Supplementary Information. Exome sequencing identifies distinct mutational patterns in liver fluke related and noninfection-related
|
|
|
- Lambert Brown
- 5 years ago
- Views:
Transcription
1 Supplementary Information Exome sequencing identifies distinct mutational patterns in liver fluke related and noninfection-related bile duct cancers Waraporn Chan-on 1,2,3*, Maarja-Liisa Nairismägi 1,2*, Choon Kiat Ong 1,2*, Weng Khong Lim 1,2*, Simona Dima 4, Chawalit Pairojkul 3, Kiat Hon Lim 5, John R. McPherson 6, Ioana Cutcutache 6, Hong Lee Heng 1,2, London Ooi 7, Alexander Chung 7, Pierce Chow 7, Peng Chung Cheow 7, Ser Yee Lee 7, Su Pin Choo 8, Iain Bee Huat Tan 8, Dan Duda 9, Anca Nastase 4, Swe Swe Myint 1,2, Bernice Huimin Wong 1,2, Anna Gan 1,2, Vikneswari Rajasegaran 1,2, Cedric Chuan Young Ng 1,2, Sanjanaa Nagarajan 1,2, Apinya Jusakul 1,2, Shenli Zhang 2, Priya Vohra 2, Willie Yu 1,2,10, DaChuan Huang 1,2, Paiboon Sithithaworn 3, Puangrat Yongvanit 3, Sopit Wongkham 3, Narong Khuntikeo 3, Vajaraphongsa Bhudhisawasdi 3, Irinel Popescu 4,#, Steven G. Rozen 6,#, Patrick Tan 2,11,12,#, Bin Tean Teh 1,2,12,# 1 NCCS-VARI Translational Research Laboratory, Division of Medical Sciences, National Cancer Centre Singapore, 11 Hospital Drive, , Singapore 2 Division of Cancer and Stem Cell Biology, Duke-NUS Graduate Medical School, 8 College Road, , Singapore 3 Liver Fluke and Cholangiocarcinoma Research Center, Faculty of Medicine, Khon Kaen University, Khon Kaen, 40002, Thailand 4 Center of Digestive Diseases and Liver Transplantation, Fundeni Clinical Institute, Bucharest, , Romania 5 Department of Pathology, Singapore General Hospital, 20 College Road, , Singapore 6 Division of Neuroscience and Behavioral Disorders, Duke-NUS Graduate Medical School, 8 College Road, , Singapore 7 Department of General Surgery, Singapore General Hospital, 20 College Road, , Singapore 8 Department of Medical Oncology, National Cancer Centre Singapore, 11 Hospital Drive, , Singapore 9 Edwin L. Steele Laboratory for Tumor Biology, Department of Radiation Oncology, Massachusetts General Hospital and Harvard Medical School, MA 02114, Boston, USA 10 National University of Singapore Graduate School for Integrative Sciences and Engineering, 28 Medical Drive, , Singapore 11 Genome Institute of Singapore, 60 Biopolis Street, Genome, , Singapore 12 Cancer Science Institute of Singapore, National University of Singapore, 28 Medical Drive, , Singapore *These authors contributed equally to the work #Co-corresponding authors Correspondence should be addressed to B.T.T. (teh.bin.tean@singhealth.com.sg), P.T. (gmstanp@duke-nus.edu.sg), S.G.R. (steve.rozen@duke-nus.edu.sg) and I.P. (irinel.popescu@icfundeni.ro). 1
2 Index of all supplementary tables and figures Supplementary Tables Supplementary Table 1 Supplementary Table 2 Supplementary Table 3 Supplementary Table 4 Supplementary Table 5 Supplementary Table 6 Supplementary Table 7 Supplementary Table 8 Supplementary Table 9 Supplementary Table 10 Supplementary Table 11 Supplementary Table 12 Supplementary Table 13 Supplementary Table 14 Supplementary Table 15 Supplementary Table 16 Supplementary Table 17 Supplementary Table 18 Clinicopathological characteristics of 101 non-o. viverriniassociated CCA cases in the discovery and the prevalence set Summary of sequencing data obtained from 10 Asian and 5 European non-o. viverrini-associated CCA exomes A list of confirmed nonsynonymous somatic mutations identified in 15 non-o. viverrini-associated samples in the discovery set Recurrently mutated genes identified in 15 samples in discovery set Clinicopathological characteristics of 108 O. viverrini-associated CCA cases in the prevalence set Serum IgG antibody against O. viverrini crude antigen Summary of mutations in KRAS, IDH1 and IDH2 identified using ultra-deep amplicon sequencing compared to Sanger sequencing Mutation frequencies of commonly mutated genes in Asian CCAs stratified by anatomical subtypes Mutation frequencies of commonly mutated genes in Asian and European non-o. viverrini-associated CCAs Frequently mutated genes identified in 101 non-o. viverriniassociated CCAs Frequently mutated genes identified in 108 O. viverriniassociated CCAs Somatic mutations in frequently mutated genes including BAP1, ARID1A, IDH1, IDH2, TP53, KRAS, SMAD4, MLL3, RNF43, GNAS and ROBO2 in non-o. viverrini-associated CCAs in discovery (N = 15) and prevalence set (N = 86) Somatic mutations in frequently mutated genes including BAP1, ARID1A, IDH1, IDH2, TP53, KRAS, SMAD4, MLL3, RNF43, GNAS and ROBO2 in 108 O. viverrini-associated CCAs Sequencing primers for the evaluation of BAP1, IDH1 and IDH2 mutations by Sanger sequencing A list of hotspots in KRAS, IDH1 and IDH2 sequenced by ultradeep amplicon sequencing Sequencing primers for the evaluation of KRAS, IDH1 and IDH2 mutations by ultra-deep amplicon sequencing A list of sirnas A list of antibodies 2
3 Supplementary Figures Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3 Supplementary Figure 4 Supplementary Figure 5 Supplementary Figure 6 Illustration of mutation types in 15 non-o. viverrini-associated CCA exomes Mutation spectra of 15 non-o. viverrini-associated CCAs compared to 8 O. viverrini-associated CCAs Analysis of copy number alterations in 15 discovery samples Enhancement in cell proliferation rates in ARID1A- and BAP1- silenced cells using multiple individual sirnas Quantification of BAP1 transcript levels in BAP1 over-expressing cells IDH1/2 mutations mediate hypermethylator phenotype in CCA tumors 3
4 Supplementary Table 1. Clinicopathological characteristics of 101 non-o. viverrini-associated CCA cases in the discovery and the prevalence set No Sample ID Sex Age Origin Race Anatomical subtype anti-o. viverrini IgG F 77 Singapore Chinese Extrahepatic N/A F 73 Singapore Chinese Intrahepatic N/A F 69 Singapore Chinese Intrahepatic Negative F 61 Singapore Chinese Intrahepatic N/A F 67 Singapore Chinese Intrahepatic N/A F 51 Singapore N/A Intrahepatic N/A M 61 Singapore Chinese Intrahepatic N/A M 51 Singapore N/A Intrahepatic N/A M 55 Singapore Chinese Intrahepatic N/A M 49 Singapore Chinese Intrahepatic N/A M 38 Singapore Chinese Intrahepatic Negative F 52 Singapore Chinese Intrahepatic N/A F 67 Singapore Chinese Intrahepatic N/A F 55 Singapore N/A Intrahepatic N/A F 26 Singapore Chinese Intrahepatic N/A M 60 Singapore Chinese Intrahepatic N/A F 60 Singapore Chinese Intrahepatic N/A F 78 Singapore Chinese Intrahepatic N/A F 61 Singapore Chinese Intrahepatic N/A M 55 Singapore N/A Intrahepatic Positive M 32 Singapore Indian Intrahepatic N/A T M 51 Singapore N/A Intrahepatic Negative M 53 Singapore Chinese Intrahepatic N/A T M 57 Singapore Malay Intrahepatic N/A F 39 Singapore N/A Intrahepatic N/A M 63 Singapore N/A Intrahepatic Negative F 60 Singapore Chinese Intrahepatic N/A M 74 Singapore Chinese Intrahepatic N/A F 74 Singapore N/A Intrahepatic N/A 30 Z321 F 59 Singapore Chinese Intrahepatic N/A 31 Z508 M 63 Singapore Chinese Intrahepatic N/A 32 Z639 M 56 Singapore Chinese Intrahepatic N/A 33 Z2403T F 77 Singapore Chinese Intrahepatic N/A 34 Z2778 M 35 Singapore N/A Intrahepatic Negative 4
5 Supplementary Table 1 (continued). Clinicopathological characteristics of 101 non-o. viverrini-associated CCA cases in the discovery and the prevalence set No Sample ID Sex Age Origin Race Anatomical subtype anti-o. viverrini IgG 35 Z2807T F 76 Singapore Chinese Intrahepatic N/A 36 Z3534T F 61 Singapore Chinese Intrahepatic Positive 37 Z3722 M 75 Singapore Chinese Intrahepatic N/A M 42 Singapore Chinese Extrahepatic N/A M 68 Singapore Chinese Extrahepatic N/A M 47 Singapore Chinese Extrahepatic N/A M 54 Singapore Chinese Extrahepatic N/A M 55 Singapore Chinese Extrahepatic N/A F 45 Singapore Chinese Extrahepatic Negative 44 Z397 M 72 Singapore Chinese Extrahepatic N/A 45 Z1326 M 62 Singapore Chinese Extrahepatic Negative 46 Z2526 F 68 Singapore Chinese Extrahepatic N/A 47 Z2911 F 59 Singapore Chinese Extrahepatic N/A 48 Z3561 F 46 Singapore N/A Extrahepatic N/A 49 Z3585 F 71 Singapore Malay Extrahepatic N/A 50 Z2293 F 70 Singapore Burmese Extrahepatic N/A M 65 Singapore Chinese Extrahepatic Negative M 64 Romania Caucasian Extrahepatic Negative M 68 Romania Caucasian Extrahepatic Negative M 70 Romania Caucasian Extrahepatic Negative F 49 Romania Caucasian Extrahepatic Negative F 69 Romania Caucasian Intrahepatic Negative M 47 Romania Caucasian Extrahepatic Negative M 40 Romania Caucasian Extrahepatic N/A M 49 Romania Caucasian Extrahepatic Negative F 54 Romania Caucasian Extrahepatic Positive M 31 Romania Caucasian Extrahepatic Negative M 44 Romania Caucasian Extrahepatic Positive F 69 Romania Caucasian Extrahepatic Negative M 61 Romania Caucasian Extrahepatic Negative M 58 Romania Caucasian Extrahepatic Negative M 72 Romania Caucasian Extrahepatic Negative M 46 Romania Caucasian Extrahepatic Positive 5
6 Supplementary Table 1 (continued). Clinicopathological characteristics of 101 non-o. viverrini-associated CCA cases in the discovery and the prevalence set No Sample ID Sex Age Origin Race Anatomical subtype anti-o. viverrini IgG M 30 Romania Caucasian Extrahepatic N/A M 51 Romania Caucasian Extrahepatic Negative F 53 Romania Caucasian Extrahepatic Positive M 59 Romania Caucasian Extrahepatic Negative F 65 Romania Caucasian Extrahepatic Negative M 61 Romania Caucasian Extrahepatic Positive F 66 Romania Caucasian Extrahepatic Negative F 58 Romania Caucasian Extrahepatic Positive F 61 Romania Caucasian Extrahepatic Negative F 58 Romania Caucasian Extrahepatic Negative M 70 Romania Caucasian Extrahepatic Negative M 68 Romania Caucasian Extrahepatic Negative F 63 Romania Caucasian Extrahepatic N/A G M 56 Romania Caucasian Extrahepatic N/A I F 50 Romania Caucasian Intrahepatic N/A P F 45 Romania Caucasian Intrahepatic Negative C F 62 Romania Caucasian Intrahepatic Negative M 45 Romania Caucasian Intrahepatic Negative M 64 Romania Caucasian Intrahepatic Negative M 65 Romania Caucasian Intrahepatic Negative F 61 Romania Caucasian Intrahepatic N/A F 49 Romania Caucasian Intrahepatic Negative F 44 Romania Caucasian Intrahepatic N/A M 50 Romania Caucasian Intrahepatic Negative M 62 Romania Caucasian Intrahepatic Negative M 69 Romania Caucasian Intrahepatic Negative F 75 Romania Caucasian Intrahepatic Positive F 66 Romania Caucasian Intrahepatic Negative F 52 Romania Caucasian Intrahepatic Negative M 66 Romania Caucasian Intrahepatic Negative M 72 Romania Caucasian Intrahepatic Negative M 49 Romania Caucasian Intrahepatic Negative M 78 Romania Caucasian Intrahepatic Negative F 75 Romania Caucasian Intrahepatic Negative Samples in the discovery set are highlighted. N/A., Not available. 6
7 Supplementary Table 2. Summary of sequencing data obtained from 10 Asian and 5 European non-o. viverrini-associated CCA exomes Pair Sample Sample Type Origin Bases in Target Region* SureSelect Capturing Kit Reads Mapped to Target Region** Ave. Depth Per Targeted Base Targeted Bases with Depth at Least 1X (%) Targeted Bases with Depth at Least 20X (%) Validated somatic mutations N Normal Singapore 37,792,881 V1 49,026, T Tumor Singapore 37,792,881 V1 53,913, N Normal Singapore 37,792,881 V1 48,913, T Tumor Singapore 37,792,881 V1 47,228, N Normal Singapore 37,792,881 V1 127,502, T Tumor Singapore 37,792,881 V1 42,091, N Normal Singapore 37,792,881 V1 55,375, T Tumor Singapore 37,792,881 V1 60,491, N Normal Singapore 37,792,881 V1 53,653, T Tumor Singapore 37,792,881 V1 59,293, N Normal Singapore 37,792,881 V1 58,741, T Tumor Singapore 37,792,881 V1 60,170, N Normal Singapore 37,792,881 V4+UTR 45,643, T Tumor Singapore 37,792,881 V4+UTR 42,266, N Normal Singapore 37,792,881 V4+UTR 47,268, T Tumor Singapore 37,792,881 V4+UTR 42,924, N Normal Singapore 37,792,881 V4+UTR 49,944, T Tumor Singapore 37,792,881 V4+UTR 49,684, N Normal Singapore 37,792,881 V4+UTR 126,824, T Tumor Singapore 37,792,881 V4+UTR 119,615,
8 Supplementary Table 2 (continued). Summary of sequencing data obtained from 10 Asian and 5 European non-o. viverriniassociated CCA exomes Pair Sample Sample Type Origin Bases in Target Region* SureSelect Capturing Kit Reads Mapped to Target Region** Ave. Depth Per Targeted Base Targeted Bases with Depth at Least 1X (%) Targeted Bases with Depth at Least 20X (%) Validated somatic mutations 0715N Normal Romania 37,792,881 V1 51,976, T Tumor Romania 37,792,881 V1 52,105, N Normal Romania 37,792,881 V1 53,318, T Tumor Romania 37,792,881 V1 52,342, N Normal Romania 37,792,881 V1 55,348, T Tumor Romania 37,792,881 V1 52,291, N Normal Romania 37,792,881 V1 51,923, T Tumor Romania 37,792,881 V1 50,976, N Normal Romania 37,792,881 V4+UTR 59,435, T Tumor Romania 37,792,881 V4+UTR 54,651, Average 37,792,881 59,164, *Sequencing data of all non-o. viverrini-associated CCA samples was mapped to the same target reference (37,792,881) regardless of exome capturing version. ** Based on the NCBI human reference genome GRCh37 (hg19) 8
