Laboratory, Division of Medical Sciences, National Cancer Centre, Singapore
|
|
|
- Amberly Cannon
- 6 years ago
- Views:
Transcription
1 Supplementary Information Exome sequencing of liver fluke-associated cholangiocarcinoma Choon Kiat Ong 1,2*, Chutima Subimerb 1,2,3*, Chawalit Pairojkul 3, Sopit Wongkham 3, Ioana Cutcutache 4, Willie Yu 1,2,5, John McPherson 4, George E. Allen 1,2, Cedric Chuan Young Ng 1,2, Bernice Huimin Wong 1,2, Swe Swe Myint 1,2, Vikneswari Rajasegaran 1,2, Hong Lee Heng 1,2, Anna Gan 1,2, Zhi Jiang Zang 6,7, Alice Yingting Wu 4, Jeanie Wu 7, Ming Hui Lee 7, DaChuan Huang 1,2, Pauline Ong 1,2, Waraporn Chan-on 1,2,3, Yun Cao 8, Chao-Nan Qian 8, Kiat Hon Lim 9, Aikseng Ooi 10, Karl Dykema 10, Kyle Furge 10, Veerapol Kukongviriyapan 3, Banchob Sripa 3, Chaisiri Wongkham 3, Puangrat Yongvanit 3, P. Andrew Futreal 11, Vajaraphongsa Bhudhisawasdi 3,#, Steve Rozen 4,#, Patrick Tan 12,7,13,#, Bin Tean Teh 1,2,# 1 National Cancer Centre Singapore-Van Andel Research Institute Translational Research Laboratory, Division of Medical Sciences, National Cancer Centre, Singapore 2 Laboratory of Cancer Therapeutics, Division of Cancer and Stem Cell Biology, Duke-NUS Graduate Medical School, Singapore 3 Liver fluke and Cholangiocarcinoma Research Center, Faculty of Medicine, Khon Kaen University, Khonkaen, Thailand 4 Division of Neuroscience and Behavioral Disorders, Duke-NUS Graduate Medical School, Singapore 5 National University of Singapore Graduate School for Integrative Sciences and Engineering, Singapore 6 Division of Cellular and Molecular Research, National Cancer Centre, Singapore 7 Division of Cancer and Stem Cell Biology, Duke-NUS Graduate Medical School, Singapore 8 Sun Yat-sen University Cancer Center, Guangzhou, P. R. China 9 Department of Pathology, Singapore General Hospital, Singapore 10 Laboratory of Computational Biology, Van Andel Research Institute, Grand Rapids, Michigan, USA 11 Cancer Genome Project, Wellcome Trust Sanger Institute, Hinxton, UK 12 Cancer Science Institute of Singapore, National University of Singapore, Centre for Life Sciences, Singapore 13 Genome Institute of Singapore, Singapore #Corresponding authors *These authors contributed equally to the work 1
2 Index of all supplementary tables, figures and notes Supplementary Tables Supplementary Table 1 Supplementary Table 2 Supplementary Table 3 Supplementary Table 4 Supplementary Table 5 Supplementary Table 6 Supplementary Table 7 Supplementary Table 8 Supplementary Table 9 Supplementary Table 10 Supplementary Table 11 Supplementary Table 12 Clinicopathological characteristics of 54 O. viverrini-associated CCA patients Sequence analysis summary of 8 O. viverrini-associated CCA exomes Non-synonymous somatic mutations identified and validated in the Discovery set of 8 tumors Somatic mutation spectra in O. viverrini-associated CCA Somatic mutations in TP53, KRAS, SMAD4, MLL3, GNAS, RNF43, ROBO2, RADIL, PEG3, NDC80, CDKN2A, PTEN, LAMA2, XIRP2 and PCDHA13 in 54 O. viverrini-associated CCA tumors Genome MuSiC analysis of mutated genes in O. viverrini-associated CCA TP53 mutations in O. viverrini-associated CCA Characteristics of O. viverrini-associated CCA patient with both SMAD4 and PTEN mutations Overall survival analysis in O. viverrini-associated CCA Mutation spectra in O. viverrini-associated Cholangiocarcinoma (CCA), Pancreatic cancer (PDAC) and Hepatocellular carcinoma (HCC) Chromosomal changes in Cholangiocarcinoma, Pancreatic cancer and Hepatocellular carcinoma Primers used for PCR amplification and Sanger sequencing Supplementary Figures Supplementary Figure 1 Distribution of mutations in 8 O. viverrini-associated CCA exomes Supplementary Figure 2 Summary of commonly mutated genes in 54 O. viverrini-associated CCA samples Supplementary Figure 3 Kaplan-Meier plots of overall survival of patients with tumors containing KRAS and RNF43 mutations Supplementary Figure 4 Analysis of LOH at ROBO2 by ASCAT (Allele-Specific Copy number Analysis of Tumors) in tumor W39, which also has an inactivating somatic mutation of ROBO2 Supplementary Figure 5 Comparisons of mutation spectra in O. viverrini-associated CCA, PDAC, and HCV-associated HCC Supplementary Notes Targeted exome capture and library preparation for massively parallel sequencing Supplementary References 2
3 Supplementary Table 1. Clinicopathological characteristics of 54 O. viverrini-associated CCA patients Code Sex Age TMN Staging Histological types Survival Status A028 F 59 T3N1M0 IVA WD tubular adenocarcinoma 474 DD A035 F 49 T3N0M0 III WD tubular adenocarcinoma 167 DD A039 F 72 T3N1M0 IVA PD tubular adenocarcinoma 874 AL A042 F 52 T3N1M0 IVA WD tubular adenocarcinoma 865 AL A043 M 45 T3N0M0 III Papillary carcinoma 426 DD A074 M 55 T4N0M0 IVA WD tubular adenocarcinoma 154 DD A105 M 53 T3N1M0 IVA WD tubular adenocarcinoma 592 DD A106 M 51 T3N1M0 IVA WD tubular adenocarcinoma 18 DD A107 M 54 T3N0M0 III WD tubular adenocarcinoma 302 DD A119 F 51 T3N0M0 III Papillary carcinoma 358 DD A120 F 53 T3N1M0 IVA Papillary carcinoma 156 DD A128 M 61 T3N1M0 IVA Papillary carcinoma 252 DD A142 F 38 T2bN1M0 IVA WD tubular adenocarcinoma 559 DD A159 M 55 T3N0M0 IIIB Papillary carcinoma 237 DD A162 M 68 T3N1M0 IVA Papillary carcinoma 617 AL B032 M 53 T3N0M0 III MD tubular adenocarcinoma 237 DD B048 F 51 T2bN0M0 IVA WD tubular adenocarcinoma 154 DD B070 M 61 T3N0M0 IIIB Papillary carcinoma 433 AL B083 F 53 T3N0M0 III WD tubular adenocarcinoma 266 DD B085 M 73 T3N0M0 III WD tubular adenocarcinoma 82 DD B087 M 56 T2bN0M0 II WD tubular adenocarcinoma 391 AL B099 M 48 T3N0M0 III WD tubular adenocarcinoma 373 AL B113 M 61 T3N1M0 IVA Papillary carcinoma 111 DD B149 M 63 T3N0M0 IIIA WD tubular adenocarcinoma 258 AL R100 F 49 T2bN1M0 IIIB WD tubular adenocarcinoma 251 DD R104 F 64 T3N0M0 III WD tubular adenocarcinoma 117 DD R134 M 50 T3N0M0 III WD tubular adenocarcinoma 222 DD R149 M 37 T4N1M0 IIIA Papillary carcinoma 475 DD T003 M 52 T4N0M0 IVA MD tubular adenocarcinoma 2253 DD T026 M 52 T3N1M0 IIIB WD tubular adenocarcinoma 307 DD 3
4 Supplementary Table 1 continued. Clinicopathological characteristics of 54 O. viverriniassociated CCA patients Code Sex Age TMN Staging Histological types Survival Status T151 M 73 T2bN1M1 IVB MD tubular adenocarcinoma 191 DD T157 M 42 T3N0M0 IIIa WD tubular adenocarcinoma 351 DD T160 M 46 T3N0M0 IIIa Papillary carcinoma 2451 AL U027 F 66 T2N1M0 IVA Papillary carcinoma 148 DD U044 M 57 T3N1M0 IIIB Papillary carcinoma 45 DD W012 F 61 T3N0M0 III WD tubular adenocarcinoma 149 DD W039 M 66 T3N0M0 III WD tubular adenocarcinoma 64 DD W040 M 56 T4N1M0 IVA WD tubular adenocarcinoma 157 DD Y002 F 45 T3N1M0 IVA Papillary carcinoma 1320 AL Y008 F 65 T3N0M0 III Papillary carcinoma 176 DD Y019 M 70 T4NXM0 IVA WD tubular adenocarcinoma 210 DD Y020 M 50 T3N1M0 IVA Papillary carcinoma 180 DD Y023 M 62 T4N0M0 IVA Papillary carcinoma 1274 AL Y032 M 69 T3N1M0 IVA Papillary carcinoma 429 DD Y033 F 51 T4N1M0 IVA WD tubular adenocarcinoma 157 DD Y035 F 64 T3N0M0 III WD tubular adenocarcinoma 754 DD Y057 M 57 T3N1M0 IVA WD tubular adenocarcinoma 416 DD Y065 M 63 T4N1M0 IVA Papillary carcinoma 558 DD Y072 M 61 T3N0M0 III Papillary carcinoma 302 DD Y074 F 40 T1N0M0 I Papillary carcinoma 1148 AL Y091 F 51 T4N0M0 IVA WD tubular adenocarcinoma 609 DD Y123 M 55 T4N0M0 IVA WD tubular adenocarcinoma 340 DD Y140 F 56 T2N0M0 II Papillary carcinoma 992 AL Y149 M 69 T4N1M0 IVA WD tubular adenocarcinoma 876 DD Highlighted in Bold = samples in Discovery set Histological types : tumor differentiation, WD = Well differentiated, MD = Moderately differentiated, PD = Poorly differentiated, PAP = Papillary Staging: TMN stage according to TMN classification, AJCC sixth edition. Survival : survival time in days. Status : follow up status ; DD = dead, AL = alive (data on 15 Aug 2011) 4
5 Supplementary Table 2. Sequence analysis summary of 8 O. viverrini-associated CCA exomes Bases in Target Region Bases Sequenced Bases Mapped to Genome * Bases After Filtering Bases Mapped to Target Region * Ave. Depth Per Targeted Base Targeted Bases with Depth at Least 1X Targeted Bases with Depth at Least 20X Somatic mutations identified in targeted region B085 B099 R104 T026 U044 W012 W039 W040 Normal 37,806,033 11,295,136,692 8,692,958,394 7,490,182,833 3,905,980, Tumor 37,806,033 11,255,244,354 8,756,261,352 7,496,830,908 3,812,200, Normal 37,806,033 7,097,490,664 5,960,436,264 5,199,247,030 2,930,117, Tumor 37,806,033 6,665,088,684 5,473,009,068 4,726,727,060 2,798,044, Normal 37,806,033 7,571,001,568 5,710,829,528 5,040,625,070 1,903,794, Tumor 37,806,033 7,418,858,456 5,981,689,212 5,293,556,256 2,668,373, Normal ,809,159,504 7,792,775,868 6,816,265,726 3,668,218, Tumor 37,806,033 8,949,919,640 7,988,945,428 6,972,740,798 3,782,109, Normal 37,806,033 7,264,404,768 6,099,026,424 5,395,414,370 2,526,282, Tumor 37,806,033 6,954,997,300 5,822,840,516 5,152,914,150 2,404,785, Normal 37,806,033 7,868,529,224 7,573,370,752 6,475,821,992 2,969,234, Tumor 37,806,033 6,755,744,456 6,473,749,104 5,451,167,598 2,519,928, Normal 37,806,033 6,349,013,668 6,118,662,324 5,167,650,066 2,428,397, Tumor 37,806,033 6,870,484,564 6,609,333,680 5,659,540,794 2,799,365, Normal 37,806,033 8,110,359,300 7,214,104,600 6,299,378,760 3,409,974, Tumor 37,806,033 8,848,149,808 7,906,602,224 6,911,891,116 3,673,858, AVERAGE 37,806,033 8,005,223,916 6,885,912,171 5,971,872,158 3,012,541, * based on the hg18 UCSC release of the human genome. 5
6 Supplementary Table 3. Non-synonymous somatic mutations identified and validated in the Discovery set of 8 tumors No. Gene Symbol Sample Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 1 ADAM12 B085 CCDS g.chr10: C>T c.2104c>t p.r702w Missense 2 AFF1 W040 CCDS g.chr4: T>C c.2282t>c p.l761s Missense 3 ALPP B085 CCDS g.chr2: C>T c.814c>t p.r272c Missense 4 ANK2 T026 CCDS g.chr4: C>T c.3466c>t p.r1156c Missense 5 ANKRD35 U044 CCDS919.1 g.chr1: G>A c.2779g>a p.e927k Missense 6 ANO4 B085 CCDS g.chr12: G>T c.790g>t p.e264x Nonsense 7 ARAF U044 CCDS g.chrx: A>T c.650a>t p.n217i Missense 8 ARID1A T026 CCDS285.1 g.chr1: C>T c.2632c>t p.q878x Nonsense 9 ARL6IP5 U044 CCDS g.chr3: T>C c.317t>c p.m106t Missense 10 ASAP3 R104 CCDS235.1 g.chr1: G>A c.992g>a p.c331y Missense 11 BBS4 W040 CCDS10246 g.chr15: delGTT c.471 to 473 delgtt fs Deletion 12 BCAS2 R104 CCDS874 g.chr1: delC c.4 delg fs Deletion 13 CALML5 T026 CCDS g.chr10: C>T c.7c>t p.g3s Missense 14 CALN1 R104 CCDS g.chr7: A>G c.380a>g p.d127g Missense 15 CCDC97 R104 CCDS g.chr19: G>T c.730g>t p.e244x Nonsense 16 CD109 B099 CCDS g.chr6: C>T c.4240c>t p.r1414c Missense 17 CDH8 U044 CCDS g.chr16: G>A c.580g>a p.a194t Missense 18 CDKN2A U044 CCDS g.chr9: C>T c.416c>t p.r139q Missense 19 CHD5 R104 CCDS57.1 g.chr1: G>A c.4330g>a p.q1444x Nonsense 20 CHRD B085 CCDS g.chr3: G>A c.1513g>a p.v505m Missense 21 CNBP W040 CCDS g.chr3: C>T c.362c>t p.s121f Missense 22 COL11A1 W040 CCDS779.1 g.chr1: G>A c.793g>a p.e255k Missense 23 COL11A2 U044 CCDS g.chr6: C>T c.2842c>t p.r948c Missense 24 CRISPLD1 B085 CCDS g.chr8: G>A c.218g>a p.r73q Missense 25 CTNNA2 W040 CCDS g.chr2: T>A c.1748t>a p.m583k Missense 26 DDX52 B085 CCDS g.chr17: G>A c.1505g>a p.r502q Missense 27 DICER1 U044 CCDS g.chr14: G>A c.4651g>a p.e1551k Missense 28 DLGAP5 B099 CCDS g.chr14: G>C c.1150g>c p.g384r Missense 29 DSP U044 CCDS g.chr6: C>T c.4763c>t p.s1588f Missense 30 DYNC1H1 W039 CCDS g.chr14: G>A c.3182g>a p.w1061x Nonsense 31 EHBP1 R104 CCDS g.chr2: G>A c.47g>a p.s16f Missense 32 EIF2C1 B085 CCDS398.1 g.chr1: G>A c.1967g>a p.r656h Missense 33 EIF3E U044 CCDS g.chr8: C>G c.80c>g p.s27c Missense 34 ENTPD8 R104 CCDS g.chr9: C>T c.178c>t p.q60x Nonsense 35 EPHA2 R104 CCDS169.1 g.chr1: G>A IVS1+1 G>A Splice site Splice site 6
