HHS Public Access Author manuscript Neuroscience. Author manuscript; available in PMC 2015 June 04.
|
|
|
- Jacob Holt
- 6 years ago
- Views:
Transcription
1 EFFECT OF POLYMORPHISM ON EXPRESSION OF THE NEUROPEPTIDE Y GENE IN INBRED ALCOHOL-PREFERRING AND -NONPREFERRING RATS J. P. SPENCE, T. LIANG, K. HABEGGER, and L. G. CARR * Departments of Medicine, Indiana University School of Medicine, Medical Research and Library Building, Room 407, 975 West Walnut Street, Indianapolis, IN 46202, USA Abstract Using animal models of alcoholism, previous studies suggest that neuropeptide Y (NPY) may be implicated in alcohol preference and consumption due to its role in the modulation of feeding and anxiety. Quantitative trait loci (QTL) analysis previously identified an interval on rat chromosome 4 that is highly associated with alcohol preference and consumption using an F2 population derived from inbred alcohol-preferring (ip) and -nonpreferring (inp) rats. NPY mapped to the peak of this QTL region and was prioritized as a candidate gene for alcohol-seeking behavior in the ip and inp rats. In order to identify a potential mechanism for reduced NPY protein levels documented in the ip rat, genetic and molecular components that influence NPY expression were analyzed between ip and inp rats. Comparing the ip rat to the inp rat, quantitative real-time polymerase chain reaction detected significantly decreased levels of NPY mrna expression in the ip rat in the six brain regions tested: nucleus accumbens, frontal cortex, amygdala, hippocampus, caudate-putamen, and hypothalamus. In addition, the functional significance of three previously identified polymorphisms was assessed using in vitro expression analysis. The polymorphism defined by microsatellite marker D4Mit7 in ip rats reduced luciferase reporter gene expression in SK-N-SH neuroblastoma cells. These results suggest that differential expression of the NPY gene resulting from the D4mit7 marker polymorphism may contribute to reduced levels of NPY in discrete brain regions in the ip rats. Keywords HHS Public Access Author manuscript Published in final edited form as: Neuroscience ; 131(4): doi: /j.neuroscience neuropeptide Y; alcohol preference; selective breeding; mrna expression; polymorphism; quantitative trait locus Alcoholism is a complex disorder influenced by the interaction between multiple genes and the environment, exhibiting a heritability that ranges from 50 to 60% in both men and women (Heath et al., 1997). Thus far, however, only the protective effects of the alcohol metabolizing enzymes have been consistently replicated (Foroud and Li, 1999; Thomasson et al., 1993). To identify genetic factors that influence alcoholism, the alcohol-preferring (P) and -nonpreferring (NP) lines were developed from a randomly bred closed colony of Wistar 2005 IBRO. Published by Elsevier Ltd. All rights reserved. * Corresponding author. Tel: ; fax: lcarr@iupui.edu (L. G. Carr).
2 SPENCE et al. Page 2 rats through bidirectional selective breeding on the basis of alcohol consumption and preference (Li et al., 1991). In this model, P rats display the characteristics that are considered necessary for an animal model of alcoholism (Cicero, 1979). Subsequently, inbred P (ip) and NP (inp) strains have been established that maintain highly discordant alcohol consumption scores. Previous studies suggest that neuropeptide Y (NPY) is implicated in the modulation of alcohol consumption in P and NP rats (Cowen et al., 2004; Thiele and Badia-Elder, 2003; Pandey et al., 2003). P rats display lower levels of NPY immunoreactivity in various regions of the brain, including the central nucleus of the amygdala, hippocampus and the frontal cortex and higher levels in the paraventricular hypothalamic nucleus and arcuate nucleus of the hypothalamus (Ehlers et al., 1998; Hwang et al., 1999). Furthermore, decreased levels of NPY are associated with increased anxiety in P rats (Colombo, 1997; Stewart et al., 1993), and i.c.v. infusion of NPY has been shown to reduce ethanol intake in the P rat (Gilpin et al., 2003). NPY is localized to an interval that is highly associated with alcohol preference and consumption, mapping to a quantitative trait locus with a lod score of 9.2 on rat chromosome 4, using an F2 population bred from ip and inp rats (Carr et al., 1998; Bice et al., 1998). In addition, mice deficient in NPY exhibit increased alcohol consumption compared with wild-type mice, and an over-expression of NPY decreases ethanol consumption in transgenic mice (Thiele et al., 1998). In humans, a polymorphism in NPY (Leu7Pro) was significantly associated with dependence in European American and Finnish alcoholics, both exhibiting an increased frequency of the Pro7 allele compared with controls (Lappalainen et al., 2002; Zhu et al., 2003). Therefore, these studies have established NPY as an excellent candidate gene for alcoholism and alcohol preference, documenting an inverse relationship between NPY levels and alcohol consumption. While a relationship between NPY and alcohol consumption has been established, the molecular and physiological mechanism responsible for this relationship remains to be elucidated. Because NPY is likely to function in memory, stress, hypertension, feeding and emotion (Heilig and Thorsell, 2002), this study was conducted to better understand the genetic and molecular components that influence NPY expression in ip and inp rats. Thus, to further characterize the relationship between NPY and alcohol preference in P and NP rats, mrna expression was evaluated in several brain regions between alcohol-naive ip and inp rats, and transient transfection assays were performed on three previously identified polymorphisms (Bice et al., 1998) to determine their functional significance. EXPERIMENTAL PROCEDURES Dissection of brain regions Five ip and four inp alcohol-naive adult male rats were killed, and the entire brain was removed and dissected using the coordinates of Paxinos and Watson (Paxinos and Watson, 1998) to produce six subregional tissue samples: 1) nucleus accumbens, 2) frontal cortex, 3) amygdala, 4) hippocampus, 5) caudate-putamen, and 6) hypothalamus. These subregions were selected because they have been implicated in the mesocorticolimbic dopamine
3 SPENCE et al. Page 3 Isolation of RNA system, reciprocally interacting with the VTA to regulate alcohol-drinking behavior (McBride and Li, 1998). The nucleus accumbens and caudate putamen are dissected from a 2 mm section generated by a coronal cut at 2 mm anterior to the optic chiasm (Bregma 1.70 mm) and a coronal cut at the optic chiasm (Bregma 0.26 mm). The nucleus accumbens is dissected bilaterally by cutting below the rhinal fissure and trimming off the olfactory tubercles and cortical tissue at the ventral and ventrolateral borders of the slice. From the remaining tissue the caudate putamen (bilateral) is dissected below the corpus callosum leaving the septum hanging in the center. The hypothalamus is dissected from the tissue posterior to the optic chiasm by making an incision 2 mm to both the right and the left of the midline and a cut in front of the mammillary bodies as the caudal limit. The amygdala is dissected by a cut at the lateral borders of the lateral hypothalamus (Bregma 2.12 mm) and ventral of the rhinal fissure, with cortical tissue then removed at the lateral edges of the dissected slice. The caudal border of the amygdalar dissection is the rostroventral border of the CA3 subfield of Ammon s horn ( 4.16 mm). The entire hippocampus is dissected from the remaining brain by a midline incision between the hemispheres and rolling the hippocampus out of the cerebral cortex. All of the experiments utilized in this study conformed to local and international guidelines on the ethical use of animals in order to minimize the number of animals used and their suffering. The microdissected tissues were snap frozen on dry ice and stored at 80 C until RNA isolation. RNA was isolated according to the RNeasy Midi manufacturer s protocol (Qiagen, Valencia, CA, USA), and the isolated RNA was resuspended in diethyl-pyrocarbonatetreated water, treated with DNase I, and stored at 80 C. Quantitative real-time RT-PCR (polymerase chain reaction) Using the ABI PRISM 7700 Sequence Detection System (PE Biosystems, Wellesley, MA, USA), the relative mrna expression levels of NPY in the ip and inp rats were determined in the nucleus accumbens, frontal cortex, amygdala, hippocampus, caudate-putamen, and hypothalamus by quantitative real-time RT-PCR (qrt-pcr). cdna was generated from the six selected brain regions of each of the five ip and four inp rats. cdna template, generated from 50 ng total RNA, was added to each PCR reaction that contained 0.1 μm forward and reverse primers and SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). Each reaction was performed in triplicate. The integrated software package accompanying the ABI PRISM 7700 was utilized to select the NPY-1F and NPY-1R primers for qrt-pcr (Table 1). The specificity of these primers was confirmed using agarose gel analysis. Each sample was amplified for 40 cycles, and the cycle threshold (Ct) was determined for each cdna template. The Ct refers to the cycle number at which the fluorescence of the amplified product reached an arbitrary threshold that was within the exponential phase of amplification. Relative values of expression were determined for each sample using the standard curve method, and these values were normalized to the Ct values of GAPDH, a standard housekeeping control gene, using glyceraldehyde phosphate dehydrogenase (GAPDH)-F and GAPDH-R primers (Table 1) for PCR amplification. t- Tests were performed to detect significant differences in expression between the ip and inp samples.