9 Supplementary Table 3. A list of confirmed nonsynonymous somatic mutations identified in 15 non-o. viverrini-associated samples in the discovery set No Gene Symbol Sample Transcript ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 1 ACCN CCDS g.chr7: G>A c.389 G>A p.g130d Missense 2 ACVR2A CCDS g.chr2: insa c.1303 insa fs Indel 3 ADAMTS CCDS g.chr9: A>G c.664 A>G p.i222v Missense 4 AGPAT CCDS g.chr8: G>A c.382 G>A p.d128n Missense 5 AGPAT CCDS g.chr8: A>G c.1220 A>G p.k407r Missense 6 AGTPBP CCDS g.chr9: C>G c.2326 G>C p.v776l Missense 7 ALDH1L CCDS g.chr3: C>T c.769 G>A p.e257k Missense 8 ANKAR CCDS g.chr2: G>T c.517 G>T p.e173x Nonsense 9 ANO CCDS g.chr12: G>A c.1913 G>A p.r638k Missense 10 AP1AR 1202 CCDS g.chr4: G>T c.395 G>T p.r132i Missense 11 APOB CCDS g.chr2: G>T c.9881 C>A p.s3294x Nonsense 12 ARHGAP CCDS g.chr10: T>C c.266 A>G p.n89s Missense 13 ARHGAP CCDS g.chr2: T>C c.533 T>C p.l178s Missense 14 ARHGEF CCDS g.chr2: G>A c.1949 G>A p.r650h Missense 15 ARID1A 3001 CCDS285.1 g.chr1: G>T c.2791 G>T p.g931x Nonsense 16 ARID1A CCDS285.1 g.chr1: insa c.3217 insa fs Indel 17 ARID1A CCDS285.1 g.chr1: G>T IVS8+1 G>T Splice site Splice site 18 ARID1A CCDS285.1 g.chr1: delc c.1558 delc fs Indel 19 ASAP1 715 CCDS g.chr8: C>G c.1483 G>C p.e495q Missense 20 ATP10A 1202 CCDS g.chr15: A>T c.3220 T>A p.l1074m Missense 21 ATP10A CCDS g.chr15: T>C c.2720 A>G p.d907g Missense 22 ATP8B4 824 CCDS g.chr15: G>C c.1588 C>G p.q530e Missense 23 ATRN CCDS g.chr20: A>C c.1118 A>C p.d373a Missense 24 BAP CCDS g.chr3: c.826_832 delcttgaga deltctcaag fs Indel 25 BAP CCDS g.chr3: delt c.1511 dela fs Indel 9
10 Supplementary Table 3 (continued). A list of confirmed nonsynonymous somatic mutations identified in 15 non-o. viverriniassociated samples in the discovery set No Gene Symbol Sample Transcript ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 26 BAP CCDS g.chr3: inst c.1360 insa fs Indel 27 BAP CCDS g.chr3: G>T c.1881 C>A p.y627x Nonsense 28 BDH CCDS g.chr3: G>A c.331 C>T p.q111x Nonsense 29 BMPR CCDS g.chr2: A>G c.526 A>G p.t176a Missense 30 BPTF CCDS g.chr17: C>T c.8572 C>T p.r2858x Nonsense 31 BRPF3 824 CCDS g.chr6: C>T c.1132 C>T p.r378c Missense 32 BRPF CCDS g.chr6: C>T c.2537 C>T p.s846f Missense 33 C11orf CCDS g.chr11: G>C c.2094 G>C p.q698h Missense 34 C12orf ENST g.chr12: T>A c.943 T>A p.c315s Missense 35 C12orf CCDS g.chr12: T>A c.865 T>A p.c289s Missense 36 C15orf CCDS g.chr15: G>T c.2363 G>T p.s788i Missense 37 C19orf ENST g.chr19: insg c.1100 insc fs Indel 38 C2orf CCDS g.chr2: G>T c.1231 G>T p.g411c Missense 39 C3orf CCDS g.chr3: G>T c.182 G>T p.s61i Missense 40 C3orf CCDS g.chr3: c.4272_4278 delctgaagt delacttcag fs Indel 41 C4A 3001 CCDS g.chr6: deltc c.3321_3322 deltc fs Indel 42 C7orf CCDS g.chr7: C>T c.671 G>A p.g224d Missense 43 CAMTA CCDS g.chr1: C>T c.3377 C>T p.a1126v Missense 44 CARD CCDS g.chr17: C>T c.451 C>T p.r151w Missense 45 CCDC CCDS g.chr15: G>A c.956 G>A p.r319h Missense 46 CCT8L CCDS g.chr22: C>T c.322 G>A p.v108m Missense 47 CCT8L CCDS g.chr22: C>T c.751 G>A p.a251t Missense 48 CDC CCDS734.1 g.chr1: C>T c.1105 C>T p.q369x Nonsense 49 CHD1L CCDS927.1 g.chr1: G>A c.1716 G>A p.m572i Missense 50 CHPF CCDS g.chr7: G>T c.2237 G>T p.r746l Missense 10
11 Supplementary Table 3 (continued). A list of confirmed nonsynonymous somatic mutations identified in 15 non-o. viverriniassociated samples in the discovery set No Gene Symbol Sample Transcript ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 51 CHST CCDS g.chr16: A>T c.859 A>T p.t287s Missense 52 CLCN7 824 CCDS g.chr16: C>T c.1307 G>A p.r436q Missense 53 CLK2 824 CCDS g.chr1: G>A c.139 C>T p.r47x Nonsense 54 CLPS 1202 CCDS g.chr6: T>A c.80 A>T p.n27i Missense 55 CLSPN CCDS396.1 g.chr1: T>C c.3305 A>G p.h1102r Missense 56 CNBP CCDS g.chr3: G>C c.477 C>G p.n159k Missense 57 CNTN6 824 CCDS g.chr3: G>A c.733 G>A p.v245i Missense 58 COL1A2 824 CCDS g.chr7: G>A c.2330 G>A p.r777h Missense 59 COLEC CCDS g.chr8: C>T c.214 C>T p.p72s Missense 60 CPXM CCDS g.chr20: C>T c.2183 G>A p.r728q Missense 61 CREB3L CCDS g.chr1: G>C c.173 G>C p.c58s Missense 62 CSRP2 824 CCDS g.chr12: G>T c.47 C>A p.t16n Missense 63 CTCF CCDS g.chr16: C>T c.1061 C>T p.s354f Missense 64 CTNND CCDS g.chr5: G>C c.517 C>G p.h173d Missense 65 CTSW 1202 CCDS g.chr11: C>A c.103 C>A p.p35t Missense 66 CWH CCDS g.chr4: A>G c.490 A>G p.t164a Missense 67 CXorf CCDS g.chrx: C>A c.443 G>T p.r148l Missense 68 CYB5R CCDS g.chr22: T>G c.732 A>C p.e244d Missense 69 CYLD 824 CCDS g.chr16: T>C IVS4-4 T>C Splice site Splice site 70 CYP4A CCDS g.chr1: C>T c.155 C>T p.p52l Missense 71 DAPK CCDS g.chr19: C>T c.1261 G>A p.e421k Missense 72 DCX CCDS g.chrx: T>C IVS2 +4 A>G Splice site Splice site 73 DDX CCDS g.chr10: G>A IVS5+1 G>A Splice site Splice site 74 DGCR6L CCDS g.chr22: G>A c.322 C>T p.r108w Missense 75 DLG CCDS g.chr3: T>C c.1859 A>G p.d620g Missense 11
12 Supplementary Table 3 (continued). A list of confirmed nonsynonymous somatic mutations identified in 15 non-o. viverriniassociated samples in the discovery set No Gene Symbol Sample Transcript ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 76 DMXL CCDS g.chr15: G>A c.8971 C>T p.r2991x Nonsense 77 DSEL 824 CCDS g.chr18: C>T c.3547 G>A p.e1183k Missense 78 DUXA CCDS g.chr19: inst c.64 insa fs Indel 79 DZIP3 824 CCDS g.chr3: A>C c.2243 A>C p.k748t Missense 80 EGR CCDS g.chr5: C>G c.56 C>G p.p19r Missense 81 EIF4E2 824 CCDS g.chr2: G>C c.443 G>C p.w148s Missense 82 ELF CCDS g.chr1: delactg c.123_126 delactg fs Indel 83 ELTD CCDS g.chr1: C>T c.592 G>A p.v198m Missense 84 ENOPH CCDS g.chr4: C>T c.340 C>T p.q114x Nonsense 85 EYA CCDS g.chr8: G>T c.144 C>A p.s48r Missense 86 F CCDS g.chrx: C>G c.4934 G>C p.w1645s Missense 87 FBN CCDS g.chr5: G>A c.1151 C>T p.t384m Missense 88 FBN CCDS g.chr19: T>A c.943 A>T p.r315w Missense 89 FBXW7 824 CCDS g.chr4: G>A c.1393 C>T p.r465c Missense 90 FGF CCDS g.chr13: C>G c.271 C>G p.r91g Missense 91 FLNA CCDS g.chrx: C>T c.973 G>A p.g325r Missense 92 FLNC CCDS g.chr7: C>T c.1208 C>T p.a403v Missense 93 FNTB 824 CCDS g.chr14: G>T c.225 G>T p.r75s Missense 94 FRMD4A 1202 CCDS g.chr10: C>T c.923 G>A p.w308x Nonsense 95 FSCN CCDS g.chr7: G>A c.341 G>A p.r114h Missense 96 FZD CCDS g.chr17: G>T c.1595 G>T p.g532v Missense 97 GAPDH CCDS g.chr12: G>T c.103 G>T p.d35y Missense 98 GCDH CCDS g.chr19: G>C c.121 G>C p.a41p Missense 99 GCFC CCDS g.chr21: deltga c.463_465 deltca In frame Indel 100 GDPD5 930 CCDS g.chr11: G>A c.700 C>T p.r234c Missense 12
13 Supplementary Table 3 (continued). A list of confirmed nonsynonymous somatic mutations identified in 15 non-o. viverriniassociated samples in the discovery set No Gene Symbol Sample Transcript ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 101 GFRA1 824 CCDS g.chr10: G>C c.1232 C>G p.t411r Missense 102 GIT CCDS g.chr12: G>A c.271 C>T p.r91c Missense 103 GLI CCDS g.chr7: G>T c.3609 C>A p.n1203k Missense 104 GLRB 1202 CCDS g.chr4: T>A c.908 T>A p.i303n Missense 105 GLYATL CCDS g.chr11: G>A IVS1+5 C>T Splice site Splice site 106 GNPAT 1202 CCDS g.chr1: delg c.780 delg fs Indel 107 GPBP1L CCDS528.1 g.chr1: T>C c.572 A>G p.q191r Missense 108 GPR CCDS g.chrx: T>A c.5320 T>A p.f1774i Missense 109 GPR CCDS g.chrx: A>G c.6719 A>G p.k2240r Missense 110 GRID2 824 CCDS g.chr4: A>G c.187 A>G p.i63v Missense 111 GRIN2A CCDS g.chr16: G>T c.3145 C>A p.p1049t Missense 112 HECTD CCDS g.chr1: C>T c.2509 G>A p.v837i Missense 113 HIF3A CCDS g.chr19: G>A c.1198 G>A p.a400t Missense 114 HMCN CCDS g.chr1: C>G c.2074 C>G p.q692e Missense 115 HMCN CCDS g.chr1: T>C c.3272 T>C p.v1091a Missense 116 HMGCR CCDS g.chr5: delc c.1723 delc fs Indel 117 HSPG CCDS g.chr1: C>A c G>T p.c3871f Missense 118 IDH CCDS g.chr2: G>A c.394 C>T p.r132c Missense 119 IDH CCDS g.chr2: G>A c.394 C>T p.r132c Missense 120 IDH CCDS g.chr2: G>A c.394 C>T p.r132c Missense 121 IGSF CCDS g.chr3: T>C c.3910 A>G p.k1304e Missense 122 IGSF CCDS g.chr1: T>C c.398 A>G p.n133s Missense 123 IGSF CCDS g.chr1: C>T c.682 G>A p.v228i Missense 124 ITGB CCDS g.chr7: G>A c.1906 G>A p.e636k Missense 125 IVNS1ABP CCDS g.chr1: T>G c.653 A>C p.e218a Missense 13
14 Supplementary Table 3 (continued). A list of confirmed nonsynonymous somatic mutations identified in 15 non-o. viverriniassociated samples in the discovery set No Gene Symbol Sample Transcript ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 126 KCNQ CCDS g.chr8: C>A c.1354 G>T p.a452s Missense 127 KLHL CCDS g.chr16: C>T c.1229 C>T p.a410v Missense 128 KRAS 930 CCDS g.chr12: C>A c.35 G>T p.g12v Missense 129 KRAS 1202 CCDS g.chr12: C>T c.35 G>A p.g12d Missense 130 KRAS CCDS g.chr12: C>T c.35 G>A p.g12d Missense 131 LMO CCDS g.chr13: G>T c.2653 G>T p.a885s Missense 132 LRCH CCDS g.chr3: delatc c.326_328 delatc In frame Indel 133 LRRIQ CCDS g.chr12: G>T IVS6-1 G>T Splice site Splice site 134 LRRIQ CCDS g.chr12: A>T c.5137 A>T p.s1713c Missense 135 LYVE CCDS g.chr11: inst c.876 insa fs Indel 136 MAGEC CCDS g.chrx: A>C c.3138 A>C p.k1046n Missense 137 MAP2K1 715 CCDS g.chr15: T>G c.357 T>G p.h119q Missense 138 MCM ENST g.chr22: A>G IVS5-2 A>G Splice site Splice site 139 MORC CCDS g.chr3: C>A c.337 G>T p.d113y Missense 140 MPO 824 CCDS g.chr17: G>T c.1078 C>A p.l360m Missense 141 MUC ENST g.chr3: G>A c.344 C>T p.a115v Missense 142 MUC CCDS g.chr7: A>G c A>G p.m4294v Missense 143 MYEOV CCDS g.chr11: G>A c.626 G>A p.g209e Missense 144 MYH CCDS g.chr14: inscg c.4085 insgc fs Indel 145 MYH CCDS g.chr14: G>A c.5134 C>T p.r1712w Missense 146 NAP1L CCDS g.chrx: G>T c.1336 C>A p.p446t Missense 147 NELL CCDS g.chr11: C>A c.1235 C>A p.t412k Missense 148 NLGN CCDS g.chr3: C>G c.450 C>G p.s150r Missense 149 NLGN4X CCDS g.chr: C>T c.1435 G>A p.e479k Missense 150 NLRP3 824 CCDS g.chr1: G>A c.1390 G>A p.a464t Missense 14
15 Supplementary Table 3 (continued). A list of confirmed nonsynonymous somatic mutations identified in 15 non-o. viverriniassociated samples in the discovery set No Gene Symbol Sample Transcript ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 151 NNT CCDS g.chr5: A>G c.2056 A>G p.i686v Missense 152 OR10J CCDS g.chr1: T>G c.604 A>C p.s202r Missense 153 OR13H CCDS g.chrx: delc c.855 delc fs Indel 154 OR13H CCDS g.chrx: C>A c.856 C>A p.p286t Missense 155 OR1C CCDS g.chr1: G>A c.166 C>T p.h56y Missense 156 OR4A5 824 CCDS g.chr11: G>A c.664 C>T p.l222f Missense 157 OR5B CCDS g.chr11: G>A c.250 C>T p.r84w Missense 158 OR8B CCDS g.chr11: C>G c.226 G>C p.v76l Missense 159 OTX CCDS g.chr2: C>T c.286 C>T p.q96x Nonsense 160 PALLD 824 CCDS g.chr4: T>C c.2635 T>C p.c879r Missense 161 PARL CCDS g.chr3: T>A c.469 A>T p.k157x Nonsense 162 PCDH CCDS g.chr5: G>A c.2405 C>T p.a802v Missense 163 PCDHA CCDS g.chr5: G>A c.1117 G>A p.v373i Missense 164 PCDHA CCDS g.chr5: G>A c.191 G>A p.r64q Missense 165 PCSK2 930 CCDS g.chr20: C>T c.844 C>T p.r282w Missense 166 PDP CCDS g.chr16: C>G c.1225 C>G p.l409v Missense 167 PITPNM CCDS g.chr11: C>G c.3423 G>C p.q1141h Missense 168 PIWIL CCDS g.chr12: C>G c.374 C>G p.p125r Missense 169 PIWIL CCDS g.chr11: T>G c.2370 T>G p.d790e Missense 170 PKNOX CCDS g.chr11: A>G c.536 A>G p.d179g Missense 171 PLCE1 824 CCDS g.chr10: G>A c.1600 G>A p.e534k Missense 172 PLXNA CCDS g.chr7: G>A c.4108 C>T p.r1370c Missense 173 POLB 1202 CCDS g.chr8: C>T c.64 C>T p.l22f Missense 174 PPFIBP1 824 CCDS g.chr12: C>T IVS16+4 C>T Splice site Splice site 175 PREX2 824 CCDS g.chr8: G>T c.3915 G>T p.r1305s Missense 15
16 Supplementary Table 3 (continued). A list of confirmed nonsynonymous somatic mutations identified in 15 non-o. viverriniassociated samples in the discovery set No Gene Symbol Sample Transcript ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 176 PRG CCDS g.chr1: A>G c.2180 A>G p.k727r Missense 177 PRMT CCDS g.chr16: G>T c.1644 G>T p.m548i Missense 178 PRR CCDS g.chr5: C>T c.503 C>T p.p168l Missense 179 PRRT CCDS g.chr6: G>A c.557 C>T p.t186m Missense 180 PTPN6 824 CCDS g.chr12: G>A c.310 G>A p.d104n Missense 181 PTPRC 1202 CCDS g.chr1: G>A c.1022 G>A p.r341k Missense 182 RABEP2 824 CCDS g.chr16: C>T c.721 G>A p.g241s Missense 183 RB CCDS g.chr13: C>T c.1654 C>T p.r552x Nonsense 184 RBM CCDS g.chrx: delg c.1482 delg fs Indel 185 RBM CCDS g.chr12: T>A IVS5-2 A>T Splice site Splice site 186 RGS CCDS g.chr1: G>C c.688 G>C p.d230h Missense 187 RGS CCDS g.chr1: C>A c.874 G>T p.e292x Nonsense 188 RHOBTB CCDS g.chr5: C>A c.144 C>A p.s48r Missense 189 RNF CCDS g.chr2: G>A c.712 C>T p.r238c Missense 190 RPL3L 3001 CCDS g.chr16: T>A c.335 A>T p.d112v Missense 191 RTEL CCDS g.chr20: G>T c.1080 G>T p.q360h Missense 192 RWDD CCDS g.chr6: G>C c.184 G>C p.d62h Missense 193 SAE CCDS g.chr19: G>A c.631 G>A p.v247m Missense 194 SBNO CCDS g.chr12: G>T c.1603 C>A p.l535m Missense 195 SCEL 824 CCDS g.chr13: A>T c.23 A>T p.k8i Missense 196 SCN11A 1202 CCDS g.chr3: T>G c.5129 A>C p.k1710t Missense 197 SCRN CCDS g.chr17: C>A c.176 G>T p.c59f Missense 198 SFSWAP CCDS g.chr12: C>T c.673 C>T p.q225x Nonsense 199 SH2D CCDS g.chr1: G>T c.417 C>A p.c139x Nonsense 200 SLC12A CCDS g.chr7: G>A c.704 G>A p.r235q Missense 16
