Apêndice A2. Lista completa na ordem crescente da conectividade intramodular
|
|
|
- Maud Phillips
- 5 years ago
- Views:
Transcription
1 Apêndice A2. Lista completa na ordem crescente da conectividade intramodular KTotal: conectividade total da rede KWithin: conectividade intramodular kout: conectividade extra modular; a diferença entre KTotal e KWithin kdiff: diferença entre KWithin e kout Gene Amostra do Array ktotal kwithin kout kdiff ACTB _x_at 13, , , , ACTB _x_at 13, , , , ACTB _x_at 13, , , , ACTB _x_at 12, , , , ACTB _x_at 12, , , , ACTB _x_at 12, , , , ACTB _x_at 11, , , , CUBN _at 11, , , , ALDOB _x_at 11, , , , PPP1R3A _at 11, , , , ALDOB _s_at 11, , , , ACTB _x_at 11, , , , ALDOB _s_at 11, , , , FN _x_at 10, , , , FN _x_at 10, , , , ATP1A _s_at 10, , , , FN _s_at 10, , , , FN _x_at 10, , , , ANGPTL _at 10, , , , PLG _at 10, , , , PTPRC _at 10, , , , SNRPB _at 10, , , , PLG _s_at 9, , , , PTPRC _s_at 9, , , , ATP1A _s_at 9, , , , G6PC _at 9, , , , SERPINE _s_at 9, , , , ALDH18A _s_at 8, , , , SORBS _s_at 8, , , , SERPINE _s_at 8, , , , ACLY _s_at 8, , , , SLC2A _at 8, , , , APOA _s_at 8, , , , RCAN _at 8, , , , LGALS _at 8, , , , APOA _at 8, , , , DRD _x_at 8, , , , BBS _x_at 8, , , , GLP1R _s_at 8, , , , MLYCD _at 8, , , ,
2 ITGB _s_at 8, , , , AKAP _at 8, , , , FABP _at 7, , , , DRD _x_at 7, , , , IGFBP _at 7, , , , RFC _s_at 7, , , , ACLY _s_at 7, , , , PRKCB _s_at 7, , , , NEIL _s_at 7, , , , TIMP _at 7, , , , CRY _at 7, , , , EXT _s_at 7, , , , PPIC _at 7, , , , ENSA _at 7, , , , RFC2 1053_at 7, , , , MCM _s_at 7, , , , ITGAL _s_at 7, , , , BAX _s_at 7, , , , ACE _at 7, , , , AXL _s_at 7, , , , TGFBI _at 7, , , , FOXM _x_at 7, , , , CD _at 7, , , , PPARGC1A _at 7, , , , CDK _s_at 7, , , , HFE _x_at 7, , , , ATP2A _s_at 6, , , , CLPS _at 6, , , , SQLE _at 6, , , , PSTPIP _s_at 6, , , , SPARC _s_at 6, , , , ACE _s_at 6, , , , FGG _s_at 6, , , , TRA2B _at 6, , , , WNT10B _at 6, , , , MFN _s_at 6, , , , CPS _s_at 6, , , , PNLIP _at 6, , , , ACTB _x_at 6, , , , PRKCB _s_at 6, , , , DCN _x_at 6, , , , LPL _s_at 6, , , , TBXA2R _x_at 6, , , , GAPDH M33197_M_at 6, , , , FGA _s_at 6, , , ,
3 CDC _at 6, , , , THBS _s_at 6, , , , DRD _s_at 6, , , , FCGR3B _s_at 6, , , , ATP2A _x_at 6, , , , PAX _s_at 6, , , , ACTB X00351_3_at 6, , , , GAPDH _x_at 6, , , , DCN _s_at 6, , , , SQSTM _at 6, , , , GH _x_at 6, , , , SORL _at 6, , , , VENTX _at 6, , , , EGFR _at 6, , , , SPARC _at 6, , , , TRA2B _s_at 6, , , , PYGM _at 6, , , , GAPDH _x_at 6, , , , ITGAM _s_at 6, , , , FN _s_at 6, , , , IL2RA _s_at 6, , , , CASQ _at 6, , , , ACP _x_at 6, , , , DCN _s_at 6, , , , FGF _at 6, , , , LMNA _s_at 6, , , , DCN _x_at 6, , , , FGF _s_at 6, , , , MTNR1B _at 6, , , , ALOX5AP _at 6, , , , BMP _at 6, , , , PPIC _s_at 6, , , , SLC5A _at 6, , , , SIRT _s_at 5, , , , AVP _at 5, , , , PLOD _at 5, , , , FGB _s_at 5, , , , PCSK1N _at 5, , , , TUB _at 5, , , , NCF _at 5, , , , NOS _at 5, , , , HSPB _s_at 5, , , , LDLR _at 5, , , , LPL _s_at 5, , , , PRLH _x_at 5, , , ,
4 MTCH _s_at 5, , , , MCM _s_at 5, , , , HADH _s_at 5, , , , MTUS _s_at 5, , , , FOXM _at 5, , , , MTUS _s_at 5, , , , HFE _x_at 5, , , , DCN _at 5, , , , MMP _s_at 5, , , , FCGR3B _at 5, , , , CD _s_at 5, , , , WDTC _at 5, , , , CD _at 5, , , , ITGA2B _at 5, , , , THBS _s_at 5, , , , PPY _at 5, , , , ACTB _x_at 5, , , , NOX _x_at 5, , , , ACLY _s_at 5, , , , BCL _at 5, , , , CTSS _s_at 5, , , , SRSF _s_at 5, , , , GAPDH M33197_3_at 5, , , , HAP _s_at 5, , , , FGF _s_at 5, , , , PTGDS _x_at 5, , , , TP _at 5, , , , MKI _s_at 5, , , , CD _s_at 5, , , , ACP _s_at 5, , , , GH _x_at 5, , , , GIPR _at 5, , , , CASP _at 5, , , , CRHR _at 5, , , , CPS _at 5, , , , SKP _s_at 5, , , , HIF1A _at 5, , , , UTS _s_at 5, , , , CLOCK _at 5, , , , RHOA _s_at 5, , , , GH _x_at 5, , , , CCKAR _at 4, , , , YY _s_at 4, , , , CCL _at 4, , , , APOC _s_at 4, , , ,
5 C1QB _at 4, , , , HMGCR _s_at 4, , , , PTGDS _x_at 4, , , , HADH _s_at 4, , , , KHDRBS _at 4, , , , GAPDH M33197_5_at 4, , , , FOXP _s_at 4, , , , DEFB _at 4, , , , AGRP _at 4, , , , GPR _at 4, , , , NPC1L _at 4, , , , C3AR _at 4, , , , TNFRSF _at 4, , , , MAS _at 4, , , , TARDBP _at 4, , , , ACAT _s_at 4, , , , CDH _x_at 4, , , , IL17A _at 4, , , , GH _x_at 4, , , , CYBB _s_at 4, , , , TNFRSF1B _at 4, , , , RB _at 4, , , , FASLG _at 4, , , , GAD _s_at 4, , , , HFE _x_at 4, , , , LTBP _at 4, , , , NLRP _s_at 4, , , , NOX _s_at 4, , , , CD _at 4, , , , PINK _s_at 4, , , , TLR _at 4, , , , FADS _s_at 4, , , , ESR _s_at 4, , , , NR1H _at 4, , , , CD _s_at 4, , , , CLC _at 4, , , , SSSCA _at 4, , , , HSD17B _at 4, , , , CYP3A _x_at 4, , , , IGF _at 4, , , , REN _at 4, , , , HTR1B _at 4, , , , CAMP _at 4, , , , CCRN4L _at 4, , , , TP _s_at 4, , , ,
6 CTSS _x_at 4, , , , GC _at 4, , , , VDR _s_at 4, , , , SIRT _s_at 4, , , , VWF _at 4, , , , GNAS _x_at 4, , , , FAIM _at 4, , , , CDH _s_at 4, , , , CDX _at 4, , , , CYBB _s_at 4, , , , PEA _s_at 4, , , , BAAT _at 4, , , , DLD _at 4, , , , TRIB _at 4, , , , MCHR _s_at 4, , , , HCRTR _at 4, , , , PTK2B _at 4, , , , SORBS _s_at 4, , , , ALDH _at 4, , , , TXK _at 4, , , , KAT2B _at 4, , , , HAP _x_at 4, , , , PKLR _at 4, , , , ATG _s_at 4, , , , CAV _at 4, , , , FN _at 4, , , , PPIL _x_at 4, , , , APOB _s_at 4, , , , KCNQ _s_at 4, , , , GNAS _x_at 4, , , , SSB _s_at 4, , , , SHBG _s_at 4, , , , IL _s_at 4, , , , GNAS _x_at 4, , , , HNF4A _s_at 4, , , , CASP _x_at 4, , , , APOA _x_at 4, , , , NLRP _s_at 4, , , , INHBA _s_at 4, , , , CDK _s_at 4, , , , CS _at 4, , , , CDKN1A _s_at 4, , , , CLEC7A _s_at 4, , , , ABCB _at 4, , , , CASP _x_at 4, , , ,
7 CEACAM _at 4, , , , PINK _s_at 4, , , , NLRP _x_at 4, , , , G6PC _at 4, , , , MPO _s_at 4, , , , CASP _s_at 4, , , , HFE _x_at 4, , , , MPO _at 4, , , , MSRB _s_at 4, , , , H6PD _s_at 3, , , , KISS _at 3, , , , GNAS _x_at 3, , , , PTPRN _x_at 3, , , , CNR _s_at 3, , , , ACTB X00351_M_at 3, , , , MOGAT _at 3, , , , COX _s_at 3, , , , NOS _s_at 3, , , , IGF _at 3, , , , FAS _x_at 3, , , , GOT _at 3, , , , AOC _s_at 3, , , , S100A _at 3, , , , LEPR _at 3, , , , TFAM _x_at 3, , , , CDH _at 3, , , , FGF _at 3, , , , EGFR _at 3, , , , HOXD _s_at 3, , , , ME _s_at 3, , , , PLAUR _s_at 3, , , , CHRNA _at 3, , , , GSK3A 632_at 3, , , , ALDOB _x_at 3, , , , SMN _s_at 3, , , , CAPN _s_at 3, , , , EPHX _at 3, , , , CCL _at 3, , , , ACAT _at 3, , , , Nnat _s_at 3, , , , LAMP _at 3, , , , LAMP _s_at 3, , , , CAV _s_at 3, , , , IL _at 3, , , , DBH _at 3, , , ,