7 Supplementary Table 3 continued. Non-synonymous somatic mutations identified and validated in the Discovery set of 8 tumors No. Gene Symbol Sample Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 36 EXOSC8 R104 CCDS g.chr13: G>A c.412g>a p.d138n Missense 37 EXTL3 W012 CCDS g.chr8: A>G c.2419a>g p.k807e Missense 38 FAM47A W040 CCDS g.chrx: G>A c.865g>a p.e289k Missense 39 FBLN1 W039 CCDS g.chr22: C>T c.1739c>t p.s580f Missense 40 FBN2 B099 CCDS g.chr5: T>A c.630t>a p.d210e Missense 41 FER W012 CCDS g.chr5: A>T c.973a>t p.n325y Missense 42 FH W039 CCDS g.chr1: C>T c.884c>t p.a295v Missense 43 FKBP3 T026 CCDS g.chr14: A>G c.235a>g p.s79g Missense 44 FLNC B085 CCDS g.chr7: G>A c.1037g>a p.g346e Missense 45 FMNL1 W039 CCDS g.chr17: G>A c.2755g>a p.v919m Missense 46 FOXR2 U044 ENST g.chrx: G>C c.775g>c p.d259h Missense 47 GATM U044 CCDS g.chr15: C>T c.565c>t p.r189c Missense 48 GDPD3 W040 CCDS10671 g.chr16: insAGCT c.938 insagct fs Insertion 49 GHRHR R104 CCDS5432 g.chr7: T>C c.346t>c p.s116p Missense 50 GHSR B085 CCDS g.chr3: G>A c.841g>a p.v281i Missense 51 GIGYF2 B099 CCDS g.chr2: G>A c.1136g>a p.r379t Missense 52 GJB6 W012 CCDS g.chr13: G>A c.380g>a p.r127q Missense 53 GLI3 T026 CCDS g.chr7: C>A c.664c>a p.l222m Missense 54 GNAS T026 CCDS g.chr20: C>T c.601c>t p.r201c Missense 55 GNAS W012 CCDS g.chr20: C>T c.601c>t p.r201c Missense 56 GPSM1 W039 CCDS48055 g.chr9: G>A c.441g>a p.r75q Missense 57 HCN4 B099 CCDS g.chr15: C>T c.1159c>t p.r387c Missense 58 HDAC2 W012 CCDS g.chr6: C>T c.514c>t p.r172w Missense 59 HDAC4 W040 CCDS g.chr2: C>T c.1633c>t p.p545s Missense 60 HECW2 R104 CCDS g.chr2: A>G c.1267a>g p.i423v Missense 61 HIST1H2AG U044 CCDS g.chr6: G>C c.364g>c p.e122q Missense 62 HOOK1 R104 CCDS612.1 g.chr1: G>T c.82g>t p.a28s Missense 63 HOXC11 U044 CCDS g.chr12: G>A c.454g>a p.d152n Missense 64 IL1RAPL1 T026 CCDS g.chrx: G>A c.277g>a p.g93r Missense 65 INHBA T026 CCDS g.chr7: C>T c.916c>t p.r306c Missense 66 IP6K2 R104 CCDS g.chr3: C>T c.37c>t p.r13c Missense 67 IQCB1 U044 CCDS g.chr3: G>T c.303g>t p.e101d Missense 68 IRX1 U044 CCDS g.chr5: G>C c.486>c p.m162i Missense 69 ITGA2B T026 CCDS g.chr17: G>A c.1708g>a p.g570r Missense 70 ITPR2 W039 CCDS g.chr12: C>T c.3766c>t p.l1256f Missense 7
8 Supplementary Table 3 continued. Non-synonymous somatic mutations identified and validated in the Discovery set of 8 tumors No. Gene Symbol Sample Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 71 JAG1 B085 CCDS g.chr20: A>T c.2315a>t p.e772v Missense 72 KCNH5 W039 CCDS g.chr14: G>A c.989g>a p.r330q Missense 73 KCNH7 B085 CCDS g.chr2: G>A c.737g>a p.r246q Missense 74 KCNJ9 W040 CCDS1194 g.chr: insT c.241 inst fs Insertion 75 KCNN4 B085 CCDS g.chr19: C>T c.1163c>t p.s388l Missense 76 KCNQ2 R104 CCDS g.chr20: C>T c.1055c>t p.s352l Missense 77 KIAA1267 U044 CCDS g.chr17: G>A IVS4+5 G>A Splice site Splice site 78 KIAA1984 W040 CCDS43906 g.chr9: delCC c.504 to 505 delcc fs Deletion 79 KLHL4 W039 CCDS g.chrx: C>T c.646c>t p.l216f Missense 80 KRAS B085 CCDS g.chr12: G>C c.35g>c p.g12a Missense 81 KRAS R104 CCDS g.chr12: C>T c.358c>t p.g13d Missense 82 KRAS W012 CCDS g.chr12: G>C c.35g>c p.g12a Missense 83 LAMA2 U044 CCDS g.chr6: C>A c.289c>a p.h97n Missense 84 LAMA2 W039 CCDS g.chr6: T>A IVS14+5 T>A Splice site Splice site 85 LMX1A U044 CCDS g.chr1: G>C c.130g>c p.d44h Missense 86 LRP1B B085 CCDS g.chr2: A>G IVS69+6 A>G Splice site Splice site 87 LRP2 W012 CCDS g.chr2: G>A c.10652g>a p.r3551h Missense 88 LRRC15 R104 CCDS g.chr3: G>A c.1162g>a p.v388i Missense 89 LRRK2 W039 CCDS g.chr12: C>T c.575c>t p.s192l Missense 90 MAGEA11 B085 CCDS14690 g.chrx: G>T c.975g>t p.e325d Missense 91 MAP2K4 W039 CCDS g.chr17: T>G c.1043t>g p.l348r Missense 92 MAP3K13 R104 CCDS g.chr3: G>A c.801g>a p.m267i Missense 93 MARK4 R104 CCDS g.chr19: T>A c.320t>a p.v107d Missense 94 MASP2 B099 CCDS123.1 g.chr1: G>C c.2000g>c p.g667a Missense 95 MEFV W012 CCDS g.chr16: G>A c.2150g>a p.r717h Missense 96 MLL3 T026 CCDS g.chr7: G>T c.14641g>t p.e4881x Nonsense 97 MLL3 W039 CCDS g.chr7: G>T c.12149g>t p.s4050x Nonsense 98 MLYCD R104 CCDS g.chr16: G>C c.646g>c p.e216q Missense 99 MXRA5 U044 CCDS g.chrx: G>A c.3725g>a p.r1242q Missense 100 MYH2 W039 CCDS g.chr17: G>A c.1334g>a p.r445h Missense 101 NDC80 W012 CCDS g.chr18: delA c.756 dela fs Deletion 102 NDC80 W039 CCDS g.chr18: G>A c.859g>a p.e287k Missense 103 NFKB1 W040 CCDS43906 g.chr4: insC c.1032 insc fs Insertion 104 NFKBIL2 W040 CCDS g.chr8: C>T c.1567c>t p.p523s Missense 105 NLGN3 B085 CCDS g.chrx: G>A c.241g>a p.e81k Missense 8
9 Supplementary Table 3 continued. Non-synonymous somatic mutations identified and validated in the Discovery set of 8 tumors No. Gene Symbol Sample Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 106 NPY5R U044 CCDS g.chr4: G>T c.250g>t p.a84s Missense 107 NUP160 R104 CCDS g.chr11: A>G c.996a>g p.k332n Missense 108 NUP160 R104 CCDS g.chr11: A>T c.994a>t p.k332e Missense 109 NUP210 W040 CCDS g.ch3r: G>A c.4054g>a p.e1352k Missense 110 ODF3 U044 CCDS g.chr11: T>G c.190t>g p.c64g Missense 111 OR51B2 T026 CCDS g.chr11: C>T c.83c>t p.p28l Missense 112 OR6Q1 R104 CCDS g.chr11: C>A c.662c>a p.s221y Missense 113 P2RY13 W012 CCDS g.chr3: C>T c.751c>t p.h251y Missense 114 P4HA3 B099 CCDS g.chr11: A>G c.377a>g p.d126g Missense 115 PAPD4 T026 CCDS g.chr5: C>T c.364c>t p.h122y Missense 116 PBX3 B099 CCDS g.chr9: A>T c.803a>t p.k268i Missense 117 PCBP3 U044 CCDS g.chr21: C>T c.512c>t p.p171l Nonsense 118 PCDH11X U044 CCDS g.chrx: G>A c.3616g>a p.a1206t Missense 119 PCDHA13 T026 CCDS g.chr5: C>T c.1985c>t p.t662m Missense 120 PCDHA13 U044 CCDS g.chr5: C>T c.1301c>t p.s434l Missense 121 PCDHA7 U044 CCDS g.chr5: G>A c.1354g>a p.a452t Missense 122 PCDHGA7 R104 ENST g.chr5: G>A c.2107g>a p.v703i Missense 123 PCNX W012 CCDS g.chr14: C>T c.4169c>t p.a1390v Missense 124 PDGFD W039 CCDS g.chr11: G>A c.970g>a p.v324m Missense 125 PEG3 W012 CCDS g.chr19: A>G c.2536a>g p.s846r Missense 126 PEG3 W039 CCDS g.chr19: G>A c.2947g>a p.d983n Missense 127 PGBD5 W039 CCDS g.chr1: C>T c.1139c>t p.t380m Missense 128 PHACTR4 B099 CCDS g.chr1: G>T c.1825g>t p.e609x Nonsense 129 POLDIP3 W040 CCDS14038 g.chr22: insC c.227 insc fs Insertion 130 POLL W039 CCDS g.chr10: G>A c.1157g>a p.r386h Missense 131 PPM1E T026 CCDS g.chr17: A>G c.1331a>g p.d444g Missense 132 PPP1R16B U044 CCDS g.chr20: A>G c.1675a>g p.k559e Missense 133 PPP2R3A W039 CCDS g.chr3: G>A c.37g>a p.d13n Missense 134 PRDM4 B099 CCDS g.chr12: T>C c.1117t>c p.f373l Missense 135 PREX2 W039 CCDS g.chr8: G>C c.4396g>c p.a1466p Missense 136 PRMT6 B085 CCDS g.chr1: A>G c.299a>g p.y100c Missense 137 PTEN R104 CCDS g.chr10: A>G c.464a>g p.y155c Missense 138 PTPRS W012 CCDS g.chr19: G>A c.230g>a p.r77h Missense 139 RAB11FIP5 R104 CCDS g.chr2: C>T c.1120c>t p.r374w Missense 140 RADIL W039 CCDS g.chr7: C>T IVS6+4 C>T Splice site Splice site 9
10 Supplementary Table 3 continued. Non-synonymous somatic mutations identified and validated in the Discovery set of 8 tumors No. Gene Symbol Sample Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 141 RADIL W040 CCDS g.chr7: G>A c.386g>a p.r129q Missense 142 RASAL2 W012 CCDS g.chr1: C>T c.3776c>t p.t1259m Missense 143 RASGRF2 W039 CCDS g.chr5: A>G IVS2-2 A>G Splice site Splice site 144 RBBP7 W040 CCDS14179 g.chrx: delGGGTCATAACCA c.98 to 109 delgggtcataacca fs Deletion 145 RBM26 B099 CCDS g.chr13: G>C c.634g>c p.e212x Nonsense 146 RGS3 R104 CCDS g.chr9: C>T c.262c>t p.a88t Missense 147 RMI1 R104 CCDS g.chr9: A>G c.554a>g p.e185g Missense 148 RNF133 B099 CCDS g.chr7: G>T c.454g>t p.v152l Missense 149 RNF43 R104 CCDS g.chr17: A>G c.355a>g p.c119r Missense 150 RNF43 U044 CCDS g.chr17: A>T c.500a>t p.n167i Missense 151 RNF43 W012 CCDS g.chr17: C>T c.21667c>t p.q723x Nonsense 152 ROBO2 T026 CCDS g.chr3: C>T c.1585cc>t p.q529x Nonsense 153 ROBO2 W039 CCDS g.chr3: insC c.3234 insc fs Insertion 154 SACS B099 CCDS g.chr13: A>T c.5660a>t p.k1887i Missense 155 SAMD7 W039 CCDS g.chr3: C>G c.25c>g p.p9a Missense 156 SCN11A R104 CCDS g.chr3: C>T c.3470c>t p.a1157v Missense 157 SEMA4F W040 CCDS g.chr2: G>C c.2067g>c p.q689h Missense 158 SEMA5A B085 CCDS g.chr5: C>A c.2170c>a p.h724n Missense 159 SEMA6D T026 CCDS g.chr15: C>T IVS16-3 C>T Splice site Splice site 160 SGSM2 B099 CCDS g.chr17: G>C c.2045g>c p.s682t Missense 161 SIGLEC12 W012 CCDS g.chr19: C>T c.197c>t p.a66v Missense 162 SLC23A1 W040 CCDS g.chr5: A>T c.227a>t p.q76l Missense 163 SLC5A5 W040 CCDS g.chr19: G>A c.1682g>a p.g561e Missense 164 SMAD4 B099 CCDS g.chr18: T>C c.1283t>c p.k428t Missense 165 SMAD4 R104 CCDS g.chr18: delAGTA c.1447 to 1450 delagta fs Deletion 166 SMAD4 U044 CCDS g.chr18: C>T c.547c>t p.q183x Nonsense 167 SMAD4 W012 CCDS g.chr18: G>T c.394g>t p.h132d Missense 168 SMARCA1 B099 CCDS g.chrx: C>A c.2594c>a p.l532m Missense 169 SMC6 W040 CCDS g.chr2: A>G c.1658a>g p.y553c Missense 170 SNAPC4 B099 CCDS g.chr9: C>T c.443c>t p.p148l Missense 171 SPAG17 U044 CCDS899.1 g.chr1: G>A IVS1+3 G>A Splice site Splice site 172 SPSB4 B099 CCDS g.chr3: G>T c.592g>t p.g198c Missense 10
11 Supplementary Table 3 continued. Non-synonymous somatic mutations identified and validated in the Discovery set of 8 tumors No. Gene Symbol Sample Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 173 STK36 T026 CCDS g.chr2: A>G c.2599a>g p.s867g Missense 174 STT3B R104 CCDS g.chr3: C>T c.398c>t p.p133l Missense 175 SYNE1 B099 CCDS g.chr6: C>T c.12953c>t p.t4318m Missense 176 SYNJ2 B085 CCDS g.chr6: C>A c.811c>a p.l271m Missense 177 TARS T026 CCDS g.chr5: C>T c.218c>t p.a73v Missense 178 TCEAL2 T026 CCDS g.chrx: C>T c.569c>t p.a190v Missense 179 TFAP2D B099 CCDS g.chr6: C>T c.1150c>t p.h384y Missense 180 THBS2 W039 CCDS g.chr6: C>T c.175c>t p.r59c Missense 181 TMCO4 W040 CCDS198.1 g.chr1: G>T c.327g>t p.l109f Missense 182 TMEM222 R104 CCDS297.2 g.chr1: G>A c.526g>a p.g176r Missense 183 TNKS2 W039 CCDS g.chr10: G>A c.2795g>a p.r932k Missense 184 TP53 T026 CCDS g.chr17: insG c.216 insg fs Insertion 185 TP53 U044 CCDS g.chr17: G>A c.310c>t p.q104x Nonsense 186 TP53 W012 CCDS g.chr17: G>A c.742g>a p.r248w Missense 187 TP53 W040 CCDS g.chr17: inst c.716 inst fs Insertion 188 TRIM28 B085 CCDS g.chr19: G>T c.1475g>t p.r492l Missense 189 TRIM48 W039 CCDS7947 g.chr11: C>T c.1048c>t p.q366x Nonsense 190 TROVE2 W040 CCDS1379 g.chr1: insTG c.1007 instg fs Insertion 191 TRPC5 B099 CCDS g.chrx: G>T c.1662g>t p.k554n Missense 192 TUBA3C U044 CCDS g.chr13: G>T c.1345g>t p.e449x Nonsense 193 UBAC1 W040 CCDS g.chr9: G>C c.484g>c p.v162m Missense 194 UTP20 W039 CCDS g.chr12: T>A IVS53+5 T>A Splice site Splice site 195 VANGL2 W040 CCDS g.chr1: T>A c.302t>a p.l101q Missense 196 VAV1 W040 CCDS g.chr19: G>A c.2383g>a p.a795t Missense 197 VCAN R104 CCDS g.chr5: G>A c.9904g>a p.v3302i Missense 198 VEGFC R104 CCDS g.chr4: A>G c.235a>g p.k79e Missense 199 VWCE T026 CCDS g.chr11: C>G c.732c>g p.f244l Missense 200 WNK3 B085 CCDS g.chrx: G>A c.5246g>a p.g1749e Missense 201 XIRP2 B085 CCDS g.chr2: inst c.9704 inst fs Insertion 202 XIRP2 T026 CCDS g.chr2: C>T c.7898c>t p.s2633l Missense 203 YSK4 W012 CCDS g.chr2: C>T c.812c>t p.s271l Missense 204 ZFY U044 CCDS g.chry: C>T c.1958c>t p.t653m Missense 205 ZNF136 W040 CCDS g.chr19: A>T c.569a>t p.h190l Missense 206 ZNF790 U044 CCDS g.chr19: G>A c.1840g>a p.e614k Missense 11
12 Supplementary Table 4. Somatic mutation spectra in O. viverrini-associated CCA Base pair change % Number Total number of substitutions Substitutions at C:G base pairs C:G to T:A C:G to G:C C:G to A:T Substitutions at T:A base pairs T:A to C:G T:A to G:C T:A to A:T Substitutions at specific dinucleotides 5'-CpG-3' '-TpC-3' NB: Mutations in this analysis include synonymous and non-synonymous coding mutations and mutations at canonical splice sites and in untranslated regions (Sanger validated and non-validated targets) for O. viverrini-associated CCA. 12
