USP19-deubiquitinating enzyme regulates levels of major myofibrillar proteins in L6 muscle cells
|
|
|
- Geraldine Carson
- 5 years ago
- Views:
Transcription
1 Am J Physiol Endocrinol Metab 297: E1283 E1290, First published September 22, 2009; doi: /ajpendo USP19-deubiquitinating enzyme regulates levels of major myofibrillar proteins in L6 muscle cells Priyanka Sundaram, 1,2 * Zhiyu Pang, 1 * Miao Miao, 1,2 *LuYu, 1 and Simon S. Wing 1,2 1 Department of Medicine and 2 Department of Biochemistry, Polypeptide Laboratory, McGill University and the McGill University Health Centre Research Institute, Montreal, Quebec, Canada Submitted 26 June 2009; accepted in final form 15 September 2009 Sundaram P, Pang Z, Miao M, Yu L, Wing SS. USP19-deubiquitinating enzyme regulates levels of major myofibrillar proteins in L6 muscle cells. Am J Physiol Endocrinol Metab 297: E1283 E1290, First published September 22, 2009; doi: /ajpendo The ubiquitin-proteasome system plays an important role in the degradation of myofibrillar proteins that occurs in muscle wasting. Many studies have demonstrated the importance of enzymes mediating conjugation of ubiquitin. However, little is known about the role of deubiquitinating enzymes. We previously showed that the USP19- deubiquitinating enzyme is induced in atrophying skeletal muscle (Combaret L, Adegoke OA, Bedard N, Baracos V, Attaix D, Wing SS. Am J Physiol Endocrinol Metab 288: E693 E700, 2005). To further explore the role of USP19, we used small interfering RNA (sirna) in L6 muscle cells. Lowering USP19 by 70 90% in myotubes resulted in a 20% decrease in the rate of proteolysis and an 18% decrease in the rate of protein synthesis, with no net change in protein content. Despite the decrease in overall synthesis, there were 1.5-fold increases in protein levels of myosin heavy chain (MHC), actin, and troponin T and a 2.5-fold increase in tropomyosin. USP19 depletion also increased MHC and tropomyosin mrna levels, suggesting that this effect is due to increased transcription. Consistent with this, USP19 depletion increased myogenin protein and mrna levels approximately twofold. Lowering myogenin using sirna prevented the increase in MHC and tropomyosin upon USP19 depletion, indicating that myogenin mediated the increase in myofibrillar proteins. Dexamethasone treatment lowered MHC and increased USP19. Depletion of USP19 reversed the dexamethasone suppression of MHC. These studies demonstrate that USP19 modulates transcription of major myofibrillar proteins and indicate that the ubiquitin system not only mediates the increased protein breakdown but is also involved in the decreased protein synthesis in atrophying skeletal muscle. ubiquitin-specific processing protease * These authors made equal contributions. Address for reprint requests and other correspondence: S. S. Wing, Polypeptide Laboratory, McGill University, Strathcona Anatomy and Dentistry Bldg., 3640 University St., Rm. W315, Montreal, QC, Canada, H3A 2B2 ( simon.wing@mcgill.ca). SKELETAL MUSCLE IS THE MOST ABUNDANT TISSUE in the human body, accounting for approximately one-half of body weight. As a major site of metabolic activity, it directly converts chemical energy into mechanical energy and heat. It is also the largest reservoir of protein in the body. During starvation, muscle protein is degraded to provide amino acids for energy and for gluconeogenesis to maintain blood glucose. Muscle protein catabolism also occurs in a variety of pathological conditions such as disuse, loss of innervation, cancer, sepsis, hypercortisolism, and uncontrolled diabetes mellitus and results in significant morbidity (reviewed in Ref. 25). Therefore, much attention has been given to elucidating the biochemical mechanisms involved in mediating protein catabolism in muscle. These studies have shown that ubiquitin-proteasome-dependent proteolysis plays an important role in this process (reviewed in Ref. 1). The highly conserved 76-amino acid peptide ubiquitin is expressed in all eukaryotic cells and exerts its cellular functions through its ability to be conjugated to other proteins. This conjugation is catalyzed sequentially by a cascade of three enzymes: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin protein ligase (E3) (reviewed in Ref. 35). When conjugated as a chain of more than four ubiquitin moieties, it can target the attached protein to the proteasome for degradation (41). Previous studies have shown that, in various catabolic conditions, steady-state levels of ubiquitinated proteins increase in the atrophying muscles (2, 26, 29, 42, 47). This suggests that there is an activation of ubiquitination that increases flux through the ubiquitin-proteasome pathway by increasing availability of ubiquitinated substrates for the proteasome. Supporting this model are many studies that have shown increased expression of many components involved in ubiquitin conjugation in various forms of muscle wasting (reviewed in Ref. 44). Increased expression in skeletal muscle of polyubiquitin genes (2, 4, 27, 40, 43, 46), ubiquitin-conjugating enzymes E2 14k, UBC-E2G, and UBC4 (7, 28, 40, 45), and ubiquitin protein ligases atrogin-1 (MAFBx), muscle RING finger-1 (MURF-1) (3, 14), and UBR2 (22) has been described in catabolic conditions. Increased expression of proteasome subunits (2, 5, 28, 36) has also been seen. Solid evidence for a role of the ubiquitin proteolytic system was confirmed with studies demonstrating that genetic inactivation in mice of the E3s MURF-1 or MAFbx/atrogin-1 (3) or of the ubiquitin-binding protein ZNF216 (18) leads to decreased loss of muscle mass following denervation. Up to now, most of the studies of the ubiquitin system in muscle wasting have focused on the enzymes involved in the conjugation of ubiquitin to proteins. However, ubiquitination can also be modulated by deubiquitinating enzymes (reviewed in Ref. 38), and therefore, these may also be involved in the wasting process (44). Structurally, deubiquitinating enzymes can be divided broadly on the basis of sequence homology into five classes (reviewed in Ref. 33). The largest class ( 75 in the human genome) are the ubiquitin-specific processing proteases (USPs). USPs are recognized by the presence of a core catalytic domain of 450 amino acids delimited by cysteine and histidine boxes. Surrounding sequences are more divergent and are likely involved in mediating substrate specificity and other protein interactions. To date, only two deubiquitinating enzymes, USP14 and USP19, have been observed to be regulated in atrophying muscle, and both were induced rather than suppressed, suggesting that they do not act as negative regu /09 $8.00 Copyright 2009 the American Physiological Society E1283
2 E1284 lators of ubiquitination of the targets of the ligases that are induced in the same catabolic muscles. Two other enzymes, USP2 and USP28, appear to be regulated by muscle stretch (39). USP14 (orthologous to yeast UBP6) was observed to be upregulated in a gene array study of muscle mrna expression in various conditions of wasting (24). This enzyme is widely expressed and associated with the proteasome (17) and appears to modulate substrate ubiquitination and proteasome activity. We previously identified USP19 as a USP that is induced in skeletal muscle atrophying in response to various catabolic conditions, including cancer, fasting, diabetes, and glucocorticoids (10). The expression of USP19 correlated inversely but very well with muscle mass, consistent with a role in atrophy (10). To further explore the potential functions of this enzyme in muscle atrophy, we determined the effects of sirnamediated depletion of this enzyme on the expression of major myofibrillar proteins in L6 myotubes. MATERIALS AND METHODS Cell culture and transfection. L6 rat muscle cells (Dr. Amira Klip, University of Toronto) were grown at 37 C with 5% CO 2 in -MEM (GIBCO) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Cells were induced to differentiate by replacing the medium with -MEM supplemented with 2% fetal bovine serum and 1% penicillin-streptomycin. L6 cells were seeded in six-well plates at a density of cells/well (for Western blot analysis) or in 60-mm plates at a density of cells/plate (for Northern blot analysis) and incubated overnight in growth medium. The next morning, the cells were 80% confluent and were transfected with 25 nm USP19 sirna or nonspecific control (CTL) sirna oligonucleotides (IDT) using Lipofectamine and Plus Reagent (Invitrogen), following the manufacturer s protocol. For myogenin and USP19 double-sirna experiments, cells were transfected with 50 nm CTL, 25 nm CTL, and 25 nm USP19 or 25 nm CTL and 25 nm myogenin sirna oligonucleotides. Three hours posttransfection, the transfection mixture was supplemented with an equal volume of -MEM containing 20% fetal bovine serum, and the cells were grown overnight at 37 C. Some samples were harvested 24 h posttransfection (day 0), whereas the remaining cells were induced to differentiate into myotubes by incubating the cells in differentiation medium. Samples were harvested at indicated time points up to day 4 of differentiation. The differentiation medium was changed every other day. To induce catabolism in the myotubes, the synthetic glucocorticoid dexamethasone (Sigma-Aldrich) was added to L6 myotubes in differentiation medium on day 2. Onday 3 the medium was replaced with fresh medium containing dexamethasone, and the cells were harvested on day 4. sirna sequences were as follows: nonspecific CTL, 5 -AAA CUC UAU CUG CAC GCU GAC-3 and 5 -GUC AGC GUG CAG AUA GAG UUU-3 ; USP19 no. 7, 5 -AAG GGU GGU CUU CUA CAG UUG-3 and 5 -CAA CUG UAG AAG ACC ACC CUU-3 ; USP19 no. 43, 5-5Phos/GGC GUG ACA AGA UCA AUG ACU UGG T-3 and 5 -ACC AAG UCA UUG AUC UUG UCA CGC CAA-3 ; myogenin, 5 -UAU CCA CUG CAA AUG CUU GTT-3 and 5 -CAA GCA UUU GCA GUG GAU ATT-3. Measurements of rates of protein turnover. Protein synthesis was measured in L6 cells on day 4 of differentiation by incubating the cells for2hindifferentiation medium containing 5 Ci of [ 3 H]tyrosine/ml and 2.0 mm nonradioactive tyrosine. The amount of labeled amino acid incorporated into protein was determined as described previously (15). To measure protein degradation, muscle cell proteins were radiolabeled after 1 day of differentiation by incubating the cells in differentiation medium containing 1 Ci of [ 3 H]tyrosine/ml and 2.0 mm nonradioactive tyrosine for 48 h. Subsequently, the monolayer of cells was rinsed three times with HBSS containing 2.0 mm nonradioactive tyrosine and then replaced with 2 ml of fresh differentiation medium containing 2.0 mm nonradioactive tyrosine. After incubation for 3 h to allow turnover of proteins with very short half-lives, the process of protein degradation was measured by collecting 0.5 ml of the cell culture medium at various time points 33 h and measuring the acid-soluble radioactivity. These subsequent steps were taken as described previously (15). Western blotting. Cells were lysed by adding 250 l of50mm Tris, ph 7.5, and 2% SDS into the wells of the six-well plate, followed by scraping to collect the cell lysate. DNA in the lysate was sheared using a 1-ml insulin syringe, and the protein concentrations of the samples were determined by the BCA Micro Protein Assay (Thermo Fisher Scientific). Samples (20 or 40 g) were subjected to SDS- PAGE, and the proteins were transferred onto m nitrocellulose membranes for Western blotting. Membranes containing 20 g of protein were probed with antibodies to myosin heavy chain (MHC; MF20, Developmental Studies Hybridoma Bank), -tubulin (Sigma Aldrich), actin (A4700; Sigma Aldrich), troponin (Sigma Aldrich), or tropomyosin (CH1; Developmental Studies Hybridoma Bank). Membranes with 40 g of protein were probed for USP19 (30), -tubulin, myogenin (F5D; Santa Cruz Biotechnology), or Myf5 (sc-302; Santa Cruz Biotechnology). Horseradish peroxidase-conjugated secondary antibodies and ECL Plus were used to detect all of the bound primary antibodies except for MF20, which was detected using an iodinated secondary antibody and a phosophoimager to avoid signal saturation. Levels of individual proteins were normalized to the level of tubulin on the same blot to control for variations in sample loading and transfer to the membrane. Northern blotting. RNA was isolated by solublizing the cells in 4 M guanidinium isothiocyanate followed by phenol-choloroform extraction (6). Northern blots were performed by electrophoresing 10 g of each RNA sample in 1% agarose gels containing formaldehyde, transferring the RNA onto nylon membranes overnight, and then cross-linking the nuclei acid to the membrane with UV light. Singlestranded 32 P-labeled probes complementary to the indicated transcripts were hybridized to the membranes at 65 C overnight and, following washing, exposed to phosphorimager screens for quantitation. Levels of individual mrna transcripts were normalized to the level of 18S ribosomal RNA on the same blot to control for variations in sample loading and transfer to the membrane. 