4 SPENCE et al. Page 4 Luciferase reporter constructs Reporter constructs were created to analyze the effects of three previously identified polymorphisms located in the 2nd intron, 3rd intron, and the 3 UTR (Bice et al., 1998; Larhammar et al., 1987) (Fig. 2). First, to amplify the regions encompassing these polymorphisms, three primer pairs were designed: Luc-1F and Luc-1R for the second intron, Luc-2F and Luc-2R for the third intron, and Luc-3F and Luc-3R for the 3 UTR (Table 1). Using ip and inp genomic DNA, six fragments were amplified and ligated into the pcr2.1- TOPO vector. Sequence analysis was then performed to confirm that there were no errors. The resulting TOPO constructs were digested with either BamHI or XbaI, depending on the desired site of insertion of each fragment into the pgl-3 Promoter vector (Promega, Madison, WI, USA). These sites were determined based on the location of each fragment in the NPY gene. The resulting fragments were gel-isolated and ligated into a pgl-3 Promoter vector that was either digested with BamHI or XbaI. Thus, the fragments encompassing the polymorphisms in the 2nd and 3rd introns were ligated into the pgl-3 Promoter vector in a site following the SV40 late poly(a) signal, while the 3 UTR fragments were ligated between the luciferase gene and the SV40 late poly(a) signal (Fig. 3). To determine fragment orientation, each fragment was amplified using either the fragment specific forward or reverse primer paired with Luc-4R, a primer specific to the pgl-3 Promoter vector. The constructs containing the correct orientation of each fragment were isolated using the EndoFree Plasmid Maxi Kit (Qiagen). Transient transfection and luciferase assays RESULTS Human neuroblastoma SK-N-SH cells were cultured in Eagle s Minimal Essential Medium containing 7.5% NaHCO 3, 2 mm Glut-max, 0.1 mm non-essential amino acids, 1 mm pyruvate, 10% FBS (Invitrogen, Carlsbad, CA, USA) and were maintained at 37 C in a humidified 5% CO 2 incubator. Twenty-four hours before transfection, cells were plated into each well of a 24 well plate; 0.5 μg of each pgl-3 luciferase test plasmid was transfected per well using Tf x 50 reagent (Promega); 2.5 ng of CMV renilla vector (prl- CMV) was cotransfected with each pgl-3-luciferase test plasmid to serve as an internal control for transfection efficiency. Cells were incubated for 48 h, washed, and harvested using passive lysis buffer. Cell extracts were assayed for firefly and Renilla luciferase activities in a TD-20/20 Luminometer, using the Dual-Luciferase Reporter Assay System (Promega). Luciferase assays were performed three to six times in triplicate, using plasmids that were independently purified at least twice. NPY exhibits reduced mrna expression in alcohol-naive ip rats compared with inp rats in multiple brain regions To assess the endogenous difference in NPY mrna expression between ip and inp rats, discrete brain regions dissected from alcohol-naive ip and inp rats were analyzed using qrt-pcr. NPY mrna expression was observed at significantly decreased levels in the nucleus accumbens (P=0.006), the frontal cortex (P=0.04), the amygdala (P=0.0004), the hippocampus (P< ), the caudate putamen (P=0.02), and the hypothalamus (P=0.02) in the ip rat (Fig. 1). The greatest difference in expression comparing inp to ip was in the
5 SPENCE et al. Page 5 hippocampus and caudate putamen (2.2-fold difference), and the smallest difference in expression was in the frontal cortex and hypothalamus (1.3-fold difference; Fig. 1). Microsatellite marker D4Mit7 polymorphism reduced luciferase activity in vitro using SK-N- SH cells DISCUSSION The microsatellite marker D4Mit7 is located in the 2nd intron of NPY. To determine the probable effects of the previously identified polymorphisms on NPY gene expression, PCR was employed to generate DNA fragments containing the polymorphisms in the 2nd intron (D4Mit7), 3rd intron and 3 UTR. The fragments containing the 2nd and 3rd intron polymorphisms were cloned into the pgl-3 Promoter luciferase vector downstream the SV40 late poly(a) signal, while the 3 UTR fragments were ligated between the luciferase gene and the SV40 late poly(a) signal (Fig. 2). All constructs were transiently transfected into SK-N-SH cells, which constitutively express the endogenous NPY gene. Luciferase expression was expressed in fold-change compared with the pgl-3 Promoter-luciferase (luc) construct (Fig. 3). The ip-(2nd intron)-luc construct defined by the D4Mit7 microsatellite marker exhibited a significant decrease in expression in SK-N-SH cells compared with the inp-(2nd intron)-luc construct (P<0.05). The 3rd intron-luc and the 3 UTR-luc vectors did not significantly alter luciferase activity in SK-N-SH cells. In this study, alcohol-naive ip rats exhibited a reduced level of NPY mrna expression compared with inp rats in the nucleus accumbens, frontal cortex, amygdala, hippocampus, caudate putamen, and hypothalamus. Three polymorphisms in the NPY gene were assessed for their functional significance in modulating NPY expression. In vitro expression analysis using neuroblastoma SK-N-SH cells yielded a significant decrease in expression of the ip- (2nd intron)-luc construct compared with the inp-(2nd intron)-luc construct (P<0.05). These results suggest that an endogenous decrease in NPY expression may be influencing the phenotypic decrease in NPY protein expression that has been previously observed in various brain regions when comparing P and NP rats. The reduced level of NPY mrna expression observed in alcohol-naive ip rats compared with inp rats corroborated the previous evidence of a reduction in NPY protein levels in central nucleus of the amygdala, hippocampus, and the frontal cortex (Ehlers et al., 1998; Hwang et al., 1999). In contrast, ip animals exhibited significantly lower levels of NPY mrna expression in the hypothalamus in the present study, whereas, in a previous study, higher levels of NPY immunoreactivity were detected in the paraventricular hypothalamic nucleus of P rats than NP rats (Hwang et al., 1999). Lack of correlation between mrna and protein levels may have resulted from procedural differences between the two studies. In our study, the entire hypothalamus was dissected, whereas a very specific region of the hypothalamus was quantified in the immunohistochemistry studies. Lower levels of NPY mrna have been documented in high alcohol drinking rats compared with the low alcohol drinking rats (Hwang et al., 1999). No difference in NPY mrna expression was observed between NP rats compared with P rats (Caberlotto et al., 2001). Together, these data suggest that the NPY expression in the hypothalamus may not play a defining role in the modulation