13 Supplementary Table 5. Somatic mutations in TP53, KRAS, SMAD4, MLL3, GNAS, RNF43, ROBO2, RADIL, PEG3, NDC80, CDKN2A, PTEN, LAMA2, XIRP2 and PCDHA13 in 54 O. viverrini-associated CCA tumors Sample Gene Symbol CCDS ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type U044 CDKN2A CCDS g.chr9: C>T c.416c>t p.r139q Missense Y091 CDKN2A CCDS g.chr9: C>T c.238 C>T p.r80x Nonsense A106 CDKN2A CCDS g.chr9: G>T c.343 G>T p.v115l Missense A074 GNAS CCDS g.chr20: G>T c.602 G>T p.r201l Missense B149 GNAS CCDS g.chr20: C>T c.601 C>T p.r201c Missense T026 GNAS CCDS g.chr20: C>T c.601 C>T p.r201c Missense W012 GNAS CCDS g.chr20: C>T c.601 C>T p.r201c Missense Y149 GNAS CCDS g.chr20: G>A c.602 G>A p.r201h Missense B085 KRAS CCDS g.chr12: G>C c.35 G>C p.g12a Missense R104 KRAS CCDS g.chr12: C>T c.38 C>T p.g13d Missense T151 KRAS CCDS g.chr12: G>A c.35 G>A p.g12d Missense U027 KRAS CCDS g.chr12: G>A c.34 G>A p.g12s Missense W12 KRAS CCDS g.chr12: G>C c.35 G>C p.g12a Missense Y008 KRAS CCDS g.chr12: G>A c.35 G>A p.g12d Missense Y019 KRAS CCDS g.chr12: G>T c.34 G>T p.g12c Missense Y020 KRAS CCDS g.chr12: G>T c.35 G>T p.g12v Missense Y149 KRAS CCDS g.chr12: G>T c.35 G>T p.g12v Missense U044 LAMA2 CCDS g.chr6: C>A c.289c>a p.h97n Missense W039 LAMA2 CCDS g.chr6: T>A IVS14+5 T>A Splice site Splice site T026 MLL3 CCDS g.chr7: G>T c G>T p.e4881x Nonsense W039 MLL3 CCDS g.chr7: G>T c G>T p.s4050x Nonsense R100 MLL3 CCDS g.chr7: insTT c.11908_11909 ins TT fs Indel R149 MLL3 CCDS g.chr7: C>T c.9934 C>T p.q3312x Nonsense Y023 MLL3 CCDS g.chr7: G>A c G>A p.w4360x Nonsense Y057 MLL3 CCDS g.chr7: A>T c.411 A>T p.q147h Missense Y091 MLL3 CCDS g.chr7: T>C c T>C p.v4527a Missense B149 MLL3 CCDS g.chr7: C>A IVS2+2 C>A Splice site Splice site U027 MLL3 CCDS g.chr7: G>C c G>C p.l4331l Synonymous W012 NDC80 CCDS g.chr18: delA c.759 dela fs Deletion W039 NDC80 CCDS g.chr18: G>A c.859 G>A p.e287k Missense W012 PEG3 CCDS g.chr19: A>G c.2536 A>C p.s846r Missense W039 PEG3 CCDS g.chr19: G>A c.2947 G>A p.d983n Missense T157 PEG3 CCDS g.chr19: C>T c.2012 C>T p.s671f Missense A142 PEG3 CCDS g.chr19: C>T c.1215 C>T ph405h Synonymous 13
14 Supplementary Table 5 continued. Somatic mutations in TP53, KRAS, SMAD4, MLL3, GNAS, RNF43, ROBO2, RADIL, PEG3, NDC80, CDKN2A, PTEN, LAMA2, XIRP2 and PCDHA13 in 54 O. viverrini-associated CCA tumors Sample Gene Symbol CCDS ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type T026 PCDHA13 CCDS g.chr5: C>T c.1985c>t p.t662m Missense U044 PCDHA13 CCDS g.chr5: C>T c.1301c>t p.s434l Missense R104 PTEN CCDS g.chr10: A>G c.464a>g p.y155c Missense A159 PTEN CCDS g.chr10: G>A c.509 G>A P.S170N Missense W040 RADIL CCDS g.chr7: G>A c.386 G>A p.r129q Missense W039 RADIL CCDS g.chr7: C>T IVS6+4 C>T Splice site Splice site A074 RNF43 CCDS g.chr17: C>G c.611 C>G p.t204r Missense A159 RNF43 CCDS g.chr17: C>T c.337 C>T p.r113x Nonsense R104 RNF43 CCDS g.chr17: A>G c.355 T>C p.c119r Missense U044 RNF43 CCDS g.chr17: A>T c.500 A>T p.n167i Missense W12 RNF43 CCDS g.chr17: C>T c.2167 C>T p.q723x Nonsense W039 ROBO2 CCDS g.chr3: insC c.3234 insc fs Insertion T026 ROBO2 CCDS g.chr3: C>T c.1585 C> T p.q529x Nonsense A159 ROBO2 CCDS g.chr3: T>A c.3999 T>A p.s1322r Missense Y020 ROBO2 CCDS g.chr3: G>T c.712 G>T p.e238x Nonsense T003 ROBO2 CCDS g.chr3: C>T c.2039 C>T p.a680v Missense A039 SMAD4 CCDS g.chr18: T>C c633 T>C p.t211t Synonymous A105 SMAD4 CCDS g.chr18: delga c.90 delga fs Deletion A159 SMAD4 CCDS g.chr18: G>A c.988 G>A p.e330k Missense A035 SMAD4 CCDS g.chr18: A>G c.1659 A>G p.x553w Missense B070 SMAD4 CCDS g.chr18: C>T c.346 C>T p.q116x Nonsense B099 SMAD4 CCDS g.chr18: T>C c.428 A>C p.k428t Missense B149 SMAD4 CCDS g.chr18: G>A c.404 G>A p.r135q Missense B149 SMAD4 CCDS g.chr18: C>T c.1081 C>T p.r361c Missense R104 SMAD4 CCDS g.chr18: delagta c.1447 delagta fs Deletion U044 SMAD4 CCDS g.chr18: C>T c.547 C>T p.q183x Nonsense W012 SMAD4 CCDS g.chr18: G>T c.394 C>G p.h132d Missense A042 TP53 CCDS g.chr17: delt c.582 delt fs Deletion A105 TP53 CCDS g.chr17: T>G c.981 T>G p.y327x Nonsense A107 TP53 CCDS g.chr17: A>G c.578 A>G p.h193r Missense A119 TP53 CCDS g.chr17: C>T c.844 C>T p.r282w Missense A120 TP53 CCDS g.chr17: G>A c.818 G>A p.r273h Missense A142 TP53 CCDS g.chr17: G>A c.673 G>A p.v225i Missense A159 TP53 CCDS g.chr17: C>T c.916 C>T p.r306x Nonsense A162 TP53 CCDS g.chr17: A>G c.395 A>G p.k132r Missense B032 TP53 CCDS g.chr17: C>G c.832 C>G p.p278s Missense 14
15 Supplementary Table 5 continued. Somatic mutations in TP53, KRAS, SMAD4, MLL3, GNAS, RNF43, ROBO2, RADIL, PEG3, NDC80, CDKN2A, PTEN, LAMA2, XIRP2 and PCDHA13 in 54 O. viverrini-associated CCA tumors Sample Gene Symbol CCDS ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type B070 TP53 CCDS g.chr17: C>T c.707 C>T p.y236c Missense B083 TP53 CCDS g.chr17: T>C c.403 T>C p.c135r Missense B099 TP53 CCDS g.chr17: G>T & inst c.686 G>T & fs Insertion ins T R134 TP53 CCDS g.chr17: G>T c.1027 G>T p.e343x Nonsense R149 TP53 CCDS g.chr17: G>A c. 853 G>A p.e285k Missense T157 TP53 CCDS g.chr17: G>A c.375 G>A p.t125t Synonymous T026 TP53 CCDS g.chr17: insG c.216 insg fs Insertion U044 TP53 CCDS g.chr17: G>A c.310 C>T p.q104x Nonsense W012 TP53 CCDS g.chr17: G>A c.742 C>T p.r248w Missense W040 TP53 CCDS g.chr17: insT c.716 ins T fs Insertion Y002 TP53 CCDS g.chr17: C>T c.586 C>T p.r196x Nonsense Y008 TP53 CCDS g.chr17: G>A c.644 G>A p.s215n Missense Y020 TP53 CCDS g.chr17: T>G c.489 T>G p.y163x Nonsense Y035 TP53 CCDS g.chr17: C>T c.817 C>T p.r273c Missense Y072 TP53 CCDS g. chr17: del c. del 750bp fs Deletion Y149 TP53 CCDS g.chr17: C>T c.817 C>T p.r273h Missense B085 XIRP2 CCDS g.chr2: inst c.9704 inst fs Insertion T026 XIRP2 CCDS g.chr2: C>T c.7898c>t p.s2633l Missense T160 XIRP2 CCDS g.chr2: C>T c.218 C>T p.s73l Missense Bold = Somatic mutation identified in discovery screening. 15
16 Supplementary Table 6. Genome MuSiC analysis of mutated genes in O. viverrini-associated CCA a) Ranking of genes according to probabilities that they harbor more non-synonymous mutations than expected by chance given the background mutation rate of the discovery set. Likelihood Ratio Test Convolution Test Rank Gene SNVs Indels P-value LRT FDR LRT P-value CT FDR CT 1 TP E E E E-06 2 SMAD E E E-05 3 KRAS E E RNF E E NDC E E GNAS E = ROBO = RPL10L = TAPBP = UFSP = PEG = XIRP = PCDHA = MLL = PTEN = CDKN2A LAMA = DNAH = RADIL We used the longest CCDS transcript (or, if no CCDS transcript was available, the longest RefSeq transcript). Rankings were computed by Genome MuSiC ( and genes are sorted by the Convolution Test FDR and then the Likelihood Ratio Test FDR. The 15 genes validated in this study (Table 1) are highlighted in orange. 16
17 b) Ranking of genes according to probabilities that they harbor more non-synonymous mutations than expected by chance given the background mutation rate, based on mutations detected in both the discovery and prevalence sets. Likelihood Ratio Test Convolution Test Rank Gene SNVs Indels P FDR P FDR 1 KRAS SMAD TP GNAS E E E E-11 5 MLL E E E E-11 6 RNF E E E E-10 7 ROBO E E E E-09 8 CDKN2A E E E E-07 9 PEG E E E E XIRP E E E E NDC E E E E PTEN E E E PCDHA LAMA RADIL c) Ranking of genes according to probabilities that they harbor more non-synonymous mutations than expected by chance given the background mutation rate, based on mutations detected in the prevalence set only. Likelihood Ratio Test Convolution Test Rank Gene SNVs Indels P-value FDR P-value FDR 1 TP KRAS E E-18 3 SMAD E E E E-14 4 GNAS E E E E-06 5 MLL E E E E-06 6 CDKN2A E E E E-05 7 ROBO E E-05 8 RNF PTEN PEG XIRP LAMA NDC PCDHA RADIL
18 Supplementary Table 7: TP53 mutations in O. viverrini-associated CCA Sample Gene Symbol CCDS ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type A042 TP53 CCDS g.chr17: delt c.582 delt fs (194) Deletion Y072 TP53 CCDS B099 TP53 CCDS g. chr17: del g.chr17: G>T & inst c. del 750bp fs (250) Deletion c.686 G>T & ins T fs (228) Insertion P53 gene structureeffected domain Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) T026 TP53 CCDS g.chr17: insG c.216 insg fs (72) Insertion Yes W040 TP53 CCDS g.chr17: inst c.716 ins T fs (238) Insertion Core domain/specific DNA binding domain ( ) A105 TP53 CCDS g.chr17: T>G c.981 T>G p.y327x Nonsense Tetramerization ( ) Yes A159 TP53 CCDS g.chr17: C>T c.916 C>T p.r306x Nonsense Yes R134 TP53 CCDS g.chr17: G>T c.1027 G>T p.e343x Nonsense Tetramerization ( ) Yes U044 TP53 CCDS g.chr17: G>A c.310 C>T p.q104x Nonsense Yes COSMIC Yes Yes Yes Yes Y002 TP53 CCDS g.chr17: C>T c.586 C>T p.r196x Nonsense Y020 TP53 CCDS g.chr17: T>G c.489 T>G p.y163x Nonsense A107 TP53 CCDS g.chr17: A>G c.578 A>G p.h193r Missense A119 TP53 CCDS g.chr17: C>T c.844 C>T p.r282w* Missense A120 TP53 CCDS g.chr17: G>A c.818 G>A p.r273h* Missense A142 TP53 CCDS g.chr17: G>A c.673 G>A p.v225i Missense A162 TP53 CCDS g.chr17: A>G c.395 A>G p.k132r Missense B032 TP53 CCDS g.chr17: C>G c.832 C>G p.p278s* Missense B070 TP53 CCDS g.chr17: C>T c.707 C>T p.y236c Missense B083 TP53 CCDS g.chr17: T>C c.403 T>C p.c135r Missense R149 TP53 CCDS g.chr17: G>A c. 853 G>A p.e285k Missense W012 TP53 CCDS g.chr17: G>A c.742 C>T p.r248w* Missense Y008 TP53 CCDS g.chr17: G>A c.644 G>A p.s215n Missense Y035 TP53 CCDS g.chr17: C>T c.817 C>T p.r273c* Missense Y149 TP53 CCDS g.chr17: C>T c.817 C>T p.r273h* Missense T157 TP53 CCDS g.chr17: G>A c.375 G>A p.t125t Synonymous Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Core domain/specific DNA binding domain ( ) Yes Yes Yes Yes Yes Yes Yes Yes Yes Yes Yes Yes Yes Yes Yes Yes Transactivation domain (1-50), Core domain/specific DNA binding domain ( ), Tetramerization ( ), Regulatory domain ( ) Reference: Monique G. C. T. van Oijen and Pieter J. Slootweg (2000) 1 * Represent reported TP53 hot spot mutation 18
19 Supplementary Table 8. Characteristics of O. viverrini-associated CCA patient with both SMAD4 and PTEN mutations Sample Age Sex Histology Stage SMAD4 mutation PTEN mutation R104 F 64 WD tubular adenocarcinoma III Frameshift Missense A159 M 55 Papillary carcinoma III Missense Missense 19
20 Supplementary Table 9: Overall survival analysis in O. viverrini-associated CCA a) Influencing mutation factors on overall survival in O. viverrini-associated CCA (n=53) Independent Variable Patient Number Univariate analysis* (P-value) TP53 mutation Absent 29 Present KRAS mutation Absent 44 Present SMAD4 mutation Absent 44 Present MLL3 mutation Absent 45 Present ROBO2 mutation Absent 48 Present GNAS mutation Absent 48 Present RNF43 mutation Absent 48 Present 5 <0.001 * Significant at a Bonferroni corrected alpha of (0.05 / 7). 20
21 b) Multivariate Cox Proportional Hazard Model for prognostic factors in O. viverriniassociated CCA patients Independent Variable Age (<56 vs >56) Sex (Female vs Male) Histology type (Papillary vs Non papillary type) Univariate Multivariate Hazard ratio analysis analysis p value* p value** (95% CI) TNM stage (I /II /III vs IIIA,B,C /IVA,B) RNF43 mutation (Absent vs Present) <0.001 < ( ) Independent Variable Univariate analysis Multivariate analysis Age (<56 vs >56) Sex (Female vs Male) Histology type (Papillary vs Non papillary type) Hazard ratio p value* p value** (95% CI) TNM stage (I /II /III vs IIIA,B,C /IVA,B) KRAS mutation (Absent vs Present) ( ) *P value < 0.05 from log rank test **P value < 0.05 from Multivariate Cox proportional hazard model analysis NB: TNM stage did not show prognostic significance because only 3 cases are of stage I and II, while the rest of the cases are of late stage disease (III and IV). 21
