Cancer Cell International. Open Access PRIMARY RESEARCH. Sandra Stempin, Anika Engel, Nora Winkler, Thorsten Buhrke * and Alfonso Lampen
|
|
|
- Janel Craig
- 5 years ago
- Views:
Transcription
1 DOI /s Cancer Cell International PRIMARY RESEARCH Open Access Morphological and molecular characterization of the human breast epithelial cell line M13SV1 and its tumorigenic derivatives M13SV1 R2 2 and M13SV1 R2 N1 Sandra Stempin, Anika Engel, Nora Winkler, Thorsten Buhrke * and Alfonso Lampen Abstract Background: The estrogen receptor-positive M13SV1 breast epithelial cell line was proposed to be a suitable in vitro model for breast cancer research since two derivatives with graduated tumorigenicity M13SV1-R2-2 and M13SV1- R2-N1 are available for this cell line. In the present study, these three cell lines were comparatively examined for their morphological and their biochemical properties on the molecular level. Methods: A transcriptomic approach (gene array analysis) was chosen to unravel differences in gene expression among the three cell lines. Network analysis was conducted to identify deregulated signaling pathways. Cellular viability was determined by impedance measurements as well as by neutral red uptake assay. Apoptosis was determined by using a caspase assay. For morphological characterization, cells were grown in three-dimensional cell culture, and cellular differentiation and spheroid formation was followed by immunofluorescence staining by using confocal laser scanning microscopy. Results: The gene array results indicated that there were only marginal differences in gene expression among the three cell lines. Network analysis predicted the R2-N1 derivative (1) to display enhanced apoptosis and (2) to have a higher migration capability compared to its parent cell line M13SV1. Enhanced apoptosis was confirmed by elevated caspase activity, and increased migration was observed in 3D culture when cells migrated out of the globular spheroids. In 3D cell culture, all three cell lines similarly formed spheroids within three days, but there was no acini formation until day 21 which is indicated by a growth arrest around day 15, cellular polarization, and the formation of hollow lumen inside the spheroids. These characteristics, however, are crucial to study, e.g., the differentiation process of breast epithelial cells in vitro. Conclusion: Due to the molecular and morphological features, the M13SV1 cell line and its tumorigenic derivatives seem to be less suitable as in vitro models than other cell lines such as the MCF-10A cell line which displays proper acini formation in 3D culture. Keywords: Human breast epithelial cell line, M13SV1, Transcriptomics, 3D cell culture, Cellular differentiation, In vitro model Background Nowadays, breast cancer research largely depends on the *Correspondence: thorsten.buhrke@bfr.bund.de Sandra Stempin and Anika Engel contributed equally to this work Department of Food Safety, Federal Institute for Risk Assessment, Max Dohrn Str. 8 10, Berlin, Germany 2015 Stempin et al. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License ( which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver ( publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
2 Page 2 of 13 availability of model systems suitable to monitor breast epithelial cell differentiation and crucial characteristics of mammary gland development in vitro. There are two major types of epithelial cells in mammary gland, namely luminal cells and basal or myoepithelial cells. These two cell types originate from the same stem cell population that is also found in the mammary epithelium [1]. For in vitro studies, numerous cell lines have been derived from mammary gland tissue in order to generate permanent cell culture models e.g., for breast cancer research [2]. Among other model systems, a protocol has been reported for the isolation of two types of human breast epithelial cells (HBEC) [3]. Type I HBEC are luminal epithelial cells with stem cell characteristics capable of e.g., differentiating into type II HBEC which have a basal epithelial cell phenotype. The M13SV1 cell line was derived from SV40-immortalized type I HBEC cells [3]. This cell line was reported to retain their stem cell characteristics and, moreover, to be positive in estrogen receptor expression [4]. On top of this, two derivatives of M13SV1 have been generated which display a graduated level of tumorigenicity. The weakly tumorigenic cell line M13SV1-R2-2 was derived from the non-tumorigenic parent cell line M13SV1 by X-ray irradiation. Subsequently, this derivative was transduced with the rat Erbb2 oncogene yielding the highly tumorigenic M13SV1-R2-N1 cell line. Tumorigenicity was evaluated according to the degree of tumor development (number and weight of tumors) in mice that had been subcutaneously inoculated with cells of these three cell lines [5]. Due to all these features, the availability of three estrogen receptor-positive type I HBECderived cell lines related to each other with a different degree of tumorigenicity was supposed to be a promising model system to be used for in vitro studies in particular with respect to estrogenic effects in breast cancer development. To further characterize this in vitro model, this study aimed on the elucidation of differences between these three cell lines on the molecular level. For this purpose, a transcriptomic approach was chosen to examine alterations in gene expression among the three cell lines under in vitro conditions in order to unravel deregulated metabolic and/or signaling pathways which may be correlated with the different degree of tumorigenicity in vivo. Moreover, the morphological characteristics of the three cell lines grown in 3D culture were compared with each other. Results A transcriptomic approach was chosen to analyze the differences between the M13SV1 cell line and its tumorigenic derivatives on the molecular level. Total RNA was prepared from the three different cell lines and subjected to gene array analysis. By using the standard settings for gene array data evaluation (fold change ± 2, p < 0.05), it turned out that only a small number of the 24,266 different transcripts represented by the gene array were differentially expressed among the three cell lines (Table 1). As an example, only 40 transcripts were differentially expressed in the weakly tumorigenic R2-2 cell line compared to its non-tumorigenic parent cell line M13SV1. The deregulation of these transcripts, however, was very significant, as still 39 of these 40 transcripts were significantly deregulated when a p-value of was applied in the analysis (Table 1). Thus, we decided also to allow more subtle deregulations and therefore to use a fold change of 1.5, but in combination with a stringent p-value of The use of these settings increased the number of deregulated genes by two- to threefold in comparison to the standard settings (Table 1) resulting in a broader data set to be used in the subsequent network analysis. The lists of deregulated genes (fold change 1.5; p < 0.001) can be found in the Additional file 1: Tables S1, Additional file 2: Table S2. The software package Ingenuity Pathway Analysis (IPA) was employed to gain insight into molecular metabolic and signaling pathways that are differentially regulated among the three cell lines. The IPA software is a literature-based database that allows a comparison of one s own experimental data with published data. In our analysis, this comparison was restricted to published experimental data obtained only with human breast cancer cell lines or with primary isolates from human mammary gland. It turned out that although 122 gene transcripts were significantly deregulated in the R2-2 cell line compared to its parent cell line M13SV1 (Table 1) there were no metabolic or signaling pathway or any biological function significantly affected according to the IPA analysis. Thus, on the molecular level these two cell lines seem to be very similar to each other. In the case of the highly tumorigenic R2-N1 cell line, the deregulation of 217 gene transcripts compared to the M13SV1 parent cell line had Table 1 Number of deregulated genes of the gene array experiment Fold change p-value R2-2 vs. M13SV1 R2-N1 vs. M13SV (15/25) 110 (71/39) (15/25) 109 (71/38) (14/25) 93 (59/34) (65/89) 418 (290/128) (65/87) 372 (250/122) (50/72) 217 (126/91) Different settings according to fold change and p-value are compared. The values in the brackets indicate the number of upreglulated genes followed by the number of downregulated genes
3 Page 3 of 13 an impact on a few signaling pathways which in turn affected a number of biological functions. IPA analysis revealed an activation of ERBB2 signaling as well as an activation of the TGFB1- and RAF1-dependent pathways, whereas WISP2 signaling was inhibited in R2-N1 cells compared to the parent cell line. Based on these findings, the IPA software predicted a decreased cellular viability corroborated by the prediction of an increased cell death and of elevated apoptosis in R2-N1 vs. M13SV1. Moreover, the software predicted an enhanced capability to migrate for the highly tumorigenic derivative R2-N1 compared to its parent cell line. The results of this analysis which are summarized in Table 2 are statistically significant (p < 0.05), however, according to the z-scores which in no case reached the threshold of ±2 it has to be stated that the predicted effects are based only on weak statistical power. By using the IPA software a molecular network was built that links the relevant deregulated genes to the upstream regulators as well as to the downstream biological functions (Fig. 1). According to this network, activation of ERBB2 and TGFB1 leads to a deregulation of a number of genes (MMP9, VIM, CDH2, WNT5A) which in turn results in an enhanced migration capability of the R2-N1 cell line compared to the parent cell line M13SV1. The predicted decreased cellular viability as well as increased cell death and apoptosis of this cell line are commonly due to the deregulation of a number of additional genes (DUSP1, PTGS2, MUC1, MAP2K6, IFI16, FOXG1, IL24) which are linked to the upstream regulators to a minor degree. Growth characteristics were examined and apoptosis assays were conducted in order to experimentally prove Table 2 Summary of the network analysis Activation status p-value z-score Upstream regulator ERBB2 Activated 1.27E RAF1 Activated 7.60E TGFB1 Activated 1.22E WISP2 Inhibited 3.29E Biological function Migration of breast cancer Activated 2.44E cell lines Cell viability of breast cancer Inhibited 1.64E cell lines Cell death of breast cancer Activated 3.52E cell lines Apoptosis of breast cancer cell lines Activated 2.87E Upstream regulators and biological functions that are activated or inhibited in the R2-N1 cell line in comparison to the parent cell line M13SV1 according to the results of the IPA analysis the predictions made by the IPA software with respect to the biological functions cell viability, cell death, and apoptosis. Growth curves obtained by continuous impedance measurement revealed that R2-N1 cells seemed to grow slightly slower than the two other cell lines (Fig. 2a), possibly pointing to a decreased cellular viability. The neutral red cytotoxicity assay, on the other hand, indicated that there were no significant differences in the viability between the three cell lines (Fig. 2b). Finally, simple trypan blue staining and counting of