In silico analysis of the grapefruit srnaome, transcriptome and gene regulation in response to CTV-CDVd co-infection
|
|
|
- Roderick Stevens
- 5 years ago
- Views:
Transcription
1 Visser et al. Virology Journal (217) 14:2 DOI /s RESEARCH Open Access In silico analysis of the grapefruit srnaome, transcriptome and gene regulation in response to CTV-CDVd co-infection Marike Visser 1, Glynnis Cook 1,2, Johan T. Burger 1 and Hans J. Maree 1,3* Abstract Background: Small RNA (srna) associated gene regulation has been shown to play a significant role during plant-pathogen interaction. In commercial citrus orchards co-infection of Citrus tristeza virus (CTV) and viroids occur naturally. Methods: A next-generation sequencing-based approach was used to study the srna and transcriptional response in grapefruit to the co-infection of CTV and Citrus dwarfing viroid. Results: The co-infection resulted in a difference in the expression of a number of srna species when comparing healthy and infected plants; the majority of these were derived from transcripts processed in a phased manner. Several RNA transcripts were also differentially expressed, including transcripts derived from two genes, predicted to be under the regulation of srnas. These genes are involved in plant hormone systems; one in the abscisic acid, and the other in the cytokinin regulatory pathway. Additional analysis of virus- and viroid-derived small-interfering RNAs (sirnas) showed areas on the pathogen genomes associated with increased sirna synthesis. Most interestingly, the starting position of the p23 silencing suppressor s subgenomic RNA generated a sirna hotspot on the CTV genome. Conclusions: This study showed the involvement of various genes, as well as endogenous and exogenous RNA-derived srna species in the plant-defence response. The results highlighted the role of srna-directed plant hormone regulation during biotic stress, as well as a counter-response of plants to virus suppressors of RNA-silencing. Keywords: Biotic stress, Citrus dwarfing viroid, Citrus tristeza virus, Citrus paradisi, High-throughput sequencing, Pathogen-response, Plant-pathogen interaction, RNA-interference, Small RNA, Transcriptome * Correspondence: hjmaree@sun.ac.za 1 Department of Genetics, Stellenbosch University, Stellenbosch, South Africa 3 Agricultural Research Council, Infruitec-Nietvoorbij: Institute for Deciduous Fruit, Vines and Wine, Stellenbosch, South Africa Full list of author information is available at the end of the article Background Plants respond to pathogen infection through a number of gene regulatory pathways. RNA-silencing is a form of regulation where double-stranded or hairpin-structured RNA precursors give rise to small RNAs (srnas), which become control elements for the expression of target genes [1 3]. Several types of srna species have been identified and characterised in plants, and their involvement in biotic stress responses have been suggested. These include micrornas (mirnas) [4 8], phasedsirnas (phasirnas) [9 12], natural-antisense transcript sirnas (nat-sirnas) [12 14], repeat-associated sirnas (rasirnas) [7] and trna-derived RNA fragments (trfs) [15, 16]. Stem pitting is a destructive symptom in fruit crops caused by various virus species. Citrus tristeza virus (CTV) is a highly destructive, phloem limited, pathogen of citrus species, causing three different disease syndromes [17] of which stem pitting currently is considered the highest threat to the industry [18]. CTV belongs to the genus Closterovirus in the family Closteroviridae [19]. The ~19,3 nt genome of CTV is organised into 12 open reading frames and represents the largest known plant virus genome [2]. The Author(s). 217 Open Access This article is distributed under the terms of the Creative Commons Attribution 4. International License ( which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver ( applies to the data made available in this article, unless otherwise stated.
2 Visser et al. Virology Journal (217) 14:2 Page 2 of 1 Due to the complex disease aetiology of CTV, which is strongly influenced by the combination of host factors and virus genotypes, the mechanism(s) behind symptom expression is poorly understood [21, 22]. A recent study, however, has suggested the involvement of different combinations of p33, p18 and p13 expression during stem pitting symptom development [18]. Three CTV RNA-silencing suppressors have been identified, namely p2, p23 and p25 (coat protein), all of which can play a role in symptom expression [23]. In addition to viruses, citrus species are also affected by viroid-infections. Citrus dwarfing viroid (CDVd), a member of the genus Apscaviroid (family Pospiviroidae), has been suggested for use in high-density orchards since it causes dwarfing of citrus varieties grafted onto Poncirus trifoliata (P. trifoliata) and its hybrids, without reducing production [24 26]. The fact that citrus species are often co-infected with CTV and viroids prompted a recent study which investigated the co-infection of CTV and CDVd in their respective indicator plants, Mexican lime and etrog citron [27]. A host-specific increase in the accumulation of CDVd was observed in the presence of CTV, along with the synthesis of CDVd-associated srnas. These observations were ascribed mainly to the involvement of the CTV-encoded silencing suppressor, p23. It is also interesting to note that the co-infection did not affect symptom expression under their experimental conditions [27]. Understanding the mechanisms involved in pathogen infection and symptom expression provides the information required to study and potentially engineer disease resistance. In this study, we used a next-generation sequencing (NGS) approach to investigate the plant responses to the co-infection of CTV and CDVd in two commercial grapefruit (Citrus paradisi) cultivars, on both the srna and transcriptome level. Our results highlighted the association of CTV-CDVd co-infection with the expression of various genes and srna species, these include srnas involved in the regulation of plant hormones. Methods Sample preparation Grapefruit plants (cultivars Marsh and Star Ruby ) on Carrizo citrange rootstocks were bark-inoculated from an asymptomatic Citrus sinensis (sweet orange) plant that was confirmed to be infected with CTV (genotype T3) and CDVd by RT-PCR using previously described protocols [28, 29]. Briefly, bark-inoculation was performed by patch-grafting two bark chips of the source plant to each grapefruit scion, once the scion was approximately 7 mm thick. All plants were inoculated at the same height. After inoculation, the scions were cut back approximately 1 cm above the inoculation point. One shoot of the new growth was allowed to grow from the top bud. Uninoculated plants served as healthy controls. Total RNA was extracted from the phloem material of three replicates of healthy and infected plants of each cultivar following an adapted CTAB method from Carra et al. [3]. Virus and viroid status was confirmed using the above-mentioned RT-PCR assays. Next-generation sequencing and data preparation Total RNA extracted from each sample was sent for sequencing on an Illumina HiSeq instrument (Fasteris, Geneva, Switzerland). Two libraries per sample were prepared and sequenced. An srna library was generated from 18 to 3 nt size-selected RNA and sequenced in a 1 5 nt run, as well as a transcriptome library generated from ribo-depleted total RNA and sequenced in a nt run. Adapter sequences were trimmed from the data using cutadapt [31]. Fastx-toolkit [32] was used to remove all low quality reads from the srna data, while Trimmomatic [33] was used to filter and trim the transcriptome data for quality. srna reads, nts in length and transcriptome reads, 2 nts and longer were retained for further analyses. Virus and viroid infection status of samples were confirmed bioinformatically with the mapping of virusderived sirna (vsirna) and viroid-derived sirna (vdsirna) derived NGS data against the respective genomes as described below. BLASTn [34] analysis of assembled contigs (described below) against NCBI s nt database was used to exclude the possible presence of any other viruses or viroids from the data. The viral status of samples were further verified using CTVspecific e-probes [35, 36]. Grapefruit transcriptome-assembly, differential expression analysis and natural-antisense transcript (NAT) identification Trinity [37, 38] was used to assemble the transcriptome data into contigs, applying default parameters. Transcript differential expression analysis was performed using the DESeq2 [39] method in Trinity. Gene ontology analyses were performed using Trinity and Blast2GO [4]. Assembled contigs were used to identify NATs by aligning contigs to each other, using BLASTn [34] to identify overlapping regions of 5 nts and longer with 1% identity. Duplex formation of the overlaps was validated with UNAfold [41]. Grapefruit srna identification, differential expression analysis and target prediction To identify novel mirnas and phased (PHAS) transcripts within grapefruit, all srna datasets were simultaneously submitted to ShortStack [42, 43], with default parameters. The assembled contigs served as
3 Visser et al. Virology Journal (217) 14:2 Page 3 of 1 template for precursor identification. DESeq2 was used to analyse the differential expression of all unique srna sequences. Differentially expressed srnas were characterised based on their comparison to different srna species as described below. The predicted novel mirnas, along with the plant entries in mirbase 21 [44 47], as well as the predicted phasirnas (srnas that fell into a dominant phasing register), served as database for the identification of differentially expressed srnas, which were mirnas or phasirnas, respectively. Differentially expressed srnas were also mapped, using Bowtie [48], onto the identified PHAS transcripts, the overlapping regions of the NATs, plant repetitive sequences in RepBase [49, 5], and the sequences of plant trnas in the PlantRNA database [51], to identify PHAS-associated srnas (not in phase with the dominant phasing register), nat-sirnas, rasir- NAs and trfs, respectively. psrnatarget [52] was used, applying default settings, to predict targets for the differentially expressed srnas using the assembled transcriptome as a list of potential targets. Pathogen-derived srna analysis vsirnas (associated with CTV) and vd-sirnas (associated with CDVd) were identified by mapping the srna reads, using Bowtie, onto the CTV-T3 (Accession No. KC525952) and CDVd (Accession No. AF184149) genomes allowing a single, or no mismatches, respectively. a Results Symptom expression in grapefruit Healthy Marsh and Star Ruby grapefruit plants were co-infected with CTV and CDVd, using an asymptomatic (CTV and CDVd infection confirmed) sweet orange plant as source. The co-infection was confirmed with RT-PCRs and supported through NGS read mapping analysis (shown below). No additional viruses or viriods were identified in the NGS data. Star Ruby plants showed more distinct leaf cupping and stem pitting symptoms than Marsh plants (Fig. 1). NGS data preparation srna and transcriptome NGS datasets were generated for each RNA sample extracted from phloem tissue. The raw data ranged from 14,74,885 to 22,862,616 srna reads and 12,726,94 to 16,41,6 transcriptome read-pairs per sample, while the high-quality datasets ranged from 8,55,133 to 12,715,167 srna reads (after quality filtering) and 12,12,34 to 15,458,59 transcriptome read-pairs (after quality filtering and trimming) per sample. Grapefruit transcriptome assembly and differential expression High-quality reads from all the transcriptome datasets were combined and de novo assembled into transcripts (contigs). Altogether 214,371 transcripts were generated, which could be grouped into 12,991, Trinity-defined, genes. b c d Fig. 1 CTV-CDVd co-infected plants. Three healthy followed by three CTV-CDVd co-infected (a) Marsh and (b) Star Ruby plants. A representative stem sample (after bark removal) is given below each plant (c and d)