22 c) Multivariate Cox Proportional Hazard Model for prognostic factors in O. viverriniassociated CCA patients (Excluding 2 cases harboring both KRAS and RNF43 mutations) Univariate Multivariate Hazard ratio Independent Variable analysis analysis p value* p value** (95% CI) Age (<56 vs >56) Sex (Female vs Male) Histology type (Papillary vs Non papillary type) TNM stage (I /II /III vs IIIA,B,C /IVA,B) RNF43 mutation (Absent vs Present) ( ) Univariate Multivariate Hazard ratio Independent Variable analysis analysis p value* p value** (95% CI) Age (<56 vs >56) Sex (Female vs Male) Histology type (Papillary vs Non papillary type) TNM stage (I /II /III vs IIIA,B,C /IVA,B) KRAS mutation (Absent vs Present) *P value < 0.05 from log rank test **P value < 0.05 from Multivariate Cox proportional hazard model analysis NB: TNM stage did not show prognostic significance because only 3 cases are of stage I and II, while the rest of the cases are of late stage disease (III and IV). 22
23 d) Multivariate Cox Proportional Hazard Model for prognostic factors including RNF43 and KRAS mutations as independent variable in O. viverrini-associated CCA patients Independent Variable Age (<56 vs >56) Sex (Female vs Male) Histology type (Papillary vs Non papillary type) Univariate Multivariate Hazard ratio analysis analysis p value* p value** (95% CI) TNM stage (I /II /III vs IIIA,B,C /IVA,B) RNF43 mutation (Absent vs Present) ( ) KRAS mutation (Absent vs Present) ( ) *P value < 0.05 from log rank test **P value < 0.05 from Multivariate Cox proportional hazard model analysis NB: TNM stage did not show prognostic significance because only 3 cases are of stage I and II, while the rest of the cases are of late stage disease (III and IV). 23
24 Supplementary Table 10. Mutation spectra in O. viverrini-associated Cholangiocarcinoma (CCA), Pancreatic cancer (PDAC) and Hepatocellular carcinoma (HCC) Mutation Category O. viverrini-associated Cholangiocarcinoma (CCA) Pancreatic ductal adenocarcinoma (PDAC)* HCV-associated Hepatocellular carcinoma (HCC) # CpG -> TpG TpC -> Tp* (not in TpCpG -> TpTpG) C:G->T:A (other than above) C:G->G:C (other than above) C:G->A:T (other than above) T:A->A:T T:A->G:C T:A->C:G Total P-values (ChiSq) $ E-16 * Data from Jones et al., Science 321, , 2008 # Data from Li et al., Nature Genetics 43, , 2011 $ P values for pairwise comparison of mutation spectrum of CCA with PDAC or HCC NB: Nine mutations in UTRs were excluded from PDAC counts. CCA mutations in this analysis include synonymous and non-synonymous coding mutations and mutations at canonical splice sites and in UTRs (Sanger validated and non-validated targets) for O. viverrini-associated CCA. 24
25 Supplementary Table 11. Chromosomal changes in Cholangiocarcinoma, Pancreatic cancer and Hepatocellular carcinoma Chromosome Chr1 Cholangiocarcinoma (CCA) 2,3,4 Pancreatic cancer (PDAC) 5,6 Hepatocellularcarcinoma (HCC) 7,8 +1q +1q +1q -1p -1p -1p Chr2 2 Chr3 +3q 3-3p -3p Chr4-4q Chr5 +5p +5 q and p Chr6-6q - 6 q and p Chr7 +7q and p +7p Chr8 +6p +8q +8q +8q -8p -8p Chr9-9p -9p Chr10 Chr11 Chr q and p Chr13-13q -13q -13q Chr14-14q Chr15 +14q Chr16-16q Chr17-17p -14q Chr18-18q -18q Chr19-6q -14q +17q +17q -17p -17p Chr20 +20q +20q Chr21 Chr22 Chr X Chr Y +Xq Yellow: Common between CCA and PDAC Green: Common between CCA and HCC Gray: Common to all 3 cancers 25
26 Supplementary Table 12. Primers used for PCR amplification and Sanger sequencing Gene Symbol Transcript IDs Coding Exon Number Genomic Region of Interest Forward primer sequence Reverse primer sequence Additional primer sequence for sequencing CDKN2A ENST Exon1 Chr9: _ GGTCCCTCCAGAGGATTTGAG GATTACAAACCCCTTCTGAAAAC CDKN2A ENST Exon2 Chr9: _ GAATGGATAGAGAACTCAAGAAGG GTGGTGGTATGCTTTGGGAAG CDKN2A ENST Exon3 Chr9: _ CCATTGCGAGAACTTTATCCATAAG CTCCAGAAAACTCCAACACAGTG GNAS ENST Exon8 and 9 Chr20: _ CAGCTTCCTGGACAAGATCGACGTG CTTGAAGAGGTTCAGAGCCTCCTGC KRAS ENST Exon2 Chr12: _ ACACGTCTGCAGTCAACTGG CATATAACTTGAAACCCAAGGTAC KRAS ENST Exon3 Chr12: _ CCTGACTATTGATGATGTTGAG CAAGTTACTCCACTGCTCTAATC MLL3 ENST Exon1 chr7: _ GAAATGCGAGAGGCTGAGC GCTCCCTCGCACTCAACTTAC GGATCCATGTGTCTTTCCTGG (F) MLL3 ENST Exon2 chr7: _ GGAGAAAATGACTGTAAACTCTG * CAAACACTAGATGGTTCATAGGCCAG MLL3 ENST Exon3 chr7: _ CGTGCATGAACTTTTCTTGTG CTAAACACAGAGGTATCAGCTTC MLL3 ENST Exon4 chr7: _ CTTATGGGTAATCACTGAATAGACTG ATTGGGTTAATGGTGAGTCAGAG MLL3 ENST Exon5 chr7: _ ACTTCCCTGTGGGTTTGGAGACTC GCATCCAGTCCGGTGGTGGTACTG CAAGGCAGATGGGAAACTTAGAGTTC [R] MLL3 ENST Exon6 chr7: _ GAAGGGCCACTTTACTGCTATCAC * CCAGATTCAAGCAAATTCTCCTG-PCR ACCATACCTGGCCTAGAATTTCTC [R] MLL3 ENST Exon7 chr7: _ CAGCGATGTGCATTTTGTAAGCACC CGATCCCTGGCAACGATCCCTCAG MLL3 ENST Exon8 chr7: _ GTCTGAAAGGTTCATGATGCAAATCC TGATGAAGTCACTGAATGAATATAGC MLL3 ENST Exon9 chr7: _ GATCAGAGGCTATTCATCTTGC GAGGGTGATGACTATGTTCATTATC CAGGGTTACCTCTGTTACTATCTGC (F) MLL3 ENST Exon10 chr7: _ GCAACAGAGTAAGAGTCCATCTC * CCTATTAGTGCTCATTTCTGATGTGG CCATCTATAACAGCAATCAGTAC [R] MLL3 ENST Exon11 chr7: _ CAACTCCTCTAACCTCTCTCCTCTC * CCATCAGATTCAGAGATGACCAAG CAGCTTTTCAGCAATGCCTTC (F) MLL3 ENST Exon12 chr7: _ CAGGCATGAACTACCACGGTTGG GATGGTAGCTGAACTACAAATACACG MLL3 ENST Exon13 chr7: _ CTTACTAATAAGTGGTACAATGTTTCATGC CCTTTCAAGAGAAACTAACCTTC MLL3 ENST Exon14-1 GTGGCTGATTCTAATCCATAAATCC GAGTCATTTCAGAGTCCATCAATCC chr7: _ MLL3 ENST Exon14-2 CAGTTGTTAGAGGAACCTGAAACAG CATTACACTGAAGTTCTATCTGAAGAC MLL3 ENST Exon15 chr7: _ GAGTCAATGTTGATGTCCTATAAACC GTGTTTCTTGGAAGCTCAATCTAG MLL3 ENST Exon16 chr7: _ CCAGGTCTCTTAATACTTATCTAAGG GGACAGTAAAATAAACATTTTGACC MLL3 ENST Exon17 chr7: _ GTCACGACTCCATTTTACGGTAG GTAATCTTCCTTCTGCTCCTTGG MLL3 ENST Exon18 chr7: _ CCTCATTGAATTCCTCCTTGCAC GTGAGCATAGAGGAGAGAGAATGC MLL3 ENST Exon19 chr7: _ GCTTCTAGGTACATAAGCACCAT GCAACTGACCCAGGAAGACTCC MLL3 ENST Exon20 chr7: _ GTTATCACACCTACTGCCTAGACC * CAAAACAGGAAATCGCTTCTGTAAG CAGAGCTTTCTCATACCACTTTGG (F) MLL3 ENST Exon21 chr7: _ CCTCTCTAATAGTTGTTCTCATTCC CAAGTTGTGATAAGTCATGTATAACC MLL3 ENST Exon22 chr7: _ TCCTCCATGACTTGAGTATTTCC * GCTAACTAGGCCGTGTTTTCTGC GTGGAAATGATTACCAACATTACAG [R] MLL3 ENST Exon23 chr7: _ CATGCAACCTTTCTGCCTGTAATCC ATGGCAAGGGTCAGTCACATCAG MLL3 ENST Exon24 chr7: _ GGAACACTGCACTAAGCAGCAC GGTTAACATTTGGGACAAGGCATTC MLL3 ENST Exon25 chr7: _ CAGGTTATTGATTTAGATTAGCACAG * GTAACAATAAGAGGCAATAAGGCTC GCAGTTTTATCCTGTGTACTGTGG (F) MLL3 ENST Exon26 chr7: _ AGTTGCCATTTCGTCATACTCATG ATGGCATTTATGATGACTAAATGAC 26
27 Supplementary Table 12 continued. Primers used for PCR amplification and Sanger sequencing Gene Symbol Transcript IDs Coding Exon Number Genomic Region of Interest Forward primer sequence Reverse primer sequence Additional primer sequence for sequencing MLL3 ENST Exon27 chr7: CTAGAAACCTTTGGTATAATTGTGTGG GCTACATCTTATACACCTGTCCAC MLL3 ENST Exon28 chr7: _ CATTAGTCACAAGACAAAATCGTCTG ATGCTGAGTCCTTTAACAAAGC MLL3 ENST Exon29 and30 chr7: _ CTGAGCGACTCTCCAAATTGATATG GCTGAGTGTTCGCAGGACTAAG MLL3 ENST Exon31 chr7: _ TATTCTAGGAGCAATTCTTGGA * CTCCCTATCAGTTTGTATACGTGG CCACTGGACATAAGGATAATTACAG [R] MLL3 ENST Exon32 chr7: _ AGTTGGTCCTCTCTATGCATGG GCAAAAGCCACAATAAACGCAA MLL3 ENST Exon33 chr7: _ GTTGCGTTTATTGTGGCTTTTGC * GAATCACATAACACGGATTTCTGC GGAATCTTCATGTTGTGGGTCATC [R] MLL3 ENST Exon34 chr7: _ CAGTGTGCCACGATCTTTGTTC CCAACTGTACCTCTGCTGAAGAG MLL3 ENST Exon35 chr7: _ GAGGTGCCGTAAGTATTTCAGC CTCATCATGGTATGTTCCATTTGTG MLL3 ENST Exon36-1 CTAATATGGGAGCAATCTGGTAGC GATGGCCTATTTGCTGTTGTCTC MLL3 ENST Exon36-2 CAGGATAGTCTTTCTCAGGCTCA ATAGGTCTTGGTGTCAAAGCAGG MLL3 ENST Exon36-3 chr7: _ CCTGTAGTTTCAGAACAAACTGC GAAATTCCAGGTCTTGGTGTTCC MLL3 ENST Exon36-4 CCAACATCTCAGGACCCATACTC CAGGAAGTTGGGAGACACCAGAG MLL3 ENST Exon36-5 GTTGGTAAGGCCACCTGATACATG CCAGACAACTATCACAAAACAAAGC MLL3 ENST Exon37 chr7: _ CTTTGAAACATTACCATAGCCAC GCAGTAACAGCTTCCTGATTAATAC MLL3 ENST Exon38-1 GCTCAGATATGGTGTTCATTATAGG * CCTCTAGACCATCAGAAGGCTGG GACAAAGGAGCTTCTGAAAATTCAC [R] MLL3 ENST Exon38-2 CTGAGACATGGAAACTTCATTCC GTGAGCAAGGAGTGTCAACATTATC CCTAATCAGCTACCTGTGCACC (F) chr7: _ MLL3 ENST Exon38-3 CAGAACTTGACATGGGAGATAAG CTGTTGATTGACCTGGAATGAGC MLL3 ENST Exon38-4 GGATCAACTCCAGTTCTCTCAAG CAAACTCCTCAAGGAGGAATAG MLL3 ENST Exon39 chr7: _ CCAGGATAAGGTTGCCATCTCAG GATCATATCGAGCTGATGCCATGA MLL3 ENST Exon40 chr7: _ GTACATGAAAGGTAGGGCTTTAGTG GATAAGGCTACTATTTAGGTTGGTG MLL3 ENST Exon41 chr7: _ GTTGCCACCACTAAATTCACTCTAC * GTGTCCTTGTCACTTAGGAAGACAG CAGTGATGTTTAATAAGACAGGAGG [R] MLL3 ENST Exon42 chr7: _ CAGTATCTCAAGAATGTGCATTCTC ACTAAGAGGTCTCAGAGCTGTCAC MLL3 ENST Exon43-1 GTGACTTTTGCAGTTACCCAAGC GTCGGAACTGTAGAAGGGAATCTG MLL3 ENST Exon43-2 CAAATGCAAATCCACAGAGTGG GATCCCGGATAACTGTGTCCATG chr7: _ MLL3 ENST Exon43-3 GACTAATGAGCGAAGGCAGGTAG GACTCTGTCTCAGCCTTTTCCAG MLL3 ENST Exon43-4 CTCCAGGCATCTCAGAAACTACC GGGTTCCAGAGAGGTAGAAATGAC MLL3 ENST Exon44 chr7: _ GCATTGACACCACTCTATCTTCC CTAGAACTCATGCCTCAAGTGATC MLL3 ENST Exon45 and 46 chr7: _ GGTGTGCAAGATACATCTTGATTAC * CCTGAAGATTATTCACCAAATTGTAC CATGAAGGTATGATTATAGCCTCC (F) MLL3 ENST Exon47 and 48 chr7: _ CATCGTATAGGTGCTGCCCAGAG GCTCTGCTCTCTGACCCTTTATCC MLL3 ENST Exon49 chr7: _ GGAATCCCTTTATGAGGATTTCTG GGTAAGTTCACCAAGGACTATACG MLL3 ENST Exon50 chr7: _ GATAATCGACTCTGTAACCATTGC * CCTTTAACAGCCTGTTGTAGCTC CAGGAACATGTGTCCATCACAC [R] MLL3 ENST Exon51 chr7: _ GAGTGAACCGTGGACCACATAG AGAGTAGCACGACTCAAGGCTCC 27
28 Supplementary Table 12 continued. Primers used for PCR amplification and Sanger sequencing Gene Symbol Transcript IDs Coding Exon Number Genomic Region of Interest Forward primer sequence Reverse primer sequence MLL3 ENST Exon53 chr7: _ GCTGCCATTTAATAAACAGGTTC CCAATGCCTTATTCCTCTAAGAG MLL3 ENST Exon54 chr7: _ CCTAAGTTTGGCTTTGAAACTG GTTCTCTTATGTAGTTGGGCACATAC MLL3 ENST Exon55 chr7: _ CTGCTAGGCAGCTTTCCATGTATG CCAAGGGACAGGATTTGATACG MLL3 ENST Exon56 chr7: _ GTAGGAATAAGCTAGTGATGGCATAG GGAATCAGTAGGTTGTGTGTGAAGC Additional primer sequence for sequencing MLL3 ENST Exon57 and 58 chr7: _ CGTGCCATGTAAATGTCATCTGTG * CATCTGGCCTATTATGGACAGAG GCTGAATGCAGCATGACAGATTTC [R] MLL3 ENST Exon59 chr7: _ GAACCTCAATAAATCAGGTCTTCC CTGCTTTAACCTAAAGGACTGAGG NDC80 ENST Exon2 chr18: _ CCTTGCAAATGAGAACTTCAGTG CAACACCTAGCAGAGTATCATGC NDC80 ENST Exon3 chr18: _ CTTATAGTACCATGTTTAGCCCATCC * CTCCACCGTGTCTGAAAGCTAGG GTGTTAGCTACACTGTCTACAGCAG [R] NDC80 ENST Exon4 and 5 chr18: _ CGTTATTTGCTCCTTTTCTCTGC TCCTACATATATTGCCCACAGTGG NDC80 ENST Exon6 chr18: _ GTATAGGTCAAGTTCTGCAGCTTCG CACTGTATTCTCCAAACTTCTTGC NDC80 ENST Exon7 chr18: _ GAGTCCATCCTTTCTAGATCCTGAAG CTATGACGTTTGGCAGGTTAAGTG NDC80 ENST Exon8 chr18: _ ATAAGCTTCCCTGTTTGTTTGACC ACTTCTCTCCACCAAAGAAACCAC NDC80 ENST Exon9 chr18: _ GAAGTACGGTGGAGTAATGGAAAG ACAAGGCTCCACAGATACCAAAC NDC80 ENST Exon10 chr18: _ GTGAAGCACTCAAACTGTTGTTG CAAACTTGTGATCATTTGTGACAGC NDC80 ENST Exon11 chr18: _ CCATTGCAAGCTAATAGGGAAATAAG * GGACTCATTTCCTCTTCTACAGAG CTGAATTGACATAAAGGGATGAGTG [R] NDC80 ENST Exon12 chr18: _ GGACTCTTATTCCAAGCTGTCAG GCAACTTGACAGTAAGGGTTAGG NDC80 ENST Exon13 chr18: _ GCAATGAATTTCCAGGATAGATGAG TGTCGAGAGCATACCCAATTCTC NDC80 ENST Exon14 chr18: _ GGACAAATGATAAGTGAGAGTCC GCAGAGAAAACTAACCCAAGATAC NDC80 ENST Exon15 chr18: _ CATAGTTGGAATTACAGGCACACACC GTATCTGTTGATAGGGAAAGTTCTCG NDC80 ENST Exon16 chr18: _ GTTACAACCCTTCGCTCCAAAGC GTTGCCACATCACTAGACAAGATTC NDC80 ENST Exon17 chr18: _ CCTCTTGCATAGTTGGAATTACAGG CAACCTGTATTTTGCCGACTTCTGG PEG3 ENST Exon4 chr19: _ CATTGTCGATGCCTCTGGTAAGG CAGCTCAATATCCCACAGTACACAGG PEG3 ENST Exon5 chr19: _ GTATGCCAGGAGTGGTTGGAAGC CTCAGATCTCTGAGTGAAGCTGACC PEG3 ENST Exon6 chr19: _ GAGCTTTGGAACTTGCTTGGAAG CTGGTGGTACCGAGTGGAACTTC PEG3 ENST Exon7 chr19: _ CTGCCACAGAATGCAGAGAAGAG GCTACTTGTCACCTAAGGGATGG PEG3 ENST Exon8 chr19: _ CGTAGGTCCCTAGTAACACTTGAC GTATCTTTACCCAAAGCCAGAGG PEG3 ENST Exon9 chr19: _ GTCTTCTGACATCCAAGGAATCAG CCAAATGTAAATACTCCCTAGTCC PEG3 ENST Exon10-1 CAACCAGTGAGATGCTATCAAGG CAATTGGCTGTGACTCGGTAAAG PEG3 ENST Exon10-2 CGCTATCATTTTGACACAGATGG TCACGTTCACGTTCATGTTCAC PEG3 ENST Exon10-3 GGCAAAGACAAATTCTACGAGTG TCGTCCTCATCACTTTCAAGAGG PEG3 ENST Exon10-4 AAGCTCAGAGCTCAGTGAGCATC TCATAGTTGCGATTCTTACTGCC chr19: _ PEG3 ENST Exon10-5 CCAAACCTCCAAGAAGTCACAATG CATGTCTGAGCCTTGAATGACAG PEG3 ENST Exon10-6 GCTACCAGCGAAGACCTCAACAC CTTCCCTATGAAGTCTCATATGCTC PEG3 ENST Exon10-7 GAATCGTGCTGCAGAGAGGAATC GCTCTGCAGCCTCTCCATCTGG PEG3 ENST Exon10-8 CACAAGTAGTTCTGAGGATTCAGG CAGTGTGGGTATTCTGGTGTCTG 28
