Clinical presentation. Duration (Month)
|
|
|
- Paul Marshall
- 5 years ago
- Views:
Transcription
1 Supplementary information, Table S1A: Clinical information for 125 PAs. No. Subtype Gender (M/F) Age at diagnosis (year) Clinical presentation Duration (Month) Maximal adenoma diameter(cm) Tumor invasion* MIB-1 (Ki-67) 1 ACTH F 36 Hypertension NO ND 2 ACTH F 38 Amenorrhea NO <1% 3 ACTH F 43 Fatigue NO 2% 4 ACTH F 32 Weight gain NO <1% 5 ACTH F 36 Peripheral edema NO 1% 6 ACTH F 43 Weight gain NO 1% 7 ACTH F 41 Acne 12 1 NO 3% 8 ACTH F 15 Amenorrhea NO 2% 9 ACTH M 25 Weight gain NO <1% 10 ACTH F 36 Weight gain 72 1 NO <1% 11 ACTH F 45 Weight gain NO 1% 12 ACTH F 46 Weight gain NO 1% 13 ACTH F 40 Moon facies 55 5 YES 1% 14 ACTH F 36 Visual loss YES 4% 15 ACTH F 34 Weight gain NO 1% 16 ACTH M 36 Weight gain NO <1% 17 ACTH M 56 Fatigue NO 2% 18 ACTH M 25 Acne NO 1% 19 ACTH F 53 Hypertension NO 1% 20 ACTH F 46 Weight gain 6 4 YES <1 21 (GH+ACTH) F 62 Peripheral edema NO 4% 22 (GH+TSH) M 33 Visual loss YES <1% 23 M 23 Acromegaly YES 2% 24 (GH+TSH) M 42 Peripheral edema YES <1% 25 F 62 Acromegaly NO 1% 26 M 46 Acromegaly NO 1% 27 M 53 Acromegaly NO <1% 28 M 47 Acromegaly NO 1% 29 F 28 Acromegaly NO 1% 30 M 60 Acromegaly NO <1% 31 F 65 Acromegaly YES 1% 32 M 32 Acromegaly YES 2% 33 (GH+ACTH) F 46 Amenorrhea YES <1% 34 M 20 Acromegaly YES <1%
2 35 M 35 Acromegaly YES <1% 36 GH M 49 Acromegaly NO 2% 37 GH M 38 Acromegaly NO <1% 38 GH F 51 Acromegaly NO <1% 39 GH M 34 Acromegaly NO ND 40 GH F 50 Acromegaly NO <1% 41 GH M 45 Acromegaly NO ND 42 GH F 41 Acromegaly 48 3 NO <1% 43 GH M 49 Diabetes mellitus NO ND 44 GH M 27 Acromegaly YES <1% 45 GH F 60 Acromegaly NO 1% 46 GH F 33 Acromegaly NO ND 47 GH F 14 Headache NO 8% 48 GH F 31 Acromegaly NO ND 49 GH F 28 Acromegaly NO ND 50 GH F 18 Amenorrhea YES 2% 51 GH M 23 Acromegaly YES <1% 52 GH F 47 Acromegaly YES 1% 53 GH M 62 Acromegaly YES 2% 54 GH F 50 Acromegaly NO 1% 55 GH M 34 Acromegaly YES 4% 56 PRL M 30 Sexual dysfunction NO 2% 57 PRL M 45 Visual loss YES ND 58 PRL M 12 Headache NO <1% 59 PRL M 60 Headache 10 2 NO <1% 60 PRL M 53 Mammoplasia NO 1% 61 PRL M 68 Epistaxis YES 1% 62 PRL F 39 Amenorrhea 48 1 NO ND 63 PRL F 13 Amenorrhea NO <1% 64 PRL M 64 Visual loss NO 1% 65 PRL M 23 Visual loss 24 5 YES 3% 66 PRL F 15 Visual loss YES 5% 67 PRL M 30 Visual loss YES <1% 68 PRL M 25 Headache 24 4 YES 3% 69 PRL M 29 Sexual dysfunction YES <1% 70 PRL F 16 Amenorrhea NO 1% 71 PRL F 27 Amenorrhea NO 2% 72 PRL F 26 Amenorrhea NO 3% 73 PRL F 24 Amenorrhea NO 1% 74 PRL F 47 Visual loss YES <1% 75 PRL F 28 Amenorrhea NO 2% 76 M 28 Visual loss YES 2% 77 M 58 Headache NO 2% 78 F 48 Visual loss YES <1% 79 F 40 Amenorrhea YES <1% 80 M 48 Visual loss YES 1%
3 81 F 60 Visual loss YES <1% 82 M 44 Visual loss YES <1% 83 M 13 Visual loss YES 2% 84 M 31 Visual loss YES <1% 85 M 25 Visual loss YES 3% 86 M 34 Visual loss YES 3% 87 F 53 Visual loss YES <1% 88 F 22 Visual loss YES 2% 89 M 70 Fatigue NO 1% 90 M 57 Headache NO 2% 91 M 47 Visual loss YES 2% 92 M 59 Dizzy NO <1% 93 M 60 Dizzy NO <1% 94 M 25 Visual loss YES 2% 95 F 28 Amenorrhea YES 2% 96 GT M 52 Visual loss YES 1% 97 GT M 38 Visual loss YES ND 98 GT M 46 Visual loss YES <1% 99 GT M 40 Visual loss YES 2% 100 GT M 29 Visual loss YES <1% 101 GT M 71 Visual loss YES <1% 102 GT M 77 Visual loss 2 4 YES 1% 103 GT M 67 Visual loss YES 2% 104 GT M 34 Visual loss NO 1% 105 GT M 56 Fatigue NO <1% 106 GT M 38 Visual loss NO <1% 107 GT M 40 Visual loss 3 3 NO 2% 108 GT M 60 Headache NO ND 109 GT M 32 Visual loss NO 1% 110 GT M 24 Visual loss YES 2% 111 GT M 72 Visual loss NO 2% 112 GT M 62 Headache NO <1% 113 GT M 57 Visual loss NO 1% 114 GT M 47 Dizzy NO <1% 115 GT M 64 Headache NO <1% 116 TSH M 27 Fatigue YES <1% 117 TSH M 32 Palpitations YES 1% 118 TSH M 48 Visual loss NO <1% 119 TSH M 48 Palpitations NO 1% 120 TSH M 57 Palpitations NO 1% 121 TSH F 16 Amenorrhea NO <1%
4 122 TSH F 28 Thyromegaly NO <1% 123 TSH F 26 Thyromegaly 4 1 NO <1% 124 TSH M 27 Palpitations NO 1% 125 TSH M 63 Palpitations NO <1% * Invasive adenomas were defined as fulfilling 1 of 2 conditions: (1) Hardy s modified classification grade III, IV and/or stage C, D and E; (2) Knosp classification grade III and IV.
5 Supplementary information, Table S1B: 412 mutations identified in exome sequencing of 125 PAs. No. Subtype 1* ACTH Function Gene symbol Genomic position Transcript Exon CDS level Protein level ANKS3 chr16:g _ NM_ exon4 c.288_306del p.a96fs 2GGCCAGCATCCGGGCTGTG>- 2* ACTH 3* ACTH 4* ACTH 5* ACTH 6* 7* ACTH ACTH 8* ACTH 9 ACTH 10 ACTH FGF4 chr11:g c>a NM_ exon1 c.g277t p.g93c nonsense IGSF10 chr3:g g>c NM_ exon4 c.c3767g p.s1256x IL18RAP chr2:g g>t NM_ exon9 c.g1006t p.v336f STAG2 chrx:g g>c NM_ exon14 c.g1363c p.g455r USP29 chr19:g a>g NM_ exon4 c.a1354g p.n452d non chr15:g _50782 USP8 NM_ CTC>exon14 c.2151_2153del p.s718del nonsense ADGRV1 chr5:g c>t NM_ exon46 c.c9877t p.r3293x CCDC159 chr19:g >c NM_ exon5 c.311dupc p.a104fs ORC4 chr2:g t>c NM_ exon7 c.a524g p.d175g PPFIA3 chr19:g g>t NM_ exon21 c.g2731t p.a911s USP8 chr15:g c>g NM_ exon14 c.c2159g p.p720r VWF chr12:g g>c NM_ exon20 c.c2634g p.f878l chr19:g _11658 non CNN1 649GAAAGGAAACAAGGTGAAC NM_ GTGGGAGTGAA>exon5 c.399_428del p.133_143del CPM chr12:g t>c NM_ exon3 c.a254g p.d85g GTF3C3 chr2:g a>g NM_ exon13 c.t1729c p.s577p NME5 chr5:g g>a NM_ exon2 c.c19t p.p7s NRP2 chr2:g g>t NM_ exon3 c.g379t p.d127y nonsense OBSL1 chr2:g g>a NM_ exon14 c.c4390t p.q1464x PRKAA2 chr1:g t>g NM_ exon7 c.t1183g p.l395v UGT8 chr4:g t>c NM_ exon1 c.t50c p.i17t URGCP chr7:g c>t NM_ exon5 c.g1279a p.g427r USP8 chr15:g t>c NM_ exon14 c.t2152c p.s718p DYNC1H1 chr14:g a>g NM_ exon12 c.a3047g p.e1016g USP8 chr15:g c>g NM_ exon14 c.c2159g p.p720r APBB1 chr11:g a>t NM_ exon11 c.t1109a p.l370h GABRE chrx:g c>a NM_ exon9 c.g1228t p.v410f USP8 chr15:g t>c NM_ exon14 c.t2152c p.s718p ZNF846 chr19:g g>c NM_ exon3 c.c139g p.l47v NRAP chr10:g a>t NM_ exon26 c.t2904a p.h968q OR7A5 chr19:g >t NM_ exon1 c.933dupa p.g312fs OR7A5 chr19:g c>g NM_ exon1 c.g930c p.m310i USP8 chr15:g t>c NM_ exon14 c.t2152c p.s718p HIST1H1B chr6:g g>t NM_ exon1 c.c225a p.d75e splicing KIR2DL5A, chr19_gl000209_random: NM_ KIR2DL5B g.95373g>a exon7 c.858+1g>a PLCL2 chr3:g g>a NM_ exon2 c.g2218a p.g740r SIPA1 chr11:g t>a NM_ exon10 c.t2442a p.d814e USP8 chr15:g t>c NM_ exon14 c.t2152c p.s718p LY75,LY75 -CD302 chr2:g c>t NM_ exon31 c.g4387a p.e1463k NUFIP1 chr13:g t>g NM_ exon3 c.a518c p.h173p SLC24A2 chr9:g g>a NM_ exon5 c.c1286t p.a429v SOX4 chr6:g a>g NM_ exon1 c.a377g p.y126c non chr15:g _50782 USP8 NM_ CTC>exon14 c.2151_2153del p.718del USP8 chr15:g c>a NM_ exon14 c.c2159a p.p720q USH2A chr1:g t>c NM_ exon13 c.a2279g p.n760s CCDC178 chr18:g g>a NM_ exon22 c.c2531t p.s844f USP8 chr15:g t>c NM_ exon14 c.t2152c p.s718p RBM12 chr20:g a>- NM_ exon2 c.831delt p.l277fs
