Single administration of recombinant IL-6 restores the gene expression of lipogenic enzymes in liver of fasting IL-6-deficient mice
|
|
|
- Elijah Stevenson
- 5 years ago
- Views:
Transcription
1 British Journal of Pharmacology RESEARCH PAPER British Journal of Pharmacology (2016) Single administration of recombinant IL-6 restores the gene expression of lipogenic enzymes in liver of fasting IL-6-deficient mice Correspondence Dr Elena Baixeras, Instituto de Investigación Biomédica de Málaga (IBIMA), Hospital Regional Universitario de Málaga, Avenida Carlos Haya no. 82, Málaga 29010, Spain. Received 30 April 2015; Revised 18 December 2015; Accepted 6 January 2016 AL Gavito 1,2,3, R Cabello 1, J Suarez 1,2,3, A Serrano 1,2,3,FJPavón 1,2,3,MVida 1,2,3,MRomero 1,2,VPardo 4,5, DBautista 6, S Arrabal 1,2,3, J Decara 1,3,ALCuesta 7, A M Valverde 4,5, F Rodríguez de Fonseca 1,2,3, *andebaixeras 1,2,3, * 1 Laboratorio de Investigación, Instituto de Investigación Biomédica de Málaga (IBIMA)/Universidad de Málaga, Málaga 29010, Spain, 2 Centro de Investigación Biomédica en Red de Fisiopatología de la Obesidad y Nutrición (CIBERobn), Instituto de Salud Carlos III (ISCIII), Madrid 28029, Spain, 3 UGC-Salud Mental, Hospital Regional Universitario de Málaga, Málaga, Spain, 4 Instituto de Investigaciones Biomédicas Alberto Sols (CSIC/UAM), Madrid 28029, Spain, 5 Ciber de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM), Instituto de Salud Carlos III, Madrid 28029, Spain, 6 Unidad de Gestión Clínica de Anatomía Patológica, Hospital Regional Universitario de Málaga, Málaga 29010, Spain, and 7 Danish Diabetes Academy, Department of Biomedical Sciences, Panum Institute, University of Copenhagen, Copenhagen, Denmark *These authors contributed equally as co-senior authors BACKGROUND AND PURPOSE Lipogenesis is intimately controlled by hormones and cytokines as well as nutritional conditions. IL-6 participates in the regulation of fatty acid metabolism in the liver. We investigated the role of IL-6 in mediating fasting/re-feeding changes in the expression of hepatic lipogenic enzymes. EXPERIMENTAL APPROACH Gene and protein expression of lipogenic enzymes were examined in livers of wild-type (WT) and IL-6-deficient (IL-6 / )mice during fasting and re-feeding conditions. Effects of exogenous IL-6 administration on gene expression of these enzymes were evaluated in vivo. The involvement of STAT3 in mediating these IL-6 responses was investigated by using sirna in human HepG2 cells. KEY RESULTS During feeding, the up-regulation in the hepatic expression of lipogenic genes presented similar time kinetics in WT and IL-6 / mice. During fasting, expression of lipogenic genes decreased gradually over time in both strains, although the initial drop was more marked in IL-6 / mice. Protein levels of hepatic lipogenic enzymes were lower in IL-6 / than in WT mice at the end of the fasting period. In WT, circulating IL-6 levels paralleled gene expression of hepatic lipogenic enzymes. IL-6 administration in vivo and in vitro showed that IL-6-mediated signalling was associated with the up-regulation of hepatic lipogenic enzyme genes. Moreover, silencing STAT3 in HepG2 cells attenuated IL-6 mediated up-regulation of lipogenic gene transcription levels. CONCLUSIONS AND IMPLICATIONS IL-6 sustains levels of hepatic lipogenic enzymes during fasting through activation of STAT3. Our findings indicate that clinical use of STAT3-associated signalling cytokines, particularly against steatosis, should be undertaken with caution. Abbreviations ACC, (Accα/β, Acaca/b) acetyl-coa carboxylase; FAS, (Fasn) fatty acid synthase; ril-6, recombinant IL-6; SCD1, (Scd1, Scd1) stearoyl-coa desaturase-1; STAT3, (Stat3, Stat3) signal transducer and activator of transcription factor 3; Srebp-1c, (Srebp-1c) sterol regulatory element-binding proteins-1c DOI: /bph The British Pharmacological Society
2 ril-6 up-regulates hepatic lipogenic enzymes BJP Tables of Links TARGETS GPCRs a Nuclear hormone receptors b Enzymes c IL-6 receptor Liver X receptor-α (Lxrα; Nrih3) Acetyl-CoA carboxylase (Acaca/b) gp130 PPARα Fatty acid synthase (Fasn) LIGANDS Acetyl-CoA Glucagon IL-6 Insulin Malonyl-CoA These Tables list key protein targets and ligands in this article which are hyperlinked to corresponding entries in guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Pawson et al., 2014) and are permanently archived in the Concise Guide to PHARMACOLOGY 2015/16 ( a,b,c Alexander et al., 2015a,b,c). Introduction IL-6 belongs to the family of IL-6-type cytokines (Heinrich et al., 2003; Qu et al., 2014). In addition to its role in the immune system, IL-6 is also involved in haematopoiesis, liver regeneration and the regulation of neural and metabolic processes (Qu et al., 2014). The IL-6 receptor is a protein complex consisting of an IL-6 binding subunit and the IL-6 transducer glycoprotein 130 (gp130). Like immune cells, endothelial cells, myocytes and adipocytes, hepatocytes also produce IL- 6, in addition to expressing high levels of the IL-6R/gp130 protein subunits in the outer membrane (Jung et al., 2000; Klein et al., 2005; Qu et al., 2014; Scheller et al., 2011).This suggests that hepatocyte production of IL-6 may be acting in both a paracrine and an autocrine manner. Several studies have shown that the liver is one of the targets for IL-6 where, in addition to other functions, it also appears to modulate lipid metabolism, although the role of IL-6 in this effect is not yet fully understood. On the one hand, chronic administration of IL-6 has been reported to exert beneficial effects on steatosis by promoting fatty acid oxidation through the stimulation of PPARα (Hong et al., 2004;ReddyandHashimoto, 2001; Vida et al., 2013). In this regard, we recently reported that recombinant IL-6 (ril-6) treatment in hepatocytes induced the transcription of the PPARα target gene for carnitine palmitoyltransferase 1 (Cpt1) whose protein promotes fatty acid β-oxidation in the mitochondria (Vida et al., 2013). On the other hand, the biological activity of IL-6 has also been associated with the promotion of lipogenesis and triglyceride secretion in hepatocytes (Brass and Vetter, 1994; Grunfeld et al., 1990; Vida et al., 2015).Moreover,andincontrastwith previous findings (Hong et al., 2004; Reddy and Hashimoto, 2001), we recently showed that chronic administration of IL-6 strikingly up-regulated the expression of the lipogenic enzymes in the liver of IL-6 / mice fed a high-fat diet, and this was associated with aggravated hepatic steatosis (Vida et al., 2015). Lipogenesis encompasses the process by which acetyl- CoA is converted to fatty acids from simple sugars such as glucose (Kersten, 2001). The lipogenic enzymes often include the acetyl-coa carboxylase isoforms (Accα/β), fatty acid synthase (FAS) and stearoyl-coa desaturase-1 (Scd1) (Strable and Ntambi, 2010). The liver X receptor-α, thesterolregulatory element-binding protein 1c (Srebp-1c) and the carbohydrate response element binding protein (Chrebp) are transcription factors particularly abundant in liver, where they play a major role in regulating the gene expression of these lipogenic enzymes (Strable and Ntambi, 2010). The Accα/β isoforms are expressed in hepatocytes and catalyse the carboxylation of acetyl-coa to malonyl-coa, which is the first step in fatty acid synthesis, followed by the action of the FAS and Scd1 enzymes. Thus, the regulation of lipogenesis occurs through changes in the abundance or activity of these enzymes that, in turn, depend on nutritional conditions (Bass and Takahashi, 2010; Fukuda et al., 1985).Indeed, lipogenesis is stimulated by feeding, and more so by a high carbohydrate diet, whereas it is inhibited by fasting and polyunsaturated fatty acids (Kersten, 2001). These effects are mediated during feeding, particularly by insulin which activates the Srebp-1c factor, and glucose which activates the Chrebp factor (Kersten, 2001). Likewise, fasting is not only associated with a significant decrease in glucose and insulin in plasma but also with increasing levels of growth hormone and glucagon, which inhibit lipogenesis (Kersten, 2001). Considering all of the above, the question arises about the real significance of IL-6 in hepatic lipogenesis, taking into account that other hormones like insulin and glucagon are crucial for the induction/inhibition of this process in the liver and, moreover, that the expression of these hormones is under the control of the feeding/fasting cycle. The IL-6 receptor complex potently activates the associated JAK2 which in turn activates the signal transducer and activator of transcription factor 3 (Stat3), the major signal transducer downstream of gp130-like receptors (Heinrich et al., 2003;Kellyet al., 2004, 2009;). The activated Stat3 translocates to the nucleus to induce the expression of specific target genes, which determine the role of IL-6 in inflammation, differentiation or development of hepatocytes (Gao, 2005). Recently, the involvement of hepatic Stat3 in the up-regulation of the genes of lipogenic enzymes has been demonstrated (Kinoshita et al., 2008). In rats, the amounts of IL-6 in serum follow a circadian rhythm, peaking at 12:00 h and reaching minimum values in the afternoon (Li et al., 2004), which parallels the feeding/fasting cycle in rodents. In humans, IL-6 is secreted in a biphasic circadian pattern, with two peaks at about 05:00 and 19:00 h (Vgontzas et al., 2005). Under normal conditions, the circulating levels of IL-6 also increase significantly during exercise, promoting fatty acid oxidation in muscle and returning to baseline levels a few hours later (Kelly et al., 2009). Under pathological conditions, permanently high British Journal of Pharmacology (2016)
3 A L Gavito et al. levels of IL-6 in plasma during an obesogenic diet are associated with insulin resistance and with steatohepatitis (Hotamisligil, 2006; Klover et al., 2003; Matthews et al., 2010; Nieto-Vazquez et al., 2008). Accordingly, these observations all support a relationship between lipid metabolism and IL-6 in both normal and pathological conditions. In this study, we further examined the significance of IL-6 in lipid metabolism, focusing our analyses on the expression of lipogenic enzymes in the liver. To do this, we used the livers of wild-type (WT) mice and mice deficient in IL-6 (IL-6 / ). First, we analysed the influence of IL-6 on the expression of genes of the lipogenic enzymes during fasting and re-feeding conditions in both mouse strains. Second, we determined the hepatic expression of the lipogenic enzymes after in vivo administration of exogenous IL-6, controlling food intake conditions, in WT and IL-6 / mice. Finally, we examined the involvement of IL-6-mediated Stat3 activation in the control of the expression of lipogenic enzymes in hepatocytes (Figure 1). Our results indicate that IL-6 induces the expression of lipogenic genes through STAT3 activation in hepatic cells. Methods Animal welfare and ethics statement Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny et al., 2010; McGrath and Lilley, 2015). Animal ethics conform to the guidance of the Spanish legislation (Royal Decree 53/2013, BOE, 34/ , 2013) and complied with the European Convention for the Protection of Vertebrate Animals used for Experimental and other Scientific Purposes (Council of Europe No. 123, Strasbourg 1985). The protocol was approved by the Ethics Committee for Animal Experiments of the University of Malaga (Permit number: A). Animals were anaesthetized with isoflurane before being