4 Visser et al. Virology Journal (217) 14:2 Page 4 of 1 Differential expression analysis was subsequently performed to identify transcripts involved in CTV- CDVd co-infection. In Marsh and Star Ruby 675 and 124 transcripts, respectively, showed altered expression (Additional file 1: Tables S1 and S2). The results also identified 154 genes for which at least one transcript were differentially expressed between healthy and infected plants, across both grapefruit varieties (Additional file 1: Table S3). According to similarity searches, these included 21 potential disease response genes, as well as 6 membrane and 1 photosystemassociated genes, highlighted through gene ontology (GO) analysis (Fig. 2, Additional file 1: Tables S4-S6 and Additional file 2: Figures S1-S3). Many transcripts were however homologous to hypothetical or uncharacterised citrus proteins, resulting in 33% of the differentially expressed genes remaining unidentified. Endogenous srna identification and regulation Combined analysis of the transcript and srna data predicted mirnas from 6 grapefruit mirna genes (MIRs) that were expressed in at least one of the samples (Additional file 3: Table S7). For 38 of these MIRs, neither of the mature mirnas predicted, were represented by any mature plant-derived mirna in mirbase. In addition to the predicted mirnas, reads with sequences identical to 216 known plant mirnas were also present in the data (Additional file 3: Table S8). Significant phasing was also seen in 7268 transcripts (called PHAS transcripts), producing 63,943 phasirnas in total, which were in phase with the dominant phasing register. To facilitate the identification of nat-sirnas, transcripts were subjected to NAT identification. The duplex formation of 25,378 transcripts, predicted to be part of one or more NAT pair, were validated in silico. The overlapping regions were extracted for subsequent nat-sirna analysis. Differential expression analysis revealed 761 srnas with altered expression levels resulting from pathogen infection (Additional file 4: Tables S9 and S1). Of these, 577 were variety-specific, while 184 showed differential expression across varieties (Additional file 4: Table S11). These srnas were characterised based on sequence homology to either the predicted, or other plant mir- NAs, predicted phasirnas, PHAS transcripts, the overlaps of NATs, as well as plant repetitive DNA-regions and trnas. While a number of srnas could be classified as nat-sirnas (22), mirnas (five), rasirnas (17) Biological process Molecular function Cellular component organic substance metabolic process primary metabolic process cellular metabolic process single-organism cellular process single-organism metabolic process nitrogen compound metabolic process regulation of cellular process biosynthetic process establishment of localization (transport) single-organism localization regulation of metabolic process cellular response to stimulus single organism signaling ion binding organic cyclic compound binding heterocyclic compound binding protein binding transferase activity small molecule binding hydrolase activity oxidoreductase activity carbohydrate derivative binding intrinsic component of membrane intracellular intracellular part membrane protein complex extrinsic component of membrane oxidoreductase complex photosynthetic membrane protein complex Number of genes Fig. 2 Gene ontology classification of differentially expressed genes. Barr-graph illustrating the number of differentially expressed genes assigned to biological process, molecular function and cellular component gene ontology terms (level 3)
5 Visser et al. Virology Journal (217) 14:2 Page 5 of 1 or trfs, the majority (59) of differentially expressed srnas were phasirnas. Some could potentially be classified into more than one srnas species, for example two srnas derived from a repetitive region and seven srnas derived from the overlapping region of a NAT that were all processed in a phased manner (Fig. 3). To determine the biological role that srnas play during CTV-CDVd co-infection, all differentially expressed srnas were subjected to in silico target prediction. Only three srnas (one uncharacterised and two derived from PHAS transcripts) showed significant inverse-regulation with respect to their predicted target transcripts (Additional file 5: Table S12), one of which was across the two varieties, and the other two specific to Star Ruby. Homology searches identified the acrossvariety srna target as a chloroplastic Magnesiumchelatase subunit ChlH (CHLH) and the Star Ruby -specific srna targets as Cytokinin dehydrogenase 6 (CKX) and a hypothetical protein. Pathogen-derived srnas Virus-derived sirnas (vsirnas) were found associated with 97% of the CTV-T3 reference genome. The majority of the vsirnas were 21 or 22 nts in length (Fig. 4). The distribution of the srna reads on the genome showed an increase in vsirnas mapping towards the 3 end of the virus (Fig. 5). A prominent hotspot for srna synthesis was observed on the negative-strand at the sub-genomic RNA initiation site of p23. In the case of CDVd, viroid-derived sirnas (vd-sir- NAs) were mostly 22 nts in length, followed by reads 21 and 24 nts in length (Fig. 4), and covered the complete viroid genome (Fig. 6). A specific area on the negativestrand of the viroid, overlapping the central and variable regions, gave rise to an abundance of srnas, indicating a potential target area. Discussion In the field, citrus plants are often subjected not only to CTV infection but also to the co-infection with viroids. It is therefore necessary to understand this combined pathogen interaction with the plant host in order to improve disease resistance strategy design. In this study, healthy Marsh and Star Ruby grapefruit plants developed leaf cupping and stem pitting symptoms after being co-infected with CTV (genotype T3) and CDVd. These phasirna 5 mirna nat-sirna trf rasirna Fig. 3 Species identification of differentially expressed srnas. Venn diagram illustrating the overlapping srna species (mirna, phasirna, nat-sirna, rasirna, trf) identities of differentially expressed srnas
6 Visser et al. Virology Journal (217) 14:2 Page 6 of 1 % of srna species vsirna vsirna (NR) vd-sirna vd-sirna (NR) length (nts) Fig. 4 Size-distribution of vsirna and vd-sirna reads. Histogram illustrating the number of vsirna and vd-sirna reads, 18 nt to 26 nt in length, from the infected samples, all (redundant) as opposed to unique (non-redundant, NR), as a percentage of the vsirna and vd-sirna reads in this size-range respectively symptoms were not present in the co-infected sweet orange plants, which served as inoculation source. Although the T3 genotype is associated with increased stem pitting, previous studies have shown that CTV isolate is not the only determinant of symptom expression, but that host species also plays a role [18, 21, 22]. The mechanism(s) that drive the severity and host-specific symptom expression remains to be elucidated. An srna and transcriptome next-generation sequencing approach was followed to study the gene-regulatory pathways involved in the CTV-CDVd co-infection of grapefruit. To compensate for the limited genomic information available for grapefruit, the transcriptome data were de novo assembled to generate a case-specific grapefruit transcriptome. Since the assembled transcripts represented both coding and non-coding transcripts, they were used to identify both the precursors and targets of srnas. As potential precursor source, the transcripts were first used for mirna discovery. The mirna registry, mirbase, currently holds no entries for grapefruit. Here we report on the identification of 6 grapefruit MIR genes along with their mature mirnas, based on in silico prediction analysis. Many of the other srna reads represented homologous plant mirnas. Once more grapefruit genome information becomes available, these homologous sequences may still prove to be true mir- NAs, expressed from grapefruit MIR genes. In addition to mirnas, phasirnas and PHAS transcripts were also identified, based on the assembled transcriptome, along with NATs that form the precursors of nat-sirnas. Many diverse genes were found differentially regulated in response CTV-CDVd co-infection across both grapefruit varieties. Similar to the results of previous studies on the infection of either CTV [53 57] or CDVd [58] in different citrus species and in P. trifoliata, grapefruit metabolic, disease response, structural and phytohormone-related pathways seem to be affected by the co-infection. Genes with known involvement in plant disease responses were mostly grouped into Replication Virion assembly and inter-cellular movement ORF 1a p33 HSP7h CPm p18 p2 ORF 1b p6 p61 CP p13 p Fig. 5 Distribution of vsirna reads along the CTV genome. vsirna profile generated from srna reads depicted as heat maps showing the reads that mapped onto the positive (+) or negative ( ) strand of CTV. A schematic representation of the genome above the heat maps illustrates the genomic position of the vsirna reads. The start of the p23 subgenomic RNA, which forms an vsirna hotspot, is indicated with an arrow
7 Visser et al. Virology Journal (217) 14:2 Page 7 of 1 T L 291/1 TCR P + - P C CCR CDVd CCR C T R V Fig. 6 Distribution of vd-sirna reads along the CDVd genome. vd-sirna profile generated from srna reads depicted as heat maps showing the reads that mapped onto the positive (+) or negative ( ) strand of CDVd. A vd-sirna hotspot was formed on the negative-strand of the viroid, overlapping the central and variable regions. C, central domain; P, pathogenic domain; TCR, terminal conserved region; CCR, central conserved region; T L, terminal left domain; T R, terminal right domain; V, variable domain leucine-rich repeat (which include tobacco mosaic virus resistance protein N-like) and ankyrin repeat-containing protein coding genes. The number of chloroplastassociated genes with altered expression supports the hypothesis that the CTV-CDVd co-infection influences the photosynthetic pathways of grapefruit, which was previously shown for CTV infection in sweet orange [56, 57] and Mexican lime [54, 59]. Many members of different endogenous srna species, such as mirnas, phasirnas, nat-sirnas, rasirnas and trfs also showed variation in expression resulting from the co-infection. Target prediction was performed to determine the biological role of the differentially expressed srnas. Despite many transcripts and srnas showing differential expression resulting from coinfection, an inverse-regulation was only seen for three srna-transcript pairs. The apparent disconnect between srnas and their predicted targets could be due to a number of factors. First, the target prediction software used, was designed specifically for mirna and phasirna target prediction and may therefore not be as effective for other srna species. Target prediction models remain to be developed for the other species, following the characterisation of their mechanism(s) of action. Second, although srna action may lead to the indirect inhibition of protein expression, the presence of any (cleaved or uncleaved) target transcript-related reads in the transcriptome dataset will count towards transcript-associated reads during differential expression analysis of the genes. Last, the relatively low read counts associated with many of the predicted target transcripts could have influenced the statistical significance of the differential expression analysis. The combined results from this study suggested the srna-directed gene regulation of plant hormone pathways during CTV-CDVd co-infection in grapefruit. The expression of chloroplastic Magnesium-chelatase subunit ChlH (CHLH) showed inverse-regulation with respect to that of its regulating srna, across both Marsh and Star Ruby plants. In Arabidopsis, CHLH plays a role in chlorophyll biosynthesis, the expression of photosynthesis-related proteins, as well as abscisic acid (ABA) signal regulation [6]. CHLH was shown to repress the expression of the disease response WRKY4 gene in the ABA signalling pathway [61]. Therefore, unsurprisingly, WRKY4 showed inverse-regulation to that of CHLH resulting from the co-infection. The involvement of the ABA pathway in plant-virus response