6 11 ACTH ACTH 20* ACTH 13* ACTH 14* ACTH 15* ACTH 16 ACTH 17 ACTH 18 ACTH ACTH (GH+ACTH) (GH+TSH) HIST1H2BA chr6:g g>a NM_ exon1 c.g202a p.v68i USP8 chr15:g c>a NM_ exon14 c.c2159a p.p720q chr2:g _23065 TRIP ACATGGCCCGTGAGCTGC NM_ CG>exon31 c.4017_4036del p.l1339fs TRIP12 chr2:g a>g NM_ exon31 c.t4016c p.l1339p MBLAC2 chr5:g a>g NM_ exon2 c.t794c p.l265p ANXA9 chr1:g a>g NM_ exon13 c.a866g p.n289s DCAF15 chr19:g c>t NM_ exon7 c.c1034t p.a345v non chr12:g _176575c IQSEC3 NM_ TCCGCCCTCGG>exon1 c.516_527del p.172_176del USP48 chr1:g c>t NM_ exon10 c.g1245a p.m415i TICRR chr15:g c>t NM_ exon20 c.c3328t p.h1110y WNK3 chrx:g a>g NM_ exon7 c.t1199c p.l400s ABCC12 chr16:g g>c NM_ exon27 c.c3787g p.q1263e BCOR chrx:g c>t NM_ exon4 c.g283a p.g95r DDX5 chr17:g t>c NM_ exon13 c.a1769g p.q590r LARP6 chr15:g c>g NM_ exon3 c.g310c p.a104p LGI1 chr10:g g>a NM_ exon4 c.g409a p.g137r MXRA5 chrx:g g>a NM_ exon4 c.c452t p.t151m POLR3B chr12:g c>- NM_ exon25 c.2787delc p.d929fs nonsense NR3C1 chr5:g g>a NM_ exon7 c.c1894t p.q632x chr5:g _14268 NR3C1 NM_ GATTTTCAGA>exon5 c.1534_1543del p.s512fs BRAF chr7:g a>t NM_ exon15 c.t1799a p.v600e FLG chr1:g c>g NM_ exon3 c.g1418c p.s473t KHDRBS2 chr6:g g>c NM_ exon6 c.c740g p.a247g NDUFV1 chr11:g a>g NM_ exon7 c.a911g p.n304s chr4:g _ YIPF7 NM_ AGGCTGCT>exon3 c.203_210del p.e68fs MAGEB10 chrx:g g>t NM_ exon3 c.g958t p.a320s HADHA chr2:g g>c NM_ exon8 c.c692g p.p231r IFT140 chr16:g >g NM_ exon19 c.2254dupc p.q752fs PHKB chr16:g c>a NM_ exon29 c.c2941a p.l981i PLIN2 chr9:g c>t NM_ exon5 c.g391a p.a131t splicing UPF1 chr19:g a>c NM_ exon11 c a>c non chrx:g _ FGD1 NM_ TCC>exon4 c.1044_1046del p.348_349del GPRIN3 chr4:g c>a NM_ exon2 c.g1828t p.d610y TAF5 chr10:g c>g NM_ exon2 c.c690g p.d230e UNKL chr16:g c>t NM_ exon3 c.g316a p.d106n AKNAD1 chr1:g a>g NM_ exon7 c.t1490c p.v497a CHD2 chr15:g a>g NM_ exon30 c.a3883g p.k1295e NR3C1 chr5:g c>g NM_ exon8 c.g2141c p.r714p non chr5:g _14267 NR3C1 NM_ ATT>exon6 c.1888_1890del p.630_630del RAP1GAP2 chr17:g g>c NM_ exon11 c.g869c p.g290a SETD2 chr3:g a>c NM_ exon4 c.t4562g p.l1521r TEDDM1 chr1:g g>c NM_ exon1 c.c813g p.s271r TADA2A chr17:g c>a NM_ exon11 c.c909a p.f303l non chr13:g _1144 GRK1 NM_ CTT>exon6 c.1275_1277del p.425_426del KIF14 chr1:g g>a NM_ exon30 c.c4615t p.r1539c MRPS9 chr2:g c>g NM_ exon2 c.c287g p.p96r SLCO1C1 chr12:g t>a NM_ exon12 c.t1594a p.c532s CCBL1 chr9:g t>c NM_ exon3 c.a104g p.q35r MAST4 chr5:g t>g NM_ exon5 c.t340g p.s114a nonsense PRAMEF19 chr1:g g>a NM_ exon3 c.c1021t p.r341x splicing SP100 chr2:g g>a NM_ exon28 c g>a SRF chr6:g g>a NM_ exon1 c.g338a p.g113e STX19 chr3:g a>t NM_ exon2 c.t530a p.m177k WDR33 chr2:g t>c NM_ exon16 c.a1834g p.m612v HNF1A chr12:g c>t NM_ exon7 c.c1424t p.p475l DYSF chr2:g g>a NM_ exon46 c.g5251a p.v1751m
7 (GH+TSH) (GH+ACTH) nonsense NEK8 chr17:g c>t NM_ exon9 c.c1297t p.q433x non PEG3 splicing USP7 chr16:g c>t NM_ exon6 c.563+1g>a ZNHIT2 chr11:g g>a NM_ exon1 c.c160t p.l54f ADRBK1 chr11:g a>c NM_ exon18 c.a1505c p.d502a ADRBK1 chr11:g a>c NM_ exon18 c.a1559c p.e520a ALDH5A1 chr6:g c>a NM_ exon6 c.c935a p.p312q nonsense CLASP2 chr3:g g>c NM_ exon15 c.c1289g p.s430x COL7A1 chr3:g c>g NM_ exon67 c.g5624c p.g1875a ENDOD1 chr11:g a>c NM_ exon2 c.a1120c p.s374r HDAC9 chr7:g g>a NM_ exon17 c.g2329a p.v777m HK2 chr2:g c>t NM_ exon12 c.c1789t p.p597s ICE1 chr5:g g>a NM_ exon13 c.g3274a p.g1092s MSANTD4 chr11:g c>t NM_ exon3 c.g472a p.e158k TCL1A chr14:g g>a NM_ exon2 c.c277t p.r93c non chr1:g _ TRNAU1AP NM_ CTT>exon2 c.51_53del p.17_18del CACNA1B chr9:g a>g NM_ exon7 c.a967g p.t323a CCDC155 chr19:g c>a NM_ exon14 c.c1060a p.l354i FOXN2 chr2:g a>g NM_ exon7 c.a1100g p.y367c IDS chrx:g g>- NM_ exon6 c.782delc p.p261fs nonsense MEN1 chr11:g c>t NM_ exon3 c.g608a p.w203x non chr7:g _ PEG10 NM_ AAA>exon2 c.27_29del p.9_10del RNF123 chr3:g g>a NM_ exon28 c.g2704a p.a902t LTBP1 chr2:g g>a NM_ exon17 c.g2296a p.g766r chr11:g _64577 MEN1 NM_ GCGGGTC>exon2 c.208_214del p.d70fs unknown KCP chr7:g c>t PAK1IP1 chr6:g t>c NM_ exon2 c.t218c p.i73t PLXNB2 chr22:g a>t NM_ exon34 c.t5219a p.l1740q SP100 chr2:g c>g NM_ exon28 c.c2532g p.h844q TBL1X chrx:g a>g NM_ exon14 c.a1216g p.i406v GNAS chr20:g a>t NM_ exon8 c.a680t p.q227l MED1 chr17:g t>c NM_ exon13 c.a1084g p.r362g MYO18B chr22:g g>a NM_ exon21 c.g3803a p.r1268h PITPNM2 chr12:g c>t NM_ exon12 c.g1684a p.v562i SOCS3 chr17:g g>a NM_ exon2 c.c491t p.a164v ADAMTSL4 chr1:g g>a NM_ exon18 c.g3047a p.r1016k DOCK8 chr9:g g>t NM_ exon12 c.g1297t p.v433f GNAS chr20:g a>t NM_ exon8 c.a680t p.q227l NFKB1 chr4:g a>g NM_ exon3 c.a73g p.i25v nonsense SLC25A48 chr5:g c>t NM_ exon4 c.c394t p.q132x GLB1L3 chr11:g t>c NM_ exon11 c.t979c p.s327p LMNTD1 chr12:g c>t NM_ exon5 c.g653a p.r218h MYH10 chr17:g t>g NM_ exon14 c.a1747c p.k583q non chr20:g _60640 TAF4 NM_ GGGCGCGGG>exon1 c.748_756del p.250_252del DESI2 chr1:g a>g NM_ exon3 c.a154g p.i52v GNAS chr20:g g>a NM_ exon8 c.g601a p.r201h MARK1 chr1:g g>t NM_ exon10 c.g1002t p.q334h MYH11 chr16:g g>a NM_ exon18 c.c2198t p.a733v OMG chr17:g c>g NM_ exon2 c.g411c p.k137n THOC6 chr16:g a>t NM_ exon6 c.a385t p.i129f XIRP2 chr2:g g>a NM_ exon7 c.g6385a p.v2129i splicing CREBBP chr16:g c>t NM_ exon23 c g>a c.4254_4255insg chr19:g >TGGCTCAGCAGCCTCCACTTC NM_ exon7 p.n1419delins AAGTGGAGGCTGCTG EVEAAEPN AGCCA RUFY2 chr10:g g>c NM_ exon18 c.c1889g p.a630g SLC9A3R1 chr17:g g>a NM_ exon3 c.g691a p.v231m
8 34 35 GNAS chr20:g g>a NM_ exon2 c.g145a p.g49r 36 GH SKIDA1 chr10:g >g NM_ exon4 c.1088dupc p.p363fs U2AF2 chr19:g c>g NM_ exon5 c.c439g p.q147e ZBP1 chr20:g a>g NM_ exon7 c.t905c p.f302s 37 GH 38 GH ADAM15 chr1:g c>t NM_ exon19 c.c2309t p.s770f 39 GH HAVCR2 chr5:g c>t NM_ exon2 c.g151a p.v51i JMJD4 chr1:g c>t NM_ exon5 c.g1036a p.e346k MGA chr15:g a>g NM_ exon19 c.a6691g p.r2231g TTC4 chr1:g t>c NM_ exon7 c.t836c p.l279p GNAS chr20:g a>t NM_ exon8 c.a680t p.q227l VPS37D chr7:g g>a NM_ exon2 c.g187a p.a63t 40 GH ZNF507 chr19:g a>- NM_ exon2 c.1775dela p.e592fs ARSD chrx:g c>t NM_ exon10 c.g1759a p.e587k 41 GH LAMA1 chr18:g c>t NM_ exon16 c.g2188a p.v730m AHCTF1 chr1:g t>c NM_ exon25 c.a3197g p.n1066s FFAR2 chr19:g g>a NM_ exon1 c.g89a p.r30q 42 GH MUC16 chr19:g g>a NM_ exon5 c.c32402t p.a10801v PARPBP chr12:g t>a NM_ exon3 c.t245a p.m82k SNX16 chr8:g g>c NM_ exon5 c.c600g p.f200l ACSS2 chr20:g a>g NM_ exon9 c.a722g p.y241c GH1 chr17:g g>- NM_ exon4 c.487delc p.l163fs 43 GH GRB10 chr7:g g>a NM_ exon10 c.c1069t p.l357f ITIH5 chr10:g g>a NM_ exon5 c.c610t p.p204s LRP10 chr14:g a>g NM_ exon5 c.a1208g p.n403s OR1S2 chr11:g g>a NM_ exon1 c.c538t p.l180f OR4A16 chr11:g g>a NM_ exon1 c.g149a p.s50n 44 GH GRB10 chr7:g t>a NM_ exon4 c.a463t p.t155s LTBP2 chr14:g c>a NM_ exon22 c.g3334t p.a1112s BCL11A chr2:g a>c NM_ exon4 c.t2323g p.c775g chr11:g _14497 COPB1 NM_ TTCT>exon13 c.1485_1488del p.k495fs ERC2 chr3:g g>a NM_ exon4 c.c1112t p.a371v LRRC7 chr1:g c>t NM_ exon19 c.c2390t p.s797l 45 GH PTPRS chr19:g c>t NM_ exon26 c.g4048a p.e1350k RAPSN chr11:g c>a NM_ exon2 c.g341t p.g114v RFX7 chr15:g >g NM_ exon9 c.3045dupc p.s1016fs SNUPN chr15:g c>t NM_ exon3 c.g202a p.d68n WIPI1 chr17:g c>t NM_ exon2 c.g133a p.v45m LXN chr3:g g>c NM_ exon5 c.c563g p.a188g 46 GH PIK3C2G chr12:g g>a NM_ exon3 c.g709a p.v237i nonsense ZNF417 chr19:g g>a NM_ exon3 c.c850t p.q284x 47 GH HECTD4 chr12:g c>t NM_ exon59 c.g8779a p.a2927t CSMD3 chr8:g a>g NM_ exon1 c.t2c p.m1t DGKZ chr11:g c>t NM_ exon5 c.c454t p.r152c 48 GH SLC22A6 chr11:g a>g NM_ exon4 c.t703c p.f235l GPN1 chr2:g g>a NM_ exon1 c.g67a p.e23k NPEPL1 chr20:g c>t NM_ exon1 c.c62t p.p21l ZNF578 chr19:g g>t NM_ exon6 c.g1116t p.k372n CASP8AP2 chr6:g t>- NM_ exon8 c.3234delt p.s1078fs 49 GH CLDN17 chr21:g a>g NM_ exon1 c.t392c p.i131t unknown SELT chr3:g a>g 50 GH MALT1 chr18:g c>g NM_ exon16 c.c2307g p.s769r MYH4 chr17:g t>c NM_ exon20 c.a2240g p.e747g