killed by decapitation in a room separate from the other experimental animals. All efforts were made to minimize animal suffering. Males from IL-6-deficient (IL-6 / ) mouse strain B6.129S2-IL-6tm1Kopf/J (SN 2650, strain/ html) and the recommended control strain C57BL/6 J (SN 0664) IL-6 +/+, henceforth referred to as WT, were purchased from Charles River Laboratories (Barcelona, Spain, S.A.). Mice were housed in groups of no more than four adults per cage and maintained in standardized conditions in animal facilities (Servicio de Estabulario, Facultad de Medicina, Universidad de Málaga) at 20 ± 2 C room temperature, 40 ± 5% relative humidity and a 12-h light/dark cycle with dawn/dusk effect, water and standard pathogen-free chow diet (Harlan Teklad, Madison, WI, USA) were provided ad libitum. The IL-6 / mice were subjected to regular genotyping by PCR following the protocol of the Jackson Laboratory for the B6.129S2-IL-6 tm1kopf /J strain mice. The mice used for these experiments were 8 12 weeks-old. For experiments in fasting conditions, the animals were maintained on nocturnal feeding prior to diurnal fasting from 08:00 h. For feeding experiments, the animals were maintained on nocturnal fasting (12 h) prior to diurnal re-feeding from 08:00 h. Samples from sera and liver tissues were obtained immediately after the animals had been killed at 08:00, 12:00, 16:00 and 20:00 h (light cycle). The number of animals per group size is indicated for each experiment. Figure 1 Diagram showing key components of the IL-6 signal transduction pathway that could account for lipogenic gene expression in hepatocytes. Signalling is initiated when IL-6 binds to its receptor IL-6R/ gp130 activating (phosphorylating) JAKs which in turn phosphorylates Stat3. Activated Stat3 dissociates from receptor, dimerizes and translocates to the nucleus where it induces transcription of target genes as well as its own Stat3 and Socs3, whose protein is rapidly induced upon IL-6 stimulation, thereby inhibiting IL-6-mediated signalling in a classic feedback loop. IL-6 also stimulates the Acc, Fasn and Scd1 gene expression in hepatocytes. To investigate the involvement of Stat3 activation in the control of the expression of the lipogenic enzymes, we knockeddown STAT3 by use of sirna. This should prevent the expression of Stat3 protein, and thereby interrupt the IL-6-mediated gene expression of lipogenic enzymes. Sample collection After the animals had been killed, liver sections were removed by surgery and immediately frozen in liquid nitrogen and stored at 80 C until the mrna or protein was isolated. Blood samples were collected in vacutainer tubes and incubated at room temperature for 1 h to allow clotting, and serum was extracted after centrifugation, aliquoted and stored at 80 C. In vivo IL-6 treatment Based on previous protocols for ril-6 administration (Vida et al., 2015; Wallenius et al., 2002), the WT and IL-6 / mice were deprived of food at 08:00 h and treated with 3.2 ngg 1 of murine ril-6 (Peprotech, Inc., Rocky Hill, NJ, USA) or vehicle alone (0.1% BSA in PBS) i.p. (n =8pergroup).Animals were killed at the times indicated in each experiment, and samples from sera and liver tissues were collected and stored British Journal of Pharmacology (2016)
4 ril-6 up-regulates hepatic lipogenic enzymes BJP IL-6 and biochemical analysis in serum IL-6 concentrations in sera were assayed in duplicate using the mouse IL-6 ELISA kit (Millipore) following the manufacturer s recommended procedures. The absorbances at 450 nm were measured in an ELISA microplate reader (VERSAmax; Molecular Devices, Sunnyvale, CA, USA), and the concentrations of IL-6 were calculated on the basis of the ODs obtained with the standards. The serum levels of triglycerides, total cholesterol and high-density lipoprotein (HDL) cholesterol were analysed using a Hitachi 737 Automatic Analyzer (Hitachi Ltd, Tokyo, Japan). HepG2 cell culture and in vitro treatments Human hepatoma cell line HepG2 was purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). HepG2 cells were grown in Eagle s minimum essential medium (ATCC) supplemented with 10% FBS (Gibco, Grand Island, NY, USA) at 37 C in 5% CO 2. For cell transfection, HepG2 cells were seeded at cells per well in a 6-well plate, and lipofectamine RNAi Max (Invitrogen, Carlsbad, CA, USA) was used for transfecting the sirnas following the manufacturer s instructions. The human STAT3 sirna (final sirna 25 pmol per well) and the corresponding Silencer Select negative control sirna n 1 (25 pmol per well) were purchased from Ambion (Thermo Fisher Scientific Inc., Austin, TX, USA). Lipid-mediated transfection efficiency was checked by oligo Block.iT Alexa Fluor Fluorescent (Invitrogen) and visualized with fluorescence microscopy. The sirna-transfected cells were incubated for 72 h at 37 C. Medium was replaced with OptiMEM (Gibco, Grand Island, NY, USA) alone or OptiMEM containing human ril-6 (30 ngml 1 ), and cells were incubated for an additional 6 h, and then collected with TRIzol (Invitrogen) or with protein sample buffer containing DTT. All assays were performed in triplicate and repeated at least twice. RNAisolationandcDNAsynthesis Total RNA from HepG2 cell cultures or from liver sections (50 mg) was extracted using TRIzol (Invitrogen) reagent according to the manufacturer s instructions. RNA concentration and purity were determined using a Nanodrop TM spectrophotometer ND-1000 (Thermo Fisher Scientific). RNA (1 μg) was reverse transcribed with the Transcriptor First Strand cdna Synthesis kit (Roche Applied Science, Mannheim, Germany). Real-time quantitative PCR The expression of genes encoding for mouse Accα (Acaca), Accβ (Acacb), FAS (Fasn), Scd1 (Scd1), Socs3 (Socs3), liver X receptor-α (Lxrα) andsrebp(srebf-1) was measured by qpcr. Normalization data were performed by the most stable pairwise combinations of the reference genes: mouse β-glucuronidase (Gusβ) and mouse glyceraldehyde-3- phosphate dehydrogenase (Gapdh). Likewise, the expression of the genes encoding for human ACC (ACC1), FAS (FASN), SCD1 (SCD1)andSOCS3(SOCS3) was measured also through qpcr. Human β-glucuronidase (GUSβ) and human β-actin (ACTINB) were used as reference genes for HepG2 hepatoma samples based on their pairwise stability. Specific oligonucleotides were designed from the Universal Probe Library (Roche Applied Science) to amplify particular regions of the genes of interest for each organism. The specificity of each primer was tested using BLAST analysis on the National Centre for Biotechnology Information database. The gene symbol, GeneID, primer sequences and amplicon length for mouse and human are listed in the Supporting Information (Tables S1 and S2). The master mix used was FastStart Universal SYBR Green Master (Rox) (Roche Applied Science). Alternatively, the expression of human STAT3 was studied by qpcr using Assays-on- Demand Gene Expression Products (TaqMan MGB probes, FAM dye-labelled) and using GAPDH and ACTINB as reference genes, all purchased from Applied Biosystems (Foster City, CA, USA) (Supporting Information Table S2). All PCR reactions were performed using a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA). Each assay included a duplicate of no-template control (water) to verify no amplification of contamination. Thermocycling parameters were the same for each amplicon, when using SYBR Green Master, following the supplier s instructions, with one cycle at 95 for 10 min, followed by 40 cycles of two steps at s and s, ending with a melting curve. Specificity was assessed with melting curves. For the TaqMan Gene Expression Assays (Applied Biosystems), the PCR reactions were subjected to one cycle at 95 C for 10 min, followed by 44 cycles of two steps at 95 C for 15 s and 60 C for 60s. Raw fluorescence data were submitted to the online Miner tool ( for calculation of quantification cycle (Cq) and efficiency values for each experimental set (Zhao and Fernald, 2005). The Cq values were converted into relative expression values taking into account amplification efficiencies and normalization factors by means of Biogazelle s qbaseplus software (Biogazelle, Zwijnaarde, Belgium) using the corresponding reference genes for human or mouse organisms (Hellemans et al., 2007). For all reference and target gene studies, two independent biological samples of each experimental condition were evaluated in technical replicates. Repeatability was accepted when the absolute difference of the Cq replicate values was 0.7 cycles. Finally, calibrated normalized relative quantity values were exported from the qbaseplus software and investigated statistically by means of the GraphPad Prism version 5.04 software program (GraphPad Software Inc., San Diego, CA, USA). Protein extraction and Western blot analysis Liver samples (50 mg) were thawed and disrupted in cold lysis buffer (50 mm Tris HCl ph 7.4, 150 mm NaCl 150, 0.5% NaDOC, 1 mm EDTA, 1% Triton, 0.1% SDS, 1 mm Na 3 VO 4, 1 mm NaF) supplemented with a cocktail of protease (Roche complete tablets). The suspension was shaken for 2 h at 4 C and centrifuged at 15300X g for 30 min at 4 C, and the soluble fraction was recovered. Protein concentration was determined by Bradford protein assay. Protein extracts were diluted 1:1 in 2X sample buffer containing DTT and boiled for 5 min before being subjected to SDS-PAGE. Samples (50 μg of total proteins each) were resolved in 4 15% Ready Gel Precast Gels (Bio-Rad) and blotted onto nitrocellulose membranes (Bio-Rad). Membranes were blocked in TBS-T (50 mm Tris HCl, ph 7.6; 200 mm NaCl and 0.1% Tween- 20) with 2% BSA. Specific proteins were detected by British Journal of Pharmacology (2016)
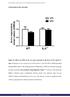
5 A L Gavito et al. incubation in TBS-T 2% BSAwith the corresponding primary antibodies: rabbit anti-accα/β, anti-fas, anti-scd1, anti-stat3 and anti-phospho-stat3, all purchased from Cell Signaling Technology Inc. (Danvers, MA, USA). Actin expression was used as loading control and was detected with rabbit antiactin (Cell Signaling Technology Inc.) antibody. When examining high MW proteins, the presence of adaptin-γ (observed size: 100 kda) was preferably used as loading control and detected with rabbit anti-adaptin-γ (Abcam Cambridge, UK). After extensive washing in TBS-T, anti-rabbit HRP-conjugated secondary antibody (Promega, Madison, MI, USA) was added for 1 h. Membranes were subjected to extensive washings in TBS-T, and the specific protein bands were revealed using the enhanced chemiluminescence detection system (Santa Cruz,BiotechnologyInc.,CA,USA),accordingtothemanufacturer s instructions, and images were visualized with an Autochemi-UVP Bioimaging System. Bands were quantified by densitometric analysis performed by ImageJ software (Rasband, W.S., ImageJ, US National Institutes of Health, Bethesda, MD, USA, ). Amounts of specific proteins were normalized to actin or adaptin levels. Statistical analysis The data and statistical analysis comply with the recommendations on experimental design and analysis in pharmacology (Curtis et al., 2015).Dataareexpressedasthe mean ± SEM of at least five to nine determinations per experimental group. All data were statistically analysed using GraphPad Prism version 5.04 software programme (GraphPad Software Inc., San Diego, CA, USA). The comparison between two groups was performed using Student s t-test. The differences between more than two groups were analysed by one-way ANOVA. In addition, the group differences using two factors or independent variables (e.g. genotype and time or