8 PEPD _at 3, , , , BRCA _s_at 3, , , , APOA _x_at 3, , , , ENSA _s_at 3, , , , CNR _x_at 3, , , , FAS _s_at 3, , , , PML _x_at 3, , , , PDX _at 3, , , , FADS _at 3, , , , CYP2C _s_at 3, , , , BCL _s_at 3, , , , ACTB X00351_5_at 3, , , , GCK _s_at 3, , , , BAX _s_at 3, , , , GSK3A _x_at 3, , , , CAV _s_at 3, , , , NPRL _s_at 3, , , , MMP _at 3, , , , CDKN1C _at 3, , , , FBP _at 3, , , , OGT _at 3, , , , SRSF _x_at 3, , , , AGAP _at 3, , , , IGFBP _at 3, , , , CRHR _s_at 3, , , , ALMS _x_at 3, , , , CCR _at 3, , , , CAPN _x_at 3, , , , CDKN1C _at 3, , , , GYS _s_at 3, , , , LYZ _s_at 3, , , , AGT _at 3, , , , PML _x_at 3, , , , VAMP _at 3, , , , PNLIP _at 3, , , , TCF7L _at 3, , , , FGF _at 3, , , , ACSM _s_at 3, , , , IRF _at 3, , , , HMGA _s_at 3, , , , RENBP _s_at 3, , , , HFE _x_at 3, , , , GCKR _at 3, , , , APOE _at 3, , , , PON _s_at 3, , , ,
9 ITGAX _at 3, , , , CDKN1C _x_at 3, , , , TJP _at 3, , , , KIR3DL _at 3, , , , TCF7L _x_at 3, , , , GJD _at 3, , , , IGF2 AS _at 3, , , , VPS26A _at 3, , , , PSEN _s_at 3, , , , PFKP _at 3, , , , NOS _s_at 3, , , , ESR _x_at 3, , , , ALDOB _s_at 3, , , , GNB _at 3, , , , HSPA _at 3, , , , SRD5A _s_at 3, , , , UCP _s_at 3, , , , CYP19A _at 3, , , , FGB _at 3, , , , CASP _s_at 3, , , , HRH _at 3, , , , SRSF _x_at 3, , , , ABCB _s_at 3, , , , PPP1R3A _s_at 3, , , , SCD _s_at 3, , , , IL1RN _s_at 3, , , , TBXA2R _x_at 3, , , , CDO _at 3, , , , NOX _at 3, , , , TCF7L _x_at 3, , , , ACSL _s_at 3, , , , PML _x_at 3, , , , GH _x_at 3, , , , SKI _s_at 3, , , , BCAT _at 3, , , , ABHD _at 3, , , , CYP2C _x_at 3, , , , CYP3A _x_at 3, , , , PLAUR _s_at 3, , , , APOBEC _at 3, , , , NBN _s_at 3, , , , UQCRC _at 3, , , , ENSA _at 3, , , , FAS _x_at 3, , , , ACSM _at 3, , , ,
10 HFE _x_at 3, , , , BCL2A _at 3, , , , AK _at 3, , , , GPRC5B _s_at 3, , , , DUSP _at 3, , , , GNAS _x_at 3, , , , CD40LG _at 3, , , , CSF _at 3, , , , CASP _at 3, , , , GH _x_at 3, , , , UBE3A _x_at 3, , , , PTK2B _s_at 3, , , , ATP2A _s_at 3, , , , CXCR _at 3, , , , CDKN1C _x_at 3, , , , CAPN _x_at 3, , , , TNFSF _s_at 3, , , , SLC15A _at 3, , , , CLTC _at 3, , , , WNT _at 3, , , , ALDOB _x_at 3, , , , NOD _s_at 3, , , , APOA _at 3, , , , SRD5A _at 2, , , , SQLE _at 2, , , , CDKN1C _x_at 2, , , , NMUR _at 2, , , , PSEN _s_at 2, , , , TBC1D _s_at 2, , , , CASP _at 2, , , , COPZ _at 2, , , , SMARCA _x_at 2, , , , DICER _at 2, , , , NTRK _at 2, , , , MYO _s_at 2, , , , CANX _s_at 2, , , , CARD _at 2, , , , OPRM _s_at 2, , , , AVPR1A _x_at 2, , , , UBE3A _s_at 2, , , , WDTC _s_at 2, , , , WDTC _x_at 2, , , , CCKAR _s_at 2, , , , PTEN _s_at 2, , , , IL6R _at 2, , , ,
11 KIR3DL _x_at 2, , , , CLCN _at 2, , , , CYP3A _x_at 2, , , , ABHD _at 2, , , , ABHD _at 2, , , , TRPC _s_at 2, , , , CPQ _at 2, , , , ATP6V1C _at 2, , , , TIMP _s_at 2, , , , IL _at 2, , , , PAX _at 2, , , , DLX _s_at 2, , , , DACT _at 2, , , , NBN _s_at 2, , , , HFE _x_at 2, , , , BCL _s_at 2, , , , TF _at 2, , , , CCR _s_at 2, , , , ACSL _at 2, , , , TRA2B _s_at 2, , , , GATA _at 2, , , , SOD _x_at 2, , , , SSB _s_at 2, , , , ROCK _at 2, , , , ATP6V1C _s_at 2, , , , TF _s_at 2, , , , GRN _x_at 2, , , , MTUS _s_at 2, , , , PPIL _x_at 2, , , , PSME _s_at 2, , , , C5AR _at 2, , , , TLR _at 2, , , , KCNQ _s_at 2, , , , HNF4A _at 2, , , , TBC1D _at 2, , , , WRN _at 2, , , , FABP _at 2, , , , BMPR1A _at 2, , , , CYP2C _x_at 2, , , , HADH _s_at 2, , , , CD _at 2, , , , EDNRA _s_at 2, , , , ASL _s_at 2, , , , LMNA _x_at 2, , , , IL1RN _s_at 2, , , ,
12 MADD 38398_at 2, , , , KIR3DL _x_at 2, , , , NOTCH _s_at 2, , , , LDLR _s_at 2, , , , GRN _x_at 2, , , , IL1RN _at 2, , , , CDK11A _x_at 2, , , , AGK _at 2, , , , ATG _s_at 2, , , , GDF _at 2, , , , ME _at 2, , , , IL _s_at 2, , , , ATP2A _s_at 2, , , , CTSL _s_at 2, , , , FAAH _s_at 2, , , , SMARCA _s_at 2, , , , CYP2C _x_at 2, , , , CD _at 2, , , , IGF2BP _at 2, , , , SLC22A _s_at 2, , , , MCCC _at 2, , , , PGD _at 2, , , , ADAM _at 2, , , , PTEN _x_at 2, , , , CRP 37020_at 2, , , , GAD _at 2, , , , IGFBP _s_at 2, , , , ABCC _at 2, , , , GAL3ST _at 2, , , , CTSL _s_at 2, , , , CDK11A _x_at 2, , , , GATA _s_at 2, , , , PDAP _at 2, , , , CAT _at 2, , , , ACACB _x_at 2, , , , STAT1 M97935_MB_at 2, , , , PPID _s_at 2, , , , IL1R _at 2, , , , ECHS _at 2, , , , PTPRF _s_at 2, , , , MAPK8IP _at 2, , , , UBE3A _s_at 2, , , , ZMPSTE _at 2, , , , CDK11A _s_at 2, , , , CFH _at 2, , , ,
13 ADM _at 2, , , , VLDLR _s_at 2, , , , ADAR _s_at 2, , , , NR3C _s_at 2, , , , SIRT _s_at 2, , , , AKAP _at 2, , , , FABP _at 2, , , , DGKZ _s_at 2, , , , ME _at 2, , , , ACADVL _at 2, , , , RRAD _s_at 2, , , , NFYC _s_at 2, , , , CYP2C _at 2, , , , CD8A _at 2, , , , PDE3A _s_at 2, , , , SLC9A _at 2, , , , TNFRSF1A _s_at 2, , , , CEL _x_at 2, , , , ESR _at 2, , , , PGM _s_at 2, , , , HNF4A _at 2, , , , UCP _at 2, , , , MAP4K _at 2, , , , PTPRF _s_at 2, , , , ITPR _at 2, , , , MFN _s_at 2, , , , BCL _at 2, , , , AANAT _at 2, , , , GRN _s_at 2, , , , AKAP _at 2, , , , NUAK _at 2, , , , HFE _x_at 2, , , , SRD5A _s_at 2, , , , PLAGL _x_at 2, , , , EDNRA _s_at 2, , , , RBMS _s_at 2, , , , NEUROD _at 2, , , , PPIL _x_at 2, , , , TLR _at 2, , , , MMP _s_at 2, , , , ARNTL _s_at 2, , , , STAT _at 2, , , , FAT _at 2, , , , AGTR _s_at 2, , , , ATM _x_at 2, , , ,
14 APP _at 2, , , , FLT _at 2, , , , EIF4A _s_at 2, , , , SRR _s_at 2, , , , RRAD _at 2, , , , TNIK _s_at 2, , , , PLAUR _at 2, , , , CRAT _x_at 2, , , , PI _at 2, , , , MGAT4A _at 2, , , , GATA _s_at 2, , , , SERPINF _at 2, , , , PDX _at 2, , , , GCG _at 2, , , , GFPT _at 2, , , , STAT1 M97935_MA_at 2, , , , MARCH _s_at 2, , , , IGF _x_at 2, , , , PYY _x_at 2, , , , OPRM _at 2, , , , PLIN _at 2, , , , MTTP _at 2, , , , SPP _s_at 2, , , , AKT _s_at 2, , , , EDNRB _at 2, , , , SCD _s_at 2, , , , ATM _s_at 2, , , , ARIH _at 2, , , , LAD _s_at 2, , , , RFC _x_at 2, , , , PON _at 2, , , , RBMS _x_at 2, , , , FOXA _at 2, , , , ITGA2B _at 2, , , , MSRB _at 2, , , , SLC2A _s_at 2, , , , RBMS _x_at 2, , , , GPT _x_at 2, , , , CX3CL _at 2, , , , ALOX _s_at 2, , , , NR3C _s_at 2, , , , IGFBP _s_at 2, , , , RAPGEF _s_at 2, , , , PRKAR2B _at 2, , , , PLAGL _s_at 2, , , ,
15 GDF _x_at 1, , , , CXCL _at 1, , , , C9orf _s_at 1, , , , NPL _s_at 1, , , , CYP2E1 1431_at 1, , , , CAPN _s_at 1, , , , PLAGL _x_at 1, , , , TPD _s_at 1, , , , GIP _at 1, , , , NFYC _s_at 1, , , , NR3C _x_at 1, , , , RFC _s_at 1, , , , OGT _x_at 1, , , , PROC _at 1, , , , SOAT _at 1, , , , DRD _x_at 1, , , , TFAP2B _at 1, , , , SMARCA _x_at 1, , , , AK _s_at 1, , , , PTPRN _s_at 1, , , , KCNJ _s_at 1, , , , TPD _s_at 1, , , , BMP _at 1, , , , OGT _s_at 1, , , , PTPRF _s_at 1, , , , PPIG _s_at 1, , , , CLEC4E _at 1, , , , ITGA2B _s_at 1, , , , CRHR _x_at 1, , , , SLC25A _at 1, , , , MARCH _x_at 1, , , , KRT _x_at 1, , , , FXN _s_at 1, , , , CXCL _s_at 1, , , , IL _x_at 1, , , , STAT1 M97935_3_at 1, , , , ARIH _at 1, , , , TRPC _at 1, , , , FSHB _at 1, , , , DOK _s_at 1, , , , RETN _at 1, , , , EDNRA _s_at 1, , , , RAPGEF _at 1, , , , SQSTM _s_at 1, , , , AR _s_at 1, , , ,
