Cdc48/p97 Ufd1 Npl4 antagonizes Aurora B during chromosome segregation in HeLa cells
|
|
|
- Sherilyn Jones
- 5 years ago
- Views:
Transcription
1 Research Article 1571 Cdc48/p97 Ufd1 Npl4 antagonizes Aurora B during chromosome segregation in HeLa cells Grzegorz Dobrynin 1,2, Oliver Popp 2, Tina Romer 2, Sebastian Bremer 1, Michael H. A. Schmitz 2, Daniel W. Gerlich 2 and Hemmo Meyer 1,2, * 1 Centre for Medical Biotechnology, University of Duisburg-Essen, Essen, Germany 2 Institute of Biochemistry, ETH Zurich, CH-8093 Zurich, Switzerland *Author for correspondence (hemmo.meyer@uni-due.de) Accepted 6 January , Published by The Company of Biologists Ltd doi: /jcs Summary During exit from mitosis in Xenopus laevis egg extracts, the AAA+ ATPase Cdc48/p97 (also known as VCP in vertebrates) and its adapter Ufd1 Npl4 remove the kinase Aurora B from chromatin to allow nucleus formation. Here, we show that in HeLa cells Ufd1 Npl4 already antagonizes Aurora B on chromosomes during earlier mitotic stages and that this is crucial for proper chromosome segregation. Depletion of Ufd1 Npl4 by small interfering RNA (sirna) caused chromosome alignment and anaphase defects resulting in missegregated chromosomes and multi-lobed nuclei. Ufd1 Npl4 depletion also led to increased levels of Aurora B on prometaphase and metaphase chromosomes. This increase was associated with higher Aurora B activity, as evidenced by the partial resistance of CENP-A phosphorylation to the Aurora B inhibitor hesperadin. Furthermore, low concentrations of hesperadin partially rescued chromosome alignment in Ufd1-depleted cells, whereas, conversely, Ufd1-depletion partially restored congression in the presence of hesperadin. These data establish Cdc48/p97 Ufd1 Npl4 as a crucial negative regulator of Aurora B early in mitosis of human somatic cells and suggest that the activity of Aurora B on chromosomes needs to be restrained to ensure faithful chromosome segregation. Key words: Mitosis, Chromosome segregation, Ubiquitin, AAA+ protein, Cdc48/p97 (VCP), Aurora B Introduction The proper segregation of the replicated and condensed chromosomes during mitosis requires correct attachment of chromatid pairs to opposite poles of the mitotic spindle and subsequent accurate separation in anaphase, followed by timely decondensation and nuclear envelope formation in telophase. The kinase Aurora B plays crucial roles in various aspects of chromosome segregation (Ducat and Zheng, 2004; Ruchaud et al., 2007). In complex with its partner proteins INCENP, survivin and borealin (also known as Dasra), it associates along chromosomes in prophase where it phosphorylates histone H3 and the centromeric histone H3 homologue CENP-A (Adams et al., 2001; Giet and Glover, 2001; Hsu et al., 2000; Murnion et al., 2001; Zeitlin et al., 2001). As mitosis progresses to metaphase, Aurora B dissociates from chromosome arms and concentrates in the centromeric region, where it controls bipolar spindle attachment to the kinetochores of sister chromatids and thus the formation of the metaphase plate (Adams et al., 2001; Cimini et al., 2006; Giet and Glover, 2001; Hauf et al., 2003; Kaitna et al., 2000; Kelly and Funabiki, 2009; Tanaka et al., 2002). Aurora B also helps to maintain the spindle checkpoint that blocks anaphase onset and exit from mitosis until all chromosomes properly attach to the spindle (Ditchfield et al., 2003; Hauf et al., 2003; Kallio et al., 2002; Musacchio and Hardwick, 2002). Upon anaphase onset, Aurora B, along with its partners, transfers to the spindle midzone where it regulates spindle dynamics and function. In telophase, Aurora B persists at the midbody where it controls cytokinesis, to ensure proper division of the daughter cells (Steigemann et al., 2009). As a consequence of its central role, RNA interference (RNAi)- mediated depletion of Aurora B, or pharmacological inhibition of its kinase activity, in mammalian tissue culture cells leads to a prominent chromosome alignment defect in prometaphase due to erroneous spindle attachment to kinetochores (Ditchfield et al., 2003; Hauf et al., 2003; Taylor and Peters, 2008). Metaphase plate formation is largely hampered, and cells display severe anaphase defects with lagging chromosomes, or they exit from mitosis before completion of anaphase with resulting micronuclei, polyploidy and multinucleated cells (Ditchfield et al., 2003; Hauf et al., 2003; Mora-Bermudez et al., 2007). How Aurora B controls and corrects spindle attachment to kinetochores and thus chromosome congression to the metaphase plate is not completely understood, but it involves regulation of a number of substrate proteins at the kinetochore with seemingly antagonizing activities (Kelly and Funabiki, 2009; Ruchaud et al., 2007). Phosphorylation of the Ndc80 (also known as Hec1) complex, the major attachment module for microtubules, is proposed to decrease its affinity for microtubules and therefore destabilizes spindle attachment (Cheeseman et al., 2006; Ciferri et al., 2008; DeLuca et al., 2006). By contrast, phosphorylation of mitotic centromere-associated kinesin (MCAK; also known as KIF2C) suppresses its microtubule-depolymerizing activity and thus might stabilize spindle attachments (Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004). The role of Aurora B in correcting spindle attachment suggests that centromeric Aurora B needs to be tightly regulated, to maintain its activity within a limited range, to allow fine-tuning of spindle attachment and consequently faithful chromatid segregation. Previously, the AAA+ ATPase Cdc48/p97 (also known as VCP in vertebrates) has been implicated in regulation of Aurora B. Cdc48/p97 is an abundant ubiquitin-dependent chaperone involved in protein quality control and signalling events in interphase and mitosis (Woodman, 2003; Ye, 2006). Cdc48/p97 associates with alternative substrate and ubiquitin adapters, including cofactor p47
2 (9) (officially known as NSFL1 cofactor p47; NSF1C) or the Ufd1 Npl4 complex, to fulfil distinct functions (Alexandru et al., 2008; Kondo et al., 1997; Meyer et al., 2000; Schuberth and Buchberger, 2008). Ufd1 and Npl4 form a stable heterodimeric complex, with both subunits containing interaction sites for Cdc48/p97, as well as binding domains for ubiquitin (Alam et al., 2004; Bruderer et al., 2004; Isaacson et al., 2007; Meyer et al., 2002). As a common activity applied in different pathways, the Cdc48/p97 Ufd1 Npl4 complex is thought to bind and extract ubiquitylated client proteins from cellular structures and to release them for either degradation or recycling (Jentsch and Rumpf, 2007; Ye, 2006). In mitosis, Cdc48/p97 Ufd1 Npl4 has been associated with different functions, such as chromosome segregation, spindle dynamics and organelle reformation, in different organisms and developmental stages (Meyer and Popp, 2008). In Xenopus laevis egg extracts, Cdc48/p97 Ufd1 Npl4 removes Aurora B from chromatin during exit from mitosis in a process that involves modification of Aurora B with ubiquitin chains (Ramadan et al., 2007). Removal of Aurora B from chromatin leads to a local decrease in activity, which then allows decondensation and nuclear envelope formation (Ramadan et al., 2007). Ufd1 Npl4 has also been associated with chromosome segregation in HeLa cells in two independent studies. RNAimediated depletion of Npl4 results in anaphase defects with entangled chromosomes (Porter et al., 2007), whereas Ufd1 depletion has been shown to cause chromosome congression defects (Vong et al., 2005). On the basis of immunohistochemistry of small interfering RNA (sirna)-treated cells, Vong and colleagues concluded that Ufd1 helps to recruit Aurora B to chromosomes. This interpretation for a positive regulation of Aurora B is in surprising contrast with our observation in Xenopus egg extracts (Ramadan et al., 2007), where Cdc48/p97 removes Aurora B from chromosomes. Two other studies using RNAi-mediated depletion of Cdc48/p97 in Caenorhabditis elegans failed to detect any role for Cdc48/p97 or Ufd1 Npl4 in mitosis (Heallen et al., 2008; Mouysset et al., 2008). The cellular relevance of the Cdc48/p97 Ufd1 Npl4 complex for regulation of mitosis and its functional relation to Aurora B has therefore remained controversial. To address this controversy, we have re-examined the relevance of Cdc48/p97 Ufd1 Npl4 for mitosis, and its functional relation to Aurora B in HeLa cells. Here, we show that Ufd1 and Npl4 cooperate in proper chromosome segregation. Moreover, we provide evidence that Ufd1 Npl4 functions by antagonizing Aurora B during segregation. Depletion of Ufd1 Npl4 resulted in increased levels of Aurora B associated with chromosomes during prometaphase and metaphase, as shown by immunofluorescence microscopy and cell fractionation. Furthermore, Ufd1 depletion increased phosphorylation of the centromeric Aurora B substrate CENP-A. Finally, and importantly, Ufd1 depletion partially rescued the rate and efficiency of chromosome congression after attenuation of Aurora B with the inhibitor hesperadin. The data establish the Cdc48/p97 Ufd1 Npl4 complex as a negative regulator of Aurora B early in mitosis. Moreover, the data imply that restraining the activity of Aurora B on chromosomes is essential for its function in controlling proper chromosome segregation. Results Ufd1 Npl4 is required for normal progression through mitosis in HeLa cells To study the role of Cdc48/p97 in regulating progression of mitosis in HeLa cells systematically, we used an sirna approach to deplete its two alternative adapters implicated in mitosis, p47 or the heterodimeric Ufd1 Npl4 complex (Kondo et al., 1997; Meyer et al., 2000; Schuberth and Buchberger, 2008), rather than Cdc48/p97 itself, in order to reduce pleiotropic effects. Depletion for 48 hours was efficient and did not affect Cdc48/p97 levels (Fig. 1A). Importantly, depletion of Npl4 also specifically destabilized Ufd1, but not vice versa, consistent with previous biochemical data showing that Ufd1 is unstable in the absence of Npl4 (Bruderer et al., 2004). We first monitored progression through mitosis in unsynchronized HeLa cells stably expressing histone-h2b mrfp by time-lapse video microscopy and measured the time between nuclear envelope breakdown (scored on the basis of the loss of a defined boundary for the chromatin regions) (Beaudouin et al., 2002) and anaphase onset (Fig. 1B,C). Ufd1- or Npl4-siRNAtreated cells displayed a delay of anaphase onset (median of ~43 minutes), compared with that of control or p47-depleted cells (median of ~30 minutes), suggesting that the spindle assembly Fig. 1. The Cdc48/p97 adapter Ufd1 Npl4 is required for normal progression through mitosis of HeLa cells. (A) Western blot analysis of HeLa Kyoto cells treated with non-silencing control (Ctrl) sirna or two independent oligonucleotides (S1 and S2) each targeting Ufd1, Npl4 or the alternative adapter p47 for 48 hours. Membranes were probed with the indicated antibodies. Note that Npl4 depletion also specifically destabilizes its binding partner Ufd1. (B) Images from representative confocal time-lapse fluorescence microscopy movies of unsynchronized HeLa Kyoto cells progressing through mitosis, after treatment with indicated sirna oligonucleotides for 48 hours. The cells are stably express histone-h2b mrfp to visualize chromatin. Time (minutes) after nuclear envelope breakdown is indicated, closed arrowheads point to anaphase onset. Note that some Npl4- sirna-treated cells did not enter anaphase (as shown in the bottom series of images). The open arrowhead indicates a chromosome bridge. Scale bars: 5 m. (C) Quantification of movies, as in B, from 5 independent experiments. The time between nuclear envelope breakdown (NEBD) and anaphase onset was determined and is represented in a box-and-whisker plot [Ctrl, median 30.0 minutes, mean 31.3 minutes (n 48); Ufd1, median: 43.3 minutes, mean: 44.9 minutes (n 28); Npl4, median: 43.3 minutes, mean: 46.3 minutes (n 12); p47, median: 29.5 minutes; mean: 31.0 minutes (n 12)]. Only Npl4-siRNAtreated cells that entered anaphase within 170 minutes were quantified. Statistical significance was tested with the Mann Whitney U-test (n.s., not significant). The sirnas Ufd1-S2, Npl4-S2 and p47-s1 were used.