genotype and treatment) were evaluated by two-way ANOVA. Bonferroni s post hoc test for multiple comparisons was performed when the main effects of factors were revealed by ANOVA through the F-statistic and the corresponding significant P-value. The level of statistical significance was set at P < Results Expression of hepatic lipogenic enzyme genes in WT and IL-6 / mice during feeding and fasting states To analyse the influence of IL-6 on the expression of lipogenic genes in the liver, and because lipogenesis is stimulated by feeding and inhibited by fasting (Kersten, 2001), we first evaluated the variations in the hepatic expression of Acaca, Acacb, Fasn and Scd1 genes during fasting and re-feeding conditions in WTand IL-6 / strains in parallel. Livers from the WT (n = 9 per group) and IL-6 / (n = 9 per group) mice exposed ad libitum to nocturnal feeding and deprived of food at 08:00 h displayed a significant gradual decrease in the diurnal expression of Acacb, Fasn and Scd1 in both strains, showing maximum levels between 08:00 and 12:00 h and minimum levels between 16:00 and 20:00 h (Figure 2A). Thus, statistical analysis rendered a significant effect of time on the expression of Acaca, Acacb, Fasn and Scd1 (Figure 2A). Also, the expression of Acaca decreased gradually in WT mice, but no significant changes were detected in its expression throughout the fasting period in IL-6-deficient mice (Figure 2A). The ANOVA indicated an effect of genotype on Acaca and Scd1 expression; interaction between genotype and time was significant only for the kinetics of Acaca expression (Figure 2A). Interestingly, further inspection of the histograms also revealed significant differences between genotypes at 12:00 h in relation to the amounts of expression of Acaca, Acacb, Fasn and Scd1 (Figure 2A). Therefore, although the expression of lipogenic genes declined with duration of fasting in both genotypes, the results indicated that the hepatic expression of these genes was sustained longer in time in WT mice than in mice lacking IL-6. Livers from WT (n = 6 per group) and IL-6 / (n = 6 per group) mice fasted overnight and re-fed at 08:00 h displayed asignificant gradual up-regulation in the expression of Acaca, Acacb and Fasn, showing maximum levels between 16:00 and 20:00h(Figure2B).Indeed,asignificant effect of time was shown for the expression of Acaca Acacb, Fasn and Scd1, and their expression were significantly increased in WT and IL- 6 / samples at 20:00 h as compared with the corresponding groups at 08:00 h (Figure 2B). Also, the analysis indicated no effect of genotype on the expression of these genes and in no case was there a significant interaction between genotype and time (Figure 2B). Therefore, these observations indicate that a deficiency in IL-6 did not have a significant effect on the expression of the genes for the hepatic lipogenic enzymes during the re-feeding period. We next investigated whether the differences in the expression of the lipogenic enzyme genes found in both strains during fasting could be associated with changes in the gene expression of their transcription factors liver X receptor or Srebp-1c. The analysis showed a significant effect of genotype and time on the expression of the gene for liver X receptor α (Lxrα, alsoknownasnr1h3), but no interaction between genotype and time (Supporting Information Figure S1). Also, a decrease in the hepatic Lxrα expression in IL-6 / mice at 20:00 h was determined compared with the samples from the 12:00 h group (Supporting Information Figure S1). The statistical analysis of Srebp-1c expression denoted no significant effect of genotype but a significant effect of time, and the interaction between genotype and time was not significant (Supporting Information Figure S1). Further analysis showed an increase in the hepatic expression of Srebp-1c at 20:00 h in WT mice and at 16:00 h in IL-6 / mice as compared with the corresponding groups at 12:00 h (Supporting Information Figure S1). Hepatic lipogenic enzyme protein expression in WT and IL-6 / mice during fasting We next assessed whether the results on the expression of the genes of lipogenic enzymes in a fasting state were also reflected at the protein level. The amounts of Accα/β, FAS and Scd1 proteins in liver tissue from fasting WT and fasting IL-6 / mice were examined by Western blot (Figure 3A). The two-way ANOVA revealed a significant effect of genotype 1074 British Journal of Pharmacology (2016)
6 ril-6 up-regulates hepatic lipogenic enzymes BJP Figure 2 Real-time PCR analysis of the gene expression of enzymes involved in lipogenesis in the liver from WT and IL-6 / mice under fasting and re-feeding conditions. (A) Expression of Acaca, Acacb, Fasn and Scd1 in liver samples from WT and IL-6 / mice (n = 9) fed overnight and deprived of food from 08:00 to 20:00 h. (B) Expression of Acaca, Acacb, Fasn and Scd1 in the liver samples from WT and IL-6 / mice (n = 6) fasted overnight and refed from 08:00 to 20:00 h. The expression of each gene was normalized with Gapdh and Gusβ as reference genes. The columns represent calibrated normalized relative quantity means ± SEM. * Denotes differences (P < 0.05) in expression versus the corresponding WT 08:00 h group. # denotes differences (P < 0.05) in expression versus the corresponding IL-6 / 08:00 h group. & denotes differences (P < 0.05) in expression between WT and IL-6 / genotypes at 12:00 h. on the expression of Accα/β with no effect on time and a significant interaction between genotype and time (Figure 3B). In WT samples, the Accα/β expression was up-regulated at 16:00 h and 20:00 h as compared with the levels observed at 12:00 h (Figure 3B). The amounts of Accα/β protein, however, remained unchanged over the fasting period in the samples from the IL-6 / mice, which resulted in significant differences in the Accα/β levels between both genotypes at 16:00 and 20:00 h (Figure 3B). The analysis of the expression of FAS revealed an effect of time, no effect of genotype and no interaction between time and genotype (Figure 3C). A modest up-regulation of FAS expression (P < 0.05) was observed at British Journal of Pharmacology (2016)
7 A L Gavito et al. Figure 3 Western blot analysis of lipogenic enzyme expression in the liver from WT and IL-6 / mice under fasting conditions. (A) The expression of Accα/β, FAS (Fas) and Scd1 proteins was determined in liver samples from WT and IL-6 / mice fasted overnight and deprived of food from 08:00 to 20:00 h. Two samples per group are shown in the corresponding blot as representative of each group. All samples were derived at the same time and processed in parallel. The adjustment to digital images did not alter the information contained therein. Densitometry of relative protein levels for Accα/β, (B) Fas (C) and Scd1 (D) were corrected for adaptin. Protein amount is expressed as fold over the WT group at 12:00 h (set at 1) for each protein. Data are the mean ± SEM. * Denotes differences (P < 0.05) in expression versus WT 12:00 h. # Denotes differences (P < 0.05) in expression versus IL-6 / 12:00 h. & Denotes differences (P < 0.05) in expression between WT and IL-6 / genotypes. 16:00 h in WTsamples (Figure 3C). Likewise, Scd1 expression showed a significant effect of genotype, no effect of time and no significant interaction between genotype and time (Figure 3D). No changes in Scd1 expression levels were observed during fasting in the WTsamples. However, in samples from IL-6 / mice a gradual but significant decrease in Scd1 levels was observed in the 20:00 h group as compared with samples from 12:00 h (Figure 3D). This effect resulted in differences (P < 0.05) in the Scd1 amounts between the genotypes at h and at 20:00 h (Figure 3D). Serum IL-6 levels in WT mice during fasting and feeding conditions NextwecomparedthepresenceofnaturallyoccurringIL-6in sera from the animals exposed to fasting and re-feeding conditions. During fasting, the mean level of IL-6 in serum of WT mice at 08:00 h was ± 2.54 pgml 1.Thesevalues gradually decreased during the fasting period. Thus, at 12:00 h, the levels of IL-6 decreased to ± 1.09 pgml 1, and at 17:00 h and 20:00 h the amounts were below the detection limit (<7.8 pgml 1 ) of the assay. Under re-feeding conditions, the levels of IL-6 were below the detection limit from 08:00 to 16:00 h, becoming detectable at 20:00 h (12 pgml 1 ± 5.18). Therefore, the levels of IL-6 paralleled those of the expression of hepatic lipogenic enzyme genes, especially during fasting conditions. Effect of a single dose of ril-6 on the levels of lipogenic enzyme genes expressed in the liver of WT and IL-6 / mice The fact that, duirng fasting, the initial drop in the expression of lipogenic genes in IL-6 / mice was more marked than that found in WT, and also that declining IL-6 levels in serum of WT paralleled the decrease in lipogenic gene expression suggests that IL-6 might play a role in sustaining the hepatic expression levels of these genes during fasting. To analyse this possibility, we studied the effects of exogenous IL-6 administration on the expression of these genes in the liver of both fasted strains. WTand IL-6 / mice were treated with a single dose of saline (n = 8) or recombinant murine IL-6 (3.2 ngg 1, 1076 British Journal of Pharmacology (2016)
8 ril-6 up-regulates hepatic lipogenic enzymes BJP ril-6; n = 8) at 12:00 h, and liver samples were extracted 1 h later. The expression of lipogenic genes after ril-6 administration was examined in both fasting strains As indicated in Figure 4A, the WT mice receiving ril-6 showed a significant increase in the gene expression of Acaca, Acacb, Scd1 and a significant up-regulation of the IL-6-inducible gene Socs3 compared with the WT group treated with vehicle alone. As for the reference genes Gusβ and Gapdh used in this analysis, no changes in the Fasn expression were observed after ril-6 treatment in these mice (Figure 4A). Examination of the expression of all these genes after administration of ril-6 for longer periods of time (up to 3 h) not only did not result in a further increase in their expression, but also no differences were observed compared with the group treated with saline. The IL-6 / mice treated with ril-6 exhibited a marked upregulation (P < 0.05) of the gene expression of Acaca, Acacb, Fasn and Scd1 as well as Socs3 compared with their corresponding group receiving vehicle alone (Figure 4B). Thus, compared with WT mice, it appeared that the IL-6-deficient mice were more receptive to exogenous IL-6. Nonetheless, it is important to note that ril-6 treatment was administered just at the initial point of depletion of the lipogenic gene Figure 4 Effect of ril-6 administration on the gene expression of enzymes involved in lipogenesis in the liver from WT (A) and IL-6 / (B) mice under fasting conditions. WT and IL-6 / mice were fasted overnight and deprived of food from 08:00 h in parallel. The ril-6 (3.2 ngg 1 ) was administered at h via i.p. route and the liver samples collected at h. The gene expression of Acaca, Acacb, Fasn and Scd1, as well as the IL-6-inducible gene Socs3 was determined and normalized with Gapdh and Gusβ as reference genes. The columns represent means ± SEM (n = 8). * Denotes significant (P < 0.05) differences in the gene expression between group treated with saline and the corresponding group treated with ril-6. Figure 5 Stat3 expression and phosphorylation status in the liver of fasted WT and IL-6 / mice treated with ril-6. WT and IL-6 / mice were fasted overnight and deprived of food from 08:00 h in parallel. The ril-6 (3.2 ngg 1 ) was administered at h via i.p. route and the liver samples collected at h. (A) A representative Western blot of the Stat3 and its phosphorylation state in the liver samples from WT and IL-6 / mice treated with saline or ril-6. Actin is shown as protein loading control. Two samples per saline-treated group and three samples per ril-6-treated group are shown in the corresponding blot as representative of each group. All samples were derived at the same time and processed in parallel. The adjustment to digital images did not alter the information contained therein. (B) Values (arbitrary units) for p-stat3/stat3 ratio densitometry. (C) Values (arbitrary units) for Stat3/actin ratio densitometry. The ratios are expressed as the fold over the WT group at 12:00 h (set at 1). Data are the mean ± SEM. * Denotes significant (P < 0.05) difference versus the corresponding group treated with vehicle. British Journal of Pharmacology (2016)