16 NFATC _at 1, , , , OXCT _at 1, , , , STAT5A _at 1, , , , CCR _at 1, , , , STAT1 M97935_5_at 1, , , , AACS _s_at 1, , , , LARS _at 1, , , , PKLR _s_at 1, , , , NBN _s_at 1, , , , NOTCH _x_at 1, , , , DLX _at 1, , , , REL _s_at 1, , , , AIM _at 1, , , , NFYC _x_at 1, , , , ESR _s_at 1, , , , SMARCA _s_at 1, , , , SDC _s_at 1, , , , CX3CR _at 1, , , , CDKN1B _at 1, , , , PML _s_at 1, , , , LGALS _s_at 1, , , , NDUFA _at 1, , , , GPRC5B _s_at 1, , , , FDFT _at 1, , , , SLC2A _s_at 1, , , , MEFV _x_at 1, , , , CDK11A _s_at 1, , , , CBL _at 1, , , , FGF _s_at 1, , , , SST _at 1, , , , PARP _at 1, , , , CTSK _s_at 1, , , , TFAM _s_at 1, , , , NEDD4L _at 1, , , , PCK _at 1, , , , CDH _s_at 1, , , , MTR _at 1, , , , STAT _s_at 1, , , , LRP _at 1, , , , AGAP _s_at 1, , , , BRCA _x_at 1, , , , SLC2A _s_at 1, , , , SLC12A _at 1, , , , SCD _x_at 1, , , , IGF _s_at 1, , , ,
17 CST _at 1, , , , DHRS _at 1, , , , GLO _at 1, , , , MAPK _x_at 1, , , , CPQ _s_at 1, , , , CYP2E _at 1, , , , NLRP _at 1, , , , GSTP _at 1, , , , CASP _s_at 1, , , , SLC25A _at 1, , , , CTDSP _at 1, , , , ADORA _s_at 1, , , , MADD _s_at 1, , , , AEBP _at 1, , , , NFYC _x_at 1, , , , PLAT _s_at 1, , , , PTBP _at 1, , , , RFC _s_at 1, , , , DEDD _x_at 1, , , , TRIM _at 1, , , , PPIL _x_at 1, , , , AXL _s_at 1, , , , RARA _s_at 1, , , , CDK11A _s_at 1, , , , SDC _at 1, , , , NR3C _s_at 1, , , , MMP _at 1, , , , RBMS _s_at 1, , , , UGCG _s_at 1, , , , APPL _s_at 1, , , , NPR _at 1, , , , ARIH _s_at 1, , , , NPAS _at 1, , , , ESR _x_at 1, , , , CYP2C _s_at 1, , , , NSFL1C _s_at 1, , , , ADCY _at 1, , , , CLOCK _at 1, , , , INSR _s_at 1, , , , EGFR _s_at 1, , , , BMPR1A _s_at 1, , , , P2RX _at 1, , , , ASIP _at 1, , , , SERPINC _at 1, , , , GALR _at 1, , , ,
18 SQSTM _s_at 1, , , , MAPK8IP _at 1, , , , ADRB _at 1, , , , BCL2L _s_at 1, , , , HMG20A _at 1, , , , PON _s_at 1, , , , MAP2K _at 1, , , , IL _s_at 1, , , , FAS _s_at 1, , , , PRKCZ _at 1, , , , FOXO _s_at 1, , , , ABCB _at 1, , , , TFAP2A _at 1, , , , BMP _at 1, , , , ZMYND _at 1, , , , FASN _s_at 1, , , , BCL2L _s_at 1, , , , NAMPT _s_at 1, , , , ARSA _at 1, , , , GHRH _at 1, , , , CXCL _x_at 1, , , , HNF1A _at 1, , , , CA _at 1, , , , SH2B _at 1, , , , CASP _s_at 1, , , , PLCG _x_at 1, , , , NRF _s_at 1, , , , OGT _s_at 1, , , , HFE _x_at 1, , , , UNC13A _at 1, , , , CYP2C _s_at 1, , , , BCL11A _s_at 1, , , , VEGFA _s_at 1, , , , ROCK _s_at 1, , , , VDR _s_at 1, , , , AKR1A _s_at 1, , , , CYP2E _s_at 1, , , , THY _x_at 1, , , , CXCL _s_at 1, , , , SLC2A _at 1, , , , LMNA _x_at 1, , , , CRH _s_at 1, , , , STK _x_at 1, , , , DYSF _at 1, , , , UBE3A _s_at 1, , , ,
19 IL _at 1, , , , AKR1B _at 1, , , , AKAP _s_at 1, , , , TNIK _at 1, , , , CANX _s_at 1, , , , TFAM _x_at 1, , , , LDLR _s_at 1, , , , NOD _at 1, , , , FOXO _s_at 1, , , , CLDN _x_at 1, , , , AR _at 1, , , , SMAD _at 1, , , , MYD _at 1, , , , ITPR _s_at 1, , , , PTPRD _at 1, , , , NLRP _at 1, , , , MYC _s_at 1, , , , SFRP _s_at 1, , , , KIR3DL _s_at 1, , , , CDKN1C _s_at 1, , , , TCF7L _s_at 1, , , , IL _at 1, , , , CLU _at 1, , , , CTNNB _at 1, , , , CPB _s_at 1, , , , ICAM _s_at 1, , , , CYP3A _at 1, , , , NPPB _at 1, , , , LDLR _s_at 1, , , , SNRPN _x_at 1, , , , TBXA2R _s_at 1, , , , PYCARD _s_at 1, , , , MYLIP _s_at 1, , , , FOXO _s_at 1, , , , ZMYND _s_at 1, , , , BCL11A _s_at 1, , , , TWIST _at 1, , , , LGALS _s_at 1, , , , NSFL1C _x_at 1, , , , ERBB _s_at 1, , , , FGFR _at 1, , , , EGFR _x_at 1, , , , AMH _at 1, , , , PIK3CB _at 1, , , , EGFR _x_at 1, , , ,
20 PML _s_at 1, , , , CDC _at 1, , , , KHDRBS _x_at 1, , , , RAB3GAP _at 1, , , , SFTPB 37004_at 1, , , , TERF2IP _s_at 1, , , , TK _at 1, , , , MAEA _s_at 1, , , , OPRM _s_at 1, , , , LIPC _at 1, , , , NPC _at 1, , , , IL1B 39402_at 1, , , , SLC17A _s_at 1, , , , CLU _s_at 1, , , , NLRP _s_at 1, , , , SFTPB _at 1, , , , MAPK _x_at 1, , , , ADORA _at 1, , , , MMP _s_at 1, , , , HIRA _s_at 1, , , , ESR _at 1, , , , LAMP _at 1, , , , CDC _at 1, , , , PML _at 1, , , , ITPR _s_at 1, , , , FOXO _s_at 1, , , , SNRNP _s_at 1, , , , FAIM _s_at 1, , , , NPAS _at 1, , , , FOXA _s_at 1, , , , F _at 1, , , , AZGP _s_at 1, , , , ATF _at 1, , , , GSK3B _s_at 1, , , , TNFAIP _s_at 1, , , , UBE3A _s_at 1, , , , KIR3DL _s_at 1, , , , CDH _s_at 1, , , , RAB3GAP _at 1, , , , FADS _s_at 1, , , , NAMPT _at 1, , , , NR0B _at 1, , , , PRKAB _at 1, , , , BMP _s_at 1, , , , ICAM _s_at 1, , , ,
Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits.
 Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits. All With Fst in the With long With SNP(s) at the 5' top 5% bracket haplotypes regulatory region
Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits. All With Fst in the With long With SNP(s) at the 5' top 5% bracket haplotypes regulatory region
Editorial Type 2 Diabetes and More Gene Panel: A Predictive Genomics Approach for a Polygenic Disease
 Cronicon OPEN ACCESS EC DIABETES AND METABOLIC RESEARCH Editorial Type 2 Diabetes and More Gene Panel: A Predictive Genomics Approach for a Polygenic Disease Amr TM Saeb* University Diabetes Center, College
Cronicon OPEN ACCESS EC DIABETES AND METABOLIC RESEARCH Editorial Type 2 Diabetes and More Gene Panel: A Predictive Genomics Approach for a Polygenic Disease Amr TM Saeb* University Diabetes Center, College
HTG EdgeSeq Immuno-Oncology Assay Gene List
 A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases
 Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
SUPPLEMENTARY FIG. S2. b-galactosidase staining of
 SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
Cardiovascular Disease Products
 Cardiovascular Disease Products For more information, visit: www.bosterbio.com Cardiovascular Disease Research Cardiovascular disease is the leading cause of death in developed nations. Boster Bio aims
Cardiovascular Disease Products For more information, visit: www.bosterbio.com Cardiovascular Disease Research Cardiovascular disease is the leading cause of death in developed nations. Boster Bio aims
Supplemental Data for Toischer et al., Cardiomyocyte proliferation prevents failure in
 Supplemental Data, Page 1 Supplemental Data for Toischer et al., Cardiomyocyte proliferation prevents failure in pressure but not volume overload. Contents: Page 3: Supplemental Figure 1 - Echocardiographic
Supplemental Data, Page 1 Supplemental Data for Toischer et al., Cardiomyocyte proliferation prevents failure in pressure but not volume overload. Contents: Page 3: Supplemental Figure 1 - Echocardiographic
Nature Genetics: doi: /ng Supplementary Figure 1
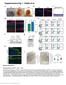 Supplementary Figure 1 Characterization of HPV Tg/+ ; Krt17 / mice. (a) Macroscopic images of HPV16 Tg/+ mouse ears at P40, P70 and P120. (b) Immunostaining of HPV16 Tg/+ ear tissue sections for K17 (red)