8 Visser et al. Virology Journal (217) 14:2 Page 8 of 1 has previously been described [62 64], and includes the restriction, to some extent, of virus movement through callose deposition [62]. In addition, ABA was shown to contribute to virus-resistance through the regulation of Argonauts [64]. The srna-directed regulation of CHLH during the CTV-CDVd co-infection could therefore potentially have a down-stream effect on virus resistance through ABA regulation. Cytokinin dehydrogenase 6 (CKX), on the other hand, is involved in the irreversible down-regulation of cytokinins [65 67], and showed inverse-regulation with respect to that of its regulating srna in Star Ruby plants. Cytokinins can stimulate plant defence response upon pathogen infection [68], and may lead to either an enhancement in resistance [69 71], or susceptibility to viral infections [72]. The observed down-regulation of CKX may lead to an increase in cytokinin levels, contributing to the grapefruit defence response. A tissue-specific up- or downregulation of CKX resulting from virus infection was also recently observed in Arabidopsis roots and shoots, respectively [73]. Pathogen-derived srna profiles have been found to vary upon different infection. The majority of the vsir- NAs, derived from CTV, were 21 or 22 nts in length, which is common for vsirnas produced by many plants, including citrus [8, 74]. The vsirnas also seem to favour the 3 end of the virus. While this increase towards the 3 end of CTV was observed before, not all genotype-host combinations showed the same pattern [8, 74]. Factors that could influence patterns of vsirna synthesis include; a host-specific response, the virus genome secondary structure, virus gene expression and host-specific suppression of virus proteins. Interestingly, when looking at the vsirna profile of CTV, a highly prominent hotspot was observed at the sub-genomic RNA initiation site of p23, associated with the negative strand, which indicates an area targeted by the host-response. p23 is known to play a major role in CTV-host interaction, especially as a suppressor to counter the host s RNA-silencing response [23]. The targeting of p23 by the host therefore adds another layer to the CTV-host interaction. Recent studies have also investigated the plant sirna response to viroid infection [75 79]. While the majority of CDVd-derived vd-sirnas were 22, 21 or 24 nts in length respectively, the observed sizedistribution may be tissue dependent [76, 77]. As was seen for CTV, an srna hotspot was observed associated with an area on the negative-strand of CDVd. A similar hotspot was previously observed for another member of the same genus, Potato spindle tuber viroid, in tomato [75, 78]. The implication of the srna targeting of this specific area of the viroid remains to be elucidated. Conclusions In silico analysis of the transcriptome and srnas generated in response to CTV and CDVd co-infection in grapefruit, suggested the involvement of srnas in the regulation of plant hormone pathways in infected plants. Further analysis revealed regions within both the CTV and CDVd genomes that form hotspots for vsirna and vd-sirna synthesis. The specific vsirna hotspot associated with p23, suggests a first report of potential vsirna counter-response by the plant to the p23-silencing suppressor of CTV. An exciting future prospect for these pathogen-derived srnas could be their application in disease resistance strategies. Additional files Additional file 1: Table S1. Results for the transcript differential expression analysis of Marsh plants. Transcripts were considered to be differentially regulated as a result of infection if a log2 fold change > =1 and padj < =.5 were observed. Table S2. Results for the transcript differential expression analysis of Star Ruby plants. Transcripts were considered to be differentially regulated as a result of infection if a log2 fold change > =1 and padj < =.5 were observed. Table S3. Results for the gene differential expression analysis across both Marsh and Star Ruby plants. Table S4. Biological process based characterisation of genes differentially expressed across both grapefruit varieties. Table S5. Molecular function based characterisation of genes differentially expressed across both grapefruit varieties. Table S6. Cellular component based characterisation of genes differentially expressed across both grapefruit varieties. (XLSX 392 kb) Additional file 2: Figure S1. Biological process based network of genes differentially expressed across both grapefruit varieties. Figure S2. Molecular function based network of genes differentially expressed across both grapefruit varieties. Figure S3. Cellular component based network of genes differentially expressed across both grapefruit varieties. (PDF 2159 kb) Additional file 3: Table S7. mirna prediction results. Table S8. Homologous plant mirna results. (XLSX 26 kb) Additional file 4: Table S9. Results for the srna differential expression analysis of Marsh plants. srnas were considered to be differentially regulated as a result of infection if a log2 fold change > =1 and padj < =.5 were observed. Table S1. Results for the srna differential expression analysis of Star Ruby plants. srnas were considered to be differentially regulated as a result of infection if a log2 fold change > =1 and padj < =.5 were observed. Table S11. Results for the srna differentially expressed across both Marsh and Star Ruby plants. srnas were considered to be differentially regulated as a result of infection if a log2 fold change > =1 and padj < =.5 were observed. (XLSX 16 kb) Additional file 5: Table S12. Differential expression analysis results showing srnas with anti-correlated expression to their targets. (XLSX 53 kb) Abbreviations ABA: abscisic acid; CDVd: Citrus dwarfing viroid; CHLH: chloroplastic magnesium-chelatase subunit ChlH; CKX: cytokinin dehydrogenase 6; CTV: Citrus tristeza virus; MIR: mirna gene; mirna: microrna; NAT: naturalantisense transcript; nat-sirna: natural-antisense transcript sirna; nts: nucleotides; P. trifoliata: Poncirus trifoliata; PHAS: phased transcript; phasirna: phased-sirna; rasirna: repeat-associated sirna; sirna: smallinterfering RNA; srna: small RNA; trf: trna-derived RNA fragment; vdsirna: viroid-derived sirna; vsirna: virus-derived sirna Acknowledgements The authors would like to acknowledge Citrus Research International (CRI), the Technology and Human Resources for Industry Programme (THRIP) and the National Research Foundation (NRF) for their financial assistance towards
9 Visser et al. Virology Journal (217) 14:2 Page 9 of 1 this research. Opinions expressed and conclusions arrived at, are those of the authors and are not necessarily to be attributed to the NRF. Funding This research project was funded by Citrus Research International (grant number 11) as well as the Technology and Human Resources for Industry Programme (grant number TP ). Availability of data and materials The datasets supporting the results of this article are available in the BioProject repository of the National Centre for Biotechnology Information, BioProject: PRJNA in Authors contributions MV participated in the design of the study, RNA samples preparation, performed data analysis and drafted the manuscript. GC participated in the design of the study, preparation of the healthy and inoculated plant material and contributed to drafting the manuscript. JTB participated in the design of the study and contributed to drafting the manuscript. HJM participated in the design of the study and contributed to drafting the manuscript. All authors read and approved the final manuscript. Ethics approval and consent to participate Not applicable. Consent for publication Not applicable. Competing interests The authors declare that they have no competing interests. Publisher s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Author details 1 Department of Genetics, Stellenbosch University, Stellenbosch, South Africa. 2 Citrus Research International, Nelspruit, South Africa. 3 Agricultural Research Council, Infruitec-Nietvoorbij: Institute for Deciduous Fruit, Vines and Wine, Stellenbosch, South Africa. Received: 1 September 217 Accepted: 16 October 217 References 1. Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286: Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by doublestranded RNA. EMBO J. 2;19: Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, et al. Widespread translational inhibition by plant mirnas and sirnas. Science. 28;32: Bester R, Burger JT, Maree HJ. Differential expression of mirnas and associated gene targets in grapevine leafroll-associated virus 3-infected plants. Arch Virol. 217;162: Gao R, Wan ZY, Wong S-M. Plant growth retardation and conserved mirnas are correlated to hibiscus chlorotic ringspot virus infection. PLoS One. 213; 8:e Naqvi AR, Haq QM, Mukherjee SK. MicroRNA profiling of tomato leaf curl new delhi virus (ToLCNDV) infected tomato leaves indicates that deregulation of mir159/319 and mir172 might be linked with leaf curl disease. Virol J. 21;7: Romanel E, Silva TF, Corrêa RL, Farinelli L, Hawkins JS, Schrago CEG, et al. Global alteration of micrornas and transposon-derived small RNAs in cotton (Gossypium Hirsutum) during cotton leafroll dwarf polerovirus (CLRDV) infection. Plant Mol Biol. 212;8: Ruiz-Ruiz S, Navarro B, Gisel A, Peña L, Navarro L, Moreno P, et al. Citrus tristeza virus infection induces the accumulation of viral small RNAs ( nt) mapping preferentially at the 3 -terminal region of the genomic RNA and affects the host small RNA profile. Plant Mol Biol. 211;75: Du P, Wu J, Zhang J, Zhao S, Zheng H, Gao G, et al. Viral infection induces expression of novel phased micrornas from conserved cellular microrna precursors. PLoS Pathog. 211;7:e Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, et al. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci. 212;19: Shivaprasad PV, Chen H-M, Patel K, Bond DM, Santos BACM, Baulcombe DC. A microrna superfamily regulates nucleotide binding site-leucine-rich repeats and other mrnas. Plant Cell. 212;24: Quintero A, Pérez-Quintero AL, López C. Identification of ta-sirnas and cisnat-sirnas in cassava and their roles in response to cassava bacterial blight. Genomics Proteomics Bioinformatics. 213;11: Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Zhu J-K, et al. A pathogen-inducible endogenous sirna in plant immunity. Proc Natl Acad Sci. 26;13: Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. A novel class of bacteriainduced small RNAs in Arabidopsis. Genes Dev. 27;21: Loss-Morais G, Waterhouse PM, Margis R. Description of plant trna-derived RNA fragments (trfs) associated with argonaute and identification of their putative targets. Biol Direct. 213;8: Visser M, Maree HJ, Rees DJ, Burger JT. High-throughput sequencing reveals small RNAs involved in ASGV infection. BMC Genomics. 214; 15: Dawson WO, Garnsey SM, Tatineni S, Folimonova SY, Harper SJ, Gowda S. Citrus tristeza virus-host interactions. Front Microbiol. 213;4 doi:1.3389/ fmicb Tatineni S, Dawson WO. Enhancement or attenuation of disease by deletion of genes from citrus tristeza virus. J Virol. 212;86: Bar-Joseph M, Garnsey SM, Gonsalves D. The Closteroviruses: a distinct group of elongated plant viruses. Adv Virus Res. 1979;25: Karasev AV, Boyko VP, Gowda S, Nikolaeva OV, Hilf ME, Koonin EV, et al. Complete sequence of the citrus tristeza virus RNA genome. Virology. 1995; 28: Garnsey SM, Civerolo EL, Gumpf DJ, Paul C, Hilf ME, Lee RF, et al. Biological characterization of an international collection of citrus tristeza virus (CTV) isolates. In: Proceedings of th 16th conference of International Organization of Citrus Virologists; 25. p publications/publication/?seqno115= Accessed 3 Mar Hilf ME, Mavrodieva VA, Garnsey SM. Genetic marker analysis of a global collection of isolates of citrus tristeza virus: characterization and distribution of CTV genotypes and association with symptoms. Phytopathology. 25;95: Lu R, Folimonov A, Shintaku M, Li W-X, Falk BW, Dawson WO, et al. Three distinct suppressors of RNA silencing encoded by a 2-kb viral RNA genome. Proc Natl Acad Sci U S A. 24;11: Hutton RJ, Broadbent P, Bevington KB. Viroid dwarfing for high density citrus plantings. In: Janick J, editor. Horticultural Reviews. Oxford: John Wiley & Sons, Inc; p doi:1.12/ ch Hardy S, Sanderson G, Barkley P, Donovan N. Dwarfing citrus trees using viroids. 74th ed. New South Wales, Australia: NSW Department of Primary Industries; orchard-management/dwarfing-viroids. Accessed 31 Mar Vidalakis G, Pagliaccia D, Bash JA, Afunian M, Semancik JS. Citrus dwarfing viroid: effects on tree size and scion performance specific to Poncirus trifoliata rootstock for high-density planting. Ann Appl Biol. 211;158: Serra P, Bani Hashemian SM, Fagoaga C, Romero J, Ruiz-Ruiz S, Gorris MT, et al. Virus-viroid interactions: citrus tristeza virus enhances the accumulation of citrus dwarfing viroid in Mexican lime via virus-encoded silencing suppressors. J Virol. 214;88: Cook G, van Vuuren SP, Breytenbach JHJ, Burger JT, Maree HJ. Expanded strain-specific RT-PCR assay for differential detection of currently known citrus tristeza virus strains: a useful screening tool. J Phytopathol. 216;164: Cook G, van Vuuren SP, Breytenbach JHJ, Steyn C, Burger JT, Maree HJ. Characterization of citrus tristeza virus single-variant sources in grapefruit in greenhouse and field trials. Plant Dis. 216;1: Carra A, Gambino G, Schubert A. A cetyltrimethylammonium bromidebased method to extract low-molecular-weight RNA from polysacchariderich plant tissues. Anal Biochem. 27;36: Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 211;17:1 2.