6 SPENCE et al. Page 6 of alcohol preference, whereas its expression in the amygdala, a region implicated in anxiety and the reinforcing properties of alcohol may be influential in the regulation of alcoholseeking behavior (Koob et al., 1998; Cowen et al., 2004). In vitro expression analysis revealed a significant decrease in luciferase expression in ip- (2nd intron)-luc construct compared with the inp-(2nd intron)-luc, providing a potential mechanism for the reduced mrna and protein expression. Furthermore, the functional insignificance of the 3 UTR polymorphism, a primary site for the regulation of mrna stability (Nair and Menon, 2000; Wang and Kiledjian, 2000; Loflin and Lever, 2001), suggests that NPY expression is regulated at the transcriptional level. Although neuroblastoma cells do not fully reflect regulatory mechanisms underlying NPY expression in neuronal cells, they show that the D4Mit7 polymorphism is functional in a cell line that expresses NPY. The reduced expression exhibited by one variant of the D4Mit7 polymorphism is consistent with the lower NPY mrna and protein levels detected in discrete brain regions of the ip strain compared with the inp strain. The polymorphism defined by the marker D4Mit7 is localized to a GC rich region, possibly involved in transcriptional regulation. Intronic polymorphisms have been associated with regulation of gene expression (Fiskerstrand et al., 1999; Arnold et al., 2000; Bream et al., 2000; Agarwal et al., 2000). Thus, polymorphic regions, previously used as genetic markers, may function in regulating gene expression (Katsuki et al., 1996). It is postulated that these alternating purine-pyrimidine tracts form Z- DNA (left-handed helices) or cruciform regions that influence gene expression, and some CA repeat sequences do exhibit moderate enhancer activities (Hamada et al., 1984). However, the difference in expression associated with the D4Mit7 polymorphism may not fully account for the observed expression difference in NPY detected between ip and inp rats. Therefore, additional studies will be necessary to define the functional significance of additional polymorphisms discovered in the NPY gene. It is also possible that sequencing additional 5 region will identify other functional polymorphisms since regulatory elements can be located many kilobases upstream in the gene. The inverse relationship between levels of NPY in various regions of the brain and alcohol consumption has been documented in previous literature (Badia-Elder et al., 2001; Thiele et al., 1998). However, the molecular mechanism of NPY remains to be elucidated even in the P and NP rats where the difference in NPY expression appears to be well defined (Ehlers et al., 1998; Hwang et al., 1999). This study has attempted to further characterize NPY expression in order to ultimately distinguish NPY expression as a primary source of the decrease in NPY protein expression observed in various brain regions between P and NP rats. In conclusion, the effects of NPY levels on alcohol preference in the P and NP rats have been established in previous studies (Badia-Elder et al., 2001). This study provided evidence of decreased mrna expression in several brain regions, including the nucleus accumbens, frontal cortex, amygdala, hippocampus, caudate putamen, and hypothalamus. The previously identified D4Mit7 marker polymorphism appeared functional, while the 3rd intron and 3 UTR polymorphisms produced no change in luc expression. All in all, these
7 SPENCE et al. Page 7 Acknowledgments Abbreviations Ct results further implicated the regulation of NPY gene expression as a likely source for the difference in NPY levels detected between ip and inp rats. U.S. Public Service grants AA10707 and AA GAPDH inp ip NP NPY P PCR References qrt-pcr cycle threshold glyceraldehyde phosphate dehydrogenase inbred alcohol-nonpreferring inbred alcohol-preferring alcohol-nonpreferring neuropeptide Y alcohol-preferring polymerase chain reaction quantitative real-time RT-PCR Agarwal AK, Giacchetti G, Lavery G, Nikkila H, Palermo M, Ricketts M, McTernan C, Bianchi G, Manunta P, Strazzullo P, Mantero F, White PC, Stewart PM. CA-Repeat polymorphism in intron 1 of HSD11B2: effects on gene expression and salt sensitivity. Hypertension. 2000; 36: [PubMed: ] Arnold R, Maueler W, Bassili G, Lutz M, Burke L, Epplen TJ, Renkawitz R. The insulator protein CTCF represses transcription on binding to the (gt)(22)(ga)(15) microsatellite in intron 2 of the HLA-DRB1(*)0401 gene. Gene. 2000; 253: [PubMed: ] Badia-Elder NE, Stewart RB, Powrozek TA, Roy KF, Murphy JM, Li TK. Effect of neuropeptide Y (NPY) on oral ethanol intake in Wistar, alcohol-preferring (P), and -nonpreferring (NP) rats. Alcohol Clin Exp Res. 2001; 25: [PubMed: ] Bice P, Foroud T, Bo R, Castelluccio P, Lumeng L, Li TK, Carr LG. Genomic screen for QTLs underlying alcohol consumption in the P and NP rat lines. Mamm Genome. 1998; 9: [PubMed: ] Bream JH, Carrington M, O Toole S, Dean M, Gerrard B, Shin HD, Kosack D, Modi W, Young HA, Smith MW. Polymorphisms of the human IFNG gene noncoding regions. Immunogenetics. 2000; 51: [PubMed: ] Caberlotto L, Thorsell A, Rimondini R, Sommer W, Hyytia P, Heilig M. Differential expression of NPY and its receptors in alcohol-preferring AA and alcohol-avoiding ANA rats. Alcohol Clin Exp Res. 2001; 25: [PubMed: ] Carr LG, Foroud T, Bice P, Gobbett T, Ivashina J, Edenberg H, Lumeng L, Li TK. A quantitative trait locus for alcohol consumption in selectively bred rat lines. Alcohol Clin Exp Res. 1998; 22: [PubMed: ] Cicero, TJ. A critique of animal analogues of alcoholism. In: Majchrowicz, E.; Noble, EP., editors. Biochemistry and pharmacology of ethanol. Vol. 2. New York: Plenum Press; p Colombo G. ESBRA-Nordmann 1996 Award Lecture: ethanol drinking behaviour in Sardinian alcohol-preferring rats. Alcohol Alcohol. 1997; 32: [PubMed: ]
8 SPENCE et al. Page 8 Cowen MS, Chen F, Lawrence AJ. Neuropeptides: implications for alcoholism. J Neurochem. 2004; 89: [PubMed: ] Ehlers CL, Li TK, Lumeng L, Hwang BH, Somes C, Jimenez P, Mathe AA. Neuropeptide Y levels in ethanol-naive alcohol-preferring and nonpreferring rats and in Wistar rats after ethanol exposure. Alcohol Clin Exp Res. 1998; 22: [PubMed: ] Fiskerstrand CE, Lovejoy E, Gerrard L, Quinn JP. An intronic domain within the rat preprotachykinin- A gene containing a CCCT repetitive motif acts as an enhancer in differentiating embryonic stem cells. Neurosci Lett. 1999; 263: [PubMed: ] Foroud T, Li TK. Genetics of alcoholism: a review of recent studies in human and animal models. Am J Addict. 1999; 8: [PubMed: ] Gilpin NW, Stewart RB, Murphy JM, Li TK, Badia-Elder NE. Neuropeptide Y reduces oral ethanol intake in alcohol-preferring (P) rats following a period of imposed ethanol abstinence. Alcohol Clin Exp Res. 2003; 27: [PubMed: ] Hamada H, Seidman M, Howard BH, Gorman CM. Enhanced gene expression by the poly(dt-dg). poly (dc-da) sequence. Mol Cell Biol. 1984; 4: [PubMed: ] Heath AC, Bucholz KK, Madden PAF, Dinwiddie SH, Slutske WS, Statham DJ, Dunne MP, Whitfield J, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997; 27: [PubMed: ] Heilig M, Thorsell. A Brain neuropeptide Y (NPY) in stress and alcohol dependence. Rev Neurosci. 