29 Supplementary Table 12 continued. Primers used for PCR amplification and Sanger sequencing Gene Symbol Transcript IDs Coding Exon Number Genomic Region of Interest Forward primer sequence Reverse primer sequence Additional primer sequence for sequencing PEG3 ENST Exon10-9 chr19: _ CCATACTATGACTGCCATGAATGC AGACTGTGGAATCTGCACATTCG PTEN ENST Exon1 chr10: _ CGTTCGGAGGATTATTCGTCTTC CTAGCAACCTGACCAGGGTTAAATG PTEN ENST Exon2 chr10: _ CCTGAATACTGTCCATGTGGAAG * GAAGTCCATTAGGTACGGTAAGCC CCTTTTATTACTCCAGCTATAGTGG (F) PTEN ENST Exon3 chr10: _ CCATAGAAGGGGTATTTGTTGG GCTCTTGGACTTCTTGACTTAATCG PTEN ENST Exon4 chr10: _ GGAAGCACCTGAATTTACAGTACTC * CCTAATTCTTAGTCTATGCTGCAC GCAATACTTTTTCCTAAAACACAACAG (F) PTEN ENST Exon5 chr10: _ CCAGTCCGTATAGCGTAAATTCC GTGAGGTGATGAATATGTTAAGTAG CCACAGTTGCACAATATCCTTTTG (F) CTCAGATCCAGGAAGAGGAAAGG [R] PTEN ENST Exon6 chr10: _ GTTCTTAAATGGCTACGACCCAG CTAGTCAGAATTGGGCTGTATTTGG CCAATGAGTTGAACAACAAAGC [R] PTEN ENST Exon7 chr10: _ CCACTAGAAGTCTAATTTTGGGAC GTCTCACCAATGCCAGAGTAAGC GATCAAGATTGCAGATACAGAATCC (F) PTEN ENST Exon8 chr10: _ GCAAATGTTTAACATAGGTGACAG CAGCAAACAACGAAGAATTAGG CTGCTACGTAAACACTGCTTCG [R] PTEN ENST Exon9 chr10: _ GCTAAGGCACGAGAATTGCTTG GGTCAGGAAAAGAGAATTGTTCC CAACAGAGCAAGACTCTTGTCTC (F) PCDHA13 ENST Exon 1-1 AAATTGGGCCTTGAGAGACAG CTTGCTTTCAGGGAATATGGG PCDHA13 ENST Exon 1-2 GGAGTGTACCATGCTGTCTTCC GATCCAGTCGGTAGGTCAATGC PCDHA13 ENST Exon 1-3 GTGAAGGTGAGGGACATTAACGAC CATATACCACTGCAGGCCATAC PCDHA13 ENST Exon 1-4 TGCAGATATTGGAGTAAACTCGG GGAGTGACAAAGAAGTGATGGTAAC PCDHA13 ENST Exon 1-5 chr5: _ TTAGAAGGCCTGTATGGCCTGC AGCAGACACCGTGAAGATGTG PCDHA13 ENST Exon 1-6 GCCCTAATCAGTGTGTCCGATC CAGCTCGCTCACTGTGCCTC PCDHA13 ENST Exon 1-7 CACATCTTCACGGTGTCTGCTC GTGCTGATCTCGCCAGTGTACAG PCDHA13 ENST Exon 1-8 AGGCACAGTGAGCGAGCTGATG CAGAACCCAGACAAGGAGGAAG PCDHA13 ENST Exon 1-9 CTGGTGGATGTCAATGTTTACTTG GTCCACAGCAGAGTTGTTAAATTCG PCDHA13 ENST Exon 2 chr5: _ CTGTGTGAGGACCCTGATGAAC CTGACCATTTGCGATCTTTCAAG PCDHA13 ENST Exon 3 chr5: _ AGCAGGCAATAAGTAAGTCAGC TCAACTCCTGAAGAGCATTAGG PCDHA13 ENST Exon 4 chr5: _ CTCAGACTTGAACAAACAACTCGTG GCAGATTTCTGAGGTCTCTAAGAGG RADIL ENST Exon2 chr7: _ TGCTCTTGGTGGTCACGATGGTTG GGAAGGCTTCAAACATCCAGGAC RADIL ENST Exon3 chr7: _ GTTTCACTGTGTTGGCCAGGCTG AATGTGGATGACTCCACATGTGC RADIL ENST Exon4-1 TGTCTCCTGGGTCTCTCGTGTG CCTTGAATAGCAGCAGGTAGTAGAG RADIL ENST Exon4-2 chr7: _ GCACATCTCCGTCAACTTCTCC GCTCAGCAGCACAGCACCGTG RADIL ENST Exon4-3 TCTCCGTCAACTTCTCCGAGGTG CAGCTGACACCTTGGGTTCTGG RADIL ENST Exon5 chr7: _ ATAGTCAGCGTCACACAGGATGC ACTCAGAGCTTCCCAGGGTGATG RADIL ENST Exon6 chr7: _ CAGGTTGATGGTTGCAGTTGTTG GCAACTGACACATGGCAGCCTAG RADIL ENST Exon7 chr7: _ TTGAGATGTTGGGTGTCTGGACC TGAAGTGGAGCTTGTCACGGTC RADIL ENST Exon8 chr7: _ ACCTCTGTGCGCCTTCTGAGTTC GGATGCGCTGTGCTGATCTGTC RADIL ENST Exon9 chr7: _ GTCATCGAGCTGTGATGTCCTCAG GGTAGCTTTGAGGTTTCCAGCC 29
30 Supplementary Table 12 continued. Primers used for PCR amplification and Sanger sequencing Gene Symbol Transcript IDs Coding Exon Number Genomic Region of Interest Forward primer sequence Reverse primer sequence Additional primer sequence for sequencing RADIL ENST Exon10 chr7: _ CCAGTAGTGGTTTCTCTGGTTGG * GAGTTTGGAACTTGGGCATGGAC TGAACTGCTGCTGTGACACCGTC (F) RADIL ENST Exon11 chr7: _ AGACACCATCAGAGACCAGTGG ACCTCGGGGCACACTGGCTG GGAATTCAGAAGGGATGAGTCC (F) RADIL ENST Exon12 chr7: _ GGAGTCCTTTGCTGACCTCTG GTCTCAGGTTGCAGGTGTCTGTG RADIL ENST Exon13 chr7: _ CACATTCTGCCTGAACAGCGTC GACAGAGGCAAGAGACAGCAGG RADIL ENST Exon14 and 15 chr7: _ GTCTCTTGCCTCTGTCCTCAAGC GCATGTTATCAACAGGCATGTCC RNF43 ENST Exon2 chr17: _ GACTTGGCTTTCTGAAACGGAAG GAATGCCAATAAGGCAGTATCTACTC RNF43 ENST Exon3 chr17: _ CAGTGGTCACTAGCTTACTAAGAGG CCACTTCTCTCAGACCAGTCATGG RNF43 ENST Exon4 and 5 chr17: _ CTTGCATAGGTAGATGTGAGTGTGC ATGAAGTTGGACTAGAGGGTTTCCAG RNF43 ENST Exon6 chr17: _ CCACTTGCAGGTTCCTCTCCATCC CTGCTGGAAATCAACAAAGACAGACG RNF43 ENST Exon7 chr17: _ GAACTCCTAAGGATGTTCAACTGG CAATAGTGCTCTTTCACTGGAAGG RNF43 ENST Exon8 chr17: _ CTGTTAAGAAAGTTGCCCAACGTC GTGAGGCACTCTGGAGCAGTGTAC RNF43 ENST Exon9-1 GAACAAAGGAGTCCCAGAGCACAG CAAAGTCACTGCTTAGGGAGCTGC RNF43 ENST Exon9-2 AGAGCAGCAGCGCCTGGCAGGAG CTGCTGAGTTGGATCTGGTGACTTGC chr17: _ RNF43 ENST Exon9-3 GTTTCCAGCCATGTCCACTACCAC TCTGGTAGCAGCCTCTTGTCCAGG RNF43 ENST Exon9-4 CAGTACCAGCAGTCTGTTCAACTTG TTGGTTGTCATCTCTGCTGTATCC RNF43 ENST Exon10 chr17: _ CCTTAGCTTTCAATCTAACCACC CACCAGTCCTCTTCCAGTGCTTCTAG ROBO2 ENST Exon1 chr3: _ GATCTTGCGATTTGCTCTTCTTGAC CTACCAACACATTCCACTTACAGCAG ROBO2 ENST Exon2 chr3: _ CAAAGGAGTTGAGATGACTTATGAG TGACCAGACTTCTCTGAAGAAGACC ROBO2 ENST Exon3 chr3: _ GGTGGATGTAGCTATGTTCTCAG GTCTCTTGTAGTCTCATCCAAATG ROBO2 ENST Exon4 chr3: _ GATCAGGAAAGAGAGGACAGCTG ACTGCTTACCATGAGTCATGAGTAAC ROBO2 ENST Exon5 chr3: _ CAGCTGAGGTACAGAGTTAGGTAC * CAAACAGTAAATATGTGGCTTGC CAAACAGTAAATATGTGGCTTGC [R] ROBO2 ENST Exon6 chr3: _ CTAAGTAATTGCACTTTGTGGCTG GATAGAGAGGACACAAAATCTAGG ROBO2 ENST Exon7 chr3: _ CTGTAGTCTCTCTCTGGCACTGAAC GCTATTGCTAGAACATCTGACTCC ROBO2 ENST Exon8 chr3: _ CTTTATCAGTGAAACCCACTGTGTG GTGGTATTCAATCAGGGAGAATTAG ROBO2 ENST Exon9 chr3: _ CAATAAATAGTATGGCCTTAGTATAG CTGTTATGAATGTGTGAGTAACG ROBO2 ENST Exon10 chr3: _ GGTAAGAGTATTATCACAGAGGAAG GTAATGGTGTGTGCAAATTCATGC ROBO2 ENST Exon11 chr3: _ GGAGAAGCAAACATTCACTTACATAC CTCTGTTCTTGAATAAAGTCAGAGAG ROBO2 ENST Exon12 chr3: _ GTGCATGGCACTTTTGTTAGGTG ACACATCTCACCAAGATAAACAGC ROBO2 ENST Exon13 chr3: _ CTTTCATTATGTCCTACCACAGTGG TGAAGTTGTTCACTTTATGGTATGC ROBO2 ENST Exon14 chr3: _ GGTTGGTATCTTAGATACTGTGAGG CATAAGATTTGGTGCAAATGGTTAC ROBO2 ENST Exon15 chr3: _ CTGTCACTGTTGAACACCGTGTG GACCTTGAACTCAGGTTCTAATACC ROBO2 ENST Exon16 chr3: _ CAGAGGTTGGGCTTTAGAAGATG CCATTTGTCAATATGATATGTGTACC ROBO2 ENST Exon17 chr3: _ GGTCAGTGTTCTTAATCACATTTGC CCTTATGGTTATAAAGCGATAGGTC ROBO2 ENST Exon18 chr3: _ CCACTGCTTGTTACTCTCTGTACAATG ATGAAACATCTGGACTTGATTCCTC 30
31 Supplementary Table 12 continued. Primers used for PCR amplification and Sanger sequencing Gene Symbol Transcript IDs Coding Exon Number Genomic Region of Interest Forward primer sequence Reverse primer sequence Additional primer sequence for sequencing ROBO2 ENST Exon19 chr3: _ GTGCCAATGTATCATCAAACCTTG * CTCAAGGTAATGAAGATACAATCG CGTCCACTGTTAATGTATGTATAGC (F) ROBO2 ENST Exon20 chr3: _ CTGTCACTGAGGCTGATAGCTTGG CTCAAGTTACTGGAAACATGCTGG ROBO2 ENST Exon21 chr3: _ CAGCTCTGCTGTTAATGCACTGG GGATTCTCAGTCCTCAATATGTGC ROBO2 ENST Exon22 chr3: _ GATTTACTGTGGTGACACCAACAGC CAGTTACAGTTACAGCTCCAGTTCC ROBO2 ENST Exon23 chr3: _ GTATTCGTAGCCATTGTTTCTGCTG GTGAACTTCTCATGAAAGCCCTAG ROBO2 ENST Exon24 chr3: _ CTACAGAGATGCACCAATATTTGG GCATGGAATGTATTTCTTGAGG ROBO2 ENST Exon25 chr3: _ GGTTCCTATTGTAGGAAATGTTGC CAATGGCCTAAGTTATGACCAAAC ROBO2 ENST Exon26 chr3: _ CACAAACTCATCTATCTGAAGACC * GAAAGAGTTAAGGTGGGTGGTGTA GCACTGTTTAGAGTCATCACTTCC [R] SMAD4 ENST Exon2 chr18: _ CAATTTTCCTTGCAACGTTAGC GCATATTTGTACAGGATGCAC SMAD4 ENST Exon3 chr18: _ GATGTATGACATGGCCAAG CAACTACAATACTCGGTTTTAGC SMAD4 ENST Exon4 chr18: _ GCTACTTCTGAATTGAAATGG CTTGTTAATGTTACTGCCTGC SMAD4 ENST Exon5 chr18: _ GACTTTTGCTGGTAAAGTAGTATGC CCTGTTTACCTATGAAAGATACTAC SMAD4 ENST Exon6 chr18: _ GATAGGCCATGGGTGAGTTACAC CAGTCCTACTTCCAGTCCAGG SMAD4 ENST Exon7 chr18: _ GCAGCAGAATGGATTTACTGG GTTAATGAAACATTAGCAACAGAG SMAD4 ENST Exon8 chr18: _ TAGCACTTGGCAGATAGCAC CATCTGAGAAGTGACCCCAT SMAD4 ENST Exon9 chr18: _ GAAGACATGGAAATTCCTACC GGAGTGCTTACAAATTCTGAG SMAD4 ENST Exon10 chr18: _ GCTCCTGACACATAGTAAGTG CCATTCCTTCCACCCAGATT SMAD4 ENST Exon11 chr18: _ CCAAGCCACCTTTCCTAACTA ATGCAAACAGGGTCATAGGC SMAD4 ENST Exon12 chr18: _ CTTCTTGGCACTTTAGCAGAG CCAGTTTCTGTCTGCTAGGAGC TP53 ENST Exon2 and 3 chr17: _ TGGAAGAGAGAATGTGAAGC GGACTGTAGATGGGTGAAAAG TP53 ENST Exon4 chr17: _ CGTTCTGGTAAGGACAAGG CAAAGGGTGAAGAGGAATC TP53 ENST Exon5 chr17: _ GTAGACGCCAACTCTCTC CTCCACACGCAAATTTCCTTC TP53 ENST Exon6 chr17: _ CATCTACAAGCAGTCACAGC ACCCATTTACTTTGCACATC TP53 ENST Exon7 chr17: _ GCACCTGTAGTCCCAGCTACTCG * TCTGGAGCCTAAGCTCCAGC TGAGAGGTGGATGGGTAGTAG [R] TP53 ENST Exon8 chr17: _ CTTAACCTGTGGCTTCTCC CATCCAGTGGTTTCTTCTTTG TP53 ENST Exon9 chr17: _ CTAAGCGAGGTAAGCAAGC GAGCCATTGTCTTTGAGG TP53 ENST Exon10 chr17: _ TAGGTACTTGAAGTGCAGTTTC CTGGGACCCAATGAGATG TP53 ENST Exon11 chr17: _ GAGGTGCTCAGTAAACATATTTGC CATCTCCCAAACATCCCTCAC 31
32 Supplementary Table 12 continued. Primers used for PCR amplification and Sanger sequencing Gene Symbol Transcript IDs Coding Exon Number Genomic Region of Interest Forward primer sequence Reverse primer sequence Additional primer sequnece for sequencing LAMA2 ENST Exon1 chr6: _ CGAGAGTGGAGGAGTGTCCT CGCTCTCATGGCATTGAACT LAMA2 ENST Exon2 chr6: _ GATGGAAGATTTAGTGGGATC GCCAGGAAGAAGCTCTTAATG LAMA2 ENST Exon3 chr6: _ GAGAATCTGAATTACTCTCCTTC ATCAGCACAGTGGGAGCAAT LAMA2 ENST Exon4 chr6: _ TGAAATGTTGCCAATGAGTT AATGTACTTTTGCTAAATCCTAT LAMA2 ENST Exon5 chr6: _ AACTAATTGGGAGAATGGGA AGCGGCACAGCTTGGATCT LAMA2 ENST Exon6 chr6: _ ACTCTTCCTTTGGGCTAAGA ACTGTGATGGCTGATTTCAA LAMA2 ENST Exon7 chr6: _ ATATACCCACTTCAAGCCTGG CATTCAGTTGAATTGGACTTC LAMA2 ENST Exon8 chr6: _ CAGCCTGGGCGACAGAGT AGTTCAACTGTTTACAGAAGC CTATTATGTATAACAGAAATGA [F] LAMA2 ENST Exon9 chr6: _ ATCTTTACTTTTGTGTAAGCATG GGAGGGAACGCTAATACACT LAMA2 ENST Exon10 chr6: _ CCCTCTACTCTTTGGTTTTAAC TGGATACTTCCAGAAGTCA LAMA2 ENST Exon11 chr6: _ AACACATAGCTTTTGAGCAG ACCTCAAAGGCTTTGAGCAA LAMA2 ENST Exon12 chr6: _ TCTAAATGTTTGTGGTTTGG GGCAATCCAATTCAGTGTGT CAAAAGTGGACACGACCAG[F] LAMA2 ENST Exon13 chr6: _ CCCTATAGAACAATAAAATTCG AGTAGTCCTGTAAGTCAGTGTCA LAMA2 ENST Exon14 chr6: _ ATAGAGCGAGACTCTGTCTCC CTGCATATTTAAAACTGTAAAG GTTCTTATCTGCTCTTTGAA [R] LAMA2 ENST Exon15 chr6: _ GGTGATTGTGAGCACTTTAGAC GCCACAGCTTTTGGTAGATA LAMA2 ENST Exon16 chr6: _ TGCCAATAACACTAAGGTCAG CTTCCTCATATTCATTAGGGAG LAMA2 ENST Exon17 chr6: _ CTCTGCTGTAAAATTGACCA TCTTTGGAATTGGTGTAGAA LAMA2 ENST Exon18 chr6: _ CTCTCCACTGGGCTATATTCAC CTTTATTTCACCTCATTGAGTG LAMA2 ENST Exon19 chr6: _ CACGTTGCGTTTGAGAAAAG GTTCTGGAATATGCAGACTCTG LAMA2 ENST Exon20 chr6: _ CCTAACAGGGCACTTAACAA GTTTAAGGTCAAATGGTAATGT LAMA2 ENST Exon21 chr6: _ TCAGTGAGGAATCCTGATAACT TACGTTGTATCAATCTGTGCTT LAMA2 ENST Exon22 chr6: _ TATTGAACAGCTCCTTTCTGA ACTTCATCATCTTTCCTACAAGT LAMA2 ENST Exon23 chr6: _ CCCAAGTGACTAAACTAGAATAGTAC CTCCATGTCTGGCAATGAGA LAMA2 ENST Exon24 chr6: _ GAGTATGCTCCCGTTATGCATT CTGTTCCTAAATGTTCACAGAGG LAMA2 ENST Exon25 chr6: _ GTGAGTTCTGTTGCTTGTGAG GATTGCATACTTGAGTTTGCC LAMA2 ENST Exon26 chr6: _ AGAGAAGATCTCCATTTGGAAC GTATATCCAAGAAGTCTTCTCGG LAMA2 ENST Exon27 chr6: _ CATTCGAGGTGGGACACC GGGAAGCCTCCATGGTTTA LAMA2 ENST Exon28 chr6: _ CAATAGAACATACTTGAGGTTGAC CTGTGTCTCACACTGTGAGTG LAMA2 ENST Exon29 chr6: _ CAGCCACTGAAGAGCTCAAG AATGCAGAATCTGTGAATATCTT LAMA2 ENST Exon30 chr6: _ AGACTGAGTTCTCTGTCAGTGAT GAACCATAGAGCATTATATACTCAC LAMA2 ENST Exon31 chr6: _ TTCATTGTGACGTCCTAGCC GACAAATTCTAAGATACACAAGTGG LAMA2 ENST Exon32 chr6: _ TCACGGCATAATCTTGATTT GGGAGAGTCACTCTTTCTCAT 32