9 51 GH CLASP1 chr2:g g>a NM_ exon13 c.c1220t p.a407v 52 GH SLC13A1 chr7:g a>g NM_ exon4 c.t383c p.m128t STAP2 chr19:g c>t NM_ exon10 c.g943a p.g315r 53 GH FRMD6 chr14:g c>t NM_ exon7 c.c649t p.l217f 54 GH CCDC160 chrx:g c>g NM_ exon2 c.c290g p.s97c 55 GH RPS3 chr11:g c>g NM_ exon2 c.c128g p.p43r 56 PRL C11orf84 chr11:g t>c NM_ exon5 c.t767c p.f256s ABCA4 chr1:g g>c NM_ exon12 c.c1660g p.p554a GABRG2 chr5:g g>t NM_ exon9 c.g1223t p.c408f 57 PRL RBL1 chr20:g c>t NM_ exon22 c.g3092a p.r1031q UPRT chrx:g g>a NM_ exon7 c.g905a p.g302e USP40 chr2:g a>t NM_ exon30 c.t3548a p.i1183n UBFD1 chr16:g g>a NM_ exon3 c.g544a p.e182k 59 PRL MGP chr12:g c>t NM_ exon4 c.g182a p.r61h LRP2 chr2:g g>a NM_ exon8 c.c803t p.p268l non BBS10 chr12:g >agc NM_ exon1 c.137_138insgct p.l46delinsll 60 PRL BRCA1 chr17:g t>c NM_ exon9 c.a3013g p.n1005d IDE chr10:g a>t NM_ exon6 c.t826a p.s276t AP2M1 chr3:g g>t NM_ exon11 c.g1274t p.r425l GORAB chr1:g c>a NM_ exon5 c.c1109a p.s370y 61 PRL IARS chr9:g c>a NM_ exon30 c.g3215t p.g1072v MAP10 chr1:g t>c NM_ exon1 c.t2279c p.v760a USP11 chrx:g a>g NM_ exon2 c.a323g p.h108r COG7 chr16:g g>t NM_ exon17 c.c2191a p.r731s FLG2 chr1:g a>c NM_ exon3 c.t6552g p.s2184r 62 PRL KIF5A chr12:g c>t NM_ exon25 c.c2813t p.t938i TLDC2 chr20:g g>c NM_ exon3 c.g259c p.g87r ZCCHC24 chr10:g g>a NM_ exon3 c.c493t p.r165c 63 PRL TBX4 chr17:g t>c NM_ exon8 c.t1594c p.y532h splicing DFNB59 chr2:g a>g NM_ exon4 c.408-2a>g 64 PRL FAM120A chr9:g g>a NM_ exon9 c.g1559a p.s520n unknown PCSK6 chr15:g g>a SLC25A16 chr10:g t>c NM_ exon7 c.a718g p.t240a 65 PRL SCN10A chr3:g c>t NM_ exon14 c.g2156a p.r719q 66 PRL ZNF674 chrx:g t>c NM_ exon6 c.a553g p.n185d PRL PRL CAMSAP3 ADAMTSL1 chr19:g c>t chr9:g g>a NM_ NM_ exon11 exon2 c.c2032t c.g95a p.r678w p.r32q TBX5 ENKD1 chr12:g g>a chr16:g g>t NM_ NM_ exon8 exon4 c.c913t c.c557a p.r305c p.t186n ECE2 chr3:g a>g NM_ exon14 c.a1612g p.t538a 69 PRL nonsense IFT52 chr20:g g>t NM_ exon14 c.g1288t p.e430x SIDT2 chr11:g c>t NM_ exon24 c.c2285t p.a762v 71 PRL nonsense NDST1 chr5:g c>t NM_ exon14 c.c2452t p.r818x unknown FOXD1 chr5:g _ GCGGCGGCGGCCTGC> 72 PRL KCTD19 chr16:g c>t NM_ exon9 c.g1337a p.r446q NLGN4X chrx:g t>a NM_ exon6 c.a2026t p.s676c TAF1 chrx:g t>c NM_ exon15 c.t2471c p.i824t 73 PRL RBFOX1 chr16:g c>g NM_ exon8 c.c763g p.q255e AGT chr1:g c>t NM_ exon5 c.g1432a p.v478m 74 PRL 75 PRL 76 HDAC4 chr2:g c>t NM_ exon4 c.g271a p.a91t HGF chr7:g c>a NM_ exon16 c.g1774t p.d592y SEC24B chr4:g t>c NM_ exon2 c.t464c p.f155s TTN chr2:g t>g NM_ exon162 c.a62084c p.y20695s FAM131A chr3:g c>a NM_ exon6 c.c715a p.p239t unknown FAM58A chrx:g c>t SYK chr9:g a>g NM_ exon4 c.a716g p.q239r TTN chr2:g g>a NM_ exon154 c.c47029t p.r15677c ALS2 chr2:g a>c NM_ exon4 c.t715g p.l239v BRIP1 chr17:g a>t NM_ exon19 c.t2726a p.l909h C17orf96 chr17:g a>g NM_ exon1 c.t2c p.m1t COL12A1 chr6:g a>g NM_ exon38 c.t4666c p.c1556r FAM170B chr10:g c>g NM_ exon2 c.g130c p.g44r HRH1 chr3:g g>c NM_ exon2 c.g214c p.a72p HRH1 chr3:g g>c NM_ exon2 c.g222c p.l74f
10 SLCO1B1 chr12:g g>a NM_ exon3 c.g173a p.r58k GNA15 chr19:g g>a NM_ exon4 c.g557a p.r186h TMEM169 chr2:g a>t NM_ exon2 c.a188t p.d63v ADAM2 chr8:g c>t NM_ exon17 c.g1874a p.c625y EBPL chr13:g a>c NM_ exon2 c.t193g p.s65a SSPO chr7:g c>t NM_ exon55 c.c8165t p.s2722f BSN chr3:g c>a NM_ exon5 c.c3703a p.r1235s SPEG chr2:g g>t NM_ exon41 c.g9740t p.r3247l 95 UHRF1BP1 chr6:g >a NM_ exon14 c.2362dupa p.l787fs BTG1 chr12:g t>c NM_ exon1 c.a43g p.i15v CFAP46 chr10:g t>c NM_ exon7 c.a668g p.h223r 96 GT KIF5A chr12:g a>g NM_ exon20 c.a2246g p.y749c SCN1A chr2:g g>a NM_ exon20 c.c3950t p.t1317i TMX1 chr14:g c>t NM_ exon1 c.c29t p.p10l 97 GT C3 chr19:g c>t NM_ exon24 c.g3125a p.r1042q CTNND1 chr11:g c>a NM_ exon2 c.c127a p.r43s FRRS1 chr1:g >t NM_ exon5 c.357_358insa p.a120fs KRT83 chr12:g g>a NM_ exon1 c.c196t p.r66c RECK chr9:g t>g NM_ exon2 c.t106g p.l36v RYR1 chr19:g g>a NM_ exon17 c.g1841a p.r614h PPAPDC1B chr8:g g>t NM_ exon1 c.c68a p.a23d TTLL11 chr9:g c>t NM_ exon1 c.g361a p.e121k ARL5A chr2:g a>c NM_ exon4 c.t161g p.v54g C20orf194 chr20:g g>a NM_ exon24 c.c2099t p.a700v DLGAP5 chr14:g t>g NM_ exon10 c.a1260c p.e420d JAGN1 chr3:g a>g NM_ exon2 c.a200g p.q67r PRMT9 chr4:g t>a NM_ exon2 c.a247t p.i83l RAD21L1 chr20:g t>c NM_ exon12 c.t1348c p.s450p TSPAN6 chrx:g g>t NM_ exon1 c.c35a p.p12q ZDBF2 chr2:g a>g NM_ exon5 c.a6637g p.i2213v ARMC8 chr3:g a>g NM_ exon16 c.a1426g p.m476v GSG2 chr17:g c>a NM_ exon1 c.c1286a p.t429k RNF216 chr7:g t>a NM_ exon7 c.a1285t p.i429f UROC1 chr3:g g>a NM_ exon14 c.c1343t p.p448l ZMYM2 chr13:g c>a NM_ exon3 c.c6a p.d2e TRIP11 chr14:g t>a NM_ exon13 c.a4778t p.d1593v TUBGCP6 chr22:g g>a NM_ exon21 c.c4744t p.l1582f OPHN1 chrx:g a>c NM_ exon5 c.t346g p.l116v COL4A3 chr2:g g>c NM_ exon25 c.g1593c p.q531h KLHL35 chr11:g c>t NM_ exon4 c.g1333a p.v445m LRP1 chr12:g g>a NM_ exon24 c.g3928a p.a1310t nonsense MAGEA3 chrx:g g>a NM_ exon3 c.c373t p.r125x OR5D16 chr11:g t>a NM_ exon1 c.t295a p.c99s SLC9A8 chr20:g g>c NM_ exon1 c.g3c p.m1i ATXN7 chr3:g a>g NM_ exon8 c.a1273g p.t425a C9orf172 chr9:g g>a NM_ exon1 c.g2242a p.g748s CAPN1 chr11:g g>a NM_ exon14 c.g1588a p.a530t FAM208A chr3:g c>t NM_ exon1 c.g155a p.s52n nonsense GPR182 chr12:g g>a NM_ exon2 c.g236a p.w79x ILF3 chr19:g a>g NM_ exon5 c.a538g p.m180v PROM2 chr2:g g>a NM_ exon2 c.g280a p.v94m RUNX2 chr6:g c>t NM_ exon6 c.c1319t p.t440i GPR132 chr14:g g>a NM_ exon3 c.c134t p.t45m PJA2 chr5:g c>t NM_ exon4 c.g1246a p.g416r RNMTL1 chr17:g t>- NM_ exon4 c.1122delt p.g374fs TRAPPC6A chr19:g g>a NM_ exon2 c.c191t p.p64l TRIP12 chr2:g g>a NM_ exon19 c.c2237t p.s746l TUBGCP5 chr15:g g>c NM_ exon6 c.g541c p.d181h ZNF334 chr20:g c>t NM_ exon5 c.g1313a p.c438y IARS chr9:g c>t NM_ exon23 c.g2423a p.r808h NOS3 chr7:g c>a NM_ exon21 c.c2521a p.p841t PEX13 chr2:g c>- NM_ exon1 c.22delc p.p8fs