Supplementary Figure 1 Characterization of HPV Tg/+ ; Krt17 / mice. (a) Macroscopic images of HPV16 Tg/+ mouse ears at P40, P70 and P120. (b) Immunostaining of HPV16 Tg/+ ear tissue sections for K17 (red)
Eosinophils! 40! 30! 20! 10! 0! NS!
 A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
Assessed Molecular Mechanism of Action of Milagro de la Selva
 Assessed Molecular Mechanism of Action of Milagro de la Selva Results A report prepared by Connexios Life Sciences Bangalore 1. Muscle - C2C12 cells. 1.1. Glucose uptake, Glycolysis and Tricarboxylic acid
Assessed Molecular Mechanism of Action of Milagro de la Selva Results A report prepared by Connexios Life Sciences Bangalore 1. Muscle - C2C12 cells. 1.1. Glucose uptake, Glycolysis and Tricarboxylic acid
antibody and ELISA experts Cancer Research Products
 Cancer Research Products WWW.BOSTERBIO.COM Cancer Research Cancer is a leading cause of death in developed nations, affecting millions of people around the world. Research into the molecular basis of these
Cancer Research Products WWW.BOSTERBIO.COM Cancer Research Cancer is a leading cause of death in developed nations, affecting millions of people around the world. Research into the molecular basis of these
Jennifer Hauenstein Oncology Cytogenetics Emory University Hospital Atlanta, GA
 Comparison of Genomic Coverage using Affymetrix OncoScan Array and Illumina TruSight Tumor 170 NGS Panel for Detection of Copy Number Abnormalities in Clinical GBM Specimens Jennifer Hauenstein Oncology
Comparison of Genomic Coverage using Affymetrix OncoScan Array and Illumina TruSight Tumor 170 NGS Panel for Detection of Copy Number Abnormalities in Clinical GBM Specimens Jennifer Hauenstein Oncology
Supplementary Figures and Tables. An inflammatory gene signature distinguishes neurofibroma Schwann cells and
 Supplementary Figures and Tables An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system Kwangmin Choi 1, Kakajan Komurov
Supplementary Figures and Tables An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system Kwangmin Choi 1, Kakajan Komurov
Precision Oncology: Experience at UW
 Precision Oncology: Experience at UW Colin Pritchard MD, PhD University of Washington, Department of Lab Medicine WSMOS Meeting November 1, 2013 Conflict of Interest Disclosures I declare the following,
Precision Oncology: Experience at UW Colin Pritchard MD, PhD University of Washington, Department of Lab Medicine WSMOS Meeting November 1, 2013 Conflict of Interest Disclosures I declare the following,
LMM / emerge III Network Reference Sequences October 2016 LMM / emerge III Network Consensus Actionable Gene List *ACMG56 gene
 LMM / emerge III Network Consensus Actionable Gene List *ACMG56 gene ACTA2* Exon 02-09 NM_001613.2 DSG2* Exon 01-15 NM_001943.3 ACTC1* Exon 01-06 NM_005159.4 DSP* Exon 01-24 NM_004415.2 APC* Exon 01-15
LMM / emerge III Network Consensus Actionable Gene List *ACMG56 gene ACTA2* Exon 02-09 NM_001613.2 DSG2* Exon 01-15 NM_001943.3 ACTC1* Exon 01-06 NM_005159.4 DSP* Exon 01-24 NM_004415.2 APC* Exon 01-15
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature15260 Supplementary Data 1: Gene expression in individual basal/stem, luminal, and luminal progenitor cells. Box plots show expression levels for each gene from the 49-gene differentiation
doi:10.1038/nature15260 Supplementary Data 1: Gene expression in individual basal/stem, luminal, and luminal progenitor cells. Box plots show expression levels for each gene from the 49-gene differentiation
Study on the Biological Basis of Hypertension and Syndrome with Liver Fire Hyperactivity Based on Data Mining Technology
 Download free from www.wjtcm.net Original Article Study on the Biological Basis of Hypertension and Syndrome with Liver Fire Hyperactivity Based on Data Mining Technology Xue-Ling Ma a, Xing Zhai b, Jing-Wei
Download free from www.wjtcm.net Original Article Study on the Biological Basis of Hypertension and Syndrome with Liver Fire Hyperactivity Based on Data Mining Technology Xue-Ling Ma a, Xing Zhai b, Jing-Wei
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study. February 2018
 Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Gene expression of muscular and neuronal pathways is cooperatively dysregulated in patients
 Gene expression of muscular and neuronal pathways is cooperatively dysregulated in patients with idiopathic achalasia Orazio Palmieri 1, Tommaso Mazza 2, Antonio Merla 1, Caterina Fusilli 2, Antonello
Gene expression of muscular and neuronal pathways is cooperatively dysregulated in patients with idiopathic achalasia Orazio Palmieri 1, Tommaso Mazza 2, Antonio Merla 1, Caterina Fusilli 2, Antonello
Transduction of lentivirus to human primary CD4+ T cells
 Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
A genome-wide association study identifies vitiligo
 A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
An Integrative Pharmacogenomic Approach Identifies Two-drug. Combination Therapies for Personalized Cancer Medicine
 An Integrative Pharmacogemic Approach Identifies Two-drug Combination Therapies for Personalized Cancer Medicine Yin Liu 1,2,3, Teng Fei 4,5, Xiaoqi Zheng 6, Myles Brown 4,5, Peng Zhang 7*, X. Shirley
An Integrative Pharmacogemic Approach Identifies Two-drug Combination Therapies for Personalized Cancer Medicine Yin Liu 1,2,3, Teng Fei 4,5, Xiaoqi Zheng 6, Myles Brown 4,5, Peng Zhang 7*, X. Shirley
Supplementary Information
 Supplementary Information Index of Supplementary Materials Supplementary Figure. Distribution of methylation levels, characteristics of cis-meqtls and association plots for meqtls in methylation-related
Supplementary Information Index of Supplementary Materials Supplementary Figure. Distribution of methylation levels, characteristics of cis-meqtls and association plots for meqtls in methylation-related
Changing the Culture of Cancer Care II. Eric Holland Fred Hutchinson Cancer Research Center University of Washington Seattle
 Changing the Culture of Cancer Care II Eric Holland Fred Hutchinson Cancer Research Center University of Washington Seattle Transforming Cancer Therapy Eric Holland Fred Hutchinson Cancer Research Center
Changing the Culture of Cancer Care II Eric Holland Fred Hutchinson Cancer Research Center University of Washington Seattle Transforming Cancer Therapy Eric Holland Fred Hutchinson Cancer Research Center
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Jocelyn Chapman, MD Division of Gynecologic Oncology. Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic
 Jocelyn Chapman, MD Division of Gynecologic Oncology Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic Genetics underlies all cancers Somatic or tumor genetics Germline or inherited genetics