2002; 13: [PubMed: ] Hwang BH, Zhang JK, Ehlers CL, Lumeng L, Li TK. Innate differences of neuropeptide Y (NPY) in hypothalamic nuclei and central nucleus of the amygdala between selectively bred rats with high and low alcohol preference. Alcohol Clin Exp Res. 1999; 23: [PubMed: ] Katsuki S, Kato J, Nakajima M, Inui N, Sasaki K, Kohgo Y, Niitsu Y. Analysis of CA repeats in first intron of class I ADH gene in Long-Evans Cinnamon rats developing fatal intoxication after ethanol intake. Alcohol Clin Exp Res. 1996; 20:33A 35A. Koob GF, Roberts AJ, Schulteis G, Parsons LH, Heyser CJ, Hyytia P, Merlo-Pich E, Weiss F. Neurocircuitry targets in ethanol reward and dependence. Alcohol Clin Exp Res. 1998; 22:3 9. [PubMed: ] Larhammar D, Ericsson A, Persson H. Structure and expression of the rat neuropeptide Y gene. Proc Natl Acad Sci USA. 1987; 84: [PubMed: ] Lappalainen J, Kranzler HR, Malison R, Price LH, Van Dyck C, Rosenheck RA, Cramer J, Southwick S, Charney D, Krystal J, Gelernter J. A functional neuropeptide Y Leu7Pro polymorphism associated with alcohol dependence in a large population sample from the United States. Arch Gen Psychiatry. 2002; 59: [PubMed: ] Li T-K, Lumeng L, Doolittle DP, Carr LG. Molecular associations of alcohol-seeking behavior in rat lines selectively bred for high and low voluntary ethanol drinking. Alcohol Alcohol. 1991; 1: Loflin P, Lever JE. HuR binds a cyclic nucleotide-dependent, stabilizing domain in the 3 untranslated region of Na (+) /glucose cotransporter (SGLT1) mrna. FEBS Lett. 2001; 509: [PubMed: ] McBride WJ, Li TK. Animal models of alcoholism: neurobiology of high alcohol-drinking behavior in rodents. Crit Rev Neurobiol. 1998; 12: [PubMed: ] Nair AK, Menon KM. Regulatory role of the 3 untranslated region of luteinizing hormone receptor: effect on mrna stability. FEBS Lett. 2000; 471: [PubMed: ] Pandey SC, Carr LG, Heilig M, Ilveskoski E, Thiele TE. Neuropeptide y and alcoholism: genetic, molecular, and pharmacological evidence. Alcohol Clin Exp Res. 2003; 27: [PubMed: ] Paxinos, G.; Watson, C. The rat brain in stereotoxic coordinates. 4. San Diego, CA: Academic Press; Stewart RB, Gatto GJ, Lumeng L, Li T-K, Murphy JM. Comparison of alcohol-preferring (P) and nonpreferring (NP) rats on tests of anxiety and for the anxiolytic effects of ethanol. Alcohol. 1993; 10:1 10. [PubMed: ]
9 SPENCE et al. Page 9 Thiele TE, Marsh DJ, Ste Marie L, Bernstein IL, Palmiter RD. Ethanol consumption and resistance are inversely related to neuropeptide Y levels. Nature. 1998; 396: [PubMed: ] Thiele TE, Badia-Elder NE. A role for neuropeptide Y in alcohol intake control: evidence from human and animal research. Physiol Behav. 2003; 79: [PubMed: ] Thomasson HR, Crabb DW, Edenberg HJ, Li TK. Alcohol and aldehyde dehydrogenase polymorphisms and alcoholism. Behav Genet. 1993; 23: [PubMed: ] Wang Z, Kiledjian M. The poly(a)-binding protein and an mrna stability protein jointly regulate an endoribonuclease activity. Mol Cell Biol. 2000; 20: [PubMed: ] Zhu G, Pollak L, Mottagui-Tabar S, Wahlestedt C, Taubman J, Virkkunen M, Goldman D, Heilig M. NPY Leu7Pro and alcohol dependence in Finnish and Swedish populations. Alcohol Clin Exp Res. 2003; 27: [PubMed: ]
10 SPENCE et al. Page 10 Fig. 1. NPY mrna expression is decreased in alcohol-naive P rats compared with NP rats. qrt- PCR analysis was utilized to compare the relative levels of NPY mrna between alcoholnaive ip and inp rats in the nucleus accumbens, the frontal cortex, the amygdala, the hippocampus, the caudate putamen, and the hypothalamus. All values were determined using the standard curve method and compared with mrna expression in the ip hypothalamus, which was arbitrarily designated 1. The graph depicts the mean±s.e.m. of the results from five independent ip and four independent inp experiments performed in triplicate, using separate preparations of cdna. Significant differences in regional NPY mrna expression between ip and inp rats was determined using the Student s t-test. * P<0.05; ** P<
11 SPENCE et al. Page 11 Fig. 2. Polymorphisms identified in the NPY gene. Sequence analysis was previously performed on the NPY gene and identified three polymorphisms in the 2nd intron, 3rd intron and 3 UTR (Bice et al., 1998). (A) The diagram depicts the NPY gene. NPY s four exons are represented with white boxes, while a black line depicts the intronic regions. The microsatellite marker D4Mit7 is represented with a black box. The three polymorphisms that have been identified in the NPY gene are labeled with arrows and their position relative to the translational start point (Tsp) that was designated +1. (B) Three polymorphisms are defined by their regional location in the NPY gene, their position relative to the Tsp (+1), and the sequence difference detected between ip and inp rats.
12 SPENCE et al. Page 12 Fig. 3. Functional importance of the NPY polymorphisms identified in the 2nd intron, 3rd intron, and 3 UTR. The Luc constructs were transiently transfected into SK-N-SH cells, and the resulting effect on luciferase expression was subsequently determined. SV40, luc+, and poly(a) denote the SV40 promoter, the luciferase gene, and the SV40 late poly(a) signal, respectively. The polymorphisms, located at +4666, +6871, and +7010, are noted in addition to the relative insertion site of each fragment into the pgl-3 Promoter vector. The activity of each construct was normalized to the internal control plasmid, prl-cmv, and was expressed as fold change compared with the activity of the pgl-3 Promoter vector, which was designated as 1. The bars and fold change show the mean±s.e.m. of the results from seven independent transfection experiments performed in triplicate, using two different plasmid preparations. The significance of differences in the resulting mean values within and between multiple constructs was analyzed using ANOVA. An asterisk denotes a significant reduction in luc expression.
13 SPENCE et al. Page 13 Table 1 List of oligos Real-time PCR NPY-1F 5 -agatactactccgctctgcga-3 NPY-1R 5 -ggcattttctgtgctttctct-3 GAPDH-F 5 -cagtcaaggctgagaatggga-3 GAPDH-R 5 -gggatctcgctcctggaag-3 Vector construction Luc-1F 5 -cggatccaactttgacttccaacag-3 Luc-1R 5 -cggatccatcaactctggaaaaac-3 Luc-2F 5 -cggatccaagtactcccttagagactg-3 Luc-2R 5 -cggatcctaaaacacaagaggcaaa-3 Luc-3F 5 -ctctagaatgaaacttgctctcctgactt-3 Luc-3R 5 -atctagatagtcacaccaggtgttcagtc-3 Luc-4R 5 -gacgatagtcatgccccgcg-3
Jonathan Kindberg BNF Molecular Biology Through Discovery. Research Proposal
 Jonathan Kindberg BNF0 491 Molecular Biology Through Discovery Research Proposal The effect of a single nucleotide polymorphism in the NPY protein and ethanol consumption in mice (08 December 2012) I.
Jonathan Kindberg BNF0 491 Molecular Biology Through Discovery Research Proposal The effect of a single nucleotide polymorphism in the NPY protein and ethanol consumption in mice (08 December 2012) I.
HHS Public Access Author manuscript Addict Biol. Author manuscript; available in PMC 2015 September 04.