3 Cdc48/p97 antagonizes Aurora B during chromosome segregation 1573 checkpoint is activated owing to compromised spindle kinetochore attachments. Indeed, inspection of the movies revealed misaligned chromosomes at the spindle pole, and a delay in congression and the formation of a metaphase plate, in Ufd1- or Ufd1 Npl4- depleted cells (Fig. 1B). When cells eventually proceeded into anaphase, we observed segregation defects, with separating chromosomes remaining entangled and delayed retraction of chromosome arms from the spindle midzone. A fraction of the Npl4-siRNA-treated cells did not enter anaphase during the course of the experiments (Fig. 1B). These findings confirm that the Cdc48/p97 Ufd1 Npl4 chaperone complex is essential for proper mitotic progression of human somatic cells. Ufd1 Npl4 depletion causes chromosome congression, as well as anaphase defects The congression defect was confirmed in fixed cells. On average 36.8% and 46.9% of cells with an apparent metaphase plate displayed misaligned chromosomes, often several at a time, upon Ufd1 or Npl4 sirna treatment, respectively, compared with 7.5% or 6.4% in control or p47-depleted cells, respectively (Fig. 2A,C). In Ufd1-depleted cells, the congression defect was largely rescued by overexpression of the mouse Ufd1 cdna that is not targeted by the sirna oligonucleotides used (Fig. 2C and supplementary material Fig. S1), showing that the effect was specific. In addition, we observed a significant (P<0.005) increase in chromosome segregation defects in anaphase from 7.8% in control and 7.7% in p47-depleted cells, to 23% and 31.4% in Ufd1- or Npl4-siRNAtreated cells, respectively (Fig. 2B,D). These defects included lagging chromosomes that probably resulted from incorrect spindle attachment. Moreover, we observed chromosome bridges between the separating chromatin masses, and often several entangled chromosomes, indicating defects in sister chromatid resolution. In particular, Npl4-siRNA-treated cells (that are depleted of the entire Ufd1 Npl4 heterodimer) displayed aberrations in nuclear morphology in interphase with micronuclei that probably originated from lagging chromosomes (Fig. 2E). Even more prominently, nuclei were often multi-lobed, a phenotype known to result from segregation errors and defective axial compaction of anaphase chromosomes upon inhibition of Aurora B (Hauf et al., 2003; Mora-Bermudez et al., 2007). These results show the shared functions of the Ufd1 and Npl4 in regulating chromosome alignment and chromosome segregation in human somatic cells. To address whether depletion of Ufd1 Npl4, in the same manner as Aurora B inhibition, causes polyploidy, we treated cells with sirna targeting Ufd1, Npl4 or Aurora B for 2 days and measured the DNA content by flow cytometry (Fig. 2F). Although inactivation of Aurora B caused an increase in the number of cells with 4N and even 8N DNA content, as previously reported (Hauf Fig. 2. Chromosome congression and segregation defects, but not multinucleation, upon Ufd1 Npl4 depletion. (A,B) Confocal fluorescence images of unsynchronized metaphase (A) and anaphase (B) HeLa Kyoto cells that had been treated for 48 hours with the indicated sirnas. Cells were fixed and stained with DAPI. Scale bar: 5 m. (C) Quantification (means+s.d.) of the experiment described in A [>200 cells from three independent experiments except for the mouse Ufd1 Myc (Ufd1:myc) overexpression, which assessed >50 cells in one experiment]. Statistical significance was analysed with Student s t-test. (D) Quantification (means+s.d.) of the experiment described in B (>200 cells from three independent experiments). Statistical significance was analysed with Student s t-test. (E) Representative fluorescence overview images of interphase cells treated for 48 hours with the indicated sirna. Cells were fixed and stained with DAPI. Note the multi-lobed nuclei and micronuclei in cells treated with the Npl4 sirna that destabilizes the entire Ufd1 Npl4 heterodimer. Scale bar: 10 m. (F) Analysis of the DNA content by flow cytometry of propidium-iodidestained cells after treatment with indicated sirnas for 48 hours.
4 (9) et al., 2003), there was no dramatic shift in the DNA content in Ufd1- or Npl4-depleted cells. Npl4 depletion did, however, cause a slight increase of cells with intermediate DNA content between 2N and 4N, at the expense of the 2N peak, which can either be explained by the aneuploidy due to the observed missegregation or by a prolonged S phase. We also counted the percentage of multinucleated cells, but did not observe any increase in Ufd1- or Npl4-siRNA-treated cells, in contrast with Aurora-B-depleted cells, indicating that cytokinesis after segregation was not affected (data not shown). Ufd1 Npl4 helps to remove a fraction of Aurora B from chromosomes as cells progress to metaphase The chromosome congression defects and the segregation errors upon Ufd1 Npl4 depletion resemble the phenotype upon inactivation of the kinase Aurora B (Ditchfield et al., 2003; Hauf et al., 2003; Mora-Bermudez et al., 2007). In apparent accordance, Vong and colleagues reported a decrease of Aurora B and survivin on chromosomes upon Ufd1 depletion (Vong et al., 2005), which would suggest a positive regulation of Aurora B by Ufd1 Npl4. By contrast, we previously showed that Cdc48/p97 Ufd1 Npl4 negatively regulates Aurora B by removing it from chromatin during exit from mitosis in embryonic systems (Ramadan et al., 2007). We therefore re-examined Aurora B and survivin localization in Ufd1 Npl4-depleted cells. To do so, we fixed unsynchronized control and sirna-treated cells and examined the localization of Aurora B and survivin by immunostaining. Wide-field fluorescence microscopy, with identical exposure settings, revealed that the levels of chromosomeassociated Aurora B increased by more than fold both in Ufd1- and in Npl4-depleted cells (Fig. 3A,B). In addition, the level of survivin on the chromosomes was increased upon Ufd1 Npl4 depletion. This finding is in sharp contrast with the observation of Vong and colleagues, who reported loss of chromatin-associated Aurora B upon Ufd1 depletion (Vong et al., 2005). We therefore examined the previously published sirna sequences (oligonucleotides V-S1 and V-S2) and compared them with a total of five Ufd1 and two Npl4 of our sirna oligonucleotides. All our sequences upregulated the localization of Aurora B to chromosomes, although to different degrees (supplementary material Fig. S2). Intriguingly, only the published V-S2 caused the reported decrease (supplementary material Fig. S2). The conflicting results were observed despite the fact that all sirna oligonucleotides efficiently depleted Ufd1 (supplementary material Fig. S2C). To solve this discrepancy, we performed restoration experiments. We introduced four silent point mutations in the mouse Ufd1 cdna to render it resistant to the published V-S2, in addition to our S2 oligonucleotide, and used it to generate a resistant Ufd1 GFP expression construct (reufd1-gfp). Overexpression of reufd1-gfp was efficient in all cell populations without affecting depletion of Ufd1 by either V-S2 or our S2 as confirmed by western blotting (supplementary material Fig. S3A). Importantly, the increased levels of chromatin-associated Aurora B in Ufd1-S2-siRNA-treated cells were reduced to almost control levels in cells overexpressing reufd1-gfp. By contrast, loss of Aurora B from chromosomes in cells treated with the V-S2 oligonucleotide was not restored (supplementary material Fig. S3B,C). Consistently, reufd1-gfp restored proper chromosome alignment in S2-siRNA-treated cells, whereas it failed to do so in V-S2-treated cells (supplementary material Fig. S3B,D). This result conclusively demonstrates that the phenotype of S2 treatment was Fig. 3. Elevated levels of Aurora B and survivin associated with chromosomes in Ufd1 Npl4-depleted cells. (A) Cells were treated with control (sictrl), Ufd1 or Npl4 sirna for 48 hours, were pre-extracted to remove soluble protein and fixed. Aurora B and survivin was detected by indirect immunofluorescence using wide-field fluorescence microscopy with identical settings. (B) The intensity of Aurora B and survivin (means+s.d.) on metaphase chromatin was determined and background-subtracted as shown in A (n>21 for each condition). Data are presented for siufd1-s2 and sinpl4-s2 (see supplementary material Fig. S2 for data from with additional oligonucleotides and supplementary material Fig. S3 for restoration experiments). (C) Western blot analysis of chromatin fractions from cells treated with control (Ctrl), Ufd1 or Aurora B (AurB) sirna. After nocodazole arrest, mitotic cells were harvested by shake-off. Mitotic indices were comparable (see supplementary material Fig. S3 for the quantification). Equal numbers of cells were lysed with detergent, and the chromatin (pellet) separated from the soluble fractions (sup) by centrifugation. The membrane was probed with antibodies against Ufd1, Aurora B, or for phosphorylated histone H3 serine 10 (ph3). Topoisomerase II alpha (TopoIIa) and GAPDH were probed, as chromatin and cytosol controls, respectively. Note the increase in chromatin-associated Aurora B in Ufd1-depleted cells. (D) Quantification of the experiment shown in C. The intensities of the Aurora B band was normalized to the TopoII signal and presented as percentage of the control intensity for the total and pellet fractions, respectively. specifically caused by Ufd1 depletion, whereas V-S2 affected mitosis indirectly through an unknown mechanism. The enhanced Aurora B association with chromosomes in Ufd1- depleted cells was observed by prophase and was even observed on segregating chromosomes in anaphase (supplementary material Fig. S4). To confirm this increase biochemically, we synchronized HeLa cells in nocodazole and harvested mitotic cells by shake-off. In such a way, we analyzed control and Ufd1-depleted cells, as well as Aurora-B-depleted cells for comparison. The mitotic index of the collected cells ranged between 90 95% in all preparations (supplementary material Fig. S5). Detergent-extracted cell lysates were separated by centrifugation into soluble and chromatin fractions. Western blotting confirmed a consistent increase of chromatin-associated Aurora B upon Ufd1 depletion, which was in part reflected in the total amount of cellular Aurora B (Fig. 3C,D). Next, we ask whether Aurora B accumulated at a specific region on the chromosome in the absence of Cdc48/p97 Ufd1 Npl4 function. Analysis of Ufd1- and Npl4-depleted cells in