9 A L Gavito et al. expression in fasting IL-6 / mice. Therefore, we wondered whether the effect of ril-6 could be more effective in fasting WT mice when administered at the initial point of the decreasing lipogenic gene expression. The results indicated that treatmentwithril-6at15:00hinfastingwtmice(n =8)and killed at 16:00 h induced a more marked increase (P < 0.05) in Acaca, Acacb, Fasn, Scd1 and Socs3 expression levels (Supporting Information Figure S2) compared with that observed at 13:00 h (Figure 4A). Examination of the status of the IL-6 signalling mediator Stat3 revealed that the Stat3 phoshorylation levels were barely detectable in the liver of both strains treated with vehicle alone, while the ril-6 treatment increased Stat3 phosphorylation in both strains to the same extent (Figure 5A). Figure 6 Inhibitory effect of STAT3 sirna on the expression of human STAT3 gene and protein in HepG2 transfected for 72 h and then treated with human ril-6. (A) Analysis of STAT3 expression in non-transfected cells (control), cells non-transfected and treated with ril-6 (30 ngml 1 ) for 6 h (ril-6), cells transfected with negative control (sirna neg) and treated with ril-6 for 6 h and cells transfected with sirna STAT3 and treated with ril-6 for 6 h. The expression of STAT3 gene was normalized using ACTINB and GAPDH as reference genes. The columns represent means ± SEM (n =3)from one of two representative experiments. * Denotes differences (P < 0.05) compared with the untreated control group. # Denotes difference (P < 0.05) between the groups treated with ril-6 as depicted. (B) Analysis of the human STAT3 and p-stat3 protein expression of the corresponding HepG2 groups defined in (A): untreated cells, cells treated with ril-6 alone, cells transfected with sirna negative control and treated with ril-6 and cells transfected with STAT3 sirna and treated with ril-6. Human ACTINB is shown as protein loading control. One sample per control or per ril-6-treated groups and two samples per sirna-transfected groups are shown in the blot as representative of each group (triplicates per group). All samples were derived at the same time and processed in parallel. The adjustment to digital images did not alter the information contained therein. (C) Densitometry values (arbitrary units) for STAT3/ACTINB and p-stat3/stat3 ratios are shown for each condition defined in (A) and (B) and as indicated in the figure. * Indicates differences (P < 0.05) compared with the untreated control group. # Indicates differences (P < 0.05) between the groups treated with ril-6 in each histogram as depicted British Journal of Pharmacology (2016)
10 ril-6 up-regulates hepatic lipogenic enzymes BJP Indeed, a thorough analysis of the results indicated a significant effect of treatment on the Stat3 phosphorylation status, with no significant differences between genotypes and no interaction between genotype and treatment (Figure 5B). Likewise, the expression of total Stat3 was up-regulated in both strains treated with ril-6, revealing a significant effect of treatment but no effect of genotype and no interaction between genotype and treatment (Figure 5C). From these observations, it can be inferred that the initial response to IL-6 is similar in both strains. The analysis of the serum lipid content of WT treated with a single dose of ril-6 revealed an increase in serum levels of cholesterol and also triglycerides compared with the group treated with saline. However, the same treatment did not appear to have any effect on the serum lipid content in IL-6 / mice, although, these mice initially showed higher levels of cholesterol and triglycerides (P < 0.05) compared with WT mice treated with saline, (Supporting Information Table S3). Effect of inhibiting Stat3 on the ability of IL-6 to increase the expression of lipogenic enzyme genes Because Stat3 signalling has been shown to be critical for IL-6- mediated responses and Stat3 has been demonstrated to be involved in the up-regulation of hepatic Acaca and Fasn enzymes without affecting Srebp-1c expression (Kinoshita et al., 2008), our observations raised the interesting possibility that IL-6 might participate in the transcription of the Figure 7 Inhibitory effect of the human STAT3 sirna on the expression of ACC1, FASN, SCD1 and SOC3 human genes mediated by IL-6 in HepG2. The gene expression of ACC1, FASN, SCD1 and SOCS3 was determined in the non-transfected group (control), group non-transfected and treated with ril-6 (30 ngml 1 ) for 6 h (ril-6), group transfected for 72 h with negative control (sirna neg) and then treated with ril-6 for 6 h and group transfected for 72 h with sirna STAT3 and then treated with ril-6 for 6 h. The expression of each gene was normalized with ACTINB (shown in the Figure) and GUSβ as reference genes. The columns represent means ± SEM (n = 3) from one of two representative experiments. * Denotes significant differences (P < 0.05) in the gene expression versus control group. # Denotes significant differences in the gene expression between groups treated with ril-6 as indicated in the figure. British Journal of Pharmacology (2016)
11 A L Gavito et al. lipogenic genes through activation of Stat3. We then hypothesized that abolishing the function or the expression of Stat3 could suppress the expression of the lipogenic enzyme genes induced by IL-6. In previous studies, we showed that lipogenic enzymes were enhanced after ril-6 treatment in the human hepatoma cell line HepG2 (Vida et al., 2015). Thus, we used this human cell line as a model to test the direct involvement of STAT3 in the transcription of the lipogenic enzymes mediated by the IL-6 response. To this aim, we applied an approach to silence the STAT3 gene by means of STAT3 sirna lipotransfection into HepG2 cells. A negative control sirna was used to test non-specific effects on gene expression. Also, transfection was monitored with the Block.iT Alexa Fluorescent oligo, which showed that 90% of the cells were efficiently transfected at 48 h (Supporting Information Figure S3). Non-transfected and transfected cells with the sirnas were stimulated with ril-6 (30 ngml 1 ) for 6 h. Expression of STAT3 was checked in all cell treatment conditions performed in triplicate. As showninfigure6a,stat3 expression was significantly upregulated in cells non-transfected or transfected with control sirna and subsequently treated with ril-6 (Figure 6A). In contrast, pretreatment with STAT3 sirna attenuated the upregulation of STAT3 mediated by ril-6 treatment, and the levels of STAT3 were even below the control group levels (Figure 6A). Notably, the levels of STAT3 in the group of cells pretreated with negative sirna control and then treated with ril-6 were even higher than those of cells treated with ril-6 alone (Figure 6A). These observations were further corroborated at protein levels (Figure 6B). In line with the gene expression data, Western blot analysis showed increased amounts of the STAT3 protein in cells treated with ril-6 alone and in cells transfected with control sirna and treated with ril-6 (Figure 6B and C). In contrast, transfection of HepG2 with STAT3 sirna markedly inhibited the expression of STAT3 even though cells were also treated with ril-6 (Figure 6B and C). Nevertheless, analysis of the p-stat3 status revealed that ril-6 was able to phoshorylate the STAT3 in all the conditions assayed(figure6bandd).indeed,eventhereducedamounts of total STAT3 in cells transfected with STAT3 sirna and treated with ril-6 showed a similar p-stat3/stat3 ratio to that of the other groups treated with ril-6 (Figure 6B and D). Next, we examined whether reducing the levels of STAT3 could affect the expression of lipogenic genes mediated by IL-6 signalling. As shown in Figure 7, the analysis of the lipogenic gene expression in cells transfected with STAT3 sirna revealed that the up-regulation of ACC1, FASN and SCD1 mediated by IL-6 stimulation was inhibited down to basal levels. In contrast, in ril-6-treated cells and those transfected with control sirna the expression levels of these genes were similarly up-regulated as those in cells treated with ril-6 alone, although in some instances, the control sirna showed a tendency to exert an additive effect on the lipogenic gene expression. This last observation was more evident when the expression of the STAT3-inducible gene SOCS3 (Auernhammer et al., 1999) was evaluated, showing that with a combination of ril-6 plus control sirna this gene was upregulated to a greater extent than with ril-6 alone. We attributed this result to a consequence of the ability of the control sirna to induce higher expression of STAT3 at gene and protein levels (Figure 6A C). Interestingly, the pretreatment of cells with STAT3 sirna attenuated the up-regulation of SOCS3 mediated by ril-6 (Figure 7). Overall, these results indicate that STAT3 is involved in the up-regulation of the lipogenic genes induced by ril-6 in hepatic cells. Discussion and conclusions The contribution of cytokines and chemokines to the regulation of metabolism in the liver and the adipose tissues is an active field of research linked to the growing epidemic of obesity. In the present study, we gained insight into how one of these signals, IL-6, by regulating the activity of the transcription factor STAT3 modifies lipid anabolic pathways in the liver. These results are important for elucidating how inflammatory processes (i.e. inflammation of the visceral adipose tissue) might contribute to the metabolic alterations observed in complicated obesity and serve as a warning about the chronic use of STAT3-associated signalling molecules (i.e. cardiotrophin, IL-6 and oncostatin) in a context where activation of lipogenesis might drive a steatotic response in the liver. Variations in the expression and activity of lipogenic enzymes in relation to diet, food intake and starving conditions have been reported previously (Fukuda and Iritani, 1991; Li et al., 2004; Morgan et al., 2008). Likewise, an IL-6 circadian rhythm in rats and humans has also been described, although its association with the feeding/fasting cycle has not been established (Li et al., 2004; Vgontzas et al., 2005). Also, the relationship between IL-6 and hepatic lipid metabolism, particularly in fatty acid oxidation and lipogenesis, has previously been demonstrated in vivo and in hepatocyte culture (Brass and Vetter, 1994; Brass and Vetter, 1995; Grunfeld et al., 1990; Hong et al., 2004; Vida et al., 2015; Vida et al., 2013). Extending previous observations found in rodents (Morgan et al., 2008), in the present study we showed that the expression of lipogenic genes Acac, Fasn and Scd1 in the liver of WT and IL-6 / mice displayed a gradual reduction during fasting conditions. In contrast, with the exception of Scd1, the expression of these genes gradually increased during re-feeding with a regular chow diet. However, the expression of Scd1 exhibited a different behaviour from that of Acac and Fasn during re-feeding, an observation in agreement with other classical reports describing the specific effects of re-feeding diets on Scd1 expression (Ntambi, 1992). In this regard, when mice were fasted and then fed a fat-free diet, the hepatic Scd1 expression was increased; however, when switched from a fat-free diet to a chow diet, Scd1 decreased to normal levels (Ntambi, 1992). This was attributed to the fat content of the chow diet being responsible for the low levels of Scd1 expressed under normal dietary conditions (Ntambi, 1992). Our findings showed that during fasting, the initial drop in the expression of lipogenic genes was more marked in the IL-6 / thaninwtmice,eventhoughtheacaca expression in the IL-6 / mice remained at low constant levels over the fasting period. This may seem a subtle difference, although enough to suggest that IL-6 might participate in sustaining the expression of these genes. Interestingly, our study showed that the circulating levels of IL-6 in WT mice peaked at 1080 British Journal of Pharmacology (2016)