 Expression Profiling and QTL Analysis: a Powerful Complementary Strategy in Drug Abuse Research JOHN P. SPENCE 1, TIEBING LIANG 1, TATIANA FOROUD 2, DAVID LO 3, and LUCINDA G. CARR 1 1 Department of Medicine,
Expression Profiling and QTL Analysis: a Powerful Complementary Strategy in Drug Abuse Research JOHN P. SPENCE 1, TIEBING LIANG 1, TATIANA FOROUD 2, DAVID LO 3, and LUCINDA G. CARR 1 1 Department of Medicine,
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Supporting Information Table of content
 Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
The levels of mrna expression in the mouse brain were measured at 52 dpi after the brains were divided into five regions: cortex, hippocampus,
 Supplemental material Supplemental method RNA extraction, reverse transcription, and real-time PCR The levels of mrna expression in the mouse brain were measured at 52 dpi after the brains were divided
Supplemental material Supplemental method RNA extraction, reverse transcription, and real-time PCR The levels of mrna expression in the mouse brain were measured at 52 dpi after the brains were divided
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Data Sheet. Notch Pathway Reporter Kit Catalog # 60509
 Data Sheet Notch Pathway Reporter Kit Catalog # 60509 6042 Cornerstone Court W, Ste B Background The Notch signaling pathway controls cell fate decisions in vertebrate and invertebrate tissues. NOTCH signaling
Data Sheet Notch Pathway Reporter Kit Catalog # 60509 6042 Cornerstone Court W, Ste B Background The Notch signaling pathway controls cell fate decisions in vertebrate and invertebrate tissues. NOTCH signaling
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Genetic Analysis of Anxiety Related Behaviors by Gene Chip and In situ Hybridization of the Hippocampus and Amygdala of C57BL/6J and AJ Mice Brains
 Genetic Analysis of Anxiety Related Behaviors by Gene Chip and In situ Hybridization of the Hippocampus and Amygdala of C57BL/6J and AJ Mice Brains INTRODUCTION To study the relationship between an animal's
Genetic Analysis of Anxiety Related Behaviors by Gene Chip and In situ Hybridization of the Hippocampus and Amygdala of C57BL/6J and AJ Mice Brains INTRODUCTION To study the relationship between an animal's
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
FOXO Reporter Kit PI3K/AKT Pathway Cat. #60643
 Data Sheet FOXO Reporter Kit PI3K/AKT Pathway Cat. #60643 Background The PI3K/AKT signaling pathway is essential for cell growth and survival. Disruption of this pathway or its regulation has been linked
Data Sheet FOXO Reporter Kit PI3K/AKT Pathway Cat. #60643 Background The PI3K/AKT signaling pathway is essential for cell growth and survival. Disruption of this pathway or its regulation has been linked
Deficits in amygdaloid camp-responsive element binding protein signaling play a role in genetic predisposition to anxiety and alcoholism
 Research article Related Commentary, page 2697 Deficits in amygdaloid camp-responsive element binding protein signaling play a role in genetic predisposition to anxiety and alcoholism Subhash C. Pandey,
Research article Related Commentary, page 2697 Deficits in amygdaloid camp-responsive element binding protein signaling play a role in genetic predisposition to anxiety and alcoholism Subhash C. Pandey,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Cell isolation. Spleen and lymph nodes (axillary, inguinal) were removed from mice
 Supplementary Methods: Cell isolation. Spleen and lymph nodes (axillary, inguinal) were removed from mice and gently meshed in DMEM containing 10% FBS to prepare for single cell suspensions. CD4 + CD25
Supplementary Methods: Cell isolation. Spleen and lymph nodes (axillary, inguinal) were removed from mice and gently meshed in DMEM containing 10% FBS to prepare for single cell suspensions. CD4 + CD25
Supplementary information
 Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
Supplemental Materials and Methods Plasmids and viruses Quantitative Reverse Transcription PCR Generation of molecular standard for quantitative PCR
 Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using
 Supplementary Information Materials and Methods RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using Trizol reagent (Invitrogen,Carlsbad, CA) according to the manufacturer's instructions.
Supplementary Information Materials and Methods RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using Trizol reagent (Invitrogen,Carlsbad, CA) according to the manufacturer's instructions.
Viral vector-induced amygdala NPYoverexpression reverses increased alcohol intake caused by repeated deprivations in Wistar rats
 Brain Advance Access published April 2, 2007 doi:10.1093/brain/awm033 Brain (2007) Page 1 of 8 Viral vector-induced amygdala NPYoverexpression reverses increased alcohol intake caused by repeated deprivations
Brain Advance Access published April 2, 2007 doi:10.1093/brain/awm033 Brain (2007) Page 1 of 8 Viral vector-induced amygdala NPYoverexpression reverses increased alcohol intake caused by repeated deprivations
In vitro DNase I foot printing. In vitro DNase I footprinting was performed as described
 Supplemental Methods In vitro DNase I foot printing. In vitro DNase I footprinting was performed as described previously 1 2 using 32P-labeled 211 bp fragment from 3 HS1. Footprinting reaction mixes contained
Supplemental Methods In vitro DNase I foot printing. In vitro DNase I footprinting was performed as described previously 1 2 using 32P-labeled 211 bp fragment from 3 HS1. Footprinting reaction mixes contained
Notch Signaling Pathway Notch CSL Reporter HEK293 Cell line Catalog #: 60652
 Notch Signaling Pathway Notch CSL Reporter HEK293 Cell line Catalog #: 60652 Background The Notch signaling pathway controls cell fate decisions in vertebrate and invertebrate tissues. Notch signaling
Notch Signaling Pathway Notch CSL Reporter HEK293 Cell line Catalog #: 60652 Background The Notch signaling pathway controls cell fate decisions in vertebrate and invertebrate tissues. Notch signaling
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
Chapter 2 Gene and Promoter Structures of the Dopamine Receptors
 Chapter 2 Gene and Promoter Structures of the Dopamine Receptors Ursula M. D Souza Abstract The dopamine receptors have been classified into two groups, the D 1 - like and D 2 -like dopamine receptors,
Chapter 2 Gene and Promoter Structures of the Dopamine Receptors Ursula M. D Souza Abstract The dopamine receptors have been classified into two groups, the D 1 - like and D 2 -like dopamine receptors,
CHAPTER 4 RESULTS. showed that all three replicates had similar growth trends (Figure 4.1) (p<0.05; p=0.0000)
 CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
Phosphate buffered saline (PBS) for washing the cells TE buffer (nuclease-free) ph 7.5 for use with the PrimePCR Reverse Transcription Control Assay
 Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation
 Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Circular RNAs (circrnas) act a stable mirna sponges
 Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Supplementary Material
 Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
 MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
LIMBIC SYSTEM. Dr. Amani A. Elfaki Associate Professor Department of Anatomy
 LIMBIC SYSTEM Dr. Amani A. Elfaki Associate Professor Department of Anatomy Learning Objectives Define the limbic system Identify the parts of the limbic system Describe the circulation of the limbic system
LIMBIC SYSTEM Dr. Amani A. Elfaki Associate Professor Department of Anatomy Learning Objectives Define the limbic system Identify the parts of the limbic system Describe the circulation of the limbic system
Single Cell Quantitative Polymer Chain Reaction (sc-qpcr)
 Single Cell Quantitative Polymer Chain Reaction (sc-qpcr) Analyzing gene expression profiles from a bulk population of cells provides an average profile which may obscure important biological differences
Single Cell Quantitative Polymer Chain Reaction (sc-qpcr) Analyzing gene expression profiles from a bulk population of cells provides an average profile which may obscure important biological differences
Department of Pharmaceutical Sciences, School of Pharmacy, Northeastern University, Boston, MA 02115, USA 2
 Pancreatic Cancer Cell Exosome-Mediated Macrophage Reprogramming and the Role of MicroRNAs 155 and 125b2 Transfection using Nanoparticle Delivery Systems Mei-Ju Su 1, Hibah Aldawsari 2, and Mansoor Amiji
Pancreatic Cancer Cell Exosome-Mediated Macrophage Reprogramming and the Role of MicroRNAs 155 and 125b2 Transfection using Nanoparticle Delivery Systems Mei-Ju Su 1, Hibah Aldawsari 2, and Mansoor Amiji
well for 2 h at rt. Each dot represents an individual mouse and bar is the mean ±
 Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
The Biology of Addiction
 The Biology of Addiction Risk factors for addiction: Biological/Genetic Family history of addiction Being male Having mental illness Exposure to substances in utero * The genes that people are born with
The Biology of Addiction Risk factors for addiction: Biological/Genetic Family history of addiction Being male Having mental illness Exposure to substances in utero * The genes that people are born with
Lack of Association between Endoplasmic Reticulum Stress Response Genes and Suicidal Victims
 Kobe J. Med. Sci., Vol. 53, No. 4, pp. 151-155, 2007 Lack of Association between Endoplasmic Reticulum Stress Response Genes and Suicidal Victims KAORU SAKURAI 1, NAOKI NISHIGUCHI 2, OSAMU SHIRAKAWA 2,
Kobe J. Med. Sci., Vol. 53, No. 4, pp. 151-155, 2007 Lack of Association between Endoplasmic Reticulum Stress Response Genes and Suicidal Victims KAORU SAKURAI 1, NAOKI NISHIGUCHI 2, OSAMU SHIRAKAWA 2,
Supplemental Figure 1
 1 Supplemental Figure 1 Effects of DATE shortening on HGF promoter activity. The HGF promoter region (-1037 to +56) containing wild-type (30As) or truncated DATE (26As, 27As, 28A, 29As) from breast cancer
1 Supplemental Figure 1 Effects of DATE shortening on HGF promoter activity. The HGF promoter region (-1037 to +56) containing wild-type (30As) or truncated DATE (26As, 27As, 28A, 29As) from breast cancer
HIF-inducible mir-191 promotes migration in breast cancer through complex regulation of TGFβ-signaling in hypoxic microenvironment.