5 Cdc48/p97 antagonizes Aurora B during chromosome segregation 1575 Fig. 4. Ufd1 Npl4 depletion leads to Aurora B spreading along chromosome arms, and its increased association at the centromere. (A) Representative confocal images of unsynchronized HeLa cells in prometaphase or metaphase after 48 hours of treatment with sirna targeting the indicated proteins. Cells were fixed and probed with DAPI, and antibodies against Aurora B (Aur B) or phosphorylated Aurora B (pt232). Images were taken with identical laser settings. Note the Aurora B localization along chromosome arms (arrowheads) in Ufd1- and Npl4-siRNA-treated cells. Scale bar: 5 m. (B) Representative chromosome spreads of prometaphase cells treated with control or Ufd1 sirna. Spreads were stained with DAPI and anti-aurora-b antibodies. The right-hand panel shows a higher magnification of a representative individual chromatid pair. Note the spreading of Aurora B along chromosome arms upon Ufd1 depletion. (C) Control-and siufd1-treated cells were fixed, immunostained with antibodies against Aurora B and the centromeric antigen CREST, and analysed by wide-field microscopy. Scale bar: 10 m. (D) Quantification (means±s.d.) of Aurora B intensities on chromosome arms and centromeres visualized as indicated in C. prometaphase and metaphase, by confocal microscopy, revealed that Aurora B localized along the entire chromosome arms, whereas it was confined to centromeric regions in control cells (Fig. 4A). Aurora B on chromosome arms was active, as it was autophosphorylated on threonine 232 (Yasui et al., 2004), which we detected with a phosphorylation-specific antibody (Fig. 4A). The distribution of Aurora B along the chromosome arms was also confirmed in chromosome spreads prepared from synchronized control- and Ufd1-depleted cells (Fig. 4B). To measure the increase, we analyzed prometaphase cells by wide-field microscopy. Fluorescence intensity measurement of centromeric regions (as defined with a CREST counterstain) and areas on distal arms revealed that Aurora B levels were significantly increased in both chromosomal regions in Ufd1-depleted cells compared with its localization in control treated cells (Fig. 4C,D). From these biochemical and morphological analyses, we conclude that impairment of Ufd1 Npl4 leads to elevated levels of Aurora B associating with chromosomes. Ufd1 Npl4 reduces chromosome-associated Aurora B activity early in mitosis To address whether the increased levels of Aurora B on chromosomal arms and centromeres also resulted in increased Aurora B activity associated with chromosomes, we monitored phosphorylation of its substrates, histone H3 and the centromeric histone H3 homologue CENP-A, by immunofluorescence microscopy. Both proteins are first phosphorylated by Aurora B in late G2 phase: histone H3 on serine 10 and CENP-A on serine 7. We did not detect differences in their steady-state phosphorylation level in control and Ufd1-depleted cells (Fig. 5C, DMSO control; data not shown), possibly because both substrates are already nearly maximally phosphorylated in control cells. We therefore asked whether a possible increase in Aurora B activity would be reflected in a partial resistance to the chemical inhibitor of Aurora B, hesperadin. To address this question, we first assessed whether phosphorylation of histone H3 or CENP-A is dynamic in prometaphase, as a result of repeated dephosphorylation and subsequent re-phosphorylation by Aurora B, which could then be probed in our analysis. To test this, we treated cells with 100 nm hesperadin for between 2 and 60 minutes, to inhibit possible rephosphorylation. We then analyzed the substrate phosphorylation state by indirect immunofluorescence with phosphorylation-specific antibodies in prometaphase and metaphase cells. Given that hesperadin treatment causes morphological changes, we confirmed the mitotic stage by assessing whether the cell had broken down the nuclear envelope but not yet degraded cyclin B1, as determined by indirect immunofluorescence (data not shown). Histone H3 phosphorylation was not affected when cells were treated for up to 15 minutes (Fig. 5A,B). However, after 30 minutes, we observed a
6 (9) Fig. 5. Ufd1 Npl4 attenuates chromosome-associated Aurora B activity towards a centromeric substrate. (A,B) Phosphorylation of CENP-A, but not of histone H3 is dynamic during prometaphase. Unsynchronized cells were treated for indicated times with 100 nm of the Aurora B inhibitor hesperadin (Hesp), fixed and phosphorylation of histone H3 (serine 10) (ph3) and CENP-A (serine 7) (pcenp-a) detected by indirect immunofluorescence with phosphorylation-specific antibodies. Cells were only assessed if they had broken down the nuclear envelope, but not yet degraded cyclin B1 (as identified with an anti-cyclin-b1 antibody). A shows fluorescence micrographs and B a quantification (means+s.d.; n 40). Note that CENP-A, but not H3 phosphorylation is sensitive to 15-minute incubations with 100 nm hesperadin. (C,D) Cells were treated with increasing concentrations of hesperadin for 15 minutes, and phosphorylation of CENP-A was detected as in A. C shows fluorescence micrographs and D a quantification (means+s.d.; n>137 from three independent experiments). Note that CENP-A phosphorylation is resistant to nm hesperadin in Ufd1-siRNAbut not control-treated cells. mixed population of cells with either fully, partially or nonphosphorylated chromatin, the latter cases most probably already reflecting inhibition of the initial phosphorylation in those cells that were in prophase during application of the inhibitor. This suggests that histone H3 phosphorylation is not dynamic and that the phosphorylation that occurs in late G2 is stable at least until anaphase onset. In marked contrast, CENP-A phosphorylation at the centromere was sensitive to a 15-minute incubation with hesperadin in almost all the sample cells, indicating that it is constantly dephosphorylated and needs to be re-phosphorylated by Aurora B to maintain its phosphorylation status during prometaphase. We therefore investigated the sensitivity of CENP-A phosphorylation to 15-minute incubations with increasing concentrations of hesperadin (from 1 to 100 nm) in control and Ufd1-depleted cells (Fig. 5C,D). We found that phosphorylation was undetectable at 20 nm in the majority of control cells, whereas phosphorylation was resistant to 50 nm hesperadin and was even partially detectable at 100 nm hesperadin. This result demonstrates that the impairment of Ufd1 Npl4 function leads to a shift towards stronger phosphorylation of CENP-A, which is consistent with an elevated Aurora B activity associated with chromosomes. Together with the observed increase in Aurora B protein, this suggests that Ufd1 Npl4 helps to restrain the activity of Aurora B early in mitosis, by removing a fraction of it from chromosomes as cells progress to metaphase. Ufd1 Npl4 antagonizes Aurora B during chromosome congression Next, we asked whether regulation of Aurora B activity by Cdc48/p97 Ufd1 Npl4 had any functional relevance. We did not observe an apparent effect on chromosome condensation in Ufd1- depleted cells, even when they were challenged with hypotonic buffer (supplementary material Fig. S6), which leads to a decompaction of unstable chromosomes (Gassmann et al., 2004). To address a possible effect on chromosome congression, we analyzed the efficiency and timing of chromosome alignment both in fixed and live cells. As a first approach, we treated control or Ufd1- depleted cells with increasing concentrations of hesperadin (from 0.7 to 100 nm), fixed the cells after 1 hour and determined the percentage of prometaphase and metaphase cells with fully aligned chromosomes (Fig. 6A,B). As expected, both depletion of Ufd1 and addition of hesperadin at concentrations equal or above 5 nm caused a marked increase in misaligned chromosomes compared with that observed in control cells. Importantly, however, chromosome alignment in Ufd1-depleted cells was partially rescued by moderate inhibition of Aurora B, with 1 nm of hesperadin, suggesting that the alignment defect in Ufd1-depleted cells was, at least in part, caused by overactivation of Aurora B. Conversely, Ufd1-depletion restored proper alignment in the presence of 5 15 nm hesperadin, showing that the overactivation of Aurora B due to impaired Ufd1 Npl4 function can overcome partial inhibition of Aurora B.
7 Cdc48/p97 antagonizes Aurora B during chromosome segregation 1577 Fig. 6. Ufd1 Npl4 antagonizes Aurora B during chromosome congression. (A,B) Unsynchronized control or Ufd1-depleted cells were treated with indicated concentrations of hesperadin (Hesp) for 1 hour, and were fixed and stained with DAPI and anti-cyclin B1 antibodies. Cells were assessed if they had broken down the nuclear envelope, but not yet degraded cyclin B1, as in Fig. 5A, and the percentage of cells with fully aligned chromosomes was determined. Data are means+s.d. from three independent experiments (n 100 for each condition). (C,D) Hela cells expressing H2B mrfp were transfected with control (Ctrl) or Ufd1 sirna. After 48 hours, cells were treated with hesperadin (50 nm), or equivalent amounts of its solvent DMSO, and imaged by confocal time-lapse microscopy as in Fig. 1B. C shows images from representative movies and D shows a quantification of the time between nuclear envelope breakdown and metaphase plate formation. Individual cells were followed in time-lapse movies and scored when they formed a distinct metaphase plate. Scored cells were plotted cumulatively for each time point. The data are from three independent experiments (sictrl, n 146; sictrl + Hesp, n 138; siufd1, n 114; siufd1 + Hesp, n 88). To confirm this result, we monitored metaphase plate formation in live cells (Fig. 6C,D). Control or Ufd1-depleted cells stably expressing histone H2B mrfp were imaged in the absence or presence of 50 nm hesperadin using time-lapse video microscopy as in Fig. 1. We determined the time from nuclear envelope breakdown to the formation of a distinct metaphase plate, when no misaligned chromosomes could be detected. We then plotted the percentage of cells that had formed metaphase plates cumulatively as a function of time (Fig. 6D). Control cells readily formed a metaphase plate within 20 minutes. In accordance with our results in Fig. 1, Ufd1-depleted cells required longer, but eventually nearly all cells completed congression. By contrast, and as expected, less than 10% of the hesperadin-treated cells managed to form a metaphase plate. Importantly, Ufd1-depletion partially restored metaphase plate formation in hesperadin-treated cells, as chromosome alignment occurred in more than 50% of cells, though at a lower rate and with metaphase plates occasionally collapsing after formation. Taken together, these results demonstrate that Ufd1 Npl4 antagonizes Aurora B during chromosome congression and suggest that misregulation of Aurora B, at least in part, accounts for the congression phenotype in Ufd1 Npl4-depleted cells. Discussion The data presented here establish that the Ufd1 Npl4 adapter of the Cdc48/p97 AAA+ ATPase is required for proper chromosome segregation in human somatic cells. Cellular depletion of Ufd1 Npl4 in HeLa cells specifically interfered with efficient chromosome alignment to the metaphase plate, delayed prometaphase and caused anaphase defects, which led to missegregation and abnormal nuclear morphology. Depletion of Ufd1 alone had a weaker effect than destabilization of the whole Ufd1 Npl4 complex after Npl4 depletion. This suggests that Npl4 alone can still partially fulfil the function of Ufd1 Npl4. Both Ufd1 and Npl4 have ubiquitin-binding activity, suggesting that one binding site might compensate for a partial depletion. Interestingly, Npl4 contains an MPN domain (residues ; Kay Hofmann, Miltenyi Biotec, personal communication), which is found in certain deubiquitinating enzymes (Ambroggio et al., 2004). Although the catalytic residues are not conserved, it is possible that this domain confers a crucial function of the Ufd1 Npl4 complex. A role for Cdc48/p97 Ufd1 Npl4 in mitosis was recently questioned, as a mitotic delay could not be observed in C. elegans embryos treated with RNAi against Cdc48/p97 or Ufd1 Npl4 (Mouysset et al., 2008). However, the RNAi in that study was fed to the animals rather than injected, which can limit efficacy. Moreover, a major delay due to chromosome misalignment is not expected in early embryos, because they have a very weak spindle assembly checkpoint under normal growth conditions, which can only delay anaphase onset moderately and only when the spindle is disrupted severely (Nystul et al., 2003). We suggest, therefore, that the evidence presented by Mouysset and colleagues (Mouysset et al., 2008) does not preclude a role for Cdc48/p97 Ufd1 Npl4 in mitosis in C. elegans. Our new results confirm the central role of Cdc48/p97 Ufd1 Npl4 in mitosis in HeLa cells. Along with previous results from C. elegans and Xenopus (Ramadan et al., 2007), as well as yeast (Cao et al., 2003; Cheng and Chen, 2010; Frohlich et al., 1991; Moir et
