Epigenetic Regulation by Chromatin Activation Mark H3K4me3 in Primate Progenitor
|
|
|
- Godfrey Melton
- 5 years ago
- Views:
Transcription
1 Epigenetic Regulation by Chromatin Activation Mark H3K4me3 in Primate Progenitor Cells within Adult Neurogenic Niche Richard S. Sandstrom 1, Michael R. Foret 2, Douglas A. Grow 2,3, Eric Haugen 1, Christopher T. Rhodes 2, Astrid E. Cardona 2, Clyde F. Phelix 2, Yufeng Wang 2, Mitchel S. Berger 4, and Chin-Hsing Annie Lin 2,3* Supplemental Figure 1. Chromatin activation mark H3K4me3 is expressed in the SVZ cell lineages of mouse brain. (A-D) Double labeling of H3K4me3 (red) and NSC marker- GFAP (A, B) as well as other lineage-specific markers showed H3K4me3 co-expressing with Mash1 (C), PSA-NCAM (E), DCX (F), and Double-labeling of EdU and H3K4me3 in SVZ (D). Double-labeled cells were marked by arrows. (G, H) Antimitotic treatment abolished most of the H3K4me3 expressing cells in adult SVZ (H), whereas H3K4me3-positive cells were retained in saline control (G). LV: lateral ventricle; CC: Corpus Callosum; Str: Striatum (12 coronal sections; 40X oil; scale bar= 50 m). Supplemental Figure 2. Network and Pathway analyses by IPA for H3K4me3 enriched genes in baboon SVZ cells. The top canonical pathways and networks were predicted by the Ingenuity Pathway Analysis software. The shaded focus genes (red highlight) were enriched with H3K4me3 identified by ChIP-Seq analysis. The node shape reflects the role of each element in the network and the direction and arrowhead shapes of each edge represent different types of interactions (see the key at left panel). (A) Post-translational modifications; (B) Network in Nervous System Development and Function; (C) Molecular transport. Supplemental Figure 3. Network and Pathway analyses by IPA for H3K4me3 enriched genes in baboon SVZ cells. (A) Metabolism. (B) RNA post-transcriptional modification
2 Supplemental Figure 4. Network and Pathway analyses by IPA for H3K4me3 enriched genes in baboon SVZ cells highlight the Pathway related to Molecular Mechanisms of Cancer. Supplemental Figure 5. Network and Pathway analyses for H3K4me3 enriched genes in baboon SVZ cells highlight the pathway of eif4 signaling and ribosomal biogenesis. Supplemental Figure 6. Venn diagram displays the differential and shared targets of H3K4me3 in the purified NSC (neural stem cell) versus NB (neuroblast) populations. The statistically overrepresented GO terms for each shared and unique targets are shown in the box. Supplemental Table 1: Summary of H3K4me3-enriched genes in the Baboon SVZ (i a-f ) NSCs; (ii a,b ) neuroblasts (NB); (iii a-f ) Total progenitors (NSC and NB); (iv a-d ) overlap comparison between NSC and NB populations; (v a,b ) Gene Ontology by DAVID for H3K4me3-enriched genes in NSCs; (vi) Gene Ontology by DAVID for H3K4me3-enriched genes in NBs. Supplemental Table 2: Summary of overlap targets and Gene Ontology by DAVID for (i a,b ) GBM1 RNA-Seq and H3K4me3 ChIP-Seq; (ii a,b ) GBM2 RNA-Seq and H3K4me3 ChIP-Seq; (iii a,b ) GBM1 RNA-Seq, GBM2 RNA-Seq, and H3K4me3 ChIP-Seq; (iv a ) Overlap genes among detectable transcripts from RNA-Seq of Baboon SVZ cells, GBM1 RNA-Seq, and GBM2 RNA-Seq; (iv b ) Overlap genes among undetectable transcripts from RNA-Seq of Baboon SVZ cells, GBM1 RNA-Seq, and GBM2 RNA-Seq. All deep sequencing data for ChIP-Seq and RNA-Seq generated during the course of this study refer to GEO files.
3 Epigenetic Regulation by Chromatin Activation Mark H3K4me3 in Primate Progenitor Cells within Adult Neurogenic Niche Richard S. Sandstrom 1, Michael R. Foret 2, Douglas A. Grow 2,3, Eric Haugen 1, Christopher T. Rhodes 2, Astrid E. Cardona 2, Clyde F. Phelix 2, Yufeng Wang 2, Mitchel S. Berger 4, and Chin-Hsing Annie Lin 2,3* Lin_Supplemental Figure 1 A B C D CC H3K4me3 GFAP DAPI LV H3K4me3 GFAP DAPI LV H3K4me3 Mash1 DAPI LV H3K4me3 EdU DAPI E F G Saline control H AraC LV LV LV LV H3K4me3 DCX DAPI H3K4me3 PSANCAM DAPI H3K4me3 DAPI H3K4me3 DAPI Str Str
4 Lin_Supplemental Figure 2 A B C
5 Lin_Supplemental Figure 3 A B
6 Lin_Supplemental Figure 4
7 Lin_Supplemental Figure 5
8 Lin_Supplemental Figure 6 NSC-specific: - Cell cycle - Cell death - Cell proliferation - DNA repair NSC NB-specific: - ribosomal biogenesis - mitochondria ribogenesis NB NSC/NB-shared pathways: - Chromatin modification - Signaling - Nervous system development and function
9 Supplemental Table 1: (i a ) H3K4me3-enriched genes in the Baboon SVZ NSC population Gene Name RefSeq ID Gene Name RefSeq ID Gene Name RefSeq ID Gene Name RefSeq ID Gene Name RefSeq ID HMX1 NM_ ATG4C NM_ MRPL24 NM_ TNR NM_ PRKCE NM_ ADD1 NM_ PGM1 NM_ PRCC NM_ RFWD2 NM_ ATP6V1E2 NM_ MAEA NM_ JAK1 NM_ CD1D NM_ ASTN1 NM_ C13H2orf61 NM_ PCGF3 NM_ LEPR NM_ CD1B NM_ PGBD2 NM_ EPCAM NM_ TTLL10 NM_ PDE4B NM_ ATP1A2 NM_ ZNF692 NM_ FOXN2 NM_ GLTPD1 NM_ SGIP1 NM_ PEA15 NM_ SH3BP5L NM_ ASB3 NM_ MRPL20 NM_ GADD45A NM_ NCSTN NM_ NLRP3 NM_ SPTBN1 NM_ VWA1 NM_ DIRAS3 NM_ F11R NM_ SMYD3 NM_ RTN4 NM_ GNB1 NM_ LRRC40 NM_ UHMK1 NM_ COX20 NM_ CCDC104 NM_ RER1 NM_ ZRANB2 NM_ TRIM11 NM_ ADSS NM_ PEX13 NM_ TPRG1L NM_ NEGR1 NM_ GUK1 NM_ AKT3 NM_ C13H2orf74 NM_ RPL22 NM_ FPGT NM_ C1H1orf35 NM_ GREM2 NM_ XPO1 NM_ PHF13 NM_ TNNI3K NM_ PSEN2 NM_ CHRM3 NM_ B3GNT2 NM_ VAMP3 NM_ RABGGTB NM_ C1H1orf95 NM_ LGALS8 NM_ EHBP1 NM_ RERE NM_ ST6GALNAC3 NM_ PYCR2 NM_ TBCE NM_ VPS54 NM_ SPSB1 NM_ PIGK NM_ EPHX1 NM_ RBM34 NM_ PELI1 NM_ CTNNBIP1 NM_ AK5 NM_ LBR NM_ KIAA1804 NM_ SERTAD2 NM_ DFFA NM_ RPF1 NM_ CAPN2 NM_ NTPCR NM_ RAB1A NM_ TNFRSF1B NM_ C1H1orf52 NM_ TLR5 NM_ DISC1 NM_ ACTR2 NM_ DHRS3 NM_ DDAH1 NM_ DUSP10 NM_ GNPAT NM_ SPRED2 NM_ EFHD2 NM_ ZNHIT6 NM_ LYPLAL1 NM_ GALNT2 NM_ C1D NM_ ZBTB17 NM_ GTF2B NM_ TGFB2 NM_ CCSAP NM_ ARHGAP25 NM_ NECAP2 NM_ LRRC8C NM_ KCTD3 NM_ PCMTD2 NM_ ANXA4 NM_ IGSF21 NM_ LRRC8D NM_ PROX1 NM_ GID8 NM_ ADD2 NM_ EMC1 NM_ ZNF326 NM_ KCNH1 NM_ LSM14B NM_ TEX261 NM_ ALPL NM_ CDC7 NM_ DIEXF NM_ ATP5E NM_ PAIP2B NM_ CELA3B NM_ RPAP2 NM_ IRF6 NM_ STX16 NM_ SPR NM_ KDM1A NM_ FAM69A NM_ TRAF3IP3 NM_ RAB22A NM_ RTKN NM_ ZNF436 NM_ DR1 NM_ PFKFB2 NM_ RBM38 NM_ TACR1 NM_ GALE NM_ CNN3 NM_ EIF2D NM_ CBLN4 NM_ CTNNA2 NM_ RHCE NM_ RWDD3 NM_ AVPR1B NM_ PFDN4 NM_ LRRTM1 NM_ TMEM57 NM_ CCDC76 NM_ NUCKS1 NM_ ZFP64 NM_ RETSAT NM_ TRIM63 NM_ DBT NM_ ELK4 NM_ SPATA2 NM_ RNF181 NM_ ZNF593 NM_ EXTL2 NM_ KLHDC8A NM_ STAU1 NM_ ST3GAL5 NM_ CD52 NM_ SLC30A7 NM_ TMCC2 NM_ CSE1L NM_ RNF103 NM_ SFN NM_ OLFM3 NM_ CNTN2 NM_ ELMO2 NM_ RPIA NM_ GPN2 NM_ PRMT6 NM_ REN NM_ MMP9 NM_ KCNIP3 NM_ TRNP1 NM_ SLC25A24 NM_ ETNK2 NM_ MYBL2 NM_ CIAO1 NM_ FGR NM_ HENMT1 NM_ ZC3H11A NM_ IFT52 NM_ SEMA4C NM_ EYA3 NM_ PSRC1 NM_ BTG2 NM_ ZHX3 NM_ ACTR1B NM_ YTHDF2 NM_ PSMA5 NM_ SYT2 NM_ TOP1 NM_ INPP4A NM_ SRSF4 NM_ GSTM3 NM_ GPR37L1 NM_ SLC32A1 NM_ MGAT4A NM_ MECR NM_ AHCYL1 NM_ TIMM17A NM_ ADIG NM_ EIF5B NM_ SNRNP40 NM_ KCNA2 NM_ CSRP1 NM_ RPRD1B NM_ CHST10 NM_ KPNA6 NM_ KCNA3 NM_ TNNT2 NM_ C10H20orf118 NM_ CREG2 NM_ ADC NM_ LRIF1 