33 Supplementary Table 12 continued. Primers used for PCR amplification and Sanger sequencing Gene Symbol Transcript IDs Coding Exon Number Genomic Region of Interest Forward primer sequence Reverse primer sequence Additional primer sequnece for sequencing LAMA2 ENST Exon33 chr6: _ CCTCAGTTTATATTATCGCCTCT AACAGTTGTGTCCCTTGATTTG LAMA2 ENST Exon34 chr6: _ CTTTGAAGGTGAGAAGGCTAAG GAAGCAGAATTAGCAAGTCAGA LAMA2 ENST Exon35 chr6: _ TTAATTGTATACGTGAGCATGG TCTCAGCAGATAATACCCTTG LAMA2 ENST Exon36 chr6: _ TTCCCAGCAGGAACACTCA AGCAAATATCTCCAGGTTGG LAMA2 ENST Exon37 chr6: _ GGTCTCAGGTACCAGTTTTCA CTTTCAGAGTTGTCACTCTTGAC LAMA2 ENST Exon38 chr6: _ CATCTAGCCACATTTCAACATG ACACTCATGTCCATTGGGAG LAMA2 ENST Exon39 chr6: _ GACAGAGCAAGACTCTGTCTCA TGTAGCCTAGGGCTATGTTCA GATGCTGCTGTAGCAAAGC [R] LAMA2 ENST Exon40 chr6: _ GGAATTGTCATAGATATATTGGC AGTGGTAGTGGGTGACTGAGA LAMA2 ENST Exon41 chr6: _ CAGTCTCCTAACTTTAACTACCGA ACTTGAAGTGAGATAGGTAGACAAC LAMA2 ENST Exon42 chr6: _ GTTCAACATGTAGAAGCATCCA AATACCATAATGCAAAGTATGGT LAMA2 ENST Exon43 chr6: _ TACACCTGAATGAGACAAAGTTC AGAAGACTTGGATTCCAGCT LAMA2 ENST Exon44 chr6: _ GGCCACTAAATTATATATTTTGTG CCACTTCGCCTGACCAAAC CTGTAACAGTGGAAAGAATAGCT[R] LAMA2 ENST Exon45 chr6: _ TTGACATTTGCCATCACACA TCCTACCCCAAATCGCTTC LAMA2 ENST Exon46-1 ACATGGAACTGTGAACAGCTC TGTAAGGTAAGACGTTGAGGTT chr6: _ LAMA2 ENST Exon46-2 GAGTATAATGGTGTCTGCATATGG ATCTGAAAGCCTACGTTAGACC LAMA2 ENST Exon47 chr6: _ GAAGCTCCTAATCATTTCACATGTC CCTCTGGTCAATACCTGGAAGG LAMA2 ENST Exon48 chr6: _ GATCAGACTCTGAGTATTGATTCG GTGTTTCCAGGTTGAATCGTC LAMA2 ENST Exon49 chr6: _ GTAGTAGCTCTAAGGATACGGACAG CAGGCAGGGACTTGAAGATTC LAMA2 ENST Exon50 chr6: _ GTAGAGATGGAGATAAGTGGAAGG CTGCCAGACAATCATCCCAG LAMA2 ENST Exon51 chr6: _ GCAATTTAGCCACAAGACCCTAAC CCCAAATCACCTATAATCTAAGCC LAMA2 ENST Exon52 chr6: _ GCTAAATATAGTGCAAGTGCTTGAG CCTGGTGTTGTTCTAACATGCAAC LAMA2 ENST Exon53 chr6: _ CCTTTGTAGATCCCTGTGAAATG CTTGCTCGTGAAACATACTATGAAC LAMA2 ENST Exon54 chr6: _ CCAGTAGACAATGGAAACCAGAGT ATGTGCTATAAAGAGCTATACATTTGG LAMA2 ENST Exon55 chr6: _ TCTTCAACTGCTCTTAAACTTTCC GGAAATATGGAATGTTCAGTCC LAMA2 ENST Exon56 chr6: _ CACTTTCCAGATCATCTCACAAGC ATGTTAAGGGTAGAACTCCTAAGG LAMA2 ENST Exon57 chr6: _ CTTAGTGTAGAATTCCTAAGCATCC GCATAGCTGTAGACAATGTAGATGC LAMA2 ENST Exon58 chr6: _ GCATCTACATTGTCTACAGCTATGC GGTAAGTCATTCAATGAAGATGTAAG LAMA2 ENST Exon59 chr6: _ GATTCACACGCACTGAGCACTC GACTTTATCAGGCTACTGATTTCC LAMA2 ENST Exon60 chr6: _ CATCTCTGAGAAGGTTTTACTCTCC GCAAACCTACACAGGTCTGAGCAC LAMA2 ENST Exon61 chr6: _ GCATACTAAGCACTGTGTACATTG CGACAGAGCGAGACTCTGTCTC LAMA2 ENST Exon62 chr6: _ CAAGTTGAGAAGCACAACACAGG AAGGATACTAGTTTAGGACTCTAGGAAC LAMA2 ENST Exon63 chr6: _ CATGATGCAGTGGTGGAAGATAG CTCCCAACAAAGAATACAGCCG LAMA2 ENST Exon64 chr6: _ GATTCTGGCTGATGTCTTTCC GGAAGCAAGTACACATAAGCACAC LAMA2 ENST Exon65 chr6: _ CAGCATTCCAATTTAATCTCAAGC GCACCATCTGAATACAGAAAGAATTC 33
34 Supplementary Table 12 continued. Primers used for PCR amplification and Sanger sequencing Gene Symbol Transcript IDs Coding Exon Number Genomic Region of Interest Forward primer sequence Reverse primer sequence Additional primer sequnece for sequencing XIRP2 ENST Exon2 chr2: _ CCAAGAGCTGGTGCTCCTAGAG CAGGACATCTGTCTAGAATACTGG XIRP2 ENST Exon3 chr2: _ CTCTTTCATTTCTGCCTTCTAACC GTCAAGTGCATTGTTTCAAACCTC XIRP2 ENST Exon4 chr2: _ GCTACAGTCCATTTTACGAGATC CACACACTATTTTTGTGCTCTTTC XIRP2 ENST Exon5 chr2: _ GACATACTGAAATAATGGTGTATGTG GCTACAGTGTTTGTTAATGAGTAATG XIRP2 ENST Exon6 chr2: _ CAGAAATTGTCACTGAAATTGCAC AGAATTCCAACTTGAAAGTGGAG XIRP2 ENST Exon7 chr2: _ GCATCTCTCAATGTACACTAAAGTC GACTGTGCTTTGGAAGTCATCC XIRP2 ENST Exon8 chr2: _ GTATTGGGATTACAGGAATGAGC GTTGAAAAGCCACCTCAAGTATTC XIRP2 ENST Exon9-1 TCACATATTGTTCCCAATGGAG CCAAGTATTCTCTTTCTGAGTTCAG XIRP2 ENST Exon9-2 CCCAAAGATGTATATTCCAAGC GAATGAAGCTGTCCAAGTTGATC XIRP2 ENST Exon9-3 CTTCTGACACTGTAGAAAATGCAG TCAATGATGACCTCTTCTGACTC XIRP2 ENST Exon9-4 GATGTAAGAACAGCACGGTGG CTCTTGTAACTCCTTCCACCTC XIRP2 ENST Exon9-5 ACAGTCCTGATATAGGAAAGCTTC CCATTTACAGGTTTTGACATCC XIRP2 ENST Exon9-6 GACCAGTTTGATGAAAGCATTC CATTGCCTTTCTGAATATCTTCAG XIRP2 ENST Exon9-7 ACAGGAGGAGATCCAAGGTGG CTGAATGTCTTCTTGTGTGACAG XIRP2 ENST Exon9-8 AGAAGAATCTGACTATATCAGCACC CTTCCATAATAGAATCGAAAGTTCG XIRP2 ENST Exon9-9 GACAAGTAGACAATTCTTTGAGTCTG ACATTTCCCTTCTCTTCTTCAC XIRP2 ENST Exon9-10 AAGAAACCTTAGAAGATCTCTACTCTC TCATTTGACTCTGTGTGGAGTTG XIRP2 ENST Exon9-11 GAAAATGATGGTGACACAATTGAGC CAATATCACCAGGGATGGCATC chr2: _ XIRP2 ENST Exon9-12 TCAGAGTACATTTGGTAAGATACCC TCACGATCTATGACAATCTTTACAG XIRP2 ENST Exon9-13 CATCAGGTTGCTGTCCAGAGG GCTTGGAGCTTTGTGTATCAG XIRP2 ENST Exon9-14 GAATCAGACAGGGCAGTGAGAG CATTAGAAGGTGGTGGAGGAGG XIRP2 ENST Exon9-15 GCAAGACACATCCAATGTAACAG GTTTGGGTTTGGGCAATGTG XIRP2 ENST Exon9-16 CCAGTAGATGAGAAATCTGAAAGAGA GGCTTTGAGTTTTAACAATGTTGTAG XIRP2 ENST Exon9-17 CTGCAAACTGTCTCTCACACACA ATGACTTTCATAGCTGTGGTCTG XIRP2 ENST Exon9-18 CATTATGCAATCCAAATCAGC GTATGCCCTTGACTTCGAGAC XIRP2 ENST Exon9-19 GGACCATCAATGATTGGTCG GCTGACATGAACATTAGATACTTTGG AGTCCCTACCACGAGTTTCTATC XIRP2 ENST Exon9-20 CAGTTCACATTGCCATGGAG AGTCCCTACCACGAGTTTCTATC XIRP2 ENST Exon9-21 GAAATTCACAGAGCAAACACTTCC CTCGCAGTTCTCTAGGATGTTTGC XIRP2 ENST Exon9-22 ACCGAACTGTTCAAATGGCTG GTTTGGTTGTTTCCACGCACTC XIRP2 ENST Exon10 chr2: _ GAGCTTTTCTACTTTAACCGCATGG GCTATACATAGAGTGCAGCTCCTG *Primer sequence used for PCR amplification only, but not for sequencing as regions of interest present high GC contents or nucleotide repeats. (F) or [R] Additional primers used for sequencing to overcome primer slippage and to obtain high coverage in region of interest.. 34
35 Supplementary Figure 1. Distribution of mutations in 8 O. viverrini-associated CCA exomes. Pie chart illustrates the types of mutations present in CCA exomes and the number of each mutation type is reflected in the table. 35
36 Code SMAD4 TP53 KRAS GNAS RNF43 ROBO2 PEG3 MLL3 CDKN2A PTEN NCD80 RADIL PCDHA13 LAMA2 XIRP2 W A U A B B Y Y Y T R A142 + W Y035 + Y002 + R134 + A162 + A107 + A042 + A119 + A120 + B032 + B083 + Y072 + R B A035 + U027 + Y019 + T151 + B T157 + Y W Y057 + Y023 + R100 + A106 + T003 + A T160 + A039 B087 Y140 Y123 Y074 Y065 Y033 Y032 A043 A028 A128 B048 B113 Supplementary Figure 2: Summary of commonly mutated genes in 54 O. viverrini-associated CCA samples. Left column indicates the patient identity and top row are genes validated in this study. Red boxes indicate genes mutated in the respective samples. Region boxed with black border indicate samples with mutated genes excluding TP53, KRAS and SMAD4. 36
37 Supplementary Figure 3: Kaplan-Meier plots of overall survival of patients with tumors containing KRAS and RNF43 mutations. (a) Five patients with RNF43 gene mutations vs. 48 patients with wildtype (P < 0.001). (b) Nine patients with KRAS gene mutation vs. 44 patients with wild-type (P = 0.014). NB: Peri-operative death cases (survival time < 30 days) were excluded from analysis. 37
38 38
Supplementary Table 2. Identified causative mutations and/or mutation candidates.
 Supplementary Table 2. Identified causative mutations and/or mutation candidates. Nonsense mutations base change aa change Average depth Result of next generation in 432 patient Hereditary form of the
Supplementary Table 2. Identified causative mutations and/or mutation candidates. Nonsense mutations base change aa change Average depth Result of next generation in 432 patient Hereditary form of the
Supplementary Information. Exome sequencing identifies distinct mutational patterns in liver fluke related and noninfection-related
 Supplementary Information Exome sequencing identifies distinct mutational patterns in liver fluke related and noninfection-related bile duct cancers Waraporn Chan-on 1,2,3*, Maarja-Liisa Nairismägi 1,2*,
Supplementary Information Exome sequencing identifies distinct mutational patterns in liver fluke related and noninfection-related bile duct cancers Waraporn Chan-on 1,2,3*, Maarja-Liisa Nairismägi 1,2*,
Whole Exome Sequenced Characteristics
 Supplementary Tables Supplementary Table 1: Patient characteristics of 45 whole exome sequenced HNSCC tumors Whole Exome Sequenced Characteristics Tumors (n=45) Age, years Median (range) 61.0 (19-90) Sex,
Supplementary Tables Supplementary Table 1: Patient characteristics of 45 whole exome sequenced HNSCC tumors Whole Exome Sequenced Characteristics Tumors (n=45) Age, years Median (range) 61.0 (19-90) Sex,
July 2015 Assay ID Assay Name Gene COSMIC ID Amino acid change Nucleotide change Wild type allele (VIC label) Mutant allele (FAM label)
 July 2015 Assay ID Assay Name Gene COSMIC ID Amino acid change Nucleotide change Wild type allele (VIC label) Mutant allele (FAM label) p AHLJ090 AKT1_33765 AKT1 33765 p.e17k c.49g>a C T 2 AHBKFRM BRAF_473
July 2015 Assay ID Assay Name Gene COSMIC ID Amino acid change Nucleotide change Wild type allele (VIC label) Mutant allele (FAM label) p AHLJ090 AKT1_33765 AKT1 33765 p.e17k c.49g>a C T 2 AHBKFRM BRAF_473
Nature Genetics: doi: /ng Supplementary Figure 1. TCGA data set on HNSCCs reanalyzed in this study.
 Supplementary Figure 1 TCGA data set on HNSCCs reanalyzed in this study. Summary of the TCGA dataset on HNSCCs re-analyzed in this study and the respective numbers of samples available within each. Supplementary
Supplementary Figure 1 TCGA data set on HNSCCs reanalyzed in this study. Summary of the TCGA dataset on HNSCCs re-analyzed in this study and the respective numbers of samples available within each. Supplementary
UW359 Ovary 3c 3 Serous Recurrent 68 BRCA1 816delGT BRCA1 del exon 1-2. UW417 Ovary 3c 3 Serous Primary 38 BRCA1 1675delA
 Supplementary Table 1. Cases with deleterious germline mutations, somatic HR mutations, and somatic PTEN mutations. ID Site Stage Grade Histology Tumor Age Germline mutation(s) a Somatic HR mutation(s)
Supplementary Table 1. Cases with deleterious germline mutations, somatic HR mutations, and somatic PTEN mutations. ID Site Stage Grade Histology Tumor Age Germline mutation(s) a Somatic HR mutation(s)
Supplementary Table 1. PIK3CA mutation in colorectal cancer
 Liao X et al. PIK3CA Mutation in Colorectal Cancer. Page 1 Supplementary Table 1. PIK3CA mutation in colorectal cancer Exon Domain Nucleotide change* Amino acid change* cases 9 Helical c.1621t>a p.e541t
Liao X et al. PIK3CA Mutation in Colorectal Cancer. Page 1 Supplementary Table 1. PIK3CA mutation in colorectal cancer Exon Domain Nucleotide change* Amino acid change* cases 9 Helical c.1621t>a p.e541t
Clinical presentation. Duration (Month)
 Supplementary information, Table S1A: Clinical information for 125 PAs. No. Subtype Gender (M/F) Age at diagnosis (year) Clinical presentation Duration (Month) Maximal adenoma diameter(cm) Tumor invasion*
Supplementary information, Table S1A: Clinical information for 125 PAs. No. Subtype Gender (M/F) Age at diagnosis (year) Clinical presentation Duration (Month) Maximal adenoma diameter(cm) Tumor invasion*
Belgian study into von Willebrand Disease (B-Will): First results
 Belgian study into von Willebrand Disease (B-Will): First results Inge Vangenechten VWD Research Unit, Antwerp University Hospital Supported by CSL Behring Chair in von Willebrand Disease, Antwerp University
Belgian study into von Willebrand Disease (B-Will): First results Inge Vangenechten VWD Research Unit, Antwerp University Hospital Supported by CSL Behring Chair in von Willebrand Disease, Antwerp University
Spectrum and Classification of ATP7B Variants in a Large Cohort of Chinese Patients with Wilson s Disease Guides Genetic Diagnosis
 Ivyspring International Publisher 638 Theranostics Research Paper 2016; 6(5): 638-649. doi: 10.7150/thno.14596 Spectrum and Classification of ATP7B Variants in a Large Cohort of Chinese Patients with Wilson
Ivyspring International Publisher 638 Theranostics Research Paper 2016; 6(5): 638-649. doi: 10.7150/thno.14596 Spectrum and Classification of ATP7B Variants in a Large Cohort of Chinese Patients with Wilson
Supplementary Online Content
 Supplementary Online Content Pearlman R, Frankel WL, Swanson B, et al. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol.
Supplementary Online Content Pearlman R, Frankel WL, Swanson B, et al. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol.