8 (9) al., 1982), they suggest that the Cdc48/p97 Ufd1 Npl4 chaperone complex has a conserved role in maintaining chromosome stability and euploidy during proliferation. Here, we also provide several lines of evidence that Cdc48/p97 Ufd1 Npl4 antagonizes Aurora B during chromosome segregation. This is supported by the observation that Ufd1 Npl4 depletion caused an increase in the levels of Aurora B associated with mitotic chromosomes, as shown by immunohistochemistry and biochemical fractionation. This notion is based on results from a total of seven Ufd1 and Npl4 sirna oligonucleotides. We note, however, that the depletion efficiencies and degree of segregation errors do not completely correlate with the various degrees of Aurora B accumulation for the different sirna oligonucleotides. Nevertheless, restoration experiments with a resistant Ufd1 cdna demonstrated that both the Aurora B accumulation and the segregation errors are a specific consequence of Ufd1 depletion. Moreover, the same experiments showed that the effect of an oligonucleotide previously reported to cause loss of Aurora B from chromosomes (Vong et al., 2005) was indirect, as it was not rescued by overexpression of the resistant Ufd1 cdna. In Xenopus egg extract, Cdc48/p97 Ufd1 Npl4 removes Aurora B from chromosomes at the end of mitosis (Ramadan et al., 2007). Although not shown directly in this manuscript, this suggests that Cdc48/p97 Ufd1 Npl4 might act in a similar manner at earlier stages of mitosis in HeLa cells and remove some Aurora B from chromosomes as cells progress from prophase (where Aurora B localizes along the entire chromosomes) to metaphase, thereby attenuating its activity. Given that Cdc48/p97 Ufd1 Npl4 function is triggered by ubiquitylation (Ramadan et al., 2007; Ye, 2006), this implies the requirement for a specific ubiquitin ligase activity early in mitosis. Interestingly, a cullin-3-based ligase has recently been shown to ubiquitylate Aurora B and to be required for Aurora B removal from chromosomes in prometaphase (Maerki et al., 2009; Sumara et al., 2007). Moreover, cullin-3 physically interacts with Cdc48/p97 (Alexandru et al., 2008) raising the possibility that the two factors cooperate to regulate chromatin-associated Aurora B. Independent of whether this holds true, both sets of findings concur that Aurora B might be actively removed from chromatin in a ubiquitin-regulated manner (Sumara et al., 2008). Other than in Xenopus egg extracts, where Aurora B remains stable, however, the available data suggests that the mobilized fraction of Aurora B in HeLa cells might, at least in part, be degraded. This hypothesis is supported by the increased total Aurora B levels in both Ufd1- depleted cells (Fig. 4C,D) and cullin-3-depleted cells (Sumara et al., 2007). Furthermore, proteasome inhibition leads to increased and persisting levels of Aurora B on chromosomes (Sumara et al., 2007). More direct evidence for the antagonizing role of Cdc48/p97 Ufd1 Npl4 with respect to Aurora B is the elevated activity towards the centromeric substrate CENP-A. Ufd1-depleted cells are less sensitive to partial inhibition of Aurora B, by addition of its inhibitor hesperadin, with respect to CENP-A phosphorylation. The central question arising from these findings is, therefore, whether the activity of Cdc48/p97 Ufd1 Npl4 in antagonizing Aurora B in prometaphase has any functional impact on chromosome segregation and hence whether excessive Aurora B activity can explain the defects caused by Ufd1 Npl4 depletion. A crucial role for Ufd1 Npl4-mediated downregulation of Aurora B in chromosome segregation is supported by the similarity of the phenotypes observed upon Ufd1 Npl4 depletion and inactivation of Aurora B (severe defects in chromosome alignment, as well as lagging and entangled chromosomes in anaphase that lead to the prominent occurrence of micronuclei and multi-lobed nuclei) (Ditchfield et al., 2003; Hauf et al., 2003; Mora-Bermudez et al., 2007; Vagnarelli and Earnshaw, 2004). Segregation defects in anaphase might also be caused indirectly through inefficient DNA replication, as observed after depletion of Cdc48/p97 or Ufd1 Npl4 in C. elegans embryos (Mouysset et al., 2008). However, although we do not exclude the possibility that problems in S phase might contribute to the observed anaphase defects, we also do not detect any major inhibition of DNA replication in Ufd1 Npl4-depleted HeLa cells (Fig. 2F), as described for C. elegans embryos (Mouysset et al., 2008). Furthermore, and more importantly, our results draw a clear link between Ufd1 Npl4 and Aurora B during mitosis, as we show that they functionally interact in regulating chromosome alignment. Attenuation of Aurora B activity, by chemical inhibition with hesperadin, improved chromosome alignment to the metaphase plate in Ufd1-depleted cells. Moreover, Ufd1- depletion partially rescued congression and metaphase plate formation in hesperadin-treated cells. This functional interaction again suggests that Cdc48/p97 Ufd1 Npl4 antagonizes Aurora B during chromosome congression and that overactivation of Aurora B accounts, at least in part, for the phenotype of Ufd1 Npl4 depletion in HeLa cells. A recent report, published during the revision of this manuscript, revealed that in Saccharomyces cerevisiae, Cdc48/p97 also counteracts Aurora (Ipl1) during chromosome bi-orientation (Cheng and Chen, 2010). Taken together with our data presented here, this suggests that the antagonizing function of Cdc48/p97 towards Aurora B is conserved during evolution. Interestingly, Cheng and Chen further suggest a role for the p47 adapter homologue Shp1. We did not find any evidence for an involvement of p47 in chromosome alignment in HeLa cells, which might point to distinct requirements for adapters in the two systems, possibly owing to differences like the open compared with the closed mitosis. The implication that excessive Aurora B activity at the centromere, upon Cdc48/p97 Ufd1 Npl4 depletion, induces congression defects is in line with the notion that both negative and positive regulation of Aurora B is crucial for its role in controlling bipolar spindle attachment (Kelly and Funabiki, 2009). Excess Aurora B activity might either overly destabilize microtubule attachments by phosphorylation of Aurora-Bcontrolled anchor points, such as Ndc80 (DeLuca et al., 2006). Alternatively, it might prevent adjustment of incorrect spindle attachment to kinetochores by phosphorylation and thus inhibition of the microtubule-depolymerizing activity of MCAK (Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004). The requirement for negative regulation to limit Aurora B activity has been shown before in the case of protein phosphatase-1, which antagonizes Aurora B at the kinetochore to ensure proper chromosome segregation (Emanuele et al., 2008; Francisco et al., 1994; Pinsky et al., 2006). It will be interesting to clarify whether phosphatases and the ubiquitin system, including Cdc48/p97, cooperate to control Aurora B activity. Materials and Methods Antibodies and plasmids Monoclonal (1:250; 5E2) and polyclonal (HME14) anti-ufd1, anti-npl4 (HME16) (all 1:500), anti-p47 (1:5000; HME22) and anti-cdc48/p97 (1:2000; HME01) antibodies were as described previously (Meyer et al., 2000). Antibodies against the following proteins were purchased and used at the indicated dilutions: Aurora B (1:500; , BD Bioscience), phosphorylated Aurora B (serine 10) (1:200; Rockland), TopoIIa (1:1000; Boehringer Ingelheim), phosphorylated histone H3
9 Cdc48/p97 antagonizes Aurora B during chromosome segregation 1579 (serine 10) (1:4000; , Upstate), GAPDH (G8795, Sigma) 1:5000, phosphorylated CENP-A (1:1000; , Millipore), cyclin B1 (1:500; sc-245, Santa Cruz Biotechnology), anti-survivin (1:500; AF886, R&D Systems) and human anti-centromere (kinetochore) CREST (1:400, Antibodies). Mouse Ufd1 cdna was cloned into pcdna3.1 (Invitrogen) with a sequence coding for a C-terminal Myc tag. Mouse Ufd1 cdna was cloned into the BamHI site of pegfp-n3 (Clontech) and the sequence stretch 5 -CTGCGGGTG - ATGGAGACCAAA-3 was mutated to 5 -CTGCGGGTAATGGAAACTAAG-3 by the Quikchange protocol to render it resistant to V-S2 Ufd1 sirna. Cell culture, establishment of stable cell lines and RNAi For routine cell culture, HeLa Kyoto cells were cultured in Dulbecco s modified Eagle s medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum and 1% penicillin/streptomycin (routine media). For RNAi, cells were plated 1 day before transfection with nm sirna (Microsynth, Switzerland) using Oligofectamine, or 10 nm sirna using Lipofectamine RNAiMAX in OptiMEM minimal medium according to the manufacturer s instructions (Invitrogen). Cells were analysed 48 hours after transfection. As a negative control in each experiment, a non-silencing sirna with the sense sequence 5 -UUCUCCGAACGUGUC - ACGUdTdT-3 (adapted from Ambion, Austin, TX) was used. The following sirnas, targeting Cdc48/p97 cofactors Ufd1, Npl4 and p47 were used (sense strand): Ufd1- S1, 5 -GGGCUACAAAGAACCCGAAdTdT-3 ; Ufd1-S2, 5 -GUG GCC ACC - UACUCCAAAUdTdT-3 ; Ufd1-S3, 5 -CTACAAAG AACCCGAAAGAdTdT-3 ; Ufd1-S4, 5 -ACAAAGAACCCGAAAGACAdTdT-3 ; Ufd1-S5, 5 -CTGG - GCTACAAAGAACCCGAAdTdT-3 ; Npl4-S1, 5 -CGUGGUGGAGGAUG - AGAUUdTdT-3 ; Npl4-S2, 5 -CAGCCUCCUCCAACAAAUCdTdT-3 ; p47-s1, 5 -AGCCAGCUCUUCCAUCUUAdTdT-3 ; and p47-s2, 5 -UCAGAG - CCUACCACAAACAdTdT-3. If not otherwise stated, Ufd1-S2, Npl4-S2 and p47-s1 were used for functional assays. Aurora B sirna, 5 -GGTGATGGAG AAT - AGCAGT-3 (Hauf et al., 2003), Ufd1-V-S1 and -V-S2 were as published previously (Vong et al., 2005). Cell extracts For western blot analysis, HeLa cells were lysed for 15 minutes on ice in 150 mm KCl, 25 mm Tris-HCl ph 7.4, 5 mm MgCl 2, 5% glycerol, 1% Triton X-100, 1 mm dithiothreitol (DTT) and EDTA-free protease inhibitor cocktail (Roche), and centrifuged at 10,000 g at 4 C for 20 minutes, with the