17 Supplementary Table 3 (continued). A list of confirmed nonsynonymous somatic mutations identified in 15 non-o. viverriniassociated samples in the discovery set No Gene Symbol Sample Transcript ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 201 SLC13A5 824 CCDS g.chr17: G>C c.1430 C>G p.a477g Missense 202 SLC18A CCDS g.chr8: C>T c.1484 G>A p.c495y Missense 203 SLC20A CCDS g.chr8: C>T c.848 G>A p.s283n Missense 204 SLC35C CCDS g.chr20: G>A c.525 C>T p.s130l Missense 205 SLC36A CCDS g.chr5: T>A c.943 A>T p.k315x Nonsense 206 SLC4A CCDS g.chr3: G>C c.1527 C>G p.d509e Missense 207 SLC5A CCDS g.chr11: A>G IVS7+2 T>C Splice site Splice site 208 SMARCA CCDS g.chr19: G>A c.92 G>A p.s31n Missense 209 SORCS1 824 CCDS g.chr10: T>A c.578 A>T p.k193m Missense 210 SPA CCDS g.chr11: G>T c.289 G>T p.e97x Nonsense 211 SPINK CCDS g.chr5: C>T c.1196 C>T p.t399i Missense 212 SPP CCDS g.chr4: A>C c.518 A>C p.k173t Missense 213 SYMPK CCDS g.chr19: T>C c.527 A>G p.n176s Missense 214 SYNE CCDS g.chr6: T>G c.9451 A>C p.k3151q Missense 215 SYNE CCDS g.chr14: G>T c G>T p.d6608y Missense 216 TAB CCDS g.chrx: G>T c.370 C>A p.p124t Missense 217 TDRD6 824 CCDS g.chr6: G>A c.440 C>T p.p147l Missense 218 TERT CCDS g.chr5: C>T c.2852 G>A p.r951q Missense 219 TGFBR CCDS g.chr9: T>G c.194 T>G p.v65g Missense 220 TLL CCDS g.chr10: G>T c.2805 C>A p.f935l Missense 221 TMEM CCDS g.chr17: G>C c.157 C>G p.l53v Missense 222 TMEM CCDS g.chr9: A>T c.3478 T>A p.s1160t Missense 223 TNIP CCDS g.chr4: T>C c.988 A>G p.k330e Missense 224 TP CCDS g.chr17: C>A c.814 G>T p.v272l Missense 225 TP CCDS g.chr17: C>T c.473 G>A p.r158h Missense 17
18 Supplementary Table 3 (continued). A list of confirmed nonsynonymous somatic mutations identified in 15 non-o. viverriniassociated samples in the discovery set No Gene Symbol Sample Transcript ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 226 TP CCDS g.chr17: delg c.80 delc fs Indel 227 TPO CCDS g.chr2: G>C IVS6+1 G>C Splice site Splice site 228 TPP CCDS g.chr13: G>T c.2215 G>T p.a739s Missense 229 TRIM CCDS376.1 g.chr1: C>T c.421 G>A p.d141n Missense 230 TRPM CCDS g.chr9: C>T c.2245 G>A p.a749t Missense 231 TSPAN CCDS g.chr10: G>T c.11 G>T p.g4v Missense 232 TSR CCDS g.chr16: A>G c.487 T>C p.s163p Missense 233 TTBK CCDS g.chr6: C>T c.772 C>T p.r258w Missense 234 UGT2B CCDS g.chr4: A>T c.593 T>A p.m198k Missense 235 UPP CCDS g.chr2: G>T c.509 G>T p.s170i Missense 236 VPS13B 824 CCDS g.chr8: C>T c.1193 C>T p.t398m Missense 237 VWF 930 CCDS g.chr12: G>A c.6908 C>T p.t2303m Missense 238 XIRP CCDS g.chr2: C>G c.9661 C>G p.l3221v Missense 239 ZBTB CCDS g.chr11: insc c.350 insg fs Indel 240 ZHX2 715 CCDS g.chr8: C>G c.2285 C>G p.a762g Missense 241 ZMYM3 930 CCDS g.chrx: C>A c.232 G>T p.e78x Nonsense 242 ZNF CCDS g.chr6: T>A c.218 A>T p.k73i Missense 243 ZNF CCDS g.chr9: G>A c.1955 G>A p.r652h Missense 244 ZNF CCDS731.1 g.chr1: T>C c.557 A>G p.n186s Missense 245 ZNF CCDS g.chr15: G>T c.1924 G>T p.d642y Missense 18
19 Supplementary Table 4. Recurrently mutated genes identified in 15 samples in discovery set No Gene Symbol Sample Transcript ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type AGPAT CCDS g.chr8: G>A c.382 G>A p.d128n Missense AGPAT CCDS g.chr8: A>G c.1220 A>G p.k407r Missense ARID1A 3001 CCDS285.1 g.chr1: G>T c.2791 G>T p.g931x Nonsense ARID1A CCDS285.1 g.chr1: delc c.1558 delc fs Indel ARID1A CCDS285.1 g.chr1: insa c.3217 insa fs Indel ARID1A CCDS285.1 g.chr1: G>T IVS8+1 G>T Splice site Splice site ATP10A 1202 CCDS g.chr15: A>T c.3220 T>A p.l1074m Missense ATP10A CCDS g.chr15: T>C c.2720 A>G p.d907g Missense BAP CCDS g.chr3: delcttgaga c.826_832 deltctcaag fs Indel BAP CCDS g.chr3: delt c.1511 dela fs Indel BAP CCDS g.chr3: inst c.1360 insa fs Indel BAP CCDS g.chr3: G>T c.1881 C>A p.y627x Nonsense BRPF3 824 CCDS g.chr6: C>T c.1132 C>T p.r378c Missense BRPF CCDS g.chr6: C>T c.2537 C>T p.s846f Missense CCT8L CCDS g.chr22: C>T c.322 G>A p.v108m Missense CCT8L CCDS g.chr22: C>T c.751 G>A p.a251t Missense GPR CCDS g.chrx: T>A c.5320 T>A p.f1774i Missense GPR CCDS g.chrx: A>G c.6719 A>G p.k2240r Missense HMCN CCDS g.chr1: C>G c.2074 C>G p.q692e Missense HMCN CCDS g.chr1: T>C c.3272 T>C p.v1091a Missense IDH CCDS g.chr2: G>A c.394 C>T p.r132c Missense IDH CCDS g.chr2: G>A c.394 C>T p.r132c Missense IDH CCDS g.chr2: G>A c.394 C>T p.r132c Missense KRAS 930 CCDS g.chr12: C>A c.35 G>T p.g12v Missense KRAS 1202 CCDS g.chr12: C>T c.35 G>A p.g12d Missense KRAS CCDS g.chr12: C>T c.35 G>A p.g12d Missense LRRIQ CCDS g.chr12: A>T c.5137 A>T p.s1713c Missense LRRIQ CCDS g.chr12: G>T IVS6-1 G>T Splice site Splice site TP CCDS g.chr17: C>A c.814 G>T p.v272l Missense TP CCDS g.chr17: delg c.80 delc fs Indel TP CCDS g.chr17: C>T c.473 G>A p.r158h Missense 19
20 Supplementary Table 5. Clinicopathological characteristics of 108 O. viverrini-associated CCA cases in the prevalence set No Sample ID Sex Age Origin Race Anatomical subtype anti-o. viverrini IgG 1 A028 F 59 Thailand Thai Intrahepatic Positive 2 A035 F 49 Thailand Thai Intrahepatic Positive 3 A039 F 72 Thailand Thai Intrahepatic Negative 4 A042 F 52 Thailand Thai Intrahepatic Positive 5 A043 M 45 Thailand Thai Intrahepatic Positive 6 A074 M 55 Thailand Thai Intrahepatic Positive 7 A105 M 53 Thailand Thai Intrahepatic Positive 8 A106 M 51 Thailand Thai Intrahepatic Positive 9 A107 M 54 Thailand Thai Intrahepatic Positive 10 A119 F 51 Thailand Thai Intrahepatic Positive 11 A120 F 53 Thailand Thai Intrahepatic Positive 12 A128 M 61 Thailand Thai Intrahepatic Positive 13 A142 F 38 Thailand Thai Intrahepatic N/A 14 A162 M 68 Thailand Thai Intrahepatic N/A 15 B032 M 53 Thailand Thai Intrahepatic N/A 16 B048 F 51 Thailand Thai Intrahepatic Positive 17 B083 F 53 Thailand Thai Intrahepatic Positive 18 B085 M 73 Thailand Thai Intrahepatic Negative 19 B087 M 56 Thailand Thai Intrahepatic Positive 20 B099 M 48 Thailand Thai Intrahepatic Positive 21 B113 M 61 Thailand Thai Intrahepatic Positive 22 R104 F 64 Thailand Thai Intrahepatic N/A 23 R134 M 50 Thailand Thai Intrahepatic N/A 24 T003 M 52 Thailand Thai Intrahepatic N/A 25 U027 F 66 Thailand Thai Intrahepatic Positive 26 W012 F 61 Thailand Thai Intrahepatic Positive 27 W039 M 66 Thailand Thai Intrahepatic N/A 28 W040 M 56 Thailand Thai Intrahepatic N/A 29 Y002 F 45 Thailand Thai Intrahepatic Positive 30 Y008 F 65 Thailand Thai Intrahepatic Positive 31 Y019 M 70 Thailand Thai Intrahepatic Negative 32 Y020 M 50 Thailand Thai Intrahepatic Positive 33 Y023 M 62 Thailand Thai Intrahepatic Positive 34 Y032 M 69 Thailand Thai Intrahepatic Positive 35 Y033 F 51 Thailand Thai Intrahepatic Positive 36 Y035 F 64 Thailand Thai Intrahepatic Positive 20
21 Supplementary Table 5 (continued). Clinicopathological characteristics of 108 O. viverriniassociated CCA cases in the prevalence set No Sample ID Sex Age Origin Race Anatomical subtype anti-o. viverrini IgG 37 Y057 M 57 Thailand Thai Intrahepatic Negative 38 Y065 M 63 Thailand Thai Intrahepatic N/A 39 Y072 M 61 Thailand Thai Intrahepatic Positive 40 Y074 F 40 Thailand Thai Intrahepatic Positive 41 Y140 F 56 Thailand Thai Intrahepatic Positive 42 Y149 M 69 Thailand Thai Intrahepatic Positive 43 C004 M 53 Thailand Thai Intrahepatic Positive 44 C008 M 47 Thailand Thai Intrahepatic N/A 45 C054 M 44 Thailand Thai Intrahepatic N/A 46 C078 M 42 Thailand Thai Intrahepatic Positive 47 C080 F 69 Thailand Thai Intrahepatic Positive 48 C084 M 62 Thailand Thai Intrahepatic Positive 49 C095 M 56 Thailand Thai Intrahepatic Positive 50 C096 F 79 Thailand Thai Intrahepatic Positive 51 C105 F 48 Thailand Thai Intrahepatic Positive 52 C107 F 49 Thailand Thai Intrahepatic Positive 53 C110 M 59 Thailand Thai Intrahepatic Negative 54 C128 F 43 Thailand Thai Intrahepatic Negative 55 C144 M 52 Thailand Thai Intrahepatic Positive 56 C151 M 73 Thailand Thai Intrahepatic Positive 57 C154 M 59 Thailand Thai Intrahepatic N/A 58 C164 F 60 Thailand Thai Intrahepatic Negative 59 C167 M 64 Thailand Thai Intrahepatic Positive 60 D017 F 52 Thailand Thai Intrahepatic Negative 61 D029 M 64 Thailand Thai Intrahepatic N/A 62 D039 F 66 Thailand Thai Intrahepatic N/A 63 A159 M 55 Thailand Thai Extrahepatic N/A 64 B070 M 61 Thailand Thai Extrahepatic Positive 65 B149 M 63 Thailand Thai Extrahepatic Positive 66 R100 F 49 Thailand Thai Extrahepatic N/A 67 R149 M 37 Thailand Thai Extrahepatic N/A 68 T026 M 52 Thailand Thai Extrahepatic N/A 69 T151 M 73 Thailand Thai Extrahepatic Positive 70 T157 M 42 Thailand Thai Extrahepatic Positive 71 T160 M 46 Thailand Thai Extrahepatic Positive 72 U044 M 57 Thailand Thai Extrahepatic N/A 21
22 Supplementary Table 5 (continued). Clinicopathological characteristics of 108 O. viverriniassociated CCA cases in the prevalence set No Sample ID Sex Age Origin Race Anatomical subtype anti-o. viverrini IgG 73 Y091 F 51 Thailand Thai Extrahepatic Positive 74 Y123 M 55 Thailand Thai Extrahepatic Positive 75 A003 F 56 Thailand Thai Extrahepatic Positive 76 A004 M 52 Thailand Thai Extrahepatic Negative 77 A011 M 53 Thailand Thai Extrahepatic Positive 78 A026 M 52 Thailand Thai Extrahepatic Positive 79 A059 M 46 Thailand Thai Extrahepatic Positive 80 A064 M 49 Thailand Thai Extrahepatic Negative 81 A072 M 52 Thailand Thai Extrahepatic Negative 82 A080 M 50 Thailand Thai Extrahepatic Positive 83 A096 F 76 Thailand Thai Extrahepatic Positive 84 A100 M 53 Thailand Thai Extrahepatic Positive 85 A114 M 76 Thailand Thai Extrahepatic Positive 86 A118 F 63 Thailand Thai Extrahepatic Positive 87 A130 M 54 Thailand Thai Extrahepatic Positive 88 A132 M 61 Thailand Thai Extrahepatic Negative 89 A150 M 64 Thailand Thai Extrahepatic Positive 90 A153 M 60 Thailand Thai Extrahepatic Positive 91 A157 M 51 Thailand Thai Extrahepatic Positive 92 A169 M 66 Thailand Thai Extrahepatic Negative 93 B005 M 50 Thailand Thai Extrahepatic Positive 94 B011 M 60 Thailand Thai Extrahepatic Positive 95 B017 M 53 Thailand Thai Extrahepatic Positive 96 B027 M 57 Thailand Thai Extrahepatic Positive 97 B103 M 63 Thailand Thai Extrahepatic Positive 98 C006 M 62 Thailand Thai Extrahepatic Positive 99 C045 M 64 Thailand Thai Extrahepatic Positive 100 C061 F 60 Thailand Thai Extrahepatic Positive 101 C085 M 52 Thailand Thai Extrahepatic Positive 102 C115 M 66 Thailand Thai Extrahepatic Positive 103 C119 F 75 Thailand Thai Extrahepatic Positive 104 C140 M 52 Thailand Thai Extrahepatic Negative 105 C150 M 57 Thailand Thai Extrahepatic Positive 106 C163 M 55 Thailand Thai Extrahepatic Positive 107 C169 F 56 Thailand Thai Extrahepatic Positive 108 C176 M 49 Thailand Thai Extrahepatic Positive Samples previously reported in Ong et al. 1 are highlighted. 22
23 Supplementary Table 6. Serum IgG antibody against O. viverrini crude antigen Cohort of CCA samples Total Positive Negative N % (N) % (N) Non-O. viverrini-cca Singapore % (2) 83.3% (10) Romania 46 15% (7) 85% (39) O. viverrini-cca Thailand % (76) 15.6% (14) 23
24 Supplementary Table 7. Summary of mutations in KRAS, IDH1 and IDH2 identified using ultra-deep amplicon sequencing compared to Sanger sequencing a) KRAS mutations in 209 cholangiocarcinoma cases Gene Sample ID Origin Sample cohort Ultra-deep amplicon sequencing Total Reads Variant Reads Variant allele Frequency (%) No. of amplicons Amino acid change cdna change Sanger validation KRAS Singapore Non-O. viverrini-cca p.g12d c.35 G>A Yes KRAS Singapore Non-O. viverrini-cca p.g12d c.35 G>A Yes KRAS Singapore Non-O. viverrini-cca p.g12v c.35 G>T Yes KRAS Z3561 Singapore Non-O. viverrini-cca p.g12d c.35 G>A Yes KRAS Singapore Non-O. viverrini-cca p.g12d c.35 G>A Yes KRAS 2143 Romania Non-O. viverrini-cca p. Q61H c.183 A>C Yes KRAS 27 Romania Non-O. viverrini-cca p. G13D c.38 G>A Yes KRAS 962 Romania Non-O. viverrini-cca p.g12v c.35 G>T Yes KRAS 1441 Romania Non-O. viverrini-cca p.g12v c.35 G>T Yes KRAS 930 Romania Non-O. viverrini-cca p. G12V c.35 G>T Yes KRAS 1202 Romania Non-O. viverrini-cca p.g12d c.35 G>A Yes KRAS R104 Thailand O. viverrini-cca p. G13D c.38 G>A Yes KRAS Y149 Thailand O. viverrini-cca p.g12v c.35 G>T Yes KRAS C154 Thailand O. viverrini-cca p.g12c c.34 G>T Yes 24
25 a) (continued). KRAS mutations in 209 cholangiocarcinoma cases Gene Sample ID Origin Sample cohort Ultra-deep amplicon sequencing Total Reads Variant Reads Variant allele Frequency (%) No. of amplicons Amino acid change cdna change Sanger validation KRAS Y020 Thailand O. viverrini-cca p.g12v c.35 G>T Yes KRAS A003 Thailand O. viverrini-cca p.g12d c.35 G>A Yes KRAS Y019 Thailand O. viverrini-cca p.g12c c.34 G>T Yes KRAS W012 Thailand O. viverrini-cca p.g12a c.35 G>C Yes KRAS C078 Thailand O. viverrini-cca p.g12v c.35 G>T Yes KRAS B085 Thailand O. viverrini-cca p.g12a c.35 G>C Yes KRAS T151 Thailand O. viverrini-cca p.g12d c.35 G>A Yes KRAS U027 Thailand O. viverrini-cca p. G12S c.34 G>A Yes KRAS Y008 Thailand O. viverrini-cca p.g12d c.35 G>A Yes KRAS Y072 Thailand O. viverrini-cca p. A146T c.436 G>A Yes KRAS 2410 Romania Non-O. viverrini-cca p.g12d c.35 G>A Yes KRAS Y033 Thailand O. viverrini-cca p. G13D c.38 G>A No KRAS T160 Thailand O. viverrini-cca p. Q61H c.183 A>C No KRAS Singapore Non-O. viverrini-cca p.g12v c.35 G>T No Yes, Mutation was consistently detected by both ultra-deep amplicon and Sanger sequencing. No, Mutation was undetectable by Sanger sequencing. Mutated samples 28/209 25/209 Mutation frequency 13.4% 12.0% 25