Jocelyn Chapman, MD Division of Gynecologic Oncology Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic Genetics underlies all cancers Somatic or tumor genetics Germline or inherited genetics
Supplemental Information. High-Throughput Microfluidic Labyrinth for the. Label-free Isolation of Circulating Tumor Cells
 Cell Systems, Volume 5 Supplemental Information High-Throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells Eric Lin, Lianette Rivera-Báez, Shamileh Fouladdel, Hyeun
Cell Systems, Volume 5 Supplemental Information High-Throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells Eric Lin, Lianette Rivera-Báez, Shamileh Fouladdel, Hyeun
Annotation SNPEff_ Annotation SNPEff_. SNPEff_. Annotation. SNPEff_. Annotation. SNPEff_. Annotation. SNPEff_. Annotation
 Chr Gene p-value Binned_ variants Phenotype Biological_Feature Note 11 DRD2 0.001179727 35 Asthma Gene 16 CES2 0.010970381 40 Asthma Gene 19 LDLR 0.023074213 43 Asthma Gene 10 ARID5B 0.025870289 44 Asthma
Chr Gene p-value Binned_ variants Phenotype Biological_Feature Note 11 DRD2 0.001179727 35 Asthma Gene 16 CES2 0.010970381 40 Asthma Gene 19 LDLR 0.023074213 43 Asthma Gene 10 ARID5B 0.025870289 44 Asthma
Supplementary Information
 Supplementary Information Title: Role of Cytochrome P450 (CYP)1A in Hyperoxic Lung Injury: Analysis of the Transcriptome and Proteome Authors: Krithika Lingappan a, Suman Maity b, Weiwu Jiang a, Lihua
Supplementary Information Title: Role of Cytochrome P450 (CYP)1A in Hyperoxic Lung Injury: Analysis of the Transcriptome and Proteome Authors: Krithika Lingappan a, Suman Maity b, Weiwu Jiang a, Lihua
APPLICATIONS OF NEXT GENERATION SEQUENCING IN SOLID TUMORS - PATHOLOGIST PROSPECTIVE
 AMP COMPANION MEETING SYMPOSIUM AT USCAP 2015 NEXT-GENERATION OF PATHOLOGY: ROLE OF PATHOLOGIST IN NGS-BASED PERSONALIZED MEDICINE APPLICATIONS OF NEXT GENERATION SEQUENCING IN SOLID TUMORS - PATHOLOGIST
AMP COMPANION MEETING SYMPOSIUM AT USCAP 2015 NEXT-GENERATION OF PATHOLOGY: ROLE OF PATHOLOGIST IN NGS-BASED PERSONALIZED MEDICINE APPLICATIONS OF NEXT GENERATION SEQUENCING IN SOLID TUMORS - PATHOLOGIST
Deconvolution of Heterogeneous Wound Tissue Samples into Relative Macrophage Phenotype Composition via Models based on Gene Expression
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2017 Supplementary Information for: Deconvolution of Heterogeneous Wound Tissue Samples into
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2017 Supplementary Information for: Deconvolution of Heterogeneous Wound Tissue Samples into
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/6/234/234ra57/dc1 Supplementary Materials for Disease Risk Factors Identified Through Shared Genetic Architecture and Electronic Medical Records Li
www.sciencetranslationalmedicine.org/cgi/content/full/6/234/234ra57/dc1 Supplementary Materials for Disease Risk Factors Identified Through Shared Genetic Architecture and Electronic Medical Records Li
Assessing the Economics of Genomic Medicine. Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center
 Assessing the Economics of Genomic Medicine Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center DISCLOSURES No conflicts Patents unenforced Consultancies unpaid
Assessing the Economics of Genomic Medicine Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center DISCLOSURES No conflicts Patents unenforced Consultancies unpaid
Figure 1: Effects of cisplatin on survival of lung cancer cells.
 Figure 1 Figure 1: Effects of cisplatin on survival of lung cancer cells. To determine the IC 50 concentration of cisplatin, cells were treated with various concentrations of cisplatin and cell survival
Figure 1 Figure 1: Effects of cisplatin on survival of lung cancer cells. To determine the IC 50 concentration of cisplatin, cells were treated with various concentrations of cisplatin and cell survival
Lions and Tigers and Bears.Oh My!
 Lions and Tigers and Bears.Oh My! TAKING THE FEAR OUT OF GENETIC TESTING Lisa Butterfield, MS, CGC Genetic Counselor Clinical Assistant Professor University of Kansas Health System Why is Genetic Testing
Lions and Tigers and Bears.Oh My! TAKING THE FEAR OUT OF GENETIC TESTING Lisa Butterfield, MS, CGC Genetic Counselor Clinical Assistant Professor University of Kansas Health System Why is Genetic Testing
qpcr-array Analysis Service
 qpcr-array Analysis Service Customer Name Institute Telephone Address E-mail PO Number Service Code Report Date Service Laboratory Department Phalanx Biotech Group, Inc 6 Floor, No.6, Technology Road 5,
qpcr-array Analysis Service Customer Name Institute Telephone Address E-mail PO Number Service Code Report Date Service Laboratory Department Phalanx Biotech Group, Inc 6 Floor, No.6, Technology Road 5,
Supplemental Figure 1
 Supplemental Figure 1 mir 26a mir 26b Supplemental Figure 1. Expression of mir-26a is higher than mir-26b in the mouse liver. Dot blot analysis of mir-26a/b expression in the livers of wild-type mice fed
Supplemental Figure 1 mir 26a mir 26b Supplemental Figure 1. Expression of mir-26a is higher than mir-26b in the mouse liver. Dot blot analysis of mir-26a/b expression in the livers of wild-type mice fed
Genomic Medicine for Physical and Mental Health
 Genomic Medicine for Physical and Mental Health Transform your health and mood with DNA-directed healthcare Learn about your DNA and what makes you unique Have you been trying to lose weight and gain muscle
Genomic Medicine for Physical and Mental Health Transform your health and mood with DNA-directed healthcare Learn about your DNA and what makes you unique Have you been trying to lose weight and gain muscle
Illumina s Cancer Research Portfolio and Dedicated Workflows
 Illumina s Cancer Research Portfolio and Dedicated Workflows Michael Sohn Clinical Sales Specialist Spain&Italy 2017 2017 Illumina, Inc. All rights reserved. Illumina, 24sure, BaseSpace, BeadArray, BlueFish,
Illumina s Cancer Research Portfolio and Dedicated Workflows Michael Sohn Clinical Sales Specialist Spain&Italy 2017 2017 Illumina, Inc. All rights reserved. Illumina, 24sure, BaseSpace, BeadArray, BlueFish,
Use of Molecular Assays to Assess Cellular Therapies
 Use of Molecular Assays to Assess Cellular Therapies D. Stroncek, P. Jin, M. Sabatino, J. Ren, L. Castiello, E. Wang, and F.M. Marincola. Cellular Therapies Section Clinical Center, NIH Methods for the
Use of Molecular Assays to Assess Cellular Therapies D. Stroncek, P. Jin, M. Sabatino, J. Ren, L. Castiello, E. Wang, and F.M. Marincola. Cellular Therapies Section Clinical Center, NIH Methods for the
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array.
 Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Sample Metrics. Allele Frequency (%) Read Depth Ploidy. Gene CDS Effect Protein Effect. LN Metastasis Tumor Purity Computational Pathology 80% 60%
 Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
B16F1 B16F10 Supplemental Figure S1
 B16F1 B16F1 Supplemental Figure S1 Representative microangiography images of B16F1 and B16F1 tumors grown in the cranial windows. FITC-dextran (2 million MW) was injected systemically to visualize blood
B16F1 B16F1 Supplemental Figure S1 Representative microangiography images of B16F1 and B16F1 tumors grown in the cranial windows. FITC-dextran (2 million MW) was injected systemically to visualize blood
Supplementary Material: Exploring wound-healing genomic machinery with a network-based approach
 Supplementary Material: Exploring wound-healing genomic machinery with a network-based approach Francesca Vitali, Simone Marini, Martina Balli, Hanne Grosemans, Maurilio Sampaolesi, Yves A. Lussier, Maria
Supplementary Material: Exploring wound-healing genomic machinery with a network-based approach Francesca Vitali, Simone Marini, Martina Balli, Hanne Grosemans, Maurilio Sampaolesi, Yves A. Lussier, Maria
103.5 Membrane metalloendopeptidase TGA GGG GTC ACG ATT TTA GG ATG ATG GTG AGG AGC AGG AC
 Supplementary Table SA Primers used for real-time RT-PCR gene regulation and their confirmation Gene Name Accession no Size Region Forward Primer Reverse Primer Efficiency% AKT v-akt oncogene homolog NM_00563
Supplementary Table SA Primers used for real-time RT-PCR gene regulation and their confirmation Gene Name Accession no Size Region Forward Primer Reverse Primer Efficiency% AKT v-akt oncogene homolog NM_00563
# 1 India s Best Selling Cancer Genomics Test Precision treatment for every Cancer Patient Patient Guide
 # 1 India s Best Selling Cancer Genomics Test Precision treatment for every Cancer Patient Patient Guide PositiveSelect is India s widely used Cancer Genomics test with every 8 out of 10 patients opting
# 1 India s Best Selling Cancer Genomics Test Precision treatment for every Cancer Patient Patient Guide PositiveSelect is India s widely used Cancer Genomics test with every 8 out of 10 patients opting
BD Rhapsody Immune Response Panel Hs
 Doc ID: 55665 Rev. 2.0 PRODUCT INSERT BD Rhapsody Immune Response Panel Hs BD Biosciences Kit Part Number: 633750 Purpose The BD Rhapsody Immune Response Panel Hs is a set of ready-to-use primer pools
Doc ID: 55665 Rev. 2.0 PRODUCT INSERT BD Rhapsody Immune Response Panel Hs BD Biosciences Kit Part Number: 633750 Purpose The BD Rhapsody Immune Response Panel Hs is a set of ready-to-use primer pools
Supplementary Table S1: Gene expression targets
 Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Exons from 137 genes were targeted for hybrid selection and massively parallel sequencing.
 SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
CD Marker Antibodies. atlasantibodies.com
 CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
q 2017 by The University of Chicago. All rights reserved. DOI: /690105
 q 2017 by The University of Chicago. All rights reserved. DOI: 10.1086/690105 Appendix from R. M. Cox et al., Hormonally Mediated Increases in Sex-Biased Gene Expression Accompany the Breakdown of Between-
q 2017 by The University of Chicago. All rights reserved. DOI: 10.1086/690105 Appendix from R. M. Cox et al., Hormonally Mediated Increases in Sex-Biased Gene Expression Accompany the Breakdown of Between-
Supplementary Figure 1. The overlap between statistically-significant and highconfidence
 Supplementary figures Supplementary Figure 1. The overlap between statistically-significant and highconfidence PPIs with previously reported PPIs in public databases. Venn diagrams showing the overlap
Supplementary figures Supplementary Figure 1. The overlap between statistically-significant and highconfidence PPIs with previously reported PPIs in public databases. Venn diagrams showing the overlap
Nature Immunology: doi: /ni.3745
 Supplementary Figure 1 Experimental settings. (a) Experimental setup and bioinformatic data analysis of transcriptomic changes induced in neonatal and adult monocytes by stimulation with 10 ng/ml LPS for
Supplementary Figure 1 Experimental settings. (a) Experimental setup and bioinformatic data analysis of transcriptomic changes induced in neonatal and adult monocytes by stimulation with 10 ng/ml LPS for
Implication of microrna regulation in para-phenylenediamine-induced cell death and senescence in normal human hair dermal papilla cells
 MOLECULAR MEDICINE REPORTS 12: 921-936, 2015 Implication of microrna regulation in para-phenylenediamine-induced cell death and senescence in normal human hair dermal papilla cells OK KYU LEE 1*, HWA JUN
MOLECULAR MEDICINE REPORTS 12: 921-936, 2015 Implication of microrna regulation in para-phenylenediamine-induced cell death and senescence in normal human hair dermal papilla cells OK KYU LEE 1*, HWA JUN
Practical implementation of personalized nutrition goals
 Practical implementation of personalized nutrition goals We all have our own nutritional needs because we are different in so many ways. Gender, age, body size, physical activity, genomic variation, gut
Practical implementation of personalized nutrition goals We all have our own nutritional needs because we are different in so many ways. Gender, age, body size, physical activity, genomic variation, gut
OncoMir Library Cancer Type Target Gene
 OncoMir Library Cancer Type Target Gene hsa-let-7a-1 Breast Cancer, Lung Cancer H RAS, HMGA2, CDK6, NRAS hsa-let-7a-2 Breast Cancer, Lung Cancer H RAS, HMGA2, CDK6, NRAS hsa-let-7a-3 Breast Cancer, Lung
OncoMir Library Cancer Type Target Gene hsa-let-7a-1 Breast Cancer, Lung Cancer H RAS, HMGA2, CDK6, NRAS hsa-let-7a-2 Breast Cancer, Lung Cancer H RAS, HMGA2, CDK6, NRAS hsa-let-7a-3 Breast Cancer, Lung
Case Study - Informatics
 bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
* Kyoto Encyclopedia of Genes and Genomes.
 Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Development of an In Vitro Human Liver System for Interrogating Non-Alcoholic. Brian R. Wamhoff, 501 Locust Ave., Charlottesville, VA 22902
 Supplemental Information Development of an In Vitro Human Liver System for Interrogating Non-Alcoholic Steatohepatitis Ryan Feaver, Banumathi K Cole, Mark J Lawson, Stephen A Hoang, Svetlana Marukian,
Supplemental Information Development of an In Vitro Human Liver System for Interrogating Non-Alcoholic Steatohepatitis Ryan Feaver, Banumathi K Cole, Mark J Lawson, Stephen A Hoang, Svetlana Marukian,
1. Jing Wang, Ph.D., Key Laboratory of Mental Health, Institute of Psychology,
 Network analysis reveals a stress-affected common gene module among seven stress-related diseases/systems which provide potential targets for mechanism research Liyuan Guo 1 *, Yang Du 1, 2 and Jing Wang
Network analysis reveals a stress-affected common gene module among seven stress-related diseases/systems which provide potential targets for mechanism research Liyuan Guo 1 *, Yang Du 1, 2 and Jing Wang
Regulation of gene expression in rats with spinal cord injury based on microarray data
 MOLECULAR MEDICINE REPORTS 12: 2465-2472, 2015 Regulation of gene expression in rats with spinal cord injury based on microarray data GUOQIANG CHEN, XIUTONG FANG and MENG YU Department of Orthopedic Surgery,
MOLECULAR MEDICINE REPORTS 12: 2465-2472, 2015 Regulation of gene expression in rats with spinal cord injury based on microarray data GUOQIANG CHEN, XIUTONG FANG and MENG YU Department of Orthopedic Surgery,
Supplemental Figure 1: Anti-GDF15 antibody blocks binding of GDF15 to hgfral.