living and dead cells revealed that there were approximately % living cells in growing cultures of the three cell lines, and there was no indication for any difference in cellular viability (data not shown). Apoptosis was examined by determining caspase activities in the three cell lines. Indeed, the derivatives R2-2 and R2-N1 displayed higher basal caspase activities compared to the parent cell line M13SV1 (Fig. 3). Moreover, induction of apoptosis by staurosporin was significantly more pronounced in the two derivatives than in the parent cell line. These findings are in line with the predictions made by the IPA software. To further characterize the molecular differences between the three cell lines in particular with respect to the increasing tumorigenic potential, qpcr experiments were conducted with a focus on genes related to breast cancer. For this purpose, the Human Breast Cancer & Estrogen Receptor Signaling RT 2 PCR Array (SABiosciences) was employed to examine the expression of 84 different breast cancer-related genes in parallel. The results of this analysis are summarized in Table 3. Again, it turned out that there are only marginal differences in gene expression between the derivatives R2-2 and R2-N1, respectively, and their parent cell line M13SV1. In the case of the weakly tumorigenic cell line R2-2, only five genes were significantly deregulated compared to M13SV1 cells; these were CCNA1, KIT, KRT19, SERPINA3, and TFF1 (standard settings, fold change ± 2, p < 0.05; see Table 3). When comparing the highly tumorigenic derivative R2-N1 with its parent cell line M13SV1, only three genes (CCNE1, COL6A1, and THBS2) were significantly deregulated (Table 3). These genes, however, were not significantly affected in the transcriptomic approach. According to the gene array data, none of these genes was deregulated by a factor of at least 1.5-fold (data not shown). On the other hand, the PCR array data were suitable to confirm the results of the gene array experiment for some of the genes depicted in Fig. 1. Four of these genes, ERBB2, GATA3, PTGS2, and MUC1, were also represented by the PCR array, and in all four cases the direction and the level of deregulation between R2-N1 and M13SV1 were comparable between the gene array and the PCR array. All in all, the gene expression data obtained with the whole genome array as
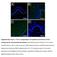
4 Page 4 of 13 Fig. 1 Network generated by the IPA software on the basis of the gene array data. The following settings were used for IPA analysis: species = human; organ = mammary gland or breast cancer cell line; only experimental data were allowed. Genes that were upregulated in R2-N1 cells compared to M13SV1 cells (fold change 1.5; p < 0.001) are highlighted in red, and downregulated genes are highlighted in green. Blue colour indicates that the IPA software predicted the respective gene to be downregulated whereas orange-coloured genes were predicted to be upregulated. The gene names are given, and the function of the respective gene products are indicated by the shape of the symbols: enzyme (vertical diamond); transcription factor (horizontal oval); cytokine (square); peptidase (horizontal diamond); growth factor (dotted square); phosphatase (triangle up); kinase (triangle down); other (circle). The relationship between two genes are indicated by a solid arrow (direct effect on gene expression), by a dotted arrow (indirect effect), or by a solid line (direct protein protein interaction). In the case of the arrows, orange colour indicates that the IPA software predicts an upregulation of the downstream gene or an activation of the downstream function whereas blue colour indicates the prediction of a downregulation or inactivation, respectively. Yellow-coloured arrows indicate inconsistency between the prediction made by IPA based on the experimental data and the literature data well as with the breast cancer PCR array lead to the conclusion, that there are only marginal differences between the three cell lines at least at the level of gene expression. In regard to the prediction that the highly tumorigenic cell line R2-N1 has an enhanced migration capability compared to the parent cell line, the growth characteristics and the morphology of the three cell lines were compared with each other. Under standard cell culture conditions, there were no apparent differences in cell growth and cell morphology when the cells grew in two dimensions on the bottom of cell culture flasks (Fig. 4a c). In the three-dimensional (3D) cell culture, all three cell lines formed spheroids within three days. These spheroids continuously grew at least until day 21, and there was no growth arrest observed for any of the cell lines (Fig. 4d f). The only difference among the three cell lines was the observation of R2-N1 cells starting to migrate out of the spheroids around day eleven (Fig. 4f), supporting the prediction that this cell line has an enhanced migration capability compared to the parent cell line. However, migrating cells leaving the spheroids were also observed to a minor degree for the other two cell lines around day 15 (data not shown), indicating that already the parent cell line M13SV1 has a migration capability. Confocal microscopy was applied to further characterize the biochemical and morphological features of
5 Page 5 of 13 a Cell Index b Absorbance Time [h] M13SV1 R2-2 R2-N1 M13SV1 R2-2 R2-N1 24 h 48 h 72 h Fig. 2 Cellular viability of the three cell lines M13SV1, R2-2, and R2-N1. a Growth curves of the three cell lines as obtained by continuous impedance measurement by using the xcelligence system (Roche); b Neutral red cytotoxicity assay. Cells of the three cell lines were seeded in 96-well plates (20,000 cells per well), and the neutral red assay was conducted after 24, 48, and 72 h, respectively. The values are the mean of three independent experiments (+SD) M13SV1 cells grown in 3D culture. Antibody staining revealed that stem cell marker such as CD24 and Oct4 were present only at day one after seeding, and that these markers were lost around day 4 in the course of spheroid growth (Fig. 5). Luminal markers (Cytokeratin 18, MUC1) were expressed right from the beginning (day one) and were present at least until day 21 whereas expression of basal markers such as Cytokeratin 14 was not detected during this period (Fig. 6). Moreover, the formation of a hollow lumen inside the spheroids due to apoptosis of the inner cells as described for other mammary epithelial cell lines such as MCF-10A [6] was not observed for the M13SV1 cell line. Within 21 days M13SV1 cells formed large spheroids of at least 100 µm in diameter that were completely filled with cells (Fig. 7), and there was no indication for any organized structure within the spheroids that would point to the formation of well-organized acini Caspase 3/7 activity [fold induction] M13SV1 R2-2 R2-N1 * * *** 0 control 0.5 µm staurosporin Fig. 3 Apoptosis as determined with a caspase 3/7 activity assay. Cells of the three cell lines were seeded in 96-well plates (15,000 cells per well). Cells were treated with 0.5 µm staurosporin to induce apoptosis (right bars). Caspase 3/7 activities were determined by using the Caspase-Glo 3/7 Assay (Promega). The values are the mean of three independent experiments (+SD) and were normalized against the value of the untreated M13SV1 cells that was set to 1. Student s t-test; *p < 0.05; **p < 0.01; ***p < as described for, e.g., the MCF-10A cell line. Accordingly, antibody staining against active Caspase-3 as a marker for apoptosis was negative (not shown). Finally, staining against specific polarization marker such as Integrin alpha 6 was also negative (not shown), indicating that M13SV1 cells do not differentiate into polarized cells as described, e.g., for MCF-10A cells [6]. Discussion The development of well-defined in vitro cell culture models for breast cancer research is a crucial issue not only for fundamental research on mammary gland carcinogenesis but also in terms of, e.g., testing of chemical substances which may have an impact on breast cancer development. With a focus on endocrine disruptors and the examination of potential carcinogenic properties of estrogenic compounds, estrogen-responsive cell lines may be preferred for the development of an in vitro test system. The M13SV1 cell line was regarded as a promising cell culture model as it was shown to be ER positive and even retained some stem cell characteristics. This feature might be regarded as an additional advantage with respect to the stem cell theory which says that numerous tumors originate from stem cells [7 9]. Indeed, M13SV1 cells have already been successfully used for toxicity testing of a number of substances. As an example, it was shown by a transcriptomic approach that treatment of the cells with 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) resulted in **
6 Page 6 of 13 Table 3 Results of the Human Breast Cancer & Estrogen Receptor Signaling RT 2 PCR Array experiment Gene name Gene symbol R2-2 vs. M13SV1 R2-N1 vs. M13SV1 Fold regulation p-value Fold regulation p-value Androgen receptor AR BCL2-associated agonist of cell death BAD BCL2-associated athanogene BAG B-cell CLL/lymphoma 2 BCL BCL2-like 2 BCL2L Complement component 3 C Cyclin A1 CCNA Cyclin A2 CCNA Cyclin D1 CCND Cyclin E1 CCNE CD44 molecule (Indian blood group) CD Cadherin 1, type 1, E-cadherin (epithelial) CDH Cyclin-dependent kinase inhibitor 1A (p21, Cip1) CDKN1A Cyclin-dependent kinase inhibitor 1B (p27, Kip1) CDKN1B Cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) CDKN2A Claudin 7 CLDN Clusterin CLU Collagen, type VI, alpha 1 COL6A Catenin (cadherin-associated protein), beta 1, 88 kda CTNNB Cathepsin B CTSB Cathepsin D CTSD Cytochrome P450, family 19, subfamily A, polypeptide 1 CYP19A Deleted in liver cancer 1 DLC Epidermal growth factor receptor EGFR V-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma ERBB derived oncogene homolog (avian) Estrogen receptor 1 ESR Estrogen receptor 2 (ER beta) ESR Fas (TNF receptor superfamily, member 6) FAS Fas ligand (TNF superfamily, member 6) FASLG Fibroblast growth factor 1 (acidic) FGF Fibronectin leucine rich transmembrane protein 1 FLRT FOS-like antigen 1 FOSL Gamma-aminobutyric acid (GABA) A receptor, pi GABRP GATA binding protein 3 GATA GNAS complex locus GNAS Gelsolin GSN High mobility group box 1 HMGB Heat shock 27 kda protein 1 HSPB Inhibitor of DNA binding 2, dominant negative helix-loop-helix protein ID Insulin-like growth factor binding protein 2, 36 kda IGFBP Interleukin 2 receptor, alpha IL2RA Interleukin 6 (interferon, beta 2) IL Interleukin 6 receptor IL6R Interleukin 6 signal transducer (gp130, oncostatin M receptor) IL6ST Integrin, alpha 6 ITGA Integrin, beta 4 ITGB Jun proto-oncogene JUN