10 Visser et al. Virology Journal (217) 14:2 Page 1 of FASTX-Toolkit. Accessed 28 Mar Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma Oxf Engl. 214;3: Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 199;215: Visser M, Burger JT, Maree HJ. Targeted virus detection in next-generation sequencing data using an automated e-probe based approach. Virology. 216;495: Jooste TL, Visser M, Cook G, Burger JT, Maree HJ. In silico probe-based detection of citrus viruses in NGS data. Phytopathol. 217;17: Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 211;29: Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with trinity. Nat Protoc. 213;8: Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 214;15: Conesa A, Gotz S. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 28;28 doi:1.1155/28/ Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol Clifton NJ. 28;453: Axtell MJ. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA. 213; doi:1.1261/rna Johnson NR, Yeoh JM, Coruh C, Axtell MJ. Improved placement of multimapping small RNAs. G3 Genes Genomes Genet. 216;6: Griffiths-Jones S, Grocock RJ, Dongen S, van Bateman A, Enright AJ. mirbase: microrna sequences, targets and gene nomenclature. Nucleic Acids Res. 26;34(suppl 1):D Griffiths-Jones S, Saini HK, Dongen S, van Enright AJ. mirbase: tools for microrna genomics. Nucleic Acids Res. 28;36(suppl 1):D Kozomara A, Griffiths-Jones S. mirbase: integrating microrna annotation and deep-sequencing data. Nucleic Acids Res. 211;39(suppl 1):D Kozomara A, Griffiths-Jones S. mirbase: annotating high confidence micrornas using deep sequencing data. Nucleic Acids Res, (Database issue):d Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 29;1:R Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 25;11: Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 28;9: Cognat V, Pawlak G, Duchene A-M, Daujat M, Gigant A, Salinas T, et al. PlantRNA, a database for trnas of photosynthetic eukaryotes. Nucleic Acids Res. 212;41:D Dai X, Zhao PX. psrnatarget: a plant small RNA target analysis server. Nucleic Acids Res. 211;39(suppl 2):W Cristofani-Yaly M, Berger IJ, Targon MLPN, Takita MA, de Dorta SO, Freitas-Astua J, et al. Differential expression of genes identified from Poncirus Trifoliata tissue inoculated with CTV through EST analysis and in silico hybridization. Genet Mol Biol. 27;3: Yang F, Wang G, Jiang B, Liu Y, Liu Y, Wu G, et al. Differentially expressed genes and temporal and spatial expression of genes during interactions between Mexican lime (Citrus aurantifolia) andasevere citrus tristeza virus isolate. Physiol Mol Plant Pathol. 213; 83(Supplement C): Cheng C, Zhang Y, Zhong Y, Yang J, Yan S. Gene expression changes in leaves of Citrus sinensis (L.) Osbeck infected by citrus tristeza virus. J Hortic Sci Biotechnol. 216;91: Laino P, Russo MP, Guardo M, Reforgiato-Recupero G, Valè G, Cattivelli L, et al. Rootstock-scion interaction affecting citrus response to CTV infection: a proteomic view. Physiol Plant. 216;156: Fu S, Shao J, Zhou C, Hartung JS. Co-infection of sweet orange with severe and mild strains of citrus tristeza virus is overwhelmingly dominated by the severe strain on both the transcriptional and biological levels. Front Plant Sci. 217;8: Tessitori M, Maria G, Capasso C, Catara G, Rizza S, De Luca V, et al. Differential display analysis of gene expression in Etrog citron leaves infected by citrus viroid III. Biochim Biophys Acta. 27;1769: Pérez-Clemente RM, Montoliu A, Vives V, López-Climent MF, Gómez-Cadenas A. Photosynthetic and antioxidant responses of Mexican lime (Citrus aurantifolia) plants to citrus tristeza virus infection. Plant Pathol. 215;64: Du S-Y, Zhang X-F, Lu Z, Xin Q, Wu Z, Jiang T, et al. Roles of the different components of magnesium chelatase in abscisic acid signal transduction. Plant Mol Biol. 212;8: Shang Y, Yan L, Liu Z-Q, Cao Z, Mei C, Xin Q, et al. The mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell. 21;22: Iriti M, Faoro F. Abscisic acid is involved in chitosan-induced resistance to tobacco necrosis virus (TNV). Plant Physiol Biochem PPB. 28;46: Alazem M, Lin K-Y, Lin N-S. The abscisic acid pathway has multifaceted effects on the accumulation of bamboo mosaic virus. Mol Plant-Microbe Interact MPMI. 214;27: Alazem M, He M-H, Moffett P, Lin N-S. Abscisic acid induces resistance against bamboo mosaic virus through Argonaute 2 and 3. Plant Physiol. 217;174: Whitty CD, Hall RH. A cytokinin oxidase in Zea mays. Can J Biochem. 1974; 52: Galuszka P, Frébort I, Sebela M, Sauer P, Jacobsen S, Pec P. Cytokinin oxidase or dehydrogenase? Mechanism of cytokinin degradation in cereals. Eur J Biochem. 21;268: Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, et al. Cytokinin oxidase regulates rice grain production. Science. 25;39: Novák J, Pavlů J, Novák O, Nožková-Hlaváčková V, Špundová M, Hlavinka J, et al. High cytokinin levels induce a hypersensitive-like response in tobacco. Ann Bot. 213;112: Masuta C, Tanaka H, Uehara K, Kuwata S, Koiwai A, Noma M. Broad resistance to plant viruses in transgenic plants conferred by antisense inhibition of a host gene essential in S-adenosylmethioninedependent transmethylation reactions. Proc Natl Acad Sci U S A. 1995;92: Clarke SF, Burritt DJ, Jameson PE, Guy PL. Influence of plant hormones on virus replication and pathogenesis-related proteins in Phaseolus vulgaris L. infected with white clover mosaic potexvirus. Physiol Mol Plant Pathol U K. 1998;53: Pogány M, Koehl J, Heiser I, Elstner EF, Barna B. Juvenility of tobacco induced by cytokinin gene introduction decreases susceptibility to tobacco necrosis virus and confers tolerance to oxidative stress. Physiol Mol Plant Pathol. 24;65: Baliji S, Lacatus G, Sunter G. The interaction between geminivirus pathogenicity proteins and adenosine kinase leads to increased expression of primary cytokinin responsive genes. Virology. 21;42: Vitti A, Nuzzaci M, Scopa A, Tataranni G, Remans T, Vangronsveld J, et al. Auxin and cytokinin metabolism and root morphological modifications in Arabidopsis thaliana seedlings infected with cucumber mosaic virus (CMV) or exposed to cadmium. Int J Mol Sci. 213;14: Licciardello G, Scuderi G, Ferraro R, Giampetruzzi A, Russo M, Lombardo A, et al. Deep sequencing and analysis of small RNAs in sweet orange grafted on sour orange infected with two citrus tristeza virus isolates prevalent in Sicily. Arch Virol. 215;16: Machida S, Yamahata N, Watanuki H, Owens RA, Sano T. Successive accumulation of two size classes of viroid-specific small RNA in potato spindle tuber viroid-infected tomato plants. J Gen Virol. 27;88: Navarro B, Pantaleo V, Gisel A, Moxon S, Dalmay T, Bisztray G, et al. Deep sequencing of viroid-derived small RNAs from grapevine provides new insights on the role of RNA silencing in plant-viroid interaction. PLoS One. 29;4:e Martinez G, Donaire L, Llave C, Pallas V, Gomez G. High-throughput sequencing of hop stunt viroid-derived small RNAs from cucumber leaves and phloem. Mol Plant Pathol. 21;11: Li R, Gao S, Hernandez AG, Wechter WP, Fei Z, Ling K-S. Deep sequencing of small RNAs in tomato for virus and viroid identification and strain differentiation. PLoS One. 212;7:e Su X, Fu S, Qian Y, Xu Y, Zhou X. Identification of hop stunt viroid infecting Citrus limon in China using small RNAs deep sequencing approach. Virol J. 215;12:1 5.