NM_ CAMSAP1L1 NM_ DLGAP4 NM_ IL1R2 NM_ SMIM12 NM_ DENND2D NM_ NEK7 NM_ SCAND1 NM_ TMEM182 NM_ NCDN NM_ WDR77 NM_ ASPM NM_ EDEM2 NM_ MRPS9 NM_ TRAPPC3 NM_ CTTNBP2NL NM_ B3GALT2 NM_ TP53INP2 NM_ C13H2orf49 NM_ LSM10 NM_ SLC16A1 NM_ RGS2 NM_ SNTA1 NM_ C13H2orf40 NM_ MEAF6 NM_ PTPN22 NM_ RGS1 NM_ KIF3B NM_ TTL NM_ MTF1 NM_ OLFML3 NM_ FAM5C NM_ DEFB129 NM_ IL1B NM_ FHL3 NM_ SIKE1 NM_ C1H1orf27 NM_ NRSN2 NM_ CXCR4 NM_ PPT1 NM_ ATP1A1 NM_ IVNS1ABP NM_ CSNK2A1 NM_ MCM6 NM_ SMAP2 NM_ CD58 NM_ EDEM3 NM_ C20orf54 NM_ DPP10 NM_ FOXJ3 NM_ CD2 NM_ C1H1orf21 NM_ PDYN NM_ INSIG2 NM_ RIMKLA NM_ TRIM45 NM_ RGL1 NM_ SNRPB NM_ C1QL2 NM_ C1H1orf50 NM_ WDR3 NM_ RNASEL NM_ ZNF343 NM_ C13H2orf76 NM_ SLC2A1 NM_ NOTCH2 NM_ STX6 NM_ IDH3B NM_ TMEM177 NM_ CDC20 NM_ SEC22B NM_ ACBD6 NM_ GNRH2 NM_ PTPN4 NM_ ELOVL1 NM_ TXNIP NM_ TOR1AIP2 NM_ FASTKD5 NM_ RALB NM_ ATP6V0B NM_ ACP6 NM_ RGS5 NM_ DDRGK1 NM_ ERCC3 NM_ EIF2B3 NM_ GNAQ NM_ NUF2 NM_ C10H20orf194 NM_ POLR2D NM_ HECTD3 NM_ FCGR1 NM_ RXRG NM_ SPEF1 NM_ MZT2B NM_ PRDX1 NM_ CA14 NM_ MGST3 NM_ MAVS NM_ IMP4 NM_ LURAP1 NM_ LASS2 NM_ UCK2 NM_ PRNP NM_ CFC1 NM_ MOB3C NM_ POGZ NM_ MAEL NM_ SLC23A1 NM_ FAM168B NM_ CMPK1 NM_ MRPL9 NM_ CD247 NM_ MCM8 NM_ CD151 NM_ SPATA6 NM_ S100A10 NM_ SFT2D2 NM_ PLCB1 NM_ CHID1 NM_ ELAVL4 NM_ SMCP NM_ XCL1 NM_ C20orf103 NM_ TOLLIP NM_ FAF1 NM_ CHTOP NM_ SLC19A2 NM_ PAK7 NM_ CTSD NM_ CDKN2C NM_ ILF2 NM_ METTL18 NM_ SNAP25 NM_ MRPL23 NM_ RNF11 NM_ CREB3L4 NM_ FMO2 NM_ MKKS NM_ CD81 NM_ GPX7 NM_ RAB13 NM_ PIGC NM_ FLRT3 NM_ TSSC4 NM_ DIO1 NM_ SHC1 NM_ FASLG NM_ PCSK2 NM_ CARS NM_ TMEM59 NM_ EFNA3 NM_ CENPL NM_ DSTN NM_ CTTN NM_ SSBP3 NM_ THBS3 NM_ SERPINC1 NM_ PET117 NM_ ORAOV1 NM_ PCSK9 NM_ RHBG NM_ CACYBP NM_ C10H20orf26 NM_ SUV420H1 NM_ JUN NM_ HAPLN2 NM_ LOC NM_ RALGAPA2 NM_ YIF1A NM_ THBD NM_ CNTN1 NM_ ISCU NM_ ICOS NM_ HS3ST3A1 NM_ NXT1 NM_ GXYLT1 NM_ ALKBH2 NM_ GPR1 NM_ COX10 NM_ CST11 NM_ YAF2 NM_ FAM222A NM_ CREB1 NM_ PMP22 NM_ APMAP NM_ ADAMTS20 NM_ GIT2 NM_ CRYGC NM_ TVP23B NM_ ABHD12 NM_ TMEM117 NM_ PPTC7 NM_ IDH1 NM_ ZNF287 NM_ NANP NM_ NELL2 NM_ PPP1CC NM_ LANCL1 NM_ ZSWIM7 NM_ CECR5 NM_ SLC38A1 NM_ BRAP NM_ IKZF2 NM_ FLCN NM_ PEX26 NM_ SLC38A2 NM_ PTPN11 NM_ MREG NM_ TOM1L2 NM_ COMT NM_ FAM113B NM_ OAS1 NM_ RPL37A NM_ ATPAF2 NM_ KLHL22 NM_ C11H12orf68 NM_ CCDC42B NM_ CXCR1 NM_ ALKBH5 NM_ SDF2L1 NM_ CACNB3 NM_ SDS NM_ ARPC2 NM_ GRAP NM_ TOP3B NM_ CCDC65 NM_ MED13L NM_ TMBIM1 NM_ B9D1 NM_ GNAZ NM_ TMBIM6 NM_ TESC NM_ CTDSP1 NM_ DHRS7B NM_ Lin_Supplemental Table 1
10 Supplemental Table 1: (i b ) H3K4me3-enriched genes in the Baboon SVZ NSC population Gene Name RefSeq ID Gene Name RefSeq ID Gene Name RefSeq ID Gene Name RefSeq ID Gene Name RefSeq ID GUCD1 NM_ COX14 NM_ RFC5 NM_ CRYBA2 NM_ MAP2K3 NM_ CRYBA4 NM_ ATF1 NM_ HSPB8 NM_ DNAJB2 NM_ WSB1 NM_ PITPNB NM_ TFCP2 NM_ PRKAB1 NM_ DES NM_ LYRM9 NM_ RASL10A NM_ C11H12orf44 NM_ RNF10 NM_ CHPF NM_ KIAA0100 NM_ GAL3ST1 NM_ EIF4B NM_ SPPL3 NM_ INHA NM_ FAM222B NM_ DUSP18 NM_ HOXC4 NM_ RNF34 NM_ EPHA4 NM_ CRYBA1 NM_ SELM NM_ OR10P1 NM_ GPR81 NM_ SCG2 NM_ NUFIP2 NM_ YWHAH NM_ CD63 NM_ ABCB9 NM_ CUL3 NM_ CORO6 NM_ TIMP3 NM_ IL23A NM_ RILPL2 NM_ KIAA1486 NM_ TMIGD1 NM_ LARGE NM_ STAT2 NM_ ZNF664 NM_ TM4SF20 NM_ OMG NM_ TOM1 NM_ HSD17B6 NM_ FAM101A NM_ CCL20 NM_ CDK5R1 NM_ RBFOX2 NM_ NAB2 NM_ PIWIL1 NM_ PID1 NM_ ASIC2 NM_ EIF3D NM_ STAT6 NM_ NOC4L NM_ CAB39 NM_ NLE1 NM_ TST NM_ STAC3 NM_ PXMP2 NM_ PSMD1 NM_ RASL10B NM_ POLR2F NM_ OS9 NM_ SPOPL NM_ EIF4E2 NM_ NFE2L1 NM_ CSNK1E NM_ AGAP2 NM_ ARHGAP15 NM_ EFHD1 NM_ HOXB8 NM_ DNAL4 NM_ CYP27B1 NM_ GTDC1 NM_ ATG16L1 NM_ ATP5G1 NM_ SMCR7L NM_ CTDSP2 NM_ MBD5 NM_ UGT1A1 NM_ NXPH3 NM_ GRAP2 NM_ USP15 NM_ RND3 NM_ SH3BP4 NM_ SPOP NM_ FAM109B NM_ C11H12orf61 NM_ CACNB4 NM_ AGAP1 NM_ NME1 NM_ TSPO NM_ C11H12orf66 NM_ ARL6IP6 NM_ CXCR7 NM_ CA10 NM_ LDOC1L NM_ WIF1 NM_ RPRM NM_ ASB1 NM_ TOM1L1 NM_ NUP50 NM_ CAND1 NM_ NR4A2 NM_ TWIST2 NM_ HLF NM_ ATXN10 NM_ DYRK2 NM_ GPD2 NM_ OTOS NM_ MMD NM_ PPARA NM_ IFNG NM_ ACVR1C NM_ GPR35 NM_ TMEM100 NM_ CDPF1 NM_ NUP107 NM_ PKP4 NM_ BOK NM_ NOG NM_ CERK NM_ SLC35E3 NM_ TANK NM_ DTYMK NM_ SCPEP1 NM_ ZBED4 NM_ MDM2 NM_ GCA NM_ ING5 NM_ AKAP1 NM_ FAM116B NM_ CNOT2 NM_ B3GALT1 NM_ MYT1L NM_ MRPS23 NM_ ADM2 NM_ THAP2 NM_ LASS6 NM_ RNASEH1 NM_ SRSF1 NM_ SLC6A12 NM_ TPH2 NM_ BBS5 NM_ IAH1 NM_ SEPT4 NM_ FBXL14 NM_ GLIPR1 NM_ PHOSPHO2 NM_ ODC1 NM_ C17orf71 NM_ LRTM2 NM_ OSBPL8 NM_ GORASP2 NM_ PDIA6 NM_ PTRH2 NM_ FKBP4 NM_ ZDHHC17 NM_ DYNC1I2 NM_ KCNF1 NM_ C6H5orf48 NM_ PARP11 NM_ PPP1R12A NM_ DLX1 NM_ ROCK2 NM_ PPM1D NM_ C11H12orf4 NM_ PPFIA2 NM_ ITGA6 NM_ E2F6 NM_ APPBP2 NM_ VWF NM_ LSM3 NM_ OLA1 NM_ FAM84A NM_ DHRS11 NM_ CD9 NM_ MRPS25 NM_ ATP5G3 NM_ DDX1 NM_ AATF NM_ MLF2 NM_ GP9 NM_ HOXD12 NM_ FAM49A NM_ C16H17orf96 NM_ C11H12orf57 NM_ RPN1 NM_ MTX2 NM_ TTC32 NM_ RPL23 NM_ SLC2A3 NM_ RUVBL1 NM_ PRKRA NM_ RHOB NM_ LASP1 NM_ PHC1 NM_ TPRA1 NM_ SESTD1 NM_ C13H2orf43 NM_ PPP1R1B NM_ KLRF1 NM_ C11H12orf29 NM_ PDE1A NM_ SF3B14 NM_ CASC3 NM_ OLR1 NM_ DUSP6 NM_ DUSP19 NM_ PTRHD1 NM_ IGFBP4 NM_ ETV6 NM_ GALNT4 NM_ NUP35 NM_ ADCY3 NM_ CCR7 NM_ LOH12CR1 NM_ POC1B NM_ ZC3H15 NM_ DNAJC27 NM_ FKBP10 NM_ CREBL2 NM_ BTG1 NM_ ITGAV NM_ KIF3C NM_ CNP NM_ HEBP1 NM_ CRADD NM_ COL3A1 NM_ EPT1 NM_ DNAJC7 NM_ WBP11 NM_ PLXNC1 NM_ ASNSD1 NM_ C13H2orf70 NM_ HCRT NM_ C11H12orf69 NM_ NDUFA12 NM_ ORMDL1 NM_ C13H2orf18 NM_ EZH1 NM_ MGP NM_ VEZT NM_ MSTN NM_ PPP1CB NM_ BECN1 NM_ RERG NM_ METAP2 NM_ NAB1 NM_ LBH NM_ HEXIM2 NM_ EPS8 NM_ ELK3 NM_ STAT1 NM_ EHD3 NM_ CRHR1 NM_ LMO3 NM_ CDK17 NM_ OBFC2A NM_ DPY30 NM_ METTL2A NM_ SLCO1A2 NM_ TMPO NM_ SDPR NM_ YIPF4 NM_ CYB561 NM_ LYRM5 NM_ FAM71C NM_ SLC39A10 NM_ RASGRP3 NM_ FTSJ3 NM_ KRAS NM_ SCYL2 NM_ GTF3C3 NM_ FAM98A NM_ DDX5 NM_ RASSF8 NM_ DRAM1 NM_ SF3B1 NM_ EIF2AK2 NM_ PRKCA NM_ SSPN NM_ IGF1 NM_ MOBKL3 NM_ CDC42EP3 NM_ PSMD12 NM_ MED21 NM_ CHST11 NM_ PLCL1 NM_ CYP1B1 NM_ C16H17orf58 NM_ STK38L NM_ C11H12orf45 NM_ C2orf60 NM_ SRSF7 NM_ MAP2K6 NM_ REP15 NM_ TCP11L2 NM_ SPATS2L NM_ SLC8A1 NM_ KCNJ2 NM_ MRPS35 NM_ C11H12orf23 NM_ FAM126B NM_ KCNG3 NM_ SOX9 NM_ PTHLH NM_ CRY1 NM_ NOP58 NM_ MTA3 NM_ COG1 NM_ FAR2 NM_ PRDM4 NM_ BMPR2 NM_ PPM1B NM_ CDC42EP4 NM_ METTL20 NM_ SART3 NM_ CD28 NM_ PREPL NM_ RAB37 NM_ QTRTD1 NM_ KCNMB2 NM_ ZNF174 NM_ RSPH1 NM_ TMEM106B NM_ IGSF11 NM_ NLGN1 NM_ CREBBP NM_ C2CD2 NM_ ICA1 NM_ B4GALT4 NM_ TNFSF10 NM_ NMRAL1 NM_ MX2 NM_ MIOS NM_ TMEM39A NM_ TNIK NM_ FAM86A NM_ LCA5L NM_ TAC1 NM_ ADPRH NM_ EIF5A2 NM_ METTL22 NM_ DYRK1A NM_ SHFM1 NM_ FSTL1 NM_ SLC7A14 NM_ CARHSP1 NM_ DSCR3 NM_ DNCI1 NM_ RABL3 NM_ CLDN11 NM_ DEXI NM_ CLDN14 NM_ COL1A2 NM_ EAF2 NM_ SEC62 NM_ PRM1 NM_ SETD4 NM_ CDK6 NM_ CSTA NM_ SERPINI1 NM_ ZC3H7A NM_ RUNX1 NM_ KRIT1 NM_ SEC22A NM_ SPTSSB NM_ BFAR NM_ ATP5O NM_ CLDN12 NM_ PTPLB NM_ SMC4 NM_ RRN3 NM_ IFNAR1 NM_ SRI NM_ ITGB5 NM_ IL12A NM_ TMC7 NM_ C3H21orf59 NM_ ABCB1 NM_ ROPN1B NM_ LXN NM_ ITPRIPL2 NM_ SOD1 NM_ C3H7orf23 NM_ SLC41A3 NM_ MLF1 NM_ IQCK NM_ GRIK1 NM_ KIAA1324L NM_ ALDH1L1 NM_ KCNAB1 NM_ THUMPD1 NM_ BACH1 NM_ SEMA3C NM_ CHST13 NM_ GMPS NM_ CDR2 NM_ USP16 NM_ CD36 NM_ CHCHD6 NM_ MME NM_ UBFD1 NM_ ADAMTS1 NM_ PHTF2 NM_ WNT7A NM_ RAP2B NM_ PRKCB NM_ GABPA NM_ TMEM60 NM_ HDAC11 NM_ P2RY1 NM_ CACNG3 NM_ C3H21orf91 NM_ DNAJC2 NM_ VGLL4 NM_ P2RY12 NM_ C20H16orf82 NM_ BTG3 NM_ ORC5 NM_ ATG7 NM_ SIAH2 NM_ TUFM NM_ HSPA13 NM_ SRPK2 NM_ SLC6A11 NM_ SELT NM_ PPP4C NM_ ABCC13 NM_ PIK3CG NM_ SEC13 NM_ COMMD2 NM_ GDPD3 NM_ COX19 NM_ PNPLA8 NM_ NFYB NM_ TM4SF18 NM_ BOLA2 NM_ ZFAND2A NM_ DNAJB9 NM_ SETD5 NM_ PLSCR1 NM_ ZNF688 NM_ FTSJ2 NM_ IMMP2L NM_ CAV3 NM_ PLSCR4 NM_ ORAI3 NM_ NUDT1 NM_ IFRD1 NM_ Lin_Supplemental Table 1