32 P-labeled probes were synthesized by PCR using myogenin, MHC, and tropomyosin cdna templates corresponding to nucleotide fragments , 5,375 5,929, and , respectively. The following reverse primer sequences were used for the PCR: myogenin, 5 -GCA- GAAGTGGTGGCGTCTGACAC-3 ; MHC (2X isoform), 5 -GGT- CACTTTCCTGCTTTGGA-3 and tropomyosin 5 -AATGCTTTCA- AGGTCTGA-3. RESULTS Depletion of USP19 modestly decreases the overall rates of protein synthesis and degradation. Since USP19 expression was previously observed to be closely but inversely correlated with muscle mass in rodents (10), we determined the effects of loss of USP19 on rates of protein turnover in L6 myotubes (Fig. 1). L6 myoblasts were transfected with USP19 or a nonspecific control sirna and induced to differentiate into myotubes for 4 days. As was previously observed in rat fibroblasts (30), the USP19 sirna oligonucleotides were effective, lowering USP19 levels by 70 90% (Fig. 2A). USP19 sirna reduced the rate of incorporation of 3 [H]tyrosine into protein by 18% compared with control sirna. We also measured the rate of proteolysis as the rate of release of acidsoluble radioactivity from cells that had proteins prelabeled for 48 h with 3 [H]tyrosine. The rate of proteolysis was 20% lower
3 Fig. 1. Depletion of ubiquitin-specific processing protease-19 (USP19) in L6 myotubes slightly decreases both rates of protein synthesis and degradation. A: L6 cells were transfected with either control (CTL) or USP19 small interfering RNA (sirna) oligonucleotides. After 4 days in differentiation medium, the cells were incubated with [ 3 H]tyrosine. Two hours later protein was isolated, and rates of incorporation of radioactive amino acid into protein were measured (shown are means SE; n 4; means are significantly different, P 0.005). B: L6 cells were transfected with either CTL or USP19 sirna oligonucleotides. After 1 day in differentiation medium, the cells were incubated with medium containing [ 3 H]tyrosine. Two days later, the medium was changed and the rate of release of free radiolabeled tyrosine into the medium measured at the indicated time points. Shown are means SE; n 4 and the rate equations obtained by linear regression analysis. CTL significantly different from USP19. P by 2-way ANOVA. in cells transfected with USP19 sirna than in control transfected cells. The similar decreases in the rates of protein synthesis and degradation indicate that overall protein turnover was probably not affected, and indeed, total protein content was unchanged by USP19 sirna (data not shown). Depletion of USP19 increases the expression of major myofibrillar proteins. Although depletion of USP19 did not affect overall protein turnover, there could still be specific effects on myofibrillar proteins, which, although they are the dominant proteins in differentiated muscle, are minor proteins in myotubes in culture. To test this hypothesis, we determined how loss of USP19 affected levels of major myofibrillar proteins in these muscle cells. Myotubes were collected after 4 days of differentiation, and the cell lysates were analyzed by Western blotting (Fig. 2). Depletion of USP19 increased levels of MHC, actin, and troponin T 1.5-fold and tropomyosin two- to threefold. There were no changes in levels of a nonmyofibrillar cytoskeletal protein, tubulin, indicating that these effects were specific. These effects were observed with two independent well-characterized sirna oligonucleotides (30), indicating E1285 that this was unlikely to be due to off-target effects. Since myofibrillar proteins are known to have very long half-lives [e.g., MHC half-life of 27 h (16)], these effects are likely due to increased synthesis of these proteins. Indeed, depletion of USP19 also increased levels of mrna transcripts of MHC (Fig. 3, A and C) and tropomyosin (Figs. 3, B and D), suggesting that USP19 acts at the level of gene transcription to increase the expression of these major muscle proteins. Depletion of USP19 increases the expression of the myogenic regulatory factor myogenin. The increase in a broad range of myofibrillar proteins suggested that USP19 may be modulating a general transcriptional pathway that controls the expression of myofibrillar proteins. Myogenesis is dependent on a family of myogenic regulatory factors consisting of MyoD, myogenin, Myf5, and Myf6 (Mrf4) (reviewed in Ref. 34). MyoD is not expressed in L6 cells, and Myf6 is expressed only in terminal stages of differentiation that are not recapitulated in the in vitro system (11). Therefore, we tested the effect of depletion of USP19 on myogenin and Myf5, which are expressed in L6 cells (Fig. 4). Transfection of the cells with USP19 sirna oligonucleotides did not affect Myf5 protein levels. However, myogenin protein levels were approximately twofold higher in cells depleted of USP19 on day 1, and they remained high in these cells until day 4, at which time they returned toward control levels. (Fig. 4, A and B). This was despite persistent low levels of USP19 on day 4 and suggests that other adaptations occur to perhaps limit differentiation to an optimal level of maturation. The overall increase in myogenin expression was associated with a slight but statistically significant increase in the extent of muscle cell fusion (defined as %cells containing 2 or more nuclei): CTL sirna, %; USP19 sirna, %. To determine whether the increased myogenin protein levels in USP19-depleted cells were due to an increase in the levels of myogenin transcript, Northern blots were performed on RNA isolated from cells collected from day 0 to day 3 (Fig. 4C). USP19 sirna resulted in an approximately twofold increase in myogenin mrna on day 1 compared with CTLtransfected cells, and this persisted to day 3 (Fig. 4D). Taken together, these results indicate that USP19 regulates the expression of the myogenic regulatory factor myogenin. USP19 regulates the expression of muscle-specific genes in a myogenin-dependent manner. To test whether USP19 depletion increases the expression of myofibrillar proteins through the increased expression of myogenin, we determined the effect of lowering myogenin levels on the ability of USP19 to modulate expression of MHC and tropomyosin (Fig. 5). We selected an sirna oligonucleotide that would not silence myogenin expression completely but would, upon concomitant treatment with myogenin and USP19 sirna oligonucleotides, result in myogenin levels similar to levels in control sirnatransfected cells. As described above, silencing of USP19 increased levels of myogenin (Fig. 5A), MHC (Fig. 5, B and C), and tropomyosin (Fig. 5, B and D). However, these increases were no longer seen when myogenin was simultaneously depleted (Fig. 5, B D). These results argue that USP19 does indeed act through myogenin to negatively regulate the expression of major muscle proteins. Depletion of USP19 can inhibit the loss of MHC induced by dexamethasone. Excess glucocorticoids are well known to activate muscle proteolysis and appear to have a preferential
4 E1286 Fig. 2. Depletion of USP19 increases the expression of major skeletal muscle myofibrillar proteins. A: L6 myoblasts were transfected with nonspecific CTL or 2 distinct USP19 sirna oligonucleotides (nos. 7 and 43) and induced to differentiate for 4 days. Western blots were performed to examine the effects of USP19 depletion on the protein levels of myosin heavy chain (MHC), actin, troponin T, and tropomyosin. B: quantitation of protein levels from A. Shown are means SE; n 7 18 for each treatment. *P compared with CTL (1-way ANOVA). effect on myofibrillar proteins (20). We observed previously that USP19 expression increases in catabolic conditions, including dexamethasone treatment (10). Other investigators have reported that incubation of L6 myotubes with dexamethasone can increase overall protein breakdown (32) and lower levels of MHC (13). Therefore, we tested whether dexamethasone can induce USP19 in muscle cells and whether lowering USP19 in muscle cells can reverse the loss of MHC induced by this catabolic stimulus (Fig. 6). We observed a small ( 20%) but reproducible increase in USP19 levels in the dexamethasone-treated myotubes and confirmed that dexamethasone is able to lower MHC levels in L6 myotubes (Fig. 6, A and C). Silencing of USP19 in these muscle cells was indeed able to reverse the loss of MHC protein induced by dexamethasone to levels similar to cells treated with control sirna but not to the higher levels seen with USP19 sirna alone (Fig. 6, A and B). DISCUSSION We have demonstrated for the first time that a deubiquitinating enzyme can regulate the expression of a panel of major myofibrillar proteins, including MHC, actin, tropomyosin, and troponin. Loss of USP19 in L6 myotubes increased the level of these proteins (Fig. 2). Although it is known that myofibrillar proteins such as MHC, myosin light chains (8, 9), and troponin I (21) are ubiquitinated in muscle, these myofibrillar proteins do not appear to be direct substrates of USP19, because sirna-mediated loss of this deubiquitinating activity would have increased their ubiquitination and degradation and therefore lowered rather than raised their levels. Instead, our data argue that USP19 negatively regulates a transcriptional program of muscle-specific genes. mrna levels of MHC and tropomyosin were found to be elevated upon USP19 depletion Fig. 3. Depletion of USP19 increases mrna levels for MHC and tropomyosin. L6 myoblasts were transfected with either nonspecific CTL or USP19 sirna oligonucleotides (nos. 7 and 43) and induced to differentiate for 3 days. RNA were prepared from the lysates and analyzed by Northern blotting to examine the effects of USP19 depletion on the mrna levels of MHC (A) and tropomyosin (B). C and D: results for MHC and tropomyosin, respectively, were quantified. Shown are means SE (n 7). *P 0.05 compared with CTL (1-way ANOVA).