Next-generation sequencing for virus detection: covering all the bases
 Visser et al. Virology Journal (2016) 13:85 DOI 10.1186/s12985-016-0539-x SHORT REPORT Open Access Next-generation sequencing for virus detection: covering all the bases Marike Visser 1,2, Rachelle Bester
Visser et al. Virology Journal (2016) 13:85 DOI 10.1186/s12985-016-0539-x SHORT REPORT Open Access Next-generation sequencing for virus detection: covering all the bases Marike Visser 1,2, Rachelle Bester
High AU content: a signature of upregulated mirna in cardiac diseases
 https://helda.helsinki.fi High AU content: a signature of upregulated mirna in cardiac diseases Gupta, Richa 2010-09-20 Gupta, R, Soni, N, Patnaik, P, Sood, I, Singh, R, Rawal, K & Rani, V 2010, ' High
https://helda.helsinki.fi High AU content: a signature of upregulated mirna in cardiac diseases Gupta, Richa 2010-09-20 Gupta, R, Soni, N, Patnaik, P, Sood, I, Singh, R, Rawal, K & Rani, V 2010, ' High
Virus-host interactions
 Virus-host interactions - Strategies viruses use to replicate their genomes in susceptible host cells replication - Strategies viruses use to move their genomes throughout susceptible host plants cell-to-cell
Virus-host interactions - Strategies viruses use to replicate their genomes in susceptible host cells replication - Strategies viruses use to move their genomes throughout susceptible host plants cell-to-cell
Identification of mirnas in Eucalyptus globulus Plant by Computational Methods
 International Journal of Pharmaceutical Science Invention ISSN (Online): 2319 6718, ISSN (Print): 2319 670X Volume 2 Issue 5 May 2013 PP.70-74 Identification of mirnas in Eucalyptus globulus Plant by Computational
International Journal of Pharmaceutical Science Invention ISSN (Online): 2319 6718, ISSN (Print): 2319 670X Volume 2 Issue 5 May 2013 PP.70-74 Identification of mirnas in Eucalyptus globulus Plant by Computational
IDENTIFICATION OF IN SILICO MIRNAS IN FOUR PLANT SPECIES FROM FABACEAE FAMILY
 Original scientific paper 10.7251/AGRENG1803122A UDC633:34+582.736.3]:577.2 IDENTIFICATION OF IN SILICO MIRNAS IN FOUR PLANT SPECIES FROM FABACEAE FAMILY Bihter AVSAR 1*, Danial ESMAEILI ALIABADI 2 1 Sabanci
Original scientific paper 10.7251/AGRENG1803122A UDC633:34+582.736.3]:577.2 IDENTIFICATION OF IN SILICO MIRNAS IN FOUR PLANT SPECIES FROM FABACEAE FAMILY Bihter AVSAR 1*, Danial ESMAEILI ALIABADI 2 1 Sabanci
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers
 Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
VirusDetect pipeline - virus detection with small RNA sequencing
 VirusDetect pipeline - virus detection with small RNA sequencing CSC webinar 16.1.2018 Eija Korpelainen, Kimmo Mattila, Maria Lehtivaara Big thanks to Jan Kreuze and Jari Valkonen! Outline Small interfering
VirusDetect pipeline - virus detection with small RNA sequencing CSC webinar 16.1.2018 Eija Korpelainen, Kimmo Mattila, Maria Lehtivaara Big thanks to Jan Kreuze and Jari Valkonen! Outline Small interfering
Supplemental Figure 1. Small RNA size distribution from different soybean tissues.
 Supplemental Figure 1. Small RNA size distribution from different soybean tissues. The size of small RNAs was plotted versus frequency (percentage) among total sequences (A, C, E and G) or distinct sequences
Supplemental Figure 1. Small RNA size distribution from different soybean tissues. The size of small RNAs was plotted versus frequency (percentage) among total sequences (A, C, E and G) or distinct sequences
Bi 8 Lecture 17. interference. Ellen Rothenberg 1 March 2016
 Bi 8 Lecture 17 REGulation by RNA interference Ellen Rothenberg 1 March 2016 Protein is not the only regulatory molecule affecting gene expression: RNA itself can be negative regulator RNA does not need
Bi 8 Lecture 17 REGulation by RNA interference Ellen Rothenberg 1 March 2016 Protein is not the only regulatory molecule affecting gene expression: RNA itself can be negative regulator RNA does not need
Proposed EPPO validation of plant viral diagnostics using next generation sequencing
 Proposed EPPO validation of plant viral diagnostics using next generation sequencing Ian Adams, Ummey Hany, Rachel Glover, Erin Lewis, Neil Boonham, Adrian Fox Adoption of Next Generation Sequencing for
Proposed EPPO validation of plant viral diagnostics using next generation sequencing Ian Adams, Ummey Hany, Rachel Glover, Erin Lewis, Neil Boonham, Adrian Fox Adoption of Next Generation Sequencing for
Small RNA and PARE sequencing in flower bud reveal the involvement of srnas in endodormancy release of Japanese pear (Pyrus pyrifolia 'Kosui')
 Bai et al. BMC Genomics (2016) 17:230 DOI 10.1186/s12864-016-2514-8 RESEARCH ARTICLE Open Access Small RNA and PARE sequencing in flower bud reveal the involvement of srnas in endodormancy release of Japanese
Bai et al. BMC Genomics (2016) 17:230 DOI 10.1186/s12864-016-2514-8 RESEARCH ARTICLE Open Access Small RNA and PARE sequencing in flower bud reveal the involvement of srnas in endodormancy release of Japanese
Hao D. H., Ma W. G., Sheng Y. L., Zhang J. B., Jin Y. F., Yang H. Q., Li Z. G., Wang S. S., GONG Ming*
 Comparison of transcriptomes and gene expression profiles of two chilling- and drought-tolerant and intolerant Nicotiana tabacum varieties under low temperature and drought stress Hao D. H., Ma W. G.,
Comparison of transcriptomes and gene expression profiles of two chilling- and drought-tolerant and intolerant Nicotiana tabacum varieties under low temperature and drought stress Hao D. H., Ma W. G.,
Leaves turn yellow: small RNAs from viruses silence plant genes such as those involved in photosynthesis. Vitantonio Pantaleo IPSP-CNR
 Consiglio Nazionale delle Ricerche Di.S.Ba Conferenza del Dipartimento di Scienze Bio-agroalimentari del CNR Accademia dei Georgofili, Firenze 24-25 Ottobre 2016 Accademia dei Georgofili Leaves turn yellow:
Consiglio Nazionale delle Ricerche Di.S.Ba Conferenza del Dipartimento di Scienze Bio-agroalimentari del CNR Accademia dei Georgofili, Firenze 24-25 Ottobre 2016 Accademia dei Georgofili Leaves turn yellow:
Complete Nucleotide Sequence of RNA1 of Cucumber Mosaic Virus Y Strain and Evolutionary Relationships among Genome RNAs of the Virus Strains
 Complete Nucleotide Sequence of RNA1 of Cucumber Mosaic Virus Y Strain and Evolutionary Relationships among Genome RNAs of the Virus Strains Jiro KATAOKA*, Chikara MASUTA* and Yoichi TAKANAMI* Abstract
Complete Nucleotide Sequence of RNA1 of Cucumber Mosaic Virus Y Strain and Evolutionary Relationships among Genome RNAs of the Virus Strains Jiro KATAOKA*, Chikara MASUTA* and Yoichi TAKANAMI* Abstract
RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays
 Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Research Article Base Composition Characteristics of Mammalian mirnas
 Journal of Nucleic Acids Volume 2013, Article ID 951570, 6 pages http://dx.doi.org/10.1155/2013/951570 Research Article Base Composition Characteristics of Mammalian mirnas Bin Wang Department of Chemistry,
Journal of Nucleic Acids Volume 2013, Article ID 951570, 6 pages http://dx.doi.org/10.1155/2013/951570 Research Article Base Composition Characteristics of Mammalian mirnas Bin Wang Department of Chemistry,
Citrus tristeza virus-host interactions
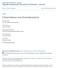 University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Papers in Plant Pathology Plant Pathology Department 5-2013 W. O. Dawson University of Florida, wodtmv@ufl.edu S. M. Garnsey
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Papers in Plant Pathology Plant Pathology Department 5-2013 W. O. Dawson University of Florida, wodtmv@ufl.edu S. M. Garnsey
Efficiency of Citrus Tristeza Virus Isolates in Inducing Early Systemic Infection in Different Citrus Hosts
 ORIGINAL SCIENTIFIC PAPER 27 Efficiency of Citrus Tristeza Virus Isolates in Inducing Early Systemic Infection in Different Citrus Hosts Katarina HANČEVIĆ ( ) Iva ŠUŠIĆ Tomislav RADIĆ Summary Citrus tristeza
ORIGINAL SCIENTIFIC PAPER 27 Efficiency of Citrus Tristeza Virus Isolates in Inducing Early Systemic Infection in Different Citrus Hosts Katarina HANČEVIĆ ( ) Iva ŠUŠIĆ Tomislav RADIĆ Summary Citrus tristeza
Bioinformation by Biomedical Informatics Publishing Group
 Predicted RNA secondary structures for the conserved regions in dengue virus Pallavi Somvanshi*, Prahlad Kishore Seth Bioinformatics Centre, Biotech Park, Sector G, Jankipuram, Lucknow 226021, Uttar Pradesh,
Predicted RNA secondary structures for the conserved regions in dengue virus Pallavi Somvanshi*, Prahlad Kishore Seth Bioinformatics Centre, Biotech Park, Sector G, Jankipuram, Lucknow 226021, Uttar Pradesh,
VIP: an integrated pipeline for metagenomics of virus
 VIP: an integrated pipeline for metagenomics of virus identification and discovery Yang Li 1, Hao Wang 2, Kai Nie 1, Chen Zhang 1, Yi Zhang 1, Ji Wang 1, Peihua Niu 1 and Xuejun Ma 1 * 1. Key Laboratory
VIP: an integrated pipeline for metagenomics of virus identification and discovery Yang Li 1, Hao Wang 2, Kai Nie 1, Chen Zhang 1, Yi Zhang 1, Ji Wang 1, Peihua Niu 1 and Xuejun Ma 1 * 1. Key Laboratory
Intrinsic cellular defenses against virus infection
 Intrinsic cellular defenses against virus infection Detection of virus infection Host cell response to virus infection Interferons: structure and synthesis Induction of antiviral activity Viral defenses
Intrinsic cellular defenses against virus infection Detection of virus infection Host cell response to virus infection Interferons: structure and synthesis Induction of antiviral activity Viral defenses
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
 Xu et al. Molecular Cancer (2019) 18:8 https://doi.org/10.1186/s12943-018-0932-8 LETTER TO THE EDITOR RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
Xu et al. Molecular Cancer (2019) 18:8 https://doi.org/10.1186/s12943-018-0932-8 LETTER TO THE EDITOR RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
MicroRNAs, RNA Modifications, RNA Editing. Bora E. Baysal MD, PhD Oncology for Scientists Lecture Tue, Oct 17, 2017, 3:30 PM - 5:00 PM
 MicroRNAs, RNA Modifications, RNA Editing Bora E. Baysal MD, PhD Oncology for Scientists Lecture Tue, Oct 17, 2017, 3:30 PM - 5:00 PM Expanding world of RNAs mrna, messenger RNA (~20,000) trna, transfer
MicroRNAs, RNA Modifications, RNA Editing Bora E. Baysal MD, PhD Oncology for Scientists Lecture Tue, Oct 17, 2017, 3:30 PM - 5:00 PM Expanding world of RNAs mrna, messenger RNA (~20,000) trna, transfer
Multiplication of RNA Plant Viruses. C.L. Mandahar
 Multiplication of RNA Plant Viruses C.L. Mandahar MULTIPLICATION OF RNA PLANT VIRUSES Multiplication of RNA Plant Viruses by C. L. MANDAHAR Botany Department, Panjab University, Chandigarh, India A C.I.P.