7 Page 7 of 13 Table 3 continued Gene name Gene symbol R2-2 vs. M13SV1 R2-N1 vs. M13SV1 Fold regulation p-value Fold regulation p-value V-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog KIT Kruppel-like factor 5 (intestinal) KLF Kallikrein-related peptidase 5 KLK Keratin 18 KRT Keratin 19 KRT Mitogen-activated protein kinase kinase 7 MAP2K Antigen identified by monoclonal antibody Ki-67 MKI Metallothionein 3 MT Mucin 1, cell surface associated MUC Nuclear transcription factor Y, beta NFYB Nerve growth factor (beta polypeptide) NGF Nerve growth factor receptor NGFR Non-metastatic cells 1, protein (NM23A) expressed in NME Pregnancy-associated plasma protein A, pappalysin 1 PAPPA Progesterone receptor PGR Plasminogen activator, urokinase PLAU Phosphatase and tensin homolog PTEN Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and PTGS cyclooxygenase) Ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding RAC protein Rac2) Ribosomal protein L27 RPL Secretoglobin, family 1D, member 2 SCGB1D Secretoglobin, family 2A, member 1 SCGB2A Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), SERPINA member 3 Serpin peptidase inhibitor, clade B (ovalbumin), member 5 SERPINB Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type SERPINE ), member 1 Solute carrier family 7 (amino acid transporter light chain, L system), member 5 SLC7A Small proline-rich protein 1B SPRR1B Stanniocalcin 2 STC Trefoil factor 1 TFF Transforming growth factor, alpha TGFA Thrombospondin 1 THBS Thrombospondin 2 THBS Tyrosine kinase with immunoglobulin-like and EGF-like domains 1 TIE Tumor necrosis factor, alpha-induced protein 2 TNFAIP Topoisomerase (DNA) II alpha 170 kda TOP2A Tumor protein p53 TP Vascular endothelial growth factor A VEGFA Akt and ERK2 activation which in turn stimulated cellular proliferation [10]. The authors concluded from their transcriptomic data that the pattern of alteration in gene expression was indicative for TCDD-induced breast tumorigenesis. The two tumorigenic derivatives of M13SV1, however, were not included in this study. In another example, it was shown that treatment of the cells with all-trans retinoic acid resulted in decreased proliferation of M13SV1 cells and of its weakly tumorigenic derivative R2-2, but proliferation was not affected in the highly tumorigenic cell line R2-N1 [11]. Finally, the molecular effects of the carcinogen 7,12-dimethylbenz(a)anthracene
8 Page 8 of 13 M13SV1 R2-2 R2-N1 a b c d e f Fig. 4 Phase-contrast microscopy. Pictures of the three cell lines as indicated above the pictures grown either in 2D culture (a c) or in 3D culture (d f; day eleven after seeding) CD24; day 1 Oct4; day 1 CD24; day 4 Oct4; day 4 a b c d e f g h Fig. 5 Expression of stem cell markers in M13SV1 cells. Cells were grown in 3D culture and stained either against the stem cell marker CD24 or Oct4, both at day 1 as well at day 4 after seeding as indicated above the pictures. a d Overlay of the phase-contrast picture and the GFP stain; e h GFP stain only (DMBA) on the three cell lines were examined, and it was shown that the non-tumorigenic cell line M13SV1 was more susceptible to the mutagen than its tumorigenic derivatives in terms of, e.g., oxidative DNA damage, formation of DNA adducts, and alteration of expression of specific cancer-related genes [12]. The aim of the present study was to compare the M13SV1 cell line with its two tumorigenic derivatives on the molecular level. Thus, the transcriptomes of the three cell lines were compared with each other in order to identify the differences in gene expression which were supposed to be related to the different tumorigenic
9 Page 9 of 13 Cytokeratin 18 MUC1 Cytokeratin 14 Fig. 6 Expression of luminal and basal epithelial cell markers in the M13SV1 cell line. Cells were grown in 3D culture and stained at day 4 after seeding against the luminal markers Cytokeratin 18 or MUC1, or against the basal marker Cytokeratin 14 as indicated above the pictures Fig. 7 Representative picture of a M13SV1 spheroid grown in 3D culture for 21 days. Nuclei were stained with SYTOX Orange Nucleic Acid Stain, and the cytoskeleton was stained with Alexa Fluor 488 Phalloidin 488. The picture was obtained with a confocal microscope and shows a cut through the middle layer of the spheroid phenotypes. It has been reported in an earlier transcriptomic study that immortalization of type I HBEC with SV40 DNA to give the M13SV1 cell line was accompanied by the deregulation of 148 genes with a fold change of at least twofold [13], however, a transcriptomic comparison of the M13SV1 cell line with its tumorigenic derivatives has not been reported so far. Although 3D cell culture undoubtedly has a number of tremendous advantages over conventional 2D-cultured cells (see below), we decided to use 2D-cultured cells for the transcriptomic approach for some practical reasons. With respect to a possible use of the cell lines for future high throughput in vitro toxicity testing, 2D culture is much cheaper and much faster than 3D-cultured cells which need approximately 20 days for full differentiation. Moreover, compared to 3D-cultured cells, 2D culture yields much more cell material for subsequent studies. Finally, Kenny and colleagues reported for a number of breast epithelial cell lines that the transcriptomes of 2D-cultures cells are very similar to the transcriptomes of the same cell line cultured under 3D conditions [14]. Our analysis, however, revealed that there are only marginal differences between the gene expression patterns of the three cell lines cultured under 2D conditions. In the case of the weakly tumorigenic cell line R2-2, only 122 out of 24,266 transcripts were significantly deregulated by a factor of at least 1.5-fold compared to its parent cell line M13SV1. The R2-2 cell line was derived from M13SV1 cells by X-ray irradiation, and the exact genetic alterations resulting from this treatment are unknown, however, the tumorigenicity which distinguishes R2-2 cells from M13SV1 cells in vivo is obviously not reflected on the transcriptomic level under in vitro conditions. Thus, post-transcriptional alterations and/or specific interactions with neighboring cells in the in vivo situation may be more relevant for the development of the tumorigenic phenotype. The R2-N1 cell line, on the other hand, was derived from R2-2 cell by a specific genetic modification, namely the introduction of the rat Erbb2 oncogene into the genome. Again, there were surprisingly few transcriptional alterations as only 217 transcripts were significantly deregulated by a factor of at least 1.5-fold between R2-N1 cells and M13SV1 cells. Indeed, the pattern of deregulated genes partly correlated with ERBB2 activation, however, the predicted alterations in biological functions such as
10 Page 10 of 13 increased apoptosis and decreased cellular viability were of only weak statistical power and were hardly reproducible with biological assays. Again, there were only marginal differences in gene expression in vitro between R2-N1 cells and the M13SV1 parent cell line which do not reflect the strong differences with respect to tumorigenicity of these two cell lines under in vivo conditions. Taken together, the transcriptomic approach revealed that there is no obvious set of biomarkers which might have been useful in regard to establish an in vitro test system based on these three cell lines. The use of a PCRarray with specific genes relevant for breast cancer and estrogen receptor signaling did also not unravel strong differences among the three cell lines, again leading to the conclusion that the three cell lines are very similar to each other on the transcriptomic level under 2D conditions. In order to establish an in vitro test system for human breast epithelial cell lines suitable to monitor substanceinduced deregulation of the differentiation process it is crucial to use a cell line which differentiates in 3D culture as similar as possible compared to the in vivo situation in regard to mammary gland formation. Kenny and colleagues have classified 25 breast cell lines into four groups according to their colony morphology [14]. Due to this classification scheme, cell lines that belong to the so-called round class seem to be most appropriate to be used for an in vitro test system. Cell lines with a round phenotype are characterized by the formation of polarized, growth-arrested colonies with many morphological features of mammary acini such as the formation of a hollow lumen due to the fact that the inner cells of the spheroids undergo apoptosis. Due to these features, cell lines with a round phenotype display the most complex differentiation program which is very similar to HBEC differentiation in vivo. The M13SV1 cell line, however, belongs to the so-called mass class according to the classification by Kenny et al. [14]. Cells with a mass phenotype still form spheroids, but there is no polarization of cells, no growth arrest, and no formation of a hollow lumen. In fact, the M13SV1 cell line as well as its two tumorigenic derivatives belong to the mass class and display very similar morphological features when grown in 3D culture. Finally, the presence of distinct stem cell markers was regarded as an advantageous feature of M13SV1 cells, however, these markers are rapidly lost in 3D culture in the course of spheroid growth. Thus, in conclusion, our analysis revealed that the M13SV1 cell line and its two tumorigenic derivatives seem to be less suitable for the development of an in vitro test system than other permanent HBEC cell lines that belong to the round class such as the MCF-12A cell line or the MPE-600 cell line [14]. Methods Cell culture The HBEC lines M13SV1, M13SV1-R2-2 (designated as R2-2 cells in the following) and M13SV1-R2-N1 (designated as R2-N1 cells in the following) were kindly provided by the working group of James E. Trosko (Department of Pediatrics/Human Development, College of Human Medicine, Michigan State University, East Lansing, USA). The cells were routinely cultured in phenol red- and hormone-free Michigan State University-1 (MSU-1) medium [3] (custom-built; Biochrom, Merck Millipore, Berlin, Germany) supplemented with 5 % charcoal stripped fetal bovine serum (PAA, Pasching, Austria), units/ml penicillin and 10 mg/ml streptomycin (PAA, Pasching, Austria). All cells were grown in a humidified atmosphere of 37 C and 5 % CO 2. For all experiments, cell cultures at a confluence of about % were washed once with phosphate buffered saline (PBS) and trypsinized (0.05 % trypsin/0.02 % EDTA). Cells were resuspended in culture medium and trypan blue staining was used to determine viability before seeding. Only cells with a viability of at least 90 % were used for experiments. Sample preparation and purification of total RNA The HBEC lines were seeded in separate cell culture flasks (25 cm 2 ). Purification of total RNA was performed using the RNeasy Mini Kit according to manufacturer s recommendations (Qiagen, Hilden, Germany). At a confluence of about % the cells were washed twice with icecold PBS, directly disrupted in lysis buffer and homogenized using QIAshredder spin columns (Qiagen, Hilden, Germany). An additional on-column DNAse digestion was performed on all samples to remove residual DNA (RNase-Free DNase Set; Qiagen, Hilden, Germany). The concentration of RNA was determined with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and integrity was assessed by using Agilent RNA 6000 Nano LabChip kit with the Agilent 2100 Bioanalyzer according to manufacturer s protocol (Agilent Technologies, Palo Alto, CA, USA). All samples for the microarray analysis as well as PCR array experiments should have a RNA Integrity Number (RIN), a measure of degradation, of 8 or higher (RIN; 1 = totally degraded, 10 = intact). Gene array analysis The gene array experiment was conducted by Eurofins Medigenomix GmbH (Ebersberg, Germany) by using Human Transcriptome 2.0 Arrays (HTA 2.0). 100 ng of total RNA was used for labelling and hybridization according to the protocols of Eurofins. Three independent biological replicates were processed for each cell line.