Supplementary Figure 1
 Supplementary Figure 1 A B HEC1A PTEN +/+ HEC1A PTEN -/- 16 HEC1A PTEN -/- 22 PTEN RAD51 % Cells with RAD51 foci 40 -IR +IR 30 20 10 * *! TUBULIN 0 HEC1A PTEN+/+ HEC1A PTEN-/- 16 HEC1A PTEN-/- 22 C D 1.0
Supplementary Figure 1 A B HEC1A PTEN +/+ HEC1A PTEN -/- 16 HEC1A PTEN -/- 22 PTEN RAD51 % Cells with RAD51 foci 40 -IR +IR 30 20 10 * *! TUBULIN 0 HEC1A PTEN+/+ HEC1A PTEN-/- 16 HEC1A PTEN-/- 22 C D 1.0
DNA Analysis in Glycogen storage disease
 DNA Analysis in Glycogen storage disease Nick Beauchamp PhD Sheffield, Sheffield Children s NHS Foundation Trust 14th October 2010 Glycogen Synthase Type 0 Glycogen Synthesis and Breakdown Type IV UDPGlucose
DNA Analysis in Glycogen storage disease Nick Beauchamp PhD Sheffield, Sheffield Children s NHS Foundation Trust 14th October 2010 Glycogen Synthase Type 0 Glycogen Synthesis and Breakdown Type IV UDPGlucose
Supplementary Information
 Supplementary Information JAK1 truncating mutations in gynecologic cancer define new role of cancerassociated protein tyrosine kinase aberrations Yuan Ren, Yonghong Zhang, Richard Z. Liu, David A. Fenstermacher,
Supplementary Information JAK1 truncating mutations in gynecologic cancer define new role of cancerassociated protein tyrosine kinase aberrations Yuan Ren, Yonghong Zhang, Richard Z. Liu, David A. Fenstermacher,
Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each
 Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each mutated gene and the panel of 125 cancer-driving genes
Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each mutated gene and the panel of 125 cancer-driving genes
Disclosure. I have nothing to disclose
 Bin T Teh MD, PhD Disclosure I have nothing to disclose Asian Cancers A Vast Unmet Clinical Need Worldwide 14 million new cancer cases and 8.2 million cancer related deaths in 2012 New cases expected to
Bin T Teh MD, PhD Disclosure I have nothing to disclose Asian Cancers A Vast Unmet Clinical Need Worldwide 14 million new cancer cases and 8.2 million cancer related deaths in 2012 New cases expected to
Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM
 Supplementary Data Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM cohort. Supplementary Table e2. Specificity, sensitivity and unadjusted ORs for glioma in participants
Supplementary Data Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM cohort. Supplementary Table e2. Specificity, sensitivity and unadjusted ORs for glioma in participants
SUPPLEMENTARY INFORMATION. Rare independent mutations in renal salt handling genes contribute to blood pressure variation
 SUPPLEMENTARY INFORMATION Rare independent mutations in renal salt handling genes contribute to blood pressure variation Weizhen Ji, Jia Nee Foo, Brian J. O Roak, Hongyu Zhao, Martin G. Larson, David B.
SUPPLEMENTARY INFORMATION Rare independent mutations in renal salt handling genes contribute to blood pressure variation Weizhen Ji, Jia Nee Foo, Brian J. O Roak, Hongyu Zhao, Martin G. Larson, David B.
Recent advances in the molecular pathology of lung cancer: role of EGFR and KRAS mutation testing in treatment selection
 ASIP 2009 USCAP Companion Meeting Molecular Pathology for the Practicing Pathologist Recent advances in the molecular pathology of lung cancer: role of EGFR and KRAS mutation testing in treatment selection
ASIP 2009 USCAP Companion Meeting Molecular Pathology for the Practicing Pathologist Recent advances in the molecular pathology of lung cancer: role of EGFR and KRAS mutation testing in treatment selection
The Role of Next Generation Sequencing in Solid Tumor Mutation Testing
 The Role of Next Generation Sequencing in Solid Tumor Mutation Testing Allie H. Grossmann MD PhD Department of Pathology, University of Utah Division of Anatomic Pathology & Oncology, ARUP Laboratories
The Role of Next Generation Sequencing in Solid Tumor Mutation Testing Allie H. Grossmann MD PhD Department of Pathology, University of Utah Division of Anatomic Pathology & Oncology, ARUP Laboratories
Familial exudative vitreoretinopathy (FEVR) is an inheritable
 Genetics Spectrum of Variants in 389 Chinese Probands With Familial Exudative Vitreoretinopathy Jia-Kai Li, 1 Yian Li, 1 Xiang Zhang, 1 Chun-Li Chen, 2 Yu-Qing Rao, 1 Ping Fei, 1 Qi Zhang, 1 Peiquan Zhao,
Genetics Spectrum of Variants in 389 Chinese Probands With Familial Exudative Vitreoretinopathy Jia-Kai Li, 1 Yian Li, 1 Xiang Zhang, 1 Chun-Li Chen, 2 Yu-Qing Rao, 1 Ping Fei, 1 Qi Zhang, 1 Peiquan Zhao,
The Personalized Cancer Medicine Initiative at Vanderbilt
 The Personalized Cancer Medicine Initiative at Vanderbilt October 26, 2009 William Pao, MD, PhD Assistant Director, Personalized Cancer Medicine Vanderbilt-Ingram Cancer Center Cancer in the United States,
The Personalized Cancer Medicine Initiative at Vanderbilt October 26, 2009 William Pao, MD, PhD Assistant Director, Personalized Cancer Medicine Vanderbilt-Ingram Cancer Center Cancer in the United States,
Belgian-Czech cooperation in the Brno-VWD study
 Inge Vangenechten I Vangenechten, P Smejkal, O Zapletal, F Bouddount, J Zavrelova, J Blatny, M Penka, JJ Michiels, A Gadisseur. Aim of the study Characterisation of VWD in South Moravia (Czech Republic)
Inge Vangenechten I Vangenechten, P Smejkal, O Zapletal, F Bouddount, J Zavrelova, J Blatny, M Penka, JJ Michiels, A Gadisseur. Aim of the study Characterisation of VWD in South Moravia (Czech Republic)
Nature Genetics: doi: /ng Supplementary Figure 1. Alternative splicing events in the 5K panel.
 Supplementary Figure 1 Alternative splicing events in the 5K panel. The majority of cryptic splicing occurred by creation of an AG or GT (Type I). While some other mutations increased the usage of a nearby
Supplementary Figure 1 Alternative splicing events in the 5K panel. The majority of cryptic splicing occurred by creation of an AG or GT (Type I). While some other mutations increased the usage of a nearby
p.r623c p.p976l p.d2847fs p.t2671 p.d2847fs p.r2922w p.r2370h p.c1201y p.a868v p.s952* RING_C BP PHD Cbp HAT_KAT11
 ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
BWA alignment to reference transcriptome and genome. Convert transcriptome mappings back to genome space
 Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
MSI positive MSI negative
 Pritchard et al. 2014 Supplementary Figure 1 MSI positive MSI negative Hypermutated Median: 673 Average: 659.2 Non-Hypermutated Median: 37.5 Average: 43.6 Supplementary Figure 1: Somatic Mutation Burden
Pritchard et al. 2014 Supplementary Figure 1 MSI positive MSI negative Hypermutated Median: 673 Average: 659.2 Non-Hypermutated Median: 37.5 Average: 43.6 Supplementary Figure 1: Somatic Mutation Burden
Supplementary appendix
 Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Wells JM, Farris RF, Gosdin TA, et al. Pulmonary
Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Wells JM, Farris RF, Gosdin TA, et al. Pulmonary
Mutational and phenotypical spectrum of phenylalanine hydroxylase deficiency in Denmark
 Clin Genet 2016: 90: 247 251 Printed in Singapore. All rights reserved Short Report 2015 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd CLINICAL GENETICS doi: 10.1111/cge.12692 Mutational and
Clin Genet 2016: 90: 247 251 Printed in Singapore. All rights reserved Short Report 2015 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd CLINICAL GENETICS doi: 10.1111/cge.12692 Mutational and
Supplementary Table 1. Information on the 174 single nucleotide variants identified by whole-exome sequencing
 Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
Nature Genetics: doi: /ng Supplementary Figure 1. TNFAIP3-associated haplotypes in family 1.
 Supplementary Figure 1 TNFAIP3-associated haplotypes in family 1. The p.leu227* mutation (shown as a star) arose de novo in the first affected member of the family (P1). Red haplotypes carry the TNFAIP3
Supplementary Figure 1 TNFAIP3-associated haplotypes in family 1. The p.leu227* mutation (shown as a star) arose de novo in the first affected member of the family (P1). Red haplotypes carry the TNFAIP3
Supplementary Document
 Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
Research Article Mutation Analysis of the Common Deafness Genes in Patients with Nonsyndromic Hearing Loss in Linyi by SNPscan Assay
 BioMed Research International Volume 2016, Article ID 1302914, 7 pages http://dx.doi.org/10.1155/2016/1302914 Research Article Mutation Analysis of the Common Deafness Genes in Patients with Nonsyndromic
BioMed Research International Volume 2016, Article ID 1302914, 7 pages http://dx.doi.org/10.1155/2016/1302914 Research Article Mutation Analysis of the Common Deafness Genes in Patients with Nonsyndromic
Supplementary Figures. Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated. for each lesion type.
 Supplementary Figures Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated for each lesion type. a b Supplementary Figure 2. Non-negative matrix factorization-derived
Supplementary Figures Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated for each lesion type. a b Supplementary Figure 2. Non-negative matrix factorization-derived
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Generation of cetuximab-resistant cells and analysis of singlecell clones. Cetuximab-sensitive cells (LIM1215 and OXCO-2) were chronically treated with cetuximab
Supplementary Figure 1 Supplementary Figure 1: Generation of cetuximab-resistant cells and analysis of singlecell clones. Cetuximab-sensitive cells (LIM1215 and OXCO-2) were chronically treated with cetuximab
b 0.50 Synonymous Subtype
 omatic mutation count Mutations per Mb a 80 * b 0.50 ynonymous 60 0.40 Non-synonymous 40 0.30 0.20 20 0.10 c 0 Fibroadenoma Phyllodes 0.00 ubtype Benign Borderline MAL upplementary Figure 1 Landscape of
omatic mutation count Mutations per Mb a 80 * b 0.50 ynonymous 60 0.40 Non-synonymous 40 0.30 0.20 20 0.10 c 0 Fibroadenoma Phyllodes 0.00 ubtype Benign Borderline MAL upplementary Figure 1 Landscape of
p.arg119gly p.arg119his p.ala179thr c.540+1g>a c.617_633+6del Prediction basis structure
 a Missense ATG p.arg119gly p.arg119his p.ala179thr p.ala189val p.gly206trp p.gly206arg p.arg251his p.ala257thr TGA 5 UTR 1 2 3 4 5 6 7 3 UTR Splice Site, Frameshift b Mutation p.gly206trp p.gly4fsx50 c.138+1g>a
a Missense ATG p.arg119gly p.arg119his p.ala179thr p.ala189val p.gly206trp p.gly206arg p.arg251his p.ala257thr TGA 5 UTR 1 2 3 4 5 6 7 3 UTR Splice Site, Frameshift b Mutation p.gly206trp p.gly4fsx50 c.138+1g>a
Importance of minor TP53 mutated clones in the clinic
 Importance of minor TP53 mutated clones in the clinic Davide Rossi, M.D., Ph.D. Hematology IOSI - Oncology Institute of Southern Switzerland IOR - Institute of Oncology Reserach Bellinzona - Switzerland
Importance of minor TP53 mutated clones in the clinic Davide Rossi, M.D., Ph.D. Hematology IOSI - Oncology Institute of Southern Switzerland IOR - Institute of Oncology Reserach Bellinzona - Switzerland
SUPPLEMENTARY INFORMATION
 Somatic ERCC2 Mutations Are Associated with a Distinct Genomic Signature in Urothelial Tumors Jaegil Kim, Kent W Mouw, Paz Polak, Lior Z Braunstein, Atanas Kamburov, Grace Tiao, David J Kwiatkowski, Jonathan
Somatic ERCC2 Mutations Are Associated with a Distinct Genomic Signature in Urothelial Tumors Jaegil Kim, Kent W Mouw, Paz Polak, Lior Z Braunstein, Atanas Kamburov, Grace Tiao, David J Kwiatkowski, Jonathan
Vertical Magnetic Separation of Circulating Tumor Cells and Somatic Genomic-Alteration Analysis in Lung Cancer Patients
 Vertical Magnetic Separation of Circulating Cells and Somatic Genomic-Alteration Analysis in Lung Cancer Patients Chang Eun Yoo 1,2#, Jong-Myeon Park 3#, Hui-Sung Moon 1,2, Je-Gun Joung 2, Dae-Soon Son
Vertical Magnetic Separation of Circulating Cells and Somatic Genomic-Alteration Analysis in Lung Cancer Patients Chang Eun Yoo 1,2#, Jong-Myeon Park 3#, Hui-Sung Moon 1,2, Je-Gun Joung 2, Dae-Soon Son
Whole exome sequencing as a first line test: Is there even a role for metabolic biochemists in the future?
 Whole exome sequencing as a first line test: Is there even a role for metabolic biochemists in the future? Tony Marinakii Purine Research Laboratory Biochemical Sciences Whole exome sequencing No doubt
Whole exome sequencing as a first line test: Is there even a role for metabolic biochemists in the future? Tony Marinakii Purine Research Laboratory Biochemical Sciences Whole exome sequencing No doubt
Table S4. List of point mutations found by WES in SMZL patients. * DNA extracted from frozen tissue *Method of detection: Variant detected using
 Table S4. List of point mutations found by WES in SMZL patients. * DNA extracted from frozen tissue *Method of detection: Variant detected using RAMSES (R) or CNAG method (C) or both (R/C) **Validation
Table S4. List of point mutations found by WES in SMZL patients. * DNA extracted from frozen tissue *Method of detection: Variant detected using RAMSES (R) or CNAG method (C) or both (R/C) **Validation
Genetic Testing and Analysis. (858) MRN: Specimen: Saliva Received: 07/26/2016 GENETIC ANALYSIS REPORT
 GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
Supplemental Table 1. The list of variants with their respective scores for each variant classifier Gene DNA Protein Align-GVGD Polyphen-2 CADD MAPP
 Supplemental Table 1. The list of variants with their respective scores for each variant classifier Gene DNA Protein Align-GVGD Polyphen-2 CADD MAPP Frequency Domain Mammals a 3 S/P b Mammals a 3 S/P b
Supplemental Table 1. The list of variants with their respective scores for each variant classifier Gene DNA Protein Align-GVGD Polyphen-2 CADD MAPP Frequency Domain Mammals a 3 S/P b Mammals a 3 S/P b
Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
 Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
Genomic Profiling on an Unselected Solid Tumor Population Reveals a Highly Mutated Wnt/β-Catenin Pathway Associated with Oncogenic EGFR Mutations
 Article Genomic Profiling on an Unselected Solid Tumor Population Reveals a Highly Mutated Wnt/β-Catenin Pathway Associated with Oncogenic EGFR Mutations Jingrui Jiang 1, *, Alexei Protopopov 2, Ruobai
Article Genomic Profiling on an Unselected Solid Tumor Population Reveals a Highly Mutated Wnt/β-Catenin Pathway Associated with Oncogenic EGFR Mutations Jingrui Jiang 1, *, Alexei Protopopov 2, Ruobai
Negative feedback-defective PRPS1 mutants drive thiopurine. resistance in relapsed childhood ALL
 SUPPLEMENTARY INFORMATION Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL Benshang Li 1,5,, Hui Li 1,5,, Yun Bai 2,, Renate Kirschner-Schwabe 3,10, Jun J
SUPPLEMENTARY INFORMATION Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL Benshang Li 1,5,, Hui Li 1,5,, Yun Bai 2,, Renate Kirschner-Schwabe 3,10, Jun J
Circulating tumour DNA in breast cancer. Kathleen Burke, PhD Bioinformatics Postdoctoral Fellow Laboratory of Dr. Jorge Reis-Filho
 Circulating tumour DNA in breast cancer Kathleen Burke, PhD Bioinformatics Postdoctoral Fellow Laboratory of Dr. Jorge Reis-Filho Conflicts of Interest I have no financial relationships to disclose I will
Circulating tumour DNA in breast cancer Kathleen Burke, PhD Bioinformatics Postdoctoral Fellow Laboratory of Dr. Jorge Reis-Filho Conflicts of Interest I have no financial relationships to disclose I will
TP53 mutational profile in CLL : A retrospective study of the FILO group.
 TP53 mutational profile in CLL : A retrospective study of the FILO group. Fanny Baran-Marszak Hopital Avicenne Bobigny France 2nd ERIC workshop on TP53 analysis in CLL, Stresa 2017 TP53 abnormalities :
TP53 mutational profile in CLL : A retrospective study of the FILO group. Fanny Baran-Marszak Hopital Avicenne Bobigny France 2nd ERIC workshop on TP53 analysis in CLL, Stresa 2017 TP53 abnormalities :
Supplementary Figure 1
 Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
Using the Bravo Liquid-Handling System for Next Generation Sequencing Sample Prep
 Using the Bravo Liquid-Handling System for Next Generation Sequencing Sample Prep Tom Walsh, PhD Division of Medical Genetics University of Washington Next generation sequencing Sanger sequencing gold
Using the Bravo Liquid-Handling System for Next Generation Sequencing Sample Prep Tom Walsh, PhD Division of Medical Genetics University of Washington Next generation sequencing Sanger sequencing gold
Unravelling the Molecular Taxonomies of Gastroesophageal Cancers
 Unravelling the Molecular Taxonomies of Gastroesophageal Cancers Patrick Tan, MD PhD Professor, Duke-NUS Medical School Deputy Executive Director, Biomedical Research Council, Agency for Science, Technology
Unravelling the Molecular Taxonomies of Gastroesophageal Cancers Patrick Tan, MD PhD Professor, Duke-NUS Medical School Deputy Executive Director, Biomedical Research Council, Agency for Science, Technology
Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases.