Multiplication of RNA Plant Viruses C.L. Mandahar MULTIPLICATION OF RNA PLANT VIRUSES Multiplication of RNA Plant Viruses by C. L. MANDAHAR Botany Department, Panjab University, Chandigarh, India A C.I.P.
Eukaryotic small RNA Small RNAseq data analysis for mirna identification
 Eukaryotic small RNA Small RNAseq data analysis for mirna identification P. Bardou, C. Gaspin, S. Maman, J. Mariette, O. Rué, M. Zytnicki INRA Sigenae Toulouse INRA MIA Toulouse GenoToul Bioinfo INRA MaIAGE
Eukaryotic small RNA Small RNAseq data analysis for mirna identification P. Bardou, C. Gaspin, S. Maman, J. Mariette, O. Rué, M. Zytnicki INRA Sigenae Toulouse INRA MIA Toulouse GenoToul Bioinfo INRA MaIAGE
MicroRNA and Male Infertility: A Potential for Diagnosis
 Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
Journal of Agricultural Technology 2012 Vol. 8(4): Journal of Agricultural
 Journal of Agricultural Technology 2012 Vol. 8(4): 1389-1395 Journal of Agricultural Available Technology online http://www.ijat-aatsea.com 2012, Vol. 8(4): 1389-1395 ISSN 1686-9141 The effect of the decreased
Journal of Agricultural Technology 2012 Vol. 8(4): 1389-1395 Journal of Agricultural Available Technology online http://www.ijat-aatsea.com 2012, Vol. 8(4): 1389-1395 ISSN 1686-9141 The effect of the decreased
Viral Genetics. BIT 220 Chapter 16
 Viral Genetics BIT 220 Chapter 16 Details of the Virus Classified According to a. DNA or RNA b. Enveloped or Non-Enveloped c. Single-stranded or double-stranded Viruses contain only a few genes Reverse
Viral Genetics BIT 220 Chapter 16 Details of the Virus Classified According to a. DNA or RNA b. Enveloped or Non-Enveloped c. Single-stranded or double-stranded Viruses contain only a few genes Reverse
In silico identification of mirnas and their targets from the expressed sequence tags of Raphanus sativus
 www.bioinformation.net Hypothesis Volume 8(2) In silico identification of mirnas and their targets from the expressed sequence tags of Raphanus sativus Charuvaka Muvva 1 *, Lata Tewari 2, Kasoju Aruna
www.bioinformation.net Hypothesis Volume 8(2) In silico identification of mirnas and their targets from the expressed sequence tags of Raphanus sativus Charuvaka Muvva 1 *, Lata Tewari 2, Kasoju Aruna
ANALYSIS OF SOYBEAN SMALL RNA POPULATIONS HLAING HLAING WIN THESIS
 ANALYSIS OF SOYBEAN SMALL RNA POPULATIONS BY HLAING HLAING WIN THESIS Submitted in partial fulfillment of the requirements for the degree of Master of Science in Bioinformatics in the Graduate College
ANALYSIS OF SOYBEAN SMALL RNA POPULATIONS BY HLAING HLAING WIN THESIS Submitted in partial fulfillment of the requirements for the degree of Master of Science in Bioinformatics in the Graduate College
Breast cancer. Risk factors you cannot change include: Treatment Plan Selection. Inferring Transcriptional Module from Breast Cancer Profile Data
 Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
Mechanisms of alternative splicing regulation
 Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
Deciphering the Role of micrornas in BRD4-NUT Fusion Gene Induced NUT Midline Carcinoma
 www.bioinformation.net Volume 13(6) Hypothesis Deciphering the Role of micrornas in BRD4-NUT Fusion Gene Induced NUT Midline Carcinoma Ekta Pathak 1, Bhavya 1, Divya Mishra 1, Neelam Atri 1, 2, Rajeev
www.bioinformation.net Volume 13(6) Hypothesis Deciphering the Role of micrornas in BRD4-NUT Fusion Gene Induced NUT Midline Carcinoma Ekta Pathak 1, Bhavya 1, Divya Mishra 1, Neelam Atri 1, 2, Rajeev
Polyomaviridae. Spring
 Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Small RNAs and Cross-Kingdom RNAi in Plant-Microbial Interaction Hailing Jin
 Small RNAs and Cross-Kingdom RNAi in Plant-Microbial Interaction Hailing Jin Department of Plant Pathology & Microbiology Institute for Integrative Genome Biology micrornas (mirnas) Small RNAs PolII PolIV
Small RNAs and Cross-Kingdom RNAi in Plant-Microbial Interaction Hailing Jin Department of Plant Pathology & Microbiology Institute for Integrative Genome Biology micrornas (mirnas) Small RNAs PolII PolIV
Fig Mosaic. Bryce W. Falk and Nida Salem Plant Pathology University of California Davis, CA
 Fig Mosaic Bryce W. Falk and Nida Salem Plant Pathology University of California Davis, CA This project was initiated in summer of 2005. We have applied for a second years funding to continue efforts along
Fig Mosaic Bryce W. Falk and Nida Salem Plant Pathology University of California Davis, CA This project was initiated in summer of 2005. We have applied for a second years funding to continue efforts along
Small RNAs and how to analyze them using sequencing
 Small RNAs and how to analyze them using sequencing RNA-seq Course November 8th 2017 Marc Friedländer ComputaAonal RNA Biology Group SciLifeLab / Stockholm University Special thanks to Jakub Westholm for
Small RNAs and how to analyze them using sequencing RNA-seq Course November 8th 2017 Marc Friedländer ComputaAonal RNA Biology Group SciLifeLab / Stockholm University Special thanks to Jakub Westholm for
Lab Tuesday: Virus Diseases
 Lab Tuesday: Virus Diseases Quiz for Bacterial Pathogens lab (pp 67-73) and Biocontrol of Crown Gall (p. 113-117), Observation of Viral Movement in Plants (p. 119), and Intro section for Viruses (pp. 75-77).
Lab Tuesday: Virus Diseases Quiz for Bacterial Pathogens lab (pp 67-73) and Biocontrol of Crown Gall (p. 113-117), Observation of Viral Movement in Plants (p. 119), and Intro section for Viruses (pp. 75-77).
AN UNUSUAL VIRUS IN TREES WITH CITRUS BLIGHT RON BRLANSKY UNIVERSITY OF FLORIDA, CREC
 AN UNUSUAL VIRUS IN TREES WITH CITRUS BLIGHT RON BRLANSKY UNIVERSITY OF FLORIDA, CREC CITRUS BLIGHT KNOWN IN FLORIDA FOR OVER 100 YEARS; FIRST DESCRIBED IN 1874 PROBLEM IN FLORIDA IN THE 1970 S WITH INCREASE
AN UNUSUAL VIRUS IN TREES WITH CITRUS BLIGHT RON BRLANSKY UNIVERSITY OF FLORIDA, CREC CITRUS BLIGHT KNOWN IN FLORIDA FOR OVER 100 YEARS; FIRST DESCRIBED IN 1874 PROBLEM IN FLORIDA IN THE 1970 S WITH INCREASE
Ch. 18 Regulation of Gene Expression
 Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
omiras: MicroRNA regulation of gene expression
 omiras: MicroRNA regulation of gene expression Sören Müller, Goethe University of Frankfurt am Main Molecular Bioinformatics Group, Institute of Computer Science Plant Molecular Biology Group, Institute
omiras: MicroRNA regulation of gene expression Sören Müller, Goethe University of Frankfurt am Main Molecular Bioinformatics Group, Institute of Computer Science Plant Molecular Biology Group, Institute
Phenomena first observed in petunia
 Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
Inferring condition-specific mirna activity from matched mirna and mrna expression data
 Inferring condition-specific mirna activity from matched mirna and mrna expression data Junpeng Zhang 1, Thuc Duy Le 2, Lin Liu 2, Bing Liu 3, Jianfeng He 4, Gregory J Goodall 5 and Jiuyong Li 2,* 1 Faculty
Inferring condition-specific mirna activity from matched mirna and mrna expression data Junpeng Zhang 1, Thuc Duy Le 2, Lin Liu 2, Bing Liu 3, Jianfeng He 4, Gregory J Goodall 5 and Jiuyong Li 2,* 1 Faculty
Arabidopsis thaliana small RNA Sequencing. Report
 Arabidopsis thaliana small RNA Sequencing Report September 2015 Project Information Client Name Client Company / Institution Macrogen Order Number Order ID Species Arabidopsis thaliana Reference UCSC hg19
Arabidopsis thaliana small RNA Sequencing Report September 2015 Project Information Client Name Client Company / Institution Macrogen Order Number Order ID Species Arabidopsis thaliana Reference UCSC hg19
Lab Tuesday: Virus Diseases
 Lab Tuesday: Virus Diseases Quiz for Bacterial Pathogens lab (pp 69-75) and Biocontrol of Crown Gall (p. 115-119), Observation of Viral Movement in Plants (p. 121), and Intro section for Viruses (pp. 77-79).
Lab Tuesday: Virus Diseases Quiz for Bacterial Pathogens lab (pp 69-75) and Biocontrol of Crown Gall (p. 115-119), Observation of Viral Movement in Plants (p. 121), and Intro section for Viruses (pp. 77-79).