11 98 GT 99 GT 100 GT 101 GT 102 GT DOCK7 chr1:g g>c NM_ exon14 c.c1667g p.p556r LPCAT2 chr16:g t>c NM_ exon5 c.t653c p.f218s UTRN chr6:g t>c NM_ exon8 c.t745c p.s249p ASH1L chr1:g c>a NM_ exon11 c.g6365t p.c2122f CCDC134 chr22:g c>t NM_ exon5 c.c476t p.s159l KIAA0368 chr9:g t>c NM_ exon43 c.a4903g p.i1635v UBASH3B chr11:g g>a NM_ exon13 c.g1703a p.g568e MAP3K6 chr1:g >g NM_ exon1 c.104dupc p.p35fs RASSF9 chr12:g c>t NM_ exon2 c.g215a p.g72e EIF4E1B chr5:g a>g NM_ exon7 c.a362g p.d121g GOLM1 chr9:g a>g NM_ exon3 c.t242c p.l81p KCNH2 chr7:g c>t NM_ exon2 c.g235a p.a79t LRFN5 chr14:g a>t NM_ exon3 c.a218t p.n73i non SKIDA1 chr10:g >ggc NM_ exon4 c.678_679insgcc p.a227delinsa A DLG1 chr3:g t>a NM_ exon12 c.a1518t p.e506d EED chr11:g t>a NM_ exon11 c.t1148a p.v383d MCOLN1 chr19:g c>g NM_ exon5 c.c631g p.l211v INPP4A chr2:g a>g NM_ exon13 c.a1097g p.h366r non chr10:g _1261 LHPP NM_ GT 77069TTC>exon3 c.390_392del p.130_131del TMEM68 chr8:g t>c NM_ exon4 c.a358g p.m120v PNMAL1 chr19:g c>t NM_ exon2 c.g1292a p.r431h 105 GT TOR1B chr9:g g>c NM_ exon2 c.g232c p.g78r GT GT GDF9 ANKRD65 chr5:g g>t chr1:g c>t NM_ NM_ exon2 exon2 c.c877a c.g34a p.q293k p.e12k OASL FAM133A chr12:g a>t chrx:g g>t NM_ NM_ exon4 exon3 c.t818a c.g5t p.i273n p.g2v 109 GT CHMP7 chr8:g a>g NM_ exon3 c.a412g p.m138v 110 GT nonsense EXOSC2 chr9:g c>t NM_ exon3 c.c259t p.r87x KLHL41 chr2:g g>c NM_ exon1 c.g668c p.r223p C2orf54 chr2:g c>g NM_ exon2 c.g703c p.g235r splicing IMPACT chr18:g a>g NM_ exon7 c.491-2a>g MAP2 chr2:g a>g NM_ exon7 c.a2083g p.i695v 111 GT nonsense NLRP11 chr19:g g>a NM_ exon4 c.c265t p.r89x RRP12 chr10:g c>a NM_ exon22 c.g2635t p.a879s WNK1 chr12:g a>g NM_ exon1 c.a266g p.n89s ZAK chr2:g g>a NM_ exon20 c.g2347a p.a783t ZNF616 chr19:g t>c NM_ exon4 c.a2330g p.n777s CFAP74 chr1:g t>- NM_ exon13 c.1445dela p.k482fs 112 GT COPRS chr17:g t>g NM_ exon4 c.a549c p.e183d TPP2 chr13:g a>g NM_ exon22 c.a2630g p.y877c 113 GT CLPB chr11:g g>a NM_ exon4 c.c604t p.r202c OR4B1 chr11:g g>a NM_ exon1 c.g771a p.m257i ACACA chr17:g t>c NM_ exon12 c.a1426g p.n476d 114 GT BRPF3 chr6:g a>g NM_ exon11 c.a3247g p.k1083e GAS2L2 chr17:g a>g NM_ exon3 c.t674c p.m225t ZFHX2 chr14:g g>a NM_ exon9 c.c5984t p.p1995l 115 GT CHRNE chr17:g c>a NM_ exon10 c.g1112t p.r371l 117 TSH ABHD17B chr9:g g>a NM_ exon4 c.c691t p.h231y 118 TSH MRPS26 chr20:g g>t NM_ exon1 c.g52t p.a18s 119 TSH C1QTNF2 chr5:g c>a NM_ exon2 c.g238t p.v80f SYNPO2 chr4:g c>a NM_ exon4 c.c2117a p.p706q GLCE chr15:g g>a NM_ exon3 c.g52a p.a18t 120 TSH PSTPIP2 chr18:g a>t NM_ exon1 c.t30a p.f10l CCDC62 chr12:g t>g NM_ exon9 c.t986g p.m329r 121 TSH nonsense MAP7D3 chrx:g a>t NM_ exon5 c.t423a p.y141x SLC33A1 chr3:g t>c NM_ exon5 c.a1294g p.i432v 122 TSH WASF2 chr1:g t>c NM_ exon8 c.a985g p.m329v 123 TSH FAM98A chr2:g g>c NM_ exon3 c.c284g p.s95c SPTA1 chr1:g a>g NM_ exon52 c.t7160c p.f2387s ADCK3 chr1:g g>a NM_ exon15 c.g1810a p.e604k 125 TSH KBTBD13 chr15:g c>- NM_ exon1 c.684delc p.g228fs SF3B6 chr2:g t>c NM_ exon4 c.a307g p.t103a * indicates the ACTH-PAs samples included in our prior study
12 Supplementary information, Table S1C: Potential Drugs in 125 PAs Accession Number Target gene Type Status DB00398 BRAF small molecule FDA approved,investigational DB08881 BRAF small molecule FDA approved DB08896 BRAF small molecule FDA approved DB08912 BRAF small molecule FDA approved DB06999 BRAF small molecule experimental DB07000 BRAF small molecule experimental DB08553 BRAF small molecule experimental DB00381 CACNA1B small molecule FDA approved DB00661 CACNA1B small molecule FDA approved DB00996 CACNA1B small molecule FDA approved,investigational DB01202 CACNA1B small molecule FDA approved,investigational DB08838 CACNA1B small molecule experimental,investigational DB00686 FGF4 small molecule FDA approved DB06176 HDAC4 small molecule FDA approved,investigational DB07879 HDAC4 small molecule experimental DB08613 HDAC4 biotech experimental DB00013 LRP2 biotech FDA approved,investigational,withdrawn DB00030 LRP2 biotech FDA approved,experimental,investigational DB00071 LRP2 small molecule FDA approved DB00798 LRP2 small molecule FDA approved DB00945 NFKB1 small molecule FDA approved DB01041 NFKB1 small molecule FDA approved,investigational,withdrawn DB01411 NFKB1 small molecule FDA approved DB08814 NFKB1 small molecule FDA approved DB00360 NOS3 small molecule FDA approved,investigational DB01110 NOS3 small molecule FDA approved,investigational DB02044 NOS3 small molecule experimental DB02141 NOS3 small molecule experimental DB02911 NOS3 small molecule experimental DB03065 NOS3 small molecule experimental DB03707 NOS3 small molecule experimental DB03910 NOS3 small molecule experimental DB04559 NOS3 small molecule experimental DB07001 NOS3 small molecule experimental DB07244 NOS3 small molecule experimental DB07388 NOS3 small molecule experimental DB08018 NOS3 small molecule experimental DB08019 NOS3 small molecule experimental DB00025 VWF small molecule FDA approved,investigational DB08655 CREBBP small molecule experimental DB02587 GNAS small molecule experimental DB06843 GNAS small molecule experimental DB08696 MED1 small molecule experimental DB06834 SYK small molecule experimental DB07159 SYK small molecule experimental DB07194 SYK small molecule experimental DB08361 SYK small molecule experimental DB08846 SYK biotech investigational
Supplementary Table 2. Identified causative mutations and/or mutation candidates.
 Supplementary Table 2. Identified causative mutations and/or mutation candidates. Nonsense mutations base change aa change Average depth Result of next generation in 432 patient Hereditary form of the
Supplementary Table 2. Identified causative mutations and/or mutation candidates. Nonsense mutations base change aa change Average depth Result of next generation in 432 patient Hereditary form of the
Whole Exome Sequenced Characteristics
 Supplementary Tables Supplementary Table 1: Patient characteristics of 45 whole exome sequenced HNSCC tumors Whole Exome Sequenced Characteristics Tumors (n=45) Age, years Median (range) 61.0 (19-90) Sex,
Supplementary Tables Supplementary Table 1: Patient characteristics of 45 whole exome sequenced HNSCC tumors Whole Exome Sequenced Characteristics Tumors (n=45) Age, years Median (range) 61.0 (19-90) Sex,
July 2015 Assay ID Assay Name Gene COSMIC ID Amino acid change Nucleotide change Wild type allele (VIC label) Mutant allele (FAM label)
 July 2015 Assay ID Assay Name Gene COSMIC ID Amino acid change Nucleotide change Wild type allele (VIC label) Mutant allele (FAM label) p AHLJ090 AKT1_33765 AKT1 33765 p.e17k c.49g>a C T 2 AHBKFRM BRAF_473
July 2015 Assay ID Assay Name Gene COSMIC ID Amino acid change Nucleotide change Wild type allele (VIC label) Mutant allele (FAM label) p AHLJ090 AKT1_33765 AKT1 33765 p.e17k c.49g>a C T 2 AHBKFRM BRAF_473
Supplementary Figure 1
 Supplementary Figure 1 A B HEC1A PTEN +/+ HEC1A PTEN -/- 16 HEC1A PTEN -/- 22 PTEN RAD51 % Cells with RAD51 foci 40 -IR +IR 30 20 10 * *! TUBULIN 0 HEC1A PTEN+/+ HEC1A PTEN-/- 16 HEC1A PTEN-/- 22 C D 1.0
Supplementary Figure 1 A B HEC1A PTEN +/+ HEC1A PTEN -/- 16 HEC1A PTEN -/- 22 PTEN RAD51 % Cells with RAD51 foci 40 -IR +IR 30 20 10 * *! TUBULIN 0 HEC1A PTEN+/+ HEC1A PTEN-/- 16 HEC1A PTEN-/- 22 C D 1.0
Supplementary Online Content
 Supplementary Online Content Pearlman R, Frankel WL, Swanson B, et al. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol.