 Supplemental Figure 1: Anti-GDF15 antibody blocks binding of GDF15 to hgfral. Anti-GDF15 antibody (mab26, a neutralizing monoclonal antibody to wtgdf15) blocks binding of GDF15 to human GFRAL in a dose-dependent
Supplemental Figure 1: Anti-GDF15 antibody blocks binding of GDF15 to hgfral. Anti-GDF15 antibody (mab26, a neutralizing monoclonal antibody to wtgdf15) blocks binding of GDF15 to human GFRAL in a dose-dependent
Supplemental Table 1: List of genes contained on the custom Steroltalk v2 microarray prepared for this study
 Supplemental Table 1: List of genes contained on the custom Steroltalk v2 microarray prepared for this study Gene GenBank No. Description Ch25h NM_009890 Cholesterol 25-hydroxylase Bile acid synthesis
Supplemental Table 1: List of genes contained on the custom Steroltalk v2 microarray prepared for this study Gene GenBank No. Description Ch25h NM_009890 Cholesterol 25-hydroxylase Bile acid synthesis
Department of International Health (Vaccine science track) Johns Hopkins University Bloomberg School of Public Health
 Acute Respiratory Distress Syndrome is a TH17 and Treg immune disease By Wan-Jiung Hu Postdoctorate Genomics Research Center Academia Sinica No 128 Academia Road section2 Nangang 115, Taipei, Taiwan Previous
Acute Respiratory Distress Syndrome is a TH17 and Treg immune disease By Wan-Jiung Hu Postdoctorate Genomics Research Center Academia Sinica No 128 Academia Road section2 Nangang 115, Taipei, Taiwan Previous
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Frampton et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing Supplementary Figure 1A Base substitution detection
SUPPLEMENTARY INFORMATION Frampton et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing Supplementary Figure 1A Base substitution detection
Genetic Testing and Analysis. (858) MRN: Specimen: Saliva Received: 07/26/2016 GENETIC ANALYSIS REPORT
 GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
The circulating T helper subsets and regulatory T cells in patients with common variable immunodeficiency with no monogenic disease
 1 Supplementary data The circulating T helper subsets and regulatory T cells in patients with common variable immunodeficiency with no monogenic disease Azizi et al. 2 Table S1. Excluded 373 genes variants
1 Supplementary data The circulating T helper subsets and regulatory T cells in patients with common variable immunodeficiency with no monogenic disease Azizi et al. 2 Table S1. Excluded 373 genes variants
We are IntechOpen, the world s leading publisher of Open Access books Built by scientists, for scientists. International authors and editors
 We are IntechOpen, the world s leading publisher of Open Access books Built by scientists, for scientists 4,000 116,000 120M Open access books available International authors and editors Downloads Our
We are IntechOpen, the world s leading publisher of Open Access books Built by scientists, for scientists 4,000 116,000 120M Open access books available International authors and editors Downloads Our
Late regulation of immune genes and micrornas in circulating leukocytes in a pig model of
 1 Supplementary material for: 2 3 4 5 6 Late regulation of immune genes and micrornas in circulating leukocytes in a pig model of influenza A (H1N2) infection Louise Brogaard, Peter M. H. Heegaard, Lars
1 Supplementary material for: 2 3 4 5 6 Late regulation of immune genes and micrornas in circulating leukocytes in a pig model of influenza A (H1N2) infection Louise Brogaard, Peter M. H. Heegaard, Lars
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature12912 Figure S1. Distribution of mutation rates and spectra across 4,742 tumor-normal (TN) pairs from 21 tumor types, as in Figure 1 from Lawrence et al. Nature
SUPPLEMENTARY INFORMATION doi:1.138/nature12912 Figure S1. Distribution of mutation rates and spectra across 4,742 tumor-normal (TN) pairs from 21 tumor types, as in Figure 1 from Lawrence et al. Nature
For focused group profiling of human HIV infection and host response genes expression
 ExProfile TM Human HIV Infection and Host Response Related Gene qpcr Array For focused group profiling of human HIV infection and host response genes expression Cat. No. QG023-A (1 x 96-well plate, Format
ExProfile TM Human HIV Infection and Host Response Related Gene qpcr Array For focused group profiling of human HIV infection and host response genes expression Cat. No. QG023-A (1 x 96-well plate, Format
Patricia Aoun MD, MPH Professor and Vice-Chair for Clinical Affairs Medical Director, Clinical Laboratories Department of Pathology City of Hope
 Patricia Aoun MD, MPH Professor and Vice-Chair for Clinical Affairs Medical Director, Clinical Laboratories Department of Pathology City of Hope National Medical Center Disclosures I have no disclosures
Patricia Aoun MD, MPH Professor and Vice-Chair for Clinical Affairs Medical Director, Clinical Laboratories Department of Pathology City of Hope National Medical Center Disclosures I have no disclosures
Supplementary Figure 1. Microarray data mining and validation
 Supplementary Figure 1. Microarray data mining and validation Microarray (>47,000 probes) Raw data Background subtraction NanoString (124 genes) Set cut-off and threshold values Raw data Normalized to
Supplementary Figure 1. Microarray data mining and validation Microarray (>47,000 probes) Raw data Background subtraction NanoString (124 genes) Set cut-off and threshold values Raw data Normalized to
BMSNF Inc., Aventura, FL 33180, USA; 4
 Int. J. Mol. Sci. 2015, 16, 25285-25322; doi:10.3390/ijms161025285 Article OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Integrated Bioinformatics, Environmental
Int. J. Mol. Sci. 2015, 16, 25285-25322; doi:10.3390/ijms161025285 Article OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Integrated Bioinformatics, Environmental
Influence of genetic ancestry and socioeconomic status on type 2 diabetes in the diverse Colombian populations of Chocó and Antioquia
 Supplementary Information For: Influence of genetic ancestry and socioeconomic status on type 2 diabetes in the diverse Colombian populations of Chocó and Antioquia Aroon T. Chande 1,2,3, Jessica Rowell
Supplementary Information For: Influence of genetic ancestry and socioeconomic status on type 2 diabetes in the diverse Colombian populations of Chocó and Antioquia Aroon T. Chande 1,2,3, Jessica Rowell
Hepatocellular Carcinoma (HCC): What Treatment Modality for Which Tumor?
 K. Rajender Reddy, MD, CG Hepatocellular Carcinoma (HCC): What Treatment Modality for Which Tumor? K. Rajender Reddy, MD, CG Ruimy Family President s Distinguished Professor of Medicine Director of Hepatology
K. Rajender Reddy, MD, CG Hepatocellular Carcinoma (HCC): What Treatment Modality for Which Tumor? K. Rajender Reddy, MD, CG Ruimy Family President s Distinguished Professor of Medicine Director of Hepatology
Inferring the Mammalian Cardiogenic Gene Regulatory Network. Jason Bazil 10/18/13 Cardiac Physiome Workshop 2013
 Inferring the Mammalian Cardiogenic Gene Regulatory Network Jason Bazil 1/18/13 Cardiac Physiome Workshop 213 Reverse Engineering Gene Regulatory Networks Two main questions need addressed: 1) What s connected
Inferring the Mammalian Cardiogenic Gene Regulatory Network Jason Bazil 1/18/13 Cardiac Physiome Workshop 213 Reverse Engineering Gene Regulatory Networks Two main questions need addressed: 1) What s connected
 Table S1. Metadata for transcriptome interaction network and pathway analysis of 5448 intracelluarly infected TEpi cells in comparison to mock TEpi cells. Gene symbol Full name Log2FC gene expression in
Table S1. Metadata for transcriptome interaction network and pathway analysis of 5448 intracelluarly infected TEpi cells in comparison to mock TEpi cells. Gene symbol Full name Log2FC gene expression in
Supporting Information
 Supporting Information Supporting Information Corrected October 10, 2013 Pawlikowski et al. 10.1073/pnas.1303491110 SI Materials and Methods Microarray. Microarray samples were prepared and processed as
Supporting Information Supporting Information Corrected October 10, 2013 Pawlikowski et al. 10.1073/pnas.1303491110 SI Materials and Methods Microarray. Microarray samples were prepared and processed as
Expression Profiling of KLF4
 Expression Profiling of KLF4 AJCR0000006 Supplemental Data Figure S1. Snapshot of enriched gene sets identified by GSEA in Klf4-null MEFs. Figure S2. Snapshot of enriched gene sets identified by GSEA in
Expression Profiling of KLF4 AJCR0000006 Supplemental Data Figure S1. Snapshot of enriched gene sets identified by GSEA in Klf4-null MEFs. Figure S2. Snapshot of enriched gene sets identified by GSEA in
An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action
 An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action Supplementary Figure : Expression levels of toll-like receptor 4 (Tlr4) in muse lung does not change throughout the
An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action Supplementary Figure : Expression levels of toll-like receptor 4 (Tlr4) in muse lung does not change throughout the
Supplemental Table 1: Enriched GO categories per cluster
 Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
The Center for PERSONALIZED DIAGNOSTICS
 The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
 Supplementary Table 1. Annotated candidate SNV and indels in melanoma tumors. Patient C1 Patient C2 Patient C3 Patient C4 Patient C5 Patient C6 Patient C7 Patient C10 Chr Position Reference Allele Sample
Supplementary Table 1. Annotated candidate SNV and indels in melanoma tumors. Patient C1 Patient C2 Patient C3 Patient C4 Patient C5 Patient C6 Patient C7 Patient C10 Chr Position Reference Allele Sample
Statistical Issues in Translational Cancer Research
 Statistical Issues in Translational Cancer Research Martin Bøgsted Department of Haematology Aalborg University Hospital and Department of Clinical Medicine Aalborg University The only useful function
Statistical Issues in Translational Cancer Research Martin Bøgsted Department of Haematology Aalborg University Hospital and Department of Clinical Medicine Aalborg University The only useful function
Nature Genetics: doi: /ng Supplementary Figure 1. Depths and coverages in whole-exome and targeted deep sequencing data.
