence of hemodynamic stress. A. Echocardiographic quantification of cardiac dimensions and
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC
 Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supplementary Materials
 Supplementary Materials Figure S1. MTT Cell viability assay. To measure the cytotoxic potential of the oxidative treatment, the MTT [3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyl tetrazolium bromide] assay
Supplementary Materials Figure S1. MTT Cell viability assay. To measure the cytotoxic potential of the oxidative treatment, the MTT [3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyl tetrazolium bromide] assay
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Nature Medicine: doi: /nm.3922
 Title: Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells Authors: Il-Kyu Kim, Byung-Seok Kim, Choong-Hyun
Title: Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells Authors: Il-Kyu Kim, Byung-Seok Kim, Choong-Hyun
JNJ : selective and slowly reversible FAAH inhibitor. Central and Peripheral PK/PD
 JNJ-42165279: selective and slowly reversible FAAH inhibitor Central and Peripheral PK/PD The Endocannabinoid System Research initiated by efforts to elucidate the active substance of Cannabis (THC in
JNJ-42165279: selective and slowly reversible FAAH inhibitor Central and Peripheral PK/PD The Endocannabinoid System Research initiated by efforts to elucidate the active substance of Cannabis (THC in
Supplementary Materials
 Supplementary Materials Supplementary Figure S1 Regulation of Ubl4A stability by its assembly partner A, The translation rate of Ubl4A is not affected in the absence of Bag6. Control, Bag6 and Ubl4A CRISPR
Supplementary Materials Supplementary Figure S1 Regulation of Ubl4A stability by its assembly partner A, The translation rate of Ubl4A is not affected in the absence of Bag6. Control, Bag6 and Ubl4A CRISPR
B220 CD4 CD8. Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN
 B220 CD4 CD8 Natarajan et al., unpublished data Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN showing B cell follicles and T cell areas. 20 µm thick. Image of magnification
B220 CD4 CD8 Natarajan et al., unpublished data Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN showing B cell follicles and T cell areas. 20 µm thick. Image of magnification
TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer
 Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. NKT ligand-loaded tumour antigen-presenting B cell- and monocyte-based vaccine induces NKT, NK and CD8 T cell responses. (A) The cytokine profiles of liver
Supplementary Figures Supplementary Figure 1. NKT ligand-loaded tumour antigen-presenting B cell- and monocyte-based vaccine induces NKT, NK and CD8 T cell responses. (A) The cytokine profiles of liver
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Edens and Levy, http://www.jcb.org/cgi/content/full/jcb.201406004/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nuclear shrinking does not depend on the cytoskeleton
Supplemental material Edens and Levy, http://www.jcb.org/cgi/content/full/jcb.201406004/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nuclear shrinking does not depend on the cytoskeleton
Cytokine Arrays Reveal Black Ops Tactics of Tumor-induced Immunosuppression
 Cytokine Arrays Reveal Black Ops Tactics of Tumor-induced Immunosuppression Jarad J Wilson, Ph.D. Technical Support & Marketing Specialist Ruo-Pan Huang, MD, Ph.D. Founder and CEO What are Antibody Arrays?
Cytokine Arrays Reveal Black Ops Tactics of Tumor-induced Immunosuppression Jarad J Wilson, Ph.D. Technical Support & Marketing Specialist Ruo-Pan Huang, MD, Ph.D. Founder and CEO What are Antibody Arrays?
Reviewers' comments: Reviewer #1 (Remarks to the Author):
 Reviewers' comments: Reviewer #1 (Remarks to the Author): The manuscript by Wu et al describes critical role of RNA binding protein CUGBP1 in the development of TGF-beta-mediated liver fibrosis. The activation
Reviewers' comments: Reviewer #1 (Remarks to the Author): The manuscript by Wu et al describes critical role of RNA binding protein CUGBP1 in the development of TGF-beta-mediated liver fibrosis. The activation
7.06 Cell Biology EXAM #3 April 24, 2003
 7.06 Spring 2003 Exam 3 Name 1 of 8 7.06 Cell Biology EXAM #3 April 24, 2003 This is an open book exam, and you are allowed access to books and notes. Please write your answers to the questions in the
7.06 Spring 2003 Exam 3 Name 1 of 8 7.06 Cell Biology EXAM #3 April 24, 2003 This is an open book exam, and you are allowed access to books and notes. Please write your answers to the questions in the
condition. Left panel, the HCT-116 cells were lysed with RIPA buffer containing 0.1%
 FIGURE LEGENDS Supplementary Fig 1 (A) sumoylation pattern detected under denaturing condition. Left panel, the HCT-116 cells were lysed with RIPA buffer containing 0.1% SDS in the presence and absence
FIGURE LEGENDS Supplementary Fig 1 (A) sumoylation pattern detected under denaturing condition. Left panel, the HCT-116 cells were lysed with RIPA buffer containing 0.1% SDS in the presence and absence
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/398/rs12/dc1 Supplementary Materials for Quantitative phosphoproteomics reveals new roles for the protein phosphatase PP6 in mitotic cells Scott F. Rusin, Kate
www.sciencesignaling.org/cgi/content/full/8/398/rs12/dc1 Supplementary Materials for Quantitative phosphoproteomics reveals new roles for the protein phosphatase PP6 in mitotic cells Scott F. Rusin, Kate
B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small cell lung cancer
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Experimental Methods Cell culture B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Experimental Methods Cell culture B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small