On the Reproducibility of TCGA Ovarian Cancer MicroRNA Profiles
 On the Reproducibility of TCGA Ovarian Cancer MicroRNA Profiles Ying-Wooi Wan 1,2,4, Claire M. Mach 2,3, Genevera I. Allen 1,7,8, Matthew L. Anderson 2,4,5 *, Zhandong Liu 1,5,6,7 * 1 Departments of Pediatrics
On the Reproducibility of TCGA Ovarian Cancer MicroRNA Profiles Ying-Wooi Wan 1,2,4, Claire M. Mach 2,3, Genevera I. Allen 1,7,8, Matthew L. Anderson 2,4,5 *, Zhandong Liu 1,5,6,7 * 1 Departments of Pediatrics
Seasonal Changes in Leaf Magnesium and Boron Contents and Their Relationships to Leaf Yellowing of Navel Orange Citrus sinensis Osbeck
 2010 37 81317 1324 Acta Horticulturae Sinica 1, 1,2,* 1 3 2 400712 400716 3 341000 6 P < 0.01 0.9082 0.9063 P > 0.05 S 666.4 A 0513-353X 2010 08-1317-08 Seasonal Changes in Leaf Magnesium and Boron Contents
2010 37 81317 1324 Acta Horticulturae Sinica 1, 1,2,* 1 3 2 400712 400716 3 341000 6 P < 0.01 0.9082 0.9063 P > 0.05 S 666.4 A 0513-353X 2010 08-1317-08 Seasonal Changes in Leaf Magnesium and Boron Contents
Supplementary Discussion 1: The mechanistic model of NIK1-mediated antiviral
 doi:10.1038/nature14171 SUPPLEMENTARY DISCUSSION Supplementary Discussion 1: The mechanistic model of NIK1-mediated antiviral signaling 1. Stress-induced oligomerization of the extracellular domain of
doi:10.1038/nature14171 SUPPLEMENTARY DISCUSSION Supplementary Discussion 1: The mechanistic model of NIK1-mediated antiviral signaling 1. Stress-induced oligomerization of the extracellular domain of
High coverage in planta RNA sequencing identifies Fusarium oxysporum effectors and Medicago truncatularesistancemechanisms
 High coverage in planta RNA sequencing identifies Fusarium oxysporum effectors and Medicago truncatularesistancemechanisms Louise Thatcher Gagan Garg, Angela Williams, Judith Lichtenzveig and Karam Singh
High coverage in planta RNA sequencing identifies Fusarium oxysporum effectors and Medicago truncatularesistancemechanisms Louise Thatcher Gagan Garg, Angela Williams, Judith Lichtenzveig and Karam Singh
RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice
 SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
MINIREVIEW. Activation and Suppression of RNA Silencing by Plant Viruses
 Virology 281, 1 5 (2001) doi:10.1006/viro.2000.0812, available online at http://www.idealibrary.com on MINIREVIEW Activation and Suppression of RNA Silencing by Plant Viruses James C. Carrington, 1 Kristin
Virology 281, 1 5 (2001) doi:10.1006/viro.2000.0812, available online at http://www.idealibrary.com on MINIREVIEW Activation and Suppression of RNA Silencing by Plant Viruses James C. Carrington, 1 Kristin
RNA interference (RNAi)
 RN interference (RNi) Natasha aplen ene Silencing Section Office of Science and Technology Partnerships Office of the Director enter for ancer Research National ancer Institute ncaplen@mail.nih.gov Plants
RN interference (RNi) Natasha aplen ene Silencing Section Office of Science and Technology Partnerships Office of the Director enter for ancer Research National ancer Institute ncaplen@mail.nih.gov Plants
Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins.
 Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins. The RNA transcribed from a complex transcription unit
Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins. The RNA transcribed from a complex transcription unit
Cross-Protection Techniques for Control of Plant Virus Diseases in the Tropics
 Symposium International Problems in Tropical Plant Pathology JOHN E. BOWMAN, PepsiCo Foods International, Dallas, TX; Chairman, Tropical Plant Pathology Committee, 1986-1987 The theme of the 1986 annual
Symposium International Problems in Tropical Plant Pathology JOHN E. BOWMAN, PepsiCo Foods International, Dallas, TX; Chairman, Tropical Plant Pathology Committee, 1986-1987 The theme of the 1986 annual
Overview: Chapter 19 Viruses: A Borrowed Life
 Overview: Chapter 19 Viruses: A Borrowed Life Viruses called bacteriophages can infect and set in motion a genetic takeover of bacteria, such as Escherichia coli Viruses lead a kind of borrowed life between
Overview: Chapter 19 Viruses: A Borrowed Life Viruses called bacteriophages can infect and set in motion a genetic takeover of bacteria, such as Escherichia coli Viruses lead a kind of borrowed life between
How a surprising lab result suggests a nutritional means to control HLB.
 How a surprising lab result suggests a nutritional means to control HLB. Prof. Eric W. Triplett, Chair Microbiology and Cell Science Department Institute of Food and Agricultural Sciences University of
How a surprising lab result suggests a nutritional means to control HLB. Prof. Eric W. Triplett, Chair Microbiology and Cell Science Department Institute of Food and Agricultural Sciences University of
Small RNA-Seq and profiling
 Small RNA-Seq and profiling Y. Hoogstrate 1,2 1 Department of Bioinformatics & Department of Urology ErasmusMC, Rotterdam 2 CTMM Translational Research IT (TraIT) BioSB: 5th RNA-seq data analysis course,
Small RNA-Seq and profiling Y. Hoogstrate 1,2 1 Department of Bioinformatics & Department of Urology ErasmusMC, Rotterdam 2 CTMM Translational Research IT (TraIT) BioSB: 5th RNA-seq data analysis course,
Profiling of the Exosomal Cargo of Bovine Milk Reveals the Presence of Immune- and Growthmodulatory Non-coding RNAs (ncrna)
 Animal Industry Report AS 664 ASL R3235 2018 Profiling of the Exosomal Cargo of Bovine Milk Reveals the Presence of Immune- and Growthmodulatory Non-coding RNAs (ncrna) Eric D. Testroet Washington State
Animal Industry Report AS 664 ASL R3235 2018 Profiling of the Exosomal Cargo of Bovine Milk Reveals the Presence of Immune- and Growthmodulatory Non-coding RNAs (ncrna) Eric D. Testroet Washington State
Computational Identification and Prediction of Tissue-Specific Alternative Splicing in H. Sapiens. Eric Van Nostrand CS229 Final Project
 Computational Identification and Prediction of Tissue-Specific Alternative Splicing in H. Sapiens. Eric Van Nostrand CS229 Final Project Introduction RNA splicing is a critical step in eukaryotic gene
Computational Identification and Prediction of Tissue-Specific Alternative Splicing in H. Sapiens. Eric Van Nostrand CS229 Final Project Introduction RNA splicing is a critical step in eukaryotic gene
Small RNAs and how to analyze them using sequencing
 Small RNAs and how to analyze them using sequencing Jakub Orzechowski Westholm (1) Long- term bioinforma=cs support, Science For Life Laboratory Stockholm (2) Department of Biophysics and Biochemistry,
Small RNAs and how to analyze them using sequencing Jakub Orzechowski Westholm (1) Long- term bioinforma=cs support, Science For Life Laboratory Stockholm (2) Department of Biophysics and Biochemistry,
WHITE PAPER. Increasing Ligation Efficiency and Discovery of mirnas for Small RNA NGS Sequencing Library Prep with Plant Samples
 WHITE PPER Increasing Ligation Efficiency and iscovery of mirns for Small RN NGS Sequencing Library Prep with Plant Samples ioo Scientific White Paper May 2017 bstract MicroRNs (mirns) are 18-22 nucleotide
WHITE PPER Increasing Ligation Efficiency and iscovery of mirns for Small RN NGS Sequencing Library Prep with Plant Samples ioo Scientific White Paper May 2017 bstract MicroRNs (mirns) are 18-22 nucleotide
York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
 MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
Identifying mirnas in RNA viruses
 1 / 13 Identifying mirnas in RNA viruses Kevin Lamkiewicz Friedrich Schiller Universität Jena 16.02.2017 32nd TBI Winterseminar Bled 2 / 13 mirnas and their function U C A C A G G U C A A G C G U U G G
1 / 13 Identifying mirnas in RNA viruses Kevin Lamkiewicz Friedrich Schiller Universität Jena 16.02.2017 32nd TBI Winterseminar Bled 2 / 13 mirnas and their function U C A C A G G U C A A G C G U U G G
Not IN Our Genes - A Different Kind of Inheritance.! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014
 Not IN Our Genes - A Different Kind of Inheritance! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014 Epigenetics in Mainstream Media Epigenetics *Current definition:
Not IN Our Genes - A Different Kind of Inheritance! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014 Epigenetics in Mainstream Media Epigenetics *Current definition:
Profiles of gene expression & diagnosis/prognosis of cancer. MCs in Advanced Genetics Ainoa Planas Riverola
 Profiles of gene expression & diagnosis/prognosis of cancer MCs in Advanced Genetics Ainoa Planas Riverola Gene expression profiles Gene expression profiling Used in molecular biology, it measures the
Profiles of gene expression & diagnosis/prognosis of cancer MCs in Advanced Genetics Ainoa Planas Riverola Gene expression profiles Gene expression profiling Used in molecular biology, it measures the
Chapter 19: Viruses. 1. Viral Structure & Reproduction. 2. Bacteriophages. 3. Animal Viruses. 4. Viroids & Prions
 Chapter 19: Viruses 1. Viral Structure & Reproduction 2. Bacteriophages 3. Animal Viruses 4. Viroids & Prions 1. Viral Structure & Reproduction Chapter Reading pp. 393-396 What exactly is a Virus? Viruses
Chapter 19: Viruses 1. Viral Structure & Reproduction 2. Bacteriophages 3. Animal Viruses 4. Viroids & Prions 1. Viral Structure & Reproduction Chapter Reading pp. 393-396 What exactly is a Virus? Viruses
Introduction retroposon
 17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
RNA-seq Introduction
 RNA-seq Introduction DNA is the same in all cells but which RNAs that is present is different in all cells There is a wide variety of different functional RNAs Which RNAs (and sometimes then translated
RNA-seq Introduction DNA is the same in all cells but which RNAs that is present is different in all cells There is a wide variety of different functional RNAs Which RNAs (and sometimes then translated
Table S1. Relative abundance of AGO1/4 proteins in different organs. Table S2. Summary of smrna datasets from various samples.
 Supplementary files Table S1. Relative abundance of AGO1/4 proteins in different organs. Table S2. Summary of smrna datasets from various samples. Table S3. Specificity of AGO1- and AGO4-preferred 24-nt
Supplementary files Table S1. Relative abundance of AGO1/4 proteins in different organs. Table S2. Summary of smrna datasets from various samples. Table S3. Specificity of AGO1- and AGO4-preferred 24-nt
Chapter 19: Viruses. 1. Viral Structure & Reproduction. What exactly is a Virus? 11/7/ Viral Structure & Reproduction. 2.