5 E1287 Fig. 4. USP19 regulates the expression of the myogenic regulatory factor myogenin. A: L6 myoblasts were transfected with nonspecific CTL or USP19 sirna oligonucleotides (nos. 7 and 43) and induced to diffentiate for the indicated times. Cell lysates were analyzed by Western blotting for Myf5 or myogenin. Representative blots on day 1 are shown. B: quantitation of myogenin protein levels during the course of differentiation. Shown are means SE (n 7). *P compared with CTL for the particular day of differentiation. C: representative Northern blots of myogenin mrna from day 0 to day 3 of differentiation. D: quantitation of myogenin mrna. Shown are means SE (n 4). *P 0.05 compared with CTL for the particular day of differentiation (2-way ANOVA). (Fig. 3). Myogenic regulatory factors are a group of musclespecific transcription factors that are master regulators of skeletal muscle development (34). We found for the first time that USP19 can modulate transcription of myogenin, a myogenic regulatory factor member that is increased during muscle differentiation, because silencing of USP19 increased levels of both myogenin mrna and protein (Fig. 4). However, USP19 does not necessarily act at the beginning of the differentiation process, because similar increases in expression of MHC could be observed when L6 cells were transfected with USP19 sirna oligonucleotides on day 2 of differentiation and harvested on day 4 (data not shown). In addition, the increase in myogenin was associated with only a small, 15% increase in the number of myotubes, suggesting that most of the stimulus of myogenic differentiation is achieved with myogenin levels present in the cells transfected with the control oligonucleotides. The mechanism by which USP19 regulates myogenin remains unclear. The simplest model is that a transcriptional repressor of myogenin transcription is a substrate of USP19 and can be destabilized upon silencing of USP19. However, at this time, we cannot exclude the possibility that USP19 acts on substrate(s) upstream in the signaling pathways that modulate the synthesis of myofibrillar proteins. Silencing of USP19 did lower the rate of overall protein degradation by 20%, and we cannot exclude that this small change contributes to the overall increase in steady-state levels
6 E1288 Fig. 5. USP19 inhibits the expression of muscle-specific genes in a myogenin-dependent manner. L6 myoblasts were transfected either singly with nonspecific CTL, USP19 (no. 7), or myogenin sirna or doubly with both myogenin and USP19 sirna oligonucleotides and induced to differentiate for 3 days. Cell lysates were analyzed by Western blotting for myogenin and USP19 (A) and MHC and tropomyosin (B). Effects of single and double sirna-mediated depletion on tropomyosin and MHC proteins were quantified in C and D, respectively (n 4). *P 0.05 compared with CTL (1-way ANOVA). of myofibrillar proteins. However, in view of the previously measured half-life of 27 h for MHC in myotubes (16), USP19 sirna would have to prolong this half-life markedly to induce a 50% increase in steady-state levels. In addition, the fact that preventing the increase in myogenin expression completely blocks the increase in myofibrillar proteins (Fig. 5) also argues that proteolysis does not play a major role in the regulation of myofibrillar protein levels by USP19. Fig. 6. Depletion of USP19 can reverse dexamethasone-induced loss of MHC. L6 myoblasts were transfected with nonspecific control (CTL) or USP19 sirna (no. 7) oligonucleotides. On day 2 of differentiation, the developing myotubes were treated with 10 M dexamethasone or vehicle. Two days later, the cells were harvested and the lysates analyzed by Western blotting for the indicated proteins. A: representative blots. B and C: quantitation of MHC and USP19 protein levels (n 3). Shown are means SE. *Means that are statistically significant with P 0.05 (1-way ANOVA).
7 As mentioned above, a study using gene arrays to identify mrna transcripts regulated in various conditions of muscle wasting identified USP14 as being induced in wasting conditions (24). This proteasome-associated deubiquitinating enzyme is orthologous to yeast UBP6, and genetic manipulation in yeast indicates that it inhibits protein degradation by deubiquitinating substrates and inhibiting proteasome activity (17). Thus, in contrast to our observations on USP19 reported here, the role of induction of this enzyme in conditions of muscle atrophy remains unclear. In our previous study, USP19 mrna levels increased by % in rat skeletal muscle atrophying in response to cancer, glucocorticoids, fasting, or diabetes. During refeeding, the increased mrna levels of USP19 in the fasted state returned to normal, coinciding with the recovery of muscle mass (10). Together with our loss of USP19 function in studies in cultured cells reported here, this suggests that USP19 has a specific role in regulating levels of myofibrillar proteins in adult differentiated muscle. Although myogenin levels are induced in adult myofibers upon denervation, studies involving other forms of muscle wasting have reported lower levels of myogenin expression in atrophying muscle (12, 37), suggesting that the mechanism that we have identified in cultured muscle cells may be relevant in vivo. Steady-state levels of muscle proteins are determined by the balance between their rates of synthesis and degradation. In many catabolic conditions, muscle atrophy arises from both a fall in the rate of synthesis and an activation of the rate of degradation (reviewed in Ref. 19). A large body of previous work has shown that components of the ubiquitin-conjugating apparatus are induced in atrophying muscles, indicating that the ubiquitin-proteasome pathway contributes to the increase in muscle protein degradation (44). A recent report indicates that the induction of the MAFbx ligase may also contribute to inhibition of protein synthesis by targeting the eif3 initiation factor for degradation (23). To our knowledge, we are showing for the first time evidence for an important role for a deubiquitinating enzyme in muscle atrophy and evidence that this enzyme may contribute to wasting by inhibiting the synthesis of major myofibrillar proteins. Our results thus expand the functions of the ubiquitin system in muscle physiology. Consistent with a potentially important role of USP19 in pathological conditions of muscle wasting, we found that depletion of USP19 reversed the loss of MHC induced by dexamethasone in L6 myotubes and that this catabolic stimulus can induce expression of USP19 (Fig. 6). Transfection with USP19 sirna in the presence of dexamethasone did not increase MHC levels to levels seen with transfection of USP19 sirna alone. This suggests that USP19 depletion cannot reverse all of the effects of glucocorticoids on MHC. Indeed, glucocorticoids promote muscle catabolism through multiple mechanisms, including decreased insulin and IGF-I signaling, increased myostatin expression, decreased translation, and activation of transcription factors that lead to induction of ubiquitin protein ligases (reviewed in Ref. 31). Therefore, our results are consistent with USP19 modulating glucocorticoid effects specifically on the transcriptional pathway that leads to synthesis of myofibrillar proteins. At this time, we cannot exclude the possibility that the sirna and glucocorticoid effects on MHC are independent of each other and additive. Further exploration of this new mechanism of muscle wasting will require the study of USP19-deficient mice. Nonetheless, our present findings suggest that inhibition of USP19 could be a novel therapeutic mechanism for the prevention and treatment of muscle protein catabolism. ACKNOWLEDGMENTS We thank Lydia Ouellet and Nathalie Bedard for technical assistance with some of the experiments. GRANTS This work