11 Lin_Supplemental Table 1 Supplemental Table 1: (i c ) H3K4me3-enriched genes in the Baboon SVZ NSC population Gene Name RefSeq ID Gene Name RefSeq ID Gene Name RefSeq ID Gene Name RefSeq ID Gene Name RefSeq ID EDEM1 NM_ C2H3orf58 NM_ COX6A2 NM_ PAPOLB NM_ GPR85 NM_ BHLHE40 NM_ CHST2 NM_ C20H16orf58 NM_ KDELR2 NM_ FOXP2 NM_ LRRN1 NM_ ZBTB38 NM_ ORC6L NM_ ZDHHC4 NM_ CAPZA2 NM_ CRBN NM_ ACPL2 NM_ C20H16orf87 NM_ BRI3 NM_ ST7 NM_ EBLN2 NM_ MRAS NM_ PHKB NM_ SMURF1 NM_ ASZ1 NM_ FOXP1 NM_ SOX14 NM_ SIAH1 NM_ FAM200A NM_ CTTNBP2 NM_ MITF NM_ NCK1 NM_ CBLN1 NM_ ZNF498 NM_ KCND2 NM_ ARL6IP5 NM_ TMEM22 NM_ SNX20 NM_ ZKSCAN1 NM_ FAM3C NM_ UBA3 NM_ PCCB NM_ RPGRIP1L NM_ C3H7orf43 NM_ RNF133 NM_ FAM19A1 NM_ MSL2 NM_ SLC6A2 NM_ CUTL1 NM_ NDUFA5 NM_ SLC25A26 NM_ PPP2R3A NM_ GNAO1 NM_ ORAI2 NM_ WASL NM_ PRICKLE2 NM_ RAB6B NM_ MT3 NM_ CCL24 NM_ GPR37 NM_ PSMD6 NM_ TMEM108 NM_ CPNE2 NM_ DNAJC30 NM_ GRM8 NM_ SYNPR NM_ ATP2C1 NM_ ARL2BP NM_ CLDN4 NM_ GCC1 NM_ C2H3orf14 NM_ METTL6 NM_ GPR56 NM_ GTF2IRD1 NM_ TNPO3 NM_ FHIT NM_ OXNAD1 NM_ ZNF319 NM_ GATSL2 NM_ UBE2H NM_ FAM107A NM_ TBC1D5 NM_ C20H16orf80 NM_ WBSCR17 NM_ C3H7orf45 NM_ SLMAP NM_ KCNH8 NM_ GINS3 NM_ TMEM248 NM_ TSGA14 NM_ PDE12 NM_ RAB5A NM_ SLC38A7 NM_ TPST1 NM_ MKLN1 NM_ SELK NM_ UBE2E2 NM_ CDH8 NM_ VKORC1L1 NM_ SLC35B4 NM_ CHDH NM_ NKIRAS1 NM_ CDH11 NM_ PHKG1 NM_ BPGM NM_ WDR82 NM_ TOP2B NM_ CKLF NM_ LANCL2 NM_ MTPN NM_ RPL29 NM_ LRRC3B NM_ NAE1 NM_ VSTM2A NM_ PTN NM_ ABHD14B NM_ CMC1 NM_ LOC NM_ GRB10 NM_ TRIM24 NM_ RBM15B NM_ AZI2 NM_ PSKH1 NM_ IGFBP3 NM_ C3H7orf55 NM_ TMEM115 NM_ TGFBR2 NM_ PSMB10 NM_ NUDCD3 NM_ RAB19 NM_ CYB561D2 NM_ CMTM7 NM_ PLA2G15 NM_ BLVRA NM_ MRPS33 NM_ TUSC2 NM_ CRTAP NM_ CYB5B NM_ HECW1 NM_ AGK NM_ GNAI2 NM_ UBP1 NM_ NOB1 NM_ PSMA2 NM_ PRSS37 NM_ CAMKV NM_ EIF4A2 NM_ WWP2 NM_ C7orf36 NM_ EPHB6 NM_ CCDC72 NM_ ST6GAL1 NM_ DHX38 NM_ SFRP4 NM_ TAS2R40 NM_ CAMP NM_ RTP4 NM_ FTSJD1 NM_ ELMO1 NM_ FAM115C NM_ CCR5 NM_ BCL6 NM_ VAC14 NM_ SEPT7 NM_ C3H7orf33 NM_ SACM1L NM_ CLDN1 NM_ ST3GAL2 NM_ NPSR1 NM_ ZNF862 NM_ TMEM42 NM_ IL1RAP NM_ FA2H NM_ RP9 NM_ GIMAP4 NM_ LOC NM_ PYDC2 NM_ CTRB1 NM_ LSM5 NM_ GIMAP2 NM_ ZNF501 NM_ FGF12 NM_ GABARAPL2 NM_ CHN2 NM_ CDK5 NM_ ABHD5 NM_ MB21D2 NM_ KARS NM_ TAX1BP1 NM_ RHEB NM_ SNRK NM_ FAM43A NM_ DYNLRB2 NM_ SKAP2 NM_ ACTR3B NM_ SEC22C NM_ ACAP2 NM_ SDR42E1 NM_ SNX10 NM_ HTR5A NM_ LYZL4 NM_ DLG1 NM_ CDH13 NM_ HNRNPA2B1 NM_ CNPY1 NM_ CTNNB1 NM_ CEP19 NM_ HSDL1 NM_ NPVF NM_ NCAPG2 NM_ ENTPD3 NM_ LOC NM_ KIAA0513 NM_ MPP6 NM_ DUSP22 NM_ MOBP NM_ PDIA2 NM_ COX4NB NM_ NPY NM_ EXOC2 NM_ CSRNP1 NM_ C20H16orf13 NM_ MAP1LC3B NM_ RAPGEF5 NM_ GMDS NM_ OXSR1 NM_ FAM195A NM_ MVD NM_ CDCA7L NM_ PSMG4 NM_ MYD88 NM_ STUB1 NM_ TRAPPC2L NM_ SP4 NM_ FAM50B NM_ TBCCD1 NM_ NARFL NM_ DPEP1 NM_ ITGB8 NM_ NRN1 NM_ C2H3orf70 NM_ GNG13 NM_ DBNDD1 NM_ TMEM196 NM_ SLC35B3 NM_ KLHL24 NM_ TRAF7 NM_ PRMT2 NM_ FERD3L NM_ C4H6orf52 NM_ SOX2 NM_ ZNF200 NM_ YBEY NM_ AGR2 NM_ C4H6orf228 NM_ DNAJC19 NM_ ZNF263 NM_ PWP2 NM_ BZW2 NM_ TMEM170B NM_ PIK3CA NM_ TIGD7 NM_ TRAPPC10 NM_ SOSTDC1 NM_ C6orf105 NM_ SLC25A31 NM_ HMGCS1 NM_ CXXC5 NM_ TMOD3 NM_ KLHL28 NM_ PHF17 NM_ CCL28 NM_ IGIP NM_ ARPP19 NM_ ARF6 NM_ SCLT1 NM_ PAIP1 NM_ PFDN1 NM_ RAB27A NM_ METTL21D NM_ C5H4orf33 NM_ FGF10 NM_ NDUFA2 NM_ MNS1 NM_ SOS2 NM_ SLC7A11 NM_ PELO NM_ PCDHA5 NM_ FAM81A NM_ ATL1 NM_ MGARP NM_ ITGA2 NM_ PCDHB1 NM_ GCNT3 NM_ SAV1 NM_ NAA15 NM_ ARL15 NM_ PCDHB8 NM_ RPS27L NM_ GNG2 NM_ RAB33B NM_ SNX18 NM_ TAF7 NM_ CA12 NM_ ERO1L NM_ SETD7 NM_ IL6ST NM_ KIAA0141 NM_ SNX1 NM_ GNPNAT1 NM_ MGST2 NM_ C5orf35 NM_ GNPDA1 NM_ SNX22 NM_ FERMT2 NM_ ELMOD2 NM_ GAPT NM_ NDFIP1 NM_ KIAA0101 NM_ CDKN3 NM_ ZNF330 NM_ RAB3C NM_ FGF1 NM_ ZNF609 NM_ CNIH NM_ IL15 NM_ DEPDC1B NM_ YIPF5 NM_ RASL12 NM_ FBXO34 NM_ USP38 NM_ NDUFAF2 NM_ KCTD16 NM_ CLPX NM_ NAA30 NM_ GAB1 NM_ KIF2A NM_ RBM27 NM_ PTPLAD1 NM_ PSMA3 NM_ SMARCA5 NM_ HTR1A NM_ JAKMIP2 NM_ RAB11A NM_ DAAM1 NM_ HHIP NM_ RGS7BP NM_ SPINK9 NM_ SNAPC5 NM_ JKAMP NM_ SMAD1 NM_ SREK1IP1 NM_ PCYOX1L NM_ RPL4 NM_ MNAT1 NM_ C5H4orf51 NM_ C6H5orf44 NM_ SLC6A7 NM_ SMAD3 NM_ TRMT5 NM_ EDNRA NM_ PIK3R1 NM_ NDST1 NM_ PIAS1 NM_ SNAPC1 NM_ TMEM184C NM_ SLC30A5 NM_ MYOZ3 NM_ FEM1B NM_ RHOJ NM_ FBXW7 NM_ GTF2H2 NM_ GPX3 NM_ GLCE NM_ ZBTB25 NM_ ARFIP1 NM_ CARTPT NM_ GLRA1 NM_ SENP8 NM_ GPX2 NM_ MND1 NM_ MAP1B NM_ SAP30L NM_ HEXA NM_ FNTB NM_ TLR2 NM_ UTP15 NM_ MRPL22 NM_ NEO1 NM_ MAX NM_ LRAT NM_ FAM169A NM_ ITK NM_ PML NM_ FAM71D NM_ NPY2R NM_ AGGF1 NM_ CYFIP2 NM_ SEMA7A NM_ TMEM229B NM_ GLRB NM_ LHFPL2 NM_ C5orf40 NM_ CSK NM_ ARG2 NM_ GRIA2 NM_ IMPACT NM_ UBLCP1 NM_ COX5A NM_ RDH12 NM_ FAM198B NM_ HOMER1 NM_ TTC1 NM_ SCAMP5 NM_ DCAF5 NM_ TMEM144 NM_ PAPD4 NM_ PTTG1 NM_ ETFA NM_ KIAA0247 NM_ C5H4orf46 NM_ FAM151B NM_ GABRB2 NM_ TSPAN3 NM_ PCNX NM_ NPY1R NM_ RPS23 NM_ GABRG2 NM_ CIB2 NM_ SIPA1L1 NM_ C5H4orf43 NM_ CCNH NM_ RARS NM_ FAH NM_ RGS6 NM_ CPE NM_ MEF2C NM_ FAM196B NM_ MESDC1 NM_ DCAF4 NM_ CBR4 NM_ CETN3 NM_ KCNMB1 NM_ IL16 NM_ PSEN1 NM_ MFAP3L NM_ ARRDC3 NM_ FBXW11 NM_ STARD5 NM_ ACOT6 NM_ SAP30 NM_ FAM172A NM_ UBTD2 NM_ C7H15orf40 NM_ ELMSAN1 NM_ GPM6A NM_ GLRX NM_ DUSP1 NM_ CPEB1 NM_ VRTN NM_
12 Lin_Supplemental Table 1 Supplemental Table 1: (i d ) H3K4me3-enriched genes in the Baboon SVZ NSC population Gene Name RefSeq ID Gene Name RefSeq ID Gene Name RefSeq ID Gene Name RefSeq ID Gene Name RefSeq ID CCDC85B NM_ ENDOD1 NM_ KLF4 NM_ TEX19 NM_ ST8SIA5 NM_ FAM89B NM_ SESN3 NM_ RAD23B NM_ FN3KRP NM_ SMAD2 NM_ FRMD8 NM_ MTMR2 NM_ TAL2 NM_ CRYL1 NM_ C18H18orf32 NM_ MRPL49 NM_ JRKL NM_ OR13C8 NM_ ZMYM2 NM_ MAPK4 NM_ DNAJC4 NM_ BIRC2 NM_ PPP3R2 NM_ PSPC1 NM_ SMAD4 NM_ MACROD1 NM_ DDI1 NM_ ZNF189 NM_ MTMR6 NM_ TXNL1 NM_ COX8A NM_ KIAA1826 NM_ ALG2 NM_ SHISA2 NM_ LMAN1 NM_ C14H11orf84 NM_ AASDHPPT NM_ GALNT12 NM_ PRHOXNB NM_ PMAIP1 NM_ C14H11orf83 NM_ ELMOD1 NM_ TMOD1 NM_ POMP NM_ RNF152 NM_ GANAB NM_ C14H11orf65 NM_ EXOSC3 NM_ SLC7A1 NM_ KDSR NM_ FADS2 NM_ C14H11orf87 NM_ ZBTB5 NM_ UBL3 NM_ SERPINB10 NM_ C14H11orf10 NM_ GPHN NM_ CCIN NM_ KATNAL1 NM_ TMX3 NM_ C14H11orf9 NM_ C14H11orf1 NM_ GLIPR2 NM_ ALOX5AP NM_ DOK6 NM_ PRPF19 NM_ HSPB2 NM_ RUSC2 NM_ RXFP2 NM_ CD226 NM_ MS4A7 NM_ C14H11orf57 NM_ FANCG NM_ STARD13 NM_ NETO1 NM_ APLNR NM_ TEX12 NM_ AQP3 NM_ RFC3 NM_ C18orf55 NM_ OR4A47 NM_ PTS NM_ SMU1 NM_ MAB21L1 NM_ CNDP2 NM_ CHRM4 NM_ ZBTB16 NM_ TMEM215 NM_ RFXAP NM_ PQLC1 NM_ PHF21A NM_ REXO2 NM_ C4H6orf203 NM_ EXOSC8 NM_ TXNL4A NM_ SLC35C1 NM_ BACE1 NM_ C15H9orf72 NM_ STOML3 NM_ RNF126 NM_ CHST1 NM_ FXYD6 NM_ MOB3B NM_ LHFP NM_ GPX4 NM_ TSPAN18 NM_ SCN2B NM_ PLAA NM_ ELF1 NM_ ATP5D NM_ ACCS NM_ ARCN1 NM_ CAAP1 NM_ C13orf15 NM_ CIRBP NM_ C14H11orf74 NM_ DPAGT1 NM_ TUSC1 NM_ C13orf31 NM_ GAMT NM_ TRIM44 NM_ USP2 NM_ ELAVL2 NM_ GTF2F2 NM_ UQCR NM_ SLC1A2 NM_ THY1 NM_ MTAP NM_ LCP1 NM_ FAM108A1 NM_ ELF5 NM_ TBCEL NM_ KLHL9 NM_ ESD NM_ SLC39A3 NM_ LMO2 NM_ SC5DL NM_ PTPLAD2 NM_ HTR2A NM_ S1PR4 NM_ CSTF3 NM_ UBASH3B NM_ RRAGA NM_ MED4 NM_ EEF2 NM_ QSER1 NM_ SCN3B NM_ SNAPC3 NM_ RB1 NM_ TNFAIP8L1 NM_ PAX6 NM_ OR4D5 NM_ CER1 NM_ LPAR6 NM_ KDM4B NM_ DNAJC24 NM_ ESAM NM_ PDCD1LG2 NM_ ARL11 NM_ VMAC NM_ MPPED2 NM_ ST3GAL4 NM_ JAK2 NM_ ALG11 NM_ SLC25A23 NM_ ARL14EP NM_ BARX2 NM_ RCL1 NM_ PCDH17 NM_ CRB3 NM_ LIN7C NM_ TMEM45B NM_ SLC1A1 NM_ PCDH9 NM_ MCOLN1 NM_ FIBIN NM_ VPS26B NM_ CBWD1 NM_ KLHL1 NM_ CD209 NM_ ANO3 NM_ ZMYND19 NM_ PTAR1 NM_ KLF12 NM_ RAB11B NM_ SVIP NM_ C15H9orf142 NM_ KLF9 NM_ TBC1D4 NM_ ACTL9 NM_ GAS2 NM_ INPP5E NM_ CYP1D1 NM_ UCHL3 NM_ OR1M1 NM_ MRGPRX2 NM_ NACC2 NM_ RORB NM_ KCTD12 NM_ OR7D4 NM_ LOC NM_ BRD3 NM_ NMRK1 NM_ FBXL3 NM_ ZNF177 NM_ RPS13 NM_ VAV2 NM_ GNA14 NM_ SLITRK1 NM_ UBL5 NM_ C14H11orf58 NM_ SURF4 NM_ TLE1 NM_ SLITRK5 NM_ PPAN NM_ SPON1 NM_ GTF3C4 NM_ GKAP1 NM_ DNAJC3 NM_ P2RY11 NM_ BTBD10 NM_ NTNG2 NM_ NTRK2 NM_ RAP2A NM_ ICAM3 NM_ ARNTL NM_ MED27 NM_ NAA35 NM_ STK24 NM_ C19H19orf52 NM_ DKK3 NM_ UCK1 NM_ DAPK1 NM_ SLC15A1 NM_ ACP5 NM_ RNF141 NM_ AIF1L NM_ CDK20 NM_ UBAC2 NM_ C19orf56 NM_ SWAP70 NM_ C15H9orf78 NM_ SHC3 NM_ TEX30 NM_ TNPO2 NM_ C11orf17 NM_ METTL11A NM_ CKS2 NM_ ARGLU1 NM_ C19H19orf43 NM_ RPL27A NM_ CRAT NM_ GADD45G NM_ ANKRD10 NM_ JUNB NM_ RIC3 NM_ SH3GLB2 NM_ NAIF1 NM_ F7 NM_ CALR NM_ OR10A4 NM_ LRRC8A NM_ FAM102A NM_ F10 NM_ STX10 NM_ TAF10 NM_ URM1 NM_ ANGPTL2 NM_ CDC16 NM_ IER2 NM_ OR52W1 NM_ PSMB7 NM_ SYK NM_ MC5R NM_ NDUFB7 NM_ TRIM22 NM_ LHX2 NM_ AUH NM_ LDLRAD4 NM_ AKAP8L NM_ HBE1 NM_ STRBP NM_ NFIL3 NM_ PSMG2 NM_ AP1M1 NM_ OR51G2 NM_ ZBTB26 NM_ CENPP NM_ CHMP1B NM_ SLC35E1 NM_ OR52M1 NM_ OR1Q1 NM_ BICD2 NM_ NAPG NM_ USE1 NM_ RHOG NM_ RBM18 NM_ NINJ1 NM_ RAB31 NM_ ABHD8 NM_ LRTOMT NM_ STOM NM_ HIATL1 NM_ NDUFV2 NM_ MRPL34 NM_ C11orf59 NM_ RAB14 NM_ FBP1 NM_ ZFP161 NM_ INSL3 NM_ PDE2A NM_ TRAF1 NM_ HSD17B3 NM_ SMCHD1 NM_ PIK3R2 NM_ FCHSD2 NM_ DBC1 NM_ GLOD4 NM_ TYMS NM_ SUGP1 NM_ FAM168A NM_ TLR4 NM_ INPP5K NM_ PHACTR1 NM_ LYRM2 NM_ PGM2L1 NM_ TRIM32 NM_ TLCD2 NM_ GFOD1 NM_ GJA10 NM_ OR2AT4 NM_ TNFSF8 NM_ CYB5D2 NM_ CD83 NM_ FUT9 NM_ SERPINH1 NM_ C15H9orf91 NM_ MED11 NM_ MYLIP NM_ KLHL32 NM_ NDUFC2 NM_ POLE3 NM_ PSMB6 NM_ CAP2 NM_ FBXL4 NM_ FAM181B NM_ HDHD3 NM_ SLC25A11 NM_ NUP153 NM_ FAXC NM_ RAB30 NM_ SLC31A1 NM_ RPAIN NM_ RNF144B NM_ MCHR2 NM_ CCDC90B NM_ KIAA1958 NM_ WSCD1 NM_ ID4 NM_ DCUN1D1 NM_ TMEM126B NM_ HSDL2 NM_ XAF1 NM_ PRL NM_ POPDC3 NM_ PICALM NM_ UGCG NM_ KCTD11 NM_ NRSN1 NM_ PREP NM_ FZD4 NM_ DNAJC25 NM_ TNFSF12 NM_ ACOT13 NM_ QRSL1 NM_ CTSC NM_ C15H9orf152 NM_ TMEM107 NM_ GMNN NM_ SOBP NM_ C14H11orf75 NM_ AKAP2 NM_ MFSD6L NM_ CMAH NM_ SEC63 NM_ FOLR4 NM_ C9orf4 NM_ RCVRN NM_ HIST1H1A NM_ GTF2H5 NM_ PIWIL4 NM_ FAM206A NM_ TMEM220 NM_ HIST1H1T NM_ ZDHHC14 NM_ SLC9A3R1 NM_ SNRPD1 NM_ LPAR2 NM_ HIST1H1D NM_ TFB1M NM_ CDR2L NM_ RBBP8 NM_ PLEKHF1 NM_ ZNF322A NM_ CNKSR3 NM_ HN1 NM_ SS18 NM_ ZNF507 NM_ HIST1H1B NM_ IPCEF1 NM_ MRPS7 NM_ AQP4 NM_ PDCD5 NM_ GPX6 NM_ MYCT1 NM_ KIAA0195 NM_ CHST9 NM_ KCTD15 NM_ GPX5 NM_ C4H6orf211 NM_ ACOX1 NM_ TTR NM_ LSM14A NM_ TRIM27 NM_ ZBTB2 NM_ SEC14L1 NM_ B4GALT6 NM_ KIAA0355 NM_ GABBR1 NM_ GINM1 NM_ TNRC6C NM_ NOL4 NM_ SCN1B NM_ MOG NM_ TAB2 NM_ SOCS3 NM_ MAPRE2 NM_ FAM187B NM_ RPP21 NM_ UST NM_ CYTH1 NM_ ZNF24 NM_ HCST NM_ NRM NM_ C6orf103 NM_ TBC1D16 NM_ GALNT1 NM_ POLR2I NM_ MDC1 NM_ RAB32 NM_ NPTX1 NM_ RPRD1A NM_ COX7AH NM_ FLOT1 NM_ FBXO30 NM_ C16H17orf70 NM_ C18orf10 NM_ ZNF790 NM_ LSM2 NM_ STX11 NM_
13 Supplemental Table 1: (i e ) H3K4me3-enriched genes in the Baboon SVZ NSC population Gene Name RefSeq ID Gene Name RefSeq ID Gene Name RefSeq ID Gene Name RefSeq ID Gene Name RefSeq ID SIRT7 NM_ RIT2 NM_ ZNF345 NM_ FKBPL NM_ PLAGL1 NM_ PYCR1 NM_ ATP5A1 NM_ SPINT2 NM_ SLC39A7 NM_ AIG1 NM_ ACTN4 NM_ CXCL13 NM_ RING1 NM_ VTA1 NM_ UBXN2B NM_ NFKBIB NM_ CCNI NM_ TAPBP NM_ ABRACL NM_ CYP7A1 NM_ MED29 NM_ STBD1 NM_ CUTA NM_ HEBP2 NM_ TOX NM_ SUPT5H NM_ PARM1 NM_ NUDT3 NM_ TNFAIP3 NM_ CA8 NM_ DYRK1B NM_ MTHFD2L NM_ RPL10A NM_ IFNGR1 NM_ RAB2A NM_ AKT2 NM_ IL8 NM_ KCTD20 NM_ TCF21 NM_ CLVS1 NM_ PLD3 NM_ ALB NM_ PI16 NM_ MOXD1 NM_ GGH NM_ CYP2B6 NM_ DCK NM_ CCDC167 NM_ MED23 NM_ PDE7A NM_ MIA NM_ IGJ NM_ GLO1 NM_ C4H6orf58 NM_ ARFGEF1 NM_ EGLN2 NM_ SULT1B1 NM_ KCNK17 NM_ CENPW NM_ RDH10 NM_ B9D2 NM_ UGT2A3 NM_ TFEB NM_ HEY2 NM_ UBE2W NM_ GSK3A NM_ STAP1 NM_ PTCRA NM_ CLVS2 NM_ PI15 NM_ PAFAH1B3 NM_ LPHN3 NM_ GTPBP2 NM_ GJA1 NM_ PKIA NM_ PRR19 NM_ POLR2B NM_ MRPS18A NM_ ASF1A NM_ IL7 NM_ ZNF428 NM_ SRP72 NM_ SLC29A1 NM_ PLN NM_ STMN2 NM_ C19orf61 NM_ PPAT NM_ SUPT3H NM_ SLC35F1 NM_ MRPS28 NM_ PVR NM_ EXOC1 NM_ ENPP4 NM_ NUS1 NM_ TPD52 NM_ GEMIN7 NM_ SRD5A3 NM_ CD2AP NM_ FAM162B NM_ ZNF704 NM_ OPA3 NM_ KIT NM_ MUT NM_ RWDD1 NM_ PMP2 NM_ SNRPD2 NM_ CHIC2 NM_ RHAG NM_ COL10A1 NM_ CHMP4C NM_ PPP5C NM_ USP46 NM_ DEFB114 NM_ NT5DC1 NM_ RALYL NM_ AP2S1 NM_ MAPK10 NM_ IL17F NM_ HS3ST5 NM_ CA2 NM_ EMP3 NM_ PPM1K NM_ TRAM2 NM_ C4H6orf225 NM_ DCAF4L2 NM_ CYTH2 NM_ NAP1L5 NM_ GSTA4 NM_ TRAF3IP2 NM_ NBN NM_ RPL13A NM_ HPGDS NM_ FBXO9 NM_ SLC16A10 NM_ LOC NM_ IZUMO2 NM_ BMPR1B NM_ GCLC NM_ RPF2 NM_ LRRC69 NM_ LRRC4B NM_ TSPAN5 NM_ LRRC1 NM_ C6orf186 NM_ TRIQK NM_ SIGLECL1 NM_ EIF4E NM_ FAM83B NM_ CDC40 NM_ FAM92A1 NM_ ZNF175 NM_ METAP1 NM_ BEND6 NM_ FIG4 NM_ TP53INP1 NM_ PPP2R1A NM_ MAPKSP1 NM_ PRIM2 NM_ SMPD2 NM_ PLEKHF2 NM_ PRPF31 NM_ DDIT4L NM_ UBE2E3 NM_ CEP57L1 NM_ C8H8orf37 NM_ RPS9 NM_ PPP3CA NM_ LMBRD1 NM_ SNX3 NM_ PTDSS1 NM_ FCAR NM_ NFKB1 NM_ C4H6orf57 NM_ NR2E1 NM_ SDC2 NM_ NLRP7 NM_ UBE2D3 NM_ KCNQ5 NM_ OSTM1 NM_ FAM188A NM_ COX6B2 NM_ TACR3 NM_ SLC17A5 NM_ SOD2 NM_ C1QL3 NM_ UBE2S NM_ INTS12 NM_ MYO6 NM_ WTAP NM_ PTPLA NM_ ZNF581 NM_ SGMS2 NM_ IRAK1BP1 NM_ PACRG NM_ ARL5B NM_ NLRP11 NM_ HADH NM_ LCA5 NM_ BRP44L NM_ DNAJC1 NM_ NLRP8 NM_ PLA2G12A NM_ ELOVL4 NM_ RNASET2 NM_ COMMD3 NM_ ESP16.9 NM_ ENPEP NM_ TPBG NM_ CCR6 NM_ THNSL1 NM_ ZNF805 NM_ NEUROG2 NM_ PRSS35 NM_ PSMB1 NM_ GAD2 NM_ ZNF304 NM_ NDST3 NM_ CYB5R4 NM_ GPR78 NM_ ARHGAP12 NM_ ZNF671 NM_ METTL14 NM_ SNX14 NM_ GRPEL1 NM_ NRP1 NM_ ZSCAN1 NM_ NDNF NM_ SYNCRIP NM_ KIAA0232 NM_ CUL2 NM_ ZBTB45 NM_ TMEM155 NM_ POLR1D NM_ MRFAP1L1 NM_ CCNY NM_ CHMP2A NM_ IL21 NM_ SLC35A1 NM_ MRFAP1 NM_ CSGALNACT2 NM_ GBE1 NM_ SPRY1 NM_ CNR1 NM_ JAKMIP1 NM_ HNRNPF NM_ CHMP2B NM_ ANKRD50 NM_ PNRC1 NM_ CYTL1 NM_ ZNF32 NM_ POU1F1 NM_ HERC4 NM_ ANKRD6 NM_ ZNF518B NM_ CXCL12 NM_ CGGBP1 NM_ LRRTM3 NM_ SPAG11B NM_ MTDH NM_ DRGX NM_ PROS1 NM_ REEP3 NM_ CTSB NM_ STK3 NM_ C10orf72 NM_ ARL6 NM_ NRBF2 NM_ FDFT1 NM_ OSR2 NM_ SNCG NM_ MINA NM_ ADO NM_ TNKS NM_ ANKRD46 NM_ BMPR1A NM_ GPR15 NM_ ZNF365 NM_ TUSC3 