supernatant recovered for analysis. For fractionation, cells were synchronized in 330 nm nocodazole for 16 hours. Mitotic cells were harvested by shake-off and the mitotic index determined. Equal numbers of cells were incubated for 15 minutes in lysis buffer (10 mm HEPES ph 7.4, 10 mm KCl, 1.5 mm MgCl 2, 340 mm sucrose, 10% glycerol, 0.25% Triton X-100, 1 mm DTT and protease inhibitors) and lysed by 20 strokes in a douncer. The chromatin was sedimented by centrifugation at 1300 g for 5 minutes and washed twice in lysis buffer; this supernatant, which contained the soluble proteins, was centrifuged at 16,000 g for 15 minutes and the supernatant harvested for analysis. Equal fractions were subjected to western blotting. Confocal live cell imaging (mitotic timing) Stable H2B mrfp HeLa Kyoto cells growing on Ibidi chambers were transfected with sirnas. Cells were cultured, for data acquisition, in minimal essential medium with Hank s F12 (MEM/F12; Gibco) supplemented with 15 mm HEPES ph 7.4. Confocal fluorescence time-lapse movies were taken with a Leica SP2 AOPS confocal microscope equipped with a NA Apofluor objective lens. Cells were kept at 37 C during imaging. For acquisition, pictures from one focal plane of living cells were taken every 1 2 minutes. Microscope settings were as follows: picture average, 6; 400 Hz scan speed; 8-bit resolution; logical size, , PMT 1: 0.7 Offset, volts; PMT2: 3.5 Offset, volts. Images were processed using Imaris (Bidplane) and ImageJ software. Indirect immunofluorescence Cells were grown on glass coverslips and transfected with sirnas as described above. HeLa cells were either first treated with pre-extraction buffer (25 mm Hepes ph 7.5, 50 mm NaCl, 1 mm EGTA, 3 mm MgCl 2, 300 mm sucrose and 0.5% Triton X-100) for 5 minutes at 4 C, or fixed directly with 4% paraformaldehyde in PBS for minutes. For hypotonic treatment, cells were incubated for 5 minutes in RSB buffer (10 mm Tris-HCl ph 7.4, 10 mm NaCl and 5 mm MgCl 2 ) before fixing. Cells were permeabilized with 0.1% Triton X-100 (Sigma) in PBS for 5 10 minutes and washed as described above. Permeabilized cells were blocked with 3% BSA in PBS for 30 minutes, followed by indirect immunostaining. Samples were mounded with Mowiol and analyzed using a Leica SP2 AOPS or a SP1 AOPS confocal microscope, or Zeiss Axiovert TV or Leica DM6000B epifluorescence microscope. Images were processed and intensities quantified with the Imaris or ImageJ software, respectively. Statistical analysis Datasets were quantified with Sigma Plot and Past ( software. Normal and non-normal distributed and datasets with equal and different sample numbers, were analyzed by Student s t-test, except for Fig. 1C where a Mann Whitney U-test was used. We thank Izabela Sumara, Matthias Peter, Monica Gotta and Patrick Meraldi for help and critical discussions. We thank Catherine Brasseur for technical help. This work was supported by a grant of the Deutscher Akademischer Austauschdienst (to O.P.), and grants of the Schweizerischer Nationalfond (3100A ), the ETH (TH ) and the Novartis Research Foundation (to H.M.). M.H.A.S. generated H2B mrfp-expressing cells and, together with D.W.G., designed quantitative live imaging assays. Supplementary material available online at References Adams, R. R., Maiato, H., Earnshaw, W. C. and Carmena, M. (2001). Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153, Alam, S. L., Sun, J., Payne, M., Welch, B. D., Blake, B. K., Davis, D. R., Meyer, H. H., Emr, S. D. and Sundquist, W. I. (2004). Ubiquitin interactions of NZF zinc fingers. EMBO J. 23, Alexandru, G., Graumann, J., Smith, G. T., Kolawa, N. J., Fang, R. and Deshaies, R. J. (2008). UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell 134, Ambroggio, X. I., Rees, D. C. and Deshaies, R. J. (2004). JAMM: a metalloproteaselike zinc site in the proteasome and signalosome. PLoS Biol. 2, E2. Andrews, P. D., Ovechkina, Y., Morrice, N., Wagenbach, M., Duncan, K., Wordeman, L. and Swedlow, J. R. (2004). Aurora B regulates MCAK at the mitotic centromere. Dev. Cell 6, Beaudouin, J., Gerlich, D., Daigle, N., Eils, R. and Ellenberg, J. (2002). Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108, Bruderer, R. M., Brasseur, C. and Meyer, H. H. (2004). The AAA ATPase p97/vcp interacts with its alternative co-factors, Ufd1 Npl4 and p47, through a common bipartite binding mechanism. J. Biol. Chem. 279, Cao, K., Nakajima, R., Meyer, H. H. and Zheng, Y. (2003). The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell 115, Cheeseman, I. M., Chappie, J. S., Wilson-Kubalek, E. M. and Desai, A. (2006). The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, Cheng, Y. L. and Chen, R. H. (2010). The AAA-ATPase Cdc48 and cofactor Shp1 promote chromosome bi-orientation by balancing Aurora B activity. J. Cell Sci. 123, Ciferri, C., Pasqualato, S., Screpanti, E., Varetti, G., Santaguida, S., Dos Reis, G., Maiolica, A., Polka, J., De Luca, J. G., De Wulf, P. et al. (2008). Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133, Cimini, D., Wan, X., Hirel, C. B. and Salmon, E. D. (2006). Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 16, DeLuca, J. G., Gall, W. E., Ciferri, C., Cimini, D., Musacchio, A. and Salmon, E. D. (2006). Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127, Ditchfield, C., Johnson, V. L., Tighe, A., Ellston, R., Haworth, C., Johnson, T., Mortlock, A., Keen, N. and Taylor, S. S. (2003). Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161, Ducat, D. and Zheng, Y. (2004). Aurora kinases in spindle assembly and chromosome segregation. Exp. Cell Res. 301, Emanuele, M. J., Lan, W., Jwa, M., Miller, S. A., Chan, C. S. and Stukenberg, P. T. (2008). Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J. Cell Biol. 181, Francisco, L., Wang, W. and Chan, C. S. (1994). Type 1 protein phosphatase acts in opposition to IpL1 protein kinase in regulating yeast chromosome segregation. Mol. Cell. Biol. 14, Frohlich, K.-U., Fries, H.-W., Rudiger, M., Erdmann, R., Botstein, D. and Mecke, D. (1991). Yeast cell cycle protein cdc48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression. J. Cell Biol. 114, Gassmann, R., Vagnarelli, P., Hudson, D. and Earnshaw, W. C. (2004). Mitotic chromosome formation and the condensin paradox. Exp. Cell Res. 296, Giet, R. and Glover, D. M. (2001). Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152, Hauf, S., Cole, R. W., LaTerra, S., Zimmer, C., Schnapp, G., Walter, R., Heckel, A., van Meel, J., Rieder, C. L. and Peters, J. M. (2003). The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161, Heallen, T. R., Adams, H. P., Furuta, T., Verbrugghe, K. J. and Schumacher, J. M. (2008). An Afg2/Spaf-related Cdc48-like AAA ATPase regulates the stability and activity of the C. elegans Aurora B kinase AIR-2. Dev. Cell 15,
Regulators of Cell Cycle Progression
 Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Posch et al., http://www.jcb.org/cgi/content/full/jcb.200912046/dc1 Figure S1. Biochemical characterization of the interaction between
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Posch et al., http://www.jcb.org/cgi/content/full/jcb.200912046/dc1 Figure S1. Biochemical characterization of the interaction between
Cell Cycle, Mitosis, and Microtubules. LS1A Final Exam Review Friday 1/12/07. Processes occurring during cell cycle
 Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Figure S1. HP1α localizes to centromeres in mitosis and interacts with INCENP. (A&B) HeLa
 SUPPLEMENTARY FIGURES Figure S1. HP1α localizes to centromeres in mitosis and interacts with INCENP. (A&B) HeLa tet-on cells that stably express HP1α-CFP, HP1β-CFP, or HP1γ-CFP were monitored with livecell
SUPPLEMENTARY FIGURES Figure S1. HP1α localizes to centromeres in mitosis and interacts with INCENP. (A&B) HeLa tet-on cells that stably express HP1α-CFP, HP1β-CFP, or HP1γ-CFP were monitored with livecell
The Cell Cycle. Packet #9. Thursday, August 20, 2015
 1 The Cell Cycle Packet #9 2 Introduction Cell Cycle An ordered sequence of events in the life of a dividing eukaryotic cell and is a cellular asexual reproduction. The contents of the parent s cell nucleus
1 The Cell Cycle Packet #9 2 Introduction Cell Cycle An ordered sequence of events in the life of a dividing eukaryotic cell and is a cellular asexual reproduction. The contents of the parent s cell nucleus
The Cell Cycle. Dr. SARRAY Sameh, Ph.D
 The Cell Cycle Dr. SARRAY Sameh, Ph.D Overview When an organism requires additional cells (either for growth or replacement of lost cells), new cells are produced by cell division (mitosis) Somatic cells
The Cell Cycle Dr. SARRAY Sameh, Ph.D Overview When an organism requires additional cells (either for growth or replacement of lost cells), new cells are produced by cell division (mitosis) Somatic cells
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Dunsch et al., http://www.jcb.org/cgi/content/full/jcb.201202112/dc1 Figure S1. Characterization of HMMR and CHICA antibodies. (A) HeLa
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Dunsch et al., http://www.jcb.org/cgi/content/full/jcb.201202112/dc1 Figure S1. Characterization of HMMR and CHICA antibodies. (A) HeLa
Mitosis and the Cell Cycle
 Mitosis and the Cell Cycle Chapter 12 The Cell Cycle: Cell Growth & Cell Division Where it all began You started as a cell smaller than a period at the end of a sentence Getting from there to here Cell
Mitosis and the Cell Cycle Chapter 12 The Cell Cycle: Cell Growth & Cell Division Where it all began You started as a cell smaller than a period at the end of a sentence Getting from there to here Cell
Origin of replication. Septum
 Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Chromatin Loop Solenoid Scaffold protein Scaffold protein Chromatin loop
Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Chromatin Loop Solenoid Scaffold protein Scaffold protein Chromatin loop
Bacterial cell. Origin of replication. Septum
 Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Scaffold protein Chromatin Loop Solenoid Scaffold protein Chromatin loop
Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Scaffold protein Chromatin Loop Solenoid Scaffold protein Chromatin loop
How Cells Divide. Chapter 10
 How Cells Divide Chapter 10 Bacterial Cell Division Bacteria divide by binary fission. -the single, circular bacterial chromosome is replicated -replication begins at the origin of replication and proceeds
How Cells Divide Chapter 10 Bacterial Cell Division Bacteria divide by binary fission. -the single, circular bacterial chromosome is replicated -replication begins at the origin of replication and proceeds
基醫所. The Cell Cycle. Chi-Wu Chiang, Ph.D. IMM, NCKU
 基醫所 The Cell Cycle Chi-Wu Chiang, Ph.D. IMM, NCKU 1 1 Introduction to cell cycle and cell cycle checkpoints 2 2 Cell cycle A cell reproduces by performing an orderly sequence of events in which it duplicates
基醫所 The Cell Cycle Chi-Wu Chiang, Ph.D. IMM, NCKU 1 1 Introduction to cell cycle and cell cycle checkpoints 2 2 Cell cycle A cell reproduces by performing an orderly sequence of events in which it duplicates
10-2 Cell Division. Chromosomes
 Cell Division In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm, is called cytokinesis.