was supported by a grant from the Canadian Institutes of Health Research (FRN 82734). P. Sundaram is the recipient of a studentship from the Natural Sciences and Engineering Research Council. S. S. Wing is the recipient of a Chercheur National salary award from the Fonds de la recherche en santé du Québec. DISCLOSURES No conflicts of interest are declared by the author(s). E1289 REFERENCES 1. Attaix D, Ventadour S, Codran A, Bechet D, Taillandier D, Combaret L. The ubiquitin-proteasome system and skeletal muscle wasting. Essays Biochem 41: , Baracos VE, DeVivo C, Hoyle DH, Goldberg AL. Activation of the ATP-ubiquitin-proteasome pathway in skeletal muscle of cachectic rats bearing a hepatoma. Am J Physiol Endocrinol Metab 268: E996 E1006, Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294: , Bossola M, Muscaritoli M, Costelli P, Bellantone R, Pacelli F, Busquets S, Argiles J, Lopez-Soriano FJ, Civello IM, Baccino FM, Rossi Fanelli F, Doglietto GB. Increased muscle ubiquitin mrna levels in gastric cancer patients. Am J Physiol Regul Integr Comp Physiol 280: R1518 R1523, Bossola M, Muscaritoli M, Costelli P, Grieco G, Bonelli G, Pacelli F, Fanelli FR, Doglietto GB, Baccino FM. Increased muscle proteasome activity correlates with disease severity in gastric cancer patients. Ann Surg 237: , Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: , Chrysis D, Underwood LE. Regulation of components of the ubiquitin system by insulin-like growth factor I and growth hormone in skeletal muscle of rats made catabolic with dexamethasone. Endocrinology 140: , Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, Rakhilin SV, Stitt TN, Patterson C, Latres E, Glass DJ. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasonetreated skeletal muscle. Cell Metab 6: , Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, Gartner C, Latres E, Goldberg AL. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol 185: , Combaret L, Adegoke OA, Bedard N, Baracos V, Attaix D, Wing SS. USP19 is a ubiquitin-specific protease regulated in rat skeletal muscle during catabolic states. Am J Physiol Endocrinol Metab 288: E693 E700, Conway K, Pin C, Kiernan JA, Merrifield P. The E protein HEB is preferentially expressed in developing muscle. Differentiation 72: , Dasarathy S, Muc S, Hisamuddin K, Edmison JM, Dodig M, McCullough AJ, Kalhan SC. Altered expression of genes regulating skeletal muscle mass in the portacaval anastamosis rat. Am J Physiol Gastrointest Liver Physiol 292: G1105 G1113, Dekelbab BH, Witchel SF, DeFranco DB. TNF-alpha and glucocorticoid receptor interaction in L6 muscle cells: a cooperative downregulation of myosin heavy chain. Steroids 72: , 2007.
8 E Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA 98: , Gulve EA, Dice JF. Regulation of protein synthesis and degradation in L8 myotubes. Biochem J 260: , Gulve EA, Mabuchi K, Dice JF. Regulation of myosin and overall protein degradation in mouse C2 skeletal myotubes. J Cell Physiol 147: 37 45, Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127: , Hishiya A, Iemura S, Natsume T, Takayama S, Ikeda K, Watanabe K. A novel ubiquitin-binding protein ZNF216 functioning in muscle atrophy. EMBO J 25: , Jackman RW, Kandarian SC. The molecular basis of skeletal muscle atrophy. Am J Physiol Cell Physiol 287: C834 C843, Kayali AG, Young VR, Goodman MN. Sensitivity of myofibrillar proteins to glucocorticoid-induced muscle proteolysis. Am J Physiol Endocrinol Metab 252: E621 E626, Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc Natl Acad Sci USA 101: , Kwak KS, Zhou X, Solomon V, Baracos VE, Davis J, Bannon AW, Boyle WJ, Lacey DL, Han HQ. Regulation of protein catabolism by muscle-specific and cytokine-inducible ubiquitin ligase E3a-II during cancer cachexia. Cancer Res 64: , Lagirand-Cantaloube J, Offner N, Csibi A, Leibovitch MP, Batonnet- Pichon S, Tintignac LA, Segura CT, Leibovitch SA. The initiation factor eif3-f is a major target for atrogin1/mafbx function in skeletal muscle atrophy. EMBO J 27: , Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J 18: 39 51, Lecker SH, Solomon V, Mitch WE, Goldberg AL. Muscle protein breakdown and the critical role of the ubiquitin-proteasome pathway in normal and disease states. J Nutr 129: 227S 237S, Llovera M, Garcia-Martinez C, Agell N, Lopez-Soriano FJ, Argiles JM. Muscle wasting associated with cancer cachexia is linked to an important activation of the ATP-dependent ubiquitin-mediated proteolysis. Int J Cancer 61: , Llovera M, Garcia-Martinez C, Agell N, Marzabal M, Lopez-Soriano FJ, Argiles JM. Ubiquitin gene expression is increased in skeletal muscle of tumour-bearing rats. FEBS Lett 338: , Lorite MJ, Smith HJ, Arnold JA, Morris A, Thompson MG, Tisdale MJ. Activation of ATP-ubiquitin-dependent proteolysis in skeletal muscle in vivo and murine myoblasts in vitro by a proteolysis-inducing factor (PIF). Br J Cancer 85: , Lorite MJ, Thompson MG, Drake JL, Carling G, Tisdale MJ. Mechanism of muscle protein degradation induced by a cancer cachectic factor. Br J Cancer 78: , Lu Y, Adegoke OA, Nepveu A, Nakayama KI, Bedard N, Cheng D, Peng J, Wing SS. USP19 deubiquitinating enzyme supports cell proliferation by stabilizing KPC1, a ubiquitin ligase for p27 Kip1. Mol Cell Biol 29: , Menconi M, Fareed M, O Neal P, Poylin V, Wei W, Hasselgren PO. Role of glucocorticoids in the molecular regulation of muscle wasting. Crit Care Med 35: S602 S608, Menconi M, Gonnella P, Petkova V, Lecker S, Hasselgren PO. Dexamethasone and corticosterone induce similar, but not identical, muscle wasting responses in cultured L6 and C2C12 myotubes. J Cell Biochem 105: , Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell 123: , Parker MH, Seale P, Rudnicki MA. Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat Rev Genet 4: , Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem 70: , Price SR, Bailey JL, Wang X, Jurkovitz C, England BK, Ding X, Phillips LS, Mitch WE. Muscle wasting in insulinopenic rats results from activation of the ATP-dependent, ubiquitin-proteasome proteolytic pathway by a mechanism including gene transcription. J Clin Invest 98: , Ramamoorthy S, Donohue M, Buck M. Decreased Jun-D and myogenin expression in muscle wasting of human cachexia. Am J Physiol Endocrinol Metab 297: E392 E401, Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 78: , Soares AG, Aoki MS, Miyabara EH, Deluca CV, Ono HY, Gomes MD, Moriscot AS. Ubiquitin-ligase and deubiquitinating gene expression in stretched rat skeletal muscle. Muscle Nerve 36: , Temparis S, Asensi M, Taillandier D, Aurousseau E, Larbaud D, Obled A, Béchet D, Ferrara M, Estrela JM, Attaix D. Increased ATP-ubiquitin-dependent proteolysis in skeletal muscles of tumor-bearing rats. Cancer Res 54: , Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J 19: , Tiao G, Fagan J, Roegner V, Lieberman M, Wang JJ, Fischer JE, Hasselgren PO. Energy-ubiquitin-dependent muscle proteolysis during sepsis in rats is regulated by glucocorticoids. J Clin Invest 97: , Tiao G, Fagan JM, Samuels N, James H, Hudson K, Lieberman M, Fisher JE, Hasselgren PO. Sepsis stimulates nonlysosomal, energydependent proteolysis and increases ubiquitin mrna levels in rat skeletal muscle. J Clin Invest 94: , Wing SS. Control of ubiquitination in skeletal muscle wasting. Int J Biochem Cell Biol 37: , Wing SS, Banville D. 14-kDa ubiquitin-conjugating enzyme: structure of the rat gene and regulation upon fasting and by insulin. Am J Physiol Endocrinol Metab 267: E39 E48, Wing SS, Goldberg AL. Glucocorticoids activate the ATP-ubiquitindependent proteolytic system in skeletal muscle during fasting. Am J Physiol Endocrinol Metab 264: E668 E676, Wing SS, Haas AL, Goldberg AL. Increase in ubiquitin-protein conjugates concomitant with the increase in proteolysis in rat skeletal muscle during starvation and denervation atrophy. Biochem J 307: , 1995.