NM_ PABPC1 NM_ GRID1 NM_ CMSS1 NM_ C9H10orf107 NM_ CSGALNACT1 NM_ NCALD NM_ TMEM254 NM_ TOMM70A NM_ RHOBTB1 NM_ FGF17 NM_ KLF10 NM_ PPIF NM_ TMEM45A NM_ CDK1 NM_ NUDT18 NM_ BAALC NM_ RPS24 NM_ FAM55C NM_ UBE2D1 NM_ REEP4 NM_ SLC25A32 NM_ KCNMA1 NM_ ALCAM NM_ MBL2 NM_ KIAA1967 NM_ LRP12 NM_ VDAC2 NM_ CD47 NM_ CSTF2T NM_ PEBP4 NM_ ANGPT1 NM_ CHCHD1 NM_ IFT57 NM_ SGMS1 NM_ NKX3-1 NM_ EMC2 NM_ ANAPC16 NM_ ABHD10 NM_ PTEN NM_ NEFL NM_ SYBU NM_ C9H10orf54 NM_ CD200 NM_ STAMBPL1 NM_ GNRH1 NM_ UTP23 NM_ LRRC20 NM_ ATP6V1A NM_ FAS NM_ BNIP3L NM_ MED30 NM_ H2AFY2 NM_ HS3ST1 NM_ PPP1R3C NM_ ADRA1A NM_ EXT1 NM_ TSPAN15 NM_ RAB28 NM_ TNKS2 NM_ TRIM35 NM_ NOV NM_ SRGN NM_ NKX3-2 NM_ CYP26A1 NM_ CCDC25 NM_ TAF2 NM_ CNOT6L NM_ CD38 NM_ LGI1 NM_ PNOC NM_ MRPL13 NM_ RGS10 NM_ PROM1 NM_ HELLS NM_ FZD3 NM_ DERL1 NM_ FAM110B NM_ LDB2 NM_ CYP2C76 NM_ LEPROTL1 NM_ FBXO32 NM_ ACBD7 NM_ LAP3 NM_ DNTT NM_ GTF2E2 NM_ RNF139 NM_ ATG4A NM_ LCORL NM_ PIK3AP1 NM_ MAK16 NM_ TRIB1 NM_ NXT2 NM_ DHX15 NM_ UBTD1 NM_ RNF122 NM_ MYC NM_ RGAG1 NM_ SOD3 NM_ AVPI1 NM_ DUSP26 NM_ FAM49B NM_ CHRDL1 NM_ ANAPC4 NM_ GOLGA7B NM_ ERLIN2 NM_ ST3GAL1 NM_ LHFPL1 NM_ TBC1D19 NM_ NDUFB8 NM_ FGFR1 NM_ CHRAC1 NM_ HNRNPH2 NM_ C5H4orf19 NM_ C9H10orf2 NM_ TM2D2 NM_ PTP4A3 NM_ PLS3 NM_ TBC1D1 NM_ LBX1 NM_ SFRP1 NM_ LYNX1 NM_ WDR44 NM_ KLF3 NM_ ELOVL3 NM_ GOLGA7 NM_ GPIHBP1 NM_ ZCCHC12 NM_ TLR10 NM_ ACTR1A NM_ GINS4 NM_ TSTA3 NM_ PGRMC1 NM_ WDR19 NM_ SFXN2 NM_ IKBKB NM_ LRRC14 NM_ ZBTB33 NM_ RHOH NM_ C9H10orf32 NM_ SLC20A2 NM_ ZNF7 NM_ LAMP2 NM_ TMEM33 NM_ NT5C2 NM_ THAP1 NM_ ZNF250 NM_ C1GALT1C1 NM_ ATP8A1 NM_ OBFC1 NM_ MCM4 NM_ ZMYND11 NM_ SH2D1A NM_ GRXCR1 NM_ GSTO1 NM_ UBE2V2 NM_ LARP4B NM_ CXHXorf64 NM_ GNPDA2 NM_ XPNPEP1 NM_ EFCAB1 NM_ WDR37 NM_ OCRL NM_ GABRA2 NM_ MXI1 NM_ SNAI2 NM_ AKR1E2 NM_ SASH3 NM_ COMMD8 NM_ PDCD4 NM_ PCMTD1 NM_ IL2RA NM_ CXHXorf57 NM_ SLAIN2 NM_ SHOC2 NM_ FAM150A NM_ PRKCQ NM_ MORC4 NM_ OCIAD2 NM_ GPAM NM_ RB1CC1 NM_ KIN NM_ TSC22D3 NM_ Lin_Supplemental Table 1
devseek (Sequence(Analysis(Panel(for(Neurodevelopmental(Disorders
 ACSL4 Mental/retardation,/XBlinked/63 XLBR ADSL Adenylosuccinase/Deficiency AR AFF2 Mental/retardation,/XBlinked,/FRAXE/type XLBR ALG6 Congenital/disorder/of/glycosylation/type/Ic AR ANK3 Mental/retardation,/autosomal/recessive/37
ACSL4 Mental/retardation,/XBlinked/63 XLBR ADSL Adenylosuccinase/Deficiency AR AFF2 Mental/retardation,/XBlinked,/FRAXE/type XLBR ALG6 Congenital/disorder/of/glycosylation/type/Ic AR ANK3 Mental/retardation,/autosomal/recessive/37
WT-TP53 DLBCL WT-TP53 GCB-DLBCL
 SUPPLEMENTAL TABLES AND FIGURES Supplemental Table S1. Summary of gene expression profiling analysis results listing counts of significant differentially expressed transcripts between defined two groups
SUPPLEMENTAL TABLES AND FIGURES Supplemental Table S1. Summary of gene expression profiling analysis results listing counts of significant differentially expressed transcripts between defined two groups
Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits.
 Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits. All With Fst in the With long With SNP(s) at the 5' top 5% bracket haplotypes regulatory region
Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits. All With Fst in the With long With SNP(s) at the 5' top 5% bracket haplotypes regulatory region
Lions and Tigers and Bears.Oh My!
 Lions and Tigers and Bears.Oh My! TAKING THE FEAR OUT OF GENETIC TESTING Lisa Butterfield, MS, CGC Genetic Counselor Clinical Assistant Professor University of Kansas Health System Why is Genetic Testing
Lions and Tigers and Bears.Oh My! TAKING THE FEAR OUT OF GENETIC TESTING Lisa Butterfield, MS, CGC Genetic Counselor Clinical Assistant Professor University of Kansas Health System Why is Genetic Testing
Quantitative Real Time PCR (RT-qPCR) Differentiation of human preadipocytes to adipocytes
 Quantitative Real Time PCR (RT-qPCR) First strand cdna was synthesized and RT-qPCR was performed using RT2 first strand kits (Cat#330401) and RT2 qpcr Master Mix (SABioscience, Frederick, MD). Experiments
Quantitative Real Time PCR (RT-qPCR) First strand cdna was synthesized and RT-qPCR was performed using RT2 first strand kits (Cat#330401) and RT2 qpcr Master Mix (SABioscience, Frederick, MD). Experiments
Regulation of eif4 and p70s6k signaling. Chondroitin and dermatan biosynthesis Interferon signaling
 Supplemental Information Table S1: Differentially regulated pathways in cortical versus cerebellar human astrocytes following treatment with PBS or IFN-β Pathway Regulation of eif4 and p70s6k signaling
Supplemental Information Table S1: Differentially regulated pathways in cortical versus cerebellar human astrocytes following treatment with PBS or IFN-β Pathway Regulation of eif4 and p70s6k signaling
The Genetics of Alcoholism. Robert Hitzemann, Ph.D. Departments of Behavioral Neuroscience & Psychiatry Oregon Health & Science University
 The Genetics of Alcoholism Robert Hitzemann, Ph.D. Departments of Behavioral Neuroscience & Psychiatry Oregon Health & Science University Genetics, Genomics and Alcoholism Robert Hitzemann, Ph.D. Departments
The Genetics of Alcoholism Robert Hitzemann, Ph.D. Departments of Behavioral Neuroscience & Psychiatry Oregon Health & Science University Genetics, Genomics and Alcoholism Robert Hitzemann, Ph.D. Departments
A genome-wide association study identifies vitiligo
 A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
Supplemental Table 1. List of genes and mature shrna sequences identified from the shrna screen in BRCA2-mutant PEO1 cells.
 Guillemette_Supplemental Table 1 Gene Mature Sequence Gene Mature Sequence ATTCATAGGATGTCAGCAG LOC157193 TATGCGTAAGTAATAAATTGCT TTAGTTCTGTCTTGGAGG LOC152225 CGCATATATGTTCTGACTT ALDH2 CACTTCAGTGTATGCCT
Guillemette_Supplemental Table 1 Gene Mature Sequence Gene Mature Sequence ATTCATAGGATGTCAGCAG LOC157193 TATGCGTAAGTAATAAATTGCT TTAGTTCTGTCTTGGAGG LOC152225 CGCATATATGTTCTGACTT ALDH2 CACTTCAGTGTATGCCT
Supplementary Table 1. mirna modulated in D HF and ND HF patients. Table shows in a linear scale the same values of Fig. 2.
 Supplementary Table 1. mirna modulated in D HF and ND HF patients. Table shows in a linear scale the same values of Fig. 2. mir D HF VS CTR ND HF VS CTR D HF VS ND HF FOLD CHANGE p FOLD CHANGE p FOLD CHANGE
Supplementary Table 1. mirna modulated in D HF and ND HF patients. Table shows in a linear scale the same values of Fig. 2. mir D HF VS CTR ND HF VS CTR D HF VS ND HF FOLD CHANGE p FOLD CHANGE p FOLD CHANGE
Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK
 Supplemental legends Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK Table SП. List of genes in cluster 1 identified using two-dimensional hierarchical clustering analysis Table SШ.
Supplemental legends Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK Table SП. List of genes in cluster 1 identified using two-dimensional hierarchical clustering analysis Table SШ.