26 b) IDH1 and IDH2 mutations in 209 cholangiocarcinoma cases Gene Sample ID Origin Sample cohort Ultra-deep amplicon sequencing Total Reads Varia nt Reads Variant allele Frequency (%) No. of amplicons Amino acid change cdna change Sanger validation IDH1 Z2778 Singapore Non-O. viverrini-cca p.r132g c.394 C>G Yes IDH Singapore Non-O. viverrini-cca p.r132c c.394 C>T Yes IDH Singapore Non-O. viverrini-cca p.r132c c.394 C>T Yes IDH Singapore Non-O. viverrini-cca p.r132c c.394 C>T Yes IDH Singapore Non-O. viverrini-cca p.r132c c.394 C>T Yes IDH Romania Non-O. viverrini-cca p.r132c c.394 C>T Yes IDH1 004C Romania Non-O. viverrini-cca p.r132c c.394 C>T Yes IDH Romania Non-O. viverrini-cca p.r132c c.394 C>T Yes IDH1 C085 Thailand O. viverrini-cca p.r132s c.394 C>A Yes IDH2 C105 Thailand O. viverrini-cca p.r172l c.515g>a Yes IDH1 A039 Thailand O. viverrini-cca p.r132v c.394_395 CG>GT Yes IDH2 Z639 Singapore Non-O. viverrini-cca p.r172w c.514 A>T Yes IDH * Singapore Non-O. viverrini-cca p.r132l c.395 G>T Yes IDH Singapore Non-O. viverrini-cca p.r132l c.395 G>T No IDH1 Z321 Singapore Non-O. viverrini-cca p.r132g c.394 C>G No Mutated samples 15/209 13/209 Mutation frequency 7.2% 6.2% Yes, Mutation was consistently detected by both ultra-deep amplicon and Sanger sequencing. No, Mutation was undetectable by Sanger sequencing. *Mutation detected by ultra-deep amplicon sequencing and later confirmed by Sanger sequencing in discovery sample but missed by whole exome sequencing. 26
27 Supplementary Table 8. Mutation frequencies a of commonly mutated genes in Asian CCAs stratified by anatomical subtypes Gene Newly identified Asian non-o. viverrini-cca b Asian O. viverrini-cca c Singapore f Thailand f Unadjusted Intrahepatic Extrahepatic P-value d FDRs e Intrahepatic Extrahepatic N=27 N=14 N=62 N=46 Unadjuste d P-value d BAP1 22.2% (6) % (2) 2.2% (1) ARID1A 7.4% (2) 7.1% (1) % (8) 23.9% (11) Differentially mutated TP53 7.4% (2) 14.2% (2) % (28) 32.6% (15) SMAD % (3) % (10) 23.9% (11) MLL % (7) 15.2% (7) GNAS % (3) 6.5% (3) IDH1/2* 22.2% (6) % (2) 2.2% (1) ROBO2 3.7% (1) 7.1% (1) % (3) 6.5% (3) RNF43 3.7% (1) 7.1% (1) % (4) 8.7% (4) KRAS* 7.4% (2) 21.4% (3) % (12) 6.5% (3) a Includes point mutations, indels and splice-site mutations. b Mutation frequencies in the independent validation samples only, excluding discovery samples. c Mutation frequencies in our previous study 1 and in new samples added in current study. d By two-sided Fisher's Exact Test. e False discovery rates (FDRs) were based on unadjusted P-values using the Benjamini-Hochberg method. f Number of samples with mutations are shown in parentheses. *Sequenced by ultra-deep amplicon sequencing (detailed in Supplementary Table 7). FDRs e 27
28 Supplementary Table 9. Mutation frequencies a of commonly mutated genes in Asian and European non-o. viverrini-associated CCAs Gene Non-O. viverrini-cca b Singapore c Romania c N=41 N=45 Unadjusted P-value d FDRs e Newly identified BAP1 14.6% (6) 6.7% (3) ARID1A 7.3% (3) 13.3% (6) Differentially mutated TP53 9.8% (4) 8.9% (4) SMAD4 7.3% (3) 4.4% (2) MLL % (3) GNAS IDH1/2* 14.6% (6) 4.4% (2) ROBO2 4.9% (2) RNF43 4.9% (2) 2.2% (1) KRAS* 12.2% (5) 11.1% (5) a Includes point mutations, indels and splice-site mutations. b Mutation frequencies in the independent validation samples only, excluding discovery samples. c Number of samples with mutations are shown in parentheses. d By two-sided Fisher's Exact Test. e False discovery rates (FDRs) were based on unadjusted P-values using the Benjamini-Hochberg method. *Sequenced by ultra-deep amplicon sequencing (detailed in Supplementary Table 7). 28
29 Supplementary Table 10. Frequently mutated genes identified in 101 non-o. viverrini-associated CCAs Gene Nonsynonymous somatic mutations Missense Nonsense, splice site or indel Synonymous Total mutations N:S ratio a Total samples harboring mutations Nonsynonymous somatic mutations Missense (N) Nonsense, splice site or indel (N) ARID1A % (13) b BAP % (2) 84.6% (11) IDH1/ % (12) 0 TP : % (7) 36.4% (4) KRAS % (13) 0 MLL : % (2) 33.3% (1) SMAD % (3) 40% (2) RNF % (3) ROBO % (2) 0 GNAS All synonymous and nonsynonymous mutations were confirmed by Sanger sequencing. Detailed information on each mutation is listed in Supplementary Table 12., no synonymous mutation was identified. a N:S, non- synonymous to synonymous mutation ratio. Nonsynonymous mutations in the N:S ratio exclude splice-site variants. b Sample that harbored both missense and indel/nonsense/splice site is classified in this group. 29
30 Supplementary Table 11. Frequently mutated genes identified in 108 O. viverrini-associated CCAs Gene ARID1A Nonsynonymous somatic mutations Missense Nonsense, splice site or indel Synonymous Total mutations N:S ratio a Total samples harboring mutations Nonsynonymous somatic mutations Missense (N) Nonsense, splice site or indel (N) % (2) 89.5% (17) b BAP % (3) IDH1/ % (3) 0 TP : % (27) 37.2% (16) KRAS % (15) 0 MLL : % (4) 71.4% (10) b SMAD : % (9) 57.1% (12) RNF % (4) 50% (4) ROBO : % (2) 66.7% (4) b GNAS % (6) 0 All synonymous and nonsynonymous mutations were confirmed by Sanger sequencing. Detailed information on BAP1 and ARID1A mutations is listed in Supplementary Table 13. Data was analyzed in the samples used in our previous study 1 and in the new samples added in the current study., no synonymous mutation was identified in this gene. a N:S, nonsynonymous to synonymous mutation ratio. Nonsynonymous mutations in the N:S ratio exclude splice-site variants. b Sample that harbored both missense and indel/nonsense/splice site is classified in this group. 30
31 Supplementary Table 12. Somatic mutations in frequently mutated genes including BAP1, ARID1A, IDH1, IDH2, TP53, KRAS, SMAD4, MLL3, RNF43, GNAS and ROBO2 in non-o. viverrini-associated CCAs in discovery (N = 15) and prevalence set (N = 86) No Gene Symbol Sample Sample Origin Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) 1 ARID1A Singapore CCDS285.1 g.chr1: delc c.1558 delc fs Indel 2 ARID1A Singapore CCDS285.1 g.chr1: insa c.3217 insa fs Indel Mutation type 3 ARID1A Singapore CCDS285.1 g.chr1: G>T IVS8+1 G>T Splice site Splice site 4 ARID1A 3001 Romania CCDS285.1 g.chr1: G>T c.2791 G>T p.g931x Nonsense 5 ARID1A Singapore CCDS285.1 g.chr1: insc c.1651 insc fs Indel 6 ARID1A Singapore CCDS285.1 g.chr1: delc c.2296 delc fs Indel 7 ARID1A * Singapore CCDS285.1 g.chr1: deltgagg c.5403_5407 deltgagg fs Indel 8 ARID1A 1774 Romania CCDS285.1 g.chr1: C>T c.4948 C>T p.q1650x Nonsense 9 ARID1A 1382 Romania CCDS285.1 g.chr1: dela c.2749 del A fs Indel 10 ARID1A 962 Romania CCDS285.1 g.chr1: deltcag c.3513_3516 del TCAG fs Indel 11 ARID1A 1178 Romania CCDS285.1 g.chr1: insgcct c.685_686 ins GCCT fs Indel 12 ARID1A 1178 Romania CCDS285.1 g.chr1: del TCCCCAGCCAGCAGACT ACAATGTATCAACAGCA c.4061_4094 del TCCCCAGCCAGCAGAC TACAATGTATCAACAGCA 13 ARID1A 004C Romania CCDS285.1 g.chr1: C>T c.4336 C>T p.r1446x Nonsense 14 ARID1A 1656 Romania CCDS285.1 g.chr1: insa c.608_609 ins A fs Indel 15 ARID1A 1656 Romania CCDS285.1 g.chr1: G>T c.6477 G>T p.k2159n Missense 16 BAP Singapore CCDS g.chr3: delcttgaga c.826_832 deltctcaag fs Indel 17 BAP Singapore CCDS g.chr3: inst c.1360 insa fs Indel 18 BAP Singapore CCDS g.chr3: delt c.1511 dela fs Indel 19 BAP Singapore CCDS g.chr3: G>T c.1881 C>A p.y627x Nonsense fs Indel 31
32 Supplementary Table 12 (continued). Somatic mutations in frequently mutated genes including BAP1, ARID1A, IDH1, IDH2, TP53, KRAS, SMAD4, MLL3, RNF43, GNAS and ROBO2 in non-o. viverrini-associated CCAs in discovery (N = 15) and prevalence set (N = 86) No Gene Symbol Sample Sample Origin Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 20 BAP1 Z639 Singapore CCDS g.chr3: insat c.1930 insta fs Indel g.chr3: c.765_ BAP * Singapore CCDS fs Indel delagcctctagt delactagaggct 22 BAP * Singapore CCDS g.chr3: C>T c.31 G>A p.d11n Missense 23 BAP1 Z508* Singapore CCDS g.chr3: C>G c.1997 G>C p.r666t Missense 24 BAP Singapore CCDS g.chr3: dela c.647 delt fs Indel 25 BAP1 Z3534 Singapore CCDS g.chr3: insa c.2085 inst fs Indel 26 BAP Romania CCDS g.chr3: G>T c.814 C>T p.q272x Nonsense 27 BAP Romania CCDS g.chr3: delt c.1269 dela fs Indel 28 BAP1 004C Romania CCDS g.chr3: G>A c.1777 C>T p.q593x Nonsense 29 IDH Singapore CCDS g.chr2: G>A c.394 C>T p.r132c Missense 30 IDH Singapore CCDS g.chr2: G>A c.394 C>T p.r132c Missense 31 IDH Romania CCDS g.chr2: G>A c.394 C>T p.r132c Missense 32 IDH Singapore CCDS g.chr2: C>A c.395 G>T p.r132l Missense 33 IDH Singapore CCDS g.chr2: G>A c.394 C>T p.r132c Missense 34 IDH Singapore CCDS g.chr2: G>A c.394 C>T p.r132c Missense 35 IDH Romania CCDS g.chr2: G>A c.394 C>T p.r132c Missense 36 IDH1 Z2778 Singapore CCDS g.chr2: G>C c.394 C>G p.r132g Missense 37 IDH1 004C Romania CCDS g.chr2: G>A c.394 C>T p.r132c Missense 38 IDH Singapore CCDS g.chr2: C>A c.395 G>T p.r132l Missense 39 IDH1 Z321 Singapore CCDS g.chr2: G>C c.394 C>G p.r132g Missense 40 IDH2 Z639 Singapore CCDS g.chr15: T>A c.514 A>T p.r172w Missense 32
33 Supplementary Table 12 (continued). Somatic mutations in frequently mutated genes including BAP1, ARID1A, IDH1, IDH2, TP53, KRAS, SMAD4, MLL3, RNF43, GNAS and ROBO2 in non-o. viverrini-associated CCAs in discovery (N = 15) and prevalence set (N = 86) No Gene Symbol Sample Sample Origin Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 41 KRAS Singapore CCDS g.chr12: C>T c.35 G>A p.g12d Missense 42 KRAS 930 Romania CCDS g.chr12: C>A c.35 G>T p.g12v Missense 43 KRAS 1202 Romania CCDS g.chr12: C>T c.35 G>A p.g12d Missense 44 KRAS Singapore CCDS g.chr12: C>A c.35 G>T p.g12v Missense 45 KRAS Singapore CCDS g.chr12: C>T c.35 G>A p.g12d Missense 46 KRAS 1441 Romania CCDS g.chr12: C>A c.35 G>T p.g12v Missense 47 KRAS Z3561 Singapore CCDS g.chr12: C>T c.35 G>A p.g12d Missense 48 KRAS Singapore CCDS g.chr12: C>T c.35 G>A p.g12d Missense 49 KRAS 27 Romania CCDS g.chr12: C>T c.35 G>A p.g12d Missense 50 KRAS 962 Romania CCDS g.chr12: C>A c.35 G>T p.g12v Missense 51 KRAS 2143 Romania CCDS g.chr12: T>G c.183 A>C p.q61h Missense 52 KRAS 2410 Romania CCDS g.chr12: C>T c.35 G>A p.g12d Missense 53 KRAS Singapore CCDS g.chr12: C>A c.35 G>T p.g12v Missense 54 MLL Romania CCDS g.chr7: insa c.4271 inst fs Indel 55 MLL3 890 Romania CCDS g.chr7: T>C c A>G p.h4339r Missense 56 MLL3 154 Romania CCDS g.chr7: G>T c.5511 C>A p.t1837t Synonymous 57 MLL Singapore CCDS g.chr7: G>A c.7540 C>T p.l2514l Synonymous 58 MLL Singapore CCDS g.chr7: T>A c.8748 A>T p.p2916p Synonymous 59 MLL Romania CCDS g.chr7: G>T c C>A p.p3513h Missense 60 RNF Singapore CCDS g.chr17: delg c.1425 del C fs Indel 33
34 Supplementary Table 12 (continued). Somatic mutations in frequently mutated genes including BAP1, ARID1A, IDH1, IDH2, TP53, KRAS, SMAD4, MLL3, RNF43, GNAS and ROBO2 in non-o. viverrini-associated CCAs in discovery (N = 15) and prevalence set (N = 86) No Gene Symbol Sample Sample Origin Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 61 RNF Singapore CCDS g.chr17: delgg c.1978_1979 delcc + fs Indel + instga instca 62 RNF Romania CCDS g.chr17: delc c.1976 delg f.s Indel 63 ROBO Singapore CCDS g.chr3: G>A c.893 G>A p.r298q Missense 64 ROBO2 Z2403 Singapore CCDS g.chr3: A>G c.1739 A>G p.y580c Missense 65 SMAD * Singapore CCDS g.chr18: c.323_334 In frame Indel delaactaaaacatg delaactaaaacatg 66 SMAD Singapore CCDS g.chr18: C>T c.1081 C>T p.r361c Missense 67 SMAD Romania CCDS g.chr18: C>T c.1333 C>T p.r445x Nonsense 68 SMAD4 Z3585 Singapore CCDS g.chr18: C>T c.1081 C>T p.r361c Missense 69 SMAD Romania CCDS g.chr18: C>T c.1081 C>T p.r361c Missense 70 TP Singapore CCDS g.chr17: C>T c.473 G>A p.r158h Missense 71 TP Singapore CCDS g.chr17: C>T c.795 G>A p.l295l Synonymous 72 TP Singapore CCDS g.chr17: delg c.80 delc fs Indel 73 TP Romania CCDS g.chr17: C>A c.814 G>T p.v272l Missense 74 TP Singapore CCDS g.chr17: C>A c.747 G>T p.r249s Missense 75 TP Singapore CCDS g.chr17: C>A c.747 G>T p.r249s Missense 76 TP Romania CCDS g.chr17: T>C c.536 A>G p.h179r Missense 77 TP Romania CCDS g.chr17: G>A c.916 C>T p.r306x Nonsense 78 TP53 Z3585 Singapore CCDS g.chr17: C>T c.713 G>A p.c238y Missense 79 TP53 Z3561 Singapore CCDS g.chr17: G>A c.844 C>T p.r282w Missense 80 TP Romania CCDS g.chr17: delg c.80 delc fs Indel 81 TP T Romania CCDS g.chr17: delg c.1024 delc fs Indel Discovery samples are highlighted. *Mutations were verified in formalin fixed paraffin embedded (FFPE) samples which had both tumor and corresponding normal DNA. 34