11 Page 11 of 13 Lists of differentially expressed genes between two cell lines were generated using the Transcriptome Analysis Console (TAC) 2.0 software (Affymetrix, Santa Clara, CA, USA). The resulting datasets were further processed by using the software Ingenuity Pathway Analysis (IPA; spring release March 2015; Qiagen, Redwood City, CA, USA) which eliminated all unmapped probe sets and which furthermore only allowed the probe set with the highest fold change for further analysis in the case that a single gene was represented by more than one probe set on the microarray. PCR array The Human Breast Cancer & Estrogen Receptor Signaling RT 2 PCR Array System (PAHS-005E) containing 84 specific primer sets was used according to the manufacturer s protocol (SABiosciences, Frederick, MD, USA). 1 μg total RNA was reverse-transcribed to cdna with the RT 2 First Strand Kit (SABiosciences, Frederick, MD, USA). The cdna was combined with RT 2 SYBR Green/ROX PCR master mix (SABiosciences, Frederick, MD, USA), and 10 µl of the mixture was added to each well of the PCR array (384-well format). The plates were run under conditions recommended by the supplier using the ABI-7900HT instrument (Applied Biosystems, Foster City, CA, USA). Data analysis was performed on four biological replicates per HBEC line using the C t method with an online analysis tool provided by the manufacturer of the PCR array (RT 2 Profiler PCR Array Data Analysis version 3.5) as described in detail elsewhere [15]. Data were normalized against a set of four endogenous control genes (B2M, HPRT1, GAPDH, and ACTB) present on the array. Resulting Ct values were converted into fold changes of tumorigenic cell lines relative to non-tumorigenic M13SV1 cells using the 2 Ct method. Genes that achieved a p-value <0.05 were considered as statistically significant for the study. Impedance measurement with the xcelligence system The xcelligence real-time cell analyzer (RTCA) system (Roche and ACEA Biosciences) is a label-free and non-invasive assay system based on a continuous measurement of electrical impedance to monitor biological processes of living cells in real time. The impedance is affected by the cells attached on the electrodes and gives information about their biological status, including cell number, viability, morphology and the degree of cell adhesion [16]. Growth of the HBEC lines was continuously monitored for at least 24 h using 96-well plates that contain microelectrodes (E-plate 96) and the RTCA SP instrument (Roche, Rotkreuz, Switzerland). The measurement of the impedance background was performed with 100 µl of cell culture medium per well. After adding the cell suspension the final volume in a single well was 200 μl (10,000 cells/ well). Electric impedance was recorded every 30 min. The impedance is displayed as Cell Index (CI), a relative and dimensionless value directly influenced by cell attachment, spreading and cell proliferation. Cytotoxicity assays The neutral red cytotoxicity assay was conducted as described in Buhrke et al. [17]. Trypan blue staining was employed to determine the ratio between living and dead cells by using the EVE Automatic cell counter (NanoEn- Tek, Seoul, Korea). Caspase 3/7 assay The activity of caspase 3 and 7 was detected using the Caspase-Glo 3/7 Assay (Promega, Mannheim, Germany). The cells were seeded in white walled 96-well plates at a density of 15,000 cells per well and allowed to attach. Twenty-four hours after plating cells were treated in triplicate with 0.5 µm staurosporin and DMSO as solvent control (0.5 %). Caspase-3 and -7 activities were determined after 12 h after staurosporin treatment according to the manufacturer s protocol. Caspase-Glo 3/7 reagent was added (25 µl/well), gently mixed, and incubated for 30 min. The assay is based on the cleavage of a proluminogenic substrate, containing a DEVD sequence, by the caspases 3 and 7 which generates a luminescent signal produced by luciferase. Luminescence, that is proportional to the amount of caspase activity present, was measured using an Infinite M200 PRO multimode reader (Tecan, Männedorf, Switzerland). Three dimensional (3D) cell culture Approximately 2000 cells per cm 2 of M13SV1, R2-2, and R2-N1 cells, respectively, were seeded on a 1 2 mm solidified layer of Growth Factor Reduced Matrigel (BD Bioscience, Heidelberg, Germany) according to the manufacturer s protocol one modification. The original matrigel was diluted with PBS to a final concentration of 5 mg/ml. The cells were grown in MSU-1 medium (see above). The medium was replaced every 2 3 days. Microscopy Light microscopic analysis of the HBEC cell lines was carried out using the Carl Zeiss inverse Axiovert 135 microscope (Zeiss, Berlin, Germany). A confocal laser scanning microscope (DMI6000, Leica, Mannheim, Germany) was used to visualize the immunofluorescent stains of cells grown in 3D culture. All primary antibodies were purchased from Abcam (Cambridge, United Kingdom): anti-cd24 (ab30350), anti-cytokeratin 14 (ab51054),
12 Page 12 of 13 anti-cytokeratin 18 (ab668), anti-muc1 (ab28081), anti- Oct4 (ab19857), anti-active Caspase-3 (ab32042) and anti- Integrin alpha 6 (ab20142). All secondary antibodies were purchased from Life Technologies (Carlsbad, CA, USA): Alexa Fluor 488 F(ab )2 Fragment of Goat Anti-Rabbit IgG (A-11070), Alexa Fluor 546 F(ab )2 Fragment of Goat Anti-Rabbit IgG (A-11070), Alexa Fluor 488 F(ab )2 fragment of goat anti-mouse IgG (A-11017) and Alexa Fluor 546 F(ab )2 fragment of goat anti-mouse IgG (A-11018). The spheroids grown in 3D culture were fixed with 3.7 % paraformaldehyde (Sigma-Aldrich, Taufkirchen, Germany) for 10 min at room temperature. After three washing steps (for 5 min each) with PBS containing 0.1 % Tween-20 (PBS-T), the spheroids were permeabilized by 0.2 % Triton X-100 for 10 min at room temperature. After another washing cycle, the spheroids were blocked by using 10 % bovine serum albumin for 15 min. Subsequently, the spheroids were incubated with the first antibody (dilution 1:100 in PBS) for 1 h at 37 C. Afterwards, the spheroids were washed again with PBS-T and then incubated with a secondary antibody (dilution 1:200 in PBS) for 20 min at 37 C. Unbound secondary antibody was removed by washing the cells three times for 5 min with PBS-T. To counterstain the nuclei and the cytoskeleton the spheroids were incubated with Alexa Fluor 488 Phalloidin (dilution 1:40 in PBS, Molecular Probes) and SYTOX Orange Nucleic Acid Stain (dilution 1:5000 in PBS, Molecular Probes) for 30 min. Statistics In the gene array experiment, statistically significant deregulated genes were identified by a one-way ANOVA test (p < 0.05). Evaluation of the gene array data with the IPA software yielded z-scores in addition to p-values with respect to the level of deregulation of molecular networks. The z-score indicates how many standard deviations the sample lies away from the mean. Networks with z-scores < 2 are regarded as significantly inactivated, and z-scores >2 indicate significant activation of the respective network. The statistical data analysis of the PCR array was performed by using the web portal software of SABiosciences. Student s t-test was used, and p-values <0.05 were considered statistically significant. The neutral red cytotoxicity data as well as the caspase 3/7 activity results were evaluated by using Student s t-test, and p < 0.05 was regarded as statistically significant. Additional files Additional file 1: Table S1. Gene expression analysis list of deregulated genes R2-2 vs. M13SV1 (fold change > 1.5; p < 0.001). Additional file 2: Table S2. Gene expression analysis list of deregulated genes R2-N1 vs. M13SV1 (fold change > 1.5; p < 0.001). Authors contributions SS carried out the PCR array analysis, the impedance measurements, and the viability assays. AE conducted the gene array experiment, and the 3D cell culture including immunofluorescence staining and microscopy. NW performed the caspase assay. TB conducted the network analysis and prepared the manuscript. AL conceived of the study, and participated in its design and helped to draft the manuscript. All authors read and approved the final manuscript. Acknowledgements We thank J.E. Trosko and C.C. Chang (Department of Pediatrics/Human Development, College of Human Medicine, Michigan State University, East Lansing, USA) for providing the cell lines M13SV1, R2-2, and R2-N1. Beatrice Rosskopp, Linda Brandenburger, and Anja Köllner are acknowledged for technical assistance. This work was funded by the Federal Institute for Risk Assessment (Project No and ). Competing interests The authors declare that they have no competing interests. Received: 2 October 2015 Accepted: 20 November 2015 References 1. Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23: Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011;13: Kao CY, Nomata K, Oakley CS, Welsch CW, Chang CC. Two types of normal human breast epithelial cells derived from reduction mammoplasty: phenotypic characterization and response to SV40 transfection. Carcinogenesis. 1995;16: Kang KS, Morita I, Cruz A, Jeon YJ, Trosko JE, Chang CC. Expression of estrogen receptors in a normal human breast epithelial cell type with luminal and stem cell characteristics and its neoplastically transformed cell lines. Carcinogenesis. 1997;18: Kang KS, Sun W, Nomata K, Morita I, Cruz A, Liu CJ, Trosko JE, Chang CC. Involvement of tyrosine phosphorylation of p185(c-erbb2/neu) in tumorigenicity induced by X-rays and the neu oncogene in human breast epithelial cells. Mol Carcinog. 