Cell Division In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm, is called cytokinesis.
Molecular Cell Biology - Problem Drill 22: The Mechanics of Cell Division
 Molecular Cell Biology - Problem Drill 22: The Mechanics of Cell Division Question No. 1 of 10 1. Which of the following statements about mitosis is correct? Question #1 (A) Mitosis involves the dividing
Molecular Cell Biology - Problem Drill 22: The Mechanics of Cell Division Question No. 1 of 10 1. Which of the following statements about mitosis is correct? Question #1 (A) Mitosis involves the dividing
Mitosis. AND Cell DiVISION
 Mitosis AND Cell DiVISION Cell Division Characteristic of living things: ability to reproduce their own kind. Cell division purpose: When unicellular organisms such as amoeba divide to form offspring reproduction
Mitosis AND Cell DiVISION Cell Division Characteristic of living things: ability to reproduce their own kind. Cell division purpose: When unicellular organisms such as amoeba divide to form offspring reproduction
10-2 Cell Division. Slide 1 of 38. End Show. Copyright Pearson Prentice Hall
 1 of 38 Cell Division In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm, is called
1 of 38 Cell Division In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm, is called
Chapter 10 Cell Growth and Division
 Chapter 10 Cell Growth and Division 10 1 Cell Growth 2 Limits to Cell Growth The larger a cell becomes, the more demands the cell places on its DNA. In addition, the cell has more trouble moving enough
Chapter 10 Cell Growth and Division 10 1 Cell Growth 2 Limits to Cell Growth The larger a cell becomes, the more demands the cell places on its DNA. In addition, the cell has more trouble moving enough
Supplementary information. The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic. Spindle Orientation
 Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
10-2 Cell Division mitosis. cytokinesis. Chromosomes chromosomes Slide 1 of 38
 In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm, is called cytokinesis. Chromosomes
In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm, is called cytokinesis. Chromosomes
Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells
 /, 2017, Vol. 8, (No. 29), pp: 46781-46800 Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells Roshan L. Shrestha 1, Grace S. Ahn 1, Mae
/, 2017, Vol. 8, (No. 29), pp: 46781-46800 Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells Roshan L. Shrestha 1, Grace S. Ahn 1, Mae
Lecture 10. G1/S Regulation and Cell Cycle Checkpoints. G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint
 Lecture 10 G1/S Regulation and Cell Cycle Checkpoints Outline: G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint Paper: The roles of Fzy/Cdc20 and Fzr/Cdh1
Lecture 10 G1/S Regulation and Cell Cycle Checkpoints Outline: G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint Paper: The roles of Fzy/Cdc20 and Fzr/Cdh1
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division Ch. 10 Where it all began You started as a cell smaller than a period
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division Ch. 10 Where it all began You started as a cell smaller than a period
Outline Interphase Mitotic Stage Cell Cycle Control Apoptosis Mitosis Mitosis in Animal Cells Cytokinesis Cancer Prokaryotic Cell Division
 The Cell Cycle and Cellular Reproduction Chapter 9 Outline Interphase Mitotic Stage Cell Cycle Control Apoptosis Mitosis Mitosis in Animal Cells Cytokinesis Cancer Prokaryotic Cell Division 1 2 Interphase
The Cell Cycle and Cellular Reproduction Chapter 9 Outline Interphase Mitotic Stage Cell Cycle Control Apoptosis Mitosis Mitosis in Animal Cells Cytokinesis Cancer Prokaryotic Cell Division 1 2 Interphase
Biology is the only subject in which multiplication is the same thing as division
 The Cell Cycle Biology is the only subject in which multiplication is the same thing as division Why do cells divide? For reproduction asexual reproduction For growth one-celled organisms from fertilized
The Cell Cycle Biology is the only subject in which multiplication is the same thing as division Why do cells divide? For reproduction asexual reproduction For growth one-celled organisms from fertilized
Prentice Hall Biology Slide 1 of 38
 Prentice Hall Biology 1 of 38 2 of 38 In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm,
Prentice Hall Biology 1 of 38 2 of 38 In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm,
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
Cell division functions in 1. reproduction, 2. growth, and 3. repair
 Cell division functions in 1. reproduction, 2. growth, and 3. repair What do you think you are looking at here??? Can something like you or I do this??? Fig. 12.1 How did you start out? How did you grow?
Cell division functions in 1. reproduction, 2. growth, and 3. repair What do you think you are looking at here??? Can something like you or I do this??? Fig. 12.1 How did you start out? How did you grow?
(a) Reproduction. (b) Growth and development. (c) Tissue renewal
 100 µm 200 µm 20 µm (a) Reproduction (b) Growth and development (c) Tissue renewal 1 20 µm 2 0.5 µm Chromosomes DNA molecules Chromosome arm Centromere Chromosome duplication (including DNA synthesis)
100 µm 200 µm 20 µm (a) Reproduction (b) Growth and development (c) Tissue renewal 1 20 µm 2 0.5 µm Chromosomes DNA molecules Chromosome arm Centromere Chromosome duplication (including DNA synthesis)
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Getting from there to here Going from egg to baby. the original
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Getting from there to here Going from egg to baby. the original
Chapter 8: Cellular Reproduction
 Chapter 8: Cellular Reproduction 1. The Cell Cycle 2. Mitosis 3. Meiosis 2 Types of Cell Division 2n 1n Mitosis: occurs in somatic cells (almost all cells of the body) generates cells identical to original
Chapter 8: Cellular Reproduction 1. The Cell Cycle 2. Mitosis 3. Meiosis 2 Types of Cell Division 2n 1n Mitosis: occurs in somatic cells (almost all cells of the body) generates cells identical to original
The Cell Cycle CHAPTER 12
 The Cell Cycle CHAPTER 12 The Key Roles of Cell Division cell division = reproduction of cells All cells come from pre-exisiting cells Omnis cellula e cellula Unicellular organisms division of 1 cell reproduces
The Cell Cycle CHAPTER 12 The Key Roles of Cell Division cell division = reproduction of cells All cells come from pre-exisiting cells Omnis cellula e cellula Unicellular organisms division of 1 cell reproduces
 Supplementary Figure Legends Supplementary Figure S1. Aurora-A is essential for SAC establishment in early mitosis. (a-c) RPE cells were treated with DMSO (a), MLN8237 (b) or BI2536 (c) for Two hours.
Supplementary Figure Legends Supplementary Figure S1. Aurora-A is essential for SAC establishment in early mitosis. (a-c) RPE cells were treated with DMSO (a), MLN8237 (b) or BI2536 (c) for Two hours.
Reproduction is a fundamental property of life. Cells are the fundamental unit of life. Reproduction occurs at the cellular level with one mother
 Cell Division ision Reproduction is a fundamental property of life. Cells are the fundamental unit of life. Reproduction occurs at the cellular level with one mother cell giving rise to two daughter cells.
Cell Division ision Reproduction is a fundamental property of life. Cells are the fundamental unit of life. Reproduction occurs at the cellular level with one mother cell giving rise to two daughter cells.
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division The Cell Cycle: Cell Growth, Cell Division 2007-2008 2007-2008 Getting from there to here Going from egg to baby. the original
Biology is the only subject in which multiplication is the same thing as division The Cell Cycle: Cell Growth, Cell Division 2007-2008 2007-2008 Getting from there to here Going from egg to baby. the original
U3.2.3: Eukaryotic chromosomes are linear DNA molecules associated with histone proteins. (Oxford Biology Course Companion page 151).
 Cell Division Study Guide U3.2.3: Eukaryotic chromosomes are linear DNA molecules associated with histone proteins. (Oxford Biology Course Companion page 151). 1. Describe the structure of eukaryotic DNA
Cell Division Study Guide U3.2.3: Eukaryotic chromosomes are linear DNA molecules associated with histone proteins. (Oxford Biology Course Companion page 151). 1. Describe the structure of eukaryotic DNA
Ploidy and Human Cell Types. Cell Cycle and Mitosis. DNA and Chromosomes. Where It All Began 11/19/2014. Chapter 12 Pg
 Ploidy and Human Cell Types Cell Cycle and Mitosis Chapter 12 Pg. 228 245 Cell Types Somatic cells (body cells) have 46 chromosomes, which is the diploid chromosome number. A diploid cell is a cell with
Ploidy and Human Cell Types Cell Cycle and Mitosis Chapter 12 Pg. 228 245 Cell Types Somatic cells (body cells) have 46 chromosomes, which is the diploid chromosome number. A diploid cell is a cell with
Unduplicated. Chromosomes. Telophase
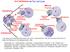 10-2 Cell Division The Cell Cycle Interphase Mitosis Prophase Cytokinesis G 1 S G 2 Chromatin in Parent Nucleus & Daughter Cells Chromatin Daughter Nuclei Telophase Mitotic Anaphase Metaphase Use what
10-2 Cell Division The Cell Cycle Interphase Mitosis Prophase Cytokinesis G 1 S G 2 Chromatin in Parent Nucleus & Daughter Cells Chromatin Daughter Nuclei Telophase Mitotic Anaphase Metaphase Use what
Creating Identical Body Cells
 Creating Identical Body Cells 5.A Students will describe the stages of the cell cycle, including DNA replication and mitosis, and the importance of the cell cycle to the growth of organisms 5.D Students
Creating Identical Body Cells 5.A Students will describe the stages of the cell cycle, including DNA replication and mitosis, and the importance of the cell cycle to the growth of organisms 5.D Students
University of Bristol - Explore Bristol Research
 Shandilya, J., Medler, K., & Roberts, S. G. E. (2016). Regulation of AURORA B function by mitotic checkpoint protein MAD2. Cell Cycle, 15(16), 2196-2201. https://doi.org/10.1080/15384101.2016.1200773 Peer
Shandilya, J., Medler, K., & Roberts, S. G. E. (2016). Regulation of AURORA B function by mitotic checkpoint protein MAD2. Cell Cycle, 15(16), 2196-2201. https://doi.org/10.1080/15384101.2016.1200773 Peer
Chapter 12. The Cell Cycle
 Chapter 12 The Cell Cycle The Key Roles of Cell Division The ability of organisms to produce more of their own kind is the one characteristic that best distinguishes living things from nonliving things.
Chapter 12 The Cell Cycle The Key Roles of Cell Division The ability of organisms to produce more of their own kind is the one characteristic that best distinguishes living things from nonliving things.