 Chapter 19: Viruses 1. Viral Structure & Reproduction 2. Bacteriophages 3. Animal Viruses 4. Viroids & Prions 1. Viral Structure & Reproduction Chapter Reading pp. 393-396 What exactly is a Virus? Viruses
Chapter 19: Viruses 1. Viral Structure & Reproduction 2. Bacteriophages 3. Animal Viruses 4. Viroids & Prions 1. Viral Structure & Reproduction Chapter Reading pp. 393-396 What exactly is a Virus? Viruses
Human breast milk mirna, maternal probiotic supplementation and atopic dermatitis in offsrping
 Human breast milk mirna, maternal probiotic supplementation and atopic dermatitis in offsrping Melanie Rae Simpson PhD candidate Department of Public Health and General Practice Norwegian University of
Human breast milk mirna, maternal probiotic supplementation and atopic dermatitis in offsrping Melanie Rae Simpson PhD candidate Department of Public Health and General Practice Norwegian University of
ESCs were lysed with Trizol reagent (Life technologies) and RNA was extracted according to
 Supplemental Methods RNA-seq ESCs were lysed with Trizol reagent (Life technologies) and RNA was extracted according to the manufacturer's instructions. RNAse free DNaseI (Sigma) was used to eliminate
Supplemental Methods RNA-seq ESCs were lysed with Trizol reagent (Life technologies) and RNA was extracted according to the manufacturer's instructions. RNAse free DNaseI (Sigma) was used to eliminate
he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003)
 MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature14008 Supplementary Figure 1. Sequence alignment of A/little yellow-shouldered bat/guatemala/060/2010 (H17N10) polymerase with that of human strain A/Victoria/3/75(H3N2). The secondary
doi:10.1038/nature14008 Supplementary Figure 1. Sequence alignment of A/little yellow-shouldered bat/guatemala/060/2010 (H17N10) polymerase with that of human strain A/Victoria/3/75(H3N2). The secondary
UC Riverside International Organization of Citrus Virologists Conference Proceedings ( )
 UC Riverside International Organization of Citrus Virologists Conference Proceedings (1957-2010) Title Reduction in Yield of Galego Lime Avoided by Preimmunization with Mild Strains of Tristeza Virus Permalink
UC Riverside International Organization of Citrus Virologists Conference Proceedings (1957-2010) Title Reduction in Yield of Galego Lime Avoided by Preimmunization with Mild Strains of Tristeza Virus Permalink
Instituto de Biociências da Universidade Estadual Paulista (Unesp), Botucatu, SP, , Brazil; 2
 Article Deep Sequencing Analysis of RNAs from Citrus Plants Grown in a Citrus Sudden Death Affected Area Reveals Diverse Known and Putative Novel Viruses Emilyn E. Matsumura 1,2, Helvécio D. Coletta Filho
Article Deep Sequencing Analysis of RNAs from Citrus Plants Grown in a Citrus Sudden Death Affected Area Reveals Diverse Known and Putative Novel Viruses Emilyn E. Matsumura 1,2, Helvécio D. Coletta Filho
Supplemental Data. Integrating omics and alternative splicing i reveals insights i into grape response to high temperature
 Supplemental Data Integrating omics and alternative splicing i reveals insights i into grape response to high temperature Jianfu Jiang 1, Xinna Liu 1, Guotian Liu, Chonghuih Liu*, Shaohuah Li*, and Lijun
Supplemental Data Integrating omics and alternative splicing i reveals insights i into grape response to high temperature Jianfu Jiang 1, Xinna Liu 1, Guotian Liu, Chonghuih Liu*, Shaohuah Li*, and Lijun
Section 6. Junaid Malek, M.D.
 Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
Gynophore mirna analysis at different developmental stages in Arachis duranensis
 Gynophore mirna analysis at different developmental stages in Arachis duranensis Y. Shen 1,2 *, Y.H. Liu 1 *, X.J. Zhang 3, Q. Sha 1 and Z.D. Chen 1 1 Institute of Industrial Crops, Jiangsu Academy of
Gynophore mirna analysis at different developmental stages in Arachis duranensis Y. Shen 1,2 *, Y.H. Liu 1 *, X.J. Zhang 3, Q. Sha 1 and Z.D. Chen 1 1 Institute of Industrial Crops, Jiangsu Academy of
A novel method for multiple water-borne plant virus detection combining Deep sequencing and Bioinformatics.
 A novel method for multiple water-borne plant virus detection combining Deep sequencing and Bioinformatics. WATER CONFERENCE OKLAHOMA CITY LIZBETH PEÑA-ZÚÑIGA 1, Jon Daniels 2, Andrés Espíndola 1, and
A novel method for multiple water-borne plant virus detection combining Deep sequencing and Bioinformatics. WATER CONFERENCE OKLAHOMA CITY LIZBETH PEÑA-ZÚÑIGA 1, Jon Daniels 2, Andrés Espíndola 1, and
A Practical Guide to Integrative Genomics by RNA-seq and ChIP-seq Analysis
 A Practical Guide to Integrative Genomics by RNA-seq and ChIP-seq Analysis Jian Xu, Ph.D. Children s Research Institute, UTSW Introduction Outline Overview of genomic and next-gen sequencing technologies
A Practical Guide to Integrative Genomics by RNA-seq and ChIP-seq Analysis Jian Xu, Ph.D. Children s Research Institute, UTSW Introduction Outline Overview of genomic and next-gen sequencing technologies
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
CRS4 Seminar series. Inferring the functional role of micrornas from gene expression data CRS4. Biomedicine. Bioinformatics. Paolo Uva July 11, 2012
 CRS4 Seminar series Inferring the functional role of micrornas from gene expression data CRS4 Biomedicine Bioinformatics Paolo Uva July 11, 2012 Partners Pharmaceutical company Fondazione San Raffaele,
CRS4 Seminar series Inferring the functional role of micrornas from gene expression data CRS4 Biomedicine Bioinformatics Paolo Uva July 11, 2012 Partners Pharmaceutical company Fondazione San Raffaele,
Marta Puerto Plasencia. microrna sponges
 Marta Puerto Plasencia microrna sponges Introduction microrna CircularRNA Publications Conclusions The most well-studied regions in the human genome belong to proteincoding genes. Coding exons are 1.5%
Marta Puerto Plasencia microrna sponges Introduction microrna CircularRNA Publications Conclusions The most well-studied regions in the human genome belong to proteincoding genes. Coding exons are 1.5%
genomics for systems biology / ISB2020 RNA sequencing (RNA-seq)
 RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
Supplementary information for: Community detection for networks with. unipartite and bipartite structure. Chang Chang 1, 2, Chao Tang 2
 Supplementary information for: Community detection for networks with unipartite and bipartite structure Chang Chang 1, 2, Chao Tang 2 1 School of Life Sciences, Peking University, Beiing 100871, China
Supplementary information for: Community detection for networks with unipartite and bipartite structure Chang Chang 1, 2, Chao Tang 2 1 School of Life Sciences, Peking University, Beiing 100871, China
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Transcription and RNA processing
 Transcription and RNA processing Lecture 7 Biology 3310/4310 Virology Spring 2018 It is possible that Nature invented DNA for the purpose of achieving regulation at the transcriptional rather than at the
Transcription and RNA processing Lecture 7 Biology 3310/4310 Virology Spring 2018 It is possible that Nature invented DNA for the purpose of achieving regulation at the transcriptional rather than at the
Obstacles and challenges in the analysis of microrna sequencing data
 Obstacles and challenges in the analysis of microrna sequencing data (mirna-seq) David Humphreys Genomics core Dr Victor Chang AC 1936-1991, Pioneering Cardiothoracic Surgeon and Humanitarian The ABCs
Obstacles and challenges in the analysis of microrna sequencing data (mirna-seq) David Humphreys Genomics core Dr Victor Chang AC 1936-1991, Pioneering Cardiothoracic Surgeon and Humanitarian The ABCs
Severe Acute Respiratory Syndrome (SARS) Coronavirus
 Severe Acute Respiratory Syndrome (SARS) Coronavirus Coronaviruses Coronaviruses are single stranded enveloped RNA viruses that have a helical geometry. Coronaviruses are the largest of RNA viruses with
Severe Acute Respiratory Syndrome (SARS) Coronavirus Coronaviruses Coronaviruses are single stranded enveloped RNA viruses that have a helical geometry. Coronaviruses are the largest of RNA viruses with
Framework for the evaluation of. and scientific impacts of plant viruses. technologies.
 Framework for the evaluation of biosecurity, commercial, regulatory and scientific impacts of plant viruses and viroids identified by NGS technologies. Sebastien Massart, Thierry Candresse, José Gil, Christophe
Framework for the evaluation of biosecurity, commercial, regulatory and scientific impacts of plant viruses and viroids identified by NGS technologies. Sebastien Massart, Thierry Candresse, José Gil, Christophe
Identification of new resistance sources for Cucumber mosaic virus new isolate-p1 (CMV-P1) in Capsicum spp.
 Absorbance at 405nm Absorbance at 405nm Absorbance at 405nm College of Agriculture and Life Sciences Identification of new resistance sources for Cucumber mosaic virus new isolate-p1 (CMV-P1) in Capsicum
Absorbance at 405nm Absorbance at 405nm Absorbance at 405nm College of Agriculture and Life Sciences Identification of new resistance sources for Cucumber mosaic virus new isolate-p1 (CMV-P1) in Capsicum
Accessing and Using ENCODE Data Dr. Peggy J. Farnham
 1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
Page 32 AP Biology: 2013 Exam Review CONCEPT 6 REGULATION
 Page 32 AP Biology: 2013 Exam Review CONCEPT 6 REGULATION 1. Feedback a. Negative feedback mechanisms maintain dynamic homeostasis for a particular condition (variable) by regulating physiological processes,
Page 32 AP Biology: 2013 Exam Review CONCEPT 6 REGULATION 1. Feedback a. Negative feedback mechanisms maintain dynamic homeostasis for a particular condition (variable) by regulating physiological processes,
Dr. Gary Mumaugh. Viruses
 Dr. Gary Mumaugh Viruses Viruses in History In 1898, Friedrich Loeffler and Paul Frosch found evidence that the cause of foot-and-mouth disease in livestock was an infectious particle smaller than any
Dr. Gary Mumaugh Viruses Viruses in History In 1898, Friedrich Loeffler and Paul Frosch found evidence that the cause of foot-and-mouth disease in livestock was an infectious particle smaller than any
EPIGENOMICS PROFILING SERVICES
 EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
Identification of mirnas and Their Targets in Cotton Inoculated with Verticillium dahliae by High-Throughput Sequencing and Degradome Analysis
 Int. J. Mol. Sci. 2015, 16, 14749-14768; doi:10.3390/ijms160714749 Article OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Identification of mirnas and
Int. J. Mol. Sci. 2015, 16, 14749-14768; doi:10.3390/ijms160714749 Article OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Identification of mirnas and
Oncolytic virus strategy
 Oncolytic viruses Oncolytic virus strategy normal tumor NO replication replication survival lysis Oncolytic virus strategy Mechanisms of tumor selectivity of several, some of them naturally, oncolytic
Oncolytic viruses Oncolytic virus strategy normal tumor NO replication replication survival lysis Oncolytic virus strategy Mechanisms of tumor selectivity of several, some of them naturally, oncolytic
MicroRNA-mediated incoherent feedforward motifs are robust
 The Second International Symposium on Optimization and Systems Biology (OSB 8) Lijiang, China, October 31 November 3, 8 Copyright 8 ORSC & APORC, pp. 62 67 MicroRNA-mediated incoherent feedforward motifs
The Second International Symposium on Optimization and Systems Biology (OSB 8) Lijiang, China, October 31 November 3, 8 Copyright 8 ORSC & APORC, pp. 62 67 MicroRNA-mediated incoherent feedforward motifs