1998;21: Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30: Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414: Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8: Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea a paradigm shift. Cancer Res. 2006;66: Ahn NS, Hu H, Park JS, Park JS, Kim JS, An S, Kong G, Aruoma OI, Lee YS, Kang KS. Molecular mechanisms of the 2,3,7,8-tetrachlorodibenzo-pdioxin-induced inverted U-shaped dose responsiveness in anchorage independent growth and cell proliferation of human breast epithelial cells with stem cell characteristics. Mutat Res. 2005;579: Ahn EH, Chang CC, Talmage DA. Loss of anti-proliferative effect of alltrans retinoic acid in advanced stage of breast carcinogenesis. Anticancer Res. 2009;29: De Flora S, Scarfi S, Izzotti A, D Agostini F, Chang CC, Bagnasco M, De Flora A, Trosko JE. Induction by 7,12-dimethylbenz(a)anthracene of molecular and biochemical alterations in transformed human mammary epithelial stem cells, and protection by N-acetylcysteine. Int J Oncol. 2006;29: Park JS, Noh DY, Kim SH, Kim SH, Kong G, Chang CC, Lee YS, Trosko JE, Kang KS. Gene expression analysis in SV40-immortalized human breast luminal epithelial cells with stem cell characteristics using a cdna microarray. Int J Oncol. 2004;24: Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, et al. The morphologies of breast
13 Page 13 of 13 cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1: Hansel NN, Cheadle C, Diette GB, Wright J, Thompson KM, Barnes KC, Georas SN. Analysis of CD4+ T-cell gene expression in allergic subjects using two different microarray platforms. Allergy. 2008;63: Kho D, MacDonald C, Johnson R, Unsworth CP, O Carroll SJ, du Mez E, Angel CE, Graham ES. Application of xcelligence RTCA biosensor technology for revealing the profile and window of drug responsiveness in real time. Biosensors (Basel). 2015;5: Buhrke T, Kibellus A, Lampen A. In vitro toxicological characterization of perfluorinated carboxylic acids with different carbon chain lengths. Toxicol Lett. 2013;218: Submit your next manuscript to BioMed Central and we will help you at every step: We accept pre-submission inquiries Our selector tool helps you to find the most relevant journal We provide round the clock customer support Convenient online submission Thorough peer review Inclusion in PubMed and all major indexing services Maximum visibility for your research Submit your manuscript at
Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3
 Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array.
 Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Focus Application. Compound-Induced Cytotoxicity
 xcelligence System Real-Time Cell Analyzer Focus Application Compound-Induced Cytotoxicity Featured Study: Using the Time Resolving Function of the xcelligence System to Optimize Endpoint Viability and
xcelligence System Real-Time Cell Analyzer Focus Application Compound-Induced Cytotoxicity Featured Study: Using the Time Resolving Function of the xcelligence System to Optimize Endpoint Viability and
Supplementary Figure 1: Neuregulin 1 increases the growth of mammary organoids compared to EGF. (a) Mammary epithelial cells were freshly isolated,
 1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1: Neuregulin 1 increases the growth of mammary organoids compared to EGF. (a) Mammary epithelial cells were freshly isolated, embedded in matrigel and exposed
1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1: Neuregulin 1 increases the growth of mammary organoids compared to EGF. (a) Mammary epithelial cells were freshly isolated, embedded in matrigel and exposed
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
Focus Application. Compound-Induced Cytotoxicity
 xcelligence System Real-Time Cell Analyzer Focus Application Compound-Induced Cytotoxicity For life science research only. Not for use in diagnostic procedures. Featured Study: Using the Time Resolving
xcelligence System Real-Time Cell Analyzer Focus Application Compound-Induced Cytotoxicity For life science research only. Not for use in diagnostic procedures. Featured Study: Using the Time Resolving
Cancer. The fundamental defect is. unregulated cell division. Properties of Cancerous Cells. Causes of Cancer. Altered growth and proliferation
 Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Cancer. The fundamental defect is. unregulated cell division. Properties of Cancerous Cells. Causes of Cancer. Altered growth and proliferation
 Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Enzyme-coupled Receptors. Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors
 Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small cell lung cancer
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Experimental Methods Cell culture B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Experimental Methods Cell culture B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small
The attached protocol was used for growing RWPE-1 cells per ATCC instructions.
 RWPE-1 (prostate epithelial transformed by HPV) From: Duke/UNC/UT/EBI ENCODE group Date: 7/7/11 ATCC: Catalog #CRL-11609 The attached protocol was used for growing RWPE-1 cells per ATCC instructions. Cell
RWPE-1 (prostate epithelial transformed by HPV) From: Duke/UNC/UT/EBI ENCODE group Date: 7/7/11 ATCC: Catalog #CRL-11609 The attached protocol was used for growing RWPE-1 cells per ATCC instructions. Cell
SUPPLEMENTARY DATA. Assessment of cell survival (MTT) Assessment of early apoptosis and cell death
 Overexpression of a functional calcium-sensing receptor dramatically increases osteolytic potential of MDA-MB-231 cells in a mouse model of bone metastasis through epiregulinmediated osteoprotegerin downregulation
Overexpression of a functional calcium-sensing receptor dramatically increases osteolytic potential of MDA-MB-231 cells in a mouse model of bone metastasis through epiregulinmediated osteoprotegerin downregulation
RNA preparation from extracted paraffin cores:
 Supplementary methods, Nielsen et al., A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor positive breast cancer.
Supplementary methods, Nielsen et al., A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor positive breast cancer.
MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands)
 Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Deregulation of signal transduction and cell cycle in Cancer
 Deregulation of signal transduction and cell cycle in Cancer Tuangporn Suthiphongchai, Ph.D. Department of Biochemistry Faculty of Science, Mahidol University Email: tuangporn.sut@mahidol.ac.th Room Pr324
Deregulation of signal transduction and cell cycle in Cancer Tuangporn Suthiphongchai, Ph.D. Department of Biochemistry Faculty of Science, Mahidol University Email: tuangporn.sut@mahidol.ac.th Room Pr324
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the
 Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening
 Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening Introduction: Vimentin (VIM) intermediate filament (IF) proteins are associated with EMT in lung cancer and its metastatic
Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening Introduction: Vimentin (VIM) intermediate filament (IF) proteins are associated with EMT in lung cancer and its metastatic
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature15260 Supplementary Data 1: Gene expression in individual basal/stem, luminal, and luminal progenitor cells. Box plots show expression levels for each gene from the 49-gene differentiation
doi:10.1038/nature15260 Supplementary Data 1: Gene expression in individual basal/stem, luminal, and luminal progenitor cells. Box plots show expression levels for each gene from the 49-gene differentiation
Genetics and Cancer Ch 20
 Genetics and Cancer Ch 20 Cancer is genetic Hereditary cancers Predisposition genes Ex. some forms of colon cancer Sporadic cancers ~90% of cancers Descendants of cancerous cells all cancerous (clonal)
Genetics and Cancer Ch 20 Cancer is genetic Hereditary cancers Predisposition genes Ex. some forms of colon cancer Sporadic cancers ~90% of cancers Descendants of cancerous cells all cancerous (clonal)
qpcr-array Analysis Service
 qpcr-array Analysis Service Customer Name Institute Telephone Address E-mail PO Number Service Code Report Date Service Laboratory Department Phalanx Biotech Group, Inc 6 Floor, No.6, Technology Road 5,
qpcr-array Analysis Service Customer Name Institute Telephone Address E-mail PO Number Service Code Report Date Service Laboratory Department Phalanx Biotech Group, Inc 6 Floor, No.6, Technology Road 5,
Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein
 Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein Lei Wang 1, Tian-Peng Zhang 1, Yuan Zhang 2, Hai-Lian
Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein Lei Wang 1, Tian-Peng Zhang 1, Yuan Zhang 2, Hai-Lian
CHAPTER VII CONCLUDING REMARKS AND FUTURE DIRECTION. Androgen deprivation therapy is the most used treatment of de novo or recurrent
 CHAPTER VII CONCLUDING REMARKS AND FUTURE DIRECTION Stathmin in Prostate Cancer Development and Progression Androgen deprivation therapy is the most used treatment of de novo or recurrent metastatic PCa.
CHAPTER VII CONCLUDING REMARKS AND FUTURE DIRECTION Stathmin in Prostate Cancer Development and Progression Androgen deprivation therapy is the most used treatment of de novo or recurrent metastatic PCa.