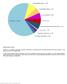 Supplementary Figure 1 Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases. A total of 85 somatically acquired
Supplementary Figure 1 Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases. A total of 85 somatically acquired
6/12/2018. Disclosures. Clinical Genomics The CLIA Lab Perspective. Outline. COH HopeSeq Heme Panels
 Clinical Genomics The CLIA Lab Perspective Disclosures Raju K. Pillai, M.D. Hematopathologist / Molecular Pathologist Director, Pathology Bioinformatics City of Hope National Medical Center, Duarte, CA
Clinical Genomics The CLIA Lab Perspective Disclosures Raju K. Pillai, M.D. Hematopathologist / Molecular Pathologist Director, Pathology Bioinformatics City of Hope National Medical Center, Duarte, CA
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma. Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute
 Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
Supporting Information
 Supporting Information Rampal et al. 10.1073/pnas.1407792111 Fig. S1. Genetic events in leukemic transformation of chronic-phase MPNs. (A) Survival of post-mpn AML patients according to mutational status
Supporting Information Rampal et al. 10.1073/pnas.1407792111 Fig. S1. Genetic events in leukemic transformation of chronic-phase MPNs. (A) Survival of post-mpn AML patients according to mutational status
MRC-Holland MLPA. Description version 05; 03 April 2019
 SALSA MLPA probemix ME012-A1 MGMT-IDH1-IDH2 Lot A1-1215. Glioblastoma, the most common malignant primary brain tumour, is characterised by aggressive behaviour and a poor survival. Hypermethylation in
SALSA MLPA probemix ME012-A1 MGMT-IDH1-IDH2 Lot A1-1215. Glioblastoma, the most common malignant primary brain tumour, is characterised by aggressive behaviour and a poor survival. Hypermethylation in
LMM / emerge III Network Reference Sequences October 2016 LMM / emerge III Network Consensus Actionable Gene List *ACMG56 gene
 LMM / emerge III Network Consensus Actionable Gene List *ACMG56 gene ACTA2* Exon 02-09 NM_001613.2 DSG2* Exon 01-15 NM_001943.3 ACTC1* Exon 01-06 NM_005159.4 DSP* Exon 01-24 NM_004415.2 APC* Exon 01-15
LMM / emerge III Network Consensus Actionable Gene List *ACMG56 gene ACTA2* Exon 02-09 NM_001613.2 DSG2* Exon 01-15 NM_001943.3 ACTC1* Exon 01-06 NM_005159.4 DSP* Exon 01-24 NM_004415.2 APC* Exon 01-15
Supplementary Information
 Supplementary Information Supplementary Figures 1-10 Supplementary Tables 1-16 Supplementary Notes Exome Sequencing of Gastric Adenocarcinoma Identifies Recurrent Somatic Mutations in Cell Adhesion and
Supplementary Information Supplementary Figures 1-10 Supplementary Tables 1-16 Supplementary Notes Exome Sequencing of Gastric Adenocarcinoma Identifies Recurrent Somatic Mutations in Cell Adhesion and
Frequency(%) KRAS G12 KRAS G13 KRAS A146 KRAS Q61 KRAS K117N PIK3CA H1047 PIK3CA E545 PIK3CA E542K PIK3CA Q546. EGFR exon19 NFS-indel EGFR L858R
 Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
GALAFOLD (migalastat) capsules, for oral use Initial U.S. Approval: 2018
 HIGHLIGHTS OF PRESCRIBING INFORMATION These highlights do not include all the information needed to use GALAFOLD safely and effectively. See full prescribing information for GALAFOLD. GALAFOLD (migalastat)
HIGHLIGHTS OF PRESCRIBING INFORMATION These highlights do not include all the information needed to use GALAFOLD safely and effectively. See full prescribing information for GALAFOLD. GALAFOLD (migalastat)
Accel-Amplicon Panels
 Accel-Amplicon Panels Amplicon sequencing has emerged as a reliable, cost-effective method for ultra-deep targeted sequencing. This highly adaptable approach is especially applicable for in-depth interrogation
Accel-Amplicon Panels Amplicon sequencing has emerged as a reliable, cost-effective method for ultra-deep targeted sequencing. This highly adaptable approach is especially applicable for in-depth interrogation
(Case 5 A ) c g>a Exon 24 splice acceptor Splicing defect - Splicing defect
 Supplementary table S1. Pathogenic potential of identified DYNC2H1 mutations Family Nucleotide Change Mutation GERP PhyloP SIFT PolyPhen-2 Mutation Taster JATD-1 c. 90443A>G p.d3015g 6.17 5.3 Tolerated
Supplementary table S1. Pathogenic potential of identified DYNC2H1 mutations Family Nucleotide Change Mutation GERP PhyloP SIFT PolyPhen-2 Mutation Taster JATD-1 c. 90443A>G p.d3015g 6.17 5.3 Tolerated
Exomes and Beyond Addressing the Genome with OneSeq. Madhuri Hegde, PhD, FACMG Emory University Emory Genetics Laboratory Atlanta, GA
 Exomes and Beyond Addressing the Genome with OneSeq Madhuri Hegde, PhD, FACMG Emory University Emory Genetics Laboratory Atlanta, GA Technologies to Detect Various Types of Mutations RESOLUTION Targeted
Exomes and Beyond Addressing the Genome with OneSeq Madhuri Hegde, PhD, FACMG Emory University Emory Genetics Laboratory Atlanta, GA Technologies to Detect Various Types of Mutations RESOLUTION Targeted
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1. Estimation of tumour content
 Supplementary Figure 1. Estimation of tumour content a, Approach used to estimate the tumour content in S13T1/T2, S6T1/T2, S3T1/T2 and S12T1/T2. Tissue and tumour areas were evaluated by two independent
Supplementary Figure 1. Estimation of tumour content a, Approach used to estimate the tumour content in S13T1/T2, S6T1/T2, S3T1/T2 and S12T1/T2. Tissue and tumour areas were evaluated by two independent
SALSA MLPA Probemix P014-B1 Chromosome 8 Lot B and B
 SALSA MLPA Probemix P014-B1 Chromosome 8 Lot B1-0916 and B1-0713. Copy number changes of the human chromosome 8 are common in many types of tumours. In most cases, losses of 8p sequences and gains of 8q
SALSA MLPA Probemix P014-B1 Chromosome 8 Lot B1-0916 and B1-0713. Copy number changes of the human chromosome 8 are common in many types of tumours. In most cases, losses of 8p sequences and gains of 8q
Supplementary Figure 1
 Supplementary Figure 1 Platelets from affected patients are comparable to controls on electron micrograph. Thin-section transmission electron micrographs of representative platelets from a normal control
Supplementary Figure 1 Platelets from affected patients are comparable to controls on electron micrograph. Thin-section transmission electron micrographs of representative platelets from a normal control
GALAFOLD (migalastat) capsules, for oral use Initial U.S. Approval: 2018
 HIGHLIGHTS OF PRESCRIBING INFORMATION These highlights do not include all the information needed to use GALAFOLD safely and effectively. See full prescribing information for GALAFOLD. GALAFOLD (migalastat)
HIGHLIGHTS OF PRESCRIBING INFORMATION These highlights do not include all the information needed to use GALAFOLD safely and effectively. See full prescribing information for GALAFOLD. GALAFOLD (migalastat)
SUPPLEMENTARY INFORMATION. Intron retention is a widespread mechanism of tumor suppressor inactivation.
 SUPPLEMENTARY INFORMATION Intron retention is a widespread mechanism of tumor suppressor inactivation. Hyunchul Jung 1,2,3, Donghoon Lee 1,4, Jongkeun Lee 1,5, Donghyun Park 2,6, Yeon Jeong Kim 2,6, Woong-Yang
SUPPLEMENTARY INFORMATION Intron retention is a widespread mechanism of tumor suppressor inactivation. Hyunchul Jung 1,2,3, Donghoon Lee 1,4, Jongkeun Lee 1,5, Donghyun Park 2,6, Yeon Jeong Kim 2,6, Woong-Yang
Characterisation of structural variation in breast. cancer genomes using paired-end sequencing on. the Illumina Genome Analyser
 Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Beta Thalassemia Case Study Introduction to Bioinformatics
 Beta Thalassemia Case Study Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu www.cs.sjsu.edu/faculty/khuri Outline v Hemoglobin v Alpha
Beta Thalassemia Case Study Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu www.cs.sjsu.edu/faculty/khuri Outline v Hemoglobin v Alpha
How has Molecular Diagnostics changed my practice? Pulmonary Pathology
 How has Molecular Diagnostics changed my practice? Pulmonary Pathology Lynette M. Sholl, M.D. Associate Pathologist Brigham and Women s Hospital Associate Director, Center for Advanced Molecular Diagnostics
How has Molecular Diagnostics changed my practice? Pulmonary Pathology Lynette M. Sholl, M.D. Associate Pathologist Brigham and Women s Hospital Associate Director, Center for Advanced Molecular Diagnostics
DYSREGULATION OF WT1 (-KTS) IS ASSOCIATED WITH THE KIDNEY-SPECIFIC EFFECTS OF THE LMX1B R246Q MUTATION
 DYSREGULATION OF WT1 (-KTS) IS ASSOCIATED WITH THE KIDNEY-SPECIFIC EFFECTS OF THE LMX1B R246Q MUTATION Gentzon Hall 1,2#, Brandon Lane 1,3#, Megan Chryst-Ladd 1,3, Guanghong Wu 1,3, Jen-Jar Lin 4, XueJun
DYSREGULATION OF WT1 (-KTS) IS ASSOCIATED WITH THE KIDNEY-SPECIFIC EFFECTS OF THE LMX1B R246Q MUTATION Gentzon Hall 1,2#, Brandon Lane 1,3#, Megan Chryst-Ladd 1,3, Guanghong Wu 1,3, Jen-Jar Lin 4, XueJun
UNIVERSITY OF TORINO DEPARTMENT OF ONCOLOGY. Giorgio V. Scagliotti University of Torino Dipartment of Oncology
 Giorgio V. Scagliotti University of Torino Dipartment of Oncology giorgio.scagliotti@unito.it Molecular landscape of MM not fully characterized to allow personalized treatment Recurrent genetic alterations
Giorgio V. Scagliotti University of Torino Dipartment of Oncology giorgio.scagliotti@unito.it Molecular landscape of MM not fully characterized to allow personalized treatment Recurrent genetic alterations
Mapping by recurrence and modelling the mutation rate
 Mapping by recurrence and modelling the mutation rate Shamil Sunyaev Broad Institute of M.I.T. and Harvard Current knowledge is from Comparative genomics Experimental systems: yeast reporter assays Potential
Mapping by recurrence and modelling the mutation rate Shamil Sunyaev Broad Institute of M.I.T. and Harvard Current knowledge is from Comparative genomics Experimental systems: yeast reporter assays Potential
CHAPTER IV RESULTS Microcephaly General description
 47 CHAPTER IV RESULTS 4.1. Microcephaly 4.1.1. General description This study found that from a previous study of 527 individuals with MR, 48 (23 female and 25 male) unrelated individuals were identified
47 CHAPTER IV RESULTS 4.1. Microcephaly 4.1.1. General description This study found that from a previous study of 527 individuals with MR, 48 (23 female and 25 male) unrelated individuals were identified
Fluxion Biosciences and Swift Biosciences Somatic variant detection from liquid biopsy samples using targeted NGS
 APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Information. PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small
 Sato T, et al. Supplementary Information PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer Teruyuki Sato 1,2, Atsushi Kaneda 1,3,4 *, Shingo Tsuji
Sato T, et al. Supplementary Information PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer Teruyuki Sato 1,2, Atsushi Kaneda 1,3,4 *, Shingo Tsuji
THE UMD TP53 MUTATION DATABASE UPDATES AND BENEFITS. Pr. Thierry Soussi
 THE UMD TP53 MUTATION DATABASE UPDATES AND BENEFITS Pr. Thierry Soussi thierry.soussi@ki.se thierry.soussi@upmc.fr TP53: 33 YEARS AND COUNTING STRUCTURE FUNCTION RELATIONSHIP OF WILD AND MUTANT TP53 1984
THE UMD TP53 MUTATION DATABASE UPDATES AND BENEFITS Pr. Thierry Soussi thierry.soussi@ki.se thierry.soussi@upmc.fr TP53: 33 YEARS AND COUNTING STRUCTURE FUNCTION RELATIONSHIP OF WILD AND MUTANT TP53 1984
Summary of Results CARRIER STATUS DNA INSIGHT. Result: Carrier, Heterozygote. Protected Health Information. Propionic Acidemia
 Test Results Reviewed & Approved by: Laboratory Director, Nilesh Dharajiya,.D. SAPLE REPORT CARRIER STATUS DNA INSIGHT PERSONAL DETAILS DOB Jan 1, 19XX ETHNICITY Asian ORDERING HEALTHCARE PROFESSIONAL
Test Results Reviewed & Approved by: Laboratory Director, Nilesh Dharajiya,.D. SAPLE REPORT CARRIER STATUS DNA INSIGHT PERSONAL DETAILS DOB Jan 1, 19XX ETHNICITY Asian ORDERING HEALTHCARE PROFESSIONAL
a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53
 1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Nangalia J, Massie CE, Baxter EJ, et al. Somatic CALR s in
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Nangalia J, Massie CE, Baxter EJ, et al. Somatic CALR s in
Reporting TP53 gene analysis results in CLL
 Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
c Tuj1(-) apoptotic live 1 DIV 2 DIV 1 DIV 2 DIV Tuj1(+) Tuj1/GFP/DAPI Tuj1 DAPI GFP
 Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Exons from 137 genes were targeted for hybrid selection and massively parallel sequencing.
 SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
NGS for Cancer Predisposition
 NGS for Cancer Predisposition Colin Pritchard MD, PhD University of Washington Dept. of Lab Medicine AMP Companion Society Meeting USCAP Boston March 22, 2015 Disclosures I am an employee of the University
NGS for Cancer Predisposition Colin Pritchard MD, PhD University of Washington Dept. of Lab Medicine AMP Companion Society Meeting USCAP Boston March 22, 2015 Disclosures I am an employee of the University
Next generation diagnostics Bringing high-throughput sequencing into clinical application
 Next generation diagnostics Bringing high-throughput sequencing into clinical application Leonardo A. Meza-Zepeda, PhD Translational Genomics Group Institute for Cancer Research Leonardo.Meza-Zepeda@rr-research.no
Next generation diagnostics Bringing high-throughput sequencing into clinical application Leonardo A. Meza-Zepeda, PhD Translational Genomics Group Institute for Cancer Research Leonardo.Meza-Zepeda@rr-research.no
Insights from Sequencing the Melanoma Exome
 Insights from Sequencing the Melanoma Exome Michael Krauthammer, MD PhD, December 2 2015 Yale University School Yof Medicine 1 2012 Exome Screens and Results Exome Sequencing of 108 sun-exposed melanomas
Insights from Sequencing the Melanoma Exome Michael Krauthammer, MD PhD, December 2 2015 Yale University School Yof Medicine 1 2012 Exome Screens and Results Exome Sequencing of 108 sun-exposed melanomas
Detection of BRCA1/2 mutations in breast cancer patients from Thailand and Pakistan
 Clin Genet 2012: 82: 594 598 Printed in Singapore. All rights reserved Letter to the Editor 2012 John Wiley & Sons A/S. Published by Blackwell Publishing Ltd CLINICAL GENETICS doi: 10.1111/j.1399-0004.2012.01869.x
Clin Genet 2012: 82: 594 598 Printed in Singapore. All rights reserved Letter to the Editor 2012 John Wiley & Sons A/S. Published by Blackwell Publishing Ltd CLINICAL GENETICS doi: 10.1111/j.1399-0004.2012.01869.x
DNA-seq Bioinformatics Analysis: Copy Number Variation
 DNA-seq Bioinformatics Analysis: Copy Number Variation Elodie Girard elodie.girard@curie.fr U900 institut Curie, INSERM, Mines ParisTech, PSL Research University Paris, France NGS Applications 5C HiC DNA-seq
DNA-seq Bioinformatics Analysis: Copy Number Variation Elodie Girard elodie.girard@curie.fr U900 institut Curie, INSERM, Mines ParisTech, PSL Research University Paris, France NGS Applications 5C HiC DNA-seq
JULY 21, Genetics 101: SCN1A. Katie Angione, MS CGC Certified Genetic Counselor CHCO Neurology
 JULY 21, 2018 Genetics 101: SCN1A Katie Angione, MS CGC Certified Genetic Counselor CHCO Neurology Disclosures: I have no financial interests or relationships to disclose. Objectives 1. Review genetic
JULY 21, 2018 Genetics 101: SCN1A Katie Angione, MS CGC Certified Genetic Counselor CHCO Neurology Disclosures: I have no financial interests or relationships to disclose. Objectives 1. Review genetic
To test the possible source of the HBV infection outside the study family, we searched the Genbank
 Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers
 Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Nature Neuroscience: doi: /nn Supplementary Figure 1. Missense damaging predictions as a function of allele frequency
 Supplementary Figure 1 Missense damaging predictions as a function of allele frequency Percentage of missense variants classified as damaging by eight different classifiers and a classifier consisting
Supplementary Figure 1 Missense damaging predictions as a function of allele frequency Percentage of missense variants classified as damaging by eight different classifiers and a classifier consisting
IntelliGENSM. Integrated Oncology is making next generation sequencing faster and more accessible to the oncology community.
 IntelliGENSM Integrated Oncology is making next generation sequencing faster and more accessible to the oncology community. NGS TRANSFORMS GENOMIC TESTING Background Cancers may emerge as a result of somatically
IntelliGENSM Integrated Oncology is making next generation sequencing faster and more accessible to the oncology community. NGS TRANSFORMS GENOMIC TESTING Background Cancers may emerge as a result of somatically
Use of panel tests in place of single gene tests in the cancer genetics clinic
 Clin Genet 2015: 88: 278 282 Printed in Singapore. All rights reserved CLINICAL GENETICS doi: 10.1111/cge.12488 Short Report se of panel tests in place of single gene tests in the cancer genetics clinic
Clin Genet 2015: 88: 278 282 Printed in Singapore. All rights reserved CLINICAL GENETICS doi: 10.1111/cge.12488 Short Report se of panel tests in place of single gene tests in the cancer genetics clinic
Variant interpretation exercise. ACGS Somatic Variant Interpretation Workshop Joanne Mason 21/09/18
 Variant interpretation exercise ACGS Somatic Variant Interpretation Workshop Joanne Mason 21/09/18 Format of exercise Compile a list of tricky variants across solid cancer and haematological malignancy.
Variant interpretation exercise ACGS Somatic Variant Interpretation Workshop Joanne Mason 21/09/18 Format of exercise Compile a list of tricky variants across solid cancer and haematological malignancy.
Supplementary Table 3. 3 UTR primer sequences. Primer sequences used to amplify and clone the 3 UTR of each indicated gene are listed.
 Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