35 Supplementary Table 13. Somatic mutations in frequently mutated genes including BAP1, ARID1A, IDH1, IDH2, TP53, KRAS, SMAD4, MLL3, RNF43, GNAS and ROBO2 in 108 O. viverrini-associated CCAs No Gene Symbol Sample Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 1 ARID1A A074 CCDS285.1 g.chr1: C>G c.1901 C>G p.s634x Nonsense 2 ARID1A R100 CCDS285.1 g.chr1: delttgaca c.5426_5441 AGCTTCCAGT delttgacaagcttccagt fs Indel 3 ARID1A R149 CCDS285.1 g.chr1: C>T c.4381 C>T p.r1461x Nonsense 4 ARID1A T026 CCDS285.1 g.chr1: C>T c.2632 C>T p.q878x Nonsense 5 ARID1A T151 CCDS285.1 g.chr1: C>A c.5729 C>A p.t1910n Missense 6 ARID1A U027 CCDS285.1 g.chr1: G>T c.6100 G>T p.e2034x Nonsense 7 ARID1A Y019 CCDS285.1 g.chr1: G>A c.6259 G>A p.g2087r Missense 8 ARID1A Y020 CCDS285.1 g.chr1: C>G c.1203 C>G p.y401x Nonsense 9 ARID1A Y057 CCDS285.1 g.chr1: deltagaggaca c.6596_6625 deltagaggacagcct GCCTTGCCGCCACACAGTTCC TGCCGCCACACAGTTCC In frame Indel 10 ARID1A Y065 CCDS285.1 g.chr1: C>A c.1820 C>A p.s607x Nonsense 11 ARID1A Y091 CCDS285.1 g.chr1: dela c.3482 dela fs Indel 12 ARID1A Y123 CCDS285.1 g.chr1: delacaaat IVS6-8 delacaaatag + c.2252_2270 Splice site fs AGGTTATATGCAGAGGAACCC delgttatatgcagaggaaccc + Indel 13 ARID1A D017 CCDS285.1 g.chr1: delg c.572 delg fs Indel 14 ARID1A C115 CCDS285.1 g.chr1: C>G c.1602 C>G p.y534x Nonsense 15 ARID1A C167 CCDS285.1 g.chr1: G>T c.1876 G>T p.e626x Nonsense 16 ARID1A C167 CCDS285.1 g.chr1: AGA>TT c.1938_1940 AGA>TT fs Indel 17 ARID1A A150 CCDS285.1 g.chr1: delg c.2245 delg fs Indel 18 ARID1A C176 CCDS285.1 g.chr1: C>T c.4270 C>T p.q1424x Nonsense 19 ARID1A A153 CCDS285.1 g.chr1: C>T c.4420 C>T p.q1474x Nonsense 20 ARID1A B011 CCDS285.1 g.chr1: G>T c.6259 G>T p.g2087x Nonsense 21 BAP1 W040 CCDS g.chr3: delgggt c.331_334 delaccc fs Indel 22 BAP1 C105 CCDS g.chr3: delag c.524_525 delct fs Indel 23 BAP1 A157 CCDS g.chr3: inst c.775 insa fs Indel 24 IDH1 A039 CCDS g.chr2: GC>AC c.394_395 CG>GT p.r132v Missense 25 IDH1 C085 CCDS g.chr2: G>T c.394 C>A p.r132s Missense 26 IDH2 C105 CCDS g.chr15: C>T c.515g>a p.r172l Missense 35
36 Supplementary Table 13 (continued). Somatic mutations in frequently mutated genes including BAP1, ARID1A, IDH1, IDH2, TP53, KRAS, SMAD4, MLL3, RNF43, GNAS and ROBO2 in 108 O. viverrini-associated CCAs No Gene Symbol Sample Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 27 KRAS C078 CCDS g.chr12: C>A c.35 G>T p.g12v Missense 28 KRAS C154 CCDS g.chr12: C>T c.34 G>T p.g12c Missense 29 KRAS A003 CCDS g.chr12: C>T c.35 G>A p.g12d Missense 30 KRAS Y072 CCDS g.chr12: C>T c.436 G>A p. A146T Missense 31 KRAS Y033 CCDS g.chr12: C>T c.38 G>A p. G13D Missense 32 KRAS T160 CCDS g.chr12: T>G c.183 A>C p. Q61H Missense 33 KRAS B085 CCDS g.chr12: G>C c.35 G>C p.g12a Missense 34 KRAS R104 CCDS g.chr12: C>T c.38 C>T p.g13d Missense 35 KRAS T151 CCDS g.chr12: G>A c.35 G>A p.g12d Missense 36 KRAS U027 CCDS g.chr12: G>A c.34 G>A p.g12s Missense 37 KRAS W12 CCDS g.chr12: G>C c.35 G>C p.g12a Missense 38 KRAS Y008 CCDS g.chr12: G>A c.35 G>A p.g12d Missense 39 KRAS Y019 CCDS g.chr12: G>T c.34 G>T p.g12c Missense 40 KRAS Y020 CCDS g.chr12: G>T c.35 G>T p.g12v Missense 41 KRAS Y149 CCDS g.chr12: G>T c.35 G>T p.g12v Missense 42 MLL3 B011 CCDS g.chr7: G>T c.734 C>A p.s245y Missense 43 MLL3 B011 CCDS g.chr7: T>G c.4126 A>C p.s1376r Missense 44 MLL3 B011 CCDS g.chr7: G>A c.4333 C>T p.l1445f Missense 45 MLL3 B011 CCDS g.chr7: G>A c.4889 C>T p.s1630l Missense 46 MLL3 B011 CCDS g.chr7: C>A c.5482 G>T p.e1828x Nonsense 47 MLL3 B011 CCDS g.chr7: A>C c.9265 T>G p.f3089v Missense 48 MLL3 B011 CCDS g.chr7: G>T c C>A p.f3689l Missense 49 MLL3 B011 CCDS g.chr7: G>A c C>T p.r3995x Nonsense 50 MLL3 B011 CCDS g.chr7: G>A c C>T R4806X Nonsense 51 MLL3 C164 CCDS g.chr7: C>T c.4385 G>A p.g1462d Missense 52 MLL3 C078 CCDS g.chr7: TT>CA c.4423_4424 AA>TG p.n1475c Missense 36
37 Supplementary Table 13 (continued). Somatic mutations in frequently mutated genes including BAP1, ARID1A, IDH1, IDH2, TP53, KRAS, SMAD4, MLL3, RNF43, GNAS and ROBO2 in 108 O. viverrini-associated CCAs No Gene Symbol Sample Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 53 MLL3 A157 CCDS g.chr7: G>A c.5614 C>T p.q1872x Nonsense 54 MLL3 A157 CCDS g.chr7: C>A c.6683 G>T p.r2228m Missense 55 MLL3 C080 CCDS g.chr7: G>A c.6082 C>T p.r2028x Nonsense 56 MLL3 A096 CCDS g.chr7: insa c.8082 inst fs Indel 57 MLL3 T026 CCDS g.chr7: G>T c G>T p.e4881x Nonsense 58 MLL3 W039 CCDS g.chr7: G>T c G>T p.s4050x Nonsense 59 MLL3 R100 CCDS g.chr7: insTT c.11908_11909 ins TT fs Indel 60 MLL3 R149 CCDS g.chr7: C>T c.9934 C>T p.q3312x Nonsense 61 MLL3 Y023 CCDS g.chr7: G>A c G>A p.w4360x Nonsense 62 MLL3 Y057 CCDS g.chr7: A>T c.411 A>T p.q147h Missense 63 MLL3 Y091 CCDS g.chr7: T>C c T>C p.v4527a Missense 64 MLL3 B149 CCDS g.chr7: C>A IVS2+2 C>A Splice site Splice site 65 MLL3 U027 CCDS g.chr7: G>C c G>C p.l4331l Synonymous 66 RNF43 A080 CCDS g.chr17: inst c.88 insa fs Indel 67 RNF43 C144 CCDS g.chr17: C>G c.619 G>C p.g207r Missense 68 RNF43 A157 CCDS g.chr17: delc c.1976 delg fs Indel 69 RNF43 A074 CCDS g.chr17: C>G c.611 C>G p.t204r Missense 70 RNF43 A159 CCDS g.chr17: C>T c.337 C>T p.r113x Nonsense 71 RNF43 R104 CCDS g.chr17: A>G c.355 T>C p.c119r Missense 72 RNF43 U044 CCDS g.chr17: A>T c.500 A>T p.n167i Missense 73 RNF43 W12 CCDS g.chr17: C>T c.2167 C>T p.q723x Nonsense 74 ROBO2 B011 CCDS g.chr3: C>A c.1370 C>A p.t457n Missense 75 ROBO2 B011 CCDS g.chr3: G>A c.2189 G>A p.r730h Missense 76 ROBO2 B011 CCDS g.chr3: C>T c.2647 C>T p.r883x Nonsense 77 ROBO2 B011 CCDS g.chr3: A>C c.2493 A>C p.831 I>I Synonymous 78 ROBO2 W039 CCDS g.chr3: insC c.3234 insc fs Indel 37
38 Supplementary Table 13 (continued). Somatic mutations in frequently mutated genes including BAP1, ARID1A, IDH1, IDH2, TP53, KRAS, SMAD4, MLL3, RNF43, GNAS and ROBO2 in 108 O. viverrini-associated CCAs No Gene Symbol Sample Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 79 ROBO2 T026 CCDS g.chr3: C>T c.1585 C> T p.q529x Nonsense 80 ROBO2 A159 CCDS g.chr3: T>A c.3999 T>A p.s1322r Missense 81 ROBO2 Y020 CCDS g.chr3: G>T c.712 G>T p.e238x Nonsense 82 ROBO2 T003 CCDS g.chr3: C>T c.2039 C>T p.a680v Missense 83 SMAD4 B103 CCDS g.chr18: G>T c.212 G>T p.c71f Missense 84 SMAD4 A064 CCDS g.chr18: delg c.568 delg fs Indel 85 SMAD4 A153 CCDS g.chr18: c.1005_1022 del delaggagagacatttaaggt AGGAGAGACATTTAAGGT In frame Indel 86 SMAD4 D017 CCDS g.chr18: C>T c.1081 C>A p.r361s Missense 87 SMAD4 A011 CCDS g.chr18: G>C c.1082 G>C p.r361p Missense 88 SMAD4 C054 CCDS g.chr18: deltcaact c.1095_1116 deltcaactc CTCCAATGTCCACAGG TCCAATGTCCACAGG fs Indel 89 SMAD4 A004 CCDS g.chr18: G>T c.1116 G>T p.r372s Missense 90 SMAD4 C164 CCDS g.chr18: C>T c.1333 C>T p.r445x Nonsense 91 SMAD4 D029 CCDS g.chr18: deltta c.1536_1538 del TTA In frame Indel 92 SMAD4 C061 CCDS g.chr18: delac c.1563_1564 del AC fs Indel 93 SMAD4 A169 CCDS g.chr18: G>T c.1612 G>T p.e538x Nonsense 94 SMAD4 C080 CCDS g.chr18: insa c.1588 ins A fs Indel 95 SMAD4 B011 CCDS g.chr18: G>A c.573 G>A p.s191s Synonymous 96 SMAD4 A080 CCDS g.chr18: T>A c.1047 T>A p.t349t Synonymous 97 SMAD4 A039 CCDS g.chr18: T>C c633 T>C p.t211t Synonymous 98 SMAD4 A105 CCDS g.chr18: delga c.90 delga fs Indel 99 SMAD4 A159 CCDS g.chr18: G>A c.988 G>A p.e330k Missense 100 SMAD4 A035 CCDS g.chr18: A>G c.1659 A>G p.x553w Missense 101 SMAD4 B070 CCDS g.chr18: C>T c.346 C>T p.q116x Nonsense 102 SMAD4 B099 CCDS g.chr18: T>C c.428 A>C p.k428t Missense 103 SMAD4 B149 CCDS g.chr18: G>A c.404 G>A p.r135q Missense 104 SMAD4 B149 CCDS g.chr18: C>T c.1081 C>T p.r361c Missense 38
39 Supplementary Table 13 (continued). Somatic mutations in frequently mutated genes including BAP1, ARID1A, IDH1, IDH2, TP53, KRAS, SMAD4, MLL3, RNF43, GNAS and ROBO2 in 108 O. viverrini-associated CCAs No Gene Symbol Sample Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 105 SMAD4 R104 CCDS g.chr18: delagta c.1447 delagta fs Indel 106 SMAD4 U044 CCDS g.chr18: C>T c.547 C>T p.q183x Nonsense 107 SMAD4 W012 CCDS g.chr18: G>T c.394 C>G p.h132d Missense 108 TP53 D029 CCDS g.chr17: insa c.211 inst fs Indel 109 TP53 A011 CCDS g.chr17: C>A c.314 G>T p.g105v Missesne 110 TP53 C164 CCDS g.chr17: C>A IVS4+1 G>T Splice site Splice site 111 TP53 C154 CCDS g.chr17: C>T IVS4-1 G>A Splice site Splice site 112 TP53 A153 CCDS g.chr17: C>T c.524 G>A p.r175h Missesne 113 TP53 C061 CCDS g.chr17: C>T c.524 G>A p.r175h Missesne 114 TP53 C110 CCDS g.chr17: G>T c.534 C>A p.h178q Missesne 115 TP53 A064 CCDS g.chr17: T>C c.614 A>G p.y205c Missesne 116 TP53 B005 CCDS g.chr17: A>C c.581 T>G p.l194r Missesne 117 TP53 C176 CCDS g.chr17: C>T c.713 G>A p.c238y Missesne 118 TP53 C008 CCDS g.chr17: c.735_749 delcatga delggcctccggttcatg ACCGGAGGCC In frame Indel 119 TP53 C045 CCDS g.chr17: inscagtgtgat c.763 insatcacactg In frame Indel 120 TP53 D017 CCDS g.chr17: C>T c.814 G>A p.v272m Missesne 121 TP53 C144 CCDS g.chr17: G>A c.817 C>T p.r273c Missesne 122 TP53 A080 CCDS g.chr17: G>A c.817 C>T p.r273c Missesne 123 TP53 C095 CCDS g.chr17: C>T c.818 G>A p.r273h Missesne 124 TP53 C080 CCDS g.chr17: C>T c.818 G>A p.r273h Missesne 125 TP53 A004 CCDS g.chr17: C>T c.853 G>A p.e285k Missesne 126 TP53 A114 CCDS g.chr17: C>T c.856 G>A p.e286k Missesne 127 TP53 A042 CCDS g.chr17: delt c.582 delt fs Indel 128 TP53 A105 CCDS g.chr17: T>G c.981 T>G p.y327x Nonsense 129 TP53 A107 CCDS g.chr17: A>G c.578 A>G p.h193r Missense 130 TP53 A119 CCDS g.chr17: C>T c.844 C>T p.r282w Missense 131 TP53 A120 CCDS g.chr17: G>A c.818 G>A p.r273h Missense 132 TP53 A142 CCDS g.chr17: G>A c.673 G>A p.v225i Missense 39
40 Supplementary Table 13 (continued). Somatic mutations in frequently mutated genes including BAP1, ARID1A, IDH1, IDH2, TP53, KRAS, SMAD4, MLL3, RNF43, GNAS and ROBO2 in 108 O. viverrini-associated CCAs No Gene Symbol Sample Transcript Accession ID Nucleotide (genomic) Nucleotide (cdna) Amino acid (protein) Mutation type 133 TP53 A159 CCDS g.chr17: C>T c.916 C>T p.r306x Nonsense 134 TP53 A162 CCDS g.chr17: A>G c.395 A>G p.k132r Missense 135 TP53 B032 CCDS g.chr17: C>G c.832 C>G p.p278s Missense 136 TP53 B070 CCDS g.chr17: C>T c.707 C>T p.y236c Missense 137 TP53 B083 CCDS g.chr17: T>C c.403 T>C p.c135r Missense 138 TP53 B099 CCDS g.chr17: G>T & inst c.686 G>T & ins T fs Indel 139 TP53 R134 CCDS g.chr17: G>T c.1027 G>T p.e343x Nonsense 140 TP53 R149 CCDS g.chr17: G>A c. 853 G>A p.e285k Missense 141 TP53 T157 CCDS g.chr17: G>A c.375 G>A p.t125t Synonymous 142 TP53 T026 CCDS g.chr17: insG c.216 insg fs Indel 143 TP53 U044 CCDS g.chr17: G>A c.310 C>T p.q104x Nonsense 144 TP53 W012 CCDS g.chr17: G>A c.742 C>T p.r248w Missense 145 TP53 W040 CCDS g.chr17: insT c.716 ins T fs Indel 146 TP53 Y002 CCDS g.chr17: C>T c.586 C>T p.r196x Nonsense 147 TP53 Y008 CCDS g.chr17: G>A c.644 G>A p.s215n Missense 148 TP53 Y020 CCDS g.chr17: T>G c.489 T>G p.y163x Nonsense 149 TP53 Y035 CCDS g.chr17: C>T c.817 C>T p.r273c Missense 150 TP53 Y072 CCDS g. chr17: _ del c. del 750bp fs Indel 151 TP53 Y149 CCDS g.chr17: C>T c.817 C>T p.r273h Missense 152 GNAS A157 CCDS g.chr20: C>T c.601 C>T p.r201c Missense 153 GNAS A074 CCDS g.chr20: G>T c.602 G>T p.r201l Missense 154 GNAS B149 CCDS g.chr20: C>T c.601 C>T p.r201c Missense 155 GNAS T026 CCDS g.chr20: C>T c.601 C>T p.r201c Missense 156 GNAS W012 CCDS g.chr20: C>T c.601 C>T p.r201c Missense 157 GNAS Y149 CCDS g.chr20: G>A c.602 G>A p.r201h Missense Data previously reported by Ong et al. 1 is highlighted. 40
41 Supplementary Table 14. Sequencing primers for the evaluation of BAP1, IDH1 and IDH2 mutations by Sanger sequencing Gene Primer Name Primer sequence (5' -> 3') Region BAP1 IDH1 IDH2 BAP1-Ex1_3-F BAP1-Ex1_3-R BAP1-Ex1-seq-R BAP1-Ex2_3-seq-F BAP1-Ex4-F BAP1-Ex4-R BAP1-Ex5-F BAP1-Ex5-R BAP1-Ex6-7-F BAP1-Ex6-7-R BAP1-Ex8-F BAP1-Ex8-R BAP1-Ex9-F BAP1-Ex9-R BAP1-Ex10-F BAP1-Ex10-R BAP1-Ex11-F BAP1-Ex11-R BAP1-Ex12-F BAP1-Ex12-R BAP1-Ex13-F BAP1-Ex13-R BAP1-Ex14-F BAP1-Ex14-R BAP1-Ex15-16-F BAP1-Ex15-16-R BAP1-Ex15-16-seq-F BAP1-Ex17-F BAP1-Ex17-R IDH1-Ex2-F IDH1-Ex2-R IDH2-Ex4-F IDH2-Ex4-R GTCCGACAGGCGGAAGACGAG CAAGGCTGCTGCTTTCTGTGAG CGAAATCTTCCACGAGCAG GAATAAGGGCTGGCTGGAG CACTTCAGACACAGTGTTGGGAAG CGTTCTGCCAGAGGATTTCTGTAG GAGTTGTCCAGATATGACTGACCTG GGAAACTCTCCTCCCTCCCTAG GTTCGTCTGTGTTCCTTCCGATTC CTGGTCGGGCAATATGGTGTAG CTACACCATATTGCCCGACCAG CTAAGCCTGATCTTGCCAGATTCAC GTGCCTGGCATGTATGGCTAGTC GTGGTTAGCTGAAGCCCAGATC CTGTGAGTGAATGGGTAGAGCCAAG CTGCTCTCCCTCTACCTTCTGAC CTTAGAGCTTGCTGACTCCCATTG CATATCAGGCAGAGGAACCTAGCAAC CTATCCAGTGTAAGTGGGTGGCAG CTCCGCAGGTGCTCAACATTATC GTTGCTTGGACCAAGTATAAGGAG CTCTGGGAAGAGAGGTCACAAG GACAGGTGGGCCTTGGACTG CAACCCAGAAAGTCTTCTGGCAC GTGCCAGAAGACTTTCTGGGTTG CAGGGCATTCCAGTTAAGACAG CTTGATAGGCATGGACTCGCTG CTGTCTTAACTGGAATGCCCTG CTGGTTCCTCCCATTCCCAG ATGAGAAGAGGGTTGAGGAGTT TGTTGAGATGGACGCCTATTTGT GTTGCTTGGGGTTCAAATTCTGG TGTCAAGGAGTGGGAAGTGTAC PCR product size (bp) Ex Ex4 430 Ex5 450 Ex Ex8 333 Ex9 410 Ex Ex Ex Ex Ex Ex Ex Ex2 393 Ex
42 Supplementary Table 15. A list of hotspots in KRAS, IDH1 and IDH2 sequenced by ultra-deep amplicon sequencing Gene Transcript ID Hotspot Region p.k5 KRAS ENST p.g10 p. A11 p.g12 p.g13 p.v14 p.l19 p.t58 Exon2 p.a59 p.g60 p.q61 p.e63 p.k117 p.a146 Exon3 Exon4 p.g70 IDH1 CCDS p.v71 p. I99M p.r100 p.r132 Exon2 IDH2 CCDS p.r140 p.r172 Exon4 42