 HIF-inducible mir-9 promotes migration in breast cancer through complex regulation of TGFβ-signaling in hypoxic microenvironment. Neha Nagpal, Hafiz M Ahmad, Shibu Chameettachal3, Durai Sundar, Sourabh
HIF-inducible mir-9 promotes migration in breast cancer through complex regulation of TGFβ-signaling in hypoxic microenvironment. Neha Nagpal, Hafiz M Ahmad, Shibu Chameettachal3, Durai Sundar, Sourabh
SOM Husse et al. Supplementary online material. Synaptotagmin10-Cre, a driver to disrupt clock genes in the SCN
 SOM Husse et al. Supplementary online material Synaptotagmin10-Cre, a driver to disrupt clock genes in the SCN Jana Husse, Xunlei Zhou, Anton Shostak, Henrik Oster and Gregor Eichele SOM Husse et al.,
SOM Husse et al. Supplementary online material Synaptotagmin10-Cre, a driver to disrupt clock genes in the SCN Jana Husse, Xunlei Zhou, Anton Shostak, Henrik Oster and Gregor Eichele SOM Husse et al.,
Islet viability assay and Glucose Stimulated Insulin Secretion assay RT-PCR and Western Blot
 Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
Doctor of Philosophy
 Regulation of Gene Expression of the 25-Hydroxyvitamin D la-hydroxylase (CYP27BI) Promoter: Study of A Transgenic Mouse Model Ivanka Hendrix School of Molecular and Biomedical Science The University of
Regulation of Gene Expression of the 25-Hydroxyvitamin D la-hydroxylase (CYP27BI) Promoter: Study of A Transgenic Mouse Model Ivanka Hendrix School of Molecular and Biomedical Science The University of
Table S1. Primers and PCR protocols for mutation screening of MN1, NF2, KREMEN1 and ZNRF3.
 Table S1. Primers and PCR protocols for mutation screening of MN1, NF2, KREMEN1 and ZNRF3. MN1 (Accession No. NM_002430) MN1-1514F 5 -GGCTGTCATGCCCTATTGAT Exon 1 MN1-1882R 5 -CTGGTGGGGATGATGACTTC Exon
Table S1. Primers and PCR protocols for mutation screening of MN1, NF2, KREMEN1 and ZNRF3. MN1 (Accession No. NM_002430) MN1-1514F 5 -GGCTGTCATGCCCTATTGAT Exon 1 MN1-1882R 5 -CTGGTGGGGATGATGACTTC Exon
SUPPLEMENTARY MATERIAL
 SYPPLEMENTARY FIGURE LEGENDS SUPPLEMENTARY MATERIAL Figure S1. Phylogenic studies of the mir-183/96/182 cluster and 3 -UTR of Casp2. (A) Genomic arrangement of the mir-183/96/182 cluster in vertebrates.
SYPPLEMENTARY FIGURE LEGENDS SUPPLEMENTARY MATERIAL Figure S1. Phylogenic studies of the mir-183/96/182 cluster and 3 -UTR of Casp2. (A) Genomic arrangement of the mir-183/96/182 cluster in vertebrates.
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated
 Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Retro-X qrt-pcr Titration Kit User Manual
 Takara Bio USA Retro-X qrt-pcr Titration Kit User Manual Cat. No. 631453 PT3952-1 (030218) 1290 Terra Bella Avenue, Mountain View, CA 94043, USA U.S. Technical Support: techus@takarabio.com United States/Canada
Takara Bio USA Retro-X qrt-pcr Titration Kit User Manual Cat. No. 631453 PT3952-1 (030218) 1290 Terra Bella Avenue, Mountain View, CA 94043, USA U.S. Technical Support: techus@takarabio.com United States/Canada
National Disease Research Interchange Annual Progress Report: 2010 Formula Grant
 National Disease Research Interchange Annual Progress Report: 2010 Formula Grant Reporting Period July 1, 2012 June 30, 2013 Formula Grant Overview The National Disease Research Interchange received $62,393
National Disease Research Interchange Annual Progress Report: 2010 Formula Grant Reporting Period July 1, 2012 June 30, 2013 Formula Grant Overview The National Disease Research Interchange received $62,393
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Behavioural effects of ketamine in non-stressed and stressed mice. Naive C57BL/6 adult male mice (n=10/group) were given a single dose of saline vehicle or ketamine (3.0 mg/kg,
Supplementary Figure 1. Behavioural effects of ketamine in non-stressed and stressed mice. Naive C57BL/6 adult male mice (n=10/group) were given a single dose of saline vehicle or ketamine (3.0 mg/kg,
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers
 Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
TITLE: A Mouse Model to Investigate the Role of DBC2 in Breast Cancer
 AD Award Number: W81XWH-04-1-0325 TITLE: A Mouse Model to Investigate the Role of DBC2 in Breast Cancer PRINCIPAL INVESTIGATOR: Valerie Boka CONTRACTING ORGANIZATION: University of Texas Health Science
AD Award Number: W81XWH-04-1-0325 TITLE: A Mouse Model to Investigate the Role of DBC2 in Breast Cancer PRINCIPAL INVESTIGATOR: Valerie Boka CONTRACTING ORGANIZATION: University of Texas Health Science
SUPPLEMENTARY INFORMATION
 -. -. SUPPLEMENTARY INFORMATION DOI: 1.1/ncb86 a WAT-1 WAT- BAT-1 BAT- sk-muscle-1 sk-muscle- mir-133b mir-133a mir-6 mir-378 mir-1 mir-85 mir-378 mir-6a mir-18 mir-133a mir- mir- mir-341 mir-196a mir-17
-. -. SUPPLEMENTARY INFORMATION DOI: 1.1/ncb86 a WAT-1 WAT- BAT-1 BAT- sk-muscle-1 sk-muscle- mir-133b mir-133a mir-6 mir-378 mir-1 mir-85 mir-378 mir-6a mir-18 mir-133a mir- mir- mir-341 mir-196a mir-17
Effects of Repeated Alcohol Deprivations on Operant Ethanol Self-Administration by Alcohol-Preferring (P) Rats
 (3) 28, 1614 1621 & 3 Nature Publishing Group All rights reserved 0893-133X/03 $25.00 www.neuropsychopharmacology.org Effects of Repeated Alcohol Deprivations on Operant Ethanol Self-Administration by
(3) 28, 1614 1621 & 3 Nature Publishing Group All rights reserved 0893-133X/03 $25.00 www.neuropsychopharmacology.org Effects of Repeated Alcohol Deprivations on Operant Ethanol Self-Administration by
SUPPLEMENTARY INFORMATION. Divergent TLR7/9 signaling and type I interferon production distinguish
 SUPPLEMENTARY INFOATION Divergent TLR7/9 signaling and type I interferon production distinguish pathogenic and non-pathogenic AIDS-virus infections Judith N. Mandl, Ashley P. Barry, Thomas H. Vanderford,
SUPPLEMENTARY INFOATION Divergent TLR7/9 signaling and type I interferon production distinguish pathogenic and non-pathogenic AIDS-virus infections Judith N. Mandl, Ashley P. Barry, Thomas H. Vanderford,
Supplementary Information
 Supplementary Information The neural correlates of subjective value during intertemporal choice Joseph W. Kable and Paul W. Glimcher a 10 0 b 10 0 10 1 10 1 Discount rate k 10 2 Discount rate k 10 2 10
Supplementary Information The neural correlates of subjective value during intertemporal choice Joseph W. Kable and Paul W. Glimcher a 10 0 b 10 0 10 1 10 1 Discount rate k 10 2 Discount rate k 10 2 10
Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples
 DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples LIBRARY
DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples LIBRARY
DNA context and promoter activity affect gene expression in lentiviral vectors
 ACTA BIOMED 2008; 79: 192-196 Mattioli 1885 O R I G I N A L A R T I C L E DNA context and promoter activity affect gene expression in lentiviral vectors Gensheng Mao 1, Francesco Marotta 2, Jia Yu 3, Liang
ACTA BIOMED 2008; 79: 192-196 Mattioli 1885 O R I G I N A L A R T I C L E DNA context and promoter activity affect gene expression in lentiviral vectors Gensheng Mao 1, Francesco Marotta 2, Jia Yu 3, Liang
Microarray profiling of gene expression after sleep deprivation and recovery sleep 03/05/2008
 Microarray profiling of gene expression after sleep deprivation and recovery sleep 03/05/2008 1. OVERVIEW...1 2. BEHAVIORAL CONDITIONS...1 3. LASER MICRODISSECTION/MICROARRAY...2 3.1. REGIONS OF INTEREST...