Biology 12 January 2004 Provincial Examination
 Biology 12 January 2004 Provincial Examination ANSWER KEY / SCORING GUIDE CURRICULUM: Organizers 1. Cell Biology 2. Cell Processes and Applications 3. Human Biology Sub-Organizers A, B, C, D E, F, G, H
Biology 12 January 2004 Provincial Examination ANSWER KEY / SCORING GUIDE CURRICULUM: Organizers 1. Cell Biology 2. Cell Processes and Applications 3. Human Biology Sub-Organizers A, B, C, D E, F, G, H
 www.lessonplansinc.com Topic: Protein Synthesis - Sentence Activity Summary: Students will simulate transcription and translation by building a sentence/polypeptide from words/amino acids. Goals & Objectives:
www.lessonplansinc.com Topic: Protein Synthesis - Sentence Activity Summary: Students will simulate transcription and translation by building a sentence/polypeptide from words/amino acids. Goals & Objectives:
Bacterial Gene Finding CMSC 423
 Bacterial Gene Finding CMSC 423 Finding Signals in DNA We just have a long string of A, C, G, Ts. How can we find the signals encoded in it? Suppose you encountered a language you didn t know. How would
Bacterial Gene Finding CMSC 423 Finding Signals in DNA We just have a long string of A, C, G, Ts. How can we find the signals encoded in it? Suppose you encountered a language you didn t know. How would
Protein sequence alignment using binary string
 Available online at www.scholarsresearchlibrary.com Scholars Research Library Der Pharmacia Lettre, 2015, 7 (5):220-225 (http://scholarsresearchlibrary.com/archive.html) ISSN 0975-5071 USA CODEN: DPLEB4
Available online at www.scholarsresearchlibrary.com Scholars Research Library Der Pharmacia Lettre, 2015, 7 (5):220-225 (http://scholarsresearchlibrary.com/archive.html) ISSN 0975-5071 USA CODEN: DPLEB4
The Cell T H E C E L L C Y C L E C A N C E R
 The Cell T H E C E L L C Y C L E C A N C E R Nuclear envelope Transcription DNA RNA Processing Pre-mRNA Translation mrna Nuclear pores Ribosome Polypeptide Transcription RNA is synthesized from DNA in
The Cell T H E C E L L C Y C L E C A N C E R Nuclear envelope Transcription DNA RNA Processing Pre-mRNA Translation mrna Nuclear pores Ribosome Polypeptide Transcription RNA is synthesized from DNA in
Figure S1. Analysis of genomic and cdna sequences of the targeted regions in WT-KI and
 Figure S1. Analysis of genomic and sequences of the targeted regions in and indicated mutant KI cells, with WT and corresponding mutant sequences underlined. (A) cells; (B) K21E-KI cells; (C) D33A-KI cells;
Figure S1. Analysis of genomic and sequences of the targeted regions in and indicated mutant KI cells, with WT and corresponding mutant sequences underlined. (A) cells; (B) K21E-KI cells; (C) D33A-KI cells;
Ernährung 2006 International Cachexia Workshop Berlin, June 2006
 Activation Peptides ATP E1 ATP Ubiquitin Proteolysis E2 Proteín Proteasome 26S E2 E3 Conjugation Ernährung 2006 International Cachexia Workshop Berlin, June 2006 Antiproteolytic strategies Prof. Dr. Josep
Activation Peptides ATP E1 ATP Ubiquitin Proteolysis E2 Proteín Proteasome 26S E2 E3 Conjugation Ernährung 2006 International Cachexia Workshop Berlin, June 2006 Antiproteolytic strategies Prof. Dr. Josep
Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy
 (Courtesy of Magda Stumpfova. Used with permission.) Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy M.D. Gomes, S.H. Lecker, R.T. Jagoe, A. Navon, and A.L. Goldberg PNAS,
(Courtesy of Magda Stumpfova. Used with permission.) Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy M.D. Gomes, S.H. Lecker, R.T. Jagoe, A. Navon, and A.L. Goldberg PNAS,
The AP-1/CJUN signaling cascade is involved in muscle differentiation: Implications in muscle wasting during cancer cachexia
 The AP-1/CJUN signaling cascade is involved in muscle differentiation: Implications in muscle wasting during cancer cachexia Rodrigo Moore-Carrasco a,b, Celia García-Martínez a,sílvia Busquets a, *, Elisabet
The AP-1/CJUN signaling cascade is involved in muscle differentiation: Implications in muscle wasting during cancer cachexia Rodrigo Moore-Carrasco a,b, Celia García-Martínez a,sílvia Busquets a, *, Elisabet
Rapid Publication. Abstract. Introduction. Methods
 Rapid Publication Muscle Wasting in Insulinopenic Rats Results from Activation of the ATP-dependent, Ubiquitin Proteasome Proteolytic Pathway by a Mechanism Including Gene Transcription S. Russ Price,
Rapid Publication Muscle Wasting in Insulinopenic Rats Results from Activation of the ATP-dependent, Ubiquitin Proteasome Proteolytic Pathway by a Mechanism Including Gene Transcription S. Russ Price,
CD31 5'-AGA GAC GGT CTT GTC GCA GT-3' 5 ' -TAC TGG GCT TCG AGA GCA GT-3'
 Table S1. The primer sets used for real-time RT-PCR analysis. Gene Forward Reverse VEGF PDGFB TGF-β MCP-1 5'-GTT GCA GCA TGA ATC TGA GG-3' 5'-GGA GAC TCT TCG AGG AGC ACT T-3' 5'-GAA TCA GGC ATC GAG AGA
Table S1. The primer sets used for real-time RT-PCR analysis. Gene Forward Reverse VEGF PDGFB TGF-β MCP-1 5'-GTT GCA GCA TGA ATC TGA GG-3' 5'-GGA GAC TCT TCG AGG AGC ACT T-3' 5'-GAA TCA GGC ATC GAG AGA
Molecular basis of diseases- muscles atrophy. Muscular System Functions. Types of Muscle
 Molecular basis of diseases- muscles atrophy Muscular System Functions Body movement (Locomotion) Maintenance of posture Respiration Diaphragm and intercostal contractions Communication (Verbal and Facial)
Molecular basis of diseases- muscles atrophy Muscular System Functions Body movement (Locomotion) Maintenance of posture Respiration Diaphragm and intercostal contractions Communication (Verbal and Facial)
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. H3F3B expression in lung cancer. a. Comparison of H3F3B expression in relapsed and non-relapsed lung cancer patients. b. Prognosis of two groups of lung cancer
Supplementary Figures Supplementary Figure 1. H3F3B expression in lung cancer. a. Comparison of H3F3B expression in relapsed and non-relapsed lung cancer patients. b. Prognosis of two groups of lung cancer
Abbreviations: P- paraffin-embedded section; C, cryosection; Bio-SA, biotin-streptavidin-conjugated fluorescein amplification.
 Supplementary Table 1. Sequence of primers for real time PCR. Gene Forward primer Reverse primer S25 5 -GTG GTC CAC ACT ACT CTC TGA GTT TC-3 5 - GAC TTT CCG GCA TCC TTC TTC-3 Mafa cds 5 -CTT CAG CAA GGA
Supplementary Table 1. Sequence of primers for real time PCR. Gene Forward primer Reverse primer S25 5 -GTG GTC CAC ACT ACT CTC TGA GTT TC-3 5 - GAC TTT CCG GCA TCC TTC TTC-3 Mafa cds 5 -CTT CAG CAA GGA
ANSC/FSTC 607 Biochemistry and Physiology of Muscle as a Food PRIMARY, SECONDARY, AND TERTIARY MYOTUBES
 ANSC/FSTC 607 Biochemistry and Physiology of Muscle as a Food PRIMARY, SECONDARY, AND TERTIARY MYOTUBES I. Satellite Cells A. Proliferative, myoblastic cells that lie in invaginations in the sarcolemma
ANSC/FSTC 607 Biochemistry and Physiology of Muscle as a Food PRIMARY, SECONDARY, AND TERTIARY MYOTUBES I. Satellite Cells A. Proliferative, myoblastic cells that lie in invaginations in the sarcolemma
Complete Student Notes for BIOL2202
 Complete Student Notes for BIOL2202 Revisiting Translation & the Genetic Code Overview How trna molecules interpret a degenerate genetic code and select the correct amino acid trna structure: modified
Complete Student Notes for BIOL2202 Revisiting Translation & the Genetic Code Overview How trna molecules interpret a degenerate genetic code and select the correct amino acid trna structure: modified
Ubiquitin-proteasome-dependent proteolysis
 Review Ubiquitin-proteasome-dependent proteolysis in skeletal muscle Didier Attaix Eveline Aurousseau Lydie Combaret, Anthony Kee Daniel Larbaud, Cécile Rallière, Bertrand Souweine Daniel Taillandier,
Review Ubiquitin-proteasome-dependent proteolysis in skeletal muscle Didier Attaix Eveline Aurousseau Lydie Combaret, Anthony Kee Daniel Larbaud, Cécile Rallière, Bertrand Souweine Daniel Taillandier,
Transcriptional Regulation of Skeletal Muscle Atrophy-Induced Gene Expression by Muscle Ring Finger-1 and Myogenic Regulatory Factors
 UNF Digital Commons UNF Theses and Dissertations Student Scholarship 2017 Transcriptional Regulation of Skeletal Muscle Atrophy-Induced Gene Expression by Muscle Ring Finger-1 and Myogenic Regulatory Factors
UNF Digital Commons UNF Theses and Dissertations Student Scholarship 2017 Transcriptional Regulation of Skeletal Muscle Atrophy-Induced Gene Expression by Muscle Ring Finger-1 and Myogenic Regulatory Factors
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
Caspase-3 Cleaves Specific 19 S Proteasome Subunits in Skeletal Muscle Stimulating Proteasome Activity
 Caspase-3 Cleaves Specific 19 S Proteasome Subunits in Skeletal Muscle Stimulating Proteasome Activity Xiaonan Wang, Emory University Liping Zhang, Baylor College of Medicine William E. Mitch, Baylor College
Caspase-3 Cleaves Specific 19 S Proteasome Subunits in Skeletal Muscle Stimulating Proteasome Activity Xiaonan Wang, Emory University Liping Zhang, Baylor College of Medicine William E. Mitch, Baylor College
Translation Activity Guide
 Translation Activity Guide Student Handout β-globin Translation Translation occurs in the cytoplasm of the cell and is defined as the synthesis of a protein (polypeptide) using information encoded in an