Dominic J Smiraglia, PhD Department of Cancer Genetics. DNA methylation in prostate cancer
 Dominic J Smiraglia, PhD Department of Cancer Genetics DNA methylation in prostate cancer Overarching theme Epigenetic regulation allows the genome to be responsive to the environment Sets the tone for
Dominic J Smiraglia, PhD Department of Cancer Genetics DNA methylation in prostate cancer Overarching theme Epigenetic regulation allows the genome to be responsive to the environment Sets the tone for
Problem Set 8 Key 1 of 8
 7.06 2003 Problem Set 8 Key 1 of 8 7.06 2003 Problem Set 8 Key 1. As a bright MD/PhD, you are interested in questions about the control of cell number in the body. Recently, you've seen three patients
7.06 2003 Problem Set 8 Key 1 of 8 7.06 2003 Problem Set 8 Key 1. As a bright MD/PhD, you are interested in questions about the control of cell number in the body. Recently, you've seen three patients
Supplementary Materials and Methods
 Supplementary Materials and Methods Immunoblotting Immunoblot analysis was performed as described previously (1). Due to high-molecular weight of MUC4 (~ 950 kda) and MUC1 (~ 250 kda) proteins, electrophoresis
Supplementary Materials and Methods Immunoblotting Immunoblot analysis was performed as described previously (1). Due to high-molecular weight of MUC4 (~ 950 kda) and MUC1 (~ 250 kda) proteins, electrophoresis
Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice
 Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice undergoing AD- and psoriasis-like disease. Immunofluorescence staining for Fn14 (green) and DAPI (blue) in skin of naïve
Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice undergoing AD- and psoriasis-like disease. Immunofluorescence staining for Fn14 (green) and DAPI (blue) in skin of naïve
Supplementary Figure (OH) 22 nanoparticles did not affect cell viability and apoposis. MDA-MB-231, MCF-7, MCF-10A and BT549 cells were
 Supplementary Figure 1. Gd@C 82 (OH) 22 nanoparticles did not affect cell viability and apoposis. MDA-MB-231, MCF-7, MCF-10A and BT549 cells were treated with PBS, Gd@C 82 (OH) 22, C 60 (OH) 22 or GdCl
Supplementary Figure 1. Gd@C 82 (OH) 22 nanoparticles did not affect cell viability and apoposis. MDA-MB-231, MCF-7, MCF-10A and BT549 cells were treated with PBS, Gd@C 82 (OH) 22, C 60 (OH) 22 or GdCl
Epstein-Barr virus driven promoter hypermethylated genes in gastric cancer
 RESEARCH FUND FOR THE CONTROL OF INFECTIOUS DISEASES Epstein-Barr virus driven promoter hypermethylated genes in gastric cancer J Yu *, KF To, QY Liang K e y M e s s a g e s 1. Somatostatin receptor 1
RESEARCH FUND FOR THE CONTROL OF INFECTIOUS DISEASES Epstein-Barr virus driven promoter hypermethylated genes in gastric cancer J Yu *, KF To, QY Liang K e y M e s s a g e s 1. Somatostatin receptor 1
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Cell cycle, signaling to cell cycle, and molecular basis of oncogenesis
 Cell cycle, signaling to cell cycle, and molecular basis of oncogenesis MUDr. Jiří Vachtenheim, CSc. CELL CYCLE - SUMMARY Basic terminology: Cyclins conserved proteins with homologous regions; their cellular
Cell cycle, signaling to cell cycle, and molecular basis of oncogenesis MUDr. Jiří Vachtenheim, CSc. CELL CYCLE - SUMMARY Basic terminology: Cyclins conserved proteins with homologous regions; their cellular
Muse Assays for Cell Analysis
 Muse Assays for Cell Analysis Multiple Assay Outputs for Cell Analysis Cell Health Cell Signalling Immunology Muse Count & Viability Kit Muse Cell Cycle Kit Muse Annexin V & Dead Cell Kit Muse Caspase
Muse Assays for Cell Analysis Multiple Assay Outputs for Cell Analysis Cell Health Cell Signalling Immunology Muse Count & Viability Kit Muse Cell Cycle Kit Muse Annexin V & Dead Cell Kit Muse Caspase
Supporting Information
 Supporting Information Identification of Novel ROS Inducers: Quinone Derivatives Tethered to Long Hydrocarbon Chains Yeonsun Hong,, Sandip Sengupta,, Wooyoung Hur, *, Taebo Sim,, * KU-KIST Graduate School
Supporting Information Identification of Novel ROS Inducers: Quinone Derivatives Tethered to Long Hydrocarbon Chains Yeonsun Hong,, Sandip Sengupta,, Wooyoung Hur, *, Taebo Sim,, * KU-KIST Graduate School
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Crosstalk between Adiponectin and IGF-IR in breast cancer. Prof. Young Jin Suh Department of Surgery The Catholic University of Korea
 Crosstalk between Adiponectin and IGF-IR in breast cancer Prof. Young Jin Suh Department of Surgery The Catholic University of Korea Obesity Chronic, multifactorial disorder Hypertrophy and hyperplasia
Crosstalk between Adiponectin and IGF-IR in breast cancer Prof. Young Jin Suh Department of Surgery The Catholic University of Korea Obesity Chronic, multifactorial disorder Hypertrophy and hyperplasia
Suppl Video: Tumor cells (green) and monocytes (white) are seeded on a confluent endothelial
 Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 )
 770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
SUPPLEMENT. Materials and methods
 SUPPLEMENT Materials and methods Cell culture and reagents Cell media and reagents were from Invitrogen unless otherwise indicated. Antibiotics and Tet-certified serum were from Clontech. In experiments
SUPPLEMENT Materials and methods Cell culture and reagents Cell media and reagents were from Invitrogen unless otherwise indicated. Antibiotics and Tet-certified serum were from Clontech. In experiments
Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h)
 Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h) after feeding. A small slice (~5-1 mm 3 ) was taken
Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h) after feeding. A small slice (~5-1 mm 3 ) was taken
* Kyoto Encyclopedia of Genes and Genomes.
 Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Erzsebet Kokovay, Susan Goderie, Yue Wang, Steve Lotz, Gang Lin, Yu Sun, Badrinath Roysam, Qin Shen,
 Cell Stem Cell, Volume 7 Supplemental Information Adult SVZ Lineage Cells Home to and Leave the Vascular Niche via Differential Responses to SDF1/CXCR4 Signaling Erzsebet Kokovay, Susan Goderie, Yue Wang,
Cell Stem Cell, Volume 7 Supplemental Information Adult SVZ Lineage Cells Home to and Leave the Vascular Niche via Differential Responses to SDF1/CXCR4 Signaling Erzsebet Kokovay, Susan Goderie, Yue Wang,
Supplementary Information. Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase
 Journal: Nature Medicine Supplementary Information Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase 1,2 Peng Wang PhD, 1,2 Juan-Carlos Alvarez-Perez
Journal: Nature Medicine Supplementary Information Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase 1,2 Peng Wang PhD, 1,2 Juan-Carlos Alvarez-Perez
Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression
 Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Figure S1. B % of Phosphorylation 32H. 32ss
 Figure S1 8H 32ss 32H 32Hc % of Phosphorylation 3 32H 2 1 32ss 1 2 3 4 Extract (μg) C % of Phosphorylation 18 12 6-32H 32Hc 8H 32ss Dbait Figure S1. List of the Dbait molecules and activation of DN-PK
Figure S1 8H 32ss 32H 32Hc % of Phosphorylation 3 32H 2 1 32ss 1 2 3 4 Extract (μg) C % of Phosphorylation 18 12 6-32H 32Hc 8H 32ss Dbait Figure S1. List of the Dbait molecules and activation of DN-PK
Assessment of pro-arrhythmic effects using Pluricyte Cardiomyocytes. on the ACEA xcelligence RTCA CardioECR
 Assessment of pro-arrhythmic effects using Pluricyte Cardiomyocytes on the ACEA xcelligence RTCA CardioECR User Guide Version 3.0 / March 2018 Contents 1. Introduction 2 2. Workflow 3 3. Important recommendations
Assessment of pro-arrhythmic effects using Pluricyte Cardiomyocytes on the ACEA xcelligence RTCA CardioECR User Guide Version 3.0 / March 2018 Contents 1. Introduction 2 2. Workflow 3 3. Important recommendations
RAS Genes. The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes.
 ۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
A549 and A549-fLuc cells were maintained in high glucose Dulbecco modified
 Cell culture and animal model A549 and A549-fLuc cells were maintained in high glucose Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum at 37 C in humidified atmosphere containing
Cell culture and animal model A549 and A549-fLuc cells were maintained in high glucose Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum at 37 C in humidified atmosphere containing
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Plate-Based Assay Methods for the Assessment of Cellular Health
 Plate-Based Assay Methods for the Assessment of Cellular Health Andrew L. Niles, Senior Research Scientist 2012, Promega Corporation. Biological Outcomes in Cell Culture Treatment -Small molecule -Bio-molecule
Plate-Based Assay Methods for the Assessment of Cellular Health Andrew L. Niles, Senior Research Scientist 2012, Promega Corporation. Biological Outcomes in Cell Culture Treatment -Small molecule -Bio-molecule
SUPPLEMENTARY INFORMATION
 b 350 300 250 200 150 100 50 0 E0 E10 E50 E0 E10 E50 E0 E10 E50 E0 E10 E50 Number of organoids per well 350 300 250 200 150 100 50 0 R0 R50 R100 R500 1st 2nd 3rd Noggin 100 ng/ml Noggin 10 ng/ml Noggin
b 350 300 250 200 150 100 50 0 E0 E10 E50 E0 E10 E50 E0 E10 E50 E0 E10 E50 Number of organoids per well 350 300 250 200 150 100 50 0 R0 R50 R100 R500 1st 2nd 3rd Noggin 100 ng/ml Noggin 10 ng/ml Noggin
Supplementary Figure 1:
 Supplementary Figure 1: (A) Whole aortic cross-sections stained with Hematoxylin and Eosin (H&E), 7 days after porcine-pancreatic-elastase (PPE)-induced AAA compared to untreated, healthy control aortas
Supplementary Figure 1: (A) Whole aortic cross-sections stained with Hematoxylin and Eosin (H&E), 7 days after porcine-pancreatic-elastase (PPE)-induced AAA compared to untreated, healthy control aortas
Serafino et al. Thymosin α1 activates complement receptor-mediated phagocytosis in human monocyte-derived macrophages. SUPPLEMENTARY FIGURES
 Supplementary Fig. S1. Evaluation of the purity and maturation of macrophage cultures tested by flow cytometry. The lymphocytic/monocytic cellular fraction was isolated from buffy coats of healthy donors
Supplementary Fig. S1. Evaluation of the purity and maturation of macrophage cultures tested by flow cytometry. The lymphocytic/monocytic cellular fraction was isolated from buffy coats of healthy donors
Notch Signaling Pathway Notch CSL Reporter HEK293 Cell line Catalog #: 60652
 Notch Signaling Pathway Notch CSL Reporter HEK293 Cell line Catalog #: 60652 Background The Notch signaling pathway controls cell fate decisions in vertebrate and invertebrate tissues. Notch signaling
Notch Signaling Pathway Notch CSL Reporter HEK293 Cell line Catalog #: 60652 Background The Notch signaling pathway controls cell fate decisions in vertebrate and invertebrate tissues. Notch signaling
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
Extended Mammosphere Culture of Human Breast Cancer Cells
 Extended Mammosphere Culture of Human Breast Cancer Cells Application Note The PromoCell 3D Tumorsphere Medium XF The PromoCell 3D Tumorsphere Medium XF has been designed to meet your requirements for
Extended Mammosphere Culture of Human Breast Cancer Cells Application Note The PromoCell 3D Tumorsphere Medium XF The PromoCell 3D Tumorsphere Medium XF has been designed to meet your requirements for
Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction. Marker Sequence (5 3 ) Accession No.
 Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.
 Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
System. xcelligence System RTCA HT Instrument. Application Note No. 14 /January Long-Term High-Throughput Cytotoxicity Profiling
 System Application Note No. 14 /January 2013 xcelligence System RTCA HT Instrument Long-Term High-Throughput Cytotoxicity Profiling For life science research only. Not for use in diagnostic procedures.
System Application Note No. 14 /January 2013 xcelligence System RTCA HT Instrument Long-Term High-Throughput Cytotoxicity Profiling For life science research only. Not for use in diagnostic procedures.
Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing)
 Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing) platform with laser manipulation to efficiently purify lung
Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing) platform with laser manipulation to efficiently purify lung
SUPPORTING MATREALS. Methods and Materials
 SUPPORTING MATREALS Methods and Materials Cell Culture MC3T3-E1 (subclone 4) cells were maintained in -MEM with 10% FBS, 1% Pen/Strep at 37ºC in a humidified incubator with 5% CO2. MC3T3 cell differentiation
SUPPORTING MATREALS Methods and Materials Cell Culture MC3T3-E1 (subclone 4) cells were maintained in -MEM with 10% FBS, 1% Pen/Strep at 37ºC in a humidified incubator with 5% CO2. MC3T3 cell differentiation
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism SUPPLEMENTARY FIGURES, LEGENDS AND METHODS
 A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
Signaling. Dr. Sujata Persad Katz Group Centre for Pharmacy & Health research
 Signaling Dr. Sujata Persad 3-020 Katz Group Centre for Pharmacy & Health research E-mail:sujata.persad@ualberta.ca 1 Growth Factor Receptors and Other Signaling Pathways What we will cover today: How
Signaling Dr. Sujata Persad 3-020 Katz Group Centre for Pharmacy & Health research E-mail:sujata.persad@ualberta.ca 1 Growth Factor Receptors and Other Signaling Pathways What we will cover today: How
VIII Curso Internacional del PIRRECV. Some molecular mechanisms of cancer
 VIII Curso Internacional del PIRRECV Some molecular mechanisms of cancer Laboratorio de Comunicaciones Celulares, Centro FONDAP Estudios Moleculares de la Celula (CEMC), ICBM, Facultad de Medicina, Universidad
VIII Curso Internacional del PIRRECV Some molecular mechanisms of cancer Laboratorio de Comunicaciones Celulares, Centro FONDAP Estudios Moleculares de la Celula (CEMC), ICBM, Facultad de Medicina, Universidad
Cancer Biology Course. Invasion and Metastasis
 Cancer Biology Course Invasion and Metastasis 2016 Lu-Hai Wang NHRI Cancer metastasis Major problem: main reason for killing cancer patients, without it cancer can be cured or controlled. Challenging questions:
Cancer Biology Course Invasion and Metastasis 2016 Lu-Hai Wang NHRI Cancer metastasis Major problem: main reason for killing cancer patients, without it cancer can be cured or controlled. Challenging questions:
Imaging of glycolytic metabolism in primary glioblastoma cells with
 63 Chapter 5 Imaging of glycolytic metabolism in primary glioblastoma cells with RIMChip 5.1. Introduction Glioblastoma(GBM) is one of the most common brain tumors 1. It is composed of heterogeneous subpopulations
63 Chapter 5 Imaging of glycolytic metabolism in primary glioblastoma cells with RIMChip 5.1. Introduction Glioblastoma(GBM) is one of the most common brain tumors 1. It is composed of heterogeneous subpopulations
Detection of Apoptosis in Primary Cells by Annexin V Binding Using the Agilent 2100 Bioanalyzer. Application Note
 Detection of Apoptosis in Primary Cells by Annexin V Binding Using the Agilent 2100 Bioanalyzer Application Note Samuel D. H. Chan Marc Valer and Tobias Preckel, Introduction The Agilent 2100 bioanalyzer
Detection of Apoptosis in Primary Cells by Annexin V Binding Using the Agilent 2100 Bioanalyzer Application Note Samuel D. H. Chan Marc Valer and Tobias Preckel, Introduction The Agilent 2100 bioanalyzer
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer. Application Note
 Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Identifying transcriptional dichotomies by "stochastic sampling"
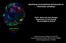 Identifying transcriptional dichotomies by "stochastic sampling" Kevin Janes and Joan Brugge Department of Cell Biology Harvard Medical School pakt (S473) keratin 10 DAPI National Cancer Institute (CA105134)
Identifying transcriptional dichotomies by "stochastic sampling" Kevin Janes and Joan Brugge Department of Cell Biology Harvard Medical School pakt (S473) keratin 10 DAPI National Cancer Institute (CA105134)
Single Cell Quantitative Polymer Chain Reaction (sc-qpcr)
 Single Cell Quantitative Polymer Chain Reaction (sc-qpcr) Analyzing gene expression profiles from a bulk population of cells provides an average profile which may obscure important biological differences
Single Cell Quantitative Polymer Chain Reaction (sc-qpcr) Analyzing gene expression profiles from a bulk population of cells provides an average profile which may obscure important biological differences
w ª wy xvwz A ª vw xvw P ª w} xvw w Æ w Æ V w,x Æ w Æ w Æ y,z Æ { Æ y,z, w w w~ w wy}æ zy Æ wyw{ xæ wz w xywæ xx Æ wv Æ } w x w x w Æ w Æ wy} zy Æ wz
 w ª wy xvwz A ª vw xvw P ª w} xvw w Æ w Æ V w,x Æ w Æ w Æ y,z Æ { Æ y,z, w w w~ w wy}æ zy Æ wyw{ xæ wz w xywæ xx Æ wv Æ } w x w x w Æ w Æ wy} zy Æ wz {w Æ Æ wyw{ x w Germ-line mutations in BRCA1 are associated
w ª wy xvwz A ª vw xvw P ª w} xvw w Æ w Æ V w,x Æ w Æ w Æ y,z Æ { Æ y,z, w w w~ w wy}æ zy Æ wyw{ xæ wz w xywæ xx Æ wv Æ } w x w x w Æ w Æ wy} zy Æ wz {w Æ Æ wyw{ x w Germ-line mutations in BRCA1 are associated
Effect of Acid and Pepsin on LPR-Sensitive Mucins In Vitro
 Novel Drug Targets for Reflux Disease Role of LPR in Injury and Disease is Controversial Nikki Johnston, Ph.D. Department of Otolaryngology and Communication Sciences Medical College of Wisconsin Milwaukee,
Novel Drug Targets for Reflux Disease Role of LPR in Injury and Disease is Controversial Nikki Johnston, Ph.D. Department of Otolaryngology and Communication Sciences Medical College of Wisconsin Milwaukee,
(A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC,
 Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Introduction: 年 Fas signal-mediated apoptosis. PI3K/Akt
 Fas-ligand (CD95-L; Fas-L) Fas (CD95) Fas (apoptosis) 年 了 不 度 Fas Fas-L 力 不 Fas/Fas-L T IL-10Fas/Fas-L 不 年 Fas signal-mediated apoptosis 度降 不 不 力 U-118, HeLa, A549, Huh-7 MCF-7, HepG2. PI3K/Akt FasPI3K/Akt
Fas-ligand (CD95-L; Fas-L) Fas (CD95) Fas (apoptosis) 年 了 不 度 Fas Fas-L 力 不 Fas/Fas-L T IL-10Fas/Fas-L 不 年 Fas signal-mediated apoptosis 度降 不 不 力 U-118, HeLa, A549, Huh-7 MCF-7, HepG2. PI3K/Akt FasPI3K/Akt
Diagnostic test Suggested website label Description Hospitals available
 Diagnostic test Suggested website label Description Hospitals available Abbott Molecular Inc, PATHVYSION HER-2 DNA Probe Kit (FISH) PathVysion kit A diagnostic tool used to determine whether a particular
Diagnostic test Suggested website label Description Hospitals available Abbott Molecular Inc, PATHVYSION HER-2 DNA Probe Kit (FISH) PathVysion kit A diagnostic tool used to determine whether a particular
Rabbit Polyclonal antibody to NFkB p65 (v-rel reticuloendotheliosis viral oncogene homolog A (avian))
 Datasheet GeneTex, Inc : Toll Free 1-877-GeneTex (1-877-436-3839) Fax:1-949-309-2888 info@genetex.com GeneTex International Corporation : Tel:886-3-6208988 Fax:886-3-6208989 infoasia@genetex.com Date :
Datasheet GeneTex, Inc : Toll Free 1-877-GeneTex (1-877-436-3839) Fax:1-949-309-2888 info@genetex.com GeneTex International Corporation : Tel:886-3-6208988 Fax:886-3-6208989 infoasia@genetex.com Date :
The Avatar System TM Yields Biologically Relevant Results
 Application Note The Avatar System TM Yields Biologically Relevant Results Liquid biopsies stand to revolutionize the cancer field, enabling early detection and noninvasive monitoring of tumors. In the
Application Note The Avatar System TM Yields Biologically Relevant Results Liquid biopsies stand to revolutionize the cancer field, enabling early detection and noninvasive monitoring of tumors. In the
Supplementary data Supplementary Figure 1 Supplementary Figure 2
 Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Case Study - Informatics
 bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
C-Phycocyanin (C-PC) is a n«sjfc&c- waefc-jduble phycobiliprotein. pigment isolated from Spirulina platensis. This water- soluble protein pigment is
 ' ^Summary C-Phycocyanin (C-PC) is a n«sjfc&c- waefc-jduble phycobiliprotein pigment isolated from Spirulina platensis. This water- soluble protein pigment is of greater importance because of its various
' ^Summary C-Phycocyanin (C-PC) is a n«sjfc&c- waefc-jduble phycobiliprotein pigment isolated from Spirulina platensis. This water- soluble protein pigment is of greater importance because of its various
Oncogenes and Tumor Suppressors MCB 5068 November 12, 2013 Jason Weber
 Oncogenes and Tumor Suppressors MCB 5068 November 12, 2013 Jason Weber jweber@dom.wustl.edu Oncogenes & Cancer DNA Tumor Viruses Simian Virus 40 p300 prb p53 Large T Antigen Human Adenovirus p300 E1A
Oncogenes and Tumor Suppressors MCB 5068 November 12, 2013 Jason Weber jweber@dom.wustl.edu Oncogenes & Cancer DNA Tumor Viruses Simian Virus 40 p300 prb p53 Large T Antigen Human Adenovirus p300 E1A
Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION
 Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION X. Shawn Liu 1, 3, Bing Song 2, 3, Bennett D. Elzey 3, 4, Timothy L. Ratliff 3, 4, Stephen F. Konieczny
Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION X. Shawn Liu 1, 3, Bing Song 2, 3, Bennett D. Elzey 3, 4, Timothy L. Ratliff 3, 4, Stephen F. Konieczny
CELL CYCLE MOLECULAR BASIS OF ONCOGENESIS
 CELL CYCLE MOLECULAR BASIS OF ONCOGENESIS Summary of the regulation of cyclin/cdk complexes during celll cycle Cell cycle phase Cyclin-cdk complex inhibitor activation Substrate(s) G1 Cyclin D/cdk 4,6
CELL CYCLE MOLECULAR BASIS OF ONCOGENESIS Summary of the regulation of cyclin/cdk complexes during celll cycle Cell cycle phase Cyclin-cdk complex inhibitor activation Substrate(s) G1 Cyclin D/cdk 4,6
Supplementary Table 1. Characterization of HNSCC PDX models established at MSKCC
 Supplementary Table 1. Characterization of HNSCC PDX models established at MSKCC Supplementary Table 2. Drug content and loading efficiency estimated with F-NMR and UV- Vis Supplementary Table 3. Complete
Supplementary Table 1. Characterization of HNSCC PDX models established at MSKCC Supplementary Table 2. Drug content and loading efficiency estimated with F-NMR and UV- Vis Supplementary Table 3. Complete
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/310/ra11/dc1 Supplementary Materials for STAT3 Induction of mir-146b Forms a Feedback Loop to Inhibit the NF-κB to IL-6 Signaling Axis and STAT3-Driven Cancer
www.sciencesignaling.org/cgi/content/full/7/310/ra11/dc1 Supplementary Materials for STAT3 Induction of mir-146b Forms a Feedback Loop to Inhibit the NF-κB to IL-6 Signaling Axis and STAT3-Driven Cancer
Protein Arrays High Throughput Protein Quantification from Total Cell Lysates
 Protein Arrays High Throughput Protein Quantification from Total Cell Lysates Dr. Markus Ehrat Mr. Markus Tobler Zeptosens A Division of Bayer (Schweiz) AG Benkenstr. 254 CH-4108 Witterswil Switzerland
Protein Arrays High Throughput Protein Quantification from Total Cell Lysates Dr. Markus Ehrat Mr. Markus Tobler Zeptosens A Division of Bayer (Schweiz) AG Benkenstr. 254 CH-4108 Witterswil Switzerland
Early cell death (FGF) B No RunX transcription factor produced Yes No differentiation
 Solution Key - Practice Questions Question 1 a) A recent publication has shown that the fat stem cells (FSC) can act as bone stem cells to repair cavities in the skull, when transplanted into immuno-compromised
Solution Key - Practice Questions Question 1 a) A recent publication has shown that the fat stem cells (FSC) can act as bone stem cells to repair cavities in the skull, when transplanted into immuno-compromised
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplemental Information. Otic Mesenchyme Cells Regulate. Spiral Ganglion Axon Fasciculation. through a Pou3f4/EphA4 Signaling Pathway
 Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
supplementary information
 DOI: 10.1038/ncb2133 Figure S1 Actomyosin organisation in human squamous cell carcinoma. (a) Three examples of actomyosin organisation around the edges of squamous cell carcinoma biopsies are shown. Myosin
DOI: 10.1038/ncb2133 Figure S1 Actomyosin organisation in human squamous cell carcinoma. (a) Three examples of actomyosin organisation around the edges of squamous cell carcinoma biopsies are shown. Myosin
Supporting Information
 Supporting Information M1 macrophage-derived nanovesicles potentiate the anticancer efficacy of immune checkpoint inhibitors Yeon Woong Choo, 1, Mikyung Kang, 2, Han Young Kim, 1 Jin Han, 1 Seokyung Kang,
Supporting Information M1 macrophage-derived nanovesicles potentiate the anticancer efficacy of immune checkpoint inhibitors Yeon Woong Choo, 1, Mikyung Kang, 2, Han Young Kim, 1 Jin Han, 1 Seokyung Kang,
Part-4. Cell cycle regulatory protein 5 (Cdk5) A novel target of ERK in Carb induced cell death
 Part-4 Cell cycle regulatory protein 5 (Cdk5) A novel target of ERK in Carb induced cell death 95 1. Introduction The process of replicating DNA and dividing cells can be described as a series of coordinated
Part-4 Cell cycle regulatory protein 5 (Cdk5) A novel target of ERK in Carb induced cell death 95 1. Introduction The process of replicating DNA and dividing cells can be described as a series of coordinated
Human Urokinase / PLAU / UPA ELISA Pair Set
 Human Urokinase / PLAU / UPA ELISA Pair Set Catalog Number : SEK10815 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized
Human Urokinase / PLAU / UPA ELISA Pair Set Catalog Number : SEK10815 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
Using Impedance-Based Approaches for Measuring Cell-Mediated Cytotoxicity; Antibody-Dependent (ADCC) and Chimeric Antigen Receptor T (CAR-T)
 Using Impedance-Based Approaches for Measuring Cell-Mediated Cytotoxicity; Antibody-Dependent (ADCC) and Chimeric Antigen Receptor T (CAR-T) Vashu Pamnani, PhD. Cambridge Bioscience T Cells Non-Adherent
Using Impedance-Based Approaches for Measuring Cell-Mediated Cytotoxicity; Antibody-Dependent (ADCC) and Chimeric Antigen Receptor T (CAR-T) Vashu Pamnani, PhD. Cambridge Bioscience T Cells Non-Adherent
FOXO Reporter Kit PI3K/AKT Pathway Cat. #60643
 Data Sheet FOXO Reporter Kit PI3K/AKT Pathway Cat. #60643 Background The PI3K/AKT signaling pathway is essential for cell growth and survival. Disruption of this pathway or its regulation has been linked
Data Sheet FOXO Reporter Kit PI3K/AKT Pathway Cat. #60643 Background The PI3K/AKT signaling pathway is essential for cell growth and survival. Disruption of this pathway or its regulation has been linked
Supplement 8: Candidate age-related genes and pathways
 Supplement 8: Candidate age-related genes and pathways Function Untreated cohort (cohort 1) Treated cohort (cohort 2) Genes Gene sets Effect of age Effect of age FDR of 2 nd Effect of age adjusted Effect
Supplement 8: Candidate age-related genes and pathways Function Untreated cohort (cohort 1) Treated cohort (cohort 2) Genes Gene sets Effect of age Effect of age FDR of 2 nd Effect of age adjusted Effect
Principles of Genetics and Molecular Biology
 Cell signaling Dr. Diala Abu-Hassan, DDS, PhD School of Medicine Dr.abuhassand@gmail.com Principles of Genetics and Molecular Biology www.cs.montana.edu Modes of cell signaling Direct interaction of a
Cell signaling Dr. Diala Abu-Hassan, DDS, PhD School of Medicine Dr.abuhassand@gmail.com Principles of Genetics and Molecular Biology www.cs.montana.edu Modes of cell signaling Direct interaction of a