Microarray profiling of gene expression after sleep deprivation and recovery sleep 03/05/2008 1. OVERVIEW...1 2. BEHAVIORAL CONDITIONS...1 3. LASER MICRODISSECTION/MICROARRAY...2 3.1. REGIONS OF INTEREST...
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Instructions for Use. RealStar Influenza Screen & Type RT-PCR Kit /2017 EN
 Instructions for Use RealStar Influenza Screen & Type RT-PCR Kit 4.0 05/2017 EN RealStar Influenza Screen & Type RT-PCR Kit 4.0 For research use only! (RUO) 164003 INS-164000-EN-S01 96 05 2017 altona
Instructions for Use RealStar Influenza Screen & Type RT-PCR Kit 4.0 05/2017 EN RealStar Influenza Screen & Type RT-PCR Kit 4.0 For research use only! (RUO) 164003 INS-164000-EN-S01 96 05 2017 altona
Electron micrograph of phosphotungstanic acid-stained exosomes derived from murine
 1 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Physical properties of murine DC-derived exosomes. a, Electron micrograph of phosphotungstanic acid-stained exosomes derived from
1 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Physical properties of murine DC-derived exosomes. a, Electron micrograph of phosphotungstanic acid-stained exosomes derived from
Supplemental information
 Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
Limbic system. Lecture 29, November 10, 2017
 Limbic system Lecture 29, November 10, 2017 Circadian rhythms (Latin, approximately a day ) Regulation of our daily rhythm Eating Sleeping Defecating Periods of activity Suprachiasmatic n. http://slideplayer.com/slide/6351731/
Limbic system Lecture 29, November 10, 2017 Circadian rhythms (Latin, approximately a day ) Regulation of our daily rhythm Eating Sleeping Defecating Periods of activity Suprachiasmatic n. http://slideplayer.com/slide/6351731/
CRISPR/Cas9 Enrichment and Long-read WGS for Structural Variant Discovery
 CRISPR/Cas9 Enrichment and Long-read WGS for Structural Variant Discovery PacBio CoLab Session October 20, 2017 For Research Use Only. Not for use in diagnostics procedures. Copyright 2017 by Pacific Biosciences
CRISPR/Cas9 Enrichment and Long-read WGS for Structural Variant Discovery PacBio CoLab Session October 20, 2017 For Research Use Only. Not for use in diagnostics procedures. Copyright 2017 by Pacific Biosciences
Supplementary Figure 1
 Supplementary Figure 1 3 3 3 1 1 Bregma -1.6mm 3 : Bregma Ref) Http://www.mbl.org/atlas165/atlas165_start.html Bregma -.18mm Supplementary Figure 1 Schematic representation of the utilized brain slice
Supplementary Figure 1 3 3 3 1 1 Bregma -1.6mm 3 : Bregma Ref) Http://www.mbl.org/atlas165/atlas165_start.html Bregma -.18mm Supplementary Figure 1 Schematic representation of the utilized brain slice
Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression
 Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
THE ROLE OF CORTICOTROPIN-RELEASING FACTOR (CRF) SIGNALING IN THE VENTRAL TEGMENTAL AREA IN THE REGULATION OF BINGE-LIKE ALCOHOL CONSUMPTION
 Gulati 1 THE ROLE OF CORTICOTROPIN-RELEASING FACTOR (CRF) SIGNALING IN THE VENTRAL TEGMENTAL AREA IN THE REGULATION OF BINGE-LIKE ALCOHOL CONSUMPTION Varun Gulati A thesis submitted to the faculty at the
Gulati 1 THE ROLE OF CORTICOTROPIN-RELEASING FACTOR (CRF) SIGNALING IN THE VENTRAL TEGMENTAL AREA IN THE REGULATION OF BINGE-LIKE ALCOHOL CONSUMPTION Varun Gulati A thesis submitted to the faculty at the
Pair-fed % inkt cells 0.5. EtOH 0.0
 MATERIALS AND METHODS Histopathological analysis Liver tissue was collected 9 h post-gavage, and the tissue samples were fixed in 1% formalin and paraffin-embedded following a standard procedure. The embedded
MATERIALS AND METHODS Histopathological analysis Liver tissue was collected 9 h post-gavage, and the tissue samples were fixed in 1% formalin and paraffin-embedded following a standard procedure. The embedded
Supplemental Data. Short Article. ATF4-Mediated Induction of 4E-BP1. Contributes to Pancreatic β Cell Survival. under Endoplasmic Reticulum Stress
 Cell Metabolism, Volume 7 Supplemental Data Short Article ATF4-Mediated Induction of 4E-BP1 Contributes to Pancreatic β Cell Survival under Endoplasmic Reticulum Stress Suguru Yamaguchi, Hisamitsu Ishihara,
Cell Metabolism, Volume 7 Supplemental Data Short Article ATF4-Mediated Induction of 4E-BP1 Contributes to Pancreatic β Cell Survival under Endoplasmic Reticulum Stress Suguru Yamaguchi, Hisamitsu Ishihara,
Oncolytic Adenovirus Complexes Coated with Lipids and Calcium Phosphate for Cancer Gene Therapy
 Oncolytic Adenovirus Complexes Coated with Lipids and Calcium Phosphate for Cancer Gene Therapy Jianhua Chen, Pei Gao, Sujing Yuan, Rongxin Li, Aimin Ni, Liang Chu, Li Ding, Ying Sun, Xin-Yuan Liu, Yourong
Oncolytic Adenovirus Complexes Coated with Lipids and Calcium Phosphate for Cancer Gene Therapy Jianhua Chen, Pei Gao, Sujing Yuan, Rongxin Li, Aimin Ni, Liang Chu, Li Ding, Ying Sun, Xin-Yuan Liu, Yourong
Supplementary Fig. S1. Schematic diagram of minigenome segments.
 open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types.
 Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
1. Intended Use New Influenza A virus real time RT-PCR Panel is used for the detection of universal influenza A virus, universal swine Influenza A vir
 New Influenza A Virus Real Time RT-PCR Kit User Manual LT028310RRFY - 1 - 1. Intended Use New Influenza A virus real time RT-PCR Panel is used for the detection of universal influenza A virus, universal
New Influenza A Virus Real Time RT-PCR Kit User Manual LT028310RRFY - 1 - 1. Intended Use New Influenza A virus real time RT-PCR Panel is used for the detection of universal influenza A virus, universal
Instructions for Use. RealStar Influenza S&T RT-PCR Kit /2017 EN
 Instructions for Use RealStar Influenza S&T RT-PCR Kit 3.0 01/2017 EN RealStar Influenza S&T RT-PCR Kit 3.0 For research use only! (RUO) 163003 INS-163000-EN-S02 96 01 2017 altona Diagnostics GmbH Mörkenstr.
Instructions for Use RealStar Influenza S&T RT-PCR Kit 3.0 01/2017 EN RealStar Influenza S&T RT-PCR Kit 3.0 For research use only! (RUO) 163003 INS-163000-EN-S02 96 01 2017 altona Diagnostics GmbH Mörkenstr.
File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables. File Name: Peer Review File Description:
 File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables File Name: Peer Review File Description: Primer Name Sequence (5'-3') AT ( C) RT-PCR USP21 F 5'-TTCCCATGGCTCCTTCCACATGAT-3'
File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables File Name: Peer Review File Description: Primer Name Sequence (5'-3') AT ( C) RT-PCR USP21 F 5'-TTCCCATGGCTCCTTCCACATGAT-3'
Nature Genetics: doi: /ng Supplementary Figure 1. Brain magnetic resonance imaging in patient 9 at age 3.6 years.