Translation Activity Guide Student Handout β-globin Translation Translation occurs in the cytoplasm of the cell and is defined as the synthesis of a protein (polypeptide) using information encoded in an
Regulation of Gene Expression in Eukaryotes
 Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
The Nobel Prize in Chemistry 2004
 The Nobel Prize in Chemistry 2004 Ubiquitous Quality Control of Life C S Karigar and K R Siddalinga Murthy The Nobel Prize in Chemistry for 2004 is shared by Aaron Ciechanover, Avram Hershko and Irwin
The Nobel Prize in Chemistry 2004 Ubiquitous Quality Control of Life C S Karigar and K R Siddalinga Murthy The Nobel Prize in Chemistry for 2004 is shared by Aaron Ciechanover, Avram Hershko and Irwin
Cardiac Atrophy Due to Cancer: Characterization, Mechanisms, and Sex Differences
 University of Colorado, Boulder CU Scholar Molecular, Cellular, and Developmental Biology Graduate Theses & Dissertations Molecular, Cellular, and Developmental Biology Spring 1-1-2011 Cardiac Atrophy
University of Colorado, Boulder CU Scholar Molecular, Cellular, and Developmental Biology Graduate Theses & Dissertations Molecular, Cellular, and Developmental Biology Spring 1-1-2011 Cardiac Atrophy
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer. Application Note
 Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Nutritional Support in Cancer
 Nutritional Support in Cancer Topic 26 Module 26.1 Molecular Mechanisms of Muscle Wasting in Cancer Cahexia Denis Guttridge Learning Objectives To understand the major mediators regulating muscle wasting
Nutritional Support in Cancer Topic 26 Module 26.1 Molecular Mechanisms of Muscle Wasting in Cancer Cahexia Denis Guttridge Learning Objectives To understand the major mediators regulating muscle wasting
Proteasomes. When Death Comes a Knock n. Warren Gallagher Chem412, Spring 2001
 Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
Plasmids Western blot analysis and immunostaining Flow Cytometry Cell surface biotinylation RNA isolation and cdna synthesis
 Plasmids psuper-retro-s100a10 shrna1 was constructed by cloning the dsdna oligo 5 -GAT CCC CGT GGG CTT CCA GAG CTT CTT TCA AGA GAA GAA GCT CTG GAA GCC CAC TTT TTA-3 and 5 -AGC TTA AAA AGT GGG CTT CCA GAG
Plasmids psuper-retro-s100a10 shrna1 was constructed by cloning the dsdna oligo 5 -GAT CCC CGT GGG CTT CCA GAG CTT CTT TCA AGA GAA GAA GCT CTG GAA GCC CAC TTT TTA-3 and 5 -AGC TTA AAA AGT GGG CTT CCA GAG
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
CHAPTER 4 RESULTS. showed that all three replicates had similar growth trends (Figure 4.1) (p<0.05; p=0.0000)
 CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
Online Data Supplement. Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2
 Online Data Supplement Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2 Yi Lin and Zhongjie Sun Department of physiology, college of
Online Data Supplement Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2 Yi Lin and Zhongjie Sun Department of physiology, college of
E3 ligase TRIM72 negatively regulates myogenesis by IRS-1 ubiquitination
 E3 ligase negatively regulates myogenesis by IRS-1 ubiquitination Young-Gyu Ko College of Life Science and Biotechnology Korea University MyHC immunofluorescence Lipid rafts are plasma membrane compartments
E3 ligase negatively regulates myogenesis by IRS-1 ubiquitination Young-Gyu Ko College of Life Science and Biotechnology Korea University MyHC immunofluorescence Lipid rafts are plasma membrane compartments
Supplemental material for Hernandez et al. Dicoumarol downregulates human PTTG1/Securin mrna expression. through inhibition of Hsp90
 Supplemental material for Hernandez et al. Dicoumarol downregulates human PTTG1/Securin mrna expression through inhibition of Hsp90 Dicoumarol-Sepharose co-precipitation. Hsp90 inhibitors can co-precipitate
Supplemental material for Hernandez et al. Dicoumarol downregulates human PTTG1/Securin mrna expression through inhibition of Hsp90 Dicoumarol-Sepharose co-precipitation. Hsp90 inhibitors can co-precipitate
All intracellular proteins and many extracellular proteins
 Review Protein Degradation by the Ubiquitin Proteasome Pathway in Normal and Disease States Stewart H. Lecker,* Alfred L. Goldberg, and William E. Mitch *Nephrology Division, Beth Israel Deaconess, and
Review Protein Degradation by the Ubiquitin Proteasome Pathway in Normal and Disease States Stewart H. Lecker,* Alfred L. Goldberg, and William E. Mitch *Nephrology Division, Beth Israel Deaconess, and
supplementary information
 DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
Skeletal Muscle 11beta-HSD1 Controls Glucocorticoid- Induced Proteolysis and Expression of E3 Ubiquitin Ligases Atrogin-1 and MuRF-1
 Skeletal Muscle 11beta-HSD1 Controls Glucocorticoid- Induced Proteolysis and Expression of E3 Ubiquitin Ligases Atrogin-1 and MuRF-1 Katrin Biedasek 1, Janin Andres 1, Knut Mai 1, Stephanie Adams 2, Simone
Skeletal Muscle 11beta-HSD1 Controls Glucocorticoid- Induced Proteolysis and Expression of E3 Ubiquitin Ligases Atrogin-1 and MuRF-1 Katrin Biedasek 1, Janin Andres 1, Knut Mai 1, Stephanie Adams 2, Simone
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism SUPPLEMENTARY FIGURES, LEGENDS AND METHODS
 A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
Neurotrophic factor GDNF and camp suppress glucocorticoid-inducible PNMT expression in a mouse pheochromocytoma model.
 161 Neurotrophic factor GDNF and camp suppress glucocorticoid-inducible PNMT expression in a mouse pheochromocytoma model. Marian J. Evinger a, James F. Powers b and Arthur S. Tischler b a. Department
161 Neurotrophic factor GDNF and camp suppress glucocorticoid-inducible PNMT expression in a mouse pheochromocytoma model. Marian J. Evinger a, James F. Powers b and Arthur S. Tischler b a. Department
Supplementary Materials
 Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
Effects of methionine-containing dipeptides on α s1 casein expression in bovine mammary epithelial cells *
 Journal of Animal and Feed Sciences, 16, Suppl. 2, 2007, 325 329 Effects of methionine-containing dipeptides on α s1 casein expression in bovine mammary epithelial cells * H.H. Wu 1, J.Y. Yang 1,2, K.
Journal of Animal and Feed Sciences, 16, Suppl. 2, 2007, 325 329 Effects of methionine-containing dipeptides on α s1 casein expression in bovine mammary epithelial cells * H.H. Wu 1, J.Y. Yang 1,2, K.
TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer
 Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Running Head: EFFECTS OF RESVERATROL ON CANCER CACHEXIA 1. Effects of resveratrol on cancer cachexia in a mouse model.
 Running Head: EFFECTS OF RESVERATROL ON CANCER CACHEXIA 1 Effects of resveratrol on cancer cachexia in a mouse model Jessica Mansfield The Ohio State University College of Nursing Running Head: EFFECTS
Running Head: EFFECTS OF RESVERATROL ON CANCER CACHEXIA 1 Effects of resveratrol on cancer cachexia in a mouse model Jessica Mansfield The Ohio State University College of Nursing Running Head: EFFECTS
Cell Quality Control. Peter Takizawa Department of Cell Biology
 Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
BMP6 treatment compensates for the molecular defect and ameliorates hemochromatosis in Hfe knockout mice
 SUPPLEMENTARY MATERIALS BMP6 treatment compensates for the molecular defect and ameliorates hemochromatosis in Hfe knockout mice Elena Corradini, Paul J. Schmidt, Delphine Meynard, Cinzia Garuti, Giuliana
SUPPLEMENTARY MATERIALS BMP6 treatment compensates for the molecular defect and ameliorates hemochromatosis in Hfe knockout mice Elena Corradini, Paul J. Schmidt, Delphine Meynard, Cinzia Garuti, Giuliana
The Regulation of Skeletal Muscle Protein Turnover During the Progression of Cancer Cachexia in the Apc Min/+ Mouse
 University of South Carolina Scholar Commons Faculty Publications Chemistry and Biochemistry, Department of 9-19-2011 The Regulation of Skeletal Muscle Protein Turnover During the Progression of Cancer
University of South Carolina Scholar Commons Faculty Publications Chemistry and Biochemistry, Department of 9-19-2011 The Regulation of Skeletal Muscle Protein Turnover During the Progression of Cancer
TFEB-mediated increase in peripheral lysosomes regulates. Store Operated Calcium Entry
 TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
C/EBPb mediates tumour-induced ubiquitin ligase atrogin1/mafbx upregulation and muscle wasting
 The EMBO Journal (2011) 30, 4323 4335 & 2011 European Molecular Biology Organization All Rights Reserved 0261-4189/11 www.embojournal.org C/EBPb mediates tumour-induced ubiquitin ligase atrogin1/mafbx
The EMBO Journal (2011) 30, 4323 4335 & 2011 European Molecular Biology Organization All Rights Reserved 0261-4189/11 www.embojournal.org C/EBPb mediates tumour-induced ubiquitin ligase atrogin1/mafbx
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
REGULATION OF ENZYME ACTIVITY. Medical Biochemistry, Lecture 25
 REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
Supplementary Table 2. Conserved regulatory elements in the promoters of CD36.