43 Supplementary Table 16. Sequencing primers for the evaluation of KRAS, IDH1 and IDH2 mutations by ultra-deep amplicon sequencing Gene Region Primer Name Primer sequence (5' -> 3') Exon2 Exon3 KRAS Exon4 KRAS-Ex2-1F-N1 TTATAAGGCCTGCTGAAAATGACTGAA KRAS-Ex2-1R-N1 CCTCTATTGTTGGATCATATTCGTCC KRAS-Ex2-2F-N1 GTGTATTAACCTTATGTGTGACATGTTCT KRAS-Ex2-2R-N1 CTGAATTAGCTGTATCGTCAAGGCAC KRAS-Ex2-3F-N1 CTGGTGGAGTATTTGATAGTGTATTAACC KRAS-Ex2-3R-N1 GTCAAGGCACTCTTGCCTACG KRAS-Ex2-4F-N1 GCCTGCTGAAAATGACTGAATATAAAC KRAS-Ex2-4R-N1 GGTCCTGCACCAGTAATATGCA KRAS-Ex3-1F-N1 TCAGGATTCCTACAGGAAGCAAGTAG KRAS-Ex3-1R-N1 TGGCAAATACACAAAGAAAGCC KRAS-Ex3-2F-N1 GTAATAATCCAGACTGTGTTTCTCCCT KRAS-Ex3-2R-N1 TCCCCAGTCCTCATGTACTGGTC KRAS-Ex3-3F-N1 GTGTTTCTCCCTTCTCAGGATTCC KRAS-Ex3-3R-N1 CAAAGAAAGCCCTCCCCAGTC KRAS-Ex4a-1F-N1 CCCAGAGAACAAATTAAAAGAGTTAAGG KRAS-Ex4a-1R-N1 CCATAACTTCTTGCTAAGTCCTGAGC KRAS-Ex4a-2F-N1 AAAGAGTTAAGGACTCTGAAGATGTACC KRAS-Ex4a-2R-N1 CCTGTCTTGTCTTTGCTGATGTTTC KRAS-Ex4a-3F-N1 CTGAAGATGTACCTATGGTCCTAGTAGG KRAS-Ex4a-3R-N1 CAGTGTTACTTACCTGTCTTGTCTTTGC KRAS-Ex4a-4F-N1 GCTCAGGACTTAGCAAGAAGTTATGG KRAS-Ex4a-4R-N1 CAGTTATGATTTTGCAGAAAACAGATC KRAS-Ex4a-5F-N1 GAACAGTAGACACAAAACAGGCTCAG KRAS-Ex4a-5R-N1 CAGTTATGATTTTGCAGAAAACAGATC KRAS-Ex4a-6F-N1 GGACTTAGCAAGAAGTTATGGAATTCC KRAS-Ex4a-6R-N1 CCTAGTATAGCATAATTGAGAGAAAAACTG PCR product size (bp)
44 Supplementary Table 16 (continued). Sequencing primers for the evaluation of KRAS, IDH1 and IDH2 mutations by ultra-deep amplicon sequencing Gene Region Primer Name Primer sequence (5' -> 3') PCR product size (bp) IDH1 IDH2 Exon2 Exon4 IDH1-G F IDH1-G R IDH1-G F IDH1-G R IDH1-G F IDH1-G R IDH1-R100-1F IDH1-R100-1R IDH1-R100-2F IDH1-R100-2R IDH1-R100-3F IDH1-R100-3R IDH1-R132-1F IDH1-R132-1R IDH1-R132-2F IDH1-R132-2R IDH1-R132-3F IDH1-R132-3R IDH2-R140-1F IDH2-R140-1R IDH2-R140-2F IDH2-R140-2R IDH2-R140-3F IDH2-R140-3R IDH2-R172-1F IDH2-R172-1R IDH2-R172-2F IDH2-R172-2R IDH2-R172-3F IDH2-R172-3R CACCAACGACCAAGTCACCAAG GAATATTTCGTATGGTGCCATTTGG AATCGTGATGCCACCAACGAC CCATTTGGTGATTTCCACATTTG GCTATGATTTAGGCATAGAGAATCGTG TGAACTCCTCAACCCTCTTCTCATC GATGAGAAGAGGGTTGAGGAGTTCA GGTTTTACCCATCCACTCACAAGC AGGAGTTCAAGTTGAAACAAATGTGG ATAAGCATGACGACCTATGATGATAGG TCAAATGTGCCACTATCACTCCTGA GCAGATAATGGCTTCTCTGAAGACC GGCACCATACGAAATATTCTGGG TGCCAACATGACTTACTTGATCCC GAAATATTCTGGGTGGCACGGTC CATGCAAAATCACATTATTGCCAAC GGTCTTCAGAGAAGCCATTATCTGC GCAAAATCACATTATTGCCAACATGAC GTTGAAAGATGGCGGCTGCA ATGGGCTCCCGGAAGACAGT TATCTCTGTCCTCACAGAGTTCAAGCT TGGTGATGGGCTTGGTCCAG CAGAGTTCAAGCTGAAGAAGATGTGG GTGCCTGCCAATGGTGATG TCATCTGCAAAAACATCCCACG AAGAGGATGGCTAGGCGAGGAG CCTAGTCCCTGGCTGGACCAA CTTGTACTGCAGAGACAAGAGGATGG GGAGCCCATCATCTGCAAAAAC AGGTCAGTGGATCCCCTCTCCA
45 Supplementary Table 17. A list of sirnas Gene sirna sirna code Vendor ARID1A Individual sirna_1 ON-TARGETplus ARID1A sirna (J ) Dharmacon, IL ARID1A Individual sirna_2 ON-TARGETplus ARID1A sirna (J ) Dharmacon, IL ARID1A Individual sirna_3 ON-TARGETplus ARID1A sirna (J ) Dharmacon, IL BAP1 Individual sirna_1 ON-TARGETplus BAP1 sirna (J ) Dharmacon, IL BAP1 Individual sirna_2 ON-TARGETplus BAP1 sirna (J ) Dharmacon, IL BAP1 Individual sirna_3 ON-TARGETplus BAP1 sirna (J ) Dharmacon, IL BAP1 SMARTpool ON-TARGETplus BAP1 sirna (L ) Dharmacon, IL Control Scrambled sirna ON-TARGETplus Non-targeting sirna #1 (D ) Dharmacon, IL 45
46 Supplementary Table 18. A list of antibodies Antibody Catalog number Type Provider Mouse Anti-Human-ARID1A sc (PSG3) Monoclonal Santa Cruz Biotechnology Mouse Anti-Human-BAP1 sc (C-4) Monoclonal Santa Cruz Biotechnology Mouse Anti-Human-β-Actin A5441 (clone AC-15) Monoclonal Sigma-Aldrich Goat Anti-Mouse IgG (H+L)-HRP HRP Conjugate Bio-Rad Goat Anti-Human IgG (H+L)-HRP HRP Conjugate Zymed 46
47 Nonsense 8.2% Indel 8.6% Splice site 4.5% Missense 78.8% Mutation Type Number Missense 193 Nonsense 20 Indel 21 Splice site 11 Total 245 Supplementary Figure 1. Illustration of mutation types in 15 non-o. viverrini-associated CCA exomes. Majority of somatic mutations identified in the tumors analyzed are missense while a relatively small fraction of mutations comprised of indels, nonsense and splice site mutations. 47
48 Supplementary Figure 2. Mutation spectra of 15 non-o. viverrini-associated CCAs compared to 8 O. viverrini-associated CCAs. Mutations belonging to the different categories are represented. (a) Non-O. viverrini-associated CCAs display a dominant C>T or G>A spectrum (left panel) that commonly occurs at CpG dinucleotides (right panel). (b) Similar mutation spectrum is observed in O. viverrini-associated CCAs. 48
49 49
50 50
51 51
52 52
53 53
54 54
55 55
56 Supplementary Figure 3. Analysis of copy number alterations in 15 discovery samples. The plots demonstrate Affymetrix SNP Array 6.0 data from paired tumor and non-tumor DNA analyzed by ASCAT 2.0 (Allele-Specific Copy number Analysis of Tumors, see Online Methods). Log R ratio (Top panel) represents the log base 2 of the ratio of total signal intensity of the tumor sample and of the matched non-tumor sample. Each red dot represents the Log R value at a heterozygous position in the non-tumor sample. The green dots show the segmentation (i.e. smoothing) of these data. B Allele Frequency (Middle panel) demonstrates the relative presence of each of the two alternative nucleotides at each SNP locus in the tumor sample, at sites that are heterozygous in the matched non-tumor sample. The green dots, which are analogous to the Log R ratio, show the segmentation of these data. The regions where the green dots are simultaneously displaced to values higher and lower than 0.5 are regions of LOH or allelic imbalance (more or fewer copies of one allele than the other). Allele Specific Copy Numbers (Bottom panel) shows the estimated genomic copy number of the two parental copies of each chromosome along the y-axis (red: major allele copy number, green: minor allele copy number). 56
57 There is a wide spread of green dots observed across multiple chromosomes in all the 16 discovery samples (a-o) including the samples that harbored 5 somatic mutations (g-j), indicating allelic imbalance or copy number alterations in these samples. Figure a-c refers to ARID1A mutated tumors ( , and 3001, respectively). Figure d-g refers to BAP1 mutated tumors ( , , and , respectively). In ARID1A mutated tumors (a-c), green dots are widely separated at the short arm of chromosome 1 (arrow). Similarly, there is a wide spread of green dots at the short arm of chromosome 3 (arrow) in BAP1 mutated tumors (d-g). These indicate allelic imbalance in both cases. Allele Specific Copy Numbers (Bottom panel) shows the estimated genomic copy number of the two parental copies of each chromosome along the y-axis (red: major allele copy number, green: minor allele copy number). ASCAT shows that one parental chromosome at chromosome 1p (including the region of ARID1A) and at chromosome 3p (including the region where BAP1 is located) is deleted. 57
58 58
59 59
60 Supplementary Figure 4. Enhancement in cell proliferation rates in ARID1A- and BAP1- silenced cells using multiple individual sirnas. Knock-down of ARID1A (a-c) and BAP1 (d-f) using non-overlapping sirnas in three cell lines consistently promoted cell proliferation. Depletion of ARID1A and BAP1 was confirmed by Western blot after 72 hours of sirna treatment. Relative proliferation rates were compared against a scrambled sirna control (sicontrol). All the results are expressed as mean ± SEM of at least three independent experiments. Error bars, SEM (standard error of the mean). P-values were computed by onesided paired t-tests on the data at day 5. * indicates P < 0.05, ** indicates P <
61 Supplementary Figure 5. Quantification of BAP1 transcript levels in BAP1 over-expressing cells. Expression of BAP1 was assessed by real-time PCR due to lack of specific antibody against truncated BAP1 mutant protein. BAP1 transcript levels in H69 (a), EGI-1 (b) and HuCCT-1 cells (c) transfected with wild-type (WT) or mutant BAP1 (K453f.s) are higher compared to the empty vector (mock) treated control cells. This indicates that both the wild-type and the mutant BAP1 were successfully expressed in the cells tested. Error bars, SEM (N = 3). 61
62 62
Supplementary Table 2. Identified causative mutations and/or mutation candidates.
 Supplementary Table 2. Identified causative mutations and/or mutation candidates. Nonsense mutations base change aa change Average depth Result of next generation in 432 patient Hereditary form of the
Supplementary Table 2. Identified causative mutations and/or mutation candidates. Nonsense mutations base change aa change Average depth Result of next generation in 432 patient Hereditary form of the
Laboratory, Division of Medical Sciences, National Cancer Centre, Singapore
 Supplementary Information Exome sequencing of liver fluke-associated cholangiocarcinoma Choon Kiat Ong 1,2*, Chutima Subimerb 1,2,3*, Chawalit Pairojkul 3, Sopit Wongkham 3, Ioana Cutcutache 4, Willie
Supplementary Information Exome sequencing of liver fluke-associated cholangiocarcinoma Choon Kiat Ong 1,2*, Chutima Subimerb 1,2,3*, Chawalit Pairojkul 3, Sopit Wongkham 3, Ioana Cutcutache 4, Willie
Whole Exome Sequenced Characteristics
 Supplementary Tables Supplementary Table 1: Patient characteristics of 45 whole exome sequenced HNSCC tumors Whole Exome Sequenced Characteristics Tumors (n=45) Age, years Median (range) 61.0 (19-90) Sex,
Supplementary Tables Supplementary Table 1: Patient characteristics of 45 whole exome sequenced HNSCC tumors Whole Exome Sequenced Characteristics Tumors (n=45) Age, years Median (range) 61.0 (19-90) Sex,
Nature Genetics: doi: /ng Supplementary Figure 1. TCGA data set on HNSCCs reanalyzed in this study.
 Supplementary Figure 1 TCGA data set on HNSCCs reanalyzed in this study. Summary of the TCGA dataset on HNSCCs re-analyzed in this study and the respective numbers of samples available within each. Supplementary
Supplementary Figure 1 TCGA data set on HNSCCs reanalyzed in this study. Summary of the TCGA dataset on HNSCCs re-analyzed in this study and the respective numbers of samples available within each. Supplementary
July 2015 Assay ID Assay Name Gene COSMIC ID Amino acid change Nucleotide change Wild type allele (VIC label) Mutant allele (FAM label)
 July 2015 Assay ID Assay Name Gene COSMIC ID Amino acid change Nucleotide change Wild type allele (VIC label) Mutant allele (FAM label) p AHLJ090 AKT1_33765 AKT1 33765 p.e17k c.49g>a C T 2 AHBKFRM BRAF_473
July 2015 Assay ID Assay Name Gene COSMIC ID Amino acid change Nucleotide change Wild type allele (VIC label) Mutant allele (FAM label) p AHLJ090 AKT1_33765 AKT1 33765 p.e17k c.49g>a C T 2 AHBKFRM BRAF_473
Clinical presentation. Duration (Month)
 Supplementary information, Table S1A: Clinical information for 125 PAs. No. Subtype Gender (M/F) Age at diagnosis (year) Clinical presentation Duration (Month) Maximal adenoma diameter(cm) Tumor invasion*
Supplementary information, Table S1A: Clinical information for 125 PAs. No. Subtype Gender (M/F) Age at diagnosis (year) Clinical presentation Duration (Month) Maximal adenoma diameter(cm) Tumor invasion*
Disclosure. I have nothing to disclose
 Bin T Teh MD, PhD Disclosure I have nothing to disclose Asian Cancers A Vast Unmet Clinical Need Worldwide 14 million new cancer cases and 8.2 million cancer related deaths in 2012 New cases expected to
Bin T Teh MD, PhD Disclosure I have nothing to disclose Asian Cancers A Vast Unmet Clinical Need Worldwide 14 million new cancer cases and 8.2 million cancer related deaths in 2012 New cases expected to
Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each
 Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each mutated gene and the panel of 125 cancer-driving genes
Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each mutated gene and the panel of 125 cancer-driving genes
Spectrum and Classification of ATP7B Variants in a Large Cohort of Chinese Patients with Wilson s Disease Guides Genetic Diagnosis
 Ivyspring International Publisher 638 Theranostics Research Paper 2016; 6(5): 638-649. doi: 10.7150/thno.14596 Spectrum and Classification of ATP7B Variants in a Large Cohort of Chinese Patients with Wilson
Ivyspring International Publisher 638 Theranostics Research Paper 2016; 6(5): 638-649. doi: 10.7150/thno.14596 Spectrum and Classification of ATP7B Variants in a Large Cohort of Chinese Patients with Wilson
X-exome Sequencing of 405 Unresolved Families Identifies Seven Novel Intellectual Disability Genes
 X-exome Sequencing of 405 Unresolved Families Identifies Seven Novel Intellectual Disability Genes Hao Hu, 1,41 Stefan A. Haas, 2,41 Jamel Chelly, 3,4 Hilde Van Esch, 5 Martine Raynaud, 6,7,8 Arjan P.M.
X-exome Sequencing of 405 Unresolved Families Identifies Seven Novel Intellectual Disability Genes Hao Hu, 1,41 Stefan A. Haas, 2,41 Jamel Chelly, 3,4 Hilde Van Esch, 5 Martine Raynaud, 6,7,8 Arjan P.M.
UW359 Ovary 3c 3 Serous Recurrent 68 BRCA1 816delGT BRCA1 del exon 1-2. UW417 Ovary 3c 3 Serous Primary 38 BRCA1 1675delA
 Supplementary Table 1. Cases with deleterious germline mutations, somatic HR mutations, and somatic PTEN mutations. ID Site Stage Grade Histology Tumor Age Germline mutation(s) a Somatic HR mutation(s)
Supplementary Table 1. Cases with deleterious germline mutations, somatic HR mutations, and somatic PTEN mutations. ID Site Stage Grade Histology Tumor Age Germline mutation(s) a Somatic HR mutation(s)
Supplementary Online Content
 Supplementary Online Content Pearlman R, Frankel WL, Swanson B, et al. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol.
Supplementary Online Content Pearlman R, Frankel WL, Swanson B, et al. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol.