Supplementary Online Content Pearlman R, Frankel WL, Swanson B, et al. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol.
Spectrum and Classification of ATP7B Variants in a Large Cohort of Chinese Patients with Wilson s Disease Guides Genetic Diagnosis
 Ivyspring International Publisher 638 Theranostics Research Paper 2016; 6(5): 638-649. doi: 10.7150/thno.14596 Spectrum and Classification of ATP7B Variants in a Large Cohort of Chinese Patients with Wilson
Ivyspring International Publisher 638 Theranostics Research Paper 2016; 6(5): 638-649. doi: 10.7150/thno.14596 Spectrum and Classification of ATP7B Variants in a Large Cohort of Chinese Patients with Wilson
DNA Analysis in Glycogen storage disease
 DNA Analysis in Glycogen storage disease Nick Beauchamp PhD Sheffield, Sheffield Children s NHS Foundation Trust 14th October 2010 Glycogen Synthase Type 0 Glycogen Synthesis and Breakdown Type IV UDPGlucose
DNA Analysis in Glycogen storage disease Nick Beauchamp PhD Sheffield, Sheffield Children s NHS Foundation Trust 14th October 2010 Glycogen Synthase Type 0 Glycogen Synthesis and Breakdown Type IV UDPGlucose
Familial exudative vitreoretinopathy (FEVR) is an inheritable
 Genetics Spectrum of Variants in 389 Chinese Probands With Familial Exudative Vitreoretinopathy Jia-Kai Li, 1 Yian Li, 1 Xiang Zhang, 1 Chun-Li Chen, 2 Yu-Qing Rao, 1 Ping Fei, 1 Qi Zhang, 1 Peiquan Zhao,
Genetics Spectrum of Variants in 389 Chinese Probands With Familial Exudative Vitreoretinopathy Jia-Kai Li, 1 Yian Li, 1 Xiang Zhang, 1 Chun-Li Chen, 2 Yu-Qing Rao, 1 Ping Fei, 1 Qi Zhang, 1 Peiquan Zhao,
Supplementary Information. Exome sequencing identifies distinct mutational patterns in liver fluke related and noninfection-related
 Supplementary Information Exome sequencing identifies distinct mutational patterns in liver fluke related and noninfection-related bile duct cancers Waraporn Chan-on 1,2,3*, Maarja-Liisa Nairismägi 1,2*,
Supplementary Information Exome sequencing identifies distinct mutational patterns in liver fluke related and noninfection-related bile duct cancers Waraporn Chan-on 1,2,3*, Maarja-Liisa Nairismägi 1,2*,
Laboratory, Division of Medical Sciences, National Cancer Centre, Singapore
 Supplementary Information Exome sequencing of liver fluke-associated cholangiocarcinoma Choon Kiat Ong 1,2*, Chutima Subimerb 1,2,3*, Chawalit Pairojkul 3, Sopit Wongkham 3, Ioana Cutcutache 4, Willie
Supplementary Information Exome sequencing of liver fluke-associated cholangiocarcinoma Choon Kiat Ong 1,2*, Chutima Subimerb 1,2,3*, Chawalit Pairojkul 3, Sopit Wongkham 3, Ioana Cutcutache 4, Willie
Belgian study into von Willebrand Disease (B-Will): First results
 Belgian study into von Willebrand Disease (B-Will): First results Inge Vangenechten VWD Research Unit, Antwerp University Hospital Supported by CSL Behring Chair in von Willebrand Disease, Antwerp University
Belgian study into von Willebrand Disease (B-Will): First results Inge Vangenechten VWD Research Unit, Antwerp University Hospital Supported by CSL Behring Chair in von Willebrand Disease, Antwerp University
Supplementary Table 1. PIK3CA mutation in colorectal cancer
 Liao X et al. PIK3CA Mutation in Colorectal Cancer. Page 1 Supplementary Table 1. PIK3CA mutation in colorectal cancer Exon Domain Nucleotide change* Amino acid change* cases 9 Helical c.1621t>a p.e541t
Liao X et al. PIK3CA Mutation in Colorectal Cancer. Page 1 Supplementary Table 1. PIK3CA mutation in colorectal cancer Exon Domain Nucleotide change* Amino acid change* cases 9 Helical c.1621t>a p.e541t
UW359 Ovary 3c 3 Serous Recurrent 68 BRCA1 816delGT BRCA1 del exon 1-2. UW417 Ovary 3c 3 Serous Primary 38 BRCA1 1675delA
 Supplementary Table 1. Cases with deleterious germline mutations, somatic HR mutations, and somatic PTEN mutations. ID Site Stage Grade Histology Tumor Age Germline mutation(s) a Somatic HR mutation(s)
Supplementary Table 1. Cases with deleterious germline mutations, somatic HR mutations, and somatic PTEN mutations. ID Site Stage Grade Histology Tumor Age Germline mutation(s) a Somatic HR mutation(s)
Table S4. List of point mutations found by WES in SMZL patients. * DNA extracted from frozen tissue *Method of detection: Variant detected using
 Table S4. List of point mutations found by WES in SMZL patients. * DNA extracted from frozen tissue *Method of detection: Variant detected using RAMSES (R) or CNAG method (C) or both (R/C) **Validation
Table S4. List of point mutations found by WES in SMZL patients. * DNA extracted from frozen tissue *Method of detection: Variant detected using RAMSES (R) or CNAG method (C) or both (R/C) **Validation
Belgian-Czech cooperation in the Brno-VWD study
 Inge Vangenechten I Vangenechten, P Smejkal, O Zapletal, F Bouddount, J Zavrelova, J Blatny, M Penka, JJ Michiels, A Gadisseur. Aim of the study Characterisation of VWD in South Moravia (Czech Republic)
Inge Vangenechten I Vangenechten, P Smejkal, O Zapletal, F Bouddount, J Zavrelova, J Blatny, M Penka, JJ Michiels, A Gadisseur. Aim of the study Characterisation of VWD in South Moravia (Czech Republic)
Nature Genetics: doi: /ng Supplementary Figure 1. TCGA data set on HNSCCs reanalyzed in this study.
 Supplementary Figure 1 TCGA data set on HNSCCs reanalyzed in this study. Summary of the TCGA dataset on HNSCCs re-analyzed in this study and the respective numbers of samples available within each. Supplementary
Supplementary Figure 1 TCGA data set on HNSCCs reanalyzed in this study. Summary of the TCGA dataset on HNSCCs re-analyzed in this study and the respective numbers of samples available within each. Supplementary
X-exome Sequencing of 405 Unresolved Families Identifies Seven Novel Intellectual Disability Genes
 X-exome Sequencing of 405 Unresolved Families Identifies Seven Novel Intellectual Disability Genes Hao Hu, 1,41 Stefan A. Haas, 2,41 Jamel Chelly, 3,4 Hilde Van Esch, 5 Martine Raynaud, 6,7,8 Arjan P.M.
X-exome Sequencing of 405 Unresolved Families Identifies Seven Novel Intellectual Disability Genes Hao Hu, 1,41 Stefan A. Haas, 2,41 Jamel Chelly, 3,4 Hilde Van Esch, 5 Martine Raynaud, 6,7,8 Arjan P.M.
GALAFOLD (migalastat) capsules, for oral use Initial U.S. Approval: 2018
 HIGHLIGHTS OF PRESCRIBING INFORMATION These highlights do not include all the information needed to use GALAFOLD safely and effectively. See full prescribing information for GALAFOLD. GALAFOLD (migalastat)
HIGHLIGHTS OF PRESCRIBING INFORMATION These highlights do not include all the information needed to use GALAFOLD safely and effectively. See full prescribing information for GALAFOLD. GALAFOLD (migalastat)
The Personalized Cancer Medicine Initiative at Vanderbilt
 The Personalized Cancer Medicine Initiative at Vanderbilt October 26, 2009 William Pao, MD, PhD Assistant Director, Personalized Cancer Medicine Vanderbilt-Ingram Cancer Center Cancer in the United States,
The Personalized Cancer Medicine Initiative at Vanderbilt October 26, 2009 William Pao, MD, PhD Assistant Director, Personalized Cancer Medicine Vanderbilt-Ingram Cancer Center Cancer in the United States,
GALAFOLD (migalastat) capsules, for oral use Initial U.S. Approval: 2018
 HIGHLIGHTS OF PRESCRIBING INFORMATION These highlights do not include all the information needed to use GALAFOLD safely and effectively. See full prescribing information for GALAFOLD. GALAFOLD (migalastat)
HIGHLIGHTS OF PRESCRIBING INFORMATION These highlights do not include all the information needed to use GALAFOLD safely and effectively. See full prescribing information for GALAFOLD. GALAFOLD (migalastat)
Nature Genetics: doi: /ng Supplementary Figure 1. Alternative splicing events in the 5K panel.