Supplementary Figures and Tables. An inflammatory gene signature distinguishes neurofibroma Schwann cells and
 Supplementary Figures and Tables An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system Kwangmin Choi 1, Kakajan Komurov
Supplementary Figures and Tables An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system Kwangmin Choi 1, Kakajan Komurov
HTG EdgeSeq Immuno-Oncology Assay Gene List
 A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
Supplementary Figure 1. Microarray data mining and validation
 Supplementary Figure 1. Microarray data mining and validation Microarray (>47,000 probes) Raw data Background subtraction NanoString (124 genes) Set cut-off and threshold values Raw data Normalized to
Supplementary Figure 1. Microarray data mining and validation Microarray (>47,000 probes) Raw data Background subtraction NanoString (124 genes) Set cut-off and threshold values Raw data Normalized to
Supplemental Data for Toischer et al., Cardiomyocyte proliferation prevents failure in
 Supplemental Data, Page 1 Supplemental Data for Toischer et al., Cardiomyocyte proliferation prevents failure in pressure but not volume overload. Contents: Page 3: Supplemental Figure 1 - Echocardiographic
Supplemental Data, Page 1 Supplemental Data for Toischer et al., Cardiomyocyte proliferation prevents failure in pressure but not volume overload. Contents: Page 3: Supplemental Figure 1 - Echocardiographic
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature12912 Figure S1. Distribution of mutation rates and spectra across 4,742 tumor-normal (TN) pairs from 21 tumor types, as in Figure 1 from Lawrence et al. Nature
SUPPLEMENTARY INFORMATION doi:1.138/nature12912 Figure S1. Distribution of mutation rates and spectra across 4,742 tumor-normal (TN) pairs from 21 tumor types, as in Figure 1 from Lawrence et al. Nature
Supplementary Materials
 Supplementary Materials Supplemental figure 1. Absolute cell count of naïve/memory compartment of CD4 and CD8 T cells in early years Absolute cell number of naïve (CD45RA+CCR7+), Stem cell memory T cell
Supplementary Materials Supplemental figure 1. Absolute cell count of naïve/memory compartment of CD4 and CD8 T cells in early years Absolute cell number of naïve (CD45RA+CCR7+), Stem cell memory T cell
Supplemental Table 1: Enriched GO categories per cluster
 Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases
 Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Integrative Radiation Biology
 Dr. Kristian Unger Integrative Biology Group Research Unit of Radiation Cytogenetics Department of Radiation Sciences Helmholtz-Zentrum München 1 Key Questions Molecular mechanisms of radiation-induced
Dr. Kristian Unger Integrative Biology Group Research Unit of Radiation Cytogenetics Department of Radiation Sciences Helmholtz-Zentrum München 1 Key Questions Molecular mechanisms of radiation-induced
Gene expression of muscular and neuronal pathways is cooperatively dysregulated in patients
 Gene expression of muscular and neuronal pathways is cooperatively dysregulated in patients with idiopathic achalasia Orazio Palmieri 1, Tommaso Mazza 2, Antonio Merla 1, Caterina Fusilli 2, Antonello
Gene expression of muscular and neuronal pathways is cooperatively dysregulated in patients with idiopathic achalasia Orazio Palmieri 1, Tommaso Mazza 2, Antonio Merla 1, Caterina Fusilli 2, Antonello
Supplementary Material. Table S1. Summary of mapping results
 Supplementary Material Table S1. Summary of mapping results Sample Total reads Mapped (%) HNE0-1 99687516 80786083 (81.0%) HNE0-2 94318720 77047792 (81.7%) HNE0-3 104033900 84348102 (81.1%) HNE15-1 94426598
Supplementary Material Table S1. Summary of mapping results Sample Total reads Mapped (%) HNE0-1 99687516 80786083 (81.0%) HNE0-2 94318720 77047792 (81.7%) HNE0-3 104033900 84348102 (81.1%) HNE15-1 94426598
Analyzing the NCI-60 Cancer Cell Lines using Data Obtained from Genome-Wide ChIP-X Experiments. Jayanth (Jay) Krishnan. Mahopac High School
 1 Analyzing the NCI-60 Cancer Cell Lines using Data Obtained from Genome-Wide ChIP-X Experiments Jayanth (Jay) Krishnan Mahopac High School January 25, 2010 2 Analyzing the NCI-60 Cancer Cell Lines using
1 Analyzing the NCI-60 Cancer Cell Lines using Data Obtained from Genome-Wide ChIP-X Experiments Jayanth (Jay) Krishnan Mahopac High School January 25, 2010 2 Analyzing the NCI-60 Cancer Cell Lines using
Supplemental Figures/Tables. Methylation forward GTCGGGGCGTATTTAGTTC. Methylation reverse AACGACGTAAACGAAAATATCG
 Supplemental Figures/Tables Supplementary Tables Table S1. Primer sequences MSP Primers SPARC2 Unmethylation forward TTTTTTAGATTGTTTGGAGAGTG Unmethylation reverse AACTAACAACATAAACAAAAATATC Methylation
Supplemental Figures/Tables Supplementary Tables Table S1. Primer sequences MSP Primers SPARC2 Unmethylation forward TTTTTTAGATTGTTTGGAGAGTG Unmethylation reverse AACTAACAACATAAACAAAAATATC Methylation
Targeted Genes and Methodology Details for Epilepsy/Seizure Genetic Panels
 Targeted s and Methodology Details for Epilepsy/Seizure tic Panels Reference transcripts based on build GRCh37 (hg19) interrogated by Epilepsy/Seizure tic Panels Epilepsy Expanded Panel Epilepsy Expanded
Targeted s and Methodology Details for Epilepsy/Seizure tic Panels Reference transcripts based on build GRCh37 (hg19) interrogated by Epilepsy/Seizure tic Panels Epilepsy Expanded Panel Epilepsy Expanded
Editorial Type 2 Diabetes and More Gene Panel: A Predictive Genomics Approach for a Polygenic Disease
 Cronicon OPEN ACCESS EC DIABETES AND METABOLIC RESEARCH Editorial Type 2 Diabetes and More Gene Panel: A Predictive Genomics Approach for a Polygenic Disease Amr TM Saeb* University Diabetes Center, College
Cronicon OPEN ACCESS EC DIABETES AND METABOLIC RESEARCH Editorial Type 2 Diabetes and More Gene Panel: A Predictive Genomics Approach for a Polygenic Disease Amr TM Saeb* University Diabetes Center, College
WERNER_FIBRO_DN LEI_MYB_REGULATED_GENES BRCA1_OVEREXP_DN SUBCLASSES_DIFF P21_P53_MIDDLE_DN
 Supplemental Table S1. Full List of Up-Regulated Conserved Biological Modules Between Zebrafish Liver Tumor with Human Liver, Lung, Gastric, and Prostate Tumors LvCMs (ZF liver cancer vs. human liver cancers
Supplemental Table S1. Full List of Up-Regulated Conserved Biological Modules Between Zebrafish Liver Tumor with Human Liver, Lung, Gastric, and Prostate Tumors LvCMs (ZF liver cancer vs. human liver cancers
Subject ID: Date Test Started: Specimen Type: Peripheral Blood Date of Report: Date Specimen Obtained:
 TEST PERFORMED Whole exome sequencing (WES) with Focused Diagnostic Panel(s): Neuropathy, Leukodystrophy, Myopathy See below for a complete list of genes analyzed. RESULTS 1 sequence variant with potential
TEST PERFORMED Whole exome sequencing (WES) with Focused Diagnostic Panel(s): Neuropathy, Leukodystrophy, Myopathy See below for a complete list of genes analyzed. RESULTS 1 sequence variant with potential
Supplementary Table 1. Information on the 174 single nucleotide variants identified by whole-exome sequencing
 Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
SCIENCE CHINA Life Sciences
 SCIENCE CHINA Life Sciences RESEARCH PAPER April 2013 Vol.56 No.4: 1 7 doi: 10.1007/s11427-013-4460-x Table S1 Human paired-end RNA-Seq data sets from the Sequence Read Archive (SRA) database Experiment
SCIENCE CHINA Life Sciences RESEARCH PAPER April 2013 Vol.56 No.4: 1 7 doi: 10.1007/s11427-013-4460-x Table S1 Human paired-end RNA-Seq data sets from the Sequence Read Archive (SRA) database Experiment
Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic
 Supplementary Information for Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic signals in endothelial cells Kazuhiro Ohkubo 1,, Yasunari Sakai 1,*,, Hirosuke Inoue 1, Satoshi
Supplementary Information for Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic signals in endothelial cells Kazuhiro Ohkubo 1,, Yasunari Sakai 1,*,, Hirosuke Inoue 1, Satoshi
Table S1. CDE Sequences and Globin Reporter mrna Half-Lives in NIH3T3 Cells, Related to Figures 1 and 2 Construct Sequence t1/2 ± SD (hrs)
 Table S1. CDE Sequences and Globin Reporter mrna Half-Lives in NIH3T3 Cells, Related to Figures 1 and 2 Construct Sequence t 1/2 ± SD (hrs) n TNF -CDE 37 -V2 AGATCCTTCAGACAGACATGTTTTCTGTGAAAACGGAGCTGAGCTAGATCT
Table S1. CDE Sequences and Globin Reporter mrna Half-Lives in NIH3T3 Cells, Related to Figures 1 and 2 Construct Sequence t 1/2 ± SD (hrs) n TNF -CDE 37 -V2 AGATCCTTCAGACAGACATGTTTTCTGTGAAAACGGAGCTGAGCTAGATCT
Supplementary Information
 1 Supplementary Information Human TSC2 Null Fibroblast-Like Cells Induce Hair Follicle Neogenesis and Hamartoma Morphogenesis Shaowei Li 1, Rajesh L. Thangapazham 1, Ji-an Wang 1, Sangeetha Rajesh 1, Tzu-Cheg
1 Supplementary Information Human TSC2 Null Fibroblast-Like Cells Induce Hair Follicle Neogenesis and Hamartoma Morphogenesis Shaowei Li 1, Rajesh L. Thangapazham 1, Ji-an Wang 1, Sangeetha Rajesh 1, Tzu-Cheg
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10860 Supplementary Discussion It remains unclear why H3K9 demethylation appeared to be more sensitive to suppression than at least some other histone methylation marks as a result of
doi:10.1038/nature10860 Supplementary Discussion It remains unclear why H3K9 demethylation appeared to be more sensitive to suppression than at least some other histone methylation marks as a result of
New network topology approaches reveal differential correlation patterns in breast cancer
 Bockmayr et al. BMC Systems Biology 2013, 7:78 RESECH TICLE Open Access New network topology approaches reveal differential correlation patterns in breast cancer Michael Bockmayr 1, Frederick Klauschen
Bockmayr et al. BMC Systems Biology 2013, 7:78 RESECH TICLE Open Access New network topology approaches reveal differential correlation patterns in breast cancer Michael Bockmayr 1, Frederick Klauschen
Jocelyn Chapman, MD Division of Gynecologic Oncology. Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic
 Jocelyn Chapman, MD Division of Gynecologic Oncology Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic Genetics underlies all cancers Somatic or tumor genetics Germline or inherited genetics
Jocelyn Chapman, MD Division of Gynecologic Oncology Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic Genetics underlies all cancers Somatic or tumor genetics Germline or inherited genetics
Precision Oncology: Experience at UW
 Precision Oncology: Experience at UW Colin Pritchard MD, PhD University of Washington, Department of Lab Medicine WSMOS Meeting November 1, 2013 Conflict of Interest Disclosures I declare the following,
Precision Oncology: Experience at UW Colin Pritchard MD, PhD University of Washington, Department of Lab Medicine WSMOS Meeting November 1, 2013 Conflict of Interest Disclosures I declare the following,
Comprehensive genetic testing for hearing and vision loss
 Comprehensive genetic testing for hearing and vision loss Hearing and vision loss can result from both genetic and non-genetic etiologies In general, there is a genetic basis for up to 50% of prelingual
Comprehensive genetic testing for hearing and vision loss Hearing and vision loss can result from both genetic and non-genetic etiologies In general, there is a genetic basis for up to 50% of prelingual
Supplementary Data to Supplementary Figure 1a to 1c
 UBC ATP13A2 UBA52 GAK HSPA4 RAB1A DNM1L SPG11 2DA2_HUMAN A30 ACMSD ADH1A ADH1B ATXN2 ATXN3 CAT CYP1B1 CYP2A6 CYP2B6 CYP2C19 CYP2J2 CYP3A43 CYP3A5 CYP4X1 FBXO7 GIGYF2 GSTO1 GSTO2 HLA-DQA2 HLA-DXA HRAS HSP90AA1
UBC ATP13A2 UBA52 GAK HSPA4 RAB1A DNM1L SPG11 2DA2_HUMAN A30 ACMSD ADH1A ADH1B ATXN2 ATXN3 CAT CYP1B1 CYP2A6 CYP2B6 CYP2C19 CYP2J2 CYP3A43 CYP3A5 CYP4X1 FBXO7 GIGYF2 GSTO1 GSTO2 HLA-DQA2 HLA-DXA HRAS HSP90AA1
Nature Genetics: doi: /ng Supplementary Figure 1. Depths and coverages in whole-exome and targeted deep sequencing data.