12 ril-6 up-regulates hepatic lipogenic enzymes BJP 08:00 h and decreased during the fasting period, and this phenomenon paralleled the drop in the expression of lipogenic genes. During the feeding period, the basal levels of IL-6 in serum of WTwere measurable only at 20:00 h, when the expression of the lipogenic genes was elevated. Considering that rodents are nocturnal and feed preferably at night, these observations are consistent with the serum IL-6 circadian rhythm found in rats, which reaches a peak at 12:00 h, decreasing progressively during the light period, thus coinciding with the fasting period (Li et al., 2004). That being so, it might appear surprising that a deficiency in IL-6 did not affect the pattern of lipogenic gene expression during the refeeding period, a condition in which lipogenesis is positively modulated. In fact, it is generally considered that insulin plays a crucial role during a fasting re-feeding programme, increasing the activity of lipogenic enzymes and also the expression of their genes (Strable and Ntambi, 2010). Therefore, the putative role of IL-6 in lipogenic gene expression during re-feeding, if any, is probably masked by the effects of insulin. In our experimental conditions, even though fasting gradually depleted the expression of the genes of lipogenic enzymes, it particularly increasedtheproteinexpressionof hepatic Accα/β in WT mice, even long after the amounts of IL-6 in serum and the expression of Acaca and Acacb were reduced. As mrna is eventually translated into protein, one might assume that there should be some correlation between the level of mrna and that of its protein. Nevertheless, according to other authors, there may not necessarily be a correlation (Reddy et al., 2006). In this regard, a transcription translation feedback loop is thought to be essential in generating circadian rhythms, particularly in the liver (Reddy et al., 2006). Thus, the Acaca/Acacb mrna could be high at one time point and then decline, while its protein follows with the appropriate lag for all the processes necessary from mrna to full protein synthesis, folding and processing. According to gene expression kinetics, although the protein levels of lipogenic enzymes were increased during fasting, this occurred at lower levels in IL-6 / than in WT mice, particularly at the end of the fasting period. The observation that the liver of IL-6 / mice showed unchanged protein amounts of Accα/β and even decreased amounts of Scd1 during fasting, thereby remaining below those found in WT mice after 8 and 12 h of food deprivation, supports the idea that IL-6 plays a role in sustaining the expression of these enzymes at the gene and protein levels in fasting conditions. In support of this notion, here, we showed that a single administration of ril-6 up-regulated the expression of Acaca/b, Fasn and Scd1 genes in WT and IL-6 / mice,eventhoughthiseffect was more apparent in IL-6 / than in WT mice. We cannot give an explanation for this difference, but it suggests that the presence of basal circulating levels of IL-6 probably primes a mechanism of signalling shutdown when levels of IL-6 rise dangerously, which is likely to be absent in IL-6- deficient mice. In this regard though, we did not observe significant differences between the two strains concerning the ability of IL-6 to stimulate the expression of natural inhibitors of Stat3, such as Socs3 (Figure 4) and, moreover, an upregulation of Stat3 or the p-stat3 status in vivo (Figure 5) was activated similarly in both WT and IL-6 / strains. Also, in line with other findings in rodents (Nonogaki et al., 1995), the administration of ril-6 increased the cholesterol and triglyceride levels in the serum of WT mice. IL-6 replacement was expected to reproduce this effect in IL-6 / mice. However, in IL-6 / mice, the basal levels of triglycerides and cholesterol were already elevated, and the single administration of IL-6 did not affect these levels. We think that this probably reflects an adaptive compensatory response in mice deficient in IL-6. Also, our recent findings have revealed that these mice display a propensity to further accumulate fat in liver when treated with ril-6 under high-fat diet nutritional conditions (Vida et al., 2015). The present results indicate that the regulatory action of IL-6 on lipogenic enzymes is dependent on the integrity of STAT3 signalling. Although the transcription of the Acaca, Acacb, Fasn and Scd1 genes is regulated mainly by the transcription factors liver X receptor-α and Srebp-1c (Strable and Ntambi, 2010), our results clearly indicate an additional role for STAT3. Both the liver X receptor-α and Srebp-1c are positively regulated by hormones like insulin after a fasting refeeding programme (Strable and Ntambi, 2010). In fact, it has been demonstrated that the liver X receptor is important in mediating the effect of insulin on Srebp-1c expression (Chen et al., 2004). To our knowledge, no evidence has been published describing how IL-6-mediated signalling might exert a direct action on the expression or activity of liver X receptor-α and Srebp-1c, although this possibility cannot be ruled out. In this context, we did not observe important differences in the gene expression profiles for Lxrα or Srebp-1c between the WT and IL-6 / genotypes during fasting conditions. Initially, it might seem surprising that the expression of Srebp-1c was increased in both strains during fasting, a condition in which insulin levels are supposed to be lower. Of note, this increase emerged earlier in the kinetics of IL-6 / mice (18:00 h) than in that of WT mice (20:00 h) and was inversely proportional to the falling expression of the target lipogenic genes, which occurred more rapidly in IL-6- deficient mice (12:00 h) than in WT mice (16:00 h). Therefore, the up-regulation of Srebp-1c might be a compensatory response to the gradual decline in the expression of the target genes. Interestingly, findings obtained in a mouse model of hepatic insulin receptor knockout (LIRKO) suggest that Srebp-1 can be stimulated by insulin-independent signalling pathways (Haas et al., 2012). Whatever the reason, it can be inferred from these observations that the expression profile of Lxrα and Srebp-1c does not explain the decreased expression of Acaca/b, Fasn and Scd1 in the fasting period. Interestingly, Stat3 has been demonstrated to have a role in the regulation of hepatic lipogenic enzymes (Kinoshita et al., 2008). The overexpression of Stat3 in the liver of insulin-resistant diabetic mice has been shown to increase the gene expression of Acaca and Fasn without affecting Srebp-1c expression (Kinoshita et al., 2008). Because Stat3 is the main transcription factor mediating IL-6 signalling, this finding raises the possibility that IL-6 participates in the induction of the gene expression of these lipogenic enzymes through its activation. In agreement with this is the observation, obtained in the present study, that declining circulating levels of IL-6 during fasting were accompanied by a gradual drop in the expression of lipogenic genes in WT mice (Figure 2A). Moreover, administration of a single dose of IL-6 restored the expression of the genes of these enzymes, particularly in IL-6 / mice (Figure 4B). Finally, the IL-6-mediated up-regulation of British Journal of Pharmacology (2016)
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Effect of a HFD on the Acox gene expression in the livers of WT and IL-6 -/- mice. Expression of Acox in the livers of WT and IL-6 -/- mice fed STD or HFD determined through
SUPPLEMENTARY FIGURES Figure S1. Effect of a HFD on the Acox gene expression in the livers of WT and IL-6 -/- mice. Expression of Acox in the livers of WT and IL-6 -/- mice fed STD or HFD determined through
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
General Laboratory methods Plasma analysis: Gene Expression Analysis: Immunoblot analysis: Immunohistochemistry:
 General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at
 Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Supporting Information Table of content
 Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination
 AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein
 Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein Lei Wang 1, Tian-Peng Zhang 1, Yuan Zhang 2, Hai-Lian
Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein Lei Wang 1, Tian-Peng Zhang 1, Yuan Zhang 2, Hai-Lian
For pair feeding, mice were fed 2.7g of HFD containing tofogliflozin
 Materials and Methods Pair Feeding Experiment For pair feeding, mice were fed 2.7g of HFD containing tofogliflozin (0.005%), which is average daily food intake of mice fed control HFD ad libitum at week
Materials and Methods Pair Feeding Experiment For pair feeding, mice were fed 2.7g of HFD containing tofogliflozin (0.005%), which is average daily food intake of mice fed control HFD ad libitum at week
TFEB-mediated increase in peripheral lysosomes regulates. Store Operated Calcium Entry
 TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
SUPPLEMENTARY DATA. Supplementary Table 1. Primers used in qpcr
 Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
Online Data Supplement. Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2
 Online Data Supplement Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2 Yi Lin and Zhongjie Sun Department of physiology, college of
Online Data Supplement Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2 Yi Lin and Zhongjie Sun Department of physiology, college of
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies
 Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Islet viability assay and Glucose Stimulated Insulin Secretion assay RT-PCR and Western Blot
 Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Supporting Information. Supporting Tables. S-Table 1 Primer pairs for RT-PCR. Product size. Gene Primer pairs
 Supporting Information Supporting Tables S-Table 1 Primer pairs for RT-PCR. Gene Primer pairs Product size (bp) FAS F: 5 TCTTGGAAGCGATGGGTA 3 429 R: 5 GGGATGTATCATTCTTGGAC 3 SREBP-1c F: 5 CGCTACCGTTCCTCTATCA