 Supplementary Figure 1 Alternative splicing events in the 5K panel. The majority of cryptic splicing occurred by creation of an AG or GT (Type I). While some other mutations increased the usage of a nearby
Supplementary Figure 1 Alternative splicing events in the 5K panel. The majority of cryptic splicing occurred by creation of an AG or GT (Type I). While some other mutations increased the usage of a nearby
Supplementary information
 Supplementary information! Recurrent activating mutation in PRKACA in cortisol-producing adrenal tumors Gerald Goh, Ute I. Scholl, James M. Healy, Murim Choi, Manju L. Prasad, Carol Nelson- Williams, John
Supplementary information! Recurrent activating mutation in PRKACA in cortisol-producing adrenal tumors Gerald Goh, Ute I. Scholl, James M. Healy, Murim Choi, Manju L. Prasad, Carol Nelson- Williams, John
Genomic Profiling on an Unselected Solid Tumor Population Reveals a Highly Mutated Wnt/β-Catenin Pathway Associated with Oncogenic EGFR Mutations
 Article Genomic Profiling on an Unselected Solid Tumor Population Reveals a Highly Mutated Wnt/β-Catenin Pathway Associated with Oncogenic EGFR Mutations Jingrui Jiang 1, *, Alexei Protopopov 2, Ruobai
Article Genomic Profiling on an Unselected Solid Tumor Population Reveals a Highly Mutated Wnt/β-Catenin Pathway Associated with Oncogenic EGFR Mutations Jingrui Jiang 1, *, Alexei Protopopov 2, Ruobai
Recent advances in the molecular pathology of lung cancer: role of EGFR and KRAS mutation testing in treatment selection
 ASIP 2009 USCAP Companion Meeting Molecular Pathology for the Practicing Pathologist Recent advances in the molecular pathology of lung cancer: role of EGFR and KRAS mutation testing in treatment selection
ASIP 2009 USCAP Companion Meeting Molecular Pathology for the Practicing Pathologist Recent advances in the molecular pathology of lung cancer: role of EGFR and KRAS mutation testing in treatment selection
Summary of Results CARRIER STATUS DNA INSIGHT. Result: Carrier, Heterozygote. Protected Health Information. Propionic Acidemia
 Test Results Reviewed & Approved by: Laboratory Director, Nilesh Dharajiya,.D. SAPLE REPORT CARRIER STATUS DNA INSIGHT PERSONAL DETAILS DOB Jan 1, 19XX ETHNICITY Asian ORDERING HEALTHCARE PROFESSIONAL
Test Results Reviewed & Approved by: Laboratory Director, Nilesh Dharajiya,.D. SAPLE REPORT CARRIER STATUS DNA INSIGHT PERSONAL DETAILS DOB Jan 1, 19XX ETHNICITY Asian ORDERING HEALTHCARE PROFESSIONAL
Negative feedback-defective PRPS1 mutants drive thiopurine. resistance in relapsed childhood ALL
 SUPPLEMENTARY INFORMATION Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL Benshang Li 1,5,, Hui Li 1,5,, Yun Bai 2,, Renate Kirschner-Schwabe 3,10, Jun J
SUPPLEMENTARY INFORMATION Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL Benshang Li 1,5,, Hui Li 1,5,, Yun Bai 2,, Renate Kirschner-Schwabe 3,10, Jun J
validated somatic mutations detected in pediatric and adult histiocytic neoplasm biopsies based
 Supplementary Figure 1. Somatic mutations detected in histiocytic neoplasms. Number of validated somatic mutations detected in pediatric and adult histiocytic neoplasm biopsies based on whole exome sequencing.
Supplementary Figure 1. Somatic mutations detected in histiocytic neoplasms. Number of validated somatic mutations detected in pediatric and adult histiocytic neoplasm biopsies based on whole exome sequencing.
Cover Page. The handle holds various files of this Leiden University dissertation.
 Cover Page The handle http://hdl.handle.net/1887/20119 holds various files of this Leiden University dissertation. Author: Lotta, Luca Andrea Title: Pathophysiology of thrombotic thrombocytopenic purpura
Cover Page The handle http://hdl.handle.net/1887/20119 holds various files of this Leiden University dissertation. Author: Lotta, Luca Andrea Title: Pathophysiology of thrombotic thrombocytopenic purpura
LMM / emerge III Network Reference Sequences October 2016 LMM / emerge III Network Consensus Actionable Gene List *ACMG56 gene
 LMM / emerge III Network Consensus Actionable Gene List *ACMG56 gene ACTA2* Exon 02-09 NM_001613.2 DSG2* Exon 01-15 NM_001943.3 ACTC1* Exon 01-06 NM_005159.4 DSP* Exon 01-24 NM_004415.2 APC* Exon 01-15
LMM / emerge III Network Consensus Actionable Gene List *ACMG56 gene ACTA2* Exon 02-09 NM_001613.2 DSG2* Exon 01-15 NM_001943.3 ACTC1* Exon 01-06 NM_005159.4 DSP* Exon 01-24 NM_004415.2 APC* Exon 01-15
Supplementary Information
 Supplementary Information JAK1 truncating mutations in gynecologic cancer define new role of cancerassociated protein tyrosine kinase aberrations Yuan Ren, Yonghong Zhang, Richard Z. Liu, David A. Fenstermacher,
Supplementary Information JAK1 truncating mutations in gynecologic cancer define new role of cancerassociated protein tyrosine kinase aberrations Yuan Ren, Yonghong Zhang, Richard Z. Liu, David A. Fenstermacher,
Supplementary Figures. Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated. for each lesion type.
 Supplementary Figures Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated for each lesion type. a b Supplementary Figure 2. Non-negative matrix factorization-derived
Supplementary Figures Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated for each lesion type. a b Supplementary Figure 2. Non-negative matrix factorization-derived
c Tuj1(-) apoptotic live 1 DIV 2 DIV 1 DIV 2 DIV Tuj1(+) Tuj1/GFP/DAPI Tuj1 DAPI GFP
 Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Subject ID: Date Test Started: Specimen Type: Peripheral Blood Date of Report: Date Specimen Obtained:
 TEST PERFORMED Whole exome sequencing (WES) with Focused Diagnostic Panel(s): Neuropathy, Leukodystrophy, Myopathy See below for a complete list of genes analyzed. RESULTS 1 sequence variant with potential
TEST PERFORMED Whole exome sequencing (WES) with Focused Diagnostic Panel(s): Neuropathy, Leukodystrophy, Myopathy See below for a complete list of genes analyzed. RESULTS 1 sequence variant with potential
Supplementary appendix
 Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Wells JM, Farris RF, Gosdin TA, et al. Pulmonary
Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Wells JM, Farris RF, Gosdin TA, et al. Pulmonary
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Nangalia J, Massie CE, Baxter EJ, et al. Somatic CALR s in
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Nangalia J, Massie CE, Baxter EJ, et al. Somatic CALR s in
A Natural History Study of Subjects with X-linked Retinoschisis in Anticipation of a Phase I/II Gene Therapy Trial
 A Natural History Study of Subjects with X-linked Retinoschisis in Anticipation of a Phase I/II Gene Therapy Trial Mark E. Pennesi, MD/PhD Associate Professor X-linked Retinoschisis Background Rare X-Linked
A Natural History Study of Subjects with X-linked Retinoschisis in Anticipation of a Phase I/II Gene Therapy Trial Mark E. Pennesi, MD/PhD Associate Professor X-linked Retinoschisis Background Rare X-Linked
Detection of BRCA1/2 mutations in breast cancer patients from Thailand and Pakistan
 Clin Genet 2012: 82: 594 598 Printed in Singapore. All rights reserved Letter to the Editor 2012 John Wiley & Sons A/S. Published by Blackwell Publishing Ltd CLINICAL GENETICS doi: 10.1111/j.1399-0004.2012.01869.x
Clin Genet 2012: 82: 594 598 Printed in Singapore. All rights reserved Letter to the Editor 2012 John Wiley & Sons A/S. Published by Blackwell Publishing Ltd CLINICAL GENETICS doi: 10.1111/j.1399-0004.2012.01869.x
Supporting Information
 Supporting Information Rampal et al. 10.1073/pnas.1407792111 Fig. S1. Genetic events in leukemic transformation of chronic-phase MPNs. (A) Survival of post-mpn AML patients according to mutational status
Supporting Information Rampal et al. 10.1073/pnas.1407792111 Fig. S1. Genetic events in leukemic transformation of chronic-phase MPNs. (A) Survival of post-mpn AML patients according to mutational status
Mutational and phenotypical spectrum of phenylalanine hydroxylase deficiency in Denmark
 Clin Genet 2016: 90: 247 251 Printed in Singapore. All rights reserved Short Report 2015 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd CLINICAL GENETICS doi: 10.1111/cge.12692 Mutational and
Clin Genet 2016: 90: 247 251 Printed in Singapore. All rights reserved Short Report 2015 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd CLINICAL GENETICS doi: 10.1111/cge.12692 Mutational and
Published Ahead of Print on March 6, 2015, as doi: /haematol Copyright 2015 Ferrata Storti Foundation.
 Published Ahead of Print on March 6, 2015, as doi:10.3324/haematol.2015.123612. Copyright 2015 Ferrata Storti Foundation. Somatic mutations of cell-free circulating DNA detected by Next Generation Sequencing
Published Ahead of Print on March 6, 2015, as doi:10.3324/haematol.2015.123612. Copyright 2015 Ferrata Storti Foundation. Somatic mutations of cell-free circulating DNA detected by Next Generation Sequencing
Supplementary Figure 1
 Supplementary Figure 1 Platelets from affected patients are comparable to controls on electron micrograph. Thin-section transmission electron micrographs of representative platelets from a normal control
Supplementary Figure 1 Platelets from affected patients are comparable to controls on electron micrograph. Thin-section transmission electron micrographs of representative platelets from a normal control
Research Article Mutation Analysis of the Common Deafness Genes in Patients with Nonsyndromic Hearing Loss in Linyi by SNPscan Assay
 BioMed Research International Volume 2016, Article ID 1302914, 7 pages http://dx.doi.org/10.1155/2016/1302914 Research Article Mutation Analysis of the Common Deafness Genes in Patients with Nonsyndromic
BioMed Research International Volume 2016, Article ID 1302914, 7 pages http://dx.doi.org/10.1155/2016/1302914 Research Article Mutation Analysis of the Common Deafness Genes in Patients with Nonsyndromic
Exomes and Beyond Addressing the Genome with OneSeq. Madhuri Hegde, PhD, FACMG Emory University Emory Genetics Laboratory Atlanta, GA
 Exomes and Beyond Addressing the Genome with OneSeq Madhuri Hegde, PhD, FACMG Emory University Emory Genetics Laboratory Atlanta, GA Technologies to Detect Various Types of Mutations RESOLUTION Targeted
Exomes and Beyond Addressing the Genome with OneSeq Madhuri Hegde, PhD, FACMG Emory University Emory Genetics Laboratory Atlanta, GA Technologies to Detect Various Types of Mutations RESOLUTION Targeted
(Case 5 A ) c g>a Exon 24 splice acceptor Splicing defect - Splicing defect
 Supplementary table S1. Pathogenic potential of identified DYNC2H1 mutations Family Nucleotide Change Mutation GERP PhyloP SIFT PolyPhen-2 Mutation Taster JATD-1 c. 90443A>G p.d3015g 6.17 5.3 Tolerated
Supplementary table S1. Pathogenic potential of identified DYNC2H1 mutations Family Nucleotide Change Mutation GERP PhyloP SIFT PolyPhen-2 Mutation Taster JATD-1 c. 90443A>G p.d3015g 6.17 5.3 Tolerated
Supplementary Document
 Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
Somatic mutations in ATP1A1 and ATP2B3 lead to aldosterone-producing adenomas and secondary hypertension
 Somatic mutations in ATP1A1 and ATP2B3 lead to aldosterone-producing adenomas and secondary hypertension Felix Beuschlein, Sheerazed Boulkroun, Andrea Osswald, Thomas Wieland, Hang N. Nielsen, Urs D. Lichtenauer,
Somatic mutations in ATP1A1 and ATP2B3 lead to aldosterone-producing adenomas and secondary hypertension Felix Beuschlein, Sheerazed Boulkroun, Andrea Osswald, Thomas Wieland, Hang N. Nielsen, Urs D. Lichtenauer,
Supplemental Data. Shin et al. Plant Cell. (2012) /tpc YFP N
 MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations
 Genomic sequencing of meningiomas identifies oncogenic SMO and AKT mutations Priscilla K. Brastianos*; Peleg M. Horowitz*; Sandro Santagata; Robert T. Jones; Aaron McKenna; Gad Getz; Keith L. Ligon; Emanuele
Genomic sequencing of meningiomas identifies oncogenic SMO and AKT mutations Priscilla K. Brastianos*; Peleg M. Horowitz*; Sandro Santagata; Robert T. Jones; Aaron McKenna; Gad Getz; Keith L. Ligon; Emanuele
Exons from 137 genes were targeted for hybrid selection and massively parallel sequencing.
 SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
A novel compound heterozygous mutation in SLC5A2 contributes to familial renal glucosuria in a Chinese family, and a review of the relevant literature
 MOLECULAR MEDICINE REPORTS A novel compound heterozygous mutation in SLC5A2 contributes to familial renal glucosuria in a Chinese family, and a review of the relevant literature SHENTANG LI 1, YEYI YANG
MOLECULAR MEDICINE REPORTS A novel compound heterozygous mutation in SLC5A2 contributes to familial renal glucosuria in a Chinese family, and a review of the relevant literature SHENTANG LI 1, YEYI YANG
Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
 Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
Distinct molecular abnormalities underlie unique clinical features of essential thrombocythemia. in children
 Distinct molecular abnormalities underlie unique clinical features of essential thrombocythemia in children Supplementary methods Patients and diagnostic criteria The study was approved by the hospital-based
Distinct molecular abnormalities underlie unique clinical features of essential thrombocythemia in children Supplementary methods Patients and diagnostic criteria The study was approved by the hospital-based
Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM
 Supplementary Data Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM cohort. Supplementary Table e2. Specificity, sensitivity and unadjusted ORs for glioma in participants
Supplementary Data Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM cohort. Supplementary Table e2. Specificity, sensitivity and unadjusted ORs for glioma in participants
b 0.50 Synonymous Subtype
 omatic mutation count Mutations per Mb a 80 * b 0.50 ynonymous 60 0.40 Non-synonymous 40 0.30 0.20 20 0.10 c 0 Fibroadenoma Phyllodes 0.00 ubtype Benign Borderline MAL upplementary Figure 1 Landscape of
omatic mutation count Mutations per Mb a 80 * b 0.50 ynonymous 60 0.40 Non-synonymous 40 0.30 0.20 20 0.10 c 0 Fibroadenoma Phyllodes 0.00 ubtype Benign Borderline MAL upplementary Figure 1 Landscape of
Supplementary Table 3. 3 UTR primer sequences. Primer sequences used to amplify and clone the 3 UTR of each indicated gene are listed.
 Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Generation of cetuximab-resistant cells and analysis of singlecell clones. Cetuximab-sensitive cells (LIM1215 and OXCO-2) were chronically treated with cetuximab
Supplementary Figure 1 Supplementary Figure 1: Generation of cetuximab-resistant cells and analysis of singlecell clones. Cetuximab-sensitive cells (LIM1215 and OXCO-2) were chronically treated with cetuximab
Whole exome sequencing as a first line test: Is there even a role for metabolic biochemists in the future?
 Whole exome sequencing as a first line test: Is there even a role for metabolic biochemists in the future? Tony Marinakii Purine Research Laboratory Biochemical Sciences Whole exome sequencing No doubt
Whole exome sequencing as a first line test: Is there even a role for metabolic biochemists in the future? Tony Marinakii Purine Research Laboratory Biochemical Sciences Whole exome sequencing No doubt
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases.
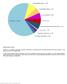 Supplementary Figure 1 Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases. A total of 85 somatically acquired
Supplementary Figure 1 Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases. A total of 85 somatically acquired
Genetic Testing and Analysis. (858) MRN: Specimen: Saliva Received: 07/26/2016 GENETIC ANALYSIS REPORT
 GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
devseek (Sequence(Analysis(Panel(for(Neurodevelopmental(Disorders
 ACSL4 Mental/retardation,/XBlinked/63 XLBR ADSL Adenylosuccinase/Deficiency AR AFF2 Mental/retardation,/XBlinked,/FRAXE/type XLBR ALG6 Congenital/disorder/of/glycosylation/type/Ic AR ANK3 Mental/retardation,/autosomal/recessive/37
ACSL4 Mental/retardation,/XBlinked/63 XLBR ADSL Adenylosuccinase/Deficiency AR AFF2 Mental/retardation,/XBlinked,/FRAXE/type XLBR ALG6 Congenital/disorder/of/glycosylation/type/Ic AR ANK3 Mental/retardation,/autosomal/recessive/37
TGF-β-SPTBN1-CTCF-regulated tumor suppression in Beckwith- Wiedemann syndrome, a human stem cell disorder
 TGF-β-SPTBN1-CTCF-regulated tumor suppression in Beckwith- Wiedemann syndrome, a human stem cell disorder Jian Chen, Zhi-Xing Yao, Jiun-Sheng Chen, Young Jin Gi, Nina M. Muñoz, Suchin Kundra, H. Franklin
TGF-β-SPTBN1-CTCF-regulated tumor suppression in Beckwith- Wiedemann syndrome, a human stem cell disorder Jian Chen, Zhi-Xing Yao, Jiun-Sheng Chen, Young Jin Gi, Nina M. Muñoz, Suchin Kundra, H. Franklin
ANNEX I SUMMARY OF PRODUCT CHARACTERISTICS
 ANNEX I SUMMARY OF PRODUCT CHARACTERISTICS 1 This medicinal product is subject to additional monitoring. This will allow quick identification of new safety information. Healthcare professionals are asked
ANNEX I SUMMARY OF PRODUCT CHARACTERISTICS 1 This medicinal product is subject to additional monitoring. This will allow quick identification of new safety information. Healthcare professionals are asked
Supplementary Table 1. Information on the 174 single nucleotide variants identified by whole-exome sequencing
 Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
Intron p Hybrid with p.leu245val p. Gly336Cys. p.leu62phe p. Ala137Val p. Asn152Thr hybrid with p.leu245val p.
 RHD negative, i.e. null Phenotype Allele name Nucleotide change Exon Predicted amino acid change D RHD*01N.01 RHD deletion 1-10 p.0 Allele name detail Comments D RHD*08N.01 RHD*Pseudogene RHD* Ψ 37- bp
RHD negative, i.e. null Phenotype Allele name Nucleotide change Exon Predicted amino acid change D RHD*01N.01 RHD deletion 1-10 p.0 Allele name detail Comments D RHD*08N.01 RHD*Pseudogene RHD* Ψ 37- bp
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature12912 Figure S1. Distribution of mutation rates and spectra across 4,742 tumor-normal (TN) pairs from 21 tumor types, as in Figure 1 from Lawrence et al. Nature
SUPPLEMENTARY INFORMATION doi:1.138/nature12912 Figure S1. Distribution of mutation rates and spectra across 4,742 tumor-normal (TN) pairs from 21 tumor types, as in Figure 1 from Lawrence et al. Nature
p.r623c p.p976l p.d2847fs p.t2671 p.d2847fs p.r2922w p.r2370h p.c1201y p.a868v p.s952* RING_C BP PHD Cbp HAT_KAT11
 ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
Hereditary hemorrhagic telangiectasia: ENG and ALK-1 mutations in Dutch patients
 Hum Genet (2005) 116: 8 16 DOI 10.1007/s00439-004-1196-5 ORIGINAL INVESTIGATION T. G. W. Letteboer Æ R. A. Zewald Æ E. J. Kamping G. de Haas Æ J. J. Mager Æ R. J. Snijder Æ D. Lindhout F. A. M. Hennekam
Hum Genet (2005) 116: 8 16 DOI 10.1007/s00439-004-1196-5 ORIGINAL INVESTIGATION T. G. W. Letteboer Æ R. A. Zewald Æ E. J. Kamping G. de Haas Æ J. J. Mager Æ R. J. Snijder Æ D. Lindhout F. A. M. Hennekam
Supplemental information
 Supplemental information Frequent reconstitution of IDH2 mutant clonal multilineage hematopoiesis following chemotherapy for acute myeloid leukemia Daniel H. Wiseman, Emma L. Williams, Deepti P. Wilks,
Supplemental information Frequent reconstitution of IDH2 mutant clonal multilineage hematopoiesis following chemotherapy for acute myeloid leukemia Daniel H. Wiseman, Emma L. Williams, Deepti P. Wilks,
ANNEX I SUMMARY OF PRODUCT CHARACTERISTICS
 ANNEX I SUMMARY OF PRODUCT CHARACTERISTICS 1 This medicinal product is subject to additional monitoring. This will allow quick identification of new safety information. Healthcare professionals are asked
ANNEX I SUMMARY OF PRODUCT CHARACTERISTICS 1 This medicinal product is subject to additional monitoring. This will allow quick identification of new safety information. Healthcare professionals are asked
SUPPLEMENTARY INFORMATION
 Somatic ERCC2 Mutations Are Associated with a Distinct Genomic Signature in Urothelial Tumors Jaegil Kim, Kent W Mouw, Paz Polak, Lior Z Braunstein, Atanas Kamburov, Grace Tiao, David J Kwiatkowski, Jonathan
Somatic ERCC2 Mutations Are Associated with a Distinct Genomic Signature in Urothelial Tumors Jaegil Kim, Kent W Mouw, Paz Polak, Lior Z Braunstein, Atanas Kamburov, Grace Tiao, David J Kwiatkowski, Jonathan
PRODUCT MONOGRAPH INCLUDING PATIENT MEDICATION INFORMATION. Migalastat Capsules. 123 mg migalastat (as migalastat hydrochloride)
 PRODUCT MONOGRAPH INCLUDING PATIENT MEDICATION INFORMATION Pr GALAFOLD TM Migalastat Capsules 123 mg migalastat (as migalastat hydrochloride) Various alimentary tract and metabolism products Amicus Therapeutics
PRODUCT MONOGRAPH INCLUDING PATIENT MEDICATION INFORMATION Pr GALAFOLD TM Migalastat Capsules 123 mg migalastat (as migalastat hydrochloride) Various alimentary tract and metabolism products Amicus Therapeutics
Detection of BRCA1 and BRCA2 germline mutations in Japanese population using next-generation sequencing
 ORIGINAL ARTICLE Detection of BRCA1 and BRCA2 germline mutations in Japanese population using next-generation sequencing Yosuke Hirotsu 1, Hiroshi Nakagomi 2, Ikuko Sakamoto 3, Kenji Amemiya 1,4, Hitoshi
ORIGINAL ARTICLE Detection of BRCA1 and BRCA2 germline mutations in Japanese population using next-generation sequencing Yosuke Hirotsu 1, Hiroshi Nakagomi 2, Ikuko Sakamoto 3, Kenji Amemiya 1,4, Hitoshi
Supplementary Figure 1 a
 Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Hereditary spastic paraplegia type 35 caused by a novel FA2H mutation
 The Turkish Journal of Pediatrics 2017; 59: 329334 DOI: 10.24953/turkjped.2017.03.016 Case Report Hereditary spastic paraplegia type 35 caused by a novel FA2H mutation Gonca Bektaş¹, Gözde Yeşil², Edibe
The Turkish Journal of Pediatrics 2017; 59: 329334 DOI: 10.24953/turkjped.2017.03.016 Case Report Hereditary spastic paraplegia type 35 caused by a novel FA2H mutation Gonca Bektaş¹, Gözde Yeşil², Edibe
Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each
 Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each mutated gene and the panel of 125 cancer-driving genes
Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each mutated gene and the panel of 125 cancer-driving genes
Supplemental Table 1. The list of variants with their respective scores for each variant classifier Gene DNA Protein Align-GVGD Polyphen-2 CADD MAPP
 Supplemental Table 1. The list of variants with their respective scores for each variant classifier Gene DNA Protein Align-GVGD Polyphen-2 CADD MAPP Frequency Domain Mammals a 3 S/P b Mammals a 3 S/P b
Supplemental Table 1. The list of variants with their respective scores for each variant classifier Gene DNA Protein Align-GVGD Polyphen-2 CADD MAPP Frequency Domain Mammals a 3 S/P b Mammals a 3 S/P b
Abstract. Introduction
 Molecular Epidemiology and Functional Assessment of Novel Allelic Variants of SLC26A4 in Non-Syndromic Hearing Loss Patients with Enlarged Vestibular Aqueduct in China Yongyi Yuan 1,4., Weiwei Guo 1.,
Molecular Epidemiology and Functional Assessment of Novel Allelic Variants of SLC26A4 in Non-Syndromic Hearing Loss Patients with Enlarged Vestibular Aqueduct in China Yongyi Yuan 1,4., Weiwei Guo 1.,
Next generation sequencing based detection of EGFR, KRAS, BRAF, NRAS, PIK3CA, Her 2 and TP53 mutations in patients with non small cell lung cancer
 MOLECULAR MEDICINE REPORTS 18: 2191-2197, 2018 Next generation sequencing based detection of EGFR, KRAS, BRAF, NRAS, PIK3CA, Her 2 and TP53 mutations in patients with non small cell lung cancer CHANGWEN
MOLECULAR MEDICINE REPORTS 18: 2191-2197, 2018 Next generation sequencing based detection of EGFR, KRAS, BRAF, NRAS, PIK3CA, Her 2 and TP53 mutations in patients with non small cell lung cancer CHANGWEN
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Lats1/2 deleted ihbs and ihps showed decreased transcripts of hepatocyte related genes (a and b) Western blots (a) and recombination PCR (b) of control and
Supplementary Figure 1 Supplementary Figure 1. Lats1/2 deleted ihbs and ihps showed decreased transcripts of hepatocyte related genes (a and b) Western blots (a) and recombination PCR (b) of control and
Names for ABO (ISBT 001) Blood Group Alleles
 Names for ABO (ISBT 001) blood group alleles v1.1 1102 Names for ABO (ISBT 001) Blood Group Alleles General description: The ABO system was discovered as in 1900 and is considered the first and clinically
Names for ABO (ISBT 001) blood group alleles v1.1 1102 Names for ABO (ISBT 001) Blood Group Alleles General description: The ABO system was discovered as in 1900 and is considered the first and clinically
Nature Genetics: doi: /ng Supplementary Figure 1. TNFAIP3-associated haplotypes in family 1.