 Supplementary Figure 1 Depths and coverages in whole-exome and targeted deep sequencing data. Depth (top) and coverage (bottom) of whole-exome sequencing for 38 independent JPN cases (mean depth = 130)
Supplementary Figure 1 Depths and coverages in whole-exome and targeted deep sequencing data. Depth (top) and coverage (bottom) of whole-exome sequencing for 38 independent JPN cases (mean depth = 130)
Nature Neuroscience: doi: /nn.4295
 Supplementary Figure 1 Gene expression profiling of EGFRvIII-expressing human brain tumor stem cells (BTSCs) and mouse astrocytes (a) The protein lysates of human BTSCs were analyzed by immunoblotting
Supplementary Figure 1 Gene expression profiling of EGFRvIII-expressing human brain tumor stem cells (BTSCs) and mouse astrocytes (a) The protein lysates of human BTSCs were analyzed by immunoblotting
Protein Symbol Full Name Patient Pre Post. Fold change (Log 2) Hrb HIV-1 Rev binding protein, mrna
 Supplementary Table 2. Upregulation of peripheral blood antibodies reactive to multiple target proteins a Protein Symbol Full Name Patient Pre Post Fold change (Log 2) Hrb HIV-1 Rev binding protein, mrna
Supplementary Table 2. Upregulation of peripheral blood antibodies reactive to multiple target proteins a Protein Symbol Full Name Patient Pre Post Fold change (Log 2) Hrb HIV-1 Rev binding protein, mrna
Category GOTERM_CC_FAT GOTERM_CC_FAT GOTERM_CC_FAT GOTERM_BP_FAT GOTERM_MF_FAT GOTERM_CC_FAT GOTERM_MF_FAT GOTERM_CC_FAT GOTERM_BP_FAT GOTERM_BP_FAT
 Category KEGG_PATHWAY KEGG_PATHWAY Term GO:0005764~lysosome GO:0000323~lytic vacuole GO:0005773~vacuole GO:0006898~receptor-mediated endocytosis GO:0019842~vitamin binding GO:0005667~transcription factor
Category KEGG_PATHWAY KEGG_PATHWAY Term GO:0005764~lysosome GO:0000323~lytic vacuole GO:0005773~vacuole GO:0006898~receptor-mediated endocytosis GO:0019842~vitamin binding GO:0005667~transcription factor
p5+mir30: AATGATACGGCGACCACCGACTAAAGTAGCCCCTTGAATTC
 SUPPORTING INFORMATION METHODS Massively parallel sequencing and shrna screen data analysis pipeline Genomic DNA extraction and purification from surviving MCF7 cells in the genome wide screen was carried
SUPPORTING INFORMATION METHODS Massively parallel sequencing and shrna screen data analysis pipeline Genomic DNA extraction and purification from surviving MCF7 cells in the genome wide screen was carried
Supplemental Table 1. 3 types of signals that are involved in T cell activation
 Supplemental Table 1. 3 types of signals that are involved in T cell activation Syn-T cells Allo-T cells CD3/28- T cells First signal MHC-TCR-CD3 pathway Not engaged MHC-alloantigen- TCR-CD3 Engaged CD3-Engaged
Supplemental Table 1. 3 types of signals that are involved in T cell activation Syn-T cells Allo-T cells CD3/28- T cells First signal MHC-TCR-CD3 pathway Not engaged MHC-alloantigen- TCR-CD3 Engaged CD3-Engaged
MathIOmica: Dynamic Transcriptome
 Printed from the Complete Wolfram Language Documentation 1 MathIOmica: Dynamic Transcriptome Loading the MathIOmica Package Classification, Clustering and Visualization of Transcriptome Time eries Importing
Printed from the Complete Wolfram Language Documentation 1 MathIOmica: Dynamic Transcriptome Loading the MathIOmica Package Classification, Clustering and Visualization of Transcriptome Time eries Importing
COMPLETE A FORM FOR EACH SAMPLE SUBMITTED ETHNIC BACKGROUND REPORTING INFORMATION
 DATE SAMPLE DRAWN: SAMPLE TYPE: BLOOD OTHER (SPECIFY): COMPLETE A FORM FOR EACH SAMPLE SUBMITTED PATIENT FIRST NAME: LAST NAME: DOB: SEX: M F UNKNOWN ADDRESS: HOME PHONE: ( ) WORK: ( ) EUROPEAN CAUCASIAN
DATE SAMPLE DRAWN: SAMPLE TYPE: BLOOD OTHER (SPECIFY): COMPLETE A FORM FOR EACH SAMPLE SUBMITTED PATIENT FIRST NAME: LAST NAME: DOB: SEX: M F UNKNOWN ADDRESS: HOME PHONE: ( ) WORK: ( ) EUROPEAN CAUCASIAN
Changing the Culture of Cancer Care II. Eric Holland Fred Hutchinson Cancer Research Center University of Washington Seattle
 Changing the Culture of Cancer Care II Eric Holland Fred Hutchinson Cancer Research Center University of Washington Seattle Transforming Cancer Therapy Eric Holland Fred Hutchinson Cancer Research Center
Changing the Culture of Cancer Care II Eric Holland Fred Hutchinson Cancer Research Center University of Washington Seattle Transforming Cancer Therapy Eric Holland Fred Hutchinson Cancer Research Center
Supplementary Figures. Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated. for each lesion type.
 Supplementary Figures Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated for each lesion type. a b Supplementary Figure 2. Non-negative matrix factorization-derived
Supplementary Figures Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated for each lesion type. a b Supplementary Figure 2. Non-negative matrix factorization-derived
Postmortem brain transcriptomes in Dup15q and idiopathic autism. T Grant Belgard, DPhil Dan Geschwind s Lab UCLA Neuropsychiatric Institute
 Postmortem brain transcriptomes in Dup15q and idiopathic autism T Grant Belgard, DPhil Dan Geschwind s Lab UCLA Neuropsychiatric Institute Many genetic causes of autism, none particularly dominant Is there
Postmortem brain transcriptomes in Dup15q and idiopathic autism T Grant Belgard, DPhil Dan Geschwind s Lab UCLA Neuropsychiatric Institute Many genetic causes of autism, none particularly dominant Is there
Supplementary Figure S1: Validation of amplicons using fluorescence in situ
 Supplementary Material Supplementary Figure S1: Validation of amplicons using fluorescence in situ hybridisation with probes for 19q13.2 (A, B), 20q13.13 (C, D) and 20q13.31-q13.32 (E, F). Ovarian clear
Supplementary Material Supplementary Figure S1: Validation of amplicons using fluorescence in situ hybridisation with probes for 19q13.2 (A, B), 20q13.13 (C, D) and 20q13.31-q13.32 (E, F). Ovarian clear
Exons from 137 genes were targeted for hybrid selection and massively parallel sequencing.
 SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
Use of Molecular Assays to Assess Cellular Therapies
 Use of Molecular Assays to Assess Cellular Therapies D. Stroncek, P. Jin, M. Sabatino, J. Ren, L. Castiello, E. Wang, and F.M. Marincola. Cellular Therapies Section Clinical Center, NIH Methods for the
Use of Molecular Assays to Assess Cellular Therapies D. Stroncek, P. Jin, M. Sabatino, J. Ren, L. Castiello, E. Wang, and F.M. Marincola. Cellular Therapies Section Clinical Center, NIH Methods for the
SUPPLEMENTAL TABLE AND FIGURES
 SUPPLEMENTAL TABLE AND FIGURES Zhang et al. (29) - Enzymes in the NAD + Salvage Pathway Regulate SIRT1 Activity at Target Gene Promoters This document contains supplemental data (1 table and 6 figures)
SUPPLEMENTAL TABLE AND FIGURES Zhang et al. (29) - Enzymes in the NAD + Salvage Pathway Regulate SIRT1 Activity at Target Gene Promoters This document contains supplemental data (1 table and 6 figures)
Supplement Results, Figures and Tables
 Supplement Results, Figures and Tables Results PET imaging The SUV was significantly higher in tumours of the parental HSC-3 cell line than tumours of the EV and the MMP-8 overexpressing cell lines (Figure
Supplement Results, Figures and Tables Results PET imaging The SUV was significantly higher in tumours of the parental HSC-3 cell line than tumours of the EV and the MMP-8 overexpressing cell lines (Figure
Nature Immunology: doi: /ni.3745
 Supplementary Figure 1 Experimental settings. (a) Experimental setup and bioinformatic data analysis of transcriptomic changes induced in neonatal and adult monocytes by stimulation with 10 ng/ml LPS for
Supplementary Figure 1 Experimental settings. (a) Experimental setup and bioinformatic data analysis of transcriptomic changes induced in neonatal and adult monocytes by stimulation with 10 ng/ml LPS for
Symbol Name Con Simva Atorva ANOVA Simvastatin - Downregulated Simvastatin - Upregulated
 Symbol Name Con Simva Atorva ANOVA Simvastatin - Downregulated Asb13 ankyrin repeat and SOCS box-containing protein 13 115 +/- 12 91 +/- 13 116 +/- 16 0.00041 Ascc3 activating signal cointegrator 1 complex
Symbol Name Con Simva Atorva ANOVA Simvastatin - Downregulated Asb13 ankyrin repeat and SOCS box-containing protein 13 115 +/- 12 91 +/- 13 116 +/- 16 0.00041 Ascc3 activating signal cointegrator 1 complex
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature11986 relative IL-6 expression Viable intracellular Bp per well 18 16 1 1 5 5 3 1 6 +DG 8 8 8 Control DG Time (h) hours B. pertussis IL-6 (pg/ml) 15 Control DG
SUPPLEMENTARY INFORMATION doi:1.138/nature11986 relative IL-6 expression Viable intracellular Bp per well 18 16 1 1 5 5 3 1 6 +DG 8 8 8 Control DG Time (h) hours B. pertussis IL-6 (pg/ml) 15 Control DG
Pushing%PET%Imaging%to%the% Cellular%Level:%Development%of%a% Radioluminescence%Microscope%
 Stanford University School of Medicine Department of Radiation Oncology Division of Radiation Physics Pushing%PET%Imaging%to%the% Cellular%Level:%Development%of%a% Radioluminescence%Microscope%! Guillem!Pratx,!PhD!
Stanford University School of Medicine Department of Radiation Oncology Division of Radiation Physics Pushing%PET%Imaging%to%the% Cellular%Level:%Development%of%a% Radioluminescence%Microscope%! Guillem!Pratx,!PhD!
Dravet Syndrome: What Warrants Further Testing
 Dravet Syndrome: What Warrants Further Testing or SCN1A in the Modern Era and SCN1A-negative Dravet syndrome Annapurna Poduri, MD, MPH Director, Epilepsy Genetics Program Division of Epilepsy & Clinical
Dravet Syndrome: What Warrants Further Testing or SCN1A in the Modern Era and SCN1A-negative Dravet syndrome Annapurna Poduri, MD, MPH Director, Epilepsy Genetics Program Division of Epilepsy & Clinical
Sample Metrics. Allele Frequency (%) Read Depth Ploidy. Gene CDS Effect Protein Effect. LN Metastasis Tumor Purity Computational Pathology 80% 60%
 Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
Palindromic amplification of the ERBB2 oncogene in human primary breast tumors. A common pattern of copy number transition of chromosome 17 in HER2-
 Supplemental Figures Table of Contents Palindromic amplification of the ERBB2 oncogene in human primary breast tumors Michael Marotta 1,5, Taku Onodera 3,5, Jeffrey Johnson 4, Thomas Budd 2, Takaaki Watanabe
Supplemental Figures Table of Contents Palindromic amplification of the ERBB2 oncogene in human primary breast tumors Michael Marotta 1,5, Taku Onodera 3,5, Jeffrey Johnson 4, Thomas Budd 2, Takaaki Watanabe
Supplementary Information
 Supplementary Information Index of Supplementary Materials Supplementary Figure. Distribution of methylation levels, characteristics of cis-meqtls and association plots for meqtls in methylation-related
Supplementary Information Index of Supplementary Materials Supplementary Figure. Distribution of methylation levels, characteristics of cis-meqtls and association plots for meqtls in methylation-related
COMPREHENSIVE DIAGNOSIS FOR MITOCHONDRIAL DISORDERS
 COMPREHENSIVE DIAGNOSIS FOR MITOCHONDRIAL DISORDERS MITOCHONDRIAL DISORDERS NGS PANELS MITOCHONDRIAL DISORDERS NGS PANELS Name Test code Gene Name Mitome200-Nuclear 2086 (164 nuclear genes) AARS2, ACACA,
COMPREHENSIVE DIAGNOSIS FOR MITOCHONDRIAL DISORDERS MITOCHONDRIAL DISORDERS NGS PANELS MITOCHONDRIAL DISORDERS NGS PANELS Name Test code Gene Name Mitome200-Nuclear 2086 (164 nuclear genes) AARS2, ACACA,
SI Table. SI Table 3. SI Table 4. SI Table 5A. SI Table 5B. SI Table 5C. SI Table 6A. SI Table 6B. SI Table 6C
 SI Table SI Table 1 SI Table 2 SI Table 3 SI Table 4 SI Table 5A SI Table 5B SI Table 5C SI Table 6A SI Table 6B SI Table 6C Title List of primers used to clone HCV genes and gene fragments HCV genotype
SI Table SI Table 1 SI Table 2 SI Table 3 SI Table 4 SI Table 5A SI Table 5B SI Table 5C SI Table 6A SI Table 6B SI Table 6C Title List of primers used to clone HCV genes and gene fragments HCV genotype
Eosinophils! 40! 30! 20! 10! 0! NS!
 A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
Supplementary Files to Töröcsik et al. Factor XIII-A is involved in the regulation of
 Supplementary Files to Töröcsik et al. Factor XIII-A is involved in the regulation of gene expression in alternatively activated human macrophages (Thromb Haemost 2010; 104.3) A Protein synthesis Cell
Supplementary Files to Töröcsik et al. Factor XIII-A is involved in the regulation of gene expression in alternatively activated human macrophages (Thromb Haemost 2010; 104.3) A Protein synthesis Cell
Supplementary Online Content
 Supplementary Online Content Lyall DM, Celis-Morales C, Ward J, et al. Association of body mass index with cardiometabolic disease in the UK Biobank: a mendelian randomization study. JAMA Cardiol. Published
Supplementary Online Content Lyall DM, Celis-Morales C, Ward J, et al. Association of body mass index with cardiometabolic disease in the UK Biobank: a mendelian randomization study. JAMA Cardiol. Published
Assessing the Economics of Genomic Medicine. Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center
 Assessing the Economics of Genomic Medicine Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center DISCLOSURES No conflicts Patents unenforced Consultancies unpaid
Assessing the Economics of Genomic Medicine Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center DISCLOSURES No conflicts Patents unenforced Consultancies unpaid
Influence of genetic ancestry and socioeconomic status on type 2 diabetes in the diverse Colombian populations of Chocó and Antioquia
 Supplementary Information For: Influence of genetic ancestry and socioeconomic status on type 2 diabetes in the diverse Colombian populations of Chocó and Antioquia Aroon T. Chande 1,2,3, Jessica Rowell
Supplementary Information For: Influence of genetic ancestry and socioeconomic status on type 2 diabetes in the diverse Colombian populations of Chocó and Antioquia Aroon T. Chande 1,2,3, Jessica Rowell
# 1 India s Best Selling Cancer Genomics Test Precision treatment for every Cancer Patient Patient Guide
 # 1 India s Best Selling Cancer Genomics Test Precision treatment for every Cancer Patient Patient Guide PositiveSelect is India s widely used Cancer Genomics test with every 8 out of 10 patients opting
# 1 India s Best Selling Cancer Genomics Test Precision treatment for every Cancer Patient Patient Guide PositiveSelect is India s widely used Cancer Genomics test with every 8 out of 10 patients opting
Supplementary information: Forstner, Hofmann et al.