Supporting Information Supporting Tables S-Table 1 Primer pairs for RT-PCR. Gene Primer pairs Product size (bp) FAS F: 5 TCTTGGAAGCGATGGGTA 3 429 R: 5 GGGATGTATCATTCTTGGAC 3 SREBP-1c F: 5 CGCTACCGTTCCTCTATCA
Rat Leptin-HS ELISA FOR LABORATORY USE ONLY YANAIHARA INSTITUTE INC AWAKURA, FUJINOMIYA - SHI SHIZUOKA, JAPAN
 YK051 Rat Leptin-HS ELISA FOR LABORATORY USE ONLY YANAIHARA INSTITUTE INC. 2480-1 AWAKURA, FUJINOMIYA - SHI SHIZUOKA, JAPAN 418 0011 Contents Introduction 2 Characteristics 3 Composition 4 Method 5-6 Notes
YK051 Rat Leptin-HS ELISA FOR LABORATORY USE ONLY YANAIHARA INSTITUTE INC. 2480-1 AWAKURA, FUJINOMIYA - SHI SHIZUOKA, JAPAN 418 0011 Contents Introduction 2 Characteristics 3 Composition 4 Method 5-6 Notes
MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands)
 Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Requires Signaling though Akt2 Independent of the. Transcription Factors FoxA2, FoxO1, and SREBP1c
 Cell Metabolism, Volume 14 Supplemental Information Postprandial Hepatic Lipid Metabolism Requires Signaling though Akt2 Independent of the Transcription Factors FoxA2, FoxO1, and SREBP1c Min Wan, Karla
Cell Metabolism, Volume 14 Supplemental Information Postprandial Hepatic Lipid Metabolism Requires Signaling though Akt2 Independent of the Transcription Factors FoxA2, FoxO1, and SREBP1c Min Wan, Karla
2.5. AMPK activity
 Supplement Fig. A 3 B phos-ampk 2.5 * Control AICAR AMPK AMPK activity (Absorbance at 45 nm) 2.5.5 Control AICAR Supplement Fig. Effects of AICAR on AMPK activation in macrophages. J774. macrophages were
Supplement Fig. A 3 B phos-ampk 2.5 * Control AICAR AMPK AMPK activity (Absorbance at 45 nm) 2.5.5 Control AICAR Supplement Fig. Effects of AICAR on AMPK activation in macrophages. J774. macrophages were
BIOL212 Biochemistry of Disease. Metabolic Disorders - Obesity
 BIOL212 Biochemistry of Disease Metabolic Disorders - Obesity Obesity Approx. 23% of adults are obese in the U.K. The number of obese children has tripled in 20 years. 10% of six year olds are obese, rising
BIOL212 Biochemistry of Disease Metabolic Disorders - Obesity Obesity Approx. 23% of adults are obese in the U.K. The number of obese children has tripled in 20 years. 10% of six year olds are obese, rising
ab SREBP-2 Translocation Assay Kit (Cell-Based)
 ab133114 SREBP-2 Translocation Assay Kit (Cell-Based) Instructions for Use For analysis of translocation of SREBP-2 into nuclei. This product is for research use only and is not intended for diagnostic
ab133114 SREBP-2 Translocation Assay Kit (Cell-Based) Instructions for Use For analysis of translocation of SREBP-2 into nuclei. This product is for research use only and is not intended for diagnostic
1. Materials and Methods 1.1 Animals experiments process The experiments were approved by the Institution Animal Ethics Committee of Jilin University
 1. Materials and Methods 1.1 Animals experiments process The experiments were approved by the Institution Animal Ethics Committee of Jilin University (Reference NO. 2015-003). 96 Kunming (KM) mice (8 weeks;
1. Materials and Methods 1.1 Animals experiments process The experiments were approved by the Institution Animal Ethics Committee of Jilin University (Reference NO. 2015-003). 96 Kunming (KM) mice (8 weeks;
Supplementary Table 1. Metabolic parameters in GFP and OGT-treated mice
 Supplementary Table 1. Metabolic parameters in GFP and OGT-treated mice Fasted Refed GFP OGT GFP OGT Liver G6P (mmol/g) 0.03±0.01 0.04±0.02 0.60±0.04 0.42±0.10 A TGs (mg/g of liver) 20.08±5.17 16.29±0.8
Supplementary Table 1. Metabolic parameters in GFP and OGT-treated mice Fasted Refed GFP OGT GFP OGT Liver G6P (mmol/g) 0.03±0.01 0.04±0.02 0.60±0.04 0.42±0.10 A TGs (mg/g of liver) 20.08±5.17 16.29±0.8
Mitochondrial Trifunctional Protein (TFP) Protein Quantity Microplate Assay Kit
 PROTOCOL Mitochondrial Trifunctional Protein (TFP) Protein Quantity Microplate Assay Kit DESCRIPTION Mitochondrial Trifunctional Protein (TFP) Protein Quantity Microplate Assay Kit Sufficient materials
PROTOCOL Mitochondrial Trifunctional Protein (TFP) Protein Quantity Microplate Assay Kit DESCRIPTION Mitochondrial Trifunctional Protein (TFP) Protein Quantity Microplate Assay Kit Sufficient materials
Supplementary Figure S1
 Lipidomic-based investigation into the regulatory effect of Schisandrin B on palmitic acid level in non-alcoholic steatotic livers Hiu Yee Kwan 1,2, Xuyan Niu 3, Wenlin Dai 4, Tiejun Tong 4, Xiaojuan Chao
Lipidomic-based investigation into the regulatory effect of Schisandrin B on palmitic acid level in non-alcoholic steatotic livers Hiu Yee Kwan 1,2, Xuyan Niu 3, Wenlin Dai 4, Tiejun Tong 4, Xiaojuan Chao
Supplementary data Supplementary Figure 1 Supplementary Figure 2
 Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary Figure S1. Effect of Glucose on Energy Balance in WT and KHK A/C KO
 Supplementary Figure S1. Effect of Glucose on Energy Balance in WT and KHK A/C KO Mice. WT mice and KHK-A/C KO mice were provided drinking water containing 10% glucose or tap water with normal chow ad
Supplementary Figure S1. Effect of Glucose on Energy Balance in WT and KHK A/C KO Mice. WT mice and KHK-A/C KO mice were provided drinking water containing 10% glucose or tap water with normal chow ad
Gene Polymorphisms and Carbohydrate Diets. James M. Ntambi Ph.D
 Gene Polymorphisms and Carbohydrate Diets James M. Ntambi Ph.D Fatty Acids that Flux into Tissue Lipids are from Dietary Sources or are Made De novo from Glucose or Fructose Glucose Fructose Acetyl-CoA
Gene Polymorphisms and Carbohydrate Diets James M. Ntambi Ph.D Fatty Acids that Flux into Tissue Lipids are from Dietary Sources or are Made De novo from Glucose or Fructose Glucose Fructose Acetyl-CoA
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original
 Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Lecithin Cholesterol Acyltransferase (LCAT) ELISA Kit
 Product Manual Lecithin Cholesterol Acyltransferase (LCAT) ELISA Kit Catalog Number STA-616 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Cholesterol is a lipid sterol
Product Manual Lecithin Cholesterol Acyltransferase (LCAT) ELISA Kit Catalog Number STA-616 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Cholesterol is a lipid sterol
ANSC/NUTR 601 GENERAL ANIMAL NUTRITION Stearoyl-CoA desaturase, VLDL metabolism, and obesity
 ANSC/NUTR 601 GENERAL ANIMAL NUTRITION Stearoyl-CoA desaturase, VLDL metabolism, and obesity I. Stearoyl coenzyme A desaturase and obesity in rodents A. Stearoyl coenzyme A desaturase (SCD) is the 9 desaturase.
ANSC/NUTR 601 GENERAL ANIMAL NUTRITION Stearoyl-CoA desaturase, VLDL metabolism, and obesity I. Stearoyl coenzyme A desaturase and obesity in rodents A. Stearoyl coenzyme A desaturase (SCD) is the 9 desaturase.
ab Adipogenesis Assay Kit (Cell-Based)
 ab133102 Adipogenesis Assay Kit (Cell-Based) Instructions for Use For the study of induction and inhibition of adipogenesis in adherent cells. This product is for research use only and is not intended
ab133102 Adipogenesis Assay Kit (Cell-Based) Instructions for Use For the study of induction and inhibition of adipogenesis in adherent cells. This product is for research use only and is not intended
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Instructions for Use. RealStar Influenza S&T RT-PCR Kit /2017 EN
 Instructions for Use RealStar Influenza S&T RT-PCR Kit 3.0 01/2017 EN RealStar Influenza S&T RT-PCR Kit 3.0 For research use only! (RUO) 163003 INS-163000-EN-S02 96 01 2017 altona Diagnostics GmbH Mörkenstr.
Instructions for Use RealStar Influenza S&T RT-PCR Kit 3.0 01/2017 EN RealStar Influenza S&T RT-PCR Kit 3.0 For research use only! (RUO) 163003 INS-163000-EN-S02 96 01 2017 altona Diagnostics GmbH Mörkenstr.
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Table 2. Plasma lipid profiles in wild type and mutant female mice submitted to a HFD for 12 weeks wt ERα -/- AF-1 0 AF-2 0
 Supplementary Table 1. List of specific primers used for gene expression analysis. Genes Primer forward Primer reverse Hprt GCAGTACAGCCCCAAAATGG AACAAAGTCTGGCCTGTATCCA Srebp-1c GGAAGCTGTCGGGGTAGCGTC CATGTCTTCAAATGTGCAATCCAT
Supplementary Table 1. List of specific primers used for gene expression analysis. Genes Primer forward Primer reverse Hprt GCAGTACAGCCCCAAAATGG AACAAAGTCTGGCCTGTATCCA Srebp-1c GGAAGCTGTCGGGGTAGCGTC CATGTCTTCAAATGTGCAATCCAT
Instructions for Use. RealStar Influenza Screen & Type RT-PCR Kit /2017 EN
 Instructions for Use RealStar Influenza Screen & Type RT-PCR Kit 4.0 05/2017 EN RealStar Influenza Screen & Type RT-PCR Kit 4.0 For research use only! (RUO) 164003 INS-164000-EN-S01 96 05 2017 altona
Instructions for Use RealStar Influenza Screen & Type RT-PCR Kit 4.0 05/2017 EN RealStar Influenza Screen & Type RT-PCR Kit 4.0 For research use only! (RUO) 164003 INS-164000-EN-S01 96 05 2017 altona
Rat Leptin ELISA FOR LABORATORY USE ONLY YANAIHARA INSTITUTE INC AWAKURA, FUJINOMIYA - SHI SHIZUOKA, JAPAN
 YK050 Rat Leptin ELISA FOR LABORATORY USE ONLY YANAIHARA INSTITUTE INC. 2480-1 AWAKURA, FUJINOMIYA - SHI SHIZUOKA, JAPAN 418-0011 Contents Introduction 2 Characteristics 3 Composition 4 Method 5-6 Notes
YK050 Rat Leptin ELISA FOR LABORATORY USE ONLY YANAIHARA INSTITUTE INC. 2480-1 AWAKURA, FUJINOMIYA - SHI SHIZUOKA, JAPAN 418-0011 Contents Introduction 2 Characteristics 3 Composition 4 Method 5-6 Notes
GPR120 *** * * Liver BAT iwat ewat mwat Ileum Colon. UCP1 mrna ***
 a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
20X Buffer (Tube1) 96-well microplate (12 strips) 1
 PROTOCOL MitoProfile Rapid Microplate Assay Kit for PDH Activity and Quantity (Combines Kit MSP18 & MSP19) 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 MSP20 Rev.1 DESCRIPTION MitoProfile Rapid Microplate
PROTOCOL MitoProfile Rapid Microplate Assay Kit for PDH Activity and Quantity (Combines Kit MSP18 & MSP19) 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 MSP20 Rev.1 DESCRIPTION MitoProfile Rapid Microplate
Phosphate buffered saline (PBS) for washing the cells TE buffer (nuclease-free) ph 7.5 for use with the PrimePCR Reverse Transcription Control Assay
 Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Human Leptin ELISA Kit
 Product Manual Human Leptin ELISA Kit Catalog Numbers MET-5057 MET-5057-5 96 assays 5 x 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Leptin is a polypeptide hormone
Product Manual Human Leptin ELISA Kit Catalog Numbers MET-5057 MET-5057-5 96 assays 5 x 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Leptin is a polypeptide hormone
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism SUPPLEMENTARY FIGURES, LEGENDS AND METHODS
 A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation
 SUPPLEMENTARY INFORMATION Materials and Methods Human cell lines and culture conditions HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation in exon 20 of BRCA1
SUPPLEMENTARY INFORMATION Materials and Methods Human cell lines and culture conditions HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation in exon 20 of BRCA1
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Human Immunodeficiency Virus type 1 (HIV-1) gp120 / Glycoprotein 120 ELISA Pair Set
 Human Immunodeficiency Virus type 1 (HIV-1) gp120 / Glycoprotein 120 ELISA Pair Set Catalog Number : SEK11233 To achieve the best assay results, this manual must be read carefully before using this product
Human Immunodeficiency Virus type 1 (HIV-1) gp120 / Glycoprotein 120 ELISA Pair Set Catalog Number : SEK11233 To achieve the best assay results, this manual must be read carefully before using this product
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.
 Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Human LDL Receptor / LDLR ELISA Pair Set
 Human LDL Receptor / LDLR ELISA Pair Set Catalog Number : SEK10231 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized in
Human LDL Receptor / LDLR ELISA Pair Set Catalog Number : SEK10231 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized in
Taylor Yohe. Project Advisor: Dr. Martha A. Belury. Department of Human Nutrition at the Ohio State University
 Atherosclerosis Development and the Inflammatory Response of Hepatocytes to Sesame Oil Supplementation Taylor Yohe Project Advisor: Dr. Martha A. Belury Department of Human Nutrition at the Ohio State
Atherosclerosis Development and the Inflammatory Response of Hepatocytes to Sesame Oil Supplementation Taylor Yohe Project Advisor: Dr. Martha A. Belury Department of Human Nutrition at the Ohio State
ab Hepatic Lipid Accumulation/ Steatosis Assay Kit
 ab133131 Hepatic Lipid Accumulation/ Steatosis Assay Kit Instructions for Use For evaluating steatosis risk of drug candidates using Oil Red O to stain neutral lipids in hepatocytes. This product is for
ab133131 Hepatic Lipid Accumulation/ Steatosis Assay Kit Instructions for Use For evaluating steatosis risk of drug candidates using Oil Red O to stain neutral lipids in hepatocytes. This product is for
Supplementary Figure S I: Effects of D4F on body weight and serum lipids in apoe -/- mice.
 Supplementary Figures: Supplementary Figure S I: Effects of D4F on body weight and serum lipids in apoe -/- mice. Male apoe -/- mice were fed a high-fat diet for 8 weeks, and given PBS (model group) or
Supplementary Figures: Supplementary Figure S I: Effects of D4F on body weight and serum lipids in apoe -/- mice. Male apoe -/- mice were fed a high-fat diet for 8 weeks, and given PBS (model group) or
GENE Forward primer Reverse primer FABP4 CCTTTGTGGGGACCTGGAAA TGACCGGATGACGACCAAGT CD68 AATGTGTCCTTCCCACAAGC GGCAGCAAGAGAGATTGGTC
 Published in "" which should be cited to refer to this work. mrna extraction and RT-PCR Total RNA from 5 15 mg of crushed white adipose tissue was isolated using the technique described by Chomczynski
Published in "" which should be cited to refer to this work. mrna extraction and RT-PCR Total RNA from 5 15 mg of crushed white adipose tissue was isolated using the technique described by Chomczynski
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
CHAPTER 4 RESULTS. showed that all three replicates had similar growth trends (Figure 4.1) (p<0.05; p=0.0000)
 CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
Free Fatty Acid Assay Kit (Fluorometric)
 Product Manual Free Fatty Acid Assay Kit (Fluorometric) Catalog Number STA-619 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Triglycerides (TAG) are a type of lipid
Product Manual Free Fatty Acid Assay Kit (Fluorometric) Catalog Number STA-619 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Triglycerides (TAG) are a type of lipid
Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression
 Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplemental Figure 1 ELISA scheme to measure plasma total, mature and furin-cleaved
 1 Supplemental Figure Legends Supplemental Figure 1 ELISA scheme to measure plasma total, mature and furin-cleaved PCSK9 concentrations. 4 Plasma mature and furin-cleaved PCSK9s were measured by a sandwich
1 Supplemental Figure Legends Supplemental Figure 1 ELISA scheme to measure plasma total, mature and furin-cleaved PCSK9 concentrations. 4 Plasma mature and furin-cleaved PCSK9s were measured by a sandwich
For in vitro Veterinary Diagnostics only. Kylt Rotavirus A. Real-Time RT-PCR Detection.
 For in vitro Veterinary Diagnostics only. Kylt Rotavirus A Real-Time RT-PCR Detection www.kylt.eu DIRECTION FOR USE Kylt Rotavirus A Real-Time RT-PCR Detection A. General Kylt Rotavirus A products are
For in vitro Veterinary Diagnostics only. Kylt Rotavirus A Real-Time RT-PCR Detection www.kylt.eu DIRECTION FOR USE Kylt Rotavirus A Real-Time RT-PCR Detection A. General Kylt Rotavirus A products are
AMPK Phosphorylation Assay Kit
 AMPK Phosphorylation Assay Kit Catalog Number KA3789 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
AMPK Phosphorylation Assay Kit Catalog Number KA3789 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
Supporting Information
 Supporting Information Pang et al. 10.1073/pnas.1322009111 SI Materials and Methods ELISAs. These assays were performed as previously described (1). ELISA plates (MaxiSorp Nunc; Thermo Fisher Scientific)
Supporting Information Pang et al. 10.1073/pnas.1322009111 SI Materials and Methods ELISAs. These assays were performed as previously described (1). ELISA plates (MaxiSorp Nunc; Thermo Fisher Scientific)
ab65336 Triglyceride Quantification Assay Kit (Colorimetric/ Fluorometric)
 Version 10 Last updated 19 December 2017 ab65336 Triglyceride Quantification Assay Kit (Colorimetric/ Fluorometric) For the measurement of triglycerides in various samples. This product is for research
Version 10 Last updated 19 December 2017 ab65336 Triglyceride Quantification Assay Kit (Colorimetric/ Fluorometric) For the measurement of triglycerides in various samples. This product is for research
Human Apolipoprotein A1 EIA Kit
 A helping hand for your research Product Manual Human Apolipoprotein A1 EIA Kit Catalog Number: 83901 96 assays 1 Table of Content Product Description 3 Assay Principle 3 Kit Components 3 Storage 4 Reagent
A helping hand for your research Product Manual Human Apolipoprotein A1 EIA Kit Catalog Number: 83901 96 assays 1 Table of Content Product Description 3 Assay Principle 3 Kit Components 3 Storage 4 Reagent
Mouse Cathepsin B ELISA Kit
 GenWay Biotech, Inc. 6777 Nancy Ridge Drive San Diego, CA 92121 Phone: 858.458.0866 Fax: 858.458.0833 Email: techline@genwaybio.com http://www.genwaybio.com Mouse Cathepsin B ELISA Kit Catalog No. GWB-ZZD154
GenWay Biotech, Inc. 6777 Nancy Ridge Drive San Diego, CA 92121 Phone: 858.458.0866 Fax: 858.458.0833 Email: techline@genwaybio.com http://www.genwaybio.com Mouse Cathepsin B ELISA Kit Catalog No. GWB-ZZD154
SUPPLEMENTAL INFORMATION
 SUPPLEMENTAL INFORMATION EXPERIMENTAL PROCEDURES Tryptic digestion protection experiments - PCSK9 with Ab-3D5 (1:1 molar ratio) in 50 mm Tris, ph 8.0, 150 mm NaCl was incubated overnight at 4 o C. The
SUPPLEMENTAL INFORMATION EXPERIMENTAL PROCEDURES Tryptic digestion protection experiments - PCSK9 with Ab-3D5 (1:1 molar ratio) in 50 mm Tris, ph 8.0, 150 mm NaCl was incubated overnight at 4 o C. The
ZL ZDF ZDF + E2 *** Visceral (g) ZDF
 Body Weight (g) 4 3 2 1 ** * ZL ZDF 6 8 1 12 14 16 Age (weeks) B * Sub-cutaneous (g) 16 12 8 4 ZL ZDF Visceral (g) 25 2 15 1 5 ZL ZDF Total fat pad weight (g) 4 3 2 1 ZDF ZL Supplemental Figure 1: Effect
Body Weight (g) 4 3 2 1 ** * ZL ZDF 6 8 1 12 14 16 Age (weeks) B * Sub-cutaneous (g) 16 12 8 4 ZL ZDF Visceral (g) 25 2 15 1 5 ZL ZDF Total fat pad weight (g) 4 3 2 1 ZDF ZL Supplemental Figure 1: Effect
YK052 Mouse Leptin ELISA
 YK052 Mouse Leptin ELISA FOR LABORATORY USE ONLY YANAIHARA INSTITUTE INC. 2480-1 AWAKURA, FUJINOMIYA-SHI SHIZUOKA, JAPAN 418-0011 Contents Ⅰ. Introduction 2 Ⅱ. Characteristics 3 Ⅲ. Composition 4 Ⅳ. Method
YK052 Mouse Leptin ELISA FOR LABORATORY USE ONLY YANAIHARA INSTITUTE INC. 2480-1 AWAKURA, FUJINOMIYA-SHI SHIZUOKA, JAPAN 418-0011 Contents Ⅰ. Introduction 2 Ⅱ. Characteristics 3 Ⅲ. Composition 4 Ⅳ. Method
Biosynthesis of Fatty Acids. By Dr.QUTAIBA A. QASIM
 Biosynthesis of Fatty Acids By Dr.QUTAIBA A. QASIM Fatty Acids Definition Fatty acids are comprised of hydrocarbon chains terminating with carboxylic acid groups. Fatty acids and their associated derivatives
Biosynthesis of Fatty Acids By Dr.QUTAIBA A. QASIM Fatty Acids Definition Fatty acids are comprised of hydrocarbon chains terminating with carboxylic acid groups. Fatty acids and their associated derivatives
Supplementary Data Table of Contents:
 Supplementary Data Table of Contents: - Supplementary Methods - Supplementary Figures S1(A-B) - Supplementary Figures S2 (A-B) - Supplementary Figures S3 - Supplementary Figures S4(A-B) - Supplementary
Supplementary Data Table of Contents: - Supplementary Methods - Supplementary Figures S1(A-B) - Supplementary Figures S2 (A-B) - Supplementary Figures S3 - Supplementary Figures S4(A-B) - Supplementary
Human Obestatin ELISA
 K-ASSAY Human Obestatin ELISA For the quantitative determination of obestatin in human serum and plasma Cat. No. KT-495 For Research Use Only. 1 Rev. 081309 K-ASSAY PRODUCT INFORMATION Human Obestatin
K-ASSAY Human Obestatin ELISA For the quantitative determination of obestatin in human serum and plasma Cat. No. KT-495 For Research Use Only. 1 Rev. 081309 K-ASSAY PRODUCT INFORMATION Human Obestatin
18s AAACGGCTACCACATCCAAG CCTCCAATGGATCCTCGTTA. 36b4 GTTCTTGCCCATCAGCACC AGATGCAGCAGATCCGCAT. Acc1 AGCAGATCCGCAGCTTG ACCTCTGCTCGCTGAGTGC
 Supplementary Table 1. Quantitative PCR primer sequences Gene symbol Sequences (5 to 3 ) Forward Reverse 18s AAACGGCTACCACATCCAAG CCTCCAATGGATCCTCGTTA 36b4 GTTCTTGCCCATCAGCACC AGATGCAGCAGATCCGCAT Acc1
Supplementary Table 1. Quantitative PCR primer sequences Gene symbol Sequences (5 to 3 ) Forward Reverse 18s AAACGGCTACCACATCCAAG CCTCCAATGGATCCTCGTTA 36b4 GTTCTTGCCCATCAGCACC AGATGCAGCAGATCCGCAT Acc1
Supplemental Figures:
 Supplemental Figures: Figure 1: Intracellular distribution of VWF by electron microscopy in human endothelial cells. a) Immunogold labeling of LC3 demonstrating an LC3-positive autophagosome (white arrow)
Supplemental Figures: Figure 1: Intracellular distribution of VWF by electron microscopy in human endothelial cells. a) Immunogold labeling of LC3 demonstrating an LC3-positive autophagosome (white arrow)
BMP6 treatment compensates for the molecular defect and ameliorates hemochromatosis in Hfe knockout mice
 SUPPLEMENTARY MATERIALS BMP6 treatment compensates for the molecular defect and ameliorates hemochromatosis in Hfe knockout mice Elena Corradini, Paul J. Schmidt, Delphine Meynard, Cinzia Garuti, Giuliana
SUPPLEMENTARY MATERIALS BMP6 treatment compensates for the molecular defect and ameliorates hemochromatosis in Hfe knockout mice Elena Corradini, Paul J. Schmidt, Delphine Meynard, Cinzia Garuti, Giuliana
ab Glucose Uptake Assay Kit (colorimetric) 1
 Version 16 Last updated 10 January 2018 ab136955 Glucose Uptake Assay Kit (Colorimetric) For the measurement of Glucose uptake in a variety of cells. This product is for research use only and is not intended
Version 16 Last updated 10 January 2018 ab136955 Glucose Uptake Assay Kit (Colorimetric) For the measurement of Glucose uptake in a variety of cells. This product is for research use only and is not intended
Influenza A H1N1 HA ELISA Pair Set
 Influenza A H1N1 HA ELISA Pair Set for H1N1 ( A/Puerto Rico/8/1934 ) HA Catalog Number : SEK11684 To achieve the best assay results, this manual must be read carefully before using this product and the
Influenza A H1N1 HA ELISA Pair Set for H1N1 ( A/Puerto Rico/8/1934 ) HA Catalog Number : SEK11684 To achieve the best assay results, this manual must be read carefully before using this product and the
Total Phosphatidic Acid Assay Kit
 Product Manual Total Phosphatidic Acid Assay Kit Catalog Number MET- 5019 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Phosphatidic Acid (PA) is a critical precursor
Product Manual Total Phosphatidic Acid Assay Kit Catalog Number MET- 5019 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Phosphatidic Acid (PA) is a critical precursor
Xenoestrogen-induced Regulation of EZH2 and Histone Methylation via Non-Genomic Estrogen
 Xenoestrogen-induced Regulation of EZH2 and Histone Methylation via Non-Genomic Estrogen Receptor Signaling to PI3K/AKT Tiffany G. Bredfeldt, Kristen L. Greathouse, Stephen H. Safe, Mien-Chie Hung, Mark
Xenoestrogen-induced Regulation of EZH2 and Histone Methylation via Non-Genomic Estrogen Receptor Signaling to PI3K/AKT Tiffany G. Bredfeldt, Kristen L. Greathouse, Stephen H. Safe, Mien-Chie Hung, Mark
Figure 1. Effects of FGF21 on adipose tissue. (A) Representative histological. findings of epididymal adipose tissue (B) mrna expression of
 SUPPLEMENTAL MATERIAL EN-12-2276 Figure 1. Effects of FGF21 on adipose tissue. (A) Representative histological findings of epididymal adipose tissue (B) mrna expression of adipocytokines in adipose tissue.