Biology is the only subject in which multiplication is the same thing as division. AP Biology
 Biology is the only subject in which multiplication is the same thing as division Chapter 12. The Cell Cycle: Cell Growth, Cell Division Where it all began You started as a cell smaller than a period at
Biology is the only subject in which multiplication is the same thing as division Chapter 12. The Cell Cycle: Cell Growth, Cell Division Where it all began You started as a cell smaller than a period at
The Cell Cycle. Chapter 12. Biology Eighth Edition Neil Campbell and Jane Reece. PowerPoint Lecture Presentations for
 Chapter 12 The Cell Cycle PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Copyright
Chapter 12 The Cell Cycle PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Copyright
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
Cellular Reproduction, Part 1: Mitosis Lecture 10 Fall 2008
 Cell Theory 1 Cellular Reproduction, Part 1: Mitosis Lecture 10 Fall 2008 Cell theory: All organisms are made of cells All cells arise from preexisting cells How do new cells arise? Cell division the reproduction
Cell Theory 1 Cellular Reproduction, Part 1: Mitosis Lecture 10 Fall 2008 Cell theory: All organisms are made of cells All cells arise from preexisting cells How do new cells arise? Cell division the reproduction
5/25/2015. Replication fork. Replication fork. Replication fork. Replication fork
 Mutations Chapter 5 Cellular Functions Lecture 3: and Cell Division Most DNA mutations alter the protein product May Make it function better (rarely) Change its function Reduce its function Make it non-functional
Mutations Chapter 5 Cellular Functions Lecture 3: and Cell Division Most DNA mutations alter the protein product May Make it function better (rarely) Change its function Reduce its function Make it non-functional
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis
 Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Chromosomes Chromosomes were first observed by the German embryologist Walther Fleming in 1882. Chromosome number varies among organisms most
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Chromosomes Chromosomes were first observed by the German embryologist Walther Fleming in 1882. Chromosome number varies among organisms most
Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion
 Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Cellular Reproduction, Part 2: Meiosis Lecture 10 Fall 2008
 Mitosis & 1 Cellular Reproduction, Part 2: Lecture 10 Fall 2008 Mitosis Form of cell division that leads to identical daughter cells with the full complement of DNA Occurs in somatic cells Cells of body
Mitosis & 1 Cellular Reproduction, Part 2: Lecture 10 Fall 2008 Mitosis Form of cell division that leads to identical daughter cells with the full complement of DNA Occurs in somatic cells Cells of body
CELL CYCLE INTRODUCTION PART I ANIMAL CELL CYCLE INTERPHASE
 CELL CYCLE INTRODUCTION The nuclei in cells of eukaryotic organisms contain chromosomes with clusters of genes, discrete units of hereditary information consisting of double-stranded DNA. Structural proteins
CELL CYCLE INTRODUCTION The nuclei in cells of eukaryotic organisms contain chromosomes with clusters of genes, discrete units of hereditary information consisting of double-stranded DNA. Structural proteins
Chapter 8 The Cell Cycle
 What molecule stores your genetic information or determines everything about you? DNA a nucleic acid How are DNA molecules arranged in the nucleus? As you can see DNA is: Chapter 8 The Cell Cycle 1. Arranged
What molecule stores your genetic information or determines everything about you? DNA a nucleic acid How are DNA molecules arranged in the nucleus? As you can see DNA is: Chapter 8 The Cell Cycle 1. Arranged
Section Cell Growth. A. Limits to Cell Growth 1. DNA Overload 2. Exchanging Materials 3. Ratio of Surface Area to Volume 4.
 Getting Through Materials move through cells by diffusion. Oxygen and food move into cells, while waste products move out of cells. How does the size of a cell affect how efficiently materials get to all
Getting Through Materials move through cells by diffusion. Oxygen and food move into cells, while waste products move out of cells. How does the size of a cell affect how efficiently materials get to all
The Cell Cycle CAMPBELL BIOLOGY IN FOCUS SECOND EDITION URRY CAIN WASSERMAN MINORSKY REECE
 CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 9 The Cell Cycle Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION Overview: The Key
CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 9 The Cell Cycle Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION Overview: The Key
Inhibitor-3 ensures bipolar mitotic spindle attachment by limiting association of SDS22 with kinetochore-bound protein phosphatase-1
 Article Inhibitor-3 ensures bipolar mitotic spindle attachment by limiting association of with kinetochore-bound protein phosphatase-1 Annika Eiteneuer 1,, Jonas Seiler 1,, Matthias Weith 1, Monique Beullens
Article Inhibitor-3 ensures bipolar mitotic spindle attachment by limiting association of with kinetochore-bound protein phosphatase-1 Annika Eiteneuer 1,, Jonas Seiler 1,, Matthias Weith 1, Monique Beullens
The Process of Cell Division
 Lesson Overview 10.2 The Process of Cell Division THINK ABOUT IT What role does cell division play in your life? Does cell division stop when you are finished growing? Chromosomes What is the role of chromosomes
Lesson Overview 10.2 The Process of Cell Division THINK ABOUT IT What role does cell division play in your life? Does cell division stop when you are finished growing? Chromosomes What is the role of chromosomes
The Cell Cycle. Chapter 12. PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece
 Chapter 12 The Cell Cycle PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Overview:
Chapter 12 The Cell Cycle PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Overview:
The Cell Cycle and How Cells Divide
 The Cell Cycle and How Cells Divide 1 Phases of the Cell Cycle The cell cycle consists of Interphase normal cell activity The mitotic phase cell divsion INTERPHASE Growth G 1 (DNA synthesis) Growth G 2
The Cell Cycle and How Cells Divide 1 Phases of the Cell Cycle The cell cycle consists of Interphase normal cell activity The mitotic phase cell divsion INTERPHASE Growth G 1 (DNA synthesis) Growth G 2
LECTURE PRESENTATIONS
 LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
Chapter 10. Cell Growth and Division
 Chapter 10 Cell Growth and Division Cell Growth A. Limits to Cell Growth 1. Two main reasons why cells divide: a. Demands on DNA as the cell get too large Cell Growth b. Moving nutrients and waste across
Chapter 10 Cell Growth and Division Cell Growth A. Limits to Cell Growth 1. Two main reasons why cells divide: a. Demands on DNA as the cell get too large Cell Growth b. Moving nutrients and waste across
TFEB-mediated increase in peripheral lysosomes regulates. Store Operated Calcium Entry
 TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
APGRU4L1 Chap 12 Extra Reading Cell Cycle and Mitosis
 APGRU4L1 Chap 12 Extra Reading Cell Cycle and Mitosis Dr. Ramesh Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008
APGRU4L1 Chap 12 Extra Reading Cell Cycle and Mitosis Dr. Ramesh Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008
BIOLOGY 4/6/2015. Cell Cycle - Mitosis. Outline. Overview: The Key Roles of Cell Division. identical daughter cells. I. Overview II.
 2 Cell Cycle - Mitosis CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Overview II. Mitotic Phase I. Prophase II. III. Telophase IV. Cytokinesis III. Binary fission
2 Cell Cycle - Mitosis CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Overview II. Mitotic Phase I. Prophase II. III. Telophase IV. Cytokinesis III. Binary fission
Cell Cycle Notes chromatin, somatic cells gametes mitosis sister chromatids, centromere cytokinesis binary fission,
 Cell Cycle Notes 1. Importance of Cell Division a. For single celled organisms, cell division increases the number of individuals. b. In a multicellular organism, cell division functions to repair and
Cell Cycle Notes 1. Importance of Cell Division a. For single celled organisms, cell division increases the number of individuals. b. In a multicellular organism, cell division functions to repair and
Campbell Biology in Focus (Urry) Chapter 9 The Cell Cycle. 9.1 Multiple-Choice Questions
 Campbell Biology in Focus (Urry) Chapter 9 The Cell Cycle 9.1 Multiple-Choice Questions 1) Starting with a fertilized egg (zygote), a series of five cell divisions would produce an early embryo with how
Campbell Biology in Focus (Urry) Chapter 9 The Cell Cycle 9.1 Multiple-Choice Questions 1) Starting with a fertilized egg (zygote), a series of five cell divisions would produce an early embryo with how
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
BIOLOGY. Cell Cycle - Mitosis. Outline. Overview: The Key Roles of Cell Division. identical daughter cells. I. Overview II.
 2 Cell Cycle - Mitosis CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Overview II. Mitotic Phase I. Prophase II. III. Telophase IV. Cytokinesis III. Binary fission
2 Cell Cycle - Mitosis CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Overview II. Mitotic Phase I. Prophase II. III. Telophase IV. Cytokinesis III. Binary fission
-The cell s hereditary endowment of DNA -Usually packaged into chromosomes for manageability
 Binary Fission-Bacterial Cell Division -Asexual reproduction of prokaryotes -No mitosis -Circular DNA and organelles replicate, the copies migrate to opposite sides of the elongating cell, and the cell
Binary Fission-Bacterial Cell Division -Asexual reproduction of prokaryotes -No mitosis -Circular DNA and organelles replicate, the copies migrate to opposite sides of the elongating cell, and the cell
Yan Ma 1.,XiYuan 1., William R. Wyatt 2, Joseph R. Pomerening 1 * Abstract. Introduction
 Expression of Constitutively Active CDK1 Stabilizes APC- Cdh1 Substrates and Potentiates Premature Spindle Assembly and Checkpoint Function in G1 Cells Yan Ma 1.,XiYuan 1., William R. Wyatt 2, Joseph R.
Expression of Constitutively Active CDK1 Stabilizes APC- Cdh1 Substrates and Potentiates Premature Spindle Assembly and Checkpoint Function in G1 Cells Yan Ma 1.,XiYuan 1., William R. Wyatt 2, Joseph R.
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis
 Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Prokaryotes Have a Simpler Cell Cycle Cell division in prokaryotes takes place in two stages, which together make up a simple cell cycle 1. Copy
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Prokaryotes Have a Simpler Cell Cycle Cell division in prokaryotes takes place in two stages, which together make up a simple cell cycle 1. Copy
Supplementary Figure 1. Mother centrioles can reduplicate while in the close association
 C1-GFP distance (nm) C1-GFP distance (nm) a arrested HeLa cell expressing C1-GFP and Plk1TD-RFP -3 s 1 2 3 4 5 6 7 8 9 11 12 13 14 16 17 18 19 2 21 22 23 24 26 27 28 29 3 b 9 8 7 6 5 4 3 2 arrested HeLa
C1-GFP distance (nm) C1-GFP distance (nm) a arrested HeLa cell expressing C1-GFP and Plk1TD-RFP -3 s 1 2 3 4 5 6 7 8 9 11 12 13 14 16 17 18 19 2 21 22 23 24 26 27 28 29 3 b 9 8 7 6 5 4 3 2 arrested HeLa
meiosis asexual reproduction CHAPTER 9 & 10 The Cell Cycle, Meiosis & Sexual Life Cycles Sexual reproduction mitosis
 meiosis asexual reproduction CHAPTER 9 & 10 The Cell Cycle, Meiosis & Sexual Sexual reproduction Life Cycles mitosis Chromosomes Consists of a long DNA molecule (represents thousands of genes) Also consists
meiosis asexual reproduction CHAPTER 9 & 10 The Cell Cycle, Meiosis & Sexual Sexual reproduction Life Cycles mitosis Chromosomes Consists of a long DNA molecule (represents thousands of genes) Also consists
BIOLOGY - CLUTCH CH.12 - CELL DIVISION.