 Supplementary Table 1. RT-qPCR primers for CD3, PPARg and CEBP. Assay Forward Primer Reverse Primer 1A CAT TTG TGG CCT TGT GCT CTT TGA TGA GTC ACA GAA AGA ATC AAT TC 1B AGG AAA TGA ACT GAT GAG TCA CAG
Supplementary Table 1. RT-qPCR primers for CD3, PPARg and CEBP. Assay Forward Primer Reverse Primer 1A CAT TTG TGG CCT TGT GCT CTT TGA TGA GTC ACA GAA AGA ATC AAT TC 1B AGG AAA TGA ACT GAT GAG TCA CAG
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Supplementary information
 Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
CASE TEACHING NOTES. Decoding the Flu INTRODUCTION / BACKGROUND CLASSROOM MANAGEMENT
 CASE TEACHING NOTES for Decoding the Flu by Norris Armstrong Department of Genetics, University of Georgia, Athens, GA INTRODUCTION / BACKGROUND This case is a clicker case. It is designed to be presented
CASE TEACHING NOTES for Decoding the Flu by Norris Armstrong Department of Genetics, University of Georgia, Athens, GA INTRODUCTION / BACKGROUND This case is a clicker case. It is designed to be presented
L I F E S C I E N C E S
 1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 OCTOBER 31, 2006
1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 OCTOBER 31, 2006
Endogenous glucocorticoids and impaired insulin signaling are both required to stimulate muscle wasting under pathophysiological conditions in mice
 Research article Endogenous glucocorticoids and impaired insulin signaling are both required to stimulate muscle wasting under pathophysiological conditions in mice Zhaoyong Hu, Huiling Wang, In Hee Lee,
Research article Endogenous glucocorticoids and impaired insulin signaling are both required to stimulate muscle wasting under pathophysiological conditions in mice Zhaoyong Hu, Huiling Wang, In Hee Lee,
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
PROVINCIAL EXAMINATION MINISTRY OF EDUCATION BIOLOGY 12 GENERAL INSTRUCTIONS
 INSERT STUDENT I.D. NUMBER (PEN) STICKER IN THIS SPACE AUGUST 1999 PROVINCIAL EXAMINATION MINISTRY OF EDUCATION BIOLOGY 12 GENERAL INSTRUCTIONS 1. Insert the stickers with your Student I.D. Number (PEN)
INSERT STUDENT I.D. NUMBER (PEN) STICKER IN THIS SPACE AUGUST 1999 PROVINCIAL EXAMINATION MINISTRY OF EDUCATION BIOLOGY 12 GENERAL INSTRUCTIONS 1. Insert the stickers with your Student I.D. Number (PEN)
The ubiquitin proteasome system in cardiovascular disease Basic mechanisms
 ECS Congress 2010 Stockholm, Sweden 28 Aug 2010 01 Sep 2010 The ubiquitin proteasome system in cardiovascular disease Basic mechanisms Saskia Schlossarek Department of Experimental and Clinical Pharmacology
ECS Congress 2010 Stockholm, Sweden 28 Aug 2010 01 Sep 2010 The ubiquitin proteasome system in cardiovascular disease Basic mechanisms Saskia Schlossarek Department of Experimental and Clinical Pharmacology
SUPPLEMENTARY INFORMATION. An orthogonal ribosome-trnas pair by the engineering of
 SUPPLEMENTARY INFORMATION An orthogonal ribosome-trnas pair by the engineering of peptidyl transferase center Naohiro Terasaka 1 *, Gosuke Hayashi 1 *, Takayuki Katoh 1, and Hiroaki Suga 1,2 1 Department
SUPPLEMENTARY INFORMATION An orthogonal ribosome-trnas pair by the engineering of peptidyl transferase center Naohiro Terasaka 1 *, Gosuke Hayashi 1 *, Takayuki Katoh 1, and Hiroaki Suga 1,2 1 Department
Biology 12 November 1999 Provincial Examination
 Biology 12 November 1999 Provincial Examination ANSWER KEY / SCORING GUIDE CURRICULUM: Organizers 1. Cell Biology 2. Cell Processes and Applications 3. Human Biology Sub-Organizers A, B, C, D E, F, G,
Biology 12 November 1999 Provincial Examination ANSWER KEY / SCORING GUIDE CURRICULUM: Organizers 1. Cell Biology 2. Cell Processes and Applications 3. Human Biology Sub-Organizers A, B, C, D E, F, G,
Spherical Nucleic Acids For Advanced Wound Healing Applications Chad A. Mirkin
 Spherical Nucleic Acids For Advanced Wound Healing Applications Chad A. Mirkin Departments of Chemistry, Infectious Disease, Materials Science & Engineering, Chemical & Biological Engineering, and Biomedical
Spherical Nucleic Acids For Advanced Wound Healing Applications Chad A. Mirkin Departments of Chemistry, Infectious Disease, Materials Science & Engineering, Chemical & Biological Engineering, and Biomedical
General aspects of bacteriology, bacterial structure and growth. Che-Hsin Lee, Ph.D. Department of Biological Sciences National Sun Yat-senUniversity
 General aspects of bacteriology, bacterial structure and growth Che-Hsin Lee, Ph.D. Department of Biological Sciences National Sun Yat-senUniversity 1 Human Microbiome Project Providing metabolic function
General aspects of bacteriology, bacterial structure and growth Che-Hsin Lee, Ph.D. Department of Biological Sciences National Sun Yat-senUniversity 1 Human Microbiome Project Providing metabolic function
This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is worth 2 points.
 MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
Supplemental Information. Inhibition of the Proteasome b2 Site Sensitizes. Triple-Negative Breast Cancer Cells
 Cell Chemical Biology, Volume 24 Supplemental Information Inhibition of the Proteasome b2 Site Sensitizes Triple-Negative Breast Cancer Cells to b5 Inhibitors and Suppresses Nrf1 Activation Emily S. Weyburne,
Cell Chemical Biology, Volume 24 Supplemental Information Inhibition of the Proteasome b2 Site Sensitizes Triple-Negative Breast Cancer Cells to b5 Inhibitors and Suppresses Nrf1 Activation Emily S. Weyburne,
Explain that each trna molecule is recognised by a trna-activating enzyme that binds a specific amino acid to the trna, using ATP for energy
 7.4 - Translation 7.4.1 - Explain that each trna molecule is recognised by a trna-activating enzyme that binds a specific amino acid to the trna, using ATP for energy Each amino acid has a specific trna-activating
7.4 - Translation 7.4.1 - Explain that each trna molecule is recognised by a trna-activating enzyme that binds a specific amino acid to the trna, using ATP for energy Each amino acid has a specific trna-activating
Aberrant Promoter CpG Methylation is a Mechanism for Lack of Hypoxic Induction of
 Aberrant Promoter CpG Methylation is a Mechanism for Lack of Hypoxic Induction of PHD3 in a Diverse Set of Malignant Cells Abstract The prolyl-hydroxylase domain family of enzymes (PHD1-3) plays an important
Aberrant Promoter CpG Methylation is a Mechanism for Lack of Hypoxic Induction of PHD3 in a Diverse Set of Malignant Cells Abstract The prolyl-hydroxylase domain family of enzymes (PHD1-3) plays an important
20S Proteasome Activity Assay Kit
 20S Proteasome Activity Assay Kit For 100 Assays Cat. No. APT280 FOR RESEARCH USE ONLY NOT FOR USE IN DIAGNOSTIC PROCEDURES USA & Canada Phone: +1(800) 437-7500 Fax: +1 (951) 676-9209 Europe +44 (0) 23
20S Proteasome Activity Assay Kit For 100 Assays Cat. No. APT280 FOR RESEARCH USE ONLY NOT FOR USE IN DIAGNOSTIC PROCEDURES USA & Canada Phone: +1(800) 437-7500 Fax: +1 (951) 676-9209 Europe +44 (0) 23
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
c Tuj1(-) apoptotic live 1 DIV 2 DIV 1 DIV 2 DIV Tuj1(+) Tuj1/GFP/DAPI Tuj1 DAPI GFP
 Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Cellular Markers of Muscle Atrophy in Chronic Obstructive Pulmonary Disease
 Cellular Markers of Muscle Atrophy in Chronic Obstructive Pulmonary Disease Pamela J. Plant 1, Dina Brooks 2, Marie Faughnan 1, Tanya Bayley 1, James Bain 3, Lianne Singer 4, Judy Correa 1, Dawn Pearce
Cellular Markers of Muscle Atrophy in Chronic Obstructive Pulmonary Disease Pamela J. Plant 1, Dina Brooks 2, Marie Faughnan 1, Tanya Bayley 1, James Bain 3, Lianne Singer 4, Judy Correa 1, Dawn Pearce
Myostatin promotes the wasting of human myoblast cultures through promoting ubiquitin-proteasome pathway-mediated loss of sarcomeric proteins
 Am J Physiol Cell Physiol 301: C1316 C1324, 2011. First published September 7, 2011; doi:10.1152/ajpcell.00114.2011. Myostatin promotes the wasting of human myoblast cultures through promoting ubiquitin-proteasome
Am J Physiol Cell Physiol 301: C1316 C1324, 2011. First published September 7, 2011; doi:10.1152/ajpcell.00114.2011. Myostatin promotes the wasting of human myoblast cultures through promoting ubiquitin-proteasome
Supplementary data Supplementary Figure 1 Supplementary Figure 2
 Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most
 Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
Nutrients, insulin and muscle wasting during critical illness
 32 nd annual meeting of the Belgian Society of Intensive Care Medicine June 15, 212 Nutrients, insulin and muscle wasting during critical illness Sarah Derde Introduction Critical illness: feeding-resistant
32 nd annual meeting of the Belgian Society of Intensive Care Medicine June 15, 212 Nutrients, insulin and muscle wasting during critical illness Sarah Derde Introduction Critical illness: feeding-resistant