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Generation of cetuximab-resistant cells and analysis of singlecell clones. Cetuximab-sensitive cells (LIM1215 and OXCO-2) were chronically treated with cetuximab
Supplementary Figure 1 Supplementary Figure 1: Generation of cetuximab-resistant cells and analysis of singlecell clones. Cetuximab-sensitive cells (LIM1215 and OXCO-2) were chronically treated with cetuximab
Supplementary Information
 Supplementary Information JAK1 truncating mutations in gynecologic cancer define new role of cancerassociated protein tyrosine kinase aberrations Yuan Ren, Yonghong Zhang, Richard Z. Liu, David A. Fenstermacher,
Supplementary Information JAK1 truncating mutations in gynecologic cancer define new role of cancerassociated protein tyrosine kinase aberrations Yuan Ren, Yonghong Zhang, Richard Z. Liu, David A. Fenstermacher,
b 0.50 Synonymous Subtype
 omatic mutation count Mutations per Mb a 80 * b 0.50 ynonymous 60 0.40 Non-synonymous 40 0.30 0.20 20 0.10 c 0 Fibroadenoma Phyllodes 0.00 ubtype Benign Borderline MAL upplementary Figure 1 Landscape of
omatic mutation count Mutations per Mb a 80 * b 0.50 ynonymous 60 0.40 Non-synonymous 40 0.30 0.20 20 0.10 c 0 Fibroadenoma Phyllodes 0.00 ubtype Benign Borderline MAL upplementary Figure 1 Landscape of
Negative feedback-defective PRPS1 mutants drive thiopurine. resistance in relapsed childhood ALL
 SUPPLEMENTARY INFORMATION Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL Benshang Li 1,5,, Hui Li 1,5,, Yun Bai 2,, Renate Kirschner-Schwabe 3,10, Jun J
SUPPLEMENTARY INFORMATION Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL Benshang Li 1,5,, Hui Li 1,5,, Yun Bai 2,, Renate Kirschner-Schwabe 3,10, Jun J
Familial exudative vitreoretinopathy (FEVR) is an inheritable
 Genetics Spectrum of Variants in 389 Chinese Probands With Familial Exudative Vitreoretinopathy Jia-Kai Li, 1 Yian Li, 1 Xiang Zhang, 1 Chun-Li Chen, 2 Yu-Qing Rao, 1 Ping Fei, 1 Qi Zhang, 1 Peiquan Zhao,
Genetics Spectrum of Variants in 389 Chinese Probands With Familial Exudative Vitreoretinopathy Jia-Kai Li, 1 Yian Li, 1 Xiang Zhang, 1 Chun-Li Chen, 2 Yu-Qing Rao, 1 Ping Fei, 1 Qi Zhang, 1 Peiquan Zhao,
DNA Analysis in Glycogen storage disease
 DNA Analysis in Glycogen storage disease Nick Beauchamp PhD Sheffield, Sheffield Children s NHS Foundation Trust 14th October 2010 Glycogen Synthase Type 0 Glycogen Synthesis and Breakdown Type IV UDPGlucose
DNA Analysis in Glycogen storage disease Nick Beauchamp PhD Sheffield, Sheffield Children s NHS Foundation Trust 14th October 2010 Glycogen Synthase Type 0 Glycogen Synthesis and Breakdown Type IV UDPGlucose
Unravelling the Molecular Taxonomies of Gastroesophageal Cancers
 Unravelling the Molecular Taxonomies of Gastroesophageal Cancers Patrick Tan, MD PhD Professor, Duke-NUS Medical School Deputy Executive Director, Biomedical Research Council, Agency for Science, Technology
Unravelling the Molecular Taxonomies of Gastroesophageal Cancers Patrick Tan, MD PhD Professor, Duke-NUS Medical School Deputy Executive Director, Biomedical Research Council, Agency for Science, Technology
Supporting Information
 Supporting Information Rampal et al. 10.1073/pnas.1407792111 Fig. S1. Genetic events in leukemic transformation of chronic-phase MPNs. (A) Survival of post-mpn AML patients according to mutational status
Supporting Information Rampal et al. 10.1073/pnas.1407792111 Fig. S1. Genetic events in leukemic transformation of chronic-phase MPNs. (A) Survival of post-mpn AML patients according to mutational status
Table S4. List of point mutations found by WES in SMZL patients. * DNA extracted from frozen tissue *Method of detection: Variant detected using
 Table S4. List of point mutations found by WES in SMZL patients. * DNA extracted from frozen tissue *Method of detection: Variant detected using RAMSES (R) or CNAG method (C) or both (R/C) **Validation
Table S4. List of point mutations found by WES in SMZL patients. * DNA extracted from frozen tissue *Method of detection: Variant detected using RAMSES (R) or CNAG method (C) or both (R/C) **Validation
Supplementary Table 1. PIK3CA mutation in colorectal cancer
 Liao X et al. PIK3CA Mutation in Colorectal Cancer. Page 1 Supplementary Table 1. PIK3CA mutation in colorectal cancer Exon Domain Nucleotide change* Amino acid change* cases 9 Helical c.1621t>a p.e541t
Liao X et al. PIK3CA Mutation in Colorectal Cancer. Page 1 Supplementary Table 1. PIK3CA mutation in colorectal cancer Exon Domain Nucleotide change* Amino acid change* cases 9 Helical c.1621t>a p.e541t
Nature Genetics: doi: /ng Supplementary Figure 1. Alternative splicing events in the 5K panel.
 Supplementary Figure 1 Alternative splicing events in the 5K panel. The majority of cryptic splicing occurred by creation of an AG or GT (Type I). While some other mutations increased the usage of a nearby
Supplementary Figure 1 Alternative splicing events in the 5K panel. The majority of cryptic splicing occurred by creation of an AG or GT (Type I). While some other mutations increased the usage of a nearby
Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases.
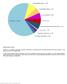 Supplementary Figure 1 Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases. A total of 85 somatically acquired
Supplementary Figure 1 Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases. A total of 85 somatically acquired
Genomic Profiling on an Unselected Solid Tumor Population Reveals a Highly Mutated Wnt/β-Catenin Pathway Associated with Oncogenic EGFR Mutations
 Article Genomic Profiling on an Unselected Solid Tumor Population Reveals a Highly Mutated Wnt/β-Catenin Pathway Associated with Oncogenic EGFR Mutations Jingrui Jiang 1, *, Alexei Protopopov 2, Ruobai
Article Genomic Profiling on an Unselected Solid Tumor Population Reveals a Highly Mutated Wnt/β-Catenin Pathway Associated with Oncogenic EGFR Mutations Jingrui Jiang 1, *, Alexei Protopopov 2, Ruobai
Belgian study into von Willebrand Disease (B-Will): First results
 Belgian study into von Willebrand Disease (B-Will): First results Inge Vangenechten VWD Research Unit, Antwerp University Hospital Supported by CSL Behring Chair in von Willebrand Disease, Antwerp University
Belgian study into von Willebrand Disease (B-Will): First results Inge Vangenechten VWD Research Unit, Antwerp University Hospital Supported by CSL Behring Chair in von Willebrand Disease, Antwerp University
Supplementary appendix
 Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Wells JM, Farris RF, Gosdin TA, et al. Pulmonary
Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Wells JM, Farris RF, Gosdin TA, et al. Pulmonary
Whole exome sequencing as a first line test: Is there even a role for metabolic biochemists in the future?
 Whole exome sequencing as a first line test: Is there even a role for metabolic biochemists in the future? Tony Marinakii Purine Research Laboratory Biochemical Sciences Whole exome sequencing No doubt
Whole exome sequencing as a first line test: Is there even a role for metabolic biochemists in the future? Tony Marinakii Purine Research Laboratory Biochemical Sciences Whole exome sequencing No doubt
Sample Metrics. Allele Frequency (%) Read Depth Ploidy. Gene CDS Effect Protein Effect. LN Metastasis Tumor Purity Computational Pathology 80% 60%
 Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
SUPPLEMENTARY INFORMATION
 Somatic ERCC2 Mutations Are Associated with a Distinct Genomic Signature in Urothelial Tumors Jaegil Kim, Kent W Mouw, Paz Polak, Lior Z Braunstein, Atanas Kamburov, Grace Tiao, David J Kwiatkowski, Jonathan
Somatic ERCC2 Mutations Are Associated with a Distinct Genomic Signature in Urothelial Tumors Jaegil Kim, Kent W Mouw, Paz Polak, Lior Z Braunstein, Atanas Kamburov, Grace Tiao, David J Kwiatkowski, Jonathan
Supplementary Table 1. Information on the 174 single nucleotide variants identified by whole-exome sequencing
 Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
Supplementary Figure 1. Estimation of tumour content
 Supplementary Figure 1. Estimation of tumour content a, Approach used to estimate the tumour content in S13T1/T2, S6T1/T2, S3T1/T2 and S12T1/T2. Tissue and tumour areas were evaluated by two independent
Supplementary Figure 1. Estimation of tumour content a, Approach used to estimate the tumour content in S13T1/T2, S6T1/T2, S3T1/T2 and S12T1/T2. Tissue and tumour areas were evaluated by two independent
Cystic fibrosis carrier screening in a North American population
 American College of Medical Genetics and Genomics Original Research Article Cystic fibrosis carrier screening in a North American population Val V. Zvereff, MD,PhD 1, Hawazin Faruki, DrPh 1, Marcia Edwards,
American College of Medical Genetics and Genomics Original Research Article Cystic fibrosis carrier screening in a North American population Val V. Zvereff, MD,PhD 1, Hawazin Faruki, DrPh 1, Marcia Edwards,
GALAFOLD (migalastat) capsules, for oral use Initial U.S. Approval: 2018
 HIGHLIGHTS OF PRESCRIBING INFORMATION These highlights do not include all the information needed to use GALAFOLD safely and effectively. See full prescribing information for GALAFOLD. GALAFOLD (migalastat)
HIGHLIGHTS OF PRESCRIBING INFORMATION These highlights do not include all the information needed to use GALAFOLD safely and effectively. See full prescribing information for GALAFOLD. GALAFOLD (migalastat)
Supplementary Figure 1
 Supplementary Figure 1 A B HEC1A PTEN +/+ HEC1A PTEN -/- 16 HEC1A PTEN -/- 22 PTEN RAD51 % Cells with RAD51 foci 40 -IR +IR 30 20 10 * *! TUBULIN 0 HEC1A PTEN+/+ HEC1A PTEN-/- 16 HEC1A PTEN-/- 22 C D 1.0
Supplementary Figure 1 A B HEC1A PTEN +/+ HEC1A PTEN -/- 16 HEC1A PTEN -/- 22 PTEN RAD51 % Cells with RAD51 foci 40 -IR +IR 30 20 10 * *! TUBULIN 0 HEC1A PTEN+/+ HEC1A PTEN-/- 16 HEC1A PTEN-/- 22 C D 1.0
Accel-Amplicon Panels
 Accel-Amplicon Panels Amplicon sequencing has emerged as a reliable, cost-effective method for ultra-deep targeted sequencing. This highly adaptable approach is especially applicable for in-depth interrogation
Accel-Amplicon Panels Amplicon sequencing has emerged as a reliable, cost-effective method for ultra-deep targeted sequencing. This highly adaptable approach is especially applicable for in-depth interrogation
Exomes and Beyond Addressing the Genome with OneSeq. Madhuri Hegde, PhD, FACMG Emory University Emory Genetics Laboratory Atlanta, GA
 Exomes and Beyond Addressing the Genome with OneSeq Madhuri Hegde, PhD, FACMG Emory University Emory Genetics Laboratory Atlanta, GA Technologies to Detect Various Types of Mutations RESOLUTION Targeted
Exomes and Beyond Addressing the Genome with OneSeq Madhuri Hegde, PhD, FACMG Emory University Emory Genetics Laboratory Atlanta, GA Technologies to Detect Various Types of Mutations RESOLUTION Targeted
NGS in Cancer Pathology After the Microscope: From Nucleic Acid to Interpretation
 NGS in Cancer Pathology After the Microscope: From Nucleic Acid to Interpretation Michael R. Rossi, PhD, FACMG Assistant Professor Division of Cancer Biology, Department of Radiation Oncology Department
NGS in Cancer Pathology After the Microscope: From Nucleic Acid to Interpretation Michael R. Rossi, PhD, FACMG Assistant Professor Division of Cancer Biology, Department of Radiation Oncology Department
Supplementary Figures. Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated. for each lesion type.
 Supplementary Figures Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated for each lesion type. a b Supplementary Figure 2. Non-negative matrix factorization-derived
Supplementary Figures Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated for each lesion type. a b Supplementary Figure 2. Non-negative matrix factorization-derived
Supplementary Figure 1
 Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
Fluxion Biosciences and Swift Biosciences Somatic variant detection from liquid biopsy samples using targeted NGS
 APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
Supplementary information
 Supplementary information! Recurrent activating mutation in PRKACA in cortisol-producing adrenal tumors Gerald Goh, Ute I. Scholl, James M. Healy, Murim Choi, Manju L. Prasad, Carol Nelson- Williams, John
Supplementary information! Recurrent activating mutation in PRKACA in cortisol-producing adrenal tumors Gerald Goh, Ute I. Scholl, James M. Healy, Murim Choi, Manju L. Prasad, Carol Nelson- Williams, John
GALAFOLD (migalastat) capsules, for oral use Initial U.S. Approval: 2018
 HIGHLIGHTS OF PRESCRIBING INFORMATION These highlights do not include all the information needed to use GALAFOLD safely and effectively. See full prescribing information for GALAFOLD. GALAFOLD (migalastat)
HIGHLIGHTS OF PRESCRIBING INFORMATION These highlights do not include all the information needed to use GALAFOLD safely and effectively. See full prescribing information for GALAFOLD. GALAFOLD (migalastat)
Next generation histopathological diagnosis for precision medicine in solid cancers
 Next generation histopathological diagnosis for precision medicine in solid cancers from genomics to clinical application Aldo Scarpa ARC-NET Applied Research on Cancer Department of Pathology and Diagnostics
Next generation histopathological diagnosis for precision medicine in solid cancers from genomics to clinical application Aldo Scarpa ARC-NET Applied Research on Cancer Department of Pathology and Diagnostics
Belgian-Czech cooperation in the Brno-VWD study
 Inge Vangenechten I Vangenechten, P Smejkal, O Zapletal, F Bouddount, J Zavrelova, J Blatny, M Penka, JJ Michiels, A Gadisseur. Aim of the study Characterisation of VWD in South Moravia (Czech Republic)
Inge Vangenechten I Vangenechten, P Smejkal, O Zapletal, F Bouddount, J Zavrelova, J Blatny, M Penka, JJ Michiels, A Gadisseur. Aim of the study Characterisation of VWD in South Moravia (Czech Republic)
Mutational and phenotypical spectrum of phenylalanine hydroxylase deficiency in Denmark
 Clin Genet 2016: 90: 247 251 Printed in Singapore. All rights reserved Short Report 2015 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd CLINICAL GENETICS doi: 10.1111/cge.12692 Mutational and
Clin Genet 2016: 90: 247 251 Printed in Singapore. All rights reserved Short Report 2015 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd CLINICAL GENETICS doi: 10.1111/cge.12692 Mutational and
Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma
 Supplementary information Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma Meng Li 1,8, Hong Zhao 2,8, Xiaosong Zhang 3, Laura D. Wood 4, Robert A. Anders 4, Michael
Supplementary information Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma Meng Li 1,8, Hong Zhao 2,8, Xiaosong Zhang 3, Laura D. Wood 4, Robert A. Anders 4, Michael
Original Article Pathological characteristics of intraductal polypoid neoplasms of bile ducts in Thailand
 Int J Clin Exp Pathol 2015;8(7):8284-8290 www.ijcep.com /ISSN:1936-2625/IJCEP0009452 Original Article Pathological characteristics of intraductal polypoid neoplasms of bile ducts in Thailand Takeo Nitta
Int J Clin Exp Pathol 2015;8(7):8284-8290 www.ijcep.com /ISSN:1936-2625/IJCEP0009452 Original Article Pathological characteristics of intraductal polypoid neoplasms of bile ducts in Thailand Takeo Nitta
SUPPLEMENTARY INFORMATION. Rare independent mutations in renal salt handling genes contribute to blood pressure variation
 SUPPLEMENTARY INFORMATION Rare independent mutations in renal salt handling genes contribute to blood pressure variation Weizhen Ji, Jia Nee Foo, Brian J. O Roak, Hongyu Zhao, Martin G. Larson, David B.
SUPPLEMENTARY INFORMATION Rare independent mutations in renal salt handling genes contribute to blood pressure variation Weizhen Ji, Jia Nee Foo, Brian J. O Roak, Hongyu Zhao, Martin G. Larson, David B.
Recent advances in the molecular pathology of lung cancer: role of EGFR and KRAS mutation testing in treatment selection
 ASIP 2009 USCAP Companion Meeting Molecular Pathology for the Practicing Pathologist Recent advances in the molecular pathology of lung cancer: role of EGFR and KRAS mutation testing in treatment selection
ASIP 2009 USCAP Companion Meeting Molecular Pathology for the Practicing Pathologist Recent advances in the molecular pathology of lung cancer: role of EGFR and KRAS mutation testing in treatment selection
Research Article Mutation Analysis of the Common Deafness Genes in Patients with Nonsyndromic Hearing Loss in Linyi by SNPscan Assay
 BioMed Research International Volume 2016, Article ID 1302914, 7 pages http://dx.doi.org/10.1155/2016/1302914 Research Article Mutation Analysis of the Common Deafness Genes in Patients with Nonsyndromic
BioMed Research International Volume 2016, Article ID 1302914, 7 pages http://dx.doi.org/10.1155/2016/1302914 Research Article Mutation Analysis of the Common Deafness Genes in Patients with Nonsyndromic
Deliverable 2.1 List of relevant genetic variants for pre-emptive PGx testing
 GA N 668353 H2020 Research and Innovation Deliverable 2.1 List of relevant genetic variants for pre-emptive PGx testing WP N and Title: WP2 - Towards shared European Guidelines for PGx Lead beneficiary:
GA N 668353 H2020 Research and Innovation Deliverable 2.1 List of relevant genetic variants for pre-emptive PGx testing WP N and Title: WP2 - Towards shared European Guidelines for PGx Lead beneficiary:
Nature Genetics: doi: /ng Supplementary Figure 1. TNFAIP3-associated haplotypes in family 1.
 Supplementary Figure 1 TNFAIP3-associated haplotypes in family 1. The p.leu227* mutation (shown as a star) arose de novo in the first affected member of the family (P1). Red haplotypes carry the TNFAIP3
Supplementary Figure 1 TNFAIP3-associated haplotypes in family 1. The p.leu227* mutation (shown as a star) arose de novo in the first affected member of the family (P1). Red haplotypes carry the TNFAIP3
Reporting TP53 gene analysis results in CLL
 Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
To test the possible source of the HBV infection outside the study family, we searched the Genbank
 Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
The Role of Next Generation Sequencing in Solid Tumor Mutation Testing
 The Role of Next Generation Sequencing in Solid Tumor Mutation Testing Allie H. Grossmann MD PhD Department of Pathology, University of Utah Division of Anatomic Pathology & Oncology, ARUP Laboratories
The Role of Next Generation Sequencing in Solid Tumor Mutation Testing Allie H. Grossmann MD PhD Department of Pathology, University of Utah Division of Anatomic Pathology & Oncology, ARUP Laboratories
Circulating tumour DNA in breast cancer. Kathleen Burke, PhD Bioinformatics Postdoctoral Fellow Laboratory of Dr. Jorge Reis-Filho
 Circulating tumour DNA in breast cancer Kathleen Burke, PhD Bioinformatics Postdoctoral Fellow Laboratory of Dr. Jorge Reis-Filho Conflicts of Interest I have no financial relationships to disclose I will
Circulating tumour DNA in breast cancer Kathleen Burke, PhD Bioinformatics Postdoctoral Fellow Laboratory of Dr. Jorge Reis-Filho Conflicts of Interest I have no financial relationships to disclose I will
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma. Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute
 Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
p.r623c p.p976l p.d2847fs p.t2671 p.d2847fs p.r2922w p.r2370h p.c1201y p.a868v p.s952* RING_C BP PHD Cbp HAT_KAT11
 ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
 Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature13898 Supplementary Information Table 1 Kras mutation status of carcinogen-induced mouse lung adenomas Tumour Treatment Strain Grade Genotype Kras status (WES)* Kras status (Sanger) 32T1
doi:10.1038/nature13898 Supplementary Information Table 1 Kras mutation status of carcinogen-induced mouse lung adenomas Tumour Treatment Strain Grade Genotype Kras status (WES)* Kras status (Sanger) 32T1
Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations
 Genomic sequencing of meningiomas identifies oncogenic SMO and AKT mutations Priscilla K. Brastianos*; Peleg M. Horowitz*; Sandro Santagata; Robert T. Jones; Aaron McKenna; Gad Getz; Keith L. Ligon; Emanuele
Genomic sequencing of meningiomas identifies oncogenic SMO and AKT mutations Priscilla K. Brastianos*; Peleg M. Horowitz*; Sandro Santagata; Robert T. Jones; Aaron McKenna; Gad Getz; Keith L. Ligon; Emanuele
HHS Public Access Author manuscript Cancer Discov. Author manuscript; available in PMC 2018 April 01.
 Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma A full list of authors and affiliations appears at the end of the article. Abstract Cholangiocarcinoma (CCA)
Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma A full list of authors and affiliations appears at the end of the article. Abstract Cholangiocarcinoma (CCA)
Genetic Testing and Analysis. (858) MRN: Specimen: Saliva Received: 07/26/2016 GENETIC ANALYSIS REPORT
 GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
Supplementary Figure 1: Number and pattern of somatic mutations in 30 LCBs. (a) Number of mutations. RK308, which had a deletion in MLH1 gene and
 Supplementary Figure 1: Number and pattern of somatic mutations in 30 LCBs. (a) Number of mutations. RK308, which had a deletion in MLH1 gene and missense mutation in MSH2 gene, had exceptionally large
Supplementary Figure 1: Number and pattern of somatic mutations in 30 LCBs. (a) Number of mutations. RK308, which had a deletion in MLH1 gene and missense mutation in MSH2 gene, had exceptionally large
Summary of Results CARRIER STATUS DNA INSIGHT. Result: Carrier, Heterozygote. Protected Health Information. Propionic Acidemia
 Test Results Reviewed & Approved by: Laboratory Director, Nilesh Dharajiya,.D. SAPLE REPORT CARRIER STATUS DNA INSIGHT PERSONAL DETAILS DOB Jan 1, 19XX ETHNICITY Asian ORDERING HEALTHCARE PROFESSIONAL
Test Results Reviewed & Approved by: Laboratory Director, Nilesh Dharajiya,.D. SAPLE REPORT CARRIER STATUS DNA INSIGHT PERSONAL DETAILS DOB Jan 1, 19XX ETHNICITY Asian ORDERING HEALTHCARE PROFESSIONAL
DYSREGULATION OF WT1 (-KTS) IS ASSOCIATED WITH THE KIDNEY-SPECIFIC EFFECTS OF THE LMX1B R246Q MUTATION
 DYSREGULATION OF WT1 (-KTS) IS ASSOCIATED WITH THE KIDNEY-SPECIFIC EFFECTS OF THE LMX1B R246Q MUTATION Gentzon Hall 1,2#, Brandon Lane 1,3#, Megan Chryst-Ladd 1,3, Guanghong Wu 1,3, Jen-Jar Lin 4, XueJun
DYSREGULATION OF WT1 (-KTS) IS ASSOCIATED WITH THE KIDNEY-SPECIFIC EFFECTS OF THE LMX1B R246Q MUTATION Gentzon Hall 1,2#, Brandon Lane 1,3#, Megan Chryst-Ladd 1,3, Guanghong Wu 1,3, Jen-Jar Lin 4, XueJun
Detection of BRCA1/2 mutations in breast cancer patients from Thailand and Pakistan
 Clin Genet 2012: 82: 594 598 Printed in Singapore. All rights reserved Letter to the Editor 2012 John Wiley & Sons A/S. Published by Blackwell Publishing Ltd CLINICAL GENETICS doi: 10.1111/j.1399-0004.2012.01869.x
Clin Genet 2012: 82: 594 598 Printed in Singapore. All rights reserved Letter to the Editor 2012 John Wiley & Sons A/S. Published by Blackwell Publishing Ltd CLINICAL GENETICS doi: 10.1111/j.1399-0004.2012.01869.x
Supplementary Text. Novel variants in NUDT15 and thiopurine intolerance in children with acute lymphoblastic leukemia from diverse ancestry
 Supplementary Text Novel variants in NUDT15 and thiopurine intolerance in children with acute lymphoblastic leukemia from diverse ancestry Takaya Moriyama a, Yung-Li Yang b, Rina Nishii a, c, Hany Ariffin
Supplementary Text Novel variants in NUDT15 and thiopurine intolerance in children with acute lymphoblastic leukemia from diverse ancestry Takaya Moriyama a, Yung-Li Yang b, Rina Nishii a, c, Hany Ariffin
AD (Leave blank) TITLE: Genomic Characterization of Brain Metastasis in Non-Small Cell Lung Cancer Patients
 AD (Leave blank) Award Number: W81XWH-12-1-0444 TITLE: Genomic Characterization of Brain Metastasis in Non-Small Cell Lung Cancer Patients PRINCIPAL INVESTIGATOR: Mark A. Watson, MD PhD CONTRACTING ORGANIZATION:
AD (Leave blank) Award Number: W81XWH-12-1-0444 TITLE: Genomic Characterization of Brain Metastasis in Non-Small Cell Lung Cancer Patients PRINCIPAL INVESTIGATOR: Mark A. Watson, MD PhD CONTRACTING ORGANIZATION:
Please Silence Your Cell Phones. Thank You
 Please Silence Your Cell Phones Thank You Utility of NGS and Comprehensive Genomic Profiling in Hematopathology Practice Maria E. Arcila M.D. Memorial Sloan Kettering Cancer Center New York, NY Disclosure
Please Silence Your Cell Phones Thank You Utility of NGS and Comprehensive Genomic Profiling in Hematopathology Practice Maria E. Arcila M.D. Memorial Sloan Kettering Cancer Center New York, NY Disclosure
Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester
 Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester dsg6@le.ac.uk CFDNA/CTDNA Circulating-free AS A LIQUID DNA BIOPSY (cfdna) Tumour Biopsy Liquid Biopsy
Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester dsg6@le.ac.uk CFDNA/CTDNA Circulating-free AS A LIQUID DNA BIOPSY (cfdna) Tumour Biopsy Liquid Biopsy
NGS in tissue and liquid biopsy
 NGS in tissue and liquid biopsy Ana Vivancos, PhD Referencias So, why NGS in the clinics? 2000 Sanger Sequencing (1977-) 2016 NGS (2006-) ABIPrism (Applied Biosystems) Up to 2304 per day (96 sequences
NGS in tissue and liquid biopsy Ana Vivancos, PhD Referencias So, why NGS in the clinics? 2000 Sanger Sequencing (1977-) 2016 NGS (2006-) ABIPrism (Applied Biosystems) Up to 2304 per day (96 sequences
Importance of minor TP53 mutated clones in the clinic
 Importance of minor TP53 mutated clones in the clinic Davide Rossi, M.D., Ph.D. Hematology IOSI - Oncology Institute of Southern Switzerland IOR - Institute of Oncology Reserach Bellinzona - Switzerland
Importance of minor TP53 mutated clones in the clinic Davide Rossi, M.D., Ph.D. Hematology IOSI - Oncology Institute of Southern Switzerland IOR - Institute of Oncology Reserach Bellinzona - Switzerland
Cancer Genomics. Nic Waddell. Winter School in Mathematical and Computational Biology. July th
 Cancer Genomics Nic Waddell Winter School in Mathematical and Computational Biology 6th July 2015 Time Line of Key Events in Cancer Genomics Michael R. Stratton Science 2011;331:1553-1558 The Cancer Genome
Cancer Genomics Nic Waddell Winter School in Mathematical and Computational Biology 6th July 2015 Time Line of Key Events in Cancer Genomics Michael R. Stratton Science 2011;331:1553-1558 The Cancer Genome
Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM
 Supplementary Data Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM cohort. Supplementary Table e2. Specificity, sensitivity and unadjusted ORs for glioma in participants
Supplementary Data Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM cohort. Supplementary Table e2. Specificity, sensitivity and unadjusted ORs for glioma in participants
Identification and clinical detection of genetic alterations of pre-neoplastic lesions Time for the PML ome? David Sidransky MD Johns Hopkins
 Identification and clinical detection of genetic alterations of pre-neoplastic lesions Time for the PML ome? David Sidransky MD Johns Hopkins February 3-5, 2016 Lansdowne Resort, Leesburg, VA Molecular
Identification and clinical detection of genetic alterations of pre-neoplastic lesions Time for the PML ome? David Sidransky MD Johns Hopkins February 3-5, 2016 Lansdowne Resort, Leesburg, VA Molecular
Insights from Sequencing the Melanoma Exome
 Insights from Sequencing the Melanoma Exome Michael Krauthammer, MD PhD, December 2 2015 Yale University School Yof Medicine 1 2012 Exome Screens and Results Exome Sequencing of 108 sun-exposed melanomas
Insights from Sequencing the Melanoma Exome Michael Krauthammer, MD PhD, December 2 2015 Yale University School Yof Medicine 1 2012 Exome Screens and Results Exome Sequencing of 108 sun-exposed melanomas
The Personalized Cancer Medicine Initiative at Vanderbilt
 The Personalized Cancer Medicine Initiative at Vanderbilt October 26, 2009 William Pao, MD, PhD Assistant Director, Personalized Cancer Medicine Vanderbilt-Ingram Cancer Center Cancer in the United States,
The Personalized Cancer Medicine Initiative at Vanderbilt October 26, 2009 William Pao, MD, PhD Assistant Director, Personalized Cancer Medicine Vanderbilt-Ingram Cancer Center Cancer in the United States,
p.arg119gly p.arg119his p.ala179thr c.540+1g>a c.617_633+6del Prediction basis structure
 a Missense ATG p.arg119gly p.arg119his p.ala179thr p.ala189val p.gly206trp p.gly206arg p.arg251his p.ala257thr TGA 5 UTR 1 2 3 4 5 6 7 3 UTR Splice Site, Frameshift b Mutation p.gly206trp p.gly4fsx50 c.138+1g>a
a Missense ATG p.arg119gly p.arg119his p.ala179thr p.ala189val p.gly206trp p.gly206arg p.arg251his p.ala257thr TGA 5 UTR 1 2 3 4 5 6 7 3 UTR Splice Site, Frameshift b Mutation p.gly206trp p.gly4fsx50 c.138+1g>a
Characterisation of structural variation in breast. cancer genomes using paired-end sequencing on. the Illumina Genome Analyser
 Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Supplementary Information
 Supplementary Information Supplementary Figures 1-10 Supplementary Tables 1-16 Supplementary Notes Exome Sequencing of Gastric Adenocarcinoma Identifies Recurrent Somatic Mutations in Cell Adhesion and
Supplementary Information Supplementary Figures 1-10 Supplementary Tables 1-16 Supplementary Notes Exome Sequencing of Gastric Adenocarcinoma Identifies Recurrent Somatic Mutations in Cell Adhesion and
Round Table: Tissue Biopsy versus Liquid Biopsy. César A. Rodríguez Hospital Universitario de Salamanca-IBSAL
 Round Table: Tissue Biopsy versus Liquid Biopsy César A. Rodríguez Hospital Universitario de Salamanca-IBSAL Introduction Classic Advantages of liquid biopsy collection over standard biopsy Standard biopsy
Round Table: Tissue Biopsy versus Liquid Biopsy César A. Rodríguez Hospital Universitario de Salamanca-IBSAL Introduction Classic Advantages of liquid biopsy collection over standard biopsy Standard biopsy
BWA alignment to reference transcriptome and genome. Convert transcriptome mappings back to genome space
 Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
Frequency(%) KRAS G12 KRAS G13 KRAS A146 KRAS Q61 KRAS K117N PIK3CA H1047 PIK3CA E545 PIK3CA E542K PIK3CA Q546. EGFR exon19 NFS-indel EGFR L858R
 Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
MRC-Holland MLPA. Description version 05; 03 April 2019
 SALSA MLPA probemix ME012-A1 MGMT-IDH1-IDH2 Lot A1-1215. Glioblastoma, the most common malignant primary brain tumour, is characterised by aggressive behaviour and a poor survival. Hypermethylation in
SALSA MLPA probemix ME012-A1 MGMT-IDH1-IDH2 Lot A1-1215. Glioblastoma, the most common malignant primary brain tumour, is characterised by aggressive behaviour and a poor survival. Hypermethylation in
Prevalence and clinical implications of BRCA1/2 germline mutations in Chinese women with breast cancer Yuntao Xie M.D., Ph.D.
 Prevalence and clinical implications of BRCA1/2 germline mutations in Chinese women with breast cancer Yuntao Xie M.D., Ph.D. Hereditary Cancer Center, Peking University Cancer Hospital 1 Breast cancer
Prevalence and clinical implications of BRCA1/2 germline mutations in Chinese women with breast cancer Yuntao Xie M.D., Ph.D. Hereditary Cancer Center, Peking University Cancer Hospital 1 Breast cancer
Supplementary Figure 1: Classification scheme for non-synonymous and nonsense germline MC1R variants. The common variants with previously established
 Supplementary Figure 1: Classification scheme for nonsynonymous and nonsense germline MC1R variants. The common variants with previously established classifications 1 3 are shown. The effect of novel missense
Supplementary Figure 1: Classification scheme for nonsynonymous and nonsense germline MC1R variants. The common variants with previously established classifications 1 3 are shown. The effect of novel missense
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Next Generation Sequencing in Haematological Malignancy: A European Perspective. Wolfgang Kern, Munich Leukemia Laboratory
 Next Generation Sequencing in Haematological Malignancy: A European Perspective Wolfgang Kern, Munich Leukemia Laboratory Diagnostic Methods Cytomorphology Cytogenetics Immunophenotype Histology FISH Molecular
Next Generation Sequencing in Haematological Malignancy: A European Perspective Wolfgang Kern, Munich Leukemia Laboratory Diagnostic Methods Cytomorphology Cytogenetics Immunophenotype Histology FISH Molecular
Supplementary Methods
 Supplementary Methods Short Read Preprocessing Reads are preprocessed differently according to how they will be used: detection of the variant in the tumor, discovery of an artifact in the normal or for
Supplementary Methods Short Read Preprocessing Reads are preprocessed differently according to how they will be used: detection of the variant in the tumor, discovery of an artifact in the normal or for
Moyamoya disease (MMD) is a vasculopathy characterized
 CLINICAL ARTICLE J Neurosurg 126:1106 1113, 2017 RNF213 as the major susceptibility gene for Chinese patients with moyamoya disease and its clinical relevance *Qian Zhang, MD, 1 Yaping Liu, PhD, 2 Dong
CLINICAL ARTICLE J Neurosurg 126:1106 1113, 2017 RNF213 as the major susceptibility gene for Chinese patients with moyamoya disease and its clinical relevance *Qian Zhang, MD, 1 Yaping Liu, PhD, 2 Dong
Supplementary Figure 1
 Supplementary Figure 1 Platelets from affected patients are comparable to controls on electron micrograph. Thin-section transmission electron micrographs of representative platelets from a normal control
Supplementary Figure 1 Platelets from affected patients are comparable to controls on electron micrograph. Thin-section transmission electron micrographs of representative platelets from a normal control
Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies
 Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies Dr. Maricel G. Kann Assistant Professor Dept of Biological Sciences UMBC 2 The term protein domain
Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies Dr. Maricel G. Kann Assistant Professor Dept of Biological Sciences UMBC 2 The term protein domain
Supplemental Table 1. The list of variants with their respective scores for each variant classifier Gene DNA Protein Align-GVGD Polyphen-2 CADD MAPP
 Supplemental Table 1. The list of variants with their respective scores for each variant classifier Gene DNA Protein Align-GVGD Polyphen-2 CADD MAPP Frequency Domain Mammals a 3 S/P b Mammals a 3 S/P b
Supplemental Table 1. The list of variants with their respective scores for each variant classifier Gene DNA Protein Align-GVGD Polyphen-2 CADD MAPP Frequency Domain Mammals a 3 S/P b Mammals a 3 S/P b
Mapping by recurrence and modelling the mutation rate
 Mapping by recurrence and modelling the mutation rate Shamil Sunyaev Broad Institute of M.I.T. and Harvard Current knowledge is from Comparative genomics Experimental systems: yeast reporter assays Potential
Mapping by recurrence and modelling the mutation rate Shamil Sunyaev Broad Institute of M.I.T. and Harvard Current knowledge is from Comparative genomics Experimental systems: yeast reporter assays Potential
IntelliGENSM. Integrated Oncology is making next generation sequencing faster and more accessible to the oncology community.
 IntelliGENSM Integrated Oncology is making next generation sequencing faster and more accessible to the oncology community. NGS TRANSFORMS GENOMIC TESTING Background Cancers may emerge as a result of somatically
IntelliGENSM Integrated Oncology is making next generation sequencing faster and more accessible to the oncology community. NGS TRANSFORMS GENOMIC TESTING Background Cancers may emerge as a result of somatically
Genetic Diagnosis of Liver Diseases
 The Hong Kong Association for the Study of Liver Diseases 27 th Annual Scientific Meeting and International Symposium on Hepatology 16 November 2014 Genetic Diagnosis of Liver Diseases Dr Chloe Mak MD,
The Hong Kong Association for the Study of Liver Diseases 27 th Annual Scientific Meeting and International Symposium on Hepatology 16 November 2014 Genetic Diagnosis of Liver Diseases Dr Chloe Mak MD,
Nature Neuroscience: doi: /nn Supplementary Figure 1. Missense damaging predictions as a function of allele frequency
 Supplementary Figure 1 Missense damaging predictions as a function of allele frequency Percentage of missense variants classified as damaging by eight different classifiers and a classifier consisting
Supplementary Figure 1 Missense damaging predictions as a function of allele frequency Percentage of missense variants classified as damaging by eight different classifiers and a classifier consisting
Nature Genetics: doi: /ng Supplementary Figure 1. Mutational signatures in BCC compared to melanoma.
 Supplementary Figure 1 Mutational signatures in BCC compared to melanoma. (a) The effect of transcription-coupled repair as a function of gene expression in BCC. Tumor type specific gene expression levels
Supplementary Figure 1 Mutational signatures in BCC compared to melanoma. (a) The effect of transcription-coupled repair as a function of gene expression in BCC. Tumor type specific gene expression levels