 Supplementary Figure 1 TNFAIP3-associated haplotypes in family 1. The p.leu227* mutation (shown as a star) arose de novo in the first affected member of the family (P1). Red haplotypes carry the TNFAIP3
Supplementary Figure 1 TNFAIP3-associated haplotypes in family 1. The p.leu227* mutation (shown as a star) arose de novo in the first affected member of the family (P1). Red haplotypes carry the TNFAIP3
Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at
 Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at different concentrations for 30 min and analyzed for
Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at different concentrations for 30 min and analyzed for
Iowa Wellstone Center Muscle Tissue and Cell Culture Repository
 Iowa Wellstone Center Muscle Tissue and Cell Culture Repository Steven A. Moore, M.D., Ph.D. The University of Iowa Department of Pathology and Iowa Wellstone Muscular Dystrophy Cooperative Research Center
Iowa Wellstone Center Muscle Tissue and Cell Culture Repository Steven A. Moore, M.D., Ph.D. The University of Iowa Department of Pathology and Iowa Wellstone Muscular Dystrophy Cooperative Research Center
stomach p63/krt5 CDH17 CEACAM5 CEACAM7 ANXA13 ANXA10 CLDN18 VSIG1 TFF2 VIL1 GDA
 Supplementary Fig. 1 a esophagus Barrett s e FABP1 Ecad stomach TFF3 b p63/krt5 CDH17 REG4 CDH17 f c signal intensity p63/krt5 EsoSC 5000 4000 3000 2000 CDH17 1500 1000 Barrett s KRT14 KRT6A KRT13 KRT5
Supplementary Fig. 1 a esophagus Barrett s e FABP1 Ecad stomach TFF3 b p63/krt5 CDH17 REG4 CDH17 f c signal intensity p63/krt5 EsoSC 5000 4000 3000 2000 CDH17 1500 1000 Barrett s KRT14 KRT6A KRT13 KRT5
Supplementary Data. Clinical Setup. References. Program for Embryo Donation
 Supplementary Data Clinical Setup Notwithstanding their value in stem cell research, the source of supernumerary human blastocysts for the derivation of new stem cell lines and the modalities of their
Supplementary Data Clinical Setup Notwithstanding their value in stem cell research, the source of supernumerary human blastocysts for the derivation of new stem cell lines and the modalities of their
Abbreviations: P- paraffin-embedded section; C, cryosection; Bio-SA, biotin-streptavidin-conjugated fluorescein amplification.
 Supplementary Table 1. Sequence of primers for real time PCR. Gene Forward primer Reverse primer S25 5 -GTG GTC CAC ACT ACT CTC TGA GTT TC-3 5 - GAC TTT CCG GCA TCC TTC TTC-3 Mafa cds 5 -CTT CAG CAA GGA
Supplementary Table 1. Sequence of primers for real time PCR. Gene Forward primer Reverse primer S25 5 -GTG GTC CAC ACT ACT CTC TGA GTT TC-3 5 - GAC TTT CCG GCA TCC TTC TTC-3 Mafa cds 5 -CTT CAG CAA GGA
Unravelling the genetic basis of simplex Retinitis Pigmentosa cases
 SUPPLEMENTARY INFORMATION Unravelling the genetic basis simplex Retinitis Pigmentosa cases Nereida Bravo-Gil 1,2#, María González-del Pozo 1,2#, Marta Martín-Sánchez 1, Cristina Méndez-Vidal 1,2, Enrique
SUPPLEMENTARY INFORMATION Unravelling the genetic basis simplex Retinitis Pigmentosa cases Nereida Bravo-Gil 1,2#, María González-del Pozo 1,2#, Marta Martín-Sánchez 1, Cristina Méndez-Vidal 1,2, Enrique
Supplemental Table 1. List of genes and mature shrna sequences identified from the shrna screen in BRCA2-mutant PEO1 cells.
 Guillemette_Supplemental Table 1 Gene Mature Sequence Gene Mature Sequence ATTCATAGGATGTCAGCAG LOC157193 TATGCGTAAGTAATAAATTGCT TTAGTTCTGTCTTGGAGG LOC152225 CGCATATATGTTCTGACTT ALDH2 CACTTCAGTGTATGCCT
Guillemette_Supplemental Table 1 Gene Mature Sequence Gene Mature Sequence ATTCATAGGATGTCAGCAG LOC157193 TATGCGTAAGTAATAAATTGCT TTAGTTCTGTCTTGGAGG LOC152225 CGCATATATGTTCTGACTT ALDH2 CACTTCAGTGTATGCCT
Cystic fibrosis carrier screening in a North American population
 American College of Medical Genetics and Genomics Original Research Article Cystic fibrosis carrier screening in a North American population Val V. Zvereff, MD,PhD 1, Hawazin Faruki, DrPh 1, Marcia Edwards,
American College of Medical Genetics and Genomics Original Research Article Cystic fibrosis carrier screening in a North American population Val V. Zvereff, MD,PhD 1, Hawazin Faruki, DrPh 1, Marcia Edwards,
A novel tumor suppressor role for the Notch pathway in bladder cancer
 A novel tumor suppressor role for the Notch pathway in bladder cancer Theodoros Rampias, Paraskevi Vgenopoulou, Margaritis Avgeris, Alexander Polyzos, Konstantinos Stravodimos, Christos Valavanis, Andreas
A novel tumor suppressor role for the Notch pathway in bladder cancer Theodoros Rampias, Paraskevi Vgenopoulou, Margaritis Avgeris, Alexander Polyzos, Konstantinos Stravodimos, Christos Valavanis, Andreas
Lions and Tigers and Bears.Oh My!
 Lions and Tigers and Bears.Oh My! TAKING THE FEAR OUT OF GENETIC TESTING Lisa Butterfield, MS, CGC Genetic Counselor Clinical Assistant Professor University of Kansas Health System Why is Genetic Testing
Lions and Tigers and Bears.Oh My! TAKING THE FEAR OUT OF GENETIC TESTING Lisa Butterfield, MS, CGC Genetic Counselor Clinical Assistant Professor University of Kansas Health System Why is Genetic Testing
Supplementary Table 1. mirna modulated in D HF and ND HF patients. Table shows in a linear scale the same values of Fig. 2.
 Supplementary Table 1. mirna modulated in D HF and ND HF patients. Table shows in a linear scale the same values of Fig. 2. mir D HF VS CTR ND HF VS CTR D HF VS ND HF FOLD CHANGE p FOLD CHANGE p FOLD CHANGE
Supplementary Table 1. mirna modulated in D HF and ND HF patients. Table shows in a linear scale the same values of Fig. 2. mir D HF VS CTR ND HF VS CTR D HF VS ND HF FOLD CHANGE p FOLD CHANGE p FOLD CHANGE
a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53
 1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
Table S1. Oligonucleotides used for the in-house RT-PCR assays targeting the M, H7 or N9. Assay (s) Target Name Sequence (5 3 ) Comments
 SUPPLEMENTAL INFORMATION 2 3 Table S. Oligonucleotides used for the in-house RT-PCR assays targeting the M, H7 or N9 genes. Assay (s) Target Name Sequence (5 3 ) Comments CDC M InfA Forward (NS), CDC M
SUPPLEMENTAL INFORMATION 2 3 Table S. Oligonucleotides used for the in-house RT-PCR assays targeting the M, H7 or N9 genes. Assay (s) Target Name Sequence (5 3 ) Comments CDC M InfA Forward (NS), CDC M
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Supplementary Materials
 Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
Genetic Alteration Panels
 TM Genetic Alteration Panels GENETIC ALTERATION PANELS Table of Contents AKT Genetic Alteration Panel (ATCC No. TCP-1029 )...1 BRAF Genetic Alteration Panel (ATCC No. TCP-1032 )...5 EGFR Genetic Alteration
TM Genetic Alteration Panels GENETIC ALTERATION PANELS Table of Contents AKT Genetic Alteration Panel (ATCC No. TCP-1029 )...1 BRAF Genetic Alteration Panel (ATCC No. TCP-1032 )...5 EGFR Genetic Alteration
Supplementary Figure 1a
 Supplementary Figure 1a Hours: E-cadherin TGF-β On TGF-β Off 0 12 24 36 48 24 48 72 Vimentin βactin Fig. S1a. Treatment of AML12 cells with TGF-β induces EMT. Treatment of AML12 cells with TGF-β results
Supplementary Figure 1a Hours: E-cadherin TGF-β On TGF-β Off 0 12 24 36 48 24 48 72 Vimentin βactin Fig. S1a. Treatment of AML12 cells with TGF-β induces EMT. Treatment of AML12 cells with TGF-β results