General Laboratory methods Plasma analysis: Gene Expression Analysis: Immunoblot analysis: Immunohistochemistry:
 General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
SUPPLEMENTAL MATERIALS AND METHODS. Puromycin-synchronized metabolic labelling - Transfected HepG2 cells were depleted of
 SUPPLEMENTAL MATERIALS AND METHODS Puromycin-synchronized metabolic labelling - Transfected HepG2 cells were depleted of cysteine and methionine and then treated with 10 μm puromycin in depletion medium
SUPPLEMENTAL MATERIALS AND METHODS Puromycin-synchronized metabolic labelling - Transfected HepG2 cells were depleted of cysteine and methionine and then treated with 10 μm puromycin in depletion medium
MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands)
 Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Key words: Branched-chain c~-keto acid dehydrogenase complex, branched-chain c~-keto acid
 Vol. 44, No. 6, May 1998 BIOCHEMISTRY and MOLECULAR BIOLOGY INTERNATIONAL Pages 1211-1216 BRANCHED-CHAIN cx-keto ACID DEHYDROGENASE KINASE CONTENT IN RAT SKELETAL MUSCLE IS DECREASED BY ENDURANCE TRAINING
Vol. 44, No. 6, May 1998 BIOCHEMISTRY and MOLECULAR BIOLOGY INTERNATIONAL Pages 1211-1216 BRANCHED-CHAIN cx-keto ACID DEHYDROGENASE KINASE CONTENT IN RAT SKELETAL MUSCLE IS DECREASED BY ENDURANCE TRAINING
Doctor of Philosophy
 Regulation of Gene Expression of the 25-Hydroxyvitamin D la-hydroxylase (CYP27BI) Promoter: Study of A Transgenic Mouse Model Ivanka Hendrix School of Molecular and Biomedical Science The University of
Regulation of Gene Expression of the 25-Hydroxyvitamin D la-hydroxylase (CYP27BI) Promoter: Study of A Transgenic Mouse Model Ivanka Hendrix School of Molecular and Biomedical Science The University of
RECAP (1)! In eukaryotes, large primary transcripts are processed to smaller, mature mrnas.! What was first evidence for this precursorproduct
 RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
Endothelial Dysfunction in Rheumatoid Arthritis: the Role of Monocyte Chemotactic
 Supplemental file Endothelial Dysfunction in Rheumatoid Arthritis: the Role of Monocyte Chemotactic Protein-1-Induced Protein Ming He, Xiao Liang, Lan He, Wen Wen, Sijia Zhao, Liang Wen, Yan Liu, John
Supplemental file Endothelial Dysfunction in Rheumatoid Arthritis: the Role of Monocyte Chemotactic Protein-1-Induced Protein Ming He, Xiao Liang, Lan He, Wen Wen, Sijia Zhao, Liang Wen, Yan Liu, John
Supplemental Information
 Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
The Synthetic Machinery of the Cell
 The Synthetic Machinery of the Cell Professor Alfred Cuschieri University of Malta Department of Anatomy Objectives State the characteristics of messenger, ribosomal and transfer RNA Distinguish between
The Synthetic Machinery of the Cell Professor Alfred Cuschieri University of Malta Department of Anatomy Objectives State the characteristics of messenger, ribosomal and transfer RNA Distinguish between
Supplementary Materials and Methods
 Supplementary Materials and Methods Immunoblotting Immunoblot analysis was performed as described previously (1). Due to high-molecular weight of MUC4 (~ 950 kda) and MUC1 (~ 250 kda) proteins, electrophoresis
Supplementary Materials and Methods Immunoblotting Immunoblot analysis was performed as described previously (1). Due to high-molecular weight of MUC4 (~ 950 kda) and MUC1 (~ 250 kda) proteins, electrophoresis
AMPK Phosphorylation Assay Kit
 AMPK Phosphorylation Assay Kit Catalog Number KA3789 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
AMPK Phosphorylation Assay Kit Catalog Number KA3789 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
Supplementary Table 3. 3 UTR primer sequences. Primer sequences used to amplify and clone the 3 UTR of each indicated gene are listed.
 Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplementary Figures
 Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
SUPPLEMENTARY INFORMATION
 -. -. SUPPLEMENTARY INFORMATION DOI: 1.1/ncb86 a WAT-1 WAT- BAT-1 BAT- sk-muscle-1 sk-muscle- mir-133b mir-133a mir-6 mir-378 mir-1 mir-85 mir-378 mir-6a mir-18 mir-133a mir- mir- mir-341 mir-196a mir-17
-. -. SUPPLEMENTARY INFORMATION DOI: 1.1/ncb86 a WAT-1 WAT- BAT-1 BAT- sk-muscle-1 sk-muscle- mir-133b mir-133a mir-6 mir-378 mir-1 mir-85 mir-378 mir-6a mir-18 mir-133a mir- mir- mir-341 mir-196a mir-17
The dynamic regulation of blood vessel caliber
 INVITED BASIC SCIENCE REVIEW The dynamic regulation of blood vessel caliber Colleen M. Brophy, MD, Augusta, Ga BACKGROUND The flow of blood to organs is regulated by changes in the diameter of the blood
INVITED BASIC SCIENCE REVIEW The dynamic regulation of blood vessel caliber Colleen M. Brophy, MD, Augusta, Ga BACKGROUND The flow of blood to organs is regulated by changes in the diameter of the blood
Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein
 Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein Lei Wang 1, Tian-Peng Zhang 1, Yuan Zhang 2, Hai-Lian
Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein Lei Wang 1, Tian-Peng Zhang 1, Yuan Zhang 2, Hai-Lian
Cellular functions of protein degradation
 Protein Degradation Cellular functions of protein degradation 1. Elimination of misfolded and damaged proteins: Environmental toxins, translation errors and genetic mutations can damage proteins. Misfolded
Protein Degradation Cellular functions of protein degradation 1. Elimination of misfolded and damaged proteins: Environmental toxins, translation errors and genetic mutations can damage proteins. Misfolded
CELLS. Cells. Basic unit of life (except virus)
 Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Using a proteomic approach to identify proteasome interacting proteins in mammalian
 Supplementary Discussion Using a proteomic approach to identify proteasome interacting proteins in mammalian cells, we describe in the present study a novel chaperone complex that plays a key role in the
Supplementary Discussion Using a proteomic approach to identify proteasome interacting proteins in mammalian cells, we describe in the present study a novel chaperone complex that plays a key role in the
FIRST BIOCHEMISTRY EXAM Tuesday 25/10/ MCQs. Location : 102, 105, 106, 301, 302
 FIRST BIOCHEMISTRY EXAM Tuesday 25/10/2016 10-11 40 MCQs. Location : 102, 105, 106, 301, 302 The Behavior of Proteins: Enzymes, Mechanisms, and Control General theory of enzyme action, by Leonor Michaelis
FIRST BIOCHEMISTRY EXAM Tuesday 25/10/2016 10-11 40 MCQs. Location : 102, 105, 106, 301, 302 The Behavior of Proteins: Enzymes, Mechanisms, and Control General theory of enzyme action, by Leonor Michaelis
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
9/16/2009. Fast and slow twitch fibres. Properties of Muscle Fiber Types Fast fibers Slow fibers
 Muscles, muscle fibres and myofibrils Fast and slow twitch fibres Rat hindlimb muscle ATPase staining at different ph and NADH Muscle fibre shortening velocity lengths/second Properties of Muscle Fiber
Muscles, muscle fibres and myofibrils Fast and slow twitch fibres Rat hindlimb muscle ATPase staining at different ph and NADH Muscle fibre shortening velocity lengths/second Properties of Muscle Fiber
Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana
 Biochemistry: Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana 38 CAMERON E. NIENABER ʻ04 Abstract Long recognized as essential plant nutrients and metabolites, inorganic and organic
Biochemistry: Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana 38 CAMERON E. NIENABER ʻ04 Abstract Long recognized as essential plant nutrients and metabolites, inorganic and organic
Insulin and IGF-1 receptors regulate FoxOmediated signaling in muscle proteostasis
 Insulin and IGF-1 receptors regulate FoxOmediated signaling in muscle proteostasis Brian T. O Neill,, K. Sreekumaran Nair, C. Ronald Kahn J Clin Invest. 2016;126(9):3433-3446. https://doi.org/10.1172/jci86522.
Insulin and IGF-1 receptors regulate FoxOmediated signaling in muscle proteostasis Brian T. O Neill,, K. Sreekumaran Nair, C. Ronald Kahn J Clin Invest. 2016;126(9):3433-3446. https://doi.org/10.1172/jci86522.
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
Boucher et al NCOMMS B
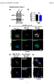 1 Supplementary Figure 1 (linked to Figure 1). mvegfr1 constitutively internalizes in endothelial cells. (a) Immunoblot of mflt1 from undifferentiated mouse embryonic stem (ES) cells with indicated genotypes;
1 Supplementary Figure 1 (linked to Figure 1). mvegfr1 constitutively internalizes in endothelial cells. (a) Immunoblot of mflt1 from undifferentiated mouse embryonic stem (ES) cells with indicated genotypes;