 Supplementary Figure 1 Depths and coverages in whole-exome and targeted deep sequencing data. Depth (top) and coverage (bottom) of whole-exome sequencing for 38 independent JPN cases (mean depth = 130)
Supplementary Figure 1 Depths and coverages in whole-exome and targeted deep sequencing data. Depth (top) and coverage (bottom) of whole-exome sequencing for 38 independent JPN cases (mean depth = 130)
Genes, Genes, Everywhere! The key to a cure for IBD
 Genes, Genes, Everywhere! The key to a cure for IBD Dermot McGovern MD, PhD, FRCP Director, Translational Medicine Professor of Medicine Joshua L and Lisa Z Greer Endowed Chair in IBD Genetics F. Widjaja
Genes, Genes, Everywhere! The key to a cure for IBD Dermot McGovern MD, PhD, FRCP Director, Translational Medicine Professor of Medicine Joshua L and Lisa Z Greer Endowed Chair in IBD Genetics F. Widjaja
Supplementary Figure 1!
 a!!gt-cre x Twist LoxP/LoxP!!GT-Cre x Twist LoxP/LoxP! x RosaLSLEYFP!!GT-Cre x Snail LoxP/LoxP!!GT-Cre x Snail LoxP/LoxP! x RosaLSLEYFP! Supplementary Figure!!GT-Cre x Twist LoxP/LoxP! x RosaLSLEYFP x
a!!gt-cre x Twist LoxP/LoxP!!GT-Cre x Twist LoxP/LoxP! x RosaLSLEYFP!!GT-Cre x Snail LoxP/LoxP!!GT-Cre x Snail LoxP/LoxP! x RosaLSLEYFP! Supplementary Figure!!GT-Cre x Twist LoxP/LoxP! x RosaLSLEYFP x
Applied Biosystems models 7500 (Fast block), 7900HT (Fast block), StepOnePlus, ViiA 7 (Fast block)
 RT² Profiler PCR Array (96-Well Format and 384-Well [4 x 96] Format) Human HIV Infection and Host Response Cat. no. 330231 PAHS-051YA For pathway expression analysis Format Format A Format C Format D Format
RT² Profiler PCR Array (96-Well Format and 384-Well [4 x 96] Format) Human HIV Infection and Host Response Cat. no. 330231 PAHS-051YA For pathway expression analysis Format Format A Format C Format D Format
The innate immune system: Contributions to Immunogenicity of Therapeutic Proteins
 The innate immune system: Contributions to Immunogenicity of Therapeutic Proteins Daniela Verthelyi, M.D., Ph.D. Laboratory of Immunology Division of Therapeu8c Proteins OBP, CDER, FDA Map to the talk:
The innate immune system: Contributions to Immunogenicity of Therapeutic Proteins Daniela Verthelyi, M.D., Ph.D. Laboratory of Immunology Division of Therapeu8c Proteins OBP, CDER, FDA Map to the talk:
SureSelect Cancer All-In-One Custom and Catalog NGS Assays
 SureSelect Cancer All-In-One Custom and Catalog NGS Assays Detect all cancer-relevant variants in a single SureSelect assay SNV Indel TL SNV Indel TL Single DNA input Single AIO assay Single data analysis
SureSelect Cancer All-In-One Custom and Catalog NGS Assays Detect all cancer-relevant variants in a single SureSelect assay SNV Indel TL SNV Indel TL Single DNA input Single AIO assay Single data analysis
A brain tumor and NGS/multiplexing
 A brain tumor and NGS/multiplexing Santiago Ramón y Cajal Jefe de Servicio. Hospital Vall d Hebron Catedrático de Anatomía Patológica U.A.B. Académico de Número de la Real Academia Nacional de Medicina
A brain tumor and NGS/multiplexing Santiago Ramón y Cajal Jefe de Servicio. Hospital Vall d Hebron Catedrático de Anatomía Patológica U.A.B. Académico de Número de la Real Academia Nacional de Medicina
SI Table. SI Table 3. SI Table 4. SI Table 5A. SI Table 5B. SI Table 5C. SI Table 6A. SI Table 6B. SI Table 6C
 SI Table SI Table 1 SI Table 2 SI Table 3 SI Table 4 SI Table 5A SI Table 5B SI Table 5C SI Table 6A SI Table 6B SI Table 6C Title List of primers used to clone HCV genes and gene fragments HCV genotype
SI Table SI Table 1 SI Table 2 SI Table 3 SI Table 4 SI Table 5A SI Table 5B SI Table 5C SI Table 6A SI Table 6B SI Table 6C Title List of primers used to clone HCV genes and gene fragments HCV genotype
Supplementary appendix
 Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Ahlqvist E, Storm P, Käräjämäki A, et al. Novel
Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Ahlqvist E, Storm P, Käräjämäki A, et al. Novel
EMT and ECM. Jesse Liang, Ph.D. Sample & Assay Technologies
 EMT and ECM Jesse Liang, Ph.D. 1 Epithelial Cells.1. Closely adjoined.2. Polarized.Epithelial Markers: E-Cadherin (adherens junctions) Claudins (tight junctions) Occludin (tight junctions) Desmoplakin
EMT and ECM Jesse Liang, Ph.D. 1 Epithelial Cells.1. Closely adjoined.2. Polarized.Epithelial Markers: E-Cadherin (adherens junctions) Claudins (tight junctions) Occludin (tight junctions) Desmoplakin
ORDERING HEALTHCARE PROFESSIONAL. Nilesh Dharajiya, M.D Nexus Center Drive San Diego, CA US
 Test Results Reviewed & Approved by: Laboratory Director, Nilesh Dharajiya, M.D. PATHWAY IT PERSONAL DETAILS DOB Jan 1, 19XX ETHNICITY Caucasian ORDERING HEALTHCARE PROESSIONAL Nilesh Dharajiya, M.D. 4755
Test Results Reviewed & Approved by: Laboratory Director, Nilesh Dharajiya, M.D. PATHWAY IT PERSONAL DETAILS DOB Jan 1, 19XX ETHNICITY Caucasian ORDERING HEALTHCARE PROESSIONAL Nilesh Dharajiya, M.D. 4755
SALSA MLPA Probemix P014-B1 Chromosome 8 Lot B and B
 SALSA MLPA Probemix P014-B1 Chromosome 8 Lot B1-0916 and B1-0713. Copy number changes of the human chromosome 8 are common in many types of tumours. In most cases, losses of 8p sequences and gains of 8q
SALSA MLPA Probemix P014-B1 Chromosome 8 Lot B1-0916 and B1-0713. Copy number changes of the human chromosome 8 are common in many types of tumours. In most cases, losses of 8p sequences and gains of 8q
Phosphorylation Site Company Cat #
 Supplemental Table 1. Antibodies used for RPPA analysis. Label Protein Phosphorylation Site Company Cat # Used on MDA_CLSS Used on MDA_Pilot 4EBP1 4EBP1 Cell Signaling 9452 No 4EBP1.pS65 4EBP1 S65 Cell
Supplemental Table 1. Antibodies used for RPPA analysis. Label Protein Phosphorylation Site Company Cat # Used on MDA_CLSS Used on MDA_Pilot 4EBP1 4EBP1 Cell Signaling 9452 No 4EBP1.pS65 4EBP1 S65 Cell
Clinically Useful Next Generation Sequencing and Molecular Testing in Gliomas MacLean P. Nasrallah, MD PhD
 Clinically Useful Next Generation Sequencing and Molecular Testing in Gliomas MacLean P. Nasrallah, MD PhD Neuropathology Fellow Division of Neuropathology Center for Personalized Diagnosis (CPD) Glial
Clinically Useful Next Generation Sequencing and Molecular Testing in Gliomas MacLean P. Nasrallah, MD PhD Neuropathology Fellow Division of Neuropathology Center for Personalized Diagnosis (CPD) Glial
Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester
 Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester dsg6@le.ac.uk CFDNA/CTDNA Circulating-free AS A LIQUID DNA BIOPSY (cfdna) Tumour Biopsy Liquid Biopsy
Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester dsg6@le.ac.uk CFDNA/CTDNA Circulating-free AS A LIQUID DNA BIOPSY (cfdna) Tumour Biopsy Liquid Biopsy