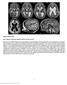 Supplementary Figure 1 Brain magnetic resonance imaging in patient 9 at age 3.6 years. (a d) Axial T2-weighted images show a marked degree of ventriculomegaly. Note the diencephalic mesencephalic junction
Supplementary Figure 1 Brain magnetic resonance imaging in patient 9 at age 3.6 years. (a d) Axial T2-weighted images show a marked degree of ventriculomegaly. Note the diencephalic mesencephalic junction
RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice
 SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
Generating Mouse Models of Pancreatic Cancer
 Generating Mouse Models of Pancreatic Cancer Aom Isbell http://www2.massgeneral.org/cancerresourceroom/types/gi/index.asp Spring/Summer 1, 2012 Alexandros Tzatsos, MD PhD Bardeesy Lab: Goals and Objectives
Generating Mouse Models of Pancreatic Cancer Aom Isbell http://www2.massgeneral.org/cancerresourceroom/types/gi/index.asp Spring/Summer 1, 2012 Alexandros Tzatsos, MD PhD Bardeesy Lab: Goals and Objectives
Chapter 2. Investigation into mir-346 Regulation of the nachr α5 Subunit
 15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep
 SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
TITLE: The Role of hcdc4 as a Tumor Suppressor Gene in Genomic Instability Underlying Prostate Cancer
 AD Award Number: TITLE: The Role of hcdc4 as a Tumor Suppressor Gene in Genomic Instability Underlying Prostate Cancer PRINCIPAL INVESTIGATOR: Audrey van Drogen, Ph.D. CONTRACTING ORGANIZATION: Sidney
AD Award Number: TITLE: The Role of hcdc4 as a Tumor Suppressor Gene in Genomic Instability Underlying Prostate Cancer PRINCIPAL INVESTIGATOR: Audrey van Drogen, Ph.D. CONTRACTING ORGANIZATION: Sidney
Neurobiology of Addiction
 Neurobiology of Addiction Domenic A. Ciraulo, MD Director of Alcohol Pharmacotherapy Research Center for Addiction Medicine Department of Psychiatry Massachusetts General Hospital Disclosure Neither I
Neurobiology of Addiction Domenic A. Ciraulo, MD Director of Alcohol Pharmacotherapy Research Center for Addiction Medicine Department of Psychiatry Massachusetts General Hospital Disclosure Neither I
Supplementary Figures
 Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
The Neuroscience of Addiction: A mini-review
 The Neuroscience of Addiction: A mini-review Jim Morrill, MD, PhD MGH Charlestown HealthCare Center Massachusetts General Hospital Disclosures Neither I nor my spouse/partner has a relevant financial relationship
The Neuroscience of Addiction: A mini-review Jim Morrill, MD, PhD MGH Charlestown HealthCare Center Massachusetts General Hospital Disclosures Neither I nor my spouse/partner has a relevant financial relationship
Breast cancer. Risk factors you cannot change include: Treatment Plan Selection. Inferring Transcriptional Module from Breast Cancer Profile Data
 Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
BIOL212 Biochemistry of Disease. Metabolic Disorders - Obesity
 BIOL212 Biochemistry of Disease Metabolic Disorders - Obesity Obesity Approx. 23% of adults are obese in the U.K. The number of obese children has tripled in 20 years. 10% of six year olds are obese, rising
BIOL212 Biochemistry of Disease Metabolic Disorders - Obesity Obesity Approx. 23% of adults are obese in the U.K. The number of obese children has tripled in 20 years. 10% of six year olds are obese, rising
Families (n=227) with at least one child with ASD were recruited by the PARIS (Paris
 Materials and Methods Families Families (n=227) with at least one child with ASD were recruited by the PARIS (Paris Autism Research International Sibpair) study at specialized clinical centers in seven
Materials and Methods Families Families (n=227) with at least one child with ASD were recruited by the PARIS (Paris Autism Research International Sibpair) study at specialized clinical centers in seven
For in vitro Veterinary Diagnostics only. Kylt Rotavirus A. Real-Time RT-PCR Detection.
 For in vitro Veterinary Diagnostics only. Kylt Rotavirus A Real-Time RT-PCR Detection www.kylt.eu DIRECTION FOR USE Kylt Rotavirus A Real-Time RT-PCR Detection A. General Kylt Rotavirus A products are
For in vitro Veterinary Diagnostics only. Kylt Rotavirus A Real-Time RT-PCR Detection www.kylt.eu DIRECTION FOR USE Kylt Rotavirus A Real-Time RT-PCR Detection A. General Kylt Rotavirus A products are
Senior Thesis. Presented to. The Faculty of the School of Arts and Sciences Brandeis University
 Greenwald 1 Mouse intercellular adhesion molecule 1 (ICAM-1) isoforms demonstrate different binding affinities to mouse macrophage-1 antigen (Mac-1) and preliminary evidence for alternatively-spliced variants
Greenwald 1 Mouse intercellular adhesion molecule 1 (ICAM-1) isoforms demonstrate different binding affinities to mouse macrophage-1 antigen (Mac-1) and preliminary evidence for alternatively-spliced variants
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Received 26 January 1996/Returned for modification 28 February 1996/Accepted 15 March 1996
 MOLECULAR AND CELLULAR BIOLOGY, June 1996, p. 3012 3022 Vol. 16, No. 6 0270-7306/96/$04.00 0 Copyright 1996, American Society for Microbiology Base Pairing at the 5 Splice Site with U1 Small Nuclear RNA
MOLECULAR AND CELLULAR BIOLOGY, June 1996, p. 3012 3022 Vol. 16, No. 6 0270-7306/96/$04.00 0 Copyright 1996, American Society for Microbiology Base Pairing at the 5 Splice Site with U1 Small Nuclear RNA
mir 375 inhibits the proliferation of gastric cancer cells by repressing ERBB2 expression
 EXPERIMENTAL AND THERAPEUTIC MEDICINE 7: 1757-1761, 2014 mir 375 inhibits the proliferation of gastric cancer cells by repressing ERBB2 expression ZHI YONG SHEN *, ZI ZHEN ZHANG *, HUA LIU, EN HAO ZHAO
EXPERIMENTAL AND THERAPEUTIC MEDICINE 7: 1757-1761, 2014 mir 375 inhibits the proliferation of gastric cancer cells by repressing ERBB2 expression ZHI YONG SHEN *, ZI ZHEN ZHANG *, HUA LIU, EN HAO ZHAO
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
LESIONS OF THE MESOLIMBIC DOPAMINE SYSTEM DISRUPT SIGNALLED ESCAPE RESPONSES IN THE RAT
 ACTA NEUROBIOL: EXP. 1988, 48: 117-121 Short communication LESIONS OF THE MESOLIMBIC DOPAMINE SYSTEM DISRUPT SIGNALLED ESCAPE RESPONSES IN THE RAT W. Jeffrey WILSON and Jennifer C. HALL Department of Psychological
ACTA NEUROBIOL: EXP. 1988, 48: 117-121 Short communication LESIONS OF THE MESOLIMBIC DOPAMINE SYSTEM DISRUPT SIGNALLED ESCAPE RESPONSES IN THE RAT W. Jeffrey WILSON and Jennifer C. HALL Department of Psychological
RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
 Xu et al. Molecular Cancer (2019) 18:8 https://doi.org/10.1186/s12943-018-0932-8 LETTER TO THE EDITOR RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
Xu et al. Molecular Cancer (2019) 18:8 https://doi.org/10.1186/s12943-018-0932-8 LETTER TO THE EDITOR RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
Product Manual. Omni-Array Sense Strand mrna Amplification Kit, 2 ng to 100 ng Version Catalog No.: Reactions
 Genetic Tools and Reagents Universal mrna amplification, sense strand amplification, antisense amplification, cdna synthesis, micro arrays, gene expression, human, mouse, rat, guinea pig, cloning Omni-Array
Genetic Tools and Reagents Universal mrna amplification, sense strand amplification, antisense amplification, cdna synthesis, micro arrays, gene expression, human, mouse, rat, guinea pig, cloning Omni-Array