SUPPLEMENTAL MATERIAL EN-12-2276 Figure 1. Effects of FGF21 on adipose tissue. (A) Representative histological findings of epididymal adipose tissue (B) mrna expression of adipocytokines in adipose tissue.
control kda ATGL ATGLi HSL 82 GAPDH * ** *** WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi iwat gwat ibat
 body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
Measurement of PDH Endogenous Activity Relative to the Fully- States
 Measurement of PDH Endogenous Activity Relative to the Fully- and Dephosphorylated Phosphorylated States Table of contents PDH Protocol #1 Measurement of PDH Endogenous Activity 1. Introduction 3 2. Regulation
Measurement of PDH Endogenous Activity Relative to the Fully- and Dephosphorylated Phosphorylated States Table of contents PDH Protocol #1 Measurement of PDH Endogenous Activity 1. Introduction 3 2. Regulation
Influenza B Hemagglutinin / HA ELISA Pair Set
 Influenza B Hemagglutinin / HA ELISA Pair Set Catalog Number : SEK11053 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized
Influenza B Hemagglutinin / HA ELISA Pair Set Catalog Number : SEK11053 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized
Supplementary Fig. 1. Identification of acetylation of K68 of SOD2
 Supplementary Fig. 1. Identification of acetylation of K68 of SOD2 A B H. sapiens 54 KHHAAYVNNLNVTEEKYQEALAK 75 M. musculus 54 KHHAAYVNNLNATEEKYHEALAK 75 X. laevis 55 KHHATYVNNLNITEEKYAEALAK 77 D. rerio
Supplementary Fig. 1. Identification of acetylation of K68 of SOD2 A B H. sapiens 54 KHHAAYVNNLNVTEEKYQEALAK 75 M. musculus 54 KHHAAYVNNLNATEEKYHEALAK 75 X. laevis 55 KHHATYVNNLNITEEKYAEALAK 77 D. rerio
Minute TM Plasma Membrane Protein Isolation and Cell Fractionation Kit User Manual (v5)
 Minute TM Plasma Membrane Protein Isolation and Cell Fractionation Kit Catalog number: SM-005 Description Minute TM plasma membrane (PM) protein isolation kit is a novel and patented native PM protein
Minute TM Plasma Membrane Protein Isolation and Cell Fractionation Kit Catalog number: SM-005 Description Minute TM plasma membrane (PM) protein isolation kit is a novel and patented native PM protein
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein
![2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein](/thumbs/86/94743397.jpg) Supplementary Information 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked amino-modification products by acrolein Ayumi Tsutsui and Katsunori Tanaka* Biofunctional Synthetic Chemistry Laboratory, RIKEN
Supplementary Information 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked amino-modification products by acrolein Ayumi Tsutsui and Katsunori Tanaka* Biofunctional Synthetic Chemistry Laboratory, RIKEN
Leptin deficiency suppresses progression of atherosclerosis in apoe-deficient mice
 Leptin deficiency suppresses progression of atherosclerosis in apoe-deficient mice Atherosclerosis, 2007 Chiba T, Shinozaki S, Nakazawa T, et al. Present by Sudaporn Pummoung Apolipoprotein E (apoe( apoe)
Leptin deficiency suppresses progression of atherosclerosis in apoe-deficient mice Atherosclerosis, 2007 Chiba T, Shinozaki S, Nakazawa T, et al. Present by Sudaporn Pummoung Apolipoprotein E (apoe( apoe)
Application Note. Introduction
 Simultaneously Measuring Oxidation of Exogenous and Endogenous Fatty Acids Using the XF Palmitate-BSA FAO Substrate with the Agilent Seahorse XF Cell Mito Stress Test Application Note Introduction The
Simultaneously Measuring Oxidation of Exogenous and Endogenous Fatty Acids Using the XF Palmitate-BSA FAO Substrate with the Agilent Seahorse XF Cell Mito Stress Test Application Note Introduction The
Supplementary Information
 Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
Human Urokinase / PLAU / UPA ELISA Pair Set
 Human Urokinase / PLAU / UPA ELISA Pair Set Catalog Number : SEK10815 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized
Human Urokinase / PLAU / UPA ELISA Pair Set Catalog Number : SEK10815 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1 Schematic depiction of the tandem Fc GDF15. Supplementary Figure 2 Supplementary Figure 2 Gfral mrna levels in the brains of both wild-type and knockout Gfral
Supplementary Figure 1 Supplementary Figure 1 Schematic depiction of the tandem Fc GDF15. Supplementary Figure 2 Supplementary Figure 2 Gfral mrna levels in the brains of both wild-type and knockout Gfral
NF-κB p65 (Phospho-Thr254)
 Assay Biotechnology Company www.assaybiotech.com Tel: 1-877-883-7988 Fax: 1-877-610-9758 NF-κB p65 (Phospho-Thr254) Colorimetric Cell-Based ELISA Kit Catalog #: OKAG02015 Please read the provided manual
Assay Biotechnology Company www.assaybiotech.com Tel: 1-877-883-7988 Fax: 1-877-610-9758 NF-κB p65 (Phospho-Thr254) Colorimetric Cell-Based ELISA Kit Catalog #: OKAG02015 Please read the provided manual
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified
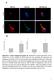 Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h)
 Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h) after feeding. A small slice (~5-1 mm 3 ) was taken
Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h) after feeding. A small slice (~5-1 mm 3 ) was taken
Serum Amyloid A3 Gene Expression in Adipocytes is an Indicator. of the Interaction with Macrophages
 Serum Amyloid A3 Gene Expression in Adipocytes is an Indicator of the Interaction with Macrophages Yohei Sanada, Takafumi Yamamoto, Rika Satake, Akiko Yamashita, Sumire Kanai, Norihisa Kato, Fons AJ van
Serum Amyloid A3 Gene Expression in Adipocytes is an Indicator of the Interaction with Macrophages Yohei Sanada, Takafumi Yamamoto, Rika Satake, Akiko Yamashita, Sumire Kanai, Norihisa Kato, Fons AJ van
ab LDL Uptake Assay Kit (Cell-Based)
 ab133127 LDL Uptake Assay Kit (Cell-Based) Instructions for Use For the detection of LDL uptake into cultured cells. This product is for research use only and is not intended for diagnostic use. Version
ab133127 LDL Uptake Assay Kit (Cell-Based) Instructions for Use For the detection of LDL uptake into cultured cells. This product is for research use only and is not intended for diagnostic use. Version
THE GLUCOSE-FATTY ACID-KETONE BODY CYCLE Role of ketone bodies as respiratory substrates and metabolic signals
 Br. J. Anaesth. (1981), 53, 131 THE GLUCOSE-FATTY ACID-KETONE BODY CYCLE Role of ketone bodies as respiratory substrates and metabolic signals J. C. STANLEY In this paper, the glucose-fatty acid cycle
Br. J. Anaesth. (1981), 53, 131 THE GLUCOSE-FATTY ACID-KETONE BODY CYCLE Role of ketone bodies as respiratory substrates and metabolic signals J. C. STANLEY In this paper, the glucose-fatty acid cycle
Supplemental Figures
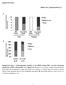 Supplemental Figures Supplemental Figure 1. Fasting-dependent regulation of the SREBP ortholog SBP-1 and lipid homeostasis mediated by the SIRT1 ortholog SIR-2.1 in C. elegans. (A) Wild-type or sir-2.1(lof)
Supplemental Figures Supplemental Figure 1. Fasting-dependent regulation of the SREBP ortholog SBP-1 and lipid homeostasis mediated by the SIRT1 ortholog SIR-2.1 in C. elegans. (A) Wild-type or sir-2.1(lof)
Supplementary Materials for
 immunology.sciencemag.org/cgi/content/full/2/16/eaan6049/dc1 Supplementary Materials for Enzymatic synthesis of core 2 O-glycans governs the tissue-trafficking potential of memory CD8 + T cells Jossef
immunology.sciencemag.org/cgi/content/full/2/16/eaan6049/dc1 Supplementary Materials for Enzymatic synthesis of core 2 O-glycans governs the tissue-trafficking potential of memory CD8 + T cells Jossef
Supplementary Information. Glycogen shortage during fasting triggers liver-brain-adipose. neurocircuitry to facilitate fat utilization
 Supplementary Information Glycogen shortage during fasting triggers liver-brain-adipose neurocircuitry to facilitate fat utilization Supplementary Figure S1. Liver-Brain-Adipose neurocircuitry Starvation
Supplementary Information Glycogen shortage during fasting triggers liver-brain-adipose neurocircuitry to facilitate fat utilization Supplementary Figure S1. Liver-Brain-Adipose neurocircuitry Starvation