 Supplementary information: Forstner, Hofmann et al., Genome-wide analysis implicates micrornas and their target genes in the development of bipolar disorder Table of contents Supplementary Figure 1: Regional
Supplementary information: Forstner, Hofmann et al., Genome-wide analysis implicates micrornas and their target genes in the development of bipolar disorder Table of contents Supplementary Figure 1: Regional
Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes
 Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes Author Hatzirodos, Nicholas, F. Irving-Rodgers, Helen, Hummitzsch, Katja, L. Harland,
Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes Author Hatzirodos, Nicholas, F. Irving-Rodgers, Helen, Hummitzsch, Katja, L. Harland,
Supplementary Table S1: Gene expression targets
 Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Supporting Information
 Supporting Information Lin et al. 10.1073/pnas.1408759111 Fig. S1. Safety and potential toxicity evaluation of M1 in immunocompetent mice. Immunocompetent BALB/c mice received two doses of i.v. infusion
Supporting Information Lin et al. 10.1073/pnas.1408759111 Fig. S1. Safety and potential toxicity evaluation of M1 in immunocompetent mice. Immunocompetent BALB/c mice received two doses of i.v. infusion
APPLICATIONS OF NEXT GENERATION SEQUENCING IN SOLID TUMORS - PATHOLOGIST PROSPECTIVE
 AMP COMPANION MEETING SYMPOSIUM AT USCAP 2015 NEXT-GENERATION OF PATHOLOGY: ROLE OF PATHOLOGIST IN NGS-BASED PERSONALIZED MEDICINE APPLICATIONS OF NEXT GENERATION SEQUENCING IN SOLID TUMORS - PATHOLOGIST
AMP COMPANION MEETING SYMPOSIUM AT USCAP 2015 NEXT-GENERATION OF PATHOLOGY: ROLE OF PATHOLOGIST IN NGS-BASED PERSONALIZED MEDICINE APPLICATIONS OF NEXT GENERATION SEQUENCING IN SOLID TUMORS - PATHOLOGIST
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study. February 2018
 Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Pol Andrés Benito 1, Jesús Moreno 1, Ester Aso 1, Mónica Povedano 2 1, 3, 4, 5. , Isidro Ferrer. Research Paper ABSTRACT INTRODUCTION
 dominant but also recessive and X-linked in some families. However, about 13% of sals cases bear a gene mutation linked to fals. Main pathological features in sals are loss of myelin and axons in the pyramidal
dominant but also recessive and X-linked in some families. However, about 13% of sals cases bear a gene mutation linked to fals. Main pathological features in sals are loss of myelin and axons in the pyramidal
SUPPLEMENTAL DATA. Table S1. Genes in each pathway selected for analysis.
 SUPPLEMENTAL DATA Table S1. Genes in each pathway selected for analysis. Gene Chr Start End bp Pathway 1 AKR1C1 10 5000044 5022159 22115 COX 2 AKR1C2 10 5029967 5060207 30240 COX 3 AKR1C3 10 5077549 5149878
SUPPLEMENTAL DATA Table S1. Genes in each pathway selected for analysis. Gene Chr Start End bp Pathway 1 AKR1C1 10 5000044 5022159 22115 COX 2 AKR1C2 10 5029967 5060207 30240 COX 3 AKR1C3 10 5077549 5149878
Supplemental Information. High-Throughput Microfluidic Labyrinth for the. Label-free Isolation of Circulating Tumor Cells
 Cell Systems, Volume 5 Supplemental Information High-Throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells Eric Lin, Lianette Rivera-Báez, Shamileh Fouladdel, Hyeun
Cell Systems, Volume 5 Supplemental Information High-Throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells Eric Lin, Lianette Rivera-Báez, Shamileh Fouladdel, Hyeun
Proteome Analysis of Human Perilymph using an Intraoperative Sampling Method
 Supporting Information Article Title Proteome Analysis of Human Perilymph using an Intraoperative Sampling Method Authors Heike A. Schmitt 1,3, Andreas Pich 2, Anke Schröder 2, Verena Scheper 1,3, Giorgio
Supporting Information Article Title Proteome Analysis of Human Perilymph using an Intraoperative Sampling Method Authors Heike A. Schmitt 1,3, Andreas Pich 2, Anke Schröder 2, Verena Scheper 1,3, Giorgio
An integrated transcriptomic and functional genomic approach identifies a. type I interferon signature as a host defense mechanism against Candida
 Supplementary information for: An integrated transcriptomic and functional genomic approach identifies a type I interferon signature as a host defense mechanism against Candida albicans in humans Sanne
Supplementary information for: An integrated transcriptomic and functional genomic approach identifies a type I interferon signature as a host defense mechanism against Candida albicans in humans Sanne
103.5 Membrane metalloendopeptidase TGA GGG GTC ACG ATT TTA GG ATG ATG GTG AGG AGC AGG AC
 Supplementary Table SA Primers used for real-time RT-PCR gene regulation and their confirmation Gene Name Accession no Size Region Forward Primer Reverse Primer Efficiency% AKT v-akt oncogene homolog NM_00563
Supplementary Table SA Primers used for real-time RT-PCR gene regulation and their confirmation Gene Name Accession no Size Region Forward Primer Reverse Primer Efficiency% AKT v-akt oncogene homolog NM_00563
Supplementary Table 1-Genes positively and negatively associated with clonogenic survival in breast cancer cell lines 67 Genes Positively Correlated
 Supplementary Table 1-Genes positively and negatively associated with clonogenic survival in breast cancer cell lines 67 Genes Positively Correlated with Radioresistance Gene Symbol Gene Name Correlation
Supplementary Table 1-Genes positively and negatively associated with clonogenic survival in breast cancer cell lines 67 Genes Positively Correlated with Radioresistance Gene Symbol Gene Name Correlation
b kb
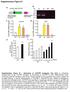 Supplementary Figure S1 COX7RP mrna/gapdh COX activity (OD 45 nm/mg protein) Relative DNA amount COX7RP mrna/gapdh a c pcxn2-flag-cox7rp CAG Flag CMV enhancer Chiken b-actin promoter 1..8.6 COX7RP Muscle
Supplementary Figure S1 COX7RP mrna/gapdh COX activity (OD 45 nm/mg protein) Relative DNA amount COX7RP mrna/gapdh a c pcxn2-flag-cox7rp CAG Flag CMV enhancer Chiken b-actin promoter 1..8.6 COX7RP Muscle
Variant prioritization
 Variant prioritization University of Cambridge Marta Bleda Latorre Cambridge, UK mb2033@cam.ac.uk 30th September 2014 Research Assistant at the Department of Medicine University of Cambridge Cambridge,
Variant prioritization University of Cambridge Marta Bleda Latorre Cambridge, UK mb2033@cam.ac.uk 30th September 2014 Research Assistant at the Department of Medicine University of Cambridge Cambridge,
g. RA Bardot et al., 2016 Supplementary Figure 1 RV LV E15.5 E6.5 E18.5 E7.5 VE RV LV E8.5 RV LV E10.5 Brightfield Foxa2Cre:YFP Brightfield
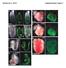 Bardot et al., 216 Supplementary Figure 1 a. VE e. RA LA LA RV LV E15.5 E6.5 b. E7.5 VE f. RA E18.5 LA c. E8.5 ML N CM g. RA RV LV LA P7 PHT RV LV d. E1.5 Brightfield Foxa2Cre:YFP H Brightfield Foxa2Cre:YFP
Bardot et al., 216 Supplementary Figure 1 a. VE e. RA LA LA RV LV E15.5 E6.5 b. E7.5 VE f. RA E18.5 LA c. E8.5 ML N CM g. RA RV LV LA P7 PHT RV LV d. E1.5 Brightfield Foxa2Cre:YFP H Brightfield Foxa2Cre:YFP
A transcriptional mirna-gene network associated with lung adenocarcinoma metastasis based on the TCGA database
 ONCOLOGY REPORTS 35: 2257-2269, 2016 A transcriptional mirna-gene network associated with lung adenocarcinoma metastasis based on the TCGA database Yubo Wang, Rui Han, Zhaojun Chen, Ming Fu, Jun Kang,
ONCOLOGY REPORTS 35: 2257-2269, 2016 A transcriptional mirna-gene network associated with lung adenocarcinoma metastasis based on the TCGA database Yubo Wang, Rui Han, Zhaojun Chen, Ming Fu, Jun Kang,
 Supplementary Table 1. Annotated candidate SNV and indels in melanoma tumors. Patient C1 Patient C2 Patient C3 Patient C4 Patient C5 Patient C6 Patient C7 Patient C10 Chr Position Reference Allele Sample
Supplementary Table 1. Annotated candidate SNV and indels in melanoma tumors. Patient C1 Patient C2 Patient C3 Patient C4 Patient C5 Patient C6 Patient C7 Patient C10 Chr Position Reference Allele Sample
RNA sequencing of cancer reveals novel splicing alterations
 RNA sequencing of cancer reveals novel splicing alterations Jeyanthy Eswaran, Anelia Horvath, Sucheta Godbole, Sirigiri Divijendra Reddy, Prakriti Mudvari, Kazufumi Ohshiro, Dinesh Cyanam, Sujit Nair,
RNA sequencing of cancer reveals novel splicing alterations Jeyanthy Eswaran, Anelia Horvath, Sucheta Godbole, Sirigiri Divijendra Reddy, Prakriti Mudvari, Kazufumi Ohshiro, Dinesh Cyanam, Sujit Nair,
Nature Immunology: doi: /ni.3494
 Supplementary Figure 1 High-throughput screen for novel inducer of T FH cell differentiation. (a) Schematic of primary screen. Purified human naïve CD4 T cells were stimulated by anti-cd3/cd28 beads on
Supplementary Figure 1 High-throughput screen for novel inducer of T FH cell differentiation. (a) Schematic of primary screen. Purified human naïve CD4 T cells were stimulated by anti-cd3/cd28 beads on
Strategy for Integrating Molecular Landscapes in Human Insulinomas
 Supplementary Figure 1 Strategy for Integrating Molecular Landscapes in Human Insulinomas 1 Supplementary Figure 2. Gene dendrogam and module colors 0.8 Height 0.9 1.00 module' The Complete Insulinoma
Supplementary Figure 1 Strategy for Integrating Molecular Landscapes in Human Insulinomas 1 Supplementary Figure 2. Gene dendrogam and module colors 0.8 Height 0.9 1.00 module' The Complete Insulinoma
Provide your cancer patients personalized treatment options with ClariFind
 ONCOLOGY ClariFind employs next-generation sequencing (NGS) for comprehensive genomic profiling of a patient s tumor sample to identify potential clinically actionable targets and associated therapies.
ONCOLOGY ClariFind employs next-generation sequencing (NGS) for comprehensive genomic profiling of a patient s tumor sample to identify potential clinically actionable targets and associated therapies.
Supplementary Data Set 5
 Supplementary Data Set 5 Selective class IIa histone deacetylase inhibition via a non-chelating zinc binding group Mercedes Lobera 1, Kevin P. Madauss 2, Denise T. Pohlhaus 2, Quentin G. Wright 1, Mark
Supplementary Data Set 5 Selective class IIa histone deacetylase inhibition via a non-chelating zinc binding group Mercedes Lobera 1, Kevin P. Madauss 2, Denise T. Pohlhaus 2, Quentin G. Wright 1, Mark
Supplementary Material. Dissected embryos were washed in cold PBS, head and entrails (heart, liver) were excised and
 Supplementary Material Supplementary Material and Methods MEFs preparation and electroporation Dissected embryos were washed in cold PBS, head and entrails (heart, liver) were excised and used for genotyping.
Supplementary Material Supplementary Material and Methods MEFs preparation and electroporation Dissected embryos were washed in cold PBS, head and entrails (heart, liver) were excised and used for genotyping.
Micro-RNAs in cancer: novel origins and sequence variation. Feng Yu
 Micro-RNAs in cancer: novel origins and sequence variation Feng Yu M.Sc., B.Sc. This thesis is submitted in fulfilment of the requirements for the Doctor of Philosophy School of Medicine The University
Micro-RNAs in cancer: novel origins and sequence variation Feng Yu M.Sc., B.Sc. This thesis is submitted in fulfilment of the requirements for the Doctor of Philosophy School of Medicine The University
UAS-SAK (M. Bettencourt-Dias), Ubiquitous-SAK-GFP (M. Bettencourt-Dias), type II
 Supplementary Materials and Methods Fly stocks and genetics The fly strains used in this paper are: ago 1 (I. Hariharan), ago 3 (I. Hariharan), apkc 06403 (C. Doe), akt 1 (H. Stocker), akt 3 (H. Stocker),
Supplementary Materials and Methods Fly stocks and genetics The fly strains used in this paper are: ago 1 (I. Hariharan), ago 3 (I. Hariharan), apkc 06403 (C. Doe), akt 1 (H. Stocker), akt 3 (H. Stocker),
Autism/ID Xpanded Panel Gene List (~2000)
 Autism/ID Xpanded Panel Gene List (~2000) Approximately 84% of the genes are covered >99% at 10X and the average coverage of a gene at 10X for this panel is >98%. A2M A2ML1 AAAS AARS AARS2 AASS ABAT ABCA1
Autism/ID Xpanded Panel Gene List (~2000) Approximately 84% of the genes are covered >99% at 10X and the average coverage of a gene at 10X for this panel is >98%. A2M A2ML1 AAAS AARS AARS2 AASS ABAT ABCA1
Inferring the Mammalian Cardiogenic Gene Regulatory Network. Jason Bazil 10/18/13 Cardiac Physiome Workshop 2013
 Inferring the Mammalian Cardiogenic Gene Regulatory Network Jason Bazil 1/18/13 Cardiac Physiome Workshop 213 Reverse Engineering Gene Regulatory Networks Two main questions need addressed: 1) What s connected
Inferring the Mammalian Cardiogenic Gene Regulatory Network Jason Bazil 1/18/13 Cardiac Physiome Workshop 213 Reverse Engineering Gene Regulatory Networks Two main questions need addressed: 1) What s connected