 !! www.clutchprep.com CONCEPT: CELL DIVISION Cell division is the process by which one cell splits into two or more daughter cells. Cell division generally requires that cells produce enough materials,
!! www.clutchprep.com CONCEPT: CELL DIVISION Cell division is the process by which one cell splits into two or more daughter cells. Cell division generally requires that cells produce enough materials,
Why do cells divide? Cells divide in order to make more cells they multiply in order to create a larger surface to volume ratio!!!
 Why do cells divide? Cells divide in order to make more cells they multiply in order to create a larger surface to volume ratio!!! Chromosomes Are made of chromatin: a mass of genetic material composed
Why do cells divide? Cells divide in order to make more cells they multiply in order to create a larger surface to volume ratio!!! Chromosomes Are made of chromatin: a mass of genetic material composed
Mitotic requirement for aurora A kinase is bypassed in the absence of aurora B kinase
 FEBS 29621 FEBS Letters 579 () 3385 3391 Mitotic requirement for aurora A kinase is bypassed in the absence of aurora B kinase Hui Yang, Teresa Burke, Jack Dempsey, Bruce Diaz, Elizabeth Collins, John
FEBS 29621 FEBS Letters 579 () 3385 3391 Mitotic requirement for aurora A kinase is bypassed in the absence of aurora B kinase Hui Yang, Teresa Burke, Jack Dempsey, Bruce Diaz, Elizabeth Collins, John
Name: Date: Block: 10-2 Cell Division Worksheet
 10-2 Cell Division Worksheet W hat do you think would happen if a cell were simple to split into two, without any advance preparation? Would each daughter cell have everything it needed to survive? Because
10-2 Cell Division Worksheet W hat do you think would happen if a cell were simple to split into two, without any advance preparation? Would each daughter cell have everything it needed to survive? Because
The Cell Cycle 4/10/12. Chapter 12. Overview: The Key Roles of Cell Division
 LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach
![klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach](/thumbs/91/104639484.jpg) DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
Unit 4: Cell Division Guided Notes
 Unit 4: Cell Division Guided Notes 1 Chromosomes are structures that contain material When Eukaryotes are not dividing, DNA and Proteins are in a mass called: When the cell divides, it condenses and becomes
Unit 4: Cell Division Guided Notes 1 Chromosomes are structures that contain material When Eukaryotes are not dividing, DNA and Proteins are in a mass called: When the cell divides, it condenses and becomes
LECTURE PRESENTATIONS
 LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
Cell Division. During interphase, a cell s DNA is in a loose form called. It condenses into tightly coiled structures called chromosomes during.
 Cell Division The is a cell s total DNA. Prokaryotes DNA is found mostly in a single called the and also in small circles called. Eukaryotes have several DNA double helices packaged into. During interphase,
Cell Division The is a cell s total DNA. Prokaryotes DNA is found mostly in a single called the and also in small circles called. Eukaryotes have several DNA double helices packaged into. During interphase,
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
Histones- protein molecules that are used to fold and package DNA into chromosomes.
 Chromosome- a portion of the DNA in a cell, a chromosome is created when the DNA segment coils around histones then twists further to create a long twisted mass. Histones- protein molecules that are used
Chromosome- a portion of the DNA in a cell, a chromosome is created when the DNA segment coils around histones then twists further to create a long twisted mass. Histones- protein molecules that are used
Chromosomes Can Congress To The Metaphase Plate Prior. To Bi-Orientation
 Chromosomes Can Congress To The Metaphase Plate Prior To Bi-Orientation Tarun M. Kapoor 1,2, Michael Lampson 1, Polla Hergert 3, Lisa Cameron 2,4, Daniela Cimini 4, E.D. Salmon 2,4, Bruce F. McEwen 3,
Chromosomes Can Congress To The Metaphase Plate Prior To Bi-Orientation Tarun M. Kapoor 1,2, Michael Lampson 1, Polla Hergert 3, Lisa Cameron 2,4, Daniela Cimini 4, E.D. Salmon 2,4, Bruce F. McEwen 3,
BIOLOGY LTF DIAGNOSTIC TEST CELL CYCLE & MITOSIS
 Biology Multiple Choice 016044 BIOLOGY LTF DIAGNOSTIC TEST CELL CYCLE & MITOSIS TEST CODE: 016044 Directions: Each of the questions or incomplete statements below is followed by five suggested answers
Biology Multiple Choice 016044 BIOLOGY LTF DIAGNOSTIC TEST CELL CYCLE & MITOSIS TEST CODE: 016044 Directions: Each of the questions or incomplete statements below is followed by five suggested answers
The Cell Cycle. Chapter 12. Biology Eighth Edition Neil Campbell and Jane Reece. PowerPoint Lecture Presentations for
 Chapter 12 The Cell Cycle PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp 1
Chapter 12 The Cell Cycle PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp 1
Cell Growth and Division *
 OpenStax-CNX module: m46034 1 Cell Growth and Division * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you will
OpenStax-CNX module: m46034 1 Cell Growth and Division * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you will
Cell Cycle. Trends in Cell Biology
 Cell Cycle Trends in Cell Biology Cell Cycle The orderly sequence of events by which a cell duplicates its contents and divides into two Daughter Cells Activities of a cell from one cell division to the
Cell Cycle Trends in Cell Biology Cell Cycle The orderly sequence of events by which a cell duplicates its contents and divides into two Daughter Cells Activities of a cell from one cell division to the
Chapter 2. Mitosis and Meiosis
 Chapter 2. Mitosis and Meiosis Chromosome Theory of Heredity What structures within cells correspond to genes? The development of genetics took a major step forward by accepting the notion that the genes
Chapter 2. Mitosis and Meiosis Chromosome Theory of Heredity What structures within cells correspond to genes? The development of genetics took a major step forward by accepting the notion that the genes
Cell Division. The Process of Cell Division Section Section 10.2: The Process of Cell Division 12/8/2010
 The Process of Cell Division Section 10.2 Biology B Section 10.2: The Process of Cell Division The student will investigate and understand common mechanisms of inheritance and protein synthesis. Key concepts
The Process of Cell Division Section 10.2 Biology B Section 10.2: The Process of Cell Division The student will investigate and understand common mechanisms of inheritance and protein synthesis. Key concepts
Mad2-independent Spindle Assembly Checkpoint Activation and Controlled Metaphase Anaphase Transition D V
 Molecular Biology of the Cell Vol. 18, 850 863, March 2007 Mad2-independent Spindle Assembly Checkpoint Activation and Controlled Metaphase Anaphase Transition D V in Drosophila S2 Cells Bernardo Orr,*
Molecular Biology of the Cell Vol. 18, 850 863, March 2007 Mad2-independent Spindle Assembly Checkpoint Activation and Controlled Metaphase Anaphase Transition D V in Drosophila S2 Cells Bernardo Orr,*
SUPPLEMENTARY INFORMATION
 DOI: 0.038/ncb33 a b c 0 min 6 min 7 min (fixed) DIC -GFP, CenpF 3 µm Nocodazole Single optical plane -GFP, CenpF Max. intensity projection d µm -GFP, CenpF, -GFP CenpF 3-D rendering e f 0 min 4 min 0
DOI: 0.038/ncb33 a b c 0 min 6 min 7 min (fixed) DIC -GFP, CenpF 3 µm Nocodazole Single optical plane -GFP, CenpF Max. intensity projection d µm -GFP, CenpF, -GFP CenpF 3-D rendering e f 0 min 4 min 0
Mitosis THE CELL CYCLE. In unicellular organisms, division of one cell reproduces the entire organism Multicellular organisms use cell division for..
 Mitosis THE CELL CYCLE In unicellular organisms, division of one cell reproduces the entire organism Multicellular organisms use cell division for.. Development from a fertilized cell Growth Repair Cell
Mitosis THE CELL CYCLE In unicellular organisms, division of one cell reproduces the entire organism Multicellular organisms use cell division for.. Development from a fertilized cell Growth Repair Cell
The questions below refer to the following terms. Each term may be used once, more than once, or not at all.
 The questions below refer to the following terms. Each term may be used once, more than once, or not at all. a) telophase b) anaphase c) prometaphase d) metaphase e) prophase 1) DNA begins to coil and
The questions below refer to the following terms. Each term may be used once, more than once, or not at all. a) telophase b) anaphase c) prometaphase d) metaphase e) prophase 1) DNA begins to coil and
8.4 The cell cycle multiplies cells. 8.4 The cell cycle multiplies cells
 8.4 The cell cycle multiplies cells! Cell division is a highly orchestrated process! The cell cycle is an ordered sequence of events that extends from the time a cell is first formed from a dividing parent
8.4 The cell cycle multiplies cells! Cell division is a highly orchestrated process! The cell cycle is an ordered sequence of events that extends from the time a cell is first formed from a dividing parent
BIOLOGY. The Cell Cycle CAMPBELL. Reece Urry Cain Wasserman Minorsky Jackson. Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick
 CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 12 The Cell Cycle Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick The Key Roles of Cell Division The ability
CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 12 The Cell Cycle Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick The Key Roles of Cell Division The ability
Chapter 12. living /non-living? growth repair renew. Reproduction. Reproduction. living /non-living. fertilized egg (zygote) next chapter
 Chapter 12 How cells divide Reproduction living /non-living? growth repair renew based on cell division first mitosis - distributes identical sets of chromosomes cell cycle (life) Cell Division in Bacteria
Chapter 12 How cells divide Reproduction living /non-living? growth repair renew based on cell division first mitosis - distributes identical sets of chromosomes cell cycle (life) Cell Division in Bacteria
CH 9: The Cell Cycle Overview. Cellular Organization of the Genetic Material. Distribution of Chromosomes During Eukaryotic Cell Division
 CH 9: The Cell Cycle Overview The ability of organisms to produce more of their own kind best distinguishes living things from nonliving matter The continuity of life is based on the reproduction of cells,
CH 9: The Cell Cycle Overview The ability of organisms to produce more of their own kind best distinguishes living things from nonliving matter The continuity of life is based on the reproduction of cells,
General Biology. Overview: The Key Roles of Cell Division. Unicellular organisms
 General Biology Course No: BNG2003 Credits: 3.00 8. The Cell Cycle Prof. Dr. Klaus Heese Overview: The Key Roles of Cell Division The continuity of life is based upon the reproduction of cells, or cell
General Biology Course No: BNG2003 Credits: 3.00 8. The Cell Cycle Prof. Dr. Klaus Heese Overview: The Key Roles of Cell Division The continuity of life is based upon the reproduction of cells, or cell
General Biology. Overview: The Key Roles of Cell Division The continuity of life is based upon the reproduction of cells, or cell division
 General Biology Course No: BNG2003" Credits: 3.00 " " " 8. The Cell Cycle Prof. Dr. Klaus Heese Overview: The Key Roles of Cell Division The continuity of life is based upon the reproduction of cells,
General Biology Course No: BNG2003" Credits: 3.00 " " " 8. The Cell Cycle Prof. Dr. Klaus Heese Overview: The Key Roles of Cell Division The continuity of life is based upon the reproduction of cells,
